Abstract
Combined BRAF and MEK inhibition is a standard of care in patients with advanced BRAF mutant melanoma but acquired resistance remains a challenge that limits response durability. Here we quantitated in vivo ERK½ activity and tumor response associated with resistance to combined BRAF and MEK inhibition in mutant BRAF xenografts. We found that ERK½ pathway re-activation preceded the growth of resistant tumors. Moreover, we detected a subset of cells that not only persisted throughout long term treatment but restored ERK½ signaling and grew upon drug removal. Cell lines derived from combination-resistant tumors (CRTs) exhibited elevated ERK½ phosphorylation, which were sensitive to ERK½ inhibition. In some CRTs, we detected a tandem duplication of the BRAF kinase domain. Monitoring ERK½ activity in vivo was efficacious in predicting tumor response during intermittent treatment. We observed maintained expression of the mitotic regulator, polo-like kinase 1 (Plk1), in melanoma resistant to BRAF and MEK inhibitors. Plk1 inhibition induced apoptosis in CRTs, leading to slowed growth of BRAF and MEK inhibitor-resistant tumors in vivo. These data demonstrate the utility of in vivo ERK½ pathway reporting as a tool to optimize clinical dosing schemes and establish suppression of Plk1 as potential salvage therapy for BRAF inhibitor and MEK inhibitor-resistant melanoma.
Keywords: melanoma, MEK, vemurafenib, resistance, BRAF
INTRODUCTION
Both targeted therapeutic strategies and immune checkpoint blockade have advanced treatment options for cutaneous melanoma patients (1). Fifty percent of melanoma harbor V600E/K mutant forms of BRAF, which encode constitutively active kinases that enhance MEK-ERK½ signaling to promote growth and survival (2,3). Mutant BRAF is druggable with RAF inhibitors including vemurafenib and dabrafenib (4) and vertical inhibition with MEK inhibitors improves response rates, progression-free survival, and overall survival (5–8). To date, the BRAF and MEK combination therapies of dabrafenib with trametinib, vemurafenib with cobimetinib, and encorafenib with binimetinib are FDA-approved for mutant BRAF V600E/K melanoma (9,10).
Despite these improvements, resistance to targeted inhibitors still develops in most patients. Mechanisms of resistance to BRAF monotherapy have been well characterized (reviewed in (11)). Studies on resistance to combined BRAF and MEK inhibitors have also implicated ERK½ re-activating alterations (12), which are driven by similar, but amplified, mechanisms of resistance to BRAF monotherapy (13). Although the importance of ERK½ signaling is established, the degree of ERK½ pathway inhibition and the timing of re-activation and signaling have not been examined during treatment and after progression during BRAF and MEK inhibition.
Here, we used an in vivo reporter system to monitor ERK½ activity throughout initial response and subsequent resistance to combined BRAF and MEK inhibitors (14). We found that combination treatment abolished ERK½ signaling, and re-activation was associated with regrowth of residual disease and acquired resistance. Given previous data and clinical interest in the use of intermittent dosing to mitigate toxicities and to delay resistance onset, we also established the utility of the reporter system as a potential tool for schedule optimization. Using a phospho-proteomics array to probe for potential salvage therapy targets, we found that expression of polo-like kinase 1 (Plk1) associated with resistance, which when inhibited reduced growth and induced cell death in combination treatment-resistant melanoma.
MATERIALS AND METHODS
Cell culture
1205Lu cells (donated by Dr. Meenhard Herlyn, Wistar Institute, Philadelphia, PA in 2005) were cultured in MCDB153 medium containing 2% FBS, 20% Leibovitz L-15 medium, with 5 μg/mL insulin. A375 cells (purchased from ATCC in 2005) were cultured in DMEM with 10% FBS. Cell lines were authenticated by sequencing at the NRAS and BRAF loci, and by STR analysis (completed July 2016). CRT cells were cultured in the same medium as parental cells with the addition of PLX4720 (1 μM) and PD0325901 (35 nM) (15,16). These levels maintain the same RAF inhibitor to MEK inhibitor ratio as used for in vivo experiments. PLX4720 was provided by Dr. Gideon Bollag (Plexxikon Inc., Berkeley, CA). PD0325901, SCH772984, GSK461364, and BI 6727 were purchased from Selleck Chemicals (Houston, TX).
Western blotting
Protein lysates were prepared in Laemmli sample buffer, separated by SDS-PAGE, and proteins were transferred to PVDF membranes. Immunoreactivity was detected using HRP-conjugated secondary antibodies (CalBioTech) and chemiluminescence substrate (Thermo Scientific) on a Chemidoc MP imaging system (BioRAD, Hercules, CA). Primary antibodies used are listed in the Supplemental Material.
Annexin V/PI analysis
Cells were treated for 72 hours at which time floating, and adherent cells were washed and incubated with 5 μL annexin V-APC (BD Biosciences) in 100 μL of binding buffer and 0.02 mg/mL propidium iodide for 15 minutes at room temperature. Cells were analyzed on the FACS Calibur or BD LSR II flow cytometers. Explanation of statistics performed can be found in the Supplemental Material.
S-phase entry analysis
S-phase incorporation was measured using the Click-it EdU Alexa Fluor 647 Flow Cytometry assay kit as per the manufacturer’s instructions (Molecular Probes, Grand Island, NY). EdU staining was quantified on the BD FacsCalibur and data were analyzed with FlowJo software.
Xenograft experiments
Human mutant BRAF melanoma cells (1205LuTR reporter and A375) were injected intradermally into athymic nude mice (1×106 cells/mouse) and allowed to grow. Size-matched tumors were placed into two treatment arms: chow laced with either vehicle or combination BRAF + MEK inhibitors (200 PPM PLX4720 plus 7 PPM PD0325901). For the Plk1 experiment, similar methods were to inject CRT 13 cells into mice on the combo chow. Formed tumors where removed from chow one day before treatment with 25 mg/kg/dose of GSK461364 or 5% DMSO PBS vehicle control by intraperitoneal injections Mice were treated every two days until the conclusion of the experiment. Luciferase activity and tumor volume were monitored 3 times a week for 21 days, described in detail in the supplemental material. GSK461364 was purchased from AdooQ BioScience,LLC (Irvine, CA) for in vivo experiments. All animal experiments were conducted in accordance with and approved by an Institutional Animal Care and Use Committee (IACUC).
IncuCyte® Live Cell Imaging Assay
IncuCyte® (Essen Bioscience, Ann Arbor, MI) imaging of Calcein AM (BD Biosciences, San Jose, CA) was used to measure viable cells. Cells were treated with indicated drugs for 72 hours, washed twice with PBS, then incubated for 5 minutes with 0.1 μM Calcein-AM PBS mixture. Green fluorescence was imaged and quantified with the IncuCyte®. Experiments were performed in triplicate, and statistical analysis performed using a two-tailed t-test assuming equal variance with error bars representing SEM.
Reverse phase protein analysis (RPPA)
Cells were seeded in 6-well plates in medium containing DMSO or 1 μM PLX4720 + 35 nM PD0325901 for 24 hours. Lysates from three independent experiments were processed and analyzed, as previously described (17).
Statistical analysis of tumor volume
Following injection, tumor volume and mouse weight were measured three times per week. In the BRAF inhibitor + MEK inhibitor experiments tumor volumes from both continuous and intermittent treatment groups were compared using the Wilcoxon rank sum with p-values adjusted for multiple testing using the Hochberg method (18). Survival curves for the Plk1 experiment were calculated with a log-rank test, using tumor volume > 1,200 mm3 as the endpoint.
Analysis of patient datasets
Analysis of normalized microarray expression data for tumors taken from patients before, during or after dabrafenib, vemurafenib or dabrafenib + trametinib treatment downloaded from NCBI’s Gene Expression Omnibus (GEO) database (accession number GSE99898) was performed using R (version 3.4.3). Data were retrieved from GEO via the GEOquery package (version 2.46.15) (19).
