Abstract
Triple negative breast cancer (TNBC) has an unusually low 5-year survival rate linked to higher metastatic rates. Our laboratory recently delineated a role for the alternative RNA splicing (AS) of cytoplasmic polyadenylation element binding protein 2 (CPEB2), via inclusion/exclusion of exon 4, in the metastasis of TNBC. In these studies, the mechanism governing the inclusion/exclusion of exon 4 was examined. Specifically, the RNA trans-factor, SRSF3, was found to be explicitly associated with CPEB2 exon 4. A SRSF3 consensus sequence was identified in exon 4, and mutation of this sequence abolished the association of SRSF3. The expression of SRSF3 was upregulated in TNBC cells upon the acquisition of anoikis resistance correlating with a reduction in the CPEB2A/B ratio. Importantly, downregulation of SRSF3 in these cells by siRNA induced the exclusion of exon 4 in cells increasing the ratio of CPEB2A (exon 4 excluded) to CPEB2B (exon 4 included). Downregulation of SRSF3 also reversed the CPEB2A/B ratio of a wild-type CPEB2 exon 4 minigene and endogenous CPEB2 pre-mRNA, but not a mutant CPEB2 minigene with the SRSF3 RNA cis-element ablated. SRSF3 downregulation ablated the anoikis resistance of TNBC cells, which was “rescued” by ectopic expression of CPEB2B. Finally, analysis of The Cancer Genome Atlas (TCGA) database showed a positive relationship between SRSF3 expression and lower CPEB2A/B ratios in aggressive breast cancers.
Implications:
These findings demonstrate that SRSF3 modulates CPEB2 AS to induce the expression of the CPEB2B isoform that drives TNBC phenotypes correlating with aggressive human breast cancer.
Keywords: CPEB2, cytoplasmic polyadenylation, epithelial to mesenchymal transition, SRSF3, trans-RNA splicing factor, exon splicing enhancer, hypoxic response, alternative RNA splicing
INTRODUCTION
Among the four main molecular subtypes of breast cancer, TNBC, which may also include the basal-like phenotype, is characterized histologically by a lack of estrogen receptor (ER), progesterone receptor (PR), and epidermal growth factor receptor 2 (ERBB2, commonly referred to as HER2) expression (1). Compared to the hormone receptor positive breast cancer subtypes, TNBC displays considerable genetic complexity and tumor heterogeneity making “targeted” therapies ineffective (2,3). The standard of care for most patients who present with TNBC is neoadjuvant chemotherapy and surgery, which, in many cases, initially provides a favorable pathologic response rate and outcome. However, the five-year progression free and survival rates for TNBC rank among the poorest of all breast cancer subtypes (4). The highly metastatic nature of TNBC and lack of known therapeutic targets make discovery of novel treatment options critical.
Increased histological and molecular complexity in TNBC suggest very early stage regulatory events in gene expression are involved in driving the underlying tumorigenic capacity of these cells (5–7). As such, the genomic alterations thought to influence breast cancer progression include alternative pre-RNA splicing (AS), which alters the coding regions of resulting proteins. While AS promotes novel and cancer specific splice variants, the upregulation of RNA splicing factors in a broad scale occurs in TNBC, and thus, provides an additional layer to the complexity of exon assembly in this paradigm (8,9). Indeed, the upregulation of specific RNA trans-activating factors alone can cause malignant transformation via dysregulated activation of downstream alternative splicing pathways (10–13).
Recently, our laboratory (2015) demonstrated that AS of cytoplasmic polyadenylation element binding protein 2 (CPEB2) affects the metastatic potential of TNBC (14). Specifically, upregulation of the CPEB2B splice variant, which incorporates exon 4 into the final transcript, produced a phenotype which was resistant to detachment-induced cell death (i.e. anoikis resistance (AnR)), and primary tumors became highly metastatic targeting the lung (14). In contrast to the CPEB2B isoform, which promoted neoplastic transformation, the CPEB2A isoform applied an opposite effect on primary tumor growth and metastasis in TNBC notably through translational repression of the DNA trans-factors TWIST1 and HIF1α (15). While the splice variant isoform ratio for CPEB2A/CPEB2B remains generally high in normal and non-tumorigenic breast tissue, the CPEB2B splice variant was found more highly expressed, reversing the A/B ratio in human TNBC and cells selected for anchorage-independent growth (14).
CPEB2 belongs to the CPEB protein family (CPEB1–4), which share a highly conserved RNA recognition motif at their C-terminal ends, however the N-terminus is highly variable among family members (16). The canonical member, CPEB1, has been shown to regulate translation of nascent mRNA transcripts through binding at the 3’ UTR and preventing extension of the polyadenylate tail (17). Much is unknown about the remaining family members, although CPEB2 has been shown to be essential for successful mitotic cell division and, by our laboratories, as required for the acquisition of anoikis resistance (18).
As to plausible regulators of CPEB2 AS, the serine/arginine (SR)-rich family is a well-known family of RNA trans-activating factors that modulate splice site selection and exon inclusion/exclusion via association of a RNA recognition motif (RRM) and RNA Binding Domain (RBD) to exonic splicing enhancers (ESE) in pre-mRNA to mediate spliceosome assembly (for reviews see (19–21)). The SR protein family member, SRSF3, is reportedly upregulated during oxidative stress and hypoxia, conditions typically found within a tumor, where the trans-RNA splicing factor can exert wide influence in genes associated with cell cycle and proliferation (22–24). In this study, our laboratory demonstrated an increased occurrence of both SRSF3 and the CPEB2B isoform in TNBC, and identified a consensus binding motif for SRSF3 in the exon promoting the CPEB2B isoform. This evidence suggested a mechanistic link to the SRSF3:CPEB2B splicing paradigm. The presented study shows that the splicing event which promotes expression of CPEB2B is driven by SRSF3 as well as linked to aggressive growth in metastatic TNBC.
MATERIALS AND METHODS
Cell culture and reagents:
MDA-MB-231 and MDA-MB-468 cells were obtained from ATCC (Manassas, VA) and maintained in RPMI (Invitrogen). The ATCC routinely authenticates cell lines using STR profiling. All cell lines were supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 1% Penicillin / Streptomycin (Invitrogen). Cells were maintained in a 95% air / 5% CO2 incubator at 37°C. Cells were passaged once every 3–5 days (~90% confluence), and all experiments were performed during the first 12 passages.
Western Blotting:
Total protein (5–20 μg) was electrophoretically separated on 7.5% - 12% SDS-polyacrylamide gels. Samples were transferred electrophoretically to PVDF membranes, then probed with the appropriate antibody as described previously (14,15,25). Antibodies were purchased from Cell Signaling with the exception of SRSF3 (ThermoFisher, Clone ID: 7B4).
Quantitative Reverse Transcriptase-Polymerase Chain Reaction (qRT-PCR):
Primer/probe sets were previously described (14). PCR was performed as described (25). cDNA was synthesized using the Superscript III kit (Life Technologies) and manufacturer’s instructions (26–28). Samples were then amplified using a BioRad CFX Connect qPCR machine and calculated using the standard curve method.
siRNA Treatment and Plasmid Transfection:
All transfections were carried out in triplicate in 6-well tissue culture dishes. Validated Silencer Select siRNA towards SRSF3 (s12732 or s12733) or non-targeting control (ThermoFisher Scientific) were utilized in this study at 30 nM concentrations and transfected using Dharmafect 4 transfection reagent (Dharmacon) as described previously and to the manufacturers specifications. Plasmid transfections were accomplished using the Effectene system (Qiagen) according to manufacturer’s instructions as described previously (14,25,29,30) and using 0.5 to 1.0 μg total DNA per well.
