Abstract Abstract
Shiraiaceae is an important family in Pleosporales (Dothideomycetes), which includes medical fungi and plant pathogens. Two hypocrellin-producing taxa, Shiraia bambusicola and a novel genus Rubroshiraiagen. nov., typified by Rubroshiraia bambusae are treated in this article. Maximum likelihood analysis, generated via RAxML (GTR+G model), using a combined SSU, LSU, TEF1 and RPB2 sequence dataset, shows that Rubroshiraia is close to Shiraia and belongs to the family Shiraiaceae. Descriptions, illustrations and a taxonomic key are provided for the genera in Shiraiaceae. Rubroshiraia morphologically differs from Shiraia in having small and dark ascostromata and filiform ascospores. Production of the ascostromatal metabolites, hypocrellin A and B, were examined by HPLC and spectrophotometer. The content of hypocrellin A and B of specimen HKAS 102255 (R. bambusae) is twice that produced by HKAS 102253 (S. bambusicola). To clarify the relationship between R. bambusae and Hypocrella bambusae, type material of the latter was examined and provided the illustration.
Keywords: HPLC, metabolite, new genus, phylogeny, taxonomy
Introduction
Liu et al. (2013) introduced the family Shiraiaceae Y.X. Liu, Zi Y. Liu & K.D. Hyde which is typified by Shiraia Henn. and placed the family in Pleosporales Luttr. ex M.E. Barr. Ariyawansa et al. (2013) accommodated Grandigallia M.E. Barr, Hanlin, Cedeño, Parra & R. Hern. in Shiraiaceae since it morphologically resembles Shiraia. Subsequent publications by Wijayawardene et al. (2014, 2017, 2018) agreed with this placement and, thus, the family currently comprises two genera.
Shiraia is typified by S. bambusicola Henn. (Hennings 1900), which is parasitic on living bamboo culms and has conspicuous large, pinkish, fleshy ascostromata with multi-locules located near the periphery, fissitunicate asci and hyaline, muriform ascospores (Liu et al. 2013). S. bambusicola has been reported from temperate regions of Asia, such as China and Japan (Table 1) (Hino 1961; Li et al. 2009; Liu et al. 2013).
Table 1.
Distribution of Shiraia bambusicola.
| Distribution | References | |
|---|---|---|
| Country | Province | |
| China | Anhui | Li et al. (2009), Lai and Fu (2000) |
| Guangxi | Li et al. (2009) | |
| Guizhou | Li et al. (2009) | |
| Henan | Li et al. (2009) | |
| Hubei | Li et al. (2009) | |
| Hunan | Li et al. (2009) | |
| Jiangsu | Zhao and Liang (2005), Li et al. (2009) | |
| Jiangxi | Li et al. (2009) | |
| Sichuan | Chen and Chen (2009), Li et al. (2009) | |
| Yunan | Fang et al. (2006), Chen et al. (2010) | |
| Zhejiang | Li et al. (2009), Liu et al. (2013) | |
| Japan | Tokyo | Hino (1961), Liu et al. (2013) |
| Osaka | Morakotkarn et al. (2007) | |
Shiraia has previously been placed in several families, depending on the opinions of authors. Hennings (1900) considered Shiraia to have unitunicate asci and treated as a member in the family Nectriaceae Tul. & C. Tul. (Hypocreales, Sordariomycetes) when he established the genus. Based on its large and fleshy fruiting bodies, Shiraia was transferred to Hypocreaceae De Not by Saccardo (1902). Amano (1980) re-examined the type specimen and regarded Shiraia as having bitunicate asci and, hence, placed the genus in Pleosporaceae Nitschke (Pleosporales, Dothideomycetes). However, it was subsequently transferred to Dothideales, genera incertae sedis by Kirk et al. (2001).
Earlier classifications of Shiraia were based on morphological characters. The first attempt of DNA-based taxonomy (Cheng et al. 2004) confirmed that Shiraia belongs in Pleosporales and was phylogenetically close to species of Phaeosphaeriaceae M.E. Barr. Thus, Cheng et al. (2004) considered Shiraia as a member in Phaeosphaeriaceae. Liu et al. (2013) carried out significant studies on Shiraia taxonomy by re-examining the holotype and carrying out phylogenetic analysis, based on LSU sequence data. Liu et al. (2013) also designated an epitype of both sexual and asexual morphs and introduced Shiraiaceae in the Pleosporales.
Shiraia bambusicola has been reported as a pathogen on various bamboo species (Table 2) or as endophyte of bamboo culms (Morakotkarn et al. 2007, 2008). The bamboo genus Brachystachyum Keng is significantly affected by S. bambusicola (Table 2; Lai and Fu 2000). The holotype of S. bambusicola was recorded from a Bambusa sp. (Liu et al. 2013). Shiraia bambusicola has also been recorded on several common bamboo genera, including Fargesia Franch., Phyllostachys Sieb. et Zucc., Pleioblastus Nakai and Indosasa Mcclure (Lai and Fu 2000; Li et al. 2009). However, these hosts need to be further verified.
Table 2.
List of bamboo hosts of Shiraia bambusicola.
| Bamboo host | References |
|---|---|
| Brachystachyum densiflorum (Rendle) Keng | Lai and Fu (2000) |
| Brachystachyum albostriatum G.H. Lai | Li et al. (2009) |
| Brachystachyum ensiflorum (Pendle) Keng | Li et al. (2009) |
| Brachystachyum yixingense | Li et al. (2009) |
| Phyllostachys nidularia Munro | GenBank |
| Phyllostachys praecox f. prevernalis S.Y. Chen & C.Y. Yao | GenBank |
| Pleioblastus amarus (Keng) Keng f. | GenBank |
Shiraia bambusicola produces hypocrellins. Four hypocrellins have been extracted from the fungal stromata (Wan and Chen 1981; Kishi et al. 1991; Chen and Chen 2009). Endophytes, named as Shiraia spp., were also shown to produce hypocrellins on media (Lu et al. 2004; Morakotkarn et al. 2008; Liang et al. 2009; Zhang et al. 2014; Tong et al. 2017). The fruiting body of “Zhuhongjun” also contains hypocrellins (Hudson et al. 1994; Huang et al. 2001). Hypocrellin seems to be an important feature when clarifying the taxa of Shiraiaceae.
A Chinese medical fungus named “Zhuhongjun” in Chinese, was identified as Hypocrella bambusae (Berk. & Broome) Sacc. by Liu (1978), based on its conspicuous and fleshy fruiting body. However, according to our knowledge, Zhuhongjun is similar to S. bambusicola and unrelated to Hypocrella. Therefore, the taxonomic status of this taxon needs to be clarified.
The monotypic genus Grandigallia, collected on Polylepis sericea Wedd. (Rosaceae), was introduced by Barr et al. (1987) with G. dictyospora M.E. Barr et al. as the type species. Grandigallia dictyospora was reported from Venezuela in a locality above 3,400 m and the fungus was found to produce large ascostromata (3–14 cm in diam.), with bitunicate asci and dictyospores (Barr et al. 1987).
In this study, ten specimens of S. bambusicola and a hypocrellin producing taxon (“Zhuhongjun” in Chinese) were collected from Yunnan Province in China. Morphological and phylogenetic studies were carried out to determine the taxonomic status of these taxa. Sequences from endophytic strains, named as Shiraia spp., were also downloaded from GenBank and included in the phylogenetic analyses. The metabolite content of hypocrellin extracted from the specimens was determined by HPLC (Chem 2012). Based on the morphology and phylogenetic analyses, the hypocrellin producing taxon “Zhuhongjun” is treated as a new genus in Shiraiaceae.
Material and methods
Collecting and examination of specimens
Bamboo culms with large, reddish to pale yellow ascostromata were collected from Yunnan, China and brought to the laboratory in 2017. Samples were examined following the methods described in Dai et al. (2017). Micro-morphological characters were examined and photographed by differential interference contrast (DIC), using a Leica DM2500 compound microscope with a Leica DMC4500 camera. Fruiting bodies were observed by stereomicroscopy using a Leica S8AP0 and photographed by HDMI 200C. Measurements were made using Tarosoft (R) Image Frame Work software. Specimens have been deposited at the herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN) and Herbarium Mycologicum, Academiae Sinicae (HMAS) in Beijing. Facesoffungi (Jayasiri et al. 2015) and Index Fungorum (Index Fungorum 2019) numbers were provided for new taxa. Type material of H. bambusae was loaned and examined from the Royal Botanic Gardens, Kew.
DNA extraction, PCR amplification and sequencing
The surface of fungal fruiting bodies was sterilised by 75% alcohol and rinsed three times in sterile water. The internal tissue with locules was cut into pieces and ground in a mortar into powder with liquid nitrogen. The powder was used to directly extract DNA with an OMEGA E.Z.N.A. Forensic DNA Kit, following the manufacturer’s instructions.
ITS5 and ITS4, NS1 and NS4 (White et al. 1990) and LROR and LR5 (Vilgalys and Hester 1990) primers were used for the amplification of internal transcribed spacers (ITS), small subunit rDNA (SSU) and large subunit rDNA (LSU), respectively. Translation elongation factor 1-α gene region (TEF 1-alpha) and RNA polymerase II second largest subunit (RPB2) genes were amplified by using EF1-983F and EF1-2218R (Rehner 2001), fRPB2-5f and fRPB2-7cr primers (Liu et al. 1999), respectively.
