Abstract
Purpose
CTG trinucleotide repeat (TNR) expansion in an intron of the TCF4 gene is the most common genetic variant associated with Fuchs' endothelial corneal dystrophy (FECD). Although several mechanisms have been implicated in the disease process, their exact pathophysiologic importance is unclear. To understand events leading from TCF4 TNR expansion to disease phenotype, we characterized splicing, gene expression, and exon sequence changes in a rare cohort of patients with TNR expansions but no phenotypic FECD (RE+/FECD−).
Methods
Corneal endothelium and blood were collected from patients undergoing endothelial keratoplasty for non-FECD corneal edema. Total RNA was isolated from corneal endothelial tissue (n = 3) and used for RNASeq. Gene splicing and expression was assessed by Mixture of Isoforms (MISO) and MAP-RSeq software. Genomic DNA was isolated from blood mononuclear cells and used for whole genome exome sequencing. Base calling was performed using Illumina's Real-Time Analysis.
Results
Three genes (MBNL1, KIF13A, AKAP13) that were previously identified as misspliced in patients with a CTG TNR expansion and FECD disease (RE+/FECD+) were found normally spliced in RE+/FECD− samples. Gene expression differences in pathways associated with the innate immune response, cell signaling (e.g., TGFβ, WNT), and cell senescence markers were also identified between RE+/FECD− and RE+/FECD+ groups. No consistent genetic variants were identified in RE+/FECD− patient exomes.
Conclusions
Identification of novel splicing patterns and differential gene expression in RE+/FECD− samples provides new insights and more relevant gene targets that may be protective against FECD disease in vulnerable patients with TCF4 CTG TNR expansions.
Keywords: Fuchs endothelial corneal dystrophy, FECD, TCF4, trinucleotide repeat expansion disease, cornea
Expansion of a normal polymorphic repeat sequence has been identified as the causative mutation for Fuchs' endothelial corneal dystrophy (FECD) and a number of other disorders including myotonic dystrophy, fragile X syndrome, and spinocerebellar ataxias.1–4 Approximately 80% of the Caucasian population with FECD have an expansion of a CTG trinucleotide repeat (TNR) within an intron (CTG18.1) of the TCF4 gene (RE+/FECD+).4 While the specific association between the CTG TNR expansion in the TCF4 gene and FECD disease pathophysiology is under investigation, studies have identified several pathogenic mechanisms directly attributable to TNR expansion and common to other TNR diseases. For example, RE+/FECD+ tissue samples contain RNA foci, sequester the splicing factor MBNL1, and have widespread changes in RNA splicing.5,6 Additionally, the presence of potentially toxic homopolymeric peptides through repeat-associated non-ATG (RAN) translation7 and the disruption of normal transcription of the TCF4 gene and some of its >80 RNA isoforms have been reported.8 However, a specific set of mechanisms responsible for pathological progression of the disease from accrual of TNR expansion to expression of the actual disease phenotype is currently unknown.
One of the limitations to studying TNR expansion diseases is the difficulty in directly associating the varied mechanisms of action from progression to disease. This is partially due to the lack of control patient groups that may help to establish disease causality. Utilizing FECD and non-FECD–based corneal transplantation procedures, we have developed a large database and tissue bank that have been analyzed for TNR expansion. Within our databases, we have identified a small cohort of elderly patients without FECD that contain pathological expansions (>40) of CTG repeats (RE+/FECD−). We performed RNA transcriptome sequencing from corneal endothelial tissue and exome sequencing from leukocyte DNA obtained from RE+/FECD− patients. Comparison of these data to those of RE+/FECD+ patients revealed significant differences in splicing and gene expression profiles between the two groups. These results provide novel insights in understanding the pathological mechanism involved in development of FECD phenotype following expansion of the CTG TNR in the TCF4 gene.
Methods
Blood and Tissue Isolation
Blood and corneal endothelial tissue were collected from patients undergoing endothelial keratoplasty for non-FECD corneal edema. Tissue samples that were utilized for RNA isolation (n = 3) were placed immediately in RNAlater ICE (ThermoFisher Scientific, Waltham, MA, USA) and stored until processed for RNASeq studies. Blood samples (n = 5) were collected and DNA was isolated from white blood cells isolated from the buffy coat. RE+/FECD+ samples utilized for comparison to RE+/FECD− samples in this study were derived from patients with a modified Krachmer scale score of either a 5 (>5-mm confluent guttae) or 6 (confluent guttae with edema), and their datasets were previously described.5,6,9 All patients whose samples were utilized in this study provided written informed consent and were enrolled in the Mayo Clinic Hereditary Eye Disease study. This research was conducted in accordance with the Declaration of Helsinki and was approved by the Mayo Clinic Institutional Review Board.
RNA Library Preparation and RNA Sequencing
Three corneal endothelial samples were collected over a 5-year period (2013–2017), and processed for RNASeq as described previously.5,6,9 Briefly, total RNA was isolated from corneal endothelial tissue samples using the RNeasy Mini QIAcube Kit (QIAGEN, Valencia, CA, USA). For this study, samples with RNA integrity number (RIN) values of ≥7.0 were depleted of ribosomal transcripts, reverse transcribed substituting deoxyuridine triphosphate (dUTP) for deoxythymidine triphosphate (dTTP), and random primed to generate the second-strand synthesis using TruSeq stranded total library preparation kit (Illumina, San Diego, CA, USA). The resulting libraries were minimally amplified and sequenced on Illumina HiSeq 4000 sequencers.
