Abstract
Many bacterial pathogens secret effectors into host cells to disable host defenses and thus promote infection. The exocyst complex functions in the transport and secretion of defense molecules, and loss of function of the EXO70B1 subunit leads to autoimmunity by activation of a truncated Toll/interleukin-1 receptor–nucleotide-binding sequence protein (TIR-NBS2; herein referred to as TN2). Here, we show that EXO70B1 is required for pathogen-associated molecular pattern-triggered immune responses in Arabidopsis thaliana. The effector AvrPtoB, an E3 ligase from Pseudomonas syringae pv. tomato (Pto) strain DC3000, associates with EXO70B1. AvrPtoB ubiquitinates EXO70B1 and mediates EXO70B1 degradation via the host’s 26S proteasome in a manner requiring E3 ligase activity. AvrPtoB enhances Pto DC3000 virulence by overcoming EXO70B1-mediated resistance. Moreover, overexpression of AvrPtoB in Arabidopsis leads to autoimmunity, which is partially dependent on TN2. Expression of TN2 in tobacco (Nicotiana tabacum and Nicotiana benthamiana) triggers strong and rapid cell death, which is suppressed by co-expression with EXO70B1 but reoccurs when co-expressed with AvrPtoB. Taken together, our data highlight that AvrPtoB targets the Arabidopsis thaliana EXO70 protein family member EXO70B1 to manipulate the defense molecule secretion machinery or immunity.
Keywords: Arabidopsis, plant immunity, EXO70B1, AvrPtoB, TIR-NBS2, exocyst complex, NLR protein
Introduction
Plants have evolved a multilayered immune system to cope with pathogens. For the initial immune response, conserved pathogen-derived molecules, termed pathogen-associated molecular patterns (PAMPs), are detected at the plant cell surface by the plasma membrane (PM)-bound receptor-like kinases (RLKs) or receptor-like proteins (RLPs), termed pattern recognition receptors (PRRs) (Wu and Zhou, 2013; Tang et al., 2017). Well-studied PRRs include the Arabidopsis leucine-rich repeat receptor kinases FLAGELLIN SENSING 2 (FLS2) and EF-TU RECEPTOR (EFR), which recognize bacterial flagellin and EF-Tu, respectively (Zipfel et al., 2004; Zipfel et al., 2006). After perception of PAMPs by the PRRs, a series of defense responses are activated, such as a burst of reactive oxygen species (ROS), an increase of Ca2+ concentration, callose deposition, mitogen-activated protein kinase (MAPK) activation, and pathogenesis-related gene expression, termed PAMP-triggered immunity (PTI) (Jones and Dangl, 2006; Tang et al., 2017). The BRASSINOSTEROID RECEPTOR-ASSOCIATED KINASE 1 (BAK1) interacts with FLS2 or EFR and functions as a co-receptor in PTI (Chinchilla et al., 2007; Sun et al., 2013).
To aid in infection, pathogens suppress PTI by secreting virulence effectors into the intercellular space or the cytoplasm of host cells and cause effector-triggered susceptibility (ETS) (Jones and Dangl, 2006). In the second layer of innate immunity, termed effector-triggered immunity (ETI), pathogen effectors are directly or indirectly recognized by the host mainly via intracellular nucleotide-binding (NB) leucine-rich repeat (LRR) domain receptors (NLRs); recognition of effectors by NLR proteins leads to robust immunity that terminates pathogen growth (Cui et al., 2015; Duxbury et al., 2016).
