Abstract
Epigenetic dysregulation plays a profound role in the pathogenesis of hematological malignancies, which is often the result of somatic mutations of chromatin regulators. Previously, these mutations were largely considered to alter gene expression in two dimensions, by activating or repressing chromatin states; however, research in the last decade has highlighted the increasing impact of the three-dimensional (3D) organization of the genome in gene regulation and disease pathogenesis. Here, we summarize the current principles of 3D chromatin organization, how the integrity of the 3D genome governs immune cell development and malignant transformation, as well as how underlying (epi-)genetic drivers of 3D chromatin alterations might act as potential novel therapeutic targets for hematological malignancies.
The advent of next-generation sequencing has led to discoveries in 3D genome formation
Research in the last decade fueled by unprecedented progress in next-generation sequencing has laid out the first maps of three-dimensional (3D) genome organization during diverse biological processes, including gametogenesis, embryonic development, stem cell differentiation and lineage commitment [1, 2]. These reports have highlighted the impact of 3D genome organization in governing long-range regulatory interactions for the precise control of gene expression [3–5]. Furthermore, such work has opened up exciting new avenues to investigate the influence of spatiotemporal changes in 3D architecture during normal development and disease. Hematopoiesis -- the process of mature blood cell and immune cell development with its well-defined cell lineages, developmental programs and cell activation stages -- has proven to be an ideal model to address novel questions in the field of 3D chromatin architecture, ranging from the compartmentalization of the genome in immune cell activation [6, 7], to mapping genome-wide cis-regulatory landscapes in immune cells [8]. In this review, we summarize the current principles and advances in 3D chromatin organization, specifically exploring the influence of 3D chromatin organization in immune cell maturation and activation following antigen stimulation. Finally, we discuss how dysregulation of 3D architecture can lead to hematological malignancies, and the implications thereafter.
Principles of 3D chromatin organization
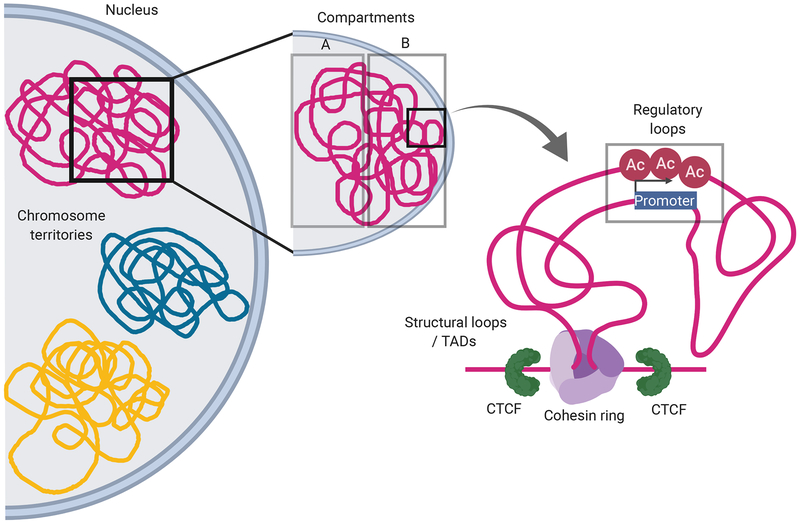
3D organization of mammalian chromatin and its role in gene regulation has been an area of significant interest over the past decade. Based on various techniques, including imaging and chromatin conformation capture approaches followed by sequencing (Box 1), the chromatin architecture can be broadly divided into multiple levels, from chromosomal territories (extensively reviewed in [9–11]), nuclear-lamina associated domains (LADs) [12], transcriptionally active (A) and inactive (B) chromatin compartments, Topologically Associated Domains (TADs), to individual promoter-enhancer contacts (amongst others) [13–16] (Figure 1). At the order of ~1–10 Mb, each chromosome can be divided into two main compartments: transcriptionally active (A), and inactive (B) compartments. These compartments are defined by mutually exclusive chromatin interaction patterns, suggesting certain nuclear co-localization for each compartment. The inactive B compartment is usually considered poor in gene content-, but enriched in repressive histone mark H3K27me3, and thus (generally) transcriptionally inert [13, 16]. Most of the B compartment is localized towards the nuclear periphery [16–18]. By contrast, the active A compartment, facing the nuclear interior, contains gene-rich regions and is considered to constitute most of a cells’ euchromatin. The enrichment of accessible chromatin and activating histone marks, such as H3K27ac, H3K4mel, H3K4me3 or H3K36me3, renders the A compartment highly transcriptionally active. Moreover, shifts from an inactive to an active compartment (or vice versa) usually coincide with cell type-specific gene expression or repression [16–18].
BOX 1-. Chromatin conformation capture (3C) techniques and beyond.
To study either local or genome-wide chromosomal architecture, various versions of proximity-based ligation assays (PLA) known as chromatin confirmation capture approaches (3C, 4C, 5C, HiC, etc) have been developed in past years (reviewed in [85, 86]). PLA use restriction enzyme digestion of crosslinked chromatin followed by ligation in diluted conditions to ligate fragments in spatial proximity.
Hi-C (High-throughput chromosome conformation capture) has enabled the genome-wide identification of chromatin-chromatin interactions, within and across the same chromosome [13]. Chemical cross-linking followed by enzymatic digestion allows the re-ligation of spatially-proximal DNA fragments which can be linearly distant. Next, a sequencing library of all such religation fragments can be prepared and sequenced with high-throughput, and thus, allow for the detection of all-to-all chromatin contacts throughout the genome. Hi-C has been used to identify the mammalian organization of chromatin into A and B compartments [13–16], as well as into smaller structures such as TADs [14, 15, 87], sub-TADs [88] and insulated neighborhoods [89].
Despite sequencing depth being a limiting factor for the resolution of chromatin contacts being studied, with deep sequencing, Hi-C can identify interactions at almost 1 kb resolution [90], thus capturing almost all possible types of 3D chromatin architecture structures known to date. In addition to the limitation of sequencing depth for defining smaller, more fine-scaled structures, Hi-C is also biased by the cross-linking and proximity ligation steps. For example, such limitations do not allow the determination of chromosome positioning within the nucleus. Another approach to overcome the limitations in sequencing depth of Hi-C experiments around specific chromatin contacts may be to identify chromatin loops associated with DNA binding proteins or histone-marks of interest. This has led to the development of conceptually similar experimental assays such as HiChIP (high-throughput chromatin immunoprécipitation (ChiP)) [91], ChlA-PET (chromatin interaction analysis of paired-end tag sequencing) [92] or PLAC-Seq (Proximity Ligation-Assisted ChlP-seq) [93], which make use of protein-specific ChIP to enrich for genomic contacts of interest. ChlA-PET utilizes ChIP followed by proximity ligation to identify chromatin interactions associated with a protein of interest. PLAC-Seq achieves proximity ligation in nuclei prior to chromatin shearing and immunoprécipitation. Although these approaches are more focused and give higher resolutions, they bear their own caveats, including the dependency on target protein abundance, antibody specificity and binding affinity to certain genomic loci, which could bias the measured contact frequency.
Figure 1: Overview of certain 3D chromatin features.
Interphase chromosomes are spatially organized in distinct chromosome territories. Each chromosome is further divided into transcriptionally active A compartment (facing the nuclear interior) and inactive B compartment (facing the nuclear lamina). Cohesin-mediated loop extrusion results in the formation of topologically associated domains (TADs) defined by structural loops enriched in CTCF binding and regulatory loops bringing promoters and enhancers into spatial proximity. The cohesin-complex, a key protein complex involved in chromatin organization, is a ring-shaped protein complex containing the four core protein subunits SMC1, SMC3, RAD21 and STAG. Red circles denoted as ‘Ac’ (acetylation) represent H3K27ac marks, indicating active enhancer elements.
The smaller chromatin structure of self-interacting TADs usually forms genomic insulated neighborhoods of ~500 kb-1,000 kb in size. The insulation refers to reduced chromatin contacts across TAD boundaries, and thus, mostly restricts chromatin contacts within TADs. As a consequence, cis-regulatory elements (herein termed ‘enhancers’) are only able to access gene promoters within the same TAD, but not across TAD boundaries [19–21]. The formation of TADs is largely invariant across cell types, and even appears highly conserved across related species [22]. TAD boundaries are highly enriched in binding of the insulator protein CCCTC-binding factor (CTCF) and strong co-localization of the cohesin complex [23–25]. TAD boundaries are predominantly formed between pairs of CTCF bindings in convergent orientation [16]. Using chromosome conformation capture approaches (e.g. Hi-C and 4C-Seq) (box 1), several groups have experimentally shown that either depletion of CTCF protein in mouse embryonic stem cells (mESCs), disruption of the CTCF binding site, or its mere switch in orientation in mESCs and human SK-/V-SH cells can lead to abrogated TAD formation [24, 26, 27]. Computational analysis of sequencing-based approaches to identify the discussed chromatin features is conceptually complex and has led to the development of various software packages to deal with individual analytic tasks (Box 2).