RESULTS
ERK½ signaling was reduced with BRAF plus MEK inhibitor treatment in vivo but was re-activated in acquired resistant xenografts
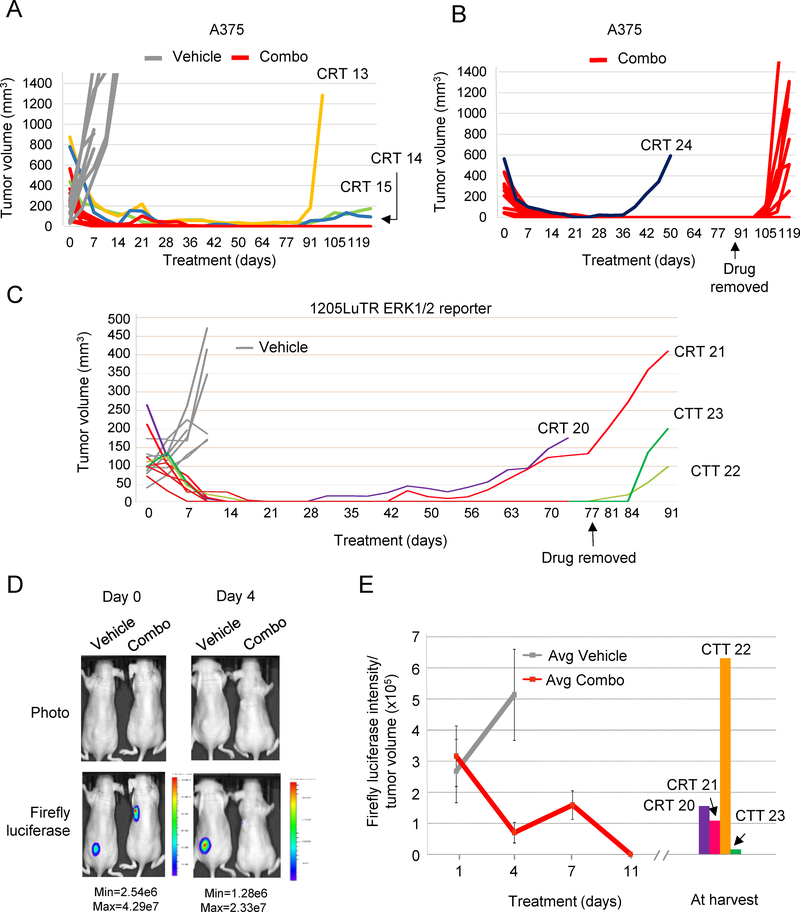
To initially develop tumors resistant to BRAF plus MEK inhibitors (combo), we used the well-characterized mutant V600E BRAF model, A375. Established A375 xenografts were treated continuously with either vehicle or combo chow. Eleven of the twelve combo-treated xenografts shrunk to a non-palpable mass and were considered to have regressed completely (Fig 1A). Prolonged treatment (>77 days) led to the development of acquired resistance with the regrowth of three combination-resistant tumors (CRT). In a second independent experiment, one A375-derived CRT (CRT 24) emerged from twelve starting tumors (Fig 1B). To test the durability of regression on the remaining 11 undetectable xenografts, we stopped treatment on day 91. Within 14 days, eight tumors re-grew indicating the presence of a subpopulation of tolerant cells that persisted during long term treatment. We termed these combination tolerant tumors (CTTs). Overall, these experiments show that combo treatment leads to drug-resistant melanoma. They also reveal that cells often persisted during long term treatment and grew after drug removal.
Fig 1. Melanoma xenografts developed resistance to prolonged combination BRAF inhibitor plus MEK inhibitor treatment in vivo.
(A) Human mutant BRAF melanoma A375 cells were injected intradermally into athymic nude mice and allowed to form palpable tumors. Mice were then given chow laced with either vehicle (n=8) or combination BRAF inhibitor plus MEK inhibitor (200 mg/kg of PLX4720 + 7 mg/kg of PD0325901; n=12). Graphed is tumor volume, as measured with digital calipers, versus time in days. Three resistant tumors arose in this cohort by day 91 (CRT 13, CRT 14, and CRT 15). (B) Tumor volumes of an independent cohort of combo-treated A375 xenografts (n=12). One CRT emerged on day 42, and the remaining 11 CTTs grew after drug removal on day 91. (C) 1205LuTR ERK½ reporter cells were used in similar experiments. Two mice developed acquired resistant tumors (CRT 20 and CRT 21). Tumor-free mice (n=6) remained on drug until day 77 at which time point drug was removed. Tumors emerged in two mice following drug removal on days 84 and 88 and were termed combination tolerant tumors (CTT 22 and CTT 23). (D) Xenograft models of 1205LuTR ERK½ reporter cells in mice that were fed either vehicle or combination drug were imaged for firefly luciferase. Representative images are shown from mice on day 0 and day 4. (E) Average ratios of firefly luciferase intensity to tumor volume. Combination-resistant and tolerant tumors were imaged for firefly luciferase on the day of tumor harvest.
To examine ERK½ activity in resistant and tolerant cells during combo treatment, we used the 1205LuTR ERK½ reporter model in which luciferase activity is driven by pathway activation (14). All established 1205LuTR reporter xenografts regressed to non-detectable levels (Fig 1C). After prolonged treatment, two out of eight tumors (CRT 20 and 21) re-grew. Cessation of therapy in the remaining six mice on day 77 led to re-growth of two CTTs. Before combo, 1205LuTR ERK½ reporter xenografts showed high levels of luciferase, which were dramatically reduced in all combo treated mice by day 11 (Fig 1D–1E). Increased ERK½ activity was observed in CRT 20 and CRT 21, as well as in the CTTs that emerged following drug removal (Supplemental Fig 1A). ERK½ signaling was restored in non-palpable residual disease on day 77 in CTT 22 and on day 83 in CTT 23. (Supplemental Fig 1B). Overall, these data show ERK½ reactivation associated with both relapse of combo-resistant tumors and re-growth of tolerant tumors after cessation of treatment.
