Abstract
Based on our previous work defining the molecular rationale for combined targeting of the PI3K and AR pathways in PTEN loss prostate cancer, the first clinical trial was recently reported demonstrating a significant benefit for combination therapy in patients with metastatic prostate cancer. In this phase II trial loss of PTEN was a biomarker predictive of response to combined AKT and AR inhibition. Given that PTEN loss prostate cancers are significantly enriched for ERG genomic rearrangements we evaluated how the aberrant expression of ERG may impact response to PI3K/AR targeted therapy. Here we show that over-expression of ERG in the setting of Pten loss promotes resistance to combined PI3K and AR pathway inhibition with associated maintenance of AR target gene expression. Importantly, following AR knock-out in the setting of ERG over-expression there is maintenance of a subset of AR lineage specific target genes making AR dispensable in this context. This has important clinical implications as even in the setting of the androgen regulated TMPRSS2:ERG genomic rearrangement, ERG expression is never abolished following AR inhibition and may allow for cell survival following AR (lineage) targeted therapies.
Introduction
Genomic loss of the tumor suppressor PTEN resulting in activation of PI3K signaling is one of the most common alterations present in metastatic castrate resistant prostate cancer (CRPC) and several studies have demonstrated that loss of PTEN promotes tumorigenesis in pre-clinical models of prostate cancer (1-3). Thus, there has been tremendous enthusiasm for targeting the PI3K pathway in prostate cancer and other malignancies harboring alterations associated with active PI3K signaling. Despite this, several clinical trials evaluating PI3K pathway inhibitors as single agents in prostate cancer have demonstrated disappointing results (4,5). We previously discovered that the PI3K and AR pathways in prostate cancer regulate one another through reciprocal feedback inhibition, such that inhibition of PI3K promotes relief of feedback resulting in activation of the AR pathway and persistent cell survival (4,6). Our model provided a molecular rationale for co-targeting these critical pathways in prostate cancers harboring loss of PTEN. Based on our work, a phase II clinical trial was performed which demonstrated a significant improvement in median radiographic progression free survival in patients with metastatic CRPC harboring loss of PTEN treated with ipatasertib (AKT inhibitor) and abiraterone (AR inhibitor) compared to abirtaterone alone (11.5 vs 4.6 months, respectively) (7). These exciting results have led to the initiation of a phase III trial evaluating ipatasertib and abiraterone in patients with metastatic CRPC which is currently accruing.
Chromosomal rearrangements leading to the aberrant expression of ETS transcription factors have been identified in over 50% of prostate cancers, with the most common alteration resulting in the over-expression of ERG (8). While many of these ETS family members are rearranged to the promoter regions of androgen regulated genes, a small subset are rearranged to constitutive promoter regions (8). In metastatic CRPC, loss of PTEN has been shown to be enriched for genomic rearrangements leading to the aberrant expression of ERG, and pre-clinical mouse modeling studies have demonstrated that ERG over-expression and loss of Pten cooperate to promote prostate tumor progression (1,9-11). Given that our previous work demonstrated the molecular rationale and therapeutic efficacy of combined PI3K and AR inhibition in ERG negative Pten loss prostate cancer model systems, we now sought to evaluate the impact of ERG on response to combination therapy in the context of loss of Pten.
Materials and Methods
GEM models:
We generated the following crosses of GEM models for our studies Pb-Cre x Ptenlox/lox and Pb-Cre x Rosa26-ERG x Ptenlox/lox. Mice were maintained and genotyped according to standard procedures and in accordance with our IACUC protocol 06–07-012.
MRI imaging:
Mouse prostate MR images were acquired on a Bruker 4.7T Biospec scanner according to our established protocol (6). Tumor volumes were reported for each mouse at initiation of study (T0) and at completion of study (T35). Changes in tumor volumes between T0 and T35 were calculated for individual mice and reported in waterfall plots.
Therapeutic agents:
The AR inhibitor MDV3100, enzalutamide, was synthesized by the MSKCC chemistry core. MDV3100 was used in our studies at an in vitro at a concentration of 10uM and in vivo with a dose of 30 mg/kg/day (6). The PI3K pathway inhibitor NVP-BEZ235 (PI3K and mTORC1/2 inhibitor) was provided by Novartis under a Materials Transfer Agreement (12). BEZ235 was dosed at 30 mg/kg/day for in vivo studies (6).
Prostate cancer organoids and cell lines:
Murine prostates were digested with Collagenase/Hyaluronidase (STEMCELL; 07912) and subsequently with TrypLE (GIBCO). Cells were cultured in suspension for 5–10 days and transferred to collagen-coated plates using established protocols. These cells were authenticated by PCR genotyping protocols established for the Ptenlox/lox and Rosa26-ERG Ptenlox/lox GEM models. The VCaP and 293FT were obtained from ATCC and validated by STR genotyping protocols. MSKPCa2 prostate cancer organoids were generated by our group from a patient with metastatic CRPC and grown using standard organoid culture methodology (13). All cell lines and prostate cancer organoids used in our studies have tested negative for mycoplasma using the MycoProbe Mycoplasma Detection Kit (R&D Systems) within one month of initiating experiments. All cell lines and organoids were freshly thawed and only passaged to achieve the number of cells required for in vitro or in vivo experiments.
