Abstract
Most of the secreted and plasma membrane proteins are synthesized on membrane-bound ribosomes on the endoplasmic reticulum (ER). They require engagement of ER-resident chaperones and foldases that assist in their folding and maturation. Since protein homeostasis in the ER is crucial for cellular function, the protein-folding status in the organelle's lumen is continually surveyed by a network of signaling pathways, collectively called the unfolded protein response (UPR). Protein-folding imbalances, or “ER stress,” are detected by highly conserved sensors that adjust the ER's protein-folding capacity according to the physiological needs of the cell. We review recent developments in the field that have provided new insights into the ER stress-sensing mechanisms used by UPR sensors and the mechanisms by which they integrate various cellular inputs to adjust the folding capacity of the organelle to accommodate to fluctuations in ER protein-folding demands.
THE ENDOPLASMIC RETICULUM AS A PROTEIN-FOLDING COMPARTMENT
All eukaryotic cells contain an endoplasmic reticulum (ER), a labyrinthine membranous network of sheets and tubules. Like all membrane-enclosed organelles, the ER allows the compartmentalization of essential cellular functions. For the ER, this includes the folding and maturation of the majority of secreted and membrane proteins. The ER is thought to have evolved by invagination of the plasma membrane, making the lumen of the ER topologically equivalent to the outside of the cell (Devos et al. 2004; Baum and Baum 2014). For this reason, the ER lumen and its proteome possess unique biochemical characteristics. The ER client and resident proteins are subjected to specific co- and posttranslational modifications, including various forms of glycosylation and disulfide bond formation (Helenius et al. 1992; Braakman and Bulleid 2011; Feige and Hendershot 2011; Gidalevitz et al. 2013; Ellgaard et al. 2016). Most of these proteins cross or become embedded in the ER membrane as they are being synthesized on the cytosolic surface of the ER by ribosomes targeted to the organelle by the signal recognition particle (Walter and Blobel 1980; Walter et al. 1982). In this cotranslational protein-targeting mechanism, the nascent polypeptides enter the ER lumen through a specialized protein-conducting channel, the Sec61 translocon, and they are folded as they grow by ER-resident chaperones (e.g., BiP and Grp94) and foldases (e.g., protein disulfide isomerases [PDIs]) that assist distinct steps in their maturation (Hammond et al. 1994; Feige and Hendershot 2011; Gidalevitz et al. 2013). Multiple maturation steps go hand-in-hand with quality-control pathways that ensure that only correctly folded client proteins reach their final destinations (Kleizen and Braakman 2004; Anelli and Sitia 2008; Barlowe and Helenius 2016).
About one-third of the human proteome consists of secreted or transmembrane proteins targeted to the ER. In steady-state conditions, the ER faces a constant influx of client proteins on the order of 0.1–1 million molecules per minute per cell (Van Cauter et al. 1992; Bromage et al. 2009; Costantini and Snapp 2013). The complex three-dimensional architecture of the ER decreases the molecule diffusion rate up to threefold relative to that of the cytoplasm (Dayel et al. 1999; Siggia et al. 2000; Sbalzarini et al. 2006). Together, these high biosynthetic rates and complex organellar architecture present a major biophysical challenge to the ER chaperones and foldases that assist the folding of client ER proteins. Therefore, to ensure protein-folding homeostasis, the ER relies on mechanisms that detect protein-folding imbalances. Such proteostasis surveillance mechanisms adjust the protein-folding capacity of the organelle to accommodate to fluctuations in protein-folding demands imposed by the physiology of cells and tissues.
THE UNFOLDED PROTEIN RESPONSE ADJUSTS THE PROTEIN-FOLDING CAPACITY OF THE ER
The protein-folding status within the ER lumen is continuously monitored by a set of evolutionarily conserved signaling pathways, collectively known as the unfolded protein response (UPR) (Cox et al. 1993; Cox and Walter 1996; Sidrauski and Walter 1997; Yoshida et al. 1998, 2001; Niwa et al. 1999; Harding et al. 2000; Tirasophon et al. 2000). When the ER protein-folding capacity is exceeded or if dysfunctional proteins that cannot be properly folded accumulate—conditions referred to as “ER stress”—the UPR is activated, allowing the cell to adjust the folding capacity of the organelle to restore homeostasis. Complementary UPR actions maintain protein-folding homeostasis in the ER. First, the UPR reduces client protein load in the ER by temporarily reducing global protein synthesis (Harding et al. 1999, 2000). Second, it enlarges the ER volume through endomembrane biosynthesis (Sriburi et al. 2004; Bommiasamy et al. 2009; Schuck et al. 2009). Third, it augments the ER-folding capacity through the up-regulation of chaperones and foldases (Lee et al. 2003; Acosta-Alvear et al. 2007). Fourth, it increases the ER protein turnover capacity through the up-regulation of ER-associated degradation (ERAD) components and ER-phagy (ER-specific autophagic mechanisms), which eliminate misfolded proteins accumulating in the ER or damaged portions of the entire organelle, respectively (Travers et al. 2000; Bernales et al. 2006; Schuck et al. 2014; Khaminets et al. 2015; Fumagalli et al. 2016; Grumati et al. 2017). Last, if homeostasis is unachievable, the UPR initiates cell-death programs that eliminate defective cells for the benefit of the organism (Lin et al. 2007; Lu et al. 2014). The life/death decision involves the interplay of molecular timers that allow cells to sense whether the defect can be fixed in an allotted time window or whether the cell poses a threat and must be eliminated (Lin et al. 2007; Lu et al. 2014). In metazoans, three unique ER-resident UPR sensors of unfolded proteins operate in parallel to offset ER stress (Cox et al. 1993; Cox and Walter 1996; Sidrauski and Walter 1997; Yoshida et al. 1998, 2001; Niwa et al. 1999; Harding et al. 2000; Tirasophon et al. 2000). These sensors are the kinase/RNase IRE1, the kinase PERK, and the membrane-tethered transcription factor ATF6. IRE1 is the most conserved sensor and exists from yeast to mammals (Mori 2009). By contrast, ATF6 and PERK are only found in metazoans. As organisms gained biological complexity, it is likely that the additional complementary ER stress-sensing pathways evolved, allowing the fine-tuning of ER homeostasis to match the specific demands of different cells and tissues, which would explain how the UPR diversified from a single IRE1-dependent ER stress-detection mechanism to multiple ones.
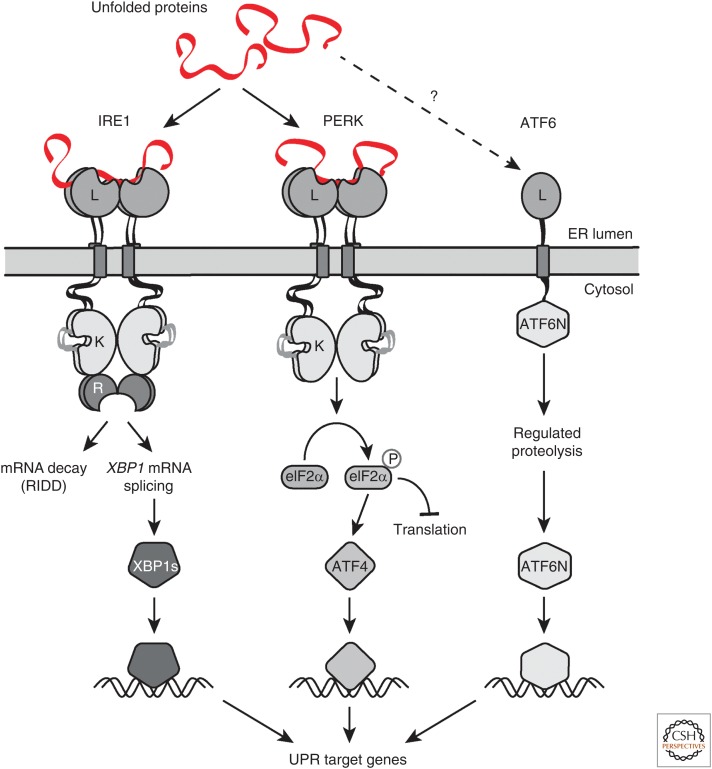
IRE1 is a single-pass transmembrane protein with an ER lumenal sensor domain and cytoplasmic effector kinase/nuclease domains (Fig. 1). There are two IRE1 paralogs in higher eukaryotes (Tirasophon et al. 1998; Wang et al. 1998). IRE1α is ubiquitously expressed and it is essential (Zhang et al. 2005), whereas IRE1β is expressed in epithelial cells that line the intestine and airways (Bertolotti et al. 2001; Martino et al. 2013; Tsuru et al. 2013). IRE1 coordinates the UPR through a well-established yet highly unconventional splicing mechanism of the messenger RNAs (mRNAs) encoding the transcription factors Hac1 in yeast and XBP1 in metazoans (Cox et al. 1993; Cox and Walter 1996; Sidrauski and Walter 1997; Yoshida et al. 1998, 2001; Calfon et al. 2002; Aragón et al. 2009; Korennykh et al. 2009; Li et al. 2010). IRE1 oligomerizes in the plane of the ER membrane in response to ER stress (Bertolotti et al. 2001; Aragón et al. 2009; Li et al. 2010; Sundaram et al. 2017). Oligomerization allows for trans-autophosphorylation and allosteric activation of its endonuclease domain, which initiates the unconventional splicing of the Hac1 or the XBP1 mRNAs (Cox et al. 1993; Cox and Walter 1996; Sidrauski and Walter 1997; Yoshida et al. 1998, 2001; Aragón et al. 2009; Korennykh et al. 2009; Li et al. 2010). Spliced XBP1 mRNA encodes the transcription factor XBP1s, which drives the expression of several target genes involved in restoring ER homeostasis, such as those encoding chaperones, foldases, lipid biosynthesis enzymes, and ERAD components (Lee et al. 2003; Acosta-Alvear et al. 2007). While the XBP1 mRNA is the only known splicing target of IRE1, active IRE1 can also cleave ER-localized mRNAs in a process known as regulated IRE1-dependent decay (RIDD), which limits the amount of client proteins entering the ER, thus helping alleviate protein-folding stress in the organelle (Fig. 1) (Hollien and Weissman 2006; Hollien et al. 2009) . In this way, IRE1 counteracts ER stress through corrective, XBP1s driven, as well as preemptive, RIDD-dependent mechanisms.
