Abstract
We present a massive investigation into the genetic basis of human lifespan. Beginning with a genome-wide association (GWA) study using a de-identified snapshot of the unique AncestryDNA database – more than 300,000 genotyped individuals linked to pedigrees of over 400,000,000 people – we mapped six genome-wide significant loci associated with parental lifespan. We compared these results to a GWA analysis of the traditional lifespan proxy trait, age, and found only one locus, APOE, to be associated with both age and lifespan. By combining the AncestryDNA results with those of an independent UK Biobank dataset, we conducted a meta-analysis of more than 650,000 individuals and identified fifteen parental lifespan-associated loci. Beyond just those significant loci, our genome-wide set of polymorphisms accounts for up to 8% of the variance in human lifespan; this value represents a large fraction of the heritability estimated from phenotypic correlations between relatives.
Keywords: GWAS, human, lifespan
A great deal of effort has been exerted to quantify the variation in human lifespan that is attributable to either genetic (Vijg and Campisi 2008) or environmental factors (Kitagawa and Hauser 1973; Crimmins and Saito 2001; Chetty et al. 2016). The magnitude of the genetic contribution to variation in human lifespan, i.e., its heritability, can be difficult to measure, and is confounded by phenomena such as assortative mating and sociocultural inheritance; nonetheless, we have previously estimated it to be under 10% (Ruby et al. 2018). Beyond genetics, numerous sociological and economic factors are known to affect life expectancy at both the level of the individual (e.g., income, education level, employment status, marital status) and the community (e.g., income inequality, access to health care) (Braveman et al. 2005; Murray et al. 2006; Jha et al. 2006; Backlund et al. 2007). Further, race as a social construct affects many of these socioeconomic factors that influence life expectancy (C. J. L. Murray et al. 2006; Backlund et al. 2007; Chetty et al. 2016). Genetic population structure and race are neither equivalent nor fully-independent of one another, confounding their effects. Similarly, the intermingling of genetic with sociocultural inheritance confounds the isolation of genetic contributions to variance in lifespan.
Our goal is to extend these efforts to find specific genetic variants affecting lifespan. The standard tool for elucidating the genetic architecture of polygenic traits is the genome-wide association (GWA) study. Usually, GWA studies test whether there exists a correlation between individuals phenotyped for the trait of interest and genotyped at a panel of common SNP variants. As a phenotype, lifespan challenges this approach: genotypes are generally gathered from living persons, whereas lifespan (total elapsed time between birth and death) is a property of deceased persons. Due to this challenge, current age has been used as a lifespan proxy trait in many human-aging GWAS (Schächter et al. 1994; Willcox et al. 2008; Newman et al. 2010; Soerensen et al. 2010; Deelen et al. 2011; Nebel et al. 2011; Beekman et al. 2013; Soerensen et al. 2013; Sebastiani et al. 2013; Deelen et al. 2014; Broer et al. 2015; Sebastiani et al. 2017). Logically, alleles that reduce lifespan should become depleted in older individuals, and vice-versa for alleles that extend lifespan. However, the retrospective sampling of young and old individuals potentially captures other differences between birth cohorts, and it is unclear whether this is an appropriate proxy for lifespan GWA analyses.
An alternative approach to investigate the genetic basis of lifespan is to prospectively measure the lifespan of deceased parents from specific birth cohorts and use the genotypes of their offspring in an association mapping analysis. The imperfect sharing of genotype between a parent and child dilutes the strength of statistical associations in such a study, thereby requiring very large sample sizes to identify high-confidence variants. This method was used in analyses of the United Kingdom Biobank (UKB) to identify loci significantly associated with parental lifespan (Pilling et al. 2016; Joshi et al. 2016; McDaid et al. 2017; Mostafavi et al. 2017; Pilling et al. 2017; Joshi et al. 2017). These candidate lifespan loci have yet to be tested in an independent cohort of significant size.
We investigated genetic factors affecting lifespan in a de-identified snapshot of more than 300,000 AncestryDNA customers that provided prior consent to participate in research and were linked to publically visible family trees. We estimated genetic ethnicity and found it to be significantly correlated with parental lifespan. To identify genetic variants that affect human longevity, we conducted GWA mapping analyses of parental lifespan and identified five loci significantly associated with paternal lifespan and one locus associated with maternal lifespan. We estimated the phenotypic variation in lifespan attributable to a genome-wide set of variants to be quite low (between 2 and 8% for different sub-cohorts) and the genetic correlation between maternal and paternal lifespan to be quite high (at least 68%). We conducted GWA analyses of the lifespan proxy trait, age, and identified a single locus, APOE, associated with both traits (age and lifespan). We also observed strong genetic correlations between the two traits (43–70%). Finally, we analyzed an independent UKB dataset in a meta-analysis of more than 650,000 individuals and corroborated the association at four of the six loci identified in the AncestryDNA analysis. In total, this meta-analysis identified eleven paternal and four maternal lifespan-associated loci, two of which have not previously been associated with parental lifespan.
Materials and Methods
Set of aggregated ancestry pedigrees provides parental lifespan data
To measure parental lifespan, we interrogated a large, non-redundant set of aggregated and anonymized pedigrees, referred to as SAP. The SAP is constructed from overlapping Ancestry customer-generated family trees designated as “public” (Ruby et al. 2018). Aggregation occurred by identifying and collapsing redundant identical ancestors and stitching them together across pedigrees. The methods used to assess data quality, compare nodes between customer-generated pedigrees and aggregate this information into the SAP used in this study are described by Ruby et al. (2018). To protect customer privacy, SAP data were de-identified with randomly generated IDs and presenting birth and death dates as years, a lower resolution than was available in the source data.
Lifespan was measured on every record that had a year of birth and death. We considered lifespan values between 40 and 120 years to reduce early-life contributions to environmental variance and remove likely errors in reporting of birth or death dates. We did not estimate the Cox proportional hazard of mortality, as in other analyses (Joshi et al. 2016; Pilling et al. 2017), because of the prevalence of missing year of death data in the SAP. Approximately 1/3 of individuals starting in 1900 and going back multiple centuries lack a death date (Ruby et al. 2018, Figure 1G) even though they are assuredly deceased. Therefore, when an individual was missing year of death data we could not determine whether: 1) the individual was still alive and legitimately right-censored, or 2) the individual was deceased and the death date was never added to the genealogy.
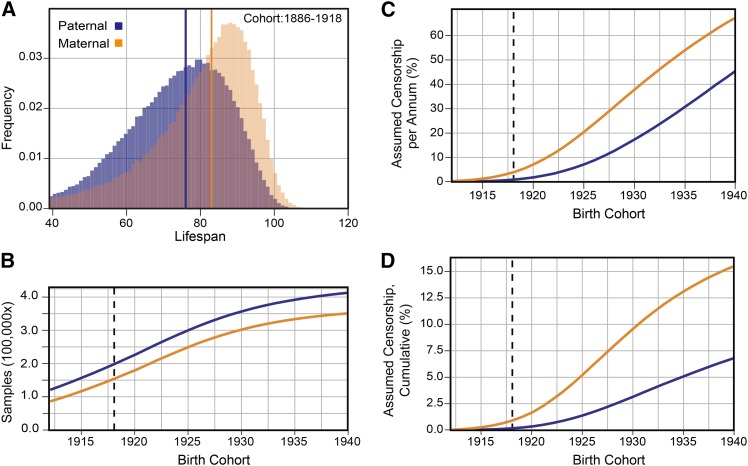
Figure 1.
(A) Distribution of parental lifespan values from the 1886-1918 birth cohort. Median values noted with colored vertical lines. (B) Cumulative number of genotyped individuals with complete lifespan data. Birth cohort span from 1886 to year on x axis. Vertical gray line denotes the 1918 cohort. (C) Estimated number of long-lived individuals assumed to be censored as fraction of annual cohort size. (D) Estimated number of long-lived individuals assumed to be censored as a fraction of cumulative cohort size. In all figure panels, paternal individuals are denoted with blue, maternal individuals with orange, and overlapping values are dark orange.
Although we were unable to determine whether any specific individual missing year of death data were alive or dead, we estimated the fraction of individuals that were alive, and assumed to be right-censored, in the 1911-1940 birth cohorts using the distribution of lifespans from the 1886-1910 cohort as a baseline. We used the lifespan distribution from 1886-1910 birth cohort as our baseline because, in the year 2016, it contained complete lifespan data for all individuals from the 1910 birth cohort with a lifespan less than or equal to 106 years, all lifespans from the 1909 birth cohort, equal to or less than 107 years, and so on. Thus, we expect few individuals to be truly censored from this baseline.
IRB statement regarding data use
All data required for this research project was collected previously with informed consent for purposes consistent with this research and was used for this research only after removing all personally identifying information. An IRB reviewed this research project and determined no further approval was required.
Genotype sample collection
Data utilized for this research project was de-identified prior to its use. The laboratory extracts DNA from a sample and genotyped it using an Illumina array (details below). In order to be included in this study, customers must: 1) activate a DNA test and agree to AncestryDNA’s terms and conditions, privacy statement, including an explicit consent to process their DNA, 2) agree to the informed consent to research agreement, and 3) provide basic personal information - including year of birth, name, and sex. Next, the customer must have associated their DNA sample to their own user-generated pedigree and made this pedigree public to other Ancestry users. This analysis was based on samples submitted between May 2012 and June 2016. The database snapshot occurred on June 19, 2016 and contained 698,812 genotyped individuals linked to the SAP.
Array based genotyping procedure
SNP variants were called by technicians at the Illumina FastTrack Microarray Services lab using the GenomeStudio platform. Genotype data were generated using an Illumina genotyping array with approximately 730,000 SNPs. During the four years of sample collection, two versions of the array were used for genotyping. All downstream analyses used the intersection of SNPs present on both versions of the chip. We included chip version as a covariate in all association mapping models.
Estimates of genetic ethnicity and test of association with parental lifespan
We used 112,909 SNPs to estimate the genetic ethnicity of de-identified genotyped individuals who consented to research and were associated with public family trees. Genetic ethnicity is the proportion of their genome that matches a reference panel of 3000 individuals from 26 ancestral populations (described in: Ball et al. 2013; Han et al. 2017). An individual’s genetic ethnicity proportions reflect ancestral admixture events between these reference populations.
We measured whether ancestral admixture proportions were independent of each other using the Pearson correlation coefficient, r. We use multivariate linear regression models to test for a correlation between percent ancestral admixture and parental lifespan. Lifespan was calculated from the mothers and fathers of genotyped individuals from the 1886-1918 birth cohort. In these models, we included all ancestral populations with a minimum of 1000 individuals which were born between 1886-1918 and possessed greater than 5% admixture assignment. The 14 ancestral populations that met these requirements were: Native American, Caucasus, Great Britain, Ireland, Europe East, Scandinavia, Italy/Greece, Europe West, Iberian Peninsula, European Jewish, Finland/NW Russia, Near East, Sub-Saharan Africa and Hispanic/Latino.
Principal component measures of population structure
In addition to genetic ethnicity, we measured population structure using a principal component analysis (PCA) of a genome-wide panel of SNPs. Given the large size of this snapshot, we estimate the first 10 principal components using the fastPCA algorithm, method: randomized, implemented in in the R package SNPRelate (Zheng et al. 2012). For this analysis, we used 50,000 randomly selected SNPs that were in approximate linkage equilibrium with each other (LD < 0.2). This procedure was implemented in PLINK v1.9 (Chang et al. 2015). We included the first 10 principal components as covariates in all association mapping models.
IBD measurement between individuals
Given the nature of the resource, it is not surprising that a significant number of closely related individuals are represented in the snapshot. To reduce the effects of shared environmental variation due to local household environment and more closely meet the assumption of genotype independence in GWA mapping analyses, we removed individuals until no pair of samples exhibit more than 300cM identity by descent (IBD). IBD was measured with a custom algorithm, J-GERMLINE (Ball et al. 2016) based upon the GERMLINE algorithm (Gusev et al. 2009). Since shorter IBD segments are difficult to accurately identify and are less informative for estimating familial relationships, we used the ‘Timber’ algorithm to filter uninformative matches (Ball et al. 2016). This caused a skew in the distribution of IBD matches less than 90 cM.
