Abstract
A diverse set of organisms has adapted to live under extreme conditions. The molecular origin of the stability is unclear, however. It is not known whether the adaptation of functional RNAs, which have intricate tertiary structures, arises from strengthening of tertiary or secondary structure. Herein we evaluate effects of sequence changes on the thermostability of tRNAphe using experimental and computational approaches. To separate out effects of secondary and tertiary structure, we modify base pairing strength in the acceptor stem, which does not participate in tertiary structure. In dilute solution conditions, strengthening secondary structure leads to non-two-state thermal denaturation curves and has small effects on thermostability, or the temperature at which tertiary structure and function is lost. In contrast, under cellular conditions with crowding and Mg2+-chelated amino acids, where two-state cooperative unfolding is maintained, strengthening secondary structure enhances thermostability. Investigation of stabilities of each tRNA stem across 44 organisms with a range of optimal growing temperatures revealed that organisms that grow in warmer environments have more stable stems. We also used Shannon entropies to identify positions of higher and lower information content, or sequence conservation, in tRNAphe and found that secondary structures have modest information content allowing them to drive thermal adaptation, while tertiary structures have maximal information content preventing them from participating in thermal adaptation. Base paired regions with no tertiary structure and modest information content thus offer a facile evolutionary route to enhancing the thermostability of functional RNA by simple molecular rules of base pairing.
Life exists at temperatures ranging from freezing to boiling water, raising questions as to the molecular mechanisms for thermal adaptation. Extensive studies have been conducted on protein folding under extreme conditions,1–3 but relatively little is known about molecular mechanisms for adaption of RNA sequence to demanding environments. Elucidation of such mechanisms can help establish how extant life has flourished on Earth, as well as provide plausible pathways for the evolution of RNA sequences on early Earth in an RNA World scenario.
Of special interest are so-called ‘functional RNAs’, which require precise tertiary structures for function. These naturally occurring RNAs include ribozymes, riboswitches, rRNA, and tRNA and are key components of an RNA World.4,5 Crystal structures reveal that tertiary contacts in functional RNAs are diverse and complex and include base triples, ribose zippers, and U-turns.6 Underlying these tertiary structures are relatively simple base paired regions that provide the structural framework of the RNA. This leads to hierarchical folding of RNA in which secondary structure formation precedes tertiary structure formation.7, 8
Given the need for functional RNAs to adapt to diverse conditions, the question arises as to whether thermostability—the temperature at which tertiary structure is lost— comes from strengthening tertiary structures or secondary structures. When tertiary structure unfolds, RNA function is lost. Therefore, it might seem that thermostability would increase by strengthening tertiary interactions rather than secondary structure. However, another mechanism is possible. Sosnick and Pan introduced the concept of functional stability, which is the difference in free energy between the fully functional state and the penultimately stable state.9 If that penultimately stable state is not secondary structure but rather unfolded RNA, then folding would be cooperative and secondary structure strength could increase thermostability.
To investigate this notion, we study the folding of a series of tRNAphe constructs with variable strength acceptor stems under diverse solution conditions including dilute buffer, crowders, and Mg2+-chelated amino acids, which we and others have found induce cooperative RNA folding.10–13 The strength of the acceptor stem was tuned by switching between G•U / AU pairs and GC pairs. We observe that strengthened tRNA mutants fold cooperatively under the most biological-like conditions of crowding in Mg2+-chelated amino acids and that such strengthening of base pairing increases thermostability. Moreover, computational analysis of a set of tRNAphe sequences from organisms that live in a wide range of temperatures reveals that base paired regions strengthen in thermophiles while tertiary structure-participating nucleotides are invariant. This suggests that in nature, tRNA adapts to harsh conditions not by strengthening tertiary interactions but by strengthening base pairing. Given the simple nature of base pairing and the large energetic effects of single nucleotide changes, this offers a simple route to adaptation of RNA and emphasizes the importance of cooperative RNA folding conditions to RNA evolution.
Materials and Methods
Chemicals.
PEG8000, HEPES, MgCl2, and sodium cacodylate were purchased from Sigma. KCl was purchased from J. T. Baker. Calf intestinal phosphatase and polynucleotide kinase were purchased from NEB.
RNA Constructs and Preparation.
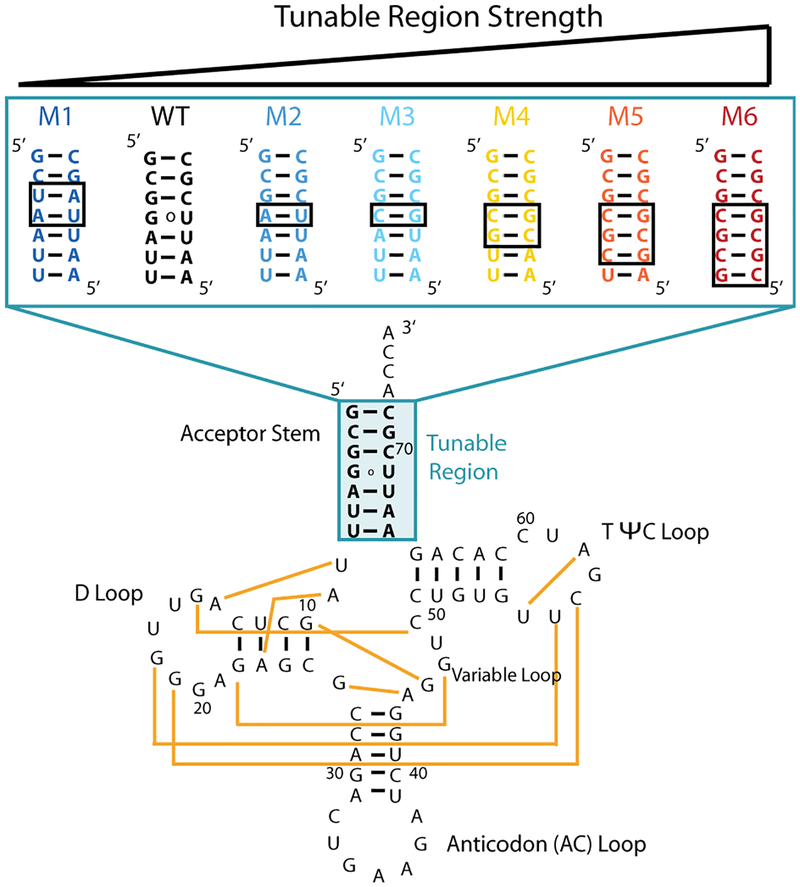
Wild-type (WT) tRNAphe and mutants (M) were transcribed with a hemi-duplex DNA template purchased from IDT that was used without further purification, as previously described11. The RNA was buffer exchanged into 10 mM sodium cacdoylate using an Amicon ultracentrifugal filter (3 kDa molecular weight cutoff). The RNA sequences are below, and nucleotides mutated from WT are in bold.