CRT-derived cell lines maintained phosphorylated ERK½ and features of resistance in vitro
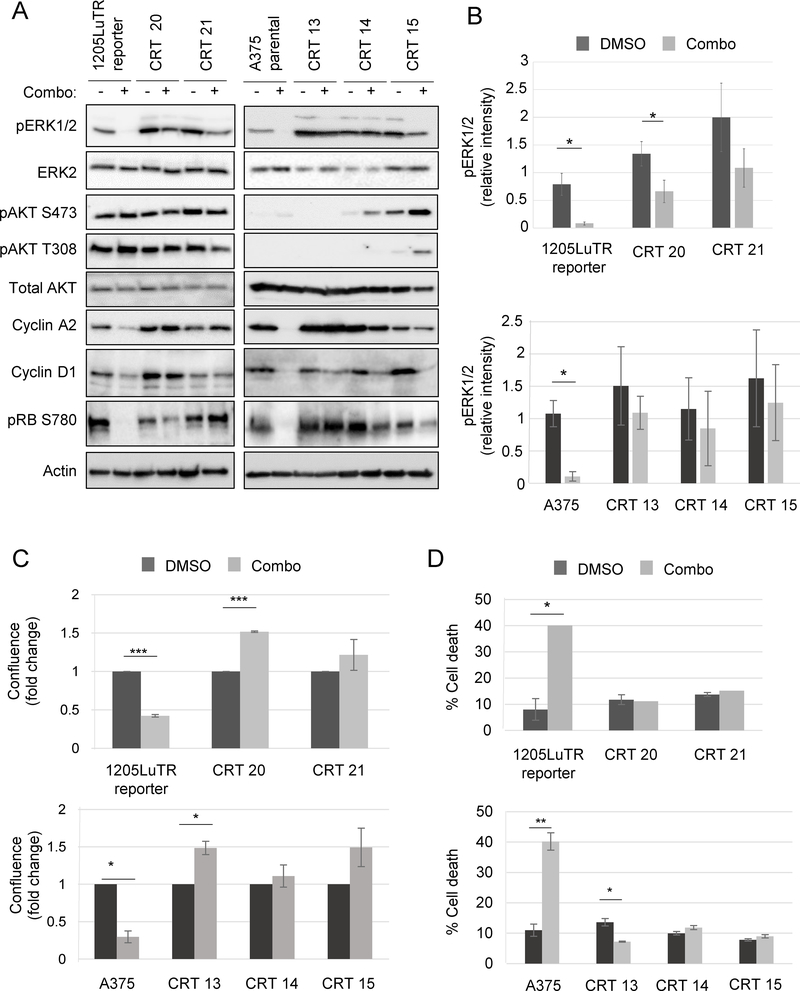
Cells lines were established from CRTs 20 and 21 (1205LuTR reporter parental) and CRTs 13, 14, and 15 (A375 parental) xenografts. As expected, combo treatment of parental cells greatly reduced ERK½ phosphorylation (Fig 2A–2B). By contrast, phosphorylated ERK½ levels were maintained in BRAF/MEK inhibitor-treated CRT-derived cell lines. While phosphorylated AKT level was unaffected by combination treatment in CRTs 20 and 21, phosphorylation increased in CRTs 14 and 15 upon combination treatment. The difference in basal AKT phosphorylation between the cell lines may reflect the PTEN-deficient status of 1205LuTR reporter compared to the PTEN-proficient A375 cells (20). Western blot analysis showed that combo treated CRT-derived lines expressed cell cycle progression markers: cyclin A2, cyclin D1, and phosphorylated RB. These findings were corroborated by functional data showing drugged CRTs progressing through G1/S phase of cell cycle (Supplemental Fig 2A–B). Combo treatment inhibited the growth of parental cells lines, whereas CRTs were either unaffected or had a growth advantage during treatment (Fig 2C). Finally, combo treatment significantly increased parental cell line death, as measured by annexin V and propidium iodide staining (Fig 2D). By contrast, combo treatment did not alter cell death in any CRTs except CRT 13, in which apoptosis was decreased by ~ 50%. These data show that cell lines derived from combo resistant melanoma maintain ERK½ phosphorylation, which associated with enhanced proliferation and abrogation of cell death during combo drug treatment.
Fig 2. Cell lines generated from combination-resistant tumors displayed ERK½ reactivation.
(A) Western blots of lysates from matched parental cell lines: 1205LuTR reporter for CRT 20 and 21; and A375 for CRT 13, 14, and 15 were blotted for signaling proteins and cell cycle progression markers as indicated (n=3). (B) Quantification of pERK½ intensity normalized to loading control in parental cells and CRTs. Shown are the mean and SEM of five independent experiments, *p<0.05. (C) Parental cells (1205LuTR and A375) and CRTs (20, 21, 13, 14, and 15) were grown for 72 hours in either vehicle (DMSO) or 1 μM PLX4720 and 35 nM PD0325901 and then assayed for 2D growth by measuring live cell counts. Shown are the mean and SEM of three independent experiments, *p<0.05, ***p<0.001 (n=3). (D) Parental cells and CRTs were grown for 72 hours in either vehicle (DMSO) or 1 μM PLX4720 and 35 nM PD0325901 and then processed for annexin V and PI staining as markers for early and late apoptosis. *p<0.05, **p<0.01 (n=3).
Combo resistant cells harbored BRAF kinase duplication and remained sensitive to direct targeting of ERK½
To identify a mechanism for ERK½ reactivation and resistance, we performed targeted sequencing on 4 vehicle-treated and 5 progressing combo-treated tumors to examine the mutational status of genes previously implicated in melanomagenesis (31 full genes and the exons of 62 other genes). Tumor-derived sequences were compared to a pool of 16 unmatched controls to filter out germline variants. Notably, the mutational status remained unchanged in KRAS, NRAS, HRAS, MEK½, and CDK4 (Table 1). ERK½ re-activation in resistant tumors has been linked to amplified mutant BRAF and aberrantly spliced mutant BRAF isoforms (21,22), but neither were detected in the CRTs (Supplemental Fig 3A). We instead observed a high molecular weight (~140 kD) form of BRAF in 3 out of 5 CRTs, present in both 1205LuTR and A375-derived lines. This BRAF protein was detected with N-terminal and C-terminal BRAF epitopes (Supplemental Figs. 3B–3C). Furthermore, all short-interfering RNAs directed at sequences across the BRAF transcript effectively knocked down expression of both 140 kD and 90 kD V600E BRAF isoforms in CRT 13 (Supplemental Fig 3D). We recently identified a tandem repeat of exons 10–18 in a patient-derived xenograft that progressed on vemurafenib and in a dabrafenib plus trametinib-resistant melanoma generated in vitro (23). To test if CRTs contained this duplicated kinase domain, we performed PCR across the non-canonical exon 18 – exon 10 junction (Supplemental Figs. 3E–3F). Sanger sequencing of the resulting PCR products confirmed the 18–10 exon fusion in CRTs 20, 21, and 13 but lack of expression in parental cells (Supplemental Fig 3G). These findings were corroborated by RNA-seq data from resistant lines (Table 2) and add to evidence linking a BRAF kinase duplication with pathway reactivation and resistance (23).
Table 1. Mutational Status of Xenografts.
Targeted sequencing on vehicle-treated and progressing combo-treated tumors to examine the mutational status of well-characterized melanoma drivers and suppressors.
| Cell line | BRAF | BRAF zyg | NRAS | HRAS | KRAS | MEK1 | MEK2 | CDK4 | CDKN2A p16INK4a) | PTEN |
|---|---|---|---|---|---|---|---|---|---|---|
| 1205LuTR Vehicle 5 | V600E | het | WT | WT | WT | WT | WT | K22Q | WT | W274X |
| 1205LuTR Vehicle 6 | V600E | het | WT | WT | WT | WT | WT | K22Q | WT | WT |
| CRT 20 | V600E | het | WT | WT | WT | WT | WT | K22Q | E61X, E69X | W274X |
| CRT 21 | V600E | het | WT | WT | WT | WT | WT | K22Q | WT | W274X |
| A375 Vehicle 7 | V600E | hom | WT | WT | WT | WT | WT | WT | E61X, E69X | WT |
| A375 Vehicle 8 | V600E | hom | WT | WT | WT | WT | WT | WT | E61X, E69X | WT |
| CRT 13 | V600E | hom | WT | WT | WT | WT | WT | WT | E61X, E69X | WT |
| CRT 14 | V600E | hom | WT | WT | WT | WT | WT | WT | E61X, E69X | WT |
| CRT 15 | V600E | hom | WT | WT | WT | WT | WT | WT | E69X | WT |
Table 2. BRAF double kinase was not detected in parental cells.
In order to count reads spanning the putative 18–10 splice junction, as well as canonical splice junctions 9–10, 18–18, and 18–19, we used BLAST to determine short sequences at the relevant end of each exon. RNA seq data from cell lines indicated that were mapped to canonical BRAF or to the BRAF kinase duplication junction.
| Total reads | 9–10 | 18–10 | 18–18 | 18–19 |
| 1205LuTR | 75 | 0 | 1 | 35 |
| CRT20 | 76 | 29 | 3 | 26 |
| CRT21 | 91 | 25 | 8 | 35 |
| Unique reads | 9–10 | 18–10 | 18–18 | 18–19 |
| 1205LuTR | 58 | 0 | 1 | 31 |
| CRT20 | 61 | 22 | 3 | 21 |
| CRT21 | 71 | 20 | 8 | 33 |
Tumors progressing on BRAF plus MEK inhibitors often reactivate the MEK-ERK½ pathway (9,24). This suggests that vertical pathway inhibition selects for ERK½-driven mechanisms of resistance, and potentially results in a larger pool of melanoma sensitive to downstream targeting of ERK½. Conversely, CRTs with increased mutant BRAF expression may augment pathway activity enough to confer some level of protection from ERK½ inhibition (24). To determine if CRTs were sensitive to targeting ERK½, we used a selective ERK½ inhibitor, SCH772984 (25). SCH772984 suppressed phosphorylation of the ERK½ target, p90RSK, and the cell cycle marker, RB, and induced cell death in all lines (Supplemental Figs. 3H–3I). We did not observe an additive effect on cell death with the triple combination except in CRT 15, suggesting that ERK½ inhibition alone may be sufficient to suppress signaling and induce apoptosis. These data show that tumors resistant to combined BRAF and MEK inhibitors treatment remain susceptible to ERK½ inhibition.