The gram-negative plant bacterial pathogen Pseudomonas syringae pv. tomato (Pto) strain DC3000 causes bacterial speck disease in tomato (Solanum lycopersicum) and Arabidopsis thaliana and injects about 30 effectors via the type III secretion system into host cells to suppress plant innate immunity (Nomura et al., 2006; Velasquez et al., 2017). AvrPto and AvrPtoB are two well-analyzed, sequence-distinct effectors from Pto DC3000. In tomato, both AvrPto and AvrPtoB interact with the tomato protein Pto, a serine/threonine protein kinase, and elicit Prf-mediated programmed cell death (PCD) and immunity (Kim et al., 2002; Xiao et al., 2007). AvrPtoB also targets a host protein kinase, Fen, to disrupt plant immunity (Rosebrock et al., 2007). In Arabidopsis, AvrPto and AvrPtoB suppress basal defense and intercept multiple PAMPs-mediated signaling upstream of MAPKKK (de Torres et al., 2006; He et al., 2006). AvrPto and AvrPtoB also can induce endogenous proteolytic activity that degrades RESISTANCE TO PSEUDOMONAS SYRINGAE PV MACULICOLA 1 (RPM1)-INTERACTING PROTEIN 4 (RIN4) in the presence of Pto and Prf (Luo et al., 2009). In Arabidopsis and tomato, AvrPto suppresses cell wall-based defenses and AvrPtoB promotes the virulence of Pto DC3000 by suppressing PCD (Abramovitch et al., 2003; Abramovitch et al., 2006; Xiang et al., 2008). AvrPto and AvrPtoB target multiple RLKs involved in the perception of PAMPs in Arabidopsis. For instance, AvrPto and AvrPtoB target BAK1 to interfere with the bacterial-induced formation of the FLS2-BAK1 receptor–signaling complex (Shan et al., 2008; Cheng et al., 2011). AvrPto targets BOTRYTIS-INDUCED KINASE 1 (BIK1), a downstream component of the FLS2 pathway, to prevent the phosphorylation of BIK1 (Xiang et al., 2011). AvrPtoB contains an N-terminal region at 1–307 amino acids (aa), which is sufficient to elicit Pto/Prf-mediated PCD (Xiao et al., 2007), and a C-terminal U-box type E3 ubiquitin ligase domain. AvrPtoB targets and degrades FLS2, EFR, and the LysM domain receptor kinase CHITIN ELICITOR RECEPTOR KINASE 1 (CERK1) to block the initiation of PTI signaling in Arabidopsis (Gohre et al., 2008; Gimenez-Ibanez et al., 2009). AvrPtoB ubiquitinates and degrades NON-EXPRESSER OF PR GENES 1 (NPR1) to block salicylic acid-dependent transcriptional reprogramming (Chen et al., 2017).
Targeted protein transport and coordinated membrane dynamics play an important role in PTI (Gu et al., 2017). These trafficking events are mediated by a large group of regulatory proteins, such as the exocyst complex, which may function in the transport of defense molecules and contributes to their secretion (Robatzek, 2007; Nathalie and Bouhidel, 2014). EXO70 is one of the eight subunits of the exocyst complex. The Arabidopsis genome encodes 23 EXO70 protein family members, some of which are involved in plant immunity (Cvrckova and Zarsky, 2013). For instance, the exo70B1 loss-of-function mutant displays enhanced resistance to the powdery mildew pathogen Golovinomyces cichoracearum, the bacterial pathogen Pto DC3000, and the oomycete pathogen Hyaloperonospora arabidopsidis Noco2 (Zhao et al., 2015). EXO70B1 is also implicated in autophagy-related transport to the vacuole, and loss of function of EXO70B1 causes reduced numbers of internalized autophagosomes inside the vacuole (Kulich et al., 2013), along with ectopic hypersensitive responses (Kulich et al., 2013; Stegmann et al., 2013; Zhao et al., 2015). In addition, EXO70B1 positively regulates light- and drought-induced stomatal movement (Hong et al., 2016; Seo et al., 2016), and EXO70B1 is co-localized with EXO84B in broad bean (Vicia faba) guard cells (Hong et al., 2016). EXO70B2 and EXO70B1 show the highest sequence similarity, and both have been reported to be necessary for PTI and resistance to pathogens (Stegmann et al., 2012; Stegmann et al., 2013). Moreover, RIN4 recruits EXO70B1 to the plant cell PM (Sabol et al., 2017). The plant U-box-type ubiquitin ligase 22 (PUB22) ubiquitinates and degrades EXO70B2, which contributes to PAMP-triggered responses (Stegmann et al., 2012).
The activated immune responses in the exo70B1 mutant require the atypical NLR protein Toll/interleukin-1 receptor–nucleotide-binding sequence protein (TIR-NBS2; herein referred to as TN2) and CALCIUM-DEPENDENT PROTEIN KINASE 5 (CPK5). Overexpression of CPK5 also leads to TN2-dependent autoimmune responses. Moreover, TN2 directly interacts with EXO70B1, and the expression level of TN2 is up-regulated in mature exo70B1-3 mutants (Zhao et al., 2015; Liu et al., 2017). TN2 belongs to the TIR-NBS (TN) family, which has 21 members in Arabidopsis ecotype Col-0 (Nandety et al., 2013; Kato et al., 2014). However, how TN2 contributes to exo70B1-activated resistance is still unclear. We proposed that EXO70B1 is a guardee of TN2 or the TN2-related complex and NLR-mediated immunity is activated when EXO70B1 is degraded (Zhao et al., 2015; Liu et al., 2017). To date, only two effectors are reported to target the components of the exocyst complex. In potato (Solanum tuberosum), Phytophthora infestans manipulates plant immunity by targeting the exocyst component Sec5 with the RXLR effector AVR1 (Du et al., 2015). In rice (Oryza sativa), the effector Avr-Pii from the rice blast fungus Magnaporthe oryzae interacts with OsEXO70F2 and OsEXO70F3, and the rice NLR protein Pii recognizes Avr-Pii and triggers OsEXO70F3-dependent immunity (Fujisaki et al., 2015).
