Osmotic stress triggers rapid reorganization of the receptor kinases QSK1 and IMK2 at plasmodesmata and induces callose-mediated plasmodesmata closure.
Abstract
Plasmodesmata act as key elements in intercellular communication, coordinating processes related to plant growth, development, and responses to environmental stresses. While many of the developmental, biotic, and abiotic signals are primarily perceived at the plasma membrane (PM) by receptor proteins, plasmodesmata also cluster receptor-like activities; whether these two pathways interact is currently unknown. Here, we show that specific PM-located Leu-rich-repeat receptor-like-kinases, Qiān Shŏu kinase (QSK1) and inflorescence meristem kinase2, which under optimal growth conditions are absent from plasmodesmata, rapidly relocate and cluster to the pores in response to osmotic stress. This process is remarkably fast, is not a general feature of PM-associated proteins, and is independent of sterol and sphingolipid membrane composition. Focusing on QSK1, previously reported to be involved in stress responses, we show that relocalization in response to mannitol depends on QSK1 phosphorylation. Loss-of-function mutation in QSK1 results in delayed lateral root (LR) development, and the mutant is affected in the root response to mannitol stress. Callose-mediated plasmodesmata regulation is known to regulate LR development. We found that callose levels are reduced in the qsk1 mutant background with a root phenotype resembling ectopic expression of PdBG1, an enzyme that degrades callose at the pores. Both the LR and callose phenotypes can be complemented by expression of wild-type and phosphomimic QSK1 variants, but not by phosphodead QSK1 mutant, which fails to relocalize at plasmodesmata. Together, the data indicate that reorganization of receptor-like-kinases to plasmodesmata is important for the regulation of callose and LR development as part of the plant response to osmotic stress.
Plasmodesmata are nanoscaled membranous pores that span the plant cell wall creating both cytoplasmic and membrane continuums between cells (Tilsner et al., 2011, 2016). By interconnecting most cells throughout the whole plant body, plasmodesmata form a symplastic network that supports and controls the movement of molecules from cell to cell, within a given tissue or organ, and long-distance transport when combined with the vasculature (Kragler et al., 1998; Corbesier et al., 2007; Liu et al., 2012; Reagan et al., 2018). Given their central function in intercellular communication, plasmodesmata orchestrate processes related to plant growth and development but also responses to pathogens and abiotic stresses (Benitez-Alfonso et al., 2010, 2013; Lee et al., 2011; Vatén et al., 2011; Liu et al., 2012; Faulkner et al., 2013; Caillaud et al., 2014; Daum et al., 2014; Gallagher et al., 2014; Cui and Lee, 2016; Lim et al., 2016; Wu et al., 2016; O’Lexy et al., 2018; Tylewicz et al., 2018; Miyashima et al., 2019). Plasmodesmata also act as specialized signaling hubs, capable of generating and/or relaying signaling from cell-to-cell through plasmodesmata-associated receptor activity (Faulkner, 2013; Stahl et al., 2013; Vaddepalli et al., 2014; Stahl and Faulkner, 2016).
Plasmodesmata specialized functions hinge on their molecular specialization (Nicolas et al., 2017). The pores are outlined by highly specialized plasma membrane (PM) microdomains that cluster a specific set of both proteins and lipids, compared to the bulk PM (Levy et al., 2007; Thomas et al., 2008; Simpson et al., 2009; Fernandez-Calvino et al., 2011; Vatén et al., 2011; Benitez-Alfonso et al., 2013; Salmon and Bayer, 2013; Grison et al., 2015a; Xu et al., 2017). Among the array of proteins that localize to plasmodesmata, receptor proteins and receptor protein kinases have recently emerged as critical players for modulating cell-to-cell signaling in response to both developmental and stress-related stimuli (Faulkner et al., 2013; Stahl and Simon, 2013; Vaddepalli et al., 2014; Stahl and Faulkner, 2016). For instance, Plasmodesmata Located Protein5 (PDLP5), a receptor-like protein, is necessary for callose induced-plasmodesmata closure in response to salicylic acid, a pivotal hormone in innate immune responses (Lee et al., 2011; Wang et al., 2013). Similarly, up-regulation of PDLP1 during mildew infection promotes down-regulation of plasmodesmata permeability (Caillaud et al., 2014). The membrane-associated receptor-like kinase (RLK) STRUBBELIG localizes at plasmodesmata, where it interacts with QUIRKY to regulate organ formation and tissue morphogenesis (Vaddepalli et al., 2014). Similarly, the receptor kinase CRINKLY4 presents dual localization at the PM and plasmodesmata and is involved in root apical meristem maintenance and columella cell identity specification (Stahl et al., 2013). CRINKLY4 forms homo- and heteromeric complexes with CLAVATA1, depending on its subcellular localization at the PM or at plasmodesmata (Stahl et al., 2013). Activation/inactivation of signaling cascades is often regulated by receptor complex association/dissociation to PM microdomains (Hofman et al., 2008). There is a high diversity of microdomains that coexist at the PM allowing the separation of different signaling pathways (Raffaele et al., 2007; Jarsch et al., 2014; Bücherl et al., 2017). For instance, in plants, the localization of FLAGELLIN SENSING2 and BRASSINOSTEROID INSENSITIVE1 in distinct microdomains enables cells to differentiate between fungus-induced immunity and steroid-mediated growth, and this is despite the fact that these two signaling cascades share common components (Bücherl et al., 2017). In mammals, the EPIDERMAL GROWTH FACTOR RECEPTOR reversibly associates and dissociates with PM microdomains, which in turn control the activation and inactivation of signaling events (Hofman et al., 2008; Bocharov et al., 2016). Spatiotemporality and dynamics of receptor complexes appear critical for regulating signaling events. In plants, both the PM and plasmodesmata pores present receptor-like activities, but at present it is not clear whether these interact.
Here, we present data revealing that the PM-located Leu rich repeat (LRR)-RLKs QSK1 (Qiān Shŏu kinase; AT3G02880) and IMK2 (inflorescence meristem kinase2; AT3G51740), which normally reside in the PM, rapidly reorganize their subcellular localization and relocate at plasmodesmata intercellular pores, upon mannitol and NaCl salt treatments. This process occurs within less than 2 min and it is not a general behavior of PM or microdomain-associated proteins. Focusing on QSK1, which has been previously shown to be involved in Suc- and abscisic acid-related responses and associated with lipid nanodomains (Wu et al., 2013, 2019; Szymanski et al., 2015; Isner et al., 2018), we show that relocalization does not depend on sterol or sphingolipid membrane composition. QSK1 is phosphorylated in response to various abiotic stresses such as salt and mannitol treatments (Hem et al., 2007; Niittylä et al., 2007; Hsu et al., 2009; Chen et al., 2010; Kline et al., 2010; Chang et al., 2012; Xue et al., 2013), and our data demonstrate that QSK1 phosphorylation is important for plasmodesmata localization in control and mannitol stress conditions. QSK1 phosphodead but not phosphomimic mutant is impaired in plasmodesmata localization upon stress. Loss-of-function in QSK1 in Arabidopsis (Arabidopsis thaliana) results in a reduction in lateral root (LR) numbers in control conditions and affects root response to mannitol treatment. These phenotypes can be complemented by QSK1 wild-type protein and QSK1 phosphomimic, but not QSK1 phosphodead protein mutant. Our data further indicate that callose deposition at plasmodesmata is modified upon mannitol stress and that phosphorylation of QSK1 is important to regulate LR response to mannitol most likely via a mechanism that modulates the levels of callose.
The work emphasizes the dynamic nature of plasmodesmata membrane domains, which can within few minutes of stimulation recruit PM-located receptor-like proteins that presumably trigger local mechanisms to regulate plasmodesmata aperture and, thereby, plant developmental response to environmental stresses.
RESULTS
The PM-Associated LRR-RLKs QSK1 and IMK2 Dynamically Associate with Plasmodesmata in Response to Mannitol and Salt Treatments
A survey of the recently published Arabidopsis plasmodesmata-proteome (Brault et al., 2019) identified several members of the RLK family present in the plasmodesmata fraction with clade III members being predominant (Supplemental Table S1). As plasmodesmata have been reported to be composed of sterol- and sphingolipid-enriched microdomains (Grison et al., 2015a; Nicolas et al., 2017), we focused on RLKs, which may preferentially associate with lipid microdomains by cross referencing the accessions with seven published detergent-resistant membrane (DRM) proteomes (Shahollari et al., 2004, 2005; Kierszniowska et al., 2009; Minami et al., 2009; Keinath et al., 2010; Demir et al., 2013; Srivastava et al., 2013; Szymanski et al., 2015). By doing so, we identified two LRR-RLKs, QSK1 (AT3G02880 also called Kinase7; Isner et al., 2018; Wu et al., 2019) and IMK2 (AT3G51740), which were relatively abundant in the plasmodesmata proteome and consistently identified in DRM fractions (Supplemental Table S2).
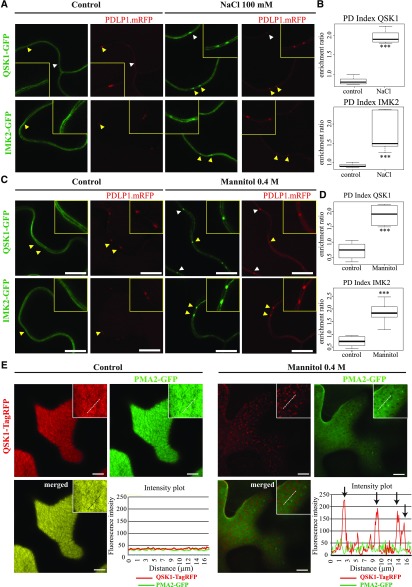
We next investigated the subcellular localization of the two LRR-RLKs, by transiently expressing the proteins as GFP fusions in Nicotiana benthamiana leaves followed by confocal imaging. Under control conditions, both QSK1 and IMK2 were found exclusively located to the PM with no specific enrichment at plasmodesmata (Fig. 1, A and C). However, when subjected to 0.4 m mannitol or 100 mm NaCl, both proteins reorganized at the cell periphery in a punctate pattern (Fig. 1, A and C, arrows). Colocalization with the plasmodesmata marker, PDLP1-mRFP (monomeric red fluorescent protein; Amari et al., 2010), revealed that the mannitol- and salt-induced peripheral dots colocalized with plasmodesmata (Fig. 1, A and C). In order to quantify plasmodesmata depletion/enrichment under control and stress conditions, we measured the plasmodesmata index, or PD index, by calculating the fluorescence intensity ratio between plasmodesmata (green signal that colocalizes with PDLP1-mRFP) versus PM (see “Materials and Methods” and Supplemental Fig. S1). In control conditions, both QSK1 and IMK2 displayed a PD index below 1 (median value), indicating no specific enrichment at plasmodesmata compared to the PM. However, upon short-term (5–30 min) mannitol or NaCl, treatment this value increased to 1.5 to 2 (Fig. 1, B and D), confirming plasmodesmata enrichment. In addition to clustering at plasmodesmata, we also observed a reorganization of the LRR-RLK QSK1 within the PM plane into microdomains at the surface of epidermal cells (Fig. 1E), from which the proton pump ATPase PMA2 (Morsomme et al., 1998) was excluded.
