Abstract
TAR DNA-binding protein 43 kDa (TDP-43), encoded by TARDBP, is an RNA-binding protein, the nuclear depletion of which is the histopathological hallmark of amyotrophic lateral sclerosis (ALS), a fatal neurodegenerative disorder affecting both upper and lower motor neurons. Besides motor symptoms, patients with ALS often develop nonneuronal signs including glucose intolerance, but the underlying pathomechanism is still controversial, i.e., whether it is impaired insulin secretion and/or insulin resistance. Here, we showed that ALS subjects reduced early-phase insulin secretion and that the nuclear localization of TDP-43 was lost in the islets of autopsied ALS pancreas. Loss of TDP-43 inhibited exocytosis by downregulating CaV1.2 calcium channels, thereby reducing early-phase insulin secretion in a cultured β cell line (MIN6) and β cell–specific Tardbp–knockout mice. Overexpression of CaV1.2 restored early-phase insulin secretion in Tardbp–knocked-down MIN6 cells. Our findings suggest that TDP-43 regulates cellular exocytosis mediated by L-type voltage–dependent calcium channels and, thus, plays an important role in the early phase of insulin secretion by pancreatic islets. Thus, nuclear loss of TDP-43 is implicated in not only the selective loss of motor neurons, but also in glucose intolerance due to impaired insulin secretion at an early stage of ALS.
Keywords: Metabolism, Neuroscience
Keywords: ALS, Calcium channels, Insulin
Introduction
Amyotrophic lateral sclerosis (ALS) is a selective motor neuron disorder leading to death in 3–5 years after the onset, due to rapid progression of muscle atrophy, including atrophy of the respiratory and bulbar muscles. TAR DNA-binding protein of 43 kDa (TDP-43) pathology is seen in 90%–95% of sporadic ALS subjects (1–4) as well as in familial ALS linked to TARDBP or C9orf72 (5, 6). Several lines of evidence indicate that the loss of nuclear TDP-43 results in motor neuron degeneration in vitro and in vivo (7–10), while the cytoplasmic aggregation of TDP-43 is also cytotoxic (3, 4, 11, 12).
In addition to their neuromuscular symptoms, ALS patients develop nonneuronal comorbidities (13). In particular, glucose intolerance and dyslipidemia are frequently reported in patients with ALS (13). Normal TDP-43 protein is abundant and ubiquitous in both neuronal and nonneuronal tissues, and it is highly expressed in the pancreas (14), suggesting the possibility that TDP-43–related pathology underlies the nonneuronal manifestation of ALS. However, the molecular mechanism underlying the extra-neural pathology of ALS is poorly understood (15–18).
The purpose of this study was to elucidate the mechanism of impaired insulin dynamics in ALS patients, with a particular focus on TDP-43 pathology in the β cells of pancreatic islets. Here we demonstrated decreased early-phase insulin secretion in patients with ALS by performing an oral glucose tolerance test (OGTT), together with the loss of nuclear TDP-43 in the pancreatic islets of autopsied cases. We further investigated the insulin secretory mechanism in Tardbp knocked-down MIN6 cells in vitro and β cell–specific Tardbp knockout mice in vivo. The results provide the novel insight that loss of TDP-43 inhibits exocytosis via the CaV1.2 calcium channel and reduces the early phase of glucose-induced insulin secretion.
Results
Patients with ALS have reduced early-phase insulin secretion and nuclear loss of TDP-43 in the islets.
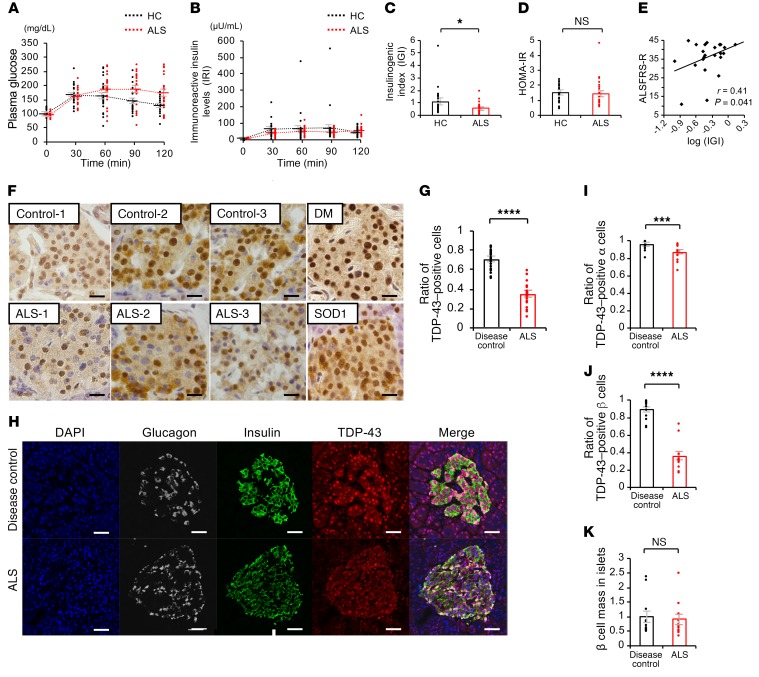
To explore the extra-neural pathogenesis, we analyzed plasma glucose levels and serum insulin using an OGTT in subjects with ALS (Table 1). Plasma glucose measured at 120 minutes after glucose load was significantly higher in ALS subjects (173 ± 59 mg/dL) than in healthy control subjects (130 ± 25 mg/dL). The immunoreactive insulin level (IRI) measured at 30 minutes was significantly lower in ALS subjects (41 ± 16 μU/mL) than in healthy control subjects (63 ± 48 μU/mL). (Figure 1, A and B). The basal insulin secretion level (fasting endogenous insulin secretion) was preserved in ALS subjects (Figure 1B). By contrast, the insulinogenic index (IGI), an index of early-phase insulin secretion, was lower in ALS subjects than in controls (Figure 1C). There were no significant differences in the homeostasis model assessment for insulin resistance (HOMA-IR), appendicular lean soft tissue mass, or creatinine among the groups (Figure 1D and Table 1). The IGI was positively correlated with the revised ALS functional rating scale (ALSFRS-R) (Figure 1E). Taken together, these results suggest that early-phase insulin secretion is decreased in the early stage of ALS and that this impairment of insulin dynamics occurs in parallel with motor dysfunction. To examine whether TDP-43 pathology is associated with the decreased insulin secretion in patients with ALS, we performed immunohistochemistry analysis of autopsied pancreas. We observed a substantial loss of nuclear TDP-43 in the islets of sporadic ALS subjects: the number of TDP-43–positive islet cells in ALS subjects (positive rate of 34% ± 2.6%) was decreased to half of that in controls (positive rate of 70% ± 2.0%) (Figure 1, F and G). Nuclear TDP-43 staining was preserved in the postmortem islets of subjects with diabetes mellitus and mutant superoxide dismutase 1–linked (SOD1-linked) familial ALS, suggesting that the pancreatic TDP-43 pathology is specific to sporadic ALS (Figure 1F). The loss of nuclear TDP-43 was prominent in the β cells compared with α cells, though the β cell mass was not significantly different between the controls and ALS patients (Figure 1, H-K). These results indicate a possible role for nuclear TDP-43 depletion in the islets in the decreased insulin secretion of patients with sporadic ALS.
Table 1. Baseline clinical and hematological features of healthy control and ALS subjects.
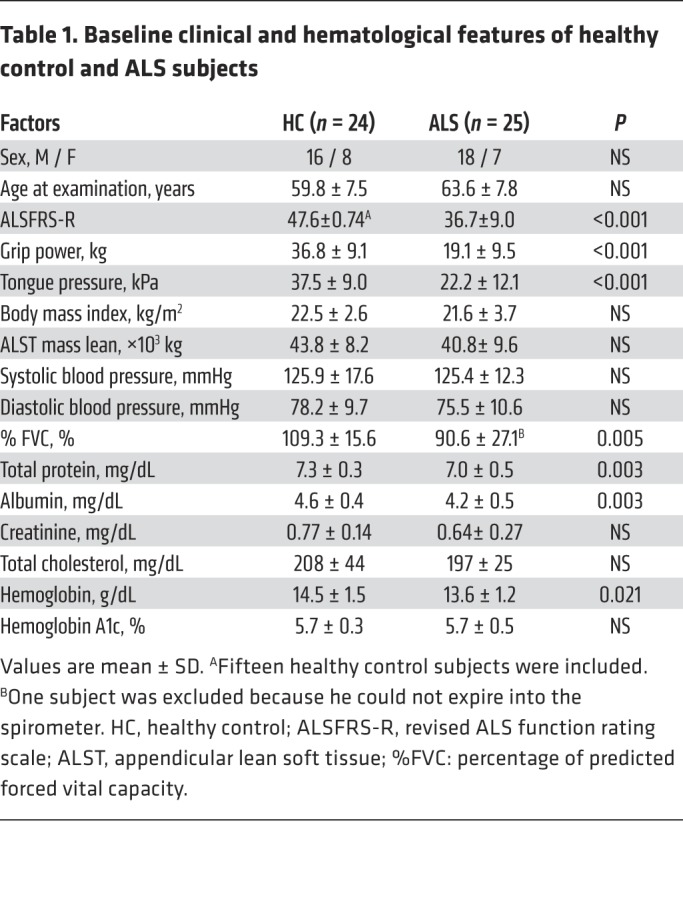
Figure 1. Decreased insulin secretion and loss of pancreatic TDP-43 in patients with early-stage ALS.
(A) Time courses of plasma glucose levels (fasting to 120 minutes). Plasma glucose measured at 120 minutes was significantly higher in ALS subjects than in healthy control subjects (P = 0.002, unpaired t test). (B) Time courses of serum insulin (IRI) (fasting to 120 minutes). IRI measured at 30 minutes was significantly lower in ALS subjects than in healthy control (HC) subjects (P = 0.03, unpaired t test). (C) Indices of glucose metabolism during an oral glucose tolerance test. IGI, an index of early-phase insulin secretion (unpaired t test). (D) Indices of insulin resistance, homeostasis model assessment for insulin resistance (HOMA-IR) (unpaired t test). (E) The base-10 logarithm of IGI plotted against the revised ALS function rating scale (ALSFRS-R) (Pearson correlation; r = 0.41, P = 0.041). For A–E, n = 24 for HC; n = 25 for ALS subjects. (F) Immunohistochemistry of autopsied pancreas in disease controls, sporadic ALS, diabetes mellitus (DM), and mutant SOD1 subjects. Scale bars: 10 μm. (G) Ratio of TDP-43–positive cells in the islets: disease control, 4 patients, 24 islets; ALS, 4 patients, 24 islets (unpaired t test). (H) Immunofluorescence of the autopsied islets. Scale bars: 10 μm. (I) Ratio of TDP-43–positive α cells (n = 12 each, unpaired t test). (J) Ratio of TDP-43–positive β cells (n = 12 each, unpaired t test). (K) β cell mass in islets (n = 12 each, unpaired t test). Values are mean ± SEM. *P < 0.05; ***P < 0.005; ****P < 0.001.
