Abstract
Background & Aims
Crohn’s disease (CD) is a debilitating inflammatory disorder that affects more than 1.6 million people in North America alone. Members of the tumor necrosis factor superfamily are key regulators of intestinal inflammation; specifically, tumor necrosis factor–like weak inducer of apoptosis (TWEAK) and its receptor, fibroblast growth factor–inducible 14 (Fn14), are involved in normal and pathologic tissue remodeling. Our aim was to determine the role of TWEAK/Fn14 in CD and a murine model of CD-like ileitis (ie, SAMP1/YitFc [SAMP] strain).
Methods
SAMP mice deficient in Fn14 (SAMP × Fn14-/-) were developed and a detailed time-course study was performed evaluating ileal tissues by histology and stereomicroscopy, as well as quantitative polymerase chain reaction and NanoString technology (Seattle, WA). Reciprocal bone marrow chimeras were generated to assess the relevance of Fn14 in hematopoietic vs nonhematopoietic compartments. Surgically resected intestinal tissues and mucosal biopsy specimens from patients with CD, ulcerative colitis, and healthy controls were analyzed for the expression of TWEAK/Fn14 by quantitative polymerase chain reaction, Western blot, immunohistochemistry, and immunofluorescence.
Results
SAMP × Fn14-/- showed a marked decrease in ileitis severity at 20 weeks of age compared with SAMP WT controls. Bone marrow chimeras showed that Fn14 was required in both hematopoietic and nonhematopoietic compartments for ileitis to develop. Transcriptome data showed multiple cellular pathways regulated by Fn14 signaling. Finally, increased expression of TWEAK and Fn14 was observed in tissue lesions from CD patients compared with ulcerative colitis and healthy controls.
Conclusions
TWEAK/Fn14 are up-regulated in CD, and also mediate experimental CD-like ileitis, by regulation of multiple innate and adaptive cellular pathways. Therefore, TWEAK/Fn14 may represent a novel therapeutic target for the treatment of small intestinal inflammation in CD.
Keywords: Gut Immunity, Intestinal Inflammation, Gene Expression, Proinflammatory Cytokines
Abbreviations used in this paper: BM, bone marrow; CD, Crohn’s disease; DSS, dextran sodium sulfate; Fn14, fibroblast growth factor–inducible 14; IBD, inflammatory bowel disease; IFNγ, interferon γ; IL, interleukin; KO, knockout; MAPK, mitogen-activated protein kinase; MLN, mesenteric lymph node; mRNA, messenger RNA; NF-κB, nuclear factor κ-light-chain-enhancer of activated B cells; qRT-PCR, quantitative real-time polymerase chain reaction; SAMP, SAMP1/YitFc; Th1, T helper cell type 1; TNF, tumor necrosis factor; TWEAK, tumor necrosis factor–like weak inducer of apoptosis; UC, ulcerative colitis; WT, wild type
Graphical abstract
Summary.
In this study, we show that tumor necrosis factor–like weak inducer of apoptosis/Fn14 is overexpressed in intestinal lesions from patients with Crohn's disease and that genetic deletion of Fn14 ameliorates the severity of ileitis in SAMP/YitFC mice by regulating critical innate and adaptive immune pathways. Therefore, tumor necrosis factor–like weak inducer of apoptosis/Fn14 may represent a novel target for the treatment of Crohn's disease.
Ulcerative colitis (UC) and Crohn’s disease (CD) are the 2 major forms of inflammatory bowel disease (IBD). CD can affect any part of the gastrointestinal tract, from the mouth to the anus. Even if the pathogenesis of CD remains incompletely understood, the disorder is thought to arise from environmental and microbial factors in genetically susceptible individuals.1 Patients affected by this condition can experience dramatic weight loss, multiple surgeries, and repeated hospital admissions. Currently, there is no cure for CD and research is focused on controlling symptoms and maintaining the disease in a phase of permanent remission. The most common treatments for CD are based on the use of steroids and monoclonal antibodies against tumor necrosis factor (TNF).2, 3, 4 Even if these immune-suppressants are effective in controlling the symptoms of CD, significant side effects have been associated with their use, including teratogenicity, hypertension, and opportunistic infections.5 Thus, there is a critical need to develop novel therapeutic strategies to treat this devastating disease.
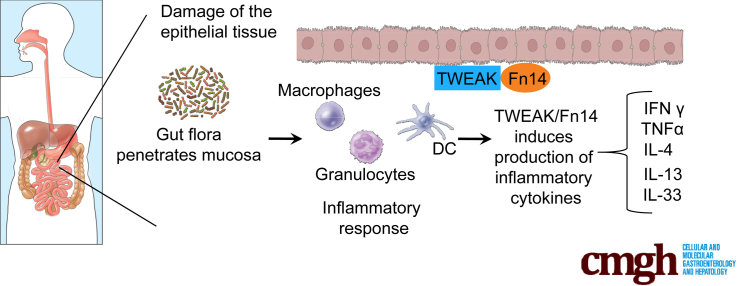
As a TNF superfamily cytokine member, tumor necrosis factor–like weak inducer of apoptosis (TWEAK) promotes its effects by binding to its specific receptor, fibroblast growth factor–inducible 14 (Fn14).6 TWEAK is widely expressed in a variety of cell types, including neurons, leukocytes, and astrocytes,7, 8 as well as in several tumor cell lines.9, 10 In healthy tissues, Fn14 is expressed at low levels. However, it can be highly induced by several growth factors in injured and diseased tissues11 on a variety of cell types, such as parenchymal, stromal, and tissue progenitor cells.12 The highly inducible pattern of Fn14 expression has been observed in mice as well as in human beings, reflecting the evolutionarily conserved nature of this pathway.13 The TWEAK/Fn14 axis has been shown to serve as a key modulator of tissue responses after acute and chronic injury, including the processes of fibrosis, malignancy, and autoimmunity.13, 14 In fact, up-regulation of Fn14 has been observed in a variety of human diseases, such as inflammatory liver diseases,15 lupus nephritis,16 and cancer.17 TWEAK/Fn14 can activate different intracellular signaling pathways, including nuclear factor-κB (NF-κB), mitogen-activated protein kinase (MAPK), and phosphatidylinositol-3-kinase and protein kinase B, as the result of an injury-induced tissue response, with different roles in acute vs chronic inflammation.18 However, the mechanism(s) by which TWEAK/Fn14 stimulate innate and adaptive immune systems has not been fully elucidated. Although several studies have confirmed that activation of TWEAK/Fn14 is beneficial during acute injury, its persistent activation during chronic inflammation can actually exacerbate damage, leading to angiogenesis, fibrosis, and pathologic hyperplasia. One of the tissue types most influenced by TWEAK/Fn14 during chronic inflammation is the intestines.19 Continued stimulation of Fn14 on intestinal epithelial cells after breakdown of the mucosal barrier may lead to excessive gut immune responses against the commensal flora. The consequences are delayed healing, dysregulated tissue repair, and fibrosis.20 Although these effects have been shown in models of colitis induced by repeated acute injury, the role of TWEAK/Fn14 in a more immunologically mediated experimental model of CD-like ileitis has yet to be investigated.
The SAMP1/YitFc (SAMP) mouse strain represents an established model of Th1/Th2-driven CD-like ileitis, with important similarities to the human condition.21 These mice develop chronic, small intestinal inflammation after 10 weeks of age, with features of cobblestone ileitis, characterized by discontinuous inflammation, fibrosis, and, in some cases, internal fistulae.22 Our previous studies have shown that several cytokines, such as TNF, interferon γ (IFNγ), and interleukin (IL)33, are important mediators of SAMP ileitis23; therefore, this model serves as an important tool for investigating the pathogenic role of novel cytokine-receptor pairs, such as TWEAK/Fn14, as possible preclinical targets for the development of novel therapies for IBD.
We report herein that genetic deletion of Fn14 in SAMP mice significantly ameliorates the severity of ileitis compared with SAMP wild-type (WT) control mice. In addition, we show that blocking TWEAK/Fn14 signaling affects a variety of intracellular and extracellular pathways, such as those involved in apoptosis, NF-κB and T-cell activation, cytokine production, and MAPK activity, which all have been shown to play a critical role in chronic intestinal inflammation. Finally, we report the novel observation that TWEAK and Fn14 are overexpressed in the ilea of CD patients with active inflammation compared with UC patients and healthy controls.
Results
Fn14 Deletion in SAMP Mice Decreases the Severity of Chronic Ileitis
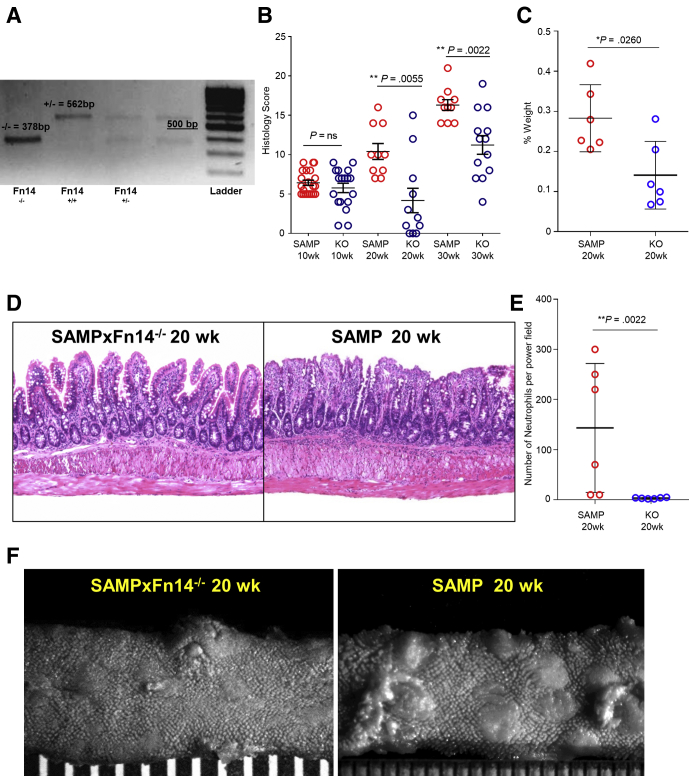
Little is known regarding the mechanisms of Fn14 signaling in modulating the chronic phase of spontaneous intestinal inflammation because most studies have been performed in murine models of chemically induced acute colitis.24 To test the role of Fn14 in chronic SAMP ileitis, we used a speed congenic approach to delete Fn14 from the SAMP genomic background (SAMP × Fn14-/-) (Figure 1A). We then performed time-course studies with histologic evaluation at early and late stages of disease (10, 20, and 30 weeks of age), and showed that SAMP × Fn14-/- have a similar severity of ileitis compared with WT littermates at 10 weeks of age. In contrast, SAMP × Fn14-/- mice showed a significant decrease in chronic ileitis compared with age-matched SAMP WT mice at both 20 and 30 weeks of age (late stage of disease) (Figure 1B and D). Mesenteric lymph nodes (MLNs) were collected on the day of death and weighed. SAMP × Fn14-/- mice presented with significantly smaller MLNs compared with WT littermates at 20 weeks of age (Figure 1C). In addition, neutrophil infiltration was decreased significantly in 20-week-old SAMP × Fn14-/- compared with age-matched SAMP littermates (Figure 1E).
Figure 1.