Figure 1.
IMK2 and QSK1 are PM-associated LRR-RLKs that reorganize at plasmodesmata upon salt and mannitol treatments. A to D, Transient expression in N. benthamiana epidermal cells of IMK2-GFP and QSK1-GFP LRR-RLKs expressed under 35S promoter and visualized by confocal microscopy. In control conditions, the two LRR-RLKs localize exclusively at the PM and present no enrichment at plasmodesmata, which are marked by PDLP1-mRFP. Upon 100 mm NaCl (A and B) or 0.4 m mannitol (C and D) treatment (5–30 min), the two LRR-RLKs relocalize to plasmodesmata (arrowheads). Yellow-boxed regions are magnification of areas indicated by yellow arrowheads. Enrichment at plasmodesmata versus the PM was quantified by the PD index, which corresponds to the fluorescence intensity ratio of the LRR-RLKs at plasmodesmata versus the PM in control and abiotic stress conditions (see “Materials and Methods” for details and Supplemental Fig. S1). n = 4 experiments, 3 plants/experiment, 10 measures/plant. Two-tailed Wilcoxon statistical analysis, *P < 0.05; **P < 0.01; ***P < 0.001. E, Transient expression in N. benthamiana epidermal cells of QSK1-TagRFP and PMA2-GFP expressed under 35S promoter and visualized by confocal microscopy. Top surface view of a leaf epidermal cell showing the uniform and smooth distribution pattern of QSK1-TagRFP and PMA2-GFP at the PM under control conditions. Mannitol treatment causes a relocalization of QSK1-TagRFP, but not of PMA2-GFP, into microdomain-like structures at the PM on the upper epidermal cell surface. Intensity plot along the white dashed line visible on the confocal images. n = 2 experiments, 3 plants/experiment. Scale bars, 10µm.
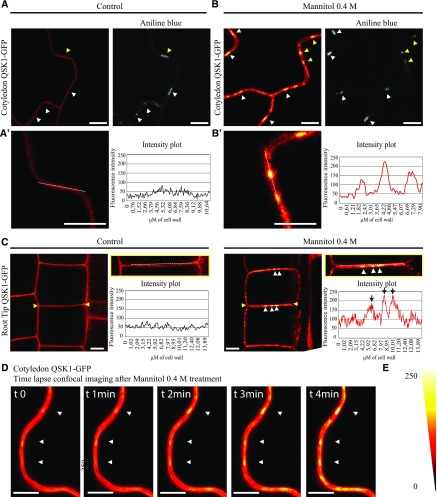
To confirm these results, we generated Arabidopsis transgenic lines expressing QSK1 tagged with GFP (Fig. 2). In control condition, QSK1 was located to the PM in both cotyledons and root tissues of 1-week-old seedlings but reorganized at the PM and relocated to plasmodesmata upon mannitol treatment (Fig. 2, A–C, arrows). Reorganization at plasmodesmata was remarkably fast and happened within 1 to 4 min posttreatment in the cotyledons (Fig. 2D; Supplemental Video S1). A similarly rapid change of localization was also observed upon NaCl (100 mm) treatment (Supplemental Fig. S2). From our data, we concluded that both QSK1 and IMK2 LRR-RLKs can rapidly modulate their subcellular localization and associate with plasmodesmata in response to osmotic stress.
Figure 2.
Reorganization of QSK1 at plasmodesmata upon abiotic stress occurs remarkably fast. Stable Arabidopsis line expressing QSK1-GFP, under 35S promoter and visualized by confocal microscopy. All images showing QSK1-GFP localization have been color-coded through a heat-map filter to highlight clustering at plasmodesmata. A and B, In control conditions, QSK1-GFP localizes exclusively at the PM in cotyledons and is not enriched at plasmodesmata (A, marked by aniline blue staining, arrowheads). A′ and B′ are magnified regions indicated by yellow arrowheads in A. Upon 0.4 m mannitol treatment, QSK1 relocalizes to plasmodesmata, where it becomes enriched (B, white arrowheads). Intensity plots along the white dashed lines are shown for QSK1-GFP localization pattern in control and mannitol conditions. C, In control condition, QSK1 localizes exclusively at the PM. Upon 0.4 m mannitol treatment, QSK1 relocalizes into dotty pattern typical from plasmodesmata. Intensity plots along the white dashed lines are shown for QSK1-GFP localization pattern in control and mannitol conditions. D, Time-lapse imaging of QSK1-GFP relocalization upon mannitol exposure. Within less than 2 min, plasmodesmata localization is already visible (white arrowhead). Please note reorganization is faster when QSK1 is stably expressed (less than 5 min when stably expressed; 5–30 min when transiently expressed). E, Color-coding bar for heat-map images. Scale bars, 10 μm.
Relocalization at Plasmodesmata Is Not a General Feature of PM or Nanodomain-Associated Proteins
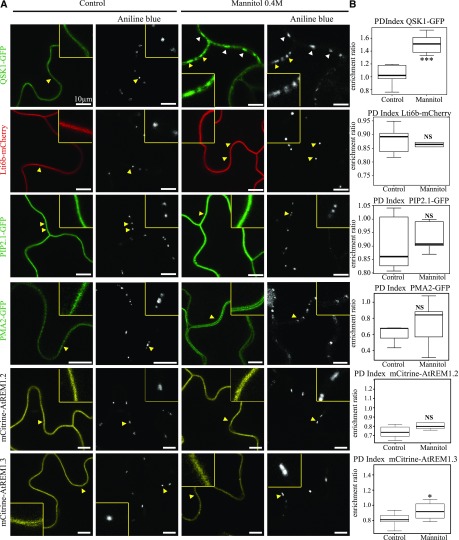
To test whether plasmodesmata association in response to osmotic stress is a common feature of PM proteins, we investigated the behavior of unrelated PM-associated proteins. We selected proteins that associate with the PM either through transmembrane domains, such as the Low Temperature Induced Protein 6B (Lti6b), the Plasma Membrane Intrinsic Protein 2;1 (PIP2;1), and PMA2 (Cutler et al., 2000; Prak et al., 2008) or through surface interaction with inner leaflet lipids such as Remorin 1.2 and 1.3, which are also well-established lipid nanodomain markers (Jarsch et al., 2014; Konrad et al., 2014; Gronnier et al., 2017). While QSK1 became significantly enriched at plasmodesmata, none of the tested PM-associated proteins displayed plasmodesmata association upon short (1–5 min) 0.4 m mannitol treatment as indicated by their PD index, which remained below 1 (Fig. 3). Altogether, our results indicate that the capacity of QSK1 and IMK2 to relocalize at plasmodesmata upon stress is not a general feature of all PM proteins.
Figure 3.
Conditional plasmodesmal association is not a general feature of PM-associated proteins. A, In control conditions, QSK1-GFP, the PM-associated proteins Lti6b-mCherry, PIP2;1-GFP, PMA2-GFP, m-Citrine-AtREM1.2, and mCitrine-AtREM1.3 show localization to the PM and are not enriched at plasmodesmata (stained with aniline blue, arrowheads). Mannitol (0.4 m) treatment (1–5 min) induces the reorganization of QSK1 at plasmodesmata, while other PM-associated proteins stay excluded from plasmodesmata. Single confocal scan images of Arabidopsis transgenic seedlings (QSK1-GFP, Lti6b-mCherry, PIP2;1-GFP, mCitrine-AtREM1.2, and mCitrine-AtREM1.3) or N. benthamiana leaves transiently expressing PMA2-GFP. Yellow boxed regions are magnifications of areas indicated by yellow arrowheads. Scale bars, 10 µm. B, PD index for each PM-associated protein tested in A in control and mannitol conditions. n = 3, 3 plants/line/experiment, 3 to 6 cells/plant, 5 to 10 regions of interest (ROI) for PM and plasmodesmata per cell. Two-tailed Wilcoxon statistical analysis: *P < 0.05; **P < 0.01; ***P < 0.001.
Changes in Sterol and Sphingolipid Composition Do Not Affect QSK1 Conditional Association with Plasmodesmata
We next decided to investigate the mechanisms underlying plasmodesmata localization of LRR-RLKs by focusing on QSK1. QSK1 has been proposed to associate with sterol- and sphingolipid-enriched PM nanodomains in plants (Shahollari et al., 2004, 2005; Kierszniowska et al., 2009; Minami et al., 2009; Keinath et al., 2010; Demir et al., 2013; Srivastava et al., 2013; Szymanski et al., 2015; Supplemental Table S2), and in animal cells, lipid-nanodomains have been reported to coalesce and form signaling platforms in a sterol-dependent manner (Rossy et al., 2014).
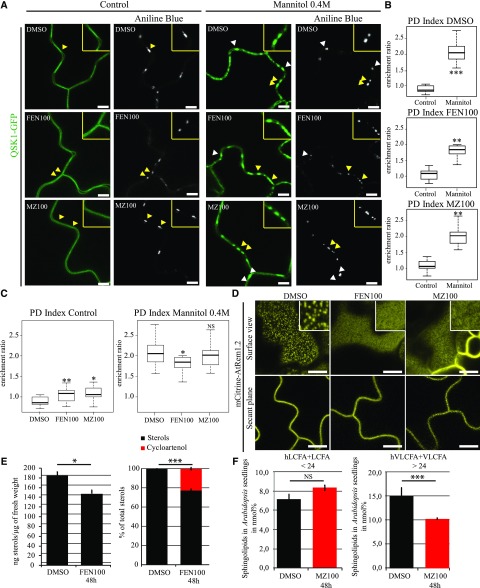
To test the importance of lipids for plasmodesmal conditional association, we used pharmacological approaches and specifically inhibited sterol and sphingolipid biosynthesis (He et al., 2003; Grison et al., 2015a; Wattelet-Boyer et al., 2016). For sterols, we used fenpropimorph (FEN100; 100 μg/mL, 48 h), which acts directly in the sterol biosynthetic pathway by inhibiting the cyclopropyl-sterol isomerase and whose effects are well characterized in Arabidopsis seedlings (Hartmann et al., 2002; He et al., 2003). For sphingolipids, we focused on glycosyl-inositol-phospho-ceramides (GIPCs), which are the main sphingolipids associated with both plasmodesmata and lipid nanodomains (Grison et al., 2015a; Cacas et al., 2016). We modulated GIPC content using metazachlor (MZ100; 100 nm/mL, 48 h), which reduces the very long chain fatty acid and hydroxylated very long chain fatty acid (VLCFA > 24C and hVLCFA > 24C) of GIPCs (Wattelet-Boyer et al., 2016). Alteration of the cellular pool of sterols and VLCFA-derived GIPCs was confirmed by gas chromatography coupled to mass spectrometry (Fig. 4, E and F). We observed a depletion of 22.6% of sterols and 30% of hVLCFA and VLCFA consistent with previous studies (Grison et al., 2015a; Wattelet-Boyer et al., 2016). Effectiveness of lipid inhibitor treatments on the PM lipid pool was also confirmed by the change of Remorin 1.2 organization at the PM surface from nanodomains to a smooth pattern (Fig. 4D).
