Abstract
Huntington’s disease (HD) is a currently incurable and, ultimately, fatal neurodegenerative disorder caused by a CAG trinucleotide repeat expansion within exon 1 of the huntingtin (HTT) gene, which results in the production of a mutant protein that forms inclusions and selectively destroys neurons in the striatum and other adjacent structures. The RNA-guided Cas9 endonuclease from CRISPR-Cas9 systems is a versatile technology for inducing DNA double-strand breaks that can stimulate the introduction of frameshift-inducing mutations and permanently disable mutant gene function. Here, we show that the Cas9 nuclease from Staphylococcus aureus, a small Cas9 ortholog that can be packaged alongside a single guide RNA into a single adeno-associated virus (AAV) vector, can be used to disrupt the expression of the mutant HTT gene in the R6/2 mouse model of HD following its in vivo delivery to the striatum. Specifically, we found that CRISPR-Cas9-mediated disruption of the mutant HTT gene resulted in a ∼50% decrease in neuronal inclusions and significantly improved lifespan and certain motor deficits. These results thus illustrate the potential for CRISPR-Cas9 technology to treat HD and other autosomal dominant neurodegenerative disorders caused by a trinucleotide repeat expansion via in vivo genome editing.
Keywords: CRISPR-Cas9, Huntington’s disease, AAV, gene therapy, genome editing
Introduction
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disorder characterized by a progressive decline in cognitive, motor, and psychiatric function.1 HD is the most common inherited neurodegenerative disease, affecting ∼1 in 10,000 individuals, and is caused by the expansion of a CAG trinucleotide repeat within exon 1 of the huntingtin (HTT) gene.2 The presence of this expansion leads to the production of a mutant protein that aggregates in the brain and disrupts important cellular functions,3, 4 such as nucleocytoplasmic transport,5 which primarily results in the loss of medium-sized spiny neurons (MSNs) and certain cortical neurons that project to the striatum.6, 7, 8 Most individuals with HD first experience disease-associated abnormalities between 35 and 45 years of age and typically perish from the disorder ∼20 years after its manifestation.9 There is no cure for HD, and currently approved therapies can only help to manage certain physical and psychiatric symptoms.10
Although the exact mechanism by which the mutant HTT protein destroys neurons remains unknown, multiple lines of evidence have suggested that deleting or reducing mutant HTT gene expression within affected areas of the brain can halt the progression of HD.11, 12 Accordingly, both antisense oligonucleotides (ASOs) and RNAi have been used to reduce mutant HTT and improve behavioral deficits in transgenic animal models of the disorder.13, 14, 15, 16, 17, 18, 19 In fact, a phase I clinical trial designed to assess the safety and tolerability of ASOs targeting SNPs associated with the mutant CAG repeat expansion (ClinicalTrials.gov: NCT03225833 and NCT03225846) is underway. Additionally, a clinical trial involving an ASO targeting both mutant and wild-type HTT mRNAs has yielded early encouraging results (ClinicalTrials.gov: NCT02519036), indicating that targeting both the native and the mutant HTT proteins could be tolerated in a clinical setting. Further, a microRNA-based gene therapy for HD20, 21, 22 is in the final stages of pre-clincial development and could soon be evaluated in patients. However, despite advances in chemistry and design, ASOs can only transiently repress the production of the HTT protein and may still require a lifetime of administrations to patients, while RNAi is prone to off-target effects.23, 24, 25 Additionally, both ASOs and RNAi are associated with incomplete knockdown and lack the capacity to correct the underlying genetic defect responsible for HD, which could ultimately limit their utility as therapeutics.
Genome editing—a method that enables the precise alteration of a targeted DNA sequence—offers an alternative approach to treat HD by providing a means to permanently disrupt the function of the HTT gene.26 CRISPR (clustered regularly interspaced short palindromic repeats)-CRISPR-associated (Cas) systems,27, 28, 29, 30, 31 in particular, have emerged as especially versatile and efficient gene-editing technologies that hold considerable potential as therapeutics.32 The CRISPR-Cas9 DNA-editing system consists of two core components: the Cas9 nuclease and a single guide RNA (sgRNA) that binds to Cas9 and directs it to a targeted genomic site via RNA-DNA base complementarity.27 Upon DNA binding, Cas9 introduces a DNA double-strand break (DSB) that stimulates non-homologous end joining (NHEJ), an error-prone DNA repair pathway that facilitates the introduction of random base insertions and deletions (indels) that can lead to a frameshift mutation33 and thereby disrupt gene expression via nonsense-mediated mRNA decay.34 As a result, CRISPR-Cas9 technology could be used to disable the production of the HTT protein and treat HD following its in vivo delivery to areas of the brain most affected by the disorder.
In the present study, we demonstrate that CRISPR-Cas9 can be deployed to the striatum via a single adeno-associated virus (AAV) vector particle to disable the expression of the human mutant HTT gene in an especially aggressive animal model of the disorder. Specifically, we show that CRISPR-Cas9-mediated disruption of the mutant HTT gene in R6/2 mice, which carry exon 1 of the human HTT gene with ∼115–150 CAG repeats, reduces the formation of neurotoxic inclusions by 2-fold, increases lifespan, and improves certain motor deficits in these same mice. Our results illustrate the potential for CRISPR-Cas9 technology to treat HD and reinforce its potential for treating other autosomal dominant neurodegenerative disorders.
Results
Using CRISPR-Cas9 to Disrupt HTT Gene Expression
Owing to its ability to induce targeted DSBs that can drive the formation of frameshift-inducing indels, we hypothesized that CRISPR-Cas9 could be used to disrupt the expression of the HTT gene following its in vivo delivery using an AAV vector, a clinically promising engineered gene delivery vehicle35, 36 capable of transducing various substructures within the brain,37 including the striatum. AAV vectors, however, have a limited carrying capacity that restricts single-particle delivery of the prototypical Cas9 nuclease from Streptococcus pyogenes (SpCas9) alongside an sgRNA expression cassette.38 Thus, in order to more effectively deliver a CRISPR-Cas9 gene-editing system in vivo, we used the Cas9 nuclease from Staphylococcus aureus (SaCas9) to target the human HTT gene.39 SaCas9 is ∼1 kb smaller than SpCas9 and can fit into a single AAV particle along with an sgRNA and a neuron-specific promoter to drive its expression in vivo.
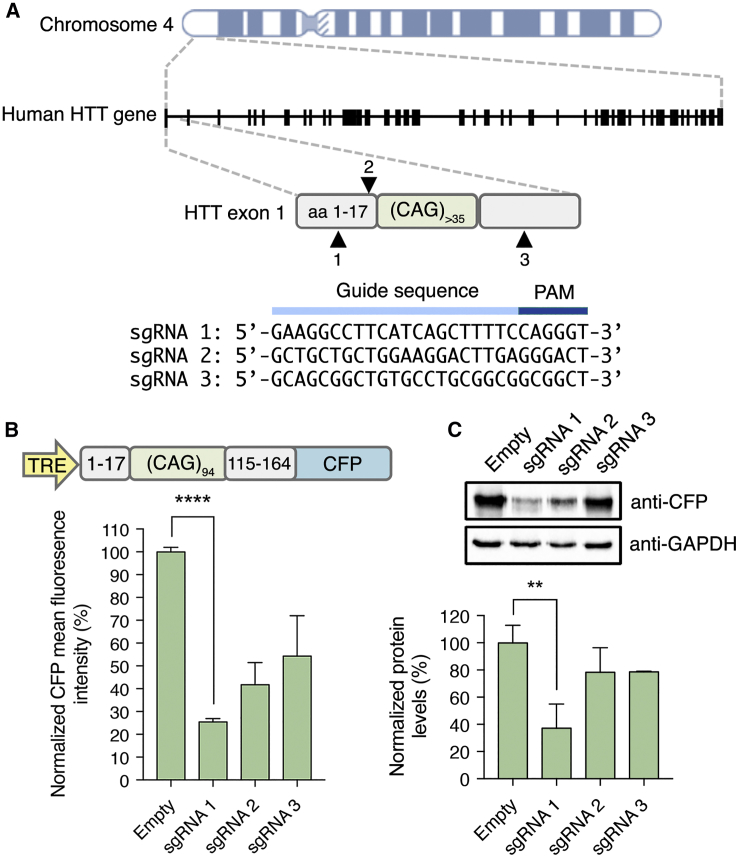
We designed several sgRNAs to target exon 1 of the human HTT gene either upstream or downstream of the CAG repeat expansion, with the expectation that SaCas9-induced indels would reduce the production of the human HTT protein (Figure 1A). Given that the CAG trinucleotide repeat, as well as the large size of the full-length HTT protein (∼350 kDa) can confound quantitative analyses, we used an established reporter that inducibly expresses exon 1 of the human HTT gene with 94 glutamines (94Q) fused to a cyan fluorescent protein (CFP) variant (Figure 1B),40 thereby linking mutant HTT gene expression to CFP fluorescence for facile evaluation of the designed sgRNAs. Following transfection into HEK293T cells, we observed that each sgRNA decreased CFP fluorescence intensity by at least 50%, with the most effective sgRNA reducing fluorescence by ∼75% (p < 0.007) (Figure 1B). These findings were corroborated by western blot, which indicated a ∼65% decrease in mutant HTT-CFP fusion protein for the most efficient sgRNA in comparison to control cells (p < 0.0001) (Figure 1C). Sanger sequencing further confirmed the presence of SaCas9-induced indels for this sgRNA within the mutant HTT transgene, though we identified mutations in only ∼10% of sequenced amplicons (Figure S1). Given the proximity of the sgRNA target site to the 3′ primer binding site used for this PCR amplification, we anticipate that SaCas9 could have mutated a fraction of these sites, thereby preventing efficient amplification of a subset of the edited transgenes. Collectively, these results indicate that SaCas9 can be used to target the human HTT gene and reduce mutant HTT protein in reporter cells.
