ABSTRACT
Background and Objectives: Dental caries is a chronic disease affecting young children and has multi-factorial risk factors. The purpose of this work was to identify sex-specific differences in the salivary microbiota within caries-active children.
Design: Saliva specimens were collected from 85 children (boys: 41; girls: 44) between the ages of 2-12 years. Salivary microbial DNA was subjected to PCR amplification using V3-V4 16S rDNA-specific primers and next-generation sequencing.
Results: Significant sex differences in salivary microbiota were found between caries-active boys versus caries-active girls. Neisseria flavescens, Rothia aeria, and Haemophilus pittmaniae were found at significantly higher levels in caries-active boys. In contrast, Lactococcus lactis, Selenomonas species HOT 126, Actinobaculum species HOT 183, Veillonella parvula, and Alloprevotella species HOT 473 were found at significantly higher levels in caries-active girls.
Conclusion: We have found the acid-generating, cariogenic Lactococcus lactis to be much more abundant in caries-active girls than caries-active boys, indicating that this microorganism may play a more significant role in shaping the cariogenic microbiome in girls. In addition, in caries-active girls, Alloprevotella species HOT 473 was the only species that exhibited both significant sex differences (4.4-fold difference; p=0.0003) as well as high abundance in numbers (1.85% of the total microbial population).
KEYWORDS: Salivary microbiome, sex differences, dental caries, children, oral microbiome
Dental caries is a common and undertreated chronic pediatric disease, exhibiting complex multifactorial etiology, lasting health effects, and serious socio-economic consequences [1–3]. The development and progression of caries are impacted by several socio-demographic risk factors, including lower education and poor oral hygiene and dietary behaviors, as well as microbiological and genetic components, including dysbiosis of the oral microbiota, altered saliva composition and flow rate, and genetic variability [1–3].
Continued cycling of enamel demineralization and remineralization is the biological basis of the caries process, with acid production by cariogenic bacteria resulting in demineralization of tooth structure, and buffering actions of saliva and fluoride promoting remineralization. An imbalance in these two processes results in caries progression and disease.
A balanced oral microbiome is a foundational element of oral health; dysbiosis and the proliferation of cariogenic microbial species result in increased tooth demineralization and cavitation [4]. Microbial species considered classically cariogenic include Streptococcus mutans and Lactobacillus [4,5]. Additional microorganisms that are associated with caries development include Scardovia wiggsiae, Firmicutes, Veillonella HOT 780, Slackia exigua, Porphyromonas, Granulicatella elegans, and Actinomyces [4–7]. These cariogenic bacteria produce an acidic environment by breaking down fermentable carbohydrates, and support the premise that diet and specific nutrients are key determinants in dental caries. The frequency of carbohydrate consumption will influence or determine the duration of acidic conditions within the oral cavity.
Recent studies demonstrate widely observed sex differences in caries prevalence, with females at higher risk to experience caries than males [8,9]. The sex gap in caries experience is reported to increase progressively with age [10]. In children, males and females express nearly equal caries rates – a pattern that shifts by the time of adolescence to females expressing higher caries susceptibility [10,11].
The higher susceptibility of females to dental caries may be due to several factors, including genetic variations, dietary factors, hormonal fluctuations, salivary differences, and social factors, all of which influence the composition of the oral microbiome [12,13]. Studies have shown differences between the sexes with lower salivary flow rates in women, in addition to lower pH and IgA levels, potentially leading to higher caries incidence [12,14,15]. Genetic variations in factors influencing taste, enamel proteins, and enamel structure are also linked to this trend [12,13,16]. Regarding patterns of tooth eruption, female teeth generally erupt at an earlier age, exposing teeth to a potential caries-inducing environment for longer durations of time [12]. Caries rates in women increase progressively throughout lifetimes, particularly during the reproductive years. Several physiological changes occur during pregnancy including hormonal fluctuations, immune suppression, dietary changes, and salivary flow [12,13]. Social factors that potentially help determine sex differences in caries experience include lower literacy rates in women globally, responsibility for food preparation, experience of domestic violence, and higher rates of eating disorders [12].
In this paper, we identified sex-specific differences in the salivary microbiome in caries-active children and describe the cariogenic salivary microbiome to gain further understanding of caries progression in females that may account for the observed global sex dissimilarity in caries experience.
Materials and methods
Participant selection and demographics
We selected study participants primarily from patients visiting the OHSU Pediatric Dental Clinic at Doernbecher Children’s Hospital, or at the OHSU School of Dentistry at the Robertson Life Sciences Building on the South Waterfront Campus. Some caries-free patients were obtained from an external practitioner’s dental clinic operated in Albany, Oregon. The study protocol was reviewed and approved by the OHSU Institutional Review Board (IRB; protocol #6535), and permissions and consent to participate were obtained from the parents or guardians of all children selected for this study. For older children, age 7 years or more, child assent and permissions were also obtained. This study is part of a larger study that examines the efficacy and use of adjunctive clinical therapies in caries-active children, and involves the use of specimens obtained before the application of any adjunctive therapies or other procedures that were applied as part of routine dental care. The inclusion parameters for study participation were children ages 2–12 years, in good general health (ASA I or II). The ASA physical status classification system is used by the American Association of Anesthesiologists to assess the fitness of patients before surgery; ASA I and ASA II designate a healthy individual, or an individual with mild systemic disease, respectively. Children excluded from the study were those who had undergone antibiotic treatment within the previous three month-period or were undergoing current orthodontic therapies, as well as children with recent exposure to topical fluoride or antiseptic mouth rinses. Eighty-five children were enrolled in this study (41 boys and 44 girls). Of the 85 children, 64 children were caries-active and exhibited DMFT scores of 1–15 at the time of appointment (DMFT average: 8.6) and 21 children were caries-free individuals, who demonstrated no apparent evidence of carious lesions. The numbers of caries-active boys, caries-active girls, caries-free boys, and caries-free girls were 28, 33, 13, and 11 individuals, respectively. Caries status and prevalence were determined by clinical examination, both visual and tactile, and with the use of the decay-missing-filled (DMFT) index and confirmatory radiographs. For children, 5 years of age or younger, tooth surfaces were examined to assess caries status.
Saliva collection, DNA extraction, and human oral microbial identification
Non-stimulated saliva specimens (1–2 ml) were collected from the oral cavity of enrolled participants using the OMNIgene-ORAL collection and stabilization kit for microbial DNA (DNA Genotek; OM-501). In some cases, especially for young children, sterile bulbs or sterile swabs were used to collect saliva from the oral cavity, prior to placement of saliva within the OMNIgene-ORAL stabilization reagent. OMNIgene-ORAL stabilizes samples at the point of collection until transport to the laboratory, and permits long-term storage up to 1 year under ambient temperatures. The OHSU Gene Profiling Shared Resources Laboratory extracted microbial genomic DNA from the saliva specimens, using kits compatible with the OMNIgene-ORAL stabilization reagent, and subsequently quantified purified DNA by absorbance at 260 nm. Microbial genomic DNA was then subjected to agarose gel electrophoresis, in the presence of ethidium bromide, and nucleic acid integrity was then documented using the Bio-Rad Gel Doc EZ Gel Documentation system (product 1,708,270; Bio-Rad Laboratories, Hercules, CA). Microbial DNA was then subjected to PCR amplification using V3-V4 16S rDNA-specific primers and next-generation sequencing with high-throughput Illumina sequencing in the Human Oral Microbial Identification Next Generation Sequencing (NGS) HOMINGS Laboratory at the Forsyth Institute (Cambridge, MA). HOMINGS as defined by the Forsyth Institute ‘permit species-level identification of nearly 600 oral bacterial taxa and genus-level identification of remaining sequences for 129 genera’. This methodology has been pioneered by Dr. Bruce J Paster, has been used extensively in microbiome studies, including oral microbiome analyses, and is described in representative publications [17–19]. Briefly, oral taxa identification and abundance of microorganisms were assessed with the ProbeSeq program, where sequence targets were matched initially with species probes and then sequentially with genus probes. Sequence targets not uniquely matched to either species or genus probes were then assessed as unmatched.
Statistical analyses
The relative abundance of oral microorganisms was compared according to caries status and by sex. All non-zero NGS count data were included in computing the relative abundance within each participant, and then averaged by the group. The differential abundance analyses of NGS count data of oral microorganisms were performed by a zero-inflated negative binomial regression model by the zinbwave and edgeR packages in the R statistical language (http://www.r-project.org) [20,21]. The ZINB-WaVE method implemented the zinbwave package and efficiently identified excess zero count data and provided microorganism and sample-specific weights. These weights were then incorporated into the dispersion estimates of the negative binomial regression model in edgeR, in order to determine or recover the power of statistical tests of relative abundance of microorganisms between caries status or by sex. Only 298 microorganisms detected in at least nine participants were kept in the analysis to stabilize dispersion parameter estimates. Two-sided p-values for testing relative abundance by caries status or by sex were adjusted by the false discovery rate (FDR) for multiple test correction [22]. Fold changes larger than 1.5, with the FDR adjusted p-value of < 0.05 were considered statistically significant [22].