FL tRNAphe:
5ʹGCGGAUUUAGCUCAGUUGGGAGAGCGCCAGACUGAAGAUCUGGAGGUCCUGUGUUCGAUCCACAGAAUUCGCACCA
Mutant 1 (M1):
5’GCUAAUUUAGCUCAGUUGGGAGAGCGCCAGACUGAAGAUCUGGAGGUCCUGUGUUCGAUCCACAGAAUUAGCACCA 3’
Mutant 2 (M2):
5’GCGAAUUUAGCUCAGUUGGGAGAGCGCCAGACUGAAGAUCUGGAGGUCCUGUGUUCGAUCCACAGAAUUCGCACCA 3’
Mutant 3 (M3):
5’GCGCAUUUAGCUCAGUUGGGAGAGCGCCAGACUGAAGAUCUGGAGGUCCUGUGUUCGAUCCACAGAAUGCGCACCA 3’
Mutant 4 (M4):
5’GCGCGUUUAGCUCAGUUGGGAGAGCGCCAGACUGAAGAUCUGGAGGUCCUGUGUUCGAUCCACAGAACGCGCACCA 3’
Mutant 5 (M5):
5’GCGCGCUUAGCUCAGUUGGGAGAGCGCCAGACUGAAGAUCUGGAGGUCCUGUGUUCGAUCCACAGAGCGCGCACCA 3’
Mutant 6 (M6):
5’GCGCGCGUAGCUCAGUUGGGAGAGCGCCAGACUGAAGAUCUGGAGGUCCUGUGUUCGAUCCACAGCGCGCGCACCA 3’
Thermal Denaturation.
RNA was renatured by denaturing at 95°C for 3 min and annealing at room temperature for 10 min in the presence of KCl and sodium cacodylate (pH 7.0). After cooling, MgCl2, polyethylene glycol (PEG) 8000, and amino acids were added to the RNA solution. The sample was then heated at 55 °C for 3 min and cooled at room temperature for 10 min. Samples were spun down at 14,000 rpm for 5 min at 4 °C to remove air bubbles and particulates. Samples were final concentrations of 0.5 μM RNA in 10 mM sodium cacodylate (pH 7.0), 140 mM KCl, 0.5–2.0 mM MgCl2, and 0–20% (w/v) PEG8000, and 0–106. mM amino acids, as indicated13. Further explanation of Mg2+-chelated samples is in Table S4. Thermal denaturation experiments were performed on an HP 8452 diode-array refurbished by OLIS, Inc with a data point collected every 0.5 °C and a data point collected ~0.5 °C/min with absorbance detection from 230–330 nm. This method is referred to as “optical melting”.
Thermal Denaturation Data Analysis
Thermal denaturation data on WT and mutant RNAs was truncated, as in Fig. 2 and Fig. S1, to remove excess baselines and fit globally from 250–290 nm using a two-state model with sloping baselines and a nonlinear Marquartdt algorithm in IgorPro14 using equation 1, where mu and mf are the slopes of the unfolded and folded baselines, bu and bf are the y-intercepts of the unfolded and folded baselines, ΔH is the enthalpy of folding, and TM is the melting temperature in Kelvin. R is the gas constant of 0.001987 kcal−1 K−1 mol−1. In global fitting the slopes and y-intercepts were allowed to vary at each wavelength, but the ΔH and TM were held constant. The traces at 260 nm were normalized for fraction unfolded using equation 2 which uses the sloping baselines from the global fits to normalize for fraction unfolded, where f is the fraction unfolded and AT is the raw absorbance at a temperature, T. There is noise in some of the polyethylene glycol data in mutant with high stem stability because the absorbance change in these transitions was low.
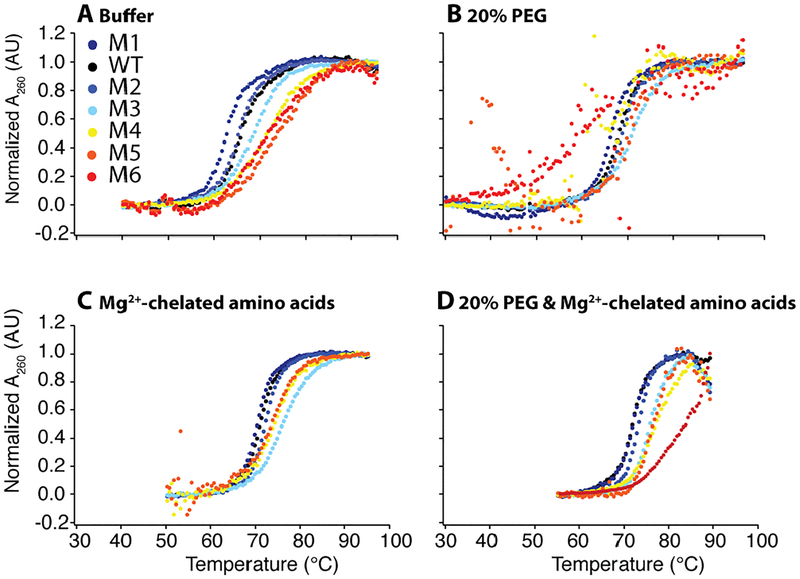
Figure 2.
WT and mutant thermal denaturation under in vivo-like solutions in the background of 2.0 mM free Mg2+. Each construct was globally fit every 2 nm between 250 and 290 nm. Thermal denaturation scans at 260 nm normalized using global fitting parameters in (A) buffer, (B) 20% PEG8000, (C) additional 14.0 mM Mg2+ weakly chelated to amino acids, and (D) 20% PEG8000 and additional 14.0 mM Mg2+ weakly chelated to amino acids. All four panels are in the background of 2.0 mM Mg2+ and 140 mM K+. Low temperature data was truncated in the fitting to avoid excess baselines, as is plotted above.
| (eq. 1) |
| (eq. 2) |
The theoretical non- and fully cooperative enthalpies of folding were calculated to compare with the experimental ΔH. The non-cooperative ΔH is the ΔHfolding derived using nearest neighbor parameters of the WT and mutant acceptor stems15. The fully cooperative ΔH is the experimentally derived ΔHfolding of WT plus or minus the nearest neighbor model ΔΔH of the mutant acceptor stems.
Small Angle X-ray Scattering Data Collection and Analysis
WT and MT tRNA constructs were transcribed, purified, and buffer exchanged as described previously11. RNA was renatured in 1× SAXS buffer by denaturing at 95 °C for 3 min and annealing at room temperature for 10 min. After cooling, MgCl2 was added to the sample which was heated at 55 °C for 3 min then cooled at room temperature for 10 min. Samples were centrifuged at 14k rpm for 10 min to minimize aggregation16.
