Abstract
Simple Summary
Src Homology 2 Domain Containing E (SHE) is a protein coding gene, and member of the SH2 domain-containing family. Sequencing revealed a 2000 bp copy number variation in the SHE gene. There is overlap between SHE copy number variation (CNV) and quantitative trait loci related to milk fat percentage and bone density. A total of 750 sheep, including Chaka sheep (CKS), Hu sheep (HS), Small Tail Han sheep (STHS) and Large Tail Han sheep (LTHS) were available to detect the CNV of the SHE gene and correlate these gene variations with economic traits. The results showed that there were more individuals with SHE copy number loss in CKS and HS than in STHS and LTHS. Association analysis showed that gain and normal copy number types performed better in body length (p < 0.05), circumference of cannon bone (p < 0.05), heart girth (p < 0.05), chest width (p < 0.05) and high at the cross (p < 0.05) in CKS, HS and STHS. Chi-square analyses found significant variation in the CNV of the SHE gene, so it varies greatly between varieties. These findings clarified the relationship between the CNV of the SHE gene and the economic traits in these four kinds of sheep, and provide a reference for sheep breeding.
Abstract
Copy number variation (CNV) caused by gene rearrangement is an important part of genomic structural variation. We found that the copy number variation region of the Src Homology 2 Domain Containing E (SHE) gene correlates with a quantitative trait locus of sheep related to milk fat percentage and bone density. The aim of our study was to detect the copy number variation of the SHE gene in four sheep breeds and to conduct a correlation analysis with economic traits, hoping to provide some reference for sheep breeding. In this study, we examined 750 sheep from four Chinese breeds: Chaka sheep (CKS), Hu sheep (HS), Large Tail Han sheep (LTHS) and Small Tail Han sheep (STHS). We used qPCR to evaluate the copy number of the SHE gene, and then used general linear models to analyze the associations between CNV and economic traits. The results showed that there were more individuals with SHE copy number loss in CKS and HS than in STHS and LTHS individuals. Association analyses showed that gain and normal copy number types were correlated to body length, circumference of cannon bone, heart girth, chest width and high at the cross in CKS, HS and STHS (p < 0.05), but this association was not observed for LTHS. Chi-square values (χ2) found prominent differences in CNV distribution among the studied breeds. Overall, the CNV of the SHE gene may be an important consideration for sheep molecular breeding.
Keywords: sheep, SHE gene, copy number variation (CNV), growth traits, association
1. Introduction
A quantitative trait locus (QTL) is a region of DNA that is associated with a particular phenotypic trait, which could be attributed to the effects of multiple genes and their environment. QTL mapping has been widely used to identify the relationships between genetic markers and quantitative traits of interest. The mapping of QTL must use genetic markers, and one seeks to locate QTLs by looking for a link between the genetic marker and the quantitative trait of interest. In other words, the genetic marker and QTL are in linkage. Copy number variation could be an abnormality caused by genome rearrangement. Abnormally sized fragments can range from 50 bp to 1 Mb, and mainly arise by genome deletion, insertion, recombination and complex mutations at multiple chromosomal sites [1]. When the function of different genes in animals is missing or altered, this can lead to changes in the animal’s phenotype and disease susceptibility. Mutations can be indels, single nucleotide polymorphisms (SNPs) or copy number variation (CNV). Indels are insertions or deletions of bases in the genome of an organism, usually less than 50 bp in length. SNPs mainly refer to DNA sequence polymorphism caused by single nucleotide variation at the genome level and are more abundant in genomic variation. CNV has a larger variation area than indels and includes a larger total number of nucleotides than the number of SNPs, so CNVs may have greater influence on genomic sequences [2].
