SUMMARY
Cryptochromes are blue light receptors that regulate various light responses in plants. Arabidopsis cryptochrome 1 (CRY1) and cryptochrome 2 (CRY2) mediate blue light inhibition of hypocotyl elongation and long-day (LD) promotion of floral initiation. It has been reported recently that two negative regulators of Arabidopsis cryptochromes, Blue light Inhibitors of Cryptochromes 1 and 2 (BIC1 and BIC2), inhibit cryptochrome function by blocking blue light-dependent cryptochrome dimerization. However, it remained unclear how cryptochromes regulate the BIC gene activity. Here we show that cryptochromes mediate light activation of transcription of the BIC genes, by suppressing the activity of CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), resulting in activation of the transcription activator ELONGATED HYPOCOTYL 5 (HY5) that is associated with chromatins of the BIC promoters. These results demonstrate a CRY–BIC negative-feedback circuitry that regulates the activity of each other. Surprisingly, phytochromes also mediate light activation of BIC transcription, suggesting a novel photoreceptor co-action mechanism to sustain blue light sensitivity of plants under the broad spectra of solar radiation in nature.
Keywords: cryptochrome (CRY), blue light inhibitors of cryptochromes (BIC), negative-feedback circuitry, Arabidopsis thaliana
INTRODUCTION
Photoreceptors are commonly controlled by negative-feedback inhibition mechanisms. For example, the blue light receptors White Collar Complex (WCC) of the filamentous fungus Neurospora mediates light-induced transcription of its negative regulator VIVID, which physically interacts with WCC to suppress the activity of WCC (Schwerdtfeger and Linden, 2003; Chen et al., 2010). Similarly, the UV-B photoreceptor of Arabidopsis, UVR8 (UV-B Resistance 8), mediates UV-B light induction of transcription of its negative regulators RUP1 and RUP2 (Repressor of UV-B Photo-morphogenesis 1 and 2), which physically interact with UVR8 to facilitate re-dimerization and inactivation of the UVR8 photoreceptor (Gruber et al., 2010; Heijde and Ulm, 2013; Findlay and Jenkins, 2016). The red/far-red light receptors phytochromes phytochrome B (phyB) appears to be regulated by its signaling protein PIF3 (Phytochrome Interacting Factor 3) via a different negative-feedback mechanism, whereby photoactivated phyB interact with PIF3 to facilitate red light-dependent phosphorylation of PIF3, resulting in not only gene expression changes in response to light but also ubiquitination and degradation of both PIF3 and phyB (Ni et al., 2014). Recurrence of negative-feedback circuitries of various photoreceptors in different evolutionary lineages is consistent with the hypothesis that negative-feedback inhibition of photoreceptors is commonly required to achieve sustained cellular photosensitivity. Although the abundance of cryptochromes are commonly regulated by the ubiquitin proteasome systems in plants and animals (Busino et al., 2007; Yu et al., 2007; Hirota et al., 2012; Xing et al., 2013), this mechanism does not seem to represent the canonical negative-feedback inhibition mechanism directly involved in the light regulation of cryptochrome activity. For example, Arabidopsis CRY2 undergoes blue light-dependent degradation that is partially dependent on the E3 ubiquitin ligase COP1 (Shalitin et al., 2002; Lin and Shalitin, 2003). However, CRY2 interacts with SPA1 to inhibit COP1 activity in response to blue light (Liu et al., 2011b; Zuo et al., 2011), which is not expected for the canonical negative-feedback inhibition mechanism.
We have recently reported that two negative regulators of cryptochromes, BIC1 and BIC2, physically interact with CRY2 to suppress photoactivation of the photoreceptor (Wang et al., 2016). However, it remained unclear how cryptochromes regulate the BIC gene activity. Here we show that cryptochromes mediate light activation of transcription of the BIC genes, by suppressing the activity of COP1 (CONSTITUTIVE PHOTOMORPHOGENIC 1), resulting in activation of the transcription activator HY5 (ELONGATED HYPOCOTYL 5) that is associated with chromatins of the BIC promoters. These results demonstrate a CRY–BIC negative-feedback circuitry that regulates the activity of each other. Surprisingly, phytochromes also mediate light activation of BIC transcription, suggesting a novel photoreceptor co-action mechanism to sustain blue light sensitivity of plants under the broad spectra of solar radiation in nature.
RESULTS AND DISCUSSION
We used an imaging-based high-resolution growth kinetics analysis to further examine the function of BIC1 antagonizing the cryptochrome-mediated blue light inhibition of hypocotyl growth (Figure 1). In this experiment, we prepared and analysed mutants and transgenic lines impaired or overexpressing the CRY or/and BIC genes (Figure 1a). To avoid post-transcriptional gene silence of the transgenes, all transgenic lines were prepared in the rdr6 mutant background defective in the RNA-dependent polymerase 6 (Peragine et al., 2004; Allen et al., 2005). We grew seedlings of these genotypes under continuous blue light (10 μmol m−2 sec−1), and measured hypocotyls lengths from the real-time images taken every 15 min between 2-day to 5-day after seed imbibition (Figure 1b). Figure 1(b) shows that the bic1bic 2 double mutant exhibited photohypersensitive phenotype resembling to that of CRY2 overexpression; that overexpression of BIC1 suppress the photohypersensitive phenotype of CRY2 overexpression; and that cry1cry2 mutations are epistatic to the bic1bic2 mutations (Figure 1b). Therefore, consistent with our previous finding based on the steady-state phenotypic analyses (Wang et al., 2016), results of the growth kinetics analyses demonstrate that BIC1 suppresses the blue light-dependent activity of plant cryptochromes. As expected, BIC1 also suppresses blue light-dependent degradation of CRY2 (Figure 1c).
Figure 1. BIC1 antagonizes the CRY-mediated blue light inhibition of hypocotyl growth.
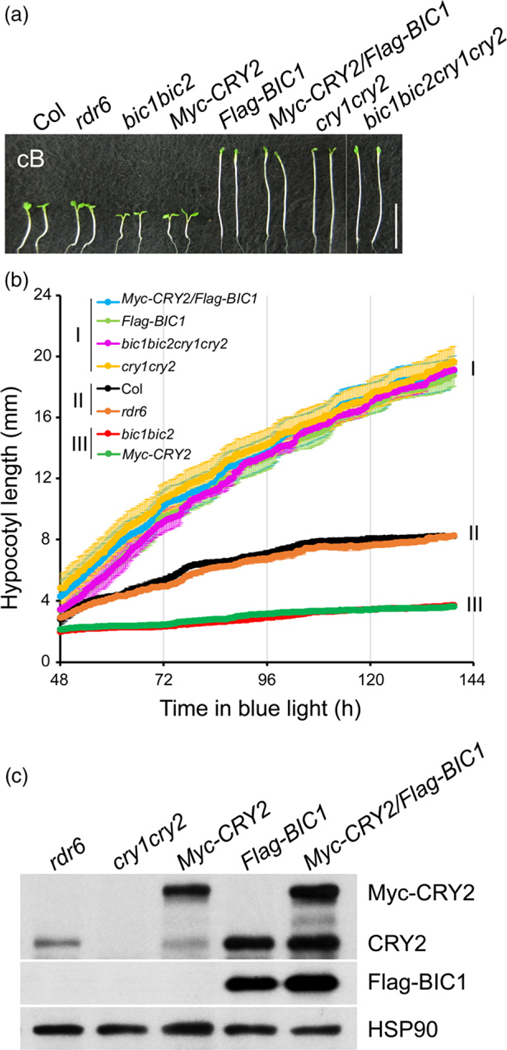
(a) The representative hypocotyl image of the WT, rdr6, bic1bic2, cry1cry2, bic1bic2cry1cry2, Myc-CRY2, Flag–BIC1, and Myc-CRY2/Flag–BIC1 grown in blue light (10 μmol m−2 sec−1) for 5 days. Scale bar = 1 cm. All transgenic lines, including Myc-CRY2, Flag–BIC1, and Myc-CRY2/Flag–BIC1 are in the rdr6 mutant that has no defects in photo-responses to avoid post-transcriptional gene silencing.