SAXS data were collected on G1 station at MacCHESS-the solution scattering beamline at the Cornell High Energy Synchrotron Source (CHESS)17 using in-line size-exclusion chromatography (SEC) to separate monomers from aggregates, as previously described11. Samples were at a final concentration of 0.2 mg/mL in buffer with 2.0 mM Mg2+. SAXS data were analyzed as previously described using the linear Gunier region to obtain structural data11. The scattering curves were compared with simulated tRNA scattering curves using the FoXS webserver18, 19.
In-line Probing
The triphosphate on the 5’ end of T7 transcripts was removed by incubation with calf intestinal phosphatase (CIP) at 37 °C for 20 min, followed by a phenol/chloroform extraction and an ethanol precipitation. This RNA was labeled on the 5’ end with [γ−32P]ATP by incubation with polynucleotide kinase at 37 °C for 30 min. The labeled RNA was purified by 10% PAGE followed by a crush and soak and ethanol precipitation procedure. 500k cpm of labeled RNA with 140 mM KCl and 20 mM Tris (pH 8.3) was renatured by denaturing at 95 °C for 1 min and annealing at room temperature for 5 min. After annealing, PEG8000, MgCl2, and amino acids were added as indicated, and the solution was heated at 55 °C for 1 min then cooled at room temperature for 5 min. The samples were incubated at 37 °C and aliquots removed at 12, 24, 36, and 48 hours, which were quenched with 50 mM Tris (pH 7.0) and 20 mM EDTA. RNA was fractionated on a 10% PAGE gel and visualized with a PhosphorImager. Lanes containing different amounts of salts and amino acids were separated on the gel to avoid running contamination. Gel data was analyzed using semi-automated footprinting analysis (SAFA) software20 to obtain reactivities of each individual band. The reactivity of each band was normalized to nucleotides 34–36 in the anticodon loop, which were single-stranded in all solution conditions. This corrects for percent reacted and loading differences.
Optimal Growth Temperature Analysis
We analyzed the strength of tRNA stems, ribosomal RNA GC percent, and genome GC percent against optimal growth temperature of organisms. The base pairs in the acceptor, D, anticodon, and TYC stem for RNA sequences were found using the Transfer RNA database21. The portions of the sequence that compose the 5’ and 3’ ends of each stem were input to the RNACofold webserver, part of the Vienna Package, to determine the ΔG of each stem. The growing temperatures of the organisms was found in the DSMZ German Collection of Microorganisms22. Genome and rRNA GC content vs. OGT was fit for a linear correlation in Igor Pro. The sequences from the organisms were binned into six temperature ranges (20–29 °C with 14 sequences, 30–40 °C with 11 sequences, 50–69 °C with 5 sequences, 70–79 °C with 4 sequences, 80–89 °C with 5 sequences, and 90–99 °C with 4 sequences), the temperature in each bin was averaged, and the five weakest ΔGfolding in each bin were averaged together. A linear fit for threshold ΔGfolding was calculated using the averaged values in Igor Pro.
Calculating Information Content
Information content was calculated for each position in tRNA to determine conservation. Nucleotides that are highly conserved have 2 bits of information, nucleotides that are not conserved have 0 bits of information, while two nucleotides that co-vary with Watson-Crick base pairing share 2 bits of information. Information content was calculated separately for the loop and stem positions, as previously described23. For the nucleotides in loops, information content was calculated by finding the Shannon uncertainty (H) from the sequence alignment using equation 4, where Pi is the probability of finding a particular base, (A, C, G, U) at position i in the sequence. The Shannon uncertainty for genomes is approximately 2, and the information content (IC) is Hgenome minus Hposition, according to equation 5.
| (eq. 4) |
| (eq. 5) |
For stem regions, covariation of base pairs was accounted for when finding information content. Equations 4 and 5 were used to find the Shannon uncertainty and the information content.
Results
Dilute Solution Conditions Lead to Non-Cooperative Folding with Stronger Base Pairing.
Folding of WT and mutant tRNAs (Fig. 1, Tables S1, S2) were first examined under dilute solution conditions using a background of 140 mM K+ 24 and either 0.5 or 2.0 mM free Mg2+, characteristic of eukaryotic and prokaryotic divalent concentrations, respectively.25–29 Data were fit according to a two-state model to calculate thermodynamic parameters and test for level of cooperativity.10 In this approach, the transition with the major absorbance change dominates the fit in a multi-transition system. The quality of the global fits can be found in Table S3.
Figure 1.
Wild-type and mutant constructs of tRNAphe. Tertiary interactions are depicted on the WT construct with yellow lines, based on PDB 1ehz. Mutant constructs (M1–M6) vary sequence and base pairing strength in the tunable region of the acceptor stem (shaded blue) and have ‘ACCA’ at the 3’ end. Base-pairing regions that are different between mutants and WT are in black boxes. Colors of WT and mutant are maintained in all display items.
In 2.0 mM Mg2+, WT tRNA unfolds in a single sharp transition and was well fit to a two-state model suggesting cooperativity, wherein secondary structure unfolds concomitant with tertiary structure (Fig. 2A). The six acceptor stem mutants (M1–M6) were fit to the same two-state model. Their apparent melting temperatures (TMs) increase monotonically over a range of 11.4 °C, reflecting increased acceptor stem base pairing strength. Folding of low stability mutants, WT and M1, in dilute buffer is cooperative as confirmed through their highly negative ΔHfolding values of ~–80 kcal/mol (Table S1). However, the three highest stability mutants, M4–M6, display a broad unfolding transition and a smaller ΔHfolding, suggesting that secondary and tertiary structure do not unfold together (Fig. 2A). In particular, these mutants have ΔHfolding values of only ~–65 to ~–40 kcal/mol, reflecting fewer bonds broken in the major unfolding transition and thus multi-state folding. Loss of cooperativity is also mirrored in relative enthalpy values between mutant and WT (ΔHM/ΔHWT), which reveals fractional values as low as 0.5 as acceptor stem base pairing strength increases (Table S1). Similar effects are found in 0.5 mM Mg2+ (Table S2, Fig. S1). As such, the apparent TM does not accurately reflect tertiary structure unfolding for the more stable mutants in dilute buffer conditions.