Figure 1.
Disruption of the Mutant HTT Gene in Reporter Cells Using CRISPR-Cas9 Nucleases
(A) Schematic representation of the human HTT locus located on chromosome 4 and the candidate sgRNA target sites. Arrowheads indicate the approximate location of the sgRNA binding site. aa, amino acids; PAM, protospacer-adjacent motif. (B) Top: graphic of the mutant HTT-CFP reporter. Bottom: the percentage of CFP-positive HEK293T cells 72 h after transfection with reporter plasmid, SaCas9, and the HTT-targeting sgRNAs or a non-targeted sgRNA (Empty) (n = 3). (C) Top: western blot of lysate from HEK293T cells 72 h after transfection with reporter plasmid, SaCas9, and the HTT-targeting sgRNAs or a non-targeted sgRNA (Empty). Bottom: quantitation of western blot results. CFP protein was normalized to GAPDH protein in each lane (n = 3). Error bars indicate SD. **p < 0.01; ***p < 0.001, unpaired t test.
CRISPR-Cas9 Reduces Mutant HTT Protein Inclusions in the R6/2 Mouse Model of HD
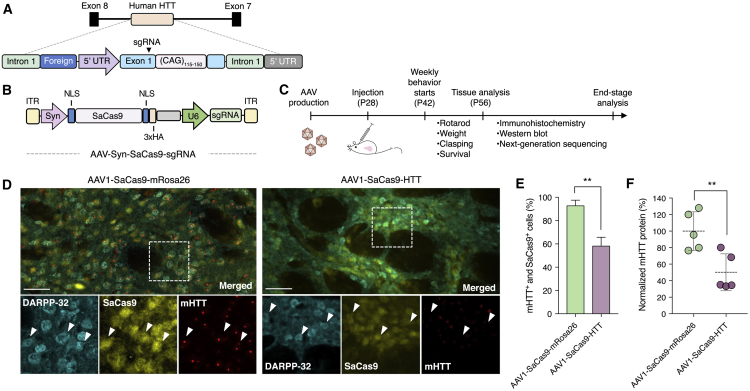
We next evaluated whether the SaCas9 nuclease could reduce mutant HTT protein in vivo following its delivery to the R6/2 mouse model of HD, a transgenic mouse strain that carries the 5′ end of the human HTT gene, which includes: (1) ∼1 kb of the 5′ UTR sequence, (2) exon 1 of the HTT gene with ∼115–150 CAG repeats, (3) the first 262 bp of intron 1, and (4) a 168-bp foreign segment from bacterial DNA41, 42 (Figure 2A). R6/2 mice develop HTT protein inclusions in the striatum and, eventually, the cortex and exhibit a progressive neurodegenerative phenotype that mimics many features of HD in humans, including weight loss, tremors, epileptic seizures, movement abnormalities, and premature death.43, 44 This strain is a well-characterized and a widely used model for studying and evaluating potential treatments for HD.
Figure 2.
In Vivo Disruption of the Mutant HTT Gene in R6/2 Mice
(A) Cartoon of the human HTT transgene located in Gm12695 (predicted gene 12695) on chromosome 4 in R6/2 mice. Arrowhead indicates approximate location of sgRNA binding site. (B) Schematic of the AAV vector. ITR, inverted terminal repeat; hSyn, human synapsin promoter; NLS, nuclear localization sequence; 3xHA, three tandem repeats of the human influenza hemagglutinin (HA) epitope tag. (C) Timeline for in vivo studies. (D) Immunofluorescent staining of striatal sections 4 weeks after R6/2 mice were injected with 6 × 1010 vector genomes of (left) AAV1-SaCas9-mRosa26 or (right) AAV1-SaCas9-HTT. Insets show high-magnification images. Arrowheads indicate representative DARPP-32+ and SaCas9+ cells with (left) high or (right) reduced mutant HTT (mHTT) protein. Images were captured using identical exposure conditions. Scale bars, 50 μm. (E) Quantitation of immunohistochemical results from R6/2 mice injected with AAV1-SaCas9-HTT (n = 3) or AAV1-SaCas9-mRosa26 (n = 3). (F) Normalized mutant HTT protein in striatal lysate 4 weeks after R6/2 mice were injected with 6 × 1010 vg of AAV1-SaCas9-mRosa26 or AAV1-SaCas9-HTT (n = 5). **p < 0.01, unpaired t test. Error bars indicate SD. All injections were performed on 28-day-old animals.
We injected the striatum of 4-week-old R6/2 mice at three depths with 2 × 1010 viral genomes (vg) of an AAV1 vector encoding SaCas9 and either the most efficient sgRNA targeting the human HTT gene (AAV1-SaCas9-HTT) or an sgRNA targeting the mouse Rosa26 locus (AAV1-SaCas9-mRosa26), a safe harbor site that can support stable transgene expression.45 Transgenic mice injected with AAV1-SaCas9-mRosa26 previously showed no signs of toxicity and displayed no changes in disease progression compared to those injected with an EGFP-encoding AAV,46 indicating the suitability of AAV1-SaCas9-mRosa26 as a negative control. Additionally, AAV1 can effectively transduce neurons in the striatum47 and has previously been used to deliver neurotrophic factors48 and engineered RNAi systems49 to HD rodent models. To ensure that SaCas9 is specifically expressed in neurons, we used the human synapsin (hSyn) promoter to drive its expression from the AAV vector (Figure 2B).
Four weeks after AAV delivery (Figure 2C), we used immunohistochemistry (IHC) to analyze brain sections from treated and untreated animals for the expression of: (1) SaCas9 via its hemagglutinin (HA) epitope tag; (2) MSNs, the major neuronal cell type of the striatum, using an antibody that targets the MSN-specific marker DARPP-32; and (3) mutant HTT, using an antibody that has been reported to efficiently recognize the mutant form of the human protein50 (Figure 2D). We observed SaCas9 expression in the central striatum (Figure S2), with quantitative analysis revealing that ∼85% of DARPP-32+ cells in the injected area expressed SaCas9 (Figure S3). Importantly, we also found that R6/2 mice infused with AAV1-SaCas9-HTT had ∼40% fewer mutant HTT protein inclusions in dual SaCas9+ and DARPP-32+ cells compared to animals injected with AAV1-SaCas9-mRosa26 (p = 0.003) (Figure 2E). Western blot analysis further revealed that mice treated by CRISPR-Cas9 had ∼50% less total mutant HTT protein in whole striatal lysate compared to control animals (p < 0.008) (Figure 2F; Figure S4).
