Abstract
Fluorogenic RNA aptamers bind and activate the fluorescence of otherwise nonfluorescent dyes. However, fluorogenic aptamers are limited by the small number of fluorogenic dyes suitable for use in live cells. Here we describe fluorogenic proteins whose fluorescence is activated by RNA aptamers. Fluorogenic proteins are highly unstable until they bind RNA aptamers inserted in mRNAs, resulting in fluorescent RNA-protein complexes that enable live imaging of mRNA in living cells.
Fluorogenic RNA aptamers are RNA aptamers that bind otherwise nonfluorescent molecules and switch them to a fluorescent form. These fluorogenic dyes can be applied to cells, enabling RNAs tagged with these fluorogenic aptamers to be imaged using fluorescence microscopy1,2. However, few fluorogenic aptamers have been developed since there are not many fluorogenic dyes that meet the criteria required for use in live cells. For example, most dyes show nonspecific fluorescence activation by cellular lipids or DNA3,4. Another problem is that the fluorogenic dyes are not genetically encoded and therefore need to be added exogenously for RNA imaging. A genetically encoded conditionally fluorescent dye would simplify the use of fluorogenic RNA aptamers.
In order to expand fluorogenic aptamer-based imaging, we sought to create a new class of fluorogenic dyes that are genetically encoded. Fluorescent proteins are particularly useful since a diverse array of spectrally distinct proteins have been described5 . However, these proteins are constitutively fluorescent. To make them dependent on RNA, we considered making them rapidly degraded in cells except when bound by a specific RNA aptamer. In this way, fluorescence would be selectively associated with RNA-protein complexes, and not with unbound fluorescent protein. This would be functionally equivalent to RNA-induced fluorescence of small molecule dyes.
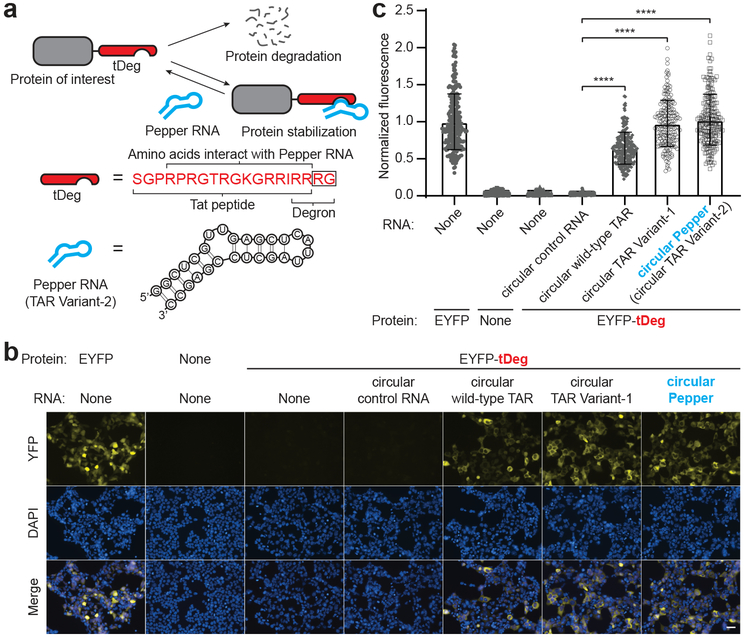
We first developed a “destabilization domain” that can be inhibited by an RNA aptamer. Previously, the Arg-Arg-Arg-Gly was described as a degron sequence when appended to the C-terminus of proteins6 . This sequence is similar to the arginine-rich RNA-binding domain of the Tat protein, which contains Arg-Arg as its last two amino acids. Therefore, we appended Arg-Gly to extend this Arg-Arg sequence so that the full Arg-Arg-Arg-Gly degron is at the C-terminus of this peptide (Fig. 1a and Supplementary Fig. 1). We termed this 19-amino acid-long bifunctional peptide “tDeg.” Tat binds a 28 nt-long RNA hairpin termed TAR7,8 , which may shield the degron and thus prevent recruitment of the proteasomal machinery needed for proteolysis (Fig. 1a and Supplementary Fig. 1).
Fig. 1 ∣. Design and optimization of an RNA-regulated protein destabilization domain.
a, Schematic drawing of a Pepper RNA-regulated protein destabilization domain, tDeg. tDeg is a bifunctional peptide that consists of the Tat peptide, which is capable of binding to the Pepper RNA aptamer, and the previously described C-terminal Arg-Arg-Arg-Gly degron6. When fused to a protein of interest, tDeg causes protein degradation. However, the protein destabilization function of tDeg is impeded when it binds to the Pepper RNA aptamer. Amino acids Arg-Gly, highlighted in a black box, are appended to the C-terminus of Tat to make the full Arg-Arg-Arg-Gly degron.
b, Pepper RNA stabilizes EYFP fused to tDeg in cells. To test whether tDeg functions as an RNA-regulated destabilization domain, we coexpressed EYFP-tDeg with different circular RNAs, and imaged the yellow fluorescence in HEK293T cells. Without circular wild-type TAR RNA or its variants, cells coexpressing EYFP-tDeg and the circular control RNA only showed minimal fluorescence above background fluorescence. Cells exhibit yellow fluorescence only when circular wild-type TAR RNA, TAR Variant-1 or TAR Vairnat-2 (named Pepper) was coexpressed. Notably, we observed higher yellow fluorescence signals in the cytosol compared to the nucleus when EYFP-tDeg was coexpressed with the circular wild-type TAR RNA or its variants. This is consistent with the cytosolic expression of small circular RNAs using the Tornado expression system 9 . All cells were stained with Hoechst dye. Scale bar, 40 μm.
c, Summary data of normalized fluorescence of untransfected HEK293T cells, or HEK293T cells expressing EYFP, or EYFP-tDeg with different RNAs as in (b). Total cellular yellow fluorescence of individual cells is plotted (n = 4 independent cell cultures). Values are means ± s.d. ****Pcircular wild-type TAR = 7.9 × 10−113; ****Pcircular TAR Variant-1 = 2.1 × 10−117; ****Pcircular TAR Variant-2 = 1.7 × 10−115 by one-way ANOVA.
