Covalently tethering RNA to hydrogel provides the prolonged presentation of RNA for locally sustained cellular gene silencing.
Abstract
Small interfering RNA (siRNA) has found many applications in tissue regeneration and disease therapeutics. Effective and localized siRNA delivery remains challenging, reducing its therapeutic potential. Here, we report a strategy to control and prolong siRNA release by directly tethering transfection-capable siRNA to photocrosslinked dextran hydrogels. siRNA release is governed via the hydrolytic degradation of ester and/or disulfide linkages between the siRNA and hydrogels, which is independent of hydrogel degradation rate. The released siRNA is shown to be bioactive by inhibiting protein expression in green fluorescent protein–expressing HeLa cells without the need of a transfection agent. This strategy provides an excellent platform for controlling nucleic acid delivery through covalent bonds with a biomaterial and regulating cellular gene expression, which has promising potential in many biomedical applications.
INTRODUCTION
RNA interference (RNAi) is a powerful therapeutic tool, which uses small interfering RNA (siRNA) and microRNA molecules to silence gene expression posttranscriptionally (1–5). These RNA molecules are capable of ameliorating a panoply of diseases and promoting tissue regeneration. For example, they can inhibit cancer-promoting genes (6) or the production of unwanted proteins that may negatively affect tissue regeneration (1). However, a number of challenges have limited their utilization as an effective therapeutic tool. Primarily, as anionic macromolecules, RNA molecules are unable to traverse the plasma membrane and transfect into cells without assistance, and they are subject to degradation by ubiquitous ribonucleases and renal clearance (7, 8). Numerous strategies have been developed to enhance cellular uptake, as well as to protect these nucleic acids from enzymatic hydrolysis, including ionic complexation with cationic polymers, such as polyethyleneimine (PEI) and others, or inclusion within nanoliposomes to form lipophilic nanoparticles (9–11). These approaches are not without drawbacks of their own; while efficacy can be greatly increased, without a targeting mechanism to reach desired cell populations, the complexed RNA can be rapidly dispersed throughout the body and give rise to undesirable side effects in nontarget cells. In addition, this may result in the need for repeated and/or high-dosage injections, which can potentially be cytotoxic (12–15). The conjugation of RNA to polymeric nanocarriers via ester or disulfide bonds has been reported to improve RNA stability and transfection efficiency (16–19). However, these systems also suffered from typical nanocarrier drawback of fast clearance. These disadvantages can severely limit the clinical applicability of RNAi. Consistent RNA delivery at target tissues is desirable to achieve prolonged or optimal cellular responses and potentially reduce unwanted side effects in nontarget tissues. Hence, there is a substantial need for development of biomaterial systems that can readily present these bioactive agents over time directly to the target tissues.
Three-dimensional biomaterials such as hydrogels (1, 20–27) and nanofiber or solid scaffolds (28–33) are a clinically valuable means of local, controlled, and/or sustained delivery of therapeutic molecular agents for a variety of tissue engineering applications and disease treatments. Notably, macroscopic biomaterial systems derived from alginate (34), dextran (DEX) (20), chitosan (35), collagen (36), poly(ethylene glycol) (1, 22, 37–41), polyphosphazenes (42, 43), and polyurethanes (29, 31) have been engineered specifically for the release of RNA to encapsulated or surrounding cells. When chemically modified, naked RNAs were physically entrapped within hydrogels, most of them were rapidly released into the surrounding medium within a few days because of their rapid diffusion throughout and from the hydrogel network (34, 35). To retard this otherwise rapid release, nanoparticles incorporating RNA were encapsulated within hydrogels with limited pore size that constrain RNA nanoparticle diffusion, thus retarding release (1, 29, 37, 38). In addition to physical entrapment of RNA nanoparticles to hinder their release, hydrogels were also chemically functionalized with cationic polymer moieties (e.g., PEI) allowing electrostatic interactions with anionic nucleic acids to retain these RNAs within the hydrogel network (20, 21, 42). These biomaterials undergo hydrolytic and/or photolytic degradation to facilitate RNA diffusion and enable release. However, some of these systems require a transfection agent or a cationic polymer, which could induce toxicity to cells. Moreover, precise spatiotemporal control of hydrogel pore size upon bulk degradation is challenging, which could additionally limit control over the RNA release.
In efforts to eliminate the use of transfection agents and obtain enhanced spatiotemporal control over RNA release that does not depend solely on RNA diffusion, RNA affinity with a delivery vehicle, and/or controlling hydrogel degradation and pore size, here, we present a simple and robust strategy to covalently tether transfection-capable siRNA to the backbone of DEX hydrogels for controlling its sustained release and regulating cellular gene expression. An advantage of covalently tethering siRNA to the hydrogel system is that it permits homogeneous and predictable distributions of a large amount of RNA within the hydrogel with minimal initial burst release. There have been no previous reports, however, of covalent conjugation of RNA into a hydrogel network nor of its use for sustained and localized RNA delivery and subsequent regulation of cellular gene expression. In addition, siRNA release from the hydrogels is primarily governed via the hydrolytic degradation of the linkages between RNA and DEX hydrogel in a controlled and sustained manner, independent from the hydrogel bulk degradation rate or RNA diffusion. This strategy may also permit spatial control over RNA distribution within the hydrogels for spatial regulation of cellular gene expression.
