Abstract
Background
Salmonella has been considered as one of the most important foodborne pathogens that threatened breeding industry and public health. To investigate the prevalence and characterization of Salmonella isolated from duck farms and a slaughterhouse in Shandong province, a total of 49 Salmonella strains were isolated from 2342 samples from four duck farms and one duck slaughterhouse in Jinan and Tai’an, Shandong province, China.
Results
Among the isolates, S. Enteritidis (20/49, 40.8%) and S. Anatum (10/49, 20.4%) were the most prevalent, and high resistance rates were detected for erythromycin (49/49, 100.0%) and nalidixic acid (47/49, 95.9%). Class I integrons were detected in 17 isolates (34.7%17/49), which contained gene cassettes aadA7 + aac3-Id(15/17) and aadA5 + dfrA17 (2/17). Eleven different kinds of resistance genes were detected while blaTEM(36/49, 73.5%) was the most prevalent, followed by sul2(14/49, 28.6%). Thirteen virulence genes were tested, and all of the strains carried invA, hilA and sipA. Multilocus sequence typing (MLST) results showed that seven sequence types (STs) were identified; ST11 was the most prevalent ST (20/49, 40.8%), followed by ST2441 (10/49, 20.4%). There was a strong correlation between STs and serovars. The results of pulsed field gel electrophoresis(PFGE) showed that 39 PFGE patterns were generated from 49 Salmonella strains. PFGE patterns were mostly diverse and revealed high similarity between the isolates from the same sampling sites.
Conclusions
The presence of Salmonella infections among duck farms revealed that ducks could also be potential reservoirs for Salmonella. The high resistance rates against commonly used antimicrobials suggested a need for more reasonable use of antimicrobials, as well as for investigating substitutes for antimicrobials.
Keywords: Salmonella, Antimicrobial resistance, Class I integrons, MLST, PFGE
Background
Salmonella has been considered to be one of the most important foodborne pathogens associated with public health worldwide, and has been frequently reported in recent years [1, 2], including topics such as its route through the food chain to humans. At present, more than 2600 different serovars have been identified and recorded [3]. In the USA, it is estimated that Salmonella causes more than 1 million infection cases annually, resulting in the loss of 365 million dollars (Centers for Disease Control and Prevention, 2014). In China, Salmonella infections are also frequently reported, especially among elderly and immunocompromised individuals [4, 5]. In addition, in China, approximately 70 to 80% of outbreaks of foodborne pathogenic diseases are caused by Salmonella [6].
In the past decades, the use of antimicrobial agents has been considered the most important way to treat and control Salmonella and other pathogens [7]. However, due to widespread utilization of antibiotics, antimicrobial-resistant and even multidrug-resistant Salmonella strains have emerged and spread worldwide, and have seriously threatened global public health [8–10].
Many molecular typing techniques are widely used in the field of microbiology and can be used to trace the origins of pathogenic bacteria [11]. Currently, the most widely used molecular typing techniques are MLST and PFGE. The MLST method is convenient and rapid; the resolution is high, and the resulting data is standardly reliable. Through the internet platform, data sharing and comparison between different laboratories is realistic and easier than ever [12]. PFGE is used by laboratories around the world for its high resolution and repeatability, and is widely considered to be the gold standard for molecular typing. However, PFGE does not have a strict, unified international naming standard, and data is not effectively communicated between different laboratories. This study combines two types of typing methods to comprehensively and systematically understand the epidemiological characteristics of Salmonella.
According to the FAO (Food and Agriculture Organization) report (2014), China is the largest producer of duck meat, producing 3 million tons annually, and consumption continues to increase every year [13]. Furthermore, Shandong province is China’s largest duck farming province, especially considering that of Tai’an and Jinan. Little information concerning the prevalence and characterization of Salmonella from ducks in farms and slaughterhouses in Shandong province is available. Therefore, this study identifies farms and a slaughterhouse as sampling points, analyzing the prevalence and drug resistance of Salmonella in these locations; furthermore, these findings may provide beneficial information for the development of the duck industry and public health.
Methods
Sample collection
A total of 2342 samples were collected between 2016 and 2017 from Tai’an and Jinan, Shandong province, including samples of duck feces, embryos, livers, intestine and leg meat, in addition to those of feed, drinking water and duck-washing pools and table surfaces (Table 3). All samples were randomly collected, according to the cluster sampling principle, from one duck farm in Jinan (n = 450), three duck farms in Tai’an (n = 1175) and one slaughterhouse in Tai’an (n = 717).In addition, the liver samples were collected from diseased ducks on the three farms, and the samples from the slaughterhouse were collected during the slaughter process. After collection, samples were immediately placed into a sterilized container, then transported to a laboratory, with ice bags, within 6 h for further bacteriological analysis; they were processed immediately upon arrival. Every sampling site was visited only once.
Table 3.
Sampling sites and isolation rates and MDR rates
| Sampling Sites | Sampling Time | Sample Amount | Positive samples | Total | MDR rate |
|---|---|---|---|---|---|
| Tai’an Farm 1 | 2016 | 72 | Embryos (11/72) | 15.3% (11/72) | 36.3% (4/11) |
| Tai’an Farm 2 | 2017 | 541 | Feces(9/466), Feed(1/41), Drinking water(0/34) | 1.9% (10/541) | 100% (10/10) |
| Tai’an Farm 3 | 2017 | 562 | Feces(4/459), Feed(0/66), Livers(4/37) | 1.4% (8/562) | 75% (6/8) |
| Jinan Farm 4 | 2016 | 450 | Feces(12/406), Livers(6/44) | 4.0% (18/450) | 83.3% (15/18) |
| Tai’an Slaughterhouse | 2017 | 717 | Leg meat(0/74), Livers(1/83), Water samples from Duck-washing pool(1/29), Cotton swabs from table surface(0/13), Intestine (0/518) | 0.3% (2/717) | 0% (0/2) |
Isolating and serotyping of Salmonella
Salmonella strains were isolated from samples using the Chinese National Standard method (GB 4789.4–2010), with some modifications. Briefly, 10.0 mL of buffered peptone water (BPW, Land Bridge Technology, Beijing, China) was added to each sample (1 g) for pre-enrichment. After incubation at 37 °C for 18 h, 1.0 mL pre-enriched culture was inoculated into 10.0 mL selenite cystine broth (SC, Land Bridge Technology) and incubated at 42 °C or 37 °C. After 24 h of incubation, a loop from each broth culture was streaked onto xylose lysinedeoxycholate medium (XLD, Land Bridge Technology) plates and incubated at 37 °C for 24 to 48 h. Next, suspected Salmonella colonies were identified by polymerase chain reaction (PCR) assays using primers invA. The invA gene was a typical violence gene of Salmonella that was able to detect and validate Salmonella strains with the inclusivity for all subspecies and exclusivity for other genera and species [14]. PCR was performed ina 25.0 μL mixture containing 12.5 μL of 2 × Taq Master Mix (Vazyme Nanjing, China), 9.5 μL ddH2O, 1.0 μL of sample DNA, and 1.0 μL of each primer.
All strains were serotyped by slide agglutination using commercial O and H antisera (Tianrun Bio-Pharmaceutical, Ningbo, China) according to the Kauffmann-White scheme.