SAMP × Fn14-/-mice show decreased severity of ileitis compared with SAMP × Fn14+/+. (A) Genotyping of Filial 1 Number of backcross generations 10 (F1N10) congenic SAMP × Fn14-/- mice confirms complete genetic deletion of Fn14 (562 bp). (B) Evaluation of disease severity by histology shows decreased ileal inflammation in SAMP × Fn14-/- mice at 20 (unpaired t test, 4.18 ± 1.55 vs 10.40 ± 1.01; P < .02; n ≥ 10/group) and 30 weeks of age (unpaired t test, 11.23 ± 1.18 vs 16.30 ± 0.68; P < .02; n ≥ 10/group), but not at 10 weeks of age (unpaired t test, 5.78 ± 0.59 vs 6.43 ± 0.33; P = NS; n ≥ 10/group) compared with SAMP × Fn14+/+ littermates. (C) MLN weight expressed as a percentage of whole-body weight; SAMP × Fn14-/- mice at 20 weeks of age possess significantly smaller MLNs compared with age-matched SAMP × Fn14+/+ littermates (unpaired t test, 0.14 ± 0.03 vs 0.28 ± 0.03; P < .05; n = 6/group). (D) Representative photomicrographs of H&E-stained ilea from 20-week-old SAMP × Fn14-/- mice show decreased immune cell infiltrates into mucosal and submucosal layers, with improved crypt-to-villous architecture compared with age-matched SAMP × Fn14+/+ littermates (left), which conversely show active severe ulcerations and an increased presence of lamina propria inflammatory cells (right). (E) Number of neutrophils per power field (40×); SAMP × Fn14-/- mice at 20 weeks of age have significantly fewer polymorphonuclear leukocytes compared with age-matched SAMP × Fn14+/+ littermates (unpaired t test: 3.3 ± 0.5 vs 143.3 ± 52.5; P < .02; n = 6/group). (F) Representative stereomicroscopic images corroborate histologic findings. Data are represented as means ± SD, and are representative of 3 independent experiments. *P < .05, **P < .02.
The mucosal surface of the ileum was examined by stereomicroscopic analysis with the 3-dimensional stereomicroscopy assessment and pattern profiling protocol to map/quantify the intestinal surface architecture.25 We found potent attenuation of ileal disease severity in SAMP × Fn14-/- compared with WT mice at 20 weeks of age. Fn14 deletion led to the generation of a population of mice with healthier intestinal 3-dimensional structure and a wider normalized distribution for the percentage of abnormal mucosa (Figure 1F). Overall, these results suggest that Fn14 plays a proinflammatory role during chronic SAMP ileitis.
SAMP × Fn14-/- Show Decreased Lymphocytic Infiltration Compared With SAMP × Fn14+/+
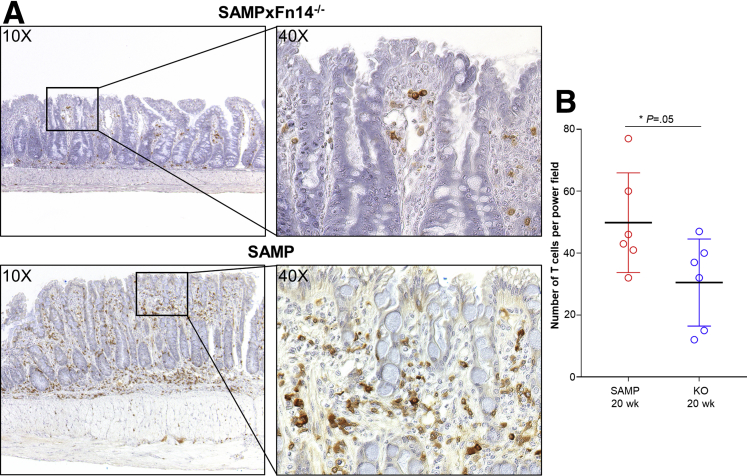
To test whether T cells play a role in mediating the significant decrease of chronic ileitis in SAMP × Fn14-/- mice, we performed immunostaining on ilea from experimental mice, targeting CD3+ cells. There was a significant decrease in the number of CD3+ cells in the subepithelial layers of ilea from 20-week-old SAMP × Fn14-/- mice compared with age-matched SAMP WT littermates (Figure 2A). Quantification of these results is shown in Figure 2B.
Figure 2.
SAMP × Fn14-/-show fewer T cells compared with SAMP × Fn14+/+. (A) Representative photomicrographs of CD3-stained ilea from 20-week-old SAMP × Fn14-/- mice highlight decreased presence of T cells in the mucosal layers (upper panel) compared with age-matched SAMP × Fn14+/+ littermates (lower panel). (B) Number of T cells per power field (40×); SAMP × Fn14-/- mice at 20 weeks of age have significantly fewer T cells compared with age-matched SAMP × Fn14+/+ littermates (unpaired t test: 30.5 ± 5.7 vs 49.8 ± 6.0; P ≤ .05; n = 6/group). Data are presented as means ± SD, and are representative of 3 independent experiments. *P ≤ .05.
Fn14 Deletion Reduces T-Helper Cell Type 1 and T-Helper Cell Type 2 Cytokine Expression During the Chronic Phase of CD-Like Ileitis
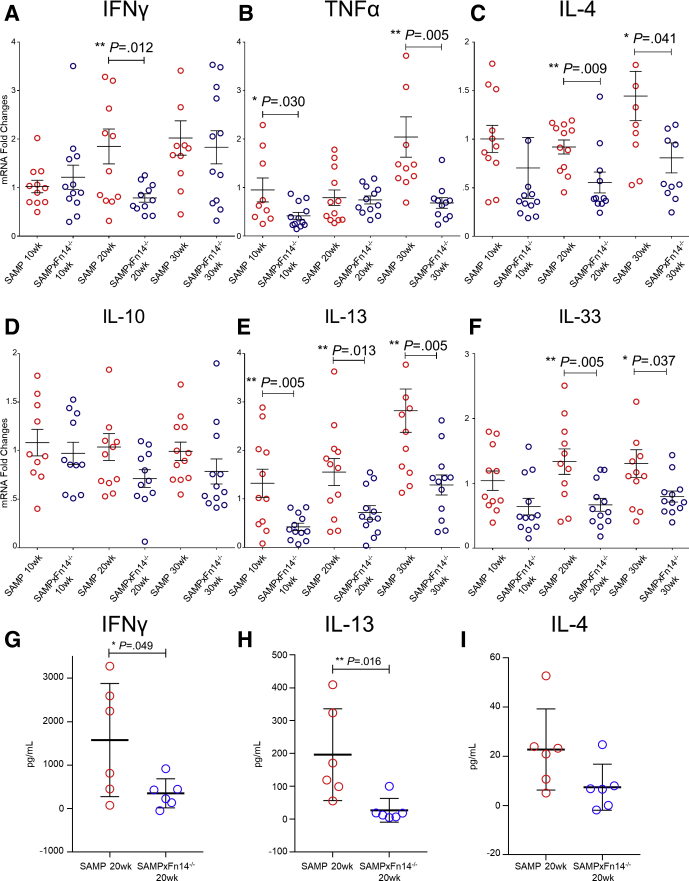
Our group previously showed that both T-helper cell type 1 (Th1) and Th2 immune responses play an important inflammatory role in the pathogenesis of SAMP ileitis.26 To explore the effects of Fn14 deletion on the types of immune responses during different stages of ileitis, we measured cytokine expression from ileal samples collected from SAMP × Fn14-/- and WT mice during the initial inflammatory phase at 10 weeks of age, and in the established chronic phase at 20 and 30 weeks of age. SAMP × Fn14-/- mice showed a significant decrease in IFNγ expression at 20 weeks of age (Figure 3A), and in TNFα (Figure 3B) and Il13 (Figure 3E) at 10 and 30 weeks of age compared with WT mice. SAMP × Fn14-/- mice also had decreased expression of Th2 cytokines at 10 and 30 weeks of age (ie, Il4, Il10, and Il33) (Figure 3C, D, and F, respectively). Altogether, these results strongly indicate that deletion of Fn14 decreases Th1- and Th2-mediated immunity during chronic SAMP ileitis and ameliorates disease severity. To confirm our quantitative reverse-transcription polymerase chain reaction (qRT-PCR) data in vitro, we isolated MLN lymphocytes from 20-week-old SAMP and SAMP × Fn14-/- mice and successively stimulated them with anti-CD3 and anti-CD28 for 72 hours. Levels of IFNγ (Figure 3G) and IL13 (Figure 3H) in the supernatants were decreased significantly in SAMP × Fn14-/- mice compared with SAMP WT littermates. Furthermore, we observed a trend in SAMP × Fn14-/- mice producing less IL4 compared with WT littermates (Figure 3I).
Figure 3.
SAMP × Fn14-/-mice show dampened Th1/Th2 immune responses compared with WT controls with established disease. Decreased expression of (A) Ifnγ in 20-week-old SAMP × Fn14-/- compared with 20-week-old SAMP (unpaired t test: 0.79- vs 1.85-fold; P < .02; N ≥ 11/group), (B) Tnf in 10-week-old (unpaired t test: 0.42- vs 0.95-fold; P < .05) and 30-week-old (unpaired t test: 0.68- vs 2.04-fold; P < .02) SAMP × Fn14-/- compared with age-matched SAMP, respectively, and (C) Il4 in 20-week-old (unpaired t test: 0.56- vs 0.92-fold; P < .02) and 30-week-old (unpaired t test: 0.81- vs 1.44-fold; P < .05) SAMP × Fn14-/- vs age-matched SAMP, respectively, with no differences observed in (D) Il10 from SAMP × Fn14-/- vs age-matched SAMP (P = NS). (E) In addition, Il13 was decreased in 10-week-old (unpaired t test: 0.42- vs 1.32-fold; P < .02), 20-week-old (unpaired t test: 0.72- vs 1.56-fold; P < .02), and 30-week-old (unpaired t test: 1.29- vs 2.82-fold; P < .02) SAMP × Fn14-/- compared with age-matched SAMP WT controls, respectively. (F) Finally, Il33 also was decreased in 20-week-old (unpaired t test: 0.67- vs 1.33-fold; P < .02) and 30-week-old (unpaired t test: 0.80- vs 1.30-fold; P < .05) SAMP × Fn14-/- compared with age-matched SAMP, respectively. (G) MLN lymphocytes isolated on day 8 of 3% DSS treatment and incubated for 72 hours with RPMI medium, anti-CD3, and anti-CD28 antibody (1 μg/mL). Cell-free supernatants were analyzed by enzyme-linked immunosorbent assay for production of IFNγ, IL13, and IL4. Twenty-week-old SAMP × Fn14-/-–derived cells produced decreased amounts of IFNγ compared with 20-week-old SAMP (unpaired t test: 351.7 ± 136.3 vs 1577.4 ± 531.7; P < .05; N = 6/group). (H) Twenty-week-old SAMP × Fn14-/-–derived cells produced decreased amounts of IL13 compared with 20-week-old SAMP (unpaired t test: 27 ± 14.7 vs 196.5 ± 6; P < .02). (I) Trend showing 20-week-old SAMP × Fn14-/-–derived cells producing decreased amounts of IL4 compared with 20-week-old SAMP (unpaired t test: 7.4 ± 3.8 vs 22.7 ± 6.7; P = NS). Data are presented as means ± SD, and are representative of 3 independent experiments. *P < .05, **P < .02.