We first asked if tDeg confers instability to proteins. We fused tDeg to the C-terminus of enhanced yellow fluorescent protein (EYFP), and expressed this fusion protein (EYFP-tDeg) in HEK293T cells. While EYFP was readily detectable, EYFP-tDeg was nearly undetectable (Fig. 1b,c). EYFP-tDeg was restored by proteasome inhibition (Supplementary Fig. 2) indicated that tDeg reduces protein stability by inducing proteasomal degradation.
We next asked if the tDeg can be regulated by the TAR RNA. We expressed the TAR RNA as a circular RNA using the Tornado ribozyme-assisted circularization approach to achieve high expression in mammalian cells9 . When TAR was expressed, EYFP-tDeg-expressing cells exhibited a 24-fold increase of fluorescence relative to control RNA (Fig. 1b,c). TAR variants that bind Tat with higher affinity, Variant-1 and Variant-210, were even more efficient at inducing EYFP-tDeg, with Variant-2 exhibiting a 38-fold increase in cellular fluorescence (Fig. 1b,c and Supplementary Fig. 3). Expression of Variant-2 induced EYFP-tDeg cellular fluorescence levels that similar to levels in cells expressing EYFP without the tDeg (Fig. 1c). Furthermore, Variant-2 induced EYFP-tDeg fluorescence in diverse cell types (Supplementary Fig. 4). Thus, the EYFP-tDeg is a fluorogenic protein that is regulated by TAR.
Because the TAR Variant-2 aptamer can control the expression of different colored fluorescent proteins, as will be described below, we named this aptamer after the multicolored vegetable Pepper, in keeping with the vegetable nomenclature system used previously for fluorogenic RNA aptamers.
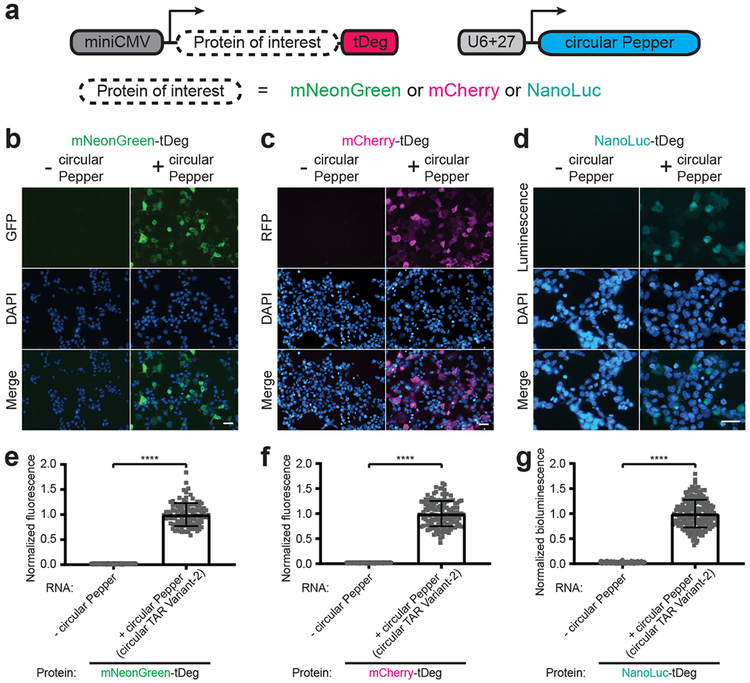
We next asked if the expression level of other proteins could be controlled by the Pepper RNA. Addition of tDeg to the C-terminus of mNeonGreen, mCherry, NanoLuc, tetracycline repressor protein (TetR), EZH2, and NF-κB, resulted in minimal or undetectable protein levels in control cells and clear induction in circular Pepper-expressing cells (Fig. 2 and Supplementary Fig. 5). Taken together, these data indicate that the tDeg tag is a versatile tag for RNA-dependent protein stabilization.
Fig. 2 ∣. tDeg confers Pepper RNA-dependent regulation to diverse proteins.
To test whether Pepper RNA stabilizes different proteins fused to tDeg, we imaged HEK293T cells expressing mNeonGreen (b, e), mCherry (c, f), and the luciferase NanoLuc (d, g) fused to a C-terminal tDeg tag with and without circular Pepper RNA (a), respectively. In each case, there was a considerable increase of fluorescence (e, f) or bioluminescence (g) of the tDeg-tagged protein only when circular Pepper RNA was coexpressed in cells. For detecting bioluminescence, cells were incubated in media with furimazine (from Promega Nano-Glo® Luciferase Assay System, diluted 100x) and imaged using a 460 ± 25 nm emission filter cube. All cells were stained with Hoechst dye. Scale bar, 40 μm. Normalized total cellular fluorescence (e and f) or bioluminescence (g) of individual cells is plotted (n = 3 independent cell cultures). Values are means ± s.d. ****PmNeonGreen-tDeg = 1.1 × 10−123; ****PmCherry-tDeg = 3.0 × 10−131; ****PNanoLuc-tDeg = 1.7 × 10−120 by unpaired two-tailed Student’s t-test.
mRNAs are commonly imaged using tethered fluorescent proteins. For example, a GFP fusion with MS2 phage coat protein (MCP) can be recruited to mRNAs contain 24–48 consecutive MS2 RNA hairpins in their 3’UTRs11. In this way, many GFPs are recruited to single mRNAs resulting in an aggregate fluorescence that can be detected by fluorescence microscopy. Typically nuclear localization elements are added to the GFP-MCP fusion to remove the unbound fluorescent protein from the cytoplasm into the nucleus11. This can reduce the fluorescence background in the cytosol, facilitating mRNA detection. However, this may introduce a potential artifact since the MS2-tagged mRNAs will contain dozens of nuclear localization sequences due to the recruited fluorescent proteins12. Fluorogenic RNA aptamers do not introduce a cellular trafficking element and may therefore bypass this concern.
