Abstract
Late embryogenesis abundant (LEA) proteins are intrinsically disordered proteins (IDPs) commonly found in anhydrobiotic organisms and are frequently correlated with desiccation tolerance. Herein we report new findings on AfrLEA6, a novel group 6 LEA protein from embryos of Artemia franciscana. Assessment of secondary structure in aqueous and dried states with circular dichroism (CD) reveals 89% random coil in the aqueous state, thus supporting classification of AfrLEA6 as an IDP. Removal of water from the protein by drying or exposure to trifluoroethanol (a chemical de-solvating agent) promotes a large gain in secondary structure of AfrLEA6, predominated by α-helix and exhibiting minimal β-sheet structure. We evaluated the impact of physiological concentrations (up to 400 mM) of the disaccharide trehalose on the folding of LEA proteins in solution. CD spectra for AfrLEA2, AfrLEA3m, and AfrLEA6 are unaffected by this organic solute noted for its ability to drive protein folding. AfrLEA6 exhibits its highest concentration in vivo during embryonic diapause, drops acutely at diapause termination, and then declines during development to undetectable values at the larval stage. Maximum cellular titer of AfrLEA6 was 10-fold lower than for AfrLEA2 or AfrLEA3, both group 3 LEA proteins. Acute termination of diapause with H2O2 (a far more effective terminator than desiccation in this Great Salt Lake, UT, population) fostered a rapid 38% decrease in AfrLEA6 content of embryos. While the ultimate mechanism of diapause termination is unknown, disruption of key macromolecules could initiate physiological signaling events necessary for resumption of development and metabolism.
Keywords: Late embryogenesis abundant proteins, Anhydrobiosis, Diapause termination, Desiccation tolerance, Secondary structure, Circular dichroism
Introduction
Anhydrobiotic animals like the brine shrimp Artemia franciscana tolerate major transitions in water content at specific points during their life cycles. During anhydrobiosis, tissue water can decrease to less than 2% (Crowe and Madin 1974), which imposes substantial challenges for defending the functionality of biological structures (Crowe et al. 1997; Crowe and Clegg 1973, 1978). One key strategy for desiccation tolerance in many anhydrobiotes is the accumulation of protective molecules such as late embryogenesis abundant (LEA) proteins, heat-shock proteins, and stabilizing organic solutes (Clegg 2005, 2011; Crowe et al. 1998, 2005; Hand et al. 2011, 2018; Tan and MacRae 2018; Tapia and Koshland 2014; Tunnacliffe and Wise 2007). The present study focuses on AfrLEA6, a group 6 LEA protein also referred to as a seed maturation protein (SMP). SMPs have previously been found in plant seeds associated with desiccation tolerance (Boucher et al. 2010; Chatelain et al. 2012). In Medicago truncatula, the SMP D34.3 accumulates in the mature seed and is correlated with long-term viability of the seed in the dried state (Chatelain et al. 2012). We have recently identified, cloned, and expressed a group 6 LEA protein from embryos of A. franciscana that exhibits strong sequence homologies to D34.3 (Hand and Menze 2015; Janis et al. 2018b; Wu et al. 2011). AfrLEA6 exhibits the lowest hydrophilicity of the LEA proteins thus far characterized in A. franciscana and more closely resembles the group 6 LEA protein MtPM25 (Boucher et al. 2010; Janis et al. 2018b). Other defining features of AfrLEA6 include two SMP domains (pfam family PF04927 with the consensus sequence kpVtpeDAaavqaAEaraageartapgGvAaaaqaAAdaNer, which contains the “pgGvA” motif specified by Jaspard et al. 2012) and a proline-rich region, which are predicted to be sites of self-interaction (Janis et al. 2018b). In this study, we investigate selected structural properties of AfrLEA6 and provide evidence for its expression in the diapause state and throughout post-diapause development of the embryo to the larval stage. The state of diapause is a programmed arrest of development that is controlled by endogenous physiological factors and may or may not involve a substantial depression of metabolism (Hand et al. 2016 and references therein). Entering diapause is a common mechanism for overwintering, because life cycle delays are beneficial for species encountering suboptimal environments.
LEA proteins are a family of intrinsically disordered proteins that are expressed in developmental and/or adult life stages of anhydrobiotic organisms and have been strongly associated with survival during water stress. Originally discovered in plants, LEA proteins have now been identified in animals such as nematodes (Browne et al. 2002; Gal et al. 2004; Solomon et al. 2000), brine shrimp (Hand et al. 2007), African chironomid insect larvae (Gusev et al. 2014; Kikawada et al. 2006), and rotifers (Denekamp et al. 2010; Tunnacliffe et al. 2005), as well as bacteria (Battista et al. 2001; Rodriguez-Salazar et al. 2017; Stacy and Aalen 1998). Similar tardigrade-specific intrinsically disordered proteins have also been described (Boothby and Pielak 2017; Boothby et al. 2017). Within these organisms, LEA proteins are present across a range of subcellular compartments including the nucleus, endoplasmic reticulum, Golgi apparatus, mitochondrion, and chloroplast (Avelange-Macherel et al. 2018; Boswell and Hand 2014; Hand et al. 2011; Tripathi et al. 2012; Tunnacliffe et al. 2010; Tunnacliffe and Wise 2007). Classification schemes place them into six groups, of which A. franciscana uniquely expresses LEA proteins from three different groups (1, 3, and 6; Hand and Menze 2015; Tunnacliffe and Wise 2007; Warner et al. 2010, 2012).
While many questions still exist in terms of the varied functions of LEA proteins, in vitro and in vivo studies have contributed insights into their roles (for reviews, see Hand et al. 2011; Hincha and Thalhammer 2012; Janis et al. 2018a; Tunnacliffe and Wise 2007). For example, LEA proteins have been shown to protect lipid bilayers of various compositions during drying and freezing (Hundertmark et al. 2011; Moore and Hand 2016; Moore et al. 2016; Navarro-Retamal et al. 2018; Steponkus et al. 1998; Thalhammer et al. 2014; Tolleter et al. 2007, 2010), preserve the activity of target enzymes (Boswell et al. 2014a; Goyal et al. 2005; Grelet et al. 2005; Popova et al. 2015), and prevent protein aggregation (“molecular shielding”; Boucher et al. 2010; Goyal et al. 2005; Yuen et al. 2019). The gain of secondary structure (α-helix, β-sheet, turns) that accompanies the dehydration of some LEA proteins may allow them to perform separate/additional roles in the dried state (Goyal et al. 2003). Finally, synergistic effects of combining trehalose, an established stabilizer of cellular components during drying and freezing (Crowe et al. 1987, 1998; Tapia and Koshland 2014), with LEA proteins have been documented (Boswell et al. 2014a; Goyal et al. 2005).
AfrLEA6 is the only group 6 LEA protein reported thus far from an animal, and our bioinformatics analyses (Janis et al. 2018b) predict distinctly different structural features compared to other LEA proteins in A. franciscana. Consequently, we evaluated secondary structural features of AfrLEA6 with circular dichroism as a function of hydration state (with and without trehalose), along with the tissue content of the protein during diapause, immediately after diapause termination, and during post-diapause development.
Methods
Collection and incubation of diapause embryos, post-diapause embryos, and larvae
Diapause embryos were collected in the hydrated state from the surface of the Great Salt Lake (UT). These embryos were verified to be in diapause by performing hatching assays as outlined in Reynolds and Hand (2004). The hatch rate of diapause embryos used in these experiments was 2.9%, indicative of a strong diapause state. Diapause embryos were rinsed in lake water and stored protected from light at ambient temperature in 1.25 M NaCl containing 200 units/ml nystatin, 50 μg/ml kanamycin, and 50 μg/ml penicillin-streptomycin. Prior to use, diapause embryos were rinsed and incubated with shaking at 110 rpm in artificial seawater (35 practical salinity units; Instant Ocean ®, Blacksburg, VA) for 4 days at room temperature in the dark to allow any embryos that had terminated diapause to hatch. Exposure to light has been associated with increased diapause breakage in the laboratory (Lavens and Sorgeloos 1987). The hatched larvae were discarded, and the remaining diapause embryos were used for experimentation.
Commercial post-diapause embryos were obtained in the dried state from Great Salt Lake Artemia, LLC (Ogden, UT; grade: laboratory reference standard) and were stored frozen at − 20 °C. Prior to use, commercial post-diapause embryos were hydrated overnight in ice-cold 0.25 M NaCl. To promote development, hydrated post-diapause embryos were incubated in 0.25 M NaCl at 23 °C with shaking, and embryos were sampled at 2, 4, 6, and 8 h. Incubation was extended through 24 h to generate free-swimming nauplius larvae. Hatched larvae were separated from embryos by phototaxis and processed separately.