(b) Growth kinetics analysis of indicated genotypes. Seedlings germinated and grown in continuous blue light (10 μmol m−2 sec−1) were imaged every 15 min, lengths of hypocotyl of three seedlings were averaged and the error bar shows the standard deviation. Three types of growth kinetics are grouped and labeled as I, II and III.
(c) Immunoblots of protein samples prepared from 6-day-old seedlings of indicated genotypes grown in continuous blue light (10 μmol m−2 sec−1) were probed with anti-CRY2, anti-Flag or anti-HSP90 antibody, respectively. The endogenous CRY2 (CRY2) is resistant to degradation in plants overexpressing BIC1.
In contrast with the ancient evolutionary origin of cryptochromes that have been found from bacteria to human, BICs appear to have emerged more recently in land plants, and their paralogs are not found in other phyla such as bacteria, fungi, or animals (Figure S1). Because BICs from different plant species possess no apparent sequence similarity to other proteins except a highly conserved cryptochrome-interacting domain among BICs (Wang et al., 2016) and that the impairment of cryptochrome-dependent blue light responses appears to be the primary phenotype of the bic1bic2 double mutant or BIC-overexpressing plants, it was speculated that BICs might be an evolutionary invention with the primary function being the antagonists of cryptochromes in land plants (Wang et al., 2016). This speculation raises a possibility that cryptochromes may positively regulate BIC genes to exert negative-feedback inhibition of the photoreceptor activity in plants. Consistent with this possibility, the BIC1 and BIC2 genes were expressed at extremely low levels in etiolated seedlings, whereas levels of the BIC mRNAs increased by up to 100-fold in response to light (Figure 2). To more thoroughly examine the blue light-induced BIC mRNA expression, we performed a two-dimensional kinetics analysis of BIC gene expression (Figure 2 and Table S1). In this experiment, 5-day-old etiolated wild-type seedlings were exposed to blue light of fluence rates ranging from 0.1 to 100 μmol m−2 sec−1 for the exposure time ranging from 20 to 120 min, and the levels of BIC1 and BIC2 mRNA was analysed by qPCR. In comparison with that of the etiolated seedlings grown in the dark, the level of BIC1 mRNA increased within 20 min of light treatment by a few folds (at relatively low fluence rates of ≤10 μmol m−2 sec−1) to >10-fold (at relatively high fluence rates of 10–100 μmol m−2 sec−1). In response to prolonged blue light exposures of 120 min, the BIC1 mRNA expression increased enormously by about 100-fold (at the lowto medium fluence rates of ≤50 μmol m−2 sec−1) to about 1000-fold (at the higher fluence rates of 50–100 μmol m−2 sec−1). The BIC2 mRNA expression also exhibited significant blue light induction, although the extent of photo-induction of BIC2 appears 3–5-fold lower than that of BIC1 (Figure 2; note the different scales used). Surprisingly, although the biochemical and physiological activities of BICs are blue light-specific (Wang et al., 2016), the light induction of BIC mRNA expression is not blue light-specific. Red light and far-red light also induced BIC1 and BIC2 mRNA expression, although the extent of red or far-red light-induced BIC1 and BIC2 mRNA expression appeared less dramatic than that caused by blue light (Figure S2b).
Figure 2. Blue light-induced mRNA expressions of the BIC1 and BIC2 genes.
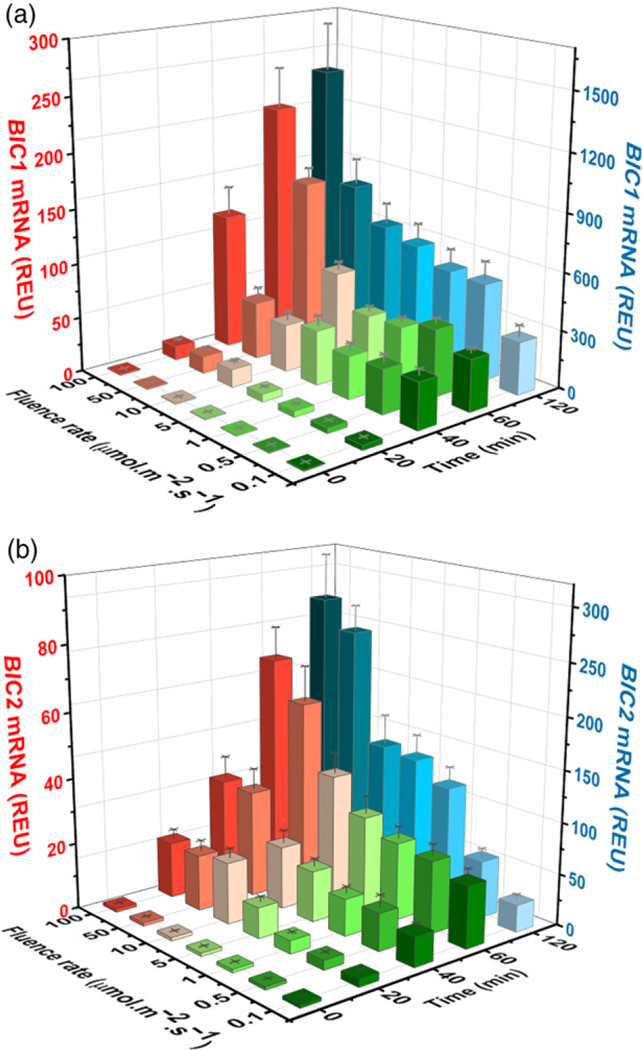
(a, b) 5-day-old etiolated wild-type seedlings were exposed to blue light with different fluence rates for the indicated time before sample collection. Relative mRNA levels of BIC1 (a) and BIC2 (b) genes are shown. The mRNA levels of seedlings exposed to blue light for 120 min are labeled by blue color (Z-axis on the right). Other colors represent mRNA levels of seedlings exposed to blue light for 0, 20, 40, or 60 min (Y-axis on the left). The relative expression unit (REU) was calculated by re-normalization of the normalized qPCR signals of samples exposed to light against the normalized qPCR signal of the sample kept in the dark. PP2A (At1g69960) was used as the reference gene for qPCR normalization.
To understand how light stimulates BIC expression, we prepared and examined the transgenic plants expressing the reporter genes, 489BIC1pro::GUS and 2100BIC2pro::GUS, which encode the reporter GUS (β-glucuronidase) under the control of the BIC1 promoter and 5’UTR (489 bp upstream of ATG), or the BIC2 promoter and 5’UTR (2100 bp upstream of ATG). Both reporter genes showed apparent increase of the GUS reporter activity in response to blue light (1 μmol m−2 sec−1) (Figures 3a and S3). These results suggest that BIC promoters are light-responsive and transcription regulation is responsible for the light-induced BIC mRNA accumulation, although a potential role of 5’UTR on stability of the BIC mRNAs cannot be completely excluded. The light-induced expression of the 489BIC1pro::GUS and 2100BIC2pro::GUS transgenes is detected in both hypocotyls and cotyledons (Figure 3a). The fact that BICs are expressed in both organs is consistent with the observation that BICs act as the general inhibitor of cryptochromes (Wang et al., 2016), and that cryptochromes mediate light inhibition of hypocotyl growth, cotyledon expansion (Lin et al., 1998), and floral initiation (Guo et al., 1998).