Mouse Xenograft procedure:
Tumor grafts measuring 1mm3 or 2.0 × 106 cells resuspended in 100 µL of 1:1 mix of growth media and Matrigel (Corning; 356237) were engrafted or injected into 6–8 weeks old CB17-SCID male mice (Taconic). Tumor size was measured weekly by Peira TM900 (Peira Scientific Instruments). The volume of tumors was calculated using the formula: Volume = Length x Width x Height. A total of 5 mice (10 tumors) per group were used to assay tumor growth in vivo in response to treatment. Treatment was initiated when tumor volumes were approximately 200mm3. All experiments were approved by our IACUC protocol 06–07-012.
Immunoblot:
Cell lysates were prepared in RIPA buffer supplemented with proteinase and phosphatase inhibitors. Proteins were resolved on NuPAGE Novex 4–12% Bis-Tris Protein Gels (Thermo Fisher Scientific) and transferred electronically onto a PVDF 0.45 μM membrane (Millipore). Membranes were blocked in 5% BSA diluted in Tris buffer saline plus 0.1% Tween 20 (TBST) for 1 hour at room temperature and were incubated with primary antibodies in 5% BSA at 4 °C overnight. After 3 washes of 10 min TBST, membranes were incubated with secondary antibodies in 5% BSA diluted in TBST for 1 hour at room temperature. After 3 washes of 10 min in TBST, membranes were developed using an Enhanced Chemiluminescence (ECL) kit (GE Healthcare Life Sciences).
Antibodies:
The following antibodies were used for Western blotting and ChIP: AR (Abcam; ab108341; 1:1,000 for Western blotting), beta-actin (Abcam; ab49900; 1:50,000 for Western blotting), ERG (Abcam; ab92513; 1:1,000 for Western blotting), phospho-AKT (Ser473) (Cell Signaling; 4060L; 1:1,000 for Western blotting), phospho-S6 Ribosomal protein (Cell Signaling; 4856S; 1:1,000 for Western blotting), PTEN (Cell Signaling; 9559L; 1:1,000 for Western blotting), AR (Abcam; ab108341; 1:1,000 for Western blotting), PSA (Cell Signaling; 5365S; 1:1,000 for Western blotting), STEAP1 (Cell Signaling; 9309S; 1:1,000 for Western blotting), and FKBP5(Cell Signaling; 12210S; 1:1,000 for Western blotting).
qRT-PCR:
For qRT-PCR expression analysis, RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (ABI). Power SYBR Master Mix (Qiagen 204057) was used to run PCR on a ViiA7 Real Time PCR System (Life Technologies).
Lentiviral CRISPR/Cas9-mediated knockout:
To knock out AR in mouse organoids, two pairs of single guide RNA (sgRNA) sequences were designed for murine AR using the design tool from the Feng Zhang Lab (MIT) and cloned into the LentiCRISPRv2 (Addgene, 52962). Lentiviruses for sgRNAs were generated in 293FT cells by standard methods using Lipofectamine 2000 (Invitrogen 11668–500). Murine prostate cells were infected with lentivirus for 48–72 hours and selected with puromycin (4 μg/mL) for 7–10 days. The target guides sequences are as follows: sgAr-1: F: CACCGGTGGAAAGTAATAGTCGAT; sgAr-1: R: AAACATCGACTATTACTTTCCACC; sgAr-2: F: CACCGGGTGGAAAGTAATAGTCGA; sgAr-2: R AAACTCGACTATTACTTTCCACCC
Stable gene expression analysis:
ERG (TMPRSS2:T4ERG) was cloned into a flag tagged retroviral vector for infection into VCaP and MSKPCa2 cells.
RNA sequencing:
QIAshredder (Qiagen; 79656) and RNeasy Mini kit (Qiagen; 74106) were used to isolate RNA from cell lines. The standard protocol of the kit was followed. RNA-seq was performed by New York genome center. RNA sequencing libraries were prepared using the TruSeq Stranded mRNA sample preparation kit in accordance with the manufacturer’s instructions. Briefly, 500ng of total RNA was used for purification and fragmentation of mRNA. Following conversion of mRNA to cDNA, DNA was adenylated, ligated to Illumina sequencing adapters, and amplified by PCR (using 10 cycles). Final libraries were quantified using the KAPA Library Quantification Kit (KAPA Biosystems), Qubit Fluorometer (Life Technologies) and Agilent 2100 BioAnalyzer and were sequenced on an Illumina HiSeq2500 sequencer (v4 chemistry) using 2 × 50bp cycles targeting 35M single-end reads per sample. RNAseq data was analyzed for differential gene expression by genotype and treatment (n=3 per condition) using DESEq2. Adjusted p-values less than 0.05 were considered significant at a fold change threshold of 2. GSEA analysis was performed to evaluate enrichment of gene sets across our comparative genotypes and a prostate luminal and basal gene set was directly compared.
Chromatin Immunoprecipitation and Sequencing:
Following established protocols, chromatin isolation from mouse organoid cell lines and immunoprecipitation using antibodies AR, ERG and H3K27ac was performed. Next generation sequencing was carried out on an Illumina HiSeq2000 platform with 50 bp or 100 bp single reads.
