Abstract
Sequence analyses of RNA virus genomes remain challenging owing to the exceptional genetic plasticity of these viruses. Because of high mutation and recombination rates, genome replication by viral RNA-dependent RNA polymerases leads to populations of closely related viruses, so-called “quasispecies.” Standard (short-read) sequencing technologies are ill-suited to reconstruct large numbers of full-length haplotypes of (1) RNA virus genomes and (2) subgenome-length (sg) RNAs composed of noncontiguous genome regions. Here, we used a full-length, direct RNA sequencing (DRS) approach based on nanopores to characterize viral RNAs produced in cells infected with a human coronavirus. By using DRS, we were able to map the longest (∼26-kb) contiguous read to the viral reference genome. By combining Illumina and Oxford Nanopore sequencing, we reconstructed a highly accurate consensus sequence of the human coronavirus (HCoV)-229E genome (27.3 kb). Furthermore, by using long reads that did not require an assembly step, we were able to identify, in infected cells, diverse and novel HCoV-229E sg RNAs that remain to be characterized. Also, the DRS approach, which circumvents reverse transcription and amplification of RNA, allowed us to detect methylation sites in viral RNAs. Our work paves the way for haplotype-based analyses of viral quasispecies by showing the feasibility of intra-sample haplotype separation. Even though several technical challenges remain to be addressed to exploit the potential of the nanopore technology fully, our work illustrates that DRS may significantly advance genomic studies of complex virus populations, including predictions on long-range interactions in individual full-length viral RNA haplotypes.
Coronaviruses (subfamily Coronavirinae, family Coronaviridae, order Nidovirales) are enveloped positive-sense (+) single-stranded (ss) RNA viruses that infect a variety of mammalian and avian hosts and are of significant medical and economic importance, as illustrated by recent zoonotic transmissions from diverse animal hosts to humans (Vijay and Perlman 2016; Menachery et al. 2017). The genome sizes of coronaviruses (∼30 kb) exceed those of most other RNA viruses. Coronaviruses use a special mechanism called discontinuous extension of minus strands (Sawicki and Sawicki 1995, 1998) to produce a nested set of 5′- and 3′-coterminal subgenomic (sg) mRNAs that carry a common 5′ leader sequence that is identical to the 5′-end of the viral genome (Zuniga et al. 2004; Sawicki et al. 2007). These sg mRNAs contain a different number of open reading frames (ORFs) that encode the viral structural proteins and several accessory proteins. With very few exceptions, only the 5′-located ORF (which is absent from the next smaller sg mRNA) is translated into protein (Fig. 1).
Figure 1.
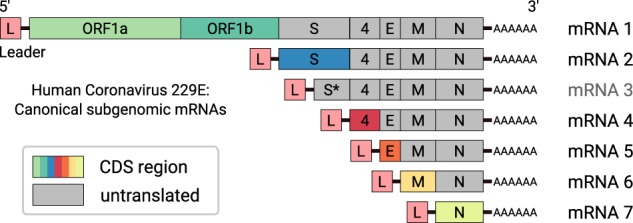
Scheme of genomic and subgenomic (sg) RNAs produced in HCoV-229E–infected cells (Raabe et al. 1990; Schreiber et al. 1989). Translation of the 5′-terminal ORF(s) of the respective mRNA gives rise to the various viral structural and nonstructural proteins (indicated by different colors). mRNA 3 is considered defective and unlikely to be translated into protein. Each mRNA has a 3′ poly(A) tail and carries a 5′-leader sequence that is identical to the 5′-end of the genome. In a process called discontinuous extension of negative strands, negative-strand RNA synthesis is attenuated at specific genomic positions. After transfer of the nascent strand to an upstream position on the template, RNA synthesis is continued by copying the 5′-leader sequence. As a result, the 3′-ends of coronavirus minus-strand mRNAs are equipped with the complement of the 5′-leader. The latter process is guided by base-pairing interactions between the transcription-regulating sequence (TRS) located immediately downstream from the leader sequence (TRS-L) at the 5′-end of the genome and the sequence complement of a TRS located upstream of one of the ORFs present in the 3′-proximal genome region (TRS-B).
In HCoV-229E–infected cells, a total of seven major viral RNAs are produced. The viral genome is also referred to as mRNA 1 because it has an mRNA function. In its 5′-terminal region, the genome RNA contains two large ORFs, 1a and 1b, that encode the viral replicase polyproteins 1a and 1ab. mRNAs 2, 4, 5, 6, and 7 are used to produce the S protein, accessory protein 4, E protein, M protein, and N protein, respectively. The 5′-region of mRNA 3 contains a truncated fragment of ORF S, which is considered defective. Although this sg RNA has been consistently identified in HCoV-229E–infected cells, its mRNA function has been disputed, and there is currently no evidence that this RNA is translated into protein (Schreiber et al. 1989; Raabe et al. 1990; Thiel et al. 2003).
Like many other +RNA viruses, coronaviruses show high rates of recombination (Lai 1992; Liao and Lai 1992; Furuya et al. 1993). In fact, the mechanism to produce 5′ leader-containing sg mRNAs represents a prime example for copy-choice RNA recombination that, in this particular case, is guided by complex RNA–RNA interactions involving the transcription-regulating sequence (TRS) core sequences and likely requires additional interactions of viral proteins with specific RNA signals. In other virus systems, RNA recombination has been shown to generate “transcriptional units” that control the expression of individual components of the genome (Holmes 2009). The mechanisms involved in viral RNA recombination are diverse and may even extend to nonreplicating systems (Gallei et al. 2004). In the vast majority of cases, recombination results in defective RNA (dRNA) copies that lack essential cis-active elements and thus cannot be replicated. In other cases, functional recombinant RNA with new properties, such as the ability to replicate in a new host, may emerge (Chang et al. 1994, 1996; Brown et al. 2007; Gustin et al. 2009). In yet other cases, defective interfering RNAs (DI-RNAs) may be produced. These defective (sg-length) RNAs contain all the cis-acting elements required for efficient replication by a helper virus polymerase and, therefore, represent parasitic RNAs that compete for components of the viral replication/transcription complex with nondefective viral RNAs (Pathak and Nagy 2009).
To elucidate the many facets of recombination and to determine full-length haplotypes of, for example, virus mutants/variants in complex viral populations (quasispecies), long-read sequencing has become the method of choice. Short-read second-generation sequencing technologies—such as Ion Torrent and Illumina—are restricted by read length (200–400 nt) (Hölzer and Marz 2017). For example, the use of highly fragmented viral RNAs considerably complicates the investigation of haplotypes (Nowak 1992; Baaijens et al. 2017). Because the nested coronavirus mRNAs are almost identical to the original genome sequence, short-read data can usually not be unambiguously assigned to specific sg RNA species.
In this study, we performed direct RNA sequencing (DRS) on an array of nanopores, as developed by Oxford Nanopore Technologies (ONT) (Garalde et al. 2018). Nanopore sequencing does not have a limited reading length but is limited only by fragmentation of the input material (Mikheyev and Tin 2014; Chua and Ng 2016; Jain et al. 2016). Further, by using DRS, we avoid several drawbacks of previous sequencing methods, in particular cDNA synthesis and amplification of the input material. Thus, for example, cDNA synthesis can create artificial RNA–RNA chimerae (Karst et al. 2018) that are difficult to discriminate from naturally occurring chimerae (such as spliced RNAs). Also, amplification before sequencing would remove all RNA modifications from the input material, whereas the nanopore sequencing technology preserves these modifications (Smith et al. 2017; Garalde et al. 2018).