Hematopoietic and Nonhematopoietic Cellular Components Participate in the TWEAK/Fn14-Mediated Proinflammatory Pathways During Chronic Ileitis
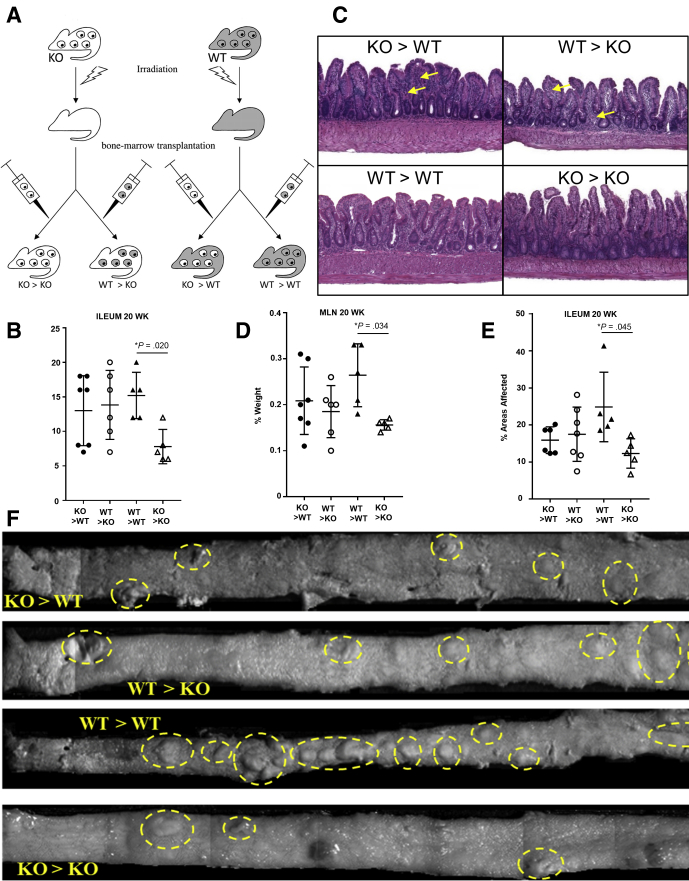
Because Fn14 is expressed in both nonhematopoietic (intestinal epithelial cells)27 and hematopoietic (immune cells)18 populations, we quantified separately the contribution of Fn14 that is derived from either cellular compartment in the development of chronic intestinal inflammation. In particular, we generated 4 groups of bone marrow (BM) chimeras that expressed either: (1) no components of the Fn14 receptor (knockout [KO] > KO mice); (2) only hematopoietic-derived Fn14 receptors (WT > KO mice); (3) only non–hematopoietic-derived Fn14 receptor (KO > WT mice); or (4) both hematopoietic and non–hematopoietic-derived Fn14 receptors (WT > WT mice) (Figure 4A). After the subsequent 8-week recovery phase, the 4 groups of mice were killed. As shown by histologic assessment of ileitis, KO > KO recipients had significantly lower inflammation compared with WT > WT mice, indicating that the anti-inflammatory role of Fn14 deletion could be owing to TWEAK/Fn14-dependent signaling mechanisms in both the hematopoietic and nonhematopoietic compartments (mean total inflammatory score: KO > KO, 8.33 ± 1.86; WT > WT, 14 ± 0.89; P < .05) (Figure 4B). No differences were observed in heterologous transplants (KO > WT and WT > KO mice), supporting the concept that Fn14 deletion in both compartments exerts the greatest anti-inflammatory effect (mean total inflammatory score: KO > WT, 12.4 ± 1.78; WT > KO, 12.71 ± 2.36; P = NS). Histology pictures show a decreased number of inflammatory cell infiltrations (yellow arrows) in the KO BM→KO recipients compared with the other 3 chimeric groups (Figure 4C). Our data are in agreement with the previous study conducted by Dohi et al19 showing that TWEAK/Fn14 pathway deficiency is protective in a γ-irradiation–induced model of intestinal epithelial damage.
Figure 4.
Both hematopoietic-derived and non–hematopoietic-derived Fn14 are essential for the development of chronic intestinal inflammation in SAMP mice. (A) SAMP × Fn14-/- and SAMP WT mice (n ≥ 7/group) were transplanted with reciprocal BM (n = 4 per group). (B) Disease severity, evaluated by histology, shows that SAMP × Fn14-/- mice transplanted with Fn14-/- BM develop less severe ileitis compared with SAMP WT transplanted with Fn14+/+ BM (unpaired t test, 8.33 ± 1.86 vs 14 ± 0.89; P < .05), but no significant difference was observed between SAMP × Fn14-/- mice transplanted with Fn14+/+ BM and SAMP WT mice transplanted with Fn14-/- BM (unpaired t test, 12.4 ± 1.78 vs 12.71 ± 2.36; P = NS). (C) Representative photomicrograph of H&E-stained ilea from SAMP × Fn14-/- mice transplanted with Fn14-/- BM show equal severity of ileitis in mucosal and submucosal layers (yellow arrows), with similar epithelial architecture when comparing SAMP WT mice transplanted with Fn14-/- BM. (D) MLN weight expressed as a percentage of whole-body weight showed no significant differences between SAMP × Fn14-/- and SAMP WT recipients receiving either Fn14+/+ or Fn14-/- BM (unpaired t test, 0.18 ± 0.03 vs 0.23 ± 0.03; P = NS), respectively, but SAMP × Fn14-/- recipients transplanted with Fn14-/- BM had significantly smaller MLNs compared with SAMP × Fn14+/+ recipients receiving Fn14+/+ BM (unpaired t test, 0.16 ± 0.01 vs 0.25 ± 0.03; P < .05). (E) Percentage of 3-dimensional abnormal ileal mucosa in SAMP × Fn14-/- recipients transplanted with Fn14+/+ BM was similar to that in SAMP WT transplanted with Fn14-/- BM (unpaired t test, 15.88 ± 1.47 vs 17.49 ± 2.77; P = NS). However, SAMP × Fn14-/- mice transplanted with Fn14-/- BM present with significantly less abnormal mucosa compared with SAMP × Fn14+/+ littermates transplanted with Fn14+/+ BM (unpaired t test, 12.25 ± 2.29 vs 24.83 ± 4.20; P < .05). (F) Representative 3-dimensional stereomicroscopic images of fixed specimens allow visualization of ileal architecture in BM transplanted mice, with yellow circles highlighting abnormal mucosa. Data are represented as means ± SD and are representative of 2 independent experiments. *P < .05.
MLNs were collected on the day of death and weighed. KO > KO recipient MLNs had significantly smaller MLNs compared with the other 3 groups (MLN weight expressed as a percentage of whole-body weight: KO > KO, 0.16 ± 0.01; WT > WT, 0.25 ± 0.03; WT > KO, 0.18 ± 0.03; KO > WT, 0.23 ± 0.03; P < .05) (Figure 4D).
Stereomicroscopic score and images (Figure 4E and F) corroborate the data obtained with histology, showing that KO > KO recipients have less percentage of abnormal mucosa compared with the other 3 chimeric groups (abnormal mucosa expressed as the percentage of the total surface of the ileum: KO > KO, 12.25 ± 2.29; WT > WT, 24.83 ± 4.20; WT > KO, 15.88 ± 1.47; KO > WT, 17.49 ± 2.77; P < .05).
Taken together, these results clearly show that the anti-inflammatory effect of Fn14 deletion in SAMP mice is the net result of a combination of effects in both hematopoietic and nonhematopoietic cells.
NanoString Data Show Mixed Temporal Expression of Proinflammatory and Anti-Inflammatory Genes
To further clarify the mechanisms that determine protection in SAMP × Fn14-/- at 20 weeks of age from ileitis, we used a Mouse Immunology and Inflammation NanoString panel (NanoString Technologies, Seattle, WA). NanoString technology is characterized by high reproducibility and sensitivity.28 As criteria of differential expression, we used the following: >2-fold change and P < .05.
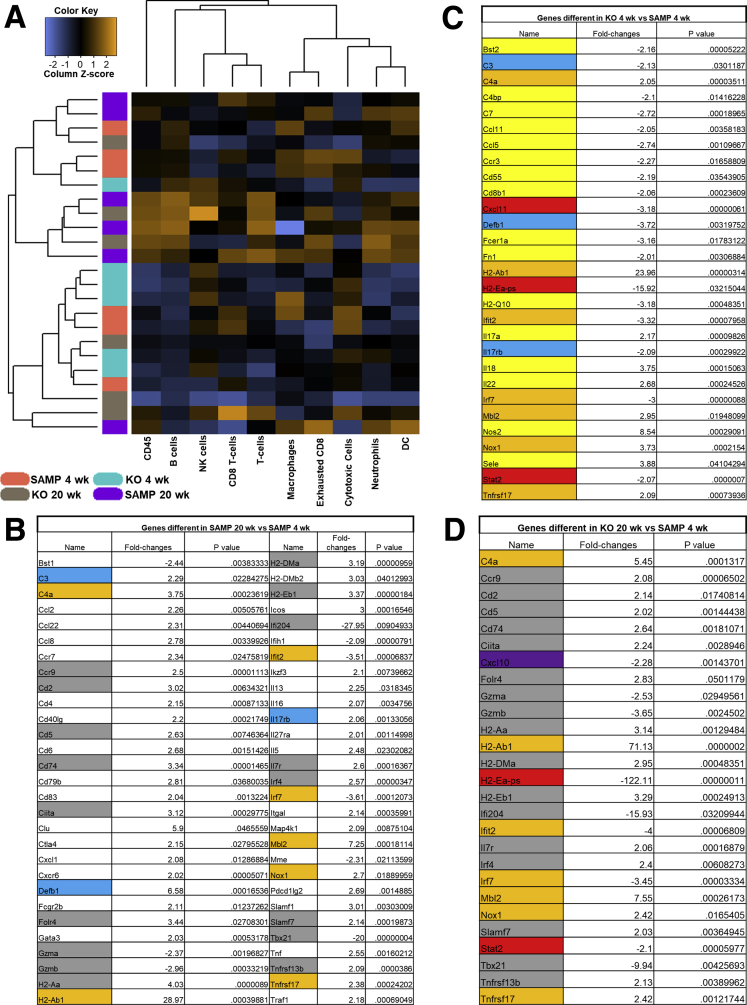
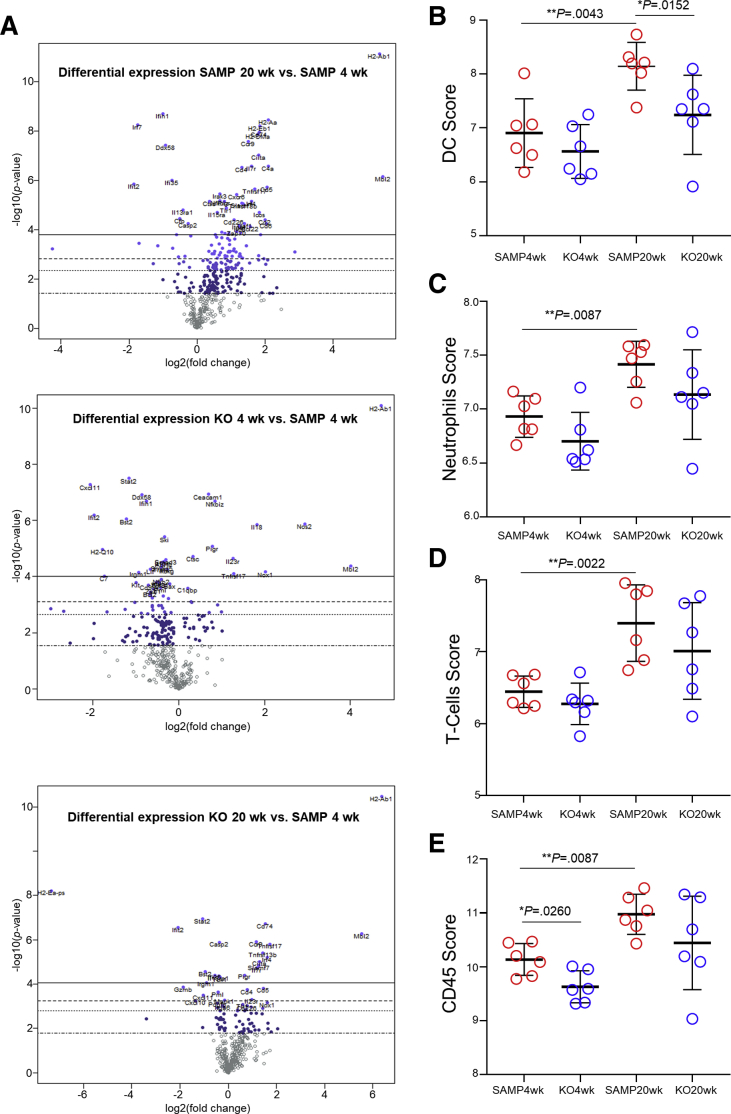
Because SAMP mice have no sign of ileitis at 4 weeks of age, and histology data showed that SAMP × Fn14-/- mice have significantly less ileitis than SAMP WT littermates at 20 weeks of age, we chose 4 weeks and 20 weeks as the ideal time points to compare SAMP × Fn14-/- and SAMP WT mice. The data obtained can be visualized in the heatmap showing the abundance of different cell types (Figure 5A). We used SAMP WT at 4 weeks of age to define baseline homeostatic expression and observed differences with SAMP at 20 weeks of age for 10.3% of the genes analyzed. Forty-nine genes were up-regulated significantly in 20-week-old SAMP WT mice and 9 were down-regulated (Figure 6A) The highest up-regulation was seen for H2-Ab1 (>20-fold compared with baseline), whereas Ifi204 and Tbx21 showed a >20-fold decrease relative to baseline 4-week-old SAMP WT expression. Among other genes significantly overexpressed in 20-week-old SAMP mice but not in the 20-week-old SAMP × Fn14-/- littermates, there are IL13 (>2-fold compared with baseline), Tnf (>2.5-fold compared with baseline), the inducible T-cell costimulatory Icos gene (>3-fold compared with baseline), and the TRAF1 gene (>2-fold compared with baseline) (Figure 5B). SAMP × Fn14-/- at 4 weeks of age showed a difference in 5.1% of the genes analyzed compared with SAMP at 4 weeks of age (Figure 5C). Interestingly, the Fn14 deletion negatively affected the expression of most of the genes altered in the 20-week-old SAMP WT and shifted their expression toward baseline of 4-week-old SAMP WT mice. In fact, SAMP × Fn14-/- at 20 weeks of age showed a very similar pattern to SAMP × Fn14 KO-/- at 4 weeks of age, with only 27 genes (Figure 5D) significantly changed in comparison with the baseline expression. Finally, we applied the same criteria of differential expression (>2-fold change and P < .05) to identify genes that were altered significantly between 20-week-old SAMP × Fn14-/- and 20-week-old SAMP WT. Conversely, only 3 genes (0.6%) were different (all of them decreased) in 20-week-old SAMP × Fn14-/- compared with 20-week-old SAMP WT. These genes were Cxcl1 (neutrophil chemotactic factor), H2-Ea-ps (antigen processing and activation of immune response), and Il22 (protein coding gene). Functional classification of the 58 genes that were different between 20-week-old SAMP WT and 4-week-old SAMP WT showed that the majority belonged to families of immunologic factors involved in adaptive and innate immune pathways. In particular, the up-regulated genes were related to the activation and activity of dendritic cells (Figure 6B), neutrophils (Figure 6C), T cells (Figure 6D), and cluster of differentiation 45(+) cells (Figure 6E).