To evaluate whether SaCas9 induced indels in vivo, we deep sequenced the human HTT transgene from whole dissociated striatal tissue, which consisted of a mixture of transduced and non-transduced cells. According to CRISPResso,51 a computational pipeline for quantifying genome-editing outcomes from sequencing data, indels were present in ∼6% of the analyzed human HTT transgenes from CRISPR-treated animals, which corresponded to a ∼12-fold increase in indels compared to that in control mice, though we observed indel frequencies up to 14% in some animals (Figure S5). To determine whether SaCas9 induced off-target effects, we deep sequenced 10 candidate off-target sites identified using the web-based tool Cas-OFFinder.52 CRISPResso analysis revealed no increase in indel formation at any of the 10 sites, including the mouse HTT gene, which deviates from the human sgRNA target site by 1 bp (Figure S6). Taken together, these results indicate that SaCas9-mediated disruption of the mutant HTT gene can reduce mutant protein in the brain.
CRISPR-Cas9-Mediated Disruption of the Mutant HTT gene Increases Survival in R6/2 Mice
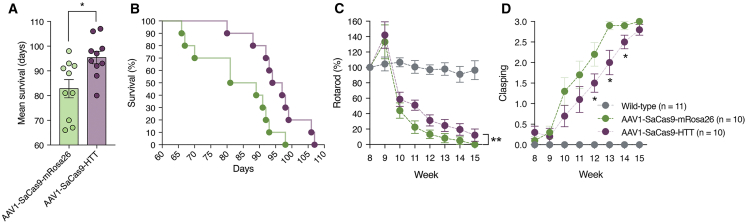
We next sought to determine whether CRISPR-Cas9-mediated disruption of the mutant HTT gene could provide therapeutic benefit to R6/2 mice, which develop a particularly aggressive and rapid form of neurodegeneration and have a shortened lifespan (typically, 16 weeks of age) compared to other transgenic models of the disorder. Starting at 4 weeks after AAV injections, we measured motor function, hindlimb clasping (an established indicator of dystonia, a clinical characteristic of HD), and weight on a weekly basis, with end-stage determined as the point at which animals either were moribund, lacked a righting reflex, or failed to respond to gentle stimulation.
R6/2 mice injected with AAV1-SaCas9-HTT displayed a ∼15% increase in mean survival compared to control animals (HTT: 95.4 ± 2.5 days; mRosa26: 82.8 ± 3.7 days; p < 0.01) (Figure 3A) and had lifespans that ranged from 80 to 106 days, compared to 66 to 98 days for control mice (p = 0.01) (Figure 3B). We also observed that R6/2 mice treated by SaCas9 gene editing had improved motor function (p < 0.01) (Figure 3C) and decreased hindlimb clasping at multiple weeks, including week 12 (p < 0.05), week 13 (p < 0.01), and week 14 (p = 0.05) (Figure 3D). In fact, we observed that more than half of all treated animals did not exhibit any signs of clasping until week 11, whereas 80% of the AAV1-SaCas9-mRosa26-treated animals had clasped by week 10. Interestingly, we observed no difference in weight (typically an indicator of disease onset) between treated and untreated mice (Figure S7), which could be due to the fact that, while HTT protein inclusions can develop as early as 2 weeks of age in R6/2 mice,53, 54 we injected AAV vector into 4-week-old animals. Thus, earlier administration of the AAV vector to this strain or delivery to a late-onset model of HD55 could shed more light on the ability of SaCas9 to slow the onset of the disease.
Figure 3.
CRISPR-Cas9-Mediated Disruption of the Mutant HTT Gene Provides Therapeutic Benefit to R6/2 Mice
(A–D) Mean survival (A), percent survival (B), normalized rotarod (C), and normalized clasping (D) scores for R6/2 mice bilaterally injected with 6 × 1010 vg of AAV1-SaCas9-HTT (n = 10) and AAV1-SaCas9-mRosa26 (n = 10). Wild-type mice (n = 11) are litter-matching B6CBAF1 mice. Values are means, and error bars indicate SEM. *p < 0.05. In (B), unpaired t test was used; in (C) and (D), a two-way ANOVA was used, followed by Tukey’s post hoc analysis.
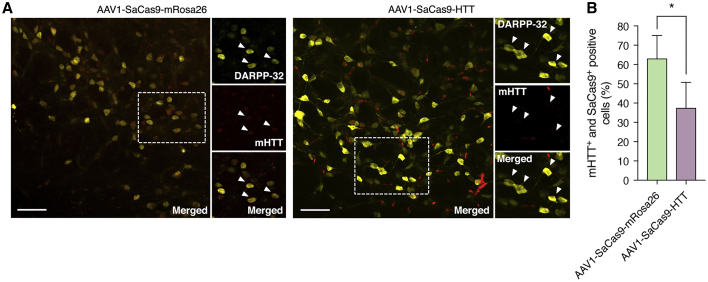
Finally, immunohistochemical analysis of striatal sections from both treated and untreated animals at end-stage revealed that gene-edited mice had ∼30% fewer mutant HTT protein inclusions in SaCas9+ cells compared to control animals (p < 0.05) (Figures 4A and 4B). Additionally, we found that end-stage mice had ∼10% more total SaCas9+ cells than animals infused with AAV1-SaCas9-mRosa26 (Figure S8). Since we observed similar transduction efficiency in animals injected with AAV1-SaCas9-HTT and AAV1-SaCas9-mRosa26 (Figure S2), these results likely indicate that CRISPR-mediated disruption of the mutant HTT gene can impart protection to some neurons from mutant HTT-induced toxicity.
Figure 4.
CRISPR-Cas9 Enhances Neuronal Survival in R6/2 Mice
(A) Immunofluorescent staining of end-stage striatal sections from R6/2 mice bilaterally injected with 6 × 1010 vector genomes of (left) AAV1-SaCas9-mRosa26 or (right) AAV1-SaCas9-HTT. Insets show high-magnification images. Arrowheads indicate representative DARPP-32+ and SaCas9+ cells with (left) high or (right) reduced mutant HTT (mHTT) protein. Images were captured using identical exposure conditions. Scale bars, 50 μm. (B) Quantitation of immunohistochemical results from R6/2 mice injected with AAV1-SaCas9-HTT (n = 4) or AAV1-SaCas9-mRosa26 (n = 3). *p < 0.05, one-way unpaired t test.
Collectively, these results establish that CRISPR-Cas9 can be used to reduce mutant HTT protein in R6/2 mice and that SaCas9-mediated disruption of the mutant HTT gene can significantly increase survival and improve motor deficits.
Discussion
HD is the most common inherited neurodegenerative disorder, affecting over 30,000 people in the United States,56 with an estimated 200,000 individuals at risk of inheriting the disease in the United States alone. There is no cure for HD, and current therapies only provide symptomatic relief. Thus, there is an urgent need for strategies that can reduce the formation of toxic HTT protein inclusions and treat the underlying cause of the disorder. Here, we show that CRISPR-Cas9—a versatile technology that we46, 57 and others58, 59, 60, 61, 62 have established can be deployed in the nervous system to facilitate genome editing—can be harnessed to disrupt the expression of the mutant HTT gene in the R6/2 mouse model of HD, one the most aggressive transgenic animal models of the disorder.63 Specifically, we demonstrate that CRISPR-Cas9 can be used to reduce neuronal protein inclusions following its delivery to the striatum, an outcome that resulted in increased lifespan and improved motor function. Our study thus illustrates the potential for genome editing to treat HD.