Figure 4.
Mannitol-induced relocalization of QSK1 is independent of sterols and sphingolipids. A to C, Stable Arabidopsis line expressing QSK1-GFP under 35S promoter and visualized by confocal microscopy after sterol- or very long chain GIPC biosynthesis inhibitor treatments and 0.4 m mannitol exposure (1–5 min). Arabidopsis seedlings were grown on normal agar plates for 5 d and then transferred to 100 µg/mL fenpropimorph (FEN100), 100 nm metazachlor (MZ100), or 3% (v/v) dimethyl sulfoxide (DMSO) agar plates for 48 h. Compared to control (DMSO) conditions, FEN100 and MZ100 induce a slight increase in plasmodesmata localization as indicated by the PD index (B and C), but QSK1-GFP was still preferentially located at the PM. Despite the lipid inhibitor treatments, QSK1-GFP was nevertheless capable of reorganizing at plasmodesmata after mannitol treatment. A, Confocal single scan images. Yellow-boxed regions are magnification of areas indicated by yellow arrowheads. B and C, PD indexes corresponding to A. n = 3 experiments, 3 plants/line/experiment, 3 to 6 cells/plant, 5 to 10 ROI for PM and plasmodesmata per cell. D, Localization pattern of Arabidopsis Remorin 1.2 (mCitrine-AtREM1.2) in Arabidopsis cotyledons after 48 h FEN100 and MZ100 treatments showing reduced lateral organization into microdomains at the epidermal cell surface upon lipid inhibitors. E, Sterol quantification after FEN100 treatment by gas chromatography coupled to mass spectrometry. Left, Arabidopsis seedlings treated with FEN100 presented a 20% decrease of the total amount of sterols after 48 h. Right, relative proportion of sterol species in Arabidopsis seedling treated with FEN100 showing cycloartenol accumulation of 22.5%. Black, “normal” sterols; red, cyloartenol (n = 3). Bars indicate SD. F, Total fatty acid methyl esters (FAMES) quantification after MZ100 treatment by gas chromatography coupled to mass spectrometry in Arabidopsis seedlings in nanomole percent (black, control condition, e.g. DMSO 48 h; red, MZ100 48 h; mean value of three replicates). VLCFA > 24 (hydroxylated and nonhydroxylated) are reduced by 30% on metazachlor (n = 3). Bars indicates SD. VLCFA, Very long chain fatty acid; LVFA, long chain fatty acid. Two-tailed Wilcoxon statistical analysis: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Scale bars, 10 µm (A and D).
Under conditions with no mannitol but sterol and sphingolipid inhibitors, we observed a minor but significant increase in the PD index of QSK1 under FEN100 and MZ100, which raised to 1.08 and 1.06, respectively, compared to DMSO control conditions with a PD index of 0.86 (Fig. 4C). The results indicate that modifying the cellular lipid pool can affect localization to plasmodesmata. However, upon mannitol treatment (0.4 m, 1–5 min), effective QSK1 relocalization to plasmodesmata was maintained in all conditions (Fig. 4, A–C). These results suggest that sterols and sphingolipids are not essential for plasmodesmata clustering of QSK1 under mannitol treatment.
QSK1 Association with Plasmodesmata Is Regulated by Phosphorylation
We next investigated whether QSK1 phosphorylation status could be involved in plasmodesmata targeting. Several phosphorylation sites have been experimentally reported for QSK1 (Supplemental Table S3). QSK1 phospho-status varies upon various abiotic stresses such as salt and mannitol treatments but also after exposure to Suc and to hormones (Hem et al., 2007; Niittylä et al., 2007; Hsu et al., 2009; Chen et al., 2010; Kline et al., 2010; Chang et al., 2012; Xue et al., 2013). In the context of this study, we focused on two phosphorylation sites (S621 and S626), which were consistently and experimentally detected in several phosphoproteomic studies, including in response to salt and mannitol exposure (Supplemental Table S3).
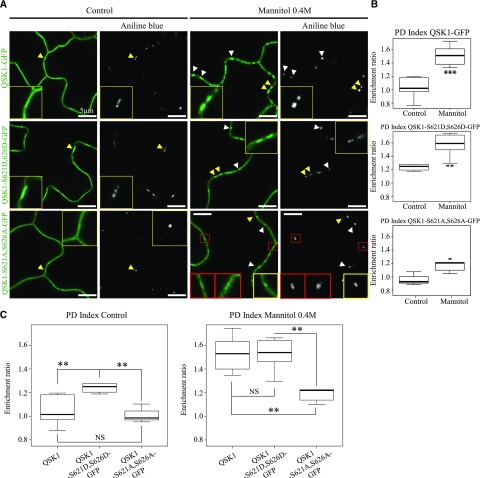
To test whether the phosphorylation of QSK1 could play a role in plasmodesmata association, we generated two QSK1 phosphomutants; the phosphomimic mutant (QSK1-S621D,S626D-GFP) and the phosphodead mutant (QSK1-S621A,S626A-GFP). Both were tagged with GFP, stably expressed under 35S in Arabidopsis, and their localization pattern analyzed along with that of the wild-type QSK1 protein (Fig. 5). Under control conditions, QSK1 and the phosphodead mutant QSK1-S621A,S626A-GFP were localized at the PM (Fig. 5A) and yielded PD indexes of 1.02 and 0.99 (median values; Fig. 5, B and C), respectively, indicating no specific plasmodesmata enrichment. By contrast QSK1-S621D,S626D-GFP displayed a significantly higher PD index of 1.24 suggesting that, in control conditions, the phosphomimic mutant can be weakly associated with plasmodesmata (Fig. 5). Mannitol exposure (0.4 m mannitol; 1–5 min treatment) triggered relocalization of all proteins to a different extent. While QSK1 and QSK1-S621D,S626D-GFP displayed a comparable PD index of 1.51 and 1.52, respectively, the phosphodead variant QSK1-S621A,S626A-GFP displayed a PD index barely reaching 1.20 (median values; Fig. 5, B and C). This suggests that QSK1-S621A,S626A-GFP ability to relocalize to plasmodesmata is diminished although not totally abolished. From these data, we concluded that QSK1 phosphorylation status influences plasmodesmata association and that mutations in the S621 and S626 phosphosites significantly alters QSK1 reorganization at the pores.
Figure 5.
QSK1 phosphorylation regulates plasmodesmata association upon mannitol treatment. A to C, Stable Arabidopsis lines expressing QSK1-GFP, QSK1-S621D,S626D-GFP (phosphomimic variant), and QSK1-S621A,S626A-GFP (phosphodead variant) under 35S promoter and visualized by confocal microscopy. Plasmodesmata were labeled by aniline blue (arrowheads). In control condition, QSK1 and the phosphodead mutant QSK1-S621A,S626A-GFP showed a “smooth” localization pattern at the PM (A) with no significant plasmodesmata association (B and C). The phosphomimic QSK1-S621D,S626D-GFP, however, displayed a weak but significant plasmodesmata localization with a shift of its PD index from 0.99 to 1.20 (A–C). After mannitol (0.4 m) exposure (1–5 min), QSK1 and QSK1-S621D,S626D-GFP similarly relocalize at plasmodesmata with a PD index of 1.52 and 1.53, respectively. Reorganization to plasmodesmata was significantly less effective for QSK1-S621A,S626A-GFP (A–C), with a PD index barely reaching 1.20 upon mannitol. For the phosphodead QSK1-S621A,S626A-GFP mutant, plasmodesmata association was not systematic as shown in red boxes in A. A, Confocal single scan images. Yellow-boxed regions are magnification of areas indicated by yellow arrowheads. B and C, PD indexes corresponding to A. n = 3 experiments, 3 plants/line/experiments, 3 to 6 cells/plants, 5 to 10 ROI for PM and PD/cells. Two-tailed Wilcoxon statistical analysis: *P < 0.05; **P < 0.01; ***P < 0.001. Scale bars, 10 µm.
QSK1 Function in Modulating Root Development and Response to Mannitol
Osmotic stress and mannitol treatments are known to affect root system architecture (Deak and Malamy, 2005; Macgregor et al., 2008; Roycewicz and Malamy, 2012; Zhou et al., 2018; Kumar et al., 2019). QSK1 localizes to plasmodesmata in response to mannitol and mutants in callose degradation and plasmodesmata transport are impaired in LR density and patterning (Benitez-Alfonso et al., 2013; Maule et al., 2013). We therefore tested QSK1 involvement in this pathway by determining its role in root development and in response to mannitol.
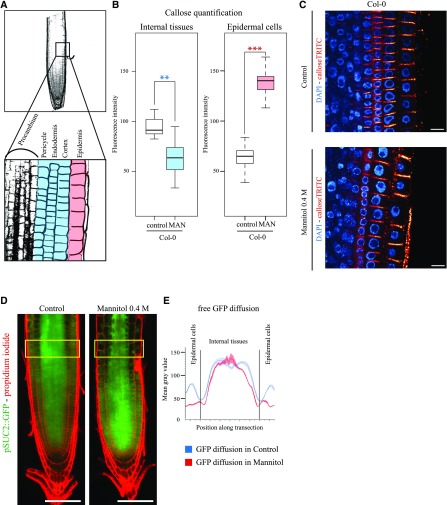
We first established the root phenotype of wild-type Col-0 seedlings in mannitol (0.4 m). After 3 d of exposure to mannitol, root length and LR number were reduced in comparison to seedlings in control media (Fig. 6A; Supplemental Fig. S3). Mannitol treatment also modified callose, which appears reduced in internal root layers and increased in the epidermal cell layer (Fig. 7, A–C) with a concomitant reduction of GFP symplastic movement into the epidermal cells when expressed under the sucrose transport protein SUC2 promoter (Fig. 7, D and E).
Figure 6.