Anoikis Resistance Assay:
Cells were transfected with the indicated siRNA and DNA plasmid using Dharmafect Duo transfection reagent (Dharmacon). After 48 hours, cells were washed, trypsinized, and added to each well of either normal or polyHEMA-coated 6-well tissue culture plates. The cells were incubated 6 hours, then collected for analysis via Western blotting.
Competitive Quantitative RT-PCR:
cDNA was synthesized as previously described (30), and then cDNA samples were subjected to competitive PCR using the following primers: endogenous CPEB2A or CPEB2B isoform amplification forward primer 5’-GCAGCAGAGGAACTCCTATAAC-3’ and reverse primer 5’-CAAAGAGTGCATATTCAAACTGTCA-3’, minigene specific CPEB2A or CPEB2B isoform amplification forward primer 5’-CAGAACAGACAACAATAGTAATACACTC-3’ and reverse primer 5’-AGGGGCAAACAACAGATGG-3’. PCR conditions for endogenous gene amplification consisted of an initial denaturing step at 98 °C for 30 seconds followed by 25 cycles of 98 °C denaturing for 10 sec., 50 °C annealing for 30 sec., 72 °C extension for 1 min., and final extension step at 72 °C for 5 mins. Minigene-specific amplification conditions were identical and used 20 cycles. All PCR reactions were amplified using standard Taq polymerase (New England Biolabs) with products run on 5% polyacrylamide-TBE and stained with SYBRgold (ThermoFisher Scientific).
RNA binding assays:
Full length Wild-type (WT) biotinylated RNA CPEB2 exon sequence (Bi:5’GUGAGAUCUAGUUUGCAGUUGCCAGCUUGGGGCUCAGAUUCACUCCAAGAUAGUUGGUGCACUGCAGCCGGAACAUCCAGAAUAGACCAG3’) or mutant sequence (MUT, see Fig.1) were incubated with recombinant SRSF3 (lsBio) and RNA-bound proteins were precipitated as described (14,15,27). Samples were subjected to immunoblotting with SRSF3 antibody.
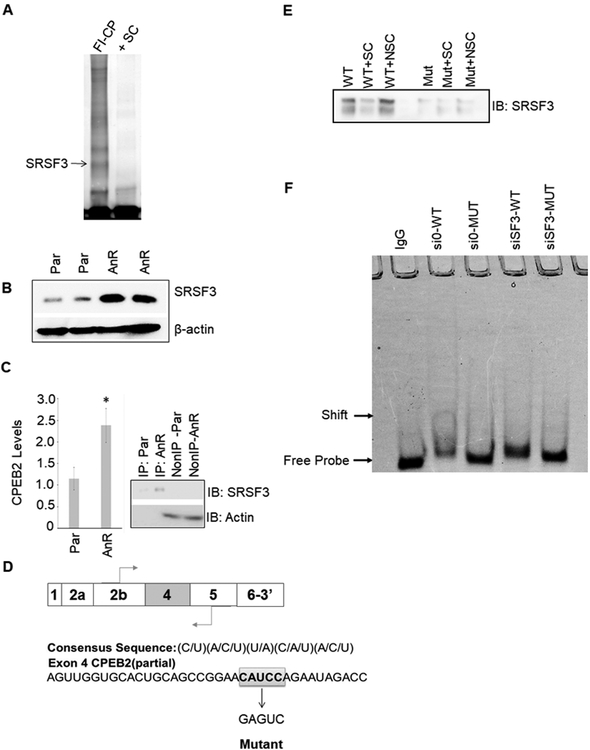
Figure 1: SRSF3/SRp20 binds specifically to exon 4 in the CPEB2 pre-mRNA.
A. MDA-MB-231 nuclear extract was incubated with either FITC-conjugated CPEB2 exon 4 sequence + “cold” nonspecific competitor (FI-CP) or pre-incubated with 100X “cold” CPEB2ex4 as a specific competitor (+SC). Samples were then electrophoresed and bound proteins were extracted and subjected to proteomic analysis. B. Lysates of Parental (Par) and anoikis resistant (AnR) MDA-MB-231 cells were immunoblotted and probed for the indicated antibodies. C. SRSF3-specific antibody was used for CLIP-qRT-PCR to detect CPEB2 levels in either MDA-MB-231 parental or AnR cells. Real-time PCR to CPEB2 at exon 4 was evaluated (data represented as n = 3 ± standard deviation (sd), * = p < 0.05). IP=immunoprecipitated fraction. Non-IP=Non-IP’d fraction. D. The consensus sequence for SRSF3 and a partial sequence of exon 4 highlighting the proposed SRSF3 binding site. E. SBAP assay was used to detect SRSF3 bound to exon 4 of CPEB2. Recombinant SRSF3 was incubated with biotinylated CPEB2 exon 4 RNA oligos with WT or mutant SRSF3 ESE cis-element. Samples were incubated with either biotin labeled CPEB2 exon 4 sequence + “cold” nonspecific competitor (NSC) or pre-incubated with 100X “cold” unlabeled CPEB2ex4 as a specific competitor (+SC). F. EMSA analysis of siRNA-depleted expression of SRSF3 in MDA-MB-231 cells. EMSA labels correspond to MDA-MB-231 cells treated with siRNA control and then total protein lysates incubated with wild type CPEB2 exon 4 ESE RNA (si0-WT), siRNA control treated cell lysates incubated with mutant CPEB2 exon 4 ESE RNA (si0-MUT), siRNA to SRSF3 treated cell lysates incubated with wild type CPEB2 exon 4 ESE RNA (siSF3-WT), or siRNA to SRSF3 treated cell lysates incubated with mutant CPEB2 exon 4 ESE RNA (siSF3-MUT). Sequences are indicated in the Materials and Methods section. Control samples were incubated with nonspecific IgG.
Electrophoretic mobility shift assay:
FITC conjugated full length (Fig.1A) or partial (Fig.1E) wild-type or mutant CPEB2 RNA sequences were subjected to EMSA as described (27).