To identify the AR and Erg binding sites on chromosome, CHIP-Seq analyses were performed on the parental cells as well as AR or ERG Crispr knockout cell lines. Sequence reads were aligned to mm10 using bowtie program, duplicated reads were excluded for subsequence analysis. MACS2 call peak program was used to call the peaks for each CHIP-Seq samples compared with input sequence using standard parameters. Peaks were annotated using Homer Annotate Peaks program with default parameters identifying promoter, gene body, and intergenic binding sites. Bigwig format files were generated by BEDTools (genomecov) with read number based normalization and presented on IGV.
Histology and immunohistochemistry:
Xenografts were fixed using 4% paraformaldehyde for 2–3 days and embedded using a Leica ASP6025 tissue processor (Leica Biosystems). 5 micron sections were stained on Ventana Discovery XT.
RNAseq and ChIPseq Accession Number:
Sequencing studies have been uploaded into GEO: RNAseq data GEO GSE112469 and ChIPseq data GEO GSE112414.
Results
Aberrant expression of ERG promotes resistance to combined PI3K and AR pathway inhibition.
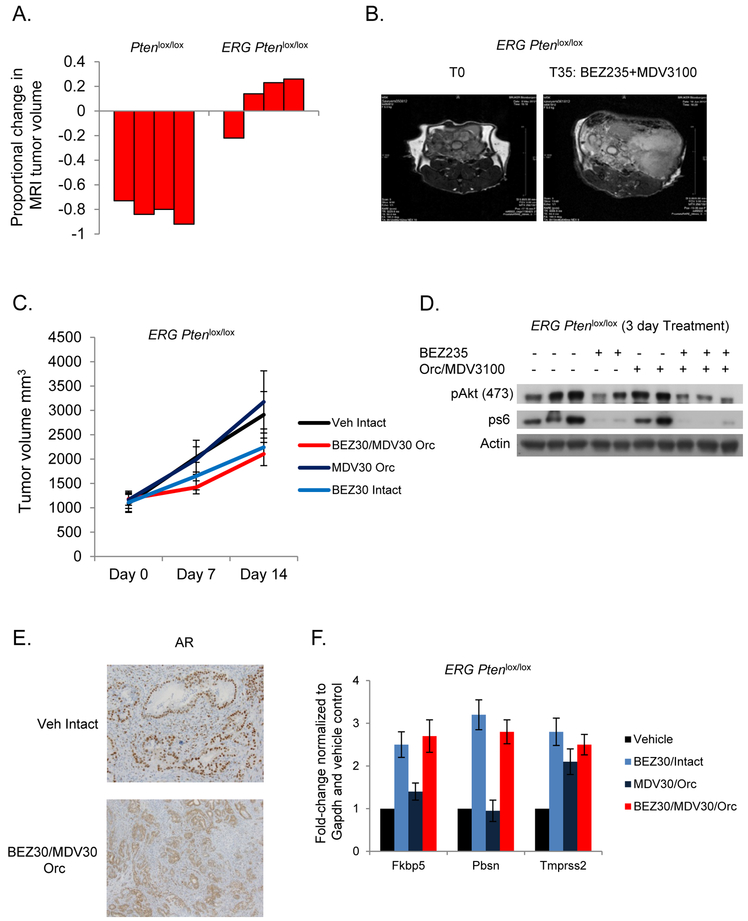
Using our Ptenlox/lox and Rosa26-ERG Ptenlox/lox GEM models (4 mice per group) we conducted a pre-clinical trial of combined PI3K and AR pathway inhibition using BEZ235 (PI3K/mTOR, 30mg/kg/day), castration, and MDV3100 (enzalutamide, AR, 30mg/kg/day) using methodology previously developed by our group (3,6,9). While Pten loss tumors demonstrated a profound response to combined PI3K/AR inhibition, the over-expression of ERG promoted resistance to combination therapy (Figure 1A, 1B). To further explore this across single agent and combination therapies, tumor grafts were established from our Rosa26-ERG Ptenlox/lox GEM model in SCID mice (10 tumors per group) and randomized to receive vehicle, castration + MDV3100, BEZ235, or castration +MDV3100 + BEZ235. Inhibition of the PI3K pathway slowed tumor growth without promoting regression (p-value 0.008), and there was no synergistic effect to combined AR inhibition (p-value = NS) (Figure 1C). Importantly, following 3 days of treatment there was significant repression of downstream PI3K signaling and sequestering of AR in the cytoplasm of cells indicating appropriate achievement of target inhibition (Figure 1D, 1E). Surprisingly however, AR target gene expression was not significantly decreased following AR targeted therapy (Figure 1F). Given the robust interaction of ERG and AR on the cistrome, we hypothesized that ERG may play a role in maintaining AR target gene expression and a luminal lineage independent of AR, thus promoting resistance to combination therapy (9,14).
Figure 1. ERG aberrant expression promotes resistance to PI3K and AR targeted therapy.
A) Waterfall plots for proportional change in tumor volume in Ptenlox/lox (n=4) and ERG Ptenlox/lox mice (n=4) treated with BEZ235 (30mg/kg/day) and MDV3100 (30mg/kg/day). B) Representative MRI images of the ERG Ptenlox/lox mice pre- and post-treatment. C) Tumor volume measurements for ERG Ptenlox/lox tumor grafts (n=10 per group) treated with vehicle, castration+MDV3100, BEZ235, and castration+MDV3100+Bez235. D) Western blot confirming PI3K pathway target inhibition following acute treatment with BEZ235. E) AR immunohistochemistry showing cytoplasmic sequestering following combined PI3K/AR inhibition. F) qRT-PCR of canonical AR target genes across the treatment groups (3 tumors per group) demonstrates maintenance of AR target gene expression despite complete androgen blockade.
