We decipher how an allosteric inhibitor activates the ClpP protease machinery by binding to the catalytic site.
Abstract
Coordinated conformational transitions in oligomeric enzymatic complexes modulate function in response to substrates and play a crucial role in enzyme inhibition and activation. Caseinolytic protease (ClpP) is a tetradecameric complex, which has emerged as a drug target against multiple pathogenic bacteria. Activation of different ClpPs by inhibitors has been independently reported from drug development efforts, but no rationale for inhibitor-induced activation has been hitherto proposed. Using an integrated approach that includes x-ray crystallography, solid- and solution-state nuclear magnetic resonance, molecular dynamics simulations, and isothermal titration calorimetry, we show that the proteasome inhibitor bortezomib binds to the ClpP active-site serine, mimicking a peptide substrate, and induces a concerted allosteric activation of the complex. The bortezomib-activated conformation also exhibits a higher affinity for its cognate unfoldase ClpX. We propose a universal allosteric mechanism, where substrate binding to a single subunit locks ClpP into an active conformation optimized for chaperone association and protein processive degradation.
INTRODUCTION
Among the most exciting antibacterial drug targets that have emerged in the past decade is the caseinolytic protease (ClpP) (1). While mostly studied in Escherichia coli (EcClpP), ClpP is present in a wide range of bacteria, as well as in mitochondria and chloroplasts. Functionally, ClpP can be defined as a cylindrical canonical serine protease composed of two stacked rings, each formed by seven subunits that encompass a large (300 kDa) proteolytic core (2). This architecture restricts access to the 14 proteolytic active sites located inside the barrel and is key for ClpP function. Although ClpP is able to rapidly hydrolyze peptides, degradation of large proteins requires the presence of an AAA+ adenosine triphosphatase (ATPase) complex, such as ClpA or ClpX in E. coli or ClpC in other species. These unfoldases provide substrate specificity not only by recognizing, unfolding, and translocating their globular protein substrates but also by opening the apical gate region of ClpP, which is otherwise blocked by flexible N-terminal tails.
ClpP protease and its associated chaperones are essential for the survival or virulence of several bacteria including pathogens such as Mycobacterium tuberculosis (Mtb) (3), Staphylococcus aureus (Sa) (4), and Listeria monocytogenes (Lm) (5). In addition, human mitochondrial ClpP was associated to human acute myeloid leukemia and obesity (6, 7). Recently, ClpP from Plasmodium falciparum (PfClpP), the parasite responsible for malaria infection, has also been proposed as a promising drug target for the control of this disease (8).
The fact that ClpP is a suitable Achilles’ heel for these organisms is further reinforced by the recent discovery of natural antibiotics that target ClpP or its co-chaperones. Efficient bacterial killing could be accomplished either by opening the axial pore of ClpP and activating unregulated cell proteolysis (9), by blocking ClpP-ATPase interaction (10), or by specifically inhibiting its chaperones, ClpX (4) or ClpC1 (11).
Despite recent advances in the field, the study and development of drugs targeting ClpP have been thwarted by the fact that ClpPs from different species show notable functional differences. For example, Mtb contains two clpP genes, clpP1 and clpP2, both of which are essential for viability and infectivity (1, 3). Although both genes encode serine proteases that form heptameric rings, first attempts to express and characterize isolated MtbClpP1 and MtbClpP2 in E. coli yielded complexes that lacked proteolytic activity (12, 13). The study of MtbClpP1P2 was not possible until the finding that a small activator, derived from classical serine protease inhibitor peptide aldehydes (3), was required to promote the formation of an active complex composed of one heptameric ring from ClpP1 and another from ClpP2 (3). Oddly enough, the small activators were found at the protein active site, and it is not clear how their presence at the active site can promote activation rather than classic competitive inhibition (14, 15). Furthermore, several structures obtained by x-ray crystallography and solution nuclear magnetic resonance (NMR) studies have suggested that ClpPs, while keeping a tetradecamer organization, can adopt several conformations (16, 17). Currently, at least three conformations have been reported: an extended state (catalytically active), a compressed state (catalytically inactive), and a compact state (catalytically inactive) (17–19). In addition, while some ClpPs are inherently active, others are purified as non-active forms (3, 12, 13, 15, 20–22).
Here, we report a study of ClpP from Thermus thermophilus (TtClpP), which is, as described for other ClpPs, purified in an inactive conformation but can be in vitro activated by small molecules. Using an integrated approach including multiple biochemical assays, x-ray crystallography, magic angle spinning (MAS) NMR and solution-state NMR, isothermal titration calorimetry (ITC), and molecular dynamics (MD) simulations, we show that bortezomib, a boronic acid previously identified as an inhibitor of MtbClpP1P2, induces a concerted conformational change in ClpP, resulting in complex activation. On the basis of our data, and building further upon previously established results, we propose a universal mechanism for ClpP activation.
RESULTS
Bortezomib in low concentrations activates TtClpP peptidase activity
Although cellular protein quality control is likely to be particularly challenging for organisms living at high temperatures, ClpP from extremophiles remains modestly documented. T. thermophilus (Tt) is a Gram-negative eubacterium used in a wide range of biotechnological applications and as a model organism for genetic manipulation, structural genomics, and systems biology. The bacterium has an optimal growth temperature of about 65°C. We obtained TtClpP by recombinant expression and purified it as a canonical tetradecamer of around 300 kDa, as monitored by size exclusion chromatography (SEC) and multi-angle laser light scattering (MALLS) (fig. S1). This tetradecamer could be reorganized into a smaller heptameric species when ammonium sulfate (300 mM) was included in the elution buffer, likely as a result of the disruption of inter-ring salt bridges (fig. S1A). As anticipated, TtClpP displayed enhanced thermostability compared to previously studied ClpPs. Using the single tryptophan of the TtClpP monomers (14 per tetradecamer) as a probe, the melting point (TM) of ClpP was determined as 84°C (fig. S1C), 30°C above the value reported for S. aureus ClpP (SaClpP) (23). The increased stability is also reflected in an elevated optimal catalytic temperature. When we measured the degradation of the green fluorescent protein GFPsrrA by TtClpP in association with the cognate TtClpX ATPase, we found maximal rates at 60°C (the highest temperature we could achieve with the fluorimeter equipment; fig. S2, A and B). However, although TtClpP is competent to degrade folded substrates in association with TtClpX, it is a rather inefficient peptidase even at 60°C. In comparison to EcClpP at 37°C, it has a 200-fold lower specific activity in degradation experiments with the short tripeptide substrate PKMamc (proline-lysine-methionine-7-amino-4-methylcoumarin; at a working concentration of 100 μM).
During attempts to identify inhibitors of TtClpP that could be used for further structural biology studies, we observed that bortezomib did not inhibit, but actually stimulated, TtClpP peptidase activity at concentrations from 3 to 100 μM (Fig. 1A and fig. S3A). Bortezomib is an N-protected dipeptide with a boronic acid instead of a carboxylic acid that forms adducts with activated threonines or serines (24). It was developed as a proteasome inhibitor targeting specifically the chymotrypsin-like sites of this complex with nanomolar affinity while exhibiting limited reactivity to the other active sites of the complex. We selected this molecule because it was recently identified in an in vivo cell screen as an inhibitor of GFPssrA degradation by the MtbClpXP1P2 complex and was proposed as a potential antituberculosis candidate (25). The bortezomib dependence of the TtClpP peptidase activity displays a bell shape, with maximum activation at 12.5 μM (about 10- to 20-fold activation), while higher drug concentrations reduced the enzymatic activity (Fig. 1A). A time-lag phase for the activation of TtClpP was consistently observed and likely resulted from the necessary fluorimeter temperature equilibrium because the assays were executed at 45° to 60°C. Bortezomib activation derives from intrinsic properties of TtClpP, and when tested against EcClpP peptidase activity, it behaved as a powerful inhibitor with an apparent IC50 (median inhibitory concentration) of 1.6 μM (fig. S3B).
Fig. 1. Bortezomib activates TtClpP for peptide and intrinsically disordered protein degradation.
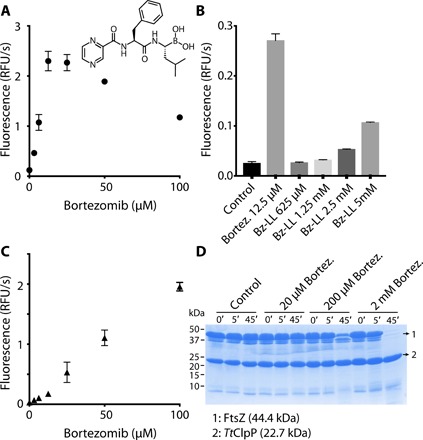
(A) TtClpP (1 μM complex) peptidase activity was measured with the substrate PKMamc (100 μM) in the presence of bortezomib at the indicated concentrations. As the peptide is cleaved, 7-amino-4-methylcoumarin is released, resulting in an increase in the measured fluorescence. Initial rates are plotted as a function of the bortezomib concentration. RFU, relative fluorescence units. (B) The activating effect of bortezomib is compared with the one observed with Bz-LL, a previously described activator of MtbClpP1P2. (C) The degradation of the unfolded protein substrate FITC-casein by TtClpP (0.1 μM complex) was measured in the presence of the indicated bortezomib concentrations. Initial degradation rates following temperature equilibration were plotted as a function of the bortezomib concentration. (D) TtClpP (1 μM complex) can degrade the intrinsically disordered E. coli FtsZ. The degradation of FtsZ by TtClpP was monitored by SDS-gel electrophoresis. While no degradation was observed with apoTtClpP, degradation of FtsZ was observed in the presence of 200 μM and 2 mM bortezomib.
Although paradoxical, activation of some ClpP complexes by inhibitors has already been reported (3, 22, 26–28). Akopian and colleagues (3) have shown that the MtbClpP complex (MtbClpP1P2), otherwise inactive, is activated by small peptide aldehydes. The most potent of these activators in Mtb, an N-terminally blocked dileucine (Bz-LL), was also capable to activate TtClpP, albeit with lower efficiency (Fig. 1B). Conversely, bortezomib could also activate the peptidase activity of the MtbClpP1P2 complex. We applied a set of biochemical, biophysical, and structural studies to understand the mechanistic and functional foundation of this inhibitor-based activation.
Bortezomib activates TtClpP unfolded protein degradation
After establishing the bortezomib-induced activation of TtClpP for degradation of small peptides, we investigated its potential to stimulate protein degradation. Contrary to small peptides, which can independently diffuse into the proteolytic chamber, the basal degradation of proteins by ClpP is low because the flexible N-terminal tails of ClpP act as a barrier to protein entry into the chamber. Our assay used as proteolytic target fluorescein isothiocyanate–labeled casein (FITC-casein), a proline-rich protein lacking stable secondary structures. Protease-catalyzed hydrolysis of FITC-casein leads to highly fluorescent dye–labeled peptides. Similarly to the peptidase activity, addition of bortezomib did not result in inhibition of FITC-casein degradation, but instead, a striking activation was observed (Fig. 1C and fig. S4A). Furthermore, contrary to what was observed with the peptidase activity, concentration dependence of this effect was linear and no reduction of the activation was observed up to 100 μM (Fig. 1C). In addition to casein, we tested whether bortezomib could also promote the degradation of other substrates. The bacterial guanosine triphosphatase (GTPase) FtsZ forms a cytokinetic ring at mid-cell, recruits the division machinery, and coordinates membrane and peptidoglycan cell wall invagination. An FtsZ monomer contains an intrinsically disordered C-terminal linker, and the degradation of FtsZ by ClpP activated by the natural antibiotic acyldepsipeptides (ADEPs) has been proposed to trigger growth inhibition (29). We therefore tested the effect of bortezomib ClpP-catalyzed proteolysis on FtsZ and found that FtsZ was, even at low temperature for TtClpP (37°C), degraded by ClpP when in the presence of bortezomib (Fig. 1D) (29). These experiments establish that bortezomib acts as a TtClpP activator for the degradation of different disordered protein substrates or small peptides.
