The short-term adaptive response to environmental cues greatly contributes to the ecological success of bacteria, and profound alterations in bacterial gene expression occur in response to physical, chemical, and nutritional stresses. Bacteria belonging to the Acinetobacter genus are ubiquitous inhabitants of soil and water though some species, such as Acinetobacter baumannii, are pathogenic and cause serious concern due to antibiotic resistance. Understanding A. baumannii pathobiology requires adequate genetic tools for gene expression analysis, and to this end we developed user-friendly shuttle vectors to probe the transcriptional responses to different environmental stresses. Vectors were constructed to overcome the problem of antibiotic selection in multidrug-resistant strains and were equipped with suitable reporter systems to facilitate signal detection. By means of these vectors, the transcriptional response of A. baumannii to DNA damage, ethanol exposure, and iron starvation was investigated both in vitro and in vivo, providing insights into A. baumannii adaptation during stress and infection.
KEYWORDS: Acinetobacter, GFP, Galleria mellonella, gene expression, in vivo monitoring, luciferase, multidrug resistance, promoter-probe, vector
ABSTRACT
The Acinetobacter genus includes species of opportunistic pathogens and harmless saprophytes. The type species, Acinetobacter baumannii, is a nosocomial pathogen renowned for being multidrug resistant (MDR). Despite the clinical relevance of infections caused by MDR A. baumannii and a few other Acinetobacter spp., the regulation of their pathogenicity remains elusive due to the scarcity of adequate genetic tools, including vectors for gene expression analysis. Here, we report the generation and testing of a series of Escherichia coli-Acinetobacter promoter-probe vectors suitable for gene expression analysis in Acinetobacter spp. These vectors, named pLPV1Z, pLPV2Z, and pLPV3Z, carry both gentamicin and zeocin resistance markers and contain lux, lacZ, and green fluorescent protein (GFP) reporter systems downstream of an extended polylinker, respectively. The presence of a toxin-antitoxin gene system and the high copy number allow pLPV plasmids to be stably maintained even without antibiotic selection. The pLPV plasmids can easily be introduced by electroporation into MDR A. baumannii belonging to the major international lineages as well as into species of the Acinetobacter calcoaceticus-A. baumannii complex. The pLPV vectors have successfully been employed to study the regulation of stress-responsive A. baumannii promoters, including the DNA damage-inducible uvrABC promoter, the ethanol-inducible adhP and yahK promoters, and the iron-repressible promoter of the acinetobactin siderophore biosynthesis gene basA. A lux-tagged A. baumannii ATCC 19606T strain, carrying the iron-responsive pLPV1Z::PbasA promoter fusion, allowed in vivo and ex vivo monitoring of the bacterial burden in the Galleria mellonella infection model.
IMPORTANCE The short-term adaptive response to environmental cues greatly contributes to the ecological success of bacteria, and profound alterations in bacterial gene expression occur in response to physical, chemical, and nutritional stresses. Bacteria belonging to the Acinetobacter genus are ubiquitous inhabitants of soil and water though some species, such as Acinetobacter baumannii, are pathogenic and cause serious concern due to antibiotic resistance. Understanding A. baumannii pathobiology requires adequate genetic tools for gene expression analysis, and to this end we developed user-friendly shuttle vectors to probe the transcriptional responses to different environmental stresses. Vectors were constructed to overcome the problem of antibiotic selection in multidrug-resistant strains and were equipped with suitable reporter systems to facilitate signal detection. By means of these vectors, the transcriptional response of A. baumannii to DNA damage, ethanol exposure, and iron starvation was investigated both in vitro and in vivo, providing insights into A. baumannii adaptation during stress and infection.
INTRODUCTION
Acinetobacter is a highly diverse bacterial genus including species which are often implicated in hospital-acquired infections (e.g., Acinetobacter baumannii, Acinetobacter dijkshoorniae, Acinetobacter nosocomialis, Acinetobacter pittii, and Acinetobacter seifertii) as well as species (e.g., Acinetobacter baylyi) which behave as environmental saprophytes and are rarely or never associated with human diseases (1). A. baumannii is the most dreaded species, showing an increasing prevalence of multidrug-resistant (MDR) (2), extensively drug-resistant (XDR) (3–5), and pandrug-resistant (PDR) (6, 7) strains, especially in health care-associated outbreaks and, occasionally, in community-acquired infections. The combination of antibiotic resistance and hypervirulent phenotypes (8) has become a major public health concern, and the World Health Organization has urged research to focus on pathogenic MDR Acinetobacter spp. as a top priority (9). It should be emphasized, however, that most Acinetobacter spp. are widely distributed in nature and commonly occur in soil and water. Some species are endowed with biotechnological potential, e.g., bioremediation of inorganic pollutants and hydrocarbons, as well as production of biopolymers, biosurfactants, and bioactive compounds (reviewed in reference 10). Despite the medical and biotechnological relevance of Acinetobacter spp., few tools for gene expression analysis are available for this genus. Real-time quantitative PCR (RT-qPCR) and comparative transcriptomics (e.g., RNA-sequencing) are in current use but have some limitations mainly related to problematic and time-consuming sample preparation which complicates time course analysis of gene expression (11–15). Integrative genetic systems based on mini-Tn7 single-copy insertion vectors have recently been employed for fluorescent protein tagging in A. baumannii, but they were not yet developed for chromosomal reporter gene fusions (16). Two promoter-probe vectors have occasionally been used for gene expression analysis in Acinetobacter spp., namely, the psychrophilic species-restricted vector pBAP1 (17) and the low-copy-number broad-host-range lacZ-based pMP220 plasmid (18). Although pMP220 made it possible to investigate iron and ethanol regulation in A. baumannii (14, 19, 20), it suffers from major limitations due to complexity of manipulation, an inadequate resistance marker (tetA) for selection in MDR Acinetobacter strains, and poor signal output. A new set of Escherichia coli-Acinetobacter species shuttle vectors has recently been generated, carrying suitable zeocin (Zeo; ble) and/or gentamicin (Gm; aacC1) resistance cassettes, and used as gene cloning vehicles in MDR, XDR, and PDR Acinetobacter spp. (21, 22). When supplied at supraphysiological concentrations, gentamicin can indeed overcome resistance displayed by clinically resistant strains (22), and zeocin resistance has never been detected in Acinetobacter spp. (21), thus ensuring in vitro selection of these markers, even upon transformation of PDR strains.
In this work, we optimized the scaffold of recently developed Escherichia coli-Acinetobacter species shuttle vectors (22) in order to generate novel promoter-probe plasmids, called pLPV, suitable for gene expression analysis in MDR, XDR, and PDR Acinetobacter spp. Luciferase (luxCDABE), β-galactosidase (lacZ), and a fast-turnover variant of the green fluorescent protein (GFPmut3) (23) were used as reporter systems. Here, the newly generated pLPV vectors have successfully been employed to investigate the transcriptional response of A. baumannii to physical (UV light), chemical, (mitomycin C [MMC]; ethanol), or nutritional (iron depletion) stress.
RESULTS
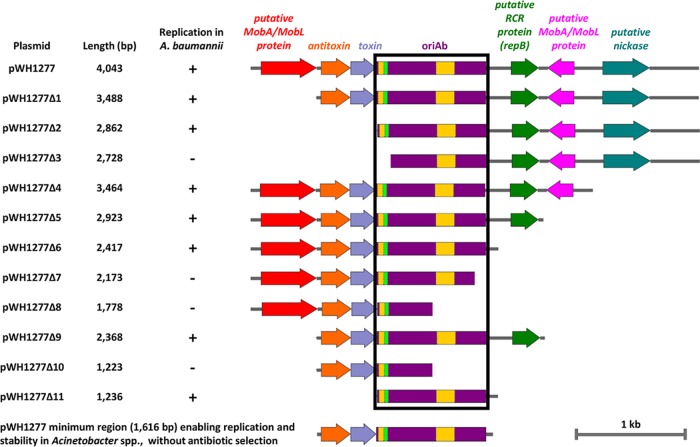
Determination of the minimal self-replicating region of pWH1277.
pWH1277 is a narrow-host-range, cryptic plasmid isolated from Acinetobacter calcoaceticus BD413, which was previously used as a scaffold for the construction of E. coli-Acinetobacter species shuttle vectors, in conjunction with a ColE1-like origin of replication (22, 24). A physical map of pWH1277 with predicted functions of annotated open reading frames (ORFs) (22) is shown in Fig. 1. To minimize the promoter-probe E. coli-Acinetobacter species shuttle vectors, the shortest DNA region enabling self-replication and autonomous maintenance of pWH1277 in A. baumannii was mapped by deletion analysis. Overlapping DNA fragments encompassing the putative replication origin of pWH1277 were generated by PCR with the primer pairs listed in Table 1, and the resulting amplicons were blunt cloned at the SmaI site of pBluescript II KS (pBS) for transformation of E. coli DH5α. After sequence verification of cloned inserts, all constructs (designated pWH1277Δ1 to pWH1277Δ11) were individually transferred by electroporation in A. baumannii ATCC 19606T to assess self-replication. Genes encoding putative MobA/MobL, nickase, RepA, and toxin-antotoxin (TA) functions could be deleted from pWH1277 without affecting the replication of recombinant constructs in A. baumannii. Conversely, short deletions affecting a 1,236-nucleotide (nt) region, designated oriAb (as in pWH1277Δ3, pWH1277Δ7, pWH1277Δ8, and pWH1277Δ10) (Fig. 1), compromised plasmid replication in A. baumannii. Interestingly, the oriAb region includes a palindromic sequence, 11 iterons with the AAAAAATAT consensus and two A+T-rich regions (22, 24) (Fig. 1), which could play a role in plasmid replication (25).
FIG 1.
Deletion analysis of pWH1277 to determine the minimal region required for autonomous plasmid replication in Acinetobacter spp. Deletion fragments of pWH1277 (24) were generated by PCR amplification with primers listed in Table 1 and blunt cloned at the SmaI site of pBS. The resulting pWH1277 deletion derivatives were introduced in A. baumannii ATCC 19606T to assess replication. The minimal self-replicating region of pWH1277 (oriAb) is boxed in black. Relevant coding regions are indicated with colors: red and pink, two putative genes coding MobA/MobL-like proteins, presumably involved in plasmid mobilization; orange, the PaaA2-like antitoxin component of the TA system involved in plasmid stability; cobalt, the ParE2-like toxin component of the TA system involved in plasmid stability; violet, the oriAb origin of replication for Acinetobacter spp., including a palindromic sequence (fluorescent green) and two A+T-rich regions (yellow); green, the gene coding for a putative RepB-like protein, presumably involved in rolling-circle replication (RCR); cyan, the gene encoding a putative nickase, likely involved in single-strand DNA nicking for rolling-circle replication. All genes are reported in scale over the total length of the vector. Images were obtained by the Snapgene software (GSL Biotech).
TABLE 1.
