Abstract
Molecular mechanisms underlying the formation of multiple classes of transport carriers or vesicles from Golgi and endosomal membranes remain poorly understood. However, one theme that has emerged over three decades is the dramatic influence of membrane lipid remodeling on transport mechanisms. A large cohort of lipid transfer proteins, lipid transporters and lipid modifying enzymes are linked to protein sorting, carrier formation and SNARE-mediated fusion events. Here we focus on one type of lipid transporter, phospholipid flippases in the type IV P-type ATPase (P4-ATPase) family, and discuss recent advances in defining P4-ATPase influences on membrane remodeling and vesicular transport.
Introduction
Transverse movement of phospholipid between leaflets of a protein-free membrane bilayer (flip-flop) is an energetically unfavorable and slow process [1]; however, rates of lipid flip-flop can be greatly accelerated by lipid transporters in biological membranes called flippases, floppases or scramblases [2]. Flippases are ATP-powered pumps from the P4-ATPase family that transport specific lipid substrates from the exofacial leaflet (extracellular leaflet of plasma membrane or luminal leaflet of Golgi/endosome membranes) to the cytosolic leaflet. Floppases are ABC transporters that catalyze lipid transport in the opposite direction and scramblases mediate energy-independent bi-directional lipid transport [2]. Most of the flippases are αβ heterodimers formed from a catalytic α subunit (the P4-ATPase) and a β subunit from the Cdc50 (TMEM30A) protein family [3]. The α subunit nomenclature is inconsistent between species, but P4-ATPases are designated ATP8A1 through ATP11C in mammals; ALA1 through ALA12 in Arabidopsis thaliani; Drs2, Neo1, Dnf1, Dnf2, Dnf3 in Saccharomyces cerevisiae; and TAT-1 through TAT-5 in C. elegans. No high-resolution structure is available for a P4-ATPase, but homology models for the catalytic subunit have been generated to provide a framework for structure/function studies into how these enzymes transport their substrate [4].
Influence of phospholipid flippases on membranes
The unidirectional transport of lipid across a membrane bilayer by a P4-ATPase has a number of consequences. Removal of phospholipid from the exofacial leaflet coupled with its addition to the cytosolic leaflet creates an imbalance in surface area between the leaflets, which imparts molecular crowding stress on the cytosolic leaflet, and a packing defect stress on the exofacial leaflet [5]. A number of P4-ATPases transport phosphatidylserine (PS), thereby displacing negative charge to the cytosolic leaflet, significantly changing the electrostatic properties of the membrane, and exacerbating the crowding stress by concentration of negatively charged lipid. The interleaflet phospholipid gradient stores potential energy that the cell can harness to drive membrane bending towards the cytosol (positive curvature), to displace lipids capable of spontaneous flip-flop (cholesterol, ceramide, diacylglycerol) to the exofacial leaflet, to facilitate extraction of lipid from the cytosolic leaflet by lipid transfer proteins, or to transduce signals by activation of a scramblase and dissipation of the gradient (Figure 1). In addition, phospholipid asymmetry generated by P4-ATPases influences the activity of other transporters and channels in the membrane, the association of soluble proteins with the membrane, and the biophysical properties of the membrane [6–9].
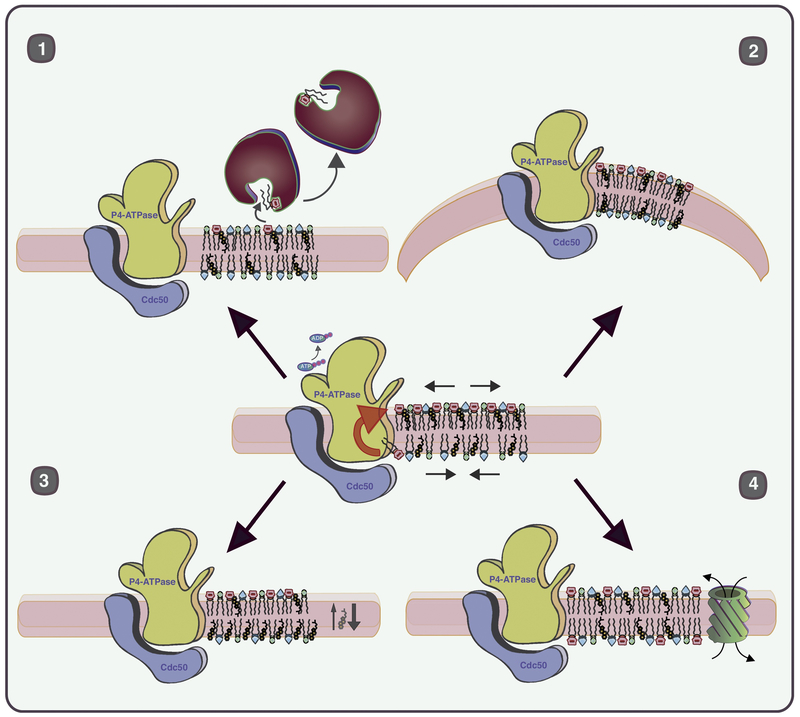
Figure 1. P4-ATPase-mediated lipid transport imparts stress on membranes that can be relieved through membrane bending, induced flop of other lipids, exchange of lipid by transfer proteins, or activation of a scramblase.