Here, we show that EXO70B1 contributes to plant immunity triggered by multiple PAMPs and AvrPtoB interacts with EXO70B1 via its N-terminal domain. AvrPtoB ubiquitinates and mediates the degradation of EXO70B1. Overexpression of AvrPtoB in Arabidopsis leads to autoimmune phenotypes, which are partially dependent on TN2. AvrPtoB contributes to the virulence of Pto DC3000 by overcoming EXO70B1-mediated resistance. AvrPtoB can rescue TN2- or TN2-TIR-triggered cell death in tobacco, which is suppressed by EXO70B1. Taken together, these findings indicate that EXO70B1 functions positively in PTI signaling and is a substrate of AvrPtoB.
Materials and Methods
Plant Materials and Growth Conditions
The Arabidopsis thaliana tn2-10 (SALK_204239C) mutant was obtained from the Arabidopsis Biological Resource Center. The exo70B1-3 mutant and the exo70B1-3 transgenic line containing a construct expressing EXO70B1-GFP under the control of the native EXO70B1 promoter were described previously (Zhao et al., 2015). Double mutants were obtained by genetic crosses and identified by polymerase chain reaction (PCR). Arabidopsis plants were grown in a growth room at 20°C to 22°C and ∼60% relative humidity with a 16/8-h day/night photoperiod for seed setting and a 9/15-h day/night photoperiod for phenotyping, with a light intensity of 7,000 to 8,000 lux. Nicotiana tabacum and Nicotiana benthamiana plants were grown under the same short-day conditions as Arabidopsis (Wu et al., 2015).
Vector Construction and Plant Transformation
The AvrPtoB sequence was amplified by PCR from Pto DC3000 genomic DNA and inserted into the pEASY vector using a pEASY Simple blunt Clonase Kit (TransGen Biotech). The AvrPtoBF479A mutant form was created using site-directed mutagenesis as described previously (Zhao et al., 2019). The AvrPtoB sequence was amplified with KpnI and SalI restriction sites and cloned into pSuper1300 with a 6 × Myc tag at the C-terminal to generate the 35S:AvrPtoB-Myc construct (Zhang et al., 2017). The Agrobacterium tumefaciens strain GV3101 carrying the construct was used to transform Arabidopsis plants by floral-dip transformation (Clough and Bent, 1998). The stably transformed plants carrying a single copy of the insert were identified in the T3 generation, and two independent stable transgenic lines AvrPtoB-Myc #2 and AvrPtoB-Myc #7 were chosen for further analysis.
ROS Assays
ROS assays were performed as described previously (Zhang et al., 2010). Leaf strips of 4-week-old plants were treated with 100 nM of flg22, 100 nM of elf18, or 0.1 mg/ml of chitin. Luminescence was detected with the GloMax 96 microplate luminometer (Promega).
Callose Deposition
Leaves from 4-week-old Arabidopsis plants were infiltrated with H2O, 20 μM of flg22, 20 μM of elf18, or 2 mg/ml of chitin in 10 mM of MgCl2 and removed 12 h after infiltration. Callose staining, image acquisition, and processing were carried out as described (Zhang et al., 2007).
Y2H Assay
The Matchmaker GAL4 Two-Hybrid System 3 (Clontech) was used for yeast two-hybrid (Y2H) assays. Bacterial effector sequences were amplified from Pto DC3000 genomic DNA and were cloned into the vector pGBKT-7. The full-length EXO70B1, EXO70B2, and EXO70A1 cDNA was cloned into pGADT-7. Different pairs of constructs were cotransformed into AH109, and the cells were grown on SD-Trp/-Leu medium. Three clones from SD-Trp/-Leu plates were transferred to SD-Trp/-Leu/-His/-Ade plates. Single clones were incubated in SD-Trp/-Leu liquid medium, and 10 μl of suspensions (diluted to OD600 = 0.5) of the overnight cultures was dropped onto different plates to test the interactions of the proteins.
RT-qPCR
Total RNA extraction and quantitative reverse transcription PCR (RT-qPCR) were performed as described previously (Shi et al., 2013).