Primers used in this study
| Primer | RSa | Sequence (5′–3′)b | Use |
|---|---|---|---|
| Ext_FW | GAAAACAGGTTGGTTTAGTGC | Deletion of pWH1277-derived portion of pVRL1 | |
| ΔMobA/MobL_FW | TGACCAAGAACAGCCTAAGC | Deletion of pWH1277-derived portion of pVRL1 | |
| ΔMobA/MobL + TA_FW | CATCGCAGGAGTGGTCATG | Deletion of pWH1277-derived portion of pVRL1 | |
| ΔMobA/MobL + TA + oriAb A+T region_FW | GGTACAGATCTTCGATACTGA | Deletion of pWH1277-derived portion of pVRL1 | |
| Δnickase + MobA/MobL + RCR + oriAb A+T region_RV | ACCAGAGGGAGGGAGCAG | Deletion of pWH1277-derived portion of pVRL1 | |
| Δnickase + MobA/MobL + RCR + oriAb partial_RV | CAAATGGTGAGGTTTTAGAGG | Deletion of pWH1277-derived portion of pVRL1 | |
| Δnickase + MobA/MobL + RCR_RV | GTAACGCTAAATTAATTACCCTG | Deletion of pWH1277-derived portion of pVRL1 | |
| Δnickase + MobA/MobL_RV | TTTCATAGGCATTAGTAGTTATAA | Deletion of pWH1277-derived portion of pVRL1 | |
| Δnickase_RV | GTTTATTAACAGCCGCCCCA | Deletion of pWH1277-derived portion of pVRL1 | |
| Ext_RV | ATGGGTTTCTTTTCATCACTTC | Deletion of pWH1277-derived portion of pVRL1 | |
| Scaffold plasmid_FW | NsiI | CCAATGCATCAGCAGCAACAGCAGCAG | Amplification of ColE1-like origin, aacC1 cassette, MCS, and lacZα from pVRL1 |
| Scaffold plasmid_RV | NsiI | CCAATGCATCAGCAGCAACAGCAGCAG | Amplification of ColE1-like origin, aacC1 cassette, MCS, and lacZα from pVRL1 |
| oriAb + TA_FW | NsiI | CCAATGCATACCAAGAACAGCCTAAGCCG | Amplification of the minimal self-replicating region (oriAb) and the TA system from pVRL1 |
| oriAb + TA_RV | NsiI | CCAATGCATCAAATGGTGAGGTTTTAGAGG | Amplification of the minimal self-replicating region (oriAb) and the TA system from pVRL1 |
| Δlac_FW | TGGGCGCTCTTCCGCTTC | Deletion of the pAb region encompassing lac promoter, CAP binding site, and the first eight codons of lacZα | |
| Δlac_RV | GCAATTAACCCTCACTAAAGGGAAC | Deletion of the pAb region encompassing lac promoter, CAP binding site, and the first eight codons of lacZα | |
| lux trascr_FW | SacI | ACCGAGCTCTAAGTAAGTAAAGAGGAGAAATTAAGCATGACTAAAAAAATTTCATTCATTATT | Amplification of luxCDABE operon from mini-CTX-lux for pLPV1 plasmid construction; this oligonucleotide presents stop codons in the three possible reading frames (5′-TAAGTAAGTAA-3′) and a canonical RBS (5′-AAAGAGGAGAAA-3′) |
| lux_RV | NcoI | CATGCCATGGAGCATTCCACTTACAATTAGGC | Amplification of luxCDABE operon from mini-CTX-lux for pLPV1 plasmid construction |
| lacZ trascr_FW | NotI | ATTAGCGGCCGCTAAGTAAGTAAAGAGGAGAAATTAAGCATGACCATGATTACGGATTCAC | Amplification of lacZ from pVRL2lacZ for pLPV2 plasmid construction; this oligonucleotide presents stop codons in the three possible reading frames (5′-TAAGTAAGTAA-3′) and a canonical RBS (5′-AAAGAGGAGAAA-3′) |
| lacZ_RV | NcoI | CATGCCATGGAGTTATTTTTGACACCAGACCAA | Amplification of lacZ from pVRL2lacZ for pLPV2 plasmid construction |
| GFP trascr_FW | NotI | ATTAGCGGCCGCTAAGTAAGTAATTAAAGAGGAGAAATTAAGCATG | Amplification of GFP gene from pVRL2gfp for pLPV3 plasmid construction; this oligonucleotide presents stop codons in the three possible reading frames (5′-TAAGTAAGTAA-3′) and a canonical RBS (5′-AAAGAGGAGAAA-3′) |
| GFP_RV | SacI | ACCGAGCTCAAGCTAGCTTGGATTCTCACC | Amplification of GFP gene from pVRL2gfp for pLPV3 plasmid construction |
| Zeo NcoI_FW | NcoI | CATGCCATGGTTCTCCTTACGCATCTGTGC | Amplification of ble cassette in order to construct pLPV1Z and pLPV2Z vectors |
| Zeo NcoI_RV | NcoI | CATGCCATGGTCATTGAGACGAGCAACAGAG | Amplification of ble cassette in order to construct pLPV1Z and pLPV2Z vectors |
| Zeo SacI_FW | SacI | ACCGAGCTCTTCTCCTTACGCATCTGTGC | Amplification of ble cassette in order to construct pLPV3Z vector |
| Zeo SacI_RV | SacI | ACCGAGCTCTCATTGAGACGAGCAACAGAG | Amplification of ble cassette in order to construct pLPV3Z vector |
| dxs Ab_FW | AGTTTGGGATGTGGGACACC | Determination of pLPV2Z vector copy number in A. baumannii; amplification from nt 195274 to nt 195387 encompassing an internal fragment (113 bp) of gene HMPREF0010_01955 | |
| dxs Ab_RV | CTTCTCTGGCTGGGAAAGCA | Determination of pLPV2Z vector copy number in A. baumannii; amplification from nt 195274 to nt 195387 encompassing an internal fragment (113 bp) of gene HMPREF0010_01955 | |
| dxs Ec_FW | CGAGAAACTGGCGATCCTTA | Determination of pLPV2Z vector copy number in E. coli; amplification from nt 436807 to nt 436694 encompassing an internal fragment (113 bp) of gene G6237 | |
| dxs Ec_RV | CTTCATCAAGCGGTTTCACA | Determination of pLPV2Z vector copy number in E. coli; amplification from nt 436807 to nt 436694 encompassing an internal fragment (113 bp) of gene G6237 | |
| aacC1 PCN FW | GATCTATATCTATGATCTCGC | Determination of pLPV2Z copy number in E. coli and A. baumannii | |
| aacC1 PCN RV | GATCACATAAGCACCAAGCG | Determination of pLPV2Z copy number in E. coli and A. baumannii | |
| PuvrA_FW | PstI | AAAACTGCAGAAGGGCATGGGCACTTTGC | Amplification and cloning of the A. baumannii DNA damage-inducible promoter region of the uvrA operon (A1S_3295) |
| PuvrA_RV | XbaI | CTAGTCTAGAAACATCTCAATTGTGTATTGATG | Amplification and cloning of the A. baumannii DNA damage-inducible promoter region of the uvrA operon (A1S_3295) |
| Petdh_FW | PstI | AAAACTGCAGTCAGGCTTCAGCCAAGGCC | Amplification and cloning of the ethanol-inducible promoter region of the A. baumannii gene encoding the ethanol dehydrogenase enzyme (HMPREF0010_01803) |
| Petdh_RV | XbaI | CTAGTCTAGAGAGTTTGATCATAACGTAAGC | Amplification and cloning of the ethanol-inducible promoter region of the A. baumannii gene encoding the ethanol dehydrogenase enzyme (HMPREF0010_01803) |
| Padh_FW | PstI | AAAACTGCAGCGGCAGAAGGACAAACGACA | Amplification and cloning of the ethanol-inducible promoter region of the A. baumannii gene encoding the aldehyde dehydrogenase enzyme (HMPREF0010_03394) |
| Padh_RV | XbaI | CTAGTCTAGATTAACTTGAGGTGCATTTTGCTT | Amplification and cloning of the ethanol-inducible promoter region of the A. baumannii gene encoding the aldehyde dehydrogenase enzyme (HMPREF0010_03394) |
| PbasA_FW | PstI | AAAACTGCAGAAGCCCGATTTTTGTACCCAC | Amplification and cloning of the A. baumannii promoter region of the iron-regulated basA gene (GI27-2634) |
| PbasA_RV | XbaI | CTAGTCTAGAGAGTCCCTACCTCAGCATAA | Amplification and cloning of the A. baumannii promoter region of the iron-regulated basA gene (GI27-2634) |
RS, restriction site in primer sequence.
GC clamps are shown in bold; restriction sites are underlined.
Although the TA system could be deleted without affecting replication in A. baumannii (as in pWH1277Δ2 and pWH1277Δ11) (Fig. 1), it was previously demonstrated that this module is essential for plasmid stability, i.e., maintenance without antibiotic selection (22). Since exposure to elevated antibiotic concentrations, such as those required for plasmid selection, can have pleiotropic effects on bacterial gene expression (26, 27), the 1,616-bp DNA fragment encompassing both oriAb and the TA gene system was regarded as the minimal region enabling plasmid replication and stability in Acinetobacter (Fig. 1).
Assembly of promoter-probe E. coli-Acinetobacter species shuttle vectors.
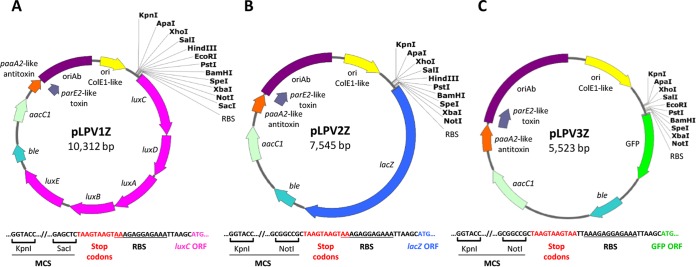
E. coli-Acinetobacter species shuttle vectors for gene expression analysis were generated by a stepwise gene cloning approach, as summarized in Fig. S1 in the supplemental material. Primers containing suitable restriction sites and template plasmids employed for PCR-based construction of the promoter-probe vectors are described in Tables 1 and 2, respectively. Using pVRL1 as the template and NsiI sites provided by the primers, the 1,634-bp region containing the oriAb and the TA modules was ligated to the 2,643-bp region encompassing the ColE1-like origin of replication, the aacC1 gene, and the multiple cloning site (MCS) within the lacZα gene fragment, generating pAb (Fig. S1B). Then, the 249-bp region containing the lac promoter (Plac), the CAP binding site, and the first 24 codons of lacZα was deleted to generate pAbΔlac (Fig. S1C). Deletion of this region was needed to avoid transcriptional interference by Plac (28) and formation of chimeric proteins between LacZα and the cloned gene products (29). Then, using appropriate primer pairs with terminal restriction sites, three different reporter gene systems were individually cloned in the distal sites of the pAbΔlac MCS, as follows: (i) the entire luxCDABE operon (abbreviated lux) from miniCTX1-lux (30), cloned at the SacI/NcoI sites to generate pLPV1 (Fig. S1D); (ii) the lacZ gene from pVRL2lacZ (22), cloned at the NotI/NcoI sites to generate pLPV2 (Fig. S1E); (iii) the GFPmut3 gene from pVRL2gfp (22), cloned at the NotI/SacI sites to generate pLPV3 (Fig. S1F). Of note, forward primers used for the amplification of the three reporter gene systems contained triplicate stop codons for all reading frames (5′-TAAGTAAGTAA-3′) to prevent possible fusion between products of cloned inserts and reporter proteins. Moreover, a canonical ribosome binding site (RBS; 5′-AAAGAGGAGAAA-3′) (29) has been placed 6 nt upstream of the start codon of each reporter gene to maximize translation efficiency (29).
TABLE 2.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Source and/or reference |
|---|---|---|
| Strains | ||
| A. baumannii | ||
| ATCC 19606T | Clinical isolate, type strain, antibiotic susceptible | 67 |
| ATCC 17978 | Clinical isolate, antibiotic susceptible | 39 |
| ACICU | MDR clinical isolate, prototype of the international clonal lineage II | 68 |
| AYE | MDR clinical isolate, prototype of the international clonal lineage I | 31 |
| A. baylyi BD413 | Naturally transformable strain | 69, 70 |
| A. dijkshoorniae 271 | Member of the ACB complex | Seifert collection (71) |
| A. nosocomialis UKK_0361 | Member of the ACB complex | Seifert collection (22) |
| A. pittii UKK_0145 | Member of the ACB complex | Seifert collection (22) |
| A. seifertii HS A23-2 | Member of the ACB complex | Seifert collection (72) |
| E. coli | ||
| DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 Nalr | 63 |
| H1717 | fhuF::λplacMu aroB araD139 ΔlacU169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR Kmr | 44 |
| Plasmids | ||
| pBluescript-II KS | E. coli cloning vector (GenBank accession number X52327.1), Apr | 73 |
| mini-CTX-lux | Integration-proficient Pseudomonas aeruginosa vector for chromosomal lux gene fusions, source of lux operon, Tcr | 30 |
| pVRL1 | E. coli-Acinetobacter species shuttle cloning vector, source of ColE1-like and oriAb origins of replication, aacC1 and TA cassette, MCS, Gmr | 22 |
| pVRL2lacZ | E. coli-Acinetobacter species shuttle plasmid for arabinose-regulated lacZ expression, source of lacZ reporter gene, Gmr | 22 |
| pVRL2gfp | E. coli-Acinetobacter species shuttle plasmid for arabinose-regulated GFP expression, source of GFP reporter gene, Gmr | 22 |
| pVRL1Z | pVRL1-derived vector carrying the ble gene, source of ble cassette, Zeor | 22 |
| pMP220 | Broad-host range, low copy number promoter-probe vector based on the lacZ reporter gene, Tcr | 18 |
| pMP220::PbasA | A. baumannii PbasA promoter cloned into pMP220, Tcr | 19 |
| pBS::PbasA | A. baumannii PbasA promoter cloned into pBluescript-II KS (pBS), Apr | 19 |
| pWH1277 | Full length pWH1277 ligated to pBluescript-II KS plasmid, Apr | This work |
| pWH1277-Δ1 | Deletion derivative of pWH1277 cloned into pBluescript-II KS, Apr | This work |
| pWH1277-Δ2 | Deletion derivative of pWH1277 cloned into pBluescript-II KS, Apr | This work |
| pWH1277-Δ3 | Deletion derivative of pWH1277 cloned into pBluescript-II KS, Apr | This work |
| pWH1277-Δ4 | Deletion derivative of pWH1277 cloned into pBluescript-II KS, Apr | This work |
| pWH1277-Δ5 | Deletion derivative of pWH1277 cloned into pBluescript-II KS, Apr | This work |
| pWH1277-Δ6 | Deletion derivative of pWH1277 cloned into pBluescript-II KS, Apr | This work |
| pWH1277-Δ7 | Deletion derivative of pWH1277 cloned into pBluescript-II KS, Apr | This work |
| pWH1277-Δ8 | Deletion derivative of pWH1277 cloned into pBluescript-II KS, Apr | This work |
| pWH1277-Δ9 | Deletion derivative of pWH1277 cloned into pBluescript-II KS, Apr | This work |
| pWH1277-Δ10 | Deletion derivative of pWH1277 cloned into pBluescript-II KS, Apr | This work |
| pWH1277-Δ11 | Deletion derivative of pWH1277 cloned into pBluescript-II KS, Apr | This work |
| pAb | E. coli-Acinetobacter species shuttle plasmid employed as scaffold vector for pLPV promoter-probe plasmid construction, Gmr | This work |
| pAbΔlac | pAb plasmid carrying a deletion of CAP binding site, lac promoter (Plac) and the first 24 codons of lacZα, Gmr | This work |
| pLPV1 | lux-based promoter-probe plasmid, Gmr | This work |
| pLPV2 | lacZ-based promoter-probe plasmid, Gmr | This work |
| pLPV3 | GFP-based promoter-probe plasmid, Gmr | This work |
| pLPV1Z | pLPV1-derived vector carrying the ble cassette, Gmr Zeor | This work |
| pLPV2Z | pLPV2-derived vector carrying the ble cassette, Gmr Zeor | This work |
| pLPV3Z | pLPV3-derived vector carrying the ble cassette, Gmr Zeor | This work |
| pLPV1Z::PuvrA | PuvrA promoter cloned in pLPV1Z, Gmr Zeor | This work |
| pLPV2Z::PuvrA | PuvrA promoter cloned in pLPV2Z, Gmr Zeor | This work |
| pLPV3Z::PuvrA | PuvrA promoter cloned in pLPV3Z, Gmr Zeor | This work |
| pLPV1Z::PadhP | PadhP promoter cloned in pLPV1Z, Gmr Zeor | This work |
| pLPV1Z::PyahK | PyahK promoter cloned in pLPV1Z, Gmr Zeor | This work |
| pLPV1Z::PbasA | PbasA promoter cloned in pLPV1Z, Gmr Zeor | This work |
| pLPV2Z::PbasA | PbasA promoter cloned in pLPV2Z, Gmr Zeor | This work |
| pLPV3Z::PbasA | PbasA promoter cloned in pLPV3Z, Gmr Zeor | This work |
Nalr, nalidixic acid resistant; Kmr, kanamycin resistant; Apr, ampicillin resistant; Tcr tetracycline resistant; Gmr, gentamicin resistant; Zeor, zeocin resistant; ACB, Acinetobacter calcoaceticus-Acinetobacter baumannii.