Expression and purification of recombinant LEA proteins from Artemia franciscana and antibody production
The original nucleic acid sequence for AfrLEA6 (GenBank accession no. MH351624) from A. franciscana embryos was amplified from our existing A. franciscana cDNA library. Primer sequences used were 5′ggcggcggccatatgATGTCTGAGAATATTGGTCATATTAACATAAATGC and 5′attagtaactagtAGTGCATCTCCCGTGATGCAGTCCATGCGGACATTCCCAAT. For the polymerase chain reaction, Q5 DNA polymerase (New England Biolabs, Ipswich, MA) was used for 25 cycles following the instructions of the manufacturer, and the PCR product obtained was cloned into pTXB1 using standard techniques and the two restriction enzymes NdeI and SpeI (New England Biolabs, Ipswich, MA). Recombinant AfrLEA6 was purified from Escherichia coli cells (BL21) designed to work with the IMPACT protein expression system (Intein-Mediated Purification with an Affinity Chitin-binding Tag; New England Biolabs Inc., Ipswich, MA). The vector (pTXB1) was used to add a chitin-binding domain and self-cleavage protein splicing elements to the expressed fusion protein. These elements permitted binding to the chitin moiety of an affinity column, and then dithiothreitol (DTT) was used to release AfrLEA6 free of any amino acid tag (see below). Bacterial cells were grown in the presence of 0.1 mM ampicillin at 37 °C with shaking until the optical density at 595 nm reached 0.6. Then protein expression was induced with 0.4 mM IPTG for 2 h at 37 °C while shaking. Cells were sedimented by centrifugation at 5000×g (4 °C) for 15 min. The cell pellets were resuspended in 40 ml of chitin column buffer (20 mM Tris, 0.5 M NaCl, pH 8.5) containing either 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich, St. Louis, MO) or one cOmplete™ protease inhibitor cocktail tablet (Roche Applied Science; Penzberg, Germany). Cells were lysed by sonication, and cellular debris was removed by centrifugation at 20,000×g (4 °C) for 30 min. The supernatant was then loaded onto a chitin resin column (S6651S; New England BioLabs Inc.; Ipswich, MA) by gravity flow (~ 1.0 ml/min). On-column cleavage was induced by incubating the column for 48 h with chitin column buffer containing 50 mM DTT at 4 °C. AfrLEA6 was eluted using chitin column buffer and dialyzed overnight against anion exchange column buffer (15 mM triethanolamine, 10 mM NaCl, pH 7.0). For further purification, the protein sample was applied to a 5-ml anion exchange column (HiTrap Q FF; GE Healthcare; Chicago, IL) using an AKTAprime plus FPLC system at a flow rate of 1.0 ml/min. After washing with 30 volumes of anion exchange column buffer, bound protein was eluted using a 50-ml linear gradient of 0–25% elution buffer (15 mM triethanolamine, 1 M NaCl, pH 7.0). The eluted protein peak was detected by absorbance at 280 nm and collected in 1 ml fractions. Pooled samples were exchanged into 10 mM potassium phosphate buffer (pH 7.5) and concentrated using Amicon Ultra-15 10K centrifugal filters (MilliporeSigma; Burlington, MA). The techniques for expressing and purifying recombinant AfrLEA2 and AfrLEA3m followed the procedures in Boswell et al. (2014b). Polyclonal antibody against the purified recombinant AfrLEA6 was raised in chicken eggs by Aves Labs, Inc. (Tigard, OR, USA).
Circular dichroism spectroscopy
Spectra were obtained with a Jasco J-815 spectropolarimeter and analyzed using Spectra Manager™ Suite (Jasco Analytical Instruments; Easton, OH). Samples were prepared in 10 mM potassium phosphate buffer, pH 7.5, at a protein concentration of 0.15 mg/ml for AfrLEA2, AfrLEA3m, AfrLEA6, and bovine serum albumin (BSA; Sigma; A6003). For aqueous protein samples, measurements were recorded between 190 and 250 nm using a quartz cuvette with a 0.1 cm path length. Dried samples were prepared by air drying a 50-μl droplet of sample containing 0.15 mg/ml protein onto one side of a demountable quartz cuvette overnight in a dry box containing desiccant (Drierite; W. A. Hammond Drierite, Xenia, OH). For conversion of spectra for dried samples to mean residue ellipticity, a path length of 0.01 cm was used. Spectra for blank samples that lacked protein were collected for each experimental condition and subtracted from the respective protein spectra. Each spectrum was then converted to mean residue ellipticity and smoothed with a convolution width of 9 (Savitzky and Golay 1964). Spectra Manager™ Suite was used to subtract blank spectra, convert values to mean residue ellipticity, and smooth the curves. Analysis of secondary structure was achieved using DICHROWEB online analysis software (Whitmore and Wallace 2004, 2008) to apply the CONTINLL algorithm (Provencher and Glockner 1981; van Stokkum et al. 1990) and SELCON3 algorithm (Sreerama et al. 1999; Sreerama and Woody 1993). Reference dataset 7 was utilized for spectra analyses as it applies to denatured proteins with measurements between 190 and 250 nm wavelengths (Sreerama et al. 1999).
Evaluation of diapause termination in the laboratory
Treatments to promote diapause termination included exposure of embryos to dehydration/rehydration cycles and to a chemical oxidant. Diapause embryos were air dried on the benchtop at room temperature and ambient humidity (50–55%) for either 24 h or 1 week. Diapause embryos were also dried for 24 h as above, rehydrated for 6 h (0 °C), and then dried again for 24 h (designated as 1.5X dehydration/rehydration cycles, 1.5X D/H). Chemical treatment involved incubating the diapause cysts in 3% H2O2 prepared in 0.25 M NaCl solution for either 15 or 30 min. When embryos were to be used for analysis of AfrLEA6 by Western blotting, embryos were rinsed with excess 0.25 M NaCl to remove any residual H2O2 prior to preparation of extracts (see below). Other experiments incorporated combinations of dehydration and chemical treatments to test for additive or sequence (order) effects.
To quantify the effectiveness of the above treatments for terminating diapause, embryos subsequently were incubated in artificial seawater for 4 days in ambient light. After the incubation, hatching assays were performed by first diluting aliquots of the treated embryos with water in a plastic petri dish (bottom marked off into squares) and adding 12 N HCl to kill any free-swimming nauplii. Then hatch counts were performed under a dissecting scope. Typically, 200–400 organisms were counted for each trial. Hatching percentages were determined by counting the total nauplii and dividing by the total count (free-swimming nauplii + unhatched cysts + E1 and E2 stages). Emergence stage E1 denotes the point at which the embryo first protrudes from its chorion (proteinaceous outer coat) but is still contained within the outer cuticular (“hatching”) membrane, while emergence stage 2 signifies that the embryo mass has protruded even further to form the shape of a parachute (cf. Neumeyer et al. 2015). All diapause and post-diapause embryos were gently blotted between sheets of filter paper to remove interstitial water as previously described (Boswell et al. 2014b; Clegg 1974; Glasheen and Hand 1989) prior to extraction for quantification of AfrLEA6 expression (see below).
