Abstract
Protein synthesis and its regulation are central to all known forms of life and impinge on biological arenas as varied as agriculture, biotechnology, and medicine. Otherwise known as translation and translational control, these processes have been investigated with increasing intensity since the middle of the 20th century, and in increasing depth with advances in molecular and cell biology. We review the origins of the field, focusing on the underlying concepts and early studies of the cellular machinery and mechanisms involved. We highlight key discoveries and events on a timeline, consider areas where current research has engendered new ideas, and conclude with some speculation on future directions for the field.
Proteins account for the largest fraction of the macromolecules in a cell, are important components of the extracellular milieu, and fulfill multiple roles—enzymatic, structural, transport, regulatory, and other—in all organisms. Their synthesis, through the translation of genetic information encoded in messenger RNA (mRNA), requires extensive biological machinery and demands delicate and sophisticated regulation (Hershey et al. 2018). Protein synthesis is modulated quantitatively, and in time and space, through a network of stimuli, responses, and interactions collectively referred to as translational control. A large proportion of the resources of cells and organisms is devoted to translation and translational control, as discussed previously in terms of genetics, bioenergetics, and cell biology (Mathews et al. 2007).
Here we summarize the beginnings of the field and outline the pathway that led to our current understanding of the processes of protein synthesis and translational control, placing landmark discoveries on a timeline (Fig. 1). The field is, and always has been, a broad one. It originated in studies of topics ranging from virus infection to embryology and development, and has grown to encompass learning, memory, and genetic disease (Tahmasebi et al. 2018), as well as therapeutic intervention, among other biomedical areas. It continues to diversify and develop with the advent of approaches of increasing depth and precision. At the same time, it continues to generate fresh concepts and present challenges to well-accepted paradigms.
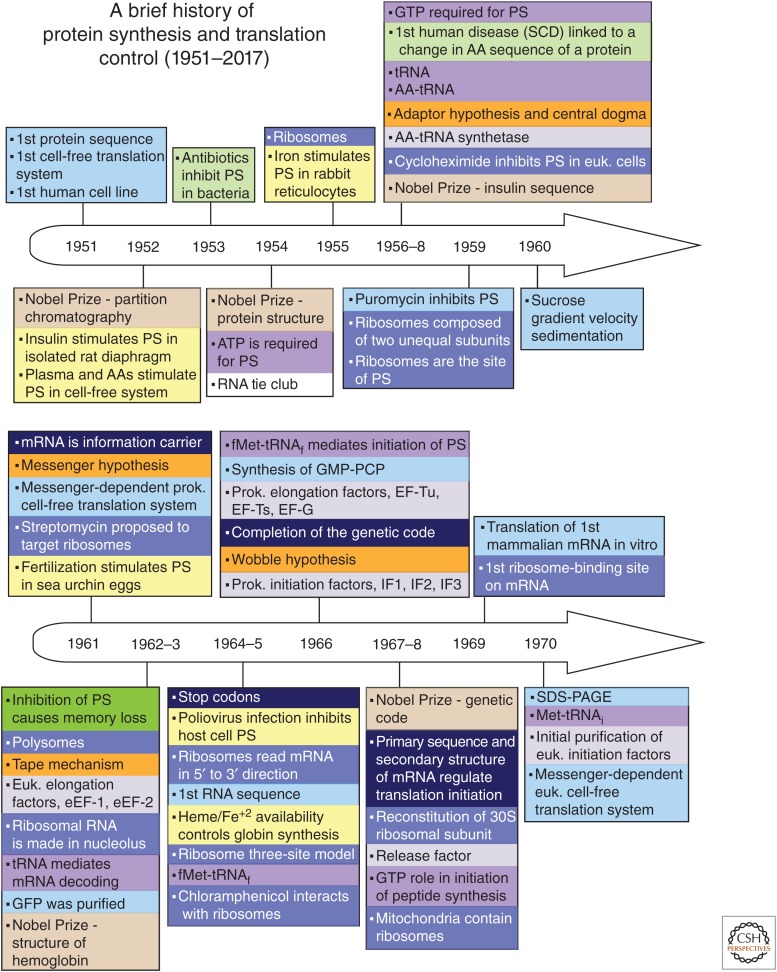
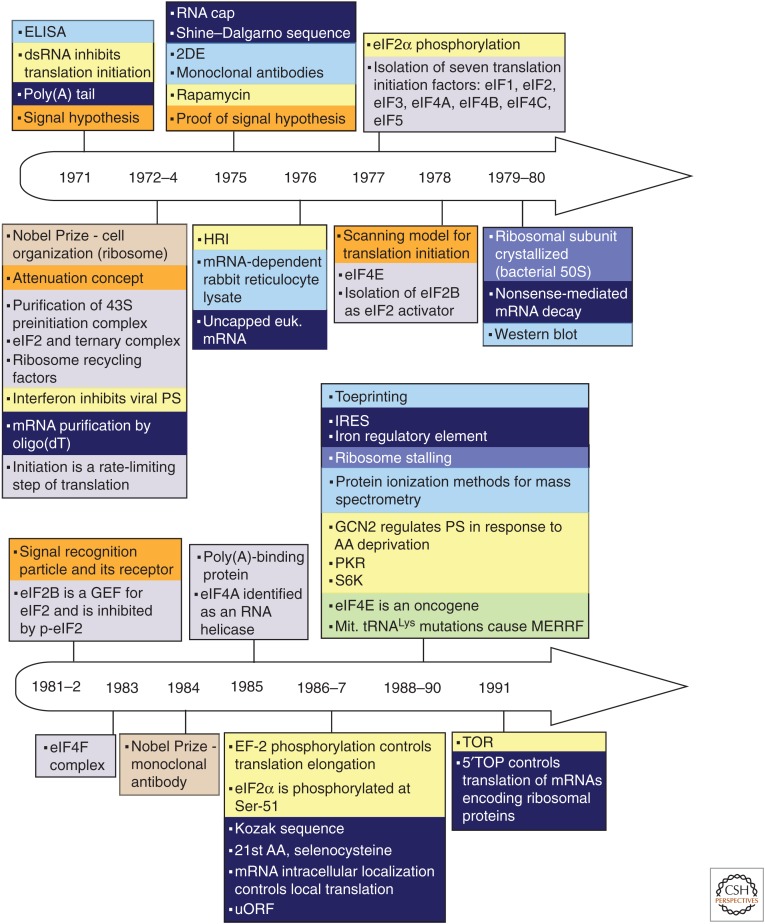
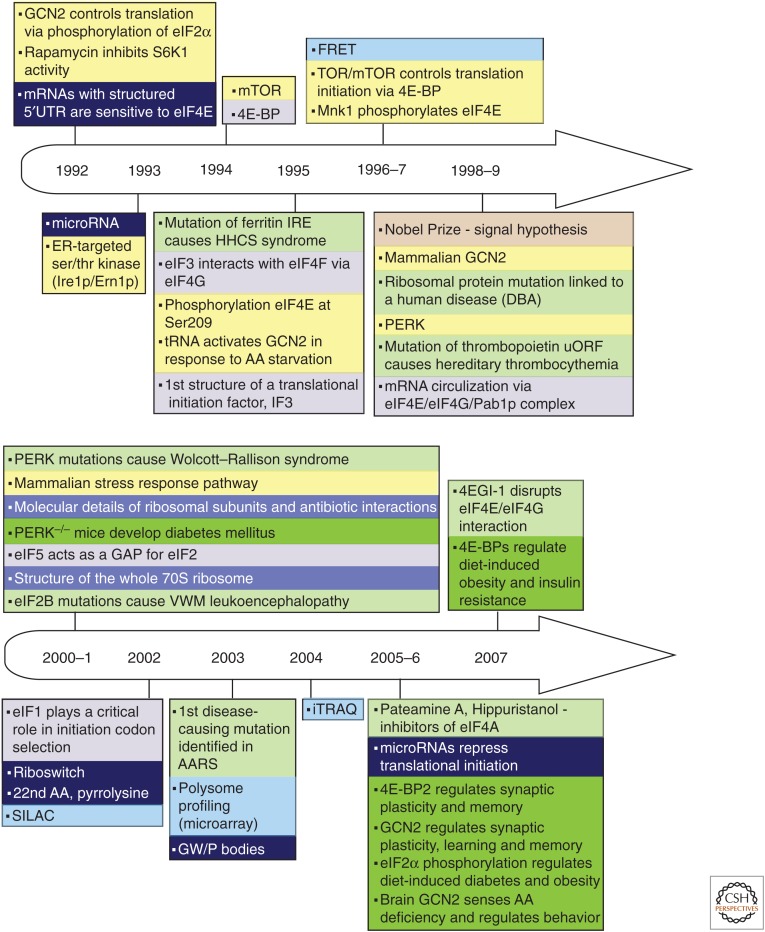
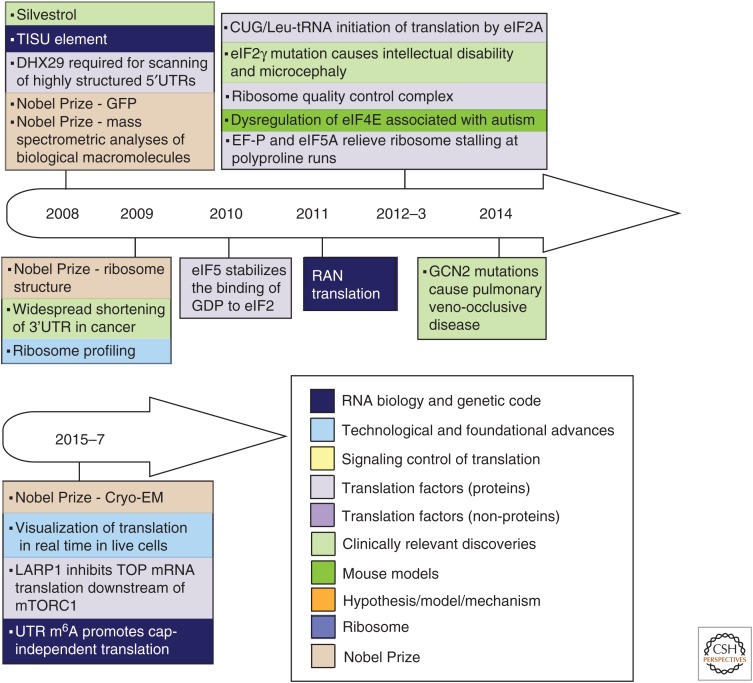
Figure 1.
Timeline of discoveries in the fields of protein synthesis (PS) and translational control (1951–2017). Principal advances are shown according to the year of publication, and are color coded by topic (color key and abbreviation definitions are at the end of the figure). Some other relevant events are also noted.
TRANSLATION TIMELINE
Before the early 1950s, most protein synthesis research addressed physiological questions and the findings were largely descriptive in nature (e.g., Daly and Mirsky 1952). Theories of protein synthesis via enzyme assembly and peptide intermediates were entertained along with template theories (Campbell and Work 1953), and no component of the translation system was known (ribosomes included [Palade 1955]). In this era, studies of protein synthesis were performed on tissues or tissue slices or in whole animals.
During the decade of the 1950s, the field underwent a transformation with the development of cell-free systems, fundamental discoveries (e.g., of transfer RNA [tRNA] and ribosomes), and crucial technical advances (such as radiolabeled amino acids, sucrose gradient centrifugation, and inhibitors). Subsequent decades were dominated by themes, concepts, and discoveries that furthered the field in different ways. Foundational discoveries (of polysomes and mRNA, for example) continued to be made in the 1960s, together with the seminal elucidation of the genetic code. The 1970s saw rapid growth of the field, including the characterization of most of the components of the eukaryotic translation system and the beginning of mechanistic studies. Mechanistic and regulatory themes dominated the 1980s, and the appreciation of regulatory pathways expanded rapidly in the 1990s. All of these themes contributed to the current activity in the first decades of the 21st century, when much research sought—and still seeks—to explain physiological and pathophysiological responses of the translation system and to develop therapeutics for treatment of genetic and acquired diseases and infections.