We therefore generated a tag for mRNA imaging consisting of consecutive Pepper aptamers. In optimization experiments, we imaged an mCherry mRNA reporter containing different 3’UTR tags comprising 10 or 20 concatenated Pepper aptamers, Pepper aptamers that were inserted into an RNA three-way junction sequence termed F30. Aptamers inserted within the F30 show improved folding13. mCherry mRNA was readily detectable as mobile fluorescent puncta in the cytoplasm when the tag contained 20 Pepper aptamers. The brightest puncta were seen when using the (F30–2xPepper)10 tag, which comprises 10 consecutive F30 sequences, with each of the two arms of F30 containing one Pepper aptamer (Supplementary Fig. 6-8 and Supplementary Video 1).
We also tested mRNA imaging using fluorogenic proteins of different brightness. These proteins comprised 2, 3, or 4 tandem mNeonGreen monomers with a C-terminal tDeg. In these experiments, a fluorogenic protein comprising four mNeonGreens provided the highest signal-to-noise ratio for imaging mRNAs (Supplementary Fig. 8). Although most fluorescent puncta were detected in the cytoplasm, occasional puncta were detected in the nucleus, potentially reflecting mRNAs prior to nuclear export (Supplementary Fig. 9).
Cellular puncta likely reflect single mRNA molecules rather than Pepper-containing mRNA fragments since northern blotting of total cellular RNA derived from cells expressing (F30–2xPepper)10-tagged mRNA, either with or without coexpression of the (mNeonGreen)4-tDeg showed mostly full-length transcripts (Supplementary Fig. 10a). Furthermore, puncta derived from mRNAs tagged with (F30–2xPepper)10 were the same size and intensity as mRNAs tagged using the PP7 fluorescent protein recruitment system, which was previously shown to reflect single mRNA molecules14 (Supplementary Fig. 10b-d).
Adding the Pepper tag to an mRNA could adversely affect mRNA fate. However, we did not find that the (F30–2xPepper)10 Pepper tag substantially altered the stability of the mCherry transcript (Supplementary Fig. 11a). Similarly, we did not see a significant difference in protein translation between the untagged and Pepper-tagged mCherry mRNA transcript (Supplementary Fig. 11b-d). Lastly, we found that expression of fluorogenic proteins did not significantly affect total cellular proteasome activity (Supplementary Fig. 11e).
We next imaged mRNAs that exhibit specific subcellular localizations. We first imaged mRNA localization to the endoplasmic reticulum (ER) using an ER-targeted reporter mRNA that encodes the first 29 amino acids of cytochrome P450, CytERM (cytoplasmic end of an endoplasmic reticulum signal-anchor membrane protein)15. This sequence tethers the mRNA to the outer ER membrane during protein translation, and restricts the mRNA’s mobility. Indeed, we observed fluorescent puncta with low mobility when this mRNA was expressed with a 3’UTR (F30–2xPepper)10 Pepper tag (Supplementary Fig. 12). Treatment with puromycin, which disrupts the ribosome and dissociates the mRNA from the nascent peptide, significantly increased puncta mobility, consistent with dissociation of the reporter mRNA from the ER (Supplementary Fig. 12 and Supplementary Video 2).
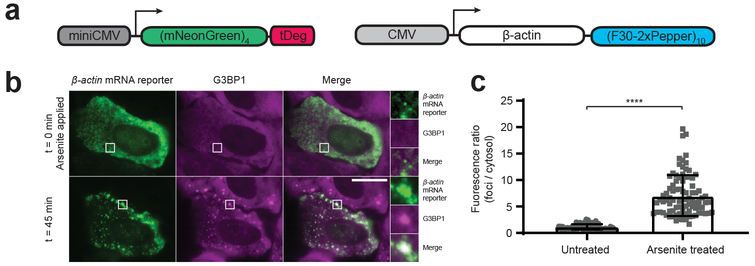
We next expressed β-actin mRNA containing a 3’UTR (F30–2xPepper)10 tag and imaged its localization in response to arsenite treatment, which induces stress granule formation16 . Upon application of 500 μM arsenite, the individual fluorescent puncta rapidly accumulated to form stress granules as evidenced by coexpression of Halo-tagged G3BP1 to label stress granules (Fig. 3 and Supplementary Fig. 13).
Fig. 3 ∣. Imaging green Pepper-tagged β-actin mRNA in live cells.
a, DNA plasmid constructs used for imaging β-actin mRNA in live cells.
b, To image β-actin mRNA localization in response to arsenite stress, we constructed a β-actin mRNA reporter containing a 3’UTR green Pepper mRNA tag, (F30-2xPepper)10. Cells coexpressing this β-actin mRNA reporter and a green fluorogenic protein, (mNeonGreen)4-tDeg were imaged before and 45 minutes after arsenite (500 μM) treatment to induce stress granules. We observed individual mRNA transcripts rapidly accumulated to form stress granules as evidenced by coexpression of tetramethylrhodamine-labeled HaloTag-G3BP1 to label stress granules. Scale bar, 20 μm.
c, Fluorescence ratio of foci / cytosol in untreated cells vs. arsenite treated cells is plotted (n = 3 independent cell cultures). Values are means ± s.d. ****P = 2.5 × 10−31 by unpaired two-tailed Student’s t-test.
To expand the color palette of RNA-regulated fluorogenic proteins, we fused two tandem copies of mVenus and two tandem copies of mCherry with a C-terminal tDeg tag to convert them into fluorogenic proteins, respectively, for imaging mRNAs. In both cases, we detected fluorescent puncta in the yellow and red fluorescence channels, respectively (Supplementary Fig. 14). Together, these data show that Pepper-tagged mRNAs can be imaged in different colors using different fluorogenic proteins.