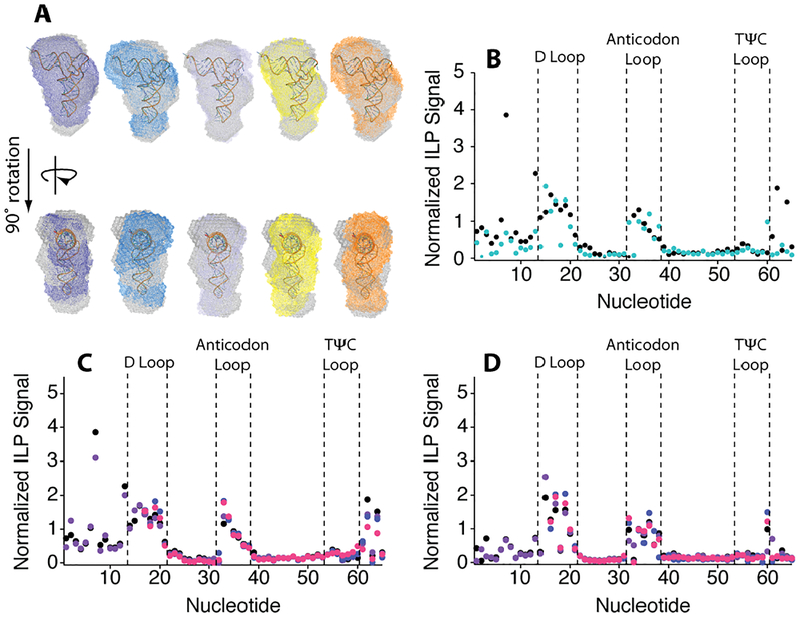
We verified that loss of cooperativity was not correlated with lack of native folding using small angle X-ray scattering (SAXS) to judge globular structure30 and in-line probing (ILP) to assess local structure31. Overlay of WT and mutant SAXS scattering envelope is excellent, supporting retention of native structure (Fig. 3A). The SAXS structural parameters of P(r), Rg, and Dmax, as well as excluded volume are similar for all constructs (Table S4, Fig. S2). Experimental scattering curves align well with the simulated scattering curve of tRNAphe (PDB 1ehz) again supporting WT-like overall structure for all constructs (Fig. S3). Moreover, the ILP profiles for WT and M5, representing low and high stability constructs, were virtually identical and consistent with native secondary structure including the critical 5’ end of the RNA where the mutations reside, supporting the same native base pairing patterns (Figs. 3B, S4). Thus, base pair changes in the acceptor stem modulate tRNA stability and folding pathway without affecting the final structure.
Figure 3.
Small angle X-ray scattering bead models of M1–M5 aligned with the WT bead model and the tRNA crystal structure in buffer with 2.0 mM Mg2+. (A) Alignment of WT (grey) and M1 (purple), M2 (blue), M3 (light blue), M4 (yellow), and M5 (orange) DAMAVER envelopes. (B) Normalized ILP signal comparing WT (black) with M5 (teal) in buffer. Normalized ILP Signal of (C) WT and (D) M5 normalized ILP signal in (black) buffer, (purple) 20% PEG8000, (blue) Mg2+-chelated amino acids and (pink) 20% PEG8000 with Mg2+-chelated amino acids in the background of 2.0 mM free Mg2+. Nucleotides 1–15 were not analyzed in samples containing Mg2+-chelated amino acids due to salt contamination.
In Vivo-Like Conditions Favor Cooperative Folding and Thermostability with Stronger Base Pairing.
We tested effects of cellular crowding in 0.5 or 2.0 mM Mg2+, using 20% w/v PEG8000, and/or cellular levels of amino acids with weakly chelated Mg2+ (aaCM).13 Previous studies from our lab revealed that the cooperativity of WT tRNA folding is enhanced equally in diverse crowders, including 20% PEG4000, PEG8000, Dextran10, Dextran70, and Ficoll70;10 20% PEG8000 is thus a representative choice. In the presence of 20% PEG8000 and 2.0 mM Mg2+, WT tRNA again unfolds in a single sharp transition, with a TM of 68.0 °C, supporting cooperative folding (Fig. 2B). Indeed, the ΔHfolding of WT tRNA is slightly larger than in dilute buffer, by almost −10 kcal/mol (Table S1). The unfolding of mutant tRNAs was sensitive to crowding agent as well (Fig. 2B). Notably, the ΔHfolding values for the WT and the mutant tRNAs are similar in crowded conditions, indicating cooperative unfolding for WT and mutants. For example, WT and M5 have ΔHfolding values of −80.4 and −62.7 kcal/mol, respectively, and ΔHM/ΔHWT range from 1.1 to 0.8. Similar effects were found in 0.5 mM Mg2+ (Table S2). However, the TMs of the mutant tRNAs are relatively unchanged in crowder. For instance, the range in TM is only 4.1 °C in crowder compared to 11.4 °C found without crowder (Table S1).
Next, we tested the stability of the tRNAs in amino acid-chelated Mg2+, which contains 2.0 mM free Mg2+ and 14.0 mM Mg2+ that is weakly chelated to a mixture of 96.0 mM L-glutamate, 4.3 mM L-aspartate, 3.8 mM L-glutamine, and 2.6 mM L-alanine (described in detail in the Materials and Methods)—the four most abundant amino acids in E. coli. This system mimics the one that is found naturally in bacteria.13 In aaCM, the observed TM and ΔH are higher in magnitude than PEG8000, by ~3 °C and ~–70 kcal/mol, respectively (Fig. 2C). Although the TMs are generally higher and range over 6 °C, as secondary strength stability increases cooperativity is lost, with ΔHmut/ΔHWT as low as 0.6. In summary, neither crowded nor aaCM solutions alone have optimal properties for thermostability: crowding has modest affects on thermostability, while aaCM loses cooperativity.
Finally, we studied the combination of aaCM and crowding to test if the favorable effects of each were additive and to more closely mimic in vivo conditions. In this combination, we observed that RNAs maintain two-state folding over a wide range of thermostability (Fig. 4A). Strikingly, large gains in thermostability and relatively high folding cooperativity are observed for all but M6, the most stable mutant (Fig. 2D). We were interested in quantifying the extent of cooperativity change for each mutant. In a cooperative system, as secondary strength increases, ΔHfolding should become larger in magnitude since more hydrogen bonds are made. We constructed theoretical fully cooperative and non-cooperative ΔHfolding limits for the most biological condition of 20% PEG8000 and aaCM (Fig. 4B). The non-cooperative limit was calculated using nearest neighbor parameters for the acceptor stems of WT and mutant tRNA32, while the fully cooperative limit was calculated as the experimentally derived ΔH of WT (Table S1) plus the nearest neighbor model ΔΔH of the mutant acceptor stems, as described in the Materials and Methods.
Figure 4.