Sheep are raised all over the world, because their temperament is timid, docile and easy to domesticate, and they can provide products such as meat and fur for people. Four Chinese breeds of sheep were studied here, Chaka sheep (CKS), Hu sheep (HS), Large Tail Han sheep (LTHS) and Small Tail Han sheep (STHS). CKS is a crossbreed between crossing local crossbred sheep and Romney sheep. The HS, STHS and the LTHS are local breeds [3]. In recent years, there have been several studies on sheep CNV. In 2016, Zhu et al. [4] found that genes such as PPARA, RXRA and KLF11 were involved in fat deposition and affected the tail type of sheep. Yang et al. [5] found that BTG3, PTGS1 and PSPH genes overlapped with sheep CNV regions which were associated with fetal muscle development, prostaglandin (PG) synthesis and bone color, respectively.
The Src Homology 2 Domain Containing E (SHE) is a protein-coding gene which is located on sheep chromosome 1 and contains 6 exons, and encodes a protein with 495 amino acids whose molecular weight is 53950 Da. The SHE gene is an important member of the SH2 domain-containing family. The SH2 (Src homology 2) domain is a structurally conserved protein domain, found in the Src oncoprotein [6] and many other intracellular signal transduction proteins [7]. Proteins containing these domains can dock to phosphorylated tyrosine residues on other proteins, and these protein–protein interactions play essential roles in cellular growth and development. Therefore, the SH2 domain can have a great effect on the economic traits of animals [8]. Studies on the SHE gene have mainly focused on diseases, such as human diseases blepharoconjunctivitis and blepharitis [9]. There have been no reports analyzing the SHE gene in sheep. However, bioinformatics analysis shows that the SHE gene of sheep has 86.2% similarity with the human SHE gene, suggesting the sheep SHE gene may also participate in disease and cellular growth and development.
In this study, our aim was to verify the CNV of the SHE gene in the four sheep breeds of CKS, HS, STHS and LTHS, and then test the association between the CNV of the SHE gene and the economic traits of these four sheep breeds. In addition, we expected to find out whether copy number variation of the SHE gene had the same effect on the growth traits of these four sheep breeds. Finally, we hoped that this work can provide useful information for sheep molecular breeding.
2. Materials and Methods
2.1. Animal Welfare
All animal experiments in the president study were performed in conformity to the applicable guidelines, animal welfare laws and policies. Moreover, all experiments were approved by the Faculty Animal Policy and Welfare Committee of Northwest A&F University (FAPWC-NWAFU, protocol number, NWAFAC1008).
2.2. Animals and Growth Traits Measurements
We selected a total of 750 sheep from four kinds of sheep to study the copy number variation of the SHE gene: CKS (n = 302, Wulan country, Qinghai Province, China), HS (n = 198, Mengjin Country, Henan Province, China), LTHS (n = 61, Yongjing Country, Gansu Province, China) and STHS (n = 189, Yongjing Country, Gansu Province, China).
We measured the economic traits of these 750 sheep. For CKS, we measured body weight, height, length and heart girth. For HS, we measured body weight, height, slanting length, heart girth, rump length and circumference of cannon bone. In the LTHS population, body weight, height, slanting length, heart girth, circumference of cannon bone, high at the cross, chest depth and chest width were measured. In the STHS population, we measured body height, slanting length, heart girth, circumference of cannon bone, high at the cross, chest depth and chest width.
2.3. Preparation of Sample and Genomic DNA
We collected all the blood samples of the sheep and the genomic DNA was extracted using the method of phenol-chloroform as described in Sonstegard et al. [10]. The quantity of the genomic DNA samples was determined using a Nanodrop 2000 Spectrophotometer. DNA samples were sequenced at a concentration of 25 ng/μL per dilution and stored at −20 °C. None of the animals included in this study showed any adverse health conditions.