Antimicrobial susceptibility testing
The Kirby-Bauer disk diffusion method, as described by the Clinical and Laboratory Standards Institute [15], was used to examine the susceptibility of Salmonella to 14 commonly used antibiotics, including ampicillin (AMP; 10 μg), ceftriaxone (CRO; 30 μg), cefotaxime (CTX; 30 μg), erythromycin (EM; 15 μg), chloramphenicol (CHL; 30 μg), florfenicol (FFN; 30 μg), gentamicin (GEN; 10 μg), streptomycin (STR; 10 μg), tetracycline (TET; 30 μg), sulfamethoxazole (SXT; 25 μg), ciprofloxacin (CIP; 5 μg), nalidixic acid (NAL; 30 μg), norfloxacin (NOR; 10 μg), polymyxin B (PB; 300 IU). Meanwhile, Escherichia coli strains ATCC 25922 and ATCC 35218 were used as control strains. Salmonellaisolates resistant to more than three classes of antimicrobials were defined as multidrug-resistance (MDR) isolates [16, 17].
Detection of antimicrobial resistance genes and virulence genes
Bacterial DNA was extracted using TIANamp Bacterial DNA Kit (TIANGEN, Beijing, China), according to the manufacturer’s instructions. After extraction of DNA, quinolone-resistance genes, including qnrA, qnrB, qnrC, qnrD, qnrS, oqxAand aac(6′)Ib-cr; β-lactamase encoding genes, including blaTEM, blaPSE, blaCMY-2, blaSHV, blaOXAand blaCTX-M; aminoglycosides-resistance genes including aaC1, aaC2,aaC3, aaC4 and Ant(2′); tetracycline-resistance genes including tetA and tetB; sulfonamides-resistance genes, including sul1, sul2 and sul3, and chloramphenicol-resistance genes, including cmlA and stcM, were detected by PCR, using previously described primers (Table 1) and procedures [18–24]. Meanwhile, 13 pairs of primers (Table 2) were used for PCR to detect the virulence genes, including invA, hilA, spvC, sipA, sopE, stnP1, pefA, rck, sipC, ssaR, ssrA, sopB and sefA [25]. All of the PCR products were sequenced (Sangon Biotech, Shanghai, China), and the resistance gene subtypes were determined for subsequent analysis.
Table 1.
Primers used to detect antimicrobial-resistance genes
| Resistance Gene Category | Resistance Gene | Primer Sequence | Reference |
|---|---|---|---|
| β-lactamase | bla TEM | F: 5′- ATAAAATTCTTGAAGACGAAA − 3′ | Ahmed et al., 2007 [18] |
| R: 5′- GACAGTTACCAATGCTTAATC − 3′ | |||
| bla SHV | F: 5′- TTATCTCCCTGTTAGCCACC − 3′ | Ahmed et al., 2007 [18] | |
| R: 5′- GATTTGCTGATTTCGCTCGG − 3′ | |||
| bla PSE | F: 5′- TAGGTGTTTCCGTTCTTG-3′ | Puah et al., 2012 [19] | |
| R: 5′- TCATTTCGCTCTTCCATT-3′ | |||
| bla OXA | F: 5′- TCAACTTTCAAGATCGCA-3′ | Ahmed et al., 2007 [18] | |
| R: 5′- GTGTGTTTAGAATGGTGA-3′ | |||
| bla CMY-2 | F: 5′- ACGGAACTGATTTCATGATG − 3′ | Ahmed et al., 2007 [18] | |
| R: 5′- GAAAGGAGGCCCAATATCCT −3′ | |||
| bla CTX-M | F: 5′- CGCTTTGCGATGTGCAG-3′ | Ahmed et al., 2007 [18] | |
| R: 5′- ACCGCGATATCGTTGGT-3′ | |||
| Quinolone | qnrA | F: 5′- ATTTCTCACGCCAGGATTTG-3′ | Ahmed et al., 2007 [18] |
| R: 5′- GATCGGCAAAGGTCAGGTCA-3′ | |||
| qnrB | F: 5′- GATCGTGAAAGCCAGAAAGG-3′ | Ahmed et al., 2007 [18] | |
| R: 5′- ACGATGCCTGGTAGTTGTCC-3′ | |||
| qnrC | F: 5′- GGTTGTACATTTATTGAATC-3′ | Ahmed et al., 2007 [18] | |
| R: 5′- TCCACTTTACGAGGTTCT −3′ | |||
| qnrD | F: 5′- AGATCAATTTACGGGGAATA-3′ | Ahmed et al., 2007 [18] | |
| R: 5′- AACAAGCTGAAGCGCCTG − 3′ | |||
| qnrS | F: 5′- ACGACATTCGTCAACTGCAA-3′ | Ahmed et al., 2007 [18] | |
| R: 5′- TAAATTGGCACCCTGTAGGC-3′ | |||
| oqxA | F: 5′- GATCAGTCAGTGGGATAGTTT-3′ | Liao et al., 2015 [20] | |
| R: 5′- TACTCGGCGTTAACTGATTA-3′ | |||
| aac(6′)-Ib-cr | F: 5′- TTGCGATGCTCTATGAGTGGCTA − 3′ | Ahmed et al., 2007 [18] | |
| R: 5′- CTCGAATGCCTGGCGTGTTT − 3′ | |||
| Aminoglycosides | aaC1 | F: ACCTACTCCCAACATCAGCC-3′ | Navajas-Benito et al., 2016 [21] |
| R: ATATAGATCTCACTACGCGC-3′ | |||
| aaC2 | F:ACTGTGATGGGATACGCGTC-3′ | Navajas-Benito et al., 2016 [21] | |
| R: CTCCGTCAGCGTTTCAGCTA-3′ | |||
| aaC3 | F: CACAAGAACGTGGTCCGCTA-3′ | Navajas-Benito et al., 2016 [21] | |
| R: AACAGGTAAGCATCCGCATC-3′ | |||
| aaC4 | F: CTTCAGGATGGCAAGTTGGT-3′ | Navajas-Benito et al., 2016 [21] | |
| R: TCATCTCGTTCTCCGCTCAT-3′ | |||
| Ant(2′) | F: ATGTTACGCAGCAGGGCAGTCG-3′ | Navajas-Benito et al., 2016 [21] | |
| R: CGTCAGATCAATATCATCGTGC-3′ | |||
| Tetracycline | tetA | F: 5′- GCGCCTTTCCTTTGGGTTCT-3′ | Navajas-Benito et al., 2016 [21] |
| R: 5′- CCACCCGTTCCACGTTGTTA-3′ | |||
| tetB | F: 5′- CATTAATAGGCGCATCGCTG-3′ | Navajas-Benito et al., 2016 [21] | |
| R: 5′- TGAAGGTCATCGATAGCAGG-3′ | |||
| Sulfonamides | sul1 | F: 5′- CTTCGATGAGAGCCGGCGGC-3′ | Aarestrup et al., 2003 [22] |
| R: 5′- GCAAGGCGGAAACCCGCGCC-3′ | |||
| sul2 | F: 5′- GCGCTCAAGGCAGATGGCATT-3′ | Aarestrup et al., 2003 [22] | |
| R: 5′- GCGTTTGATACCGGCACCCGT-3′ | |||
| sul3 | F: 5′- AGATGTGATTGATTTGGGAGC-3′ | Zhang et al., 2009 [23] | |
| R: 5′- TAGTTGTTTCTGGATTAGAGCCT-3′ | |||
| Chloramphenicol | cmlA | F: 5′- TGTCATTTACGGCATACTCG-3′ | Guerra et al., 2001 [24] |
| R: 5′- ATCAGGCATCCCATTCCCAT-3′ | |||
| stcM | F: 5′- CACGTTGAGCCTCTATATGG-3′ | Guerra et al., 2001 [24] | |
| R: 5′- ATGCAGAAGTAGAACGCGAC-3′ |
Table 2.