Insulin secretion is impaired in Tardbp knocked-down MIN6 cells.
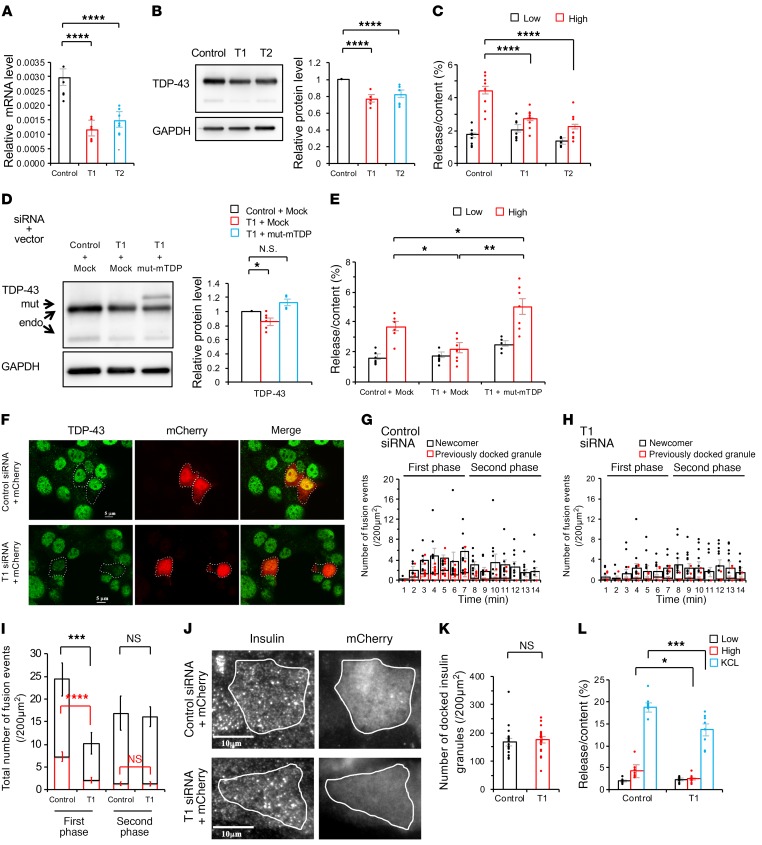
We examined the hypothesis that nuclear TDP-43 regulates early-phase insulin secretion in MIN6 cells by performing in vitro knockdown experiments. The efficiency of Tardbp siRNA knockdown (set1, T1; set2, T2) was confirmed by quantitative real-time polymerase chain reaction (qRT-PCR) and immunoblotting (Figure 2, A and B). Both sets of Tardbp siRNA significantly decreased glucose-induced insulin secretion in MIN6 cells (Figure 2C). The siRNA-resistant form of TDP-43 (a plasmid of mutant [mut]-mTDP-43) recovered glucose-induced early-phase insulin secretion (Figure 2, D and E). Total internal reflection fluorescence (TIRF) imaging was used to confirm the decrease of early-phase insulin secretion due to Tardbp knockdown. First, we confirmed the transfection of control or T1 siRNA into MIN6 cells with mCherry (Figure 2F). We then labeled the insulin granules with GFP, and then analyzed the real-time imaging of the insulin-GFP signal, which was released from the mCherry-positive MIN6 cells with glucose stimulation. The results showed that insulin-GFP signaling was decreased in Tardbp knocked-down MIN6 cells, which revealed an impaired insulin secretion (Supplemental Videos 1 and 2). Quantitative analysis revealed that the glucose-stimulated, first-phase insulin secretion was inhibited in Tardbp knocked-down MIN6 cells (Figure 2, G–I), although the number of insulin granules morphologically docked to the plasma membrane was not affected (Figure 2, J and K). KCL-induced insulin secretion was also significantly decreased in Tardbp knocked-down cells (Figure 2L). Collectively, these findings demonstrate the dysfunction in the first phase of insulin secretion in Tardbp knocked-down MIN6 cells.
Figure 2. Endogenous TDP-43 depletion suppresses the first phase of insulin secretion in MIN6 cells.
(A) mRNA expression levels of Tardbp, encoding TDP-43 (control siRNA, Control; Tardbp siRNA, T1 or T2) (n = 7 each, 1-way ANOVA). (B) Immunoblotting of MIN6 cells transfected with control, T1, or T2 (control, n = 10; T1, n = 5; T2, n = 7, 1-way ANOVA). (C) The insulin assay with low (2.8 mmol/L) and high glucose (16.7 mmol/L) (control, n = 12; T1, n = 12; T2, n = 7, 1-way ANOVA). (D) TDP-43 rescue experiments with mock plasmid or a siRNA-resistant form of murine TDP-43 (mut-mTDP) (n = 6 each, 1-way ANOVA). (E) The insulin assay with low and high glucose. mut-mTDP-43 recovered the glucose-induced insulin secretion (n = 8 each, 1-way ANOVA). (F) T1 siRNA against Tardbp effectively reduced the protein level of TDP-43 (green). Scale bars: 5 μm. (G and H) Histogram showing the number of fusion events from GFP-tagged granules in control siRNA (G) and T1 siRNA–treated MIN6 cells (H) (per 200 μm2) at 1-minute intervals after glucose stimulation and measured by TIRF microscopy (control, n = 11 cells; T1, n = 15 cells, unpaired t test). (I) Quantitative analysis of the total number of exocytotic events detected during the first phase (0–7 minutes) or second phase (>7 minutes). (J) TIRF imaging of insulin granules morphologically docked to the plasma membrane. Scale bar: 10 μm. (K) Number of docked insulin granules (per 200 μm2, control, n = 20; T1, n = 23 cells, unpaired t test). (L) The insulin assay with low glucose, high glucose, and KCL stimulation (n = 8 each, 1-way ANOVA). Values are mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
Loss of TDP-43 reduces insulin secretion by downregulating CaV1.2 calcium channel expression in MIN6 cells.
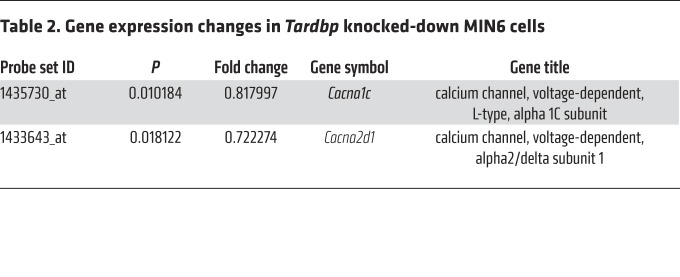
To identify the gene expression changes that mediate Tardbp knockdown–induced impairment of early-phase insulin secretion, we performed microarray analysis on the MIN6 cells treated with control siRNA or T1 siRNA against Tardbp. In this analysis, we identified genes with altered expression: the expression of 338 genes was increased (>120%; Supplemental Table 1), while the expression of 611 genes was decreased (<80%; Supplemental Table 2), in the T1 siRNA–treated MIN6 cells compared with control siRNA–treated MIN6 cells (Gene Expression Omnibus database, accession number GSE125424). These contained 2 voltage-dependent calcium channels, Cacna1c and Cacna2d1 (Table 2). We confirmed that T1 siRNA decreased Cacna1c and Cacna2d1 mRNA levels as well as CaV1.2 and Ca2d1 protein levels (Figure 3, A and B). However, T2 siRNA against Tardbp did not decrease the mRNA levels of Cacna2d1, indicating an off-target effect (Figure 3, A and B). Cotransfection of Tardbp siRNA and a CaV1.2 plasmid recovered glucose-induced insulin secretion (Figure 3, C and D), confirming that TDP-43 regulates early-phase insulin secretion via CaV1.2. In situ hybridization showed that human CACNA1C mRNA staining was substantially attenuated in the islets of patients with sporadic ALS (Figure 3E). As CACNA1D and CACNA1A are abundantly expressed in islets, we also examined the mRNA staining of those genes. CACNA1D mRNA staining was attenuated but CACNA1A expression was enhanced in the islets of patients with sporadic ALS (Figure 3F). In Fura-2 imaging, Ca2+ influx was decreased in response to 30 mmol/L KCl and 50 mmol/L glucose in Tardbp knocked-down MIN6 cells (Figure 4, A–H). Furthermore, in whole-cell patch-clamp recordings of the Ca2+ current measurements, the voltage-dependent calcium channel inward currents were significantly smaller in T1 siRNA–treated MIN6 cells than in control siRNA–treated MIN6 cells (Figure 4I). Collectively, these findings demonstrate that the dysfunction in the first phase of insulin secretion in Tardbp knocked-down MIN6 cells was due to the inhibition of CaV1.2 and eventual impairment of calcium influx.
Table 2. Gene expression changes in Tardbp knocked-down MIN6 cells.
Figure 3. CaV1.2 downregulation mediates impairment of the first phase of insulin secretion in Tardbp knockdown MIN6 cells.
(A) mRNA expression levels of Cacna1c and Cacna2d1 that encode CaV1.2 and Ca2d1, respectively (control siRNA, Control; Tardbp siRNA, T1 or T2) (Cacna1c, n = 6; Cacna2d1, n = 12 each, 1-way ANOVA). (B) Immunoblotting of MIN6 cells transfected with control, T1, or T2 (n = 6 each, one-way ANOVA). (C) Mock or CaV1.2 plasmid was cotransfected into MIN6 cells with control or T1 siRNA (n = 4 each, 1-way ANOVA). (D) The insulin assay with low and high glucose (n = 6 each, 1-way ANOVA). (E) In situ hybridization of the islets of disease control and ALS subjects with a human CACNA1C mRNA antisense probe. Scale bars: 20 μm. (F) Quantitative analysis of in situ hybridization of human CACNA1C, CACNA1D, and CACNA1A (n = 4 patients each, unpaired t test). Values are mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
Figure 4. Fura-2 imaging and whole-cell patch-clamp recordings of Tardbp knocked-down MIN6 cells.
