Significance
Mechanisms to balance the acquisition of sufficient Fe while preventing a toxic overload differ in bacteria, fungi, animals, and plants. Identification of specific E3 ligases acting directly on a key transcription factor for Fe uptake in Arabidopsis indicates how this balance is regulated in dicotyledonous plants. The domain structure and function of the E3 ligases show interesting parallels to a distantly related protein regulating Fe homeostasis in mammals. Moreover, the accumulation of Fe in weaker mutant alleles of the E3 ligases could be exploited for biofortification of crops.
Keywords: iron, ubiquitin, bHLH transcription factor, dicotyledon, micronutrient
Abstract
Organisms need to balance sufficient uptake of iron (Fe) with possible toxicity. In plant roots, a regulon of uptake genes is transcriptionally activated under Fe deficiency, but it is unknown how this response is inactivated when Fe becomes available. Here we describe the function of 2 partially redundant E3 ubiquitin ligases, BRUTUS-LIKE1 (BTSL1) and BTSL2, in Arabidopsis thaliana and provide evidence that they target the transcription factor FIT, a key regulator of Fe uptake, for degradation. The btsl double mutant failed to effectively down-regulate the transcription of genes controlled by FIT, and accumulated toxic levels of Fe in roots and leaves. The C-terminal domains of BTSL1 and BTSL2 exhibited E3 ligase activity, and interacted with FIT but not its dimeric partner bHLH39. The BTSL proteins were able to poly-ubiquitinate FIT in vitro and promote FIT degradation in vivo. Thus, posttranslational control of FIT is critical to prevent excess Fe uptake.
Iron (Fe) is the fourth most abundant element in the Earth’s crust, but its bioavailability is greatly limited by the insolubility of Fe hydroxides. High-affinity uptake mechanisms are therefore essential for most organisms, from prokaryotes to multicellular species. Plants have developed 2 molecular strategies for Fe uptake, historically divided into Strategy I (reductive strategy) present in dicotyledonous plants, and Strategy II (chelating strategy) in grasses (1, 2). The reductive strategy in plants such as Arabidopsis thaliana involves a ferric reductive oxidase, FRO2, to reduce Fe3+ to Fe2+, which is then taken up by the iron-regulated transporter IRT1. A key regulator of Fe uptake in dicot plants is the basic helix–loop–helix (bHLH) transcription factor FIT (FER-like iron deficiency-induced transcription factor) (3). FIT forms heterodimers with 1 of 4 bHLH proteins from subgroup Ib; namely, bHLH38, bHLH39, bHLH100, and bHLH101. Mutant studies combined with transcriptomics have identified more than 400 genes that are controlled by FIT (4, 5). These include FRO2 and IRT1 for which direct promoter binding by the FIT-bHLH Ib dimer has been shown (6, 7).
Fe is essential as a cofactor for many enzymes, but in its free form it is toxic. Redox chemistry of Fe2+/Fe3+ catalyzes the production of oxygen radicals, known as the Fenton reaction. Therefore, the uptake of Fe, storage mechanisms, and Fe cofactor biosynthesis must be tightly regulated (8). In their natural environment, plants are generally Fe starved, and the Fe deficiency response is engaged to maximize uptake. However, when Fe becomes available through new root growth or changes in the environment, uptake needs to be switched off immediately to avoid Fe overload. In addition to internalization of IRT1 (9–11), the activity of upstream transcription factors is down-regulated. FIT protein levels are controlled by 26S-proteasome-dependent turnover, which, paradoxically, is initiated during Fe deficiency while FIT is transcriptionally up-regulated (12, 13). It has been proposed that a short-lived pool of FIT is important to anticipate a sudden increase in Fe in the environment. However, the E3 ligase(s) that may facilitate the turnover of FIT have so far not been identified.
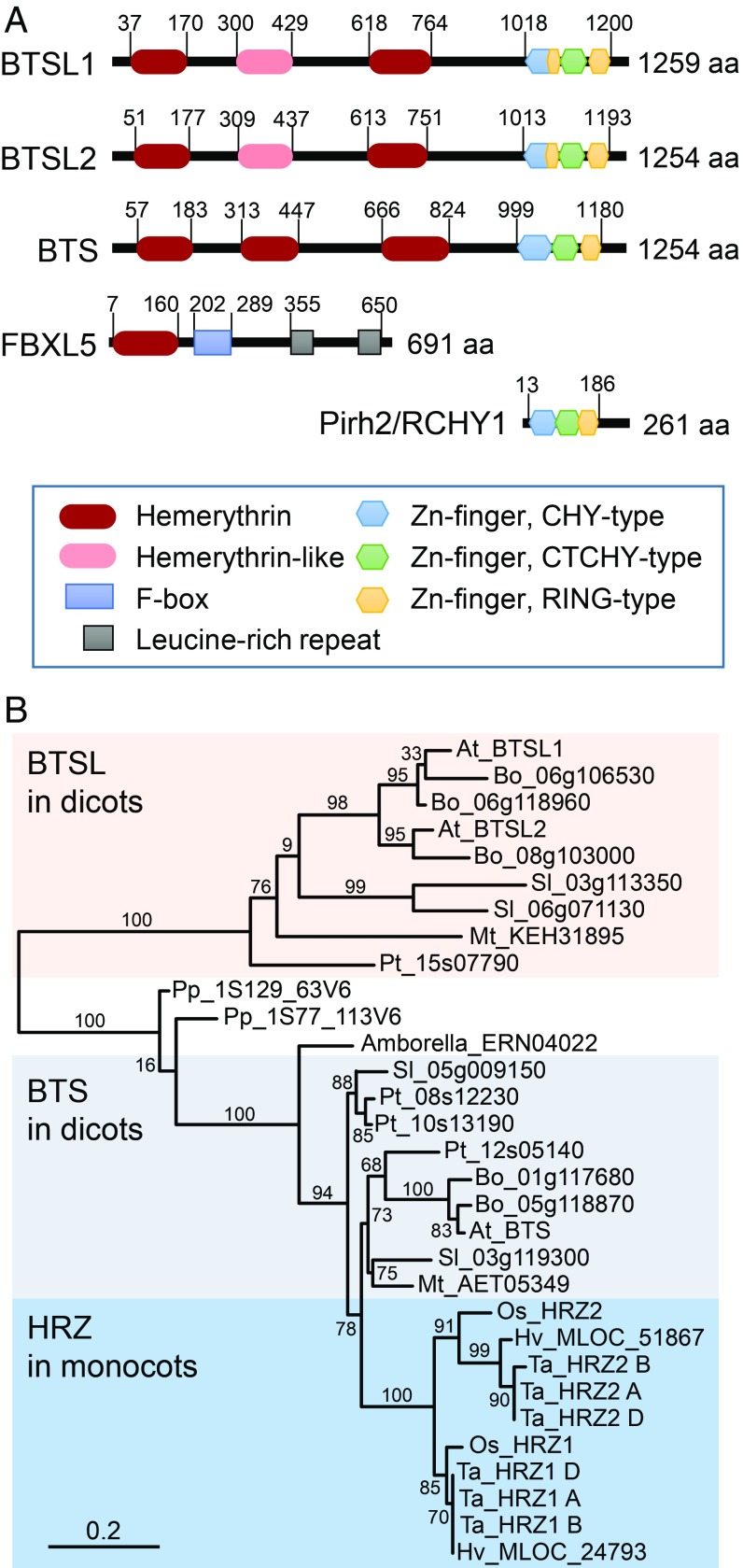
Among the 20 different E3 ligases that are up-regulated during Fe deficiency (4, 14, 15) is a small gene family encoding E3 ubiquitin ligases with N-terminal hemerythrin motifs. These include BRUTUS (BTS) in Arabidopsis (16, 17); and the hemerythrin motif-containing RING- and zinc (Zn)-finger proteins (HRZ1, HRZ2) in rice (18). The BTS and HRZ proteins have been characterized as negative regulators of Fe homeostasis, since bts and hrz mutant lines accumulated Fe and exhibited increased tolerance to Fe deficiency. The combination of an Fe-binding hemerythrin motif associated with an E3 ligase is also found in mammalian FBXL5 (F-box/LRR protein 5, Fig. 1A). The stability of FBXL5 is regulated by Fe and oxygen, providing a switch to promote the ubiquitination and degradation of IRP1 and IRP2, iron regulatory proteins 1 and 2 (19–21). However, there are no functional homologs of IRP1 and IRP2 in plants; therefore, the BTS/HRZ proteins must have different targets. Their E3 ligase domain shows striking homology to the mammalian Pirh2/RCHY1 E3 ligase (ref. 22 and Fig. 1A), which regulates the levels of p53 transcription factor (23, 24). In Arabidopsis, BTS was shown to interact with, and affect the stability of, the transcription factors bHLH105 (ILR3) and bHLH115, which are involved in Fe signaling (17). In rice, HRZ1 targets PRI1, the homolog of ILR3, for degradation (25). Two genes homologous to BTS are found in the Arabidopsis genome, named BRUTUS-LIKE1 (AT1G74770) and BRUTUS-LIKE2 (AT1G18910). The btsl1 btsl2 double knockout mutant accumulated Fe and was more tolerant to Fe deficiency (26). A triple mutant of btsl1, btsl2, and bts had an enhanced phenotype; therefore, it was suggested that the function of BTSLs is redundant with BTS.