We have previously reported that plasmid pVRL1, which served as a scaffold for the generation of all pLPV plasmids, can be selected by supraphysiological Gm concentrations (up to 512 μg/ml) even in A. baumannii isolates which are clinically resistant to Gm (22). This holds true also for the whole pLPV plasmid series although a few exceptions exist, e.g., strain AYE (31) which is hyperresistant to Gm (>512 μg/ml) due to the presence of multiple aminoglycoside-detoxifying mechanisms (Table 3). To overcome this limitation, the ble cassette from pVRL1Z (22) was cloned in the unique NcoI or SacI sites of pLPV vectors to obtain the Zeo-resistant derivatives pLPV1Z, pLPV2Z, and pLPV3Z (Fig. 2 and Fig. S1). Briefly, convenient features of pLPV promoter-probe vectors are the possibility of selection even in PDR strains, the relatively small size (from 5.5 to 10.3 kb, depending on the reporter system), and the presence of an extended polylinker with ≥10 unique restriction sites flanked by annealing regions for universal sequencing oligonucleotides (i.e., the T3, pBS KS, and pBS SK primers), as illustrated in Fig. 2.
TABLE 3.
Transformation efficiency and stability of pLPV plasmids and antibiotic concentrations required for their selection
| Strain (plasmid) | TE (CFU/μg DNA)a | Stability (NAnt/N0)b | Selected by: |
|
|---|---|---|---|---|
| Gm (μg/ml) | Zeo (μg/ml) | |||
| E. coli DH5α(pLPV1Z) | (1.16 ± 0.38) × 105 | 1.07 ± 0.09 | 10 | 25 |
| E. coli DH5α(pLPV2Z) | (1.38 ± 0.31) × 105 | 1.12 ± 0.14 | 10 | 25 |
| E. coli DH5α(pLPV3Z) | (1.80 ± 0.01) × 105 | 1.13 ± 0.11 | 10 | 25 |
| E. coli DH5α(pVRL1) | (1.02 ± 0.21) × 105 | 1.00 ± 0.33 | 10 | |
| A. baumannii ATCC 19606T(pLPV1Z) | (1.56 ± 0.06) × 102 | 0.80 ± 0.23 | 100 | 250 |
| A. baumannii ATCC 19606T(pLPV2Z) | (2.87 ± 0.93) × 102 | 0.83 ± 0.10 | 100 | 250 |
| A. baumannii ATCC 19606T(pLPV3Z) | (4.57 ± 0.29) × 102 | 0.81 ± 0.09 | 100 | 250 |
| A. baumannii ATCC 19606T(pVRL1) | (6.95 ± 0.21) × 102 | 0.92 ± 0.29 | 100 | |
| A. baumannii ATCC17978(pLPV1Z) | (4.37 ± 0.07) × 101 | 0.85 ± 0.14 | 100 | 250 |
| A. baumannii ATCC17978(pLPV2Z) | (5.01 ± 5.01) × 101 | 0.78 ± 0.21 | 100 | 250 |
| A. baumannii ATCC17978(pLPV3Z) | (3.89 ± 0.29) × 101 | 0.75 ± 0.17 | 100 | 250 |
| A. baumannii ATCC17978(pVRL1) | (4.95 ± 0.21) × 101 | 0.92 ± 0.29 | 100 | |
| A. baumannii ACICU(pLPV1Z) | (1.06 ± 0.87) × 102 | 0.74 ± 0.09 | 200 | 250 |
| A. baumannii ACICU(pLPV2Z) | (9.12 ± 0.74) × 103 | 0.73 ± 0.13 | 200 | 250 |
| A. baumannii ACICU(pLPV3Z) | (9.49 ± 1.25) × 103 | 0.71 ± 0.22 | 200 | 250 |
| A. baumannii ACICU(pVRL1) | (1.78 ± 0.21) × 103 | 0.83 ± 0.34 | 200 | |
| A. baumannii AYE(pLPV1Z) | (1.02 ± 1.05) × 103 | 0.92 ± 0.08 | NS | 250 |
| A. baumannii AYE(pLPV2Z) | (1.19 ± 0.98) × 103 | 1.02 ± 0.12 | NS | 250 |
| A. baumannii AYE(pLPV3Z) | (2.84 ± 0.64) × 103 | 0.90 ± 0.11 | NS | 250 |
| A. baumannii AYE(pVRL1Z) | (1.02 ± 0.21) × 103 | 0.95 ± 0.01 | NT | 250 |
| A. pittii UKK_0145(pLPV1Z) | (1.81 ± 0.76) × 104 | 0.67 ± 0.13 | 50 | 250 |
| A. pittii UKK_0145(pLPV2Z) | (1.97 ± 0.04) × 104 | 0.77 ± 0.20 | 50 | 250 |
| A. pittii UKK_0145(pLPV3Z) | (2.84 ± 0.64) × 104 | 0.87 ± 0.17 | 50 | 250 |
| A. pittii UKK_0145(pVRL1) | (1.55 ± 0.05) × 104 | 0.79 ± 0.25 | 50 | |
| A. baylyi BD413(pLPV1Z) | (1.16 ± 0.03) × 104 | 0.64 ± 0.25 | 10 | 25 |
| A. baylyi BD413(pLPV2Z) | (1.38 ± 0.02) × 104 | 0.62 ± 0.27 | 10 | 25 |
| A. baylyi BD413(pLPV3Z) | (1.80 ± 0.01) × 104 | 0.79 ± 0.32 | 10 | 25 |
| A. baylyi BD413(pVRL1) | (9.5 ± 0.75) × 103 | 0.99 ± 0.17 | 10 | 25 |
TE, transformation efficiency. E. coli chemically competent cells were transformed by a heat shock protocol (63); A. baumannii and A. pittii electrocompetent cells were transformed by electroporation (74); A. baylyi naturally competent cells were transformed as previously reported (64). Data are the means ± standard deviations from three independent experiments.
In vitro stability in the absence of antibiotic selection is expressed as the ratio between the number of bacterial colonies grown on LB agar supplemented with the indicated antibiotic concentrations (NAnt) and the number of bacterial colonies grown on LB agar without antibiotic (N0). Data are the means ± standard deviations from three independent experiments. NS, not selectable (Gm MIC > 512 μg/ml); NT, not tested.
FIG 2.
Physical and functional map of pLPV1Z (A), pLPV2Z (B), and pLPV3Z (C) promoter-probe vectors. The nucleotide sequence upstream of the reporter gene is shown for each vector. In all vectors, the ribosome binding site (RBS) is provided by the canonical sequence 5′-AAAGAGGAGAAA-3′, preceded by stop codons in the three reading frames (red). Unique cutter restriction enzymes are indicated in bold. Nomenclature: ble, Zeo resistance gene; aacC1, Gm resistance gene; ori ColE1-like, ColE1-derived origin of replication for E. coli; oriAb, origin of replication for Acinetobacter spp. All genes are reported in scale over the total length of the vector. Images were obtained by Snapgene software (GSL Biotech).
Copy number, transformation efficiency, host range, and stability of pLPV plasmids.
The plasmid copy number (PCN) per cell of pLPV2, taken as an example of the pLPV vector series, was determined by real-time quantitative PCR as outlined in Materials and Methods (22), using E. coli DH5α and A. baumannii ATCC 19606T as host strains. The PCNs of pLPV2 were 75 ± 10 per E. coli DH5α cell and 57 ± 12 per A. baumannii ATCC 19606T cell and, thus, very similar to the PCN of the ancestor plasmid pVRL1 (22) (Table S1). Therefore, pLPV vectors can be considered high-copy-number plasmids according to the conventional classification (32). Accordingly, all pLPV plasmids could be extracted with good yields from early-stationary-phase E. coli DH5α and A. baumannii ATCC 19606T cells, attaining up to 1.65 and 0.45 μg of DNA per milliliter of culture at an optical density at 600 nm (OD600) of 1 (Table S1).
The pLPV vectors could be introduced by electroporation in A. baumannii strains belonging to different lineages, e.g., A. baumannii AYE, ACICU, and ATCC 17978 (33), in addition to the type strain ATCC 19606T, as well as in Acinetobacter baylyi BD413 and Acinetobacter pittii UKK_0145 (Table 3). However, the transformation efficiencies (TE) varied greatly between species and strains, ranging from ca. 105 CFU/μg DNA for E. coli DH5α to ca. 50 CFU/μg DNA for A. baumannii ATCC 17978 (Table 3).
To confirm the stability of pLPV vectors in both E. coli and Acinetobacter spp., individual strains carrying the different pLPV constructs were grown for ca. 40 generations, without antibiotic, before being plated on selective medium supplemented with the appropriate antibiotic. Plasmid stability was expressed as the NAnt/N0 ratio, where NAnt and N0 are the numbers of CFU grown with and without antibiotic, respectively. Elevated segregational stability was observed for all pLPV vectors in all tested species, with little or no plasmid loss after ca. 40 generations (Table 3).
Transcriptional response of the uvrABC promoter to DNA damage.
Uvr-dependent excision repair is the main system for the correction of UV-induced pyrimidine dimers and a variety of other forms of DNA damage in E. coli. UvrA, UvrB, and UvrC proteins act in concert to introduce a nick at the end of a DNA lesion to initiate the repair process. Expression of the uvrABC operon in E. coli is controlled by RecA and LexA transcriptional regulators and is inducible by DNA damage following exposure to UV light or other DNA-damaging agents, such as mitomycin C (MMC) (34). It was observed that there is no Uvr-dependent response to DNA lesions in Acinetobacter spp., with the remarkable exception of A. baumannii and a few other species (15, 35) in which different genes, including the nucleotide excision repair component uvrA, are upregulated in response to the DNA-damaging agent MMC (15).