Pull-Down Assay
The pull-down assay was performed as described previously (Yu et al., 2016). The AvrPtoB and AvrPtoB1–410 sequences were amplified from AvrPtoB-pEASY, and the AvrPtoBF479A sequence was amplified from AvrPtoBF479A-pEASY with EcoRI and NotI restriction sites and were cloned into the prokaryotic expression vector pGEX-4T-1. The positive clones were selected by sequencing. The GST-AvrPtoB, GST-AvrPtoBF479A, and GST-AvrPtoB1–410 fusion proteins were expressed in Escherichia coli as described previously (Xie et al., 1999). The EXO70B1 sequence was amplified from the EXO70B1-AD plasmid with KpnI and SalI restriction sites and cloned into pSuper1300 with a GFP tag at the C-terminal. EXO70B1-GFP was transiently expressed in N. benthamiana leaves by Agrobacterium tumefaciens-mediated infiltration at OD600 = 1.5, and samples were collected after 2 days. EXO70B1-GFP was extracted in native extraction buffer as described previously (Shi et al., 2013). Equimolar amounts of glutathione S-transferase (GST), GST-AvrPtoB, GST-AvrPtoBF479A, and GST-AvrPtoB1–410 were incubated with equal amounts of EXO70B1-GFP extracts at 4°C for 4 h. After incubation, the GST binding beads (Novagen) were added to incubate for another 2 h. Then, the mix was washed three times using GST washing buffer (40 mM of Na2HPO4, 8.5 mM of KH2PO4, 1.4 M of NaCl, 27 mM of KCl [pH 7.3], and 5 mM of dithiothreitol [DTT]). After the last centrifugation, the GST washing buffer was removed. Eighty microliters of GST washing buffer and 20 μl of 5 × SDS sample buffer were added, and the beads were boiled for 10 min. The proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and then detected using anti-GST (Abmart) and anti-GFP (Abmart) antibodies.
Co-IP Assay in N. Benthamiana
Coimmunoprecipitation (Co-IP) assays were performed as described previously, with minor modifications (Shi et al., 2013). Two days after infiltration, leaves were syringe infiltrated with or without 50 μM of MG132 for 3 h before freezing in liquid nitrogen. For anti-Myc immunoprecipitation, protein extracts were incubated with agarose-conjugated anti-Myc antibody (MBL) for 4 h at 4°C with gentle rotation. The agarose beads were washed and resuspended in 80 μl of PBS and 20 μl of 5 × SDS sample buffer and boiled for 10 min. Protein samples were detected with anti-GFP (Abmart) and anti-Myc (Abmart) immunoblot.
Cell-Free and In Vivo Protein Degradation Assay
The 35S:AvrPtoB-Myc construct or an empty vector control construct was transiently expressed in N. benthamiana leaves by Agrobacterium tumefaciens-mediated infiltration at OD600 = 1.0, and samples were collected after 2 days. Total leaf proteins were extracted in native extraction buffer. The EXO70B1 sequence was amplified from the EXO70B1-AD plasmid with BamHI and SalI restriction sites and were cloned into the prokaryotic expression vector pMAL-c2g-1, and the MBP-EXO70B1 fusion protein was expressed in E. coli. Maltose-binding protein (MBP) and the MBP-EXO70B1 recombinant protein were incubated with a cell-free crude extract for 1.5 and 3 h in the presence or absence of 50 μM of MG132 (AG Scientific). The reactions were terminated by adding 5 × SDS sample buffer. The protein degradation patterns were analyzed by immunoblotting with anti-MBP (New England Biolabs) antibody, and AvrPtoB-Myc protein was detected by anti-Myc (Abmart) antibody.
For the in vivo protein degradation assay, the EXO70B1 coding sequence was inserted into the PUC19 vector with a HA tag using SacI and SalI restriction sites. The 35S:EXO70B1-HA and 35S:GFP plasmids were cotransfected with the 35S:AvrPtoB-Myc or 35S:AvrPtoBF479A-Myc construct into protoplasts prepared from rosette leaves of wild-type Arabidopsis plants using the polyethylene glycol (PEG) method (Ryu et al., 2010). After incubation for 12 h, the protoplasts were treated with MG132 (50 μM) or dimethyl sulfoxide (DMSO) for 3 h. The total proteins were extracted in native extraction buffer, and then 5 × SDS sample buffer was added. The patterns of protein degradation were analyzed by immunoblot analysis with anti-HA (Abmart) and anti-GFP (Abmart) antibodies. AvrPtoB-Myc was detected by anti-Myc (Abmart) antibody.
In Vitro Ubiquitination
In vitro ubiquitination was carried out as described (Gimenez-Ibanez et al., 2009; Stegmann et al., 2012). In brief, each reaction of 60 μl of final volume contained ubiquitination buffer (50 mM of Tris–HCl [pH 7.5], 5 mM of MgCl2, 2 mM of DTT, 4 mM of ATP, and 1 × protease inhibitor cocktail) and 1 μg of MBP-EXO70B1 or MBP-EXO70B2 in the presence or absence of 15 μg of ubiquitin, 50 ng of human E1 (UBE1), 200 ng of Arabidopsis E2 His-UBC8, and 500 ng of GST-AvrPtoB or GST-AvrPtoBF479A. The reactions were incubated at 30°C for 3 h and stopped by adding 5 × SDS sample buffer and boiling for 10 min. Samples were separated by an 8% SDS-PAGE gel followed by detection of ubiquitinated substrate by immunoblotting using anti-MBP (New England Biolabs) and anti-GST (Abmart) antibodies.