Fig. 1.
BTSL are uniquely found in dicotyledonous plants. (A) Domain organization of the BTSL and BTS proteins and closest mammalian homologs. (B) Phylogenetic tree of BTS homologs from selected plant species. Sequences were found by BLASTing the amino acid sequences of Arabidopsis BTS, BTSL1, BTSL2, and rice HRZ1 in Ensembl Plants (http://plants.ensembl.org). Species used: Amborella, Amborella trichocarpa; At, A. thaliana; Bo, Brassica oleracea; Hv, Hordeum vulgare; Mt, Medicago truncatula; Os, Oryza sativa; Pp, Physcomitrella patens; Pt, Populus trichocarpa; Sl, Solanum lycopersicum; Ta, Triticum aestivum. Numbers next to branches indicate bootstrapping values for 100 replications. The scale bar indicates the rate of evolutionary change expressed as number of amino acid substitutions per site.
Here we show that the function of the BTSL proteins differs from BTS in their tissue-specific expression and ubiquitination targets. BTSL1 and BTSL2 are expressed predominantly in the root epidermis and cortex and are coregulated with Fe uptake genes, whereas BTS is coexpressed with other Fe homeostasis genes. On standard medium, btsl1 btsl2 double mutants accumulated moderate amounts of Fe, but when challenged with Fe resupply after a period of deficiency, large amounts of Fe accumulated as the mutant plants failed to rapidly switch off the transcription of FRO2 and IRT1. Using a range of assays, we show that the E3 ligase domains of BTSL1 and BTSL2 were able to ubiquitinate FIT in vitro and target the transcription factor for degradation.
Results
BTSL1 and BTSL2 Proteins Are Unique to Dicotyledonous Plants.
To study the evolutionary relationship between hemerythrin E3 ligases in plants, we performed a phylogenetic analysis of homologous protein sequences across the plant kingdom. Interestingly, Arabidopsis BTSL1 and BTSL2 are in a separate clade from BTS, and the BTSL clade is unique to dicotyledonous species (Fig. 1B).
In terms of domain organization, the Arabidopsis BTSL1/2 proteins have 2 predicted hemerythrin motifs compared to 3 motifs in BTS, and a C-terminal CHY/RING Zn-finger domain (Fig. 1A). The 2 hemerythrin motifs of BTSL1/2 correspond to the first and third motif in BTS. Further inspection revealed a degenerate hemerythrin sequence in BTSL1/2, not recognized by motif searching algorithms, corresponding to the second hemerythrin motif of BTS. Although the 4 conserved histidine residues found in canonical hemerythrin proteins are lacking, the sequence is predicted to form a 4 α-helical bundle typical of hemerythrins (SI Appendix, Fig. S1).
The C-terminal Zn-finger domain has 80% amino acid identity between BTSL1 and BTSL2, and 65% identity with BTS. This particular type of CHY/RING Zn-finger domain is also found in 4 other Arabidopsis E3 ligases such as MIEL1 (27), and in 1 mammalian E3 ligase, Pirh2/RCHY1 (23). Structural studies of Pirh2 using NMR showed a unique arrangement of Zn-fingers, with a total of 9 Zn-binding sites. The first 2 Zn-fingers (CHY- and CTCHY-type) form a “tweezer” together with the C-terminal amino acids for interaction with its target, the transcription factor p53. The RING Zn-finger domain provides the interaction platform for the E2 ligase (24). It is interesting to note that BTSL proteins have an extra RING Zn-finger motif inserted between the CHY and CTHY Zn-finger motifs (Fig. 1A).
BTSL1 and BTSL2 Promoters Are Activated by Fe Deficiency in Specific Root Tissues.