Quantification of AfrLEA6 expression by Western blot analysis
Diapause embryos, post-diapause embryos (0, 2, 4, 6, and 8 h), and 24 h free-swimming nauplii were used to analyze AfrLEA6 across development. Extracts were prepared by homogenizing 100 mg of embryos or larvae in 1.9 ml of 1X Laemmli buffer with a ground-glass homogenizer. Samples were heat treated at 95 °C for 10 min after homogenization and then centrifuged at 20,000×g for 20 min to remove insoluble cell debris. The supernatant was collected and frozen at − 20 °C prior to analysis. Total protein was measured with a modified Lowry assay as described by Peterson (1977). Equal protein (40 μg) was loaded for each time point onto an SDS polyacrylamide gel (4% stacking gel, 11% resolving gel). A standard curve was prepared with 0–40 ng of purified, recombinant AfrLEA6 to quantify AfrLEA6 across the developmental stages. Electrophoresis was performed at 125 V for 80 min in a Bio-Rad Mini Protean 3 cell. Following electrophoresis, samples were transferred to a nitrocellulose membrane at 80 V for 60 min in Towbin’s buffer (25 mM tris, 192 mM glycine, 20% v/v methanol, 0.025% SDS). The nitrocellulose membranes were then blocked for 1 h in 5% fat-free dry milk prepared in TBS-T (20 mM Tris, 500 mM NaCl, 0.1% Tween 20, pH 7.6) at room temperature with rocking. The blots were then incubated overnight at 4 °C with AfrLEA6 polyclonal antibody (raised in chicken eggs, Aves Labs Inc.) diluted 1:50,000 in 5% dry milk prepared in TBS-T. Because α-tubulin served as a loading control, blots were incubated simultaneously with α-tubulin primary antibody raised in rabbits (Cell Signaling Technology, Danvers, MA) at a 1:1000 dilution. Afterwards, membranes were given three 15-min rinses in TBS-T with shaking. The blots were incubated at room temperature for 1 h with two horseradish peroxide-labeled (HRP) secondary antibodies: goat anti-chicken (Aves Labs Inc., Tigard, OR) at a 1:10,000 dilution and mouse anti-rabbit (Cell Signaling Technologies; Danvers, MA) at a 1:1000 dilution. The blots were developed using an Amersham ECL Prime Western Blotting Detection Kit (GE Healthcare; Chicago, IL) and imaged with a Bio-Rad ChemiDoc™ XRS+ Imager. Bio-Rad Image Lab™ Software was used for quantifying bands. Values for AfrLEA6 expression were converted to mg AfrLEA6 protein per ml embryo water by calculating the total mg of AfrLEA6 present per gram embryo (hydrated and blotted as above) and then dividing by the corrected water content of metabolically active embryos (0.573, i.e., 57.3% embryo water) as published in Glasheen and Hand (1989). Because preliminary bioinformatic analysis and histochemical localization (B. LeBlanc and S. Hand, unpublished) indicate AfrLEA6 is a cytoplasmic protein, this concentration unit is appropriate for estimating its effective titer in the cell.
Statistical analyses
Analyses were performed using IBM SPSS Statistics for Windows, version 22.0 (IBM Corp., Armonk, New York, USA). Levene’s test was performed to check for equal variance among data sets. To determine significant differences among treatments, one-way ANOVA was applied for data sets with equal variance, and Welch’s ANOVA was used for data sets with unequal variance. For post hoc tests used in pair-wise comparisons, Tukey’s honestly significant difference (HSD) was chosen for data sets with equal variance, and the Games-Howell test was applied for data sets with unequal variances (Games and Howell 1976; McDonald 2014).
Results
Purification of recombinant AfrLEA6
Fractions containing protein eluted from the chitin column after treatment with DTT were analyzed via SDS-PAGE to estimate purity of recombinant AfrLEA6, which has a calculated molecular mass of 27.0 kDa. Several contaminating proteins were present in the eluent with estimated molecular masses of ~ 50 kDa, ~ 60 kDa, and ~ 20 kDa. Heat treatment and anion exchange chromatography were investigated as techniques to remove the contaminating proteins and further enrich AfrLEA6. Heat treatment followed by centrifugation was ineffective at precipitating these contaminating proteins (Fig. 1a). Anion exchange chromatography with a 0–250 mM NaCl gradient eluted a protein peak between 130 and 180 mM NaCl. The leading edge of the peak (130–150 mM NaCl) contained AfrLEA6 without visible contaminating bands; the purity of the pooled fractions was sufficient for production of AfrLEA6 primary antibody (Fig. 1a). The trailing edge of the peak at about 160–180 mM NaCl (Fig. 1a) showed contamination. The removed contaminants likely included the intact fusion protein composed of AfrLEA6 bound to the intein (22 kDa) and the chitin-binding domain protein (CBD, 6 kDa), along with free intein and intein fused with the CBD. A Western blot of purified AfrLEA6 versus the native protein in an embryo extract documented equivalent migration at approximately 27 kDa (Fig. 1b).
Fig. 1.
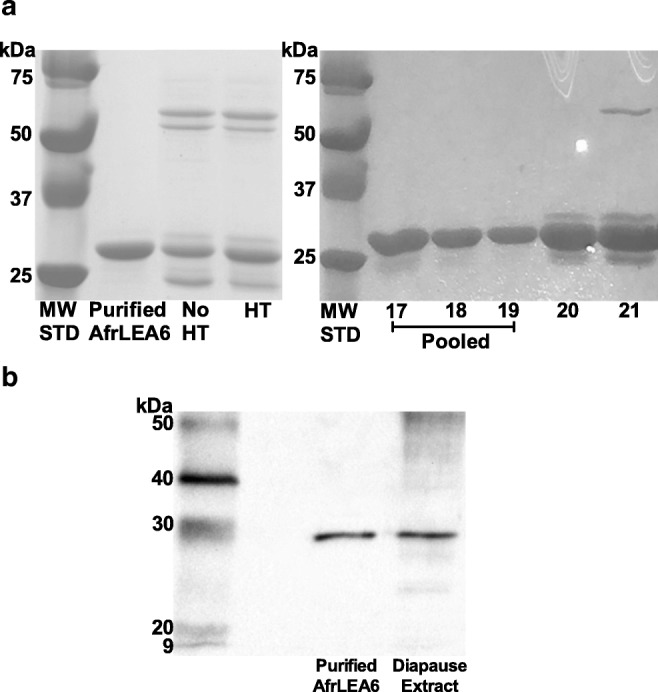
Purification of recombinant AfrLEA6. aLeft panel: SDS-PAGE analysis of purified protein after IMPACT column chromatography combined with anion exchange chromatography. Evidence that heat treatment (HT) is ineffective for removing contaminants present after IMPACT chromatography alone. Total protein loaded per lane was 40 μg. MW STD, ladder of molecular weight standards. Right panel: SDS-PAGE analysis of 1-ml fractions eluted from the anion exchange column (HiTrap Q FF resin) that contained AfrLEA6. Contamination is present in the trailing edge of the peak, which was not pooled. Each lane was loaded with 10 μl of eluant. b Western blot of purified, recombinant AfrLEA6 versus the native protein in a diapause embryo extract shows equivalent migration. Approximately 40 ng of purified AfrLEA6 and 40 μg total protein from an embryo extract prepared in Laemmli buffer were loaded in respective lanes. Ladder of MW standards shown at left
Secondary structure of LEA proteins
The secondary structure of AfrLEA6 was investigated under various solution conditions and in the dried state. AfrLEA6 exists as an intrinsically disordered protein in solution with a minimum ellipticity of 200 nm that is characteristic of disordered, random coil proteins (Fig. 2a). When interactions with water are decreased by drying or by the addition of 70% trifluoroethanol (TFE; a chemical de-solvating agent), AfrLEA6 gains significant secondary structure (α-helix, β-sheet, turns). A similar outcome was promoted by adding 2% SDS. A substantial increase in the proportion of α-helix was indicated in the CD spectra by double minima at 208 and 222 nm and a maximum at 191 nm (Fig. 2a). The proportion of α-helices increased from 3.9% in the aqueous state to 47.1% in the dried state (Fig. 2b). Similarly, the proportion of α-helix structure increased from 3.9 to 34% and 44.3% after the additions of 2% SDS and 70% TFE, respectively (Fig. 2b). These data were substantiated by comparisons to bovine serum albumin (BSA), which is a typical globular protein known to exhibit secondary structure in solution and to AfrLEA2, which is a positive control previously shown to behave as an intrinsically disordered protein (Boswell et al. 2014a). As expected, the structure of BSA was predominantly α-helix in aqueous solution and did not substantially change after treatment with 2% SDS or 70% TFE (Fig. 3) (also see Boswell et al. 2014a; Takeda et al. 1987). In aqueous solution, there were noteworthy differences in secondary structure between AfrLEA6 and AfrLEA2 (Fig. 3). Overall, AfrLEA6 exhibited a lower percentage of secondary structure (11%) when compared to AfrLEA2 (26.7%), the major difference being the reduced content of β-sheet for AfrLEA6 (0.5%) versus AfrLEA2 (15.6%). AfrLEA6 gained more total secondary structure in 70% TFE (71.1%) than in 2% SDS (61.1%), whereas the percentages for AfrLEA2 were the same across the two treatments (70 versus 71%).
Fig. 2.