BOX 2-. Computational analysis of Hi-C data.
The computational demand for the analysis of complex data is rising, and includes genome-wide chromatin conformation capture techniques followed by sequencing, requiring thorough and robust assessment of the results. The concepts and potential pitfalls of the computational analysis of Hi-C data and related assays has been extensively reviewed [94–96], but shortly summarized here. Allowing users to analyze raw data by guiding the user through each necessary step from read alignment and normalization to downstream analyses and visualization, computational pipelines have been developed previously, such as hic-bench [97] juicer/juice-box [98], HiC-Pro [99] and HiC-explorer [100], to name a few. The identification of compartments was first described using eigenvector analysis to define two distinct classes of mutual exclusive interactions [13], and implemented for example in the c-score tool [101]. This computational analysis using eigenvector decomposition of Hi-C contact matrices to define chromatin compartmentalization. Other algorithms, such as hic-ratio [97], TopDom [102], TADtree [103] and Amartus [104], were developed to reveal TADs, using similar approaches of defining genome-wide ‘insulation scores’ to detect local maxima capable of defining a TAD boundary. Insulation scores are computational measurements of the capacity of a single genomic position to insulate its upstream area from its downstream area in 3D space. Within TADs, the detection of individual loops can also be achieved with tools such as mango [105] or FitHiC [106], with various specializations being developed for other techniques such as HiChIP or ChlA-PET. Other applications for the analysis of Hi-C involve the detection of structural variants such as copy-number variants [107] or chromosomal translocations [84].
Loop domains are formed via loop extrusion
Individual contacts between any two genomic positions in the range of ~10 kb up to ~1 Mb are often referred to as chromatin ‘loops’. These loops can be broadly divided into two categories, the structural loops (sometimes referred to as ‘domain loops’) and regulatory loops. Domain loops demarcate the contact between two TAD boundaries due to the strong binding of CTCF proteins [13, 25, 28]. Regulatory loops facilitate contacts between gene promoters and cis-regulatory elements, such as typical enhancers or super-enhancers, which can also be strengthened by additional structural factors such as eukaryotic YY1 [29]. YY1 binds both promoters and active enhancers and shows similar properties as CTCF, including the ability to dimerize. YY1 was thus implicated in the formation of individual promoter-enhancer contacts; CRISPR-mediated deletion of YY1 binding motifs in enhancers led to reduced promoter-looping in mESCs relative to controls [29]. Currently, the widely-accepted model for loop formation is the ‘loop extrusion model’. Therein, a single cohesin ring is loaded onto DNA, which is actively extruded through the cohesin ring in an ATP-dependent manner [28], As a result, a DNA loop is formed to bring two linearly distant DNA elements into direct contact at the site of the cohesin ring [30–32]. Extrusion via cohesin can be blocked through oriented binding of structural proteins, such as CTCF and YY1, and thus form a temporarily stable contact between the two DNA elements encircled by cohesin [30–32]. Further Hi-C evidence supporting this model has shown that deletion of cohesin release factor, WAPL, diminishes local intra-TAD contacts and thus dynamic loop formation in human FIAP1 cells, resulting in an enrichment of nearly all, long-range loops [33]. In addition, other studies have shown that complete loss of loop extrusion via deletion of cohesin subunit RAD21 in human colorectal carcinoma cell line FICT116 results in little to no detection of structural loops in Hi-C data [25]. These reports together support loop extrusion as a primary mechanism of loop formation that contributes to modulating chromatin organization and transcriptional events.
Phase-separated condensates around super-enhancers significantly boost transcription
Complementary to loop extrusion as a mechanism of establishing genomic interactions critical for the regulation of gene expression, others have described a synergistic formation of condensates established by liquid phase-separation, especially around highly regulatory ‘superenhancers’ [34–36]. This model proposes the formation of highly dense conglomerates of transcription factors, nucleic acids, and co-activators such as MED1 or BRD4, displaying a collective binding affinity beyond what would be expected from interactions simply driven by DNA-sequence motif recognition and the orderly recruitment of such factors [34]. This idea is consistent with the formation of processing ribonucleoprotein complexes (p-bodies) and stress granules, which also act to compartmentalize critical regulatory processes into biochemically-distinct condensates [37, 38]. Specifically, the co-factors MEDI and BRD4 share large domains called ‘intrinsically disordered regions’ (IDRs) with other proteins known to phase-separate. Thus, MEDI and BRD4 themselves have been hypothesized to harbor the potential of forming nuclear condensates around super-enhancers via liquid phase-separation [35, 36]. These reports have led to new models aiming to explain different aspects of super-enhancer formation, activation, and sensitivity to perturbations of super-enhancer high-density coactivators [34].
3D architecture in immune cell maturation and leukemia development
The immune system has emerged as an ideal system to study 3D chromosomal landscape alterations as it includes multiple well-defined lineages, extensively characterized developmental programs, and a large variety of activation stages. Also, the abundance of immune cells in peripheral blood, bone marrow and peripheral lymphoid organs, allows for easy access to experimental samples from both animal models (e.g. rodents) and humans. Finally, the immune system can lend ‘a ground’ for the development of a large number of neoplasms, including leukemias and lymphomas. These are clonal hematopoietic neoplasms characterized by the proliferation and accumulation of hematopoietic progenitor cells (FIPCs) that impede normal blood production and major organ function [39]. Gene discovery studies have identified a number of recurrent and targetable genetic and epigenetic lesions contributing to disease pathogenesis. These genetic lesions affect both the normal differentiation of hematopoietic progenitors into mature lineages, while endowing these progenitor cells with uncontrolled pro-survival and pro-proliferative capacity. For example, activating mutations in transmembrane protein NOTCH1 and its signaling pathway (targeting for example FBXW7) can result in T cell maturation arrest leading to continuous proliferation of immature lymphocytes, ultimately causing T cell acute lymphoblastic leukemia (T-ALL) [40, 41]. Flence, to understand the role of chromatin architecture in hematological malignancies, it is essential to first understand the changes in the 3D landscape occurring throughout lineage differentiation -- from healthy hematopoietic stem and progenitor cells, to functional immune cells.
A role for 3D genome organization in hematopoiesis and lymphocyte maturation
Recent reports have addressed how the 3D chromatin landscape changes specifically during T cell and B cell developmental programs [42, 43]. The development of a competent and diverse T cell receptor repertoire involves a complex, step-wise process whereby bone marrow hematopoietic progenitors must egress to the thymus to commit to the T cell lineage. These committed thymocytes undergo multiple rounds of genomic deletion and proliferation in order to differentiate into mature T cells bearing a fully-functional T cell receptor (TCR) [44, 45]. Using the well-characterized αβ T cell differentiation program in WT (WT) C57BL/6 mice as a model system, 3D chromatin changes were examined along eight intermediary states; from hematopoietic stem cells (FISCs), to double negative (DN) CD4− CD8− thymocytes, and then to TCR-expressing CD4+CD8+ double-positive (DP) thymocytes [42] (Figure 2a). The authors of this study observed that the most significant changes in 3D organization occurred at the transition stage of DN2-DN3 (the point of T cell lineage commitment), and at the DN4-DP transition (a key step for (β-chain selection and completion of TCRβ gene rearrangement). Specifically, chromatin reorganization during T cell development was found to lag behind transcriptional responses; this is relevant as it might suggest that 3D chromatin architectural changes are not drivers of gene expression changes, but rather, that they ensure lineage commitment, preventing cells from reverting back to immature states, or from being redirected to alternate lineages. Alternatively, as these populations are heterogeneous and pass through various transitions with different kinetics, it is possible that only singe-cell Hi-C approaches could correctly estimate the impact of 3D changes on transcriptional programs [46]. However, robust testing is warranted to fully distinguish these possibilities.