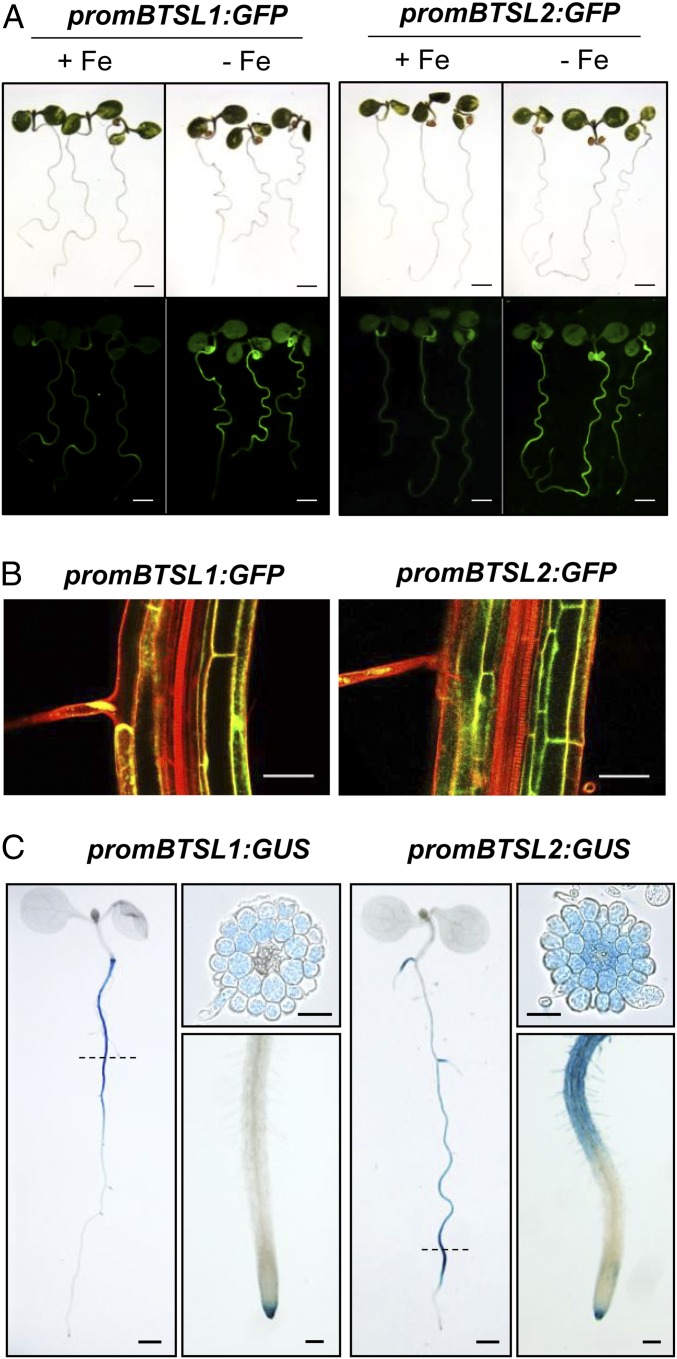
BTSL1 and BTSL2 are expressed predominantly in roots under Fe deficiency (26, 28). To investigate where in the roots and in which cell types BTSL1 and BTSL2 are expressed, the promoter regions of these genes were cloned upstream of an eGFP-GUS reporter gene (29). Seeds were germinated on agar plates with minimum salts (SI Appendix, Materials and Methods) supplemented either with 50 µM FeEDTA (+Fe) or with 100 µM ferrozine to effectively deplete Fe (−Fe). After 5 d of growth, GFP was highly induced in roots −Fe, whereas no GFP fluorescence was observed +Fe, except for auto-fluorescence in the seed coat (Fig. 2A). In older seedlings which had developed lateral roots, the expression pattern of the BTSL1 and BTSL2 promoters started to differentiate from each other: GUS staining showed that BTSL1 is expressed in the upper half of the root under Fe deficiency, whereas BTSL2 is expressed in the lower half of the root, predominantly in the differentiation zone with root hairs (Fig. 2C). BTSL1 and BTSL2 promoter-GUS activity was also found in the root cap (columella), but not in the root meristem or elongation zone. Cross-sections of the GUS-stained roots (Fig. 2C) and imaging of GFP expression in the longitudinal plane (Fig. 2B) showed that both promoters are induced in the epidermis, including root hairs, and cortex cells. BTSL2 was also expressed in the endodermis and stele in the differentiation zone (Fig. 2 B and C). The expression patterns of BTSL1 and BTSL2 contrast with the pattern observed for BTS, which has little promoter activity in the epidermis and cortex, but was strongly up-regulated in the stele in response to Fe deficiency (17). Moreover, BTS is expressed in leaves and in embryos (17) which lack expression of BTSL1 and BTSL2 (SI Appendix, Fig. S2).
Fig. 2.
Promoter activity of BTSL1 and BTSL2. Promoter sequences of BTSL1 (−880 nt) and BTSL2 (−2097 nt) were inserted upstream of eGFP-GUS and the constructs were stably expressed in wild-type Arabidopsis. (A) GFP fluorescence in seedlings grown on medium with 50 μM FeEDTA (+Fe) or with 100 µM ferrozine to deplete Fe (−Fe) for 6 d. (Scale bar, 1 mm.) (B) GFP fluorescence in optical transverse sections of roots in 6-d-old seedlings grown −Fe. Cell walls were stained with propidium iodide (red). (Scale bar, 50 μm.) (C) GUS activity staining in seedlings grown −Fe for 8 d. Scale bar is 0.5 mm for whole seedlings, 100 μm for a close-up of the root tip, and 50 μm for root cross-sections. Images are representative of 3 independent lines for each promoter construct.
Using publicly available microarray data, we built coexpression networks around BTSL1, BTSL2, and BTS, then filtered for Fe-regulated genes (30). First, we found that BTSL1 and BTSL2 are coregulated with each other but not with BTS (SI Appendix, Fig. S3 and Table S1). Second, we noticed that a large network could be built around BTSL1 and BTSL2 using microarray data from root samples, but correlations with BTS were only found when using data sets from shoots. BTSL1 and BTSL2 are coregulated with the Fe transporter gene IRT1 and genes for the biosynthesis and export of coumarin-derived Fe chelators (4CL2, F6’H1 and PDR9). Also coexpressed with BTSL1 and BTSL2 are the transcription factor FIT and its partner bHLH39. Basically, the entire root Fe uptake regulon is found, except for bHLH38 and FRO2, but these genes are not present on the microarray chip that is most commonly used. The BTS network includes the transcription factor POPEYE (PYE), as previously documented (16), and the ferric reductase FRO3 (SI Appendix, Fig. S3B and Table S1). The different coexpression networks of BTSL1/2 and BTS correlate well with their tissue-specific expression patterns.
BTSL1 and BTSL2 Prevent Excess Fe Uptake under Fluctuating Levels.
To further compare the functions of BTSL1, BTSL2, and BTS, we obtained T-DNA insertion lines. Three independent mutant lines were selected for BTSL1 (btsl1-1, btsl1-2, and btsl1-3), but only 1 T-DNA insertion line was available for BTSL2 (btsl2-2) (SI Appendix, Fig. S4A). The btsl1-1 allele has been described as btsl1 in ref. 26. The expression of BTSL1 was virtually abolished in the btsl1-1 and btsl1-3 alleles, but residual expression remained in btsl1-2 (SI Appendix, Fig. S4B). The btsl2-2 mutant lacked detectable levels of BTSL2 transcript, while BTSL1 expression was ∼3-fold higher than in wild type −Fe. A double knockout line was produced by crossing btsl1-1 with the btsl2-2 line. The btsl1-1 btsl2-2 double mutant was genetically complemented by either BTSL1:YFP or BTSL2:GFP (see below).
Mutant lines were tested for phenotypes on a range of Fe concentrations, from 0 to 500 µM FeEDTA (SI Appendix, Fig. S5A). Single insertion lines showed no obvious growth phenotype as previously reported (26). On the other hand, the btsl1-1 btsl2-2 double mutant line showed subtle but noticeable phenotypes in response to both Fe deficiency and Fe excess. After 4 d without Fe, the btsl double mutant appeared less chlorotic than wild type, which was confirmed by chlorophyll measurements (SI Appendix, Fig. S5 A and B). The btsl2-2 mutant allele also retained some chlorophyll, suggesting that BTSL1 and BTSL2 are not fully redundant. In the presence of excess Fe, growth of the btsl double mutant was significantly impaired compared to wild type or the single mutants, associated with a 2-fold accumulation of Fe in the shoots (SI Appendix, Fig. S5 A and C).
