Abstract
In the last decade, extended-spectrum cephalosporin and carbapenem resistant Gram-negative bacilli (GNB) have been extensively reported in the literature as being disseminated in humans but also in animals and the environment. These resistant organisms often cause treatment challenges due to their wide spectrum of antibiotic resistance. With the emergence of colistin resistance in animals and its subsequent detection in humans, the situation has worsened. Several studies reported the transmission of resistant organisms from animals to humans. Studies from the middle east highlight the spread of resistant organisms in hospitals and to a lesser extent in livestock and the environment. In view of the recent socio-economical conflicts that these countries are facing in addition to the constant population mobilization; we attempt in this review to highlight the gaps of the prevalence of resistance, antibiotic consumption reports, infection control measures and other risk factors contributing in particular to the spread of resistance in these countries. In hospitals, carbapenemases producers appear to be dominant. In contrast, extended spectrum beta lactamases (ESBL) and colistin resistance are becoming a serious problem in animals. This is mainly due to the continuous use of colistin in veterinary medicine even though it is now abandoned in the human sphere. In the environment, despite the small number of reports, ESBL and carbapenemases producers were both detected. This highlights the importance of the latter as a bridge between humans and animals in the transmission chain. In this review, we note that in the majority of the Middle Eastern area, little is known about the level of antibiotic consumption especially in the community and animal farms. Furthermore, some countries are currently facing issues with immigrants, poverty and poor living conditions which has been imposed by the civil war crisis. This all greatly facilitates the dissemination of resistance in all environments. In the one health concept, this work re-emphasizes the need to have global intervention measures to avoid dissemination of antibiotic resistance in humans, animals and the environment in Middle Eastern countries.
Keywords: colistin, ESBL, carbapenemases, one health, MDROs
Introduction
In the 1940s, the discovery of antibiotics was seen as one of medicine’s major achievements that saved millions of lives (van Hoek et al., 2011). However, in the last decade antimicrobial resistance has significantly increased in bacteria and reduced the effectiveness of many clinically important antibiotics (Seiffert et al., 2013). Gram-negative bacilli (GNB) are among the most common causative agents of infectious diseases (Tian et al., 2012). Members of this family are ubiquitous, i.e., can be found in humans and animals’ intestinal microflora, but also in the environment (Verraes et al., 2013). Among other resistant organisms, GNB are distinct in view of their complex mechanisms of resistance. These are mainly mediated via the production of extended spectrum beta lactamases (ESBL), AmpC and carbapenemases (Schill et al., 2017). These hydrolyzing enzymes confer bacterium resistance toward the most common class of antibiotics prescribed nowadays in clinical settings: beta lactams (Ruppe et al., 2015). Furthermore, resistance genes of these enzymes are often located on plasmids harboring resistance determinants to other classes of antibiotics; thus challenging therapeutic options when infectious diseases do occur (Ruppe et al., 2015). The dissemination GNB resistant to extended spectrum cephalosporins and carbapenem, necessitates the re-use of colistin (a polymyxin E antibiotic) previously abandoned due to its toxicity and side effects (Olaitan et al., 2014b). The re-introduction of colistin in recent years has also seen the emergence of resistance, further complicating the clinical situation. Colistin resistance occurs either via chromosomal mutations that mediates the alteration of the lipid A moiety of the lipopolysaccharide chain (Baron et al., 2016); or via the acquisition of an mcr gene (Olaitan et al., 2016a).
Previously known to be confined to the hospital settings, multi-drug resistant organisms (MDROs) are nowadays widely spread in animals and the environment (Rafei et al., 2015a). Dandachi et al. (2018a) reported the wide dissemination of ESBL producers as well as colistin resistant GNB in poultry, cattle, swine and companion animals in Mediterranean countries. For instance, several studies have shown that multi-drug resistance (MDR) can be readily transferred from one ecosystem to another via direct/indirect contact with contaminated animals and/or animal products (Huijbers et al., 2014), dust (Blaak et al., 2015), air (von Salviati et al., 2015), contaminated wastewaters (Guenther et al., 2011), and soil fertilized with animal manure (Laube et al., 2014). Humans, animals, and the environment together therefore form an interconnected system that should be carefully addressed in terms of bacterial resistance, antibiotic stewardship, and infection control measures.
In this context, Middle Eastern countries are thus of special interest. The dissemination of MDROs in this region of the world involves an interplay of over/misuse of antibiotics in humans and animals, the absence of infection control measures and most importantly the recent continuous population mobilization due to socio-economic conflicts and multiple war crises. In this review, our aim is to describe the epidemiology of extended spectrum cephalosporin, carbapenem and colistin resistant GNB in humans, animals and the environment in the Middle Eastern area. The Middle East includes 15 countries: Bahrain, Egypt, Iraq, Iran, Jordan, the Kingdom of Saudi Arabia (KSA), Kuwait, Lebanon, Palestine, Qatar, Sultanate Oman, Syria, Turkey, the United Arab Emirates (UAE), and Yemen. Our attempt is to highlight the gaps in bacterial resistance reports, antibiotic consumption data as well as infection control measures in this distinct area of the world.
Distribution of Multi-Drug Resistant Organisms in Humans
Infections With ESBL/AmpC Producers
Extended spectrum cephalosporins and penicillin’s have been widely used in clinical settings due to their wide spectrum of activity as well as their negligible toxicity compared to other antimicrobial agents (Bush and Bradford, 2016). Their un-restricted use by physicians, in addition to their purchasing ease, without medical prescription in the community pharmacies, plays an important role in the emergence of bacteria resistant to these antimicrobial agents (Figure 1).
FIGURE 1.
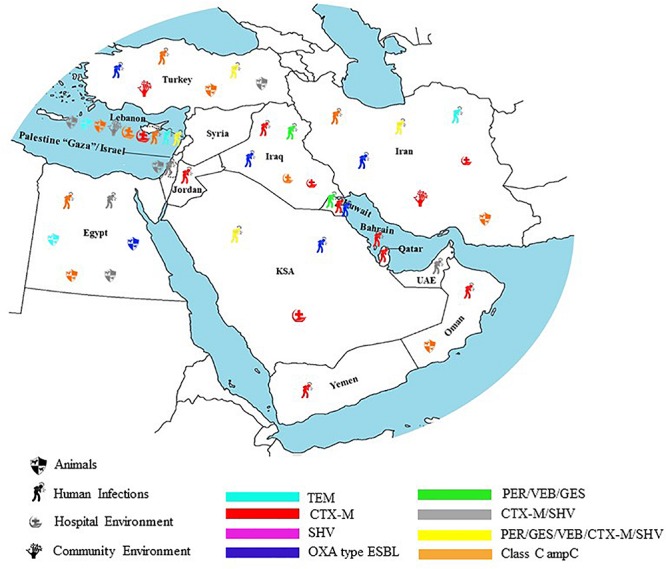
Geographical distribution of ESBL in humans, animals, hospital, and community environment in the Middle East.
In Iran, studies have shown that in Klebsiella spp., resistance to extended spectrum beta lactams is mainly mediated via the production of CTX-M variants (CTX-M-15, CTX-M-3, CTX-M-8, CTX-M-1, and CTX-M-2) (Feizabadi et al., 2010; Peerayeh et al., 2014; Bialvaei et al., 2016; Akya et al., 2018; Dehshiri et al., 2018) and to a lesser degree via SHV (SHV-12, SHV-11, SHV-5) (Feizabadi et al., 2010; Shakib et al., 2018), TEM genes (Gholipour et al., 2014; Maleki et al., 2018) and others (VEB, PER, and GES) (Sedighi et al., 2017). pAmpC beta lactamase genes were reported by two studies in clinical isolates of Klebsiella pneumoniae (Mansouri et al., 2012; Kiaei et al., 2018). PFGE and ERIC-PCR analysis in these studies showed the presence of different clones in each clinical center (Akya et al., 2018; Hashemizadeh et al., 2018; Kiaei et al., 2018; Maleki et al., 2018). This is with the exception of one study where high clonal relatedness among ESBL K. pneumoniae strains was reported (Ghaffarian et al., 2018). Mehrgan et al. (2010) showed that intensive care unit (ICU) or medical ward stays are significantly associated with the acquisition of ESBL Klebsiella spp. Indeed, these resistant organisms are often found in patients who are very young and who have not yet developed full immunity, thus making them susceptible to opportunistic pathogen infections (Mehrgan et al., 2010). Similarly, to Klebsiella spp., CTX-M-15 followed by SHV, TEM and to a lesser extent CIT, were the most common beta lactamase genes detected in clinical strains of Escherichia coli (Gholipour et al., 2014; Rezai et al., 2015; Shayan and Bokaeian, 2015; Bialvaei et al., 2016). It is worth noting the detection of CTX-M, TEM, and CIT beta lactamases in diarrheagenic E. coli strains: Enteroaggregative and Enteropathogenic ones (Heidary et al., 2014; Aminshahidi et al., 2017). These E. coli patotypes are always pathogenic when present in human intestines (Fratamico et al., 2016). Moreover, one recent study reported the isolation of CTX-M-15 extraintestinal pathogenic E. coli ST131 from inpatients and outpatients in Iran (Namaei et al., 2017). Statistical analysis indicated that ESBL producing ST131 E. coli strains were positively associated with CTX-M variants, CTX-M-15, and TEM beta lactamases (Namaei et al., 2017). Moreover, strictly pathogenic species producing ESBL were also detected: CTX-M-1/CTX-M-15 Salmonella spp. and CTX-M-15/CMY-2 Shigella spp. (Salimian Rizi et al., 2015; Bialvaei et al., 2016; Aminshahidi et al., 2017). The high incidence of cephalosporins resistance in pathogenic bacteria in this country may be attributed in part to their inappropriate and high use in clinical settings (Amin et al., 2018); this is in addition to their extensive utilization in the Iranian community via self-medication (SM) (Zamanlou et al., 2018). Other ESBL producers that have been detected in clinical settings in Iran include CTX-M-15/TEM-169, SHV-12 producing Enterobacter cloacae (Peymani et al., 2014). On the other hand, in Pseudomonas aeruginosa, the major ESBL types were OXA-10/OXA-4, PER-1, VEB-1, and GES-1 (Mirsalehian et al., 2010; Alikhani et al., 2014; Farshadzadeh et al., 2014; Emami et al., 2015; Davodian et al., 2016; Amirkamali et al., 2017). This is followed by CTX-M, TEM-116, SHV-12, DHA and hyper-produced ADC enzymes (Bokaeian et al., 2014; Fazeli and Momtaz, 2014; Rafiee et al., 2014). Only one study in Iran revealed 13 distinct profiles among 100 ESBL/carbapeneme resistant P. aeruginosa isolated from burn patients via RAPD analysis. A dominant RAPD type was observed consisting of 80 isolates, thus revealing the possible existence of endemic clones circulating among patients (Neyestanaki et al., 2014).
In Turkey, ESBL production in E. coli and K. pneumoniae is mainly mediated via CTX-M-group1 (CTX-M-15 and CTX-M-1) and CTX-M-group2. Others include PER and OXA-10 in E. coli (Elaldi et al., 2013; Gorgec et al., 2015; Iraz et al., 2015). Furthermore, CMY-2, CIT, MOX, EBC, FOX, and ACT-1 have been detected in E. coli and Klebsiella spp., respectively (Demirbakan et al., 2008; Sari et al., 2013; Yilmaz et al., 2013). PFGE analysis showed no major clonal relationship per species in each clinical center (Durmaz et al., 2015; Gorgec et al., 2015). Multivariate analysis showed that urinary catheter insertion was a common risk factor for acquiring an infection with an ESBL quinolone resistant E. coli strain in inpatients and outpatients alike (Durmaz et al., 2015). Moreover, in two other studies the risk factors for the development of an ESBL K. pneumoniae blood stream infections were high, with the duration of hospitalization being a common factor (Serefhanoglu et al., 2009). Other factors included prior antibiotic use and the use of aminoglycosides (Tanir Basaranoglu et al., 2017). Other ESBL producing Enterobacteriaceae detected in Turkey include CTX-M/TEM/SHV/qnrA aac(6′)-ib Enterobacter spp. and VEB-1/qnrA1 Providencia stuartii (Nazik et al., 2009, 2011; Erdem et al., 2018). Interestingly, Agin et al. (2011) reported an outbreak of Salmonella enterica serovar typhimurium producing SHV-12 and CTX-M-3 ESBLs. In view of this, handwashing and disinfection procedures in addition to the establishment of an active surveillance program were initiated. These infection control measures led to the containment of the outbreak after 2 years. As for non-fermenters, PER-1 was the main ESBL type detected in P. aeruginosa and Acinetobacter spp. alike (Atilla et al., 2012; Erac et al., 2013; Keskin et al., 2014). In Pseudomonas spp. additional types were also detected such as OXA-10, OXA-14, and GES-1 (Aktas et al., 2012; Er et al., 2015).
In Lebanon, clinical epidemiological studies showed the predominance of CTX-M-15 and SHV-5a in E. coli and K. pneumoniae (Charrouf et al., 2014; Daoud et al., 2017). Furthermore, one report described the presence of SHV-11/CTX-M-15/acc(6′)-lb-cr/qnrb1 producing ST336 K. pneumoniae (Tokajian et al., 2015). PFGE analysis revealed clonal diversity among ESBL producing E. coli and K. pneumoniae (Daoud et al., 2017). As for the effect of antibiotic prescription and its correlation with the level of bacterial resistance, Daoud et al. (2017) found a significant association between aztreonam resistance and the use of penicillin’s, and between cefuroxime, ceftazidime, cefoxitin, ciprofloxacin resistance and 3rd/4th generation cephalosporins use in Klebsiella spp. Moreover, one study reported the detection of four unrelated ESBL producing Shigella sonnei isolated from the stool samples of patients admitted for bacillary dysentery. These isolates harbored the CTX-M-15 gene on the plasmid and were flanked by ISEcp1 (Sabra et al., 2009).
In Israel, one study found low prevalence of ESBL producers in a clinical center (Chazan et al., 2009). The authors suggest that one of the reasons for this finding is the strict supervision of antibiotic prescription applied in their hospital; in addition to the limited use of ceftazidime (Chazan et al., 2009). Another study in the same country, argued that recent hospitalization, urinary tract infection (UTI) prophylaxis and Klebsiella spp. UTI are risk factors for the development of community acquired ESBL UTI (Dayan et al., 2013). Another has found that prior antipseudomonal therapy and empirical cephalosporin therapy are independent risk factors for UTI, caused by an ESBL producing Proteus mirabilis (Cohen-Nahum et al., 2010). As for the underlying genes of resistance, one study showed the presence of CTX-M-2, CTX-M-15, and CTX-M-14 in predominantly ST131 E. coli strains (Karfunkel et al., 2013). In this study, 93 and 51% of the isolates were co-resistant to fluoroquinolones and gentamicin, respectively. Transformation experiments suggest that aminoglycosides resistance is co-carried on the same plasmid harboring the CTX-M gene (Karfunkel et al., 2013). Other studies in Palestine have found clonal diversity among ESBL producing E. coli clinical strains (Adwan et al., 2014; Tayh et al., 2016). On the other hand, in Israel, Karfunkel et al. (2013) reported the dominance of the ST131 among 41 CTX-M positive E. coli strains isolated for community onset bacteremia (COBSI) at Tel Aviv Sourasky Medical Center. In this center, the incidence of COBSI has increased 2.7-fold over a 7 year period. This increase appears to be correlated with the clonal expansion of ST131 E. coli strains carrying the blaCTX-M-14 and blaCX-M-15 genes (Karfunkel et al., 2013). ESBL production by K. pneumoniae in clinical settings was reported, whereby CTX-M-15, CTX-M-14a, CTX-M-3, SHV-12, SHV-5, and SHV-33 were detected (Tayh et al., 2017). In Jordan, very few studies have addressed the prevalence of ESBL producers in clinical settings. However, blaCTX-M (CTX-M-15, CTX-M-1, and CTX-M-9) was the only ESBL type detected in Enterobacteriaceae notably E. coli and ST131 K. pneumoniae strains (Hayajneh et al., 2015; Aqel et al., 2017; Nairoukh et al., 2018).
In Iraq, CTX-M-1, SHV, TEM producing E. coli strains were reported in recurrent UTI patients. In this report, MDR was significantly higher in ESBL E. coli versus non-ESBL ones (Al-Mayahie and Al Kuriashy, 2016). Similar results were obtained in a study addressing ESBL producers in pregnant/non-pregnant women with symptomatic genital tract infection. It is worth mentioning that ESBL producers co-resistant to non-beta lactam antibiotics is of special interest in this category; this is in view of the narrow choice of antibiotics that could be used in this category of patients (Al-Mayahie, 2013). Furthermore, in this country, CTX-M, SHV, TEM, and OXA ESBLs were described in clinical isolates of Morganella morganii with high resistance toward minocycline, trimethoprim- sulfamethoxazole and ciprofloxacin (Al-Muhanna et al., 2016). In parallel, VEB, PER, and OXA-10 were detected in high risk strains of P. aeruginosa: ST244, ST235, ST308, and ST654 (van Burgh et al., 2018).
In Kuwait, diverse genetic profiles of ESBL producing E. coli strains were detected in inpatients and outpatients alike (Dashti et al., 2014). CTX-M-15 followed by SHV-12, CMY-2, CTX-M-14, CTX-M-56, and CTX-M-2 are the most common ESBL types detected (Dashti et al., 2014; Jamal et al., 2015). In contrast to E. coli, one study in Kuwait reported identical PFGE profiles of K. pneumoniae SHV-112 positive strains isolated from blood and urine specimens of ICU patients (Dashti et al., 2010a). Another study however, reported different sequence types of K. pneumoniae detected in hospitalized patients: ST677, ST16, ST107, and ST485 producing CTX-M-15, SHV-11, and CTX-M-14 beta lactamases (Jamal et al., 2015).
In KSA, ST131 followed by ST38 E. coli strains producing ESBL appears to be predominant in clinical settings (Alghoribi et al., 2015; Alyamani et al., 2017; Yasir et al., 2018). In these, the main ESBL types detected were: CTX-M-15, CTX-M-9, CTX-M-1, CTX-M-8/25, CTX-M-2, CTX-M-14, SHV-12, and SHV-5 (Shibl et al., 2012; Al Sheikh et al., 2014; Alyamani et al., 2017; Yasir et al., 2018). Indeed, one study has shown that ESBL producers were significantly more resistant to aminoglycosides, ciprofloxacin and trimethoprim-sulfamethoxazole (Hassan and Abdalhamid, 2014). Al-Otaibi and Bukhari (2013) found that recurrent UTIs, surgical intervention, children with vesicoureteric reflux and patients who had underlying renal transplant and renal disease are all possible risk factors for the acquisition of an ESBL UTI E. coli strain.
As for ESBL K. pneumoniae, the situation appears to be similar to their E. coli counterparts (Ahmad et al., 2009; Hassan and Abdalhamid, 2014; Somily et al., 2014). This is with the exception to the additional detection of other CTX-M variants such as CTX-M-3, CTX-M-82, CTX-M-57, and CTX-M-27 in K. pneumoniae as compared to E. coli strains (Al-Qahtani et al., 2014). In addition, in view of the wide diversity of ESBL K. pneumoniae isolates, it seems that clonal spread plays a negligible role in the dissemination of these strains (Al-Qahtani et al., 2014). Moreover, one study reported the detection of CTX-M-14 and SHV-12 in clinical isolates of Citrobacter freundii and Enterobacter spp. (Al Sheikh et al., 2014). SHV-5, CMY-2, and DHA-1 were also detected in Enterobacter spp. isolated from clinical settings in KSA (Abdalhamid et al., 2017a). On the other hand, VEB, GES, and OXA-10 were detected in P. aeruginosa clinical strains (Al-Agamy et al., 2012; Tawfik et al., 2012).
ESBL production in Acinetobacter baumannii on the other hand, was meditated via CTX-M and GES variants (Alyamani et al., 2015; Al-Agamy et al., 2017). Similarly, to other ESBL producing GNB in KSA, MLST analysis revealed the presence of a wide variety of sequence types in ESBL A. baumannii strains (Alyamani et al., 2015; Al-Agamy et al., 2017) (Table 1). Moreover, one study addressing the hajj pilgrims of Marseille, reported the detection of 2 CTX-M-2 producing Salmonella spp. Both strains were gentamicin and colistin resistant, in addition, they belonged to the epidemic Newport serotype ST45 (Olaitan et al., 2015). This finding calls for improved public health surveillance during the Hajj period in order to prevent the dissemination of MDROs in KSA and worldwide (Olaitan et al., 2015).
TABLE 1.
Sequence and plasmid types associated with ESBL genes in humans, animals, and environment in the Middle East.
| Country | ESBL gene | Reservoir | Species | Sequence type/phylogroup | Plasmid type |
| Iran | CTX-M | Humans | E. coli | ||
| K. pneumoniae | |||||
| E. cloacae | |||||
| Salmonella spp. | |||||
| Shigella spp. | |||||
| Hospital environment | E. coli | ||||
| Community environment | A. baumannii | ||||
| TEM | Humans | E. coli | |||
| K. pneumoniae | |||||
| E. cloacae | |||||
| A. baumannii | |||||
| P. aeruginosa | |||||
| Hospital environment | E. coli | ||||
| P. aeruginosa | |||||
| Community environment | A. baumannii | ||||
| SHV | Humans | E. coli | |||
| K. pneumoniae | |||||
| E. cloacae | |||||
| P. aeruginosa | |||||
| Animals | E. coli | ||||
| Hospital environment | E. coli | ||||
| P. aeruginosa | |||||
| OXA | Humans | E. coli | |||
| P. aeruginosa | |||||
| Hospital environment | E. coli | ||||
| GES | Humans | K. pneumoniae | |||
| P. aeruginosa | |||||
| VEB | Humans | E. coli | |||
| K. pneumoniae | |||||
| P. aeruginosa | |||||
| PER | Humans | K. pneumoniae | |||
| A. baumannii | |||||
| P. aeruginosa | |||||
| Turkey | CTX-M | Humans/community environment | E. coli | – | |
| Animals | E. coli | A, D, B1, B2 | |||
| Humans | K. pneumoniae | ||||
| Humans | Enterobacter spp. | ||||
| TEM | Humans/animals | E. coli | |||
| Humans | K. pneumoniae | ||||
| Humans | Enterobacter spp. | ||||
| SHV | Humans | E. coli | |||
| Enterobacter spp. | |||||
| S. paratyphi | |||||
| Animals | E. coli | A, D, B1, B2 | |||
| Community environment | E. coli | ||||
| OXA | Humans | E. coli | |||
| P. aeruginosa | |||||
| Community environment | E. coli | ||||
| GES | Humans | P. aeruginosa | |||
| VEB | Humans | K. pneumoniae | |||
| PER | Humans | E. coli | |||
| P. aeruginosa | |||||
| A. baumannii | |||||
| Lebanon | CTX-M | Humans | E. coli | ||
| K. pneumoniae | |||||
| Shigella spp. | |||||
| Animals | E. coli | ST10, ST617, ST58, ST69, ST1303, ST156, ST5470, ST354, ST155, ST3224 | |||
| Community environment | E. coli | ST328, ST405, ST34, ST48, ST131, ST120, ST2067, ST10, ST38, ST410 | IncFII | ||
| ST38, ST1431, ST46 | IncFIA | ||||
| ST212 | IncY | ||||
| ST617, ST4144, ST6470, ST6222, ST90, ST38, ST4608, ST6894, ST127 | |||||
| K. pneumoniae | ST22, ST336, ST15, ST16 | IncFIIk | |||
| C. freundii | IncY | ||||
| C. braakii | IncY | ||||
| Hospital environment | E. coli | ||||
| K. pneumoniae | |||||
| TEM | Humans | E. coli | |||
| K. pneumoniae | |||||
| Animals | E. coli | ||||
| K. pneumoniae | |||||
| Hospital environment | E. coli | ||||
| K. pneumoniae | |||||
| SHV | Humans | E. coli | |||
| K. pneumoniae | |||||
| Animals | E. coli | ||||
| K. pneumoniae | |||||
| Community environment | E. coli | ST46 | IncFII | ||
| ST617, ST6470, ST90, ST4608, ST6480, ST4144 | |||||
| SHV | Hospital environment | E. coli | |||
| K. pneumoniae | |||||
| VEB | Humans | K. pneumoniae | |||
| Palestine/Israel | CTX-M | Humans | E. coli | ST1, ST2, ST39, ST131, ST472, ST473, ST474, ST475, ST476, ST477 | |
| K. pneumoniae | |||||
| Animals | E. coli | ||||
| TEM | Humans | E. coli | |||
| SHV | Humans | K. pneumoniae | |||
| Animals | E. coli | ||||
| Jordan | CTX-M | Humans | E. coli | ST131 | |
| K. pneumoniae | A2, C | ||||
| Iraq | CTX-M | Humans | E. coli | ||
| M. morganii | |||||
| TEM | Humans | M. morganii | |||
| SHV | Humans | E. coli | |||
| M. morganii | |||||
| OXA | Humans | P. aeruginosa | ST244, ST308 | ||
| VEB | Humans | P. aeruginosa | ST235 | ||
| PER | Humans | P. aeruginosa | |||
| Bahrain | CTX-M | Humans | E. coli | ||
| K. pneumoniae | |||||
| TEM | Humans | E. coli | |||
| K. pneumoniae | |||||
| SHV | Humans | E. coli | |||
| Qatar | CTX-M | Humans | E. coli | ||
| K. pneumoniae | |||||
| TEM | Humans | E. coli | |||
| K. pneumoniae | |||||
| SHV | Humans | E. coli | |||
| United Arab Emirates | CTX-M | Humans | E. coli | ST131 | IncFII-FIA-FIB |
| ST131 | |||||
| SHV | Humans | K. pneumoniae | |||
| Oman | CTX-M | Humans | E. coli | ||
| Yemen | CTX-M | Humans | E. coli | ST131 | |
| K. pneumoniae | ST1399, ST340, ST405, ST147, ST29 | ||||
| SHV | Humans | K. pneumoniae | ST1399, ST340, ST405, ST147 | ||
| Kuwait | CTX-M | Humans | E. coli | ST131, ST405, ST38 | |
| K. pneumoniae | |||||
| SHV | Humans | E. coli | ST131 | ||
| K. pneumoniae | |||||
| KSA | CTX-M | Humans | E. coli | ST131, ST493, ST73, ST2346, ST1193, ST92, ST421, ST636, ST410, ST3268, ST120, ST602, | |
| ST224, ST1196, ST2852, ST58, ST1011, ST6438, | |||||
| ST167, ST8162, ST4981, ST1284, ST648, ST117, ST457, ST394, ST69, ST38, ST10 | |||||
| K. pneumoniae | |||||
| C. freundii | |||||
| Enterobacter spp. | |||||
| A. baumannii | ST195 | ||||
| Community environment | E. cloacae | ||||
| TEM | Humans | E. coli | ST493, ST73, ST131, ST2346, ST1193, ST92, ST421, ST636, ST410, ST4040, ST46, ST38, ST3268, ST405 | ||
| ST648, ST117, ST394, ST69, ST8162, ST4981, ST1284 | |||||
| A. baumannii | ST195 | ||||
| Animals | E. coli | ||||
| SHV | Humans | E. coli | |||
| K. pneumoniae | |||||
| C. freundii | |||||
| Enterobacter spp. | |||||
| Animals | E. coli | ||||
| OXA | Humans | E. coli | |||
| P. aeruginosa | |||||
| GES | Humans | A. baumannii | ST1, ST2, ST15, ST17, ST113, ST114, ST115, ST116 | ||
| P. aeruginosa | |||||
| VEB | Humans | P. aeruginosa | |||
| Egypt | CTX-M | Humans | E. coli | ST405, ST68, ST131, ST648; Phylogroup A, B1, B2, D | |
| K. pneumoniae | |||||
| Enterobacter spp. | |||||
| Salmonella spp. | |||||
| P. mirabilis | |||||
| C. freundii | |||||
| S. marcescens | |||||
| A. baumannii | |||||
| P. aeruginosa | |||||
| Animals | E. coli | ST131 | IncFII | ||
| D | |||||
| TEM | Humans | K. pneumoniae | |||
| A. baumannii | |||||
| Animals | E. coli | B1, C, D | |||
| Salmonella spp. | |||||
| SHV | Humans | E. coli | ST68, Phylogroups A and D | ||
| A. baumannii | |||||
| P. aeruginosa | |||||
| Animals | E. coli | D | |||
| OXA | Animals | E. coli | C. B2 |
References are cited in the main text.
In Bahrain, CTX-M-grp1 and CTX-M-grp9 with high resistance to ciprofloxacin, nitrofurantoin, and trimethoprim-sulfamethoxazole have been described as the predominant ESBL types detected in E. coli and K. pneumoniae clinical strains (Bindayna and Murtadha, 2011; Shahid et al., 2014; Zowawi et al., 2014). In Qatar, CTX-M-group1, CTX-M-group9, TEM and SHV dominated the E. coli and K. pneumoniae clinical isolates (Sid Ahmed S.S. et al., 2016; Eltai et al., 2018b). In United Arab Emirates, CTX-M-15 and SHV-258 were detected in K. pneumoniae isolated from inpatients (Alfaresi et al., 2011). In parallel, CTX-M-15, CTX-M-3, and CTX-M-14 producing ST131 E. coli strains were also reported (Peirano et al., 2014). This same E. coli sequence type was isolated recently from the urine sample of a 76-year-old male patient. This isolate harbored the blaCTX-M-27 gene carried on an IncFII-FIA-FIB plasmid along with aminoglycosides (aadA5, strA, and strB), sulfonamide (sul1 and sul2), TET [tet(A)], macrolides (mphA), and trimethoprim (dfrA17), resistance determinants (Mutti et al., 2018). Ranjan Dash et al. (2018) found that age, gender, recurrent UTI and catheterization are significant factors for developing an ESBL UTI in United Arab Emirates.
In the Sultanate of Oman, the main risk factors for ESBL infections in children was suggested to include being female, severe illness, prolonged hospital stays and previous exposure to antimicrobials (Al Muharrmi et al., 2008). As for the ESBL types detected, only one study showed the presence of CTX-M-15 producing a clinical E. coli strain (Zowawi et al., 2014).
Last, but not least in the gulf region, in Yemen, CTX-M-15, SHV-11, SHV-76, and SHV-184 were detected in clonally diverse K. pneumoniae clinical isolates (Gharout-Sait et al., 2014). On the other hand, CTX-M-15 was observed in ST131 E. coli strains (Alsharapy et al., 2018). As it becomes evident, ST131 is highly associated with ESBL production in the Middle Eastern region as well as other countries across the world: Israel (Karfunkel et al., 2013), KSA (Alghoribi et al., 2015; Alyamani et al., 2017; Yasir et al., 2018), Iran (Moghanni et al., 2018), Bulgaria (Markovska et al., 2012), Ecuador (Zurita et al., 2019), and Spain (Merino et al., 2016).
In Egypt, ESBL producers are widely spread in hospitals. One recent study showed a significant association between 3rd generation cephalosporins and resistance fluoroquinolones, gentamicin and tetracycline in hospital acquired infections (Galal et al., 2018). CTX-M-1, CTX-M-9, CTX-M-15, CTX-M-14, and SHV-12 were reported in E. coli strains isolated from different clinical origins (Hassan et al., 2012; Abdelaziz et al., 2013a; Abdallah et al., 2015b; Helmy and Kashef, 2017).
Additionally, TEM and SHV variants were also reported by El-Badawy et al. (2017), who found that among 61 clinical isolates of E. coli producing ESBL, SHV-11, and TEM-214 were predominant followed by others such as SHV-48, TEM-206, TEM-57, TEM-135, TEM-207, TEM-34, and TEM-176. This study was the first to report the detection of GES in E. coli strains isolated from Egyptian patients. A total of 92.30% ESBL E. coli isolates belonged to the ST131 clone and 45.83% of them belonged to the O25b serotype (El-Badawy et al., 2017). The association of E. coli ST131 with high antimicrobial resistance and virulence was previously reported in the literature (Can et al., 2015). On the other hand, CTX-M-15, CTX-M-14, SHV-11, and SHV-12 were detected in ESBL positive K. pneumoniae strains (Abdelaziz et al., 2013a; Abdallah et al., 2015b). Considerable resistance against aminoglycosides, fluoroquinolones and trimethoprim-sulfamethoxazole was observed in these isolates (Abdallah et al., 2017). Other ESBL producers detected in Egyptian hospitals include CTX-M-14/CTX-M-15 Enterobacter spp. (Abdallah et al., 2015b; Galal et al., 2018), CTX-M-15/SHV P. mirabilis, CTX-M-15/SHV C. freundii, CTX-M-14 Serratia marcescens (Helmy and Kashef, 2017; Galal et al., 2018) and CTX-M Salmonella spp. (Abdallah et al., 2017). CTX-M-14, CTX-M-15, and SHV ESBL types were detected in P. aeruginosa and A. baumannii (Abdelkader et al., 2017; Alkasaby and El Sayed Zaki, 2017; Helmy and Kashef, 2017). Furthermore, as for AmpC production, CMY variants (CMY-2, CMY-42, CMY-102), DHA-1, EBC, FOX, and MOX were detected in clinical isolates of Enterobacteriaceae such as E. coli, K. pneumoniae, and P. mirabilis (Abdelaziz et al., 2013a; Helmy and Wasfi, 2014; Wassef et al., 2015).
