Abstract
We examined the ability of neuronal ensembles from rat motor cortex to predict behavioral performance during a reaction time task. We found that neurons that were the best individual predictors of task performance were not necessarily the neurons that contributed the most predictive information to an ensemble of neurons. To understand this result, we applied a framework for quantifying statistical relationships between neurons (Schneidman et al., 2003) to all possible combinations of neurons within our ensembles. We found that almost all neurons (96%) contributed redundant predictive information to the ensembles. This redundancy resulted in the maintenance of predictive information despite the removal of many neurons from each ensemble. Moreover, the balance of synergistic and redundant interactions depended on the number of neurons in the ensemble. Small ensembles could exhibit synergistic interactions (e.g., 23 ± 9% of ensembles with two neurons were synergistic). In contrast, larger ensembles exhibited mostly redundant interactions (e.g., 99 ± 0.1% of ensembles with eight neurons were redundant). We discuss these results with regard to constraints on interpreting neuronal ensemble data and with respect to motor cortex involvement in reaction time performance.
Keywords: motor cortex, population coding, neuronal ensembles, reaction time, synergy, redundancy
Introduction
How do ensembles of cortical neurons work together to encode information? Pioneering studies of motor cortex by Georgopoulos et al. (1982, 1988) have suggested that “population vectors,” constructed from weighted averages of the responses of single neurons, can accurately predict behavioral variables, such as movement direction. This approach has been used to study population coding in a number of cortical systems (Gochin et al., 1994; Britten et al., 1996; Priebe and Lisberger, 2004; Romo et al., 2004). These studies have led to the view that cortical neurons act as independent processors of information (Gochin et al., 1994; Rolls et al., 1997, 2004; Reich et al., 2001; Purushothaman and Bradley, 2005). Although correlations between neurons may be important factors for neural coding (Zohary et al., 1994; Vaadia et al., 1995; Dan et al., 1998; Abbott and Dayan, 1999; Bair et al., 2001; Nirenberg and Latham, 2003; Averbeck and Lee, 2004; Johnson, 2004), few studies have demonstrated that considering neuronal interactions increases predictive information over that contained in individual neurons (Riehle et al., 1997; Hatsopoulos et al., 1998; Gat and Tishby, 1998; Maynard et al., 1999; Averbeck et al., 2003; Samonds et al., 2004).
Gawne and Richmond (1993) and, more recently, Schneidman et al. (2003) have proposed a theoretical framework for interpreting neuronal ensemble activity in which information encoded by an ensemble of neurons is compared with the sum of information encoded by the component neurons. In this framework, an independent coding scheme suggests that predictive information encoded by an ensemble of neurons should approximately equal the sum of its individual parts. However, if ensemble interactions exist between recorded neurons, interactions can be either synergistic (i.e., predictive information encoded by the ensemble of neurons is greater than the sum of the information of the individual neurons) or redundant (i.e., the predictive information encoded by the ensemble is less than the sum of information of individual neurons). With redundancy, a hallmark of distributed processing, the removal of individual neurons from an ensemble does not affect overall system performance. Redundant systems are thus resistant to error and can have improved signal-to-noise (Barlow, 2001).
In the present study, we investigated the relationship between ensembles of simultaneously recorded cortical neurons and their component neurons by using statistical classifiers to estimate predictive information at the individual-neuron and ensemble levels. We found that relationships between the activity of multiple neurons (that is, joint activity of neuronal ensembles) are important factors for the performance of statistical classifiers in predicting behavioral outcomes. We report three results: (1) the predictive information of a single neuron is not related to its informational contribution to a neuronal ensemble; (2) this effect is attributable to high levels of redundancy; and (3) the extent of redundant or synergistic interactions depends on the size of the neuronal ensemble. Ensembles of more than a few neurons were almost entirely redundant. Smaller ensembles of neurons, especially pairs or triplets, could be either redundant or synergistic.
Materials and Methods
Behavioral and electrophysiological methods
Data were collected from the motor cortex of awake rats performing a simple reaction time task (Laubach et al., 2000). Adult, male rats (Long-Evans) were trained to perform a simple reaction time task (Fig. 1). In this task, the rats pressed a lever to initiate a trial and maintained the lever press over a variable delay period of 400-800 ms. At the end of the delay, a trigger stimulus was presented (tone or vibration of the lever), and the rats released the lever within 1 s of stimulus onset to obtain a reward (∼0.02 ml of water). “Conditioned” responses in the task occurred when the rats successfully maintained the lever press for the full delay period and released the lever within 1 s of the trigger stimulus. “Premature” responses occurred when the lever was released before the trigger stimulus. “Late” responses occurred when the lever was released >1 s after the trigger stimulus and were infrequent after initial training sessions (<10% of trials). In the present study, neuronal ensemble activity was compared for trials with conditioned and premature responses. Data analysis was performed for two sessions from each of three rats. These data were collected after several weeks of training, i.e., some time after the data described in the study by Laubach et al. (2000).
Figure 1.
The simple reaction time task, in which rats were trained to initiate a trial by pressing a lever, which they held for a variable delay lasting 400-800 ms. The rats received a vibrotactile or an auditory stimulus. A, On conditioned release trials, rats performed the task appropriately and released the lever within 1 s of the conditioning stimulus to receive a water reward. B, On premature release trials, rats released the lever prematurely before they received the vibrotactile or auditory stimulus. If the response was premature, there was a 2 s timeout. Note that the stimulus (represented in gray) did not occur on premature trials.
Procedures for chronically implanting arrays of microwire electrodes (NB Labs, Denison, TX) are described in detail previously (Nicolelis, 1998). Two chronic microwire arrays (50 μm stainless steel wire, Teflon insulation to a blunt tip, arranged in 2 × 8 matrices with individual wires spaced by 250 μm) were stereotaxically implanted in rat motor cortex (caudal array: -0.5-1.0 mm anteroposterior, ±2.5-3.5 mm mediolateral, -1.5 mm dorsoventral at -20° anterior to the frontal plane; rostral array: +2.0-3.5 mm, ±2.0-3.0 mm, -1.5 mm at +20° posterior to the frontal plane; coordinates from bregma) under aseptic conditions. The locations of microwires were reconstructed from 50-mm-thick, thionin-stained, frontal sections after the completion of all experiments. All rats were treated in accordance with National Institutes of Health guidelines.
Neuronal ensemble recordings were made using a many-neuron acquisition program (MNAP; Plexon, Dallas, TX). Single neuronal units were identified off-line by the following criteria: consistent waveform shapes, an average waveform amplitude at least three times larger than background activity, and a consistent refractory period of at least 2 ms in interspike interval histograms. Only units whose activity was modulated around the time of lever release were analyzed, as assessed visually by using cumulative density plots (Stranger; Biographics, Winston-Salem, NC).
The animals' behavior was recorded on videotape and examined to ensure that it was consistent for all trials included in a given analysis. Additional control data were collected in one animal in which neuronal ensemble activity was recorded together with electromyographical (EMG) data (agonist: triceps brachii, which extends the forelimb; antagonist: trapezius, which rotates and pulls the shoulder blade toward the spinal column). Additionally, an electrolytic tile sensor (Advanced Orientation Systems, Linden, NJ) was mounted on the response lever and recorded the angle of the lever. Signals from the EMG electrodes and the positional sensor were amplified and filtered at 20-500 Hz (SA Instrumentation, Encinitas, CA) and sampled at 1 kHz. As described by Laubach et al. (2000), there were no significant differences between the evoked EMG or lever-position data that accompanied lever releases on trials with conditioned versus premature responses.