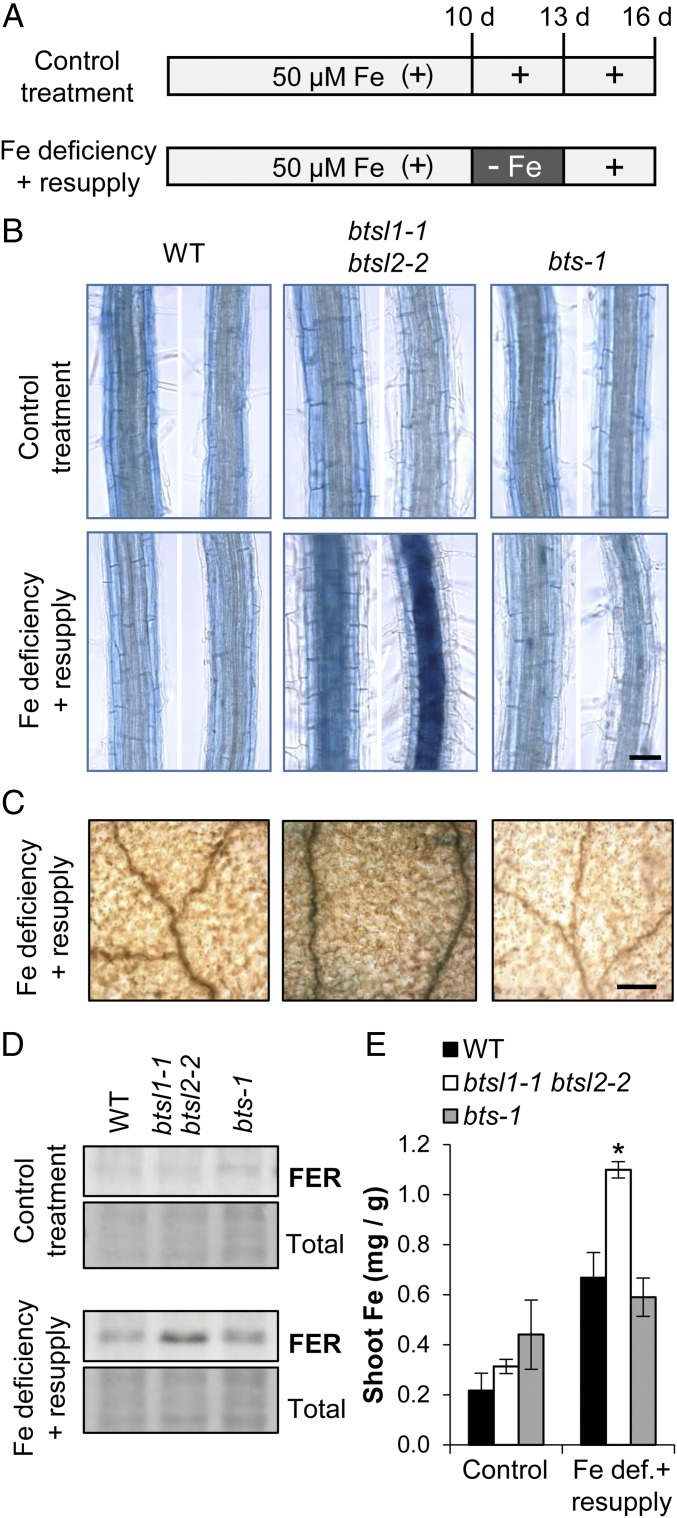
We tested whether the BTSL genes have a specific function in the transition from Fe deficiency to Fe sufficiency. Wild-type seedlings and btsl double mutants were grown up with 50 μM FeEDTA (+Fe) for 10 d and then transferred to −Fe medium for 3 d to induce Fe deficiency. After that, plants were resupplied with Fe and sampled after 3 d (Fig. 3A). While there were no obvious growth phenotypes, the btsl double mutant accumulated large amounts of Fe in the central cylinder of the root and in leaf veins as shown by Perls’ staining (Fig. 3 B and C). Ferritin as a marker for Fe status accumulated in the roots of the btsl double mutant (Fig. 3D), and the shoots contained 4-fold more Fe (Fig. 3E). Iron did not accumulate when btsl double mutants were grown continuously on 50 μM Fe. For comparison, we also grew the viable brutus-1 (bts-1) mutant (17) under the same conditions. The bts-1 behaved as wild type and did not show increased Perls’ staining in any part of the root or leaves after the Fe deficiency-resupply treatment. Interestingly, we found that wild-type plants, when subjected to deficiency and resupply, contained 3-fold more Fe compared to continuous growth on 50 µM Fe (Fig. 3E). It is likely that up-regulation of the Fe uptake machinery during Fe deficiency leads to a sudden influx of Fe when this is resupplied, but to levels that can be controlled by redistribution in wild-type plants.
Fig. 3.
The btsl double mutant hyper-accumulates Fe after a period of Fe deficiency. (A) Diagram of the Fe treatments used in this study. Seedlings were germinated on medium with 50 μM FeEDTA. On day 10, seedlings were transferred to a new plate (control treatment) or to medium depleted of Fe (100 μM ferrozine). After 3 d, seedlings were transferred back to medium with 50 μM FeEDTA for another 3 d. (B) Perls’ Prussian Blue staining for Fe in roots in wild type, btsl double mutant and bts-1 seedlings following control (Top) and Fe deficiency-resupply treatments (Bottom) according to the diagram in A. The images show a section of the differentiation zone. (Scale bar, 50 μm.) (C) Perls’ Prussian Blue staining of leaves after the Fe deficiency-resupply treatment. The images are a close-up of the adaxial leaf surface with veins. (Scale bar, 0.2 mm.) (D) Immunoblot of ferritin (FER) protein in roots and (E) Fe concentrations in shoots of wild type, btsl double mutant, and bts-1 seedlings after control and Fe deficiency-resupply treatments. Bars represent the mean of 3 biological replicates of 10 pooled plants each ±SD (*P < 0.05 using a 2-tailed t test).
In summary, our data show that the btsl double mutant is unable to limit Fe uptake when this is supplied after a period of Fe deficiency. Moreover, lack of Fe accumulation in the bts-1 mutant under these conditions suggest that BTSLs and BTS have different functions.
Transcriptional Down-Regulation of Fe Uptake Genes Is Delayed in the btsl1-1 btsl2-2 Double Mutant.
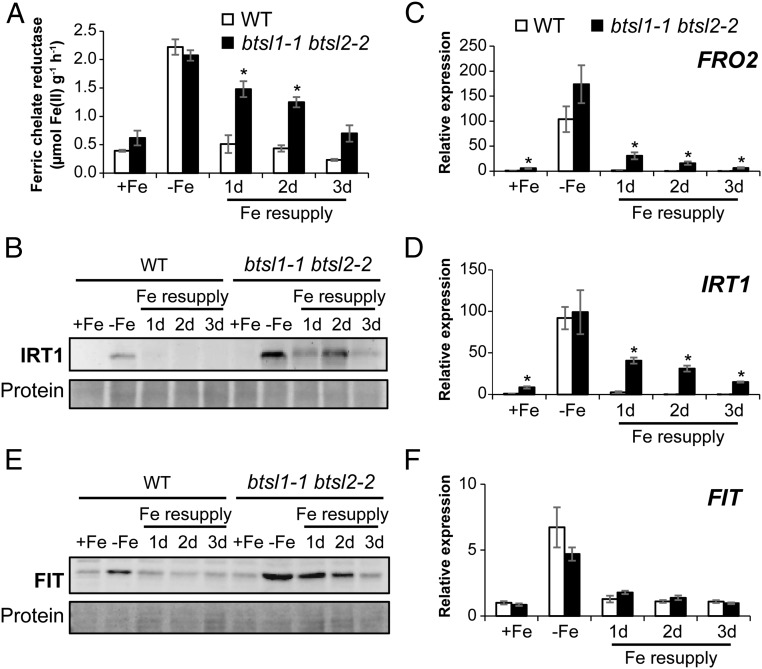
Next, we investigated the levels of FRO2 and IRT1, 2 key players in Fe uptake, during Fe deficiency and resupply. Wild-type seedlings showed a ∼6-fold induction in FRO2 activity in response to Fe deficiency, and down to basal levels 1 d after Fe resupply (Fig. 4A). In the btsl double mutant, FRO2 enzyme activity remained high after Fe resupply: the activity was 4-fold induced after 1 d, 3-fold after 2 d, and it was still nearly double the basal wild-type levels after 3 d (Fig. 4A). Of the single mutant lines, btsl1-1 behaved like wild type, but btsl2-2 showed a slight delay in switching off FRO2 activity (SI Appendix, Fig. S6A). The sustained induction of FRO2 activity in the btsl double mutant was restored by expressing either 35S:BTSL1-YFP or 35S:BTSL2-GFP (SI Appendix, Fig. S6B).
