Abstract
The G-protein-coupled μ-opioid receptor (μ-OR) mediates the majority of analgesia effects for morphine and other pain relievers. Despite extensive studies of its structure and activation mechanisms, the inherently low maturation efficiency of μ-OR represents a major hurdle to understanding its function. Here we computationally designed μ-OR mutants with altered stability to probe the relationship between cell-surface targeting, signal transduction, and agonist efficacy. The stabilizing mutation T315Y enhanced μ-OR trafficking to the plasma membrane and significantly promoted the morphine- mediated inhibition of downstream signaling. In contrast, the destabilizing mutation R165Y led to intracellular retention of μ-OR and reduced the response to morphine stimulation. These findings suggest that μ-OR stability is an important factor in regulating receptor signaling and provide a viable avenue to improve the efficacy of analgesics.
Keywords: opioid receptor, protein engineering, GPCR, maturation, signal transduction
Introduction
The human μ-OR constitutes the main target for opiate analgesics and endogenous neuropeptides, including morphine, codeine and P-endorphin 1–3. Activation of μ-OR promotes the signals transmission through the Gi/adenylyl cyclase pathway and regulation through the P-arrestin pathway, resulting in analgesics, respiratory suppression, opioid dependence, and tolerance 4–6. The efficacy of signal transduction for μ-OR and other G protein-coupled receptors (GPCR) critically depends on the surface expression and trafficking to the plasma membrane 7,8. Incorrectly folded nascent receptors are excluded from endoplasmic reticulum (ER)-derived trafficking vesicles or retained in pre-Golgi compartments 9,10. For example, only 40% of newly synthesized δOR (a close family member of μ-OR) could reach to the plasma membrane, representing a hurdle to biogenesis of functional GPCRs 11–14 The ER-retained mutants can also exert suppression on the wild-type receptor maturation, as observed for rhodopsin and β2 adrenoreceptor 15,16. Thus, impaired cell-surface targeting can lead to defects in GPCR signaling and reduced pharmacological efficacy of corresponding agonists.
The quantity of GPCR targeting to cell surface as the functional form is determined by cumulative effects of GPCR maturation, internalization, recycling and degradation 7,17 These processes critically depend on the rate of protein folding, interaction with endogenous molecular chaperones, temperature, as well as binding to small molecules (e.g. pharmacological chaperones) 9. Specifically, pharmacological chaperones are cell permeable ligands that stabilize the newly synthesized receptor and restore protein mislocalization or misfolding 11,18. For example, incubation with the μ-OR agonist etorphine or the opioid antagonist naloxone could rescue the traffic-deficient mutants of μ-OR 19. The sustained treatment of morphine also enhanced the δOR targeting to neuronal membranes and, consequently, its antinociceptive potency 20. However, the capability of morphine and other pharmacological chaperones to stabilize the correctly folded protein typically coupled with their agonist or antagonist effects 21, thus hindering a clear understanding of the rescue mechanisms (signaling vs. chaperoning). A lack of direct observation on the relationship between μ-OR stability, trafficking, and signal transmission also encumber the drug development of more efficient analgesics. Hence here we examined whether the modulation of μ-OR stability could directly affect μ-OR cellular localization and signaling efficiency.
Materials and Methods
Computational protein design
We estimate the change of protein free energy upon mutation using our in house-developed software Eris 22,23. Eris evaluates protein stability by re-packing the side chains of all other amino acids surrounding (within 10 Å) the mutation site using a Monte Carlo simulated annealing procedure 24,25. The free energies were calculated as a weighted sum of van der Waals forces, implicit solvation model, hydrogen bonding, and backbone-dependent statistical energies. The change of the protein free energy of folding upon mutation is estimated by the thermodynamic stability of the mutant protein, evaluated as ΔΔG value (ΔGMut - ΔGWt, wherein the reference state for ΔG in the unfolded protein) using Medusa force field. The models of inactive state and active state μ-OR were built from the crystal structure (PDB 4DKL and 5C1M, respectively) of murine μ-OR (93% sequence homology to the human μ-OR), we performed a systematic mutation of 170 receptor amino acids.
We excluded from our analysis all of the amino acids implicated in receptor dimerization, G protein interaction and opiates binding (orthosteric site). These residues were identified in previous studies 4,26. Residues potentially implicated in the receptor dimerization are: M65, V66, T67, I69, M72, S76, I77, V80 (TM1); P122, V126, L129, M130 (TM2); W226, Y227, N230, L231, I234, F237, I238, F241, I242, V245, L246, T249, Y252, M255, I256, L257 (TM5); T279, R280, L283, I290, T294, I298, I301, I302, L305, I306, T307 (TM6); F347, C351 (TM8).