2.4. Candidate Gene Identification and Primer Design
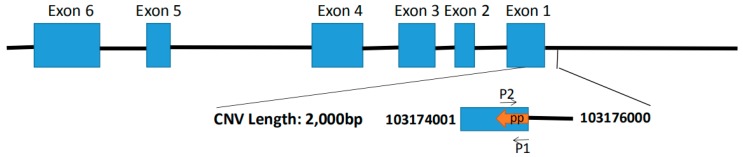
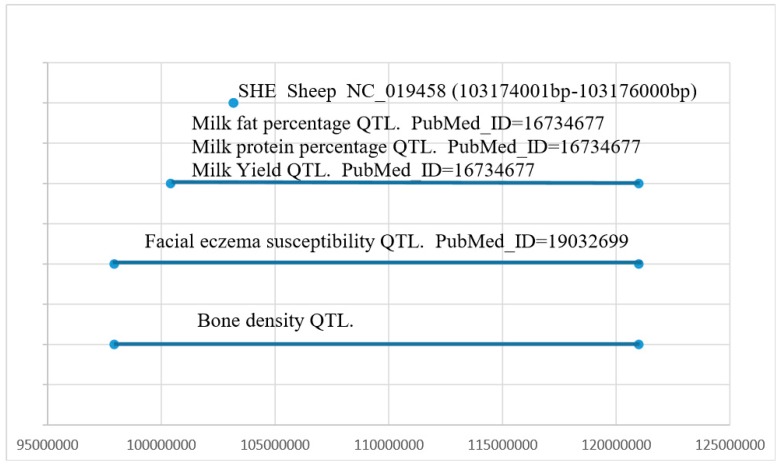
We found that the SHE gene was located in QTLs related to milk fat percentage and bone density, and we identified a 2000 bp (NC_019458.2 103174001 bp–103176000 bp) copy number variation in the SHE gene (Figure 1) (Huang et al., unpublished data). This CNV overlaps with the QTL associated with sheep milk fat percentage and bone density (Figure 2). The ANKRD1 gene appeared stable with two copies (Huang et al., unpublished data). We also found that the ANKRD1 gene is present in two copies in both cattle and sheep according to the animal omics database, so we selected to use the ANKRD1 gene as a diploid internal reference. All primers were designed by NCBI primer blast and are listed in Table 1.
Figure 1.
The region of the SHE gene’s copy number variation (CNV) in sheep breeds. The blue boxes indicate the coding area. The CNV region starts at 103174001 to 103176000 in Chr1 (Huang et al.). pp. (primer pair-CNV): 103174905 to 103175078, the detection sequence size is 174 bp.
Figure 2.
The CNV region of the SHE gene overlaps with the QTL of the sheep.
Table 1.
The primer information used in this study.
| Gene | Primer Pairs Sequences (5′–3′) | Amplification Length (bp) | Tm (°C) |
|---|---|---|---|
| SHE | 1F: AACAAAGCGCATTTAGGGCA | 174 | 60 |
| 1R: ACGTCATGATCCAGCGATAGT | 174 | 60 | |
| ANKRD1 | 2F: TGGGCACCACGAAATTCTCA | 143 | 60 |
| 2R: TGGCAGAAATGTGCGAACG | 143 | 60 |
F: forward primer; R: reverse primer.
2.5. Copy Number Variation and Gene Expression Analysis of the SHE Gene
We used quantitative polymerase chain reaction (qPCR) to determine the copy number of potential CNVs. Genomic DNA qPCR experiments were conducted using SYBR Green in triplicate reactions, each with a reaction volume of 10 μL, which contained 25 ng of genomic DNA, 5 μL of SYBR Premix Ex TaqII (GenStar, Beijing, China) and 0.5 μL of primers. Thermal cycling conditions were as follows: 95 °C for 10 min followed by 39 cycles of 95 °C for 15 s and 60 °C for 40 s. Melting curve analysis was completed at the end of the amplification with the following conditions: From 65 °C to 95 °C, 0.5 °C per 0.05 s. The copy number of the SHE gene was determined based on the assumption that the DNA fragment from the DNA of the calibrated animal has two copies.