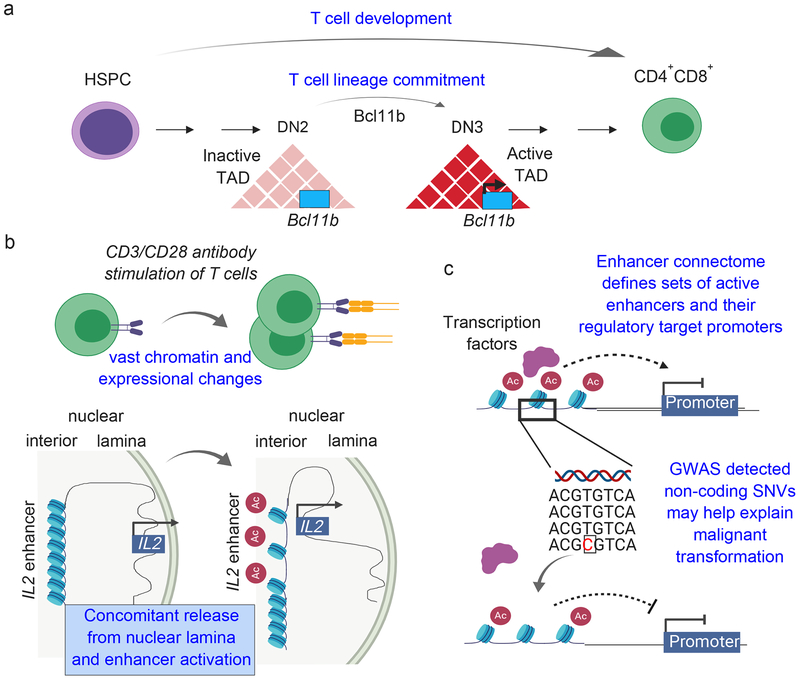
Figure 2: 3D architectural changes during human and mouse normal hematopoiesis and immunity.
a) T cell maturation is associated with widespread changes from hematopoietic stem and progenitor cells (HSPC) to mature T cells, but especially during the double-negative (DN) CD4 CD8’ DN2 to DN3 transition. Specifically, a topologically associated domain (TAD) harboring T cell lineage commitment factor Bc/11b gains TAD connectivity during this transition, and is associated with transcriptional activation of Bc/11b [42]. Red triangles and rectangles represent 3D chromatin interactions between the linear genome represented as a black line, b) T cell activation via transmembrane antibody stimulation results in rapid cell proliferation, and requires alterations in chromatin accessibility, enhancer activity and chromatin loops. For example, in human Jurkat T cells, positioning of IL2 and its enhancer element locus, transitions from a highly repressive environment at the nuclear periphery into the nuclear interior, along with enhancer activation, due to more accessible chromatin. Such rewiring allows upregulation of IL2 during T cell activation [7]. c) The genome-wide enhancer-promoter interaction landscape in human T cell subsets can be defined using H3K27ac HiChIP (red circles denoted with ‘Ac’ represent H3K27ac marks). In this diagram, specific enhancer sequences are overlaid with noncoding mutations associated with certain autoimmune diseases, and tested for expressional changes upon CRISPR-Cas9-mediated silencing in corresponding T cells from healthy individuals [8]. Such non-coding mutations might help explain their putative involvement in gene expression and malignant transformation. SNV: single nucleotide variant
Furthermore, when describing a role for lineage defining transcription factors in the regulation of the 3D nucleome, T cell lineage commitment transcription factor, Bc|11b, was also shown to modulate 3D organization in mice during the αβ T cell differentiation program [42]. Specifically, a TAD harboring Bc|11b itself, along with TADs displaying high Bc|11b binding at the DN3 stage gained more intra-TAD interactions at the DN2-DN3 transition relative to other T cell maturation stages; this suggested a specific role for Bc|11b in regulating the chromatin interaction landscape of thymocytes in C57BL/6 mice during T-cell lineage commitment (Figure 2a) [42]. This increase in chromatin interactions correlated with increased expression of Bc|11b and its targets in the DN3 stage, relative to other distinct stages of T cell maturation. Bc|11b transcription itself was also previously reported as being regulated at the DN2 stage by large-scale changes in the nuclear architecture, including the repositioning of a Bc/11b enhancer from the nuclear lamina to the interior in mice during T cell development [47]. This repositioning of the enhancer element regulating Bc|llb expression is orchestrated by c/s transcription of a long non-coding RNA, ThymoD [47], ThymoD transcription favored binding of cohesin and CTCF proteins that allowed the juxtaposition of the Bc/11b enhancer and promoter into a single loop domain to initiate transcription of Bc|11b. To elucidate the mechanism by which ThymoD transcription promoted CTCF binding, DN2 cells were isolated from WT ThymoD+/+ and ThymoDp(A)/p(A) mice, where insertion of a poly A site interfered with ThymoD transcription [47]. DN2 cells from ThymoDp(A)/p(A) mice (the latter frequently developing T cell leukemias and lymphomas), harbored several hypermethylated CpG residues within the CTCF binding sites in the Bc/11b locus relative to DN2 cells from WT ThymoD mice. The findings also suggested that ThymoD anti-sense transcription was associated with demethylation events essential for the recruitment of CTCF and cohesin to the Bc/11b enhancer region, thus facilitating Bc/11b enhancer-promoter looping, and the subsequent initiation of Bc/11b transcription in mouse T cells, which would be relevant for the potential initiation of lymphomagenesis [47].
Further supporting a role for lineage-defining transcription factors in the establishment and maintenance of genome organization, another recent report identified transcription factor Paired Box 5 (Pax5) as being critical for the global lineage-specific architecture of B cells [43]. Pax5 is a well-known B cell commitment transcription factor that activates B cell specific genes and represses genes promoting other immune cell lineages [48]. By correlating differential interactions detected by in situ Hi-C with Pax5 binding data, along key distinct stages of B cell development in C57BL/6 mice, a clear role for Pax5 in transcriptional control via 3D interactions at the pro-B cell stage was elucidated. The authors found that most interactions associated with binding of Pax5 specifically in pro-B cells were weakened during B cell differentiation. Furthermore, comparison of WT and Pax5−/− pro-B cells identified differential 3D contacts predominantly in the immunoglobulin heavy chain (Igh) locus. This report corroborates previous findings describing a role for Pax5 in the reorganization of the Igh locus in pro-B cells, promoting chromatin looping over long distances to facilitate locus contraction in order to regulate target gene expression [43]. Indeed, locus contraction is the process where for example, immunoglobulin (or TCR) V (variable) genes move closer to the J (joining) genes, providing spatial proximity leading to VD(J) recombination (D; diversity), an essential step for the generation of a diverse antibody repertoire [49, 50]. Collectively, these studies validate the concept that specific changes in 3D architecture during hematopoiesis and lymphocyte maturation also require lineage-defining transcription factors such as Bc|11b and Pax5 to modulate gene transcription and mediate B and T cell lineage commitment (extensively reviewed in [51]). However, of note, these master transcription factors do not possess intrinsic chromatin remodeling activity and hence, understanding the underlying mechanisms by which lineage commitment factors influence 3D organization remains an interesting future question.
Examples of chromatin changes during adaptive immunity
Germinal Center Responses
3D chromatin changes have also been observed during adaptive immune responses. Widespread genomic architectural changes are observed in germinal center B cells, arising upon activation of naive B cells. These 3D chromatin changes occur amongst genes that specify the germinal center B cell phenotype [6], For instance, the genomic locus on human chromosome 3 encoding the B cell lymphoma 6 transcription factor (BCL6) -- a master regulator of the germinal center (GC) B cell program -- is governed by multiple layers of 3D reorganization in human GC B cells relative to naive B cells, including increased promoter connectivity, increased interactions with an active enhancer/locus control region (LCR) located upstream of the BCL6 gene, and 5´ to 3´ gene looping (measured by Hi-C) [6]. Indeed, several genes such as CADPS, LRCH3, GPR160, LPP, BFSP2 and MME that gain interactions with the BCL6 promoter or LCR have been described as genes enriched in GC survival pathways [6]. Moreover, CRISPR-Cas9-mediated deletion of a syntenic LCR in the germline of C57BL/6 mice located in chromosome 16, 165 kb upstream of the Bc/6 gene, resulted in failed GC formation (as evidenced from immunohisto-chemical staining of Bcl6 and Ki-67, following T-cell dependent immune activation with sheep red blood cells in vitro; these findings highlighted the importance of this LCR as a hub for chromatin interactions that could contribute to establishing the GC B cell program in humans and mice [6].