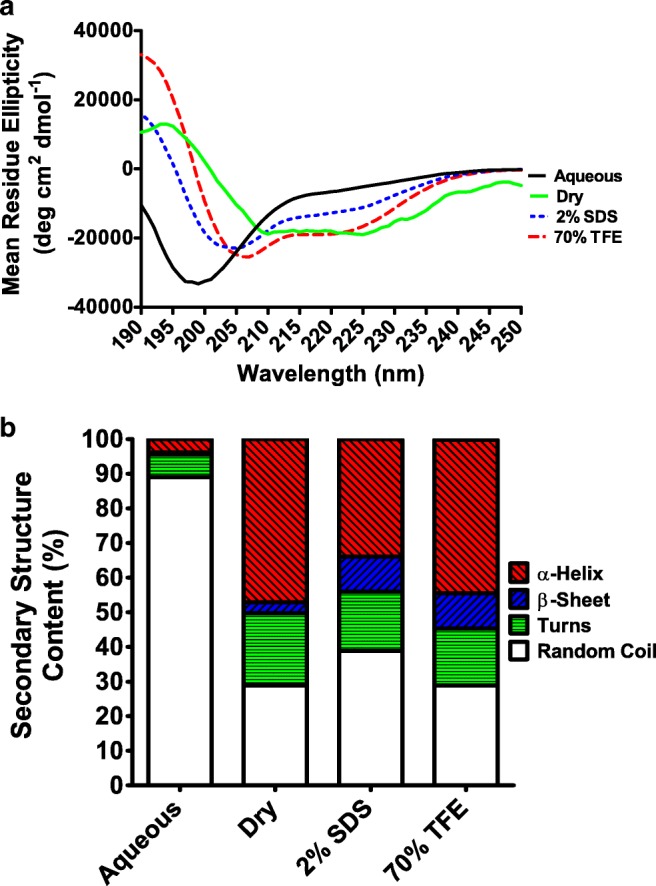
a Circular dichroism (CD) spectra of 0.15 mg/ml AfrLEA6 in the aqueous and dried state as well as in the presence of 2% SDS and 70% TFE. Lines smoothed via the Savitzky-Golay method with convolution width 9. b Dichroweb analysis of secondary structures in CD spectra of AfrLEA6 using the Contin-LL method (see text)
Fig. 3.
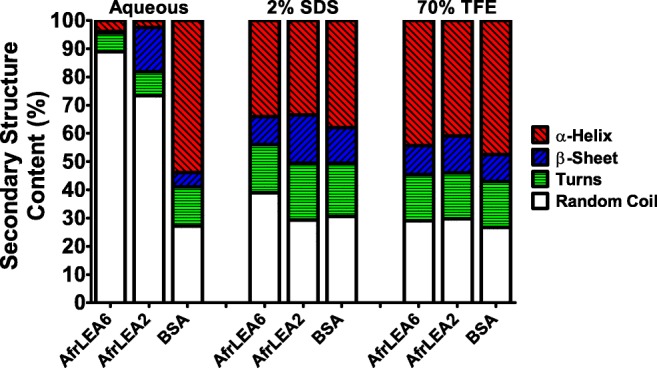
Comparison of secondary structure content for AfrLEA6, AfrLEA2, and bovine serum albumin (BSA) based on CD spectra recorded in the aqueous state, presence of 2% SDS, and presence of 70% TFE. All proteins were measured at a concentration of 0.15 mg/ml. Analysis of secondary structure was performed using the Dichroweb server with the Contin-LL method (see text)
Secondary structures of AfrLEA2, AfrLEA3m, and AfrLEA6 were also investigated in the aqueous state in the presence of trehalose. In embryos of A. franciscana, trehalose concentrations can reach at least 340 mM (Glasheen and Hand 1989) or higher, so it was of interest to evaluate whether this stabilizing organic solute might promote gain of structure in LEA proteins. In all cases, the structure of LEA proteins was unaffected by the presence of trehalose (Fig. 4). The CD spectra of LEA proteins remained indicative of a disordered, random coil structure. The α-helix structure of BSA remained unchanged as well.
Fig. 4.

Comparison of secondary structure content for AfrLEA2, AfrLEA3m, and AfrLEA6 based on CD spectra recorded in the presence of various concentrations of trehalose in the aqueous state. All proteins were measured at a concentration of 0.15 mg/ml. Analysis of secondary structure was performed using the Dichroweb server with the Contin-LL method (see text)
Protein expression of AfrLEA6 during development
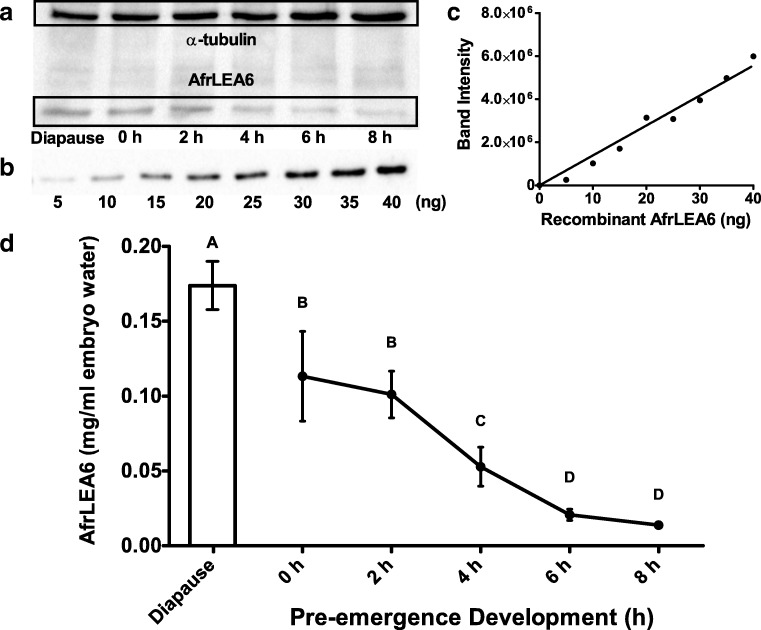
The quantity of AfrLEA6 present in each developmental sample was determined by comparing band intensities to a standard curve of purified, recombinant AfrLEA6 (Fig. 5a–c). These quantities were converted to units of mg LEA protein per ml embryo water. The in vivo titer of AfrLEA6 was highest during diapause and decreased throughout pre-emergence development (Fig. 5d) until it was no longer quantifiable in 24 h nauplii (data not shown). AfrLEA6 was present in diapause embryos at a concentration of 0.173 ± 0.016 (mean ± SD; n = 3) mg per ml embryo water and decreased significantly to a concentration of 0.113 ± 0.03 (mean ± SD; n = 3) mg per ml embryo water by 0 h of pre-emergence development—a reduction of 35% (Fig. 5d). By 8 h of pre-emergence development, the titer of AfrLEA6 had been reduced to 8% of the diapause values, corresponding to 0.014 ± 0.0025 (mean ± SD; n = 3) mg per ml embryo water.
Fig. 5.
Quantification of AfrLEA6 in extracts of A. franciscana by Western blot analysis using an AfrLEA6 polyclonal antibody. a AfrLEA6 was quantified in extracts from diapause embryos and post-diapause embryos through 8 h of pre-emergence development. Total protein loaded per lane was 40 μg. α-Tubulin is included as a loading control for each time point. AfrLEA6 was too low in 24-h free-swimming nauplii to reliably quantify (data not shown). b, c Quantification of AfrLEA6 in extracts was based on a standard curve with 5–40 ng of purified recombinant AfrLEA6. Bio-Rad Image Lab™ software was used to compare band intensities. d AfrLEA6 concentrations from diapause throughout 8 h of pre-emergence development. All values were normalized to α-tubulin and then expressed per ml embryo water as described in “Methods.” Data are presented as means ± SD. Different letters above means indicate statistically significant differences (one-way ANOVA, Tukey, p < 0.01, n = 3)
Diapause termination under defined conditions
We were surprised by the drop in AFrLEA6 between diapause and post-diapause (0 h) embryos (Fig. 5d), an observation based in part on measurements for post-diapause embryos obtained from commercial sources. Consequently, trials were conducted to terminate diapause directly under controlled conditions to more rigorously evaluate this state-specific change in AfrLEA6. Two termination cues were used to break the diapause state in the laboratory—desiccation and incubation in hydrogen peroxide. As shown in Fig. 6a, H2O2 was by far the more effective treatment. Treatment of hydrated diapause embryos with the 3% H2O2 solution for 15 min and 30 min resulted in 75.9 ± 7.0% and 72.6 ± 4.9% hatching (means ± SD; n = 3), respectively; the two results were statistically identical (p = 0.998). Further studies with H2O2 involving time courses, additional concentrations, and other Artemia species and populations are available (Lavens et al. 1986; Lavens and Sorgeloos 1987; Van Stappen et al. 1998). Drying the diapause embryos for 24 h versus applying 1.5X D/H cycles promoted 1.7 ± 0.39% and 7.7 ± 1.2% hatching (means ± SD; n = 3), respectively (Fig. 6a). The hatching percentage promoted by 1.5X D/H cycles was significantly higher than for one 24-h drying bout (p = 0.048), but the difference was modest. Drying for 1 week was no more effective for diapause termination than drying for 24 h (p = 1.000; Fig. 6b). Combining drying with 3% H2O2 (in either order, Fig. 6b) did not improve diapause termination compared to H2O2 alone (all p values > 0.05).