Figure 3. The promoter-dependent transcription activation of expression of the BIC mRNA and protein.
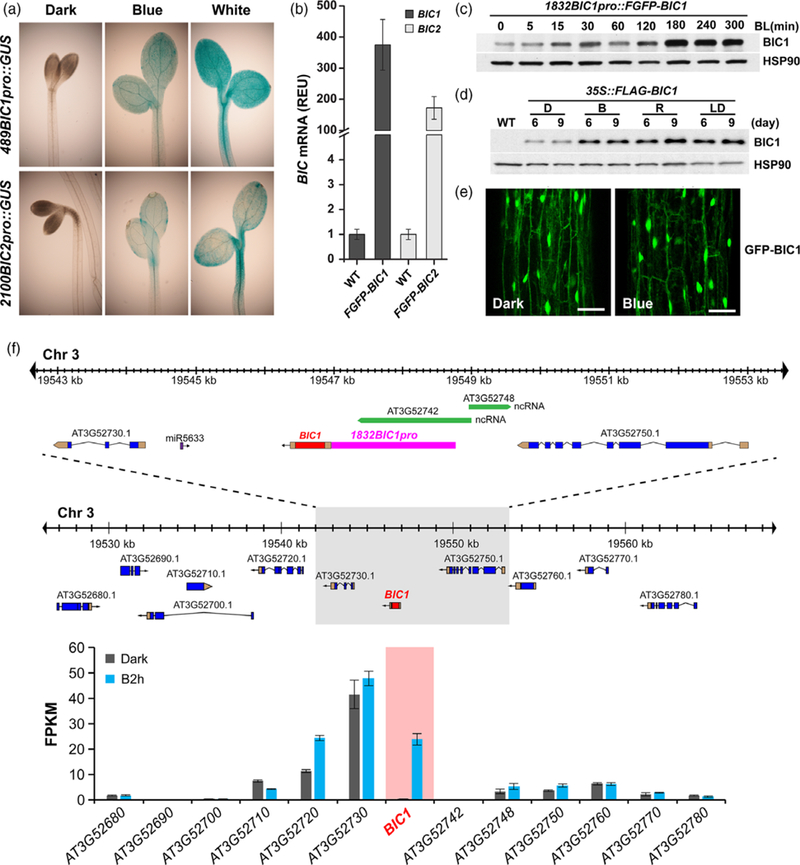
(a) Histochemical GUS staining of 4-day-old etiolated seedlings expressing the indicated reporter genes. Seedlings were left in the dark (Dark) or exposed to blue (Blue, 1 μmol m−2 sec−1) and white light (White) for 20 h. The promoter regions of BIC1 (489 bp upstream from the ATG codon) or BIC2 (2100 bp upstream from the ATG codon) genes were used to prepare the 489BIC1pro::GUS and 2100BIC2pro::GUS reporters.
(b) Genome browser view of the BIC1 locus on chromosome 3 in A. thaliana accession Col. The 10-kb region (top) enlarged from the ~40-kb (middle) areas around the BIC1 gene and all genes within this ~40-kb region are shown. Blue and golden boxes represent exons and untranslated regions are shown. BIC1 has no intron and it is highlighted by red color. Green boxes represent non-protein-coding RNAs genes. Purple boxes represent microRNA genes. Arrows indicate the direction of transcription. The promoter sequences used for making transgenic lines in Figure 3(c) is highlighted by the pink box. The relative levels of mRNA expression derived from a RNA-seq experiment for all the genes in the ~40-kb region are shown (bottom). In this RNA-seq experiment, etiolated wild-type seedlings were kept in the dark (Dark) or exposed to blue light (20 μmol m−2 sec−1) for 2 h (B2 h) were analysed. The gene expression levels were shown as the mean value of Fragments Per Kilobase of transcript per Million mapped reads (FPKM) from three biological repeats and the error bar indicates the standard deviation. Levels of BIC1 mRNA expression is highlighted, the absolute values of BIC1 mRNA expression is included in Figure S2(a).
(c) Immunoblots showing the level of the Flag–GFP–BIC1 fusion protein (FGFP–BIC1) driven by the BIC1 promoter (1832 bp upstream from the ATG codon). 7-day-old etiolated seedlings expressing the 1832BIC1pro::FGFP–BIC1 transgene were either kept in the dark (0) or exposed to blue (30 μmol m−2 sec−1) for indicated time (BL(min)). Immunoblots were probed with the anti-Flag antibody or the anti-HSP90 antibody, respectively.
(d) Immunoblots showing the level of FLAG–BIC1 fusion protein driven by the constitutive 35S promoter. Transgenic seedlings expressing the 35S::Flag–BIC1 transgene were grown in the dark, continuous blue light (30 μmol m−2 sec−1), continuous red light (30 μmol m−2 sec−1), or LD photoperiod (white light) for 6 days (6) or 9 days (9). The recombinant BIC1 protein was detected by immunoblot probed with the anti-Flag antibody. HSP90 was used as the loading control.
(e) Lack of light effect on BIC1 subcellular localization. 3-day-old etiolated seedlings expressing the pACT2::GFP–BIC1 transgene driven by the constitutive ACT2 promoter were either kept in the dark (Dark) or exposed to blue light (10 μmol m−2 sec−1) for 3 h (Blue). Subcellular distributions of the GFP–BIC1 fusion protein in hypocotyl were observed under a confocal microscope. Scale bar = 50 μm.
(f) Relative levels of mRNA expression of the endogenous BICs genes or the transgenes 1832BIC1pro::FGFP–BIC1 (FGFP–BIC1) or 1822BIC2pro::FGFP–BIC2 (FGFP–BIC2, see Figure S4) in the respective transgenic plants grown in the dark. The BIC-specific PCR primers or the GFP–specific PCR primers were used to detect endogenous BICs genes in wild-type plants or the FGFP–BICs transgenes, respectively, in the qPCR assays. All qPCR signals are normalized by that of the PP2A gene (At1g69960). The relative expression unit (REU) of the endogenous BIC1 or BIC2 genes (WT) are set to 1, the REU values of the 1832BIC1pro::FGFP–BIC1 (FGFP–BIC1) or 1822BIC2pro::FGFP–BIC2 (FGFP–BIC2) transgenes are calculated by dividing the normalized qPCR signals of the transgenes with that of the endogenous BIC1 or BIC2 genes, respectively. Three biological repeats were performed and the error bars indicate the standard error of three repeats.