Pathogen Infection Assays
Bacterial strains were grown overnight at 28°C in King’s B medium with appropriate antibiotics. Seedling infection assays were performed as described previously (Lu et al., 2010). In brief, seedlings were grown in 1/2 MS liquid medium for 10 days in a 12-well tissue culture plate. Bacteria were added at the concentration of OD600 = 0.2. Bacterial counting was performed 3 days after inoculation.
For powdery mildew infection, Golovinomyces cichoracearum strain UCSC1 was used to infect Arabidopsis plants at a high spore density (Adam and Somerville, 1996). The infected leaves were stained with Trypan Blue at 8 days postinfection to visualize hyphae and dead cells (Frye and Innes, 1998).
Gene Expression
For gene expression analysis, Arabidopsis leaves of Col-0 plants were hand-infiltrated with Pto DC3000 or the Pto ΔAvrPtoB mutant at OD600 = 0.2 in 10 mM of MgCl2. Total RNA extraction and RT-qPCR were performed as described above.
Hypersensitive Response Assay
Agrobacterium strain GV3101 was injected into 4-week-old N. benthamiana or N. tabacum leaves. The injected leaves were removed at 4 days postinoculation and photographed, then stained using Trypan Blue to examine cell death, and photographed.
Oligonucleotide Sequences
The primers used in this study are listed in Supplementary Table 1 .
Results
EXO70B1 Associates With AvrPtoB
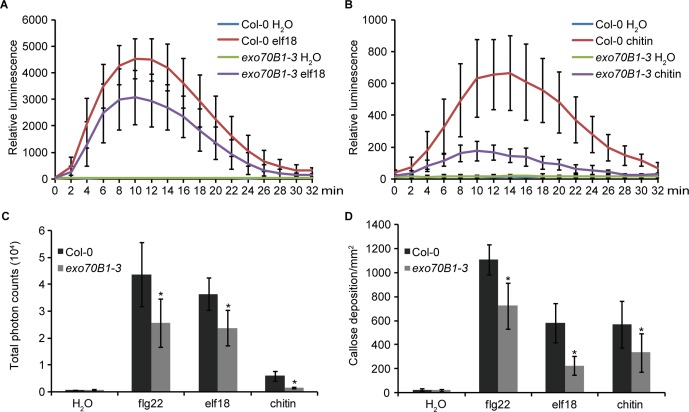
EXO70B1 is required for bacterial flagellin peptide 22 (flg22)-triggered immunity in plants (Stegmann et al., 2012). To further investigate the role of EXO70B1 in PTI, we examined elf18- and chitin-induced ROS production and callose deposition in the exo70B1-3 mutant. When treated with elf18 or chitin, the exo70B1-3 mutant showed reduced production of ROS than did the wild type Col-0 ( Figures 1A–C ). Similarly, flg22-, elf18-, and chitin-induced callose deposition was also significantly reduced in the exo70B1-3 mutant ( Figure 1D ), indicating that EXO70B1 functions positively in multiple PAMPs-triggered responses in Arabidopsis.
Figure 1.
EXO70B1 is required for responses triggered by multiple PAMPs. (A) and (B) Luminescence-based assays of ROS production in leaves of four-week-old Col-0 and exo70B1-3 Arabidopsis plants following treatment with 100 nM of elf18 (A) or 0.1 mg/ml of chitin (B). The means and standard deviations were determined using 12 independent biological samples. (C) Flg22-, elf18-, chitin-, and control (H2O)-induced ROS production over 30 min, presented as total photon counts in Col-0 and exo70B1-3 mutants. Data represent the mean and standard deviation of eight independent biological samples. Asterisk indicates a statistically significant difference between Col-0 and exo70B1-3 mutants (P < 0.05; Student’s t-test). (D) Flg22-, elf18-, chitin-, and control (H2O)-induced callose deposition in Col-0 and exo70B1-3 mutants. Data represent the mean and standard deviation of 27 independent biological samples. Asterisk indicates a statistically significant difference between Col-0 and exo70B1-3 mutants (P < 0.05; Student’s t-test). These experiments were repeated three times with similar results.