Analysis of Differentially Spliced Genes
RNASeq datasets were analyzed using MAP-RSeq10 and MISO (Mixture of Isoforms)11 software as previously described.6 MAP-RSeq was utilized to obtain in-depth quality control data, transcriptome read alignment, and abundance of gene and exon expression. Binary Alignment Map (BAM) files obtained by MAP-RSeq were analyzed by MISO to identify and quantify expression levels of alternatively spliced genes between RE+/FECD− and RE+/FECD+ samples. To identify differentially spliced genes, MISO was used to calculate Bayesian probabilities and provide Percent Spliced In (PSI) values for every skipped exon event (range from 0 to 1). A value of 1 indicates that the exon in question is uniformly included in the final transcript. In contrast, a value of 0 indicates that the exon was skipped and is not represented in the final transcript. The PSI value provided a quantifiable measurement that was used to identify differences in splicing between exons in RE+/FECD−, RE+/FECD+, and RE−/FECD− samples. Previously reported datasets generated for RE+/FECD+ and RE−/FECD− samples (i.e., Gene Expression Omnibus [GEO] under accession number GSE112201) and filtering criteria as previously reported were used in RE+/FECD− comparisons.5,6,9 Genes whose PSI values in all three samples that were within 1 standard deviation (SD) of the PSI mean for RE−/FECD− or RE+/FECD+ were placed in that category.
Analysis of Differential Gene Expression
Raw gene counts of all samples were obtained by featureCounts12 using the Subread package (v1.4.6) through the MAPR-Seq pipeline. Differential gene expression analysis between the RE+/FECD− group and RE+/FECD+ group was performed using the edgeR package,13 and the batch effect across the flow cells was corrected as a confounding factor in the model. Log2-transformed normalized gene expression in counts per million (CPM) was obtained through edgeR. Differentially expressed genes that are reported as significant were selected by requiring both adjusted P value ≤ 0.05 and fold change ≥ 2.0 or ≤ −2.0. Supplementary Tables include genes that had a mean average of ≥30 transcripts for RE+/FECD+ and RE+/FECD− groups.
Pathway Analysis Using PANTHER
RNASeq results were analyzed using the PANTHER web portal (http://www.pantherdb.org/; in the public domain). Overrepresentation analysis of genes in specific PANTHER families and pathways was identified following false discovery rate correction for multiple testing.
Whole Exome Sequencing
Genomic DNA was isolated from peripheral blood mononuclear cells obtained from consenting patients (n = 5) using QIAGEN FlexiGene chemistry on an AutoGen FLEX-STAR (Hollister, MA, USA). Briefly, red blood cells were lysed; white cells were pelleted by centrifugation, lysed, and treated with protease. DNA was precipitated with isopropanol, washed with 70% ethanol, and suspended in Tris-EDTA (TE) buffer. Paired-end libraries were prepared using the Bravo liquid handler (Agilent, Santa Clara, CA, USA) and whole exome capture was performed using an Agilent V4+UTR or Agilent V5+UTR kit. Concentration and size distribution of the libraries were determined on Qubit (Invitrogen, Carlsbad, CA, USA) and Agilent Bioanalyzer DNA 1000 chips. Exome libraries were loaded onto TruSeq Rapid run paired end flow cells and sequenced on an Illumina HiSeq 2500 sequencer using TruSeq Rapid SBS kit version 1. Base calling was performed using Illumina's RTA version 1.17.21.3. Filtering for variants predicted to be deleterious (missense, indel, stop codon change or changes at splice junctions) was accomplished using Ingenuity Variant Analysis version 5.2 settings for dominant variants. Variants with a call quality of at least 100 for cases and 50 for controls, outside the top 5% of exonically variable 100 base windows in healthy public genomes, and with an allele frequency of 0.5% or less in NHLBI exomes, ExAC database, or gnomAD were kept.
DNA Isolation and TNR Characterization
TCF4 TNR length was determined as described previously.4,6 Briefly, genomic DNA was isolated from leukocytes using QIAGEN AutoGen FlexiGene and suspended in TE buffer at a final concentration of 250 ng/μL. TCF4 TNR regions were PCR amplified using 100 ng genomic DNA and 10 pmol oligonucleotide primers 5-TCF-Fuchs and 3-TCF-Fuchs1 in the presence of Invitrogen Platinum PCR Super Mix High Fidelity. PCR program consisted of a single cycle at 95°C for 6 minutes followed by 35 cycles at 95°C for 1 minute, 62°C for 1 minute, and 68°C for 3 minutes. Final 68°C incubation for 7 minutes was performed, at which time the sample was refrigerated at 4°C. Primer sequences for 5-TCF-Fuchs and 3-TCF-Fuchs1 have been described previously.4
For Short Tandem Repeat analysis, a 5′ FAM primer was used in place of 5-TCF-Fuchs and PCR was performed as described above. After PCR amplification, 2 μL DNA was mixed with 12 μL diluted Map Marker 1000 (BioVentures, Inc., Murfreesboro, TN, USA) and TCF4 TNR determination was performed using GeneScan on an ABI 3730XL DNA Analyzer (Foster City, CA, USA). 5′ FAM primer sequence was described previously.4
Results
Our cohort of RE+/FECD− phenotype comprises five elderly Caucasian patients (78.4 ± 7.6; age range, 66–86 years) with RE+ (small allele = range 12–27 repeats; large allele = range 67–83 repeats) who did not have clinical FECD (Table 1). These patients had unilateral pseudophakic corneal edema, did not have guttae in the study eye or the contralateral eye, and were undergoing endothelial keratoplasty for complications due to prior glaucoma and/or cataract surgery. We performed RNASeq analysis on three corneal endothelial samples and exome sequencing on all five patients.