Primers used to detect virulence genes
| Virulence Gene | Primer Sequence | Reference |
|---|---|---|
| hilA | F: 5′- CGTGAAGGGATTATCGCAGT −3′ | Fardsanei et al., 2017 [25] |
| R: 5′- GTCCGGGAATACATCTGAGC −3′ | ||
| invA | F: 5′- ACAGTGCTCGTTTACGACCTGAAT − 3′ | Fardsanei et al., 2017 [25] |
| R: 5′- AGACGACTGGTACTGATCGATAAT − 3′ | ||
| pefA | F: 5′- TTGCACTGGGTGGTTCTGG − 3′ | Fardsanei et al., 2017 [25] |
| R: 5′- TGTAACCCACTGCGAAAG − 3′ | ||
| rck | F: 5′- AACGGACGGAACACAGAGTC − 3′ | Fardsanei et al., 2017 [25] |
| R: 5′- TGTCCTGACGAAAGTGCATC − 3′ | ||
| sefA | F: 5′- GCAGCGGTTACTATTGCAGC − 3′ | Fardsanei et al., 2017 [25] |
| R: 5′- TGTGACAGGGACATTTAGCG − 3′ | ||
| sipA | F: 5′- CCATTCGACTAACAGCAGCA − 3′ | Fardsanei et al., 2017 [25] |
| R: 5′- CGGTCGTACCGGCTTTATTA − 3′ | ||
| sipC | F: 5′- AGACAGCTTCGCAATCCGTT − 3′ | Fardsanei et al., 2017 [25] |
| R: 5′- ATTCATCCCTTCGCGCATCA − 3′ | ||
| sopB | F: 5′- CCTCAAGACTCAAGATG − 3′ | Fardsanei et al., 2017 [25] |
| R: 5′- TACGCAGGAGTAAATCGGTG − 3′ | ||
| sopE | F: 5′- CGAGTAAAGACCCCGCATAC − 3′ | Fardsanei et al., 2017 [25] |
| R: 5′- GAGTCGGCATAGCACACTCA − 3′ | ||
| spvC | F: 5′- ACTCCTTGCACAACCAAATGCGGA − 3′ | Fardsanei et al., 2017 [25] |
| R: 5′- TGTCTCTGCATTTCGCCACCATCA − 3′ | ||
| ssaR | F: 5′- GTTCGGATTTGCTTCGG − 3′ | Fardsanei et al., 2017 [25] |
| R: 5′- TCTCCAGTGACTAACCCTAACCAA − 3′ | ||
| ssrA | F: 5′- CTTACGATTACGCCATTTACGG − 3′ | Fardsanei et al., 2017 [25] |
| R: 5′- ATTTGGTGGAGCTGGCGGGACT − 3′ | ||
| stnP1 | F: 5′- TTGTCTCGCTATCACTGGCAACC − 3′ | Fardsanei et al., 2017 [25] |
| R: 5′- ATTCGTAACCCGCTCTCGTCC − 3′ |
Detection of class I integrons
To investigate the presence of class I integrons, a 25 μL total reaction volume, consisting of 12.5 μL 2 × Taq Master Mix (Vazyme Nanjing, China), 9.5 μL ddH2O, 1.0 μL of sample DNA, and 1.0 μL of each pair of primers was prepared for PCR (White et al., 2000). PCR products were purified by a purification Kit and then sequenced (Sangon Biotech, Shanghai, China).
Multilocus sequence typing (MLST)
Seven housekeeping genes (aroC, dnaN, hemD, hisD, purE, sucA, and thrA) were selected for molecular typing of Salmonella strains according to the instructions from the University of Warwick (http://mlst.warwick.ac.uk/mlst/). The provided protocols from the MLST homepage were used including PCR conditions and primers (http://mlst.warwick.ac.uk/mlst/dbs/Senterica). PCR products were sequenced by Sangon Biotech (Shanghai, China) and then compared with provided housekeeping genes by Blast+ (Basic Local Alignment Search Tool) [11].
Pulsed-field gel electrophoresis (PFGE)
PFGE was performed based on the protocol of the Centers for Disease Control and Prevention (CDC) with minor modifications [26]. In brief, isolates were streaked on Luria-Bertani (LB) plates and incubated at 37 °C for 24 h, and Salmonella were collected and suspended in a cell suspension buffer (CSB). Subsequently,400.0 μL of cell suspension was transferred to a sterile tube and mixed with 20.0 μL of proteinase K (20.0 mg/mL).400.0 μL of melted 1.0% SeaKem Gold agarose (Lonza, Morristown, NJ, USA) with 1.0% sodium dodecyl sulfate (SDS) was then added to 400.0 μL of the cell suspension. The mixture was poured into the plug molds, cooled down, and then transferred to the lysis solution. After the 2-h lysis, the agarose-embedded DNA was stored in 0.5 × Tris-Borate-EDTA (TBE) at 4 °C. The bacterial cells in the agarose plugs were digested with 50 U of XbaI (TaKaRa, Dalian, China) for 2 h at 37 °C. Digested fragments were resolved in 1.0%SeaKem Gold agarose gel in 0.5× TBE using a ChefMapper electrophoresis system (Bio-Rad, Hercules, CA, USA). After performing electrophoresis at 14 °C for 19 h, the gel was stained with Gel-Red (TIANGEN, Beijing, China), the gel images were obtained by UV trans-illumination (Bio-Rad) and the fingerprinting profiles were analyzed by the BioNumerics Software (Applied Maths, Kortrijk, Belgium). According to the manufacturer’s instructions, the unweighted pair-group method (UPGMA) was performed to generate the dendrogram, with settings of 1.5% position tolerance and 0.5% optimization.
Results
Isolation and serotyping of Salmonella
A total of 49 Salmonella strains were isolated from 2342 samples obtained from four large-scaleduck farms and one slaughterhouse, having an isolation rate of 2.1%. Eleven strains were collected from Farm1 in Tai’an (No.1–11), 10 strains were collected from Farm2 in Tai’an (No.12–21), 8 strains were collected from Farm3 in Tai’an (No.22–29), 18 strains were collected from Farm 4 in Jinan (No. 30–47) and 2 strains were collected from the slaughterhouse in Tai’an (No. 48–49). The prevalence of positive samples was 15.3, 1.9, 1.4, 4.0 and 0.3% in farms 1, 2, 3 and 4 and in the slaughterhouse, respectfully (Table 3).The positive rate for Salmonella in duck livers from the diseased duck farms (12.3%, 10/81) was higher than that in duck livers from the slaughterhouse (1.2%, 1/83).In addition, a Salmonella strain was also isolated from duck feed.
Forty-nine Salmonella isolates were divided into 6 serotypes (Table 4), including S. Enteritidis (n = 20), S. Anatum (n = 10), S. Typhimurium (n = 8), S.Kentucky (n = 5), S. Indiana (n = 5) and S. Montevideo (n = 1), while S. Enteritidis (40.8%, 20/49) was the predominant one, followed by S. Anatum (20.4%, 10/49). The Salmonella serotype of Farm 1 was relatively singular, being primarily S. Anatum (90.9%). In the four duck farms and the slaughterhouse, S. Enteritidis (3/5), S. Typhimurium (3/5) and S. Indiana (3/5) were widely prevalent serotypes. Among the 6 serotypes in this study, we found that all S. Indiana from the three farms were multi-drug resistant, being resistant to at least 12 antibiotics, and also contained the most detected type of resistance genes including blaTEM, blaOXA, blaCTX-M, sul1, sul2and aaC4.