Minigene Construct and Plasmids:
The genomic region spanning exons 3,4, and 5 of the CPEB2 gene were analyzed in this study. Template DNA was amplified from the RPCI-11 HS BAC Clone (ThermoFisher, Clone ID: 629A7) in two different 1.7 kilobase fragments. PCR reactions used forward and reverse primers to amplify exon 3, intron 3, exon 4, and a partial segment of intron 4 with sequences 5’-AAACGGGCCCTCTAGATTTCCCTAGCCTCTTCTGA-3’ and 5’-GGAAGGAATGCTAGATGACTAACGGTTTCTCCATA-3’. The second fragment of DNA was amplified using forward and reverse primers to amplify a region of intron 4 directly upstream exon 5 and all of exon 5 except the last three codons. Forward and reverse PCR primers consisted of sequences 5’-TCTAGCATTCCTTCCGTCA-3’ and 5’-TACCGAGCTCGGATCCGGATCATGCTCTGCTCTC-3’. Genomic DNA fragments were amplified using standard PCR conditions and proofreading Taq polymerase (Phusion High Fidelity DNA Polymerase, New England Biolabs). Standard PCR conditions consisted of an initial denaturing step at 98 °C for 30 seconds followed by 30 cycles of 98 °C denaturing for 10 sec., 60 °C annealing for 30 sec., 72 °C extension for 30 sec., and final extension step at 72 °C for 10 mins. Fusion of amplified genomic material with the pcDNA3.1(−) mammalian expression vector (Invitrogen) was carried out using the In-Fusion HD cloning reaction (Clontech). At each step of PCR amplification and plasmid generation, the CPEB2 minigene sequence was verified by Sanger dideoxy method (GenScript). The CPEB2 minigene insert was designed to retain the XbaI and BamHI restriction sites at the 5’ and 3’ ends respectively. All primers used in cloning and analysis were synthesized by Integrated DNA Technologies.
Site-directed mutagenesis was carried out using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent) according to the manufacturer’s instructions and primers containing the mutated CPEB2 ESE with forward and reverse sequences 5’-GGTGCACTGCAGCCGGAAGAGTCAGAATAGACCAGGTAGG-3’ and 5’- CCTACCTGGTCTATTCTGACTCTTCCGGCTGCAGTGCACC-3’.
The CPEB2B-flag plasmid construct was previously reported (14).
Biostatistics:
Biostatistical analyses were performed using either SPSS or R. Statistical tests used include one-way ANOVA/ pooled t-test (in the case of only two samples), ANOVA (in the case of multiple samples) and an FDR-adjusted p-value with Tukey HSD posthoc calculation.
RESULTS
Unbiased proteomic analysis identified SRSF3 associated with exon 4 of CPEB2.
Previously, our laboratory reported that the acquisition of anoikis by TNBC cells required the inclusion of exon 4 into the mature CPEB2 mRNA to produce the CPEB2B isoform (14,15). These findings led to the investigation of the mechanism governing the inclusion of exon 4. promoting mRNA maturation towards the CPEB2B isoform. To this end, electrophoretic mobility shift assays (EMSA) analysis were employed using nuclear extract from MDA-MB-231 cells to examine protein complexes associated with exon 4 of CPEB2. Unbiased proteomic analysis of complexes bound to CPEB2 exon 4 identified SRSF3, hnRNP hnRNP F, and hnRNP H1 bound to exon 4 of CPEB2 (Fig. 1A; Table 1). Of the four RNA trans-factors associated with exon 4, SRSF3 was validated using an SRSF3-specific antibody for cross-linking immunoprecipitation coupled with quantitative real-time RT-PCR (CLIP-qRT-PCR, Fig 1C). SRSF3 was shown to cross-link total RNA in either MDA-MB-231 parental (Par) or AnR cells via immunoprecipitation, and was shown to interact with Exon 4 of CPEB2 in both Par versus AnR cells through qRT-PCR amplification specific to CPEB2 Exon 4. A significant increase in RNA association of SRSF3 with CPEB2 Exon 4 was also observed in the AnR cells (Fig. 1B). Importantly, the levels of SRSF3 were increased in TNBC cells that acquire AnR (Fig. 1B).
Table 1.
RNA trans-splicing factor candidate screen with siRNA for CPEB2A/B Protein Ratio
| Protein | CPEB2A/B ratio |
|---|---|
| No Treatment | 7.40 |
| Non-targeting control | 6.96 |
| SRSF3 | 19.0 |
| hnRNP H1 | 9.78 |
| hnRNP F | 7.06 |
| hnRNP H1/F | 3.58 |
Examination of exon 4 elucidated a consensus sequence for SRSF3 (C/U)(A/C/U)(U/A)(C/A/U)(A/C/U) (Fig. 1C), and to examine the association of SRSF3 with this sequence, a streptavidin-biotin affinity purification (SBAP) assay was developed using CPEB2 exon 4 RNA as “bait” and combined with recombinant SRSF3. mutation of this sequence abolished the association of SRSF3. This assay revealed that SRSF3 specifically binds to CPEB2 exon 4, with competition for SRSF3 binding achieved with unlabeled CPEB2 exon 4 RNA at excess concentration (100X); while non-specific competitor RNA showed no effect on the RNA:protein complex (Fig. 1C–E). Importantly, mutation of the SRSF3 consensus sequence (CAUCC → GAGUC) abolished the association of SRSF3 (Fig. 1C–E). Importantly, reduction of the levels of SRSF3 induced a dramatic decrease in the amount of SRSF3 bound to the SRSF3 consensus sequence in TNBC cells (Fig. 1F). These data demonstrate that SRSF3 associates with exon 4 of CPEB2 in a specific manner via the SRSF3 consensus sequence, CAUCC.
Downregulation of SRSF3 reduced the inclusion of exon 4 into the mature CPEB2 mRNA transcript.
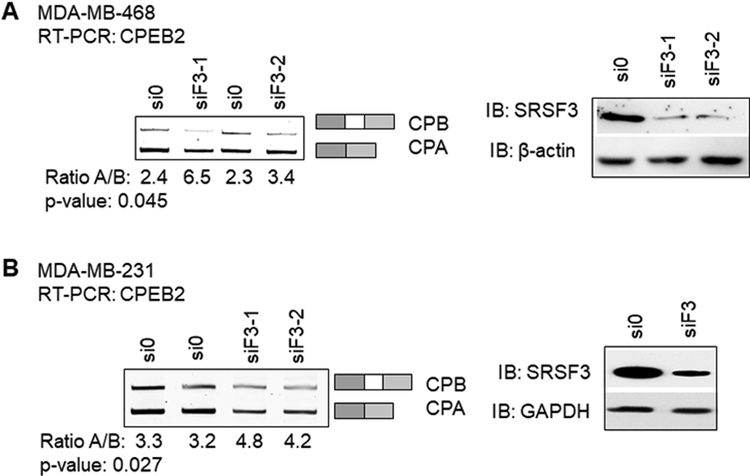
To determine whether SRSF3 regulated the inclusion/exclusion of CPEB2 exon 4, multiple siRNA sequences were utilized to downregulate SRSF3 as well as the other siRNA trans-factors identified in the unbiased proteomic screen (Fig. 2, Table 1). Downregulation of ≥ 75% of each protein was achieved in MDA-MB-468 and MDA-MB-231 cells (Table 2, Fig. 2) using SRSF3-specific siRNA as compared to siRNA controls. Downregulation of SRSF3 induced a significant increase in the CPEB2A/B mRNA and protein ratio demonstrating a decrease in the inclusion of exon 4 into the mature CPEB2 mRNA transcript (Fig. 2A & 2B). Reducing the SRSF3 levels in MDA-MB-468 cell line, which inherently expresses more of the CPEB2B isoform versus MDA-MB-231 (ratio CPA/CPB 2.4 versus 3.3, respectively), also increased the CPA/CPB ratio (Fig. 2A & 2B). These data demonstrate that SRSF3 is an RNA trans-factor that functions to enhance the inclusion of exon 4 into the mature mRNA transcript.