ClpX enhances peptidase activity of TtClpP
Because ClpP acts in association with specific chaperones in vivo (ClpX, ClpA, or ClpC), we proceeded by assessing the effect of TtClpX association on TtClpP peptidase activity. When we measured the degradation of PKMamc by TtClpP in the presence of TtClpX, we observed a marked increase in peptidase activity that was dependent on TtClpX concentration (Fig. 2, A and B). Similarly to TtClpP activation by bortezomib, a lag phase was observed, which could result either from temperature equilibrium or from oligomerization of TtClpX hexamers from monomers in the presence of adenosine 5′-triphosphate (ATP).
Fig. 2. TtClpX activates TtClpP peptidase activity.
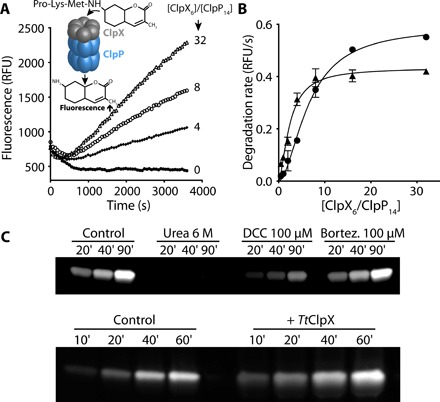
(A) TtClpP (0.1 μM complex) peptidase activity was measured in the presence of TtClpX at the indicated ClpX6/ClpP14 ratios. (B) The activation of TtClpP peptidase activity in the absence (circles) or presence (triangles) of bortezomib (10 μM) is plotted as a function of the molar ClpX6/ClpP14 ratio. Half-maximal peptidase activity was obtained at 6.5 ± 0.5 and 2.4 ± 0.3 ClpX6/ClpP14 ratio for the apo and bortezomib-loaded TtClpP, respectively. (C) Activation of the TtClpP active site by bortezomib (100 μM) and TtClpX (1 μM), monitored by labeling with TAMRA-FP. TAMRA-FP (5 μM) was incubated with TtClpP (1 mg/ml), and aliquots of the reaction mixture were removed at the indicated time points. Labeling of the active-site serines is reflected by the increase in fluorescence observed.
Because bortezomib and ClpX are both able to activate TtClpP, we tested whether the effects of bortezomib and ClpX were synergistic. Bortezomib (10 μM) did not induce further activation but instead acted as an inhibitor (fig. S4B) in the presence of ClpX. However, in the presence of bortezomib, a marked feature of the ClpX activation curve was a certain level of cooperativity, i.e., less TtClpX concentration was required to achieve maximal activity, suggesting that bortezomib binding can modulate the affinity of ClpP for ClpX (Fig. 2B). In the presence of bortezomib, half-maximal activation was achieved with a TtClpX6/ClpP14 ratio of 2.4, corresponding to 120 nM TtClpX6. This value for TtClpX-TtClpP association is on the same order of magnitude as the one reported for E. coli using an indirect ATPase assay (27).
The large activation of ClpP-catalyzed degradation of small peptides by ClpX was unexpected, because auxiliary chaperones are required for unfolding, translocation, and opening of the N-terminal pores facilitating entry of protein substrates into the ClpP proteolytic chamber, while small peptides can just freely diffuse into the proteolytic chamber. The hydrolysis rate of small peptides (dipeptides) in EcClpP has been independently shown to be unaffected by ClpX association (30, 31). Activation by EcClpX is, however, essential for bigger peptides and proteins, and ClpX activation increases as a function of peptide molecular weight. ClpX binding has also been shown to stimulate active-site modification of ClpP by fluorophosphates, but only to the level predicted from faster diffusion of the inhibitor into the ClpP chamber (30). Nonetheless, it was recently proposed that, in addition to the opening of the ClpP gate, chaperones can also allosterically activate the ClpP catalytic sites by inducing a shift of ClpP from the inactive compressed state, in which the catalytic triad is distorted, to the extended state, in which the catalytic residues are positioned for enzymatic action (23).
Because the small molecular weight of the substrate used in our assays is not an obstacle to its free diffusion into the proteolytic chamber (molecular weight of 574 Da) (30), we tested whether TtClpX binding to TtClpP could affect the protease active sites. To confirm the effect of co-chaperone or bortezomib on the active site, we used an assay based on fluorophosphonate (FP) conjugated tetramethylrhodamine (TAMRA-FP, molecular weight of 680 Da), which specifically and covalently labels activated Ser active sites with a fluorescent dye. TAMRA-FP can be used to quantify the reactivity of a given protease active site, e.g., if the catalytic triad is in the correct geometry for proper serine activation. The presence of an activated Ser can thus be detected using fluorescent gel imaging. We incubated TtClpP and TAMRA-FP with either urea, bortezomib, dichloroisocoumarin (DCC; an inhibitor of serine proteases), or TtClpX. While urea (by unfolding the complex) and DCC (by irreversibly blocking the active site) reduced the amount of TAMRA bound to ClpP, ClpX association resulted in a significant increase of active-site acetylation—a clear indication that ClpX association activates the ClpP active site. Although bortezomib also leads to an activation of ClpP, we failed to observe any increase of TAMRA-FP labeling (Fig. 2C). This could result from competition between the two inhibitors (bortezomib and TAMRA-FP). Therefore, we cannot exclude that TAMRA-FP binding also results in TtClpP activation. However, contrary to bortezomib, TAMRA-FP forms irreversible adducts with TtClpP, thereby preventing activity measurements.
Bortezomib binds to the serine of ClpP active site
To understand bortezomib’s mechanism of action, we examined its binding site using NMR spectroscopy. With a molecular weight of 300 kDa, NMR studies of TtClpP are challenging. In solution, the slow tumbling leads to severe line broadening, to the extent that standard solution NMR studies are not feasible. The most sensitive route for observing solution NMR signals of proteins of the size of ClpP is the specific labeling of methyl groups in an otherwise deuterated background (32). Although being a very powerful approach for probing interactions and dynamics, methyl-directed NMR is able to observe only the subset of methyl-bearing amino acid types, and the sequence-specific assignment of NMR resonances is challenging, generally requiring multiple mutants.
MAS solid-state NMR does not suffer from this inherent size limitation and is able to detect all amino acid types. We have used samples of TtClpP that were either obtained from sedimenting ClpP from solution into MAS NMR sample tubes (1.3-mm rotors) or obtained by addition of methyl-pentane-diol, an oft-used precipitant in crystallography. Spectra acquired from these two preparations are very similar. We have used proton-detected three-dimensional (3D) and 4D MAS NMR approaches (33) to obtain the sequence-specific resonance assignment for (1HN, 15N, 13Cα, 13CO) nuclei of 97 residues spread throughout the molecule, including the active site and the central helix αE, which undergoes most changes in the compressed-to-extended transition. The N-terminal gate region was not assigned, presumably due to its structural heterogeneity and dynamics, which lead to inefficient NMR coherence transfers and line broadening (16). Figure 3A shows the residue-wise secondary structure obtained from these MAS NMR resonance assignments. As expected, the location of secondary structures matches well the information inferred from crystallography (see below).
Fig. 3. Bortezomib binding to TtClpP investigated by MAS NMR and solution NMR.
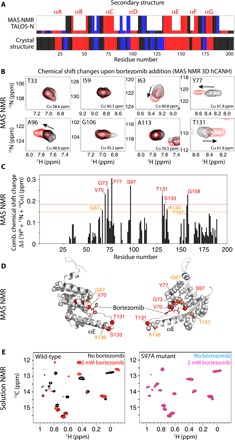
(A) Secondary structures (loop, black; α-helix, red; β-strand, blue) as a function of sequence number obtained from the resonance assignments obtained by MAS NMR and analysis with TALOS-N (24) (tall bars). For residues for which no assignment has been obtained, the small bars are obtained by TALOS-N from a database approach. The bottom panel shows the comparison with the secondary structure obtained from the crystal structure (see Fig. 5). (B) Zooms onto 2D 1H-15N excerpts from 3D hCANH MAS NMR correlation spectra of TtClpP sedimented in the presence of 10 mM bortezomib (red) or without bortezomib (black). Chemical shift changes are indicated with arrows. Note that the CSPs are relatively small, presumably due to the difficulty of saturating the binding site in a sedimented sample, given the high Kd value, resulting in incompletely occupied binding sites. ppm, parts per million. (C) Combined CSPs upon addition of bortezomib from the added peak shifts in 1H, 13Cα, and 15N dimensions obtained from the spectra shown in (B). (D) Location of residues with substantial CSPs mapped onto the x-ray structure of the TtClpP:bortezomib complex obtained from the data shown in (C). (E) Solution-state NMR spectra of wild-type (left) and S97A (right) TtClpP in the absence and presence of bortezomib, showing large spectra changes for the former due to binding, but no effect on the mutant.
3D spectra correlating intra-residue 1H-13Cα-15N frequencies (referred to as hCANH) of sedimented TtClpP with and without bortezomib are reported in Fig. 3 (B and C). The observed changes are modest, which is likely linked to the relatively low affinity of bortezomib, making it difficult to saturate the protein in the highly concentrated sedimented sample. Nevertheless, chemical shift perturbations (CSPs) are observed for a number of residues in the vicinity of the active site, including Val70, Gly73, Tyr77, and the catalytic Ser97, as well as residues in the neighboring helix αE (Fig. 3D). These data reveal that bortezomib interacts with the active site, and suggest that the interaction induces also small changes in the structure and the dynamics of helix αE, which is involved in mediating the compressed-to-extended transition.
To independently confirm the direct involvement of the active site for bortezomib binding in solution, we have performed additional NMR experiments, making use of perdeuterated, Ile-δ1-CH3–labeled TtClpP. Spectra of apo TtClpP, and of the latter in the presence of 2 mM bortezomib, are shown in Fig. 3E (left). Although we did not attempt to assign the resonances of the 18 isoleucines per ClpP subunit, the clear differences in the spectra without or with bortezomib unambiguously confirms binding. In a control experiment performed with the catalytic S97A mutant, no spectral changes were observed, corroborating that the bortezomib interaction requires the presence of the catalytic serine (Fig. 3E, right). The absence of any substantial CSP in the S97A mutant demonstrates that all spectral changes are a result of the direct binding of bortezomib to the active site and its associated conformational changes—not from nonspecific binding to other parts of the protein.
Together, MAS NMR and solution NMR provide direct evidence that bortezomib binds to the catalytic site. CSPs outside the active site (particularly in helix αE) suggest that binding may lead to further structural changes, possibly connected to the observed TtClpP activation.
Structural basis for bortezomib activation
To understand the structural details of the TtClpP-bortezomib interaction initially characterized by NMR, we used x-ray crystallography and determined the crystal structures of TtClpP in the absence and presence of bortezomib at a resolution of 1.95 and 2.7 Å, respectively (Fig. 4).
Fig. 4. Structure and dynamics of TtClpP from x-ray crystallography, MAS NMR, and MD simulations.
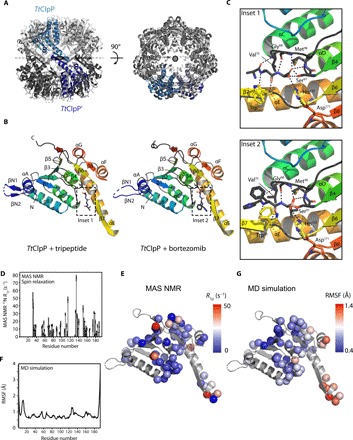
(A) Side and top views of the TtClpP 14-mer. One TtClpP monomer per heptameric ring (light and dark gray) is highlighted in light and dark blue, respectively. (B) Cartoon representation of the TtClpP monomer in peptide-bound (left) and bortezomib-bound (right) states. Helices are named by letters, and strands are indicated by numbers. A zoom of the ligands present in the active sites (dashed boxes) is shown in (C). (C) Substrate-binding pocket of TtClpP. The residues involved in the binding to the model peptide (inset 1) and bortezomib (inset 2) are shown as sticks. (D and E) Residue-wise MAS NMR amide 15N R1α relaxation rate constants. High R1α rate constants point to enhanced nanosecond-to-millisecond motions and are found primarily in loop regions and in helix αE, as shown in (C). (F and G) MD-derived root mean square fluctuations (RMSFs) over the 1-μs-long MD trajectory of the assembled 14-mer ClpP.