Table 1.
Demographics of RE+/FECD− Samples
|
Sample Name |
Sex |
Age |
Race |
Small Allele* |
Large Allele* |
Krachmer Grade |
Clinical Characteristics of Study Eye |
| 4583 | Female | 66 | Caucasian | 27 | 74 | 0 | Dry AMD, cataracts, PEX syndrome without glaucoma, pseudophakia |
| 4575 | Male | 77 | Caucasian | 18 | 83 | 0 | Dry AMD, unilateral macular hole, pseudophakia |
| RNA95 | Female | 82 | Caucasian | 17 | 82 | 0 | Chronic corneal edema, PEX glaucoma, pseudophakia |
| RNA191 | Male | 86 | Caucasian | 16 | 83 | 0 | Chronic corneal edema, PEX glaucoma, pseudophakia |
| RNA184 | Female | 81 | Caucasian | 12 | 67 | 0 | Progressive corneal edema, pseudophakia |
AMD, age-related macular degeneration; PEX, pseudoexfoliation.
Represents repeat number.
Qualitative Splicing Differences in RE+/FECD− Gene Expression
Previous studies on corneal endothelial tissue obtained from RE+/FECD+ patients showed consistent mRNA missplicing patterns in 24 genes.5,6,9 This was attributed to sequestration of MBNL1 and MBNL2 in RNA foci produced by expression of the CTG TNR expansion in TCF4. To determine if these missplicing events were also found in RE+/FECD− samples, we performed RNASeq and compared the transcriptome of RE+/FECD− samples (n = 3) to previously reported datasets of both RE+/FECD+ and RE−/FECD− (GEO accession number GSE112201).5,6,9 Analysis of the 24 missplicing events in samples from RE+/FECD− patients showed traits of both RE+/FECD+ and RE−/FECD− (Table 2). Splicing patterns for three genes, MBNL1, KIF13A, and AKAP13, were consistent with RE−/FECD− tissue as PSI values for all three samples were within 1 SD of the mean RE−/FECD− values. The MISO-calculated PSI values for the MBNL1 alternative splicing event in RE+/FECD− patients was 0.41, 0.49, and 0.28 (0.39 ± 0.11; mean ± SD), similar to the mean of 0.38 for RE−/FECD−. Similarly, KIF13A and AKAP13 also showed PSI values for all three samples consistent with RE−/FECD− samples (Table 2). Due to low RNA yields from corneal endothelial tissue and scarcity of RE+/FECD− samples, we were not able to orthogonally validate these splicing results. However, in previous work we found excellent concordance between RNASeq and real-time PCR findings.5,6
Table 2.
Missplicing Events in RE+/FECD− Samples
|
Gene |
Event Name |
Average PSI Values, Mean ± SD |
||
|
RE−/FECD−,
N
= 12 |
RE+/FECD+,
N
= 18 |
RE+/FECD−,
N
= 3 |
||
| Splicing patterns associating with RE−/FECD− | ||||
| MBNL1 | chr3:152163071:152163328:+@chr3:152164493:152164546:+@chr3:152165409:152165562:+ | 0.38 ± 0.18* | 0.85 ± 0.06 | 0.39 ± 0.11* |
| KIF13A | chr6:17772139:17772290:−@chr6:17771345:17771449:−@chr6:17763924:17765177:− | 0.20 ± 0.15* | 0.47 ± 0.08 | 0.22 ± 0.08* |
| AKAP13 | chr15:86198648:86199018:+@chr15:86201768:86201821:+@chr15:86207794:86207986:+ | 0.77 ± 0.09* | 0.54 ± 0.06 | 0.77 ± 0.13* |
| Splicing patterns associating with RE+/FECD+ | ||||
| CLASP1 | chr2:122204913:122205083:−@chr2:122203025:122203072:−@chr2:122187649:122187753:− | 0.84 ± 0.13 | 0.28 ± 0.18† | 0.28 ± 0.10† |
| GOLGA2 | chr9:131036129:131036251:−@chr9:131035064:131035144:−@chr9:131030699:131030803:− | 0.47 ± 0.17 | 0.11 ± 0.10† | 0.03 ± 0.04† |
| MYO6 | chr6:76618213:76618344:+@chr6:76621389:76621415:+@chr6:76623780:76623998:+ | 0.37 ± 0.21 | 0.14 ± 0.09† | 0.13 ± 0.06† |
| NHSL1 | chr6:138768138:138768330:−@chr6:138763120:138763251:−@chr6:138751530:138754817:− | 0.42 ± 0.20 | 0.77 ± 0.13† | 0.83 ± 0.15† |
| Splicing patterns not associating with either RE−/FECD− or RE+/FECD+ | ||||