Principal discoveries are recorded chronologically and thematically (distinguished by color coding) in Figure 1. The timeline illustrates the fact that in this field, as in others, research did not always progress in a systematic and orderly fashion. Some discoveries developed slowly or even lay fallow for many years; the roles of eukaryotic initiation factors (eIFs) afforded several examples of this. Other discoveries, such as the poly(A) tail of eukaryotic mRNA, immediately spawned far-reaching advances in multiple areas of mRNA characterization, isolation, metabolism, and translation. Many advances benefitted from studies in greater depth that were enabled by new techniques (e.g., gel electrophoresis, blotting, toeprinting, ribosome profiling) and technological developments in related fields (cloning, reverse genetics, X-ray crystallography). These drove research forward, providing fresh insights and mechanistic understanding of increasing clarity and detail. Striking examples came from structural investigations of ribosomes and components of the translation system with which they interact (Jobe et al. 2018). Concurrently, investigations sometimes in related fields of study, led to a steady stream of unforeseen observations, including unorthodox initiation mechanisms (Meyer et al. 2015; Zhou et al. 2015; Kwan and Thompson 2018; Zu et al. 2018) and novel concepts such as ribosomal heterogeneity (Sauert et al. 2015; Genuth and Barna 2018) and the ribosomal concentration model (Mills and Green 2017; Khajuria et al. 2018), which expanded the scope of the field.
DEVELOPMENT OF THE PROTEIN SYNTHESIS FIELD
Biochemical investigations of protein synthesis began when concepts that are now nearly axiomatic were still uncertain. The view of proteins as unique linear arrays of just 20 amino acid residues was about to be established with the publication of the first protein sequence (the insulin B chain [Sanger and Tuppy 1951]), and mechanisms of protein synthesis involving the reversal of proteolysis or phosphorylated intermediates were entertained (Zamecnik 1969). Radioactive isotopes had begun to revolutionize many areas of biomedical science in the late 1940s, and radiolabeled amino acids came into use as tracers around 1950. Researchers synthesized them from simple labeled compounds such as formaldehyde or sodium cyanide as a first step in their experiments (e.g., Borsook et al. 1952), until they became commercially available in the latter part of the decade. Enabled by this profound technical advance, investigations proliferated rapidly and biochemistry ran ahead of genetics until the advent of cloning and the systematic exploitation of the yeast system that began to make their mark in the 1980s.
Siekevitz and Zamecnik (1951) produced a cell-free preparation from rat liver that incorporated amino acids into protein and showed that energy was required in the form of ATP and GTP (Zamecnik and Keller 1954; Keller and Zamecnik 1956). The translation system was refined by stages and resolved into subfractions including a microsomal fraction that contained ribosomes attached to intracellular membrane fragments (Zamecnik 1960). Pulse-chase experiments showed that ribosomes are the site of protein synthesis, not an easy task in bacterial cells where protein synthesis is very rapid: the assembly of a protein chain on a ribosome was estimated to take only 5–10 seconds (McQuillen et al. 1959). It is salutary to recall that this was accomplished in advance of an understanding of the central role of RNA in the flow of genetic information to protein, before the visualization of polysomes, and well before the first RNA sequence was completed (Holley et al. 1965). Amid many concepts (Crick 1959), one idea posited that each ribosome is dedicated to the synthesis of a single protein, the “one gene–one ribosome–one protein” hypothesis. Early in the 1960s, however, polysomes were observed and their function appreciated in light of the messenger hypothesis and the “tape mechanism” of translation discussed below (Marks et al. 1962; Warner et al. 1962, 1963; Arlinghaus and Schweet 1963; Gierer 1963; Goodman and Rich 1963; Nakamoto et al. 1963; Noll and Wettstein 1963; Wettstein et al. 1963). Technical advances in electron microscopy (EM) and high-speed centrifugation made vital contributions during this phase of the field’s development.