The overall structure of TtClpP is essentially identical to the “extended” state (34–37) of ClpP described for other species {root mean square deviation (RMSD) to EcClpP tetradecamer [Protein Data Bank (PDB): 6MT6] = 2.115 Å for 1451 aligned Cα atoms and RMSD to EcClpP monomer = 0.73 Å for 176 aligned Cα atoms}, which is in agreement with its high sequence similarity to ClpP orthologs (% identity to EcClpP = 58.25). The structure of one TtClpP monomer is presented in Fig. 4B and depicts an α/β-fold composed of two domains: a head domain comprising six repeats of α/β units (helices A to D, F, and G and strands 1 to 5, 7, and 8) and a handle domain (helix E and strand 6). The N terminus of TtClpP forms a short antiparallel β-hairpin that protrudes from the apical surface of the barrel. This N-terminal β-hairpin is constituted of residues 5 to 8 and residues 15 to 18 of TtClpP, with missing electron density for residues 9 to 14. While the overall monomer structure of ClpP is highly conserved among the structures solved to date, a high degree of variability exists in the conformation of the N-terminal regions. Several ClpP crystal structures feature N-terminal β-hairpin structures of varying lengths and degrees of flexibility, some of which appear to be stabilized by crystal-packing contacts (38). Solution NMR studies have demonstrated that the N termini of ClpP are structurally heterogeneous and adopt multiple conformations (16, 39), which might explain the missing density near the end of the N-terminal β-hairpins in the TtClpP structure.
Unexpectedly, in the TtClpP crystals without added bortezomib, we consistently observe a continuous stretch of electron density near the active-site serine (Ser97) of each TtClpP monomer, which, based on its shape, suggests the presence of a tripeptide (Fig. 4C and fig. S5). Because no peptide was added, the nature of this molecule was unknown. We, therefore, built it in the electron density as a tri-l-alanine (see also omit and polder maps presented in fig. S5). The presence of short peptides in Helicobacter pylori ClpP structures resulting from proteolysis during crystallization has been previously described (40). While in that particular case the observed tri- and tetrapeptides are caused by proteolysis of added heptapeptides before crystallization, the origin of the tripeptides observed in our TtClpP crystals remains unclear. Matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) measurements exclude the presence of these peptides in fresh purified TtClpP. We speculate that they are the result of autoproteolysis of unfolded or partially unfolded TtClpP in the crystallization drops followed by the formation of TtClpP:peptide crystals. The serendipitous presence of peptides near the active-site Ser97 provides valuable insights into how TtClpP binds its substrates (Fig. 4C, inset 1). Apart from the active-site Ser97, the terminal carboxy group of the peptide (residue P3) is further stabilized by hydrogen bonds with the Nε2 atom of His122 (part of the ClpP catalytic triad) and the backbone amide groups of Met98 and Gly68. The Nδ1 atom of His122 on its turn forms an additional hydrogen bond with Asp171, the third residue of the ClpP catalytic triad. The amino groups of peptide residues P2 and P3 are stabilized by backbone interactions with Gly68 and Trp125, respectively, while the carbonyl groups are stabilized by interactions with amino groups of Trp125 and Val70 (Fig. 4C, inset 1). The geometry and distances between the catalytic triad residues (Ser97, His122, and Asp171) are consistent with a functional TtClpP catalytic triad because their distances are in the range of the ones observed in classical serine proteases and active ClpPs (40). Figure 4B displays the 2.7-Å resolution structure of the bortezomib-bound state (see also fig. S5). Similarly to the peptides present in the TtClpP:peptide structure, bortezomib tightly fits between β-strand 6 and a small β-turn formed by Gly68 and Val70, thereby forming an antiparallel β-sheet. This conformation is characterized by a hydrogen bond network almost identical to the one observed in the TtClpP:peptide interaction (Fig. 4C, inset 2). Unlike the TtClpP:peptide complex, where Ser97 is forming hydrogen bonds with the terminal carboxy group of the peptide, bortezomib seems to be covalently bound to the active-site Ser97 via its boron atom. Other significant differences are observed in the conformation of Trp125, which is displaced toward bortezomib and forms an aromatic interaction with the bortezomib phenylalanyl group. In addition, the nearby Thr131 residue, which in peptide-bound TtClpP interacts with Arg170 of a neighboring ClpP monomer, now forms a hydrogen bond with Gln123 of an opposing ClpP monomer in the other heptameric ring. Despite the rearrangement of the Trp125 side chain, the catalytic triad geometry is again consistent with a fully active protease.
To probe the flexibility of TtClpP, we used MAS NMR spin relaxation experiments and MD simulations. Site-specific amide 15N R1ρ experiments probe the local amplitudes and time scales of backbone motion and are particularly sensitive to nanosecond-to-millisecond motions (41). The sites with highest 15N R1ρ rate constants are found not only in the loop regions but also at the tip of helix αE (Fig. 4, D and E). One-microsecond-long MD simulations of the TtClpP tetradecamer in the apo and peptide-bound states corroborate this observation, albeit with a greater flexibility at the tip of the helix (Fig. 4, F and G). Enhanced flexibility in this helix has been reported also in EcClpP by methyl-directed solution-state NMR and suggested by other methodologies (16–19, 34, 35). Because of the lack of reliable parametrization of the boronic acid moiety of bortezomib, we focused on simulating tripeptides bound to the active site. It is noteworthy that over the length of three independent simulations of ClpP initially loaded with tri-alanine at all active sites, only a fourth of the peptides remain associated to their designated active sites (fig. S6), consistent with the idea that the products of TtClpP are necessarily weak binders, which ought to be released rapidly to leave the catalytic sites ready for the next degradation reaction.
Bortezomib induces the transition from a tense state to a relaxed state with higher affinity for the substrate
Having identified the bortezomib-binding site of TtClpP by NMR and x-ray crystallography, we turned to ITC to characterize thermodynamically the interaction at different temperatures ranging from 25° to 45°C. At 25°C, far from the ideal catalytic temperature of TtClpP (65°C), only a moderate affinity and symmetric binding isotherm was observed. The binding enthalpy was negative and rather small (see Table 1), thus indicating that binding was mainly entropy-driven. However, when the temperature was increased to 35° and 45°C, considerable changes were observed in the binding isotherm. The asymmetry of the curve and the appearance of two different phases in the binding isotherm along the titration, within the low ligand saturation region, were clear indicators of positive cooperativity (Fig. 5A). Because of the architecture of TtClpP, comprising two heptameric rings forming a tetradecameric assembly, the ITC data were fitted to the Monod-Wyman-Changeux (MWC) model (Fig. 5B), which considers two intrinsically noncooperative conformations, with low and high binding affinity, denoted as tense (T) state and relaxed (R) state, respectively. In the MWC model, ligand binding shifts the equilibrium toward the high-affinity conformation (R) in a concerted cooperative way (i.e., all subunits change their conformation simultaneously; Table 1). This model has been previously transposed to other enzymatic systems, where activation at low ligand concentration and inactivation at high ligand concentration were also observed (42).
Table 1. Thermodynamic parameters of bortezomib binding from ITC and fitting to the MWC model.
KR and KT refer to the bortezomib association constants for the relaxed and tense state. ΔHR, ΔHT, and ΔHγ refer to the binding enthalpy of the R state, T state, and conformational enthalpy between R and T conformations. γ is the equilibrium constant between R and T conformations in the absence of the ligand. N is the fraction of active or binding-competent protein. Relative errors in equilibrium constants are 30%. Absolute error in enthalpies is 0.3 kcal/mol.
| T (°C) | N |
KR (M−1) |
ΔHR (kcal/mol) |
KT (M−1) |
ΔHT (kcal/mol) |
γ |
ΔHγ (kcal/mol) |
| 25°C | 0.97 | 4.5 × 104 | −0.5 | No apparent cooperativity | |||
| 35°C | 0.97 | 8.0 × 104 | −0.9 | 2.8 × 104 | −0.1 | 3.3 | −0.3 |
| 45°C | 1.00 | 6.5 × 104 | −1.5 | 2.5 × 104 | −0.3 | 3.2 | −0.2 |
Fig. 5. Cooperative bortezomib binding detected by ITC experiments.
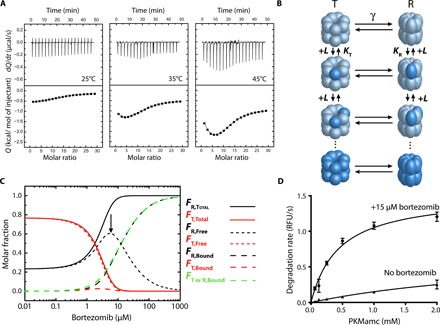
(A) Calorimetric titrations for the interaction of TtClpP (10 μM in the calorimetric cell) with bortezomib (1.4 mM in the injecting syringe) in 50 mM Hepes (pH 7.6) and 50 mM NaCl. Experiments were performed at three different temperatures: 25°, 35°, and 45°C. Thermograms (thermal power as a function of time) are displayed in the upper plots, and binding isotherms (ligand-normalized heat effects per injection as a function of the molar ratio, [L]T/[P]T) are displayed in the lower plots. The binding isotherms were analyzed with the MWC model for TtClpP, consisting of 14 identical subunits, each one containing a single ligand-binding site. Nonlinear least squares regression analysis allows the determination of the following binding parameters (see Table 1): association constants for the R and T states (KR, KT), binding enthalpies to the R and T states (ΔHR, ΔHT), conformational equilibrium constant and conformational enthalpy change between states R and T (γ, ΔHγ), and fraction of active protein (N). (B) MWC model for a 14-mer oligomeric protein. The protein can populate only two conformational states in equilibrium (with equilibrium constant γ): all subunits in R (relaxed, ellipsoidal shape) conformation or all subunits in T (tense, spherical shape) conformation. Subunits have a single ligand-binding site, exhibiting the R conformation a higher binding affinity (KR > KT). Ligand binding occurs through an independent (noncooperative) fashion within an oligomer (ligand-free subunits in light blue and ligand-bound subunits in dark blue). T conformation is favored at low ligand concentration; the higher ligand-binding affinity for R conformation promotes a highly cooperative compulsory concerted conformational change driven by ligand binding and involving all subunits within an oligomer, displacing the equilibrium toward the R conformation. (C) Molar fraction of the different protein species (total, ligand-free, and ligand-bound R and T conformations) as a function of ligand concentration: total fraction of subunits in R conformation (continuous black line), total fraction of subunits in T conformation (continuous red line), fraction of subunits in ligand-bound R conformation (dashed black line), fraction of subunits in ligand-bound T conformation (dashed red line), fraction of ligand-free subunits in R conformation (dotted black line, highlighted with an arrow), and fraction of ligand-free subunits in T conformation (dotted red line). In addition, the fraction of ligand-bound subunits in either R or T conformation is shown (dashed green line). It is obvious that the contribution of subunits in T conformation to the ligand binding is very small (dashed red line). At very low ligand concentration, 77% of the protein subunits are in T conformation and 23% are in R conformation, according to the value of the equilibrium constant γ equal to 3.3. At low ligand concentration, the total fraction of subunits in R conformation increases, while the total fraction of subunits in T conformation decreases due to the T→R conversion. However, both the fractions of ligand-bound subunits in R and T conformation increase due to ligand binding (although the increment in ligand-bound subunits in T conformation is negligible). At high ligand concentration, the total fraction of subunits in R conformation further increases, while that of total subunits in T conformation decreases, and the fraction of ligand-bound subunits in R conformation further increases due to ligand binding, but the fraction of ligand-bound subunits in T conformation decreases due to the T→R conversion. The fraction of ligand-free subunits in R conformation dominates this region, with a maximal population of 60%. This is due to the concerted conversion of subunits within a given protein oligomer (all subunits within an oligomer undergo the conformational change T→R, although not all of the subunits bind a ligand). At very high ligand concentration, the fraction of ligand-bound subunits in R conformation dominates the conformational ensemble, reaching a maximal population of 100%. The R↔T equilibrium shows a switchover or crossover point (R and T are equally populated) at around 2 μM free bortezomib concentration. (D) TtClpP (1 μM complex) peptidase activity was measured as a function of PKMamc concentration in the presence (15 μM) and absence of bortezomib.