Since R6/2 mice harbor the 5′ end of exon 1 of the human HTT gene with an expansion of ∼115–150 CAG trinucleotide repeats, we designed sgRNAs to target exon 1 of the human HTT allele. As a result, this therapeutic genome-editing strategy is not allele specific and unable to discriminate between the mutant and wild-type forms of the HTT gene in an HD patient. However, while disrupting the wild-type HTT gene can affect early neurological development and lead to embryonic lethality or cause certain side effects in young mice,64, 65, 66 several studies have suggested that reducing wild-type and mutant HTT protein in adult mice18, 66 and larger animals21 is well tolerated. In fact, a clinical trial aimed at accessing the safety of an ASOs targeting both the mutant and wild-type HTT mRNA is underway (ClinicalTrials.gov: NCT02519036), with more in the pipeline,20, 21, 22 underscoring the therapeutic feasibility of a non-allele-specific targeting approach, though additional studies are still needed to firmly establish this. In the case that specific disruption of the mutant HTT gene is required, CRISPR-Cas9 could be deployed to target distinct heterozygous SNPs associated with the CAG repeat, which was demonstrated as feasible in previous studies.16, 19, 60 However, since many of these SNPs vary among the patient population, this approach would likely require using different specialized CRISPR-Cas9 systems to target each mutation, which could make its implementation challenging.
To date, several genome-editing and targeted gene-regulating technologies have been used to reduce the expression of the mutant HTT gene, including an engineered zinc-finger repressor protein that targeted the CAG repeat expansion67 and also CRISPR-Cas9, which has been used to excise an expanded CAG repeat59 and disrupt the expression of the HTT gene.60, 61 However, to our knowledge, no other study has demonstrated that CRISPR-Cas9 technology can increase survival in an HD animal model, an important benchmark for a HD therapeutic. In particular, while Yang et al.59 used the SpCas9 nuclease to delete the CAG repeat expansion in HD140Q-knockin mice (which express exon 1 of the human mutant HTT gene in place of exon 1 of the mouse HTT gene), they did not analyze survival and reported that only early neuropathology effects were attenuated by gene editing. Similarly, while Monteys et al.60 reported that CRISPR-Cas9 can be used to selectively target mutant HTT-associated SNPs, they also did not analyze whether gene editing could rescue motor deficits in an animal model of the disorder. Importantly, our results establish that CRISPR-Cas9-mediated disruption of the mutant HTT gene can significantly increase lifespan in R6/2 mice, which aggressively and rapidly manifest HD-like symptoms, and that gene editing can confer protection to neurons from striatal degeneration. Our results also demonstrate that CRISPR-Cas9 can provide a therapeutic benefit that is on par with those previously observed with an HTT-targeting ASO that was administered to the cerebrospinal fluid of R6/2 mice.18 Finally, compared to synthetic zinc-finger repressors, which were used to improve rotarod function and reduce clasping in R6/2 mice at 5 and 7 weeks of age,67 we observed improved motor performance in mice from 10 to 14 weeks of age following Cas9-mediated gene editing.
Of note, past therapeutic gene-editing studies for HD have relied on the SpCas9 nuclease to facilitate modification of the HTT gene.59, 60, 61 Due to its relatively large size, the SpCas9 nuclease typically requires the use of two AAV particles for delivery with its sgRNA, a requirement that could reduce in vivo editing efficiency and limit therapeutic effectiveness. Our results show that SaCas9, a smaller Cas9 ortholog that can fit into a single AAV vector alongside an sgRNA and a promoter to drive its expression, can facilitate efficient disruption of the HTT gene, indicating that alternate CRISPR-Cas9 systems that are more accommodating to AAV-mediated delivery could be used to treat HD.
To effectively treat HD, CRISPR-Cas9 or any other gene-editing cargo must be efficiently delivered to the cell types affected by the disorder. Our immunohistochemical results indicated that ∼85% of DARPP-32+ cells analyzed in the striatum expressed SaCas9 4 weeks after AAV delivery, indicating efficient transduction to the targeted area. However, other neuronal populations, including certain large pyramidal projection neurons,68 are also vulnerable to HD-associated toxicity. Thus, directed evolution or other protein engineering strategies could be used to create specialized AAV capsid variants69 that can facilitate enhanced delivery to all or most of the cell populations susceptible to HD. AAV vectors, in particular, hold tremendous potential as therapeutic gene delivery vehicles, as they have demonstrated efficacy in several clinical trials.70, 71, 72, 73, 74 In fact, a retina-directed AAV-based therapy earned regulatory approval by the U.S. Food and Drug Administration in 2017, the first such endorsement for an AAV therapy in the United States, with more potentially on the horizon. Additionally, while numerous advances have been made to help minimize the frequency that CRISPR-Cas9 nucleases introduce off-target mutations in cells,75, 76, 77, 78, 79 the continuous expression of Cas9 or any other gene-editing nuclease from an AAV vector could, nonetheless, cause off-target effects to accumulate. Self-inactivating gene-editing technologies61, 80, 81 such as the KamiCas9 system,61 which has been used to target the HTT gene in an HD mouse model, could be harnessed to limit the duration that a nuclease is exposed within a cell, though it remains to be seen whether such an approach can be integrated into an AAV vector. Further, to assess the safety of this therapeutic gene-editing strategy in a genetic background relevant to HD, unbiased methods for identifying Cas9-induced DSBs39, 82, 83 should be performed in neurons differentiated from human induced pluripotent stem cells derived from HD patient cells in order to comprehensively curate sgRNA specificity. Though our analysis revealed no increase in indels at any analyzed candidate off-target cleavage site, the use of Cas9 variants with enhanced targeting specificity84 could be used to help minimize the potential for off-target effects.
Finally, we note that, while treatment with CRISPR-Cas9 increased lifespan in R6/2 mice and protected some striatal neurons from death, the therapeutic genome-editing strategy described here is unable to restore lost cells. However, we85 and others86 have demonstrated that transplanted neural progenitors can innervate within a host tissue and form new synaptic connections with endogenous neurons.85 Thus, in the future, CRISPR-Cas9-mediated gene editing could be used in combination with cell replacement therapy to treat HD via a two-step process that first involves CRISPR-mediated disruption of the endogenous mutant HTT gene to reduce its neurodegenerative effect and then involves the integration of a functional striatal graft to replace lost cells, thereby unifying the benefits of both approaches to treat HD in a potentially more effective manner.
In conclusion, we show that CRISPR-Cas9-mediated gene editing can reduce the formation of mutant HTT protein inclusions and provide therapeutic benefit to a mouse model of HD. Our results lay a foundation for using gene editing to treat HD and suggest that CRISPR-Cas9 and other emerging gene-editing technologies87 have broad potential to treat neurodegenerative disorders.
Materials and Methods
Plasmid Construction
The hSyn promoter was PCR amplified from pAAV-hSyn-mCherry (kindly provided by Dr. John Flannery) and cloned into the SpeI and AgeI sites of pAAV-CMV-SaCas9-U6-sgRNA (Addgene, #61591). Exon 1 of the human HTT gene was searched for the motif 5′-(N)20–21-NNGRRT-3′ (where N = A, T, C, or G, and R = A or G) to identify potential SaCas9 cleavage sites. Oligonucleotides encoding the identified sgRNA targeting sequences were synthesized (Elim Biopharmaceuticals), phosphorylated by T4 polynucleotide kinase (New England Biolabs), annealed, and then ligated into the BsaI restriction sites of pAAV-hSyn-SaCas9-U6-sgRNA. Plasmid sequences were verified by DNA sequencing (UC Berkeley DNA Sequencing Facility) using the oligonucleotides described in Table S1.