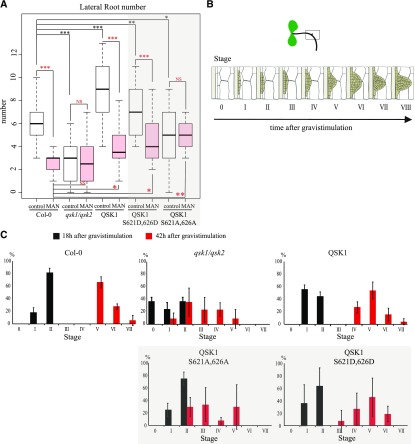
QSK1 is involved in root development and response to mannitol. A, LR number in wild-type Col-0, qsk1.qsk2 double mutant, qsk1.qsk2 double mutant expressing QSK1-GFP, QSK1-S621D,S626D-GFP, QSK1-S621A,S626A-GFP under 35S promoter. Arabidopsis lines were grown for 9 d on Murashige and Skoog plates for control conditions or 6 d then transferred to MS plate containing 0.4 m mannitol before root phenotyping. LR number is represented by white and red box plots for control and mannitol treatment, respectively. In control conditions, the qsk1.qsk2 double mutant displays a decrease of LR number compared to the wild type. Overexpression of QSK1 and the phosphomimic QSK1-S621D,S626D-GFP reverse this phenotype with more LR. Overexpression of QSK1-S621A,S626A-GFP phosphodead only partially rescues the qsk1.qsk2 double mutant LR number phenotype. In response to mannitol treatment, Col-0 wild-type and Arabidopsis seedlings overexpressing QSK1 and QSK1-S621D,S626D-GFP in the qsk1.qsk2 double mutant background all showed a decrease in LR number, whereas the qsk1.qsk2 double mutant and the qsk1.qsk2 double mutant overexpressing QSK1-S621A,S2626A-GFP display the same number of LR as in control conditions. n = 2 experiments, 10 plants/line/experiments. Two-tailed Wilcoxon statistical analysis, *P < 0.05; **P < 0.01; ***P < 0.001. Scale bars, 10 µm. (B and C) Graphical summary of the gravistimulation and the development stages of the LR primordia (B) adapted from Péret et al. (2012). The LR primordium stages were determined 18 h and 42 h after gravistimulation and are color-coded respectively in black and red (C). At 18 h, the qsk1.qsk2 double mutant display a delay in LR primordium initiation with the absence of LR primordium initiation (stage 0) in 35% of the plants observed. At 42 h, both the qsk1.qsk2 double mutant and QSK1-S621A,S2626A-GFP-expressing lines showed a delay in LR primordium compared to other lines, with no stage VI or VII LR primordium.
Figure 7.
Callose and plasmodesmata trafficking is modulated upon mannitol treatment. A, Square indicates the region where the callose quantification was done (top). High magnification of region similar to A top square with the different cell layers (bottom). Epidermal cells are colored in red, and “internal layers” are colored in blue. The same color code has been conserved in the box plot representation to facilitate the comprehension of the figure. B, Callose level quantifications; upon mannitol treatment (3 h, 0.4 m mannitol), callose levels are down-regulated in internal layers (blue) of the root while being up-regulated in the epidermis (red). C, Representative confocal images of callose immunofluorescence (red) in wild-type Col-0 Arabidopsis roots in control and mannitol treatment. DAPI staining of DNA (blue) was performed to highlight the cellular organization of root tissues. Scale bars, 10 μm. n = 2 experiments, 10 roots/experiments, 5 to 10 cell wall/cell lineage/roots. Two-tailed Wilcoxon statistical analysis, *P < 0.05; **P < 0.01; ***P < 0.001. D, Arabidopsis seedlings expressing pSUC2::GFP in under control and mannitol treatment (16 h, 0.4 m mannitol). Scale bars, 50 μm. E, GFP symplastic unloading from the phloem to surrounding tissues is modified under mannitol treatment. We observed a reduction of GFP diffusion in epidermal cells, which showed increased callose levels at plasmodesmata. n = 3.
Next, we compared the root phenotype of the wild-type Col-0 and loss-of-function qsk1 Arabidopsis mutant grown in parallel. Since QSK1 shares more than 90% similarity at the amino acid level to the LRR-RLK QSK2 (AT5G16590) and these proteins also display very similar expression profiles (Supplemental Figs. S4 and S5), we generated a double loss-of-function mutant named qsk1.qsk2 double mutant. The qsk1.qsk2 double mutant and the overexpressor line 35S::QSK1-GFP in the mutant background were grown in Murashige and Skoog control media and root phenotype was analyzed 9 d after germination. We found that the primary root length was not significantly different between all the lines in both conditions (Supplemental Fig. S3, A and B). LR development, on the other hand, was significantly affected in the qsk1.qsk2 double mutant and the QSK1 overexpressing line in control conditions, with qsk1.qsk2 double mutant displaying a reduced number of LR and QSK1 overexpressor showing the opposite phenotype with an increase in LR number in comparison to wild type (Fig. 6A, white box plots).
To further dissect this phenotype, we examined the different stages of LR formation by subjecting the seedlings to a 90° gravitropic stimulus, which triggers LR initiation in a very synchronized manner at the outer edge of the bend root (Péret et al., 2012). LR initiation and outgrow was observed at 18 h and 42 h postgravitropic stimuli (Fig. 6, B–C). LR initiation was impaired in the qsk1.qsk2 Arabidopsis double mutant as 35% of the bend roots did not display LR primordium 18 h after gravistimulation and no stage VI and VII primordia were found after 42 h. Overexpression of QSK1, on the other hand, resulted in only a slight delay in LR development.
We also tested the response of the qsk1.qsk2 double mutant and QSK1 overexpressing line to mannitol treatment. While Col-0 wild type showed a reduced number of LR in mannitol compared to control growth conditions, qsk1.qsk2 double mutant was not significantly affected (Fig. 6A, compare white and red box plots). Hence, in qsk1.qsk2 double mutant, the number of LR was not reduced further by mannitol exposure in comparison to control growth conditions. Expression of QSK1 in qsk1.qsk2 double mutant background complemented the phenotype restoring LR response (reduced LR number) to mannitol (Fig. 6A). In summary, LR development and response to mannitol is significantly affected by mutation in QSK1.
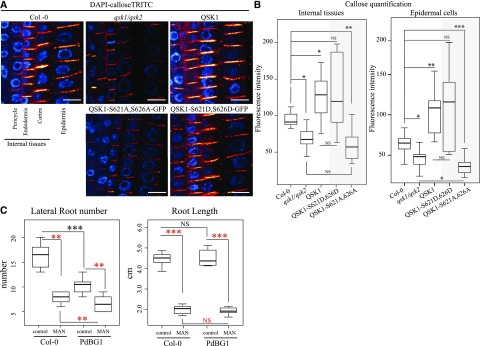
Since mannitol induces changes in callose deposition (Fig. 7), we used immunolocalization to detect callose levels in qsk1.qsk2 double mutant and QSK1 overexpressor line (Fig. 8). The qsk1.qsk2 double mutant showed reduced callose levels compared to wild-type seedlings, while the overexpressing QSK1 lines appear to accumulate more callose (Fig. 8, A and B). These results suggest that callose down-regulation may be accountable for qsk1.qsk2 double mutant LR phenotype. To test this hypothesis, we studied the root phenotype in a line ectopically expressing PdBG1, a plasmodesmata associated β1-3 glucanase (AT3G13560) that degrades callose (Benitez-Alfonso et al., 2013; Maule et al., 2013). Similar to the qsk1.qsk2 double mutant, overexpression of PdBG1 did not affect primary root length, but LR number was reduced compared to Col-0 in control conditions (Fig. 8C). After mannitol treatment, changes in LR number were reduced in the PdBG1 overexpressor to a lesser extent than wild type, partially resembling the qsk1.qsk2 double-mutant response. This suggests that ectopic callose degradation is, at least partly, related to the LR response in control and mannitol growth conditions. Taken together, these results suggest that QSK1 is necessary to regulate LR development and response to mannitol, and our results highlight a link between callose deposition and the observed phenotypes.
Figure 8.
QSK1 is involved in callose regulation at plasmodesmata, which depends on QSK1 phosphorylation status. (A and B) Quantification of callose levels in Col-0, qsk1.qsk2 double mutant, qsk1.qsk2 double mutant overexpressing QSK1-GFP, QSK1-S621D,S626D-GFP, or QSK1-S621A,S626A-GFP Arabidopsis roots. Seedlings were grown for 6 d on Murashige and Skoog plates. Both the qsk1.qsk2 double mutant and the qsk1.qsk2 double mutant expressing QSK1-S621A,S626A-GFP present a defect in callose deposition with reduced levels internal tissues and in epidermal cells compared to the Col-0. In the opposite way, overexpression of QSK1 and QSK1-S621D,S626D-GFP phosphomimic induces an increase in callose deposition. A, Representative confocal images of callose immunofluorescence (red) in roots. DAPI staining of DNA (blue) was performed to highlight the cellular organization of root tissues. Scale bars, 10 μm. B, Callose quantifications in “internal” root cell layers and epidermal cells. n = 3 experiments, 10 roots/experiments, 5 to 10 cell wall/cell lineage/roots. Two-tailed Wilcoxon statistical analysis, *P < 0.05; **P < 0.01; ***P < 0.001. C, LR number in wild-type Col-0 and PdBG1 overexpressing line. Arabidopsis lines were grown for 9 d on Murashige and Skoog plates for control conditions or 6 d then transferred to Murashige and Skoog plate containing 0.4 m mannitol before root phenotyping. LR number is represented by white and red box plots for control and mannitol treatment, respectively. In control conditions, PdBG1 overexpressor displays a decrease of LR number compared to the wild type. In response to mannitol treatment, Col-0 wild type and Arabidopsis seedlings overexpressing PdBG1 showed a decrease in LR number. The primary root length was measured in parallel to the LR (A) using FIJI software. None of the lines tested presented a significant root length difference compare to Col-0 in control conditions (white box plot). After mannitol treatment, all the lines were similarly affected with a reduction of the primary root length (red box plot). n = 3 experiments, 5 to 10 roots/lines. Two-tailed Wilcoxon statistical analysis: *P < 0.05; **P < 0.01; ***P < 0.001.
QSK1 Plasmodesmata Localization Is Required to Regulate Callose and the Root Response to Mannitol
Changes in QSK1 phosphorylation were found to be necessary for localization of the protein at plasmodesmata in response to mannitol. To investigate the implications of QSK1 phosphorylation for LR response to mannitol, we tested complementation of the qsk1.qsk2 double-mutant phenotype with both the QSK1 phosphomimic (QSK1-S621D,S626D-GFP) and the phosphodead (QSK1-S621A,S626A-GFP) mutant variants. Under control conditions, overexpression of both QSK1-S621D,S626D-GFP and QSK1-S621A,S626A-GFP variants in the qsk1.qsk2 double mutant background did not affect root length (Supplemental Fig. S3). Reduced LR phenotype in the qsk1.qsk2 double mutant was fully restored by expression of QSK1-S621D,S626D-GFP and only partially by expression of QSK1-S621A,S626A-GFP (Fig. 6A, white boxes). Concomitantly, lines expressing QSK1-S621A,S626A-GFP variant displayed a delay in LR primordium development with no stage VI and VII primordia at 42 h after gravistimulation, a phenotype resembling the qsk1.qsk2 double mutant (Fig. 6C). Next, we tested the phenotype of these lines in mannitol. As in wild-type Col-0, LR number was reduced in response to mannitol in qsk1.qsk2 double mutants expressing the phosphomimic but not the phosphodead QSK1 variant, suggesting that QSK1 phosphorylation is important for LR response to mannitol (Fig. 6A, compare white to red boxes).
