Abstract
The enzyme soluble guanylyl cyclase (sGC) is a heterodimer composed of an α subunit and a heme-containing β subunit. It participates in signaling by generating cGMP in response to nitric oxide (NO). Heme insertion into the β1 subunit of sGC (sGCβ) is critical for function, and heat shock protein 90 (HSP90) associates with heme-free sGCβ (apo-sGCβ) to drive its heme insertion. Here, we tested the accuracy and relevance of a modeled apo-sGCβ–HSP90 complex by constructing sGCβ variants predicted to have an impaired interaction with HSP90. Using site-directed mutagenesis, purified recombinant proteins, mammalian cell expression, and fluorescence approaches, we found that (i) three regions in apo-sGCβ predicted by the model mediate direct complex formation with HSP90 both in vitro and in mammalian cells; (ii) such HSP90 complex formation directly correlates with the extent of heme insertion into apo-sGCβ and with cyclase activity; and (iii) apo-sGCβ mutants possessing an HSP90-binding defect instead bind to sGCα in cells and form inactive, heme-free sGC heterodimers. Our findings uncover the molecular features of the cellular apo-sGCβ–HSP90 complex and reveal its dual importance in enabling heme insertion while preventing inactive heterodimer formation during sGC maturation.
Keywords: heat shock protein 90 (HSP90), chaperone, heme, structural model, protein-protein interaction, protein folding, cell signaling, client protein
Introduction
The enzyme soluble guanylyl cyclase (sGC,3 EC 4.6.1.2) participates in numerous biologic signaling cascades by generating cGMP in response to NO (1–3). Disruption of this pathway contributes to a wide spectrum of pulmonary and cardiovascular diseases (4–10). Mature, active sGC is a heterodimer composed of an α-subunit and a β-subunit (11–13). Both subunits are modular and contain an N-terminal H-NOX domain, a middle Per-Arnt-Sim like domain (PAS domain), a coiled-coil domain (CC domain), and a C-terminal catalytic cyclase domain (11–13). The H-NOX domain of sGCβ, but not that of sGCα, contains a binding site for iron protoporphyrin IX (heme) (14, 15). When loaded onto sGCβ, the heme functions to bind NO and as a nexus for consequent protein structural changes that activate catalysis in the sGC heterodimer (1–3).
Whereas sGC has been studied extensively for decades, relatively little is known about the processes that form mature, functional sGCα/β heterodimers in mammalian cells. Several studies established that the chaperone heat shock protein 90 (HSP90) plays an important role in sGC maturation and function (6, 16–18). Antibody pulldown studies first showed that sGC associates with HSP90 in cells, although the reasons for this association were not clear (19–21). More recent studies showed that HSP90 only associates with the heme-free (apo) form of the sGCβ subunit (17), thereby helping to drive heme insertion into apo-sGCβ in an ATP-dependent process (16, 17). Once the sGCβ subunit acquires heme, it dissociates from HSP90 and associates instead with an sGCα subunit partner to form a mature sGCα/β heterodimer (16, 17). Although clarifying, these cell-based studies did not define the nature of the HSP90-apo-sGCβ association and thus provided no molecular-level information.
Our subsequent study with purified apo-sGCβ and HSP90 proteins revealed that they directly form a complex with low micromolar affinity, in which the middle domain of HSP90 interacts with the PAS domain of apo-sGCβ (22). This information allowed us to build an energy-minimized structural model of a directly interacting complex (18, 22). However, this complex model has not been tested with regard to its accuracy, its formation in cells, or its relevance for heme insertion into apo-sGCβ. To address these gaps, we generated a series of sGCβ variants that contain model-based point and deletion mutations designed to disrupt the apo-sGCβ–HSP90 interaction. We investigated the ability of each variant to interact with HSP90 both in vitro and in mammalian cells, to undergo heme insertion, to form a heterodimer with sGCα, and to display cyclase activity. Our findings show that our model for the HSP90–apo-sGCβ complex is valid, that the complex forms in live cells, and that HSP90 both drives heme insertion into apo-sGCβ and prevents premature association of the sGCα subunit during the heme insertion process. This improves our molecular-level understanding of sGC heterodimer maturation and further defines the key roles played by the HSP90 chaperone.
Results
Generation and general properties of sGCβ mutant proteins
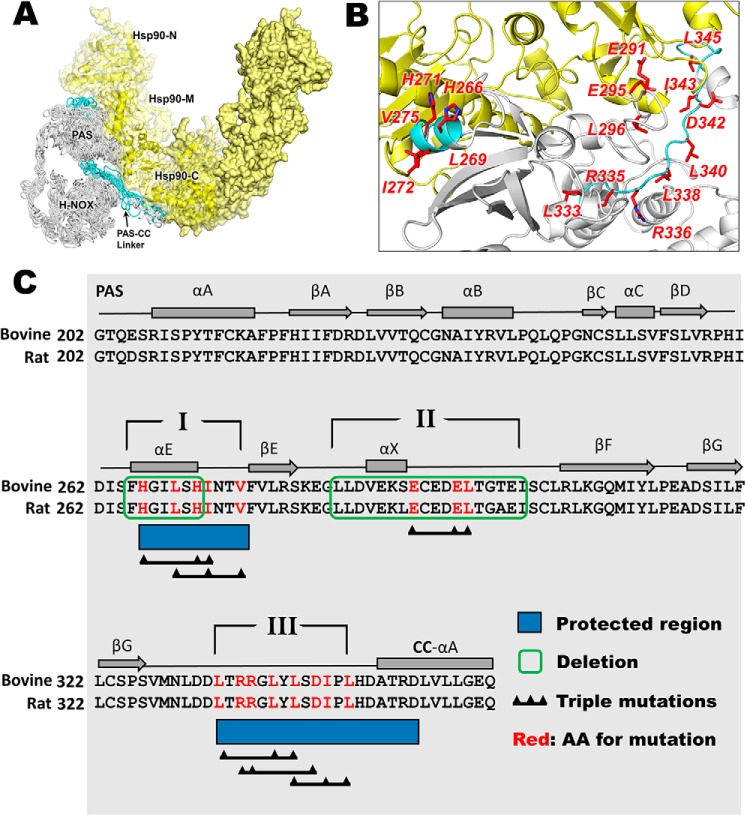
The deletions and amino acid substitutions that we incorporated into our bovine sGCβ1(1–358) bacterial expression construct are listed in Table S1. All of the sequence changes were located in the C-terminal region that contains the PAS domain, a downstream linker element, and an N-terminal portion of the CC domain (Fig. 1). The deletions and substitutions cluster into three regions. We hypothesized that these regions may interact directly with HSP90, based on our previous hydrogen–deuterium exchange MS study of HSP90–sGCβ interaction (22) (regions I and III) or on the model that we built of the HSP90–sGCβ complex (22) (region II).
Figure 1.
Model structure of the apo-sGCβ–HSP90 complex, the protein–protein interface, and the sGCβ residues targeted for mutagenesis. A, sGCβ(1–358)–HSP90 interaction model. Adapted from Ref. 22. This research was originally published in the Journal of Biological Chemistry. Sarkar, A., Dai, Y., Haque, M. M., Seeger, F., Ghosh, A., Garcin, E. D., Montfort, W. R., Hazen, S. L., Misra, S., and Stuehr, D. J. Heat shock protein 90 associates with the Per-Arnt-Sim domain of heme-free soluble guanylate cyclase: implications for enzyme maturation. J. Biol. Chem. 2015; 290:21615–21628. © the American Society for Biochemistry and Molecular Biology. The H-NOX, PAS, and PAS-linker domains of sGCβ are colored white or teal, with teal indicating regions reported to become protected upon HSP90 binding. The HSP90 homodimer is colored yellow with the N-terminal (N), middle (M), and C-terminal (C) domains of one protomer indicated. B, view of the sGCβ-PAS (white and teal) and HSP90 M (yellow) interacting region. The red highlighted residues are those mutated in the current study. C, portion of the primary sequence and secondary structure of sGCβ that we targeted for mutagenesis. Dark blue rectangles, sequences that become protected by HSP90 complex formation. The three regions (indicated by Roman numerals) and residues within them targeted for deletion or point mutation are indicated.
