Abstract
The circadian oscillator is a complex network of interconnected feedback loops that regulates a wide range of physiological processes. Indeed, variation in clock genes has been implicated in an array of plant environmental adaptations, including growth regulation, photoperiodic control of flowering, and responses to abiotic and biotic stress. Although the clock is buffered against the environment, maintaining roughly 24-h rhythms across a wide range of conditions, it can also be reset by environmental cues such as acute changes in light or temperature. These competing demands may help explain the complexity of the links between the circadian clock network and environmental response pathways. Here, we discuss our current understanding of the clock and its interactions with light and temperature-signaling pathways. We also describe different clock gene alleles that have been implicated in the domestication of important staple crops.
Through the day, all organisms are exposed to diel environmental rhythms such as the daily transition from light to dark and the daily fluctuation of temperature. Organisms have evolved light and temperature sensors, which enable them to sense and respond to these changes, maintaining homeostatic balance (Larner et al. 2018). Although responding to environmental changes to maintain homeostasis is important, it is also beneficial for organisms to anticipate daily changes and prepare for them beforehand. To this end, most organisms, including plants, have developed an internal timing mechanism known as the circadian clock that enables them to anticipate and align internal biological processes with these daily rhythms (for review, see Harmer 2009; Millar 2016). Circadian clocks are cell autonomous and each cell maintains its own 24-h rhythm, which allows multicellular organisms to maintain tissue and organ-specific clocks (Endo 2016). The main components of a circadian system are the input signals from the environment that reset the clock, the central oscillator that maintains a roughly 24-h rhythm even in the absence of input signals, and the output signals that generate daily rhythms in physiology.
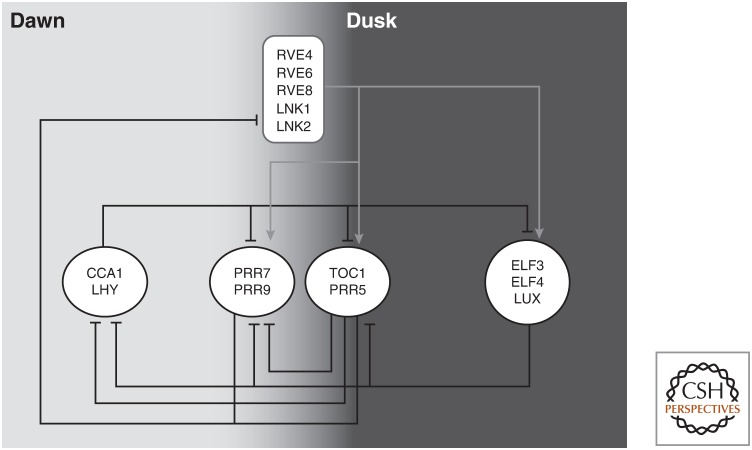
In plants, the central oscillator is a complex gene regulatory network of repressors and activators that form multiple interlocking feedback loops (Fig. 1). These clock genes are expressed at specific times of the day and in addition to regulating each other's expression they also influence multiple physiological processes. Clock-regulated pathways may show rhythmicity, with peak activity at distinct times of day, and in addition may be “gated” by the clock such that they are more responsive to environmental stimuli at specific times of day (Greenham and McClung 2015). This mechanism ensures that a plant is most responsive to light during daylight hours, to growth hormones during the night, and to environmental stresses at times when adverse conditions are most likely (Covington and Harmer 2007; Arana et al. 2011; Zhu et al. 2016). To appropriately gate plant responses to external factors, the clock is directly linked with the light and temperature-signaling pathways, which also ensures synchronicity between the external and internal rhythms (Casal and Qüesta 2018). The cross talk between these regulatory pathways also provides seasonal information to the plant, allowing for example the determination of day length for the appropriate control of the transition to flowering (Song et al. 2015). Although the clock can be reset by temperature and light, it would be detrimental to a plant if the clock were sensitive to minor intermittent fluctuations of temperatures throughout the day. To counter this, the clock is not only reset by large temperature changes but is also buffered against ambient changes in a mechanism known as temperature compensation, in which the clock maintains an approximately 24-h period even when temperatures are fluctuating over time (Gil and Park 2019).
Figure 1.
A highly simplified representation of the plant circadian regulatory network. Similar genes operating at similar times during the day in a similar manner are grouped together in white circles. Black lines with blunt ends indicate genes function as repressors in the negative feedback loops. Gray lines and arrows indicate genes acting as activators in the regulatory network.
Given the central role of the circadian clock in modulating plant responses to environmental cues, it is not surprising that selection of circadian clock variants has been implicated in the adaptation and domestication of many agriculturally important species (Bendix et al. 2015; Blümel et al. 2015). In this review, we discuss our current understanding of the circadian clock network and how environmental cues are integrated into this complex regulatory system. We also discuss the role of the clock in the adaptation of crop species to different latitudes and to distinct biotic and abiotic stresses.
THE PLANT CIRCADIAN CLOCK
The plant circadian clock is a complex network of intertwined feedback loops comprised of repressor and activator transcription factors (Harmer 2009; Hsu and Harmer 2014; Huang and Nusinow 2016). Levels of these proteins are in constant flux, each peaking at a specific time of day and feeding back to regulate each other's expression. The morning expressed MYB-like transcription factors CCA1 (CIRCADIAN CLOCK ASSOCIATED1) and LHY (LATE ELOGATED HYPOCOTYL) repress the afternoon expressed PSEUDO-RESPONSE REGULATOR (PRR) genes, including PRR1/TOC1 (TIMING OF CAB EXPRESSION1), PRR5, PRR7, and PRR9 (Alabadí et al. 2001; Farré et al. 2005; Kamioka et al. 2016). TOC1, along with the other PRR proteins, in turn, repress the expression of CCA1 and LHY, closing this feedback loop (Alabadí et al. 2001; Nakamichi et al. 2010). CCA1/LHY are themselves primarily repressors of transcription and bind to a cis-motif termed the evening element (EE) found in the regulatory regions of many clock genes, including the PRRs. Other direct targets of CCA1/LHY activity include genes that encode members of the transcriptional regulatory evening complex, ELF3 (EARLY FLOWERING3), ELF4, and LUX (LUX ARRHYTHMO). These three genes are expressed at night, at which time the evening complex feeds back to repress multiple morning- and afternoon-expressed genes to complete another feedback loop in the network (Fig. 1; Huang and Nusinow 2016).
The circadian regulatory network in plants is not only comprised of negative feedback loops: a second set of midday-expressed MYB-like transcription factors, REVEILLE4 (RVE4), RVE6, and RVE8, has been shown to activate expression of several clock genes including TOC1, the PRRs and the evening complex genes (Farinas and Mas 2011; Rawat et al. 2011; Hsu et al. 2013). To activate gene expression, RVE8 forms a complex with LNK1 (NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED1) and LNK2 and associates with the promoters of TOC1 and PRR5 (Xie et al. 2014). The RVE activator proteins are not simply a second layer of regulation on top of the core circadian clock but are connected and embedded into the clock regulatory network (Fig. 1). It has previously been shown that RVE8 expression is repressed by TOC1 and the PRRs, forming yet another negative feedback loop in this network (Rawat et al. 2011; Hsu et al. 2013). Interestingly, the CCA1 and LHY repressors and the RVE activators both have highly similar DNA-binding domains and can bind the same EE-binding site in similar sets of promotors (Harmer and Kay 2005; Rawat et al. 2011). A recent study showed that the balance between the expression levels of the activating and repressing MYB-like factors is more important in regulation of circadian period than the presence or absence of any specific factor (Shalit-Kaneh et al. 2018).
INTEGRATING LIGHT AND TEMPERATURE CUES INTO THE CIRCADIAN CLOCK REGULATORY NETWORK
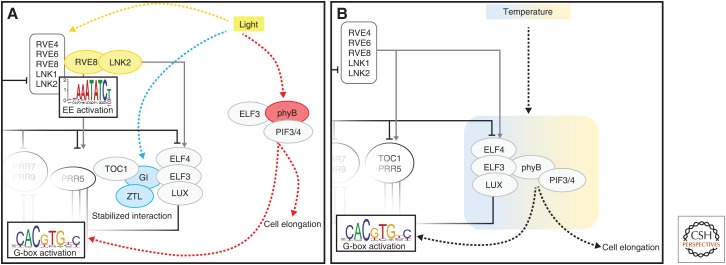
Circadian clocks must be continually adjusted by environmental cues so that the processes they control are appropriately timed even as temperature and day-length change with the seasons. For this reason, the plant clock uses multiple mechanisms to sense and integrate external signals into the feedback loops described above (Fig. 2). The phytochrome-signaling pathway is one of the main mechanisms through which plants sense and respond to changes in red light availability and is directly linked to the clock regulatory network (Oakenfull and Davis 2017). Of the phytochrome receptors, phytochrome B (phyB) is the main red-light receptor and its effects on plant growth and development have been extensively studied (Larner et al. 2018). phyB photoconverts from an inactive form (Pr) to an active form (Pfr) on absorption of red light (Viczián et al. 2017). Pfr interacts with the PHYTOCHROME-INTERACTING FACTORS (PIFs) and targets these transcription factors for degradation during the day to limit cell elongation to the night time hours (Seluzicki et al. 2017). PIF proteins have been established as transcriptional regulators of morning-expressed LHY and CCA1, directly linking the light and clock regulatory networks (Martínez-García et al. 2000). Recently, PIFs have also been shown to mediate metabolic signals to the circadian oscillator (Shor et al. 2017). Another link between the clock- and light-signaling pathways occurs via interactions between phyB and the evening complex protein ELF3 (Liu et al. 2001; Huang et al. 2016). ELF3 also binds to PIF4 independently of the other evening complex components to repress PIF4 function, thus regulating a light-signaling component controlling hypocotyl elongation (Nieto et al. 2015). Similarly, TOC1 and other PRR proteins have been shown to bind directly to PIF3 and PIF4 and inhibit their ability to activate transcription. Thus, the association of PRR factors with PIFs on the G-box elements of target promoters serves to limit PIF transactivation function to the predawn hours (Liu et al. 2016; Soy et al. 2016; Zhu et al. 2016). The PRRs and the evening complex have also been shown to regulate transcription of PIF genes (Nusinow et al. 2011; Nakamichi et al. 2012; Liu et al. 2016). Thus, both ELF3 and the PRR proteins limit the function and expression of the important growth regulatory PIF factors and provide further links between clock and light regulation of growth (Fig. 2A).
