Abstract
The balance between self-renewal and differentiation is crucial to ensure the homeostasis of the hematopoietic system, and is a hallmark of hematopoietic stem cells. However, the underlying molecular pathways, including the role of micro-RNA, are not completely understood. To assess the contribution of micro-RNA, we performed micro-RNA profiling of hematopoietic stem cells and their immediate downstream progeny multi-potent progenitors from wild-type control and Pbx1-conditional knockout mice, whose stem cells display a profound self-renewal defect. Unsupervised hierarchical cluster analysis separated stem cells from multi-potent progenitors, suggesting that micro-RNA might regulate the first transition step in the adult hematopoietic development. Notably, Pbx1-deficient and wild-type cells clustered separately, linking micro-RNAs to self-renewal impairment. Differential expression analysis of micro-RNA in the physiological stem cell-to-multi-potent progenitor transition and in Pbx1-deficient stem cells compared to control stem cells revealed miR-127-3p as the most differentially expressed. Furthermore, miR-127-3p was strongly stem cell-specific, being quickly down-regulated upon differentiation and not re-expressed further downstream in the bone marrow hematopoietic hierarchy. Inhibition of miR-127-3p function in Lineage-negative cells, achieved through a lentiviral-sponge vector, led to severe stem cell depletion, as assessed with serial transplantation assays. miR-127-3p-sponged stem cells displayed accelerated differentiation, which was uncoupled from proliferation, accounting for the observed stem cell reduction. miR-127-3p overexpression in Lineage-negative cells did not alter stem cell pool size, but gave rise to lymphopenia, likely due to lack of miR-127-3p physiological downregulation beyond the stem cell stage. Thus, tight regulation of miR-127-3p is crucial to preserve the self-renewing stem cell pool and homeostasis of the hematopoietic system.
Introduction
Hematopoietic stem cells (HSC) are characterized by their ability to give rise to all blood lineages for the entire lifespan of an individual. In order to preserve this capability throughout life, a reservoir of quiescent stem cells is maintained in the bone marrow (BM) microenvironment.1 This occurs through transcriptional and epigenetic mechanisms that actively repress cell cycling, differentiation, apoptosis, and senescence, or protect from oxidative stress. To generate the progenitors that are constantly required to replenish differentiated blood cells characterized by high turnover or to respond to peripheral injuries such as bleeding, a portion of HSC must awake from their dormant state and re-activate proliferation and differentiation programs.2 Thanks to their ability to correctly balance self-renewal and multi-potent differentiation the HSC pool size is maintained. However, the molecular pathways underlying this regulation are not completely understood, including the role of micro-RNA (miRNA). These are evolutionary conserved small non-coding RNA (ncRNA) that regulate mRNA stability and translation at the post-transcriptional level through non-perfect binding to target sequences.3
To study the potential role for miRNA in HSC self-renewal, we took advantage of the Pbx1 conditional knockout (Pbx1-cKO) mouse model. Pbx1 is a homeodomain transcription factor that positively regulates HSC quiescence.4 Its absence in post-natal HSC causes an excessive proliferation that ultimately leads to their exhaustion, indicating a profound self-renewal defect, and a premature myeloid differentiation at the expense of lymphoid differentiation.5 Therefore, the study of Pbx1-deficient HSC provides the opportunity to identify miRNA involved in the maintenance of HSC identity.
We employed Pbx1-cKO mice (and controls) to perform miRNA profiling of HSC and their immediate downstream progeny multi-potent progenitors (MPP) characterized by the absence of the Flk2 marker on their cell surface (Flk2-MPP, previously known as short-term HSC), representing one of the very first differentiation steps from HSC, with similar multi-potent differentiation capacity but reduced self-renewal. This approach allowed the identification of an HSC-specific miRNA, miR-127-3p. By modulating its activity through lentiviral vectors we demonstrate that miR-127 acts as a novel brake on differentiation that HSC employ to maintain their pool.
Methods
Mice
Tie2Cre+.Pbx1−/f and Mx1Cre+.Pbx1−/f mice have been described.4,6 Briefly, Tie2Cre+.Pbx1+/− and Mx1Cre+.Pbx1+/f mice were bred with Pbx1f/f mice to obtain Tie2Cre+.Pbx1−/f or MxCre+.Pbx1f/f Pbx1-cKO and their littermate controls.
miRNA profile and bioinformatic analysis
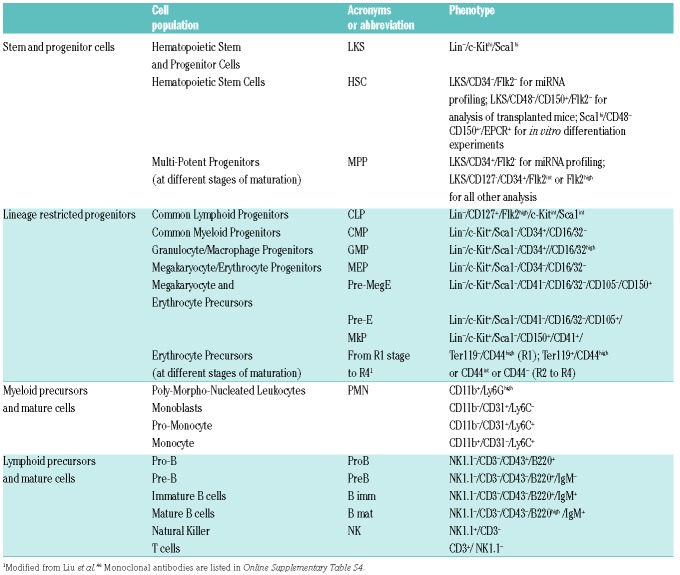
RNA was extracted with MirVana isolation kit (Ambion, ThermoFisher Scientific) from 1.4-2×103 HSC and 2-6×103 Flk2-MPPs (Table 1) sorted from the BM of Polyinosinic-polycytidylic acid [poly(I:C)] (InvivoGen)-treated Mx1Cre+Pbx1f/f and Mx1Cre-Pbx1f/f control mice (four experimental groups, 4-5 samples from individual mice/group). miRNA profiling was performed using the Megaplex TaqMan Assay system coupled with PreAmplification step (Rodent Pool A, Applied Biosystems, ThermoFisher Scientific). The expression level of 376 miRNA plus eight controls was obtained. Non-expressed miRNA (Ct level ≥35) were filtered out, whereas miRNA expressed in at least one of the four groups were further analyzed. Several normalization strategies were applied (quantile, median and endogenous normalization strategy) and quantile normalization was primarily chosen due to its smallest co-efficient of variation among replicates (cv=sdev/mean).7
Table 1.
Analyzed and/or sorted cell populations.
Differential expression (DE) analysis was performed by Statistical Analysis of Microarray8 (SAM) (FDR with q-value <0.1).