Because expression of the GUS reporter cannot be used to accurately estimate the extent of light induction of the BIC genes, we further analysed the protein and mRNA expression of transgenic line expressing the 1832BIC1pro::FGFP–BIC1 (1832 bp upstream of ATG) and 1822BIC2pro::FGFP–BIC2 (1822 bp upstream of ATG) minigenes, which are also physiologically active (Wang et al., 2016). The 1832BIC1pro::FGFP–BIC1 and 1822BIC2pro::FGFP–BIC2 minigenes encode the Flag–GFP–BIC1 or Flag–GFP–BIC2 fusion proteins driven by the BIC1 promoter and 5’UTR or the BIC2 promoter and 5’UTR, respectively (Figures 3b and S4). As expected, levels of the FGFP–BIC1 or FGFP–BIC2 recombinant proteins increased in etiolated seedlings exposed to blue light or red light (Figures 3c, S5 and S6). We noticed that levels of the FGFP–BIC1 or FGFP–BIC2 recombinant proteins expressed from the 1832BIC1pro::FGFP–BIC1 and 1822BIC2pro::FGFP–BIC2 minigenes increased by less than 10-fold in response to light (Figures 3c, S5a, b and S6a, b). Similarly, mRNA expression of the 1832BIC1pro::FGFP–BIC1 and 1822BIC2pro::FGFP–BIC2 minigenes also did not exceed 10-fold photo-induction (Figure S6c, d). These results indicate that the range of photo-induction of the BIC minigenes (<10-fold) is markedly lower than the range of photo-induction of the BIC native genes (≥100-fold) (Figure 2). One possible explanation of this observation would be that transcription of the BIC native genes is suppressed in the dark but the BIC minigenes somehow lost such control. Therefore, we compared the levels of mRNA of the endogenous BIC genes and that of the BIC minigenes in etiolated seedlings (Figure 3f). Figure 3(f) shows that, in etiolated seedlings grown in the dark, the BIC minigenes accumulated >102 folds more mRNA than that of the BIC native genes. This result appears to argue that the higher amplitude photo-induction of the BIC native genes is primarily due to their extremely low level of mRNA expression in etiolated seedlings, whereas the hampered photo-induction of the 1832BIC1pro::FGFP–BIC1 and 1822BIC2pro::FGFP–BIC2 minigenes is most likely due to the loss of dark-suppression of transcription of these minigenes. It is conceivable that the intergenic regions of the BIC native genes may contain transcriptional silencers, or DNA elements that suppress transcription of the BIC native genes in the dark, which are located beyond the sequences included in the minigenes (Figure 3b). Alternatively, the genomic regions surrounding the BIC native genes may possess heterochromatin-like structures that suppress transcription in the dark. However, analyses of the RNA-seq data (Wang et al., 2016) of the 12 neighboring genes in the genomic regions (~40 kb) of the BIC1 gene indicate that BIC1 is the only gene that exhibits both extremely low transcription in the dark and the strong light induction of transcription (Figure 3b). Similarly, BIC2 is also the only gene in the surrounding ~40 kb region that showed both extremely low transcription in the dark and markedly induced transcription in response to light (Figure S4). Therefore, there is presently little evidence supporting the hypothesis that a heterochromatin-like chromatin structure around the BIC genes suppresses transcription in the dark. Nevertheless, results of our experiments argue strongly that light stimulation of transcriptional activity of the BIC promoters is the primary mechanism responsible for the light-induced BIC mRNA expression, whereas the molecular nature of the putative ‘dark silencers’ of the BIC genes remain to be further investigated in the future.
To examine whether light also affect post-transcriptional regulation of the BIC genes and BIC proteins, we analysed transgenic plants expressing the constitutively transcribed 35S::Flag–BIC1 and 35S::Flag–BIC2 transgenes, which, like the BIC minigenes, abolished cryptochrome activities in transgenic plants (Wang et al., 2016). Immunoblot analyses showed that levels of the recombinant BIC proteins increased slightly in 6-day-old or 9-day-old seedlings grown under continuous blue, red, or white light, or long-day (LD) photoperiodic conditions, in comparison to that grown in the dark (Figures 3d and S5e). Similarly, a slight increase of the constitutively transcribed BIC recombinant proteins was also detected in etiolated seedlings exposed to blue light (Figure S5c) or red light (Figure S5d). BIC proteins were detected in both nucleus and cytosol as reported previously (Wang et al., 2016) (Figures 3e and S5f), but light treatment showed little effect on the relative distribution of the GFP–BIC1 or BIC2–GFP proteins in the nucleus and cytoplasm (Figures 3e and S5f). We conclude that wavelength-independent light activation of transcription is the primary mechanism responsible for the light-regulated expression of the BIC genes, although light may also slightly increase stability of the BIC proteins under the conditions tested.
Given that blue, red and far-red light could stimulate mRNA expression of the BIC genes, we hypothesize that both phytochromes and cryptochromes may mediate light induction of BIC transcription. It is well established that phytochromes and cryptochromes can both suppress activity of the SPA1/COP1 E3 ubiquitin ligase to stabilize light signaling transcription factors, such as HY5, which activates transcription of light-induced genes (Lian et al., 2011; Liu et al., 2011a; Zuo et al., 2011; Huang et al., 2014). Indeed, light-induced BIC mRNA ed (P < 0.01) in the cry1-cry2 mutant (in blue light), the phyAphyB mutant (in red light or far-red light), and the hy5 mutant (in all lights) (Figure 4a–f). In contrast, higher level of the BIC1 and BIC2 mRNA were detected in the cop1 mutant comparing to that of the wild-type plants (Figure 4a–f). These results demonstrate that cryptochromes and phytochromes mediate light suppression of COP1 activity, resulting in increased HY5 activity and induced BIC mRNA expression. To further test possible physiological roles of light-induced BIC mRNA expression, we examined whether pre-treatment by far-red light may affect blue light-dependent CRY2 degradation. We have previously shown that BIC inhibits blue light-induced CRY2 dimerization and consequently the blue light-induced CRY2 degradation (Wang et al., 2016). If the light-induced BIC expression affects the CRY2 activity, one might expect pre-treatment of a long-wavelength light, such as far-red light, that is not absorbed by CRY2 but induces BIC mRNA expression, would suppressed CRY2 degradation in seedlings subsequently treated with blue light. Indeed, as shown in Figure 4, pre-treatment of seedlings with far-red light moderately suppresses the subsequent blue light-induced CRY2 degradation in wild-type (WT) seedlings (Figure 4g, h). This result clearly demonstrates that the light-induced BIC mRNA expression is physiologically relevant to at least the blue light regulation of CRY2 protein stability. As expected, this far-red light effect was weakened in the bic1bic2 mutant seedlings (Figure 4g, h). However, as reported previously (Wang et al., 2016), elimination of the BIC proteins by the bic1bic2 double mutation only resulted in a modest enhancement of CRY2 degradation (Figure 4), although overexpression of BIC almost completely suppressed CRY2 degradation (Wang et al., 2016). Because the BIC1 and BIC2 are expressed at extremely low levels in etiolated seedlings (Figures 2 and 3), the modest effect of FR-pre-treatment on CRY2 degradation under the conditions tested may not be surprising. A relatively long delay of BIC mRNA translation may partially explain the contradiction between the significant FR induction of BIC mRNA expression (Figure 4c, f) and the lack of significant effect of FR-pre-treatment on CRY2 degradation (Figure 4g, h). Alternatively, a relatively mild effect of the light-induced BIC on the function or regulation of CRY2 during early de-etiolation, such as that mimicked by our experimental conditions, may be necessary to ensure high sensitivity of the cryptochrome photosensory system when seedlings first emerge from soil, whereas prolonged light induction of BIC expression may be needed in late seedling development to prevent oversensitive light responses once de-etiolation is established. Additional kinetics and genetics analyses are needed to further clarify how phytochrome-mediated light induction of BIC mRNA expression affect CRY2 function.
Figure 4. Cryptochromes and phytochromes mediate light activation of the BIC transcription via the COP1/SPA1/HY5 pathway.