Fitting of the ITC data revealed that in the absence of the ligand, 76% of TtClpP exists in the T state, which has a lower affinity for bortezomib (Kd,T = KT−1 = 40 μM). Bortezomib binding induces a concerted transition from the T state to the R state, with a higher affinity for substrate (Kd,R = KR−1 = 15 μM). A model in which we assumed that there is no ligand binding to the subunits in the T conformation (KT = 0, ΔHT = 0) can be ruled out based on significantly worse fitting. The higher affinity of the ligand for the R conformation drives the conversion of T subunits into R subunits. In addition, the compulsory concerted conformational change within a given protein oligomer (i.e., all subunits in a given protein oligomer must undergo the conformational change at once, even if not all of them are occupied by ligand) will further promote the conversion of T subunits into R subunits, thereby increasing the total fraction of R subunits. The Hill coefficient [maximal slope in the log(nLB/(14 − nLB)) versus log[ligand] plot] of TtClpP is 1.3, which is rather small for a tetradecameric protein but was expected considering the small value for the conformational equilibrium constant γ.
Although the MWC model was used 40 years ago for the calorimetric study of the interaction of trout hemoglobin with carbon monoxide using a gas-liquid reaction calorimeter (43), the present work represents the first detailed mathematical description and implementation of the MWC model in ITC, for which the experimental methodology and the mathematical formalism are not the same as those outlined in the aforementioned seminal work.
The MWC model provides a rationale for TtClpP activation by bortezomib. Activation of TtClpP by the inhibitor bortezomib at moderate concentrations occurs as a consequence of the concerted conformational change within the protein tetradecamer, which makes the total fraction of subunits in the enzymatically active R conformation (FR,Total) much larger than the fraction of ligand-bound subunits in R conformation (FR,Bound), i.e., there is no proportionality between FR,Total and FR,Bound (Fig. 5C). At intermediate bortezomib concentration, the fraction of inhibitor-free (active) subunits in an R conformation is maximal and predominates in the speciation distribution (Fig. 5C). Thus, the extents of cooperativity for binding and cooperativity for activity are different [a phenomenon previously reported for allosteric proteins in (44)]. If the conformational changes were not concerted, then the activity of TtClpP would continuously decrease with increasing bortezomib concentration, with no maximal activity at intermediate bortezomib concentration. The increase in the fraction of inhibitor-free R subunits at moderate inhibitor concentration is the molecular event underlying the increase in activity. Similarly, the decrease in the fraction of inhibitor-free R subunits at high inhibitor concentration is the molecular event underlying the decrease in activity at high inhibitor concentration.
The ITC-derived model predicts accurately the activity data. According to the peptide degradation activity measurements (Fig. 1A), the maximal TtClpP activity is found around 12.5 μM of total bortezomib. The ITC-derived MWC model predicts that the maximum fraction of inhibitor-free TtClpP in the R conformation (FR,Free) should be around 6 μM of free bortezomib (Fig. 5C). Considering the population of bound TtClpP at 6 μM free bortezomib concentration (FTorR,Bound = 0.3; Fig. 5C), and taking into account the TtClpP subunit concentration of 10 μM (used in our activity measurements), one can calculate that the concentration of bortezomib bound to TtClpP is 3 μM. Consequently, the total concentration of bortezomib for achieving 6 μM free bortezomib (i.e., the concentration for maximal bortezomib-free subunits in R conformation) is around 9 μM, in excellent agreement with the activity measurements reporting a maximum activity for 12.5 μM bortezomib concentration (Fig. 1A).
Furthermore, to verify that the bortezomib-induced R state has a higher affinity for the substrate, we measured peptide hydrolysis as a function of substrate concentration in the apo state and in the presence of nonsaturating concentrations of bortezomib (15 μM). While there are differences between Kd, a thermodynamic equilibrium constant, versus a purely kinetic constant as KM, in the presence of bortezomib, TtClpP displayed an apparent KM around 400 μM. In its absence, no indications of substrate saturation were observed (Fig. 5D), reflecting the major differences between the bortezomib-bound and apo states. Put together, the activity measurements and ITC data reveal that activation of TtClpP by an inhibitor is achieved by shifting the conformational population toward a higher-affinity R state.
Off-equilibrium MD simulations provide rationale for stabilization of extended state
Activity measurements, ITC data, x-ray structures, and NMR spectroscopic data have established that, by binding to a subset of TtClpP active sites, bortezomib and peptide substrates favor a catalytically more active particle by keeping both the bound and free subunits in an active state, with the catalytic triad poised for catalysis. We attempted to bridge the functional and thermodynamic information from biochemistry and ITC with the structural view by performing in silico experiments. In a nutshell, in these MD simulations, we investigated the energetics of the transition from the extended (active) to the compressed (inactive) state and interrogated whether the presence of a molecule bound to the active site alters the relative stability of the two states. Near-equilibrium MD experiments were performed, in which the two heptameric rings were slowly brought from the extended state to the compressed state using the centers of mass of the two rings as single variable for the steered MD protocol (see Materials and Methods). Figure 6 (A and B) displays the force required for this extended-to-compressed transition, obtained from five independent in silico experiments, as a function of the distance of the two half-rings. Figure 6B shows the work (i.e., the integrated force) for this process. The present simulations have been performed for both the apo state (gray) and a substoichiometrically peptide-loaded state in which only 2 or 3 of the 14 binding sites were occupied with tri-alanine peptide. During the compression process, none of the initially bound peptides were expulsed from their target catalytic sites, and all the secondary structure elements were preserved (Fig. 6, C to E). Compressing ClpP from the extended conformation requires a stronger force in the presence of a substoichiometric amount of alanine tripeptides compared to the empty protein (Fig. 6, A and B).
Fig. 6. In silico pulling experiment of TtClpP from the extended state to the compressed state.
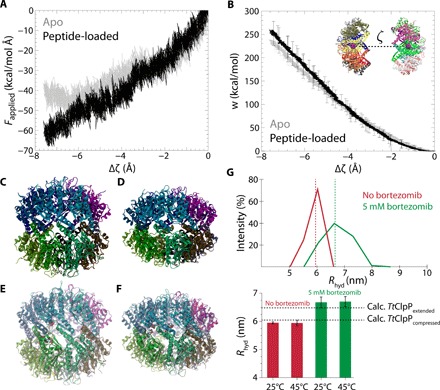
(A and B) Profile of the force (A) and work (B) required for compressing the two rings toward each other as a function of the distance ζ between the rings (shown in the inset, with exaggerated distance for better visibility). Three independent 200-ns simulations were performed for apo TtClpP or TtClpP, in which 2 or 3 of the 14 binding sites were occupied with trialanine peptide (see Materials and Methods). (C and D) Conformations of TtClpP extended (C) and SaClpP compressed (PDB: 3QWD) (D) states observed in crystal structures. (E) Starting conformation of TtClpP in the steered MD simulations (shown as sphere representation). (F) Final conformation of the compressed state at the conclusion of the steered MD. Note that the peptides are still bound and that the structure is in good overall agreement with the compressed state from x-ray diffraction shown in (D). (G) DLS analysis of TtClpP (3 μM) supplemented without or with 5 mM bortezomib. The upper graph shows one representative curve of three replicates measured at 25°C for TtClpP without (red curve) or with 5 mM bortezomib (green curve), with the relative intensity (in %) plotted versus particle size [hydrodynamic radius, Rhyd (in nm)]. The average of the three replicates is indicated using a dotted line. The lower graph depicts average hydrodynamic radii (Rhyd) calculated from three independent experiments measured at 25° and 45°C (0 mM bortezomib, 25°C: 5.946 ± 0.036 nm; 0 mM bortezomib, 45°C: 5.940 ± 0.102 nm; 5 mM bortezomib, 25°C: 6.673 ± 0.192 nm; 5 mM bortezomib, 45°C: 6.702 ± 0.176 nm). Theoretical calculated hydrodynamic radii of compressed TtClpP (6.05 nm) and the extended TtClpP:bortezomib complex (6.47 nm) are indicated using a dotted black line (see also Materials and Methods).
Together, these data provide a rationale for the enhanced stability of the extended conformation in the presence of a ligand: Even substoichiometric amounts of the ligand, i.e., only partially occupied sites, lead to an overall stabilization of all subunits in the active state. The simulations were performed with peptides rather than bortezomib due to the lack of reliable force field parameters for boronic acid and reveal the substrate-induced stabilization of the active extended state. It can be expected that this stability difference of extended over compressed state is even more pronounced with the covalently bound bortezomib.
The MD simulations support the idea that bound ligand, even at substoichiometric amounts, favors a stable extended form, which we believe is the R state modeled in the ITC data, while the apo state is a compressed state. We attempted to validate the compressed-to-extended state transition observed from the MD experiments by performing dynamic light scattering (DLS). Figure 6G shows the hydrodynamic radii (Rhyd) of TtClpP in the absence and presence of 5 mM bortezomib, revealing that addition of bortezomib causes an increase of approximately 8 Å in Rhyd (or 16 Å in diameter) of TtClpP. The experimentally obtained hydrodynamic radii for TtClpP without and with 5 mM bortezomib (5.95 and 6.67 nm, respectively) are in excellent agreement with the theoretical hydrodynamic radii obtained for compressed and extended states of ClpP, supporting the view that the T-to-R transition modeled from the ITC data corresponds to a compressed-to-extended transition.
DISCUSSION
Activation of different ClpPs by previously identified inhibitors has been independently reported (3, 22), but to date, no mechanism for these paradoxical results has been proposed. Bortezomib, just like N-blocked peptide aldehyde Mtb activators reported previously, does not follow the canonical definition for an allosteric activator, i.e., they bind to the protease active site (14, 15) and their effect is not universal to all ClpP homologs. For instance, we show here that although bortezomib activates TtClpP (up to 100 μM), it is, in the same concentration range, a good inhibitor of EcClpP. This result is rather puzzling, considering the high sequence and structural similarity between the two proteins (58% sequence identity).
We show in the present contribution that ClpP activation by inhibitors derives from the intrinsically cooperative character of the ClpP tetradecameric structure. Bortezomib, although blocking a number of active sites, can overall increase the catalytic rate by promoting a concerted transition in the complex, stabilizing the active-state conformation. In line with this observation, the near-equilibrium MD simulations performed in our study suggest that compression of the extended-active conformation of TtClpP is hampered when a fraction of the catalytic sites is occupied by unspecific tripeptide substrates. In the presence of a stronger specific binder such as bortezomib, it could be reasonably anticipated that compression toward the inactive conformation is even more hindered. A comparable mechanism likely underlies MtbClpP1P2 activation, because it has been shown that the peptide activator would sterically clash with a non-active ClpP1 complex, thereby forcing the complex into an active conformation (22).
The similarity between the bortezomib TtClpP structure and the structure of TtClpP with peptides in the active site strongly suggests that our findings reflect an inherent substrate-dependent activation mechanism of the enzyme. Through this mechanism, the presence of the substrate in the ClpP active site promotes the activation of the remaining subunits, allowing efficient and processive protein degradation. Although we were unable to obtain a crystal structure of apo TtClpP, the DLS experiments performed here are consistent with a bortezomib-induced transition from a compressed or compacted state to an extended TtClpP conformation. Addition of bortezomib altered chemical shifts in MAS NMR experiments, possibly pointing also toward the changes in the equatorial region of ClpP, expected for a compressed/extended transition (Fig. 3D).
Our results also support the previously described bidirectional cross-talk between chaperone binding and protease active sites (14, 22, 23, 26–28), because bortezomib is no longer an activator in the presence of the co-chaperone TtClpX and TtClpP partially saturated with bortezomib binds strongly to TtClpX. Significant differences were observed between the activation of TtClpP peptidase activity and proteolytic activity by bortezomib. While peptidase activation displayed a bell-shaped curve, protease activation showed a linear rise in the range of the used bortezomib concentration. Although the process underlying bortezomib activation in the two activities should be common, important differences exist between the two assays. Protein degradation is strictly dependent on substrate access to the proteolytic chamber, because larger substrates cannot diffuse freely toward the proteolytic chamber. Our results showing protein degradation activated by bortezomib demonstrate that modulation of protein entry can also be achieved via binding to the ClpP active site and not exclusively by ClpX or ADEP binding. Protein access to the proteolytic chamber leads to high local concentrations of the substrate present at the ClpP active sites. Although bortezomib activates protein degradation at low concentrations, it must compete with high concentrations of substrate in the proteolytic activity assay, thereby rationalizing the requirement for higher concentrations of drug to achieve activation. The range of bortezomib concentrations used in our proteolytic activity assay likely corresponds to the rising part of the bell curve monitored in the peptidase activity assay and also explains why several ClpP inhibitors are 10-fold more potent in peptidase assays compared with protease assays (45).