All of the His6-tagged sGCβ1(1–358) constructs that were generated, including WT, expressed well in Escherichia coli. Following purification, the recombinant proteins were predominantly heme-free, with residual heme content ranging from 0.03 to 0.5 mol of heme/mol of protein (Fig. S1). Adding heme to the purified proteins followed by filtration through a desalting column increased their heme contents, ranging from 0.64 to 0.99 mol of heme/mol of protein. Each heme-reconstituted protein exhibited a predominant Soret peak at 431 nm (Fig. S1) and visible spectral features corresponding to predominantly ferrous and five-coordinate heme, as is present in native sGC (23, 24). This shows that none of the bacterially expressed and purified apo-sGCβ1(1–358) proteins had a significantly compromised or altered heme binding, consistent with the mutations being located outside of the heme-binding domain.
Effect of mutations on apo-sGCβ association with HSP90
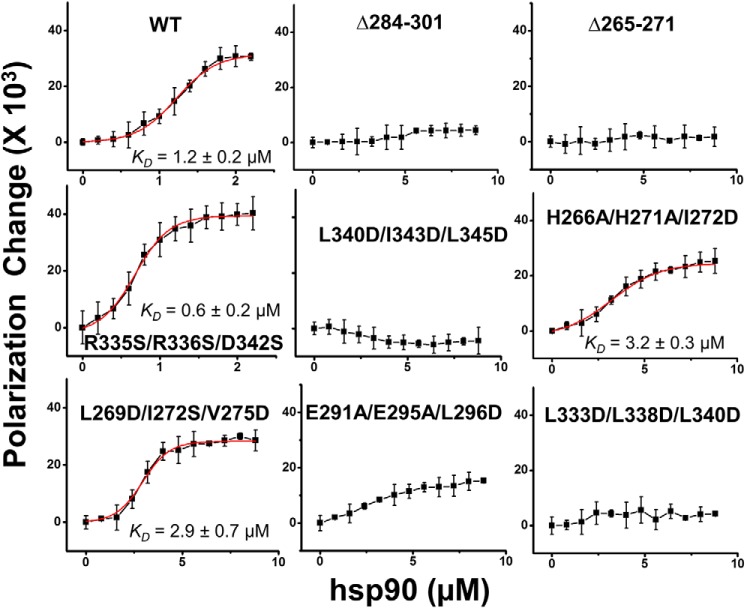
We utilized fluorescence polarization to monitor the binding of purified HSP90 with each purified WT or mutant apo-sGCβ1(1–358) protein (22). Fig. 2 shows residual fluorescence polarization traces for each FITC-labeled apo-sGCβ1(1–358) protein upon titration with increasing concentrations of HSP90. The proteins displayed a range of HSP90 interaction capabilities, with R335S/R336S/D342S showing saturation binding and an estimated affinity as good as or slightly better than WT. Two other mutants (L269D/I272S/V275D and H266A/H271A/I272D) showed saturation binding but somewhat poorer estimated affinities, and the other five mutants had HSP90-binding affinities that were either significantly diminished (E291A/E295A/L296D) or undetectable by this method. To exclude labeling artifacts, we repeated the binding experiment using FITC-labeled HSP90 and unlabeled apo-sGCβ1(1–358) proteins. The results were similar and are shown in Fig. S2. Thus, amino acid residues within each of the three proposed binding regions in apo-sGCβ1(1–358) influence its affinity toward HSP90 in the purified protein system.
Figure 2.
Binding interactions of FITC-labeled sGCβ1(1–358) mutant proteins with HSP90. Each indicated FITC-labeled sGCβ protein (0.5 μm) was titrated with increasing concentrations of HSP90, and the residual polarization change was recorded and plotted. The points depict mean ± S.D. (error bars) for three samples and are representative of two independent experiments. Fit lines are red, and the resulting binding constant estimates are indicated.
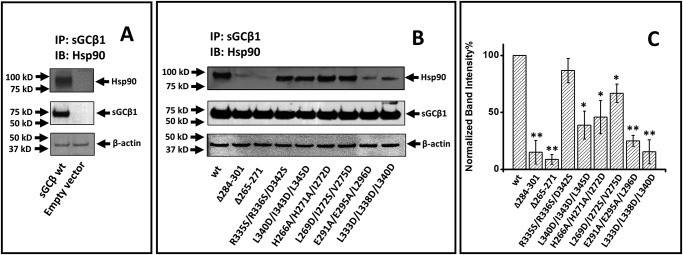
We next introduced the same mutations into a mammalian expression vector that encoded full-length rat sGCβ1(1–619) (17). We transiently expressed each construct in COS-7 cells under heme-deficient conditions, which support the maximum levels of apo-sGCβ–HSP90 interaction (17). We analyzed cell supernatants to assess the expression level and degree of HSP90 association of each mutant. All of the sGCβ proteins showed similar expression levels but displayed different degrees of HSP90 association (Fig. 3). In general, the rank order of the in-cell HSP90 associations mimicked the HSP90-binding behaviors that we observed for the purified sGCβ mutant proteins as described in Fig. 2. The good correlation is consistent with apo-sGCβ directly associating with HSP90 in the cells through an interaction that involves an interface similar to the one depicted in our structural model of the apo-sGCβ–HSP90 complex.
Figure 3.
Association of sGCβ1(1–619) mutants with HSP90 in mammalian cells. Heme-deficient COS-7 cells were transfected to express each sGCβ1(1–619) mutant protein. After 48 h, supernatants (equal protein) were immunoprecipitated with an anti-sGCβ antibody followed by SDS-PAGE and Western blot analysis (IB) with anti-HSP90 and sGCβ antibodies. A, representative Western blotting indicating HSP90 pulled down with WT sGCβ1 and an empty vector as negative control. B, representative Western blotting indicating the HSP90 pulled down with each sGCβ1 protein on the beads. C, densitometric quantification of HSP90 binding, with the values being the mean ± S.D. (error bars) of three independent experiments. The normalized band intensity of each sGC mutants is compared with WT for significance determination. **, p < 0.01; *, 0.01 < p < 0.05.
HSP90 interaction is needed for heme insertion into apo-sGCβ in cells
To assess how HSP90 binding correlates with sGCβ1 heme insertion in live cells, we utilized a construct of rat sGCβ1(1–619) originally developed by Hoffmann et al. (25), which incorporates a tetra-Cys motif (TC) at a specific location near the sGCβ heme-binding site. Once the TC motif binds the indicator dye FlAsH (FlAsH-TC-sGCβ), it becomes fluorescent and in turn undergoes quenching when heme binds. Such FlAsH-TC-sGCβ constructs have been used to study heme content and conformational changes in sGCβ (25, 26).