In vivo ERK½ reporting predicted tumor response to intermittent treatment
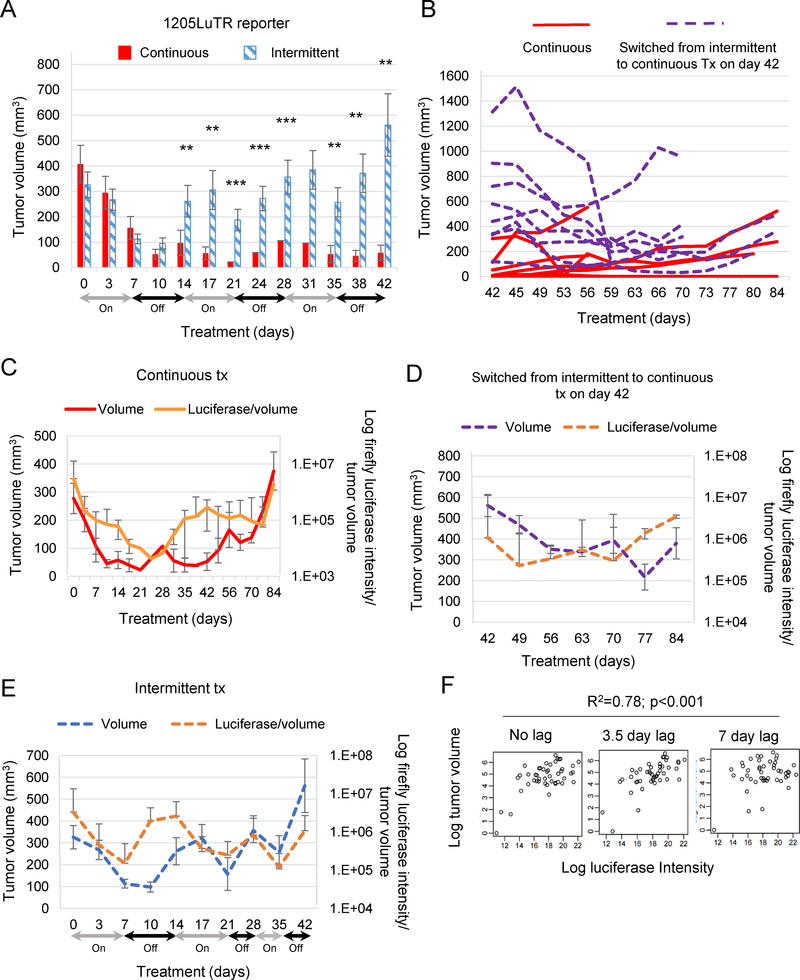
Despite improved efficacy, toxicities from inhibiting BRAF, MEK½, and ERK½ (either as monotherapies or in combination) have led to clinical testing of intermittent dosing (26). Since preclinical models of these efforts are limited, we used a characterized in vivo reporter system to monitor ERK½ activity and tumor response during intermittent combo treatment. Established 1205LuTR reporter tumors were treated either continuously or intermittently using a 1 week on, 1 week off schedule. Complete and durable responses were observed in 5 out of 9 consistently treated xenografts, while the other 4 xenografts relapsed between days 28–49 (Supplemental Fig 4A). By contrast, 1 of 10 xenografts in the intermittent dosing arm regressed completely and durably (Fig 3A). Intermittently treated tumors were significantly larger than those in the continuous group on day 42, at which time they were switched to constant dosing (Fig 3B). Responses were mixed in tumors that changed treatment schedules, 4 of which were reduced by over 50%, while tumor regression was less robust in the other 5, with the earliest relapse observed on day 53. All 8 switched xenografts re-grew during treatment.
Fig 3. Modeling of tumor volume using in vivo ERK½ pathway activation.
(A-B) 1205LuTR reporter xenografts were treated continuously (n=9) for 84 days or intermittently (n=10) on a one week on, one week off cycle until day 42. Intermittently treated mice were switched to continuous dosing on day 42. Bar graph represents average tumor volume up to day 42. After day 42 data, are presented as a line graph for individual mice. (C-E) Average tumor volumes and luciferase intensity normalized to tumor volume during continuous (red), intermittent (blue), or intermittent switched to continuous (purple) treatment schemes. (F) Regression analysis of intermittent dosing as quantification for the association between firefly expression and tumor volume.
Continuous treatment lowered firefly luciferase intensity and tumor volume (Figs. 3C–D). Ultimately, progression was associated with re-activation of ERK½ signaling. In the intermittent treatment arm, ERK½ activity closely tracked treatment status, with a delay between changes in luciferase expression induced by switching to and from the ‘on-drug’ and ‘off drug’ states. Tumor volume lagged but followed the ERK½ reporter changes. The residual effects of adding or removing drug on tumor size persisted from 3.5 to 7 days after treatment change (Fig 3E). To maximize resolution while modeling intermittent dosing, luciferase and tumor volumes were obtained 3 times a week from day 0 to day 21. Accounting for the lag in tumor volume changes, we modeled luciferase as a predictor of tumor volume to determine the utility of the reporter system in this setting. The luciferase expression 0, 3.5, or 7 days prior to tumor measurement significantly predicted tumor volume during intermittent dosing (Fig 3E). Taken together, luciferase expression data accounted for 78% of changes in tumor volume (multiple R-squared = 0.78; Fig 3F). When characterizing cell lines derived from these resistant tumors, we found variable re-activation of ERK½ signaling (Supplemental Fig 4B); the tandem duplication of the BRAF kinase domain was not detected in these lines (Supplemental Fig 4C). Collectively, these data show that non-invasive monitoring of ERK½ activity quantitatively predicts tumor response to intermittent combo treatment regimens and that resistance to intermittent treatment associated with re-activation of ERK½.