Fig. 6.
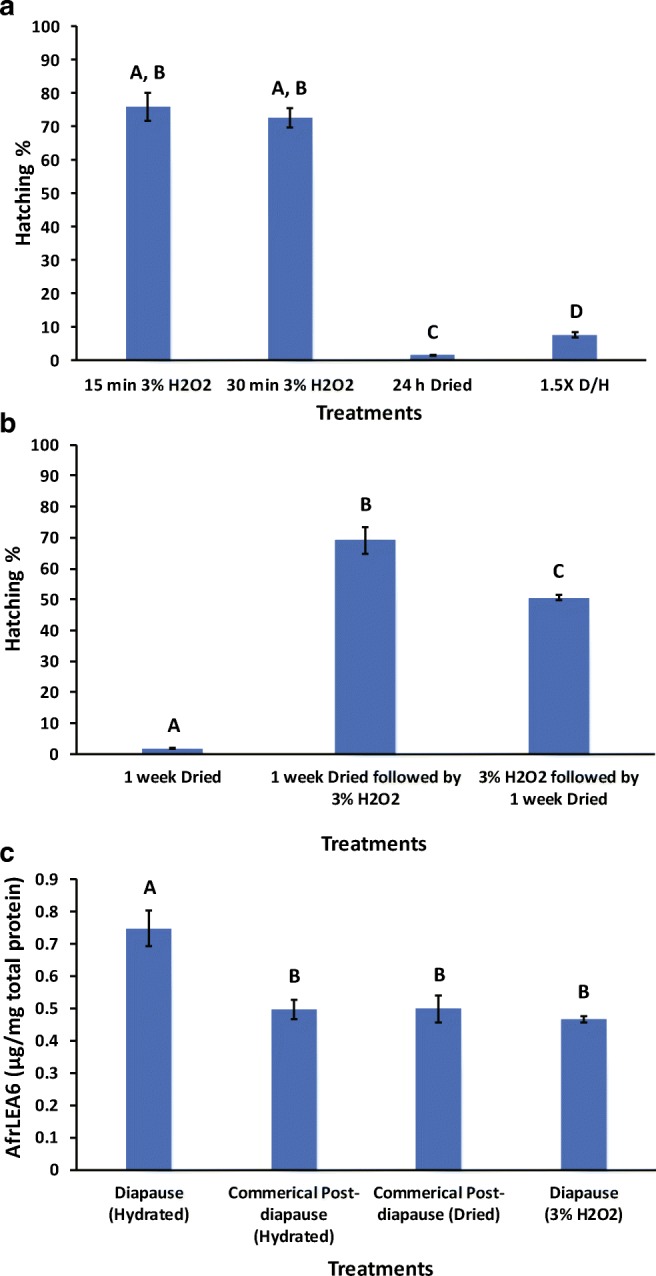
Hatching percentages of diapause embryos from A. franciscana after H2O2 and drying treatments. All treatments were performed at ambient humidity and at room temperature. a 1.5X D/H is defined as drying hydrated diapause embryos for 24 h, rehydrating for 6 h, and drying again (24 h) prior to the 4-day incubation to promote hatching. Bars represent means ± SD. Different letters above bars indicate statistically significant differences among means (Levene’s F (9, 20) = 5.742, p < 0.05. Welch’s ANOVA, Games-Howell, p < 0.05, n = 3). b Hatch percentages after combined treatments with H2O2 and 1-week drying. Bars represent means ± SD (Levene’s F (9, 20) = 5.742, p < 0.05. Welch’s ANOVA, Games-Howell, p < 0.05, n = 3). c AfrLEA6 contents for diapause embryos not treated and treated with H2O2 and for commercially obtained post-diapause embryos of A. franciscana. AfrLEA6 was quantified by Western blotting. Bars represent means ± SD. Different letters above bars indicate statistically significant differences among means (Levene’s F (3, 16) = 3.635, p < 0.05, Welch’s ANOVA, Games-Howell, p < 0.05; from left to right, n = 8, n = 6, n = 3, n = 3)
Consequently, the 15-min treatment with H2O2 was used to terminate diapause and quantify the impact on the AfrLEA6 content of embryos. There was a significant 38% drop in AfrLEA6 content in embryos released from diapause with H2O2 compared to the untreated diapause embryos (Fig. 6c). Further, the AfrLEA6 content in these H2O2-treated embryos was statistically identical to that measured for the commercial post-diapause embryos (Fig. 6c). Thus, the method for diapause termination used commercially (proprietary) promoted the same content of AfrLEA6.
Discussion
Despite sequence and bioinformatic evidence for ArfLEA6 indicating substantially less hydrophilicity than other LEA proteins from A. franciscana, results presented here demonstrate that AfrLEA6 is indeed intrinsically disordered in aqueous solutions and gains structure as water is removed. Further, we show for the first time that the very high physiological concentration of trehalose known to exist in embryos of A. franciscana is insufficient to promote the gain of secondary structure for any of the three LEA proteins (AfrLEA2, AfrLEA3m, AfrLEA6) evaluated in aqueous solution. One might predict that the reduced hydrophilic poise of AfrLEA6 would render it more likely to initiate folding, particularly considering the strong ability of trehalose to force protein assembly/folding through preferential exclusion from the peptide backbone (Street et al. 2006; Xie and Timasheff 1997). AfrLEA6 exhibits its highest concentration in vivo during the diapause state, drops acutely at diapause termination, and then the content declines progressively during development to the larval stage. After confirming the capacity of H2O2 to quickly terminate diapause, we document that the state-dependent, step change in AfrLEA6 at diapause breakage can be produced in the laboratory under controlled conditions.
Our CD studies indicate that AfrLEA6 is clearly intrinsically disordered in solution based on an estimated 89% content of random coil. Further, the α-helix content of AfrLEA6 increased from 4% in solution to 47% when dried, 44% in TFE, and 34% in SDS. Solutions containing 2% SDS and 70% TFE have been documented to induce α-helix conformation in LEA proteins (Ismail et al. 1999; Kentsis and Sosnick 1998; Shih et al. 2004, 2012; Tolleter et al. 2007). In the presence of either solution, AfrLEA6 adopts a majority of α-helix conformation. By comparison to group 3 LEA proteins, Boswell et al. (2014a) estimated the total α-helix content of AfrLEA3m in the aqueous state was 2%, which increased to 41% and 36% after treatment with 2% SDS or 70% TFE, respectively. In the dried state, however, α-helix content only increased to 18%. Gains of structure were also seen for AfrLEA2 under identical conditions. However, note that recombinant AfrLEA2 and AfrLEA3m in that study contained up to 10% sequence that was atypical of the mature protein (hexa-histidine tag, amphipathic mitochondrial targeting sequence, etc.), so precise comparisons of percentages to AfrLEA6 (all native sequence due to purification with the IMPACT expression system/chitin-binding column) should be limited. Recent bioinformatics predictions place AfrLEA6 at the top of the most hydrophobic LEA proteins found in A. franciscana, with AfrLEA2 being second. Despite its high hydrophobicity, the DisEMBL prediction of overall disorder for AfrLEA6 was 80.9% (Janis et al. 2018b). In summary, both experimental data and bioinformatics strongly support a classification of ArfLEA6 as an intrinsically disordered protein (IDP).
As mentioned earlier, trehalose is a stabilizing organic solute that is present in embryonic stages of A. franciscana. At concentrations that are physiologically relevant to these embryos in the hydrated state (≥ 340 mM), trehalose has been shown to drive folding equilibrium towards the native state of globular proteins (Auton et al. 2011; Xie and Timasheff 1997). LEA protein folding in the presence of molecular crowding agents at high concentration (≥ 2 M) has been successfully demonstrated (Bremer et al. 2017; Navarro-Retamal et al. 2018). However, our CD spectra gathered for AfrLEA2, AfrLEA3m, and AfrLEA6 in solutions of trehalose across physiological concentrations indicate no impact of trehalose on the structure of LEA proteins or BSA. If trehalose were to play a role in promoting a gain of secondary structure in LEA proteins, it likely would be seen at higher concentrations that are approached as cellular water is removed during desiccation. Further investigations into the secondary structure of LEA proteins at intermediate hydration values, with and without trehalose, could be warranted.