Recently, nanopore sequencing has been used for metagenomic forays into the virosphere (Warwick-Dugdale et al. 2019) and studies focusing on transmission routes (Quick et al. 2016; Faria et al. 2017). Furthermore, viral transcriptomes have been investigated using nanopore sequencing of cDNA (Moldován et al. 2017, 2018a,b; Tombácz et al. 2017), being subject to bias from reverse transcription and amplification. Other studies used DRS to study the human poly(A) transcriptome (Workman et al. 2018) and the transcriptome of DNA viruses such as HSV (Depledge et al. 2018). Furthermore, the genome of influenza A virus has been completely sequenced in its original form using DRS (Keller et al. 2018).
In the present study, we sequenced one of the largest known RNA genomes, that of HCoV-229E, a member of the genus Alphacoronavirus, with a genome size of ∼27,300 nt, in order to assess the complex architectural details for viral sg RNAs produced in cells infected with recombinant HCoV-229E. By using DRS, we aim to capture complete viral mRNAs, including the full coronavirus genome, in single contiguous reads. Sequence analysis of thousands of full-length sg RNAs will allow us to determine the architectures (including leader–body junction sites) of the major viral mRNAs. In addition, this approach provides insight into the diversity of additional HCoV-229E sg RNAs, probably including DI-RNAs. Further, we aim to assess whether RNA modifications can be called directly from the raw nanopore signal of viral molecules without prior in vitro treatment, as has been shown for DNA (Stoiber et al. 2016; McIntyre et al. 2019).
Results
Full-genome sequencing without amplification
We sequenced total RNA samples obtained from Huh7 cells infected with serially passaged recombinant human coronaviruses: wild-type (WT) HCoV-229E, HCoV-229E_SL2-SARS-CoV, and HCoV-229E_SL2-BCoV, respectively. In the latter two viruses, a conserved stem–loop structure (SL2) residing in the HCoV-229E 5′ UTR was replaced with the equivalent SL2 element from SARS-CoV and BCoV, respectively (Madhugiri et al. 2018). Total RNA samples obtained for the latter two (chimeric) viruses were pooled before sequence analysis. Hereafter, we refer to the first sample as WT RNA and to the second (pooled) sample as SL2 RNA (see Methods).
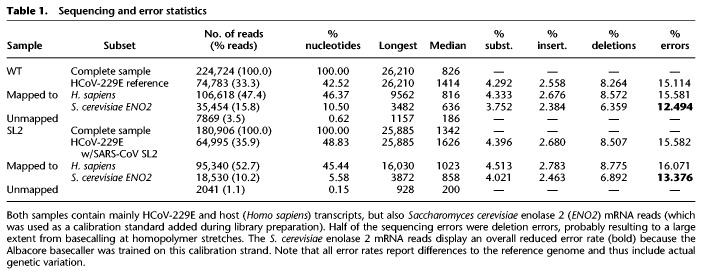
We performed two DRS runs (one per sample) on a MinION Nanopore sequencer. As shown in Table 1, we achieved a throughput of 0.237 and 0.282 Gb with 225,000 and 181,000 reads for the WT and SL2 sample, respectively. See Supplemental Figure S1A for an overview of the read length distribution. For the WT and SL2 samples, 33.3% and 35.9% of the reads mapped to the reference HCoV-229E sequences, respectively; 15.8% and 10.2%, respectively, mapped to the yeast enolase 2 mRNA sequence, a calibration strand added during the library preparation, whereas 47.4% and 52.7% could be attributed to human host cell RNA. Minimap2 (Li 2018) did not align the remaining 3.50% and 1.11% of reads. Using BLAST (Altschul et al. 1990) against the nucleotide database, 18.1% and 20.7% of these reads can be attributed again to HCoV, human or yeast. As reads that were not aligned by minimap2 were mostly very short (median ≤200 nt), of poor basecalling quality and represented only 0.62% and 0.15% of total nucleotides, respectively, we decided to only use the higher quality reads that minimap2 could align (for detailed statistics, see Supplemental Fig. S2).
Table 1.
Sequencing and error statistics
The visualized raw voltage signal of a nanopore read is commonly called “squiggle” (see Supplemental Fig. S3). Different from all previous sequencing technologies, nanopore sequencing preserves the information about base modifications in the raw signal (Garalde et al. 2018). However, one of the biggest challenges is the accurate mapping of the raw voltage signal to bases (“basecalling”).
As expected for nanopore DRS (Garalde et al. 2018; Keller et al. 2018), reads had a median uncorrected error rate of ∼15% for human and virus reads, whereas basecalling errors were reduced for yeast ENO2 mRNA reads, as the basecaller was trained on this calibration strand (see Table 1). This included gaps but omitted discontinuous sites >6 nt because they indicated recombination. Half of all errors were deletions. In addition, we found that more than half of all single-nucleotide deletions occur in homopolymers, and most of these stretches that coincide with a deletion are ≥3 nt long (see Supplemental Fig. S4). A quarter of the errors were substitutions, which we argue are largely because of modified bases that impede the basecaller's ability to assign bases correctly.
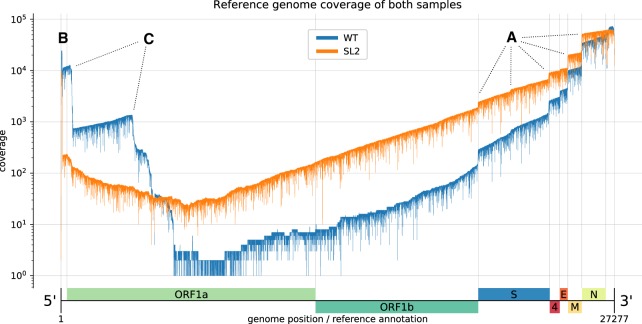
The HCoV-229E genome was 99.86% covered, with a large coverage bias toward both ends (see Figs. 1, 2). The high coverage of the 3′-end reflects the higher abundance of mRNAs produced from the 3′-terminal genome regions and is a result of the discontinuous transcription mechanism used by coronaviruses and several other nidoviruses (Pasternak et al. 2006; Sawicki et al. 2007; Sola et al. 2015). The 3′-coverage is further increased by the directional sequencing that starts from the mRNA 3′-terminal poly(A) tail. Also, the observed coverage bias for the very 5′-end results from the coronavirus-specific transcription mechanism because all viral mRNAs are equipped with the 65-nt 5′-leader sequence derived from the 5′-end of the genome. The remainder of the high 5′-coverage bias likely reflects the presence of high numbers of DI-RNAs in which 5′- and 3′-proximal genomic sequences were fused, probably resulting from illegitimate recombination events as shown previously for other coronaviruses (Liao and Lai 1992; Furuya et al. 1993; Luytjes et al. 1996). For the WT and SL2 samples, 38.37% and 16.32% were split-mapped, respectively. Of these, only 278 and 181 had multiple splits. The considerably larger fraction of split reads in the WT sample is explained by the high abundance of potential DI-RNA molecules (see Figure 2C).
Figure 2.
Reference genome coverage of the HCoV-229E WT sample (blue) and the SL2 sample (orange) based on alignments with minimap2. There is an inverse correlation between sg RNA abundance and length. (A) Notable vertical “steps” in the coverage correspond to borders expected for the canonical sg RNAs (see Fig. 1). (B) The presence of the leader sequence (∼65 nt) in canonical sg RNAs gives rise to the sharp coverage peak at the 5′-end. (C) We also observed unexpected “steps,” especially in the WT sample (blue). We hypothesize that the sequences correspond to DI-RNA molecules that may arise by recombination at TRS-like sequence motifs as well as other sites displaying sequence similarities that are sufficient to support illegitimate recombination events (see Fig. 3). We attribute the difference in the observed (noncanonical) recombination sites between the two samples to biological factors that we either did not control for or do not know (see also legend to Fig. 3).
