Abstract
For a safe and sustainable environment, effective microbes as biocontrol agents are in high demand. We have isolated a new Bacillus velezensis strain DTU001, investigated its antifungal spectrum, sequenced its genome, and uncovered the production of lipopeptides in HPLC-HRMS analysis. To test the antifungal efficacy, extracts of B. velezensis DTU001 was tested against a range of twenty human or plant pathogenic fungi. We demonstrate that inhibitory potential of B. velezensis DTU001 against selected fungi is superior in comparison to single lipopeptide, either iturin or fengycin. The isolate showed analogous biofilm formation to other closely related Bacilli. To further support the biocontrol properties of the isolate, coculture with Candida albicans demonstrated that B. velezensis DTU001 exhibited excellent antiproliferation effect against C. albicans. In summary, the described isolate is a potential antifungal agent with a broad antifungal spectrum that might assist our aims to avoid hazardous pathogenic fungi and provide alternative to toxicity caused by chemicals.
Keywords: Bacillus velezensis, Candida albicans, Secondary metabolites, Biocontrol, Biofilm
1. Introduction
The prerequisite for benign and effective antifungal agents increases in parallel with the existing and escalating emergence of fungal pathogens resistant to current therapies [1], being relevant to both therapeutics and biocontrol. While infectious diseases impose serious public health burden and often have devastating consequences, plant diseases constitute an emerging threat to global food security. Additionally, extensive use of chemicals to control plant diseases has distressed the ecological balance of microbes inhabiting soil. This in turn has led to the appearance of resistant pathogenic strains, groundwater contamination, and obvious health risks to human beings. Thus, the identification and development of environmental friendly alternatives to the currently used chemical pesticides for combating a variety of crop diseases is nowadays one of the biggest ecological challenges [2]. It is therefore vital to obtain a broad range of antifungal agents that are non-hazardous to human health as well as to the environment.
Several microorganisms have so far been discovered for the development of biopesticides at commercial level. Among the various microorganisms, Bacillus spp., Pseudomonas spp., and some mycorrhizal fungi are widely used groups for biocontrol [3]. Especially, the Bacillus genus is proven to be source an excellent source of antifungal agents and thus Bacilli are widely used as biocontrol agents [4,5]. The US Food and Drug Administration (US FDA) declared Bacillus subtilis as GRAS (generally recognized as safe) organisms for its use in food processing industries [6]. Members of the B. subtilis groups were found to produce numerous highly active antifungal compounds [7]. The B. subtilis group contains the “Operational Group Bacillus amyloliquefaciens” that includes B. amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis [8]. Among these, B. velezensis FZB42 (formerly B. amyloliquefaciens subsp. plantarum FZB42) is the most researched commercially used Bacillus-based biofertilizer and biocontrol agent in agriculture [9].
Antifungal cyclopeptides appeared to play a major role in biological control of plant pathogens [4,10]. Composed of a peptide and a fatty acyl moiety, these amphiphilic metabolites have consolidated their position as potent therapeutic compounds, such as daptomycin, a lipopeptide antibiotic used in human medicine [11]. Antibacterial, -viral, -fungal, -tumor activity and immune-modulatory effects are some of their proven and emerging properties [12]. In addition, secondary metabolites seem to play an important role in bacterial-fungal interaction of Bacilli [13] and other bacterial life styles, including surface spreading [[14], [15], [16]].
Here, we describe a new Bacillus isolate DTU001, which exhibited a broad and strong antifungal spectrum against different pathogenic fungi ranging from plant pathogens to human pathogens. Using in silico DNA hybridization, it was identified as B. velenzensis. To dissect the genetic potential as an antifungal agent, we genome-mined BGCs in 31 Bacillus species and compared their biosynthetic potential with a special focus on commercially applied strains. To detect the synthesized secondary metabolites of B. velezensis DTU001, the lipopetide profile of the strain's crude extract was investigated.
Lipopeptides produced by Bacilli, such as iturin, fengycin and surfactin are believed to contribute to their antifungal potential. When new isolates from the B. subtilis species complex are evaluated, the role of single lipopeptide in antifungal activity is scrutinized lacking systematic in vitro evaluation of the antifungal efficacy of the combination of these secondary metabolites. To fill in this gap, we also studied the efficacy of extracted iturin and fengycin in comparison to the crude extract of B. velezensis DTU001 using twenty critical human or plant pathogens. In this report, we outline the phylogeny, genomics, metabolomics, biofilm formation, and antifungal effects of B. velezensis DTU001.
2. Materials and methods
2.1. Bacterial and fungal strains
B. velezensis DTU001 is an airborne bacterium, isolated indoor at the DTU campus, Kongens Lyngby Denmark. Twenty selected plant/human pathogens (Table 1) covering five genera Aspergillus, Penicillium, Talaromyces, Cryptococcus and Candida were obtained from the DTU Bioengineering IBT fungal collection. All microbes are maintained at the department.
Table 1.