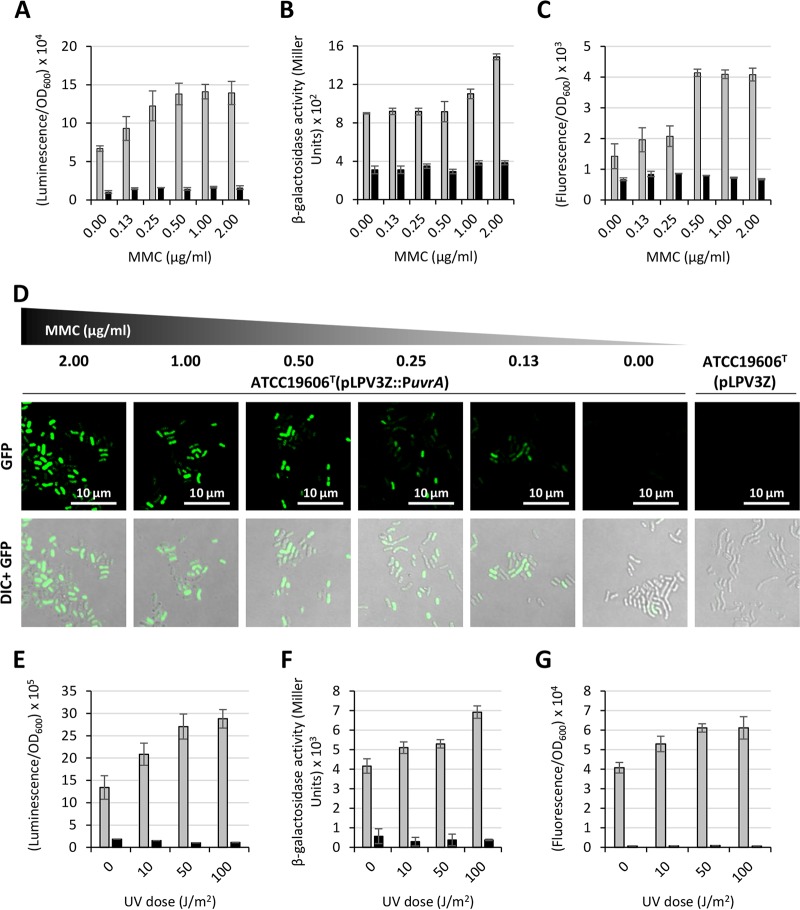
For a preliminary proficiency testing of the performances of pLPV vectors, we investigated the regulation of the DNA damage-inducible uvrABC operon in A. baumannii ATCC 19606T. To this purpose, a 572-bp DNA fragment encompassing the predicted promoter region of the uvrABC operon (PuvrA) was cloned upstream of lux, lacZ, and GFP genes, yielding pLPV1Z::PuvrA, pLPV2Z::PuvrA, and pLPV3Z::PuvrA, respectively. A. baumannii ATCC 19606T transformants were challenged with different MMC concentrations, as indicated in Fig. 3. Significant induction of reporter gene expression was observed for all constructs upon exposure of bacteria to MMC though different dose-response profiles were observed, depending on the reporter system. The most sensitive and dose-responsive systems were pLPV1Z::PuvrA and pLPV3Z::PuvrA, showing significant PuvrA induction even at low MMC concentrations (Fig. 3A and C). It should be pointed out that the pLPV3Z::PuvrA (GFP-based) fusion had to be tested in M9 minimal medium supplemented with 20 mM sodium succinate (M9-S medium) instead of Luria-Bertani (LB) medium due to the high fluorescent background of LB medium upon excitation at 475 nm. Laser scanning confocal microscopy provided visual evidence of the dose-dependent increase of GFP expression in A. baumannii ATCC 19606T(pLPV3Z::PuvrA) exposed to MMC (Fig. 3D). Intriguingly, few GFP-producing cells were also detected without MMC induction, suggesting that the uvrABC operon is expressed by a minority of the bacterial population even under noninductive conditions.
FIG 3.
Regulation of the uvrABC operon upon exposure of A. baumannii to DNA-damaging agents MMC and UV light. Overnight cultures of A. baumannii ATCC 19606T carrying any of the three PuvrA fusions (pLPV1Z::PuvrA or pLPV2Z::PuvrA or pLPV3Z::PuvrA) or the corresponding promoterless vectors were subcultured (1:100 dilution) in LB broth (pLPV1Z::PuvrA or pLPV2Z::PuvrA) or M9-S medium (pLPV3Z::PuvrA) and incubated at 37°C until the culture reached the mid-exponential phase (OD600 of 0.6). Cultures were treated with different MMC concentrations, as indicated, and luminescence (A), β-galactosidase activity (B), and fluorescence emission (C) were measured after 16 h. GFP-producing cells were also visualized using a Leica SP5 confocal laser scanning microscope equipped with a 63× oil immersion objective (D). Representative images of either GFP or GFP and differential interference contrast (DIC)-merged channels are shown. Scale bar, 10 μm. Overnight cultures were also suspended in M9-S medium at an OD600 of 1.0, and 5 ml of each suspension was irradiated with 0, 10, 50, and 100 J/m2 UV light at 300 nm. Luminescence (E), β-galactosidase activity (F), and fluorescence emission (G) were measured after a 2-h rescue at 37°C. Gray and black histograms indicate strains carrying individual PuvrA fusions and the promoterless vectors, respectively. Data are the means ± standard deviations from three independent experiments.
To confirm the uvrA transcriptional response to DNA damage, A. baumannii ATCC 19606T carrying any of the three PuvrA promoter fusions was suspended in M9-S medium and exposed to increasing doses of UV light (i.e., 0, 10, 50, and 100 J/m2; λ = 300 nm). UV rays are known to cause the formation of pyrimidine dimers which can be repaired by photolyase reactivation or a Uvr-dependent nucleotide excision repair mechanism. Notably, the PuvrA promoter showed a dose-dependent response to UV light induction, even at the lowest dose, in all three pLPV reporter gene fusions (Fig. 3E to G). These experiments provide new evidence of UV light-dependent activation of uvrA in A. baumannii.
Transcriptional response of ethanol-inducible promoters.
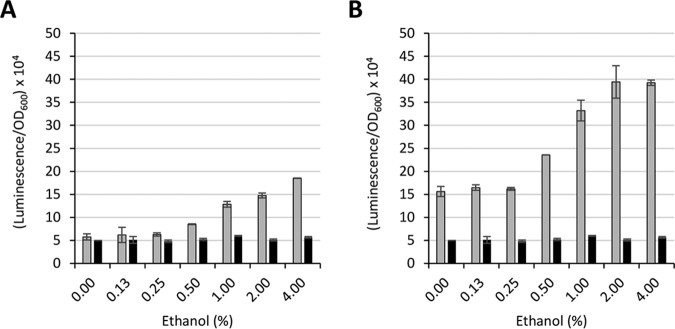
Ethanol has pleiotropic effects on A. baumannii physiology; it stimulates bacterial growth yields and increases salt tolerance and pathogenicity in different animal models (36–38). Moreover, ethanol affects the expression of several regulatory genes, suggesting that it could play global regulatory functions (14, 39). Prominent genes which were reported to be induced by ethanol are those coding for ethanol dehydrogenase (adhP) and aldehyde dehydrogenase (yahK), due to their implication in ethanol detoxification (14). To further validate pLPV vectors, DNA fragments encompassing the predicted promoter region of the adhP and yahK genes (1,073 bp and 1,080 bp, respectively) were cloned in pLPV1Z to generate pLPV1Z::PadhP and pLPV1Z::PyahK, respectively, and then introduced by electroporation in A. baumannii strains ATCC 19606T, ATCC 17978, and AYE. Transformants were challenged with up to 4% ethanol concentrations for quantification of bioluminescent emission at 15 min postexposure. Significant induction of both PyahK and PadhP promoters was observed at >0.5% ethanol, with a nearly linear dose-response effect up to 2 to 4% ethanol in A. baumannii ATCC 19606T (Fig. 4A and B). Ethanol induction of both PyahK and PadhP was also observed in A. baumannii ATCC 17978 and AYE although to a lesser extent (Fig. S2A and B). While differences in the expression levels of individual promoter fusions were observed between different strains, pLPV1Z::PyahK invariably produced higher bioluminescence than pLPV1Z::PadhP (Fig. 4 and Fig. S2). This might correlate with previous evidence that yahK transcripts are more abundant than adhP transcripts in ethanol-exposed A. baumannii (14). In all the tested strains, the luminescent signal rapidly decreased 15 min after induction and almost completely disappeared after 1 h (data not shown). There are no obvious explanations for this observation; it could be due to ethanol toxicity, alcohol evaporation, decreased reporter protein stability/activity in the presence of ethanol, or other reasons. Repeated attempts to detect ethanol induction of PyahK and PadhP using lacZ- or GFP-based fusions failed, probably due to a delayed response of these reporter systems (data not shown).
FIG 4.
Regulation of the A. baumannii ethanol-inducible adhP (A) and yahK (B) genes. A. baumannii ATCC 19606T carrying either pLPV1Z::PadhP or pLPV1Z::PyahK was cultured overnight in LB broth, diluted at an OD600 of 0.1 in LB broth, and challenged with different ethanol concentrations, as indicated. The luminescence emission was recorded at 15 min postinduction. Gray and black histograms indicate pLPV1Z::PadhP (A) or pLPV1Z::PyahK (B) fusions and the promoterless pLPV1Z vector, respectively. Data are the means ± standard deviations from three independent experiments.
Iron-regulated gene expression.
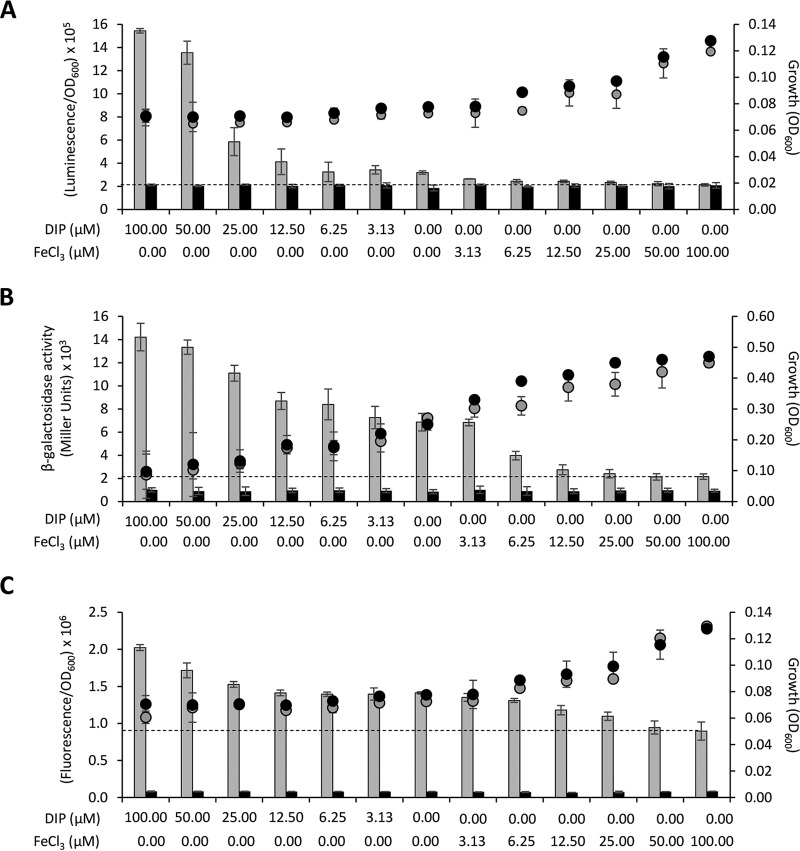
A. baumannii requires iron for growth, and in order to acquire iron from the environment, it has evolved multiple iron uptake strategies (reviewed in references 11, 40, and 41). Iron uptake must be finely tuned to prevent toxicity due to iron overload, and the Fur repressor protein acts as the global regulator of iron homeostasis in Gram-negative bacteria. In cells containing sufficient iron levels, the Fur-Fe2+ complex represses transcription arising from iron-regulated promoters, which are transcribed only under conditions of iron scarcity (42). The acinetobactin siderophore-mediated iron uptake system is the best studied among A. baumannii iron transport systems, and it involves both siderophore synthesis and uptake functions encoded by bas and bau genes, respectively (reviewed in references. 11, 40, and 41). To assess the usefulness of pLPV vectors as tools to investigate iron regulation in A. baumannii, the Fur-Fe2+-repressible promoter of the basA gene (PbasA) (43), implicated in acinetobactin biosynthesis, was cloned in pLPV vectors to obtain pLPV1Z::PbasA, pLPV2Z::PbasA, and pLPV3Z::PbasA.
Since Fur-dependent regulation of PbasA has previously been demonstrated (19, 20, 43), we initially wondered if pLPV plasmids could directly be employed for the genetic screening of Fur-repressible promoters by a Fur titration assay (FURTA) (44). For this purpose, E. coli H1717 carrying either pLPV1Z::PbasA or pLPV3Z::PbasA was grown on MacConkey agar plates supplemented with increasing Fe2+ concentrations, and β-galactosidase expression directed by the chromosomal fhuA::lacZ fusion was recorded after overnight incubation at 37°C (44). Remarkably, both pLPV1Z::PbasA and pLPV3Z::PbasA showed the same lacZ expression profile as the positive-control pBS::PbasA (19), indicating that pLPV plasmids have sufficient copy numbers to be directly used for the genetic screening of Fur-controlled promoters (Fig. S3).