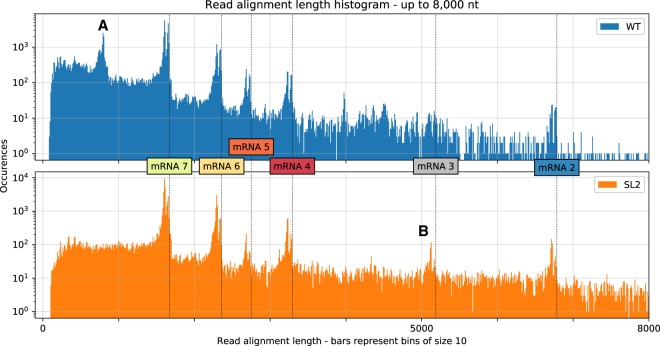
An alignment of the longest reads from both samples to the HCoV-229E reference indicates that they represent near complete virus genomes (Supplemental Fig. S1B). The observed peaks in the aligned reads length distribution (see Fig. 4, below) corresponded very well with the abundances of the known mRNAs produced in HCoV-229E–infected cells (see Fig. 1; Schreiber et al. 1989; Raabe et al. 1990; Thiel et al. 2003). Alignment of the reads to these canonical mRNA sequences confirmed these observed abundances (Supplemental Fig. S5).
Figure 4.
Distribution of aligned read lengths up to 8000 nt for both samples based on alignments with minimap2. We observed clusters that correspond well with the transcript lengths expected for canonical mRNAs (vertical dotted lines). Alignment of the reads to these canonical mRNA sequences confirmed these observed abundances (Supplemental Fig. S5). The distribution shows double peaks at the cluster positions because reads corresponding to mRNAs often miss the leader sequence, possibly owing to basecalling or mapping errors. We also observed additional clusters that likely correspond to highly abundant DI-RNAs. (A) A cluster in the WT sample that represents chimeric ∼820-nt sg RNAs composed of both 5′- and 3′-terminal genome regions (∼540 and ∼280 nt, respectively). We propose the template switch for this transcript occurs around position 27,000 and RNA synthesis resumes at around position 540 (see corresponding peaks in Fig. 3, track B). (B) A cluster from the SL2 sample that contains transcripts with an approximate length of 5150 nt, which represents mRNA 3 (see Fig. 1). These transcripts are probably formed owing to a transcription stop at a TRS motif around position 22,150 (see corresponding peak in Fig. 3, track C).
The median read length for the combined set of reads from both samples was 826 nt, with a maximum of 26,210 nt, covering 99.86% of the 27,276-nt-long virus genome, missing only 21 nt at the 5′-end, 15 nt at the 3′-end, and those nucleotides that correspond to the skewed error distribution, with 5.7 percentage points more deletions than insertions (see Table 1). The median read length might sound short; however, most of the viral RNAs (including many DI-RNAs) identified in HCoV-229E–infected cells were <2000 nt in length. Furthermore, this number nearly doubles the longest read length that can be obtained with short-read sequencing methods. We observed an abundance of very short reads, representing the 3′ (poly[A]) end of the genome. This could be an artifact of RNA degradation, although we cannot estimate the exact fraction of affected transcripts. Because sequencing starts at the poly(A) tail, fragmented RNA will not be sequenced beyond any 3′ break point. It is thus best to minimize handling time during RNA extraction and library preparation. Innovations in these fields will directly translate into larger median read lengths.
We obtained 99.15% and 98.79% identity in both samples (WT, SL2), respectively, with the help of the consensus caller Ococo (Břinda et al. 2017) using the reference genomes and all reads mapping to it. We attempted a standard long-read assembly using Canu (Koren et al. 2017), which yielded unusable results (WT: 389 contigs, longest 13 kb, all other <4 kb; SL2: 517 contigs, all <6 kb). We think that current nanopore-only assembly tools are not equipped to handle special read data sets such as those originating from a small RNA virus genome. In addition, we assembled WT and SL2 consensus sequences using Nanopore and Illumina data with HG-CoLoR (Morisse et al. 2018) in an approach that uses long nanopore reads to traverse an assembly graph constructed from short Illumina reads. We thereby recovered 99.57% of the reference genome in a single contiguous sequence at 99.90% sequence identity to reference using this approach with the single longest read from the SL2 sample. This hybrid approach illustrates how short- and long-read technologies can be combined to reconstruct long transcripts accurately, which will greatly facilitate studies of haplotypes.
Uncharacterized sg and DI-RNAs
In addition to the leader-to-body junctions expected for the canonical sg mRNAs 2, 4, 5, 6, and 7, we observed a high number of recombination sites (Fig. 3) that were consistently found in our samples but have not been described previously (Fig. 3). In this study, we defined a recombination site as any site that flanks more than 100 consecutive gaps, as determined in a discontinuous mapping (“spliced” mapping). Although there is currently no consensus on how to define such sites, we believe this to be a conservative definition, as this type of pattern is unlikely to result from, for example, miscalled homopolymer runs that, in our experience, typically affect less than 10 consecutive bases. We observe all known canonical HCoV-229E mRNAs at their expected lengths, including the (presumably) noncoding mRNA 3 (Fig. 4).
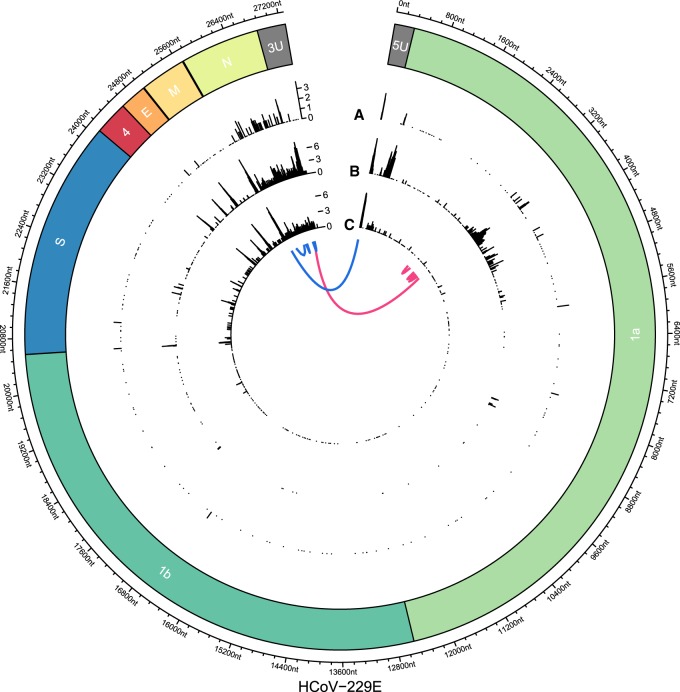
Figure 3.