Cell Culture and Flow Cytometry
HEK293T cells were cultured in a 5% CO2 atmosphere at 37°C and kept in DMEM (Corning) supplemented with 10% (v/v) fetal bovine serum (FBS; Life Technologies) and 1% (v/v) Antibiotic-Antimycotic (Anti-Anti; Life Technologies). For transfections, HEK293T cells were seeded onto 24-well plates at a density of 3 × 105 cells per well. At 16 h after seeding, cells were transfected with 100 ng pTreTight-HTT94Q-CFP (Addgene, #23966), 100 ng tTA/TRE-mCherry, and 800 ng pAAV-CMV-SaCas9-U6-sgRNA using 3 μL polyethylenimine (1 μg/μL), as described elsewhere.88 At 72 h after transfection, cells were washed once with PBS, and mCherry and CFP fluorescence was evaluated by flow cytometry using a BD LSRFortessa X-20 cytometer (UC Berkeley Flow Cytometry Core Facility). For each sample, 50,000 live events were collected, and data were analyzed using FlowJo v10 (Tree Star)
Western Blot
Homogenized HEK293T cells transfected with 100 ng pTreTight-HTT94Q-CFP, 100 ng tTA/TRE-mCherry, and 800 ng pAAV-CMV-SaCas9-U6-sgRNA were lysed using RIPA buffer (50 mM Tris, 150 mM NaCl, 0.2% SDS, 0.5% deoxycholate, 1% NP-40, 1 mM EDTA [pH 8.0]) containing protease inhibitor cocktail (Sigma-Aldrich) for 30 min on ice and then centrifuged at 14,000 × g for 5 min at 4°C. Supernatants were collected, and protein concentration was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) before 15 μg protein was electrophoresed by SDS-PAGE and transferred onto a nitrocellulose membrane in transfer buffer (20 mM Tris-HCl, 150 mM glycine, and 20% [v/v] methanol) for 1 h at 160 V. Membranes were blocked with 5% (v/v) Blotting-Grade Blocker (Bio-Rad) in Tris-buffered saline (TBS) (20 mM Tris-HCl, 150 mM NaCl, [pH 7.5]) with 0.05% (v/v) Tween-20 (TBST) for 1 h and incubated overnight at 4°C with primary antibodies in blocking solution. The following antibodies were used: rabbit anti-GFP (Abcam, #ab6556) and rabbit anti-GAPDH, clone 14C10 (Cell Signaling, #2118). Membranes were washed three times with TBST and incubated with goat anti-rabbit secondary antibody horseradish peroxidase conjugate (Thermo Fisher Scientific, #65-6120) in blocking solution for 1 h at room temperature. After three washes with TBST, blots were developed using the SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific) and visualized by automated chemiluminescence using the Gel Doc XR Imaging System (Bio-Rad). Band intensity was quantitated using Image Lab (Bio-Rad). Total mutant HTT protein was normalized to GAPDH protein in each lane.
Harvested striatal tissue was lysed using RIPA buffer with 2% SDS, electrophoresed on a NuPAGE 4%–12% Bis-Tris Protein Gel (Thermo-Fisher Scientific), and transferred onto an Odyssey Nitrocellulose Membrane (Li-COR Biosciences). Membranes were incubated overnight at 4°C with the following primary antibodies: mouse anti-HTT, clone mEM48 (Millipore-Sigma, #MAB5374); and rabbit anti-GAPDH (Sigma-Aldrich). Membranes were washed three times with TBST and incubated with either biotinylated goat anti-rabbit immunoglobulin G (IgG) (Abcam, #ab64256) or biotinylated goat anti-mouse IgG (Abcam, #ab64255) in blocking solution for 1 h at room temperature. After three washes with TBST, blots were incubated with a streptavidin-Alexa Fluor 700 conjugate (Thermo Fisher Scientific, #S21383) for 1 h at room temperature and visualized using an Odyssey Imaging System. Band intensity was quantitated using Image Lab (Bio-Rad), and mutant HTT protein was normalized to GAPDH control protein in each lane.
Sanger Sequencing
HEK293T cells were seeded onto 24-well plates at a density of 3 × 105 cells per well and transfected with 100 ng tTA/TRE-mCherry and 800 ng pAAV-CMV-SaCas9-U6-sgRNA. At 72 h after transfection, mCherry+ cells were isolated using fluorescence-activated cell sorting (FACS) (BD FACSAria Fusion; UC Berkeley Flow Cytometry Core Facility), and genomic DNA was isolated using QuickExtract DNA Extraction Solution (Epicenter), according to the manufacturer’s instructions. The mutant HTT transgene was PCR amplified using the primers EcoRI-HTT-Exon 1-Fwd and XbaI-HTT-Exon 1-Rev and cloned into the EcoRI and XbaI restriction sites in pcDNA 3.1(+). Individual colonies were mini-prepped and sequenced using the primers EcoRI-HTT-Exon 1-Fwd and XbaI-HTT-Exon 1-Rev.
AAV Vector Production
AAV was manufactured as described elsewhere.88 Briefly, HEK293T cells were seeded onto 15-cm plates at a density of 4 × 107 cells per plate in DMEM with 10% (v/v) FBS and 1% Anti-Anti. At 16 h after seeding, cells were transfected with 15 μg pHelper, 15 μg AAV1, and 15 μg either pAAV-hSyn-SaCas9-U6-sgRNA-HTT or pAAV-hSyn-SaCas9-U6-sgRNA-mRosa25 using 135 μL polyethylamine (1 μg/μL). At 48 h after transfection, cells were harvested and centrifuged at 4,000 × g for 5 min at room temperature and freeze-thawed three times in lysis buffer (50 mM Tris-HCl, 150 mM NaCl [pH 8.0]). Cell lysate was incubated with 10 U benzonase nuclease (Sigma-Aldrich) for 30 min at 37°C and centrifuged at 10,000 × g for 10 min at room temperature. Supernatant was collected and laid over an iodixanol gradient and centrifuged at 42,000 × g for 2 h at 18°C. AAV was extracted from the iodixanol gradient and buffer exchanged with PBS with 0.001% Tween-20 using an Ultra-15 Centrifugal Filter Unit (Amicon) at 4,000 × g and concentrated to ∼200 μL. Virus was stored at 4°C, and the genomic titer was determined by qRT-PCR using SYBR Green (Sigma-Aldrich), as described elsewhere.88
Injections
Animal procedures were approved by the Office of Laboratory Animal Care at UC Berkeley and conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals. Four-week-old R6/2 mice bred from 8-week-old R6/2 mice (B6CBA-Tg(HDexon1)62Gpb/3J; Jackson Laboratory, stock #006494) and 8-week-old female B6CBAF1 mice (Jackson Laboratory, stock #100011) were bilaterally injected with 2 × 1010 vector genomes of AAV1-hSyn-SaCas9-HTT or AAV1-hSyn-SaCas9-mRosa26 in 2 μL PBS with 0.001% Tween-20 at stereotaxic coordinates anterior-posterior (AP) = 0.50 mm; medial-lateral (ML) = ±1.75 mm; and dorsal-ventral (DV) = −2.0 mm, −1.5 mm, and −1 mm using a 10-μL syringe with a 22G Point Style 4 needle with a 30° angle (Hamilton). Before injections, animals were genotyped by PCR using genomic DNA purified from an ear clip.
Behavior
Starting 2 weeks after injections, treated and control R6/2 mice were monitored weekly with the rotarod assay, clasping test, and weight measurements. Treatment groups were gender balanced, and all measurements were performed in a blinded manner. For the rotarod, mice were placed onto a Rotamex-5 rotarod (Columbus Instruments), and the latency to fall (measured in seconds) was recorded for each animal. Each session consisted of three trials, with the rotarod programmed to accelerate from 4 to 40 rpm in 180 s. For the clasping test, mice were suspended by the tail for 30 s and scored from 0 to 3: 0 = no clasping was observed; 1 = mice clasped hindlimbs within 30 s; 2 = mice clasped hindlimbs but recovered within 5 s when released; and 3 = mice clasped hindlimbs but failed to recover within 5 s when released. All data were normalized to values determined at week 8 (i.e., 4 weeks after injection). End-stage was defined as the point at which animals were moribund, lacked a righting reflex, failed to respond to gentle stimulation, or decreased to 80% of their peak weight.
Striatal Tissue Harvesting
Mice were anesthetized by intraperitoneal injection of ketamine (100 mg/kg) and xylazine (10 mg/kg) and transcardially perfused with 0.9% saline. Brains were harvested, and the striatum was isolated by manual dissection on a pre-chilled surface. Tissue was homogenized by incubation in trypsin for 90 min at 37°C with constant CO2 equilibration and gentle perturbation every 15 min. Cells were centrifuged at 2,000 × g for 5 min at room temperature, washed once with PBS, and then centrifuged for an additional 2,000 × g for 5 min at room temperature.
