Abstract
NMR-based quantification of human lipoprotein (sub)classes is a powerful high-throughput method for medical diagnostics. We evaluated select proton NMR signals of serum lipoproteins for elucidating the physicochemical features and the absolute NMR visibility of their lipids. We separated human lipoproteins of different subclasses by ultracentrifugation and analyzed them by 1H NMR spectroscopy at different temperatures (283–323 K) and pressures (0.1–200 MPa). In parallel, we determined the total lipid content by extraction with chloroform/methanol. The visibility of different lipids in the 1H NMR spectra strongly depends on temperature and pressure: it increases with increasing temperatures but decreases with increasing pressures. Even at 313 K, only part of the lipoprotein is detected quantitatively. In LDL and in HDL subclasses HDL2 and HDL3, only 39%, 62%, and 90% of the total cholesterol and only 73%, 70%, and 87% of the FAs are detected, respectively. The choline head groups show visibilities of 43%, 75%, and 87% for LDL, HDL2, and HDL3, respectively. The description of the NMR visibility of lipid signals requires a minimum model of three different compartments, A, B, and C. The thermodynamic analysis of compartment B leads to melting temperatures between 282 K and 308 K and to enthalpy differences that vary for the different lipoproteins as well as for the reporter groups selected. In summary, we describe differences in NMR visibility of lipoproteins and variations in biophysical responses of functional groups that are crucial for the accuracy of absolute NMR quantification.
Keywords: metabolomics, serum lipids, intermediate density lipoprotein, very low density lipoprotein, low density lipoprotein, high density lipoprotein, high pressure nuclear magnetic resonance
Lipoprotein particles of different size and composition mediate lipid metabolism by transporting them to their place of destination via the blood or the lymphatic system (1, 2). The outer layer of all lipoprotein classes is covered by hydrophilic moieties of apolipoproteins and of amphiphilic lipids, such as free unesterified cholesterol, phospholipids (PLs), and SM, in order to guarantee water solubility of the assembly. The core of the particles is mainly composed of cholesteryl esters (CEs), triacylglycerides (TGs), and also free cholesterols. The lipoprotein composition directly affects the molecular dynamics of the lipids and may alter the conformation of apolipoproteins, which has direct influence on metabolic processes like receptor binding or cholesterol exchange (3–5).
With rising temperature, lipid assemblies usually are subjected to a phase transition from a solid-like gel state to a liquid-crystalline-like state with higher internal mobility. The core lipids of LDLs undergo a broad phase transition from a smectic-like crystal state to a liquid-like state with increased fluidity at body temperature primarily due to a molecular rearrangement of the CEs (6–8). Above a minimum relative concentration of TGs, CEs are melting cooperatively with TG, as it is the case in larger ApoB-containing particles. CEs do not accumulate in segregated microdomains but are equally distributed within the TGs (9). In fact, the melting temperature Tm was shown to correlate inversely with the CE/TG ratio (10) and indicates a discontinuity at a critical CE/TG ratio of about 7:1 (11). On the other hand, it was possible to detect phase transitions in CE-rich VLDLs from rabbits that were fed a hypercholesterolemic diet (12), but not in HDL particles whose rather small TG content should actually indicate a rearrangement in CEs (13, 14). A proposed explanation for this behavior is the limited space and the high curvature of the small particles, which constrains the CE molecules from arranging in ordered layers (15).
NMR spectroscopy, in contrast to common calorimetric methods, has been shown to be a powerful method in detecting thermal transitions not only for core CEs, but also for specific lipid resonances of all lipid classes (7, 14, 16–20). Strong signals of specific lipid groups were used for the characterization of lipoproteins in these NMR studies. Especially the FA methyl and methylene groups, as well as the protons in or close to double bonds, give rise to characteristic NMR signals. In addition, the choline head group incorporated in phosphatidylcholines (PCs) and SMs shows a sharp singlet NMR signal (21, 22) and can be used to characterize the membranes of lipoprotein particles.
Decreasing temperature leads to restriction of the lipid mobility and hence to loss of signal intensity and/or significant line broadening and a partial disappearance of the signal in the baseline noise level (7, 18). By plotting the signal amplitudes, linewidths, or peak areas against the measuring temperature, sigmoidal curve shapes were obtained, whose points of inflection were highly correlated with the phase transition temperature Tm of the internal lipids (17). Thus, it is possible to detect structural rearrangements not only concerning the core lipids of LDL, but also of the lipids within the membrane of LDL and HDL (14). The cholesterol spine caused a rather broad background signal, which was therefore difficult to access and overlaid especially with the desired methyl and methylene groups of the lipids.
The thermodynamics of biomolecular assemblies can be studied by using temperature T and/or pressure P perturbations of the system, influencing the free Gibbs energies ΔG and hence the thermodynamic equilibrium. According to Heremans and Smeller (23), the free energy difference at temperature T and pressure P can be expressed as a Taylor series around T0 and P0 as
| (Eq. 1) |
The dependence of ΔG on pressure and temperature can be used to extract the thermodynamic quantities ΔG0 (free energy), ΔS0 (entropy), ΔCp (isobaric heat capacity), ΔV0 (volume), Δκ (compressibility), and Δα (thermal expansibility) for obtaining a full description of the process.
Some high pressure studies of artificial lipid systems were reported in the past (for a review, see Ref. 24) but are still rather rare compared with applications to proteins. Especially high-pressure NMR spectroscopy is mainly used to study proteins, where it is applied to folding/unfolding events, in the study of conformational transitions and in drug design (see, e.g., Refs. 25–28).
Various NMR studies have been carried out with the objective to quantify not only the amount of lipoprotein lipids, like cholesterol or triglycerides, but also the distribution of the different lipoprotein classes and subclasses and even the amount of apolipoproteins via line-shape fitting models (29–32), neural networks (33–35), multivariate statistics (35), computational algorithms (36, 37), or specific NMR methods based on wavelet transformation (38) or diffusion-edited NMR spectroscopy (39, 40). All of these methods revealed very good agreement of the obtained lipoprotein subclass distributions compared with conventional biochemical assays.
In medical diagnostics, probably the most important high-throughput application of NMR spectroscopy is the determination of particle numbers of lipoproteins of different subclasses in human serum first introduced by Otvos et al. (30, 31). Many other groups including several analytical companies (see, e.g., Refs. 32–40) are now involved in these studies because it turned out the NMR derived particle numbers are better correlated to human diseases such as coronary heart disease (41, 42); for a review see, e.g., Ref. 43) as lipoprotein concentrations determined by classical biochemical methods.
Here, we present an elaborate evaluation of selected lipoprotein NMR signals (including the terminal methyl group of FAs, choline, and of the cholesterol spine) of isolated lipoproteins at different temperatures ranging from 283 to 323 K and quantitatively compare them to NMR spectra of lipids extracted from the same batch of lipoproteins. As an alternative physical method for the study of phase transitions in lipoproteins, we used high-pressure NMR spectroscopy. These findings reveal new structural insights into lipid mobility within lipoprotein particles and provide important information concerning the accuracy of absolute NMR quantification.
MATERIALS AND METHODS
Lipoprotein preparation
Initially, experiments were performed with sera of different donors for establishing the methods and for optimizing the experimental and computational setup. However, in this publication, we restricted to a single healthy donor for getting a coherent picture because otherwise the data could not be interpreted quantitatively in detail. Data shown are from a healthy donor (male, 48 years old, BMI 27.4) after an overnight fasting and stored at −20°C until its utilization. Freezing and storage of the serum at −20°C could potentially lead to irreversible deterioration of the samples. By NMR, we do not see any spectral differences between freshly drawn samples and samples that where repeatedly frozen, stored at −20°C, and thawed. An example is shown in supplemental Figs. S1 and S2. In our context, we only compare the lipid visibility in serum fractions of the same sample; small changes of lipid contents caused by freezing and thawing would not be relevant here.
Typical concentrations of total cholesterol, total triglycerides, LDL cholesterol, and HDL cholesterol were 217, 103, 147, and 48.8 mg/dl, respectively. The main thermodynamics analysis of the protein was performed from a single large serum sample of the donor, because otherwise the data of different fraction would not fit together.
Lipoproteins were isolated by sequential ultracentrifugation using KBr to establish the desired density of 1.006, 1.019, 1.063, 1.125, and 1.210 g/ml for VLDL, IDL, LDL, HDL2, and HDL3, respectively (44). Each 8–10 ml of the samples was centrifuged in a Beckman Type 75 Ti fixed angle titanium rotor at 235,469 g for 24 h (4°C) within a Beckman L-70 ultracentrifuge. The upper lipoprotein fraction was taken. The lower residue was adjusted to the next higher density and diluted with an appropriate KBr solution to undergo further separation steps leaving only the lipoprotein-deficient serum (LPDS). LPDS was withdrawn in two fractions. The upper layer was called LPDS1 and the lower LPDS2. LPDS was also obtained directly from serum by establishing the density for HDL3 in the samples.
Using Vivaspin concentrators (molecular mass cutoff 10 kDa), all samples were transferred into a quasiphysiological extracellular NMR buffer described by Freund and Kalbitzer (45) by at least three concentration/dilution cycles. At the end of these procedure, the original buffer was diluted by a factor > 150. Even at the highest density used in the ultracentrifugation (1.2 kg/l), the KBr concentration was less than 20 mM after dilution, substantially lower than the salt concentration in the physiological buffer. The buffer contained 2.2 mM Na2HPO4, 0.4 mM Na2SO4, 94.6 mM NaC1, 5.0 mM KC1, 0.85 mM MgC12, 17.0 mM Na2CO3, 8.0 mM NaHCO3, and 21.1 mM CD3COOD in bidest. H2O. The pH was adjusted to 7.4 with NaOH. The buffer has an ionic strength of 153 mM and an ionic equivalent of 150 mM. A quantity of 0.02% of NaN3 was added. For NMR measurements, each 475 µl of the samples were mixed with 25 µl of a 2 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) solution in D2O. Alternatively, as additional references, 25 µl of a solution containing 1.9 mM DSS, 1.9 mM 1,4-dioxane, and 3.8 mM pyrazine in 5 vol% D2O/95 vol% H2O were added.
Lipid extraction
To extract the lipids from the lipoproteins, a modified protocol of Srivastava et al. (55) was used. A total of 90 µl of lyophilized lipoproteins were treated with 700 µl of a mixture of CDCl3 [including 0.03 vol% TMS and 14.11 mM pyrazine] and methanol-d4 (2:1, by volume) and were sonicated for 3 × 5 min within an ice bucket to obtain a homogeneous suspension. The solid matter was separated by centrifugation at 2,375 g for 5 min (277 K), and 500 µl of the supernatant was transferred directly into a 5 mm NMR tube and measured immediately. All extractions steps were carried out using glass tubes and instruments in order to avoid contamination due to instability of plastic containers.
NMR spectroscopy
If not stated otherwise 1H NMR experiments were performed under identical conditions on a Bruker Avance 600 MHz spectrometer with a TXI cryoprobe. The high-pressure experiments were performed on a Bruker Avance 800 MHz spectrometer with a TCI cryoprobe. The temperature was stabilized by a variable temperature unit, which was calibrated using the signal of methanol contained in a capillary. The samples were contained in high-quality 5 mm NMR tubes. For quantitative measurements, the sample was tuned/matched and shimmed, and the 90 degree pulse was readjusted after the thermal equilibrium was established.
Solutions in organic solvents were measured at 293 K using the Bruker zg pulse sequence with a delay of 30 s (10 s for the lipid standards), an acquisition time of 2.28 s, and a spectral width of 11.9725 ppm. Accurate water suppression for aqueous solutions was achieved using the Bruker noesygppr1d pulse sequence with spoil gradients, a mixing time of 10 ms, and presaturation during a relaxation delay of 2 s. An additional relaxation delay of 13 s was used to assure sufficient relaxation of the lipoprotein signals. Acquisition time and spectral width were identical to those used for organic solutions, and the numbers of scans were adapted as necessary. Spectra recorded in organic solvents were referenced to TMS, spectra in aqueous directly to internal DSS, or indirectly to DSS using internal pyrazine, 1,4-dioxane, or lactate. Transverse relaxation times were measured with a Carr-Purcell-Meiboom-Gill (CPMG)-type sequence (46, 47) with the Bruker pulse program cpmgpr. The distance between the hard π-pulses was set to 1 ms, and the repetition time was 14 s. Spectra were not corrected for the chemical shift difference of −0.094 ppm between TMS in chloroform and D2O [the value measured in D2O should be larger by 0.094 ppm and for TMS and DSS in D2O by 0.02 ppm (48)]. This would lead to a correction of the chemical shifts measured relative to the reference TMS in CDCl3 by 0.074 ppm when measured in D2O at 303 K relative to the reference DSS. Note that DSS is interacting with lipoproteins and serum proteins. In blood serum, the DSS shift is concentration-dependent. At 310 K, the signal of 0.1 mM DSS is shifted by approximately 0.029 ppm upfield from its unperturbed position. In spectra of serum fractions (except HDL), such a shift was not observed. The lactate methyl signal is usually used as secondary standard in spectroscopy of blood serum. Under our conditions, its value was 1.321 ppm relative to internal DSS.
For lipid signal assignment, 2D total correlation spectroscopy (TOCSY) measurements were performed utilizing a broad-band spin-lock sequence MLEV-17 and a mixing time of 60 ms (49). T1 relaxation times were obtained using a Bruker t1ir pulse sequence with and without presaturation. T1 relaxation times of pyrazine and 1,4-dioxane were extremely long in aqueous solutions compared with the lipoprotein relaxation times, especially at higher temperatures. Thus, the measuring time was shortened up to the time mentioned above where all lipoprotein signals experienced full relaxation. The signal intensities of the internal standards were corrected afterwards for different repetition times by dividing the obtained intensities of the reference compounds by 1 − exp(−Trep/T1), which led to very good agreement of the standard signal intensities among one another. As control of this correction for series of the temperature-dependent measurements, intensities of lipoprotein signals were also corrected using the changes of the 90 degree pulse lengths and the Boltzmann factor. Within the limits of error, the same values were obtained.