Prostate cancer organoids derived from our GEM models recapitulate the molecular and therapeutic phenotypes.
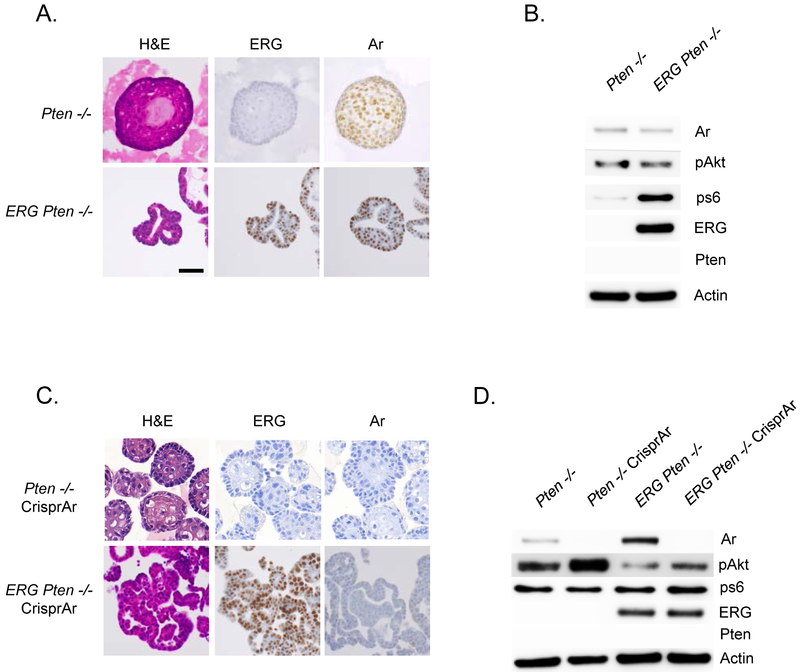
To evaluate the interaction of ERG and AR in established prostate cancer, prostate cancer organoids were derived from Ptenlox/lox and Rosa26-ERG Ptenlox/lox mice (Figure 2A) (13,15). Western blot analysis of these organoids confirmed loss of Pten, activation of pAkt, and over-expression of ERG accordingly (Figure 2B). These prostate cancer organoids recapitulate the histologic and molecular phenotypes of the GEM models they were derived from. Importantly, the injection of ERG Pten−/− organoids into the flank of SCID mice (10 tumors per group), demonstrate similar histologic findings as that observed in our GEM model, and were resistant to combined PI3K/AR pathway inhibition (Supplementary Figure 1A, 1B). To further evaluate the interaction of ERG and AR, Crispr knock-out of AR was performed in our Pten−/− and ERG Pten−/− prostate cancer organoids (Figure 2C). AR knock-out was confirmed by western blot analysis, and as previously reported based on our model of reciprocal feed-back regulation of AR and PI3K signaling, pAkt levels increased following AR knock-out in the Pten−/− organoids (Figure 2D).
Figure 2. Establishment and characterization of Pten−/− and ERG Pten−/− prostate cancer organoids.
A) Tumors derived from our Ptenlox/lox and ERG Ptenlox/lox GEM models were used to establish prostate cancer organoids (3 clones for each genotype) and characterized in 3-D culture conditions for histology, and immunohistochemistry was performed for ERG and AR. B) Western blotting confirming loss of Pten, activation of PI3K pathway, and ERG over-expression. C) Pten−/− and ERG Pten−/− organoids underwent AR Crispr (3 individual clones for each genotype) and were characterized in 3-D culture conditions for histology, and immunohistochemistry was performed for ERG and AR. D) Western blotting confirming loss of Pten, activation of PI3K pathway, ERG over-expression, and loss of AR following AR Crispr.
A subset of AR target genes maintain expression following knock-out of AR in ERG Pten−/− prostate cancer organoids.