Based on data presented, AfrLEA6 protein content is highest during the diapause state and undetectable in free-swimming nauplii. This pattern parallels the loss of desiccation tolerance seen at the larval stages through adults, which is consistent with a protective role of LEA proteins during times of desiccation. However, there is an acute 38% decrease in AfrLEA6 during the termination of diapause with H2O2 that was unanticipated, and the reason for this drop remains an open question. This observation suggests that H2O2 triggers an immediate degradation of selected proteins, which apparently includes AfrLEA6 [H2O2 treatment of commercial post-diapause embryos has less impact (29% decrease); M. Le, B. LeBlanc, S Hand, unpublished]. Disruption of key macromolecules could initiate physiological signaling events necessary for resumption of development and metabolism. For example, it has been speculated that the small heat-shock protein p26 in A. franciscana embryos might bind proteins critical to the maintenance of diapause, or perhaps sequester a signaling protein important for diapause termination (King and MacRae 2012). Ultimately, the mechanism explaining termination of diapause with hydrogen peroxide, or any other release cues (e.g., cold temperature, desiccation), is currently unknown (cf. Hand et al. 2016). Population-specific differences exist in the efficacy of termination cues (Lavens and Sorgeloos 1987). As documented here (Fig. 6a, b), desiccation for 24 h up to 1 week is largely ineffective at diapause release for Great Salt Lake embryos, whereas drying works well for embryos from the San Francisco Bay (cf. Clegg et al. 1996). Until definitive mechanisms of diapause termination in A. franciscana become available, these interesting population differences will remain unexplained (cf. Hand et al. 2016).
Finally, considering that the cellular titer of AfrLEA6 in diapause and post-diapause embryos is approximately 10-fold lower than for AfrLEA2 and AfrLEA3m (cf. Boswell et al. 2014b), AfrLEA6 apparently promotes its functions in A. franciscana at a much lower content than these group 3 LEA proteins. Based on sequence homologies between AfrLEA6 and SMPs that are associated with desiccation tolerance in plants (Boucher et al. 2010; Chatelain et al. 2012), potential functions of AfrLEA6 may involve the prevention and dispersion of protein aggregates during desiccation and/or their rapid dissolution during rehydration similar to MtPM25. Contribution to a glassy state during desiccation as with pollen D-7 LEA protein (Wolkers et al. 2001) is also possible. The amino acid sequence of AfrLEA6 contains features associated with proteins known to undergo liquid-liquid phase separations in the hydrated state under certain conditions (Janis et al. 2018a, b). An attractive hypothesis may be that the protein is involved in the partitioning of signaling peptides or other biomolecules that regulate diapause in the cyst. Due to the increasing concentration of solutes in the drying cell, AfrLEA6 may precipitate from solution into another liquid phase that partitions such biomolecules at moderate to low water content and releases them as it dissolves upon rehydration. Experiments to test this hypothesis are currently underway. In any case, our results underscore the concept that multiple LEA proteins exist at different amounts and within different cellular compartments within a single organism and may act together or separately to protect the organism during events of water stress.
Acknowledgments
Thanks are extended to Dr. Brad Marden (Research Division, Great Salt Lake Artemia, Ogden, UT) for his help in collecting diapause embryos of A. franciscana from the Great Salt Lake.
Funding information
We recognize the National Science Foundation grant IOS-1457061/IOS-1659970 for support of this work.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Blase M. LeBlanc, Email: blebl61@lsu.edu
Mike T. Le, Email: mle12@lsu.edu
Brett Janis, Email: brett.janis@louisville.edu.
Michael A. Menze, Email: michael.menze@louisville.edu
Steven C. Hand, Email: shand@lsu.edu
References
- Auton M, Rosgen J, Sinev M, Holthauzen LM, Bolen DW. Osmolyte effects on protein stability and solubility: a balancing act between backbone and side-chains. Biophys Chem. 2011;159:90–99. doi: 10.1016/j.bpc.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avelange-Macherel MH, Candat A, Neveu M, Tolleter D, Macherel D. Decoding the divergent subcellular location of two highly similar paralogous LEA proteins. Int J Mol Sci. 2018;19:1620. doi: 10.3390/ijms19061620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battista JR, Park MJ, McLemore AE. Inactivation of two homologues of proteins presumed to be involved in the desiccation tolerance of plants sensitizes Deinococcus radiodurans R1 to desiccation. Cryobiology. 2001;43:133–139. doi: 10.1006/cryo.2001.2357. [DOI] [PubMed] [Google Scholar]
- Boothby TC, Pielak GJ. Intrinsically disordered proteins and desiccation tolerance: elucidating functional and mechanistic underpinnings of anhydrobiosis. Bioessays. 2017;39:1700119. doi: 10.1002/bies.201700119. [DOI] [PubMed] [Google Scholar]
- Boothby TC, Tapia H, Brozena AH, Piszkiewicz S, Smith AE, Giovannini I, Rebecchi L, Pielak GJ, Koshland D, Goldstein B. Tardigrades use intrinsically disordered proteins to survive desiccation. Mol Cell. 2017;65:975–984 e975. doi: 10.1016/j.molcel.2017.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boswell LC, Hand SC. Intracellular localization of group 3 LEA proteins in embryos of Artemia franciscana. Tissue Cell. 2014;46:514–519. doi: 10.1016/j.tice.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Boswell LC, Menze MA, Hand SC. Group 3 late embryogenesis abundant proteins from embryos of Artemia franciscana: structural properties and protective abilities during desiccation. Physiol Biochem Zool. 2014;87:640–651. doi: 10.1086/676936. [DOI] [PubMed] [Google Scholar]
- Boswell LC, Moore DS, Hand SC. Quantification of cellular protein expression and molecular features of group 3 LEA proteins from embryos of Artemia franciscana. Cell Stress Chaperones. 2014;19:329–341. doi: 10.1007/s12192-013-0458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher V, et al. MtPM25 is an atypical hydrophobic late embryogenesis-abundant protein that dissociates cold and desiccation-aggregated proteins. Plant Cell Environ. 2010;33:418–430. doi: 10.1111/j.1365-3040.2009.02093.x. [DOI] [PubMed] [Google Scholar]
- Bremer A, Wolff M, Thalhammer A, Hincha DK. Folding of intrinsically disordered plant LEA proteins is driven by glycerol-induced crowding and the presence of membranes. FEBS J. 2017;284:919–936. doi: 10.1111/febs.14023. [DOI] [PubMed] [Google Scholar]
- Browne J, Tunnacliffe A, Burnell A. Anhydrobiosis: plant desiccation gene found in a nematode. Nature. 2002;416:38. doi: 10.1038/416038a. [DOI] [PubMed] [Google Scholar]
- Chatelain E, et al. Temporal profiling of the heat-stable proteome during late maturation of Medicago truncatula seeds identifies a restricted subset of late embryogenesis abundant proteins associated with longevity. Plant Cell Environ. 2012;35:1440–1455. doi: 10.1111/j.1365-3040.2012.02501.x. [DOI] [PubMed] [Google Scholar]
- Clegg JS. Interrelationships between water and metabolism in Artemia salina cysts—hydration-dehydration from liquid and vapor phases. J Exp Biol. 1974;61:291–308. doi: 10.1242/jeb.61.2.291. [DOI] [PubMed] [Google Scholar]
- Clegg JS. Desiccation tolerance in encysted embryos of the animal extremophile, Artemia. Integr Comp Biol. 2005;45:715–724. doi: 10.1093/icb/45.5.715. [DOI] [PubMed] [Google Scholar]
- Clegg JS. Stress-related proteins compared in diapause and in activated, anoxic encysted embryos of the animal extremophile, Artemia franciscana. J Insect Physiol. 2011;57:660–664. doi: 10.1016/j.jinsphys.2010.11.023. [DOI] [PubMed] [Google Scholar]
- Clegg JS, Drinkwater LE, Sorgeloos P. The metabolic status of diapause embryos of Artemia franciscana (SFB) Physiol Zool. 1996;69:49–66. doi: 10.1086/physzool.69.1.30164200. [DOI] [Google Scholar]
