In rice, OsSHOC1 and OsPTD constitute an XPF-ERCC1-like complex and collaborate with OsMSH5, OsHEI10, and OsZIP4 to control class I meiotic crossover formation.
Abstract
In most eukaryotes, a set of conserved proteins that are collectively termed ZMM proteins (named for molecular zipper 1 [ZIP1], ZIP2, ZIP3, and ZIP4, MutS homologue 4 [MSH4] and MSH5, meiotic recombination 3, and sporulation 16 [SPO16] in yeast [Saccharomyces cerevisiae]) are essential for the formation of the majority of meiotic crossovers (COs). Recent reports indicated that ZIP2 acts together with SPO16 and ZIP4 to control CO formation through recognizing and stabilizing early recombination intermediates in budding yeast. However, whether this mechanism is conserved in plants is not clear. Here, we characterized the functions of SHORTAGE OF CHIASMATA 1 (OsSHOC1; ZIP2 ortholog) and PARTING DANCERS (OsPTD; SPO16 ortholog) and their interactions with other ZMM proteins in rice (Oryza sativa). We demonstrated that disruption of OsSHOC1 caused a reduction of CO numbers to ∼83% of wild-type CO numbers, whereas synapsis and early meiotic recombination steps were not affected. Furthermore, OsSHOC1 interacts with OsPTD, which is responsible for the same set of CO formations as OsSHOC1. In addition, OsSHOC1 and OsPTD are required for the normal loading of other ZMM proteins, and conversely, the localizations of OsSHOC1 and OsPTD were also affected by the absence of OsZIP4 and human enhancer of invasion 10 in rice (OsHEI10). OsSHOC1 interacts with OsZIP4 and OsMSH5, and OsPTD interacts with OsHEI10. Furthermore, bimolecular fluorescence complementation and yeast-three hybrid assays demonstrated that OsSHOC1, OsPTD, OsHEI10, and OsZIP4 were able to form various combinations of heterotrimers. Moreover, statistical and genetic analysis indicated that OsSHOC1 and OsPTD are epistatic to OsHEI10 and OsZIP4 in meiotic CO formation. Taken together, we propose that OsSHOC1, OsPTD, OsHEI10, and OsZIP4 form multiple protein complexes that have conserved functions in promoting class I CO formation.
Meiosis is a special cell division employed in organisms undergoing sexual reproduction. In one meiotic cycle, a single round of DNA replication is followed by two rounds of nuclear division separating homologous chromosomes (meiosis I) and then sister chromatids (meiosis II), and finally produces four daughter cells with half the number of chromosomes. Accurate separation of chromosomes depends on the successful completion of homologous recombination and formation of meiotic crossovers (COs). COs establish physical links between homologs, which play a vital role in balancing the opposite pulling force of the spindle at metaphase I (Wang and Copenhaver, 2018).
Homologous recombination is initiated by the induction of programmed double-strand breaks (DSBs). DSBs are generated by a widely conserved sporulation 11 (SPO11)-meiotic topoisomerase VIB-like (MTOPVIB) protein complex (Fu et al., 2016; Robert et al., 2016; Vrielynck et al., 2016; Xue et al., 2016). DSBs are processed by the radiation-sensitive 50 (RAD50)-meiotic recombination 11-X-ray sensitive 2 complex to produce 3′-single-stranded DNA (ssDNA) tails that are bound sequentially by replication protein A and recombinases RAD51 and DNA meiotic recombinase 1 (DMC1). Subsequently, RAD51 and DMC1 mediate invasion of these nucleoprotein filaments into the intact homologous DNA duplexes to form stable single-end invasion (SEI) intermediates (Neale and Keeney, 2006). With capturing of the 3′-end on the other side of the break, a recombination intermediate termed the double Holliday junction (dHj) is formed. The differential resolution of dHjs may produce COs or non-COs (Roeder, 1997; Börner et al., 2004; Gerton and Hawley, 2005). COs are generated via two pathways in the vast majority of organisms (Higgins et al., 2004). Class I COs ensure formation of the obligate CO, so that each homologous chromosome pair receives at least one CO (Osman et al., 2011). The class I pathway is interference sensitive, whereby one CO reduces the probability of a second CO occurring in an adjacent interval and is responsible for >75% of COs in most eukaryotes. The class II, interference-insensitive pathway produces randomly distributed COs and accounts for <25% of COs (Börner et al., 2004; Hollingsworth and Brill, 2004).
In budding yeast (Saccharomyces cerevisiae), synaptonemal complex (SC) assembly and interference-sensitive CO formation rely on a group of functionally related proteins consisting of molecular zipper 1 (ZIP1), ZIP2, ZIP3, ZIP4, MutS homologue 4 (MSH4), MSH5, meiotic recombination 3, and sporulation 16 (SPO16; ZMM proteins; Lynn et al., 2007; Shinohara et al., 2008). It has been demonstrated that ZMM orthologs are evolutionarily conserved in many other eukaryotes, including the model plants Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa; Wang and Copenhaver, 2018). ZIP1 encodes the transverse filament protein of the SC. Synapsis and recombination are functionally linked in budding yeast and in most animal and plant species. Failure in SC assembly causes defects in CO formation. Arabidopsis AtZYP1 and rice OsZEP1 also play essential roles in synapsis and CO formation. Contrary to ZIP1 and AtZYP1 promoting CO formation, OsZEP1 blocks CO formation in rice (Sym et al., 1993; Higgins et al., 2005; Wang et al., 2010). Except for MER3, disruption of the other six orthologs leads to a reduction of CO numbers to ∼85% of the wild-type level in Arabidopsis, while no obvious, or only mild, synapsis defects were observed in their mutants (Higgins et al., 2004, 2008; Chen et al., 2005; Mercier et al., 2005 Chelysheva et al., 2007, 2012; Macaisne et al., 2008). However, mutations of ZIP3/ZIP4/MER3 orthologs are merely responsible for ∼70% of COs in wild-type rice (Wang et al., 2009; Shen et al., 2012). Meiosis-specific mismatch repair family proteins MSH4 and MSH5 form heterodimers that stabilize dHJs and promote class I COs (Snowden et al., 2004; Higgins et al., 2008). In rice, osmsh4-osmsh5 mutants have the greatest reduction in Class I COs of the ZMMs accompanied by complete SC installation and may be required for normal localization of other ZMM proteins on chromosomes, indicating an upstream function (Luo et al., 2013; Zhang et al., 2014).
In yeast, ZIP2 together with ZIP3 and ZIP4 serve as the key components of the synapsis initiation complex. This complex promotes initiation of synapsis via localization to interhomolog recombination sites (Chua and Roeder, 1998; Tsubouchi et al., 2006). ZIP2 orthologs, which were named SHORTAGE IN CHIASMATA (SHOC1), were later identified in Arabidopsis, human, mouse, and rice. Similar to yeast, mutations in human (Homo sapiens) and mouse (Mus musculus) excision repair cross complementation group 4 (ERCC4)-helix-hairpin-helix (HhH)2-like-domain–containing protein SHOC1 disturb SC formation and were described to ensure CO formation by stabilizing CO recombination intermediates (De Muyt et al., 2018; Guiraldelli et al., 2018). In Arabidopsis, AtSHOC1 belongs to the same epistatic group as AtMSH5 and AtZIP4, controlling ∼85% of CO formation (Macaisne et al., 2008). Moreover, Arabidopsis ERCC1-like PARTING DANCERS (AtPTD) can interact with AtSHOC1 to specifically control interference-sensitive CO formation (Wijeratne et al., 2006; Macaisne et al., 2011). More recently, Ren et al. (2019) found that OsSHOC1 and its partner, OsPTD, are both required for CO formation in rice, but how they control CO formation and their relationships with other ZMM proteins remain unclear.
Xeroderma pigmentosa complementation group F (XPF)/methyl methansulfonate and ultraviolet-sensitive gene clone 81 (MUS81) family proteins have a similar catalytic domain followed by a tandem (HhH)2 motif (within the XPF protein family) or are flanked by single HhH motifs (within the MUS81 protein family), and exert their normal functions as homodimers (archaea) or heterodimers (eukaryotes; Ciccia et al., 2008). A common feature of ZIP2 and SHOC1 proteins is that they all possess a predicted XPF-like domain. Proteins belonging to the XPF/MUS81 family are conserved endonucleases that have essential roles in DNA replication and DNA damage repair. During evolution, XPF/MUS81 ancestors have diverged from a homodimeric complex to several types of heterodimers, including well-characterized XPF-ERCC1, MUS81-essential meiotic endonuclease 1 (EME1), MUS81-EME2, and defective in FA complementation group M (FANCM)-Fanconianemia-associated protein 24 kDa (FAAP24) in eukaryotes (Ciccia et al., 2008). It has been shown that ZIP2 and SPO16, AtSHOC1 and AtPTD, and SHOC1 and SPO16 form three meiosis-specific XPF-ERCC1-like complexes in budding yeast, Arabidopsis, and mammals, respectively (Macaisne et al., 2011; De Muyt et al., 2018; Zhang et al., 2019). Normally a heterodimer consists of a catalytic and a noncatalytic subunit. FANCM-FAAP24 heterodimers are one exception and have no detectable endonuclease activity, indicating that they may have noncatalytic roles, such as ssDNA binding or targeting endonucleases to defined DNA structures (Ciccia et al., 2008). ZIP2 and SHOC1 contain a typical (HhH)2 motif. However, compared with the canonical XPF endonucleases, the characteristic GDXnERKX3D active site of the nuclease domain is highly diverged or missing, suggesting that, similar to FANCM-FAAP24, they behave as DNA structure recognition modules rather than endonucleases. Recent studies demonstrated that the ZIP2-SPO16 heterodimer and human SHOC1 possess branched DNA binding activity (De Muyt et al., 2018; Guiraldelli et al., 2018). ZIP2-SPO16 and ZIP4 form a stable ZIP2-ZIP4-SPO16 (ZZS) complex that is able to stabilize meiotic early recombination intermediates to regulate CO formation (De Muyt et al., 2018).
Although ZIP2 homologs have conserved functions in CO formation, some functional differentiation is still clearly visible in different species. For example, both ZIP2/SHOC1 and SPO16 are indispensable for synapsis in yeast and mammals, but AtSHOC1 and AtPTD are not required for SC formation in Arabidopsis. Although their homologs in rice have been identified, their roles in homologous pairing and synapsis are unknown. For relatively well elucidated ZMM proteins in yeast, their role in plants is less understood, particularly how they cooperate to control CO formation. Here, we report the functional characterization of OsSHOC1 and its partner OsPTD in rice. Our results showed that both OsSHOC1 and OsPTD are dispensable for chromosome synapsis but are specifically required for the formation of interference-sensitive COs. OsSHOC1 and OsPTD form an XPF-ERCC1-like complex, and both are involved in forming heterotrimers with OsZIP4 and OsHEI10. In addition, OsZIP4 can mediate the interaction of OsSHOC1 and OsHEI10. Considering the relationship of localization interdependence among these four proteins (OsSHOC1, OsPTD, OsHEI10, and OsZIP4), we proposed that these ZMM proteins form heterotrimeric or heterotetrametric complexes to ensure formation of the obligate class I COs in rice.
RESULTS
Identification of osshoc1-2
OsSHOC1 has been reported recently to be involved in meiotic CO formation (Ren et al., 2019). Here, we isolated another allele of osshoc1 from a rice (9522) mutant library (Chen et al., 2006). The mutant exhibited no obvious defects at the vegetative development stage and showed normal panicle morphology, while bearing no seeds at maturity (Supplemental Fig. S1, A and B). A closer observation showed that the mutant spikelets developed normally, except that they contained slightly smaller and pale yellow anthers (Supplemental Fig. S1, C–F), which produced no viable pollen grains at the heading stage (Supplemental Fig. S1, G and H). When pollinated with wild-type pollen grains, the mutant still could not produce seeds, suggesting that it was defective in both male and female gamete development.