Data analysis
A statistical pattern recognition approach was used to quantify the predictive relationship between neuronal activity and the behavioral outcome of each trial (i.e., conditioned vs premature response). This approach involves three basic steps that are described below: (1) preprocessing, (2) dimension reduction, and (3) prediction of behavior using statistical classifiers.
Preprocessing. Neural activity was collected from 400 ms before to 100 ms after lever release. For each neuron, average spiking responses for conditioned and premature trials were computed using a bin size of 1 ms. Single-trial perievent time histograms of neural activity were then convolved over 5 ms epochs using low-pass filtering (Richmond and Optican, 1987) and decimation (for additional details, see Laubach, 2004). This low-pass filtering was performed on the raw perievent histograms before the wavelet packet analysis below.
Dimension reduction. A wavelet-based method, discriminant pursuit (Buckheit and Donoho, 1995; Laubach, 2004), was used to reduce the dimensionality of the neuronal data before analysis with statistical classifiers. This was performed in four steps, as follows. First, a difference vector was computed for each neuron by subtracting the mean response on different types of trials (i.e., conditioned vs premature response). Second, the difference vector was decomposed with the wavelet packet transform using the Daubechies four-tap wavelet. The result of the wavelet packet transform is several components, analogous to components in principal component analysis. Each wavelet component represents variance in the original data and corresponds to a “feature” in the firing pattern of a single neuron, such as an increase in firing rate preceding movement on conditioned trials. Third, wavelet components were ranked by the between-groups variance that they accounted for in the original spike trains. Finally, a reduced set of components was then chosen for subsequent analysis by using only those features from a given neuron that accounted for more variance than features extracted from random data; that is, the amplitudes of features extracted from neural data were compared with those extracted from 1000 “random” data sets, in which the outcome of each trial was assigned pseudorandomly. This approach is an alternative to using an arbitrary criterion for determining the number of features (cf. Laubach, 2004). In the present study, neurons with task-related features had an average of 5.5 ± 2.5 features. Interactions between neurons were not examined at this stage of processing. Instead, neuronal interactions were evaluated using statistical classifiers described below.
Statistical classification. Features obtained with the wavelet packet analysis were subsequently used as input for a variety of statistical classifiers. Exploratory analysis of statistical classifiers was conducted in R (Venables and Ripley, 2002; Venables et al., 2004) and in WEKA (http://www.cs.waikato.ac.nz/~ml/weka/; WEKA is a data mining tool that enables rapid processing of data and visualization of results with a multitude of classifiers). We selected three classifiers for additional analysis: learning vector quantization (LVQ) (Kohonen, 2000) (we chose LVQ because previous studies have used it; see below), penalized discriminant analysis (PDA) (Hastie et al., 1995) (we chose PDA because it is a linear method), and support vector machines (SVMs) (Hastie et al., 2001; Hsu et al., 2003) (we chose SVMs because they performed well). LVQ was implemented using the “class” library for R. An algorithm for SVMs (libSVM) with a sigmoid transfer function was implemented using the “e1071” library for R, and PDA, which involves the use of ridge regression to define discriminant functions that are subsequently analyzed with a Gaussian mixture model, was implemented using the “mda” library for R (of special note, the algorithms for these implementations in R used original source code developed by the authors of each algorithm.) No effort was made to optimize parameters of any statistical classifiers in this study. All results were confirmed with multiple classifiers; however, for the purposes of comparison with previously published papers (Nicolelis et al., 1998a; Ghazanfar et al., 2000; Laubach et al., 2000; Furukawa and Middlebrooks, 2001; Middlebrooks and Bierer, 2002), the results in this manuscript were described using LVQ, except when explicitly reported.
The statistical classifiers were trained on subsets of trials (i.e., training data), and predictions were made for trials not included in the training data (i.e., testing data). To generate stable estimates of predictive information, 10-fold cross validation was used; that is, each trial was a member of one testing data set and a member of nine training data sets. The result of each statistical classifier is a “classification matrix” (also called a confusion matrix); that is, a table of correct and error predictions by the classifier for each class on the testing data.
For every value of information and accuracy reported, error bars were estimated by performing 10 10-fold cross validations (100 classifications). Chance levels of classification and confidence intervals were estimated by permutation tests (Good, 2000) in which the behavioral labels for all trials were randomly permuted, and classification analysis was repeated 1000 times with a unique random seed for each iteration.
All analysis described in this manuscript was performed using scripts written for R, WEKA, and Matlab (MathWorks, Natick, MA). These analyses were run over a cluster of 16 Linux workstations via batch scripts in the Bash shell. All analysis and clustering scripts used in this study are available on request from the corresponding author.
Limitations of statistical classifiers. In this study, statistical classifiers were used to predict a behavioral condition (i.e., conditioned vs premature response) from neural data as represented by features from multiple neurons. Information contained in the neural data were estimated by applying information theory to confusion matrices for each classifier and each data set (see below). It is important to point out that this approach cannot estimate the true mutual information contained in the neuronal data because the informational estimates depend on the specific classifier used for prediction (for review, see Ripley, 1996; Duda et al., 2001; Hastie et al., 2001). Consequently, several different statistical classifiers were used when estimating neural information. However, it is always possible that another statistical classifier exists that will perform better on our data. Even if such a classifier exists, there is no way to verify that rules used by a statistical classifier have any relationship to the true mutual information or rules used by the animal. For these reasons, and according to the data processing inequality (Cover and Thomas, 1991; Schneidman et al., 2003), we and others (Averbeck et al., 2003; Foffani et al., 2004) can only estimate a lower bound of mutual information between neural data and behavioral outcomes. Nevertheless, we believe that the statistical pattern recognition approach is a practical solution to the problem of interpreting high-dimensional neuronal ensemble data. This approach has been used for a variety of real-world datasets (Ripley, 1996; Witten and Frank, 2000; Hastie et al., 2001) and is able to quantify relationships between neurons and animal behavior on a trial-by-trial basis (compared with information theoretic methods, which are average measures).
Assessment of classifier performance. Statistical classifier performance can be assessed in several ways. One index of statistical classifier performance is “error rate,” or the number of trials classified incorrectly divided by the total trials. For clarity, “accuracy” (1 - error rate) is reported in this manuscript. Note that the chance accuracy is not 50%; rather, it is the percentage of trials in the larger class.
One method of assessing statistical classifier performance that allows us to compare our results with other neuroscientific studies is to calculate mutual information (Gawne and Richmond, 1993; Rolls et al., 1997; Ghazanfar et al., 2000; Laubach et al., 2000; Reich et al., 2001; Schneidman et al., 2003). We calculate the mutual information from the classification matrix according to the following equation (Cover and Thomas, 1991):
 |
(1) |
where S is the actual outcome, and R is the predicted outcome. In general, measures of statistical classifier performance are highly correlated. Accuracy and mutual information for individual neurons were correlated at R = 0.94 ± 0.04 (t ≫ 10; p ≪ 0.001).