- Crowe J, Crowe L, Carpenter J, Prestrelski S, Hoekstra F. Anhydrobiosis: cellular adaptations to extreme dehydration. In: Dantzler W, editor. Handbook of physiology section 13, comparative physiology. Oxford: Oxford University Press; 1997. pp. 1445–1477. [Google Scholar]
- Crowe JH, Carpenter JF, Crowe LM. The role of vitrification in anhydrobiosis. Annu Rev Physiol. 1998;60:73–103. doi: 10.1146/annurev.physiol.60.1.73. [DOI] [PubMed] [Google Scholar]
- Crowe JH, Clegg JS. Anhydrobiosis. Stroudsburg: Dowden, Hutchinson & Ross; 1973. [Google Scholar]
- Crowe JH, Clegg JS. Dry biological systems. New York: Academic Press; 1978. [Google Scholar]
- Crowe JH, Crowe LM, Carpenter JF, Aurell Wistrom C. Stabilization of dry phospholipid bilayers and proteins by sugars. Biochem J. 1987;242:1–10. doi: 10.1042/bj2420001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe JH, Crowe LM, Wolkers WF, Oliver AE, Ma X, Auh JH, Tang M, Zhu S, Norris J, Tablin F. Stabilization of dry mammalian cells: lessons from nature. Integr Comp Biol. 2005;45:810–820. doi: 10.1093/icb/45.5.810. [DOI] [PubMed] [Google Scholar]
- Crowe JH, Madin KA. Anhydrobiosis in tardigrades and nematodes. Trans Am Microsc Soc. 1974;93:513–524. doi: 10.2307/3225155. [DOI] [Google Scholar]
- Denekamp NY, Reinhardt R, Kube M, Lubzens E. Late embryogenesis abundant (LEA) proteins in nondesiccated, encysted, and diapausing embryos of rotifers. Biol Reprod. 2010;82:714–724. doi: 10.1095/biolreprod.109.081091. [DOI] [PubMed] [Google Scholar]
- Gal TZ, Glazer I, Koltai H. An LEA group 3 family member is involved in survival of C. elegans during exposure to stress. FEBS Lett. 2004;577:21–26. doi: 10.1016/j.febslet.2004.09.049. [DOI] [PubMed] [Google Scholar]
- Games PA, Howell JF. Pairwise multiple comparison procedures with unequal n’s and/or variances: a Monte Carlo study. J Educ Stat. 1976;1:113–125. [Google Scholar]
- Glasheen JS, Hand SC. Metabolic heat dissipation and internal solute levels of Artemia embryos during changes in cell associated water. J Exp Biol. 1989;145:263–282. [Google Scholar]
- Goyal K, Tisi L, Basran A, Browne J, Burnell A, Zurdo J, Tunnacliffe A. Transition from natively unfolded to folded state induced by desiccation in an anhydrobiotic nematode protein. J Biol Chem. 2003;278:12977–12984. doi: 10.1074/jbc.M212007200. [DOI] [PubMed] [Google Scholar]
- Goyal K, Walton LJ, Tunnacliffe A. LEA proteins prevent protein aggregation due to water stress. Biochem J. 2005;388:151–157. doi: 10.1042/BJ20041931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelet J, Benamar A, Teyssier E, Avelange-Macherel MH, Grunwald D, Macherel D. Identification in pea seed mitochondria of a late-embryogenesis abundant protein able to protect enzymes from drying. Plant Physiol. 2005;137:157–167. doi: 10.1104/pp.104.052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev O, Suetsugu Y, Cornette R, Kawashima T, Logacheva MD, Kondrashov AS, Penin AA, Hatanaka R, Kikuta S, Shimura S, Kanamori H, Katayose Y, Matsumoto T, Shagimardanova E, Alexeev D, Govorun V, Wisecaver J, Mikheyev A, Koyanagi R, Fujie M, Nishiyama T, Shigenobu S, Shibata TF, Golygina V, Hasebe M, Okuda T, Satoh N, Kikawada T. Comparative genome sequencing reveals genomic signature of extreme desiccation tolerance in the anhydrobiotic midge. Nat Commun. 2014;5:4784. doi: 10.1038/ncomms5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand SC, Denlinger DL, Podrabsky JE, Roy R. Mechanisms of animal diapause: recent developments from nematodes, crustaceans, insects, and fish. Am J Phys Regul Integr Comp Phys. 2016;310:R1193–R1211. doi: 10.1152/ajpregu.00250.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand SC, Jones D, Menze MA, Witt TL. Life without water: expression of plant LEA genes by an anhydrobiotic arthropod. J Exp Zool A Ecol Genet Physiol. 2007;307:62–66. doi: 10.1002/jez.a.343. [DOI] [PubMed] [Google Scholar]
- Hand SC, Menze MA. Molecular approaches for improving desiccation tolerance: insights from the brine shrimp Artemia franciscana. Planta. 2015;242:379–388. doi: 10.1007/s00425-015-2281-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand SC, Menze MA, Toner M, Boswell L, Moore D. LEA proteins during water stress: not just for plants anymore. Annu Rev Physiol. 2011;73:115–134. doi: 10.1146/annurev-physiol-012110-142203. [DOI] [PubMed] [Google Scholar]
- Hand SC, Moore DS, Patil Y. Challenges during diapause and anhydrobiosis: mitochondrial bioenergetics and desiccation tolerance. IUBMB Life. 2018;70:1251–1259. doi: 10.1002/iub.1953. [DOI] [PubMed] [Google Scholar]
- Hincha DK, Thalhammer A. LEA proteins: IDPs with versatile functions in cellular dehydration tolerance. Biochem Soc Trans. 2012;40:1000–1003. doi: 10.1042/BST20120109. [DOI] [PubMed] [Google Scholar]
- Hundertmark M, Dimova R, Lengefeld J, Seckler R, Hincha DK. The intrinsically disordered late embryogenesis abundant protein LEA18 from Arabidopsis thaliana modulates membrane stability through binding and folding. Biochim Biophys Acta. 2011;1808:446–453. doi: 10.1016/j.bbamem.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Ismail AM, Hall AE, Close TJ. Purification and partial characterization of a dehydrin involved in chilling tolerance during seedling emergence of cowpea. Plant Physiol. 1999;120:237–244. doi: 10.1104/pp.120.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janis B, Belott C, Menze MA. Role of intrinsic disorder in animal desiccation tolerance. Proteomics. 2018;18:e1800067. doi: 10.1002/pmic.201800067. [DOI] [PubMed] [Google Scholar]
- Janis B, Uversky VN, Menze MA. Potential functions of LEA proteins from the brine shrimp Artemia franciscana—anhydrobiosis meets bioinformatics. J Biomol Struct Dyn. 2018;36:3291–3309. doi: 10.1080/07391102.2017.1387177. [DOI] [PubMed] [Google Scholar]
- Jaspard E, Macherel D, Hunault G. Computational and statistical analyses of amino acid usage and physico-chemical properties of the twelve late embryogenesis abundant protein classes. PLoS One. 2012;7(5):e36968. doi: 10.1371/journal.pone.0036968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentsis A, Sosnick TR. Trifluoroethanol promotes helix formation by destabilizing backbone exposure: desolvation rather than native hydrogen bonding defines the kinetic pathway of dimeric coiled coil folding. Biochemistry. 1998;37:14613–14622. doi: 10.1021/bi981641y. [DOI] [PubMed] [Google Scholar]
- Kikawada T, Nakahara Y, Kanamori Y, Iwata KI, Watanabe M, McGee B, Tunnacliffe A, Okuda T. Dehydration-induced expression of LEA proteins in an anhydrobiotic chironomid. Biochem Biophys Res Commun. 2006;348:56–61. doi: 10.1016/j.bbrc.2006.07.003. [DOI] [PubMed] [Google Scholar]
- King AM, MacRae TH. The small heat shock protein p26 aids development of encysting Artemia embryos, prevents spontaneous diapause termination and protects against stress. PLoS One. 2012;7:e43723. doi: 10.1371/journal.pone.0043723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavens P, Sorgeloos P. The cryptobiotic state of Artemia cysts, its diapause deactivation and hatching: a review. In: Sorgeloos P, Bengtson DA, Decleir W, Jaspers E, editors. Artemia research and its applications. Wetteren: Universa Press; 1987. pp. 27–63. [Google Scholar]
- Lavens P, Tackaert W, Sorgeloos P. International study on Artemia. XLI. Influence of culture conditions and specific diapause deactivation methods on the hatchability of Artemia cysts produced in a standard culture system. Mar Ecol Prog Ser. 1986;31:197–203. doi: 10.3354/meps031197. [DOI] [Google Scholar]
- McDonald J. Handbook of biological statistics. 3. Baltimore: Sparky House Publishing; 2014. [Google Scholar]
- Moore DS, Hand SC. Cryopreservation of lipid bilayers by LEA proteins from Artemia franciscana and trehalose. Cryobiology. 2016;73:240–247. doi: 10.1016/j.cryobiol.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Moore DS, Hansen R, Hand SC. Liposomes with diverse compositions are protected during desiccation by LEA proteins from Artemia franciscana and trehalose. Biochim Biophys Acta. 2016;1858:104–115. doi: 10.1016/j.bbamem.2015.10.019. [DOI] [PubMed] [Google Scholar]