The recorded data were zero-filled and multiplied by an exponential line-broadening function of 0.3 or 2.0 Hz for the lipid and lipoprotein samples, respectively. After Fourier transformation and phase correction, the baseline of each spectrum was adjusted manually using a 2nd degree polynomial function to the entire frequency range. Additional baseline correction was carried out for separate signal quantification, paying attention to frame the integral limits into local minima next to the peaks. Concerning the CH3 group of FAs and the C-18 methyl cholesterol signal, an artificially broadened cholesterol spectrum was scaled to quantify and subtract the cholesterol signal contribution (see also supplemental Fig. S6). Since the cholesterol linewidths are temperature-dependent, different line broadenings had to be used for this correction.
All NMR spectra to be quantified were corrected according to receiver gain, number of scans, dilution, and internal standard concentration. Peak areas of individual NMR signals were obtained by adding all intensities within the desired frequency range. We will always refer to peak areas defined in this way whenever we talk about signal intensities or integrals. As far as possible, a mixed deconvolution function integrated in TOPSPIN 2.1 (mdcon) was used to fit the line shape of the selected peaks exactly by picking several peaks within one single singlet.
Lipid signal quantification
Quantitative analysis of the lipid signals within NMR spectra of the intact lipoproteins as well as the extracted lipids was performed in TOPSPIN 2.1 using standardized integration (intrng) of predefined spectral ranges given in supplemental Fig. S3 and supplemental Table S3, as well as in Table 4. All spectral ranges were chosen to terminate in local minima. Areas next to predominant signals were avoided. Slight baseline offsets were corrected manually via the TOPSPIN integrated baseline functionality. Cholesterol quantification in lipoproteins is described in detail in supplemental Fig. S6.
TABLE 4.
Thermodynamic parameters for VLDL, IDL, LDL, HDL2, and HDL3
| Frequency Range (ppm) | Group | Parameter | VLDL | IDL | LDL | HDL2 | HDL3 |
| 0.1–3.0 | CH2/CH3 region | ΔH(Tm) | −49 ± 17 | −80 ± 4 | −108 ± 12 | −43 ± 14 | −61 ± 21 |
| [kJ/mol] | |||||||
| Tm [K] | 314 ± 7 | 288.0 ± 0.5 | 299.2 ± 0.7 | 291.4 ± 6.3 | 295.3 ± 4.1 | ||
| A [%] | 40 ± 7.2 | 14.6 ± 6 | 11.0 ± 1.5 | 9.4 ± 16.8 | 44.3 ± 9.6 | ||
| B [%] | 60 ± 15 | 83.9 ± 10 | 39.1 ± 2.2 | 67 ± 22 | 46 ± 13 | ||
| C [%] | 0a | 0a | 49.9 ± 3.7 | 24 ± 39 | 10 ± 23 | ||
| 0.62–0.78 | Cholesterol C-18 | ΔH(Tm) | −166 ± 8 | −138 ± 30 | −123.0 ± 14.3 | −29.9 ± 3.9 | −74.0 ± 29.5 |
| [kJ/mol] | |||||||
| Tm [K] | 314 ± 7 | 288 ± 1 | 301.0 ± 0.7 | 285.6 ± 2.9 | 304.0 ± 2.9 | ||
| A [%] | 62 ± 3.3 | 40.2 ± 4.8 | 12.8 ± 0.1 | 0.0 ± 0.0 | 68.6 ± 4.4 | ||
| B [%] | 38 ± 8.3 | 59.8 ± 6.4. | 32.9 ± 0.2 | 85.8 ± 5.7 | 34.9 ± 9.4 | ||
| C [%] | 0a | 0a | 54.3 ± 0.3 | 14.2 ± 5.7 | 0 ± 13.8 | ||
| 0.7–1.0 | FAMethyl | ΔH(Tm) | −114 ± 14 | −114 ± 16 | −131.4 ± 6.0 | −56 ± 10 | −49 ± 11 |
| [kJ/mol] | |||||||
| Tm [K] | 319 ± 3 | 290 ± 1 | 296.6 ± 0.3 | 291.9 ± 2.9 | 282.0 ± 1.6 | ||
| A [%] | 67.6 ± 0.7 | 45.5 ± 4.4 | 16.8 ± 1.0 | 10.5 ± 10.4 | 0.0 ± 0.0 | ||
| B [%] | 32.4 ± 4.7 | 54.5 ± 5.2 | 62.6 ± 1.4 | 76.0 ± 13.1 | 100.5 ± 5.9 | ||
| C [%] | 0a | 0a | 21.6 ± 2.4 | 13.5 ± 23.5 | 0 ± 5.9 | ||
| 2.6–2.8 | Bis allylic Methylene | ΔH(Tm) | −118 ± 100 | −286 ± 200 | −79 ± 20 | −38.4 ± 4.1 | −27.5 ± 7.0 |
| [kJ/mol] | |||||||
| Tm [K] | 310 ± 4 | 291.2 ± 3 | 302.7 ± 3.8 | 300.5 ± 3.8 | 309 ± 18 | ||
| A [%] | 74.5 ± 3.5 | 13 ± 21 | 0.0 ± 0.0 | 0.0 ± 0.0 | (0) | ||
| B [%] | 25.5 ± 5.5 | 87 ± 24 | 62.0 ± 9.1 | 85.3 ± 7.4 | (100) | ||
| C [%] | 0a | 0a | 38.0 ± 9.1 | 14.7 ± 7.4 | (0) | ||
| 3.0–3.4 | Choline -N+(CH3)3 | ΔH(Tm) | −103 ± 94 | −138 ± 130 | −104 ± 23 | −45 ± 15 | −135 ± 43 |
| [kJ/mol] | |||||||
| Tm [K] | 301 ± 12 | 295 ± 16 | 304.4 ± 1.4 | 295.8 ± 4.5 | 304.4 ± 1.8 | ||
| A [%] | 37.9 ± 13.1 | 85 ± 8 | 24.9 ± 1.6 | 38 ± 12 | 67 ± 19 | ||
| B [%] | 62.1 ± 20.2 | 14 ± 9 | 27.9 ± 3.3 | 54 ± 18 | 26.7 ± 3.8 | ||
| C [%] | 0a | 0a | 45.2 ± 4.9 | 8 ± 30 | 6.3 ± 23 | ||
| Mean | All 4 reporter groups | Tm [K] | 311 ± 7 | 290 ± 4.3 | 301.2 ± 0.1 | 293.5 ± 2.2 | 299.9 ± 4.3 |
Parameters were obtained by fitting the data shown in Figs. 7 and 8 with equation 8. A, B, and C correspond to the fractions of lipids that are visible at all temperatures, get visible at increasing temperatures, and are never visible, respectively. ΔH(Tm) is the difference of enthalpies at the melting temperature for compartment B. For values in parentheses, no stable fit to equation 8 could be obtained from our data; however, 100% of the signal is observed at high temperature (Fig. 8).
Value could not be determined experimentally but was assumed to be 0%.
Data evaluation within a thermodynamic model
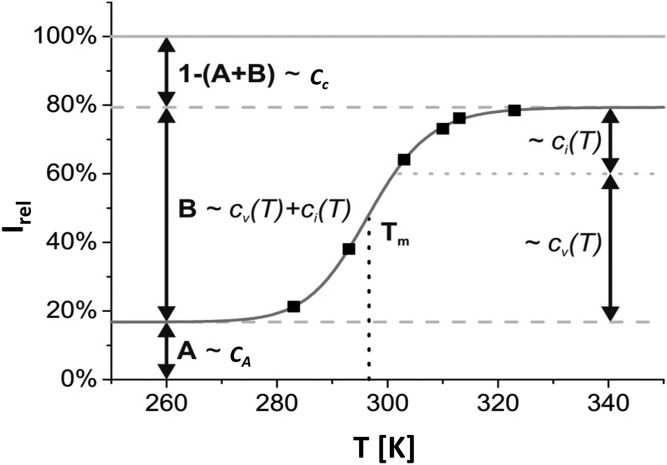
It turned out that a satisfying fit of the temperature dependence of the lipoprotein spectra required the assumption of at least three compartments: a compartment A characterized by lipid signals visible at any temperature; a compartment B characterized by a phase transition between an immobilized state and a state with high (internal) mobility; and a compartment C containing lipids that were not observable at any temperature studied by solution NMR (Fig. 1). In this model, the concentrations of spins (lipid groups) with resonance frequencies in the spectral range under consideration are temperature-independent with cA, cB, and cC their concentrations in the compartments A, B, and C, respectively. The concentrations cv and ci describe the NMR-visible and the invisible components in compartment B, respectively. The equilibrium constant K between these two states is given by equation 2 with the Gibbs free energy difference ΔG, the gas constant R, and the temperature T.
Fig. 1.
Schematic representation of the three-compartment model. The relative intensity Irel is defined as the ratio between the lipid peak areas ILP obtained by adding up all intensities within a selected frequency range divided by the expected total signal ILipid of the lipoprotein if all signals would be visible. cv and ci describe the temperature-dependent concentrations of the lipoprotein signal, which is visible or invisible for NMR detection. The sum of these two variables cB is constant, but the ratio varies with the temperature. cA refers to the concentration of lipoprotein signal, which is always detectable by NMR, and cC represents the concentration of lipoprotein in compartment C, which is never visible.
| (Eq. 2) |
The difference of the Gibbs free energies of the two states ΔG contains two contributions, the differences of the enthalpies ΔH and the entropies ΔS,
| (Eq. 3) |
At constant pressure, ΔG can be expressed in terms of the heat capacity Cp and the melting (phase transition) temperature Tm defined by ΔG(Tm) = 0
| (Eq. 4) |
In order to quantify Irel, the lipoprotein NMR signal intensity ILP observable in the native lipoproteins was divided by the appropriately normalized signal intensity ILipid of the lipids extracted from the same sample
| (Eq. 5) |
Two temperature-independent parameters A and B can be defined by
| (Eq. 6) |
leading to a simple expression for Irel
| (Eq. 7) |
Substituting equations 2 and 4 into equation 7 leads to the final equation
| (Eq. 8) |
that is used for fitting the experimental data. The difference in heat capacity in lipids is usually rather small and was set to zero in the fit of data.
High-pressure system
A homebuilt online-pressure system according to Yamada-method (50) was used. Pressure produced by a homemade manually operated piston compressor was transmitted via a high-pressure line (High Pressure Equipment Co., Linden, PA) by methylcyclohexane to the high-pressure ceramic cell (an outer diameter 5 mm, inner diameter of 3 mm) from Daedalus Innovations LLC (Aston, PA). A polyethylene membrane acts as a flexible separator between the pressure fluid und the aqueous sample. To reduce the volume of the ceramic cell a cylindric polyether-ether-ketone displacement body was used. A titanium autoclave connects the ceramic cell with the closed pressure line. It is similar to the original autoclave provided by Daedalus Innovations, but is produced from titanium and contains a safety valve that closes rapidly when the cell should brake.
RESULTS
Separation and reconstitution of spectra from different molecular constituents contained in human blood serum
Blood serum is a mixture of many different molecular constituents, mainly proteins, lipoproteins, and small metabolites. In the NMR spectra of blood serum usually used for lipoprotein analysis, they contribute differently, depending on their molecular mass and their internal mobility. Low-molecular-mass compounds are characterized by very narrow lines and can easily be recognized. These signals can be suppressed by applying magnetic field gradients (see e.g., Ref. 40). At relatively small rotational correlation times (in proton NMR corresponding to a molecular mass of a rigid macromolecule of up to approximately 40 kDa) all molecules can be detected in solution via 1H NMR spectroscopy. Typical 1H linewidths at 40 kDa are 30 Hz (at 600 MHz corresponding to 0.05 ppm). Because for rigid molecular complexes the rotational correlation times and the linewidths increase proportional to the molecular masses, the lines for very large molecular complexes are too broad to be separated from background noise and baseline variations. However, also, larger molecular complexes can contribute to the visible spectrum in solution if the effective rotational correlation time of its components is decreased by internal motions. Such an effect is clearly observed for lipoproteins that are well visible in 1H NMR spectra.
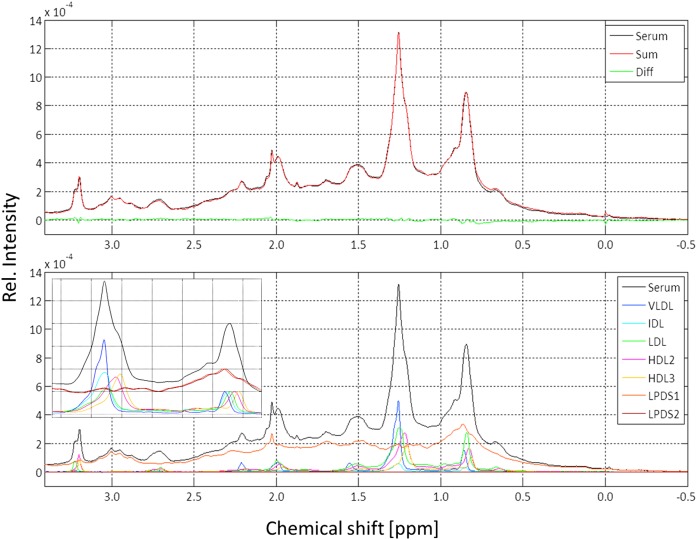
An important test for the validity of the separation techniques is the possibility to reconstruct the original serum spectrum from the individual spectra of all fractions isolated by sequential ultracentrifugation (Fig. 2). For obtaining meaningful results, fractionation of the serum and the spectral analysis was performed from a serum sample taken from a single healthy donor. The serum was separated in seven different fractions, the lipoprotein fractions VLDL, IDL, LDL, HDL2, HDL3, and two serum protein fractions LPDS1 and LPDS2. All fractions as well as the original serum sample were transferred into a synthetic “physiological” extracellular buffer (45, 51) that does not contain components visible by 1H NMR spectroscopy (see Materials and Methods). By this treatment, the low-molecular-mass compounds originally contained in the serum were also removed. The spectra were normalized to the same integral in the range from 3.2 to –0.5 ppm and thus correspond to the same number of proton spins in this range. On the basis of NMR-visible protons in the spectral range shown, an optimal fit of the serum spectrum could be obtained with a negligible residual between the fit and the original serum spectrum (Fig. 2). The corresponding weighting factors are given in Table 1. The largest contribution to the spectrum in the spectral range given comes from the serum proteins (64%, LPDS1 and LPDS2 in Fig. 2), not from the lipoproteins, and with 12%, the second largest contribution comes from the LDLs.