Joining of noncontiguous genome sequences in sg RNAs identified in HCoV-229E–infected cells. On the circular axis, the annotations of the reference genome (including 5′ UTR [5U] and 3′ UTR [3U]) are shown. Genomic positions of “discontinuous sites” identified in Illumina reads (A; outer track), nanopore reads of sample HCoV-229E WT (B; middle track), and nanopore reads of SL2 sample (C; inner track) reveal multiple recombination sites across the whole genome. An aggregation of recombination sites can be observed in the region that encodes the viral N protein. Furthermore, clear recombination sites can be seen at intergenic boundaries and at the 5′ and 3′ UTRs, with the former corresponding to the boundary between the leader sequence and the rest of the genome. Another prominent cluster can be observed in ORF1a in the WT nanopore sample but not in SL2. This cluster is supported by the WT Illumina data, excluding sequencing bias as a potential source of error. We hypothesize that because samples WT and SL2 were obtained from nonplaque-purified serially passaged virus populations derived from in vitro transcribed genome RNAs transfected into Huh7 cells, differences in the proportion of full-length transcripts versus abortive transcripts could translate into different patterns of recombination. Generally, nanopore-based sequencing allows more detailed analysis of recombination events owing to the long read length. Even complex isoforms such as two exemplary reads, each with four discontinuous segments, can be observed (blue and pink).
The aligned reads distribution revealed clusters for all known mRNAs which closely fit the expected molecule lengths (Fig. 4). The cluster positions show double peaks with a consistent distance of ∼65 nt, that is, the length of the leader sequence. We observed that the 5′-end of reads has larger-than-average error rates and is often missing nucleotides (for detailed statistics, see Supplemental Fig. S8). This might be because of a bias of the basecaller toward the end of reads. This is plausible because the underlying classification algorithm is a bidirectional (i.e., forward and backward looking) long-short-term memory neural network (LSTM). The mapping algorithm was often unable to align these erroneous 5′-ends, leading to soft-clipped bases. Thus, for many reads representing canonical mRNAs, the included leader sequence was not aligned, which gives rise to the secondary peak at each cluster position. We also observed additional clusters that likely correspond to highly abundant dRNAs (Fig. 4).
We also observed several unexpected recombination sites, for example, at positions 3000–4000 (within ORF1a, see Fig. 3). These sites were confirmed by both Nanopore and Illumina sequencing. They had a high read support and defined margins, suggesting a specific synthesis/amplification of these sg RNAs that, most likely, represent DI-RNAs. Because DI-RNAs are byproducts of viral replication and transcription, they present a larger diversity than the canonical viral mRNAs (Penzes et al. 1994, 1996; Méndez et al. 1996; Brian and Spaan 1997; Izeta et al. 1999).
Nanopore sequencing captures recombination events far better than Illumina, which allowed us to identify even complex sg RNAs (composed of sequences derived from more than two noncontiguous genome regions) at much higher resolution: For example, we found sg RNAs with up to four recombination sites in the 5′- and 3′-terminal genome regions (Fig. 3).
Consistent 5mC methylation signatures of coronavirus RNA
Nanopore sequencing preserves information on nucleotide modifications. By using a trained model, DNA and RNA modifications such as 5mC methylation could be identified (Fig. 5). To assess the false-positive rate (FPR) of the methylation calling, we used an unmethylated RNA calibration standard (RCS) as a negative control that was added in the standard library preparation protocol for DRS. We considered a position to be methylated if at least 90% of the reads showed a methylation signal for this particular position. By using this threshold, the estimated FPR was calculated to be <5%. Our experimental setup did not include a positive methylation control.
Figure 5.
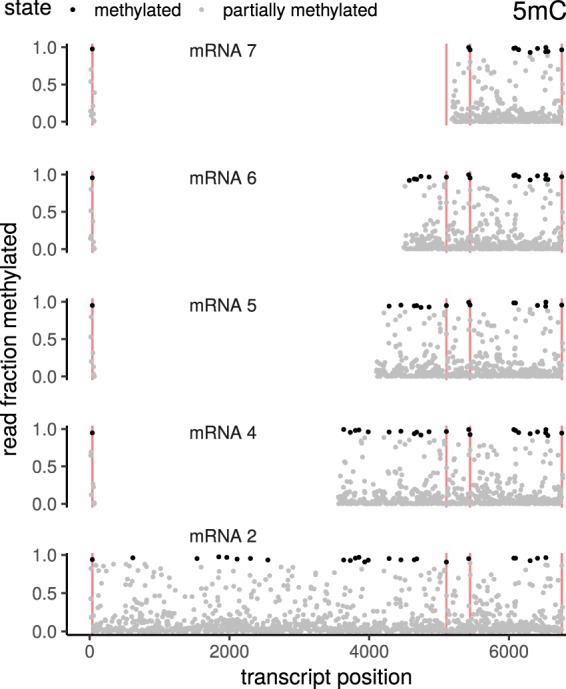
5mC methylation of various annotated coronavirus mRNAs. The transcripts have been aligned such that corresponding genomic positions can be found in the same vertical column across facets. Both the leader sequence (including the TRS) and the nested sg sequences show consistent patterns of methylation across transcripts. Exemplary positions that display consistent methylation across all investigated transcripts are marked as red vertical lines. Note that although coronavirus recombination uses two TRSs, the resulting transcript has only one TRS, because of self-similarity-based pairing of the TRS. Positions have been labeled “methylated” if at least 90% of the reads show a methylation signal. Using this threshold, the estimated FPR is <5%.
When analyzing 5mC methylation across various RNAs, we observed consistent patterns (Fig. 5) that were reproducible for the corresponding genomic positions in different RNAs, suggesting that the methylation of coronavirus RNAs is sequence-specific and/or controlled by RNA structural elements. Methylated nucleotides could be identified across the genome, both in the leader sequence and in the body regions of viral mRNAs.
Although the overall methylation pattern looks similar between sg RNAs and the negative control (see Supplemental Fig. S9), we nevertheless find consistent methylation across different sg RNA “types”; that is, methylated positions of mRNA 2 are mirrored in mRNA 4 etc.
Discussion
We identified highly diverse sg RNAs in coronavirus-infected cells, with many sg RNAs not corresponding to the known canonical mRNAs. These “noncanonical” sg RNAs had abundant read support, and full-length sequences could be obtained for most of these RNAs.
As indicated above, only 12% of the sg RNAs were found to conform to our current understanding of discontinuous mRNA transcription in coronaviruses, resulting in mRNAs that (all) carry an identical 5′-leader sequence that is fused to the 3′-coding (“body”) sequence of the respective mRNA. We, however, believe that 12% represents an underestimate because a large number of sg RNAs were probably omitted from the analysis: First, RNA molecules degrade rapidly under laboratory conditions, even when handled carefully. The resulting fragments will only be sequenced if they contain a poly(A) tail. Second, the high sequencing error may introduce mismappings, especially for low-quality reads. These reads would not be assigned to the canonical model under our assumptions because of the high number of mismatches. However, we think the associated bias is low, because minimap2 is very robust against high error rates and because the reads are very long, thus ensuring that the mapper has sufficient aggregate information on a given read to position it very reliably on a reference. Third, the library preparation protocol for DRS includes the ligation of adapters via a T4 ligase. Any ligase could potentially introduce artificial chimera, although we did not investigate this systematically. Again, we think that this does not affect our results substantially: (1) This bias is random, and it seems unlikely that we would observe the very same RNA “isoform” many times if it was created by random ligation; (2) many “isoforms” that we observed only once (e.g., those colored pink and blue in Fig. 3) were structured plausibly: They contained a leader sequence and had recombined at expected (self-similar) sites corresponding to putative or validated TRSs, with downstream sequences being arranged in a linear 5′-3′-order. Fourth, finally, it is important to note that the RNA used for DRS was isolated from cells infected with a serially passaged pool of recombinant viruses rescued after transfection of in vitro transcribed genome-length (27.3-kb) RNAs. Transfection of preparations of in vitro transcribed RNA of this large size likely included a significant proportion of abortive transcripts that lacked varying parts of the 3′ genome regions, rendering them dysfunctional. It is reasonable to suggest that the presence of replication-incompetent RNAs lacking essential 3′-terminal genome regions may have triggered recombination events resulting in the emergence of DI-RNAs that contained all the 5′- and 3′-terminal cis-active elements required for RNA replication but lacked nonessential internal genome regions. Upon serial passaging of the cell culture supernatants for 21 times, DI-RNAs may have been enriched, especially in the HCoV-229E (WT) sample (Fig. 3). Comparative DRS analyses of RNA obtained from cells infected with (1) plaque-purified HCoV-229E and (2) newly rescued recombinant HCoV-229E (without prior plaque purification), respectively, would help to address the possible role of prematurely terminated in vitro transcripts produced from full-length cDNA in triggering the large number of DI-RNAs observed in our study.