Figure 1.
Three branches of the unfolded protein response (UPR). Three endoplasmic reticulum (ER) stress sensors, IRE1, PERK, and ATF6, monitor the protein-folding conditions in the ER lumen. Each pathway uses a unique mechanism of signal transduction that results in the activation of specialized transcription factors that drive transcription of target genes that alleviate ER stress. The IRE1 and PERK branches also reduce the ER-folding load by impeding further client protein load in the ER either by degrading ER-targeted messenger RNAs (mRNAs) or by negatively regulating translation, respectively.
The second branch of the UPR is mediated by the ER-resident transmembrane serine/threonine kinase PERK (Fig. 1; Harding et al. 1999). Upon ER stress, and similar to IRE1, PERK oligomerizes in the plane of the ER membrane, leading to its autophosphorylation and activation (Harding et al. 1999, 2002; Bertolotti et al. 2000; Marciniak et al. 2006). Active PERK phosphorylates the α subunit of the translation initiation factor 2 (eIF2), a key regulator of protein synthesis (Shi et al. 1998; Harding et al. 1999, 2002). Phosphorylation of eIF2α inhibits global translation, reducing the influx of proteins entering the ER, and hence alleviates ER stress akin to the reduction in protein-folding load afforded by RIDD. Paradoxically, the phosphorylation of eIF2α also leads to preferential translation of a few mRNAs containing short open reading frames in their 5′-UTRs (5′ untranslated regions), an unfortunate misnomer. One of these mRNAs encodes for the transcription factor ATF4, which, similar to XBP1, regulates the expression of target genes that help alleviate ER stress by increasing the biosynthetic capacity of the cell, such as those encoding amino acid importers and redox homeostasis regulators (Harding et al. 2003). Like IRE1, PERK counteracts ER stress through corrective, ATF4-driven, and preemptive phospho-eIF2α-dependent mechanisms.
The third branch of the UPR is regulated by ATF6 (Fig. 1). Like IRE1, ATF6 has two homologs in vertebrates, ATF6β and ATF6α, which are type II transmembrane proteins with cytoplasmic transcription factor domains that become severed from the membrane upon ER stress (Haze et al. 1999, 2001). Neither protein is essential for normal development in the mouse, but their combined deletion is embryonic lethal, suggesting they have highly overlapping functions (Yamamoto et al. 2007). Accumulation of unfolded proteins causes ATF6 to be exported to the Golgi apparatus by a still-ill-defined mechanism (Haze et al. 1999). Upon reaching the Golgi apparatus, ATF6 is sequentially processed by S1P and S2P (site 1 and site 2 proteases), which remove their lumenal domain (LD) and transmembrane anchor, respectively (Haze et al. 1999; Ye et al. 2000). This regulated intramembrane proteolysis mechanism releases the cytosolic transcription factor portion, ATF6(N), which is then imported into the nucleus to activate UPR target genes, sometimes together with XBP1 as ATF6(N) and XBP1s can heterodimerize (Yamamoto et al. 2007). Unlike PERK and IRE1, ATF6 does not prevent client protein loading into the ER. Rather, it increases the ER volume and its protein-processing and degradative capacities (Yoshida et al. 1998; Wang et al. 2000; Nadanaka et al. 2006b; Adachi et al. 2008; Bommiasamy et al. 2009). Thus, it is likely that the coordinated actions of IRE1, PERK, and ATF6 serve overlapping regulatory functions, with preemptive measures that kick in simultaneously with corrective ones that, together, maintain ER-folding homeostasis.
As discussed before, the UPR does not only safeguard cellular health by adapting the folding capacity of the ER to cell's needs, but it can also drive cell death. IRE1, PERK, and ATF6 detect the same problem, namely, imbalances in the folding capacity of the ER, yet coordinate different outputs—adaptation or death—computed from the intensity and time-dependent activity of each UPR sensor (Lu et al. 2014). UPR signaling output is likely to be coordinated in a condition- and cell-type-specific manner to accommodate to the physiological state of cells and tissues. Because the UPR induces both prosurvival and proapoptotic pathways, it sits at the center of life-or-death decisions that can affect the progression of numerous diseases, including neuropathologies, cancer, diabetes, atherosclerosis, and infection by bacterial and viral pathogens (Bi et al. 2005; Feldman et al. 2005; Lin et al. 2007; Mulvey et al. 2007; Zhang and Kaufman 2008; Sung et al. 2009; Behrman et al. 2011; Vidal et al. 2012; Stahl et al. 2013; Lu et al. 2014; Treacy-Abarca and Mukherjee 2015; Tufanli et al. 2017). The double-edged sword behavior of the UPR requires robust and precise mechanisms that allow the exquisitely sensitive detection of protein-folding perturbations in the ER. Recent data suggest that a complex and intricate interaction network—formed among chaperones, unfolded proteins, and the UPR sensors—tunes the activity of the UPR to allow the reinstatement of homeostasis while avoiding premature cell death.
SIGNALING THE ER PROTEIN-FOLDING STATUS ACROSS THE MEMBRANE
The first step in resolving ER stress is transmitting information on the protein-folding status in the ER lumen across the ER membrane and into the cell's nucleus by activation of UPR transcription factors. For each of the UPR sensors, the transmission of this information relies on changes in their oligomerization state. IRE1 and PERK, which share similar structural features in their ER-lumenal sensor domains, form higher-order oligomers during ER stress. This type of self-association juxtaposes the cytosolic effector domains of either protein for their activation (Bertolotti et al. 2000; Aragón et al. 2009; Li et al. 2010). Impairing ER-lumenal oligomerization of either IRE1 or PERK negatively impacts their activity in the cell, underlining the importance of oligomerization for their function (Credle et al. 2005; Carrara et al. 2015a; Karagöz et al. 2017). ATF6 activation is the antipode of IRE1 and PERK activation. Inactive ATF6 is held in the ER in the form of disulfide-linked oligomers, which are reduced before leaving the ER en route to the Golgi apparatus for activation (Mori and Nadanaka 2003; Nadanaka et al. 2006a,b, 2007). The precise mechanism by which ATF6 senses unfolded proteins in the ER lumen prior to its transport to the Golgi apparatus remains to be defined. Whereas changes in the oligomerization status of ER stress sensors are evidently important for activity, the initial step in resolving ER stress is detecting unfolded proteins in the ER lumen. The mechanisms by which the UPR sensors detect ER stress are becoming rapidly elucidated.
STRESS-SENSING MECHANISMS—EARLY MODELS
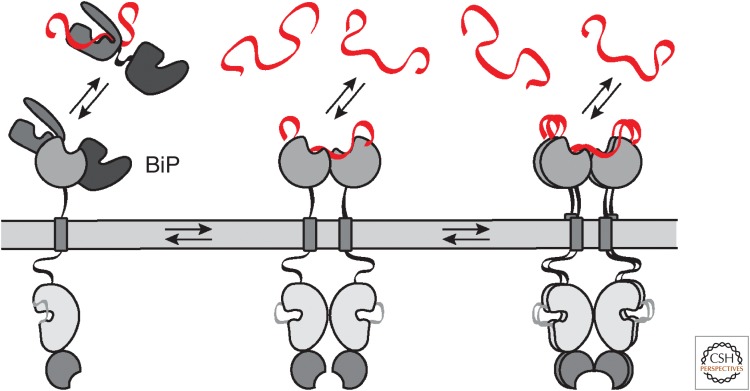
The evolutionary conservation of IRE1 makes it the most mechanistically understood branch of the UPR. Two models can describe how IRE1's LD senses ER stress: an earlier model in which the reversible dissociation of the ER Hsp-70 type chaperone BiP from the IRE1 LD is responsible for activation/deactivation (Bertolotti et al. 2000; Okamura et al. 2000; Zhou et al. 2006), and a revised model in which unfolded proteins act as direct ligands for IRE1's sensor domain (Fig. 2; Credle et al. 2005).
Figure 2.
IRE1's endoplasmic reticulum (ER) protein-folding stress-sensing mechanism. In steady-state conditions, the ER-resident chaperone BiP binds IRE1's sensor domain and shifts IRE1 to a monomeric inactive state, buffering IRE1's activity. During ER stress, BiP is titrated to unfolded proteins accumulating in the ER, relieving its buffering effect on IRE1. Simultaneously, unfolded proteins are directly recognized by IRE1's sensor lumenal domain. These unfolded proteins serve as activating ligands that drive IRE1 oligomerization and activation.