P4-ATPases convert the chemical potential energy in ATP to potential energy stored in a lipid gradient within the membrane. In the example shown, the flippase-catalyzed unidirectional transport of PS creates molecular crowding stress in the cytosolic leaflet, packing defect stress in the luminal leaflet, and a charge difference between the leaflets. The potential energy in the membrane can be used by the cell to drive at least four different processes. 1) Exchange of cytosolic leaflet lipids with a bulky headgroup (such as phosphatidylinositol-4-phosphate or PS) for sterol or ceramide by lipid transfer proteins can relieve molecular crowding stress and facilitate membrane remodeling in the Golgi (sphingolipid synthesis and sterol loading). 2) Membrane bending to support budding or scission of protein transport carriers. 3) Displacement of lipids capable of spontaneous flip-flop to the luminal leaflet. Cholesterol is an example of a lipid that flip-flops rapidly between leaflets in an energy-independent manner. Coupling of PS flip to the cytosolic leaflet with cholesterol flop to the luminal leaflet could play an important role in lateral segregation of sterol, sphingolipid and protein in raft-like structures for sorting into exocytic carriers. 4) Rapid exposure of PS in the outer leaflet of cells through activation of a scramblase. This signaling event plays a critical role in apoptosis, blood clotting, immune system suppression, fusion of myoblasts and bone formation.
Several observations support the biological significance of these effects of P4-ATPase catalyzed lipid transport on vesicular transport. Golgi cisternae and endosomes are highly curved tubular-vesicular structures in budding yeast, but drs2 and neo1 mutants display a loss of Golgi curvature and protein transport defects [10,11]. In addition, the PS flippase activity of Drs2 is required to generate the curvature and anionic surface charge on these membranes needed for membrane recruitment of an ArfGAP (Gcs1) via its curvature and charge-sensitive ALPS (ArfGAP Lipid Packing Sensor) motif [9]. The PS flippase activity of ATP8A1 and ATP8A2, mammalian orthologs of Drs2, is similarly required to recruit EHD1 (Eps15 Homology Domain-containing protein 1) to recycling endosomes [12]. TAT-5 deficiency in C. elegans causes substantial shedding of plasma membrane derived extracellular vesicles [13,14]. This observation suggests that TAT-5 flippase activity normally counterbalances a force on the plasma membrane that promotes outward bending (negative curvature). P4-ATPases are required to support COPI-[11,15], clathrin adaptor- [10,16–19], and retromer-dependent trafficking routes [20–22], suggesting the coat proteins are taking advantage of flippase membrane-bending power to facilitate carrier formation.
Many different transport factors are argued to drive the membrane curvature needed for carrier formation (such as coat proteins, small GTPases and BAR-domain proteins [23]) and so it is difficult to tease out the precise contributions of any one protein to membrane deformation. For example, P4-ATPase loss of function trafficking defects could be a direct result of a loss of curvature in the donor membrane, or a secondary effect of perturbing the activity of other membrane-bending, cargo sorting, or membrane scission factors. However, a recent study demonstrates a gain-of-function condition where a P4-ATPase activity induces membrane curvature [24]. Expression the phosphatidylcholine (PC) flippase ATP10A in cells that do not normally express this P4-ATPase induces tubulation of the plasma membrane. PC is normally enriched in the plasma membrane outer leaflet of cells and its redistribution to the cytosolic leaflet should impart stress on the membrane to drive curvature. It remains to be determined how ATP10A influences membrane architecture and endocytosis in cells where it is normally expressed. In contrast, several other P4-ATPases are known to play critical roles in protein trafficking and are functionally intertwined with well-characterized transport machineries.
Trafficking pathways requiring ATP8A1, ATP8A2 and TAT-1
The group of orthologous PS/phosphatidylethanolamine (PE) flippases have been linked to endosomal recycling pathways in mammalian cells [12] and C. elegans [25,26](Figure 2). Depending on the cell type, ATP8A1 and ATP8A2 localize to endosomes, Golgi or the plasma membrane, although the primary site of residence appears to be Rab11-positive recycling endosomes (REs) [3,12]. Depletion of ATP8A1 in COS-1 cells results in a defect in transferrin recycling and its accumulation in REs, and a defect in shiga toxin transport from REs to the Golgi [12]. Morphologically, REs accumulates long thin tubules in ATP8A1 depleted cells and phenocopies cells depleted for EHD1, which is thought to facilitate fission of tubular carriers. In fact, EHD1 is a PS-binding protein with preference for curved membranes and its recruitment to the RE requires ATP8A1 activity [12]. These observations are consistent with prior reports showing a critical role for TAT-1 in RME-1 (EHD1 ortholog)-dependent endosomal protein recycling in C. elegans, although RME-1 recruitment to tat-1 endosomes appeared normal [25–27]. ATP8A2 is expressed in neurons, but it can replace the endocytic recycling function of ATP8A1 in COS-1 cells and primary neurons from Atp8a2−/− mice accumulate transferrin receptors internally [12]. Thus, ATP8A1, ATP8A2 and TAT-1 likely function similarly to support endocytic recycling by recruiting EHD1/RME-1 to help drive scission of tubular carriers.