Infections With Carbapenemase Producers
In Iran, K. pneumoniae is the most common carbapenemase producing Enterobacteriaceae clinical isolate. Carbapenem resistance in these is often mediated by NDM-1 and OXA-48 production followed by KPC (KPC-2), VIM (VIM-1 and VIM-4), and IMP (Rastegar Lari et al., 2013; Azimi et al., 2014; Nobari et al., 2014; Fazeli et al., 2015; Rajabnia et al., 2015; Firoozeh et al., 2016, 2017; Sedighi et al., 2017; Shahcheraghi et al., 2017; Armin et al., 2018; Ghotaslou et al., 2018; Hosseinzadeh et al., 2018; Moghadampour et al., 2018a). The majority of the studies reported no clonal relatedness among isolated carbapenem resistant K. pneumoniae in each center (Shahcheraghi et al., 2013; Jafari et al., 2018; Kiaei et al., 2018). This is with the exception of three centers where an identical genotype was observed for VIM-1 producers (Nobari et al., 2014), for NDM-1 producers the strains were distributed into two major clonal complexes including ST13 and ST392 (Shoja et al., 2018) and among NDM-1 and/or OXA-48 positive ones the predominant cluster/pulsotype was associated to ST11 and ST893 (Solgi et al., 2018). In this latter study, OXA-48 and NDM-1 genes were located on IncL/M and IncFII plasmids, respectively. These transferable plasmids are known as potent contributors to the dissemination of resistance genes including NDM-1, OXA-48, and CTX-M-15 among enterobacterial species (Solgi et al., 2018). OXA-48 and NDM (NDM-1, NDM-7) were dominant in carbapenem resistant E. coli strains (Hojabri et al., 2017; Solgi et al., 2017b). In one study, isolated strains belonged to the ST131 (Hojabri et al., 2017) whereas in the other one, carbapenemase producing E. coli strains were distributed into 18 different sequence types with the predominance of ST648 and ST167 (Solgi et al., 2017b). Interestingly, in one of the aforementioned studies, OXA-48 was found on the same transferable plasmid type IncL/M that was previously detected in K. pneumoniae (Solgi et al., 2017b) (Table 2). This finding emphasizes the role of the IncL/M incompatibility group in the horizontal gene transfer of the OXA-48 gene among Enterobacteriaceae. In contrast, in the same study, NDM-1 was detected on an IncA/C plasmid type. In this study, ST648 and ST167 were dominant in NDM-1 and/or OXA-48 producing E. coli strains (Solgi et al., 2017b). Concerning Salmonella, two VIM positive strains were reported in Iran (Shahcheraghi et al., 2017). In A. baumannii, OXA-23 were dominant in all studies addressing carbapenem resistant Acinetobacter spp. in clinical settings (Azizi et al., 2015; Zanganeh and Eftekhar, 2015; Shoja et al., 2016, 2017; Mohajeri et al., 2017; Sarikhani et al., 2017; Zafari et al., 2017; Rezaei et al., 2018; Shirmohammadlou et al., 2018). Other carbapenem resistance genes included OXA-24, OXA-58, IMP, VIM, KPC, GIM, SIM, and SPM (Peymani et al., 2011; Azimi et al., 2015; Bagheri Josheghani et al., 2015; Aghamiri et al., 2016; Maspi et al., 2016; Moghadam et al., 2016; Khorvash et al., 2017; Armin et al., 2018; Soltani et al., 2018). Isolated strains of carbapenem resistant A. baumannii are genetically diverse with the predominance of International clone I and II (Peymani et al., 2011; Savari et al., 2017; Mahdian et al., 2015; Sarhaddi et al., 2017). The rapid evolution of bacterial resistance in Acinetobacter spp. could be attributed to its genome plasticity that allows the acquisition and loss of mobile genetic elements (plasmids, transposons) that modifies the organism’s genomic structure (Savari et al., 2017). As for non baumannii species, only one study reported the detection of OXA-23 and SPM producing Acinetobacter nosocomialis in patients with blood infections (Pourabbas et al., 2016). In P. aeruginosa, MBLs were the most common carbapenemases including: IMP (IMP-1 and IMP-55) and VIM variants (VIM-1, VIM-2) (Abiri et al., 2015; Lari et al., 2015; Mirbagheri et al., 2015; Moosavian and Rahimzadeh, 2015; Azizi et al., 2016; Saffari et al., 2016; Kazeminezhad et al., 2017; Dogonchi et al., 2018; Pournajaf et al., 2018; Rostami et al., 2018). Only two studies reported the detection of OXA-23 and SPM-1 in Iranian clinical isolates of P. aeruginosa (Ostad Asadolah-Malayeri et al., 2016; Azimi et al., 2018). Akhi et al. (2018) found that the main risk factor for acquiring an MBL infection is non-intensive wards hospitalization. Whether the dissemination of carbapenem resistant P. aeruginosa in Iran is polyclonal or not, cannot be assumed. This is because the genetic relatedness was investigated in only two studies; in one of these different genotypes (Akhi et al., 2018) were detected whereas in the other, the strains were distributed into three distinct genotypes (Azimi et al., 2018).
TABLE 2.
Sequence and plasmid types associated with carbapenemase genes in humans, animals, and environment in the Middle East.
| Country | Carbapenemase gene | Reservoir | Species | Sequence type/phylogroup | Plasmid type |
| Iran | Class D oxacillinases | Humans | E. coli | ST131, ST167, ST1431, ST5005, ST889, ST10, ST3737, ST410, ST5114, ST5164, ST315, | |
| ST6350, ST648, ST167, ST410, ST431, ST3134 | IncL/M | ||||
| ST178, ST6826 | IncA/C | ||||
| K. pneumoniae | A, B | ||||
| A. baumannii | ST2 | ||||
| P. aeruginosa | |||||
| Hospital environment | K. pneumoniae | ||||
| A. baumannii | |||||
| KPC | Humans | K. pneumoniae | B | ||
| NDM | Humans | E. coli | C,D, ST167, ST131 | ||
| ST3044, ST167, ST178, ST6826 | IncA/C | ||||
| ST315 | IncL/M | ||||
| K. pneumoniae | C, D | ||||
| ST11, ST893 | IncFII | ||||
| Enterobacter spp. | |||||
| A. baumannii | |||||
| P. aeruginosa | |||||
| Community environment | P. aeruginosa | ||||
| Hospital environment | K. pneumoniae | ||||
| VIM | Humans | K. pneumoniae | ST13, ST392 | ||
| Salmonella spp. | |||||
| A. baumannii | |||||
| P. aeruginosa | |||||
| Community environment | P. aeruginosa | ||||
| IMP | Humans | K. pneumoniae | |||
| A. baumannii | |||||
| P. aeruginosa | |||||
| Community environment | P. aeruginosa | ||||
| Hospital environment | K. pneumoniae | ||||
| Turkey | Class D oxacillinases | Humans | E. coli | ||
| K. pneumoniae | ST11, ST258, ST16 | ||||
| K. oxytoca | |||||
| E. cloacae | |||||
| E. aerogenes | |||||
| P. mirabilis | |||||
| P. stuartii | |||||
| P. rettgeri | |||||
| C. freundii | |||||
| S. marcescens | |||||
| M. morganii | |||||
| R. planticola | |||||
| A. baumannii | ST2, ST15, ST157, ST10, ST158 | ||||
| P. aeruginosa | |||||
| NDM | Humans | E. coli | |||
| K. pneumoniae | |||||
| E. cloacae | |||||
| P. rettgeri | |||||
| VIM | Humans | E. coli | |||
| K. pneumoniae | |||||
| E. cloacae | |||||
| P. aeruginosa | |||||
| IMP | Humans | K. pneumoniae | |||
| E. cloacae | |||||
| P. aeruginosa | |||||
| Palestine/Israel | Class D oxacillinases | Humans | P. mirabilis | IncL/M | |
| KPC | Humans | E. coli | ST512 | ||
| K. pneumoniae | ST340, ST277, ST376 | ||||
| ST327, ST512, ST258 | pKpQIL | ||||
| Hospital and community environment | Enterobacteriaceae | ||||
| NDM | Humans | A. baumannii | ST103, ST2, ST183 | ||
| Hospital and community environment | Enterobacteriaceae | ||||
| VIM | Humans | P. aeruginosa | ST1562, ST1563, ST1564 | ||
| Lebanon | Class D oxacillinases | Humans | E. coli | ST405 | IncL/M |
| K. pneumoniae | IncL/M | ||||
| E. cloacae | IncL/M | ||||
| S. marcescens | IncL/M | ||||
| M. morganii | IncL/M | ||||
| R. ornithinolytica | |||||
| A. baumannii | ST2, ST25, ST1, ST570, ST85, ST600 | ||||
| A. pittii | |||||
| Animals | E. coli | ST38 | |||
| A. baumannii | ST2, ST491, ST493, ST20, ST492 | ||||
| Community environment | E. coli | ST38 | IncHI2 | ||
| – | IncL | ||||
| K. pneumoniae | ST16 | IncL | |||
| A. baumannii | |||||
| NDM | Humans | K. pneumoniae | ST14, ST25 | ||
| Hospital environment | E. cloacae | ||||
| VIM | Humans | P. aeruginosa | ST233, ST699, ST654 | ||
| Animals | P. aeruginosa | ST1759, ST1760, ST1761, ST1762 | |||
| IMP | Humans | P. aeruginosa | ST446, ST654 | ||
| Syria | Class D oxacillinases | Humans | E. coli | ST38 | |
| K. pneumoniae | ST101, ST1633 | ||||
| NDM | Humans | E. coli | |||
| K. pneumoniae | ST278 | ||||
| E. cloacae | |||||
| P. rettgeri | |||||
| C. braakii | |||||
| A. baumannii | ST85 | ||||
| Jordan | Class D oxacillinase | Humans | K. pneumoniae | A1, A3, B | IncL/M |
| Hospital environment | A. baumannii | ||||
| NDM | Humans | E. coli | |||
| K. pneumoniae | A2, C | ||||
| IncFIB, IncFII | |||||
| E. cloacae | |||||
| VIM | Humans | E. cloacae | |||
| Iraq | Class D oxacillinases | Humans | A. baumannii | ST2, ST136, ST194, ST623, ST792, ST793 | |
| NDM | Humans | K. pneumoniae | |||
| P. aeruginosa | |||||
| IMP | Humans | K. pneumoniae | |||
| P. aeruginosa | |||||
| Kuwait | Class D oxacillinases | Humans | E. coli | ST405, ST38 | |
| K. pneumoniae | ST677, ST16, ST107, ST485 | ||||
| E. cloacae | |||||
| M. morganii | |||||
| A. baumannii | ST158 | pK50a | |||
| ST195, ST208, ST229, ST436, ST450, ST452, ST499 | |||||
| NDM | Humans | E. coli | ST448 | IncX3 | |
| K. pneumoniae | |||||
| E. cloacae | |||||
| P. stuartii | |||||
| M. morganii | |||||
| VIM | Humans | E. coli | IncA/C | ||
| K. pneumoniae | IncA/C | ||||
| K. oxytoca | |||||
| E. cloacae | |||||
| P. aeruginosa | |||||
| IMP | Humans | A. baumannii | |||
| KSA | Class D oxacillinases | Humans | E. coli | ||
| K. pneumoniae | ST29, ST709, ST37, ST111, ST974, ST29, ST199, ST48, ST348, ST152, ST340, ST17, ST298, ST16, | ||||
| ST11, ST353 | |||||
| E. cloacae | |||||
| E. aerogenes | |||||
| A. baumannii | ST2, ST19, ST20, ST25, ST236, ST1096, ST945, ST208, | ||||
| ST450, ST452, ST499, ST195, ST229, ST436, | |||||
| NDM | Humans | K. pneumoniae | ST152, ST348, ST199, ST1045 | ||
| E. cloacae | |||||
| E. aerogenes | |||||
| A. baumannii | |||||
| Community environment | E. coli | ||||
| VIM | Humans | K. pneumoniae | |||
| K. oxytoca | |||||
| E. cloacae | |||||
| A. baumannii | ST1, ST2, ST195, ST196, ST487, ST489, ST20 | ||||
| IMP | Humans | A. baumannii | ST1, ST2, ST195, ST196, ST487, ST489, ST20 | ||
| Bahrain | Class D oxacillinases | Humans | A. baumannii | ST208, ST452 | |
| VIM | Humans | P. aeruginosa | |||
| Qatar | Class D oxacillinases | Humans | E. coli | ||
| K. pneumoniae | |||||
| A. baumannii | ST229 | ||||
| NDM | Humans | K. pneumoniae | |||
| VIM | Humans | P. aeruginosa | |||
| United Arab Emirates | Class D oxacillinases | Humans | E. coli | ||
| K. pneumoniae | ST14, ST15, ST23, ST48 | ||||
| Citrobacter spp. | |||||
| S. marcescens | |||||
| KPC | Humans | K. pneumoniae | ST14 | IncX3 | |
| NDM | Humans | E. coli | |||
| K. pneumoniae | ST14 | HI1b | |||
| ST147 | IncFII | ||||
| E. cloacae | |||||
| VIM | Humans | K. pneumoniae | |||
| Oman | Class D oxacillinases | Humans | E. coli | ST648 | IncL/M |
| ST138 | |||||
| K. pneumoniae | ST753, ST754 | IncL/M | |||
| A. baumannii | ST195 | ||||
| NDM | Humans | E. coli | ST2527 | IncF | |
| ST4107 | IncX3 | ||||
| K. pneumoniae | ST15, ST147 | IncH1B | |||
| ST372 | IncFII | ||||
| ST11 | IncL/M, IncH1B | ||||
| VIM | Humans | K. pneumoniae | |||
| Yemen | Class D Oxacillinase | Humans | A. baumannii | ST2 | |
| NDM | Humans | K. pneumoniae | ST1399, ST147, ST29, ST405, ST340 | ||
| E. cloacae | |||||
| Egypt | Class D oxacillinases | Humans | E. coli | ST101, ST648 | |
| K. pneumoniae | |||||
| Salmonella spp. | |||||
| S. marcescens | |||||
| A. baumannii | ST331, ST410, ST208, ST108, ST411, ST409, ST236 | ||||
| P. aeruginosa | |||||
| Animals | K. pneumoniae | ||||
| KPC | Humans | K. pneumoniae | |||
| E. cloacae | |||||
| A. baumannii | |||||
| P. aeruginosa | |||||
| Animals | K. pneumoniae | ||||
| NDM | Humans | E. coli | ST5018 | Incl1-ly | |
| K. pneumoniae | ST147, ST11, ST17 | IncR | |||
| E. cloacae | |||||
| S. marcescens | |||||
| A. baumannii | ST103 | ||||
| P. aeruginosa | ST233 | ||||
| Animals | K. pneumoniae | ||||
| VIM | Humans | E. coli | A, B | ||
| K. pneumoniae | |||||
| S. maltophilia | |||||
| A. baumannii | |||||
| P. aeruginosa | ST233, ST198, ST303, ST629, ST507, ST406, ST274, ST990, ST683, ST884, ST738 |
References are cited in the main text.
In Turkey, K. pneumoniae were the most common GNB resistant to carbapenems (Us et al., 2010). Resistance to these agents is mainly mediated via the production of OXA-48 carbapenemases (Nazik et al., 2012, 2014; Iraz et al., 2015; Baran and Aksu, 2016; Haciseyitoglu et al., 2017; Kutlu et al., 2018). Some studies found a clonal relationship between isolated OXA-48 strains in the clinical center investigated (Castanheira et al., 2014; Karabay et al., 2016; Haciseyitoglu et al., 2017) while others did not (Kilic et al., 2011; Nazik et al., 2012; Iraz et al., 2015). Interestingly, in one of the studies, post the detection of a clonal spread of OXA-48 K. pneumoniae in a tertiary care center, infection control measures including hand washing, high level surface disinfection, screening of colonization before admission were strictly followed (Ece et al., 2018). Later on, this resulted in a significant decrease of the rate of carbapenem resistant K. pneumoniae (Ece et al., 2018). Other types of carbapenemases were also detected in Turkish isolates of K. pneumoniae: NDM, VIM and IMP (Balkan et al., 2014; Candan and Aksoz, 2015; Cakar et al., 2016; Guven Gokmen et al., 2016). In E. coli strains, OXA-48, NDM, and VIM were detected (Gulmez et al., 2008; Carrer et al., 2010; Kilic et al., 2011; Nazik et al., 2012; Baron et al., 2016; Cakar et al., 2016; Kuskucu et al., 2016; Kutlu et al., 2018). Other carbapenemases producing Enterobacteriaceae include: OXA-48 and MBL Enterobacter species (Carrer et al., 2010; Baron et al., 2016; Haciseyitoglu et al., 2017), OXA-48 C. freundii, S. marcescens, P. mirabilis, M. morganii, Raoultella planticola, P. stuartii, and Providencia rettgeri (Carrer et al., 2010; Baron et al., 2016). Otlu et al. (2018) reported the detection of two genetically indistinguishable OXA-48/NDM-1 producing P. rettgeri isolates. These strains were isolated from two different patients about 7 months apart in the same unit. These data show that Turkey is an endemic area of OXA-48 producers, thus warranting the urgent implementation of infection control measures as well as antibiotic stewardship programs (ASP). In non-fermenters, OXA-23, OXA-24, and OXA-58 were detected in A. baumannii with no evidence of clonal dissemination (Kulah et al., 2010; Ciftci et al., 2013; Metan et al., 2013; Castanheira et al., 2014; Cicek et al., 2014; Aksoy et al., 2015; Ahmed S.S. et al., 2016; Direkel et al., 2016). In two studies, co-resistance to colistin was detected (Ergin et al., 2013; Keskin et al., 2014); this fact is threatening and should be taken into real consideration since colistin is currently a last resort therapeutic agent against carbapenem resistant organisms (Olaitan et al., 2014b). On the other hand, MBL (VIM-1, VIM-2, VIM-38, IMP-1, and IMP-9) (Iraz et al., 2014; Yilmaz et al., 2014; Er et al., 2015; Malkocoglu et al., 2017) and OXA-23/OXA-58 were detected in clonaly diverse P. aeruginosa (Tasbent and Ozdemir, 2015).
In Syria, the epidemiology of MDR is unknown due to the civil war crisis. However, injured Syrian refuges are considered a source of MDROs in the country they are residing in Peretz et al. (2014b), Reinheimer et al. (2016). Indeed, recent studies showed the introduction of ST85 NDM-1 positive A. baumannii into Lebanese clinical settings from wounded Syrian refugees (Rafei et al., 2014b, 2015b). Subsequently, NDM-1 positive Acinetobacter spp. were isolated from Lebanese patients (Rafei et al., 2015b). Similarly, NDM/OXA-48 K. pneumoniae and E. coli and NDM producing E. cloacae, P. rettgeri, and Citrobacter braakii were isolated from Syrian refugees in North Palestine (Lerner et al., 2016). In both reports, the origin of the detected isolates could not be determined; the infection might have been acquired on the battlefield from environmental sources, during the patients stay in Syrian hospitals or during evacuation from Syria to another territory (Rafei et al., 2014b). Instead, what is sure is that screening of refugees arriving from countries with unknown epidemiology of carbapenem resistance, upon hospital admission, is a must and is crucial in order to contain the dissemination of these highly resistant MDROs (Lerner et al., 2016).
In Lebanon, in early 2012, OXA-48 and OXA-48/NDM-1 positive E. coli and K. pneumoniae, respectively, were isolated from the blood and urine cultures of Iraqi patients (El-Herte et al., 2012). Indeed, the most common carbapenemases detected in Enterobacteriaceae isolated from Lebanese hospitals are class D oxacillinases; these include OXA-48/OXA-232 E. coli, ST14 NDM-1 K. pneumoniae, OXA-48, OXA-162, OXA-232 K. pneumoniae, OXA-48/OXA-232 E. cloacae and OXA-48 producing S. marcescens, M. morganii, and Raoultella ornithinolytica (Hammoudi et al., 2015b; Al-Bayssari et al., 2016; Tokajian et al., 2016; Hammoudi Halat et al., 2017; Alousi et al., 2018). In two studies, the OXA-48 gene was located on the same plasmid IncL/M in E. coli and K. pneumoniae alike (Hammoudi Halat et al., 2017; Alousi et al., 2018). This emphasizes the crucial role that mobile genetic elements play in the spread of resistance determinants between different species. On the other hand, within the same species, no clonal relatedness was observed among carbapenem resistant K. pneumoniae strains (Baroud et al., 2013). NDM-1 producing K. pneumoniae belonging to ST14 was reported by Alousi et al. (2018). Furthermore, an NDM-1 ST15 K. pneumoniae strain was isolated from the urine sample of an old Syrian refugee in Lebanon (Salloum et al., 2017). ST15 is heavily reported in hospitals worldwide such as Nepal (Stoesser et al., 2014), Vietnam (Tada et al., 2017), Thailand (Netikul et al., 2014), and China (Hu et al., 2013). The successful dissemination of ST15 K. pneumoniae could be attributed to its ability to acquire several resistance genes with no fitness cost (Toth et al., 2014). On the other hand, bacterial resistance in A. baumannii have largely evolved in Lebanon since its first detection by Matar et al. (1992). OXA-58 were at first the most common carbapenemase detected in clinical isolates of A. baumannii (Giannouli et al., 2009; Di Popolo et al., 2011), thereafter, OXA-23 and OXA-24 dominated (Rafei et al., 2014a; Hammoudi et al., 2015a, b; Hammoudi Halat et al., 2017). The dissemination of carbapenem resistant A. baumannii in Lebanese hospitals appears to be mainly mediated via the international clone II (Al Atrouni et al., 2016; Dahdouh et al., 2016; Hajjar Soudeiha et al., 2018). However, horizontal gene transfer has also played a major role. This is illustrated in a study conducted by Kanj et al. (2018) who found that the prevalence of OXA-23 positive A. baumannii have significantly increased between 2007, 2008, and 2013. Molecular analysis revealed only a 22% genomic relatedness among isolated strains. This emphasizes the role of horizontal gene transfer in the dissemination of resistance determinants among A. baumannii in Lebanese hospitals. Only one study reported the detection of non baumannii Acinetobacter species: Acinetobacter pittii producing NDM-1 and OXA-72 carbapenemases; these strains were isolated from the urine culture of a 4-month-old child and from a febrile gastroenteritis infected patient, respectively (Al Atrouni et al., 2016a). As for P. aeruginosa isolated from Lebanese hospitals, only MBLs were detected: IMP-1, IMP-2, IMP-15, and VIM-2 (Al Bayssari et al., 2014; Hammoudi et al., 2015b; Hammoudi Halat et al., 2017).
In Israel, KPC-3 and to a lesser extent KPC-2 producing K. pneumoniae appear to be endemic (Navon-Venezia et al., 2009; Leavitt et al., 2010a, b; Warburg et al., 2012; Castanheira et al., 2014). PFGE analysis showed that isolated KPC-3 K. pneumoniae belonged to the same genetic clone (Navon-Venezia et al., 2009). On the other hand, MLST analysis in three other studies showed the predominance of ST258 (Leavitt et al., 2010a; Warburg et al., 2012; Castanheira et al., 2014). The fact that KPC was also detected in other enterobacterial species such as E. coli (KPC-2) (Goren et al., 2010a, b), Enterobacter spp. (Lazarovitch et al., 2015), and C. freundii (Castanheira et al., 2014) suggests a possible monoclonal spread of KPC in K. pneumoniae and its subsequent successful horizontal gene transfer to other species. Other carbapenemase producing GNB detected in Palestine involve: OXA-48 located in the IncL/M plasmid in P. mirabilis (Chen et al., 2015), VIM-2/VIM-4 P. aeruginosa in Palestine (Sjolander et al., 2014) and NDM-2/OXA-23/OXA-24 A. baumannii in both Israel and Palestine (Castanheira et al., 2014; Sjolander et al., 2014).
In Jordan, NDM-1 and NDM-1/VIM-4 were detected in E. coli and E. cloacae clinical isolates, respectively (Aqel et al., 2018). Furthermore, two studies reported the detection of NDM and OXA-48 in genetically diverse K. pneumoniae strains isolated from clinical specimens (Aqel et al., 2017, 2018). In one study, NDM-1 and OXA-48 were located on FII(K)/FIB and IncL/M, respectively (Aqel et al., 2018). The interesting finding in the second study is that distinct NDM-1 positive K. pneumoniae were isolated from a Yemeni patient and a native Jordanian without a history of travel, hospitalized at the same time period. In the same report, distinct OXA-48 K. pneumoniae was isolated from a Yemeni and also a native Jordanian treated in the same ward with specimens 12 days apart (Aqel et al., 2017). Altogether, these data highlight the importance of horizontal gene transfer and the absence of effective infection control measures in the dissemination of carbapenem resistance genes in Jordanian hospitals.
In Iraq, the most common carbapenemase producers are the non-fermenters including OXA-23/OXA-24 A. baumannii (Kusradze et al., 2011; Ganjo et al., 2016) and NDM (NDM-1, NDM-2), IMP and SPM P. aeruginosa (Al-Charrakh et al., 2016; Ismail and Mahmoud, 2018). According to Al-Charrakh et al. (2016), the high resistance of carbapenem resistant P. aeruginosa to non-beta lactams such as ciprofloxacin and gentamicin, can be attributed to the over-use of these antimicrobial agents in Iraqi clinical practices. In Enterobacteriaceae, only NDM-1 and SPM K. pneumoniae strains were detected (Hussein, 2018).
In Kuwait, VIM-4, NDM (NDM-1, NDM-7), and OXA-48 carbapenemases were detected in clinical isolates of clonally unrelated E. coli and K. pneumoniae strains (Jamal et al., 2013, 2015, 2016; Pal et al., 2017). In one study, the blaVIM gene was located on the same plasmid type IncA/C both in E. coli and K. pneumoniae (Sonnevend et al., 2017b). Moreover, several reports described the detection of NDM-1 P. stuartii, OXA-48, NDM-1 M. morganii and VIM (VIM-4), NDM-1, and OXA-48 E. cloacae in Kuwaiti hospitals (Jamal et al., 2013, 2015, 2016; Sonnevend et al., 2015a, 2017b). In A. baumannii, OXA-23 were mainly detected followed by IMP-1, VIM (VIM-1 and VIM-2) (Jamal et al., 2009; Al-Sweih et al., 2012; Zowawi et al., 2015; Wibberg et al., 2018). On the other hand, only one study reported the detection of VIM positive P. aeruginosa clinical strains (Zowawi et al., 2018). The P. aeruginosa strains were distributed into 14 sequence type clusters with some of them being recognized as highly disseminated international clones such as ST111, ST235, ST357, and ST654 (Zowawi et al., 2018). In fact, according to one report, it has been suggested that the dissemination of carbapenem resistance in the clinical settings of Kuwait appears to be promoted by immigration, in-sufficient infection control measures, environmental spread, and antibiotic misuse (Jamal et al., 2016).
In KSA, OXA-48, and MBL (NDM-1, VIM-4, and VIM-29) are the most common carbapenem resistance genes detected in Enterobacteriaceae (Memish et al., 2015; Sonnevend et al., 2015b; Algowaihi et al., 2016; Alotaibi et al., 2017; Al-Zahrani and Alsiri, 2018; Zaman et al., 2018). These isolates included mainly K. pneumoniae and others (E. coli, E. cloacae, and Enterobacter aerogenes) (Al-Agamy et al., 2013; Shibl et al., 2013; Uz Zaman et al., 2014; Memish et al., 2015; Abdalhamid et al., 2017b). In fact, several studies reported a clonal relatedness among carbapenemase producing K. pneumoniae in each clinical center (Balkhy et al., 2012; Uz Zaman et al., 2014; Abdalhamid et al., 2017b). In one study, the clonal relatedness of carbapenem resistant K. pneumoniae was 93.2% (Abdalhamid et al., 2017b) whereas in other studies, MLST analysis revealed the predominance of certain sequence types such as ST29, ST199, and ST152 (Uz Zaman et al., 2014; Zaman et al., 2018). Furthermore, in one of the reports, the IncL/M plasmid type was predominant in OXA-48 Klebsiella spp. (Zaman et al., 2018). Indeed, one explanation for the MBL and OXA-48 predominance in the clinical isolates of Enterobacteriaceae in KSA is the big number of migrant workers and visitors coming from endemic areas such as India, Pakistan and Turkey (Al-Zahrani and Alsiri, 2018). Moreover, one study found that most of the patients infected with a carbapenem resistant K. pneumoniae had prolonged hospital stays, indwelling devices, surgical procedures, carbapenem usage and infection/carriage with MDROs (Balkhy et al., 2012). On the other hand, class D oxacillinase (OXA-23, OXA-24, and OXA-58) predominate in A. baumannii followed by NDM, VIM, and IMP (Alsultan et al., 2013; Elabd et al., 2015; Aly et al., 2016; Al-Agamy et al., 2017; Alhaddad et al., 2018). Clonal diversity revealed by different sequence types as well as PFGE patterns among isolated strains was reported in all the studies (Aly et al., 2014; Lopes et al., 2015; Zowawi et al., 2015; El-Mahdy et al., 2017). In one study, Aljindan et al. (2015) found that carbapenem resistant A. baumannii were more resistant to gentamicin, amikacin, ciprofloxacin, and tigecycline compared to the susceptible ones. In P. aeruginosa with high clonal diversity, VIM, IMP, VIM-1, VIM-2, VIM-4, VIM-11, VIM-28 were detected (Al-Agamy et al., 2012, 2016; Tawfik et al., 2012).
In Bahrain, VIM and class D oxacillinases (OXA-23, OXA-58, OXA-72, and OXA-40) were detected in genetically variant P. aeruginosa and A. baumannii, respectively (Mugnier et al., 2009; Zowawi et al., 2015, 2018). In Qatar, OXA-48 E. coli, NDM/OXA-48 K. pneumoniae, OXA-23 A. baumannii, and VIM P. aeruginosa were reported in clinical settings (Zowawi et al., 2014, 2015, 2018; Rolain et al., 2016).
In United Arab Emirates, NDM (NDM-1 and NDM-5), OXA-48 and to a lesser degree KPC, are the predominant carbapenemases detected in clinical isolates of K. pneumoniae (Dash et al., 2014; Sonnevend et al., 2015a, 2017a). MLST analysis revealed different sequence types with the most common being ST11, ST14, and ST147 (Sonnevend et al., 2013, 2015a, 2017a; Moubareck et al., 2018). ST147 is of special interest since in their study, Sonnevend et al. (2017a) reported a multi-hospital occurrence of a pan-resistant ST147 K. pneumoniae isolated from four patients over a 1 year period. The strains had highly similar genotypes and PFGE patterns. Furthermore, with more deep genetic analysis, extensive similarities (backbone and resistance islands) were found between these strains and the ST147 K. pneumoniae strains isolated in South Korea. Interestingly, one of the Korean isolates was from a patient transferred from the United Arab Emirates. This reveals the huge capacity of the ST147 K. pneumoniae clone in maintaining itself over a long period of time in addition to its ability to be transmitted internationally (Sonnevend et al., 2017a). Similarly, NDM and OXA-48 were also found in other GNB in the Imarati hospitals including E. coli, E. cloacae, Citrobacter spp., S. marcescens, and A. baumannii (Ghazawi et al., 2012; Sonnevend et al., 2013, 2015b). In two studies, the NDM gene was located on an IncX3 plasmid (Sonnevend et al., 2013; Pal et al., 2017; Moubareck et al., 2018). According to Sonnevend et al. (2013), the Middle East is the second region where IncX3 plasmids with very similar structures that carry the blaNDM-1 were detected; found in different species, this emphasizes the role of this plasmid type on the inter-generic dissemination of this MBL gene.
In Oman, carbapenem resistance in Enterobacteriaceae (E. coli and K. pneumoniae) is mediated via the production of NDM (NDM-1 and NDM-7) and OXA-48 carbapenemases (Dortet et al., 2012; Zowawi et al., 2014). Reported sequence types for K. pneumoniae include ST14, ST340, ST11, and ST147 (Poirel et al., 2011a; Potron et al., 2011; Sonnevend et al., 2015a). Furthermore, as reported in United Arab Emirates, NDM-7 in E. coli was located on the epidemiologically important IncX3 plasmid (Pal et al., 2017). On the other hand, OXA-23 was detected in A. baumannii whereas in P. aeruginosa VIM and IMP were found (Zowawi et al., 2015, 2018). In the gulf, Zowawi et al. (2015) found that several clusters of indistinguishable OXA-23 A. baumannii strains are circulating. These include ST208 and ST195 that belong to the clonal complex 92, which is internationally disseminated (Chen et al., 2017; Rieber et al., 2017).
In Yemen, OXA-23 producing ST2 A. baumannii were isolated from clinical settings (Bakour et al., 2014); this is in addition to clonally un-related NDM-1 K. pneumoniae (ST1399, ST147, ST29, ST405, and ST340) and E. cloacae strains (Gharout-Sait et al., 2014).
In Egyptian hospitals, KPC, VIM (VIM-1, VIM-2, and VIM-29), NDM (NDM-1 and NDM-5), and OXA-48 are the predominant carbapenamases detected in Enterobacteriaceae (Abdelaziz et al., 2013b; Metwally et al., 2013; Hamdy Mohammed el et al., 2016; Abdallah et al., 2017; Barwa and Shaaban, 2017; Khalifa et al., 2017; Khalil et al., 2017; Abdulall et al., 2018; Kamel et al., 2018). Molecular analysis revealed that no clonal relationship was observed among carbapenem resistant E. coli and K. pneumoniae strains (Abdelaziz et al., 2013b; Khalifa et al., 2017; Khalil et al., 2017; Abdulall et al., 2018). The polyclonal spread of carbapenem resistant K. pneumoniae in Egypt is further documented in a study conducted in Italy. In this study, two NDM producing K. pneumoniae were isolated from unrelated patients with recent hospitalization in an Egyptian hospital (Principe et al., 2017). Isolated strains belonged to different sequence types. ST15 which was previously reported in Africa (Poirel et al., 2011b) and other Middle Eastern countries such as Lebanon (Salloum et al., 2017); and ST11 which is the sequence type to which the first NDM-1 K. pneumoniae strain isolated from Egypt belonged to (Abdelaziz et al., 2013b; Gamal et al., 2016). Polyclonal and horizontal gene transfer via mobile genetic elements appears to play an important role in the spread of carbapenemase producers in Egyptian clinical settings. However, more genetic analyses (MLST, plasmid typing) are needed to confirm this assumption. Other carbapenemase producers detected in Egyptian clinical settings include: VIM, KPC, NDM E. cloacae, OXA-48 M. morganii and Salmonella, OXA-48, NDM-1 S. marcescens and VIM Stenotrophomonas maltophilia (Hamdy Mohammed el et al., 2016; Khalifa et al., 2017; Abdulall et al., 2018; Kamel et al., 2018). In non-fermenters, carbapenem resistance A. baumannii was mediated mainly via OXA-23, OXA-24, OXA-58 followed by NDM (NDM-1 and NDM-2), VIM (VIM-1 and VIM-2), IMP, SIM, and GIM (Kaase et al., 2011; Fouad et al., 2013; El-Ageery and Al-Hazmi, 2014; Lopes et al., 2015; Hamdy Mohammed el et al., 2016; Alkasaby and El Sayed Zaki, 2017; Ghaith et al., 2017; Abdulall et al., 2018; Abdulzahra et al., 2018; Kamel et al., 2018; Ramadan R.A. et al., 2018). High genetic diversity was observed among isolated strains (Al-Hassan et al., 2013; Ghaith et al., 2017; El Bannah et al., 2018). As for associated risk factors, one study showed that the empirical intake of carbapenem 1 month ago is significantly associated with the development of a carbapenem resistance caused infection (ElMahallawy et al., 2018). On the other hand, VIM (VIM-2, VIM-28, and VIM-1-like), NDM (NDM-1), IMP, and OXA-48 genes were reported in P. aeruginosa (El-Mahdy, 2014; Zafer et al., 2014, 2015; Khalifa et al., 2017). The majority of isolated strains were genetically diverse with different sequence types including ST233, ST303, ST198, ST629, and ST507 (Zafer et al., 2014, 2015; Khalifa et al., 2017).