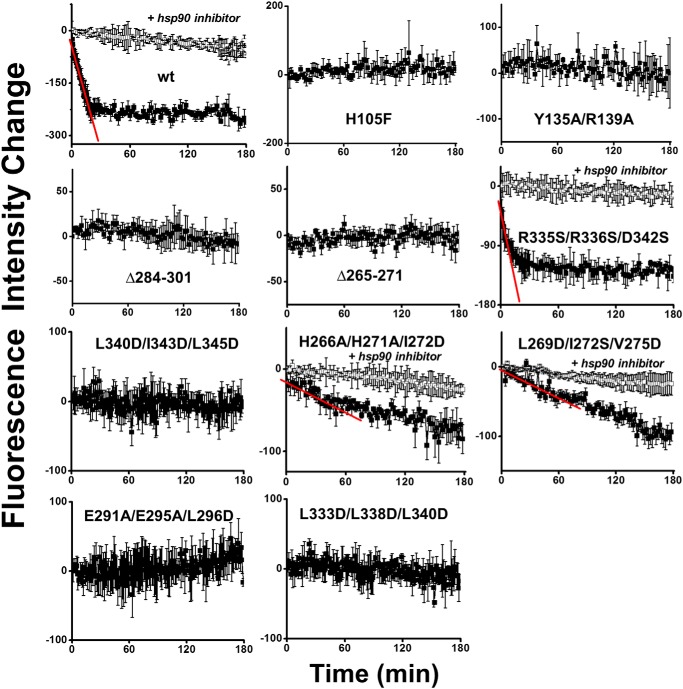
To validate the method, we expressed WT TC-sGCβ1(1–619) and either of two mutant constructs that are defective in heme-binding, TC-sGCβ1(1–619) H105F (17, 27) and TC-sGCβ1(1–619) Y135A/R139A (27), in heme-deficient COS-7 cells. We bound FlAsH to their TC motifs and determined how heme addition to the cells impacted their FlAsH fluorescence. Fig. 4 shows that adding heme to cells expressing the WT FlAsH-TC-apo-sGCβ1(1–619) caused a fluorescence decrease that was time-dependent, saturable, and sensitive to HSP90 inhibition, with an initial rate of fluorescence decrease estimated to be −11.4 ± 0.9 min−1. In contrast, adding heme had no effect on the fluorescence signal intensity from cells expressing either of the sGCβ heme-binding mutants. We thus went on to use the same strategy to evaluate heme insertion into the various apo-TC-sGCβ1(1–619) mutants in the live cells. Fig. 4 shows that the mutant proteins exhibited a range of heme insertion capacities relative to WT, which were also sensitive to HSP90 inhibition. The R335S/R336S/D342S mutant showed heme insertion kinetics similar to or slightly faster than WT, as judged from a fit of its initial fluorescence decrease rate (−13.1 ± 2.2 min−1), whereas the L269D/I272S/V275D and H266A/H271A/I272D mutants had detectable but 10-fold slower initial rates of heme insertion, as estimated from fits of their fluorescence decreases (−0.9 ± 0.1 and −1.0 ± 0.2 min−1, respectively). Heme binding to the other five mutant proteins was essentially undetectable by this method. In general, heme insertion into the various sGCβ proteins expressed in cells correlated well with their different HSP90-binding capabilities, with the exception of the L269D/I272S/V275D mutant, whose heme insertion was somewhat less than what one might expect based on its HSP90 binding capacity. Overall, the results imply that heme insertion into apo-sGCβ in cells depends on, and is likely limited by, the ability of HSP90 to directly bind to apo-sGCβ regions I, II, and III.
Figure 4.
Kinetics of heme insertion into FlAsH-TC-sGCβ1(1–619) WT, heme binding–deficient mutants, and HSP90-binding mutant proteins in mammalian cells. TC-sGCβ1(1–619) WT and mutants were expressed in heme-deficient COS-7 cells. The proteins were FlAsH-labeled, and then their fluorescence was monitored with time after adding 5 μm heme to the cell cultures. The fluorescence decrease is proportional to the heme insertion into the TC-sGCβ1(1–619) proteins over time. In some cases, the initial rates of fluorescence decrease were estimated by fitting (red lines). Curve points show the mean ± S.D. (error bars) from three wells and are representative of three independent experiments.
HSP90 interaction is critical for development of heme-dependent sGCβ catalytic activity
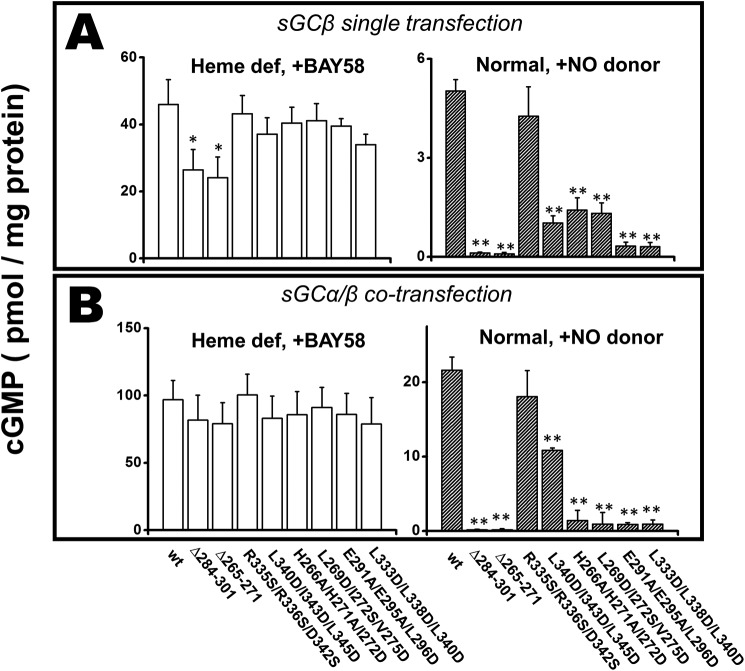
To independently test the importance of HSP90 complex formation, we compared the ability of each sGCβ mutant to become catalytically active in cells. sGCβ can become active in cells in the absence or presence of an sGCα partner, although the activity achieved is greater in the latter case (16, 17). We first determined the ability of BAY58-2667 (4-[[(4-carboxybutyl)[2-[2-[[4-(2-phenylethyl)-phenyl]methoxy]phenyl]ethyl]amino]methyl]benzoic acid hydrochloride; BAY58) to activate catalysis by each mutant when it was expressed in heme-deficient cells with or without sGCα co-expression. Under these circumstances, BAY58 binds within the heme pocket of apo-sGCβ (28), inducing structural changes that activate catalysis independent of heme or HSP90. Fig. 5 (A and B, left panels) shows the guanylate cyclase activities we obtained when the cell supernatants were given BAY58. All displayed activities approached the level for WT, with the exception of the supernatants containing the two apo-sGCβ deletion mutants, which were about 50–60% as active in the absence of sGCα co-expression, respectively. All of the sGCβ proteins expressed in the heme-deficient cells showed similar expression levels (Fig. S3), and as expected, they all had a very poor GTP cyclase activity in response to the NO donor NOC-18 (Fig. S4). The results establish that each sGCβ mutant protein maintains a robust intrinsic catalytic activity when it is elicited in a heme- and HSP90-independent manner by BAY58.
Figure 5.
Guanylyl cyclase activity of the sGCβ1(1–619) proteins in cell supernatants in response to an sGC activator drug or to an NO donor. COS-7 cells were cultured in normal medium (normal) or cultured in heme-depleted conditions (heme def) and transfected to express each sGCβ1(1–619) protein alone or along with sGCα. Their supernatants were collected and used for GTP cyclase activity measurements in response to BAY 58 or to the NO donor NOC-18, as indicated. A, activities of the singly transfected sGCβ1(1–619) proteins; B, activity of the sGCβ1(1–619) proteins when co-transfected with sGCα. Values are the mean ± S.D. (error bars) of three measurements and are representative of two experiments each. The activity of each sGC mutant is compared with WT for significance determination. **, p < 0.01; *, 0.01 < p < 0.05.
The GTP cyclase activities of supernatants from similarly transfected cells that were grown in normal medium and in response to the NO donor NOC-18 are shown in the right panels of Fig. 5 (A and B). In this circumstance, the level of cyclase activity depends completely on heme insertion having occurred in sGCβ. The activities varied greatly in response to the NO donor, most were lower than WT, and their rank order was similar with or without the sGCα co-expression. In general, the NO-dependent activities of the mutants corresponded well with their capacities for HSP90 complex formation and for achieving heme insertion (as determined above). Thus, heme insertion into sGCβ and its consequent catalytic activity requires that HSP90 bind to PAS domain regions I, II, and III.
