Abstract
In this study we investigated the influence of oxygen availability on a phenotypic microtiter screen to identify new, natural product inhibitors of growth for the bovine mastitis-causing microorganisms; Streptococcus uberis, Staphylococcus aureus, and Escherichia coli. Mastitis is a common disease in dairy cattle worldwide and is a major cause of reduced milk yield and antibiotic usage in dairy herds. Prevention of bovine mastitis commonly relies on the application of teat disinfectants that contain either iodine or chlorhexidine. These compounds are used extensively in human clinical settings and increased tolerance to chlorhexidine has been reported in both Gram-positive and Gram-negative microorganisms. As such new, non-human use alternatives are required for the agricultural industry. Our screening was conducted under normoxic (20% oxygen) and hypoxic (<1% oxygen) conditions to mimic the conditions on teat skin and within the mammary gland respectively, against two natural compound libraries. No compounds inhibited E. coli under either oxygen condition. Against the Gram-positive microorganisms, 12 inhibitory compounds were identified under normoxic conditions, and 10 under hypoxic conditions. Data revealed a clear oxygen-dependency amongst compounds inhibiting growth, with only partial overlap between oxygen conditions. The oxygen-dependent inhibitory activity of a naturally occurring quinone, β-lapachone, against S. uberis was subsequently investigated and we demonstrated that this compound is only active under normoxic conditions with a minimum inhibitory concentration and minimum bactericidal concentration of 32 μM and kills via a reactive oxygen species-dependent mechanism as has been demonstrated in other microorganisms. These results demonstrate the importance of considering oxygen-availability in high-throughput inhibitor discovery.
Keywords: mastitis, oxygen-dependent, Streptococcus uberis, Staphylococcus aureus, antimicrobial, natural product inhibitors, β-lapachone
Introduction
Bovine mastitis is the most common reason for antibiotic use in the dairy industry worldwide (Mcdougall, 2014; Gomes and Henriques, 2016). Application of teat antiseptics commonly containing either chlorhexidine or iodine is highly effective in reducing transmission of bacteria hence controlling the risk of mastitis (Pankey et al., 1984; Williamson and Lacy-Hulbert, 2013). Globally, use of chlorhexidine as a teat disinfectant accounts for 10–25% of the market (Hemling, 2002) and increased tolerance to chlorhexidine has been reported in both Gram-positive (Alotaibi et al., 2017; Hardy et al., 2018) and Gram-negative hospital-associated microorganisms (Guo et al., 2015).
As antibiotic use globally continues to rise against a backdrop of spreading antimicrobial resistance, the use of medically relevant human-use antimicrobials in agriculture is coming under increasing scrutiny (O’Neill, 2014; Klein et al., 2018). Additionally, antibiotic therapy of mastitis may increase the risk of selection of resistant bacteria, and as some of the same classes of medically important antibiotics used in agriculture (e.g., β-lactams) are also used in human medicine, the emergence of resistant bacteria is concerning. Discovery of antimicrobial alternatives for animal, but not human use, so-called ‘Green Antimicrobials’ (Toutain et al., 2016), for the prevention, or treatment of bacterial infections, such as bovine mastitis, would greatly assist in reducing the global usage of medically important human-use antimicrobials.
Many bacterial species have been associated with bovine mastitis, but Streptococcus uberis, Staphylococcus aureus, and Escherichia coli are amongst the most common. Environmental bacteria are a major cause of disease in agriculture and are, by necessity, capable of growth and survival under a range of conditions. Streptococcus uberis, a Gram-positive environmental pathogen, is the main cause of clinical and subclinical bovine mastitis cases in the early dry period, and around calving in New Zealand (Williamson et al., 1995; Pankey et al., 1996; Mcdougall, 1998) and is a cause of bovine mastitis throughout the world (Hogan et al., 1989; Wilson et al., 1997; Phuektes et al., 2001). S. uberis colonizes cows and their environment and has been isolated from the skin, lips, tonsils, gut, genital tract, teat orifice and canal, infected udders of cows as well as in bedding and pasture from the dairy herd environment (King, 1981). These diverse locations require the bacterium to not only tolerate variations in conditions for environmental growth and persistence, but also in their disease-causing location, the mammary gland, where during infection low oxygen conditions are prevalent (Mayer et al., 1988; Lopez-Benavides et al., 2007). Thus, pathogens are subjected to changes in growth conditions depending on their location (Kromker et al., 2014) and these diverse conditions are rarely, if ever, optimal for growth. The last two decades have revealed conventional antimicrobial discovery approaches, e.g., target-based and phenotypic HTS, to be largely unsuccessful (Payne et al., 2007; Tommasi et al., 2015). Bacteria are routinely grown, and antimicrobial screening conducted, under conditions optimized for growth in the laboratory (oxygenation, temperature, pH, nutrient availability), even though conditions under which the target organisms cause disease may differ considerably from laboratory conditions and in fact, may be ‘sub-optimal’ for growth (Cooper, 2013). Variations in screening conditions have been shown to identify different inhibitors both in target-based and phenotypic screens (Miller et al., 2009; Dunn et al., 2015). Furthermore, it has recently been shown that apparently simple antimicrobial killing assays are very sensitive to variations in culture conditions and bacterial growth phase (Harms et al., 2017). Anti-infectives, therefore, must also be effective under the same diverse range of conditions to prevent or treat disease (Nathan et al., 2008). In the case of oxygenation this is particularly significant, as antimicrobial efficacy has recently been directly linked to bacterial respiration and energy production (Lobritz et al., 2015) and reduced oxygen concentrations have been demonstrated within the mammary gland of cows with mastitis (Mayer et al., 1988).