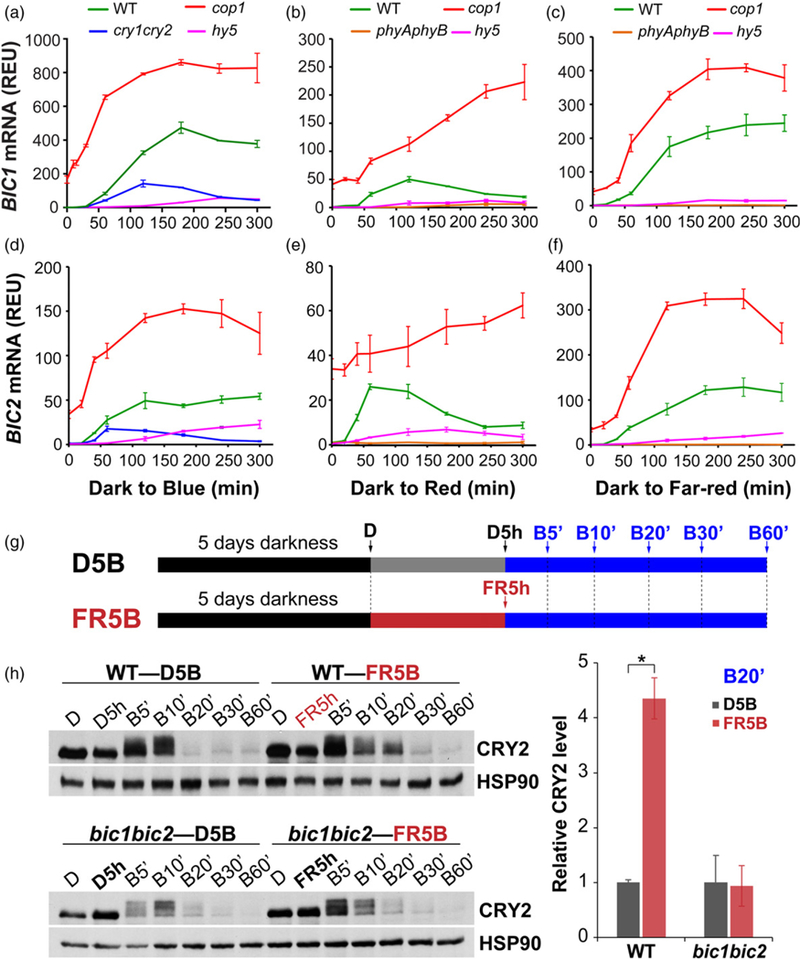
(a–f) qPCR showing light induction of BIC1 (a–c) and BIC2 (d–f) mRNA expressions in 5-day-old etiolated seedlings of indicated genotypes (wild-type (WT), cry1-cry2, phyAphyB, cop1, hy5) exposed to blue light (a, d; 10 μmol m−2 sec−1), red light (b, e; 10 μmol m−2 sec−1) or far-red light (c, f; 5 μmol m−2 sec−1) for the indicated time (min). The relative expression unit (REU) was calculated by re-normalization of the normalized qPCR signal of the BIC genes in plants (of the indicated genotypes) exposed to light against the normalized qPCR signals of the BIC genes in plants (of the wild-type plants) grown in the dark. Three biological repeats were performed and the error bars indicate the standard error of three repeats.
(g) A schematic of two light treatment conditions (D5B and FR5B) of seedlings used for the immunoblot analyses. The time points for sampling are indicated by arrows. Five-day-old etiolated seedlings (D) of WT and bic1bic2 were either irradiated with far-red light (10 μmol m−2 sec−1; FR5 h) or kept in the dark (D5 h) for 5 h, then exposed to blue light (30 μmol m−2 sec−1) for indicated times (5, 10, 20, 30 and 60 min).
(h) CRY2 degradation in response to blue light in WT and bic1bic2 seedlings. CRY2 was detected by immunoblot probed with the anti-CRY2 antibody. HSP90 was used as the loading control. The band intensities of CRY2 at B20’ were quantified and normalized against that of HSP90, then re-normalized to the value of sample D5B (i.e. CRY2D5B/CRY2D5B and CRY2FR5B/CRY2D5B) to calculate the ‘Relative CRY2 level’ in the indicated genotypes. Three biological repeats have been used for CRY2 level quantification and the error bar indicates the standard error of three repeats (*P < 0.05).
Because HY5 activates BICs expression, we investigated whether HY5 is associated with chromatins of the BIC genes by the chromatin immunoprecipitation (ChIP) assay, using a previously reported protocol that minimizes the change of protein stability of HY5 in response to a short illumination (Lee et al., 2007; Zhang et al., 2011). In this experiment, transgenic seedlings expressing the pHY5::HY5–GFP transgene were grown in LD photoperiod for 5 days, transferred to the dark for 8 h, and exposed to blue light (30 μmol m−2 sec−1) for 3 h before harvest. This experimental condition of relatively short light exposure was specifically designed to avoid light-induced increase of the HY5 protein level, such that to minimize effects of the change of cellular levels of the HY5 protein on the ChIP-PCR results. As previously reported (Lee et al., 2007; Zhang et al., 2011), the level of HY5 exhibited little change under this short light exposure (Figure 5a, insert). Figure 5(a) shows clearly that HY5 interacts with the chromatin regions that contain the HY5-binding DNA elements, such as G-Box (CACGTG) and CT-Box (GACGTT). This result is consistent with HY5 being a transcription regulator directly activating BIC transcription. However, we detected no significant change of the interaction between HY5 and chromatins of the promoter regions of both BIC1 and BIC2 genes (Figure 5a). This result may not be surprising because the genome-wide studies of the HY5-chromatin interaction also showed little light-induced change of HY5- chromatin interaction on other light- and HY5-regulated genes (Lee et al., 2007; Zhang et al., 2011). Given that HY5 can activate BIC transcription in the absence of apparent change of the HY5 protein level or alteration of the HY5-chromatin association, exactly how HY5 activates BIC mRNA expression remains to be further studied.
Figure 5. HY5 associates with BIC chromatin regions in plants.