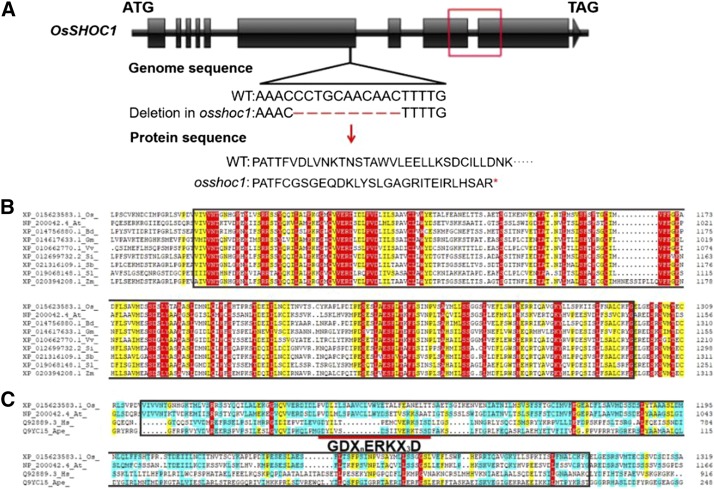
Sequence analysis indicated that an 11-bp deletion occurred within the sixth exon of OsSHOC1, causing premature translational termination after 849 amino acids and disrupting the ERCC4-(HhH)2-like domain (Fig. 1A). Homologous alignment revealed that the ERCC4-(HhH)2-like domain is highly conserved in plants (Fig. 1B). However, similar to AtSHOC1, an active GDXnERKX3D motif within the ERCC4-(HhH)2-like domain was not conserved in OsSHOC1 when compared with the active motif in canonical human and archaeal XPFs (Fig. 1C). For the complementation test, a genomic fragment of OsSHOC1 and a genomic fragment translationally fused with the enhanced GFP (eGFP) reporter gene were introduced into the mutant. Both constructs were able to fully restore the fertility of the mutant (Supplemental Fig. S2). These results confirmed that the sterility was caused by the mutation in OsSHOC1.
Figure 1.
OsSHOC1 encodes an XPF-like protein in rice. A, Structure of the OsSHOC1 gene. OsSHOC1 consists of 10 exons (black blocks) and 9 introns (black lines). An 11-bp deletion (red shot lines) causes a downstream frame shift and premature translational termination, which abolishes a conserved region (red box) in the predicted protein sequence. B, Protein sequence alignment of the conserved region in OsSHOC1 and its homologs, including the ERCC4-(HhH)2 like domain (black outline; 1070–1298 in OsSHOC1). Conserved amino acids are highlighted in red. At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Gm, Glycine max; Os, Oryza sativa; Sb, Sorghum bicolor; Si, Setaria italica; Sl, Solanum lycopersicum; Vv, Vitis vinifera; Zm, Zea mays. C, Protein sequence alignment of rice and Arabidopsis SHOC1 ERCC4-(HhH)2-like domain (black outline) with canonical XFP proteins HsXPF and ApeXPF. Ape, Aeropyrum pernix; At, Arabidopsis thaliana; Hs, Homo sapiens; Os, Oryza sativa. The short red line indicates the highly conserved active site GDXnERKX3D motif in HsXPF and ApeXPF.
The Number of Interference-Sensitive COs Is Dramatically Decreased in osshoc1-2
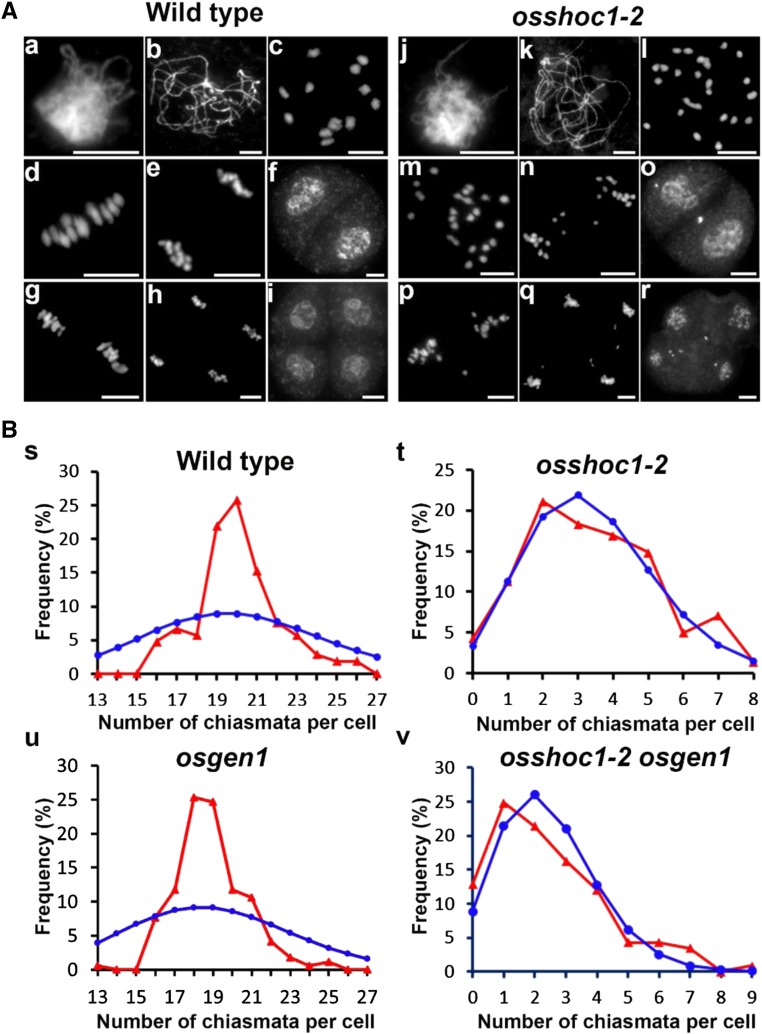
It has been reported that OsSHOC1 functions in the formation of meiotic COs, however, it is not yet clear whether OsSHOC1 controls class I COs in rice (Ren et al., 2019). We first analyzed wild-type and osshoc1-2 chromosome behaviors using 4',6-diamidino-2-phenylindole (DAPI) staining. Similar to the recent report Ren et al. (2019), no visible chromosome abnormalities were observed in the mutant during the zygotene or the pachytene when compared with the wild type (Fig. 2A, a, b, j, and k). At diakinesis, 12 bivalents could be clearly distinguished in wild-type meiocytes (Fig. 2A, c), whereas a mixture of univalents and bivalents were found in osshoc1-2 (Fig. 2A, l). At metaphase I, 12 further condensed bivalents were aligned on the equatorial plate (Fig. 2A, d). Subsequently, dyads and tetrads with equal chromosome numbers were produced at the end of meiosis I and II, respectively (Fig. 2A, e–i). Conversely, in osshoc1-2 meiocytes, univalents displayed a random distribution and separation at anaphase I (Fig. 2A, m and n), leading to the formation of a dyad with an unequal number of chromosomes in two daughter cells (Fig. 2A, o). At meiosis II, sister chromatids underwent similar random separation, and finally a tetrad with multiple micronuclei was produced after the second division (Fig. 2A, p–r).
Figure 2.
Meiosis progression and the chiasma frequency of the wild type and osshoc1-2 in rice. A, Chromosome behaviors of male meiocytes in the wild type (a–i) and osshoc1-2 (j–r) at the different stages of meiosis: zygotene (a and j), pachytene (b and k), diakinesis (c and l). metaphase I (d and m), anaphase I (e and n), dyad (f and o), metaphase II (g and p), telophase II (h and q), and tetrad (i and r). Bars = 5 µm. B, The observed (red triangles) and predicted (blue circles) Poisson distributions of chiasmata in wild-type (n = 105; s), osshoc1-2 (n = 142; t), osgen1 (n = 170; u), and osshoc1-2 osgen1 (n = 117; v) PMCs are shown.
To further understand how OsSHOC1 affected CO formation in rice, we calculated the frequency and distribution of chiasmata in wild-type and osshoc1-2 pollen mother cells (PMCs) by studying the shape of the bivalents at diakinesis and metaphase I, as previously described (Sanchez Moran et al., 2001). Compared with chiasmata per cell in the wild type (20.1 ± 2.10, n = 105), chiasma frequency dropped to 3.40 ± 1.89 (n = 142) per cell in osshoc1-2, an ∼83% reduction. In addition, chiasmata in the wild type were highly regulated in number as their distribution severely deviated from the predicted Poisson distribution that would reflect a numerically random distribution (Kolmogorov-Smirnov test, P < 0.01; Fig. 2B, s). However, the distribution of the residual chiasmata in osshoc1-2 was consistent with predicted Poisson distributions (Kolmogorov-Smirnov test, P > 0.05; Fig. 2B, t).
OsGEN1 is considered to function in the interference-insensitive CO pathway in rice (Wang et al., 2017). A significant reduction in the chiasma frequency was detected in the osshoc1-2 osgen1 double mutant (2.43 ± 1.71, n = 117) compared with that in the osshoc1-2 single mutant (3.40 ± 1.89, n = 142), and the distribution of residual chiasmata in osshoc1-2 osgen1 was consistent with a Poisson distribution (Kolmogorov-Smirnov test, P > 0.05) compared with that in the osgen1 single mutant (Kolmogorov-Smirnov test, P < 0.01; Fig. 2B, u and v; Table 1). These results clearly showed that OsSHOC1 and OsGEN1 function in two different CO-formation pathways. Taken together, we concluded that OsSHOC1 is an essential component in the class I CO pathway in rice.
Table 1. Chiasma frequency in different genotypes.
Multigroup comparisons of the means were carried out by one-way ANOVA test with post hoc contrasts by Newman-Keuls multiple-comparisons test (P < 0.01). The groups showing significant differences from each other are indicated by lowercase letters a through e. All values are represented as the mean ± sd. n, number of meiocytes observed.
| Genotype | Chiasma per Cell | Reference |
|---|---|---|
| Wild type | 20.1 ± 2.10 (n = 105)a | This study |
| osshoc1-2 | 3.40 ± 1.89 (n = 142)b | This study |
| osptd-1 | 3.12 ± 1.76 (n = 113)b | This study |
| oshei10 | 6.06 ± 2.86 (n = 167)c | (Wang et al., 2017) |
| oszip4 | 6.51 ± 2.56 (n = 47)c | This study |
| osgen1 | 18.9 ± 1.84 (n = 170)d | (Wang et al., 2017) |
| osshoc1-2 osptd-1 | 3.26 ± 1.84 (n = 109)b | This study |
| osshoc1-2 oshei10 | 3.36 ± 1.90 (n = 94)b | This study |
| osshoc1-2 oszip4 | 3.38 ± 1.78 (n = 122)b | This study |
| osshoc1-2 osgen1 | 2.43 ± 1.71 (n = 117)e | This study |
The osshoc1 Mutation Does Not Affect Early Steps of Homologous Chromosome Recombination
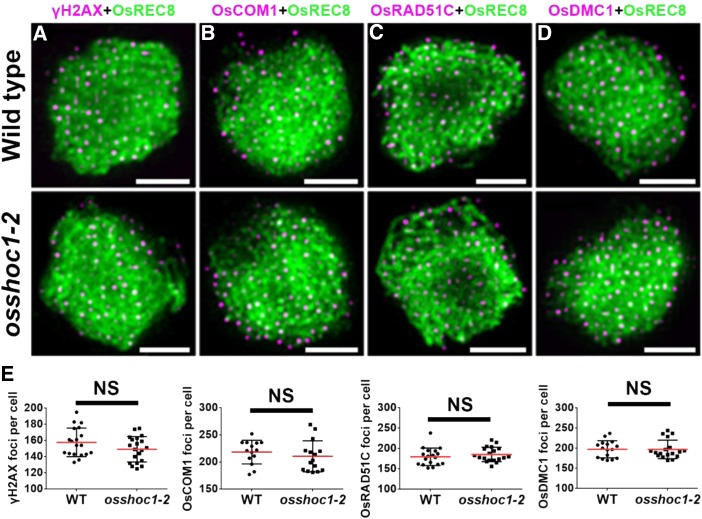
Meiotic recombination is initiated by DSB formation. The histone H2A variant, H2AX, is rapidly phosphorylated to γH2AX at DSB sites; thus, γH2AX foci are used as an indirect marker of DSBs (Hunter et al., 2001). To investigate whether the reduction of chiasmata in osshoc1-2 was caused by defects in DSB formation, we monitored numbers of γH2AX foci in wild-type and mutant meiocytes. OsREC8 is a sister-chromatid cohesion protein that has been used as a marker for early prophase I chromosomes (Shao et al., 2011). Normal loading of OsREC8 on meiotic chromosomes was demonstrated in osshoc1-2 compared with the wild type (Fig. 3). There were no obvious differences in numbers of γH2AX foci between wild type (158.5, n = 19) and osshoc1-2 (149.2, n = 20) at the zygotene (Fig. 3, A, top and bottom, and E), indicating that DSB formation is normal in the mutant.