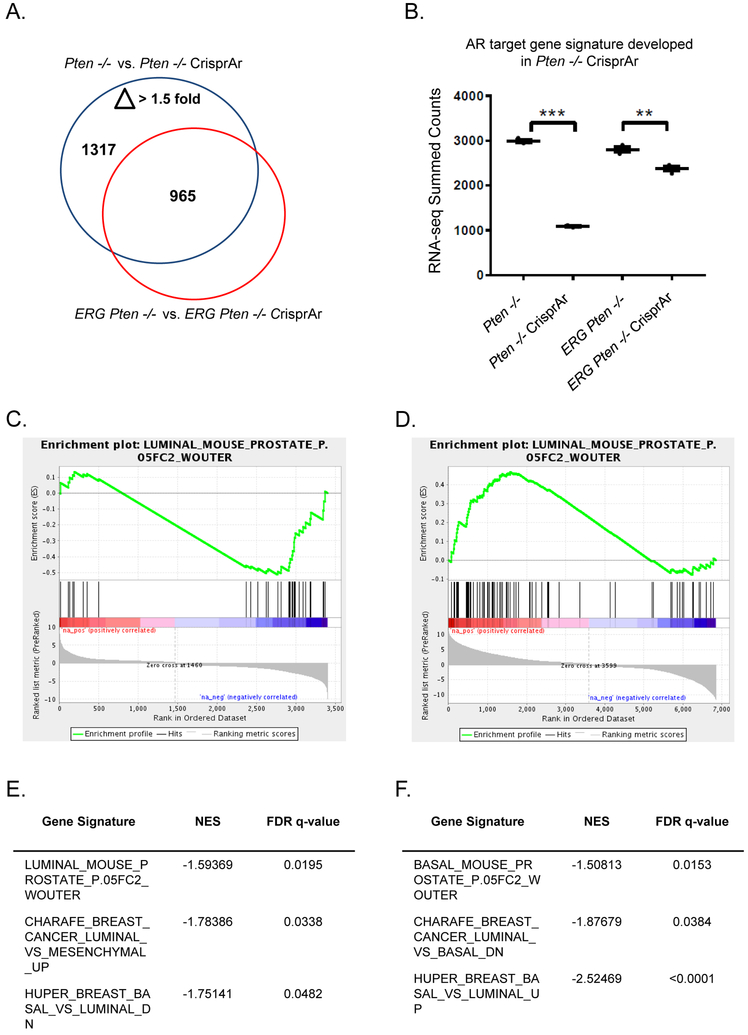
To evaluate the impact of AR knock-out in our Pten−/− and ERG Pten−/− organoids RNAseq analysis was performed. Using our Pten−/− organoids as our baseline, we discovered 1317 differentially expressed transcripts (p<0.05, fold-change > 1.5) following knock-out of AR. We then analyzed the expression of these transcripts in our ERG Pten−/− organoids following AR knock-out and discovered that 352 transcripts (331 known genes) showed no change in expression and many others demonstrated a less significant change (Figure 3A, 3B). Furthermore gene-set enrichment analysis revealed that knock-out of AR in the Pten−/− organoids enriched for the loss of luminal signatures (AR lineage), while knock-out of AR in ERG Pten−/− organoids did not (Figure 3C, 3E). Furthermore, comparisons between AR knock-out amongst Pten−/− and ERG Pten−/− demonstrated loss of luminal signatures and enrichment of basal signatures in the Pten−/− organoids (Figure 3D, 3F). Collectively, our data demonstrates that in the setting of ERG aberrant expression, despite AR inhibition through knock-out, a subset of AR target genes are preserved, particularly those genes represented in luminal signatures.
Figure 3. AR target and luminal gene expression is maintained in the presence of ERG following AR knock-out.
A) RNAseq expression profiling (3 clones for each genotype) for genes differentially expressed greater than 1.5 fold following AR Crispr in Pten−/− and ERG Pten−/− organoids shows substantial overlap in AR targets and a subset of genes that are not differentially regulated in the context of ERG. B) AR gene signature was developed in the Pten−/− organoids following AR knock-out and then applied across our organoid models. C) Gene set enrichment analysis was performed revealing loss of enrichment for the AR regulated luminal gene set in Pten−/− organoids following AR knock-out. D) Gene set enrichment analysis was performed comparing Pten−/− and ERG Pten−/− following AR knock-out which demonstrated enrichment for the luminal signature in the ERG Pten−/− AR Crispr organoids. E) Gene set enrichment analysis revealed loss of enrichment for luminal gene sets in our Pten−/− organoids following AR knock-out. F) Gene set enrichment analysis demonstrating enrichment of basal signatures (loss of luminal signatures) in the Pten−/− versus ERG Pten−/− following AR Crispr.
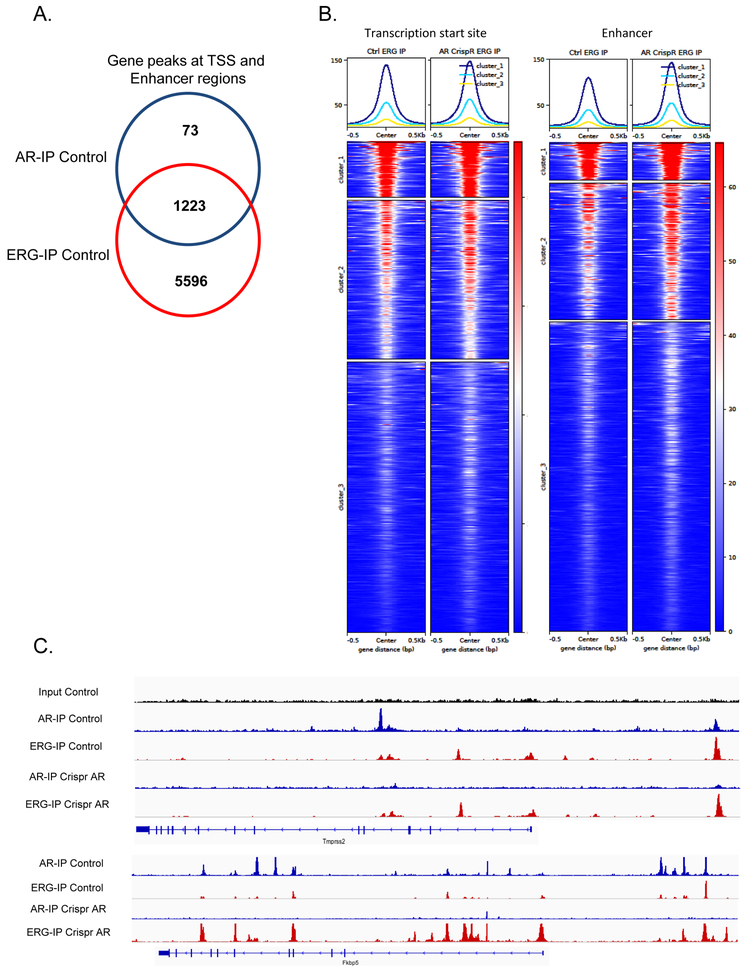
ERG binding increases at enhancer regions following AR inhibition in established prostate cancer.