The role of aminoacyl-tRNA was established in the late 1950s. An intermediate, activated state of amino acids was first detected (Hultin and Beskow 1956), then characterized (Hoagland et al. 1958, 1959) and recognized as the physical manifestation of the adaptor RNA predicted on theoretical grounds (Crick 1958). Once its function had been realized, the name transfer RNA replaced the term “soluble” RNA (sRNA). Chemical modification of the amino acid moiety of cysteine-charged tRNACys (to alanine) confirmed that the RNA component is responsible for decoding the template (Chapeville et al. 1962). Thus, fidelity of information transfer from nucleic acid to protein rests in part on the aminoacyl-tRNA synthetases. One of these, the valine-specific enzyme from Escherichia coli, was arguably the first macromolecular component of the protein synthetic apparatus to be characterized (Berg and Ofengand 1958), and additional synthetases soon followed.
Numerous enzymes catalyzing and facilitating the several steps in protein synthesis were steadily purified over the years, with an intense burst of activity in the 1960s and 1970s. In advance of full authentication of their purity and function, these proteins were provisionally called “factors,” a term that has stuck. Although many of the factors have been known for almost half a century, the activities of some of them remained obscure or debatable until recently (e.g., EF-P and its homolog eIF5A) (Kang and Hershey 1994; Aoki et al. 1997; Doerfel et al. 2013; Ude et al. 2013), whereas others are still emerging (e.g., eIF2A and eIF2D) (Komar et al. 2005; Ventoso et al. 2006; Dmitriev et al. 2010; Starck et al. 2012; Kearse and Wilusz 2017), and new ones with specialized functions, such as the internal ribosome entry site (IRES) trans-acting factors (ITAFs), are being discovered (King et al. 2010; Lee et al. 2017).
The messenger RNA concept revolutionized thinking about gene expression in all cells. Genetics and bacteriophage biology, as well as biochemistry, played key parts in the genesis and confirmation of the messenger hypothesis (see, for example, Cobb 2015). Jacob and Monod (1961) hypothesized the existence of an unstable intermediate between the DNA of the gene and the ribosome, which could be related to the RNA produced in phage T2-infected cells (Volkin and Astrachan 1956; Nomura et al. 1960). On this view, the ribosome and other components of the protein synthesis machinery constitute a relatively stable decoding and synthetic apparatus that is programmed by unstable mRNA. This was soon confirmed in bacteria (Brenner et al. 1961; Gros et al. 1961) and bacterial cell-free systems. The discovery that poly(U) can direct the synthesis of polyphenylalanine in vitro (Nirenberg and Matthaei 1961) was a transformative event, spearheading the elucidation of the genetic code by the mid-1960s. The wobble hypothesis, which rationalizes features of the code’s redundancy and its decoding by tRNAs, was published by Crick (1966). In higher cells, the existence of a class of rapidly labeled RNA, heterogeneous in size and with distinct chromatographic properties, was recognized. Its essential feature as an informational intermediary were confirmed and messenger-dependent eukaryotic cell-free translation systems appeared at the end of the decade (Laycock and Hunt 1969; Lockard and Lingrel 1969; Mathews and Korner 1970).
Building on these foundations, mechanisms explaining important aspects of translation in both prokaryotes and eukaryotes emerged in the 1970s. The question of initiation site selection was largely accounted for in prokaryotes by the Shine–Dalgarno sequence base-pairing with 16S ribosomal RNA (Shine and Dalgarno 1975). A solution to the problem in eukaryotes came later, in the 1980s, with recognition of cap-dependent scanning and the Kozak consensus sequence (Kozak 1978, 1986, 1987; Kozak and Shatkin 1978), followed by identification of IRES-dependent mechanisms (Jang et al. 1988; Pelletier and Sonenberg 1988). The optimal codon hypothesis explained how a bias in the usage of synonymous codons can influence the production levels of individual genes (Ikemura 1981) and the signal hypothesis (discussed below) accounted for protein transport into the endoplasmic reticulum (ER). Investigations of mRNA translation and metabolism were greatly facilitated by the discovery in the early 1970s of the terminal hallmarks found on most eukaryotic mRNAs, 5′ caps (Adams and Cory 1975; Both et al. 1975; Furuichi and Miura 1975; Furuichi et al. 1975a,b; Perry and Kelley 1975; Wei and Moss 1975) and 3′ poly(A) tails (Darnell et al. 1971a,b; Edmonds et al. 1971; Lee et al. 1971).
By the end of the 1980s, as a result of the identification and purification of most of the components of the translation system and the reconstitution of their activities in vitro, the pathway of protein synthesis had been defined and was well understood in outline (Hershey et al. 2018). Subsequent detailed analyses led to an in-depth understanding of many of the mechanisms of initiation, elongation, and termination in bacteria (Rodnina 2018) and in eukaryotic cells, with indispensable contributions from yeast genetics and biochemistry (Dever et al. 2018; Hellen 2018; Merrick and Pavitt 2018). This has allowed questions of regulation to be addressed at ever-increasing levels of sophistication.
ORIGINS OF TRANSLATIONAL CONTROL
The idea of regulation at the transcriptional level flowed naturally from the messenger concept. Jacob and Monod (1961) wrote that “the synthesis of individual proteins may be provoked or suppressed within a cell, under the influence of specific external agents, and … the relative rates at which different proteins are synthesized may be profoundly altered, depending on external conditions.” They recognized that such regulation “is absolutely essential to the survival of the cell,” although the notion that it could be exerted at the translational level was not a principal focus in the bacterial field. Still, the seeds of the concept that gene expression can be regulated by the efficiency of protein synthesis emerged early, and some from work in bacterial systems.
Among the first observations of regulation at this level of gene expression were those made in rabbit reticulocytes (Borsook et al. 1952; Kruh and Borsook 1956). The term “translational control” itself was used in 1963 with respect to the differential expression of proteins from the RNA genome of MS2 phage in an E. coli cell-free translation system (Ohtaka and Spiegelman 1963). The concept spread rapidly into other areas of research, to the extent that less than 10 years later the presence of translationally silent mRNA that is activated on fertilization of sea urchin eggs was referred to as a “classical conclusion” (Humphreys 1971). After the early studies on phages, much of the focus was on eukaryotic systems. A virtue of translation as a site of regulation is that it affords a rapid response to external stimuli without invoking nuclear pathways for mRNA synthesis, processing, and transport. Correspondingly, the first cases recognized were mostly ones in which it was evident or simple to establish that transcription and other nuclear events were not responsible. To illustrate how the evidence for translational control arose, we briefly describe four paradigms, together with an early example of translational control at the level of elongation.
Sea Urchin Eggs
The eggs of sea urchins and other invertebrates synthesize protein at a very low rate but are triggered to incorporate amino acids within a few minutes of fertilization with little or no concomitant RNA synthesis (Hultin 1961; Nemer 1962; Gross et al. 1964). The first wave of increased translation, lasting several hours, is not blocked by inhibiting transcription (Gross et al. 1964) because the eggs contain preexisting mRNAs in a masked form that are not translated until fertilization. In principle, the limitation could be caused by a deficiency in the translational machinery, but there is little evidence to support this possibility (Humphreys 1969). For example, egg ribosomes can translate added poly(U) even though they display little intrinsic protein synthetic activity (Nemer 1962; Wilt and Hultin 1962). Deproteinized egg RNA can be translated in a cell-free system (Maggio et al. 1964; Monroy et al. 1965) and cytoplasmic messenger ribonucleoprotein (mRNP) particles were observed (Spirin and Nemer 1965). Because the assembly of masked mRNP complexes must take place during oogenesis, the sea urchin system exemplifies a reversible process of mRNA repression and activation. Current understanding of the diverse translational control processes operative during embryonic development and stem-cell differentiation in the adult are described by Teixeira and Lehmann (2018).