Figure 3.
OsSHOC1 is not required for early homologous recombination repair in rice. Dual immunolocalization of OsREC8 (green) with γH2AX (A), OsCOM1 (B), OsRAD51C (C), and OsDMC1 (D; magenta) in wild-type and osshoc1-2 zygotene meiocytes. (E) Statistical analysis of the numbers of γH2AX, OsCOM1, OsRAD51C, and OsDMC1 foci revealed no significant differences (NS) between the wild-type (n = 19, 14, 18, and 16, respectively) and osshoc1-2 zygotene cells (n = 20, 15, 19, and 18, respectively). All values represent the mean ± sd (P > 0.05, Student’s t tests). Bars = 2 µm.
In rice, following DSB formation, OsCOM1 functions in DSB end resection to generate 3′-ssDNA tails (Ji et al., 2012). Subsequently, the rice recombinase A homolog OsDMC1 binds to the ssDNA and mediates invasion into the intact homologous duplexes (Wang et al., 2016b). Another rice recombinase A homolog, OsRAD51C, is important for meiotic DSB repair (Tang et al., 2014). As shown in Figure 3, the localizations of OsCOM1, OsRAD51C, and OsDMC1 foci were indistinguishable from those in wild-type meiocytes. The average numbers of OsCOM1, OsRAD51C, and OsDMC1 foci in osshoc1-2 (210.5 [n = 15], 185.5 [n = 19], and 197.2 [n = 18], respectively) were comparable to those in the wild type (218.4 [n = 14], 179.7 [n = 18], and 197.5 [n = 16], respectively; Fig. 3E). These observations indicated that early meiotic recombination steps involving DSB formation, processing, and single-strand invasion are not altered in the osshoc1-2 mutant.
Homologous Chromosome Pairing and Synapsis Occur Normally in osshoc1-2
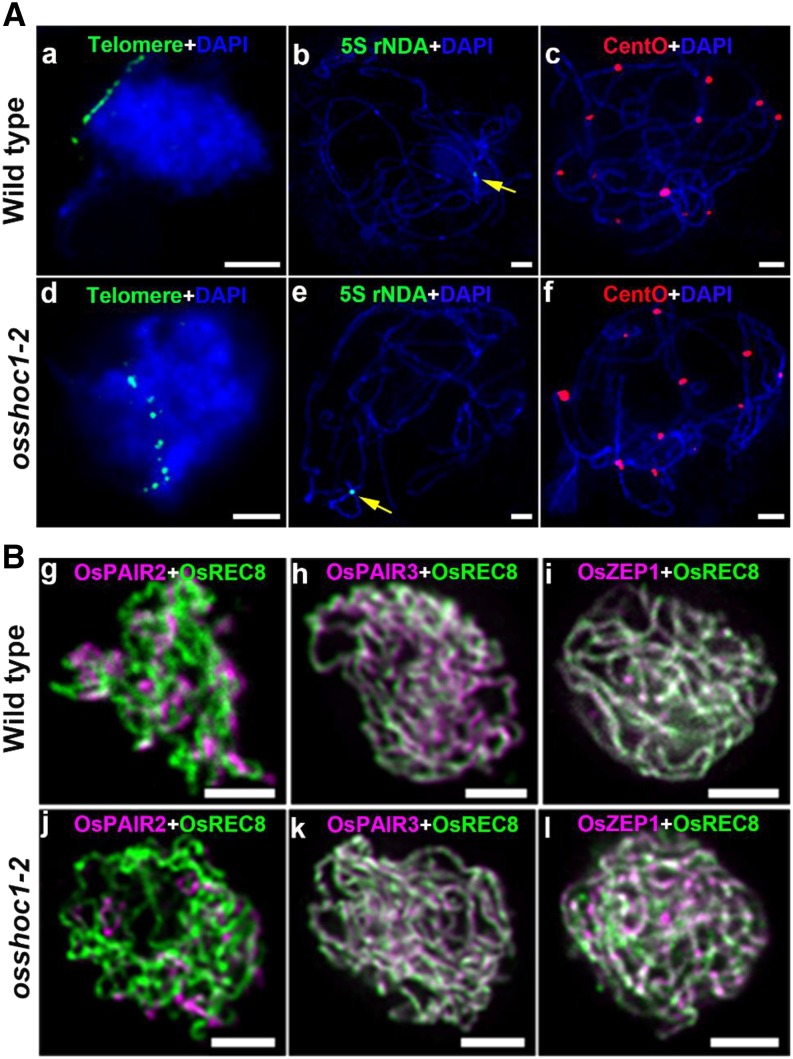
In plants, aberrations in homologous chromosome pairing and SC formation generally impair normal CO formation. We first investigated the impact of OsSHOC1 disruption on homologous chromosome pairing. The telomere bouquet is a highly conserved configuration that efficiently facilitates homology searching and pairing, thus influencing the subsequent homology synapsis and recombination in meiotic prophase I (Harper et al., 2004). Fluorescent in situ hybridization (FISH) analysis using fluorescein isothiocyanate probe-labeled telomere-specific sequences showed that ∼78.6% (n = 42) and ∼78.3% (n = 60) of early-zygotene meiocytes were observed to have telomere signals clustering in the wild type and osshoc1-2, respectively (Fig. 4A, a and d). To further investigate the progression of homologous pairing, we conducted FISH analysis with a 5S repetitive DNA (rDNA) probe located on the short arm of chromosome 11 and a centromere-specific tandem repeat of O. sativa (CentO) probe, which is specific to centromeric regions on all chromosomes. When PMCs enter into pachytene, full-length homologous pairing has been achieved. Only one 5S rDNA signal and 12 CentO signals were detected on the wild-type (n = 20) and osshoc1-2 (n = 28) pachytene chromosomes, indicative of paired homologs in osshoc1-2 (Fig. 4A, b, c, e, and f).
Figure 4.
OsSHOC1 is dispensable for homologous pairing and SC formation in rice. A, Bouquet formation and homologous pairing analysis in the wild type and the osshoc1-2 mutant. Telomere-specific probes (green) were used to check telomere clustering in wild-type (a) and osshoc1-2 (d) male meiocytes. Homologous pairing was revealed by 5S rDNA probe (yellow arrows) and CentO probe (red) on wild-type (b and c) and osshoc1-2 (e and f) chromosomes, respectively. Meiotic chromosomes are indicated by DAPI staining (blue). Bars = 5 μm. B, Synaptonemal complex assembly in the wild type and osshoc1-2. Dual immunolocalization of REC8 (green) with PAIR2, PAIR3, and ZEP1 proteins (magenta) in wild-type (g–i) and osshoc1-2 (j–l) pachytene meiocytes respectively. Bars = 2 μm.
SC formation occurs following homologous chromosome pairing. In rice, PAIR2 and PAIR3 serve as two parallel axial elements (Nonomura et al., 2006; Wang et al., 2011). A transverse filament protein, ZEP1, serves as the central element of SC in rice (Wang et al., 2010). We investigated a possible role for OsSHOC1 in SC assembly by performing dual-immunolocalization assays of REC8 with PAIR2, PAIR3, and ZEP1 in the wild type and osshoc1-2. In osshoc1-2 meiocytes, PAIR2 and PAIR3 loaded onto the chromosome axes as in the wild type (Fig. 4B, g, h, j, and k). The linear signals of ZEP1 along the pachytene chromosomes were observed in wild-type (n = 10) and osshoc1-2 (n = 12) meiocytes with a similar pattern (Fig. 4B, i and l). Altogether, mutation of osshoc1 does not appear to affect homologous pairing or SC assembly.
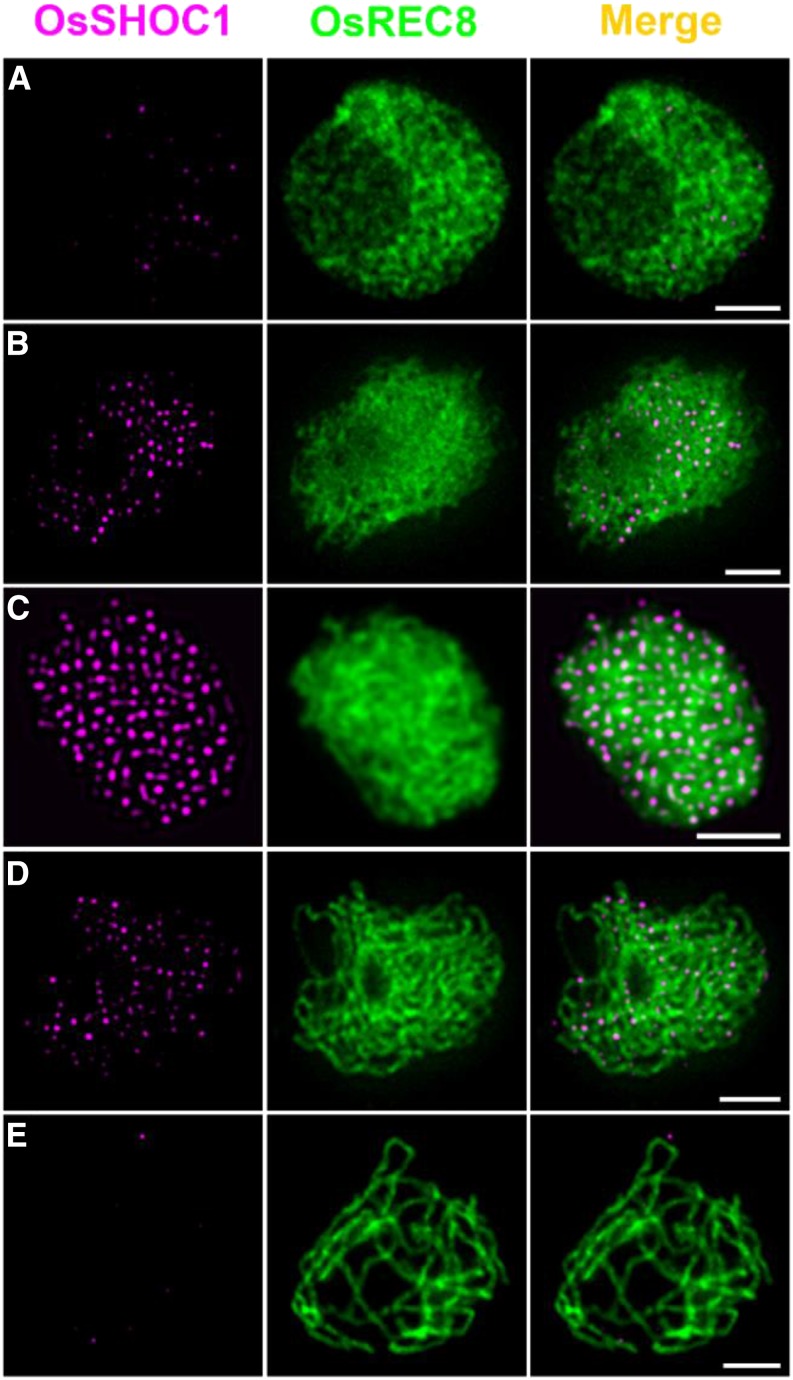
A Dynamic Localization Pattern of OsSHOC1 onto the Meiotic Chromosomes
To understand the dynamics of OsSHOC1 association and dissociation with meiotic chromosomes, we conducted dual-immunolocalization assays of OsSHOC1 and OsREC8. At early leptotene, a few bright OsSHOC1 foci were first observed, followed by a rapid increase (Fig. 5, A and B). The maximal number of OsSHOC1 foci was observed at zygotene (average 197.4, n = 16, range 171–226) and decreased thereafter at pachytene (Fig. 5, C and D). Almost no visible foci could be detected at late pachytene (Fig. 5E). By contrast, no signals were observed when the anti-OsSHOC1 antibody was applied in osshoc1-2 meiocytes (Supplemental Fig. S3). This specific localization pattern was further confirmed by detecting OsSHOC1-GFP recombinant protein distribution in male meiocytes of osshoc1-2/proOsSHOC1:OsSHOC1:eGFP complementation lines (Supplemental Fig. S2).