It is unclear why the action of bortezomib and activators is not uniformly conserved between different ClpPs. Differences in enzymatic activity are generally more difficult to decode than major structural differences because allosteric pathways often result from subtle changes in conformational dynamics that affect population distributions, which are not readily observed from comparisons of structures. For example, single mutations or truncations in EcClpP N-terminal have been shown to induce reversible inactivation of ClpP (16), and using an indirect methodology based on the association of an EcClpP E14W mutant with EcClpA, the equilibrium constant between an EcClpP active state (R) and inactive state (T) was previously quantified as Keq = 7.5 (37°C) (46), i.e., a value similar to the one we describe here for TtClpP Keq = 3.3 (45°C). This finding is particularly interesting, as it suggests that in the absence of mutations, EcClpP preferentially populates active states, while TtClpP is preferentially inactive (16). The energy barrier between the two states is, however, rather small, with an energy gap of 0.7 kcal/mol between the R and the T conformations in TtClpP. Given the high sequence identity between EcClpP and TtClpP, the key to the difference in the equilibria between active and non-active states must necessarily result from small sequence changes. EcClpP and SaClpP contain a conserved Q131XT/S133 motif at the tip of the turn between helix αE and the β-strand 6 in the handle domain (fig. S7). Here, Gln131 engages in hydrogen bonds with the η1 and η2 nitrogens of Arg170 on a neighboring subunit of the same heptamer ring (intra-ring) and with the backbone oxygen of Glu169 and ε1 oxygen of Gln123 on a monomer of the other ring (inter-ring). The side chain of the conserved Thr133 forms hydrogen bonds with the nitrogen atoms of Gln123 and Lys146 (inter-ring). Gln123 is part of the conserved His-Gln-Pro (HQP) motif, which includes the catalytic triad His122, while Arg170 is the vicinal residue of the catalytic triad Asp171. It is clear that this extended hydrogen bond network restricts the degrees of freedom of the aforementioned catalytic triad residues. It is noteworthy that these residues are conserved neither in TtClpP nor in MtbClpP1. In TtClpP, Gln131 is replaced by a threonine (Thr131), which interacts either with the ε1 nitrogen of Arg170 (intra-ring) or with Gln123 (inter-ring) in the TtClpP:peptide or TtClpP:bortezomib structures, respectively. Ser133 of TtClpP contacts Gln123 and Lys146 (inter-ring) in a similar way as EcClpP Thr133 (fig. S7). The replacement of the conserved glutamine Q131 by the shorter threonine in TtClpP or serine in MtbClpP1 leads to modifications in the hydrogen network that normally sustains the extended helix αE. Structural and MD studies in SaClpP have shown that these key residues are essential for keeping the long helix αE in a straight conformation. In TtClpP, the exclusive hydrogen bond acceptor character and shorter side chain of Thr131 does not allow the formation of a network that simultaneously coordinates two of the members of the catalytic triad. This reduced H-bonding capacity is expected to result in an increased flexibility in that region and likely to an equilibrium shift toward nonfunctional conformations, supported by the flexibility measurements reported here by MAS NMR and MD simulations (Fig. 5). We propose that bortezomib binding through preferential stabilization of the active site (R form) shifts the entire population toward a fully active enzyme.
A different hydrogen bond network is also observed for MtbClp1. In this case, Gln131 is replaced by serine, while Thr133 is replaced by alanine. When purified alone, ClpP1 is an inactive enzyme, and the x-ray structure of the ClpP1 tetradecamer shows that the catalytic triad is not in the active state (13). However, in complex with ClpP2, ClpP1 has been shown to be fully active, likely because MtbClpP2, which contains the conserved QFT motif at the tip of the helix αE, can stabilize ClpP1 by forming a similar network, as has been observed in EcClpP (3).
Cooperativity allows multimeric systems to adapt rapidly to environmental changes by more efficiently converting a biochemical input, i.e., the substrate/ligand concentration, into a biochemical response. Cooperative response to incoming substrates has been reported for other machineries of the protein quality control system. Because protein degradation is key to cell homeostasis, several levels of regulation have evolved to prevent uncontrolled proteolysis that would be deleterious to the cell. ClpP, as well as other complexes like the 20S proteasome, hides their active sites inside an inaccessible catalytic chamber to prevent uncontrolled proteolysis. This architecture is fine-tuned by ClpP-specific co-chaperones, which add another layer of regulation in protease control, either by “opening” the axial pores of the cylinder and translocating unfolded substrates into the chamber or by directly activating the ClpP active sites (2). Several studies have reported that ClpX is able to revert the conformational inhibition of ClpP induced by chemical compounds (14) or even by mutations known to promote ClpP inactive states (23). The fact that one of these inactive-state promoting mutations, namely, the Asp172Asn SaClpP mutant, corresponds to the native form of LmClpP1 and several other ClpPs suggests that this mechanism could be relevant for in vivo ClpP regulation in these species (47). Our data support a similar mechanism of conformational control for TtClpP. Our observation that TtClpX and EcClpX (data not shown) can activate TtClpP further suggests that this enzyme is conformationally attenuated, i.e., in the absence of the chaperone, it is in a latent inactive state. The nonphysiological activation observed with bortezomib is both a consequence of the inherent cooperativity of the protease and the fact that, in the absence of activators, its predominant state is the non-active T state (Fig. 5C). The small molecule–induced conformational shift opens new possibilities for drug development, particularly as not only peptide degradation but also protein degradation is stimulated, as we report here (Fig. 1C).
In summary, using a host of biochemical, biophysical, and structural approaches, we have proposed a mechanism for the activation of ClpP by inhibitors with a key element, namely, a concerted activating conformational change in the supramolecular ClpP tetradecameric structure elicited by the binding of inhibitors at subsaturating concentrations. Hints to a similar allosteric behavior have been reported for the proteasome in the presence of bortezomib (47). While bortezomib inhibits efficiently the chymotrypsin site of the proteasome, it has been observed that it induces a paradoxical activation of the trypsin site of the complex (24).
MATERIALS AND METHODS
Cloning and mutagenesis
TtClpP DNA was directly amplified from T. thermophilus HB8 DNA and cloned in to a pET41c (Novagen) expression vector using Ned I and Xho I restriction enzymes. The expressed gene contained the additional residues LEHHHHHHHH at its C terminus to allow affinity chromatography purification. Full-length ClpX DNA was first cloned into a pET28 vector using Ned I and Hind III restriction enzymes, resulting in residues MGSSHHHHHHSSGLVPRGSH added to the N terminus. While full-length ClpX was insoluble, similar to ClpX orthologs from other species, removal of its N-terminal domain (residues 1 to 54) allowed expression of a soluble ClpX construct (∆TtClpX) (48). Removal of the TtClpX N terminus was performed by GeneCust using Ned I and Hind III as restriction enzymes. TtClpP and ∆TtClpX were expressed in E. coli Bl21RIL cells following overnight expression at 20°C after induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). A TtClpP mutant containing an S97A mutation was constructed using the QuikChange II Site-Directed Mutagenesis Kit from Thermo Fisher Scientific.
Protein purification
GFPssrA was purified as previously described (49). Casein FITC from bovine milk was obtained from Sigma (C0528) and loaded onto a G25 column (GE Healthcare) to remove free fluorescein (49). TtClpP and ∆TtClpX were purified using native NiNTA (nickel-charged nitrilotriacetic acid) affinity chromatography followed by SEC using a 16/600 S200 200 pg Superdex column and a final SEC buffer containing 100 mM tris (pH 8.5), 100 mM NaCl, and 5% glycerol. U-[2H,15N],Ile-δ1-[13CH3]TtClpP was expressed as described (33). When expressed in D2O, TtClpP was insoluble and the protein was recovered from E. coli inclusion bodies using denaturing conditions. Briefly, cells were harvested and resuspended in denaturing buffer containing 8 M urea followed by sonication and centrifugation to remove cell debris. The resulting supernatant was loaded onto a NiNTA column and washed several times with denaturing buffer. The column was then equilibrated in refolding buffer [100 mM sodium phosphate (NaPi) (pH 8), 5% glycerol, and 100 mM NaCl] followed by elution with elution buffer [100 mM NaPi (pH 8), 5% glycerol, 100 mM NaCl, and 400 mM imidazole]. The eluted fractions were loaded into a 16/600 S200 200 pg Superdex column, and the fractions corresponding to native ClpP were pulled. EcClpP was purified as described in (50). FtsZ was a gift of T. Hosek and was purified as previously described (50).
Biochemical assays
Concentrations of ClpP and ClpX were calculated using the molecular weight of a tetradecamer and a hexamer, respectively. PKMamc degradation by TtClpP and EcClpP was measured as described (45). Briefly, TtClpP in Hepes buffer [50 mM Hepes (pH 7.6) and 50 mM NaCl] was mixed with indicated concentrations of PKMamc and bortezomib/Bz-LL [or respective dimethyl sulfoxide (DMSO) volume] at 60°C (or indicated temperature), and fluorescence (340-nm excitation, 462-nm emission) was measured at defined time intervals. FITC-casein degradation by TtClpP was measured as described (45); in a typical assay, FITC-casein (0.15 μM) was mixed with TtClpP14 (0.1 μM) and fluorescence increase (440-nm excitation, 509-nm emission, 495-nm cutoff) was monitored. GFPssrA (0.5 μM) degradation by TtClpX6 (0.2 μM) and TtClpP14 (0.1 μM) in the presence of ATP (10 mM) was monitored, taking advantage of the intrinsic fluorescence of GFPssra (440-nm excitation, 509-nm emission, 495-nm cutoff) at the indicated temperatures. FtsZ degradation by TtClpP was measured at 37°C. Briefly, TtClpP14 (1 μM) was mixed with purified FtsZ (20 μM) in the presence of the indicated concentrations of bortezomib. Aliquots were removed at the indicated time points, and the samples were loaded in a 15% SDS agarose gel. It is important to note the substantial differences in TtClpP concentration used for the different techniques, with 10-fold more protease being used for the FtsZ protein degradation study compared with FITC-casein degradation, as a consequence of the lower sensitivity of SDS–polyacrylamide gel electrophoresis staining versus fluorescein fluorescence methods. TtClpP reaction with ActivX TAMRA-FP was executed according to the manufacturer’s instructions (Thermo Fisher Scientific). TtClpP14 (1 mg/ml) was incubated in the indicated conditions with 1 μl of TAMRA-FR reagent (final concentration of 5 μM). At defined time intervals, aliquots were removed and loaded in a 4 to 20% SDS gel. Gels were visualized in a Bio-Rad ChemiDoc XRS system. TtClpP (20 μM) unfolding was measured in a Varian Cary Eclipse spectrofluorimeter monitoring tryptophan intrinsic fluorescence (280-nm excitation, 350-nm emission). TtClpP14 analytical SEC was performed using a Superdex 200 10/300 GL column.
NMR experiments
MAS solid-state NMR experiments were performed on a Bruker Avance III spectrometer with a 1.3-mm MAS NMR probe tuned to 1H, 13C, 15N and 2H frequencies at a MAS frequency of 55 kHz and an effective sample temperature of 30°C. Backbone resonance assignment was achieved with proton-detected triple-resonance experiments described earlier (33). The assignment was obtained for 97 residues; many more spin systems were observable in the spectra, but spectral overlap challenged further assignment, which likely requires additional high-dimensional experiments. Given that in the present context we were interested in probing interactions and dynamics, the current extent of assignment was sufficient here.