Results
Abundance of salivary microorganisms within caries-active boys versus caries-active girls
Table 1 ranks the abundance of microorganisms in caries-active girls, caries-active boys, caries-free girls, caries-free boys, caries-active children, and caries-free children; using box representations, Figures 1 and 2 display the abundance of the top 20 microorganisms differentiated by caries status and sex, respectively. Tables 2 (caries-active boys versus caries-active girls) and 3 (caries-free boys versus caries-free girls) compares the fold changes for each microorganism between boys versus girls and ranks the fold changes with high positive numbers representing higher proportional representation of microorganisms in boys and extremely low negative values representing higher proportional representation of microorganisms in girls. The abundance list in Table 1 is sorted according to rankings of microorganisms observed in caries-active children and then applied in the same sequence for all remaining groups, and lists only the top 100 microorganisms. In Tables 2 and 3, microorganisms found in significantly-higher proportions in boys versus girls are shaded in gray, and in general, only lists microorganisms with statistically significant differences according to sex. In Table 1, Streptococci are the most abundant microorganisms observed in all groups. Prevotella melaninogenica is the second most abundant microorganism in caries-active children while it is the fourth most abundant in caries-free children. In caries-active boys, Neisseria flavescens and Haemophilus pittmaniae were the only two microorganisms (shaded in gray in Table 1) that exhibited both significant sex differences (> 1.5-fold changes with FDR adjusted p < 0.05) and abundance comprising > 0.5% of the microbial population (Tables 1 and 2). N. flavescens and H. pittmaniae were abundant in caries-active boys, comprising 3.92% and 1.53%, respectively, of the salivary microbial population. In caries-active girls, Alloprevotella species HOT 473 was the only species that exhibited significant sex differences (4.4-fold difference between caries-active girls and caries-active boys; p = 0.0003) and high abundance in numbers (1.85% of the total microbial population in caries-active girls).
Table 1.
Abundance of salivary microorganisms found in caries-active and caries-free children.
| Probe ID | Microorganism | Caries-Active Girls | Caries-Active Boys | Caries-Free Girls | Caries-Free Boys | Caries-Active Children | Caries-Free Children |
|---|---|---|---|---|---|---|---|
| GP-081 | Streptococcus_Genus_probe_4 | 25.49% | 21.76% | 30.93% | 21.28% | 23.78% | 25.70% |
| PR-14 | Prevotella_melaninogenica | 9.26% | 8.06% | 6.50% | 4.84% | 8.71% | 5.60% |
| HA-05 | Haemophilus_parainfluenzae | 6.21% | 8.13% | 7.69% | 12.72% | 7.09% | 10.42% |
| RO-03 | Rothia_mucilaginosa | 5.89% | 8.08% | 5.36% | 11.55% | 6.90% | 8.71% |
| GE-02 | Gemella_haemolysans | 4.56% | 5.07% | 5.02% | 4.16% | 4.79% | 4.56% |
| GP-060 | Neisseria_Genus_probe_2 | 4.60% | 4.87% | 3.43% | 6.68% | 4.72% | 5.19% |
| GR-02 | Granulicatella_elegans | 4.47% | 4.12% | 3.16% | 3.29% | 4.31% | 3.23% |
| FU-10 | Fusobacterium_periodonticum | 2.09% | 2.91% | 2.11% | 4.10% | 2.47% | 3.18% |
| NE-03 | Neisseria_flavescens | 1.10% | 3.92% | 2.15% | 2.26% | 2.39% | 2.21% |
| PO-09 | Porphyromonas_pasteri | 2.31% | 2.05% | 1.14% | 2.18% | 2.19% | 1.70% |
| GP-040 | Haemophilus_Genus_probe_3 | 1.81% | 2.28% | 0.82% | 1.31% | 2.02% | 1.09% |
| BE-02 | Bergeyella_sp_HOT_322 | 2.20% | 1.18% | 4.05% | 0.84% | 1.74% | 2.31% |
| AL-04 | Alloprevotella_sp_HOT_473 | 1.85% | 0.43% | 0.40% | 1.05% | 1.20% | 0.75% |
| PR-79 | Prevotella_nanceiensis | 1.19% | 1.10% | 1.30% | 1.20% | 1.15% | 1.25% |
| GR-01 | Granulicatella_adiacens | 1.17% | 1.00% | 1.48% | 1.49% | 1.09% | 1.49% |
| HA-07 | Haemophilus_pittmaniae | 0.57% | 1.53% | 1.61% | 1.43% | 1.01% | 1.51% |
| ST-20 | Streptococcus_sanguinis | 0.93% | 1.07% | 0.99% | 0.88% | 1.00% | 0.94% |
| GP-038 | Fusobacterium_Genus_probe_4 | 0.89% | 0.93% | 0.63% | 1.40% | 0.91% | 1.05% |
| GP-089 | Veillonella_Genus_probe_2 | 1.08% | 0.68% | 0.67% | 0.83% | 0.90% | 0.76% |
| PR-09 | Prevotella_histicola | 1.19% | 0.35% | 0.39% | 0.06% | 0.80% | 0.21% |
| PO-24 | Porphyromonas_pasteri | 0.65% | 0.78% | 0.77% | 0.89% | 0.71% | 0.84% |
| PR-25 | Prevotella_salivae | 0.86% | 0.51% | 0.76% | 0.06% | 0.70% | 0.38% |
| GP-039 | Gemella_Genus_probe | 0.65% | 0.67% | 0.78% | 0.57% | 0.66% | 0.66% |
| GP-126 | Streptococcus_Genus_probe_1 | 0.51% | 0.62% | 0.45% | 0.28% | 0.56% | 0.36% |
| LE-16 | Leptotrichia_sp_HOT_417 | 0.65% | 0.45% | 0.26% | 0.08% | 0.56% | 0.16% |
| VE-08 | Veillonella_sp_HOT_780 | 0.67% | 0.32% | 0.32% | 0.41% | 0.51% | 0.37% |
| LE-06 | Leptotrichia_hongkongensis | 0.43% | 0.52% | 0.40% | 0.13% | 0.47% | 0.25% |
| PR-22 | Prevotella_pallens | 0.42% | 0.50% | 0.25% | 0.05% | 0.46% | 0.14% |
| LE-07 | Leptotrichia_shahii | 0.28% | 0.64% | 0.39% | 0.17% | 0.44% | 0.27% |
| GP-019 | Campylobacter_Genus_probe_2 | 0.46% | 0.42% | 0.19% | 0.20% | 0.44% | 0.19% |
| NE-19 | Neisseria_subflava | 0.20% | 0.62% | 0.53% | 0.32% | 0.39% | 0.41% |
| GE-04 | Gemella_sanguinis | 0.33% | 0.44% | 0.67% | 0.66% | 0.38% | 0.67% |
| VE-21 | Veillonella_atypica | 0.48% | 0.26% | 0.60% | 0.07% | 0.38% | 0.31% |
| GP-073 | Rothia_Genus_probe | 0.32% | 0.43% | 0.27% | 0.38% | 0.37% | 0.33% |
| RO-01 | Rothia_aeria | 0.20% | 0.55% | 0.22% | 0.20% | 0.36% | 0.21% |