We next investigated the effect of QSK1 phosphomimic and phosphodead variants on the callose mutant phenotype. We used immunolocalization to compare callose levels in wild type and in the qsk1.qsk2 double mutant expressing either QSK1-S621A,S626A-GFP or QSK1- S621Q,S626Q-GFP (Fig. 8, A and B). While callose levels in the qsk1.qsk2 double mutant expressing the phosphomimic version were comparable to QSK1 overexpressing line, the phosphodead variant displayed a reduction of callose levels comparable to the qsk1.qsk2 double mutant (Fig. 8, A and B). To summarize, expression and phosphorylation-dependent relocalization of QSK1 is important to regulate LR response to mannitol via a mechanism that modulates the levels of callose.
DISCUSSION
In this study, we report the rapid change of location of two PM-located LRR-RLKs in response to osmotic stress. Under standard growth conditions, both QSK1 and IMK2 show an exclusive PM localization, but exposure to salt or mannitol triggered their relocalization to plasmodesmata. This rearrangement happens remarkably fast, within the first 2 min after stimulation, suggesting that this process may be either posttranscriptionally or posttranslationally regulated. Both QSK1 and IMK2 were originally identified as enriched in our plasmodesmata-fraction, but they show no particular plasmodesmata association in Arabidopsis seedlings grown under normal condition when subcellular localization was examined by confocal. An early step of the plasmodesmata purification process includes a short plasmolysis treatment with 0.45 m mannitol, which aims at retracting the protoplast from the wall and minimizing contamination with the PM (Grison et al., 2015b). Most likely during this step, QSK1 and IMK2 relocate to plasmodesmata, which explain why they are found enriched in our proteomic screen while being in fact excluded from the pores in normal growth condition.
Dynamic plasmodesmal association is neither a general feature of PM-associated proteins nor of microdomain-associated proteins, such as REM1.2 and 1.3, whose localizations remain “static.” So far, receptor-like proteins that associate with plasmodesmata have been reported to be spatially and stably confined to the PM microdomain lining the pores (Thomas et al., 2008; Faulkner et al., 2013; Stahl et al., 2013; Caillaud et al., 2014; Vaddepalli et al., 2014; Carella et al., 2015; Lim et al., 2016). Conditional association with plasmodesmata has been reported for the endoplasmic reticulum-PM membrane contacts site protein Synaptotagmin SYTA, which within a few days postviral infection is recruited by Tobamovirus viral movement protein to plasmodesmata active in cell-to-cell spread (Levy et al., 2015). Recently, the group of M. Wrzaczek demonstrated that another class of RLK, the Cys-rich receptor-like kinase CRK2, reorganizes from the PM to plasmodesmata in response to salt stress in a phospholipase D-dependent manner (Hunter et al., 2019). Taken together, this suggests that plasmodesmata molecular composition is more dynamic than previously thought and changes in response to biotic and abiotic stimuli.
An important feature of the PM, which acts at the interface between the apoplastic and symplastic compartment, is its ability to respond to external and internal stimuli by remodeling its molecular organization. This process takes many forms from the association/dissociation of proteins with nanodomains and complexes, through protein/protein and protein/lipid interactions, through the modification of endoplasmic reticulum-PM contacts, or posttranslational modification such as phosphorylation or ubiquitination (Demir et al., 2013; Gronnier et al., 2017; Dubeaux et al., 2018; Perraki et al., 2018; Lee et al., 2019). This most likely also applies to plasmodesmata, which need to quickly integrate development and biotic/abiotic stimuli to regulate their aperture. Spatiotemporal rearrangement of RLKs from the bulk PM to plasmodesmata may provide a different membrane environment and protein partners, which in turn could modify the protein function. In line with that, the RLK CRINKLY4 is known to interact with CLAVATA1, and the heteromer displays different composition at the PM and at plasmodesmata, indicating that local territory indeed modifies receptor activity/function (Stahl et al., 2013).
In plants, protein mobility within the plane of the PM is restricted by the cell wall and appears to be rather slow compared to animal cells (Martinière et al., 2012). Rapid rearrangement of QSK1 within the plane of the PM was therefore unexpected. This pushed us to investigate the molecular determinants controlling plasmodesmata association. Our group previously showed that the specialized PM domain of plasmodesmata is enriched in sterols and sphingolipids. Altering the membrane sterol pool lead to plasmodesmata protein mislocalization and defects in callose-mediated cell-to-cell trafficking (Grison et al., 2015a). Both QSK1 and IMK2 were reported to associate with DRM (Shahollari et al., 2005; Kierszniowska et al., 2009; Keinath et al., 2010; Demir et al., 2013; Srivastava et al., 2013; Szymanski et al., 2015), hence supposedly sterol- and sphingolipid-enriched PM nanodomains. However, inhibiting sterol- and VLCFA-sphingolipid synthesis had no effect on QSK1 relocalization to plasmodesmata upon stress conditions (Kierszniowska et al., 2009; Demir et al., 2013).
Protein phosphorylation has been reported as one of the early posttranslational responses to osmotic stress (Nikonorova et al., 2018), and QSK1 has multiple phosphorylation sites and is phosphorylated in response to abiotic stress (Niittylä et al., 2007; Chang et al., 2012). Using phosphomutants of QSK1, we showed that the phosphorylation status of QSK1 is important for subcellular localization with the QSK1-S621D,S626D-GFP phosphomimic mutant partially associating with plasmodesmata even in control conditions, while the QSK1-S621A,S626A-GFP phosphodead mutant was significantly affected in its capacity to localize to plasmodesmata after mannitol treatment. Having said that, QSK1-S621A,S626A-GFP mutant is still able to partially localize to the pores after stress (PD index of 1.2), indicating that other factors may be important to control this process. For QSK1, localization to the PM microdomains was previously shown to depend on cytoskeletal integrity (Szymanski et al., 2015), and involvement of cytoskeletal components in reorganization to plasmodesmata should be investigated in further studies.
An explanation for why QSK1 and IMK2 cluster at plasmodesmata in response to mannitol and NaCl and how this exactly impacts on plasmodesmata function remains to be determined. We postulate that our mannitol treatment induces a change in plasmodesmata permeability through callose deposition or removal as it has been observed for cold, oxidative, nutrient, and biotic stresses (Sivaguru et al., 2000; Bilska and Sowinski, 2010; Benitez-Alfonso et al., 2011; Zavaliev et al., 2011; Faulkner et al., 2013; Cui and Lee, 2016; O’Lexy et al., 2018). Callose is a well-established regulator of plasmodesmata-mediated cell-to-cell communication, and modifying callose deposition at the pores has a strong impact on numerous developmental programs, including LR formation (Benitez-Alfonso et al., 2013; Maule et al., 2013; Otero et al., 2016). The balance between callose synthesis and degradation is tightly regulated through a set of callose-related enzymes. The plasmodesmata-associated β1-3 glucanase PdBG1 (AT3G13560) is involved in modulating plasmodesmata aperture through callose degradation and has been implicated in LR formation and patterning (Benitez-Alfonso et al., 2013; Maule et al., 2013). Our data indicate that the QSK1-induced LR response in control and mannitol stress condition is likely to involve callose. Modifying plasmodesmata permeability by overexpressing PdBG1 affects LR phenotype and resembles that of the qsk1.qsk2 double mutant and the qsk1.qsk2 double mutant overexpressing QSK1-S621A,S626A-GFP lines, which are also defective in callose regulation. However, at this stage it is unclear why fast QSK1 relocalization would be required to control a slow process such LR formation. Possibly, QSK1 reorganization to plasmodesmata is part of an early stress response, and other partners are likely to act in combination to incorporate and sustain stress response at the pores.
To conclude, our work highlights the complex and dynamic regulation of symplastic intercellular communication in response to osmotic stress, a situation that plants are often confronted with in their environment. We propose that reorganization of PM-located RLKs to plasmodesmata is an ingenious mechanism that combines “stress sensing” at the bulk PM and modulation of cell-to-cell trafficking at plasmodesmata.
MATERIALS AND METHODS
Proteomic Analyses
We used the label-free plasmodesmata proteomic analysis of Brault et al. (2019) to select RLK candidates. For that, all members of the LRR-RLK family that displayed with a significant fold change (plasmodesmata/PM enrichment ratio > 2) were selected (Supplemental Table S1) and cross referenced with DRM proteomic studies (Supplemental Table S2).
Cloning
IMK2 and QSK1 were cloned using classical gateway system with p221 as DNR plasmid and pGBW661 or pGBW641 as Destination plasmid comprising 35S promoter and C-terminal tag GFP and TagRFP, respectively. Amplifications were run on plasmid containing the full-length cDNA (U12366 TAIR), purified with QIAquick gel extraction kit, and inserted into p221 DNR (see Supplemental Table S4 for primer details) and then inserted into Destination plasmid for stable expression in Arabidopsis (Arabidopsis thaliana) or for transient expression in Nicotiana benthamiana.
Plant Material and Growth Conditions
The following Arabidopsis transgenic lines were used: p35S:Lti6b-mCherry, p35S::PIP2;1-GFP, pREM1.2:REM1.2-YFP (yellow fluorescent protein), pREM1.3:REM1.3-YFP, p35S::mCitrine-PdBG1, p35S:QSK1-GFP, p35S:QSK1-S621A,S626A-GFP, p35S:QSK1-S621D,S626D-GFP, and the mutant line qsk1/qsk2 (Cutler et al., 2000; Prak et al., 2008; Benitez-Alfonso et al., 2013; Jarsch et al., 2014; Szymanski et al., 2015; Wu et al., 2019).
For confocal microscopy, Arabidopsis seedlings were grown 6 d on agar (8 g/L) plates containing Murashige and Skoog salts including 2.2 g/L vitamins, 10 g/L Suc, and 0.5 g/L MES at pH 5.8 in a culture room at 22°C in long-day light conditions (150 µE/m2/s) followed by treatment with NaCl or mannitol (see section “Mannitol and NaCl treatments”).
For LR phenotyping, Arabidopsis seedlings were grown 9 d on agar (8 g/L) plates containing Murashige and Skoog salts including 2.2 g/L vitamins, 10 g/L Suc, and 0.5 g/L MES at pH 5.8 in a culture room at 22°C in long-day light conditions (150 µE/m2/s) for control conditions or 6 d then transferred to the same media supplemented with 0.4 m mannitol for another 3 d.
Mannitol and NaCl Treatments
For short-term treatment, mannitol (0.4 m solution) or NaCl (100 mm solution) were infiltrated in Arabidopsis cotyledons (for stable expression) or N. benthamiana leaves (for transient expression), and samples were immediately observed by confocal microscopy. For Arabidopsis roots, seedlings were grown for 6 d on half-strength Murashige and Skoog 1% (w/v) Suc agar plates in long-day conditions then transferred in liquid half-strength Murashige and Skoog 1% (w/v) Suc media containing 0.4 m mannitol for 3 h before analysis (confocal live imaging or immunolocalization against callose on whole-mount tissues). For control conditions, leaves/cotyledons were infiltrated with water and Arabidopsis roots incubated in half-strength Murashige and Skoog 1% (w/v) Suc media without mannitol.