Fig. 2.
Reconstruction of a serum spectrum from its fractions obtained by sequential ultracentrifugation. All samples were transferred into extracellular buffer (45) as described in Materials and Methods. Only the highfield part of the 600 MHz 1H NMR spectrum is shown. Temperature was 310 K. Lines were broadened by applying an artificial line broadening of 2 Hz. Upper: Original serum spectrum (black) and synthetic spectrum calculated adding the appropriately weighted experimental spectra of the lipoprotein subfractions obtained by sequential ultracentrifugation of the same serum sample (red) and residual (green). Lower: All individual spectra that were added are shown with a color code indicated. (Insert) Methyl-methylene region of different subfractions. The intensity of the methyl signal was normalized here always to the same value. Diff., difference; Rel. relative.
TABLE 1.
Relative number of spins contributing to the 1H NMR serum spectrum
| Frequency Range (ppm]) | Group | Relative Proton Concentration (%) | ||||||
| VLDL | IDL | LDL | HDL2 | HDL3 | LPDS1 | LPDS2 | ||
| −0.5 to 3.5 | 7.2 | 1.5 | 12.0 | 9.6 | 5.6 | 57.6 | 6.7 | |
| 0.75 to 1.0 | CH3 | 6.3 | 1.7 | 16.3 | 11.6 | 6.7 | 52.5 | 6.4 |
| 1.1 to 1.35 | CH2 | 18.1 | 3.1 | 20.3 | 17.2 | 9.8 | 28.6 | 3.2 |
| 1.4 - 1.6 | CH2 in β-position to carboxyl | 6.3 | 1.5 | 11.9 | 8.2 | 4.6 | 59.8 | 6.3 |
| 1.9 - 2.1 | CH2 adjacent to double bonds | 6.9 | 1.4 | 10.4 | 7.9 | 4.9 | 56.9 | 9.7 |
| 2.1 - 2.3 | CH2 in α-position to carboxyl | 7.2 | 1.2 | 7.0 | 6.8 | 4.5 | 64.3 | 6.8 |
| 2.6 - 2.8 | CH2 between double bonds | 4.7 | 1.1 | 9.8 | 9.4 | 6.1 | 57.0 | 6.8 |
| 3.18 to 3.22 | N(CH3)3 of choline | 1.6 | 1.0 | 10.3 | 29.7 | 19.4 | 34.6 | 2.3 |
The 1H NMR signals in the given ranges between are quantified at 310 K. For details see Fig. 2. Chemical group assignments apply only for lipids, not for the LPDS-fractions.
Spectral features of the LPDS fractions
The signals of the serum proteins and the lipoproteins overlap in most regions; however, there are some positions in the spectral range considered where characteristic protein signals can be observed almost unperturbed. A well-defined singlet resonance at 2.05 ppm and a linewidth of approximately 5 Hz is only contained in the LPDS fraction. It corresponds to methyl signals of N-acetyl-glucosamine groups of the proinflammatory acute phase proteins α1-acid glycoprotein, haptoglobin, α1-antitrypsin, α1-antichymotrypsin, and transferrin (52, 53). This group of signals is called GlycA and is located at 2.0 ppm in the reference system of Otvos et al. (52). The chemical shift difference to our data is explained by the use of a somewhat different referencing that was shifted upfield by approximately 0.04 ppm relative to internal DSS (52). The triplet signal at 3.016 ppm only contained in the serum protein fractions could correspond to the ε-methylene groups of lysine residues in protein(s) [expectation value in proteins 3.004 ppm (54)]. Note that small molecules as isolated amino acids were already removed by the separation procedure. Protein signals in the range from 2.8 to 3.2 ppm probably correspond to β-methylene groups of histidine, phenylalanine, tyrosine, tryptophan, aspartate, asparagine, and cysteine methylene groups (54). The region downfield from the water signal of LPDS is dominated by the albumin signal.
Visibility of lipid signals from lipoproteins
The spectra of the lipoprotein fractions are dominated by the lipids contained in the lipoproteins but also contain weak apolipoprotein signals. These resonances of aromatic amino acids and amide protons of parts with high internal mobility are visible in the downfield part of the spectra (data not shown). Although the serum NMR spectrum can be reconstituted almost perfectly from the spectra of the fractions obtained by ultracentrifugation (Fig. 2), it does not imply that all constituents are completely visible. Large rotational correlation times lead to an extensive line broadening that finally results in the disappearance of parts of the expected signals in the background noise. Large rotational correlation times are usually found for high effective molecular masses and high local viscosities. Low temperatures and high pressures also have similar effects. Even at quite high temperatures, as 310 K, it cannot be assumed that the resonances of all molecules contained in the sample are visible in the solution NMR spectrum. For a quantitative analysis of lipoprotein concentrations in the native serum, it is mandatory to know what proportion of the lipid signals is actually observable in the spectra under different experimental conditions.
Lipid extraction and signal quantification in the extracts
In order to establish not only a relative but also an absolute quantification of all lipids, we extracted all lipids from the lipoprotein particles and performed 1H NMR spectroscopy directly within the extraction solvent. We chose an extraction procedure based on a similar protocol reported earlier by Srivastava et al. (55). By using deuterated chloroform and methanol as extracting agents (2:1 by volume), it was possible to extract all lipids in a single step without subsequent drying and redissolving in deuterated solvents. Pyrazine and TMS were also contained in the extraction solution in well-defined quantities as internal standard for quantification. Only the protein precipitate was removed to ensure homogeneity of the solution. Thus, sample loss and chemical modifications due to air oxidation were minimized. Because in the organic solvent, macromolecular assemblies are destroyed, the signals of all lipids can be observed and can serve as a measure for the absolute quantity of lipids contained in the native fractions. Always two aliquots of each sample were treated in parallel.
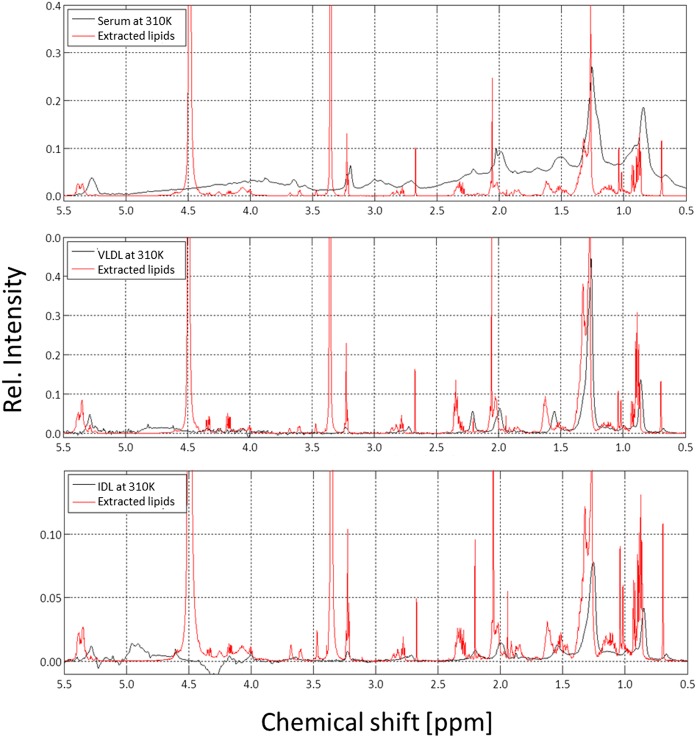
1H NMR spectra of the lipids extracted from human blood serum and different lipoprotein fractions are presented together with the spectra of the corresponding intact lipoprotein particles in Figs. 3 and 4. As expected, the signals of the extracted lipids of different lipoprotein fractions and the chemical shifts of their molecular components do not vary (see also supplemental Fig. S3, where the different extracts are directly compared); only the signal intensities of their constituents vary due to their variable concentrations. Signal assignment was based on published data (21) and was verified by the measurement of standard lipids (cholesterol; cholesteryl palmitate; 1,2-dioctyl-sn-glycero-3-phosphocholine; and 1,2-dipalmitoyl-sn-glycerol). In addition, assignments of the resonances were confirmed by 2D NMR spectroscopy (TOCSY) of the extracts. As an example, the TOCSY spectrum of LDL is shown in supplemental Fig. S4, together with typical coupling patterns of FAs, cholesterol, CEs, sphingosine, and lipid head groups. Except for minor differences, the line shapes and chemical shifts of the observed signals were identical to those obtained in rat liver extracts (20) and are given in supplemental Table S1.
Fig. 3.
1H NMR spectra of intact human serum, VLDL, IDL, and of the corresponding extracted lipids. The 600 MHz 1H NMR spectra of human blood serum (top), VLDL (middle), and IDL (bottom) together with the lipids extracted from the same samples. Samples were contained in synthetic extracellular buffer and measured at 310 K; extracted lipids were dissolved in CDCl3/methanol-d4 (2:1) and measured at 293 K. The spectra were referred to DSS or TMS, respectively, as internal standards. Rel., relative.
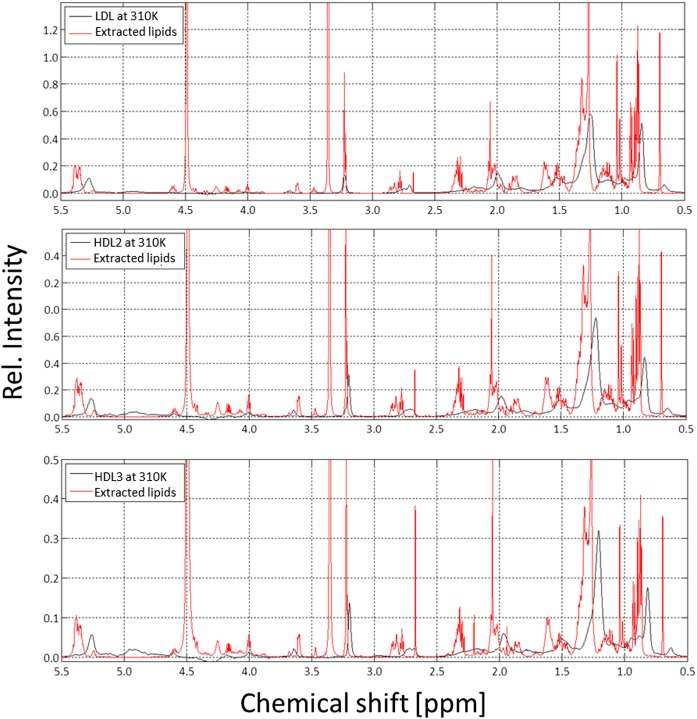
Fig. 4.
1H NMR spectra of human LDL, HDL2, HDL3, and of the corresponding extracted lipids. The 600 MHz 1H NMR spectra of human LDL (top), HDL2 (middle), and HDL3 (bottom) together with the lipids extracted from the same samples. Samples were contained in synthetic extracellular buffer and measured at 310 K; extracted lipids were dissolved in CDCl3/methanol-d4 (2:1) and measured at 293 K. The spectra were referenced to DSS or TMS, respectively, as internal standards. Rel., relative.
The main contribution to the spectra of the extracts are provided by the FA chains with signals of the terminal methyl (CH3) groups (0.75–1.0 ppm), of methylene (CH2) groups adjacent to other methylene groups (1.1–1.35 ppm), CH2 groups adjacent to or between double bonds (1.9–2.1/2.6–2.8 ppm), and CH2 groups in α- or β-position to carboxyl groups (2.1–2.3/1.4–1.6 ppm).
A characteristic contribution comes from the choline head groups found in PC and SM with sharp singlet signals at approximately 3.2 ppm. Additionally, cholesterol and CE show a variety of different small signals which cause a characteristic background signal. Two signals of cholesterol CH3 groups should be mentioned in particular, namely, C-18 methyl (0.7 ppm), which makes it easy to quantify the total amount of cholesterol, and C-19 methyl (1.02–1.04 ppm), which permits quantitative distinction between free and esterified cholesterol. An example for these assignments and the integration areas used in extracts of HDL is shown in supplemental Fig. S5.
As shown above, in native human blood serum, most of these signals can also be identified by comparing the characteristic spectra of the extracts with the untreated serum (Figs. 3 and 4). However, one has to take into account the chemical shift difference of TMS in chloroform-methanol and DSS in aqueous solution (see Materials and Methods) and the additional shifts depending on the particle size.
Accurate NMR quantification requires the use of internal standards. Thus, for intact lipoproteins in aqueous solution, we added pyrazine and 1,4-dioxane, which both give rise to singlets at 8.64 and 3.75 ppm, respectively. DSS was not a suitable standard for quantification because it appeared to interact with the lipoprotein particles and experienced considerable signal loss. In organic solutions, we used pyrazine and TMS. For the analysis of the absolute lipid concentrations in extracts, NMR experiments have only to be performed at one temperature. We selected 293 K because chloroform and methanol are volatile solvents. In these organic solvents, all lipids are completely dissolved at such a temperature.