Mammalian Reticulocytes
It was taken for granted that protein synthesis (mainly hemoglobin) in mammalian reticulocytes, which are enucleate immature red blood cells, would be regulated at the translational level. In the intact rabbit reticulocyte, the synthesis of heme parallels that of globin (Kruh and Borsook 1956) and globin synthesis is controlled by the availability of heme or ferrous ions (Bruns and London 1965). Regulation by heme occurs in the reticulocyte lysate (Lamfrom and Knopf 1964), the forerunner of the messenger-dependent translation system of Pelham and Jackson (1976) and some coupled transcription-translation systems. When globin synthesis is inhibited in cells or extracts, the polysomes dissociate to monosomes (Hardesty et al. 1963; Waxman and Rabinowitz 1966), arguing that regulation impacts translation initiation. The effects of heme deprivation are mediated by the protein kinase HRI (heme-regulated inhibitor, EIF2AK1) and are mimicked by unrelated stimuli, including addition of glutathione disulfide (Kosower et al. 1971) or double-stranded RNA mediated by PKR (protein kinase R, EIF2AK2) (Ehrenfeld and Hunt 1971; Kosower et al. 1971). Regulation extends to all mRNAs in the reticulocyte lysate (Mathews et al. 1973), implying that a general mechanism of translational control is being invoked. This mechanism centers on the phosphorylation of the α subunit of eIF2, which results in reduced levels of ternary complex (eIF2•GTP•Met-tRNAi) and impaired loading of the 40S ribosomal subunit with Met-tRNAi (Farrell et al. 1977). Considerable attention has been given to the family of eIF2 kinases, which confer sensitivity to a wide range of stimuli. In addition to HRI and PKR, PERK (PKR-like ER kinase, EIF2AK3) and GCN2 (general control nonderepressible 2, EIF2AK4) are activated by ER stress and uncharged tRNA, respectively (Merrick and Pavitt 2018; Wek 2018). PKZ, a PKR-like eIF2α kinase in fish, is activated by Z-DNA and can also inhibit translation (Bergan et al. 2008; Liu et al. 2013; Taghavi and Samuel 2013).
Physiological Stimuli
Cells and tissues of higher organisms regulate the expression of individual genes or classes of genes at the translational level in response to a wide variety of stimuli and conditions. Examples include responses to hormones (Eboué-Bonis et al. 1963; Garren et al. 1964; Martin and Young 1965; Tomkins et al. 1965) and ions (Drysdale and Munro 1965); changes in cell state, such as mitosis (Steward et al. 1968; Hodge et al. 1969; Fan and Penman 1970) and differentiation (Heywood 1970); and stress resulting from heat shock (McCormick and Penman 1969), treatment with noxious substances, and the incorporation of amino acid analogs (Thomas and Mathews 1984). Although these findings strengthened the view that such control is widespread and important, proof that it was exerted at the translational level was sometimes challenging in nucleated cells, let alone in tissues and whole organisms. One approach to this issue took advantage of selective inhibitors of transcription or translation, such as actinomycin D and cycloheximide, but the results were liable to be complicated by indirect or side effects of the drugs in complex systems. The rapidity of a response could also provide suggestive evidence for an effect at the translational level. Compelling data often came from investigations of the underlying biochemical processes, for example, by demonstrating changes in polysome profiles or initiation factor phosphorylation states. Several methods can provide rigorous evidence (Hershey et al. 2018) and ribosome profiling is a powerful and increasingly popular modern approach (Ingolia et al. 2009, 2018).
Cell growth is dependent on protein synthesis and translational control mediated by the mammalian or mechanistic target of rapamycin (mTOR), a protein kinase that lies at the nexus of numerous regulatory pathways. In the early 1990s, genetic screening for rapamycin-resistant genes in budding yeast uncovered TOR as a major regulator of cell growth (Heitman et al. 1991). A few years later, it was established that protein synthesis is a major downstream target (Barbet et al. 1996), and that mTOR controls translation initiation through phosphorylation of eIF4E-binding proteins (4E-BPs) (Beretta et al. 1996). eIF4E is the mRNA cap-binding protein (Sonenberg et al. 1979) required for cap-dependent initiation, and its activity is prevented by dephosphorylated 4E-BP. mTOR phosphorylates 4E-BP, releasing eIF4E, and allowing cap-dependent translation. The control of several other translation factors and regulators is also linked to mTOR activity (Proud 2018). mRNAs that harbor a 5′-terminal oligopyrimidine (TOP) motif (Levy et al. 1991) were the first ones found to be translationally suppressed by rapamycin (Jefferies et al. 1994; Terada et al. 1994). The TOP mRNA class includes those encoding ribosomal proteins and elongation factors, consistent with the importance of mTOR in ribosome biogenesis, cell growth, and cancer (Proud 2018; Robichaud et al. 2018). The exact mechanism of TOP mRNA translation regulation remained elusive for many years, but in recent years the LARP1 protein has been shown to mediate this effect (Fonseca et al. 2015; Lahr et al. 2017).
Virus-Infected Cells
The small RNA phages, MS2 and its relatives, provided some of the first evidence for translational control, as well as the first clear case of a mechanism specific for the synthesis of an individual protein. The phage genome encodes four polypeptides (the maturation, coat, and lysis proteins and RNA replicase) that are initiated individually and are produced at dissimilar rates. Several regulatory interactions among them are now known. One was revealed by the observation that a nonsense mutation early in the cistron coding for phage coat protein down-regulates replicase synthesis (Lodish and Zinder 1966); passage of ribosomes through a critical region of the coat protein cistron melts the long-range RNA structure and allows replicase translation. A second nonsense mutation leads to overproduction of the replicase because the coat protein acts as a repressor of replicase translation, and the binding of phage coat protein to the hairpin structure containing the replicase AUG is a well-characterized RNA–protein interaction (Witherell et al. 1991). Subsequent studies have disclosed numerous translational control mechanisms in phages and in viruses infecting eukaryotes (Breaker 2018; Stern-Ginossar et al. 2018).
Cellular mRNAs are also subject to translational control during infection with many viruses (Stern-Ginossar et al. 2018). Inhibition of cellular mRNA translation, an aspect of host cell shutoff, may begin before the onset of viral protein synthesis and without any apparent interference with cellular mRNA production or stability. In poliovirus infection, the shutoff of host-cell translation can be complete within 2 hours after infection and is followed by a wave of viral protein synthesis (Summers et al. 1965). In the first phase, polysomes break down without any effect on translation elongation or termination (Penman and Summers 1965; Summers and Maizel 1967). In the second phase, virus-specific polysomes form (Penman et al. 1963). The cellular mRNA remains intact and translatable in vitro (Leibowitz and Penman 1971), evidence that initiation has become selective for viral mRNA. Translational inhibition extends to mRNAs produced by other viruses in a double infection (Ehrenfeld and Lund 1977), indicative of a general effect that later work ascribed to modification of the cap-binding complex, eIF4F. Cleavage of the eIF4G subunit of this complex prevents cap-dependent initiation on cellular mRNAs but does not interfere with initiation on the viral mRNA, which occurs by internal ribosome entry (Kwan and Thompson 2018). Viruses have evolved many such specialized mechanisms (Stern-Ginossar et al. 2018), some of which have led to the identification of parallel mechanisms in uninfected cells themselves.