T Cell Activation
Similarly, the ability of T cells to rapidly respond to antigen stimulation with significant expressional changes and proliferation has raised questions about the impact of chromatin alterations in stimulated T cells. For instance, heterochromatin-rich regions of total resting T cells isolated from C57BL/6 mice splenocytes, were reorganized following activation with anti-CD3 antibodies, rendering chromatin more accessible to transcriptional activation, thus enabling rapid gene expression [52] (Figure 2b). Moreover, in human naive Jurkat T cells, despite the fact that numerous immediate-responding genes already reside in active compartment A, other immune response genes, such as IL2 or NFkbIZ, found in proximity to the chromatin-repressive nuclear lamina, can be released from released from this compartment following activation with human Raji B cells. [7] (Figure 2b). Indeed, the spatial localization of these genes transitioned from the nuclear lamina in the periphery, towards the nuclear interior, subsequently becoming accessible (permissive) for transcription factor binding and transcription initiation [7]. In this study, due to the loss of repressive histone marks in repositioned LADs towards the nuclear interior, 513 out of 974 enhancer clusters -- previously assumed to be functional during human T cell activation [53] – were indeed activated [7]. The authors suggested that late-stage enhancer activation might potentially allow for chromatin contacts with nearby genes, such as BTLA, a gene responding to T cell activation with increased expression, and previously reported to be involved in attenuating T cell activation and immune responses [54]. In line with this report, fine-layered promoter-enhancer interactions are extensively rewired during T cell stimulation, as evidenced from the Trac-looping method [55]. Contrary to Hi-C, Trac-looping is independent of cross-linking, due to the use of transposable linker sequences that specifically insert into accessible chromatin via Tn5 transposase [55]. Because of the use of bivalent linker sequences, these will interact at genomic loci that are of spatial proximity, allowing for paired-end sequencing of chromatin interactions at a genome-wide level [55]. An example for coinciding chromatin accessibility gain and chromatin looping in anti-CD3/CD28 activated human CD4+ T cells relative to naive human CD4+ T cells is focused around the IL2 locus, which can also be released from repressive nuclear lamina during activation of human Jurkat T cells [7] –suggestive of a potentially more accessible environment in activated T cells. In this study, multiple distal enhancers for IL2 gained chromatin accessibility, and looping changes were confirmed by various techniques (Figure 2b); however, no functional link has been demonstrated between IL2 enhancer activity and T cell responses. The authors also investigated whether specific DNA binding protein motifs were enriched in the anchors of chromatin interactions gained during T cell activation, based on the observation of an association between an open chromatin state and looping (using Trac-looping) [7]. The motifs were associated with binding of FOS transcription factor family members, which were also transcriptionally upregulated in anti-CD3/CD28 activated human CD4+ T cells relative to naive human CD4+ T cells; these motifs were specifically enriched in gained chromatin loops in activated T cells, suggesting an involvement of FOS factors in looping changes during T cell activation [55]. However, whether FOS family proteins can act as DNA binding factors capable of opening chromatin to recruit additional transcription factors and to induce DNA looping, or whether additional co-factors are required for such changes, remains an open question.
Assessing T Cell Subsets
To dissect different processes of enhancer looping in human naive T cells, regulatory T cells (Treg) and T helper 17 (Thl7) cells, a study used FI3K27ac HiChIP, to specifically define chromatin interactions of active enhancers and promoters [8]. Despite focusing on genes exclusively expressed in specific cell types, the data suggested that events called ‘enhancer skipping’ or ‘enhancer switching’ were occurring; namely, in the former scenario, an enhancer preferentially loops with a more distant promoter, while for enhancer switching, one gene remains highly active but its expression is mediated by another set of enhancers [8]. Using the enhancer connectome map as a reference for human naive, regulatory and Thl7 T cells, the authors linked single nucleotide variants (SNVs) -- from genome-wide association studies (GWAS) – first, with enhancer activity, and then used the chromatin interactions to associate these enhancers with regulated genes such as IL18RAP, IL18R1, IL1RL1, and IL1RL2 (Figure 2c). SNVs located in non-coding regions that were prevalent in patients with 21 different autoimmune diseases (including Type-1 diabetes (T1D), celiac disease, multiple sclerosis, rheumatoid arthritis and other) were found to be ‘rewired’ in a cell-type specific manner in human naive, regulatory and Thl7 cells (as evidenced from enhancer contacts). This rewiring suggested a possible role for chromatin remodeling and gene expression in the pathogenesis of autoimmune diseases originating from distinct T cell subsets, such as Tregs or Thl7 cells [8]. However, a direct link between the complex interplay of SNVs, cell-type specific enhancer looping in T cell subsets, associated genes and respective autoimmune diseases remains to be assessed. Nevertheless, as an example, a specific SNV located within the genomic sequence of a Thl7-specific enhancer of ID2 was associated with T1D. CRISPR-Cas9-mediated silencing of this enhancer in a human My-La CD4+ cell line using complementary guide RNAs to the enhancer sequence, confirmed a loss of expression of the target gene ID2 [8]. This finding suggested that the TID-associated non-coding SNV might potentially modulate ID2 expression by inactivating the enhancer activity and thus, potentially be implicated in the development or progression of human T1D, although this remains highly speculative at present. Nevertheless, taken together, these reports highlight the complex interplay of epigenetic and 3D chromatin changes that are necessary to regulate gene expression, and to potentially mount an effective immune response -- however, the latter remains to be demonstrated by functional experiments.
The role for 3D architecture in immune memory constitutes an area of current and future investigations. For instance, comparisons between human naive and memory CD8+ T cells have identified distinct epigenetic profiles associated with chromatin accessibility, as well as histone and DNA modification patterns, correlating with the transcriptional profiles and primed states of memory CD8+ T cells, as evidenced from integrated ATAC-Seq, RNA-Seq and ChlP-Seq datasets [56–59]. Thus, a significant question is whether these epigenetic differences are accompanied by changes in 3D genome architecture, and whether these might be related to the intrinsic ability of memory CD8+ T cells to rapidly reactivate upon challenge, and in a context-dependent manner. Preliminary 3D maps using promoter capture Hi-C in 17 primary human hematopoietic cell types, including human naive CD8+ T cells and a heterogeneous pool of total human CD8+T cells, have identified cell type-specific promoter-interacting regions [60], albeit lacking in-depth functional characterization of these differences. Indeed, high resolution genome-wide chromatin interaction maps of specific CD8+ (or CD4+) T cell subsets will be invaluable in accurately identifying and interpreting enhancer activities and gene expression, and their effects on memory cell priming and responses in the context of health and disease.
The Importance of the 3D architectural alterations in hematopoietic malignancies
Given the importance of structural chromatin rearrangements in immune cells during normal hematopoiesis and antigen-activation, disruption of the 3D chromatin landscape under certain circumstances, has been postulated to lead or contribute to, impaired hematopoiesis, differentiation and activation/function. Thus, these changes have also been implicated in the initiation and progression of certain hematological malignancies. For instance, a primary report identified recurrent microdeletions in T-ALL genomes spanning boundary regions of insulated neighborhoods containing prominent oncogenes associated with T-ALL pathogenesis such as TALI and LMO2, thus suggesting that significant changes in 3D chromatin structure might also constitute a potential mechanism for promoting leukemia [61]. The authors recapitulated some of these deletions in vitro in human embryonic kidney fibroblasts by CRISPR-Cas9-mediated genome editing; these deletions led to new long-range regulatory gene interactions and subsequent upregulation of the proto-oncogenes TALI and LMO2 [61], suggesting that genetic alterations that disrupt insulated neighborhoods can lead to oncogene activation in malignant cells.