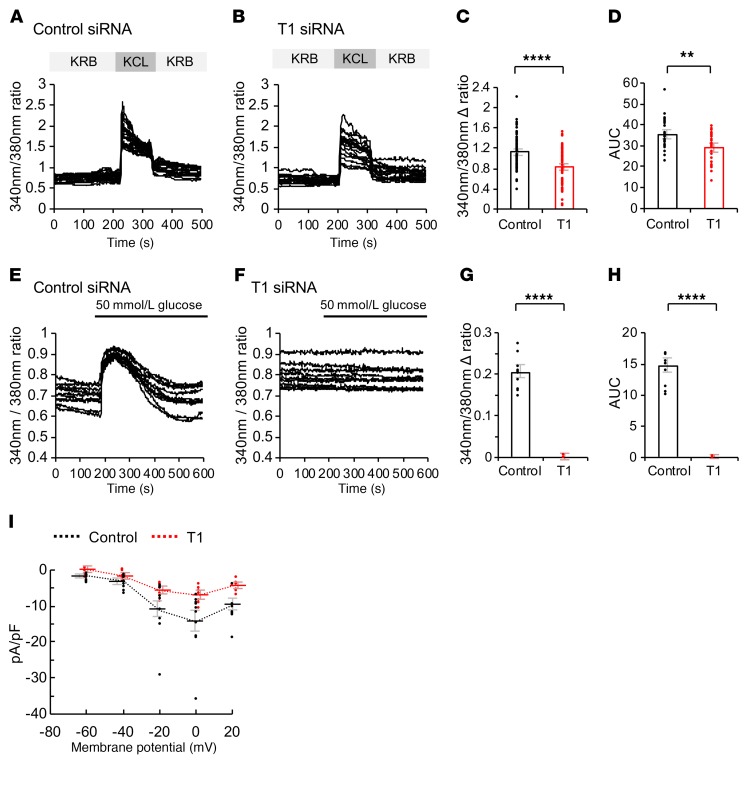
(A and B) Fura-2 imaging of control (A) or T1 (B) siRNA–treated MIN6 cells with KCL stimulation (control, n = 11; T1, n = 14). (C) The F340/F380 fluorescence ratio, after KCL (30 mmol/L) stimulation to KRB buffer (n = 65 each, unpaired t test). (D) The AUC of the F340/F380 fluorescence from 0 to 200 seconds after KCL stimulation (n = 30 each, unpaired t test). (E and F) Fura-2 imaging of control (E) or T1 (F) siRNA–treated MIN6 cells with glucose stimulation (n = 10 each). (G) The F340/F380 fluorescence ratio, after 50 mmol/L glucose stimulation to base line (n = 10 each, unpaired t test). (H) The AUC of the F340/F380 fluorescence from 0 to 200 seconds after 50 mmol/L glucose stimulation (n = 10 each, unpaired t test). (I) Charge (pA/pF)—voltage (V) relationships recorded in control or T1 siRNA–treated MIN6 cells (control, n = 10; T1, n = 7; mean ± SEM; P < 0.05, 2-way ANOVA). Values are mean ± SEM. **P < 0.01; ****P < 0.001.
TDP-43 regulates the transcriptional activity of CaV1.2 calcium channel.
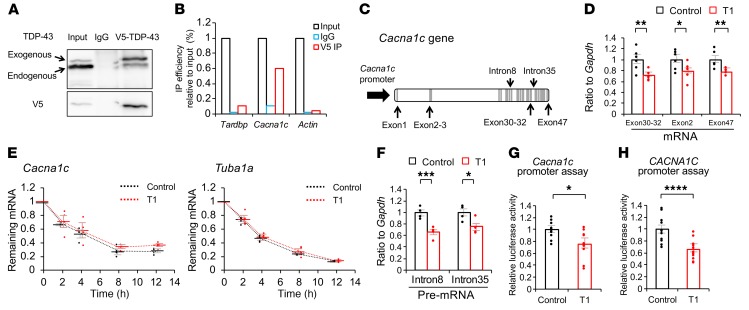
As TDP-43 is a nuclear protein that has a variety of roles in RNA metabolism, we investigated how TDP-43 controls the mRNA expression of Cacna1c. RNA immunoprecipitation analysis showed that mouse WT TDP-43 binds to Cacna1c (Figure 5, A and B), and that Tardbp knockdown reduces mature Cacna1c mRNA levels in MIN6 cells (Figure 5, C and D). To confirm the nonsense-mediated decay of TDP-43 as RNA binding protein, we performed RNA stability. The results showed that there was no difference in mRNA stability of Cacna1c between the MIN6 cells treated with control siRNA and those treated with T1 Tardbp siRNA (Figure 5E). Instead, the amount of premature RNA was reduced in Tardbp knocked-down MIN6 cells (Figure 5F). Furthermore, luciferase assays showed that Tardbp knockdown reduces the promoter activity of both mouse Cacna1c and human CACNA1C (Figure 5, G and H). In silico analysis showed that the TDP-43 protein bound to the promoter region of CACNA1C according to TARDBP chromatin immunoprecipitation sequencing (ChIP-seq) data (ENCODE phase3 data base, accession number ENCSR753GIA, https://www.encodeproject.org/experiments/ENCSR753GIA/). These results indicate that TDP-43 depletion downregulates CaV1.2 in MIN6 cells by decreasing gene transcripts and not by increasing RNA processing.
Figure 5. TDP-43 regulates the transcription of CaV1.2 calcium channels.
(A) Immunoprecipitation (IP) of lysates from MIN6 cells overexpressing V5-tagged TDP-43 with rabbit IgG or an anti-V5 antibody (IP-V5), analyzed with immunoblotting with the indicated antibody. (B) RNA-IP of V5 in cultured MIN6 cells. Bound RNA was analyzed with qRT-PCR using the indicated primers. IP efficiency was calculated relative to input. Representative data from triplicate experiments are shown. (C) Schematics of mature mRNA (exon), premature mRNA (intron), and putative promoter are shown over the Cacna1c gene. (D) Each mature mRNA (exon) were analyzed with qRT-PCR using the indicated primers (n = 6 each, unpaired t test). (E) mRNA stability was measured in MIN6 cells (control, T1) treated with 10 mg/mL actinomycin D with qRT-PCR using the indicated Cacna1c and Tuba1a primers (n = 4 each, 2-way ANOVA). The mRNA levels relative to pretreatment were plotted against time after treatment. Residual mRNA levels were compared at 2, 4, 8, and 12 hours after treatment. (F) Each premature mRNA (intron) were analyzed with qRT-PCR using the indicated primers (n = 6 each, unpaired t test). (G) Luciferase reporter assay of the –1542/+257 mouse Cacna1c promoter (n = 10 each, unpaired t test). (H) Human CACNA1C promoter luciferase reporter assay using the LightSwitch promoter reporter GoClone collection (n = 11 each, unpaired t test). Values are mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
Early-phase insulin secretion and CaV1.2 expression are reduced in pancreas-specific Tardbp knockout mice.
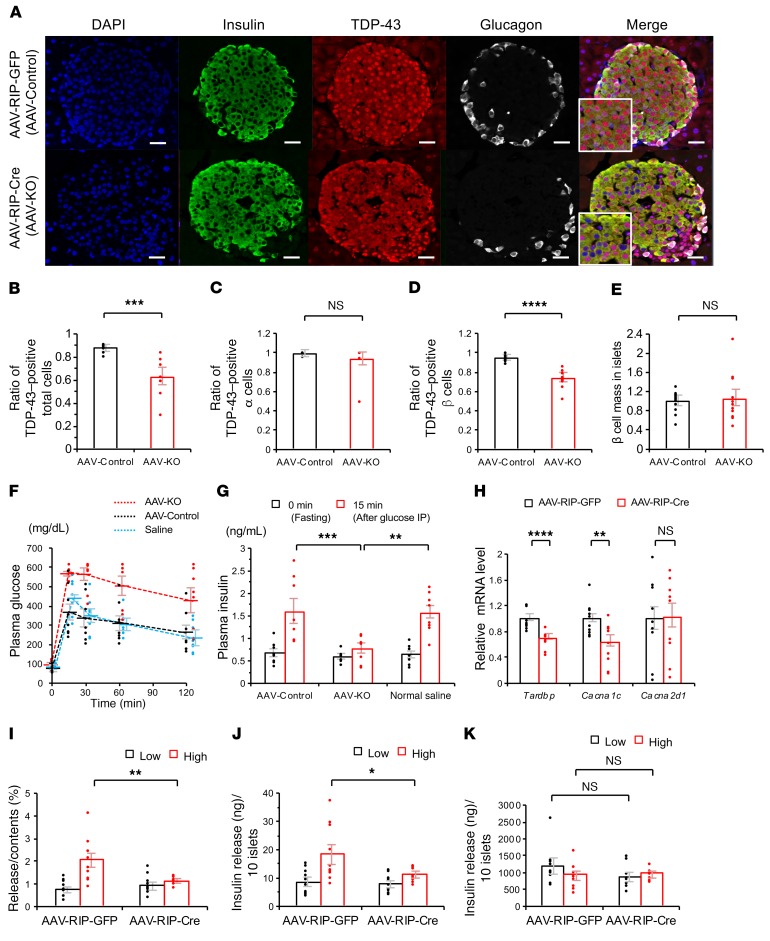
To confirm the role of TDP-43 in early-phase insulin secretion in vivo, we created an AAV-mediated β cell–specific Tardbp knockout (AAV-KO) mouse by intraperitoneally administrating AAV8 carrying Cre recombinase driven by rat insulin promoter (AAV-RIP-Cre) to Tardbpfl/fl mice. TDP-43 expression was attenuated in the islets of AAV-KO mice by about 30%, but not in Tardbpfl/fl mice treated with AAV-Control (Figure 6, A and B). A loss of nuclear TDP-43 was seen in the β cells but not in the α cells, though the β cell mass was not significantly different between AAV-Control and AAV-KO (Figure 6, B–E). IPGTT showed that the plasma glucose at 120 minutes was higher in AAV-KO mice than in control groups (Figure 6F). Insulin secretion at 15 minutes of IGTT was decreased in AAV-KO mice (Figure 6G). In situ hybridization showed that Cacna1c mRNA staining was substantially attenuated in the islets of AAV-KO mice (Supplemental Figure 1, A and B). We also infected primary islets extracted from Tardbpfl/fl mice with AAV-RIP-Cre. In those primary islets, the mRNA levels of Tardbp and Cacna1c were reduced by 30% and 37%, respectively, but Cacna2d1 was not downregulated (Figure 6H). The islets infected with AAV-RIP-Cre had a significantly decreased glucose-induced insulin secretion compared with the control AAV-infected islets (Figure 6, I–K).
Figure 6. AAV vector–mediated pancreatic-specific Tardbp knockout in mice.