Infections With Colistin Resistant Gram-Negative Bacilli
In Egypt, the first mcr-1 producing E. coli isolated from a clinical setting occurred in 2016. This strain co-produced the CTX-M-15 and had a sequence type of ST1011 which was previously detected in an avian E. coli strain within this same country. This finding could be a direct manifestation of a zoonotic transmission of mcr-1 from animals to humans (Elnahriry et al., 2016). Another study conducted on carbapenem resistant A. baumannii revealed substitutional mutations in the pmrA/B genes and subsequent colistin resistance. A. baumannii is considered an opportunistic pathogen and is usually treated with colistin if found to be carbapenem resistant. This association of colistin resistance with resistance to other antimicrobials is thus especially worrisome (Abdulzahra et al., 2018).
In Lebanon, Okdah et al. (2017) reported the detection of colistin resistance in three unrelated K. pneumoniae strains (ST268, ST2296, and ST348) with mutations in the mgrB, phoQ, pmrA/B genes in a hospital in Beirut. In Israel, one study reported the case of an Israeli patient with prior colistin administration during hospitalization and subsequent isolation of colistin resistant K. pneumoniae from his stool, supporting the theory of colistin resistance emergence as a result of antibiotic overuse in hospitals (Olaitan et al., 2014a). Lalaoui et al. (2019), reported the detection of colistin resistance in NDM-1 and KPC-3 harboring K. pneumoniae strains isolated from a medical center in Jerusalem. Resistance to colistin in these isolates was mediated by inactivation of the mgrB gene via an IS5-like insertion sequence (Lalaoui et al., 2019). Similarly, to nearby countries, colistin resistant K. pneumoniae strains in Israel are genetically diverse with different sequence types including ST16, ST76, ST258, and ST512 (Lalaoui et al., 2019). Indeed, this country lacks quantitative investigation of the dosages and/or duration of colistin administration that significantly increase the risk of development of colistin resistance in a strain or a patient (Drozdinsky et al., 2018).
In Jordan, Nazer et al. (2015) conducted a study where they focused on critically ill cancer patients with carbapenem resistant A. baumannii and the adverse effects of colistin as choice of treatment. In the latter, despite 66% of the patients being cleared of their respiratory infections with colistin resistant A. baumannii, nephrotoxicity and even mortality were significantly associated with this therapeutic regimen. This warrants quantitative studies that are not necessarily targeted at determining doses and frequency that lead to emergence of A. baumannii colistin resistant strains, but are rather targeted at finding treatments for different types of infections in different populations (critically ill cancer patients for example) with minimal side effects and optimal outcomes (Nazer et al., 2015).
In the region of the Arabian Peninsula, colistin resistance is a public health challenge that is worth addressing. In the United Arab Emirates for instance, K. pneumoniae strains were isolated from different hospitals in different emirates. ST147 K. pneumoniae was isolated from a hospital in Abu Dhabi as well as from two different hospitals in Um al Quwain (Sonnevend et al., 2017a). This strain was not only carbapenem resistant through the blaOXA-181 gene but was also colistin resistant through an insertion in its mgrB gene. Interestingly enough, the insertion into the mgrB gene which resulted in colistin resistance was in fact the functional blaOXA-181 gene (Sonnevend et al., 2017a). Those findings imply that not only is there a spread of this ST over a large geographic area, but also that this strain is one of many that have developed resistance to both carbapenems and colistin and therefore has the potential to cause epidemics (Sonnevend et al., 2017a). Moreover, a study conducted in Dubai, on clinical isolates from hospitals with the broadest medical and surgical exposure in the country to assess resistance to carbapenems as well as to colistin, found that 31.4% of the carbapenem resistant K. pneumoniae strains isolated were also colistin resistant (Moubareck et al., 2018). The mechanism of colistin resistance was not identified but was confirmed not to be the mcr plasmid mediated gene. While 40% of the K. pneumoniae isolates that were both colistin and carbapenem resistant were sporadic cases, 31.4% were associated with the K. pneumoniae ST14 clone, which is locally prevalent. Along with the fact that Dubai is a major economical, touristic, and medical city in the region, the above information showcases the potential of Dubai playing an important role in the spread of colistin resistance from a One Health Concept perspective (Moubareck et al., 2018). Indeed, in the UUnited Arab EmiratesAE, only one ST131 E. coli strain harboring the mcr-1 gene was reported (Sonnevend et al., 2016).
In Qatar, a colistin resistant clinical E. coli strain positive for the mcr-1 gene was recently reported. This isolate belongs to ST95, known to cause meningitis in humans as well as severe avian infections. It is worth mentioning that this strain had an ISApl1 element in the same plasmid carrying the mcr-1 gene and the pap2-like phosphatase gene (Forde et al., 2018). The PAP2-like phosphatase can potentially contribute to colistin resistance by modifying the lipid A of the GNB outer membranes. The extent to which this gene contributes to colistin resistance in bacteria remains unknown but is worth investigating (Forde et al., 2018). Similarly, in Bahrain, the clinical colistin resistant E. coli strains (ST648 and ST224) were associated with the mcr-1 gene being on an Incl2 plasmid type (Sonnevend et al., 2016).
Additionally, in Oman, a clinical isolate of E. coli carrying mcr-1 was isolated in 2016. This strain belongs to ST10 and also harbors a plasmid of the IncI2 type. The detection of colistin resistance in ST10 E. coli is worrisome given that this clonal group has been known to mediate the spread of ESBL and quinolone resistance genes globally (Mohsin et al., 2018). In Kuwait, the development of colistin resistance in Acinetobacter spp. was evaluated in 2011. Of a total of 250 strains collected from eight governmental hospitals, 12% were found to be resistant to colistin. Compared to 0% in 2009, this significant increase prevalence could be attributed to the sudden increase in colistin prescription due to the global emergence of MDR infections (Al-Sweih et al., 2011).
In the KSA, a study by Mirnejad et al. (2018), focused on resistance to polymyxin B rather than colistin (polymyxin E). Those two antibiotics however, cover the same spectrum of organisms and can be used interchangeably as they have very similar mechanisms of action (Mirnejad et al., 2018). It was found that 13.2% of A. baumannii strains collected were resistant to polymyxin B (Memish et al., 2012). Another study found that the rate of resistance to colistin among A. baumannii in the KSA increased from 2.6 to 4.7% over the course of 2 years between 2010 and 2011 (Baadani et al., 2013). The danger that accompanies the appearance of colistin resistant strains in this country was embodied in a study in which two out of seven patients involved died due to colistin resistance. In that study, there was a history of colistin use reported in all patients except for one, suggesting that sporadic emergence rather than horizontal transmission of resistance might have played a more important role in the rise of colistin resistance in the isolated strains (Garbati et al., 2013). Moreover, sporadic cases of mcr-1 in hospitals in the KSA has previously been reported (Sonnevend et al., 2016). However, most of the studies conducted demonstrated chromosomal mutations (mgrB and phoP) responsible for colistin resistance (Uz Zaman et al., 2018). In all of these studies, no clonal relatedness was observed among isolated colistin GNB strains (Sonnevend et al., 2016; Uz Zaman et al., 2018). The polyclonal spread of colistin resistance questions the level of colistin use in hospitals of the Arabian peninsula.
While addressing the topic of colistin resistance and the One Health Concept in the KSA, it is very important to mention the yearly Muslim pilgrimage, Hajj, that takes place in the city of Mecca. Plasmid mediated mcr-1 carrying strains of predominantly genetically diverse E. coli strains and to a lesser extent K. pneumoniae have previously been isolated from patients during the Hajj. Pilgrims arrive from different countries, different occupations, and therefore with different sources of colistin resistance acquisition. These sources might be from the environment, food, animals, or from other humans (Leangapichart et al., 2016b).
In Turkey, colistin resistance has raised great concern as it has been associated with poor prognosis (Yilmaz et al., 2016). A study done by Cizmeci et al. (2017), found that six out of eight patients with K. pneumoniae that are resistant to both carbapenems and colistin ended up dying when all treatment options failed. Carbapenem resistant isolates positive for the NDM-1 gene have been found to have a higher rate of concomitant colistin resistance than isolates positive for the OXA-48 gene (Cizmeci et al., 2017). Furthermore, not only is the potential for colistin resistant infections to be fatal worrisome in Turkey, their potential to cause epidemics is also worrisome; the isolation of identical colistin resistant strains circulating in the country over short periods of time validate those concerns (Metan et al., 2017).
In Iran, one study reported the isolation of two colistin resistant P. aeruginosa from university teaching hospitals. The two isolates presented with different sequence types and more importantly were isolated from patients with no history of colistin consumption. The mechanisms of colistin resistance in both isolates was the overexpression of MexB and MexY genes which code for MexAB-OprM and MexXY-OprM efflux pumps. Despite colistin not being a specific substrate for those efflux pumps, the over expression of the MexAB-OprM and MexXY-OprM efflux was suspected to have played a role in the development of colistin resistance (Goli et al., 2016). This theory is partly supported by the fact that the over expression of MexAB-OprM and MexXY-OprM efflux pumps has already been linked to resistance in P. aeruginosa in multiple antimicrobial agents such as aminoglycosides (Hocquet et al., 2003). One interesting study done by Bahador et al. (2018) found that resistance to colistin in A. baumannii isolates is linked to the increase in virulence factors such as biofilm formation in burn patients. This renders the treatment of such MDR more challenging, as both resistance to colistin and virulence factors must be tackled at once (Bahador et al., 2018). Furthermore, two studies reported the isolation of colistin resistant A. baumannii and K. pneumoniae with mutations in the pmrB and mgrB genes, respectively (Haeili et al., 2017, 2018) (Table 3).
TABLE 3.
Mechanisms of colistin resistance described in GNB in the Middle East.
| Country | Origin | Species | Sequence type | Mechanism of colistin resistance | References |
| Iran | Clinical samples | A. baumannii | pmrB∗ | Haeili et al., 2018 | |
| A. baumannii | pmrA/B∗ | Sepahvand et al., 2016 | |||
| K. pneumoniae | mgrB∗ | Haeili et al., 2017 | |||
| P. aeruginosa | MexAB-OprM/MexXY-OprM∗∗ | Goli et al., 2016 | |||
| Lebanon | Clinical samples | K. pneumoniae | ST268 | mgrB∗ | Okdah et al., 2017 |
| K. pneumoniae | ST2296 | mgrB∗, PhoQ∗ | |||
| K. pneumoniae | ST348 | pmrA/B∗ | |||
| Poultry | E. coli | ST515 | mcr-1 | Dandachi et al., 2018c | |
| Swine | E. coli | mcr-1 | Dandachi et al., 2018b | ||
| Palestine | Clinical samples | K. pneumoniae | mgrB∗ | Olaitan et al., 2014a | |
| K. pneumoniae | ST512, ST76 | mgrB∗ | Lalaoui et al., 2019 | ||
| Bahrain | Clinical samples | E. coli | ST648, ST224 | mcr-1 on IncI2 | Sonnevend et al., 2016 |
| Qatar | Clinical samples | E. coli | ST95 | mcr-1 on IncHI2 | Forde et al., 2018 |
| United Arab Emirates | Clinical samples | K. pneumoniae | ST147 | mgrB∗ | Sonnevend et al., 2017a |
| E. coli | ST131 | mcr-1 on IncI2 | Sonnevend et al., 2016 | ||
| Oman | Clinical samples | E. coli | ST10 | mcr-1 on IncI2 | Mohsin et al., 2018 |
| KSA | Clinical samples | K. pneumoniae | ST974, ST37, ST709, ST348, ST37 | PhoP∗ | Uz Zaman et al., 2018 |
| K. pneumoniae | ST14, ST15, ST16, ST22, ST48, ST101, ST152, ST307 | mgrB∗ | |||
| K. pneumoniae | ST15 | mgrB∗, phoP∗ | |||
| E. coli | ST68 | mcr-1 on IncHI2 | Sonnevend et al., 2016 | ||
| Egypt | Clinical samples | A. baumannii | pmrCAB∗ | Abdulzahra et al., 2018 | |
| Animal | E. coli | ST10 | mcr-1 | Khalifa et al., 2016 | |
| E. coli O157 | mcr-1 | Lima Barbieri et al., 2017 | |||
| E. coli O158 | mcr-1 |
∗Mutations. ∗∗Efflux pump over-expression.
Carriage of ESBL/Carbapenemase Producers
The main concern of MDROs intestinal carriage is the acquisition of MDRO caused infections with limited therapeutic options (Magwenzi et al., 2017). In addition, as the carriage can last from months to years, the asymptomatic colonization of MDROs constitute a potent reservoir for transmission and dissemination (Decker et al., 2018).
In Iran, one study reported an 18.3% rectal carriage rate of ESBL K. pneumoniae among ICU patients and outpatients. The main mechanism of resistance was the production of CTX-M-15 detected in 86.3% of isolated strains (Aghamohammad et al., 2018). MLST analysis revealed that isolates of K. pneumoniae belonged to 16 different STs with a predominance of ST15, ST147, and ST16 (Aghamohammad et al., 2018). ST15 K. pneumoniae is widely associated worldwide with the production of CTX-M-15 (Lee et al., 2011; Rodrigues et al., 2014; Caneiras et al., 2019). On the other hand, carbapenem resistant Enterobacteriaceae (CRE) colonization in Iranian inpatients was associated with 3rd generation cephalosporins, meropenem, colistin, and vancomycin exposure. This is in addition to ICU hospitalization, urinary catheter, mechanical ventilation, recent surgery, patient transfers from another hospital/unit and being male (Solgi et al., 2017a). Isolated CRE included, NDM and OXA-48 producing K. pneumoniae, E. coli, E. cloacae, and P. mirabilis (Solgi et al., 2017a).
In Turkey, the rectal carriage of CRE (OXA-48, NDM-1, and IMP K. pneumoniae) as well as carbapenem resistant non-fermenters (CR-NF) was reported in several studies (Alp et al., 2013; Karaaslan et al., 2016). In one study, only carbapenem intake was associated with OXA-48/IMP producing K. pneumoniae infections (Zarakolu et al., 2016). An interesting clinical experience was the one reported by Poirel et al. (2014) when an outbreak of carbapenem resistance was suspected with the first isolation of two similar carbapenem resistant E. cloacae from two patients’ residing in the neonatal ICU (NICU). Accordingly, nasal and rectal screening was performed for all NICU patients. In addition, contact isolation precautions were implemented as well as an intensive infection control program was performed for all staff personnel. Subsequently, after 1 month, no infection/colonization with CRE was observed (Poirel et al., 2014). This emphasizes the significance of the microbiology laboratory and infection control unit’s cooperation in preventing the dissemination of CRE (Poirel et al., 2014). A more recent study, also conducted in a Turkish NICU, showed that ages less than 1 year, carbapenem administration, presence of underlying diseases, urinary catheterization, and nasogastric tube placement were independent risk factors for CRE colonization. In this study, CRE included OXA-48, IMP, NDM K. pneumoniae, E. coli, and E. cloacae (Karaaslan et al., 2016). On the other hand, CR-NF carriage (NDM, IMP-1, OXA-23, OXA-24, and OXA-58 producing A. baumannii) was correlated to an ICU stay, ampicillin carbapenem use, mean daily antibiotic use, presence of underlying diseases, surgical intervention and nasogastric tube placement (Karaaslan et al., 2016).
In Lebanon, two studies addressed the rectal carriage of ESBL producing Enterobacteriaceae in nursing home residents in Beirut and Tripoli in the north (Jallad et al., 2015). In Beirut, constipation and antibiotic intake were independent risk factors for ESBL carriage (Jallad et al., 2015); whereas in Tripoli, only antibiotic administration was found (Dandachi et al., 2016). Nursing homes are community facilities where MDROs can easily emerge and spread due to the uncontrolled or unprofessional prescription of antibiotics and inadequate environmental decontamination, waste disposal, and hygiene practices (Dandachi et al., 2016). Another study conducted in healthy infants showed that CTX-M-15, CTX-M-9, and CTX-M-2 positive Enterobacteriaceae are prevalent in the Lebanese community (Hijazi et al., 2016). Hospital birth, cesarean delivery, being formula-fed and being male are important risk factors for ESBL colonization in this category. In this report, proper hygiene was associated with a colonization rate decrease (Hijazi et al., 2016). On the other hand, Christophy et al. (2017) reported a high prevalence of carbapenem resistance fecal carriage in cancer patients undergoing chemotherapy. The carbapenem resistant strains included mainly OXA-48/CTX-M E. coli, OXA-48 E. cloacae and VIM Pseudomonas stutzeri.
In Israel, a study conducted at a rehabilitation center revealed the patient’s rectal carriage of CTX-M-27, CTX-M-15, CTX-M-14, CTX-M-39, CTX-M-55, SHV-5, SHV-12, and CMY-4 and CMY-2 producing E. coli strains of diverse sequence types including ST131 (Izdebski et al., 2013). In parallel, among patients admitted to a teaching hospital in one study, 8% were carriers of ESBL. The risk factors for this colonization were female sex and recent antibiotic intake. On the other hand, 21% of admitted patients acquired ESBL carriage. The latter was significantly associated with being older than 65 years and having an extended spectrum beta lactam antibiotic intake (Friedmann et al., 2009). Additionally, in clinical settings, one study raised concern about the real efficiency of antibiotic prophylaxis post-bowel surgery on ESBL carriage and subsequent infection. This is because in this report, immunosuppressive therapy and antibiotic use in the previous 3 months were independent risk factors for ESBL rectal carriage in this patients’ category (Pfeffer et al., 2016). Moreover, in this country, the carriage of carbapenem resistance was significantly higher to ESBL positive Enterobacteriaceae in view of the number of reports. In one study, the carriage of CR-KP was significantly associated with a prolonged hospital stay, room sharing with a previously known carrier and residency in a high carrier ward (Ben-David et al., 2011). In fact, in their study, Wiener-Well et al. (2010), found that during the surveillance of CR-KP carriage in hospitalized patients, isolated strains had identical PFGE patterns showing a clonal origin. The authors argued that strict isolation of carriers might help reduce the transmission of the CR-GNB from one patient to another (Wiener-Well et al., 2010). In another study, the CR-KP carriage was dependent on recent surgery and a sequential organ failure assessment (SOFA) score (Debby et al., 2012). Other described risk factors for CR-KP intestinal carriage include diaper use, length of hospital stay and vancomycin use (Wiener-Well et al., 2010). Adler et al. (2015) reported an increase in the rectal carriage of KPC producing K. pneumoniae (represented by ST258) in a post-acute care hospital (PACH) from 65% in 2008 to 80% in 2013. The acquisition source of more than 50% of the carriers was the PACH itself (Adler et al., 2015). Moreover, one report showed that the duration of CRE carriage can last for 3 months, 6 months and even up to 1 year. The carriage duration was affected mainly by repeated hospitalization and the isolation of a clinical and not surveillance positive culture (Zimmerman et al., 2013). One explanation for this finding is that recurrent hospitalization often represents re-infection and flags more severely ill people who need more time to eradicate CRE (Zimmerman et al., 2013). Furthermore, CRE infected patients might have larger loads of CRE compared to those who are only colonized with; contributing subsequently to longer periods of continuous carriage (Zimmerman et al., 2013). Interestingly, one study assessed the risk factors responsible for the development of CRE infection after CRE colonization. These latter included ICU admission, antibiotic intake (especially fluoroquinolones and metronidazole), diabetes mellitus and central venous catheter insertion (Schechner et al., 2013). The identification of these factors are important in order to predict CRE infections and direct accordingly antibiotic empirical therapy (Schechner et al., 2013). Other carbapenem resistant species detected in Palestinian colonizers include VIM (VIM-1, VIM-35) producing Aeromonas species and NDM-1/OXA-10 positive P. rettgeri (Adler et al., 2014; Olaitan et al., 2016b). In Jordan, one study reported the rectal carriage of CTX-M-15, CTX-M-2, and CTX-M-1 E. coli in infants less than 1 year of age to the Pediatric unit in a hospital in Amman (Badran et al., 2016).
In the Gulf region, Dashti et al. (2010b) reported the detection of a single ESBL producing E. coli clone in blood cultures of neonates and health care workers’ (HCW) hand in a Kuwaiti hospital. This highlight the important role that the health care personnel can play as vectors and reservoirs from which bacterial resistance can spread. This is especially true when non-adherence to proper sanitation and hand hygiene occur. In Qatar, only one study reported the fecal carriage of MDR ESBL E. coli in food handlers (Eltai et al., 2018c). This finding is of public health concern, since MDROs can be silently transmitted to the general community via contaminated food, contributing thus further to the dissemination of bacterial resistance (Eltai et al., 2018c).
In KSA, only two studies reported the rectal carriage of MDROs in the clinical settings. These latter included ICU patients carrying of highly diverse OXA-23 A. baumannii, CTX-M-15 K. pneumoniae, and NDM, VIM producing P. aeruginosa (Aljindan et al., 2015; Abdalhamid et al., 2016). Indeed, the rectal colonization of MDROs was mostly addressed in the Hajj period. One study reported in 2013 a significant CTX-M intestinal carriage in pilgrims with the rate of the latter increasing from 10.08% before Hajj to 32.56% post Hajj (Leangapichart et al., 2016a). In the same context, the same author reported similar findings in 2014. The acquisition rate of ESBL producers did not significantly differed between the 2 years (Leangapichart et al., 2017). Indeed, Leangapichart et al. also found that there was a difference not only at the level of intestinal colonization rate but also at the level of the bacterial diversity detected. For instance, A. baumannii strains were isolated from 26 rectal specimens and 16 pharyngeal one’s post Hajj while none was detected in the samples collected prior to Hajj travel (Leangapichart et al., 2016c). It is worth mentioning the detection of one A. baumannii strain that is carbapenem resistant and produced the OXA-72 carbapenemase post Hajj. Likewise, an E. coli positive for blaNDM-5, blaCTX-M-15 was also detected after Hajj travel (Leangapichart et al., 2016c). These data emphasize the role of this season as a mediator of bacterial resistance dissemination in the KSA and worldwide. More effort is warranted for the improvement of the public health conditions during this period of the year. Moreover, recent Hajj travel should be taken into consideration when pilgrim patients are admitted to hospitals in their hometown in order to control for the introduction of new MDROs to clinical settings. However, more studies are needed in order to characterize “recent Hajj travel” as a risk factor for MDROs fecal carriage.
In Egypt, fecal carriage of ESBL producing GNB was detected in hospitalized patients. These included CTX-M (CTX-M-15, CTX-M-14, CTX-M-2, and CTX-M-grp9) SHV and TEM ESBL type (Khalaf et al., 2009; Fam et al., 2015). Fouda et al. (2016) found that ESBL carriage was associated with increased mortality in ICU admitted patients. In the same context, two studies reported the intestinal carriage of ESBL and AmpC beta lactamases in HCW in two hospitals (Abdel Rahman et al., 2010; Bassyouni et al., 2015). As already mentioned, HCW constitute a potent reservoir of bacterial resistance when infection control measures and proper hand hygiene are lacking in a clinical center (Bassyouni et al., 2015). Furthermore, NDM-1 positive ST267 A. baumannii were isolated from hospitalized patients during a rectal screening surveillance in this same country (Krahn et al., 2016).
Distribution of Multi-Drug Resistant Organisms in Animals
ESBL/AmpC Producers
In the Middle East, studies from Egypt reported the detection of TEM, SHV, CTX-M-9, CTX-M-15, and OXA-7 producing E. coli strains in broiler farms. The plasmid mediated AmpC beta lactamase genes blaCMY-2 and blaDHA-1 were also observed (Moawad et al., 2018). Furthermore, studies done on poultry hatcheries revealed similar results where blaTEM, followed by blaSHV, blaMOX-like, blaCIT-like, and blaFOX were the most common beta lactamase genes detected (Osman et al., 2018). BlaCTX-M-15 has also been reported in Egyptian poultry with other β-lactamase-encoding genes such as blaTEM-104, blaCMY-2, and blaOXA-30 in E. coli strains including the sequence type ST131 (Ahmed et al., 2013; Abdallah et al., 2015a; Ramadan H.H. et al., 2018). Multidrug-resistant E. coli O25b:H4 ST131 has been reported to be spread worldwide in humans, companion animals and livestock (Ahmed et al., 2013). Another study on chickens in Egypt, reported other ESBL types including blaTEM-57, blaSHV-12, blaCTX-M-14 (El-Shazly et al., 2017). As for pathogenic bacteria, two studies reported the detection of TEM ESBL type in Salmonella species isolated from chicken meat as well as from pigeons (Ahmed H.A. et al., 2016; Abdeen et al., 2018). Indeed, one of the main contributors to this high prevalence of ESBL/AmpC producers in the Egyptian poultry sector is the misuse of antibiotics. According to El-Shazly et al. (2017), many farmers in Egypt tend to use cefotaxime injections (a 3rd generation cephalosporin banned in poultry) to treat diseases in chicken (such as colibacillosis) after the failure of other antimicrobial treatments like fluoroquinolone and aminoglycosides. In addition, due to the low cost of antibiotics, many veterinarians still over-use antibiotics such as tetracycline, quinolone and beta lactams to treat and prevent zoonotic diseases and growth promotion (Braun et al., 2016; Hakim et al., 2017). Moreover, in pets, blaTEM along with blaSHV, blaPSE-1 and blaCTX-M were detected in E. coli strains isolated from dogs in in the same country (Aly et al., 2012). In cattle, TEM, SHV (SHV-11, SHV-27), and CTX-M-15 were detected in E. coli and K. pneumoniae strains (Hammad and Shimamoto, 2011; Braun et al., 2016). Another report on dairy calves, reported the detection of blaCMY-2 and blaSHV-12 genes in Salmonella spp. including S. enterica serovars enteritidis and S. typhimurium (Ahmed et al., 2009). Other studies in the Egyptian dairy products revealed the presence of CTX-M-variants (CTX-M-15, CTX-M-104, CTX-M-3), TEM-52, SHV-12, and CMY-2 producing E. coli strains (Ahmed and Shimamoto, 2015; Ombarak et al., 2018). Other ESBL (including OXA-10 and SHV-28) and AmpC producing GNB detected in the Egyptian bovine sector include Klebsiella oxytoca and C. freundii (Ahmed and Shimamoto, 2011).
In Palestine, blaCTX-M (including CTX-M-1, CTX-M-9) and SHV-12 were the only ESBL types detected in E. coli strains isolated in Chicken (Qabajah et al., 2014). Similarly, these ESBL types were detected in cattle in Israel (Adler et al., 2016). In Lebanon, a recent nationwide study conducted in chicken farms, found a considerable number of ESBL and AmpC producing GNB. These included mainly blaCTX-M, blaTEM and blaCMY genes (Dandachi et al., 2018). On the other hand, Diab et al. (2016) reported the dissemination of CTX-M-15 producing E. coli in Lebanese cattle. One more recent study conducted by Dandachi et al. (2018b) found that CTX-M followed by CMY are the most common beta lactamases detected in E. coli strains isolated from Swine farms. Both in cattle and poultry, MLST analysis revealed high variety of sequence types in isolated E. coli strains with some of them previously described in the literature as being common to animals as well as to humans (ST10, ST617, ST58, ST69, ST155, and ST156) (Diab et al., 2016; Dandachi et al., 2018). This emphasizes the role of livestock in the dissemination of MDROs in the one health concept.
In Turkey, CTX-M-15 was detected in E. coli strains belonging to the B1 phylogenetic group isolated from cattle with bovine mastitis (Pehlivanoglu et al., 2016). Furthermore, one study targeting MDROs in dogs reported the detection of CTX-M-15, BlaCMY-2, blaCTX-M-3, blaCTX-M-1, and blaSHV-12 in E. coli isolates with A1 and D2 being the most common phylogenetic groups identified. In this report, ST131/B2 E. coli positive for CTX-M-15 were detected. This clone is a human pandemic one that can possibly be transmitted to humans via direct or indirect contact with companion animals (Aslantas and Yilmaz, 2017).
In the gulf and specifically in the KSA, blaSHV and blaTEM were reported in E. coli strains isolated from poultry (Altalhi et al., 2010; Abo-Amer et al., 2018). Furthermore, ESBL and AmpC producers were detected in the Qatari chickens and green turtles in Oman, respectively (Al-Bahry et al., 2012; Eltai et al., 2018a). In Iran, E. coli strains producing blaSHV were isolated from raw milk and dairy products across the country (Ranjbar et al., 2018). Furthermore, this same gene was detected in Uropathogenic E. coli strains isolated from dogs (Yousefi and Torkan, 2017).
Carbapenem and Colistin Resistance
Unlike ESBL and AmpC producers, carbapenemase producing GNB are not widely spread in animals of the Middle East (Figure 2). Al Bayssari et al. (2015b) reported the isolation of ST38 E. coli positive for the blaOXA-48 from fowl in Lebanon. Furthermore, they detected VIM-2 carbapenemase in P. aeruginosa and blaOXA-23/blaOXA-58 genes A. baumannii strains isolated from cattle, swine and fowl (Al Bayssari et al., 2015a). Furthermore, Rafei et al. (2015a) reported the isolation of OXA-143 A. baumannii and OXA-24 A. pittii from a horse and a rabbit oral cavity, respectively. In Egypt, carbapenem resistant K. pneumoniae (CR-KP) have been isolated from broilers, drinking water and workers in chicken farms (Abdallah et al., 2015a). The genes responsible for resistance were blaKPC, blaOXA-48, and blaNDM. In cattle, OXA-48 and OXA-181 producing E. coli were detected (Braun et al., 2016).
FIGURE 2.

Geographical distribution of carbapenemases in humans, animals, hospital, and community environment in the Middle East.
As for colistin resistance (Figure 3), in Egypt, it is known that colistin is used in animal husbandry in farms, calves, poultry, and rabbits (Lima Barbieri et al., 2017). In poultry for example, colistin has been used for colibacillosis. Colistin resistant avian isolates of E. coli that have been found in Egyptian farms imply that the overuse of colistin in the farming industry can indeed have participated in the emergence of colistin resistance in Egypt (Lima Barbieri et al., 2017). Indeed, in samples collected from both poultry and cattle, the mcr-1 gene was detected (Khalifa et al., 2016; Lima Barbieri et al., 2017). In cattle, mcr-1 was harbored by an ST10 E. coli strain (Khalifa et al., 2016). In the light of the One Health concept, those resistant strains can potentially enter the human food chain and result in treatment challenging infections that pose a serious threat to the Egyptian population. This is especially relevant in a country like Egypt which is known to struggle with infectious diseases and poor control of antibiotic use (Khalifa et al., 2016). On the other hand, Lebanon is considered one of the more recent countries in which colistin resistance has emerged. Dandachi et al. (2018c) reported the first detection of ESBL/mcr-1 ST515 E. coli strain isolated from chicken in the South of Lebanon. mcr-1 E. coli strains were also detected in Lebanese pigs (Dandachi et al., 2018b).
FIGURE 3.
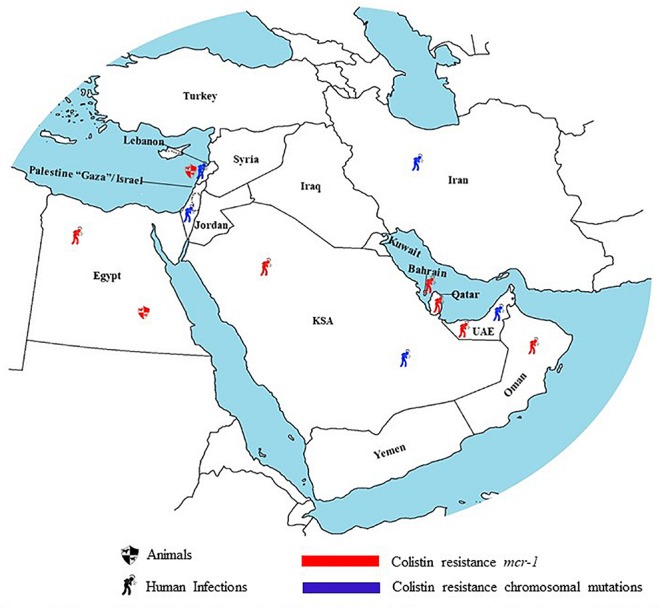
Geographical distribution of colistin resistance in humans and animals in the Middle East.
Distribution of Multi-Drug Resistant Organisms in the Environment
Community Environment
In Iran, a study addressing MDROs in surface water resources found a high prevalence of CTX-M, TEM, SHV, and OXA genes among isolated E. coli strains (Ranjbar and Sami, 2017). In another study conducted in community water filtering systems, P. aeruginosa producing blaVIM-1, blaNDM, and blaIMP-1 were detected (Mombini et al., 2019). In Turkey, ESBL producing E. coli strains were detected in samples collected from Orontes River. The most common ESBL type was CTX-M-15 with the majority co-harboring the sulfonamide resistance gene sul (Kurekci et al., 2017).