Then, A. baumannii ATCC 19606T carrying any of the three Fur-controlled gene fusions (pLPV1Z::PbasA or pLPV2Z::PbasA or pLPV3Z::PbasA) was grown in M9-S medium supplemented with increasing concentrations (0 to 100 μM) of either FeCl3 or the iron-chelator 2,2′-dipyridyl (DIP), and reporter gene expression was monitored for up to 6 h (Fig. 5). Of note, induction times were preliminarily set by testing the response of individual PbasA fusions at 2, 4, and 6 h postinoculation (data not shown). As expected, downregulation of all three fusions was observed in the presence of exogenously added FeCl3, as opposed to upregulation with increasing DIP concentrations (Fig. 5). Although a dose-response effect was evident for all fusions, different levels of responsiveness were observed among reporter systems. Considering 100 μM FeCl3 as the baseline (complete PbasA repression), nearly linear responses were observed for the lacZ fusion in pLPV2Z::PbasA after a 6-h induction (Fig. 5B) and for the GFP fusion in pLPV3Z::PbasA after a 2-h induction (Fig. 5C), while the lux-based system in pLPV1Z::PbasA showed a nearly linear response up to 6.25 μM DIP, followed by an exponential increase at higher DIP concentrations, after a 2-h induction (Fig. 5A). The link between upregulation of GFP expression and decreased iron availability was confirmed by laser scanning confocal microscopy analysis of A. baumannii ATCC 19606T(pLPV3Z::PbasA) cells (Fig. S4). The number of fluorescent cells increased in parallel with the extent of iron limitation. Intriguingly, fluorescent cells were occasionally detected also in the presence of exogenously added FeCl3, possibly accounting for the elevated fluorescent background of the pLPV3Z::PbasA fusion (Fig. 5C).
FIG 5.
Regulation of the basA promoter in response to iron availability. A. baumannii ATCC 19606T carrying any of the three PbasA fusions, namely pLPV1Z::PbasA (A), pLPV2Z::PbasA (B), or pLPV3Z::PbasA (C), or the corresponding promoterless vectors was cultured overnight in LB broth, washed in saline, and suspended at an OD600 of 0.1 in M9-S medium supplemented with different concentrations of DIP or FeCl3, as indicated on the abscissa. Luminescence (A), β-galactosidase activity (B), and fluorescence emission (C) were recorded after a 2-h (A and C) or 6-h (B) incubation at 37°C. Gray and black histograms indicate the PbasA fusions and the corresponding promoterless vectors, respectively. Dotted lines denote the baseline PbasA activity under maximum repression (100 μM FeCl3). Circles indicate bacterial growth (OD600) values without optical path adjustment; the optical path ratio between A or C and B is 0.30. Data are the means ± standard deviations from three independent experiments.
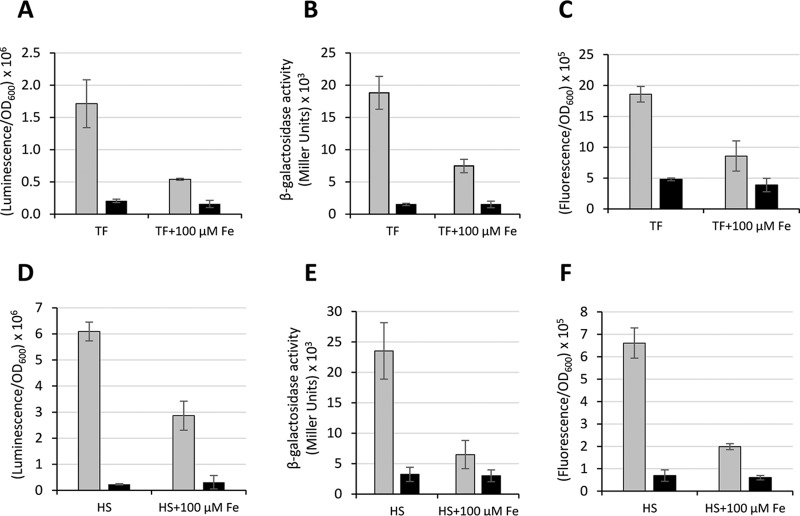
Probing iron regulation in biological fluids and in vivo.
During infection, bacteria must face a severe nutritional stress imposed by the iron-withholding response of the host (45). The iron binding proteins transferrin (TF) in serum and lactoferrin in mucosal secretions play a crucial role in the competition for iron with invading pathogens (46). On this basis, the response of the three PbasA fusions was tested in A. baumannii ATCC 19606T grown in M9-S medium supplemented with 2.5 mg/ml TF or in heat-inactivated (complement-free) human serum (HS), with or without 100 μM FeCl3. In both media, clear repression of the PbasA promoter carried by any of the three vectors (pLPV1Z::PbasA, pLPV2Z::PbasA, and pLPV3Z::PbasA) was observed upon addition of 100 μM FeCl3 due to complete saturation of TF iron binding sites in both TF-supplemented M9-S medium and HS (Fig. 6). These observations indicate that the PbasA-based transcriptional fusions can be used to monitor the iron starvation response of A. baumannii under conditions which mimic in vivo growth.
FIG 6.
Regulation of the basA promoter in TF-supplemented medium and in HS. A. baumannii ATCC 19606T carrying any of the three PbasA fusions (pLPV1Z::PbasA or pLPV2Z::PbasA or pLPV3Z::PbasA) or the corresponding promoterless vector was grown overnight in LB broth, washed with saline, and inoculated (OD600 of 0.1) in either M9-S medium supplemented with 2.5 mg/ml TF (TF) or in HS (HS). As a control, 100 μM FeCl3 was added to TF (TF + 100 μM Fe) or HS (HS + 100 μM Fe) to saturate the TF iron binding capacity. Luminescence (A), β-galactosidase activity (B), and fluorescence emission (C) were recorded after a 4-h (A to C) or 6-h (D to F) incubation at 37°C. Gray and black histograms indicate the PbasA fusions and the corresponding promoterless vectors, respectively. Data are the means ± standard deviations from three independent experiments.
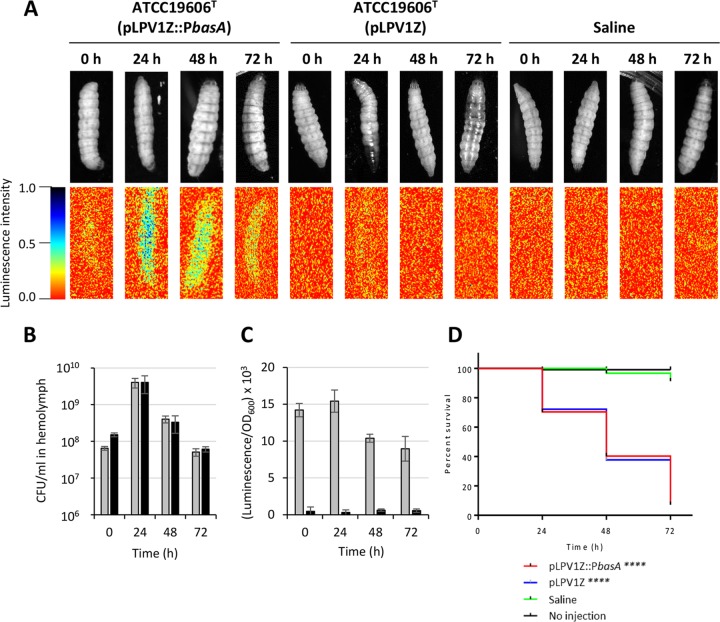
Since the basA gene is upregulated in response to iron starvation, we wondered if the lux-based pLPV1Z::PbasA transcriptional fusion could emit a detectable signal under the iron-limited conditions of Galleria mellonella caterpillar hemocoel, with the aim of real-time monitoring A. baumannii ATCC 19606T(pLPV1Z::PbasA) iron-regulated gene expression and growth in vivo. G. mellonella caterpillars are a suitable nonmammalian infection model that has extensively been employed to study host-pathogen interactions in many bacterial species, including A. baumannii (47, 48). The caterpillar hemocoel cavity contains a fluid, called hemolymph, which is functionally comparable to blood (49). Interesting similarities with the human immune system can also be observed for the insect cellular (phagocytic cell-mediated) and humoral immune responses even if insects lack adaptive immunity (50). Moreover, G. mellonella caterpillars can be maintained at 37°C, enabling the expression of bacterial virulence factors at the temperature of the human body.
About 106 viable cells of A. baumannii ATCC 19606T(pLPV1Z::PbasA) were injected in the second-to-last left proleg of G. mellonella caterpillars and maintained at 37°C for up to 72 h postinfection. A. baumannii CFU counts, luminescent emission by both intact larvae and hemolymph, and larval survival were daily monitored. It was observed that the progression of infection was accompanied by an increasing number of melanized insects (Fig. 7A), indicating that A. baumannii triggers the host innate immune response, resulting in the activation of the prophenoloxidase cascade (51). A dramatic increase of caterpillar-associated bioluminescence became detectable at 24 h postinfection (Fig. 7A), concomitant with an increase of bacterial counts (Fig. 7B) and luminescent emission by the hemolymph (Fig. 7C). While bacterial viability and luminescence gradually decrease over time, larval killing was delayed with respect to the maximum bacterial burden, culminating with 80% of dead caterpillars at 72 h postinfection (Fig. 7D). Notably, the in vivo stability of pLPV1Z::PbasA in A. baumannii ATCC 19606T cells, calculated at the end of the experiment, was 0.87 ± 0.16. Overall, these results indicate that A. baumannii is faced with iron scarcity during G. mellonella infection, paving the way for the use of pLPV-based gene fusions for probing the A. baumannii transcriptional response to environmental stressors also in vivo.
FIG 7.
In vivo and ex vivo monitoring of A. baumannii iron-regulated gene expression during G. mellonella infection. Larvae were injected with ca. 106 cells of A. baumannii ATCC 19606T carrying either pLPV1Z::PbasA or the promoterless pLPV1Z vector and monitored daily for up to 72 h by visual inspection and luminescence imaging of the intact animals (A). Saline injection was used as a control of G. mellonella viability. Luminescence emission by the larvae is shown below the corresponding caterpillars. The hemolymph was sampled from live or agonic caterpillars at 24-h intervals, diluted in saline, and used to determine bacterial viability (B) or luminescence emission (C). Gray and black histograms indicate pLPV1Z::PbasA and the corresponding promoterless pLPV1Z vector, respectively. Data are the means ± standard deviations from 5 larvae. (D) Kaplan-Meier curves of the caterpillar population. A log rank test revealed no statistically significant differences between survival rates of larvae injected with A. baumannii ATCC 19606T carrying either pLPV1Z::PbasA or the empty vector, while statistically significant differences were observed between saline- and A. baumannii-injected larvae (****, P < 0.0001).
DISCUSSION
The availability of user-friendly tools for bacterial gene expression analysis, both in vitro and in natural environments, has become a fundamental requirement for understanding environment-responsive regulatory circuitry. Almost 10% of the A. baumannii coding capacity consists of putative or confirmed regulatory genes (39, 52, 53), but still little is known about the regulation of gene expression in this species. Indeed, several studies have investigated A. baumannii virulence-related traits (reviewed in reference 41), but very few have focused on their regulation (reviewed in references 54 and 55).
Motivated by the need of user-friendly tools for gene expression analysis in Acinetobacter spp., we have developed different promoter-probe plasmid vectors which can be used to generate episomal transcriptional fusions. These vectors can replicate in both E. coli and Acinetobacter spp. thanks to the presence of ColE1-like and oriAb replication origins, both derived from the ancestor plasmid pVRL1 (22). To minimize plasmid size without affecting stability and antibiotic selection, only a small region of the pVRL1 ancestor was incorporated in pLPV vectors, including the aacC1 and ble antibiotic resistance cassettes, encoding Gm and Zeo resistance, respectively, for selection in MDR A. baumannii strains, and the TA system which is essential for plasmid maintenance in the absence of antibiotic selection (22, 56). Vectors were then equipped with three alternative reporter systems, namely the lux operon, the lacZ gene, or the GFP gene, downstream of an extended polylinker.