2.6. Statistical Analysis
The copy number of the SHE gene was calculated using 2 × 2−∆Ct [11]. Three types of copy number (gain, loss and normal) were classified as >2, <2 or 2 copies. We used a general linear model in SPSS v19.0 software (SPSS, Inc., Chicago, IL, USA) to examine the associations of SHE CNV with growth traits within CKS, HS, LTHS and STHS breeds, where age and sex were considered fixed factors. Individuals of each breed were unrelated and from the same farm and the phenotypes were measured in the same season [12]. We used the following model to analyze the CNV effects on phenotypic traits: Yijk = μ + Ai + Sj + CNVk + eijk, where Yijk is the observation of growth traits; μ is the overall mean of each trait; Ai is the effect due to age; Sj is effect due to sex; CNVk is the effect of the SHE gene’s CNV type; and eijk is the random residual error. Finally, the variance between varieties was tested by chi-square [13]. The additive and dominance effect of the SHE gene CNV were calculated by the method of Falconer et al. [14] and Riollet et al. [15].
3. Results
3.1. The Distribution of SHE Gene’s CNV in Four Sheep Breeds
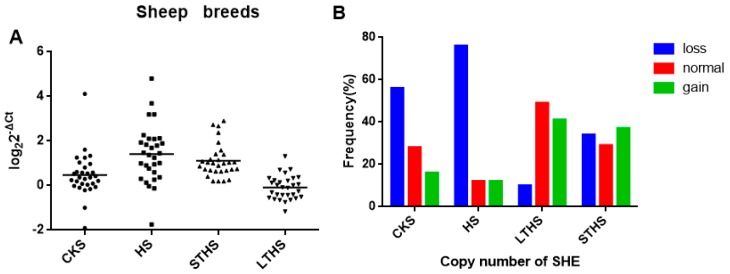
Available sequencing data revealed a 2000 bp copy number variation in the SHE gene. In order to study the distribution of this CNV in different sheep breeds, we examined CKS, HS, LTHS and STHS. The specific primers used are shown in Figure 3. Three types of copy number (gain, loss and normal) were classified as >2, <2 and 2 copies, and were calculated according to 2 × 2−∆Ct. As displayed in Figure 4A, the results revealed copy number variation of the SHE gene in the four breeds. The frequencies of copy number polymorphisms in the four sheep breeds revealed that most showed a loss copy number type, and this phenomenon was more pronounced in CKS and HS (Figure 4B).
Figure 3.
The specific primers of the SHE gene and the ANKRD1 gene.
Figure 4.
(A) The distribution of the SHE gene’s CNV in four sheep breeds. (B) The frequency of the copy numbers of the SHE gene’s CNV in the four sheep breeds.
3.2. Differences in CNV Distribution of the SHE Gene among Four Sheep Breeds
Based on the above findings, we calculated the frequencies of CNV types of the SHE gene in the four sheep breeds. The proportion of loss in CKS and HS breeds exceeded 50%, the proportion of loss in LTHS was only 10%, and there were relatively similar proportions of gain, loss and normal in STHS (Table 2). Chi-square values showed that a significantly different CNV distribution among the four sheep breeds exist (Table 3), indicating that the SHE CNV may be breed-specific.
Table 2.
Frequency of different CNV types in four sheep breeds.
| Breeds | Loss | Normal | Gain |
|---|---|---|---|
| CKS | 0.56 | 0.28 | 0.16 |
| HS | 0.76 | 0.12 | 0.12 |
| STHS | 0.34 | 0.29 | 0.37 |
| LTHS | 0.10 | 0.49 | 0.41 |
Numbers represent the proportion of different CNV types in this breed.
Table 3.
The chi-square test of CNV types among Chinese sheep breeds.
| Breed | CKS | HS | STHS | LTHS |
|---|---|---|---|---|
| CKS | 22.886 (p = 1.07 × 10−5) | 45.736 (p = 1.17 × 10−10) | 33.844 (p = 4.47 × 10−8) | |
| HS | 84.897 (p = 3.67 × 10−19) | 67.481 (p = 2.22 × 10−15) | ||
| STHS | 15.809 (p = 3.69 × 10−4) | |||
| LTHS |
Chi-square values (χ2) for differences in CNV between two breeds.