RESULTS AND DISCUSSION
siRNA targeting the green fluorescent protein (GFP) was synthesized and functionalized with a thiol group and cholesterol moiety at the 5′ and 3′ ends of the sense strand, respectively (denoted “siGFP-SH”; detailed information is provided in table S1 and fig. S1A), that allows its delivery to cells in the absence of transfection reagents and covalent tethering to mono(2-acryloyloxyethyl) succinate–modified DEX (DEX-MAES) photocrosslinked hydrogels. siGFP was selected because of the ease in monitoring its ability to silence cellular GFP expression. DEX-MAES (fig. S2) containing three hydrolytically degradable ester linkages between the DEX and acrylate groups was synthesized via an esterification reaction between DEX hydroxyl groups and the carboxylic acids of MAES (44). siGFP-SH was synthesized using standard solid phase synthesis and was shown to silence cellular GFP expression without a transfection agent (fig. S1B). siGFP-SH reacts with acrylate groups of DEX-MAES via a Michael-type addition to form hydrolytically degradable β-thioether ester linkages (Fig. 1A). The remaining acrylate groups of DEX-MAES were then photopolymerized, in the presence of a photoinitiator (PI), to form DEX hydrogels containing covalently tethered siRNA (fig. S3). When one of the three ester groups between siRNA and DEX hydrogels hydrolytically degrades, siRNA containing a carboxylic acid (─COOH) group is decoupled from the hydrogels and can diffuse out (Fig. 1B and fig. S3).
Fig. 1. Tethering siRNA to the hydrogel via Michael-addition chemistry and its release.
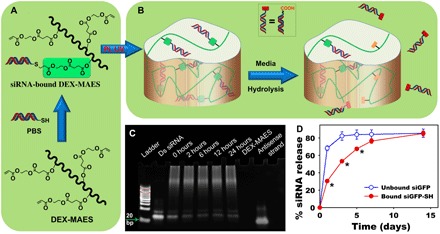
Schematic illustration of (A) siRNA conjugation to DEX-MAES and (B) hydrogel formation via photocrosslinking and siRNA release upon hydrolytic degradation. (C) Acrylamide gel showing conjugation of siRNA-SH to DEX-MAES over time. (D) Release profiles of unbound (physically trapped siGFP, “unbound siGFP”) and covalently tethered siGFP-SH (“bound siGFP-SH”) from 10% (w/w) DEX hydrogels (20 μg of RNA per 50 μl of gel) (*P < 0.001 compared with “unbound siGFP” at the same time point). UV, ultraviolet; PBS, phosphate-buffered saline; Ds siRNA, double-stranded siRNA; bp, base pairs.
Acrylamide gel analysis has demonstrated high conjugation of the synthesized siGFP-SH to the DEX-MAES macromer (Fig. 1C), which could help to prevent initial diffusion and control siRNA release from the fabricated hydrogels. Densitometric calculation from the acrylamide gel indicated that 71, 85, 87, 86, and 85% of siRNA was conjugated to DEX-MAES after 0-min (right after mixing) and 2-, 6-, 12-, and 24-hour reactions, respectively. siRNA (71%) was conjugated at 0 hours, implying that the coupling reaction occurred rapidly and shortly after mixing. This conjugation further delayed the release of covalently tethered siGFP-SH (denoted as bound siGFP-SH) from DEX photocrosslinked hydrogels compared to that of unconjugated siGFP control that lacked the terminal thiol (denoted as unbound siGFP) (Fig. 1D). Specifically, while most of unbound siGFP was released after 3 days, it took up to 14 days to release most bound siGFP-SH, as a result of the covalent tethering. Changing the hydrogel concentration between 6, 8, and 10% resulted in differences in hydrogel persistence time but had little effect on siRNA release behavior (fig. S4A). This finding indicates that the release of tethered siRNA was governed via hydrolysis of ester linkages between siRNA and the hydrogel network and was independent of potential differences in hydrogel pore size or bulk degradation.
Retaining bioactivity of released siRNA is an important factor of a biomaterial delivery system. Therefore, the capacity of released siGFP from the DEX hydrogels to silence cellular GFP expression was examined via culturing with destabilized GFP-expressed HeLa cells without additional transfection agents, followed by quantification of GFP expression via flow cytometry. Unfortunately, the cellular GFP expression could not be suppressed by released siGFP after 2 days of culture (fig. S4B), which could be attributed to the generation of ─COOH groups after the siRNA was released (Fig. 1B) that may prevent the cell internalization of released siRNA. To confirm this, the released siGFP was complexed with Lipofectamine, a commercial transfection reagent, and then treated with destabilized GFP-expressed HeLa cells. By complexation with Lipofectamine, the released siGFP from both “unbound” and “bound” hydrogels silenced cellular GFP expression at significant levels compared to the “no treatment” and nontargeted RNA groups at both tested siRNA concentrations, in which lower siGFP concentration showed significantly decreased siGFP knockdown extent (fig. S5). Both original (“fresh”) and released thiolated siGFP were less effective in silencing cellular GFP expression compared to corresponding nonthiolated siGFP, which is likely due to the thiol modification. These observations confirmed that the ─COOH groups on the released siRNA (Fig. 1B) eliminated its ability to transfect cells by itself, which was recovered by the addition of a transfection reagent. In addition, fluorescence confocal images confirmed that when complexes of siGFP or siGFP-SH with PEI were encapsulated into the DEX-hydrogels, the GFP expression of coencapsulated HeLa cells was significantly reduced compared to no RNA group (no treatment; fig. S6, A and B), while high cell viability was maintained (fig. S6C).