(A) Immunofluorescence of the islets of Tardbp flox mice treated with intraperitoneal AAV-RIP-GFP (AAV-Control) or AAV-RIP-Cre (Tardbp-knockout; AAV-KO). Scale bars: 10 μm. (B) Ratio of TDP-43–positive total cells (n = 8 each, unpaired t test). (C) Ratio of TDP-43–positive α cells (n = 10 each, unpaired t test). (D) Ratio of TDP-43–positive β cells (n = 10 each, unpaired t test). (E) β cell mass in islets (n = 12 each, unpaired t test). (F) Intraperitoneal glucose tolerance test (IPGTT). After fasting for 16 hours (17:00–09:00), 2.0 g/kg body weight D-glucose was injected intraperitoneally into the normal saline–treated, AAV-Control, and AAV-KO mice. Plasma glucose measured at 120 minutes was significantly higher in AAV-KO mice than in AAV-Control mice (n = 8 each, P = 0.015, ANOVA). (G) Insulin measurement at 0 minutes (fasting) and 15 minutes after intraperitoneal glucose load in normal saline–treated, AAV-Control, and AAV-KO mice (n = 8, 1-way ANOVA). (H) mRNA expression levels of Tardbp, Cacna1c, and Cacna2d1 in primary islets infected with AAV-RIP-GFP and AAV-RIP-Cre (Tardbp and Cacna1c, n = 10 each; Cacna2d1, n = 9 each, unpaired t test). (I–K) The insulin assay with low and high glucose in primary islets infected with AAV-RIP-GFP and AAV-RIP-Cre (I) Release/Content (%) (n = 10 each, unpaired t test). (J) Insulin release (ng)/10 islets (n = 10 each, unpaired t test). (K) Insulin content (ng)/10 islets (n = 10 each, unpaired t test). Values are mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
Similarly, Tardbpfl/fl mice were mated with RIP-Cre transgenic mice to produce another type of β cell–specific Tardbp knockout (CKO-TDP) mouse. The CKO-TDP mice showed glucose intolerance at 120 minutes with impaired insulin secretion at 15 minutes (Figure 7, A–G). The severity of glucose intolerance was greater in CKO-TDP than in AAV-KO, reflecting the fact that the efficacy of Tardbp knockdown is higher in CKO-TDP mice (Figure 7, F and G). To examine the time course of the insulin secretion response to 16.7 mmol/L glucose, perfusion experiments were performed. In CKO-TDP mice, the amount of secreted insulin (AUCinsulin) after glucose stimulation (from 5 to 10 minutes) was significantly less than that of Tardbpfl/fl mice (Figure 7, H and I). The Cacna1c mRNA levels were also reduced in the islets of CKO-TDP (Supplemental Figure 2, A and B). By contrast, nuclear TDP-43 staining was preserved in the islets of NOD mice, an animal model of diabetes mellitus (Supplemental Figure 2C). Cre recombinase driven by the rat insulin promoter has been shown to be expressed in the hypothalamus in addition to the islet (19). In the present study, Cre recombinase was barely expressed in the hypothalamus of AAV-KO mice, but it was strongly expressed in the hypothalamus of CKO-TDP mice (Figure 8, A–C). This difference appears to underlie the observation that CKO-TDP mice had a greater impairment of glucose tolerance than AAV-KO mice. The CaV1.2 protein was not downregulated in the hippocampus of neuron-specific Tardbp knockout mice (Figure 8, D–F). Taken together, these findings suggest that TDP-43 regulates early-phase insulin secretion from islet β cells in vivo.
Figure 7. Insulin secretion is suppressed in Tardbp flox mouse mated with RIP-Cre mouse (CKO-TDP).
(A) Immunofluorescence of the islets in Tardbpfl/fl and CKO-TDP mice. Scale bars: 10 μm. (B) Ratio of TDP-43–positive total cells (n = 8 each, unpaired t test). (C) Ratio of TDP-43-positive α cells (n = 10 each, unpaired t test). (D) Ratio of TDP-43–positive β cells (n = 10 each, unpaired t test). (E) β cell mass in islets (n = 12 each, unpaired t test). (F) IPGTT showed that plasma glucose measured at 120 minutes was significantly higher in CKO-TDP mice than in Tardbpfl/fl mice (n = 10 each, P < 0.0001, ANOVA). (G) Insulin measurement at 0 minutes (fasting) and 15 minutes after intraperitoneal glucose load (n = 10 each, unpaired t test). (H) Insulin secretion in perfused pancreas in response to high glucose (16.7 mmol/L) (n = 4 each). (I) The amounts of secreted insulin of Tardbpfl/fl and CKO-TDP mice after glucose stimulation were expressed as the AUCinsulin from 5 to 10 minutes (n = 4 each, unpaired t test). Values are mean ± SEM. *P < 0.05; ****P < 0.001.
Figure 8. Brain expression driven by the RIP-Cre promoter in pancreatic-specific and neuron-specific Tardbp KO mice.
(A) X-gal staining of the islets of the CKO-TDP and AAV-KO mice. Scale bar, 20 μm. (B) X-gal staining of brain sections including the hypothalamus in CKO-TDP and AAV-KO mice. Scale bars: 500 μm. (C) Immunohistochemistry of the hypothalamus in Tardbpfl/fl, CKO-TDP, AAV-Control, and AAV-KO mice with an anti–TDP-43 antibody. Nuclear TDP-43 staining was lost only in the hypothalamic neurons of CKO-TDP mice, Scale bars: 20 μm. (D) Immunohistochemistry of the hippocampus in Tardbpfl/fl and neuron-specific Tardbp knockout mouse with an anti–TDP-43 antibody. Nuclear TDP-43 staining was lost in the neuron of cortices and CA1 in neuron-specific Tardbp knockout mouse. Scale bars: 20 μm. (E and F) Immunoblotting of cortices and CA1 in Tardbpfl/fl and neuron-specific Tardbp knockout mice, together with quantitation of the bands in a ratio to GAPDH (n = 3 each, P < 0.05, unpaired t test). Values are mean ± SEM. ***P < 0.005.
Discussion
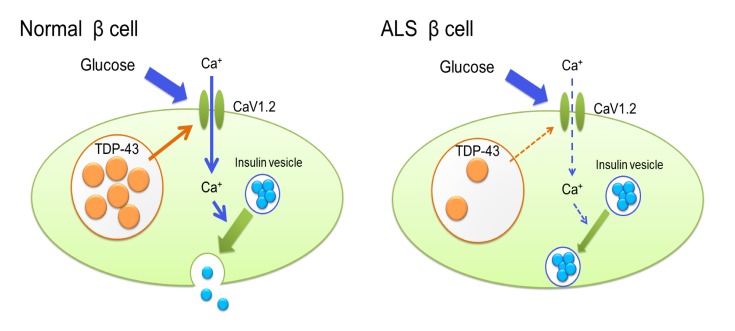
TDP-43 is a nuclear protein which possesses a variety of functions in RNA metabolism (transcription, splicing, and transport), synapse (synaptic vesicle transport, neurotransmitter secretion, and synaptic transmission), development, and cell morphology (3, 4, 11, 12). TDP-43 is normally localized in the nucleus and histopathology of ALS is characterized by the loss of TDP-43 from the nucleus as well as the presence of cytoplasmic protein aggregates containing TDP-43. Neuro-specific depletion of TDP-43 leads to a progressive neurodegenerative phenotype similar to human ALS, suggesting that TDP-43 is required for the maintenance of neuronal integrity (3, 8–10). However, the precise pathomechanism of neurodegeneration due to loss of TDP-43 is elusive. Here we revealed a novel function of TDP-43 as a regulator of insulin secretion. Insulin secretion consists of 2 phases, early and late, which are regulated differently. Early-phase insulin secretion is instigated by glucose uptake via glucose transporters and facilitated by the activation of potassium channels followed by Ca2+ influx via calcium channels. This process releases the insulin pooled in vesicles via exocytosis. Our results indicate that TDP-43 facilitates insulin secretion via the transcriptional regulation of the L-type calcium channel CaV1.2, which is required for early-phase insulin release and the maintenance of systemic glucose tolerance (20, 21). The plasma glucose values during the IPGTT in β cell–specific CaV1.2 knockout mice were shown to be similar to those of our β cell–specific Tardbp knockout mice (20). Thus, the present findings suggest that TDP-43 is a novel regulator of insulin secretion, providing a molecular basis for insulin intolerance in early-stage ALS subjects.
TDP-43 is a global regulator of gene expression involved in regulation of transcription, as well as a DNA- and RNA-binding protein involved in RNA processing (4, 11, 22). The TDP-43 exonic targets include RNA metabolism, synapse, development, cell morphology, and cell signaling, among others (22). Particularly, there is increasing evidence that TDP-43 controls synaptic protein expression (11, 22), although the precise mechanism has yet to be elucidated. TDP-43 overexpression impaired presynaptic integrity by downregulating soluble N-ethylmaleimide–sensitive factor (NSF) attachment receptors (SNARE) proteins including synapsin I and synaptotagmin, resulting in altered glutamate metabolism together with dysregulated excitatory-inhibitory neurotransmitter balance and synchrony (23). The pathogenic mutant TDP-43 upregulates HDAC6 with Bruchpilot deacetylase, leading to synaptic and locomotion defects in flies (24). The present study thus provides what we believe is a novel insight into the function of TDP-43 in vesicle exocytosis.
L-type calcium channels play an important role not only in the pancreas but also in the nervous system (25, 26). The association of TDP-43 with CaV1.2 is shown by the basis of cross-linking, immunoprecipitation, and high-throughput sequencing (CLIP-seq or HITS-CLIP) experiments in the mouse brain (11). In Drosophila, lack of neuronal TDP-43 results in locomotion deficits, which are restored by the exogenous expression of a voltage-gated calcium channel (26). Furthermore, L-type calcium channel agonists ameliorate motor impairment in zebrafish larvae bearing a TDP-43 mutation (25). Given that subtle synaptic dysfunction has been postulated to be a primary event in the initial pathophysiology of ALS (9, 27), L-type calcium channels may also be affected by the loss of TDP-43 in motor neurons, though further work is needed to verify this hypothesis. Although the possibility that there is a CaV1.2-independent pathway in the TDP-43–mediated regulation of insulin secretion cannot be excluded, the decreased Ca2+ entry upon KCL stimulation by Tardbp knockdown and rescue of insulin secretion by exogenous CaV1.2 strongly indicate the primary roles of calcium channels in the regulation of insulin secretion by TDP-43. This view is further supported by physiology experiments using MIN6 cells.
Glucose intolerance in patients with ALS has been repeatedly reported, but the pathophysiologic mechanisms have been elusive (13, 15–18, 28, 29). Glucose intolerance may result from insulin resistance with a marked loss of skeletal muscle (17, 18, 29) and/or impaired insulin secretion (15, 16, 28). Our results suggest that the loss of nuclear TDP-43 underlies the impaired early-phase secretion of insulin in early-stage ALS subjects. Our observation that the IGI, an index of insulin secretion, correlated with motor function measured with the ALSFRS-R further implies that motor neuronal damage occurs in parallel with β cell dysfunction in patients with ALS. This view is likely supported by the fact that islet β cells and motor neurons share key molecules, such as islet1 and HB9, during their development (30, 31).
Methods
ALS patients and oral glucose tolerance test.
We enrolled 25 consecutive Japanese ALS patients without diabetes mellitus or family history of ALS (Table 1). All patients were diagnosed as definite, probable, or probable laboratory-supported ALS under the El Escorial and the revised Airlie House diagnostic criteria. We compared these ALS patients with 24 age- and sex-matched Japanese volunteers without neurological disease and diabetes mellitus. All patients were followed in Nagoya University Hospital. The data were collected from May 2013 and August 2014.