Figure 5.
Dual immunolocalization of OsREC8 (green) and OsSHOC1 (magenta) in rice meiocyte cells in the early leptotene (A) late leptotene (B), zygotene (C), early pachytene (D), and late pachytene (E) stages. Meiotic chromosomes are indicated by anti-OsREC8 antibody. Bars = 2 μm.
OsPTD Functions in the Formation of Class I COs
In Arabidopsis, SHOC1 and PTD can form an XPF-ERCC1-like complex that is essential for class I CO formation (Macaisne et al., 2011) and in yeast/mammalian ZIP2/SHOC1 interacts with the ERCC1-like protein SPO16 to regulate CO formation (De Muyt et al., 2018; Zhang et al., 2019). Recently, it has been reported that OsSHOC1 interacts with OsPTD in rice (Ren et al., 2019). To further understand the role of OsPTD in meiotic CO formation, we generated two alleles of osptd using the clustered regularly interspaced short palindromic repeats(CRISPR)/CRISPR associated protein 9 (Cas9) system. Sequencing of two mutant alleles revealed a 1-bp (T) insertion in the second exon of osptd-1 and a 1-bp (A) insertion in the seventh exon of osptd-2 (Supplemental Fig. S4A). Both alleles produced sterile plants with similar meiotic defects (Supplemental Fig. S4, B and C). The osptd-1 mutant allele was used for observation and data generation throughout our research.
The meiotic chromosome behavior of osptd-1 PMCs was investigated by DAPI staining. No apparent defects were seen until diakinesis, when a mixture of univalents and bivalents was observed, as described by Ren et al. (2019). The univalents segregated randomly during meiosis II in osptd-1, leading to the production of abnormal tetrads (Supplemental Fig. S4C). Moreover, the chiasma frequency was reduced to 3.12 ± 1.76 (n = 113), and the residual chiasmata showed a Poisson-like distribution in osptd-1 (Kolmogorov-Smirnov test, P > 0.05; Supplemental Fig. S4D).
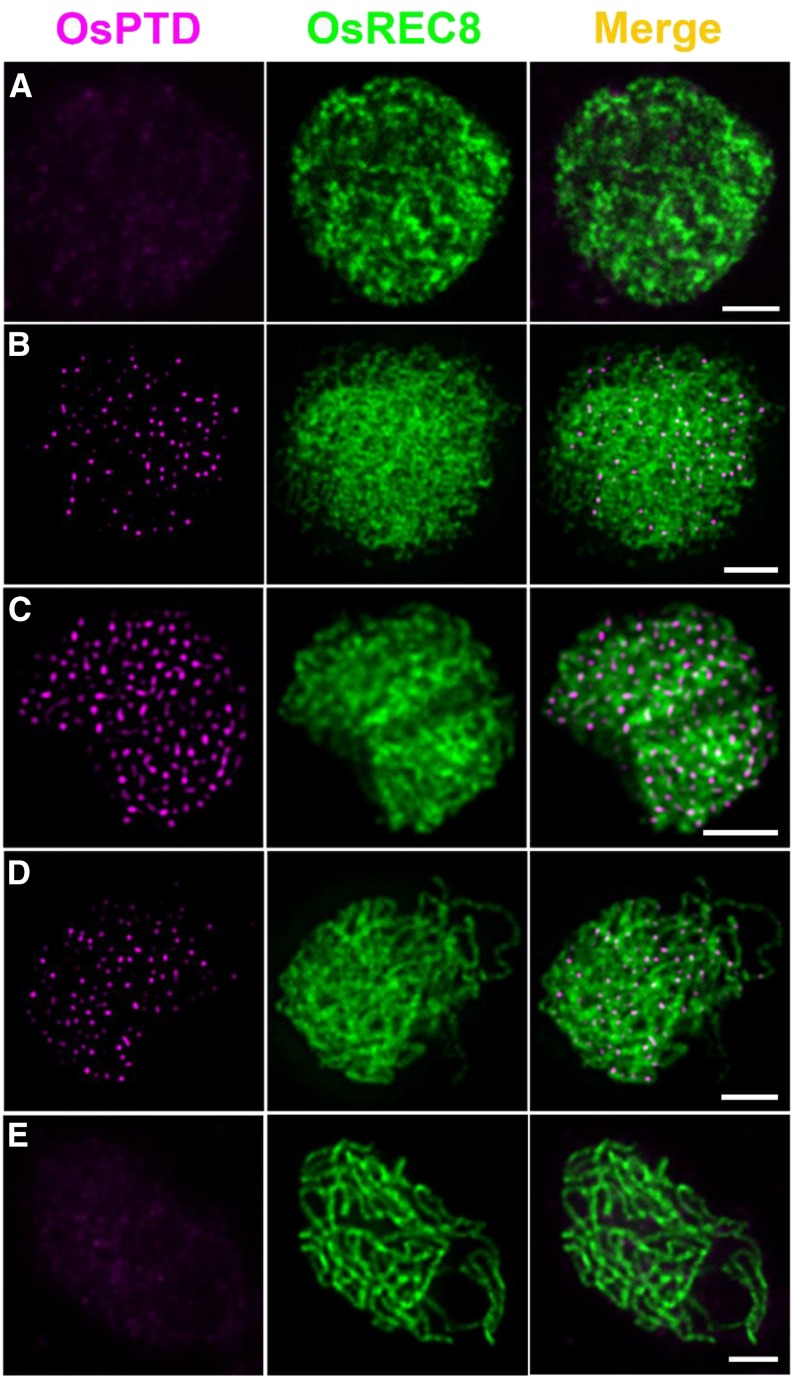
We raised an OsPTD-specific antibody to reveal the temporal and spatial distribution of OsPTD on meiotic chromosomes. Dual-immunodetection assays showed that visible OsPTD foci first appeared at leptotene, that maximum levels occurred at zygotene (average 204.7, n = 12, range 170–244), and none were detected at late pachytene (Fig. 6). No visible foci could be detected in osptd-1 meiocytes (Supplemental Fig. S3D).
Figure 6.
Dual immunolocalization of OsREC8 (green) and OsPTD (magenta) in rice meiocyte cells at the interphase (A), leptotene (B), zygotene (C), early pachytene (D), and late pachytene stages (E). Meiotic chromosomes are indicated by anti-OsREC8 antibody. Bars = 2 μm.
The Interdependent Loading Pattern between OsSHOC1 and OsPTD
To investigate the relationship between OsSHOC1 and OsPTD, we first conducted yeast two-hybrid (Y2H) assays. Similar to the findings of Ren et al. (2019), OsSHOC1 displayed a bidirectional interaction with OsPTD (Supplemental Fig. S5A). This interaction was further confirmed in tobacco cells by bimolecular fluorescence complementation (BiFC) assays (Supplemental Fig. S5B).
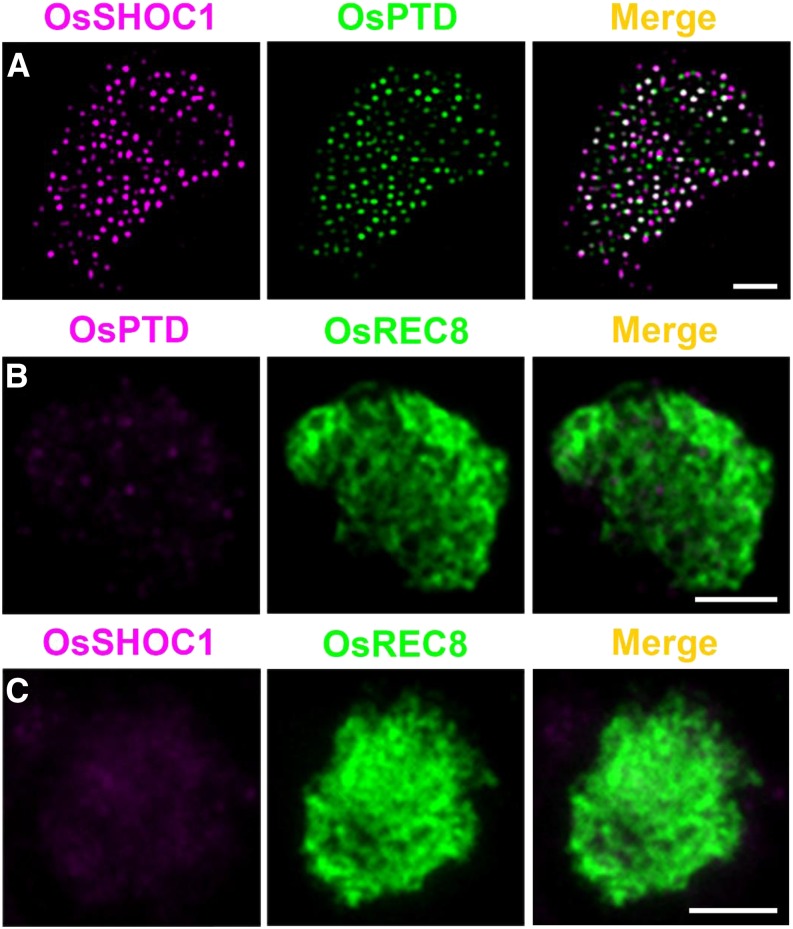
To further demonstrate the direct interaction of OsSHOC1 and OsPTD in vivo, we performed dual-immunolocalization assays using polyclonal antibodies against OsPTD and the GFP tag. As shown in Figure 7A, the visual colocalization patterns between OsSHOC1 and OsPTD were captured in male zygotene osshoc1-2/proOsSHOC1:OsSHOC1:eGFP transgenic lines (n = 8). In addition, the cytofluorogram result also suggested a close association between OsSHOC1 and OsPTD (Supplemental Fig. S6A). When combined with the further intensity correlation analysis and colocalization parameter results, we observed an extensive colocalization between OsSHOC1 and OsPTD in meiocyte chromosomes (Supplemental Fig. S6, B–D). Moreover, no OsPTD signals were detected in osshoc1-2 meiocytes (Fig. 7B), and no visible OsSHOC1 signals were observed in 54% of osptd-1 meiotic cells (n = 214), while the remaining ones had substantially decreased numbers of OsSHOC1 foci (Fig. 7C).
Figure 7.
Colocalization of OsSHOC1 and OsPTD. A, Dual immunolocalization of OsSHOC1 (magenta) and OsPTD (green). Bars = 2 mm. B, OsPTD foci (magenta) in osshoc1-2 zygotene meiocytes. C, OsSHOC1 foci (magenta) in osptd-1 zygotene meiocytes. Meiotic chromosomes are indicated by anti-OsREC8 antibody (green). Bars = 2 μm.