In Lebanon, Diab et al. (2018) targeted the issue of MDROs contamination in three water sources: estuaries, wells and spring water (Diab et al., 2018). It was found that in estuaries, 17 ESBL and four carbapenemase producing GNB were isolated. The most prevalent ESBL gene was the bla-CTX-M-15 followed by CTX-M-55, CTX-M-14, and SHV-12. IncF type plasmid was the most common plasmid type detected among ESBL. In parallel, carbapenem resistance was mediated by the presence of bla-OXA-48 carried by the IncL plasmid and the bla-OXA-244 carried by the IncHl2 plasmid. On the other hand, only ESBL producers were detected in wells and spring water. These included the blaCTX-M-15 gene located on an IncF plasmid (Diab et al., 2018). In another study, ESBL producing E. coli were addressed in a refugee camp and from river effluents (Tokajian et al., 2018). The prevalence of ESBL was similar in both groups (53.11% versus 49.1%). However, the latter presented with different phylogroups and sequence types; in addition, refugee camp isolated strains had higher resistance rates toward aminoglycosides, fluoroquinolones and trimethoprim-sulfamethoxazole (Tokajian et al., 2018). In this study, the most common ESBL types detected in both sources were CTX-M-15, CTX-M-27, CTX-M-14, and CTX-M-9 (Tokajian et al., 2018). Furthermore, it is worth noting the detection of OXA-72 producing Acinetobacter calcoaceticus in vegetables in Lebanon. The presence of MDROs in vegetables could be the result of direct animal contamination or indirect environmental contamination with soil or irrigation water (Al Atrouni et al., 2015, 2016b). Altogether, due to the lack of proper water treatment systems in Lebanon, water sources have become major environmental reservoirs for MDROs. In Israel, one study reported the detection of blaKPC and blaNDM-1 producing Enterobacteriaceae (mainly K. pneumoniae and E. cloacae) in sewage systems (Meir-Gruber et al., 2016).
In the KSA, blaNDM-1 ST101 E. coli was isolated from wastewater (Mantilla-Calderon et al., 2016). In addition, Alghoribi et al. (2015) reported the detection of CTX-M producing E. cloacae from a community sewage.
Hospital Environment
In Iran, TEM and SHV producing P. aeruginosa were detected in hospital environmental samples (Gholami et al., 2017). Another study, in which the samples were taken from an ICU setting, revealed that among the Klebsiella species isolated, the majority were carbapenem resistant K. pneumoniae. The ICU’s contaminated areas are a great source for the spread of MDROs in their surroundings (Moghadampour et al., 2018b). In isolated K. pneumoniae strains, the highest resistance rate was observed for β-lactam antibiotics and the lowest resistance rate was toward tigecycline. In this study, blaOXA-48 was the most prevalent carbapenemase detected, followed by blaNDM (Moghadampour et al., 2018b). Furthermore, various hospitals have been suffering from antibiotic resistant A. baumannii. In one study, several isolates where taken from different parts of the hospital with the greatest number of A. baumannii coming from the ICU. These strains carried the OXA-23 and OXA-24 genes (Shamsizadeh et al., 2017). The prevalence of CRKP has been another problem in Iran, especially on hospital equipment where in one study 34 CRKP were isolated (Moghadampour et al., 2018b). Aliramezani et al. (2016) also reported the detection of carbapenem resistant GNB (OXA-23 A. baumannii) in instruments that are frequently used for the care of patients such as dressing sets, suction tubes, hand-washing sinks and faucets. The occurrence of MDROs in the aforementioned surfaces raise the chance of acquisition and HGT of resistance genes among patients, health care personnel and visitors, creating thus a significant source of hospital acquired infections (Aliramezani et al., 2016). Moreover, other studies in Iran reported the isolation of carbapenemase producing A. baumannii (OXA-23, OXA-24, and OXA-58) from hospital environmental samples (Kulah et al., 2010; Salehi et al., 2018). Of special interest is the detection of OXA-23 and CTX-M-32 genes in air samples collected from operating theaters, ICUs, surgery and internal medicine wards (Mirhoseini et al., 2016). This study unveiled another source of the environmental route of MDROs transmission. Not only is area disinfection warranted but the application of efficient working ventilation and air quality monitoring systems is also highly needed (Mirhoseini et al., 2016).
In Turkey, only one study conducted in hospitals reported the detection of ESBL K. pneumoniae in broiler distilled water in a NICU (Hosbul et al., 2012). In Israel, a study conducted inside a general hospital with emphasis on the bacteria present on wheelchairs, found that P. aeruginosa followed by A. baumannii were the most common MDROs detected and exhibited resistance to all antibiotics tested, especially in the samples taken from wheelchairs in the surgery department (Peretz et al., 2014a). Interestingly, one study assessed the prevalence of environmental contamination of CRE in the vicinity of 34 carriers. Among these 26 were spreaders with a group of six being responsible for 79% of the environmental CRE detected. Statistical analysis revealed that fecal continence was the sole independent factor associated with CRE non-spread. On the other hand, high rectal colonization with these MDROs in addition to being admitted with a respiratory disease were the only independent risk factors for CRE shedding (Lerner et al., 2015). Therefore, imminent protocols must be set to minimize contamination and spread of infections in all hospital settings.
In Lebanon, the most discussed source of MDROs contamination are of a human source whether directly or indirectly through hospital sewage. In addition to that, water is considered a pivotal driver of contamination because it acts as a reservoir for MDROs and receives them from multiple sources. Furthermore, Daoud et al. (2018) found that isolates of E. coli from the hospital wastewater produced CTX-M ESBL at a rate of 81.5% in one hospital and 94.4% in another. SHV beta-lactamases were produced by 55.6 and 44.4% of the isolates in each hospital, respectively. In the same context, Daoud et al. (2017), addressed the MDROs in two hospital sewage treatment plants. In this latter, ESBL and AmpC producers including E. coli, E. cloacae, Klebsiella spp., and Serratia odorifera were isolated. The most common ESBL types found were CTX-M followed by SHV, TEM, and OXA. Furthermore, only one CRE was detected (Daoud et al., 2017).
In Iraq, only one study revealed the detection of ESBL Klebsiella spp. and E. coli producing CTX-M-15, AmpC beta lactamases and SHV ESBL types, respectively, in hospital environmental samples (Huang et al., 2012). Interestingly, Obeidat et al. (2014) reported the isolation of carbapenem resistant A. baumannii (OXA-23 and OXA-24 producing) from a hospital environment as well as from patients respiratory tracts; the high similarity of MDR patterns suggest the persistence of these MDRO in the environment is responsible for their high colonization rates detected in the respiratory tracts of ICU patients.
In the gulf, one study in the KSA found ESBL producing GNB in hospital sewage. The sewage tank might play a significant role in the dissemination of MDROs, especially if it enters the sea and beach recreational activity areas, subsequently affecting the community population (Alghoribi et al., 2015).
Antibiotic Use in the Middle East
Clinical Setting and Community
Nowadays, antibiotics are among the most common drugs prescribed worldwide. Between 2000 and 2010, antibiotic consumption increased by 20 billion standard units (Auta et al., 2019). The growing use of antibiotics through prescriptions or non-prescriptions is linked to the spread of MDROs, therefore causing a global public health concern (Auta et al., 2019).
In the Middle East, studies have shown that SM is highly prevalent. In Iran, SM ranges from 35.4 to 83%, 32 to 42% in Lebanon, 32 to 62% in Jordan, 98% in Palestine, 85% in Syria. In the Gulf, SM rates were as high as 89.2% in the United Arab Emirates, 48% in the KSA and 60% in Yemen (Khalifeh et al., 2017). This is unlike European countries where “over-the-counter” access to antibiotics is strictly regulated, thus resulting in SM prevalence rates ranging from 1 to 4% only (Alhomoud et al., 2017). The major types of antibiotics sold over-the-counter in the Middle East include penicillin, macrolides, cephalosporins, fluoroquinolones, and tetracycline (Alhomoud et al., 2017). The main reason behind the common practice of SM in Middle Eastern countries is the lack of strict policies controlling the sale of antibiotics without a prescription from pharmacies. This is in addition to the low economic status and lack of health care insurance that push individuals to retrieve medications from pharmacists to avoid consultations costs (Alhomoud et al., 2017).
Besides SM, inappropriate use of antibiotics in hospitals is another reason behind the dissemination of MDROs in the Middle East. Indeed, ASP, although implemented in some hospitals in several countries such as Lebanon, Jordan, Palestine, the KSA, the United Arab Emirates, Bahrain, Qatar, and Oman; these are still in their infancy (Nasr et al., 2017). Barriers for the implementation of ASP in the Middle East are divided mainly into two levels: individual and hospital barriers. Individually speaking, physicians often lack up-to-date knowledge for appropriate antibiotic use and resistance, reluctance for antibiotic prescription other than the usual and fear of patient complications especially in very sick patients are other individual barriers (Alghamdi et al., 2018). On the other hand, lack of expertise, unavailability of some antibiotics, lack of education/training for appropriate usage of antibiotics and antimicrobial resistance as well as a lack of financial, administrative and management support are all barriers against the implementation of ASP at the hospital level (Alghamdi et al., 2018).
Further research assessing the knowledge, attitude and practices of antibiotic prescription among expatriates is crucial for the adoption of successful programs, in order to promote the rational use of antimicrobial agents in the Middle East (Alhomoud et al., 2017). Furthermore, hospital leadership is paramount to ensure policies’ enforcement, in collaboration with physicians and other stakeholders (Alghamdi et al., 2018). On the other hand, as for SM, enforcing regulatory measures that restrict antibiotic access to “prescribed-only,” developing national resistance as well as antibiotic consumption surveillance systems can all help in reducing the rates of SM (Alhomoud et al., 2017). This is definitely in addition to public awareness campaigns addressing the proper use of antibiotics in addition to the dangers of their inappropriate use and over-intake (Alhomoud et al., 2017).
Animals and Environment
Unfortunately, it is evident that MDROs are nowadays disseminated in animals and the environment as it has been reported worldwide (Rizzo et al., 2013; Alonso et al., 2017; Dandachi et al., 2018a). Similar to humans, among other factors, the un-regulated use of antibiotics in veterinary medicine is the main cause for MDRO dissemination (Guerra et al., 2014). Besides treatment, in animals, antibiotics are also given as growth promoters and for prophylaxis. As growth promoters, this practice is no longer applied in the European Union, but it persists in North America and other countries (Economou and Gousia, 2015). MDROs in animals can be transmitted to humans via direct/indirect contact or via the surrounding environment (Pomba et al., 2017). Despite their importance in the transmission chain, surveillance studies addressing MDROs in these two ecosystems in the Middle East are scarce. As shown in Figures 1–3, epidemiological studies describing the dissemination of MDR in animals and the environment were conducted in only six out of the 15 countries. The level of antibiotic consumption in livestock is unknown and thus policies to control the misuse and overuse of antimicrobial agents in veterinary medicine are not yet in place. Furthermore, the role of the environment in the transmission route is also unknown in this region of the world. In the environment, resistant bacteria can spread either due to the shedding of MDROs from human/animal waste or via the antibiotic selective pressure created by antimicrobial release in livestock and human waste streams (Dar et al., 2016). In the one health concept “the health of people is connected to the health of animals and the environment” (Centers for Disease Control and Prevention [CDC], 2018). Researchers in Middle Eastern countries are therefore recruited to conduct studies to fill the gaps of epidemiological distribution of MDROs as well as antibiotic consumption in ecosystems other than humans. Furthermore, the implementation of an integrated human-animal surveillance system where samples are obtained from both humans, livestock and the environment and then processed with a synchronized monitoring system can assist these speculations (Manyi-Loh et al., 2018). The first worldwide system integrating humans and animals was the “DANMAP” (Danish Integrated Antimicrobial Monitoring and Resistance Program) which addresses the problem of MDR in livestock, food of animal origin and people (Dar et al., 2016).
Conclusion
This review shows the extensive dissemination of ESBL and carbapenemase producing GNB in Middle Eastern hospitals. The prevalence of these MDROs is less well documented in animals and the environment. However, studies reported that ESBL is common in livestock whereas carbapenemases are scarce. In the environment, to some extent both groups (ESBL and carbapenemases) were reported equally. This emphasizes that the environment plays a double route in the transmission of resistant organisms from humans to animals and vice versa. In some countries especially in the gulf, nothing is known about the spread of MDROs in animals nor the environment; therefore, a clear conclusion cannot be drawn. One major mediator of MDROs spread in the Middle East is the recent population mobilization due to the socio-economic crisis and the Syrian war. This conflict promotes the introduction of resistance genes not previously reported in those countries. The emergence of colistin resistance is another major issue. In most of the epidemiological studies, colistin susceptibility is assessed by the Kirby-Bauer technique. This method is unreliable and might underestimate the real prevalence of colistin resistance in all ecological niches.
Author Contributions
ID, AC, JH, and JM wrote the manuscript. ZD corrected the manuscript. All authors approved and revised the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Abdalhamid B., Albunayan S., Shaikh A., Elhadi N., Aljindan R. (2017a). Prevalence study of plasmid-mediated AmpC beta-lactamases in enterobacteriaceae lacking inducible ampC from Saudi hospitals. J. Med. Microbiol. 66 1286–1290. 10.1099/jmm.0.000504 [DOI] [PubMed] [Google Scholar]
- Abdalhamid B., Elhadi N., Albunayan S., Alsamman K., Aljindan R. (2017b). First description of methyltransferases in extensively drug-resistant Klebsiella pneumoniae isolates from Saudi Arabia. J. Med. Microbiol. 66 859–863. 10.1099/jmm.0.000480 [DOI] [PubMed] [Google Scholar]
- Abdalhamid B., Elhadi N., Alabdulqader N., Alsamman K., Aljindan R. (2016). Rates of gastrointestinal tract colonization of carbapenem-resistant enterobacteriaceae and Pseudomonas aeruginosa in hospitals in Saudi Arabia. New Microbes New Infect. 10 77–83. 10.1016/j.nmni.2016.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah H. M., Alnaiemi N., Reuland E. A., Wintermans B. B., Koek A., Abdelwahab A. M., et al. (2017). Fecal carriage of extended-spectrum beta-lactamase- and carbapenemase-producing enterobacteriaceae in Egyptian patients with community-onset gastrointestinal complaints: a hospital -based cross-sectional study. Antimicrob. Resist. Infect. Control 6:62. 10.1186/s13756-017-0219-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah H. M., Reuland E. A., Wintermans B. B., Al Naiemi N., Koek A., Abdelwahab A. M., et al. (2015a). Extended-spectrum beta-lactamases and/or carbapenemases-producing enterobacteriaceae isolated from retail chicken meat in Zagazig, Egypt. PLoS One 10:e0136052. 10.1371/journal.pone.0136052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah H. M., Wintermans B. B., Reuland E. A., Koek A., al Naiemi N., Ammar A. M., et al. (2015b). Extended-spectrum beta-lactamase- and carbapenemase-producing enterobacteriaceae isolated from Egyptian patients with suspected blood stream infection. PLoS One 10:e0128120. 10.1371/journal.pone.0128120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdeen E., Elmonir W., Suelam I. I. A., Mousa W. S. (2018). Antibiogram and genetic diversity of Salmonella enterica with zoonotic potential isolated from morbid native chickens and pigeons in Egypt. J. Appl. Microbiol. 124 1265–1273. 10.1111/jam.13697 [DOI] [PubMed] [Google Scholar]
- Abdel Rahman A. T., Hafez S. F., Abdelhakam S. M., Ali-Eldin Z. A., Esmat I. M., Elsayed M. S., et al. (2010). Antimicrobial resistant bacteria among health care workers in intensive care units at ain shams university hospitals. J. Egypt Soc. Parasitol. 40 71–83. [PubMed] [Google Scholar]
- Abdelaziz M. O., Bonura C., Aleo A., Fasciana T., Cala C., Mammina C. (2013a). Cephalosporin resistant Escherichia coli from cancer patients in Cairo. Egypt Microbiol. Immunol. 57 391–395. 10.1111/1348-0421.12046 [DOI] [PubMed] [Google Scholar]
- Abdelaziz M. O., Bonura C., Aleo A., Fasciana T., Mammina C. (2013b). NDM-1- and OXA-163-producing Klebsiella pneumoniae isolates in Cairo, Egypt, 2012. J. Glob. Antimicrob. Resist. 1 213–215. 10.1016/j.jgar.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Abdelkader M. M., Aboshanab K. M., El-Ashry M. A., Aboulwafa M. M. (2017). Prevalence of MDR pathogens of bacterial meningitis in Egypt and new synergistic antibiotic combinations. PLoS One 12:e0171349. 10.1371/journal.pone.0171349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdulall A. K., Tawfick M. M., El Manakhly A. R., El Kholy A. (2018). Carbapenem-resistant gram-negative bacteria associated with catheter-related bloodstream infections in three intensive care units in Egypt. Eur. J. Clin. Microbiol. Infect. Dis. 37 1647–1652. 10.1007/s10096-018-3294-7 [DOI] [PubMed] [Google Scholar]
- Abdulzahra A. T., Khalil M. A. F., Elkhatib W. F. (2018). First report of colistin resistance among carbapenem-resistant Acinetobacter baumannii isolates recovered from hospitalized patients in Egypt. New Microbes New Infect. 26 53–58. 10.1016/j.nmni.2018.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abiri R., Mohammadi P., Shavani N., Rezaei M. (2015). Detection and genetic characterization of metallo-beta-lactamase IMP-1 and VIM-2 in Pseudomonas aeruginosa strains from different hospitals in Kermanshah, Iran. Jundishapur J. Microbiol. 8:e22582. 10.5812/jjm.22582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abo-Amer A. E., Shobrak M. Y., Altalhi A. D. (2018). Isolation and antimicrobial resistance of Escherichia coli isolated from farm chickens in Taif, Saudi Arabia. J. Glob. Antimicrob. Resist. 15 65–68. 10.1016/j.jgar.2018.05.020 [DOI] [PubMed] [Google Scholar]
- Adler A., Assous M. V., Paikin S., Shulman A., Miller-Roll T., Hillel S., et al. (2014). Emergence of VIM-producing Aeromonas caviae in Israeli hospitals. J. Antimicrob. Chemother. 69 1211–1214. 10.1093/jac/dkt505 [DOI] [PubMed] [Google Scholar]
- Adler A., Hussein O., Ben-David D., Masarwa S., Navon-Venezia S., Schwaber M. J., et al. (2015). Persistence of Klebsiella pneumoniae ST258 as the predominant clone of carbapenemase-producing enterobacteriaceae in post-acute-care hospitals in Israel, 2008-13. J. Antimicrob. Chemother. 70 89–92. 10.1093/jac/dku333 [DOI] [PubMed] [Google Scholar]
- Adler M., Anjum M., Andersson D. I., Sandegren L. (2016). Combinations of mutations in envZ, ftsI, mrdA, acrB and acrR can cause high-level carbapenem resistance in Escherichia coli. J. Antimicrob. Chemother. 71 1188–1198. 10.1093/jac/dkv475 [DOI] [PubMed] [Google Scholar]
- Adwan K., Jarrar N., Abu-Hijleh A., Adwan G., Awwad E. (2014). Molecular characterization of Escherichia coli isolates from patients with urinary tract infections in Palestine. J. Med. Microbiol. 63(Pt 2), 229–234. 10.1099/jmm.0.067140-0 [DOI] [PubMed] [Google Scholar]
- Aghamiri S., Amirmozafari N., Fallah Mehrabadi J., Fouladtan B., Hanafi Abdar M. (2016). Antibiotic resistance patterns and a survey of metallo-beta-lactamase genes including bla-IMP and bla-VIM types in Acinetobacter baumannii isolated from hospital patients in Tehran. Chemotherapy 61 275–280. 10.1159/000443825 [DOI] [PubMed] [Google Scholar]
- Aghamohammad S., Badmasti F., Solgi H., Aminzadeh Z., Khodabandelo Z., Shahcheraghi F. (2018). First report of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae among fecal carriage in Iran: high diversity of clonal relatedness and virulence factor profiles. Microb. Drug Resist. 10.1371/journal.pone.0173510 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Agin H., Ayhan F. Y., Gulay Z., Gulfidan G., Yasar N., Erac B., et al. (2011). The evaluation of clusters of hospital infections due to multidrug-resistant Salmonella enterica serovar typhimurium in the neonatal unit: a two-year experience. Turk. J. Pediatr. 53 517–521. [PubMed] [Google Scholar]
- Ahmad S., Al-Juaid N. F., Alenzi F. Q., Mattar E. H., Bakheet O. (2009). Prevalence, antibiotic susceptibility pattern and production of extended-spectrum beta-lactamases amongst clinical isolates of Klebsiella pneumoniae at armed forces hospital in Saudi Arabia. J. Coll. Physicians. Surg. Pak. 19 264–265. [PubMed] [Google Scholar]
- Ahmed A. M., Shimamoto T. (2011). Molecular characterization of antimicrobial resistance in gram-negative bacteria isolated from bovine mastitis in Egypt. Microbiol. Immunol. 55 318–327. 10.1111/j.1348-0421.2011.00323.x [DOI] [PubMed] [Google Scholar]
- Ahmed A. M., Shimamoto T. (2015). Molecular analysis of multidrug resistance in shiga toxin-producing Escherichia coli O157:H7 isolated from meat and dairy products. Int. J. Food Microbiol. 193 68–73. 10.1016/j.ijfoodmicro.2014.10.014 [DOI] [PubMed] [Google Scholar]
- Ahmed A. M., Shimamoto T., Shimamoto T. (2013). Molecular characterization of multidrug-resistant avian pathogenic Escherichia coli isolated from septicemic broilers. Int. J. Med. Microbiol. 303 475–483. 10.1016/j.ijmm.2013.06.009 [DOI] [PubMed] [Google Scholar]
- Ahmed A. M., Younis E. E., Osman S. A., Ishida Y., El-Khodery S. A., Shimamoto T. (2009). Genetic analysis of antimicrobial resistance in Escherichia coli isolated from diarrheic neonatal calves. Vet. Microbiol. 136 397–402. 10.1016/j.vetmic.2008.11.021 [DOI] [PubMed] [Google Scholar]
- Ahmed H. A., El-Hofy F. I., Shafik S. M., Abdelrahman M. A., Elsaid G. A. (2016). Characterization of virulence-associated genes, antimicrobial resistance genes, and class 1 integrons in Salmonella enterica serovar typhimurium isolates from chicken meat and humans in Egypt. Foodborne Pathog. Dis. 13 281–288. 10.1089/fpd.2015.2097 [DOI] [PubMed] [Google Scholar]
- Ahmed S. S., Alp E., Ulu-Kilic A., Dinc G., Aktas Z., Ada B., et al. (2016). Spread of carbapenem-resistant international clones of Acinetobacter baumannii in Turkey and Azerbaijan: a collaborative study. Eur. J. Clin. Microbiol. Infect. Dis. 35 1463–1468. 10.1007/s10096-016-2685-x [DOI] [PubMed] [Google Scholar]
- Akhi M. T., Khalili Y., Ghotaslou R., Yousefi S., Kafil H. S., Naghili B., et al. (2018). Evaluation of carbapenem resistance mechanisms and its association with Pseudomonas aeruginosa infections in the northwest of Iran. Microb. Drug Resist. 24 126–135. 10.1089/mdr.2016.0310 [DOI] [PubMed] [Google Scholar]
- Aksoy M. D., Cavuslu S., Tugrul H. M. (2015). Investigation of metallo beta lactamases and oxacilinases in carbapenem resistant Acinetobacter baumannii strains isolated from inpatients. Balkan Med. J. 32 79–83. 10.5152/balkanmedj.2015.15302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktas Z., Satana D., Kayacan C., Can B., Gonullu N., Kucukbasmaci O. (2012). Antibiotic susceptibility rates and beta-lactam resistance mechanisms of Pseudomonas aeruginosa strains. Mikrobiyol. Bul. 46 386–397. [PubMed] [Google Scholar]
- Akya A., Elahi A., Chegenelorestani R., Rezaee M. (2018). Dissemination of multidrug-resistant, class I and II integrons and molecular typing of CTX-M-producing Klebsiella pneumoniae. Int. J. Appl. Basic Med. Res. 8 100–105. 10.4103/ijabmr.IJABMR_333_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Atrouni A., Hamze M., Jisr T., Lemarie C., Eveillard M., Joly-Guillou M. L., et al. (2016). Wide spread of OXA-23-producing carbapenem-resistant Acinetobacter baumannii belonging to clonal complex II in different hospitals in Lebanon. Int. J. Infect. Dis. 52 29–36. 10.1016/j.ijid.2016.09.017 [DOI] [PubMed] [Google Scholar]
- Al Atrouni A., Joly-Guillou M. L., Hamze M., Kempf M. (2016a). Emergence of NDM-1 and OXA-72 producing Acinetobacter pittii clinical isolates in Lebanon. New Microbes New Infect. 12 43–44. 10.1016/j.nmni.2016.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Atrouni A., Hamze M., Rafei R., Eveillard M., Joly-Guillou M. L., Kempf M. (2016b). Diversity of Acinetobacter species isolated from different environments in Lebanon: a nationwide study. Future Microbiol. 11 1147–1156. 10.2217/fmb-2016-0082 [DOI] [PubMed] [Google Scholar]
- Al Atrouni A., Kempf M., Eveillard M., Rafei R., Hamze M., Joly-Guillou M. L. (2015). First report of oxa-72-producing Acinetobacter calcoaceticus in Lebanon. New Microbes New Infect. 9 11–12. 10.1016/j.nmni.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Bayssari C., Dabboussi F., Hamze M., Rolain J. M. (2015a). Emergence of carbapenemase-producing Pseudomonas aeruginosa and Acinetobacter baumannii in livestock animals in Lebanon. J. Antimicrob. Chemother. 70 950–951. 10.1093/jac/dku469 [DOI] [PubMed] [Google Scholar]
- Al Bayssari C., Olaitan A. O., Dabboussi F., Hamze M., Rolain J. M. (2015b). Emergence of OXA-48-producing Escherichia coli clone ST38 in fowl. Antimicrob. Agents Chemother. 59 745–746. 10.1128/aac.03552-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Bayssari C., Diene S. M., Loucif L., Gupta S. K., Dabboussi F., Mallat H., et al. (2014). Emergence of VIM-2 and IMP-15 carbapenemases and inactivation of oprD gene in carbapenem-resistant Pseudomonas aeruginosa clinical isolates from Lebanon. Antimicrob. Agents Chemother. 58 4966–4970. 10.1128/AAC.02523-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Muharrmi Z., Rafay A. M., Balkhair A., Al-Tamemi S., Al Mawali A., Al Sadiri H. (2008). Extended-spectrum beta-lactamase (ESBL) in omani children: study of prevalence, risk factors and clinical outcomes at sultan qaboos university hospital, sultanate of Oman. Sultan Qaboos Univ. Med. J. 8 171–177. [PMC free article] [PubMed] [Google Scholar]
- Al Sheikh Y. A., Marie M. A., John J., Krishnappa L. G., Dabwab K. H. (2014). Prevalence of 16S rRNA methylase genes among beta-lactamase-producing enterobacteriaceae clinical isolates in Saudi Arabia. Libyan J. Med. 9:24432. 10.3402/ljm.v9.24432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Agamy M. H., Jeannot K., El-Mahdy T. S., Samaha H. A., Shibl A. M., Plesiat P., et al. (2016). Diversity of molecular mechanisms conferring carbapenem resistance to Pseudomonas aeruginosa isolates from Saudi Arabia. Can. J. Infect. Dis. Med. Microbiol. 2016:4379686. 10.1155/2016/4379686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Agamy M. H., Jeannot K., El-Mahdy T. S., Shibl A. M., Kattan W., Plesiat P., et al. (2017). First detection of GES-5 carbapenemase-producing Acinetobacter baumannii isolate. Microb. Drug Resist. 23 556–562. 10.1089/mdr.2016.0152 [DOI] [PubMed] [Google Scholar]
- Al-Agamy M. H., Shibl A. M., Elkhizzi N. A., Meunier D., Turton J. F., Livermore D. M. (2013). Persistence of Klebsiella pneumoniae clones with OXA-48 or NDM carbapenemases causing bacteraemias in a Riyadh hospital. Diagn. Microbiol. Infect. Dis 76 214–216. 10.1016/j.diagmicrobio.2013.02.006 [DOI] [PubMed] [Google Scholar]
- Al-Agamy M. H., Shibl A. M., Tawfik A. F., Elkhizzi N. A., Livermore D. M. (2012). Extended-spectrum and metallo-beta-lactamases among ceftazidime-resistant Pseudomonas aeruginosa in Riyadh, Saudi Arabia. J. Chemother. 24 97–100. 10.1179/1120009X12Z.00000000015 [DOI] [PubMed] [Google Scholar]
- Al-Bahry S. N., Al-Zadjali M. A., Mahmoud I. Y., Elshafie A. E. (2012). Biomonitoring marine habitats in reference to antibiotic resistant bacteria and ampicillin resistance determinants from oviductal fluid of the nesting green sea turtle, chelonia mydas. Chemosphere 87 1308–1315. 10.1016/j.chemosphere.2012.01.051 [DOI] [PubMed] [Google Scholar]
- Al-Bayssari C., Olaitan A. O., Leangapichart T., Okdah L., Dabboussi F., Hamze M., et al. (2016). Whole-genome sequence of a blaOXA-48-harboring Raoultella ornithinolytica clinical isolate from Lebanon. Antimicrob. Agents Chemother. 60 2548–2550. 10.1128/AAC.02773-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Charrakh A. H., Al-Awadi S. J., Mohammed A. S. (2016). Detection of metallo-beta-lactamase producing Pseudomonas aeruginosa isolated from public and private hospitals in Baghdad, Iraq. Acta Med. Iran 54 107–113. [PubMed] [Google Scholar]
- Alfaresi M. S., Elkoush A. A., Alshehhi H. M., Abdulsalam A. I. (2011). Molecular characterization and epidemiology of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates in the United Arab Emirates. Med. Princ. Pract. 20 177–180. 10.1159/000319912 [DOI] [PubMed] [Google Scholar]
- Alghamdi S., Shebl N. A., Aslanpour Z., Shibl A., Berrou I. (2018). Hospital adoption of antimicrobial stewardship programmes in gulf cooperation council countries: a review of existing evidence. J. Glob. Antimicrob. Resist. 15 196–209. 10.1016/j.jgar.2018.07.014 [DOI] [PubMed] [Google Scholar]
- Alghoribi M. F., Gibreel T. M., Farnham G., Al Johani S. M., Balkhy H. H., Upton M. (2015). Antibiotic-resistant ST38, ST131 and ST405 strains are the leading uropathogenic Escherichia coli clones in riyadh, Saudi Arabia. J. Antimicrob. Chemother. 70 2757–2762. 10.1093/jac/dkv188 [DOI] [PubMed] [Google Scholar]