Bias and variance. There are two sources of error in estimates of mutual information from the statistical pattern recognition approach: “bias” and “variance” (Ripley, 1996; Hastie et al., 2001). Bias refers to error between estimates of predictive information and true information. For statistical classifiers, there are three sources of bias. First, a statistical classifier might have a “language bias” based on its representational ability; that is, some algorithms may be better able to capture certain types of data as a function of the way they internally represent that data. Second, statistical classifiers might suffer from a “search bias” in the heuristic rules that they use to fit the data. If the rules are inappropriate, inaccurate conclusions may be reached. Finally, statistical classifiers can suffer from an “overfitting bias,” or tendency to find patterns that do not exist in the data. For an additional discussion of these topics, see Witten and Frank (2000) or Hastie et al. (2001). The first two sources of bias were addressed by using multiple statistical classifiers with distinct internal representations and decision rules. It is always possible that there exists another classifier that has an internal representation that better matches our data or that has more appropriate decision rules. The last source of bias, attributable to overfitting, was addressed directly in two ways. First, for every dataset and every number of neurons, the statistical pattern recognition approach was applied to data in which the behavioral labels for all trials were randomly permuted (see Results). Second, statistical classifiers were applied to benchmark data with a known Bayesian error rate (the “waveform” benchmark data, available in the “mlbench” library for R; 86% error rate), and it was found that these classifiers did not greatly overfit or underfit the benchmark data (generally within ±1%) (see Table 3, row 1). These tests lent confidence to our interpretations of the results of this study. The second source of error in classification was variance, i.e., errors in classification attributable to the sample chosen from which mutual information was predicted. Variance in our data were accounted for by repeating classifications several times.
Table 3.
Statistical classifier performance across datasets
|
Dataset |
Trials |
Behavioral performance |
|
Individual neuron |
Chance |
PDA |
Chance |
SVM |
Chance |
LVQ |
PEnsemble |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benchmark | 3000 | Acc | 86 ± 1% | 86 ± 1% | 87 ± 3% | % | ||||||
| Info | 0.87 ± 0.00 | 0.85 ± 0.00 | 0.89 ± 0.00 | Bits | ||||||||
| A1 | 173 | 58% | Acc | 64 ± 3% | 54 | 86 ± 1% | 56 | 89 ± 2% | 56 | 79 ± 2% | % | |
| Info | 0.05 ± 0.03 | 0.01 | 0.39 ± 0.03 | 0.01 | 0.50 ± 0.05 | 0.01 | 0.23 ± 0.04 | −0.43 | Bits | |||
| A2 | 163 | 63% | Acc | 65 ± 2% | 52 | 77 ± 1% | 60 | 91 ± 1% | 57 | 79 ± 1% | % | |
| Info | 0.03 ± 0.02 | 0.01 | 0.19 ± 0.02 | 0.01 | 0.52 ± 0.04 | 0.01 | 0.22 ± 0.02 | −0.31 | Bits | |||
| B1 | 171 | 76% | Acc | 77 ± 1% | 64 | 81 ± 2% | 75 | 84 ± 1% | 75 | 79 ± 1% | % | |
| Info | 0.03 ± 0.01 | 0.00 | 0.13 ± 0.03 | 0.00 | 0.18 ± 0.03 | 0.01 | 0.07 ± 0.02 | −0.14 | Bits | |||
| B2 | 244 | 72% | Acc | 73 ± 1% | 62 | 87 ± 1% | 72 | 87 ± 1% | 70 | 80 ± 2% | % | |
| Info | 0.02 ± 0.02 | 0.00 | 0.30 ± 0.02 | 0.00 | 0.28 ± 0.02 | 0.00 | 0.13 ± 0.03 | −0.12 | Bits | |||
| C1 | 229 | 59% | Acc | 63 ± 5% | 51 | 76 ± 1% | 54 | 79 ± 1% | 55 | 75 ± 1% | % | |
| Info | 0.04 ± 0.04 | 0.00 | 0.19 ± 0.01 | 0.00 | 0.24 ± 0.02 | 0.00 | 0.16 ± 0.01 | −0.38 | Bits | |||
| C2 | 188 | 70% | Acc | 71 ± 2% | 59 | 85 ± 2% | 70 | 89 ± 2% | 68 | 82 ± 1% | % | |
|
|
|
|
Info |
0.03 ± 0.03 |
0.00
|
0.30 ± 0.04 |
0.00
|
0.37 ± 0.06 |
0.00
|
0.19 ± 0.03 |
−0.12 |
Bits |
Accuracy (Acc) and information (Info) rates of three different statistical classifiers (PDA, SVM, and LVQ) for all six datasets and one benchmark dataset are shown. For each dataset (labeled in the leftmost column), accuracy and information rates are each presented on a separate row (labeled in the rightmost column). Chance levels for each classifier are listed to the left of the results for each classifier in italics. For reference, the number of trials, the behavioral performance of the animal for each session, and the average predictive information for individual neurons within a particular dataset are listed in the first three columns. The redundancy for each dataset (PEnsemble) is listed in the last column.
Synergy and redundancy
To assess the contributions of each neuron to ensemble information, features for each neuron were removed from each ensemble, and the predictive information from the remaining neurons was estimated (i.e., “neuron dropping”) (Nicolelis et al., 1998b). The information contributed to the ensemble by a given neuron (IContrib) was defined as the difference between the predictive information of the full ensemble (IEnsemble with neuron) and the predictive information of a subensemble without a given neuron (IEnsemble without neuron):
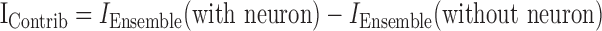 |
(2) |
Because a given neuron can participate in several subensembles of the original ensemble [for instance, for a three-neuron ensemble, neuron 3 participates in subensembles (2, 3), (1, 3), and (1, 2, 3)], IContrib was calculated for each of these subensembles that a given neuron participated in (for an illustration of the computation, see Table 1) by applying Equation 2 to all possible subensembles including a given neuron. Although computationally intensive (e.g., a data set with 17 neurons has 131,071 subensembles, requiring 182 h of CPU time to analyze all combinations of neurons using a single AMD 1800 XP processor; this same data set could be analyzed in 13 h using a cluster with workstations with 16 processors), this analysis provided a robust estimate of IContrib. Within each ensemble, IContrib was compared only with ensembles of similar size (i.e., an average IContrib for 2, 3, 4... n neuron ensembles was calculated separately).
Table 1.
Illustration of combinations for three neuron ensemble
|
Ensembles with neuron 3 |
|
Ensembles without neuron 3 |
|---|---|---|
| 3 2 1 | → | 2 1 |
| 3 2 | → | 2 |
| 3 1 | → | 1 |
| 3 |
→ |
N/A |
To measure how a particular neuron interacts with the rest of the ensemble, we defined a notion of information contributed by single neurons to the ensemble, PNeuron. This value was calculated from the contribution of a neuron to ensemble predictive information subtracted from the individual predictive information of that neuron:
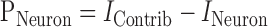 |
(3) |
In Equation 3, IContrib is the contribution of a particular neuron to an ensemble as measured by the predictive information lost after removing that neuron from an ensemble as calculated by Equation 2, and INeuron is the predictive information (in bits) encoded by that neuron.
This equation answers the following question: how does removing a neuron from an ensemble change ensemble predictive information? There are three possible results for removing a neuron from an ensemble that define the type of net contribution made by a neuron to information at the ensemble level. If PNeuron is zero (i.e., if IContrib ≈ INeuron), then the information lost by removing a neuron is equal to the individual information encoded by that neuron. Such a neuron contributes “independent” information to the ensemble. Second, if PNeuron is negative (if IContrib < INeuron), the information lost by removing a neuron is less than the individual information encoded by that neuron. Such a neuron contributes “redundant” information to the ensemble. Third, if PNeuron is positive (if IContrib > INeuron), the information lost by removing a neuron is greater than the individual information encoded by that neuron. Such a neuron contributes “synergistic” information to the ensemble.
Interactions at the ensemble level were defined as PEnsemble based on the ensemble information and the sum of the individual information of its component neurons:
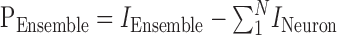 |
(4) |
where IEnsemble is the predictive information of the ensemble, and INeuron, as above, is the predictive information of individual neurons in the ensemble.