Melting temperature and enthalpy of folding of WT and mutant tRNAs in 2.0 mM free Mg2+ with 20% PEG8000 with Mg2+-chelated amino acids. (A) Melting temperature and (B) ΔHfolding of tRNA and mutants in 20% PEG8000 and additional Mg2+-chelated amino acids. See Materials and Methods for the calculation of (pink) non-cooperative ΔH and (purple) fully cooperative ΔH limits.
As depicted in Figure 4, constructs M1, WT, and M2 fit to a ΔHfolding very close to the fully cooperative limit. However, as acceptor stem strength increases the ΔH becomes somewhat less cooperative (Fig. 4). For M3–M5, maintain partial cooperative behavior. However, the strongest mutant, M6, unfolds in an almost fully non-cooperative manner, presumably because its all-GC acceptor stem unfolds subsequent to tertiary structure. Nonetheless, it is clear that strengthening base pairing in the acceptor stem leads to greater thermal stability for WT and M1–M5. When we look at less cellular solution conditions, cooperativity is lost at much lower secondary structure strength (Fig. S5). Finally, we note that the various crowded and aaCM conditions did not affect the global tRNA structure, as the ILP profiles for WT and M5 were unaffected by buffer, 20% PEG8000, aaCM, or a combination 20%PEG8000 and aaCM (Fig. 3C, D).
Evidence that Nature Selects for Thermostable tRNAs Through Strengthening Secondary Structure not Tertiary Structure.
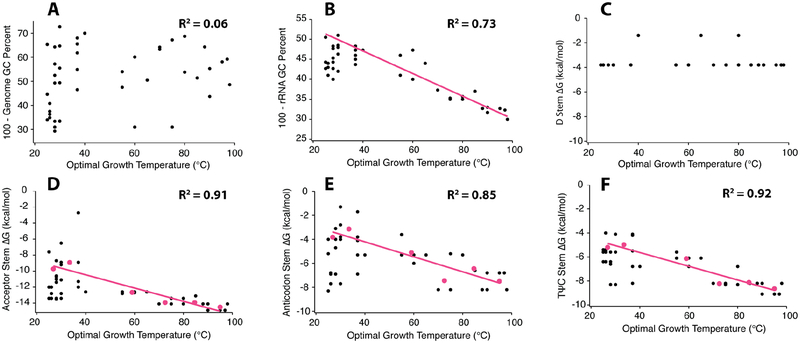
Given that our above results show that increased secondary structure strength drives increased functional stability under biological conditions, we hypothesized that strong secondary structures may be found in functional RNAs from thermophiles owing to natural selection. We analyzed the following characteristics in a series of organisms with a wide range of optimal growth temperatures (OGT): genomic and rRNA GC percentage, ΔG of each stem in tRNAphe, and average ΔG (ΔGavg) of the four stems in tRNAphe.
No observable trend was found between OGT and genome GC percent, where the R2 of a linear fit is 0.063 (plotted as 100–GC percentage in Fig. 5A). This indicates that there is no underlying GC bias to the genome of thermophilic organisms. However, a strong positive linear correlation, with an R2 of 0.73, exists between rRNA GC percent and OGT (plotted as 100–GC percentage in Fig 5B); clearly, organisms that grow optimally at higher temperatures have higher GC content in their rRNA, similar to previous reports.33–35 Strikingly, linear trends also exist between OGT and a threshold ΔGfolding for tRNA acceptor, anticodon, and TΨC stems, as well as ΔGavg; the R2 for linear fits for these four plots are all at or above 0.85 (Fig. 5D–F, Fig. S6). In these plots, the term “threshold ΔGfolding” means that we average the five weakest ΔGfolding values within a given temperature bin, as defined in the Materials and Methods, which represent a minimal stability needed to maintain function (see Discussion). For the D stem, no correlation of ΔG and OGT was found; rather, we observed two sequence clusters, each of which is fairly weak (Fig. 5C). Notably, the D stem is the only stem in tRNAphe that participates in tertiary interactions (Fig. 1), with three of its four base pairs engaged in such interactions. We hypothesize that the highly conserved tertiary contacts preclude variation of these nucleotides.
Figure 5.
Analysis of tRNA sequences from organisms with a large range of optimal growing temperatures. A comparison between optimal growth temperature and (A) whole genome GC percent, (B) ribosomal RNA GC percent, and the stability of the (C) D stem, (D) acceptor stem, (E) anticodon stem, (F) TψC stem. In panels B, D-F there are sequence bins based on OGT (pink dots) and threshold DGfolding lines (pink lines) for stem stability as a function of growth temperature, see Materials and Methods for further details.
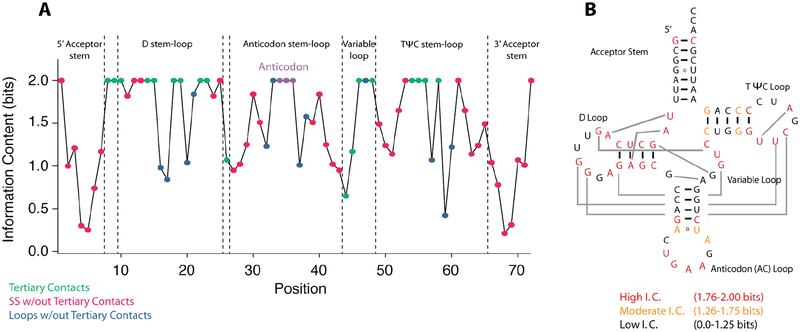
To look for variable residues, tRNAphe sequences from 44 organisms were aligned and the information content, a measure of conservation, of each position in the RNA was calculated according to Shannon entropies.6, 36, 37 As described in the Materials and Methods, nucleotides that are highly conserved have 2 bits of information, nucleotides that are not conserved have 0 bits of information, while two nucleotides that co-vary with Watson-Crick base pairing share 2 bits of information. Strikingly, nucleotides that are involved in tertiary contacts have an information content close to 2 (Fig. 6A). There are eighteen nucleotides that participate in tertiary contacts and fifteen of these have 2 bits of information each. This suggests that strengthening of tertiary structure is not a route to thermal stability in tRNA.
Figure 6.
Information content of each position in tRNAphe. (A) The information bits for each position of tRNAphe. Positions are color coded as follows: tertiary contacts (green), secondary structure without tertiary contacts (pink), loops without tertiary contacts (blue), and the anticodon stem (purple). (B) Information content mapped onto the secondary structure of tRNA. Regions of high information content (1.76–2.00 bits), moderate information content (1.26–1.75 bits) and low information content (0.00–1.25 bits) are colored in red, orange, and black respectively. Joining lines between the D and TYC loops are passing behind the AC stem.