For the measurement of CSPs upon addition of bortezomib, we prepared two samples of U-[2H,13C,15N]–labeled ClpP in H2O-based buffer [100 mM tris (pH 8.5), 5% glycerol, and 100 mM NaCl] containing either 10 mM bortezomib and 5% DMSO or only 5% DMSO. In both cases, the protein (10 mg/ml) was sedimented into a 1.3-mm Bruker MAS NMR rotor, and hCANH spectra were collected in ca. 2.5 days per spectrum. The peak positions in the two spectra were extracted for all assigned backbone sites using CCPN (Collaborative Computing Project for NMR) software (51).
Spin relaxation measurements (15N R1ρ) were performed with proton-detected 3D hCANH experiments. The 3D approach was chosen to resolve sites that would be overlapped in 2D 1H-15N correlation spectra. The experiment comprised a 15N spin-lock element of 15-kHz radiofrequency field strength and delays of 2 and 55 ms. Total experimental times were 3 days per 3D experiment. All spectra were acquired with TopSpin v3.5 and analyzed with CCPN. Relaxation rate constants were obtained using in-house written Python scripts using the peak intensities extracted from the 3D spectra, and error bars were determined using Monte Carlo analysis based on the spectral noise level (determined in CCPN).
Solution-state NMR experiments were performed on a Bruker Avance III spectrometer, equipped with cryogenically cooled TCI probeheads, operating at a magnetic field strength corresponding to 1H Larmor frequencies of 700 MHz. The sample temperature was set to 60°C, and 2D SOFAST-methyl-TROSY NMR experiments were recorded with an adjusted duration depending on the final concentration of the proteins (experimental time ranging from 10 to 120 min per sample) (52). The angle of the proton excitation pulse was set to 30°, and the recycling delay was optimized to 0.6 s to achieve the highest sensitivity (51). U-[2H,15N],Ile-δ1-[13CH3]TtClpP and U-[2H,15N],Ile-δ1-[13CH3]S97ATtClpP (250 μM) were mixed with 2 mM bortezomib (or DMSO equivalent) in tris buffer [100 mM tris (pH 8.5), 100 mM NaCl, and 5% glycerol].
Crystallization of TtClpP and the TtClpP:bortezomib complex
Crystallization trials were set up manually in a sitting drop vapor diffusion setup using 24-well sitting drop plates (Hampton Research) with drop volumes of 1 μl (0.5 μl of protein solution + 0.5 μl of reservoir solution). Purified TtClpP was concentrated to 9 mg/ml. Approximately 1 week after setting up crystal plates at 293 K, large cube-shaped crystals were obtained in a condition containing 0.1 M sodium acetate (pH 4.8) and 30% polyethylene glycol 400 (PEG-400).
To obtain crystals of the TtClpP:bortezomib complex before setting up crystal plates, 5 μl of a 100 mM bortezomib solution dissolved in DMSO was added to 45 μl of TtClpP solution (9 mg/ml), resulting in a final concentration of 10 mM bortezomib and 8.1 mg/ml of TtClpP. Large brick-shaped crystals of the TtClpP:bortezomib complex were obtained at 293 K in the same condition as TtClpP [0.1 M sodium acetate (pH 4.8) and 30% PEG-400]. TtClpP crystals were scooped, transferred to a solution containing 32% PEG-400 and 15% glycerol, and subsequently flash-cooled in liquid nitrogen. Crystals of the TtClpP:bortezomib complex were scooped directly from the drops and flash-cooled in liquid nitrogen. The diffraction data of TtClpP (1.95 Å) and the TtClpP:bortezomib complex (2.70 Å) were collected at the ID30A-MASSIF (TtClpP) and ID23-1 MX (TtClpP:bortezomib) beamlines at the European Synchrotron Radiation Facility (ESRF), Grenoble, France. The obtained datasets were processed using the XDS package (53).
Structure determination of TtClpP and the TtClpP:bortezomib complex
Diffraction data for TtClpP were processed in space group C2 (a = 105.98 Å, b = 162.79 Å, c = 107.95 Å, α = γ = 90°, β = 116.34°). The structure was solved by maximum-likelihood molecular replacement (MR) using Phaser (54) in Phenix (55). The x-ray structure of EcClpP in complex with ADEP1 was used as a search model [PDB: 3KTI; (56)]. Before running Phaser, side chains of the search model were trimmed using the Schwarzenbacher (57) side-chain pruning method in Sculptor (Phenix). The resulting MR solution was refined in Phenix using positional (XYZ) and real-space refinement with noncrystallographic symmetry (NCS) restraints, occupancy refinement, individual ADP (atomic displacement parameter) refinement with TLS (translation liberation screw), and optimized x-ray/stereochemistry and x-ray/ADP weights. Multiple rounds of refinement in Phenix were followed by additional manual model building carried out in Coot (58).
Diffraction data for TtClpP:bortezomib were processed in space group C2221 (a = 135.14 Å, b = 168.74 Å, c = 166.08 Å, α = β = γ = 90°). The structure was solved by maximum-likelihood MR using Phaser in Phenix, taking the refined 1.95-Å TtClpP structure as a search model. Refinement of the TtClpP:bortezomib structure in Phenix was carried out using the same strategy as for TtClpP (positional and real-space refinement with NCS restraints, occupancy refinement, individual ADP refinement with TLS, and optimized x-ray/stereochemistry and x-ray/ADP weights, followed by additional manual model building in Coot).
Isothermal titration calorimetry
The interaction between TtClpP and bortezomib was assessed by ITC using an Auto-iTC200 microcalorimeter (MicroCal-Malvern Panalytical, Malvern, UK). Calorimetric titrations were performed with a 1.4 mM bortezomib solution in the injecting syringe and a 10 μM ClpP solution in the calorimetric cell in 50 mM Hepes (pH 7.6) and 50 mM NaCl. All solutions were properly degassed to avoid bubble formation during stirring. In each titration, a sequence of 19 2-μl injections was programmed, with reference power of 10 μcal/s, initial delay of 60 s, spacing between injections of 150 s, and stirring speed of 750 rpm.
The heat evolved after each ligand injection was obtained from the integral of the calorimetric signal. Experiments were performed in replicates, and data were analyzed using in-house developed software implemented in Origin 7.0 (OriginLab, Northampton, MA). The binding isotherms (ligand-normalized heat as a function of the molar ratio) were analyzed considering the MWC model for ClpP, an oligomeric macromolecule consisting of 14 identical subunits containing a single ligand-binding site each (52). Nonlinear least squares regression allows the determination of the following binding parameters: equilibrium constants (KR, KT, γ), enthalpies (ΔHR, ΔHT, ΔHγ), and fraction of active protein (N). A fitting routine was implemented in Origin 7.0 (OriginLab).
MWC model implemented in ITC
The MWC model considers an oligomeric macromolecule consisting of n identical subunits containing a single ligand-binding site each. Those subunits may adopt two conformations, R (relaxed) and T (tense), with different ligand-binding affinities (KR > KT). Within a given oligomer, all subunits exhibit the same conformation and ligand binding occurs to any subunit independently. However, although T state predominates initially, as a consequence of the higher ligand-binding affinity for R conformation, ligand binding will shift the conformational equilibrium by indirectly eliciting a concerted conformational change of all subunits toward R conformation within the same oligomer at once, resulting in a cooperative binding behavior. The MWC model can only reproduce positive cooperativity, while the Koshland-Nemethy-Filmer (KNF) model can reproduce both negative and positive cooperativities. Both models, MWC and KNF, represent reduced and limited versions of a general allosteric model (44, 59) that takes into account all possible conformational states (e.g., mixed conformations within a protein oligomer) and all possible liganded states, but, in practice, they are considerably more useful and manageable than the general allosteric model.
The binding polynomial for a macromolecule with n subunits that can exist, all at once within a given oligomer, in two different conformations, R and T, and each subunit with a single ligand-binding site, is given by
| (1) |
where PLi represents the protein complex with i ligand molecules bound, and RLi and TLi refer to the complexes of each oligomeric conformational state with i binding sites occupied by ligand molecules. In terms of site-specific binding parameters, the MWC binding polynomial is written as follows
| (2) |
where KR and KT are the site-specific microscopic intrinsic association constants for a binding site in a protein subunit in the R and T states, respectively, with KR > KT, and γ is the equilibrium constant for the conformational equilibrium between the T and R oligomers (γ = [Tn]/[Rn]). Because initially the oligomer in T conformation predominates, the equilibrium constant γ is larger than 1. Each term represents the binding subpolynomial considering n independent ligand-binding sites in each conformational state, R and T.
Valuable information can be extracted from the binding polynomial. In particular, the molar fraction of each liganded species
| (3) |
The two first derivatives of the binding polynomial relative to ligand concentration and temperature provide two fundamental quantities, the average number of ligand molecules bound per oligomeric macromolecule
| (4) |
that can be expressed more conveniently as
| (5) |
and the average molar excess binding enthalpy
| (6) |
that can also be expressed more conveniently as
| (7) |
where ∆HR and ∆HT are the site-specific microscopic intrinsic ligand-binding enthalpies for a binding site in a protein subunit in the R and T states, respectively, and ∆Hγ is the enthalpy associated to the concerted conformational change between the R and T states.
The binding equations corresponding to the binding equilibrium derive from the mass conservation and the chemical equilibrium
| (8) |
The last equation can be transformed into a (n + 1)th-degree polynomial equation in [L] (in this case, a 15th degree polynomial equation) with coefficients that are functions of KR, KT, γ, [P]T, and [L]T and that can be solved numerically (e.g., Newton-Raphson method) for the unknown [L]. Equation 8 must be solved for each experimental point in the calorimetric titration (i.e., each ligand injection j), for which the total concentrations of protein and ligand after each injection j are calculated as follows
| (9) |
where [P]0 is the initial macromolecule concentration in the cell, [L]0 is the ligand concentration in the syringe, V0 is the calorimetric cell volume, and v is the injection volume. Once the free ligand concentration is known, the concentration of each complex after each injection can be calculated (subscript j omitted for the sake of clarity)
| (10) |
The heat effect, qj, associated with each injection j is calculated, considering that it reflects the change in the average excess molar binding enthalpy or in the concentration of all complexes in the calorimetric cell between injection j and j − 1
| (11) |
Last, qj is normalized by the amount of the ligand injected during each injection, and an adjustable parameter qd accounting for the background injection heat (due to solution mismatch, turbulence, etc.) is also included
| (12) |
In addition, a normalizing parameter N is included in Eq. 9 by multiplying [P]0 to account for the active or binding-competent fraction of macromolecule (percentage of protein able to bind ligand).
Nonlinear least squares regression allows the determination of the binding parameters KR, KT, γ, ∆HR, ∆HT, ∆Hγ, N, and qd. A fitting routine was implemented in Origin 7.0 (OriginLab).
For the sake of simplicity, a non-normalized binding polynomial has been used in Eq. 2, where only the ligand-free R state has been taken as the reference state. A renormalized binding polynomial can be constructed taking the subensemble of ligand-free states, R and T, as the reference (ensemble) state, resulting in a standard binding polynomial with a leading term equal to 1 when grouping terms according to powers in [L]. The new binding polynomial is given by
| (13) |
and the only difference in the subsequent development is that a different reference value for the excess average ligand-binding enthalpy is used
| (14) |
The second term in the right-hand side of Eq. 14 after the minus sign is a constant value equal to the average excess enthalpy of the non-liganded fraction of protein (i.e., ligand-free R and T states)
| (15) |
Thus, that term introduces a shift in the enthalpy scale, and an additional constant term in the average excess molar enthalpy makes no difference when calculating the heat effect for each injection, as can be easily proven
| (16) |
A nested MWC cooperativity model for the behavior of ClpP in which each heptamer might undergo a specific concerted conformational change could have been applied to ClpP but that would have added three additional equilibrium constants and three additional enthalpy changes, which would result in overparameterization in the fitting function and correlation/dependency between fitting parameters. In addition, the KNF cooperativity model in which sequential conformational changes are occurring as the oligomer is being occupied by the ligand could have been applied to ClpP, but, again, more parameters should have been considered in the fitting function, resulting in overparameterization. Besides, the KNF model cannot reproduce the activation effect induced by bortezomib because, in the KNF model, the conformational change induced by the ligand is specifically restricted to those subunits binding the ligand, with no possibility of ligand-free subunits undergoing the activating conformational change. Thus, the MWC cooperativity model is the minimal model able to reproduce the behavior of ClpP.