Although, for the above reasons, the low percentage of canonical mRNAs (12%) in our samples likely represents an underestimate, our study may stimulate additional studies, for example, to revisit the production of mRNAs from noncanonical templates (Wu and Brian 2010; Sola et al. 2011). Also, it is worth mentioning in this context that, for several other nidoviruses, such as murine hepatitis virus (MHV), bovine coronavirus (BCoV), and arteriviruses, evidence has been obtained that sg RNA transcription may also involve noncanonical TRS motifs (Joo and Makino 1992; Lai and Cavanagh 1997; Lai 1998; Ozdarendeli et al. 2001; Alonso et al. 2002).
The majority of sg RNAs (other than mRNAs) we found in our samples likely represent DI-RNAs, which are a common occurrence in coronavirus in vitro studies (Pathak and Nagy 2009).
To our knowledge, this study is the first to perform RNA modification calling without prior treatment of the input sample. It only relies on the raw nanopore signal. Although DNA modifications such as 5mC methylation have been explored extensively (Breiling and Lyko 2015), less is known about RNA modifications (Roundtree et al. 2017), the importance of which is debated (Grozhik and Jaffrey 2017). We found consistent 5mC methylation patterns across viral RNAs when tested at a FPR <5%. We were not able to assess the sensitivity and specificity of the methylation calling owing to the absence of a positive control group, which was beyond the scope of this study.
RNA is known to have many different modifications, and we expect the presence of these on coronavirus sg RNAs (Machnicka et al. 2013) too. However, to our knowledge no comprehensive data exist on prior expectations for such modifications in coronaviruses, which might or might not correspond to those observed in for example, humans.
In addition, we observed that the software used (Stoiber et al. 2016) will likely present high error rates in regions of low coverage or where the underlying reference assembly is erroneous. This is because the resquiggle algorithm—upon which this method is based—has to align the raw nanopore read signal to the basecalled read sequence (see Supplemental Fig. S6). This is necessary to test the raw signal against learned modification models, of which at the time of manuscript preparation (May 2019) only 5mC was implemented for RNA. Nevertheless, new options to call these modifications at an acceptable error rate without any RNA pretreatment is a powerful method.
The validity of the methylation signal should be confirmed in future studies using, for example, bisulfite sequencing. Ideally, this validation should start from in vitro synthetic transcripts in which modified bases have been inserted in known positions. Furthermore, RNA modification detection from single-molecule sequencing is a current bioinformatic frontier, and algorithms and tools are under active development. We showed that consistent 5mC methylation patterns were seen across different sg RNAs. However, the overall pattern of the methylation calls between sg RNAs and the negative control was very similar. At a FPR of 5%, the RNA modifications we identified are supported by their consistent occurrence. However, we cannot rule out that instead, the observed pattern might be caused by an alignment artifact. In the used methylation calling algorithm, the raw signal is aligned to the nucleotide sequence after basecalling. If there is a systematic bias in this alignment and certain sequence motifs cause a consistent mapping mismatch, this mismatch could lead to false-positive methylated sites. This is because in these positions the signal would deviate from the expected one owing to the misalignment and not owing to methylation. In future experiments, this can be decided using a positive control in the form of an RNA transcript with known 5mC methylated sites. However, even if we are in its early stages, the reading of RNA modifications from the read signal has great potential to elucidate viral biology.
We were able to reconstruct accurate consensus sequences, for both the Illumina and Nanopore data. We also showed that individual transcripts can be characterized. More problematic was the resolution of quasispecies in our experimental setup. Although DRS allowed us to confirm the presence of each of the two heterologous SL2 structures present in the SL2 sample, this was only possible for sg-length (DI-) RNAs. It appears that the high error rate of >10% was a critical limitation when analyzing the SL2 region located at the extreme 5′-terminal end of the 27.3-kb genome RNA. This high error rate made variant calling difficult, particularly under low-coverage conditions. The current generation of long-read assemblers is not well suited to reconstruct many viral genome architectures, such as nested ones. The development of specialized assemblers would be of great help in virology projects.
We used a hybrid error correction method (HG-CoLoR) (Morisse et al. 2018) that uses Illumina data to correct read-level errors. However, it remains questionable whether the corrected read sequence is truly representative of the ground truth read sequence. Signal-based correction methods such as Nanopolish (Loman et al. 2015) may be more promising; however at the time of manuscript writing (May 2019), correction on direct RNA data has not been implemented. We expect this to become available in the near future. Combined with the ever-increasing accuracy of the nanopore technology, we think this method might be able to study quasispecies soon.
There are recombination events observed in the Illumina data that were not detected in the nanopore data. These are likely caused by misalignment of the short single-end reads (50 nt). A minimum of only 10 nt was required for mapping on either end of the gapped alignment. This was a trade-off between sensitivity to identify recombination sites and unspecific mapping.
In this work, we showed the potential of long-read data as produced by nanopore sequencing. We were able to directly sequence the RNA molecules of two different samples of one of the largest RNA virus genomes known to date. We showed how very large RNA genomes and a diverse set of sg RNAs with complex structures can be investigated at high resolution without the need for a prior assembly step and without the bias introduced by cDNA synthesis that is typically required for transcriptome studies.
The detail and quality of the available data still require significant bioinformatic expertise as the available tooling is still at an early development stage. However, the technological potential of nanopore sequencing for new insights into different aspects of viral replication and evolution is very promising.
Future studies should investigate both strands of the coronavirus transcriptome. Studies focusing on RNA modifications need to use well-defined positive and negative controls to assess the error rate of the current software alternatives. Also, the DRS method will be extremely powerful if it comes to analyzing the nature and dynamics of specific haplotypes in coronavirus populations under specific selection pressures, for example, mutations and/or drugs affecting replication efficiency and others.
Our work also serves as a proof of concept showing that consistent RNA modifications can be detected using nanopore DRS.
To fully exploit the potential of DRS, several improvements are needed: First and foremost, a significant reduction of the currently very high per-read error rate is crucial. This is especially problematic in studies focusing on intra-sample heterogeneity and haplotypes. Second, protocols that limit RNA degradation during library preparation would be of great value. This could be achieved by shortening the library protocol. To limit the cost of DRS, barcoded adapters would be desirable. On the bioinformatics side, the basecaller for DRS data is still at an early stage and, for example, cannot accurately call the poly(A) regions as well as the RNA-DNA-hybrid adapter sequences. Further basecalling errors likely result from RNA modifications, which need to be modeled more accurately. However, once these limitations will be fixed, the use of nanopore-based DRS can be expected to greatly advance our understanding of the genomics of virus populations and their multiple haplotypes.