Residues that are part of intracellular or extracellular loops: Y96, T97, K98, M99, K100, T101 (ICL1); H171, P172, V173, K174, A175, L176, D177, F178, R179, T180, P181 (ICL2); L259, K260, S261, V262, R263, E270, K271, D272 (ICL3); G131, T132, W133, P134, F135, G136, N137 (ECL1); M205, A206, T207, T208, K209, Y210, R211, Q212, G213, S214, I215, D216, C217, T218, L219, T220, F221, S222, H223, P224 (ECL2); I306, T307, I308, P309, E310, T311 (ECL3)
Residues that are part of the orthosteric-binding site (within 5 Å of β-FNA in 4DKL crystal structure): D147, Y148, M151, E229, K233, V236, W293, I296, H297, V300, K303, W318, I322, G324, Y325
Focusing on the remaining 170 residues, for each one of them, we performed an exhaustive scanning of all 19 non-native variants by performing three rounds of calculations: 1) one Monte Carlo simulated annealing for all 170 residues; 2) twenty Monte Carlo simulated annealing for all mutations with a negative ΔΔG; and 3) 2500 independent Monte Carlo simulated annealing to obtain converged ΔΔG and standard errors. We performed simulated annealing only with the side chains and kept the protein backbone constrained. Additionally, side chains repacking was allowed only within a 10 A radius to avoid perturbation of protein structure. Our criteria of choosing the mutations include: 1) large positive or negative ΔΔG values; 2) the significance of the calculation, as characterized by Z-score; 3) the location of residues (polar vs. hydrophobic interior part of μ-OR). We retrieved mutations for which ΔΔG values deviate of three standard deviations from the mean value computed over all mutated residues (i.e., more than 99.73% chance of rejecting the possibility of a randomly identified mutation, Z-Score < −3), and ultimately retrieved the stabilizing mutation T315Y and the destabilizing mutation R165Y.
Plasmids construct
The WT μ-OR plasmids (human pIRES-EGFP-Myc-7TM) were described previously 27 μ-OR mutants R165Y and T315Y were generated by overlapping polymerase chain reaction with Q5® Site-Directed Mutagenesis Kit (NEB) following the manufacture’s instruction.
Immunocytochemistry in HEK293 and HEK293T cells
HEK293 and HEK293T cell lines were purchased from ATCC, plated on Poly-D-Lysine coated coverslips, and cultured in high glucose (4.5 g/L) Dulbecco’s Modified Eagle’s Medium (Gibco) containing 10% (v/v) fetal bovine serum (Gibco). All media used for cell cultured were supplemented with 50 units/ml of Penicillin and 50 μg/ml streptomycin, and all cells were maintained with 5% CO2 in a 37°C incubator. Transfection (2 μg cDNA) was performed with Lipofectamine™ 2000 Reagent (Invitrogen) at 80% confluency and the transfected cells were cultured in the same growth medium for 48 h. Then the coverslips were washed and fixed with 4% PFA for 15 min (fixed cells were permeabilized with Triton X-100), washed and incubated with anti-Myc primary antibody (millipore, rabbit, 1:100) at 4°C for 1 hour. The cells on coverslips were incubated with secondary antibodies conjugated with cy3 (1:100; Jackson ImmunoResearch, West Grove, PA) and examined under a Leica SP5 inverted confocal microscope.
The immunofluorescent intensities of μ-OR on the cell surface and the whole cell were quantified with Image Pro Plus 5 (Media Cybernetics, Inc.), and indicated as the percentage of μ-OR distribution on the cell surface versus the intracellular compartment. For quantitative analysis, 5–7 microscope fields of view from 3 separate transfections were selected, GFP positive cells in each microscope fields of view were selected, analyzed, and the percentage of μ-OR distribution on the cell surface versus the intracellular compartment within one field were averaged. One-Way ANOVA was used for statistical analysis, **P<0.05, vs. WT, ***P<0.001, vs. WT, 5–7 repeats/group, number of cells analyzed (HEK293T) = 76, (HEK293) = 73.