- Algowaihi R., Ashgar S., Sirag B., Shalam S., Nassir A., Ahmed A. (2016). Draft genome sequence of a multidrug-resistant Klebsiella pneumoniae strain isolated from King abdullah medical city, Makkah, Saudi Arabia. Genome Announc. 4 e375–16. 10.1128/genomeA.00375-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhaddad M. S., AlBarjas A. K., Alhammar L. E., Al Rashed A. S., Badger-Emeka L. I. (2018). Molecular characterization and antibiotic susceptibility pattern of Acinetobacter baumannii isolated in intensive care unit patients in Al-hassa, Kingdom of Saudi Arabia. Int. J. Appl. Basic Med. Res. 8 19–23. 10.4103/ijabmr.IJABMR_91_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hassan L., El Mehallawy H., Amyes S. G. (2013). Diversity in Acinetobacter baumannii isolates from paediatric cancer patients in Egypt. Clin. Microbiol. Infect. 19 1082–1088. 10.1111/1469-0691.12143 [DOI] [PubMed] [Google Scholar]
- Alhomoud F., Aljamea Z., Almahasnah R., Alkhalifah K., Basalelah L., Alhomoud F. K. (2017). Self-medication and self-prescription with antibiotics in the middle east-do they really happen? A systematic review of the prevalence, possible reasons, and outcomes. Int. J. Infect. Dis. 57 3–12. 10.1016/j.ijid.2017.01.014 [DOI] [PubMed] [Google Scholar]
- Alikhani M. Y., Karimi Tabar Z., Mihani F., Kalantar E., Karami P., Sadeghi M., et al. (2014). Antimicrobial resistance patterns and prevalence of blaPER-1 and blaVEB-1 genes among ESBL-producing Pseudomonas aeruginosa isolates in west of Iran. Jundishapur J. Microbiol. 7:e8888. 10.5812/jjm.8888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliramezani A., Douraghi M., Hajihasani A., Mohammadzadeh M., Rahbar M. (2016). Clonal relatedness and biofilm formation of OXA-23-producing carbapenem resistant Acinetobacter baumannii isolates from hospital environment. Microb. Pathog. 99 204–208. 10.1016/j.micpath.2016.08.034 [DOI] [PubMed] [Google Scholar]
- Aljindan R., Bukharie H., Alomar A., Abdalhamid B. (2015). Prevalence of digestive tract colonization of carbapenem-resistant Acinetobacter baumannii in hospitals in Saudi Arabia. J. Med. Microbiol. 64(Pt 4), 400–406. 10.1099/jmm.0.000033 [DOI] [PubMed] [Google Scholar]
- Alkasaby N. M., El Sayed Zaki M. (2017). Molecular study of Acinetobacter baumannii isolates for metallo-beta-lactamases and extended-spectrum-beta-lactamases genes in intensive care unit, Mansoura university hospital, Egypt. Int. J. Microbiol. 2017:3925868. 10.1155/2017/3925868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mayahie S., Al Kuriashy J. J. (2016). Distribution of ESBLs among Escherichia coli isolates from outpatients with recurrent UTIs and their antimicrobial resistance. J. Infect. Dev. Ctries 10 575–583. 10.3855/jidc.6661 [DOI] [PubMed] [Google Scholar]
- Al-Mayahie S. M. (2013). Phenotypic and genotypic comparison of ESBL production by vaginal Escherichia coli isolates from pregnant and non-pregnant women. Ann. Clin. Microbiol. Antimicrob. 12:7. 10.1186/1476-0711-12-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Muhanna A. S., Al-Muhanna S., Alzuhairi M. A. (2016). Molecular investigation of extended-spectrum beta-lactamase genes and potential drug resistance in clinical isolates of Morganella morganii. Ann. Saudi Med. 36 223–228. 10.5144/0256-4947.2016.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso C. A., Zarazaga M., Ben Sallem R., Jouini A., Ben Slama K., Torres C. (2017). Antibiotic resistance in Escherichia coli in husbandry animals: the african perspective. Lett. Appl. Microbiol. 64 318–334. 10.1111/lam.12724 [DOI] [PubMed] [Google Scholar]
- Al-Otaibi F. E., Bukhari E. E. (2013). Clinical and laboratory profiles of urinary tract infections caused by extended-spectrum beta-lactamase-producing Escherichia coli in a tertiary care center in central Saudi Arabia. Saudi Med. J. 34 171–176. [PubMed] [Google Scholar]
- Alotaibi F. E., Bukhari E. E., Al-Mohizea M. M., Hafiz T., Essa E. B., AlTokhais Y. I. (2017). Emergence of carbapenem-resistant enterobacteriaceae isolated from patients in a university hospital in Saudi Arabia. epidemiology, clinical profiles and outcomes. J. Infect. Public Health 10 667–673. 10.1016/j.jiph.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alousi S., Salloum T., Arabaghian H., Matar G. M., Araj G. F., Tokajian S. T. (2018). Genomic characterization of MDR Escherichia coli harboring blaOXA-48 on the IncL/M-type plasmid isolated from blood stream infection. Biomed. Res. Int. 2018:3036143. 10.1155/2018/3036143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alp E., Percin D., Colakoglu S., Durmaz S., Kurkcu C. A., Ekincioglu P., et al. (2013). Molecular characterization of carbapenem-resistant Klebsiella pneumoniae in a tertiary university hospital in Turkey. J. Hosp. Infect. 84 178–180. 10.1016/j.jhin.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Al-Qahtani A. A., Al-Agamy M. H., Ali M. S., Al-Ahdal M. N., Aljohi M. A., Shibl A. M. (2014). Characterization of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae from Riyadh, Saudi Arabia. J. Chemother. 26 139–145. 10.1179/1973947813Y.0000000124 [DOI] [PubMed] [Google Scholar]
- Alsharapy S. A., Yanat B., Lopez-Cerero L., Nasher S. S., Diaz-De-Alba P., Pascual A., et al. (2018). Prevalence of ST131 clone producing both ESBL CTX-M-15 and AAC(6’)ib-cr among ciprofloxacin-resistant Escherichia coli isolates from Yemen. Microb. Drug Resist. 10.1016/j.jgar.2016.02.001 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Alsultan A. A., Evans B. A., Elsayed E. A., Al-Thawadi S. I., Al-Taher A. Y., Amyes S. G., et al. (2013). High frequency of carbapenem-resistant Acinetobacter baumannii in patients with diabetes mellitus in Saudi Arabia. J. Med. Microbiol. 62(Pt 6), 885–888. 10.1099/jmm.0.057216-0 [DOI] [PubMed] [Google Scholar]
- Al-Sweih N. A., Al-Hubail M., Rotimi V. O. (2012). Three distinct clones of carbapenem-resistant Acinetobacter baumannii with high diversity of carbapenemases isolated from patients in two hospitals in Kuwait. J. Infect. Public Health 5 102–108. 10.1016/j.jiph.2011.11.004 [DOI] [PubMed] [Google Scholar]
- Al-Sweih N. A., Al-Hubail M. A., Rotimi V. O. (2011). Emergence of tigecycline and colistin resistance in Acinetobacter species isolated from patients in Kuwait hospitals. J. Chemother. 23 13–16. 10.1179/joc.2011.23.1.13 [DOI] [PubMed] [Google Scholar]
- Altalhi A. D., Gherbawy Y. A., Hassan S. A. (2010). Antibiotic resistance in Escherichia coli isolated from retail raw chicken meat in Taif, Saudi Arabia. Foodborne Pathog. Dis. 7 281–285. 10.1089/fpd.2009.0365 [DOI] [PubMed] [Google Scholar]
- Aly M., Tayeb H. T., Al Johani S. M., Alyamani E. J., Aldughaishem F., Alabdulkarim I., et al. (2014). Genetic diversity of OXA-51-like genes among multidrug-resistant Acinetobacter baumannii in Riyadh, Saudi Arabia. Eur. J. Clin. Microbiol. Infect. Dis. 33 1223–1228. 10.1007/s10096-014-2068-0 [DOI] [PubMed] [Google Scholar]
- Aly M. M., Abu Alsoud N. M., Elrobh M. S., Al Johani S. M., Balkhy H. H. (2016). High prevalence of the PER-1 gene among carbapenem-resistant Acinetobacter baumannii in Riyadh, Saudi Arabia. Eur. J. Clin. Microbiol. Infect. Dis. 35 1759–1766. 10.1007/s10096-016-2723-8 [DOI] [PubMed] [Google Scholar]
- Aly S. A., Debavalya N., Suh S. J., Oryazabal O. A., Boothe D. M. (2012). Molecular mechanisms of antimicrobial resistance in fecal Escherichia coli of healthy dogs after enrofloxacin or amoxicillin administration. Can. J. Microbiol. 58 1288–1294. 10.1139/w2012-105 [DOI] [PubMed] [Google Scholar]
- Alyamani E. J., Khiyami A. M., Booq R. Y., Majrashi M. A., Bahwerth F. S., Rechkina E. (2017). The occurrence of ESBL-producing Escherichia coli carrying aminoglycoside resistance genes in urinary tract infections in Saudi Arabia. Ann. Clin. Microbiol. Antimicrob. 16:1. 10.1186/s12941-016-0177-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alyamani E. J., Khiyami M. A., Booq R. Y., Alnafjan B. M., Altammami M. A., Bahwerth F. S. (2015). Molecular characterization of extended-spectrum beta-lactamases (ESBLs) produced by clinical isolates of Acinetobacter baumannii in Saudi Arabia. Ann. Clin. Microbiol. Antimicrob. 14:38. 10.1186/s12941-015-0098-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Zahrani I. A., Alsiri B. A. (2018). The emergence of carbapenem-resistant Klebsiella pneumoniae isolates producing OXA-48 and NDM in the southern (Asir) province, Saudi Arabia. Saudi Med. J. 39 23–30. 10.15537/smj.2018.1.21094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin M., Sirous M., Javaherizadeh H., Motamedifar M., Saki M., Veisi H., et al. (2018). Antibiotic resistance pattern and molecular characterization of extended-spectrum beta-lactamase producing enteroaggregative Escherichia coli isolates in children from southwest Iran. Infect. Drug Resist. 11 1097–1104. 10.2147/IDR.S167271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminshahidi M., Arastehfar A., Pouladfar G., Arman E., Fani F. (2017). Diarrheagenic Escherichia coli and shigella with high rate of extended-spectrum beta-lactamase production: two predominant etiological agents of acute diarrhea in Shiraz, Iran. Microb. Drug Resist. 23 1037–1044. 10.1089/mdr.2017.0204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirkamali S., Naserpour-Farivar T., Azarhoosh K., Peymani A. (2017). Distribution of the bla OXA, bla VEB-1, and bla GES-1 genes and resistance patterns of ESBL-producing Pseudomonas aeruginosa isolated from hospitals in Tehran and Qazvin, Iran. Rev. Soc. Bras. Med. Trop. 50 315–320. 10.1590/0037-8682-0478-2016 [DOI] [PubMed] [Google Scholar]
- Aqel A. A., Findlay J., Al-Maayteh M., Al-Kaabneh A., Hopkins K. L., Alzoubi H., et al. (2018). Characterization of carbapenemase-producing enterobacteriaceae from patients in Amman, Jordan. Microb. Drug Resist. 24 1121–1127. 10.1089/mdr.2017.0238 [DOI] [PubMed] [Google Scholar]
- Aqel A. A., Giakkoupi P., Alzoubi H., Masalha I., Ellington M. J., Vatopoulos A. (2017). Detection of OXA-48-like and NDM carbapenemases producing Klebsiella pneumoniae in Jordan: a pilot study. J. Infect. Public Health 10 150–155. 10.1016/j.jiph.2016.02.002 [DOI] [PubMed] [Google Scholar]
- Armin S., Fallah F., Azimi L., Samadi Kafil H., Ghazvini K., Hasanzadeh S., et al. (2018). Warning: spread of NDM-1 in two border towns of Iran. Cell. Mol. Biol. 64 125–129. [PubMed] [Google Scholar]
- Aslantas O., Yilmaz E. S. (2017). Prevalence and molecular characterization of extended-spectrum beta-lactamase (ESBL) and plasmidic AmpC beta-lactamase (pAmpC) producing Escherichia coli in dogs. J. Vet. Med. Sci. 79 1024–1030. 10.1292/jvms.16-0432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atilla A., Eroglu C., Esen S., Sunbul M., Leblebicioglu H. (2012). Investigation of the frequency of PER-1 type beta-lactamase and antimicrobial resistance rates in nosocomial isolates of Pseudomonas aeruginosa. Mikrobiyol. Bul. 46 1–8. [PubMed] [Google Scholar]
- Auta A., Hadi M. A., Oga E., Adewuyi E. O., Abdu-Aguye S. N., Adeloye D., et al. (2019). Global access to antibiotics without prescription in community pharmacies: a systematic review and meta-analysis. J. Infect. 78 8–18. 10.1016/j.jinf.2018.07.001 [DOI] [PubMed] [Google Scholar]
- Azimi A., Peymani A., Pour P. K. (2018). Phenotypic and molecular detection of metallo-beta-lactamase-producing Pseudomonas aeruginosa isolates from patients with burns in Tehran, Iran. Rev. Soc. Bras. Med. Trop. 51 610–615. 10.1590/0037-8682-0174-2017 [DOI] [PubMed] [Google Scholar]
- Azimi L., Nordmann P., Lari A. R., Bonnin R. A. (2014). First report of OXA-48-producing Klebsiella pneumoniae strains in Iran. GMS Hyg. Infect. Control 9:Doc07. 10.3205/dgkh000227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimi L., Talebi M., Pourshafie M. R., Owlia P., Rastegar Lari A. (2015). Characterization of carbapenemases in extensively drug resistance Acinetobacter baumannii in a burn care center in Iran. Int. J. Mol. Cell. Med. 4 46–53. [PMC free article] [PubMed] [Google Scholar]
- Azizi O., Shakibaie M. R., Badmasti F., Modarresi F., Ramazanzadeh R., Mansouri S., et al. (2016). Class 1 integrons in non-clonal multidrug-resistant Acinetobacter baumannii from Iran, description of the new blaIMP-55 allele in In1243. J. Med. Microbiol. 65 928–936. 10.1099/jmm.0.000315 [DOI] [PubMed] [Google Scholar]
- Azizi O., Shakibaie M. R., Modarresi F., Shahcheraghi F. (2015). Molecular detection of class-D OXA carbapenemase genes in biofilm and non-biofilm forming clinical isolates of Acinetobacter baumannii. Jundishapur J. Microbiol. 8:e21042. 10.5812/jjm.21042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baadani A. M., Thawadi S. I., El-Khizzi N. A., Omrani A. S. (2013). Prevalence of colistin and tigecycline resistance in Acinetobacter baumannii clinical isolates from 2 hospitals in Riyadh region over a 2-year period. Saudi Med. J. 34 248–253. [PubMed] [Google Scholar]
- Badran E. F., Qamer Din R. A., Shehabi A. A. (2016). Low intestinal colonization of Escherichia coli clone ST131 producing CTX-M-15 in Jordanian infants. J. Med. Microbiol. 65 137–141. 10.1099/jmm.0.000210 [DOI] [PubMed] [Google Scholar]
- Bagheri Josheghani S., Moniri R., Firoozeh F., Sehat M., Dasteh Goli Y. (2015). Susceptibility pattern and distribution of oxacillinases and bla PER-1 genes among multidrug resistant Acinetobacter baumannii in a teaching hospital in Iran. J. Pathog. 2015:957259. 10.1155/2015/957259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahador A., Farshadzadeh Z., Raoofian R., Mokhtaran M., Pourakbari B., Pourhajibagher M., et al. (2018). Association of virulence gene expression with colistin-resistance in Acinetobacter baumannii: analysis of genotype, antimicrobial susceptibility, and biofilm formation. Ann. Clin. Microbiol. Antimicrob. 17:24. 10.1186/s12941-018-0277-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakour S., Alsharapy S. A., Touati A., Rolain J. M. (2014). Characterization of Acinetobacter baumannii clinical isolates carrying bla(OXA-23) carbapenemase and 16S rRNA methylase armA genes in Yemen. Microb. Drug Resist. 20 604–609. 10.1089/mdr.2014.0018 [DOI] [PubMed] [Google Scholar]
- Balkan I. I., Aygun G., Aydin S., Mutcali S. I., Kara Z., Kuskucu M., et al. (2014). Blood stream infections due to OXA-48-like carbapenemase-producing enterobacteriaceae: treatment and survival. Int. J. Infect. Dis. 26 51–56. 10.1016/j.ijid.2014.05.012 [DOI] [PubMed] [Google Scholar]
- Balkhy H. H., El-Saed A., Al Johani S. M., Francis C., Al-Qahtani A. A., Al-Ahdal M. N., et al. (2012). The epidemiology of the first described carbapenem-resistant Klebsiella pneumoniae outbreak in a tertiary care hospital in Saudi Arabia: how far do we go? Eur. J. Clin. Microbiol. Infect. Dis. 31 1901–1909. 10.1007/s10096-011-1519-0 [DOI] [PubMed] [Google Scholar]
- Baran I., Aksu N. (2016). Phenotypic and genotypic characteristics of carbapenem-resistant enterobacteriaceae in a tertiary-level reference hospital in Turkey. Ann. Clin. Microbiol. Antimicrob. 15:20. 10.1186/s12941-016-0136-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron S., Hadjadj L., Rolain J. M., Olaitan A. O. (2016). Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int. J. Antimicrob. Agents 48 583–591. 10.1016/j.ijantimicag.2016.06.023 [DOI] [PubMed] [Google Scholar]
- Baroud M., Dandache I., Araj G. F., Wakim R., Kanj S., Kanafani Z., et al. (2013). Underlying mechanisms of carbapenem resistance in extended-spectrum beta-lactamase-producing Klebsiella pneumoniae and Escherichia coli isolates at a tertiary care centre in Lebanon: role of OXA-48 and NDM-1 carbapenemases. Int. J. Antimicrob. Agents 41 75–79. 10.1016/j.ijantimicag.2012.08.010 [DOI] [PubMed] [Google Scholar]
- Barwa R., Shaaban M. (2017). Molecular characterization of Klebsiella pneumoniae clinical isolates with elevated resistance to carbapenems. Open Microbiol. J. 11 152–159. 10.2174/1874285801711010152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassyouni R. H., Gaber S. N., Wegdan A. A. (2015). Fecal carriage of extended-spectrum beta-lactamase- and AmpC- producing Escherichia coli among healthcare workers. J. Infect. Dev. Ctries 9 304–308. 10.3855/jidc.5633 [DOI] [PubMed] [Google Scholar]
- Ben-David D., Masarwa S., Navon-Venezia S., Mishali H., Fridental I., Rubinovitch B., et al. (2011). Carbapenem-resistant Klebsiella pneumoniae in post-acute-care facilities in Israel. Infect. Control Hosp. Epidemiol. 32 845–853. 10.1086/661279 [DOI] [PubMed] [Google Scholar]
- Bialvaei A. Z., Kafil H. S., Asgharzadeh M., Aghazadeh M., Yousefi M. (2016). CTX-M extended-spectrum beta-lactamase-producing Klebsiella spp, Salmonella spp, Shigella spp and Escherichia coli isolates in Iranian hospitals. Braz. J. Microbiol. 47 706–711. 10.1016/j.bjm.2016.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindayna K. M., Murtadha M. (2011). High prevalence of blaCTX-M in enterobacteriaceae isolates from the Kingdom of Bahrain. Asian Pac. J. Trop. Med. 4 937–940. 10.1016/S1995-7645(11)60222-8 [DOI] [PubMed] [Google Scholar]
- Blaak H., van Hoek A. H., Hamidjaja R. A., van der Plaats R. Q., Kerkhof-de Heer L., de Roda Husman A. M., et al. (2015). Distribution, numbers, and diversity of ESBL-producing E. coli in the poultry farm environment. PLoS One 10:e0135402. 10.1371/journal.pone.0135402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokaeian M., Zahedani S. S., BajgIran M. S., Moghaddam A. A. (2014). Frequency of PER, VEB, SHV, TEM and CTX-M genes in resistant strains of Pseudomonas aeruginosa producing extended spectrum beta-lactamases. Jundishapur J. Microbiol. 8:e13783. 10.5812/jjm.13783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S. D., Ahmed M. F., El-Adawy H., Hotzel H., Engelmann I., Weiss D., et al. (2016). Surveillance of extended-spectrum beta-lactamase-producing Escherichia coli in dairy cattle farms in the nile delta, Egypt. Front. Microbiol. 7:1020. 10.3389/fmicb.2016.01020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K., Bradford P. A. (2016). Beta-lactams and beta-lactamase inhibitors: an overview. Cold Spring Harb. Perspect. Med. 6:a025247. 10.1101/cshperspect.a025247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakar A., Akyon Y., Gur D., Karatuna O., Ogunc D., Ozhak Baysan B., et al. (2016). Investigation of carbapenemases in carbapenem-resistant Escherichia coli and Klebsiella pneumoniae strains isolated in (2014) in Turkey. Mikrobiyol. Bul. 50 21–33. 10.5578/mb.10695 [DOI] [PubMed] [Google Scholar]
- Can F., Azap O. K., Seref C., Ispir P., Arslan H., Ergonul O. (2015). Emerging Escherichia coli O25b/ST131 clone predicts treatment failure in urinary tract infections. Clin. Infect. Dis 60 523–527. 10.1093/cid/ciu864 [DOI] [PubMed] [Google Scholar]
- Candan E. D., Aksoz N. (2015). Klebsiella pneumoniae: Characteristics of carbapenem resistance and virulence factors. Acta Biochim. Pol. 62 867–874. 10.18388/abp.2015_1148 [DOI] [PubMed] [Google Scholar]
- Caneiras C., Lito L., Melo-Cristino J., Duarte A. (2019). Community- and hospital-acquired Klebsiella pneumoniae urinary tract infections in portugal: virulence and antibiotic resistance. Microorganisms 7:E138. 10.3390/microorganisms7050138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrer A., Poirel L., Yilmaz M., Akan O. A., Feriha C., Cuzon G., et al. (2010). Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob. Agents Chemother. 54 1369–1373. 10.1128/AAC.01312-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanheira M., Costello S. E., Woosley L. N., Deshpande L. M., Davies T. A., Jones R. N. (2014). Evaluation of clonality and carbapenem resistance mechanisms among Acinetobacter baumannii-Acinetobacter calcoaceticus complex and enterobacteriaceae isolates collected in european and mediterranean countries and detection of two novel beta-lactamases, GES-22 and VIM-35. Antimicrob. Agents Chemother. 58 7358–7366. 10.1128/AAC.03930-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention [CDC], (2018). One Health Basics. Available at: https://www.cdc.gov/onehealth/basics/index.html (accessed May 14, 2019). [Google Scholar]
- Charrouf F. O., Hamze M., Mallat H., Achkar M., Dabboussia F. (2014). Characterization of resistance genes in 68 ESBL-producing Klebsiella pneumonia in Lebanon. Med. Mal. Infect. 44 535–538. 10.1016/j.medmal.2014.10.001 [DOI] [PubMed] [Google Scholar]
- Chazan B., Raz R., Teitler N., Nitzan O., Edelstein H., Colodner R. (2009). Epidemiology and susceptibility to antimicrobials in community, hospital and long-term care facility bacteremia in northern Israel: a 6 year surveillance. Isr. Med. Assoc. J. 11 592–597. [PubMed] [Google Scholar]
- Chen L., Al Laham N., Chavda K. D., Mediavilla J. R., Jacobs M. R., Bonomo R. A., et al. (2015). First report of an OXA-48-producing multidrug-resistant Proteus mirabilis strain from Gaza, Palestine. Antimicrob. Agents Chemother. 59 4305–4307. 10.1128/AAC.00565-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Gao J., Zhang H., Ying C. (2017). Spread of the blaOXA-23-containing Tn2008 in carbapenem-resistant Acinetobacter baumannii isolates grouped in CC92 from China. Front. Microbiol. 8:163. 10.3389/fmicb.2017.00163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophy R., Osman M., Mallat H., Achkar M., Ziedeh A., Moukaddem W., et al. (2017). Prevalence, antibiotic susceptibility and characterization of antibiotic resistant genes among carbapenem-resistant gram-negative bacilli and yeast in intestinal flora of cancer patients in north Lebanon. J. Infect. Public Health 10 716–720. 10.1016/j.jiph.2016.10.009 [DOI] [PubMed] [Google Scholar]
- Cicek A. C., Saral A., Iraz M., Ceylan A., Duzgun A. O., Peleg A. Y., et al. (2014). OXA- and GES-type beta-lactamases predominate in extensively drug-resistant Acinetobacter baumannii isolates from a Turkish university hospital. Clin. Microbiol. Infect. 20 410–415. 10.1111/1469-0691.12338 [DOI] [PubMed] [Google Scholar]
- Ciftci I. H., Asik G., Karakece E., Oksuz L., Yagci S., Sesli Cetin E., et al. (2013). Distribution of blaOXA genes in Acinetobacter baumannii strains: a multicenter study. Mikrobiyol. Bul. 47 592–602. 10.5578/mb.6388 [DOI] [PubMed] [Google Scholar]
- Cizmeci Z., Aktas E., Otlu B., Acikgoz O., Ordekci S. (2017). Molecular characterization of carbapenem- resistant enterobacteriaceae yields increasing rates of NDM-1 carbapenemases and colistin resistance in an OXA-48- endemic area. J. Chemother. 29 344–350. 10.1080/1120009X.2017.1323149 [DOI] [PubMed] [Google Scholar]
- Cohen-Nahum K., Saidel-Odes L., Riesenberg K., Schlaeffer F., Borer A. (2010). Urinary tract infections caused by multi-drug resistant Proteus mirabilis: risk factors and clinical outcomes. Infection 38 41–46. 10.1007/s15010-009-8460-5 [DOI] [PubMed] [Google Scholar]
- Dahdouh E., Hajjar M., Suarez M., Daoud Z. (2016). Acinetobacter baumannii isolated from lebanese patients: phenotypes and genotypes of resistance, clonality, and determinants of pathogenicity. Front. Cell Infect. Microbiol. 6:163. 10.3389/fcimb.2016.00163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandachi I., Chabou S., Daoud Z., Rolain J. M. (2018a). Prevalence and emergence of extended-spectrum cephalosporin-, carbapenem- and colistin-resistant gram negative bacteria of animal origin in the mediterranean basin. Front. Microbiol. 9:2299. 10.3389/fmicb.2018.02299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandachi I., Fayad E., El-Bazzal B., Daoud Z., Rolain J. M. (2018b). Prevalence of extended-spectrum beta-lactamase-producing gram-negative bacilli and emergence of mcr-1 colistin resistance gene in lebanese swine farms. Microb. Drug Resist. 25 233–240. 10.1089/mdr.2018.0110 [DOI] [PubMed] [Google Scholar]
- Dandachi I., Leangapichart T., Daoud Z., Rolain J. M. (2018c). First detection of mcr-1 plasmid mediated colistin resistant E. coli in lebanese poultry. J. Glob. Antimicrob. Resist. 12 137–138. 10.1016/j.jgar.2018.01.004 [DOI] [PubMed] [Google Scholar]
- Dandachi I., Salem Sokhn E., Najem E., Azar E., Daoud Z. (2016). Carriage of beta-lactamase-producing enterobacteriaceae among nursing home residents in north Lebanon. Int. J. Infect. Dis. 45 24–31. 10.1016/j.ijid.2016.02.007 [DOI] [PubMed] [Google Scholar]
- Dandachi I., Sokhn E. S., Dahdouh E. A., Azar E., El-Bazzal B., Rolain J. M., et al. (2018). Prevalence and characterization of multi-drug-resistant gram-negative bacilli isolated from Lebanese poultry: a nationwide study. Front. Microbiol. 9:550. 10.3389/fmicb.2018.00550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daoud Z., Farah J., Sokhn E. S., El Kfoury K., Dahdouh E., Masri K., et al. (2018). Multidrug-resistant enterobacteriaceae in Lebanese hospital wastewater: implication in the one health concept. Microb. Drug Resist. 24 166–174. 10.1089/mdr.2017.0090 [DOI] [PubMed] [Google Scholar]
- Daoud Z., Salem-Sokhn E., Dahdouh E., Irani J., Matar G. M., Doron S. (2017). Resistance and clonality in Escherichia coli and Klebsiella spp. and relationship with antibiotic consumption in major Lebanese hospitals. J. Glob. Antimicrob. Resist. 11 45–51. 10.1016/j.jgar.2017.07.011 [DOI] [PubMed] [Google Scholar]
- Dar O. A., Hasan R., Schlundt J., Harbarth S., Caleo G., Dar F. K., et al. (2016). Exploring the evidence base for national and regional policy interventions to combat resistance. Lancet 387 285–295. 10.1016/S0140-6736(15)00520-6 [DOI] [PubMed] [Google Scholar]
- Dash N., Panigrahi D., Zarouni M. A., Darwish D., Ghazawi A., Sonnevend A., et al. (2014). High incidence of new delhi metallo-beta-lactamase-producing Klebsiella pneumoniae isolates in Sharjah, United Arab Emirates. Microb. Drug Resist. 20 52–56. 10.1089/mdr.2013.0040 [DOI] [PubMed] [Google Scholar]
- Dashti A. A., Jadaon M. M., Amyes S. G. (2010a). Retrospective study of an outbreak in a Kuwaiti hospital of multidrug-resistant Klebsiella pneumoniae possessing the new SHV-112 extended-spectrum beta-lactamase. J. Chemother. 22 335–338. 10.1179/joc.2010.22.5.335 [DOI] [PubMed] [Google Scholar]
- Dashti A. A., Jadaon M. M., Gomaa H. H., Noronha B., Udo E. E. (2010b). Transmission of a Klebsiella pneumoniae clone harbouring genes for CTX-M-15-like and SHV-112 enzymes in a neonatal intensive care unit of a Kuwaiti hospital. J. Med. Microbiol. 59(Pt 6), 687–692. 10.1099/jmm.0.019208-0 [DOI] [PubMed] [Google Scholar]
- Dashti A. A., Vali L., El-Shazly S., Jadaon M. M. (2014). The characterization and antibiotic resistance profiles of clinical Escherichia coli O25b-B2-ST131 isolates in Kuwait. BMC Microbiol. 14:214. 10.1186/s12866-014-0214-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davodian E., Sadeghifard N., Ghasemian A., Noorbakhsh S. (2016). Presence of blaPER-1 and blaVEB-1 beta-lactamase genes among isolates of Pseudomonas aeruginosa from south west of Iran. J. Epidemiol. Glob. Health 6 211–213. 10.1016/j.jegh.2016.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayan N., Dabbah H., Weissman I., Aga I., Even L., Glikman D. (2013). Urinary tract infections caused by community-acquired extended-spectrum beta-lactamase-producing and nonproducing bacteria: a comparative study. J. Pediatr. 163 1417–1421. 10.1016/j.jpeds.2013.06.078 [DOI] [PubMed] [Google Scholar]
- Debby B. D., Ganor O., Yasmin M., David L., Nathan K., Ilana T., et al. (2012). Epidemiology of carbapenem resistant Klebsiella pneumoniae colonization in an intensive care unit. Eur. J. Clin. Microbiol. Infect. Dis. 31 1811–1817. 10.1007/s10096-011-1506-5 [DOI] [PubMed] [Google Scholar]
- Decker B. K., Lau A. F., Dekker J. P., Spalding C. D., Sinaii N., Conlan S., et al. (2018). Healthcare personnel intestinal colonization with multidrug-resistant organisms. Clin. Microbiol. Infect 24 82.e1–82.e4. 10.1016/j.cmi.2017.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehshiri M., Khoramrooz S. S., Zoladl M., Khosravani S. A., Parhizgari N., Motazedian M. H., et al. (2018). The frequency of Klebsiella pneumonia encoding genes for CTX-M, TEM-1 and SHV-1 extended-spectrum beta lactamases enzymes isolated from urinary tract infection. Ann. Clin. Microbiol. Antimicrob. 17:4. 10.1186/s12941-018-0256-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirbakan H., Midilli K., Ogunc D., Ozen N., Ongut G., Daglar D., et al. (2008). Investigation of plasmid-mediated AmpC beta-lactamase types in Klebsiella spp. and Escherichia coli isolates resistant or intermediate to cefoxitin. Mikrobiyol. Bul. 42 545–551. [PubMed] [Google Scholar]
- Di Popolo A., Giannouli M., Triassi M., Brisse S., Zarrilli R. (2011). Molecular epidemiological investigation of multidrug-resistant Acinetobacter baumannii strains in four mediterranean countries with a multilocus sequence typing scheme. Clin. Microbiol. Infect. 17 197–201. 10.1111/j.1469-0691.2010.03254.x [DOI] [PubMed] [Google Scholar]
- Diab M., Hamze M., Bonnet R., Saras E., Madec J. Y., Haenni M. (2018). Extended-spectrum beta-lactamase (ESBL)- and carbapenemase-producing enterobacteriaceae in water sources in Lebanon. Vet. Microbiol. 217 97–103. 10.1016/j.vetmic.2018.03.007 [DOI] [PubMed] [Google Scholar]
- Diab M., Hamze M., Madec J. Y., Haenni M. (2016). High prevalence of non-ST131 CTX-M-15-producing Escherichia coli in healthy cattle in Lebanon. Microb. Drug Resist. 23 261–266. 10.1089/mdr.2016.0019 [DOI] [PubMed] [Google Scholar]
- Direkel S., Copur Cicek A., Karagoz A., Aydogan Ejder N., Oktay E., Delialioglu N., et al. (2016). Antimicrobial susceptibility and molecular characterization of multidrug-resistant Acinetobacter baumannii isolated in an university hospital. Mikrobiyol. Bul. 50 522–534. 10.5578/mb.34158 [DOI] [PubMed] [Google Scholar]
- Dogonchi A. A., Ghaemi E. A., Ardebili A., Yazdansetad S., Pournajaf A. (2018). Metallo-beta-lactamase-mediated resistance among clinical carbapenem-resistant Pseudomonas aeruginosa isolates in northern Iran: a potential threat to clinical therapeutics. Ci Ji Yi Xue Za Zhi 30 90–96. 10.4103/tcmj.tcmj_101_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortet L., Poirel L., Al Yaqoubi F., Nordmann P. (2012). NDM-1, OXA-48 and OXA-181 carbapenemase-producing enterobacteriaceae in Sultanate of Oman. Clin. Microbiol. Infect. 18 E144–E148. 10.1111/j.1469-0691.2012.03796.x [DOI] [PubMed] [Google Scholar]