Secretory Pathway
Protein synthesis is regulated predominantly at the level of initiation, consistent with the principle that it is more efficient to govern a pathway at its outset than to interrupt it midstream with the ensuing accumulation of intermediates and logjam of recyclable components. Nevertheless, well-characterized cases do occur later in the translational pathway, at the elongation and termination level (Dever et al. 2018). Proteins destined for secretion or retention in cell membranes are made on polysomes attached to the ER. In the early 1970s, it began to seem likely that ribosomes become associated with cell membranes only after protein synthesis has been initiated (Lisowska-Bernstein et al. 1970; Rosbash 1972) and what came to be called a signal peptide was found on secreted proteins (Milstein et al. 1972; Devilliers-Thiery et al. 1975). These findings lent substance to the signal hypothesis that proposed that an amino-terminal sequence might be responsible for secretion (Blobel and Sabatini 1971). The development of cell-free systems enabled biochemical dissection of the secretory pathway (Blobel and Dobberstein 1975) leading to the discovery of the signal recognition particle (SRP), a ribonucleoprotein, and its receptor on the ER (Walter and Blobel 1981; Walter et al. 1981; Gilmore et al. 1982a,b; Meyer et al. 1982). The SRP also interacts with the ribosome such that the binding of the SRP to a nascent signal peptide causes translational arrest that is relieved when the ribosome docks with its ER receptor. This mechanism ensures cotranslational protein export and prevents the accumulation of secretory proteins in an improper subcellular compartment (the cytosol). From another perspective, this mechanism also represents an example of mRNA localization achieved by controlling its translation, distinct from several other methods used by cells to compartmentalize translation (Buxbaum et al. 2015; Biswas et al. 2018).
WHAT OF THE FUTURE?
In the past seven decades, combined genetic, biochemical, cell biological, pharmacological and structural approaches have uncovered the major components involved in protein synthesis, their interactions, and many of the sophisticated regulatory processes that adjust protein synthesis to developmental and environmental demands. Technological improvements such as cryoelectron microscopy (cryo-EM), real-time single-molecule microscopy, DNA and RNA sequencing, mass spectrometry, and rapid kinetic analysis now provide the opportunity to interrogate translation on spatial and temporal scales in ways that were not possible before. This increased resolution promises to bring the field from studies of cells in bulk to organelle-specific and even single-mRNA levels in vivo. Together with the increasingly detailed understanding of the role of translation in physiology and disease pathogenesis (Tahmasebi et al. 2018), there is optimism that therapeutic relief from acquired and genetic diseases may be on the horizon.
Yet the immense complexity of the translation system continues to pose new challenges and many uncharted areas remain. Little is known, for example, about protein synthesis and translational control in archaea, or in chloroplasts and mitochondria, even though mitochondrial ribosomes were reported half a century ago (Kuntzel and Noll 1967). Novel mechanisms can be confidently predicted to be uncovered by study of these organisms and organelles, as well, perhaps, from the giant viruses of Acanthamoeba that encode components of the translation apparatus (Bekliz et al. 2018; Stern-Ginossar et al. 2018). Even in well-studied areas, growing appreciation of the flexibility and complexity of the translational apparatus points to the likelihood that more mechanistic variety will be found than is currently appreciated. Discoveries such as repeat-associated non-ATG (RAN) translation (Zu et al. 2011, 2018) and ribosomal heterogeneity (Kondrashov et al. 2011; Robichaud et al. 2018) exemplify how much is still to be learned. Although it would be rash to be specific, advances in RNA biology—including base modifications and epitranscriptomics (Peer et al. 2018), and regulation by circular and noncoding RNAs (Chekulaeva and Rajewsky 2018) and microRNAs (Duchaine and Fabian 2018), as well as antisense and interfering RNAs—present new avenues for basic research and open fresh possibilities for bench-to-bedside translation.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Adams JM, Cory S. 1975. Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature 255: 28–33. [DOI] [PubMed] [Google Scholar]
- Aoki H, Dekany K, Adams SL, Ganoza MC. 1997. The gene encoding the elongation factor P protein is essential for viability and is required for protein synthesis. J Biol Chem 272: 32254–32259. [DOI] [PubMed] [Google Scholar]
- Arlinghaus RF, Schweet R. 1963. A ribosome-bound intermediate in polypeptide synthesis. Biochem Biophys Res Commun 11: 92–96. [Google Scholar]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell 7: 25–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekliz M, Azza S, Seligmann H, Decloquement P, Raoult D, La Scola B. 2018. Experimental analysis of mimivirus translation initiation factor 4a reveals its importance in viral protein translation during infection of Acanthamoeba polyphaga. J Virol 92: e00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. 1996. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J 15: 658–664. [PMC free article] [PubMed] [Google Scholar]
- Berg P, Ofengand EJ. 1958. An enzymatic mechanism for linking amino acids to RNA. Proc Natl Acad Sci 44: 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergan V, Jagus R, Lauksund S, Kileng O, Robertsen B. 2008. The Atlantic salmon Z-DNA binding protein kinase phosphorylates translation initiation factor 2α and constitutes a unique orthologue to the mammalian dsRNA-activated protein kinase R. FEBS J 275: 184–197. [DOI] [PubMed] [Google Scholar]
- *.Biswas J, Liu Y, Singer RH, Wu B. 2018. Fluorescence imaging methods to investigate translation in single cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B. 1975. Transfer of proteins across membranes. II: Reconstitution of functional rough microsomes from heterologous components. J Cell Biol 67: 852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Sabatini DD. 1971. Ribosome-membrane interaction in eukaryotic cells. In Biomembranes (ed. Manson LA), pp. 193–195. Plenum, New York. [Google Scholar]
- Borsook H, Deasy CL, Haagensmit AJ, Keighley G, Lowy PH. 1952. Incorporation in vitro of labeled amino acids into proteins of rabbit reticulocytes. J Biol Chem 196: 669–694. [PubMed] [Google Scholar]
- Both GW, Furuichi Y, Muthukrishnan S, Shatkin AJ. 1975. Ribosome binding to reovirus mRNA in protein synthesis requires 5′ terminal 7-methylguanosine. Cell 6: 185–195. [DOI] [PubMed] [Google Scholar]
- *.Breaker RR. 2018. Riboswitches and translation control. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Jacob F, Meselson M. 1961. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature 190: 576–581. [DOI] [PubMed] [Google Scholar]
- Bruns GP, London IM. 1965. The effect of hemin on the synthesis of globin. Biochem Biophys Res Commun 18: 236–242. [DOI] [PubMed] [Google Scholar]
- Buxbaum AR, Haimovich G, Singer RH. 2015. In the right place at the right time: Visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol 16: 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PN, Work TS. 1953. Biosynthesis of proteins. Nature 171: 997–1001. [DOI] [PubMed] [Google Scholar]
- Chapeville F, Lipmann F, von Ehrenstein G, Weisblum B, Ray WJ, Benzer S. 1962. On the role of soluble ribonucleic acid in coding for amino acids. Proc Natl Acad Sci 48: 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Chekulaeva M, Rajewsky N. 2018. Roles of long noncoding RNAs and circular RNAs in translation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb M. 2015. Who discovered messenger RNA? Curr Biol 25: R526–R532. [DOI] [PubMed] [Google Scholar]
- Crick FHC. 1958. On protein synthesis. Symp Soc Exp Biol 12: 138–163. [PubMed] [Google Scholar]