cAMP assay
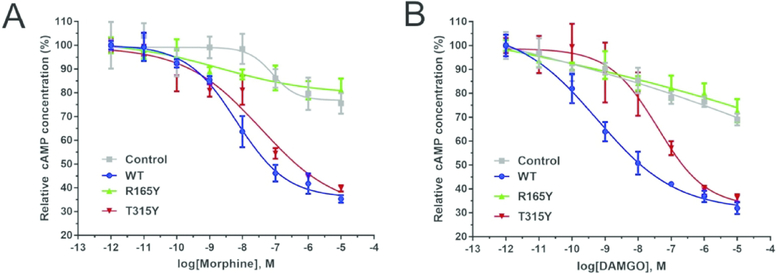
HEK293 cells were grown in DMEM supplemented with 10% FBS (Gibco, Invitrogen). For monitoring cAMP levels, the GloSensor cAMP-sensitive luciferase reporter assay (Promega) was performed, as previously described 6,28. Briefly, cells were transiently co-transfected with GloSensor-22F cAMP-sensitive construct and either of the following expressing constructs: pIRES-EGFP-μ-OR (WT μ-OR), μ-OR mutants: R165Y or T315Y, or empty vector (Control) 27 The day after transfection (24 hr), cells were seeded in 384-well plates; the following day cells were pretreated with 100 nM isoproterenol and challenged with morphine or DAMGO (Sigma Aldrich) at different concentrations. For this assay and cell line, the use of isoproterenol, instead of forskolin, was designed and optimized. Luminescence was recorded on a PHERAstar plate reader (BMG LABTECH), repeated three times, and analyzed with GraphPad Prism 6.0. The basal cAMP levels (cellular cAMP concentrations before adding agonists) were normalized as 100% and the activity of μ-OR was recorded as the percentage of cAMP signal reduction compared to the basal levels (Figure. 3).
Figure 3. Measurement of μ-OR activity in HEK293 cells.
HEK293 cells transiently transfected with vectors containing μ-OR (WT, R165Y, T315Y), or empty vectors (Control). Cells were then treated with μ-OR agonist (A) morphine or (B) DAMGO at concentrations of 10−12 to 10−4 M. The activities were measured with the cAMP-sensitive luciferase reporter assay. The stabilizing mutant T315Y exhibited elevated efficacy to both agonists, in comparison to the destabilizing mutant R165Y. Error bar: S.E.M., n = 3.
Results
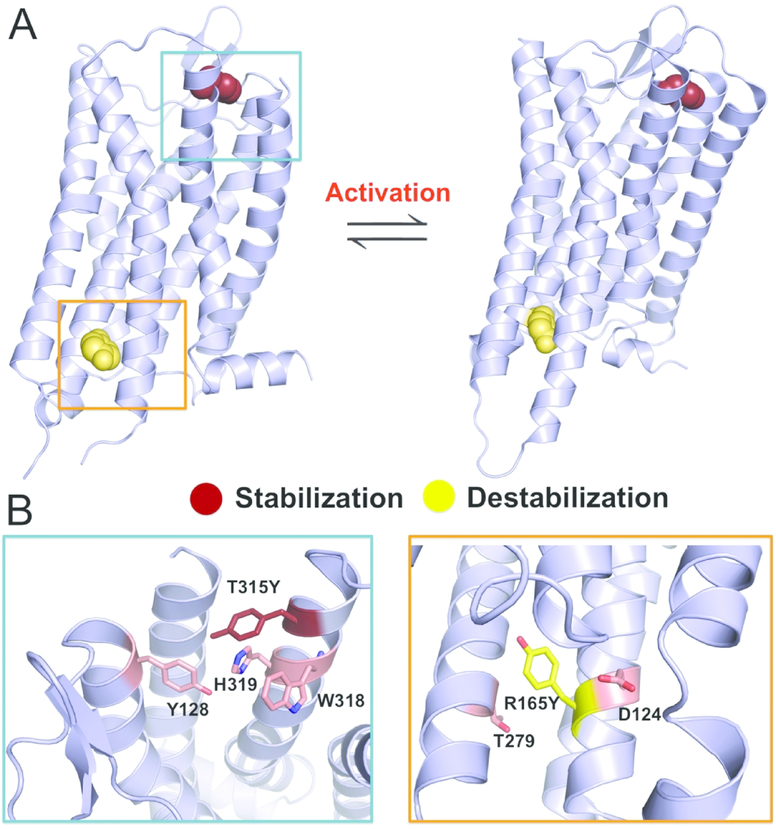
We addressed the question of μ-OR stability and signaling through a combination of computational protein design, cell imaging, and μ-OR activity assays. First, we identified μ-OR mutants with altered thermodynamic stability through the molecular design suite Eris 22,23. The change of folding free energies upon mutation was estimated by ΔΔG (AGmut - AGwt, as a computational prediction) wherein a negative ΔΔG indicates the stabilizing effect. To avoid the impact on μ-OR biological functions, we kept intact the μ-OR residues implicated in receptor dimerization, G protein interaction, and orthosteric ligand binding (see Methods), thus scanning the rest 170 residues in Eris. Based on the crystal structure of μ-OR (PDB 4DKL, the conformation before agonist stimulation) 26, we found two mutations: R165Y and T315Y (numbered as in PDB 4DKL). R165Y destabilizes μ-OR (ΔΔG = 7.9 ± 0.4 kcal/mol) while T315Y stabilizes its conformation (ΔΔG = −6.4 ± 0.5 kcal/mol) (Figure. 1A, Figure S1, and Table 1). R165 (R3.50 in Ballesteros-Weinstein numbers) resides on the transmembrane helix 3 (TM3) and constitute the DRY motif, a conserved motif in class A GPCRs implicated in stabilizing the receptor in the inactive state 29,30. Mutation to tyrosine likely abolishes the salt bridge between R165 and D164 and the polar interaction to T279, leading to destabilization (Figure. 1B). T315Y (T7.32) resides on TM7 and mutation to tyrosine could facilitate the formation of hydrogen-bond network between Y128 and H319, as well as aromatic packing with W318 (Figure. 1B), thus stabilizing μ-OR.
