Abstract
Paternal mitochondria are eliminated following fertilization by selective autophagy, but the mechanisms that restrict this process to sperm-derived organelles are not well understood. FUNDC1 (FUN14 domain containing1) is a mammalian mitophagy receptor expressed on the mitochondrial outer membrane that contributes to mitochondrial quality control following hypoxic stress. Like FUNDC1, the C. elegans ortholog FNDC-1 is widely expressed in somatic tissues and mediates hypoxic mitophagy. Here, we report that FNDC-1 is strongly expressed in sperm but not oocytes and contributes to paternal mitochondria elimination. Paternal mitochondrial DNA is normally undetectable in wildtype larva, but can be detected in the cross-progeny of fndc-1 mutant males. Moreover, loss of fndc-1 retards the rate of paternal mitochondria degradation, but not that of membranous organelles, a nematode specific membrane compartment whose fusion is required for sperm motility. This is the first example of a ubiquitin-independent mitophagy receptor playing a role in the selective degradation of sperm mitochondria.
Keywords: autophagy, mitochondria, mitophagy, maternal inheritance, C. elegans
INTRODUCTION
Maternal mitochondrial inheritance is widely-conserved among metazoans. However, our understanding of the molecular processes that specify paternal mitochondria elimination (PME) is incomplete. Mitophagy is a process in which mitochondria are engulfed by autophagosomes and delivered to lysosomes for destruction. PME was first reported to occur through mitophagy in the genetic model organism C. elegans [1–3], and mitophagy has recently been shown to underlie PME in mouse embryos [4]. Nevertheless, the signal that initiates selective degradation of paternal mitochondria following fertilization is unknown.
One form of mitophagy is triggered by the loss of mitochondrial membrane potential (ΔΨ) in a pathway that requires the E3 kinase PINK1 and E3 ubiquitin ligase Parkin [5]. In somatic cells, these proteins contribute to mitochondria quality control (MQC) through the selective recognition of damaged mitochondria. Indeed, a loss of membrane potential has been reported to occur in mammalian sperm mitochondria within 36 hours of fertilization, and PME engages PINK1, Parkin, and other MQC players to facilitate mitochondrial degradation [4]. However, PINK1 mostly functions as a sentinel that recognizes mitochondrial damage and is not causative for mitophagy in-and-of itself. Furthermore, the deletion mutants in the PINK1-Parkin pathway in C. elegans are not affected for the degradation of sperm mitochondria nor the nematode specific membranous organelles (MOs) [6].
The identification of new molecules that contribute to PME may help to broaden our understanding of underlying mechanisms and signaling pathways that target sperm mitochondria specifically for degradation. For example, phb-2 (prohibitin 2) was recently demonstrated to form a paternal mitophagy receptor at the mitochondrial inner membrane in C. elegans [7]. It is notable however that outer membrane rupture is necessary to unveil the PHB-2 receptor, making this an unlikely candidate for an initial selectivity filter.
Alternative modes of mitophagy involve mitochondrial outer membrane receptors that can recruit autophagosomes through direct binding to microtubule-associated protein light chain 3 (LC3), including BNIP3, NIX, and FUNDC1 [8]. This hypothesis is an attractive alternative to other autophagy pathways since paternal mitochondria in C. elegans do not seem to be strongly ubiquitinated [6,9]. These receptors are thought to operate independent of PINK1 and Parkin in somatic cells, but it is currently unknown whether any of these proteins play a role in PME, which is temporally constrained to a very short period and results in removal of the entire paternal mitochondrial pool.
Moreover, mining of bioinformatics databases suggested that FUNDC1 expression in testes responded to a genetic perturbation expected to impact MQC differently than nearly all somatic tissues, making it an attractive candidate to test (NCBI GEO GDS4791 / 1424215_PM_at “Mitochondrial protease Clpp knockout effect on various tissues”).
FUNDC1 (FUN14 domain containing1) was identified based upon its ability to facilitate clearance of mitochondria damaged by exposure to hypoxia and selectively associates with LC3B [10]. LC3 binding is regulated by reversible phosphorylation via ULK1 and PGAM5 [11,12]. FUNDC1 is localized to the mitochondria outer membrane at sites of contact with ER [10], where it interacts with both the IP3 receptor and components of the mitochondrial fission-fusion machinery [13]. In mice, FUNDC1 contributes to cardiac health and modulates ischemia-reperfusion injury [13,14].
We recently found that the C. elegans T06d8.7 / fndc-1 gene product, an ortholog to human FUNDC1, is broadly expressed across many somatic tissues where it localizes to the mitochondria and contributes to hypoxic mitophagy (manuscript in preparation). Our results here demonstrate that FNDC-1 is also expressed strongly in the male germ line, where it localizes in sperm mitochondria and acts as a paternal mitophagy receptor that contributes to the uniparental inheritance of mitochondria.