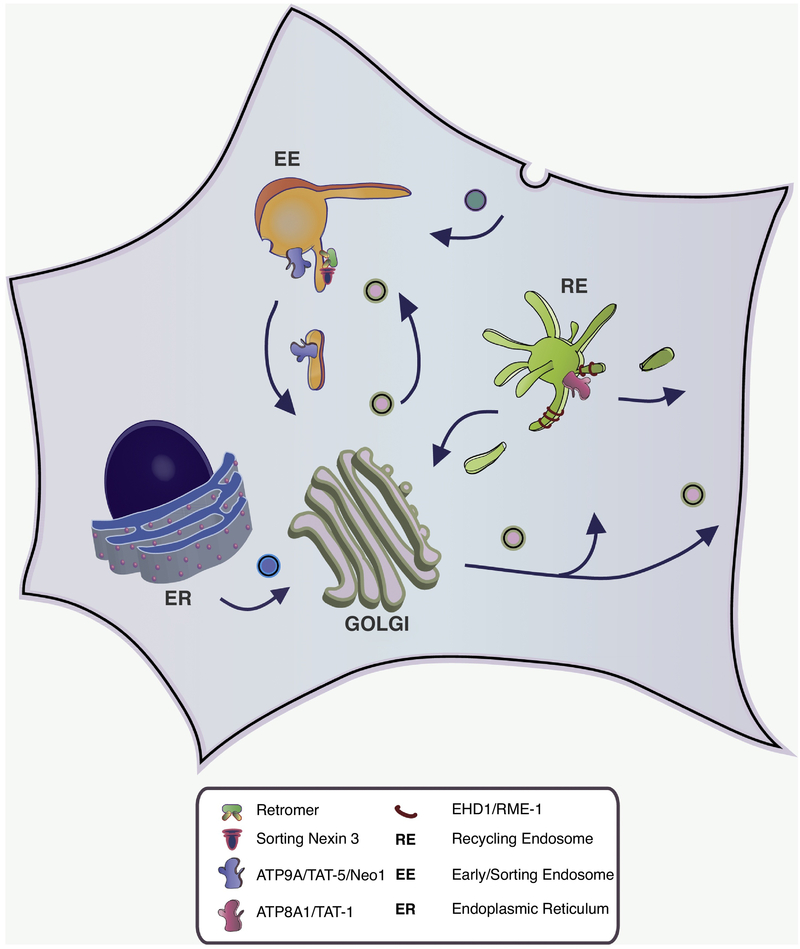
Figure 2. Roles for ATP9A and ATP8A1 in endosomal recycling pathways.
ATP8A1 (TAT-1 in C. elegans) and EHD1 (RME-1 in C. elegans) are required for transport of endocytosed proteins from recycling endosomes to the Golgi and for return back to the plasma membrane. Transport of PS to the cytosolic leaflet by ATP8A1 helps recruit EHD1 to recycling endosomes (RE) to potentially drive scission of tubular transport carriers. ATP9A (TAT-5 in C. elegans and Neo1 in S. cerevisiae) and Snx3-retromer is required for protein transport from early endosomes (EE) to the Golgi. MON2 and DOPEY2 are also linked to the function of ATP9A in this pathway.
Deficiency of ATP8A2 causes severe neurological disease that might be attributable to defects in endosomal recycling. An ATP8A2 mutation causes a striking disease in humans called cerebellar ataxia, mental retardation, disequilibrium syndrome (CAMRQ) where affected individuals walk on all fours [28]. In addition, the wabbler-lethal mouse carries a mutation Atp8a2−/− and this mouse displays progressive motor neuron degeneration and early death [29]. Interestingly, the phenotypically similar wobbler mouse, a model for amyotrophic lateral sclerosis, harbors a mutation in Vps54, a subunit of the Golgi-Associated Retrograde Protein (GARP) complex involved in tethering endosome-derived carriers to the Golgi [30]. Mutations in four different genes (VLDLR, WDR81, CA8 and ATP8A2) cause CAMRQ and WDR81 function is also linked to endosomal trafficking [31,32]. WDR81 is a BEACH (Beige and Chediak Higashi) domain and WD40 repeat protein (SORF-1 in C. elegans) that regulates phosphatidylinositol-3-phosphate (PI3P) levels on endosomes [31]. In brain, the very low density lipoprotein receptor (VLDLR) is a receptor for reelin, a secreted patterning factor involved in neuronal migration and synaptic plasticity [33]. A speculative model is that ATP8A2 and WDR81 mutations perturb recycling of VLDLR (and other neuronal membrane proteins) through the endosomal system to cause CAMRQ. The human, mouse and C. elegans genetics support an endosomal recycling role for ATP8A/TAT-1, consistent with foundational studies implicating their budding yeast ortholog, Drs2, in protein trafficking [9,10,15,19,34,35].