Figure 1. Rational design of μ-OR stabilizing and destabilizing mutants.
(A) Eris scan identifies a stabilizing mutation (T315Y) and destabilizing mutation (R165Y) using μ-OR structural models (Left: inactive state, Right: active state). The mutated tyrosine residues were shown as spheres.
(B) T315Y can mediate hydrogen bond network (Y128-H319-T315Y) and aromatic interactions (T315Y-W318) in the design model, while R165Y disrupts polar interactions with D124 and T279 of μ-OR.
Table 1.
Results of Eris calculation
| Mutations† | ΔΔG (kcal/mol) | ΔΔG (kcal/mol) |
|---|---|---|
| Inactive State | Active State | |
| R165Y | 7.99 ± 0.44 | −1.41 ± 0.49 |
| T315Y | −6.44 ± 0.54 | 3.54 ± 0.75 |
| N40D | 2.98 ± 0.34 | 0.82 ± 0.32 |
| R260H | 3.18 ± 0.37 | 1.60 ± 0.33 |
| S268P | 0.49 ± 0.52 | 0.21 ± 0.06 |
Two computationally designed μ-OR mutations: R165Y and T315Y; three naturally occurred μ-OR mutations: N40D, R260H, and S268P
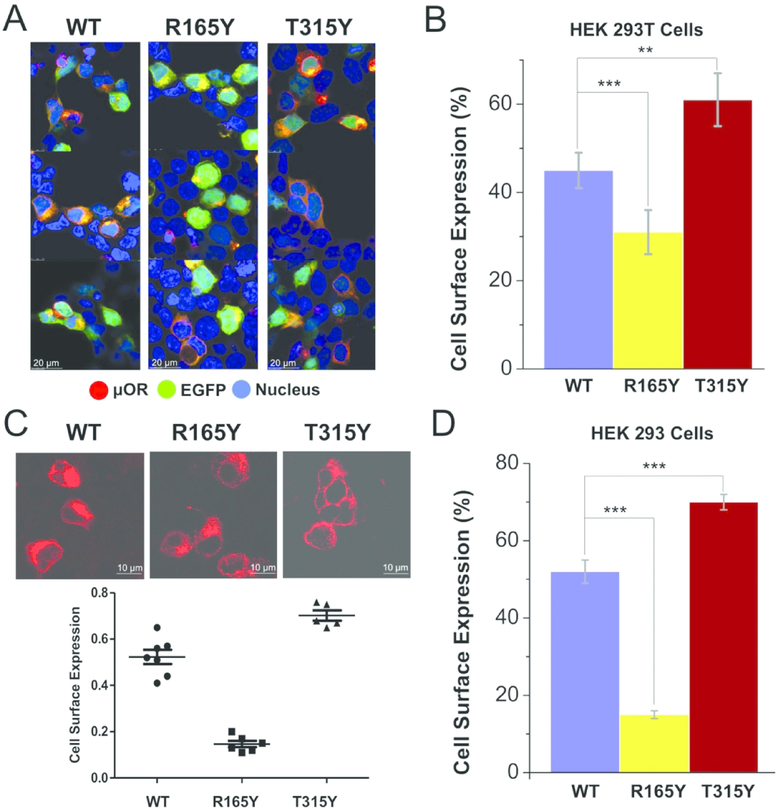
We then examined the cellular distribution of three μ-OR proteins (WT, R165Y, and T315Y) using fluorescence imaging. Expression of μ-ORs was accomplished by transfection of pIRES-EGFP-Myc-7TM construct in HEK293T cell line (μ-OR and EGFP expressed separately). Expression of EGFP (green fluorescence, Figure. 2A) indicates successful transfection of the vector as well as the cytoplasmic location. Expression of μ-ORs was detected by anti-Myc antibodies (red fluorescence). The large overlap between μ-OR and EGFP (yellow fluorescence) suggests intracellular retention of μ-OR. Compared to WT μ-OR (45% cell-surface expression), the destabilizing mutation R165Y results in a reduction in the membrane expression of μ-OR (31%), while T315Y accumulated more on the membrane (61%) (Figure. 2B), suggesting that μ-OR stabilization can facilitate its exportation from the ER lumen and trafficking to the plasma membrane.