This equation answers the following question: how do neurons work together to encode predictive information? As above, there are three possible coding schemes. First, if PEnsemble is zero, ensemble predictive information is the same as the sum of predictive information from neurons in the ensemble. As a whole, this ensemble is neither redundant nor synergistic. Second, if PEnsemble is negative, ensemble predictive information is the less than the sum of predictive information from neurons in the ensemble. As a whole, this ensemble is redundant. Third, if PEnsemble is positive, ensemble predictive information is greater than the sum of predictive information from neurons in the ensemble. As a whole, this ensemble is synergistic (Fig. 2).
Figure 2.
Because neurons are added to an independent ensemble, predictive information increases proportionally to the number of neurons in the ensemble. If the ensemble is redundant, predictive information increases slower than the number of neurons added to the ensemble. If the ensemble is synergistic, predictive information increases faster than the number of neurons added to the ensemble.
In this manuscript, the quantities IContrib and PEnsemble were calculated across all possible subensembles for a given data set (Table 2). Because these values were predictive information from classification, their units were expressed in bits. Also, note that Equation 2 assumes that the estimated informational values of each neuron in a given ensemble did not depend on each other. Although our estimates of predictive information were generated from statistical classifiers instead of from direct information theoretic measures, our results are analogous to those obtained in previous studies (Gawne and Richmond, 1993; Maynard et al., 1999; Averbeck et al., 2003; Schneidman et al., 2003; Samonds et al., 2004).
Table 2.
Ensemble combinations
|
Dataset |
Neurons |
Features |
Combinations |
|---|---|---|---|
| A1 | 16 | 84 | 65,535 |
| A2 | 17 | 102 | 131,071 |
| B1 | 8 | 57 | 255 |
| B2 | 11 | 51 | 2047 |
| C1 | 13 | 50 | 8191 |
| C2 |
13 |
76 |
8191 |
Results
Neuronal and ensemble predictive information
We analyzed ensemble neuronal data from three awake, behaving rats performing a simple reaction time task (Laubach et al., 2000). Recording sessions (two from each rat) yielded an average of 23.5 ± 5.2 (mean ± SD) neurons (range of 17-27 neurons, 140 neurons total, 32 electrodes per animal performing 195 ± 33 trials per session). Of recorded neurons, 55.5% (78 of 140 recorded neurons, or an average of 13 neurons per session) had task-related features, i.e., had more features than would be expected from random data (see Materials and Methods) (Fig. 3).
Figure 3.
Perievent rasters and time histograms for an ensemble of 16 neurons with task-related features recorded from the motor cortex of one animal (dataset A1) while performing a simple reaction time task. A time interval from 400 ms before the lever release to 100 ms after (period shown in gray, release indicated by dashed vertical line) was used for the analyses reported in this study. Neurons 1-13 are from the caudal motor cortex, whereas neurons 14-16 are from the rostral motor cortex.
Analysis using statistical pattern recognition (preprocessing/discriminant pursuit/LVQ, unless otherwise stated) revealed that the single neurons could be used to discriminate between conditioned and premature trials with an average accuracy of 68 ± 3% (accuracy for individual neurons with randomly permuted behavioral labels was 64 ± 8%). Using LVQ, the average single neuron encoded 0.034 ± 0.03 bits of predictive information (ÎNeuron) of a possible 0.90 ± 0.02 bits of predictive information [note that the possible information is <1 because, in every dataset, there are more conditioned vs premature trials (Table 3)] or 4 ± 3% of the possible 0.90 ± 0.02 bits of predictive information (Fig. 4). Using SVM for classification, the average single neuron encoded 0.025 ± 0.03 bits of predictive information, and, using PDA for classification, the average single neuron encoded 0.046 ± 0.04 bits of predictive information.
Figure 4.
The individual predictive information of neurons and their contributions to ensemble predictive information. The individual predictive information of each neuron (positive values, INeuron) was plotted above the predictive information lost when that neuron is removed from the ensemble (negative values, IContrib or ensemble contribution) for all datasets (A1, A2, B1, B2, C1, and C2). Rasters of neurons from dataset A1 are shown in Figure 3. The 95% confidence level for chance is drawn as a light gray line at 0.016 bits.
To determine chance levels of significant predictive information, we calculated predictive information from neural datasets with randomly permuted behavioral labels (i.e., random data). Across datasets, the 95% confidence interval of predictive information of random data fell at 0.016 ± 0.003 bits (average predictive information was 0.004 ± 0.005 bits). We interpreted values of predictive information above this value as significant. We found that 58% (45 of 78) of neurons with task-related features had predictive information above the 95% confidence intervals of predictive information from random data (INeuron > 0.016) and that the mean predictive information of individual neurons in our data were twice this value.
Ensembles of neurons could be used to discriminate between conditioned and premature trials with an average accuracy of 82.5 ± 4.9%. Accuracy for datasets with randomly permuted behavioral labels was 61.7 ± 8.1%. Therefore, the improvement over chance levels of classification was >20%. Ensembles encoded an average of 0.16 ± 0.06 bits (ÎEnsemble) of predictive information with LVQ and as much as 0.31 ± 0.12 bits of predictive information with SVM (Table 3); that is, ensembles encoded from 18 ± 7% [with LVQ (Table 3)] to 34 ± 13% [with SVM (Table 3)] of the possible predictive information. Ensembles encoded significantly more predictive information than single neurons (t = 7.34; p ≪ 0.001). Larger ensembles tended to encode more predictive information (R = 0.85; p < 0.03). Ensembles with pseudorandomly assigned behavioral labels provided 0.004 ± 0.005 bits of predictive information, significantly less than ensembles with appropriately assigned behavioral labels (t = 6.64; p ≪ 0.001).
Importantly, we performed all of the analysis above with multiple statistical classifiers in both WEKA and R. Results of analysis on ensemble data with three different statistical classifiers are presented in Table 3. Although some classifiers (i.e., SVM) performed better than others (i.e., LVQ), all statistical classifiers indicated that the ensembles had significantly higher predictive information than random data (t = 6.12; p ≪ 0.001) and had significantly higher predictive information than single neurons (t = 8.98; p ≪ 0.001). Across datasets, information values calculated by different statistical classifiers were correlated for ensembles (LVQ vs SVM, R = 0.92; SVM vs PDA, R = 0.53; PDA vs LVQ, R = 0.59) and for individual neurons (LVQ vs SVM, R = 0.80; SVM vs PDA, R = 0.60; PDA vs LVQ, R = 0.69). Additionally, redundancy values of different classifiers were highly correlated (LVQ vs SVM, R = 0.94; SVM vs PDA, R = 0.76; PDA vs LVQ, R = 0.88). These results indicate that the different statistical classifiers used in this study generated primarily convergent estimates of predictive information and redundancy.
Single neurons contribute redundant information
To estimate the contributions of individual neurons to an ensemble (IContrib), we calculated how much predictive information was lost when a neuron was removed from all ensembles in which a neuron might participate (see Materials and Methods) (Table 1). Removing a single neuron from an ensemble resulted in an average decrease in predictive information of 0.016 ± 0.05 bits (ÎContrib). Importantly, IContrib for 87% of neurons (68 of 78) statistically included zero (Fig. 4, note large error bars for IContrib), indicating that removing a neuron from an ensemble often did not greatly change the predictive information of the ensemble.
We found that information lost with neuron removal was significantly less than the information provided by the neurons themselves (i.e., IContrib < INeuron; paired t = 4.32; p ≪ 0.001), and there was no obvious relationship between these informational values (R = 0.11; p < 0.17) (Fig. 4). These results suggest that the information contributed by single neurons to the ensembles was not independent.