Fig. 4.
The btsl double mutant has delayed down-regulation of FRO2, IRT1, and FIT upon Fe resupply. (A) Ferric chelate reductase activity as a measure for FRO2 protein activity in roots of wild type and btsl1-1 btsl2-2 double mutants. Bars represent the mean of 3 biological replicates (n = 5 seedlings/assay) ± SE (*P < 0.05 using a 2-tailed t test). (B) Immunoblot of IRT1 protein levels. (C, D, and F) Expression of FRO2, IRT1, and FIT, respectively, determined by quantitative RT-qPCR in roots of wild type and btsl1-1 btsl2-2 mutants. All values are relative to wild type +Fe and the mean of 3 biological replicates ±SE (*P < 0.05 using a 2-tailed t test). (E) Immunoblot of FIT protein. Quantification of 3 independent immunoblots can be found in SI Appendix, Fig. S7A.
IRT1 protein levels in wild-type plants were strongly increased under Fe deficiency, and down after Fe resupply. In the btsl double mutant, IRT1 was not detected under standard conditions (+Fe), but, after 3 d of Fe deficiency, was present at much higher levels than in wild-type roots. Upon Fe resupply, the IRT1 protein levels remained high (Fig. 4B). The sustained presence of FRO2 and IRT1 would explain the increased Fe accumulation in the btsl double mutant following the Fe deficiency-resupply treatment.
To investigate which step of FRO2 and IRT1 expression is misregulated, we analyzed transcript levels of FRO2 and IRT1 by RT-qPCR. Interestingly, under control conditions (+Fe), FRO2 and IRT1 transcript levels were already increased 5- to 8-fold, respectively, in the btsl double mutant compared to wild type (Fig. 4 C and D). After 3 d of Fe deficiency, transcription of FRO2 and IRT1 was strongly up-regulated by a similar magnitude in both mutant and in wild type. However, upon Fe resupply, FRO2 and IRT1 transcript levels remained high in the btsl double mutant (Fig. 4 C and D), matching the sustained FRO2 activity and IRT1 protein levels, respectively.
The transcription of FRO2 and IRT1 is directly regulated by FIT together with 1 of 4 partially redundant bHLH transcription factors, bHLH38, bHLH39, bHLH100, or bHLH101 (6, 7). To investigate the levels of FIT protein, specific antibodies were raised against recombinantly expressed protein. FIT has a predicted molecular weight of 35.5 kDa, but on SDS/PAGE it migrates at 55 kDa, as previously shown (12). In wild-type seedlings, FIT was increased under Fe deficiency and decreased after resupply, as expected (Fig. 4E and SI Appendix, Fig. S7). In the btsl double mutant –Fe, the levels of FIT protein were 2-fold higher than in wild type, and only slowly diminished after Fe resupply. Strikingly, there was no significant change in FIT transcript levels in btsl1 btsl2 compared to wild type in any of the tested Fe conditions (Fig. 4F). These findings indicate that FIT protein synthesis or turnover are misregulated in the btsl mutant. In contrast, the levels of FIT protein in the bts-1 mutant resemble those of wild type, and FRO2 and IRT1 expression are normal under changing Fe conditions (SI Appendix, Fig. S7). Thus, it is likely that BTSL and BTS differ in their downstream targets.
The E3 Ligase Domains of BTSL1 and BTSL2 Interact with FIT.
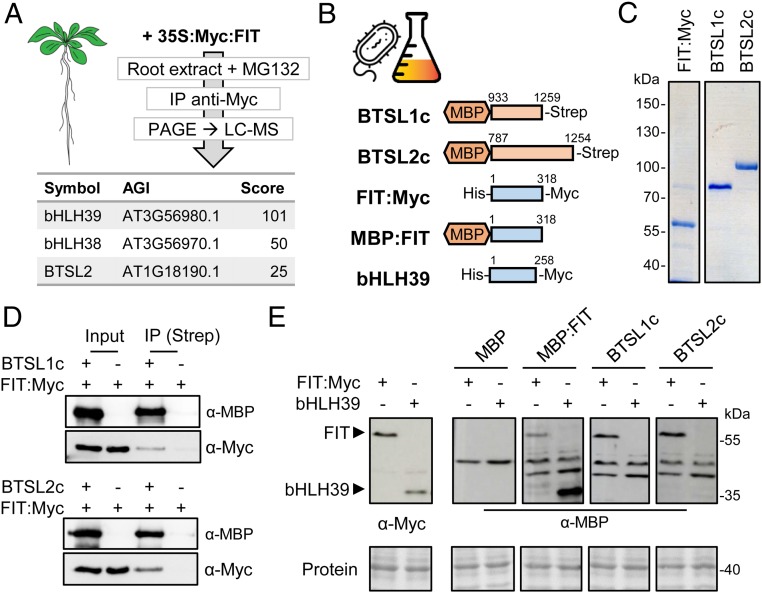
To investigate if FIT is a direct target of the BTSL E3 ubiquitin ligases, we tested for protein interaction. Because BTSL protein levels are very low even when expressed using the 35S promoter (SI Appendix, Fig. S8), we used FIT as a bait to pull down interacting proteins. Plants expressing 35S:Myc-FIT were grown and roots were harvested for protein extraction in the presence of the proteasomal inhibitor MG132. Proteins bound to Myc affinity resin were separated by SDS/PAGE gel, followed by mass spectrometry for identification. Among the proteins pulled down with Myc-FIT were bHLH38 and bHLH39 (Fig. 5A), which are known to form stable heterodimers with FIT for transcriptional activity (6). In addition, we detected peptides matching BTSL2 (P < 0.05) in pull-downs from Myc-FIT expressing plants, but not in control plants (Fig. 5A). Confirmation of the interaction by yeast 2-hybrid assays was hampered by auto-activation. As an alternative approach, protein interactions were validated in vitro using recombinant proteins expressed in Escherichia coli. The C-terminal half of BTSL1 (residues 933–1259) and BTSL2 (residues 787–1254), encompassing the predicted binding sites for E2 protein and putative target proteins, were fused to maltose-binding protein (MBP) at the N terminus to improve protein stability (Fig. 5B). A C-terminal Strep-tag was added to the BTSLc proteins for affinity purification. FIT was expressed with His and Myc-tags at the N- and C-termini, respectively (Fig. 5B). The FIT and BTSLc proteins were purified by affinity resins and the purity of the recombinant proteins was verified by SDS/PAGE and Coomassie staining (Fig. 5C). FIT and BTSLc proteins were mixed and BTSL1c or BTSL2c were recaptured using Strep-tactin resin. FIT was also pulled down, but not when BTSL was omitted (Fig. 5D).
Fig. 5.
The C-terminal domain of BTSL1 or BTSL2 interacts with FIT. (A) BTSL2 coimmunoprecipitated (IP) with FIT from extracts of Arabidopsis roots. Proteins were separated by SDS/PAGE and identified by LC-MS. (B) Diagram of the recombinant proteins produced in E. coli for in vitro analyses. (C) Purified FIT, BTSL1c, and BTSL2c proteins stained with Coomassie blue. (D) Interaction between the C-terminal E3 ligase domain of BTSL proteins and FIT shown by immunoprecipitation. (E) Far-Western blot analysis to compare the interaction of BTSL proteins with FIT and its partner bHLH39. Protein extracts of bacteria producing FIT:Myc or bHLH39:Myc were separated by SDS/PAGE, blotted, and incubated with the indicated proteins, followed by immunodetection of MBP. Ponceau staining shows equal protein loading.