| GP-112 | Haemophilus_Genus_probe_2 | 0.39% | 0.32% | 0.18% | 0.31% | 0.36% | 0.25% |
| VE-03 | Veillonella_dispar | 0.49% | 0.20% | 0.22% | 0.11% | 0.36% | 0.16% |
| AG-06 | Aggregatibacter_sp_HOT_513 | 0.33% | 0.36% | 0.03% | 0.09% | 0.35% | 0.06% |
| GP-099 | Aggregatibacter_Genus_probe_1 | 0.27% | 0.44% | 0.18% | 0.26% | 0.34% | 0.22% |
| AB-01 | Abiotrophia_defectiva | 0.44% | 0.22% | 0.19% | 0.46% | 0.34% | 0.34% |
| NE-16 | Neisseria_flavescens | 0.03% | 0.64% | 0.00% | 0.02% | 0.31% | 0.01% |
| VE-20 | Veillonella_atypica | 0.40% | 0.21% | 0.26% | 0.04% | 0.31% | 0.14% |
| LA-29 | Lautropia_mirabilis | 0.26% | 0.35% | 0.84% | 0.77% | 0.30% | 0.80% |
| CL-03 | Ruminococcaceae[G-1]_sp_HOT_075 | 0.33% | 0.25% | 0.08% | 0.07% | 0.29% | 0.07% |
| RO-02 | Rothia_dentocariosa | 0.35% | 0.20% | 0.31% | 0.32% | 0.28% | 0.32% |
| NE-08 | Neisseria_pharyngis | 0.13% | 0.41% | 0.04% | 0.07% | 0.26% | 0.06% |
| OR-01 | Oribacterium_sinus | 0.21% | 0.28% | 0.21% | 0.59% | 0.24% | 0.42% |
| LE-13 | Leptotrichia_sp_HOT_221 | 0.32% | 0.12% | 0.05% | 0.05% | 0.23% | 0.05% |
| GP-049 | Leptotrichia_Genus_probe_3 | 0.27% | 0.16% | 0.06% | 0.02% | 0.22% | 0.04% |
| GP-050 | Leptotrichia_Genus_probe_4 | 0.18% | 0.27% | 0.12% | 0.29% | 0.22% | 0.21% |
| GE-07 | Gemella_morbillorum | 0.20% | 0.23% | 0.04% | 0.21% | 0.21% | 0.13% |
| AL-11 | Alloprevotella_sp_HOT_914 | 0.22% | 0.20% | 0.07% | 0.19% | 0.21% | 0.14% |
| VE-07 | Veillonella_rogosae | 0.11% | 0.33% | 0.40% | 0.67% | 0.21% | 0.54% |
| LE-11 | Leptotrichia_sp_HOT_218 | 0.31% | 0.09% | 0.09% | 0.00% | 0.21% | 0.04% |
| FU-12 | Fusobacterium_nucleatum_subsp_animalis | 0.11% | 0.28% | 0.02% | 0.01% | 0.18% | 0.02% |
| PR-18 | Prevotella_nigrescens | 0.16% | 0.21% | 0.19% | 0.03% | 0.18% | 0.10% |
| PR-51 | Prevotella_veroralis | 0.28% | 0.06% | 0.07% | 0.01% | 0.18% | 0.04% |
| TM-05 | TM7[G-1]_sp_HOT_352 | 0.13% | 0.19% | 0.17% | 0.12% | 0.16% | 0.15% |
| LE-26 | Leptotrichia_sp_HOT_215 | 0.15% | 0.16% | 0.13% | 0.08% | 0.15% | 0.10% |
| GP-005 | Alloprevotella_Genus_probe | 0.20% | 0.09% | 0.08% | 0.15% | 0.15% | 0.12% |
| GP-100 | Aggregatibacter_Genus_probe_2 | 0.09% | 0.21% | 0.03% | 0.08% | 0.15% | 0.06% |
| NE-18 | Neisseria_oralis | 0.11% | 0.18% | 0.17% | 0.06% | 0.14% | 0.11% |
| GP-004 | Actinomyces_Genus_probe_4 | 0.16% | 0.12% | 0.18% | 0.13% | 0.14% | 0.15% |
| PR-26 | Prevotella_scopos | 0.09% | 0.19% | 0.06% | 0.07% | 0.13% | 0.07% |
| GE-05 | Gemella_morbillorum | 0.10% | 0.15% | 0.03% | 0.11% | 0.12% | 0.07% |
| TM-01 | TM7[G-1]_sp_HOT_346 | 0.20% | 0.02% | 0.00% | 0.07% | 0.12% | 0.04% |
| CO-03 | Corynebacterium_matruchotii | 0.13% | 0.09% | 0.54% | 0.20% | 0.11% | 0.36% |
| EU-03 | Peptostreptococcaceae[XI][G-1][Eubacterium]_sulci | 0.09% | 0.14% | 0.07% | 0.04% | 0.11% | 0.05% |
| ME-01 | Megasphaera_micronuciformis | 0.17% | 0.03% | 0.13% | 0.01% | 0.11% | 0.06% |
| PR-70 | Prevotella_pallens | 0.14% | 0.07% | 0.04% | 0.03% | 0.11% | 0.03% |
| LE-22 | Leptotrichia_wadei | 0.13% | 0.08% | 0.12% | 0.00% | 0.11% | 0.06% |
| VE-06 | Veillonella_parvula | 0.18% | 0.02% | 0.02% | 0.01% | 0.11% | 0.02% |
| LA-28 | Lactococcus_lactis | 0.19% | 0.00% | 0.00% | 0.00% | 0.10% | 0.00% |
| LA-06 | Lachnoanaerobaculum_umeaense | 0.11% | 0.08% | 0.11% | 0.16% | 0.10% | 0.13% |
| PR-20 | Prevotella_oris | 0.11% | 0.08% | 0.17% | 0.02% | 0.10% | 0.09% |
| KI-03 | Kingella_oralis | 0.13% | 0.06% | 0.05% | 0.02% | 0.10% | 0.03% |
| GP-067 | Porphyromonas_Genus_probe_2 | 0.08% | 0.12% | 0.02% | 0.12% | 0.10% | 0.07% |
| GP-118 | Moraxella_Genus_probe_1 | 0.00% | 0.22% | 0.00% | 0.00% | 0.10% | 0.00% |
| PR-21 | Prevotella_oulorum | 0.12% | 0.07% | 0.06% | 0.01% | 0.10% | 0.03% |
| GP-069 | Prevotella_Genus_probe_2 | 0.10% | 0.09% | 0.05% | 0.07% | 0.10% | 0.06% |
| SO-01 | Solobacterium_moorei | 0.08% | 0.11% | 0.11% | 0.07% | 0.10% | 0.09% |
| CL-04 | Ruminococcaceae[G-2]_sp_HOT_085 | 0.11% | 0.07% | 0.04% | 0.02% | 0.09% | 0.03% |
| GE-03 | Gemella_morbillorum | 0.08% | 0.10% | 0.02% | 0.09% | 0.09% | 0.06% |
| GP-096 | Fusobacterium_Genus_probe_3 | 0.11% | 0.07% | 0.04% | 0.12% | 0.09% | 0.09% |
| PE-21 | Peptostreptococcus_stomatis | 0.08% | 0.10% | 0.06% | 0.08% | 0.09% | 0.07% |
| PO-26 | Porphyromonas_sp_HOT_930 | 0.09% | 0.08% | 0.00% | 0.00% | 0.09% | 0.00% |
| AC-20 | Actinomyces_sp_HOT_172 | 0.10% | 0.07% | 0.16% | 0.07% | 0.09% | 0.11% |
| OR-06 | Oribacterium_asaccharolyticum | 0.09% | 0.08% | 0.06% | 0.06% | 0.08% | 0.06% |
| GP-130 | Veillonella_Genus_probe_1 | 0.11% | 0.05% | 0.14% | 0.03% | 0.08% | 0.08% |
| PR-06 | Prevotella_denticola | 0.07% | 0.09% | 0.01% | 0.00% | 0.08% | 0.00% |
| CA-37 | Catonella_morbi | 0.06% | 0.09% | 0.05% | 0.13% | 0.08% | 0.09% |
| AL-09 | Alloprevotella_tannerae | 0.07% | 0.08% | 0.01% | 0.01% | 0.07% | 0.01% |
| PR-39 | Prevotella_sp_HOT_309 | 0.09% | 0.06% | 0.04% | 0.01% | 0.07% | 0.03% |
| GP-113 | Kingella_Genus_probe_1 | 0.07% | 0.08% | 0.05% | 0.08% | 0.07% | 0.06% |
| PO-03 | Porphyromonas_endodontalis | 0.06% | 0.08% | 0.00% | 0.00% | 0.07% | 0.00% |
| GP-076 | Selenomonas_&_Centipeda_Genus_probe | 0.12% | 0.02% | 0.21% | 0.34% | 0.07% | 0.28% |
| NE-02 | Neisseria_elongata | 0.07% | 0.07% | 0.05% | 0.13% | 0.07% | 0.09% |
Figure 1.
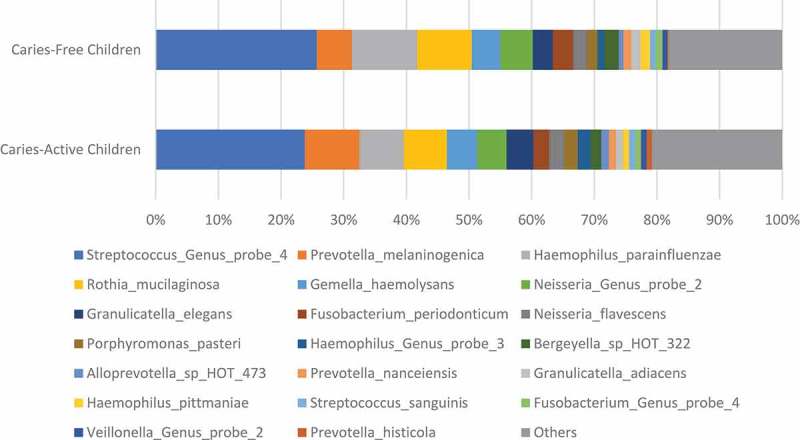
Abundance of top 20 salivary microorganisms found in caries-active and caries-free children.
Plots were calculated based on select information displayed in Table 1.
Figure 2.