To determine whether interactions between the neurons were synergistic or redundant, we calculated PNeuron according to Equation 3 for each neuron in each ensemble. The average value of PNeuron for all neurons was -0.016 ± 0.03 bits, which suggests average redundancy. Most neurons contributed redundant information to the ensembles (43 of 45 neurons, or 96%, of neurons with significant information were redundant). Only neurons 1 and 6 in dataset B1 (2 of 45 neurons, or 4% of neurons with significant information) had synergistic ensemble interactions.
Because individual neurons encoded a small fraction of the possible information (4 ± 3%), redundant information contributed by individual neurons was unlikely to be a result of information saturation by high information rates of individual neurons; that is, if a few neurons were highly informative, any additional neurons would by definition provide redundant information. In our data, we found that the of removal of highly informative neurons did not greatly impact ensemble performance (Fig. 4, neuron 8 in dataset A1, neurons 3 and 8 in dataset C1), once again suggesting redundant interactions between neurons at the ensemble level.
Neuronal ensembles are highly redundant
To determine whether ensembles as a whole were redundant or synergistic, we calculated the value PEnsemble according to Equation 4, which represents the difference between the predictive information of an ensemble and the sum of the predictive information of the component neurons. All six ensembles were redundant (P̂Ensemble = -0.24 ± 0.14 bits) (Table 3).
A redundant ensemble should maintain performance as it is degraded. To test this property, we progressively removed neuronal data from each ensemble and estimated accuracy and predictive information. Data from all animals suggested that, regardless of classifier, accuracy of prediction decreased slowly as neurons were removed when ensembles were larger than a few neurons and decreased rapidly as neurons were removed when ensembles were small (Fig. 5A,B). Predictive information increased much less slowly than would be predicted with an independent coding scheme (in which each neuron encoded an average of 0.034 bits) (Fig. 5C,E). As neurons were excluded, the remaining ensembles maintained accuracy (across datasets, 80 ± 12% of ensemble information was maintained with only 50% of neurons) (Fig. 5A,B). However, predictive information declined rapidly after <30% of the neurons remained in the ensemble (Fig. 5C,E). For small ensembles of neurons, the average value of PEnsemble was near zero (e.g., 0.006 ± 0.012 for pairs) (Fig. 5D,F). For larger ensembles, PEnsemble became more negative (e.g., -0.13 ± 0.07 for 8 neuron ensembles) (Fig. 5D,F), signifying a net increase in redundancy across datasets (t = 3.67; p < 0.006) with an increase in ensemble size across datasets.
Figure 5.

Analysis of ensemble predictive information as neurons are added/removed reveals high levels of redundancy. Accuracy of ensemble prediction as a function of the number of neurons for dataset A1 (A) and for dataset C1 (B), as quantified by three different statistical classifiers (SVM, PDA, and LVQ) for all possible combinations of neurons. SD plotted as lighter lines. C, Predictive information as a function of the number of neurons in the ensemble for dataset C1 with three different statistical classifiers (SVM, PDA, and LVQ; colors as in A). The dotted gray line represents the increase in predictive information that would be expected from an independent coding scheme if each neuron encoded an average of 0.034 bits, the average predictive information for individual neurons in our datasets. Compare with Figure 2. D, PEnsemble as a function of the number of neurons in the ensemble for dataset C1 with three different statistical classifiers (SVM, PDA, and LVQ; colors as in A). E, Predictive information as a function of the number of neurons in the ensemble for all datasets; datasets are labeled on the figure. F, PEnsemble as a function of the number of neurons in the ensemble for all datasets; datasets are labeled on the figure.
Across our datasets, we found that PEnsemble was indeed negatively correlated with predictive information (mean R = -0.78; p ≪ 0.001 with exponentially transformed predictive information) (Fig. 5E). We also found that PEnsemble was negatively correlated with the number of neurons in the ensemble (across datasets, mean R = -0.85 ± 0.02; p ≪ 0.001) (Fig. 5F). A multiple linear regression revealed that 73% of the variance of PEnsemble was explained by predictive information and ensemble size (R2 of ensemble size = 0.45, R2 of exponentially transformed predictive information = 0.28; R2 = 0.73; Fmodel = 209; p ≪ 0.001). These data indicate that redundancy increases (i.e., PEnsemble becomes more negative) as a function of both predictive information and ensemble size.
Finally, we investigated whether the redundancy we observed was a result of a “ceiling effect,” i.e., a saturation of predictive information attributable to similarities in neuronal response properties (Rolls et al., 1997). This effect is unlikely for three reasons. First, individual neurons provided little predictive information (i.e., 4 ± 3%). Second, removing any particular neuron had a small effect on ensemble predictive information (Figs. 4, 5). Third, trial shuffling (i.e., randomizing the trial orders within each trial type for individual neurons) significantly increased ensemble predictive information across datasets (increase in accuracy, 3.5 ± 1.9%; paired t = 4.6; p < 0.006).
Redundancy and synergy
Our experimental data revealed redundant interactions within large ensembles of weakly coding neurons. However, in our data from the rat motor cortex, we found that small ensembles (less than six neurons) exhibited rapid increases in predictive information as neurons were added to the ensembles (Fig. 5C,E). This data suggested a nonredundant coding scheme (Fig. 2), such as synergy. To assess the extent of synergy in our data, we plotted the percentage of significant synergistic ensembles as a function of the ensemble size and found that the fraction of significant synergistic interactions was inversely related to the number of neurons in the ensemble (Fig. 6A). Significance levels were determined by calculating PEnsemble for 1000 two-neuron ensembles with randomly permuted behavioral labels. The 95% confidence interval for PEnsemble was between 0.016 and -0.032. We interpreted PEnsemble values outside this range to be significant. We found declining synergy with increasing ensemble size in every dataset and with every statistical classifier we used, although some statistical classifiers found higher levels of synergy (i.e., SVM) than others (i.e., LVQ).
Figure 6.
Synergy among ensemble data. A, Fraction of synergistic ensembles as a function of number of neurons in ensembles. B, PEnsemble for pairs of neurons plotted against distance between microelectrodes from which neurons were recorded.
Finally, we examined whether there were any spatial determinants for synergistic or redundant neuronal interactions. We determined the distance between each neuron across the microelectrode arrays used for the neuronal recordings and plotted those distances against PEnsemble for pairs of neurons. We observed no obvious relationship between PEnsemble and the distance between electrodes from which neurons were recorded (R = 0.08; p < 0.12) (Fig. 6B).
Discussion
We studied how interactions among neuronal ensembles in the rat motor cortex influence predictions of behavioral performance in a simple reaction time task. We used statistical pattern recognition methods to quantify predictive information conveyed by single neurons and groups of neurons of varying complexity. Using a cluster of computer workstations, we were able to account for interactions between all possible combinations of neurons (involving many thousands of combinations; Table 2), something that, to our knowledge, has not been accomplished previously. We report three main findings. First, the predictive information conveyed by single cortical neurons is not related to the contribution that neuron makes to information conveyed by neuronal ensemble. Second, ensembles of cortical neurons tend to be highly redundant. Finally, although large ensembles are highly redundant, significant synergistic interactions exist in smaller ensembles.