Simulation of GWA analysis of parental traits using offspring genotypes
In order to estimate the statistical power of our mapping populations, we simulated a GWA mapping analysis in which the phenotypes and genotypes were measured in offset generations. Briefly, we modeled a situation in which a single, additive allele effects our focal trait and segregates in a large, random mating population at Hardy-Weinberg equilibrium. We randomly assigned genotypes in the parental generation. Next, we calculated the parental trait value given their genotype and environmental variance, modeled with a random-normal distribution. The offspring genotype was calculated using the genotype of a parent where the trait value was known and a second parent, randomly sampled from the population. We assumed perfect Mendelian segregation of alleles between generations. Finally, we regressed the offspring’s genotype against a single parental phenotype.
Simulations were run varying the effect allele frequency (from 0.001 to 0.5) and the additive effect size of a single copy of the allele (from 0.06 to 3.8 years). We simulated mapping populations of 80,000 to 480,000 individuals. Fifty replicate simulations were run for each parameter combination. Power was measured as the number of replicate simulations which detected a significant effect (P value less than 5.0e-8) of the focal allele vs. the total number of simulations for each parameter combination. For each simulation we also calculated the allelic effect inferred from the genotypes of the parents vs. genotypes from the offspring.
Parental lifespan GWA mapping analyses
We conducted quantitative GWA mapping analyses of parental lifespan using PLINK v1.9 (Chang et al. 2015). We conducted separate analyses for paternal and maternal lifespans. The majority of genotyped individuals were linked to two parental lifespans and we did not wish to test the same genotype against multiple phenotypes in a single GWA analysis. We further sub-divided our cohort and ran separate GWA analyses on the three broad population groups: European, Sub-Saharan African, and East Asian/Native American. For each mapping population, we identified close genetic relatives (IBD greater than 300 cM) and randomly removed one of the individuals from the pair. We examined the genetic basis of lifespan of parents born in both the 1886-1918 and 1886-1940 cohorts.
We applied the following genotype filters: minor allele frequency (MAF) threshold of 0.5%, variant missingness threshold of 20%, an individual missingness threshold of 2%, and 1% of SNPs with strongest deviation from Hardy-Weinberg Equilibrium. Additionally, we used the ‘–indep-pairwise’ function in Plink v1.9 to filter sites in which linkage disequilibrium with an adjacent site exceeded 0.9. After applying these filters, between 540,852 and 541,614 SNPs were included in any one GWA mapping analysis. We included the mitochondria and Y chromosomes in our initial analyses but found no effect (results not shown) and removed them from all subsequent analyses. The array version and first ten PCs were added as covariates to the association mapping models.
To identify lifespan-associated variants, we controlled for the genomic variance inflation factor, λ, and used a Bonferroni correction to establish a threshold of statistical significance at P = 9.2e-8. For each lifespan associated variant, we ensured that missingness was less than 5%. Next, we estimated the phase of each individual genotype for 5.0 megabase (Mb) regions surrounding an associated variant using the AncestryDNA algorithm, Underdog (Ball et al. 2016). We additionally imputed all cataloged variants from each 5.0 Mb region using the 1000 Genomes (1000G) phase 3 reference panel (The 1000 Genomes Project Consortium et al. 2015). Imputation was performed using the default parameters in minimac3 (Das et al. 2016).
Genetic heritability analyses
We used LD Score Regression v1.0.0 (Finucane et al. 2015) to estimate the heritability, hg2, of lifespan using genotyping array variants that passed all quality control filters. For all heritability estimates, we used LD scores measured using the AncestryDNA, European population group, rather than the 1000G European (1000G,EUR) population LD score estimates included with the program. For this analysis, we included 482,066 male and female individuals; this sample contained no relatives with pairwise IBD scores greater than 300cM. We estimated the LD scores of 542,710 variants in 1 cM windows. Our analyses with the AncestryDNA LD scores produced h2g intercept values close to 1.0 and ratio values lower than the 1000G,EUR LD scores, which suggested that these scores adequately captured the LD structure of the AncestryDNA dataset.
GWA analysis of lifespan proxy trait: age
We conducted GWA analyses on the age of genotyped individuals from the European population group. We removed close genetic relatives (IBD greater than 300 cM) from this mapping population. Age, at a resolution of years, was calculated by subtracting 2016 from an individual’s birth year. We assumed all of these individuals were still alive, because all individuals recently provided a saliva sample in the four years since the launch of AncestryDNA (http://www.ancestry.com/corporate/about-ancestry/company-facts) and the time of this snapshot. In contrast to the lifespan phenotype, right-censorship of lifespan is an explicit assumption of the age trait and therefore addressed a potential confounder.
We conducted GWA analyses using both the case/control as well as quantitative trait frameworks. In the case/control analyses, the “cases” were individuals above a specified age, and the “controls” were individuals below another specified age, with a gap existing between the two age thresholds. The age threshold for the control population was set to the median age: 65 years for men and 64 years for women. We investigated two thresholds for the case population: 2.5% (male: 91 years, N = 5,860; female: 92 years, N = 6,803) and 5% (male: 86 years, N = 12,288; female: 87 years, N = 13,265). We reported results from the 5% case threshold. No additional loci were identified in the 2.5% case threshold GWA analysis (data not shown). We conducted GWA analyses for each gender separately (male N= 117,396, female N= 140,017) as well as together (N = 257,413) and included gender as a covariate in the association mapping model. To increase the mapping population size, we also conducted a quantitative trait GWA mapping analysis with all individuals of both genders between the ages of 40 and 120 years into a single analysis (N= 482,066).
Genetic correlation analyses
We used LD Score regression (Finucane et al. 2015) to estimate the genetic correlation, rg, between: 1) maternal and paternal lifespan; and 2) the age of genotyped individuals and parental lifespan. To estimate the genetic correlation between traits, we constrained the h2g intercept to 1.0 for both traits and the genetic covariance intercept to: rp*N12 / sqrt(N1*N2) where rp is the phenotypic correlation, N1 is the number of samples in study 1, N2 is the number of samples in study 2, and N12 is the sample overlap (Bulik-Sullivan et al. 2015).
For our estimate of the genetic correlation between age and lifespan, we required all samples to have both age and parental lifespan measurements within each gender specific comparison. Second, we aimed to minimize biases associated with younger individuals having a higher rate of parental lifespan censorship compared to older individuals, by restricting the birth cohort of genotyped individuals included in the analysis to be between the years 1916 and 1956. This resulted in an age distribution of genotyped individuals to be between 60 and 100 years. Parental lifespan was examined for the 1886-1940 birth cohort, and the distribution of lifespans examined were between 40 and 120 years.
Cohort specific effects of lifespan associated loci
To investigate variation in the association of SNP variants and lifespan between birth cohorts, we ran separate association mapping analyses for 4 – 8 year slices spanning the 1886-1940 cohort. The cohorts interrogated were: 1886-1894, 1894-1902, 1902-1908, 1908-1912, 1912-1916, 1916-1920, 1920-1924, 1924-1928, 1928-1932, 1932-1936, 1935-1940. The number of individuals in the narrowly defined birth cohort slices range from 10722 – 23211 for paternal lifespan and 6764 – 22469 for maternal lifespan. We also measured the change in MAF between cohorts for variants at each lifespan associated locus. For APOE, the one locus that exhibited a change in MAF between cohorts, we also measured the allele frequency in samples binned by lifespan value.
Comparison of AncestryDNA and UK Biobank parental lifespan analyses
We compared the AncestryDNA GWA of parental lifespan from the broadly defined birth cohort (1886-1940) to the largest UKB association analysis of parental lifespan published to date: Pilling et al. (2017). The focal trait in the UKB analysis was attained parental age, which was calculated using the Cox proportional hazard model and includes our focal trait, complete lifespan, as well as the current age of parents which were not deceased (Pilling et al. 2017). We downloaded GWA summary statistics from the UKB analysis of attained parents age from: https://figshare.com/articles/Plling_et_al_2017_UKB_parents_attained_age_GWAS/5439382/2.
We conducted a meta-analysis of more than 658,000 individuals using the sample size based approach implemented in METAL (Willer, Li, and Abecasis 2010). We included SNP variants that were common to both studies, possessed a cohort-specific MAF of greater than or equal to 0.5% and had matching alleles. With these requirements, we interrogated 458,525 and 458,865 variants in our meta-analyses of maternal and paternal lifespan. This sub-set of SNPs meant that we were only able to include variants that were found to be significant in the UKB analysis at 10 of 14 paternal attained-age loci and four of six maternal attained-age loci. We controlled for the genomic inflation factor in each dataset within METAL and established a P value of 5.0e-8 as our significance threshold.
We measured the genetic correlation between AncestryDNA and UKB GWA analyses using LD Score Regression (Finucane et al. 2015). We ran analyses with LD scores from both the AncestryDNA European population group and the 1000G,EUR population. We reported results from the AncestryDNA LD scores analysis because the h2 ratio values were lower and the h2g intercept values were closer to 1.0, suggesting that these LD scores are a better match than the 1000G,EUR to the genetic structure of the UKB mapping population (Finucane et al. 2015). In contrast to the LD score analyses of solely the AncestryDNA GWA results, we did not constrain the h2g or genetic covariance intercepts in these analyses because we could not estimate the phenotypic correlation and amount of sample overlap between the two datasets. The GWA meta-analyses and LD score analyses used z-score transformed effect sizes from the AncestryDNA and UKB GWA mapping results. In order to make the effect sizes consistent with the values reported in the UKB analysis, we used effect sizes from the AncestryDNA analysis calculated with offspring genotypes -rather than effects sizes re-scaled to reflect allelic dosage in the parental generation.
Privacy considerations & data availability
Supplemental Figures and Tables, with their Legends, are provided as a single PDF document: Supplemental Tables & Figures. In addition, Supplemental Tables 8 and 9 are provided as Excel documents and contain SNP-by-SNP summary statistics from each presented GWAS for representative SNPs with genomic control adjusted P values less than 3.0e-3 in the AncestryDNA cohorts (Supplemental Table 8) or 2.5e-3 in the AncestryDNA-UKB meta analyses (Supplemental Table 9).
The growing literature and commentary on genomic privacy guided our decision to release summary statistics for only a subset of SNPs in this instance. The power to use an individual’s genome-wide variant data to infer whether that individual was present in or absent in a GWA cohort using association mapping summary statistics has not yet been exhaustively explored, but has nonetheless been considered from both theoretical (Visscher and Hill 2009; Sankararaman et al. 2009) and practical perspectives (Homer et al. 2008; Im et al. 2012; Gymrek et al. 2013; Cai et al. 2015). These studies consistently identify the ratio of reported SNP statistics to cohort size as the key factor governing identifiability. This relationship is typically described as m independent SNP markers, n individuals, with the power to identify individuals is proportional to sqrt(m/n) (Equation 1; Sankararaman et al. 2009). As a result, we err on the side of caution in protecting Ancestry customers’ privacy.
In our Supplemental Tables 8 and 9, we provide abbreviated summary statistics for seven GWA mapping analyses (for a total of 10,171 unique SNPs). Our basis for this was inclusion of all SNPs with P values less than 3.0e-3 for the five AncestryDNA specific analyses and 2.5e-3 for the two AncestryDNA-UKB meta analyses. This standard ensured that all significant and near-significant results were reported, and also that all other SNPs were known by the reader to be non-significant. For our smallest-cohort analysis (maternal lifespan for the 1886-1918 birth cohort: 133,203 individuals), we report 1,471 snps, which is an m/n ratio of 0.011, substantially below the ratios analyzed in the privacy literature noted above and therefore presumably safe. In contrast, the community standard of GWAS data disclosure – provision of summary statistics for all 540,852 analyzed variants – would produce an m/n ratio greater than 4.0. Re-identification power for such a high ratio is plotted by Sankararaman et al. (2009; Figure 1; m/n = 1.0, plotted for m = 1,000). While low, the risk of re-identification in this scenario is not negligible.