In contrast, nucleotides involved in secondary structure have moderate (1.26–1.75 bits) or low (0.00–1.25 bits) information content. This suggests that secondary structure base pairs tend to co-vary to maintain base pairing. When mapped onto the tRNA secondary structure, positions of lower information content and conservation cluster in the acceptor, anticodon, and TYC stems, with lowest information content in the acceptor stem where we made the mutants that were tested experimentally (Fig. 6B). In summary, bases with lower information content are found in the stems and tend towards higher GC content in thermophiles, while bases with higher information content are found in tertiary interactions and are not routes to thermal stability.
Discussion
RNA can have diverse functions including catalysis in ribozymes and small molecule binding in riboswitches to regulate gene expression. In addition, certain RNAs such as tRNA and rRNA have extensive tertiary structures and mediate protein expression. The pathways for adaption of these functional RNA to extreme temperatures has remained elusive. Since loss of tertiary structure leads to loss of function, one route to thermal stability might be strengthening of tertiary interactions. This could occur, for example, through enhanced metal ion interactions and long-range interactions. Such a means for stability was found in a SELEX experiment on a group I intron,38 but the question remains as to how thermal stability is obtained during natural selection.
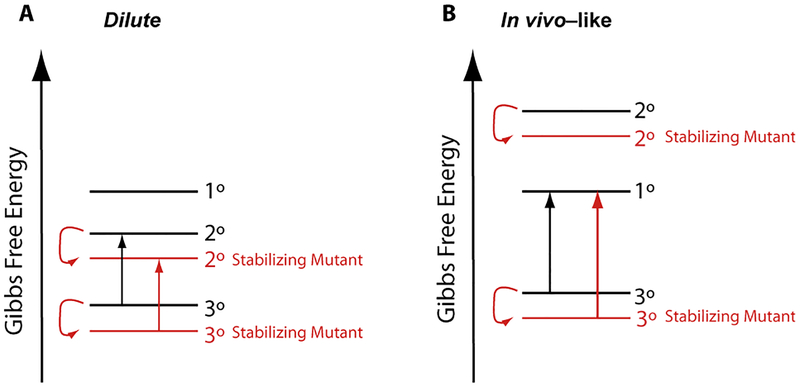
Proteins are known to fold cooperatively in nature, especially small compact proteins.39,40 Less is known about whether RNAs fold cooperatively. Crothers and co-workers showed that tRNA can fold cooperatively in high Mg2+ conditions,41 and we found that yeast tRNAphe folds cooperatively in crowded conditions that arises by both stabilization of tertiary structure and destabilization of secondary structure.10, 11 When model RNAs unfold cooperatively, strengthening of base pairing, even in positions without tertiary interactions, can contribute to thermostability.42 We thus entertained the notion that variations in stems apart from tertiary interactions could tune the temperature at which tertiary interactions in natural RNAs melt. Indeed, we found this to be the case. We found that strengthening the acceptor stem by changing G•U and AU pairs to GC pairs led to greater thermal stability, but only when folding was cooperative, i.e. under cellular crowded conditions containing Mg2+-chelated amino acids. Figure 7 provides a conceptual framework for this observation using the Pan and Sosnick concept of functional free energy.9 Under dilute conditions, where folding is non-cooperative, strengthening of secondary structure leads to no change in the functional free energy since secondary structure is the penultimately stable state and also contained in the fully folded RNA. However, under in vivo-like conditions, where folding is cooperative, strengthening of secondary structure enhances the functional free energy since the unfolded RNA is the penultimately stable state.
Figure 7.
Conceptual free energy diagram of WT and stabilizing mutants under dilute and in vivo-like conditions. (A) In dilute solution conditions folding is non-cooperative, and stabilization of secondary structure does not increase cooperativity because the penultimate state is also stabilized. (B) In more in vivo-like conditions folding is cooperative, and stabilization of secondary structure leads to an increase in cooperativity because the penultimate state is primary structure, which does not change in stability.
These findings were corroborated by analysis of a database we constructed of tRNAphe from 44 organisms that live at a broad range of temperatures. This analysis revealed that long-range tertiary interacting nucleotides have maximal information content and thus cannot change. With the exception of a single GC pair fixed toward the end of each helix, presumably present to stabilize the helix ends,43 the regions of moderate information content were largely found to be in the stems (Fig. 6). Indeed, in nature the stems strengthen with growing temperature, suggesting that they provide a route to stability. Such base pairing changes can lead to several kcal/mol gain in free energy from just a single base change.44 Thus, strengthening secondary structure of a cooperatively folding natural RNA provides a facile route to large effects in stability, albeit the sequence may be subject to other selection pressures such as serving as an identity element for aminoacylation. Moreover, observation that secondary structure increases with (OGT) in nature supports that tRNAs fold cooperatively in vivo in a diverse set of organisms.
Changing the tertiary structure of the RNA, rather than secondary structure, could be detrimental, given the generally greater molecular complexity of tertiary structure. For instance, new tertiary interactions in tRNA might interfere with the translation machinery. Additionally, observation that nucleotides involved in tertiary structure do not change with elevated growth temperature suggests that population of tertiary structure is relatively temperature independent or even endothermic; this contrasts sharply with secondary structure formation, which is strongly exothermic.6 Given that formation of tertiary structure is generally accompanied by water release,45, 46 tertiary structure formation may be entropically driven. Indeed, early studies show that metal binding to ATP is endothermic, presumably driven by the entropy gain from water release.47 In addition, docking of the P1 helix into the catalytic core of the Tetrahymena ribozyme, which is mediated by 2’ hydroxyl tertiary interactions, is entropically favored with a ΔS ranging from +37 to +62 eu and a ΔH ranging from +8.5 to +19 kcal/mol.48, 49 The origin of this effect has been attributed to water release. The Woodson lab reported that the entropy of the unfolded state of Azoarcus ribozyme is decreased in the presence of crowding agents, which decreases the entropy loss upon tertiary structure docking.12, 50 In other cases, folding of other RNA tertiary interactions have been reported to be modestly enthalpically favored, indicating that the actual results will depend on the details of number and type of tertiary interactions.51,52
The effects observed herein were lost with the most stable stem of all GC base pairs in M6. However, our experiments were conducted on tRNAs without the natural modifications. It is well established that modifications strengthen tertiary structure,53, 54 thus it is possible that even this mutant would fold cooperatively in this background. Hyperthermophiles tend to have more modified bases than mesophiles. These modifications are found in positions that participate in both secondary and tertiary structure, often strengthening contacts or increasing structure rigidity, suggesting that nature has multiple methods of selecting for RNA thermostability.55, 56 Thermostable proteins use several methods to improve stability, including more charged interactions, more disulfide bonds, and increasing rigidity.57
In this study, we introduced the notion of threshold free energy, which we define as the minimal free energy needed to keep a helix folded. In other words, it is possible to have a helix with greater stability than the threshold but not less. A similar notion was used by Szostak and co-workers in describing the threshold information content needed to attain an RNA with a given function, such as catalytic rate.23 Inspection of Figure 5 suggests that the threshold is approached by most sequences in all but the lowest growth temperatures presumably because of the entropy gain from lower information content. It remains to be explored whether similar principles apply to other functional RNAs. Our results suggest a simple mechanism for functional RNAs to adapt to higher temperatures by increasing the stability of secondary structure regions with low to moderate information content while maintaining tertiary contacts, structure, and stability.