Figure 2: Downregulation of SRSF3 decreases exon 4 inclusion in endogenous CPEB2 transcript and correlates to loss of CPEB2B protein expression.
A. MDA-MB-468 cells were subjected to nonspecific siRNA treatment (si0) or two different siRNA sequences specific to SRSF3 (siF3–1 and siF3–2), and endogenous levels of either CPEB2A (CPA) or CPEB2B (CPB) mature RNA transcript were detected by RT-PCR with primers spanning exon 4 (left gel). Western blot analysis of the siRNA treated MDA-MB-468 cells are shown to indicate expression of SRSF3 in siRNA depleted samples (right blot). B. After validating siRNA sequences in the MDA-MB-468 cell line, MDA-MB-231 cells were treated with a combination of the two siRNA sequences (equal molar concentrations) in duplicate. RT-PCR products for endogenous CPEB2 isoform mRNA were quantified via densitometry (right gel). Western blot analysis of the siRNA treated samples in MDA-MB-231 cells are shown to indicate expression of SRSF3 in siRNA depleted samples.
Table 2.
Percent “knock-down” and siRNA IDs for proteins identified in screen
| Protein | %of control signal | Thermo Fisher siRNA ID# |
|---|---|---|
| No Treatment | NA | NA |
| Non-targeting control | NA | 4404020 |
| SRSF3 | 0.13±0.09 | s1273, s12732, s12733 (pooled) |
| hnRNP H1 | 0.11±0.06 | s6728, s6729, s6730 (pooled) |
| hnRNP F | 0.20±0.11 | s6725, s6727 s230280 (pooled) |
| hnRNP H1/F | (see above) | (see above) |
The consensus cis-element for SRSF3 is essential for the inclusion of CPEB2 exon 4.
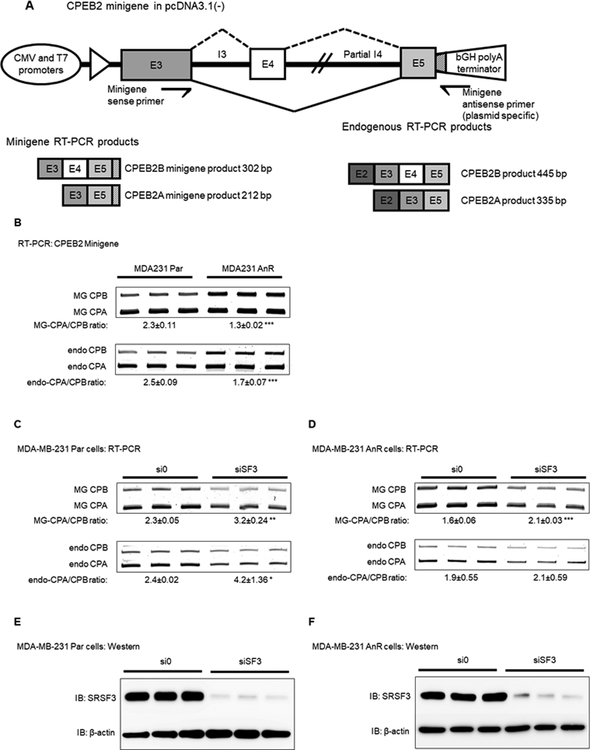
The consensus pentamer nucleotide motif for SRSF3 association is contained within CPEB2 exon 4, and this motif has previously been reported to promote alternative splicing of target pre-RNAs including coding and non-coding species (31). To interrogate the SRSF3 binding site in CPEB2 exon 4 for regulation of CPEB2 AS, a minigene reporter system was generated for mutational analysis of this RNA cis-element (Fig. 3A) (26,32,33). Specifically, CPEB2 exon 3, intron 3, exon 4, part of intron 4, and most of exon 5 were cloned into a mammalian expression vector under a CMV promoter (pcDNA3.1(–)). Utilizing competitive RT-PCR the inclusion/exclusion of exon 4 into the minigene mRNA was assayed with a plasmid-specific, reverse primer to ensure the assay remains reporter specific rather than amplifying endogenous splicing events (Fig. 3A). Both MDA-MB-231 Par and AnR cells were evaluated for their minigene splicing levels to test whether increased SRSF3 expression in AnR cells directly correlated with CPEB2 splicing in TNBC. Importantly, expression of the CPEB2 minigene in TNBC cells demonstrated a similar CPEB2 A/B mRNA ratio as endogenous CPEB2 mRNA (2.3±0.11 for the MG vs 2.5±0.09 for endogenous) in MDA-MB-231 Par cells (Fig. 3B). While wild type CPEB2 minigene splicing in MDA-MB-231 AnR cells followed a similar trend (1.3±0.02 for the MG vs 1.7±0.07 for endogenous) (Fig. 3B).
Figure 3: Modulating levels of SRSF3 in TNBC cells affects the CPEB2 splice isoform ratio at exon 4.
A. Schematic of CPEB2 exon 3–4/5 minigene. Genomic DNA was amplified from RPCI-11 Hs BAC Clone using primers which spanned the entirety of exon 3, 4, and the majority of exon 5. The complete sequence for intron 3 was included, and partial sequence of intron 4 was included. Primers specific to the minigene were used to detect splicing events in RT-PCR analysis. B. MDA-MB-231 Par cells were compared to MDA-MB-231 AnR cells for basal levels of minigene splicing for the MG-specific CPA/CPB (MG) ratio and compared to endogenous CPEB2 splicing (endo). C-D. MDA-MB-231 Par (C) and MDA-MB-231 AnR (D) cells were treated with a combination of two siRNA sequences targeting SRSF3 (shown in Fig.1). CPEB2 minigene splicing and endogenous CPEB2 splicing was detected via RT-PCR. E-F. MDA-MB-231 Par (E) and MDA-MB-231 AnR (F) SRSF3 protein levels were detected after siRNA treatment as indicated by Western blot. Representative images from 3 independent experiments are shown, and for all quantitation n = 3 ± standard deviation (sd) via densitometry. Statistical significance is reported as p-value from oneway ANOVA pooled t-test of the MG or endo CPA/CPB ratio. (* = p-value < 0.05, ** = p-value < 0.01, *** = p-value < 0.001).
To determine whether SRSF3 expression affected the inclusion/exclusion of CPEB2 exon 4, siRNA downregulation of SRSF3 using siRNA induced a significant increase in the CPEB2A/B minigene mRNA ratio mimicking the effects observed on endogenous levels (Fig. 3C–F). Reductions in CPEB2B splicing were significantly different in siRNA treatment to SRSF3 compared to control siRNA and was observed to be approximately 1.3 – 1.4 fold less in both the parental and AnR cell lines (p-value = 0.0023 for Par and p-value = 0.0003 for AnR cells).