Motor cortex and simple reaction time tasks
In this study, spike trains were collected from chronically implanted arrays of electrodes in rat motor cortex during performance of a simple reaction time task with a variable foreperiod. In such a task, there is an increasing likelihood that the trigger stimulus will occur as time passes during the foreperiod (Naatanen, 1970). Prompt reaction times are thought to depend on processes of time estimation and stimulus detection that converge on a “response generator,” e.g., motor cortex (Ollman and Billington, 1972; Kornblum, 1973). The motor cortex should thus be responsive to temporal and sensory factors during the foreperiod. However, processes related to stimulus detection should only occur on trials with conditioned (correct) responses. There are several large-scale brain potentials that become active during the foreperiod of simple reaction time tasks (for review, see Brunia, 1999). One of them, the contingent negative variation (CNV), is evoked by the trigger stimulus. If rats are capable of generating this potential (Pirch, 1980), then a CNV-like process would differentiate neuronal processing on the two types of trials examined in this study. The lack of activation of motor cortex by a CNV-like process may have resulted in reduced levels of excitability in motor cortex, as is apparent in the perievent histograms in Figure 3. This effect might also result in quantitative differences in neuronal ensemble level activity on conditioned and premature responses, and these effects might be manifest in redundant interactions among neurons.
The shortest observed RTs may represent anticipatory responses initiated before the trigger stimulus. If neuronal stimulus relationships were different on such anticipatory trials, then removing such trials should improve classification of conditioned versus premature trials. However, when we removed trials with very short reaction times, we observed equivalent or slightly decreased classification of the remaining conditioned responses versus premature responses (data not shown). These results suggest that motor cortex was influenced more by the absence of the trigger stimulus than by anticipatory responses on the premature response trials.
Functional interpretation of synergy and redundancy
Our results suggest that single neurons in the rat motor cortex do not function as independent encoders of behavioral performance in a simple reaction time task. Instead, the neurons are highly redundant. As was pointed out by Meijers and Eijkman (1974), redundancy may exist in the motor cortex to overcome the large amount of variability in the onset latencies of corticospinal neurons (Evarts, 1966) relative to onset latencies in muscle (Botwinick and Thompson, 1966). Redundancy might also be attributable to activations of functionally related groups of muscles (Donoghue et al., 1992; Plautz et al., 2000; Holdefer and Miller, 2002; Jackson et al., 2003; Kargo and Nitz, 2003; Morrow and Miller, 2003) that are used to control the response lever and maintain the rat's posture during the trial. Redundancy in motor cortex would allow for increased efficiency in controlling functionally related sets of muscles (Barlow, 2001), or, more practically, to changes that are known to occur with motor skill learning in which accessory muscles tend to change with learning more than primary mover muscles (Heise, 1995). Indeed, increases in correlated firing in motor cortex, an indirect measure of redundant processing, have been reported recently to accompany the learning of sensorimotor tasks (Gemba and Sasaki, 1984, 1988; Laubach et al., 2000; Cohen and Nicolelis, 2004; Kargo and Nitz, 2004; Paz and Vaadia, 2004). More generally, the advantages of redundancy in cortical neurons may offset the advantages of channel capacity reduction (Shannon and Weaver, 1949) and economy of energy (Allman, 1990) because of greatly enhanced signal-to-noise during goal-oriented behavior (Barlow, 2001).
Distributed coding in neuronal ensembles
Distributed coding has been examined in a relatively small number of studies. Humphrey et al. (1970) found that small sets of neurons could accurately predict several motor variables. More recently, Wessberg et al. (2000) performed a similar analysis in ensembles with hundreds of neurons in multiple parts of primate cortex to predict hand position during two- and three-dimensional arm movement tasks and found that ensembles performed at high levels (prediction above 90% of the variance of hand trajectory) despite removal of up to one-third of the neurons in the ensemble. Similarly, Averbeck et al. (2003) found evidence for high levels of redundancy while recording from ensembles up to 22 neurons in primate prefrontal cortex during a shape-copying task. In the rat primary somatosensory cortex, Ghazanfar et al. (2000) found graceful degradation of cortical ensembles when discriminating among sites of stimulation on the whisker pad. These results are consistent with our results and further suggest that redundancy is a major factor in information processing in multiple cortical systems.
Studies that have investigated how small cortical ensembles work together to encode information have found less redundancy than reported here. Gawne and Richmond (1993) studied responses of pairs of neurons in the inferior temporal cortex to complex visual stimuli and found that 20% of all pairwise neuronal interactions were redundant. Rolls et al. (2004) report lower levels of redundancy (4-10%) in inferior temporal cortex when discriminating between 20 faces. In the primate motor cortex, Maynard et al. (1999) found only ∼20% of all pairwise neuronal interactions were redundant in the motor cortex during directional limb movements, whereas the remainder were synergistic. Reich et al. (2001) found entirely independent interactions between small ensembles (less than six neurons) of simultaneously recorded neurons in primary visual cortex. Samonds et al. (2004) found high levels of synergy for small ensembles (up to six neurons) when recording from cat V1 while a drifting sinusoidal grating was presented.
In contrast, in the present study, we found that most (99.3 ± 0.01%) of large ensembles (more than eight neurons) are entirely redundant. However, just 36 ± 12% of neuron pairs were redundant, with 23 ± 9% exhibiting synergistic interactions (the remainder of pairs did not exhibit significant synergy or redundancy). One explanation for the high redundancy we find is that larger ensembles may mask synergy present in smaller ensembles. Thus, the low levels of redundancy observed in small ensembles in previous studies (Gawne and Richmond, 1993; Maynard et al., 1999; Reich et al., 2001; Rolls et al., 2004; Samonds et al., 2004; Purushothaman and Bradley, 2005) are entirely compatible with our results, because we find little redundancy in small ensembles. An alternative explanation, however, is that the sensory mapping studies above used many classes of stimuli, whereas our study had just two outcomes of task performance. This issue will require additional experimental studies to be resolved.
Limitations of the statistical pattern recognition approach
Our estimates of mutual information depend to some extent on the specific algorithm used for classification. As a result, our estimates may underestimate limits on mutual information for encoding behavioral performance (see Materials and Methods). Furthermore, we calculated synergy and redundancy based on estimates generated by statistical classifiers. It is possible that the classifiers have sources of bias that direct estimates of mutual information would avoid (see Materials and Methods) (Schneidman et al., 2003). We used different classifiers on benchmark data and random neuronal data to examine this issue. However, we cannot rule out bias in our measures of PEnsemble because we know of no practical method to directly measure mutual information for large ensembles of neurons. Despite these concerns, we believe that our approach is a feasible way to analyze interactions among ensembles of neurons. The high levels of redundancy we found in all datasets using multiple statistical classifiers lends confidence to our interpretation. We hope that future studies will consider these issues. However, we believe the ultimate test of the value of synergistic and redundant neuronal interactions for encoding behaviorally relevant information will be through experiments, perhaps involving real-time analyses of spike trains recorded in awake, behaving animals (Humphrey et al., 1970; Wessberg et al., 2000; Serruya et al., 2002; Taylor et al., 2002; Ben-Shaul et al., 2003; Carmena et al., 2003; Lebedev et al., 2004).
Footnotes
This work was supported by the National Institutes of Health, Defense Advanced Research Projects Agency, The John B. Pierce Laboratory, and a training grant from the National Institutes of Health through the Yale Medical Scientist Training Program (N.S.N., E.Y.K.). The physiological data presented here were collected in the laboratory of Miguel A. L. Nicolelis at Duke University (Durham, NC). We thank him for support and feedback on this manuscript. We also thank Brian Dobbins (Yale Mechanical Engineering) for help with implementing efficient methods for analyzing subensemble interactions using a cluster of Linux workstations in our laboratory. Finally, we thank two anonymous reviewers for insightful suggestions. The classifiers and software used in this study are freely available (http://spikelab.jbpierce.org), and we invite groups to use this approach to test their respective neural systems. This manuscript is dedicated to the memories of Tilghman E. Laubach and William M. Laubach.