Bioactivity of iturin (1 mg/mL), fengycin (1mg/mL), and crude B. velezensis extract (1mg/mL) against 16 human- and plant-pathogenic filamentous fungi and 4 yeasts. No inhibition is denoted with "-", while faint inhibition is labeled "+".
| Name of the fungi | Strain information | Zone of inhibition (mm) |
||
|---|---|---|---|---|
| Iturin | Fengycin | B. velezensis supernatant | ||
| A. niger IBT27878 | Harvest decay of fresh and dried fruits and certain vegetables, nuts, beans, and cereals | 27 | – | 25 |
| A. terreus IBT22089 | Opportunistic human pathogen; crops pathogen, e.g. wheat, ryegrass, and potatoes | 20 | – | 19 |
| A. felis IBT34215 | Causative agent of Aspergillosis in humans, dogs and cats | – | – | – |
| A. flavus IBT32105 | 30 | – | 26 | |
| T. amestolkiae IBT23485 | Common cosmopolitan species that has been cultured from indoor house dust, sputum and lungs from cystic fibrosis patients, indoor air, wheat, soil, pineapple, sculptures and manure | 26 | 20 | 24 |
| T. atroroseus IBT11181 | 29 | 20 | 30 | |
| P. expansum IBT34672 | 24 | – | 20 | |
| P. frequentans IBT34879 | 25 | – | 23 | |
| P. italicum IBT34168 | 30 | – | 33 | |
| P. digitatum IBT34217 | + | – | + | |
| P. ulaiense IBT23038 | 30 | – | 30 | |
| P. arizonense IBT12857 | + | – | + | |
| P. solitum IBT34928 | 33 | – | 33 | |
| P. sclerotigenum IBT30034 | Opportunistic human infection, associated with blue mold of yam in Korea | – | – | 32 |
| A. uvarum IBT32575 | Contamination of several agricultural products, including grape-derived products, coffee and cocoa | 25 | – | 35 |
| A. calidoustus IBT28322 | Opportunistic human infection | – | – | 32 |
| C. albicans IBT654 | Superficial and serious systemic diseases | 21 | – | 19 |
| C. albicans IBT657 | 24 | – | 20 | |
| C. neoformans IBT10488 | 30 | – | 31 | |
| C. neoformans IBT10496 | 26 | – | 29 | |
2.2. Genome sequencing
Genomic DNA was isolated from B. velezensis DTU001 using Bacterial and Yeast Genomic DNA kit (EURx, Poland). Genome sequencing has been performed on PacBio RSII platform, de novo assembly and annotation were done using the RS HGAO Assembly (v3.0) and Prokka (v1.12b) software, respectively. The complete sequence has been deposited in NCBI (accession number CP035533) followed by automatic annotation via NCBI PGAP [17].
2.3. Chemicals
Solvents used in this study were LC–MS grade, and all other chemicals were analytical grade. Iturin A, fengycin and solvents as well were from Sigma-Aldrich (Steinheim, Germany) unless otherwise stated. Water was purified using a Milli-Q system (Millipore, Bedford, MA). ESI–TOF tune mix was purchased from Agilent Technologies (Torrance, CA, USA).
2.4. Digital DNA–DNA hybridization (dDDH)
The genome-to-genome-distance calculator (GGDC) version 2.1 provided by DSMZ (http://ggdc.dsmz.de/) was used for genome-based species delineation [18] and genome-based subspecies delineation [19]. Distances were calculated by (i) comparing two genomes using the chosen program to obtain HSPs/MUMs and (ii) inferring distances from the set of HSPs/MUMs using three distinct formulas. Next, the distances were transformed to values analogous to DDH. The DDH estimates were based on an empirical reference dataset comprising real DDH values and genome sequences. The DDH estimate resulted from a generalized linear model (GLM) which also provided the estimate's confidence interval. Three formulas are available for the calculation: (1) HSP length/total length; (2) identities/HSP length; and (3) identities/total length. Formula 2, which is especially appropriate to analyze draft genomes, was used in this study.
2.5. Construction of the evolutionary tree
An initial whole genome phylogeny was inferred using the automlst tool (http://automlst.ziemertlab.com; [38]). This automatically identifies related strains based on a seed collection of sequences obtained from NCBI Genbank (B. velezensis DTU001, B. thuringiensis serovar Berliner ATCC 10792, B. cereus 03BB102, B. megaterium DSM319, B. velezensis FZB42), using a MASH-based algorithm, that extracts 80 phylogenetically relevant genes and builds a MLST phylogeny out of these data. The sequence alignment obtained from automlst was then manually refined (removal of very similar species). To infer the best phylogenetic model, jModeltest (v.2.1.10) was used with default parameters [20]. In all three evaluation criteria (Akaike Information Criterion AIC, Bayesian Information Criterion BIC and the Decision Therory Performance-based Selection), the GTR + I + G model was deemed most suitable. Thus, a maximum-likelihood tree was constructed in PhyML (v3.3.20180621) [21] using the parameters suggested by jModeltest. The tree then was checked with Dendroscope (v3.5.10) [22] and plotted using the ETE3 toolkit [23].
2.6. Genome mining of secondary metabolites
B. velezensis DTU001 and other 32 Bacillus strains were mined for the biosynthetic gene clusters using the antiSMASH platform [24] and BiG-SCAPE [25].