- Drozdinsky G., Ben-Zvi H., Kushnir S., Leibovici L., Yahav D. (2018). Colistin exposure as a risk factor for infections caused by inherently colistin resistant enterobacteriaceae-a case-control study. Clin. Microbiol. Infect. 24 896–899. 10.1016/j.cmi.2017.11.022 [DOI] [PubMed] [Google Scholar]
- Durmaz S., Percin D., Ercal B. D. (2015). Molecular epidemiology of quinolon resistant strains of extended spectrum beta-lactamase producing Escherichia coli. Pak. J. Med. Sci. 31 1067–1072. 10.12669/pjms.315.8186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ece G., Tunc E., Otlu B., Aslan D., Ece C. (2018). Detection of blaOXA-48 and clonal relationship in carbapenem resistant K. pneumoniae isolates at a tertiary care center in western Turkey. J. Infect. Public Health 11 640–642. 10.1016/j.jiph.2018.04.003 [DOI] [PubMed] [Google Scholar]
- Economou V., Gousia P. (2015). Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect. Drug Resist. 8 49–61. 10.2147/IDR.S55778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bannah A. M. S., Nawar N. N., Hassan R. M. M., Salem S. T. B. (2018). Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii in a tertiary care hospital in Egypt: clonal spread of blaOXA-23. Microb. Drug Resist. 24 269–277. 10.1089/mdr.2017.0057 [DOI] [PubMed] [Google Scholar]
- Elabd F. M., Al-Ayed M. S., Asaad A. M., Alsareii S. A., Qureshi M. A., Musa H. A. (2015). Molecular characterization of oxacillinases among carbapenem-resistant Acinetobacter baumannii nosocomial isolates in a Saudi hospital. J. Infect. Public Health 8 242–247. 10.1016/j.jiph.2014.10.002 [DOI] [PubMed] [Google Scholar]
- El-Ageery S. M., Al-Hazmi S. S. (2014). Microbiological and molecular detection of VIM-1 metallo beta lactamase-producing Acinetobacter baumannii. Eur. Rev. Med. Pharmacol. Sci. 18 965–970. [PubMed] [Google Scholar]
- Elaldi N., Gozel M. G., Kolayli F., Engin A., Celik C., Bakici M. Z., et al. (2013). Community-acquired CTX-M-15-type ESBL-producing Escherichia coli meningitis: a case report and literature review. J. Infect. Dev. Ctries 7 424–431. 10.3855/jidc.2820 [DOI] [PubMed] [Google Scholar]
- El-Badawy M. F., Tawakol W. M., Maghrabi I. A., Mansy M. S., Shohayeb M. M., Ashour M. S. (2017). Iodometric and molecular detection of ESBL production among clinical isolates of E. coli fingerprinted by ERIC-PCR: the first Egyptian report declares the emergence of E. coli O25b-ST131clone harboring blaGES. Microb. Drug Resist. 23 703–717. 10.1089/mdr.2016.0181 [DOI] [PubMed] [Google Scholar]
- El-Herte R. I., Araj G. F., Matar G. M., Baroud M., Kanafani Z. A., Kanj S. S. (2012). Detection of carbapenem-resistant Escherichia coli and Klebsiella pneumoniae producing NDM-1 in Lebanon. J. Infect. Dev. Ctries 6 457–461. 10.3855/jidc.2340 [DOI] [PubMed] [Google Scholar]
- ElMahallawy H. A., Zafer M. M., Amin M. A., Ragab M. M., Al-Agamy M. H. (2018). Spread of carbapenem resistant enterobacteriaceae at tertiary care cancer hospital in Egypt. Infect. Dis. 50 560–564. 10.1080/23744235.2018.1428824 [DOI] [PubMed] [Google Scholar]
- El-Mahdy T. S. (2014). Identification of a novel metallo-beta-lactamase VIM-28 located within unusual arrangement of class 1 integron structure in Pseudomonas aeruginosa isolates from Egypt. Jpn. J. Infect. Dis. 67 382–384. 10.7883/yoken.67.382 [DOI] [PubMed] [Google Scholar]
- El-Mahdy T. S., Al-Agamy M. H., Al-Qahtani A. A., Shibl A. M. (2017). Detection of blaOXA-23-like and blaNDM-1 in Acinetobacter baumannii from the eastern region, Saudi Arabia. Microb. Drug Resist. 23 115–121. 10.1089/mdr.2015.0304 [DOI] [PubMed] [Google Scholar]
- Elnahriry S. S., Khalifa H. O., Soliman A. M., Ahmed A. M., Hussein A. M., Shimamoto T. (2016). Emergence of plasmid-mediated colistin resistance gene mcr-1 in a clinical Escherichia coli isolate from Egypt. Antimicrob. Agents Chemother. 60 3249–3250. 10.1128/aac.00269-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shazly D. A., Nasef S. A., Mahmoud F. F., Jonas D. (2017). Expanded spectrum beta-lactamase producing Escherichia coli isolated from chickens with colibacillosis in Egypt. Poult. Sci. 96 2375–2384. 10.3382/ps/pew493 [DOI] [PubMed] [Google Scholar]
- Eltai N. O., Abdfarag E. A., Al-Romaihi H., Wehedy E., Mahmoud M. H., Alawad O. K., et al. (2018a). Antibiotic resistance profile of commensal Escherichia coli isolated from broiler chickens in Qatar. J. Food Prot. 81 302–307. 10.4315/0362-028X.JFP-17-191 [DOI] [PubMed] [Google Scholar]
- Eltai N. O., Al Thani A. A., Al-Ansari K., Deshmukh A. S., Wehedy E., Al-Hadidi S. H., et al. (2018b). Molecular characterization of extended spectrum beta -lactamases enterobacteriaceae causing lower urinary tract infection among pediatric population. Antimicrob. Resist. Infect. Control 7:90. 10.1186/s13756-018-0381-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltai N. O., Yassine H. M., Al Thani A. A., Abu Madi M. A., Ismail A., Ibrahim E., et al. (2018c). Prevalence of antibiotic resistant Escherichia coli isolates from fecal samples of food handlers in Qatar. Antimicrob. Resist. Infect. Control 7:78. 10.1186/s13756-018-0369-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami A., Bazargani A., Mohammadi A. A., Zardosht M., Seyed Jafari S. M. (2015). Detection of blaPER-1 & blaOxa10 among imipenem resistant isolates of Pseudomonas aeruginosa isolated from burn patients hospitalized in Shiraz burn hospital. Iran J. Microbiol. 7 7–11. [PMC free article] [PubMed] [Google Scholar]
- Er H., Altindis M., Asik G., Demir C. (2015). Molecular epidemiology of beta-lactamases in ceftazidime-resistant Pseudomonas aeruginosa isolates. Mikrobiyol. Bul. 49 156–165. 10.5578/mb.8901 [DOI] [PubMed] [Google Scholar]
- Erac B., Hosgor-Limoncu M., Ermertcan S., Tasli H., Aydemir S. (2013). Prevalence of blaPER-1 and integrons in ceftazidime-resistant gram-negative bacteria at a university hospital in Turkey. Jpn. J. Infect. Dis. 66 146–148. 10.7883/yoken.66.146 [DOI] [PubMed] [Google Scholar]
- Erdem I., Kara Ali R., Ardic E., Elbasan Omar S., Mutlu R., Topkaya A. E. (2018). Community-acquired lower urinary tract infections: etiology, antimicrobial resistance, and treatment results in female patients. J. Glob. Infect. Dis. 10 129–132. 10.4103/jgid.jgid_86_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergin A., Hascelik G., Eser O. K. (2013). Molecular characterization of oxacillinases and genotyping of invasive Acinetobacter baumannii isolates using repetitive extragenic palindromic sequence-based polymerase chain reaction in ankara between 2004 and 2010. Scand. J. Infect. Dis. 45 26–31. 10.3109/00365548.2012.708782 [DOI] [PubMed] [Google Scholar]
- Fam N. S., Defasque S., Bert F., Leflon-Guibout V., El-Ray A., El-Ghannam M., et al. (2015). Faecal carriage of extended-spectrum beta-lactamase (ESBL)-producing enterobacteria in liver disease patients from two hospitals in Egypt and france: a comparative epidemiological study. Epidemiol. Infect. 143 1247–1255. 10.1017/S0950268814001812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farshadzadeh Z., Khosravi A. D., Alavi S. M., Parhizgari N., Hoveizavi H. (2014). Spread of extended-spectrum beta-lactamase genes of blaOXA-10, blaPER-1 and blaCTX-M in Pseudomonas aeruginosa strains isolated from burn patients. Burns 40 1575–1580. 10.1016/j.burns.2014.02.008 [DOI] [PubMed] [Google Scholar]
- Fazeli H., Norouzi-Barough M., Ahadi A. M., Shokri D., Solgi H. (2015). Detection of new delhi metallo-beta-lactamase-1 (NDM-1) in carbapenem- resistant Klebsiella pneumoniae isolated from a university hospital in Iran. Hippokratia 19 205–209. [PMC free article] [PubMed] [Google Scholar]
- Fazeli N., Momtaz H. (2014). Virulence gene profiles of multidrug-resistant Pseudomonas aeruginosa isolated from Iranian hospital infections. Iran Red. Crescent Med. J. 16 e15722. 10.5812/ircmj.15722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feizabadi M. M., Delfani S., Raji N., Majnooni A., Aligholi M., Shahcheraghi F., et al. (2010). Distribution of bla(TEM), bla(SHV), bla(CTX-M) genes among clinical isolates of Klebsiella pneumoniae at Labbafinejad hospital, Tehran, Iran. Microb. Drug Resist. 16 49–53. 10.1089/mdr.2009.0096 [DOI] [PubMed] [Google Scholar]
- Firoozeh F., Aghaseyed-Hosseini M., Zibaei M., Piroozmand A. (2016). Detection of blaKPC and blaGES carbapenemase genes in Klebsiella pneumoniae isolated from hospitalized patients in Kashan, Iran. Recent Pat. Antiinfect. Drug Discov. 11 183–188. 10.2174/1574891X11666160813192556 [DOI] [PubMed] [Google Scholar]
- Firoozeh F., Mahluji Z., Shams E., Khorshidi A., Zibaei M. (2017). New delhi metallo-beta-lactamase-1-producing Klebsiella pneumoniae isolates in hospitalized patients in Kashan, Iran. Iran J. Microbiol. 9 283–287. [PMC free article] [PubMed] [Google Scholar]
- Forde B. M., Zowawi H. M., Harris P. N. A., Roberts L., Ibrahim E., Shaikh N., et al. (2018). Discovery of mcr-1-mediated colistin resistance in a highly virulent Escherichia coli lineage. mSphere 3 e486–18. 10.1128/mSphere.00486-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouad M., Attia A. S., Tawakkol W. M., Hashem A. M. (2013). Emergence of carbapenem-resistant Acinetobacter baumannii harboring the OXA-23 carbapenemase in intensive care units of Egyptian hospitals. Int. J. Infect. Dis. 17 e1252–e1254. 10.1016/j.ijid.2013.07.012 [DOI] [PubMed] [Google Scholar]
- Fouda R., Soliman M. S., ElAnany M. G., Abadeer M., Soliman G. (2016). Prevalence and risk factors of MRSA, ESBL and MDR bacterial colonization upon admission to an Egyptian medical ICU. J. Infect. Dev. Ctries 10 329–336. 10.3855/jidc.6798 [DOI] [PubMed] [Google Scholar]
- Fratamico P. M., DebRoy C., Liu Y., Needleman D. S., Baranzoni G. M., Feng P. (2016). Advances in molecular serotyping and subtyping of Escherichia coli. Front. Microbiol. 7:644. 10.3389/fmicb.2016.00644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann R., Raveh D., Zartzer E., Rudensky B., Broide E., Attias D., et al. (2009). Prospective evaluation of colonization with extended-spectrum beta-lactamase (ESBL)-producing enterobacteriaceae among patients at hospital admission and of subsequent colonization with ESBL-producing enterobacteriaceae among patients during hospitalization. Infect. Control Hosp. Epidemiol. 30 534–542. 10.1086/597505 [DOI] [PubMed] [Google Scholar]
- Galal L., Abdel Aziz N. A., Hassan W. M. (2018). Defining the relationship between phenotypic and genotypic resistance profiles of multidrug-resistant Enterobacterial clinical isolates. Adv. Exp. Med. Biol. 10.1007/5584_2018_208 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Gamal D., Fernandez-Martinez M., Salem D., El-Defrawy I., Montes L. A., Ocampo-Sosa A. A., et al. (2016). Carbapenem-resistant Klebsiella pneumoniae isolates from Egypt containing blaNDM-1 on IncR plasmids and its association with rmtF. Int. J. Infect. Dis. 43 17–20. 10.1016/j.ijid.2015.12.003 [DOI] [PubMed] [Google Scholar]
- Ganjo A. R., Maghdid D. M., Mansoor I. Y., Kok D. J., Severin J. A., Verbrugh H. A., et al. (2016). OXA-carbapenemases present in clinical Acinetobacter baumannii-calcoaceticus complex isolates from patients in Kurdistan region, Iraq. Microb. Drug Resist. 22 627–637. 10.1089/mdr.2015.0060 [DOI] [PubMed] [Google Scholar]
- Garbati M. A., Bin Abdulhak A., Baba K., Sakkijha H. (2013). Infection due to colistin-resistant Enterobacteriacae in critically-ill patients. J. Infect. Dev. Ctries 7 713–719. 10.3855/jidc.2851 [DOI] [PubMed] [Google Scholar]
- Ghaffarian F., Hedayati M., Sedigh Ebrahim-Saraie H., Atrkar Roushan Z., Mojtahedi A. (2018). Molecular epidemiology of ESBL-producing Klebsiella pneumoniae isolates in intensive care units of a tertiary care hospital, north of Iran. Cell. Mol. Biol. 64 75–79. [PubMed] [Google Scholar]
- Ghaith D. M., Zafer M. M., Al-Agamy M. H., Alyamani E. J., Booq R. Y., Almoazzamy O. (2017). The emergence of a novel sequence type of MDR Acinetobacter baumannii from the intensive care unit of an Egyptian tertiary care hospital. Ann. Clin. Microbiol. Antimicrob. 16:34. 10.1186/s12941-017-0208-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharout-Sait A., Alsharapy S. A., Brasme L., Touati A., Kermas R., Bakour S., et al. (2014). enterobacteriaceae isolates carrying the new delhi metallo-beta-lactamase gene in Yemen. J. Med. Microbiol. 63(Pt 10), 1316–1323. 10.1099/jmm.0.073767-0 [DOI] [PubMed] [Google Scholar]
- Ghazawi A., Sonnevend A., Bonnin R. A., Poirel L., Nordmann P., Hashmey R., et al. (2012). NDM-2 carbapenemase-producing Acinetobacter baumannii in the United Arab Emirates. Clin. Microbiol. Infect. 18 E34–E36. 10.1111/j.1469-0691.2011.03726.x [DOI] [PubMed] [Google Scholar]
- Gholami S., Tabatabaei M., Sohrabi N. (2017). Comparison of biofilm formation and antibiotic resistance pattern of Pseudomonas aeruginosa in human and environmental isolates. Microb. Pathog. 109 94–98. 10.1016/j.micpath.2017.05.004 [DOI] [PubMed] [Google Scholar]
- Gholipour A., Soleimani N., Shokri D., Mobasherizadeh S., Kardi M., Baradaran A. (2014). Phenotypic and molecular characterization of extended-spectrum beta-lactamase produced by Escherichia coli, and Klebsiella pneumoniae isolates in an educational hospital. Jundishapur J. Microbiol. 7:e11758. 10.5812/jjm.11758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghotaslou R., Sadeghi M. R., Akhi M. T., Hasani A., Asgharzadeh M. (2018). Prevalence and antimicrobial susceptibility patterns of ESBL, AmpC and carbapenemase-producing enterobactericeae isolated from hospitalized patients in Azerbaijan, Iran. Iran J. Pharm. Res. 17(Suppl.), 79–88. [PMC free article] [PubMed] [Google Scholar]
- Giannouli M., Tomasone F., Agodi A., Vahaboglu H., Daoud Z., Triassi M., et al. (2009). Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii strains in intensive care units of multiple mediterranean hospitals. J. Antimicrob. Chemother. 63 828–830. 10.1093/jac/dkp032 [DOI] [PubMed] [Google Scholar]
- Goli H. R., Nahaei M. R., Ahangarzadeh Rezaee M., Hasani A., Samadi Kafil H., Aghazadeh M. (2016). Emergence of colistin resistant Pseudomonas aeruginosa at Tabriz hospitals, Iran. Iran J. Microbiol. 8 62–69. [PMC free article] [PubMed] [Google Scholar]
- Goren M. G., Carmeli Y., Schwaber M. J., Chmelnitsky I., Schechner V., Navon-Venezia S. (2010a). Transfer of carbapenem-resistant plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in patient. Emerg. Infect. Dis. 16 1014–1017. 10.3201/eid1606.091671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren M. G., Navon-Venezia S., Chmelnitsky I., Carmeli Y. (2010b). Carbapenem-resistant KPC-2-producing Escherichia coli in a Tel aviv medical center, 2005 to 2008. Antimicrob. Agents Chemother. 54 2687–2691. 10.1128/AAC.01359-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgec S., Kuzucu C., Otlu B., Yetkin F., Ersoy Y. (2015). Investigation of beta-lactamase genes and clonal relationship among the extended-spectrum beta-lactamase producing nosocomial Escherichia coli isolates. Mikrobiyol. Bul. 49 15–25. 10.5578/mb.8437 [DOI] [PubMed] [Google Scholar]
- Guenther S., Ewers C., Wieler L. H. (2011). Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front. Microbiol. 2:246. 10.3389/fmicb.2011.00246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra B., Fischer J., Helmuth R. (2014). An emerging public health problem: acquired carbapenemase-producing microorganisms are present in food-producing animals, their environment, companion animals and wild birds. Vet. Microbiol. 171 290–297. 10.1016/j.vetmic.2014.02.001 [DOI] [PubMed] [Google Scholar]
- Gulmez D., Woodford N., Palepou M. F., Mushtaq S., Metan G., Yakupogullari Y., et al. (2008). Carbapenem-resistant Escherichia coli and Klebsiella pneumoniae isolates from Turkey with OXA-48-like carbapenemases and outer membrane protein loss. Int. J. Antimicrob. Agents 31 523–526. 10.1016/j.ijantimicag.2008.01.017 [DOI] [PubMed] [Google Scholar]
- Guven Gokmen T., Nagiyev T., Meral M., Onlen C., Heydari F., Koksal F. (2016). NDM-1 and rmtC-producing Klebsiella pneumoniae isolates in Turkey. Jundishapur J. Microbiol. 9:e33990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haciseyitoglu D., Dokutan A., Abulaila A., Erdem F., Cag Y., Ozer S., et al. (2017). The first Enterobacter cloacae co-producing NDM and OXA-48 carbapenemases and interhospital spread of OXA-48 and NDM-producing Klebsiella pneumoniae in Turkey. Clin. Lab. 63 1213–1222. 10.7754/Clin.Lab.2017.170120 [DOI] [PubMed] [Google Scholar]
- Haeili M., Javani A., Moradi J., Jafari Z., Feizabadi M. M., Babaei E. (2017). MgrB alterations mediate colistin resistance in Klebsiella pneumoniae isolates from Iran. Front. Microbiol. 8:2470. 10.3389/fmicb.2017.02470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeili M., Kafshdouz M., Feizabadi M. M. (2018). Molecular mechanisms of colistin resistance among pandrug-resistant isolates of Acinetobacter baumannii with high case-fatality rate in intensive care unit patients. Microb. Drug Resist. 24 1271–1276. 10.1089/mdr.2017.0397 [DOI] [PubMed] [Google Scholar]
- Hajjar Soudeiha M., Dahdouh E., Daoud Z., Sarkis D. K. (2018). Phenotypic and genotypic detection of beta-lactamases in Acinetobacter spp. isolates recovered from Lebanese patients over a 1-year period. J. Glob. Antimicrob. Resist. 12 107–112. 10.1016/j.jgar.2017.09.016 [DOI] [PubMed] [Google Scholar]
- Hakim A. S., Omara S. T., Syame S. M., Fouad E. A. (2017). Serotyping, antibiotic susceptibility, and virulence genes screening of Escherichia coli isolates obtained from diarrheic buffalo calves in Egyptian farms. Vet. World 10 769–773. 10.14202/vetworld.2017.769-773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdy Mohammed el S., Elsadek Fakhr A., Mohammed El Sayed H., Al Johery S. A., Abdel Ghani Hassanein W. (2016). Spread of TEM, VIM, SHV, and CTX-M beta-lactamases in imipenem-resistant gram-negative bacilli isolated from Egyptian hospitals. Int. J. Microbiol. 2016:8382605. 10.1155/2016/8382605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad A. M., Shimamoto T. (2011). Asymptomatic intramammary infection with multidrug-resistant gram-negative bacteria in a research dairy farm: incidence and genetic basis of resistance. J. Vet. Med. Sci. 73 1089–1092. 10.1292/jvms.10-0361 [DOI] [PubMed] [Google Scholar]
- Hammoudi D., Moubareck C. A., Hakime N., Houmani M., Barakat A., Najjar Z., et al. (2015a). Spread of imipenem-resistant Acinetobacter baumannii co-expressing OXA-23 and GES-11 carbapenemases in Lebanon. Int. J. Infect. Dis 36 56–61. 10.1016/j.ijid.2015.05.015 [DOI] [PubMed] [Google Scholar]
- Hammoudi D., Moubareck C. A., Kanso A., Nordmann P., Sarkis D. K. (2015b). SURVEILLANCE of CARBAPENEM NON-SUSCEPTIBLE GRAM NEGATIVE STRAINS and CHARACTERIZATION of CARBAPENEMASES of CLASSES A, B and D in a LEBANESE HOSPITAL. J. Med. Liban. 63 66–73. 10.12816/0012553 [DOI] [PubMed] [Google Scholar]
- Hammoudi Halat D., Moubareck C. A., Sarkis D. K. (2017). Heterogeneity of carbapenem resistance mechanisms among gram-negative pathogens in Lebanon: results of the first cross-sectional countrywide study. Microb. Drug Resist. 23 733–743. 10.1089/mdr.2016.0077 [DOI] [PubMed] [Google Scholar]
- Hashemizadeh Z., Kalantar-Neyestanaki D., Mansouri S. (2018). Clonal relationships, antimicrobial susceptibilities, and molecular characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolates from urinary tract infections and fecal samples in southeast Iran. Rev. Soc. Bras. Med. Trop. 51 44–51. 10.1590/0037-8682-0080-2017 [DOI] [PubMed] [Google Scholar]
- Hassan H., Abdalhamid B. (2014). Molecular characterization of extended-spectrum beta-lactamase producing enterobacteriaceae in a Saudi Arabian tertiary hospital. J. Infect. Dev. Ctries 8 282–288. 10.3855/jidc.3809 [DOI] [PubMed] [Google Scholar]
- Hassan W. M., Hashim A., Domany R. (2012). Plasmid mediated quinolone resistance determinants qnr, aac(6’)-ib-cr, and qep in ESBL-producing Escherichia coli clinical isolates from Egypt. Indian J. Med. Microbiol. 30 442–447. 10.4103/0255-0857.103766 [DOI] [PubMed] [Google Scholar]
- Hayajneh W. A., Hajj A., Hulliel F., Sarkis D. K., Irani-Hakimeh N., Kazan L., et al. (2015). Susceptibility trends and molecular characterization of gram-negative bacilli associated with urinary tract and intra-abdominal infections in Jordan and Lebanon: SMART 2011-2013. Int. J. Infect. Dis 35 56–61. 10.1016/j.ijid.2015.04.011 [DOI] [PubMed] [Google Scholar]
- Heidary M., Momtaz H., Madani M. (2014). Characterization of diarrheagenic antimicrobial resistant Escherichia coli isolated from pediatric patients in Tehran, Iran. Iran Red Crescent Med. J. 16:e12329. 10.5812/ircmj.12329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmy M. M., Wasfi R. (2014). Phenotypic and molecular characterization of plasmid mediated AmpC beta-lactamases among Escherichia coli, Klebsiella spp., and Proteus mirabilis isolated from urinary tract infections in Egyptian hospitals. Biomed. Res. Int. 2014:171548. 10.1155/2014/171548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmy O. M., Kashef M. T. (2017). Different phenotypic and molecular mechanisms associated with multidrug resistance in gram-negative clinical isolates from Egypt. Infect. Drug Resist. 10 479–498. 10.2147/IDR.S147192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijazi S. M., Fawzi M. A., Ali F. M., Abd El Galil K. H. (2016). Multidrug-resistant ESBL-producing enterobacteriaceae and associated risk factors in community infants in Lebanon. J. Infect. Dev. Ctries 10 947–955. 10.3855/jidc.7593 [DOI] [PubMed] [Google Scholar]
- Hocquet D., Vogne C., El Garch F., Vejux A., Gotoh N., Lee A., et al. (2003). MexXY-OprM efflux pump is necessary for an adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 47 1371–1375. 10.1128/aac.47.4.1371-1375.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojabri Z., Mirmohammadkhani M., Kamali F., Ghassemi K., Taghavipour S., Pajand O. (2017). Molecular epidemiology of Escherichia coli sequence type 131 and its H30/H30-rx subclones recovered from extra-intestinal infections: first report of OXA-48 producing ST131 clone from Iran. Eur. J. Clin. Microbiol. Infect. Dis. 36 1859–1866. 10.1007/s10096-017-3021-9 [DOI] [PubMed] [Google Scholar]
- Hosbul T., Ozyurt M., Karademir F., Suleymanoglu S., Haznedaroglu T. (2012). Investigation of a nosocomial outbreak caused by ESBL positive Klebsiella pneumoniae in neonatal intensive care unit by AP-PCR. Mikrobiyol. Bul. 46 101–105. [PubMed] [Google Scholar]
- Hosseinzadeh Z., Sedigh Ebrahim-Saraie H., Sarvari J., Mardaneh J., Dehghani B., Rokni-Hosseini S. M. H., et al. (2018). Emerge of blaNDM-1 and blaOXA-48-like harboring carbapenem-resistant Klebsiella pneumoniae isolates from hospitalized patients in southwestern Iran. J. Chin. Med. Assoc. 81 536–540. 10.1016/j.jcma.2017.08.015 [DOI] [PubMed] [Google Scholar]
- Hu L., Zhong Q., Tu J., Xu Y., Qin Z., Parsons C., et al. (2013). Emergence of blaNDM-1 among Klebsiella pneumoniae ST15 and novel ST1031 clinical isolates in China. Diagn. Microbiol. Infect. Dis. 75 373–376. 10.1016/j.diagmicrobio.2013.01.006 [DOI] [PubMed] [Google Scholar]
- Huang X. Z., Frye J. G., Chahine M. A., Glenn L. M., Ake J. A., Su W., et al. (2012). Characteristics of plasmids in multi-drug-resistant enterobacteriaceae isolated during prospective surveillance of a newly opened hospital in Iraq. PLoS One 7:e40360. 10.1371/journal.pone.0040360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers P. M., Graat E. A., Haenen A. P., van Santen M. G., van Essen-Zandbergen A., Mevius D. J., et al. (2014). Extended-spectrum and AmpC beta-lactamase-producing Escherichia coli in broilers and people living and/or working on broiler farms: prevalence, risk factors and molecular characteristics. J. Antimicrob. Chemother. 69 2669–2675. 10.1093/jac/dku178 [DOI] [PubMed] [Google Scholar]
- Hussein N. H. (2018). Emergence of NDM-1 among carbapenem-resistant Klebsiella pneumoniae in Iraqi hospitals. Acta Microbiol. Immunol. Hung. 65 211–227. 10.1556/030.64.2017.026 [DOI] [PubMed] [Google Scholar]
- Iraz M., Duzgun A. O., Cicek A. C., Bonnin R. A., Ceylan A., Saral A., et al. (2014). Characterization of novel VIM carbapenemase, VIM-38, and first detection of GES-5 carbapenem-hydrolyzing beta-lactamases in Pseudomonas aeruginosa in Turkey. Diagn. Microbiol. Infect. Dis. 78 292–294. 10.1016/j.diagmicrobio.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Iraz M., Ozad Duzgun A., Sandalli C., Doymaz M. Z., Akkoyunlu Y., Saral A., et al. (2015). Distribution of beta-lactamase genes among carbapenem-resistant Klebsiella pneumoniae strains isolated from patients in Turkey. Ann. Lab. Med. 35 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail S. J., Mahmoud S. S. (2018). First detection of new delhi metallo-beta-lactamases variants (NDM-1, NDM-2) among Pseudomonas aeruginosa isolated from Iraqi hospitals. Iran J. Microbiol. 10 98–103. [PMC free article] [PubMed] [Google Scholar]
- Izdebski R., Baraniak A., Fiett J., Adler A., Kazma M., Salomon J., et al. (2013). Clonal structure, extended-spectrum beta-lactamases, and acquired AmpC-type cephalosporinases of Escherichia coli populations colonizing patients in rehabilitation centers in four countries. Antimicrob. Agents Chemother. 57 309–316. 10.1128/AAC.01656-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari Z., Harati A. A., Haeili M., Kardan-Yamchi J., Jafari S., Jabalameli F., et al. (2018). Molecular epidemiology and drug resistance pattern of carbapenem-resistant Klebsiella pneumoniae isolates from Iran. Microb. Drug Resist. 25 336–343. 10.1089/mdr.2017.0404 [DOI] [PubMed] [Google Scholar]
- Jallad M. A., Naoufal R., Irani J., Azar E. (2015). Extended spectrum beta-lactamase carriage state among elderly nursing home residents in beirut. ScientificWorldJournal 2015:987580. 10.1155/2015/987580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal W., Rotimi V. O., Albert M. J., Khodakhast F., Nordmann P., Poirel L. (2013). High prevalence of VIM-4 and NDM-1 metallo-beta-lactamase among carbapenem-resistant enterobacteriaceae. J. Med. Microbiol. 62(Pt 8), 1239–1244. 10.1099/jmm.0.059915-0 [DOI] [PubMed] [Google Scholar]
- Jamal W., Salama M., Dehrab N., Al Hashem G., Shahin M., Rotimi V. O. (2009). Role of tigecycline in the control of a carbapenem-resistant Acinetobacter baumannii outbreak in an intensive care unit. J. Hosp. Infect. 72 234–242. 10.1016/j.jhin.2009.03.023 [DOI] [PubMed] [Google Scholar]
- Jamal W. Y., Albert M. J., Khodakhast F., Poirel L., Rotimi V. O. (2015). Emergence of new sequence type OXA-48 carbapenemase-producing enterobacteriaceae in Kuwait. Microb. Drug Resist 21 329–334. 10.1089/mdr.2014.0123 [DOI] [PubMed] [Google Scholar]
- Jamal W. Y., Albert M. J., Rotimi V. O. (2016). High prevalence of new delhi metallo-beta-lactamase-1 (NDM-1) producers among carbapenem-resistant enterobacteriaceae in Kuwait. PLoS One 11:e0152638. 10.1371/journal.pone.0152638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaase M., Nordmann P., Wichelhaus T. A., Gatermann S. G., Bonnin R. A., Poirel L. (2011). NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J. Antimicrob. Chemother. 66 1260–1262. 10.1093/jac/dkr135 [DOI] [PubMed] [Google Scholar]
- Kamel N. A., El-Tayeb W. N., El-Ansary M. R., Mansour M. T., Aboshanab K. M. (2018). Phenotypic screening and molecular characterization of carbapenemase-producing gram-negative bacilli recovered from febrile neutropenic pediatric cancer patients in Egypt. PLoS One 13:e0202119. 10.1371/journal.pone.0202119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanj S., Tayyar R., Shehab M., El-Hafi B., Rasheed S., Kissoyan K., et al. (2018). Increase of blaOXA-23-like in Acinetobacter baumannii at a tertiary care center in Lebanon between 2007 and 2013. J. Infect. Dev. Ctries 12 228–234. 10.3855/jidc.9642 [DOI] [PubMed] [Google Scholar]
- Karaaslan A., Soysal A., Altinkanat Gelmez G., Kepenekli Kadayifci E., Soyletir G., Bakir M. (2016). Molecular characterization and risk factors for carbapenem-resistant gram-negative bacilli colonization in children: emergence of NDM-producing Acinetobacter baumannii in a newborn intensive care unit in Turkey. J. Hosp. Infect. 92 67–72. 10.1016/j.jhin.2015.09.011 [DOI] [PubMed] [Google Scholar]
- Karabay O., Altindis M., Koroglu M., Karatuna O., Aydemir O. A., Erdem A. F. (2016). The carbapenem-resistant enterobacteriaceae threat is growing: NDM-1 epidemic at a training hospital in Turkey. Ann. Clin. Microbiol. Antimicrob. 15:6. 10.1186/s12941-016-0118-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karfunkel D., Carmeli Y., Chmelnitsky I., Kotlovsky T., Navon-Venezia S. (2013). The emergence and dissemination of CTX-M-producing Escherichia coli sequence type 131 causing community-onset bacteremia in Israel. Eur. J. Clin. Microbiol. Infect. Dis. 32 513–521. 10.1007/s10096-012-1765-9 [DOI] [PubMed] [Google Scholar]
- Kazeminezhad B., Bostanmanesh, Rad A., Gharib A., Zahedifard S. (2017). blaVIM and blaIMP genes detection in isolates of carbapenem resistant P. aeruginosa of hospitalized patients in two hospitals in Iran. Iran. J. Pathol. 12 392–396. [PMC free article] [PubMed] [Google Scholar]
- Keskin H., Tekeli A., Dolapci I., Ocal D. (2014). Molecular characterization of beta-lactamase-associated resistance in Acinetobacter baumannii strains isolated from clinical samples. Mikrobiyol. Bul. 48 365–376. 10.5578/mb.7796 [DOI] [PubMed] [Google Scholar]
- Khalaf N. G., Eletreby M. M., Hanson N. D. (2009). Characterization of CTX-M ESBLs in Enterobacter cloacae, Escherichia coli and Klebsiella pneumoniae clinical isolates from cairo, Egypt. BMC Infect. Dis 9:84. 10.1186/1471-2334-9-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifa H. O., Ahmed A. M., Oreiby A. F., Eid A. M., Shimamoto T. (2016). Characterisation of the plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli isolated from animals in Egypt. Int. J. Antimicrob. Agents 47 413–414. 10.1016/j.ijantimicag.2016.02.011 [DOI] [PubMed] [Google Scholar]
- Khalifa H. O., Soliman A. M., Ahmed A. M., Shimamoto T., Hara T., Ikeda M., et al. (2017). High carbapenem resistance in clinical gram-negative pathogens isolated in Egypt. Microb. Drug Resist. 23 838–844. 10.1089/mdr.2015.0339 [DOI] [PubMed] [Google Scholar]