HSP90 prevents sGCα binding to apo-sGCβ during heme insertion
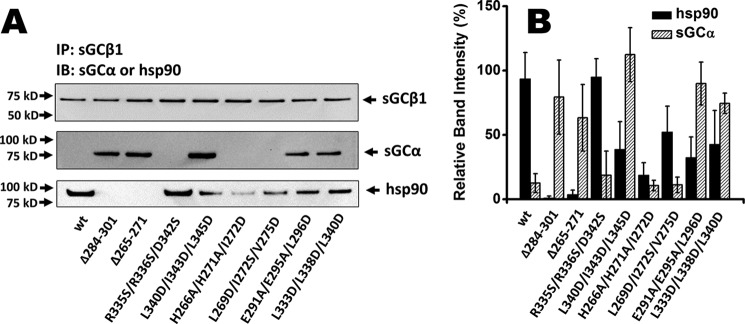
Co-IP studies suggest that sGCα/β heterodimer formation occurs after apo-sGCβ acquires heme and dissociates its HSP90 chaperone (16, 17). Our current study allowed us to test the relative importance of HSP90 association versus heme insertion in governing sGCβ subunit interaction with an sGCα partner. We co-transfected heme-depleted COS-7 cells with sGCα and each of our sGCβ1(1–619) mutants and then analyzed their associations with sGCα versus with HSP90 by performing co-IP on the cell supernatants. Fig. 6A shows that WT apo-sGCβ associated strongly with HSP90 but minimally with sGCα, as established previously (16). In comparison, five of the seven apo-sGCβ mutants with completely or partially impaired HSP90 binding were found to associate much more strongly with sGCα. The sGCα-binding capacities of these five mutants correlated inversely with their HSP90 association capacities (Fig. 6B). The remaining two sGCβ1(1–619) mutants with partially impaired HSP90 binding (H266A/H271A/I272D and L269D/I272S/V275D) associated poorly with sGCα in the heme-deficient cells, revealing that they are also sGCα-binding mutants under this circumstance. Co-expression of sGCα did not significantly alter the levels of HSP90 association with any of the sGCβ1(1–619) mutants (Fig. S5), suggesting that their heme content is the major factor that determines their HSP90 interaction. Overall, our findings reveal that sGCβ and sGCα subunits have an intrinsic binding affinity toward one another, even when the sGCβ subunit is heme-free, but sGCα can only bind to apo-sGCβ when it has defective HSP90 binding. Thus, besides enabling heme insertion, HSP90 may also competitively block sGCα from binding to apo-sGCβ in cells during sGC maturation.
Figure 6.
Association of each sGCβ protein with sGCα or HSP90 in heme-deficient mammalian cells. Heme-deficient COS-7 cells were transfected to express each sGCβ1(1–619) mutant and sGCα for 48 h. Supernatant aliquots (equal protein) were immunoprecipitated with an anti-sGCβ antibody followed by SDS-PAGE and Western blot analysis (IB) with anti-HSP90 and sGCα antibodies. A, representative Western blot analysis of bound sGCα, HSP90, and sGCα retained on the beads. B, densitometric quantifications of HSP90 or sGCα associated with each sGCα protein. Results are the mean ± S.D. (error bars) of three independent experiments.
Discussion
Heme insertion during sGC maturation ultimately leads to formation of an NO-responsive heterodimer, but questions remain about the mechanism and regulation of this process. Early studies established that sGC associates with HSP90 in cells and tissues under a variety of conditions. Their association usually correlated with an increase in sGC activity, and pulldown and column binding experiments implicated the sGCβ PAS domain in enabling the HSP90 association (20, 21). More recently, we determined that (i) in cells and tissues, HSP90 associates with only the heme-free form of sGCβ, and not with holo-sGCβ, sGCα, or the sGC heterodimer (16, 17), and (ii) purified apo-sGCβ and HSP90 interact directly to form a complex (22). Using interaction mapping based on hydrogen–deuterium exchange MS and biophysical experiments, existing structural data, and computer-assisted docking, we built a partial model of an sGCβ–HSP90 interaction complex (18, 22) (as in Fig. 1). In the present study, we sought to determine whether a complex corresponding to our model actually forms in mammalian cells and, if so, to define its nature and its biological relevance for sGCβ heme insertion, sGC heterodimer formation, and catalytic activity. Our findings suggest that a direct complex does form between apo-sGCβ and HSP90 in cells that is similar to our model structure, because model-guided deletions and mutations successfully disrupted the sGCβ–HSP90 complex. Moreover, we found that the complex must form in mammalian cells to drive heme insertion into apo-sGCβ during maturation. Our study also shows that HSP90, by virtue of binding to apo-sGCβ, prevents premature association of apo-sGCβ with an sGCα partner subunit. In this way, HSP90 prevents formation of a heme-free sGC heterodimer in cells.
To interpret the results of our study, it was important to determine whether the mutations caused unintended disruptions to sGCβ behavior, such as the ability of its H-NOX domain to bind heme properly or to impact the catalytic activity of sGC. Two lines of evidence suggest that our designed mutations had a negligible impact. (i) All of the purified apo-sGCβ mutants bound exogenously added heme to generate a common species with spectral features characteristic of native sGC. (ii) When each of the full-length sGCβ mutants was expressed in heme-deficient mammalian cells, they displayed good catalytic activity upon the addition of BAY58, which binds and activates apo-sGCβ independent of HSP90 or NO (29, 30). Thus, the effects of the mutations appeared to be restricted to PAS domain functions, and their impacts on HSP90 binding, sGCβ heme acquisition, and NO-dependent catalytic activity in mammalian cells could be clearly interpreted.
Structural findings
The residues that we selected for mutagenesis or deletion were clustered in three regions of apo-sGCβ that form the HSP90 complex interface in our model structure. Mutant proteins from all three regions either partly or completely impaired HSP90 interaction when tested either as purified constructs or when expressed as full-length sGCβ proteins in mammalian cells.
Region I residues Phe265–His271 consist of a surface α-helix (αE) that is a common structural element in PAS domain folds (31), followed by a proximal C-terminal loop, which interacts directly with the HSP90 N-M domain interface in our model. Deletion of region I eliminated HSP90 binding and allowed sGCα interaction with the heme-free sGCβ. It is unlikely that the deletion grossly perturbed the folding of sGCβ, as the expression level was normal and the purified protein could correctly bind the provided heme. The triple substitutions within region I of surface-exposed His to Ala or of surface-exposed aliphatic side chains to Asp/Ser also induced a partial loss in HSP90 affinity, suggesting that region I of the PAS domain engages in a relatively distributed interaction with HSP90. Interestingly, the heme-free triple mutants were both defective in binding an sGCα partner subunit, implying that region I also enables the intrasubunit PAS–PAS domain interactions that are needed for sGC heterodimer formation.
Region II contains an α-helix insertion that is not typical of PAS domains (31) along with a relatively extended loop. We designed mutations in this region solely guided by our model of the complex, which suggests that region II contributes to an interaction surface distinct from that of region I and adjacent to that of region III. We found that determinants in region II are also important for HSP90–sGCβ interaction. Deletion of the entire region abolished the interaction, whereas the E291A/E295A/L296D mutant showed substantially weaker affinity toward HSP90. Their location within the loop of region II suggests that this flexible segment is important for the HSP90 interaction while not excluding the possibility that the upstream helical portion also contributes to the binding interface.
Region III corresponds primarily to an unstructured linker region that connects the PAS and CC domains. Mutating aliphatic residues within region III, specifically substituting Leu residues with acidic residues, often but not always abrogated HSP90 binding to apo-sGCβ1(1–358). The HSP90-binding defect was observed for the L333D/L338D/L340D mutant. The L340D/I343D/L345D mutant also showed no detectable HSP90 binding in pure form but did exhibit partial HSP90 binding when expressed in mammalian cells and exhibited some NO-dependent catalytic activity, particularly when co-expressed with sGCα. In comparison, the region III mutant R335S/R336S/D342S bound well to HSP90 and had normal heme insertion. This is consistent with these particular charged side chains pointing away from the HSP90 interface, as they do in our model complex.
A common feature among all three regions is that surface-exposed aliphatic residues (e.g. Leu/Ile) make important contributions to the HSP90 interaction. This agrees with the concept that HSP90 prefers to bind flexible, conformation-reporting epitopes that may otherwise be buried in intraprotein or protein–protein interactions (32).
It remains to be elucidated how residues within the three regions may contribute to domain–domain interactions within sGCβ or between the subunits in the sGC heterodimer. We found that only substitutions within region I antagonized the natural binding affinity that exists between the sGCβ and sGCα subunits. In this regard, it is noteworthy that deleting regions I or II only partially diminished sGC heterodimer activation by BAY58 (Fig. 5B). This suggests that, despite the region I and II deletions, the conformational changes induced by BAY58 binding are for the most part still relayed from the HNOX domain to the catalytic domains of the sGC heterodimer. A conservative interpretation is that interdomain contacts that involve regions I or II help to support, but are not essential for, signal transmission. These concepts can now be further investigated.