- Crick FH. 1959. Biochemical activities of nucleic acids. The present position of the coding problem. Brookhaven Symp Biol 12: 35–39. [PubMed] [Google Scholar]
- Crick FH. 1966. Codon–anticodon pairing: The wobble hypothesis. J Mol Biol 19: 548–555. [DOI] [PubMed] [Google Scholar]
- Daly MM, Mirsky AE. 1952. Formation of protein in the pancreas. J Gen Physiol 36: 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE, Philipson L, Wall R, Adesnik M. 1971a. Polyadenylic acid sequences: Role in conversion of nuclear RNA into messenger RNA. Science 174: 507–510. [DOI] [PubMed] [Google Scholar]
- Darnell JE, Wall R, Tushinski RJ. 1971b. An adenylic acid-rich sequence in messenger RNA of HeLa cells and its possible relationship to reiterated sites in DNA. Proc Natl Acad Sci 68: 1321–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dever TE, Dinman JD, Green R. 2018. Translation elongation and recoding in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devilliers-Thiery A, Kindt T, Scheele G, Blobel G. 1975. Homology in amino-terminal sequence of precursors to pancreatic secretory proteins. Proc Natl Acad Sci 72: 5016–5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN. 2010. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem 285: 26779–26787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. 2013. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339: 85–88. [DOI] [PubMed] [Google Scholar]
- Drysdale JW, Munro HN. 1965. Failure of actinomycin D to prevent induction of liver apofentin after iron administration. Biochim Biophys Acta 103: 185–188. [DOI] [PubMed] [Google Scholar]
- *.Duchaine TF, Fabian MR. 2018. Mechanistic insights into microRNA-mediated gene silencing. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eboué-Bonis D, Chambaut AM, Volfin P, Clauser H. 1963. Action of insulin on the isolated rat diaphragm in the presence of actinomycin D and puromycin. Nature 199: 1183–1184. [DOI] [PubMed] [Google Scholar]
- Edmonds M, Vaughan MH Jr, Nakazato H. 1971. Polyadenylic acid sequences in the heterogeneous nuclear RNA and rapidly-labeled polyribosomal RNA of HeLa cells: Possible evidence for a precursor relationship. Proc Natl Acad Sci 68: 1336–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenfeld E, Hunt T. 1971. Double-stranded poliovirus RNA inhibits initiation of protein synthesis by reticulocyte lysates. Proc Natl Acad Sci 68: 1075–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenfeld E, Lund H. 1977. Untranslated vesicular stomatitis virus messenger RNA after poliovirus infection. Virology 80: 297–308. [DOI] [PubMed] [Google Scholar]
- Fan H, Penman S. 1970. Regulation of protein synthesis in mammalian cells. II: Inhibition of protein synthesis at the level of initiation during mitosis. J Mol Biol 50: 655–670. [DOI] [PubMed] [Google Scholar]
- Farrell PJ, Balkow K, Hunt T, Jackson RJ, Trachsel H. 1977. Phosphorylation of initiation factor eIF-2 and the control of reticulocyte protein synthesis. Cell 11: 187–200. [DOI] [PubMed] [Google Scholar]
- Fonseca BD, Zakaria C, Jia JJ, Graber TE, Svitkin Y, Tahmasebi S, Healy D, Hoang HD, Jensen JM, Diao IT, et al. 2015. La-related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1). J Biol Chem 290: 15996–16020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y, Miura K. 1975. A blocked structure at the 5′ terminus of mRNA from cytoplasmic polyhedrosis virus. Nature 253: 374–375. [DOI] [PubMed] [Google Scholar]
- Furuichi Y, Morgan M, Muthukrishnan S, Shatkin AJ. 1975a. Reovirus messenger RNA contains a methylated, blocked 5′-terminal structure: m7G(5′)ppp(5′)G-MpCp. Proc Natl Acad Sci 72: 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y, Morgan M, Shatkin AJ, Jelinek W, Salditt-Georgieff M, Darnell JE. 1975b. Methylated, blocked 5 termini in HeLa cell mRNA. Proc Natl Acad Sci 72: 1904–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garren LD, Howell RR, Tomkins GM, Crocco RM. 1964. A paradoxical effect of actinomycin D: The mechanism of regulation of enzyme synthesis by hydrocortisone. Proc Natl Acad Sci 52: 1121–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genuth NR, Barna M. 2018. Heterogeneity and specialized functions of translation machinery: From genes to organisms. Nat Rev Genet 10.1038/s41576-018-0008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer A. 1963. Function of aggregated reticulocyte ribosomes in protein synthesis. J Mol Biol 6: 148–157. [DOI] [PubMed] [Google Scholar]
- Gilmore R, Blobel G, Walter P. 1982a. Protein translocation across the endoplasmic reticulum. I: Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol 95: 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore R, Walter P, Blobel G. 1982b. Protein translocation across the endoplasmic reticulum. II: Isolation and characterization of the signal recognition particle receptor. J Cell Biol 95: 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman HM, Rich A. 1963. Mechanism of polyribosome action during protein synthesis. Nature 199: 318–322. [DOI] [PubMed] [Google Scholar]
- Gros F, Hiatt H, Gilbert W, Kurland GG, Risebrough RW, Watson JD. 1961. Unstable ribonucleic acid revealed by pulse labelling of Escherichia coli. Nature 190: 581–585. [DOI] [PubMed] [Google Scholar]
- Gross PR, Malkin LI, Moyer WA. 1964. Templates for the first proteins of embryonic development. Proc Natl Acad Sci 51: 407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardesty B, Miller R, Schweet R. 1963. Polyribosome breakdown and hemoglobin synthesis. Proc Natl Acad Sci 50: 924–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253: 905–909. [DOI] [PubMed] [Google Scholar]
- *.Hellen CUT. 2018. Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hershey JWB, Sonenberg N, Mathews MB. 2018. Principles of translational control. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heywood SM. 1970. Specificity of mRNA binding factor in eukaryotes. Proc Natl Acad Sci 67: 1782–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland MB, Stephenson ML, Scott JF, Hecht LI, Zamecnik PC. 1958. A soluble ribonucleic acid intermediate in protein synthesis. J Biol Chem 231: 241–257. [PubMed] [Google Scholar]
- Hoagland MB, Zamecnik PC, Stephenson ML. 1959. A hypothesis concerning the roles of particulate and soluble ribonucleic acids in protein synthesis. In A symposium on molecular biology (ed. Zirkle RE). University of Chicago, Chicago, IL. [Google Scholar]
- Hodge LD, Robbins E, Scharff MD. 1969. Persistence of messenger RNA through mitosis in HeLa cells. J Cell Biol 40: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, Merrill SH, Penswick JR, Zamir A. 1965. Structure of a Ribonucleic Acid. Science 147: 1462–1465. [DOI] [PubMed] [Google Scholar]
- Hultin T. 1961. Activation of ribosomes in sea urchin eggs in response to fertilization. Exp Cell Res 25: 405–417. [DOI] [PubMed] [Google Scholar]
- Hultin T, Beskow G. 1956. The incorporation of 14C-L-leucine into rat liver proteins in vitro visualized as a two-step reaction. Exp Cell Res 11: 664–666. [DOI] [PubMed] [Google Scholar]
- Humphreys T. 1969. Efficiency of translation of messenger-RNA before and after fertilization in sea urchins. Dev Biol 20: 435–458. [DOI] [PubMed] [Google Scholar]
- Humphreys T. 1971. Measurements of messenger RNA entering polysomes upon fertilization of sea urchin eggs. Dev Biol 26: 201–208. [DOI] [PubMed] [Google Scholar]
- Ikemura T. 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: A proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol 151: 389–409. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Ingolia NT, Hussmann JA, Weissman JS. 2018. Ribosome profiling: Global views of translation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Monod J. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol 3: 318–356. [DOI] [PubMed] [Google Scholar]
- Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 62: 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies HB, Reinhard C, Kozma SC, Thomas G. 1994. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc Natl Acad Sci 91: 4441–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Jobe A, Liu Z, Gutierrez-Vargas C, Frank J. 2018. New insights into ribosome structure and function. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HA, Hershey JW. 1994. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J Biol Chem 269: 3934–3940. [PubMed] [Google Scholar]