The aforedescribed results demonstrate that the Michael-addition chemistry successfully conjugated thiolated siRNA to DEX-MAES and delayed siRNA release. However, the ─COOH group on the released siRNA might prevent it from entering cells, although the released bound siGFP-SH could silence cellular GFP expression when complexed with a transfection reagent. To retain the capacity of the released siRNA to transfect cells without a transfection reagent, we therefore used an alternative chemistry to covalently tether thiolated siRNA to DEX-MAES hydrogels to permit siRNA release through hydrolytic degradation without bearing a ─COOH group. In this strategy, named the “methacrylation approach,” siGFP methacrylate (siGFP-MA) was synthesized by conjugating siGFP-SH to a pyridyldithiol group of 2-aminoethyl methacrylate conjugated pyridyldithiol (AEMA–DP) via disulfide bond formation (Fig. 2A). The resulting siGFP-MA was then co-photopolymerized with DEX-MAES in the presence of PI to construct siGFP-tethered hydrogels (fig. S7). When the ester or disulfide bonds between the siGFP and hydrogels degrade, siGFP will be released as hydroxyl-terminated siGFP or its original siGFP-SH form (Fig. 2B and fig. S7), which was shown to effectively knock down GFP expression without a transfection reagent before conjugating to the hydrogels (fig. S1B). In addition, the disulfide bond between nucleic acid and residual polymer molecules can also be intracellularly cleaved by redox enzymes (16–19). Acrylamide gel results (Fig. 2C) showed that siRNA-MA was not covalently conjugated to DEX after being mixed with 2% (w/w) DEX-MAES without ultraviolet (UV) application, as indicated by a strong RNA band (lane 3) at the same height as control siRNA (lane 2). When UV light was applied, a large fraction of siRNA-MA was tethered to DEX, as indicated by a weak siRNA band (lane 4) at the same height of control siRNA (lane 2). At a higher DEX-MAES concentration [8% (w/w)], the sample existed as an siRNA-MA–bound hydrogel disk after photocrosslinking, and therefore, there was only one siRNA band observed at the acrylamide gel well position where the DEX hydrogel was initially placed (lane 5). As controls, siRNA without SH and AEMA modification showed no conjugation to 2% (w/v) DEX-MAES without (lane 6) or with (lane 7) UV application. With 8% DEX-MAES, siRNA without SH and AEMA conjugation was only physically trapped within the hydrogel formed via photocrosslinking, and thus, a large fraction of the siRNA ran out of the DEX hydrogel and into the acrylamide gel when a voltage was applied (lane 8).
Fig. 2. Tethering siRNA to the hydrogel via UV conjugation.
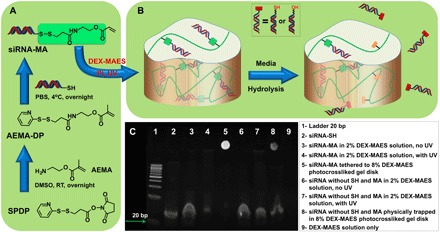
Schematic illustration of (A) siRNA-MA synthesis and (B) conjugation of siRNA-MA to DEX-MAES via photopolymerization and siRNA release upon hydrolytic degradation of ester and/or disulfide bonds within the DEX hydrogel. (C) Polyacrylamide gels confirming conjugation of siRNA-MA to DEX-MAES after application of UV light. SPDP, succinimidyl 3-(2-pyridyldithio) propionate; DMSO, dimethyl sulfoxide; RT, room temperature.
After confirming the conjugation of siRNA-MA to DEX hydrogels via photopolymerization, we examined whether this methacrylation approach could also permit sustained siRNA release. As shown in Fig. 3A, the release of siGFP-MA covalently tethered to the hydrogels (bound siGFP-MA) was delayed and prolonged up to 10 days compared to the rapid 1-day release of siGFP lacking SH and AEMA conjugation (unbound siGFP). Specifically, after 1 day, 80.6% unbound siGFP was released, while only 45.3% bound siGFP-MA was released as a result of covalent tethering. Similar results were also obtained with longer sampling intervals (fig. S8), indicating the reproducibility of this approach. To demonstrate that the methacrylation approach preserves siRNA bioactivity after being covalently conjugated to and released from the hydrogels, bioactivity of “released bound siGFP-MA” and “released unbound siGFP” was assessed in Dulbecco’s modified Eagle’s medium–high glucose (DMEM-HG). When the same volume of releasates containing different concentrations of released siRNA was used to transfect the cells (Fig. 3B), released unbound siGFP, which was only physically trapped within the hydrogels, significantly inhibited 85.4 and 68.5% GFP expression at 3- and 12-hour release time points, respectively. The GFP silencing level returned to 15.8 and 0% at the 1- and 2-day release time points, respectively, as most of the siRNA was released within the first 12 hours. In contrast, released bound siGFP-MA sustained GFP silencing up to 14 days as a result of prolonged siGFP release (Fig. 3, B and C). Covalent tethering of siRNA to the hydrogel system resulted in sustained retention and release from the hydrogels with high siRNA concentrations (>100 nM) in releasates for up to 10 days compared to only 12 hours for unbound, physically trapped siRNA (Fig. 3C). High concentrations of released siRNA in releasates from bound siGFP-MA (Fig. 3C) permitted significant knockdown of gene expression for up to 10 days, whereas the unbound siGFP group achieved significant knockdown for only 1 day (Fig. 3B). The duration of RNA release and gene knockdown may potentially be varied and extended by changing parameters in the system such as the amount of tethered siRNA in the hydrogels and the degradable functional group(s) between the RNA and hydrogel backbone.
Fig. 3. Release of phototethered siRNA from the hydrogels and RNA bioactivity.
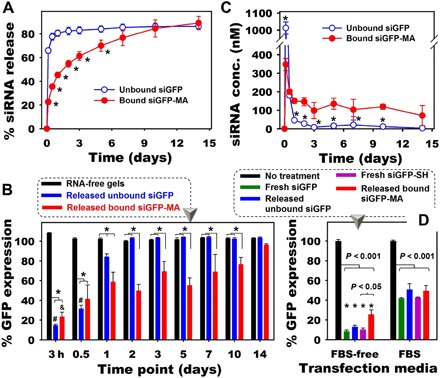
(A) Release profiles of siRNA (10 μg per 50 μl of gel) from 10% (w/w) DEX hydrogels into phenol red–free DMEM-HG (*P < 0.001 compared with unbound siGFP at the same time point). (B) GFP expression of HeLa cells treated with the same volume of releasates from different groups without the addition of transfection reagent and in the absence of FBS for 2 days (*P < 0.05; #P < 0.05 and &P < 0.05 compared to different time points of the same hydrogels). (C) Concentration of siRNA in releasate at different time points, which were used as transfection media to obtain (B) (*P < 0.05 compared with “bound siGFP-MA” at the same time point). (D) Bioactivity of released siGFP at the same siRNA concentration (350 nM) performed in DMEM-HG containing 0% FBS (“FBS-free”) or 2.5% (v/v) FBS [*P < 0.001 compared to the corresponding (color) FBS groups].