A direct HSP90–apo-sGCβ interaction drives heme insertion
We were able to monitor heme insertion into FlAsH-TC-apo-sGCβ1(1–619) proteins in live cells to determine whether HSP90 binding corresponded to heme acquisition. Overall, we observed that the two processes correlate well. WT apo-sGCβ incorporated heme over a 15–30-min period, and a mutant with slightly improved HSP90-binding affinity (R335S/R336S/D342S) showed a slightly faster heme incorporation, suggesting that it is a gain-of-function mutant. In contrast, the four PAS domain mutants with the poorest HSP90 binding affinities (TC-sGCβ1(1–619) Δ265–271, Δ284–301, E291A/E295A/L296D, and L333D/L338D/L340D) showed no heme insertion over a 90–180-min period. Two PAS domain mutants with partly compromised HSP90 binding (L269D/I272S/V275D and H266A/H271A/I272D) showed detectable heme incorporation, but at rates slower than the WT. The acquisition of NO-stimulated sGCβ catalytic activity, which is a heme-dependent function (1), generally correlated well with mutant protein heme insertion and with HSP90-binding capabilities. The correlations between HSP90 binding, heme insertion, and gain of heme-dependent function argue strongly that direct interaction of HSP90 with apo-sGCβ drives the heme insertion, rather than any other HSP90 interactions that could conceivably be present within a multiprotein heme insertion complex.
Possible mechanism of HSP90 action in heme insertion
We currently hypothesize that HSP90 functions in the heme insertion through its conformational control of the apo-sGCβ client. If one adds heme to the cells, they will incorporate the heme into the HSP90-bound apo-sGCβ over a 15–30 min period, and inhibiting HSP90 ATPase activity during this period blocked the heme insertion. Thus, one can imagine HSP90 helping to form and stabilize heme-accepting protein conformations of apo-sGCβ and facilitating heme insertion in an ATP-dependent manner or, similarly, HSP90 facilitating shifts between protein conformations (partial folding/unfolding) that ultimately facilitate heme insertion in an ATP-dependent manner. In this way, we propose that HSP90 drives heme insertion into apo-sGCβ through an ATP-dependent process.
Besides sGCβ, it is intriguing to speculate that heme insertion into other known HSP90 clients, such as globins (33) and NO synthases (34), may also involve a direct complex formation with HSP90. These clients do not contain PAS domains, so their interactions with HSP90 would need to involve distinct protein regions. Our current study provides a template to investigate how HSP90 interaction facilitates heme insertion at the molecular level and how this process may be regulated by co-chaperones or post-translational modifications.
HSP90 controls two steps of sGC maturation
Heterodimer formation is the final step in sGC maturation and is critical because both the α and β subunits provide residues that form the active site and the substrate-binding site (11, 12). Heterodimer formation also permits cross-subunit communication that regulates catalytic activity in response to NO binding, S-nitrosation, and other signaling mechanisms within cells (12, 21, 35–37). Our previous work confirmed that two sGCβ subpopulations (heme-free and heme-bound) exist in cells and revealed that each has a distinct protein binding partner: apo-sGCβ is bound to HSP90, whereas holo-sGCβ either self-associates or forms a heterodimer with a sGCα subunit partner (17). Our current study clarifies how sGCβ interactions with HSP90 versus sGCα are governed.
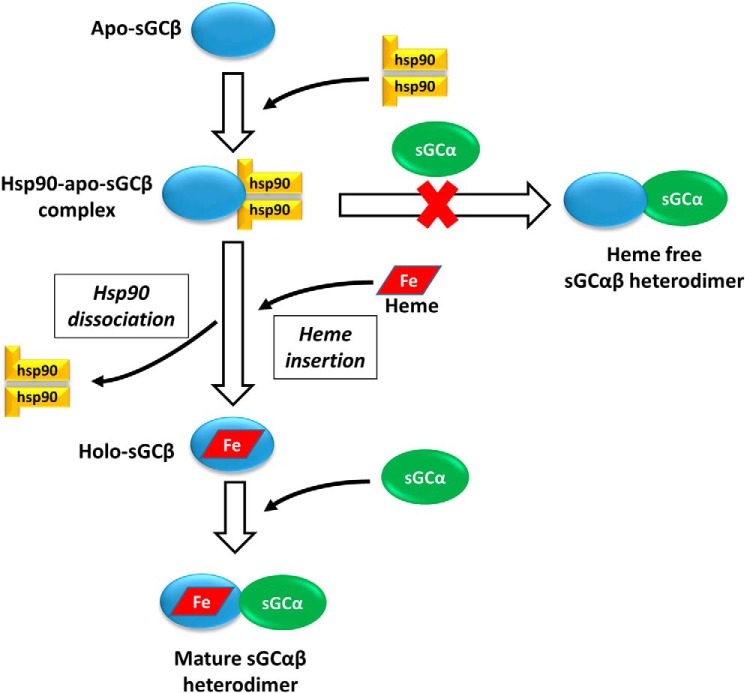
Surprisingly, most of the apo-sGCβ mutant proteins with defective HSP90 binding now associated with sGCα in cells to form heme-free heterodimers. This behavior has not been observed for WT apo-sGCβ (17) and implies that (i) unlike many other HSP90 client proteins (38–41), apo-sGCβ does not strictly require HSP90 binding to stabilize it or protect it from degradation in mammalian cells, and (ii) apo-sGCβ is inherently capable of binding with sGCα but does not do so in mammalian cells because it is instead sequestered by HSP90. Thus, in the context of sGC maturation, a heterodimer forms after heme insertion not because heme binding itself enables sGCβ to bind sGCα, but because HSP90 blocks premature heterodimer formation until heme is inserted into the apo-sGCβ. It is likely that heme insertion promotes conformational changes in sGCβ that favor HSP90 dissociation. The ability of HSP90 to block sGCα–sGCβ heterodimerization is consistent with it interacting directly with the sGCβ PAS domain, because the PAS domains of sGCβ and sGCα interact to form the heterodimer (42, 43). Thus, HSP90 and sGCα may compete for binding to the PAS domain of sGCβ. The fact that HSP90 also drives heme insertion into apo-sGCβ means that it governs sGC maturation in two ways: HSP90 blocks formation of a heme-free heterodimer and drives the heme insertion reaction of apo-sGCβ. This presents new regulatory points to modulate the total cellular pool of active sGC. A model for sGC maturation that incorporates these concepts is shown in Fig. 7.
Figure 7.
Bimodal regulation of sGC maturation by HSP90 in mammalian cells. When HSP90 binds to apo-sGCβ, it drives heme insertion into apo-sGCβ while also preventing its premature interaction with sGCα to form a heme-free, nonfunctional sGC heterodimer. Once heme insertion is complete, HSP90 dissociates from sGCβ, and this allows it to partner with sGCα to form a mature and functional sGC heterodimer.
Our work raises questions regarding the possible functions of apo-sGC in vivo. Prior studies showed that heme could be removed from purified sGC heterodimer with a weak detergent and that the basal activity of the heme-free sGC was greater than its heme-bound counterpart (23, 44, 45). Our work questions whether such heme-free sGC heterodimers could accumulate in mammalian cells, as we always find that apo-sGCβ is complexed with HSP90 and unable to associate with sGCα. When the H105F sGCβ heme-binding mutant was expressed in mammalian cells, it was found in complex with HSP90 and remained so even after the cells were provided heme (17). Tissues from knock-in mice bred to express the H105F sGCβ mutant in place of the WT exhibited a basal sGC activity that was higher than that of WT tissues, consistent with expression of heme-free sGCβ (46). Unfortunately, it was not determined whether the H105F sGCβ in the tissues was present as a homodimer, as a heterodimer, or in complex with HSP90. Of note, our previous study (17) showed that sGC activators like BAY58, which fill the heme-binding site, drive sGC homodimer or heterodimer formation in cells independent of heme or of HSP90. This also appeared to occur in the H105F sGCβ knock-in mice, as judged by an observed increase in tissue sGC activity upon treatment with BAY58 (46). These considerations provide a strong incentive for further studies of the biological nature and behaviors of apo-sGCβ in cells, tissues, animal models, and human physiology.