Lentiviral constructs
Design and production of lentiviral vectors for stable ectopic miRNA-overexpression (OE vector) or functional downregulation (DR vector) were as previously described.9,10 Briefly, both vectors exploit the spleen focus forming virus (SFFV) promoter and couple miR-127-3p up- or downregulation with the expression of a co-transcribed fluorescence reporter protein (mOrange fluorescent protein, OFP, for OE vector and destabilized Green Fluorescence Protein, dGFP, for DR vector respectively). For the OE vector, a 274bp fragment containing murine pre-mir-127 was PCR-amplified and cloned into the XhoI and MluI sites inside the EF1a intron of lentiviral transfer plasmid #1394 (SFFV.EFintron.OFP; described by Lechman et al.11). In the DR or ‘sponge’ vector, four tandem copies of an imperfectly complementary miR-127-3p target sequence were synthesized as described by Gentner et al.10 and cloned into the 3′ untranslated region (XbaI-XmaI sites) of the #1031scrT (LV.SFFV.dGFP) lentiviral backbone.11 Third-generation lentiviral vector particles pseudotyped with VSV-G were generated as described.12,13
Ethics
The study was approved by the Humanitas Clinical and Research Center - IRCCS Institutional Ethical Committee (prot. n. CE Humanitas, as per Ministerial Decree 127/14 of 8/2/2013).
Statistical analysis
Data are represented as mean ± Standard Error (SE) when n>3. The significance of differences was determined by two-tailed Mann-Whitney unpaired test unless otherwise stated. P<0.05 was considered statistically significant (ns: not significant; * P<0.05; ** P<0.005; ***P<0.0005; ****P<0.0001). Statistical analyses were performed with GraphPad Prism (GraphPad Software).
Additional methods are presented in the Online Supplementary Methods.
Results
Pbx1-cKO mouse as a model to identify candidate miRNA regulating hematopoietic stem cell self-renewal
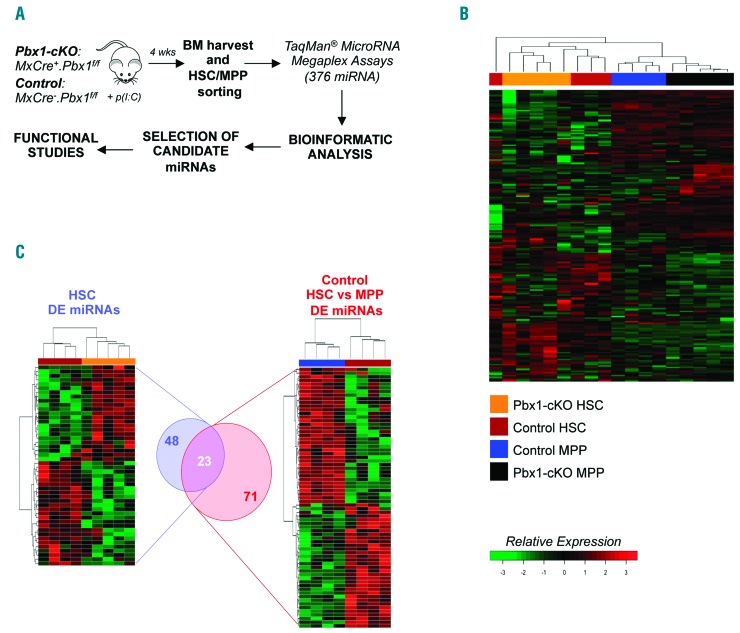
Hematopoietic stem cells and Flk2−MPP were prospectively isolated from the BM of five Pbx1-cKO (Mx1Cre+.Pbx1f/f) and four control (Mx1Cre−.Pbx1f/f) individual mice. The expression level of 376 different miRNA was obtained using a multiplexed Taq-Man-based real-time stem-loop PCR method14 and subjected to normalization, filtering and analysis (Figure 1A). A global analysis through unsupervised hierarchical clustering indicated a clear distinction between HSC and Flk2-MPP at the level of miRNA expression (Figure 1B), suggesting that miRNA regulate the first transition step in adult hematopoietic development. Within each group, Pbx1-deficient and control cells clustered separately, linking miRNA to self-renewal impairment. Differential expression analysis indicated that 71 miRNA are differentially expressed (DE) during the physiological HSC-to-MPP transition (Figure 1C, right, and Online Supplementary Table S1). A similar analysis on Pbx1-deficient HSC revealed 48 miRNA DE compared to control HSC (Figure 1C, left), half of which (n=23) are concordantly DE in the HSC-to-MPP list (Online Supplementary Table S2), in accordance with the hypothesis that miRNA are involved in HSC self-renewal. This result is similar to that obtained by analyzing mRNA, with Pbx1-deficient HSC exhibiting a transcriptional profile typical of their immediate downstream progeny.4 Importantly, the expression of most of the DE miRNA changed more than 2-fold. Of note, the most up-regulated miRNA in the normal HSC-to-MPP transition is miR-221 (Online Supplementary Figure S1). This represents a positive control of our analysis, since miR-221 has been predicted to be an important regulator of HSC maturation due to its ability to repress cKit.15 We compared the lists of DE miRNA with those of DE transcripts obtained by microarray.4 None of the DE miRNA between mutant HSC or Flk2−MPP and corresponding controls are located within a transcript detected in the microarray, indicating that their different level of expression is not the result of the level of expression of a host gene.
Figure 1.
Strategy for the selection of candidate miRNA regulating hematopoietic stem cell (HSC) self-renewal. (A) Experimental workflow. (B) Heat map shows the unsupervised hierarchical clustering of relative miRNA expression in HSC and multi-potent progenitors (MPP) characterized by the absence of the Flk2 marker on their cell surface (Flk2−MPP). The color-scale represents Z-score transformed signal intensity. (C) Hierarchical cluster dendrogram is shown for relative expression of differential expression (DE) miRNA in mutant HSC compared to wild-type (wt) HSC (left) and in the normal HSC-to-MPP transition (right). Venn diagram shows the overlap of deregulated miRNA in the two analyses. The significance of overlap was computed by hypergeometric test (P-value 5×10−5). BM: bone marrow.
Among the 23 over-lapping miRNA (Figure 1C), we applied stringent criteria to select a few candidates for subsequent studies. We considered only miRNA that: 1) were DE also by applying a different normalization method; 2) had a fold difference higher than five; 3) had anti-correlated predicted targets (PT) among previously described DE mRNA in Pbx1-deficient HSC;4 4) were DE also with independent real-time PCR in wild-type (wt) samples. Only three miRNA candidates fulfilled each condition, all evolutionary conserved and all down-regulated (miR-127-3p, miR-411-5p, miR-34b-3p). Among these, the most DE both in Pbx1-deficient HSC and in the normal HSC-to-MPP transition is miR-127-3p (Online Supplementary Table S3). This miRNA was therefore chosen as a candidate HSC− self-renewal regulator.