Our report of summary statistics across seven GWA analyses increases the risk of re-identification. The work of Im et al. (2012) clearly identifies multi-phenotype GWAS as adding tremendous power to re-identification (presented in their Figure 6). Such added power derives from the ability to regress the same SNPs across multiple phenotypes; in contrast, our presentation of summary statistics for the most-significant SNPs for each analysis resulted in largely non-overlapping sets of SNPs, thereby negating the additional re-identification power that multiple GWAS provide. Had we reported summary statistics across all SNPs, then the SNPs would have been totally consistent between phenotypes. The additional power gained from the multiple phenotypes – together with the added power provided by the larger SNP set – would have profoundly facilitated re-identification, far beyond what the increased m/n ratio would imply by itself.
The GWAS literature from another commercial entity with legal obligations to the members of its study population consistently reports the 10,000 most-significant SNPs (Jansen et al. 2019; Shaffer et al. 2017; Hu et al. 2016; Jones et al. 2017; Aberg et al. 2018; Huusko et al. 2018; Lee et al. 2018; Jones et al. 2019). For our complete study, 10,171 unique SNPs provided a level of disclosure that permits scientific scrutiny while comfortably maintaining the anonymity of our users.
Ancestry’s highest priority is protecting our customers’ privacy and being good stewards of their data; that starts with the basic belief that customers should always maintain ownership and control over their own data. We have provided access to the significant findings that would allow for a level of replication in the supplemental files. We collaborate with scientific researchers on a case-by-case basis to advance science in specific areas while using research methodologies that protect consumers when appropriate. Examples to be considered in these requests include but are not limited to:
Technical and scientific feasibility
Level of detail required from the underlying data (including its sensitivity)
Novelty/potential impact of the proposed work
Academic institution (in good standing and acting in good faith while implementing appropriate technical, administrative, and physical measures to protect the privacy of the participants and the security of the data)
Amount of resource allocation for Ancestry required to support the collaboration
Correspondence and requests should be addressed to Catherine Ball (cball@ancestry.com). Supplemental material available at Figshare: https://doi.org/10.25387/g3.8479022.
Results
The Ancestry pedigree facilitated GWA mapping analysis of human lifespan
To measure parental lifespan, we interrogated a large, non-redundant set of aggregated and anonymized pedigrees, referred to as SAP, generated by stitching together overlapping customer-generated family trees designated as “public” (Ruby et al. 2018). The June 2016 snapshot of AncestryDNA customers contained more than 680,000 individuals who provided prior consent to research, were linked to the SAP and had at least one parent with complete lifespan information, i.e., recorded year of birth and death. The majority the parents with lifespan information (76.0% of fathers and 77.7% of mothers) were born in the 1900-1930 cohort. For this cohort, nearly 80% of these individuals were born in the United States, ∼10% were born in Europe and the remaining ∼10% born outside of the US and Europe (Ruby et al. 2018, Figure 1I). We considered lifespan values between 40 and 120 years to reduce early-life contributions to environmental variance and remove potential errors in the recording of birth or death dates (Figure 1A).
To maximize sample size and power in the association mapping analysis, we sought to include individuals from a wide range of birth cohorts (Figure 1B). However, the addition of younger birth cohorts increased the systematic removal of long-lived, right-censored individuals, because they lack a year of death. We estimated the fraction of individuals that were assumed to be censored (i.e., were missing a year of death and were presumably alive in the 1911-1940 birth cohorts) using the lifespan distribution from 1886-1910 birth cohort as a baseline (Figure 1C, D). We found the 1886-1918 cohort possessed a large sample size with minimal assumed censorship: for fathers, N = 208,707, assumed censorship = 0.3%; for mothers, N= 162,659, assumed censorship = 1.3% (Figure 1B, D). Median paternal lifespan ranged from 75 to 77 years when comparing the 1886-1894 cohort (N = 10,721) to the 1916-1918 cohort (N = 20,858; Figure S1A), whereas maternal lifespan ranged from 81-83 years across the same timespan (N= 6,764 for 1886-1894; N = 19,239 for 1916-1918; Figure S1B). A wider range of birth cohorts, 1886-1940, increased the number of parents with lifespan information (fathers: 423,817 and mothers: 359,719, Figure 1B) and increased the fraction of missing long-lived individuals, estimated to be 6.8% for fathers and 15.5% for mothers (Figure 1D).
Given the constraint of a single-generation offset between measures of phenotype and genotype, we estimated our power to detect genetic associations of various effect sizes in datasets of this scale. The power to identify an allele with an additive effect size of 0.75 years at 20% minor allele frequency in the population would be 0.90 in a cohort of 170,000 individuals and nearly 1.0 in cohorts exceeding 230,000 individuals (Figure S2). We concluded that for our snapshot, there was sufficient power to identify a genetic variant at moderate allele frequency with an effect size of 0.75 years or greater.
Genetic measures of ethnicity were significantly associated with lifespan
A non-random distribution of lifespans with respect to the underlying genetic structure in the mapping population can cause false positives in a GWA mapping. Previous research demonstrates the association between race, defined as a socially constructed categorical variable maintained by dominant social groups (Mills 1997; Haslanger 2008), and life expectancy (C. J. L. Murray et al. 2006; Backlund et al. 2007; Chetty et al. 2016). We sought to determine whether there was a similar association in this snapshot, however we have no knowledge of individuals’ race, thus we instead estimated their genetic ethnicity and compared this to the lifespan of their parents.
We estimated the genetic ethnicity as the proportion of the genome that is assigned to a reference panel of 26 ancestral populations for de-identified genotyped individuals who have previously consented to research and were associated with public family trees (described in: Ball et al. 2013; Han et al. 2017). We found that genetic ethnicity proportions, which represent ancestral admixture events between these reference populations, were generally uncorrelated with each other in this snapshot (Figure 2A), except in the case of the eight Sub-Saharan African ethnicities (r > 0.38), which we combined into a single Sub-Saharan African group; and the Native American and Iberian ancestral admixture (r = 0.44), which, for individuals exhibiting evidence of admixture (Han et al. 2017), were combined into a single Latino group.
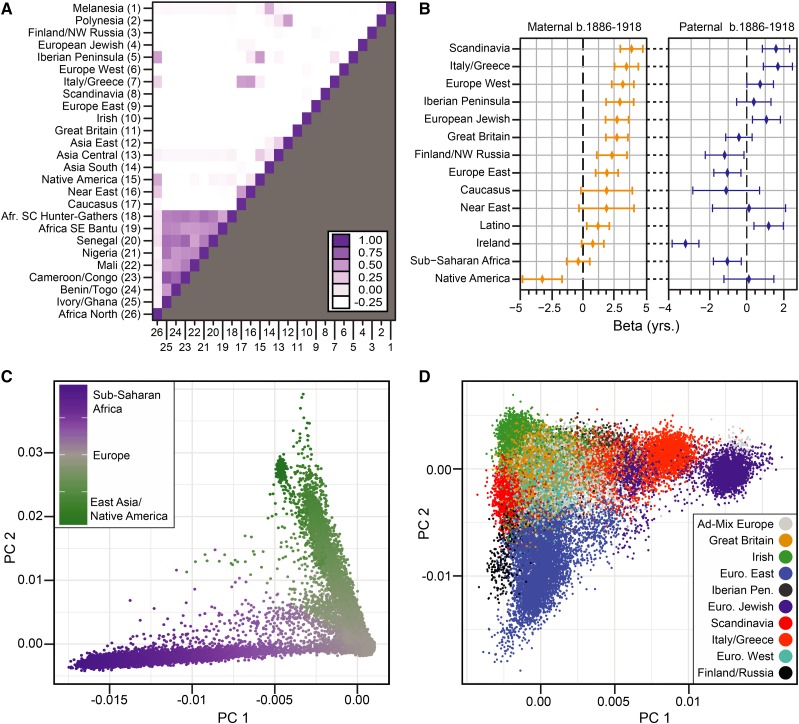
Figure 2.
(A) Pairwise correlation between ancestral admixture proportions of genotyped individuals from analysis of paternal lifespan, 1886-1918 birth cohort. (B) Linear regression coefficient (+/−SE) for admixture population and paternal or maternal lifespan. The dashed line at zero represents the mean lifespan for all individuals in the analysis. (C) Population structure, measured with principal component analysis, of individuals from analysis of paternal lifespan, 1886-1918 birth cohort. Each circle is a single individual, colored according to percent composition of Sub-Saharan Africa and Native American / East Asian admixture populations. (D) Population structure within individuals of European descent, after filtering Sub-Saharan Africa, Native American/East Asian groups. Colors depict individuals with majority ethnicity assignment in one of nine European populations.
Using multivariate, linear regression models we found that ancestral admixture proportions were significantly associated with both maternal and paternal lifespan (P < 2e-16) although the magnitude of the correlations were not large (maternal lifespan r = 0.047, paternal lifespan r = 0.056). We found significant, positive correlations with maternal lifespan and genetic ethnicity assignment to the following populations: Europe West, European Jewish, Scandinavia, Italy/Greece, and Great Britain (Figure 2B, Table S1). Conversely, there was a negative correlation between maternal lifespan and ancestral admixture with the Native American population, although this was not significant after multiple test correction (Table S1). Paternal lifespan was positively correlated with ancestral admixture with Scandinavian and Italian/Greek populations, although this was not significant after multiple test correction (Figure 2B, Table S2). Ancestral admixture with a single population, Ireland, exhibited a significant, negative correlation with paternal lifespan (Table S2). Given the relationship between race and life-expectancy (C. Murray et al. 1998; Backlund et al. 2007; Chetty et al. 2016), we inferred that non-genetic, social factors contributed to the correlation between lifespan and genetic ethnicity.
We attempted to control for these factors in our association analyses by calculating genetic differentiation using principal component analysis (PCA) and splitting this snapshot of de-identified genotyped individuals who provided prior consent to research and were associated with public family trees into one of three broad population groups: Sub-Saharan African, Native American/East Asian or European (Figure 2C). We applied a conservative minimum threshold of 5% ancestral admixture assignment inclusion in the Sub-Saharan African and Native American/East Asian groups; these samples comprised a small proportion of the snapshot. All remaining individuals were placed into the European population group. Next, we re-calculated genetic PCs for each group and used these as covariates in the association mapping analyses. GWA analyses focused on the European population group (Figure 2D) because it contained the majority of samples.
GWA mapping revealed three loci to be significantly associated with parental lifespan in the 1886-1918 birth cohort
We first investigated the genetic basis of maternal and paternal lifespan in the 1886-1918 birth cohort. This mapping population was of primarily European descent and contained no close genetic relatives (IBD threshold 300 cM; Figure S3). The number of individuals in maternal and paternal analyses was 133,203 and 167,179, respectively. After quality control filtering of array genotyping variants, the minimum number of variants tested in each analysis was 540,852 (Table S3).
We found genetic variants at two loci associated with paternal lifespan (Figure 3A) and no variants associated with maternal lifespan (Figure 3B) in the European population group at a Bonferroni-corrected P value less than 0.05, which corresponded to observed P value less than 9.24e-8 (Table 1). The genotyping chip version covariate was not significant in these analyses (Table S4; Figure S4). These variants remained significant after controlling for the genomic variance inflation factors: λPaternal = 1.084, λMaternal = 1.030 (Table S3). We inferred that most of this inflation was due to polygenicity, rather than unaccounted population structure, because the LD score regression intercepts were nearly 1.0 and the mean LD score regression (LDSC) χ2 estimates were similar to the genomic variance inflation estimates: 1.085 and 1.033 for paternal and maternal lifespan, respectively (Table S3; Finucane et al., 2015).