The final volume of the polymerase chain reaction (PCR) was prepared following Dai et al. (2017). The PCR thermal cycle programme of ITS, SSU, LSU, RPB2 and TEF 1-alpha genes amplifications were run under the same conditions as described in Dai et al. (2017). The quality of PCR products was checked by 1% Biowest agarose gel electrophoresis. Amplified PCR fragments were sequenced at Shanghai Majorbio Bio-Pharm Technology Co., Ltd. and BGI Tech Solutions Co., Ltd. (BGI-Tech), P.R. China. Generated new sequences of ITS, LSU, SSU, Rpb2 and TEF1 regions are deposited in GenBank (Table 4).
Table 4.
List of newly generated sequences with their culture collection numbers and GenBank accession numbers.
The holotype specimen is highlighted in bold. Abbreviations: HKAS: herbarium of Kunming Institute of Botany, Chinese Academy of Sciences.
Phylogenetic analysis
The BLAST searches in GenBank, using LSU and ITS sequence data were carried out to obtain the close strains. Additional sequences were downloaded from GenBank based on recent publications (Liu et al. 2017).
Single gene sequence alignments were carried out with MAFFT v. 7.215 (Katoh and Standley 2013, http://mafft.cbrc.jp/alignment/server/index.html) and edited manually when necessary in BioEdit v. 7.0 (Hall 2004). The alignments of LSU, SSU, Rpb2 and TEF1 regions were combined in MEGA6 version 6.0 (Tamura et al. 2013).
Maximum-likelihood (ML) analyses, including 1000 bootstrap replicates, were run using RAxMLGUI v.1.0. (Stamatakis 2006; Silvestro and Michalak 2011). Alignments in PHYLIP format were exchanged and loaded from the website (http://sing.ei.uvigo.es/ALTER/). The online tool Findmodel (http://www.hiv.lanl.gov/content/sequence/findmodel/findmodel.html) was used to determine the best nucleotide substitution model for each partition data.
Maximum-parsimony (MP) analyses were carried out in PAUP v. 4.0b10 (Swofford 2002) with 1000 replications. Maxtrees were set to 1000, branches of zero length were collapsed and all multiple equally most parsimonious trees were saved. The robustness of the most parsimonious trees was evaluated from 1000 bootstrap replications (Phillips et al. 2013).
Bayesian analyses were performed using MrBayes v. 3.0b4 (Ronquist and Huelsenbeck 2003). The model of evolution was performed using MrModeltest v. 2.2 (Nylander 2004). Posterior Probabilities (PP) (Rannala and Yang 1996; Zhaxybayeva and Gogarten 2002) were determined by Markov Chain Monte Carlo sampling (MCMC) in MrBayes v. 3.0b4 (Huelsenbeck and Ronquist 2001). Six simultaneous Markov chains were run for 1,000,000 generations and trees were sampled every 100th generation. The burn-in was set to 0.25 and the run was automatically stopped when the average standard deviation of split frequencies reached below 0.01 (Maharachchikumbura et al. 2015).
Trees were visualised with TreeView (Page 1996) or FigTree v. 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/) and, additionally, layouts were done with Adobe Illustrator CS v. 5. Maximum-likelihood bootstrap values (MLBP) and Maximum-parsimony bootstrap values (MPBP) equal to or greater than 50% are given for each tree. Bayesian posterior probabilities (BYPP) > 0.90 are indicated as thickened lines. The sequences used in this study are listed in Table 1. The combined alignment and phylogenetic tree were submitted at TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S24345).
HPLC profiling
Standards of hypocrellin A and hypocrellin B were purchased from Shanghai Tauto Biotech CO., Ltd. (http://www.tautobiotech.com) and used as received. Their purity is ≥ 98% (HPLC) and their structures are redrawn based on references (Wan and Chen 1981; Morakotkarn et al. 2008) and shown in Figure 1. The dry powder of ascostromata of S. bambusicola (HKAS102266) and “Zhuhongjun” (HKAS102270) was extracted followed the methods described by Stadler et al. (2001) and accurately weighed to 0.5 g and added to 25 ml of methanol and sonicated for 30 min. Semi-preparative HPLC was performed on an Agilent 1260 apparatus equipped with a UV detector and a CAPCELL PAK C18 (Agilent, 4.6 mm × 25 cm, 5 µm) column, with 38% solvent A: H2O + 0.5% formic acid; 62% solvent B: acetonitrile, isocratic elution, UV/Vis the detection in the range of 265 nm (Table 3). The UV-Vis spectra were recorded at room temperature on a Perkin-Elmer Lambda 900 spectrophotometer (Fig. 5).
Figure 1.
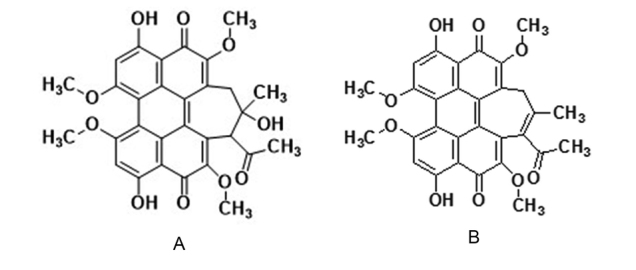
Chemical structures of hypocrellin A and hypocrellin B. A hypocrellin A B hypocrellin B.
Table 3.
HPLC condition used in this study.
| Instrument | Condition |
|---|---|
| Reverse phase-column | CAPCELL PAK C18 (4.6 mm × 25 cm, 5 µm) |
| Oven temp. (°C) | 35 |
| Flow rate (ml/min) | 1 |
| Mobile phase (%) | 38% solvent A: H2O + 0.5% formic acid; 62% solvent B: acetonitrile |
| UV Absorbance (nm) | 265 |
| Gradient elution | isocratic elution |
| Run time (min) | 30–40 |
Figure 5.
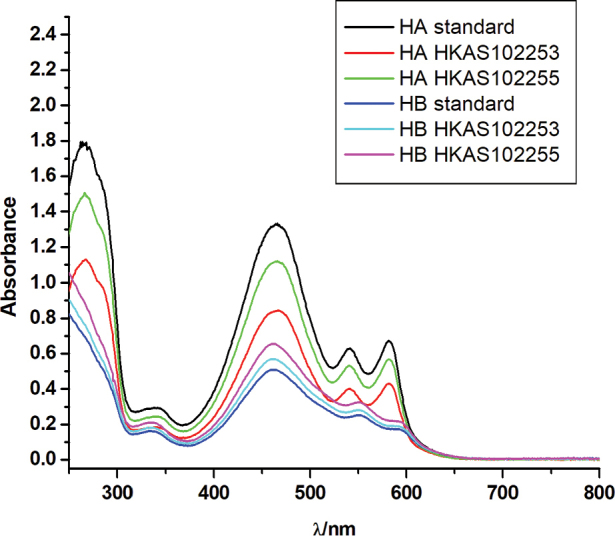
The UV spectrum of the standards and of hypocrellin A and B from the samples (Shiraia bambusicolaHKAS 102253 and Rubroshiraia bambusaeHKAS 102255) were recorded in alcohol at room temperature. HA: hypocrellin A, HB: hypocrellin B.
Results
Phylogeny
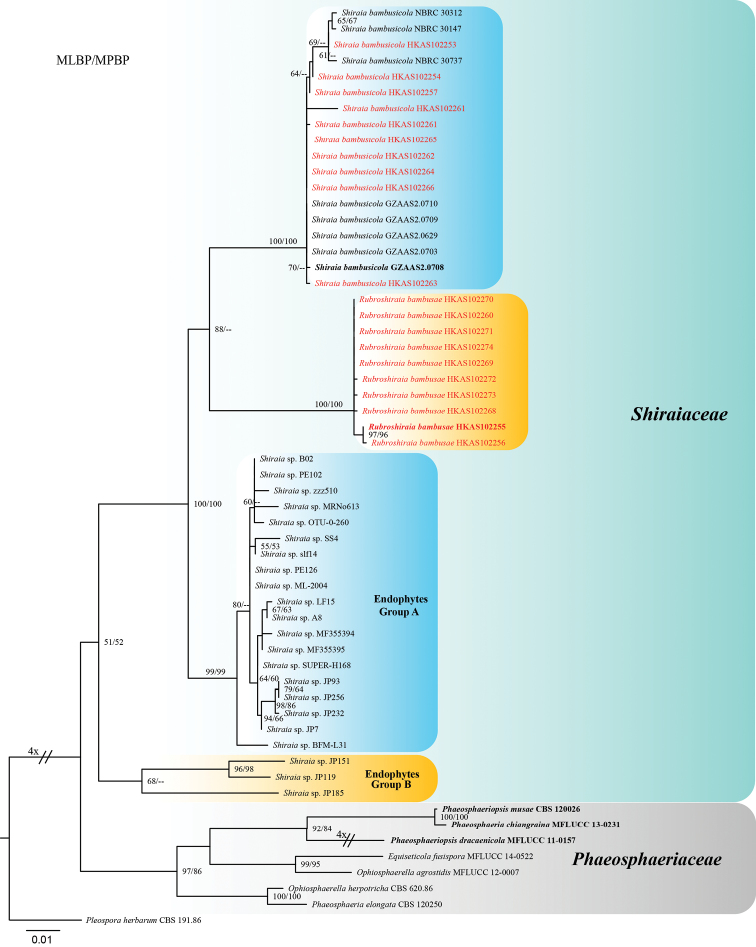
To clarify the family placement of newly established taxa, maximum likelihood phylogenetic analysis was generated from RAxML (GTR+G model), based on combined SSU, LSU, TEF1 and RPB2 sequences data (Fig. 2). The combined alignment comprised 4025 characters including gaps for 127 ingroup taxa and one outgroup taxon Dothidea insculpta (CBS 189.58). Based on the phylogenetic tree in Fig. 2, the new collections cluster within family Shiraiaceae with high bootstrap support (96/1.00 MLBS/BSPP) and emerge as two groups, which are S. bambusicola lineage and a new clade named as R. bambusae in this paper. Shiraia and Rubroshiraia have more or less similar ascostromata and both of them can produce the metabolite hypocrellins. However, they can be phylogenetically distinguished with high bootstrap support (100/1.00 MLBS/BSPP) (Fig. 2). Grandigallia has not been included in phylogenetic analysis as it is lacking sequences in the GenBank. However, the new taxa can be morphologically distinguished from it. Shiraiaceae is phylogenetically close with family Phaeosphaeriaceae in Pleosporales and this has been confirmed by Liu et al. (2013).