miR-127-3p is an HSC-specific miRNA
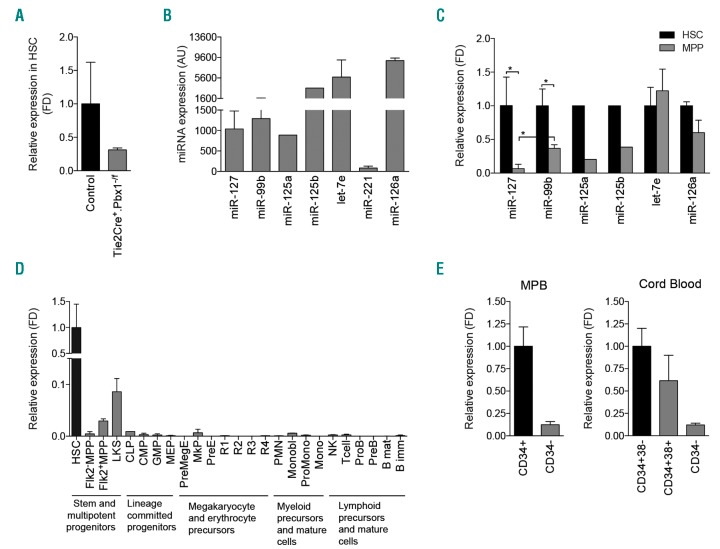
As further proof that miR-127-3p downregulation correlates with loss of self-renewal, we confirmed its DE in phenotypically-defined HSC from a different previously described Pbx1-cKO mouse model (Tie2Cre+Pbx1−/f)4,5 (Figure 2A).
Figure 2.
Expression of miR-127-3p. (A) Histogram shows fold difference (FD) of miR-127-3p expression in hematopoietic stem cell (HSC) from 3-4-week old Tie2Cre+Pbx1−/f mice relative to HSC from Tie2Cre-Pbx1+/f littermate controls, as measured by quantitative real-time polymerase chain reaction (qRT-PCR). Due to the extreme paucity of stem cells from the Tie2Cre+Pbx1−/f model,4 HSC from nine mice were grouped in two pools to perform qRT-PCR; bars indicate the range. (B) Expression of the indicated miRNA in HSC from 4-12-week old wild-type (wt) mice as measured by qRT-PCR and expressed in arbitrary units (AU). (C) Expression of the indicated miRNA in multi-potent progenitors (MPP) from wt mice relative to their expression level in HSC. (B and C) miR127-3p and let-7e, n=4; miR-99b, n=8; miR-125a and miR-125b, n=1; miR-221, n=6; miR-126a, n=2; n: number of pools (2-5 mice/pool). For miR-126, bars indicate the range. (D) miR-127-3p expression level in steady state bone marrow cell subpopulations from two pools of wt mice (2-3 mice/pool). (E) qRT-PCR analysis of miR-127 expression in human hematopoietic mature CD34− cells (or in CD34+CD38+ progenitors) relative to the CD34+ mobilized peripheral blood (MPB) or CD34+CD38− cord blood (CB) compartments. N=2 healthy donors for each source, bars indicate the range. All donors signed informed consent. When miRNA expression is indicated as FD, black bars indicate the sample relative to which FD is calculated.
We then prospectively isolated HSC and Flk2−MPP from wt mice and compared the expression level of miR-127-3p to that of other miRNA previously associated to HSC, such as miR-99b, miR-125, let-7, miR-221 and miR-126.11,15–19 We found that miR-127-3p expression in HSC is not particularly high, being similar to that of other miRNA already reported to play a role in HSC biology (Figure 2B). We confirmed the recently reported downregulation of miR-99b in Flk2−MPP20 (Figure 2C). However, miR-127-3p showed the highest downregulation (>100-fold; miR-99b 3-fold) in the first step of hematopoietic differentiation, whereas the majority of the other miRNA are still expressed at the progenitor level and have only a modest reduction compared to HSC. We then used real-time PCR to investigate whether miR-127-3p is expressed in other hematopoietic cell subpopulations other than HSC within the BM in steady-state conditions. This issue was addressed by analyzing miR-127-3p expression from purified populations including different lineage committed progenitors, megakaryocyte and erythrocyte precursors, myeloid precursors and mature cells, and various lymphoid subsets (Table 1 and Figure 2D), since expression in only one subgroup would not be detectable in pooled populations. Impressively, miR-127-3p was highly HSC-specific, being quickly down-regulated in the earliest step of hematopoietic differentiation, where loss of self-renewal occurs. Furthermore, we confirmed its expression in human HSC-enriched CD34+ cells from different sources (Figure 2E).
Inhibition of miR-127-3p function severely impairs HSC self-renewal
To investigate the role of miR-127-3p in HSC we transplanted lineage-negative (Lin-) cells from CD45.2+ wt mice into lethally irradiated syngeneic CD45.1+ recipients, after permanently inhibiting binding to its targets through a lentiviral sponge vector carrying the reporter protein dGFP, as previously described for other miRNA.11 Since miR-127-3p targets in HSC are not known, we assessed effective downregulation of miR-127-3p activity on K562 cells. Upregulation of XBP-1 and BLIMP-1, two miR-127-3p targets,21 indicated that the sponge vector worked as expected (Online Supplementary Figure S2).
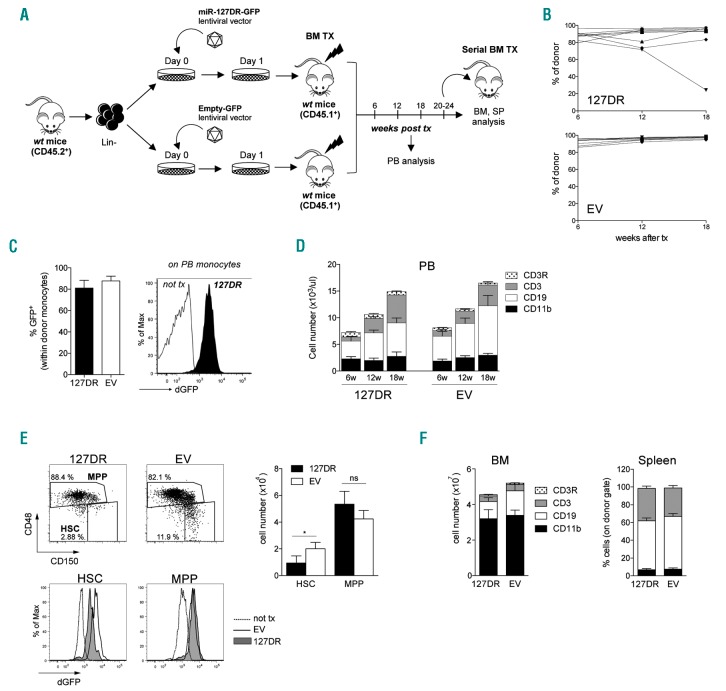
Engraftment and multipotent reconstitution by Lintransduced cells was then monitored through periodic blood sampling of transplanted mice and at necropsy several weeks after transplant. The BM of some of the reconstituted mice was transplanted into secondary recipients to assess if, in the absence of miR-127-3p activity, HSC maintain their self-renewal ability (Figure 3A).
Figure 3.