Figure 2.
Schematic representation of how light and temperature-signaling pathways integrate with the circadian clock regulatory network. The underlying clock network is the same as Figure 1 with either the light-signaling pathway (A) or the temperature-signaling pathway (B) linking to points in the circadian regulatory network. (A) Red and blue pathways indicate how these different wavelengths of light are integrated into the clock at different points via independent pathways. (B) Different temperatures influence the same pathway (blue to orange shaded box), with cooler temperatures stabilizing complex formation and warmer temperatures releasing growth factors such as PIFs.
The clock protein ZEITLUPE (ZTL) is unique in being both a component of the plant clock and a blue-light photoreceptor. ZTL interacts directly with GIGANTEA (GI), another clock component, and this interaction is stabilized by blue light via the photosensory LOV domain of ZTL. This ZTL–GI complex can maintain circadian rhythms by influencing the stability of both TOC1 and GI proteins (Más et al. 2003; Kim et al. 2007, 2013). GI stability is also affected by a second protein complex consisting of ELF3 and COP1 (CONSTITUTIVE PHOTOMORPHOGENIC1) that acts downstream from the blue light photoreceptor CRY2 (Yu et al. 2008). The ELF3–COP1 complex targets GI for degradation and represents yet another point at which light signals are integrated into the circadian clock network. The LNK2 and RVE8 complex also appears to play a role in the integration of the clock and light-signaling pathways (Fig. 2A). It is possible that clock entrainment relies on the induction of LNK expression by phytochromes in conjunction with the early morning expression of CCA1 and LHY (Wang and Tobin 1998; Kim et al. 2003; Rugnone et al. 2013).
Temperature is another external signal that is integrated into the clock network (Fig. 2B). Recent research has revealed that plant photoreceptors can also function as temperature receptors (Delker et al. 2017; Casal and Qüesta 2018). One such receptor is phyB, with the rate of reversion from the active Pfr form to the inactive Pr form increasing at higher temperatures (Jung et al. 2016; Legris et al. 2016). Given the multiple links between phytochrome-signaling components and clock proteins described above, temperature regulation of phytochrome function is a likely point of temperature integration into the clock. PIF4 has also been shown to respond to changes in temperature to alter plant development and morphology (Paik et al. 2017). Because degradation of PIF4 is promoted by Pfr, the increased rate of Pfr to Pr reversion at higher temperatures may account for the warm temperature-induced posttranscriptional accumulation of PIF4 protein (Foreman et al. 2011; Zhu et al. 2016).
Temperature has also been shown to regulate the activity of the evening complex. At higher temperatures, association of ELF3 with target promoters is reduced via an unknown mechanism (Box et al. 2015; Mizuno et al. 2015; Ezer et al. 2017). Thus, in warm conditions, evening-complex mediated repression of targets such as the clock genes PRR7, PRR9, GI, LUX, and the growth regulating PIF4 is relieved, leading to elevated levels of these transcripts during warm nights. Integration of cold temperature cues into the clock network can occur via CBF1/DREB1a (COLD-INDUCIBLE C-REPEAT/DROUGHT-RESPONSIVE ELEMENT-BINDING FACTOR). CBF1 expression is highly induced by cold, and this factor has been shown to bind directly to the promoter of the evening complex component LUX and promote its high-amplitude rhythmic expression at cold temperatures (Chow et al. 2014). Intriguingly, phyB and PIF proteins have been reported to repress CBF1 expression (Kidokoro et al. 2009; Lee and Thomashow 2012), suggesting links between distinct temperature-sensing pathways. Finally, expression of CBF1 is regulated by a number of clock components including the PRRs, the evening complex, and CCA1/LHY (Kinmonth-Schultz et al. 2013), providing yet another example of the ubiquitous feedback loops found in the circadian system.
CLOCK REGULATION OF GROWTH PATHWAYS
Plant growth is regulated by both environmental and internal cues, and therefore plant growth pathways are highly interconnected with the circadian clock regulatory network as well as with light- and temperature-signaling pathways (Nozue and Maloof 2006; Farré 2012; Kinmonth-Schultz et al. 2013; Henriques et al. 2018). The best-studied example of plant growth is the elongation of Arabidopsis hypocotyls, which is driven by anisotropic growth of cells formed during embryo development. In short-day conditions, hypocotyl elongation is rhythmic, with peak growth occurring at the end of the night/beginning of dawn. However, in long-day or constant-light conditions, the peak phase of growth is shifted to midmorning or end of the subjective day, respectively. These findings show that hypocotyl growth is regulated both by light and the circadian clock (Nozue et al. 2007).
The PIF transcriptional activators are key mediators of this and other growth rhythms, and are important integrators of clock, light, and temperature signals (Legris et al. 2016; Paik et al. 2017; Pham et al. 2018). Clock and light regulation of PIF activity restricts hypocotyl elongation to the end of the night in short-day conditions by the following mechanism. First, PIF4 and PIF5 expression is limited to the day and predawn hours because of the direct binding of the repressive evening complex to the promoters of these genes (Nusinow et al. 2011). However, during the day, active phyB (Pfr) sequesters PIF proteins and targets them for degradation, preventing PIF protein accumulation and inhibiting cell elongation (Nozue et al. 2007; Park et al. 2012; Soy et al. 2012). During the night, Pfr converts back to Pr and PIF degradation is relieved, allowing PIF protein accumulation and promoting hypocotyl elongation near dawn (Nozue et al. 2007). The circadian clock also regulates hypocotyl elongation via control of PIF transactivation activity through other core clock components. TOC1 and the other repressive PRR proteins physically interact with the transactivating PIF proteins to inhibit the induction of growth-promoting genes (Liu et al. 2016; Soy et al. 2016; Zhu et al. 2016; Martín et al. 2018). Thus, the circadian clock and lightsignaling pathways facilitate late-night/early-day phased hypocotyl elongation via multiple mechanisms. Growth in the morning is speculated to ensure that expansion coincides with maximal water availability for increased turgor pressure and higher availability of carbon for cell wall remodeling (Nozue et al. 2007; Robertson et al. 2009).
New findings suggest that the circadian clock can regulate growth not only on a whole-plant or organ level but can differentially regulate growth in a subset of cells within a specific organ (Atamian et al. 2016; Endo 2016; Apelt et al. 2017; Ke et al. 2018). The first written record of diel rhythms was the observation in the fourth century BC that a number of plants show daily rhythms in leaf movement (McClung, 2006). Some plants have specialized motor cells, called pulvini, which undergo rapid changes in turgor pressure to facilitate such movement (Whippo and Hangarter 2009). However, most species lack pulvini and leaf movements are thought to rely on differential expansion of cells on the adaxial and abaxial sides of petioles (Polko et al. 2012; Rauf et al. 2013). Recently, this differential growth and leaf movement in Arabidopsis was found to depend on the PRR clock components, with a prr7prr9 double mutant displaying poor leaf movements compared with wild-type plants (Apelt et al. 2017). These results suggest that the circadian clock can regulate the differential expansion of specific cell layers to mediate leaf movement.
A similar growth regulatory mechanism is thought to underlie the daily movement of the stems of juvenile sunflowers. Although their ability to bend from east to west each day to track the apparent movement of the sun is well known, it is less recognized that they bend back from west to east each night in anticipation of the coming dawn. These rhythmic back-and-forth movements persist for several days when plants are moved to constant environmental conditions, suggesting involvement of the circadian clock in these heliotropic movements (Atamian et al. 2016). This tracking motion in juvenile sunflowers is caused by differential growth on opposite sides of the stem, and indeed signaling genes of the growth hormone, auxin, are differentially expressed on the east and west sides of solar tracking stems. Disruption of tracking movements causes a reduction in leaf area and biomass, perhaps caused by a decrease in leaf photon capture (Atamian et al. 2016). Similarly, a study on diel flower opening in waterlily found that movement was initiated by differential expansion of cells only in a specific region of the petal above its base. This cell expansion and the petal movement was found to be regulated by light signals that are thought to activate downstream auxin signaling and cell wall remodeling pathways (Ke et al. 2018). Taken together, these findings suggest that there are further regulatory networks to be explored that limit clock and environmental effects to specific cells, which fine-tune plant adaption to environmental challenges.