Because of the positive role of EXO70B1 in PTI, we hypothesized that EXO70B1 may be targeted by pathogen effectors. To validate this hypothesis, we performed a Y2H screen to identify bacterial effectors that interact with EXO70B1. We identified several effectors from Pto DC3000 that interact with EXO70B1, including HopH1 (Wei et al., 2007), HopK1 (He et al., 2004), HopO1-1 (Guo et al., 2005), HopY1 (Guttman et al., 2002), AvrPto (Kim et al., 2002; He et al., 2006), and AvrPtoBF479A, the E3 ligase-deficient mutant of AvrPtoB (Gohre et al., 2008; Gimenez-Ibanez et al., 2009) ( Table 1 ). According to our hypothesis, TN2 recognizes effectors by monitoring the status of EXO70B1, and the TN2-mediated resistance is activated when EXO70B1 disappeared (Zhao et al., 2015; Liu et al., 2017). Then, we focused on AvrPtoB as AvrPtoB is a well-characterized effector, which contains an E3 ligase domain in its C-terminal. EXO70B1 interacted with the E3 ligase-deficient mutant form of AvrPtoB only, indicating that EXO70B1 may be a substrate of wild-type AvrPtoB.
Table 1.
EXO70B1 interacting effectors from Pto DC3000 identified by a Y2H screening.
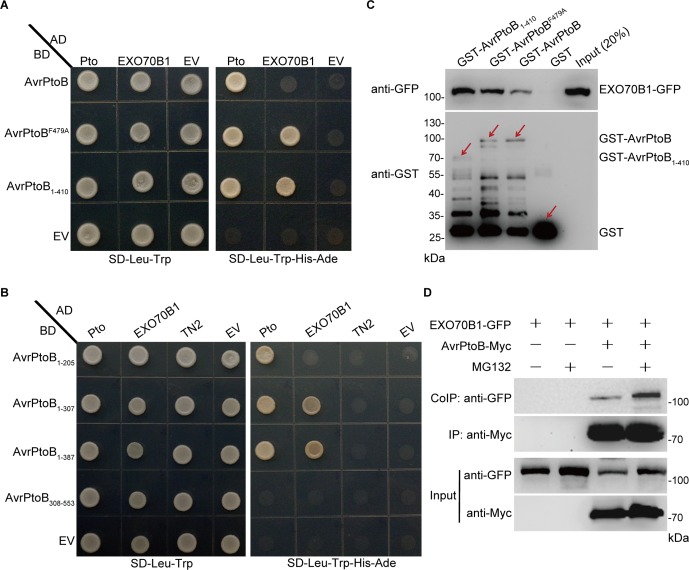
AvrPtoB contains two major domains and several specific domains in the N-terminus ( Supplementary Figure S1A ). AvrPtoB interacts with several target proteins through its N-terminal domains and degrades them via E3 ubiquitin ligase activity (Gohre et al., 2008; Gimenez-Ibanez et al., 2009; Chen et al., 2017). To test whether AvrPtoB interacts with EXO70B1 via its N-terminal domains, we introduced constructs encoding the N-terminal region lacking the E3 ligase domain of AvrPtoB (AvrPtoB1–410) as well as the E3-ligase-deficient mutant AvrPtoBF479A into yeast (Saccharomyces cerevisiae) along with constructs encoding EXO70B1. Both forms of AvrPtoB interacted with EXO70B1 ( Figure 2A ), indicating that AvrPtoB maybe associated with EXO70B1 via one of the protein interacting domains in its N-terminal region. We did not observe that the full-length AvrPtoB interacts with EXO70B1 in Y2H assay, maybe because of the AvrPtoB E3 ligase activity, which may affect protein interaction in yeast.
Figure 2.
EXO70B1 associates with AvrPtoB. (A) EXO70B1 interacts with AvrPtoB in a Y2H assay. The coding sequences of AvrPtoB, AvrPtoBF479A, and AvrPtoB1–410 were fused to the Gal4 DNA binding domain (BD), and the coding sequence of EXO70B1 was fused to the Gal4 transactivation domain (AD). As a positive control, tomato Pto was also fused to the AD. Different pairs of constructs were cotransformed into AH109. A 10-μl suspension (OD600 = 0.5) of each cotransformant was dropped onto synthetic dropout (SD) medium lacking Leu and Trp and SD medium lacking Ade, His, Leu, and Trp. Photographs were taken after 5 days of incubation. (B) Amino acids 1–307 of AvrPtoB are necessary for the interaction with EXO70B1 in a Y2H assay. Different truncated fragments of AvrPtoB (1–205, 1–307, 1–387, and 308–553) were fused to the BD, and EXO70B1 and TN2 were fused to the AD. Different pairs of constructs were cotransformed into AH109. Yeast cells containing the indicated plasmids were spotted onto SD-Leu-Trp and SD-Ade-His-Leu-Trp. Photographs were taken after 5 days of incubation. (C) EXO70B1 interacts with AvrPtoB in a GST-pull-down assay. E. coli-expressed GST, GST-AvrPtoB, GST-AvrPtoBF479A, and GST-AvrPtoB1–410 were incubated with plant-expressed EXO70B1-GFP in pull-down assays. After incubation for 4 h at 4°C, the beads were washed three times, and 5 × SDS sample buffer was added. Then, 20 μl of the samples was utilized to detect the precipitated EXO70B1-GFP by anti-GFP antibody, and 10 μl of the samples was used to detect the levels of GST, GST-AvrPtoB, GST-AvrPtoBF479A, and GST-AvrPtoB1–410 by anti-GST antibody. Arrows indicate the positions of the corresponding proteins. (D) EXO70B1 interacts with AvrPtoB in a Co-IP assay. EXO70B1-GFP was co-expressed with AvrPtoB-Myc in N. benthamiana leaves. After 48 h, leaves were treated with or without 50 μM of MG132 for 3 h. Total protein was extracted and subjected to immunoprecipitation of AvrPtoB by anti-Myc antibody. Proteins were analyzed in an immunoblot using anti-GFP and anti-Myc antibodies. These experiments were repeated three times with similar results.