Plk1 inhibition was effective against combined BRAF and MEK inhibitor-refractory melanoma in vitro and in vivo
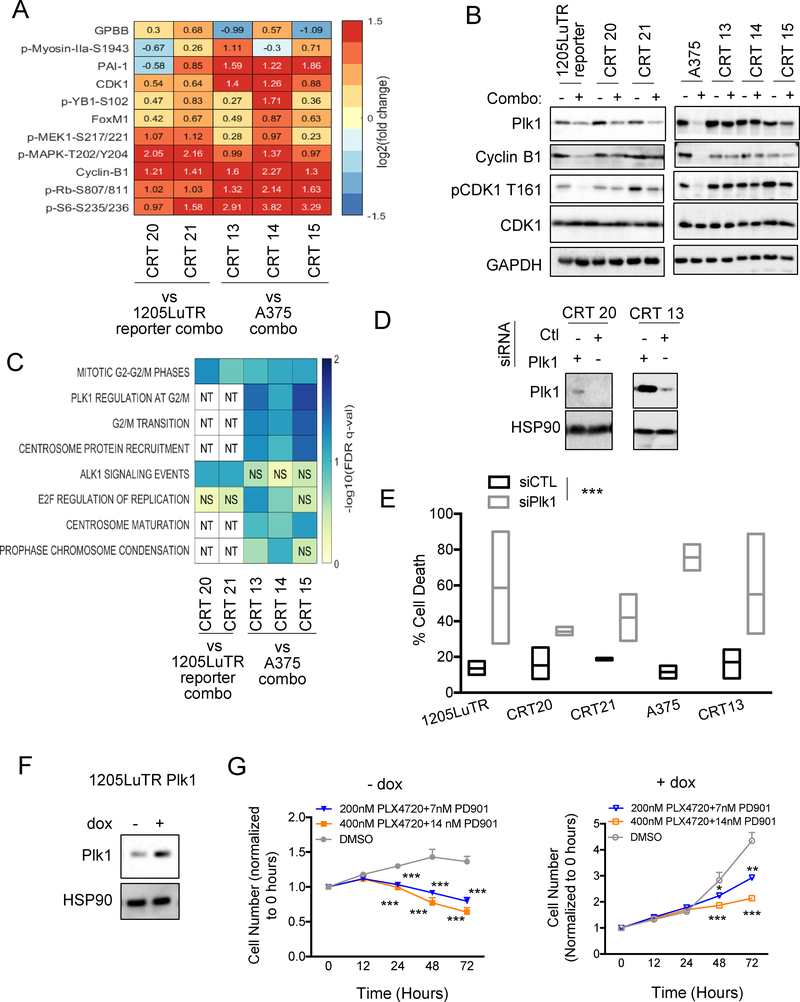
Persistent re-activation of ERK½ associated with resistance to direct targeting of mutant BRAF, MEK, or ERK½ (27) implies the need to identify actionable downstream pathways for salvage therapy. To identify such pathways, we used a reverse phase protein array (RPPA) analysis to evaluate phospho-proteomic changes in the CRTs. Forty-seven proteins and phosphoproteins were significantly altered in at least one CRT compared to parental lines (Supplemental Fig 5A). Eleven proteins were significantly modified in all treated CRTs including increased expression of positive cell cycle regulators, CDK1 and cyclin B1 (Fig 4A). We validated these data via Western blot analysis and found maintained expression of phospho-CDK1 and Cyclin B1 in combo-treated resistant cell lines (Fig 4B), which was consistent EdU data showing cell cycle progression while on treatment (Supplemental Fig 2A–2B).
Fig 4. Plk1 is involved in resistance to BRAF and MEK inhibition.
(A). Heatmap showing the significant fold change for proteins in all CRTs relative to parental. (B) Western blot analysis of cell cycle progression proteins and Plk1 expression in combo. (C) Heatmap of pathways found to be significantly enriched in at least 2 of the comparisons. ‘NS’ represents pathways that were not found to be significant, and ‘NT’ represents pathways that were not tested due to the comparator having less than the number of proteins needed to represent the pathway). (D) Western blot analysis of Plk1 in CRTs after siRNA knockdown of Plk1 (n=3). (E) Percent cell death, as determined by annexin and PI staining, in parental and CRT cell lines after siRNA knockdown of Plk1, ***p<0.001 (n=2–4). (F) Western blot analysis of Plk1 in 1205LuTR Plk1 after 24 hours of doxycycline (dox) treatment, *p<0.05 (n=3). (G) Cell growth of 1205LuTR Plk1 cells +/− dox (24 hours pretreatment) treated with PLX4720 and PD’901 represented as normalized cell number over time, *p<0.05, **p<0.01, ***p<0.001 (n=3).
Pathway analysis of combo-altered proteins revealed significant enrichment of the mitotic G2-G2/M phases pathway in all CRTs (Fig 4C). A375-derived CRTs were also enriched for additional pathways including Plk1 regulation at G2/M, G2/M transition, centrosome protein recruitment, and centrosome maturation. Since Plk1 is central to all processes mentioned above, we examined its expression in treated parental and combination-resistant cell lines (28–31). Plk1 expression appeared independent of resistance status in 1205LuTR cells and their combo-resistant counterparts since they were similarly modulated in response to combo. Conversely, combo abolished Plk1 expression in parental A375s, but was largely maintained in CRTs 13 and 14, and was moderately reduced in CRT 15 (Fig 4B).
To determine how Plk1 expression affects tumor growth and response to combo, we modulated Plk1 gene expression in CRTs and parental cell lines. Knockdown of Plk1 by siRNA induced cell death in parental and CRT cell lines (Fig 4D–4E; Supplemental Fig 5B), suggesting Plk1 expression is involved in tumor cell survival. To directly test whether Plk1 could confer resistance to combo, we generated 1205LuTR Plk1 cells which inducibly expressed exogenous Plk1 when treated with doxycycline (Fig 4F). Exponential growth rate analysis indicated that whereas treatment with combo abrogated growth rate of controls, Plk1 overexpression reversed the effect at comparative doses (Fig 4G and Supplemental Fig 5C). These data suggest that Plk1 expression is directly involved in the growth of melanoma tumor cells and resistance to combo therapy.
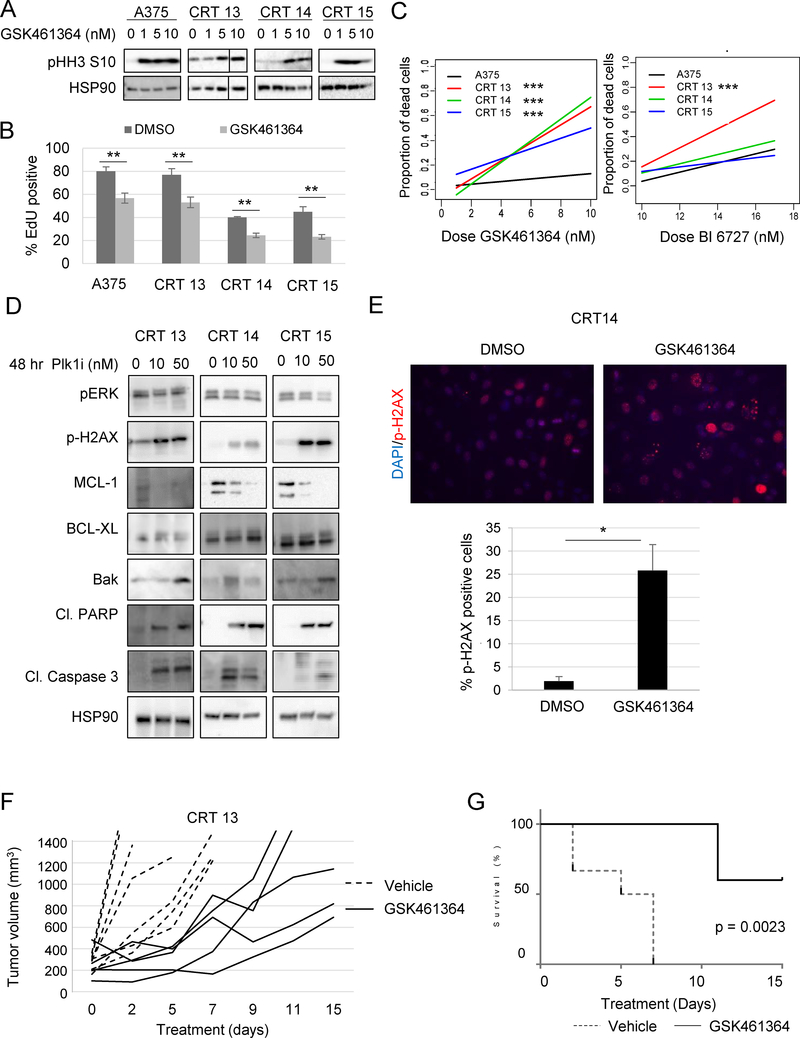
Given the apparent difference in Plk1 expression between treated A375 parental cells and their combo-resistant counterparts, we used CRTs 13, 14, and 15 to test whether pharmacologic Plk1 inhibition could be an effective second-line therapy for combo resistant tumors. We used GSK461364, a targeted inhibitor with > 390-fold selectivity for Plk1 relative to Plk2 and Plk3 (32). Treatment increased expression of phospho-histone H3 (pHH3), a mitotic arrest marker and biochemical readout for Plk1 inhibition (33). GSK461364 treatment decreased cell count and S-phase progression (Fig 5A–5B; Supplemental Fig 5D). Next, we treated primary human melanocytes to test toxicity in healthy tissue and observed no effect of Plk1 inhibition on cell count (Supplemental Fig 5D). Cell line sensitivity to GSK461364 mirrored Plk1 changes during combo treatment; minimal death of A375 cells, robust killing of CRTs 13 and 14, and a moderate response in CRT 15 (Fig 5C; Supplemental Fig 5E). Comparison of the fitted slopes of dead cells in increasing drug concentrations confirmed enhanced killing in CRTs compared to A375 (Fig 5C). Similar analysis was performed using BI 6727, a less potent Plk1 inhibitor exhibiting 6- and 65-fold superior activity against Plk2 and Plk3 over Plk1 (34). BI 6727 strongly induced death in CRTs 13 and 14 and a moderate response in A375 and CRT 15 (Supplemental Fig 5F). Together, these data suggest that pharmacologic Plk1 inhibition induces robust cell death of CRT cell lines.