All patients underwent a 75-gram OGTT after a 12-hour overnight fast. The plasma glucose and immunoreactive insulin levels were measured before glucose loading and every 30 minutes until 120 minutes after glucose loading. OGTT was performed via feeding tubes in the 2 ALS patients with gastrostomy tubes and orally in the remaining 23 ALS patients and in 24 healthy controls. In an effort to examine glucose metabolic pathology, we calculated several indices of insulin secretion and resistance by measuring C-peptide immunoreactivity, fasting and GTT plasma glucose, and IRI. The IGI, a measure of early-phase insulin secretion, defined as the ratio of the increment of insulin to that of plasma glucose 30 minutes after a glucose load, was calculated with the formula: IGI = [(IRI30 – IRI0) / (PG30 – PG0)], where IRI0 is fasting plasma insulin (μU/mL); IRI30 is insulin 30 minutes after glucose load (μU/mL); PG0 is fasting plasma glucose (mg/dL); and PG30 is plasma glucose 30 minutes after glucose load (mg/dL) (32, 33). Insulin secretory insufficiency was defined as an IGI <0.4 (34). The serum C-peptide immunoreactivity index (CPI), an index of fasting endogenous insulin secretion, was calculated by the formula: CPI = [fasting serum C-peptide levels (ng/mL) × 100 / PG0 (mg/dL)] (35). The HOMA-IR, which primarily reflects hepatic insulin resistance, was calculated by the formula: HOMA-IR = [PG0 × IRI0 / 405] (36). The measured biochemical parameters included serum creatinine and appendicular lean soft tissue (ALST) mass as indices of skeletal muscle mass (37).
Immunocytochemistry for human autopsy samples.
We analyzed autopsied pancreatic islet specimens from patients with ALS and from age-matched disease control subjects. Pathological diagnoses of the disease controls were hepatic failure, Parkinson’s disease, and diabetes mellitus. The ALS pancreatic islets were excised at autopsy and fixed immediately in a 10% buffered formalin solution. Sections (3 μm) were deparaffinized, heated in a microwave for 15 minutes in 10 mM citrate buffer (pH 6.0), and incubated overnight with an anti–TDP-43 rabbit polyclonal antibody (1:6000; 10782-2-AP, Proteintech). Subsequent staining procedures were performed using the DAKO EnVision+ HRP System (Dako) and photographed with an optical microscope (BZ-X710; Keyence) as previously described (38, 39).
Immunofluorescence for human autopsy samples.
The ALS pancreatic islets were excised at autopsy and fixed immediately in a 10% buffered formalin solution. Sections (3 μm) were deparaffinized, heated in a microwave for 15 minutes in 10 mM citrate buffer (pH 6.0), and incubated overnight with the following primary antibodies: anti–TDP-43 rabbit polyclonal (1:1000; 10782-2-AP, Proteintech), anti-insulin pig (1:200; ab7842, Abcam), and anti-glucagon mouse (1:500; ab10988, Abcam). After washing, the samples were incubated with Alexa-488–conjugated donkey anti-pig IgG (1:1000; A11073, Invitrogen), Alexa-488–conjugated donkey anti-mouse IgG (1:1000; R37114, Invitrogen), Alexa-546–conjugated donkey anti-rabbit IgG (1:1000; A10040, Invitrogen), and Alexa-647–conjugated donkey anti-mouse IgG (1:1000; A31571, Invitrogen) for 1 hour, mounted using ProLong Gold with DAPI (P36935, Thermo Fisher Scientific), then imaged with a laser confocal microscope (LSM880; Zeiss).
Cell culture and transfection.
MIN6 β cells, a line derived from mouse insulinoma at passage 20–30, were a gift from S. Seino (Kobe University, Kobe, Japan) and J. Miyazaki (Osaka University, Osaka, Japan). The cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% (vol/vol) FBS (10270, Life Technologies) with 5% penicillin/streptomycin (15140-122, Wako) and β-mercaptoethanol (M6250, Sigma-Aldrich) at 37°C and 5% CO2. MIN6 cells seeded at a density of 3.0 × 105 cells on a 12-well plate were electroporated with 100 pmol siRNA (Control, TDP-43 set1, and TDP-43 set2) using the NEON Transfection System (electroporation: 1200 V pulse, 20 ms width; Thermo Fisher Scientific) according the manufacturer’s procedure, unless otherwise mentioned. Oligonucleotide siRNA duplexes were synthesized by Sigma-Aldrich with the following sequences: control-siRNA, 5′-GAAUCAGAUGCACAUGAGUTT-3′; Tardbp siRNA-set1-T1, 5′-GAACGAUGAACCCAUUGAATT-3′; and Tardbp siRNA-set2-T2, 5′-GUUCUUAUGGUUCAGGUCATT-3′. Unless otherwise mentioned, set1 siRNA (T1) was used for Tardbp knockdown throughout the experiments. The siRNA-resistant form of the TDP-43 gene (mut-mTDP-43) was generated as described elsewhere (7). In the TDP-43 and CaV1.2 rescue experiments, MIN6 cells seeded at a density of 3.0 × 105 cells on a 12-well plate were electroporated with 100 pmol control siRNA, 2 μg mock plasmid, 100 pmol T1-siRNA, and either 2 μg mut-mTDP-43 or 50 ng v5-His-CaV1.2 plasmid (as appropriate), using the NEON Transfection System as described above. The pCDNA6/V5-His-CaV1.2 (v5-His-CaV1.2) construct was purchased from Addgene (plasmid 26572). At 48 hours after transfection, MIN6 cells were collected and used for qRT-PCR, immunoblotting, and measurement of insulin secretion.
RNA extraction and qRT-PCR.
mRNA levels were measured by qRT-PCR as previously described (38, 39). RNA was prepared from MIN6 cells using an RNeasy Mini Kit (74106, Qiagen) according to the manufacturer’s instructions. Total RNA (500 ng) was used as a template for reverse transcription using ImProm-II Reverse Transcriptase (A3800, Promega). qRT-PCR was performed using the primers listed in Table 3 and KAPA SYBR FAST qPCR Master mix (KK4602, KAPA Biosystems) according to the manufacturer’s instructions. Data were shown as the ratio of the mRNA level to that of Gapdh.
Table 3. Primer sequences.

Immunoblotting.
MIN6 cells were scraped from the culture dish and homogenized with RIPA buffer (182-02451, Wako) containing 1% protease inhibitors (4693116001, Roche) and phosphatase inhibitors (1862495, Thermo Fisher Scientific). Homogenates were incubated on ice for 20 minutes and centrifuged at 13,000g for 10 minutes at 4°C. The lysates were mixed with 4 times NuPAGE LDS sample buffer (NP0008, Novex) and heated at 95°C for 5 minutes. After denaturation, each cell lysate was separated by SDS-PAGE (5%–20% gradient gel) and analyzed by immunoblotting with ECL Plus detection reagents (NEL121001EA, PerkinElmer) using the following primary antibodies: anti–TDP-43 rabbit polyclonal (1:3000, 10782-2-AP; Proteintech), anti-GAPDH mouse monoclonal (1:1000; M171-3, MBL International Corporation), anti-CaV1.2 rabbit polyclonal (1:2000; ACC-003, Alomone Labs), anti-Ca2d1 (E10) mouse monoclonal (1:100, sc-271697; Santa Cruz Biotechnology), and anti–v5-Tag (D3H8Q) rabbit polyclonal (1:1000, 13151; Cell Signaling Technology). Band intensity was quantified by Multi Gauge 3.0 software (Fujifilm).
Insulin secretion in MIN6 cells following glucose stimulation.
MIN6 cells were cultured at a density of 3.0 × 105 cells on a 12-well plate for 48 hours after electroporation (MPK10025, Thermo Fisher Scientific) as described above. The cells were preincubated with a 2.8 mmol/L (low glucose), Krebs-Ringer Modified Buffer (KRB) containing 10 mM HEPES (pH 7.4), 110 mM NaCl, 4.4 mM KCl, 1.45 mM KH2PO4, 1.2 mM MgSO4, 2.3 mM calcium gluconate, 4.8 mM NaHCO3, 4 mM glucose, and 0.3% bovine serum albumin for 30 minutes, and stimulated with 16.7 mmol/L (high glucose) glucose and 30 mmol/L KCL for 30 minutes. Insulin levels in the supernatant (release) and acid ethanol extraction (content) were measured using an Insulin Ultra-sensitive Assay kit (62IN2PEG, CisBio). The amount of secreted insulin was calculated as a ratio of release to content.
Microarray analysis of MIN6 cells transfected with control and Tardbp siRNA.
We analyzed alterations in gene expression in MIN6 cells transfected with control and Tardbp siRNA, using GeneChip Mouse Genome 430 2.0 array (Affymetrix). Total RNA was isolated from the MIN6 cells treated by control or Tardbp T1 siRNA according to the manufacturer’s instructions (RNeasy, Qiagen). The cDNA preparation, hybridization process, and microarray data analysis were performed at RIKEN Genesis. Biotinylated cRNA was prepared using GeneChip 3′IVT PLUS Reagent Kit (Thermo Fisher Scientific) according to the manufacturer’s standard protocol from 100 ng total RNA. Following fragmentation, 12.5 μg cRNA was hybridized for 16 hours at 45°C on a GeneChip Mouse Genome 430 2.0 Array. GeneChips were washed and stained in the GeneChip Fluidics Station 450. GeneChips were scanned using the GeneChip Scanner 3000 7G. The data were analyzed with Microarray Suite version 5.0 (MAS 5.0) using Affymetrix Expression Console Software (Thermo Fisher Scientific) according to the default analysis settings. The criteria used to detect the differences in gene expression were a 1.2-fold change in MIN6 cells treated with control siRNA compared with Tardbp siRNA. The microarray profiling data were deposited in the NCBI’s Gene Expression Omnibus database (GSE125424).
TIRF imaging for insulin secretion by MIN6 cells transfected with control or Tardbp siRNA.
MIN6 cells were plated on fibronectin-coated high-refractive-index glass for imaging by TIRF microscopy or on fibronectin-coated normal coverslips for immunostaining (40). After 18–24 hours, the cells were transfected with 50 nM various vectors (insulin-GFP, ref. 41), mCherry (42), and siRNAs (control or Tardbp siRNAs) using Lipofectamine 2000 (11668-019, Invitrogen) in the presence of serum, according to the manufacturer’s instructions. The experiments were performed 3 days after transfection. The Olympus total internal reflection system was used with a high-aperture objective lens (Apo ×100 OHR, numerical aperture 1.65; Olympus) essentially as previously described (43). For real-time imaging of the insulin granules labeled by insulin-GFP under TIRF microscopy, MIN6 cells were incubated for 20 minutes at 37°C in KRB. The cells were then transferred to the stage of a TIRF microscope and glucose stimulation was achieved by the addition of 40 mM glucose in KRB into the chamber (final concentration 22 mM glucose).