Correspondence should be addressed to Dr. Mark Laubach, The John B. Pierce Laboratory, 290 Congress Avenue, New Haven, CT 06519. E-mail: mark.laubach@yale.edu.
Copyright © 2005 Society for Neuroscience 0270-6474/05/254207-10$15.00/0
References
- Abbott LF, Dayan P (1999) The effect of correlated variability on the accuracy of a population code. Neural Comput 11: 91-101. [DOI] [PubMed] [Google Scholar]
- Allman J (1990) The origin of the neocortex. Neurosciences 2: 257-262. [Google Scholar]
- Averbeck BB, Lee D (2004) Coding and transmission of information by neural ensembles. Trends Neurosci 27: 225-230. [DOI] [PubMed] [Google Scholar]
- Averbeck BB, Crowe DA, Chafee MV, Georgopoulos AP (2003) Neural activity in prefrontal cortex during copying geometrical shapes. II. Decoding shape segments from neural ensembles. Exp Brain Res 150: 142-153. [DOI] [PubMed] [Google Scholar]
- Bair W, Zohary E, Newsome WT (2001) Correlated firing in macaque visual area MT: time scales and relationship to behavior. J Neurosci 21: 1676-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow H (2001) Redundancy reduction revisited. Network 12: 241-253. [PubMed] [Google Scholar]
- Ben-Shaul Y, Drori R, Asher I, Stark E, Nadasdy Z, Abeles M (2003) Neuronal activity in motor cortical areas reflects the sequential context of movement. J Neurophysiol 89: 1136-1142. [DOI] [PubMed] [Google Scholar]
- Botwinick J, Thompson LW (1966) Premotor and motor components of reaction time. J Exp Psychol 71: 9-15. [DOI] [PubMed] [Google Scholar]
- Britten KH, Newsome WT, Shadlen MN, Celebrini S, Movshon JA (1996) A relationship between behavioral choice and the visual responses of neurons in macaque MT. Vis Neurosci 13: 87-100. [DOI] [PubMed] [Google Scholar]
- Brunia CH (1999) Neural aspects of anticipatory behavior. Acta Psychol 101: 213-242. [DOI] [PubMed] [Google Scholar]
- Buckheit J, Donoho DL (1995) Improved linear discrimination using time-frequency dictionaries. Proc SPIE 2569: 540-551. [Google Scholar]
- Carmena JM, Lebedev MA, Crist RE, O'Doherty JE, Santucci DM, Dimitrov D, Patil PG, Henriquez CS, Nicolelis MA (2003) Learning to control a brain-machine interface for reaching and grasping by primates. PLoS Biol 1: E42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Nicolelis MA (2004) Reduction of single-neuron firing uncertainty by cortical ensembles during motor skill learning. J Neurosci 24: 3574-3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover TM, Thomas JA (1991) Elements of information theory. Hoboken, NJ: Wiley-Interscience.
- Dan Y, Alonso JM, Usrey WM, Reid RC (1998) Coding of visual information by precisely correlated spikes in the lateral geniculate nucleus. Nat Neurosci 1: 501-507. [DOI] [PubMed] [Google Scholar]
- Donoghue JP, Leibovic S, Sanes JN (1992) Organization of the forelimb area in squirrel monkey motor cortex: representation of digit, wrist, and elbow muscles. Exp Brain Res 89: 1-19. [DOI] [PubMed] [Google Scholar]
- Duda RO, Hart PE, Stork DG (2001) Pattern classification. New York: Wiley.
- Evarts EV (1966) Pyramidal tract activity associated with a conditioned hand movement in the monkey. J Neurophysiol 29: 1011-1027. [DOI] [PubMed] [Google Scholar]
- Foffani G, Tutunculer B, Moxon KA (2004) Role of spike timing in the forelimb somatosensory cortex of the rat. J Neurosci 24: 9266-9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa S, Middlebrooks JC (2001) Sensitivity of auditory cortical neurons to locations of signals and competing noise sources. J Neurophysiol 86: 226-240. [DOI] [PubMed] [Google Scholar]
- Gat I, Tishby N (1998) Synergy and redundancy among brain cells of behaving monkeys. In: Advances in neural information processing systems: Proceedings of the 1998 Conference (Kearns MS, Solla SA, Cohn DA, eds), pp 111-117. Cambridge, MA: MIT.
- Gawne TJ, Richmond BJ (1993) How independent are the messages carried by adjacent inferior temporal cortical neurons? J Neurosci 13: 2758-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemba H, Sasaki K (1984) Studies on cortical field potentials recorded during learning processes of visually initiated hand movements in monkeys. Exp Brain Res 55: 26-32. [DOI] [PubMed] [Google Scholar]
- Gemba H, Sasaki K (1988) Changes in cortical field potentials associated with learning processes of audio-initiated hand movements in monkeys. Exp Brain Res 70: 43-49. [DOI] [PubMed] [Google Scholar]
- Georgopoulos AP, Kalaska JF, Caminiti R, Massey JT (1982) On the relations between the direction of two-dimensional arm movements and cell discharge in primate motor cortex. J Neurosci 2: 1527-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos AP, Kettner RE, Schwartz AB (1988) Primate motor cortex and free arm movements to visual targets in three-dimensional space. II. Coding of the direction of movement by a neuronal population. J Neurosci 8: 2928-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazanfar AA, Stambaugh CR, Nicolelis MA (2000) Encoding of tactile stimulus location by somatosensory thalamocortical ensembles. J Neurosci 20: 3761-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gochin PM, Colombo M, Dorfman GA, Gerstein GL, Gross CG (1994) Neural ensemble coding in inferior temporal cortex. J Neurophysiol 71: 2325-2337. [DOI] [PubMed] [Google Scholar]
- Good PI (2000) Permutation tests: a practical guide to resampling methods for testing hypotheses. New York: Springer.
- Hastie T, Tibshirani R, Buja A (1995) Flexible discriminant analysis and mixture models. In: Neural networks and statistics (Kay J, Titterington D, eds). Oxford, UK: Oxford UP.
- Hastie T, Tibshirani R, Friedman J (2001) The elements of statistical learning. New York: Springer.
- Hatsopoulos NG, Ojakangas CL, Paninski L, Donoghue JP (1998) Information about movement direction obtained from synchronous activity of motor cortical neurons. Proc Natl Acad Sci USA 95: 15706-15711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise GD (1995) Changes in agonist muscles during practice of a multijoint throwing skill. J Electromyogr Kinisiol 5: 81-94. [DOI] [PubMed] [Google Scholar]
- Holdefer RN, Miller LE (2002) Primary motor cortical neurons encode functional muscle synergies. Exp Brain Res 146: 233-243. [DOI] [PubMed] [Google Scholar]
- Humphrey DR, Schmidt EM, Thompson WD (1970) Predicting measures of motor performance from multiple cortical spike trains. Science 170: 758-762. [DOI] [PubMed] [Google Scholar]
- Hsu CW, Chang CC, Lin CJ (2003) A practical guide to support vector classification. Available at http://www.csie.ntu.edu.tw/~cjlin/papers/guide/guide.pdf.