Figure 2. Cellular distribution of WT μ-OR and two mutants.
(A) Expression of μ-OR was visualized by immunocytochemistry (blue: Hoechst stain; red: μ-OR expression; green: EGFP expression; yellow: overlap between red and green fluorescence, showing intracellular retention of μ-OR).
(B) Quantified cell surface expression levels for WT, R165Y, and T315Y indicates that increasing μ-OR stability facilitates its maturation in HEK293T cells. ** (P<0.05), *** (P<0.001). 5–7 repeats/group.
(C) R165Y was located mainly intracellularly, while the majority of T315Y translocated to the plasma membrane in HEK293 cells. *** (P<0.001). 5–7 repeats/group..
(D) Quantified cell surface expression levels for WT, R165Y, and T315Y in HEK 293 cells.
We also confirmed the effect of μ-OR stabilization in a different cell line (HEK293), which possesses smaller nucleus content. Using confocal fluorescence microscopy, we could better detect the signals from μ-OR (red fluorescence, Figure. 2C and D) and distinguish the cytoplasmic contents. In agreement with the measurement made in HEK293T cells, destabilizing μ-OR significantly impaired its export ability (cell surface expression of R165Y: 15%, T315Y: 70%, WT: 52%).
We have previously optimized the μ-OR functional assay in human HEK293 cells, measuring the downstream adenylate cyclase signals 6,28,31. Upon μ-OR activation, Gi protein is released from the plasma membrane and subsequently inhibits the adenylate cyclase activity, leading to reduction of cyclic adenosine monophosphate (cAMP) levels and suppressed neuronal activity 32,33. We investigated the dose-dependent responses of the three μ-ORs to morphine treatment (Figure. 3A). For R165Y, incubation with morphine resulted in 28% reduction of cAMP levels. In contrast, morphine exerted significantly elevated efficacy on T315Y, leading to more than 74% cAMP reduction when the agonist concentrations changes from 10−12 to 10−4 M. Although T315Y enhanced the surface expression in comparison to WT μ-OR (Figure. 2), the differences between their responses to agonists were not significant, likely due to a destabilizing effect of T315Y on the μ-OR active conformation (ΔΔG = 3.5 ± 0.7 kcal/mol, Table 1). Interestingly, the EC50 of morphine to all three μ-ORs remained similar (Log[EC50]: WT −8.2 ± 0.1, R165Y −8.6 ± 0.8, T315Y −7.8 ± 0.5), suggesting that the mutations did not affect ligand binding capacity of the mutants. This observation corroborates our design strategy of keeping the overall fold and orthosteric ligand binding sites intact. We also repeated the assay with DAMGO (an analog to endogenous opioid peptides and a selective μ-OR binder) (Figure. 3B). Similarly, the elevated efficacy of the ligand is correlated with an increase surface expression and μ-OR stability.
Discussion
Naturally occurring mutations and single nucleotide polymorphism (SNP) in the messenger RNA of μ-OR may also affect cellular μ-OR production and, consequently, μ-OR associated physiology or drug treatment 34–38. One SNP (rs10485058) in the 3′-untranslated region leads to reduced μ-OR availability. The patients bearing this single variation response poorly to the methadone treatment for opioid dependence, due to reduced population of μ-OR 39. The most relevant natural mutations of μ-OR also include N40D, R260H, and S268P, all destabilizing based on our calculation (Table 1). Based on a meta-analysis of 5,902 patients, N40D (10.5–18.8% allelic frequency) causes more opioids consumption for analgesia and impaired responsiveness of morphine 37 R260H (< 1% allelic frequency) result in reduced spontaneous receptor signaling 35. Neither N40D nor R260H is close to the μ-OR ligand binding site or Gi protein interaction site. Hence, their impacts on physiology may be induced by the overall destabilizing of μ-OR. The effects of S268P (< 1% allelic frequency) on μ-OR stability is not significant (ΔΔG < 1 kcal/mol), but its location at the third intracellular loop means S268P may impair agonist-induced receptor signaling, resulting in a 75% reduction of opioid efficacy 36. These observations, along with our results of designed mOR mutants, suggest that defects of μ-OR maturation impair μ-OR signaling.