To further understand the relationship between OsSHOC1 and OsPTD, we calculated the chiasma frequency in the osshoc1-2 osptd-1 double mutant (Table 1). The mean chiasma frequency was 3.26 ± 1.84 (n = 109) per cell, which was comparable to frequencies in the osshoc1-2 (3.40 ± 1.89, n = 142) and osptd-1 (3.12 ± 1.76, n = 113) single mutants, suggesting that OsSHOC1 and OsPTD function in the same pathway for class I CO formation. The interaction and interdependent localization between OsSHOC1 and OsPTD, together with similar meiotic chromosome defects conferred by their single and double mutations, suggest that SHOC1-PTD forms an XPF-ERCC1-like complex required to promote class I COs.
The Interrelation between OsSHOC1, OsPTD, and Rice ZMM Proteins
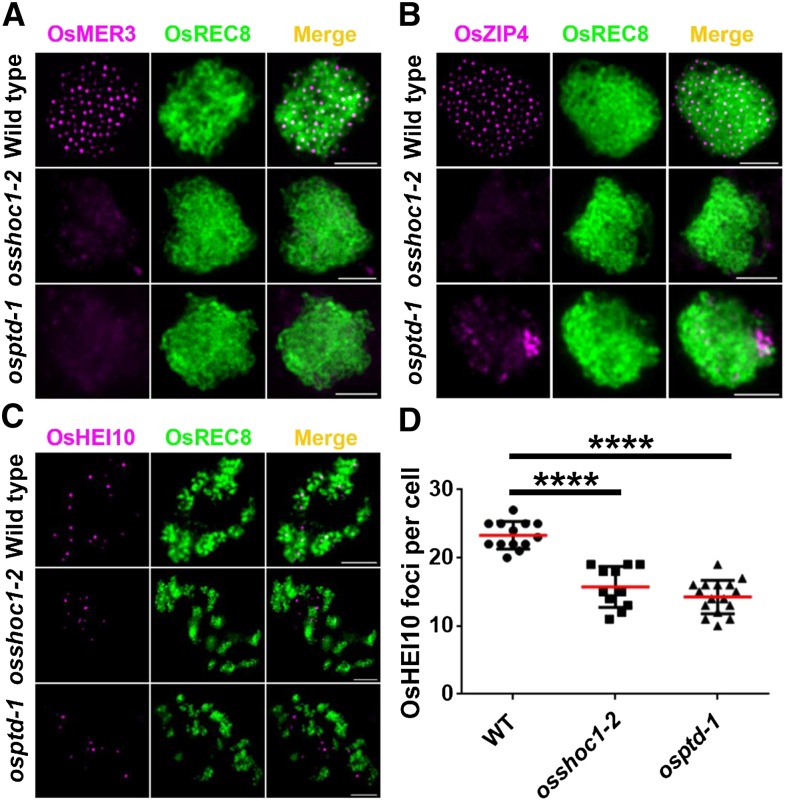
Displacement loops, SEIs, and dHJs are important recombination intermediates generated progressively following DSB formation. It has been suggested that ZMM proteins play a crucial role in stabilizing and protecting these intermediates toward CO formation (De Muyt et al., 2012; Kaur et al., 2015). So far, homologous ZMM proteins in rice, including OsMSH4, OsMSH5, OsZEP1, OsHEI10, OsZIP4, and OsMER3, have been characterized. In all of these proteins except OsZEP1, loss of function causes a substantial decrease in the number of class I COs, which is very similar to what is observed in osshoc1-2 and osptd-1 (Wang et al., 2009, 2010, 2012, 2016a; Shen et al., 2012; Luo et al., 2013; Zhang et al., 2014). To understand the relationship of OsSHOC1 and OsPTD with other ZMM components, we investigated the localization of these proteins in osshoc1-2 and osptd-1 male meiotic chromosomes. During zygotene, OsMER3 was not detected in 66% (n = 483) of osshoc1-2 cells and 58% (n = 370) of osptd-1 cells, and the remaining cells had decreased OsMER3 foci (Fig. 8A). OsZIP4 signals were not detected in osshoc1-2, and OsZIP4 foci aggregated abnormally in osptd-1 (Fig. 8B). In rice, OsHEI10 is used as a marker for Class I COs during late prophase I (Wang et al., 2012). Compared to HEI10 foci in the wild type at diakinesis (23.3, n = 13, range 20–27), fewer foci were observed in osshoc1-2 (15.7, n = 11, range 11–19) and osptd-1 (14.3, n = 16, range 10–19; Fig. 8, C and D). These results suggest that OsSHOC1 and OsPTD are required for normal localization of OsMER3, OsZIP4, and OsHEI10.
Figure 8.
Immunodetection analysis of OsMER3, OsZIP4, and OsHEI10 (magenta) in the wild type, osshoc1-2, and osptd-1 mutants in rice. A, OsMER3 foci in wild-type, osshoc1-2, and osptd-1 zygotene meiocytes. B, OsZIP4 foci in wild-type, osshoc1-2, and osptd-1 zygotene meiocytes. C, OsHEI10 foci in wild-type, osshoc1-2, and osptd-1 diakinesis meiocytes. D, Statistical analysis of the numbers of bright OsHEI10 foci in wild-type (n = 13), osshoc1-2 (n = 11), and osptd-1 (n = 16) diakinesis cells. All values represent the mean ± sd. The asterisks indicate statistical significance between the wild type and the mutants, as determined by one-way ANOVA test with post hoc contrasts by the Newman-Keuls multiple-comparisons test (****P < 0.0001). Meiotic chromosomes are indicated by anti-OsREC8 antibody (green). Bars = 2 μm.
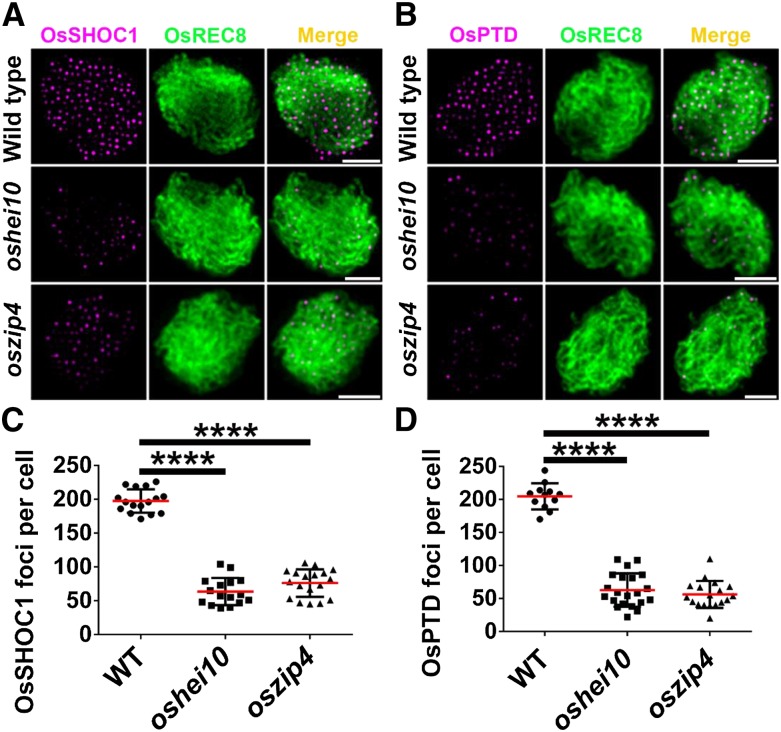
To further characterize the relationship among these proteins, we assessed the immunolocalization of OsSHOC1 and OsPTD proteins in oshei10 and oszip4 mutants. The results showed that the average number of OsSHOC1 foci was reduced by 69–79% in oshei10 (43.3, n = 30, range 16–103) and oszip4 (61.4, n = 43, range 37–106), respectively. The number of OsPTD foci was reduced to a similar extent in oshei10 (51.6, n = 35, range 18–116) and oszip4 (76.7, n = 40, range 37–117), respectively (Fig. 9). In conclusion, one of the four proteins (OsSHOC1, OsHEI10, OsZIP4, and OsPTD) impacts the normal loading of the other three proteins but to varying degrees.
Figure 9.
Immunodetection analysis of OsSHOC1 and OsPTD (magenta) in the wild type and the oszip4 and oshei10 mutants. A, OsSHOC1 foci in wild-type, oshei10, and oszip4 zygotene meiocytes. B, OsPTD foci in wild-type, oshei10, and oszip4 zygotene meiocytes. C, Statistical analysis of numbers of OsSHOC1 foci in wild-type (n = 16), oshei10 (n = 30), and oszip4 (n = 43) zygotene cells. D, Statistical analysis of numbers of OsPTD foci in wild-type (n = 12), oshei10 (n = 35), and oszip4 (n = 40) zygotene cells. All values represent the mean ± sd. The asterisks indicate statistical significance between the wild type and the mutants, as determined by one-way ANOVA test with post hoc contrasts by the Newman-Keuls multiple-comparisons test (****P < 0.0001). Meiotic chromosomes are indicated by anti-OsREC8 antibody (green). Bars = 2 μm.
The hypothesized upstream function of OsSHOC1 in rice CO formation in relation to other ZMM proteins was investigated by genetic analysis. The mean chiasma frequency in oshei10 (6.06 ± 2.86, n = 167), as determined by Wang et al. (2017), was much higher than that in osshoc1-2 (3.40 ± 1.89, n = 142) or osptd-1 (3.12 ± 1.76, n = 113). Moreover, the mean chiasma frequency in osshoc1-2 oshei10 double mutants (3.36 ± 1.90, n = 94) was similar to that in osshoc1-2, osptd-1, and osshoc1-2 osptd-1 (Table 1). Consistent with that report (Shen et al., 2012), there was an ∼70% (6.51 ± 2.56, n = 47) decrease in chiasma frequency in oszip4 generated by CRISPR-Cas9 (Supplemental Fig. S7). However, a significant reduction in frequency happened in the osshoc1-2 oszip4 double mutant (3.38 ± 1.78, n = 122) compared with the oszip4 single mutant (Table 1). Altogether, these results showed that OsSHOC1 and OsPTD function upstream of OsHEI10 and OsZIP4 and genetically control the same set of class I COs.
Protein Interactions between Rice ZMM Proteins
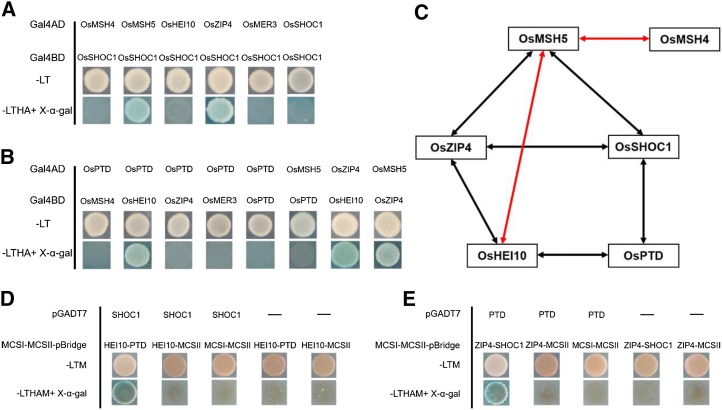
It has been reported that in rice, OsMSH5 can interact with OsMSH4 and OsHEI10 (Zhang et al., 2014; Wang et al., 2016a). However, the interactions among most rice ZMM proteins are yet unknown. Recent reports indicate that ZIP4 interacts with ZIP2, ZIP3, MSH5, and SPO16 (De Muyt et al., 2018). We first investigated the interactions between OsSHOC1, OsPTD, OsHEI10, and OsZIP4 using Y2H assays. As shown in Figure 10 and Supplemental Figure S8, OsSHOC1 interacts with OsZIP4, OsPTD interacts with OsHEI10, and OsHEI10 can interact with OsZIP4. BiFC assays were used to further confirm these interactions (Supplemental Fig. S9A). Considering that archaea XPF protein can form a functional homodimer (Dehé and Gaillard, 2017), we also tested whether XPF-like protein OsSHOC1 acts as a homodimer. The results showed that OsSHOC1 was unable to form a homodimer (Fig. 10A). Similarly, OsPTD also did not function as a homodimer (Fig. 10B). In addition, interactions between OsSHOC1, OsPTD, and the other three ZMM members (OsMSH4, OsMSH5, and OsMER3) were investigated. The results showed that OsSHOC1 can interact with OsMSH5 (Fig. 10A; Supplemental Fig. S8). However, OsPTD does not interact with any of these proteins (Fig. 10B). Regardless, the investigation of interactions between OsZIP4 and other ZMM proteins indicated that OsZIP4 can interact with OsMSH5 (Fig. 10B; Supplemental Fig. S8).