| TSPOAP1 | chr17:56387328:56387519:−@chr17:56385902:56386741:−@chr17:56385203:56385302:− | 0.91 ± 0.08 | 0.28 ± 0.12 | 0.42 ± 0.40‡ |
| NUMA1 | chr11:71723941:71727306:−@chr11:71723447:71723488:−@chr11:71721832:71721900:− | 0.74 ± 0.06 | 0.27 ± 0.08 | 0.54 ± 0.10‡ |
| PPFIBP1 | chr12:27829361:27829532:+@chr12:27829997:27830029:+@chr12:27832422:27832572:+ | 0.67 ± 0.15 | 0.14 ± 0.06 | 0.38 ± 0.25‡ |
| SYNE1 | chr6:152469180:152469513:−@chr6:152466622:152466690:−@chr6:152464758:152464900:− | 0.72 ± 0.16 | 0.27 ± 0.09 | 0.51 ± 0.26‡ |
| MBNL2 | chr13:97999058:97999321:+@chr13:98009050:98009103:+@chr13:98009736:98009889:+ | 0.07 ± 0.06 | 0.58 ± 0.19 | 0.34 ± 0.19‡ |
| KIF13A | chr6:17794480:17794626:−@chr6:17790103:17790141:−@chr6:17788007:17788106:− | 0.74 ± 0.17 | 0.13 ± 0.08 | 0.45 ± 0.44‡ |
| ITGA6 | chr2:173362703:173362828:+@chr2:173366500:173366629:+@chr2:173368819:173371181:+ | 0.72 ± 0.13 | 0.18 ± 0.10 | 0.49 ± 0.14‡ |
| ABI1 | chr10:27065994:27066170:−@chr10:27060004:27060018:−@chr10:27059174:27059274:− | 0.76 ± 0.16 | 0.35 ± 0.13 | 0.54 ± 0.10‡ |
| ADD3 | chr10:111890121:111890244:+@chr10:111892063:111892158:+@chr10:111893084:111895323:+ | 0.05 ± 0.03 | 0.41 ± 0.11 | 0.23 ± 0.08‡ |
| COPZ2 | chr17:46105838:46105876:−@chr17:46105042:46105155:−@chr17:46103533:46103841:− | 0.08 ± 0.04 | 0.35 ± 0.13 | 0.12 ± 0.05‡ |
| SCARB1 | chr12:125270903:125271049:−@chr12:125267229:125267357:−@chr12:125262174:125263132:− | 0.70 ± 0.16 | 0.30 ± 0.14 | 0.44 ± 0.29‡ |
| INF2 | chr14:105180540:105181193:+@chr14:105181621:105181677:+@chr14:105185132:105185947:+ | 0.86 ± 0.14 | 0.24 ± 0.10 | 0.22 ± 0.32‡ |
| CD46 | chr1:207958964:207959027:+@chr1:207963598:207963690:+@chr1:207966864:207968861:+ | 0.59 ± 0.07 | 0.33 ± 0.09 | 0.42 ± 0.03‡ |
| VEGFA | chr6:43746626:43746655:+@chr6:43749693:43749824:+@chr6:43752278:43754223:+ | 0.46 ± 0.17 | 0.77 ± 0.07 | 0.57 ± 0.17‡ |
| FGFR1 | chr8:38314874:38315052:−@chr8:38287200:38287466:−@chr8:38285864:38285953:− | 0.40 ± 0.21 | 0.77 ± 0.23 | 0.27 ± 0.28‡ |
| PLEKHM2 | chr1:16046229:16046415:+@chr1:16047824:16047883:+@chr1:16051812:16052040:+ | 0.68 ± 0.13 | 0.28 ± 0.09 | 0.55 ± 0.35‡ |
| EXOC1 | chr4:56749989:56750094:+@chr4:56755054:56755098:+@chr4:56756389:56756552:+ | 0.24 ± 0.18 | 0.80 ± 0.10 | 0.41 ± 0.24‡ |
RE−/FECD−, no TCF4 trinucleotide repeat expansion and no Fuchs' endothelial corneal dystrophy; RE+/FECD+, TCF4 trinucleotide repeat expansion and Fuchs' endothelial corneal dystrophy; RE+/FECD−; TCF4 trinucleotide repeat expansion and no Fuchs' endothelial corneal dystrophy.
PSI values that are similar between RE+/FECD− and RE−/FECD−.
PSI values that are similar between RE+/FECD− and RE+/FECD+.
PSI values for RE+/FECD− that do not show similarity to RE−/FECD− or RE+/FECD+.
In addition to genes whose splicing patterns associated with RE−/FECD− samples, our analysis of missplicing in RE+/FECD− samples identified four genes (CLASP1, GOLGA2, MYO6, NHSL1) whose PSI values were within 1 SD from the mean of RE+/FECD+ values (Table 2). For example, in RE+/FECD− samples, CLASP1 PSI values were 0.37, 0.31, and 0.17, all within 1 SD of RE+/FECD+ patients (0.28 ± 0.18). Similar results were identified for GOLGA2, MYO6, and NHSL1.
For the remaining 17 genes, the PSI values for at least one RE+/FECD− sample fell between those recorded for RE+/FECD+ and the RE−/FECD− samples (Table 2). Due to the small sample size, we could not specifically identify whether their splicing patterns were more similar to RE−/FECD− or RE+/FECD+ samples, or had splicing patterns consistently between those reported for RE−/FECD− and RE+/FECD+ samples.