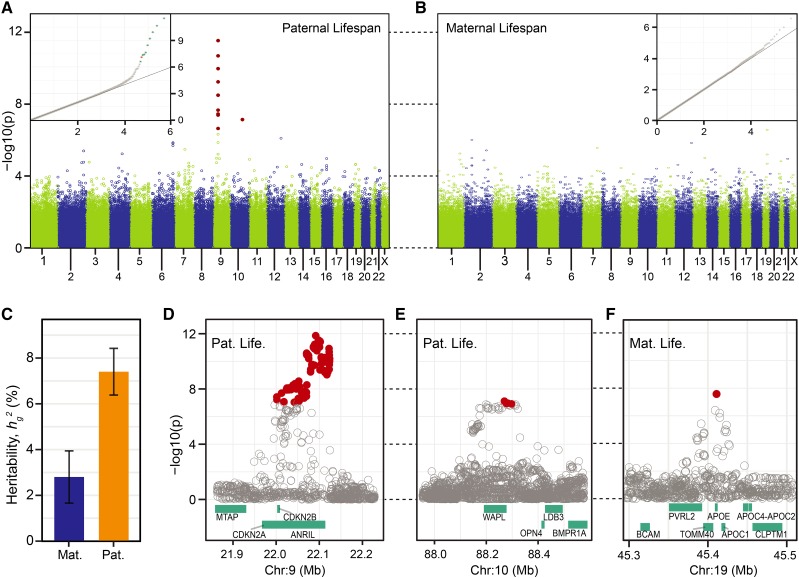
Figure 3.
Manhattan plot of GWA results of (A) paternal and (B) maternal lifespan, cohort 1886-1918. Each circle is a single genotyped SNP, solid red circles denote SNPs with Bonferroni corrected P values less than 0.05. Inset: Q-Q plot with expected (x-axis) vs. observed (y-axis) –log10 P values. Non-gray points denote SNPs with Bonferroni corrected P values less than 0.05. Association analysis of candidate loci with imputed data: (C) Heritability of lifespan, estimate (+/−SE) with genome-wide panel of SNPs. (D) Chrm9: ANRIL, paternal lifespan (E) Chrm10: WAPL, paternal lifespan (F) Chrm19: APOE, maternal lifespan. Solid red circle are SNPs with Bonferroni corrected P values less than 0.05. Cyan bars: hg19 gene models.
Table 1. Candidate paternal and maternal lifespan loci. For each locus, we list the lead genotyped variants, and for SRRM3 and APOE we also list the lead imputed variant, indicated with SNP ID prefix: bp. The additive effect of each allele at the candidate locus is given in years, scaled to the expected allelic dosage in the parental generation. h2snp is the fraction of variance in lifespan explained per snp. The statistical significance of the association at each candidate variant is given as an uncorrected (PUC) value and genomic control corrected (PGC) value. Significance of PGC after Bonferroni multiple test correction: *** PBonf < 0.001; * PBonf < 0.05; - PBonf > 0.05.
| Lifespan | Locus | RSID | Chr | Effect Allele | Effect Allele Freq. | 1886 - 1918 | 1886 - 1940 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect (years) | SE (years) | h2snp (%) | PUC | PGC | Effect (years) | SE (years) | h2snp (%) | PUC | PGC | ||||||||
| Paternal | ANRIL | rs1333042 | 9 | A | 0.49 | 0.65 | 0.09 | 0.063 | 4.01e-13 | 3.21e-12 | *** | 0.51 | 0.07 | 0.039 | 7.00e-15 | 8.13e-13 | *** |
| WAPL | rs10887623 | 10 | G | 0.10 | −0.84 | 0.15 | 0.037 | 2.26e-08 | 7.89e-08 | * | −0.51 | 0.11 | 0.014 | 3.36e-06 | 1.92e-05 | — | |
| LPA | rs9457925 | 6 | A | 0.02 | −1.66 | 0.33 | 0.030 | 5.49e-07 | 1.51e-06 | — | −1.71 | 0.25 | 0.031 | 4.14e-12 | 1.83e-10 | *** | |
| SRRM3 | rs17685 | 7 | A | 0.28 | 0.38 | 0.10 | 0.018 | 1.13e-04 | 2.08e-04 | — | 0.40 | 0.07 | 0.019 | 5.41e-08 | 5.73e-07 | — | |
| bp75763624 | 7 | A | 0.32 | 0.42 | 0.07 | 0.023 | 2.64e-09 | 4.39e-08 | * | ||||||||
| CHRNA3/5 | rs931794 | 15 | G | 0.35 | −0.34 | 0.09 | 0.017 | 1.92e-04 | 3.40e-04 | — | −0.41 | 0.07 | 0.023 | 3.19e-09 | 5.17e-08 | * | |
| — | |||||||||||||||||
| Maternal | APOE | rs4420638 | 19 | G | 0.18 | −0.60 | 0.13 | 0.033 | 4.83e-06 | 6.60e-06 | — | −0.49 | 0.09 | 0.022 | 5.94e-08 | 2.76e-07 | — |
| bp45411941 | 19 | C | 0.14 | −0.83 | 0.15 | 0.048 | 1.86e-08 | 2.96e-08 | * | −0.6 | 0.1 | 0.026 | 2.53e-09 | 1.61e-08 | * | ||
To estimate the additive effect of variants on lifespan, we doubled the values measured in our association mapping analyses – which used offspring genotypes – because the observed allelic dose in the genotyped offspring is half of the expected allelic dose in the parents (Figure S5; Joshi et al., 2016). All effect sizes reported for the AncestryDNA specific analyses are scaled to the allelic dose in the parental generation (Table 1).
We estimated the fraction of phenotypic variance in parental lifespan explained by genotyping array variants, hg2, to be 7.38% (SE = 1.02%; Nsnps = 390,234) and 2.78% (SE = 1.14%; Nsnps = 385,494) for paternal and maternal lifespan, respectively, using LD score regression (Finucane et al. 2015) (Figure 3C). As with our variant effect size estimates, these hg2 values have been re-scaled to the allelic dose in the parental generation (Figure S5; Joshi et al., 2016).
Next, we examined whether the non-normal distribution of lifespan values (Figure 1A) contributed to genomic variance inflation with a permutation analysis of lifespan values from this dataset. We found that the distribution of λ values was centered at 1.00, and did not overlap the observed λ values (Figure S6). We also considered whether the increase in median lifespan between the years of 1886 and 1918 (Figure S1) may have altered these results. Re-running these analyses with an additional median lifespan per cohort covariate did not significantly change the distribution of P values across the genome (Figure S7), nor at the lifespan associated variants (Table S5).
To define each region of association more precisely, we imputed a 5.0 Mb block of sequence using the 1000G, phase 3 reference panel. The chromosome 9 locus, which exhibited the strongest association with paternal lifespan, included two annotated protein-coding genes (CDKN2A/B) and one annotated long non-coding RNA (lncRNA; ANRIL). Hereafter, we refer to this locus as ANRIL because the lead variants, defined by the strength of the statistical association, were in the lncRNA, rather than either of the two protein-coding genes (Figure 3D). The second locus associated with paternal lifespan included a single annotated, protein-coding gene: WAPL (Figure 3E), and, to our knowledge, has not been previously associated with any life-shortening diseases or lifespan. Although no variants achieved genome-wide significance in our analysis of maternal lifespan significant, the SNP with the lowest P value was located near APOE, a locus previously associated with survivorship to advanced age (Deelen et al. 2011; Nebel et al. 2011; Sebastiani et al. 2017) and parental lifespan (Joshi et al. 2016; Pilling et al. 2017). We imputed SNPs at this locus and identified a single variant which was significantly associated with maternal lifespan (Table 1; Figure 3F). This association was gender-specific: no imputed variants at the APOE locus were significantly associated with paternal lifespan (Figure S8A). In summary, we identified a total of three loci associated with parental lifespan in this narrowly defined birth cohort.
We found no variants that were significantly associated with maternal or paternal lifespan in the Sub-Saharan African or Native American/East Asian population groups (Figure S9A-D). With fewer than 10,000 individuals, these analyses were underpowered and this negative result was expected (Figure S2).
Increased GWA mapping power in an expanded birth cohort identified one maternal and four paternal lifespan associated loci
To increase the power of our association mapping analysis, at the cost of increased censorship of long-lived individuals, we expanded the analysis to the 1886-1940 birth cohort. The European population group from this birth cohort contained, after filtering closely related individuals, 270,548 and 309,383 samples with maternal and paternal lifespans (Table S3).
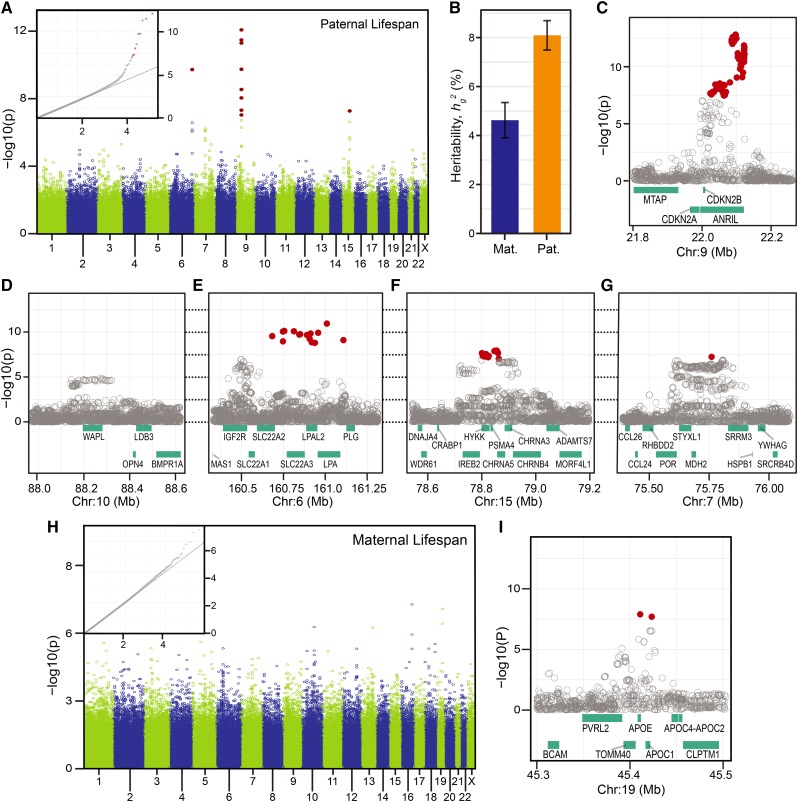
In the expanded cohort, variants at four loci (ANRIL, LPA, CHRNA3/5, and SRRM3; Table 1; Figure 4A) were significantly associated with paternal lifespan after controlling for genomic variance inflation (λPaternal =1.182; Table S3). Similar to the narrowly defined cohort, we interpreted most of this inflation was due to polygenicity, LDSCχ2 = 1.161 (Table S3), and hg2 was 8.08%, SE = 0.60% (Figure 4B). ANRIL was the one locus significantly associated with lifespan in both the narrow and expanded birth cohorts (Figure 4C). In contrast, WAPL did not achieve genome-wide significance in the broader birth cohort (Figure 4D). We identified significantly associated variants at two loci, LPA and CHRNA3/5 (Figure 4E,F), that were previously associated with paternal lifespan (Joshi et al. 2016; Pilling et al. 2017). The fourth locus, SRRM3 (Figure 4G), was defined by a single imputed SNP (genotyping array variants were near the threshold of significance; Table 1) which, to our knowledge has not been previously associated with any life-shortening diseases or lifespan.
Figure 4.
Manhattan plot of association test statistics from analyses of (A) paternal and (H) maternal lifespan, cohort 1886-1940. (B) Heritability of lifespan, estimate (+/−SE) with genotyped variants. Association analyses with imputed genotypes at candidate loci: (C) ANRIL, (D) WAPL, (E) LPA, (F) CHRNA3/5, (G) SRRM3, (I) APOE. Annotation details are the same as in Figure 3.