For long-term mannitol treatment, seedlings were grown for 6 d on half-strength Murashige and Skoog 1% (w/v) Suc agar plates in long-day conditions, then transferred on half-strength Murashige and Skoog 1% (w/v) Suc agar plates containing 0.4 m mannitol for 3 d, before analysis.
Confocal Live Imaging
For transient expression in N. benthamiana, leaves of 3-week-old plants were pressure-infiltrated with GV3101 agrobacterium strains, previously electroporated with the relevant binary plasmids. Prior to infiltration, Agrobacteria cultures were grown in Luria and Bertani medium with appropriate antibiotics at 28°C for 1 d then diluted to one tenth and grown until the culture reached an OD600 of about 0.8. Bacteria were then pelleted and resuspended in water at a final OD600 of 0.3 for individual constructs, 0.2 each for the combination of two. Agroinfiltrated N. benthamiana leaves were imaged 3 d postinfiltration at room temperature using a confocal laser scanning microscope Zeiss LSM 880 using X63 oil lens. Immediately before imaging leaves were infiltrated with water 0.4 m mannitol or 100 mm NaCl solutions supplemented with 20 µg/mL aniline blue (Biosupplies) for plasmodesmata colocalization and PD index, ∼0.5-cm leaf pieces were cut out and mounted with the lower epidermis facing up onto glass microscope slides.
For Arabidopsis lines, seedlings were grown for 6 d on half-strength Murashige and Skoog 1% (w/v) Suc agar plate prior to treatment. For cotyledon observation, seedlings were vacuum infiltrated with water or 0.4 m mannitol treatment supplemented with 20 µg/mL aniline blue and immediately mounted onto glass microscope slides with the lower epidermis facing up for confocal observation. For roots, seedlings were incubated for 3 h with appropriate solution before observation.
For time-lapse imaging, QSK1-expressing Arabidopsis cotyledons were cut in half and dry mounted onto microscope glass and cover slip, and 0.4 m mannitol solution was gently injected between glass and cover slip and immediately followed by imaging.
For GFP and YFP imaging, excitation was performed with 2% to 8% of 488-nm laser power and fluorescence emission collected at 505 to 550 nm and 520 to 580 nm, respectively. For mRFP imaging, excitation was achieved with 2% to 5% of 561-nm laser power and fluorescence emission collected at 580 to 630 nm. For aniline blue imaging, excitation was performed with 0.5% to 6% of 405-nm laser power and fluorescence emission collected at 420 to 480 nm. For colocalization, sequential scanning was systematically used.
PD Index
Plasmodesmata depletion or enrichment was assessed by calculating the fluorescence intensity ratio between the GFP/YFP/mRFP/mCherry-tagged protein intensity at plasmodesmata (indicated PDLP1-mRFP or aniline blue) versus the PM outside plasmodesmata. Confocal images of leaf/cotyledon or roots epidermal cells (N. benthamiana or Arabidopsis) were acquired by sequential scanning of PDLP1-mRFP or aniline blue (as plasmodesmata markers) and GFP/YFP/mRFP/mCherry-tagged (see section "Confocal Live Imaging" for details). About 30 images of leaf epidermis cells were acquired with a minimum of three biological replicates. Individual images were then processed using Fiji by defining five to 20 regions of interest (ROIs) at plasmodesmata (using plasmodesmata marker to define the ROI) and five to 20 ROIs outside plasmodesmata. The ROI size and imaging condition were kept the same. The GFP/YFP/mRFP/mCherry-tagged protein mean intensity was measured for each ROI then averaged for single image. The PD index corresponds to intensity ratio between fluorescence intensity of proteins at plasmodesmata versus outside the pores. (see Supplemental Fig. S1).
Callose Quantification in Arabidopsis Roots by Whole-Mount Immunolocalization
Arabidopsis seedlings were grown on half-strength Murashige and Skoog 1% (w/v) Suc agar plate for 6 d then incubated 3 h in half-strength Murashige and Skoog 1% (w/v) Suc liquid media for control condition or half-strength Murashige and Skoog 1% (w/v) Suc liquid media containing 0.4 m mannitol, prior to fixation. The immunolocalization procedure was done according to Boutté and Grebe (2014). The callose antibody (Australia Biosupplies) was diluted to 1/300 in MTSB (microtubule stabilizing buffer) containing 5% (v/v) of neutral donkey serum. The secondary anti-mouse antibody coupled to tetramethylrhodamine was diluted to 1/300 in MTSB buffer containing 5% (v/v) of neutral donkey serum. Nuclei were stained using 4′,6-diamidino-2-phenylindole diluted to 1/200 in MTSB buffer for 20 min. Samples were then imaged with a Zeiss LSM 880 using X40 oil lens. 4′,6-diamidino-2-phenylindole excitation was performed using 0.5% of 405 laser power, and fluorescence collected at 420 to 480 nm; GFP excitation was performed using 5% of 488-nm laser power and fluorescence emission collected at 505 to 550 nm; tetramethylrhodamine excitation was performed with 5% of 561-nm power and fluorescence collected at 569 to 590 nm. All parameters were kept consistent between experiments to allow quantifications.
Callose deposition was then quantified using Fiji software. Callose fluorescence intensity was measured at the apicobasal cell walls of epidermal cells and internal layers endodermal and cortex cells for the “inner tissues.” A total of 20 cell wall intensity were measured per cell lineage (e.g. 20 epidermal; 20 endodermal + 20 cortex) per roots, 10 roots per transgenic lines. Two biological replicates were performed.
LR Number and LR Primordium Developmental Stage Quantifications
Arabidopsis seedlings were grown 9 d on half-strength Murashige and Skoog 1% (w/v) Suc agar plates for control or 6 d on half-strength Murashige and Skoog 1% (w/v) Suc agar plates then transferred for 3 d on half-strength Murashige and Skoog 1% (w/v) Suc agar plates supplemented with 0.4 m mannitol. The number of emerged LRs and LR primordia (from stage 2) was imaged and quantified using a macroscope Axiozoom Leica with a 150× magnification. LR primordium stages were analyzed according to Péret et al. (2012).
Root length was measured by using Image J software after taking pictures of the plates with Bio-Rad Chemidoc.
Sterol and Sphingolipid Inhibitor Treatments
For sterols and sphingolipids inhibitor experiments, 5-d-old seedlings were transferred to Murashige and Skoog agar plates containing 100 μg/mL Fenpropimorph (stock solution 100 mg/mL in DMSO) or 100 nm Metazachlor (stock solution 1 mm in DMSO). Control plates contained an equal amount of 0.1% (v/v) DMSO solvent. Seedlings were observed by confocal microscopy 48 h after treatment and lipid analysis was performed in parallel (see below for details).
Lipid Analysis
For the analysis of total fatty acids by gas chromatography mass spectrometry (GC-MS; FAMES), Arabidopsis seedlings were harvested 48 h after transfer on Murashige and Skoog plates containing 100 nm Metazachlor or 0.1% (v/v) DMSO. Transmethylation and trimethylsilylation of fatty acids from 150 mg of fresh material was performed as described in Grison et al. (2015a). An HP-5MS capillary column (5% [w/v] phenyl-methyl-siloxane, 30-m, 250-mm, and 0.25-mm film thickness; Agilent) was used with helium carrier gas at 2 mL/min; injection was done in splitless mode; injector and mass spectrometry detector temperatures were set to 250°C; the oven temperature was held at 50°C for 1 min, then programmed with a 25°C/min ramp to 150°C (2-min hold) and a 10°C/min ramp to 320°C (6-min hold). Quantification of nonhydroxylated and hydroxylated fatty acids was based on peak areas that were derived from the total ion current.
For sterol analysis by GC-MS, Arabidopsis seedlings were harvested 48 h after transfer on Murashige and Skoog plates containing 100 µg/mL Fenpropimorph or 0.1% (v/v) DMSO. A saponification of 150 mg of fresh material was performed by adding 1 mL of ethanol containing the internal standard α-cholestanol (25 µg/mL) and 100 mL of 11 n KOH and incubating it for 1 h at 80°C. After the addition of 1 mL of hexane and 2 mL of water, the sterol-containing upper phase was recovered and evaporated under an N2 gas stream. Sterols were derivatized by (N, O-Bis(trimethylsily)trifluoroacetamide) as described for FAMES and resuspended in 100 µL of hexane before analysis by GC-MS (Grison et al., 2015a).
Phylogenetic Tree Construction
Sequence alignment and phylogenetic tree building were performed with SeaView version 4 multiplatform program. Alignment algorithm chosen was ClustalW and PhyML version 3 was used to reconstruct maximum-likelihood tree of 34 clade III LRR-RLKs (ten Hove et al., 2011).
Statistical Analysis
Statistical analyses were done using “R” software (https://www.r-project.org). For all analyses, we applied “two-tailed Wilcoxon rank-sum test,” which is a nonparametrical statistical test commonly used for a small range number of replicate (e.g. n < 20).
Accession Numbers
Sequence data from this article can be found at https://www.arabidopsis.org under accession numbers: QSK1: AT3G02880, IMK2: AT3G51740, PdBG1: AT3G57270, PMA2: AT4G30190.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. PD index calculation.
Supplemental Figure S2. Reorganization of QSK1 at plasmodesmata upon salt stress.
Supplemental Figure S3. Root length measurement.
Supplemental Figure S4. Phylogenetic tree of clade III LRR-RLKs.
Supplemental Figure S5. Expression pattern of QSK1 and QSK2.
Supplemental Table S1. List of LRR-RLKs extracted from the label-free Arabidopsis plasmodesmata proteome from Brault et al. (2019).
Supplemental Table S2. RLKs associated with lipid microdomains according to seven DRM proteomic studies.
Supplemental Table S3. QSK1 phosphorylation sites.
Supplemental Table S4. List of primers used in this work.
Supplemental Video S1. Time-lapse confocal movie showing the rapid relocalization of QSK1-GFP immediately after mannitol treatment.
Footnotes
This work was supported by the National Agency for Research (grant ANR-14-CE19-0006-01 to E.M.B.), Osez l’interdisciplinarité CNRS OSEZ-2017-BRIDGING CNRS program to E.M.B., the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme (grant agreement no. 772103-BRIDGING to E.M.B.). P.K. was supported by a BBSRC DTP (BB/M011151/1). Y.B.-A.’s lab work is supported by research grants from the Leverhulme Trust (RPG-2016-136). Work in W.X.S.’s lab was funded by the Deutsche Forschungsgemeinschaft, grant SCHU1533/9-1 to W.X.S. and X.N.W.
Articles can be viewed without a subscription.