Figure 10.
Interactions among rice ZMM proteins. A, Analysis of interactions between OsSHOC1 and ZMM proteins using the Y2H assay. B, Analysis of interactions between ZMM proteins using the Y2H assay. Full-length OsSHOC1, OsPTD, and ZMM protein coding sequences were constructed into both pGADT7 (Gal4AD) and PGBKT7 (Gal4BD) vectors. Selective-medium -LT (SD-Leu/-Trp), and -LTHA (SD-Leu/-Trp/-His/-Ade) with X-α-gal, were used to test interactions in the Y2H system. C, Diagram summarizing the network of interactions among ZMM proteins identified by Y2H system. Two-way arrows indicate interactions between rice ZMM proteins identified in this study (black) and those in previous reports (red). D and E, Y3H analysis among OsSHOC1, OsPTD, OsHEI10, and OsZIP4. Full-length OsSHOC1 and OsPTD protein coding sequences were constructed into pGADT7 vectors, and OsZIP4, OsHEI10, OsSHOC1, and OsPTD were separately cloned into multiple cloning sites I (MCSI) and II (MCSII) of pBridge vectors. Yeast cells were grown on -LTM (SD-Leu/-Trp/-Met) selection agar plates or -LTHAM (SD-Leu/-Trp/-His/-Ade/-Met) agar plates with X-α-gal for interaction detection.
Based on the fact that localizations of OsSHOC1, OsPTD, HEI10, and OsZIP4 proteins on the chromosomes were interdependent, we proposed that they may form multiprotein complexes, although no direct interactions between OsHEI10 and OsSHOC1, OsPTD, and OsZIP4 were detected by Y2H assay (Fig. 10). To test this possibility, full-length OsSHOC1 and OsHEI10 proteins fused with the N-terminal and C-terminal halves, respectively, of YFP, together with OsZIP4 or OsPTD proteins fused with FLAG-tag (FLG), were simultaneously expressed in tobacco cells. The results showed that the presence of OsZIP4 or OsPTD can mediate the interaction of OsSHOC1 and OsHEI10 (Supplemental Fig. S9B). To confirm the reliability of this result, we detected the expression of OsZIP4 and OsPTD proteins by western blot assay (Supplemental Fig. S10). Moreover, the yeast three-hybrid (Y3H) assay was also employed to screen “a third protein” whose presence is required for the interactions between OsHEI10 and OsSHOC1, and OsPTD and OsZIP4. The results also showed that OsSHOC1 could interact with OsHEI10 when OsPTD was present (Fig. 10D). Additionally, the presence of OsSHOC1 mediated the interaction between OsPTD and OsZIP4 (Fig. 10E). Based on the protein-protein interactions and mutual dependence of protein localization among these four proteins, we proposed that they may form heterotrimeric or heterotetrametric complexes in vivo, perhaps transiently, to promote class I COs in rice.
DISCUSSION
OsSHOC1-OsPTD Complex Is Indispensable for Rice CO Formation
In yeast, ZIP2 acts together with other ZMM proteins, including ZIP1, ZIP3, ZIP4, MSH4, MSH5, MER3, and SPO16 to control class I CO formation (Lynn et al., 2007; Shinohara et al., 2008). Recent reports indicated that orthologs of ZIP2 in Arabidopsis (AtSHOC1) and mammals (chromsome 9 open reading frame 84, SHOC1, and mammalian ortholog of ZIP2) also play essential roles in meiotic recombination (Macaisne et al., 2008; Guiraldelli et al., 2018; Zhang et al., 2018). In this study, we showed that mutations in OsSHOC1, the rice ZIP2 ortholog, led to the loss of ∼83% of total meiotic COs, which was higher than that in previous studies (∼78%; Ren et al., 2019). The residual chiasmata in osshoc1-2 were randomly distributed, indicating that OsSHOC1 is involved in the formation of class I COs. Consistently, a further decrease in the number of COs was observed in the double mutant of OsSHOC1 and OsGEN1, the latter of which encodes a resolvase required for class II CO formation (Wang et al., 2017).
ZIP2 and SHOC1 show weak homology to a structure-specific endonuclease XPF, which plays important roles in DNA lesions repair (Ciccia et al., 2008). In archaebacteria, XPFs are active as homodimers, while in eukaryotes ERCC1-like proteins are usually required for XPF-like proteins to fulfill their normal functions (Dehé and Gaillard, 2017). In agreement, it has been shown that ZIP2 and SPO16, AtSHOC1 and AtPTD, and SHOC1 and SPO16 form functional XPF-ERCC1-like complexes in yeast, Arabidopsis, and mammals (Macaisne et al., 2011; De Muyt et al., 2018; Zhang et al., 2019). Our study demonstrated that OsSHOC1 did not homodimerize and was able to directly interact in vivo with a rice ERCC1-like protein, OsPTD. In meiotic cells, the localization of OsSHOC1 and OsPTD on chromosomes was mutually dependent. Furthermore, loss of function of OsPTD also resulted in a substantial reduction in the number of class I COs. Statistical analysis of data indicated that the number of residual chiasmata in osshoc1-2, osptd-1, and osshoc1-2 osptd-1 was not different, indicating that the functions of these two proteins are epistatic. These results suggest that OsSHOC1 and OsPTD form a meiotic XPF-ERCC1-like complex to promote formation of class I COs. However, a recent report claimed that the residual chiasma frequency in the osptd1 mutants was significantly higher than that in osshoc1-2 mutants (Ren et al., 2019). Interestingly, very close mutation sites were created in osptd-1 and ko-2-osptd1 mutants, suggesting that the differences may be due to different genetic backgrounds and/or potential cognitive and statistical bias. Our results, together with those from previous studies (Shen et al., 2012; Zhang et al., 2014), demonstrate that all eight of the ZMM homologs in rice—OsMSH4, OsMSH5, OsMER3, OsZEP1, OsSHOC1, OsHEI10, OsZIP4, and OsPTD—have evolutionarily conserved functions in class I CO formation.
Yeast SPO16 and ZIP2, and mammalian SPO16, SHOC1, and mammalian ortholog of ZIP2 have an additional role in synapsis (Chua and Roeder, 1998; Shinohara et al., 2008; Guiraldelli et al., 2018; Zhang et al., 2018). Asynapsis was also observed in other budding yeast or mammalian ZMM mutants. By contrast, no obvious synapsis defects occurred in reported plant ZMM mutants. Consistent with previous reports, synapsis was not affected in osshoc1-2 and osptd-1 mutants. Similarly, synaptonemal complexes were well assembled in the absence of Caenorhabditis elegans ZMM homologs (Lynn et al., 2007). The reason for the distinct requirement for ZMM proteins in SC formation in different species remains elusive.
OsSHOC1-OsPTD Complex May Function in the Post-Strand Invasion Step
Immunological detection showed that absence of OsSHOC1 had no effects on DSB formation, implying that homologous recombination initiated normally in osshoc1-2. In rice, disruption of the DSB end resection protein OsCOM1 or strand invasion proteins OsRAD51C and OsDMC1 leads to serious consequences, such as entangled chromosomes and fragmented chromosomes and asynapsis (Ji et al., 2012; Tang et al., 2014; Wang et al., 2016b). These defects were not observed in osshoc1-2, osptd-1, and osshoc1-2 osptd-1 mutants. This is consistent with the observation that the loading of OsCOM1, OsRAD51C, and OsDMC1 is not affected in osshoc1-2. Normal loading of strand-invasion proteins in zmm mutants has also been reported in Arabidopsis and C. elegans (Higgins et al., 2004; Jantsch et al., 2004; Mercier et al., 2005; Macaisne et al., 2008; Chelysheva et al., 2012). Considering these findings along with the dynamic localization of OsSHOC1 and OsPTD, we propose that the OsSHOC1-OsPTD complex may function primarily after the initiation of single-strand invasion during the homologous recombination process.
The GDXnERKX3D catalytic site found in canonical XPF endonucleases is not conserved in ZIP2 and mammalian SHOC1s, and it is totally missing in Arabidopsis and rice SHOC1s (Macaisne et al., 2008; this study), suggesting that the SHOC1-PTD complex does not possess intrinsic nuclease activity. Recent studies indicate that the ZIP2-SPO16 complex and recombinant mammalian SHOC1 preferentially bind branched DNA structures that are similar to meiotic recombination intermediates, such as D-loops and dHJs (De Muyt et al., 2018; Guiraldelli et al., 2018). Combined with the findings that ZIP2 and SHOC1 are not able to cleave these DNA structures and the number of recombination intermediates produced later (SEIs and dHJs) is dramatically reduced, the authors proposed that these proteins function as a DNA recognition module that binds and stabilizes D-loops rather than being a DNA endonuclease or resolvase. Based on prior works and the current study, we suggest that the OsSHOC1-OsPTD complex promotes class I CO formation by binding and stabilizing early recombination intermediates, protecting them against anti-CO activities.
How Rice ZMM Proteins Coordinately Control Class I CO Formation
Most ZMM proteins in rice have been characterized, but how they act together to control class I CO formation is largely unknown. In yeast, each ZMM member is epistatic in regard to CO number. Mutations of any ZMM protein cause similar reductions in the chiasma frequency (Börner et al., 2004). Similar observations were also reported in C. elegans, in which MSH4, MSH5, and ZIP3-homologous protein are required for all class I COs (Zalevsky et al., 1999; Kelly et al., 2000; Jantsch et al., 2004). In contrast, plant ZMMs are divided into different epistasis groups. In Arabidopsis, AtSHOC1 and AtPTD belong to the same epistasis group as AtMSH4, AtMSH5, AtZIP4, and AtHEI10, and all six are required for ∼85% of COs (Higgins et al., 2004; Chen et al., 2005; Chelysheva et al., 2007; Macaisne et al., 2008). The CO number is less affected in the two other Arabidopsis zmm mutants, atmlh3 and rck/atmer3 (40% and 25%–32%, respectively, of the wild-type CO number; Chen et al., 2005; Mercier et al., 2005; Jackson et al., 2006). In rice, OsMSH4 and OsMSH5 are responsible for ∼90% of COs, which is much higher than the number of COs attributed to OsHEI10, OsZIP4, and OsMER3 (Zhang et al., 2014). Our study showed that the number of COs eliminated in osshoc1-2 oshei10 and osshoc1-2 oszip4 double mutants (∼83%) was higher than that in oshei10 and oszip4 single mutants (∼70%) but similar to those in osshoc1-2 and osptd-1 single mutants. Thus, we propose that OsSHOC1, OsPTD, OsZIP4, and OsHEI10 function in the same CO formation process, but OsSHOC1 and OsPTD function upstream of OsHEI10, OsZIP4, and OsMER3.