Overall, our studies describe how constitutively fluorescent proteins can be converted to fluorogenic proteins that are regulated by RNA aptamers. We confer fluorogenicity to a protein by making its stability controlled by an RNA aptamer, Pepper. In this way, unbound fluorogenic protein is rapidly degraded, but the fluorogenic protein bound to Pepper remains stable. Thus, these Pepper-regulated fluorogenic proteins are functionally analogous to RNA-regulated fluorogenic dyes. This system has the advantage of being able to use diverse fluorogenic proteins with diverse spectral properties. Additionally, unlike our previous Spinach system1, the fluorogenic system described here is fully genetically encoded.
Fluorophore maturation kinetics may also contribute to the low fluorescence of the Pepper system. Since the tDeg tag is highly efficient, it is possible that newly synthesized mNeonGreen is degraded prior to chromophore maturation. mNeonGreen that is bound to the RNA may persist for a sufficiently long time to mature to a fluorescent form while bound to RNA. This may further contribute to the low background fluorescence in cells.
Unlike previous mRNA imaging systems, no nuclear localization elements are added to fluorescent proteins to lower cytosolic background fluorescence. Instead, low background fluorescence is achieved by the highly efficient degradation of the unbound fluorogenic protein. The simplicity of this system should simplify mRNA imaging.
An important question is whether the tagged mRNA faithfully recapitulates behavior of the endogenous mRNA. The Pepper tag did not substantially affect the stability, translation, and localization of the specific mRNAs we tested. Nevertheless, imaging tags are best used when comparing two mRNAs that differ by a single sequence alteration, or the same mRNA compared in two different conditions. In this way the role of a putative functional RNA element or RNA-regulatory pathway can be inferred and then validated with the endogenous mRNA.
Although we used the RNA-regulated destabilization domains to create fluorogenic proteins for RNA imaging, the ability to control protein expression levels through the Pepper aptamer can potentially enable novel synthetic biology applications. For these applications, Pepper can be expressed on its own, rather than part of an mRNA. By expressing tDeg-tagged proteins, diverse types of protein functions can be regulated by RNA aptamer expression levels.
ONLINE METHODS
General methods and materials.
Single stranded synthetic DNA oligonucleotides for PCR were purchased from Integrated DNA Technologies. Phusion® High-Fidelity DNA Polymerase (NEB M0530) were used for routine PCR amplifications. PCR products were run on 1% TAE agarose gels. PCR products with correct size were then excised and purified with the Qiaquick Gel Extraction kit (Qiagen 28704). Restriction endonucleases used for restriction digest were purchased from New England Biolabs, and used according to the manufacturer’s recommended protocol. DNA ligation reactions were carried out using the Quick Ligation™ Kit (NEB M2200L). DNA plasmids were propagated using chemically competent E. coli (Agilent 200314). The QIAprep Spin Plasmid Miniprep Kit (Qiagen 27106) was used for DNA plasmid extraction and purification from E. coli. DNA sequencing (GENEWIZ) was used to verify the inserted gene sequences.
Cell culture and transfection.
HEK293T/17 (ATCC CRL-11268), U2OS (ATCC HTB-96), COS-7 (ATCC CRL-1651), and HeLa (ATCC CCL-2) cells were cultured in DMEM (Thermo Fisher Scientific 11995–065) supplemented with 10% fetal bovine serum (Corning 35–010-CV), 100 U ml−1 penicillin and 100 μg ml−1 of streptomycin (Thermo Fisher Scientific 15140122) under 37°C with 5% CO2. TrypLE Express (Thermo Fisher Scientific 12604013) was used for detaching cells from culture flasks during cell passage. All cell lines used in this study were transfected using FuGENE HD (Promega 2311) according to the manufacturer’s instructions. Prior to live-cell imaging, cells were changed to imaging media: phenol red-free DMEM (Thermo Fisher Scientific 31053–028) supplemented with 10% fetal bovine serum (Corning 35–010-CV), 100 U ml−1 penicillin and 100 μg ml−1 of streptomycin (Thermo Fisher Scientific 15140122), 1x GlutaMAX™(Thermo Fisher Scientific 35050–061), and 1 mM sodium pyruvate (Thermo Fisher Scientific 11360–070).
Fluorescence and bioluminescence imaging of tDeg-tagged proteins.
To construct an expression vector for EYFP, EYFP-tDeg, mNeonGreen-tDeg, mCherry-tDeg, NanoLuc-tDeg, EGFP-TetR-tDeg, EGFP-EZH2-tDeg, or mCherry-NF-κB-tDeg, a pcDNA3.1(+) vector was digested by MluI and XbaI and ligated to an insert comprising a miniCMV promoter (5’-GGTAGGCGTGTACGGT GGGAGGCCTATATAAGCAGAGCT-3’), a HindIII restriction site, a Kozak sequence (5’-GCCACC-3’), and the gene encoding EYFP, EYFP, mNeonGreen, mCherry, NanoLuc, EGFP-TetR, EGFP-EZH2, or mCherry-NF-κB, respectively, fused with tDeg. These expression vectors were called miniCMV-EYFP, miniCMV-EYFP-tDeg, miniCMV-mNeonGreen-tDeg, miniCMV-mCherry-tDeg, miniCMV-NanoLuc-tDeg, miniCMV-EGFP-TetR-tDeg, miniCMV-EGFP-EZH2-tDeg, and miniCMV-mCherry-NF-κB-tDeg respectively. For control constructs of miniCMV-EGFP-TetR, miniCMV-EGFP-EZH2, and miniCMV-mCherry-NF-κB, a stop codon was inserted on the immediate upstream of the coding sequence of tDeg using QuikChange Site-Directed Mutagenesis Kits (Agilent).