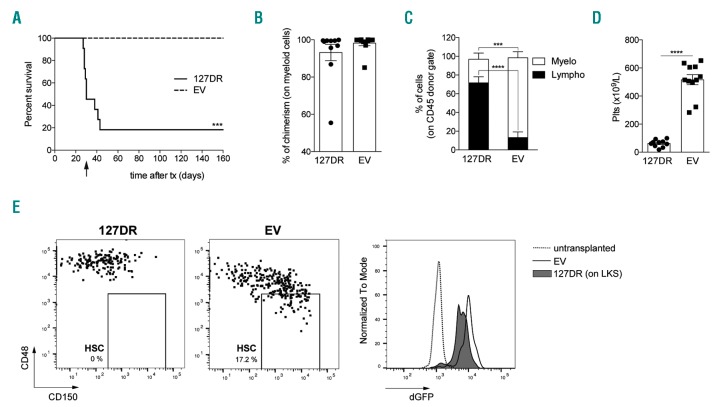
Inhibition of miR-127-3p function leads to hematopoietic stem cell (HSC) depletion. (A) Experimental workflow. (B) Level of engraftment of transduced cells over time, measured as percentage of cells expressing the donor marker CD45.2 within total peripheral blood (PB) CD45+ cells. Graphs show engraftment in individual mice transplanted with Lin− cells transduced with 127DR (top, n=11) or empty vector (EV) (bottom, n=9). Tx: transplant. (C) dGFP expression in PB donor monocytes measured by FACS analysis 18 weeks after transplant. (Left) Average of eight mice; (right) representative FACS histogram. (D) Kinetics of multi-lineage differentiation of transduced cells during PB sampling (6-12-18 weeks). Histograms show the absolute cell number, determined by hemocytometer analysis, and stacked columns represent the relative abundance of each population (CD11b+ myeloid cells, CD19+ B cells, CD3+ T cells; CD3R: T cells from recipient), determined by FACS analysis (n=6-8). (E) FACS analysis of 127DR and EV stem and progenitor cells in the bone marrow (BM) of transplanted mice. (Left) Representative FACS analysis of HSC and multi-potent progenitors (MPP) gated on LKS (dot plots) and of their dGFP expression (bottom histograms) compared to that of a representative non-transplanted mouse. (Right) Average of HSC and MPP absolute numbers (n=6-7). (F) Central and peripheral multi-lineage differentiation by transplanted 127DR-and EV-transduced cells in primary recipients. Histograms show absolute BM cell numbers (left) and percentage of donor cells in the spleen (right). Stacked columns represent relative abundance of each lineage (n=6-8). All graphs summarize results from two independent experiments, and BM cells counts are relative to one femur and one tibia.
In primary recipients, survival curves were similar for all transplanted mice (Online Supplementary Figure S2B). Similarly, the level of engraftment by sponge-transduced HSC (named 127DR for miR-127-3p functional downregulation), measured as the proportion of cells expressing the donor marker CD45.2 over time, was comparable to that of HSC transduced with a control vector carrying only dGFP (named EV for empty vector) (Figure 3B) or with untransduced cells (data not shown), with the exception of one mouse out of eleven. Sponge and control vectors express dGFP under the SFFV promoter, which is highly active in HSC, progenitors and myeloid cells, particularly monocytes. Due to its rapid turnover, dGFP is only detectable when expressed at very high levels. Therefore, even though the SFFV promoter has good and modest activity in B and T lymphocytes, respectively, the dGFP marker is hardly detectable in these lineages.22 For this reason, we followed by FACS analysis dGFP expression in monocytes; this also served as an indirect measure of dGFP expression in HSC since, due to their high turnover, myeloid cells are continuously generated from HSC. Virtually all monocytes were GFP+, indicating a very efficient transduction of long-term reconstituting cells (Figure 3C). Importantly, GFP-negative or low B and T lymphocytes isolated from the spleen of transplanted mice contained vector sequences (Online Supplementary Figure S2C), indicating that they derived from transduced cells. We therefore safely monitored the presence of transduced cells in vivo through the donor marker CD45.2, since we could not rely on dGFP expression in all cells.
The kinetics of peripheral myeloid, B- and T-cell reconstitution from donor cells was similar in mice transplanted with 127DR- or EV-transduced HSC, as well as the proportion and number of the different cell types (Figure 3D), suggesting that suppression of miR-127-3p activity did not affect multi-lineage reconstitution. However, FACS analysis of the BM at necropsy revealed a significant depletion of donor-derived phenotypically defined HSC, which were GFP+ (Figure 3E), without affecting their output of multi-potent progenitors and their lymphoid and myeloid differentiation potential, as confirmed on BM and spleen (Figure 3F). None of the transplanted animals displayed extramedullary hematopoiesis (data not shown), indicating that the observed HSC reduction was not due to abnormal egress from the BM.
To evaluate whether there was a decrease in functional HSC independently of the immunophenotype used to define them, secondary transplants were performed by injecting a high dose of total BM cells from two individual primary recipients in lethally irradiated CD45.1+ secondary recipient mice. The number of transplanted donor BM cells was sufficient for allowing recovery after irradiation; however, most of the mice transplanted with cells derived from 127DR BM succumbed 4-6 weeks after transplant, soon after the challenge of the first blood withdrawal, suggesting a severe HSC defect (Figure 4A). Four weeks after transplant, donor myeloid chimerism was very high in all mice, without significant differences among the groups of mice transplanted with 127DR or EV transduced BM (Figure 4B), indicating that the early mortality of 127DR BM-transplanted mice was likely not due to homing or engraftment impairment. However, sponging miR127-3p severely compromised myeloid cell and platelet production in secondary recipients (Figure 4C and D), in accordance with a stem cell defect.
Figure 4.
Inhibition of miR-127-3p function severely impairs hematopoietic stem cell (HSC) self-renewal. (A) Kaplan-Mayer survival curves of secondary recipients (n=10-11 from two independent experiments; Log-rank test) of 127DR- and empty vector (EV)-transduced cells. The arrow below the horizontal axis indicates the time of first peripheral blood (PB) withdrawal (4 weeks after transplant). (B-D) PB analysis four weeks after transplant (n=10-11 from two independent experiments). (B) FACS analysis for myeloid chimerism, measured as percentage of donor CD45.2+ cells within CD11b+ cells. (C) Percentage of myeloid and lymphoid cells within CD45+ donor gate determined by FACS analysis. (D) Platelet counts determined by hemocytometer analysis. (E) FACS analysis of 127DR stem and progenitor cells in one of the two secondary recipients that survived throughout the experiment. Dot plots are gated on LKS population and a representative sample is shown for the EV group. Histogram overlay (right) shows dGFP expression within the HSC (non-transplanted and EV) or the LKS (127DR) gate.