Table 4.
Serotype distribution of duck Salmonella isolates
| Serotype | No. of isolates (%) | Total (n = 49) | ||||
|---|---|---|---|---|---|---|
| Farm 1 (n = 11) | Farm 2 (n = 10) | Farm 3 (n = 8) | Farm 4 (n = 18) | Slaughterhouse (n = 2) | ||
| S. Enteritidis | 0 | 0 | 4 (50.0) | 14 (77.8) | 2 (100.0) | 20 (40.8) |
| S. Anatum | 10 (90.9) | 0 | 0 | 0 | 0 | 10 (20.4) |
| S. Typhimurium | 0 | 3 (30.0) | 2 (25.0) | 3 (16.7) | 0 | 8 (16.3) |
| S. Kentucky | 0 | 5 (50.0) | 0 | 0 | 0 | 5 (10.2) |
| S. Indiana | 0 | 2 (20.0) | 2 (25.0) | 1 (5.6) | 0 | 5 (10.2) |
| S. Montevideo | 1 (9.1) | 0 | 0 | 0 | 0 | 1 (2.0) |
Antimicrobial susceptibility testing
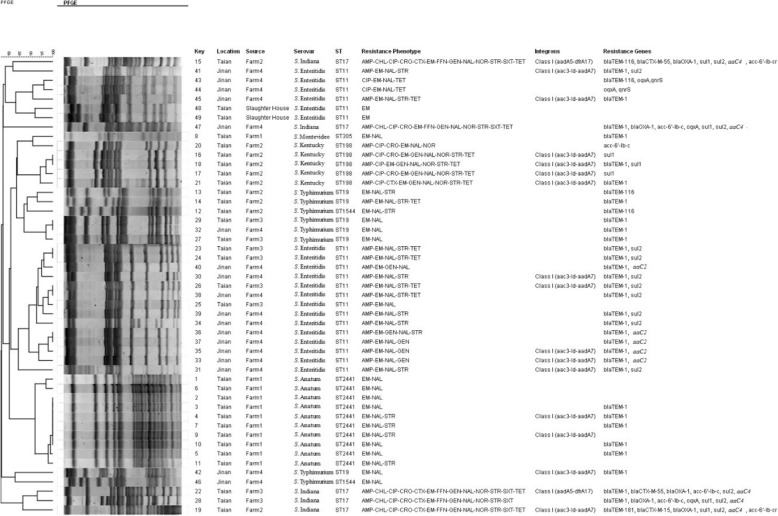
All of the 49 isolated Salmonella strains were tested for susceptibility against 14 antimicrobial agents; the results are listed in Table 5. It is noteworthy that 100.0 and 95.9% of those strains were resistant against EM and NAL, respectively, while all of the isolates were sensitive to PB. Salmonella isolated from the slaughterhouse were only resistant to EM, compared to Salmonella isolated from the farms (Table 6). The most common drug resistance spectrum was EM-NAL (n = 12). Among all of the isolates, 35 isolates exhibited multidrug-resistance (MDR), yielding the high rate of 71.4% (Fig. 1).
Table 5.
Antimicrobial resistance rates for 49 Salmonella serovars
| Drugs | Enteritidis (n = 20) | Anatum (n = 10) | Typhimurium (n = 8) | Kentucky (n = 5) | Indiana (n = 5) | Montevideo (n = 1) |
|---|---|---|---|---|---|---|
| AMP | 80.0 | 0 | 12.5 | 100.0 | 100.0 | 0 |
| CRO | 0 | 0 | 0 | 60.0 | 100.0 | 0 |
| CTX | 0 | 0 | 0 | 20.0 | 80.0 | 0 |
| EM | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| GEN | 25.0 | 0 | 0 | 80.0 | 100.0 | 0 |
| STR | 55.0 | 40.0 | 37.5 | 80.0 | 100.0 | 0 |
| TET | 35.0 | 0 | 12.5 | 80.0 | 80.0 | 0 |
| SXT | 0 | 0 | 0 | 0 | 100.0 | 0 |
| NOR | 0 | 0 | 0 | 100.0 | 100.0 | 0 |
| CIP | 10.0 | 0 | 0 | 100.0 | 100.0 | 0 |
| NAL | 90.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| CHL | 0 | 0 | 0 | 0 | 100.0 | 0 |
| FFN | 0 | 0 | 0 | 0 | 100.0 | 0 |
| PB | 0 | 0 | 0 | 0 | 0 | 0 |
Table 6.
Antimicrobial resistance phenotypes of 49 Salmonella isolates
| Antibacterial agents | Number of resistant isolates (%) | Total (n = 49) |
||||
|---|---|---|---|---|---|---|
| Sample from | ||||||
| Farm 1 (n = 11) |
Farm 2 (n = 10) |
Farm 3 (n = 8) |
Farm 4 (n = 18) |
Slaughterhouse (n = 2) |
||
| β-Lactams | ||||||
| AMP | 0 | 8 (80.0) | 6 (75.0) | 13(72.2) | 0 | 27 (55.1) |
| CRO | 0 | 5 (50.0) | 2 (25.0) | 1 (5.6) | 0 | 8 (16.3) |
| CTX | 0 | 3 (30.0) | 2 (25.0) | 0 | 0 | 5 (10.2) |
| Macrolides | ||||||
| EM | 11 (100.0) | 10 (100.0) | 8 (100.0) | 18 (100.0) | 2 (100.0) | 49 (100.0) |
| Aminoglycosides | ||||||
| GEN | 0 | 6 (60.0) | 2 (25.0) | 6 (33.3) | 0 | 14 (28.6) |
| STR | 4 (36.4) | 9 (90.0) | 5 (62.5) | 9 (50.0) | 0 | 27 (55.1) |
| Tetracyclines | ||||||
| TET | 0 | 7 (70.0) | 4 (50.0) | 5 (27.8) | 0 | 16 (32.7) |
| Sulfonamides | ||||||
| SXT | 0 | 2 (20.0) | 2 (25.0) | 1 (5.6) | 0 | 5 (10.2) |
| Quinolones | ||||||
| NOR | 0 | 7 (70.0) | 2 (25.0) | 1 (5.6) | 0 | 10 (20.5) |
| CIP | 0 | 7 (70.0) | 2 (25.0) | 3 (16.7) | 0 | 12 (24.5) |
| NAL | 11 (100.0) | 10 (100.0) | 8 (100.0) | 18 (100.0) | 0 | 47 (95.9) |
| Amphenicols | ||||||
| CHL | 0 | 2 (20.0) | 2 (25.0) | 1 (5.6) | 0 | 5 (10.2) |
| FFN | 0 | 2 (20.0) | 2 (25.0) | 1 (5.6) | 0 | 5 (10.2) |
| Polypeptide | ||||||
| PB | 0 | 0 | 0 | 0 | 0 | 0 |
Fig. 1.