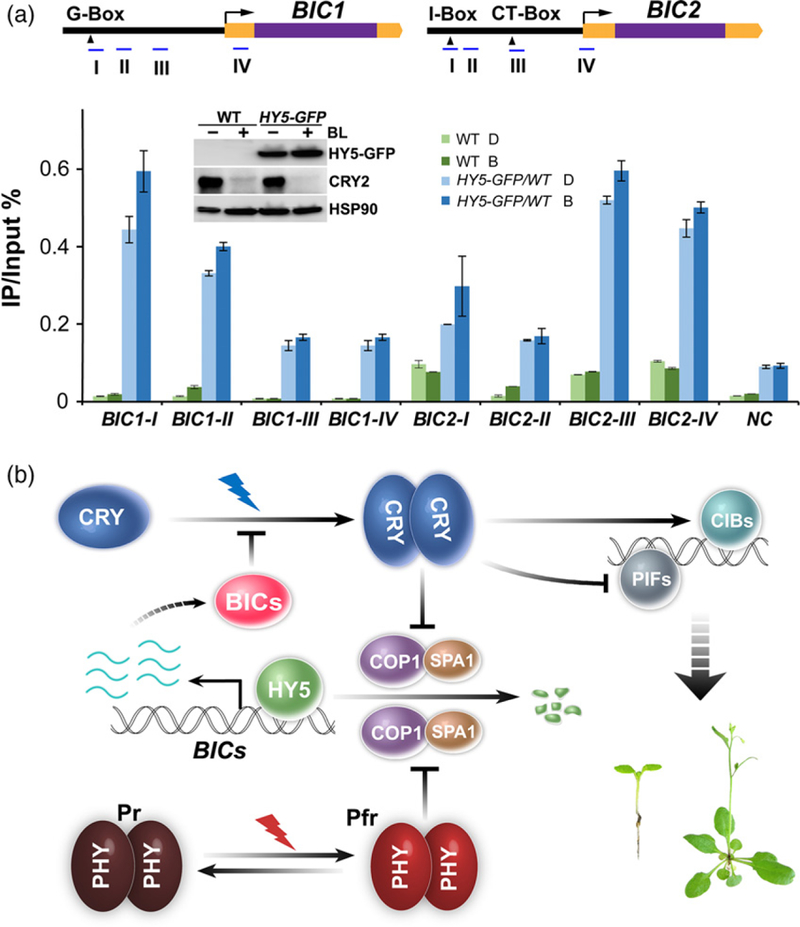
(a) Results of the ChlP-qPCR experiments showing interaction of HY5 with genomic regions of BIC1 and BIC2. Five-day-old LD-grown seedlings of wild-type (WT) and pHY5::HY5-GFP (HY5–GFP) transgenic lines were adapted to the dark (D) for 8 h, then exposed to blue light (30 μmol m−2 sec−1; B) for 3 h. ChIP was performed using the GFP-trap beads. The upper panels show the structures of BIC1 and BIC2 genomic loci. Blue lines indicate genomic regions amplified by the ChlP-qPCR. Yellow and purple boxes indicate UTRs and exons, respectively. The black arrowheads indicate locations of G-Box (CACGTG), I-Box (TAGATAACC), and CT-Box (GACGTT). A genomic region of At4G26900 was amplified as the negative control (NC), representing the background signals of GFP–trap pull-down experiments. The immunoblot inside the bar graph shows the lack of dramatic change in the HY5 protein expression under the condition used to perform the ChlP-qPCR assay. Two biological repeats were performed for the ChlP-qPCR assay. Two repeats showed similar results and only one experiment was presented in this figure. The error bar indicates the standard deviation of four technical repeats for each sample. Protein levels of HY5–GFP, CRY2 and HSP90 were detected by immunoblots probed with the anti-GFP, anti-CRY2 and anti-HSP90 antibodies respectively. Blue light-dependent CRY2 degradation was used to assess the blue light treatment, HSP90 was used as the loading control.
(b) A hypothetic model depicting the CRY2-BlCs negative-feedback circuitry. CRY2 exists as inactive monomers in the dark. Photoexcited CRY2 becomes homodimers or oligomers, which interact with CRY-signaling proteins (such as SPA1, PlFs and ClBs) to activate photomorphogenic development. Cryptochromes and phytochromes mediate blue light or red/far-red light inhibition of the activity of COP1/SPA1 E3 ligase, resulting in activation of HY5. The HY5 protein binds to the promoter of BIC genes to active transcription of the BIC genes in response to light. The BIC proteins bind to CRY2 to inhibit CRY2 dimerization and activation.
Based on results presented in this and previous studies (Wang et al., 2016), we proposed a CRY–BIC negative-feedback circuitry regulating the activity of cryptochromes in response to light (Figure 5b). According to this model, cryptochromes become photoactivated by blue light-dependent homodimerization; the photoactivated cryptochromes mediate blue light-dependent change of transcription of many genes, including the BIC genes; increased expression of the BIC proteins inhibits cryptochrome dimerization to complete the negative-feedback circuitry.
Co-action between different photoreceptors is a well known phenomenon in plant light responses (Casal, 2000; Su et al., 2017). Common partners associated with different photoreceptors represent one possible mechanism underlying the phytochrome and cryptochrome co-action. For example, the phytochrome signaling proteins, PIF4 (Phytochrome Interacting Factor 4) and PIF5, interact with CRY1 and CRY2 to control hypocotyl growth under LBL (Low Blue Light) or elevated temperature (Ma et al., 2016; Pedmale et al., 2016), respectively, leading to the convergence of diverse light signaling pathways mediated by PHYs and CRYs. It has also been reported recently that Photoregulatory Protein Kinases (PPKs) interact with not only CRY2 to catalyze CRY2 phosphorylation (Liu et al., 2017), but also PIF3and phyBto promote the light-induced phosphorylation and degradation of PIF3 (Ni et al., 2017). Our finding reported here that both cryptochromes and phytochromes mediate light stimulation of BIC transcription represents a distinct mechanism for the co-action of cryptochromes and phytochromes, whereby the two types of photoreceptors coordinately elicit the BIC negative-feedback circuitry to suppress cryptochromes in response to the broad spectra of light in nature. It would be interesting to further explore how these two different mechanisms may interplay to coordinate co-action of the respective photoreceptors.
EXPERIMENTAL PROCEDURES
Plant materials and growth conditions
All Arabidopsis plants used in this work are of the Columbia (Col) accession. The bic1bic2, cry1cry2, bic1bic2cry1cry2, pACT2::Myc-CRY2, 35S::Flag–BIC1, 35S::Flag–BIC2, 35S::BIC2–GFP, pACT2::FGFP–BIC1, pBIC1::FGFP–BIC1 and pBIC2::FGFP–BIC2 are described previously (Wang et al., 2016). The double transgenic plant expressing both Myc-CRY2 and Flag–BIC1 were prepared by co-transforming pACT2::Myc-CRY2 and 35S::Flag–BIC1 constructs into rdr6–11 allele, which suppresses gene silencing, using the standard floral-dip method. The transgenic T1 populations were screened on MS agar medium containing 25 mg L−1 hygromycin and 25 mg L−1 glufosinate-ammonium.
For the light-response experiments, light-emitting diode (LED) was used to obtain monochromatic blue (peak 450 nm; half-bandwidth of 20 nm), red (peak 660 nm; half-bandwidth of 20 nm) or far-red (peak 730 nm; half-bandwidth of 20 nm) light. Cool white fluorescence tubes were used for the white light.
Image-based growth kinetics analyses
For the image-based growth kinetics analysis, Arabidopsis seeds were ethanol-sterilized and plated onto 0.8% agar MS plates. After imbibition in the dark at 4°C for 3 days, the plates were placed vertically under continuous blue light (10 μmol m−1 sec−2) for imaging. Seedlings were imaged by time-lapse photography, using a CCD camera (Jinghang JHSM500B) equipped with a prime macro lens. Images were taken every 15 min from 48 h after imbibition for the next 4 days. Image acquisition was controlled by a custom-designed software. Hypocotyl length of each image was measured by a semi-automated MATLAB script modified from that provided by Dr Christian Fankhauser (Kohnen et al., 2016). Three seedlings of each genotype were measured for time-lapsed imaging. Two independent biological repeats showed the similar result, and the results of one experiment was represented in the Figure 1(b).