THE ROLE OF THE PLANT CLOCK IN PHOTOPERIODIC REGULATION OF FLOWERING
It is important for plants to detect seasonal changes so that biological processes such as flowering, dormancy, and budbreak can be aligned with the appropriate season. Links between the clock and photoperiod-mediated regulation of the transition between vegetative and reproductive growth have been particularly well studied. Integration of information from the light- and thermosensory pathways into the photoperiodic flowering time network via the clock helps ensure plants reproduce in the appropriate season, maximizing plant fitness (Blümel et al. 2015; Song et al. 2015). Many plant species are photoperiodic, with time to flowering hastened either by long days in the case of long-day plants or by short days in the case of short-day plants. These traits are associated with distinct reproductive strategies; for example, many short-day plants use the shortening days of fall as a cue to produce flowers and seeds before the onset of winter, whereas many long-day plants use the lengthening days of spring as a cue to reproduce before the onset of a hot and dry summer. However, many crop cultivars have been selected for day neutrality, with time to flowering independent of day length. Although some regulators of flowering vary across species, promotion of flowering in response to accumulation of homologs of the FT (FLOWERING LOCUS T) protein in the shoot apex is highly conserved (Andrés and Coupland 2012).
In the long-day plant, Arabidopsis thaliana, expression of FT is activated in leaf vasculature by the CO (CONSTANS) transcription factor, which increases FT expression via a feed-forward mechanism (An et al. 2004). CO transcript abundance is negatively regulated by the clock-controlled CDF (CYCLING DOF FACTOR) proteins (Fornara et al. 2009). The CDF proteins are degraded by a protein complex comprised of GI and a ZTL-related protein, FKF1 (FLAVIN-BINDING, KELCH REPEAT, F-BOX1). This complex is stabilized by blue light, in a similar manner to the GI–ZTL complex, and degrades the CDF proteins to promote the transition to flowering (Imaizumi et al. 2005; Song et al. 2015). There is also evidence that GI can directly bind to the FT promoter to regulate flowering independently of CO, suggesting GI is a central factor regulating flowering time (Sawa and Kay 2011).
In the short-day plant rice, the CO and FT homologs Hd1 (HEADING DATE1) and Hd3a also play key roles in the photoperiodic control of flowering. As is also true in Arabidopsis, the rice homolog of GI promotes expression of the CO homolog Hd1, and an ELF3 homolog has also been implicated in photoperiodic control of flowering (Hori et al. 2016). However, although CO promotes flowering in Arabidopsis in long days, the rice CO homolog Hd1 promotes flowering in short days and inhibits it in long days (Izawa 2007). Thus, despite important differences in the molecular circuitry controlling the transition to flowering, clock components play key roles in relaying environmental information to the photoperiodic flowering pathways in both short- and long-day plants.
SELECTION OF CLOCK GENE VARIANTS FOR FLOWERING TIME ADAPTATION
The highly integrated nature of the circadian clock with light and temperature response networks suggest that these genes and pathways play a central role in the ability of plants to adapt to their environment. Indeed, recent studies have found that natural variation in circadian clock genes has facilitated the migration and domestication of many different plant species (Bendix et al. 2015; Blümel et al. 2015). Genetic variation in a number of clock-related genes across different crop species have been valuable in expanding their growth range and yield (Bendix et al. 2015; Blümel et al. 2015). Many of these genes result in altered flowering time and photoperiod sensitivity (Table 1). In general, crops that originated in the tropics such as rice, sorghum, and maize are ancestrally short-day plants with flowering inhibited by long days. Many of these crops have been adapted to the long summer days at higher latitudes by breeding for photoperiod-insensitive variants that flower earlier under long days than ancestral genotypes (Hung et al. 2012). Soybean is also a short-day plant but was originally adapted to a limited latitudinal range. Expansion of soybean cultivation to higher latitudes has been enabled by selection for varieties that flower early in long days (Weller and Ortega 2015). Conversely, adaptation to more equatorial regions has been achieved by selection for cultivars that are less responsive to inductive short days, allowing more vegetative growth before the transition to flowering and thus increasing biomass and yield (Lu et al. 2017b). Many long-day crops such as wheat, barley, pea, and lentil are from temperate regions, and in the ancestral state flowering is promoted by long days (Cockram et al. 2007; Weller et al. 2012). In cereals, like wheat and barley, the long-day growth habit ensures the plants that germinate in the fall will flower as days lengthen in the spring, allowing for grain filling during the wet season and harvest before the hot dry summers. Selection for short rotation varieties that can be sown in the spring and harvested soon thereafter has enabled production of two successive crops each year, an innovation instrumental in the green revolution (Borlaug 1983; Cockram et al. 2007).
Table 1.
Clock alleles influencing important agronomic traits in different crop species grouped together by phenotype and allele
| Phenotype | Agriculturally important alleles of clock genes | Crop species | References |
|---|---|---|---|
| Early flowering | ELF3-like | Hordeum vulgare | Faure et al. 2012; Zakhrabekova et al. 2012 |
| Triticum aestivum | Wang et al. 2016a; Zikhali et al. 2016 | ||
| Triticum monococcum | Alvarez et al. 2016 | ||
| Pisum sativum | |||
| Lens culinaris | Weller et al. 2012 | ||
| ELF4-like | Pisum sativum | Liew et al. 2009 | |
| Zea mays | Li et al. 2016 | ||
| PRR-like | Sorghum bicolor | Murphy et al. 2011 | |
| Oryza sativa | Koo et al. 2013 | ||
| Triticum aestivum | Beales et al. 2007 | ||
| GI-like | Glycine max | Watanabe et al. 2011; Wang et al. 2016b | |
| Oryza sativa | Hayama et al. 2003 | ||
| Zea mays | Bendix et al. 2013 | ||
| LUX-like | Hordeum vulgare | Campoli et al. 2013 | |
| Triticum monococcum | Gawroński et al. 2014; Nishiura et al. 2018 | ||
| Pisum sativum | Liew et al. 2014 | ||
| Delayed flowering | ELF3-like | Glycine max | Lu et al. 2017a |
| Triticum monocuccum | Alvarez et al. 2016 | ||
| Oryza sativa | Saito et al. 2012; Zhao et al. 2012; Yang et al. 2013 | ||
| PRR-like | Oryza sativa | Murakami et al. 2005; Yan et al. 2013 | |
| Hordeum vulgare | Turner et al. 2005 | ||
| GI-like | Pisum sativum | Hecht et al. 2007 | |
| CCA1-like | Brassica rapa | Yi et al. 2017 | |
| LNK2-like | Solanum lycopersicum | Müller et al. 2018 | |
| Water stress | ELF3-like | Hordeum vulgare | Habte et al. 2014 |
| PRR-like | Hordeum vulgare | Habte et al. 2014 | |
| Glycine max | Syed et al. 2015 | ||
| LUX-like | Glycine max | Syed et al. 2015 | |
| Temperature stress | GI-like | Brassica rapa | Xie et al. 2015; Kim et al. 2016 |
| LUX-like | Triticum monococcum | Gawroński et al. 2014 | |
| CCA1-like | Brassica oleracea | Song et al. 2018 | |
| Biennial growth | PRR-like | Beta vulgaris | Pin et al. 2012 |
PRR Gene Variation Alters Photoperiodic Flowering and Expands Growth Range
In the model plant Arabidopsis, mutations in most of the PRR genes delay flowering in long days, the inductive photoperiodic condition (Nakamichi et al. 2007). PRR genes have been characterized in many monocot crop species (including rice, wheat, barley, sorghum, and maize); however, these genes have undergone independent duplications in the cereals leading to ambiguity in the evolutionary relationships between these genes across different species (Brambilla et al. 2017; Li and Xu 2017). To reflect this ambiguity, the PRR genes in cereals have been named PRR1, PRR37, PRR59, PRR73, and PRR95, indicating the closest Arabidopsis relative(s) for each gene (Campoli et al. 2012). Rice PRR genes are expressed in a sequential manner throughout the day and function as core components of the rice circadian clock in a similar manner to the PRR genes in Arabidopsis (Murakami et al. 2003, 2007).
In rice, analysis of the progeny resulting from a cross between cultivars with different photoperiodic sensitivity led to the identification of several heading date (HD) QTLs (quantitative trait loci). Two of these loci map near PRR-like genes (Murakami et al. 2005). One such locus is Ghd7.1/Hd2, which corresponds to the OsPRR37 gene. Although a functional version of this gene contributes to delayed flowering in long days, multiple nonfunctional alleles of this locus are present in Asian and European cultivars and are thought to contribute to adaptation of rice cultivation to higher latitudes (Koo et al. 2013; Yan et al. 2013). A knockout allele, with a T-DNA insertion within the OsPRR37 locus, causes early flowering owing to an increase in Hd3a (FT homolog) expression in long days (Koo et al. 2013). In barley, ancestrally a long-day plant, delayed flowering in normally inductive photoperiods was associated with genetic variation at the Ppd-H1 locus (also known as eam1) and was shown to be caused by a loss-of-function mutation in HvPRR73 that caused a decrease in HvFT expression (Turner et al. 2005), a phenotype similar to that seen in Arabidopsis prr7 mutants (Yamamoto et al. 2003). Conversely, in wheat, spring varieties have been selected that are photoperiod insensitive because of variation in an allele of PRR37 (the Ppd-D1a locus) that causes an increase in TaFT expression and early flowering in short days (Beales et al. 2007). A variant at the sorghum Ma1 locus, also a PRR37 homolog, was also found to significantly advance flowering time in normally noninductive long days (Murphy et al. 2011). How these different PRR alleles function on a molecular level in different crops is still under investigation but it is clear that this clock-related gene family has played an important role in extending the growth range of several important staple crop species.