Trafficking pathways requiring Drs2
Drs2 localizes to the TGN/endosomal system and its flippase activity is acutely required for sorting of AP-1/clathrin cargos [10,18]. Drs2 also contributes to trafficking pathways mediated by AP-3 and GGA adaptor proteins, but does so redundantly with Dnf1, Dnf2 and Dnf3 P4-ATPases [19]. The specific requirement for Drs2 in AP-1 function can be attributed to its ability to flip PS because Drs2 mutations that perturb recognition pf PS, but not PE, cause these trafficking defects [9]. Moreover, gain of function mutations in Dnf1 that greatly increase its ability to flip PS allow these Dnf1PS variants to replace Drs2’s trafficking functions [9]. Surprisingly, AP-1 appears to be recruited normally to the trans-Golgi network (TGN) in drs2 mutants, but is nonfunctional [18]. This result implies that AP-1/clathrin coat assembly does not provide adequate energy to bend the Golgi membranes in the absence of the flippase. Moreover, fungal tetrameric adaptors lack clathrin binding motifs found in metazoan adaptors, and AP-2 and AP-3 can function independently of clathrin in yeast [17,36]. In addition, all of the yeast adaptors, including AP-2, are functionally linked to flippases [17–19,37]. We speculate that cells compensate for weak (or lost) interactions between fungal tetrameric adaptor proteins and clathrin heavy chain by using flippases to drive curvature of the transport carriers. The observed loss of AP-1 function does not explain all of the drs2 trafficking defects because endocytic recycling of an exocytic v-SNARE, Snc1, back to the plasma membrane requires Drs2 but is unaffected in clathrin adaptor or retromer mutants [19,34,38,39].
We recently discovered a surprising role for COPI in the Drs2-dependent recycling of Snc1 (Figure 3) [15]. Snc1 is ubiquitinated on several lysine residues within the SNARE motif and mutation of the preferred lysine, or attachment of a potent deubiquitinase to GFP-Snc1, disrupts its recycling [15,40]. Ubiquitin is well known to form a sorting signal on membrane proteins for endocytosis and sorting of protein into intraluminal vesicles [41], but these studies implicate ubiquitin as a recycling sorting signal and COPI as the coat acting in this pathway [15,40]. The clathrin-like α and β’ subunits of COPI contain WD40 repeats that form twin β-propeller structures, which bind to polyubiquitin with specificity for the chain length (3 or more) and linkage (K63)[15]. Deletion of a single propeller from β’COP perturbs Snc1 recycling, but replacing this propeller with a small domain that binds specifically to K63-linked polyubiquitin (the NZF domain from Tab2) fully restores Snc1 recycling [15]. Rcy1 is an F-box protein (potential substrate adaptor for SCF ubiquitin ligases) needed for Snc1 recycling [42,43], but Rcy1 may function in this pathway through binding and activation of Drs2 rather than as a ubiquitin ligase [15,39]. The role of Drs2 PS flippase activity in this pathway is only partially understood. Drs2 helps establish the negative charge and curvature required to recruit Gcs1 to TGN/EE membranes, and this ArfGAP binds to both COPI and Snc1 [44,45]. However, the gcs1 mutant displays a weaker Snc1 recycling defect than drs2 or COPI mutants [9], and so it is likely that additional Drs2 effectors are acting in this pathway. Whether or not flippases, polyubiquitin and COPI contribute to SNARE (e.g. VAMP2) recycling in animal cells remains to be determined.
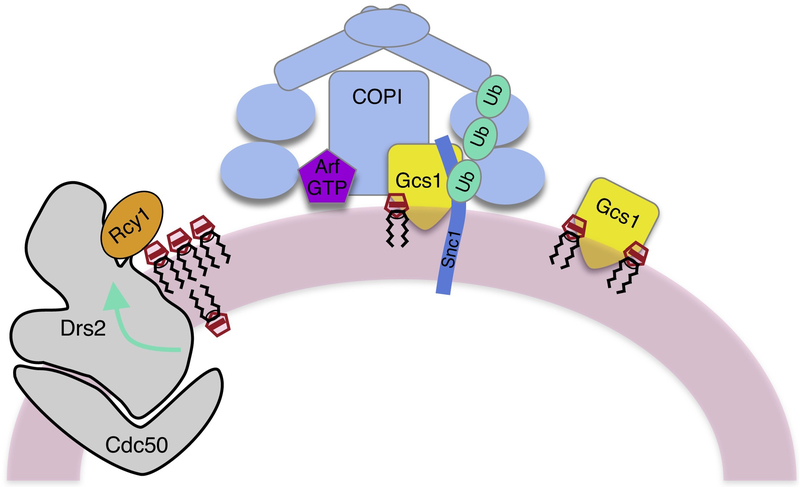
Figure 3. Model for how a PS flippase (Drs2-Cdc50) contributes to the polyubiquitin- and COPI-dependent recycling of an exocytic SNARE.
Snc1 is a t-SNARE that functions in the fusion of exocytic carriers with the plasma membrane. After endocytosis, Snc1 recycles through the endosomal system back to the Golgi for re-packaging into exocytic carriers. This recycling process requires Drs2-Cdc50, Rcy1, the ArfGAP Gcs1, COPI, and modification of Snc1 by K63-linked polyubiquitin. Rcy1 binds to a C-terminal regulatory domain on Drs2 and likely stimulates flippase activity. PS flip to the cytosolic leaflet increases membrane curvature and negative charge to recruit Gcs1, which binds to both COPI and Snc1. In addition, the twin β propellers of α and β’ COP bind directly to K63-linked polyubiquitin and this interaction is essential for Snc1 recycling.