Deep Sequencing
Candidate OT sites were identified using Cas-OFFinder, as described elsewhere.46 Briefly, the mm10 mouse reference genome was searched for sites with, at most, four mismatches (up to two nucleotide mismatches and up to two DNA or sgRNA bulges) from the sgRNA target site in the human HTT gene. The 10 most similar sites were chosen for analysis. Striatal tissue from R6/2 mice injected with AAV1-SaCas9-HTT or AAV1-SaCas9-mRosa26 was harvested, and genomic DNA was extracted using the DNeasy Blood & Tissue Kit (QIAGEN). The sgRNA target site in the human HTT gene and the candidate off-target cleavage site in the mouse HTT gene were amplified (Table S1) using a touchdown PCR with Taq DNA Polymerase (New England Biolabs). The remaining candidate off-target sites were amplified (Table S1) using Phusion High-Fidelity DNA Polymerase (New England Biolabs). Mouse-specific barcode sequences were then incorporated using an additional round of PCR. All barcoded amplicons were quantified using PicoGreen (Thermo Fisher Scientific) and purified using AMPure PCR purification (Beckman) at the UC Berkeley Functional Genomics Laboratory. Barcoded amplicons were pooled together, and 150-bp single-ended reads were generated using the HiSeq 4000 System (Illumina; QB3 Vincent J. Coates Genomics Sequencing Laboratory). Samples were demultiplexed based on their index, and adaptor sequences were trimmed from the reads. Using CRISPResso, sequences were filtered for >99% confidence (phred33 threshold > 20) and aligned using EMBOSS Needle. Indel frequency was quantified within a 25-bp window of the cleavage site (substitutions were ignored). Samples with fewer than 1,000 reads post-analysis were removed from statistical analysis.
Immunofluorescence Staining
Mice were anesthetized by intraperitoneal injection of ketamine (100 mg/kg) and xylazine (10 mg/kg) and transcardially perfused with 0.9% saline followed by 4% paraformaldehyde. Brains were harvested and post-fixed in 4% paraformaldehyde for 48 h at 4°C and then transferred to a 30% (w/v) sucrose solution. Brains were cut to 40-μm coronal sections and stored in cytoprotectant at −20°C. Sections were then washed three times with PBS, incubated with blocking solution (PBS with 2% [v/v] BSA, 5% [v/v] donkey serum, and 0.2% [v/v] Triton X-100) at room temperature for 2 h, and stained with primary antibodies in blocking solution for 48 h at 4°C. Sections were washed four times with PBS and incubated with secondary antibodies in blocking solution for 2 h followed by a 10-min incubation with DAPI nuclear stain. After staining, sections were washed four times with PBS, mounted onto slides, and visualized using a Zeiss LSM 710 AxioObserver confocal microscope (UC Berkeley Molecular Imaging Center). Image analysis was performed using ImageJ (https://imagej.nih.gov/ij/).
The following antibodies were used for brain sections: mouse anti-HTT, clone mEM48 (Millipore-Sigma, #MAB5374); goat anti-DARPP-32 (R&D Systems, #AF6259); and rabbit anti-HA, clone C29F4 (Cell Signaling Technology, #3724). The following secondary antibodies were used: donkey anti-goat Alexa Fluor 488 (Jackson ImmunoResearch Laboratories, #703-545-155), donkey anti-rabbit Alexa Fluor 555 (Thermo-Fisher Scientific, #A-31572), and donkey anti-mouse Alexa Fluor 647 (Thermo-Fisher Scientific, #A-31571).
Statistics
Statistical analysis was performed using Prism 7 (GraphPad Software). Mutant HTT protein, mean lifespan, and neuron survival were compared using an unpaired t test. Rotarod times, weight loss, and clasping scores were compared using a one-way ANOVA followed by Tukey’s post hoc analysis. Kaplan-Meier plots were analyzed using the log-rank test.
Author Contributions
T.G. and D.V.S. conceived of the study. F.K.E. designed constructs; performed molecular biology; packaged the AAV vector; performed injections, behavior studies, and immunofluorescent staining; and analyzed the data. D.S.O. performed molecular biology and animal studies. M.M.A. performed injections and immunofluorescent staining. P.A.L. performed next-generation sequencing and western blot. T.G. designed the experiments, performed molecular biology studies, and analyzed the data. T.G. and F.K.E. wrote the manuscript, with contributions from all authors.
Conflicts of Interest
D.V.S. is an inventor on patents related to AAV vectors and cofounder of 4D Molecular Therapeutics, a company focused on the clinical development of gene therapies for recessive diseases using engineered AAV variants. D.V.S. is also a member of the board of directors of uniQure, a company focused on clinical AAV gene therapy. D.S.O. is now an employee of Sangamo Therapeutics. T.G. is a consultant for 4D Molecular Therapeutics. F.K.E., M.M.A., and P.A.L. declare no competing interests.
Acknowledgments
We thank J.G. Flannery for providing space for behavioral studies, A. Dillin for providing the rotarod, S. Kumar for access to the Odyssey Imaging system, and G.N. Ramadoss for technical assistance. T.G. was supported by a Ruth L. Kirschstein National Research Service Award (NRSA) (F32GM113446) and by the Judith and Jean Pape Adams Charitable Foundation. D.S.O. was supported by an NSF Graduate Research Fellowship and a UC Berkeley Dissertation-Year Fellowship. D.V.S. was supported by the NIH (R01EY022975) and a gift from D. Chan.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.07.009.
Contributor Information
David V. Schaffer, Email: schaffer@berkeley.edu.
Thomas Gaj, Email: gaj@illinois.edu.
Supplemental Information
References
- 1.Bates G.P., Dorsey R., Gusella J.F., Hayden M.R., Kay C., Leavitt B.R., Nance M., Ross C.A., Scahill R.I., Wetzel R. Huntington disease. Nat. Rev. Dis. Primers. 2015;1:15005. doi: 10.1038/nrdp.2015.5. [DOI] [PubMed] [Google Scholar]
- 2.MacDonald M.E., Ambrose C.M., Duyao M.P., Myers R.H., Lin C., Srinidhi L., Barnes G., Taylor S.A., James M., Groot N. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 3.Gutekunst C.A., Li S.H., Yi H., Mulroy J.S., Kuemmerle S., Jones R., Rye D., Ferrante R.J., Hersch S.M., Li X.J. Nuclear and neuropil aggregates in Huntington’s disease: relationship to neuropathology. J. Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim Y.J., Yi Y., Sapp E., Wang Y., Cuiffo B., Kegel K.B., Qin Z.H., Aronin N., DiFiglia M. Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington’s disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc. Natl. Acad. Sci. USA. 2001;98:12784–12789. doi: 10.1073/pnas.221451398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grima J.C., Daigle J.G., Arbez N., Cunningham K.C., Zhang K., Ochaba J., Geater C., Morozko E., Stocksdale J., Glatzer J.C. Mutant huntingtin disrupts the nuclear pore complex. Neuron. 2017;94:93–107.e6. doi: 10.1016/j.neuron.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 7.Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G.P., Davies S.W., Lehrach H., Wanker E.E. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 8.Waldvogel H.J., Kim E.H., Tippett L.J., Vonsattel J.P., Faull R.L. The neuropathology of Huntington’s disease. Curr. Top. Behav. Neurosci. 2015;22:33–80. doi: 10.1007/7854_2014_354. [DOI] [PubMed] [Google Scholar]