Methods
RNA virus samples
The two total RNA samples used in this study for DRS (ONT MinION) and Illumina sequencing were prepared at 24 h post infection from Huh7 cells infected at an MOI of three with recombinant HCoV-229E WT, HCoV-229E_SL2-SARS-CoV, and HCoV-229E_SL2-BCoV, respectively (Madhugiri et al. 2018). Before sequence analysis, the two RNA samples obtained from HCoV- 229E_SL2-SARS-CoV–infected and HCoV-229E_SL2-BCoV–infected cells were pooled (SL2 sample) (see Supplemental Fig. S7).
Generation of recombinant viruses and total RNA isolation were performed as described previously (Madhugiri et al. 2018). Briefly, full-length cDNA copies of the genomes of HCoV-229E (GenBank accession number NC_002645), HCoV-229E_SL2- SARS-CoV, and HCoV-229E_SL2-BCoV, respectively, were engineered into recombinant vaccinia viruses using previously described methods (Thiel et al. 2001; Hertzig et al. 2004; Thiel and Siddell 2005). Next, full-length genomic RNAs of HCoV-229E, HCoV-229E_SL2-SARS-CoV, and HCoV-229E_SL2-BCoV, respectively, were transcribed in vitro using purified ClaI-digested genomic DNA of the corresponding recombinant vaccinia virus as a template; 1.5 µg of full-length viral genome RNA, along with 0.75 µg of in vitro transcribed HCoV-229E nucleocapsid protein mRNA, was used to transfect 1 × 106 Huh7 cells using the TransIT-mRNA transfection kit according to the manufacturer's instructions (Mirus Bio). At 72 h post transfection (p.t.), cell culture supernatants were collected and serially passaged in Huh7 cells for 21 (WT) or 12 times (HCoV-229E_SL2-SARS-CoV and HCoV-229E_SL2-BCoV), respectively.
Nanopore sequencing and long-read assessment
For nanopore sequencing, 1 µg of RNA in 9 µL was carried into the library preparation with the Oxford Nanopore DRS protocol (SQK-RNA001). All steps were followed according to the manufacturer's specifications. The library was then loaded on an R9.4 flow cell and sequenced on a MinION device (ONT). The sequencing run was terminated after 48 h.
The raw signal data were basecalled using Albacore (v2.2.7; available through the Oxford Nanopore community forum).
Although it is customary to remove adapters after DNA sequencing experiments, we did not perform this preprocessing step. The reason is that the sequenced RNA is attached to the adapter molecule via a DNA linker, effectively creating a DNA–RNA chimera. The current basecaller, being trained on RNA, is not able to reliably translate the DNA part of the sequence into base space, which makes adapter trimming based on sequence distance unreliable. However, we found that the subsequent mapping is very robust against these adapter sequences. All mappings were performed with minimap2 (v2.8-r672) (Li 2018) using the “spliced” preset without observing the canonical GULAG splicing motif (parameter -u n), and k-mer size set to 14 (-k 14).
Raw reads coverage and sequence identity to the HCoV-229E reference genome (WT: GenBank, NC_002645.1; SL2: stem loop 2 sequence replaced with SARS-CoV SL2 sequence) were determined from mappings to the references produced by minimap2. Read origin and sequencing error statistics were assessed by mapping the reads simultaneously with minimap2 to a concatenated mock-genome consisting of HCoV-229E (WT and SL2 variants, respectively), yeast enolase 2 mRNA (calibration strand, GenBank, NP_012044.1), and the human genome (GRCh38). Identity and error rates are the number of matching nucleotides (or number of nucleotide substitutions, insertions, or deletions) divided by the total length of the alignment including gaps from indels.
Consensus calling of nanopore reads was performed with Ococo (v0.1.2.6) (Břinda et al. 2017). The minimum required base quality was set to zero in order to avoid gaps in low coverage domains.
We used the hybrid error correction tool HG-CoLoR (Morisse et al. 2018) in conjunction with the Illumina HiSeq short-read data sets of both samples to reduce errors in all reads that exceed 20,000 nt in length. The program builds a de Bruijn graph from the near noise-free short-read data and then substitutes fragments of the noisy long reads with paths found in the graph that correspond to that same fragment of the sequence. HG-CoLoR was run with default parameters except for the maximum order of the de Bruijn graph, which was set to 50 in order to fit the length of the short reads.
Illumina HiSeq sequencing and assembly
Illumina short-read sequencing was performed using the TruSeq RNA v2 kit to obtain RNA from species with poly(A) tails and without any strand information. The three samples (WT, SL2_SARS-CoV, SL2_BCoV) selected for this study were prepared on a HiSeq 2500 lane and sequenced with 51 cycles. After demultiplexing, 23.2, 22.0, and 23.8 million single-end reads were obtained for the WT and the two SL2 samples, respectively. The raw sequencing data were deposited at the OSF (doi:10.17605/OSF.IO/UP7B4) and ENA (PRJEB33797).
Characterization of transcript isoforms and sg RNAs
We first defined TRS as an 8-mer with a maximum Hamming distance of two from the motif UCUCAACU. We then searched the HCoV-229E reference genome (GenBank, NC_002645.1) for all matching 8-mers. We then synthesized sg RNAs in silico as follows: For each pair of complementary 8-mers (5′-TRS, 3′-TRS), we accepted at most one mismatch to simulate base-pairing under a stable energy state. We then joined two reference subsequences for each pair or TRS: first, the 5′-end up to but not including the 5′-TRS and, second, the 3′-end of the reference genome including the 3′-TRS and excluding the poly(A) tail.
This way we obtained about 5000 candidate sg RNAs. To validate them, we mapped the nanopore reads to these “mock” sg RNAs in a nondiscontinuous manner; that is, all reads had to map consecutively without large gaps. To count as a putative hit, 95% of the read length had to uniquely map to a given mock transcript, and the mock transcript could not be >5% longer than the read.
We only considered putative hits as plausible if they had a read support of at least five. With this threshold, we aim to balance the sensitivity of finding plausible novel transcripts with a need to control the number of false positives.
Identification of 5mC methylation
We used Tombo (v1.3, default parameters) (Stoiber et al. 2016) to identify signal level changes corresponding to 5mC methylation (see Supplemental Fig. S6).
To assess the FPR of the methylation calling, we used an RCS as a negative control. It is added in the standard library preparation protocol for DRS. This mRNA standard is derived from the yeast enolase II (YHR174W) gene (Engel et al. 2014) and is produced using an in vitro transcription system. As a consequence, the mRNA standard is not methylated.
For a conservative resquiggle match score of 1.3 (part of the Tombo algorithm, default setting for RNA) and a methylation threshold of 0.9, the FPR was 4.67%, which met our requirement that the FPR be <5%. Our experimental setup did not include a positive methylation control.
Data access
Both the short-read cDNA (Illumina) and the basecalled long-read RNA (ONT) data from this study have been submitted to the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) under accession number PRJEB33797, as well as to the Open Science Framework repository UP7B4. Raw long-read data in fast5 format have also been submitted to this OSF repository. All analysis code has been submitted to the same OSF repository and is also available in Supplemental_Code.zip from the Supplemental Material.
Supplementary Material
Acknowledgments
We sincerely thank Celia Diezel for technical assistance in nanopore sequencing. We thank Ivonne Görlich and Marco Groth from the Core Facility DNA sequencing of the Leibniz Institute on Aging–Fritz Lipmann Institute in Jena for their help with Illumina sequencing. We also thank Nadja Karl (Medical Virology, Giessen) for excellent technical assistance. M.H. appreciates the support of the Joachim Herz Foundation by the add-on fellowship for interdisciplinary life science. This work was supported by BMBF–InfectControl 2020 (03ZZ0820A; K.L., M.M.) and is part of the Collaborative Research Centre AquaDiva (CRC 1076 A06 AquaDiva) of the Friedrich Schiller University Jena, funded by the Deutsche Forschungsgemeinschaft (DFG), supporting M.H. The study is further supported by DFG TRR 124 “FungiNet,” INST 275/365-1, B05 (M.M.). The work of J.Z. was supported by the DFG (SFB 1021-A01 and KFO 309-P3). We used Inkscape version 0.92.1 (available from inkscape.org) to finalize our figures for publication.