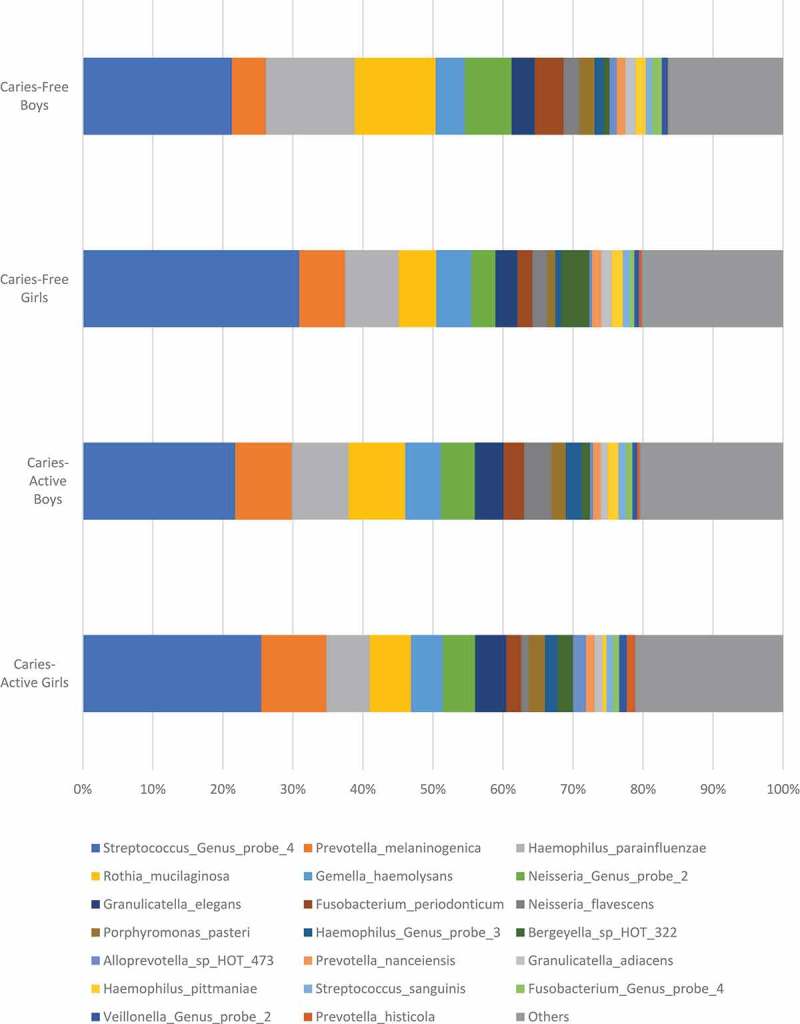
Abundance of top 20 salivary microorganisms found in caries-active and caries-free children differentiated by sex.
Plots were calculated based on select information displayed in Table 1.
Table 2.
Significant differences in salivary microorganisms found in caries-active boys versus caries-active girls.
| Probe ID | Microorganism | Average Counts per Million | Fold Changes | p-value | FDR p-value |
|---|---|---|---|---|---|
| GP-057 | Moraxella_Genus_probe_2 | 339.5 | 171.0 | 0.0000 | 0.0004 |
| GP-127 | Streptococcus_Genus_probe_2 | 434.2 | 26.4 | 0.0000 | 0.0004 |
| NE-16 | Neisseria_flavescens | 2056.0 | 20.7 | 0.0011 | 0.0287 |
| RO-01 | Rothia_aeria | 3918.0 | 3.9 | 0.0001 | 0.0114 |
| HA-07 | Haemophilus_pittmaniae | 15,037.3 | 3.8 | 0.0004 | 0.0177 |
| AL-04 | Alloprevotella_sp_HOT_473 | 10,366.4 | −4.4 | 0.0003 | 0.0169 |
| VE-06 | Veillonella_parvula | 676.9 | −5.4 | 0.0005 | 0.0194 |
| PR-41 | Prevotella_sp_HOT_315 | 54.4 | −7.3 | 0.0007 | 0.0271 |
| LA-03 | Lachnoanaerobaculum_sp_HOT_083 | 75.9 | −8.9 | 0.0012 | 0.0295 |
| AC-03 | Actinobaculum_sp_HOT_183 | 266.6 | −9.8 | 0.0010 | 0.0287 |
| SE-37 | Selenomonas_sp_HOT_126 | 209.9 | −21.5 | 0.0009 | 0.0283 |
| LA-28 | Lactococcus_lactis | 498.9 | −92.2 | 0.0002 | 0.0169 |
Microorganisms exhibiting positive value fold changes (shaded in gray) indicate higher prevalence in caries-active boys compared to caries-active girls.Microorganisms exhibiting negative value fold changes (unshaded) indicate higher prevalence in caries-active girls compared to caries-active boys.
Table 3.
Significant differences in salivary microorganisms found in caries-free boys versus caries-free girls.
| Probe ID | Microorganism | Average Counts per Million | Fold Changes | p-value | FDR p-value |
|---|---|---|---|---|---|
| LA-31 | Lachnospiraceae[G-3]_sp_HOT_100 | 527.2 | 39.5 | 0.0023 | 0.0341 |
| BE-03 | Bergeyella_sp_HOT_900 | 313.2 | 21.8 | 0.0032 | 0.0427 |
| VE-21 | Veillonella_atypica | 3025.7 | −6.2 | 0.0020 | 0.0314 |
| BE-02 | Bergeyella_sp_HOT_322 | 24,673.6 | −6.2 | 0.0029 | 0.0412 |
| PR-25 | Prevotella_salivae | 4980.9 | −6.8 | 0.0033 | 0.0427 |
| GP-095 | Fusobacterium_Genus_probe_2 | 412.7 | −7.1 | 0.0008 | 0.0173 |
| ME-01 | Megasphaera_micronuciformis | 842.4 | −11.5 | 0.0006 | 0.0144 |
| PR-20 | Prevotella_oris | 952.2 | −11.7 | 0.0003 | 0.0118 |
| AC-36 | Actinomyces_naeslundii | 347.0 | −16.9 | 0.0003 | 0.0118 |
| SE-18 | Selenomonas_sputigena | 126.9 | −26.8 | 0.0015 | 0.0263 |
| GP-022 | Cardiobacterium_Genus_probe | 1195.8 | −28.6 | 0.0001 | 0.0085 |
| PR-23 | Prevotella_pleuritidis | 338.1 | −28.7 | 0.0014 | 0.0261 |
| LE-22 | Leptotrichia_wadei | 865.2 | −37.4 | 0.0006 | 0.0144 |
| ST-09 | Streptococcus_anginosus | 185.0 | −49.3 | 0.0002 | 0.0111 |
| AC-19 | Actinomyces_sp_HOT_171 | 166.9 | −69.4 | 0.0001 | 0.0085 |
| LE-19 | Leptotrichia_sp_HOT_498 | 398.4 | −78.7 | 0.0001 | 0.0085 |
| OR-08 | Oribacterium_sp_HOT_078 | 386.3 | −87.3 | 0.0008 | 0.0180 |
| LE-11 | Leptotrichia_sp_HOT_218 | 1106.9 | −102.0 | 0.0019 | 0.0311 |
| AG-10 | Aggregatibacter_sp_HOT_949 | 117.6 | −137.3 | 0.0013 | 0.0250 |
| AC-18 | Actinomyces_sp_HOT_170 | 636.4 | −159.1 | 0.0002 | 0.0111 |
| PR-19 | Prevotella_oralis | 172.0 | −206.2 | 0.0004 | 0.0129 |
| PR-15 | Prevotella_micans | 687.0 | −298.6 | 0.0006 | 0.0144 |
| SE-05 | Selenomonas_noxia | 2956.5 | −357.0 | 0.0000 | 0.0060 |
Microorganisms exhibiting positive value fold changes (shaded in gray) indicate higher prevalence in caries-active boys compared to caries-active girls. Microorganisms exhibiting negative value fold changes (unshaded) indicate higher prevalence in caries-active girls compared to caries-active boys.
Significant sex-specific differences in salivary microbiota in caries-active and caries-free children
Moraxella genus probe 2, Streptococcus genus probe 2, N. flavescens, Rothia aeria, and H. pittmaniae were found at significantly higher levels in caries-active boys than caries-active girls, exhibiting 171-, 26.4-, 20.7-, 3.9- and 3.8-fold differences, respectively (Table 2). Alternately, Lactococcus lactis, Selenomonas species HOT 126, Actinobaculum species HOT 183, Lachnoanaerobaculum species HOT 083, Prevotella species HOT 315, Veillonella parvula, and Alloprevotella species HOT 473 were found at significantly higher levels in caries-active girls than caries-active boys, exhibiting 92.2-, 21.5-, 9.8-, 8.9-, 7.3-, 5.4- and 4.4-fold differences, respectively (Table 2).
Conversely, for caries-free children, Lachnospiraceae [G-3] species HOT 100 and Bergeyella species HOT 900 were found in significantly higher levels in caries-free boys than caries-free girls, exhibiting 39.5 and 21.8-fold differences, respectively (Table 3). In addition, Selenomonas noxia, Prevotella micans, Prevotella oralis, Actinomyces species HOT 170, Aggregatibacter species HOT 949, Leptotrichia species HOT 218, Oribacterium species HOT 078, Leptotrichia species HOT 498, Actinomyces species HOT 171, Streptococcus anginosos, Leptotrichia wadei, Prevotella pleuritidis, Cardiobacterium genus probe, Selenomonas sputigena, and Actinomyces naeslundii were found at significantly higher levels in caries-free girls than caries-free boys, exhibiting 357-, 299-, 206-, 159-, 137-, 102-, 87.3-, 78.7-, 69.4-, 49.4-, 37.4-, 28.7-, 28.6-, 26.8-, and 16.9-fold differences, respectively (Table 3).