3.3. Association between the Copy Number Variation and Growth Traits
Many recent studies found correlations of animal economic traits to copy number variation. Here, we analyzed the association of the SHE gene CNV copy number types with the economic traits of four sheep breeds using a general linear model. The association showed that the CNV of the SHE gene in CKS had a significant effect on body length (p < 0.05). In HS, CNV had a significant effect on the circumference of cannon bone and heart girth (p < 0.05). In STHS, the CNV of the SHE gene was related to chest width and high at the cross (p < 0.05). No effect was observed for LTHS (Table 4, Supplementary Table S1). Sheep with copy number gain had a significantly higher circumference of cannon bone and heart girth than those with loss or normal copy types in HS (p < 0.05). Consistently, gain and normal copy number subjects also had high body length, chest width and high at the cross than loss copy number groups in CKS and STHS, with a significance of p < 0.05. What is more, the phenotypic variance explained by this CNV is shown in Supplementary Table S2, and the additive, dominance genetic effects of this CNV are shown in Supplementary Table S3.
Table 4.
Association analysis of the SHE gene CNV with growth traits in four sheep breeds.
| Breed | Growth Traits | CNV Type (Mean ± SE) | p | ||
|---|---|---|---|---|---|
| Loss | Normal | Gain | |||
| CKS | Body length (cm) | 71.66 ± 1.60 b | 74.22 ± 1.588 a | 73.33 ± 1.59 a,b | 0.028 |
| HS | Circumference of cannon bone (cm) | 76.39 ± 0.36 c | 76.40 ± 0.89 a,b | 78.77 ± 0.89 a | 0.046 |
| STHS | Heart girth (cm) | 7.03 ± 0.04 b | 7.29 ± 0.11 a,b | 7.31 ± 0.11 a | 0.012 |
| Chest width (cm) | 19.58 ± 0.39 a,b | 20.26 ± 0.44 a | 18.60 ± 0.38 b | 0.019 | |
| High at the cross (cm) | 62.06 ± 0.51 B | 62.02 ± 0.58 B,C | 64.20 ± 0.50 A | 0.004 | |
a,b Values that differ significantly at p < 0.05; A,B,C Values that differ significantly at p < 0.01. CKS: loss (169), normal (86), gain (47). HS: loss (150), normal (24), gain (24). STHS: loss (65), normal (54), gain (70).
4. Discussion
CNVs are caused by the insertion, deletion or replication of large fragments. As the main genetic form of submicroscopic structural variation, CNVs are widely distributed in the human genome, and affect gene expression, phenotypic variation and adaptation by interfering with genes and changing gene dosage [16,17,18,19]. Recent studies have confirmed that CNVs are not only ubiquitous in humans, but are also widely interspersed in mammals and plants, including domestic cattle [20], dogs [21], chickens [22], pigs [23], goats [24], sheep [25] and rabbits [26]. The concept of QTL was proposed by Geldermann in 1975 [27] and refers to the localization of multiple genes that collectively control a trait to one or more fragments on a chromosome. For example, the CNV of the bovine GBP4 gene is part of a QTL that determines its effect on the height of adult cattle [6].
As an SH2 domain-containing proteins, most studies of the SHE gene have focused on human diseases. The SH2 (Src Homology 2) domain is a structurally conserved protein domain present in the Src oncoprotein [7] and in many other intracellular signal-transducing proteins [28]. The function of this domain is to recognize the phosphorylated state of tyrosine residues, and initiates a series of events, which ultimately results in altered patterns of gene expression or other cellular responses. Therefore, the SHE gene in sheep is a promising candidate with an important function for sheep breeding.