For accurate quantification of the 1H NMR signals, relaxation effects on the signal intensity have to be minimized. Ideally, the repetition time (in our experiments 30 s) should be much larger than the relevant T1 times. This was clearly true for the lipid resonances in extracts and serum with T1 values. The largest value in extracts was found for methyl groups of FAs with 3.16 s. In serum lipoproteins, these values were substantially smaller (supplemental Table S2). For the internal standard pyrazine, T1 was 6.85 s at 293 K in the extract and in serum 6.92 s, and increased in serum at 323 K to 12.36 s (supplemental Table S3). For 1,4-dioxane, the relaxation properties were somewhat more favorable. Here, the T1 time was 7.0 s at the highest temperature. Details of the correction of (small) relaxation effects are given in Materials and Methods.
The extracts of the different fractions can be analyzed and quantified with respect to their main constituents. Although the general composition is quite similar, the relative concentrations of its components vary largely. Qualitatively, this can be seen in the overlay of the 1H NMR spectra of different lipoprotein fractions with their corresponding extracts presented in Figs. 3 and 4. As an example, the results obtained from a sample of a healthy human donor are presented in Tables 2 and 3.
TABLE 2.
Typical composition of a different lipoprotein fractions of a healthy donor obtained from the analysis of the 1D spectra
| Serum | VLDL | IDL | LDL | HDL2 | HDL3 | |
| Relative Molar Ratio [%] | ||||||
| Total cholesterol | 54.7 | 35.6 | 57.4 | 67.1 | 54.5 | 46.4 |
| Cholesterol | 14.3 ± 0.9 | 13.5 ± 2.1 | 19.2 ± 3.4 | 18.6 ± 0.1 | 14.4 ± 0.3 | 7.0 ± 0.1 |
| CE | 40.5 ± 0.9 | 22.0 ± 2.1 | 38.3 ± 3.4 | 48.4 ± 0.1 | 40.1 ± 0.3 | 39.3 ± 0.1 |
| Ratio CE/cholesterol | 2.84 | 1.63 | 1.99 | 2.60 | 2.78 | 5.61 |
| SLs | 6.9 ± 2.3 | 4.0 ± 0.9 | 6.5 ± 0.8 | 6.0 ± 1.8 | 5.9 ± 2.4 | 8.0 ± 0.4 |
| PGs | 25.2 | 21.4 | 21.9 | 22.0 | 33.8 | 38.4 |
| Ratio PG/SL | 3.6 | 5.4 | 3.4 | 3.7 | 5.8 | 4.8 |
| TG | 9.0 | 37.8 | 13.2 | 4.1 | 3.5 | 4.9 |
| Plasmalogenes | 4.1 | 1.4 | 0.9 | 0.8 | 2.3 | 2.4 |
| Choline | 24.0 ± 0.1 | 19.0 ± 5.3 | 18.9 ± 5.0 | 21.1 ± 4.7 | 32.3 ± 3.8 | 36.5 ± 2.5 |
| Ethanolamine | 0.9 | 1.1 | 1.4 | 0.4 | 0.1 | 0.4 |
| Inositol | 0.4 | 1.2 | 1.6 | 0.5 | 1.4 | 1.5 |
| Ratio CE/TG | 4.5 | 0.6 | 2.9 | 11.7 | 11.4 | 8.0 |
| FAs | 110.3 | 177.4 | 113.2 | 92.1 | 106.9 | 115.6 |
| Saturated FAs | 31.7 | 40.3 | 45.4 | 39.2 | 45.4 | 49.2 |
| Average chain length | 16.5 | 16.4 | 17.0 | 17.9 | 17.7 | 17.1 |
| Unsaturated FAs | 68.3 | 59.6 | 54.6 | 60.8 | 54.6 | 50.8 |
| Monounsaturated | 24.1 | 28.5 | 13.7 | 12.4 | 7.0 | 2.2 |
| Diunsaturated | 31.9 | 22.8 | 30.3 | 37.0 | 34.4 | 33.6 |
| Triunsaturated | 12.3 | 8.3 | 10.6 | 11.5 | 13.2 | 15.0 |
Values given correspond to the molar ratio relative to the sum of total cholesterol, TGs, and PLs and were calculated from the NMR spectra of the lipid extracts.
TABLE 3.
Relative visibility Irel of all observed signals shown in given spectral ranges
| Signal | T (K) | Relative Visibility Irel (%) | ||
| LDL | HDL2 | HDL3 | ||
| CH2/CH3 region (0.1–3.0 ppm) | 283 | 14.1 | 34.3 | 55.9 |
| 293 | 21.9 | 44.4 | 65.2 | |
| 303 | 36.3 | 53.9 | 73.8 | |
| 313 | 42.4 | 58.4 | 78.6 | |
| 323 | 45.1 | 61.3 | 82.0 | |
| Total cholesterol C-18H3 (0.62–0.78 ppm) | 283 | 14.3 | 40.0 | 72.0 |
| 293 | 19.4 | 50.4 | 77.7 | |
| 303 | 32.3 | 57.8 | 84.9 | |
| 313 | 38.6 | 61.8 | 89.8 | |
| 323 | 41.8 | 64.4 | 94.2 | |
| CH3 FAs (0.7–1.0 ppm) | 283 | 21.3 | 35.3 | 51.9 |
| 293 | 38.1 | 50.0 | 70.1 | |
| 303 | 64.1 | 63.7 | 79.5 | |
| 313 | 73.1 | 70.1 | 86.9 | |
| 323 | 76.2 | 73.7 | 91.7 | |
| Bis-allylic-methylene (2.6–2.8 ppm) | 283 | 6.2 | 23.0 | 41.4 |
| 293 | 13.9 | 35.6 | 56.2 | |
| 303 | 36.1 | 44.8 | 71.4 | |
| 313 | 40.4 | 52.3 | 76.8 | |
| 323 | 43.1 | 55.1 | 82.6 | |
| Choline N+(CH3)3 (3.0–3.4 ppm) | 283 | 26.2 | 53.9 | 66.7 |
| 293 | 29.1 | 62.3 | 71.5 | |
| 303 | 38.1 | 70.1 | 77.5 | |
| 313 | 42.8 | 74.7 | 87.1 | |
| 323 | 46.4 | 77.6 | 89.2 | |
The relative visibilities Irel are calculated from the normalized individual peak areas in the lipoprotein spectra ILP divided by the corresponding peak areas Ilipid of the extracted lipids. The given values are the average of the two samples of the same healthy donor. The ppm values given correspond to the approximate limits of the peaks in native lipoprotein preparations. The error estimated from the two parallel estimations is about 5%. The intensity of the noncholesterol methyl groups is obtained as described in Materials and Methods. Bold type indicates the recommended experimental temperature.
The highest relative molar concentrations of total cholesterol and CEs in all lipoproteins are found in LDL. Also, the highest ratio of CE to TG is observed here. The CE to cholesterol ratio is highest in HDL3. In HDL3, the relative concentrations of typical membrane lipids (PG, SL) are higher than in other lipoproteins. A corresponding marker is also the choline head group. In addition, the relative number of double bonds in FAs can be determined from the signals. The different lipids contained in the extracts can be quantitatively analyzed by selecting resonances of characteristic groups. Details on the quantification method are given in Materials and Methods. As an example, the total cholesterol concentration can be determined from the C-18 methyl resonance at 0.69 ppm, and cholesterol and CEs can be distinguished by their C-19 methyl resonances at 1.02 and 1.04 ppm. Triacylglycerol can be recognized by signals in the range between 4.30 and 4.38 ppm, phosphatidylinositol by signals between 3.74 and 3.82 ppm, PCs and SMs by signals between 3.20 and 3.26 ppm, sphingolipids (SLs) including their main component SM alone in the range between 5.50 and 5.76 ppm, plasmalogens by signals between 5.89 to 5.96 ppm, and ethanolamine containing lipids by the signal of the methylene group at 3.102 ppm (3.07–3.13 ppm). Monoacylglycerides and diacylglycerides were neglected. A distinction between ceramide and lyso-compounds was not established (supplemental Table S4).
A detailed analysis of the FA composition is impossible using only 1H NMR spectra because the spin system of the FA chains and the lipid head groups was disturbed by the ester group of the lipids. However, the intensity ratios of the FA signals were used to estimate a rough FA characterization. All FAs contain one terminal methyl group, which can be identified by signals in the range of 0.73–0.95 ppm. Quantification, however, is imprecise because the contribution of methyl groups from other sources is necessary (see above). Signals between 1.22 and 1.42 ppm depict methylene group signals, which provide insight into the saturation degree of the FAs. The more double bonds a FA has, the fewer methylene groups are present while maintaining the chain length. Signals between 2.24 and 2.44 ppm and between 1.55 and 1.77 ppm reflect methylene groups in α- or β-position to the carboxyl group functionality, respectively. These, in contrast to the methyl groups, are preferred for quantification since superposition is reduced and both groups only exist once in a FA. Unsaturated FA can be identified at different positions as well. Signals between 1.95 and 2.44 ppm, 2.73 and 2.80 ppm, and 2.80 and 2.93 ppm reflect protons next to, between two, or between more than two double bonds, respectively. Signals in the range between 5.16 and 5.53 ppm correspond directly to the double bonds in methine groups.
The average chain-length distribution can be determined from the ratio of the relative concentration of FA methyl groups to FA methylene and methine groups. However, details of the distribution itself cannot be obtained from the 1D spectra without additional information. A multi parameter fit of the spectral data assuming a continuous length distribution gives reasonable results (see supplemental Table S5).
Quantitative analysis of intact lipoproteins with their broad lines was more difficult than the analysis of lipid components with narrow lines. In extracts, most lines are well separated. In intact lipoprotein samples, the exact peak area of more complex but clearly separated signals like the choline head group at 3.2 ppm or the bis-allylic methylene group at 2.8 ppm could be determined by simple integration without line shape fitting. FA methyl signals at 0.8 ppm, however, were highly overlaid by signals of cholesterol backbone methyl groups. For separating the signals of noncholesterol methyl groups from cholesterol signals, we subtracted a suitably broadened, weighted neat cholesterol NMR spectrum. The weighting factor was determined from the intensity of the C-18H3 signal. Thus, we obtained a reduced spectrum that contains only the methyl groups of the FAs for integration (supplemental Fig. S6).
Temperature-dependent visibility and thermodynamic analysis of lipoprotein signals
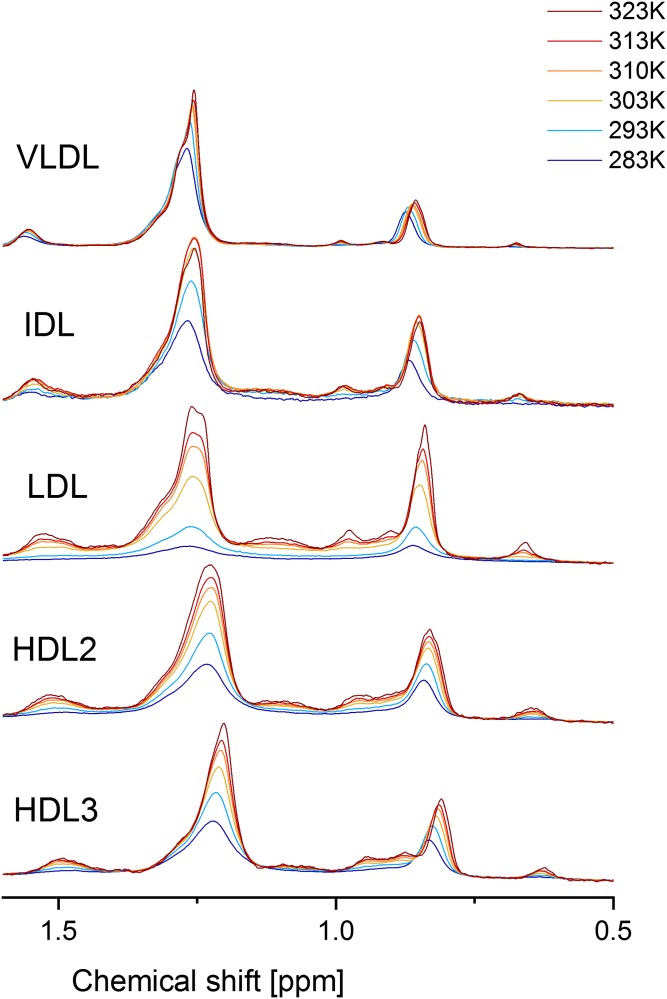
A series of 1H NMR spectra of serum, VLDL, IDL, LDL, HDL2, and HDL3 were recorded in steps of 10 K in the range of 283 K up to 323 K. The methyl and methylene regions of the corresponding spectra at these temperatures are depicted in Fig. 6. We corrected all determined signal intensities of the different lipoproteins using the signals of the internal standards in order to minimize external influences. All lipoproteins show characteristic 1H NMR signal increases and line shape variances due to higher lipid mobility, phase transitions, and lipid rearrangement during the heating process. The VLDL and IDL sample preparations were not stable at high temperatures for longer times. This can clearly be seen for IDL in Fig. 6, where the intensities of the methylene and methyl resonances decrease at 323 K. This is indicative for a denaturation/precipitation of particles. Consequently, this temperature point had to be omitted in the quantitative analysis of VLDL and IDL spectra.
Fig. 6.
Temperature-dependent 1H NMR spectra of human VLDL, IDL, LDL, HDL2, and HDL3 from 283 K up to 323 K. All spectra were referred to DSS, and temperature-dependent sensitivity variations are corrected by internal standards.