- 9.Vonsattel J.P., Myers R.H., Stevens T.J., Ferrante R.J., Bird E.D., Richardson E.P., Jr. Neuropathological classification of Huntington’s disease. J. Neuropathol. Exp. Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 10.Ross C.A., Aylward E.H., Wild E.J., Langbehn D.R., Long J.D., Warner J.H., Scahill R.I., Leavitt B.R., Stout J.C., Paulsen J.S. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat. Rev. Neurol. 2014;10:204–216. doi: 10.1038/nrneurol.2014.24. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto A., Lucas J.J., Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 12.Díaz-Hernández M., Torres-Peraza J., Salvatori-Abarca A., Morán M.A., Gómez-Ramos P., Alberch J., Lucas J.J. Full motor recovery despite striatal neuron loss and formation of irreversible amyloid-like inclusions in a conditional mouse model of Huntington’s disease. J. Neurosci. 2005;25:9773–9781. doi: 10.1523/JNEUROSCI.3183-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harper S.Q., Staber P.D., He X., Eliason S.L., Martins I.H., Mao Q., Yang L., Kotin R.M., Paulson H.L., Davidson B.L. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc. Natl. Acad. Sci. USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boudreau R.L., McBride J.L., Martins I., Shen S., Xing Y., Carter B.J., Davidson B.L. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol. Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drouet V., Perrin V., Hassig R., Dufour N., Auregan G., Alves S., Bonvento G., Brouillet E., Luthi-Carter R., Hantraye P., Déglon N. Sustained effects of nonallele-specific Huntingtin silencing. Ann. Neurol. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- 16.Carroll J.B., Warby S.C., Southwell A.L., Doty C.N., Greenlee S., Skotte N., Hung G., Bennett C.F., Freier S.M., Hayden M.R. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin. Mol. Ther. 2011;19:2178–2185. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Southwell A.L., Skotte N.H., Bennett C.F., Hayden M.R. Antisense oligonucleotide therapeutics for inherited neurodegenerative diseases. Trends Mol. Med. 2012;18:634–643. doi: 10.1016/j.molmed.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Kordasiewicz H.B., Stanek L.M., Wancewicz E.V., Mazur C., McAlonis M.M., Pytel K.A., Artates J.W., Weiss A., Cheng S.H., Shihabuddin L.S. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drouet V., Ruiz M., Zala D., Feyeux M., Auregan G., Cambon K., Troquier L., Carpentier J., Aubert S., Merienne N. Allele-specific silencing of mutant huntingtin in rodent brain and human stem cells. PLoS ONE. 2014;9:e99341. doi: 10.1371/journal.pone.0099341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miniarikova J., Zimmer V., Martier R., Brouwers C.C., Pythoud C., Richetin K., Rey M., Lubelski J., Evers M.M., van Deventer S.J. AAV5-miHTT gene therapy demonstrates suppression of mutant huntingtin aggregation and neuronal dysfunction in a rat model of Huntington’s disease. Gene Ther. 2017;24:630–639. doi: 10.1038/gt.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evers M.M., Miniarikova J., Juhas S., Vallès A., Bohuslavova B., Juhasova J., Skalnikova H.K., Vodicka P., Valekova I., Brouwers C. AAV5-miHTT gene therapy demonstrates broad distribution and strong human mutant huntingtin lowering in a Huntington’s sisease minipig model. Mol. Ther. 2018;26:2163–2177. doi: 10.1016/j.ymthe.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miniarikova J., Zanella I., Huseinovic A., van der Zon T., Hanemaaijer E., Martier R., Koornneef A., Southwell A.L., Hayden M.R., van Deventer S.J. Design, characterization, and lead selection of therapeutic miRNAs targeting huntingtin for development of gene therapy for huntington’s disease. Mol. Ther. Nucleic Acids. 2016;5:e297. doi: 10.1038/mtna.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 24.Jackson A.L., Burchard J., Schelter J., Chau B.N., Cleary M., Lim L., Linsley P.S. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh S., Narang A.S., Mahato R.I. Subcellular fate and off-target effects of siRNA, shRNA, and miRNA. Pharm. Res. 2011;28:2996–3015. doi: 10.1007/s11095-011-0608-1. [DOI] [PubMed] [Google Scholar]
- 26.Gaj T., Sirk S.J., Shui S.L., Liu J. Genome-editing technologies: principles and applications. Cold Spring Harb. Perspect. Biol. 2016;8:a023754. doi: 10.1101/cshperspect.a023754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho S.W., Kim S., Kim J.M., Kim J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 32.Maeder M.L., Gersbach C.A. Genome-editing technologies for gene and cell therapy. Mol. Ther. 2016;24:430–446. doi: 10.1038/mt.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santiago Y., Chan E., Liu P.Q., Orlando S., Zhang L., Urnov F.D., Holmes M.C., Guschin D., Waite A., Miller J.C. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. USA. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hentze M.W., Kulozik A.E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 35.Ojala D.S., Amara D.P., Schaffer D.V. Adeno-associated virus vectors and neurological gene therapy. Neuroscientist. 2015;21:84–98. doi: 10.1177/1073858414521870. [DOI] [PubMed] [Google Scholar]
- 36.Lentz T.B., Gray S.J., Samulski R.J. Viral vectors for gene delivery to the central nervous system. Neurobiol. Dis. 2012;48:179–188. doi: 10.1016/j.nbd.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aschauer D.F., Kreuz S., Rumpel S. Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PLoS ONE. 2013;8:e76310. doi: 10.1371/journal.pone.0076310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaj T., Epstein B.E., Schaffer D.V. Genome engineering using adeno-associated virus: basic and clinical research applications. Mol. Ther. 2016;24:458–464. doi: 10.1038/mt.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maynard C.J., Böttcher C., Ortega Z., Smith R., Florea B.I., Díaz-Hernández M., Brundin P., Overkleeft H.S., Li J.Y., Lucas J.J., Dantuma N.P. Accumulation of ubiquitin conjugates in a polyglutamine disease model occurs without global ubiquitin/proteasome system impairment. Proc. Natl. Acad. Sci. USA. 2009;106:13986–13991. doi: 10.1073/pnas.0906463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., Bates G.P. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 42.Chiang C., Jacobsen J.C., Ernst C., Hanscom C., Heilbut A., Blumenthal I., Mills R.E., Kirby A., Lindgren A.M., Rudiger S.R. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat. Genet. 2012;44:390–397. doi: 10.1038/ng.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menalled L., El-Khodor B.F., Patry M., Suárez-Fariñas M., Orenstein S.J., Zahasky B., Leahy C., Wheeler V., Yang X.W., MacDonald M. Systematic behavioral evaluation of Huntington’s disease transgenic and knock-in mouse models. Neurobiol. Dis. 2009;35:319–336. doi: 10.1016/j.nbd.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cummings D.M., Alaghband Y., Hickey M.A., Joshi P.R., Hong S.C., Zhu C., Ando T.K., André V.M., Cepeda C., Watson J.B., Levine M.S. A critical window of CAG repeat-length correlates with phenotype severity in the R6/2 mouse model of Huntington’s disease. J. Neurophysiol. 2012;107:677–691. doi: 10.1152/jn.00762.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irion S., Luche H., Gadue P., Fehling H.J., Kennedy M., Keller G. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat. Biotechnol. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- 46.Gaj T., Ojala D.S., Ekman F.K., Byrne L.C., Limsirichai P., Schaffer D.V. In vivo genome editing improves motor function and extends survival in a mouse model of ALS. Sci. Adv. 2017;3:eaar3952. doi: 10.1126/sciadv.aar3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hadaczek P., Stanek L., Ciesielska A., Sudhakar V., Samaranch L., Pivirotto P., Bringas J., O’Riordan C., Mastis B., San Sebastian W. Widespread AAV1- and AAV2-mediated transgene expression in the nonhuman primate brain: implications for Huntington’s disease. Mol. Ther. Methods Clin. Dev. 2016;3:16037. doi: 10.1038/mtm.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Connor B., Sun Y., von Hieber D., Tang S.K., Jones K.S., Maucksch C. AAV1/2-mediated BDNF gene therapy in a transgenic rat model of Huntington’s disease. Gene Ther. 2016;23:283–295. doi: 10.1038/gt.2015.113. [DOI] [PubMed] [Google Scholar]