RESULTS
Expression of fndc-1 in C. elegans sperm mitochondria
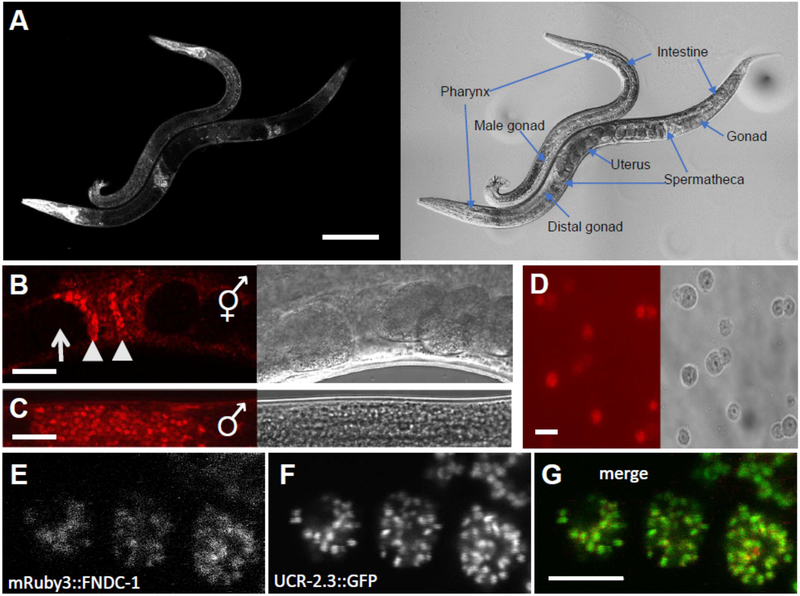
The predicted gene T06D8.7 / fndc-1 codes for a protein that shares 32% identity and 50% similarity to human FUNDC1 and is the second gene in a C. elegans operon that includes a non-ATPase subunit of the 26S proteasome’s 19S regulatory particle base subcomplex rpn-9, cytochrome c-type heme lyase cclh-1, and cytochrome c oxidase assembly factor cox-15. As might be expected based upon the other genes in this operon, a CRISPR-Cas9 fusion [15] between mRuby3 and fndc-1 is broadly expressed in many somatic tissues (Fig 1A), consistent with its expression in mice [16]. Robust levels of mRuby3 fluorescence were also observed in both the hermaphrodite spermatheca (Fig 1B) and the male gonad (Fig 1C). In dissected male gonads, the mRuby3::FNDC-1 fusion protein were found to label the spermatids (Fig 1D). Likewise, closer examination of the hermaphrodites revealed brightly labelled sperm surrounded by dimmer surrounding somatic tissue, and oocytes which lacked appreciable fndc-1 expression entirely. Finally, the mRuby3::FNDC-1 fusion protein appeared to co-localize in spermatids with a CRISPR-Cas9 mediated insertion of GFP coding sequencing to ucr-2.3, coding for the complex III ubiquinol-cytochrome c reductase core protein 2 of the mitochondrial electron transport chain (Fig 1E), and localized in mature spermatazoa to the cell body and not to the pseudopod (data not shown), consistent with mitochondrial targeting.
Figure 1 – FNDC-1 is expressed in spermatids and targeted to mitochondria.
mRuby3 red fluorescent protein sequence was inserted into the N-terminal genomic coding region of fndc-1 using CRISPR-Cas9 editing and HDR.
Images in panels A - D contain both fluorescence (left) and differential contrast interference (right).
A Intestine, body wall muscles, and pharynx of males and hermaphrodites, as labeled. Scale bar: 100 μm.
B Hermaphrodite spermatheca. Note the lack of expression in oocytes (arrows) compared to spermatids (arrowheads). Scale bar: 20 μm.
C Male gonad. Scale bar: 20 μm.
D Isolated spermatids from males expressing mRuby3::FNDC-1. Scale bar: 5 μm.
E Higher magnification image of mRuby3::FNDC-1 in spermatids.
F Genomic single copy CRISPR-Cas9 modified ucr-2.3::GFP in the same spermatids as panel E.
G Overlay of mRuby3::FNDC-1 (red) and ucr-2.3::GFP (green). Scale bar: 5 μm.