PFGE Dendrogram of 49 Salmonella isolates from duck farms and a slaughterhouse in Shandong Province, China
Detection of antimicrobial resistance genes and virulence genes
Among the 49 Salmonella isolates, 11 kinds of resistance genes were detected (Fig. 1).It is noteworthy that there were 36 strains carrying blaTEM(73.5%, 36/49), including blaTEM-1 (n = 31), blaTEM-116 (n = 4), and blaTEM-181 (n = 1), and 14 isolates carrying sul2 (28.6%, 14/49).None of the 49 Salmonella strains displayed the tetracycline resistance gene and the chloramphenicol resistance gene. Moreover, there are nine Salmonella isolates in which resistance genes were not detected, of which five were collected from Farm1 (Fig. 1). Thirteen virulence genes including invA, hilA, spvC, sipA, sopE, stnP1, pefA, rck, sipC, ssaR, ssrA, sopB and sefA were also detected (Table 7).We found that all of the isolates(100.0%, 49/49) carried invA, hilA and sipA, followed by stnP1 (91.8%, 45/49) and ssrA (91.8%, 45/49); only a few isolates carried the ssaR (16.3%, 8/49) and sopE (16.3%, 8/49) genes.
Table 7.
Results of detection of virulence genes
| Virulence genes | Farm 1 (n = 11) | Farm 2 (n = 10) | Farm 3 (n = 8) | Farm 4 (n = 18) | Slaughterhouse (n = 2) | Total (n = 49) |
|---|---|---|---|---|---|---|
| invA | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| hilA | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| sipA | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| stnP1 | 100.0 | 100.0 | 87.5 | 94.4 | 0 | 91.8 |
| ssrA | 100.0 | 100.0 | 87.5 | 88.9 | 50.0 | 91.8 |
| sipC | 100.0 | 90.0 | 50.0 | 38.9 | 0 | 63.3 |
| spvC | 0 | 30.0 | 50.0 | 77.8 | 0 | 42.9 |
| rck | 0 | 30.0 | 62.5 | 61.1 | 0 | 38.8 |
| sefA | 0 | 0 | 50.0 | 77.8 | 50.0 | 38.8 |
| pefA | 0 | 30.0 | 50.0 | 66.7 | 0 | 38.8 |
| sopB | 45.5 | 50.0 | 50.0 | 22.2 | 0 | 36.7 |
| ssaR | 0 | 20.0 | 25.0 | 22.2 | 0 | 16.3 |
| sopE | 0 | 0 | 25.0 | 33.3 | 0 | 16.3 |
Detection of class I integrons
Among all of the 49 Salmonella strains, 17 strains were found to be carrying class I integrons (Fig. 1), yielding at detection rate of 34.7%. Two kinds of gene cassettes were found in those class Iintegrons, which were aadA7 + aac3-Id (15/17, 88.2%) and aadA5 + dfrA17 (2/17, 11.8%). Furthermore, 94% of Salmonella carrying class I integrons were multi-drug resistant in our study.
MLST
The 49 Salmonella isolates were classified into 7 STs (Table 8). The dominant ST isST11 (20/49, 40.8%), followed by ST2441 (10/49, 20.4%), ST19 (6/49, 12.2%) and ST17 (5/49, 10.2%) (Table 8). The STs identified in the present study show the following correlations with Salmonella serovars: ST11 with S. Enteritidis, ST19 with S. Typhimurium, ST17 with S. Indiana and ST198 with S. Kentucky.
Table 8.
Prevalence of sequence types (STs) for the Salmonella isolates
| STs | Serovars | Allelic type | No. of isolates | ||||||
|---|---|---|---|---|---|---|---|---|---|
| aroC | dnaN | hemD | hisD | purE | sucA | thrA | |||
| ST11 | Enteritidis | 5 | 2 | 3 | 7 | 6 | 6 | 11 | 19 |
| ST11 | Enteritidis | 5 | 2 | 3 | 7 | 6 | 6 | 67 | 1 |
| ST2441 | Anatum | 10 | 500 | 15 | 31 | 25 | 20 | 33 | 6 |
| ST2441 | Anatum | 76 | 500 | 15 | 31 | 25 | 20 | 33 | 3 |
| ST2441 | Anatum | 10 | 500 | 47 | 31 | 25 | 20 | 33 | 1 |
| ST19 | Typhimurium | 10 | 7 | 12 | 9 | 5 | 9 | 2 | 5 |
| ST1544 | Typhimurium | 10 | 7 | 12 | 230 | 5 | 9 | 2 | 2 |
| ST19 | Typhimurium | 10 | 7 | 12 | 9 | 5 | 6 | 2 | 1 |
| ST198 | Kentucky | 10 | 14 | 3 | 77 | 64 | 64 | 67 | 3 |
| ST198 | Kentucky | 8 | 14 | 11 | 77 | 64 | 64 | 67 | 1 |
| ST198 | Kentucky | 76 | 14 | 3 | 77 | 64 | 64 | 67 | 1 |
| ST17 | Indiana | 8 | 8 | 11 | 11 | 5 | 11 | 15 | 5 |
| ST305 | Montevideo | 43 | 41 | 16 | 42 | 34 | 13 | 23 | 1 |
PFGE
As shown in Fig. 1, the PFGE patterns were generally diverse between different sampling sites, and showed similarity values of 80–100% among all of the strains. The 49 Salmonella isolates were divided into 39 PFGE patterns, which grouped into ten clusters. Most strains of the same serotype have similar PFGE patterns. However, there are also a few strains of the same serotype that have very different PFGE patterns, for example S. Indiana (Key 47 and Key 15). In addition, it is noteworthy that the strains derived from the same farm or slaughterhouse exhibited high similarity, and that some of the isolates from different districts have the same PFGE patterns, as exemplified by two S. Typhimurium isolates, Key 29 and Key 32; Key 29 is from Tai’an and Key 32 is from Jinan.
Discussion
In this study, the total isolation rate of Salmonella strains was 2.1% (49/2342), which is significantly less than that (12.2%) in conventional farms in Sichuan province in China [16] and the isolation rates, 2.4 and 7.5%, respectively, in Shandong province, as reported by a study conducted from 2009 to 2012 [10]. However, the isolation rate of Salmonella in duck embryos (15.3%) from the diseased duck farms was relatively high. This result was similar to other reports [27], which found the positive rate of Salmonella to be 21.1% in dead embryos. The isolation rate of Salmonella in duck feces samples (1.8%) collected from the diseased duck farms was lower than that (12.3%) in liver samples, probably due to intermittent detoxification of Salmonella; even when the duck is infected with Salmonella, the pathogen may not be detectable in the collected fecal samples. In addition, different regions, environmental climates and seasons may also cause differing rates of Salmonella isolation.
In our study, the predominant serotype was S. Enteritidis (40.8%, 20/49), which is consistent with a study [28], conducted throughout twelve provinces in China, that found the most prevalent serotype among duck farms to be S. Enteritidis (36.6%, 15/41). This report differs from those of other countries [1, 8], however, which report that S. Typhimurium is the most prevalent serotype in Penang, Malaysia and Korea. The underlying reason may be differences in geography and species isolation among farms. In addition, S.Kentucky has been rarely reported in ducks; however, it has been reported in other animals, such as chicken [29, 30], pork [31], beef [32] and rabbit [33]. In this study, we found that S. Indiana from the three farms showed a high MDR rate (100%, 5/5) and that our findings concerning the phenomenon of particularly serious drug resistance are similar the corresponding report in China [34]. This may be related to the characteristics of S. Indiana itself; most of the S. Indiana isolates were resistant to many antibiotics, including streptomycin, tetracycline, chloramphenicol and fluoroquinolones, etc. We have detected Class I integrons and related gene cassettes in 3 out of 5 S.indiana isolates, which may be responsible for the MDR (Fig. 1.). Generally, the integrons were located at the conjugative plasmids or at the chromosome within the Salmonella Genome island 1 (SGI1) [35]. However, it has been reported that the S. Indiana lacks of SGI1, which was supported by whole genome and PCR analyses [36]. At this stage, the exact mechanisms underlying the antimicrobial resistance of the S.indiana may remained to be further studied.