We found that the CNV region of the SHE gene overlaps with a QTL region correlated with sheep milk fat percentage and bone density. Previous research reported that, in chickens, some QTL that correlated with CNV fragments are related to body weight, carcass weight and breast muscle mass [28,29]. Therefore, we speculated that the copy number variation of the SHE gene may be related to the economic traits of sheep, so four sheep (CKS, HS, STHS and LTHS) breeds were used in this study to analyze the CNV of the SHE gene. We found the CNV of the SHE gene existed in all four sheep breeds, and have associations with economic traits in CKS, HS and STHS. No association was observed in the LTHS, possibly due to the diversity across breeds [30]. For body length of CKS, the normal type was significantly better than the loss and gain types, and this difference was significant (p < 0.05). Notably, the gain type of HS associated with increased circumference of cannon bone and heart girth, and the normal and gain types were related with better chest width and high at the cross in STHS. Thus, we can conclude that the normal and gain CNV types of the SHE gene are related to better phenotypes in these four sheep breeds. We found different frequencies of the three CNV types in the four sheep breeds. Importantly, the dominant genotypes of these four sheep were not the same, which may be attributed to the diverse breeding backgrounds [31]. Gain and normal as the dominant CNV types of the SHE gene may be beneficial to the meat production and wool production traits of sheep, and further exploration is warranted.
CKS is a crossbred with little genomics studied. The meat value of CKS may be higher than the other local varieties. HS offers good quality lamb skin. LTHS have big tails, but poor development of the forequarters. STHS grow faster and are “high leg sheep” because of their tall and thin legs. The different characteristics of these four sheep breeds may be partially related to the CNV of the SHE gene.
In conclusion, this is the first detection and validation of CNVs of the SHE gene in four Chinese sheep breeds. The frequency of loss in CKS and HS exceeded 50%, the proportion of that in LTHS was only 10% and the proportion of the three copy number types in STHS was relatively balanced. Normal and gain copy number were associated with better economic traits than loss in CKS, HS and STHS, but had no difference in LTHS. The results suggested that the CNV of the SHE gene participates in economic traits and may be a molecular marker for sheep breeding.
Acknowledgments
We would like to thank the Chaka sheep breeding farm (Wulan, Qinghai, China), Hu sheep breeding farm (Mengjin, Henan, China), Large Tail Han sheep breeding farm (Yongjing, Gansu, China) and Small Tail Han sheep breeding farm (Yongjing, Gansu, China) for providing us with sampling sites.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2615/9/8/531/s1, Table S1: Association of the SHE gene CNV with growth traits in four sheep breeds, Table S2: The phenotypic variance explained by the CNV of the SHE gene, Table S3: The additive and dominance effect of the SHE gene CNV.
Author Contributions
Conceptualization, R.J., H.C., and L.-Y.H.; formal analysis, R.J., J.C., X.-K.C., and Y.-L.M.; funding acquisition, B.C., Y.-Z.H., X.-Y.L., C.-Z.L., L.-Y.H., and H.C.; investigation, R.J., X.-K.C., and Y.-L.M.; methodology, R.J., J.C., X.-K.C., and Y.-L.M.; project administration, Y.-Z.H., X.-Y.L., C.-Z.L., L.-Y.H., and H.C.; resources, L.-Y.H. and H.C.; software, R.J., J.C., and Y.-L.M.; supervision, Y.-Z.H. and X.-Y.L.; validation, L.-Y.H., and H.C; visualization, Y.-Z.H. and X.-Y.L.; writing—original draft writing, R.J. and J.C.; writing—review and editing, R.J., J.C., X.-Y.L., and B.C.