Paternal mtDNA are detectable in the cross-progeny of fndc-1(lf) males
The differential expression of fndc-1 in male and female germ cells motivated the question of whether FNDC-1 might also play a role in PME. uaDf5 is a mitochondrial haplotype represented by a large, stable deletion in the mitochondrial genome [17]. Mitochondrial haplotypes are not inherited through Mendelian genetics, but their abundance can be represented through heteroplasmy, which compares the relative amount of mutant to wildtype mitochondrial haplotypes. Within uaDf5 strains, uaDf5 generally contributes ~70% of the mitochondrial pool in a non-mutant nuclear genome background [17]. Most importantly here, uaDf5 can be distinguished from wildtype mtDNA via PCR [18]. When a uaDf5 male is mated with a fem-3(e2006) hermaphrodite that lacks sperm production under non-permissive conditions, it is possible to monitor the removal (or retention) of the male sperm mtDNA by tracking uaDf5 haplotype by PCR in their progeny.
In the background of a wildtype nuclear genome, residual uaDf5 mtDNA can be detected by PCR up to but not beyond the 64-cell stage of embryonic development [2]. However, previous studies have shown that uaDf5 mtDNA can be detected in the fully-developed cross-progeny of a uaDf5 male and an atg-7(bp422) autophagy mutant hermaphrodite [19]; since the worm atg8/LC3 homologs LGGs are necessary to remove paternal mtDNA, this cross results in a similar residual, but detectable level of uaDf5 in cross-progeny of uaDf5 males and mutant hermaphrodites [1,2,20].
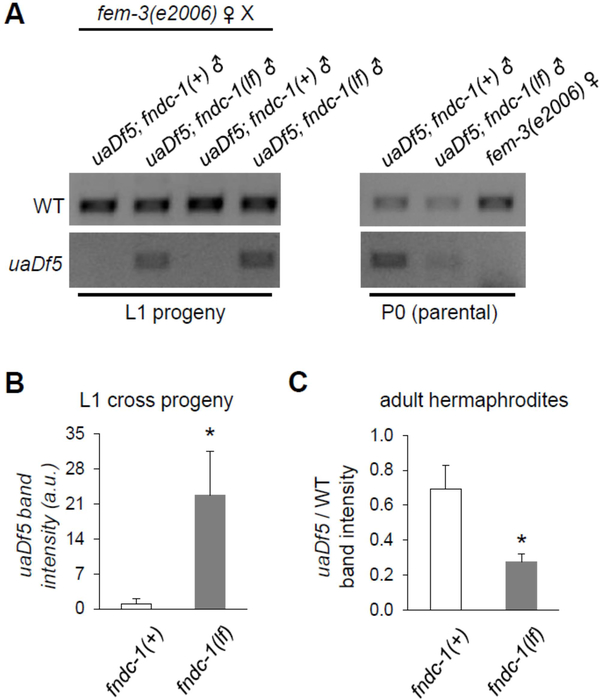
Here, either fndc-1(+); uaDf5 males or fndc-1(rny14); uaDf5 males were crossed to fem-3(e2006) hermaphrodites, and the abundance of the uaDf5 haplotype was assessed by PCR in recently-hatched L1 cross-progeny. In contrast to fndc-1(+) controls, where uaDf5 mtDNA was minimal, uaDf5 mtDNA could be amplified from the cross-progeny of fndc-1(lf) males (Fig 2A and B). This result was specific to the male germ line, as maternal fndc-1(lf) had no impact on PME following mating by a fndc-1(+); uaDf5 male (data not shown).
Figure 2 -. The paternal mitochondrial genome is detectable in cross progeny of fndc-1(lf) males.
uaDf5 is a mitochondrial haplotype with a large deletion in the mitochondrial genome that can be detected by PCR.
A The first four lanes contain ethidium bromide stained gel-separated PCR products from ~100 L1 cross-progeny of fem-3(e2006) females that had been mated to either fndc-1(+) or fndc-1(lf) males containing uaDf5 mtDNA. These data are two replicates of six indepenent experiments that were performed. The second set of three lanes are similar PCR reactions from the paternal and maternal worms that were used to create the cross-progeny. Note that amplification cycles (generally 25–35) have been optimized for each of the primer combinations, whether used as pairs or in triplicate, for L1 versus P0 generations, and for multiple versus single worms (see Methods). As such, band intensities are only directly comparable within the boundaries of each image.
B uaDf5 in L1 cross-progeny from fndc-1(+) or fndc-1(lf) males and fem-3(e2006) hermaphrodites. The band intensity for the uaDf5 PCR product was determined and normalized to the wildtype PCR product to control for total mtDNA content. The data represents the average values from six independent experiments (mean ± SEM; *p < 0.05; t-test).
C Relative uaDf5 abundance in fndc-1(+) and fndc-1(lf) adult hermaphrodites. The data (for which the PCR reactions are not shown) represents the average values from three independent experiments (mean ± SEM; *p < 0.05; t-test).