Figure 2.
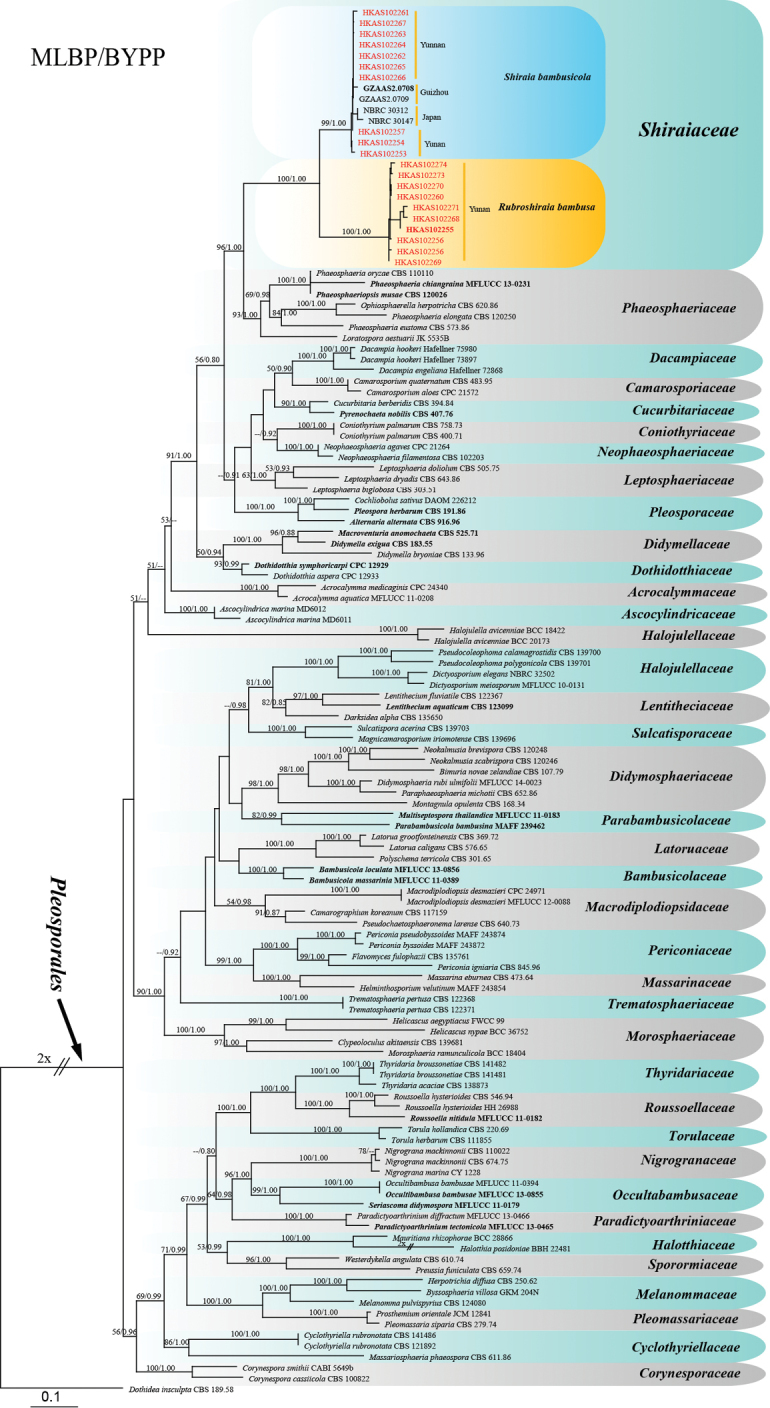
Maximum likelihood phylogenetic tree generated from RAxML (GTR+G model), based on combined LSU, SSU, TEF1 and RPB2 sequences data. ML values (MLBP) (> 50%), resulting from 1000 bootstrap replicates and Bayesian posterior probabilities (BYPP) greater than 0.90, are given at the nodes. The original isolate numbers’ codes are noted after the species names. The tree is rooted to Dothidea insculpta (CBS 189.58). Ex-type or ex-epitype strains are in bold. Newly generated strains are in red and the new genus is in yellow background.
To clarify the relationship between endophytic strains named as shiraia-like (Shiraia spp.) and Shiraiaceae, a phylogenetic tree was constructed (RAxML (GTR+G model), based on combined LSU and ITS sequences data and compared. The combined alignment comprises 1442 characters including gaps for 57 ingroup taxa and one outgroup taxon Pleospora herbarum (CBS 191.86). Of the 1442 characters of the combined matrix, 1116 were constant and 220 were parsimony informative. The endophytic strains separated into two lineages (Group A and group B) forming at the base clade of Shiraiaceae (Fig. 3). Several strains in group A ca. JP7, JP93, JP232, JP256, SUPER-H168, A8 and ML-2004, isolated from bamboo tissue can produce hypocrellins in media (Lu et al. 2004; Morakotkarn et al. 2008; Liang et al. 2009; Cai et al. 2011; Zhang et al. 2014). However, no hypocrellins were detected from Group B, which included three Japanese strains viz. JP119, JP151 and JP185 (Morakotkarn et al. 2008).
Figure 3.
Maximum likelihood phylogenetic tree generated from RAxML (GTR+G model), based on combined LSU and ITS sequences data. ML and MP values (MLBP/MPBP) (> 50%), resulting from 1000 bootstrap replicates, are given at the nodes. The original isolate numbers’ codes are noted after the species names. The tree is rooted to Pleospora herbarum (CBS 191.86). Ex-type or ex-epitype strains are in bold. Newly generated strains are in red.
Metabolites production
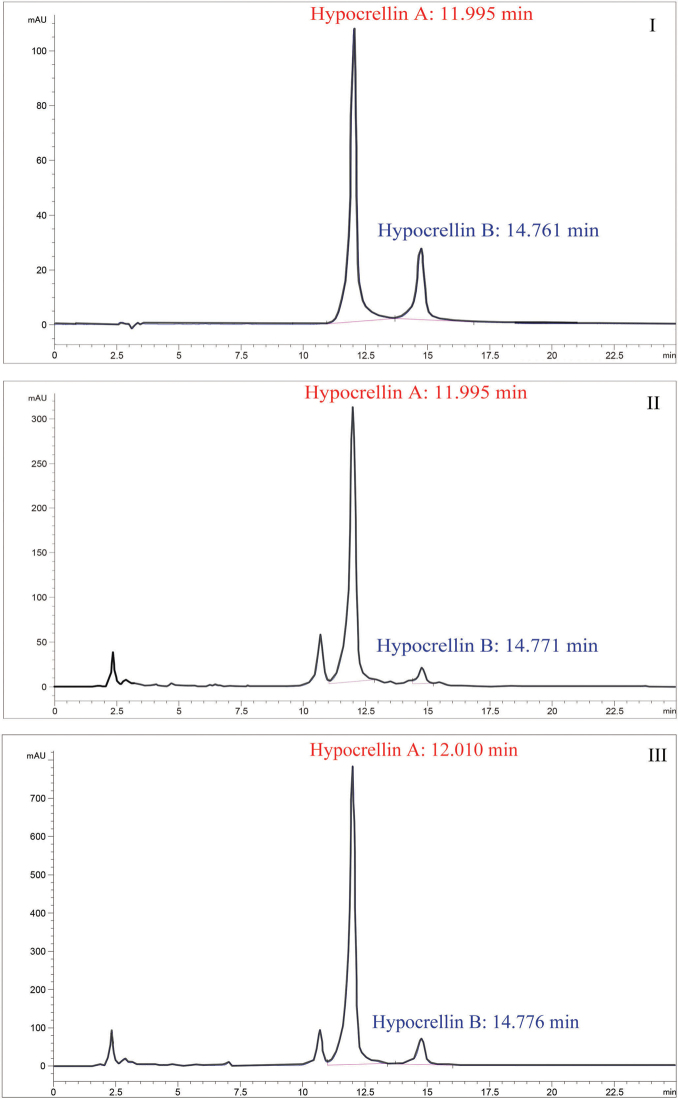
Stromatal extracts from specimens of S. bambusicola (HKAS102266) and R. bambusae (HKAS102270) contained high quantities of hypocrellin A (304.03 ng/ul and 790.86 ng/ul, respectively). Stromatal extracts from specimens of S. bambusicola contained 42.55 ng/ul hypocrellin B, whereas R. bambusae produces a higher quantity (204.60 ng/ul). The HPLC profiles of S. bambusicola and R. bambusae are depicted in Figure 4. The UV spectrum of the standards and of hypocrellin A and B from the samples (S. bambusicolaHKAS 102253 and R. bambusaeHKAS 102255) were recorded in alcohol and shown in Figure 5.