In budding yeast, both ZIP2 and SPO16 can interact with ZIP4 to form a stable ZZS complex that is essential for stabilizing and promoting the resolution of recombination intermediates (De Muyt et al., 2018). In our study, we verified that OsSHOC1 interacted with OsPTD and OsZIP4 in vivo, respectively. Consistent with Arora and Corbett (2019), who did not identify an interaction between SPO16 with ZIP4, we also did not detect a direct interaction between OsPTD and OsZIP4, but Y3H results suggested that OsSHOC1, OsZIP4, and OsPTD could form a ZZS-like heterotrimer. In addition, the localization of these proteins on the meiotic chromosome was mutually dependent. The TPR-containing protein ZIP4 has been suggested to function as a scaffold protein with primary roles in mediating protein interactions (Perry et al., 2005; Zeytuni and Zarivach, 2012). We showed that OsZIP4 mediated the interaction between OsHEI10 and OsSHOC1. Additionally, this interaction could also be induced by the presence of OsPTD. The formation of different heterotrimers in yeast implied that OsSHOC1, OsPTD, OsZIP4, and OsHEI10 may likely form different complexes, including heterotrimers and even a heterotetramer, in vivo. Previous studies indicated that the human hMSH4-hMSH5 heterodimer could stabilize D-loops by forming a sliding clamp that embraces homologous chromosomes to promote stable dHJ formation (Snowden et al., 2004). Our results indicate that both OsSHOC1 and OsZIP4 interact with OsMSH5. We suggest that the OsSHOC1-OsPTD-OsZIP4 complex acts together with the OsMSH4-OsMSH5 heterodimer to stabilize early recombination intermediates, and that the XPF-ERCC1-like DNA binding complex may represent a conserved feature in eukaryotic meiosis.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Cloning of OsSHOC1
Rice (Oryza sativa) plants were all grown in the paddy field at Shanghai Jiao Tong University during the natural growing season. To identify OsSHOC1, a F2 genetic segregating population was generated from a cross between heterozygous plants of osshoc1-2 (O. sativa ssp. japonica) and Guang Lu Ai4 (O. sativa ssp. indica). From primary mapping, 70 sterile plants were collected for genotyping. The mutation locus was narrowed between two insertion-deletion markers (6712 and 7690) on the long arm of chromosome 2. By high-throughput sequencing, an 11-bp deletion was detected within the coding region of LOC_Os02g42910. osgen1 and oshei10 were described previously (Wang et al., 2017). Four double mutants, osshoc1-2 oshei10, osshoc1-2 oszip4, osshoc1-2 osptd-1, and osshoc1-2 osgen1, were generated by crossing heterozygous plants of osshoc1-2 with the other heterozygous plants, respectively, and were identified from F2 populations. Nicotiana benthamiana plants were grown in the greenhouse under 16 h/8 h light/dark conditions.
Complementation of osshoc1-2
For the complementation test, the genomic sequence of OsSHOC1 containing 2505 bp upstream of the start codon, the coding region, and 131 bp downstream of the stop codon was cloned into the binary vector pCAMBIA1301. To construct the pCAMBIA1301-OsSHOC1-eGFP plasmid, 2505 bp upstream of the start codon and the coding region without the stop codon were fused with the eGFP reporter gene. Two plasmids were independently transformed into homozygous callus induced from young panicles of the homozygous osshoc1-2 plants. Primers used in this assay are shown in Supplemental Table S1.
Obtaining CRISPR Knockout Mutants
CRISPR-Cas9 mutants were obtained using methods previously described (Wang et al., 2017). The primers for constructing the single-guide RNA vectors and verifying the transgenic plants are listed in Supplemental Table S1.
Antibody Production
Rabbit polyclonal antibodies against OsSHOC1, OsPTD, OsZIP4, and OsMER3 were prepared by ABclonal. To obtain these antibodies, 151, 248, 200, and 193 amino acid peptides of OsSHOC1, OsPTD, OsZIP4, and OsMER3 (residues 91–241, 1–248, 1–200, and 950–1142, respectively) were expressed in Rosetta (DE3; TIANGEN) using pGEX-4T-1 expression vectors. Antisera were prepared and purified by ABclonal. The specificity of the antibodies is shown in Supplemental Figure S4. The anti-GFP antibody was obtained from Abcam (catalog no. ab6673). Antibodies against other rice meiotic proteins (REC8, PAIR2, PAIR3, ZEP1, COM1, RAD51C, DMC1, and HEI10) and H2AX were generated as described previously (Fu et al., 2016; He et al., 2016; Wang et al., 2017).
Microscopy Analysis
Anthers were collected from flowering spikelets and stained with 1% (w/v) I2-KI solution to determine the pollen viability of the mutants and wild type. For DAPI staining and FISH assays, young spikelets at male meiotic stages were collected and fixed with Carnoy’s solution (ethanol:glacial acetic 3:1 [v/v]), and then prepared as described previously (Cheng, 2013). The sequences of the telomere and centromere FISH probes were determined according to previous studies (Mizuno et al., 2006; Yang et al., 2016). The sequence of the 5S rDNA FISH probe labeled with fluorescein isothiocyanate is listed in Supplemental Table S1. For immunolocalization assays, fresh young panicles were fixed in 4% (w/v) paraformaldehyde for 20–30 min at room temperature and then prepared as described previously (Cheng, 2013). Different antibody combinations were diluted (1:50–1:500, depending on the antibody titer) in TNB buffer (0.1 m Tris-HCl, pH 7.5, 0.15 m NaCl, and 0.5% [w/v] blocking reagent). After three rounds of washing in phosphate-buffered saline, donkey anti-goat immunoglobulin G (heavy and light chain; Alexa Fluor 555; Abcam; 1:200), Alex 555-conjugated goat anti-rabbit antibody (Life Technologies; 1:200), and DyLight 488-conjugated goat anti-rat/rabbit antibody (Abbkine; 1:200) were added to the slides. After counterstaining with DAPI, the slides were photographed using an Eclipse Ni-E microscope (Nikon), and analysis was performed using NIS-Elements Advanced Research software. Image deconvolution was carried out using the function “Mexican Hat.” The graphs and parameters of colocalization analysis were generated from deconvolution images using JACoP plugins (https://imagej.nih.gov/ij/plugins/track/jacop.html) in ImageJ 1.52a software (Li et al., 2004; Bolte and Cordelières, 2006).
Y2H and Y3H Assays
For the Y2H assay, the coding sequences of ZMM proteins were cloned into pGADT7 and pGBKT7, then transformed into the yeast (Saccharomyces cerevisiae) strain AH109. The Y2H assays were performed according to the manufacturer’s user manual (Clontech). For the Y3H assay, full-length OsSHOC1, OsPTD, OsHEI10, and OsZIP4 coding sequences were cloned into the pGADT7 vector; MCSI was fused with the GAL4 binding domain or MCSII was driven by a Met-responsive promoter, Met-25, of the pBridge vector (Clontech); then, both vectors were cotransformed into the yeast strain AH109. As described by Maruta et al. (2016), transformed yeast cells were selected on synthetic dropout (SD/-Leu/-Trp/-Met) nutrient media, and positive clones were transferred and grown on a synthetic dropout (SD/-Leu/-Trp/-His/-Ade/-Met) nutrient medium with X-α-gal to test the interaction. Primers used in this assay are shown in Supplemental Table S1.
BiFC Assay
The coding sequences of the ZMM proteins were cloned into PXY104-C-terminal yellow fluorescent protein and PXY106-N-terminal yellow fluorescent protein plasmids. The pGreenII0000-FLG vector was used to express OsZIP4-FLG and OsPTD-FLG recombinant proteins in N. benthamiana cells. Constructs were transformed into Agrobacterium tumefaciens strain EHA105, and then infiltrated into young N. benthamiana leaves. After 36 h dark incubation, leaves were excised and visualized using a TCS SP5 confocal laser scanning microscope (Leica). Primers used in this assay are shown in Supplemental Table S1.
Multiple Alignments and Statistical Analysis
For the multiple sequence alignment, DNAMAN software (version 6.0) was applied using homologous protein sequences obtained from PSI-BLAST (National Center for Biotechnology Information; https://www.ncbi.nlm.nih.gov/). The gene structure was drawn using IBS software (version 6.0; http://ibs.biocuckoo.org/). Graphs were generated using GraphPad Prism 6 software. Statistical significance was carried out using the function “Unpaired two-tailed t test” and “one-way ANOVA test with post hoc contrasts by Newman-Keuls Multiple comparisons test” in GraphPad Prism 6 software (http://www.graphpad.com/). Kolmogorov-Smirnov tests were carried out using IBM SPSS Statistics (version 20.0).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: OsSHOC1 (Os02g0642600), OsPTD (Os05g0588200), OsMSH4 (Os07g0486000), OsMSH5 (Os05g0498300), OsHEI10 (Os02g0232100), OsZIP4 (Os01g0890900), OsMER3 (Os02g0617500), OsREC8 (Os05g0580500), PAIR2 (Os09g0506800), PAIR3 (Os10g0405500), ZEP1 (Os04g0452500), OsCOM1 (Os06g0613400), DMC1 (Os11g0146800, Os12g0143800), RAD51C (Os01g0578000), OsGEN1 (Os09g0521900), H2AX (Os03g0721900).
Supplemental Materials
The following supplemental materials are available.
Supplemental Figure S1. Morphological analyses of the wild type and osshoc1-2 mutant in rice.
Supplemental Figure S2. Complementation of rice osshoc1-2 lines and immunodetection of GFP protein in complementation lines.
Supplemental Figure S3. Dual immunodetection analysis of OsREC8 (green) and OsSHOC1, OsHEI10, OsZIP4, and OsPTD (magenta) in rice osshoc1-2, oshei10, oszip4, and osptd-1 mutants, respectively.
Supplemental Figure S4. Meiotic defects of rice osptd.
Supplemental Figure S5. OsSHOC1 interacts with OsPTD in rice.
Supplemental Figure S6. Colocalization analysis between OsSHOC1 and OsPTD in osshoc1-2 transgenic complementary lines.
Supplemental Figure S7. Meiotic defects of rice OsZIP4 CRISPR lines.
Supplemental Figure S8. Interaction analysis between rice ZMM proteins by Y2H assay.
Supplemental Figure S9. Interactions among rice ZMM proteins by BiFC.
Supplemental Figure S10. Western blot assay of rice OsZIP4 and OsPTD expression in Nicotiana tabacum (tobacco) cells
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank Mingjiao Chen and Zhijing Luo for mutant screening, generation of the F2 population, and mapping. We thank Jie Wang for gene map-based cloning.
Footnotes
This work was supported by the National Key Research and Development Program of China (grant no. 2016YFD0100903), by the Programme of Introducing Talents of Discipline to Universities (111 Project, grant no. B14016), and in part by the Next-Generation BioGreen 21 Program (grant no. PJ01369001 to K.-H.J.).
Articles can be viewed without a subscription.