Figure 5.
Difference in gene expression between 4-week-old SAMP mice and 20-week-old SAMP, and 4-week-old SAMP × Fn14-/-and 20-week-old SAMP × Fn14-/-. (A) Heatmap of normalized data, highlighting associations between different immune cell populations in 4- and 20-week-old SAMP WT and age-matched SAMP × Fn14-/- mice. Data are plotted by z-score and provide potential associations to covariates of interest (blue, low expression; black, average expression; orange, high expression). Each row represents a single probe, and each column represents a single sample, and plot is scaled with relation to average probe performance across samples to give all genes equal mean and variance. Hierarchical clustering is used to generate dendrograms. (B) List of genes with significantly different expression between SAMP WT at 20 weeks of age and SAMP WT at 4 weeks of age. A total of 10.3% of the genes analyzed were different. Forty-nine genes were up-regulated significantly in 20-week-old SAMP WT mice and 9 were down-regulated. (C) List of genes with significantly different expression between SAMP × Fn14-/- at 4 weeks of age and SAMP WT at 4 weeks of age. SAMP × Fn14-/- at 4 weeks of age showed a difference in 29 genes (5.1%) analyzed compared with SAMP at 4 weeks of age. (D) List of genes with significantly different expression between SAMP × Fn14-/- at 20 weeks of age and SAMP WT at 4 weeks of age. SAMP × Fn14-/- at 20 weeks of age have only 27 genes (4.8%) that were significantly changed in comparison with SAMP WT at 4 weeks of age (N = 6/group). *P < .05 and fold-changes > 2. Yellow, difference between 4-week-old SAMP × Fn14-/- and baseline; purple, difference between 20-week-old SAMP × Fn14-/- and baseline; white, difference between 20-week-old SAMP WT and baseline; blue, difference in 4-week-old SAMP × Fn14-/- and 20-week-old SAMP WT compared with baseline; orange, difference in 4-week-old SAMP × Fn14-/-, 20-week-old SAMP × Fn14-/-, and 20-week-old SAMP WT compared with baseline; red, difference in 4-week-old and 20-week-old SAMP × Fn14-/- compared with baseline; and grey, difference in 20-week-old SAMP × Fn14-/- and 20-week-old SAMP WT compared with baseline. DC, dendritic cell; NK, natural killer.
Figure 6.
Differential expression profiles of immune cell populations in young vs old SAMP, and in SAMP WT vs age-matched SAMP × Fn14-/-mice. (A) Volcano plots expressing NanoString data for 561 genes were obtained from nSolver software and showed that SAMP WT at 20 weeks of age have a higher number of genes differentially expressed compared with either 4-week-old SAMP WT, and 4- and 20-week-old SAMP × Fn14-/- mice. When comparing 20- vs 4-week-old SAMP WT mice, genes related to the development of (B) dendritic cells (DCs) (1-way analysis of variance; DC score: 8.144 ± 0.1814 vs 6.908 ± 0.26; P < .02), (C) neutrophils (1-way analysis of variance; neutrophil score: 7.415 ± 0.08741 vs 6.93 ± .0788; P < .02), (D) T cells (1-way analysis of variance; T-cell score: 7.398 ± 0.218 vs 6.445 ± 0.090; P < .02), and (E) CD45+ immune cells (1-way analysis of variance; CD45 score: 10.98 ± 0.15 vs 10.14 ± 0.12; P < .02) all are increased significantly, although no significant differences in the aforementioned groups were detected when comparing 20-week-old SAMP × Fn14-/- with 4-week-old SAMP WT mice (P = NS). Data are represented as means ± SD (N = 6/group). **P < .02.
Taken together, these data show that Fn14 in SAMP mice decreases the expression of several proinflammatory genes and shifts the phenotype to the homeostasis position, reducing the severity of chronic intestinal inflammation.
Multiple Cellular Pathways Are Suppressed With Fn14 Deletion
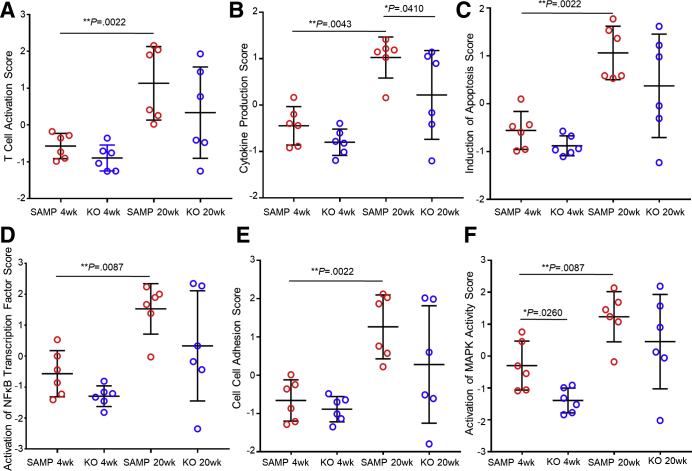
We used the first principal component of the expression of groups of genes involved in specific pathways to extract pathway-level information from our 4 experimental groups. This particular approach was established by Tomfohr et al.29 Each experimental group was composed of 6 mice. The results obtained show that several pathways were up-regulated in SAMP WT mice at 20 weeks of age compared with SAMP WT mice at 4 weeks of age: T-cell activation (1.132 ± 0.4078 vs -0.5716 ± 0.1403; P < .02) (Figure 7A); cytokine production (1.026 ± 0.1806 vs -0.4455 ± 0.169; P < .02) (Figure 7B); induction of apoptosis (1.063 ± 0.2277 vs -0.5579 ± 0.1624; P < .02) (Figure 7C); activation of NF-κB transcription factor (1.529 ± 0.3332 vs -0.5671 ± 0.304; P < .02) (Figure 7D); cell–cell adhesion (1.263 ± 0.3413 vs -0.6576 ± 0.22; P < .02) (Figure 7E); and activation of MAPK activity (1.235 ± 0.3205 vs -0.2972 ± 0.313; P < .02) (Figure 7F). The same pathways, on the contrary, were not significantly different between SAMP × Fn14-/- at 4 weeks of age and SAMP WT at 4 weeks of age and between SAMP × Fn14-/- at 20 weeks of age and SAMP WT at 4 weeks of ages (P = NS). One-way analysis of variance was used to determine if there were any statistically significant differences between the means of the 4 experimental groups.
Figure 7.
Suppression of multiple cellular pathways after Fn14 deletion. When comparing 20- vs 4-week-old SAMP WT mice, activation of (A) T cells (1-way analysis of variance; P < .02), (B) cytokines (1-way analysis of variance; P < .02), (C) apoptosis (1-way analysis of variance; P < .02), (D) NF-κB (1-way analysis of variance; P < .02), (E) cell–cell adhesion (1-way analysis of variance; P < .02), and (F) MAPK activity (1-way analysis of variance; P < .02) was observed, although no significant differences between 20-week-old SAMP × Fn14-/- and 4-week-old SAMP were measured for any of the aforementioned parameters (1-way analysis of variance; P = NS); N = 6/group. Data are represented as means ± SD (N = 6/group). *P< .05, **P < .02.
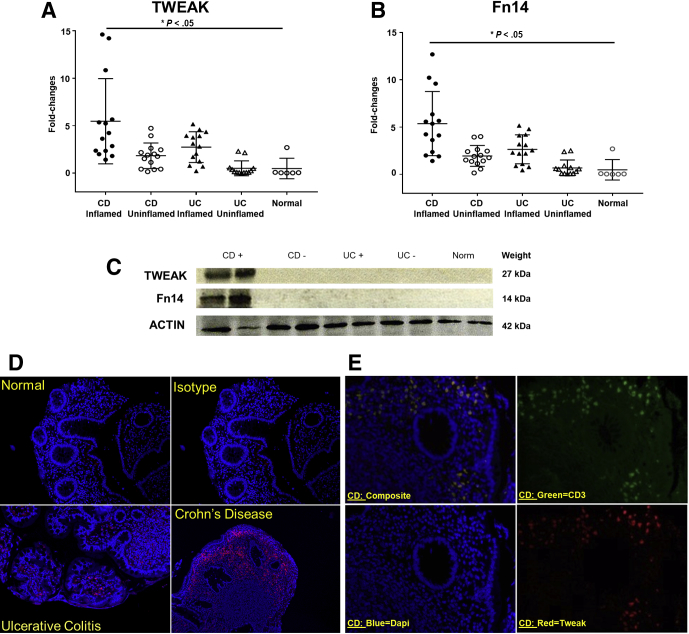
TWEAK and Fn14 Are Overexpressed in CD Patients During Active Disease
To confirm the translational significance of our findings in SAMP mice, we then measured the expression of TWEAK and Fn14 in ileal tissue samples from patients with CD and UC, with or without active inflammation, compared with healthy controls. Real-time RT-PCR analyses showed that TWEAK (Figure 8A) and Fn14 (Figure 8B) were up-regulated significantly in CD patients during active inflammation by 5.1-fold and 5.9-fold, respectively (N = 15; P < .05). As a control, we used normal ileal tissue samples from patients who underwent surgery for reasons not related to IBD (N = 6). By comparison, there was no significant difference in terms of TWEAK/Fn14 expression between CD patients without inflammation, UC patients, and normal tissue specimens. These results were confirmed by Western blot analyses showing increased protein expression of TWEAK/Fn14 in CD inflamed samples (Figure 8C).
Figure 8.
TWEAK and Fn14 are overexpressed in CD patients during active disease. (A) TWEAK and (B) Fn14 in ileal tissues from patients with CD and UC, with or without active inflammation, vs healthy controls (N ≥ 6/group) show up-regulation in CD patients during active inflammation compared with patients who underwent surgery for reasons not related to IBD (by 5.1- and 5.9-fold, respectively, both P < .05). (C) Representative Western blot of TWEAK and Fn14 in CD(+) inflamed, CD(-) uninflamed, UC(+) inflamed, UC(-) uninflamed, and healthy patients (N ≥ 6/group), with actin as an internal control, corroborates results obtained by qPCR. (D) Representative immunofluorescence shows that TWEAK is more expressed and distributed in CD patients compared with UC and normal patients (N ≥ 6/group). Isotype antibodies were used as a negative control. (E) Representative immunofluorescence double staining with anti-TWEAK and anti-CD3 antibodies shows that TWEAK is expressed by T lymphocytes in the subepithelial layer (N ≥ 6/group). Isotype antibodies were used as negative control. Data are represented as means ± SD, and results are representative of 2 independent experiments. *P < .05.