To evaluate the number of docked insulin granules by TIRF microscopy, MIN6 cells transfected with mCherry and control or Tardbp siRNA were cultured on high-refractive-index glass, fixed, permeabilized with 4% paraformaldehyde/0.1% Triton X-100, and processed for immunohistochemistry. The cells were labeled with an anti–insulin antibody (I2018, Sigma-Aldrich) and processed with goat anti-mouse IgG conjugated to Alexa Fluor 488 (A-11001, Invitrogen). For the TIRF imaging of Alexa Fluor 488–labeled insulin granules in cells expressing mCherry, we used the 488-nm and 561-nm laser line for excitation and an image splitter (W-view Gemini; Hamamatsu Photonics) that divided the green and red components of the images with a 565-nm dichroic mirror (Q565; Chroma Technology Corporation), passing the green component through a 530 nm ± 15 nm bandpass filter (HQ530/30 m; Chroma Technology Corporation) and the red component through a 630 nm ± 25 nm bandpass filter (HQ630/50 m; Chroma Technology Corporation) (43).
Measurement of intracellular Ca2+ concentration.
Intracellular free Ca2+ concentration ([Ca2+]i) was estimated by microfluorometry in MIN6 cells loaded with Fura-2 AM (4987481523647, Dojindo Laboratories) as previously described (44). MIN6 cells, electroporated with control siRNA or Tardbp siRNA (T1), were incubated for 60 minutes at room temperature with the acetoxymethyl ester Fura-2 (10 μM) in the standard KRB solution (2.8 mmol/L). The Fura-2–loaded MIN6 cells were attached to glass coverslips, which were then covered with 500 μL KRB for 2–4 minutes, KCL (30 mmol/L) for 1 minute, and KRB for 2–4 minutes at 37°C on the stage of an inverted microscope (Olympus IX). Alternatively, the experiment was performed with 500 μL KRB for 2–4 minutes and 50 mM glucose for 7 minutes at 37°C. Microfluorometry was performed on 10–20 MIN6 cells illuminated alternately at 340 and 380 nm. Fluorescence intensities (F340 and F380) were measured at 510 nm. Changes in [Ca2+]i are presented as changes in the F340/F380 fluorescence ratio.
Whole-cell voltage-clamp recordings.
MIN6 cells were transfected with 500 pmol/2 mL siRNA using Lipofectamine 3000 (L3000015, Invitrogen). The cells were used 48–72 hours after transfection. The bath solution contained 140 mM NaCl, 5 mM KCl, 5 mM EGTA, 6.9 mM BaCl2, (approximately 2 mM free Ba2+ calculated by the MAXC program, Stanford University), 2 mM MgCl2, 10 mM glucose, and 10 mM HEPES, pH 7.40, adjusted with NaOH. The pipette solution contained 140 mM NMDG-Cl, 5 mM EGTA, and 10 mM HEPES, pH 7.30, adjusted with NMDG. Pipette resistances were 3 ± 1 MΩ. The currents were recorded with step pulses from –60 mV to +20 mV in 20-mV increments from a –90 mV holding potential using an Axopatch 200B amplifier (Molecular Devices), filtered at 5 kHz with a low-pass filter, and digitized with Digidata 1440A (Axon Instruments). Data were acquired with pCLAMP 10 (Axon Instruments).
Immunoprecipitation.
The pcDNA3.1/V5-His-mouse TDP-43 (GenBank accession number NM_145556, v5-mTDP-43-WT) construct was produced as previously described (7). Total lysates of MIN6 cells transfected with 2 μg v5-mTDP-43-WT by electroporation (1.0 × 106 cells, 1200 V, 20 ms, 2 pulses) on a 6-well plate were mixed with 2 μg rabbit IgG as control or 2 μg anti–v5-Tag (D3H8Q) rabbit polyclonal antibody (13151, Cell Signaling Technology), which binds to WT mTDP-43, and rotated at 4°C for 1 hour. Protein G Dynabeads (10004D, Invitrogen) were added to the mixture and further incubated at 4°C for 2 hours. The beads were washed 4 times with RIPA buffer (182-02451, Wako) containing 1% protease inhibitors (4693116001, Roche) and phosphatase inhibitors (1862495, Thermo Fisher Scientific). The beads were mixed directly with 2× NuPAGE LDS-PAGE sample buffer (NP0008, Novex) containing β-mercaptoethanol, heated at 70°C for 10 minutes, and analyzed by immunoblotting using an anti–TDP-43 rabbit polyclonal antibody (1:3000, 10782-2-AP; Proteintech) and anti-GAPDH mouse monoclonal antibody (1:1000; M171-3, MBL International Corporation).
Ribonucleoprotein immunoprecipitation.
Ribonucleoprotein immunoprecipitation was performed using the RIP Assay Kit (RN1001, MBL International Corporation) according to the manufacturer’s instructions. RNA concentrations were measured with the NanoDrop system (Thermo Fisher Scientific). RNA obtained from immunoprecipitates was reverse transcribed into first-strand cDNA using ImProm-II Reverse Transcriptase (A3800, Promega). qRT-PCR was performed using the primers listed in Table 3 with KAPA SYBR FAST qPCR Master mix (KK4602, KAPA Biosystems).
Luciferase assay for mouse Cacna1c promoter.
To analyze the putative promoter activity of mouse Cacna1c (CaV1.2, gene ID 12288), a series of reporter plasmid constructs were made using the backbone of the pGL3-Basic reporter vector and pRL-TK vector (E2231, Promega). To clone the 5′-flanking region of mouse Cacna1c as a putative promoter region, the following 2 primers were used: 5′-GTAGAGCTCCACAGTGCCA-3′, which contained a 10-bp SacI DNA fragment (–1542 to –1533), and 5′-GGACTCGAGTGTCTGGTTG-3′, which contained a 10-bp XhoI Cacna1c gene fragment (+257 to +248). The Cacna1c promoter was amplified by PCR from a BAC clone (RP23-123O15, ID 638275; Advanced GenoTechs), and contained a 1799-bp SacI/XhoI insertion (–1542 to +257). Amplified DNA was ligated into the SacI and XhoI sites of the pGL3-Basic-reporter vector (Promega), generating a 1799-bp fragment (–1542 to +257) of the CaV1.2 promoter in pGL3-Basic. The construct was verified by sequencing. MIN6 cells seeded at 9.0 × 103 cells on a 96-well plate were transfected with 100 pmol siRNA (Control, T1), 10 ng pGL3-Basic-CaV1.2 promoter, and 5 ng pRL/SV40 (Promega) using the NEON Transfection System (Thermo Fisher Scientific) as described above. At 48 hours after transfection, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (E1910, Promega) according to the manufacturer’s instructions.
Human CACNA1C promoter reporter assay.
HEK293T cells seeded at 9.0 × 103 cells on a 96-well plate were transfected with 100 pmol siRNA (Control, T1) and 50 ng GoClone plasmid DNA (human CACNA1C promoter, ID S71093) using Lipofectamine 2000 (11668-019, Invitrogen) according to the manufacturer’s instructions. At 48 hours after transfection, luciferase activity was measured using the LightSwitch Assay Reagent (LS010, SwitchGear Genomics) according to the manufacturer’s instructions.
RNA stability.
Actinomycin D (final concentration 10 μg/mL), a potent inhibitor of mRNA synthesis, was added to MIN6 cells (3.0 × 105 cells) in a 12-well plate at 48 hours after electroporation of control or T1 siRNA. Total RNA was extracted at 0, 2, 4, 8, and 12 hours after treatment, and the RNA was then subjected to qRT-PCR as described above. The data are presented as values relative to the expression levels detected at 0 hours of actinomycin D treatment.
In silico analysis.
We extracted the TARDBP ChIP-seq data from the NCBI Gene Expression Omnibus database (ENCSR753GIA, ENCODE phase3, https://www.encodeproject.org/experiments/ENCSR753GIA/). Using the UCSC genome browser, we confirmed the presence of a binding site for TADBP protein in CACNA1C promoter region (human CACNA1C promoter, ID S71093, LightSwitch Assay; Active Motif).
Animals.
All mice (background C57BL/6J) were maintained at 25°C in a light-controlled (12-hour light-dark cycle) environment (3–5 mice per cage) with food and water provided ad libitum. Tardbpfl/fl mice were generated using the Cre/LoxP recombination system as previously described (9). RIP-Cre transgenic mice (B6.Cg-Tg[Ins2-cre]25Mgn/J, 3573) were purchased from The Jackson Laboratory. NOD female 10-week-old mice (NOD/Shi JIC) were purchased from CLEA-Japan.
Recombinant AAV production.
Modified pBluescript II KS/RIP-Cre-ER (DM 265) was purchased from Addgene (plasmid 15029). The AAV vector plasmids contained an expression cassette consisting of a RIP, Cre recombinase, and poly A tail (RIP-Cre), and RIP-GFP between the inverted terminal repeats of the AAV3 genome. The recombinant AAV vectors were produced by transient transfection of HEK293 cells with the vector plasmid, an AAV8 vp expression plasmid, and an adenoviral helper plasmid (pHelper, Agilent Technologies), as previously described (45). The recombinant viruses were purified by isolation from 2 sequential continuous CsCl gradients, and viral titers were determined by qPCR.
Pancreas-specific Tardbp knockout mice.
Male Tardbpfl/fl mice (8–10 weeks old) were injected intraperitoneally with normal saline, 4.0 × 1011 vector genomes (vg)/mouse AAV8-RIP-GFP (AAV-Control), or 4.0 × 1011 vg/mouse AAV8-RIP-Cre (AAV-KO) as Tardbp knockout mice. The IPGTT and insulin test were performed 2 weeks after injection. RIP-Cre mice were purchased from The Jackson Laboratory (strain B6.Cg-Tg[Ins2-cre]25Mgn/J, 3573). Pancreas-specific Tardbp knockout mice were generated by mating Tardbpfl/fl mice with RIP-Cre mice (CKO-TDP). In Tardbpfl/fl and CKO-TDP mice without AAV injection, the IPGTT and insulin test were performed in 6-week-old male mice.
Insulin and glucose tolerance tests in mice.
After fasting for 16 hours (17:00–9:00), 2.0 g/kg bodyweight D-glucose was injected intraperitoneally into the mice. Plasma glucose levels were measured after 0, 15, 30, 60, and 120 minutes using the Glucogard G Sensor (Arkray). Plasma insulin levels were measured after 0 and 15 minutes using a mouse insulin enzyme-linked immunosorbent assay kit (Morinaga Institute of Biological Science).
Immunofluorescence for islets.
Pancreatic islets were excised from mice with perfusion fixation using 4% paraformaldehyde, and fixed immediately in a 10% buffered formalin solution. Sections (3 μm) were deparaffinized and heated in a microwave for 15 minutes in 10 mM citrate buffer (pH 6.0). The following primary and secondary antibodies were the same as used for human samples: anti–TDP-43 rabbit polyclonal, anti-insulin pig, and anti-glucagon mouse.
Pancreatic islet isolation.