The concentration (and the evolution along a titration) of the different protein species (R and T conformations) can be readily calculated from the previous equations. The total fractions of protein subunits in the conformation R or T, FR,T and FT,T, respectively, are given by
| (17) |
and the fractions of protein subunits in the conformation R or T that are bound to the ligand, FR,B and FT,B, respectively, are given by
| (18) |
The fractions of ligand-free protein subunits in the conformation R or T, FR,F and FT,F, respectively, can be calculated as the following differences
| (19) |
MD simulations
Computational assays
The 3D structure used in the simulations was obtained from TtClpP:peptide, which corresponds to an extended conformation of the protein, bereft of substrate. The missing N-terminal β-hairpin residues were added using Modeller 9.17 (60). Positioning of the seven loops was done concomitantly for each of the upper and the lower hemispheres of the protein. For the protein with bound peptide substrates, use was made of the TtClpP:peptide structure featuring stretches of electron density characteristic of short peptides, over which zwitterionic alanine tripeptides were superimposed. The computational assays consisted of the extended TtClpP, with or devoid of alanine tripeptides bound to the 14 serine active sites of the protein, immersed in a bath of 75,307 water molecules, with a 0.16 M NaCl concentration corresponding to a cubic simulation cell of length equal to 138 Å at thermodynamic equilibrium.
MD protocol
All the MD simulations reported in this study were performed using NAMD 2.12 (61), with the CHARMM36 (62) force field for proteins and lipids, and the TIP3P water model. A Langevin thermostat with a damping coefficient of 1 p s−1 maintained the temperature at 27°C. The Langevin piston method was used to keep the computational assay at a nominal pressure of 1 atm. Covalent bonds involving hydrogen atoms were constrained to their equilibrium length by the Rattle algorithm. The Settle algorithm was used to constrain water molecules to their equilibrium geometry. Long-range electrostatic forces were evaluated using the particle mesh Ewald algorithm with a grid spacing of 1.2 Å, while a smoothed 12-Å spherical cutoff was applied to truncate short-range van der Waals and electrostatic interactions. The r-RESPA multiple time-stepping algorithm combined with a mass-repartitioning scheme was used to integrate the equations of motion with an effective time step of 4 fs for short-range interactions and of 8 fs for long-range electrostatic interactions.
Simulation design
The extended apo-TtClpP was probed in a 1-μs MD simulation. In addition, the computational assay consisting of the protein with zwitterionic alanine tripeptides bound to its 14 serine active sites was examined in three independent, 1-μs MD simulations, using distinct initial conditions. These simulations are aimed at exploring how the presence of the substrate affects the flexibility of TtClpP, alongside the relevance of the alanine tripeptide as a proxy for the peptide chain that binds to the Ser97 active site.
Near-equilibrium pulling experiments
To address how the occupancy of the serine active sites of TtClpP affects the compressibility of TtClpP, we have examined the conformational transition of the protein from its extended state to its compressed state by means of near-equilibrium pulling experiments. Toward this end, a steered MD protocol was used, pulling at constant velocity a stiff spring of force constant equal to 100 kcal/mol Å. The transition coordinate was defined as the Euclidian distance separating the center of mass of the upper hemisphere to that of the lower hemisphere computed from the position of all α-carbon atoms. The length of the transition coordinate was determined on the basis of apo-TtClpP and the structure of the compressed conformation (PDB: 3QWD), and amounts to 8 Å. Three independent, 200-ns-long realizations were performed for the empty protein and three trajectories for ClpP containing either two (in one trajectory) or three (in the other trajectories) zwitterionic alanine tripeptides, pulling irreversibly the two lobes of the protein from an extended state to a compressed one.
Multi-angle laser light scattering
SEC-MALLS experiments were conducted at room temperature on a high-performance liquid chromatography (HPLC) system consisting of a DGU-20 AD degasser, an LC-20 AD pump, a SIL20-ACHT autosampler, a CBM-20A communication interface, an SPD-M20A UV-Vis detector (Shimadzu, Kyoto, Japan), an XL-Therm column oven (WynSep, Sainte Foy d’Aigrefeuille, France), and static light scattering miniDAWN Treos, DLS DynaPro NANOSTAR, and refractive index Optilab rEX detectors. Data were analyzed using version 5.4.3.20 of the ASTRA software (Wyatt Technology, Santa Barbara, USA). TtClpP (1 mg/ml, 100 μl) was injected at 0.5 ml/min on a Superdex 200 10/300 GL column (GE Healthcare), equilibrated with a buffer containing 100 mM tris (pH 8.5), 100 mM NaCl, and 5% glycerol. Bovine serum albumin at 2 mg/ml in phosphate-buffered saline (PBS) buffer was injected as a control in each experiment. The extinction coefficient and refractive index increments for the proteins were calculated from the amino acid sequences using the program SEDFIT.
Dynamic light scattering
TtClpP (3 μM) in a buffer containing 100 mM tris (pH 8.5), 100 mM NaCl, and 5% glycerol without or with 5 mM added bortezomib was measured using a DynaPro NANOSTAR (Wyatt, Santa Barbara, USA) instrument operated at 25° or 45°C and analyzed with the associated DYNAMICS software. Each experiment was performed in triplicate (three times for each of two TtClpP samples, both measured at 25° and 45°C), and hydrodynamic radii were calculated as the average of these replicates. Theoretical hydrodynamic radii of compressed TtClpP [based on PDB: 3QWD (18)] and extended TtClpP (using the TtClpP:bortezomib structure), after modeling of missing residues in the N-terminal β-hairpins and the N and C termini using SWISS-MODEL (63), were calculated using Hydropro10 (64) using a solvent viscosity and a solvent density of 0.0010387 poise and 1.0114 g/cm3, respectively (5% glycerol, 25°C), or 0.00068656 poise and 1.0043 g/cm3, respectively (5% glycerol, 45°C).
Supplementary Material
Acknowledgments
Funding: This work used the platforms of the Grenoble Instruct center (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from INSTRUCT (“Innovative EM/NMR approach for the characterization of the drug target ClpP APPID: 301“), FRISBI (ANR-10-INSB-05-02), and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB). We thank A. Le Roi and C. Ebel for assistance with MALLS and DLS data acquisition. Special thanks to T. Hosek (Institute Biologie Structurale) for FtsZ and T. Akopian and A. Goldberg (Harvard School of Public Health, USA) for testing peptide substrates. Activation of MtbClpP1P2 by bortezomib was first observed by T. Akopian and A. Goldberg. We thank J. Boisbouvier (IBS Grenoble) for hosting H.F. in his team for a related (previously published) work. We thank the ESRF for beamtime at ID30A and ID23-1. Funding: This work was supported by Spanish Ministerio de Economia y Competitividad (BFU2016-78232-P) and Instituto de Salud Carlos III co-funded by European Union (PI15/00663 and PI18/00349, ERDF/ESF, “Investing in your future”). This work was financially supported by the European Research Council (ERC-Stg-2012-311318 to P.S.). J.F. is supported by an EMBO long-term post-doctoral fellowship (ALTF441-2017). Author contributions: J.F. prepared ClpP + inhibitor crystals, processed x-ray diffraction data, refined and analyzed crystal structures, performed MALLS and DLS experiments, prepared figures, and wrote the manuscript. K.W. performed solution NMR experiments, discussed results, and prepared figures. C.C. and F.D. designed, performed, and analyzed the MD simulations and prepared figures. A.H. performed site-directed mutagenesis and protein sample preparation. D.F.G. performed modeling and computational simulations proof of concepts and prepared figures. C.M. prepared ClpP crystals and collected x-ray diffraction data. O.A. performed and analyzed ITC experiments. I.G. designed research and discussed results. A.V.-C. performed and analyzed ITC experiments, prepared figures, discussed results, and wrote the manuscript. P.S. performed and analyzed NMR experiments, designed research, managed and prepared figures, discussed results, and wrote the manuscript. H.F. performed biochemical experiments and sample preparation for NMR and x-ray, managed and prepared figures, discussed results, and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Coordinates and structure factors for TtClpP in complex with peptide or bortezomib have been deposited in the PDB with accession codes 6HWM and 6HWN, respectively.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/9/eaaw3818/DC1
Fig. S1. Biochemical characterization of TtClpP.
Fig. S2. Characterization of GFPssrA degradation by TtClpXP.
Fig. S3. Characterization of TtClpP peptidase activity in the absence and presence of bortezomib.
Fig. S4. Characterization of FITC-casein degradation by TtClpP in the absence and presence of bortezomib and/or TtClpX.
Fig. S5. Ligand validation by 2mFo-DFc, omit, and polder maps.
Fig. S6. Distance RMSD of the 14 alanine tripeptide ligands with respect to their initial bound state in TtClpP for three independent, 1-μs-long MD simulations.
Fig. S7. Analysis of structural differences between TtClpP and EcClpP.
Table S1. Crystallographic data collection and refinement statistics.