- Kearse MG, Wilusz JE. 2017. Non-AUG translation: A new start for protein synthesis in eukaryotes. Genes Dev 31: 1717–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller EB, Zamecnik PC. 1956. The effect of guanosine diphosphate and triphosphate on the incorporation of labeled amino acids into proteins. J Biol Chem 221: 45–59. [PubMed] [Google Scholar]
- Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SK, Abdulhay NJ, Specht H, Keshishian H, Mani DR, et al. 2018. Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell 173: 90–103.e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King HA, Cobbold LC, Willis AE. 2010. The role of IRES trans-acting factors in regulating translation initiation. Biochem Soc Trans 38: 1581–1586. [DOI] [PubMed] [Google Scholar]
- Komar AA, Gross SR, Barth-Baus D, Strachan R, Hensold JO, Goss Kinzy T, Merrick WC. 2005. Novel characteristics of the biological properties of the yeast Saccharomyces cerevisiae eukaryotic initiation factor 2A. J Biol Chem 280: 15601–15611. [DOI] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Ishijima J, Shiroishi T, Barna M. 2011. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145: 383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosower NS, Vanderhoff GA, Benerofe B, Hunt T, Kosower EM. 1971. Inhibition of protein synthesis by glutathione disulfide in the presence of glutathione. Biochem Biophys Res Commun 45: 816–821. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1978. How do eukaryotic ribosomes select initiation regions in messenger RNA. Cell 15: 1109–1123. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44: 283–292. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15: 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M, Shatkin AJ. 1978. Migration of 40 S ribosomal subunits on messenger RNA in the presence of edeine. J Biol Chem 253: 6568–6577. [PubMed] [Google Scholar]
- Kruh J, Borsook H. 1956. Hemoglobin synthesis in rabbit reticulocytes in vitro. J Biol Chem 220: 905–915. [PubMed] [Google Scholar]
- Kuntzel H, Noll H. 1967. Mitochondrial and cytoplasmic polysomes from Neurospora crassa. Nature 215: 1340–1345. [DOI] [PubMed] [Google Scholar]
- *.Kwan T, Thompson SR. 2018. Noncanonical translation initiation in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr RM, Fonseca BD, Ciotti GE, Al-Ashtal HA, Jia JJ, Niklaus MR, Blagden SP, Alain T, Berman AJ. 2017. La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. eLife 6: e24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamfrom H, Knopf PM. 1964. Initiation of haemoglobin synthesis in cell-free systems. J Mol Biol 9: 558–575. [DOI] [PubMed] [Google Scholar]
- Laycock DG, Hunt JA. 1969. Synthesis of rabbit globin by a bacterial cell free system. Nature 221: 1118–1122. [DOI] [PubMed] [Google Scholar]
- Lee SY, Mendecki J, Brawerman G. 1971. A polynucleotide segment rich in adenylic acid in the rapidly-labeled polyribosomal RNA component of mouse sarcoma 180 ascites cells. Proc Natl Acad Sci 68: 1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Chen CJ, Shih SR. 2017. Regulation mechanisms of viral IRES-driven translation. Trends Microbiol 25: 546–561. [DOI] [PubMed] [Google Scholar]
- Leibowitz R, Penman S. 1971. Regulation of protein synthesis in HeLa cells. III: Inhibition during poliovirus infection. J Virol 8: 661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Avni D, Hariharan N, Perry RP, Meyuhas O. 1991. Oligopyrimidine tract at the 5′ end of mammalian ribosomal protein mRNAs is required for their translational control. Proc Natl Acad Sci 88: 3319–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisowska-Bernstein B, Lamm ME, Vassalli P. 1970. Synthesis of immunoglobulin heavy and light chains by the free ribosomes of a mouse plasma cell tumor. Proc Natl Acad Sci 66: 425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZY, Jia KT, Li C, Weng SP, Guo CJ, He JG. 2013. A truncated Danio rerio PKZ isoform functionally interacts with eIF2α and inhibits protein synthesis. Gene 527: 292–300. [DOI] [PubMed] [Google Scholar]
- Lockard RE, Lingrel JB. 1969. The synthesis of mouse hemoglobin β-chains in a rabbit reticulocyte cell-free system programmed with mouse reticulocyte 9S RNA. Biochem Biophys Res Commun 37: 204–212. [DOI] [PubMed] [Google Scholar]
- Lodish HF, Zinder ND. 1966. Mutants of the bacteriophage f2. VIII: Control mechanisms for phage-specific syntheses. J Mol Biol 19: 333–348. [DOI] [PubMed] [Google Scholar]
- Maggio R, Vittorelli ML, Rinaldi AM, Monroy A. 1964. In vitro incorporation of amino acids into proteins stimulated by RNA from unfertilized sea urchin eggs. Biochem Biophys Res Commun 15: 436–441. [DOI] [PubMed] [Google Scholar]
- Marks PA, Burka ER, Schlessinger D. 1962. Protein synthesis in erythroid cells. I. Reticulocyte ribosomes active in stimulating amino acid incorporation. Proc Natl Acad Sci 48: 2163–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TE, Young FG. 1965. An in vitro action of human growth hormone in the presence of actinomycin D. Nature 208: 684–685. [Google Scholar]
- Mathews M, Korner A. 1970. Mammalian cell-free protein synthesis directed by viral ribonucleic acid. Eur J Biochem 17: 328–338. [DOI] [PubMed] [Google Scholar]
- Mathews MB, Hunt T, Brayley A. 1973. Specificity of the control of protein synthesis by haemin. Mammalian messenger RNA. Nat New Biol 243: 230–233. [DOI] [PubMed] [Google Scholar]
- Mathews MB, Sonenberg N, Hershey JW. 2007. Origins and principles of translational control. In Translational control in biology and medicine (ed. Mathews MB, Sonenberg N, Hershey JWB). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- McCormick W, Penman S. 1969. Regulation of protein synthesis in HeLa cells: Translation at elevated temperatures. J Mol Biol 39: 315–333. [DOI] [PubMed] [Google Scholar]
- McQuillen K, Roberts RB, Britten RJ. 1959. Synthesis of nascent protein by ribosomes in Escherichia coli. Proc Natl Acad Sci 45: 1437–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Merrick WC, Pavitt GD. 2018. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DI, Krause E, Dobberstein B. 1982. Secretory protein translocation across membranes-the role of the “docking protein.” Nature 297: 647–650. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 2015. 5′ UTR m6A promotes cap-independent translation. Cell 163: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EW, Green R. 2017. Ribosomopathies: There’s strength in numbers. Science 358: eaan2755. [DOI] [PubMed] [Google Scholar]
- Milstein C, Brownlee GG, Harrison TM, Mathews MB. 1972. A possible precursor of immunoglobulin light chains. Nat New Biol 239: 117–120. [DOI] [PubMed] [Google Scholar]
- Monroy A, Maggio R, Rinaldi AM. 1965. Experimentally induced activation of the ribosomes of the unfertilized sea urchin egg. Proc Natl Acad Sci 54: 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto TC, Conway TW, Allende JE, Spyrides G, Lipmann F. 1963. Formation of peptide bonds. I: Peptide formation from aminoacyl-S-RNA. Cold Spring Harb Quant Biol 28: 227. [Google Scholar]
- Nemer M. 1962. Interrelation of messenger polyribonucleotides and ribosomes in the sea urchin egg during embryonic development. Biochem Biophys Res Commun 8: 511–515. [DOI] [PubMed] [Google Scholar]
- Nirenberg MW, Matthaei JH. 1961. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc Natl Acad Sci 47: 1588–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll HS, Wettstein T. 1963. Ribosomal aggregates engaged in protein synthesis: Ergosome breakdown and messenger ribonucleic acid transport. Nature 198: 632–638. [Google Scholar]
- Nomura MA, Hall BD, Spiegelman BM. 1960. Characterization of RNA synthesized in Escherichia coli after bacteriophage T2 infection. J Mol Biol 2: 306–326. [Google Scholar]