Conclusions
We tested the reliability of an HSP90–sGCβ model complex that could become useful to study structure–function aspects of heme insertion into apo-sGCβ. Further, we determined that when the complex forms in cells, it drives heme insertion while actively preventing sGCα from binding to apo-sGCβ during sGC maturation until heme has been inserted. In this way, HSP90 may directly prevent the formation of NO-unresponsive, heme-free sGC heterodimers and further regulate the NO and cGMP signaling pathways.
Experimental procedures
General methods and materials
The TC-FlAsH In-Cell Tetracysteine Tag Detection Kit was obtained from Invitrogen. All other reagents and materials were obtained from sources reported elsewhere (17).
Antibodies
Rat polyclonal sGCβ1 and sGCα1 antibodies were obtained from Cayman Chemicals (Ann Arbor, MI) and Novus Biologicals (Centennial, CO), respectively. Rabbit polyclonal HSP90 antibody was purchased from Cell Signaling Technology (Danvers, MA).
Molecular biology
Bovine sGCβ1(1–358) DNA (which codes for the H-NOX domain, the PAS domain, and the N-terminal portion of the coiled-coil domain) (22) was subcloned into pMAL-c2X vector on the N-terminal side of maltose-binding protein (MBP) for bacterial expression. pET20b vector containing rat sGCβ1(1–385) with a C-terminal His6 tag was a generous gift from Dr. Michael Marletta (University of California, Berkeley). pCMV5 mammalian expression plasmid containing full-length rat sGCα1(1–690) or rat sGCβ1(1–619) (H-NOX domain, PAS domain, coiled-coil domain, and catalytic domain) was described previously (17). The bovine and rat sGCβ1 amino acid sequences are 98% identical, and none of the residues we modified for our study differ between the bovine and rat sequences. pCMV5 mammalian expression plasmid containing rat sGCβ1(1–690) with a CCPGCC sequence in residues 239–244 (TC-sGCβ1) was made by Genscript (Piscataway, NJ). Designed mutations listed in Table S1 were prepared by site-directed mutagenesis using primers (Invitrogen) with Q5 DNA polymerase (New England Biolabs, Ipswich, MA) on pMAL-sGCβ1(1–358), pET20b-sGCβ1(1–385), pCMV5-sGCβ1(1–619), and pCMV5-TC-sGCβ1(1–619). The sequences of mutations were confirmed by DNA sequencing at the Cleveland Clinic Genomics Core Facility. The pET15MHL plasmid containing full-length human HSP90β(1–724) was described previously (22).
Expression and purification of WT and mutant sGCβ1(1–358), sGCβ1(1–385), and HSP90β in E. coli
pMAL-sGCβ1(1–358) WT and mutants were each transformed into E. coli BL21 (DE3) for expression and purified by a method described previously with modifications (22). Briefly, sGCβ-MBP recombinant proteins were first purified on an amylose column using a method described elsewhere (47), and their purity was checked by SDS-PAGE. In some cases, further anion-exchange chromatography on a Q-Sepharose column with a liner gradient of 50 mm to 1 m NaCl in 40 mm EPPS, 10% glycerol, pH 8.5, buffer was used to further purify sGCβ-MBP and mutant proteins (22). pET20b-sGCβ1(1–385) was grown and purified using a method reported previously (24) with modifications. Briefly, WT and mutant sGCβ1(1–385) constructs were each transformed into BL21(DE3)-Rosetta cells for expression. Overnight cultures were grown at 37 °C in Luria broth and used to inoculate 500-ml expression cultures grown in Terrific broth. Expression cultures were grown until A600 ∼0.5 and subsequently cold-shocked for 10 min in an ice bath. Protein expression was induced with 250 μm isopropyl-d-1-thiogalactopyranoside for 20 h at 25 °C at 250 rpm. Cells were harvested by centrifugation and then resuspended in 40 mm EPPS, 150 mm NaCl, 10% glycerol, pH 7.5, buffer supplemented with protease inhibitors, benzonase, and lysozyme. Resuspended cells were lysed by sonication and clarified by centrifugation. Lysate was then purified with a gravity nickel-nitrilotriacetic acid column with a method described previously (22). Human HSP90β(1–724) with an N-terminal histidine tag was transformed into BL21 colon+ cells for expression and purified with a gravity column with nickel-nitrilotriacetic acid–agarose as described previously (22).
Fluorescence polarization measurements
FITC labeling of purified sGCβ1(1–358) proteins and fluorescence polarization measurements were performed using a method described previously (22). The resulting curves of WT, R335S/R336S/D342S, H266A/H271A/I272D, and L269D/I272S/V275D were fit in Origin Lab 8.0 (Origin Lab Corp., Northampton, MA) to calculate binding affinities.
Cell culture and transient transfection of cells
COS-7 cells were grown in DMEM + 10% FBS on fluorescence 96-well plates or on 10-cm dishes. Cultures (50–60% confluent) of COS-7 cells were transfected with expression constructs of rat sGCα1(1–690), rat sGCβ1(1–619), or TC-sGCβ1(1–619) by a method described previously (17). After 24 or 48 h of expression, cells were either harvested for supernatant production using a method described previously (17) or kept in 96-well plates. In some cases, a 400 μm concentration of the heme biosynthesis inhibitor succinyl acetone was added to COS-7 cells 72 h prior to transfection to enable production of apo-sGCβ1(1–619) (17).
Western blotting and immunoprecipitation
Western blotting and immunoprecipitation were performed by methods described previously (16). For immunoprecipitation, 500 μg of total cell supernatant protein was precleared with Protein G beads (20 μl) (16).
In vivo labeling of TC-sGCβ with FlAsH
TC-sGCβ1(1–619) expressed in COS-7 cells was labeled with FlAsH using a method described elsewhere (25) with modifications. Briefly, transfected cells grown in fluorescence 96-well plates were washed three times with phenol red–free DMEM containing 1 g/liter glucose. Cells were then incubated with a mixture of FlAsH/1,2-ethanedithiol made in Opti-MEM (final concentration 0.5 μm/12.5 μm) for 1 h at room temperature. Afterward, cells were washed three times with 250 μm 1,2-ethanedithiol in phenol red-free DMEM + 10% FBS and then three times with phenol red–free DMEM + 10% FBS. Phenol red–free DMEM + 10% serum and 5 μm heme was then added to the cells for monitoring heme insertion.
Measurement of fluorescence of FlAsH-TC-sGC in cells
We transiently transfected COS-7 cells that grew in fluorescence 96-well plates in heme-depleted medium in the presence of succinyl acetone to enable the production of mutant and WT TC-apo-sGCβ1(1–619). Two heme-binding mutants, TC-sGCβ1(1–619) H105F (16) and TC-sGCβ1(1–619) Y135A/R139A (27), were also used as negative controls. After 24 h of transfection, we labeled the TC-sGC1(1–619) proteins with FlAsH, added heme, and monitored the heme insertion over 3 h by following the fluorescence intensity of each FlAsH-TC-sGC protein with a Flexstation 3 microplate reader (Molecular Devices, San Jose, CA). FlAsH-TC-sGC proteins were excited at 488 nm, and emission was detected at 528 nm. 100 readings were taken for signal integration every 1 or 2 min.
cGMP enzyme–linked immunosorbent assay
cGMP formation and its concentration in cell supernatant assays was determined by a method described previously (17) with modifications. Briefly, immediately after lysing COS-7 cells, different amounts of cell supernatant were added to a reaction mixture containing 500 μm GTP alone or with 30 μm NO donor NOC-18, and these mixtures were incubated for 30 min at 37 °C. In some groups, 10 μm BAY58 was added in place of NO donor. The cGMP in the cell supernatant reactions was then quantified with a cGMP ELISA kit (Cell Signaling Technology, Danvers, MA).