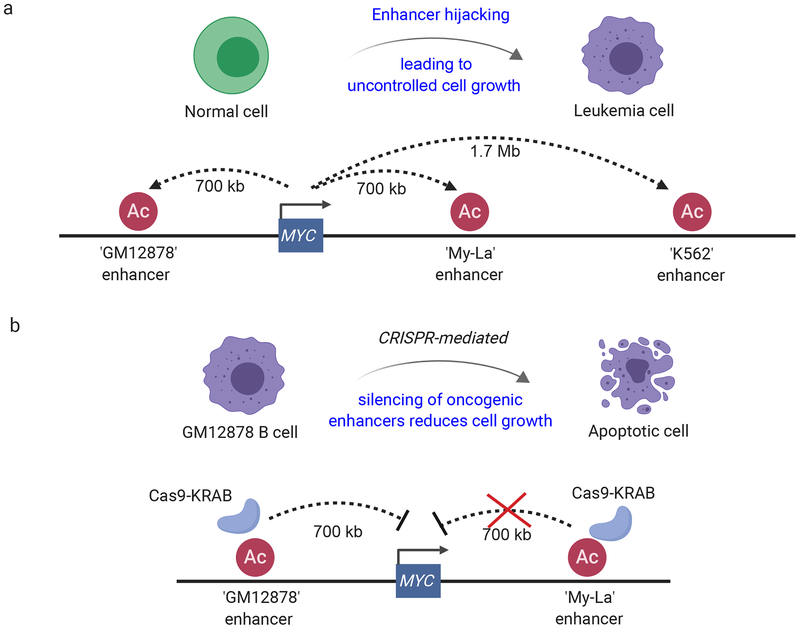
Another relevant example is that of the proto-oncogene MYC, a prevalent driver of proliferation in various cancer sub-types in mammals [62], MYC has a multitude of known proximal and distal enhancer elements that regulate MYC expression in various cancers in a cell type specific manner [63]. For example, the MYC promoter loops with a downstream enhancer cluster approximately 1.4 Mb to 1.7 Mb downstream of the MYC promoter in mice and humans [64–66] (Figure 3a). This cluster of enhancers was described as important for normal hematopoiesis, as well as in various leukemia mouse models, including acute myeloid leukemia (AML), chronic myeloid leukemia (CML) and T-ALL [64–66]. Specifically, upon partial or full deletion of the enhancer cluster in a C57BL/6 mice background, flow cytometric analysis identified an accumulation of multipotent progenitor cells, and depletion of differentiated hematopoietic cells, pheno-copied a murine knock-out of MYC in hematopoietic stem cells [64–66]. Thus, hijacking the MYC enhancer cluster (or any element within) can upregulate MYC expression and increase predisposition to hematopoietic malignancies, at least in mice [64–66] (Figure 3a). A recent study demonstrated cell-type specific looping of MYC in human cell lines K562 (CML), My-La (CD4+) and GM12878 (B cell lymphoblast) using H3K27ac HiChIP, further supporting a cell type specific enhancer landscape capable of regulating MYC expression [8], By performing virtual 4C analysis on the FI3K27 HiChIP data, the authors confirmed robust looping of the MYC promoter with the 3’ super-enhancer element ~1.7 Mb distally in K562, as well as more proximal downstream looping of the MYC promoter in My-La CD4+ T cells [8]. Contrary to these observations, the MYC promoter in GM12878 displayed robust interactions with an upstream enhancer that was spatially located in a different TAD. Furthermore, the use of CRISPR-Cas9-mediated silencing of the enhancer in GM12878 cells -- but not the enhancer in My-La cells --led to significant down-regulation of MYC expression and reduced proliferation relative to WT GM12878 cells, comparable to targeting the MYC promoter directly in GM12878 cells. These findings validated the regulatory nature of these enhancer-promoter interactions due to a lack of transcriptional inhibition of MYC, when silencing enhancer elements not in spatial contact with the promoter (Figure 3b) [8]. These reports collectively suggest that targeting cell-type specific enhancer elements can constitute a putative mechanism to perturb malignant transformation in a cell type specific manner (e.g., of one but not another cell type).
Figure 3: Complex regulation of proto-oncogene expression can be hijacked in human leukemogenesis.
a) The proto-oncogene MYC is an important driver of proliferation in various leukemias. Its transcriptional regulation is mediated by a multitude of enhancers, represented by H3K27ac HiChIP data from human cell lines K562 (CML), My-La (CD4+) and GM12878 (B cell lymphoblast), which are often ‘hijacked’ in tumors, leading to uncontrolled MYC expression and thus, cell proliferation [8], A distal enhancer cluster on chromosome 8 ~1.7 Mb downstream of the MYC promoter is known to be important during normal hematopoiesis and has been associated with MYC overexpression in human and mouse AML, T-ALL and CML [66]. b) CRISPR-Cas9-mediated silencing using a Cas9-fusion with transcriptional inhibitor domain of KRAB of a GM 12878-specific enhancer but not My-La specific enhancers in GM12878 cells, has been shown to result in reduced MYC expression leading to increased cell death (apoptotic cell) relative to controls [8].
Mutations in structural proteins driving 3D changes in leukemia
Given the importance of structural proteins such as CTCF and cohesin in the formation of 3D chromatin structures, it is not surprising that many malignancies have been linked to various mutations in both CTCF and cohesin [67–69]. For instance, during hematopoiesis, loss-of- function mutations of cohesin have proven to impair differentiation and increase self-renewal and proliferation of human and mouse hematopoietic stem and progenitor cells relative to WT cells [70–72]. Specifically, as demonstrated by these studies, impaired cohesin function accelerated the initiation of myeloproliferative-like disorders with increased extramedullary hematopoiesis and splenomegaly in vivo in mouse models in the background of inducible knockdown of cohesin subunits Rad21, Smcla, Stag2 and conditional knockout of Smc3 relative to controls, suggesting a tumor suppressive role for cohesin in the myeloid lineage [70–72] (Figure 4a). Despite evidence of little impact on overall transcription and chromatin accessibility in conditional cohesin knock-out systems in mice and humans [25, 73], one study reported a link between specific transcriptional responses to inflammation and cohesin mutations in mouse macrophages [73]. Myeloid TLR4-dependent cell activation of mouse macrophages via bacterial cell wall component lipopolysaccharide (LPS) treatment was directly impaired by cohesin mutations compared to WT mouse macrophages, mainly due to a lack of enhancer reorganization around IFN signaling genes, such as Egr2, Ifnarl and Cebpb, as evidenced from ATAC-Seq, FI3K27ac and FI3K4mel ChlP-Seq [73]. The lack of IFN production in cohesin-deficient mouse macrophages led to failed signaling and expression of inducible genes relative to WT cells; this response was partially rescued by direct IFN-beta treatment in cohesin-deficient cells [73]. Moreover, expression data from the TCGA database for AML patients [69] aligned with these findings; indeed, a subgroup of TCGA-profiled AML patients with cohesin mutations lacked expression of inflammatory genes, in particular IFN-pathway genes relative to healthy individuals [73]. As stated, the cohesin complex and the insulator protein CTCF are important players in the 3D nucleome, and an aberrant function of these is therefore likely involved in disease pathogenesis. Thus, the impairment of a proper immune response in the myeloid lineage upon cohesin loss-of-function mutations might contribute to explaining the reduced differentiation potential of monocytes and, presumably, a predisposition for leukemogenesis in AML, although this possibility warrants robust testing.
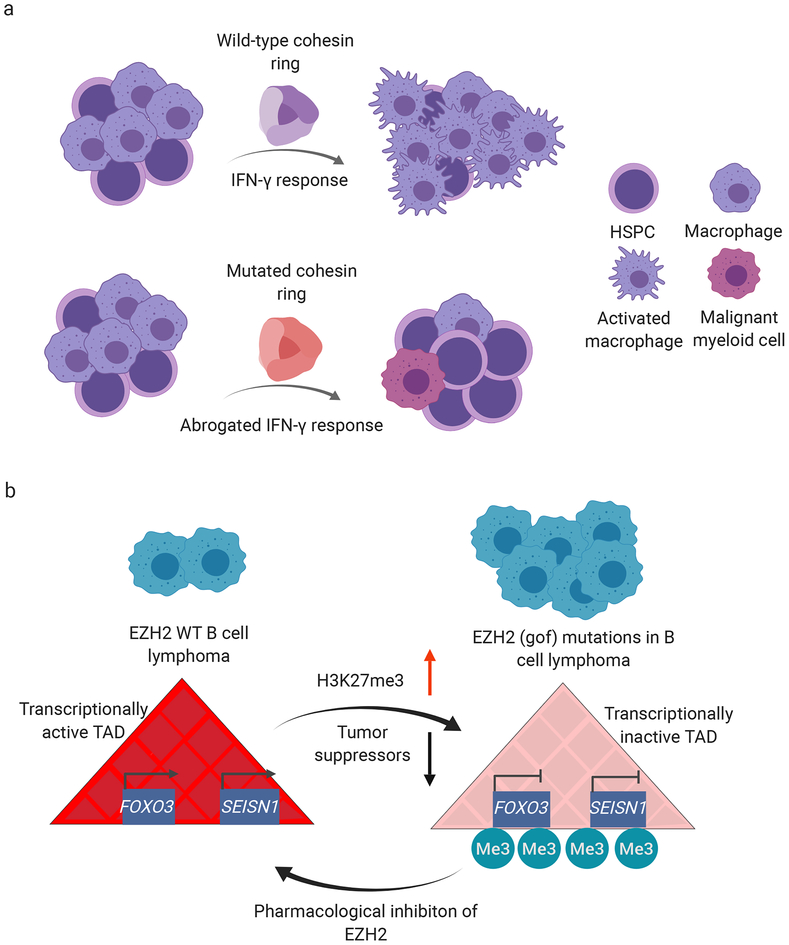
Figure 4: Mutations in key epigenetic regulators can promote human hematopoietic malignancies.