It is possible that the BTSL proteins primarily interact with bHLH38 and bHLH39, which would result in coimmunoprecipitation of FIT. To test this, bHLH39 was expressed in E. coli, but because of poor protein stability, far-Western blot analysis was used to probe the interaction with BTSL E3 ligase domains. Immunodetection of the Myc-tag confirmed the expression of FIT and bHLH39 (Fig. 5 E, Left). Purified MBP, MBP:FIT or MBP:BTSLc were incubated with the membrane, followed by antibody detection of MBP. MBP:FIT bound strongly to bHLH39 and weakly to itself, as expected. The immuno-signals were clearly separated from an aspecific band in the MBP control. Both MBP:BTSL1c and MBP:BTSL2c bound to FIT, but not to bHLH39 (Fig. 5E). Thus, bHLH39 is unlikely to be a target of BTSL1 or BTSL2, in agreement with RT-qPCR analysis showing that misregulation of bHLH39 is at the transcriptional level (SI Appendix, Fig. S9).
The BTSL Proteins Display E3 Ligase Activity and Promote Degradation of FIT.
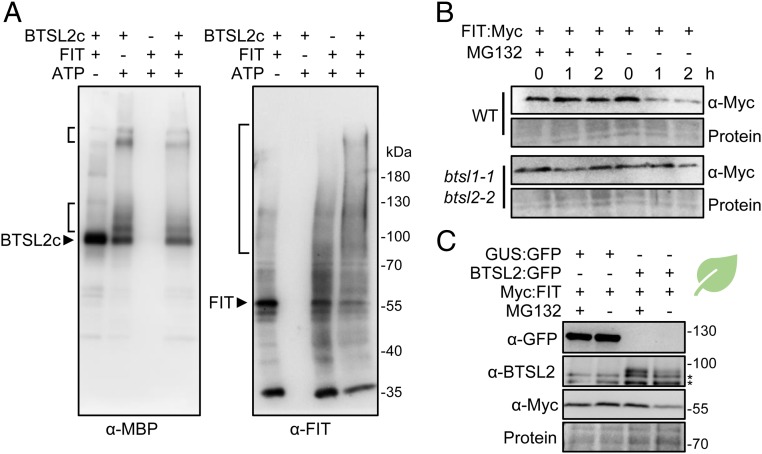
To test if the BTSL proteins are functional E3 ligases, purified MBP:BTSL1c or MBP:BTSL2c were incubated with E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme (UbcH5c), ubiquitin, and ATP. Immunoblot analysis using antibodies against MBP showed additional higher molecular weight forms of BTSL1c and BTSL2c when ATP was present (Fig. 6 A, Left and SI Appendix, Fig. S10). These are likely to be poly-ubiquitinated forms, showing that the E3 ligase domain has self-ubiquitination activity, which is commonly observed for these enzymes (31). Next, FIT was added to the reaction mixture as a potential substrate of BTSL2, which is more active than BTSL1 (SI Appendix, Fig. S10) and therefore used in subsequent experiments. Immunodetection of FIT revealed higher molecular weight forms typical of ubiquitinated protein products in the presence of BTSL2 but not in control reactions (Fig. 6 A, Right).
Fig. 6.
The C-terminal domain of BTSL2 catalyzes FIT ubiquitination and degradation. (A) In vitro ubiquitination activity of MBP:BTSL2c in the presence of [E1 + E2] and ATP. Proteins were separated by SDS/PAGE and higher molecular weight protein products were detected by Western blot analysis using anti-MBP antibodies for MBP:BTSL2c and purified anti-FIT antibodies for FIT. (B) Cell-free degradation of recombinant FIT in Arabidopsis root extracts. Protein extracts from 10-d-old wild-type (WT) and btsl double mutant seedlings were spiked with recombinant FIT:Myc protein and incubated in the presence of either MG132 (proteasome inhibitor) or DMSO (solvent) for the indicated length of time. (C) Steady state protein levels present in extracts from N. benthamiana leaves transiently expressing Myc:FIT and BTSL2c:GFP or GUS:GFP in the presence or absence of MG132. BTSL2c:GFP could be detected with anti-BTSL2c antibodies but not with commercial anti-GFP. Aspecific bands are marked (*). Ponceau staining shows equal loading.
To test if BTSL2 is able to promote the proteasomal degradation of FIT in vivo we performed cell-free degradation assays. First, protein extracts from wild-type Arabidopsis seedlings and the btsl double mutant were incubated with recombinant FIT protein. In the absence of the proteasomal inhibitor MG132, FIT protein levels were decreased within 1 h of incubation in wild-type extracts, but not in extracts from btsl double mutants (Fig. 6B). The cell-free degradation assay was also performed using Nicotiana benthamiana leaves transiently expressing Myc:FIT and BTSL2c:GFP and showed similar results (SI Appendix, Fig. S10). To further show support for the BTSL-dependent degradation of FIT, the proteins were expressed in N. benthamiana leaves followed by Western blot detection. In the absence of MG132, Myc:FIT protein levels were diminished when BTSL2c:GFP was coexpressed, but not when GUS:GFP was coexpressed (Fig. 6C). These results confirm that BTSL2 is implicated in FIT proteasomal degradation. Interestingly, BTSL2c:GFP protein was also stabilized by MG132, highlighting again its self-ubiquitination activity and tight regulation of its abundance, which is extremely low, even when expressed using strong constitutive promoters.
Collectively, our data show that the E3 ligase domain of either BTSL1 or BTSL2 can interact with FIT, and that BTSL2 is able to poly-ubiquitinate FIT in vitro and promote the proteasomal degradation of this transcription factor.
Discussion
Despite substantial differences in Fe homeostasis mechanisms, plants and animals employ hemerythrin E3 ubiquitin ligases for posttranslational regulation of Fe uptake. Functional comparison of the hemerythrin E3 ligases in Arabidopsis showed that the BTSL1 and BTSL2 genes have a distinct function from BTS, and are not redundant paralogs as previously proposed (26). BTSL genes have evolved specifically in dicotyledonous plants (Fig. 1B) and are coregulated with the Fe uptake machinery in the root (SI Appendix, Fig. S3). The different expression patterns of BTSL1/2 and BTS (Fig. 2 and SI Appendix, Fig. S2) suggest that the E3 ligases target different proteins for degradation. Indeed, we found that the partially redundant BTSL1 and BTSL2 target the bHLH transcription factor FIT for degradation (Figs. 4–6), whereas BTS has previously been found to interact with and induce the degradation of the bHLH proteins ILR3 and bHLH115 (17). In rice, mutant lines of HRZ1 and HRZ2 showed misregulation of Fe-regulated transcripts including OsIRO2 and OsIRO3 (18). More recently, it was shown that HRZ1 poly-ubiquitinates OsPRI1, the homolog of ILR3 and a transcriptional regulator of OsIRO2 (25). The target of HRZ2 has not yet been identified.
Arabidopsis BTSL1 and BTSL2 are closely related in sequence and, based on phylogeny, result from an ancient chromosome segment duplication (Fig. 1B). Although mutant analysis showed that the function of the 2 genes is overlapping, BTSL2 appears to be the dominant paralog. Comparing full knockout alleles of BTSL1 and BTSL2, the btsl2-2 mutant has a weak phenotype on its own, including residual chlorophyll in Fe-deficient seedlings (SI Appendix, Fig. S5). These phenotypic observations are in agreement with residual FRO2 activity in btsl2-2, but not btsl1-1, upon Fe resupply (SI Appendix, Fig. S6). Of the 2 BTSL genes, the expression pattern of BTSL2 is more similar to that of FIT, with both transcripts being produced in the differentiation zone with root hairs where nutrients are taken up (Fig. 2) (4, 32). Either BTSL1 is slowly degenerating and becoming obsolete, or it is taking on a different function, with different ubiquitination targets. For both BTSL1 and BTSL2, it will be interesting to explore whether they have other targets besides FIT. In particular, the misregulation of bHLH38 and bHLH39 transcripts in the btsl double mutant (SI Appendix, Fig. S9) may be explained by this possibility if 1 of the BTSLs were to target ILR3 for degradation (33).