2.7. Agar-diffusion assay
Agar-diffusion assay was carried to test the anti-fungal activities. B. velezensis DTU001 grown on agar plates was extracted by ethyl acetate, concentrated and re-dissolved in methanol (1 mg/mL). DMSO was used as a negative control. Initial screening of the bacterial extract was done by standard agar disc diffusion method [26] and the zone of inhibition was noted. The 16 pathogenic filamentous fungi were grown on sterile MEAOX plates and the 4 yeasts were cultivated on PDA at 25 °C. 5-days old fungal plates and 2-days old yeast plates were taken for further investigation. Spore suspensions harvested in tween 20 from MEAOX medium plates with a final cell concentration of 5 × 105 cfu/mL of each microbial culture (fungi and yeast) were maintained for the disc diffusion assay. Plugs were harvested from 48 h grown B. velezensis DTU001 from MEAOX plates and extracted in isopropanol-ethyl acetate-1% formic acid followed by 1 h ultra-sonication. The supernatant was collected and dried utilizing nitrogen evaporator. The test sample (B. velezensis extract) and the standard compounds (iturin and fengycin) were dissolved in 0.5% DMSO to a concentration of 1 mg/mL each. Sterile Muller Hinton Agar plates were inoculated with the test organisms using lawn culture method. Discs of 6 mm were cut at the centre of the sterile plates using a sterile cork borer. The discs were loaded with 50 μl of each test sample. The plates were maintained at 25 ± 2 °C for 5 days and observed for clear zone of inhibition. Iturin and fengycin standards were maintained as positive control. Blank assays in DMSO were applied to avoid any possible effect of the solvent. Each experiment was conducted in triplicates for further confirmation and reproducibility of results.
2.8. Anti-Candida effects under planktonic and biofilm conditions
The impact of B. velezensis DTU001 on C. albicans SC5314 reference strain was assayed. C. albicans and B. velezensis were subcultured on Sabouraud dextrose agar and lysogeny broth (LB) agar, respectively. The growth of the individual strains in both monoculture and coculture was evaluated in order to examine the effect of B. velezensis on C. albicans planktonic growth. The final cell concentrations were adjusted to 2 × 105 cells/ml in RPMI-1640 broth + LB broth (50:50 v/v %) both for C. albicans and B. velezensis. Test tubes were incubated with agitation in darkness at 35 °C. 100 μl culture was removed at 0, 2, 4, 6, 8, 12 and 24 h, serially diluted tenfold in sterile physiological saline then plated onto either Sabouraud dextrose agar or LB agar. Plates were incubated for 24 and 48 h at 35 °C for B. velezensis and C. albicans, respectively. Results are representative of three independent experiments and are expressed as mean ± SD (error bars), which was presented using GraphPad Prism 6.05.
For the biofilm assays, polystyrene flat-bottom 96-well microtiter plates (TPP, Trasadingen, Switzerland) were used for the formation of single and mixed species biofilms of C. albicans and B. velezensis. A volume of 200 μl of cell suspension (1 × 106 cells/mL in RPMI-1640 broth + LB broth (50:50 v/v%) for C. albicans and 1 × 106 cells/mL in RPMI-1640 broth + LB broth (50:50 v/v%) for B. velezensis) was pipetted to each well for single biofilms, while 100 μL from each suspension (2 × 106 cells/mL for C. albicans with 2 × 106 cells/ml for B. velezensis) was added for mixed species biofilms to wells assigned to endpoints 2, 4, 6, 8, 12 and 24 h. The plates were incubated in darkness with static conditions. After 2, 4, 6, 8, 12 and 24 h of incubation, the corresponding wells were washed three times with sterile physiological saline then 200 μL sterile saline was added into wells, scraped off the adhered cells from surfaces; afterwards, aliquots of 100 μL were removed. Cells were released by mild sonication and vortexing then diluted serially ten-fold and plated onto either Sabouraud dextrose agar or LB agar. Plates were incubated for 24 and 48 h at 35 °C for B. velezensis and C. albicans, respectively. Results are representative of three independent experiments and are expressed as mean ± SD (error bars), which was presented using GraphPad Prism 6.05.
2.9. Bacterial biofilm formation
Bacterial strains (B. velezensis DTU001, B. subtilis NCBI 3610 [28], B. subtilis ATCC6633 (ATCC collection), B. subtilis PS216 [27], B. velezensis FZB42 (obtained from Bacillus Genetic Stock Centre), and B. methylotrophicus NCCB100236 (obtained from Bacillus Genetic Stock Centre)) were inoculated in lysogeny broth (Lenox LB, Carl Roth, Germany) and grown overnight for 16 h at 37 °C at 225 rpm. For pellicle development 20 μl was inoculated in 2 mL MSgg medium and placed in a 24-well plate, while 5 μL spotted on MSgg medium solified with 1,5% agar that was prepared as described previously [8,28]. Both pellicle and colony biofilms were incubated at incubated at 30 °C static conditions and imaged after 3 days.
2.10. Secondary metabolites identification
A UHPLC–DAD–QTOF method was set up for screening, with typical injection volumes of 0.1–2 μL extract. Separation was performed on a Dionex Ultimate 3000 UHPLC system (Thermo Scientific, Dionex, Sunnyvale, California, USA) equipped with a 100 × 2.1 mm, 2.6 μm, Kinetex C18 column, held at a temperature of 40 °C, and using a linear gradient system composed of A: 20 mmol L−1 formic acid in water, and B: 20 mmol L−1 formic acid in acetonitrile. The flow was 0.4 mL min−1, 90% A graduating to 100% B in 10 min, 100% B 10–13 min, and 90% A 13.1–15 min.