As with the narrowly defined birth cohort, our analysis of the expanded birth cohort identified a single locus, APOE, which was significantly associated with maternal lifespan (Table 1). After controlling for variance inflation, λMaternal = 1.113, none of the genotyped variants were significant (Figure 4H); however, imputed SNPs at APOE achieved genome-wide significance (Figure 4I). We estimated the LDSCχ2 to be 1.097 (Table S3) and heritability of maternal lifespan to be 4.62%, SE =0.72% (Figure 4B). Consistent with our findings from the narrowly defined birth cohort, APOE was not associated with paternal lifespan in the broadly defined cohort (Figure S8B).
Table 3. AncestryDNA-UKB GWA meta-analysis of parental lifespan. The P value is given of the lead variant at each locus in the test of association with parental lifespan in the AncestryDNA, UKB and combined cohorts. The z-score transformed variant effect sizes and standard errors are listed for the AncestryDNA and UKB analyses. Variants which were significantly associated with lifespan in either the AncestryDNA or UKB cohorts, and were not significant in the meta-analysis, are provided at the bottom of the table.
| Meta-Analysis | AncestryDNA | UKB | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Locus | rsid | chr | bp | P | P | Effect (z-score) | SE | P | Effect (z-score) | SE |
| Paternal | CLESR2..PSRC1 | rs599839 | 1 | 109822166 | 4.43e-09 | 1.66e-03 | 0.010 | 0.003 | 4.00e-09 | 0.015 | 0.003 |
| Lifespan | MICA..MICB | rs3131621 | 6 | 31425499 | 8.86e-09 | 7.72e-04 | −0.011 | 0.003 | 3.60e-08 | −0.012 | 0.002 |
| LPA | rs140570886 | 6 | 161013013 | 1.13e-24 | 1.84e-13 | −0.080 | 0.011 | 1.10e-17 | −0.077 | 0.009 | |
| LPL | rs15285 | 8 | 19824667 | 8.46e-10 | 2.44e-05 | 0.012 | 0.003 | 1.20e-07 | 0.013 | 0.002 | |
| EPHX2..CLU | rs7844965 | 8 | 27442064 | 5.79e-10 | 9.06e-06 | 0.013 | 0.003 | 2.20e-07 | 0.013 | 0.003 | |
| ANRIL | rs1556516 | 9 | 22100176 | 1.57e-28 | 2.99e-15 | 0.020 | 0.003 | 1.30e-20 | 0.020 | 0.002 | |
| ATXN2 | rs10774625 | 12 | 111910219 | 1.39e-08 | 9.97e-02 | 0.004 | 0.003 | 2.80e-12 | 0.015 | 0.002 | |
| SEMA6D | rs4774495 | 15 | 47651362 | 4.92e-08 | 4.29e-03 | −0.011 | 0.004 | 3.30e-08 | −0.019 | 0.003 | |
| CHRNA3/5 | rs9788721 | 15 | 78802869 | 1.10e-31 | 8.49e-10 | −0.016 | 0.003 | 1.40e-32 | −0.027 | 0.002 | |
| LDLR | rs6511720 | 19 | 11202306 | 2.73e-09 | 6.78e-05 | 0.016 | 0.004 | 1.70e-07 | 0.018 | 0.003 | |
| APOE | rs769449 | 19 | 45410002 | 1.86e-20 | 4.31e-05 | −0.016 | 0.004 | 5.60e-24 | −0.033 | 0.003 | |
| Maternal | IP6K1 | rs9872864 | 3 | 49792023 | 6.90e-10 | 5.61e-05 | −0.011 | 0.003 | 1.30e-07 | −0.011 | 0.002 |
| Lifespan | LPA | rs186696265 | 6 | 161111700 | 8.43e-09 | 3.33e-03 | −0.034 | 0.012 | 1.60e-08 | −0.051 | 0.009 |
| CHRNA3/5 | rs4887072 | 15 | 78925435 | 1.92e-09 | 3.82e-05 | 0.014 | 0.003 | 6.80e-07 | 0.013 | 0.003 | |
| APOE | rs429358 | 19 | 45411941 | 4.37e-57 | 2.53e-09 | −0.023 | 0.004 | 1.30e-68 | −0.053 | 0.003 | |
| Not Replicated | |||||||||||
| Paternal | SRMM3 | rs28689051 | 7 | 75763624 | 3.41e-03 | 2.64e-09 | 0.016 | 0.003 | 2.60e-01 | −0.003 | 0.002 |
| Lifespan | WAPL | rs10887623 | 10 | 88315181 | 3.32e-03 | 3.36e-06 | −0.020 | 0.004 | 9.20e-01 | 0.000 | 0.004 |
| ZW10 | rs17613838 | 11 | 113638177 | 1.92e-04 | 8.11e-01 | 0.001 | 0.003 | 6.10e-09 | −0.016 | 0.003 | |
| C20orf187 | rs6077996 | 20 | 10972839 | 2.28e-04 | 7.21e-01 | 0.001 | 0.003 | 4.60e-09 | −0.013 | 0.002 | |
| Maternal Lifespan | PSORS1C3 | rs1265159 | 6 | 31140047 | 6.43e-07 | 3.43e-01 | −0.004 | 0.004 | 3.60e-10 | −0.016 | 0.003 |
We found no variants that were significantly associated with maternal or paternal lifespan in the expanded birth cohort analyses of the Sub-Saharan African or Native American/East Asian population groups (Figure S9E-H). With fewer than 20,000 individuals, these analyses were underpowered and this negative result was expected (Figure S2).
Age was strongly associated with the APOE locus
We next sought to compare the lifespan GWA results to the commonly used proxy trait: age. We evaluated age in both the quantitative trait framework, including all individuals in a cohort (Figure S10A), and in the routinely employed case/control framework (e.g., Deelen et al. 2011; Deelen et al. 2014; Broer et al. 2015), wherein the ‘cases’ were individuals above a specific age (e.g., 5% tail: 86 years for males and 87 years for females) and the ‘controls’ were individuals below a certain age (e.g., median age: 65 years for males and 64 years for females). The GWA mapping population included the European population group; individuals with close relatives in this population (IBD threshold of 300cM) were removed. In agreement with many previous studies, we identified genome-wide significant SNPs at the APOE locus in analyses of males, females, and combining samples from both genders (Table S6; Figure S10B-F). Additionally, we identified a SNP at the MAP2K6 gene, which narrowly achieved genome wide significance in the case/control analysis of female age (Figure S10C).
Genetic correlations between paternal lifespan and age
We measured the genetic correlation, rg, between the maternal and paternal lifespan and between age and lifespan using LD Score regression (Finucane et al. 2015; Bulik-Sullivan et al. 2015). We estimated the genetic correlation between maternal and paternal lifespan to be quite high: rg = 0.65 (SE = 0.09) for the 1886-1918 cohort and rg = 0.94 (SE = 0.04) in the 1886-1940 cohort (Table 2). The high genetic correlations suggest that the majority of significant allelic effects on lifespan will be similar in magnitude and direction between men and women, with a minority of alleles effecting lifespan in a gender-specific manner.
Table 2. Phenotypic and genetic correlations for maternal vs. paternal lifespan and age vs. lifespan. N- Overlap is the number of samples which have phenotypes measured for both traits. N - SNPs is the number of variants used in LD Score regression analysis. Phenotypic variance (PVAR), the slope of the correlation (beta), and the Pearson correlation coefficient (rp). Genetic heritability (h2g) and genetic correlation (rg).
| Traits | Phenotypic Correlation | Genetic Correlation | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Lifespan | Lifespan | N - Overlap | PVAR Mat. | PVAR Pat. | rp | N - SNPs | %h2g Maternal (SE) | %h2g Paternal (SE) | rg (SE) |
| Maternal_1918 | Paternal_1918 | 112498 | 161.53 | 166.20 | 0.068 | 385494 | 3.66 (0.64) | 7.54 (0.58) | 0.651 (0.090) |
| Maternal_1940 | Paternal_1940 | 219514 | 153.42 | 160.27 | 0.095 | 386724 | 5.34 (0.36) | 7.62 (0.32) | 0.937 (0.041) |
| Age | Lifespan | N - Overlap | PVAR Age | PVAR Lifespan | rp | N - SNPs | %h2g Age (SE) | %h2g Lifespan (SE) | rg (SE) |
| Male | Maternal | 90496 | 62.03 | 153.94 | 0.095 | 385974 | 3.04 (0.42) | 4.78 (0.92) | 0.700 (0.127) |
| Male | Paternal | 98313 | 59.88 | 162.09 | 0.000 | 386086 | 2.68 (0.37) | 7.04 (0.86) | 0.432 (0.097) |
| Female | Maternal | 109571 | 66.67 | 153.84 | 0.098 | 386209 | 3.69 (0.44) | 4.88 (0.76) | 0.693 (0.094) |
| Female | Paternal | 115806 | 59.86 | 161.66 | 0.004 | 386293 | 2.61 (0.40) | 7.54 (0.80) | 0.527 (0.094) |
Next, we estimated within and between gender genetic correlations between age and parental lifespan. Similar to the parental lifespan analysis, the genetic correlations between these traits were high: maternal lifespan compared to female and male age was 0.69 (SE = 0.09) and 0.70 (SE = 0.13); paternal lifespan compared to male and female age was 0.43 (SE = 0.10) and 0.53 (SE = 0.09) (Table 2). In summary: hg2 for each trait was quite low (between 2 and 8% for different sub-cohorts), but the variants that underlie maternal vs. paternal lifespan or parental lifespan vs. age had substantially correlated effects.
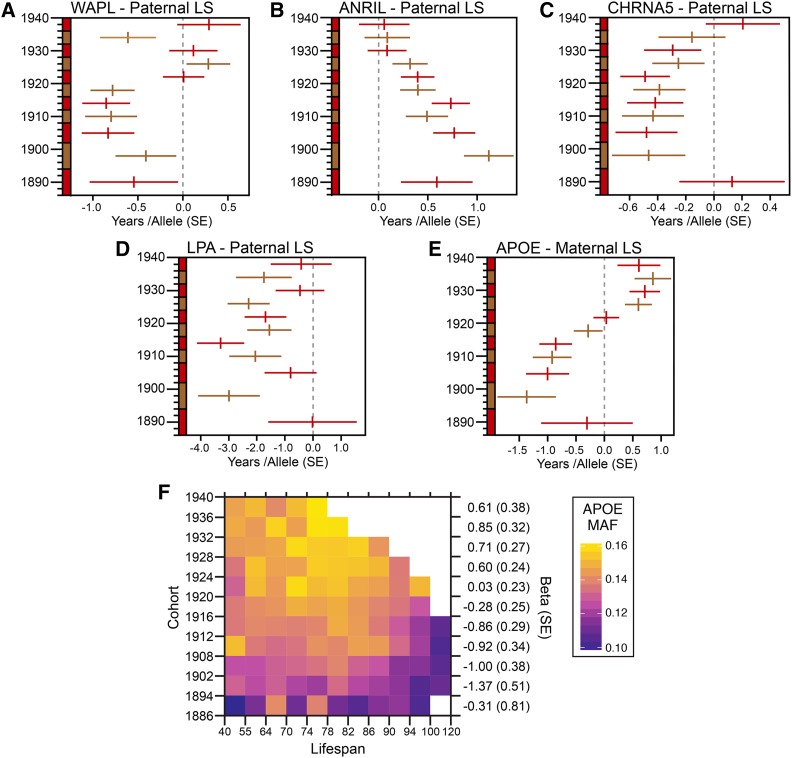
Effects of WAPL and APOE on parental lifespan varied across birth cohorts
We investigated whether lifespan determining variants exhibited birth-cohort-specific effects because of the inconsistent WAPL results between the 1886-1918 (Figure 3D) and 1886-1940 (Figure 4D) cohorts: loss of significance despite increased sample size. We found that variants at WAPL had a mean effect of 0.70 (SE = 0.07) years in the 1886-1920 cohorts, and in all later cohorts, save 1928-1932, it was not significantly different from zero (Figure 5A). For comparison, ANRIL, CHRNA3/5, and LPA had significantly affected lifespan through 1928 (Figure 5B-D), after which we observed a decrease in mean effect size, likely due to increased censorship in the younger cohorts (assumed to be 18.5–40.9% for men in 1928-1940 cohorts; Figure 1D). In contrast, the youngest cohort in which WAPL had no significant effect, 1920-1924, had an assumed censorship of only 4.1% (Figure 1D). The shift in effect size could not be attributed to variation in the MAF – the standard deviation of MAF estimates, normalized to the mean, was 1.4%.