For the actual study presented in the following, we prepared two sets of lipoprotein fractions taken from the same donor at the same time and extracted the lipids from defined aliquots of these lipoprotein preparations. The two data were evaluated separately for obtaining an estimate for the experimental error resulting from the sample preparation and data evaluation. The 1H NMR spectra of the corresponding extracts and the original preparations were recorded and intensity was corrected as described above. The signal shape did not vary considerably in all spectra at matching temperatures. Thus, the spectra of the two lipoprotein isolations could be compared with the temperature-dependent spectra just shown. By comparing the integrals of the corresponding frequency ranges, we defined a relative visibility index Irel by calculating the integrals in predefined spectral ranges in the lipoproteins spectra and the extract spectra, which gave us information about how many lipids are absolutely visible in an intact lipoprotein particle observed with NMR spectroscopy at a chosen temperature (Table 4). The integrals were appropriately normalized as described in Materials and Methods. Besides slight deviations, we achieved very good agreement of both datasets.
For a fit of the data of LDL and HDL, we used the three-compartment model described above. For VLDL and IDL experiments, absolute quantification of lipids failed, and therefore the total visibility could not be determined. Therefore, the data could only be analyzed with the plausible assumption (see Discussion) that compartment C is very small and can be set to 0% in the evaluation.
Applying equation 8 to our data, we found that a stable solution was only obtainable when assuming (as usually done in literature) that the change of the heat capacity can be neglected (ΔCp = 0) and defining the compartments to be positive (A, B, C ≥ 0). In fact, in datasets that can be fitted acceptably without neglecting ΔCp, ΔCp is relatively small and does not affect the values of the other parameters significantly. The parameters defined by the fit function are the transition temperature Tm of the system and the change of enthalpy ΔH during the phase transition. The parameters A and C reveal the relative amounts of lipoprotein signals that are always detectable by solution NMR or are not detectable at all (in a reasonable temperature range).
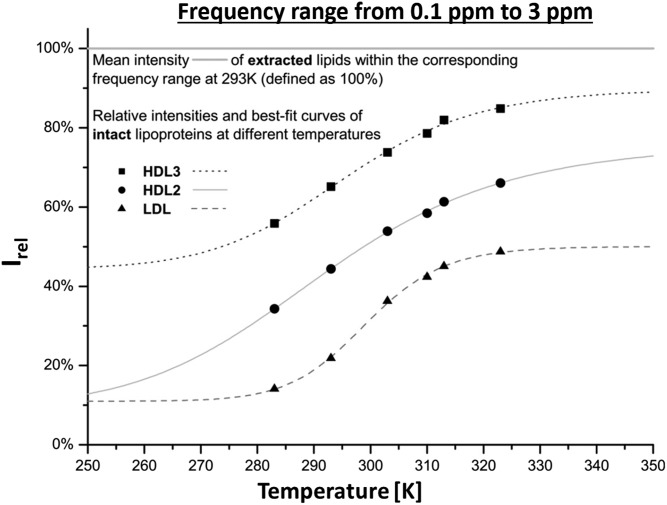
First, we examined the frequency range from 0.0 to 3.0 ppm that contains the majority of methyl and methylene groups of FAs and most of the cholesterol signals to get an overview of more or less all lipids present in the particle. Of course, all DSS signals were excluded. The resulting temperature dependence of the main lipid constituents shows typical sigmoidal characteristics, which is most prominent for LDL data (Fig. 5). The obtained melting temperatures were highest for VLDL with Tm = 314 K, followed by 299 K, 295 K, 291 K, and 288 K for LDL, HDL3, HDL2, and IDL, respectively (Table 4). The same order was also obtained for the melting temperatures averaged over all signals analyzed. The enthalpy difference of LDL was also highest with ΔH(Tm) = −107.8 kJ/mol compared with −60.8 kJ/mol (HDL3) and −42.6 kJ/mol (HDL2) (Table 4). The overall NMR visibility of LDL was very low, so that even at high temperature, not even 50% of the lipid intensity could be detected. On the contrary, HDL3, which is significantly smaller than LDL, revealed almost 100% at high temperature but also nearly 50% of the signal intensity was visible at low temperature. HDL2 showed average visibility but a less pronounced sigmoidal curve. For VLDL and LDL, compartment A was rather large, indicating that even at the lowest temperature, a substantial part of the signal intensity can be observed.
Fig. 5.
1H NMR signal enhancement as a function of temperature. The integrals Irel = ILP/ILipid from 0.1 to 3.0 ppm of the 1H NMR spectra of LDL, HDL2, and HDL3 are plotted in relation to the corresponding extracted lipid signal, which was defined as 100%. All lipoprotein subclasses show successive intensity increase along with the temperature, but do not reach the absolute lipid intensity of 100%. HDL3 gains about 90%, HDL2 75%, and LDL merely 50%. Irel was fitted with equation 8. The fit values are given in Table 4.
As can be seen in Fig. 6, not only the intensity increased with increasing temperature in all lipoproteins including VLDL and IDL (neglecting precipitation of IDL at 323K), but also the line shape varied, indicating that functional groups and lipids were affected by the temperature in different ways. Specific functional groups gave rise to clearly separated peaks, which makes them available for integration. We chose the broad methyl group signal at about 0.8 ppm, the cholesterol C-18 signal at 0.7 ppm, the choline head group signal at 3.2 ppm, and the bis-allylic methylene signal at 2.8 ppm. The quantification strategy has already been discussed in the previous section. For all individual functional groups, the corresponding intensities of the lipid extracts were used to calculate the relative intensity. The obtained data and the corresponding values are shown in Figs. 5–7, respectively.
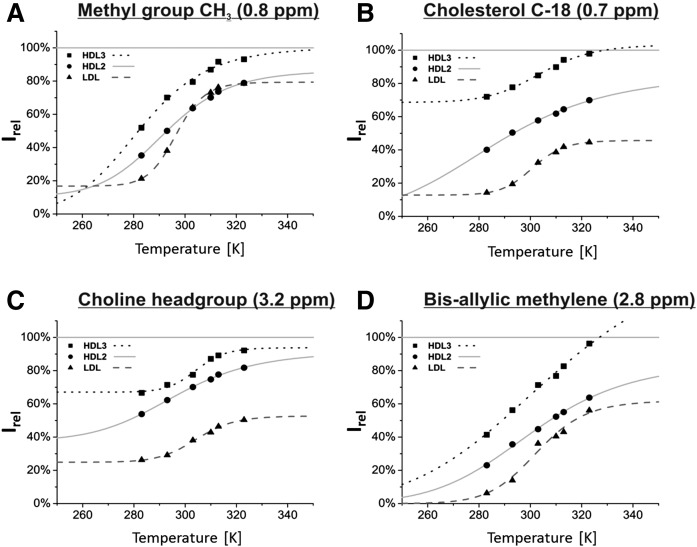
Fig. 7.
Temperature dependence of the lipid visibility Irel of specific functional groups. Some well-separated lipid NMR signals were evaluated. To improve accuracy, the baseline around each signal was adjusted separately, and the integral limits were framed into local minima next to the peaks. Note that the temperature dependence of the methyl signal (A) represents the FAs only since the cholesterol signal was first subtracted as described before (see also Fig. 6). The overlaying methanol signal next to the choline head group [C, -N(CH3)3, ∼3.2 ppm] in extracts was subtracted by deconvoluting the spectra, as was discussed earlier. The resulting integrals were compared with the corresponding integrals of the extracted lipids, respectively. The temperature dependency of Irel was fitted with equation 8; the fit parameters are listed in Table 4.
Compared with the overall frequency range just discussed, the general shape of the intensity gain with temperature of the chosen signals is quite similar. The bis-allylic methylene signal reveals a rather poor signal-to-noise ratio and can therefore not be fitted very accurately. Except of VLDL, it shows a low NMR visibility at low temperatures so that for all lipoprotein subclasses the curve approaches almost zero. The choline head group on the contrary, shows in general a high basic visibility (parameter A), especially regarding HDL. The relative intensities of cholesterol differ highly referring to the individual lipoprotein subclasses. Whereas LDL cholesterol tended to be rather invisible at all times, HDL3 showed a visibility of 70% even at low temperatures. The terminal methyl groups of FAs, on the other hand, behaved quite similarly in all lipoproteins. Note that the contributions of cholesterol were removed by our procedure in Fig. 7, but not in Fig. 5, where a large spectral range was used. Apart from the more distinct sigmoidal curve in LDL, all lipoprotein subclasses showed an intensity gain that covered a wide range of relative intensities, meaning that within the limits of accuracy, the terminal methyl groups from FAs altered from complete invisibility to complete visibility for HDL but not in LDL (Table 4).
Transverse relaxation times of serum lipoprotein signals
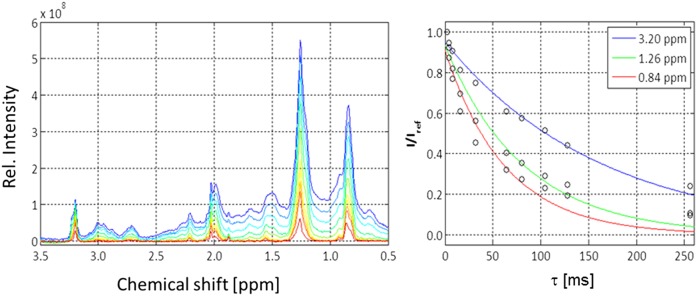
The signals of the different lipid groups of serum lipoproteins appear to be rather broad. The apparent transverse relaxation times T2,app calculated from the linewidth Δν1/2 at half-maximum were correspondingly small (Fig. 8 and Table 5). However, for inhomogeneous lines, this does not give the intrinsic transverse relaxation times caused by the physical relaxation processes. Therefore, we determined the relaxation times of different samples by a CPMG spin-echo sequence. Fig. 8 shows the NMR signals at different waiting times τ. As to be expected, the signals decreased continuously with the waiting time τ. Signal positions at 0.84, 1.26, and 3.2 ppm were selected that corresponded to the maximum of the inhomogeneous resonance lines of the lipid methyl, lipid methylene, and choline N+(CH3)3 groups. The low-molecular-mass compounds were removed by exchange with the physiological buffer. As to be expected, we observed a multiexponential decay because signals of different lipoproteins as well as protein signals were superposed. The protein signal strongly contributed to the signals at 0.84 and 1.26 ppm that clearly showed a slower-decaying component. By measuring separately, the T2 relaxation times of the serum-protein fraction LPDS one obtains for these signals a transverse relaxation time of the order of 20 ms, explaining the slow components of the signal decays. However, for an approximate estimate of the average relaxation the data were fitted by a monoexponential function. The values are given in Table 5 together with the apparent relaxation times. Clearly the homogeneous linewidths are much smaller than the inhomogeneous linewidths. The inhomogeneous linewidths of the methyl and methylene signals were larger by a factor of 17 and 20 than the intrinsic linewidths, respectively. This indicates that the inhomogeneous linewidth is mainly determined by the size distributions of the particles, not the intrinsic linewidths. For completeness, also the corresponding longitudinal relaxation times discussed earlier are given. As to be expected for higher rotational correlation times, they are substantially larger by approximately one order of magnitude than the transverse relaxation times, although they were measured at a lower temperature (293 K) than the T2 times. At 310 K, they should be even larger.
Fig. 8.
T2 relaxation of serum lipoproteins. T2 relaxation times of serum in the physiological extracellular buffer were measured with a CPMG-pulse sequence at 310 K. Left: NMR spectra recorded with different total waiting times τ, with τ = 2 ms (blue) and τ = 256 ms (red). The color code of the other spectra corresponds to an increase of the τ values. The actual τ values were 2, 4, 8, 16, 32, 64, 80, 104, 128, and 256 ms. Right: Plot of the signal intensities at 3.2, 1.26, and 0.84 ppm corresponding to the signals of choline-N(CH3)3, FA methylene groups, and methyl groups of FA and cholesterol as a function of τ. Rel., relative.
TABLE 5.
T2-relaxation times of lipid signals of human blood serum
| FA-CH3 (0.84 ppm) | FA-CH2 (1.26 ppm) | Choline -N(CH3)3 (3.25 ppm) | |
| T1 [s] | 0.71 ± 0.01 | 0.66 ± 0.01 | 0.64 ± 0.02 |
| T2 [ms] | 63 ± 20 | 81 ± 18 | 163 ± 25 |
| Δν1/2 [Hz] | 5.1 | 3.9 | 2.0 |
| T2,app [ms] | 3.7 | 4.0 | 11.8 |
| Δν1/2, app [Hz] | 86 | 79 | 27 |
The relaxation times T1, T2, and the full linewidth at half height were obtained at 310 K from a fit of the data presented in Fig. 6. The apparent relaxation times T2, app was calculated from the linewidths at half height of the corresponding in homogeneously broadened peaks.
Pressure dependence of the lipid signals of human blood serum
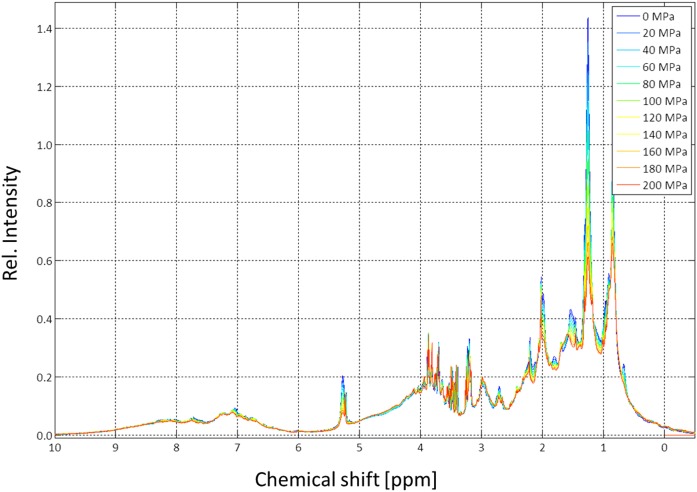
We applied pressures between 0.1 and 200 MPa with a home-built system (see Materials and Methods) at a 1H resonance frequency of 800.2 MHz to human serum. In contrast to the former samples, the actual sample still contained all small, natural metabolites. When pressure was applied to human serum proteins, the signal intensities of the lipoproteins decreased substantially, whereas the signals of the low-molecular-mass compounds slightly increased, and the signals of soluble serum proteins remained almost unaffected. The behavior of the low-molecular-mass components is to be expected because the pressure does not have an effect on their structure, but the compression of the solvent leads to an additional small increase of their concentrations.