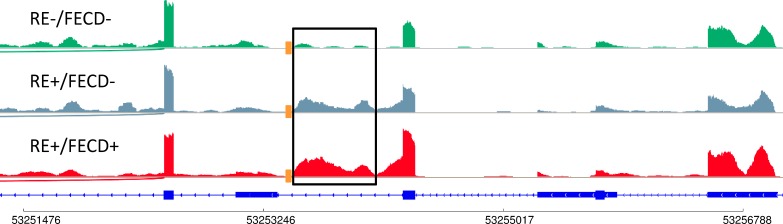
In addition to the gene set described above, the impact of the TNR expansion on splicing in the TCF4 gene was examined. We and others had previously reported that the intron just upstream of the TNR expansion in TCF4 is preferentially retained in RE+/FECD+ samples5,14 but was absent in FECD samples that lacked a TNR expansion. Examining this region in the RE+/FECD− samples revealed that this intron is also retained, similar to RE+/FECD+ samples even in the absence of clinical FECD (Figure). This suggests that the retention of this intron might be a reliable marker for identifying the presence of a TNR expansion, but is not a reliable marker for FECD status.
Figure.
Effect of TNR expansion on splicing in the TCF4 gene. RNASeq analysis of the TCF4 gene shows that in RE+/FECD− and RE+/FECD+ patients, missplicing occurs in the intron region (outlined in black) immediately downstream from the location of the TNR expansion (orange square). This additional sequence is not present in TCF4 transcripts obtained from RE−/FECD− patients.
Effect of TNR Expansion on Gene Expression in Patients With or Without FECD
To identify genes that link TNR expansion to FECD pathophysiology, we compared gene expression profiles between RE+/FECD+ and RE+/FECD− individuals. A total of 810 genes with at least a 2-fold higher expression in RE+/FECD+ compared to RE+/FECD− samples were identified (Supplementary Table S1). This included SLC4A11, a gene that has previously been implicated in the pathogenesis of FECD (Table 3).15 Several genes involved in mRNA splicing (ARVCF, CELF5, KHDRBS2, and RBFOX1) and Wnt signaling were also increased in RE+/FECD+ samples (Table 3). Analysis with PANTHER revealed that RE+/FECD+ samples had increased expression of molecules involved in ion transport and metabolic systems.
Table 3.
Selected Genes With Increased Expression in RE+/FECD+
|
Gene Name |
Log2 Fold Change |
Description |
| mRNA splicing | ||
| ARVCF | 2.42 | Armadillo repeat protein deleted in Velocardiofacial syndrome |
| CELF5 | 4.92 | CUGBP Elav-like family member 5 |
| KHDRBS2 | 4.34 | KH RNA binding domain containing, signal transduction associated 2 |
| RBFOX1 | 2.35 | RNA binding Fox-1 homolog 1 |
| Wnt signaling | ||
| DKK4 | 3.22 | Dickkopf WNT signaling pathway inhibitor 4 |
| DKKL1 | 8.38 | Dickkopf-like acrosomal protein 1 |
| ERBB4 | 3.70 | Erb-B2 receptor tyrosine kinase 4 |
| FRZB | 2.94 | Frizzled related protein |
| KDR | 3.17 | Vascular endothelial growth factor receptor 2 |
| MST1R | 2.48 | Macrophage stimulating 1 receptor |
| RNF43 | 2.00 | Ring finger protein 43 |
| SFRP1 | 3.34 | Secreted frizzled-related protein 1 |
In addition to genes showing increased expression in RE+/FECD+ samples, we also identified 1372 genes that have more than 2-fold lower expression in RE+/FECD+ samples when compared to RE+/FECD− (Supplementary Table S2). Analysis of this decreased expression gene set with PANTHER revealed significant reduction for intracellular signaling pathways. This included decreased expression of Toll-like receptor signaling, TGFβ superfamily members, and genes involved in TGFβ2 expression and activation (Table 4). This gene set also identified a number of downregulated molecules in the RE+/FECD+ group whose expression is associated with cell senescence.
Table 4.