To construct an expression vector for different circular RNAs, the Tornado expression plasmid9 containing an F30 scaffold was digested, then ligated to inserts encoding the following sequences, respectively: wild-type TAR RNA (5’-GGCTCGTGTAGCTCATTAGCTCCGAGCC-3’), TAR Variant-1 (5’-GGCTCGTCTGAGCTCATTAGCTCCGAGCC-3’), Pepper (TAR Variant-2) (5’-GGCTCGTTGAGCTCATTAGCTCCGAGCC-3’), or a control RNA, the MS2 hairpin (5’-ACATGAGGATCACCCATGT-3’). We called these vectors: U6+27-tnd-wildtype TAR, TAR Variant-1, Pepper (TAR Variant-2), control RNA, respectively.
For live-cell imagining experiments with HEK293T cells, HEK293T cells were seeded into 12-well flat bottom cell culture plates (Corning™ 3513) with 2 × 105 cells per well, and were cultured overnight. On the next day, cells were transfected using FuGENE HD (Promega 2311) according to the manufacturer’s instructions. Specifically, for imaging experiments in Figure 1, we cotransfected 550 ng of miniCMV-EYFP-tDeg with 550 ng of U6+27-tnd-wildtype TAR, TAR Variant-1, Pepper (TAR Variant-2), or control RNA, respectively. In the case of EYFP, we transfected 550 ng of miniCMV-EYFP with 550 ng of diluent DNA (pUC19 plasmid) to maintain 1.1 μg of total plasmid DNA per well. For imaging experiments in Figure 2 and Supplementary Figure 5, we cotransfected 550 ng of miniCMV-protein X-tDeg (protein X = mNeonGreen, mCherry, NanoLuc, EGFP-TetR, EGFP-EZH2, or mCherry-NF-κB) with 550 ng of circular Pepper (TAR Variant-2) or with 550 ng of diluent DNA (pUC19 plasmid). At 24 hours after transfection, cells were subcultured into 35 mm imaging dishes precoated with poly-D-lysine (Mattek Corporation P35GC-1.5–14C) and mouse laminin I (Cultrex® 3401–010-02) in culture media. Cells were then cultured overnight. Cell culture media was changed imaging media prior to fluorescence or bioluminescence live-cell imaging.
For live-cell imagining experiments in Supplementary Figures 4, U2OS cells, COS-7 cells, or HeLa cells were seeded into 35 mm imaging dishes precoated with poly-D-lysine (Mattek Corporation P35GC-1.5–14C) with 2 × 105 cells per dish, respectively. On the next day, cells were transfected using FuGENE HD (Promega 2311) according to the manufacturer’s instructions. Specifically, we cotransfected 1.4 μg of miniCMV-EYFP-tDeg with 1.4 μg of circular Pepper (TAR Variant-2) or 1.4 μg of diluent DNA (pUC19 plasmid). At 48 hours after transfection, cell culture media was changed imaging media prior to fluorescence live-cell imaging.
Prior to live-cell fluorescence or bioluminescence imaging, 1 μL of Hoechst 33342 (Thermo Fisher Scientific H3570) per 2 ml of imaging media was added to the cells. In the case of proteasome inhibitor treatment, cells were treated with either DMSO or 10 μM (final concentration in the media) MG132 for 7 hours prior to live-cell imaging. In the case of bioluminescence imaging of NanoLuc, 20 μL of furimazine (Promega Nano-Glo® Luciferase Assay System) per 2 ml of imaging media was added to the cells prior to bioluminescence imaging.
For live-cell fluorescence or bioluminescence imaging, we used an epifluorescence inverted microscope (Nikon Eclipse TE2000-E) equipped with a CoolSnap HQ2 CCD camera and a 130-W Nikon mercury lamp. We used the NIS-Elements Advanced Research software (Nikon) to control the microscope and camera. Cells were imaged with a 20×/0.75-NA (numerical aperture) or a 40×/0.75-NA air objective (Nikon) at 37°C. A FITC filter cube (with excitation filter 470 ± 20 nm, dichroic mirror 495 nm (long pass), and emission filter 525 ± 25 nm) was used for detecting EGFP-TetR-tDeg or EGFP-EZH2-tDeg with an exposure time of 500 msec. A YFP filter cube (with excitation filter 500±12 nm, dichroic mirror 520 nm (long pass), and emission filter 542±13.5 nm) was used for detecting EYFP, EYFP-tDeg, or mNeonGreen-tDeg with an exposure time of 500 msec. A TRITC filter cube (with excitation filter 560 ± 20 nm, dichroic mirror 585 nm (long pass), and emission filter 630 ± 37.5nm) was used for detecting mCherry-tDeg, or mCherry-NF-κB-tDeg with an exposure time of 500 msec. A filter cube (with emission filter 460 ± 25 nm) was used for detecting the bioluminescence of NanoLuc with an exposure time of 3 minutes. A DAPI filter cube (with 350 ± 25 nm excitation filter, 400 nm (long pass) dichroic mirror, and 460 ± 25 nm emission filter) was used for detecting the Hoechst-stained nuclei in cells with an exposure time of 100–500 msec. All filters used in these filter cubes are purchased from Chroma Technology. Cell fluorescence/bioluminescence was calculated using ImageJ by measuring the mean fluorescence/bioluminescence signal in a cell’s area and subtracting background based on average signal of culture media. Normalized fluorescence/bioluminescence was calculated by dividing the cell fluorescence/bioluminescence intensity of each cell to the averaged cell fluorescence/bioluminescence of the whole cell population.