Trafficking pathways requiring ATP9A, ATP9B, TAT-5, and Neo1
These orthologous P4-ATPases are also linked to protein trafficking in the Golgi and endosomal system, and are unusual because they do not require a β subunit from the Cdc50 family to function [11,20–22,46–48]. In addition, phospholipid flippase activity has not yet been definitely demonstrated for any of these proteins through purification and reconstitution. However, knockdown or mutations in TAT-5 and Neo1 cause a loss of PE asymmetry in C. elegans [13], and both PE and PS asymmetry in budding yeast [49]. Thus, it is likely that these proteins are PE or PE/PS flippases. Neo1, ATP9A and ATP9B localize to the Golgi and endosomal system, and although TAT-5 localizes to the plasma membrane, it requires endosomal recycling to maintain cell surface localization [14,20,46]. Neo1, ATP9A and TAT-5 engage in a conserved set of interactions with Snx3, DOPEY2 (Dopl in yeast, PAD-1 in C. elegans) and MON2 (an ArfGEF-related protein), and are functionally linked to retromer [21,22,50].
The core retromer heterotrimer (Vps26, Vps29, Vps35) forms two distinct complexes with sorting nexins, either a SNX-BAR heterodimer or a PX-SNX (Snx3), and the retromer-Snx3 complex specifically retrieves Neo1 from endosomes back to the Golgi [22]. In addition, mutation of the sorting signal in Neo1 required for interaction with Snx3 not only disrupts Neo1 trafficking, but another retromer-Snx3 cargo as well. Thus, it appears Neo1 is needed to form the retromer-Snx3 carriers [22]. SNX3 and ATP9A/TAT-5 are also required for retromer-dependent retrieval of Wntless from endosomes to the Golgi so it can be reused for Wnt secretion in animal cells [21]. SNX3 interacts with the DOPEY-MON2 complex, and depletion of ATP9A/TAT-5, DOPEY2 or MON2 disrupts Wntless trafficking and Wnt signaling. This is consistent with prior studies demonstrating functionally important interactions between Dopl, Mon2 and Neo1 in budding yeast [22,50]. In C. elegans, MON-2 plays a role in TAT-5 recycling from the endosomal system redundantly with sorting nexins, although curiously independent of core retromer subunits, and it appears the DOPEY (PAD-1) interaction with TAT-5 regulates its PE flippase activity. Like tat-5, pad-1 and mutants defective in TAT-5 recycling excessively shed extracellular vesicles [14].
Neo1 and Dop1 are essential for viability in budding yeast [11,51], in contrast to the near normal growth of retromer mutants [52]. Therefore, Neo1 and Dop1 must have other, retromer-independent functions necessary for growth. Conditional neo1 mutants display several Golgi-associated defects, including mislocalization of COPI-dependent retrograde cargos targeted to the ER [11]. Thus, the remaining four P4-ATPases do not normally compensate for loss of Neo1. However, neo1Δ, dop1Δ or mon2Δ cells grow well when ANY1, encoding a conserved PQ-loop membrane protein, is knocked out [53]. Growth of neo1Δ any1Δ cells requires Drs2 and it appears that Drs2 can carry out Neo1’s essential function these cells [53]. How Anyl segregates Neo1 and Drs2 functions is still unclear, although it has been suggested that Any1 could be a scramblase that dissipates the phospholipid gradients formed by Drs2 and Neo1 in the Golgi [53].
Concluding remarks
Membrane remodeling by P4-ATPases plays a crucial role in protein trafficking, although there are many gaps in our understanding of the molecular basis for this role. The activity of ATP9A, TAT-5 and Neo1 in facilitating retromer-Snx3 carrier formation (or cargo sorting) from endosome for delivery to the Golgi is a great example of a highly conserved P4-ATPase function in a specific trafficking pathway [21,22]. However, there is no direct evidence that these P4-ATPases are bona fide flippases, comparable to studies with Drs2 or ATP8A1 [54,55]. The biochemical functions of MON2, DOPEY, and Any1 “regulators” of ATP9A/TAT-5/Neo1 remain obscure, as does the essential function of Neo1 and TAT-5 and their relationship to COPI-dependent transport from Golgi cisternae. The most clearly conserved function of ATP8A/TAT-1/Drs2 is in endosomal recycling. Drs2 has been linked to COPI and AP-1 function for trafficking between the Golgi and endosomes, but the nature of the donor compartment (TGN, recycling endosome, early endosome?) and directionality of transport is not clear. The tubular carriers ATP8A1/ATP8A2/TAT-1 help generate from recycling endosomes in animal cells are linked to EHD1 (RME-1) activity, but the precise function of this ATPase and how cargo is sorted into these carriers is unknown. While recent advances in the field have been impressive, we have a long way to go before the final chapter on phospholipid flippases in protein transport can be written.