Supplementary Material
Acknowledgments
We thank Dr. Richard Gillian and Dr. Jesse Hopkins for help with small-angle X-ray scattering experiments. This work was supported by U.S. National Institutes of Health Grant R01-GM110237 (P.C.B.). Experiments conducted at the Cornell High Energy Synchrotron Source (CHESS) were supported by the National Science Foundation and the National Institutes of Health/National Institute of General Medical Sciences under NSF award DMR-0936384, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by GM-103485 from the National Institutes of Health, through its National Institute of General Medical Sciences. We thank Elizabeth Jolley, Raghav Poudyal, Laura Ritchey, Jacob Sieg, and Ryota Yamagami and for helpful comments and discussions on the manuscript.
Footnotes
Supporting Information
The supporting information contains additional tables with the thermodynamics parameters derived from global fitting, global fit quality and errors, and SAXS parameters of WT and mutant constructs. There is a table describing the concentrations of each species in solutions with amino acid chelated-Mg2+. There are additional figures with the thermal denaturation curves of WT and mutant constructs in 0.5 mM Mg2+ with and without cellular additives, figures containing supporting SAXS data, ILP gel images, enthalpy models of WT and, and average tRNA stem free energy compared with growth temperature.
References
- (1).Berezovsky IN, and Shakhnovich EI (2005) Physics and evolution of thermophilic adaptation, Proc Natl Acad Sci USA 102, 12742–12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Greaves RB, and Warwicker J (2007) Mechanisms for stabilisation and the maintenance of solubility in proteins from thermophiles, BMC Struct Bio 7, 18–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Bae E, and Phillips GN (2004) Structures and analysis of highly homologous psychrophilic, mesophilic, and thermophilic adenylate kinases, J Biol Chem 279, 28202–28208. [DOI] [PubMed] [Google Scholar]
- (4).Trevino SG, Zhang N, Elenko MP, Lupták A, and Szostak JW (2011) Evolution of functional nucleic acids in the presence of nonheritable backbone heterogeneity, Proc Natl Acad Sci USA 108, 13492–13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Chen X, Li N, and Ellington AD (2007) Ribozyme catalysis of metabolism in the RNA world, Chem Biodivers 4, 633–655. [DOI] [PubMed] [Google Scholar]
- (6).Bloomfield VA, Crothers DM, and Tinoco I (2000) Nucleic Acids: Structures, Properties, and Functions, University Science Books, Sausalito, California. [Google Scholar]
- (7).Tinoco I, and Bustamante C (1999) How RNA folds, J Mol Biol 293, 271–281. [DOI] [PubMed] [Google Scholar]
- (8).Brion P, and Westhof E (1997) Hierarchy and dynamics of RNA folding, Annual Review of Biophysics and Biomolecular Structure 26, 113–137. [DOI] [PubMed] [Google Scholar]
- (9).Fang XW, Golden BL, Littrell K, Shelton V, Thiyagarajan P, Pan T, and Sosnick TR (2001) The thermodynamic origin of the stability of a thermophilic ribozyme, Proc Natl Acad Sci USA 98, 4355–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Strulson CA, Boyer JA, Whitman EE, and Bevilacqua PC (2014) Molecular crowders and cosolutes promote folding cooperativity of RNA under physiological ionic conditions, RNA 20, 331–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Leamy KA, Yennawar NH, and Bevilacqua PC (2017) Cooperative RNA folding under cellular conditions arises rrom both tertiary structure stabilization and secondary structure destabilization, Biochemistry 56, 3422–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kilburn D, Roh JH, Behrouzi R, Briber RM, and Woodson SA (2013) Crowders perturb the entropy of RNA energy landscapes to favor folding, J Am Chem Soc 135, 10055–10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Yamagami R, Bingaman JL, Frankel EA, and Bevilacqua PC (2018) Cellular conditions of weakly chelated magnesium ions strongly promote RNA folding, stability, and catalysis, Nat Comm, Under Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Siegfried NA, and Bevilacqua PC (2009) Thinking inside the box: designing, implementing, and interpreting thermodynamic cycles to dissect cooperativity in RNA and DNA folding, Methods Enzymol 455, 365–393. [DOI] [PubMed] [Google Scholar]
- (15).Serra MJ, and Turner DH (1995) Predicting thermodynamic properties of RNA, Methods Enzymol 259, 242–261. [DOI] [PubMed] [Google Scholar]
- (16).Skou S, Gillilan RE, and Ando N (2014) Synchrotron-based small-angle X-ray scattering (SAXS) of proteins in solution, Nat Protoc 9, 1727–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Acerbo AS, Cook MJ, and Gillilan RE (2015) Upgrade of MacCHESS facility for X-ray scattering of biological macromolecules in solution, J Synchrotron Radiat 22, 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Schneidman-Duhovny D, Hammel M, Tainer John A., and Sali A (2013) Accurate SAXS profile computation and its assessment by contrast variation experiments, Biophys J 105, 962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Schneidman-Duhovny D, Hammel M, Tainer JA, and Sali A (2016) FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles, Nucleic Acids Res 44, W424–W429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Das R, Laederach A, Pearlman SM, Herschlag D, and Altman RB (2005) SAFA: Semiautomated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments, RNA 11, 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, and Pütz J (2009) tRNAdb 2009: compilation of tRNA sequences and tRNA genes, Nucleic Acids Res 37, D159–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Söhngen C, Podstawka A, Bunk B, Gleim D, Vetcininova A, Reimer LC, Ebeling C, Pendarovski C, and Overmann J (2016) BacDive – The bacterial diversity metadatabase in 2016, Nucleic Acids Res 44, D581–D585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Carothers JM, Oestreich SC, Davis JH, and Szostak JW (2004) Informational complexity and functional activity of RNA structures, J Am Chem Soc 126, 5130–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Feig AL, and Uhlenbeck OC (1999) The role of metal ions in RNA biochemistry, In The RNA World, 2nd Ed (Gesteland RF, Cech TR, and Atkins JF, Eds.), pp 287–320, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- (25).Lusk JE, Williams RJ, and Kennedy EP (1968) Magnesium and the growth of Escherichia coli, J Biol Chem 243, 2618–2624. [PubMed] [Google Scholar]