- Ohtaka Y, Spiegelman S. 1963. Translational control of protein synthesis in a cell-free system directed by a polycistronic viral RNA. Science 142: 493–497. [DOI] [PubMed] [Google Scholar]
- Palade GE. 1955. A small particulate component of the cytoplasm. J Biophys Biochem Cytol 1: 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Peer Y, Moshitch-Moshkovitz S, Rechavi G, Dominissini D. 2018. The epitranscriptome in translation regulation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HRB, Jackson RJ. 1976. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem 67: 247–256. [DOI] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334: 320–325. [DOI] [PubMed] [Google Scholar]
- Penman S, Summers D. 1965. Effects on host cell metabolism following synchronous infection with poliovirus. Virol 27: 614–620. [DOI] [PubMed] [Google Scholar]
- Penman S, Scherrer K, Becker Y, Darnell JE. 1963. Polyribosomes in normal and poliovirus-infected HeLa cells and their relationship to messenger RNA. Proc Natl Acad Sci 49: 654–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RP, Kelley DE. 1975. Methylated constituents of heterogeneous nuclear RNA: Presence in blocked 5′ terminal structures. Cell 6: 13–19. [DOI] [PubMed] [Google Scholar]
- *.Proud CG. 2018. Phosphorylation and signal transduction pathways in translational control. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Robichaud N, Sonenberg N, Ruggero D, Schneider RJ. 2018. Translational control in cancer. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Rodnina MV. 2018. Translation in prokaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M. 1972. Formation of membrane-bound polyribosomes. J Mol Biol 65: 413–422. [DOI] [PubMed] [Google Scholar]
- Sanger F, Tuppy H. 1951. The amino-acid sequence in the phenylalanyl chain of insulin. 2. The investigation of peptides from enzymic hydrolysates. Biochem J 49: 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauert M, Temmel H, Moll I. 2015. Heterogeneity of the translational machinery: Variations on a common theme. Biochimie 114: 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J, Dalgarno L. 1975. Determinant of cistron specificity in bacterial ribosomes. Nature 254: 34–38. [DOI] [PubMed] [Google Scholar]
- Siekevitz P, Zamecnik PC. 1951. In vitro incorporation of 1-14C-DL-alanine into proteins of rat-liver granular fractions. Fed Proc 10: 246–247. [Google Scholar]
- Sonenberg N, Rupprecht KM, Hecht SM, Shatkin AJ. 1979. Eukaryotic mRNA cap binding protein: Purification by affinity chromatography on sepharose-coupled m7GDP. Proc Natl Acad Sci 76: 4345–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin AS, Nemer M. 1965. Messenger RNA in early sea-urchin embryos: Cytoplasmic particles. Science 150: 214–217. [DOI] [PubMed] [Google Scholar]
- Starck SR, Jiang V, Pavon-Eternod M, Prasad S, McCarthy B, Pan T, Shastri N. 2012. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science 336: 1719–1723. [DOI] [PubMed] [Google Scholar]
- *.Stern-Ginossar N, Thompson SR, Mathews MB, Mohr I. 2018. Translational control in virus-infected cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward DL, Shaeffer JR, Humphrey RM. 1968. Breakdown and assembly of polyribosomes in synchronized Chinese hamster cells. Science 161: 791–793. [DOI] [PubMed] [Google Scholar]
- Summers DF, Maizel JV. 1967. Disaggregation of HeLa cell polysomes after infection with poliovirus. Virol 31: 550–552. [DOI] [PubMed] [Google Scholar]
- Summers DF, Maizel JV, Darnell JE. 1965. Evidence for virus-specific noncapsid proteins in poliovirus-infected HeLa cells. Proc Natl Acad Sci 54: 505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghavi N, Samuel CE. 2013. RNA-dependent protein kinase PKR and the Z-DNA binding orthologue PKZ differ in their capacity to mediate initiation factor eIF2α-dependent inhibition of protein synthesis and virus-induced stress granule formation. Virology 443: 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahmasebi S, Khoutorsky A, Mathews MB, Sonenberg N. 2018. Translation deregulation in human disease. Nat Rev Mol Cell Biol 10.1038/s41580-018-0034-x. [DOI] [PubMed] [Google Scholar]
- *.Teixeira FK, Lehmann R. 2018. Translational control during development transitions. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada N, Patel HR, Takase K, Kohno K, Nairn AC, Gelfand EW. 1994. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc Natl Acad Sci 91: 11477–11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GP, Mathews MB. 1984. Alterations of transcription and translation in HeLa cells exposed to amino acid analogs. Mol Cell Biol 4: 1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkins GM, Garren LD, Howell RR, Peterkofsky B. 1965. The regulation of enzyme synthesis by steroid hormones: the role of translation. J Cell Comp Physiol 66: 137–151. [DOI] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. 2013. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339: 82–85. [DOI] [PubMed] [Google Scholar]
- Ventoso I, Sanz MA, Molina S, Berlanga JJ, Carrasco L, Esteban M. 2006. Translational resistance of late α virus mRNA to eIF2α phosphorylation: A strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev 20: 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkin E, Astrachan L. 1956. Phosphorus incorporation in Escherichia coli ribonucleic acid after infection with bacteriophage T2. Virology 2: 149–161. [DOI] [PubMed] [Google Scholar]
- Walter P, Blobel G. 1981. Translocation of proteins across the endoplasmic reticulum. II: Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol 91: 551–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ibrahimi I, Blobel G. 1981. Translocation of proteins across the endoplasmic reticulum. I: Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol 91: 545–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, Rich A, Hall CE. 1962. Electron microscope studies of ribosomal clusters synthesizing hemoglobin. Science 138: 1399–1403. [DOI] [PubMed] [Google Scholar]
- Warner JR, Knopf PM, Rich A. 1963. A multiple ribosomal structure in protein synthesis. Proc Natl Acad Sci 49: 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman HS, Rabinowitz M. 1966. Control of reticulocyte polyribosome content and hemoglobin synthesis by heme. Biochim Biophys Acta 129: 369–379. [Google Scholar]
- Wei CM, Moss B. 1975. Methylated nucleotides block 5′-terminus of vaccinia virus messenger RNA. Proc Natl Acad Sci 72: 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wek RC. 2018. Role of eIF2α kinases in translational control and adaptation to cellular stresses. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettstein FO, Staehelin T, Noll H. 1963. Ribosomal aggregate engaged in protein synthesis: characterization of the ergosome. Nature 197: 430–435. [DOI] [PubMed] [Google Scholar]
- Wilt FH, Hultin T. 1962. Stimulation of phenylalanine incorporation by polyuridylic acid in homogenates of sea urchin eggs. Biochem Biophys Res Commun 9: 313–317. [DOI] [PubMed] [Google Scholar]
- Witherell GW, Gott JM, Uhlenbeck OC. 1991. Specific interaction between RNA phage coat proteins and RNA. Prog Nucleic Acid Res Mol Biol 40: 185–220. [DOI] [PubMed] [Google Scholar]
- Zamecnik PC. 1960. Historical and current aspects of the problem of protein synthesis. Harvey Lectures 54: 256–281. [Google Scholar]
- Zamecnik PC. 1969. An historical account of protein synthesis, with current overtones—A personalized view. Cold Spring Harb Symp Quant Biol 34: 1–16. [DOI] [PubMed] [Google Scholar]
- Zamecnik PC, Keller EB. 1954. Relation between phosphate energy donors and incorporation of labeled amino acids into proteins. J Biol Chem 209: 337–354. [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. 2015. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526: 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, et al. 2011. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci 108: 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Zu T, Pattamatta A, Ranum LPW. 2018. RAN translation in neurological diseases. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033019. [DOI] [PMC free article] [PubMed] [Google Scholar]