Previously, we showed that EXO70B1 associates with TN2, and CPK5 phosphorylates EXO70B1 (Zhao et al., 2015; Liu et al., 2017). To examine whether AvrPtoB also interacts with TN2 or CPK5, we introduced constructs encoding AvrPtoB, AvrPtoB1–410, and AvrPtoBF479A into yeast along with TN2 or CPK5, with Pto as a positive control. None of these constructs interacted with TN2 or CPK5 ( Supplementary Figure S1B ). Taken together, these data indicated that AvrPtoB interacts with EXO70B1, but not TN2 or CPK5, via its N-terminal region, and the E3 ligase domain of AvrPtoB is dispensable for the interaction.
To identify the specific domains of AvrPtoB that interact with EXO70B1 and to further confirm that there is no interaction between AvrPtoB and TN2, we generated a series of AvrPtoB truncations, including AvrPtoB1–205, AvrPtoB1–307, AvrPtoB1–387, and AvrPtoB308–553 ( Supplementary Figure S1A ). Two truncated proteins, AvrPtoB1–307 and AvrPtoB1–387, interacted with EXO70B1, and none of the deletion mutants interacted with TN2 ( Figure 2B ). As a positive control, Pto did interact with AvrPtoB1–205, which indicated that amino acid residues 1–307 of AvrPtoB are sufficient for the interaction between AvrPtoB and EXO70B1.
To test biochemically whether AvrPtoB interacts with EXO70B1, we performed GST-pull-down assays. The 35S:EXO70B1-green fluorescent protein (GFP) vector was transiently expressed in Nicotiana benthamiana leaves by Agrobacterium tumefaciens-mediated infiltration. Total leaf proteins were extracted, and EXO70B1-GFP was detected by immunoblotting using anti-GFP antibody. Bacterially expressed GST, GST-AvrPtoB, GST-AvrPtoBF479A, and GST-AvrPtoB1–410 recombinant proteins were incubated with total proteins prepared from N. benthamiana leaves transiently expressing EXO70B1-GFP. GST protein was used as the negative control. GST-AvrPtoB, GST-AvrPtoBF479A, and GST-AvrPtoB1–410 could pull down EXO70B1-GFP, and the EXO70B1-GFP precipitated by GST-AvrPtoBF479A and GST-AvrPtoB1–410 is increased possibly because of the degradation activity of wild-type AvrPtoB ( Figure 2C ).
To further test the interaction between AvrPtoB and EXO70B1 in planta, we performed Co-IP assays by transiently expressing EXO70B1-GFP and AvrPtoB-Myc in N. benthamiana leaves, using EXO70B1-GFP alone as a negative control. AvrPtoB was immunoprecipitated using anti-Myc antibody, and EXO70B1 was detected with anti-GFP antibody only in the precipitate from the leaves that co-expressed both EXO70B1-GFP and AvrPtoB-Myc, but not from the negative control leaves that only expressed EXO70B1-GFP. Interestingly, the protein level of EXO70B1 and the association of EXO70B1 and AvrPtoB were markedly increased upon treatment with MG132, an inhibitor of the proteasome complex (Bogyo et al., 1997) ( Figure 2D ). These results indicated that EXO70B1 and AvrPtoB form a protein complex in N. benthamiana, and AvrPtoB may mediate the degradation of EXO70B1.