Most Salmonella isolates identified in our study showed high resistance to NAL (95.9%) and AMP (55.1%), having resistance rates slightly higher than those seen in the study on ducks in the Sichuan province of China [16], which were reported to be 69.6 and 34.8% to NAL and AMP, respectively. Our corresponding results were also higher than another study conducted in South Korea [37] that reported resistance rates to NAL (73.6%) and AMP (24.0%), suggesting that these drugs may have been widely used in ducks during disease control efforts or prevention. In this study, NAL (95.9%) was obviously higher than CIP (24.5%) in quinolone antibiotic. This may be caused by a significantly greater use of NAL than that of CIP. This result was consistent with a study on ducks in Sichuan province in China [16]. The resistance rate of third-generation cephalosporins to CRO was 16.3%, and that to CTX was 10.2%, which are similar to the reports of another study on ducks in Penang, Malaysia [8]. Our study indirectly proved that third-generation cephalosporins have become the primary drugs for the treatment of Salmonella, which is in agreement with another study [38]. In our study, MDR isolate rate of Salmonella (71.4%) was similar to another study (73.9%) on ducks in Sichuan province in China [16], but higher than a study (50.5%) on ducks from South Korea [36]. MDR Salmonella isolates were frequently observed among the farms in this study; notably, S. Indiana isolates were resistant to at least 12 antimicrobials, posing a great risk to public health, should these MDR Salmonella isolates be transferable to humans via duck or duck-derived products. Reducing the use of antibiotics in ducks is especially important to limiting the emergence of super-MDR organisms and to maintaining good public health, as well as for other animals.
Concerning the detection of antimicrobial resistance genes, the most prevalent β-lactamase-resistance gene was blaTEM (36/49, 73.47%), which is different than reports from Sichuan province that state that the most commonly isolated β-lactamase-resistance gene was blaOXA [16].The most common quinolone-resistance genes were aac(6′)-Ib-cr (10.2%, 5/49), followed by oqxA (8.2%, 4/49); these reports differ from those in Xinjiang province, China that report the most prevalent to be qnrB (34.3%) [39]. Those differences may be due to the usage of antimicrobials in different areas of China. Meanwhile, in the present study, there was no tetracycline-resistance gene detected among all strains, while 34.7% (17/49) of strains showed resistance to tetracycline; this could be due to mutations of the resistance strains. Furthermore, there were only a few antimicrobial resistance genes and antibiotics detected in isolates from Farm 1, probably due to fewer parental breeding duck resistance genes and to rational use of antibiotics on this farm. In this study, Class I integrons were detected in 17 Salmonella strains out of 49 strains (34.7%), which provided a supplement to the data of Salmonella in ducks and was similar to the percentage (31.5%) presented in a study on chicken [40]. Class I integrons were associated with MDR Salmonella isolates, a finding which is consistent with other reports [16, 41].
All of the strains carried invA, hilA and sipA, in this study; the high detection rates of these virulence genes have also been observed by other researchers. For instance, it was reported that all 34 S. Enteritidis strains (100.0%) isolated in Iran harbored invA, hilA and sipA genes [25]. Generally, a high detection rate of virulence genes emphasizes the pathogenic potential of these isolates, which may foreshadow severe Salmonellosis and threats to public health [25]. Moreover, Salmonella isolated from Farm 1 and the slaughterhouse had multiple virulence genes not detected. This phenomenon is similar to patterns in Salmonella resistance and antimicrobial resistance genes detected in our study. We suspect that there may be some connection between them, and propose that further study is necessary.
MLST results revealed that 7 STs were identified in the duck farm and slaughterhouse isolates. Among them, ST11 was the most prevalent ST, which is in agreement with the results of a previous study on ducks [37]. Additionally, the result was in concordance with other reports of ST11 being the predominant ST among Salmonella isolates from human and food-producing animals in China [42]; our corroboration of these results further highlight the prevalence of ST11 strains in China. Furthermore, we noticed that no ST40 was detected in our study, which is different from reports of previous studies that found ST40 to be significantly prevalent in slaughterhouse and retail markets in Yangzhou, China. It has also been widely detected in the pig industries of the United States and Europe [6, 16]. This may be due to geographical and environmental differences. In this study, the following correlations between STs and Salmonella serovars were founded: ST11 with S. Enteritidis, ST2441 with S. Anatum, ST19 with S. Typhimurium, ST17 with S. Indiana and ST198 with S. Kentucky. These correlations are consistent with those observed in a previously reported study that found STs and serovars to be tightly connected [6].
With regard to other genotyping methods, whole genome sequencing (WGS) is recently considered as one of the most powerful method to differentiate foodborne microorganisms and determine the genetic relatedness of Salmonella isolates, and now WGS is growingly being used by Food Safety Authorities worldwide for outbreak investigation and surveillance [43]. Compared to WGS, PFGE, which used to be known as the golden standard of genotyping, are no longer considered cutting edge but still have been efficient in detecting, investigating and control of foodborne infection outbreaks in the past two decades due to its discriminatory power and reproducibility [44]. In the present study, the phenomenon that the strains derived from the same sampling site exhibited high similarity suggests the possibility of clonal spread; for example, isolates collected from Farm 1 exhibited high genotype similarity. However, the strains isolated from different areas, such as Key 26 (Tai’an Farm 3) and Key 38 (Jinan Farm 4), showed the same PFGE pattern, ST, serovar and resistance profiles; this demonstrates that it is vital to take surveillance and controlling measures to prevent the dissemination of Salmonella clones. These results suggest the possibility that Salmonella can be transmitted between different farms, a conclusion similar to one proposed in a previous study [45]. Furthermore, we have observed that Salmonella isolates with the same serotypes have higher PFGE pattern similarity, that PFGE may be more suitable for Salmonella genotyping of the same serotype and that PFGE showed greater power in molecular typing, compared with MLST. For instance, the 49 Salmonella isolates were divided into 39 patterns and only 7 STs. However, MLST can be used for typing between different serotypes of Salmonella. Therefore, the two types of methods can complement each other, and we can use two types of typing methods for bacterial typing in order to analyze their genetic relationships.
Conclusions
The prevalence and antimicrobial resistance of Salmonella in duck farms are potential risks to public health. The data presented in our study illustrated that several duck farms and a slaughterhouse in the Tai’an and Jinan areas of China are contaminated with Salmonella and that antimicrobial resistance and MDR is widespread among the strains. PFGE results revealed that the Salmonella strains may have the ability to spread among different areas, as well as the ability to cause clonal spread. It is still necessary and critical to reinforce the surveillance and control of Salmonella and to search for a substitution for antimicrobials.
Acknowledgments
Not Applicable.
Abbreviations
- AMP
Ampicillin
- BPW
Buffered peptone water
- CDCP
Centers for Disease Control and Prevention
- CHL
Chloramphenicol
- CIP
Ciprofloxacin
- CLSI
Clinical and Laboratory Standards Institute
- CRO
Ceftriaxone
- CSB
Cell suspension buffer
- CTX
Cefotaxime
- EM
Erythromycin
- FAO
Food and Agriculture Organization
- FFN
Florfenicol
- GEN
Gentamicin
- MDR
Multidrug-resistance
- MLST
Multilocus sequence typing
- NAL
Nalidixic acid
- NOR
Norfloxacin
- PB
Polymyxin B
- PCR
Polymerase chain reaction
- PFGE
Pulsed field gel electrophoresis
- SC
Selenite cystine broth
- SDS
Sodium dodecyl sulfate
- STR
Streptomycin
- STs
Sequence types
- SXT
Sulfamethoxazole
- TBE
Tris-Borate-EDTA
- TET
Tetracycline
- UPGMA
Unweighted pair-group method
- XLD
Xylose lysinedeoxycholate medium
Authors’ contributions
SS, JY and ZJ conceived the project and designed the study. All authors performed the experiments and analyzed the results. JY and ZJ wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National key R&D project (2016YFD0501608) and (2016 YFD0500510); Taishan Scholar Program (201511023); Funds of Shandong “Double Tops” program.