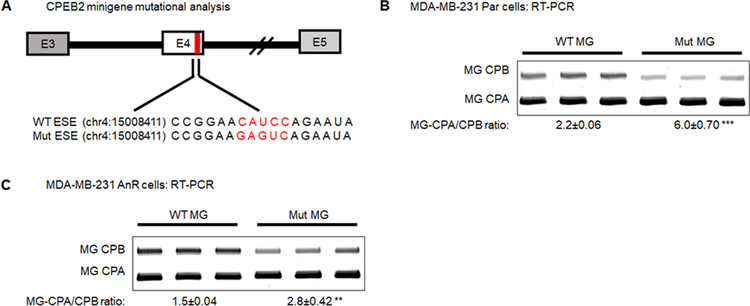
To show SRSF3 binding to CPEB2 exon 4 is required to mediate enhancement of exon 4 inclusion, site-directed mutagenesis was performed on the putative SRSF3 ESE site in exon 4 (Fig. 4A) (31,34). Mutations using nucleotide substitution with residues predicted to ablate binding (mutations utilized in Fig. 1C and D) were tested. Mutation of the SRSF3 consensus sequence reduced the CPEB2A/B minigene mRNA ratio basally in MDA-MB-231 Par and AnR cells, although not to equal amounts (6.0±0.70 and 2.8±0.42, respectively, Fig. 4B & 4C). Mutating the ESE cis-acting RNA element specific for SRSF3 reduced splicing of the CPEB2B isoform significantly in both parental and AnR cells (p-value = 0.0007 and 0.0053, respectively). These data demonstrate that SRSF3 regulates the inclusion of CPEB2 exon 4 via association with the CAUCC pentameric sequence.
Figure 4: Mutational analysis of SRSF3 RNA cis-element indicates the pentameric ESE is essential for CPEB2 exon 4 inclusion.
A. Schematic representation of the mutant ESE minigene. Red bar indicates the position of the RNA cis-element is located near the 5’ splice site. Genomic coordinates indicate the first nucleotide base in the RNA cis-element and were extracted from Genome Reference Consortium Human Build 38 (GRCh38.p12) B. RT-PCR analysis of MDA-MB-231 Par cells for the wild type (WT) CPEB2 ESE compared to mutant (Mut) MG-specific CPA/CPB ratio. C. RT-PCR analysis of MDA-MB-231 AnR cells for the WT CPEB2 ESE compared to Mut MG-specific CPA/CPB ratio. Representative images from 3 independent experiments are shown. All quantitation shown as n = 3 ± standard deviation (sd) via densitometry. Statistical significance is reported as p-value from oneway ANOVA pooled t-test of the MG CPA/CPB ratio. (* = p-value < 0.05, ** = p-value < 0.01, *** = p-value < 0.001).
SRSF3 regulates the resistance of TNBC cells to anoikis by enhancing the inclusion of exon 4 in CPEB2 mRNA.
To determine whether SRSF3 was a key mediator of biological functions linked to CPEB2 AS (e.g., anoikis), SRSF3 was downregulated in both parental and anoikis resistant TNBC cells (MDA-MB-231 AnR cells). Interestingly, downregulation of SRSF3 induced an increase in both basal apoptosis as well as detachment induced cell death in both AnR cell lines (Fig. 5A and data not shown). Conversely, SRSF3 downregulation did not, by itself, induce apoptosis in the parental line (Fig. 5B). Importantly, ectopic expression of the CPEB2B isoform (exon 4 included) blocked the effect of SRSF3 downregulation for cleaved Caspase-3 (but, interestingly, not PARP cleavage in the AnR line) (Fig. 5A). These data provide a biological link between the SRSF3 RNA trans-factor, its splicing target, and subsequent phenotypic presentation (i.e. anoikis sensitivity). Taken together, this study shows clear evidence of the SRSF3/CPEB2 AS axis regulating anoikis sensitivity by inducing the expression of the CPEB2B isoform.
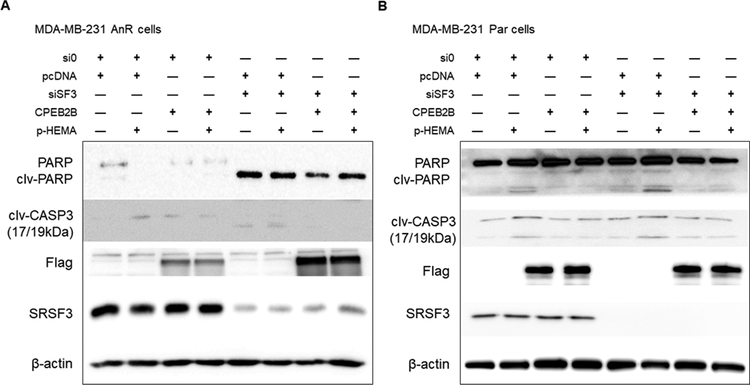
Figure 5: SRSF3 modulates TNBC cells’ sensitivity to cell death due to anoikis via expression of CPEB2B.
siRNA treatment was applied for a 48-hour cycle then incubated for 6 hours on adherent substrate or poly-HEMA substrate which forced cells into suspension. After incubation, early stage apoptosis was evaluated using Western blot to probe for cleaved PARP (clv-PARP) and cleaved Caspase 3 (clv-CASP3). Antibodies for apoptotic markers detected full-size PARP (116 kDa) and cleaved PARP (89 kDa), and both large fragments of activated cleaved Caspase 3 (17/19 kDa doublet). A. MDA-MB-231 AnR cells were treated as indicated in the plus/minus graphical organizer with non-specific siRNA control (si0), pcDNA3.1(–) empty vector (pcDNA), siRNA to SRSF3 (siSF3), CPEB2B-Flag overexpression plasmid (CPEB2B), and poly-HEMA coated substrate (p-HEMA). Samples shown representative of experiments done in triplicate for each treatment. B. MDA-MB-231 Par cells were treated identically to the cells described in panel A.
Expression of SRSF3 is enhanced in human TNBC and basal-like breast cancer.
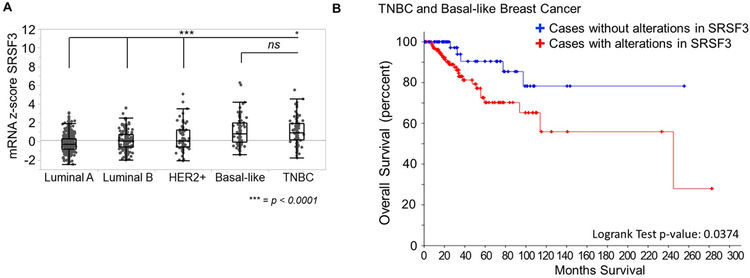
This mechanistic study linking SRSF3 with CPEB2B expression coupled to our reports that CPEB2B is highly expressed in human TNBC and regulates TNBC metastasis, forms the premise that SRSF3 expression will correlate with human TNBC and the aggressiveness of human breast cancers. To examine the validity of this premise, sequence data contained in TCGA Breast Invasive Carcinoma (BRCA) dataset of unique patient cases with both RNAseq and clinical data associated were evaluated for mRNA levels of SRSF3 (35). The mRNA z-scores were extracted for each PAM50 gene-expression based subtyping. TNBC and basal-like subtypes showed the highest expression of SRSF3, while Luminal A, Luminal B, and HER2+ subtypes all expressed significantly less (Fig. 6A). Using the R coding platform to interrogate the TCGA survival data (36), we observed stratifying patients by SRSF3 expression was predictive of survival probability (Fig. 6B). These data support the prediction that in TNBC with high SRSF3 expression, the CPEB2A/B isoform ratio will be low, and provides further insight into the powerful role of dysregulated AS in cancer progression and metastasis.
Figure 6: SRSF3 is over represented in the most aggressive and metastatic breast cancers in The Cancer Genome Atlas.