To examine the possible effects of the hydrogel preparation process, the release experiment itself, and the presence of fetal bovine serum (FBS) in the release microenvironment on the bioactivity of released siRNA, transfection was then performed using the same concentration (350 nM) of released or fresh siRNA in the presence or absence of FBS [2.5% (v/v)]. Note that this differs from the previous experiment in which the releasate volume was kept constant. When performing the transfection in DMEM-HG without FBS, HeLa cells without siRNA treatment (no treatment) served as a control with 100% GFP expression, and treatment with siLuc (“fresh siLuc”) as a negative control exhibited 101.6% GFP expression. At the same siRNA concentration of 350 nM, fresh non-thiol–AEMA siGFP (“fresh siGFP”) and thiolated siGFP (“fresh siGFP-SH”) silenced 91.5 and 89.7% of the GFP-expressing cells, respectively, while released unbound siGFP lacking SH and AEMA conjugation (released unbound siGFP) and released bound siGFP-MA silenced expression to 87.1 and 74.5%, respectively (Fig. 3D). When the transfection was performed in DMEM-HG containing 2.5% FBS, 58.1, 49.2, 57.3, and 50.5% GFP was silenced when the cells were treated with 350 nM fresh siGFP, released unbound siGFP, fresh siGFP-SH, and released bound siGFP-MA, respectively (Fig. 3D), indicating that this strategy may be implemented in serum-containing conditions. A similar trend was also obtained, however, with significantly lower GFP reduction when the cells were transfected with a lower siRNA concentration (200 nM; fig. S9).
It has been shown that cholesterol-modified RNA offers many advantages compared to unmodified RNA, such as enhancement of tissue targeting, circulation half-life, endosomal escape, target binding specificity, and nuclease resistance, along with cell internalization without the need of an additional transfection agent (45, 46). Conjugation of siRNA to a hydrogel network not only provides localized and sustained RNA presentation but also offers many other advantages, such as improved RNA stability against nucleases and enzymes, which may be present in inflammatory environments when the hydrogel constructs are implanted (18). After conjugation into the hydrogel network, the increase in steric hindrance of RNA molecules against degradative factors acts as another protective mechanism. In addition, the hydrogel network may also act as an additional protective layer shielding the tethered siRNA from degradation by molecules that must diffuse into the gel network. After being released from the hydrogels, cholesterol-modified siRNA is rather stable in the cellular microenvironment as demonstrated in the literature (18, 45, 46). This strategy could be used to provide prolonged presentation of siRNA from hydrogels and subsequent gene silencing that could strongly benefit tissue regeneration and disease therapeutics.
CONCLUSIONS
We developed strategies for controlling siRNA release via covalently tethering siRNA to a hydrogel. Synthesized, cholesterol-modified siGFP-SH inhibited GFP expression in constitutively GFP-expressing HeLa cells without the need of a transfection reagent. The release rate of covalently tethered siRNA was delayed compared to the nontethered siRNA and independent from polymer concentrations. Although the Michael-addition chemistry could be used to tether siRNA to the hydrogel for delayed release, the released siRNA lost its ability to inhibit cellular gene expression without a transfection reagent. The methacrylation approach, in which siRNA-SH was methacrylated before phototethering to the hydrogel, not only allowed for sustained siRNA release but also resulted in transfection without a transfection reagent to knockdown cellular gene expression regardless of the presence of FBS in the media. This approach provides a promising biomaterial platform for controlling nucleic acid delivery for tissue regeneration and disease therapeutics.
MATERIALS AND METHODS
Synthesis and characterization of RNA oligomers
Phosphoramidite monomers (2′OH A/G, 2′-fluoro–modified C/U, and 2′-o-methyl C/U) were purchased from ChemGenes (Wilmington, MA). The 3′ cholesterol–modified controlled pore glass (3′-Cholesterol SynBase CPG 1000/110) was purchased from Link Technologies Ltd. (Lanarkshire, Scotland). The 5′ thiol modification was introduced using a 5′ C6 S-S thiol modifier (ChemGenes). All other RNA synthesis reagents were purchased form Glen Research (Sterling, VA). Positions containing phosphorothioates were introduced by replacing the oxidizer (I2/H20/pyridine) with 1,2,4-dithiazole-5-thione in a mixture of 40% pyridine in acetonitrile. A time of 120 s was used for sulfurization.
All siRNAs were synthesized via solid-phase synthesis on an Expedite 8909 DNA synthesizer (Biolytic Lab Performance, Fremont, CA) at the 1-μmole scale with the final dimethoxytrityl (DMT) group left on to facilitate purification. Support cleavage and base deprotection was achieved by treatment with a 50/50 mixture of concentrated ammonium hydroxide (~30% in H2O) and methylammonium hydroxide (40% in H2O) for 10 min at 65°C. Following a lyophilization step, 2′TOM protecting groups on A and G residues were removed by treatment with 250 μl of mixture of N-methyl-2-pyrrolidone, triethylamine, and triethylamine trihydrofluoride [1.5:0.75:1.0 (v/v/v)] for 2.5 hours at 65°C. The deprotected RNA was recovered by precipitation following the addition of 550 μl of trimethylmethoxysilane. All synthesized oligonucleotides were purified using a reversed-phase HPLC with an Xbridge C18 column (10 mm by 50 mm; Waters, Milford, MA) and a linear gradient of acetonitrile in 0.1 M triethylammonium acetate at pH 7.5. The DMT groups were removed by treatment with 20% acetic acid for 30 min at room temperature, followed by extraction with ethyl acetate. The identity of all oligonucleotides was confirmed using an Agilent 1200/6130 liquid chromatography–mass spectrometry. For analysis, oligonucleotides were desalted using a linear gradient of MeOH in 400 mM hexafluoroisopropanol/8 mM triethylamine using a Waters Xbridge C18 column (2.1 mm by 50 mm). The sequence of the sense (S) and antisense (AS) siRNA oligonucleotides is provided in table S1.