Immunoblot analysis
To prepare protein extracts, seedlings were ground in liquid N2 and boiled in Protein Extraction Buffer (120 mM Tris–HCl pH 6.8; 100 mM EDTA; 4% SDS; 10% β-ME; 5% glycerol and 0.05% bromophenol blue) for 10 min. The protein extracts were separated by 10% SDS-PAGE and transferred to Pure Nitrocellulose Blotting Membrane (BioTrace NT, Pall Life Sciences). The membrane was stained with Ponceau S Red and blocked with 5% skimmed milk in PBST solution. After probed with primary and secondary antibodies, the membrane was incubated in the home-made ECL [Solution A: 100 mM Tris–HCl pH 8.5; 0.2 mM coumaric acid; Solution B: 100 mM Tris–HCl pH 8.5; 1.25 mM luminol; Before use, mix 3 ml Solution A with 3 ml Solution B and add 2 μl 30% H2O2] and exposed to an X-ray film to detect the signals. The primary anti-bodies used in this study included: anti-CRY2 (1:3000) (Wang et al., 2016), anti-HSP90 (1:1000, sc-33755, Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and anti-FLAG (1:3000, F3165, Sigma-Aldrich Corp., St. Louis, MO, USA). Rabbit IgG HRP-linked F(ab’)2 fragment (from donkey) (NA9340–1ML, GE Healthcare, Chicago, IL, USA) was used as the secondary antibody (1:10 000) to detect anti-CRY2 and anti-HSP90 antibodies. Mouse IgG HRP-Linked F (ab’)2 fragment (from sheep) (NA9310–1ML, GE Healthcare) was used (1:10 000) to detect the anti-FLAG antibody.
Confocal microscopy
To determine the subcellular localization of GFP–BIC1 and BIC2–GFP proteins, seedlings of pACT2::GFP–BIC1 and 35S::BIC2–GFP were grown in the dark or blue light (10 μmol m−2 sec−1), fixed in 1% formaldehyde in phosphate-buffered saline (PBS) solution for 15 min, washed twice with PBS and examined by a confocal laser scanning microscope (LSM 810, Carl Zeiss).
Chromatin immunoprecipitation (ChIP) assays
The ChIP experiments were performed using 5-day-old LD-grown seedlings adapted in the dark for 8 h and exposed to blue light for 3 h before harvesting. Approximately 4 g seedlings were ground in liquid nitrogen and homogenized in 40 mL nuclei isolation and X-linking buffer [0.4 m sucrose, 10 mM HEPEs (pH 8.0), 5 mM KCl, 5 mM MgCl2, 0.5% Triton X-100, 1 mM PMSF, 1× Protease inhibitor cocktail (Roche) and 1% formaldehyde] for 20 min at room temperature. Formaldehyde cross-linking was stopped by addition of 2.5 ml 2 M glycine at room temperature for 10 min. Homogenized slurry was filtered through Miracloth and the filtrate was centrifuged at 2500 g for 10 min at 4°C. The pellet was suspended in 2 ml extraction buffer 2 [0.25 M sucrose, 10 mM Tris–HCl pH8.0, 10 mM MgCl2, 1% Triton X-100, 1 mM DTT and 1× Protease inhibitor cocktail (Roche)], centrifuged at 2500 g for 10 min at 4°C. The pellet was suspended in 400 μl of nuclei lysis buffer [50 mM Tris pH 8.0, 10 mM EDTA, 1% SDS, 1 mM PMSF and 1× Protease inhibitor cocktail], and the nuclei was lysed by pipetting up and down slowly at room temperature. The nuclear lysate was mixed with ChIP dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris pH 8.0, 167 mM NaCl, 1 mM PMSF and 1× Protease inhibitor cocktail) to a final volume of 1200 μl, treated in Bioruptor for 30 min (30 sec high, 30 sec off) in the ice-water bath, centrifuged at 14 000 g for 10 min at 4°C. After centrifugation, the supernatants were diluted with ChIP dilution buffer to a final volume of 4 ml, of which 100 μl was saved as Input. Approximately 4 ml chromatin solution was pre-cleared by addition of 20 μl ProtA/G-agarose beads (Pierce) and incubated at 4°C for 30 min. Beads were removed by centrifugation. The supernatant was mixed with 100 L GFP–trap agarose beads, incubated at 4°C overnight with rotation. Beads were washed once with low-salt wash buffer (150 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA and 20 mM Tris–HCl, pH 8.0), high-salt wash buffer (500 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA and 20 mM Tris–HCl, pH 8.0) and LiCl wash buffer (0.25 m LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mM EDTA and 10 mM Tris–HCl, pH 8.0), respectively, washed twice with TE buffer (10 mM Tris–HCl, pH 8.0, and 1 mM EDTA). The immunoprecipitated protein-DNA complexes were eluted from the GFP–trap beads with 1% SDS and 0.1 M NaHCO3. After reverse-crosslinking, the immunoprecipitated DNA was purified using the QIAquick kit (Qiagen, Hilden, German) according to the manufacturer’s instructions, and quantified by qPCR analysis. ChIP-qPCR values for each set of primers were normalized to that of the Input (IP/Input). A genomic region of At4G26900 was amplified as the negative control (NC) of HY5-binding sites (Lee et al., 2007; Zhang et al., 2011).
Histochemical GUS analysis
To investigate the expression patterns of BIC1 and BIC2 in plants, the upstream regions of BIC1 (489 bp including 5’-UTR) and BIC2 (2100 bp including 5’-UTR) were PCR-amplified with the following primers; 5’-CCCAAGCTTGGATCCTCTAGTTGAGTTTGGTCAC-3’ (forward) and 5’-GCTCTAGAGATGACACAATAGTAAAGCAGATTCAG-3’ (reverse) for BIC1; 5’-CCCAAGCTTGTTTTTCCGGTTTAG-TAGTTGTTTG-3’ (forward) and 5’-CGGGATCCTTTGAACTCTTTTCTTTATTTTTACTTTTG-3’ (reverse) for BIC2.
To generate GUS expression vectors controlled by the promoters of BICs, each PCR product was subcloned into pBI101 (Jefferson et al., 1987) with HindIII and XbaI for BIC1, and with HindIII and BamHI for BIC2. The resulting vectors 489BIC1pro::GUS and 2100BIC2pro::GUS were then introduced into Arabidopsis thaliana (Col). Transgenic lines segregating ~3:1 for antibiotic resistance in the T2 generation were selected, and the T3 or T4 homozygous seeds were used for subsequent analyses.
To detect the histochemical BICs expression pattern, the seedlings of 489BIC1pro::GUS and 2100BIC2pro::GUS were grown in “MS for 5 days under various light conditions. For histochemical GUS assays, the seedlings were incubated in GUS staining buffer (80 mM sodium phosphate, pH 7.0, 0.4 mM potassium ferricyanide, 0.4 mM potassium ferrocyanide, 8 mM sodium-EDTA, 0.01% Triton X-100, 0.8 mg ml−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, and 20% methanol) for 8 h in the dark and washed with absolute ethanol to remove chlorophylls with gentle agitation.
RNA analyses
Total RNA from the seedlings was extracted using RNeasy Plant Mini Kit with DNase I digestion (QIAGEN). Single-stranded cDNA was synthesized from 3 μg of total RNA using Superscript III (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) and oligo-dT primer following the manufacturer’s protocol. Quantitative PCR reactions were performed with gene specific primers and the SYBR Premix Ex Taq (TaKaRa) on Mx3005P™ Real-Time PCR System (Stratagene, Santa Clara, CA, USA). The qPCR signals were normalized to that of the reference gene PP2A (At1g69960) using the ΔCT method. Transcriptomes were analysed by RNA-seq experiments as described previously (Wang et al., 2016), and the results of BIC1 and BIC2 mRNA expression derived from the previous experiment are presented in Figure S2 as an additional supporting information.