Microorganisms were also compared between caries-active children as a combined cohort compared to microorganisms contained within caries-free children (Table 4). Fold changes were also determined for each microorganism contained within caries-active children versus caries-free children and then ranked from high positive values (indicating a high prevalence of specific microorganisms within caries-active children) and extremely low negative values (indicating a high prevalence of specific microorganisms within caries-free children). Microorganisms displaying statistically significant fold changes (> 1.5 fold changes with FDR adjusted p < 0.05) are displayed in Table 4; gray shading indicates a high prevalence of microorganisms found in caries-active children compared to caries-free children. This table lists only those microorganisms displaying significant fold changes according to caries status, and omits the large majority of species (hundreds) that exhibit nominal or minor fold changes between caries-active children and caries-free children.
Table 4.
Ranking of salivary microorganisms found in caries-active children versus caries-free children.
| Probe ID | Microorganism | Average Counts per Million | Fold Changes | p-value | FDR p-value |
|---|---|---|---|---|---|
| BA-17 | Bacteroidetes[G-5]_sp_HOT_511 | 115.3 | 531.4 | 0.0004 | 0.0031 |
| PR-23 | Prevotella_pleuritidis | 338.1 | 504.9 | 0.0001 | 0.0010 |
| TR-02 | Treponema_denticola | 93.9 | 370.2 | 0.0001 | 0.0009 |
| ST-21 | Streptococcus_sobrinus | 151.1 | 331.3 | 0.0036 | 0.0171 |
| LE-19 | Leptotrichia_sp_HOT_498 | 398.4 | 241.0 | 0.0001 | 0.0010 |
| LA-28 | Lactococcus_lactis | 498.9 | 209.7 | 0.0144 | 0.0455 |
| GP-057 | Moraxella_Genus_probe_2 | 339.5 | 189.7 | 0.0055 | 0.0223 |
| LE-11 | Leptotrichia_sp_HOT_218 | 1106.9 | 187.4 | 0.0038 | 0.0178 |
| NE-16 | Neisseria_flavescens | 2056.0 | 171.4 | 0.0082 | 0.0299 |
| PO-26 | Porphyromonas_sp_HOT_930 | 791.7 | 132.8 | 0.0023 | 0.0120 |
| AG-01 | Aggregatibacter_actinomycetemcomitans | 85.8 | 127.9 | 0.0031 | 0.0158 |
| GP-012 | Bacteroidetes[G-5]_Genus_probe | 66.3 | 96.5 | 0.0004 | 0.0035 |
| PR-06 | Prevotella_denticola | 520.6 | 92.0 | 0.0040 | 0.0184 |
| GP-127 | Streptococcus_Genus_probe_2 | 434.2 | 70.3 | 0.0020 | 0.0110 |
| EU-02 | Peptostreptococcaceae[XI][G-1][Eubacterium]_infirmum | 64.7 | 65.5 | 0.0007 | 0.0051 |
| TR-04 | Treponema_lecithinolyticum | 72.3 | 63.4 | 0.0055 | 0.0223 |
| CA-11 | Capnocytophaga_haemolytica | 73.3 | 56.7 | 0.0050 | 0.0214 |
| TR-33 | Treponema_sp_HOT_257 | 64.5 | 55.9 | 0.0045 | 0.0199 |
| GP-086 | Treponema_Genus_probe_4 | 77.0 | 54.2 | 0.0058 | 0.0228 |
| MY-10 | Mycoplasma_salivarium | 81.8 | 50.6 | 0.0168 | 0.0497 |
| LA-08 | Lachnospiraceae[G-2]_sp_HOT_096 | 276.9 | 39.3 | 0.0088 | 0.0313 |
| TR-58 | Treponema_sp_HOT_237 | 62.1 | 35.1 | 0.0034 | 0.0168 |
| LA-11 | Butyrivibrio_sp_HOT_455 | 369.3 | 32.7 | 0.0055 | 0.0223 |
| CA-28 | Capnocytophaga_sp_HOT_902 | 73.8 | 32.4 | 0.0048 | 0.0207 |
| PE-01 | Peptococcus_sp_HOT_167 | 61.1 | 29.7 | 0.0070 | 0.0262 |
| AL-09 | Alloprevotella_tannerae | 482.6 | 27.4 | 0.0093 | 0.0317 |
| FU-12 | Fusobacterium_nucleatum_subsp_animalis | 1129.1 | 25.2 | 0.0006 | 0.0045 |
| TR-05 | Treponema_maltophilum | 54.0 | 14.5 | 0.0156 | 0.0476 |
| PR-51 | Prevotella_veroralis | 1406.4 | 13.8 | 0.0140 | 0.0449 |
| AC-07 | Actinomyces_gerencseriae | 67.6 | 13.1 | 0.0148 | 0.0464 |
| GP-049 | Leptotrichia_Genus_probe_3 | 1238.6 | 8.8 | 0.0059 | 0.0229 |
| PO-09 | Porphyromonas_pasteri | 26,451.6 | −4.1 | 0.0114 | 0.0377 |
| GR-02 | Granulicatella_elegans | 49,220.8 | −4.6 | 0.0091 | 0.0316 |
| RO-03 | Rothia_mucilaginosa | 99,788.6 | −4.8 | 0.0089 | 0.0313 |
| GP-038 | Fusobacterium_Genus_probe_4 | 9703.2 | −5.1 | 0.0008 | 0.0056 |
| GP-112 | Haemophilus_Genus_probe_2 | 4763.9 | −5.3 | 0.0168 | 0.0497 |
| HA-05 | Haemophilus_parainfluenzae | 101,160.9 | −5.4 | 0.0011 | 0.0070 |
| FU-10 | Fusobacterium_periodonticum | 29,184.7 | −5.6 | 0.0035 | 0.0171 |
| CA-04 | Campylobacter_gracilis | 635.3 | −6.3 | 0.0161 | 0.0484 |
| GP-050 | Leptotrichia_Genus_probe_4 | 2282.6 | −6.3 | 0.0030 | 0.0158 |
| CA-31 | Cardiobacterium_hominis | 590.2 | −6.6 | 0.0039 | 0.0180 |
| GP-039 | Gemella_Genus_probe | 8474.1 | −6.7 | 0.0016 | 0.0091 |
| GP-025 | Corynebacterium_Genus_probe | 360.7 | −7.2 | 0.0041 | 0.0185 |
| PO-24 | Porphyromonas_pasteri | 8037.8 | −7.4 | 0.0008 | 0.0053 |
| ST-29 | Stenotrophomonas_maltophilia | 51.4 | −7.5 | 0.0069 | 0.0259 |
| LA-06 | Lachnoanaerobaculum_umeaense | 1228.0 | −7.5 | 0.0004 | 0.0033 |
| RO-02 | Rothia_dentocariosa | 3995.8 | −7.8 | 0.0018 | 0.0101 |
| GP-046 | Lactobacillus_Genus_probe_3 | 74.2 | −7.8 | 0.0047 | 0.0206 |
| GR-01 | Granulicatella_adiacens | 14,304.5 | −7.9 | 0.0000 | 0.0006 |
| HA-07 | Haemophilus_pittmaniae | 15,037.3 | −8.0 | 0.0014 | 0.0082 |
| CA-30 | Capnocytophaga_sputigena | 766.6 | −8.1 | 0.0005 | 0.0043 |
| GP-023 | Catonella_Genus_probe | 354.0 | −8.3 | 0.0085 | 0.0305 |
| OR-01 | Oribacterium_sinus | 3558.9 | −8.7 | 0.0003 | 0.0031 |
| LE-15 | Leptotrichia_sp_HOT_392 | 620.5 | −8.7 | 0.0067 | 0.0255 |
| CA-12 | Capnocytophaga_leadbetteri | 864.1 | −11.3 | 0.0003 | 0.0031 |
| GE-04 | Gemella_sanguinis | 5988.5 | −11.7 | 0.0001 | 0.0009 |
| GE-02 | Gemella_haemolysans | 75,652.7 | −11.9 | 0.0000 | 0.0005 |
| ST-09 | Streptococcus_anginosus | 185.0 | −13.4 | 0.0152 | 0.0473 |
| PO-27 | Porphyromonas_sp_HOT_284 | 472.5 | −13.7 | 0.0010 | 0.0063 |
| GP-003 | Actinomyces_Genus_probe_3 | 990.3 | −13.8 | 0.0001 | 0.0015 |
| GP-081 | Streptococcus_Genus_probe_4 | 374,334.7 | −14.1 | 0.0000 | 0.0000 |
| GP-083 | TM7_Genus_probe | 789.1 | −14.1 | 0.0001 | 0.0014 |
| CA-09 | Capnocytophaga_gingivalis | 447.6 | −15.0 | 0.0001 | 0.0015 |
| PR-73 | Prevotella_sp_HOT_317 | 211.0 | −15.2 | 0.0127 | 0.0410 |
| VE-07 | Veillonella_rogosae | 3703.2 | −16.1 | 0.0001 | 0.0016 |
| CA-32 | Cardiobacterium_valvarum | 116.1 | −16.3 | 0.0010 | 0.0063 |
| GP-020 | Capnocytophaga_Genus_probe_2 | 406.4 | −16.9 | 0.0000 | 0.0002 |
| AC-24 | Actinomyces_lingnae | 868.9 | −17.0 | 0.0001 | 0.0013 |
| CA-01 | Campylobacter_concisus | 216.9 | −17.3 | 0.0003 | 0.0031 |
| LE-25 | Leptotrichia_sp_HOT_223 | 197.4 | −17.4 | 0.0058 | 0.0228 |
| AC-30 | Actinomyces_sp_HOT_877 | 81.4 | −17.8 | 0.0120 | 0.0392 |
| SE-05 | Selenomonas_noxia | 2956.5 | −19.6 | 0.0095 | 0.0320 |
| PE-28 | Peptostreptococcaceae[XI][G-7]_sp_HOT_922 | 188.2 | −20.9 | 0.0096 | 0.0320 |
| TM-14 | TM7[G-1]_sp_HOT_952 | 227.5 | −21.1 | 0.0006 | 0.0046 |
| CO-02 | Corynebacterium_durum | 1140.3 | −23.9 | 0.0000 | 0.0000 |
| PR-42 | Prevotella_sp_HOT_317 | 288.0 | −25.3 | 0.0012 | 0.0074 |
| CA-17 | Capnocytophaga_sp_HOT_332 | 223.8 | −26.2 | 0.0016 | 0.0091 |
| AC-43 | Actinomyces_odontolyticus | 87.5 | −27.0 | 0.0016 | 0.0091 |
| SR-01 | SR1[G-1]_sp_HOT_345 | 321.9 | −28.6 | 0.0003 | 0.0031 |
| AC-22 | Actinomyces_sp_HOT_178 | 100.7 | −29.9 | 0.0076 | 0.0281 |
| PR-15 | Prevotella_micans | 687.0 | −33.9 | 0.0154 | 0.0473 |