Time-of-flight detection was performed using a maXis 3G QTOF orthogonal mass spectrometer (Bruker Daltonics, Bremen, Germany) operated at a resolving power of ∼50000 full width at half maximum (FWHM). The instrument was equipped with an orthogonal electrospray ionization source, and mass spectra were recorded in the range m/z 100–1500 as centroid spectra, with five scans per second. For calibration, 1 μL 10 mmol L−1 sodium formate was injected at the beginning of each chromatographic run, using the divert valve (0.3–0.4 min). Data files were calibrated post-run on the average spectrum from this time segment, using the Bruker HPC (high-precision calibration) algorithm.
For ESI+ the capillary voltage was maintained at 4200 V, the gas flow to the nebulizer was set to 2.4 bar, the drying temperature was 220 °C, and the drying gas flow was 12.0 L min−1. Transfer optics (ion-funnel energies, quadrupole energy) were tuned on HT-2 toxin to minimize fragmentation. For ESI− the settings were the same, except that the capillary voltage was maintained at −2500 V. Unless otherwise stated, ion-cooler settings were: transfer time 50 μs, radio frequency (RF) 55 V peak-to-peak (Vpp), and pre-pulse storage time 5 μs.
3. Results and discussion
3.1. The genome of the indoor airborne isolate B. velezensis DTU001 encodes numerous secondary metabolites
B. velezensis DTU001 was isolated indoor at the DTU campus in Denmark and selected for detailed analysis based on its potential antimicrobial activity. The complete genome sequence of B. velezensis DTU001 consisted of a circular 3.927.210 bp chromosome with a GC content of 46.6%. Its 16S rRNA gene sequence exhibited 99% similarity to B. velezensis and B. amyloliquefaciens in genome database.
The phylogenetic affiliation of DTU001 isolate was further investigated using digital DNA:DNA-hybridization (dDDH) similarities and differences in genomic G + C content. (https://ggdc.dsmz.de/). These distances are transformed to values analogous to DDH using a generalized linear model (GLM) inferred from an empirical reference dataset comprising real DDH values and genome sequences. Compared to B. velezensis FZB42 (NC-009725.1), DDH value of 80.3%, difference in % G + C: 0.10. Meanwhile, there is a DDH analogous value of B. amyloliquefaciens (FN597644.1) 55.4%, difference in % G + C: 0.51. This result clearly supports that DTU001 is closer related to B. velezensis, where the DDH value exceeded the threshold of 70%.
A phylogenetic analysis with automlst (http://automlst.ziemertlab.com) was carried out, which automatically extracts 80 conserved genes, concatenates them, computes a multiple sequence alignment and builds a phylogenetic tree. The sequence alignment was manually checked, and a maximum-likelihood tree refined with jModelTest2 [30] and PhyML [21]. According to the MLST analysis, B. velezensis DTU1 forms a clade with other Bacilli that were recently proposed to be assigned the new species Bacillus lentimorphus [29].
To determine the genetic potential of B. velezensis DTU001 as biocontrol strain, the complete genome was surveyed for biosynthetic gene clusters that identified nine gene clusters involved in nonribosomal synthesis of lipopeptides, polyketides, and siderophore bacillibactin. About 15.4% of the whole DTU001 genome was devoted to secondary metabolites synthesis of nonribosomal synthesis of antimicrobial compounds and siderophores. In addition, genes for synthesis of the highly bioactive extracellular alkaline guanyl-preferring ribonuclease, previously detected in B. amyloliquefaciens [30] was also identified in the genome of DTU001.
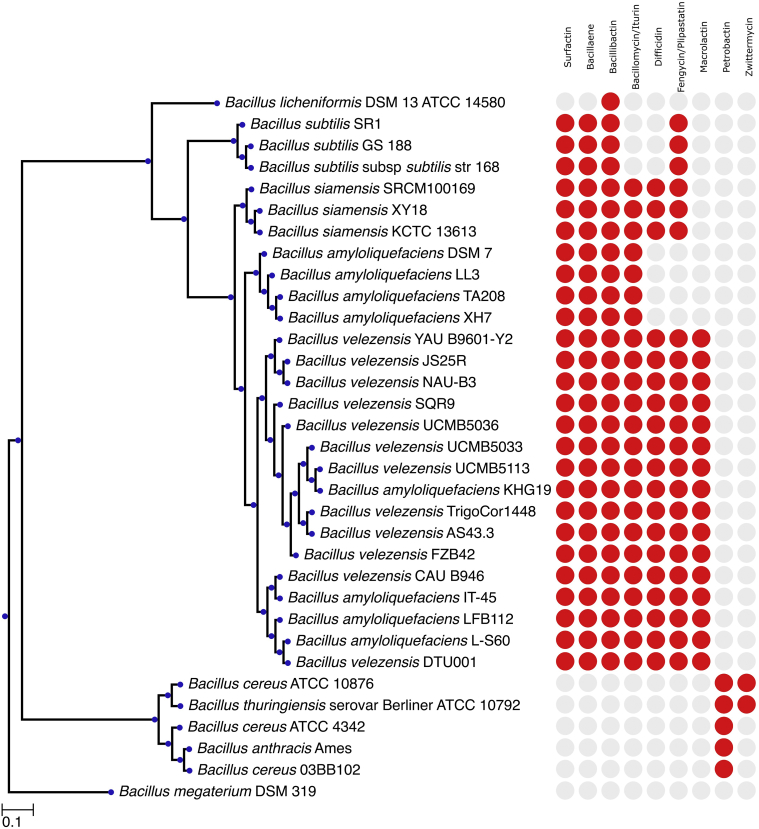
In order to compare the biosynthetic potentials between different Bacilli, B. velezensis together with other 17 species were mined for the genetic potential of secondary metabolite production using antiSMASH and BiG-SCAPE. All Bacilli related to B. velezensis DTU001 have the genetic potential to utilize bacillibactin as a siderophore to acquire irons from the environments (Fig. 1). All selected B. amyloliquefaciens, B. velezensis, and B. subtilis strains showed high similarities in BGC potentials and are considered as talented strains producing various lipopetides or polyketides. More distantly related Bacillus strains, such as members of the Bacillus cereus group showed a different secondary metabolite production genotype. All of these strains (B. cereus, B. anthracis, B. thuringiensis) seem to be unable of producing lipopeptides of the surfactin, iturin or fengycin families.