A. mRNA z-scores were derived from patient samples with both clinical breast cancer subtype based on PAM50 and histopathology and RNASeq data in the the Breast Invasive Carcinoma (TCGA, Cell 2015) data set containing 1105 patient samples. SRSF3 expression data across the main breast cancer subtypes was mined from cBioPortal and evaluated using ANOVA with post-hoc Tukey HSD (** = p-value < 0.01, *** = p-value < 0.0001). B. Clinical overall survival data for BRCA data set was extracted from TCGA for a combined Basal-like and TNBC cohort consisting of 131 patients and survival probability based on SRSF3 expression levels were plotted on Kaplan-Meyer curve using cBioPortal. Cases with alterations were defined as deviating from the mRNA z-score in the upper or lower quantiles of the population. Statistical significance is reported as logrank test analysis (p-value = 0.0323).
DISCUSSION
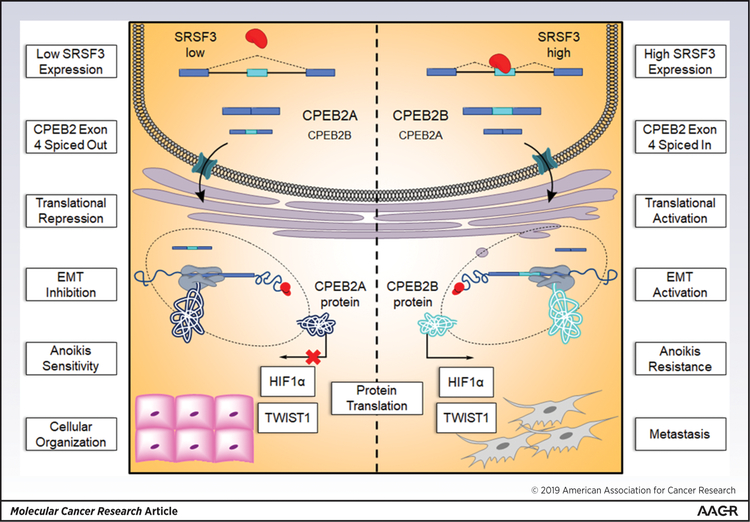
Herein, our laboratory provides evidence which describes the mechanism for AS of CPEB2 driven through the trans-RNA splicing factor SRSF3 in TNBC. Specifically, SRSF3 expression is upregulated in hypoxic conditions found in the tumor microenvironment typical of solid and malignant breast tumors. Previously we reported two isoforms of CPEB2, CPEB2A and CPEB2B, exert opposing roles in the transformation from primary to metastatic phenotype in TNBC through acquisition of AnR (15). For the first time, the proto-oncogene, SRSF3, is shown to promote the pro-metastatic CPEB2B isoform. We have identified AS of CPEB2 precursor-mRNA is facilitated through SRSF3 by binding to a pyrimidine-rich, pentameric ESE found at the distal 3’ end of coding exon 4. This interaction promotes inclusion of the characteristically weak CPEB2B coding exon 4 into the mature transcript and leads to activation of signaling pathways, which initiate epithelial to mesenchymal transition (EMT) pathology (Fig. 7). Importantly, SRSF3-mediated splicing of CPEB2 exon 4 is dependent on the fidelity of its RNA cis-element ESE in exon 4. Indeed, we observe a reversal of the CPEB2A/B ratio by substitution mutation in the CPEB2 minigene reporter at the SRSF3 ESE. Finally, modulating the expression of SRSF3 through siRNA to deplete expressed protein levels led to a decrease in CPEB2B transcript levels, thus reversing the low CPA/CPB ratio to the higher ratio observed in cells sensitive to anoikis. Reversing this ratio also caused increased apoptosis in the biological phenotype, especially for the non-AnR cell line. The biological phenotype observed did not, however, show robust rescue by forced CPEB2B expression in the AnR cells as predicted in our original hypothesis. Levels of cleaved PARP were not attenuated by circumventing SRSF3 activity with the CPEB2B transient transfection in the splicing factor depleted group. These data suggest that the maintenance of survival after detachment is dependent on SRSF3 expression.
Figure 7.
Proposed pathway for the mechanism of SRSF3 and CPEB2 splicing which partially contributes to the metastasis of TNBC.
SRSF3 plays a substantial role in regulation of AS, and increasing levels of SR proteins bound to ESE prevents exon skipping (37). Recent studies reveal that SRSF3 may play an antagonistic role compared to other SR-family proteins where SRSF3 acts to promote exon inclusion leading to tumor initiation, progression, and/or resistance (10,22,38). Indeed, data suggests SRSF3 is a potential master regulator of the transcriptome by the observation that it binds multiple SR-family proteins and promotes “poison cassette exon” autoregulatory feedback loops through nonsense mediated decay (NMD) (31). While the current research shows CPEB2 AS is tightly connected to SRSF3 binding at the ESE of exon 4 in precursor mRNA, this alone may not be the sole regulatory mechanism for CPEB2 splicing. For example, we observed short-term transient overexpression of SRSF3 cDNA in MDA-MB-231 cells did not lead to increases in CPEB2B levels as would be predicted (data not shown). In fact, protein levels of SRSF3 may be increased due to chromosomal amplifications in the MDA-MB-231 cell line which is shown to be pentameric at the SRSF3 locus 6p21.1 (MDA-MB-231 SKY/M-FISH SKYGRAM, (39)). One possible explanation suggests alternative oncogenic pathways involving extensive post-translational modifications (PTMs) which both hyperactivate SRSF3 and provide resistance to proteolytic degradation are at work in the splicing regimen of TNBC cells. Further investigation into the activity of SRSF3 under various post-translational modifications may uncover the influence this splicing factor exerts in driving the CPEB2A/B isoform ratio lower seen in AnR and TNBC.
In regard to PTMs of SRSF3, NEDDylation, a modification covalently linking the small ubiquitin-like protein NEDD8 to Lys85 of SRSF3 has been shown to promote stress granule (SG) assembly during times of oxidative stress (40). This metabolically efficient strategy is co-opted by tumor cells experiencing persistent stress to keep nascent mRNA and proteins compartmentalized and ready to regulate signaling pathway activity (41). Not only does SRSF3 localize to SG, but both CPEB2A and B proteins contain a low complexity domain (LCD) immediately distal to the amino acids encoded by exon 4 in the primary amino acid sequence of CPEB2B. The LCD of CPEB2 and other RNA binding proteins contribute dynamically to the formation of ordered sub-organelle compartments such as the SG (42). Furthermore, extensive phosphorylation of SRSF3 at the RS domain is crucial for spliceosomal assembly, while AS catalysis is dependent on subsequent dephosphorylation. The cycle of phosphorylation/dephosphorylation also allows SRSF3 to interact with the nuclear export factor, NXF1 to couple AS and polyadenylation during NXF1-mediated mRNA export (43). In the context of TNBC where SRSF3 levels are inherently high, one can hypothesize that the metastatic potential of primary tumors may be activated by CPEB2 precursor mRNA bound by highly phosphorylated SRSF3 at exon 4 and shuttled to SG in the cytoplasm, thereby increasing cytoplasmic abundance of the CPEB2B transcript via promotion of exon inclusion and activating the EMT pathway DNA trans-factors TWIST1 and HIF1α (Fig. 7).