Availability of data and materials
The data used and analyzed during the present study are accessible from the corresponding author on request.
Ethics approval and consent to participate
Verbal consent for all the sampling procedures was obtained from the owners of the animals. All procedures were approved by the Animal Care and Use of Shandong Agricultural University (SDAUA-2018-027).
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jie Yang and Zijing Ju are co-first author and contributed equally to this work.
Contributor Information
Jie Yang, Email: snyangjie@yeah.net.
Zijing Ju, Email: jzjsdau@163.com.
Yi Yang, Email: yangyi1987512@163.com.
Xiaonan Zhao, Email: 605376729@qq.com.
Zhiyu Jiang, Email: 380402876@qq.com.
Shuhong Sun, Email: ssh6811@163.com.
References
- 1.Bae DH, Dessie HK, Baek HJ, Kim SG, Lee HS, Lee YJ. Prevalence and characteristics of Salmonella spp. isolated from poultry slaughterhouses in Korea. J Vet Med Sci. 2013;75:1193–1200. doi: 10.1292/jvms.13-0093. [DOI] [PubMed] [Google Scholar]
- 2.Shao D, Shi Z, Wei J, Ma Z. A brief review of foodborne zoonoses in China. Epidemiol Infect. 2011;139:1497–1504. doi: 10.1017/S0950268811000872. [DOI] [PubMed] [Google Scholar]
- 3.Guibourdenche M, Roggentin P, Mikoletit M, Fields PI, Bockemuhi J, Grimont PA, et al. Supplement 2003-2007 (no.47) to the white-Kauffmann-le minor scheme. Res Microbiol. 2010;161:26–29. doi: 10.1016/j.resmic.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Lee LA, Puhr ND, Maloney EK, Bean NH, Tauxe RV. Increase in antimicrobial-resistant Salmonella infections in the United States, 1989-1990. J Infect Dis. 1994;170:128–134. doi: 10.1093/infdis/170.1.128. [DOI] [PubMed] [Google Scholar]
- 5.Liang Z, Ke BX, Deng XL, Liang JH, Ran L, Lu LG. Serotypes, seasonal trends, and antibiotic resistance of non-typhoidal Salmonella from human patients in Guangdong Province, China, 2009-2012. BMC Infect Dis. 2015;15:53. doi: 10.1186/s12879-015-0784-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai Y, Tao J, Jiao Y, Fei X, Zhou L, Wang Y, et al. Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou, China. Int J Food Microbiol. 2016;222:56–64. doi: 10.1016/j.ijfoodmicro.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 7.Kuang X, Hao H, Dai M, Wang Y, Ahmad I, Liu Z, et al. Serotypes and antimicrobial susceptibility of Salmonella spp. isolated from farm animals in China. Front Microbiol. 2015;6:602. doi: 10.3389/fmicb.2015.00602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adzitey F, Rusul G, Huda N. Prevalence and antibiotic resistance of Salmonella serovars in ducks, duck rearing and processing environments in Penang, Malaysia. Food Res Int. 2012;45:947–952. doi: 10.1016/j.foodres.2011.02.051. [DOI] [PubMed] [Google Scholar]
- 9.Gong JS, Xu M, Zhu CH, Miao JF, Liu XX, Xu B, et al. Antimicrobial resistance, presence of integrons and biofilm formation of Salmonella Pullorum isolates from eastern China (1962-2010) Avian Pathol. 2013;42:290–294. doi: 10.1080/03079457.2013.788129. [DOI] [PubMed] [Google Scholar]
- 10.Lai J, Wu CM, Wu CB, Qi J, Wang Y, Wang HY, et al. Serotype distribution and antibiotic resistance of Salmonella in food-producing animals in Shandong province of China, 2009 and 2012. Int J Food Microbiol. 2014;180:30–38. doi: 10.1016/j.ijfoodmicro.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 11.Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 2012;8:e1002776. doi: 10.1371/journal.ppat.1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bilhere E, Lucas PM, Claisse O, Lonvaudfunel A. Multilocus sequence typing of Oenococcus oeni: detection of two subpopulations shaped by intergenic recombination. Appl Environ Microb. 2009;75:1291–1300. doi: 10.1128/AEM.02563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang CB, Chen QM, Zhang C, Yang J, Lu ZX, Lu FX, et al. Characterization of a broad host-spectrum virulent Salmonella bacteriophagefmb-p1 and its application on duck meat. Virus Res. 2017;236:14–23. doi: 10.1016/j.virusres.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Malorny B, Hoorfar J, Bunge C, Helmuth R. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl Environ Microb. 2003;28:290–296. doi: 10.1128/AEM.69.1.290-296.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CLSI . Performance standards for antimicrobial susceptibility testing: twentieth-third informational supplement M100-S23. Wayne: Clinical and Laboratory Standards Institute; 2013. [Google Scholar]
- 16.Li RC, Lai J, Wang Y, Liu SL, Li Y, Liu KY, et al. Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan Province, China. Int J Food Microbiol. 2013;163:14–18. doi: 10.1016/j.ijfoodmicro.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 17.Pokharel BM, Koirala J, Dahal RK, Mishra SK, Khadga PK, Tuladhar N. Multidrug-resistant and extended-spectrum beta-lactamase (ESBL)-producing Salmonella enterica (serotypes Typhi and Paratyphi A) from blood isolates in Nepal: surveillance of resistance and a search for newer alternatives. Int J Infect Dis. 2006;10:434–438. doi: 10.1016/j.ijid.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Ahmed AM, Motoi Y, Sato M, Maruyama A, Watanabe H, Fukumoto Y, et al. Zoo animals as reservoirs of gram-negative bacteria harboring integrons and antimicrobial resistance genes. Appl Environ Microbiol. 2007;73:6686–6690. doi: 10.1128/AEM.01054-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puah SM, Puthucheary SD, Liew FY, Chua KH. Aeromonas aquariorum clinical isolates: antimicrobial profiles, plasmids and genetic determinants. Int J Antimicrob Ag. 2012;41:281–284. doi: 10.1016/j.ijantimicag.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Liao XP, Xia J, Yang L, Li L, Sun J, Liu YH, et al. Characterization of CTX-M-14-producing Escherichia coli from food-producing animals. Front Microbiol. 2015;6:1136. doi: 10.3389/fmicb.2015.01136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navajas-Benito EV, Alonso CA, Sanz S, Olarte C, Martinezolarte R, Hidalgosanz S, et al. Molecular characterization of antibiotic resistance in Escherichia coli strains from a dairy cattle farm and its surroundings. J Sci Food Agr. 2016;97:362–365. doi: 10.1002/jsfa.7709. [DOI] [PubMed] [Google Scholar]
- 22.Aarestrup FM, Lertworapreecha M, Evans MC, Bangtrakulnonth A, Chalermchaikit T, Hendriksen RS, et al. Antimicrobial susceptibility and occurrence of resistance genes among Salmonella enterica serovar Weltevreden from different countries. J Antimicrob Chemother. 2003;52:715–718. doi: 10.1093/jac/dkg426. [DOI] [PubMed] [Google Scholar]
- 23.Zhang AY, Wang HN, Tian GB, Zhang Y, Yang X, Xia QQ, et al. Phenotypic and genotypic characterisation of antimicrobial resistance in faecal bacteria from 30 Giant pandas. Int J Antimicrob Agents. 2009;33:456–460. doi: 10.1016/j.ijantimicag.2008.10.030. [DOI] [PubMed] [Google Scholar]
- 24.Guerra B, Soto SM, Arguelles JM, Mendoza MC. Multidrug resistance is mediated by largeplasmids carrying a class I integron in the emergent Salmonella enterica serotype [4,5, 12:I:-] Antimicrob Agents Chemother. 2001;5:1305–1308. doi: 10.1128/AAC.45.4.1305-1308.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fardsanei F, Soltan DMM, Douraghi M, Zahraei ST, Mahmoodi M, Memariani H, et al. Genetic diversity and virulence genes of Salmonella enteric subspecies enterica serotype Enteritidis isolated from meats and eggs. Microb Pathog. 2017;107:451–456. doi: 10.1016/j.micpath.2017.04.026. [DOI] [PubMed] [Google Scholar]
- 26.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 27.Ha PDH, Phan TT, Hanh TX, Le TMK, Cuong NV, Binh NX. Investigation of Salmonella and Escherichia coli infections of ducks in Long An province, Vietnam. Hoc Ky Thuat Thu Y. 2000;7(4):29-34.