- (26).Truong DM, Sidote DJ, Russell R, and Lambowitz AM (2013) Enhanced group II intron retrohoming in magnesium-deficient Escherichia coli via selection of mutations in the ribozyme core, Proc Natl Acad Sci USA 110, E3800–E3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Alberts B, Bray D, Lewis J, Roberts K, and Watson JD (1994) Molecular biology of the cell 3rd ed. [Google Scholar]
- (28).London RE (1991) Methods for measurement of intracellular magnesium: NMR and fluorescence, Annual Reviews of Physiology 53, 241–258. [DOI] [PubMed] [Google Scholar]
- (29).Romani AM (2007) Magnesium homeostasis in mammalian cells, Frontiers in Bioscience 12, 308–331. [DOI] [PubMed] [Google Scholar]
- (30).Pollack L (2011) Time resolved SAXS and RNA folding, Biopolymers 95, 543–549. [DOI] [PubMed] [Google Scholar]
- (31).Regulski EE, and Breaker RR (2008) In-line probing analysis of riboswitches, In Post-Transcriptional Gene Regulation (Wilusz J, Ed.), pp 53–67, Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- (32).Serra MJ, and Turner DH (1995) Predicting thermodynamic properties of RNA, In Methods Enzymol, pp 242–261, Academic Press. [DOI] [PubMed] [Google Scholar]
- (33).Wang H.-c., and Hickey DA (2002) Evidence for strong selective constraint acting on the nucleotide composition of 16S ribosomal RNA genes, Nucleic Acids Res 30, 2501–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Jegousse C, Yang Y, Zhan J, Wang J, and Zhou Y (2017) Structural signatures of thermal adaptation of bacterial ribosomal RNA, transfer RNA, and messenger RNA, PLOS ONE 12, e0184722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Galtier N, and Lobry JR (1997) Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes, J Mol Evol 44, 632–636. [DOI] [PubMed] [Google Scholar]
- (36).Shannon CE (1948) A mathematical theory of communication, Bell Syst Tech J 27, 379–423. [Google Scholar]
- (37).Schneider TD, Stormo GD, Gold L, and Ehrenfeucht A (1986) Information content of binding sites on nucleotide sequences, J Mol Biol 188, 415–431. [DOI] [PubMed] [Google Scholar]
- (38).Juneau K, and Cech TR (1999) In vitro selection of RNAs with increased tertiary structure stability, RNA 5, 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Malhotra P, and Udgaonkar JB (2016) How cooperative are protein folding and unfolding transitions?, Protein Sci 25, 1924–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Portman JJ (2010) Cooperativity and protein folding rates, Curr. Opin. Struct. Biol 20, 11–15. [DOI] [PubMed] [Google Scholar]
- (41).Stein A, and Crothers DM (1976) Conformational changes of transfer RNA. The role of magnesium(II), Biochemistry 15, 160–168. [DOI] [PubMed] [Google Scholar]
- (42).Blose JM, Silverman SK, and Bevilacqua PC (2007) A simple molecular model for thermophilic adaptation of functional nucleic acids, Biochemistry 46, 4232–4240. [DOI] [PubMed] [Google Scholar]
- (43).Xia T, SantaLucia J, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, and Turner DH (1998) Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs, Biochemistry 37, 14719–14735. [DOI] [PubMed] [Google Scholar]
- (44).Blose JM, Manni ML, Klapec KA, Stranger-Jones Y, Zyra AC, Sim V, Griffith CA, Long JD, and Serra MJ (2007) Non-nearest-neighbor dependence of the stability for RNA bulge loops based on the complete set of group I single-nucleotide bulge loops, Biochemistry 46, 15123–15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Miyoshi D, Karimata H, and Sugimoto N (2006) Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions, J Am Chem Soc 128, 7957–7963. [DOI] [PubMed] [Google Scholar]
- (46).Nakano S. i., Karimata HT, Kitagawa Y, and Sugimoto N (2009) Facilitation of RNA enzyme activity in the molecular crowding media of cosolutes, J Am Chem Soc 131, 16881–16888. [DOI] [PubMed] [Google Scholar]
- (47).Banyasz JL, and Stuehr JE (1973) Interactions of divalent metal ions with inorganic and nucleoside phosphates. III. Temperature dependence of the magnesium(II)--adenosine 5’-triphosphate, --adenosine 5’-diphosphate, and --cytidine 5’-diphosphate systems, J Am Chem Soc 95, 7226–7231. [DOI] [PubMed] [Google Scholar]
- (48).Li Y, Bevilacqua PC, Mathews D, and Turner DH (1995) Thermodynamics and activation parameters for binding of pyrene-labeled substrate by the Tetrahymena ribozyme: Docking is not diffusion-controlled and is driven by a favorable entropy change, Biochemistry 34, 14394–14399. [DOI] [PubMed] [Google Scholar]
- (49).Narlikar GJ, and Herschlag D (1996) Isolation of a local tertiary folding transition in the context of a globally folded RNA, Nat Struct Mol Biol 3, 701. [DOI] [PubMed] [Google Scholar]
- (50).Kilburn D, Behrouzi R, Lee H-T, Sarkar K, Briber RM, and Woodson SA (2016) Entropic stabilization of folded RNA in crowded solutions measured by SAXS, Nucleic Acids Res 44, 9452–9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Crothers DM, Cole PE, Hilbers CW, and Shulman RG (1974) The molecular mechanism of thermal unfolding of Escherichia coli formylmethionine transfer RNA, J Mol Biol 87, 63–88. [DOI] [PubMed] [Google Scholar]
- (52).Szewczak AA, Podell ER, Bevilacqua PC, and Cech TR (1998) Thermodynamic stability of the P4−P6 domain RNA tertiary structure measured by temperature gradient gel electrophoresis, Biochemistry 37, 11162–11170. [DOI] [PubMed] [Google Scholar]
- (53).Helm M (2006) Post-transcriptional nucleotide modification and alternative folding of RNA, Nucleic Acids Res 34, 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Nobles KN, Yarian CS, Liu G, Guenther RH, and Agris PF (2002) Highly conserved modified nucleosides influence Mg2+-dependent tRNA folding, Nucleic Acids Res 30, 4751–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Kowalak JA, Dalluge JJ, McCloskey JA, and Stetter KO (1994) The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles, Biochemistry 33, 7869–7876. [DOI] [PubMed] [Google Scholar]
- (56).Lorenz C, Lünse CE, and Mörl M (2017) tRNA modifications: Impact on structure and thermal adaptation, Biomolecules 7, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Pucci F, and Rooman M (2017) Physical and molecular bases of protein thermal stability and cold adaptation, Curr Opin Struct Biol 42, 117–128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.