The pLPV vectors are high-copy-number plasmids, and their host range is limited to A. baumannii, including MDR isolates, and a few other Acinetobacter spp., e.g., A. baylyi and A. pittii (Table 3 and data not shown). Despite many attempts to introduce pLPV vectors in other members of the Acinetobacter calcoaceticus-Acinetobacter baumannii (ACB) complex, no transformants were obtained for Acinetobacter dijkshoorniae 271, Acinetobacter nosocomialis UKK_0361, and Acinetobacter seifertii HS A23-2 (data not shown) while these species could, indeed, be transformed by the ancestor plasmid pVRL1 (22). Probably, deletion of some ORFs from pVRL1 has narrowed the pLPV host range. These features, together with the absence of putative mobilization functions, would prevent undesired spread of Zeo and Gm resistance genes to neighbor species.
A major shortcoming of plasmid-based transcriptional fusions is the need for antibiotic selection to ensure plasmid maintenance. However, exposure to antibiotics, even at subinhibitory concentrations, has a profound impact on the bacterial transcriptome and can cause a bias in gene expression analysis (26, 27). Since the three pLPV vectors proved to be stably maintained in different Acinetobacter species and strains for up to ca. 40 generations, they could safely be used to analyze gene expression without the need of antibiotic pressure. Indeed, the duration of our in vitro gene expression assays never exceeded 16 h (ca. 10 generations), and in vivo assays provided evidence of elevated pLPV1Z::PbasA stability for up to 72 h during G. mellonella infection.
To test the proficiency of pLPV vectors and to gain insight into the transcriptional regulation of well-characterized A. baumannii genes in response to major environmental stressors, transcriptional fusions carrying the promoter regions of DNA damage-, ethanol-, and iron starvation-inducible genes were generated in the three pLPV plasmids. Upon appropriate vector selection, the predicted regulation was confirmed for all promoter fusions, with a clear difference between inducing and noninducing conditions. For instance, it was possible to demonstrate clear induction of the PuvrA promoter in response to MMC and UV light exposure. Moreover, thanks to the rapid response and strong signal of the lux-based pLPV1Z vector, ethanol-inducible expression of adhP and yahK genes was observed in A. baumannii. Despite strain-dependent variability in the expression levels of the adhP and yahK genes, clear induction of ethanol-responsive gene expression was shown for isolates belonging to different lineages, suggesting that ethanol regulation is a universal feature of A. baumannii.
Then, iron-regulated gene fusions were generated by cloning the promoter region of the basA gene, implicated in acinetobactin synthesis, in all three pLPV plasmids. Since basA expression is controlled by the Fur repressor protein, both pLPV1Z::PbasA and pLPV3Z::PbasA were directly employed to demonstrate the presence of functional Fur binding sequences in PbasA, bypassing tedious promoter subcloning into E. coli high-copy-number vectors, as recommended in the FURTA (44). Accordingly, growth of A. baumannii under conditions of increasing iron deficiency (M9-S medium supplemented with different concentrations of either FeCl3 or DIP) showed progressive upregulation of the PbasA promoter with decreasing iron availability. The lacZ-based pLPV2Z::PbasA fusion provided a more robust and linear response to iron starvation than pLPV1Z::PbasA and pLPV3Z::PbasA and overall higher β-galactosidase levels than with the low-copy-number pMP220::PbasA construct, which was previously employed by our group to probe the iron starvation response in A. baumannii (19, 20) (see Fig. S5 in the supplemental material). Moreover, all three PbasA fusions provided clear evidence of upregulation under conditions which mimic bacterial growth in vivo, i.e., TF-supplemented M9-S medium and HS.
Since in vivo bioluminescence imaging has emerged as a powerful technique to monitor the progression of bacterial infection in different animal models (57–59), PbasA::lux-tagged A. baumannii was employed to infect G. mellonella larvae. An increase of bioluminescence produced by A. baumannii ATCC 19606T(pLPV1Z::PbasA) was observed at 24 h postinfection, followed by a gradual decrease of bioluminescence over time, which correlated with ex vivo bacterial cell counts, suggesting that pLPV1Z provides a suitable promoter-probe system for real-time monitoring of gene expression and growth in vivo. Bioluminescence-based analysis of bacterial burden in vivo is a simple and rapid method for noninvasive monitoring of A. baumannii infection in intact animals, with the potential to assess the expression pattern of virulence genes. Coherent with our findings, it has been demonstrated that active iron uptake is essential for A. baumannii pathogenicity in different animals, including G. mellonella larvae (19, 60). Furthermore, bioluminescence monitoring positively contributes to the implementation of two of the three Rs (replacement, reduction, and refinement) of ethical principles in animal experimentation (61). Refinement and reduction are promoted by the noninvasive nature of the method, given that photon emission can be used to quantify bacterial growth and/or gene expression within animal tissues (62). Remarkably, our findings also demonstrated that lux expression in vivo does not affect A. baumannii lethality in G. mellonella since nearly identical killing plots were observed for the basA::lux fusion and the promoterless vector (Fig. 7D).
In conclusion, three promoter-probe vectors for gene expression analysis in MDR A. baumannii have been developed and proficiency tested. Overall, the lux-based fusion (pLPV1Z) provided faster detectable signal than lacZ- and gfp-based fusions and did not require sample processing as it was strictly dependent on intracellular ATP content. These characteristics make the lux-based reporter system suitable for detection of early regulatory responses and for the assessment of spatiotemporal dynamics of A. baumannii infection in living organisms. On the other hand, the lacZ reporter-based system (pLPV2Z) was more appropriate under some in vitro conditions, but a reporter gene assay is complex and time-consuming. The GFP-based reporter system (pLPV3Z) does not need exogenous substrates for signal emission and is suited for A. baumannii since this species is not intrinsically fluorescent (data not shown) although the use of this system could be hampered by the autofluorescence of some culture media (e.g., LB medium and HS). Notably, the GFPmut3b protein used as reporter in pLPV3 is endowed with a short half-life as it is susceptible to the activity of endogenous proteases, which provides an advantage for in situ studies of temporal gene expression due to limited GFP intracellular accumulation (23). The pLPV vectors will be made freely available to the scientific community with the hope that they will help provide more insights into the regulation of gene expression in Acinetobacter spp.
MATERIALS AND METHODS
Bacterial strains and culture media.
Bacterial strains used in this study are listed in Table 2. All bacterial strains were routinely grown in Luria-Bertani (LB) broth or on LB agar plates at 37°C. Ampicillin (Ap), carbenicillin (Cb), and tetracycline (Tc) were added at the following concentrations: for E. coli, 100 μg/ml Ap and 12.5 μg/ml Tc; for A. baumannii ATCC 19606 T, 250 μg/ml Cb and 50 μg/ml Tc. The gentamicin (Gm) and zeocin (Zeo) concentrations used are indicated in Table 3. Zeo selection was performed on low-salt LB agar (10 g/liter tryptone, 0.5 g/liter NaCl, and 5 g/liter yeast extract) to avoid the inhibition of Zeo activity due to the high ionic strength of LB agar (21). Where specified in the text, bacteria were grown in M9 minimal medium containing 20 mM sodium succinate as a carbon source (M9-S medium) (63).
Preparation of Acinetobacter spp. and E. coli competent cells.
Electrocompetent cells of Acinetobacter spp. were prepared as previously described (22). Briefly, bacteria were grown in LB broth for 18 h at 37°C. Bacterial cultures were refreshed 1:100 into 50 ml of prewarmed LB broth and incubated for 24 h at 37°C with vigorous shaking. Cells were harvested by centrifugation (3,000 × g, 15 min), washed twice with 25 ml of 10% glycerol at room temperature, and suspended in 1.5 ml of 10% glycerol. Eighty-microliter aliquots of competent cells were stored at −80°C. Electroporation was performed using 500 ng of plasmid in 0.2-cm electroporation cuvettes (Gene Pulser; Bio-Rad). After pulsing (2.5 kV/cm, 200 Ohm, 25 μF), cells were recovered in 1 ml of prewarmed SOC medium (2% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM glucose) and incubated at 37°C for 1 h. Transformants were selected on LB agar or low-salt LB agar with the appropriate antibiotic concentration. Competent E. coli cells were prepared by the rubidium-calcium chloride method and transformed according to a heat shock protocol (63).
Plasmid construction.
The plasmid construction strategy has been illustrated in Results, according to established protocols (63). All plasmids used in this study and their characteristics are listed in Table 2.
Transformation efficiency, in vitro stability, and plasmid extraction yield.
Transformation efficiency (TE) was determined by transformation of 75 ng and 500 ng of plasmid DNA in calcium-competent E. coli and electrocompetent Acinetobacter species cells, respectively. Naturally competent A. baylyi BD413 was transformed with 150 ng of plasmid, as reported previously (64). Transformants were plated on LB agar supplemented with the appropriate concentration of Gm. Bacterial colonies grown on Gm-supplemented plates were streaked on low-salt LB agar plates containing the appropriate Zeo concentration. TE was expressed as the ratio between CFU number and amount of plasmid DNA (in micrograms) used for transformation. Plasmid stability was assessed in E. coli DH5α and Acinetobacter spp. Bacterial strains were preliminarily grown for 18 h in LB broth with the appropriate antibiotic concentration and then washed and diluted 1,000-fold in LB broth without antibiotics. Bacterial cultures were refreshed (1:1,000) every 12 h for 48 h. Bacterial colony counts were determined on LB agar or low-salt LB agar (N0) and on LB agar or low-salt LB agar supplemented with Gm or Zeo at the appropriate concentrations (NAnt). Plasmid stability is expressed as the NAnt/N0 ratio (24).
To evaluate the extraction yield, bacterial cultures were grown for 18 h at 37°C in LB broth supplemented with the appropriate antibiotic concentration. Cultures were diluted to an OD600 of 1, and plasmids were extracted using a Wizard Plus SV Miniprep DNA purification system (Promega) according to the manufacturer’s instructions. Plasmid yields were expressed as micrograms of DNA per milliliter of culture.
Determination of plasmid copy number.
The plasmid copy number (PCN) of pLPV plasmids in A. baumannii ATCC 19606T and E. coli DH5α was determined by real-time quantitative PCR (RT-qPCR), as previously described (22). All of the following experimental procedures were applied also to determine the PCN of pVRL1, employed as a reference plasmid in which the PCN has previously been determined (22). pLPV2Z was chosen as a representative pLPV plasmid for PCN determination because it was comparable in size to pVRL1 (7,545 bp versus 7,276 bp, respectively). Three primer pairs for the amplification of the Gm resistance gene (aacC1) and of the d-1-deoxyxylulose 5-phosphate synthase gene (dxs) of A. baumannii ATCC 19606T (locus tag HMPREF0010_02600) or E. coli DH5α (EcoCyc database accession number G6237) were used (Table 1). The aacC1 gene is present in single copy in the pLPV and pVRL1 vectors, while it is absent in A. baumannii ATCC 19606T and E. coli DH5α chromosomes, and dxs is a single-copy gene in both A. baumannii ATCC 19606T and E. coli DH5α. Therefore, RT-qPCR quantification of aacC1 and dxs in samples containing both plasmid and A. baumannii ATCC 19606T or E. coli DH5α chromosomes, relative to samples containing known amounts of plasmid or A. baumannii ATCC 19606T or E. coli DH5α chromosomal DNA alone, makes it possible to extrapolate the PCN of pLPV vectors in these bacteria. RT-qPCR was performed using an AriaMX real-time PCR system (Agilent) with software (version 1.1). Reactions were performed in a 20-μl total volume, containing 1× iTaq Universal SYBR Green Supermix (Bio-Rad), 0.4 μM each 10 μM concentrated primer, and 2 μl of 2-fold-diluted template DNA (final DNA amount ranging from 0.05 ng to 50 ng per sample). Separate reaction mixtures were prepared for detection of chromosomal or plasmid-specific amplicons for each template DNA concentration. The thermal cycling protocol was as follows: initial denaturation of 4 min at 95°C, followed by 40 cycles of denaturation for 15 s at 95°C and annealing/extension for 45 s at 60°C. Cycle threshold (CT) values were determined after automatic fine-tuning of the baseline and manual adjustment of the fluorescence threshold. Standard curves (R2 ≥ 0.99) from five independent samples containing A. baumannii ATCC 19606T or E. coli DH5α chromosomal DNA (chromosome) or containing plasmid alone were generated, placing the log value of the amount of template DNA (determined according to dilution) on the x axis and the average CT value on the y axis. Standard curves were used to extrapolate the copy number of pLPV vectors in samples containing both chromosomal and plasmid DNA.
DNA manipulations.
Genomic DNA was extracted using a QIAamp DNA Mini kit (Qiagen), and plasmid DNA was purified from bacterial cultures using a Wizard Plus SV Miniprep DNA purification system (Promega Corporation), according to the manufacturer’s instructions. PCRs were performed using Thermo Scientific Phusion high-fidelity DNA polymerase and primers listed in Table 1. FastDigest restriction enzymes were purchased from Thermo Fisher Scientific. DNA sequencing was performed using an ABI3730 sequencer (service by Bio-fab Research, Rome, Italy).
Deletion analysis of pWH1277 and generation of promoter fusions.