The aim of this investigation was to determine the effect of oxygen, the levels of which affect cellular energy production, on microtiter screening assays to identify new antimicrobials for agricultural usage. Our results demonstrated that there was a strong oxygen dependency on the inhibitory profiles of various antimicrobial compounds against S. uberis. To determine the underlying molecular basis of the oxygen dependency identified by our screening approach, we selected the compound β-lapachone which was inhibitory under normal oxygen concentrations, but not inhibitory under low oxygen concentrations, for further investigation.
Materials and Methods
Bacterial Strains and Growth Conditions
All growth media and reagents were purchased from Sigma-Aldrich (New Zealand) unless otherwise stated and were prepared according to the manufacturers’ specifications. The S. uberis strain used in this investigation was a clinical bovine mastitis isolate from the Manawatu-Wanganui region of New Zealand and was kindly provided by Dr. Olaf Bork (Mastaplex, New Zealand). Recently the genome of this isolate has been sequenced and deposited under Accession No. CP022435.1 and designated as S. uberis NZ01 (Taiaroa et al., 2018). S. uberis was routinely maintained on THBA and grown in THB at 37°C with agitation (200 rpm). Staphylococcus aureus BB255 (Berger-Bächi, 1983) and E. coli MG1655 (Bachmann, 1996) were grown under the same conditions in BHI and LB, respectively. 96-well microtiter plates were inoculated with an OD600 of 0.005 in a final volume of 200 μL of either THB (S. uberis), BHI (S. aureus) or LB (E. coli) and incubated overnight at 37°C with agitation (200 rpm). Low-oxygen growth conditions for microtiter 96-well plates were established using AnaeroPack 2.5 L containers and AnaeroGen sachets (Thermo Fisher Scientific) according to the manufacturer’s instructions. Growth was monitored via OD600 using a Varioskan Flash plate reader.
Phenotypic High-Throughput Screening Assays
Mastitis-causing bacteria were screened for inhibition of growth against the NPSII (NCI, Bethesda, MD, United States) library and the commercially available NPL (Selleckchem, United States). Library compounds, dissolved in DMSO, were added at a final concentration of 10 μM (or 20 μM for E. coli) to 96-well polystyrene microtiter plates (Thermo Fisher Scientific, New Zealand) inoculated with either Streptococcus uberis, S. aureus, or E. coli (final volume 200 μL). The concentration of DMSO did not exceed 2% (vol/vol) and control wells containing 2% (vol/vol) DMSO were included in each plate. Additional control wells included: un-inoculated medium (THB, BHI, or LB), and chlorhexidine gluconate (35 μM). All controls were included in technical triplicate. Plates were incubated overnight at 37°C with agitation (200 rpm) and OD600 were recorded using a Thermo Scientific Varioskan Flash plate reader. Assay performance was assessed by the statistical parameters Z and Z′, which take account of both data variability and signal window (Zhang et al., 1999). Z′ is a measure of the suitability of the assay set up and takes in to account the separation between negative and positive controls, Z-factor assess the impact of the screening library on the assay (Zhang et al., 1999). Statistical analysis was conducted in GraphPad Prism 8.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed in biological triplicate by measuring MICs by broth microdilution in polystyrene 96-well flat-bottom microtiter plates. MICs of β-lapachone, oleanolic acid, and chlorhexidine against S. uberis were determined. S. uberis cells grown overnight were diluted to an OD600 of 0.05 in THB and dispensed into microtiter wells. Test compounds were added to starting wells and two-fold serial dilutions were undertaken to generate a range of inhibitor concentrations in a final volume of 200 μL. Media, compound-free (untreated), and DMSO vehicle controls were included in each microtiter plate in triplicate. After 24 h incubation at 37°C and 200 rpm, the OD600 of wells were read using a Varioskan Flash plate reader. The MIC was reported as the lowest concentration of the test compound for which no growth occurred. To determine the MBC inhibited cells from the MIC assay were subsequently diluted 10-fold to 1 × 10–6, and 20 μL of each 10-fold dilution was spotted onto THBA plates.
Cell Killing Assays Against Exponentially Growing S. uberis
To investigate the kinetics of cell death induced by β-lapachone, cell killing assays were conducted with S. uberis, using the Miles-Misra drop-plate method to measure the viability of cells in response to β-lapachone challenge against both growing and stationary phase cells (Miles et al., 1938). To establish the cell killing kinetics of β-lapachone against growing cells, an overnight culture of stationary phase S. uberis cells (OD600 ∼ 2.0) was diluted in THB to an OD600 of 0.05, and grown to mid-exponential phase (OD600 0.5) at 37°C, with shaking (200 rpm). Once cells had reached mid-exponential phase the culture was split into 2 mL aliquots in a 7 mL glass bijou vial, and challenged with ×1 (32 μM), ×4 (128 μM), or ×8 (256 μM) the MIC of β-lapachone. An untreated control containing 1.28% (vol/vol) DMSO, equivalent to the amount of DMSO present in the ×8 sample, was also included. Following addition of the test compound, samples were obtained at 0.5, 1, 2, 4, and 6 h post-treatment. For all treatment groups, time point samples were serially diluted 10-fold in PBS, and 20 μL of each dilution was dropped in technical triplicate onto THBA plates. Plates were incubated at 37°C for 18 h. Following incubation, colonies were counted for each technical triplicate, and the mean cfu/mL was determined from at least two independent experiments.