As a homeostatic mechanism to modulate receptor sensitization, μ-OR translocation and maturation represent the major factors affecting the strength of analgesia-related signal transduction and the efficacy of opioid alkaloids. Newly synthesized membrane proteins are packaged into COPII-coated vesicles in the ER and recruited to the plasma membrane through the secretory pathway of pre-Golgi compartments 40. This process critically relies on the intrinsic stability of target proteins, as the failure to attain native conformations often lead to exclusion from transport and elimination through ER quality control 9. To improve the stability of GPCR native conformations, researchers have applied means of directed evolution and engineering intermolecular disulfide bonds 41,42. The stabilized GPCRs not only provide ideal model systems for drug developments, but also facilitate its crystallization and structural elucidation. In current work, we designed two μ-OR mutants with altered stabilities and demonstrated their impacts on the efficiency of cell-surface targeting. The destabilizing mutation R165Y caused intracellular accumulation, while the majority of T315Y, a stabilizing mutant, insert into the plasma membrane as functional forms. Importantly, our results suggest that inefficient folding and cellular mislocalization, as observed in R165Y, severely impair the μ-OR signaling through cAMP pathway, a consequence at the root of many GPCR-associated diseases 43. Hence, protein stabilization provides a viable approach to circumvent difficulties in GPCR maturation and may alleviate loss-of-function diseases.
Conclusion
Here, we demonstrated a rational protein design method that can specifically stabilize or destabilize the μ-OR inactive state, thus regulating the pharmacological efficacy of μ-OR agonists. The ability to modulate protein stabilities should make possible the identification of rescue mutants for misfolded protein 44, or the study of transient species in protein conformational changes 45–48, thus providing broad applications to diverse biosystems.
Supplementary Material
Acknowledgments
We thank Oskar Laur for the generation of receptor mutants and Stephen L. Upton for assistance in performing mutagenesis. This work was supported by NIH grants R01GM114015 and GM123247 (to N.V.D.), 1R41DA032293 (to L.D., N.V.D.), and CIHR grant CERC09 (to L.D.).
Footnotes
Conflict of Interest Statement
The authors declare that they have no competing interests.
References
- 1.Matthes HWD, Maldonado R, Simonin F, et al. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu-opioid-receptor gene. Nature. 1996;383:819–823. [DOI] [PubMed] [Google Scholar]
- 2.Law PY, Wong YH, Loh HH. Molecular mechanisms and regulation of opioid receptor signaling. Annu Rev Pharmacol. 2000;40:389–430. [DOI] [PubMed] [Google Scholar]
- 3.Chan H, McCarthy D, Li J, Palczewski K, Yuan S. Designing Safer Analgesics via μ-Opioid Receptor Pathways. Trends in Pharmacological Sciences 2017;38:1016–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang WJ, Manglik A, Venkatakrishnan AJ, et al. Structural insights into mu-opioid receptor activation. Nature. 2015;524:315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Childers SR. Opioid Receptor-Coupled 2nd Messenger Systems. Life Sci 1991;48:1991–2003. [DOI] [PubMed] [Google Scholar]
- 6.Serohijos AWR, Yin SY, Ding F, et al. Structural Basis for mu-Opioid Receptor Binding and Activation. Structure. 2011;19:1683–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong CM, Filipeanu CM, Duvernay MT, Wu GY. Regulation of G protein-coupled receptor export trafficking. Bba-Biomembranes. 2007;1768:853–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan CM, Brady AE, Nickols HH, Wang Q, Limbird LE. Membrane trafficking of G protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2004;44:559–609. [DOI] [PubMed] [Google Scholar]
- 9.Hegde RS, Ploegh HL. Quality and quantity control at the endoplasmic reticulum. Curr Opin Cell Biol 2010;22:437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teasdale RD, Jackson MR. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu Rev Cell Dev Biol 1996;12:27–54. [DOI] [PubMed] [Google Scholar]