The Role of the Evening Complex in Modification of Photoperiodic Flowering Requirements
Given the central role of ELF3 and the evening complex in the circadian clock, light-signaling, temperature-sensing, and photoperiodic flowering pathways (Figs. 1, 2), it is not surprising that evening complex genes have played an important role in the domestication of plants. In Arabidopsis, elf3 mutants were first identified based on their day-neutral flowering phenotype, flowering significantly earlier than wild-type in short days (Zagotta et al. 1996). Later studies provided evidence that mutations in any one of the three evening complex genes not only have major effects on clock function but also result in an early flowering phenotype (Hicks et al. 2001; Doyle et al. 2002; Hazen et al. 2005). Alleles of evening complex genes have been identified as the underlying cause for variation in photoperiod sensitivity and flowering time in multiple crop species (Table 1).
Variation of several genetic loci have been associated with differences in the photoperiodic flowering of barley, with some primarily affecting flowering time in long-day conditions and others primarily affecting flowering time in short-day conditions, such as eam 7 to 10 and Ppd-H2 (Boyd et al. 2003). eam8, which induces early flowering in short-day conditions, was identified as a homolog of Arabidopsis ELF3. Mutations in this gene disrupt clock function and increase HvFT expression, resulting in an early flowering phenotype in short days that is advantageous in regions with short growing seasons, such as Scandinavia (Faure et al. 2012; Zakhrabekova et al. 2012). Interestingly, although HvPRR37/ppd-H1 expression levels are elevated in eam8 mutants, early flowering is maintained in plants with both the eam8 and ppd-H1 alleles (Faure et al. 2012). These data suggest that although HvELF3 regulates HvPRR37 expression in barley, there is also an alternate pathway for its regulation of HvFT expression and flowering time.
An ELF3 homolog in pea (HR locus) and lentil has also been identified as a genetic factor underlying variation in flowering time (Weller and Ortega 2015). Similarly, in wheat, one of the genes underlying an earliness per se locus that regulates flowering time has been identified as an ELF3 homolog (Alvarez et al. 2016; Wang et al. 2016a). Most of these alleles confer the expected early flowering phenotype; however, the Eps-Am1-l allele appears to confer a late flowering phenotype (Alvarez et al. 2016). The different effects of these alleles appear to be light and temperature sensitive (Lewis et al. 2008), in keeping with the central role of ELF3 in these signaling pathways as described above. Similarly, in the short-day crop soybean, an ELF3 homolog has been identified at the J locus, an important regulator of flowering time. Loss-of-function alleles of j flower late in short days caused by the loss of inhibition of expression of the legume-specific E1 gene, which encodes a repressor of FT gene expression (Lu et al. 2017a). Importantly, the j allele has allowed the expansion of soybean cultivation to equatorial regions by extending the vegetative phase of development in short days. Finally, in rice, two orthologs of the Arabidopsis ELF3 gene have been identified. Mutation in the OsELF3-1 gene leads to a delay in flowering in both long and short days, although mutations in the duplicate gene have little or no effect on flowering (Saito et al. 2012; Zhao et al. 2012).
Unlike ELF3, there are very few ELF4 homologs characterized in crop species and this gene may not be present in all angiosperms. For example, there does not appear to be an ELF4 ortholog in rice (Izawa et al. 2003). In maize, a member of the DUF1313 protein family was found to have sequence similarity to ELF4 and this gene appeared to be a good marker for days to silking, suggesting it may be involved in photoperiodic flowering (Li et al. 2016). In pea, the DNE locus is homologous to Arabidopsis ELF4 and mutations in this locus result in early flowering in normally noninductive short days (Liew et al. 2009). Interestingly, although the dne allele causes early flowering similar to that seen in Arabidopsis elf4 mutants, clock function appears largely intact in dne plants, thus it is not clear whether DNE is a core part of the pea circadian clock (Liew et al. 2009).
A few homologs and allelic variants of LUX, the final evening complex component, have been identified in barley, wheat, and pea (Table 1). Campoli et al. (2013) found that the gene underlying the barley eam10 locus is a homolog of LUX and that this mutation disrupted the expression of core clock genes including the PRRs and CCA1. The eam10 region in barley and the Eps-3Am locus in spring wheat are syntenic and have been conserved across species (Gawroński et al. 2014). Eps-3Am was identified as a causal factor in a very early flowering wheat mutant with disrupted circadian rhythms and high TaFT expression. In pea, the STERILE NODES (SN) locus was identified as a LUX homolog and knockdown mutations in this gene produces an early flowering phenotype in short-day conditions (Liew et al. 2014). Genetic interactions between the SN (LUX), DNE (ELF4), and HR (ELF3) loci in pea suggest that the role of the evening complex is well conserved between pea and Arabidopsis (Liew et al. 2014). These findings indicate that alterations in the evening complex of the circadian clock have played a central role in the adaptation of crop species to wide latitudinal distributions.
Other Clock Gene Variants Influencing Photoperiodic Flowering in Crop Species
Alleles of GI have also played an important role in the development of crops that flower in a photoperiod-insensitive manner. Mutations in GI result in a delayed flowering phenotype in Arabidopsis under long days but cause no phenotype under short days (Araki and Komeda 1993). Similarly, in a pea mutant screen under long-day conditions, a delayed flowering allele (LATE BLOOMER 1) was identified and associated with a mutation in a pea GI homolog. This mutation drastically decreases the expression of the FT homolog PsFTL and thus delays flowering in a clock and light-dependent manner (Hecht et al. 2007). Rice mutant for GI also shows a late-flowering phenotype specifically in inductive short-day photoperiods (Izawa et al. 2011). Although maize, like rice, is ancestrally a short-day plant, GI mutants in this species display an early flowering phenotype under long days but no phenotype in short days (Bendix et al. 2013). This advance in flowering is the result of an early accumulation of conz1 (the maize CO homolog) and increased expression of zcn8 (a maize FT homolog) in these mutants, suggesting that GI acts to repress conz1 under long days (Bendix et al. 2013). A loss-of-function allele in a soybean GI homolog was identified as the gene underlying the e2 QTL that causes early flowering in field-grown plants (Watanabe et al. 2011). Taken together, these results indicate that GI plays an important role in the ability of plants to distinguish between long and short days and adapt to these growth conditions.
There are fewer reports of allelic variation in other clock genes such as LHY, CCA1, TOC1, PRR5, PRR9, RVEs, and LNK1 or LNK2 leading to photoperiod adaptation. In the Chinese cabbage and leafy varieties of Brassica rapa, early flowering generally leads to decreased productivity and yield. A recent study found that B. rapa cultivars with variable photoperiod sensitivities contained a great deal of sequence variation in the BrCCA1 homolog and several of these variations could be associated with a delayed flowering phenotype, suggesting that CCA1 is a good candidate for marker-assisted breeding in Brassica (Yi et al. 2017). In tomato, deletion of the LNK2 homolog likely enabled cultivation of this crop beyond its natural range to higher latitudes, perhaps by lengthening the period of the circadian clock (Müller et al. 2018). It will be interesting to see whether other clock genes have also played roles in major crop domestication events or whether these genes could be used to drive domestication in the future.
VARIATION IN CLOCK GENES ENABLES PLANT ADAPTATION TO STRESS
Recently, there have been reports of variation in clock genes playing a role in the ability of plants to respond to abiotic stress (Table 1). In natural populations of the annual plant, Mimulus guttatus, leaf movement rhythms were assessed to monitor clock function and revealed that clock period is correlated with latitude. Mimulus populations derived from more northerly latitudes tend to have longer periods than their southern counterparts; these altered rhythms are thought to promote local adaptation to the environment (Greenham et al. 2017). In barley, cultivars with variation in the HvPRR73 (Ppd-H1) and HvELF3 genes were subjected to osmotic stress. It was found that mutations in HvELF3 changed the phase and waveform of expression of stress-response genes, whereas HvPRR73 alleles affected the overall levels at which stress-response genes were expressed (Habte et al. 2014). A comparison of drought-tolerant and drought-susceptible soybean cultivars under drought conditions revealed differences in LUX gene expression. The tolerant cultivar showed a significant decrease in LUX expression during drought and reverted to normal levels on watering (Syed et al. 2015). In the same study, the investigators found that TOC1 and PRR7 homologs were phase shifted under drought and flooding conditions and a PRR3 homolog underwent significant alternate splicing during these stress events. Freezing tolerance in Brassica oleracea was associated with two BoCCA1 alleles, in which BoCCA1-1 was associated with freezing tolerance and BoCCA1-2 was linked to freezing susceptibility (Song et al. 2018). In wheat, cultivars from warmer climates have a higher degree of sequence variation within a LUX homolog than those from cooler regions, suggesting alterations in this locus may help adapt temperate cereals to warmer climes (Gawroński et al. 2014). Anthocyanins are also well known to play a role in abiotic stress responses, and the circadian clock pathway has long been linked to anthocyanin biosynthesis (Harmer et al. 2000). It has recently been reported that the LNK2 and RVE8 transcriptional regulators directly control expression of anthocyanin biosynthetic genes (Pérez-García et al. 2015). To date, no natural variation in these genes has been associated with anthocyanin biosynthesis and stress response, but this is an interesting area for future research.