Based on our results showing BTSL-dependent modification of FIT in vitro and previous studies on FIT protein dynamics (12), we propose that BTSLs are required for constant recycling of FIT, which helps to rapidly switch off its transcriptional activity when Fe is resupplied (Fig. 7A). A parallel can be drawn with the role of RNF in mouse, an E3 ligase that turns over the transcription factor STAT1 for instant down-regulation of the inflammation response (34). In Fe-deficient plants, inhibition of the 26S-proteasome by MG132 resulted in a lower transcriptional activity of FIT under Fe deficiency (12), presumably because promoters are populated with ubiquitinated, inactive FIT which cannot be removed. In contrast, there was no difference in transcriptional activity of FIT under Fe deficiency in the btsl double mutant compared to wild type (Fig. 4A), which could be explained by FIT remaining active when not ubiquitinated in the btsl mutant.
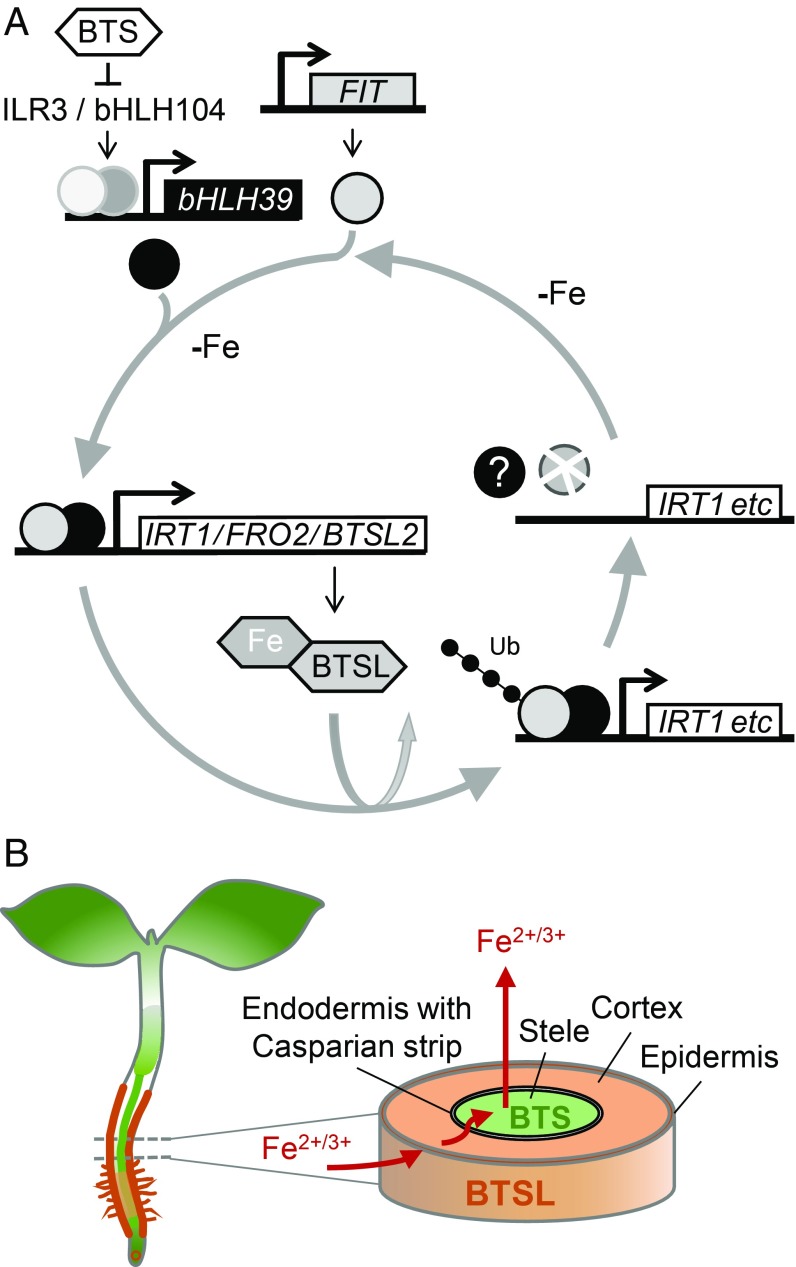
Fig. 7.
Proposed modes of action of BTSL and BTS in Fe homeostasis. (A) The transcription of FIT is up-regulated under Fe deficiency (top), resulting in increased levels of FIT protein, which forms a dimer with bHLH39 (or bHLH38). The FIT + bHLH39 dimer up-regulates the transcription of genes for Fe uptake, including IRT1 and FRO2. The transcript levels of BTSL2 are also controlled by FIT. The BTSL proteins act as E3 ubiquitin ligases and promote the degradation of FIT. Iron binding to the N-terminal hemerythrin domains of BTSL is likely to affect their stability and forms another layer of regulation. The constant turnover of FIT is proposed to facilitate rapid down-regulation of the Fe deficiency response when Fe becomes available. (B) BTSL1 and BTSL2 (orange) are expressed primarily in the epidermis and cortex cells, where they regulate the level of FIT protein. BTS (green) is predominantly expressed in the stele and shoot, and regulates the levels of ILR3. Thus, BTSL proteins provide a first protection mechanism against Fe overload outside the endodermis and Casparian strip, whereas BTS protects against Fe overload inside this barrier for nutrients.
Because BTSL2 is under transcriptional control by FIT (4, 32), a negative feedback loop operates continuously to regulate FIT protein levels (Fig. 7A). Another layer of regulation is likely to be provided by Fe binding to the N-terminal hemerythrin domain of BTSL proteins, similar to BTS/HRZ and the mammalian FBXL5 protein (35). Whereas FBXL5 protein is stabilized by Fe binding (19, 20), the Arabidopsis BTS protein appears to be destabilized by Fe (17). The effect of Fe on BTSL protein stability remains to be investigated. Furthermore, our results suggest that the heterodimeric partner protein bHLH39 is not targeted by BTSLs. Indeed, targeting FIT rather than bHLH38/39/100/101 would be more effective, as FIT is nonredundant, and many signaling pathways converge on this protein to regulate Fe homeostasis (36).
A closer look at the expression patterns of the BTSL1, BTSL2, and BTS genes suggests a demarcation by the endodermis: the 2 BTSL genes are predominantly expressed in the root epidermis and cortex, whereas BTS is expressed in the root stele and in the shoot. The endodermal Casparian strip, when made impermeable by suberin, is well known to affect metal and nutrient distribution, providing an effective barrier against nutrient overload in the leaves (37, 38). We propose a model where BTSL proteins act as a primary defense mechanism against excess Fe uptake in the root. A second defense mechanism against Fe overload is then regulated by the BTS protein in the stele and leaf tissues, behind the Casparian strip (Fig. 7B). This 2-step mechanism would explain why the double knockout btsl mutant has a mild phenotype, but a bts knockout is not viable. Although the btsl mutant takes up excess Fe in the root, BTS is able to regulate Fe redistribution in the rest of the plant. If BTSLs are functional, but BTS is not, the accumulation of toxic levels of Fe causes embryo lethality (17). The expression of BTSL2 and BTS may overlap in the stele of the uptake zone where the Casparian strip is not fully suberinized (Fig. 2). It will be interesting to study the cell-specific turnover of BTSL and BTS targets, by using constitutively expressed FIT-GFP and ILR3-GFP, respectively. Moreover, the model could help explain the observed local and systemic control of Fe homeostasis (39–41).