Author contributions: A.V. developed the experimental design for sequencing with nanopores. A.V., S.K., and K.L. analyzed and interpreted the data. J.Z., R.M., M.H., and M.M. were major contributors for discussion and in writing the manuscript. All authors wrote, commented, edited, and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.247064.118.
References
- Alonso S, Izeta A, Sola I, Enjuanes L. 2002. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J Virol 76: 1293–1308. 10.1128/JVI.76.3.1293-1308.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Baaijens JA, Aabidine AZE, Rivals E, Schönhuth A. 2017. De novo assembly of viral quasispecies using overlap graphs. Genome Res 27: 835–848. 10.1101/gr.215038.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiling A, Lyko F. 2015. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics Chromatin 8: 24 10.1186/s13072-015-0016-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian DA, Spaan WJ. 1997. Recombination and coronavirus defective interfering RNAs. Semin Virol 8: 101–111. 10.1006/smvy.1997.0109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Břinda K, Boeva V, Kucherov G. 2017. Ococo: an online consensus caller. arXiv:1712.01146 [q-bio.GN].
- Brown CG, Nixon KS, Senanayake SD, Brian DA. 2007. An RNA stem-loop within the bovine coronavirus nsp1 coding region is a cis-acting element in defective interfering RNA replication. J Virol 81: 7716–7724. 10.1128/JVI.00549-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R-Y, Hofmann MA, Sethna PB, Brian DA. 1994. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J Virol 68: 8223–8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R-Y, Krishnan R, Brian DA. 1996. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J Virol 70: 2720–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua EW, Ng PY. 2016. MinION: a novel tool for predicting drug hypersensitivity? Front Pharmacol 7: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depledge DP, Puthankalam SK, Sadaoka T, Beady D, Mori Y, Placantonakis DG, Mohr I, Wilson AC. 2018. Native RNA sequencing on nanopore arrays redefines the transcriptional complexity of a viral pathogen. bioRxiv 10.1101/373522 [DOI] [PMC free article] [PubMed]
- Engel SR, Dietrich FS, Fisk DG, Binkley G, Balakrishnan R, Costanzo MC, Dwight SS, Hitz BC, Karra K, Nash RS, et al. 2014. The reference genome sequence of Saccharomyces cerevisiae: then and now. G3 (Bethesda) 4: 389–398. 10.1534/g3.113.008995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria NR, Quick J, Claro I, Theze J, de Jesus JG, Giovanetti M, Kraemer MUG, Hill SC, Black A, da Costa AC, et al. 2017. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature 546: 406–410. 10.1038/nature22401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya T, Macnaughton TB, La Monica N, Lai MM. 1993. Natural evolution of coronavirus defective-interfering RNA involves RNA recombination. Virology 194: 408–413. 10.1006/viro.1993.1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallei A, Pankraz A, Thiel H-J, Becher P. 2004. RNA recombination in vivo in the absence of viral replication. J Virol 78: 6271–6281. 10.1128/JVI.78.12.6271-6281.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A, et al. 2018. Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods 15: 201–206. 10.1038/nmeth.4577 [DOI] [PubMed] [Google Scholar]
- Grozhik AV, Jaffrey SR. 2017. Epitranscriptomics: shrinking maps of RNA modifications. Nature 551: 174–176. 10.1038/nature24156 [DOI] [PubMed] [Google Scholar]
- Gustin KM, Guan B-J, Dziduszko A, Brian DA. 2009. Bovine coronavirus nonstructural protein 1 (p28) is an RNA binding protein that binds terminal genomic cis-replication elements. J Virol 83: 6087–6097. 10.1128/JVI.00160-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzig T, Scandella E, Schelle B, Ziebuhr J, Siddell SG, Ludewig B, Thiel V. 2004. Rapid identification of coronavirus replicase inhibitors using a selectable replicon RNA. J Gen Virol 85: 1717–1725. 10.1099/vir.0.80044-0 [DOI] [PubMed] [Google Scholar]
- Holmes EC. 2009. The evolution and emergence of RNA viruses. Oxford University Press, Oxford, New York. [Google Scholar]
- Hölzer M, Marz M. 2017. Software dedicated to virus sequence analysis “bioinformatics goes viral.” Adv Virus Res 99: 233–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izeta A, Smerdou C, Alonso S, Penzes Z, Mendez A, Plana-Durán J, Enjuanes L. 1999. Replication and packaging of transmissible gastroenteritis coronavirus-derived synthetic minigenomes. J Virol 73: 1535–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Olsen HE, Paten B, Akeson M. 2016. The oxford nanopore minION: delivery of nanopore sequencing to the genomics community. Genome Biol 17: 239 10.1186/s13059-016-1103-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo M, Makino S. 1992. Mutagenic analysis of the coronavirus intergenic consensus sequence. J Virol 66: 6330–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst SM, Dueholm MS, McIlroy SJ, Kirkegaard RH, Nielsen PH, Albertsen M. 2018. Retrieval of a million high-quality, full-length microbial 16S and 18S rRNA gene sequences without primer bias. Nat Biotechnol 36: 190–195. 10.1038/nbt.4045 [DOI] [PubMed] [Google Scholar]
- Keller MW, Rambo-Martin BL, Wilson MM, Ridenour CA, Shepard SS, Stark TJ, Neuhaus EB, Dugan VG, Wentworth DE, Barnes JR. 2018. Direct RNA sequencing of the coding complete influenza A virus genome. Sci Rep 8: 14408 10.1038/s41598-018-32615-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27: 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MM. 1992. RNA recombination in animal and plant viruses. Microbiol Rev 56: 61–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MM. 1998. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology 244: 1–12. 10.1006/viro.1998.9098 [DOI] [PubMed] [Google Scholar]
- Lai MM, Cavanagh D. 1997. The molecular biology of coronaviruses. Adv Virus Res 48: 1–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CL, Lai MM. 1992. RNA recombination in a coronavirus: recombination between viral genomic RNA and transfected RNA fragments. J Virol 66: 6117–6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman NJ, Quick J, Simpson JT. 2015. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods 12: 733–735. 10.1038/nmeth.3444 [DOI] [PubMed] [Google Scholar]
- Luytjes W, Gerritsma H, Spaan WJ. 1996. Replication of synthetic defective interfering RNAs derived from coronavirus mouse hepatitis virus-A59. Virology 216: 174–183. 10.1006/viro.1996.0044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. 2013. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res 41: D262–D267. 10.1093/nar/gks1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhugiri R, Karl N, Petersen D, Lamkiewicz K, Fricke M, Wend U, Scheuer R, Marz M, Ziebuhr J. 2018. Structural and functional conservation of cis-acting RNA elements in coronavirus 5′-terminal genome regions. Virology 517: 44–55. 10.1016/j.virol.2017.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre ABR, Alexander N, Grigorev K, Bezdan D, Sichtig H, Chiu CY, Mason CE. 2019. Single-molecule sequencing detection of N6-methyladenine in microbial reference materials. Nat Commun 10: 579 10.1038/s41467-019-08289-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery VD, Graham RL, Baric RS. 2017. Jumping species: a mechanism for coronavirus persistence and survival. Curr Opin Virol 23: 1–7. 10.1016/j.coviro.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez A, Smerdou C, Izeta A, Gebauer F, Enjuanes L. 1996. Molecular characterization of transmissible gastroenteritis coronavirus defective interfering genomes: packaging and heterogeneity. Virology 217: 495–507. 10.1006/viro.1996.0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheyev AS, Tin MM. 2014. A first look at the Oxford Nanopore MinION sequencer. Mol Ecol Resour 14: 1097–1102. 10.1111/1755-0998.12324 [DOI] [PubMed] [Google Scholar]