Primers used in this study
| Primer Name |
Primer Sequence | Assay |
|---|---|---|
| BIC1-I-F | 5’-CAACACCGAATCTCTCAACACAAAC-3’ | ChIP-qPCR |
| BIC1-I-R | 5’-TAAAGCAGATTCAGATTCTTGCAGG-3’ | |
| BIC1-II-F | 5’-TCAAAATGCCCGTTTTCTCTCTT-3’ | |
| BIC1-II-R | 5’-TGTGACCAAACTCAACTAGAGGAT-3’ | |
| BIC1-III-F | 5’-CCGATGTATCCCCACCTAAACA-3’ | |
| BIC1-III-R | 5’-AGCATTCAAAGCGGTTGGATTTT-3’ | |
| BIC1-IV-F | 5’-AAAGGGAAACGAATTTGATCGAAGG-3’ | |
| BIC1-IV-R | 5’-TAAAAGTACTTTCTCACGTGGCCGT-3’ | |
| BIC2-I-F | 5’-TCGACTAGACGTTAGATGCTCATGC-3’ | |
| BIC2-I-R | 5’-TCGTTGACGTTTCTACGTCTCATGG-3’ | |
| BIC2-II-F | 5’-ACACTCTCCTTCGTTTTCAACCTTG-3’ | |
| BIC2-II-R | 5’-GGCTTCGACGTGTGTGTTGTATATA-3’ | |
| BIC2-III-F | 5’-CCGATCAATAGCGATTAGAAGAAGC-3’ | |
| BIC2-III-R | 5’-TTATAGGTGGCGGGCAACAA-3’ | |
| BIC2-IV-F | 5’-CTTTTAGAGGGGAAGGGGGC-3’ | |
| BIC2-IV-R | 5’-TGCGAAGCGGTTAAAGATTACT-3’ | |
| NC-F | 5’-ATCGGAGCTCCAATAGGTCTG-3’ | |
| NC-R | 5’-AATAAGATCTAGACGAGAGAG-3’ | |
| BIC1-QPCR-F | 5’-TGGACACTGGGAGAGAGAGG-3’ | RT-qPCR |
| BIC1-QPCR-R | 5’-AGCAGTACGTGCAGACGAGA-3’ | |
| BIC2-QPCR-F | 5’-GTTCCTATACCGGATAGTTGGGG-3’ | |
| BIC2-QPCR-R | 5’- CGCAGCTCGAGCAGAGACAATCTGG-3’ | |
| GFP-QPCR-F | 5’-GGACGACGGCAACTACAAGA-3’ | |
| GFP-QPCR-R | 5’-TGAAGTCGATGCCCTTCAGC-3’ | |
| PP2A-QPCR-F | 5’-TATCGGATGACGATTCTTCGTGCAG-3’ | |
| PP2A-QPCR-R | 5’- GCTTGGTCGACTATCGGAATGAGAG-3’ | |
ACCESSION NUMBERS
CRY1 (AT4G08920), CRY2 (AT1G04400), BIC1 (AT3G52740), BIC2 (AT3G44450), HY5 (AT5G11260), COP1 (AT2G32950), PhyA (AT1G09570), PhyB (AT2G18790).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Markus Kohnen and Christian Fankhauser for providing the MATLAB script for the image-based growth kinetics analysis. This work is supported in part by the National Institute of Health (GM56265 to CL), National Science Foundation of China (31500991 to QW, 31650110478 to YO, 31422041 to BL, and 31371411 to ZZ), Fujian-Taiwan Joint Innovative Center for Germplasm Resources and cultivation of crop (Fujian 2011 Program, [2015] 75), and Rural Development Administration of Republic of Korea (PJ01104001 to JIK). The authors thank the UCLA-FAFU (Fujian Agriculture and Forestry University) Joint Research Center on Plant Proteomics, Haixia Institute of Science and Technology, and Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology for institutional supports.
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- Allen E, Xie Z, Gustafson AM and Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell, 121,207–221. [DOI] [PubMed] [Google Scholar]
- Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF and Pagano M (2007) SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science, 316, 900–904. [DOI] [PubMed] [Google Scholar]
- Casal JJ (2000) Phytochromes, cryptochromes, phototropin: photoreceptor interactions in plants. Photochem. Photobiol. 71,1–11. [DOI] [PubMed] [Google Scholar]
- Chen C-H, DeMay BS, Gladfelter AS, Dunlap JC and Loros JJ (2010) Physical interaction between VIVID and white collar complex regulates photoadaptation in Neurospora. Proc. Natl Acad. Sci. USA, 107, 16715–16720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay KMW and Jenkins GI (2016) Regulation of UVR8 photoreceptor dimer/monomer photo-equilibrium in Arabidopsis plants grown under photoperiodic conditions. Plant Cell Environ. 39,1706–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK and Ulm R (2010) Negative-feedback regulation of UV-B–induced photomorphogenesis and stress acclimation in Arabidopsis. Proc. Natl Acad. Sci. USA, 107, 20132–20137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC and Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science, 279, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Heijde M and Ulm R (2013) Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc. Natl Acad. Sci. USA, 110,1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lee JW, John PC et al. (2012) Identification of small molecule activators of cryptochrome. Science, 337,1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Ouyang X and Deng XW (2014) Beyond repression of photomorphogenesis: role switching of COP/DET/FUS in light signaling. Curr. Opin. Plant Biol. 21, 96–103. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA and bevan MW (1987) GUS fusions: β-glu-coronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohnen MV, Schmid-Siegert E, Trevisan M, Petrolati LA, Sénéchal F, Müller-Moulé P, Maloof J, Xenarios I and Fankhauser C (2016) Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell, 28, 2889–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I and Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell, 19, 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L and Yang HQ (2011) Blue light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25, 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C and Shalitin D (2003) Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 54, 469–496. [DOI] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J and Cashmore AR (1998) Enhancement of blue light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA, 95, 2686–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X and Lin C (2011a) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25, 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Liu B, Zhao C, Pepper M and Lin C (2011b) The action mechanisms of plant cryptochromes. Trends Plant Sci. 16, 684–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang Q, Deng W et al. (2017) Molecular basis for blue light-dependent phosphorylation of Arabidopsis cryptochrome 2. Nat. Commun. 8, 15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP and Liu H (2016) Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl Acad. Sci. USA, 113, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu S-L, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang Z-Y and Quail PH (2014) A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science, 344,1160–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Xu3 S-L, Chalkley RJ, Huhmer AF, Burlingame AL, Wang4 Z-Y and Quail PH (2017) PPKs mediate signal transduction from phytochrome photoreceptors to bHLH factor PIF3. Nat. Commun. 8, https://org/10.1038/ncomms15236 (2017). In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale UV, Huang SSC, Zander M et al. (2016) Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164, 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL and Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18, 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerdtfeger C and Linden H (2003) VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 22, 4846–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC and Lin C (2002) Regulation of Arabidopsis cryptochrome 2 by blue light-dependent phosphorylation. Nature, 417, 763–767. [DOI] [PubMed] [Google Scholar]
- Su J, Liu B, Liao J, Yang Z, Lin C and Oka Y (2017) Coordination of cryptochrome and phytochrome signals in the regulation of plant light responses. Agronomy, 7, 25. [Google Scholar]
- Wang Q, Zuo Z, Wang X et al. (2016) Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science, 354, 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W, Busino L, Hinds TR, Marionni ST, Saifee NH, Bush MF, Pagano M and Zheng N (2013) SCFFBXL3 ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature, 496, 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, Yang H, Lee J, Liu X, Lopez J and Lin C (2007) Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell, 19, 3146–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, He H, Wang X, Wang X, Yang X, Li L and Deng XW (2011) Genome-wide mapping of the HY5-mediated genenetworks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65, 346–358. [DOI] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X and Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 21, 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