Cell Killing Assays Against Stationary Phase S. uberis
To investigate the impact of β-lapachone on stationary phase S. uberis cells, an overnight culture of S. uberis stationary phase cells (OD600 ∼ 2.0) was washed twice in PBS by centrifugation at 10,000 × g, 25°C, for 5 min. Following washing, cells were resuspended in PBS to an OD600 of 0.05. 1 mL of the washed cells was used per challenge group (×1, ×4, or ×8 the MIC of β-lapachone). An untreated control containing 1.28% (vol/vol) DMSO, equivalent to the amount of DMSO present in the ×8 sample, was also included. The antioxidant L-Glutathione (50 mM) was prepared as a neutralized stock solution in distilled-H2O and was pre-incubated for 10-min in the cell suspensions, prior to the addition of β-lapachone. Cell killing kinetics were determined as previously described.
Results and Discussion
Growth of S. uberis, S. aureus, and E. coli for HTS was readily achieved in microtiter 96-well plates under both normoxic and hypoxic conditions, where hypoxic conditions are defined as <1% oxygen. Microtiter cultures of S. uberis, S. aureus, and E. coli typically reached an OD600 of 0.53, 1.2, and 0.9 respectively under normoxic conditions, and 0.33, 0.3–0.5, and 0.4 respectively under hypoxic conditions. The MICs of chlorhexidine gluconate against S. uberis and S. aureus displayed only minor variations between normal- and low-oxygen conditions: Supplementary Table S1. Under low-oxygen conditions, chlorhexidine MICs were approximately two- and four-fold higher for S. uberis and S. aureus, respectively. The MIC for E. coli (14.3 μg/mL) was several times higher than that of the Gram-positive organisms, but was not affected by oxygen levels.
To determine the effect of oxygen availability on the antimicrobial profile of these libraries, screening was conducted in microtiter plates, in duplicate, under both normoxic and hypoxic conditions. S. uberis, S. aureus and E. coli were screened against the well-characterized, NPSII library (120 compounds) and the commercially available NPL (131 compounds). The initial screening concentration for the library compounds was 20 μM and Z′ values (derived from positive and negative control wells) were 0.6–0.9 for all organisms under both low and normal-oxygen conditions, indicating excellent assay performance. However, we observed that the NPSII library contained many highly-colored compounds which, at the initial screening concentration of 20 μM, had an undesirable effect on the sample OD600 absorbance signals for S. uberis and S. aureus, though not E. coli. This was confirmed by statistical analysis as the Z-factors (<0) indicated a low separation band due to sample and control signal variation bands touching/overlapping (Zhang et al., 1999). To address the low Z-factor observed screening Gram-positives with the NPSII, we re-screened S. aureus under normal-oxygen conditions, as this assay had previously returned the lowest Z-factor, with a reduced compound concentration (10 μM). The reduced screening concentration had a marked effect on the sample signal distribution such that the Z-factor was improved from <0, indicating almost no confidence in the data, to a positive value (0.1), demonstrating that this assay was now suitable for HTS. This was not observed with the NPL screened against the Gram-positive organisms, where good to excellent mean Z′ and Z-factors were returned (e.g., S. aureus Z′ = 0.78, Z-factor = 0.70). The decision was then taken that all remaining Gram-positive organism screens would be conducted at a library compound concentration of 10 μM.