Funding
This work was supported by the Qinghai Province major science and technology projects (2017-NK-153).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Mills R.E., Walter K., Stewart C., Handsaker R.E., Chen K., Alkan C., Abyzov A., Yoon S.C., Ye K., Cheetham R.K., et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henrichsen C.N., Vinckenbosch N., Zöllner S., Chaignat E., Pradervand S., Schütz F., Ruedi M., Kaessmann H., Reymond A. Segmental copy number variation shapes tissue transcriptomes. Nat. Genet. 2009;41:424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- 3.Ma Q., Liu X., Pan J., Ma L., Ma Y., He X., Zhao Q., Pu Y., Li Y., Jiang L. Genome-wide detection of copy number variation in Chinese indigenous sheep using an ovine high-density 600 K SNP array. Sci. Rep. 2017;7:912. doi: 10.1038/s41598-017-00847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu C., Fan H., Yuan Z., Hu S., Ma X., Xuan J., Wang H., Zhang L., Wei C., Zhang Q., et al. Genome-wide detection of CNVs in Chinese indigenous sheep with different types of tails using ovine high-density 600K SNP arrays. Sci. Rep. 2016;6:27822. doi: 10.1038/srep27822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang L., Xu L., Zhou Y., Liu M., Wang L., Kijas J.W., Zhang H., Li L., Liu G.E. Diversity of copy number variation in a worldwide population of sheep. Genomics. 2018;110:143–148. doi: 10.1016/j.ygeno.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Sadowski I., Stone J.C., Pawson T. A noncatalytic domain conserved among cytoplasmic protein-tyrosine kinases modifies the kinase function and transforming activity of Fujinami sarcoma virus P130gag-fps. Mol. Cell. Biol. 1986;6:4396–4408. doi: 10.1128/MCB.6.12.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russell R.B., Breed J., Barton G.J. Conservation analysis and structure prediction of the SH2 family of phosphotyrosine binding domains. FEBS Lett. 1992;304:15–20. doi: 10.1016/0014-5793(92)80579-6. [DOI] [PubMed] [Google Scholar]
- 8.Hosoe Y., Numoto N., Inaba S., Ogawa S., Morii H., Abe R., Ito N., Oda M. Structural and functional properties of Grb2 SH2 dimer in CD28 binding. Biophys. Physicobiol. 2019;16:80–88. doi: 10.2142/biophysico.16.0_80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherif N., D’Hermies F., Barraco P., Elmaleh C., Renard G., Pouliquen Y. Meibomian adenocarcinoma in its blepharo-conjunctival form. Apropos of a case. J. Fr. Ophtalmol. 1997;20:293–296. [PubMed] [Google Scholar]
- 10.Sonstegard T.S., Garrett W.M., Ashwell M.S., Bennett G.L., Kappes S.M., Van Tassell C.P. Comparative map alignment of BTA27 and HSA4 and 8 to identify conserved segments of genome containing fat deposition QTL. Mamm. Genome. 2000;11:682–688. doi: 10.1007/s003350010130. [DOI] [PubMed] [Google Scholar]
- 11.Bae J.S., Cheong H.S., Kim L.H., NamGung S., Park T.J., Chun J.Y., Kim J.Y., Pasaje C.F., Lee J.S., Shin H.D. Identification of copy number variations and common deletion polymorphisms in cattle. BMC Genomics. 2010;11:232. doi: 10.1186/1471-2164-11-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J., Zhu X., Ma L., Xu H., Cao X., Luo R., Chen H., Sun X., Cai Y., Lan X. Detection of a new 20-bp insertion/deletion (indel) within sheep PRND gene using mathematical expectation (ME) method. Prion. 2017;11:143–150. doi: 10.1080/19336896.2017.1300740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benjamini Y., Hochberg Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 14.Falconer D.S., Mackay T.F.C. Introduction to quantitative genetics, 4th ed. Am. J. Hum. Genet. 1996;46:1231. [Google Scholar]