a) Cohesin is important in mediating an effective immune response within the myeloid lineage, leading to activation of macrophages in response to extracellular stimuli. Loss-of-function mutations in cohesin can specifically inhibit IFN responses and myeloid activation, which may promote a predisposed environment for leukemogenesis due to uncontrolled growth of progenitor cells, b) EZH2 gain-of-function (gof) mutations affecting amino acid Tyr646 (e.g. EZH2y646x) are highly prevalent in B cell lymphomas and are known to vastly increase H3K27me3 marks genome-wide. Such hyper-methylation phenotype interferes with transcriptional output of certain topologically associated domains (TAD), for example simultaneously inhibiting tumor suppressors FOXN3 and SESN1, which reside in the same TAD, in EZH2Y646X mutant lymphoma cell lines Karas422 and WSU-DLCL2 compared to EZH2 WT lymphoma cell line OCI-Lyl9. Pharmacological inhibition of the methyltransferase activity of EZH2 in EZH2Y646X lymphoma cell lines with GSK126 and EPZ6438 can revert both H3K27me3 as well as transcriptional inhibition within TADs. Red arrow indicates increase of repressive H3K27me3 marks, black arrow indicates reduction of tumor suppressor expression when comparing EZH2 WT with EZH2Y646X lymphoma cells [74]. Me3: tri-methylation
Mutations in epigenetic players driving 3D changes in hematopoietic malignancies and solid tumors
In addition to chromatin structural variations, proteins that regulate the epigenetic landscape can also alter the nature of 3D chromatin architecture, for instance, by altering histone modifications, DNA accessibility or DNA methylation. This was recently documented in human non-Hodgkin lymphoma cell lines Karpas422 and WSU-DLCL2, harboring gain-of-function mutations affecting Tyr646 in the catalytic domain of the zeste homolog 2 gene (EZH2) [74]. EZH2 is the catalytic methyltransferase component of the Polycomb repressive complex (PRC2). Gain-of-function mutations in EZH2 can lead to a hyperactive methyltransferase enzyme in lymphomas, in turn leading to genome-wide increases in repressive Histone-3 K27 trimethylation (H3K27me3) and depletion of active chromatin marks (Figure 4b) [74]. This study correlated widespread changes in H3K27me3 with 3D chromatin architecture (specifically TADs), as evidenced from genome-wide Hi-C data comparing data from human B cell lymphoma cell line OCI-Lyl9 (EZH2 WT) with EZH2Y646X gain-of-function mutant human B cell lymphoma cell lines Karpas422, WSU-DLCL2. The authors reported that gain-of-function mutations of EZH2 correlated with silencing of entire TAD domains in terms of gene transcription, and hypothesized that this could potentially synergistically inactivate multiple tumor suppressors in non-Hodgkin lymphoma [74], Furthermore, silencing of TAD domains was associated with changes in intra-TAD gene promoter interactions. As an example of tumor suppressor silencing, the authors reported that a single TAD domain containing F0X03 and SESN1 -- among other genes -- was partially inhibited by H3K27me3 in EZH2Y646X mutant cell lines relative to EZH2 WT cell line OCI-Lyl9. This also led to reduced long-range chromatin interactions between the promoters of FOXO3 and SESN1 within the reported TAD (Figure 4b). Moreover, individual as well as combined knock-down of Foxo3 and Sesnl in mouse HPCs isolated from Eμ-myc mice, and re-transplanted into WT mice, accelerated lymphomagenesis in vivo and decreased recipient animal survival significantly when compared to WT animals [74]. In addition, pharmacological inhibition of methyltransferase activity of hyperactive EZH2 with GSK126 or EPZ6438 restored chromatin interactions and gene expression to WT levels, suggesting that the underlying chromatin reorganization might contribute to the disease phenotype in this model (Figure 4b) [74].
Other examples of epigenetic players include mutations in genes DNMT3A, TET2, IDH1 and IDH2 (among others) that regulate DNA methylation at CpG dinucleotides; indeed, these are frequent initiating events of AML [69]. DNMT3A is a de novo methyltransferase whereas TET2 catalyzes the conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosines (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) in the genome, and these modifications are considered as key intermediates in the process of active DNA demethylation [75, 76]. Gain-of-function mutations in the gene encoding iscocitrate dehydrogenase (IDH) in gliomas have been shown to lead to the production of the oncometabolite 2-hydroxyglutarate which inhibits hydroxylases -- including TET enzymes -- by competing with α-ketoglutarate, a cofactor of these proteins [77, 78], This can result in a hyper-methylated phenotype in AML and glioma samples harboring IDH mutations [78], Mutations in IDH and TET genes are mutually exclusive, and hyper-methylation of cytosines within CTCF biding motifs can disrupt binding of the insulator protein CTCF [79–81], suggesting that changes in DNA methylation might directly alter TAD boundaries and loop formation. In support of this hypothesis, increased DNA methylation at the CTCF site and reduced CTCF binding to hypermethylated CTCF sites resulted in partial disruption of a TAD structure in IDH mutant human gliomas relative to gliomas with WT IDH1 [4]. In this study, TAD disruption led to the activation of oncogenic driver PDGFRA (encoding this receptor tyrosine kinase) by enhancers located outside the normal TAD of PDGFRA, resulting in increased expression of PDGFRA in IDH mutant gliomaspheres relative to controls. However, it remains to be investigated whether similar mechanisms of tumorigenesis can be observed in leukemias. In summary, these studies have laid the groundwork for concepts of how genetic aberrations in epigenetic players, such as EZH2 or TET enzymes, can lead to disruption of the 3D genome, resulting in malignant cell transformation due to an inability to control tumor suppressor and/or oncogene expression.
Concluding Remarks
Since the advent of high-throughput assays for the analysis of chromatin conformation during the past decade, numerous studies have confirmed various roles for epigenetic and chromatin regulators in normal hematopoiesis and malignant transformation. These epigenetic and chromatin regulators are mainly implicated in the transcriptional control of cell lineage determining factors or can act as initiating events in tumorigenesis. Genetic mutations targeting such regulators can render tumor suppressor genes inactive, e.g. via uncontrolled spread of H3K27me3 in the case of EZH2-mutant lymphomas, and/or, can contribute to reducing adequate immune responses, e.g in the case of cohesin-deficient AML. However, few reports to date have addressed the direct involvement of such aberrations in the specific regulation of key oncogenes during tumorigenesis. Recent pan-cancer analyses on primary cancer patient samples using a novel computational framework termed cis expression structural alteration mapping (CESAM) has identified that several proto-oncogenes are potentially activated by reorganization of TAD structures, such as IRS4 in sarcoma and squamous cancer, IGF2 in colorectal cancer, PRDM6, ERBB2, ETV1, ETV4, MYC and TERT in medulloblastoma and other cancer subtypes [82–84]. These reports propose that there may be a significant role for 3D chromatin architectural changes in tumor initiation, development and relapse. Such methods --together with accessible primary tumor cells from peripheral blood and bone marrow aspirates, in conjunction with an increased understanding of human leukemia subtypes -- may offer ideal models to evaluate the broad impact of 3D chromatin alterations in tumorigenesis. Comprehending the nature of structural 3D reorganization and epigenetic dysregulation might enable the identification of cell-type specific host events that could be hijacked to promote malignant cell transformation and/or other events (see Outstanding Questions). Further research in this area might also offer valuable information to understand the roles of 3D chromosomal structures in immunity, e.g. by mapping DNA looping events relevant to B and T cell differentiation, activation, or the acquisition of memory phenotypes. Such advances may have an important impact and increase our knowledge of mechanisms of host defense, immunosurveillance and anti-tumor immunity.
Outstanding questions.
What is the impact of genomewide changes in epigenetic/3D chromatin landscape for malignant transformation relative to known mutations targeting individual oncogenes and tumor suppressors?
Can epigenetics and 3D architecture, e.g. assayed via simplified blood tests, help in diagnostic and prognostic of hematological malignancies in future?
Can the information on 3D architecture be used to identify targets to induce differentiation of immature leukemic blasts as a therapeutic avenue?
Is the epigenetic status and changes in 3D chromatin architecture associated with potential resistance mechanisms to targeted therapy?
Despite the immense progress in immune and gene targeted therapies, we have only begun to explore possible initiation events for malignant transformation, which, if followed up, can broaden therapeutic applications by a multitude.
Highlights.
The 3D chromatin architecture is emerging as an important player in the pathogenesis of hematological malignancies.