The former model, which suggested that BiP is the sole regulator of the UPR, was based on the main observations that BiP overexpression attenuated UPR signaling and that depleting BiP activated the UPR (Dorner et al. 1992; Ng et al. 1992; Kohno et al. 1993). The model gained further traction when additional studies showed that BiP binds to IRE1, PERK, and ATF6 in unstressed cells, and that it dissociates from them during acute ER stress (Bertolotti et al. 2000; Okamura et al. 2000; Shen et al. 2002). Together, these observations supported that the titration of BiP from IRE1 by unfolded proteins provided the molecular switch for IRE1 activation. According to this view, BiP binding sequesters the UPR sensors in an inactive state and its dissociation upon ER stress licenses their activation (Bertolotti et al. 2000; Zhou et al. 2006; Oikawa et al. 2009; Carrara et al. 2015b).
This simple view has been experimentally refuted as it became evident that BiP is not the primary regulator of IRE1 activity (Kimata et al. 2004; Pincus et al. 2010). A mutational analysis of IRE1's LD in yeast mapped the BiP-binding site to the juxtamembrane segment (Kimata et al. 2004; Pincus et al. 2010). Removal of this region abolished ER stress-regulated BiP binding but it did not lead to unrestrained activation of IRE1. Instead, removing the BiP-binding site in IRE1 resulted in delayed deactivation kinetics, suggesting that BiP association is crucial for buffering IRE1 activity (Fig. 2; Pincus et al. 2010). Together, these data suggest that, at least in yeast, the release of BiP cannot be the primary trigger for IRE1 activation.
STRUCTURAL INSIGHTS INTO STRESS-SENSING MECHANISM—DIRECT ACTIVATION MODEL
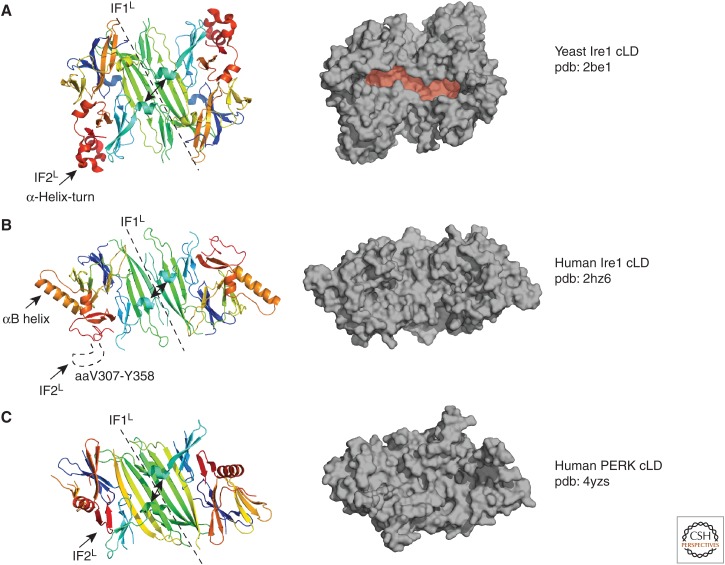
A more recent model poses that IRE1 directly senses ER stress. This model emerged from the crystal structure of the conserved core region of the LD (core lumenal domain [cLD]) of yeast IRE1, which shows architectural similarities with the major histocompatibility complexes (MHCs) (Credle et al. 2005). Yeast IRE1 cLD structure displayed a deep pocket that extends across a dimerization interface reminiscent of the peptide-binding groove of the MHCs (Fig. 3A; Credle et al. 2005). Based on the architectural similarity of the yeast IRE1 cLD to that of MHCs, it was proposed that misfolded proteins directly interact with the IRE1 cLD, inducing the formation of active IRE1 oligomers. Mutation of the amino acids lining the bottom of the peptide-binding groove abolished IRE1 signaling in cells, providing functional evidence for the importance of these structural elements for IRE1 activation (Credle et al. 2005). A follow-up study showed that yeast IRE1 selectively binds misfolded ER-lumenal proteins in vivo and that purified yeast IRE1 cLD directly interacts with peptides (used experimentally as misfolded protein surrogates) in vitro, leading to its oligomerization (Gardner and Walter 2011). Peptide tiling arrays revealed that IRE1 cLD recognizes peptides with a distinct amino acid composition bias, consistent with it recognizing exposed polypeptide stretches that would normally be buried inside folded proteins (Gardner and Walter 2011). Together, these observations revealed that IRE1 can sense ER stress directly, where unfolded proteins act as agonists inducing its oligomerization and activation (Fig. 2).
Figure 3.
Comparison between the crystal structures of the sensor domains of IRE1 and PERK. Crystal structures of the core lumenal domains (cLDs) of yeast (A), and human IRE1 (B), as well as that of the human PERK cLD (C) reveal that key structural elements forming the dimerization interface (endoplasmic reticulum [ER]-lumenal interface 1, IF1L; indicated by dashed lines) are highly conserved among yeast and mammalian IRE1 as well as in the mammalian PERK cLD. By contrast, the α-helix turn forming oligomerization interface 2 (IF2L, indicated by the black arrow) in the yeast IRE1 cLD is not conserved in the mammalian ER stress sensors. The structures are shown in colored ribbon diagram representations on the left and as surface representations in grey on the right. The major histocompatibility complex (MHC)-like groove in the yeast IRE1 cLD structure is shaded in red. The αB helix found only in human IRE1α cLD is indicated with an arrow. The distance between the helices surrounding the groove is depicted with double-pointed black arrows.
STRESS SENSING THROUGH DIRECT RECOGNITION OF UNFOLDED PROTEINS—NOVEL INSIGHTS
Differences between the crystal structures of human and yeast IRE1 cLDs challenge the direct activation model. Although the crystal structure of human IRE1α cLD displays conserved structural elements in the core of the MHC-like fold, there are three notable differences between the crystal structures of human and yeast IRE1 cLDs known to date. First, the helices flanking the putative peptide-binding groove are too closely juxtaposed in the human structure to allow peptide binding as has been proposed and modeled in the yeast protein (Zhou et al. 2006). Second, the mammalian cLD lacked a second interface, present in the yeast structure (ER-lumenal interface 2, or IF2L) that provides contacts for higher-order oligomerization and is indispensable for IRE1 activation in yeast (Credle et al. 2005). Third, a prominent α-helix (“αB helix”; residues V245-I263) in mammalian cLD that is absent in the yeast IRE1 cLD would prevent formation of the equivalent of IF2L due to steric hindrance (Fig. 3B; Zhou et al. 2006). Recent crystal structures of PERK cLD further complicate the picture as the oligomerization interface in PERK cLD diverges from both the human and the yeast IRE1 cLDs. Similar to the mammalian IRE1 cLD, the helices forming the putative peptide-binding groove in yeast IRE1 cLD are too close to each other in the PERK dimer structure to accommodate peptides in the groove (Fig. 3C).
IRE1 represents the most conserved component in the UPR network. Thus, rather than proposing alternative mechanisms for IRE1 activation in different species, it seems more plausible that the structures of the cLDs of mammalian IRE1 and PERK represent different states in a spectrum of possible conformational states that the cLDs from any species could assume. After all, the structure adopted by a protein in a crystal lattice represents a singular snapshot of one of many possible conformational states. In this notion, the crystal structures of mammalian IRE1α or PERK cLDs would represent a “closed” conformation that can shift toward an “open” state to allow peptide binding in the MHC-like groove that is clearly apparent and functionally validated in the structure of the yeast ortholog (Gardner and Walter 2011; Gardner et al. 2013). Therefore, it remains entirely plausible that the mammalian ER stress sensors IRE1/PERK and the yeast IRE1 cLD use a common mechanism of activation that hinges on direct recognition of unfolded protein ligands. Indeed, replacing the yeast IRE1 LD with mammalian IRE1 or PERK LDs, as well as exchanging the LDs of IRE1 and PERK, results in functional UPR signaling in yeast and mammalian cells, respectively, underscoring the existence of a conserved stress-sensing mechanism (Bertolotti et al. 2000; Liu et al. 2000; Mai et al. 2018).
Recent biochemical and structural analyses on the human IRE1α activation mechanism (Karagöz et al. 2017) showed that, similar to yeast IRE1, human IRE1α cLD recognizes unfolded proteins as activating ligands. These studies revealed that the human IRE1α cLD directly binds select peptides and unfolded proteins (Karagöz et al. 2017). Notably, peptide arrays revealed similarities in the peptide recognition principles of human and yeast IRE1 (Gardner and Walter 2011; Karagöz et al. 2017). Mammalian IRE1 cLD's affinity for peptides varied between 5 and 30 µM, which is in the same order of magnitude as those reported for most chaperones (Marcinowski et al. 2011; Street et al. 2011; Karagöz et al. 2014), and peptide binding induces the formation of IRE1 cLD oligomers as assessed by analytical ultracentrifugation analyses (Karagöz et al. 2017). Together, these observations support the notion that the driving force for IRE1 oligomerization/activation is the recognition and binding of unfolded protein ligands, that a biochemical signature of unfolded peptide ligands is recognized by IRE1, and that such signature is conserved from yeast to mammals (Gardner and Walter 2011; Karagöz et al. 2017).