Author contributions
Y. D. and D. J. S. conceptualization; Y. D., S. S., M. M. H., and S. M. data curation; Y. D., S. S., S. M., and D. J. S. formal analysis; Y. D. validation; Y. D. investigation; Y. D. and D. J. S. methodology; Y. D. writing-original draft; Y. D., S. M., and D. J. S. writing-review and editing; S. M. software; D. J. S. supervision; D. J. S. funding acquisition; D. J. S. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Andreas Papapetropoulos (University of Patras, Patras, Greece) for sharing the pCMV5-sGCα1 and pCMV5-sGCβ1 constructs, and we thank Dr. Michael Marletta (University of California, Berkeley) and Dr. Elsa Garcin (University of Maryland Baltimore County) for the sGCβ bacterial expression constructs. We also thank all members of the Stuehr laboratory for helpful advice.
This work was supported by National Institutes of Health Grants P01HL081064 and P01HL103453 (to D. J. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1 and Figs. S1–S5.
- sGC
- soluble guanylyl cyclase
- HSP90
- heat shock protein 90
- PAS
- Per-Arnt-Sim–like domain
- CC domain
- coiled-coil domain
- TC
- tetracysteine motif
- BAY58
- BAY58-2667 (4-[[(4-carboxybutyl)[2-[2-[[4-(2-phenylethyl)-phenyl]methoxy]phenyl]ethyl]amino]methyl]benzoic acid hydrochloride)
- EPPS
- 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid
- NOC-18
- 2,2′-(hydroxynitrosohydrazino)bis-ethanamine
- DMEM
- Dulbecco's modified Eagle's medium
- IP
- immunoprecipitation
- MBP
- maltose-binding protein
- FBS
- fetal bovine serum.
References
- 1. Denninger J. W., and Marletta M. A. (1999) Guanylate cyclase and the •NO/cGMP signaling pathway. Biochim. Biophys. Acta 1411, 334–350 10.1016/S0005-2728(99)00024-9 [DOI] [PubMed] [Google Scholar]
- 2. Martin E., Berka V., Tsai A. L., and Murad F. (2005) Soluble guanylyl cyclase: the nitric oxide receptor. Methods Enzymol. 396, 478–492 10.1016/S0076-6879(05)96040-0 [DOI] [PubMed] [Google Scholar]
- 3. Lucas K. A., Pitari G. M., Kazerounian S., Ruiz-Stewart I., Park J., Schulz S., Chepenik K. P., and Waldman S. A. (2000) Guanylyl cyclases and signaling by cyclic GMP. Pharmacol. Rev. 52, 375–414 [PubMed] [Google Scholar]
- 4. Dasgupta A., Bowman L., D'Arsigny C. L., and Archer S. L. (2015) Soluble guanylate cyclase: a new therapeutic target for pulmonary arterial hypertension and chronic thromboembolic pulmonary hypertension. Clin. Pharmacol. Ther. 97, 88–102 10.1002/cpt.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dupont L. L., Glynos C., Bracke K. R., Brouckaert P., and Brusselle G. G. (2014) Role of the nitric oxide-soluble guanylyl cyclase pathway in obstructive airway diseases. Pulm. Pharmacol. Ther. 29, 1–6 10.1016/j.pupt.2014.07.004 [DOI] [PubMed] [Google Scholar]
- 6. Ghosh A., Koziol-White C. J., Asosingh K., Cheng G., Ruple L., Groneberg D., Friebe A., Comhair S. A., Stasch J. P., Panettieri R. A. Jr, Aronica M. A., Erzurum S. C., and Stuehr D. J. (2016) Soluble guanylate cyclase as an alternative target for bronchodilator therapy in asthma. Proc. Natl. Acad. Sci. U.S.A. 113, E2355–E2362 10.1073/pnas.1524398113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stasch J. P., Pacher P., and Evgenov O. V. (2011) Soluble guanylate cyclase as an emerging therapeutic target in cardiopulmonary disease. Circulation 123, 2263–2273 10.1161/CIRCULATIONAHA.110.981738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. West A. R., and Tseng K. Y. (2011) Nitric oxide-soluble guanylyl cyclase-cyclic GMP signaling in the striatum: new targets for the treatment of Parkinson's disease? Front. Syst. Neurosci. 5, 55 10.3389/fnsys.2011.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wagenaar G. T., Hiemstra P. S., and Gosens R. (2015) Therapeutic potential of soluble guanylate cyclase modulators in neonatal chronic lung disease. Am. J. Physiol. Lung Cell Mol. Physiol. 309, L1037–L1040 10.1152/ajplung.00333.2015 [DOI] [PubMed] [Google Scholar]
- 10. Tabima D. M., Frizzell S., and Gladwin M. T. (2012) Reactive oxygen and nitrogen species in pulmonary hypertension. Free Radic. Biol. Med. 52, 1970–1986 10.1016/j.freeradbiomed.2012.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Derbyshire E. R., and Marletta M. A. (2012) Structure and regulation of soluble guanylate cyclase. Annu. Rev. Biochem. 81, 533–559 10.1146/annurev-biochem-050410-100030 [DOI] [PubMed] [Google Scholar]
- 12. Potter L. R. (2011) Guanylyl cyclase structure, function and regulation. Cell. Signal. 23, 1921–1926 10.1016/j.cellsig.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Montfort W. R., Wales J. A., and Weichsel A. (2017) Structure and activation of soluble guanylyl cyclase, the nitric oxide sensor. Antioxid. Redox Signal. 26, 107–121 10.1089/ars.2016.6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pellicena P., Karow D. S., Boon E. M., Marletta M. A., and Kuriyan J. (2004) Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases. Proc. Natl. Acad. Sci. U.S.A. 101, 12854–12859 10.1073/pnas.0405188101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Purohit R., Weichsel A., and Montfort W. R. (2013) Crystal structure of the α subunit PAS domain from soluble guanylyl cyclase. Protein Sci. 22, 1439–1444 10.1002/pro.2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ghosh A., and Stuehr D. J. (2012) Soluble guanylyl cyclase requires heat shock protein 90 for heme insertion during maturation of the NO-active enzyme. Proc. Natl. Acad. Sci. U.S.A. 109, 12998–13003 10.1073/pnas.1205854109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ghosh A., Stasch J. P., Papapetropoulos A., and Stuehr D. J. (2014) Nitric oxide and heat shock protein 90 activate soluble guanylate cyclase by driving rapid change in its subunit interactions and heme content. J. Biol. Chem. 289, 15259–15271 10.1074/jbc.M114.559393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ghosh A., and Stuehr D. J. (2017) Regulation of sGC via hsp90, cellular heme, sGC agonists, and NO: new pathways and clinical perspectives. Antioxid. Redox Signal. 26, 182–190 10.1089/ars.2016.6690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nedvetsky P. I., Meurer S., Opitz N., Nedvetskaya T. Y., Müller H., and Schmidt H. H. (2008) Heat shock protein 90 regulates stabilization rather than activation of soluble guanylate cyclase. FEBS Lett. 582, 327–331 10.1016/j.febslet.2007.12.025 [DOI] [PubMed] [Google Scholar]
- 20. Yetik-Anacak G., Xia T., Dimitropoulou C., Venema R. C., and Catravas J. D. (2006) Effects of hsp90 binding inhibitors on sGC-mediated vascular relaxation. Am. J. Physiol. Heart Circ. Physiol. 291, H260–H268 10.1152/ajpheart.01027.2005 [DOI] [PubMed] [Google Scholar]
- 21. Papapetropoulos A., Zhou Z., Gerassimou C., Yetik G., Venema R. C., Roussos C., Sessa W. C., and Catravas J. D. (2005) Interaction between the 90-kDa heat shock protein and soluble guanylyl cyclase: physiological significance and mapping of the domains mediating binding. Mol. Pharmacol. 68, 1133–1141 10.1124/mol.105.012682 [DOI] [PubMed] [Google Scholar]