- Jackson A, Gee VJ, Baker SN, Lemon RN (2003) Synchrony between neurons with similar muscle fields in monkey motor cortex. Neuron 38: 115-125. [DOI] [PubMed] [Google Scholar]
- Johnson DH (2004) Neural population structures and consequences for neural coding. J Comput Neurosci 16: 69-80. [DOI] [PubMed] [Google Scholar]
- Kargo WJ, Nitz DA (2003) Early skill learning is expressed through selection and tuning of cortically represented muscle synergies. J Neurosci 23: 11255-11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargo WJ, Nitz DA (2004) Improvements in the signal-to-noise ratio of motor cortex cells distinguish early versus late phases of motor skill learning. J Neurosci 24: 5560-5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohonen T (2000) Self-organizing maps. New York: Springer.
- Kornblum S (1973) Simple reaction times as a race between signal detection and time estimation: a paradigm and a model. Percept Psychophy 13: 109-112. [Google Scholar]
- Laubach M (2004) Wavelet-based processing of neuronal spike trains prior to discriminant analysis. J Neurosci Methods 134: 159-168. [DOI] [PubMed] [Google Scholar]
- Laubach M, Wessberg J, Nicolelis MA (2000) Cortical ensemble activity increasingly predicts behavior outcomes during learning of a motor task. Nature 405: 567-571. [DOI] [PubMed] [Google Scholar]
- Lebedev MA, Messinger A, Kralik JD, Wise SP (2004) Representation of attended versus remembered locations in prefrontal cortex. PLoS Biol 2: e365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard EM, Hatsopoulos NG, Ojakangas CL, Acuna BD, Sanes JN, Normann RA, Donoghue JP (1999) Neuronal interactions improve cortical population coding of movement direction. J Neurosci 19: 8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijers LMM, Eijkman EGJ (1974) The motor system in simple reaction time experiments. Acta Psychol (Amst) 38: 367-377. [DOI] [PubMed] [Google Scholar]
- Middlebrooks JC, Bierer JA (2002) Auditory cortical images of cochlearimplant stimuli: coding of stimulus channel and current level. J Neurophysiol 87: 493-507. [DOI] [PubMed] [Google Scholar]
- Morrow MM, Miller LE (2003) Prediction of muscle activity by populations of sequentially recorded primary motor cortex neurons. J Neurophysiol 89: 2279-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naatanen R (1970) The diminishing time uncertainty with the lapse of time after the warning signal in reaction-time experiments with varying foreperiods. Acta Psychologia 34: 399-419. [DOI] [PubMed] [Google Scholar]
- Nicolelis MA, ed (1998) Methods in neuronal ensemble recording. Boca Raton, FL: CRC.
- Nicolelis MA, Ghazanfar AA, Stambaugh CR, Oliveira LM, Laubach M, Chapin JK, Nelson RJ, Kaas JH (1998a) Simultaneous encoding of tactile information by three primate cortical areas. Nat Neurosci 1: 621-630. [DOI] [PubMed] [Google Scholar]
- Nicolelis MA, Stambaugh CR, Brisben A, Laubach M (1998b) Methods for simultaneous multisite neural ensemble recordings in behaving primates. In: Methods for neuronal ensemble recording (Nicolelis MA, ed). Boca Raton, FL: CRC.
- Nirenberg S, Latham PE (2003) Decoding neuronal spike trains: how important are correlations? Proc Natl Acad Sci USA 100: 7348-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollman R, Billington MJ (1972) The deadline model for simple reaction times. Cognit Psychol 3: 311-336. [Google Scholar]
- Paz R, Vaadia E (2004) Learning-induced improvement in encoding and decoding of specific movement directions by neurons in the primary motor cortex. PLoS Biol 2: 264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirch JH (1980) Event-related slow potentials in rat cortex during a reaction time task: cortical area differences. Brain Res Bull 5: 199-201. [DOI] [PubMed] [Google Scholar]
- Plautz EJ, Milliken GW, Nudo RJ (2000) Effects of repetitive motor training on movement representations in adult squirrel monkeys: role of use versus learning. Neurobiol Learn Mem 74: 27-55. [DOI] [PubMed] [Google Scholar]
- Priebe NJ, Lisberger SG (2004) Estimating target speed from the population response in visual area MT. J Neurosci 24: 1907-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushothaman G, Bradley DC (2005) Neural population code for fine perception in area MT. Nat Neurosci 8: 99-106. [DOI] [PubMed] [Google Scholar]
- Reich DS, Mechler F, Victor JD (2001) Independent and redundant information in nearby cortical neurons. Science 294: 2566-2568. [DOI] [PubMed] [Google Scholar]
- Richmond BJ, Optican LM (1987) Temporal encoding of two-dimensional patterns by single units in primate inferior temporal cortex. II. Quantification of response waveform. J Neurophysiol 57: 147-161. [DOI] [PubMed] [Google Scholar]
- Riehle A, Grun S, Diesmann M, Aertsen A (1997) Spike synchronization and rate modulation differentially involved in motor cortical function. Science 278: 1950-1953. [DOI] [PubMed] [Google Scholar]
- Ripley BD (1996) Pattern recognition and neural networks. New York: Cambridge.
- Rolls ET, Treves A, Tovee MJ (1997) The representational capacity of the distributed encoding of information provided by populations of neurons in primate temporal visual cortex. Exp Brain Res 114: 149-162. [DOI] [PubMed] [Google Scholar]
- Rolls ET, Aggelopoulos NC, Franco L, Treves A (2004) Information encoding in the inferior temporal visual cortex: contributions of the firing rates and the correlations between the firing of neurons. Biol Cybern 90: 19-32. [DOI] [PubMed] [Google Scholar]
- Romo R, Hernandez A, Zainos A (2004) Neuronal correlates of a perceptual decision in ventral premotor cortex. Neuron 41: 165-173. [DOI] [PubMed] [Google Scholar]
- Samonds JM, Allison JD, Brown HA, Bonds AB (2004) Cooperative synchronized assemblies enhance orientation discrimination. Proc Natl Acad Sci USA 101: 6722-6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman E, Bialek W, Berry II MJ (2003) Synergy, redundancy, and independence in population codes. J Neurosci 23: 11539-11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serruya MD, Hatsopoulos NG, Paninski L, Fellows MR, Donoghue JP (2002) Instant neural control of a movement signal. Nature 416: 141-142. [DOI] [PubMed] [Google Scholar]
- Shannon CE, Weaver W (1949) The mathematical theory of communication. Urbana, Il: University of Illinois.
- Taylor DM, Tillery SI, Schwartz AB (2002) Direct cortical control of 3D neuroprosthetic devices. Science 296: 1829-1832. [DOI] [PubMed] [Google Scholar]
- Vaadia E, Haalman I, Abeles M, Bergman H, Prut Y, Slovin H, Aertsen A (1995) Dynamics of neuronal interactions in monkey cortex in relation to behavioral events. Nature 373: 515-518. [DOI] [PubMed] [Google Scholar]
- Venables WN, Ripley BD (2002) Modern applied statistics with S. New York: Springer.
- Venables WN, Smith DM, R Development Core Team (2004) An introduction to R. Bristol, UK: Network Theory.
- Wessberg J, Stambaugh CR, Kralik JD, Beck PD, Laubach M, Chapin JK, Kim J, Biggs SJ, Srinivasan MA, Nicolelis MA (2000) Real-time prediction of hand trajectory by ensembles of cortical neurons in primates. Nature 408: 361-365. [DOI] [PubMed] [Google Scholar]
- Witten I, Frank E (2000) Data mining. San Diego: Academic.
- Zohary E, Shadlen MN, Newsome WT (1994) Correlated neuronal discharge rate and its implications for psychophysical performance. Nature 370: 140-143. [DOI] [PubMed] [Google Scholar]