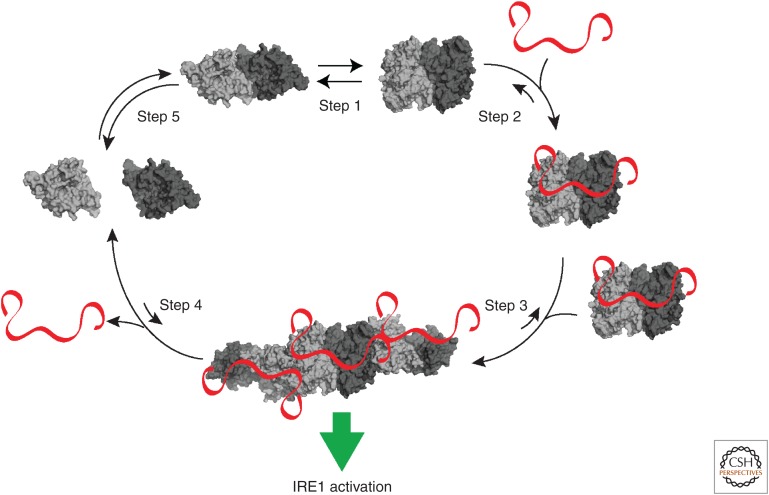
Further support for the peptide ligand-driven activation model of IRE1 comes from nuclear magnetic resonance (NMR) spectroscopy experiments that provide atomic resolution information on dynamic protein complexes. Such studies revealed that the human IRE1α cLD scans various conformational states in solution, which can explain its plasticity and promiscuity in recognizing unfolded proteins (Karagöz et al. 2017). NMR experiments also showed that peptide binding maps to the center of the MHC-like fold in the human IRE1α cLD and that it induces conformational changes in regions of the protein that were thought to sterically block IRE1α cLD's oligomerization (Karagöz et al. 2017). These data support the model that peptide binding induces an allosteric conformational switch that leads to the formation of a functional oligomerization interface. From solution dynamics of the human IRE1 cLD, a new picture emerges that supports, first, that the crystal structures are indeed likely snapshots of possible conformational states, and second, that IRE1-peptide interactions allosterically license the first step in IRE1 activation: its oligomerization (Fig. 4).
Figure 4.
Apo-human IRE1 lumenal domain (LD) dimers are found in equilibrium between closed and open conformations (step 1). Upon endoplasmic reticulum (ER) stress, unfolded proteins accumulating in the ER lumen bind the IRE1 LD, stabilizing the sensor domain in its open conformation. Peptide binding also induces a conformational change in the αB helix and the neighboring structural elements (steps 2 and 3), that facilitate activation by allowing the formation of an IF2L-like interface in the protein's LD (step 4). When protein-folding homeostasis is achieved, the dynamic IRE1 LD oligomers re-adopt their inactive conformation (step 5).
The MHC-like fold in the human IRE1α cLD is enriched in aromatic residues and displays a negatively charged surface that chemically complements IRE1α-binding peptides. IRE1-binding peptides have moderate hydrophobicity, suggesting that IRE1 recognizes peptides with lower aggregation propensity compared to those typically recognized by ER chaperones and cochaperones, such as BiP and the J proteins (Flynn et al. 1991; Blond-Elguindi et al. 1993; Behnke et al. 2016). Supporting this notion, human and yeast IRE1 cLD-binding peptides only partially overlap with those bound by BiP, suggesting that the UPR sensors do not compete with highly abundant BiP for binding sites in unfolded proteins (Gardner and Walter 2011; Karagöz et al. 2017). Moreover, human IRE1α interacts with unfolded proteins in cells (remarkable considering that BiP exists in levels that are orders of magnitude higher than those of IRE1), further corroborating these in vitro observations (Sundaram et al. 2018).
Recently, it was found that similar to human IRE1 LD, the structurally related PERK LD could also recognize unfolded polypeptides. PERK LD binds polypeptides displayed in a peptide tiling array (Dalton et al. 2018), and a phage display strategy showed it specifically interacts with unfolded polypeptides (Wang et al. 2018). Moreover, PERK displays aggregation prevention properties similar to those of yeast IRE1 cLD and chaperones (Kimata et al. 2007; Wang et al. 2018). These findings suggest that PERK and IRE1 use a common stress-sensing mechanism that relies on direct recognition of unfolded proteins.
To date, only one peptide has been cocrystalized with the PERK cLD. This crystal structure displayed the bound peptide on a cleft that is involved in the formation of a novel tetramerization interface that was mapped based on an earlier PERK cLD crystal structure (Carrara et al. 2015a; Wang et al. 2016, 2018). Notably, in the human IRE1 cLD crystal structure, the corresponding cleft is occupied by a carboxy-terminal segment that plays a role in IRE1 LD oligomerization, as assessed by mutational analyses in vitro and in vivo (Karagöz et al. 2017). Importantly, this segment was truncated in the PERK cLD. The amino-terminal flexible segment in yeast IRE1 fine-tunes its response, as it allows it to compete with unfolded polypeptides (Mathuranyanon et al. 2015). Therefore, it is plausible that different peptides might occupy different pockets in the UPR sensors, promoting either assembly or disassembly of higher-order oligomers. In this way, different ligands could act as either agonists or antagonists of UPR signaling. Thus, and even though we currently lack the direct comparison of peptide recognition principles of mammalian IRE1 and PERK lumenal domains, it is attractive to speculate that different unfolded polypeptides may regulate the different ER stress sensors to tip the UPR outcome toward life or death.
In these proposed mechanisms, the UPR sensors do not principally depend on saturation of BiP by unfolded substrate proteins to engage with IRE1. Although it currently remains unknown whether BiP release is a prerequisite for IRE1 to become receptive to peptide binding, direct activation by unfolded proteins would allow the UPR sensors to detect the ER stress dynamically.
FINE-TUNING THE UPR THROUGH ER STRESS SENSOR PROTEIN INTERACTORS
Two Distinct Modes of BiP Interaction with IRE1
Although the ER-chaperone BiP has long been proposed to be a regulator of the UPR, we only recently obtained a detailed in vitro characterization of BiP's interaction with the UPR sensors. BiP consists of an amino-terminal nucleotide-binding domain (NBD) and a carboxy-terminal substrate-binding domain (SBD). BiP binds to unfolded proteins by recognizing hydrophobic patches (Flynn et al. 1991; Blond-Elguindi et al. 1993) through its SBD (Gething 1999). The ATPase-coupled conformational changes in BiP allow binding and release of its substrates. Recent lines of evidence support that BiP can fine-tune IRE1's activity through two different modes of action: (1) a novel nucleotide-independent interaction involving BiP's NBD (Carrara et al. 2015b), and (2) by a substrate recognition-like interaction assisted by the cochaperone ERDJ4 (Amin-Wetzel et al. 2017). A recent study that used primarily an in vitro reconstitution strategy suggested noncanonical interactions between BiP-NBD and IRE1/PERK LDs (Carrara et al. 2015b). A similar interaction was suggested earlier for yeast IRE1 and BiP using an in vivo mapping strategy (Todd-Corlett et al. 2007). Moreover, binding of unfolded proteins to BiP's SBD relieved the interaction between BiP and IRE1/PERK LD, suggesting a novel allosteric model for UPR induction (Carrara et al. 2015b; Kopp et al. 2018). The precise mechanism behind such allosteric control (i.e., the release of BiP from IRE1's cLD upon substrate binding) remains to be investigated. Yet, the selective binding of BiP to ER stress sensors and unfolded proteins through two different sites in BiP provides an attractive model in which BiP could facilitate engaging unfolded proteins with ER stress sensors. Subsequent conformational changes may allow BiP-release and handover of the unfolded proteins to the ER stress sensors for their activation.
By itself, BiP is a poor ATPase, and it relies on J-protein cochaperones to enhance its ATPase activity (Marcinowski et al. 2011; Behnke et al. 2015). New data suggest a role of the ER-lumenal cochaperone ERDJ4 in regulating IRE1's activity through BiP (Amin-Wetzel et al. 2017). In cells, ERDJ4 depletion increased XBP1 mRNA splicing, suggesting that ERDJ4 behaves as an IRE1 repressor. In vitro, ERDJ4 promoted the formation of a complex between BiP and IRE1 LD by breaking IRE1 dimers. These observations support a scenario in which ERDJ4 associates with IRE1's LD to recruit BiP through the stimulation of ATP hydrolysis. Interestingly, depleting ERDJ4 only resulted in partial activation of IRE1 in cells, suggesting that the surmised silencing of IRE1 activity by ERDJ4 is not absolute. Considering that ERDJ4 has a role in ERAD—it interacts with the ERAD component Derlin-1 (Lai et al. 2012)—it remains possible that the ERDJ4-dependent BiP action results in partial unfolding and degradation of IRE1. IRE1 regulates the expression of the ERDJ4 gene downstream of XBP1 (Lee et al. 2003; Acosta-Alvear et al. 2007; Adamson et al. 2016). Thus, it is possible that the BiP/ERDJ4 interaction is crucial for shutting off IRE1 signaling, as previously shown for the role of the BiP/IRE1 interaction in yeast (Kimata et al. 2004; Pincus et al. 2010). Mammals possess eight different ERDJ proteins (ERDJ1-8) that recognize various clients. Therefore, it is conceivable that competition for unfolded proteins between IRE1 and ERDJ4 (and perhaps others), allows fine-tuning IRE1 activity in higher eukaryotes to match tissue-specific folding demands.