Annealing of siRNA
Previously lyophilized modified sense or antisense RNA oligomers were reconstituted in distilled nuclease-free water (diH2O) to a concentration of 200 μM. To remove protecting trityl groups from the sense strand RNA oligomer, the strand was treated with tris(carboxyethyl)phosphine (TCEP; Sigma-Aldrich, St. Louis, MO) to obtain a 20 mM TCEP (Thermo Fisher Scientific, Pittsburgh, PA) and 0.1 M triethyl ammonium acetate (TEAA; Thermo Fisher Scientific) solution and a final RNA concentration of 100 μM. This mixture was placed in a thermocycler (Genius, Techne Ltd., Cambridge, UK) at 70°C for 3 min and subsequently returned to ambient temperature for 1 hour. The solution was then centrifuged in Microspin 6 desalting columns (Bio-Rad, Hercules, CA), according to the procedure delineated by the manufacturer, to remove the TCEP, TEAA, and protecting group salts. The concentration of RNA in solutions was then quantified using a NanoDrop (Thermo Fisher Scientific, Wilmington, DE). The two strand solutions were combined in equal single-oligomer mole proportion and diluted with phosphate-buffered saline (PBS) to afford a 50 μM solution of double-stranded siRNA. The resulting solution was heated to 70°C in the thermocycler for 3 min and was then removed from the thermocycler and allowed to cool to ambient temperature for 15 min. The annealed, reduced siRNA (annealed siRNA) solution was then frozen at −20°C, and siRNA concentration was quantified using the NanoDrop.
Polyacrylamide gel analysis was performed to confirm the annealing of the siRNA. Precast 15% acrylamide gels (Bio-Rad) were loaded with a solution containing 4 μl of annealed double-stranded siRNA or single-stranded RNA oligomers and 4 μl of loading buffer containing 50 mM tris-HCl, 5 mM EDTA, 25% glycerol, 0.2% bromophenol blue (Thermo Fisher Scientific), and 0.2% xylene cyanole FF (Sigma-Aldrich). These gels were run in tris-borate EDTA (TBE) buffer at 100 V for 90 min. The gels were stained with SYBR Gold (Thermo Fisher Scientific) according to the manufacturer’s instructions for 5 min with agitation. The gels were imaged with a ChemiDoc imaging system (Bio-Rad) (fig. S1A).
Evaluating the bioactivity of synthesized siRNA
Destabilized GFP (deGFP) HeLa cells used for all cell experiments were expanded in DMEM-HG (Sigma-Aldrich), containing 5% FBS and 0.5% G418 (Invitrogen) in a humidified 37°C incubator with 5% CO2. To evaluate the bioactivity of synthesized siGFP and siGFP-SH, 24-well plates were seeded with 50,000 cells per well and grown for at least 16 hours before treating with 0.5 ml of DMEM-HG without FBS containing 0.35 μM siRNA without the addition of transfection agents for 2 days before they were harvested for GFP silencing determination using a flow cytometer (EPICS XLMCL, Beckman Coulter, Fullerton, CA) (n = 3). Accell siGFP (Dharmacon, Lafayette, CO) was used as a positive control, and siLuc-SH served as a negative control. Cells cultured with media only served as a control with 100% GFP expression (no treatment; 100% GFP expression), and all other groups were normalized to the no treatment group.
Synthesis of DEX-MAES
DEX-MAES was synthesized via an esterification reaction between the hydroxyl groups of DEX (Sigma-Aldrich) and the carboxylic acids of MAES (TIC America, Portland, OR), as described in our previous report (44). The actual degree of MAES modification was 16%, calculated using proton nuclear magnetic resonance spectra.
Michael-addition chemistry for siRNA tethering and release from hydrogels
To confirm the conjugation of siGFP-SH to DEX-MAES macromer via a Michael-addition reaction, polyacrylamide gel analysis was performed. A solution of 10 μg of annealed siGFP-SH was mixed with 10% (w/w) DEX-MAES solution in nuclease-free PBS (pH 8.0) containing 0.05% (w/w) Irgacure D-2959 photoinitiator (PI) (Sigma-Aldrich) to a total volume of 50 μl, and the mixture was allowed to react under ambient conditions in darkness. After 0 min (right after mixing) and 2, 6, 12, or 24 hours, the samples (10 μl) were collected and frozen in −80°C at the desired time points to halt the reaction and to be thawed immediately before running acrylamide gels (described in the previous section). A standard of 1 μl of antisense strand siRNA, which did not contain a thiol group to bind to the DEX, and a standard of RNA-free DEX were both added as controls. The antisense RNA was also used to standardize the densitometric calculations, on which the estimates of reaction completion were based.
To examine the release of Michael-addition tethered siRNA from DEX hydrogels, 50 μl of each DEX-siRNA conjugate solution was added to wells of a clear plastic 96-well plate. The wells were irradiated with UV light (OmniCure S1000 UV Spot Cure System, Lumen Dynamics Group, Mississauga, Ontario, Canada) for 150.0 s at the intensity of 2.5 mW/cm2 to cross-link the hydrogels. To fabricate the hydrogels with different macromer concentrations [6, 8, and 10% (w/w)] while keeping the total RNA amount constant, the same amount of RNA (10 μg) was conjugated with the same volume of macromer solutions with different DEX-MAES concentrations before cross-linking. The cross-linked gels were then gently moved to nuclease-free Eppendorf tubes containing 1.0 ml of nuclease-free PBS (pH 7.4). The tubes were placed in a 37°C incubator for subsequent siRNA release. The PBS was periodically carefully removed from the Eppendorf tubes and subsequently frozen. One milliliter of fresh nuclease-free PBS (pH 7.4) was then added, and the gels were allowed to continue degrading. In this manner, the amount of siRNA released from the gels could be detected within the PBS at each time point to monitor the degradation of the hydrogels. Once complete degradation occurred, the frozen aliquots were thawed and their siRNA contents were determined by a RiboGreen (Thermo Fisher Scientific) assay, as per the manufacturer’s procedure. The assay was performed using a plate reader (VersaMax, Molecular Devices, Sunnyvale, CA) with an excitation of 485 nm and an emission of 520 nm. Standards were made of fresh corresponding annealed double-stranded siRNA to establish standard curves to calculate the concentration of released RNAs. Assayed values from degraded RNA-free hydrogels were then subtracted from the experimental samples to take into account any potential assay interference from DEX (n = 3). To examine the bioactivity of released siRNA, the releasates containing released siGFP from days 1 to 10 were pooled and used to treat monolayer cultured HeLa cells for 2 days, followed by quantification of cellular GFP expression via flow cytometry (fig. S4B).