Two out of eleven mice transplanted with 127DR cells survived throughout the experiment, with very different levels of donor chimerism (Online Supplementary Figure S3A). Several weeks after transplant, we analyzed the most primitive hematopoietic compartment within the BM of the two secondary sponge recipients that were still alive. This analysis revealed that HSC were either absent (Figure 4E) or belonged to the recipient (Online Supplementary Figure S3B). LKS multipotent progenitors were donor-derived GFP+, indicating that HSC had originally engrafted at the time of the secondary transplant and were able to generate differentiated progeny throughout time; however, they failed to self-renew. Therefore, miR-127-3p activity is crucial for the maintenance of phenotypically and functionally defined HSC.
miR-127-3p prevents premature differentiation
To discern the biological mechanism at the basis of HSC loss, we tested if miR-127-3p is involved in regulating HSC quiescence, metabolic properties, survival, or prevention of premature differentiation, all necessary to preserve self-renewal. All these features were initially analyzed on Lin− cells isolated ex vivo from transplanted mice, as opposed to newly transduced cells, in order to avoid potential influences of cell culture.
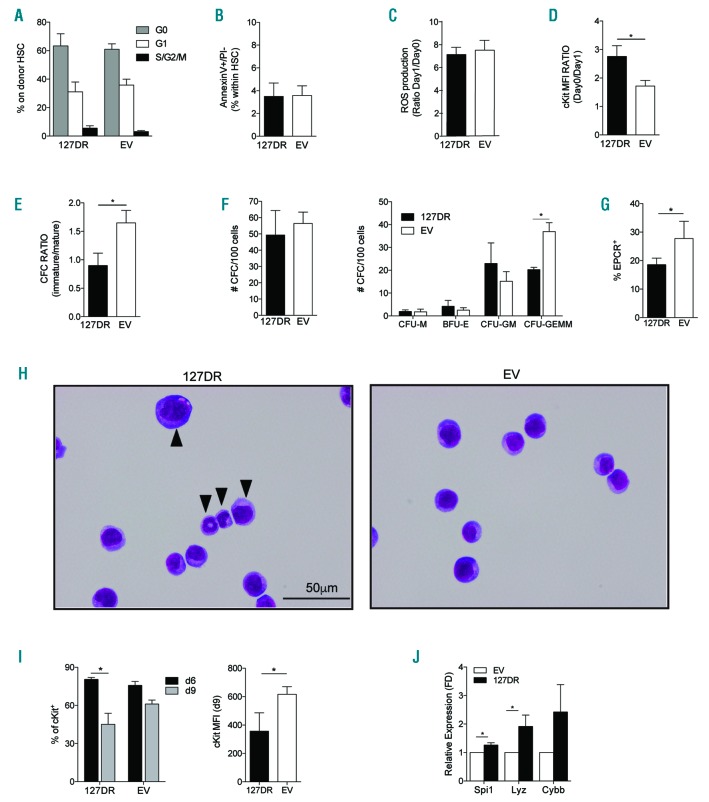
The proportion of cycling and quiescent HSC was similar in 127DR- and EV-transduced BM (Figure 5A), as well as the proportion of apoptotic cells (Figure 5B), indicating that miR127-3p downregulation does not alter HSC cell cycle or apoptosis.
Figure 5.
miR-127-3p prevents premature differentiation. (A) Cell cycle status of 127DR- and empty vector (EV)-transduced hematopoietic stem cell (HSC) was determined by FACS analysis with Ki67/Sytox staining on the bone marrow (BM) of primary recipients (n= 4 and 5, respectively). (B-D) FACS analysis of 127DR- and EV-transduced Lin− cells isolated ex vivo from the BM of primary recipients (n= 5 and 6). (B) Analysis of early apoptosis (AnnexinV+PI−) in 127DR- and EV-transduced HSC identified by FACS within the Lin− cell population. (C) Reactive oxygen species (ROS) production in 127DR- and EV-transduced HSC in response to short (24 hours) cytokine stimulation in culture with StemSpan medium supplemented with SCF, TPO, Flt-3 Ligand and IL-3, measured by FACS after incubation with cellROX reagent. Vertical axis shows the ratio of the mean fluorescence intensity (MFI) at day (d) 1 versus mean fluorescence intensity (MFI) immediately after Lin− isolation (d0). (D) Early differentiation of ex vivo isolated 127DR and EV-transduced Lin− cells measured as reduction of cKit expression after one day of culture as in (C) (MFI ratio, d0/d1). (E) Colony forming cell (CFC) assay performed on ex vivo isolated 127DR- and EV-transduced Lin− cells (n=6). Colonies were scored as immature (CFU-GM and CFU-GEMM) and mature (CFU-M and CFU-G) at d9. Vertical axis shows the ratio between immature and mature colonies. (F) CFC assay performed on wild-type (wt) HSC two days after lentiviral transduction (d3 of culture, n=4 individual mice). (Left) Total number of colonies; (right) number of each different type of colonies. (G) FACS analysis for endothelial protein C receptor (EPCR) expression on GFP+cKit+CD48−CD150+ cells (n=4). (H) Representative cytospin preparations stained with May-Grunwald and Giemsa of 127DR-and EV-transduced HSC at d6 of culture. (I) FACS analysis for cKit expression at d6 and d9 (n=4). (J) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of the expression of the indicated genes in transduced HSC at d9 of culture relative to d9 non-transduced cells (n=3).
Since oxidative stress has been described to negatively affect HSC function, we measured reactive oxygen species (ROS) production in response to cytokine stimulation, as described.23 ROS production induced upon short in vitro cytokine exposure was similar in HSC within Lin−cells isolated from 127DR or EV recipients (Figure 5C), suggesting that miR-127-3p downregulation does not lead to an increase in oxidative stress. However, after only one day of culture, a reduced expression level (measured by MFI) of the stem-cell associated cKit marker was observed in 127DR cells (Figure 5D), suggesting that miR-127-3p downregulation leads to accelerated differentiation. Lin− cells isolated from 127DR or EV recipients were also subjected to a standard colony assay (Figure 5E). 127DR stem and progenitor cells generated a significantly lower proportion of immature colonies compared to mature ones, in accordance with the hypothesis that they are more prone to differentiate. We confirmed this phenotype using freshly isolated HSC cultured in vitro. HSC were sorted from wt mice using the EPCR marker,24,25 transduced and analyzed at different time points. Single-cell colony assay performed early after transduction revealed that, despite the fact that the total number of colonies was comparable in the 127DR and EV groups, sponged cells generated fewer CFU-GEMM colonies, which originate from the most immature cells (Figure 5F). FACS analysis did not show any differences in the two groups when the conventional CD48, CD150 staining was performed (Online Supplementary Figure S4A); however, at day 6, the proportion of HSC expressing EPCR was reduced (Figure 5G and Online Supplementary Figure S4B). Moreover, a higher proportion of cells displayed early signs of differentiation in cytospin preparation (Figure 5H). The proportion of cKit+ within dGFP+ cells was very high in both experimental groups at day 6. However, at day 9, it dropped significantly in cultures derived from sponged HSC, while this decrease was less dramatic in the EV group. Accordingly, cKit expression level was significantly lower at day 9 in the 127DR group (Figure 5I). Finally, the expression of genes with myeloid expression pattern (Gene Expression Commons26) was higher in sponged cells from day 9 cultures compared to cells transduced with EV (Figure 5J).