Pancreatic islets were isolated from Tardbpfl/fl mice at 8–10 weeks of age by modifying a previously described procedure (46). Briefly, the pancreas of an anesthetized mouse was distended via the pancreatic duct with 3 ml RPMI 1640 (11875119, Thermo Fisher Scientific) containing 1.5 mg/mL collagenase (C7657, Sigma-Aldrich). The distended pancreas was then excised and digested in a water bath at 37°C for 11.5 minutes, and then shaken manually 5 times in sets of 7. The digest was washed 3 times with 25 mL RPMI 1640 containing 10% (vol/vol) FBS (10270, Life Technologies), and then filtered through a tea strainer. The filtered tissue was centrifuged (1000g for 2 minutes), and the pellet resuspended with 10 mL Ficoll-Paque PLUS (17144002, GE Healthcare) in a 50-mL conical tube. Next, it was overlayed slowly with 10 mL RPMI 1640 containing 10% (vol/vol) FBS, which was centrifuged (1000g for 20 minutes at 4°C). Isolated islets appeared at the intermediate phase and were moved on the cell culture dish. The islets were isolated by hand-picking using a pipette with RPMI 1640 containing 10% (vol/vol) FBS, and incubated for 2 hours in 5% CO2 incubator at 37°C.
Infection of isolated islets by AAV.
The isolated islets were cultured with 1.2 × 1011 vg/mL AAV8-RIP-GFP or AAV8-RIP-Cre for 36 hours. Then mRNA levels were measured with qRT-PCR as described above for MIN6 cells, and the insulin secretion of infected islets was measured with an Insulin Ultra-Sensitive Assay kit (62IN2PEG, CisBio) as described above for MIN6 cells. The 10 isolated islets were preincubated on 96-well plate with a 2.8 mmol/L glucose (low glucose) for 30 minutes, and stimulated with 16.7 mmol/L glucose (high glucose) for 30 minutes.
Perfusion experiments of Tardbpfl/fl and CKO-TDP mice.
Perfusion experiments were performed as previously described (47, 48). Briefly, 10- to 12-week-old male mice were fasted overnight (16 hours). The perfusion protocol began with a 10-minute equilibration period with the same buffer used in initial step (1–5 minutes; 2.8 mM glucose) and glucose loading step (5–25 minutes, 16.7 mM glucose) (Figure 6). The insulin levels in the perfusate were measured with an Insulin Ultra-Sensitive Assay kit (62IN2PEG, CisBio).
In situ hybridization.
We purchased 4 probes against mouse Cacna1c (NM_009781.4, bp4846–6605, ACD445451), human CACNA1C (NM_199460.3, bp501–2144, ACD460061), human CACNA1D (NM_000720.3, bp544-1957, ACD544881), and human CACNA1A (NM_023035.2, bp646-1855, made-to-order ACD320269) from Advanced Cell Diagnostics, and applied them to 3-μm sections of paraffin-embedded pancreas samples according to the RNAscope Sample Preparation and Pretreatment Guide for FFPE Tissue Part 1 (catalog 320511). We used the single-plex RNAscope assay according to the RNAscope 2.5 HD Detection Kit (BROWN) User Manual Part 2 (catalog 320497), containing a number of channel 1 probes against mouse Cacna1c, human CACNA1C, human CACNA1D, and human CACNA1A molecules detected with DAB. A semiquantitative scoring scale was used according to the manufacturer’s guidelines to evaluate the staining results: 0, no staining or less than 1 dot per 10 cells; 1, 1–3 dots per cell; 2, 4–9 dots per cell with no or very few dot clusters; and 3, 10–15 dots per cell and fewer than 10% dots are in clusters. Scoring was performed at ×60 magnification.
X-gal staining for Cre recombinase driven by the RIP promoter.
ROSA26 mice were injected intraperitoneally with 4.0 × 1011 vg/mouse AAV8-RIP-Cre (AAV-KO) or mated with RIP-Cre mice (CKO-TDP). Pancreatic islets and brains of male AAV-KO ROSA26 mice and CKO-TDP ROSA26 mice were analyzed at 8–10 weeks of age. The pancreatic islets and brains were excised with perfusion fixation using 4% paraformaldehyde, and fixed immediately in 4% paraformaldehyde for 2 hours. Then, the brains were implanted in 3% gel at 4°C overnight. Horizontal slices of the hippocampus (50 μm) were cut on a vibratome (Leica VT 1200 S) and stained using an X-gal staining kit (MIR 2600; Mirus Bio LLC). After fixation in 4% paraformaldehyde, pancreatic tissues were embedded in Tissue-Tek compound and snap-frozen. Sections (10 μm) were cut with a Leica CM3050 S Cryostat and stained with the same method as for the brains (MIR 2600; Mirus Bio LLC).
Neuron-specific Tardbp knockout mice.
We crossed Tardbpfl/fl mice with CamkII-Cre mice (49) (gift from Tsuyoshi Miyakawa, Fujita Health University, Toyoake, Japan) and generated Tardbpfl/fl/CamKII-Cre+ mice as neuron-specific Tardbp knockout mice. The brains were excised from the neuron-specific Tardbp knockout mice at 12 weeks of age with perfusion fixation using 4% paraformaldehyde, and fixed immediately in a 10% buffered formalin solution. Sections (3 μm) were deparaffinized and heated in a microwave for 15 minutes in 10 mM citrate buffer (pH 6.0). The anti–TDP-43 rabbit polyclonal was the same as for human samples. Horizontal slices of the hippocampus were excised and homogenized in RIPA buffer (182-02451, Wako) containing 1% protease inhibitors (4693116001, Roche) and phosphatase inhibitors (1862495, Thermo Fisher Scientific) with a Nippi Biomasher II homogenizer. Homogenates were incubated on ice for 20 minutes and centrifuged at 13,000g for 10 minutes at 4°C. The lysates were mixed with 4× NuPAGE LDS sample buffer (NP0008, Novex) and heated at 95°C for 5 minutes. After denaturation, each cell lysate was separated by SDS-PAGE (5%–20% gradient gel) and analyzed by immunoblotting with ECL Plus detection reagents (NEL104001EA, PerkinElmer) using the following primary antibodies: anti–TDP-43 rabbit polyclonal (1:3000; 10782-2-AP, Proteintech), GAPDH mouse monoclonal (1:1000; MBL International Corporation), and anti-CaV1.2 rabbit polyclonal (1:2000; ACC-003, Alomone Labs). Band intensity was quantified by Multi Gauge 3.0 software (Fujifilm).
Statistics.
For qRT-PCR and immunoblotting in vitro, the data were obtained from at least 3 independent experiments and are presented as relative value to the control. For the IPGTT and insulin test in vivo, at least 3 animals were in each group. Statistical analysis throughout the study was performed by applying a 2-tailed Student’s t test for 2-group comparison and 1-way ANOVA with Turkey post hoc test for multi-group comparison. In all experiments and for clinical studies, graph and other data are expressed as mean ± SEM. Significance was defined at P less than 0.05 using the SPSS 23.0J statistical software package. In clinical studies, intergroup differences in categorical and continuous variables were assessed using unpaired t test, and correlation coefficients were assessed using the Pearson correlation. For experiments using cell cultures and mice, the number of experiments or animals is stated in the figure legends.
Study approval.
This human study adhered to the ethics guidelines for human genome/gene analysis research and those for medical and health research involving human subjects endorsed by the Japanese government, and was approved by the ethics review committees of Nagoya University Graduate School of Medicine. All participants provided written informed consent. Experimental procedures involving human subjects were conducted according to the principles expressed in the Declaration of Helsinki. Animal protocols were approved by the IACUC at Nagoya University.
Author contributions
MK, KA, and AA contributed to the study concept and design. KA, AA, DH, TI, A Hashizume, YH, SY, YI, A Hara, KI, KK, SI, YN, ST, YS, AY, YT, SH, MT, MOI, AS, HI, AE, MY, HA, SM, GS, and MK conducted experiments and acquired data. KA, AA, MI, and MK drafted the manuscript and figures.
Supplementary Material
Acknowledgments
We thank Mika Ito and Naomi Takino (Jichi Medical University) for their help with the production of the AAV vectors. This work was supported by Grants-in-Aid (KAKENHI) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (grants 16K09742, 16K15480, 17H04195, and 17K08547 to MK; grants 16K09392 and 16K09393 to HI). This work was partly supported by grants from the Japan Agency of Medical Research and Development (AMED) (grants JP17dm0107105h0002 and JP16kk0205009h0001 to MY). This work was also supported by Grants-in-Aid from the Research Committee of CNS Degenerative Diseases, Research on Policy Planning and Evaluation for Rare and Intractable Diseases, Health, Labor and Welfare Sciences Research Grants, the Ministry of Health, Labor and Welfare, Japan (to MY), a grant from the Naito Foundation (to MK), and a grant from the Hori Sciences & Arts Foundation (to MK).
Version 1. 07/29/2019
Electronic publication
Version 2. 09/03/2019
Print issue publication
Footnotes
Conflict of interest: SM owns equity in Gene Therapy Research Institution, a gene therapy company that commercializes the use of AAV vectors for gene therapy applications.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(9):3578–3593.https://doi.org/10.1172/JCI124481.
Contributor Information
Kunihiko Araki, Email: karaki@med.nagoya-u.ac.jp.
Amane Araki, Email: amane@nagoya2.jrc.or.jp.
Daiyu Honda, Email: thisisabook0922@gmail.com.
Takako Izumoto, Email: izumoto@med.nagoya-u.ac.jp.
Atsushi Hashizume, Email: hassy0707@med.nagoya-u.ac.jp.
Yasuhiro Hijikata, Email: hijikata_yasuhiro@yahoo.co.jp.
Shinichiro Yamada, Email: s.yamada@med.nagoya-u.ac.jp.
Yohei Iguchi, Email: iguyo@med.nagoya-u.ac.jp.
Akitoshi Hara, Email: a.hara0621@med.nagoya-u.ac.jp.
Kazuhiro Ikumi, Email: ikumikazuhiro@gmail.com.
Kaori Kawai, Email: kawaik@med.nagoya-u.ac.jp.
Shinsuke Ishigaki, Email: shinsuke.ishigaki@gmail.com.
Yoko Nakamichi, Email: nakamiti@ks.kyorin-u.ac.jp.
Shin Tsunekawa, Email: tsune87@med.nagoya-u.ac.jp.
Yusuke Seino, Email: yusuke@med.nagoya-u.ac.jp.
Akiko Yamamoto, Email: akikoy@htc.nagoya-u.ac.jp.
Yasunori Takayama, Email: takayama@nips.ac.jp.
Shihomi Hidaka, Email: sakai220@fujita-hu.ac.jp.
Makoto Tominaga, Email: tominaga@nips.ac.jp.
Mica Ohara-Imaizumi, Email: mimaizu@ks.kyorin-u.ac.jp.
Atsushi Suzuki, Email: aslapin@fujita-hu.ac.jp.
Hiroshi Ishiguro, Email: ishiguro@htc.nagoya-u.ac.jp.
Atsushi Enomoto, Email: enomoto@iar.nagoya-u.ac.jp.
Mari Yoshida, Email: myoshida@aichi-med-u.ac.jp.
Hiroshi Arima, Email: arima105@med.nagoya-u.ac.jp.