Fig 5. Plk1 inhibition induces cell death, leading to tumor growth delay of CRTs.
(A) Western blot analysis of cell lines treated for 24 hours with the indicated dose and probed for readout of Plk1 inhibition (n=3). (B) EdU incorporation of cells treated with Plk1 inhibitor (GSK461364; 50 nM) for 72 hours, **p<0.01 (n=3). (C) Slopes of the proportion of dead cells after 72 hours of treatment with indicated doses of GSK461364, ***p<0.001. (D) Western blot analysis of pERK½, p-H2AX, MCL-1, BCL-XL, Bak, cleaved PARP, and cleaved-caspase 3 after treatment with Plk1 inhibitor (GSK461364, n=3). (E) Immunofluorescent staining of p-H2AX in CRT14 after treatment with 20 nM GSK461364, *p<0.05 (n=3). (F) Tumor growth, indicated by change in volume over time, of CRT 13 tumors treated with Plk1 inhibitor (GSK461364; 25 mg/kg/dose) in nude mice. (G) Kaplan Meier survival curve of F, ***p<0.001.
Next, using Western blot analysis, we investigated the apoptotic mechanism by which Plk1 inhibition caused CRT death (Fig 5D). Plk1 treatment increased expression of the apoptotic effector, Bak, and augmented PARP and caspase 3 cleavage. Both Bcl-XL and Mcl-1 inhibit apoptosis by sequestering Bak (35). We did not detect any changes in Bcl-XL levels; however, the expression of Mcl-1 decreased in a dose-dependent manner. Since Plk1 is a member of the mitotic DNA damage checkpoint (36) and reduced Mcl-1 has been linked to the impaired repair of damaged DNA (37), we also tested for expression pH2AX as a read-out for DNA damage (Fig 5E). Plk1 treatment significantly intensified DNA damage as shown by Western blot and immunofluorescence analysis of pH2AX foci (Fig 5E). These data imply Plk1 inhibition promotes induction of the intrinsic apoptotic pathway in CRTs via Mcl-1 degradation which correlated with increased Bak protein and DNA damage.
To establish Plk1 inhibitor efficacy in resistant tumors in vivo, we implanted combo treated mice with CRT 13 cells and allowed them to grow. Pre-formed xenografts were then taken off combo one day before treatment with either vehicle or GSK461364. All vehicle-treated mice had to be sacrificed due to tumor burden by day 7, whereas GSK461364 delayed tumor growth (Fig. 5F). Survival analysis showed that GSK461364 treatment significantly extended mouse survival compared to vehicle treatment-treated mice (Fig. 5G). Overall these data suggest that Plk1 expression is a driver of BRAF mutant melanoma resistant to BRAF and MEK inhibitor combinations and that targeting Plk1 in vitro and in vivo is a possible salvage treatment strategy.
Plk1 expression was elevated in patients progressing on BRAF or BRAF plus MEK inhibitors
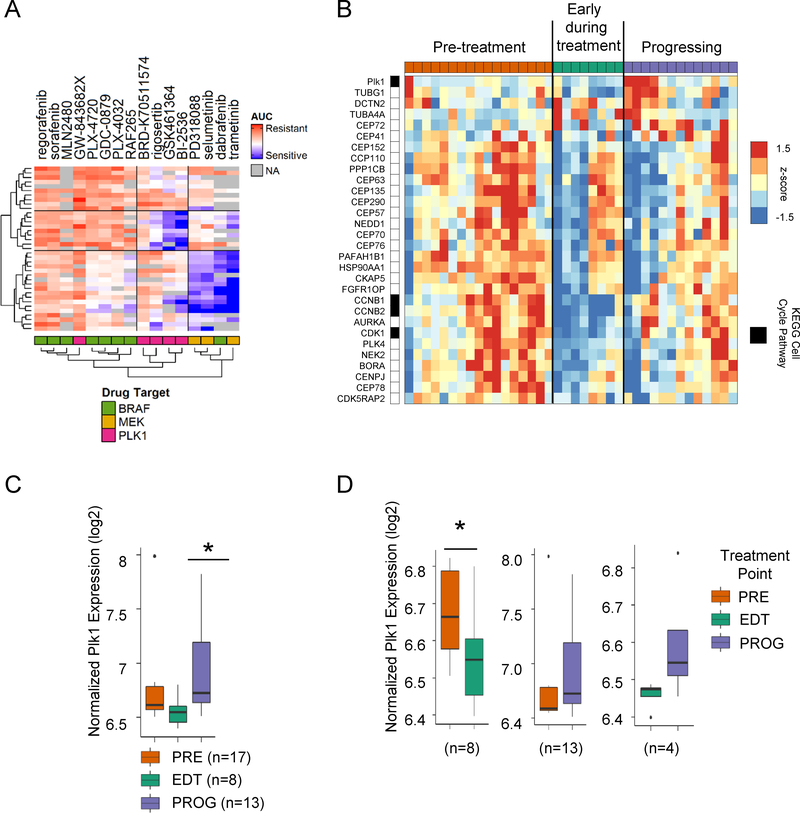
To analyze Plk1 expression in large data sets, we used the Cancer Cell Line Encyclopedia (38) and the Cancer Therapeutics Response Portal (39) and cross-referenced the viability of mutant BRAF melanoma (n=37) with response to Plk1, BRAF, and MEK inhibitors. Area under the curve (AUC) analysis revealed a subset of cell lines that were resistant to BRAF inhibitors but were sensitive to Plk1 targeting agents suggesting Plk1 efficacy in an expanded set of BRAF mutant human melanoma (Fig 6A). To corroborate with patient data, we analyzed a microarray dataset (GSE99898) of tumors taken from patients before treatment (PRE), early during treatment (EDT), or after resistance developed (PROG) to BRAF or BRAF+MEK inhibitors for changes in expression of genes involved in the regulation of Plk1 activity at G2M transition (40). Using a paired sample analysis, we observed that 30 out of 79 genes belonging to this pathway were differentially expressed in at least one time point (Fig 6B). When analyzed in aggregate, we detected significantly higher Plk1 levels in the progressing tumors compared to those early during treatment (Fig 6C). To examine Plk1 expression changes in individual patients, we analyzed matched tumors samples throughout the treatment course. In line with our in vitro data, Plk1 decreased compared to baseline upon treatment, but was unchanged in progressing tumors (Fig 6D). Enhanced Plk1 expression was observed in advancing tumors compared to tumors responding to treatment; however, it did not reach significance. This may be attributed to the small number of matched tumors that existed in these two time points. These patient data reinforce our findings and highlight the role and clinical relevance of Plk1 in the context of drug resistance in mutant BRAF melanoma.
Fig 6. Plk1 expression changes in patients throughout stages of treatment.