References
- Amari K, Boutant E, Hofmann C, Schmitt-Keichinger C, Fernandez-Calvino L, Didier P, Lerich A, Mutterer J, Thomas CL, Heinlein M, et al. (2010) A family of plasmodesmal proteins with receptor-like properties for plant viral movement proteins. PLoS Pathog 6: e1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Faulkner C, Ritzenthaler C, Maule AJ (2010) Plasmodesmata: Gateways to local and systemic virus infection. Mol Plant Microbe Interact 23: 1403–1412 [DOI] [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Jackson D, Maule A (2011) Redox regulation of intercellular transport. Protoplasma 248: 131–140 [DOI] [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Faulkner C, Pendle A, Miyashima S, Helariutta Y, Maule A (2013) Symplastic intercellular connectivity regulates lateral root patterning. Dev Cell 26: 136–147 [DOI] [PubMed] [Google Scholar]
- Bilska A, Sowinski P (2010) Closure of plasmodesmata in maize (Zea mays) at low temperature: A new mechanism for inhibition of photosynthesis. Ann Bot 106: 675–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocharov EV, Lesovoy DM, Pavlov KV, Pustovalova YE, Bocharova OV, Arseniev AS (2016) Alternative packing of EGFR transmembrane domain suggests that protein-lipid interactions underlie signal conduction across membrane. Biochim Biophys Acta 1858: 1254–1261 [DOI] [PubMed] [Google Scholar]
- Boutté Y, Grebe M (2014) Immunocytochemical fluorescent in situ visualization of proteins in Arabidopsis. Methods Mol Biol 1062: 453–472 [DOI] [PubMed] [Google Scholar]
- Brault ML, Petit JD, Immel F, Nicolas WJ, Glavier M, Brocard L, Gaston A, Fouché M, Hawkins TJ, Crowet J-M, et al. (2019) Multiple C2 domains and transmembrane region proteins (MCTPs) tether membranes at plasmodesmata. EMBO Rep e47182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bücherl CA, Jarsch IK, Schudoma C, Segonzac C, Mbengue M, Robatzek S, MacLean D, Ott T, Zipfel C (2017) Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife 6: 25114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacas J-L, Buré C, Grosjean K, Gerbeau-Pissot P, Lherminier J, Rombouts Y, Maes E, Bossard C, Gronnier J, Furt F, et al. (2016) Revisiting plant plasma membrane lipids in tobacco: A focus on sphingolipids. Plant Physiol 170: 367–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud M-C, Wirthmueller L, Sklenar J, Findlay K, Piquerez SJM, Jones AME, Robatzek S, Jones JDG, Faulkner C (2014) The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog 10: e1004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carella P, Isaacs M, Cameron RK (2015) Plasmodesmata-located protein overexpression negatively impacts the manifestation of systemic acquired resistance and the long-distance movement of Defective in Induced Resistance1 in Arabidopsis. Plant Biol (Stuttg) 17: 395–401 [DOI] [PubMed] [Google Scholar]
- Chang IF, Hsu JL, Hsu PH, Sheng WA, Lai SJ, Lee C, Chen CW, Hsu JC, Wang SY, Wang LY, et al. (2012) Comparative phosphoproteomic analysis of microsomal fractions of Arabidopsis thaliana and Oryza sativa subjected to high salinity. Plant Sci 185-186: 131–142 [DOI] [PubMed] [Google Scholar]
- Chen Y, Hoehenwarter W, Weckwerth W (2010) Comparative analysis of phytohormone-responsive phosphoproteins in Arabidopsis thaliana using TiO2-phosphopeptide enrichment and mass accuracy precursor alignment. Plant J 63: 1–17 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distrance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Cui W, Lee J-Y (2016) Arabidopsis callose synthases CalS1/8 regulate plasmodesmal permeability during stress. Nat Plants 2: 16034. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Medzihradszky A, Suzaki T, Lohmann JU (2014) A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc Natl Acad Sci USA 111: 14619–14624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak KI, Malamy J (2005) Osmotic regulation of root system architecture. Plant J 43: 17–28 [DOI] [PubMed] [Google Scholar]
- Demir F, Horntrich C, Blachutzik JO, Scherzer S, Reinders Y, Kierszniowska S, Schulze WX, Harms GS, Hedrich R, Geiger D, et al. (2013) Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proc Natl Acad Sci USA 110: 8296–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubeaux G, Neveu J, Zelazny E, Vert G (2018) Metal sensing by the IRT1 transporter-receptor orchestrates its own degradation and plant metal nutrition. Mol Cell 69: 953–964.e5 [DOI] [PubMed] [Google Scholar]
- Faulkner C. (2013) Receptor-mediated signaling at plasmodesmata. Front Plant Sci 4: 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C, Petutschnig E, Benitez-Alfonso Y, Beck M, Robatzek S, Lipka V, Maule AJ (2013) LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc Natl Acad Sci USA 110: 9166–9170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Calvino L, Faulkner C, Walshaw J, Saalbach G, Bayer E, Benitez-Alfonso Y, Maule A (2011) Arabidopsis plasmodesmal proteome. PLoS One 6: e18880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher KL, Sozzani R, Lee C-M (2014) Intercellular protein movement: Deciphering the language of development. Annu Rev Cell Dev Biol 30: 207–233 [DOI] [PubMed] [Google Scholar]
- Grison MS, Brocard L, Fouillen L, Nicolas W, Wewer V, Dörmann P, Nacir H, Benitez-Alfonso Y, Claverol S, Germain V, et al. (2015a) Specific membrane lipid composition is important for plasmodesmata function in Arabidopsis. Plant Cell 27: 1228–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grison MS, Fernandez-Calvino L, Mongrand S, Bayer EMF (2015b) Isolation of plasmodesmata from Arabidopsis suspension culture cells. Methods Mol Biol 1217: 83–93 [DOI] [PubMed] [Google Scholar]
- Gronnier J, Crowet J-M, Habenstein B, Nasir MN, Bayle V, Hosy E, Platre MP, Gouguet P, Raffaele S, Martinez D, et al. (2017) Structural basis for plant plasma membrane protein dynamics and organization into functional nanodomains. eLife 6: e26404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann MA, Perret AM, Carde JP, Cassagne C, Moreau P (2002) Inhibition of the sterol pathway in leek seedlings impairs phosphatidylserine and glucosylceramide synthesis but triggers an accumulation of triacylglycerols. Biochim Biophys Acta 1583: 285–296 [DOI] [PubMed] [Google Scholar]
- He J-X, Fujioka S, Li T-C, Kang SG, Seto H, Takatsuto S, Yoshida S, Jang J-C (2003) Sterols regulate development and gene expression in Arabidopsis. Plant Physiol 131: 1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hem S, Rofidal V, Sommerer N, Rossignol M (2007) Novel subsets of the Arabidopsis plasmalemma phosphoproteome identify phosphorylation sites in secondary active transporters. Biochem Biophys Res Commun 363: 375–380 [DOI] [PubMed] [Google Scholar]
- Hofman EG, Ruonala MO, Bader AN, van den Heuvel D, Voortman J, Roovers RC, Verkleij AJ, Gerritsen HC, van Bergen En Henegouwen PMP (2008) EGF induces coalescence of different lipid rafts. J Cell Sci 121: 2519–2528 [DOI] [PubMed] [Google Scholar]
- Hsu JL, Wang LY, Wang SY, Lin CH, Ho KC, Shi FK, Chang IF (2009) Functional phosphoproteomic profiling of phosphorylation sites in membrane fractions of salt-stressed Arabidopsis thaliana. Proteome Sci 7: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter K, Kimura S, Rokka A, Tran HC, Toyota M, Kukkonen JP, Wrzaczek M (2019) CRK2 enhances salt tolerance by regulating callose deposition in connection with PLDα1. Plant Physiol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isner JC, Begum A, Nuehse T, Hetherington AM, Maathuis FJM (2018) KIN7 kinase regulates the vacuolar TPK1 K+ channel during stomatal closure. Curr Biol 28: 466–472.e4 [DOI] [PubMed] [Google Scholar]
- Jarsch IK, Konrad SSA, Stratil TF, Urbanus SL, Szymanski W, Braun P, Braun K-HH, Ott T (2014) Plasma membranes are subcompartmentalized into a plethora of coexisting and diverse microdomains in Arabidopsis and Nicotiana benthamiana. Plant Cell 26: 1698–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato-Asano H, Grossniklaus U, Schulze WX, Robatzek S, Panstruga R (2010) PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J Biol Chem 285: 39140–39149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszniowska S, Seiwert B, Schulze WX (2009) Definition of Arabidopsis sterol-rich membrane microdomains by differential treatment with methyl-beta-cyclodextrin and quantitative proteomics. Mol Cell Proteomics 8: 612–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline KG, Barrett-Wilt GA, Sussman MR (2010) In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc Natl Acad Sci USA 107: 15986–15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad SSA, Popp C, Stratil TF, Jarsch IK, Thallmair V, Folgmann J, Marín M, Ott T (2014) S-acylation anchors remorin proteins to the plasma membrane but does not primarily determine their localization in membrane microdomains. New Phytol 203: 758–769 [DOI] [PubMed] [Google Scholar]
- Kragler F, Monzer J, Shash K, Xoconostle-Cázares B, Lucas WJ (1998) Cell-to-cell transport of proteins: Requirement for unfolding and characterization of binding to a putative plasmodesmal receptor. Plant J 15: 367–381 [Google Scholar]
- Kumar M, Yusuf MA, Yadav P, Narayan S, Kumar M (2019) Overexpression of chickpea defensin gene confers tolerance to water-deficit stress in Arabidopsis thaliana. Front Plant Sci 10: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Vanneste S, Pérez-Sancho J, Benitez-Fuente F, Strelau M, Macho AP, Botella MA, Friml J, Rosado A (2019) Ionic stress enhances ER-PM connectivity via phosphoinositide-associated SYT1 contact site expansion in Arabidopsis. Proc Natl Acad Sci USA 116: 1420–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Wang X, Cui W, Sager R, Modla S, Czymmek K, Zybaliov B, van Wijk K, Zhang C, Lu H, et al. (2011) A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 23: 3353–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Erlanger M, Rosenthal M, Epel BL (2007) A plasmodesmata-associated beta-1,3-glucanase in Arabidopsis. Plant J 49: 669–682 [DOI] [PubMed] [Google Scholar]
- Levy A, Zheng JY, Lazarowitz SG (2015) Synaptotagmin SYTA forms ER-plasma membrane junctions that are recruited to plasmodesmata for plant virus movement. Curr Biol 25: 2018–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim GH, Shine MB, de Lorenzo L, Yu K, Cui W, Navarre D, Hunt AG, Lee JY, Kachroo A, Kachroo P (2016) Plasmodesmata localizing proteins regulate transport and signaling during systemic acquired immunity in plants. Cell Host Microbe 19: 541–549 [DOI] [PubMed] [Google Scholar]