- 22. Sarkar A., Dai Y., Haque M. M., Seeger F., Ghosh A., Garcin E. D., Montfort W. R., Hazen S. L., Misra S., and Stuehr D. J. (2015) Heat shock protein 90 associates with the Per-Arnt-Sim domain of heme-free soluble guanylate cyclase: implications for enzyme maturation. J. Biol. Chem. 290, 21615–21628 10.1074/jbc.M115.645515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin E., Sharina I., Kots A., and Murad F. (2003) A constitutively activated mutant of human soluble guanylyl cyclase (sGC): implication for the mechanism of sGC activation. Proc. Natl. Acad. Sci. U.S.A. 100, 9208–9213 10.1073/pnas.1633590100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Underbakke E. S., Iavarone A. T., and Marletta M. A. (2013) Higher-order interactions bridge the nitric oxide receptor and catalytic domains of soluble guanylate cyclase. Proc. Natl. Acad. Sci. U.S.A. 110, 6777–6782 10.1073/pnas.1301934110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoffmann L. S., Schmidt P. M., Keim Y., Hoffmann C., Schmidt H. H., and Stasch J. P. (2011) Fluorescence dequenching makes haem-free soluble guanylate cyclase detectable in living cells. PLoS One 6, e23596 10.1371/journal.pone.0023596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pan J., Yuan H., Zhang X., Zhang H., Xu Q., Zhou Y., Tan L., Nagawa S., Huang Z. X., and Tan X. (2017) Probing the molecular mechanism of human soluble guanylate cyclase activation by NO in vitro and in vivo. Sci. Rep. 7, 43112 10.1038/srep43112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schmidt P. M., Schramm M., Schröder H., Wunder F., and Stasch J. P. (2004) Identification of residues crucially involved in the binding of the heme moiety of soluble guanylate cyclase. J. Biol. Chem. 279, 3025–3032 10.1074/jbc.M310141200 [DOI] [PubMed] [Google Scholar]
- 28. Martin F., Baskaran P., Ma X., Dunten P. W., Schaefer M., Stasch J. P., Beuve A., and van den Akker F. (2010) Structure of cinaciguat (BAY 58–2667) bound to Nostoc H-NOX domain reveals insights into heme-mimetic activation of the soluble guanylyl cyclase. J. Biol. Chem. 285, 22651–22657 10.1074/jbc.M110.111559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krieg T., Liu Y., Rütz T., Methner C., Yang X. M., Dost T., Felix S. B., Stasch J. P., Cohen M. V., and Downey J. M. (2009) BAY 58–2667, a nitric oxide-independent guanylyl cyclase activator, pharmacologically post-conditions rabbit and rat hearts. Eur. Heart J. 30, 1607–1613 10.1093/eurheartj/ehp143 [DOI] [PubMed] [Google Scholar]
- 30. Nossaman B., Pankey E., and Kadowitz P. (2012) Stimulators and activators of soluble guanylate cyclase: review and potential therapeutic indications. Crit. Care Res. Pract. 2012, 290805 10.1155/2012/290805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Möglich A., Ayers R. A., and Moffat K. (2009) Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17, 1282–1294 10.1016/j.str.2009.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karagöz G. E., and Rüdiger S. G. (2015) Hsp90 interaction with clients. Trends Biochem. Sci. 40, 117–125 10.1016/j.tibs.2014.12.002 [DOI] [PubMed] [Google Scholar]
- 33. Ghosh A., Garee G., Sweeny E. A., Nakamura Y., and Stuehr D. J. (2018) Hsp90 chaperones hemoglobin maturation in erythroid and nonerythroid cells. Proc. Natl. Acad. Sci. U.S.A. 115, E1117–E1126 10.1073/pnas.1717993115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Harris M. B., Mitchell B. M., Sood S. G., Webb R. C., and Venema R. C. (2008) Increased nitric oxide synthase activity and Hsp90 association in skeletal muscle following chronic exercise. Eur. J. Appl. Physiol. 104, 795–802 10.1007/s00421-008-0833-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fernhoff N. B., Derbyshire E. R., Underbakke E. S., and Marletta M. A. (2012) Heme-assisted S-nitrosation desensitizes ferric soluble guanylate cyclase to nitric oxide. J. Biol. Chem. 287, 43053–43062 10.1074/jbc.M112.393892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beuve A. (2017) Thiol-based redox modulation of soluble guanylyl cyclase, the nitric oxide receptor. Antioxid. Redox Signal. 26, 137–149 10.1089/ars.2015.6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kollau A., Gesslbauer B., Russwurm M., Koesling D., Gorren A. C. F., Schrammel A., and Mayer B. (2018) Modulation of nitric oxide-stimulated soluble guanylyl cyclase activity by cytoskeleton-associated proteins in vascular smooth muscle. Biochem. Pharmacol. 156, 168–176 10.1016/j.bcp.2018.08.009 [DOI] [PubMed] [Google Scholar]
- 38. Sato S., Fujita N., and Tsuruo T. (2000) Modulation of Akt kinase activity by binding to Hsp90. Proc. Natl. Acad. Sci. U.S.A. 97, 10832–10837 10.1073/pnas.170276797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Müller L., Schaupp A., Walerych D., Wegele H., and Buchner J. (2004) Hsp90 regulates the activity of wild type p53 under physiological and elevated temperatures. J. Biol. Chem. 279, 48846–48854 10.1074/jbc.M407687200 [DOI] [PubMed] [Google Scholar]
- 40. Minet E., Mottet D., Michel G., Roland I., Raes M., Remacle J., and Michiels C. (1999) Hypoxia-induced activation of HIF-1: role of HIF-1α-Hsp90 interaction. FEBS Lett. 460, 251–256 10.1016/S0014-5793(99)01359-9 [DOI] [PubMed] [Google Scholar]
- 41. Lewis J., Devin A., Miller A., Lin Y., Rodriguez Y., Neckers L., and Liu Z. G. (2000) Disruption of hsp90 function results in degradation of the death domain kinase, receptor-interacting protein (RIP), and blockage of tumor necrosis factor-induced nuclear factor-κB activation. J. Biol. Chem. 275, 10519–10526 10.1074/jbc.275.14.10519 [DOI] [PubMed] [Google Scholar]
- 42. Rothkegel C., Schmidt P. M., Atkins D. J., Hoffmann L. S., Schmidt H. H., Schröder H., and Stasch J. P. (2007) Dimerization region of soluble guanylate cyclase characterized by bimolecular fluorescence complementation in vivo. Mol. Pharmacol. 72, 1181–1190 10.1124/mol.107.036368 [DOI] [PubMed] [Google Scholar]
- 43. Ma X., Sayed N., Baskaran P., Beuve A., and van den Akker F. (2008) PAS-mediated dimerization of soluble guanylyl cyclase revealed by signal transduction histidine kinase domain crystal structure. J. Biol. Chem. 283, 1167–1178 10.1074/jbc.M706218200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Foerster J., Harteneck C., Malkewitz J., Schultz G., and Koesling D. (1996) A functional heme-binding site of soluble guanylyl cyclase requires intact N-termini of α1 and β1 subunits. Eur. J. Biochem. 240, 380–386 10.1111/j.1432-1033.1996.0380h.x [DOI] [PubMed] [Google Scholar]
- 45. Ignarro L. J., Degnan J. N., Baricos W. H., Kadowitz P. J., and Wolin M. S. (1982) Activation of purified guanylate cyclase by nitric oxide requires heme: comparison of heme-deficient, heme-reconstituted and heme-containing forms of soluble enzyme from bovine lung. Biochim. Biophys. Acta 718, 49–59 10.1016/0304-4165(82)90008-3 [DOI] [PubMed] [Google Scholar]
- 46. Thoonen R., Cauwels A., Decaluwe K., Geschka S., Tainsh R. E., Delanghe J., Hochepied T., De Cauwer L., Rogge E., Voet S., Sips P., Karas R. H., Bloch K. D., Vuylsteke M., Stasch J. P., et al. (2015) Cardiovascular and pharmacological implications of haem-deficient NO-unresponsive soluble guanylate cyclase knock-in mice. Nat. Commun. 6, 8482 10.1038/ncomms9482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maina C. V., Riggs P. D., Grandea A. G. 3rd, Slatko B. E., Moran L. S., Tagliamonte J. A., McReynolds L. A., and Guan C. D. (1988) An Escherichia coli vector to express and purify foreign proteins by fusion to and separation from maltose-binding protein. Gene 74, 365–373 10.1016/0378-1119(88)90170-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.