Tethering siGFP to DEX-MAES hydrogels via UV conjugation
Succinimidyl 3-(2-pyridyldithio) propionate (SPDP) (Fisher) and AEMA (Polysciences Inc., Warrington, PA) were dissolved in dimethyl sulfoxide (Sigma-Aldrich) at concentrations of 20 and 75 mM, respectively. SPDP and AEMA solutions were then mixed together [3:1 (v/v)] and incubated at room temperature overnight to obtain a 15 mM AEMA-DP solution (Fig. 2A and fig. S7A). To conjugate the linker AEMA-DP to siGFP-SH to create siGFP-MA, a solution containing siGFP-SH (15 μM) and AEMA-SPDP (1.5 mM) in PBS was prepared and incubated at 4°C overnight. The siGFP-MA was then mixed with DEX-MAES solution in 0.05% PI followed by application of UV light (150.0 s, 2.5 mW/cm2) to fabricate an siGFP-bound hydrogel network (Fig. 2A and fig. S7). To examine the conjugation of siGFP-MA to DEX-MAES, solutions (20 μl) containing siGFP-MA (300 ng), loading buffer (4 μl), and DEX-MAES [final concentration, 2 or 8% (w/w)] before and after UV application were examined on an electrophoresis gel. Precast 15% acrylamide gels (Bio-Rad) were loaded with 20 μl of sample solutions or gels (for samples with 8% DEX-MAES after UV application). These acrylamide gels were run in TBE buffer at 100 V for 90 min (as described in previous section). The gels were stained with ethidium bromide (1 μg/ml; Fisher) for 30 min and imaged with a ChemiDoc imaging system.
To perform siRNA release from photoconjugated DEX-MAES hydrogels (Fig. 2 and fig. S7), 50-μl solutions of 10% (w/w) DEX-MAES containing 10 μg of siGFP-MA were used, and the experiment was performed in a similar manner to the Michael-addition siRNA-conjugated hydrogels with a slight modification. Phenol red–free DMEM-HG (0.5 ml; Sigma-Aldrich) was used as release media instead of 1.0 ml of nuclease-free PBS.
For examination, the bioactivity of released siRNA phototethered DEX-MAES hydrogels, three different types of transfection solutions were used (using the previous protocol) to test three different questions: (i) To examine the effect of tethering on siRNA release rate and then its ability to control cellular gene expression, 0.25 ml of pooled releasates from three hydrogels at specific time points were used as transfection solutions; (ii) to examine whether the hydrogel preparation and/or release process have negative effects on the bioactivity of released siRNA, DMEM-HG containing the same concentration of released or fresh siRNA were used as transfection solutions; and (iii) to examine the effect of FBS on the bioactivity of released siRNA, transfection solutions containing the same concentration of released and fresh siRNA with and without FBS have been used. The cells were then cultured in transfection solutions for two more days and harvested for flow cytometry to quantify the degree of GFP silencing. Cells cultured with media only served as a control with 100% GFP expression (“Ctrl”), and all other groups were normalized to the “Ctrl.”
Statistical analysis
Data are presented as means ± SD. Statistical analysis was performed with Tukey-Kramer multiple comparisons test with one-way analysis of variance (ANOVA) using InStat software (GraphPad Software, La Jolla, CA). P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
Funding: The authors acknowledge funding from the NIH National Institute of Dental and Craniofacial Research (R56DE022376 to E.A.), the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR069564 and R01AR066193 to E.A.), and National Cancer Institute (R21CA182330 to M.L.), the Department of Defense Congressionally Directed Medical Research Programs (OR110196 to E.A.), and a Stand Up To Cancer (SU2C) Innovative Research Grant (IRG) (to M.L.). Author contributions: M.K.N., C.T.H., M.L., and E.A. designed the research strategy and experiments; M.K.N., C.T.H., A.G., S.E.W., K.E.M., N.K., and E.A. performed experiments and/or analyzed the data; M.K.N., C.T.H., A.G., M.L., and E.A. wrote the paper; C.T.H., M.L., and E.A. revised the paper; and M.L. and E.A. supervised the whole process. Competing interests: E.A. is an inventor on a pending patent related to this work filed by Case Western Reserve University (no. 12/191,034, filed on 13 August 2008). E.A., M.K.N., C.T.H., A.G., and M.L. are inventors on another pending patent related to this work filed by Case Western Reserve University (no. 14/435,378, filed on 15 October 2013). The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/8/eaax0801/DC1
Table S1. Sequence of the synthesized oligonucleotides.
Fig. S1. Annealing and gene silencing ability of synthesized siRNA.
Fig. S2. Chemical structure of DEX-MAES.
Fig. S3. Schematic illustration of siRNA conjugation to hydrogels via Michael-addition reaction and its release.
Fig. S4. Effect of hydrogel concentration on the tethered siRNA release behavior and bioactivity of released siRNA.
Fig. S5. Lipofectamine-mediated bioactivity of released siRNA from siRNA-tethered hydrogels.
Fig. S6. Transfection reagent-mediated bioactivity of siRNA loaded or tethered in the hydrogels.
Fig. S7. Schematic illustration of siRNA conjugation to hydrogels via photoconjugation and its release.
Fig. S8. Additional release profiles of tethered siGFP from hydrogels with longer sampling intervals compared to Fig. 3A.
Fig. S9. Additional bioactivity data of released siGFP (200 and 350 nM RNA treatment).