In addition to the role of the clock in response to abiotic stress, the circadian system also affects plant–pathogen and plant–pest interactions (Seo and Mas 2015; Lu et al. 2017b). Plant susceptibility to a variety of insects and microbial pathogens is gated by the clock, presumably due, at least in part, to circadian regulation of the defense hormones salicylic and jasmonic acid (Wang et al. 2011; Goodspeed et al. 2012; Korneli et al. 2014; Ingle et al. 2015; Lu et al. 2017b). Genetic analyses have also shown links between clock genes and plant defenses, with perturbation of expression of the clock genes CCA1, LHY, or ELF3 in Arabidopsis, increasing susceptibility to bacterial, fungal, and oomycete attack (Bhardwaj et al. 2011; Wang et al. 2011; Zhang et al. 2013; Lu et al. 2017b) and the silencing of ZTL expression rendering wild tobacco more susceptible to a generalist herbivore (Li et al. 2018). Genome-wide association mapping in Arabidopsis identified LHY and LUX alleles as associated with Botrytis cinera infection traits such as lesion eccentricity and size (Corwin et al. 2016; Fordyce et al. 2018). Despite these clear links between the clock and immune responses, whether allelic variation in clock genes of cultivated plants affects biotic stress responses remains to be determined.
CONCLUDING REMARKS
This review has touched on several aspects of plant physiology that are known to be circadian regulated, but with a third of all Arabidopsis transcripts being circadian regulated it is likely there are many other ecologically and agronomically important processes regulated by the clock (Covington et al. 2008). It would be interesting to assess crop cultivars with known clock gene variants under different nutrient and environmental stresses to get a fuller picture of the role of the clock in response to different environmental challenges. A relatively unexplored area for future research is how clock-regulated processes in plants affect and are affected by clock-regulated processes in other organisms during plant–pathogen, plant–pollinator, and plant–microbiome interactions, and what the implications are for adaptation and domestication (Hevia et al. 2015; Yon et al. 2017; Fenske et al. 2018; Hubbard et al. 2018). It is clear that the plant circadian clock has a central role to play in adapting crops to the ever-changing environment; however, there remains a great deal we do not yet know about circadian clocks in different plant species and their roles in distinct environments. Finally, given the central role of the circadian clock in environmental signal perception and response, it is important to understand the trade-offs between different pathways when selecting for or targeting specific clock-related traits.
ACKNOWLEDGMENTS
This work was supported by awards from the National Institutes of Health (R01 GM069418), the National Science Foundation (IOS 1238040), and the U.S. Department of Agriculture–National Institute of Food and Agriculture (CA-D-PLB-2259-H).
Footnotes
Editor: Pamela C. Ronald
Additional Perspectives on Engineering Plants for Agriculture available at www.cshperspectives.org
REFERENCES
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. 2001. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883. 10.1126/science.1061320 [DOI] [PubMed] [Google Scholar]
- Alvarez MA, Tranquilli G, Lewis S, Kippes N, Dubcovsky J. 2016. Genetic and physical mapping of the earliness per se locus Eps-Am1 in Triticum monococcum identifies EARLY FLOWERING 3 (ELF3) as a candidate gene. Funct Integr Genomics 16: 365–382. 10.1007/s10142-016-0490-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C. 2004. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626. 10.1242/dev.01231 [DOI] [PubMed] [Google Scholar]
- Andrés F, Coupland G. 2012. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639. 10.1038/nrg3291 [DOI] [PubMed] [Google Scholar]
- Apelt F, Breuer D, Olas JJ, Annunziata MG, Flis A, Nikoloski Z, Kragler F, Stitt M. 2017. Circadian, carbon, and light control of expansion growth and leaf movement. Plant Physiol 174: 1949–1968. 10.1104/pp.17.00503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, Komeda Y. 1993. Analysis of the role of the late-flowering locus, GI, in the flowering of Arabidopsis thaliana. Plant J 3: 231–239. 10.1046/j.1365-313X.1993.t01-15-00999.x [DOI] [Google Scholar]
- Arana MV, Marín-de la Rosa N, Maloof JN, Blázquez MA, Alabadí D. 2011. Circadian oscillation of gibberellin signaling in Arabidopsis. Proc Natl Acad Sci 108: 9292–9297. 10.1073/pnas.1101050108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamian HS, Creux NM, Brown EA, Garner AG, Blackman BK, Harmer SL. 2016. Circadian regulation of sunflower heliotropism, floral orientation, and pollinator visits. Science 353: 587–590. 10.1126/science.aaf9793 [DOI] [PubMed] [Google Scholar]
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. 2007. A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115: 721–733. 10.1007/s00122-007-0603-4 [DOI] [PubMed] [Google Scholar]
- Bendix C, Mendoza JM, Stanley DN, Meeley R, Harmon FG. 2013. The circadian clock-associated gene gigantea1 affects maize developmental transitions. Plant Cell Environ 36: 1379–1390. 10.1111/pce.12067 [DOI] [PubMed] [Google Scholar]
- Bendix C, Marshall CM, Harmon FG. 2015. Circadian clock genes universally control key agricultural traits. Mol Plant 8: 1135–1152. 10.1016/j.molp.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Bhardwaj V, Meier S, Petersen LN, Ingle RA, Roden LC. 2011. Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS ONE 6: e26968 10.1371/journal.pone.0026968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blümel M, Dally N, Jung C. 2015. Flowering time regulation in crops—What did we learn from Arabidopsis? Curr Opin Biotechnol 32: 121–129. 10.1016/j.copbio.2014.11.023 [DOI] [PubMed] [Google Scholar]
- Borlaug NE. 1983. Contributions of conventional plant breeding to food production. Science 219: 689–693. 10.1126/science.219.4585.689 [DOI] [PubMed] [Google Scholar]
- Box MS, Huang BE, Domijan M, Jaeger KE, Khattak AK, Yoo SJ, Sedivy EL, Jones DM, Hearn TJ, Webb AA. 2015. ELF3 controls thermoresponsive growth in Arabidopsis. Curr Biol 25: 194–199. 10.1016/j.cub.2014.10.076 [DOI] [PubMed] [Google Scholar]
- Boyd WJR, Li CD, Grime CR, Cakir M, Potipibool S, Kaveeta L, Men S, Kamali MJ, Barr AR, Moody DB. 2003. Conventional and molecular genetic analysis of factors contributing to variation in the timing of heading among spring barley (Hordeum vulgare L.) genotypes grown over a mild winter growing season. Aust J Agric Res 54: 1277–1301. 10.1071/AR03014 [DOI] [Google Scholar]
- Brambilla V, Gomez-Ariza J, Cerise M, Fornara F. 2017. The importance of being on time: Regulatory networks controlling photoperiodic flowering in cereals. Front Plant Sci 8: 665 10.3389/fpls.2017.00665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli C, Shtaya M, Davis SJ, von Korff M. 2012. Expression conservation within the circadian clock of a monocot: Natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biol 12: 97 10.1186/1471-2229-12-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli C, Pankin A, Drosse B, Casao CM, Davis SJ, von Korff M. 2013. HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: Phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol 199: 1045–1059. 10.1111/nph.12346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Qüesta JI. 2018. Light and temperature cues: Multitasking receptors and transcriptional integrators. New Phytol 217: 1029–1034. 10.1111/nph.14890 [DOI] [PubMed] [Google Scholar]
- Chow BY, Sanchez SE, Breton G, Pruneda-Paz JL, Krogan NT, Kay SA. 2014. Transcriptional regulation of LUX by CBF1 mediates cold input to the circadian clock in Arabidopsis. Curr Biol 24: 1518–1524. 10.1016/j.cub.2014.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockram J, Jones H, Leigh FJ, O'Sullivan D, Powell W, Laurie DA, Greenland AJ. 2007. Control of flowering time in temperate cereals: Genes, domestication, and sustainable productivity. J Exp Bot 58: 1231–1244. 10.1093/jxb/erm042 [DOI] [PubMed] [Google Scholar]
- Corwin JA, Copeland D, Feusier J, Subedy A, Eshbaugh R, Palmer C, Maloof J, Kliebenstein DJ. 2016. The quantitative basis of the Arabidopsis innate immune system to endemic pathogens depends on pathogen genetics. PLoS Genet 12: e1005789 10.1371/journal.pgen.1005789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Harmer SL. 2007. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5: e222 10.1371/journal.pbio.0050222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. 2008. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9: R130 10.1186/gb-2008-9-8-r130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C, van Zanten M, Quint M. 2017. Thermosensing enlightened. Trends Plant Sci 22: 185–187. 10.1016/j.tplants.2017.01.007 [DOI] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognár L, Nagy F, Millar AJ, Amasino RM. 2002. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77. 10.1038/nature00954 [DOI] [PubMed] [Google Scholar]
- Endo M. 2016. Tissue-specific circadian clocks in plants. Curr Opin Plant Biol 29: 44–49. 10.1016/j.pbi.2015.11.003 [DOI] [PubMed] [Google Scholar]