| LA-03 | Lachnoanaerobaculum_sp_HOT_083 | 75.9 | −40.1 | 0.0004 | 0.0031 |
| GP-072 | Pseudomonas_Genus_probe | 113.8 | −45.0 | 0.0053 | 0.0221 |
| CA-03 | Campylobacter_curvus | 71.9 | −48.4 | 0.0032 | 0.0159 |
| LE-12 | Leptotrichia_sp_HOT_219 | 530.0 | −49.4 | 0.0001 | 0.0009 |
| LA-29 | Lautropia_mirabilis | 8113.6 | −55.9 | 0.0000 | 0.0000 |
| PR-71 | Prevotella_saccharolytica | 214.3 | −60.6 | 0.0003 | 70.0031 |
| CO-03 | Corynebacterium_matruchotii | 3137.4 | −61.6 | 0.0000 | 0.0000 |
| PR-72 | Prevotella_saccharolytica | 169.6 | −71.0 | 0.0011 | 0.0070 |
| TA-02 | Tannerella_sp_HOT_286 | 1137.6 | −91.4 | 0.0000 | 0.0000 |
| PO-16 | Porphyromonas_catoniae | 329.7 | −91.9 | 0.0005 | 0.0043 |
| BA-03 | Bacteroidales[G-2]_sp_HOT_274 | 1761.5 | −99.3 | 0.0000 | 0.0006 |
| AC-10 | Actinomyces_johnsonii | 67.7 | −100.5 | 0.0001 | 0.0010 |
| LA-31 | Lachnospiraceae[G-3]_sp_HOT_100 | 527.2 | −102.8 | 0.0000 | 0.0008 |
| GP-022 | Cardiobacterium_Genus_probe | 1195.8 | −110.2 | 0.0000 | 0.0000 |
| AG-03 | Aggregatibacter_paraphrophilus | 286.5 | −112.5 | 0.0006 | 0.0046 |
| GP-076 | Selenomonas_&_Centipeda_Genus_probe | 1541.7 | −155.9 | 0.0000 | 0.0000 |
| AC-11 | Actinomyces_massiliensis | 186.1 | −201.7 | 0.0000 | 0.0000 |
| LA-09 | Lachnospiraceae[G-3]_sp_HOT_100 | 242.2 | −215.3 | 0.0000 | 0.0000 |
| AC-18 | Actinomyces_sp_HOT_170 | 636.4 | −389.8 | 0.0000 | 0.0000 |
| TM-13 | TM7[G-1]_sp_HOT_348 | 2241.3 | −514.4 | 0.0000 | 0.0000 |
Microorganisms exhibiting positive value fold changes (shaded in gray) indicate higher prevalence in caries-active children compared to caries-free children.
Microorganisms exhibiting negative value fold changes (unshaded) indicate higher prevalence in caries-free children compared to caries-active children.
Discussion
Salivary microbiota and influence on oral and systemic health and disease
The salivary microbiota has direct and significant implications on oral and systemic health, and may influence the development of dental caries and other dental diseases, including endodontic infections and periodontal disease. Salivary microorganisms commonly found to be associated with caries include Atopobium, Streptococcus, and Veillonella [23–25]. Other salivary microorganisms associated with caries also include members from Actinobaculum, Aggregatibacter, Rothia, Granulicatella, Gemella, Actinomyces, Selenomonas, Haemophilus, Megaspora, Prevotella, Bacteroides, and Bifidobacterium [23–27]. Caries-associated bacteria within the Prevotella genus include Prevotella denticola and Prevotella histicola [28,29]. Conversely, a number of salivary microorganisms are associated with caries-free status, including Porphyromonas pasteri, Streptococcus Genus Probe 4, Bergeyella species HOT 322, and Corynebacterium durum [23,30,31]. Microbiota identified within caries-free individuals also include Actinomyces, Bergeyella, Campylobacter, Granulicatella, Kingella, Leptotrichia, Shuttleworthia satelles, Rothia, Treponema, Peptococcus, Porphyromonas catoniae, and N. flavescens [23,25,30,32]. Microorganisms associated with endodontic infections include P. denticola, Granulicatella adiacens, and Capnocytophaga sputigena [28,33–36].
Numerous members of the salivary microbiome are associated with periodontitis in varying levels of severity. SR1 [G-1] species HOT 345 and P. denticola and Prevotella species HOT 317 were found to be associated with periodontitis or serve as risk indicators for periodontitis [37–39]. TM7 Genus Probe was associated with generalized gingivitis [40]. Microorganisms implicated in refractory periodontitis include G. adiacens, Gemella sanguinis, and S. noxia [41–43]. Those microorganisms associated with generalized aggressive periodontitis include G. adiacens, S. noxia, Bacteroidales [G-2] species HOT 274, and Cardiobacterium Genus Probe [37,40,41,44,45]. S. noxia and Bacteroidales [G-2] species HOT 274 are also associated with chronic periodontitis [45,46]. Conversely, C. sputigena was associated with periodontal health [43,47].
Rationale for examining the salivary microbiome and influence of saliva on the oral microbiome and dental caries
The oral microbiome encompasses heterogeneous microorganisms from different sites of the mouth, including periodontal sulcus, dental plaque, tongue, buccal mucosa and saliva (48). Saliva contains little if any indigenous microbiota, instead acquiring bacteria shed from desquamating oral epithelial surfaces and oral biofilms, including dental plaque. Dental plaque contains Streptococcus mutans, one significant microbial contributor to dental caries and tooth decay, as well as other adherent microorganisms. Therefore, the salivary microbiome is considered to be an important reservoir that contains microorganisms found throughout the oral cavity [48,49], and upon interaction with the gut microbiome, may influence the onset of systemic diseases including inflammatory bowel disease. Thus, defining the factors underlying the constituency of the salivary microbiome are critical in understanding the complete oral microbiome and its influence in oral and systemic health and disease.
Sex differences in the salivary microbiome of caries-active and caries-free children
Our data demonstrate significant differences in the salivary microbiome between caries-active and caries-free children, consistent with the known shifts in acidity of the oral cavity in the development of dental caries. For caries-active boys, Streptococci may represent more important determinants in the onset of caries, while for caries-active girls, Veillonella may be more important factors. The high prevalence of Actinomyces, of which some species are known to have cariogenic potential, in caries-free girls (Table 3), indicates that this microorganism may not be a major influential determinant in dental caries. The detection of several periodontal microorganisms, including S. sputigena, S. noxia, and Aggregatibacter species HOT 949, in caries-free girls, is consistent with the more neutral pH found in the non-cariogenic oral cavity. In caries-active girls, Alloprevotella species HOT 473 was the only species that exhibited significant sex differences and moderate abundance within the total microbial population (Tables 1 and 2).
Other non-microbial factors influencing sex differences in dental caries
Shaffer et al. [1] have indicated that genetics may significantly influence variability in dental caries experience between individuals. In genome-wide scans, the genetic region signifying the potential for ‘higher caries experience’ was located at 14q24.3, which is in close proximity to ESRRB (estrogen-related receptor beta) [16]. ESRRB is similar to the estrogen receptor and contributes to various biological functions in physiology and pathology [50]. High expression of ESRRB in females may lead to higher caries rates due to increased enamel surface sensitivity towards demineralization in the acidic oral environment [51].