RT-qPCR
Total RNA was isolated from cells using Trizol according to the manufacturer’s instruction. To remove residual DNA contaminations, the purified RNA was treated with DNaseI (Thermo-Fisher) according to the manufacturer’s instructions. The same amount of DNaseI-treated RNA was reverse transcribed to cDNA using SuperScript IV First-Strand kit (Invitrogen) with random hexamers according to the manufacturer’s instructions. To measure relative expression levels of the RNAs of interest, qPCR measurements were performed using the iQ SYBR Green Supermix with 0.250 ng of cDNA in the final reaction mix. For the amplification, the following protocol was used: 98°C for 2 minutes, 40 cycles of 95°C for 10 seconds, 60°C for 40 seconds. Primer sets for amplifying the cDNA of EYFP and mCherry are listed in Supplementary Table 2. Every primer set was tested for its efficiency. To test primer specificity, melting curves were performed at the end of the 40 cycles of amplification. In the case of mCherry quantification, an untrasfected sample was added as additional negative control. Relative measurements (2^-ΔCq) of mCherry, EYFP were performed using GAPDH and RPS18 as housekeeping genes. Biological replicates were tested.
Gel staining
Total RNA was isolated from cells using Trizol according to the manufacturer’s instruction. Then, 2.5 μg of isolated total RNA was separated using a precast 6% TBE-Urea Gel (Life Technologies EC68655). This gel was run at 200 V in TBE buffer until completion, and stained with SYBR Gold (ThermoFisher S11494) diluted 1:10,000 in TBE buffer for 15 minutes. After SYBR Gold staining, RNA bands were imaged on a ChemiDoc XRS+ system (Bio-Rad).
mRNA imaging using tDeg and Pepper.
To construct an expression vector for fluorogenic proteins used in mRNA imaging, a pcDNA3.1(+) vector was digested by MluI and XbaI and ligated to an insert comprising a miniCMV promoter (5’-GGTAGGCGTGTACGGT GGGAGGCCTATATAAGCAGAGCT-3’), a HindIII restriction site, a Kozak sequence (5’-GCCACC-3’), and the gene encoding tandem copies of mNeonGreen, mVenus, or mCherry, respectively. To construct an expression vector for an mCherry mRNA reporter containing different 3’UTR tags comprising 10 or 20 concatenated Pepper aptamers, a pcDNA3.1(+) vector was first digested by HindIII and XbaI and ligated to an insert encoding the gene of mCherry followed by XhoI after its stop codon. We called this vector CMV-mCherry. CMV-mCherry was then digested XhoI and XbaI, and ligated to different Pepper tags, respectively. All the Pepper tags were synthesized by GenScript.
U2OS cells were seeded into 35 mm imaging dishes precoated with poly-D-lysine (Mattek Corporation P35GC-1.5–14C) with 2 × 105 cells per dish. On the next day, cells were transfected using FuGENE HD (Promega 2311) according to the manufacturer’s instructions. Specifically, we cotransfected 1.4 μg of fluorogenic protein plasmids with 1.4 μg of mRNA reporter plasmids. At 48 hours after transfection, cell culture media was changed to imaging media prior to imaging experiments.
For mRNA imaging experiments, we used an epifluorescence inverted microscope (Olympus IX-70) equipped with a Evolve® 512 EMCCD OEM camera (Photometrics) and an Insight SSI 7 color solid state illumination system (Applied Precision). We used the Resolve3D softWoRx-Acquire Version: 6.5.2 to control the microscope and camera. Cells were imaged with a 100×/1.4-NA oil objective at 37°C, with N = 1.520 immersion oil (Applied Precision). A FITC filter cube (with excitation filter 475 ± 14 nm, dichroic mirror with a reflection band of 481–502 nm, and a transmission band of 506–543 nm), and emission filter 525 ± 25 nm) was used for detecting mNeonGreen with an exposure time of 50 msec. A YFP filter cube (with excitation filter 513 ± 8.5 nm, dichroic mirror with a reflection band of 496–528 nm, and a transmission band of 537–550 nm, and emission filter 559 ± 19 nm) was used for detecting mVenus with an exposure time of 100 msec. A TRITC filter cube (with excitation filter 542 ± 13.5 nm, dichroic mirror with a reflection band of 547–565 nm, and a transmission band of 576–630 nm, and emission filter 594 ± 22.5nm) was used for detecting reporter plasmids encoding mCherry with an exposure time of 10–100 msec. Signal-to-noise ratio of the fluorescent puncta was calculated by the mean fluorescence intensity of each mRNA puncta divided by the mean fluorescence intensity of the adjacent cytosolic background fluorescence.
Northern blot.
HEK293T cells were seeded into 10 cm culture dish with 3 × 106 cells per dish. On the next day, cells were cotransfected with CMV-mCherry-(F30–2xPepper)10 and miniCMV-(mNeonGreen)4-tDeg or pUC19, respectively. A total amount of 19 μg plasmid DNA was used for each culture dish, and pUC19 vector was used here as a diluent DNA to ensure the same amount of plasmid DNA transfected to the cells. All transfections were performed using FuGENE HD (Promega 2311) according to the manufacturer’s instructions. Cells were harvested after 48 hours of transfection. Total RNA was extracted with TRIzol (Thermo Fisher Scientific 15596026) followed by isopropanol precipitation. The purified total RNA was then subjected to RNase-free DNase I (Thermo Fisher Scientific AM2224) digestion at 37°C for 1 hour. After digestion, the RNA was subjected to phenol-chloroform (Thermo Fisher Scientific AM9720) extraction and ethanol purification.
For gel electrophoresis, we used a 1.5% agarose/formaldehyde gel (20 mM MOPS, 5 mM sodium acetate, 1 mM EDTA, 1.5% w/v agarose, 2% formaldehyde). We loaded 20 μg of total RNA in each lane. The RNA was resuspended in 20 μL of RNA sample buffer (20 mM MOPS, 5 mM sodium acetate, 1 mM EDTA, 50% v/v formamide, 3.7% formaldehyde). The RNA samples were heated at 70°C for 10 minutes, and then chilled on ice for more than 1 minute. Before loading the RNA samples into the gel, we mixed the RNA samples with 2 μL of loading buffer (50% glycerol, 5 mM EDTA, 0.4% bromophenol blue, 0.4% xylene cyanol). The gel was run at 70 V for 2 hours. After electrophoresis, the gel was stained with 1x SYBR™ Gold Nucleic Acid Gel Stain (Thermo Fisher Scientific S11494) to assess the quality of the RNA and check for separation. All solutions mentioned above were made in diethylpyrocarbonate (DEPC)-treated water.