Colonic biopsy specimens also were collected from the same experimental groups for immunofluorescence to localize the distribution of TWEAK and CD3. Confocal images showed that TWEAK was highly expressed in the ileum of CD patients, compared with UC and healthy controls (Figure 8D). Furthermore, double staining of TWEAK and CD3 indicated that the TWEAK-positive cells most likely are intestinal lymphocytes (Figure 8E).
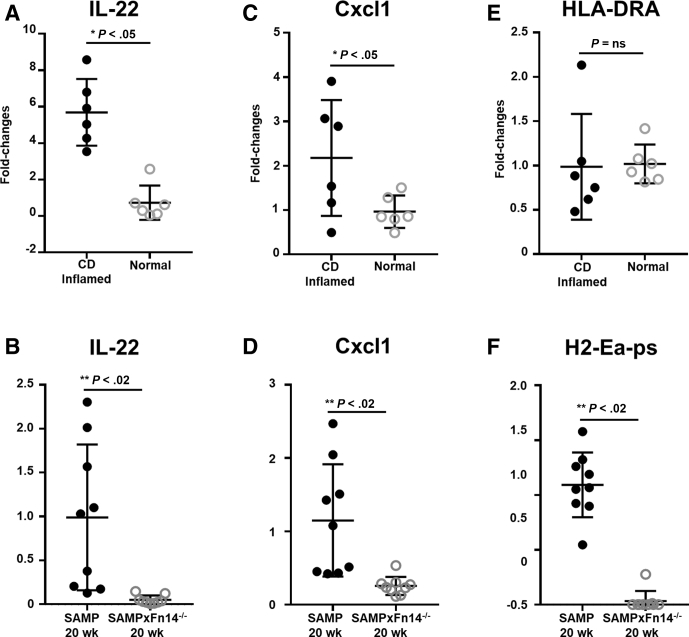
To further corroborate the translational relevance of the data obtained in mouse and human samples, we analyzed by qRT-PCR the expression of the 3 most significant genes in the Nanostring analysis (ie, Il22, Cxcl1, and H2-Ea-ps). Our results confirm that Il22, Cxcl1, and H2-Ea-ps were decreased in 20-week-old SAMP × Fn14-/- compared with WT SAMP littermates. Remarkably, qRT-PCR data in CD patients resemble the same gene expression patterns observed in the SAMP model. Specifically, CD patients and 20-week-old SAMP mice showed a significantly increased expression of Il22 (Figure 9A and B) and Cxcl1 (Figure 9C and D) compared with healthy controls and 20-week-old SAMP × Fn14-/-, respectively. Because H2-Ea-ps is a mouse-specific gene, we measured the expression of the correspondent ortholog human gene HLA-DRA. SAMP mice showed increased expression of H2-Ea-ps compared with the SAMP × Fn14-/- littermates (Figure 9F), but there was no difference in the expression of HLA-DRA between CD patients and healthy controls (Figure 9E).
Figure 9.
CD patients show the same gene expression patterns highlighted by qRT-PCR mouse data. (A) Increased expression of Il22 in CD inflamed patients compared with healthy controls (unpaired t test: 5.69 ± 0.75 vs 0.73 ± 0.38; P < .001; N = 6/group). (B) Decreased expression of Il22 in 20-week-old SAMP × Fn14-/- compared with 20-week-old SAMP (unpaired t test: 0.05 ± 0.02 vs 0.99 ± 0.28; P < .02; N = 9/group). (C) Increased expression of Cxcl1 in CD inflamed patients compared with healthy controls (unpaired t test: 2.17 ± 0.53 vs 0.96 ± 0.15; P ≤ .05; N = 6/group). (D) Decreased expression of Cxcl1 in 20-week-old SAMP × Fn14-/- compared with 20-week-old SAMP (unpaired t test: 0.26 ± 0.04 vs 1.15 ± 0.26; P < .02; N = 9/group). (E) No significant change in expression of HLA-DRA in CD patients compared with healthy controls (unpaired t test: 0.99 ± 0.24 vs 1.02 ± 0.09; P = NS; N = 6/group). (F) Decreased expression of H2-Ea-ps in 20-week-old SAMP × Fn14-/- compared with 20-week-old SAMP (unpaired t test: 0.03 ± 0.03 vs 1.09 ± 0.09; P < .001; N = 9/group). Data are presented as means ± SD, and are representative of 3 independent experiments. *P ≤ .05, **P < .02, and ***P < .001.
Discussion
In the present study, we showed that deletion of TWEAK’s cognate receptor, Fn14, significantly improves chronic CD-like ileitis in SAMP mice through down-regulation of several inflammatory genes. Furthermore, we determined that the anti-inflammatory effects of Fn14 deletion involve suppression of Th1- and Th2-type immune responses. Although the role of Fn14 as a proinflammatory cytokine and related Th2 responses have been described previously in a model of colitis induced by repeated rectal administration of a hapten,30 our data were generated using SAMP mice, a well-characterized model of CD, in which the main pathology is a spontaneous ileitis that is remarkably comparable with the human condition, with skip lesions, stricture formation, and transmural inflammation. In support of the translational relevance of these findings, we showed that CD patients with active inflammation preferentially overexpressed TWEAK and Fn14 compared with UC and noninflamed control patients. Furthermore, 2 of the 3 genes (ie, Il22, Cxcl1) that showed the highest expression in ilea of 20-week-old SAMP WT vs age-matched Fn14-/- mice also were found to be overexpressed in CD patients compared with healthy controls. These results further confirm the striking resemblance between the intestinal pathology observed in CD and in SAMP mice. We therefore suggest that increased expression of TWEAK and Fn14 may represent a key mechanism by which inflammation is exacerbated in CD patients with chronic active inflammation. Our group recently reported that Fn14 deletion in mice on an intestinal disease-free genetic background (eg, C57BL/6J strain) aggravates the severity of acute colitis induced by dextran sodium sulfate (DSS) administration.31 In fact, Fn14 deletion increases the susceptibility of C57BL/6J mice to DSS-induced acute colitis compared with WT mice, suggesting that in normal immunocompetent mice, TWEAK/Fn14 plays a protective role after an acute challenge. In contrast, the present study shows that Fn14 deletion significantly attenuates the severity of inflammation in adult ileitis-prone SAMP mice, particularly in the late stages of disease (age, 20–30 wk). Taken together, these results show a dichotomous role of TWEAK/Fn14 in acute intestinal injury compared with chronic intestinal inflammation, similar to that observed with other TNF and IL1 family members, including TNF, IL1, IL23, IL33, and IL36.32, 33
Our data show that several critical proinflammatory cytokines involved with regulating key signaling pathways of the mucosal immune system are affected by genetic deletion of Fn14. In fact, we showed that the absence of Fn14 was associated with inhibition of the prototypic Th1 cytokines, TNF and IFNγ, which are important mediators in killing intracellular parasites and perpetuating autoimmune responses.34, 35 Hence, because TNF and IFNγ also notably are increased in CD patients,36, 37 Fn14 may represent a novel cellular target for therapeutic intervention in this condition. The absence of Fn14 also abolishes the expression of the Th2 cytokines, IL4, IL10, and IL13, which normally are associated with Th2 immune responses and fibrosis.38 Interestingly, TNF and IL13 were suppressed significantly in SAMP × Fn14-/- mice at 10 weeks of age, preceding the decrease in disease severity observed at 20 and 30 weeks. These data suggest that these 2 cytokines may be key downstream mediators of Fn14 signaling, early during the development of ileitis.39 This is of particular importance because TNF and IL13 are considered to be critical mediators in the pathogenesis of IBD.40 Altogether, these results show that the anti-inflammatory effects of Fn14 deletion in CD-like ileitis are mediated by key proinflammatory cytokines relevant to the human condition.
Next, we expanded our gene expression analysis to 571 inflammatory and immunologic genes using NanoString technology. We identified 58 inflammatory genes that show significantly altered expression in 20-week-old SAMP WT mice with established disease compared with noninflamed 4-week-old SAMP WT, before the onset of ileitis. Interestingly, only 27 genes were altered significantly in 20-week-old SAMP × Fn14-/- mice, in which inflammation was considerably dampened, compared with 4-week-old SAMP WT controls. In short, NanoString analysis confirmed that both Th1 and Th2 responses were inhibited in 20-week-old SAMP × Fn14-/- mice. Interestingly, none of the genes involved in Th17 differentiation were found to be significantly changed in 20- compared with 4-week-old SAMP mice. These data are consistent with recent clinical trials in CD patients showing that blockade of IL17 was ineffective in this patient population.41, 42 Furthermore, Fn14 deletion on the SAMP background inhibited overexpression of several other genes encoding proinflammatory cytokines, receptors, and intracellular molecules involved in activation and differentiation of Th1 and Th2 cells, such as Il27ra (which promotes the differentiation of CD4+ cells into Th1 cells), Traf1 (which is required for TNF-mediated activation of NF-κB), Gata3 (important regulator of Th2 development), and Il16 (a modulator of T-cell activation). Taken together, our data strongly suggest that Fn14 signaling functions as a key upstream mediator of Th1 and Th2 responses during chronic intestinal inflammation.
We further analyzed and interpreted the high-throughput NanoString gene expression data using statistical gene set–enrichment techniques in which groups of genes correlating with a specific cellular pathway were cross-referenced with differentially expressed genes.43 The results showed that the NF-κB and MAPK signaling pathways, both activated by the Fn14 receptor,44 play pivotal roles in establishing the CD-like ileitis observed in SAMP WT mice at 20 weeks of age. In addition, the observed decrease in NF-κB and MAPK activation correlated with significant improvement in the severity of ileitis in SAMP × Fn14-/- mice at the same age. As part of our investigation, because engagement of TWEAK/Fn14 leads to activation of apoptosis, we also hypothesized that one effect of Fn14 deficiency could be the reduction of apoptosis, which may result in decreased intestinal inflammation. In support of this hypothesis, the NanoString data showed that induction of apoptosis activity was reduced dramatically in 20-week-old SAMP × Fn14-/- mice compared with age-matched SAMP WT mice.
Furthermore, we found that Fn14 requires both hematopoietic and nonhematopoietic compartments to completely exert its proinflammatory effects within the intestinal mucosa, as supported by the results from the bone marrow chimera experiments. These data confirm previously published results by our laboratory, showing that Fn14 requires both hematopoietic and nonhematopoietic compartments to exert its protective role during acute DSS-induced colitis.31 As such, these findings were somehow predictable because Fn14 is expressed in multiple cell types, such as mesenchymal cells, epithelial cells, granulocytes, and macrophages, as previously reported.18, 20 These results provide further support that TWEAK/Fn14 represents a critical cytokine pathway that regulates intestinal inflammation by acting on both hematopoietic and nonhematopoietic cells, reducing lymphocytic and neutrophilic infiltration.
Finally, to confirm the translational relevance of our findings, we analyzed the expression of TWEAK and Fn14 in ileal tissues derived from IBD patients and patients not affected by IBD. These data showed that TWEAK and Fn14 were overexpressed significantly in ileal biopsy specimens from CD patients during active inflammation compared with those from UC and patients who underwent surgery for reasons not related to IBD. Interestingly, TWEAK and Fn14 were not overexpressed in CD patients during the remission phase. These data support our hypothesis that TWEAK/Fn14 signaling pathways are up-regulated during acute and chronic inflammation as a defensive mechanism, but their excessive activation in IBD may lead to exaggerated gut immune responses against gut commensal flora. The consequences are delayed healing, dysregulated tissue repair, and fibrosis.45, 46 Our results are different from those reported by Kawashima et al,20 showing that TWEAK/Fn14 were increased in patients with active UC, but not in CD patients and healthy controls. Several points may explain this difference. First, in the aforementioned study, only messenger RNA (mRNA) levels were reported, whereas in our study, we evaluated both mRNA by qPCR as well as protein expression by Western blot and immunofluorescence. Second, only 4 patients with CD were included in the Kawashima et al20 studies, compared with 25 in this report. Finally, our analyses primarily included tissue specimens from the small intestine, whereas the former only studied colonic specimens. Findings within the present study reported TWEAK/Fn14 overexpression in tissue lesions obtained from patients with active CD. Furthermore, our mouse and human data related to IL22 and Cxcl1 are in agreement with the studies by Brand et al47 and Kim et al,48 highlighting the proinflammatory role(s) of these cytokines in active CD. In addition, the discovery that Fn14 activation causes significant overexpression of both Il22 and Cxcl1 lays the groundwork for developing novel therapeutic strategies to prevent flares in CD patients by blocking Fn14.