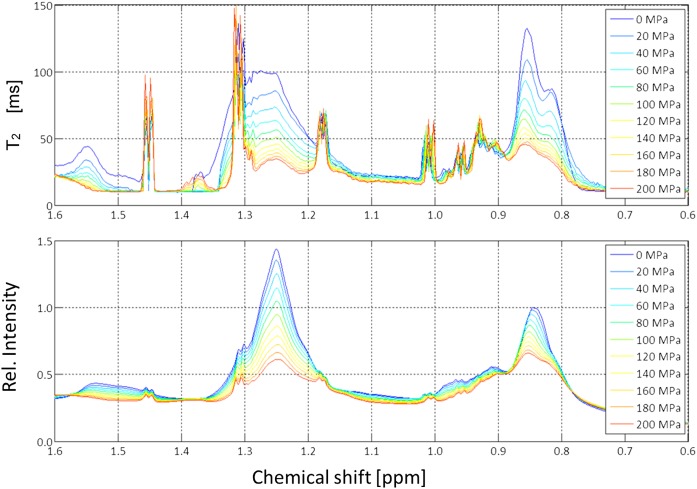
Because pressure has an influence on phase transitions as well as on viscosity, transverse relaxation times T2 were measured with a CPMG-pulse sequence at different pressures (Fig. 9). Only the high field region was depicted in more detail in Fig. 10 together with the relaxation times obtained for different pressures by fitting the data locally with an exponential function. The obtained values correspond to the weighted average of all signals contained in a slot of 0.003 ppm and thus were determined by the compound with the highest signal amplitude in the given slot. Note that the T2 values shown here at ambient pressure are somewhat higher than those given in Table 5. The small differences can be explained by the fact that the temperature in the measurement depicted in Fig. 10 was higher, and, probably more important, that the data were taken from different serum preparations. In Fig. 8, serum was exchanged with physiological extracellular buffer, and in Figs. 9 and 10, only 10% D2O was added to the native sample for providing a lock signal. The linewidths increased rapidly with increasing pressure; at 200 MPa the intrinsic transverse relaxation times of the lipid signals decreased by approximately 60%. This indicates that, as to be expected, the local effective viscosity inside the particles was increasing with increasing pressure.
Fig. 9.
Pressure dependence of lipid signals in human blood serum. The 800 MHz spectra of human blood serum were recorded at 313 K. The signal amplitude of the methyl group at 0.1 MPa was set to 1. Rel., relative.
Fig. 10.
Pressure dependence of the transverse relaxation times of lipid signals in human blood serum. Upfield part of the 800 MHz spectrum of human blood serum recorded at 313 K. Same sample as shown in Fig. 9. Bottom: The 1D NMR spectra at different pressures. Top: T2 values measured with a CPMG-pulse sequence at various pressures. The T2-values were calculated for small slots of 0.003 ppm and depicted as a function of chemical shifts. Rel., relative.
DISCUSSION
In this article, we investigated 1) what proportion of the expected lipid signals for different lipids in different lipoprotein (sub)classes can be observed at different temperatures by 1H NMR spectroscopy; and 2) characterized these effects by a thermodynamic analysis of the temperature (and pressure)-dependent changes of spectral shapes and intensities.
Contribution of protein signals to the lipoprotein signals
For obtaining a consistent set of observations for the main analysis of the visibility of lipids in different lipoprotein subclasses and their thermodynamics, it was mandatory for this study to prepare all lipoprotein fractions from the single large blood sample of the same donor. Because of the relatively time-consuming experiments the sample had to be frozen and stored at 253 K before being used. In agreement with literature (see, e.g., Refs. 56–59) and as discussed in Materials and Methods, only negligible effects are to be expected when native or frozen serum is studied. The possible variations of the observed features in different individuals are not topic of this paper, but variations can be expected when the lipid composition is different.
One-dimensional 1H NMR spectroscopy is the method of choice for high-throughput analysis of lipoprotein subclasses in clinical medicine (e.g., Refs. 29–43). From the NMR point of view, it is not clear what macromolecular components of the serum are really visible in the spectra and how well they can be directly quantified. In the serum itself, the protein fractions LPDS1 and LPDS2 significantly contributed to the total NMR signal intensity (number of protons observable), ranging from 59% in the methyl region to 32% in the methylene region of the spectra at 310 K (Table 1). In the spectral range from −0.5 to 3.5 ppm, it was 64.2%. Omitting this contribution would give much too high values of the lipoprotein concentrations; care has to be taken to handle this contribution properly.
In the main part of our study, we used purified lipoprotein fractions. Because here these proteins were not present, only signals of the apolipoproteins could add up to the lipid signals and influence the analysis.
Mobile regions of the apolipoproteins could contribute to the “lipid” signals, an increase of the apparent visibility would result, the size of compartment A would increase, and the size of compartment C would decrease and potentially would be negative. Using the lipid composition determined in our sample (Table 2), the number of methyl groups in each lipid (supplemental Table S1) and an average of 3,000 and 200 lipid molecules per LDL and HDL particle, respectively (60), we could calculate the maximum number of lipid protons in the methyl region (0.7 to 1.0 ppm) of the spectrum as approximately 43,000 (8,300 in FAs only) in LDL and 2,500 (700 in FAs only) in HDL. In comparison, the signals of all methyl groups of leucine, isoleucine, and valine of apoB-100 in LDL could contribute at most 6,379 methyl protons. For being visible in the methyl region, all amino acids of the protein would have to be in highly mobile regions of the protein, a property that is very unlikely. In this unlikely case, about 13% of all signals in the methyl region could come from the protein. HDL particles of different subclasses contain different numbers of apoA-I and apoA-II. Assuming that large HDL contains four copies of apoA-I, we would expect 1,248 protein protons in the methyl region, corresponding to 33% of all signals in the methyl region expected. This means that even when all protein signals can be observed, their contribution to the methyl signal between 0.7 and 1.0 ppm can be almost neglected in LDL, but not in HDL. In practice, only mobile, unstructured parts of the apolipoproteins should contribute in the rather narrow range from 0.7 to 1.0 ppm, because folding leads to additional shifts and immobilization in the membrane to severe line broadening. It is not unlikely that compartment A contains protein signals while the basic signal intensity is approximately independent of temperature. The predicted maximum protein signal contribution of 33% could also partly explain the relative high values obtained for compartment A in HDL2 of 10.5% of the signal intensity (Table 4). Even for LDL, the maximum protein signal contribution of 13% is close to the experimental value of 16.8% in compartment A.
Anyway, an enhanced total visibility toward smaller particle sizes remains (Tables 3 and 4) for LDL and HDL. If the signal contribution of apolipoproteins can be neglected, an alternative explanation would be a looser packing of lipids and thus higher mobility due to the higher curvature of smaller particles.
NMR visibility of lipid signals of lipoprotein particles
The compartments A, B, and C are primarily introduced for describing the experimental data, namely, that for a given reporter group (e.g. the methyl group of FAs), some signals are always observable (A), some show a typical two-states phase-transition behavior going from undetectable (very broad) lines to lines with a sufficiently narrow linewidth (B), and some predicted to be never visible from the melting curve in temperature range studied (C). This model neither requires that the reporter groups are located in a well-defined closed spatial region nor that different reporter groups reflect the same spatial regions. Nevertheless, a simplified structural model for a lipoprotein such as been proposed, e.g., by Hevonoja et al. (60) for LDL would make sense for model building and discussion. This model has also three compartments, but now defined by their location and composition in the particle, an outer membrane shell, followed by a shell with rather low density and a more densely packed core. At low temperature, the outer shell mainly consists of CEs, the core of TGs. At high temperature, the TGs and CEs are not separated anymore. More recent cryo-electron microscopy (cryo-EM) studies at 279 K reveal planar CE layers that are 3 nm apart. It is assumed that the cholesterol ring structures of CEs constitute these denser regions and the FA (tails) the less dense regions (61).
At 310 K, in LDL, about 55% of all expected lipid signals in the range from 0.1 to 3.0 ppm (Table 3) cannot be observed, whereas in HDL with 82%, almost all expected lipid signals can be observed in the limits of error at high temperatures (Table 4). However, when protein signals would significantly contribute to the proton NMR signal (compartment A), also in HDL not all expected lipid signals would be observable.
Compartment B is characterized by a strong temperature dependence of the signal intensity that can be described by a phase transition with melting temperatures between 282 and 319 K for the different lipoprotein fractions and reporter groups (Table 4). In this model, the region with immobilized lipids may shrink with temperature without changing the local lipid composition, or different lipids may be redistributed, creating regions with higher mobility.
Compartment C contains reporter groups in an environment where either their linewidths are very large because of low mobility (large rotational correlation times) or large chemical shift dispersion. These do not significantly contribute to the observed signals. In a rigid-body approximation, one would expect linewidths of the order of 60–300 Hz (0.1–0.5 ppm at 600 MHz) for HDL3 particles, but much larger linewidths for the other particles. In contrast, the observed lipid resonances in serum have rather small intrinsic linewidths between 2 Hz [N+(CH3)3] and 5 Hz (CH3) at 310 K, as determined by CPMG experiments (Table 5).
Different reporter groups reflect different local environments
The positively charged choline trimethylamine group should be preferably found at the outer surface of the membrane and thus would be a marker for that structural compartment. For the two HDLs, HDL2 and HDL3, the compartment C is approximately zero within the limits of error that is the choline groups can be detected completely at elevated temperatures (Table 4). In contrast, in LDL, about 50% of the choline head groups cannot be observed even at 325 K.
This means that in the LDL membrane, the choline head groups are highly mobile in about 50% of the lipids, but 50% are immobilized. The highly mobile groups are probably located in regions of the membrane in the liquid crystalline state. For an immobilization of a lipid group in the membrane, there are two plausible reasons, an interaction with proteins or the formation of more rigid lipid raft-like clusters with high viscosity.
The signals of methyl groups of lipid FAs were found to vary from nearly complete invisibility to almost complete visibility when passing from low to high temperature for LDL and HDL. The intrinsic linewidths of the methyl as well as methylene groups of FAs were again unexpectedly small, almost as small as those observed for small molecules such as lactate (Figs. 8 and 10; Table 5). This means that, surprisingly, the local viscosity of the lipid phase in these particles was very low and comparable to that of the aqueous phase. The spin-spin-relaxation rates and thus the individual linewidths of the “visible” signals was increasing with increasing pressure and decreasing temperature. However, as Fig. 10 shows, this increase was rather moderate and would not explain the intensity reduction by pressure (or by temperature). Again, only a phase transition to a phase with much higher viscosity and thus much lower mobility can explain the observations.
The visibility of the FA methyl groups at high temperatures was rather high; compartment C varied from 22% for LDL to 0% for HDL3 (Table 4). Other FA groups showed a somewhat smaller visibility than methyl groups, even the bis-allylic methylene group as a reporter for unsaturated FA chains. Using the cholesterol C-18 methyl group as reporter for the maximum visibility of the rigid cholesterol ring gives an extremely low visibility in LDL even at high temperatures of 46%, but a complete visibility in HDL3. For VLDL and IDL, no absolute analysis of the maximum relative visibility was available because we did not measure the total concentration. However, it is plausible that, again, compartment C is very small.
Transition temperatures
The temperature at which LDL undergoes cooperative melting varies among individual donors and with differing lipid composition (10, 11). Hence, diseases that affect the lipid composition have a great influence on the phase-transition temperature Tm as well. In this study, we chose the sample of one healthy donor in order to obtain a homogeneous data set for the different studied particles, which is not averaged due to their origin from different donors.
We found that transition temperatures averaged over all reporter groups between 290 K (IDL) and 311 K (VLDL). For the FA methyl groups, the melting temperatures varied between 282 K (HDL3) and 319 ± 3 K (VLDL). Except for VLDL, the transition temperatures were below the body temperature of 310 K in all observed lipoproteins (Table 4; Figs. 5–7). This suggests that the transition temperature in VLDL describes a different process than in the other lipoproteins.
The transition temperatures in the observed frequency range from 0.1 to 3.0 ppm constituted a mean for the different contributing reporter groups. The melting point dropped from 314 K in VLDL to 288 K in IDL, with LDL, HDL3, and HDL2 in between. However, the rather large calculated standard errors reduced the significance of this statement (Table 4). As expected, the transition temperatures were somewhat different for different reporter groups analyzed because their direct environments were influenced by their location in the particles determining the temperature dependence. In IDL and HDL2, the lowest melting point was found for cholesterol; in LDL and HDL3, methyl groups of FAs; and in VLDL, in choline.
A possible explanation described in literature would be that the smectic to liquid phase transition temperature caused by the CEs within the particle core is rising with an increasing ratio of CEs/TGs (10, 11, 13–15). However, in our case, this seems not to be the dominant factor because the CE/TG ratio determined in our particles does not strictly correlate well with the observed melting temperatures Tm (Tables 2 and 4).
The signal of the trimethylamine group is a reporter group for the membrane surface. It shows somewhat higher melting temperatures with 301 K (VLDL), 295 K (IDL), 304 K (LDL), 296 K (HDL2), and 304 K (HDL3) than the core lipids (Table 4). The phosphoglyceride (PG)/SL ratios in Table 2 revealed a maximum for HDL2 particles of 5.8 (VLDL, 5.4; IDL, 3.4; LDL, 3.7; HDL3, 4.8). Again, a clear correlation with the melting temperature cannot be observed.