- Ezer D, Jung JH, Lan H, Biswas S, Gregoire L, Box MS, Charoensawan V, Cortijo S, Lai X, Stöckle D. 2017. The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat Plants 3: 17087 10.1038/nplants.2017.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinas B, Mas P. 2011. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J 66: 318–329. 10.1111/j.1365-313X.2011.04484.x [DOI] [PubMed] [Google Scholar]
- Farré EM. 2012. The regulation of plant growth by the circadian clock. Plant Biol 14: 401–410. 10.1111/j.1438-8677.2011.00548.x [DOI] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. 2005. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54. 10.1016/j.cub.2004.12.067 [DOI] [PubMed] [Google Scholar]
- Faure S, Turner AS, Gruszka D, Christodoulou V, Davis SJ, von Korff M, Laurie DA. 2012. Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc Natl Acad Sci 109: 8328–8333. 10.1073/pnas.1120496109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenske MP, Nguyen LP, Horn EK, Riffell JA, Imaizumi T. 2018. Circadian clocks of both plants and pollinators influence flower seeking behavior of the pollinator hawkmoth Manduca sexta. Sci Rep 8: 2842 10.1038/s41598-018-21251-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordyce RF, Soltis NE, Caseys C, Gwinner R, Corwin JA, Atwell S, Copeland D, Feusier J, Subedy A, Eshbaugh R, et al. 2018. Digital imaging combined with genome-wide association mapping links loci to plant–pathogen interaction traits. Plant Physiol 178: 1406–1422. 10.1104/pp.18.00851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J, Johansson H, Hornitschek P, Josse EM, Fankhauser C, Halliday KJ. 2011. Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J 65: 441–452. 10.1111/j.1365-313X.2010.04434.x [DOI] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G. 2009. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17: 75–86. 10.1016/j.devcel.2009.06.015 [DOI] [PubMed] [Google Scholar]
- Gawroński P, Ariyadasa R, Himmelbach A, Poursarebani N, Kilian B, Stein N, Steuernagel B, Hensel G, Kumlehn J, Sehgal SK. 2014. A distorted circadian clock causes early flowering and temperature-dependent variation in spike development in the Eps-3Am mutant of einkorn wheat. Genetics 113: 158444 10.1534/genetics.113.158444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil KE, Park CM. 2019. Thermal adaptation and plasticity of the plant circadian clock. New Phytol 221: 1215–1229. 10.1111/nph.15518 [DOI] [PubMed] [Google Scholar]
- Goodspeed D, Chehab EW, Min-Venditti A, Braam J, Covington MF. 2012. Cozzarelli Prize Winner: Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc Natl Acad Sci 109: 4674–4677. 10.1073/pnas.1116368109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenham K, McClung CR. 2015. Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet 16: 598–610. 10.1038/nrg3976 [DOI] [PubMed] [Google Scholar]
- Greenham K, Lou P, Puzey JR, Kumar G, Arnevik C, Farid H, Willis JH, McClung CR. 2017. Geographic variation of plant circadian clock function in natural and agricultural settings. J Biol Rhythms 32: 26–34. 10.1177/0748730416679307 [DOI] [PubMed] [Google Scholar]
- Habte E, Müller LM, Shtaya M, Davis SJ, von Korff M. 2014. Osmotic stress at the barley root affects expression of circadian clock genes in the shoot. Plant Cell Environ 37: 1321–1337. 10.1111/pce.12242 [DOI] [PubMed] [Google Scholar]
- Harmer SL. 2009. The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377. 10.1146/annurev.arplant.043008.092054 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Kay SA. 2005. Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17: 1926–1940. 10.1105/tpc.105.033035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. 2000. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113. 10.1126/science.290.5499.2110 [DOI] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. 2003. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722. 10.1038/nature01549 [DOI] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA. 2005. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci 102: 10387–10392. 10.1073/pnas.0503029102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Knowles CL, Vander Schoor JK, Liew LC, Jones SE, Lambert MJ, Weller JL. 2007. Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol 144: 648–661. 10.1104/pp.107.096818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Papdi C, Ahmad Z, Bögre L. 2018. Circadian regulation of plant growth. In Annual plant reviews online (ed. Roberts JA). Wiley, Hoboken, NJ: 10.1002/9781119312994.apr0655 [DOI] [Google Scholar]
- Hevia MA, Canessa P, Müller-Esparza H, Larrondo LF. 2015. A circadian oscillator in the fungus Botrytis cinerea regulates virulence when infecting Arabidopsis thaliana. Proc Natl Acad Sci 112: 8744–8749. 10.1073/pnas.1508432112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Albertson TM, Wagner DR. 2001. EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13: 1281–1292. 10.1105/tpc.13.6.1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Matsubara K, Yano M. 2016. Genetic control of flowering time in rice: Integration of Mendelian genetics and genomics. Theor Appl Genet 129: 2241–2252. 10.1007/s00122-016-2773-4 [DOI] [PubMed] [Google Scholar]
- Hsu PY, Harmer SL. 2014. Wheels within wheels: The plant circadian system. Trends Plant Sci 19: 240–249. 10.1016/j.tplants.2013.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Devisetty UK, Harmer SL. 2013. Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2: e00473 10.7554/eLife.00473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Nusinow DA. 2016. Into the evening: Complex interactions in the Arabidopsis circadian clock. Trends Genet 32: 674–686. 10.1016/j.tig.2016.08.002 [DOI] [PubMed] [Google Scholar]
- Huang H, Alvarez S, Bindbeutel R, Shen Z, Naldrett MJ, Evans BS, Briggs SP, Hicks LM, Kay SA, Nusinow DA. 2016. Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol Cell Proteomics 15: 201–217. 10.1074/mcp.M115.054064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard CJ, Brock MT, van Diepen LT, Maignien L, Ewers BE, Weinig C. 2018. The plant circadian clock influences rhizosphere community structure and function. ISME J 12: 400–410. 10.1038/ismej.2017.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung HY, Shannon LM, Tian F, Bradbury PJ, Chen C, Flint-Garcia SA, McMullen MD, Ware D, Buckler ES, Doebley JF, et al. 2012. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc Natl Acad Sci 109: E1913–E1921. 10.1073/pnas.1203189109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. 2005. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297. 10.1126/science.1110586 [DOI] [PubMed] [Google Scholar]
- Ingle RA, Stoker C, Stone W, Adams N, Smith R, Grant M, Carré I, Roden LC, Denby KJ. 2015. Jasmonate signalling drives time-of-day differences in susceptibility of Arabidopsis to the fungal pathogen Botrytis cinerea. Plant J 84: 937–948. 10.1111/tpj.13050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T. 2007. Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J Exp Bot 58: 3091–3097. 10.1093/jxb/erm159 [DOI] [PubMed] [Google Scholar]
- Izawa T, Takahashi Y, Yano M. 2003. Comparative biology comes into bloom: Genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr Opin Plant Biol 6: 113–120. 10.1016/S1369-5266(03)00014-1 [DOI] [PubMed] [Google Scholar]
- Izawa T, Mihara M, Suzuki Y, Gupta M, Itoh H, Nagano AJ, Motoyama R, Sawada Y, Yano M, Hirai MY. 2011. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell 23: 1741–1755. 10.1105/tpc.111.083238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S. 2016. Phytochromes function as thermosensors in Arabidopsis. Science 354: 886–889. 10.1126/science.aaf6005 [DOI] [PubMed] [Google Scholar]
- Kamioka M, Takao S, Suzuki T, Taki K, Higashiyama T, Kinoshita T, Nakamichi N. 2016. Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED 1 in Arabidopsis circadian clock. Plant Cell 28: 696–711. 10.1105/tpc.15.00737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke M, Gao Z, Chen J, Qiu Y, Zhang L, Chen X. 2018. Auxin controls circadian flower opening and closure in the waterlily. BMC Plant Biol 18: 143 10.1186/s12870-018-1357-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro S, Maruyama K, Nakashima K, Imura Y, Narusaka Y, Shinwari ZK, Osakabe Y, Fujita Y, Mizoi J, Shinozaki K, et al. 2009. The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol 151: 2046–2057. 10.1104/pp.109.147033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Song HR, Taylor BL, Carré IA. 2003. Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J 22: 935–944. 10.1093/emboj/cdg075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE. 2007. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360. 10.1038/nature06132 [DOI] [PubMed] [Google Scholar]
- Kim J, Geng R, Gallenstein RA, Somers DE. 2013. The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development 140: 4060–4069. 10.1242/dev.096651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JA, Jung H, Hong JK, Hermand V, McClung CR, Lee YH, Kim JY, Lee SI, Jeong MJ, Kim J. 2016. Reduction of GIGANTEA expression in transgenic Brassica rapa enhances salt tolerance. Plant Cell Rep 35: 1943–1954. 10.1007/s00299-016-2008-9 [DOI] [PubMed] [Google Scholar]