- Khalifeh M. M., Moore N. D., Salameh P. R. (2017). Self-medication misuse in the middle east: a systematic literature review. Pharmacol. Res. Perspect. 5:e00323. 10.1002/prp2.323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil M. A. F., Elgaml A., El-Mowafy M. (2017). Emergence of multidrug-resistant new delhi metallo-beta-lactamase-1-producing Klebsiella pneumoniae in Egypt. Microb. Drug Resist. 23 480–487. 10.1089/mdr.2016.0003 [DOI] [PubMed] [Google Scholar]
- Khorvash F., Yazdani M., Shabani S., Soudi A. (2017). Pseudomonas aeruginosa-producing metallo-beta-lactamases (VIM, IMP, SME, and AIM) in the clinical isolates of intensive care units, a university hospital in isfahan, Iran. Adv. Biomed. Res. 6:147. 10.4103/2277-9175.219412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiaei S., Moradi M., Hosseini Nave H., Hashemizadeh Z., Taati-Moghadam M., Kalantar-Neyestanaki D. (2018). Emergence of co-existence of blaNDM with rmtC and qnrB genes in clinical carbapenem-resistant Klebsiella pneumoniae isolates in burning center from southeast of Iran. Folia Microbiol. 64 55–62. 10.1007/s12223-018-0630-3 [DOI] [PubMed] [Google Scholar]
- Kilic A., Aktas Z., Bedir O., Gumral R., Bulut Y., Stratton C., et al. (2011). Identification and characterization of OXA-48 producing, carbapenem-resistant enterobacteriaceae isolates in Turkey. Ann. Clin. Lab. Sci. 41 161–166. [PubMed] [Google Scholar]
- Krahn T., Wibberg D., Maus I., Winkler A., Bontron S., Sczyrba A., et al. (2016). Intraspecies transfer of the chromosomal Acinetobacter baumannii blaNDM-1 carbapenemase gene. Antimicrob. Agents Chemother. 60 3032–3040. 10.1128/AAC.00124-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulah C., Mooij M. J., Comert F., Aktas E., Celebi G., Ozlu N., et al. (2010). Characterisation of carbapenem-resistant Acinetobacter baumannii outbreak strains producing OXA-58 in Turkey. Int. J. Antimicrob. Agents 36 114–118. 10.1016/j.ijantimicag.2010.03.017 [DOI] [PubMed] [Google Scholar]
- Kurekci C., Aydin M., Yipel M., Katouli M., Gundogdu A. (2017). Characterization of extended spectrum beta-lactamase (ESBL)-producing Escherichia coli in asi (orontes) river in Turkey. J. Water Health 15 788–798. 10.2166/wh.2017.257 [DOI] [PubMed] [Google Scholar]
- Kuskucu M. A., Karakullukcu A., Ailiken M., Otlu B., Mete B., Aygun G. (2016). Investigation of carbapenem resistance and the first identification of Klebsiella pneumoniae carbapenemase (KPC) enzyme among Escherichia coli isolates in Turkey: a prospective study. Travel Med. Infect. Dis. 14 572–576. 10.1016/j.tmaid.2016.11.006 [DOI] [PubMed] [Google Scholar]
- Kusradze I., Diene S. M., Goderdzishvili M., Rolain J. M. (2011). Molecular detection of OXA carbapenemase genes in multidrug-resistant Acinetobacter baumannii isolates from Iraq and Georgia. Int. J. Antimicrob. Agents 38 164–168. 10.1016/j.ijantimicag.2011.03.021 [DOI] [PubMed] [Google Scholar]
- Kutlu H. H., Us E., Tekeli A. (2018). Investigation of carbapenemase genes and molecular epidemiology of enterobacteriaceae strains isolated between 2010-2014 in a university hospital. Mikrobiyol. Bul. 52 1–12. 10.5578/mb.66156 [DOI] [PubMed] [Google Scholar]
- Lalaoui R., Bakour S., Livnat K., Assous M. V., Diene S. M., Rolain J. M. (2019). Spread of carbapenem and colistin-resistant Klebsiella pneumoniae ST512 clinical isolates in israel: a cause for vigilance. Microb. Drug Resist. 25 63–71. 10.1089/mdr.2018.0014 [DOI] [PubMed] [Google Scholar]
- Lari A. R., Azimi L., Soroush S., Taherikalani M. (2015). Low prevalence of metallo-beta-lactamase in Pseudomonas aeruginosa isolated from a tertiary burn care center in Tehran. Int. J. Immunopathol. Pharmacol. 28 384–389. 10.1177/0394632015578343 [DOI] [PubMed] [Google Scholar]
- Laube H., Friese A., von Salviati C., Guerra B., Rosler U. (2014). Transmission of ESBL/AmpC-producing Escherichia coli from broiler chicken farms to surrounding areas. Vet. Microbiol. 172 519–527. 10.1016/j.vetmic.2014.06.008 [DOI] [PubMed] [Google Scholar]
- Lazarovitch T., Amity K., Coyle J. R., Ackerman B., Tal-Jasper R., Ofer-Friedman H., et al. (2015). The complex epidemiology of carbapenem-resistant enterobacter infections: a multicenter descriptive analysis. Infect. Control Hosp. Epidemiol. 36 1283–1291. 10.1017/ice.2015.186 [DOI] [PubMed] [Google Scholar]
- Leangapichart T., Dia N. M., Olaitan A. O., Gautret P., Brouqui P., Rolain J. M. (2016a). Acquisition of extended-spectrum beta-lactamases by Escherichia coli and Klebsiella pneumoniae in gut microbiota of pilgrims during the hajj pilgrimage of 2013. Antimicrob. Agents Chemother. 60 3222–3226. 10.1128/AAC.02396-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leangapichart T., Gautret P., Brouqui P., Memish Z. A., Raoult D., Rolain J. M. (2016b). Acquisition of mcr-1 plasmid-mediated colistin resistance in Escherichia coli and Klebsiella pneumoniae during hajj (2013) and (2014). Antimicrob. Agents Chemother. 60 6998–6999. 10.1128/aac.01486-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leangapichart T., Gautret P., Griffiths K., Belhouchat K., Memish Z., Raoult D., et al. (2016c). Acquisition of a high diversity of bacteria during the hajj pilgrimage, including Acinetobacter baumannii with blaOXA-72 and Escherichia coli with blaNDM-5 carbapenemase genes. Antimicrob. Agents Chemother. 60 5942–5948. 10.1128/AAC.00669-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leangapichart T., Tissot-Dupont H., Raoult D., Memish Z. A., Rolain J. M., Gautret P. (2017). Risk factors for acquisition of CTX-M genes in pilgrims during hajj 2013 and 2014. J. Antimicrob. Chemother. 72 2627–2635. 10.1093/jac/dkx155 [DOI] [PubMed] [Google Scholar]
- Leavitt A., Carmeli Y., Chmelnitsky I., Goren M. G., Ofek I., Navon-Venezia S. (2010a). Molecular epidemiology, sequence types, and plasmid analyses of KPC-producing Klebsiella pneumoniae strains in Israel. Antimicrob. Agents Chemother. 54 3002–3006. 10.1128/AAC.01818-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt A., Chmelnitsky I., Ofek I., Carmeli Y., Navon-Venezia S. (2010b). Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug-resistant epidemic Klebsiella pneumoniae strain. J. Antimicrob. Chemother. 65 243–248. 10.1093/jac/dkp417 [DOI] [PubMed] [Google Scholar]
- Lee M. Y., Ko K. S., Kang C. I., Chung D. R., Peck K. R., Song J. H. (2011). High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: diverse clones and clonal dissemination. Int. J. Antimicrob. Agents 38 160–163. 10.1016/j.ijantimicag.2011.03.020 [DOI] [PubMed] [Google Scholar]
- Lerner A., Adler A., Abu-Hanna J., Cohen Percia S., Kazma Matalon M., Carmeli Y. (2015). Spread of KPC-producing carbapenem-resistant enterobacteriaceae: the importance of super-spreaders and rectal KPC concentration. Clin. Microbiol. Infect. 21 470.e1–7. 10.1016/j.cmi.2014.12.015 [DOI] [PubMed] [Google Scholar]
- Lerner A., Solter E., Rachi E., Adler A., Rechnitzer H., Miron D., et al. (2016). Detection and characterization of carbapenemase-producing enterobacteriaceae in wounded Syrian patients admitted to hospitals in northern Israel. Eur. J. Clin. Microbiol. Infect. Dis. 35 149–154. 10.1007/s10096-015-2520-9 [DOI] [PubMed] [Google Scholar]
- Lima Barbieri N., Nielsen D. W., Wannemuehler Y., Cavender T., Hussein A., Yan S. G., et al. (2017). Mcr-1 identified in avian pathogenic Escherichia coli (APEC). PLoS One 12:e0172997. 10.1371/journal.pone.0172997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes B. S., Al-Agamy M. H., Ismail M. A., Shibl A. M., Al-Qahtani A. A., Al-Ahdal M. N., et al. (2015). The transferability of blaOXA-23 gene in multidrug-resistant Acinetobacter baumannii isolates from Saudi Arabia and Egypt. Int. J. Med. Microbiol. 305 581–588. 10.1016/j.ijmm.2015.07.007 [DOI] [PubMed] [Google Scholar]
- Magwenzi M. T., Gudza-Mugabe M., Mujuru H. A., Dangarembizi-Bwakura M., Robertson V., Aiken A. M. (2017). Carriage of antibiotic-resistant enterobacteriaceae in hospitalised children in tertiary hospitals in Harare, Zimbabwe. Antimicrob. Resist. Infect. Control 6:10. 10.1186/s13756-016-0155-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdian S., Sadeghifard N., Pakzad I., Ghanbari F., Soroush S., Azimi L., et al. (2015). Acinetobacter baumannii clonal lineages I and II harboring different carbapenem-hydrolyzing-beta-lactamase genes are widespread among hospitalized burn patients in Tehran. J. Infect. Public Health 8 533–542. 10.1016/j.jiph.2015.04.030 [DOI] [PubMed] [Google Scholar]
- Maleki N., Tahanasab Z., Mobasherizadeh S., Rezaei A., Faghri J. (2018). Prevalence of CTX-M and TEM beta-lactamases in Klebsiella pneumoniae isolates from patients with urinary tract infection, al-zahra hospital, Isfahan, Iran. Adv. Biomed. Res. 7:10. 10.4103/abr.abr_17_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkocoglu G., Aktas E., Bayraktar B., Otlu B., Bulut M. E. (2017). VIM-1, VIM-2, and GES-5 carbapenemases among Pseudomonas aeruginosa isolates at a tertiary hospital in Istanbul, Turkey. Microb. Drug Resist. 23 328–334. 10.1089/mdr.2016.0012 [DOI] [PubMed] [Google Scholar]
- Mansouri S., Kalantar D., Asadollahi P., Taherikalani M., Emaneini M. (2012). Characterization of Klebsiella pneumoniae strains producing extended spectrum beta-lactamases and AMPC type beta-lactamases isolated from hospitalized patients in Kerman, Iran. Roum. Arch. Microbiol. Immunol. 71 81–86. [PubMed] [Google Scholar]
- Mantilla-Calderon D., Jumat M. R., Wang T., Ganesan P., Al-Jassim N., Hong P. Y. (2016). Isolation and characterization of NDM-positive Escherichia coli from municipal wastewater in jeddah, Saudi Arabia. Antimicrob. Agents Chemother. 60 5223–5231. 10.1128/AAC.00236-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manyi-Loh C., Mamphweli S., Meyer E., Okoh A. (2018). Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules 23:E795. 10.3390/molecules23040795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovska R., Schneider I., Ivanova D., Keuleyan E., Stoeva T., Sredkova M., et al. (2012). High prevalence of CTX-M-15-producing O25b-ST131 Escherichia coli clone in Bulgarian hospitals. Microb. Drug Resist. 18 390–395. 10.1089/mdr.2011.0186 [DOI] [PubMed] [Google Scholar]
- Maspi H., Mahmoodzadeh Hosseini H., Amin M., Imani Fooladi A. A. (2016). High prevalence of extensively drug-resistant and metallo beta-lactamase-producing clinical Acinetobacter baumannii in Iran. Microb. Pathog. 98 155–159. 10.1016/j.micpath.2016.07.011 [DOI] [PubMed] [Google Scholar]
- Matar G. M., Gay E., Cooksey R. C., Elliott J. A., Heneine W. M., Uwaydah M. M., et al. (1992). Identification of an epidemic strain of Acinetobacter baumannii using electrophoretic typing methods. Eur. J. Epidemiol. 8 9–14. 10.1007/bf02427385 [DOI] [PubMed] [Google Scholar]
- Mehrgan H., Rahbar M., Arab-Halvaii Z. (2010). High prevalence of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in a tertiary care hospital in Tehran, Iran. J. Infect. Dev. Ctries 4 132–138. 10.3855/jidc.488 [DOI] [PubMed] [Google Scholar]
- Meir-Gruber L., Manor Y., Gefen-Halevi S., Hindiyeh M. Y., Mileguir F., Azar R., et al. (2016). Population screening using sewage reveals pan-resistant bacteria in hospital and community samples. PLoS One 11:e0164873. 10.1371/journal.pone.0164873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z. A., Assiri A., Almasri M., Roshdy H., Hathout H., Kaase M., et al. (2015). Molecular characterization of carbapenemase production among gram-negative bacteria in Saudi Arabia. Microb. Drug Resist. 21 307–314. 10.1089/mdr.2014.0121 [DOI] [PubMed] [Google Scholar]
- Memish Z. A., Shibl A. M., Kambal A. M., Ohaly Y. A., Ishaq A., Livermore D. M. (2012). Antimicrobial resistance among non-fermenting gram-negative bacteria in Saudi Arabia. J. Antimicrob. Chemother. 67 1701–1705. 10.1093/jac/dks091 [DOI] [PubMed] [Google Scholar]
- Merino I., Shaw E., Horcajada J. P., Cercenado E., Mirelis B., Pallares M. A., et al. (2016). CTX-M-15-H30Rx-ST131 subclone is one of the main causes of healthcare-associated ESBL-producing Escherichia coli bacteraemia of urinary origin in Spain. J. Antimicrob. Chemother. 71 2125–2130. 10.1093/jac/dkw133 [DOI] [PubMed] [Google Scholar]
- Metan G., Ilbay A., Eser O. K., Unal S., Zarakolu P. (2017). A silent epidemic of colistin- and carbapenem-resistant enterobacteriaceae at a turkish university hospital. Infect. Control Hosp. Epidemiol. 38 254–257. 10.1017/ice.2016.255 [DOI] [PubMed] [Google Scholar]
- Metan G., Sariguzel F., Sumerkan B., Reijden T., Dijkshoorn L. (2013). Clonal diversity and high prevalence of OXA-58 among Acinetobacter baumannii isolates from blood cultures in a tertiary care centre in Turkey. Infect. Genet. Evol. 14 92–97. 10.1016/j.meegid.2012.11.003 [DOI] [PubMed] [Google Scholar]
- Metwally L., Gomaa N., Attallah M., Kamel N. (2013). High prevalence of Klebsiella pneumoniae carbapenemase-mediated resistance in K. pneumoniae isolates from Egypt. East Mediterr. Health J. 19 947–952. 10.26719/2013.19.11.947 [DOI] [PubMed] [Google Scholar]
- Mirbagheri S. Z., Meshkat Z., Naderinasab M., Rostami S., Nabavinia M. S., Rahmati M. (2015). Study on imipenem resistance and prevalence of blaVIM1 and blaVIM2 metallo-beta lactamases among clinical isolates of Pseudomonas aeruginosa from Mashhad, northeast of Iran. Iran J. Microbiol. 7 72–78. [PMC free article] [PubMed] [Google Scholar]
- Mirhoseini S. H., Nikaeen M., Shamsizadeh Z., Khanahmad H. (2016). Hospital air: a potential route for transmission of infections caused by beta-lactam-resistant bacteria. Am. J. Infect. Control 44 898–904. 10.1016/j.ajic.2016.01.041 [DOI] [PubMed] [Google Scholar]
- Mirnejad R., Heidary M., Bahramian A., Goudarzi M., Pournajaf A. (2018). Evaluation of polymyxin B susceptibility profile and detection of drug resistance genes among Acinetobacter baumannii clinical isolates in Tehran, Iran during 2015-2016. Mediterr. J. Hematol. Infect. Dis. 10:e2018044. 10.4084/MJHID.2018.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsalehian A., Feizabadi M., Nakhjavani F. A., Jabalameli F., Goli H., Kalantari N. (2010). Detection of VEB-1, OXA-10 and PER-1 genotypes in extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patients. Burns 36 70–74. 10.1016/j.burns.2009.01.015 [DOI] [PubMed] [Google Scholar]
- Moawad A. A., Hotzel H., Neubauer H., Ehricht R., Monecke S., Tomaso H., et al. (2018). Antimicrobial resistance in enterobacteriaceae from healthy broilers in Egypt: emergence of colistin-resistant and extended-spectrum beta-lactamase-producing Escherichia coli. Gut Pathog. 10:39. 10.1186/s13099-018-0266-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadam M. N., Motamedifar M., Sarvari J., Sedigh E. S., Mousavi S. M., Moghadam F. N. (2016). Emergence of multidrug resistance and metallo-beta-lactamase producing Acinetobacter baumannii isolated from patients in shiraz, Iran. Ann. Med. Health Sci. Res. 6 162–167. 10.4103/2141-9248.183946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadampour M., Rezaei A., Faghri J. (2018a). The emergence of blaOXA-48 and blaNDM among ESBL-producing Klebsiella pneumoniae in clinical isolates of a tertiary hospital in Iran. Acta Microbiol. Immunol. Hung. 65 335–344. 10.1556/030.65.2018.034 [DOI] [PubMed] [Google Scholar]
- Moghadampour M., Salari-Jazi A., Faghri J. (2018b). High rate of carbapenem-resistant Klebsiella pneumoniae detected from hospital equipments in Iran. Acta Microbiol. Immunol. Hung. 65 529–538. 10.1556/030.65.2018.039 [DOI] [PubMed] [Google Scholar]
- Moghanni M., Ghazvini K., Farsiani H., Namaei M. H., Derakhshan M., Yousefi M., et al. (2018). High prevalence of sequence type 131 isolates producing CTX-M-15 among extended-spectrum beta-lactamase-producing Escherichia coli strains in northeast Iran. J. Glob. Antimicrob. Resist. 15 74–78. 10.1016/j.jgar.2018.05.016 [DOI] [PubMed] [Google Scholar]
- Mohajeri P., Farahani A., Mehrabzadeh R. S. (2017). Molecular characterization of multidrug resistant strains of Acinetobacter baumannii isolated from intensive care units in west of Iran. J. Clin. Diagn. Res. 11 DC20–DC22. 10.7860/JCDR/2017/21156.9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsin J., Pal T., Petersen J. E., Darwish D., Ghazawi A., Ashraf T., et al. (2018). Plasmid-mediated colistin resistance gene mcr-1 in an Escherichia coli ST10 bloodstream isolate in the Sultanate of Oman. Microb. Drug Resist. 24 278–282. 10.1089/mdr.2017.0131 [DOI] [PubMed] [Google Scholar]
- Mombini S., Rezatofighi S. E., Kiyani L., Motamedi H. (2019). Diversity and metallo-beta-lactamase-producing genes in Pseudomonas aeruginosa strains isolated from filters of household water treatment systems. J. Environ. Manage. 231 413–418. 10.1016/j.jenvman.2018.10.068 [DOI] [PubMed] [Google Scholar]
- Moosavian M., Rahimzadeh M. (2015). Molecular detection of metallo-beta-lactamase genes, bla IMP-1, bla VIM-2 and bla SPM-1 in imipenem resistant Pseudomonas aeruginosa isolated from clinical specimens in teaching hospitals of ahvaz, Iran. Iran J. Microbiol. 7 2–6. [PMC free article] [PubMed] [Google Scholar]
- Moubareck C. A., Mouftah S. F., Pal T., Ghazawi A., Halat D. H., Nabi A., et al. (2018). Clonal emergence of Klebsiella pneumoniae ST14 co-producing OXA-48-type and NDM carbapenemases with high rate of colistin resistance in Dubai, United Arab Emirates. Int. J. Antimicrob. Agents 52 90–95. 10.1016/j.ijantimicag.2018.03.003 [DOI] [PubMed] [Google Scholar]
- Mugnier P. D., Bindayna K. M., Poirel L., Nordmann P. (2009). Diversity of plasmid-mediated carbapenem-hydrolysing oxacillinases among carbapenem-resistant Acinetobacter baumannii isolates from Kingdom of Bahrain. J. Antimicrob. Chemother. 63 1071–1073. 10.1093/jac/dkp052 [DOI] [PubMed] [Google Scholar]
- Mutti M., Sonnevend A., Pal T., Junttila S., Ekker H., Galik B., et al. (2018). Complete genome sequence of Escherichia coli 81009, a representative of the sequence type 131 C1-M27 clade with a multidrug-resistant phenotype. Genome Announc. 6 e56–18. 10.1128/genomeA.00056-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairoukh Y. R., Mahafzah A. M., Irshaid A., Shehabi A. A. (2018). Molecular characterization of multidrug resistant uropathogenic E. coli isolates from Jordanian patients. Open Microbiol. J. 12 1–7. 10.2174/1874285801812010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namaei M. H., Yousefi M., Ziaee M., Salehabadi A., Ghannadkafi M., Amini E., et al. (2017). First report of prevalence of CTX-M-15-producing Escherichia coli O25b/ST131 from Iran. Microb. Drug Resist. 23 879–884. 10.1089/mdr.2016.0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasr Z., Paravattil B., Wilby K. J. (2017). The impact of antimicrobial stewardship strategies on antibiotic appropriateness and prescribing behaviours in selected countries in the middle east: a systematic review. East. Mediterr. Health J. 23 430–440. 10.26719/2017.23.6.430 [DOI] [PubMed] [Google Scholar]
- Navon-Venezia S., Leavitt A., Schwaber M. J., Rasheed J. K., Srinivasan A., Patel J. B., et al. (2009). First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob. Agents Chemother. 53 818–820. 10.1128/AAC.00987-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazer L. H., Rihani S., Hawari F. I., Le J. (2015). High-dose colistin for microbiologically documented serious respiratory infections associated with carbapenem-resistant Acinetobacter baummannii in critically ill cancer patients: a retrospective cohort study. Infect. Dis. 47 755–760. 10.3109/23744235.2015.1055586 [DOI] [PubMed] [Google Scholar]
- Nazik H., Aydin S., Albayrak R., Bilgi E. A., Yildiz I., Kuvat N., et al. (2014). Detection and spread of oxa-48-producing Klebsiella oxytoca isolates in Istanbul, Turkey. Southeast Asian J. Trop. Med. Public Health 45 123–129. [PubMed] [Google Scholar]
- Nazik H., Bektore B., Ongen B., Ozyurt M., Baylan O., Haznedaroglu T. (2011). Co-expression of plasmid-mediated quinolone resistance-qnrA1 and blaVEB-1 gene in a Providencia stuartii strain. New Microbiol. 34 225–228. [PubMed] [Google Scholar]
- Nazik H., Ilktac M., Ongen B. (2009). Prevalence of qnrA, qnrB, qnrS and aac(6’)-ib-cr (in qnr-positive isolates) among ESBL-positive and/or ciprofloxacin-resistant isolates in Turkey. J. Chemother. 21 219–221. 10.1179/joc.2009.21.2.219 [DOI] [PubMed] [Google Scholar]
- Nazik H., Ongen B., Ilktac M., Aydin S., Kuvat N., Sahin A., et al. (2012). Carbapenem resistance due to bla(OXA-48) among ESBL-producing Escherichia coli and Klebsiella pneumoniae isolates in a univesity hospital, Turkey. Southeast Asian J. Trop. Med. Public Health 43 1178–1185. [PubMed] [Google Scholar]
- Netikul T., Sidjabat H. E., Paterson D. L., Kamolvit W., Tantisiriwat W., Steen J. A., et al. (2014). Characterization of an IncN2-type blaNDM-(1)-carrying plasmid in Escherichia coli ST131 and Klebsiella pneumoniae ST11 and ST15 isolates in Thailand. J. Antimicrob. Chemother. 69 3161–3163. 10.1093/jac/dku275 [DOI] [PubMed] [Google Scholar]
- Neyestanaki D. K., Mirsalehian A., Rezagholizadeh F., Jabalameli F., Taherikalani M., Emaneini M. (2014). Determination of extended spectrum beta-lactamases, metallo-beta-lactamases and AmpC-beta-lactamases among carbapenem resistant Pseudomonas aeruginosa isolated from burn patients. Burns 40 1556–1561. 10.1016/j.burns.2014.02.010 [DOI] [PubMed] [Google Scholar]
- Nobari S., Shahcheraghi F., Rahmati Ghezelgeh F., Valizadeh B. (2014). Molecular characterization of carbapenem-resistant strains of Klebsiella pneumoniae isolated from Iranian patients: first identification of blaKPC gene in Iran. Microb. Drug Resist. 20 285–293. 10.1089/mdr.2013.0074 [DOI] [PubMed] [Google Scholar]
- Obeidat N., Jawdat F., Al-Bakri A. G., Shehabi A. A. (2014). Major biologic characteristics of Acinetobacter baumannii isolates from hospital environmental and patients’ respiratory tract sources. Am. J. Infect. Control 42 401–404. 10.1016/j.ajic.2013.10.010 [DOI] [PubMed] [Google Scholar]
- Okdah L., Leangapichart T., Hadjadj L., Olaitan A. O., Al-Bayssari C., Rizk R., et al. (2017). First report of colistin-resistant Klebsiella pneumoniae clinical isolates in Lebanon. J. Glob. Antimicrob. Resist. 9 15–16. 10.1016/j.jgar.2017.02.004 [DOI] [PubMed] [Google Scholar]
- Olaitan A. O., Dia N. M., Gautret P., Benkouiten S., Belhouchat K., Drali T., et al. (2015). Acquisition of extended-spectrum cephalosporin- and colistin-resistant Salmonella enterica subsp. enterica serotype newport by pilgrims during hajj. Int. J. Antimicrob. Agents 45 600–604. 10.1016/j.ijantimicag.2015.01.010 [DOI] [PubMed] [Google Scholar]
- Olaitan A. O., Chabou S., Okdah L., Morand S., Rolain J. M. (2016a). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16 147–149. [DOI] [PubMed] [Google Scholar]
- Olaitan A. O., Diene S. M., Assous M. V., Rolain J. M. (2016b). Genomic plasticity of multidrug-resistant NDM-1 positive clinical isolate of Providencia rettgeri. Genome Biol. Evol. 8 723–728. 10.1093/gbe/evv195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaitan A. O., Diene S. M., Kempf M., Berrazeg M., Bakour S., Gupta S. K., et al. (2014a). Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in lao PDR, thailand, Israel, nigeria and france owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int. J. Antimicrob. Agents 44 500–507. 10.1016/j.ijantimicag.2014.07.020 [DOI] [PubMed] [Google Scholar]
- Olaitan A. O., Morand S., Rolain J. M. (2014b). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643 10.3389/fmicb.2014.00643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ombarak R. A., Hinenoya A., Elbagory A. M., Yamasaki S. (2018). Prevalence and molecular characterization of antimicrobial resistance in Escherichia coli isolated from raw milk and raw milk cheese in Egypt. J. Food Prot. 81 226–232. 10.4315/0362-028X.JFP-17-277 [DOI] [PubMed] [Google Scholar]
- Osman K. M., Kappell A. D., Elhadidy M., ElMougy F., El-Ghany W. A. A., Orabi A., et al. (2018). Poultry hatcheries as potential reservoirs for antimicrobial-resistant Escherichia coli: a risk to public health and food safety. Sci. Rep. 8:5859. 10.1038/s41598-018-23962-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostad Asadolah-Malayeri H., Hakemi-Vala M., Davari K. (2016). Role of aders and OXA23 genes among imipenem resistant Acinetobacter baumannii isolates from two hospitals of Tehran, Iran. Iran J. Pathol. 11 345–353. [PMC free article] [PubMed] [Google Scholar]
- Otlu B., Yakupogullari Y., Gursoy N. C., Duman Y., Bayindir Y., Tekerekoglu M. S., et al. (2018). Co-production of OXA-48 and NDM-1 carbapenemases in Providencia rettgeri: the first report. Mikrobiyol. Bul. 52 300–307. 10.5578/mb.67153 [DOI] [PubMed] [Google Scholar]
- Pal T., Ghazawi A., Darwish D., Villa L., Carattoli A., Hashmey R., et al. (2017). Characterization of NDM-7 carbapenemase-producing Escherichia coli isolates in the arabian peninsula. Microb. Drug Resist. 23 871–878. 10.1089/mdr.2016.0216 [DOI] [PubMed] [Google Scholar]
- Peerayeh S. N., Rostami E., Siadat S. D., Derakhshan S. (2014). High rate of aminoglycoside resistance in CTX-M-15 producing Klebsiella pneumoniae isolates in Tehran, Iran. Lab. Med. 45 231–237. 10.1309/LMDQQW246NYAHHAD [DOI] [PubMed] [Google Scholar]
- Pehlivanoglu F., Turutoglu H., Ozturk D. (2016). CTX-M-15-type extended-spectrum beta-lactamase-producing Escherichia coli as causative agent of bovine mastitis. Foodborne Pathog. Dis. 13 477–482. 10.1089/fpd.2015.2114 [DOI] [PubMed] [Google Scholar]
- Peirano G., van der Bij A. K., Freeman J. L., Poirel L., Nordmann P., Costello M., et al. (2014). Characteristics of Escherichia coli sequence type 131 isolates that produce extended-spectrum beta-lactamases: global distribution of the H30-rx sublineage. Antimicrob. Agents Chemother. 58 3762–3767. 10.1128/AAC.02428-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretz A., Koiefman A., Dinisman E., Brodsky D., Labay K. (2014a). Do wheelchairs spread pathogenic bacteria within hospital walls? World J. Microbiol. Biotechnol. 30 385–387. 10.1007/s11274-013-1458-2 [DOI] [PubMed] [Google Scholar]
- Peretz A., Labay K., Zonis Z., Glikman D. (2014b). Disengagement does not apply to bacteria: a high carriage rate of antibiotic-resistant pathogens among syrian civilians treated in Israeli hospitals. Clin. Infect. Dis. 59 753–754. 10.1093/cid/ciu374 [DOI] [PubMed] [Google Scholar]
- Peymani A., Farivar T. N., Sanikhani R., Javadi A., Najafipour R. (2014). Emergence of TEM, SHV, and CTX-M-extended spectrum beta-lactamases and class 1 integron among enterobacter cloacae isolates collected from hospitals of Tehran and Qazvin, Iran. Microb. Drug Resist. 20 424–430. 10.1089/mdr.2013.0191 [DOI] [PubMed] [Google Scholar]
- Peymani A., Nahaei M. R., Farajnia S., Hasani A., Mirsalehian A., Sohrabi N., et al. (2011). High prevalence of metallo-beta-lactamase-producing Acinetobacter baumannii in a teaching hospital in Tabriz, Iran. Jpn. J. Infect. Dis. 64 69–71. [PubMed] [Google Scholar]
- Pfeffer I., Zemel M., Kariv Y., Mishali H., Adler A., Braun T., et al. (2016). Prevalence and risk factors for carriage of extended-spectrum beta-lactamase-producing enterobacteriaceae among patients prior to bowel surgery. Diagn. Microbiol. Infect. Dis. 85 377–380. 10.1016/j.diagmicrobio.2016.04.002 [DOI] [PubMed] [Google Scholar]
- Poirel L., Al Maskari Z., Al Rashdi F., Bernabeu S., Nordmann P. (2011a). NDM-1-producing Klebsiella pneumoniae isolated in the Sultanate of Oman. J. Antimicrob. Chemother. 66 304–306. 10.1093/jac/dkq428 [DOI] [PubMed] [Google Scholar]
- Poirel L., Benouda A., Hays C., Nordmann P. (2011b). Emergence of NDM-1-producing Klebsiella pneumoniae in morocco. J. Antimicrob. Chemother. 66 2781–2783. 10.1093/jac/dkr384 [DOI] [PubMed] [Google Scholar]
- Poirel L., Yilmaz M., Istanbullu A., Arslan F., Mert A., Bernabeu S., et al. (2014). Spread of NDM-1-producing enterobacteriaceae in a neonatal intensive care unit in Istanbul, Turkey. Antimicrob. Agents Chemother. 58 2929–2933. 10.1128/AAC.02047-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomba C., Rantala M., Greko C., Baptiste K. E., Catry B., van Duijkeren E., et al. (2017). Public health risk of antimicrobial resistance transfer from companion animals. J. Antimicrob. Chemother. 72 957–968. 10.1093/jac/dkw481 [DOI] [PubMed] [Google Scholar]
- Potron A., Nordmann P., Lafeuille E., Al Maskari Z., Al Rashdi F., Poirel L. (2011). Characterization of OXA-181, a carbapenem-hydrolyzing class D beta-lactamase from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 55 4896–4899. 10.1128/AAC.00481-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourabbas B., Firouzi R., Pouladfar G. (2016). Characterization of carbapenem-resistant Acinetobacter calcoaceticus-baumannii complex isolates from nosocomial bloodstream infections in southern Iran. J. Med. Microbiol. 65 235–239. 10.1099/jmm.0.000219 [DOI] [PubMed] [Google Scholar]
- Pournajaf A., Razavi S., Irajian G., Ardebili A., Erfani Y., Solgi S., et al. (2018). Integron types, antimicrobial resistance genes, virulence gene profile, alginate production and biofilm formation in Iranian cystic fibrosis Pseudomonas aeruginosa isolates. Infez. Med. 26 226–236. [PubMed] [Google Scholar]
- Principe L., Mauri C., Conte V., Pini B., Giani T., Rossolini G. M., et al. (2017). First report of NDM-1-producing Klebsiella pneumoniae imported from Africa to Italy: evidence of the need for continuous surveillance. J. Glob. Antimicrob. Resist. 8 23–27. 10.1016/j.jgar.2016.10.004 [DOI] [PubMed] [Google Scholar]
- Qabajah M., Awwad E., Ashhab Y. (2014). Molecular characterisation of Escherichia coli from dead broiler chickens with signs of colibacillosis and ready-to-market chicken meat in the west bank. Br. Poult. Sci. 55 442–451. 10.1080/00071668.2014.935998 [DOI] [PubMed] [Google Scholar]
- Rafei R., Dabboussi F., Hamze M., Eveillard M., Lemarie C., Gaultier M. P., et al. (2014a). Molecular analysis of Acinetobacter baumannii strains isolated in Lebanon using four different typing methods. PLoS One 9:e115969. 10.1371/journal.pone.0115969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafei R., Dabboussi F., Hamze M., Eveillard M., Lemarie C., Mallat H., et al. (2014b). First report of blaNDM-1-producing Acinetobacter baumannii isolated in Lebanon from civilians wounded during the Syrian war. Int. J. Infect. Dis. 21 21–23. 10.1016/j.ijid.2014.01.004 [DOI] [PubMed] [Google Scholar]