Shin-ichi Muramatsu, Email: muramats@jichi.ac.jp.
Gen Sobue, Email: sobueg@med.nagoya-u.ac.jp.
Masahisa Katsuno, Email: ka2no@med.nagoya-u.ac.jp.
References
- 1.Arai T, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351(3):602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 2.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 3.Wegorzewska I, Baloh RH. TDP-43-based animal models of neurodegeneration: new insights into ALS pathology and pathophysiology. Neurodegener Dis. 2011;8(4):262–274. doi: 10.1159/000321547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scotter EL, Chen HJ, Shaw CE. TDP-43 proteinopathy and ALS: insights into disease mechanisms and therapeutic targets. Neurotherapeutics. 2015;12(2):352–363. doi: 10.1007/s13311-015-0338-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iguchi Y, et al. TDP-43 depletion induces neuronal cell damage through dysregulation of Rho family GTPases. J Biol Chem. 2009;284(33):22059–22066. doi: 10.1074/jbc.M109.012195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu LS, Cheng WC, Shen CK. Targeted depletion of TDP-43 expression in the spinal cord motor neurons leads to the development of amyotrophic lateral sclerosis-like phenotypes in mice. J Biol Chem. 2012;287(33):27335–27344. doi: 10.1074/jbc.M112.359000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iguchi Y, et al. Loss of TDP-43 causes age-dependent progressive motor neuron degeneration. Brain. 2013;136(Pt 5):1371–1382. doi: 10.1093/brain/awt029. [DOI] [PubMed] [Google Scholar]
- 10.Yang C, et al. Partial loss of TDP-43 function causes phenotypes of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2014;111(12):E1121–E1129. doi: 10.1073/pnas.1322641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14(4):459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor JP, Brown RH, Cleveland DW. Decoding ALS: from genes to mechanism. Nature. 2016;539(7628):197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupuis L, Pradat PF, Ludolph AC, Loeffler JP. Energy metabolism in amyotrophic lateral sclerosis. Lancet Neurol. 2011;10(1):75–82. doi: 10.1016/S1474-4422(10)70224-6. [DOI] [PubMed] [Google Scholar]
- 14.Buratti E, Dörk T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20(7):1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quick DT, Greer M. Pancreatic dysfunction in patients with amyotrophic lateral sclerosis. Neurology. 1967;17(2):112–116. doi: 10.1212/WNL.17.2.112. [DOI] [PubMed] [Google Scholar]
- 16.Goto F, Kitamura A, Koto A, Kataoka K, Atsuji H. Abnormal insulin secretion in amyotrophic lateral sclerosis. J Neurol Sci. 1972;16(2):201–207. doi: 10.1016/0022-510X(72)90089-5. [DOI] [PubMed] [Google Scholar]
- 17.Reyes ET, Perurena OH, Festoff BW, Jorgensen R, Moore WV. Insulin resistance in amyotrophic lateral sclerosis. J Neurol Sci. 1984;63(3):317–324. doi: 10.1016/0022-510X(84)90154-0. [DOI] [PubMed] [Google Scholar]
- 18.Pradat PF, et al. Impaired glucose tolerance in patients with amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2010;11(1-2):166–171. doi: 10.3109/17482960902822960. [DOI] [PubMed] [Google Scholar]
- 19.Song J, Xu Y, Hu X, Choi B, Tong Q. Brain expression of Cre recombinase driven by pancreas-specific promoters. Genesis. 2010;48(11):628–634. doi: 10.1002/dvg.20672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulla V, et al. Impaired insulin secretion and glucose tolerance in beta cell-selective Ca(v)1.2 Ca2+ channel null mice. EMBO J. 2003;22(15):3844–3854. doi: 10.1093/emboj/cdg389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor JT, Huang L, Keyser BM, Zhuang H, Clarkson CW, Li M. Role of high-voltage-activated calcium channels in glucose-regulated beta-cell calcium homeostasis and insulin release. Am J Physiol Endocrinol Metab. 2005;289(5):E900–E908. doi: 10.1152/ajpendo.00101.2005. [DOI] [PubMed] [Google Scholar]
- 22.Sephton CF, et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem. 2011;286(2):1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heyburn L, Moussa CE. TDP-43 overexpression impairs presynaptic integrity. Neural Regen Res. 2016;11(12):1910–1911. doi: 10.4103/1673-5374.195272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miskiewicz K, et al. HDAC6 is a Bruchpilot deacetylase that facilitates neurotransmitter release. Cell Rep. 2014;8(1):94–102. doi: 10.1016/j.celrep.2014.05.051. [DOI] [PubMed] [Google Scholar]
- 25.Armstrong GA, Drapeau P. Calcium channel agonists protect against neuromuscular dysfunction in a genetic model of TDP-43 mutation in ALS. J Neurosci. 2013;33(4):1741–1752. doi: 10.1523/JNEUROSCI.4003-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang JC, Hazelett DJ, Stewart JA, Morton DB. Motor neuron expression of the voltage-gated calcium channel cacophony restores locomotion defects in a Drosophila, TDP-43 loss of function model of ALS. Brain Res. 2014;1584:39–51. doi: 10.1016/j.brainres.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Zundert B, Izaurieta P, Fritz E, Alvarez FJ. Early pathogenesis in the adult-onset neurodegenerative disease amyotrophic lateral sclerosis. J Cell Biochem. 2012;113(11):3301–3312. doi: 10.1002/jcb.24234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saffer D, Morley J, Bill PL. Carbohydrate metabolism in motor neurone disease. J Neurol Neurosurg Psychiatry. 1977;40(6):533–537. doi: 10.1136/jnnp.40.6.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perurena OH, Festoff BW. Reduction in insulin receptors in amyotrophic lateral sclerosis correlates with reduced insulin sensitivity. Neurology. 1987;37(8):1375–1379. doi: 10.1212/WNL.37.8.1375. [DOI] [PubMed] [Google Scholar]
- 30.Nakano T, Windrem M, Zappavigna V, Goldman SA. Identification of a conserved 125 base-pair Hb9 enhancer that specifies gene expression to spinal motor neurons. Dev Biol. 2005;283(2):474–485. doi: 10.1016/j.ydbio.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 31.Lee H, et al. Slit and Semaphorin signaling governed by Islet transcription factors positions motor neuron somata within the neural tube. Exp Neurol. 2015;269:17–27. doi: 10.1016/j.expneurol.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips DI, Clark PM, Hales CN, Osmond C. Understanding oral glucose tolerance: comparison of glucose or insulin measurements during the oral glucose tolerance test with specific measurements of insulin resistance and insulin secretion. Diabet Med. 1994;11(3):286–292. doi: 10.1111/j.1464-5491.1994.tb00273.x. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto K, et al. Glucose tolerance, insulin secretion, and insulin sensitivity in nonobese and obese Japanese subjects. Diabetes Care. 1997;20(10):1562–1568. doi: 10.2337/diacare.20.10.1562. [DOI] [PubMed] [Google Scholar]
- 34.Matsuda A, Kuzuya T. The prevalence of low insulin responders to oral glucose load among groups with various patterns of family history of diabetes. Diabet Med. 1996;13(9 Suppl 6):S59–S62. [PubMed] [Google Scholar]
- 35.Funakoshi S, et al. Utility of indices using C-peptide levels for indication of insulin therapy to achieve good glycemic control in Japanese patients with type 2 diabetes. J Diabetes Investig. 2011;2(4):297–303. doi: 10.1111/j.2040-1124.2010.00096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 37.Hijikata Y, et al. Impaired muscle uptake of creatine in spinal and bulbar muscular atrophy. Ann Clin Transl Neurol. 2016;3(7):537–546. doi: 10.1002/acn3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Udagawa T, et al. FUS regulates AMPA receptor function and FTLD/ALS-associated behaviour via GluA1 mRNA stabilization. Nat Commun. 2015;6:7098. doi: 10.1038/ncomms8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokoi S, et al. 3’UTR length-dependent control of SynGAP isoform α2 mRNA by FUS and ELAV-like proteins promotes dendritic spine maturation and cognitive function. Cell Rep. 2017;20(13):3071–3084. doi: 10.1016/j.celrep.2017.08.100. [DOI] [PubMed] [Google Scholar]
- 40.Ohara-Imaizumi M, et al. ELKS, a protein structurally related to the active zone-associated protein CAST, is expressed in pancreatic beta cells and functions in insulin exocytosis: interaction of ELKS with exocytotic machinery analyzed by total internal reflection fluorescence microscopy. Mol Biol Cell. 2005;16(7):3289–3300. doi: 10.1091/mbc.e04-09-0816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohara-Imaizumi M, Nakamichi Y, Tanaka T, Ishida H, Nagamatsu S. Imaging exocytosis of single insulin secretory granules with evanescent wave microscopy: distinct behavior of granule motion in biphasic insulin release. J Biol Chem. 2002;277(6):3805–3808. doi: 10.1074/jbc.C100712200. [DOI] [PubMed] [Google Scholar]
- 42.Aoyagi K, Ohara-Imaizumi M, Nishiwaki C, Nakamichi Y, Nagamatsu S. Insulin/phosphoinositide 3-kinase pathway accelerates the glucose-induced first-phase insulin secretion through TrpV2 recruitment in pancreatic β-cells. Biochem J. 2010;432(2):375–386. doi: 10.1042/BJ20100864. [DOI] [PubMed] [Google Scholar]
- 43.Ohara-Imaizumi M, et al. Imaging analysis reveals mechanistic differences between first- and second-phase insulin exocytosis. J Cell Biol. 2007;177(4):695–705. doi: 10.1083/jcb.200608132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto A, et al. Ethanol induces fluid hypersecretion from guinea-pig pancreatic duct cells. J Physiol (Lond) 2003;551(Pt 3):917–926. doi: 10.1113/jphysiol.2003.048827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li XG, et al. Viral-mediated temporally controlled dopamine production in a rat model of Parkinson disease. Mol Ther. 2006;13(1):160–166. doi: 10.1016/j.ymthe.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 46.Rhodes CJ, Halban PA. Newly synthesized proinsulin/insulin and stored insulin are released from pancreatic B cells predominantly via a regulated, rather than a constitutive, pathway. J Cell Biol. 1987;105(1):145–153. doi: 10.1083/jcb.105.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miki T, et al. Distinct effects of glucose-dependent insulinotropic polypeptide and glucagon-like peptide-1 on insulin secretion and gut motility. Diabetes. 2005;54(4):1056–1063. doi: 10.2337/diabetes.54.4.1056. [DOI] [PubMed] [Google Scholar]
- 48.Shibasaki T, et al. Essential role of Epac2/Rap1 signaling in regulation of insulin granule dynamics by cAMP. Proc Natl Acad Sci U S A. 2007;104(49):19333–19338. doi: 10.1073/pnas.0707054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsien JZ, Huerta PT, Tonegawa S. The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell. 1996;87(7):1327–1338. doi: 10.1016/S0092-8674(00)81827-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.