Overall, these data suggest that a faster differentiation to the next maturation step is the underlying cause of the observed HSC loss in vivo when miR-127-3p is not active.
Forced miR-127-3p expression does not affect hematopoietic stem cell maintenance
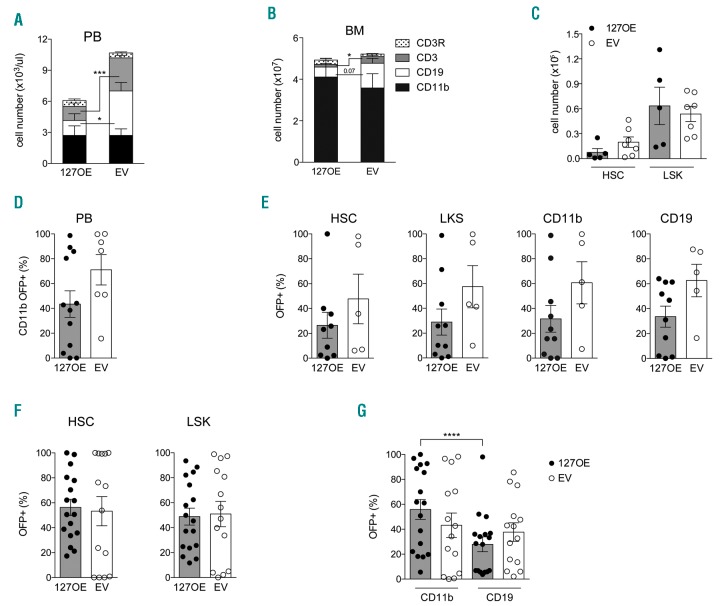
We next asked whether increased miR-127-3p levels affect HSC function in vivo. Lentiviral-mediated miR-127 overexpression was performed in wt CD45.2+ Lin− cells, followed by transplantation in CD45.1+ recipient mice (Online Supplementary Figure S5A). miR-127-overexpression (127OE) vector carried the Orange Fluorescent Protein (OFP) as reporter gene and the corresponding empty vector (EV), carrying only the reporter protein, was used as control. We obtained very high transduction efficiency with both vectors (data not shown). Expression analysis through real-time PCR before and after transduction confirmed that the progeny of infected cells expressed miR-127-3p, reaching levels similar to those of miR-16, which is present in all hematopoietic cells (Online Supplementary Figure S5B). Periodic PB analysis revealed that 127OE Lin− cells were able to engraft and undergo multi-lineage differentiation, similar to Lin− cells transduced with the EV (data not shown). However, several weeks after transplant, peripheral B- and T-cell counts were significantly lower in mice transplanted with miR-127 over-expressing cells, compared to mice transplanted with the EV (Figure 6A). These differences were present also in BM (Figure 6B) and spleen (data not shown), and are consistent with the reported suppression of germinal center regulators by human miR-127,21,27 likely reflecting the lack of miR-127 downregulation from the MPP stage onward rather than upregulation in HSC. In mice transplanted with 127OE cells, the level of miR-127-3p expression in the spleen several weeks after transplantation was similar to that of cells recently infected, and inferior to that of miR-16 (Online Supplementary Figure S5C), thus excluding effects due to supra-physiological expression.
Figure 6.
Forced miR-127 expression does not affect hematopoietic stem cell (HSC) maintenance. (A and B) FACS analysis of multi-lineage differentiation in peripheral blood (PB) 18 weeks after transplant (A) and in bone marrow (BM) (B) of 127OE and empty vector (EV)-recipient mice. Histograms show absolute cell numbers and stacked columns show the relative abundance of each lineage determined by FACS analysis. (A) PB absolute cell number derives from hemocytometer analysis (127OE: n=6; EV: n=9; two independent experiments). (B) BM counts are relative to one femur and one tibia (127OE: n=5; EV: n=7; two independent experiments). (C) FACS analysis of 127OE and EV stem and progenitor cells in the BM of primary recipient mice. Cell numbers are relative to one femur and one tibia. (D-G) FACS analysis of 127OE and EV Lin− transduced cells transplanted in competition with non-transduced cells. Analyses are relative to primary (D and E) and secondary (F and G) recipient mice. (D) Percentage of OFP+ cells within donor CD11b+ cells in PB 18 weeks after transplant of transduced Lin−cells injected in competition with non-transduced cells in a 9:1 ratio (P=0.24). (E) Frequency of OFP+ cells within the indicated BM populations of primary recipients. (F) Frequency of OFP+ cells within the indicated BM populations of secondary recipients. (G) Percentage of OFP+ cells within donor CD11b+ and CD19+ BM cells of secondary recipients (****P<0.0001; Wilcoxon paired test).
The observed lymphopenia was not accompanied by myeloproliferation or splenomegaly, as expected if 127OE were to lead to HSC expansion. Indeed, 127OE did not affect maintenance of stem and progenitor cells (Figure 6C). This is similar to what has been described for miR-99, which did not cause HSC defects when over-expressed, despite its function in regulating HSC self-renewal being revealed after downregulation.20
To test whether miR-127OE HSC show an advantage when subjected to challenging conditions, transduced Lin-cells were transplanted in competition with un-transduced cells at different ratios, and the presence of OFP+ donor cells was monitored over time on myeloid cells in the PB, as described for the sponge experiments, as well as in BM and spleen at necropsy (Online Supplementary Figure S5D). 127OE OFP+ cells did not expand in primary recipients (Figure 6D and E). Donor-derived OFP+ stem cells were functionally competent, since they engrafted, expanded and gave rise to multi-lineage progeny in secondary recipients (Figure 6F and G), independently from the overexpression of miR-127. However, lower lymphoid reconstitution was detected in secondary recipients (Figure 6G).
Taken together, these results suggest that miR-127-3p maintains the HSC pool by limiting premature differentiation, and must be down-regulated upon HSC maturation to maintain correct homeostasis of the hematopoietic system.
Discussion
In this report we demonstrate that miR-127-3p is an important novel regulator of the balance between self-renewal and differentiation in HSC.