Figure 5.
(A-E) Forest plots of allelic effect in years (+/−SE) of lead SNP at each candidate locus in 1886-1940 birth cohorts. Alternating red and brown lines mark successive cohorts. (F) Heat map of APOE minor allele frequency in the 1886-1940 birth cohorts, binned into lifespan phenotype classes spanning 40 – 120 years. White squares are missing data- birth cohort, phenotype combinations less than 500 individuals.
Next, we examined birth-cohort-specific effects at APOE, which exhibited significant negative effects on lifespan in the 1894-1920 birth cohorts and consistently positive effects in birth cohorts after 1923 (Figure 5E). Measurement of the frequency of the APOE allele as a function of both phenotypic class and birth cohort (Figure 5F) revealed that: 1) the MAF increased from 11.38% in older birth cohorts to 15.15% in younger cohorts; and 2) the increase was not consistent across phenotypic classes, but was most pronounced for intermediate lifespan values (74-86 years). Thus, the APOE allele was most strongly associated with intermediate lifespans, suggesting it may be beneficial early in life and detrimental later in life (Figure 5F). The apparent positive effect of the APOE allele on lifespan in cohorts after 1923 (displayed as an increase in MAF at intermediate lifespan values, 74-86 years, relative to shorter lifespan values, 40-74 years in Figure 5F) was actually due to systematic censorship of longer lifespans in younger cohorts (shown as empty squares in top right corner of Figure 5F). In summary: the minor-frequency APOE allele was associated with intermediate lifespan values, and this association was stronger in younger cohorts.
GWA meta-analysis of parental lifespan with UKB dataset identified 15 lifespan associated variants
The only GWA studies of parental lifespan comparable to this analysis are derived from the UKB (Pilling et al. 2016; Joshi et al. 2016; McDaid et al. 2017; Joshi et al. 2017; Mostafavi et al. 2017; Pilling et al. 2017). Of these studies, the analysis with the largest sample size, N = 389,166 (Pilling et al. 2017) identified six and fourteen loci associated with maternal and paternal attained-age, respectively. The trait investigated in that analysis is hazard of survival, a derived variable that is inclusive of both lifespan and age. We estimated the same-gender genetic correlation between the lifespan and attained-age across the two analyses and found that these values were close to 1.0 (paternal rg = 0.98, SE = 0.06; maternal rg = 1.02, SE = 0.11; S Table 7). We then estimated the across-gender genetic correlations for the two analyses and found that rg ranged from 0.88 (SE = 0.08) to 0.90 (SE = 0.08); these values were similar to those observed for the AncestryDNA specific analyses (Table 2; Table S7). Next, we tested whether the AncestryDNA lifespan associated loci were also significant in a meta-analysis of results from the UKB attained-age GWA.
We conducted the AncestryDNA-UKB meta-analysis of more than 658,000 individuals using METAL (Willer, Li, and Abecasis 2010). We included AncestryDNA GWA results from the broadly defined birth cohort (1886-1940; the larger of the two cohorts we investigated). Only variants that were common to both studies were included in the analysis; this requirement removed some variants that were found to be significant in the UKB study (details provided below).
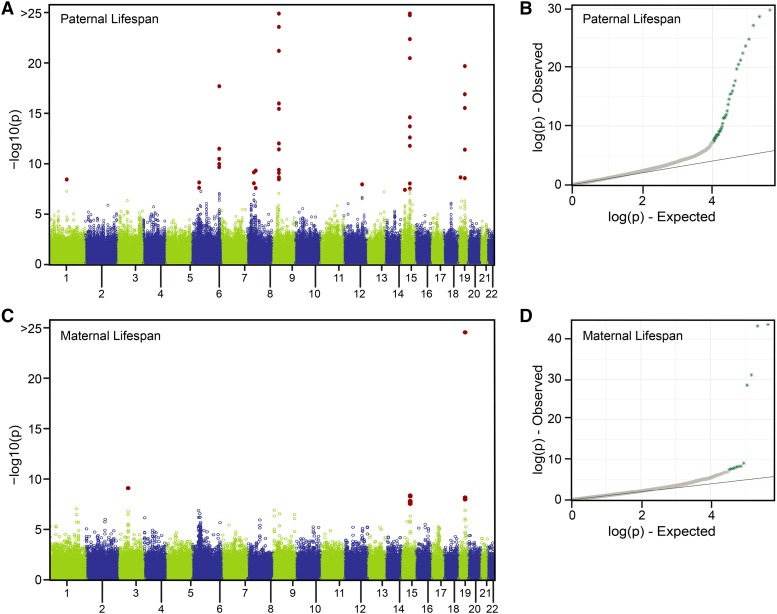
We found a total of 11 loci significantly associated with paternal lifespan (P < 5e-8) in the meta-analysis (Figure 6A,B). Of the five AncestryDNA paternal lifespan associated loci, three (LPA, ANRIL, and CHRNA3/5) maintained genome-wide significance in the meta-analysis, while WAPL and SRRM3 failed to do so (Table 3). Of the 14 loci associated with attained-age of fathers in the UKB GWA (Pilling et al. 2017) genetic variants from ten loci were included in this meta-analysis (see Methods). Eight of those reached statistical significance (Table 3). In addition to the previously mentioned LPA, ANRIL, CHRNA3/5 we found variants at: APOE, CLESR2-PSRC1, ATXN2, SEMA6D, and MICA-MICB were significant in the meta-analysis (Table 3). Two attained-age associated loci from the UKB analysis, ZW10 and C20orf187, had P values greater than 5e-8 in the meta-analysis (Table 3). Finally, we identified significant associations at three loci, LPL, EPHX2/CLU, and LDLR (Table 3), which were marginally associated with paternal lifespan in the either the AncestryDNA or UKB studies.
Figure 6.
AncestryDNA-UKB meta-analysis results for maternal and paternal lifespan. Manhattan plots (A,C) where each circle shows the P value association test statistic for each genotyped SNP and solid red circles denote SNPs with Bonferroni corrected P values less than 0.05. QQ plots (B, D) of association test statistics, aquamarine colored points denote SNPs with Bonferroni corrected P values less than 0.05.
We found a total of four loci significantly associated with maternal lifespan in the meta-analysis, including APOE, the one locus significantly associated with maternal lifespan in the AncestryDNA dataset (Table 3; Figure 6C,D). Six loci were associated with maternal attained-age in the UKB GWA, and three of them, including APOE, were analyzed in the AncestryDNA-UKB meta-analysis. One locus, LPA, achieved statistical significance, whereas PSORS1C3 failed to do so (Table 3). Finally, we identified two loci which, to our knowledge, have not been previously associated with maternal lifespan: IP6K1 and CHRNA3/5 (Table 3).
Discussion
The AncestryDNA-UKB meta-analysis interrogated more than 658,000 samples and identified 11 loci significantly associated with paternal lifespan and four loci significantly associated with maternal lifespan. This analysis was the first large-scale, independent comparison of UKB parental lifespan GWA results.
Many lifespan-associated loci were also linked with life-shortening diseases
We found variants at five loci (ANRIL, LPA, APOE, LDLR, LPL) that we refer to as candidate lifespan loci to emphasize that we identified a significant statistical association, while acknowledging that we have no functional-mechanistic data on how these genetic variants may affect lifespan. However, we suspect these loci to affect lifespan via increased risk for cardiovascular disease because they were previously associated with this disease (Nikpay et al. 2015; van der Harst and Verweij 2018) and it was the leading cause of death in the United States from 1980 to 2014 (Dwyer-Lindgren et al. 2016). ANRIL, LPA and APOE were significantly associated with parental lifespan in both the individual AncestryDNA and UKB analyses as well as the meta-analysis (Table 3). In contrast, LDLR and LPL were only significantly associated with lifespan in the large meta-analysis and marginally associated with lifespan in the individual AncestryDNA and UKB analyses (Table 3).
The CHRN3/5 locus has been linked to smoking behavior and incidence of chronic obstructive pulmonary disease (Hobbs et al. 2017), another leading life-threatening disease in the United States (Dwyer-Lindgren et al. 2016). This locus was significantly associated with paternal lifespan in both the AncestryDNA and UKB independent analyses. In the meta-analysis, the combined power of both cohorts also revealed this locus to also be significantly associated with maternal lifespan (Table 3).
We found two lifespan candidate loci which were previously associated with Alzheimer’s disease (Lambert et al. 2013): the newly identified EPHX2/CLU and, perhaps the most widely reported candidate lifespan locus: APOE (Deelen et al. 2011; Deelen et al. 2014; Broer et al. 2015). Cohort-specific analysis of APOE revealed two curious results: 1) APOE exhibited a negative effect on lifespan in older cohorts and a positive effect in younger cohorts (Figure 5E) and 2) the minor allele frequency increased from 11 to 15% between the 1886 and 1940 birth cohorts (Figure 5G). The minor allele at APOE was at highest frequency for intermediate lifespan values (74-86 years). This pattern was most pronounced in the younger birth cohorts, and it suggested that this allele (or a linked allele or alleles) confers a survival benefit early in life but a survival detriment later in life. While it may be tempting to invoke the antagonistic pleiotropy theory of aging to explain such a result (Williams 1957), two variants in tight linkage disequilibrium, which could even modify the action of two different genes in this gene-dense region (S Figure 8), would equally explain this result.
Finally, three loci had no known disease associations. WAPL and SRRM3, identified in the AncestryDNA analysis (Table 1) and IP6K1, identified in the meta-analysis (Table 3) have, to our knowledge, not been associated with any life-shortening diseases.
Heterogeneity of GWA results across analyses
Several loci that were significantly associated with lifespan in either the AncestryDNA analysis (WAPL and SRRM3) or the UKB analysis (maternal lifespan: PSORS1C3; paternal lifespan: ZW10 and C20orf187) did not achieve significance in the meta-analysis. One explanation is that these loci do not affect parental lifespan. This could be due to underlying population structure that was not fully accounted for in the GWA mapping models. If so, the signal of association would not be replicated in the comparison between the primarily-British vs. primarily-American cohorts. Alternatively, the candidate loci may directly affect parental lifespan, but distinct population structure, environmental hazards, or phenotypic definitions between the two studies may have resulted in a failure to replicate across independent cohorts.
A second factor to consider is that the focal trait examined in our AncestryDNA study was lifespan and only included deceased parents, whereas the UKB study examined the attained-age of both living and deceased parents. We reiterate that we did not include parents lacking a death date in the AncestryDNA analysis because we could not determine whether any specific individual was in fact alive or deceased and their date of death was never recorded (Ruby et al. 2018, Figure 1G). The difference between focal traits implies that the UKB population included younger individuals from more recent birth cohorts compared to AncestryDNA, and differences in the environment or population sub-structure between historical eras may have contributed to the lack a replication between studies.
Differences in the genetic architecture of lifespan vs. age
The difficulty of obtaining genetic information from deceased individuals has motivated investigators to use age as a proxy phenotype for human lifespan (Schächter et al. 1994; Willcox et al. 2008; Newman et al. 2010; Soerensen et al. 2010; Deelen et al. 2011; Nebel et al. 2011; Beekman et al. 2013; Soerensen et al. 2013; Sebastiani et al. 2013; Deelen et al. 2014; Broer et al. 2015; Sebastiani et al. 2017). We applied that strategy in this study. However, we also addressed this issue using a second strategy: we examined the complete lifespans of parents and the genotypes of their offspring (similar to the approach formalized in Liu et al. (2017)). The genetic correlations between the outcomes of these two strategies was relatively high, ranging from 0.43 to 0.70 in gender specific comparisons (Table 2). Nonetheless, of the several loci that were associated with either age or parental lifespan to a statistically significant degree, only the APOE locus was associated with both (Figure 4I; Figure S10). These results suggested that the use age as a proxy for lifespan in a GWA analysis is informative but not comprehensive.