DNA fragments of pWH1277 were amplified by PCR using pVRL1 as the template (22) with the primers listed in Table 1. The amplicons were blunt cloned into the unique SmaI site of the pBS plasmid, and the resulting constructs were introduced by electroporation in A. baumannii ATCC 19606T. Transformants were selected on LB agar with 250 μg/ml Cb.
The promoter region of the uvrABC operon (PuvrA; locus tag A1S_3295) was amplified by PCR from A. baumannii ATCC 17978. The promoter regions of the adhP (PadhP; locus tag HMPREF0010_01803) and yahK (PyahK; locus tag HMPREF0010_03394) genes were amplified from A. baumannii ATCC 19606T. The promoter region of basA (PbasA) was amplified by PCR from A. baumannii ACICU (BioCyc database accession number ACICU_02587). Primers used to generate the PuvrA (572 bp), PadhP (1,073 bp), PyahK (1,080 bp), and PbasA (938 bp) amplicons are listed in Table 1. The PCR products were directionally ligated to the unique PstI/XbaI sites of pLPV1Z, pLPV2Z, and pLPV3Z.
Analysis of the PuvrA promoter activity.
The DNA-damaging agent MMC was used to induce the transcription of the PuvrA promoter, as previously described (15), with minor modifications. Briefly, A. baumannii ATCC 19606T strains harboring pLPV1Z::PuvrA, pLPV2Z::PuvrA, pLPV3Z::PuvrA, or the corresponding promoterless vectors were grown overnight in LB broth supplemented with 100 μg/ml Gm, diluted 1:100 in LB broth, and incubated at 37°C under vigorous shaking. At the mid-exponential phase (OD600 of 0.6), cultures were treated with increasing concentrations of MMC (from 0.1 to 2.0 μg/ml) for 16 h. The OD600 value and luminescence or fluorescence were recorded using a Tecan Spark 10M microtiter reader, while β-galactosidase activity was determined as previously described (65). Twenty microliters of A. baumannii ATCC 19606T cultures carrying the pLPV3Z::PuvrA or pLPV3Z plasmid was also mounted on a glass slide covered with 0.5% agarose and visualized using a laser scanning confocal microscope (Leica SP5 mounting a HCX PL APO lambda blue 63×/1.40 immersion oil objective; excitation λ [λEx] of 488 nm; emission λ [λEm] of 500 nm to 600 nm).
For UV light induction, A. baumannii ATCC 19606T strains harboring pLPV1Z::PuvrA, pLPV2Z::PuvrA, pLPV3Z::PuvrA, or the corresponding promoterless vectors were grown overnight in LB broth with the appropriate antibiotic concentration. Bacterial cultures were washed and suspended in M9-S medium to an OD600 of 1. Five milliliters of each bacterial suspension was irradiated with 0, 10, 50, and 100 J/m2 UV light at 300 nm by using a UV Transilluminator 2000 (Bio-Rad) equipped with a UV light meter (Sper Scientific). Irradiated cultures were rescued for 2 h at 37°C before bioluminescence, β-galactosidase activity, and fluorescence emission were determined.
Analysis of PadhP and PyahK promoter activity.
A. baumannii ATCC 19606T, ATCC 17978, and AYE carrying pLPV1Z::PadhP, pLPV1Z::PyahK, or pLPV1Z were grown overnight in LB broth supplemented with the appropriate antibiotic concentration. Bacterial cultures were washed and diluted to an OD600 of 0.1 in LB broth supplemented with increasing ethanol concentrations (from 0.13% to 4.00%). Bioluminescence and the OD600 value were recorded 15 min after ethanol exposure by using a Tecan Spark 10M microtiter reader.
Analysis of PbasA promoter activity.
A. baumannii ATCC 19606T carrying pLPV1Z::PbasA, pLPV2Z::PbasA, pLPV3Z::PbasA, pMP220::PbasA, or the corresponding promoterless vectors was grown overnight in LB broth supplemented with the appropriate antibiotic concentration, washed with M9-S medium, and diluted to an OD600 of 0.1 in M9-S medium or in M9-S medium supplemented with increasing concentrations of 2,2′-dipyridyl (DIP) or FeCl3 (from 3.1 μM to 100 μM). The OD600 value and bioluminescence or fluorescence emission were recorded after a 2-h incubation at 37°C using a Tecan Spark 10M microtiter reader. Twenty microliters of the A. baumannii ATCC 19606T(pLPV3Z::PbasA) culture was also mounted on a glass slide covered with 0.5% agarose and visualized using a laser scanning confocal microscope (Leica SP5 mounting a HCX PL APO lambda blue 63×/1.40 immersion oil objective; λEx of 488 nm; λEm of 500 nm to 600 nm). β-Galactosidase activity expressed by A. baumannii ATCC 19606T carrying pLPV2Z::PbasA, pLPV2Z, pMP220::PbasA, and pMP220 vectors was evaluated after 6 h of incubation, as previously described (65).
LB broth-grown overnight cultures of A. baumannii ATCC 19606T carrying pLPV1Z::PbasA, pLPV2Z::PbasA, pLPV3Z::PbasA, or the corresponding promoterless vectors were diluted to an OD600 of 0.1 also in HS or in M9-S medium supplemented with 2.5 mg/ml TF (Sigma-Aldrich). The mean TF concentration in healthy human patients is ca. 2.5 mg/ml (66). Where indicated in the text, 100 μM FeCl3 was added to both media. Bioluminescence, β-galactosidase activity, and fluorescence emission were recorded after a 4-h or 6-h incubation at 37°C in cultures grown in M9-S medium supplemented with TF or in HS, respectively. HS was collected from 30 healthy donors, pooled, filtered, and inactivated (30 min, 56°C), as previously described (19, 20). Bulk serum chemistry was as follows: total serum proteins, 80 mg/ml; total iron, 0.70 μg/ml; ferritin, 0.243 μg/ml; TF, 2.63 mg/ml; total iron binding capacity, 4.27 mg/ml (20% TF saturation).
Fur titration assay.
A Fur titration assay (FURTA) was performed as previously described (44). Briefly, the plasmids pLPV1Z::PbasA and pLPV3Z::PbasA and the corresponding promoterless vectors (pLPV1Z and pLPV3Z) were individually introduced into E. coli H1717. E. coli H1717(pBSPbasA) and E. coli H1717(pBS) were employed as controls (19, 20). Briefly, 1-ml bacterial cultures grown overnight at 37°C in LB broth were washed twice with saline and diluted to obtain ca. 5 × 108 cells/ml. Ten microliters of the bacterial suspensions was deposited on MacConkey agar plates supplemented with different concentrations of Fe(NH4)2(SO4)2 (100, 50, 25, 10, 5, and 0 μM). Plates were incubated at 37°C for 24 h before visual inspection.
In vivo and ex vivo monitoring of PbasA promoter activity.
Overnight cultures of A. baumannii ATCC 19606T harboring pLPV1Z::PbasA or pLPV1Z were harvested by centrifugation (5 min at 5,000 × g). Bacterial cells were washed twice with saline and suspended to an OD600 of 1 in sterile saline. A cohort of 240 larvae was divided into four experimental groups: 90 larvae were injected with A. baumannii ATCC 19606T(pLPV1Z::PbasA), 90 larvae were injected with A. baumannii ATCC 19606T(pLPV1Z), 30 larvae were injected with sterile saline, and 30 larvae were not injected. In more detail, G. mellonella fifth-instar larvae were injected with 10 μl of bacterial cultures (corresponding to ≈ 6 × 106 CFU) through the second-to-last left proleg into the hemocoel using a BD Plastipak insulin syringe with a 0.3-mm needle, mounted on a Tridak stepper pipette. G. mellonella larvae were incubated at 37°C, and their viability and luminescence were monitored every 24 h for 3 days. The bioluminescent emission by viable or agonic larvae was recorded with a ChemiDoc XRS+ imaging system (Bio-Rad), with a 30-s exposition time. Five viable caterpillars injected with bacterial culture were sacrificed at each time point, t (0, 24, 48, and 72 h postinfection), to extract the hemolymph. The extracted hemolymph was serially diluted 1:10 in saline in a black, clear-bottom 96-well microplate plate (Greiner), and luminescence was recorded using a Tecan Spark 10M microtiter reader. Serial dilutions were then plated on LB agar, with or without 100 μg/ml Gm, and incubated at 37°C for 18 h. CFU counts and light emission were determined in the resulting colonies to verify in vivo plasmid stability. Viability of A. baumannii ATCC 19606T was expressed as the number of CFU per milliliter of hemolymph.
Statistical analysis.
Statistical analysis was performed with GraphPad Instat software. G. mellonella survival curves were generated by the Kaplan-Meier method and analyzed by a log rank test. Differences having a P value of ≤0.05 were considered statistically significant.
Data availability.
The full-length sequences of pLPV1Z, pLPV2Z, and pLPV3Z plasmids have been deposited in the GenBank database under accession numbers MK681506, MK681507, and MK681508, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Excellence Departments grant from the Italian Ministry of Education, University and Research (MIUR, Italy) (Art. 1, commi 314-337 Legge 232/2016) to the Department of Science, Roma Tre University, and by the PRIN 2017 grant protocol 20177J5Y3P from MIUR to F.I. and P.V.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01334-19.
REFERENCES
- 1.Turton JF, Shah J, Ozongwu C, Pike R. 2010. Incidence of Acinetobacter species other than A. baumannii among clinical isolates of Acinetobacter: evidence for emerging species. J Clin Microbiol 48:1445–1449. doi: 10.1128/JCM.02467-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manchanda V, Sanchaita S, Singh N. 2010. Multidrug resistant Acinetobacter. J Glob Infect Dis 2:291–304. doi: 10.4103/0974-777X.68538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. 2017. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev 30:409–447. doi: 10.1128/CMR.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsueh P-R, Teng L-J, Chen C-Y, Chen W-H, Ho S-W, Luh K-T. 2002. Pandrug-resistant Acinetobacter baumannii causing nosocomial infections in a university hospital. Emerg Infect Dis 8:827–832. doi: 10.3201/eid0808.020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nowak J, Zander E, Stefanik D, Higgins PG, Roca I, Vila J, McConnell MJ, Cisneros JM, Seifert H. 2017. High incidence of pandrug-resistant Acinetobacter baumannii isolates collected from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J Antimicrob Chemother 72:3277–3282. doi: 10.1093/jac/dkx322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ou H-Y, Kuang SN, He X, Molgora BM, Ewing PJ, Deng Z, Osby M, Chen W, Xu HH. 2015. Complete genome sequence of hypervirulent and outbreak-associated Acinetobacter baumannii strain LAC-4: epidemiology, resistance genetic determinants and potential virulence factors. Sci Rep 5:8643. doi: 10.1038/srep08643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf?ua=1.