In conclusion, our results show significant TWEAK/Fn14 overexpression in intestinal tissue lesions from patients with CD, and mechanistically show a critical role of Fn14 signaling in the pathogenesis of experimental CD-like ileitis. With the availability of novel drugs that target TWEAK/Fn14,49 our data indicate that potential pharmacologic inhibition of Fn14 signaling may be a feasible and beneficial strategy to prevent Th1- and Th2-driven intestinal inflammation in patients with CD.
Materials and Methods
Experimental Animals
SAMP mice were obtained from the Mouse Models Core of the National Institutes of Health P30 Cleveland Digestive Disease Research Core Center. A marker-assisted selection protocol was used to generate congenic SAMP mice carrying deletion of the Fn14 gene: using a marker-assisted speed congenic approach, we generated a group of 96 microsatellite genotyping markers to characterize and separate SAMP and C57/BL6 strains. We then created 10 generations of Fn14+/- mice crossing the male Fn14+/- mice of each generation with female SAMP mice. After generation N10, we were able to introduce single Fn14 deletions onto the SAMP background. Genotyping analysis of these mice confirm that the Fn14 gene is completely knocked out compared with WT SAMP littermates. SAMP WT littermates were used as controls. All mice used in this study were between 4 and 30 weeks of age and were age-matched among experimental groups. An equal number of male and female mice were used. SAMP WT and SAMP × Fn14-/- mice were maintained under specific pathogen–free conditions, fed standard chow (Harlan Teklad, Indianapolis, IN), and kept on 12-hour light/dark cycles. All experiments were conducted in a blinded manner, without prior knowledge of treatments and mouse groups by the experimenter. Mice were randomized to different interventions using a progressive numeric number. The code for each mouse was known only to the animal caretaker and was shown at the end of the study. All procedures performed were approved by the Institutional Animal Care and Use Committee at Case Western Reserve University and followed the American Association for Laboratory Animal Care guidelines.
Histology
Ilea from SAMP WT mice and SAMP × Fn14-/- mice were removed, flushed of fecal contents, opened longitudinally, and placed in Bouin’s fixative. Tissues were embedded in paraffin and stained with H&E. Inflammation was evaluated by a trained pathologist in a blinded fashion using a previously described scoring system.50 Scores were given for 3 histologic changes: (1) active inflammation (neutrophil infiltration of tissue); (2) chronic inflammation (lymphocytes, plasma cells, and macrophages in the mucosa and submucosa); and (3) villus distortion (flattening and/or widening of normal mucosal villous architecture). Scores ranged from 0 to 3, with 0 showing normal histology and 1–3 showing incremental severity of histologic changes based on the presence of scattered polymorphonuclear leukocytes in the lamina propria and/or confluent areas of crypt abscesses and mucosal erosion, increase of mononuclear inflammatory cells in lamina propria (based primarily on the increased cell number below the base of the crypts), height of the villi, and percentage of cross-sectional area. In addition, the presence or absence of granuloma formation (aggregates of epithelioid histiocytes), multinucleated histiocytic giant cell formation, and hypertrophy of the intestinal muscular layer were noted.
Neutrophil Count
Neutrophils were counted by a trained pathologist on tissue stained with H&E with a magnification of 40×. The neutrophils were identified by a characteristic multilobated nucleus, usually 3–5 lobes that are connected by thin joined strands of chromatin. The cytoplasm of neutrophils contains numerous azurophilic granules and secondary granules. Three pictures were taken at the distal, middle, and proximal side for each section, and the average number of neutrophils in the 3 pictures was calculated.
Stereomicroscopy 3-Dimensional Pattern Profiling of Intestinal Inflammation
The 3-dimensional stereomicroscopy assessment and pattern profiling protocol was used to map and quantify the intestinal health in SAMP mice and SAMP × Fn14-/- and to determine the extent of mucosal involvement that occurs with chronic inflammation as previously described.25 To analyze the variable patterns of stereomicroscopic abnormalities, stereomicroscopy photographs were analyzed using ImageJ (National Institutes of Health, Bethesda, MD) public domain software (imagej.nih.gov/ij/).
Bone Marrow Transfer Experiments
Before transplant, SAMP WT and SAMP × Fn14-/- recipient mice were irradiated twice (600 rad), with a 6-hour interval between the first and the second dose. BM then was harvested from femurs and tibias of 12-week-old SAMP × Fn14-/- or SAMP × Fn14+/+ mice in RPMI medium (10% fetal calf serum), and cell suspensions were washed and diluted to a concentration of 30 × 106 cells/mL in Hank's balanced salt solution. Immediately after the second dose of 600 rad, a total of 7.5 × 106 cells/250 μL then were injected intravenously into the lateral tail veins of recipient SAMP × Fn14-/- or SAMP WT mice, resulting in 4 different groups of chimeric mice, as follows: (1) Fn14-/- BM→ Fn14-/- mice, (2) Fn14+/+ BM→ Fn14-/- mice, (3) Fn14-/- BM→ Fn14+/+ mice, and (4) Fn14+/+ BM→ Fn14+/+ mice. BM chimera recipient mice were placed on antibiotic water (0.7 mmol/L neomycin sulfate, 80 mmol/L sulfamethoxazole, and 0.37 mmol/L trimethoprim) for 2 weeks after irradiation, and then given autoclaved water for 6 weeks to reconstitute normal gut microbiota.
qRT-PCR
Ilea samples from each mouse were collected and subjected to physical homogenization (with 100 mg of 1.4-mm ceramic beads; 4000 rpm), and total RNA was isolated using an RNeasy Mini Kit (Qiagen, Hilden, Germany). Reverse transcription was performed using the High-Capacity RNA-to-Complementary DNA Kit (Applied Biosystems, Carlsbad, CA). qPCR amplification of complementary DNA samples was performed using clear 96-well plates (Roche, Branchburg, NJ) and the Roche 480 LightCycler SYBR Green (Roche) run template settings (hot start 95°C, 10 min, 40 amplification cycles; 95°C, 15 s; 60°C, 30 s; 72°C, 30 s); β-actin was used as a housekeeping gene.
Primers for qRT-PCR
The following primers were custom designed and synthesized by Integrated DNA Technology. For mouse work, we used the following: B actin: forward primer: 5’CAGGGTGTGATGGTGGGAATG3’; reverse primer: 5’GTAGAAGGTGTGGTGCCAGATC3’; IL22: forward primer: 5’TCGTCAACCGCACCTTTATG3’; reverse primer: 5’CCCGATGAGCCGGACAT3’; Cxcl1: forward primer: 5’TGTTGTGCGAAAAGAAGTGC3’; reverse primer: 5’CGAGACGAGACCAGGAGAAA3’; H2-Ea-ps: forward primer: 5’CGTGTGTGCTCTTGGGTTGTTTGT3’; reverse primer: 5’GTCGGCGTTCTACAACATTGCGTT3’; Il4: forward primer: 5’GCTAGTTGTCATCCTGCTCTTC-3’; reverse primer: 5’-GGCGTCCCTTCTCCTGTG3’; Il33: forward primer: 5’TCCTTGCTTGGCAGTATCCA3’, reverse primer: 5’TGCTCAATGTGTCAACAGACG3’; TNFα: forward primer: 5’GCGGTGCCTATGTCTCAG3’; reverse primer: 5’GCCATTTGGGAACTTCTCATC3’; Il13: forward primer: 5’GGGAGTCTGGTCTTGTGTGATG3’; reverse primer: 5’TTGCTTGCCTTGGTGGTCTC3’; IFNγ: forward primer: 5’TGAGCTCATTGAATGCTTGG3’; reverse primer: 5’ACAGCAAGGCGAA AAAGGAT3’; Il10: forward primer: 5’TGGCCCAGAAATCAAGGAGC3’; reverse primer: 5’CAGCAGACTCAATACACACT3’. For human work, we used the following: β actin: forward primer: 5’AGAGCTACGAGCTGCCTGAC3’; reverse primer: 5’AGCACTGTGTTGGCGTACAG3’; Il22: forward primer: 5’GCAGGCTTGACAAGTCCAACT3’; reverse primer: 5’GCCTCCTTAGCCAGCATGAA3’; Cxcl1: forward primer: AACCGAAGTCATAGCCACAC; reverse primer: 5’GTTGGATTTGTCACTGTTCAGC3’; HLA-DRA: forward primer: 5’AGACAAGTTCACCCCACCAG3’; reverse primer: 5’AGCATCAAACTCCCAGTGC3’.
Cell Isolation and Cytokine Enzyme-Linked Immunosorbent Assay
MLNs were removed aseptically and pressed gently against a 70-μm cell strainer to obtain single-cell suspensions. Resulting cells were cultured (1 × 106 /mL) in RPMI 1640 with 10% fetal bovine serum and 1% penicillin/streptomycin for 72 hours in the presence of αCD3/αCD28 monoclonal antibodies (1 μg/mL) as previously described.51 After the incubation period, supernatants were collected for TNFα, IFNγ, IL4, and IL13 analysis by enzyme-linked immunosorbent assay (Thermo Fisher Scientific, Waltham, MA). All enzyme-linked immunosorbent assays were performed according to the manufacturer’s instructions.
NanoString nCounter Gene Expression Analysis
Total RNA was extracted from ilea of 4-week-old and 20-week-old SAMP × Fn14-/- and SAMP WT mice using the RNeasy Mini Kit (Qiagen, Hilden, Germany). Extracted RNA was incubated with a custom panel of 560 bar-coded probes (NanoString Technologies) specific for genes associated with mouse immunology and inflammation. Six biological replicates per condition were evaluated. Standard NanoString protocols were followed.52 Reporter probes, hybridization solution, sample, and capture probes were mixed together and hybridized overnight at 65°C. After hybridization, samples were transferred and processed in the NanoString nCounter Prep Station. The Prep Station washed away excess probes and purified the target/probe complexes using magnetic beads. Briefly, the hybridization solution containing the target-probe complexes is mixed with magnetic beads complementary to sequences on the capture probe. This process is followed by a wash step to remove the excess reporter probes. The target-probe complexes then are hybridized to magnetic beads complementary to sequences on the reporter probe. A final wash step is performed to remove excess capture probes. The purified target/probe complexes were deposited in a cartridge, laid flat, and immobilized for data collection.