ER-Resident Chaperones Fine-Tune IRE1 Activity
Fine-tuning the UPR goes beyond interactions with the highly conserved chaperone BiP. As organisms evolved and acquired more complex proteomes, so did their chaperone repertoire to accommodate to cell- and tissue-specific protein-folding demands. Recently, a more complex interaction network of the UPR sensors with chaperones has been identified in higher eukaryotes. The ER-resident chaperones PDIA6 and Hsp47 were found to fine-tune the UPR by directly interacting with the UPR sensors. On one hand, the disulfide isomerase PDIA6 regulates attenuation of the UPR by interacting with both IRE1 and PERK LDs and breaking stress-induced disulfide-stabilized oligomers (Eletto et al. 2014, 2016). On the other hand, the ER chaperone Hsp47 was suggested to enhance IRE1 activation by competing with BiP in the initial stages of ER stress (Sepulveda et al. 2018). These findings suggest that a complex network of ER chaperones adjusts the activation/deactivation dynamics of the UPR sensors in cells and, moreover, could account for differences in UPR activity in different tissues. Since Hsp47 is a specialized chaperone for collagen (Ito and Nagata 2017), it is attractive to contemplate that the interaction between IRE1 and Hsp47 evolved to regulate IRE1 activity in cells specialized for collagen secretion, such as osteoblasts, tenocytes, or chondrocytes. By contrast, PDIA6 might regulate the duration of IRE1 activity through competition with select unfolded protein substrates for IRE1 LD binding. Together, these findings underline the existence of a multilayered regulation system that balances the magnitude and duration of the UPR according to the physiological burden of the cell experiencing ER stress.
Translocon Interaction Tunes IRE1 Oligomerization
Nascent polypeptides enter the ER through the Sec61 translocon. Recent work emphasized the importance interactions between the translocon and IRE1 in regulating IRE1's activity. Disruption of the Sec61-IRE1 complex deregulates IRE1 signaling (Plumb et al. 2015; Adamson et al. 2016; Sundaram et al. 2017, 2018), suggesting that such interaction, much like the interactions with the chaperone network, fine-tunes IRE1's activity. It is notable that translocons are highly abundant, present at levels at least an order of magnitude higher than those of IRE1 (Wang et al. 2015). Such disparate stoichiometry suggests that IRE1 associates with specialized translocon complexes that might be involved in the translocation of select ER clients. By physically engaging with the translocon, the cell would ensure that IRE1 is located at the spot where the action happens, allowing it to monitor protein-folding fidelity as the incoming ER-folding clients are cotranslationally inserted in the ER. In such a scenario, when the chaperones are exhausted during protein-folding stress, IRE1 could bind to nascent chains that cannot fold properly, allowing it to sense local changes in protein-folding load and perhaps single out the mRNAs that are the source of the problem, which would then be degraded by RIDD.
UPR SENSORS MONITOR THE HEALTH OF THE ER MEMBRANE
Besides sensing protein-folding problems in the lumen of the ER, the ER stress sensors also act as surveyors of membrane composition. Aberrant lipid compositions of the ER membrane, referred to as lipid bilayer stress, can activate the UPR independently of unfolded protein accumulation in the ER lumen (Kimata et al. 2007; Promlek et al. 2011; Volmer et al. 2013; Halbleib et al. 2017; Kono et al. 2017). Yeast IRE1 uses an amphipathic helix juxtaposed to its transmembrane domain to sense and respond to aberrant physical properties of the ER membrane. During lipid bilayer stress, these amphipathic helices bend the membrane, thermodynamically facilitating IRE1 oligomerization (Halbleib et al. 2017). A similar amphipathic juxtamembrane helix exists in PERK (Halbleib et al. 2017), suggesting the mechanism to sense lipid bilayer stress may be conserved among ER stress sensors. In this way, ER stress sensors could detect physicochemical properties of the ER membrane environment, which may be detrimental for membrane protein folding or for trafficking through the secretory pathway and, as such, could negatively impact cell health. Together, the evidence to date indicates that UPR sensors integrate various signals to sense and respond to perturbations in ER homeostasis.
CONCLUDING REMARKS: THE UPR PROVIDES UNIQUE SOLUTIONS FOR UNIQUE PROTEOMES
Various physiological and pathological conditions that overwhelm the protein-folding capacity of the ER activate the UPR. It has become clear that the UPR is indispensable for ensuring development, proper function, and survival of professional secretory cells, including antibody-secreting plasma cells, collagen-secreting osteoblasts, hepatocytes, and cells within endocrine/exocrine tissues (Reimold et al. 2000, 2001; Harding et al. 2001; Gass et al. 2002; Zhang et al. 2002, 2005, 2006; Iwakoshi et al. 2003b; Shaffer et al. 2004; Lee et al. 2005, 2008; Kaser et al. 2008; Wei et al. 2008; Murakami et al. 2009; Iwawaki et al. 2010; Saito et al. 2011). Additionally, emerging evidence suggests that the UPR serves physiological roles that are crucial for the development and maintenance of tissues that are not classically thought of as “secretory.” Specifically, the UPR seems to be essential for neuronal development, differentiation, and maturation, as well as for maintenance of mature neurons (Cho et al. 2009; Kawada et al. 2014; Tekko et al. 2014; Martinez et al. 2016; Murao and Nishitoh 2017). It is intriguing that different branches of the UPR appear to be required by different tissues to address their physiological needs. For instance, the PERK branch of the UPR participates in olfactory receptor (OR) choice though a feedback process in which unfolded ORs entering the ER trigger PERK activation (Dalton et al. 2013). RTP1, an OR-specific chaperone that is a target of the PERK branch of the UPR, is required for proper trafficking of ORs and is part of the feedback signal that ascertains only a single one of the ∼1000 loci that encode ORs in the genome is selected for expression (Dalton et al. 2013; Sharma et al. 2017). Whereas IRE1/XBP1 are required for plasma cell differentiation (Reimold et al. 2000; Iwakoshi et al. 2003a; Zhang et al. 2005), PERK signaling is suppressed in the process (Ma et al. 2010), underscoring the notion that in specialized tissues, the UPR sensors and ER clients might have coevolved to sensitize/desensitize different branches of the UPR in recognition of misfolded stretches of specific client proteins exposed upon misfolding (Sung et al. 2009; Sharma et al. 2017).
Dysregulation of the UPR contributes to neuropathologies, diabetes, cancer, metabolic disease, atherosclerosis, as well as bacterial and viral infections (for review, see Lin et al. 2008). In these instances, the different branches of the UPR are differentially regulated. For example, multiple myeloma, a cancer of plasma cells, relies on IRE1/XBP1 (Carrasco et al. 2007), and pharmacologically blocking IRE1 shows antimyeloma activity (Papandreou et al. 2011; Mimura et al. 2012). Active IRE1/XBP1 is also observed in mouse models of insulin resistance and of atherosclerosis (Ozcan et al. 2004; Tufanli et al. 2017), making the therapeutic targeting of this pathway attractive for multiple disease states (Hotamisligil 2010; Vidal et al. 2012; Martinez et al. 2016; Tufanli et al. 2017). Similarly, the foreign proteomes and high biosynthetic demands of viruses and bacterial pathogens differentially turn on or suppress individual UPR pathways (Mulvey et al. 2007; Sung et al. 2009; Stahl et al. 2013; Treacy-Abarca and Mukherjee 2015). Altogether, converging lines of evidence suggest that the UPR gets activated during physiological and pathological conditions, in which changes in the proteome result in fluctuations of ER protein-folding demands. As accumulating data indicate, the activation profile of the UPR signaling is unique in every cell and relies on activation of different UPR branches. It is likely that the shifting composition of different proteomes and/or the expression of UPR modulators, many of which likely to still be discovered, allow presentation of different unfolded polypeptide ligands for the UPR sensors as well as the chaperone network to differentially activate each one of the UPR branches. Such differential activation of the UPR remains an exciting and still largely unexplored area of research.