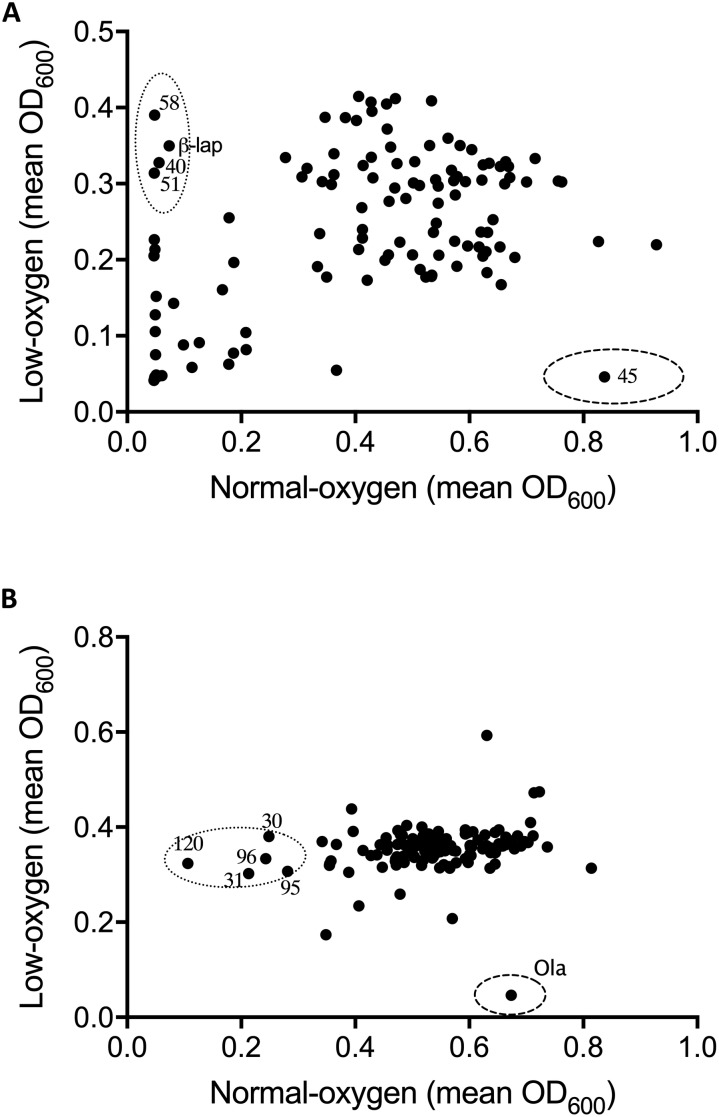
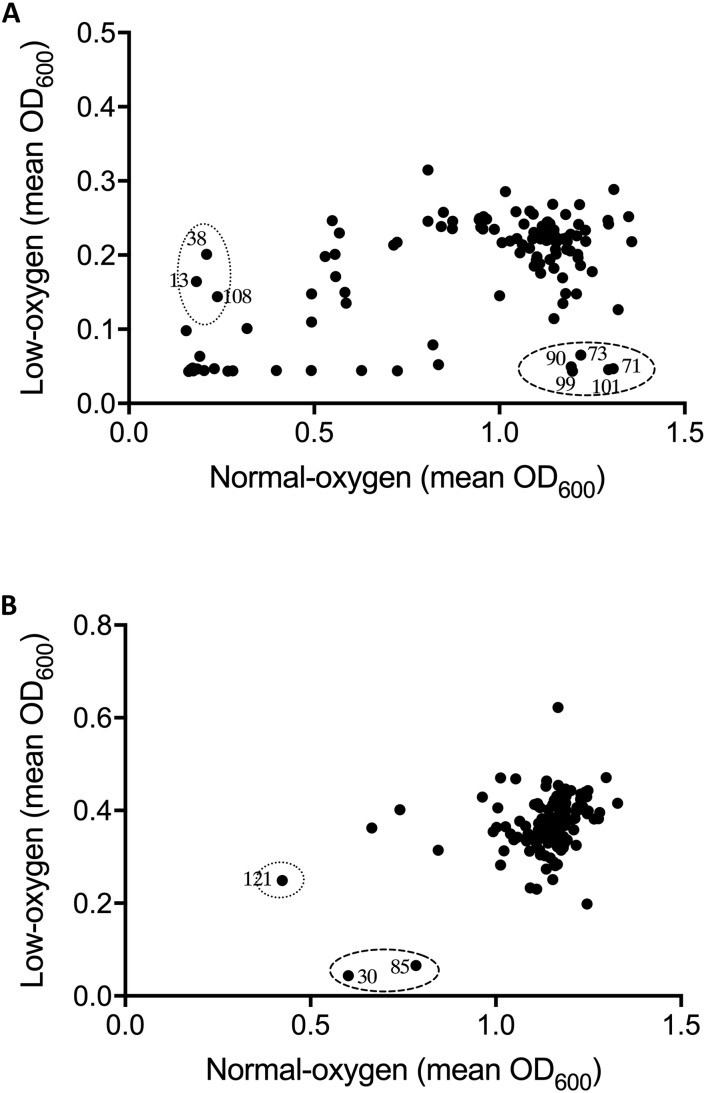
Following optimization of screening conditions S. uberis and S. aureus were then re-screened, under both oxygen conditions, against NPSII and NPL at 10 μM, whereas E. coli was screened at a final compound library concentration of 20 μM. Results from screening assays were qualitatively assessed in terms of the inhibition of growth of mastitis causing bacteria under normoxic conditions, hypoxic conditions, or both. Neither of the libraries screened inhibited the growth of E. coli under either oxygen condition. However, an oxygen-dependent inhibitory effect was observed with S. uberis and S. aureus (Figures 1, 2, respectively. A number of library compounds were only capable of inhibiting growth under normoxic conditions, while others only inhibited growth under hypoxic conditions (contained within the indicative dotted or dashed lines Figures 1, 2, respectively). Furthermore, a collection of compounds which appeared to be capable of working under both hypoxic and normoxic conditions were identified from the NPSII, but not the NPL library against S. uberis (Figure 1A) and S. aureus (Figure 2B). Compounds which were found to be active under both oxygen conditions against S. uberis and S. aureus included the antibiotics; Antibiotic X-536A (Bovatec R), Bactobolin, and Siomycin A, as well as the toxin 4-Ipomeanol. Several other inhibitory compounds were found to be active against S. uberis and S. aureus individually, however, compounds which were inhibitory under both oxygen conditions were not examined further in this study.
FIGURE 1.
Effect of oxygen availability on the antimicrobial profile of the natural product libraries NPSII (A), and NPL (B) against Streptococcus uberis NZ01. β-Lapachone (β-lap) and oleanolic acid (Ola) are indicated. Compounds that inhibited growth under normoxic conditions only are within dotted line, those inhibiting growth under hypoxic conditions are within dashed line. Results are the mean of two independent experiments.
FIGURE 2.
Effect of oxygen availability on the antimicrobial profile of the natural product libraries NPSII (A), and NPL (B) against Staphylococcus aureus BB255. Compounds which inhibited growth under normoxic conditions only are within dotted line, those inhibiting growth under hypoxic conditions are within the dashed line. Results are the mean of two independent experiments.
Oxygen-dependent inhibition of growth was clearly observed for S. uberis for some compounds in the NPSII (Figure 1A) library and fewer compounds in the NPL (Figure 1B) library. Though less pronounced than for S. uberis, inhibition of S. aureus was also observed for some compounds in the NPSII library, but few compounds in the NPL library were inhibitory: Supplementary Tables S2–S5. The different compositions of these two commercially available libraries clearly contributed to the oxygen-dependency of growth inhibition. The variation observed in the compounds that inhibited growth of S. uberis or S. aureus under different oxygen conditions also suggests that the composition of libraries for antimicrobial screening should be carefully considered, based on both the final usage conditions and key target pathogens.