Selected Genes With Decreased Expression in RE+/FECD+
|
Gene Name |
Log2 Fold Change |
Description |
| Innate immune system | ||
| TLR4 | −3.57 | Toll like receptor 4 |
| TLR5 | −2.88 | Toll like receptor 5 |
| TLR6 | −2.93 | Toll like receptor 6 |
| TLR7 | −3.97 | Toll like receptor 7 |
| TLR8 | −3.27 | Toll like receptor 8 |
| TLR10 | −2.87 | Toll like receptor 10 |
| CD14 | −2.36 | CD14 antigen |
| TGFβ superfamily members | ||
| BMP2 | −3.17 | Bone morphogenetic protein 2 |
| BMP4 | −4.20 | Bone morphogenetic protein 4 |
| BMP6 | −4.93 | Bone morphogenetic protein 6 |
| GDF7 | −2.78 | Growth differentiation factor 7 |
| GDF15 | −2.06 | Growth differentiation factor 15 |
| GDNF | −6.56 | Glial cell-derived neurotrophic factor |
| INHBA | −5.60 | Inhibin subunit beta A |
| LEFTY2 | −4.85 | Left-right determination factor 2 |
| TGFβ2 | −2.13 | Transforming growth factor beta 2 |
| TGFβ activation/signaling | ||
| ADAMTS2 | −2.81 | ADAM metallopeptidase with thrombospondin type 1 motif 2 |
| ADAMTS10 | −3.40 | ADAM metallopeptidase with thrombospondin type 1 motif 10 |
| ELN | −4.61 | Elastin |
| FBLN2 | −2.89 | Fibulin 2 |
| FBLN5 | −3.27 | Fibulin 5 |
| FBN1 | −2.82 | Fibrillin 1 |
| FBN2 | −5.31 | Fibrillin 2 |
| LTBP2 | −2.10 | Latent transforming growth factor beta binding protein 2 |
| miR21 | −3.56 | MicroRNA 21 |
| MMP2 | −2.13 | Matrix metalloproteinase 2 |
| TGFβ2 | −4.28 | Transforming growth factor beta 2 |
| VCAN | −5.66 | Versican |
| Senescence markers | ||
| CDKN1A | −2.83 | Cyclin dependent kinase inhibitor 1A |
| CDKN2A | −4.08 | Cyclin dependent kinase inhibitor 2A |
| CDKN2B | −3.53 | Cyclin dependent kinase inhibitor 2B |
| SERPINE1 | −4.05 | Serpin family E member 1 |
| TAGLN | −4.14 | Transgelin |
Identification of Genetic Variants in RE+/FECD−
The apparent disruption of multiple splicing and gene expression changes in RE+/FECD− samples could reflect underlying changes in the DNA sequence. To identify genetic variants, exome sequencing was performed on leukocyte DNA obtained from the 5 RE+/FECD− patients (Table 1). Analysis of the RE+/FECD− samples did not identify any variants that were common to all five samples, but did identify 94 genes that had uncommon variants in at least two of the RE+/FECD− samples (Supplementary Table S3). Comparison of the exome sequencing results to those obtained by RNASeq showed that 64 of the 94 genes were expressed in the corneal endothelium. Analysis of the 64 genes by PANTHER did not reveal any statistically significant enrichment for molecular function, biological function, or cellular component association.
Discussion
To understand the molecular events leading from TNR expansion to expression of FECD disease phenotype, we analyzed a cohort of elderly patients with CTG TNR expansion in the pathological range but not showing any signs of phenotypic FECD (RE+/FECD−). By performing RNASeq and exome sequencing on corneal endothelial tissue and blood mononuclear cells, respectively, from these patients and comparing them to RE+/FECD+ and RE−/FECD− datasets, we identified a marked reduction in TNR expansion-driven missplicing in MBNL1, KIF13A, and AKAP13 and decreased expression of several known splicing factors in RE+/FECD− samples. This is consistent with previous reports stating that splicing changes in RE+/FECD+ patients have significant implication in the pathogenesis of FECD.5,6,9 Additionally, gene expression analysis of RE+/FECD− patients identified several important signaling pathways that have been implicated in FECD.
In RE+/FECD− samples, only 4 of the 24 previously described6 most common misspliced genes (CLASP1, NHSL1, MYO6, and GOLGA2) associated with TNR expansion and FECD were identified with a splicing signature consistent with RE+/FECD+ samples. This was somewhat of a surprise given that RE+/FECD− and RE+/FECD+ samples contain TNR expansions in the TCF4 gene. In contrast, the pattern of splicing of MBNL1, KIF13A, and AKAP13 was consistent with RE−/FECD− samples. Of particular interest is MBNL1. Missplicing of MBNL1 has been identified as a key event in the pathogenesis of type I myotonic dystrophy, another TNR expansion disease.16,17 Given that the same MBNL1 missplicing event is found in both FECD and myotonic dystrophy, it is reasonable to hypothesize that the absence of MBNL1 missplicing in RE+/FECD− samples is a key driver for the absence of FECD in these individuals. The work presented here does not address this important question experimentally, so additional studies will be required to rigorously assess this possibility. Nevertheless, it is clear that some disruption of TNR expansion-induced missplicing events occurs in RE+/FECD− patients, reinforcing a prominent role of MBNL1-mediated mechanisms in expression of FECD phenotype.
The observation of decreased mRNA levels for four genes known to be involved in alternative splicing events (ARVCF, CELF5, KHDRBS2, and RBFOX1; Table 3) in the RE+/FECD− samples compared to the RE+/FECD+ samples also merits consideration in FECD disease pathogenesis. Previous data have shown that RBFOX1 and MBNL1 cooperate to regulate the splicing of many transcripts in myotonic dystrophy patients and, in some cell types, RBFOX1 knockdown has been shown to influence the splicing pattern of MBNL1 transcripts.18 One hypothesis for the lack of FECD in RE+/FECD− patients could be that altered activity of some splicing factors may negate some missplicing events, counteracting the influence of TNR expansion-induced missplicing in these patients.
The increased expression of 810 genes, including SLC4A11 (Supplementary Table S1), in RE+/FECD+ samples compared to RE+/FECD− samples provides a window into more global differences in gene expression associated with the development and progression of FECD. In addition to the increased expression of alternative splicing factors discussed above, higher expression of proteins involved in WNT signaling was identified. While several of these molecules act as antagonists to Wnt signaling (i.e., DKK4, DKKL1, SFRP1), and with decreased expression of WNT2, WNT7a, and WNT11, it is clear that normal WNT signaling is altered in RE+/FECD+ compared to RE+/FECD−. The overall effect on cell health and disease causality based on these findings is unclear, but endothelial-to-mesenchymal transition, a process controlled by WNT signaling, has been implicated as a possible factor in FECD.19 Further studies to determine the effect these modifications to WNT signaling have on corneal endothelial cellular physiology in FECD are warranted.