- Kinmonth-Schultz HA, Golembeski GS, Imaizumi T. 2013. Circadian clock-regulated physiological outputs: dynamic responses in nature. Semin Cell Dev Biol 24: 407–413. 10.1016/j.semcdb.2013.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BH, Yoo SC, Park JW, Kwon CT, Lee BD, An G, Zhang Z, Li J, Li Z, Paek NC. 2013. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol Plant 6: 1877–1888. 10.1093/mp/sst088 [DOI] [PubMed] [Google Scholar]
- Korneli C, Danisman S, Staiger D. 2014. Differential control of pre-invasive and post-invasive antibacterial defense by the Arabidopsis circadian clock. Plant Cell Physiol 55: 1613–1622. 10.1093/pcp/pcu092 [DOI] [PubMed] [Google Scholar]
- Larner VS, Franklin KA, Whitelam GC. 2018. Photoreceptors and light signalling pathways in plants. In Annual plant reviews online (ed. Roberts JA). Wiley, Hoboken, NJ: 10.1002/9781119312994 [DOI] [Google Scholar]
- Lee CM, Thomashow MF. 2012. Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc Natl Acad Sci 109: 15054–15059. 10.1073/pnas.1211295109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie ES, Rojas CCR, Neme M, Hiltbrunner A, Wigge PA, Schäfer E, Vierstra RD, Casal JJ. 2016. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354: 897–900. 10.1126/science.aaf5656 [DOI] [PubMed] [Google Scholar]
- Lewis S, Faricelli ME, Appendino ML, Valárik M, Dubcovsky J. 2008. The chromosome region including the earliness per se locus Eps-Am1 affects the duration of early developmental phases and spikelet number in diploid wheat. J Exp Bot 59: 3595–3607. 10.1093/jxb/ern209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu M. 2017. CCT family genes in cereal crops: A current overview. Crop J 5: 449–458. 10.1016/j.cj.2017.07.001 [DOI] [Google Scholar]
- Li J, Hu E, Chen X, Xu J, Lan H, Li C, Hu Y, Lu Y. 2016. Evolution of DUF1313 family members across plant species and their association with maize photoperiod sensitivity. Genomics 107: 199–207. 10.1016/j.ygeno.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Li R, Llorca LC, Schuman MC, Wang Y, Wang L, Joo Y, Wang M, Vassão DG, Baldwin IT. 2018. ZEITLUPE in the roots of wild tobacco regulates jasmonate-mediated nicotine biosynthesis and resistance to a generalist herbivore. Plant Physiol 177: 833–846. 10.1104/pp.18.00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew LC, Hecht V, Laurie RE, Knowles CL, Vander Schoor JK, Macknight RC, Weller JL. 2009. DIE NEUTRALIS and LATE BLOOMER 1 contribute to regulation of the pea circadian clock. Plant Cell 21: 3198–3211. 10.1105/tpc.109.067223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew LC, Hecht V, Sussmilch FC, Weller JL. 2014. The pea photoperiod response gene STERILE NODES is an ortholog of LUX ARRHYTHMO. Plant Physiol 165: 648–657. 10.1104/pp.114.237008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR. 2001. ELF3 encodes a circadian clock–regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304. 10.1105/tpc.13.6.1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TL, Newton L, Liu MJ, Shiu SH, Farré EM. 2016. A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol 170: 528–539. 10.1104/pp.15.01562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Zhao X, Hu Y, Liu S, Nan H, Li X, Fang C, Cao D, Shi X, Kong L. 2017a. Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat Genet 49: 773–779. 10.1038/ng.3819 [DOI] [PubMed] [Google Scholar]
- Lu H, McClung CR, Zhang C. 2017b. Tick tock: Circadian regulation of plant innate immunity. Annu Rev Phytopathol 55: 287–311. 10.1146/annurev-phyto-080516-035451 [DOI] [PubMed] [Google Scholar]
- Martín G, Rovira A, Veciana N, Soy J, Toledo-Ortiz G, Gommers CM, Boix M, Henriques R, Minguet EG, Alabadí D, et al. 2018. Circadian waves of transcriptional repression shape PIF-regulated photoperiod-responsive growth in Arabidopsis. Curr Biol 28: 311–318.e5. 10.1016/j.cub.2017.12.021 [DOI] [PubMed] [Google Scholar]
- Martínez-García JF, Huq E, Quail PH. 2000. Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863. 10.1126/science.288.5467.859 [DOI] [PubMed] [Google Scholar]
- Más P, Kim WY, Somers DE, Kay SA. 2003. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570. 10.1038/nature02163 [DOI] [PubMed] [Google Scholar]
- McClung CR. 2006. Plant circadian rhythms. Plant Cell 18: 792–803. 10.1105/tpc.106.040980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ. 2016. The intracellular dynamics of circadian clocks reach for the light of ecology and evolution. Annu Rev Plant Biol 67: 595–618. 10.1146/annurev-arplant-043014-115619 [DOI] [PubMed] [Google Scholar]
- Mizuno T, Kitayama M, Takayama C, Yamashino T. 2015. Insight into a physiological role for the EC night-time repressor in the Arabidopsis circadian clock. Plant Cell Physiol 56: 1738–1747. 10.1093/pcp/pcv094 [DOI] [PubMed] [Google Scholar]
- Müller NA, Zhang L, Koornneef M, Jiménez-Gómez JM. 2018. Mutations in EID1 and LNK2 caused light-conditional clock deceleration during tomato domestication. Proc Natl Acad Sci 115: 7135–7140. 10.1073/pnas.1801862115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Ashikari M, Miura K, Yamashino T, Mizuno T. 2003. The evolutionarily conserved OsPRR quintet: Rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol 44: 1229–1236. 10.1093/pcp/pcg135 [DOI] [PubMed] [Google Scholar]
- Murakami M, Matsushika A, Ashikari M, Yamashino T, Mizuno T. 2005. Circadian-associated rice pseudo response regulators (OsPRRs): Insight into the control of flowering time. Biosci Biotechnol Biochem 69: 410–414. 10.1271/bbb.69.410 [DOI] [PubMed] [Google Scholar]
- Murakami M, Tago Y, Yamashino T, Mizuno T. 2007. Characterization of the rice circadian clock-associated pseudo-response regulators in Arabidopsis thaliana. Biosci Biotechnol Biochem 71: 1107–1110. 10.1271/bbb.70048 [DOI] [PubMed] [Google Scholar]
- Murphy RL, Klein RR, Morishige DT, Brady JA, Rooney WL, Miller FR, Dugas DV, Klein PE, Mullet JE. 2011. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc Natl Acad Sci 108: 16469–16474. 10.1073/pnas.1106212108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Niinuma K, Ito S, Yamashino T, Mizoguchi T, Mizuno T. 2007. Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol 48: 822–832 10.1093/pcp/pcm056 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H. 2010. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605. 10.1105/tpc.109.072892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T. 2012. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci 109: 17123–17128. 10.1073/pnas.1205156109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C, López-Salmerón V, Davière JM, Prat S. 2015. ELF3–PIF4 interaction regulates plant growth independently of the evening complex. Curr Biol 25: 187–193. 10.1016/j.cub.2014.10.070 [DOI] [PubMed] [Google Scholar]
- Nishiura A, Kitagawa S, Matsumura M, Kazama Y, Abe T, Mizuno N, Nasuda S, Murai K. 2018. An early-flowering einkorn wheat mutant with deletions of PHYTOCLOCK 1/LUX ARRHYTHMO and VERNALIZATION 2 exhibits a high level of VERNALIZATION 1 expression induced by vernalization. J Plant Physiol 222: 28–38. 10.1016/j.jplph.2018.01.002 [DOI] [PubMed] [Google Scholar]
- Nozue K, Maloof JN. 2006. Diurnal regulation of plant growth. Plant Cell Environ 29: 396–408. 10.1111/j.1365-3040.2005.01489.x [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. 2007. Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361. 10.1038/nature05946 [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA. 2011. The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402. 10.1038/nature10182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakenfull RJ, Davis SJ. 2017. Shining a light on the Arabidopsis circadian clock. Plant Cell Environ 40: 2571–2585. 10.1111/pce.13033 [DOI] [PubMed] [Google Scholar]
- Paik I, Kathare PK, Kim JI, Huq E. 2017. Expanding roles of PIFs in signal integration from multiple processes. Mol Plant 10: 1035–1046. 10.1016/j.molp.2017.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Park J, Kim J, Nagatani A, Lagarias JC, Choi G. 2012. Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J 72: 537–546. 10.1111/j.1365-313X.2012.05114.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-García P, Ma Y, Yanovsky MJ, Mas P. 2015. Time-dependent sequestration of RVE8 by LNK proteins shapes the diurnal oscillation of anthocyanin biosynthesis. Proc Natl Acad Sci 112: 5249–5253. 10.1073/pnas.1420792112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VN, Kathare PK, Huq E. 2018. Phytochromes and phytochrome interacting factors. Plant Physiol 176: 1025–1038. 10.1104/pp.17.01384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin PA, Zhang W, Vogt SH, Dally N, Büttner B, Schulze-Buxloh G, Jelly NS, Chia TY, Mutasa-Göttgens ES, Dohm JC. 2012. The role of a pseudo-response regulator gene in life cycle adaptation and domestication of beet. Curr Biol 22: 1095–1101. 10.1016/j.cub.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Polko JK, van Zanten M, van Rooij JA, Marée AF, Voesenek LA, Peeters AJ, Pierik R. 2012. Ethylene-induced differential petiole growth in Arabidopsis thaliana involves local microtubule reorientation and cell expansion. New Phytol 193: 339–348. 10.1111/j.1469-8137.2011.03920.x [DOI] [PubMed] [Google Scholar]