- 10.Doughari HJ, Ndakidemi PA, Human IS, Benade S. 2011. The ecology, biology and pathogenesis of Acinetobacter spp.: an overview. Microbes Environ 26:101–112. doi: 10.1264/jsme2.me10179. [DOI] [PubMed] [Google Scholar]
- 11.Zimbler DL, Arivett BA, Beckett AC, Menke SM, Actis LA. 2013. Functional features of TonB energy transduction systems of Acinetobacter baumannii. Infect Immun 81:3382–3394. doi: 10.1128/IAI.00540-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lannan FM, O'conor DK, Broderick JC, Tate JF, Scoggin JT, Moran NA, Husson CM, Hegeman EM, Ogrydziak CE, Singh SA, Vafides AG, Brinkley CC, Goodin JL. 2016. Evaluation of virulence gene expression patterns in Acinetobacter baumannii using quantitative real-time polymerase chain reaction array. Mil Med 181:1108–1113. doi: 10.7205/MILMED-D-15-00437. [DOI] [PubMed] [Google Scholar]
- 13.Sepahvand S, Davarpanah MA, Roudgari A, Bahador A, Karbasizade V, Kargar Jahromi Z. 2017. Molecular evaluation of colistin-resistant gene expression changes in Acinetobacter baumannii with real-time polymerase chain reaction. Infect Drug Resist 10:455–462. doi: 10.2147/IDR.S141196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camarena L, Bruno V, Euskirchen G, Poggio S, Snyder M. 2010. Molecular mechanisms of ethanol-induced pathogenesis revealed by RNA-sequencing. PLoS Pathog 6:e1000834. doi: 10.1371/journal.ppat.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aranda J, Poza M, Shingu-Vázquez M, Cortés P, Boyce JD, Adler B, Barbé J, Bou G. 2013. Identification of a DNA-damage-inducible regulon in Acinetobacter baumannii. J Bacteriol 195:5577–5582. doi: 10.1128/JB.00853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ducas-Mowchun K, De Silva PM, Patidar R, Schweizer HP, Kumar A. 2019. Tn7-based single-copy insertion vectors for Acinetobacter baumannii. Methods Mol Biol 1946:135–150. doi: 10.1007/978-1-4939-9118-1_13. [DOI] [PubMed] [Google Scholar]
- 17.Wei Y-L, Lin L-B, Ji X-L, Jing S-R. 2007. Construction of promoter probe vector for a cold-adapted bacterium, Acinetobacter sp. DWC6. Sheng Wu Gong Cheng Xue Bao 23:530–534. (In Chinese.) [PubMed] [Google Scholar]
- 18.Spaink HP, Okker RJ, Wijffelman CA, Pees E, Lugtenberg BJ. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol Biol 9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 19.Antunes LCS, Imperi F, Minandri F, Visca P. 2012. In vitro and in vivo antimicrobial activities of gallium nitrate against multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 56:5961–5970. doi: 10.1128/AAC.01519-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Runci F, Gentile V, Frangipani E, Rampioni G, Leoni L, Lucidi M, Visaggio D, Harris G, Chen W, Stahl J, Averhoff B, Visca P. 2019. The contribution of active iron-uptake to Acinetobacter baumannii pathogenicity. Infect Immun 87:e00755-18. doi: 10.1128/IAI.00755-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luna BM, Ulhaq A, Yan J, Pantapalangkoor P, Nielsen TB, Davies BW, Actis LA, Spellberg B. 2017. Selectable markers for use in genetic manipulation of extensively drug-resistant (XDR) Acinetobacter baumannii HUMC1. mSphere 2:e00140-17. doi: 10.1128/mSphere.00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lucidi M, Runci F, Rampioni G, Frangipani E, Leoni L, Visca P. 2018. New shuttle vectors for gene cloning and expression in multidrug-resistant Acinetobacter species. Antimicrob Agents Chemother 62:e02480-17. doi: 10.1128/AAC.02480-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol 64:2240–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunger M, Schmucker R, Kishan V, Hillen W. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45–51. doi: 10.1016/0378-1119(90)90494-c. [DOI] [PubMed] [Google Scholar]
- 25.Khan SA. 2000. Plasmid rolling-circle replication: recent developments. Mol Microbiol 37:477–484. [DOI] [PubMed] [Google Scholar]
- 26.Davies J. 2009. Everything depends on everything else. Clin Microbiol Infect 15:1–4. doi: 10.1111/j.1469-0691.2008.02682.x. [DOI] [PubMed] [Google Scholar]
- 27.Heo A, Jang H-J, Sung J-S, Park W. 2014. Global transcriptome and physiological responses of Acinetobacter oleivorans DR1 exposed to distinct classes of antibiotics. PLoS One 9:e110215. doi: 10.1371/journal.pone.0110215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friesen JD, An G, Fiil N. 1983. The lethal effect of a plasmid resulting from transcriptional readthrough of rplJ from the rplKA operon in Escherichia coli. Mol Gen Genet 189:275–281. doi: 10.1007/BF00337817. [DOI] [PubMed] [Google Scholar]
- 29.Makrides SC. 1996. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol Rev 60:512–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becher A, Schweizer HP. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 29:948–952. doi: 10.2144/00295bm04. [DOI] [PubMed] [Google Scholar]
- 31.Fournier P-E, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie J-M. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayer MP. 1995. A new set of useful cloning and expression vectors derived from pBlueScript. Gene 163:41–46. doi: 10.1016/0378-1119(95)00389-n. [DOI] [PubMed] [Google Scholar]
- 33.Wen H, Wang K, Liu Y, Tay M, Lauro FM, Huang H, Wu H, Liang H, Ding Y, Givskov M, Chen Y, Yang L. 2014. Population dynamics of an Acinetobacter baumannii clonal complex during colonization of patients. J Clin Microbiol 52:3200–3208. doi: 10.1128/JCM.00921-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kenyon CJ, Walker GC. 1981. Expression of the Escherichia coli uvrA gene is inducible. Nature 289:808–810. doi: 10.1038/289808a0. [DOI] [PubMed] [Google Scholar]
- 35.Norton MD, Spilkia AJ, Godoy VG. 2013. Antibiotic resistance acquired through a DNA damage-inducible response in Acinetobacter baumannii. J Bacteriol 195:1335–1345. doi: 10.1128/JB.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith MG, Des Etages SG, Snyder M. 2004. Microbial synergy via an ethanol-triggered pathway. Mol Cell Biol 24:3874–3884. doi: 10.1128/mcb.24.9.3874-3884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nwugo CC, Arivett BA, Zimbler DL, Gaddy JA, Richards AM, Actis LA. 2012. Effect of ethanol on differential protein production and expression of potential virulence functions in the opportunistic pathogen Acinetobacter baumannii. PLoS One 7:e51936. doi: 10.1371/journal.pone.0051936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gandhi JA, Ekhar VV, Asplund MB, Abdulkareem AF, Ahmadi M, Coelho C, Martinez LR. 2014. Alcohol enhances Acinetobacter baumannii-associated pneumonia and systemic dissemination by impairing neutrophil antimicrobial activity in a murine model of infection. PLoS One 9:e95707. doi: 10.1371/journal.pone.0095707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, Snyder M. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev 21:601–614. doi: 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McConnell MJ, Actis L, Pachón J. 2013. Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol Rev 37:130–155. doi: 10.1111/j.1574-6976.2012.00344.x. [DOI] [PubMed] [Google Scholar]
- 41.Harding CM, Hennon SW, Feldman MF. 2018. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol 16:91–102. doi: 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J-W, Helmann JD. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- 43.Eijkelkamp BA, Hassan KA, Paulsen IT, Brown MH. 2011. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics 12:126. doi: 10.1186/1471-2164-12-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stojiljkovic I, Bäumler AJ, Hantke K. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J Mol Biol 236:531–545. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- 45.Weinberg ED. 2009. Iron availability and infection. Biochim Biophys Acta 1790:600–605. doi: 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Barber MF, Elde NC. 2015. Buried treasure: evolutionary perspectives on microbial iron piracy. Trends Genet 31:627–636. doi: 10.1016/j.tig.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peleg AY, Jara S, Monga D, Eliopoulos GM, Moellering RC, Mylonakis E. 2009. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob Agents Chemother 53:2605–2609. doi: 10.1128/AAC.01533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wand ME, Bock LJ, Turton JF, Nugent PG, Sutton JM. 2012. Acinetobacter baumannii virulence is enhanced in Galleria mellonella following biofilm adaptation. J Med Microbiol 61:470–477. doi: 10.1099/jmm.0.037523-0. [DOI] [PubMed] [Google Scholar]
- 49.Olsen RJ, Watkins ME, Cantu CC, Beres SB, Musser JM. 2011. Virulence of serotype M3 group A Streptococcus strains in wax worms (Galleria mellonella larvae). Virulence 2:111–119. doi: 10.4161/viru.2.2.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai C-Y, Loh JMS, Proft T. 2016. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7:214–229. doi: 10.1080/21505594.2015.1135289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu A, Zhang Q, Zhang J, Yang B, Wu K, Xie W, Luan Y-X, Ling E. 2014. Insect prophenoloxidase: the view beyond immunity. Front Physiol 5:252. doi: 10.3389/fphys.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiss A, Broach WH, Lee MC, Shaw LN. 2016. Towards the complete small RNome of Acinetobacter baumannii. Microb Genom 2:e000045. doi: 10.1099/mgen.0.000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Casella LG, Weiss A, Pérez-Rueda E, Ibarra JA, Shaw LN. 2017. Towards the complete proteinaceous regulome of Acinetobacter baumannii. Microb Genom 3:mgen000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kröger C, Kary SC, Schauer K, Cameron AD. 2016. Genetic regulation of virulence and antibiotic resistance in Acinetobacter baumannii. Genes 8:12. doi: 10.3390/genes8010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eze EC, Chenia HY, El Zowalaty ME. 2018. Acinetobacter baumannii biofilms: effects of physicochemical factors, virulence, antibiotic resistance determinants, gene regulation, and future antimicrobial treatments. Infect Drug Resist 11:2277–2299. doi: 10.2147/IDR.S169894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Unterholzner SJ, Poppenberger B, Rozhon W. 2013. Toxin-antitoxin systems: biology, identification, and application. Mob Genet Elements 3:e26219. doi: 10.4161/mge.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuklin NA, Pancari GD, Tobery TW, Cope L, Jackson J, Gill C, Overbye K, Francis KP, Yu J, Montgomery D, Anderson AS, McClements W, Jansen KU. 2003. Real-time monitoring of bacterial infection in vivo: development of bioluminescent staphylococcal foreign-body and deep-thigh-wound mouse infection models. Antimicrob Agents Chemother 47:2740–2748. doi: 10.1128/aac.47.9.2740-2748.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosa SLL, Solheim M, Diep DB, Nes IF, Brede DA. 2015. Bioluminescence based biosensors for quantitative detection of enterococcal peptide–pheromone activity reveal inter-strain telesensing in vivo during polymicrobial systemic infection. Sci Rep 5:8339. doi: 10.1038/srep08339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Avci P, Karimi M, Sadasivam M, Antunes-Melo WC, Carrasco E, Hamblin MR. 2018. In-vivo monitoring of infectious diseases in living animals using bioluminescence imaging. Virulence 9:28–63. doi: 10.1080/21505594.2017.1371897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaddy JA, Arivett BA, McConnell MJ, López-Rojas R, Pachón J, Actis LA. 2012. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun 80:1015–1024. doi: 10.1128/IAI.06279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tannenbaum J, Bennett BT. 2015. Russell and Burch’s 3Rs then and now: the need for clarity in definition and purpose. J Am Assoc Lab Anim Sci 54:120–132. [PMC free article] [PubMed] [Google Scholar]
- 62.Francis KP, Joh D, Bellinger-Kawahara C, Hawkinson MJ, Purchio TF, Contag PR. 2000. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect Immun 68:3594–3600. doi: 10.1128/iai.68.6.3594-3600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 64.Renda BA, Chan C, Parent KN, Barrick JE. 2016. Emergence of a competence-reducing filamentous phage from the genome of Acinetobacter baylyi ADP1. J Bacteriol 198:3209–3219. doi: 10.1128/JB.00424-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 66.Stites SW, Nelson ME, Wesselius LJ. 1995. Transferrin concentrations in serum and lower respiratory tract fluid of mechanically ventilated patients with COPD or ARDS. Chest 107:1681–1685. doi: 10.1378/chest.107.6.1681. [DOI] [PubMed] [Google Scholar]
- 67.Janssen P, Maquelin K, Coopman R, Tjernberg I, Bouvet P, Kersters K, Dijkshoorn L. 1997. Discrimination of Acinetobacter genomic species by AFLP fingerprinting. Int J Syst Bacteriol 47:1179–1187. doi: 10.1099/00207713-47-4-1179. [DOI] [PubMed] [Google Scholar]
- 68.Iacono M, Villa L, Fortini D, Bordoni R, Imperi F, Bonnal RJP, Sicheritz-Ponten T, De Bellis G, Visca P, Cassone A, Carattoli A. 2008. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob Agents Chemother 52:2616–2625. doi: 10.1128/AAC.01643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Juni E, Janik A. 1969. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum). J Bacteriol 98:281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vaneechoutte M, Young DM, Ornston LN, De Baere T, Nemec A, Van Der Reijden T, Carr E, Tjernberg I, Dijkshoorn L. 2006. Naturally transformable Acinetobacter sp. strain ADP1 belongs to the newly described species Acinetobacter baylyi. Appl Environ Microbiol 72:932–936. doi: 10.1128/AEM.72.1.932-936.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cosgaya C, Marí-Almirall M, Van Assche A, Fernández-Orth D, Mosqueda N, Telli M, Huys G, Higgins PG, Seifert H, Lievens B, Roca I, Vila J. 2016. Acinetobacter dijkshoorniae sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex mainly recovered from clinical samples in different countries. Int J Syst Evol Microbiol 66:4105–4111. doi: 10.1099/ijsem.0.001318. [DOI] [PubMed] [Google Scholar]
- 72.Nemec A, Krizova L, Maixnerova M, Sedo O, Brisse S, Higgins PG. 2015. Acinetobacter seifertii sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex isolated from human clinical specimens. Int J Syst Evol Microbiol 65:934–942. doi: 10.1099/ijs.0.000043. [DOI] [PubMed] [Google Scholar]
- 73.Alting-Mees MA, Short JM. 1989. pBluescript II: gene mapping vectors. Nucleic Acids Res 17:9494. doi: 10.1093/nar/17.22.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thompson MG, Yildirim S. 2019. Transformation of Acinetobacter baumannii: electroporation. Methods Mol Biol 1946:69–74. doi: 10.1007/978-1-4939-9118-1_7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full-length sequences of pLPV1Z, pLPV2Z, and pLPV3Z plasmids have been deposited in the GenBank database under accession numbers MK681506, MK681507, and MK681508, respectively.