Footnotes
Editors: Richard I. Morimoto, F. Ulrich Hartl, and Jeffery W. Kelly
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. 2007. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell 27: 53–66. 10.1016/j.molcel.2007.06.011 [DOI] [PubMed] [Google Scholar]
- Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. 2008. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct 33: 75–89. 10.1247/csf.07044 [DOI] [PubMed] [Google Scholar]
- Adamson B, Norman TM, Jost M, Cho MY, Nunez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et al. 2016. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167: 1867–1882.e21. 10.1016/j.cell.2016.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin-Wetzel N, Saunders RA, Kamphuis MJ, Rato C, Preissler S, Harding HP, Ron D. 2017. A J-protein cochaperone recruits BiP to monomerize IRE1 and repress the unfolded protein response. Cell 171: 1625–1637.e13. 10.1016/j.cell.2017.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli T, Sitia R. 2008. Protein quality control in the early secretory pathway. EMBO J 27: 315–327. 10.1038/sj.emboj.7601974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragón T, van Anken E, Pincus D, Serafimova IM, Korennykh AV, Rubio CA, Walter P. 2009. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature 457: 736–740. 10.1038/nature07641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C, Helenius A. 2016. Cargo capture and bulk flow in the early secretory pathway. Annu Rev Cell Dev Biol 32: 197–222. 10.1146/annurev-cellbio-111315-125016 [DOI] [PubMed] [Google Scholar]
- Baum DA, Baum B. 2014. An inside-out origin for the eukaryotic cell. BMC Biol 12: 76 10.1186/s12915-014-0076-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke J, Feige MJ, Hendershot LM. 2015. BiP and its nucleotide exchange factors Grp170 and Sil1: Mechanisms of action and biological functions. J Mol Biol 427: 1589–1608. 10.1016/j.jmb.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke J, Mann MJ, Scruggs FL, Feige MJ, Hendershot LM. 2016. Members of the Hsp70 family recognize distinct types of sequences to execute ER quality control. Mol Cell 63: 739–752. 10.1016/j.molcel.2016.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrman S, Acosta-Alvear D, Walter P. 2011. A CHOP-regulated microRNA controls rhodopsin expression. J Cell Biol 192: 919–927. 10.1083/jcb.201010055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. 2006. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol 4: e423 10.1371/journal.pbio.0040423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol 2: 326–332. 10.1038/35014014 [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Wang X, Novoa I, Jungreis R, Schlessinger K, Cho JH, West AB, Ron D. 2001. Increased sensitivity to dextran sodium sulfate colitis in IRE1β-deficient mice. J Clin Invest 107: 585–593. 10.1172/JCI11476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, et al. 2005. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J 24: 3470–3481. 10.1038/sj.emboj.7600777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething MJ. 1993. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75: 717–728. 10.1016/0092-8674(93)90492-9 [DOI] [PubMed] [Google Scholar]
- Bommiasamy H, Back SH, Fagone P, Lee K, Meshinchi S, Vink E, Sriburi R, Frank M, Jackowski S, Kaufman RJ, et al. 2009. ATF6α induces XBP1-independent expansion of the endoplasmic reticulum. J Cell Sci 122: 1626–1636. 10.1242/jcs.045625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Bulleid NJ. 2011. Protein folding and modification in the mammalian endoplasmic reticulum. Annu Rev Biochem 80: 71–99. 10.1146/annurev-biochem-062209-093836 [DOI] [PubMed] [Google Scholar]
- Bromage E, Stephens R, Hassoun L. 2009. The third dimension of ELISPOTs: quantifying antibody secretion from individual plasma cells. J Immunol Methods 346: 75–79. 10.1016/j.jim.2009.05.005 [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415: 92–96. 10.1038/415092a [DOI] [PubMed] [Google Scholar]
- Carrara M, Prischi F, Nowak PR, Ali MM. 2015a. Crystal structures reveal transient PERK luminal domain tetramerization in endoplasmic reticulum stress signaling. EMBO J 34: 1589–1600. 10.15252/embj.201489183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrara M, Prischi F, Nowak PR, Kopp MC, Ali MM. 2015b. Noncanonical binding of BiP ATPase domain to Ire1 and Perk is dissociated by unfolded protein CH1 to initiate ER stress signaling. eLife 4: e03522 10.7554/eLife.03522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco DR, Sukhdeo K, Protopopova M, Sinha R, Enos M, Carrasco DE, Zheng M, Mani M, Henderson J, Pinkus GS, et al. 2007. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell 11: 349–360. 10.1016/j.ccr.2007.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YM, Jang YS, Jang YM, Chung SM, Kim HS, Lee JH, Jeong SW, Kim IK, Kim JJ, Kim KS, et al. 2009. Induction of unfolded protein response during neuronal induction of rat bone marrow stromal cells and mouse embryonic stem cells. Exp Mol Med 41: 440–452. 10.3858/emm.2009.41.6.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini L, Snapp E. 2013. Probing endoplasmic reticulum dynamics using fluorescence imaging and photobleaching techniques. Curr Protoc Cell Biol 60: Unit 21 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Walter P. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87: 391–404. 10.1016/S0092-8674(00)81360-4 [DOI] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73: 1197–1206. 10.1016/0092-8674(93)90648-A [DOI] [PubMed] [Google Scholar]
- Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. 2005. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci 102: 18773–18784. 10.1073/pnas.0509487102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton RP, Lyons DB, Lomvardas S. 2013. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell 155: 321–332. 10.1016/j.cell.2013.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton RP, Karagöz GE, Kahiapo J, Sharma R, Bashkirova L, Lyons DB, Matsunami H, Walter P. 2018. Olfactory and vomeronasal receptor feedback employ divergent mechanisms of PERK activation. bioRxiv. [Google Scholar]
- Dayel MJ, Hom EF, Verkman AS. 1999. Diffusion of green fluorescent protein in the aqueous-phase lumen of endoplasmic reticulum. Biophys J 76: 2843–2851. 10.1016/S0006-3495(99)77438-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. 2004. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol 2: e380 10.1371/journal.pbio.0020380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner AJ, Wasley LC, Kaufman RJ. 1992. Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO J 11: 1563–1571. 10.1002/j.1460-2075.1992.tb05201.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eletto D, Eletto D, Dersh D, Gidalevitz T, Argon Y. 2014. Protein disulfide isomerase A6 controls the decay of IRE1α signaling via disulfide-dependent association. Mol Cell 53: 562–576. 10.1016/j.molcel.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eletto D, Eletto D, Boyle S, Argon Y. 2016. PDIA6 regulates insulin secretion by selectively inhibiting the RIDD activity of IRE1. FASEB J 30: 653–665. 10.1096/fj.15-275883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L, McCaul N, Chatsisvili A, Braakman I. 2016. Co- and post-translational protein folding in the ER. Traffic 17: 615–638. 10.1111/tra.12392 [DOI] [PubMed] [Google Scholar]
- Feige MJ, Hendershot LM. 2011. Disulfide bonds in ER protein folding and homeostasis. Curr Opin Cell Biol 23: 167–175. 10.1016/j.ceb.2010.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman DE, Chauhan V, Koong AC. 2005. The unfolded protein response: a novel component of the hypoxic stress response in tumors. Mol Cancer Res 3: 597–605. 10.1158/1541-7786.MCR-05-0221 [DOI] [PubMed] [Google Scholar]
- Flynn GC, Pohl J, Flocco MT, Rothman JE. 1991. Peptide-binding specificity of the molecular chaperone BiP. Nature 353: 726–730. 10.1038/353726a0 [DOI] [PubMed] [Google Scholar]
- Fumagalli F, Noack J, Bergmann TJ, Cebollero E, Pisoni GB, Fasana E, Fregno I, Galli C, Loi M, Solda T, et al. 2016. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol 18: 1173–1184. 10.1038/ncb3423 [DOI] [PubMed] [Google Scholar]
- Gardner BM, Walter P. 2011. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 333: 1891–1894. 10.1126/science.1209126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P. 2013. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol 5: a013169 10.1101/cshperspect.a013169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass JN, Gifford NM, Brewer JW. 2002. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem 277: 49047–49054. 10.1074/jbc.M205011200 [DOI] [PubMed] [Google Scholar]
- Gething MJ. 1999. Role and regulation of the ER chaperone BiP. Semin Cell Dev Biol 10: 465–472. 10.1006/scdb.1999.0318 [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Stevens F, Argon Y. 2013. Orchestration of secretory protein folding by ER chaperones. Biochim Biophys Acta 1833: 2410–2424. 10.1016/j.bbamcr.2013.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P, Morozzi G, Holper S, Mari M, Harwardt MI, Yan R, Muller S, Reggiori F, Heilemann M, Dikic I. 2017. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife 6: e25555 10.7554/eLife.25555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbleib K, Pesek K, Covino R, Hofbauer HF, Wunnicke D, Hänelt I, Hummer G, Ernst R. 2017. Activation of the unfolded protein response by lipid bilayer stress. Mol Cell 67: 673–684.e8. 10.1016/j.molcel.2017.06.012 [DOI] [PubMed] [Google Scholar]
- Hammond C, Braakman I, Helenius A. 1994. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc Natl Acad Sci 91: 913–917. 10.1073/pnas.91.3.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397: 271–274. 10.1038/16729 [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell 5: 897–904. 10.1016/S1097-2765(00)80330-5 [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Bertolotti A, Zeng H, Zhang Y, Urano F, Jousse C, Ron D. 2001. Translational regulation in the cellular response to biosynthetic load on the endoplasmic reticulum. Cold Spring Harb Symp Quant Biol 66: 499–508. 10.1101/sqb.2001.66.499 [DOI] [PubMed] [Google Scholar]
- Harding HP, Calfon M, Urano F, Novoa I, Ron D. 2002. Transcriptional and translational control in the Mammalian unfolded protein response. Annu Rev Cell Dev Biol 18: 575–599. 10.1146/annurev.cellbio.18.011402.160624 [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11: 619–633. 10.1016/S1097-2765(03)00105-9 [DOI] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell 10: 3787–3799. 10.1091/mbc.10.11.3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze K, Okada T, Yoshida H, Yanagi H, Yura T, Negishi M, Mori K. 2001. Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem J 355: 19–28. 10.1042/bj3550019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Marquardt T, Braakman I. 1992. The endoplasmic reticulum as a protein-folding compartment. Trends Cell Biol 2: 227–231. 10.1016/0962-8924(92)90309-B [DOI] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. 2006. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313: 104–107. 10.1126/science.1129631 [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. 2009. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 186: 323–331. 10.1083/jcb.200903014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS. 2010. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 140: 900–917. 10.1016/j.cell.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Nagata K. 2017. Biology of Hsp47 (Serpin H1), a collagen-specific molecular chaperone. Semin Cell Dev Biol 62: 142–151. 10.1016/j.semcdb.2016.11.005 [DOI] [PubMed] [Google Scholar]