- Liu L, Liu C, Hou X, Xi W, Shen L, Tao Z, Wang Y, Yu H (2012) FTIP1 is an essential regulator required for florigen transport. PLoS Biol 10: e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor DR, Deak KI, Ingram PA, Malamy JE (2008) Root system architecture in Arabidopsis grown in culture is regulated by sucrose uptake in the aerial tissues. Plant Cell 20: 2643–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinière A, Lavagi I, Nageswaran G, Rolfe DJ, Maneta-Peyret L, Luu D-T, Botchway SW, Webb SED, Mongrand S, Maurel C, et al. (2012) Cell wall constrains lateral diffusion of plant plasma-membrane proteins. Proc Natl Acad Sci USA 109: 12805–12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule AJ, Gaudioso-Pedraza R, Benitez-Alfonso Y (2013) Callose deposition and symplastic connectivity are regulated prior to lateral root emergence. Commun Integr Biol 6: e26531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami A, Fujiwara M, Furuto A, Fukao Y, Yamashita T, Kamo M, Kawamura Y, Uemura M (2009) Alterations in detergent-resistant plasma membrane microdomains in Arabidopsis thaliana during cold acclimation. Plant Cell Physiol 50: 341–359 [DOI] [PubMed] [Google Scholar]
- Miyashima S, Roszak P, Sevilem I, Toyokura K, Blob B, Heo JO, Mellor N, Help-Rinta-Rahko H, Otero S, Smet W, et al. (2019) Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature 565: 490–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsomme P, Dambly S, Maudoux O, Boutry M (1998) Single point mutations distributed in 10 soluble and membrane regions of the Nicotiana plumbaginifolia plasma membrane PMA2 H+-ATPase activate the enzyme and modify the structure of the C-terminal region. J Biol Chem 273: 34837–34842 [DOI] [PubMed] [Google Scholar]
- Nicolas WJ, Grison MS, Bayer EMF (2017) Shaping intercellular channels of plasmodesmata: The structure-to-function missing link. J Exp Bot 69: 91–103 [DOI] [PubMed] [Google Scholar]
- Niittylä T, Fuglsang AT, Palmgren MG, Frommer WB, Schulze WX (2007) Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol Cell Proteomics 6: 1711–1726 [DOI] [PubMed] [Google Scholar]
- Nikonorova N, Van den Broeck L, Zhu S, van de Cotte B, Dubois M, Gevaert K, Inzé D, De Smet I (2018) Early mannitol-triggered changes in the Arabidopsis leaf (phospho)proteome reveal growth regulators. J Exp Bot 69: 4591–4607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Lexy R, Kasai K, Clark N, Fujiwara T, Sozzani R, Gallagher KL (2018) Exposure to heavy metal stress triggers changes in plasmodesmatal permeability via deposition and breakdown of callose. 69: 3715–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero S, Helariutta Y, Benitez-Alfonso Y (2016) Symplastic communication in organ formation and tissue patterning. Curr Opin Plant Biol 29: 21–28 [DOI] [PubMed] [Google Scholar]
- Péret B, Li G, Zhao J, Band LR, Voß U, Postaire O, Luu DT, Da Ines O, Casimiro I, Lucas M, et al. (2012) Auxin regulates aquaporin function to facilitate lateral root emergence. Nat Cell Biol 14: 991–998 [DOI] [PubMed] [Google Scholar]
- Perraki A, Gronnier J, Gouguet P, Boudsocq M, Deroubaix A-F, Simon V, German-Retana S, Legrand A, Habenstein B, Zipfel C, et al. (2018) REM1.3’s phospho-status defines its plasma membrane nanodomain organization and activity in restricting PVX cell-to-cell movement. PLoS Pathog 14: e1007378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prak S, Hem S, Boudet J, Viennois G, Sommerer N, Rossignol M, Maurel C, Santoni V (2008) Multiple phosphorylations in the C-terminal tail of plant plasma membrane aquaporins: Role in subcellular trafficking of AtPIP2;1 in response to salt stress. Mol Cell Proteomics 7: 1019–1030 [DOI] [PubMed] [Google Scholar]
- Raffaele S, Mongrand S, Gamas P, Niebel A, Ott T (2007) Genome-wide annotation of remorins, a plant-specific protein family: Evolutionary and functional perspectives. Plant Physiol 145: 593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagan BC, Ganusova EE, Fernandez JC, McCray TN, Burch-Smith TM (2018) RNA on the move: The plasmodesmata perspective. Plant Sci 275: 1–10 [DOI] [PubMed] [Google Scholar]
- Rossy J, Ma Y, Gaus K (2014) The organisation of the cell membrane: Do proteins rule lipids? Curr Opin Chem Biol 20: 54–59 [DOI] [PubMed] [Google Scholar]
- Roycewicz P, Malamy JE (2012) Dissecting the effects of nitrate, sucrose and osmotic potential on Arabidopsis root and shoot system growth in laboratory assays. Philos Trans R Soc Lond B Biol Sci 367: 1489–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon MS, Bayer EM (2013) Dissecting plasmodesmata molecular composition by mass spectrometry-based proteomics. Front Plant Sci 3: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahollari B, Peskan-Berghöfer T, Oelmüller R (2004) Receptor kinases with leucine-rich repeats are enriched in Triton X-100 insoluble plasma membrane microdomains from plants. Physiol Plant 122: 397–403 [Google Scholar]
- Shahollari B, Varma A, Oelmüller R (2005) Expression of a receptor kinase in Arabidopsis roots is stimulated by the basidiomycete Piriformospora indica and the protein accumulates in Triton X-100 insoluble plasma membrane microdomains. J Plant Physiol 162: 945–958 [DOI] [PubMed] [Google Scholar]
- Simpson C, Thomas C, Findlay K, Bayer E, Maule AJ (2009) An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21: 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaguru M, Fujiwara T, Samaj J, Baluska F, Yang Z, Osawa H, Maeda T, Mori T, Volkmann D, Matsumoto H (2000) Aluminum-induced 1→3-β-d-glucan inhibits cell-to-cell trafficking of molecules through plasmodesmata. A new mechanism of aluminum toxicity in plants. Plant Physiol 124: 991–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V, Malm E, Sundqvist G, Bulone V (2013) Quantitative proteomics reveals that plasma membrane microdomains from poplar cell suspension cultures are enriched in markers of signal transduction, molecular transport, and callose biosynthesis. Mol Cell Proteomics 12: 3874–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y, Faulkner C (2016) Receptor complex mediated regulation of symplastic traffic. Trends Plant Sci 21: 450–459 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Simon R (2013) Gated communities: Apoplastic and symplastic signals converge at plasmodesmata to control cell fates. J Exp Bot 64: 5237–5241 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Grabowski S, Bleckmann A, Kühnemuth R, Weidtkamp-Peters S, Pinto KG, Kirschner GK, Schmid JB, Wink RH, Hülsewede A, et al. (2013) Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr Biol 23: 362–371 [DOI] [PubMed] [Google Scholar]
- Szymanski WG, Zauber H, Erban A, Gorka M, Wu XN, Schulze WX (2015) Cytoskeletal components define protein location to membrane microdomains. Mol Cell Proteomics 14: 2493–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hove CA, Bochdanovits Z, Jansweijer VMA, Koning FG, Berke L, Sanchez-Perez GF, Scheres B, Heidstra R (2011) Probing the roles of LRR RLK genes in Arabidopsis thaliana roots using a custom T-DNA insertion set. Plant Mol Biol 76: 69–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CL, Bayer EM, Ritzenthaler C, Fernandez-Calvino L, Maule AJ (2008) Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol 6: 0060007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J, Amari K, Torrance L (2011) Plasmodesmata viewed as specialised membrane adhesion sites. Protoplasma 248: 39–60 [DOI] [PubMed] [Google Scholar]
- Tilsner J, Nicolas W, Rosado A, Bayer EM (2016) Staying tight: Plasmodesmal membrane contact sites and the control of cell-to-cell connectivity in plants. Annu Rev Plant Biol 67: 337–364 [DOI] [PubMed] [Google Scholar]
- Tylewicz S, Petterle A, Marttila S, Miskolczi P, Azeez A, Singh RK, Immanen J, Mähler N, Hvidsten TR, Eklund DM, et al. (2018) Photoperiodic control of seasonal growth is mediated by ABA acting on cell-cell communication. Science 360: 212–215 [DOI] [PubMed] [Google Scholar]
- Vaddepalli P, Herrmann A, Fulton L, Oelschner M, Hillmer S, Stratil TF, Fastner A, Hammes UZ, Ott T, Robinson DG, et al. (2014) The C2-domain protein QUIRKY and the receptor-like kinase STRUBBELIG localize to plasmodesmata and mediate tissue morphogenesis in Arabidopsis thaliana. Development 141: 4139–4148 [DOI] [PubMed] [Google Scholar]
- Vatén A, Dettmer J, Wu S, Stierhof YD, Miyashima S, Yadav SR, Roberts CJ, Campilho A, Bulone V, Lichtenberger R, et al. (2011) Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell 21: 1144–1155 [DOI] [PubMed] [Google Scholar]
- Wang X, Sager R, Cui W, Zhang C, Lu H, Lee J-Y (2013) Salicylic acid regulates Plasmodesmata closure during innate immune responses in Arabidopsis. Plant Cell 25: 2315–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattelet-Boyer V, Brocard L, Jonsson K, Esnay N, Joubès J, Domergue F, Mongrand S, Raikhel N, Bhalerao RP, Moreau P, et al. (2016) Enrichment of hydroxylated C24- and C26-acyl-chain sphingolipids mediates PIN2 apical sorting at trans-Golgi network subdomains. Nat Commun 7: 12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, O’Lexy R, Xu M, Sang Y, Chen X, Yu Q, Gallagher KL (2016) Symplastic signaling instructs cell division, cell expansion, and cell polarity in the ground tissue of Arabidopsis thaliana roots. Proc Natl Acad Sci USA 113: 11621–11626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XN, Sanchez Rodriguez C, Pertl-Obermeyer H, Obermeyer G, Schulze WX (2013) Sucrose-induced receptor kinase SIRK1 regulates a plasma membrane aquaporin in Arabidopsis. Mol Cell Proteomics 12: 2856–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Chu L, Xi L, Pertl-Obermeyer H, Li Z, Sklodowski K, Sanchez-Rodriguez C, Obermeyer G, Schulze WX (2019) Sucrose-Induced Receptor Kinase 1 is modulated by an interacting kinase with short extracellular domain. Mol Cell Proteomics 18: 1556–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Cheval C, Laohavisit A, Hocking B, Chiasson D, Olsson TSG, Shirasu K, Faulkner C, Gilliham M (2017) A calmodulin-like protein regulates plasmodesmal closure during bacterial immune responses. New Phytol 215: 77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Wang P, Wang L, Renzi E, Radivojac P, Tang H, Arnold R, Zhu JK, Tao WA (2013) Quantitative measurement of phosphoproteome response to osmotic stress in arabidopsis based on Library-Assisted eXtracted Ion Chromatogram (LAXIC). Mol Cell Proteomics 12: 2354–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavaliev R, Ueki S, Epel BL, Citovsky V (2011) Biology of callose (β-1,3-glucan) turnover at plasmodesmata. Protoplasma 248: 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Ma H, Feng S, Gong S, Wang J (2018) A novel sugar transporter from Dianthus spiculifolius, DsSWEET12, affects sugar metabolism and confers osmotic and oxidative stress tolerance in Arabidopsis. Int J Mol Sci 19: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]