We have previously shown that ERG over-expression in the prostate prior to tumor development dramatically increases the landscape of AR binding, however little is known how ERG and AR interact at the chromatin level during tumor progression (9). To evaluate the interplay of ERG and AR in established prostate cancer, we performed AR and ERG ChIP seq across our ERG Pten−/− organoids with or without AR and ERG Crispr knock-out. As previously reported there was significant overlap of ERG and AR binding in genes at both enhancer and transcription start site regions (Figure 4A). In fact, the majority of AR gene peaks demonstrated ERG binding peaks as well. While AR knock-out did not impact ERG binding at transcription start sites, at enhancer regions a subset of genes surprisingly demonstrated an increase in ERG binding following knock-out of AR (Figure 4B). For example, at the TMPRSS2 enhancer region we observe both ERG and AR binding at consensus sites and following AR knock-out ERG binding persists, while at the FKBP5 enhancer region, we observe increased ERG binding following AR knock-out (Figure 4C). This data demonstrates that the chromatin interplay between ERG and AR may be more complex in established tumors where ERG demonstrates compensatory binding at a subset of target genes following loss of AR. Our data also suggests that ERG may play a role in regulating its own expression when translocated to the TMPRSS2 locus.
Figure 4. Interaction of ERG and AR on transcription start site and enhancer regions.
A) ChIP seq analyses for ERG and AR shows that in ERG Pten−/− organoids there is significant overlap of AR and ERG binding peaks at transcription start site and enhancer regions. B) AR knock-out revealed an increase in ERG binding at enhancer regions in a subset of genes. C) Integrated genomics view of AR and ERG chromatin binding at the enhancer regions of Tmprss2 and Fkbp5.
Maintenance of ERG expression promotes resistance to combined PI3K/AR inhibition in an ERG dependent prostate cancer model.
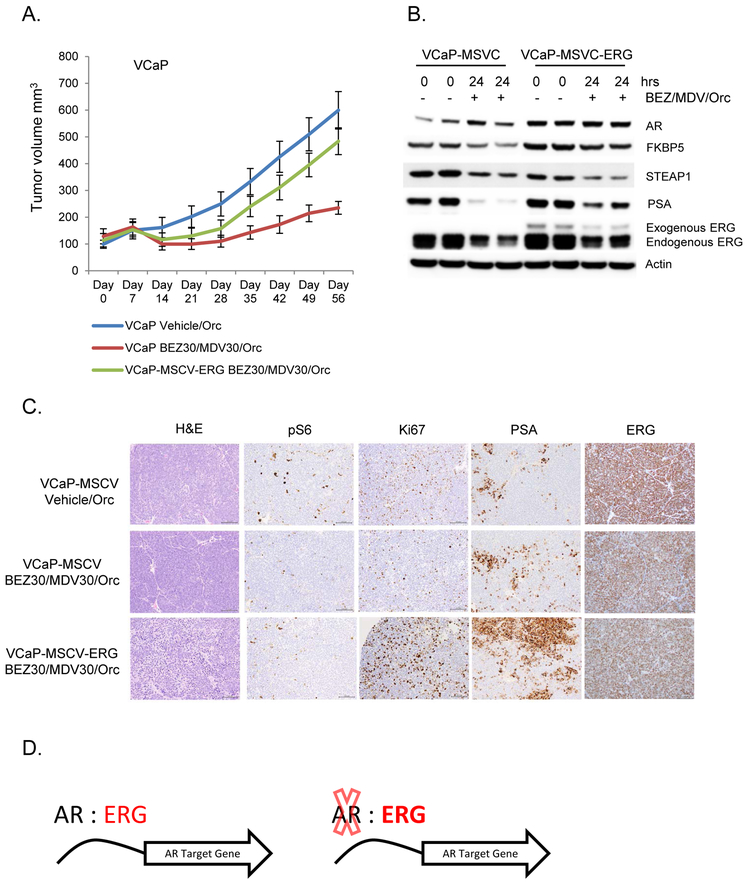
To evaluate the impact of ERG expression on promoting resistance to combined PI3K/AR pathway inhibition in human prostate cancer model systems, we constitutively over-expressed Flag tagged ERG or vector control in the VCaP cell line which harbors the TMPRSS2:ERG genomic rearrangement and established xenograft models (10 tumors per group) for pre-clinical therapeutic studies (Supplementary Figure 1C). Similar to that observed in our GEM model studies, the over-expression of ERG, even at modestly low levels, promoted resistance to combined PI3K and AR inhibition (p-value <0.001) (Figure 5A). Interestingly, over-expression of ERG promoted increased expression of the AR target genes PSA and FKBP5 which remained persistently higher following acute inhibition of PI3K and AR, highlighting the interplay of ERG and AR on driving AR target gene expression (Figure 5B). The expression of exogenous ERG promoted the increased expression of endogenous TMPRSS2:ERG pre- and post-treatment compared to vector control cells. We have shown that both AR and ERG bind at the enhancer region of TMPRSS2 and thus ERG may play a role in supporting it’s own transcription from the TMPRSS2 translocated enhancer region (Figure 4C). Furthermore, in the vector control cells, complete androgen blockade reduced but did not abolish the AR regulated expression of TMPRSS2:ERG and all end of study tumors across the different cohorts displayed diffuse ERG expression by IHC (Figure 5C). While pS6 levels were repressed in the PI3K/AR treatment groups, Ki67 and PSA staining was notably increased in the tumors with exogenous ERG over-expression despite combined PI3K/AR targeted therapy (Figure 5C). Furthermore, over-expression of ERG in the androgen dependent prostate cancer organoid MSKPCa2 promoted enhanced cell proliferation in the presence of enzalutamide (MDV3100) compared to vector control (Supplementary Figure 2) (13). Importantly, over-expression of ERG also resulted in maintenance of the AR target genes PSA and STEAP1 in the MSKPCa2 prostate cancer organoids in the presence of enzalutamide (Supplementary Figure 2).