- 11.Petaja-Repo UE, Hogue M, Bhalla S, Laperriere A, Morello JP, Bouvier M. Ligands act as pharmacological chaperones and increase the efficiency of delta opioid receptor maturation. Embo J 2002;21:1628–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petaja-Repo UE, Hogue M, Laperriere A, Bhalla S, Walker P, Bouvier M. Newly synthesized human delta opioid receptors retained in the endoplasmic reticulum are retrotranslocated to the cytosol, deglycosylated, ubiquitinated, and degraded by the proteasome. J Biol Chem 2001;276:4416–4423. [DOI] [PubMed] [Google Scholar]
- 13.Petaja-Repo UE, Hogue M, Laperriere A, Bhalla S, Walker P, Bouvier M. Newly synthesized human delta opioid receptors retained in the endoplasmic reticulum are retrotranslocated to the cytosol, deglycosylated, ubiquitinated, and degraded by the proteasome. J Biol Chem 2001;276:4416–4423. [DOI] [PubMed] [Google Scholar]
- 14.Petaja-Repo UE, Hogue M, Laperriere A, Walker P, Bouvier M. Export from the endoplasmic reticulum represents the limiting step in the maturation and cell surface expression of the human delta opioid receptor. J Biol Chem 2000;275:13727–13736. [DOI] [PubMed] [Google Scholar]
- 15.Salahpour A, Angers S, Mercier JF, Lagace M, Marullo S, Bouvier M. Homodimerization of the beta 2-adrenergic receptor as a prerequisite for cell surface targeting. JBiol Chem 2004;279:33390–33397. [DOI] [PubMed] [Google Scholar]
- 16.Rajan RS, Kopito RR. Suppression of wild-type rhodopsin maturation by mutants linked to autosomal dominant retinitis pigmentosa. J Biol Chem 2005;280:1284–1291. [DOI] [PubMed] [Google Scholar]
- 17.Tao YX, Conn PM. Chaperoning G Protein-Coupled Receptors: From Cell Biology to Therapeutics. EndocrRev. 2014;35:602–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Convertino M, Das J, Dokholyan NV. Pharmacological Chaperones: Design and Development of New Therapeutic Strategies for the Treatment of Conformational Diseases. Acs Chem Biol 2016;11:1471–1489. [DOI] [PubMed] [Google Scholar]
- 19.Chaipatikul V, Erickson-Herbrandson LJ, Loh HH, Law PY. Rescuing the traffic-deficient mutants of rat mu-opioid receptors with hydrophobic ligands. Mol Pharmacol. 2003;64:32–41. [DOI] [PubMed] [Google Scholar]
- 20.Cahill CM, Morinville A, Lee MC, Vincent JP, Collier B, Beaudet A. Prolonged morphine treatment targets delta opioid receptors to neuronal plasma membranes and enhances delta-mediated antinociception. J Neuro Sci 2001;21:7598–7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wuller S, Wiesner B, Loffler A, et al. Pharmacochaperones post-translationally enhance cell surface expression by increasing conformational stability of wild-type and mutant vasopressin V2 receptors. J Biol Chem 2004;279:47254–47263. [DOI] [PubMed] [Google Scholar]
- 22.Yin S, Ding F, Dokholyan NV. Eris: an automated estimator of protein stability. Nature methods. 2007;4:466–467. [DOI] [PubMed] [Google Scholar]
- 23.Yin S, Ding F, Dokholyan NV. Modeling backbone flexibility improves protein stability estimation. Structure. 2007;15:1567–1576. [DOI] [PubMed] [Google Scholar]
- 24.Ding F, Dokholyan NV. Emergence of protein fold families through rational design, PLoS Comput Biol 2006;2:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu C, Mowrey DD, Dokholyan NV. Computational Protein Design Through Grafting and Stabilization. Methods in molecular biology. 2017;1529:227–241. [DOI] [PubMed] [Google Scholar]
- 26.Manglik A, Kruse AC, Kobilka TS, et al. Crystal structure of the mu-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gris P, Gauthier J, Cheng P, et al. A novel alternatively spliced isoform of the μ-opioid receptor: functional antagonism. Molecular Pain. 2010;6:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Convertino M, Samoshkin A, Viet CT, et al. Differential Regulation of 6-and 7-Transmembrane Helix Variants of mu-Opioid Receptor in Response to Morphine Stimulation. Plos One. 2015;10: e0142826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rovati GE, Capra V, Neubig RR. The highly conserved DRY motif of class A G protein-coupled receptors: Beyond the ground state. Mol Pharmacol. 2007;71:959–964. [DOI] [PubMed] [Google Scholar]