Acknowledgements
This project was supported by a grant from the National Institutes of Health (GM118452) to TRG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
References and Recommended reading
Papers of particular interest, published within the period of review, have been highlightes as”
• of special interest
•• of outstanding interest
- 1.Kornberg RD, McConnell HM: Inside-outside transitions of phospholipids in vesicle membranes. Biochemistry 1971, 10:1111–1120. [DOI] [PubMed] [Google Scholar]
- 2.Hankins HM, Baldridge RD, Xu P, Graham TR: Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution. Traffic 2015, 16:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shin HW, Takatsu H: Substrates of P4-ATPases: beyond aminophospholipids (phosphatidylserine and phosphatidylethanolamine). FASEB J 2018:fj201801873R. [DOI] [PubMed] [Google Scholar]
- 4.Roland BP, Graham TR: Decoding P4-ATPase substrate interactions. Crit Rev Biochem Mol Biol 2016, 51:513–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham TR, Kozlov MM: Interplay of proteins and lipids in generating membrane curvature. Curr Opin Cell Biol 2010, 22:430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S: Membrane phosphatidylserine regulates surface charge and protein localization. Science 2008, 319:210–213. [DOI] [PubMed] [Google Scholar]
- 7.Alexander RT, Jaumouille V, Yeung T, Furuya W, Peltekova I, Boucher A, Zasloff M, Orlowski J, Grinstein S: Membrane surface charge dictates the structure and function of the epithelial Na+/H+ exchanger. EMBO J 2011, 30:679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •8.Tsuchiya M, Hara Y, Okuda M, Itoh K, Nishioka R, Shiomi A, Nagao K, Mori M, Mori Y, Ikenouchi J, et al. : Cell surface flip-flop of phosphatidylserine is critical for PIEZO1-mediated myotube formation. Nat Commun 2018, 9:2049. [DOI] [PMC free article] [PubMed] [Google Scholar]; Regulated cell surface exposure of PS was known to play a crucial role in myoblast fusion during skeletal muscle development. This paper shows that the transbilayer distribution of PS regulates PIEZO1, a mechanosensitive Ca++ channel, to control polarized membrane fusion. PS in the outer leaflet of the plasma membrane inhibits Ca++ influx and its subsequent flip to the cytosolic leaflet activates PIEZO1. This work provides a nice example of how phospholipid asymmetry can regulate the activity of a membrane protein.
- 9.Xu P, Baldridge RD, Chi RJ, Burd CG, Graham TR: Phosphatidylserine flipping enhances membrane curvature and negative charge required for vesicular transport. J Cell Biol 2013, 202:875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen CY, Ingram MF, Rosal PH, Graham TR: Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J Cell Biol 1999, 147:1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hua Z, Graham TR: Requirement for Neo1p in retrograde transport from the Golgi complex to the endoplasmic reticulum. Mol Biol Cell 2003, 14:4971–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S, Uchida Y, Wang J, Matsudaira T, Nakagawa T, Kishimoto T, Mukai K, Inaba T, Kobayashi T, Molday RS, et al. : Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase. EMBO J 2015, 34:669–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wehman AM, Poggioli C, Schweinsberg P, Grant BD, Nance J: The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr Biol 2011, 21:1951–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •14.Beer KB, Rivas-Castillo J, Kuhn K, Fazeli G, Karmann B, Nance JF, Stigloher C, Wehman AM: Extracellular vesicle budding is inhibited by redundant regulators of TAT-5 flippase localization and phospholipid asymmetry. Proc Natl Acad Sci U S A 2018, 115:E1127–E1136. [DOI] [PMC free article] [PubMed] [Google Scholar]; TAT-5 activity is required to inhibit plasma membrane deformation required for extracellular vesicle release (ectocytosis) from embryonic cells [13]. This study demonstrates the importance of recycling TAT-5 from endosomes back to the plasma membrane for repression of extocytosis. In addition, MON-2 and PAD-1 (MON2 and DOPEY orthologs) are shown to be critical for TAT-5 recycling and/or activity. Several different sorting nexins contribute to TAT-5 recycling, including SNX-3, but in this case retromer does not appear to play a role in contrast to the observations with mammalian cells [21] and yeast [22].
- ••15.Xu P, Hankins HM, MacDonald C, Erlinger SJ, Frazier MN, Diab NS, Piper RC, Jackson LP, MacGurn JA, Graham TR: COPI mediates recycling of an exocytic SNARE by recognition of a ubiquitin sorting signal. Elife 2017, 6:e28342. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mechanisms for how the v-SNARE Snc1 is recycled from the plasma membrane back to the Golgi for re-incorporation into exocytic vesicles were unclear. This study, along with [40], provides evidence that ubiquitination of Snc1 is critical for its recycling. In addition, the COPI coat was shown to bind K63-linked polyubiquitin for the first time and that this interaction is required for Snc1 recycling. Along with [9], these studies implicate Drs2 PS flippase activity in budding COPI-coated vesicles used to recycle Snc1.
- 16.Schultzhaus Z, Yan H, Shaw BD: Aspergillus nidulans flippase DnfA is cargo of the endocytic collar and plays complementary roles in growth and phosphatidylserine asymmetry with another flippase, DnfB. Mol Microbiol 2015, 97:18–32. [DOI] [PubMed] [Google Scholar]
- ••17.Martzoukou O, Amillis S, Zervakou A, Christoforidis S, Diallinas G: The AP-2 complex has a specialized clathrin-independent role in apical endocytosis and polar growth in fungi. Elife 2017, 6:e20083. [DOI] [PMC free article] [PubMed] [Google Scholar]; A phylogenetic analysis of tetrameric adaptor proteins showed a loss of clathrin-binding motifs in the C-terminal hinge and ear domain of AP-1, AP-2 and AP-3 β subunits in most fungi. This study further shows a clathrin-independent role for Aspergillus nidulans AP-2 in endocytosis and imply that P4-ATPases (Dnf1 and Drs2 orthologs) are crucial cargos of AP-2 (also see [16]). Flippase and AP-2 mutants display comparable defects in cell polarity and polarized growth and these proteins may function together to drive carrier budding.