Heteroplasmy is also subject to genetic control, and heteroplasmic tolerance can differ in mutant backgrounds, particularly those that impact MQC. Hence, it was intriguing that the relative abundance of uaDf5 to wildtype mtDNA was decreased in the paternal fndc-1(lf) males (Fig 2A). A similarly reduced heteroplasmic level was observed in fndc-1(lf) hermaphrodites (Fig 2C). This is akin to the effect reported in other MQC mutants [21]. However, it is important to note that the decrease in uaDf5 load in the paternal fndc-1(lf) males is likely not responsible for the increased amount of uaDf5 observed in their progeny. In fact, a reduced amount of uaDf5 haplotype contributed at fertilization would be expected to reduce, not extend, the time-to-clearance.
Removal of paternal mitochondria but not membranous organelles is facilitated by fndc-1
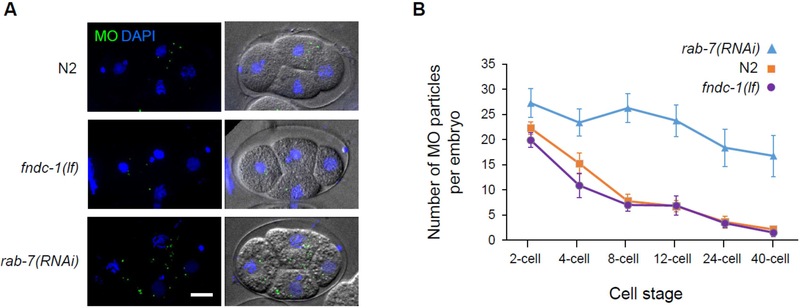
Sperm-derived MOs in C. elegans function to deliver cargo to the plasma membrane of mature spermatozoon [22] and are removed following fertilization through autophagy [1,2,20]. RAB-7 is a small GTPase that regulates membrane traffic into and between early-to-late endosomes and its loss has been shown to delay MO clearance in worms [23]. We also found that depletion of oocyte rab-7 by RNAi delayed MO clearance in the resulting embryos; however, the absence of FNDC-1 in the sperm did not (Fig 3). The absence of paternal FNDC-1 did result in a transient and reproducible delay in PME (Fig. 4). In agreement with previous results obtained for mtDNA, this result confirms that paternal fndc-1 was required for normal sperm mitochondria elimination. This delay being specific to sperm mitochondria degradation reinforces the conclusion that MO removal and early removal of paternal mitochondria occurs via distinct and separate ubiquitin-dependent process (Molina et al., in this issue).
Figure 3 – Membranous organelles are effectively cleared in the absence of paternal FNDC-1.
A No defect in MOs elimination is observed in the absence of FNDC-1 compared to wildtype (N2) embryos. A delay in MOs elimination can be observed in the rab-7(RNAi) embryos. Maximum-intensity Z-stacks projections of fixed embryos. Wildtype (N2), fndc-1(lf) and rab-7(RNAi) 4-cell stage embryos labeled for MOs (green) and stained for DNA (blue) (left panels) and DIC images (right panels) are shown. Scale bar: 10μm.
B Quantification of MO particles per cell stage in wildtype (N2) (purple), fndc-1(lf) (orange) and rab-7(RNAi) (blue) embryos (n=8 embryos per condition). Data shown in the graph corresponds to the mean ± SEM of MOs per embryo.
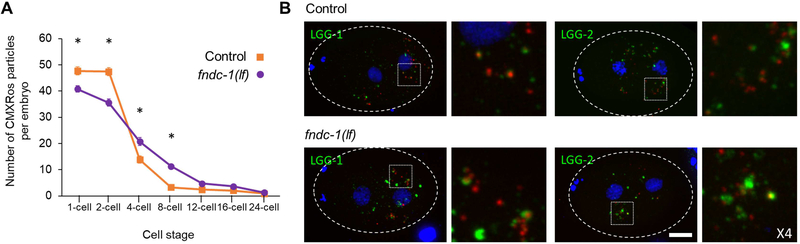
Figure 4 – Paternal FNDC-1 facilitates mitophagy at fertilization.
A Quantification of CMXRos stained sperm mitochondria per cell stage in embryos from control (purple) and fndc-1(lf) (purple) male matings (n = 5 separate experiments with n≥15 embryos analyzed for each stage). Data shown in the graph corresponds to the mean ± SEM of CMXRos particles per embryo. *p < 0.05; t-test (p-values: 1-cell, 8.8E-04; 2-cell, 1.6E-06; 4-cell, 1.9E-03; 8-cell, 1.2E-09).