Figure 4.
Hypocrellin A and hypocrellin B HPLC-UV profiles (265 nm) of standards and stromatal HPLC-UV profiles (265 nm) of specimens of Shiraia bambusicola (HKAS 102253) (II) and Rubroshiraia bambusae (HKAS 102255) (III) and DAD spectra of major metabolites.
Taxonomy
Shiraiaceae
Y.X. Liu, Zi Y. Liu & K.D. Hyde, Phytotaxa 103(1): 53 (2013)
FE829B88F4F85051A601B10DCC0C5256
Notes.
The family Shiraiaceae was introduced by Liu et al. (2013) with a single genus and later Grandigallia was added to this family by Ariyawansa et al. (2013). In previous studies, Shiraiaceae was closely related with Phaeosphaeriaceae and their distinction was questionable (Cheng et al. 2004, Liu et al. 2013). However, our multi-gene analyses (Fig. 2) clearly indicate that Shiraiaceae and Phaeosphaeriaceae are distinct. Evidence is also borne out by the fact the Phaeosphaeriaceae have single ascostromata (Phookamsak et al. 2014), while in Shiraiaceae, ascostromata have multiple ascomata. Moreover, Shiraiaceae produces a high quantity of hypocrellins and no such metabolites, secreted by Phaeosphaeriaceae, were reported as far as we know (Phookamsak et al. 2014). In this study, the third genus (i.e. Rubroshiraia) is introduced to the family and produces hypocrellins. The endophytic strains in the phylogenetic tree in Figure (2) probably can be named as new genera, once the types are selected. Thus, currently three genera are placed in Shiraiaceae.
Type genus.
Shiraia Henn., Bot. Jb. 28(3): 274 (1900).
Type species.
S. bambusicola Henn., Bot. Jb. 28(3): 274 (1900).
Shiraia bambusicola
Henn., Bot. Jb. 28(3): 274 (1900)
981E92E478AF50D18ABCF92E6A692755
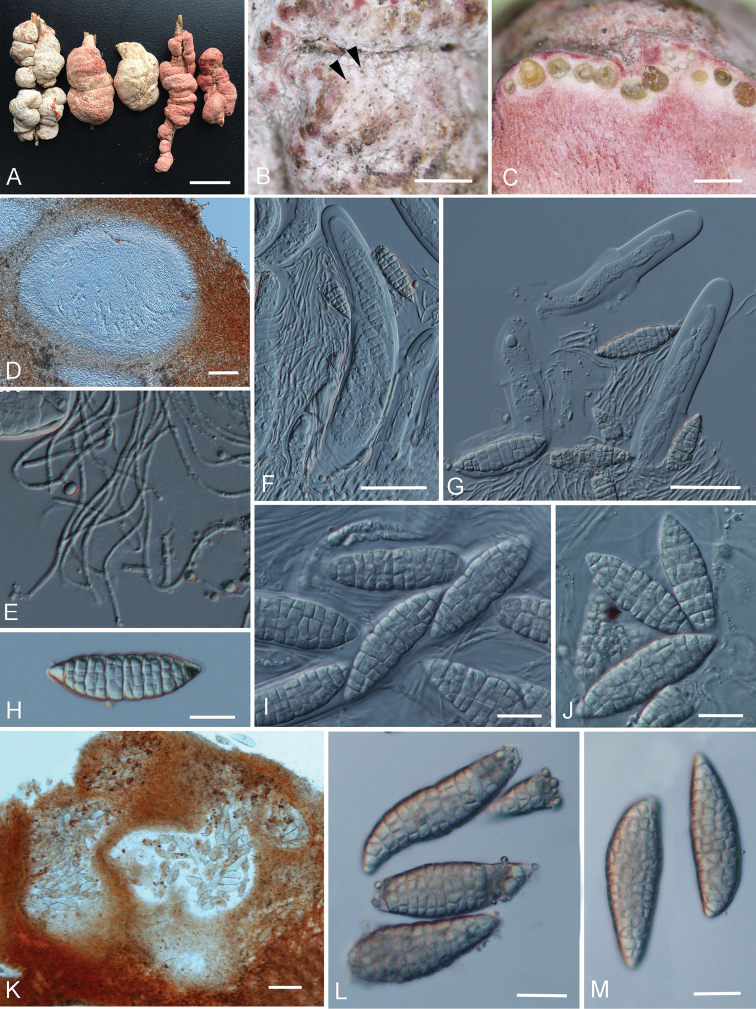
Figure 6.
Shiraia bambusicolaA–J sexual morph A fruiting bodies (HKAS102253, HKAS102254, HKAS102257, HKAS102261, HKAS102262) B–J photographs from material HKAS102253 B Surface of ascostromata showing the dark openings of ostiole C vertical section of ascostromata D vertical section of locule E pseudoparaphyses F, G asci (G Showing the fissitunicate asci) H–J ascospores K–M asexual morph K vertical section of asexual locules L–M conidia. Scale bars: 2 cm (A), 5 mm (B), 1 mm (C), 100 μm (D, K), 50 μm (F, G), 20 μm (H–J, L, M).
Description.
Parasitic on living branches of bamboo. Sexual morph: Ascostromata 1–6 cm long × 1–4 cm wide, solitary, superficial, subglobose, long ellipsoid to irregular, tuberculate, fleshy, white to pinkish, with locules lining the periphery, with dark ostiolate points appearing on surface. Ascostromatic tissue thick, pinkish, composed of wide, woven hyphae of textura intricata. Locules in vertical section 370–700 µm high × 370–700 µm diam. (x̄ = 541 × 513 µm, n = 20), globose to subglobose, immersed in the peripheral layer of ascostromata, with 100–200 µm wide ostioles. Peridium 20–45 µm thick, composed of several layers of hyaline to light brown, small cells of textura angularis to textura intricata. Hamathecium composed of interthecial, hyaline septate, branched pseudoparaphyses, 1–2.5 µm wide. Asci 200–370 × 20–35 µm (x̄ = 291.6 × 26.6 µm, n = 20), 4–6-spored, thick-walled, bitunicate, fissitunicate, cylindrical, short-pedicellate, with an ocular chamber. Ascospores 50–77 × 15–24 µm (x̄ = 62.3 × 18.1 µm, n = 20), 1-seriate, overlapped, fusiform, muriform, hyaline, with 7 transverse septa, constricted at the septum, smooth-walled. Asexual morph: Conidiomata 200–500 µm high, 300–400 µm wide, loculate, forming within ascostromata, globose to subglobose or irregular. Wall of locules 20–40 µm thick, composed of several layers of hyaline to light brown, small cells of textura intricata. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 3–6 × 2–3 µm (x̄ = 4.7 × 2.1 µm, n = 10), blastic, cylindrical, hyaline, smooth-walled. Conidia 60–80 × 19–25 µm (x̄ = 75.4 × 23.1 µm, n = 20), fusiform, muriform, hyaline, with irregularly transverse and longitudinal septa, straight to curved, smooth-walled.
Culture characters.
Colonies growing slowly, attaining 30 mm diam. after 2 weeks at 27 °C under dark, circular, with even margin, floccose at the centre, drift white at margin, light greenish at centre, dark from below.
Material examined.
CHINA, Yunnan province, Lijiang, on living branches of Brachystachyum densiflorum (Rendle) Keng, 3 May 2017, Dong-Qin Dai, DDQ00409 (HKAS102253), Ibid. (duplicate specimen deposited in HMAS 290446), Ibid. DDQ00410 (HKAS102254), Ibid. DDQ00413 (HKAS102257), Ibid. 10 June 2017, Dong-Qin Dai, DDQ00418 (HKAS102261), Ibid. DDQ00419 (HKAS102262), Ibid. DDQ00420 (HKAS102263), Ibid. DDQ00421 (HKAS102264), Ibid. DDQ00422 (HKAS102265), Ibid. DDQ00423 (HKAS102266), Ibid. DDQ00424 (HKAS102267).
Notes.
Shiraia bambusicola was erected by Hennings (1900), based on a collection from Japan. Liu et al. (2013) re-examined the holotype with 1–2.5 cm wide ascostromata, which is smaller than the new collections (1–4 cm wide in ascostromata) in China. The holotype has large ascospores compared with the new specimens in this study (75–125 × 23–47 µm vs. 50–77 × 15–24 µm). The epitype designated by Liu et al. (2013) which has similar-sized (50–77 × 15–24 µm) ascospores and similar ITS sequence, as in our new collections.
Other genera included
Grandigallia
M.E. Barr et al., Mycotaxon 29: 196. 1987.
779F11C3FA33521CA77BAA085CE562DD
Description.