The differences in GWA mapping results between age and lifespan were likely due, at least in part, to the fundamental nature of the two traits. Lifespan is a property of deceased individuals and was measured prospectively on individuals from the same birth cohort. Age is a property of the living and GWA analyses of this trait are retrospective comparisons between differentially aged individuals from distinct birth cohorts. Prospective GWA analysis of lifespan potentially avoids problems that are specific to retrospective studies: recruitment bias of long-lived individuals and establishment of an appropriate youthful control population (Clayton and McKeigue 2001; Manolio, Bailey-Wilson, and Collins 2006). Recruitment biases may explain why the association between APOE and current age is much stronger than with lifespan; variants at this locus significantly increases the incidence of Alzheimer’s disease in old age, and may inhibit participation in web-based genealogy services and in providing informed consent to research.
Little heritability is “missing” from the SAP cohort
We estimated hg2 for paternal and maternal lifespan to be between 2 and 8%, depending on the cohort examined (Figure 3F, 4B). For many phenotypes, it has been noted that estimated hg2 values, measured using GWA methods, are generally much less than h2 values measured based on familial phenotypic correlations; this recurring discrepancy is known as the “missing heritability” problem (Manolio et al. 2009). While our hg2 values could potentially have been increased through the evaluation of genome-wide imputed variants (Yang et al. 2010; Yang et al. 2015), our previously published measure of lifespan heritability derived from familial correlations in the same SAP used for this analysis (Ruby et al. 2018) produced a range of h2 values, all below 10%. While our estimate of hg2 was slightly lower than our estimate of h2, the values reported in each study cover the same range, giving the impression that there was little heritability remaining to be explained.
In comparison to previously published estimates of h2 for human lifespan, our estimate of hg2 was low, and would have been interpreted as consistent with the “missing heritability” problem (Manolio et al. 2009). For instance, another large-pedigree study recently estimated h2 to be between 16 and 18% (Kaplanis et al. 2018), and multiple smaller studies estimated h2 to be in the range of 15–30% (Philippe 1978; Mayer 1991; Herskind et al. 1996; Ljungquist et al. 1998; Gavrilov et al. 1998; Mitchell et al. 2001; Kerber et al. 2001; Gögele et al. 2011). Had those estimates of heritability been used, the apparently “missing” component following our present analysis would have been quite large. Importantly, our own estimates of under 10% overall heritability for lifespan differed from the precedent literature not by the familial correlations, but rather by our accounting for considerable assortative mating (Ruby et al. 2018). Therefore, our observation that the small difference between h2 and hg2 is due to the consideration of assortative mating is an alternative hypothesis to the often invoked explanations: rare variants, structural variation, or epistatic interactions (Manolio et al. 2009). The extent of assortative mating, either by primary or secondary means, has not been thoroughly explored for many other phenotypes. It is worth considering that the inflation of h2 estimates due to assortative mating may be more generally responsible for the recurrence of sizable “missing heritability” gaps.
Acknowledgments
We gratefully acknowledge the AncestryDNA customers who consented to participate in research. This manuscript was greatly improved by comments from Anastasia Baryshnikova, Madeleine Cule, Nicholas Eriksson, Eugene Melamud, Leland Taylor, and David Botstein at Calico Life Sciences LLC. In memory of Natalie Myres, whose dedicated work has made this manuscript a reality.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.8479022.
Communicating editor: B. Andrews
Literature Cited
- Aberg K. A., Shabalin A. A., Chan R. F., Zhao M., Kumar G. et al. , 2018. Convergence of Evidence from a Methylome-Wide CpG-SNP Association Study and GWAS of Major Depressive Disorder. Translational Psychiatry 8(1): 162. 10.1038/s41398-018-0205-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backlund E., Rowe G., Lynch J., Wolfson M. C., Kaplan G. A. et al. , 2007. Income inequality and mortality: a multilevel prospective study of 521 248 individuals in 50 US states. Int. J. Epidemiol. 36: 590–596. 10.1093/ije/dym012 [DOI] [PubMed] [Google Scholar]
- Ball C., Barber M., Byrnes J., Callaway J., Chahine K. et al. , 2013. Ethnicity Estimate White Paper. https://www.ancestry.com/dna/resource/whitePaper/AncestryDNA-Ethnicity-White-Paper.pdf.
- Ball C., Barber M., Byrnes J., Carbonetto P., Chahine K. et al. , 2016. Ancestry DNA Matching White Paper. https://www.ancestry.com/corporate/sites/default/files/AncestryDNA-Matching-White-Paper.pdf.
- Beekman M., Blanché H., Perola M., Hervonen A., Bezrukov V. et al. , 2013. Genome-Wide Linkage Analysis for Human Longevity: Genetics of Healthy Aging Study. Aging Cell 12: 184–193. 10.1111/acel.12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braveman P. A., Cubbin C., Egerter S., Chideya S., Marchi K. S. et al. , 2005. Socioeconomic Status in Health Research. JAMA 294(22): 2879–2888. 10.1001/jama.294.22.2879 [DOI] [PubMed] [Google Scholar]
- Broer L., Buchman A. S., Deelen J., Evans D. S., Faul J. D. et al. , 2015. GWAS of Longevity in CHARGE Consortium Confirms APOE and FOXO3 Candidacy. J. Gerontol. A Biol. Sci. Med. Sci. 70: 110–118. 10.1093/gerona/glu166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B., Finucane H. K., Anttila V., Gusev A., Day F. R. et al. , 2015. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47: 1236–1241. 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai R., Hao Z., Winslett M., Xiao X., Yang Y. et al. , 2015. Deterministic identification of specific individuals from GWAS results. Bioinformatics 31: 1701–1707. 10.1093/bioinformatics/btv018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. C., Chow C. C., Tellier L. C. A. M., Vattikuti S., Purcell S. M. et al. 2015. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience 4: 1. 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed]
- Chetty R., Stepner M., Abraham S., Lin S., Scuderi B. et al. , 2016. The association between income and life expectancy in the United States, 2001–2014. JAMA 315: 1750–1766. Erratum: 317: 90. 10.1001/jama.2016.4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D., and McKeigue P. M., 2001. Epidemiological Methods for Studying Genes and Environmental Factors in Complex Diseases. The Lancet 358(9290): 1356–1360. 10.1016/S0140-6736(01)06418-2 [DOI] [PubMed]
- Crimmins E. M., and Saito Y., 2001. Trends in Healthy Life Expectancy in the United States, 1970–1990: Gender, Racial, and Educational Differences. Soc. Sci. Med. 52: 1629–1641. 10.1016/S0277-9536(00)00273-2 [DOI] [PubMed] [Google Scholar]
- Das S., Forer L., Schönherr S., Sidore C., Locke A. E. et al. , 2016. Next-generation genotype imputation service and methods. Nat. Genet. 48: 1284–1287. 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen J., Beekman M., Uh H. W., Broer L., Ayers K. L. et al. , 2014. Genome-Wide Association Meta-Analysis of Human Longevity Identifies a Novel Locus Conferring Survival beyond 90 Years of Age. Hum. Mol. Genet. 23: 4420–4432. 10.1093/hmg/ddu139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen J., Beekman M., Uh H. W., Helmer Q., Kuningas M. et al. , 2011. Genome-Wide Association Study Identifies a Single Major Locus Contributing to Survival into Old Age; the APOE Locus Revisited. Aging Cell 10: 686–698. 10.1111/j.1474-9726.2011.00705.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer-Lindgren L., Bertozzi-Villa A., Stubbs R. W., Morozoff C., Kutz M. J. et al. , 2016. US County-Level Trends in Mortality Rates for Major Causes of Death, 1980–2014. JAMA 316(22): 2385–2401. 10.1001/jama.2016.13645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane H. K., Brendan B.-S., Gusev A., Trynka G., Reshef Y., Loh P.-R., Anttila V. et al. , 2015. Partitioning Heritability by Functional Annotation Using Genome-Wide Association Summary Statistics. Nature Genetics 47 (11): 1228–1235. 10.1038/ng.3404 [DOI] [PMC free article] [PubMed]
- Gavrilov N. S., Gavrilov L. A., Evdokushkina G. N., Semyonova V. G., Gavrilova A. L. et al. , 1998. Evolution, mutations, and human longevity: European royal and noble families. Hum Biol. 70: 799–804. [PubMed] [Google Scholar]
- Gögele M., Pattaro C., Fuchsberger C., Minelli C., Pramstaller P. P. et al. , 2011. Heritability analysis of life span in a semi-isolated population followed across four centuries reveals the presence of pleiotropy between life span and reproduction. J. Gerontol. A Biol. Sci. Med. Sci. 66: 26–37. 10.1093/gerona/glq163 [DOI] [PubMed] [Google Scholar]
- Gusev A., Lowe J. K., Stoffel M., Daly M. J., Altshuler D. et al. , 2009. Whole population, genome-wide mapping of hidden relatedness. Genome Res. 19: 318–326. 10.1101/gr.081398.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymrek M., McGuire A., Golan D., Halperin E., and Erlich Y., 2013. Identifying Personal Genomes by Surname Inference. Science 339 (6117). American Association for the Advancement of Science. 10.1126/science.1229566 [DOI] [PubMed]
- Han E., Carbonetto P., Curtis R. E., Wang Y., Granka J. M. et al. , 2017. Clustering of 770,000 genomes reveals post-colonial population structure of North America. Nat. Commun. 8: 14238 10.1038/ncomms14238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Harst P., and Verweij N., 2018. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ. Res. 122: 433–443. 10.1161/CIRCRESAHA.117.312086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslanger S., 2008. A Social Constructionist Analysis of Race, pp. 56–69 in Revisiting Race in a Genomic Age, edited by Koenig B. A., Lee S. S. J., and Richardson S. S.. Rutgers University Press, New Brunswick, NJ. [Google Scholar]
- Herskind A. M., McGue M., Holm N. V., Sørensen T. I. A., Harvald B. et al. 1996. The Heritability of Human Longevity: A Population-Based Study of 2872 Danish Twin Pairs Born 1870–1900. Human Genetics 97(3): 319–323. 10.1007/BF02185763 [DOI] [PubMed]
- Hobbs B. D., de Jong K., Lamontagne M., Bossé Y., Shrine N. et al. , 2017. Genetic Loci Associated with Chronic Obstructive Pulmonary Disease Overlap with Loci for Lung Function and Pulmonary Fibrosis. Nature Genetics 49(3): 426–32. 10.1038/ng.3752 [DOI] [PMC free article] [PubMed]
- Homer N., Szelinger S., Redman M., Duggan D., Tembe W. et al. , 2008. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genetics 4: e1000167 10.1371/journal.pgen.1000167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Shmygelska A., Tran D., Eriksson N., Tung J. Y. et al. , 2016. GWAS of 89,283 Individuals Identifies Genetic Variants Associated with Self-Reporting of Being a Morning Person. Nature Communications 7(1): 10448. 10.1038/ncomms10448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huusko J. M., Karjalainen M. K., Graham B. E., Zhang G., Farrow E. G. et al. , 2018. Whole exome sequencing reveals HSPA1L as a genetic risk factor for spontaneous preterm birth. PLoS Genetics 14: e1007394 10.1371/journal.pgen.1007394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im Hae Kyung, Gamazon Eric R., Nicolae Dan L., and Cox Nancy J., 2012. On Sharing Quantitative Trait GWAS Results in an Era of Multiple-Omics Data and the Limits of Genomic Privacy. The American Journal of Human Genetics 90(4): 591–598. 10.1016/j.ajhg.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen P. R., Kyoko W., Stringer S., Skene N., Bryois J. et al. , 2019. Genome-Wide Analysis of Insomnia in 1,331,010 Individuals Identifies New Risk Loci and Functional Pathways. Nature Genetics 51(3): 394–403. 10.1038/s41588-018-0333-3 [DOI] [PubMed]