Results were analyzed using the nSolver Analysis Software 4.0 (NanoString). Each sample was normalized first to the geometric mean of positive controls, and then to the geometric mean of reference genes. Differences in gene expression were considered significant when there was a fold-change greater than 2 times compared with the control group and the P value for comparison between target and control was less than .05. We used the method described by Danaher et al53 to measure the abundance of various cell populations. The method quantifies cell populations using marker genes that are expressed stably and specifically in given cell types. These marker genes act as reference genes specific to individual cell types because they are expressed only in their nominal cell type, at the same level in each cell. The closer the biomarker genes defined in the probe annotation are to this ideal scenario, the more reliable the scores. A cell type’s abundance can be measured as the average log-scale expression of its characteristic genes. The algorithm used to identify appropriate marker genes and exclude badly behaving cell-type–specific genes from estimates of cell type abundance is detailed later, as is the permutation test used to derive a P value assessing a cell type’s marker genes. First, a similarity metric between 2 candidate cell type–specific genes was defined. Under the assumption that both genes are specific to the same cell type and consistently expressed within it, they will be highly correlated with a slope of 1. To measure 2 gene’s adherence to this pattern, a slightly modified version of the Pearson correlation metric was used:
where x and y are the vectors of log-transformed, normalized expression values of the 2 genes, ® and ® are their sample means, and var(x) and var(y) are their sample variances. The similarity() function equals 1 when the 2 genes are perfectly correlated with a slope of 1 and decreases for gene pairs with low correlation or a slope diverging from 1. Our gene selection algorithm is as follows. Assume there are p genes and n samples. Use the similarity function to compute a p*p similarity matrix among the genes. Each gene has a similarity of 1 with itself. Label all gene pairs with similarity below 0.2 as discordant. Iteratively remove genes: although there are more than 2 genes remaining and although at least 1 discordant pair of genes remains: count the number of discordant pairs each gene participates in, call the maximum of these counts n-discord; and identify the genes with n.discord instances of discordance with another gene. Of these genes, remove the single gene with the lowest average similarity to the other remaining genes. This process is similar to the geNorm algorithm.54 This similarity is not a coincidence because cell type marker genes can be thought of cell type–specific housekeeper genes. We tested the null hypothesis that the given gene set shows no greater cell type–specific–like behavior than a randomly selected gene set of similar size. First, we required a metric of a gene set’s adherence to the assumption of cell type–specific and consistent expression.
where X is the matrix of log-transformed, normalized expression values of the gene set, and where p is the number of genes. The concordance() function evaluates at 1 if all genes are perfectly correlated with a slope of 1, and degrades to 0 as this pattern weakens. We performed our permutation test as follows. Assume the given gene set has p genes, of which p0 survived the iterative gene selection procedure. Call the data from the gene set X, and the data from the reduced gene set X0. Compute concordance(X0). Choose 1000 random genes sets of size p. Denote the data from a random gene set X’. For each gene set, apply the criteria of the gene selection algorithm to reduce X’ to only its best p0 genes. Call the data from this reduced random gene set X0’, and compute concordance(X0’). Return a p value equal to the proportion of concordance(X0’) values greater than concordance(X0).
Patients
In the present study we included a total of 45 IBD and 10 control patients. All intestinal surgical specimens and endoscopic biopsy specimens were obtained in an unidentified manner (institutional review board exempt) from the Biorepository Core of the National Institutes of Health Cleveland Digestive Diseases Research Core Center. For Western blot or immunofluorescence, surgical tissue specimens were obtained from CD or UC patients who underwent therapeutic bowel resection at Cleveland Medical Center or control patients receiving a bowel resection for noninflammatory, malignant, and nonmalignant conditions. All surgical specimens were collected immediately after resection, opened longitudinally, rinsed, and examined for gross pathologic changes, and representative full-thickness samples were obtained from areas of active disease (involved) as well as macroscopically normal (uninvolved) areas from the same patient. For mRNA studies, endoscopic biopsy specimens were obtained from CD and UC patients from areas of active inflammation, before the initiation of biologic therapies, or after a wash-out period of 3 weeks or more after biologic therapies. Biopsy specimens from patients undergoing colonoscopy for diagnostic or surveillance purposes were used as controls. The mean ages were 59 ± 5 years (CD), 55 ± 5 years (UC), and 50 ± 7 years (control). All diagnoses were confirmed by accepted clinical, radiologic, endoscopic, and histologic criteria. Informed consent was obtained from each patient. All protocols were approved by the Institutional Review Board of University Hospitals Cleveland Medical Center (Cleveland, OH).
Immunohistochemistry
Ileal samples (n = 6/group) prepared in Bouin’s fixative were embedded in paraffin and cut. Furthermore, we performed immunohistochemistry for detecting CD3. The sections obtained were stained with anti-CD3 antibody (Ab11089; Abcam, Cambridge, United Kingdom) and then assessed by confocal microscopy to characterize the distribution of CD3 in mucosal and submucosal layers. CD3+ cells have been counted by a trained pathologist on tissue counterstained with hematoxylin with a magnification of 40×. Three pictures were taken at distal, middle, and proximal side for each section, and the average of number of CD3+ cells in the 3 pictures was calculated.
Immunofluorescence
Immunofluorescence was performed to detect TWEAK and CD3 in colon tissues of patients with CD, UC, and normal patients (n = 10/group) with anti-TWEAK (Ab73170; Abcam) at a concentration of 20 μg/mL and anti-CD3 antibody (Ab699; Abcam) 1/40 dilution. An isotype antibody was used as a negative control.
Western Blot Analysis
Colonic and ileal biopsy specimens of IBD patients with and without active inflammation, and control patients who underwent intestinal surgery for reasons not related to IBD (n = 10/group), were lysed and centrifuged. Supernatants were standardized through the Bradford assay (BioRad, Hercules, CA), and Western blot was completed on 0.45-μm pore size nitrocellulose membranes (Thermo Scientific, Rockford, IL) and blotted with anti-TWEAK (Ab73170; Abcam) with a concentration of 1 μg/mL and anti-Fn14 (4403S; Cell Signaling, Danvers, MA) with a dilution of 1:1000. Actin was used as a loading control. Immunoreactive bands were shown with an enhanced chemiluminescent substrate for detection of horseradish peroxidase (Thermo Scientific, Waltham, MA), and blots were exposed to blue basic autorad films (Bioexpress, Kaysville, UT).
Statistical Analysis
Experiments were conducted at a minimum in duplicate. Univariate and multivariate analyses were conducted using the collective data from replicated experiments. When the data fulfilled the assumptions for parametric statistics, comparisons of continuous data across experimental groups were conducted using the Student t test. Alternative nonparametric tests were used for data with unfulfilled distributional assumptions regarding the normality of the data. Data were expressed as SEMs, and 95% CIs were reported when appropriate. An α level of .05 was considered significant. All analyses were conducted with STATA (Statacorp, College Station, TX) and GraphPad software (San Diego, CA).
Acknowledgments
The authors thank Giorgos Bamias and Kristen O. Arseneau for critical discussion of the manuscript. The authors also thank the core services provided by the Cleveland Digestive Research Core Center.
Footnotes
Author contributions Luca Di Martino was responsible for the study concept, design, and acquisition of murine data; Abdullah Osme analyzed and interpreted the histologic data; Sarah Kossak-Gupta acquired human data; Theresa T. Pizarro was responsible for the study concept, design, obtained funding, and critical revision of the manuscript for important intellectual content; and Fabio Cominelli was responsible for the study concept, design, obtained funding, and study supervision.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by National Institutes of Health grants DK055812 (F.C.), DK042191 (F.C. and T.T.P.), DK091222 (F.C. and T.T.P.), DK056762 (T.T.P.), and the Crohn's and Colitis Foundation grant RFA481439 (L.D.M.). Also supported by the Histology/Imaging and Biorepository Cores of the Cleveland Digestive Disease Research Core Center (DK097948 to F.C.).
Supplementary Material
References
- 1.Xavier R.J., Podolsky D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 2.Feldman P.A., Wolfson D., Barkin J.S. Medical management of Crohn's disease. Clin Colon Rectal Surg. 2007;20:269–281. doi: 10.1055/s-2007-991026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanauer S.B., Feagan B.G., Lichtenstein G.R., Mayer L.F., Schreiber S., Colombel J.F., Rachmilewitz D., Wolf D.C., Olson A., Bao W., Rutgeerts P., ACCENT I Study Group Maintenance infliximab for Crohn's disease: the ACCENT I randomised trial. Lancet. 2002;359:1541–1549. doi: 10.1016/S0140-6736(02)08512-4. [DOI] [PubMed] [Google Scholar]
- 4.Lirhus S.S., Hoivik M.L., Moum B., Melberg H.O. Regional differences in anti-TNF-alpha therapy and surgery in the treatment of inflammatory bowel disease patients: a Norwegian nationwide cohort study. Scand J Gastroenterol. 2018;53:952–957. doi: 10.1080/00365521.2018.1495258. [DOI] [PubMed] [Google Scholar]
- 5.Sandborn W.J., Feagan B.G., Lichtenstein G.R. Medical management of mild to moderate Crohn's disease: evidence-based treatment algorithms for induction and maintenance of remission. Aliment Pharmacol Ther. 2007;26:987–1003. doi: 10.1111/j.1365-2036.2007.03455.x. [DOI] [PubMed] [Google Scholar]
- 6.Burkly L.C. TWEAK/Fn14 axis: the current paradigm of tissue injury-inducible function in the midst of complexities. Semin Immunol. 2014;26:229–236. doi: 10.1016/j.smim.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Nakayama M., Harada N., Okumura K., Yagita H. Characterization of murine TWEAK and its receptor (Fn14) by monoclonal antibodies. Biochem Biophys Res Commun. 2003;306:819–825. doi: 10.1016/s0006-291x(03)01051-9. [DOI] [PubMed] [Google Scholar]
- 8.Yepes M., Brown S.A., Moore E.G., Smith E.P., Lawrence D.A., Winkles J.A. A soluble Fn14-Fc decoy receptor reduces infarct volume in a murine model of cerebral ischemia. Am J Pathol. 2005;166:511–520. doi: 10.1016/S0002-9440(10)62273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawakita T., Shiraki K., Yamanaka Y., Yamaguchi Y., Saitou Y., Enokimura N., Yamamoto N., Okano H., Sugimoto K., Murata K., Nakano T. Functional expression of TWEAK in human hepatocellular carcinoma: possible implication in cell proliferation and tumor angiogenesis. Biochem Biophys Res Commun. 2004;318:726–733. doi: 10.1016/j.bbrc.2004.04.084. [DOI] [PubMed] [Google Scholar]
- 10.Tran N.L., McDonough W.S., Donohue P.J., Winkles J.A., Berens T.J., Ross K.R., Hoelzinger D.B., Beaudry C., Coons S.W., Berens M.E. The human Fn14 receptor gene is up-regulated in migrating glioma cells in vitro and overexpressed in advanced glial tumors. Am J Pathol. 2003;162:1313–1321. doi: 10.1016/S0002-9440(10)63927-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burkly L.C., Michaelson J.S., Hahm K., Jakubowski A., Zheng T.S. TWEAKing tissue remodeling by a multifunctional cytokine: role of TWEAK/Fn14 pathway in health and disease. Cytokine. 2007;40:1–16. doi: 10.1016/j.cyto.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Scholzke M.N., Rottinger A., Murikinati S., Gehrig N., Leib C., Schwaninger M. TWEAK regulates proliferation and differentiation of adult neural progenitor cells. Mol Cell Neurosci. 2011;46:325–332. doi: 10.1016/j.mcn.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Zheng T.S., Burkly L.C. No end in site: TWEAK/Fn14 activation and autoimmunity associated- end-organ pathologies. J Leukoc Biol. 2008;84:338–347. doi: 10.1189/jlb.0308165. [DOI] [PubMed] [Google Scholar]