- 18.Liu K, Surendhran K, Nothwehr SF, Graham TR: P4-ATPase requirement for AP-1/clathrin function in protein transport from the trans-Golgi network and early endosomes. Mol Biol Cell 2008, 19:3526–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua Z, Fatheddin P, Graham TR: An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol Biol Cell 2002, 13:3162–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y, Takar M, Cuentas-Condori AA, Graham TR: Neo1 and phosphatidylethanolamine contribute to vacuole membrane fusion in Saccharomyces cerevisiae. Cell Logist 2016, 6:e1228791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••21.McGough IJ, de Groot REA, Jellett AP, Betist MC, Varandas KC, Danson CM, Heesom KJ, Korswagen HC, Cullen PJ: SNX3-retromer requires an evolutionary conserved MON2:DOPEY2:ATP9A complex to mediate Wntless sorting and Wnt secretion. Nat Commun 2018, 9:3737. [DOI] [PMC free article] [PubMed] [Google Scholar]; While ATP9A had been linked to endosomal recycling previously [48], this paper provides the first demonstration that ATP9A is required for Snx3-retromer function in mammalian cells. Moreover, the biological significance of the MON2, DOPEY2 and ATP9A (TAT-5) interactions with SNX3-retromer in endosome to Golgi trafficking of Wntless is demonstrated using C. elegans and mammalian cells. Deficiency of any of these components leads to a failure to retrieve Wntless from endosomes and a defect in Wnt secretion.
- ••22.Dalton LE, Bean BDM, Davey M, Conibear E: Quantitative high-content imaging identifies novel regulators of Neo1 trafficking at endosomes. Mol Biol Cell 2017, 28:1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]; A prior study had found that Neo1 requires retromer for its retrieval from endosomes in budding yeast [20], and this paper went on to show it is specifically the Snx3-retromer complex that sorts Neo1. A sorting signal within Neo1 that binds Snx3 was identified and remarkably, mutation of this sorting signal disrupted trafficking of another Snx3-retromer cargo. This observation suggests that Neo1’s flippase activity is required for the formation of Snx3-retromer carriers.
- 23.Jarsch IK, Daste F, Gallop JL: Membrane curvature in cell biology: An integration of molecular mechanisms. J Cell Biol 2016, 214:375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••24.Takada N, Naito T, Inoue T, Nakayama K, Takatsu H, Shin HW: Phospholipid-flipping activity of P4-ATPase drives membrane curvature. EMBO J 2018, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]; Prior studies linking P4-ATPase function to protein transport and membrane curvature were primarily performed using loss-of-function mutations in flippases (e.g. [9, 10, 12, 26]). The authors of this paper discovered a gain-of-function membrane curvature phenotype associated with overexpression of ATP10A, a PC flippase. Co-expression of ATP10A with a BAR domain that can bind and stabilize curved membranes but cannot induce curvature on it own, causes substantial tubulation of the plasma membrane. This study nicely emphasizes the membranebending power of phospholipid flippases.
- 25.Chen B, Jiang Y, Zeng S, Yan J, Li X, Zhang Y, Zou W, Wang X: Endocytic sorting and recycling require membrane phosphatidylserine asymmetry maintained by TAT-1/CHAT-1. PLoS Genet 2010, 6:e1001235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nilsson L, Jonsson E, Tuck S: Caenorhabditis elegans Numb Inhibits Endocytic Recycling by Binding TAT-1 Aminophospholipid Translocase. Traffic 2011, 12:1839–1849. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Chen B, Yoshina S, Cai T, Yang F, Mitani S, Wang X: Inactivation of Caenorhabditis elegans aminopeptidase DNPP-1 restores endocytic sorting and recycling in tat-1 mutants. Mol Biol Cell 2013, 24:1163–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emre Onat O, Gulsuner S, Bilguvar K, Nazli Basak A, Topaloglu H, Tan M, Tan U, Gunel M, Ozcelik T: Missense mutation in the ATPase, aminophospholipid transporter protein ATP8A2 is associated with cerebellar atrophy and quadrupedal locomotion. Eur J Hum Genet 2012, 21:281–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu X, Libby RT, de Vries WN, Smith RS, Wright DL, Bronson RT, Seburn KL, John SW: Mutations in a p-type ATPase gene cause axonal degeneration. PLoS Genet 2012, 8:e1002853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitt-John T: VPS54 and the wobbler mouse. Front Neurosci 2015, 9:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu K, Jian Y, Sun X, Yang C, Gao Z, Zhang Z, Liu X, Li Y, Xu J, Jing Y, et al. : Negative regulation of phosphatidylinositol 3-phosphate levels in early-to-late endosome conversion. J Cell Biol 2016, 212:181–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rapiteanu R, Davis LJ, Williamson JC, Timms RT, Paul Luzio J, Lehner PJ: A Genetic Screen Identifies a Critical Role for the WDR81-WDR91 Complex in the Trafficking and Degradation of Tetherin. Traffic 2016, 17:940–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dlugosz P, Nimpf J: The Reelin Receptors Apolipoprotein E receptor 2 (ApoER2) and VLDL Receptor. Int J Mol Sci 2018, 19:E3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakane H, Yamamoto T, Tanaka K: The functional relationship between the Cdc50p-Drs2p putative aminophospholipid translocase and the Arf GAP Gcs1p in vesicle formation in the retrieval pathway from yeast early endosomes to the TGN. Cell Struct Funct 2006, 31:87–108. [DOI] [PubMed] [Google Scholar]