- Jha P., Peto R., Zatonski W., Boreham J., and Jarvis M. J. et al. , 2006. Social Inequalities in Male Mortality, and in Male Mortality from Smoking: Indirect Estimation from National Death Rates in England and Wales, Poland, and North America. Lancet 368: 367–370. 10.1016/S0140-6736(06)68975-7 [DOI] [PubMed] [Google Scholar]
- Jones A. V., Tilley M., Gutteridge A., Hyde C., Nagle M. et al. , 2017. GWAS of self-reported mosquito bite size, itch intensity and attractiveness to mosquitoes implicates immune-related predisposition loci. Hum. Mol. Genet. 26: 1391–1406. 10.1093/hmg/ddx036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. E., Lane J. M., Wood A. R., van Hees V. T., Tyrrell J. et al. , 2019. Genome-Wide Association Analyses of Chronotype in 697,828 Individuals Provides Insights into Circadian Rhythms. Nature Communications 10(1): 343. 10.1038/s41467-018-08259-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi P. K., Fischer K., Schraut K. E., Campbell H., Esko T. et al. , 2016. Variants near CHRNA3/5 and APOE Have Age- and Sex-Related Effects on Human Lifespan. Nat. Commun. 7: 11174 10.1038/ncomms11174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi P. K., Pirastu N., Kentistou K. A., Fischer K., Hofer E. et al. , 2017. Genome-Wide Meta-Analysis Associates HLA- DQA1/DRB1 and LPA and Lifestyle Factors with Human Longevity. Nat. Commun. 8: 910 10.1038/s41467-017-00934-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplanis J., Gordon A., Shor T., Weissbrod O., Geiger D. et al. , 2018. Quantitative Analysis of Population-Scale Family Trees with Millions of Relatives. Science (New York, N.Y.) 360(6385): 171–175. 10.1126/science.aam9309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerber R. A., O’Brien E., Smith K. R., and Cawthon R. M., 2001. Familial excess longevity in Utah genealogies. J. Gerontol. A Biol. Sci. Med. Sci. 56: B130–B139. 10.1093/gerona/56.3.B130 [DOI] [PubMed] [Google Scholar]
- Kitagawa E. M., and Hauser P. M., 1973. Differential Mortality in the United States, Harvard University Press, Cambridge, MA and London, England: 10.4159/harvard.9780674188471 [DOI] [Google Scholar]
- Lambert J.-C., Ibrahim-Verbaas C. A., Harold D., Naj A. C., Sims R. et al. 2013. Meta-Analysis of 74,046 Individuals Identifies 11 New Susceptibility Loci for Alzheimer’s Disease. Nature Genetics 45: 1452–1458. 10.1038/ng.2802 [DOI] [PMC free article] [PubMed]
- Lee J. J., Wedow R., Okbay A., Kong E., Maghzian O. et al. , 2018. Gene Discovery and Polygenic Prediction from a Genome-Wide Association Study of Educational Attainment in 1.1 Million Individuals. Nature Genetics 50(8): 1112–1121. 10.1038/s41588-018-0147-3 [DOI] [PMC free article] [PubMed]
- Liu J. Z, Erlich Y., and Pickrell J. K.. 2017. Case–Control Association Mapping by Proxy Using Family History of Disease. Nature Genetics 49(3): 325–331. 10.1038/ng.3766 [DOI] [PubMed]
- Ljungquist B., Berg S., Lanke J., McClearn G. E., and Pedersen N. L., 1998. The effect of genetic factors for longevity: a comparison of identical and fraternal twins in the Swedish twin registry. J. Gerontol. A Biol. Sci. Med. Sci. 53: M441–M446. 10.1093/gerona/53A.6.M441 [DOI] [PubMed] [Google Scholar]
- Manolio T. A., Bailey-Wilson J. E., and Collins F. S., 2006. Genes, Environment and the Value of Prospective Cohort Studies. Nature Reviews Genetics 7(10): 812–820. 10.1038/nrg1919 [DOI] [PubMed]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A. et al. 2009. Finding the Missing Heritability of Complex Diseases. Nature 461(7265): 747–753. 10.1038/nature08494 [DOI] [PMC free article] [PubMed]
- Mayer P. J. 1991. Inheritance of Longevity Evinces No Secular Trend among Members of Six New England Families Born 1650–1874. American Journal of Human Biology 3(1): 49–58. 10.1002/ajhb.1310030109 [DOI] [PubMed]
- McDaid A. F., Joshi P. K., Porcu E., Komljenovic A., Li H. et al. , 2017. Bayesian Association Scan Reveals Loci Associated with Human Lifespan and Linked Biomarkers. Nature Communications 8 (July). Nature Publishing Group 15842 10.1038/ncomms15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills C. W., 1997. The Racial Contract, Cornell University Press, Ithaca. [Google Scholar]
- Mitchell B. D., Hsueh W.-C., King T. M., Pollin T. I., and Sorkin J.. et al, 2001. Heritability of Life Span in the Old Order Amish. American Journal of Medical Genetics 102(4): 346–352. 10.1002/ajmg.1483 [DOI] [PubMed] [Google Scholar]
- Mostafavi H., Berisa T., Day F. R., Perry J. R. B., Przeworski M. et al. , 2017. Identifying genetic variants that affect viability in large cohorts. PLoS Biol. 15: e2002458 10.1371/journal.pbio.2002458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C. J., Kulkarni S. C., Michaud C., Tomijima N., Bulzacchelli M. T. et al. , 2006. Eight Americas: investigating mortality disparities across races, counties, and race-counties in the United States. PLoS Med. 3: e260 10.1371/journal.pmed.0030260 Erratum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C. J., Michaud C. M., McKenna M. T., and Marks J. S., 1998. US patterns of mortality by county and race: 1965–1994. Cambridge: Harvard Center for Population and Development Studies.
- Nebel A., Kleindorp R., Caliebe A., Nothnagel M., Blanché H. et al. , 2011. A Genome-Wide Association Study Confirms APOE as the Major Gene Influencing Survival in Long-Lived Individuals. Mech. Ageing Dev. 132: 324–330. 10.1016/j.mad.2011.06.008 [DOI] [PubMed] [Google Scholar]
- Newman A. B., Walter S., Lunetta K. L., Garcia M. E., Slagboom P. E. et al. , 2010. A meta-analysis of four genome-wide association studies of survival to age 90 years or older: the cohorts for heart and aging research in genomic epidemiology consortium. J. Gerontol. A Biol. Sci. Med. Sci. 65: 478–487. 10.1093/gerona/glq028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikpay M., Goel A., Won H.-H., Hall L. M., Willenborg C. et al. , 2015. A Comprehensive 1000 Genomes–Based Genome-Wide Association Meta-Analysis of Coronary Artery Disease. Nature Genetics 47(10): 1121–1130. 10.1038/ng.3396 [DOI] [PMC free article] [PubMed]
- Philippe P. 1978. Familial Correlations of Longevity: An Isolate-Based Study. American Journal of Medical Genetics 2(2): 121–129. 10.1002/ajmg.1320020203 [DOI] [PubMed] [Google Scholar]
- Pilling L. C., Kuo C.-L., Sicinski K., Tamosauskaite J., Kuchel G. A. et al. , 2017. Human Longevity: 25 Genetic Loci Associated in 389,166 UK Biobank Participants. Aging Impact Journals, LLC. http://www.aging-us.com/article/101334/text [DOI] [PMC free article] [PubMed]
- Pilling L. C., Atkins J. L., Bowman K., Jones S. E., Tyrrell J. et al. , 2016. Human longevity is influenced by many genetic variants: evidence from 75,000 UK biobank participants. Aging (Albany N.Y.) 8: 547–560. 10.18632/aging.100930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J., Wright K. M., Rand K. A., Kermany A., Noto K. et al. , 2018. Estimates of the heritability of human longevity are substantially inflated due to assortative mating. Genetics 210: 1109–1124. 10.1534/genetics.118.301613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S., Obozinski G., Jordan M. I., and Halperin E., 2009. Genomic privacy and limits of individual detection in a pool. Nat. Genet. 41: 965–967. 10.1038/ng.436 [DOI] [PubMed] [Google Scholar]
- Schächter F., Faure-Delanef L., Guénot F., Rouger H., Froguel P. et al. , 1994. Genetic Associations with Human Longevity at the APOE and ACE Loci. Nature Genetics 6(1): 29–32. http://www.nature.com/doifinder/10.1038/ng0194-29 [DOI] [PubMed] [Google Scholar]
- Sebastiani P., Bae H., Sun F. X., Andersen S. L., Daw E. W. et al. , 2013. Meta‐analysis of genetic variants associated with human exceptional longevity. Impact Journals 5: 653–661. 10.18632/aging.100594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P., Gurinovich A., Bae H., Andersen S., Malovini A. et al. , 2017. Four genome-wide association studies identify new extreme longevity variants. J. Gerontol. A Biol. Sci. Med. Sci. 72: 1453–1464. 10.1093/gerona/glx027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer J. R., Li J., Lee M. K., Roosenboom J., Orlova E. et al. 2017. Multiethnic GWAS Reveals Polygenic Architecture of Earlobe Attachment. The American Journal of Human Genetics 101: 913–924. 10.1016/j.ajhg.2017.10.001 [DOI] [PMC free article] [PubMed]
- Soerensen M., Dato S., Christensen K., McGue M., Stevnsner T., 2010. Replication of an Association of Variation in the FOXO3A Gene with Human Longevity Using Both Case-Control and Longitudinal Data. Aging Cell 9(6): 1010–1017. 10.1111/j.1474-9726.2010.00627.x [DOI] [PMC free article] [PubMed]
- Soerensen M., Dato S., Tan Q., Thinggaard Mikael, Kleindorp R. et al. , 2013. Evidence from Case–Control and Longitudinal Studies Supports Associations of Genetic Variation in APOE, CETP, and IL6 with Human Longevity. AGE 35(2): 487–500. 10.1007/s11357-011-9373-7 [DOI] [PMC free article] [PubMed]
- The 1000 Genomes Project Consortium, Auton A., Abecasis G. R., Altshuler D. M., Durbin R. M., Abecasis G. R. et al. , 2015. A Global Reference for Human Genetic Variation. Nature 526(7571): 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed]
- Vijg J. and Campisi J., 2008. Puzzles, Promises and a Cure for Ageing. Nature 454(7208): 1065–1071. 10.1038/nature07216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P. M. and Hill W. G., 2009. The limits of individual identification from sample allele frequencies: theory and statistical analysis. PLoS Genet. 5: e1000628 10.1371/journal.pgen.1000628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox B. J., Donlon Q. H. T. A., Chen R., Grove J. S., Yano K. et al. , 2008. FOXO3A Genotype Is Strongly Associated with Human Longevity. Proceedings of the National Academy of Sciences of the United States of America 105 (37). National Academy of Sciences: 13987–13992. 10.1073/pnas.0801030105 [DOI] [PMC free article] [PubMed]
- Willer C. J., Li Y., and Abecasis G. R., 2010. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191. 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G. C. 1957. Pleiotropy, Natural Selection, and the Evolution of Senescence. Evolution 11(4): 398–411. 10.2307/2406060 [DOI]
- Yang J., Bakshi A., Zhu Z., Hemani G., Vinkhuyzen A. A. E. et al. , 2015. Genetic Variance Estimation with Imputed Variants Finds Negligible Missing Heritability for Human Height and Body Mass Index. Nature Genetics 47(10): 1114–1120. 10.1038/ng.3390 [DOI] [PMC free article] [PubMed]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K. et al. , 2010. Common SNPs Explain a Large Proportion of the Heritability for Human Height. Nat. Genet. 42: 565–569. 10.1038/ng.608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Levine D., Shen J., Gogarten S. M., Laurie C. et al. , 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28: 3326–3328. 10.1093/bioinformatics/bts606 [DOI] [PMC free article] [PubMed] [Google Scholar]