Similar considerations apply to the 1372 genes (Supplementary Table S2) that are downregulated in RE+/FECD+ samples relative to the RE+/FECD− samples. PANTHER overrepresentation analysis for this group of genes identified downregulation of several key intracellular pathways (e.g., Toll-like receptor, TGFβ2 signaling). Toll-like receptors recognize pathogens and respond to cell and tissue injury by activating the innate immune system.20 Decreased expression of TOLL-like receptors in FECD suggests a reduced ability of these cells to respond to pathogenic changes in the microenvironment. Because individuals with FECD may have increased levels of inflammation,21 it is feasible that part of this response may be due to changes in immune defense within the corneal endothelium.
In addition to Toll-like receptors, we also found decreased expression of second messenger signaling pathways including TGFβ superfamily members (Table 4) in RE+/FECD+ compared to RE+/FECD−. This general reduction in cell signaling cascades may be a reflection on the altered health of RE+/FECD+ cells and its environment compared to RE+/FECD−. For example, TGFβ2 is secreted from cells in a pro-protein form that binds to latent transforming growth factor binding proteins (LTBP), fibrillins (FBN), and fibulins (FBLN), which enables TGFβ2 to localize in the extracellular matrix. From here, active TGFβ2 levels are regulated by several proteases such as ADAM metallopeptidases (ADAMTS) and matrix metalloproteases (MMP) that respond to cues from the cell and extracellular matrix for proper expression. In RE+/FECD+ samples, this process appears to be downregulated. In addition to reduced levels of TGFβ2 mRNA, these cells also have reduced expression of TGFβ-associated proteins LTBP2, ADAMTS2, ADAMTS10, FBN1, FBN2, FBLN2, FBLN5, and MMP2. Because TGFβ2 signaling is important for expression of a variety of extracellular matrix proteins, this downregulation may alter their levels of expression, which are observed in these cells (i.e., VCAN, ELN, and a variety of collagen subunits including COL1A1, COL4A1, and COL4A2; Table 4; Supplementary Table S2). This hypothesis seems to be in contrast to that previously reported by Okumura et al.22 In their study, they found elevated levels of TGFβ2-induced expression of several extracellular matrix proteins that triggered the intrinsic apoptotic pathway through the unfolded protein response in a cell model of FECD. While differences in model systems (cell culture versus tissue), controls (normal corneal cells versus non-FECD tissue), and endpoint parameters (protein versus RNA) can substantiate the differences in results obtained in the two studies, it is clear that both studies show involvement of TGFβ2 signaling and FECD pathophysiology. While our analysis is speculative at this point, it will be necessary to evaluate changes in specific proteins to determine the important role our findings have on FECD pathogenesis and progression.
While the missplicing and global changes in expression between RE+/FECD− and RE+/FECD+ samples are of significant interest as they relate to the influence of a TCF4 TNR expansion sequence in patients with and without FECD, we cannot rule out the influence of other non-FECD disease processes in the RE+/FECD− group. Although all three subjects were noted clinically to be free of guttae in both the study and contralateral eyes, the tissue samples were “abnormal” because they were obtained from patients during endothelial keratoplasty for endothelial dysfunction after glaucoma and/or cataract surgery. Given this limitation, it will be important to identify additional RE+/FECD− samples to continue exploring differences in missplicing and gene expression in non-FECD patients who have a TNR expansion. Likewise, the repeat expansion length measured in leukocytes might not reflect repeat length in endothelial cells due to somatic instability of the expanded repeat sequence. While this is important information, current technology limits our ability to accurately measure repeat length in these relatively hypocellular endothelial samples.
The exome sequencing work presented here revealed no clear “protective” mutations in single genes or pathways that might account for the phenotypic absence of FECD in RE+/FECD− elderly patients. Given the biological complexity of any disease process, it is understandable that no single event is responsible for FECD disease progression. Because our sequencing efforts were limited to exons, it remains possible that additional variants in noncoding regulatory regions of critical genes may be present and might help to explain the absence of FECD in this cohort of RE+ patients.
While the generalizability of our conclusions is limited by the low numbers of RE+/FECD− samples, our findings provide important insights toward understanding the underlying mechanism of FECD pathogenesis. The association of TNR expansions of CTG18.1 and FECD is very strong and has been replicated many times, so assembling even a small cohort of RE+/FECD− DNA and tissue samples is a challenge. The magnitude of this challenge is emphasized by the absence of extensive data on expansion-positive, disease-negative samples in other repeat expansion disorders like late-onset neurologic and neuromuscular disorders that are all very rare. For most reported TNR disorders, expansion of the repeat leads to disease. However, there are rare instances of individuals with CAG TNRs in the HTT gene that fall within the pathological range but who do not demonstrate the clinical phenotypes of Huntington's disease.23 Because FECD is much more common than any other RE+ disease and phenotypically normal TCF4 RE+ individuals are more easily categorized, additional studies on such RE+/FECD− patients are feasible. Insights gained from such work will prove useful in identifying and confirming molecular targets that are protective in these patients and may serve as a platform to develop productive approaches to therapy across the spectrum of RE+ diseases.