- 15.Riollet C., Rainard P., Poutrel B. Differential induction of complement fragment C5a and inflammatory cytokines during intramammary infections with Escherichia coli and Staphylococcus aureus. Clin. Diagn. Lab. Immunol. 2000;7:161–167. doi: 10.1128/CDLI.7.2.161-167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feuk L., Carson A.R., Scherer S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 17.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conrad D.F., Pinto D., Redon R., Feuk L., Gokcumen O., Zhang Y., Aerts J., Andrews T.D., Barnes C., Campbell P., et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang F., Gu W., Hurles M.E., Lupski J.R. Copy Number Variation in Human Health, Disease, and Evolution. Annu. Rev. Genom. Hum. Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu G.E., Hou Y., Zhu B., Cardone M.F., Jiang L., Cellamare A., Mitra A., Alexander L.J., Coutinho L.L., Dell’Aquila M.E., et al. Analysis of copy number variations among diverse cattle breeds. Genome Res. 2010;20:693–703. doi: 10.1101/gr.105403.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X., Nahashon S., Feaster T.K., Bohannon-Stewart A., Adefope N. An initial map of chromosomal segmental copy number variations in the chicken. BMC Genomics. 2010;11:351. doi: 10.1186/1471-2164-11-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fadista J., Nygaard M., Holm L.-E., Thomsen B., Bendixen C. A Snapshot of CNVs in the Pig Genome. PLoS ONE. 2008;3:e3916. doi: 10.1371/journal.pone.0003916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fontanesi L., Martelli P.L., Beretti F., Riggio V., Dall’Olio S., Colombo M., Casadio R., Russo V., Portolano B.L. An initial comparative map of copy number variations in the goat (Capra hircus) genome. BMC Genomics. 2010;11:639. doi: 10.1186/1471-2164-11-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fontanesi L., Beretti F., Martelli P., Colombo M., Dall’Olio S., Occidente M., Portolano B., Casadio R., Matassino D., Russo V. A first comparative map of copy number variations in the sheep genome. Genomics. 2011;97:158–165. doi: 10.1016/j.ygeno.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Fontanesi L., Martelli P.L., Scotti E., Russo V., Rogel-Gaillard C., Casadio R., Vernesi C. Exploring copy number variation in the rabbit (Oryctolagus cuniculus) genome by array comparative genome hybridization. Genomics. 2012;100:245–251. doi: 10.1016/j.ygeno.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Geldermann H. Investigations on inheritance of quantitative characters in animals by gene markers I. Methods. Theor. Appl. Genet. 1975;46:319–330. doi: 10.1007/BF00281673. [DOI] [PubMed] [Google Scholar]
- 27.Cao X.K., Huang Y.Z., Ma Y.L., Cheng J., Qu Z.X., Ma Y., Bai Y.Y., Tian F., Lin F.P., Ma Y.L., et al. Integrating CNVs into meta-QTL identified GBP4 as positional candidate for adult cattle stature. Funct. Integr. Genomics. 2018;18:559–567. doi: 10.1007/s10142-018-0613-0. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., Gu X., Feng C., Song C., Hu X., Li N. A genome-wide survey of copy number variation regions in various chicken breeds by array comparative genomic hybridization method. Anim. Genet. 2012;43:282–289. doi: 10.1111/j.1365-2052.2011.02308.x. [DOI] [PubMed] [Google Scholar]
- 29.Shi T., Xu Y., Yang M., Huang Y., Lan X., Lei C., Qi X., Yang X., Chen H. Copy number variations at LEPR gene locus associated with gene expression and phenotypic traits in Chinese cattle. Anim. Sci. J. 2016;87:336–343. doi: 10.1111/asj.12531. [DOI] [PubMed] [Google Scholar]
- 30.Lehnert S.A., Reverter A., Byrne K.A., Wang Y., Nattrass G.S., Hudson N.J., Greenwood P.L. Gene expression studies of developing bovine longissimus muscle from two different beef cattle breeds. BMC Dev. Biol. 2007;7:95. doi: 10.1186/1471-213X-7-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng Z., Yuan Y., Li Y.J., Wang H.X., Shi J., Cao W.X., Luo H.W., Deng J.R., Feng W.L. Targeting BCR tyrosine177 site with novel SH2-DED causes selective leukemia cell death in vitro and in vivo. Int. J. Biochem. Cell Biol. 2012;44:861–868. doi: 10.1016/j.biocel.2012.02.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.