To further investigate the oxygen-dependency observed in our screen, we selected two inhibitory compounds (β-lapachone and oleanolic acid) that were active against S. uberis under differing oxygen availability. β-Lapachone is an ortho-naphthoquinone with antitumor (Silvers et al., 2017), antibacterial (Cruz et al., 1978; Pereira et al., 2006), antiparasitic (Salas et al., 2008; Dos Anjos et al., 2016), and antimalarial (De Andrade-Neto et al., 2004) properties dependent on the activity of NAD(P)H:quinone oxidoreductase (Pink et al., 2000; Bey et al., 2007). In Bacillus subtilis and B. stearothermophilus β-lapachone increases the generation of a superoxide anion and hydrogen peroxide (H2O2) (Cruz et al., 1978). Based on these previous studies it is therefore likely that β-lapachone also exerts its bactericidal effects against S. uberis by a ROS-dependent mechanism. Consistent with this mechanism we observed that β-lapachone exerts its inhibitory and bactericidal activity against S. uberis growing under normoxic conditions with a MIC and MBC of 32 μM, comparable to the screening concentration used of 10 μM. To demonstrate the strict oxygen requirement for the inhibitory effect of β-lapachone, MIC assays were conducted under low-oxygen conditions. The resulting hypoxic MIC of >256 μM is in agreement with the lack of observed activity in the hypoxic screen. Further investigation of the oxygen-dependent antimicrobial activity of β-lapachone was undertaken against the beta-hemolytic mastitis-causing pathogens Streptococcus dysgalactiae and Streptococcus agalactiae. As we observed for S. uberis, β-lapachone inhibited S. dysagalactiae and S. agalactiae under normoxic conditions with MIC values of 32 and 64 μM respectively. However, under hypoxic conditions no (≤256 μM) β-lapachone associated antimicrobial activity was observed (Table 1). Oleanolic acid, a pentacyclic triterpenoid, inhibited S. uberis growth only under low-oxygen conditions, with a MIC of 128 μM, far higher than the 10 μM used in the compound library screen. The reason for this discrepancy between the measured MIC, and the concentration used in the library screen is not clear, but demonstrates the need to further validate initial results from HTS using MIC and MBC assays. Given the high MIC against S. uberis and that the mechanism of action of oleanolic acid against Streptococcus mutans UA159 (Park et al., 2017) has already been characterized, we did not investigate this compound further.
TABLE 1.
Effect of oxygen on the minimum inhibitory, and bactericidal concentration of β-lapachone against three mastitis-causing streptococci.
| β-Lapachone | ||||
| MIC (μM) | MBC (μM) | |||
| Normoxic | Hypoxic | Normoxic | Hypoxic | |
| Streptococcus uberis | 32 | >256 | 32 | >256 |
| Streptococcus dysgalactiae | 32 | >256 | 64 | >256 |
| Streptococcus agalactiae | 64 | 256 | 64 | 256 |
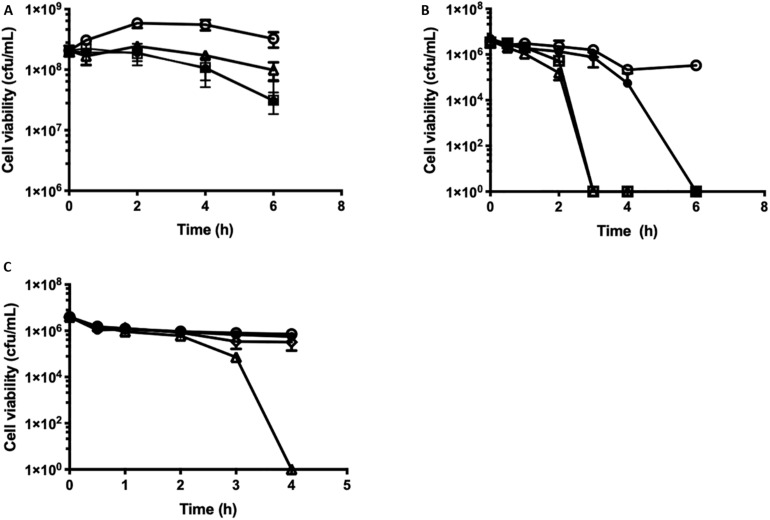
The kinetics of β-lapachone mediated cell killing were investigated by time-dependent cell viability assays against S. uberis. Growing cells, OD600 0.5 (2 × 108 cfu/mL) were challenged with increasing concentrations of β-lapachone, and the viability of the cells was determined (Figure 3A). The addition of ×1–8 MIC (32–256 μM) of β-lapachone did not induce cell lysis, as observed by the lack of change in the optical density readings between challenged and untreated cells. At ×8 MIC (256 μM), β-lapachone challenge exerted a bacteriostatic effect (Figure 3A) against exponentially growing cells with cell numbers reduced by 1-log10 in 6 h. No viable cells were detected 24 h post-treatment, indicating that the bactericidal activity of β-lapachone against growing cells is dependent upon duration of exposure. However, when PBS-washed stationary phase cells (4 × 106 cfu/mL) were treated with β-lapachone a more pronounced bactericidal effect was observed (Figure 3B). Six hours post-treatment, ×1 MIC (32 μM) killed all viable cells, and 3 h post-treatment ×4 MIC (128 μM) killed all viable cells, indicating that stationary phase S. uberis cells in PBS are sensitive to the action of β-lapachone and are unable to respond to challenge by ROS. The bactericidal activity observed with β-lapachone against stationary phase cells could be prevented by the addition of the antioxidant glutathione (50 mM), confirming a ROS-dependent mechanism of killing as has been previously described for β-lapachone (Figure 3C) (Cruz et al., 1978). Protection against β-lapachone-induced ROS by glutathione has also been demonstrated in cancer cells (Park et al., 2014).
FIGURE 3.