REFERENCES AND NOTES
- 1.Nguyen M. K., Jeon O., Krebs M. D., Schapira D., Alsberg E., Sustained localized presentation of RNA interfering molecules from in situ forming hydrogels to guide stem cell osteogenic differentiation. Biomaterials 35, 6278–6286 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krebs M. D., Alsberg E., Localized, targeted, and sustained siRNA delivery. Chemistry 17, 3054–3062 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitehead K. A., Langer R., Anderson D. G., Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov. 8, 129–138 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gori M., Trombetta M., Santini D., Rainer A., Tissue engineering and microRNAs: future perspectives in regenerative medicine. Expert. Opin. Biol. Ther. 15, 1601–1622 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Saraswathy M., Gong S., Recent developments in the co-delivery of siRNA and small molecule anticancer drugs for cancer treatment. Mater. Today 17, 298–306 (2014). [Google Scholar]
- 6.Xu C.-f., Wang J., Delivery systems for siRNA drug development in cancer therapy. Asian J. Pharm. Sci. 10, 1–12 (2015). [Google Scholar]
- 7.Leung A. K., Tam Y. Y., Cullis P. R., Lipid nanoparticles for short interfering RNA delivery. Adv. Genet. 88, 71–110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Draz M. S., Fang B. A., Zhang P., Hu Z., Gu S., Weng K. C., Gray J. W., Chen F. F., Nanoparticle-mediated systemic delivery of siRNA for treatment of cancers and viral infections. Theranostics 4, 872–892 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urban-Klein B., Werth S., Abuharbeid S., Czubayko F., Aigner A., RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 12, 461–466 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Kim J., Sunshine J. C., Green J. J., Differential polymer structure tunes mechanism of cellular uptake and transfection routes of poly(β-amino ester) polyplexes in human breast cancer cells. Bioconjug. Chem. 25, 43–51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patil M. L., Zhang M., Betigeri S., Taratula O., He H., Minko T., Surface-modified and internally cationic polyamidoamine dendrimers for efficient siRNA delivery. Bioconjug. Chem. 19, 1396–1403 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Tamura A., Nagasaki Y., Smart siRNA delivery systems based on polymeric nanoassemblies and nanoparticles. Nanomedicine 5, 1089–1102 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Xue H. Y., Liu S., Wong H. L., Nanotoxicity: A key obstacle to clinical translation of siRNA-based nanomedicine. Nanomedicine 9, 295–312 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrissey D. V., Lockridge J. A., Shaw L., Blanchard K., Jensen K., Breen W., Hartsough K., Machemer L., Radka S., Jadhav V., Vaish N., Zinnen S., Vargeese C., Bowman K., Shaffer C. S., Jeffs L. B., Judge A., MacLachlan I., Polisky B., Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 23, 1002–1007 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Hu-Lieskovan S., Heidel J. D., Bartlett D. W., Davis M. E., Triche T. J., Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing's sarcoma. Cancer Res. 65, 8984–8992 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Park K., Yang J.-A., Lee M.-Y., Lee H., Hahn S. K., Reducible hyaluronic acid-siRNA conjugate for target specific gene silencing. Bioconjug. Chem. 24, 1201–1209 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Rozema D. B., Lewis D. L., Wakefield D. H., Wong S. C., Klein J. J., Roesch P. L., Bertin S. L., Reppen T. W., Chu Q., Blokhin A. V., Hagstrom J. E., Wolff J. A., Dynamic polyconjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 104, 12982–12987 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J. S., Oh M. H., Park J. Y., Park T. G., Nam Y. S., Protein-resistant, reductively dissociable polyplexes for in vivo systemic delivery and tumor-targeting of siRNA. Biomaterials 34, 2370–2379 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Namgung R., Kim W. J., A highly entangled polymeric nanoconstruct assembled by siRNA and its reduction-triggered siRNA release for gene silencing. Small 8, 3209–3219 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Nguyen K., Dang P. N., Alsberg E., Functionalized, biodegradable hydrogels for control over sustained and localized siRNA delivery to incorporated and surrounding cells. Acta Biomater. 9, 4487–4495 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huynh C. T., Nguyen M. K., Tonga G. Y., Longé L., Rotello V. M., Alsberg E., Photocleavable hydrogels for light-triggered siRNA release. Adv. Healthc. Mater. 5, 305–310 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen M. K., Jeon O., Dang P. N., Huynh C. T., Varghai D., Riazi H., McMillan A., Herberg S., Alsberg E., RNA interfering molecule delivery from in situ forming biodegradable hydrogels for enhancement of bone formation in rat calvarial bone defects. Acta Biomater. 75, 105–114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huynh C. T., Nguyen Q. V., Lym J. S., Kim B. S., Huynh D. P., Jae H. J., Kim Y. I., Lee D. S., Intraarterial gelation of injectable cationic pH/temperature-sensitive radiopaque embolic hydrogels in a rabbit hepatic tumor model and their potential application for liver cancer treatment. RSC Adv. 6, 47687–47697 (2016). [Google Scholar]