- Navarro-Retamal C, Bremer A, Ingólfsson HI, Alzate-Morales J, Caballero J, Thalhammer A, González W, Hincha DK. Folding and lipid composition determine membrane interaction of the disordered protein COR15A. Biophys J. 2018;115:968–980. doi: 10.1016/j.bpj.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumeyer CH, Gerlach JL, Ruggiero KM, Covi JA. A novel model of early development in the brine shrimp, Artemia franciscana, and its use in assessing the effects of environmental variables on development, emergence, and hatching. J Morphol. 2015;276:342–360. doi: 10.1002/jmor.20344. [DOI] [PubMed] [Google Scholar]
- Peterson GL. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Popova AV, Rausch S, Hundertmark M, Gibon Y, Hincha DK. The intrinsically disordered protein LEA7 from Arabidopsis thaliana protects the isolated enzyme lactate dehydrogenase and enzymes in a soluble leaf proteome during freezing and drying. Biochim Biophys Acta. 2015;1854:1517–1525. doi: 10.1016/j.bbapap.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Provencher SW, Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- Reynolds JA, Hand SC. Differences in isolated mitochondria are insufficient to account for respiratory depression during diapause in Artemia franciscana embryos. Physiol Biochem Zool. 2004;77:366–377. doi: 10.1086/420950. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Salazar J, Moreno S, Espin G. LEA proteins are involved in cyst desiccation resistance and other abiotic stresses in Azotobacter vinelandii. Cell Stress Chaperones. 2017;22:397–408. doi: 10.1007/s12192-017-0781-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitzky A, Golay MJE. Smoothing + differentiation of data by simplified least squares procedures. Anal Chem. 1964;36:1627. doi: 10.1021/ac60214a047. [DOI] [Google Scholar]
- Shih MD, Hsieh TY, Jian WT, Wu MT, Yang SJ, Hoekstra FA, Hsing YI. Functional studies of soybean (Glycine max L.) seed LEA proteins GmPM6, GmPM11, and GmPM30 by CD and FTIR spectroscopy. Plant Sci. 2012;196:152–159. doi: 10.1016/j.plantsci.2012.07.012. [DOI] [PubMed] [Google Scholar]
- Shih MD, Lin SC, Hsieh JS, Tsou CH, Chow TY, Lin TP, Hsing YI. Gene cloning and characterization of a soybean (Glycine max L.) LEA protein, GmPM16. Plant Mol Biol. 2004;56:689–703. doi: 10.1007/s11103-004-4680-3. [DOI] [PubMed] [Google Scholar]
- Solomon A, Salomon R, Paperna I, Glazer I. Desiccation stress of entomopathogenic nematodes induces the accumulation of a novel heat-stable protein. Parasitology. 2000;121(Pt 4):409–416. doi: 10.1017/S0031182099006563. [DOI] [PubMed] [Google Scholar]
- Sreerama N, Venyaminov SY, Woody RW. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999;8:370–380. doi: 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama N, Woody RW. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal Biochem. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- Stacy RA, Aalen RB. Identification of sequence homology between the internal hydrophilic repeated motifs of group 1 late-embryogenesis-abundant proteins in plants and hydrophilic repeats of the general stress protein GsiB of Bacillus subtilis. Planta. 1998;206:476–478. doi: 10.1007/s004250050424. [DOI] [PubMed] [Google Scholar]
- Steponkus PL, Uemura M, Joseph RA, Gilmour SJ, Thomashow MF. Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1998;95:14570–14575. doi: 10.1073/pnas.95.24.14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street TO, Bolen DW, Rose GD. A molecular mechanism for osmolyte-induced protein stability. Proc Natl Acad Sci U S A. 2006;103:13997–14002. doi: 10.1073/pnas.0606236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Shigeta M, Aoki K. Secondary structures of bovine serum albumin in anionic and cationic surfactant solutions. J Colloid Interface Sci. 1987;117:120–126. doi: 10.1016/0021-9797(87)90174-3. [DOI] [Google Scholar]
- Tan J, MacRae TH. Stress tolerance in diapausing embryos of Artemia franciscana is dependent on heat shock factor 1 (Hsf1) PLoS One. 2018;13:e0200153. doi: 10.1371/journal.pone.0200153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia H, Koshland DE. Trehalose is a versatile and long-lived chaperone for desiccation tolerance. Curr Biol. 2014;24:2758–2766. doi: 10.1016/j.cub.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Thalhammer A, Bryant G, Sulpice R, Hincha DK. Disordered cold regulated15 proteins protect chloroplast membranes during freezing through binding and folding, but do not stabilize chloroplast enzymes in vivo. Plant Physiol. 2014;166:190–201. doi: 10.1104/pp.114.245399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolleter D, Hincha DK, Macherel D. A mitochondrial late embryogenesis abundant protein stabilizes model membranes in the dry state. Biochim Biophys Acta. 2010;1798:1926–1933. doi: 10.1016/j.bbamem.2010.06.029. [DOI] [PubMed] [Google Scholar]
- Tolleter D, Jaquinod M, Mangavel C, Passirani C, Saulnier P, Manon S, Teyssier E, Payet N, Avelange-Macherel MH, Macherel D. Structure and function of a mitochondrial late embryogenesis abundant protein are revealed by desiccation. Plant Cell. 2007;19:1580–1589. doi: 10.1105/tpc.107.050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi R, Boschetti C, McGee B, Tunnacliffe A. Trafficking of bdelloid rotifer late embryogenesis abundant proteins. J Exp Biol. 2012;215:2786–2794. doi: 10.1242/jeb.071647. [DOI] [PubMed] [Google Scholar]
- Tunnacliffe A, Hincha DK, Leprince O, Macherel D. LEA proteins: versatility of form and function. In: Lubzens E, Cerda J, Clark M, editors. Dormancy and resistance in harsh environments. Berlin: Springer; 2010. pp. 91–108. [Google Scholar]
- Tunnacliffe A, Lapinski J, McGee B. A putative LEA protein, but no trehalose, is present in anhydrobiotic bdelloid rotifers. Hydrobiologia. 2005;546:315–321. doi: 10.1007/s10750-005-4239-6. [DOI] [Google Scholar]
- Tunnacliffe A, Wise MJ. The continuing conundrum of the LEA proteins. Naturwissenschaften. 2007;94:791–812. doi: 10.1007/s00114-007-0254-y. [DOI] [PubMed] [Google Scholar]
- Van Stappen G, Lavens P, Sorgeloos P. Effects of hydrogen peroxide treatment in Artemia cysts of different geographical origin. Arch Hydrobiol Spec Issues Advanc Limnol. 1998;52:281–296. [Google Scholar]
- van Stokkum IH, Spoelder HJ, Bloemendal M, van Grondelle R, Groen FC. Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal Biochem. 1990;191:110–118. doi: 10.1016/0003-2697(90)90396-Q. [DOI] [PubMed] [Google Scholar]
- Warner AH, Chakrabortee S, Tunnacliffe A, Clegg JS. Complexity of the heat-soluble LEA proteome in Artemia species. Comp Biochem Physiol D Genomics Proteomics. 2012;7:260–267. doi: 10.1016/j.cbd.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Warner AH, Miroshnychenko O, Kozarova A, Vacratsis PO, MacRae TH, Kim J, Clegg JS. Evidence for multiple group 1 late embryogenesis abundant proteins in encysted embryos of Artemia and their organelles. J Biochem. 2010;148:581–592. doi: 10.1093/jb/mvq091. [DOI] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- Wolkers WF, McCready S, Brandt WF, Lindsey GG, Hoekstra FA. Isolation and characterization of a D-7 LEA protein from pollen that stabilizes glasses in vitro. Biochim Biophys Acta Protein Struct Mol Enzymol. 2001;1544:196–206. doi: 10.1016/S0167-4838(00)00220-X. [DOI] [PubMed] [Google Scholar]
- Wu G, Zhang H, Sun J, Liu F, Ge X, Chen WH, Yu J, Wang W. Diverse LEA (late embryogenesis abundant) and LEA-like genes and their responses to hypersaline stress in post-diapause embryonic development of Artemia franciscana. Comp Biochem Physiol B Biochem Mol Biol. 2011;160:32–39. doi: 10.1016/j.cbpb.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Xie G, Timasheff SN. The thermodynamic mechanism of protein stabilization by trehalose. Biophys Chem. 1997;64:25–43. doi: 10.1016/S0301-4622(96)02222-3. [DOI] [PubMed] [Google Scholar]
- Yuen F, Watson M, Barker R, Grillo I, Heenan RK, Tunnacliffe A, Routh AF. Preferential adsorption to air-water interfaces: a novel cryoprotective mechanism for LEA proteins. Biochem J. 2019;476:1121–1135. doi: 10.1042/BCJ20180901. [DOI] [PMC free article] [PubMed] [Google Scholar]