- 49.Franich N.R., Fitzsimons H.L., Fong D.M., Klugmann M., During M.J., Young D. AAV vector-mediated RNAi of mutant huntingtin expression is neuroprotective in a novel genetic rat model of Huntington’s disease. Mol. Ther. 2008;16:947–956. doi: 10.1038/mt.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang C.E., Zhou H., McGuire J.R., Cerullo V., Lee B., Li S.H., Li X.J. Suppression of neuropil aggregates and neurological symptoms by an intracellular antibody implicates the cytoplasmic toxicity of mutant huntingtin. J. Cell Biol. 2008;181:803–816. doi: 10.1083/jcb.200710158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinello L., Canver M.C., Hoban M.D., Orkin S.H., Kohn D.B., Bauer D.E., Yuan G.C. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016;34:695–697. doi: 10.1038/nbt.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morton A.J., Lagan M.A., Skepper J.N., Dunnett S.B. Progressive formation of inclusions in the striatum and hippocampus of mice transgenic for the human Huntington’s disease mutation. J. Neurocytol. 2000;29:679–702. doi: 10.1023/a:1010887421592. [DOI] [PubMed] [Google Scholar]
- 54.Meade C.A., Deng Y.P., Fusco F.R., Del Mar N., Hersch S., Goldowitz D., Reiner A. Cellular localization and development of neuronal intranuclear inclusions in striatal and cortical neurons in R6/2 transgenic mice. J. Comp. Neurol. 2002;449:241–269. doi: 10.1002/cne.10295. [DOI] [PubMed] [Google Scholar]
- 55.Ehrnhoefer D.E., Butland S.L., Pouladi M.A., Hayden M.R. Mouse models of Huntington disease: variations on a theme. Dis. Model. Mech. 2009;2:123–129. doi: 10.1242/dmm.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rawlins M.D., Wexler N.S., Wexler A.R., Tabrizi S.J., Douglas I., Evans S.J., Smeeth L. The prevalence of Huntington’s disease. Neuroepidemiology. 2016;46:144–153. doi: 10.1159/000443738. [DOI] [PubMed] [Google Scholar]
- 57.Tervo D.G., Hwang B.Y., Viswanathan S., Gaj T., Lavzin M., Ritola K.D., Lindo S., Michael S., Kuleshova E., Ojala D. A designer AAV variant permits efficient retrograde access to projection neurons. Neuron. 2016;92:372–382. doi: 10.1016/j.neuron.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swiech L., Heidenreich M., Banerjee A., Habib N., Li Y., Trombetta J., Sur M., Zhang F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang S., Chang R., Yang H., Zhao T., Hong Y., Kong H.E., Sun X., Qin Z., Jin P., Li S., Li X.J. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J. Clin. Invest. 2017;127:2719–2724. doi: 10.1172/JCI92087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Monteys A.M., Ebanks S.A., Keiser M.S., Davidson B.L. CRISPR/Cas9 editing of the mutant huntingtin allele in vitro and in vivo. Mol. Ther. 2017;25:12–23. doi: 10.1016/j.ymthe.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Merienne N., Vachey G., de Longprez L., Meunier C., Zimmer V., Perriard G., Canales M., Mathias A., Herrgott L., Beltraminelli T. The self-inactivating KamiCas9 system for the editing of CNS disease genes. Cell Rep. 2017;20:2980–2991. doi: 10.1016/j.celrep.2017.08.075. [DOI] [PubMed] [Google Scholar]
- 62.Suzuki K., Tsunekawa Y., Hernandez-Benitez R., Wu J., Zhu J., Kim E.J., Hatanaka F., Yamamoto M., Araoka T., Li Z. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferrante R.J. Mouse models of Huntington’s disease and methodological considerations for therapeutic trials. Biochim. Biophys. Acta. 2009;1792:506–520. doi: 10.1016/j.bbadis.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nasir J., Floresco S.B., O’Kusky J.R., Diewert V.M., Richman J.M., Zeisler J., Borowski A., Marth J.D., Phillips A.G., Hayden M.R. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 65.Duyao M.P., Auerbach A.B., Ryan A., Persichetti F., Barnes G.T., McNeil S.M., Ge P., Vonsattel J.P., Gusella J.F., Joyner A.L. Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- 66.Wang G., Liu X., Gaertig M.A., Li S., Li X.J. Ablation of huntingtin in adult neurons is nondeleterious but its depletion in young mice causes acute pancreatitis. Proc. Natl. Acad. Sci. USA. 2016;113:3359–3364. doi: 10.1073/pnas.1524575113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garriga-Canut M., Agustín-Pavón C., Herrmann F., Sánchez A., Dierssen M., Fillat C., Isalan M. Synthetic zinc finger repressors reduce mutant huntingtin expression in the brain of R6/2 mice. Proc. Natl. Acad. Sci. USA. 2012;109:E3136–E3145. doi: 10.1073/pnas.1206506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han I., You Y., Kordower J.H., Brady S.T., Morfini G.A. Differential vulnerability of neurons in Huntington’s disease: the role of cell type-specific features. J. Neurochem. 2010;113:1073–1091. doi: 10.1111/j.1471-4159.2010.06672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kotterman M.A., Schaffer D.V. Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nathwani A.C., Reiss U.M., Tuddenham E.G., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.MacLaren R.E., Groppe M., Barnard A.R., Cottriall C.L., Tolmachova T., Seymour L., Clark K.R., During M.J., Cremers F.P., Black G.C. Retinal gene therapy in patients with choroideremia: initial findings from a phase 1/2 clinical trial. Lancet. 2014;383:1129–1137. doi: 10.1016/S0140-6736(13)62117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stroes E.S., Nierman M.C., Meulenberg J.J., Franssen R., Twisk J., Henny C.P., Maas M.M., Zwinderman A.H., Ross C., Aronica E. Intramuscular administration of AAV1-lipoprotein lipase S447X lowers triglycerides in lipoprotein lipase-deficient patients. Arterioscler. Thromb. Vasc. Biol. 2008;28:2303–2304. doi: 10.1161/ATVBAHA.108.175620. [DOI] [PubMed] [Google Scholar]
- 73.Carpentier A.C., Frisch F., Labbé S.M., Gagnon R., de Wal J., Greentree S., Petry H., Twisk J., Brisson D., Gaudet D. Effect of alipogene tiparvovec (AAV1-LPL(S447X)) on postprandial chylomicron metabolism in lipoprotein lipase-deficient patients. J. Clin. Endocrinol. Metab. 2012;97:1635–1644. doi: 10.1210/jc.2011-3002. [DOI] [PubMed] [Google Scholar]
- 74.Bainbridge J.W., Smith A.J., Barker S.S., Robbie S., Henderson R., Balaggan K., Viswanathan A., Holder G.E., Stockman A., Tyler N. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N. Engl. J. Med. 2008;358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 75.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guilinger J.P., Thompson D.B., Liu D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsai S.Q., Wyvekens N., Khayter C., Foden J.A., Thapar V., Reyon D., Goodwin M.J., Aryee M.J., Joung J.K. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen Y., Liu X., Zhang Y., Wang H., Ying H., Liu M., Li D., Lui K.O., Ding Q. A self-restricted CRISPR system to reduce off-target effects. Mol. Ther. 2016;24:1508–1510. doi: 10.1038/mt.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Petris G., Casini A., Montagna C., Lorenzin F., Prandi D., Romanel A., Zasso J., Conti L., Demichelis F., Cereseto A. Hit and go CAS9 delivered through a lentiviral based self-limiting circuit. Nat. Commun. 2017;8:15334. doi: 10.1038/ncomms15334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R., Hwang J., Kim J.I., Kim J.S. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015;12 doi: 10.1038/nmeth.3284. 237–243, 1, 243. [DOI] [PubMed] [Google Scholar]
- 84.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Adil M.M., Gaj T., Rao A.T., Kulkarni R.U., Fuentes C.M., Ramadoss G.N., Ekman F.K., Miller E.W., Schaffer D.V. hPSC-derived striatal cells generated using a scalable 3D hydrogel promote recovery in a Huntington disease mouse model. Stem Cell Reports. 2018;10:1481–1491. doi: 10.1016/j.stemcr.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reidling J.C., Relaño-Ginés A., Holley S.M., Ochaba J., Moore C., Fury B., Lau A., Tran A.H., Yeung S., Salamati D. Human neural stem cell transplantation rescues functional deficits in R6/2 and Q140 Huntington’s disease mice. Stem Cell Reports. 2018;10:58–72. doi: 10.1016/j.stemcr.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brooks A.K., Gaj T. Innovations in CRISPR technology. Curr. Opin. Biotechnol. 2018;52:95–101. doi: 10.1016/j.copbio.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 88.Gaj T., Schaffer D.V. Adeno-associated virus-mediated delivery of CRISPR-Cas systems for genome engineering in mammalian cells. Cold Spring Harb. Protoc. 2016;2016 doi: 10.1101/pdb.prot086868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.