References
- Arora K, Corbett KD (2019) The conserved XPF:ERCC1-like Zip2:Spo16 complex controls meiotic crossover formation through structure-specific DNA binding. Nucleic Acids Res 47: 2365–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Cordelières FP (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224: 213–232 [DOI] [PubMed] [Google Scholar]
- Börner GV, Kleckner N, Hunter N (2004) Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45 [DOI] [PubMed] [Google Scholar]
- Chelysheva L, Gendrot G, Vezon D, Doutriaux MP, Mercier R, Grelon M (2007) Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet 3: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L, Vezon D, Chambon A, Gendrot G, Pereira L, Lemhemdi A, Vrielynck N, Le Guin S, Novatchkova M, Grelon M (2012) The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet 8: e1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhang W, Timofejeva L, Gerardin Y, Ma H (2005) The Arabidopsis ROCK-N-ROLLERS gene encodes a homolog of the yeast ATP-dependent DNA helicase MER3 and is required for normal meiotic crossover formation. Plant J 43: 321–334 [DOI] [PubMed] [Google Scholar]
- Chen L, Chu H, Yuan Z, Pan Ah, Liang W, Huang H, Shen M, Zhang D, Chen L (2006) Isolation and genetic analysis for rice mutants treated with 60 Co γ-Ray. J. Xiamen Univ 45: 82–85 [Google Scholar]
- Cheng Z. (2013) Analyzing meiotic chromosomes in rice. Methods Mol Biol 990: 125–134 [DOI] [PubMed] [Google Scholar]
- Chua PR, Roeder GS (1998) Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell 93: 349–359 [DOI] [PubMed] [Google Scholar]
- Ciccia A, McDonald N, West SC (2008) Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem 77: 259–287 [DOI] [PubMed] [Google Scholar]
- Dehé PM, Gaillard PH (2017) Control of structure-specific endonucleases to maintain genome stability. Nat Rev Mol Cell Biol 18: 315–330 [DOI] [PubMed] [Google Scholar]
- De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, Lichten M (2012) BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol Cell 46: 43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Pyatnitskaya A, Andréani J, Ranjha L, Ramus C, Laureau R, Fernandez-Vega A, Holoch D, Girard E, Govin J, et al. (2018) A meiotic XPF-ERCC1-like complex recognizes joint molecule recombination intermediates to promote crossover formation. Genes Dev 32: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Wang C, Xue F, Higgins J, Chen M, Zhang D, Liang W (2016) The DNA topoisomerase VI-B subunit OsMTOPVIB is essential for meiotic recombination initiation in rice. Mol Plant 9: 1539–1541 [DOI] [PubMed] [Google Scholar]
- Gerton JL, Hawley RS (2005) Homologous chromosome interactions in meiosis: Diversity amidst conservation. Nat Rev Genet 6: 477–487 [DOI] [PubMed] [Google Scholar]
- Guiraldelli MF, Felberg A, Almeida LP, Parikh A, de Castro RO, Pezza RJ (2018) SHOC1 is a ERCC4-(HhH)2-like protein, integral to the formation of crossover recombination intermediates during mammalian meiosis. PLoS Genet 14: e1007381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper L, Golubovskaya I, Cande WZ (2004) A bouquet of chromosomes. J Cell Sci 117: 4025–4032 [DOI] [PubMed] [Google Scholar]
- He Y, Wang C, Higgins JD, Yu J, Zong J, Lu P, Zhang D, Liang W (2016) MEIOTIC F-BOX is essential for male meiotic DNA double-strand break repair in rice. Plant Cell 28: 1879–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Armstrong SJ, Franklin FCH, Jones GH (2004) The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes Dev 18: 2557–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FCH (2005) The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev 19: 2488–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Vignard J, Mercier R, Pugh AG, Franklin FCH, Jones GH (2008) AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J 55: 28–39 [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Brill SJ (2004) The Mus81 solution to resolution: Generating meiotic crossovers without Holliday junctions. Genes Dev 18: 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N, Börner GV, Lichten M, Kleckner N (2001) Gamma-H2AX illuminates meiosis. Nat Genet 27: 236–238 [DOI] [PubMed] [Google Scholar]
- Jackson N, Sanchez-Moran E, Buckling E, Armstrong SJ, Jones GH, Franklin FCH (2006) Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. EMBO J 25: 1315–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch V, Pasierbek P, Mueller MM, Schweizer D, Jantsch M, Loidl J (2004) Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol Cell Biol 24: 7998–8006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Tang D, Wang K, Wang M, Che L, Li M, Cheng Z (2012) The role of OsCOM1 in homologous chromosome synapsis and recombination in rice meiosis. Plant J 72: 18–30 [DOI] [PubMed] [Google Scholar]
- Kaur H, De Muyt A, Lichten M (2015) Top3-Rmi1 DNA single-strand decatenase is integral to the formation and resolution of meiotic recombination intermediates. Mol Cell 57: 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly KO, Dernburg AF, Stanfield GM, Villeneuve AM (2000) Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lau A, Morris TJ, Guo L, Fordyce CB, Stanley EF (2004) A syntaxin 1, GαO, and N-type calcium channel complex at a presynaptic nerve terminal: Analysis by quantitative immunocolocalization. J Neurosci 24: 4070–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q, Tang D, Wang M, Luo W, Zhang L, Qin B, Shen Y, Wang K, Li Y, Cheng Z (2013) The role of OsMSH5 in crossover formation during rice meiosis. Mol Plant 6: 729–742 [DOI] [PubMed] [Google Scholar]
- Lynn A, Soucek R, Börner GV (2007) ZMM proteins during meiosis: Crossover artists at work. Chromosome Res 15: 591–605 [DOI] [PubMed] [Google Scholar]
- Macaisne N, Novatchkova M, Peirera L, Vezon D, Jolivet S, Froger N, Chelysheva L, Grelon M, Mercier R (2008) SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr Biol 18: 1432–1437 [DOI] [PubMed] [Google Scholar]
- Macaisne N, Vignard J, Mercier R (2011) SHOC1 and PTD form an XPF-ERCC1-like complex that is required for formation of class I crossovers. J Cell Sci 124: 2687–2691 [DOI] [PubMed] [Google Scholar]
- Maruta N, Trusov Y, Botella JR (2016) Yeast three-hybrid system for the detection of protein-protein interactions. Methods Mol Biol 1363: 145–154 [DOI] [PubMed] [Google Scholar]
- Mercier R, Jolivet S, Vezon D, Huppe E, Chelysheva L, Giovanni M, Nogué F, Doutriaux MP, Horlow C, Grelon M, et al. (2005) Two meiotic crossover classes cohabit in Arabidopsis: One is dependent on MER3, whereas the other one is not. Curr Biol 15: 692–701 [DOI] [PubMed] [Google Scholar]
- Mizuno H, Wu J, Kanamori H, Fujisawa M, Namiki N, Saji S, Katagiri S, Katayose Y, Sasaki T, Matsumoto T (2006) Sequencing and characterization of telomere and subtelomere regions on rice chromosomes 1S, 2S, 2L, 6L, 7S, 7L and 8S. Plant J 46: 206–217 [DOI] [PubMed] [Google Scholar]
- Neale MJ, Keeney S (2006) Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Nakano M, Eiguchi M, Suzuki T, Kurata N (2006) PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J Cell Sci 119: 217–225 [DOI] [PubMed] [Google Scholar]
- Osman K, Higgins JD, Sanchez-Moran E, Armstrong SJ, Franklin FCH (2011) Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol 190: 523–544 [DOI] [PubMed] [Google Scholar]
- Perry J, Kleckner N, Börner GV (2005) Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. Proc Natl Acad Sci USA 102: 17594–17599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Chen D, Li W, Zhou D, Luo T, Yuan G, Zeng J, Cao Y, He Z, Zou T, et al. (2019) OsSHOC1 and OsPTD1 are essential for crossover formation during rice meiosis. Plant J 98: 315–328 [DOI] [PubMed] [Google Scholar]
- Robert T, Nore A, Brun C, Maffre C, Crimi B, Guichard V, Bourbon HM, de Massy B (2016) The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science 351: 943–949 [DOI] [PubMed] [Google Scholar]
- Roeder GS. (1997) Meiotic chromosomes: It takes two to tango. Genes Dev 11: 2600–2621 [DOI] [PubMed] [Google Scholar]
- Sanchez Moran E, Armstrong SJ, Santos JL, Franklin FCH, Jones GH (2001) Chiasma formation in Arabidopsis thaliana accession Wassileskija and in two meiotic mutants. Chromosome Res 9: 121–128 [DOI] [PubMed] [Google Scholar]
- Shao T, Tang D, Wang K, Wang M, Che L, Qin B, Yu H, Li M, Gu M, Cheng Z (2011) OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol 156: 1386–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Tang D, Wang K, Wang M, Huang J, Luo W, Luo Q, Hong L, Li M, Cheng Z (2012) ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J Cell Sci 125: 2581–2591 [DOI] [PubMed] [Google Scholar]
- Shinohara M, Oh SD, Hunter N, Shinohara A (2008) Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat Genet 40: 299–309 [DOI] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R (2004) hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell 15: 437–451 [DOI] [PubMed] [Google Scholar]
- Sym M, Engebrecht JA, Roeder GS (1993) ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72: 365–378 [DOI] [PubMed] [Google Scholar]
- Tang D, Miao C, Li Y, Wang H, Liu X, Yu H, Cheng Z (2014) OsRAD51C is essential for double-strand break repair in rice meiosis. Front Plant Sci 5: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T, Zhao H, Roeder GS (2006) The meiosis-specific zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with zip2. Dev Cell 10: 809–819 [DOI] [PubMed] [Google Scholar]
- Vrielynck N, Chambon A, Vezon D, Pereira L, Chelysheva L, De Muyt A, Mézard C, Mayer C, Grelon M (2016) A DNA topoisomerase VI-like complex initiates meiotic recombination. Science 351: 939–943 [DOI] [PubMed] [Google Scholar]
- Wang Y, Copenhaver GP (2018) Meiotic recombination: Mixing it up in plants. Annu Rev Plant Biol 69: 577–609 [DOI] [PubMed] [Google Scholar]
- Wang C, Wang Y, Cheng Z, Zhao Z, Chen J, Sheng P, Yu Y, Ma W, Duan E, Wu F, et al. (2016a) The role of OsMSH4 in male and female gamete development in rice meiosis. J Exp Bot 67: 1447–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Higgins JD, He Y, Lu P, Zhang D, Liang W (2017) Resolvase OsGEN1 mediates DNA repair by homologous recombination. Plant Physiol 173: 1316–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hu Q, Tang D, Liu X, Du G, Shen Y, Li Y, Cheng Z (2016b) OsDMC1 is not required for homologous pairing in rice meiosis. Plant Physiol 171: 230–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Tang D, Wang M, Lu J, Yu H, Liu J, Qian B, Gong Z, Wang X, Chen J, et al. (2009) MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J Cell Sci 122: 2055–2063 [DOI] [PubMed] [Google Scholar]
- Wang K, Wang M, Tang D, Shen Y, Qin B, Li M, Cheng Z (2011) PAIR3, an axis-associated protein, is essential for the recruitment of recombination elements onto meiotic chromosomes in rice. Mol Biol Cell 22: 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang M, Tang D, Shen Y, Miao C, Hu Q, Lu T, Cheng Z (2012) The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet 8: e1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang K, Tang D, Wei C, Li M, Shen Y, Chi Z, Gu M, Cheng Z (2010) The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22: 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeratne AJ, Chen C, Zhang W, Timofejeva L, Ma H (2006) The Arabidopsis thaliana PARTING DANCERS gene encoding a novel protein is required for normal meiotic homologous recombination. Mol Biol Cell 17: 1331–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Li Y, Zhang L, Shi W, Zhang C, Feng M, Zhang F, Tang D, Yu H, Gu M, et al. (2016) OsMTOPVIB promotes meiotic DNA double-strand break formation in rice. Mol Plant 9: 1535–1538 [DOI] [PubMed] [Google Scholar]
- Yang R, Li Y, Su Y, Shen Y, Tang D, Luo Q, Cheng Z (2016) A functional centromere lacking CentO sequences in a newly formed ring chromosome in rice. J Genet Genomics 43: 694–701 [DOI] [PubMed] [Google Scholar]
- Zalevsky J, MacQueen AJ, Duffy JB, Kemphues KJ, Villeneuve AM (1999) Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153: 1271–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeytuni N, Zarivach R (2012) Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20: 397–405 [DOI] [PubMed] [Google Scholar]
- Zhang L, Tang D, Luo Q, Chen X, Wang H, Li Y, Cheng Z (2014) Crossover formation during rice meiosis relies on interaction of OsMSH4 and OsMSH5. Genetics 198: 1447–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shao J, Fan HY, Yu C (2018) Evolutionarily-conserved MZIP2 is essential for crossover formation in mammalian meiosis. Commun Biol 1: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QT, Ji SY, Busayavalasa K, Yu C (2019) SPO16 binds SHOC1 to promote homologous recombination and crossing-over in meiotic prophase I. Sci Adv 5: eaau9780. [DOI] [PMC free article] [PubMed] [Google Scholar]