The miR-127 gene is encoded in a large evolutionary conserved miRNA gene cluster located within an imprinted domain (14q32 in human, 12qF in mouse),28 the Dlk1-Gtl2 locus. On the maternally imprinted allele, this locus encodes multiple long ncRNA, snoRNA and a mega-cluster of miRNA organized in two groups, among which the miR-127/miR-136 cluster composed of 7 miRNA genes. RNA-seq data showed that the entire locus, which is regulated by a common ciselement, is specifically transcribed in CD49blo long-term HSC compared to all other BM progenitor and mature cells.28 However, according to our profiling, of the miR-127/miR-136 cluster, only miR-127 and miR-434 are expressed as mature forms in adult HSC. Similarly, of all the other miRNA included in the Dlk1-Gtl2 locus beside the miR-127/miR-136 cluster, only miR-411 is expressed in HSC. Of note, three out of the four top miRNA in our HSC-to-MPP list are part of this locus (Online Supplementary Table S2), in line with previous data showing that ncRNA within the Dlk1-Gtl2 locus can be considered HSC markers.28 The miR-127 gene is embedded in a CpG island; therefore, it is conceivable that the rapid downregulation in the earliest step of hematopoietic differentiation is due to epigenetic silencing. Indeed, the expression of miR-127 was experimentally induced in cancer cell lines by chromatin-modifying drugs, which did not affect expression of the other members of the miRNA gene cluster.29
The entire locus has been inactivated in the maternally- derived allele.30,31 Due to embryonic lethality, the consequences on hematopoiesis were studied using fetal liver stem and progenitor cells. The phenotype observed presented features similar to what we observed in mice transplanted with 127DR Lin− cells, such as reduction of HSC despite normal progenitor and lineage cell counts, and defective long-term reconstitution capacity.28 Altered PI3K-mTOR pathway affecting mitochondrial metabolism explained this phenotype,28 although other molecular mechanisms cannot be excluded. Our data on ROS production and apoptosis do not support the hypothesis that miR-127-3p is involved in regulating oxidative stress in HSC, and indeed miR-127-3p was not one of the miRNA for which a target within the PI3K-mTOR pathway was confirmed.28 A mouse model with specific deletion of miR-127 was generated;32 however, the effects on hematopoiesis have not been investigated.
MiR-127 is aberrantly expressed in the context of several solid tumors, potentially acting as either an oncogene (oncomiR)33–35 or as a tumor suppressor.36–41 Within hematopoietic malignancies, miR-127 was found up-regulated, together with other miRNA included within the 14q32 domain, in the acute promyelocytic leukemia (APML) due to PML-RARa translocation.42 It would be worth investigating whether miR-127 aberrant expression contributes to the differentiation block in APML myeloid progenitors. Although miR-127-3p overexpression did not lead to myelo-or lymphoproliferation, we cannot exclude that, in the presence of other genetic abnormalities, miR-127-3p may act as an oncomiR.
A number of miR-127-3p targets have been found in cancer cells,21,33,35–39,41 in other cell lines,43,44 or during development,32 but they are either not expressed in hematopoietic progenitors, or their expression does not change in the HSC-to-MPP transition. Our search for predicted miR-127-3p targets within DE mRNA in the HSC-to-MPP transition and in Pbx1-deficient HSC revealed two potential targets, Gp1bb and Nek2 (Online Supplementary Table S3). We confirmed by qRT-PCR their upregulation in the physiological HSC-to-MPP transition (data not shown); however, the increase in Gp1bb expression was extremely mild, whereas Nek2 seed-sequence is located within an alternatively spliced, non-coding portion of the transcript. It is, therefore, unlikely that Nek2 protein expression is directly regulated by miR-127-3p. Since miR-127-3p binding to its target mRNA might result in translation inhibition rather than mRNA degradation, a global proteomic approach would be required to gain insights on miR-127-3p functional targets mediating the observed phenotype, although protein analysis on rare primary HSC is technically challenging, especially in a model in which stem cells are reduced.
The physiological downregulation of miR-127-3p observed in the transition from HSC to MPP coincides with the loss of self-renewal ability that is known to occur during this differentiation step. Moreover, miR-127-3p is absent in HSC obtained from Pbx1-cKO mice, which display a profound self-renewal defect.4 This stimulates future studies to investigate if Pbx1, together with its homeobox partners, directly regulates miR-127 expression, or if miR-127 OE can rescue part of the severe Pbx1-cKO phenotype. Our data strongly suggest that HSC must maintain proper levels of miR-127-3p to preserve their long-term self-renewal capacity. We also found that miR-29a, miR-99 and miR-126a are expressed in HSC but also present in Flk2−MPP although at lower levels (Figure 2B and C and Online Supplementary Table S1). However: 1) none of them was DE in HSC from our self-renewal defective mouse model; or 2) the downregulation was less than 3-fold. Among other miRNA whose function in the hematopoietic system has been previously established, miR-29a and miR-126 were shown to play a role in HSC self-renewal and to be differentially expressed in HSC compared to lineage-committed progenitors, but still highly expressed at the MPP stage,11,45 whereas miR-99 was recently found to be DE in the HSC-to-MPP transition.20 Interestingly, these miRNA regulate HSC pool maintenance through different biological mechanisms. miR-126 restrains HSC cell cycle entry, while miR-99 and miR-29a limit HSC maturation, similar to miR-127. However, the accelerated differentiation displayed by miR-127DR cells was not associated to increased apoptosis or altered cell cycle. This might explain why the effect of miR-127DR was only observed with secondary transplants, since HSC depletion was likely due to a gradual exit of HSC from the self-renewing pool. Likewise, it is not surprising that the accelerated differentiation did not result in an increased differentiated progeny in vivo, since: a) differentiation was not associated with increased proliferation; and b) miR-127 is not normally expressed beyond the HSC stage, therefore an effect of miR-127DR in more mature cell population is not expected.
In conclusion, we identified miR-127 as novel player for preserving the HSC pool. Our discovery has potential implications for several translational aspects of experimental hematology, including hematopoietic malignancies, BM transplantation, and regenerative medicine.
Acknowledgments
The authors would like to thank Dario Strina, Stefano Mantero and Lucia Susani for technical assistance, Massimiliano Mirolo and Ciro Menale for technical suggestions, and Paolo Vezzoni for helpful discussions and for critically reading the manuscript.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/9/1744
Funding
We acknowledge support from Marie Curie IRG 256452, from Ricerca Finalizzata GR 2010-2307975 and from AIRC-Fondazione Cariplo (TRIDEO 15882) to FF, from CNR National Program Aging Project to AV, and NIH grant CA116606 to ML. LC was recipient of a fellowship from Fondazione Nicola del Roscio; SM was recipient of a fellowship from Fondazione Damiano per l’Ematologia.