Genes expressed in enamel mineralization, AMELX (amelogenin) and AMNB, as well as ESRRB were found to be associated with calcium levels in saliva. Genetic mutations in enamel mineralization are directly responsible for X-linked Amelogenesis Imperfecta, which supports their contribution in enamel matrix formation and caries vulnerability [52]. Deeley et al. [53] found that ‘higher caries experience’ was associated with irregular amelogenin allele markers. This may occur in the presence of X-inactivation and mosaicism and may contribute to female caries susceptibility [53].
Depending on the type and frequency of sugar introduced in the diet, a cariogenic oral environment may also develop via fermentation of sugars by acidogenic microorganisms, resulting in enamel decalcification [12]. Vegetarian diets, which contain sugar-containing fruits and berries, have been shown to increase caries incidence [54,55]. Higher caries prevalence was found in participants with vegetarian diets, while protein-meat consumption was suggested to be caries-protective because of the development of a more alkaline oral environment due to protein metabolism [56]. Females were noted to engage more in vegetarianism than males [57], with western societies containing more vegetarian women than vegetarian men, and women consuming substantially less meat in comparison to men [54].
Throughout the female lifespan, various biological stages are influenced by reproductive hormone fluctuations that coincide with changes in body structure. Estrogen fluctuations that occur during pregnancy influence suppression of the immune system and have known impact on salivary composition and flow rate contributing to a less protective oral environment and greater caries susceptibility. The parotid, buccal and labial salivary glands demonstrated lower flow rates in females than males, with elderly participants demonstrating substantial differences [15,58]. The minor salivary gland in women consistently contained substantially lower IgA concentrations among all tested salivary glands, and the higher IgA concentration found in men may provide greater protective anti-cariogenic effects in the oral cavity when compared to women [15].
Oral health disparities in men and women can be influenced by cultural and social differences [59]. Overall, dental caries unequally affects ethnic minorities and low socioeconomic status populations, with women in these categories being more affected than men [60]. Female-inclined behaviors, such as labor and domestic role in food preparation, may contribute to increased caries rates known to be associated with women. This role provides easy access to food supplies and a greater tendency towards frequent snacking [61]. Some countries exhibit this trend in caries incidence due to social and religious involvement, including son preference, fasting, and pregnancy-related dietary restrictions [62].
Limitations of the study and concluding remarks
One limitation of the study concerns the difficulty in obtaining caries-free children, which was primarily due to the referral-based, region-wide patient acquisition system at the OHSU School of Dentistry. This referral system resulted in the predominant availability of more difficult and problematic dental cases, including advanced caries, and very few caries-free individuals. We supplemented the caries-free cohort with additional patients obtained from an external practitioner’s clinic, but we do acknowledge that increasing the caries-free control population may increase the statistical power of the study, and would have the potential to further discriminate borderline differences in microbial species between the sexes. Another limitation was the use of participants in different developmental dental stages – individuals with primary dentition or mixed dentition. We initially started the study attempting to identify only young individuals – ages 2–6 – with only primary dentition, but were not able to identify sufficient numbers of participants, and later expanded our age range to age 12 years. The majority of participants in this study were ages 2–6 years (30 boys and 34 girls) and the smaller remainder (11 boys and 13 girls) were within the 7–12 years age range, many with mixed dentition and eruption of permanent central and lateral incisors, first molars and cuspids. The limitations of different developmental stages were best mitigated with the use of saliva specimens instead of plaque specimens from specific teeth, many of which were missing or replaced with permanent teeth. The use of saliva also facilitated investigation of the salivary microbiome, perhaps the most reflective of the entire repertoire of microorganisms contained within the oral cavity. Any potential limitations were also mitigated with the use of our advanced multi-stage statistical analyses, which use the criterion of abundance and statistical differences in fold changes to identify the most relevant microbial species based on caries-status and by sex.
As the primary finding in our studies, in caries-active boys, N. flavescens and H. pittmaniae were the only two microorganisms (shaded in gray in Table 1) that exhibited both significant sex differences (> 1.5-fold changes with FDR adjusted p < 0.05) and abundance comprising > 0.5% of the microbial population (Tables 1 and 2). In caries-active girls, Alloprevotella species HOT 473 was the only species that exhibited both significant sex differences (4.4-fold difference between caries-active girls and caries-active boys; p = 0.0003) as well as high abundance in numbers (1.85% of the total microbial population). Alloprevotella species are associated with neutral pH in the oral cavity and has been found in cases of caries progression, while Neisseria species are considered to be acid producers and are more conducive to the presence within a cariogenic environment. Determining the causative reason why Alloprevotella species are found more predominantly in caries-active girls, or conversely why N. flavescens and H. pittmaniae are found more predominantly in caries-active boys, is beyond the scope of this study, and could be potentially dictated by genetic, nutritional, hormonal or immune factors or mechanisms, all of which could represent avenues for future investigation.
Funding Statement
This work was supported by the NIH - NIDCR [NIH DE024317].
Acknowledgments
SO, EH and CL are equal contributors to this work. EH is a recipient of the Dean’s Student Research Fellowship Award from the OHSU School of Dentistry, and was the OHSU-SCADA student representative at the 2019 IADR Meeting in Vancouver, BC, Canada (June 19-22, 2019). KR is also the current recipient of the AADR Student Research Day Award, supporting travel to the 2020 IADR Meeting in Washington, DC (March 18-21, 2020). SO, EH, KR, BK, and MS are students in the academic DMD degree program at the OHSU School of Dentistry. CL is the Clinical Study Coordinator for our project and AP is a graduating resident in the Pediatric Dentistry program. TM and CM receive salary support from the OHSU School of Dentistry and AF has an adjunct appointment at the School of Dentistry. This project and CM were also supported in part by NIH DE024317. We thank Dr. Kim Kutsch, who provided several caries-free study participants from his private dental practice in Albany, Oregon, and for establishing and supporting with Carifree (Albany, Oregon), the Bob Bowers Memorial Award for Student Research in Dental Caries. The Bob Bowers Memorial Award helped support SO for travel to present research at the 2019 IADR Meeting. We also thank Dr. Bruce J Paster and the HOMINGS Laboratory for providing commentary on next generation sequencing procedures, and Dr. Elizabeth Palmer for assistance in patient recruitment, and Drs. Tom Shearer, Bill Knight, Bob Steelman, John Engle, John Peterson, and Dena Fischer (NIDCR) for many discussions and support.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Shaffer JR, Wang X, McNeil DW, et al. Genetic susceptibility to dental caries differs between the sexes: A family-based study. Caries Res. 2015;49(2):133–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hakan Ç, Dülgergil Ç, Dalli M, et al. Early childhood caries update: A review of causes, diagnoses, and treatments. J Nat Sci Bio Med. 2013;4(1):29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Featherstone JDB. Dental caries: A dynamic disease process. Aust Dent J. 2008;53(3):286–291. [DOI] [PubMed] [Google Scholar]
- [4].Fakhruddin KS, Ngo HC, Samaranayake LP. Cariogenic microbiome and microbiota of the early primary dentition: A contemporary overview. Oral Dis. 2019;25(4):982–995. [DOI] [PubMed] [Google Scholar]
- [5].Ma C, Chen F, Zhang Y, et al. Comparison of oral microbial profiles between children with severe early childhood caries and caries-free children using the human oral microbe identification microarray. PLoS One. 2015;10(3):e0122075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jiang W, Zhang J, Chen H. 2013. Pyrosequencing analysis of oral microbiota in children with severe early childhood dental caries. Curr Microbiol 2013;67(5):537–542. [DOI] [PubMed] [Google Scholar]
- [7].Agnello M, Marques J, Cen L, et al. Microbiome associated with severe caries in Canadian first nations children. J Dent Res. 2017;96(12):1378–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ramezani GH, Norozi A, Valael N. The prevalence of nursing caries in 18 to 60 months old children in Qazvin. J Indian Soc Predod Prev Dent. 2003;21(1):19–26. [PubMed] [Google Scholar]
- [9].Madléna M, Hermann P, Jáhn M, et al. Caries prevalence and tooth loss in Hungarian adult population: Results of a national survey. BMC Public Health. 2008;8:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mangi SL. The effect of pregnancy on the incidence of dental caries in Indian women. J All India Dent Assoc. 1954;26:1–4. [PubMed] [Google Scholar]
- [11].Saravanan S, Anuradha KP, Bhaskar DJ. Prevalence of dental caries and treatment needs among school going children of Pondicherry, India. J Indian Soc Pedod Dent. 2003;21(1):1–12. [PubMed] [Google Scholar]