Regarding other proteins in the SR family, it is well established in the literature that these trans-factors regulate a variety of apoptosis/survival proteins and thereby hallmarks of cancer (reviewed in 44). For example, SRSF1 is involved in both breast and lung cancer pathogenesis and SRSF6 has been shown to regulate Bim expression in melanoma (44). Hence, our data will not “rule out” other SR-dependent pathways which may induce breast cancer metastasis.
Defective AS frequently impacts dysregulation in many of the hallmarks of cancer which have potential to promote primary tumor metastasis (45). In the case of TNBC, the higher level of genetic heterogeneity may be attributed to defects in pre-mRNA splicing and the resulting structural protein variants which contribute to therapeutic resistance (46). Increased interest in targeting not only the products of aberrant splicing in cancer, but also trans-RNA splicing factors, namely SRSF3, using antisense oligonucleotide (ASO)-mediated inhibition has emerged and appears to be a promising step in development of novel therapeutic targets in cancer (47,48). Certainly, the feasibility of targeting the SRSF3:CPEB2B splicing paradigm is worth pursuing in the context of TNBC.
In summary, we have identified SRSF3 as the trans-RNA splicing factor responsible for mediating AS of CPEB2 through inclusion of exon 4 via binding at the cis-RNA pentameric ESE. Our study demonstrates that the SRSF3:CPEB2B splicing paradigm is essential for the acquisition of AnR in TNBC. We also show that siRNA-mediated depletion of SRSF3 results in a loss of resistance to anchorage independent growth and reverts the CPEB2A/ B ratio to that of nontumorigenic breast tissue. Further studies will be needed to derive the activation of this novel splicing ontology and its potential to translate into a tangible clinical target for human breast carcinoma.
ACKNOWLEDGEMENTS
The authors would like to heartily thank Dr. Binks Wattenberg for his assistance and critical review in this effort.
GRANT SUPPORT
This work was supported by research grants from the American Cancer Society (ACS), IRG-17-173-22 (MAP), U.S. Department of Veterans Affairs (VA Merit Review, I BX001792 (CEC) and a Research Career Scientist Award, 13F-RCS-002 (CEC); from the National Institutes of Health via TR000057-04 (MAP), 1U01HD087198-01 (CEC), CA117950 (CEC), CA154314 (CEC) and AI139072 (CEC). The contents of this manuscript do not represent the views of the Department of Veterans Affairs or the United States Government.
Footnotes
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article
References Cited
- 1.Schmadeka R, Harmon BE, and Singh M (2014) Triple-negative breast carcinoma: current and emerging concepts. Am J Clin Pathol 141, 462–477 [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas N (2012) Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Zhang T, Kwiatkowski N, Abraham BJ, Lee TI, Xie S, Yuzugullu H, Von T, Li H, Lin Z, Stover DG, Lim E, Wang ZC, Iglehart JD, Young RA, Gray NS, and Zhao JJ (2015) CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell 163, 174–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, and Narod SA (2007) Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 13, 4429–4434 [DOI] [PubMed] [Google Scholar]
- 5.Pradella D, Naro C, Sette C, and Ghigna C (2017) EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer 16, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pareja F, Geyer FC, Marchio C, Burke KA, Weigelt B, and Reis-Filho JS (2016) Triple-negative breast cancer: the importance of molecular and histologic subtyping, and recognition of low-grade variants. NPJ Breast Cancer 2, 16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, and Pietenpol JA (2011) Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121, 2750–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eswaran J, Horvath A, Godbole S, Reddy SD, Mudvari P, Ohshiro K, Cyanam D, Nair S, Fuqua SA, Polyak K, Florea LD, and Kumar R (2013) RNA sequencing of cancer reveals novel splicing alterations. Sci Rep 3, 1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, and Gertler FB (2011) An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet 7, e1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gautrey H, Jackson C, Dittrich AL, Browell D, Lennard T, and Tyson-Capper A (2015) SRSF3 and hnRNP H1 regulate a splicing hotspot of HER2 in breast cancer cells. RNA Biol 12, 1139–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anczukow O, Akerman M, Clery A, Wu J, Shen C, Shirole NH, Raimer A, Sun S, Jensen MA, Hua Y, Allain FH, and Krainer AR (2015) SRSF1-Regulated Alternative Splicing in Breast Cancer. Mol Cell 60, 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ke H, Zhao L, Zhang H, Feng X, Xu H, Hao J, Wang S, Yang Q, Zou L, Su X, Wang L, Wu C, Wang Y, Nie J, and Jiao B (2018) Loss of TDP43 inhibits progression of triple-negative breast cancer in coordination with SRSF3. Proc Natl Acad Sci U S A 115, E3426–E3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silipo M, Gautrey H, and Tyson-Capper A (2015) Deregulation of splicing factors and breast cancer development. J Mol Cell Biol 7, 388–401 [DOI] [PubMed] [Google Scholar]
- 14.Johnson RM, Vu NT, Griffin BP, Gentry AE, Archer KJ, Chalfant CE, and Park MA (2015) The Alternative Splicing of Cytoplasmic Polyadenylation Element Binding Protein 2 Drives Anoikis Resistance and the Metastasis of Triple Negative Breast Cancer. J Biol Chem 290, 25717–25727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLigio JT, Lin G, Chalfant CE, and Park MA (2017) Splice variants of cytosolic polyadenylation element-binding protein 2 (CPEB2) differentially regulate pathways linked to cancer metastasis. J Biol Chem 292, 17909–17918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez-Miranda G, and Mendez R (2012) The CPEB-family of proteins, translational control in senescence and cancer. Ageing Res Rev 11, 460–472 [DOI] [PubMed] [Google Scholar]
- 17.D’Ambrogio A, Nagaoka K, and Richter JD (2013) Translational control of cell growth and malignancy by the CPEBs. Nat Rev Cancer 13, 283–290 [DOI] [PubMed] [Google Scholar]
- 18.Giangarra V, Igea A, Castellazzi CL, Bava FA, and Mendez R (2015) Global Analysis of CPEBs Reveals Sequential and Non-Redundant Functions in Mitotic Cell Cycle. PLoS One 10, e0138794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin S, and Fu XD (2007) SR proteins and related factors in alternative splicing. Adv Exp Med Biol 623, 107–122 [DOI] [PubMed] [Google Scholar]
- 20.Long JC, and Caceres JF (2009) The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417, 15–27 [DOI] [PubMed] [Google Scholar]
- 21.Anko ML (2014) Regulation of gene expression programmes by serine-arginine rich splicing factors. Semin Cell Dev Biol 32, 11–21 [DOI] [PubMed] [Google Scholar]