Supplementary Material
Acknowledgments
Supported by National Eye Institute Grants EY21727 and EY26490, Robert Waller Career Development Award, and the Mayo Foundation.
Disclosure: E.D. Wieben, None; K.H. Baratz, None; R.A. Aleff, None; K.R. Kalari, None; X. Tang, None; L.J. Maguire, None; S.V. Patel, None; M.P. Fautsch, None
References
- 1.Kremer EJ, Pritchard M, Lynch M, et al. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991;252:1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- 2.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 3.Nelson DL, Orr HT, Warren ST. The unstable repeats--three evolving faces of neurological disease. Neuron. 2013;77:825–843. doi: 10.1016/j.neuron.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wieben ED, Aleff RA, Tosakulwong N, et al. A common trinucleotide repeat expansion within the transcription factor 4 (TCF4, E2-2) gene predicts Fuchs corneal dystrophy. PLoS One. 2012;7:e49083. doi: 10.1371/journal.pone.0049083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du J, Aleff RA, Soragni E, et al. RNA toxicity and missplicing in the common eye disease fuchs endothelial corneal dystrophy. J Biol Chem. 2015;290:5979–5990. doi: 10.1074/jbc.M114.621607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wieben ED, Aleff RA, Tang X, et al. Trinucleotide repeat expansion in the transcription factor 4 (TCF4) gene leads to widespread mRNA splicing changes in Fuchs' endothelial corneal dystrophy. Invest Ophthalmol Vis Sci. 2017;58:343–352. doi: 10.1167/iovs.16-20900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soragni E, Petrosyan L, Rinkoski TA, et al. Repeat-associated non-ATG (RAN) translation in Fuchs' endothelial corneal dystrophy. Invest Ophthalmol Vis Sci. 2018;59:1888–1896. doi: 10.1167/iovs.17-23265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sepp M, Kannike K, Eesmaa A, Urb M, Timmusk T. Functional diversity of human basic helix-loop-helix transcription factor TCF4 isoforms generated by alternative 5′ exon usage and splicing. PLoS One. 2011;6:e22138. doi: 10.1371/journal.pone.0022138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wieben ED, Aleff RA, Tang X, et al. Gene expression in the corneal endothelium of Fuchs endothelial corneal dystrophy patients with and without expansion of a trinucleotide repeat in TCF4. PLoS One. 2018;13:e0200005. doi: 10.1371/journal.pone.0200005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalari KR, Nair AA, Bhavsar JD, et al. MAP-RSeq: Mayo analysis pipeline for RNA sequencing. BMC Bioinformatics. 2014;15:224. doi: 10.1186/1471-2105-15-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao Y, Smyth GK. Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 13.Robinson MD, McCarthy DJ. Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sznajder LJ, Thomas JD, Carrell EM, et al. Intron retention induced by microsatellite expansions as a disease biomarker. Proc Natl Acad Sci U S A. 2018;115:4234–4239. doi: 10.1073/pnas.1716617115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iliff BW, Riazuddin SA, Gottsch JD. The genetics of Fuchs' corneal dystrophy. Expert Rev Ophthalmol. 2012;7:363–375. doi: 10.1586/eop.12.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chamberlain CM, Ranum LP. Mouse model of muscleblind-like 1 overexpression: skeletal muscle effects and therapeutic promise. Hum Mol Genet. 2012;21:4645–4654. doi: 10.1093/hmg/dds306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang ET, Cody NA, Jog S, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klinck R, Fourrier A, Thibault P, et al. RBFOX1 cooperates with MBNL1 to control splicing in muscle, including events altered in myotonic dystrophy type 1. PLoS One. 2014;9:e107324. doi: 10.1371/journal.pone.0107324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baratz KH, Tosakulwong N, Ryu E, et al. E2-2 protein and Fuch's corneal dystrophy. N Engl J Med. 2010;363:1016–1024. doi: 10.1056/NEJMoa1007064. [DOI] [PubMed] [Google Scholar]
- 20.Vidya MK, Kumar VG, Sejian V, Bagath M, Krishnan G, Bhatta R. Toll-like receptors: significance, ligands, signaling pathways, and functions in mammals. Int Rev Immunol. 2018;37:20–36. doi: 10.1080/08830185.2017.1380200. [DOI] [PubMed] [Google Scholar]
- 21.Aggarwal S, Cavalcanti BM, Regali L, et al. In vivo confocal microscopy shows alterations in nerve density and dendritiform cell density in Fuchs' endothelial corneal dystrophy. Am J Ophthalmol. 2018;196:136–144. doi: 10.1016/j.ajo.2018.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okumura N, Hashimoto K, Kitahara M, et al. Activation of the TGF-β signaling induces cell death via the unfolded protein response in Fuchs endothelial corneal dystrophy. Sci Rep. 2017;7:6801. doi: 10.1038/s41598-017-06924-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kremer B, Goldberg P, Andrew SE, et al. A worldwide study of the Huntington's disease mutation. The sensitivity and specificity of measuring CAG repeats. N Engl J Med. 1994;330:1401–1406. doi: 10.1056/NEJM199405193302001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.