- 28.Gong JS, Zhang JQ, Xu M, Zhu CH, Yu Y, Liu XX, et al. Prevalence and Fimbrial genotype distribution of poultry Salmonella isolates in China (2006 to 2012) Appl Environ Microb. 2014;80:687–693. doi: 10.1128/AEM.03223-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aslam M, Checkley S, Avery B, Chalmers G, Bohaychuk V, Gensler G, et al. Phenotypic and genetic characterization of antimicrobial resistance in Salmonella serovars isolated from retail meats in Alberta, Canada. Food Microbiol. 2012;32:110–117. doi: 10.1016/j.fm.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 30.Guran HS, Mann D, Alali WQ. Salmonella prevalence associated with chicken parts with and without skin from retail establishments in Atlanta metropolitan area, Georgia. Food Control. 2017;73:462–467. doi: 10.1016/j.foodcont.2016.08.038. [DOI] [Google Scholar]
- 31.Hernandez M, Gomez-Laguna J, Luque I, Herrera-Leon S, Maldonado A, Reguillo L, et al. Salmonella prevalence and characterization in a free-range pig processing plant: tracking in trucks, lairage, slaughter line and quartering. Int J Food Microbiol. 2012;162:48–54. doi: 10.1016/j.ijfoodmicro.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 32.Stevens A, Kabore Y, Perrier JD, Millemann Y, Brisabois A, Catteau M, et al. Prevalence and antibiotic-resistance of Salmonella isolated from beef sampled from the slaughterhouse and from retailers in Dakar (Senegal) Int J Food Microbiol. 2006;110:178–186. doi: 10.1016/j.ijfoodmicro.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 33.Kylie J, McEwen SA, Boerlin P, Reid-Smith RJ, Weese JS, Turner PV. Prevalence of antimicrobial resistance in fecal Escherichia coli and Salmonella enterica in Canadian commercial meat, companion, laboratory, and shelter rabbits (Oryctolagus cuniculus) and its association with routine antimicrobial use in commercial meat rabbits. Prev Vet Med. 2017;147:53–57. doi: 10.1016/j.prevetmed.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Lu Y, Wu CM, Wu GJ, Zhao HY, He T, Cao XY, et al. Prevalence of antimicrobial resistance among Salmonella from chicken in China. Foodborne Pathog Dis. 2011;8:45–53. doi: 10.1089/fpd.2010.0605. [DOI] [PubMed] [Google Scholar]
- 35.Fluit AC. Towards more virulent and antibiotic-resistant Salmonella? FEMS Immunol Med Microbiol. 2005;43:1–11. doi: 10.1016/j.femsim.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 36.Gong JS, Wang C, Shi S, Bao H, Zhu C, Kelly P, et al. Highly drug-resistant Salmonella enterica Serovar Indiana clinical isolates recovered from broilers and poultry workers with diarrhea in China. Antimicrob Agents Chemother. 2016;60:1943–1947. doi: 10.1128/AAC.03009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cha SY, Kang M, Yoon RH, Park CK, Moon OK, Jang HK. Prevalence and antimicrobial susceptibility of Salmonella isolates in Pekin ducks from South Korea. Comp Immunol Microb. 2013;36:473–479. doi: 10.1016/j.cimid.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 38.Zhao X, Yang J, Zhang B, Sun S, Chang W. Characterization of integrons and resistance genes in Salmonella isolates from farm animals in Shandong Province. China Front Microbiol. 2017;8:1300. doi: 10.3389/fmicb.2017.01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin M, Yang B, Wu Y, Wang L, Wu H, Zhang T, et al. Prevalence and characterisation of Salmonellaenterica serovar in retail meats in market place in Uighur, Xinjiang, China. Food Control. 2016;64:165–172. doi: 10.1016/j.foodcont.2015.12.029. [DOI] [Google Scholar]
- 40.Wang HS, Hu RP, Gao YM, Yang Z, Deng XL, Zhang JY, et al. Study on molecular characterization of class I integron and integron-associated antimicrobial resistance in Escherichia coli from beef cattle. China Anim Hus Vet Med. 2014;41:63–67. [Google Scholar]
- 41.Firoozeh F, Zahraei-Salehi T, Shahcheraghi F, Karimi V, Aslani MM. Characterization of class I integrons among Salmonella enterica serovar Enteritidis isolated from humans and poultry. FEMS Immunol Med Microbiol. 2012;64:237–243. doi: 10.1111/j.1574-695X.2011.00883.x. [DOI] [PubMed] [Google Scholar]
- 42.Chao GX, Wang C, Wu TQ, Zhang XR, Chen JH, Qi XX, et al. Molecular epidemiology and antibiotic resistance phenotypes and genotypes of Salmonella from food supply chains in China. Food Control. 2017;77:32–40. doi: 10.1016/j.foodcont.2017.01.022. [DOI] [Google Scholar]
- 43.Nadon C, Van Walle I, Gerner-Smidt P, Campos J, Chinen I, Concepcion-Acevedo J, et al. PulseNet international: vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Euro Surveill. 2017;22. 10.2807/1560-7917.ES.2017.22.23.30544. [DOI] [PMC free article] [PubMed]
- 44.Rumore JL, Tschetter L, Nadon C. The impact of multilocusvariable-number tandem-repeat analysis on PulseNet Canada Escherichia coli O157:H7 laboratory surveillance and outbreak support, 2008-2012. Foodborne Pathog Dis. 2016;13:255–261. doi: 10.1089/fpd.2015.2066. [DOI] [PubMed] [Google Scholar]
- 45.Ni P, Xu Q, Yin Y, Liu D, Zhang J, Wu Q, et al. Prevalence and characterization of Salmonella serovars isolated from farm products in Shanghai. Food Control. 2018;85:269–275. doi: 10.1016/j.foodcont.2017.10.009. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used and analyzed during the present study are accessible from the corresponding author on request.