Normal hematopoiesis and immune cell maturation are accompanied by widespread changes in chromatin compartments, topologically associated domains and epigenetic features to ensure lineage commitment.
Chromatin architectural changes driven by either genetic or epigenetic dysregulation can activate proto-oncogenes by altering regulatory interactions, contributing to tumorigenesis.
Genetic mutations in key epigenetic players can drive cancer development by impacting 3D chromatin architecture.
Acknowledgements
The authors acknowledge Matthew Witkowski and Sofia Nomikou for critical reading of the manuscript and helpful discussions. All images were created with BioRender. Related research was supported by the NIH/NCI (RO1CA194923, RO1CA216421, RO1CA228135, PO1CA229086) and the NY State Department of Health (NYSTEM, C32563GG and C32587GG, to IA), the AACR Leukemia Incyte Fellowship (to PT), and the American Cancer Society (RSG-15-189-01-RMC), the Leukemia and Lymphoma Society and the St. Baldrick’s Foundation (to AT). A.T. is a Scientific Advisor to Intelligence.Al.
Glossary
- Chromosomal territories
Individual chromosomes in interphase that occupy a defined 3D space in the nucleus
- Chromatin compartments
Transcriptionally active (A) and repressive (B) compartments are defined by mutually exclusive chromatin interactions in Hi-C contact maps
- LADs
Genomic regions in close contact with nuclear lamina in the nuclear periphery
- TADs
Genomic sequences in the range of Mb in length; physically interact with themselves more frequently than with the rest of the genome
- Euchromatin
Uncoiled loosely packed chromatin occupying the nuclear interior and enriched in actively transcribed genes
- Heterochromatin
Highly condensed chromatin (showing dark staining to DAPI); present in the nuclear periphery and associated with transcriptionally repressed genes
- Cohesin complex
Ring-shaped protein complex containing four core protein subunits, SMC1, SMC3, RAD21 and STAG
- Super-enhancers
Large clusters of enhancers occupied by a high density of interacting factors controlling genes and oncogenes, and involved in defining lineage specificity
- Liquid liquid phase-separation
dense liquid-like droplet phase within a more dilute liquid phase. The dense liquid phase is usually formed by proteins that exhibit multivalent interactions among each other
- p-bodies
Cytoplasmic granules containing mRNAs associated with translational repressors and mRNA decay machinery
- Stress granules
Cytoplasmic conserved ribonucleoprotein granules formed by a group of mRNAs stalled in translation initiation; they contain various translation initiation factors, RNA binding proteins and non RNA-binding proteins
- IDRs Intrinsically disordered regions (IDRs)
polypeptide segments in a protein that are not likely to form a defined 3D structure
- DN2-DN3
Commitment to the T cell lineage occurs during the transition from the CD4 CD8” double negative thymocyte development stage DN2 to DN3
- DN4-DP
Transition of developing thymocytes from CD4 CD8” double negative cells to CD4+CD8+ double positive cells
- TCRβ gene rearrangement
enables developing thymocytes to express TCRβ with an invariant pre-Tα
- 3D nucleome
Three-dimensional organization of chromatin within the nucleus
- Hypermethylated CpG residues
DNA regions of higher than average CG dinucleotide content that are extensively methylated at cytosine residues
- CRISPR-Cas9
RNA guided DNA cleaving genome editing system
- Syntenic region
Genomic blocks that share homology between two species (conserved order of genes)
- LCR locus control region
long-range cis-regulatory element enhancing the expression of linked genes at distal chromatin sites
- Immediate-responding genes
upregulated expression immediately following stimulation (e.g. antigen stimulation of CD4+T cells)
- Trac-looping
Transposase-dependent assay used to study chromatin looping without chromatin fragmentation and proximity-based ligation
- Thl7 helper T cells
Subset of T helper cells that produce of IL-17A, IL-17F, and IL-22; one function is to protect mucosa from bacterial and fungal infections
- Single nucleotide variants
Somatic variation of a single base-pair compared to the germline genome
- GWAS Genome-wide association studies
investigate whether a genetic variant is associated with a phenotypic trait
- Memory CD8+ T cells
have the ability to mount a rapid immune response to a previously encountered antigen
- TET (Ten-eleven-translocation) enzymes
sequentially oxidize 5-methylcytosines to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxycytosine (5caC)
- CESAM Cis-expression structural alteration mapping
computational framework developed to detect cancer related gene overexpression resulting from reorganization of TAD structures
- Hi-C
Proximity ligation assay and high-throughput sequencing to identify all pairwise interactions throughout the genome
- HiChIP
Combination of Hi-C and ChlP-Seq (achieves proximity ligation in nuclei prior to chromatin shearing and immunoprécipitation)
- AML
Acute leukemia with an immature myeloid phenotype
- CML
Chronic leukemia with a mature myeloid phenotype
- T-ALL
Acute leukemia with an immature T cell phenotype
- Insulated neighborhoods
DNA chromatin loop structures that hold genes and their regulatory regions together
- Enhancer skipping
when a promoter preferentially loops to a more distant enhancer than the proximal enhancer
- Enhancer switching
Specific usage of different enhancer elements for a particular gene (e.g. in the direct comparison of two cell types)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hug CB and Vaquerizas JM (2018) The Birth of the 3D Genome during Early Embryonic Development. Trends in genetics : TIG 34, 903–914 [DOI] [PubMed] [Google Scholar]
- 2.Zheng H and Xie W (2019) The role of 3D genome organization in development and cell differentiation. Nature reviews. Molecular cell biology [DOI] [PubMed] [Google Scholar]
- 3.Lupiáñez Darío G. , et al. Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell 161,1012–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flavahan WA, et al. (2016) Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529,110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spielmann Mv et al. (2018) Structural variation in the 3D genome. Nat Rev Genet [DOI] [PubMed] [Google Scholar]
- 6.Bunting KL, et al. (2016) Multi-tiered Reorganization of the Genome during B Cell Affinity Maturation Anchored by a Germinal Center-Specific Locus Control Region. Immunity 45, 497–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robson MI, et al. (2017) Constrained release of lamina-associated enhancers and genes from the nuclear envelope during T-cell activation facilitates their association in chromosome compartments. Genome Res 27,1126–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mumbach MR, et al. (2017) Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat Genet 49,1602–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cremer T and Cremer C (2001) Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nature reviews genetics 2, 292. [DOI] [PubMed] [Google Scholar]
- 10.Cremer T and Cremer M (2010) Chromosome territories. Cold Spring Harbor perspectives in biology 2, a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider R and Grosschedl R (2007) Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes & development 21, 3027–3043 [DOI] [PubMed] [Google Scholar]
- 12.Guelen L , et al. (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948. [DOI] [PubMed] [Google Scholar]
- 13.Lieberman-Aiden E, et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science (New York, N.Y.) 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sexton T, et al. (2012) Three-dimensional folding and functional organization principles of the drosophila genome. Cell 148. [DOI] [PubMed] [Google Scholar]
- 15.Dixon JR, et al. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao SSP, et al. (2014) A 3D Map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peric-Hupkes D , et al. (2010) Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Molecular cell 38, 603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robson MI, et al. (2016) Tissue-specific gene repositioning by muscle nuclear membrane proteins enhances repression of critical developmental genes during myogenesis. Molecular cell 62, 834–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sexton T and Cavalli G (2015) The role of chromosome domains in shaping the functional genome. Cell 160,1049–1059 [DOI] [PubMed] [Google Scholar]
- 20.Dixon JR, et al. (2016) Chromatin Domains: The Unit of Chromosome Organization. Molecular cell 62, 668–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dekker J and Mirny L (2016) The 3D Genome as Moderator of Chromosomal Communication. Cell 164,1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon JR, et al. (2016) Chromatin Domains: The Unit of Chromosome Organization. Molecular cell 62, 668–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuin J , et al. (2014) Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proceedings of the National Academy of Sciences 111, 996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nora EP, et al. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 169, 930–944.e922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao SSP, et al. (2017) Cohesin Loss Eliminates All Loop Domains. Cell 171, 305–320.e324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Y , et al. (2015) CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell 162, 900–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Wit E, et al. (2015) CTCF Binding Polarity Determines Chromatin Looping. Molecular cell 60, 676–684 [DOI] [PubMed] [Google Scholar]
- 28.Vian L , et al. (2018) The Energetics and Physiological Impact of Cohesin Extrusion. Cell 173, 1165–1178.ell20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weintraub AS, et al. (2017) YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 171,1573–1588.el528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanborn AL, et al. (2015) Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proceedings of the National Academy of Sciences of the United States of America 112, E6456–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fudenberg G , et al. (2016) Formation of Chromosomal Domains by Loop Extrusion. Cell Rep 15, 2038–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganji M , et al. (2018) Real-time imaging of DNA loop extrusion by condensin. Science (New York, N.Y.) 360, 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flaarhuis JHI, et al. (2017) The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell 169, 693–707.e614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hnisz D , et al. (2017) A Phase Separation Model for Transcriptional Control. Cell 169,13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabari BR, et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science (New York, N.Y.) 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boija A , et al. (2018) Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brangwynne CP, et al. (2009) Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science (New York, N.Y.) 324,1729–1732 [DOI] [PubMed] [Google Scholar]