- 35.Furuta N, Fujimura-Kamada K, Saito K, Yamamoto T, Tanaka K: Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol Biol Cell 2007, 18:295–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vowels JJ, Payne GS: A dileucine-like sorting signal directs transport into an AP-3-dependent, clathrin-independent pathway to the yeast vacuole. EMBO J 1998, 17:2482–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, et al. : Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell 2006, 126:611–625. [DOI] [PubMed] [Google Scholar]
- 38.Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HR: Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell 2000, 11:23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanamatsu H, Fujimura-Kamada K, Yamamoto T, Furuta N, Tanaka K: Interaction of the phospholipid flippase Drs2p with the F-box protein Rcy1p plays an important role in early endosome to trans-Golgi network vesicle transport in yeast. J Biochem 2014, 155:51–62. [DOI] [PubMed] [Google Scholar]
- 40.Chen SH, Shah AH, Segev N: Ypt31/32 GTPases and their F-Box effector Rcy1 regulate ubiquitination of recycling proteins. Cell Logist 2011, 1:21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacGurn JA, Hsu PC, Emr SD: Ubiquitin and membrane protein turnover: from cradle to grave. Annu Rev Biochem 2012, 81:231–259. [DOI] [PubMed] [Google Scholar]
- 42.Wiederkehr A, Avaro S, Prescianotto-Baschong C, Haguenauer-Tsapis R, Riezman H: The Fbox protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J Cell Biol 2000, 149:397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen SH, Chen S, Tokarev AA, Liu F, Jedd G, Segev N: Ypt31/32 GTPases and their novel Fbox effector protein Rcy1 regulate protein recycling. Mol Biol Cell 2005, 16:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson M, Poon PP, Schindler C, Murray LE, Kama R, Gabriely G, Singer RA, Spang A, Johnston GC, Gerst JE: The Gcs1 Arf-GAP mediates Snc1,2 v-SNARE retrieval to the Golgi in yeast. Mol Biol Cell 2006, 17:1845–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suckling RJ, Poon PP, Travis SM, Majoul IV, Hughson FM, Evans PR, Duden R, Owen DJ: Structural basis for the binding of tryptophan-based motifs by delta-COP. Proc Natl Acad Sci U S A 2015, 112:14242–14247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takatsu H, Baba K, Shima T, Umino H, Kato U, Umeda M, Nakayama K, Shin HW: ATP9B, a P4-ATPase (a putative aminophospholipid translocase), localizes to the trans-Golgi network in a CDC50-independent manner. J Biol Chem 2011, 286:38159–38167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wicky S, Schwarz H, Singer-Kruger B: Molecular interactions of yeast Neo1p, an essential member of the Drs2 family of aminophospholipid translocases, and its role in membrane trafficking within the endomembrane system. Mol Cell Biol 2004, 24:7402–7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka Y, Ono N, Shima T, Tanaka G, Katoh Y, Nakayama K, Takatsu H, Shin HW: The phospholipid flippase ATP9A is required for the recycling pathway from the endosomes to the plasma membrane. Mol Biol Cell 2016, 27:3883–3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takar M, Wu Y, Graham TR: The Essential Neo1 Protein from Budding Yeast Plays a Role in Establishing Aminophospholipid Asymmetry of the Plasma Membrane. J Biol Chem 2016, 291:15727–15739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barbosa S, Pratte D, Schwarz H, Pipkorn R, Singer-Kruger B: Oligomeric Dop1p is part of the endosomal Neo1p-Ysl2p-Arl1p membrane remodeling complex. Traffic 2010, 11:1092–1106. [DOI] [PubMed] [Google Scholar]
- 51.Gillingham AK, Whyte JR, Panic B, Munro S: Mon2, a relative of large Arf exchange factors, recruits Dop1 to the Golgi apparatus. J Biol Chem 2006, 281:2273–2280. [DOI] [PubMed] [Google Scholar]
- 52.Seaman MN, Marcusson EG, Cereghino JL, Emr SD: Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J Cell Biol 1997, 137:79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Leeuwen J, Pons C, Mellor JC, Yamaguchi TN, Friesen H, Koschwanez J, Usaj MM, Pechlaner M, Takar M, Usaj M, et al. : Exploring genetic suppression interactions on a global scale. Science 2016, 354:aag0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou X, Graham TR: Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc Natl Acad Sci U S A 2009, 106:16586–16591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coleman JA, Kwok MC, Molday RS: Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes. J Biol Chem 2009, 284:32670–32679. [DOI] [PMC free article] [PubMed] [Google Scholar]