Time-dependent cell killing assays against growing and stationary phase cells of S. uberis in response to β-lapachone. (A) Growing Streptococcus uberis NZ01 cells at OD600 0.5 (2 × 108 cfu/mL) or (B) PBS-washed stationary phase cells at OD600 0.05 (4 × 106 cfu/mL) were challenged as follows with: ×1 (closed circles), ×4 (open squares), and ×8 (open triangles) MIC of β-lapachone, or untreated (open circles). (C) PBS-washed stationary phase S. uberis cells at OD600 0.05 (3.93 × 106 cfu/mL) were challenged as follows with: glutathione (50 mM) (closed circles), β-lapachone (×8 MIC) (open triangles), and β-lapachone (×8 MIC) with glutathione (50 mM) (open diamonds), or untreated (open circles). Error bars represent the standard deviation of the mean from three biological replicates. Where no error bars are visible, error bars are smaller than the symbol size. All cfu/mL determinations were performed in technical triplicate.
Examination of the S. uberis NZ01 genome (Accession No. CP022435.1), reveals that the organism has genes for detoxification of ROS, including one annotated superoxide dismutase for the conversion of to H2O2 and O2, as well as AhpCF alkyl hydroperoxide reductase (Taiaroa et al., 2018), though the activity of these enzymes in S. uberis NZ01 have not yet been investigated. Analysis of the S. uberis NZ01 genome also reveals that to mitigate the H2O2 produced by superoxide dismutase, NZ01 encodes a gene for the flavoprotein NADH oxidase. NADH oxidase is essential in the normoxic metabolism of S. pyogenes (Gibson et al., 2000), and plays a key role in the oxidative stress tolerance of S. suis (Zheng et al., 2017).
Conclusion
Growth and screening conditions both impact the susceptibility of microorganisms to antimicrobials (Payne et al., 2007; Tommasi et al., 2015; Harms et al., 2017). Here, we show that oxygen levels in phenotypic screening assays had a profound effect on the ability of natural products to inhibit the growth of two mastitis-causing organisms S. uberis and S. aureus. β-Lapachone was identified as an oxygen-dependent inhibitor of S. uberis, which exerted a time-dependent bactericidal effect, most likely mediated by accumulation of ROS. This work demonstrates that in the search for new antimicrobials exclusively for use in agriculture and with no human application, researchers should carefully consider both screening and end-usage conditions.
Data Availability
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
SF, GC, AH, and KB conceived and designed the research. GC and AH supervised the project. SF, AM, EVZ, and C-YC performed the research. SF, DR, MB, SM, GC, and AH analyzed the data. SF and AH wrote the manuscript. All authors contributed to the manuscript revision, read, and approved the submitted version.
Conflict of Interest Statement
KB is CEO of Deosan Ltd., manufacturers of TeatX, a chlorhexidine-based teat disinfectant. This work was, in part, funded by Deosan Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dave Malcolm for his assistance with the manuscript and to Alex Krause for the technical assistance. Streptococcus uberis NZ01 was kindly supplied by Dr. Olaf Bork (Mastaplex, New Zealand).
Abbreviations
- BHI
brain heart infusion
- DMSO
dimethyl sulfoxide
- HTS
high-throughput screening
- LB
Luria-Bertani
- MBC
minimum bactericidal concentration
- MIC
minimum inhibitory concentration
- NPL
Natural Product Library
- NPSII
Natural Products Set II
- OD600
optical density 600 nm
- PBS
phosphate buffered saline
- ROS
reactive oxygen species
- THBA
Todd-Hewitt agar
- THB
Todd-Hewitt Broth
- vol/vol
volume/volume
Footnotes
Funding. This work was supported by the Agricultural and Marketing Research Development Trust (New Zealand) Agribusiness (grant number A16012) and Deosan, Ltd. (Waharoa, New Zealand).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01995/full#supplementary-material
References
- Alotaibi S. M. I., Ayibiekea A., Pedersen A. F., Jakobsen L., Pinholt M., Gumpert H., et al. (2017). Susceptibility of vancomycin-resistant and -sensitive Enterococcus faecium obtained from Danish hospitals to benzalkonium chloride, chlorhexidine and hydrogen peroxide biocides. J. Med. Microbiol. 66 1744–1751. 10.1099/jmm.0.000642 [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. (1996). “Derivations and Genotypes of Some Mutant Derivatives of Escherichia coli K-12,” in Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, ed. Neidhardt F. C. (Washington, DC: ASM Press; ), 2460–2488. [Google Scholar]
- Berger-Bächi B. (1983). Insertional inactivation of staphylococcal methicillin resistance by Tn551. J. Bacteriol. 154 479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bey E. A., Bentle M. S., Reinicke K. E., Dong Y., Yang C.-R., Girard L., et al. (2007). An NQO1- and PARP-1-mediated cell death pathway induced in non-small-cell lung cancer cells by β-lapachone. Proc. Natl. Acad. Sci. U.S.A. 104 11832–11837. 10.1073/pnas.0702176104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper C. B. (2013). Development of Mycobacterium tuberculosis whole cell screening hits as potential antituberculosis agents. J. Med. Chem. 56 7755–7760. 10.1021/jm400381v [DOI] [PubMed] [Google Scholar]