- Moldován N, Balázs Z, Tombácz D, Csabai Z, Szűcs A, Snyder M, Boldogkői Z. 2017. Multi-platform analysis reveals a complex transcriptome architecture of a circovirus. Virus Res 237: 37–46. 10.1016/j.virusres.2017.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldován N, Tombácz D, Szűcs A, Csabai Z, Balázs Z, Kis E, Molnár J, Boldogkői Z. 2018a. Third-generation sequencing reveals extensive polycistronism and transcriptional overlapping in a baculovirus. Sci Rep 8: 8604 10.1038/s41598-018-26955-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldován N, Tombácz D, Szűcs A, Csabai Z, Snyder M, Boldogkői Z. 2018b. Multi-platform sequencing approach reveals a novel transcriptome profile in pseudorabies virus. Front Microbiol 8: 2708 10.3389/fmicb.2017.02708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisse P, Lecroq T, Lefebvre A. 2018. Hybrid correction of highly noisy long reads using a variable-order de Bruijn graph. Bioinformatics 34: 4213–4222. [DOI] [PubMed] [Google Scholar]
- Nowak MA. 1992. What is a quasispecies? Trends Ecol Evol 7: 118–121. 10.1016/0169-5347(92)90145-2 [DOI] [PubMed] [Google Scholar]
- Ozdarendeli A, Ku S, Rochat S, Williams GD, Senanayake SD, Brian DA. 2001. Downstream sequences influence the choice between a naturally occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J Virol 75: 7362–7374. 10.1128/JVI.75.16.7362-7374.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak AO, Spaan WJM, Snijder EJ. 2006. Nidovirus transcription: how to make sense…? J Gen Virol 87: 1403–1421. 10.1099/vir.0.81611-0 [DOI] [PubMed] [Google Scholar]
- Pathak KB, Nagy PD. 2009. Defective interfering RNAs: foes of viruses and friends of virologists. Viruses 1: 895–919. 10.3390/v1030895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes Z, Tibbles K, Shaw K, Britton P, Brown TD, Cavanagh D. 1994. Characterization of a replicating and packaged defective RNA of avian coronavirus infectious bronchitis virus. Virology 203: 286–293. 10.1006/viro.1994.1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes Z, Wroe C, Brown TD, Britton P, Cavanagh D. 1996. Replication and packaging of coronavirus infectious bronchitis virus defective RNAs lacking a long open reading frame. J Virol 70: 8660–8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick J, Loman NJ, Duraffour S, Simpson JT, Severi E, Cowley L, Bore JA, Koundouno R, Dudas G, Mikhail A, et al. 2016. Real-time, portable genome sequencing for ebola surveillance. Nature 530: 228–232. 10.1038/nature16996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raabe T, Schelle-Prinz B, Siddell SG. 1990. Nucleotide sequence of the gene encoding the spike glycoprotein of human coronavirus HCV 229E. J Gen Virol 71(Pt 5): 1065–1073. 10.1099/0022-1317-71-5-1065 [DOI] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, He C. 2017. Dynamic RNA modifications in gene expression regulation. Cell 169: 1187–1200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki SG, Sawicki DL. 1995. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv Exp Med Biol 380: 499–506. 10.1007/978-1-4615-1899-0_79 [DOI] [PubMed] [Google Scholar]
- Sawicki SG, Sawicki DL. 1998. A new model for coronavirus transcription. Adv Exp Med Biol 440: 215–219. 10.1007/978-1-4615-5331-1_26 [DOI] [PubMed] [Google Scholar]
- Sawicki SG, Sawicki DL, Siddell SG. 2007. A contemporary view of coronavirus transcription. J Virol 81: 20–29. 10.1128/JVI.01358-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SS, Kamahora T, Lai MM. 1989. Sequence analysis of the nucleocapsid protein gene of human coronavirus 229E. Virology 169: 142–151. 10.1016/0042-6822(89)90050-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Jain M, Mulroney L, Garalde DR, Akeson M. 2017. Reading canonical and modified nucleotides in 16S ribosomal RNA using nanopore direct RNA sequencing. bioRxiv 10.1101/132274 [DOI] [PMC free article] [PubMed]
- Sola I, Mateos-Gomez PA, Almazan F, Zuñiga S, Enjuanes L. 2011. RNA-RNA and RNA-protein interactions in coronavirus replication and transcription. RNA Biol 8: 237–248. 10.4161/rna.8.2.14991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I, Almazan F, Zúñiga S, Enjuanes L. 2015. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol 2: 265–288. 10.1146/annurev-virology-100114-055218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoiber MH, Quick J, Egan R, Lee JE, Celniker SE, Neely RK, Loman N, Pennacchio LA, Brown J. 2016. De novo identification of DNA modifications enabled by genome-guided nanopore signal processing. bioRxiv 10.1101/094672 [DOI]
- Thiel V, Siddell SG. 2005. Reverse genetics of coronaviruses using vaccinia virus vectors. Curr Top Microbiol Immunol 287: 199–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V, Herold J, Schelle B, Siddell SG. 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J Gen Virol 82: 1273–1281. 10.1099/0022-1317-82-6-1273 [DOI] [PubMed] [Google Scholar]
- Thiel V, Ivanov KA, Putics A, Hertzig T, Schelle B, Bayer S, Weissbrich B, Snijder EJ, Rabenau H, Doerr HW, et al. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol 84: 2305–2315. 10.1099/vir.0.19424-0 [DOI] [PubMed] [Google Scholar]
- Tombácz D, Csabai Z, Szűcs A, Balázs Z, Moldován N, Sharon D, Snyder M, Boldogkői Z. 2017. Long-read isoform sequencing reveals a hidden complexity of the transcriptional landscape of herpes simplex virus type 1. Front Microbiol 8: 1079 10.3389/fmicb.2017.01079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay R, Perlman S. 2016. Middle east respiratory syndrome and severe acute respiratory syndrome. Curr Opin Virol 16: 70–76. 10.1016/j.coviro.2016.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwick-Dugdale J, Solonenko N, Moore K, Chittick L, Gregory AC, Allen MJ, Sullivan MB, Temperton B. 2019. Long-read viral metagenomics captures abundant and microdiverse viral populations and their niche-defining genomic islands. PeerJ 7: e6800. 10.7717/peerj.6800 [DOI] [PMC free article] [PubMed]
- Workman RE, Tang A, Tang PS, Jain M, Tyson JR, Zuzarte PC, Gilpatrick T, Razaghi R, Quick J, Sadowski N. 2018. Nanopore native RNA sequencing of a human poly(A) transcriptome. bioRxiv 10.1101/459529 [DOI] [PMC free article] [PubMed]
- Wu H-Y, Brian DA. 2010. Subgenomic messenger RNA amplification in coronaviruses. Proc Natl Acad Sci 107: 12257–12262. 10.1073/pnas.1000378107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga S, Sola I, Alonso S, Enjuanes L. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J Virol 78: 980–994. 10.1128/JVI.78.2.980-994.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.