- [12].Ferraro M, Vieira AR. Explaining gender differences in caries: A multifactorial approach to a multifactorial disease. Int J Dent. 2010;2010:649643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lukacs JR. Sex differences in dental caries experience: Clinical evidence, complex etiology. Clin Oral Investig. 2011;15(5):649–656. [DOI] [PubMed] [Google Scholar]
- [14].Galvão-Moreira LV, de Andrade CM, de Oliveira JFF, et al. Sex differences in salivary parameters of caries susceptibility in healthy individuals. Oral Hlth Prev Dent. 2018;16(1):71–77. [DOI] [PubMed] [Google Scholar]
- [15].Eliasson L, Birkhed D, Osterberg T, et al Minor salivary gland secretion rates and immunoglobulin A in adults and the elderly. Eur J Oral Sci. 2006;114(6):494–499. [DOI] [PubMed] [Google Scholar]
- [16].Vieira AR, Marazita ML, Goldstein-McHenry T. Genome-wide scan finds suggestive caries loci. J Dent Res. 2008;87(5):435–439. [DOI] [PubMed] [Google Scholar]
- [17].Palmer RJ Jr, Cotton SL, Kokaras AS, et al. Analysis of oral bacterial communities: Comparison of HOMINGS with a tree-based approach implemented in QIIME. J Oral Microbiol. 2019;11(1):1586413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Belstrom D, Grande MA, Sembler-Moller ML, et al. Influence of periodontal treatment on subgingival and salivary microbiotas. J Periodontol. 2018;89(5):531–539. [DOI] [PubMed] [Google Scholar]
- [19].Mitwali H, Mourao MDA, Dennison J, et al. Effect of silver diamine fluoride treatment on microbial profiles of plaque biofilms from root/cervical caries lesions. Caries Res. 2019;53(5):555–566. [DOI] [PubMed] [Google Scholar]
- [20].Risso D, Perraudeau F, Gribkova S, et al. A general and flexible method for signal extraction from single-cell RNA-seq data. Nat Commun. 2018;9(1):284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57(1):289–300. [Google Scholar]
- [23].Lif Holgerson P, Öhman C, Rönnlund A, et al. Maturation of oral microbiota in children with or without dental caries. PLoS One. 2015;10(5):e0128534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jagathrakshakan SN, Sethumadhava RJ, Mehta DT, et al. 16S rRNA gene-based metagenomics analysis identifies a novel bacterial co-prevalence pattern in dental caries. Eur J Dent. 2015;9(1):127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Xu L, Chen X, Wang Y, et al. Dynamic alterations in salivary microbiota related to dental caries and age in preschool children with deciduous dentition: A 2-year follow-up study. Front Physiol. 2018;9:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yang F, Zeng X, Ning K, et al. Saliva microbiomes distinguish caries-active from healthy human populations. Isme J. 2012;6(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kaur R, Gilbert SC, Sheehy EC, et al. Salivary levels of Bifidobacteria in caries-free and caries-active children. Int J Paediatr Dent. 2013;23(1):32–38. [DOI] [PubMed] [Google Scholar]
- [28].Wade WG, Weightman AJ. Specificity of the oral microflora in dentinal caries, endodontic infections and periodontitis. Int Congr Ser. 2005;1284:150–157. [Google Scholar]
- [29].Asakawa M, Takeshita T, Furata M, et al. Tongue microbiota and oral health status in community-dwelling elderly adults. mSphere. 2018;3(4):e00332–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yasunaga H, Takeshita T, Shibata Y, et al. Exploration of bacterial species associated with the salivary microbiome of individuals with low susceptibility to dental caries. Clin Oral Invest. 2017;21(8):2399–2406. [DOI] [PubMed] [Google Scholar]
- [31].Richards VP, Alvarez AJ, Luce AR, et al. Microbiomes of site-specific dental plaques from children with different caries status. Infect Immun. 2017;85(8):e00106–00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Crielaard W, Zaura E, Schuller AA, et al. Exploring the oral microbiota of children at various development stages of their dentition in the relation to their oral health. BMC Med Genomics. 2011;4:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sakamoto M, Rôças IN, Siqueira JF, et al. Molecular analysis of bacteria in asymptomatic and symptomatic endodontic infections. Oral Microbiol Immun. 2006;21(2):112–122. [DOI] [PubMed] [Google Scholar]
- [34].Hsiao WL, Li KL, Liu Z, et al. Microbial transformation from normal oral microbiota to acute endodontic infections. BMC Genomics. 2012;13:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Siqueira JF, Rôças IN. Catonella morbi and Granulicatella adiacens: New species in endodontic infections. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;102(2):259–264. [DOI] [PubMed] [Google Scholar]
- [36].Didilescu AC, Rusu D, Anghel A, et al. Investigation of six selected bacterial species in endo-periodontal lesions. Int Endo J. 2012;45(3):282–293. [DOI] [PubMed] [Google Scholar]
- [37].Lourco TG, Heller D, Silva-Boghossian CM, et al. Microbial signature profiles of periodontally healthy and diseased patients. J Clin Periodontol. 2014;41(11):1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Pérez-Chaparro PJ, Gonçalves C, Figueiredo LC, et al. Newly identified pathogens associated with periodontitis: A systematic review. J Dent Res. 2014;93(9):846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Nadkarni MA, Chhour KL, Browne GV, et al. Age-dependent changes in Porphyromonas gingivalis and Prevotella species/phylotypes in healthy gingiva and inflamed/diseased sub-gingival sites. Clin Oral Invest. 2015;19(4):911–919. [DOI] [PubMed] [Google Scholar]
- [40].Huang S, Yang F, Zeng X, et al. Preliminary characterization of the oral microbiota of Chinese adults with and without gingivitis. BMC Oral Health. 2011;11(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kuboniwa M, Inaba H, Amano A. Genotyping to distinguish microbial pathogenicity in periodontitis. Periodontol 2000. 2010;54(1):136–159. [DOI] [PubMed] [Google Scholar]
- [42].Colombo AP, Bennet S, Cotton SL, et al. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: Comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J Periodontol. 2012;83(10):1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Colombo AP, Boches SK, Cotton SL, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80(9):1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gonçalves LF, Fermiano D, Feres M, et al. Levels of Selenomonas species in generalized aggressive periodontitis. J Periodontal Res. 2012;47(6):711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Oliverira RR, Fermiano D, Feres M, et al. Levels of candidate periodontal pathogens in subgingival biofilm. J Dent Res. 2016;95(6):711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Teles RP, Bogren A, Patel M, et al. A three-year prospective study of adult subjects with gingivitis II: Microbiological parameters. J Clin Periodontol. 2007;34(1):7–17. [DOI] [PubMed] [Google Scholar]
- [47].Martínez-Hernández M, Olivares-Navarrete R, Almaguer-Flores A. Influence of the periodontal status on the initial-biofilm formation on titanium surfaces. Clin Implant Dent Relat Res. 2016;18(1):174–181. [DOI] [PubMed] [Google Scholar]
- [48].Shaw L, Ribeiro ALR, Levine AP, et al. The human salivary microbiome is shaped by shared environment rather than genetics: Evidence from a large family of closely related individuals. mBio. 2017;8(5):e01237–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Costalonga M, Herzberg MC. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett. 2014;162(2 Pt A):22–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhou W, Liu Z, Wu J, et al. Identification and characterization of two novel splicing isoforms of human estrogen-related receptor beta. J Clin Endocr Metab. 2006;91(2):569–579. [DOI] [PubMed] [Google Scholar]
- [51].Weber ML, Hsin HY, Kalay E, et al. Role of estrogen related receptor beta (ESRRB) in DFN35B hearing impairment and dental decay. BMC Med Genet. 2014;15:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kuchler EC, Pecharki GD, Castro ML, et al. Genes involved in the enamel development are associated with calcium and phosphorus level in saliva. Caries Res. 2017;51(3):225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Deeley K, Letra A, Rose EK, et al. Possible association of amelogenin to high caries experience in a Guatemalan-Mayan population. Caries Res. 2008;42(1):8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ruby MB. Vegetarianism. A blossoming field of study. Appetite. 2012;58(1):141–150. [DOI] [PubMed] [Google Scholar]
- [55].Love HJ, Sulikowski D. Of meat and men: Sex differences in implicit and explicit attitudes toward meat. Front Psychol. 2018;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Khan AA, Jain SK, Shrivastav A. Prevalence of dental caries among the population of Gwalior (India) in relation of different associated factors. Eur J Dent. 2008;2(2):81–85. [PMC free article] [PubMed] [Google Scholar]
- [57].Shah N. Gender issues and oral health in elderly Indians. Int Dent J. 2003;53(6):475–484. [DOI] [PubMed] [Google Scholar]
- [58].Percival RS, Challacombe SJ, Marsh PD. Flow rates of resting whole and stimulated parotid saliva in relation to age and gender. J Dent Res. 1994;73(8):1416–1420. [DOI] [PubMed] [Google Scholar]
- [59].Martinez-Mier EA, Zandona AF. The impact of gender on caries prevalence and risk assessment. Dent Clin N Am. 2013;57(2):301–315. [DOI] [PubMed] [Google Scholar]
- [60].Petersen PE. Sociobehavioural risk factors in dental caries – international perspectives. Community Dent Oral Epidemiol. 2005;33(4):274–279. [DOI] [PubMed] [Google Scholar]
- [61].Lukacs JR, Largaespada LL. Explaining sex differences in dental caries prevalence: Saliva, hormones, and “life-history” etiologies. Am J Hum Biol. 2006;18(4):540–555. [DOI] [PubMed] [Google Scholar]
- [62].Lukacs JR. Gender differences in oral health in South Asia: Metadata imply multifactorial biological and cultural causes. Am J Hum Biol. 2011;23(3):398–411. [DOI] [PubMed] [Google Scholar]