- 24.Lym J. S., Nguyen Q. V., Ahn D. W., Huynh C. T., Jae H. J., Kim Y. I., Lee D. S., Sulfamethazine-based pH-sensitive hydrogels with potential application for transcatheter arterial chemoembolization therapy. Acta Biomater. 41, 253–263 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Harding J. L., Osmond M. J., Krebs M. D., Engineering osteoinductive biomaterials by bioinspired synthesis of apatite coatings on collagen hydrogels with varied pore microarchitectures. Tissue Eng. Part A 23, 1452–1465 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Kowalczewski C. J., Saul J. M., Surface-mediated delivery of siRNA from fibrin hydrogels for knockdown of the BMP-2 binding antagonist noggin. Acta Biomater. 25, 109–120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segovia N., Pont M., Oliva N., Ramos V., Borrós S., Artzi N., Hydrogel doped with nanoparticles for local sustained release of siRNA in breast cancer. Adv. Healthc. Mater. 4, 271–280 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Chen M., Gao S., Dong M., Song J., Yang C., Howard K. A., Kjems J., Besenbacher F., Chitosan/siRNA nanoparticles encapsulated in PLGA nanofibers for siRNA delivery. ACS Nano 6, 4835–4844 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Nelson C. E., Kim A. J., Adolph E. J., Gupta M. K., Yu F., Hocking K. M., Davidson J. M., Guelcher S. A., Duvall C. L., Tunable delivery of siRNA from a biodegradable scaffold to promote angiogenesis in vivo. Adv. Mater. 26, 607–614 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen L. H., Gao M., Lin J., Wu W., Wang J., Chew S. Y., Three-dimensional aligned nanofibers-hydrogel scaffold for controlled non-viral drug/gene delivery to direct axon regeneration in spinal cord injury treatment. Sci. Rep. 7, 42212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin J. R., Nelson C. E., Gupta M. K., Yu F., Sarett S. M., Hocking K. M., Pollins A. C., Nanney L. B., Davidson J. M., Guelcher S. A., Duvall C. L., Local delivery of PHD2 siRNA from ROS-degradable scaffolds to promote diabetic wound healing. Adv. Healthc. Mater. 5, 2751–2757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mencia Castaño I., Curtin C. M., Shaw G., Murphy J. M., Duffy G. P., O’Brien F. J., A novel collagen-nanohydroxyapatite microRNA-activated scaffold for tissue engineering applications capable of efficient delivery of both miR-mimics and antagomiRs to human mesenchymal stem cells. J. Control. Release 200, 42–51 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Elangovan S., Khorsand B., Do A. V., Hong L., Dewerth A., Kormann M., Ross R. D., Sumner D. R., Allamargot C., Salem A. K., Chemically modified RNA activated matrices enhance bone regeneration. J. Control. Release 218, 22–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krebs M. D., Jeon O., Alsberg E., Localized and sustained delivery of silencing RNA from macroscopic biopolymer hydrogels. J. Am. Chem. Soc. 131, 9204–9206 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Han H. D., Mora E. M., Roh J. W., Nishimura M., Lee S. J., Stone R. L., Bar-Eli M., Lopez-Berestein G., Sood A. K., Chitosan hydrogel for localized gene silencing. Cancer Biol. Ther. 11, 839–845 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng H., Yang H., Song L., Zhou Z., Sun J., Du Y., Lu K., Li T., Yin A., Xu J., Wei S., Sustained delivery of siRNA/PEI complex from in situ forming hydrogels potently inhibits the proliferation of gastric cancer. J. Exp. Clin. Cancer Res. 35, 57 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huynh C. T., Nguyen M. K., Naris M., Tonga G. Y., Rotello V. M., Alsberg E., Light-triggered RNA release and induction of hMSC osteogenesis via photodegradable, dual-crosslinked hydrogels. Nanomedicine 11, 1535–1550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huynh C. T., Zheng Z., Nguyen M. K., McMillan A., Tonga G. Y., Rotello V. M., Alsberg E., Cytocompatible catalyst-free photodegradable hydrogels for light-mediated RNA release to induce hMSC osteogenisis. ACS Biomater Sci. Eng. 3, 2011–2023 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Huynh C. T., Liu F., Cheng Y., Coughlin K. A., Alsberg E., Thiol-epoxy “Click” chemistry to engineer cytocompatible PEG-based hydrogel for siRNA-mediated osteogenesis of hMSCs. ACS Appl. Mater. Interfaces 10, 25936–25942 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., Malcolm D. W., Benoit D. S. W., Controlled and sustained delivery of siRNA/NPs from hydrogels expedites bone fracture healing. Biomaterials 139, 127–138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L. L., Sloand J. N., Gaffey A. C., Venkataraman C. M., Wang Z., Trubelja A., Hammer D. A., Atluri P., Burdick J. A., Injectable, guest-host assembled polyethylenimine hydrogel for siRNA delivery. Biomacromolecules 18, 77–86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim Y.-M., Park M.-R., Song S.-C., Injectable polyplex hydrogel for localized and long-term delivery of siRNA. ACS Nano 6, 5757–5766 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Kim Y. M., Song S. C., Targetable micelleplex hydrogel for long-term, effective, and systemic siRNA delivery. Biomaterials 35, 7970–7977 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Nguyen M. K., McMillan A., Huynh C. T., Schapira D. S., Alsberg E., Photocrosslinkable, biodegradable hydrogels with controlled cell adhesivity for prolonged siRNA delivery to hMSCs to enhance their osteogenic differentiation. J. Mater. Chem. B 5, 485–495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S. H., Kang Y. Y., Jang H.-E., Mok H., Current preclinical small interfering RNA (siRNA)-based conjugate systems for RNA therapeutics. Adv. Drug Deliv. Rev. 104, 78–92 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Benizri S., Gissot A., Martin A., Vialet B., Grinstaff M. W., Barthelemy P., Bioconjugated oligonucleotides: Recent developments and therapeutic applications. Bioconjug. Chem. 30, 366–383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/8/eaax0801/DC1
Table S1. Sequence of the synthesized oligonucleotides.
Fig. S1. Annealing and gene silencing ability of synthesized siRNA.
Fig. S2. Chemical structure of DEX-MAES.
Fig. S3. Schematic illustration of siRNA conjugation to hydrogels via Michael-addition reaction and its release.
Fig. S4. Effect of hydrogel concentration on the tethered siRNA release behavior and bioactivity of released siRNA.
Fig. S5. Lipofectamine-mediated bioactivity of released siRNA from siRNA-tethered hydrogels.
Fig. S6. Transfection reagent-mediated bioactivity of siRNA loaded or tethered in the hydrogels.
Fig. S7. Schematic illustration of siRNA conjugation to hydrogels via photoconjugation and its release.
Fig. S8. Additional release profiles of tethered siGFP from hydrogels with longer sampling intervals compared to Fig. 3A.
Fig. S9. Additional bioactivity data of released siGFP (200 and 350 nM RNA treatment).