B Recruitment of autophagosome markers, LGG-1 and LGG-2, around sperm-labeled mitochondria can be observed in the absence of FNDC-1. Maximum-intensity Z-stacks projections of cross-fertilized 2-cell stage embryos from wildtype unlabeled hermaphrodites and either fndc-1(lf) or control CMXRos labeled males. Embryos are labeled for LGG-1 (green; top panels) and LGG-2 (green; bottom panels); CMXRos was used to label sperm mitochondria (red) and DAPI to visualize DNA (blue). For clarity, 4-fold magnifications of the highlighted areas are shown in the right panels. However, the entire embryo was analyzed to generate the quantitative data presented in panel A. Scale bar: 10μm.
Autophagy is required for PME in C. elegans, with Atg8/LC3 ubiquitin-like proteins LGG-1 and LGG-2 being recruited around sperm mitochondria [1–3]. LGG-1 is required for autophagosome formation while LGG-2 participates in microtubule-dependent transport toward the pericentrosomal area prior to acidification [23]. Given that mammalian FUNDC1 binds directly to LC3, we reasoned that recruitment of these factors to paternal mitochondria may be impacted by fndc-1(lf). However, both endogenous LGG-1 and LGG-2 (expressed maternally) appeared to localize normally around sperm mitochondria in 2-cell embryos following fertilization by fndc-1(lf) mutant males (Fig 4B). We hypothesize that this result reflects functional redundancy in LGG recruitment, perhaps through other ubiquitin-independent mitophagy receptors.
DISCUSSION
The asymmetric expression of FNDC-1 on sperm mitochondria and the delayed PME in the fndc-1(lf) mutant suggest an attractive idea: that ubiquitin-independent mitophagy receptors may contribute to the selectivity filter that restricts mitochodrial degradation to the male germ line derived organelles. CPS-6, a mitochondrial endonuclease G, serves as the sole example of a paternal mitochondrial factor that is critical for PME [24]. Other molecules such as the core autophagy machinery that regulate PME appear to be contributed through the maternal germline [25] and are required for both MO clearance and mitophagy. In contrast, we have shown that FNDC-1 is contributed through the paternal germ line, is specific to PME, and has no role in the clearance of MO. Also, FNDC-1 is not required for recruitment of either LGG-1 or LGG-2 to sperm derived organelles. However, we have not ruled out a role for FNDC-1 in contributing to the stabilization of these proteins following their initial recruitment or that FNDC-1 may act at a later step in autophagosome formation. Of course, FNDC-1 could also act redundantly with other ubiquitin independent mitophagy receptors. To distinguish between these possibilities, it may be necessary to develop reagents that are better able to probe the discreet steps involved in PME downstream of atg-8/LC3 protein recruitment.
We note that there is an apparent discrepancy between the persistence of uaDf5 mtDNA in fndc-1(lf) mutant L1 larva (Fig 2) versus the “delay” observed in PME as assessed by CMXRos labeled sperm mitochondria (Fig 4). It is known that CMXRos staining disappears more quickly than genomic material, with published data showing that uaDf5 can be detected up to the 64-cell stage in WT worms, even though CMXRos is entirely gone well prior to that [2]. This could illustrate the sensitivity of PCR over conventional fluorescent imaging approaches, although it is also possible that mitochondrial fusion/fission events could enable mtDNA from the sperm to be transferred to a maternal mitochondria. It is important that this effect appears to be exacerbated in the fndc-1(lf) mutant.
In this study, we have shown that the FNDC-1 is strongly expressed in the male germ line and contributes to PME assurance. However, we have not interrogated the mechanisms coordinating FNDC-1 activity with fertilization. In somatic cells, FUNDC1 helps coordinate the signaling pathway mediating mitochondrial fission and fusion by interacting with MARCH5, OPA1, calnexin, and DRP1 at the mitochondrion-associated membrane [26], and mitochondrial dynamics are important for PME [19]. Whether the same signaling pathways and interactions that have been defined in mammals act in a similar fashion in worms and can be applied to fertilization remains to be determined.
It is notable however that calcium signaling at fertilization is well-established in both worms and mammals [27,28] and that calcium-induced reactive oxygen species (ROS) produced at fertilization have recently been shown to control the early embryonic cell cycle in Xenopus [29]. Hence, it would not be difficult to envision an elegant process involving hypoxic stimulation of ROS generation to link the somatic and germ cell functions of FNDC-1.
In summary, our results demonstrating paternal germline expression of FNDC-1 and its role in PME should motivate further examination of ubiquitin-independent mitophagy receptors in the early embryo.