After electrophoresis, the RNA was transferred to Amersham Hybond-N+ nylon membrane (GE Healthcare Life Sciences RPN203B) using the VacuGene XL Vacuum Blotting System (GE Healthcare Life Sciences) according to the manufacturer’s instructions. The RNA was then UV crosslinked to the nylon membrane. The membrane was washed with NorthernMax® Prehybridization/Hybridization Buffer (Thermo Fisher Scientific AM8677) at 42°C for at least 30 minutes. Biotinylated (at 5’) single-stranded DNA probes (Integrated DNA Technologies) as shown in Supplementary Table 1 were mixed with NorthernMax® Prehybridization/Hybridization Buffer and incubated with the membrane at 42°C overnight. On the following day, the membrane was washed in 50 mL of wash buffer 1 (2xSSC, 0.1% SDS) twice at 42°C for 10 minutes each time, and then washed with wash buffer 2 (0.1xSSC, 0,1% SDS) twice at 42°C for 15 minutes. The membrane was visualized by Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Fisher Scientific 89880).
Imaging membrane-tethered mRNA.
U2OS cells were seeded 72 hours before imaging in 96-well glass bottom dishes (Matriplates, Brooks Life Science Systems) at 40% confluency. Cells were transfected with DNA plasmids that encode miniCMV-(mNeonGreen)4-tDeg, PCP-3xmCherry-CAAX and the mRNA reporter 48 hours before imaging using 0.5 μl FuGENE 6 (Promega) and 200–300 ng DNA per well. The transfection mix was prepared in OptiMEM (Sigma-Aldrich) and added to the cells in a total volume 150–200 μl of medium.
Twenty-four hours prior to imaging, transcription of the reporters was induced by addition of doxycycline (1 μg/ml) (Sigma-Aldrich). Thirty minutes before imaging, the cell culture medium was replaced with pre-warmed CO2-independent Leibovitz’s-15 medium (Gibco) with doxycycline. Images were acquired using a Nikon TI inverted microscope with perfect focus system equipped with a Yokagawa CSU-X1 spinning disc, a 100× 1.49 NA objective and an iXon Ultra 897 EMCCD camera (Andor) and was controlled by NIS software (Nikon). During the experiment, cells were maintained at a constant temperature of 37°C. Single Z-plane images were acquired, with the bottom plasma membrane of the cell in the focal plane. Camera exposure times of 500 ms were used for both mNeonGreen and mCherry.
To determine the fluorescence intensity of mRNA foci, mean spot intensities were measured in Image J in a region of interest (ROI) 0.53 × 0.53 μm in size. For each spot, local background fluorescence intensity was measured in a ROI (0.53 × 0.53 μm in size) directly next to the spot of interest, and mean background fluorescence intensities were subtracted from the mean spot intensity. Cells with very high number of mRNAs (more than ~50) were excluded from the analysis.
Western Blotting.
Cells were lysed in whole cell lysis buffer (10 mM Tris-HCl pH 7.4, 10 mM EDTA, 50 mM NaCl, 1% Triton X-100, 0.1% SDS) containing 1X protease and phosphatase inhibitor (Pierce, 78440). Lysates were cleared by centrifugation (12,000g for 10 minutes). Protein quantification was performed using the Pierce BCA protein assay kit according to the manufacturer’s instruction (Thermo Fisher Scientific, 23227). Equal quantities of proteins were mixed with loading dye, and incubated at 95°C for 5 minutes before they were separated on 4–12% Bis–Tris gels (Invitrogen) and transferred onto a PVDF membrane at constant 350 mA at 4°C for 1 hour. Membranes were blocked by incubation in 5% milk for 1 hour at room temperature under agitation and then incubated with the following primary antibodies: mouse anti-GAPDH (Santa Cruz) with a 1:5000 dilution in 1%milk overnight, or rabbit anti-mCherry (Abcam, ab167453) with a 1:1000 dilution in 1%milk overnight, or rabbit anti-ubiquitin (Abcam, ab19247) with a 1:1000 dilution in 1%milk overnight. After incubation with the appropriate secondary antibodies conjugated to HRP and extensive washing, blots were imaged on a ChemiDoc XRS+ system (Bio-Rad).
Imaging ER-targeting mRNA.
To construct an expression vector for an ER-targeting mRNA reporter, DNA sequence that encodes the first 29 amino acids of cytochrome p450, CytERM, and a linker sequence (MDPVVVLGLCLSCLLLLSLWKQSYGGGKLGGSGGTGGSGTSGG) was cloned into the upstream of the mCherry sequence of the CMV-mCherry-(F30–2xPepper)10 plasmid to make CMV-CytERM-mCherry-(F30–2xPepper)10. To construct the plasmid that encodes the fluorogenic protein used in this experiment, we replaced the miniCMV promoter sequence in miniCMV-(mNeonGreen)4-tDeg to the human ubiquitin C promoter sequence to make UbC-(mNeonGreen)4-tDeg.
U2OS cells were seeded into 35 mm imaging dishes precoated with poly-D-lysine (Mattek Corporation P35GC-1.5–14C) with 2 × 105 cells per dish. On the following day, cells were cotransfected with 1.4 μg of CMV-CytERM-mCherry-(F30–2xPepper)10, 0.28 μg of UbC-(mNeonGreen)4-tDeg, and 1.12 μg of pUC19 (as a diluent DNA) using FuGENE HD (Promega 2311) according to the manufacturer’s instructions. At 48 hours after transfection, cell culture media was changed to imaging media prior to imaging experiments. This imaging setup for these experiments are the same as the one used for mRNA imaging using tDeg and Pepper.
Imaging β-actin mRNA after arsenite stress.