Fig. 1.
Evolutionary relationships of taxa between B. velezensis DTU001 and other Bacilli. A maximum-likelihood phylogenetic tree based on 80 conserved genes of selected strains of Bacilli is shown; B. megaterium DSM 319 was selected as outgroup. The presence/absence of typical Bacillus BGCs is depicted on the right.
3.2. Antifungal characterization of B. velezensis
Iturins are well-known secondary metabolites important for biocontrol against plant pathogens such as Xanthomonas campestris subsp. Cucurbitae, Pectobacterium carotovorum subsp. Carotovorum, Rhizoctonia solani, Fusarium graminearum etc. [31,32]. However, the impact of iturins against medically-relevant fungal pathogens is less explored. Therefore, we tested the efficacy and target specificity of DTU001 against selected important plant and human pathogenic fungi. In addition, we compared the effectiveness of the DTU001 crude extract and the purified lipopeptides, iturin and fengycin. We have selected 17 important plant and human pathogenic filamentous fungi (Table 1) ranging from the genera Penicillium, Aspergillus, Talaromyces, Candida to Cryptococcus. Besides those notorious plant pathogens such as Aspergillus uvarum infecting grapes, citrus rotting Penicillium ulaiense and post-harvest pathogen Penicillium expansum infecting apples, several important human pathogens were tested, such as Aspergillus niger and Candida.
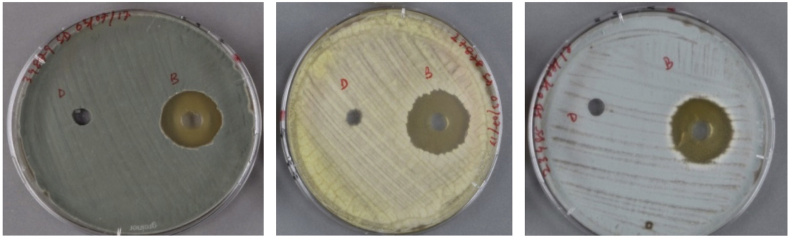
The organic extracts of B. velezensis DTU001 displayed strong antifungal effects against A. uvarum, P. ulaiense and P. expansum (Table 1 and Fig. 2). Due to the susceptibility to infection of mature and overripe fruit, post-harvest treatment of fruit with fungicides has traditionally been the most common method of combating P. expansum. Also, bio-fungicides using active ingredients such as bacteria and yeast have been successful in preventing infection but are ineffective against existing infections [33]. Considering the size of the apple product industry and the large number of people that may come into contact with infected fruits, control of the P. expansum is vitally important. Interestingly both Talaromyces atroroseus and Talaromyces amestolkiae were inhibited by both fengycin (20 mm) and iturin (29, 26 mm) as well as the crude extract (Table 1 and Fig. 2). All the tested organisms showed less or no susceptibility towards fengycin.
Fig. 2.
Extracts of B. velezensis DTU001 against fungi showing clear inhibition zones. Images left to right: P. frequentans, A. niger and T. amestolkiae. DMSO control loaded to the left hole, while the crude extract was applied to the right hole demonstrating the specific inhibition zone against the fungi.
In summary, the agar diffusion test showed that iturin and fengycin displayed selective anti-fungal activities against certain fungal pathogens, which means that some strains are insensitive to the addition of a single purified compound. For example, neither iturin nor fengycin displayed activities against Penicillium sclerotigenum and Aspergillus calidoustus. Overall, iturin displayed stronger and broader activities compared to fengycin. Compared to iturin and fengycin standards, extracts of B. velezensis DTU001 exhibited stronger inhibition activities and displayed a broader spectrum against test fungi including P. sclerotigenum and A. calidoustus. One explanation might be that B. velezensis produced a cocktail mixture which functions in a synergistic and more effective manner or additional secondary metabolite plays a role in the inhibition. However, the combined impact of the lipopeptides could avoid fast resistance evolution and help the bacterium in the environmental survival. It seemed that Aspergillus felis harbored certain mechanisms to avoid anti-fungal effects from B. velezensis DTU001 as neither lipopeptide standards nor the bacterial crude extract displayed inhibition activities against this fungus.