- 14.Ucero A.C., Benito-Martin A., Fuentes-Calvo I., Santamaria B., Blanco J., Lopez-Novoa J.M., Ruiz-Ortega M., Egido J., Burkly L.C., Martinez-Salgado C., Ortiz A. TNF-related weak inducer of apoptosis (TWEAK) promotes kidney fibrosis and Ras-dependent proliferation of cultured renal fibroblast. Biochim Biophys Acta. 2013;1832:1744–1755. doi: 10.1016/j.bbadis.2013.05.032. [DOI] [PubMed] [Google Scholar]
- 15.Jakubowski A., Ambrose C., Parr M., Lincecum J.M., Wang M.Z., Zheng T.S., Browning B., Michaelson J.S., Baetscher M., Wang B., Bissell D.M., Burkly L.C. TWEAK induces liver progenitor cell proliferation. J Clin Invest. 2005;115:2330–2340. doi: 10.1172/JCI23486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J., Kwan B.C., Lai F.M., Choi P.C., Tam L.S., Li E.K., Chow K.M., Wang G., Li P.K., Szeto C.C. Gene expression of TWEAK/Fn14 and IP-10/CXCR3 in glomerulus and tubulointerstitium of patients with lupus nephritis. Nephrology (Carlton) 2011;16:426–432. doi: 10.1111/j.1440-1797.2011.01449.x. [DOI] [PubMed] [Google Scholar]
- 17.Michaelson J.S., Burkly L.C. Therapeutic targeting of TWEAK/Fnl4 in cancer: exploiting the intrinsic tumor cell killing capacity of the pathway. Results Probl Cell Differ. 2009;49:145–160. doi: 10.1007/400_2008_18. [DOI] [PubMed] [Google Scholar]
- 18.Burkly L.C., Michaelson J.S., Zheng T.S. TWEAK/Fn14 pathway: an immunological switch for shaping tissue responses. Immunol Rev. 2011;244:99–114. doi: 10.1111/j.1600-065X.2011.01054.x. [DOI] [PubMed] [Google Scholar]
- 19.Dohi T., Borodovsky A., Wu P., Shearstone J.R., Kawashima R., Runkel L., Rajman L., Dong X., Scott M.L., Michaelson J.S., Jakubowski A., Burkly L.C. TWEAK/Fn14 pathway: a nonredundant role in intestinal damage in mice through a TWEAK/intestinal epithelial cell axis. Gastroenterology. 2009;136:912–923. doi: 10.1053/j.gastro.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 20.Kawashima R., Kawamura Y.I., Oshio T., Son A., Yamazaki M., Hagiwara T., Okada T., Inagaki-Ohara K., Wu P., Szak S., Kawamura Y.J., Konishi F., Miyake O., Yano H., Saito Y., Burkly L.C., Dohi T. Interleukin-13 damages intestinal mucosa via TWEAK and Fn14 in mice-a pathway associated with ulcerative colitis. Gastroenterology. 2011;141:2119–2129 e8. doi: 10.1053/j.gastro.2011.08.040. [DOI] [PubMed] [Google Scholar]
- 21.Pizarro T.T., Pastorelli L., Bamias G., Garg R.R., Reuter B.K., Mercado J.R., Chieppa M., Arseneau K.O., Ley K., Cominelli F. SAMP1/YitFc mouse strain: a spontaneous model of Crohn's disease-like ileitis. Inflamm Bowel Dis. 2011;17:2566–2584. doi: 10.1002/ibd.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cominelli F., Arseneau K.O., Rodriguez-Palacios A., Pizarro T.T. Uncovering pathogenic mechanisms of inflammatory bowel disease using mouse models of Crohn's disease-like ileitis: what is the right model? Cell Mol Gastroenterol Hepatol. 2017;4:19–32. doi: 10.1016/j.jcmgh.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pastorelli L., De Salvo C., Vecchi M., Pizarro T.T. The role of IL-33 in gut mucosal inflammation. Mediators Inflamm. 2013;2013:608187. doi: 10.1155/2013/608187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dohi T., Kawashima R., Kawamura Y.I., Otsubo T., Hagiwara T., Amatucci A., Michaelson J., Burkly L.C. Pathological activation of canonical nuclear-factor kappaB by synergy of tumor necrosis factor alpha and TNF-like weak inducer of apoptosis in mouse acute colitis. Cytokine. 2014;69:14–21. doi: 10.1016/j.cyto.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Palacios A., Kodani T., Kaydo L., Pietropaoli D., Corridoni D., Howell S., Katz J., Xin W., Pizarro T.T., Cominelli F. Stereomicroscopic 3D-pattern profiling of murine and human intestinal inflammation reveals unique structural phenotypes. Nat Commun. 2015;6:7577. doi: 10.1038/ncomms8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bamias G., Martin C., Mishina M., Ross W.G., Rivera-Nieves J., Marini M., Cominelli F. Proinflammatory effects of TH2 cytokines in a murine model of chronic small intestinal inflammation. Gastroenterology. 2005;128:654–666. doi: 10.1053/j.gastro.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 27.Qi X., Qin L., Du R., Chen Y., Lei M., Deng M., Wang J. Lipopolysaccharide upregulated intestinal epithelial cell expression of Fn14 and activation of Fn14 signaling amplify intestinal TLR4-mediated inflammation. Front Cell Infect Microbiol. 2017;7:315. doi: 10.3389/fcimb.2017.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veldman-Jones M.H., Brant R., Rooney C., Geh C., Emery H., Harbron C.G., Wappett M., Sharpe A., Dymond M., Barrett J.C., Harrington E.A., Marshall G. Evaluating robustness and sensitivity of the nanostring technologies ncounter platform to enable multiplexed gene expression analysis of clinical samples. Cancer Res. 2015;75:2587–2593. doi: 10.1158/0008-5472.CAN-15-0262. [DOI] [PubMed] [Google Scholar]
- 29.Tomfohr J., Lu J., Kepler T.B. Pathway level analysis of gene expression using singular value decomposition. BMC Bioinformatics. 2005;6:225. doi: 10.1186/1471-2105-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Son A., Oshio T., Kawamura Y.I., Hagiwara T., Yamazaki M., Inagaki-Ohara K., Okada T., Wu P., Iseki M., Takaki S., Burkly L.C., Dohi T. TWEAK/Fn14 pathway promotes a T helper 2-type chronic colitis with fibrosis in mice. Mucosal Immunol. 2013;6:1131–1142. doi: 10.1038/mi.2013.10. [DOI] [PubMed] [Google Scholar]
- 31.Di Martino L., Dave M., Menghini P., Xin W., Arseneau K.O., Pizarro T.T., Cominelli F. Protective role for TWEAK/Fn14 in regulating acute intestinal inflammation and colitis-associated tumorigenesis. Cancer Res. 2016;76:6533–6542. doi: 10.1158/0008-5472.CAN-16-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bamias G., Corridoni D., Pizarro T.T., Cominelli F. New insights into the dichotomous role of innate cytokines in gut homeostasis and inflammation. Cytokine. 2012;59:451–459. doi: 10.1016/j.cyto.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ngo V.L., Abo H., Maxim E., Harusato A., Geem D., Medina-Contreras O., Merlin D., Gewirtz A.T., Nusrat A., Denning T.L. A cytokine network involving IL-36gamma, IL-23, and IL-22 promotes antimicrobial defense and recovery from intestinal barrier damage. Proc Natl Acad Sci U S A. 2018;115:E5076–E5085. doi: 10.1073/pnas.1718902115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kassiotis G., Kollias G. TNF and receptors in organ-specific autoimmune disease: multi-layered functioning mirrored in animal models. J Clin Invest. 2001;107:1507–1508. doi: 10.1172/JCI13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollard K.M., Cauvi D.M., Toomey C.B., Morris K.V., Kono D.H. Interferon-gamma and systemic autoimmunity. Discov Med. 2013;16:123–131. [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Borra J., Lopez-Larrea C., Gonzalez S., Fuentes D., Dieguez A., Deschamps E.M., Perez-Pariente J.M., Lopez-Vazquez A., de Francisco R., Rodrigo L. High serum tumor necrosis factor-alpha levels are associated with lack of response to infliximab in fistulizing Crohn's disease. Am J Gastroenterol. 2002;97:2350–2356. doi: 10.1111/j.1572-0241.2002.05990.x. [DOI] [PubMed] [Google Scholar]
- 37.Noguchi M., Hiwatashi N., Liu Z., Toyota T. Enhanced interferon-gamma production and B7-2 expression in isolated intestinal mononuclear cells from patients with Crohn's disease. J Gastroenterol. 1995;30(Suppl 8):52–55. [PubMed] [Google Scholar]
- 38.Morishima Y., Ishii Y. Targeting Th2 cytokines in fibrotic diseases. Curr Opin Investig Drugs. 2010;11:1229–1238. [PubMed] [Google Scholar]
- 39.Bamias G., Arseneau K.O., Cominelli F. Cytokines and mucosal immunity. Curr Opin Gastroenterol. 2014;30:547–552. doi: 10.1097/MOG.0000000000000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abraham C., Dulai P.S., Vermeire S., Sandborn W.J. Lessons learned from trials targeting cytokine pathways in patients with inflammatory bowel diseases. Gastroenterology. 2017;152:374–388 e4. doi: 10.1053/j.gastro.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hueber W., Sands B.E., Lewitzky S., Vandemeulebroecke M., Reinisch W., Higgins P.D., Wehkamp J., Feagan B.G., Yao M.D., Karczewski M., Karczewski J., Pezous N., Bek S., Bruin G., Mellgard B., Berger C., Londei M., Bertolino A.P., Tougas G., Travis S.P. Secukinumab in Crohn's Disease Study Group. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn's disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut. 2012;61:1693–1700. doi: 10.1136/gutjnl-2011-301668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mozaffari S., Nikfar S., Abdollahi M. Inflammatory bowel disease therapies discontinued between 2009 and 2014. Expert Opin Investig Drugs. 2015;24:949–956. doi: 10.1517/13543784.2015.1035432. [DOI] [PubMed] [Google Scholar]
- 43.Tsang H.F., Xue V.W., Koh S.P., Chiu Y.M., Ng L.P., Wong S.C. NanoString, a novel digital color-coded barcode technology: current and future applications in molecular diagnostics. Expert Rev Mol Diagn. 2017;17:95–103. doi: 10.1080/14737159.2017.1268533. [DOI] [PubMed] [Google Scholar]
- 44.Tajrishi M.M., Zheng T.S., Burkly L.C., Kumar A. The TWEAK-Fn14 pathway: a potent regulator of skeletal muscle biology in health and disease. Cytokine Growth Factor Rev. 2014;25:215–225. doi: 10.1016/j.cytogfr.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogler G., Hausmann M. Factors promoting development of fibrosis in Crohn's disease. Front Med (Lausanne) 2017;4:96. doi: 10.3389/fmed.2017.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rieder F., Brenmoehl J., Leeb S., Scholmerich J., Rogler G. Wound healing and fibrosis in intestinal disease. Gut. 2007;56:130–139. doi: 10.1136/gut.2006.090456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brand S., Beigel F., Olszak T., Zitzmann K., Eichhorst S.T., Otte J.M., Diepolder H., Marquardt A., Jagla W., Popp A., Leclair S., Herrmann K., Seiderer J., Ochsenkuhn T., Goke B., Auernhammer C.J., Dambacher J. IL-22 is increased in active Crohn's disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am J Physiol Gastrointest Liver Physiol. 2006;290:G827–G838. doi: 10.1152/ajpgi.00513.2005. [DOI] [PubMed] [Google Scholar]
- 48.Kim T.J., Koo J.S., Kim S.J., Hong S.N., Kim Y.S., Yang S.K., Kim Y.H. Role of IL-1ra and Granzyme B as biomarkers in active Crohn's disease patients. Biomarkers. 2018;23:161–166. doi: 10.1080/1354750X.2017.1387933. [DOI] [PubMed] [Google Scholar]
- 49.Wajant H. The TWEAK-Fn14 system as a potential drug target. Br J Pharmacol. 2013;170:748–764. doi: 10.1111/bph.12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corridoni D., Kodani T., Rodriguez-Palacios A., Pizarro T.T., Xin W., Nickerson K.P., McDonald C., Ley K.F., Abbott D.W., Cominelli F. Dysregulated NOD2 predisposes SAMP1/YitFc mice to chronic intestinal inflammation. Proc Natl Acad Sci U S A. 2013;110:16999–17004. doi: 10.1073/pnas.1311657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kosiewicz M.M., Nast C.C., Krishnan A., Rivera-Nieves J., Moskaluk C.A., Matsumoto S., Kozaiwa K., Cominelli F. Th1-type responses mediate spontaneous ileitis in a novel murine model of Crohn's disease. J Clin Invest. 2001;107:695–702. doi: 10.1172/JCI10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geiss G.K., Bumgarner R.E., Birditt B., Dahl T., Dowidar N., Dunaway D.L., Fell H.P., Ferree S., George R.D., Grogan T., James J.J., Maysuria M., Mitton J.D., Oliveri P., Osborn J.L., Peng T., Ratcliffe A.L., Webster P.J., Davidson E.H., Hood L., Dimitrov K. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 53.Danaher P., Warren S., Dennis L., D'Amico L., White A., Disis M.L., Geller M.A., Odunsi K., Beechem J., Fling S.P. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer. 2017;5:18. doi: 10.1186/s40425-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schlotter Y.M., Veenhof E.Z., Brinkhof B., Rutten V.P., Spee B., Willemse T., Penning L.C. A GeNorm algorithm-based selection of reference genes for quantitative real-time PCR in skin biopsies of healthy dogs and dogs with atopic dermatitis. Vet Immunol Immunopathol. 2009;129:115–118. doi: 10.1016/j.vetimm.2008.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.