To construct an expression vector for a β-actin mRNA reporter containing a (F30–2xPepper)10 tag, the full length β-actin gene (from Addgene Plasmid #27123) was amplified by PCR and digested by XhoI and HindIII, and then ligated to a vector from CMV-mcherry-(F30–2xPepper)10 digested by the same restriction endonucleases to cut out the gene encoding mCherry. This expression vector was called CMV-β-actin-(F30–2xPepper)10.
U2OS cells stably expresses Halo-G3BP1 were seeded into 35 mm imaging dishes precoated with poly-D-lysine (Mattek Corporation P35GC-1.5–14C) with 2 × 105 cells per dish. On the following day, cells were cotransfected with 1.4 μg of miniCMV-(mNeonGreen)4-tDeg with 1.4 μg of CMV-β-actin-(F30–2xPepper)10 using FuGENE HD (Promega 2311) according to the manufacturer’s instructions. For control experiments, 1.4 μg of miniCMV-(mNeonGreen)4-tDeg with 1.4 μg of U6+27-tnd-Pepper was used following the same transfection protocol. At ~40 hours after transfection, cell culture media was changed to imaging media with the HaloTag® TMRDirect™ Ligand (Promega G2991) for 5 hours. Cells were then rinsed with 1xPBS (Thermo Fisher Scientific 10010049) and incubated in imaging media prior to imaging experiments. We used the same microscope setup as in the above mRNA imaging experiments. To induce stress granule formation, 1 mL of imaging media supplemented with 1000 μM of sodium arsenite was added to the cells cultured in 1 mL of imaging media to reach a final concentration of 500 μM of sodium arsenite.
Statistical analysis.
All data are expressed as means ± s.d. with sample sizes (n) listed for each experiment. Statistical analyses were performed using Excel (Microsoft) and Prism (Graphpad). For different circular TAR variants’ inhibition of tDeg’s destabilizing effect, and optimization of the number of fluorogenic mNeonGreen monomers in the fluorogenic protein for imaging mRNA in live cells, one-way ANOVA was used to analyze significant differences between group means. For Pepper RNA-dependent regulation of protein stability, imaging green Pepper-tagged β-actin mRNA, proteasomal inhibition, imaging membrane-tethered mRNA, two tailed Student’s t-tests were used to analyze significant differences between group means. P values were reported for each experiment.
Supplementary Material
Acknowledgements
This work was supported by NIH grant R01NS056306 (S.R.J.), a FRAXA Postdoctoral Fellowship (J.W.). M.E.T. and D.K. were supported by the Oncode Institute that is partly funded by the Dutch Cancer Society (KWF). We thank L. Mei and V. Despic for technical support, the Bio-Imaging Resource Center at Rockefeller University for technical support on imaging experiments, and members of the S.R.J. laboratory and especially X. Li, J. Litke, and J.D. Moon for useful comments and suggestions.
Footnotes
Accessions
PRIMARY ACCESSIONS
NCBI Reference Sequence
Data Availability
The sequence of mammalian expression plasmids used in this study are available on GenBank: CMV-mCherry-(F30–2xPepper)10 (accession number: MN052904), miniCMV-(mNeonGreen)4-tDeg (accession number: MN052905), CMV-CytERM-mCherry-(F30–2xPepper)10 (accession number: MN052906), UbC-(mNeonGreen)4-tDeg (accession number: MN052907), pAV-U6+27-Tornado-F30-Pepper(TAR Variant-2) (accession number: MN052908), pAV-U6+27-Tornado-F30-TAR Variant-1 (accession number: MN052909). These plasmids are available via Addgene according to the terms of the Uniform Biological Material Transfer Agreement. Source Data for Figures 1, 2, 3 and Supplementary Figures 2, 3, 4, 5, 8, 9, 10, 11, and 12 are available with the paper online.
Supplementary Information is linked to the online version of the paper.
Competing Financial Interests
The authors declare no competing financial interests.
REFERENCES
- 1.Paige JS, Wu KY & Jaffrey SR RNA mimics of green fluorescent protein. Science 333, 642–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braselmann E et al. A multicolor riboswitch-based platform for imaging of RNA in live mammalian cells. Nat. Chem. Biol 14, 964–971 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo̊ber G The fluorescence of dye-nucleic acid complexes. Journal of Luminescence 22, 221–265 (1981). [Google Scholar]
- 4.Fam TK, Klymchenko AS & Collot M Recent Advances in Fluorescent Probes for Lipid Droplets. Materials (Basel) 11, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez EA et al. The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins. Trends Biochem. Sci 42, 111–129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonger KM, Chen LC, Liu CW & Wandless TJ Small-molecule displacement of a cryptic degron causes conditional protein degradation. Nat. Chem. Biol 7, 531–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye X, Kumar RA & Patel DJ Molecular recognition in the bovine immunodeficiency virus Tat peptide-TAR RNA complex. Chem. Biol 2, 827–40 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Puglisi JD, Chen L, Blanchard S & Frankel AD Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex. Science 270, 1200–3 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Litke JL & Jaffrey SR Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith CA, Crotty S, Harada Y & Frankel AD Altering the context of an RNA bulge switches the binding specificities of two viral Tat proteins. Biochemistry 37, 10808–14 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Bertrand E et al. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–45 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Tyagi S Imaging intracellular RNA distribution and dynamics in living cells. Nat. Methods 6, 331–8 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Filonov GS, Kam CW, Song W & Jaffrey SR In-gel imaging of RNA processing using broccoli reveals optimal aptamer expression strategies. Chem. Biol 22, 649–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan X, Hoek TA, Vale RD & Tanenbaum ME Dynamics of Translation of Single mRNA Molecules In Vivo. Cell 165, 976–89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costantini LM, Fossati M, Francolini M & Snapp EL Assessing the tendency of fluorescent proteins to oligomerize under physiologic conditions. Traffic 13, 643–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tourrière H et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol 160, 823–31 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.