3.3. B. velezensis DTU001 is able to weaken biofilms of C. albicans
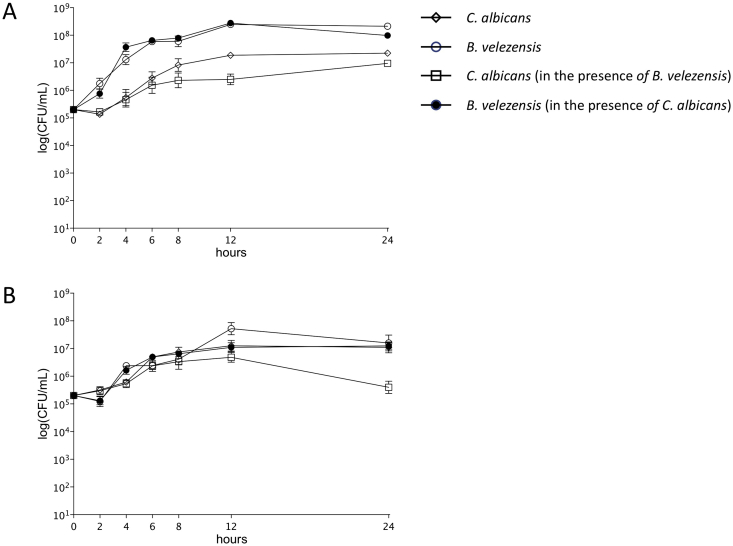
As B. velezensis DTU001 showed broad anti-fungal activity spectrum, we further evaluated if the bacterial isolate can directly reduce the growth of the human pathogen fungus, C. albicans under planktonic conditions and during biofilm development. First, the growth of the fungus and bacterium was monitored in planktonic co-cultures and compared to the mono-cultures (Fig. 3). Importantly, the B. velezensis cell count in the coculture with C. albicans did not differ significantly compared to the monoculture. In contrast, C. albicans planktonic cells between 6 and 24 h diminished in the presence of B. velezensis DTU001, therefore the bacterial isolate was able to inhibit the proliferation of the fungus in the co-cultures. Similarly, B. velezensis DTU001 was able to reduce the biofilms of C. albicans when co-cultured. The inhibitory effect was even higher that under planktonic conditions after 24 h (2-log decrease in fungal cell count). These experiments further support the applicability of B. velezensis DTU001 as biocontrol agent even in the presence of the target organism.
Fig. 3.
Planktonic (A) and biofilm (B) cultivation of C. albicans SC5314 and B. velezensis DTU001. Colony forming units were recorded at the indicated time points for C. albicans (open rhombus indicates C. albicans alone, open square denotes C. albicans in the presence of B. velezensis) and B. velezensis (open circle indicates B. velezensis alone, filled circle denotes B. velezensis in the presence of C. albicans).
3.4. B. velezensis DTU001 is a potent biofilm former
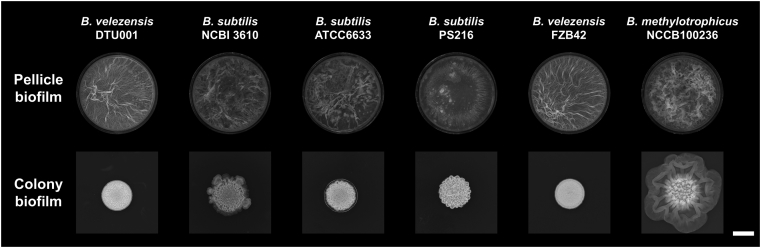
The probiotic effects of Bacilli depend on the ability to form biofilm and production of a matrix, that acts as an immune system stimulator [34], protects from environmental stress [35], and is required for plant colonization [36,37]. Therefore, we have compared the biofilm formation of B. velezensis DTU001 to other Bacillus species known for biofilm formation. Both architecturally complex colonies on agar medium and air-liquid localized pellicles were tested. The biofilm development can be qualitatively monitored using these models: the more complex structures and enhanced wrinkles produced, the increased the biofilm development of Bacilli [28,37]. In addition, pellicle formation strongly depends on the matrix components, without which cell are unable to float on the air-medium interface [8]. Both the architecturally complex biofilm colonies and the pellicles showed robust biofilm formation (i.e. wrinkled pellicles and colonies) by B. velezensis DTU001 that was comparable to other closely related isolates from the B. subtilis species complex (Fig. 4).
Fig. 4.
Pellicle (above) and colony biofilms (below) of selected strains from the B. subtilis species complex. The ability of biofilm formation is indicated by the development of wrinkles on the top of the pellicles and colonies. Scale bar indicates 5 mm.
3.5. Secondary metabolites characterization of B. velezensis DTU001
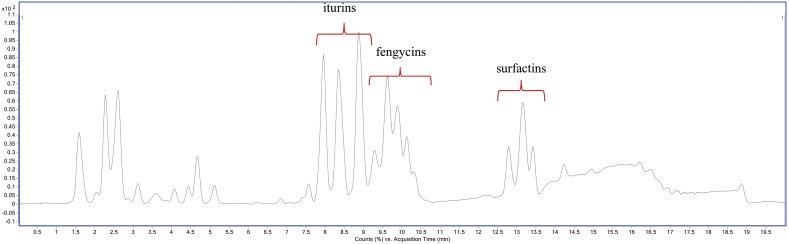
Based on the genome sequences and antifungal activity, B. velezensis DTU001 was predicted to produce various lipopeptides. To specifically demonstrate these secondary metabolites, LC-HRMS analysis was performed on ethyl acetate extracts harvested from agar grown B. velezensis DTU001. Indeed, chemical analysis along with purified standards detected three groups of bioactive lipopeptides: iturins, fengycins, and surfactins (Fig. 5, Figs. S1, S2, S3 and S4 in the supplemental data).
Fig. 5.