The melting temperature of 301 K determined here for LDL (Table 4) by using cholesterol as NMR reporter group is close to 303 K, a value also observed earlier by differential scanning calorimetry (DSC) (6). Similar results were also reported by Kroon (17) from NMP spectroscopy and DSC from LDL, where a Tm of 304 K was found. More recent DSC and pressure perturbation calorimetry studies (61) display this transition that is supposed to correspond to the reversible smectic-to-disorder phase transition in the core CEs in LDL. However, with FA methyl groups as reporter group, we found a significant lower melting temperature of 297 K. Similar results were also reported by Kroon (17) from NMR spectroscopy and DSC of LDL, where a Tm of 304 K was found. An analysis of the 1H NMR signals of methylene groups by Ala-Korperela et al. (14) identified two transitions at 292 and 301 K. The mean of these two values (297 K) is identical to the value observed by us assuming for the description of the temperature dependence a two-state model. However, the NMR visible transitions for HDL2 and HDL3 were not observable by DSC, supposedly because the HDL core was too small to form a larger smectic phase. For VLDL, again DSC did not find any transition for the lipids in the temperature range from 283 to 318 K (9). The data were interpreted by a disordered TG-rich homogeneous core. In agreement with these observations, we found a large portion of 62% of lipids that showed no temperature dependence. However, a smaller portion of the lipids showed a clear transition with a transition temperature of 319 ± 3 K (Table 4) as detected by their FA methyl group signal. This transition was accompanied by characteristic line-shape changes of the methylene signal (Fig. 6). The phase transition may not have been observed by DSC because it comprised a relatively small part of the lipids and was on the border of the reported temperature range. It may represent the lipid shell that is close to the membrane and their embedded proteins. The IDL particles behave more than the LDL particles concerning their melting temperatures, but still show a rather large compartment A of always visible signals (Table 4).
The data from high-pressure NMR spectroscopy complement nicely the temperature dependence because changing the pressure leads also to strong effects on the visibility of the lipid signals and thus confirms the phase transitions in the lipids of the lipoproteins (Figs. 9 and 10). As to be expected, at higher pressures, the size of the smectic phase is increasing and the signal intensity decreases. In contrast, the intensity of the protein signals (well visible in the downfield part of the spectra) do only show small pressure effects in comparison to the lipid signals. This is also true for the marker signal for proinflammatory acute-phase proteins at 2.05 ppm. Interestingly, the pressure-induced phase transition is observed earlier for the upfield shifted lipid signals characteristic for smaller particles.
Thermodynamics of lipoprotein particles
The enthalpy changes determined by using cholesterol in the core of LDL as reporter group are −123 kJ/mol (Table 5), very different to the value of 2.11 MJ/mol obtained by DSC (61). This indicates that the two methods characterize different physical processes. From a cryo-EM reconstruction of LDL at 279 K, it is known that the lipid core transition to higher disorder is accompanied by a change of the LDL structural appearance from a slight discoid structure to a more spheroidal structure (cryo-EM reconstruction of LDL at 310 K) (62, 63). The temperature-dependent lipid core phase transition is complete within less than 10 ms (64). The activation energies for slow kinetically controlled transitions measured by CD spectroscopy using an Arrhenius plot of the data are in the range of 250 kJ/mol (65, 66), but for a transition that occurred at much higher temperature of 355 K and led to strong partly irreversible structural changes and fusions of the particles. The observed enthalpy difference of −123 kJ/mol at the melting temperature is in the range of kinetically controlled transitions and might reflect the sensitivity of NMR relaxation properties (i.e., the visibility) to the orientational structure factor of the lipoproteins and the effective correlation times τc of the observed groups. This could include conformational changes and lipid-binding properties of ApoB-100, as well as increasing fluidity of the membrane with increasing temperature when the choline group is concerned with an even smaller ΔH value of −104 kJ/mol.
Smaller enthalpy changes were obtained for cholesterol C-18H as reporter group, with −30 and −74 kJ/mol for HDL2 and HDL3, respectively (Table 4). Both values are again much too small for classical lipid phase transitions, which were never observed for HDL in calorimetric studies. Instead, the stability of human plasma HDL seems to be stabilized by kinetic factors. Mehta et al. (67) observed in chemical denaturation experiments of intact HDL two kinetic phases and in their heating experiments a scan-rate dependence of the melting curves indicative of a high enthalpic barrier for particle rupture. Gursky and coworkers (68) discussed the importance of structural local disorder for the reverse cholesterol transport. Their model contains discoidal nascent HDL, small mature spherical HDL3, and large mature spherical HDL2. They suggest that at least two kinetic steps are required and that in spherical HDLs, the second step involves protein dissociation, particle rupture, and release of apolar core lipids. Generally, the energy barriers between the different steps are of the order of 80 kJ/mol for, e.g., particle rupture. The enthalpy differences observed in our temperature range from 283 to 323 K for different reporter groups describe the increase of local and structural disorder and dynamics, which is important for the physiological function of HDLs. The model of Guha et al. (68) suggests that in a first step, structural disorder facilitates efflux of lipids from the plasma membrane to apolipoproteins; in a second step, local disorder in nascent discoidal HDLs facilitates cholesterol insertion and esterification; and in a third step, the destabilization of spherical HDLs promotes their metabolic remodeling and fusion. In a study investigating the effects of salt on the thermal stability of human HDL, Jayaraman et al. (69) detected two kinetic phases in HDL protein unfolding in 150 mM NaCl: a faster phase with small inactivation energies below 60 kJ/mol and a slower phase with activation energies around 220 kJ/mol. Their gel electrophoresis and electron microscopic data suggested that the faster phase involves partial protein unfolding but no significant protein dissociation or changes in HDL size. The slower phase is characterized by complete protein unfolding, partial protein dissociation, and HDL fusion. The fast phase with energies of less than 60 kJ/mol agrees very well with our observed enthalpy changes for HDL2 of −30 kJ/mol and HDL3 of −74 kJ/mol. The first study on the thermostability of VLDL were reported by Guha et al. (70). Their data analysis revealed two kinetic phases with activation energies around −220 kJ/mol, which correspond to distinct morphological transitions observable by electron microscopy. Their conclusion was that, similar to HDL and LDL, also VLDL is stabilized by kinetic barriers. These barriers then prevent particle fusion and rupture and, in addition, might decelerate spontaneous interconversion among lipoprotein classes and subclasses. In our study, again, two states are observed, but with lower values of −80 kJ/mol (Table 5).
CONCLUSION
Thermodynamic analysis of a heterogeneous, multicomponent system that possibly also contains different microenvironments with temperature-dependent sizes is very difficult; the quantitative results may depend on the experimental methods and the models used for their description. It is not surprising that values reported in literature and their interpretation are sometimes contradictory (see above). Only rather detailed structural data may be help to solve some of the problems. The three-compartment model we used here is a simple descriptive model that mainly serves for the description of the temperature-dependent visibility of different lipids in the lipoproteins. Here, the atomic resolution of NMR providing different chemical reporter groups adds at least some additional information for modeling and interpretation.
The presented data have practical consequences when NMR is used to analyze lipoproteins. The signal intensities are strongly temperature dependent, and the temperature dependence is different for different lipoproteins and different lipids. For obtaining data that are not dependent on individual melting points, temperatures clearly above the melting points of all lipids would have to be used. Our data indicate that temperatures ≥ 323 K would be optimal in this respect. A problem is that at high temperatures, especially larger particles such as IDL and VLDL, appear to be less stable, and thus errors in quantification may occur. At 323 K, all lipids of the HDL particles can be quantified correctly. However, when using the methyl peak for quantification of the LDL particles, about 50% of cholesterol signals and 20% of the FA signals remain invisible. Therefore, for the lipoprotein quantification, the obtained LDL data have to be renormalized suitably.
More information can be obtained by also using other resonances of lipids. This would have the advantage that the protein background may be weaker, and additional information about the lipoprotein composition can be obtained. Because of the visibility differences between lipids in different lipoproteins, additional valuable information could be obtained by furthermore using the simple extraction procedure described here and quantifying the lipids by NMR or mass spectrometry.
The background signal of the serum proteins contributes strongly to the signal intensities observed for the signal around 0.8 ppm, usually used for the analysis of the lipoproteins. The actual influence of the serum protein signals could be approximated from the signal intensity in the downfield part of the spectrum. A correspondingly weighted protein spectrum could be subtracted from the obtained spectrum before evaluation of the lipoproteins.
The 1H NMR visibility of lipid signals is strongly influenced by the intrinsic transverse relaxation times that are strongly influenced by the external physical parameters pressure and temperature. NMR pressure data were presented here for the first time. In principle, the dependence of the transverse relaxation times of different groups on temperature and pressure can provide additional information about specific lipoproteins and may also be characteristic for different individuals and their specific (risk of) diseases. This additional information (also the relative visibility) is not used routinely and is neglected in biomedical studies. It may be worthwhile to include that in metabolomic studies in future.
Supplementary Material
Footnotes
Abbreviations:
- CE
- cholesteryl ester
- CPMG
- Carr-Purcell-Meiboom-Gill
- cryo-EM
- cryo-electron microscopy
- DSC
- differential scanning calorimetry
- DSS
- 4,4-dimethyl-4-silapentane-1-sulfonic acid
- LPDS
- lipoprotein-deficient serum
- PC
- phosphatidylcholine
- PG
- phosphoglyceride
- PL
- phospholipid
- SL
- sphingolipid
- TG
- triacylglyceride
- TOCSY
- total correlation spectroscopy
This work was supported by the Verband der Chemischen Industrie, the Deutsche Forschungsgemeinschaft, and the Bayerische Forschungsstiftung.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Eisenberg S. 1983. Lipoproteins and lipoprotein metabolism. Klin. Wochenschr. 61: 119–132. [DOI] [PubMed] [Google Scholar]
- 2.Dolphin P. J. 1985. Lipoprotein metabolism and the role of apolipoproteins as metabolic programmers. Can. J. Biochem. Cell Biol. 63: 850–869. [DOI] [PubMed] [Google Scholar]
- 3.Schroeder F., and Goh E. H.. 1979. Regulation of very low density lipoprotein interior core lipid physicochemical properties. J. Biol. Chem. 254: 2464–2470. [PubMed] [Google Scholar]
- 4.Aviram M., Lund-Katz S., Phillips M. C., and Chait A.. 1988. The influence of the triglyceride content of low density lipoprotein on the interaction of apolipoprotein B-100 with cells. J. Biol. Chem. 263: 16842–16848. [PubMed] [Google Scholar]
- 5.Ibdah J. A., Lund-Katz S., and Phillips M. C.. 1989. Molecular packing of high-density and low-density lipoprotein surface lipids and Apolipoprotein A-I binding. Biochemistry. 28: 1126–1133. [DOI] [PubMed] [Google Scholar]
- 6.Deckelbaum R.J., Shipley G.G., Small D.M., Lees R.S., and George P.K.. 1975. Thermal transitions in human plasma low density lipoproteins. Science. 190: 392–394. [DOI] [PubMed] [Google Scholar]
- 7.Sears B., Deckelbaum R. J., Janiak M. J., Shipley G. G., and Small D. M.. 1976. Temperature-dependent Carbon-13 nuclear magnetic resonance studies of human serum low density lipoproteins. Biochemistry. 15: 4151–4157. [DOI] [PubMed] [Google Scholar]
- 8.Atkinson D., Deckelbaum R. J., Small D. M., and Shipley G. G.. 1977. Structure of human plasma low-density lipoproteins: molecular organization of the central core. Proc. Natl. Acad. Sci. USA. 74: 1042–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deckelbaum R. J., Tall A. R., and Small D. M.. 1977. Interaction of cholesterol ester and triglyceride in human plasma very low density lipoprotein. J. Lipid Res. 18: 164–168. [PubMed] [Google Scholar]
- 10.Deckelbaum, R. J., G. G. Shipley, and D. M. Small. 1977. Structure and interactions of lipids in human plasma low density lipoproteins. J. Biol. Chem. 252: 744–754. [PubMed] [Google Scholar]