Figure 5. Over-expression of ERG promotes resistance to combined PI3K and AR inhibition in a TMPRSS2:ERG pre-clinical model.
A) VCaP cells harboring the TMPRSS2: ERG genomic rearrangement display resistance to PI3K (BEZ235 30mg/kg/day) and AR (castration+enzalutamide 30mg/kg/day) pathway inhibition following modest exogenous over expression of ERG (n=10 tumors per group). B) Western blot following acute in vivo treatment with PI3K and AR pathway inhibition. C) Hisotlogic and immunohistochemical analyses of VCaP tumors. D) Model of ERG and AR interaction in established prostate cancers.
Our data provides a framework where in established prostate cancers harboring ERG genomic rearrangements, ERG and AR chromatin binding significantly overlaps and ERG is capable of maintaining a subset of AR target genes in the absence of AR to maintain luminal lineage and promote resistance to AR targeted therapies (Figure 5D).
Discussion
Loss of the tumor suppressor PTEN, resulting in activation of the PI3K pathway, has been shown to promote resistance to androgen ablation and AR targeted strategies in a variety of pre-clinical models, but the implications of this in patients with metastatic prostate cancer has been less consistent (6,16). Several studies have demonstrated that loss of PTEN is enriched in metastatic CRPC compared to primary prostate cancers, and loss of PTEN has been associated with less favorable response to second-generation AR pathway targeted therapies (7,17,18). Collectively, this data has led to recent clinical trials investigating the role of combined PI3K and AR pathway inhibition in metastatic prostate cancers demonstrating loss of PTEN. However, it has not been well studied how ERG genomic rearrangements and the complex interaction of ERG and PTEN, which are concomitantly enriched in prostate cancer, impact the response to AR or PI3K targeted therapies. Here we show in established pre-clinical model systems that the expression of ERG in the background of Pten loss promotes resistance to combined PI3K and AR inhibition in a dose dependent manner. Our data is directly applicable to the small subset of prostate cancers where ERG is rearranged to non-androgen regulated gene promoter regions, leading to constitutive over-expression. However, even in the setting of the most common TMPRSS2:ERG genomic rearrangement, we show that ERG expression is not abolished by second-generation AR targeted therapies, even modest over-expression of ERG is sufficient to promote resistance, and ERG expression is restored as resistance develops. Furthermore, we show that ERG and AR significantly overlap at the chromatin level and that in the absence of AR, ERG maintains the expression of a subset of AR target genes, especially those enriched in luminal signatures, thus negating the global phenotype of AR inhibition.
The results of our study are in slight contrast to the data reported by Blee et al, which found that ERG promoted modest sensitivity to enzalutamide in the context of Pten and Tp53 loss (14). However, these results may be explained by the fact that in their studies, ERG was expressed under the control of the ARR2PB promoter, which is significantly more androgen dependent compared to the endogenous TMPRSS2 promoter that ERG is commonly rearranged to. Thus, in the context of ERG dependency, loss of ERG expression as driven by ARR2PB may be a driving force in the modest sensitivity of enzalutamide in this model system. Importantly, these tumors still displayed growth throughout treatment and following 3 weeks of therapy were found to express ERG in the growing population of cells, and similar to our findings, these investigators also found that ERG promoted a luminal phenotype (14). Additionally, a recent study by Knuuttila et al, reported that ERG positive prostate cancers had enhanced androgen-regulated gene expression possibly through activation of testosterone independent DHT synthesis (19). Thus in TMPRSS2:ERG rearranged tumors activation of AR target genes and maintenance of ERG expression is crucial.