- Iwakoshi NN, Lee AH, Glimcher LH. 2003a. The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol Rev 194: 29–38. 10.1034/j.1600-065X.2003.00057.x [DOI] [PubMed] [Google Scholar]
- Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. 2003b. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol 4: 321–329. 10.1038/ni907 [DOI] [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Kohno K. 2010. IRE1α disruption causes histological abnormality of exocrine tissues, increase of blood glucose level, and decrease of serum immunoglobulin level. PLoS ONE 5: e13052 10.1371/journal.pone.0013052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagöz GE, Duarte AM, Akoury E, Ippel H, Biernat J, Moran Luengo T, Radli M, Didenko T, Nordhues BA, Veprintsev DB, et al. 2014. Hsp90-Tau complex reveals molecular basis for specificity in chaperone action. Cell 156: 963–974. 10.1016/j.cell.2014.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagöz GE, Acosta-Alvear D, Nguyen HT, Lee CP, Chu F, Walter P. 2017. An unfolded protein-induced conformational switch activates mammalian IRE1. eLife 6: e30700 10.7554/eLife.30700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, et al. 2008. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134: 743–756. 10.1016/j.cell.2008.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada K, Iekumo T, Saito R, Kaneko M, Mimori S, Nomura Y, Okuma Y. 2014. Aberrant neuronal differentiation and inhibition of dendrite outgrowth resulting from endoplasmic reticulum stress. J Neurosci Res 92: 1122–1133. 10.1002/jnr.23389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, et al. 2015. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522: 354–358. 10.1038/nature14498 [DOI] [PubMed] [Google Scholar]
- Kimata Y, Oikawa D, Shimizu Y, Ishiwata-Kimata Y, Kohno K. 2004. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J Cell Biol 167: 445–456. 10.1083/jcb.200405153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Ishiwata-Kimata Y, Ito T, Hirata A, Suzuki T, Oikawa D, Takeuchi M, Kohno K. 2007. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J Cell Biol 179: 75–86. 10.1083/jcb.200704166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleizen B, Braakman I. 2004. Protein folding and quality control in the endoplasmic reticulum. Curr Opin Cell Biol 16: 343–349. 10.1016/j.ceb.2004.06.012 [DOI] [PubMed] [Google Scholar]
- Kohno K, Normington K, Sambrook J, Gething MJ, Mori K. 1993. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol 13: 877–890. 10.1128/MCB.13.2.877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono N, Amin-Wetzel N, Ron D. 2017. Generic membrane-spanning features endow IRE1α with responsiveness to membrane aberrancy. Mol Biol Cell 28: 2318–2332. 10.1091/mbc.e17-03-0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp MC, Nowak PR, Larburu N, Adams CJ, Ali MM. 2018. In vitro FRET analysis of IRE1 and BiP association and dissociation upon endoplasmic reticulum stress. eLife 7: e30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Zhang C, Shokat KM, Stroud RM, Walter P. 2009. The unfolded protein response signals through high-order assembly of Ire1. Nature 457: 687–693. 10.1038/nature07661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CW, Otero JH, Hendershot LM, Snapp E. 2012. ERdj4 protein is a soluble endoplasmic reticulum (ER) DnaJ family protein that interacts with ER-associated degradation machinery. J Biol Chem 287: 7969–7978. 10.1074/jbc.M111.311290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23: 7448–7459. 10.1128/MCB.23.21.7448-7459.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. 2005. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J 24: 4368–4380. 10.1038/sj.emboj.7600903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH. 2008. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320: 1492–1496. 10.1126/science.c1158042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Korennykh AV, Behrman SL, Walter P. 2010. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc Natl Acad Sci 107: 16113–16118. 10.1073/pnas.1010580107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. 2007. IRE1 signaling affects cell fate during the unfolded protein response. Science 318: 944–949. 10.1126/science.1146361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Walter P, Yen TS. 2008. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol 3: 399–425. 10.1146/annurev.pathmechdis.3.121806.151434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Schröder M, Kaufman RJ. 2000. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem 275: 24881–24885. 10.1074/jbc.M004454200 [DOI] [PubMed] [Google Scholar]
- Lu M, Lawrence DA, Marsters S, Acosta-Alvear D, Kimmig P, Mendez AS, Paton AW, Paton JC, Walter P, Ashkenazi A. 2014. Cell death. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science 345: 98–101. 10.1126/science.1254312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Shimizu Y, Mann MJ, Jin Y, Hendershot LM. 2010. Plasma cell differentiation initiates a limited ER stress response by specifically suppressing the PERK-dependent branch of the unfolded protein response. Cell Stress Chaperones 15: 281–293. 10.1007/s12192-009-0142-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai TC, Munakata T, Tran DM, Takagi H, Kimata Y. 2018. A chimeric mutant analysis in yeast cells suggests BiP independent regulation of the mammalian endoplasmic reticulum-stress sensor IRE1α. Biosci Biotechnol Biochem 82: 1527–1530. [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Garcia-Bonilla L, Hu J, Harding HP, Ron D. 2006. Activation-dependent substrate recruitment by the eukaryotic translation initiation factor 2 kinase PERK. J Cell Biol 172: 201–209. 10.1083/jcb.200508099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcinowski M, Höller M, Feige MJ, Baerend D, Lamb DC, Buchner J. 2011. Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat Struct Mol Biol 18: 150–158. 10.1038/nsmb.1970 [DOI] [PubMed] [Google Scholar]
- Martinez G, Vidal RL, Mardones P, Serrano FG, Ardiles AO, Wirth C, Valdes P, Thielen P, Schneider BL, Kerr B, et al. 2016. Regulation of memory formation by the transcription factor XBP1. Cell Rep 14: 1382–1394. 10.1016/j.celrep.2016.01.028 [DOI] [PubMed] [Google Scholar]
- Martino MB, Jones L, Brighton B, Ehre C, Abdulah L, Davis CW, Ron D, O'Neal WK, Ribeiro CM. 2013. The ER stress transducer IRE1β is required for airway epithelial mucin production. Mucosal Immunol 6: 639–654. 10.1038/mi.2012.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathuranyanon R, Tsukamoto T, Takeuchi A, Ishiwata- Kimata Y, Tsuchiya Y, Kohno K, Kimata Y. 2015. Tight regulation of the unfolded protein sensor Ire1 by its intramolecularly antagonizing subdomain. J Cell Sci 128: 1762–1772. 10.1242/jcs.164111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura N, Fulciniti M, Gorgun G, Tai YT, Cirstea D, Santo L, Hu Y, Fabre C, Minami J, Ohguchi H, et al. 2012. Blockade of XBP1 splicing by inhibition of IRE1α is a promising therapeutic option in multiple myeloma. Blood 119: 5772–5781. 10.1182/blood-2011-07-366633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K. 2009. Signalling pathways in the unfolded protein response: Development from yeast to mammals. J Biochem 146: 743–750. 10.1093/jb/mvp166 [DOI] [PubMed] [Google Scholar]
- Mori K, Nadanaka S. 2003. Intracellular traffic of membrane-bound transcription factors. Seikagaku 75: 506–511. [PubMed] [Google Scholar]
- Mulvey M, Arias C, Mohr I. 2007. Maintenance of endoplasmic reticulum (ER) homeostasis in herpes simplex virus type 1-infected cells through the association of a viral glycoprotein with PERK, a cellular ER stress sensor. J Virol 81: 3377–3390. 10.1128/JVI.02191-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Saito A, Hino S, Kondo S, Kanemoto S, Chihara K, Sekiya H, Tsumagari K, Ochiai K, Yoshinaga K, et al. 2009. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat Cell Biol 11: 1205–1211. 10.1038/ncb1963 [DOI] [PubMed] [Google Scholar]
- Murao N, Nishitoh H. 2017. Role of the unfolded protein response in the development of central nervous system. J Biochem 162: 155–162. 10.1093/jb/mvx047 [DOI] [PubMed] [Google Scholar]
- Nadanaka S, Yoshida H, Mori K. 2006a. Reduction of disulfide bridges in the lumenal domain of ATF6 in response to glucose starvation. Cell Struct Funct 31: 127–134. 10.1247/csf.06024 [DOI] [PubMed] [Google Scholar]
- Nadanaka S, Yoshida H, Sato R, Mori K. 2006b. Analysis of ATF6 activation in Site-2 protease-deficient Chinese hamster ovary cells. Cell Struct Funct 31: 109–116. 10.1247/csf.06015 [DOI] [PubMed] [Google Scholar]
- Nadanaka S, Okada T, Yoshida H, Mori K. 2007. Role of disulfide bridges formed in the luminal domain of ATF6 in sensing endoplasmic reticulum stress. Mol Cell Biol 27: 1027–1043. 10.1128/MCB.00408-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DT, Watowich SS, Lamb RA. 1992. Analysis in vivo of GRP78-BiP/substrate interactions and their role in induction of the GRP78-BiP gene. Mol Biol Cell 3: 143–155. 10.1091/mbc.3.2.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Sidrauski C, Kaufman RJ, Walter P. 1999. A role for presenilin-1 in nuclear accumulation of Ire1 fragments and induction of the mammalian unfolded protein response. Cell 99: 691–702. 10.1016/S0092-8674(00)81667-0 [DOI] [PubMed] [Google Scholar]
- Oikawa D, Kimata Y, Kohno K, Iwawaki T. 2009. Activation of mammalian IRE1α upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp Cell Res 315: 2496–2504. 10.1016/j.yexcr.2009.06.009 [DOI] [PubMed] [Google Scholar]
- Okamura K, Kimata Y, Higashio H, Tsuru A, Kohno K. 2000. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem Biophys Res Commun 279: 445–450. 10.1006/bbrc.2000.3987 [DOI] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS. 2004. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 306: 457–461. 10.1126/science.1103160 [DOI] [PubMed] [Google Scholar]