Type species.
Grandigallia dictyospora M.E. Barr et al., Mycotaxon 29: 196 (1987)
Notes.
The monotypic genus Grandigallia was introduced by Barr (1987) and is typified by G. dictyospora. The fungus infects branches of Polylepis sericea Wedd. (Rosaceae) and produces conspicuous (3–14 cm in diam.) and black ascostromata. Grandigallia closely resembles Shiraia in having muriform ascospores, however, it differs by its black and larger ascostromata. Kirk et al. (2008) and Lumbsch and Huhndorf (2010) placed Grandigallia in Dothideomycetes, genera incertae sedis.Ariyawansa et al. (2013) re-examined the type material and transferred it to Shiraiaceae in Pleosporales. Wijayawardene et al. (2014, 2017, 2018) accepted this placement.
Rubroshiraia
D.Q. Dai & K.D. Hyde gen. nov.
D006E91FAED454F682DB6F5C97D99A97
Etymology.
The epithet “Rubro” means red colour referring to reddish ascotromata similar to the genus Shiraia.
Description.
Parasitic on living branches of bamboo. Sexual morph: Ascostromata solitary, superficial, globose to subglobose, fleshy, reddish, with locules lining the periphery, with dark ostiolate tips appearing on surface. Ascostromatic tissue thick, pinkish, composed of wider woven hyphae of textura intricata. Locules globose to subglobose, immersed in the peripheral layer of ascostromata, with narrow ostiolate openings. Peridium composed of several layers of hyaline to dark brown, small cells of textura angularis to textura intricata. Hamathecium of interthecial, hyaline, septate, branched pseudoparaphyses above asci. Asci 8-spored, thick-walled, bitunicate, fissitunicate, cylindrical, short-pedicellate, with an ocular chamber. Ascospores spirally arranged in asci, filiform, hyaline, with transverse septa, smooth-walled. Asexual morph: Undetermined.
Type species.
R. bambusae D.Q. Dai & K.D. Hyde.
Notes.
The hypocrellin-producing fungus R. bambusae is a well-known taxon used in Chinese traditional medicine which is called “Zhuhongjun” or “Zhuxiaorouzhuojun” in Chinese. However, without molecular data, it was wrongly named as H. bambusae (Liu 1978).
Hypocrella bambusae was combined by Saccardo (1878), based on its linear asci and filiform ascospores. Index Fungorum (2019) lists its basionym as Hypocrea bambusae Berk. & Broome, which was collected on the inflorescences of bamboo in Sir Lanka and had linear asci and filiform ascospores (Berkeley and Broome 1875). Liu (1978) recorded a well-known Chinese medicinal ascomycete, producing 0.7–1.5 mm diam., hemispheric and reddish stromata with multi-locules, cylindrical asci and filiform ascospores which are spirally arranged and more than 250 μm long on bamboo culms. Liu (1978) identified this fungus as H. bambusae, probably based on its cylindrical asci and filiform ascospores. In addition, species of Hypocrella usually produce perithecial ascomata (Saccardo 1878). To our knowledge, no fungal records or herbal medicine like that described in Liu (1978) occur in Sir Lanka. Moreover, based on the examination of type material of Hypocrea bambusae, it has smaller (0.1 cm vs. 0.7–1.5 mm in diam.) and black stromata, unitunicate asci and ascospores are in a single fascicle but not significantly helically coiled (Fig. 7). Hence, we conclude that Liu (1978) made a wrong identification.
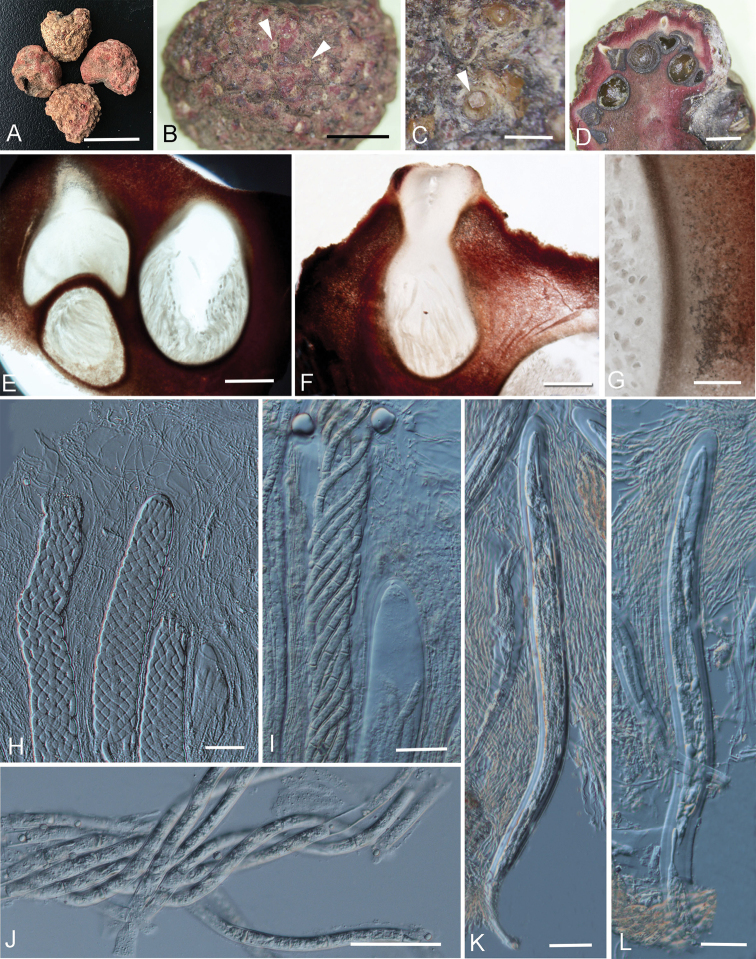
Figure 7.
Rubroshiraia bambusae (HKAS102255, holotype) A fruiting bodies B, C surface of ascostromata showing the openings of ostiole D vertical section of ascostromata E, F vertical section of locule G peridium of locule H asci and pseudoparaphyses I asci and asci ocular chamber J ascospores K, L immature asci. Scale bars: 1 cm (A), 25 mm (B), 2 mm (C, D), 500 μm (E, F), 200 μm (G), 50 μm (H–L).
New collections of “Zhuhongjun” were collected and sequenced. The phylogenetic analyses showed it belongs to Shiraiaceae and is separate from Shiraia with high bootstrap support (100/1.00 MLBS/BSPP) (Fig. 2). Grandigallia has not been included in the phylogenetic tree as it is lacking gene sequences in the GenBank (retrieved date: 13 May 2019). However, Grandigallia can be morphologically distinguished from the new taxon in having black ascostromata and muriform ascospores (Barr 1987; Ariyawansa et al. 2013). Thus, this fungus is introduced as R. bambusae gen. et sp. nov in this study.
Rubroshiraia bambusae is often confused with S. bambusicola by Chinese traditional folk residents, probably because of the similarity of their ascostromata, parasitism on bamboo host and similar efficacy of medical treatment. However, it differs from S. bambusicola by its smaller sized ascostromata (0.7–1.2 cm long × 0.7–1 cm wide vs. 1–6 cm long × 1–4 cm wide) and distinct ascospores (filiform ascospores vs. fusiform and muriform ones). Both of the above species can produce the metabolites hypocrellin A and B, whereas R. bambusae contains almost double the content compared to S. bambusicola (Fig. 4).
Rubroshiraia bambusae
D.Q. Dai & K.D. Hyde sp. nov.
B8CC7A3EDFC3517F8169748C3DCE32BA
Etymology.
Refers the bamboo host.
Holotype.
HKAS102255.
Description.
Parasitic on living branches of bamboo. Sexual morph: Ascostromata 0.7–1.5 cm long × 0.7–1.3 cm wide, solitary, superficial, globose to subglobose, fleshy, reddish, with locules lining the periphery, with dark ostiolate points appearing on the surface. Ascostromatic tissue thick, pinkish, composed of wider woven hyphae of textura intricata. Locules in vertical section 800–1800 µm high × 1000–2000 µm diam. (x̄ = 1289.4 ×1368.8 µm, n = 20), globose to subglobose, immersed in the periphery layer of ascostromata, with 250–500 µm wide × 450–550 µm high ostioles. Peridium 20–35 µm thick, composed of several layers of hyaline to dark brown, small cells of textura angularis to textura intricata. Hamathecium of interthecial, hyaline septate, branched pseudoparaphyses, 1–3 µm wide. Asci 660–800 × 45–55 µm (x̄ = 751.6 × 49.5 µm, n = 20), 8-spored, thick-walled, bitunicate, fissitunicate, cylindrical, short-pedicellate, with an ocular chamber. Ascospores 600–750 × 5.5–11 µm (x̄ = 728.8 × 9.1 µm, n = 20), spirally arranged in asci, filiform, hyaline, with 15–18 transverse septa, smooth-walled. Asexual morph: Undetermined.
Material examined.