References
- 1.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson A, Laurenti E, Trumpp A. Balancing dormant and self-renewing hematopoietic stem cells. Curr Opin Genet Dev. 2009;19(5):461–468. [DOI] [PubMed] [Google Scholar]
- 3.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Bio. 2014;15(8):509–524. [DOI] [PubMed] [Google Scholar]
- 4.Ficara F, Murphy MJ, Lin M, Cleary ML. Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell. 2008;2(5):484–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ficara F, Crisafulli L, Lin C, et al. Pbx1 restrains myeloid maturation while preserving lymphoid potential in hematopoietic progenitors. J Cell Sci. 2013; 126(Pt14):3181–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koss M, Bolze A, Brendolan A, et al. Congenital asplenia in mice and humans with mutations in a Pbx/Nkx2-5/p15 module. Dev Cell. 2012;22(5):913–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deo A, Carlsson J, Lindlof A. How to choose a normalization strategy for miRNA quantitative real-time (qPCR) arrays. J Bionif Comput Biol. 2011;9(6):795–812. [DOI] [PubMed] [Google Scholar]
- 8.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amendola M, Passerini L, Pucci F, Gentner B, Bacchetta R, Naldini L. Regulated and multiple miRNA and siRNA delivery into primary cells by a lentiviral platform. Mol Ther. 2009;17(6):1039–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gentner B, Schira G, Giustacchini A, et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods. 2009;6(1):63–66. [DOI] [PubMed] [Google Scholar]
- 11.Lechman ER, Gentner B, van Galen P, et al. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell. 2012;11(6):799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet. 2000;25(2):217–222. [DOI] [PubMed] [Google Scholar]
- 13.Guenechea G, Gan OI, Inamitsu T, et al. Transduction of human CD34+ CD38-bone marrow and cord blood-derived SCID-repopulating cells with third-generation lentiviral vectors. Mol Ther. 2000;1(6):566–573. [DOI] [PubMed] [Google Scholar]
- 14.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felli N, Fontana L, Pelosi E, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005; 102(50):18081–18086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerrits A, Walasek MA, Olthof S, et al. Genetic screen identifies microRNA cluster 99b/let-7e/125a as a regulator of primitive hematopoietic cells. Blood. 2012; 119(2):377–387. [DOI] [PubMed] [Google Scholar]
- 17.Guo S, Lu J, Schlanger R, et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci U S A. 2010; 107(32):14229–14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Connell RM, Chaudhuri AA, Rao DS, Gibson WS, Balazs AB, Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci U S A. 2010;107(32):14235–14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ooi AG, Sahoo D, Adorno M, Wang Y, Weissman IL, Park CY. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci U S A. 2010;107(50):21505–21510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khalaj M, Woolthuis CM, Hu W, et al. miR-99 regulates normal and malignant hematopoietic stem cell self-renewal. J Exp Med. 2017;214(8):2453–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leucci E, Onnis A, Cocco M, et al. B-cell differentiation in EBV-positive Burkitt lymphoma is impaired at posttranscriptional level by miRNA-altered expression. Int J Cancer. 2010;126(6):1316–1326. [DOI] [PubMed] [Google Scholar]
- 22.Zonari E, Pucci F, Saini M, et al. A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood. 2013;122(2):243–252. [DOI] [PubMed] [Google Scholar]
- 23.Ito K, Hirao A, Arai F, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431(7011):997–1002. [DOI] [PubMed] [Google Scholar]
- 24.Kent DG, Copley MR, Benz C, et al. Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood. 2009;113(25):6342–6350. [DOI] [PubMed] [Google Scholar]
- 25.Kent DG, Dykstra BJ, Eaves CJ. Isolation and Assessment of Single Long-Term Reconstituting Hematopoietic Stem Cells from Adult Mouse Bone Marrow. Curr Protoc Stem Cell Biol. 2016;38:2A.4.1–2A.4.24. [DOI] [PubMed] [Google Scholar]
- 26.Seita J, Sahoo D, Rossi DJ, et al. Gene Expression Commons: an open platform for absolute gene expression profiling. PloS One. 2012;7(7):e40321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Wang M, Guo M, Xie Y, Cong YS. miR-127 regulates cell proliferation and senescence by targeting BCL6. PloS One. 2013;8(11):e80266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian P, He XC, Paulson A, et al. The Dlk1-Gtl2 Locus Preserves LT-HSC Function by Inhibiting the PI3K-mTOR Pathway to Restrict Mitochondrial Metabolism. Cell Stem Cell. 2016;18(2):214–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with down-regulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9(6):435–443. [DOI] [PubMed] [Google Scholar]
- 30.Lin SP, Youngson N, Takada S, et al. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35(1):97–102. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Cheunsuchon P, Nakayama Y, et al. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development. 2010; 137(16):2643–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito M, Sferruzzi-Perri AN, Edwards CA, et al. A trans-homologue interaction between reciprocally imprinted miR-127 and Rtl1 regulates placenta development. Developmen. 2015;142(14):2425–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang H, Hua D, Zhang J, et al. MicroRNA-127-3p promotes glioblastoma cell migration and invasion by targeting the tumor-suppressor gene SEPT7. Oncol Rep. 2014;31(5):2261–2269. [DOI] [PubMed] [Google Scholar]
- 34.Shi L, Wang Y, Lu Z, et al. miR-127 promotes EMT and stem-like traits in lung cancer through a feed-forward regulatory loop. Oncogene. 2017;36(12):1631–1643. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Kong D. Knockdown of lncRNA MEG3 inhibits viability, migration, and invasion and promotes apoptosis by sponging miR-127 in osteosarcoma cell. Cell Biochem. 2018;119(1):669–679. [DOI] [PubMed] [Google Scholar]
- 36.Bi L, Yang Q, Yuan J, et al. MicroRNA-127-3p acts as a tumor suppressor in epithelial ovarian cancer by regulating the BAG5 gene. Oncol Rep. 2016;36(5):2563–2570. [DOI] [PubMed] [Google Scholar]
- 37.Gao X, Wang X, Cai K, et al. MicroRNA-127 is a tumor suppressor in human esophageal squamous cell carcinoma through the regulation of oncogene FMNL3. Eur J Pharmacol. 2016;791:603–610. [DOI] [PubMed] [Google Scholar]
- 38.Herr I, Sahr H, Zhao Z, et al. MiR-127 and miR-376a act as tumor suppressors by in vivo targeting of COA1 and PDIA6 in giant cell tumor of bone. Cancer Lett. 2017;409:49–55. [DOI] [PubMed] [Google Scholar]
- 39.Wang D, Tang L, Wu H, Wang K, Gu D. MiR-127-3p inhibits cell growth and invasiveness by targeting ITGA6 in human osteosarcoma. IUBMB Life. 2018;70(5):411–419. [DOI] [PubMed] [Google Scholar]
- 40.Yu Y, Liu L, Ma R, Gong H, Xu P, Wang C. MicroRNA-127 is aberrantly downregulated and acted as a functional tumor suppressor in human pancreatic cancer. Tumor Biol. 2016;37(10):14249–14257. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Hou W, Chai M, et al. MicroRNA-127-3p inhibits proliferation and invasion by targeting SETD8 in human osteosarcoma cells. Biochem Biophys Res Commun. 2016;469(4):1006–1611. [DOI] [PubMed] [Google Scholar]
- 42.Dixon-McIver A, East P, Mein CA, et al. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PloS One. 2008; 3(5):e2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma H, Lin Y, Zhao ZA, et al. MicroRNA-127 Promotes Mesendoderm Differentiation of Mouse Embryonic Stem Cells by Targeting Left-Right Determination Factor 2. J Biol Chem. 2016;291(23):12126–12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhai L, Wu R, Han W, Zhang Y, Zhu D. miR-127 enhances myogenic cell differentiation by targeting S1PR3. Cell Death Dis. 2017;8(3):e2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han YC, Park CY, Bhagat G, et al. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med. 2010;207(3):475–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Zhang J, Ginzburg Y, et al. Quantitative analysis of murine terminal erythroid differentiation in vivo: novel method to study normal and disordered erythropoiesis. Blood. 2013;121(8):e43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]