Reference (65)
REFERENCES AND NOTES
- 1.Raju R. M., Goldberg A. L., Rubin E. J., Bacterial proteolytic complexes as therapeutic targets. Nat. Rev. Drug Discov. 11, 777–789 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Wang J., Hartling J. A., Flanagan J. M., The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell 91, 447–456 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Akopian T., Kandror O., Raju R. M., UnniKrishnan M., Rubin E. J., Goldberg A. L., The active ClpP protease from M. tuberculosis is a complex composed of a heptameric ClpP1 and a ClpP2 ring. EMBO J. 31, 1529–1541 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fetzer C., Korotkov V. S., Thänert R., Lee K. M., Neuenschwander M., von Kries J. P., Medina E., Sieber S. A., A chemical disruptor of the ClpX chaperone complex attenuates the virulence of multidrug-resistant Staphylococcus aureus. Angew. Chem. Int. Ed. Engl. 56, 15746–15750 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Gaillot O., Pellegrini E., Bregenholt S., Nair S., Berche P., The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35, 1286–1294 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Bhaskaran S., Pharaoh G., Ranjit R., Murphy A., Matsuzaki S., Nair B. C., Forbes B., Gispert S., Auburger G., Humphries K. M., Kinter M., Griffin T. M., Deepa S. S., Loss of mitochondrial protease ClpP protects mice from diet-induced obesity and insulin resistance. EMBO Rep. 19, e45009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole A., Wang Z., Coyaud E., Voisin V., Gronda M., Jitkova Y., Mattson R., Hurren R., Babovic S., Maclean N., Restall I., Wang X., Jeyaraju D. V., Sukhai M. A., Prabha S., Bashir S., Ramakrishnan A., Leung E., Qia Y. H., Zhang N., Combes K. R., Ketela T., Lin F., Houry W. A., Aman A., Al-awar R., Zheng W., Wienholds E., Xu C. J., Dick J., Wang J. C. Y., Moffat J., Minden M. D., Eaves C. J., Bader G. D., Hao Z., Kornblau S. M., Raught B., Schimmer A. D., Inhibition of the mitochondrial protease ClpP as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 27, 864–876 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rathore S., Sinha D., Asad M., Böttcher T., Afrin F., Chauhan V. S., Gupta D., Sieber S. A., Mohmmed A., A cyanobacterial serine protease of Plasmodium falciparum is targeted to the apicoplast and plays an important role in its growth and development. Mol. Microbiol. 77, 873–890 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Brötz-Oesterhelt H., Beyer D., Kroll H.-P., Endermann R., Ladel C., Schroeder W., Hinzen B., Raddatz S., Paulsen H., Henninger K., Bandow J. E., Sahl H.-G., Labischinski H., Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 11, 1082–1087 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Famulla K., Sass P., Malik I., Akopian T., Kandror O., Alber M., Hinzen B., Ruebsamen-Schaeff H., Kalscheuer R., Goldberg A. L., Brötz-Oesterhelt H., Acyldepsipeptide antibiotics kill mycobacteria by preventing the physiological functions of the ClpP1P2 protease. Mol. Microbiol. 101, 194–209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinhäupl K., Brennich M., Kazmaier U., Lelievre J., Ballell L., Goldberg A., Schanda P., Fraga H., The antibiotic cyclomarin blocks arginine-phosphate-induced millisecond dynamics in the N-terminal domain of ClpC1 from Mycobacterium tuberculosis. J. Biol. Chem. 293, 8379–8393 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benaroudj N., Raynal B., Miot M., Ortiz-Lombardia M., Assembly and proteolytic processing of mycobacterial ClpP1 and ClpP2. BMC Biochem. 12, 61 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingvarsson H., Maté M. J., Högbom M., Portnoï D., Benaroudj N., Alzari P. M., Ortiz-Lombardía M., Unge T., Insights into the inter-ring plasticity of caseinolytic proteases from the X-ray structure of Mycobacterium tuberculosis ClpP1. Acta Crystallogr. D 63, 249–259 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Li M., Kandror O., Akopian T., Dharkar P., Wlodawer A., Maurizi M. R., Goldberg A. L., Structure and functional properties of the active form of the proteolytic complex, ClpP1P2, from Mycobacterium tuberculosis. J. Biol. Chem. 291, 7465–7476 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitz K. R., Carney D. W., Sello J. K., Sauer R. T., Crystal structure of Mycobacterium tuberculosis ClpP1P2 suggests a model for peptidase activation by AAA+ partner binding and substrate delivery. Proc. Natl. Acad. Sci. U.S.A. 111, E4587–E4595 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vahidi S., Ripstein Z. A., Bonomi M., Yuwen T., Mabanglo M. F., Juravsky J. B., Rizzolo K., Velyvis A., Houry W. A., Vendruscolo M., Rubinstein J. L., Kay L. E., Reversible inhibition of the ClpP protease via an N-terminal conformational switch. Proc. Natl. Acad. Sci. U.S.A. 115, E6447–E6456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sprangers R., Gribun A., Hwang P. M., Houry W. A., Kay L. E., Quantitative NMR spectroscopy of supramolecular complexes: Dynamic side pores in ClpP are important for product release. Proc. Natl. Acad. Sci. U.S.A. 102, 16678–16683 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geiger S. R., Böttcher T., Sieber S. A., Cramer P., A conformational switch underlies ClpP protease function. Angew. Chem. Int. Ed. Engl. 50, 5749–5752 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Zhang J., Ye F., Lan L., Jiang H., Luo C., Yang C. G., Structural switching of Staphylococcus aureus Clp protease. J. Biol. Chem. 286, 37590–37601 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood N. A., Chung K. Y., Blocker A. M., Rodrigues de Almeida N., Conda-Sheridan M., Fisher D. J., Ouellette S. P., Initial characterization of the two ClpP paralogs of Chlamydia trachomatis suggests unique functionality for each. J. Bacteriol. 201, e00635-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahmen M., Vielberg M.-T., Groll M., Sieber S. A., Structure and mechanism of the caseinolytic protease ClpP1/2 heterocomplex from Listeria monocytogenes. Angew. Chem. Int. Ed. 54, 3598–3602 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Schmitz K. R., Sauer R. T., Substrate delivery by the AAA+ ClpX and ClpC1 unfoldases activates the mycobacterial ClpP1P2 peptidase. Mol. Microbiol. 93, 617–628 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gersch M., Famulla K., Dahmen M., Göbl C., Malik I., Richter K., Korotkov V. S., Sass P., Rübsamen-Schaeff H., Madl T., Brötz-Oesterhelt H., Sieber S. A., AAA+ chaperones and acyldepsipeptides activate the ClpP protease via conformational control. Nat. Commun. 6, 6320 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Groll M., Berkers C. R., Ploegh H. L., Ovaa H., Crystal structure of the boronic acid-based proteasome inhibitor bortezomib in complex with the yeast 20S proteasome. Structure 14, 451–456 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Moreira W., Santhanakrishnan S., Dymock B. W., Dick T., Bortezomib warhead-switch confers dual activity against mycobacterial caseinolytic protease and proteasome and selectivity against human proteasome. Front. Microbiol. 8, 333–336 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balogh D., Dahmen M., Stahl M., Poreba M., Gersch M., Drag M., Sieber S. A., Insights into ClpXP proteolysis: Heterooligomerization and partial deactivation enhance chaperone affinity and substrate turnover in Listeria monocytogenes. Chem. Sci. 8, 1592–1600 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joshi S. A., Hersch G. L., Baker T. A., Sauer R. T., Communication between ClpX and ClpP during substrate processing and degradation. Nat. Struct. Mol. Biol. 11, 404–411 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Singh S. K., Guo F., Maurizi M. R., ClpA and ClpP remain associated during multiple rounds of ATP-dependent protein degradation by ClpAP protease. Biochemistry 38, 14906–14915 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Sass P., Josten M., Famulla K., Schiffer G., Sahl H. G., Hamoen L., Brotz-Oesterhelt H., Antibiotic acyldepsipeptides activate ClpP peptidase to degrade the cell division protein FtsZ. Proc. Natl. Acad. Sci. U.S.A. 108, 17474–17479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee M. E., Baker T. A., Sauer R. T., Control of substrate gating and translocation into ClpP by channel residues and ClpX binding. J. Mol. Biol. 399, 707–718 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson M. W., Singh S. K., Maurizi M. R., Processive degradation of proteins by the ATP-dependent Clp protease from Escherichia coli. Requirement for the multiple array of active sites in ClpP but not ATP hydrolysis. J. Biol. Chem. 269, 18209–18215 (1994). [PubMed] [Google Scholar]
- 32.Vitali Tugarinov P. M., Hwang A., Jason E., Ollerenshaw L. E. K., Cross-Correlated Relaxation Enhanced 1H−13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J. Am. Chem. Soc. 125, 10420–10428 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Fraga H., Arnaud C.-A., Gauto D. F., Audin M., Kurauskas V., Macek P., Krichel C., Guan J.-Y., Boisbouvier J., Sprangers R., Breyton C., Schanda P., Solid-State NMR H-N-(C)-H and H-N-C-C 3D/4D correlation experiments for resonance assignment of large proteins. ChemPhysChem 18, 2697–2703 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye F., Zhang J., Liu H., Hilgenfeld R., Zhang R., Kong X., Li L., Lu J., Zhang X., Li D., Jiang H., Yang C.-G., Luo C., Helix unfolding/refolding characterizes the functional dynamics of Staphylococcus aureus Clp protease. J. Biol. Chem. 288, 17643–17653 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimber M. S., Yu A. Y. H., Borg M., Leung E., Chan H. S., Houry W. A., Structural and theoretical studies indicate that the cylindrical protease ClpP samples extended and compact conformations. Structure 18, 798–808 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Li D. H. S., Chung Y. S., Gloyd M., Joseph E., Ghirlando R., Wright G. D., Cheng Y.-Q., Maurizi M. R., Guarné A., Ortega J., Acyldepsipeptide antibiotics induce the formation of a structured axial channel in ClpP: A model for the ClpX/ClpA-bound state of ClpP. Chem. Biol. 17, 959–969 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szyk A., Maurizi M. R., Crystal structure at 1.9 Å of E. coli ClpP with a peptide covalently bound at the active site. J. Struct. Biol. 156, 165–174 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Alexopoulos J. A., Guarné A., Ortega J., ClpP: A structurally dynamic protease regulated by AAA+ proteins. J. Struct. Biol. 179, 202–210 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Religa T. L., Ruschak A. M., Rosenzweig R., Kay L. E., Site-directed methyl group labeling as an NMR probe of structure and dynamics in supramolecular protein systems: Applications to the proteasome and to the ClpP protease. J. Am. Chem. Soc. 133, 9063–9068 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Kim D. Y., Kim K. K., The structural basis for the activation and peptide recognition of bacterial ClpP. J. Mol. Biol. 379, 760–771 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Schanda P., Ernst M., Studying dynamics by magic-angle spinning solid-state NMR spectroscopy: Principles and applications to biomolecules. Prog. Nucl. Magn. Reson. Spectrosc. 96, 1–46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerhart J. C., Pardee A. B., The effect of the feedback inhibitor CTP, on subunit interactions in aspartate transcarbamylase. Cold Spring Harb. Symp. Quant. Biol. 28, 491–496 (1963). [Google Scholar]
- 43.Barisas B. G., Gill S. J., Thermodynamic analysis of carbon monoxide binding by hemoglobin trout I. Biophys. Chem. 9, 235–244 (1979). [DOI] [PubMed] [Google Scholar]
- 44.Monod J., Wyman J., Changeux J.-P., On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 12, 88–118 (1965). [DOI] [PubMed] [Google Scholar]
- 45.Akopian T., Kandror O., Tsu C., Lai J. H., Wu W., Liu Y., Zhao P., Park A., Wolf L., Dick L. R., Rubin E. J., Bachovchin W., Goldberg A. L., Cleavage Specificity of Mycobacterium tuberculosis ClpP1P2 protease and identification of novel peptide substrates and boronate inhibitors with anti-bacterial activity. J. Biol. Chem. 290, 11008–11020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kress W., Mutschler H., Weber-Ban E., Assembly pathway of an AAA+ protein: Tracking ClpA and ClpAP complex formation in real time. Biochemistry 46, 6183–6193 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Zeiler E., List A., Alte F., Gersch M., Wachtel R., Poreba M., Drag M., Groll M., Sieber S. A., Structural and functional insights into caseinolytic proteases reveal an unprecedented regulation principle of their catalytic triad. Proc. Natl. Acad. Sci. U.S.A. 110, 11302–11307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wojtyra U. A., Thibault G., Tuite A., Houry W. A., The N-terminal zinc binding domain of ClpX is a dimerization domain that modulates the chaperone function. J. Biol. Chem. 278, 48981–48990 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Smith D. M., Fraga H., Reis C., Kafri G., Goldberg A. L., ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell 144, 526–538 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leung E., Datti A., Cossette M., Goodreid J., McCaw S. E., Mah M., Nakhamchik A., Ogata K., el Bakkouri M., Cheng Y.-Q., Wodak S. J., Eger B. T., Pai E. F., Liu J., Gray-Owen S., Batey R. A., Houry W. A., Activators of cylindrical proteases as antimicrobials: Identification and development of small molecule activators of ClpP protease. Chem. Biol. 18, 1167–1178 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D., The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 59, 687–696 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Amero C., Schanda P., Durá M. A., Ayala I., Marion D., Franzetti B., Brutscher B., Boisbouvier J., Fast two-dimensional NMR spectroscopy of high molecular weight protein assemblies. J. Am. Chem. Soc. 131, 3448–3449 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee B.-G., Park E. Y., Lee K.-E., Jeon H., Sung K. H., Paulsen H., Rübsamen-Schaeff H., Brötz-Oesterhelt H., Song H. K., Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism. Nat. Struct. Mol. Biol. 17, 471–478 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Schwarzenbacher R., Godzik A., Grzechnik S. K., Jaroszewski L., The importance of alignment accuracy for molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 60, 1229–1236 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.J. Wyman, S. J. Gill, Binding and Linkage: Functional Chemistry of Biological Macromolecules (University Science Books, 1990). [Google Scholar]
- 60.Šali A., Blundell T. L., Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993). [DOI] [PubMed] [Google Scholar]
- 61.Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K., Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MacKerell A. D., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiórkiewicz-Kuczera J., Yin D., Karplus M., All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998). [DOI] [PubMed] [Google Scholar]
- 63.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F. T., de Beer T. A. P., Rempfer C., Bordoli L., Lepore R., Schwede T., SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ortega A., Amorós D., García de la Torre J., Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J. 101, 892–898 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karplus P. A., Diederichs K., Linking crystallographic model and data quality. Science 336, 1030–1033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/9/eaaw3818/DC1
Fig. S1. Biochemical characterization of TtClpP.
Fig. S2. Characterization of GFPssrA degradation by TtClpXP.
Fig. S3. Characterization of TtClpP peptidase activity in the absence and presence of bortezomib.
Fig. S4. Characterization of FITC-casein degradation by TtClpP in the absence and presence of bortezomib and/or TtClpX.
Fig. S5. Ligand validation by 2mFo-DFc, omit, and polder maps.
Fig. S6. Distance RMSD of the 14 alanine tripeptide ligands with respect to their initial bound state in TtClpP for three independent, 1-μs-long MD simulations.
Fig. S7. Analysis of structural differences between TtClpP and EcClpP.
Table S1. Crystallographic data collection and refinement statistics.
Reference (65)