(A) Heat map of the area under drug response dose curve (AUC) data for mutant BRAF melanoma cell lines (n=37), for BRAF, MEK and Plk1 inhibitors from the Cancer Therapeutic Response Portal (CTRP v2.1) data set. Missing drug sensitivity data are colored in gray. (B) Heat map of median-centered expression data for significantly altered genes (>50% fold-change) in the Regulation of Plk1 Activity at G/2M Transition gene set for pre-treatment (PRE), early-during treatment (EDT) and progressing on treatment (PROG) samples taken from patient tumors before, during or after dabrafenib, vemurafenib or dabrafenib plus trametinib treatment. Samples were separated based on treatment time points (row 1). KEGG cell cycle genes are indexed in black next to the gene name. Box plot showing normalized log2 Plk1 expression in (C) all samples and (D) matched samples from patients with PRE, EDT and PROG tumors. A linear model was used for statistical analysis of the aggregate (non-paired) Plk1 tumors and paired samples. Letters in the brackets identify which sub-image accurately represents the samples used.
DISCUSSION
Re-activation of MEK-ERK½ signaling is a prevalent mechanism of resistance to BRAF monotherapy and combined BRAF and MEK inhibition (12–14,41), but efforts characterizing the degree of response or addressing challenges of vertical pathway inhibition are lacking. Here we provide a preclinical model to study ERK½ activity and resistance to combined BRAF and MEK inhibition in vivo. We detected both drug resistance and tolerance to BRAF and MEK inhibitors. ERK½ signaling was re-activated during acquired resistance and after treatment cessation of tolerant xenografts. Cell lines generated from resistant tumors remained sensitive to direct ERK½ inhibition, which induced apoptosis. We used our reporter system to temporally and quantitatively model response to intermittent dosing of combined BRAF and MEK inhibitors in mutant BRAF xenografts. Lastly, we used a high-throughput protein array and identified Plk1 as a potential therapeutic target for combination-resistant melanoma.
Tolerance to targeted inhibitors is attributed to non-genetic changes in phenotype allowing tumors to persist until they acquire permanent resistance mechanisms (32). Previous work identified altered cell cycling concurrent with de-differentiation (33), upregulated receptor tyrosine kinase expression and G-coupled protein signaling (34–36), and changes in chromatin remodeling (37, 38) as being associated with tolerance to short term MEK-ERK½ pathway targeted inhibitor treatment. A recent report examining cancers that withstood treatment showed a selective dependence on the lipid hydroperoxidase, GPX4, for survival, which was shared across a range of cancers and targeted therapies (42). Consistent with the existence of a pool of drug-tolerant persister cells, we observed re-growth of residual disease after removal of sustained treatment. Given the clinical implications, defining and targeting the mechanism underlying CTT resilience could be a fruitful area for future research.
One recent study in a patient-derived xenograft model demonstrated that an intermittent schedule of combined BRAF, MEK, and, ERK½ inhibitors was superior to sequential administration of three monotherapies (43). They proposed a model whereby the triple combination stifled selection of BRAF amplified clones whose increased basal ERK½ activity conferred resistance to the next treatment. While the extent of inhibition has been previously associated with response and resistance (2, 3), these findings emphasize the importance of characterizing the kinetics and degree of ERK½ pathway activation in vivo. When examining the utility of ERK½ reporting in this context, we found ERK½ signaling was low in tolerant cells, and drug removal led to pathway reactivation followed by tumor growth. In contrast, ERK½ activity was increased in nearly all acquired resistant tumors, which is consistent with current literature (12,13,21,44). We did not detect a difference in the onset of resistance between continuous and weekly on-off dosing schemes. Importantly, we selected a weekly design as a baseline to generate a model, rather than to evaluate a specific schedule. Changes in ERK½ activity during intermittent dosing significantly predicted tumor volume, providing proof-of-concept that quantitative pathway monitoring can be used as a tool to optimize future clinical trials.
Using a high throughput protein expression array, we detected increased expression of the mitotic regulator, Plk1, in combo treated resistant cell lines. Targeting Plk reduced expression of phospho ERK½ in CRT 15 but remained mostly unchanged in CRTs 13 and 14, suggesting that Plk1 inhibitor-mediated anti-tumor effects may be independent of the ERK½ signaling. Plk1 inhibitor treatment of CRTs reduced expression of pro-survival molecule MCL-1 and increased levels of the DNA damage marker, p-H2AX. We hypothesize that Plk1 inhibition lead to intrinsic apoptosis via Mcl-1 loss, DNA damage and cell death. These data correlated to a significant increase in the survival of CRT tumor bearing animals treated with Plk1 inhibitor as a salvage therapy. Several preclinical studies have demonstrated the efficacy of Plk1 inhibition as a first line treatment for melanoma (45–47). Our findings, which are supported by patient data, provide new evidence for a role for Plk1 in therapeutic resistance and suggest that targeting Plk1 may be a valuable salvage therapy for patients progressing on the combination of BRAF plus MEK inhibitors.
Vertically targeting multiple nodes along the same pathway, while more efficacious, can be limited by toxicities. Here we show monitoring ERK½ in vivo during intermittent dosing is predictive of tumor response, which may be used to design treatment schemes optimized for safety and efficacy. We also demonstrate the selective killing of resistant cells by Plk1 inhibitors. Together, our data provide insight on strategic and therapeutic avenues that may be employed in melanoma during treatment or after progression on BRAF plus MEK inhibitors.
Supplementary Material
Acknowledgments
Financial Support: A.E. Aplin is supported by NIH R01 awards CA160495 and CA182365 and by Dr. Miriam and Sheldon G. Adelson Medical Research Foundation. I. Sanchez is supported by a diversity supplement to CA160495. The Sidney Kimmel Cancer Center Flow Cytometry, Translational Pathology, and Meta-Omics core facilities are supported by NIH/National Cancer Institute Support Grant (P30 CA056036). The RPPA studies were performed at the Functional Proteomics Core Facility at The University of Texas MD Anderson Cancer Center, which is supported by an NCI Cancer Center Support Grant (CA16672).
Conflict of Interest: A.E. Aplin has received funding from Pfizer. M.A. Davies is on advisory boards for Novartis, Roche/Genentech, GSK, Sanofi-Aventis, BMS and has received institutional grants from GSK, Roche/Genentech, Sanofi-Aventis, and Oncothyeon.