The androgen receptor plays a dominant role in prostate cancer and is a driver luminal differentiation (20,21). Previous studies have demonstrated that inhibition of AR promotes loss of a luminal phenotype with associated dysregulation of luminal associated genes, and therapeutic efficacy for androgen dependent prostate cancers (20). Furthermore, as androgen dependent prostate cancers evolve in the setting of AR targeted therapies, a subset will dedifferentiate to an AR negative neuroendocrine or double negative phenotype as a mechanism of resistance (22,23). Here we show that in addition to AR, ERG is capable of maintaining expression of genes associated with luminal signatures independent of AR and promoting resistance to AR lineage targeted therapies, this may explain in part the low frequency of ERG genomic rearrangements in AR negative neuroendocrine prostate cancers (1).
Based on the exciting results of the ipatasertib and abiraterone phase II clinical trial there is tremendous enthusiasm for combining PI3K and AR pathway inhibitors in patients with prostate cancers harboring loss of PTEN (7). Our data would suggest that ERG positive prostate cancers harboring loss of PTEN will be less responsive to combination therapy, and as the clinical trials mature this can be directly addressed in patients. Furthermore, our data suggests the optimal way of inhibiting ERG activity may not be through AR targeted strategies, highlighting the importance of further understanding the molecular biology of ERG in prostate cancer and discovering more potent ways of targeting ERG therapeutically.
Supplementary Material
Acknowledgements
A special thanks to members of the Chen, Sawyers, and Rosen labs for providing informative discussion. B.S.C. and N.M. designed the study. B.S.C., N.M., and D.C. performed the mouse experiments. B.S.C., N.M., D.G., and Y.S.L. performed the in vitro experiments. H.H. performed the RNAseq analyses. W.H. performed the ChIPseq analyses. B.S.C. and N.M. prepared the manuscript. This work was funded in part through: NIH/NCI Prostate SPORE P50-CA092629–14, NIH/NCIR01-CA182503–01A1 (B. Carver, N. Mao, YS. Lee.), PCF Challenge Award (B. Carver, Y. Chen) and the MSKCC NIH/NCI Cancer Center Support Grant P30 CA008748. Funding through the STARR Cancer Consortium (Y.Chen, B. Carver) allowed for establishment of a prostate organoid core to assist in our experiments. All authors approved of the final manuscript.
Footnotes
Declaration of Interests: No authors have any conflict of interest
References
- 1.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell 2015;162:454. [DOI] [PubMed] [Google Scholar]
- 2.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18:11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol 2003;1:E59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rathkopf DE, Larson SM, Anand A, Morris MJ, Slovin SF, Shaffer DR, et al. Everolimus combined with gefitinib in patients with metastatic castration-resistant prostate cancer: Phase 1/2 results and signaling pathway implications. Cancer 2015;121:3853–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham L, Banda K, Torres A, Carver BS, Chen Y, Pisano K, et al. A phase II study of the dual mTOR inhibitor MLN0128 in patients with metastatic castration resistant prostate cancer. Invest New Drugs 2018;36:458–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 2011;19:575–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bono JS, De Giorgi U, Nava Rodrigues D, Massard C, Bracarda S, Font A, et al. Randomized Phase II Study of Akt Blockade With or Without Ipatasertib in Abiraterone-Treated Patients With Metastatic Prostate Cancer With and Without PTEN Loss. Clin Cancer Res 2018 [DOI] [PubMed] [Google Scholar]
- 8.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 2007;448:595–9 [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med 2013;19:1023–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet 2009;41:524–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet 2009;41:619–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maira SM, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther 2008;7:1851–63 [DOI] [PubMed] [Google Scholar]
- 13.Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014;159:176–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blee AM, He Y, Yang Y, Ye Z, Yan Y, Pan Y, et al. TMPRSS2-ERG Controls Luminal Epithelial Lineage and Antiandrogen Sensitivity in PTEN and TP53-Mutated Prostate Cancer. Clin Cancer Res 2018;24:4551–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 2014;159:163–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sackmann Sala L, Boutillon F, Menara G, De Goyon-Pelard A, Leprevost M, Codzamanian J, et al. A rare castration-resistant progenitor cell population is highly enriched in Pten-null prostate tumours. J Pathol 2017;243:51–64 [DOI] [PubMed] [Google Scholar]
- 17.Abida W, Armenia J, Gopalan A, Brennan R, Walsh M, Barron D, et al. Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making. JCO Precis Oncol 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Attard G, Swennenhuis JF, Olmos D, Reid AH, Vickers E, A’Hern R, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res 2009;69:2912–8 [DOI] [PubMed] [Google Scholar]
- 19.Knuuttila M, Mehmood A, Maki-Jouppila J, Ryberg H, Taimen P, Knaapila J, et al. Intratumoral androgen levels are linked to TMPRSS2-ERG fusion in prostate cancer. Endocr Relat Cancer 2018;25:807–19 [DOI] [PubMed] [Google Scholar]
- 20.Wu CT, Altuwaijri S, Ricke WA, Huang SP, Yeh S, Zhang C, et al. Increased prostate cell proliferation and loss of cell differentiation in mice lacking prostate epithelial androgen receptor. Proc Natl Acad Sci U S A 2007;104:12679–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipianskaya J, Cohen A, Chen CJ, Hsia E, Squires J, Li Z, et al. Androgen-deprivation therapy-induced aggressive prostate cancer with neuroendocrine differentiation. Asian J Androl 2014;16:541–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov 2011;1:487–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R, et al. Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell 2017;32:474–89 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.