- 11.Pregetter M., Prassl R., Schuster B., Kriechbaum M., Nigon F., Chapman J., and Laggner P.. 1999. Microphase separation in low density lipoproteins evidence for a fluid triglyceride core below the lipid melting transition. J. Biol. Chem. 274: 1334–13411.. [DOI] [PubMed] [Google Scholar]
- 12.Morrisett J. D., Gaubatz J. W., Tarver A. P., Allen J. K., Pownall H. J., Laggner P., and Hamilton J. A.. 1984. Thermotropic properties and molecular dynamics of cholesteryl ester rich very low density lipoproteins: effect of hydrophobic core on polar surface. Biochemistry. 23: 5343–5352. [DOI] [PubMed] [Google Scholar]
- 13.Tall A. R., Deckelbaum R. J., Small D. M., and Shipley G. G.. 1977. Thermal behavior of human plasma high density lipoprotein. Biochim. Biophys. Acta 487: 145–153. [DOI] [PubMed] [Google Scholar]
- 14.Ala-Korpela M., Oja J., Lounila J., Jokisaari J., Savolainen M. J., and Kesäniemi Y. A.. 1995. Structural changes of lipoprotein lipids by 1H NMR. Chem. Phys. Lett. 242: 95–100. [Google Scholar]
- 15.Tall A. R. 1980. Structure of plasma lipoproteins: view from calorimetric studies. Ann. N. Y. Acad. Sci. 348: 335–351. [DOI] [PubMed] [Google Scholar]
- 16.Kroon P. A., and Krieger M.. 1981. The mobility of cholesteryl esters in native and reconstituted low density lipoprotein as monitored by nuclear magnetic resonance spectroscopy. J. Biol. Chem. 256: 5340–5344. [PubMed] [Google Scholar]
- 17.Kroon P. A. 1981. The order-disorder transition of the core cholesteryl esters of human plasma low density lipoprotein. a proton nuclear magnetic resonance study. J. Biol. Chem. 256: 5332–5339. [PubMed] [Google Scholar]
- 18.Ginsburg G. S., Small D. M., and Hamilton J. A.. 1982. Temperature-dependent molecular motions of cholesterol esters: a carbon-13 nuclear magnetic resonance study. Biochemistry. 21: 6857–6867. [DOI] [PubMed] [Google Scholar]
- 19.Hamilton J. A., Small D. M., and Parks J. S.. 1983. 1H NMR studies of lymph chylomicra and very low density lipoproteins from nonhuman primates. J. Biol. Chem. 258: 1172–1179. [PubMed] [Google Scholar]
- 20.Parks J. S., and Hauser H.. 1996. Low density lipoprotein particle size and core cholesteryl ester physical state affect the proton NMR magnetic environment of fatty acid methylene and methyl nuclei. J. Lipid Res. 37: 1289–1297. [PubMed] [Google Scholar]
- 21.Casu M., Anderson G. J., Choi G., and Gibbons W. A.. 1991. NMR lipid profiles of cells, tissues and body fluids. 1D and 2D proton NMR of lipids from rat liver. Magn. Reson. Chem. 29: 594–602. [Google Scholar]
- 22.Nicholson J. K., Foxall P. J. D., Spraul M., Farrant R. D., and Lindon J. C.. 1995. 750 MHz 1H and 1H–13C NMR spectroscopy of human blood plasma. Anal. Chem. 67: 793–811. [DOI] [PubMed] [Google Scholar]
- 23.Heremans K., and Smeller L.. 1998. Protein structure and dynamics at high pressure. Biochim. Biophys. Acta. 1386: 353–370. [DOI] [PubMed] [Google Scholar]
- 24.Winter R. 2015. Pressure effects on artificial and cellular membranes. In High Pressure Bioscience. K. Akasaka and H. Matsuki, editors. Springer, Heidelberg. 345–370. [DOI] [PubMed] [Google Scholar]
- 25.Akasaka K. 2006. Probing conformational fluctuation of proteins by pressure perturbation. Chem. Rev. 106: 1814–1835. [DOI] [PubMed] [Google Scholar]
- 26.Kremer W. 2006. High-pressure NMR studies in proteins. Annu. Rep. NMR Spectrosc. 57: 177–203. [Google Scholar]
- 27.Kitahara R., Hata K., Li H., Williamson M. P., and Akasaka K.. 2013. Pressure induced chemical shifts as probes for conformational fluctuations in proteins. Prog. Nucl. Magn. Reson. Spectrosc. 71: 35–58. [DOI] [PubMed] [Google Scholar]
- 28.Kalbitzer H. R., Rosnizeck I. C., Munte C. E., Puthenpurackal Narayanan S., Kropf V., and Spoerner M.. 2013. Intrinsic allosteric inhibition of signaling proteins by targeting rare interaction states detected by high-pressure NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 52: 14242–14246. [DOI] [PubMed] [Google Scholar]
- 29.Hiltunen Y., Ala-Korpela M., Jokisaari J., Eskelinen S., Kiviniitty K., Savolainen M., and Kesäniemi Y. A.. 1991. A lineshape fitting model for 1H NMR spectra of human blood plasma. Magn. Reson. Med. 21: 222–232. [DOI] [PubMed] [Google Scholar]
- 30.Otvos J. D., Jeyarajah E. J., and Bennett D. W.. 1991. Quantification of plasma lipoproteins by proton nuclear magnetic resonance spectroscopy. Clin. Chem. 37: 377–386. [PubMed] [Google Scholar]
- 31.Otvos J. D., Jeyarajah E. J., Bennett D. W., and Krauss R. M.. 1992. Development of a proton nuclear magnetic resonance spectroscopic method for determining plasma lipoprotein concentrations and subspecies distributions from a single, rapid measurement. Clin. Chem. 38: 1632–1638. [PubMed] [Google Scholar]
- 32.Ala-Korpela M., Korhonen A., Keisala J., Hörkkö S., Korpi P., Ingman L. P., Jokisaari J., Savolainen M. J., and Kesäniemi Y. A.. 1994. 1H NMR-based absolute quantitation of human lipoproteins and their lipid contents directly from plasma. J. Lipid Res. 35: 2292–2304. [PubMed] [Google Scholar]
- 33.Hiltunen Y., Heiniemi E., and Alakorpela M.. 1995. Lipoprotein-lipid quantification by neural-network analysis of 1H-NMR data from human blood plasma. J. Magn. Reson. B. 106: 191–194. [DOI] [PubMed] [Google Scholar]
- 34.Ala-Korpela M., Hiltunen Y., and Bell J. D.. 1995b. Quantification of biomedical NMR data using artificial neural network analysis: lipoprotein lipid profiles from 1H NMR data of human plasma. NMR Biomed. 8: 235–244. [DOI] [PubMed] [Google Scholar]
- 35.Bathen T. F., Krane J., Engan T., Bjerve K. S., and Axelson D.. 2000. Quantification of plasma lipids and apolipoproteins by use of proton NMR spectroscopy, multivariate and neural network analysis. NMR Biomed. 13: 271–288. [DOI] [PubMed] [Google Scholar]
- 36.Vehtari A., Mäkinen V-P., Soininen P., Ingman P., Mäkelä S., Savolainen M., Hannuksela M., Kaski K., and Ala-Korpela M.. 2007. A novel Bayesian approach to quantify clinical variables and to determine their spectroscopic counterparts in 1H NMR metabonomic data. BMC Informatics. 8: S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalbitzer H. R., Lang E., Huber F., Kremer W., and Theis F.. 2009. Method for determining lipoprotein components in a lipoprotein mixture to be analyzed and data processing system. Patent application WO2009152805A1. [Google Scholar]
- 38.Serrai H., Nadal L., Leray G., Leroy B., Delplanque B., and de Certaines J. D.. 1998. Quantification of plasma lipoprotein fractions by wavelet transform time-domain data processing of the proton nuclear magnetic resonance methylene spectral region. NMR Biomed. 11: 273–280. [DOI] [PubMed] [Google Scholar]
- 39.Kremer W., Kalbitzer H. R., and Huber F.. 2005, 2011. Process for the determination of Lipoproteins in body fluids. Patents DE 10 2004 026 903 B4, AU 2005250571 B2, CA 2568705 A1, and US 7927878. [Google Scholar]
- 40.Dyrby M., Petersen M., Whittaker A. K., Lambert L., Nørgaard L., Bro R., and Engelsen S. B.. 2005. Analysis of lipoproteins using 2D diffusion-edited NMR spectroscopy and multi-way chemometrics. Anal. Chim. Acta 531: 209–216. [Google Scholar]
- 41.Otvos J. D., Jeyarajah E. J., and Cromwell W. C.. 2002. Measurement issues related to lipoprotein heterogeneity. Am. J. Cardiol. 90(suppl): 22i–29i. [DOI] [PubMed] [Google Scholar]
- 42.Freedman D. S., Otvos J. D., Jeyarajah E. J., Shalaurova I., Cupples L. A., Parise H., D’Agostino R. B., Wilson P. W. F., and Schaefer E. J.. 2004. Sex and age differences in lipoprotein subclasses measured by nuclear magnetic resonance spectroscopy: The Framingham Study. Clin. Chem. 50: 1189–1200. [DOI] [PubMed] [Google Scholar]
- 43.Kontush A. 2015. HDL particle number and size as predictors of cardiovascular disease. Front. Pharmacol. 6: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Havel R. J., Eder H. A., and Bragdon J. H.. 1955. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J. Clin. Invest. 34: 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freund J., and Kalbitzer H. R.. 1995. Physiological buffers for NMR spectroscopy. J. Biomol. NMR. 5: 321–322. [DOI] [PubMed] [Google Scholar]
- 46.Carr H. Y., and Purcell E. M.. 1954. Effects of diffusion on free precession in nuclear magnetic resonance experiments. Phys. Rev. 54: 630–638. [Google Scholar]
- 47.Meiboom S., and Gill D.. 1958. Modified spin echo method for measuring nuclear relaxation times. Rev. Sci. Instrum. 29: 688–691. [Google Scholar]
- 48.Hoffman R. E. 2006. Standardization of chemical shifts of TMS and solvent signals in NMR solvents. Magn. Reson. Chem. 44: 606–616. [DOI] [PubMed] [Google Scholar]
- 49.Bax A., and Davis D. G.. 1985. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 65: 355–360. [Google Scholar]
- 50.Yamada H. 1974. Pressure-resisting glass cell for high pressure, high resolution NMR measurement. Rev. Sci. Instrum. 45: 5. [Google Scholar]
- 51.Kremer W., and Kalbitzer H. R.. 2001. Physiological conditions and practicality for protein nuclear magnetic resonance spectroscopy: experimental methodologies and theoretical background. Methods. Enzymol. 339: 3–19. [DOI] [PubMed] [Google Scholar]
- 52.Otvos J. D., Shalaurova I., Wolak-Dinsmore J., Connelly M. A., Mackey R. H., Stein J. H., and Tracy R. P.. 2015. GlycA: a composite nuclear magnetic resonance biomarker of systemic inflammation. Clin. Chem. 61: 714–723. [DOI] [PubMed] [Google Scholar]
- 53.Duprez D. A., Otvos J., Sanchez O. A., Mackey R. A., Tracy R., and Jacobs D. R.. 2016. Comparison of the predictive value of GlycA and other biomarkers of inflammation for total death, incident cardiovascular events, noncardiovascular and noncancer inflammatory-related events, and total cancer events. Clin. Chem. 62: 1020–1031. [DOI] [PubMed] [Google Scholar]
- 54.Groß K-H., and Kalbitzer H. R.. 1988. Distribution of chemical shifts in 1H nuclear magnetic resonance spectra of proteins. J. Magn. Reson. 76: 87–99. [Google Scholar]
- 55.Srivastava N. K., Pradhan S., Mittal B., Kumar R., and Nagana Gowda G. A.. 2006. An improved, single step standardized method of lipid extraction from human skeletal muscle tissue. Anal. Lett. 39: 297–315. [Google Scholar]
- 56.Kronenberg F., Lobentam E-V., Konig P., Utermann G., and Dieplinger H.. 1994. Effect of sample storage on the measurement of lipoprotein[a], apolipoproteins B and A-IV, total and high density lipoprotein cholesterol and triglycerides. J. Lipid Res. 35: 1318–1328. [PubMed] [Google Scholar]
- 57.Zivkovic A. M., Wiest M. M., Nguyen U. T., Davis R., Watkins S. M., and German J. B.. 2009. Effects of sample handling and storage on quantitative lipid analysis in human serum. Metabolomics. 5: 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cuhadar S., Koseoglu M., Atay A., and Dirican A.. 2013. The effect of storage time and freeze-thaw cycles on the stability of serum samples. Biochem. Med. (Zagreb). 23: 70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jobard E., Trédan O., Postoly D., André F., Martin A., Elena-Herrmann B., and Boyault S.. 2016. A systematic evaluation of blood serum and plasma pre-analytics for metabolomics cohort studies. Int. J. Mol. Sci. 17: 2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hevonoja T., Pentikainen M. O., Hyvonen M. T., Kovanen P. T., and Ala-Korpela M.. 2000. Structure of low density lipoprotein (LDL) particles: basis for understanding molecular changes in modified LDL. Biochim. Biophys. Acta. 1488: 189–210. [DOI] [PubMed] [Google Scholar]
- 61.Kumar V., Butcher S. J., Öörrni K., Engelhardt P., Heikkonen J., Kaski K., Ala-Korpela M., and Kovane P. T.. 2011. Three-dimensional cryoEM reconstruction of native LDL particles to 16Å resolution at physiological body temperature. PLoS One. 6: e18841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jayaraman S., Jasuja R., Zakharov M. N., and Gursky O.. 2011. Pressure perturbation calorimetry of lipoproteins reveals an endothermic transition without detectable volume changes. Implications for adsorption of apolipoprotein to a phospholipid surface. Biochemistry. 50: 3919–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y., Luo D., and Atkinson D.. 2011. Human LDL core cholesterol ester packing: three-dimensional image reconstruction and SAXS simulation studies. J. Lipid Res. 52: 256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prassl R., Pregetter M., Amenitsch H., Kriechbaum M., Schwarzenbacher R., Chapman J. M., and Laggneret P.. 2008. Low density lipoproteins as circulating fast temperature sensors. PLoS One. 3: e4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jayaraman S., Gantz D. L., and Gursky O.. 2005. Structural basis for thermal stability of human low-density lipoprotein. Biochemistry. 44: 3965–3971. [DOI] [PubMed] [Google Scholar]
- 66.Gursky O., and Gantz D. L.. 2002. Complex of human Apolipoprotein C-1 with phospholipid: thermodynamic or kinetic stability? Biochemistry. 41: 7373–7384. [DOI] [PubMed] [Google Scholar]
- 67.Mehta R., Gantz D. L., and Gursky O.. 2003. Human plasma high-density lipoproteins are stabilized by kinetic factors. J. Mol. Biol. 328: 183–192. [DOI] [PubMed] [Google Scholar]
- 68.Guha M., Gao X., Jayaraman S., and Gursky O.. 2008. Correlation of structural stability with functional remodeling of high-density lipoproteins: the importance of being disordered. Biochemistry. 47: 11393–11397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jayaraman S., Gantz D. L., and Gursky O.. 2006. Effects of salt on the thermal stability of human plasma high-density lipoprotein. Biochemistry. 45: 4620–4628. [DOI] [PubMed] [Google Scholar]
- 70.Guha M., England C., Herscovitz H., and Gursky O.. 2007. Thermal transitions in human very-low-density lipoprotein: fusion, rupture, and dissociation of HDL-like particles. Biochemistry. 46: 6043–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.