- 38.Molliex A , et al. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163,123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawyers CL, et al. (1991) Leukemia and the disruption of normal hematopoiesis. Cell 64, 337–350 [DOI] [PubMed] [Google Scholar]
- 40.Kourtis N , et al. (2015) Emerging roles for the FBXW7 ubiquitin ligase in leukemia and beyond. Current opinion in cell biology 37, 28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belver L and Ferrando A (2016) The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nature reviews. Cancer 16, 494–507 [DOI] [PubMed] [Google Scholar]
- 42.Hu G , et al. (2018) Transformation of Accessible Chromatin and 3D Nucleome Underlies Lineage Commitment of Early T Cells. Immunity 48, 227–242.e228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johanson TM, et al. (2018) Transcription-factor-mediated supervision of global genome architecture maintains B cell identity. Nature immunology 19,1257–1264 [DOI] [PubMed] [Google Scholar]
- 44.Yui MA and Rothenberg EV (2014) Developmental gene networks: a triathlon on the course to T cell identity. Nature Reviews Immunology 14, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.von Boehmer H , et al. (1999) The impact of pre-T-cell receptor signals on gene expression in developing T cells. Cold Spring Harbor symposia on quantitative biology 64, 283–289 [DOI] [PubMed] [Google Scholar]
- 46.Nagano T , et al. (2013) Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Isoda T , et al. (2017) Non-coding Transcription Instructs Chromatin Folding and Compartmentalization to Dictate Enhancer-Promoter Communication and T Cell Fate. Cell 171, 103–119.ell8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nutt SL, et al. (1999) Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401, 556. [DOI] [PubMed] [Google Scholar]
- 49.Montefiori L , et al. (2016) Extremely Long-Range Chromatin Loops Link Topological Domains to Facilitate a Diverse Antibody Repertoire. Cell Rep 14, 896–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barajas-Mora EM, et al. (2019) A B-Cell-Specific Enhancer Orchestrates Nuclear Architecture to Generate a Diverse Antigen Receptor Repertoire. Molecular cell 73, 48–60.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Proudhon C , et al. (2015) Long-Range Regulation of V(D)J Recombination. Advances in immunology 128,123–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rawlings JS, et al. (2011) Chromatin condensation via the condensin II complex is required for peripheral T-cell quiescence. The EMBOjournal 30, 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hnisz D , et al. (2013) Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe N , et al. (2003) BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nature immunology 4, 670. [DOI] [PubMed] [Google Scholar]
- 55.Lai B , et al. (2018) Trac-looping measures genome structure and chromatin accessibility. Nature methods 15, 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Araki Y , et al. (2009) Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity 30, 912–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Russ BE, et al. (2014) Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8(+) T cell differentiation. Immunity 41, 853–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moskowitz DM, et al. (2017) Epigenomics of human CD8T cell differentiation and aging. Science immunology 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Henning AN, et al. (2018) Epigenetic control of CD8(+) T cell differentiation. Nature reviews. Immunology 18, 340–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Javierre BM, et al. (2016) Lineage-Specific Genome Architecture Links Enhancers and Noncoding Disease Variants to Target Gene Promoters. Cell 167,1369–1384.el319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hnisz D , et al. (2016) Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science (New York, N.Y.) 351,1454–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dang CV (2012) MYC on the path to cancer. Cell 149, 22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lancho O and Herranz D (2018) The MYC Enhancer-ome: Long-Range Transcriptional Regulation of MYC in Cancer. Trends in Cancer 4, 810–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Herranz D , et al. (2014) A NOTCHl-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med 20,1130–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yashiro-Ohtani Y , et al. (2014) Long-range enhancer activity determines Myc sensitivity to Notch inhibitors in T cell leukemia. Proceedings of the National Academy of Sciences of the United States of America 111, E4946–4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bahr C , et al. (2018) A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature 553, 515–520 [DOI] [PubMed] [Google Scholar]
- 67.Zhang J , et al. (2012) The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. 481,157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Y , et al. (2017) The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. 49,1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ley TJ, et al. (2013) Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. The New England journal of medicine 368, 2059–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mullenders J, et al. (2015) Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. The Journal of experimental medicine 212,1833–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Viny AD, et al. (2015) Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. The Journal of experimental medicine 212,1819–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mazumdar C, et al. (2015) Leukemia-Associated Cohesin Mutants Dominantly Enforce Stem Cell Programs and Impair Human Hematopoietic Progenitor Differentiation. Cell Stem Cell 17, 675–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cuartero S, et al. (2018) Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nature immunology 19, 932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Donaldson-Collier MC, et al. (2019) EZH2 oncogenic mutations drive epigenetic, transcriptional, and structural changes within chromatin domains. Nature Genetics 51, 517–528 [DOI] [PubMed] [Google Scholar]
- 75.Tahiliani M , et al. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science (New York, N.Y.) 324, 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cimmino Lv et al. (2011) TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell 9,193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dang L , et al. (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu W , et al. (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 19,17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bell AC and Felsenfeld G (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482. [DOI] [PubMed] [Google Scholar]
- 80.Hark AT, et al. (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/lgf2 locus. Nature 405, 486. [DOI] [PubMed] [Google Scholar]
- 81.Wang H , et al. (2012) Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res 22,1680–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weischenfeldt J , et al. (2017) Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat Genet 49, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Northcott PA, et al. (2017) The whole-genome landscape of medulloblastoma subtypes. Nature 547, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dixon JR, et al. (2018) Integrative detection and analysis of structural variation in cancer genomes. Nature Genetics 50,1388–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu M and Ren B (2017) The Three-Dimensional Organization of Mammalian Genomes. Annual review of cell and developmental biology 33, 265–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Denker A and de Laat W (2016) The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev 30,1357–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nora EP, et al. (2012) Spatial partitioning of the regulatory landscape of the X-inactivation centre. 485, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Phillips-Cremins JE, et al. (2013) Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153,1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dowen JM, et al. (2014) Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159, 374–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bonev B , et al. (2017) Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 171, 557–572.e524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mumbach MR, et al. (2016) HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nature methods 13, 919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li G , et al. (2010) ChlA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol 11, R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fang R , et al. (2016) Mapping of long-range chromatin interactions by proximity ligation-assisted ChlP-seq. Cell Research 26,1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lajoie BR, et al. (2015) The Hitchhiker’s guide to Hi-C analysis: practical guidelines. Methods 72, 65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schmitt AD, et al. (2016) Genome-wide mapping and analysis of chromosome architecture. Nature reviews Molecular cell biology 17, 743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pal K , et al. (2019) Hi-C analysis: from data generation to integration. Biophysical reviews 11, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lazaris C , et al. (2017) HiC-bench: comprehensive and reproducible Hi-C data analysis designed for parameter exploration and benchmarking. BMC Genomics 18, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Durand NC, et al. (2016) Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell systems 3, 95–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Servant N , et al. (2015) HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wolff J , et al. (2018) Galaxy HiCExplorer: a web server for reproducible Hi-C data analysis, quality control and visualization. Nucleic Acids Res 46, Wll–wl6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zheng X and Zheng Y (2018) CscoreTool: fast Hi-C compartment analysis at high resolution. Bioinformatics 34,1568–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shin H , et al. (2016) TopDom: an efficient and deterministic method for identifying topological domains in genomes. Nucleic Acids Res 44, e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Weinreb C and Raphael BJ (2016) Identification of hierarchical chromatin domains. Bioinformatics 32,1601–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Filippova D , et al. (2014) Identification of alternative topological domains in chromatin. Algorithms Mol Biol 9,14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Phanstiel DH, et al. (2015) Mango: a bias-correcting ChlA-PET analysis pipeline. Bioinformatics 31, 3092–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ay F , et al. (2014) Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res 24, 999–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chakraborty A and Ay F (2017) Identification of copy number variations and translocations in cancer cells from Hi-C data. Bioinformatics [DOI] [PMC free article] [PubMed] [Google Scholar]