Materials and Methods
C. elegans strains and maintenance
Strains were maintained under standard conditions at 20°C on the E. coli OP50 seeded NGM plates. The bacterial clone for rab-7 RNAi came from the J. Ahringer library and empty vector L4440 was used for control RNAi feeding experiments [27].
Strains used in this study were the parental N2 Bristol strain and the following:
APW202 ucr-2.3(jbm39 [ucr-2.3::link::GFP])III
CB3844 fem-3(e2006)IV
CU607 smIs23[pdk-2::gfp]II; him-5(e1490)V
KWN638 fndc-1(rny15 [mRuby3::fndc-1])II
KWN703 fndc-1(rny14)II
KWN706 fndc-1(rny14)II; him-5(e1490)V; uaDf5/+
KWN708; him-5(e1490)V; uaDf5/+
KWN722 fndc-1(rny15 [mRuby3::fndc-1])II; him-5(e1490)V
KWN774 ucr-2.3(jbm39); fndc-1(rny15 [mRuby3::fndc-1])II
KWN775 fndc-1(rny14); him-5(e1490)V
Generation of the fndc-1 transgenic worms
fndc-1 deletion and transgenic worm lines were generated via CRISPR Cas9-triggered homologous recombination following a previously described method [15]. crRNAs were obtained from Dharmacon, Inc, Lafayette, CO and 6xHis-Cas9 were purified by conventional nickel column chromatography. A crRNA containing a guide sequence with an adjacent protospacer motif (PAM) near a ClaI site in the first exon of fndc-1 (GUAGUAGAGGACUGUAUCGATGG) was injected with purified Cas9 enzyme and dpy-10 crRNA as a co-injection marker. The deletion mutant was identified by single worm genomic PCR of dpy-10 rol mutants using primers flanking the targeting site and screening of the PCR product by ClaI digest, as the ClaI site was disrupted by non-homologous end joining during the repair process. The rny15 allele was created through homology-directed repair using a crRNA guide sequence where the PAM was within the fndc-1 start codon (CUGUGAUUGUUCCAGCCATGG) and a PCR repair template containing the mRuby3 coding sequence with 35 nt arms homologous to the genome on either side of the cleavage site that disrupted the PAM. All CRISPR clones were fully sequenced within several hundred nucleotides of the modification site and outcrossed to the ancestral N2 strain prior to use.
Detection of uaDf5 mtDNA
To assess uaDf5 and wildtype mtDNA abundance in adult worms, lysis and PCR was performed as described [18]. Lysis was performed for 60 minutes at 65°C in 10 to 30μl of lysis buffer (50mM KCl, 10mM Tris, 20mM MgCl2, 0.45% NP40, 0.45% Tween-20, 0.01% gelatin) containing 1mg/ml proteinase K, followed by 15 minutes of inactivation at 95°C. PCR was performed using three primers in combination: a mutant-specific forward primer 5’-CCATCCGTGCTAGAAGACAA-3’ with the wildtype-specific forward primer 5’-TTGGTGTTACAGGGGCAACA-3’ (in the region spanning the deletion), and reverse primer 5’-CTTCTACAGTGCATTGACCTAGTC-3’ common to both mutant and wildtype mtDNA. PCR products were separated on a 2% agarose gel, stained with ethidium bromide, and photographed under UV illumination. Band intensities were measured using Image Lab software (BioRad). To quantify PME efficiency, relative abundances in cross-progeny, 30–40 young adult males heteroplasmic for uaDf5 were mated with 12 feminized fem-3(e2006) L4 stage worms at 20°C for 16 hr. After mating, gravid females were transferred to new NGM plate to obtain eggs for three hours. The females were removed from the plates, which were then incubated at 20°C overnight. The next day, 100 L1 progeny were harvested from each of the plates, lysed and analyzed as described above, with the following modification: seperate PCR reactions were used to detect the wildtype and uaDf5 mtDNA with the appropriate primer pairs. This was necessary due to the low abundcance of uaDf5 in the L1 cross-progeny. Two experimental replicates each consisting of three sets of independent matings were performed on different days.
MitoTracker CMXRos staining to monitor PME
To stain mitochondria in male sperm, L4 stage male worms were transferred to NGM plates containing Mitotracker Red CMXRos (1μg/ml, Molecular Probes) for 12 hr in the dark. Prior to microscopy, the worms were placed onto new unlabeled NGM plates for one hour. Sperm were isolated from male worms according to a previously reported protocol [30]. To monitor PME, labeled males were crossed with wildtype unlabeled young adult hermaphrodites overnight at 15°C. Hermaphrodites with labeled spermatheca, indicating cross-fertilization, were picked under a fluorescence binocular dissecting microscope.