- Rauf M, Arif M, Fisahn J, Xue G-P, Balazadeh S, Mueller-Roeber B. 2013. NAC transcription factor SPEEDY HYPONASTIC GROWTH regulates flooding-induced leaf movement in Arabidopsis. Plant Cell 25: 4941–4955. 10.1105/tpc.113.117861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, Phinney BS, Harmer SL. 2011. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet 7: e1001350 10.1371/journal.pgen.1001350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson FC, Skeffington AW, Gardner MJ, Webb AAR. 2009. Interactions between circadian and hormonal signalling in plants. Plant Mol Biol 69: 419–427. 10.1007/s11103-008-9407-4 [DOI] [PubMed] [Google Scholar]
- Rugnone ML, Soverna AF, Sanchez SE, Schlaen RG, Hernando CE, Seymour DK, Mancini E, Chernomoretz A, Weigel D, Más P. 2013. LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc Natl Acad Sci 110: 12120–12125. 10.1073/pnas.1302170110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Ogiso-Tanaka E, Okumoto Y, Yoshitake Y, Izumi H, Yokoo T, Matsubara K, Hori K, Yano M, Inoue H, et al. 2012. Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short- and long-day conditions. Plant Cell Physiol 53: 717–728. 10.1093/pcp/pcs029 [DOI] [PubMed] [Google Scholar]
- Sawa M, Kay SA. 2011. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc Natl Acad Sci 108: 11698–11703. 10.1073/pnas.1106771108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluzicki A, Burko Y, Chory J. 2017. Dancing in the dark: Darkness as a signal in plants. Plant Cell Environ 40: 2487–2501. 10.1111/pce.12900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Mas P. 2015. STRESSing the role of the plant circadian clock. Trends Plant Sci 20: 230–237. 10.1016/j.tplants.2015.01.001 [DOI] [PubMed] [Google Scholar]
- Shalit-Kaneh A, Kumimoto RW, Filkov V, Harmer SL. 2018. Multiple feedback loops of the Arabidopsis circadian clock provide rhythmic robustness across environmental conditions. Proc Natl Acad Sci 115: 7147-7152. 10.1073/pnas.1805524115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E, Paik I, Kangisser S, Green R, Huq E. 2017. PHYTOCHROME INTERACTING FACTORS mediate metabolic control of the circadian system in Arabidopsis. New Phytol 215: 217–228. 10.1111/nph.14579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T. 2015. Photoperiodic flowering: Time measurement mechanisms in leaves. Annu Rev Plant Biol 66: 441–464. 10.1146/annurev-arplant-043014-115555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Yi H, Han CT, Park JI, Hur Y. 2018. Allelic variation in Brassica oleracea CIRCADIAN CLOCK ASSOCIATED 1 (BoCCA1) is associated with freezing tolerance. Hortic Environ Biotechnol 59: 423–434. 10.1007/s13580-018-0045-8 [DOI] [Google Scholar]
- Soy J, Leivar P, González-Schain N, Sentandreu M, Prat S, Quail PH, Monte E. 2012. Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J 71: 390–401. 10.1111/j.1365-313x.2012.04992.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J, Leivar P, González-Schain N, Martín G, Diaz C, Sentandreu M, Al-Sady B, Quail PH, Monte E. 2016. Molecular convergence of clock and photosensory pathways through PIF3–TOC1 interaction and co-occupancy of target promoters. Proc Natl Acad Sci 113: 4870–4875. 10.1073/pnas.1603745113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed NH, Prince SJ, Mutava RN, Patil G, Li S, Chen W, Babu V, Joshi T, Khan S, Nguyen HT. 2015. Core clock, SUB1, and ABAR genes mediate flooding and drought responses via alternative splicing in soybean. J Exp Bot 66: 7129–7149. 10.1093/jxb/erv407 [DOI] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. 2005. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034. 10.1126/science.1117619 [DOI] [PubMed] [Google Scholar]
- Viczián A, Klose C, Ádám É, Nagy F. 2017. New insights of red light-induced development. Plant Cell Environ 40: 2457–2468. 10.1111/pce.12880 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. 1998. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217. 10.1016/S0092-8674(00)81464-6 [DOI] [PubMed] [Google Scholar]
- Wang W, Barnaby JY, Tada Y, Li H, Tör M, Caldelari D, Lee D, Fu XD, Dong X. 2011. Timing of plant immune responses by a central circadian regulator. Nature 470: 110–114. 10.1038/nature09766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wen W, Hanif M, Xia X, Wang H, Liu S, Liu J, Yang L, Cao S, He Z. 2016a. TaELF3-1DL, a homolog of ELF3, is associated with heading date in bread wheat. Mol Breed 36: 161 10.1007/s11032-016-0585-5 [DOI] [Google Scholar]
- Wang Y, Gu Y, Gao H, Qiu L, Chang R, Chen S, He C. 2016b. Molecular and geographic evolutionary support for the essential role of GIGANTEAa in soybean domestication of flowering time. BMC Evol Biol 16: 79 10.1186/s12862-016-0653-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Xia Z, Hideshima R, Tsubokura Y, Sato S, Yamanaka N, Takahashi R, Anai T, Tabata S, Kitamura K. 2011. A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188: 395–407. 10.1534/genetics.110.125062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Ortega R. 2015. Genetic control of flowering time in legumes. Front Plant Sci 6: 207 10.3389/fpls.2015.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Liew LC, Hecht VF, Rajandran V, Laurie RE, Ridge S, Wenden B, Vander Schoor JK, Jaminon O, Blassiau C. 2012. A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc Natl Acad Sci 109: 21158–21163. 10.1073/pnas.1207943110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whippo CW, Hangarter RP. 2009. The “sensational” power of movement in plants: A Darwinian system for studying the evolution of behavior. Am J Bot 96: 2115–2127. 10.3732/ajb.0900220 [DOI] [PubMed] [Google Scholar]
- Xie Q, Wang P, Liu X, Yuan L, Wang L, Zhang C, Li Y, Xing H, Zhi L, Yue Z. 2014. LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 26: 2843–2857. 10.1105/tpc.114.126573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Lou P, Hermand V, Aman R, Park HJ, Yun DJ, Kim WY, Salmela MJ, Ewers BE, Weinig C. 2015. Allelic polymorphism of GIGANTEA is responsible for naturally occurring variation in circadian period in Brassica rapa. Proc Natl Acad Sci 112: 3829–3834. 10.1073/pnas.1421803112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Sato E, Shimizu T, Nakamich N, Sato S, Kato T, Tabata S, Nagatani A, Yamashino T, Mizuno T. 2003. Comparative genetic studies on the APRR5 and APRR7 genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis. Plant Cell Physiol 44: 1119–1130. 10.1093/pcp/pcg148 [DOI] [PubMed] [Google Scholar]
- Yan W, Liu H, Zhou X, Li Q, Zhang J, Lu L, Liu T, Liu H, Zhang C, Zhang Z. 2013. Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res 23: 969–971. 10.1038/cr.2013.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Peng Q, Chen GX, Li XH, Wu CY. 2013. OsELF3 is involved in circadian clock regulation for promoting flowering under long-day conditions in rice. Mol Plant 6: 202–215. 10.1093/mp/sss062 [DOI] [PubMed] [Google Scholar]
- Yi H, Li X, Lee SH, Nou IS, Lim YP, Hur Y. 2017. Natural variation in CIRCADIAN CLOCK ASSOCIATED 1 is associated with flowering time in Brassica rapa. Genome 60: 402–413. 10.1139/gen-2016-0052 [DOI] [PubMed] [Google Scholar]
- Yon F, Kessler D, Joo Y, Kim SG, Baldwin IT. 2017. Fitness consequences of a clock pollinator filter in Nicotiana attenuata flowers in nature. J Integr Plant Biol 59: 805–809. 10.1111/jipb.12579 [DOI] [PubMed] [Google Scholar]
- Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML, Sullivan JA. 2008. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32: 617–630. 10.1016/j.molcel.2008.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, Meeks-Wagner DR. 1996. The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J 10: 691–702. 10.1046/j.1365-313X.1996.10040691.x [DOI] [PubMed] [Google Scholar]
- Zakhrabekova S, Gough SP, Braumann I, Müller AH, Lundqvist J, Ahmann K, Dockter C, Matyszczak I, Kurowska M, Druka A, et al. 2012. Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc Natl Acad Sci 109: 4326–4331. 10.1073/pnas.1113009109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Xie Q, Anderson RG, Ng G, Seitz NC, Peterson T, McClung CR, McDowell JM, Kong D, Kwak JM, et al. 2013. Crosstalk between the circadian clock and innate immunity in Arabidopsis. PLoS Pathog 9: e1003370 10.1371/journal.ppat.1003370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Huang X, Ouyang X, Chen W, Du A, Zhu L, Wang S, Deng XW, Li S. 2012. OsELF3-1, an ortholog of Arabidopsis early flowering 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS ONE 7: e43705 10.1371/journal.pone.0043705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Oh E, Wang T, Wang ZY. 2016. TOC1–PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat Commun 7: 13692 10.1038/ncomms13692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zikhali M, Wingen LU, Griffiths S. 2016. Delimitation of the Earliness per se D1 (Eps-D1) flowering gene to a subtelomeric chromosomal deletion in bread wheat (Triticum aestivum). J Exp Bot 67: 287–299. 10.1093/jxb/erv458 [DOI] [PMC free article] [PubMed] [Google Scholar]