- Rafei R., Hamze M., Pailhories H., Eveillard M., Marsollier L., Joly-Guillou M. L., et al. (2015a). Extrahuman epidemiology of Acinetobacter baumannii in Lebanon. Appl. Environ. Microbiol. 81 2359–2367. 10.1128/AEM.03824-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafei R., Pailhories H., Hamze M., Eveillard M., Mallat H., Dabboussi F., et al. (2015b). Molecular epidemiology of Acinetobacter baumannii in different hospitals in Tripoli, Lebanon using bla OXA-51-like sequence based typing. BMC Microbiol. 15:103. 10.1186/s12866-015-0441-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiee R., Eftekhar F., Tabatabaei S. A., Minaee Tehrani D. (2014). Prevalence of extended-spectrum and metallo beta-lactamase production in AmpC beta-lactamase producing Pseudomonas aeruginosa isolates from burns. Jundishapur J. Microbiol. 7:e16436. 10.5812/jjm.16436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajabnia R., Asgharpour F., Ferdosi Shahandashti E., Moulana Z. (2015). Nosocomial emerging of (VIM1) carbapenemase-producing isolates of Klebsiella pneumoniae in north of Iran. Iran J. Microbiol. 7 88–93. [PMC free article] [PubMed] [Google Scholar]
- Ramadan H. H., Jackson C. R., Taha S. A., Moawad A. A., Barrett J. B., Woodley T. A. (2018). Contribution of healthy chickens to antimicrobial-resistant Escherichia coli associated with human extraintestinal infections in Egypt. Vector Borne Zoonotic Dis. 18 408–416. 10.1089/vbz.2017.2237 [DOI] [PubMed] [Google Scholar]
- Ramadan R. A., Gebriel M. G., Kadry H. M., Mosallem A. (2018). Carbapenem-resistant Acinetobacter baumannii and Pseudomonas aeruginosa: characterization of carbapenemase genes and E-test evaluation of colistin-based combinations. Infect. Drug Resist. 11 1261–1269. 10.2147/IDR.S170233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan Dash N., Albataineh M. T., Alhourani N., Khoudeir A. M., Ghanim M., Wasim M., et al. (2018). Community-acquired urinary tract infections due to extended-spectrum beta -lactamase-producing organisms in United Arab Emirates. Travel Med. Infect. Dis. 22 46–50. 10.1016/j.tmaid.2018.01.007 [DOI] [PubMed] [Google Scholar]
- Ranjbar R., Safarpoor Dehkordi F., Sakhaei Shahreza M. H., Rahimi E. (2018). Prevalence, identification of virulence factors, O-serogroups and antibiotic resistance properties of shiga-toxin producing Escherichia coli strains isolated from raw milk and traditional dairy products. Antimicrob. Resist. Infect. Control 7:53. 10.1186/s13756-018-0345-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjbar R., Sami M. (2017). Genetic investigation of beta-lactam associated antibiotic resistance among Escherichia coli strains isolated from water sources. Open Microbiol. J. 11 203–210. 10.2174/1874285801711010203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastegar Lari A., Azimi L., Rahbar M., Fallah F., Alaghehbandan R. (2013). Phenotypic detection of Klebsiella pneumoniae carbapenemase among burns patients: first report from Iran. Burns 39 174–176. 10.1016/j.burns.2012.02.025 [DOI] [PubMed] [Google Scholar]
- Reinheimer C., Kempf V. A., Gottig S., Hogardt M., Wichelhaus T. A., O’Rourke F., et al. (2016). Multidrug-resistant organisms detected in refugee patients admitted to a university hospital, Germany June December(2015). Euro. Surveill. 21:30110. [DOI] [PubMed] [Google Scholar]
- Rezaei A., Fazeli H., Moghadampour M., Halaji M., Faghri J. (2018). Determination of antibiotic resistance pattern and prevalence of OXA-type carbapenemases among Acinetobacter baumannii clinical isolates from inpatients in Isfahan, central Iran. Infez. Med. 26 61–66. [PubMed] [Google Scholar]
- Rezai M. S., Salehifar E., Rafiei A., Langaee T., Rafati M., Shafahi K., et al. (2015). Characterization of multidrug resistant extended-spectrum beta-lactamase-producing Escherichia coli among uropathogens of pediatrics in north of Iran. Biomed. Res. Int. 2015:309478. 10.1155/2015/309478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieber H., Frontzek A., Pfeifer Y. (2017). Molecular investigation of carbapenem-resistant Acinetobacter spp. from hospitals in north rhine-westphalia, Germany. Microb. Drug Resist. 23 25–31. 10.1089/mdr.2016.0035 [DOI] [PubMed] [Google Scholar]
- Rizzo L., Manaia C., Merlin C., Schwartz T., Dagot C., Ploy M. C., et al. (2013). Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci. Total Environ. 447 345–360. 10.1016/j.scitotenv.2013.01.032 [DOI] [PubMed] [Google Scholar]
- Rodrigues C., Machado E., Ramos H., Peixe L., Novais A. (2014). Expansion of ESBL-producing Klebsiella pneumoniae in hospitalized patients: a successful story of international clones (ST15, ST147, ST336) and epidemic plasmids (IncR, IncFIIK). Int. J. Med. Microbiol. 304 1100–1108. 10.1016/j.ijmm.2014.08.003 [DOI] [PubMed] [Google Scholar]
- Rolain J. M., Loucif L., Al-Maslamani M., Elmagboul E., Al-Ansari N., Taj-Aldeen S., et al. (2016). Emergence of multidrug-resistant Acinetobacter baumannii producing OXA-23 carbapenemase in Qatar. New Microbes New Infect. 11 47–51. 10.1016/j.nmni.2016.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostami S., Farajzadeh Sheikh A., Shoja S., Farahani A., Tabatabaiefar M. A., Jolodar A., et al. (2018). Investigating of four main carbapenem-resistance mechanisms in high-level carbapenem resistant Pseudomonas aeruginosa isolated from burn patients. J. Chin. Med. Assoc. 81 127–132. 10.1016/j.jcma.2017.08.016 [DOI] [PubMed] [Google Scholar]
- Ruppe E., Woerther P. L., Barbier F. (2015). Mechanisms of antimicrobial resistance in gram-negative bacilli. Ann. Intensive Care 5 21. 10.1186/s13613-015-0061-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabra A. H., Araj G. F., Kattar M. M., Abi-Rached R. Y., Khairallah M. T., Klena J. D., et al. (2009). Molecular characterization of ESBL-producing Shigella sonnei isolates from patients with bacilliary dysentery in Lebanon. J. Infect. Dev. Ctries 3 300–305. [DOI] [PubMed] [Google Scholar]
- Saffari M., Firoozeh F., Pourbabaee M., Zibaei M. (2016). Evaluation of metallo-beta-lactamase-production and carriage of bla-VIM genes in Pseudomonas aeruginosa isolated from burn wound infections in isfahan. Arch. Trauma Res. 5:e34343. 10.5812/atr.34343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi B., Goudarzi H., Nikmanesh B., Houri H., Alavi-Moghaddam M., Ghalavand Z. (2018). Emergence and characterization of nosocomial multidrug-resistant and extensively drug-resistant Acinetobacter baumannii isolates in Tehran, Iran. J. Infect. Chemother. 24 515–523. 10.1016/j.jiac.2018.02.009 [DOI] [PubMed] [Google Scholar]
- Salimian Rizi K., Najar Peerayeh S., Bakhshi B., Rahbar M. (2015). Prevalence of the bla CTX-M-1 group and their transferability in resistant clinical isolates of Salmonella serogroups from several hospitals of Tehran. Iran J. Microbiol. 7 203–207. [PMC free article] [PubMed] [Google Scholar]
- Salloum T., Arabaghian H., Alousi S., Abboud E., Tokajian S. (2017). Genome sequencing and comparative analysis of an NDM-1-producing Klebsiella pneumoniae ST15 isolated from a refugee patient. Pathog. Glob. Health 111 166–175. 10.1080/20477724.2017.1314069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarhaddi N., Soleimanpour S., Farsiani H., Mosavat A., Dolatabadi S., Salimizand H., et al. (2017). Elevated prevalence of multidrug-resistant Acinetobacter baumannii with extensive genetic diversity in the largest burn centre of northeast Iran. J. Glob. Antimicrob. Resist. 8 60–66. 10.1016/j.jgar.2016.10.009 [DOI] [PubMed] [Google Scholar]
- Sari A. N., Bicmen M., Gulay Z. (2013). Investigation of plasmid mediated AmpC beta-lactamases among Escherichia coli and Klebsiella pneumoniae isolated from blood cultures. Mikrobiyol. Bul. 47 582–591. 10.5578/mb.6240 [DOI] [PubMed] [Google Scholar]
- Sarikhani Z., Nazari R., Nateghi Rostami M. (2017). First report of OXA-143-lactamase producing Acinetobacter baumannii in qom, Iran. Iran J. Basic Med. Sci. 20 1282–1286. 10.22038/IJBMS.2017.9490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savari M., Ekrami A., Shoja S., Bahador A. (2017). Plasmid borne carbapenem-hydrolyzing class D beta-lactamases (CHDLs) and AdeABC efflux pump conferring carbapenem-tigecycline resistance among Acinetobacter baumannii isolates harboring TnAbaRs. Microb. Pathog. 104 310–317. 10.1016/j.micpath.2017.01.045 [DOI] [PubMed] [Google Scholar]
- Schechner V., Kotlovsky T., Kazma M., Mishali H., Schwartz D., Navon-Venezia S., et al. (2013). Asymptomatic rectal carriage of blaKPC producing carbapenem-resistant enterobacteriaceae: who is prone to become clinically infected? Clin. Microbiol. Infect. 19 451–456. 10.1111/j.1469-0691.2012.03888.x [DOI] [PubMed] [Google Scholar]
- Schill F., Abdulmawjood A., Klein G., Reich F. (2017). Prevalence and characterization of extended-spectrum beta-lactamase (ESBL) and AmpC beta-lactamase producing enterobacteriaceae in fresh pork meat at processing level in Germany. Int. J. Food Microbiol. 257 58–66. 10.1016/j.ijfoodmicro.2017.06.010 [DOI] [PubMed] [Google Scholar]
- Sedighi M., Halajzadeh M., Ramazanzadeh R., Amirmozafari N., Heidary M., Pirouzi S. (2017). Molecular detection of beta-lactamase and integron genes in clinical strains of Klebsiella pneumoniae by multiplex polymerase chain reaction. Rev. Soc. Bras. Med. Trop. 50 321–328. 10.1590/0037-8682-0001-2017 [DOI] [PubMed] [Google Scholar]
- Seiffert S. N., Hilty M., Perreten V., Endimiani A. (2013). Extended-spectrum cephalosporin-resistant gram-negative organisms in livestock: an emerging problem for human health? Drug Resist. Updat. 16 22–45. 10.1016/j.drup.2012.12.001 [DOI] [PubMed] [Google Scholar]
- Sepahvand S., Doudi M., Davarpanah M. A., Bahador A., Ahmadi M. (2016). Analyzing pmrA and pmrB genes in Acinetobacter baumannii resistant to colistin in shahid rajai shiraz, Iran hospital by PCR: first report in Iran. Pak. J. Pharm. Sci. 29(4 Suppl.), 1401–1406. [PubMed] [Google Scholar]
- Serefhanoglu K., Turan H., Timurkaynak F. E., Arslan H. (2009). Bloodstream infections caused by ESBL-producing E. coli and K. pneumoniae: risk factors for multidrug-resistance. Braz. J. Infect. Dis. 13 403–407. 10.1590/s1413-86702009000600003 [DOI] [PubMed] [Google Scholar]
- Shahcheraghi F., Aslani M. M., Mahmoudi H., Karimitabar Z., Solgi H., Bahador A., et al. (2017). Molecular study of carbapenemase genes in clinical isolates of enterobacteriaceae resistant to carbapenems and determining their clonal relationship using pulsed-field gel electrophoresis. J. Med. Microbiol. 66 570–576. 10.1099/jmm.0.000467 [DOI] [PubMed] [Google Scholar]
- Shahcheraghi F., Nobari S., Rahmati Ghezelgeh F., Nasiri S., Owlia P., Nikbin V. S., et al. (2013). First report of new delhi metallo-beta-lactamase-1-producing Klebsiella pneumoniae in Iran. Microb. Drug Resist. 19 30–36. 10.1089/mdr.2012.0078 [DOI] [PubMed] [Google Scholar]
- Shahid M., Al-Mahmeed A., Murtadha M. M., Qareeballa A., Eltahir M. A., Tabbara K. S., et al. (2014). Characterization of cephalosporin-resistant clinical enterobacteriaceae for CTX-M ESBLs in Bahrain. Asian Pac. J. Trop. Med. 7S1 S212–S216. 10.1016/S1995-7645(14)60234-0 [DOI] [PubMed] [Google Scholar]
- Shakib P., Ramazanzadeh R., Taherikalani M., Nouri B. (2018). Detection of extended-spectrum beta-lactamases (ESBLs) and antibiotic susceptibility patterns in Klebsiella pneumoniae in western, Iran. Infect. Disord. Drug Targets 18 156–163. 10.2174/1871526517666170713101734 [DOI] [PubMed] [Google Scholar]
- Shamsizadeh Z., Nikaeen M., Nasr Esfahani B., Mirhoseini S. H., Hatamzadeh M., Hassanzadeh A. (2017). Detection of antibiotic resistant Acinetobacter baumannii in various hospital environments: potential sources for transmission of Acinetobacter infections. Environ. Health Prev. Med. 22:44. 10.1186/s12199-017-0653-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayan S., Bokaeian M. (2015). Detection of ESBL- and AmpC-producing E. coli isolates from urinary tract infections. Adv. Biomed. Res. 4:220. 10.4103/2277-9175.166643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibl A., Al-Agamy M., Memish Z., Senok A., Khader S. A., Assiri A. (2013). The emergence of OXA-48- and NDM-1-positive Klebsiella pneumoniae in Riyadh, Saudi Arabia. Int. J. Infect. Dis. 17 e1130–e1133. 10.1016/j.ijid.2013.06.016 [DOI] [PubMed] [Google Scholar]
- Shibl A. M., Al-Agamy M. H., Khubnani H., Senok A. C., Tawfik A. F., Livermore D. M. (2012). High prevalence of acquired quinolone-resistance genes among enterobacteriaceae from Saudi Arabia with CTX-M-15 beta-lactamase. Diagn. Microbiol. Infect. Dis 73 350–353. 10.1016/j.diagmicrobio.2012.04.005 [DOI] [PubMed] [Google Scholar]
- Shirmohammadlou N., Zeighami H., Haghi F., Kashefieh M. (2018). Resistance pattern and distribution of carbapenemase and antiseptic resistance genes among multidrug-resistant Acinetobacter baumannii isolated from intensive care unit patients. J. Med. Microbiol. 67 1467–1473. 10.1099/jmm.0.000826 [DOI] [PubMed] [Google Scholar]
- Shoja S., Ansari M., Faridi F., Azad M., Davoodian P., Javadpour S., et al. (2018). Identification of carbapenem-resistant Klebsiella pneumoniae with emphasis on new delhi metallo-beta-lactamase-1 (blaNDM-1) in bandar abbas, south of Iran. Microb. Drug Resist. 24 447–454. 10.1089/mdr.2017.0058 [DOI] [PubMed] [Google Scholar]
- Shoja S., Moosavian M., Rostami S., Abbasi F., Tabatabaiefar M. A., Peymani A. (2016). Characterization of oxacillinase and metallo-beta-lactamas genes and molecular typing of clinical isolates of Acinetobacter baumannii in ahvaz, south-west of Iran. Jundishapur J. Microbiol. 9:e32388. 10.5812/jjm.32388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoja S., Moosavian M., Rostami S., Farahani A., Peymani A., Ahmadi K., et al. (2017). Dissemination of carbapenem-resistant Acinetobacter baumannii in patients with burn injuries. J. Chin. Med. Assoc. 80 245–252. 10.1016/j.jcma.2016.10.013 [DOI] [PubMed] [Google Scholar]
- Sid Ahmed M. A., Bansal D., Acharya A., Elmi A. A., Hamid J. M., Sid Ahmed A. M., et al. (2016). Antimicrobial susceptibility and molecular epidemiology of extended-spectrum beta-lactamase-producing enterobacteriaceae from intensive care units at hamad medical corporation, Qatar. Antimicrob. Resist. Infect. Control 5:4. 10.1186/s13756-016-0103-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjolander I., Hansen F., Elmanama A., Khayyat R., Abu-Zant A., Hussein A., et al. (2014). Detection of NDM-2-producing Acinetobacter baumannii and VIM-producing Pseudomonas aeruginosa in Palestine. J. Glob. Antimicrob. Resist. 2 93–97. 10.1016/j.jgar.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Solgi H., Badmasti F., Giske C. G., Aghamohammad S., Shahcheraghi F. (2018). Molecular epidemiology of NDM-1- and OXA-48-producing Klebsiella pneumoniae in an Iranian hospital: clonal dissemination of ST11 and ST893. J. Antimicrob. Chemother. 73 1517–1524. 10.1093/jac/dky081 [DOI] [PubMed] [Google Scholar]
- Solgi H., Badmasti F., Aminzadeh Z., Giske C. G., Pourahmad M., Vaziri F., et al. (2017a). Molecular characterization of intestinal carriage of carbapenem-resistant enterobacteriaceae among inpatients at two Iranian university hospitals: first report of co-production of bla NDM-7 and bla OXA-48. Eur. J. Clin. Microbiol. Infect. Dis. 36 2127–2135. 10.1007/s10096-017-3035-3 [DOI] [PubMed] [Google Scholar]
- Solgi H., Giske C. G., Badmasti F., Aghamohammad S., Havaei S. A., Sabeti S., et al. (2017b). Emergence of carbapenem resistant Escherichia coli isolates producing blaNDM and blaOXA-48-like carried on IncA/C and IncL/M plasmids at two Iranian university hospitals. Infect. Genet. Evol. 55 318–323. 10.1016/j.meegid.2017.10.003 [DOI] [PubMed] [Google Scholar]
- Soltani B., Heidari H., Ebrahim-Saraie H. S., Hadi N., Mardaneh J., Motamedifar M. (2018). Molecular characteristics of multiple and extensive drug-resistant Acinetobacter baumannii isolates obtained from hospitalized patients in southwestern Iran. Infez. Med. 26 67–76. [PubMed] [Google Scholar]
- Somily A. M., Habib H. A., Absar M. M., Arshad M. Z., Manneh K., Al Subaie S. S., et al. (2014). ESBL-producing Escherichia coli and Klebsiella pneumoniae at a tertiary care hospital in Saudi Arabia. J. Infect. Dev. Ctries 8 1129–1136. 10.3855/jidc.4292 [DOI] [PubMed] [Google Scholar]
- Sonnevend A., Al Baloushi A., Ghazawi A., Hashmey R., Girgis S., Hamadeh M. B., et al. (2013). Emergence and spread of NDM-1 producer enterobacteriaceae with contribution of IncX3 plasmids in the United Arab Emirates. J. Med. Microbiol. 62(Pt 7), 1044–1050. 10.1099/jmm.0.059014-0 [DOI] [PubMed] [Google Scholar]
- Sonnevend A., Ghazawi A., Alqahtani M., Shibl A., Jamal W., Hashmey R., et al. (2016). Plasmid-mediated colistin resistance in Escherichia coli from the Arabian peninsula. Int. J. Infect. Dis. 50 85–90. 10.1016/j.ijid.2016.07.007 [DOI] [PubMed] [Google Scholar]
- Sonnevend A., Ghazawi A., Darwish D., AlDeesi Z., Kadhum A. F., Pal T. (2015a). Characterization of KPC-type carbapenemase-producing Klebsiella pneumoniae strains isolated in the Arabian peninsula. J. Antimicrob. Chemother. 70 1592–1593. [DOI] [PubMed] [Google Scholar]
- Sonnevend A., Ghazawi A. A., Hashmey R., Jamal W., Rotimi V. O., Shibl A. M., et al. (2015b). Characterization of carbapenem-resistant enterobacteriaceae with high rate of autochthonous transmission in the Arabian peninsula. PLoS One 10:e0131372. 10.1371/journal.pone.0131372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnevend A., Ghazawi A., Hashmey R., Haidermota A., Girgis S., Alfaresi M., et al. (2017a). Multihospital occurrence of pan-resistant Klebsiella pneumoniae sequence type 147 with an ISEcp1-directed blaOXA-181 insertion in the mgrB gene in the United Arab Emirates. Antimicrob. Agents Chemother. 61 e418–17. 10.1128/AAC.00418-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnevend A., Yahfoufi N., Ghazawi A., Jamal W., Rotimi V., Pal T. (2017b). Contribution of horizontal gene transfer to the emergence of VIM-4 carbapenemase producer enterobacteriaceae in Kuwait. Infect. Drug Resist. 10 469–478. 10.2147/IDR.S149321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoesser N., Giess A., Batty E. M., Sheppard A. E., Walker A. S., Wilson D. J., et al. (2014). Genome sequencing of an extended series of NDM-producing Klebsiella pneumoniae isolates from neonatal infections in a Nepali hospital characterizes the extent of community- versus hospital-associated transmission in an endemic setting. Antimicrob. Agents Chemother. 58 7347–7357. 10.1128/AAC.03900-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Tsuchiya M., Shimada K., Nga T. T. T., Thu L. T. A., Phu T. T., et al. (2017). Dissemination of carbapenem-resistant Klebsiella pneumoniae clinical isolates with various combinations of carbapenemases (KPC-2, NDM-1, NDM-4, and OXA-48) and 16S rRNA methylases (RmtB and RmtC) in Vietnam. BMC Infect. Dis 17:467. 10.1186/s12879-017-2570-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanir Basaranoglu S., Ozsurekci Y., Aykac K., Karadag Oncel E., Bicakcigil A., Sancak B., et al. (2017). A comparison of blood stream infections with extended spectrum beta-lactamase-producing and non-producing Klebsiella pneumoniae in pediatric patients. Ital. J. Pediatr. 43:79. 10.1186/s13052-017-0398-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasbent E. F., Ozdemir M. (2015). The presence of OXA type carbapenemases in Pseudomonas strains: first report from Turkey. Mikrobiyol. Bul. 49 26–34. 10.5578/mb.8563 [DOI] [PubMed] [Google Scholar]
- Tawfik A. F., Shibl A. M., Aljohi M. A., Altammami M. A., Al-Agamy M. H. (2012). Distribution of ambler class A, B and D beta-lactamases among Pseudomonas aeruginosa isolates. Burns 38 855–860. 10.1016/j.burns.2012.01.005 [DOI] [PubMed] [Google Scholar]
- Tayh G., Ben Sallem R., Ben Yahia H., Gharsa H., Klibi N., Boudabous A., et al. (2016). First report of extended-spectrum beta-lactamases among clinical isolates of Escherichia coli in gaza strip, Palestine. J. Glob. Antimicrob. Resist. 6 17–21. 10.1016/j.jgar.2016.01.013 [DOI] [PubMed] [Google Scholar]
- Tayh G., Ben Sallem R., Ben Yahia H., Gharsa H., Klibi N., Boudabous A., et al. (2017). First report of extended-spectrum beta-lactamases among clinical isolates of Klebsiella pneumoniae in gaza strip, Palestine. Microb. Drug Resist. 23 169–176. 10.1089/mdr.2016.0089 [DOI] [PubMed] [Google Scholar]
- Tian G. B., Wang H. N., Zhang A. Y., Zhang Y., Fan W. Q., Xu C. W., et al. (2012). Detection of clinically important beta-lactamases in commensal Escherichia coli of human and swine origin in western China. J. Med. Microbiol. 61(Pt 2), 233–238. 10.1099/jmm.0.036806-0 [DOI] [PubMed] [Google Scholar]
- Tokajian S., Eisen J. A., Jospin G., Farra A., Coil D. A. (2015). Whole genome sequencing of extended-spectrum beta-lactamase producing Klebsiella pneumoniae isolated from a patient in Lebanon. Front. Cell. Infect. Microbiol. 5:32 10.3389/fcimb.2015.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokajian S., Eisen J. A., Jospin G., Matar G., Araj G. F., Coil D. A. (2016). Draft genome sequence of Klebsiella pneumoniae KGM-IMP216 harboring blaCTX-M-15, blaDHA-1, blaTEM-1B, blaNDM-1, blaSHV-28, and blaOXA-1, isolated from a patient in Lebanon. Genome Announc. 4 e1632–15. 10.1128/genomeA.01632-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokajian S., Moghnieh R., Salloum T., Arabaghian H., Alousi S., Moussa J., et al. (2018). Extended-spectrum beta-lactamase-producing Escherichia coli in wastewaters and refugee camp in Lebanon. Future Microbiol. 13 81–95. 10.2217/fmb-2017-0093 [DOI] [PubMed] [Google Scholar]
- Toth A., Kocsis B., Damjanova I., Kristof K., Janvari L., Paszti J., et al. (2014). Fitness cost associated with resistance to fluoroquinolones is diverse across clones of Klebsiella pneumoniae and may select for CTX-M-15 type extended-spectrum beta-lactamase. Eur. J. Clin. Microbiol. Infect. Dis. 33 837–843. 10.1007/s10096-013-2022-6 [DOI] [PubMed] [Google Scholar]
- Us E., Tekeli A., Arikan Akan O., Dolapci I., Sahin F., Karahan Z. C. (2010). Molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae strains isolated between 2004-2007 in Ankara university hospital, Turkey. Mikrobiyol. Bul. 44 1–10. [PubMed] [Google Scholar]
- Uz Zaman T., Albladi M., Siddique M. I., Aljohani S. M., Balkhy H. H. (2018). Insertion element mediated mgrB disruption and presence of ISKpn28 in colistin-resistant Klebsiella pneumoniae isolates from Saudi Arabia. Infect. Drug Resist. 11 1183–1187. 10.2147/IDR.S161146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uz Zaman T., Aldrees M., Al Johani S. M., Alrodayyan M., Aldughashem F. A., Balkhy H. H. (2014). Multi-drug carbapenem-resistant Klebsiella pneumoniae infection carrying the OXA-48 gene and showing variations in outer membrane protein 36 causing an outbreak in a tertiary care hospital in Riyadh, Saudi Arabia. Int. J. Infect. Dis. 28 186–192. 10.1016/j.ijid.2014.05.021 [DOI] [PubMed] [Google Scholar]
- van Burgh S., Maghdid D. M., Ganjo A. R., Mansoor I. Y., Kok D. J., Fatah M. H., et al. (2018). PME and other ESBL-positive multiresistant Pseudomonas aeruginosa isolated from hospitalized patients in the region of Kurdistan, Iraq. Microb. Drug Resist. 25 32–38. 10.1089/mdr.2018.0036 [DOI] [PubMed] [Google Scholar]
- van Hoek A. H., Mevius D., Guerra B., Mullany P., Roberts A. P., Aarts H. J. (2011). Acquired antibiotic resistance genes: an overview. Front. Microbiol. 2:203. 10.3389/fmicb.2011.00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verraes C., Van Boxstael S., Van Meervenne E., Van Coillie E., Butaye P., Catry B., et al. (2013). Antimicrobial resistance in the food chain: a review. Int. J. Environ. Res. Public Health 10 2643–2669. 10.3390/ijerph10072643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Salviati C., Laube H., Guerra B., Roesler U., Friese A. (2015). Emission of ESBL/AmpC-producing Escherichia coli from pig fattening farms to surrounding areas. Vet. Microbiol. 175 77–84. 10.1016/j.vetmic.2014.10.010 [DOI] [PubMed] [Google Scholar]
- Warburg G., Hidalgo-Grass C., Partridge S. R., Tolmasky M. E., Temper V., Moses A. E., et al. (2012). A carbapenem-resistant Klebsiella pneumoniae epidemic clone in Jerusalem: sequence type 512 carrying a plasmid encoding aac(6’)-ib. J. Antimicrob. Chemother. 67 898–901. 10.1093/jac/dkr552 [DOI] [PubMed] [Google Scholar]
- Wassef M., Abdelhaleim M., AbdulRahman E., Ghaith D. (2015). The role of OmpK35, OmpK36 porins, and production of beta-lactamases on imipenem susceptibility in Klebsiella pneumoniae clinical isolates, Cairo, Egypt. Microb. Drug Resist. 21 577–580. 10.1089/mdr.2014.0226 [DOI] [PubMed] [Google Scholar]
- Wibberg D., Salto I. P., Eikmeyer F. G., Maus I., Winkler A., Nordmann P., et al. (2018). Complete genome sequencing of Acinetobacter baumannii strain K50 discloses the large conjugative plasmid pK50a encoding carbapenemase OXA-23 and extended-spectrum beta-lactamase GES-11. Antimicrob. Agents Chemother. 62 e212–18. 10.1128/AAC.00212-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener-Well Y., Rudensky B., Yinnon A. M., Kopuit P., Schlesinger Y., Broide E., et al. (2010). Carriage rate of carbapenem-resistant Klebsiella pneumoniae in hospitalised patients during a national outbreak. J. Hosp. Infect. 74 344–349. 10.1016/j.jhin.2009.07.022 [DOI] [PubMed] [Google Scholar]
- Yasir M., Ajlan A. M., Shakil S., Jiman-Fatani A. A., AlmaSaudi S. B., Farman M., et al. (2018). Molecular characterization, antimicrobial resistance and clinico-bioinformatics approaches to address the problem of extended-spectrum beta-lactamase-producing Escherichia coli in western Saudi Arabia. Sci. Rep. 8:14847. 10.1038/s41598-018-33093-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz G. R., Dizbay M., Guven T., Pullukcu H., Tasbakan M., Guzel O. T., et al. (2016). Risk factors for infection with colistin-resistant gram-negative microorganisms: a multicenter study. Ann. Saudi Med. 36 216–222. 10.5144/0256-4947.2016.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz N. O., Agus N., Bozcal E., Oner O., Uzel A. (2013). Detection of plasmid-mediated AmpC beta-lactamase in Escherichia coli and Klebsiella pneumoniae. Indian J. Med. Microbiol. 31 53–59. 10.4103/0255-0857.108723 [DOI] [PubMed] [Google Scholar]
- Yilmaz N. O., Agus N., Bozcal E., Uzel A. (2014). Prevalence and molecular characterisation of metallo-beta-lactamase producing strains of imipenem-resistant Pseudomonas aeruginosa in Turkey. Indian J. Med. Microbiol. 32 349–350. [DOI] [PubMed] [Google Scholar]
- Yousefi A., Torkan S. (2017). Uropathogenic Escherichia coli in the urine samples of Iranian dogs: antimicrobial resistance pattern and distribution of antibiotic resistance genes. Biomed Res. Int. 2017:4180490. 10.1155/2017/4180490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafari M., Feizabadi M. M., Jafari S., Abdollahi A., Sabokbar A. (2017). High prevalence of OXA-type carbapenemases among Acinetobacter baumannii strains in a teaching hospital of Tehran. Acta Microbiol. Immunol. Hung. 64 385–394. 10.1556/030.64.2017.031 [DOI] [PubMed] [Google Scholar]
- Zafer M. M., Al-Agamy M. H., El-Mahallawy H. A., Amin M. A., El Din Ashour S. (2015). Dissemination of VIM-2 producing Pseudomonas aeruginosa ST233 at tertiary care hospitals in Egypt. BMC Infect. Dis. 15:122. 10.1186/s12879-015-0861-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafer M. M., Amin M., El Mahallawy H., Ashour M. S., Al Agamy M. (2014). First report of NDM-1-producing Pseudomonas aeruginosa in Egypt. Int. J. Infect. Dis. 29 80–81. 10.1016/j.ijid.2014.07.008 [DOI] [PubMed] [Google Scholar]
- Zaman T. U., Alrodayyan M., Albladi M., Aldrees M., Siddique M. I., Aljohani S., et al. (2018). Clonal diversity and genetic profiling of antibiotic resistance among multidrug/carbapenem-resistant Klebsiella pneumoniae isolates from a tertiary care hospital in Saudi Arabia. BMC Infect. Dis. 18:205. 10.1186/s12879-018-3114-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanlou S., Ahangarzadeh Rezaee M., Aghazadeh M., Ghotaslou R., Babaie F., Khalili Y. (2018). Characterization of integrons, extended-spectrum beta-lactamases, AmpC cephalosporinase, quinolone resistance, and molecular typing of Shigella spp. from Iran. Infect. Dis. 50 616–624. 10.1080/23744235.2018.1455222 [DOI] [PubMed] [Google Scholar]
- Zanganeh Z., Eftekhar F. (2015). Correlation of oxacillinase gene carriage with the genetic fingerprints of imipenem-resistant clinical isolates of Acinetobacter baumannii. Jundishapur J. Microbiol. 8:e26545. 10.5812/jjm.26545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarakolu P., Eser O. K., Aladag E., Al-Zahrani I. A., Day K. M., Atmaca O., et al. (2016). Epidemiology of carbapenem-resistant Klebsiella pneumoniae colonization: a surveillance study at a turkish university hospital from 2009 to 2013. Diagn. Microbiol. Infect. Dis. 85 466–470. 10.1016/j.diagmicrobio.2016.05.012 [DOI] [PubMed] [Google Scholar]
- Zimmerman F. S., Assous M. V., Bdolah-Abram T., Lachish T., Yinnon A. M., Wiener-Well Y. (2013). Duration of carriage of carbapenem-resistant enterobacteriaceae following hospital discharge. Am. J. Infect. Control 41 190–194. 10.1016/j.ajic.2012.09.020 [DOI] [PubMed] [Google Scholar]
- Zowawi H. M., Sartor A. L., Balkhy H. H., Walsh T. R., Al Johani S. M., AlJindan R. Y., et al. (2014). Molecular characterization of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the countries of the gulf cooperation council: dominance of OXA-48 and NDM producers. Antimicrob. Agents Chemother. 58 3085–3090. 10.1128/AAC.02050-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zowawi H. M., Sartor A. L., Sidjabat H. E., Balkhy H. H., Walsh T. R., Al Johani S. M., et al. (2015). Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates in the gulf cooperation council states: dominance of OXA-23-type producers. J. Clin. Microbiol. 53 896–903. 10.1128/JCM.02784-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zowawi H. M., Syrmis M. W., Kidd T. J., Balkhy H. H., Walsh T. R., Al Johani S. M., et al. (2018). Identification of carbapenem-resistant Pseudomonas aeruginosa in selected hospitals of the gulf cooperation council states: dominance of high-risk clones in the region. J. Med. Microbiol. 67 846–853. 10.1099/jmm.0.000730 [DOI] [PubMed] [Google Scholar]
- Zurita J., Solis M. B., Ortega-Paredes D., Barba P., Paz Y., Mino A., et al. (2019). High prevalence of B2-ST131 clonal group among ESBL-producing Escherichia coli isolated from bloodstream infections in Quito, Ecuador. J. Glob. Antimicrob. Resist. 10.1016/j.jgar.2019.04.019 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]