- Papandreou I, Denko NC, Olson M, Van Melckebeke H, Lust S, Tam A, Solow-Cordero DE, Bouley DM, Offner F, Niwa M, et al. 2011. Identification of an Ire1α endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood 117: 1311–1314. 10.1182/blood-2010-08-303099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus D, Chevalier MW, Aragon T, van Anken E, Vidal SE, El-Samad H, Walter P. 2010. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol 8: e1000415 10.1371/journal.pbio.1000415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumb R, Zhang ZR, Appathurai S, Mariappan M. 2015. A functional link between the co-translational protein translocation pathway and the UPR. eLife 4 10.7554/eLife.07426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promlek T, Ishiwata-Kimata Y, Shido M, Sakuramoto M, Kohno K, Kimata Y. 2011. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol Biol Cell 22: 3520–3532. 10.1091/mbc.e11-04-0295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, et al. 2000. An essential role in liver development for transcription factor XBP-1. Genes Dev 14: 152–157. [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. 2001. Plasma cell differentiation requires the transcription factor XBP-1. Nature 412: 300–307. 10.1038/35085509 [DOI] [PubMed] [Google Scholar]
- Saito A, Ochiai K, Kondo S, Tsumagari K, Murakami T, Cavener DR, Imaizumi K. 2011. Endoplasmic reticulum stress response mediated by the PERK-eIF2α-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. J Biol Chem 286: 4809–4818. 10.1074/jbc.M110.152900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbalzarini IF, Hayer A, Helenius A, Koumoutsakos P. 2006. Simulations of (an)isotropic diffusion on curved biological surfaces. Biophys J 90: 878–885. 10.1529/biophysj.105.073809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. 2009. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol 187: 525–536. 10.1083/jcb.200907074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, Gallagher CM, Walter P. 2014. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J Cell Sci 127: 4078–4088. 10.1242/jcs.154716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda D, Rojas-Rivera D, Rodriguez DA, Groenendyk J, Kohler A, Lebeaupin C, Ito S, Urra H, Carreras-Sureda A, Hazari Y, et al. 2018. Interactome screening identifies the ER luminal chaperone Hsp47 as a regulator of the unfolded protein response transducer IRE1α. Mol Cell 69: 238–252.e7. 10.1016/j.molcel.2017.12.028 [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, et al. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21: 81–93. 10.1016/j.immuni.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Sharma R, Ishimaru Y, Davison I, Ikegami K, Chien MS, You H, Chi Q, Kubota M, Yohda M, Ehlers M, et al. 2017. Olfactory receptor accessory proteins play crucial roles in receptor function and gene choice. eLife 6: e21895 10.7554/eLife.21895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R. 2002. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell 3: 99–111. 10.1016/S1534-5807(02)00203-4 [DOI] [PubMed] [Google Scholar]
- Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC. 1998. Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translational control. Mol Cell Biol 18: 7499–7509. 10.1128/MCB.18.12.7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Walter P. 1997. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90: 1031–1039. 10.1016/S0092-8674(00)80369-4 [DOI] [PubMed] [Google Scholar]
- Siggia ED, Lippincott-Schwartz J, Bekiranov S. 2000. Diffusion in inhomogeneous media: Theory and simulations applied to whole cell photobleach recovery. Biophys J 79: 1761–1770. 10.1016/S0006-3495(00)76428-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriburi R, Jackowski S, Mori K, Brewer JW. 2004. XBP1: A link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol 167: 35–41. 10.1083/jcb.200406136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl S, Burkhart JM, Hinte F, Tirosh B, Mohr H, Zahedi RP, Sickmann A, Ruzsics Z, Budt M, Brune W. 2013. Cytomegalovirus downregulates IRE1 to repress the unfolded protein response. PLoS Pathog 9: e1003544 10.1371/journal.ppat.1003544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street TO, Lavery LA, Agard DA. 2011. Substrate binding drives large-scale conformational changes in the Hsp90 molecular chaperone. Mol Cell 42: 96–105. 10.1016/j.molcel.2011.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram A, Plumb R, Appathurai S, Mariappan M. 2017. The Sec61 translocon limits IRE1α signaling during the unfolded protein response. eLife 6: e27187 10.7554/eLife.27187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram A, Appathurai S, Plumb R, Mariappan M. 2018. Dynamic changes in complexes of IRE1α, PERK, and ATF6α during endoplasmic reticulum stress. Mol Biol Cell 29: 1376–1388. 10.1091/mbc.E17-10-0594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung SC, Chao CY, Jeng KS, Yang JY, Lai MM. 2009. The 8ab protein of SARS-CoV is a luminal ER membrane-associated protein and induces the activation of ATF6. Virology 387: 402–413. 10.1016/j.virol.2009.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekko T, Lillevali K, Luuk H, Sutt S, Truu L, Ord T, Mols M, Vasar E. 2014. Initiation and developmental dynamics of Wfs1 expression in the context of neural differentiation and ER stress in mouse forebrain. Int J Dev Neurosci 35: 80–88. 10.1016/j.ijdevneu.2014.03.009 [DOI] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ. 1998. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev 12: 1812–1824. 10.1101/gad.12.12.1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ. 2000. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev 14: 2725–2736. 10.1101/gad.839400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd-Corlett A, Jones E, Seghers C, Gething MJ. 2007. Lobe IB of the ATPase domain of Kar2p/BiP interacts with Ire1p to negatively regulate the unfolded protein response in Saccharomyces cerevisiae. J Mol Biol 367: 770–787. 10.1016/j.jmb.2007.01.009 [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258. 10.1016/S0092-8674(00)80835-1 [DOI] [PubMed] [Google Scholar]
- Treacy-Abarca S, Mukherjee S. 2015. Legionella suppresses the host unfolded protein response via multiple mechanisms. Nat Commun 6: 7887 10.1038/ncomms8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuru A, Fujimoto N, Takahashi S, Saito M, Nakamura D, Iwano M, Iwawaki T, Kadokura H, Ron D, Kohno K. 2013. Negative feedback by IRE1β optimizes mucin production in goblet cells. Proc Natl Acad Sci 110: 2864–2869. 10.1073/pnas.1212484110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufanli O, Telkoparan Akillilar P, Acosta-Alvear D, Kocaturk B, Onat UI, Hamid SM, Çimen I, Walter P, Weber C, Erbay E. 2017. Targeting IRE1 with small molecules counteracts progression of atherosclerosis. Proc Natl Acad Sci 114: E1395–E1404. 10.1073/pnas.1621188114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cauter E, Mestrez F, Sturis J, Polonsky KS. 1992. Estimation of insulin secretion rates from C-peptide levels. Comparison of individual and standard kinetic parameters for C-peptide clearance. Diabetes 41: 368–377. 10.2337/diabetes.41.3.368 [DOI] [PubMed] [Google Scholar]
- Vidal RL, Figueroa A, Court FA, Thielen P, Molina C, Wirth C, Caballero B, Kiffin R, Segura-Aguilar J, Cuervo AM, et al. 2012. Targeting the UPR transcription factor XBP1 protects against Huntington's disease through the regulation of FoxO1 and autophagy. Hum Mol Genet 21: 2245–2262. 10.1093/hmg/dds040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volmer R, van der Ploeg K, Ron D. 2013. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc Natl Acad Sci 110: 4628–4633. 10.1073/pnas.1217611110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G. 1980. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc Natl Acad Sci 77: 7112–7116. 10.1073/pnas.77.12.7112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Gilmore R, Muller M, Blobel G. 1982. The protein translocation machinery of the endoplasmic reticulum. Philos Trans R Soc Lond B Biol Sci 300: 225–228. 10.1098/rstb.1982.0168 [DOI] [PubMed] [Google Scholar]
- Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. 1998. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J 17: 5708–5717. 10.1093/emboj/17.19.5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. 2000. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem 275: 27013–27020. [DOI] [PubMed] [Google Scholar]
- Wang M, Herrmann CJ, Simonovic M, Szklarczyk D, von Mering C. 2015. Version 4.0 of PaxDb: Protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics 15: 3163–3168. 10.1002/pmic.201400441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Li J, Sha B. 2016. The ER stress sensor PERK luminal domain functions as a molecular chaperone to interact with misfolded proteins. Acta Crystallogr D Struct Biol 72: 1290–1297. 10.1107/S2059798316018064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Li J, Tao J, Sha B. 2018. The luminal domain of the ER stress sensor protein PERK binds misfolded proteins and thereby triggers PERK oligomerization. J Biol Chem 293: 4110–4121. 10.1074/jbc.RA117.001294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Sheng X, Feng D, McGrath B, Cavener DR. 2008. PERK is essential for neonatal skeletal development to regulate osteoblast proliferation and differentiation. J Cell Physiol 217: 693–707. 10.1002/jcp.21543 [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. 2007. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev Cell 13: 365–376. 10.1016/j.devcel.2007.07.018 [DOI] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Davé UP, Prywes R, Brown MS, Goldstein JL. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell 6: 1355–1364. 10.1016/S1097-2765(00)00133-7 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Haze K, Yanagi H, Yura T, Mori K. 1998. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem 273: 33741–33749. 10.1074/jbc.273.50.33741 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107: 881–891. 10.1016/S0092-8674(01)00611-0 [DOI] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ. 2008. From endoplasmic-reticulum stress to the inflammatory response. Nature 454: 455–462. 10.1038/nature07203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Cavener DR. 2002. The PERK eukaryotic initiation factor 2α kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol 22: 3864–3874. 10.1128/MCB.22.11.3864-3874.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. 2005. The unfolded protein response sensor IRE1α is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest 115: 268–281. 10.1172/JCI200521848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Feng D, Li Y, Iida K, McGrath B, Cavener DR. 2006. PERK EIF2AK3 control of pancreatic β cell differentiation and proliferation is required for postnatal glucose homeostasis. Cell Metab 4: 491–497. 10.1016/j.cmet.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Zhou J, Liu CY, Back SH, Clark RL, Peisach D, Xu Z, Kaufman RJ. 2006. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci 103: 14343–14348. 10.1073/pnas.0606480103 [DOI] [PMC free article] [PubMed] [Google Scholar]