REFERENCES
- 1.Wargo JA, Cooper ZA, Flaherty KT. Universes collide: combining immunotherapy with targeted therapy for cancer. Cancer Dis 2014;4:1377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conner SR, Scott G, Aplin AE. Adhesion-dependent activation of the ERK½ cascade is by-passed in melanoma cells. J Biol Chem 2003;278:34548–54. [DOI] [PubMed] [Google Scholar]
- 3.Hingorani SR, Jacobetz MA, Robertson GP, Herlyn M, Tuveson DA. Suppression of BRAF(V599E) in human melanoma abrogates transformation. Cancer research 2003;63:5198–202. [PubMed] [Google Scholar]
- 4.Malissen N, Grob JJ. Metastatic Melanoma: Recent Therapeutic Progress and Future Perspectives. Drugs 2018;78:1197–209 doi 10.1007/s40265-018-0945-z. [DOI] [PubMed] [Google Scholar]
- 5.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. The New England journal of medicine 2012;367:107–14. [DOI] [PubMed] [Google Scholar]
- 6.Ribas A, Gonzalez R, Pavlick A, Hamid O, Gajewski TF, Daud A, et al. Combination of vemurafenib and cobimetinib in patients with advanced BRAF(V600)-mutated melanoma: a phase 1b study. Lancet Oncol 2014;15:954–65 doi 10.1016/S1470-2045(14)70301-8. [DOI] [PubMed] [Google Scholar]
- 7.Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. The New England journal of medicine 2015;372:30–9. [DOI] [PubMed] [Google Scholar]
- 8.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med 2014;371:1877–88 doi 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 9.Eroglu Z, Ribas A. Combination therapy with BRAF and MEK inhibitors for melanoma: latest evidence and place in therapy. Ther Adv Med Oncol 2016;8:48–56 doi 10.1177/1758834015616934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shirley M Encorafenib and Binimetinib: First Global Approvals. Drugs 2018;78:1277–84 doi 10.1007/s40265-018-0963-x. [DOI] [PubMed] [Google Scholar]
- 11.Hartsough E, Shao Y, Aplin AE. Resistance to RAF inhibitors revisited. The Journal of investigative dermatology 2014;134:319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagle N, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer discovery 2014;4:61–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moriceau G, Hugo W, Hong A, Shi H, Kong X, Yu CC, et al. Tunable-combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction. Cancer Cell 2015;27:240–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abel EV, Basile KJ, Kugel CH, Witkiewicz AK, Le K, Amaravadi RK, et al. Melanoma adapts to RAF/MEK inhibitors through FOXD3-mediated upregulation of ERBB3. The Journal of clinical investigation 2013;123:2155–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A 2008;105:3041–6 doi 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett SD, Bridges AJ, Dudley DT, Saltiel AR, Fergus JH, Flamme CM, et al. The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorg Med Chem Lett 2008;18:6501–4 doi 10.1016/j.bmcl.2008.10.054. [DOI] [PubMed] [Google Scholar]
- 17.Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther 2006;5:2512–21. [DOI] [PubMed] [Google Scholar]
- 18.Hochberg Y A sharper Bonferroni procedure for multiple tests of significance. Biometrika 1988. [Google Scholar]
- 19.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007;23:1846–7 doi 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 20.Spofford LS, Abel EV, Boisvert-Adamo K, Aplin AE. Cyclin D3 expression in melanoma cells is regulated by adhesion-dependent phosphatidylinositol 3-kinase signaling and contributes to G1-S progression. J Biol Chem 2006;281:25644–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011;480:387–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basile KJ, Abel EV, Dadpey N, Hartsough EJ, Fortina P, Aplin AE. In vivo MAPK reporting reveals the heterogeneity in tumoral selection of resistance to RAF inhibitors. Cancer Research 2013;73:7101–10 doi 10.1158/0008-5472.CAN-13-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kemper K, Krijgsman O, Kong X, Cornelissen-Steijger P, Shahrabi A, Weeber F, et al. BRAF(V600E) Kinase Domain Duplication Identified in Therapy-Refractory Melanoma Patient-Derived Xenografts. Cell Rep 2016;16:263–77 doi 10.1016/j.celrep.2016.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med 2014;371:1867–76 doi 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 25.Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov 2013;3:742–50 doi 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 26.Emmons MF, Faiao-Flores F, Smalley KS. The role of phenotypic plasticity in the escape of cancer cells from targeted therapy. Biochem Pharmacol 2016;122:1–9 doi 10.1016/j.bcp.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jha S, Morris EJ, Hruza A, Mansueto MS, Schroeder GK, Arbanas J, et al. Dissecting Therapeutic Resistance to ERK Inhibition. Mol Cancer Ther 2016;15:548–59 doi 10.1158/1535-7163.MCT-15-0172. [DOI] [PubMed] [Google Scholar]
- 28.Wakida T, Ikura M, Kuriya K, Ito S, Shiroiwa Y, Habu T, et al. The CDK-PLK1 axis targets the DNA damage checkpoint sensor protein RAD9 to promote cell proliferation and tolerance to genotoxic stress. Elife 2017;6 doi 10.7554/eLife.29953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruinsma W, Macurek L, Freire R, Lindqvist A, Medema RH. Bora and Aurora-A continue to activate Plk1 in mitosis. J Cell Sci 2014;127:801–11 doi 10.1242/jcs.137216. [DOI] [PubMed] [Google Scholar]
- 30.Malumbres M, Barbacid M. Cell cycle kinases in cancer. Curr Opin Genet Dev 2007;17:60–5 doi 10.1016/j.gde.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z, Sun Q, Wang X. PLK1, A Potential Target for Cancer Therapy. Transl Oncol 2017;10:22–32 doi 10.1016/j.tranon.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hornberger KR, Badiang JG, Salovich JM, Kuntz KW, Emmitte KA, Cheung M. Regioselective synthesis of benzimidazole thiophene inhibitors of polo-like kinase 1. Tetrahedron Lett 2008;49:6348–51 doi 10.1016/j.tetlet.2008.08.077. [DOI] [Google Scholar]
- 33.Li J, Hong MJ, Chow JP, Man WY, Mak JP, Ma HT, et al. Co-inhibition of polo-like kinase 1 and Aurora kinases promotes mitotic catastrophe. Oncotarget 2015;6:9327–40 doi 10.18632/oncotarget.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudolph D, Steegmaier M, Hoffmann M, Grauert M, Baum A, Quant J, et al. BI 6727, a Polo-like kinase inhibitor with improved pharmacokinetic profile and broad antitumor activity. Clin Cancer Res 2009;15:3094–102 doi 10.1158/1078-0432.ccr-08-2445. [DOI] [PubMed] [Google Scholar]
- 35.Hockings C, Alsop AE, Fennell SC, Lee EF, Fairlie WD, Dewson G, et al. Mcl-1 and Bcl-xL sequestration of Bak confers differential resistance to BH3-only proteins. Cell Death Differ 2018;25:719–32 doi 10.1038/s41418-017-0010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyun SY, Hwang HI, Jang YJ. Polo-like kinase-1 in DNA damage response. BMB Rep 2014;47:249–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattoo AR, Pandita RK, Chakraborty S, Charaka V, Mujoo K, Hunt CR, et al. MCL-1 Depletion Impairs DNA Double-Strand Break Repair and Reinitiation of Stalled DNA Replication Forks. Mol Cell Biol 2017;37 doi 10.1128/mcb.00535-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603–7 doi 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seashore-Ludlow B, Rees MG, Cheah JH, Cokol M, Price EV, Coletti ME, et al. Harnessing Connectivity in a Large-Scale Small-Molecule Sensitivity Dataset. Cancer Discov 2015;5:1210–23 doi 10.1158/2159-8290.cd-15-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kakavand H, Rawson RV, Pupo GM, Yang JYH, Menzies AM, Carlino MS, et al. PD-L1 Expression and Immune Escape in Melanoma Resistance to MAPK Inhibitors. Clin Cancer Res 2017;23:6054–61 doi 10.1158/1078-0432.CCR-16-1688. [DOI] [PubMed] [Google Scholar]
- 41.Gardner PP, Eldai H. Annotating RNA motifs in sequences and alignments. Nucleic Acids Res 2015;43:691–8 doi 10.1093/nar/gku1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hangauer MJ, Viswanathan VS, Ryan MJ, Bole D, Eaton JK, Matov A, et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 2017;551:247–50 doi 10.1038/nature24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue Y, Martelotto L, Baslan T, Vides A, Solomon M, Mai TT, et al. An approach to suppress the evolution of resistance in BRAFV600E-mutant cancer. Nat Med 2017;23:929–37 doi 10.1038/nm.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat comm 2012;3:724 doi 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Posch C, Cholewa BD, Vujic I, Sanlorenzo M, Ma J, Kim ST, et al. Combined Inhibition of MEK and Plk1 Has Synergistic Antitumor Activity in NRAS Mutant Melanoma. J Invest Dermatol 2015;135:2475–83 doi 10.1038/jid.2015.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Oliveira JC, Brassesco MS, Pezuk JA, Morales AG, Valera ET, Montaldi AP, et al. In vitro PLK1 inhibition by BI 2536 decreases proliferation and induces cell-cycle arrest in melanoma cells. J Drugs Dermatol 2012;11:587–92. [PubMed] [Google Scholar]
- 47.Schmit TL, Zhong W, Setaluri V, Spiegelman VS, Ahmad N. Targeted depletion of Polo-like kinase (Plk) 1 through lentiviral shRNA or a small-molecule inhibitor causes mitotic catastrophe and induction of apoptosis in human melanoma cells. J Invest Dermatol 2009;129:2843–53 doi 10.1038/jid.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.