CHINA, Yunnan, Dali, on living branches of Fargesia spathacea Franch, 13 May 2017, Dong-Qin Dai, DDQ00411 (HKAS102255, holotype), Ibid. (HMAS 290447, isotype), Ibid. DDQ00412 (HKAS102256), Ibid. DDQ00416 (HKAS102260), Ibid. 20 June 2017, Dong-Qin Dai, DDQ00425 (HKAS102268), Ibid. DDQ00426 (HKAS102269), Ibid. DDQ00427 (HKAS102270), Ibid. DDQ00428 (HKAS102271), Ibid. DDQ00429 (HKAS102272), Ibid. DDQ00430 (HKAS102273), Ibid. DDQ00431 (HKAS102274).
Key for distinguishing genera in Shiraiaceae
| 1 | Parasitising bamboo branches, ascostromata are white to reddish | 2 |
| – | Parasitising Rosaceae branches, ascostromata are black | Grandigallia |
| 2 | Ascospores muriform | Shiraia |
| – | Ascospores filiform | Rubroshiraia |
Since the familial placement of H. bambusae is controversial in different studies (Berkeley and Broome 1875, Saccardo 1878, Liu 1978), we re-studied the isotype. Based on morphology, we conclude that it has unitunicate asci thus related to Sordariomycetes.
Hypocrella bambusae
(Berk. & Broome) Sacc. 1878
C63A90F9A247512CBE1F4AC911BD2CD0
Figure 8.
Hypocrella bambusae (K(M)52469, isotype, images are accredited to the Royal Botanic Gardens, Kew) A, C fruiting bodies on inflorescence of bamboo B vertical section of stromata showing the perithecia locating D herbarium envelope E filiform ascospores F asci with caps (Staining by cotton blue). Scale bars: 5 mm (A), 200 μm (B), 2 mm (C), 20 μm (E, F).
Basionym.
Hypocrea bambusae Berk. & Broome, 1873
Description.
Parasitic on living inflorescence of bamboo. Sexual morph: Stromata around 0.14 cm diam., 0.06 cm high, solitary, superficial, subglobose, fleshy to coriaceous, black, with around 20 perithecia lining the periphery, with ostioles slightly raised above stroma surface. Stromatic tissue thick, brown to dark brown. Perithecia in vertical section around 100 µm diam., 200 µm high, pyriform, immersed in the periphery layer of stromata. Asci more than 220 µm long, 5–6 µm diam., 8-spored, unitunicate, cylindrical, with a glassy refractive cap around 3 µm from apex to base. Ascospores around 180 µm long, 1–1.5 µm diam., in a single fascicle but not significantly helically coiled, filiform, hyaline, with 9–10 transverse septa, with rounded ends, smooth-walled. Asexual morph: Undetermined.
Material examined.
SRI LANKA, on inflorescence of bamboo, January 1855, G.H.K. Thwaites s.n. (ex herb. M.J. Berkeley), K(M)52469, isotype.
Notes.
This taxon has typical morphology of the Clavicipitaceae, which is pyriform perithecia with a gradually tapering upper part and cylindrical asci with a glassy refractive cap. New collections are required and need to be sequenced to clarify its placement.
Discussion
Members of the family Shiraiaceae are distributed from Asia to South America but so far reported only from three countries, viz. China, Japan and Venezuela (Barr et al. 1987; Liu et al. 2013). The family comprises three genera, i.e. Grandigallia, Rubroshiraia and Shiraia wherein the former genus is lacking DNA sequences and, thus in here, we did not include it in the molecular analyses (Figs 2 and 3). These genera show the typical characters of Shiraiaceae, viz. conspicuous large, tuberculate, fleshy and multi-loculate ascostromata producing bitunicate asci. Shiraia bambusicola has various types of ascostromata, such as subglobose to tuberculate with white to pinkish colours (Fig. 6). However, the phylogenetic analysis shows these specimens with different types of ascostromata belong to same species (Figs 2 and 3). Thus, we assume that the different shapes of ascostromata are because of the host and different environment conditions.
Stromatal methanol extracts of Rubroshiraia and Shiraia contain Hypocrellins (Fig. 4). However, so far no extracts have been reported from Grandigallia. Fresh material of Grandigallia is essential to determine the metabolites. Rubroshiraia has darker reddish ascostromata compared with Shiraia, probably because its stromatal methanol extracts contain larger quantity of hypocrellins. Some endophytes, named as Shiraia-like fungi, are known to produce hypocrellins on media. They were isolated from different parts of bamboo, such as seeds, nods and internodes (Lu et al. 2004; Morakotkarn et al. 2008; Liang et al. 2009; Cai et al. 2011; Zhang et al. 2014). Other Shiraia-like endophytes, isolated from the rhizome of Gastrodia, leaves of Huperzia serrata and from Triticum aestivum, phylogenetically cluster within the former group (Fig. 3). However, no hypocrellins were produced from their mycelium (Zhu et al. 2010; Wang et al. 2011, 2016). The bamboo tissue may be providing the needful substances for fungi to produce hypocrellins. The endophytic Shiraia-like taxa (Fig. 3) appear as a distinct genus in Shiraiaceae. The nomination will be made once the type material is available.
Shiraia bambusicola has been used as a Chinese traditional folk-medicine, in curing rheumatoid arthritis, infantile convulsion and pertussis etc. for more than 400 years, because of its stromatal metabolites (Huang et al. 2001; Shen et al. 2002). Japanese scientists first obtained three perylenequinones from air-dried ascostromata of S. bambusicola and named them as hypocrellin A, B and C (Kishi et al. 1991). However, hypocrellin A was originally discovered by Wan and Chen (1981) from a different fungus on bamboo which was called as “Zhuhongjun” in Chinese and was erroneously identified as H. bambusae (Liu 1978). Later the fourth hypocrellin analogue (hypocrellin D) was named by Fang et al. (2006). Therefore, in total, four hypocrellins have so far been named. Hypocrellins are types of biologically active compounds and naturally occurring perylenequinones with photodynamic activity (Wan and Chen 1981; Kishi et al. 1991; Chowdhury et al. 2002; Liang et al. 2009; Liu et al. 2013). These secondary metabolites have gained much attention owing to their light-induced anti-tumour, anti-fungal and anti-viral activities (Wan and Chen 1981; Liang et al. 2009; Li et al. 2000a, b). In clinical trials, hypocrellin shows promising treatment for various skin diseases, such as skin cancer and white lesions of the vulva (Wan and Chen 1981; Li et al. 2000b). In China, a costly medicinal unguent named Bamboo Parasitic Fungus Ointment is made of hypocrellin B (Dai et al. 2018). Interestingly, it was proved that hypocrellin has bactericidal activities which inhibit various bacteria, such as Bacillus subtilis Ehrenberg and Micrococcus luteus Schroeter (Chen et al. 2010). In addition, hypocrellin A has an antiviral activity against human immunodeficiency virus (HIV-1) (Hudson et al. 1994) and is promising as a new-fashioned photoelectric conversion material (Li et al. 2000a).
Hypocrellin has wide application prospects, but it was earlier only found existing in ascostromata of S. bambusicola and “Zhuhongjun” (R. bambusae in this paper) (Wan and Chen 1981; Kishi et al. 1991). For gaining a high yield of Hypocrellin, scientists devoted themselves to looking for strains that can produce hypocrellin through fermentation production (Liang et al. 2009). Numerous endophytes, isolated from bamboo tissue such as culms, leaves, nodes and seeds, were published (Lu et al. 2004; Morakotkarn et al. 2007, 2008; Liang et al. 2009; Cai et al. 2011; Shen et al. 2012, 2014; Zhang et al. 2014), several of which had the potential for hypocrellin production (Lu et al. 2004; Morakotkarn et al. 2008; Liang et al. 2009; Zhang et al. 2014; Tong et al. 2017). However, the strains with promising industrial fermentation were identified as Shiraia sp. based on the blast search in GenBank by ITS sequences. More endophytes producing biologically active compounds, such as huperzine, isolated from the plant Huperzia serrata (Thunb. ex Murray) Trev., were also named as Shiraia sp. (Wang et al. 2011, 2016; Zhu et al. 2010). These strains usually have around 80%–90% ITS similarity with S. bambusicola, which also shows that they are phylogenetically close with members of Shiraiaceae. In this study, these endophytes are placed in Shiraiaceae, based on the phylogenetic analyses (Fig. 3).
According to Deng et al. (2017), polyketide synthase (SbaPKS) is involved in hypocrellin biosynthesis, based on the methods of CRISPR/Cas9 genome editing. It provides evidence for decoding the hypocrellin pathway (Deng et al. 2017). This pathway has the potential for producing high quality hypocrellins.
Supplementary Material
Acknowledgements
This work was supported by the Key Laboratory of Yunnan Province Universities of the Diversity and Ecological Adaptive Evolution for Animals and plants on Yun-Gui Plateau, the National Natural Science Foundation of China (No. NSFC 31760013, 31950410558, 31260087, 31460561, 31860005, 31460179 and 31860057) and the Scientific Research Foundation of Yunnan Provincial Department of Education (2017ZZX186). Dong-Qin Dai would like to thank Yunnan Province Universities of the Science and Technology Innovation Team for the exploitation and utilisation of endophytes and the Thousand Talents Plan, Youth Project of Yunnan Provinces for support. Chao Liu thanks the Yunnan Local Colleges Applied Basic Research Projects (2017FH001-034). Dong-Qin Dai would like to thank Xiu Gao (Qujing Normal University) for the help with drawing chemical structures of hypocrellins and is grateful to Dr. Joanne E. Taylor for the help of the loaned herbarium.