Materials and Methods
Plant Material, Cultivation, and Phenotyping.
A. thaliana wild-type and mutant lines, growth conditions, Fe treatments, and analysis of chlorophyll and Fe concentrations are described in detail in SI Appendix, SI Materials and Methods.
Promoter Activity and Gene Expression Analysis.
The promoter:GUS/GFP constructs and quantitative reverse transcriptase PCR are described in SI Appendix, SI Materials and Methods.
Protein Production and Protein Interaction Studies.
Details of the constructs for protein expression in E. coli, protein purification, and interaction studies by co-IP and far-Western blot analysis are provided in SI Appendix, SI Materials and Methods.
All other methods, such as in-vitro ubiquitination and western-blot analysis are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Huilan Wu for help with western blot analysis of FIT and IRT1; Benjamin Planterose for assistance with cloning of the BTSL:GFP constructs; Abigail Turner and Jed de Ruiter-Swain for cloning and protein expression; and the Bioimaging platform at the John Innes Centre for sectioning and confocal microscopy. This work was funded by a Marie Sklodowska Curie fellowship form the European Union Horizon 2020 programme, Ref. 655043 (J.R.-C.); the Biotechnology and Biological Sciences Research Council, grant awards BB/N001079/1, BB/P012523/1, and BB/J004561/1 (J.R.-C., J.M.C., I.K., R.T.G., M.F., and J.B.); the John Innes Centre/Chinese Academy of Sciences Center of Excellence for Plant and Microbial Sciences (J.R.-C., Y.C., H.-Q.L., and J.B.); grant award 2016ZX08009003-005 from the Ministry of Agriculture of China (H.-Q.L); and grants from the Academia Sinica (Y.-T.C. and K.-C.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Arabidopsis gene functions have been deposited in The Arabidopsis Information Resource (TAIR) database, https://www.arabidopsis.org/ (accession nos. AT1G74770 for BTSL1 and AT1G18910 for BTSL2).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907971116/-/DCSupplemental.
References
- 1.Römheld V., Marschner H., Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol. 80, 175–180 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai H.-H., Schmidt W., Mobilization of iron by plant-borne coumarins. Trends Plant Sci. 22, 538–548 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Bauer P., Ling H. Q., Guerinot M. L., FIT, the FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR in Arabidopsis. Plant Physiol. Biochem. 45, 260–261 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Colangelo E. P., Guerinot M. L., The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16, 3400–3412 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mai H. J., Pateyron S., Bauer P., Iron homeostasis in Arabidopsis thaliana: Transcriptomic analyses reveal novel FIT-regulated genes, iron deficiency marker genes and functional gene networks. BMC Plant Biol. 16, 211–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan Y., et al. , FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 18, 385–397 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Wang N., et al. , Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol. Plant 6, 503–513 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Connorton J. M., Balk J., Rodríguez-Celma J., Iron homeostasis in plants–A brief overview. Metallomics 9, 813–823 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barberon M., et al. , Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. U.S.A. 108, E450–E458 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin L. J., et al. , IRT1 degradation factor1, a ring E3 ubiquitin ligase, regulates the degradation of iron-regulated transporter1 in Arabidopsis. Plant Cell 25, 3039–3051 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubeaux G., Neveu J., Zelazny E., Vert G., Metal sensing by the IRT1 transporter-receptor orchestrates its own degradation and plant metal nutrition. Mol. Cell 69, 953–964.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Sivitz A., Grinvalds C., Barberon M., Curie C., Vert G., Proteasome-mediated turnover of the transcriptional activator FIT is required for plant iron-deficiency responses. Plant J. 66, 1044–1052 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Meiser J., Lingam S., Bauer P., Posttranslational regulation of the iron deficiency basic helix-loop-helix transcription factor FIT is affected by iron and nitric oxide. Plant Physiol. 157, 2154–2166 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinneny J. R., et al. , Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320, 942–945 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Buckhout T. J., Yang T. J. W., Schmidt W., Early iron-deficiency-induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genomics 10, 147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long T. A., et al. , The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22, 2219–2236 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selote D., Samira R., Matthiadis A., Gillikin J. W., Long T. A., Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol. 167, 273–286 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi T., et al. , Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat. Commun. 4, 2792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vashisht A. A., et al. , Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326, 718–721 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salahudeen A. A., et al. , An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326, 722–726 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Outten C. E., Checks and balances for the iron bank. J. Biol. Chem. 292, 15990–15991 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthiadis A., Long T. A., Further insight into BRUTUS domain composition and functionality. Plant Signal. Behav. 11, e1204508 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leng R. P., et al. , Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112, 779–791 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Sheng Y., et al. , Molecular basis of Pirh2-mediated p53 ubiquitylation. Nat. Struct. Mol. Biol. 15, 1334–1342 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H. M., Li Y., Yao X. N., Liang G., Yu D., POSITIVE REGULATOR OF IRON HOMEOSTASIS1, OsPRI1, facilitates iron homeostasis. Plant Physiol. 175, 543–554 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hindt M. N., et al. , BRUTUS and its paralogs, BTS LIKE1 and BTS LIKE2, encode important negative regulators of the iron deficiency response in Arabidopsis thaliana. Metallomics 9, 876–890 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H. G., Seo P. J., The Arabidopsis MIEL1 E3 ligase negatively regulates ABA signalling by promoting protein turnover of MYB96. Nat. Commun. 7, 12525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodríguez-Celma J., et al. , The transcriptional response of Arabidopsis leaves to Fe deficiency. Front. Plant Sci. 4, 276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karimi M., Inzé D., Depicker A., GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Schmidt W., Buckhout T. J., A hitchhiker’s guide to the Arabidopsis ferrome. Plant Physiol. Biochem. 49, 462–470 (2011). [DOI] [PubMed] [Google Scholar]
- 31.de Bie P., Ciechanover A., Ubiquitination of E3 ligases: Self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ. 18, 1393–1402 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakoby M., Wang H. Y., Reidt W., Weisshaar B., Bauer P., FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett. 577, 528–534 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Zhang J., et al. , The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 27, 787–805 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S., et al. , Nuclear RNF2 inhibits interferon function by promoting K33-linked STAT1 disassociation from DNA. Nat. Immunol. 19, 41–52 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Rodríguez-Celma J., Chou H., Kobayashi T., Long T. A., Balk J., Hemerythrin E3 ubiquitin ligases as negative regulators of iron homeostasis in plants. Front. Plant Sci. 10, 98 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi T., Understanding the complexity of iron sensing and signaling cascades in plants. Plant Cell Physiol. 60, 1440–1446 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Naseer S., et al. , Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc. Natl. Acad. Sci. U.S.A. 109, 10101–10106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barberon M., et al. , Adaptation of root function by nutrient-induced plasticity of endodermal differentiation. Cell 164, 447–459 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Grusak M. A., Pezeshgi S., Shoot-to-root signal transmission regulates root Fe(III) reductase activity in the dgl mutant of pea. Plant Physiol. 110, 329–334 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schikora A., Schmidt W., Iron stress-induced changes in root epidermal cell fate are regulated independently from physiological responses to low iron availability. Plant Physiol. 125, 1679–1687 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vert G. A., Briat J.-F., Curie C., Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiol. 132, 796–804 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.