- 22.Jia R, Li C, McCoy JP, Deng CX, and Zheng ZM (2010) SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int J Biol Sci 6, 806–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kano S, Nishida K, Kurebe H, Nishiyama C, Kita K, Akaike Y, Kajita K, Kurokawa K, Masuda K, Kuwano Y, Tanahashi T, and Rokutan K (2014) Oxidative stress-inducible truncated serine/arginine-rich splicing factor 3 regulates interleukin-8 production in human colon cancer cells. Am J Physiol Cell Physiol 306, C250–262 [DOI] [PubMed] [Google Scholar]
- 24.Ajiro M, Jia R, Yang Y, Zhu J, and Zheng ZM (2016) A genome landscape of SRSF3-regulated splicing events and gene expression in human osteosarcoma U2OS cells. Nucleic Acids Res 44, 1854–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vu NT, Park MA, Shultz MD, Bulut GB, Ladd AC, and Chalfant CE (2016) Caspase-9b Interacts Directly with cIAP1 to Drive Agonist-Independent Activation of NF-kappaB and Lung Tumorigenesis. Cancer Res 76, 2977–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vu NT, Park MA, Shultz JC, Goehe RW, Hoeferlin LA, Shultz MD, Smith SA, Lynch KW, and Chalfant CE (2013) hnRNP U enhances caspase-9 splicing and is modulated by AKT-dependent phosphorylation of hnRNP L. J Biol Chem 288, 8575–8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shultz JC, Goehe RW, Wijesinghe DS, Murudkar C, Hawkins AJ, Shay JW, Minna JD, and Chalfant CE (2010) Alternative splicing of caspase 9 is modulated by the phosphoinositide 3-kinase/Akt pathway via phosphorylation of SRp30a. Cancer Res 70, 9185–9196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbour SE, Nguyen PT, Park M, Emani B, Lei X, Kambalapalli M, Shultz JC, Wijesinghe D, Chalfant CE, and Ramanadham S (2015) Group VIA Phospholipase A2 (iPLA2beta) Modulates Bcl-x 5’-Splice Site Selection and Suppresses Anti-apoptotic Bcl-x(L) in beta-Cells. J Biol Chem 290, 11021–11031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goehe RW, Shultz JC, Murudkar C, Usanovic S, Lamour NF, Massey DH, Zhang L, Camidge DR, Shay JW, Minna JD, and Chalfant CE (2010) hnRNP L regulates the tumorigenic capacity of lung cancer xenografts in mice via caspase-9 pre-mRNA processing. J Clin Invest 120, 3923–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shapiro BA, Vu NT, Shultz MD, Shultz JC, Mietla JA, Gouda MM, Yacoub A, Dent P, Fisher PB, Park MA, and Chalfant CE (2016) Melanoma Differentiation-associated Gene 7/IL-24 Exerts Cytotoxic Effects by Altering the Alternative Splicing of Bcl-x Pre-mRNA via the SRC/PKCdelta Signaling Axis. J Biol Chem 291, 21669–21681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anko ML, Muller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, and Neugebauer KM (2012) The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol 13, R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massiello A, Salas A, Pinkerman RL, Roddy P, Roesser JR, and Chalfant CE (2004) Identification of two RNA cis-elements that function to regulate the 5’ splice site selection of Bcl-x pre-mRNA in response to ceramide. J Biol Chem 279, 15799–15804 [DOI] [PubMed] [Google Scholar]
- 33.Patel NA, Apostolatos HS, Mebert K, Chalfant CE, Watson JE, Pillay TS, Sparks J, and Cooper DR (2004) Insulin regulates protein kinase CbetaII alternative splicing in multiple target tissues: development of a hormonally responsive heterologous minigene. Mol Endocrinol 18, 899–911 [DOI] [PubMed] [Google Scholar]
- 34.Akerman M, David-Eden H, Pinter RY, and Mandel-Gutfreund Y (2009) A computational approach for genome-wide mapping of splicing factor binding sites. Genome Biol 10, R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, Bowlby R, Shen H, Hayat S, Fieldhouse R, Lester SC, Tse GM, Factor RE, Collins LC, Allison KH, Chen YY, Jensen K, Johnson NB, Oesterreich S, Mills GB, Cherniack AD, Robertson G, Benz C, Sander C, Laird PW, Hoadley KA, King TA, Network TR, and Perou CM (2015) Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 163, 506–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, Ceccarelli M, Bontempi G, and Noushmehr H (2016) TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 44, e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ibrahim EC, Schaal TD, Hertel KJ, Reed R, and Maniatis T (2005) Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc Natl Acad Sci U S A 102, 5002–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sotillo E, Barrett DM, Black KL, Bagashev A, Oldridge D, Wu G, Sussman R, Lanauze C, Ruella M, Gazzara MR, Martinez NM, Harrington CT, Chung EY, Perazzelli J, Hofmann TJ, Maude SL, Raman P, Barrera A, Gill S, Lacey SF, Melenhorst JJ, Allman D, Jacoby E, Fry T, Mackall C, Barash Y, Lynch KW, Maris JM, Grupp SA, and Thomas-Tikhonenko A (2015) Convergence of Acquired Mutations and Alternative Splicing of CD19 Enables Resistance to CART-19 Immunotherapy. Cancer Discov 5, 1282–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knutsen T, Gobu V, Knaus R, Padilla-Nash H, Augustus M, Strausberg RL, Kirsch IR, Sirotkin K, and Ried T (2005) The interactive online SKY/M-FISH & CGH database and the Entrez cancer chromosomes search database: linkage of chromosomal aberrations with the genome sequence. Genes Chromosomes Cancer 44, 52–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jayabalan AK, Sanchez A, Park RY, Yoon SP, Kang GY, Baek JH, Anderson P, Kee Y, and Ohn T (2016) NEDDylation promotes stress granule assembly. Nat Commun 7, 12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grabocka E, and Bar-Sagi D (2016) Mutant KRAS Enhances Tumor Cell Fitness by Upregulating Stress Granules. Cell 167, 1803–1813 e1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R, and McKnight SL (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller-McNicoll M, Botti V, de Jesus Domingues AM, Brandl H, Schwich OD, Steiner MC, Curk T, Poser I, Zarnack K, and Neugebauer KM (2016) SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev 30, 553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kędzierska H, and Piekiełko-Witkowska A (2017) Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer. Cancer Lett. 396, 53–65 [DOI] [PubMed] [Google Scholar]
- 45.Sveen A, Kilpinen S, Ruusulehto A, Lothe RA, and Skotheim RI (2016) Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 35, 2413–2427 [DOI] [PubMed] [Google Scholar]
- 46.Lawrence RT, Perez EM, Hernandez D, Miller CP, Haas KM, Irie HY, Lee SI, Blau CA, and Villen J (2015) The Proteomic Landscape of Triple-Negative Breast Cancer. Cell Rep 11, 990. [DOI] [PubMed] [Google Scholar]
- 47.Dewaele M, Tabaglio T, Willekens K, Bezzi M, Teo SX, Low DH, Koh CM, Rambow F, Fiers M, Rogiers A, Radaelli E, Al-Haddawi M, Tan SY, Hermans E, Amant F, Yan H, Lakshmanan M, Koumar RC, Lim ST, Derheimer FA, Campbell RM, Bonday Z, Tergaonkar V, Shackleton M, Blattner C, Marine JC, and Guccione E (2016) Antisense oligonucleotide-mediated MDM4 exon 6 skipping impairs tumor growth. J Clin Invest 126, 68–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo J, Che X, Wang X, Jia R (2018) Inhibition of the expression of oncogene SRSF3 by blocking an exonic splicing suppressor with antisense oligonucloetides. RSC Adv 8, 7159–7163 [DOI] [PMC free article] [PubMed] [Google Scholar]