Base peak chromatogram showing the presence of iturin, fengycin and surfactin produced by B. velezensis DTU001.
4. Conclusions
Through genomics and metabolomics investigation, we demonstrated that B. velezensis DTU001 harbored the capacity to produce bioactive lipopeptides, comparable to the commercially utilized B. amyloliquefaciens and B. velezensis strains. Production of bioactive iturin, fengycin and surfactin by this strain was confirmed by HPLC-HRMS. Phylogenetic distance between DTU001 and those commercially utilized B. amyloliquefaciens together with their shared chemistry makes DTU001 as an attractive biocontrol option. Additionally, our targeted comparative genomics study revealed the versatility of secondary metabolite production by the selected Bacilli.
Either iturin or fengycin were previously proposed to be responsible for the antifungal activities of isolates within the B. subtilis species complex, however, we propose here that a cocktail anti-fungal agent derived from the crude extract of B. velezensis DTU001 functions more efficiently than single components. In addition, it will be interesting to examine whether purified surfactin also impacts the inhibitory potential of DTU001. In summary, this newly described isolate can be utilized as a biocontrol agent with a broad antifungal spectrum and provides a sustainable alternative for replacement of hazardous chemicals.
Declaration of interests
None.
Acknowledgements
This work was supported by the Danish National Research Foundation (DNRF137) for the Center for Microbial Secondary Metabolites (CeMiSt). SD was supported by a Novozymes and Henning Holck Larsen fellowship during her stay at DTU. TW was supported by a grant from Novo Nordisk Foundation (NNF10CC1016517).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2019.08.002.
Contributor Information
Ákos T. Kovács, Email: atkovacs@dtu.dk.
Ling Ding, Email: lidi@dtu.dk.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.De Lucca A.J., Walsh T.J. Antifungal peptides: novel therapeutic compounds against emerging pathogens. Antimicrob Agents Chemother. 1999;43:1–11. doi: 10.1128/aac.43.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ongena M., Jacques P. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 2008;16:115–125. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Rahman S.F.S.A., Singh E., Pieterse C.M.J., Schenk P.M. Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 2018;2:102–111. doi: 10.1016/j.plantsci.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Chae G.P., Shoda M., Kubota H. Suppressive effect of Bacillus subtilis and its products on phytopathogenic microorganisms. J Ferment Bioeng. 1990;69:1–7. [Google Scholar]
- 5.Shafi J., Tian H., Ji M.S. Bacillus species as versatile weapons for plant pathogens: a review. Biotechnol Biotechnol Equip. 2017;31:446-459. [Google Scholar]
- 6.Denner W.G.T. Stockton Press; New York: 1996. The legislative aspects of the use of industrial enzymes in the manufacture of food and food ingredients. Industrial enzymology; p. 6. [Google Scholar]
- 7.Grubbs K.J., Bleich R.M., Santa Maria K.C., Allen S.E., Farag S., AgBiome T., Shank E.A., Bowers A.A. Large-scale bioinformatics analysis of Bacillus genomes uncovers conserved roles of natural products in bacterial physiology. mSystems. 2017;2 doi: 10.1128/mSystems.00040-17. e00040-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan B., Blom J., Klenk H.P., Borriss R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an "operational group B. amyloliquefaciens" within the B. subtilis species complex. Front Microbiol. 2017;8:22. doi: 10.3389/fmicb.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan B., Wang C., Ding X., Zhu B., Song X., Borriss R. AmyloWiki: an integrated database for Bacillus velezensis FZB42, the model strain for plant growth-promoting Bacilli. Database. 2019;1:baz071. doi: 10.1093/database/baz071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verschuere L., Rombaut G., Sorgeloos P., Verstraete W. Probiotic bacteria as biological control agents in aquaculture. Microbiol Mol Biol Rev. 2000;64:655–671. doi: 10.1128/mmbr.64.4.655-671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanafani Z.A., Corey G.R. Daptomycin: a rapidly bactericidal lipopeptide for the treatment of Gram-positive infections. Expert Rev Anti Infect Ther. 2007;5:177–184. doi: 10.1586/14787210.5.2.177. [DOI] [PubMed] [Google Scholar]
- 12.Meena K.R., Kanwar K.S. Lipopeptides as the antifungal and antibacterial agents: applications in food safety and therapeutics. BioMed Res Int. 2015:473050. doi: 10.1155/2015/473050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benoit I., van den Esker M.H., Patyshakuliyeva A., Mattern D.J., Blei F., Zhou M., Dijksterhuis J., Brakhage A.A., Kuipers O.P., de Vries R.P., Kovács Á.T. Bacillus subtilis attachment to Aspergillus niger hyphae results in mutually altered metabolism. Environ Microbiol. 2015;17:2099–2113. doi: 10.1111/1462-2920.12564. [DOI] [PubMed] [Google Scholar]