Citation
Dai D-Q, Wijayawardene NN, Tang L-Z, Liu C, Han L-H, Chu H-L, Wang H-B, Liao C-F, Yang E-F, Xu R-F, Li Y-M, Hyde KD, Bhat DJ, Cannon PF (2019) Rubroshiraia gen. nov., a second hypocrellin-producing genus in Shiraiaceae (Pleosporales). MycoKeys 58: 1–26. https://doi.org/10.3897/mycokeys.58.36723
Funding Statement
National Natural Science Foundation of China
Contributor Information
Li-Zhou Tang, Email: biologytang@163.com.
Chao Liu, Email: liuchao_80@163.com.
References
- Amano N. (1980) Studies on the Japanese Loculoascomycetes. II. Taxonomic position of the genus Shiraia. Bulletin of the National Science Museum 6: 55–60. [Google Scholar]
- Ariyawansa HA, Kang JC, Alias SA, Chukeatirote E, Hyde KD. (2013) Towards a natural classification of Dothideomycetes: The genera Dermatodothella, Dothideopsella, Grandigallia, Hysteropeltella and Gloeodiscus (Dothideomycetesincertae sedis). Phytotaxa 147: 35–47. 10.11646/phytotaxa.147.2.1 [DOI] [Google Scholar]
- Barr ME, Boise JR, Hanlin RT. (1987) A spectacular Loculoascomycete from Venezuela. Mycotaxon 29: 195–198. [Google Scholar]
- Berkeley MJ, Broome CE. (1875) Enumeration of the fungi of Ceylon. Part II. Botanical Journal of the Linnean Society 14: 29–141. 10.1111/j.1095-8339.1873.tb00301.x [DOI] [Google Scholar]
- Cai Y, Liao X, Liang X, Ding Y, Sun J, Zhang D. (2011) Induction of hypocrellin production by Triton X-100 under submerged fermentation with Shiraia sp. SUPER-H168. New Biotechnology 28: 588–592. 10.1016/j.nbt.2011.02.001 [DOI] [PubMed] [Google Scholar]
- Chem A. (2012) Practical HPLC method development. Carbohydrate Polymers 16: 338–338. 10.1016/0144-8617(91)90119-W [DOI] [Google Scholar]
- Chen H, Chen WQ. (2009) The analysis and comparison of bioactive ingredients in Shiraia bambusicola from different regions. Journal of Zhejiang Shuren University 9: 17–19. [Google Scholar]
- Chen YJ, Zhong WW, Yang SY. (2010) Study on the Antibacterial Activity of Shiraia bambusicola Henn. Journal of Yunnan University of Nationalities (Natural Sciences Edition) 19: 154–156. [Google Scholar]
- Cheng TF, Jia XM, Ma XH, Lin HP, Zhao YH. (2004) Phylogenetic study on Shiraia bambusicola by rDNA sequence analyses. Journal of Basic Microbiology 44: 339–350. 10.1002/jobm.200410434 [DOI] [PubMed] [Google Scholar]
- Chowdhury PK, Das K, Datta A, Liu WZ, Zhang HY, Petrich JW. (2002) A comparison of the excited-state processes of nearly symmetrical perylene quinones: hypocrellin A and hypomycin B. Journal of Photochemistry & Photobiology A: Chemistry 154: 107–116. 10.1016/S1010-6030(02)00309-X [DOI] [Google Scholar]
- Dai DQ, Phookamsak R, Wijayawardene NN, Li WJ, Bhat DJ, Mortimer PE, Xu JC, Taylor JE, Hyde KD, Chukeatirote E. (2017) Bambusicolous fungi. Fungal Diversity 82: 1–105. 10.1007/s13225-016-0367-8 [DOI] [Google Scholar]
- Dai DQ, Tang LZ, Wang HB. (2018) Review of bambusicolous ascomycetes. Bamboo: Current and Future Prospects 2018: 165. 10.5772/intechopen.76463 [DOI]
- Deng HX, Gao RJ, Liao XG, Cai YJ. (2017) Genome editing in Shiraia bambusicola using CRISPR-Cas9 system. Journal of Biotechnology 259: 228–234. 10.1016/j.jbiotec.2017.06.1204 [DOI] [PubMed] [Google Scholar]
- Fang LZ, Qing C, Shao HJ, Yang YD, Dong ZJ, Wang F, Zhao W, Yang WQ, Liu JK. (2006) Hypocrellin D, a cytotoxic fungal pigment from fruiting bodies of the ascomycete Shiraia bambusicola. Journal of Antibiotics 59: 351–354. 10.1038/ja.2006.49 [DOI] [PubMed] [Google Scholar]
- Hall T. (2004) BioEdit. Ibis Therapeutics, Carlsbad. http://www.mbio.ncsu.edu/BioEdit/bioedit.html [18 Mar 2005]
- Hennings P. (1900) Fungi Japonici. Botanische Jahrbücher für Systematik, Pflanzengeschichte und Pflanzengeographie 28: 259–280. [Google Scholar]
- Hino I. (1961) Icones fungorum bambusicolorum japonicorum. The Fuji Bamboo Garden, Kobe.
- Huang TK, Ding ZZ, Zhao SX, Yan YQ, Xu GJ, Chen L, Yu CL, Gao XL, Zhang ZD. (2001) Xian Dai Ben Cao Gang Mu. China Medical Science Press, Beijing. [In Chinese]
- Hudson JB, Zhou J, Chen J, Harris L, Towers GH. (1994) Hypocrellin, from Hypocrella bambusae, is phototoxic to human immunodeficiency virus. Photochem Photobiol 60: 253–255. 10.1111/j.1751-1097.1994.tb05100.x [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Index Fungorum (2019) Index Fungorum. http://www.indexfungorum.org/names/Names.asp
- Jayasiri SC, Hyde KD, Ariyawansa HA, Bhat DJ, Buyck B, Cai L, Dai YC, Abd-Elsalam KA, Ertz D, Hidayat I, Jeewon R, Jones EBG, Bahkali AH, Karunarathna SC, Liu JK, Luangsa-ard JJ, Lumbsch HT, Maharachchikumbura SSN, McKenzie EHC, Moncalvo JM, Ghobad-Nejhad M, Nilsson H, Pang KL, Pereira OL, Phillips AJL, Raspé O, Rollins AW, Romero AI, Etayo J, Selçuk F, Stephenson SL, Suetrong S, Taylor JE, Tsui CKM, Vizzini A, Abdel-Wahab MA, Wen TC, Boonmee S, Dai DQ, Daranagama DA, Dissanayake AJ, Ekanayaka AH, Fryar SC, Hongsanan S, Jayawardena RS, Li WJ, Perera RH, Phookamsak R, de Silva NI, Thambugala KM, Tian Q, Wijayawardene NN, Zhao RL, Zhao Q, Kang JC, Promputtha I. (2015) The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Diversity 74: 3–18. 10.1007/s13225-015-0351-8 [DOI] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk PM, Cannon PF, David JC, Stalpers JA. (2001) Ainsworth and Bisby’s Dictionary of the Fungi (9th edn). CAB International, Wallingford, 655 pp. [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, Stalpers JA. (2008) Dictionary of the Fungi (10th edn). CABI, Wallingford, 784 pp. [Google Scholar]
- Kishi T, Tahara S, Taniguchi N, Tsuda M, Tanaka C, Takahashi S. (1991) New perylenequinones from Shiraia bambusicola. Planta Medica 57: 376–379. 10.1055/s-2006-960121 [DOI] [PubMed] [Google Scholar]
- Lai GH, Fu LY. (2000) Study on main host plants of Shiraia bambusicola. Chinese Wild Plant Resources 1: 8–11. [In Chinese] [Google Scholar]
- Li C, Chen YT, Lin NY, Wang HQ, Liu WZ, Xie JL. (2000a) Analysis on the chemical components of a fungus producing perylenequinones photosensitive compounds. Mycosystema 19: 122–127. [Google Scholar]
- Li C, Wang HQ, Xie JL, Lin NY, Dai WH, Chen YT. (2000b) Analysis and comparisons of compounds among three medical fungi of Hypocreaceae. Chinese Traditional and Herbal Drugs 31: 250–251. [Google Scholar]
- Li XM, Gao J, Yue YD, Hou CL. (2009) Studies on Systematics, Biology and Bioactive Substance of Shiraia bambusicola. Forest Research 22: 279–284. [Google Scholar]
- Liang XH, Cai YJ, Liao XR, Wu K, Wang L, Zhang DB, Meng Q. (2009) Isolation and identification of a new hypocrellin A-producing strain Shiraia sp. SUPER-H168. Microbiological Research 164: 9–17. 10.1016/j.micres.2008.08.004 [DOI] [PubMed] [Google Scholar]
- Liu B. (1978) Chinese medicinal fungi (2nd edn). Shanxi people’s publishing house, Taiyuan. [In Chinese]
- Liu YJ, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Liu YX, Hyde KD, Ariyawansa HA, Li WJ, Zhou DQ, Yang YL, Chen YM, Liu ZY. (2013) Shiraiaceae, new family of Pleosporales (Dothideomycetes, Ascomycota). Phytotaxa 103: 51–60. 10.11646/phytotaxa.103.1.4 [DOI] [Google Scholar]
- Liu JK, Hyde KD, Jeewon R, Phillips AL, Maharachchikumbura SSN, Ryberg M, Liu ZY, Zhao Q. (2017) Ranking higher taxa using divergence times: a case study in Dothideomycetes. Fungal Diversity 84: 75–99. 10.1007/s13225-017-0385-1 [DOI] [Google Scholar]