Fixation of embryos
Hermaphrodites were dissected directly on poly-l-lysine–coated glass slides. Embryos’ eggshell was opened by freeze cracking and immediately fixed in pre-cooled methanol for 20 min at −20°C. Slides were rehydrated in phosphate-buffered saline (PBS) with 0.1% Tween 20 and blocked with 5% milk PBS-T. Embryos wereincubated with primary antibodies for 2 hr at room temperature and then washed two times for 30 min in PBS with 0.1% Tween 20. Secondary antibodies’ incubation was done for 1h at room temperature. The embryos were washed as before and mounted with VECTASHIELD® Antifade Mounting Medium with DAPI (Vector Laboratories).
Antibodies
An affinity-purified rabbit polyclonal antibodies against LGG-1 [3], a mouse monoclonal antibody against peptides VPSFKERRPFHERQ and NSMSMSNLYSQERD of LGG-2, and the SP56 monoclonal antibody (generous gift from S. Strome; [25]) were used to immunolocalize LGG-1 (1:200), LGG-2 (1:25) and MO (1:25) respectively in fixed embryos. Fluorescent secondary antibodies (1:500) were from Invitrogen and included anti-mouse Alexa-488, anti-rabbit Alexa 568 and anti-rabbit Alexa 488.
Fluorescent imaging
Paralyzed (with 0.1% tetramisole) worms or isolated sperm were mounted on 2% agarose pads on glass slides for microscopic examination using an Olympus FV1000 confocal microscope and FV10-ASW version 4 software (University of Rochester Light Microscopy Core). For the quantification of the number MOs and sperm mitochondria in fixed embryos, all embryos were imaged as multi-channel stacks of images every 0.4um (close to theoretical 0.29 optimal resolution) on a Zeiss Cell Observer spinning disk microscope and maximum intensity projection were generated using Image J and Zen 2012 (blue edition) in order to count the number of particles. Embryonic stages were established by counting the number of DAPI labelled nuclei.
Statistical analyses
Since most of our datasets satisfied the conditions of normality, equal variance, and independence, two-sample unpaired t-tests were used to assess significance between genotypes.
HIGHLIGHTS.
C. elegans selectively express the mitophagy receptor FNDC-1 on sperm mitochondria
Loss of fndc-1 results in delayed paternal mitochondria elimination
Loss of fndc-1 does not impact the removal of nematode-specific membranous organelles
ACKNOWLEDGMENTS
Strains were provided by the Caenorhabditis Genetic Center and by Dr. Andrew Wojtovich whom we thank in particular for the ucr-2.3::GFP strain. Technical assistance provided by Teresa Sherman was greatly appreciated. This work was supported by R01 HL127891 (KN, PSB, YI, PS), NIH R15 HD083882 (PM), the Fondation pour la Recherche Medicale (Equipe FRM DEQ20160334874) and the European COST Program (BM1408 GENiE) (VG and KR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Sato M, Sato K (2011) Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 334: 1141–1144. [DOI] [PubMed] [Google Scholar]
- 2.Zhou Q, Li H, Xue D (2011) Elimination of paternal mitochondria through the lysosomal degradation pathway in C. elegans. Cell Res 21: 1662–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al Rawi S, Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V (2011) Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 334: 1144–1147. [DOI] [PubMed] [Google Scholar]
- 4.Rojansky R, Cha M-Y, Chan DC (2016) Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. Elife 5:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitworth AJ, Pallanck LJ (2017) PINK1/Parkin mitophagy and neurodegeneration-what do we really know in vivo? Curr Opin Genet Dev 44: 47–53. [DOI] [PubMed] [Google Scholar]
- 6.Sato M, Sato K, Tomura K, Kosako H, Sato K (2018) The autophagy receptor ALLO-1 and the IKKE-1 kinase control clearance of paternal mitochondria in Caenorhabditis elegans. Nat Cell Biol 20: 81–91. [DOI] [PubMed] [Google Scholar]