- 14.Grau R.R., de Ona P., Kunert M., Lenini C., Gallegos-Monterrosa R., Mhatre E., Vileta D., Donato V., Holscher T., Boland W., Kuipers O.P., Kovács Á.T. A duo of potassium-responsive histidine kinases govern the multicellular destiny of Bacillus subtilis. mBio. 2015;6 doi: 10.1128/mBio.00581-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kearns D.B. A field guide to bacterial swarming motility. Nat Rev Microbiol. 2010;8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hölscher T., Kovács Á.T. Sliding on the surface: bacterial spreading without an active motor. Environ Microbiol. 2017;19:2537–2545. doi: 10.1111/1462-2920.13741. [DOI] [PubMed] [Google Scholar]
- 17.Haft D.H., DiCuccio M., Badretdin A., Brover V., Chetvernin V., O'Neill K., Li W., Chitsaz F., Derbyshire M.K., Gonzales N.R., Gwadz M., Lu F., Marchler G.H., Song J.S., Thanki N., Yamashita R.A., Zheng C., Thibaud-Nissen F., Geer L.Y., Marchler-Bauer A., Pruitt K.D. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018;46:D851–D860. doi: 10.1093/nar/gkx1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meier-Kolthoff J.P., Auch A.F., Klenk H.P., Goker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meier-Kolthoff J.P., Hahnke R.L., Petersen J., Scheuner C., Michael V., Fiebig A., Rohde C., Rohde M., Fartmann B., Goodwin L.A., Chertkov O., Reddy T., Pati A., Ivanova N.N., Markowitz V., Kyrpides N.C., Woyke T., Goker M., Klenk H.P. Complete genome sequence of DSM 30083(T), the type strain (U5/41(T)) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand Genomic Sci. 2014;9:2. doi: 10.1186/1944-3277-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 22.Huson D.H., Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 23.Huerta-Cepas J., Serra F., Bork P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber T., Blin K., Duddela S., Krug D., Kim H.U., Bruccoleri R., Lee S.Y., Fischbach M.A., Muller R., Wohlleben W., Breitling R., Takano E., Medema M.H. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Navarro-Muñoz J., Selem-Mojica N., Mullowney M., Kautsar S., Tryon J., Parkinson E., De Los Santos E., Yeong M., Cruz-Morales P., Abubucker S., Roeters A., Lokhorst W., Fernandez-Guerra A., Teresa Dias Cappelini L., Thomson R., Metcalf W., Kelleher N., Barona-Gomez F., Medema M.H. A computational framework for systematic exploration of biosynthetic diversity from large-scale genomic data. bioRxiv. 2018:445270. doi: 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bauer A.W., Kirby W.M., Sherris J.C., Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- 27.Stefanic P., Mandic-Mulec I. Social interactions and distribution of Bacillus subtilis pherotypes at microscale. J Bacteriol. 2009;191:1756–1764. doi: 10.1128/JB.01290-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Branda S.S., Gonzalez-Pastor J.E., Ben-Yehuda S., Losick R., Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parks D.H., Chuvochina M., Waite D.W., Rinke C., Skarshewski A., Chaumeil P.A., Hugenholtz P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 30.Hartley R.W., Rogersen D.L., Jr., Smeaton J.R. Production and purification of the extracellular ribonuclease of Bacillus amyloliquefaciens (barnase) and its intracellular inhibitor (barstar). II. Barstar. Prep Biochem. 1972;2:243–250. doi: 10.1080/00327487208061474. [DOI] [PubMed] [Google Scholar]
- 31.Gong A.D., Li H.P., Yuan Q.S., Song X.S., Yao W., He W.J., Zhang J.B., Liao Y.C. Antagonistic mechanism of iturin A and plipastatin A from Bacillus amyloliquefaciens S76-3 from wheat spikes against Fusarium graminearum. PLoS One. 2015;10 doi: 10.1371/journal.pone.0116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeriouh H., Romero D., Garcia-Gutierrez L., Cazorla F.M., de Vicente A., Perez-Garcia A. The iturin-like lipopeptides are essential components in the biological control arsenal of Bacillus subtilis against bacterial diseases of cucurbits. Mol Plant Microbe Interact. 2011;24:1540–1552. doi: 10.1094/MPMI-06-11-0162. [DOI] [PubMed] [Google Scholar]
- 33.Konstantinou S., Karaoglanidis G.S.K., Bardas G.A. Postharvest fruit rots of apple in Greece: pathogen incidence and relationships between fruit quality parameters, cultivar susceptibility, and patulin production. Plant Dis. 2011;95:7. doi: 10.1094/PDIS-11-10-0856. [DOI] [PubMed] [Google Scholar]
- 34.Jones S.E., Paynich M.L., Kearns D.B., Knight K.L. Protection from intestinal inflammation by bacterial exopolysaccharides. J Immunol. 2014;192:4813–4820. doi: 10.4049/jimmunol.1303369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovács Á.T., van Gestel J., Kuipers O.P. The protective layer of biofilm: a repellent function for a new class of amphiphilic proteins. Mol Microbiol. 2012;85:8–11. doi: 10.1111/j.1365-2958.2012.08101.x. [DOI] [PubMed] [Google Scholar]
- 36.Beauregard P.B., Chai Y., Vlamakis H., Losick R., Kolter R. Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci. 2013;110:E1621–E1630. doi: 10.1073/pnas.1218984110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallegos-Monterrosa R., Mhatre E., Kovács Á.T. Specific Bacillus subtilis 168 variants form biofilms on nutrient-rich medium. An Microbiol. 2016;162:1922–1932. doi: 10.1099/mic.0.000371. [DOI] [PubMed] [Google Scholar]
- 38.Alanjary M., Steinke K., Ziemert N. AutoMLST: an automated web server for generating multi-locus species trees highlighting natural product potential. Nucleic Acids Res. 2019;47:W276–W282. doi: 10.1093/nar/gkz282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.