- Cruz F. S., Docampo R., Boveris A. (1978). Generation of superoxide anions and hydrogen peroxide from β-lapachone in bacteria. Antimicrob. Agents Chemother. 14 630–633. 10.1128/aac.14.4.630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Andrade-Neto V. F., Goulart M. O., Da Silva Filho J. F., Da Silva M. J., Pinto Mdo C., Pinto A. V., et al. (2004). Antimalarial activity of phenazines from lapachol, β-lapachone and its derivatives against Plasmodium falciparum in vitro and Plasmodium berghei in vivo. Bioorg. Med. Chem. Lett. 14 1145–1149. 10.1016/j.bmcl.2003.12.069 [DOI] [PubMed] [Google Scholar]
- Dos Anjos D. O., Sobral Alves E. S., Goncalves V. T., Fontes S. S., Nogueira M. L., Suarez-Fontes A. M., et al. (2016). Effects of a novel β-lapachone derivative on Trypanosoma cruzi: parasite death involving apoptosis, autophagy and necrosis. Int. J. Parasitol. Drugs Drug Resist. 6 207–219. 10.1016/j.ijpddr.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn E. A., Cook G. M., Heikal A. (2015). Comparison of lipid and detergent enzyme environments for identifying inhibitors of membrane-bound energy-transducing proteins. J. Microbiol. Methods 120 41–43. 10.1016/j.mimet.2015.11.012 [DOI] [PubMed] [Google Scholar]
- Gibson C. M., Mallett T. C., Claiborne A., Caparon M. G. (2000). Contribution of NADH oxidase to aerobic metabolism of Streptococcus pyogenes. J. Bacteriol. 182 448–455. 10.1128/jb.182.2.448-455.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes F., Henriques M. (2016). Control of bovine mastitis: old and recent therapeutic approaches. Curr. Microbiol. 72 377–382. 10.1007/s00284-015-0958-8 [DOI] [PubMed] [Google Scholar]
- Guo W., Shan K., Xu B., Li J. (2015). Determining the resistance of carbapenem-resistant Klebsiella pneumoniae to common disinfectants and elucidating the underlying resistance mechanisms. Pathog. Glob. Health 109 184–192. 10.1179/2047773215Y.0000000022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy K., Sunnucks K., Gil H., Shabir S., Trampari E., Hawkely P., et al. (2018). Increased usage of antiseptics is associated with reduced susceptibility in clinical isolates of Staphylococcus aureus. mBio 9 e894–18. 10.1128/mBio.00894-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A., Fino C., Sorensen M. A., Semsey S., Gerdes K. (2017). Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. mBio 8 e1964–17. 10.1128/mBio.01964-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemling T. C. (2002). “Teat Condition - Prevention and cure through teat dips,” in Proceedings of the British Mastitis Conference, (Brockworth: Institute for Animal Health/Milk Development Council; ), 1–14. [Google Scholar]
- Hogan J. S., Smith K. L., Hoblet K. H., Schoenberger P. S., Todhunter D. A., Hueston W. D., et al. (1989). Field survey of clinical mastitis in low somatic cell count herds. J. Dairy Sci. 72 1547–1556. 10.3168/jds.s0022-0302(89)79266-3 [DOI] [PubMed] [Google Scholar]
- King J. S. (1981). Streptococcus uberis: a review of its role as a causative organism of bovine mastitis 1. characteristics of the organism. Br. Vet. J. 137 36–52. 10.1016/s0007-1935(17)31786-4 [DOI] [PubMed] [Google Scholar]
- Klein E. Y., Van Boeckel T. P., Martinez E. M., Pant S., Gandra S., Levin S. A., et al. (2018). Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. U.S.A. 115 E3463–E3470. 10.1073/pnas.1717295115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kromker V., Friederike R., Jan-Hendrik P., Grabowski N. (2014). Bovine Streptococcus uberis intramammary infections and mastitis. Clin. Microbiol. 3:4. [Google Scholar]
- Lobritz M. A., Belenky P., Porter C. B. M., Gutierrez A., Yang J. H., Schwarz E. G., et al. (2015). Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. U.S.A. 112 8173–8180. 10.1073/pnas.1509743112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Benavides M. G., Williamson J. H., Pullinger G. D., Lacy-Hulbert S. J., Cursons R. T., Leigh J. A. (2007). Field observations on the variation of Streptococcus uberis populations in a pasture-based dairy farm. J. Dairy Sci. 90 5558–5566. 10.3168/jds.2007-0194 [DOI] [PubMed] [Google Scholar]
- Mayer S. J., Waterman A. E., Keen P. M., Craven N., Bourne F. J. (1988). Oxygen concentration in milk of healthy and mastitic cows and implications of low oxygen tension for the killing of Staphylococcus aureus by bovine neutrophils. J. Dairy Res. 55 513–519. 10.1017/s0022029900033288 [DOI] [PubMed] [Google Scholar]
- Mcdougall S. (1998). Efficacy of two antibiotic treatments in curing clinical and subclinical mastitis in lactating dairy cows. N. Z. Vet. J. 46 226–232. 10.1080/00480169.1998.36094 [DOI] [PubMed] [Google Scholar]
- Mcdougall S. (2014). “Current prescribing for mastitis,” in Proceedings of the Society of Dairy Cattle Veterinarians of the NZVA Annual Conference, (Hamilton: VetLearn Foundation; ). [Google Scholar]
- Miles A. A., Misra S. S., Irwin J. O. (1938). The estimation of the bactericidal power of the blood. J. Hyg. 38 732–749. 10.1017/s002217240001158x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. H., Nisa S., Dempsey S., Jack C., O’toole R. (2009). Modifying culture conditions in chemical library screening identifies alternative inhibitors of mycobacteria. Antimicrob. Agents Chemother. 53 5279–5283. 10.1128/AAC.00803-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C., Gold B., Lin G., Stegman M., De Carvalho L. P. S., Vandal O., et al. (2008). A philosophy of anti-infectives as a guide in the search for new drugs for tuberculosis. Tuberculosis 88 S25–S33. 10.1016/S1472-9792(08)70034-9 [DOI] [PubMed] [Google Scholar]