- 7.Wei Y, Chiang W-C, Sumpter R, Mishra P, Levine B (2017) Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell 168: 224–238.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoo S-M, Jung Y-K (2018) A Molecular Approach to Mitophagy and Mitochondrial Dynamics. Mol Cells 41: 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hajjar C, Sampuda KM, Boyd L (2014) Dual roles for ubiquitination in the processing of sperm organelles after fertilization. BMC Dev Biol 14: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu W, Lin C, Wu K, Jiang L, Wang X, Li W, Zhuang H, Zhang X, Chen H, Li S, et al. (2016) FUNDC1 regulates mitochondrial dynamics at the ER-mitochondrial contact site under hypoxic conditions. EMBO J 35: 1368–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu H, Xue D, Chen G, Han Z, Huang L, Zhu C, Wang X, Jin H, Wang J, Zhu Y, et al. (2014) The BCL2L1 and PGAM5 axis defines hypoxia-induced receptor-mediated mitophagy. Autophagy 10: 1712–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu W, Tian W, Hu Z, Chen G, Huang L, Li W, Zhang X, Xue P, Zhou C, Liu L, et al. (2014) ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep 15: 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu S, Lu Q, Wang Q, Ding Y, Ma Z, Mao X, Huang K, Xie Z, Zou M-HH (2017) Binding of FUN14 Domain Containing 1 With Inositol 1,4,5-Trisphosphate Receptor in Mitochondria-Associated Endoplasmic Reticulum Membranes Maintains Mitochondrial Dynamics and Function in Hearts in Vivo. Circulation 136: 2248–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W, Ren H, Xu C, Zhu C, Wu H, Liu D, Wang J, Liu L, Li W, Ma QQ, et al. (2016) Hypoxic mitophagy regulates mitochondrial quality and platelet activation and determines severity of I/R heart injury. Elife 5: 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paix A, Folkmann A, Seydoux G (2017) Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods 121–122: 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, et al. (2012) Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol 14: 177–185. [DOI] [PubMed] [Google Scholar]
- 17.Tsang WY, Lemire BD (2002) Stable heteroplasmy but differential inheritance of a large mitochondrial DNA deletion in nematodes. Biochem Cell Biol 80: 645–654. [DOI] [PubMed] [Google Scholar]
- 18.Gitschlag BL, Kirby CS, Samuels DC, Gangula RD, Mallal SA, Patel MR (2016) Homeostatic Responses Regulate Selfish Mitochondrial Genome Dynamics in C. elegans. Cell Metab 24: 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Zhang Y, Chen L, Liang Q, Yin X-MM, Miao L, Kang B-HH, Xue D (2016) Kinetics and specificity of paternal mitochondrial elimination in Caenorhabditis elegans. Nat Commun 7: 12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al Rawi S, Louvet-Valle, Djeddi A, Sachese M, CUletto E, Hajar C, Boyd L, Legouis R, Galy V (2011) Postfertilization Autophagy of Sperm. Science (80- ) 334: 1144–1147. [DOI] [PubMed] [Google Scholar]
- 21.Lin Y-F, Schulz AM, Pellegrino MW, Lu Y, Shaham S, Haynes CM (2016) Maintenance and propagation of a deleterious mitochondrial genome by the mitochondrial unfolded protein response. Nature 533: 416–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shakes DC, Ward S (1989) Mutations that disrupt the morphogenesis and localization of a sperm-specific organelle in Caenorhabditis elegans. Dev Biol 134: 307–316. [DOI] [PubMed] [Google Scholar]
- 23.Djeddi A, Al Rawi S, Deuve JL, Perrois C, Liu Y-Y, Russeau M, Sachse M, Galy V (2015) Sperm-inherited organelle clearance in C. elegans relies on LC3-dependent autophagosome targeting to the pericentrosomal area. Development 142: 1705–1716. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Q, Li H, Li H, Nakagawa A, Lin JLJ, Lee E-S, Harry BL, Skeen-Gaar RR, Suehiro Y, William D, et al. (2016) Mitochondrial endonuclease G mediates breakdown of paternal mitochondria upon fertilization. Science 353: 394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward S (1986) The asymmetric localization of gene products during the development of Caenorhabditis elegans spermatozoa In, Gametogenesis and the Early Embryo, Gall J, ed. (New York: A.R. Liss; ), pp. 55–75. pp 55–75. [Google Scholar]
- 26.Wu X, Wu F-H, Wu Q, Zhang S, Chen S, Sima M (2017) Phylogenetic and Molecular Evolutionary Analysis of Mitophagy Receptors under Hypoxic Conditions. Front Physiol 8: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samuel ADT, Murthy VN, Hengartner MO (2001) Calcium dynamics during fertilization in C. elegans. BMC Dev Biol 1: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miao Y-L, Williams CJ (2012) Calcium signaling in mammalian egg activation and embryo development: the influence of subcellular localization. Mol Reprod Dev 79: 742–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han Y, Ishibashi S, Iglesias-Gonzalez J, Chen Y, Love NR, Amaya E (2018) Ca2+-Induced Mitochondrial ROS Regulate the Early Embryonic Cell Cycle. Cell Rep 22: 218–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singaravelu G, Chatterjee I, Marcello MR, Singson A (2011) Isolation and in vitro activation of Caenorhabditis elegans sperm. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]