- Lu MF, Huang YB, Zhang HY, Wang HN, Zhang YJ. (2004) Cloning and sequencing of the ITS in rDNA Gene of a Fungus Producing Perylenequinones Deriveration. Journal of Sichuan Agricultural University 22: 138–141. 10.1300/J064v24n01_09 [DOI] [Google Scholar]
- Lumbsch HT, Huhndorf SM. (2010) Myconet Volume 14. Part one. Outline of Ascomycota-2009. Part Two. Notes on Ascomycete Systematics. Nos. 4751–5113. Fieldiana Life and Earth Sciences 1: 1–64. 10.3158/1557.1 [DOI] [Google Scholar]
- Maharachchikumbura SS, Hyde KD, Jones EBG, McKenzie EHC, Huang SK, Abdel-Wahab MA, Daranagama DA, Dayarathne M, D’souza MJ, Goonasekara ID, Hongsanan S, Jayawardena RS, Kirk PM, Konta S, Liu JK, Liu ZY, Norphanphoun C, Pang KL, Perera RH, Senanayake IC, Shang Q, Shenoy BD, Xiao YP, Bahkali AH, Kang JC, Somrothipol S, Suetrong S, Wen TC, Xu JC. (2015) Towards a natural classification and backbone tree for Sordariomycetes. Fungal Diversity 72: 199–301. 10.1007/s13225-015-0331-z [DOI] [Google Scholar]
- Morakotkarn D, Kawasaki H, Seki T. (2007) Molecular diversity of bamboo-associated fungi isolated from Japan. FEMS Microbiology Letters 266: 10–19. 10.1111/j.1574-6968.2006.00489.x [DOI] [PubMed] [Google Scholar]
- Morakotkarn D, Kawasaki H, Tanaka K, Okane I, Seki T. (2008) Taxonomic characterization of Shiraia-like fungi isolated from bamboos in Japan. Mycoscience 49: 258–265. 10.1007/S10267-008-0419-3 [DOI] [Google Scholar]
- Nylander JAA. (2004) MrModeltest 2.0. Program distributed by the author. Evolutionary Biology Centre, Uppsala University
- Page RDM. (1996) TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12: 357–358. 10.1093/bioinformatics/12.4.357 [DOI] [PubMed] [Google Scholar]
- Phillips AJL, Alves A, Abdollahzadeh J, Slippers B, Wingfield MJ, Groenewald JZ, Crous PW. (2013) The Botryosphaeriaceae: genera and species known from culture. Studies in Mycology 76: 51–167. 10.3114/sim0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phookamsak R, Liu JK, McKenzie EHC, Manamgoda DS, Ariyawansa H, Thambugala KM, Dai DQ, Camporesi E, Chukeatirote E, Wijayawardene NN, Bahkali AH, Mortimer PE, Xu JC, Hyde KD. (2014) Revision of Phaeosphaeriaceae. Fungal Diversity 68: 159–238. 10.1007/s13225-014-0308-3 [DOI] [Google Scholar]
- Rannala B, Yang Z. (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of molecular evolution 43: 304–311. 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- Rehner S. (2001) Primers for elongation factor 1-α (EF1-α). http://ocid.NACSE.ORG/research/deephyphae/EF1primer.pdf
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Saccardo PA. (1878) Enumeratio pyrenomycetum Hpocreaceorum hucusque cognitorum systemate carpologico dispositorum. Michelia 1: 277–325. [Google Scholar]
- Saccardo PA. (1902) Sylloge Fungorum. Omnium hucosque cognitorum. Supplementum universale XVI: 421
- Shen YX, Rong XG, Gao ZH. (2002) Studies on the chemical constituents of Shiraia bambusicola. China Journal of Chinese Materia Medica 27: 674–677. [PubMed] [Google Scholar]
- Shen X, Zheng D, Gao J, Hou CL. (2012) Isolation and evaluation of endophytic fungi with antimicrobial ability from Phyllostachys edulis. Bangladesh Journal of Pharmacology 7: 249–257. 10.3329/bjp.v7i4.12068 [DOI] [Google Scholar]
- Shen XY, Cheng YL, Cai CJ, Fan L, Gao J, Hou CL. (2014) Diversity and Antimicrobial Activity of Culturable Endophytic Fungi Isolated from Moso Bamboo Seeds. PloS one 9: e95838. 10.1371/journal.pone.0095838 [DOI] [PMC free article] [PubMed]
- Silvestro D, Michalak I. (2011) raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution 12: 335–337. 10.1093/bioinformatics/btg180 [DOI] [Google Scholar]
- Stadler M, Wollweber H, Mühlbauer A, Henkel T, Asakawa Y, Hashimoto T, Rogers JD, Ju YM, Wetzstein HG, Tichy HV. (2001) Secondary metabolite profiles, genetic fingerprints and taxonomy of Daldinia and allies. Mycotaxon 77: 379–429. [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Swofford DL. (2002) PAUP: phylogenetic analysis using parsimony, version 4.0 b10. Sinauer Associates, Sunderland. 10.1002/0471650129.dob0522 [DOI]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZW, Mao L, Liang H, Zhang Z, Wang Y, Yan R, Zhu D. (2017) Simultaneous Determination of Six Perylenequinones in Shiraiaia sp. Slf14 by HPLC. Journal of Liquid Chromatography & Related Technologies 40: 536–540. 10.1080/10826076.2017.1331172 [DOI] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan XY, Chen YT. (1981) Hypocrellin A-A new drug for photochemotherapy. Chinese Science Bulletin 26: 1040–1042. [Google Scholar]
- Wang Y, Zeng QG, Zhang ZB, Yan RM, Wang LY, Zhu D. (2011) Isolation and characterization of endophytic huperzine A-producing fungi from Huperzia serrata. Journal of Industrial Microbiology & Biotechnology 38: 1267–1278. 10.1007/s10295-010-0905-4 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zheng L, Li XX, Yan RM, Zhang ZB, Yang HL, Zhu D. (2016) Isolation, diversity and acetylcholinesterase inhibitory activity of the culturable endophytic fungi harboured in Huperzia serrata from Jinggang Mountain, China. World Journal of Microbiology & Biotechnology 32: 1–23. 10.1007/s11274-015-1966-3 [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (Eds) PCR protocols: a guide to methods and applications, Academic, San Diego, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Wijayawardene NN, Crous PW, Kirk PM, Hawksworth DL, Boonmee S, Braun U, Chomnunti P, Dai DQ, D’souza MJ, Diederich P, Dissanayake A, Doilom M, Hongsanan S, Jones EBG, Groenewald JZ, Jayawardena R, Lawrey JD, Liu JK, Lücking R, Madrid H, Manamgoda DS, Muggia L, Nelsen MP, Phookamsak R, Suetrong S, Tanaka K, Thambugala KM, Wikee S, Zhang Y, Aptroot A, Ariyawansa HA, Bahkali AH, Bhat JD, Gueidan C, De Hoog GS, Knudsen K, McKenzie EHC, Miller AN, Mortimer PE, Wanasinghe DN, Phillips AJL, Raja HA, Slippers B, Shivas RS, Taylor JE, Wang Y, Woudenberg JHC, Piątek M, Cai L, Jaklitsch WM, Hyde KD. (2014) Naming and outline of Dothideomycetes. (2014) including proposals for the protection or suppression of generic names. Fungal Diversity 69: 1–55. 10.1007/s13225-014-0309-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayawardene NN, Papizadeh M, Phillips AJL, Wanasinghe DN, Bhat DJ, Weerahewa HLD, Shenoy BD, Wang Y, Huang YQ. (2017) Mycosphere Essays 19: Recent advances and future challenges in taxonomy of coelomycetous fungi. Mycosphere 8: 934–950. 10.5943/mycosphere/8/7/9 [DOI] [Google Scholar]
- Wijayawardene NN, Hyde KD, Lumbsch HT, Liu JK, Maharachchikumbura SSN, Ekanayaka AH, Tian Q, Phookamsak R. (2018) Outline of Ascomycota: 2017. Fungal Diversity 88: 167–263. 10.1007/s13225-018-0394-8 [DOI] [Google Scholar]
- Zhang M, Pang W, Wang J. (2014) Effect of oxidative stress on hypocrellin A yield in submerged cultures of endophytic Shiraia sp. A8. Planta Med 80: P1N2. 10.1055/s-0034-1394593 [DOI]
- Zhaxybayeva O, Gogarten JP. (2002) Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC Genomics 3: 4. 10.1186/1471-2164-3-4 [DOI] [PMC free article] [PubMed]
- Zhao D, Liang ZQ. (2005) Reviews of Studies on Isolation and Culture of Shiraia bambusicola Henn. Journal of Fungal Research 3: 53–57. [Google Scholar]
- Zhu D, Wang JX, Zeng QG, Zhang ZB, Yan RM. (2010) A novel endophytic huperzine A-producing fungus, Shiraia sp. Slf14, isolated from Huperzia serrata. Journal of Applied Microbiology 109: 1469–1478. 10.1111/j.1365-2672.2010.04777.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.