- O’Neill J. (2014). Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. Review on Antimicrobial Resistance. Available at: https://amr-review.org/Publications.html (accessed April 8, 2019). [Google Scholar]
- Pankey J. W., Eberhart R. J., Cuming A. L., Daggett R. D., Farnsworth R. J., Mcduff C. K. (1984). Uptake on postmilking teat antisepsis. J. Dairy Sci. 67 1336–1353. 10.3168/jds.s0022-0302(84)81443-5 [DOI] [PubMed] [Google Scholar]
- Pankey J. W., Pankey P. B., Barker R. M., Williamson J. H., Woolford M. W. (1996). The prevalence of mastitis in primiparous heifers in eleven Waikato dairy herds. N. Z. Vet. J. 44 41–44. 10.1080/00480169.1996.35932 [DOI] [PubMed] [Google Scholar]
- Park E. J., Min K. J., Lee T. J., Yoo Y. H., Kim Y. S., Kwon T. K. (2014). β-Lapachone induces programmed necrosis through the RIP1-PARP-AIF-dependent pathway in human hepatocellular carcinoma SK-Hep1 cells. Cell Death Dis. 5:e1230. 10.1038/cddis.2014.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-N., Lim Y. K., Choi M.-H., Cho E., Bang I. S., Kim J. M., et al. (2017). Antimicrobial mechanism of oleanolic and ursolic acids on Streptococcus mutans UA159. Curr. Microbiol. 75 11–19. 10.1007/s00284-017-1344-5 [DOI] [PubMed] [Google Scholar]
- Payne D. J., Gwynn M. N., Holmes D. J., Pompliano D. L. (2007). Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6 29–40. 10.1038/nrd2201 [DOI] [PubMed] [Google Scholar]
- Pereira E. M., Machado Tde B., Leal I. C., Jesus D. M., Damaso C. R., Pinto A. V., et al. (2006). Tabebuia avellanedae naphthoquinones: activity against methicillin-resistant staphylococcal strains, cytotoxic activity and in vivo dermal irritability analysis. Ann. Clin. Microbiol. Antimicrob. 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuektes P., Mansell P. D., Dyson R. S., Hooper N. D., Dick J. S., Browning G. F. (2001). Molecular epidemiology of Streptococcus uberis isolates from dairy cows with mastitis. J. Clin. Microbiol. 39 1460–1466. 10.1128/jcm.39.4.1460-1466.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pink J. J., Planchon S. M., Tagliarino C., Varnes M. E., Siegel D., Boothman D. A. (2000). NAD(P)H:Quinone oxidoreductase activity is the principal determinant of beta-lapachone cytotoxicity. J. Biol. Chem. 275 5416–5424. 10.1074/jbc.275.8.5416 [DOI] [PubMed] [Google Scholar]
- Salas C., Tapia R. A., Ciudad K., Armstrong V., Orellana M., Kemmerling U., et al. (2008). Trypanosoma cruzi: activities of lapachol and α- and β-lapachone derivatives against epimastigote and trypomastigote forms. Bioorg. Med. Chem. 16 668–674. 10.1016/j.bmc.2007.10.038 [DOI] [PubMed] [Google Scholar]
- Silvers M. A., Deja S., Singh N., Egnatchik R. A., Sudderth J., Luo X., et al. (2017). The NQO1 bioactivatable drug, β-lapachone, alters the redox state of NQO1+ pancreatic cancer cells, causing perturbation in central carbon metabolism. J. Biol. Chem. 292 18203–18216. 10.1074/jbc.m117.813923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiaroa G., Harbison-Price N., Ferguson S. A., Carter G. P., Williamson D. A., Macknight R. C., et al. (2018). Complete genome sequence of a New Zealand isolate of the bovine pathogen Streptococcus uberis. Genome Announc. 6 e119–18. 10.1128/genomeA.00119-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasi R., Brown D. G., Walkup G. K., Manchester J. I., Miller A. A. (2015). ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 14 529–542. 10.1038/nrd4572 [DOI] [PubMed] [Google Scholar]
- Toutain P. L., Ferran A. A., Bousquet-Melou A., Pelligand L., Lees P. (2016). Veterinary medicine needs new green antimicrobial drugs. Front. Microbiol. 7:1196. 10.3389/fmicb.2016.01196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson J. H., Lacy-Hulbert S. J. (2013). Effect of disinfecting teats post-milking or pre- and post-milking on intramammary infection and somatic cell count. N. Z. Vet. J. 61 262–268. 10.1080/00480169.2012.751576 [DOI] [PubMed] [Google Scholar]
- Williamson J. H., Woolford M. W., Day A. M. (1995). The prophylactic effect of a dry-cow antibiotic against Streptococcus uberis. N. Z. Vet. J. 43 228–234. 10.1080/00480169.1995.35898 [DOI] [PubMed] [Google Scholar]
- Wilson D. J., Gonzalez R. N., Das H. H. (1997). Bovine mastitis pathogens in New York and Pennsylvania: prevalence and effects on somatic cell count and milk production. J. Dairy Sci. 80 2592–2598. 10.3168/jds.s0022-0302(97)76215-5 [DOI] [PubMed] [Google Scholar]
- Zhang J. H., Chung T. D. Y., Oldenburg K. R. (1999). A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomolecular Screen. 4 67–73. 10.1177/108705719900400206 [DOI] [PubMed] [Google Scholar]
- Zheng C., Ren S., Xu J., Zhao X., Shi G., Wu J., et al. (2017). Contribution of NADH oxidase to oxidative stress tolerance and virulence of Streptococcus suis serotype 2. Virulence 8 53–65. 10.1080/21505594.2016.1201256 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.