- 30.Valentin-Hansen L, Groenen M, Nygaard R, Frimurer TM, Holliday ND, Schwartz TW. The Arginine of the DRY Motif in Transmembrane Segment III Functions as a Balancing Micro-switch in the Activation of the beta 2-Adrenergic Receptor. J Biol Chem 2012;287:31973–31982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samoshkin A, Convertino M, Viet CT, et al. Structural and functional interactions between six-transmembrane mu-opioid receptors and beta(2)-adrenoreceptors modulate opioid signaling. Sci Rep. 2015;5:18198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crain SM, Shen KF. Antagonists of excitatory opioid receptor functions enhance morphine’s analgesic potency and attenuate opioid tolerance/dependence liability. Pain. 2000;84:121–131. [DOI] [PubMed] [Google Scholar]
- 33.Inturrisi CE. Clinical pharmacology of opioids for pain. Clin J Pain. 2002;18:S3–S13. [DOI] [PubMed] [Google Scholar]
- 34.Lotsch J, Geisslinger G. Are mu-opioid receptor polymorphisms important for clinical opioid therapy? Trends Mol Med 2005;11:82–89. [DOI] [PubMed] [Google Scholar]
- 35.Wang D, Quillan JM, Winans K, Lucas JL, Sadee W. Single nucleotide polymorphisms in the human mu opioid receptor gene alter basal G protein coupling and calmodulin binding. JBiol Chem 2001;276:34624–34630. [DOI] [PubMed] [Google Scholar]
- 36.Befort K, Filliol D, Decaillot FM, Gaveriaux-Ruff C, Hoehe MR, Kieffer BL. A single nucleotide polymorphic mutation in the human mu-opioid receptor severely impairs receptor signaling. J Biol Chem 2001;276:3130–3137. [DOI] [PubMed] [Google Scholar]
- 37.Beyer A, Koch T, Schroder H, Schulz S, Hollt V. Effect of the A118G polymorphism on binding affinity, potency and agonist-mediated endocytosis, desensitization, and resensitization of the human mu-opioid receptor. J NeuroChem 2004;89:553–560. [DOI] [PubMed] [Google Scholar]
- 38.Redler RL, Das J, Diaz JR, Dokholyan NV. Protein destabilization as a common factor in diverse inherited disorders, J Mol Evol. 2016;82:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crist RC, Doyle GA, Nelson EC, et al. A polymorphism in the OPRM1 3′-untranslated region is associated with methadone efficacy in treating opioid dependence. Pharmacogenomics J 2018;18:173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brandizzi F, Barlowe C. Organization of the ER-Golgi interface for membrane traffic control. Nat Rev Mol Cell Bio. 2013;14:382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott DJ, Pluckthun A. Direct Molecular Evolution of Detergent-Stable G Protein-Coupled Receptors Using Polymer Encapsulated Cells. J Mol Biol 2013;425:662–677. [DOI] [PubMed] [Google Scholar]
- 42.Standfuss J, Xie GF, Edwards PC, Burghammer M, Oprian DD, Schertler GFX. Crystal structure of a thermally stable rhodopsin mutant. J Mol Biol 2007;372:1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spiegel AM, Weinstein LS. Inherited diseases involving G proteins and G protein-coupled receptors. Annu Rev Med 2004;55:27–39. [DOI] [PubMed] [Google Scholar]
- 44.Proctor EA, Kota P, Aleksandrov AA, He LH, Riordan JR, Dokholyan NV. Rational coupled dynamics network manipulation rescues disease-relevant mutant cystic fibrosis transmembrane conductance regulator. Chem Sci 2015;6:1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu C, Beck MV, Griffith JD, Deshmukh M, Dokholyan NV. Large SOD1 aggregates, unlike trimeric SOD1, do not impact cell viability in a model of amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2018;115:4661–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding F, Dokholyan NV. Simple but predictive protein models. Trends Biotechnol. 2005;23:450–455. [DOI] [PubMed] [Google Scholar]
- 47.Dokholyan NV. Studies of folding and misfolding using simplified models. Curr Opin Struct Biol 2006;16:79–85 [DOI] [PubMed] [Google Scholar]
- 48.Chen Y, Ding F, Nie H, Serohijos AWR, Sharma S, Wilcox KC, Yin S, Dokholyan NV. Protein folding: then and now. Arch Biochem Biophys. 2008;469:4–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.