AvrPtoB Degrades EXO70B1 in a Proteasome-Dependent Manner
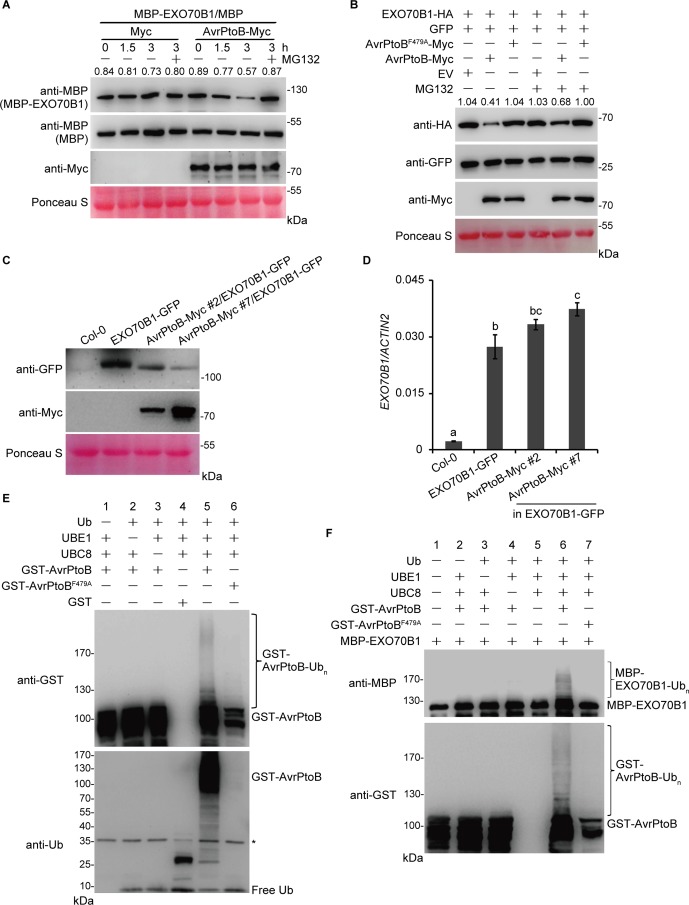
The interaction between EXO70B1 and AvrPtoB suggests that EXO70B1 may be a substrate of AvrPtoB and may be degraded by AvrPtoB. To investigate this possibility, we first conducted a cell-free degradation assay. The 35S:AvrPtoB-Myc or an empty vector control construct was transiently expressed in N. benthamiana leaves. Total leaf proteins were extracted, and AvrPtoB-Myc was identified by immunoblotting using anti-Myc antibody. Bacterially expressed MBP or MBP-EXO70B1 recombinant proteins were incubated with total proteins prepared from N. benthamiana leaves transiently expressing AvrPtoB-Myc or Myc with or without MG132 and were subjected to immunoblot analysis with anti-MBP antibody at the indicated time. After 1.5 h of incubation, the level of MBP-EXO70B1 was slightly reduced in the AvrPtoB-Myc cell-free extracts. After 3 h of incubation, the level of MBP-EXO70B1 was substantially reduced in the AvrPtoB-Myc cell-free extracts. By contrast, MBP-EXO70B1 remained stable in the Myc cell-free extracts and in the presence of MG132 ( Figure 3A ), indicating that MBP-EXO70B1 is degraded in a proteasome-dependent manner in the presence of AvrPtoB-Myc. The MBP protein, as a negative control, remained unaffected in both extracts.
Figure 3.
The degradation and ubiquitination of EXO70B1 by AvrPtoB. (A) Cell-free degradation assay of EXO70B1 by AvrPtoB. Bacterially expressed MBP or MBP-EXO70B1 was incubated with a cell-free crude extract prepared from N. benthamiana leaves transiently expressing AvrPtoB-Myc or Myc, in the presence or absence of 50 μM of MG132. The time-dependent changes of protein levels were monitored by immunoblotting with anti-MBP antibody. Numbers indicate the protein levels of MBP-EXO70B1 normalized to MBP using ImageJ software. The levels of Rubisco are shown as an equal loading control of cell-free extracts. (B) In vivo degradation assay of EXO70B1 by AvrPtoB in protoplasts. The 35S:EXO70B1-HA and 35S:GFP constructs were cotransfected with the 35S:Myc, 35S:AvrPtoB-Myc, or 35S:AvrPtoBF479A-Myc constructs into protoplasts freshly isolated from wild-type Arabidopsis leaf tissues using PEG-mediated transformation. After overnight transfection, protoplasts were incubated with or without 50 μM of MG132 for 3 h. Total proteins were extracted and examined by immunoblot analysis with anti-HA, anti-GFP, and anti-Myc antibodies. The levels of GFP show the transfection efficiency of protoplasts. Numbers indicate the protein levels of EXO70B1-HA normalized to GFP using ImageJ software. The levels of Rubisco are shown as an equal loading control. (C) EXO70B1-GFP and AvrPtoB-Myc levels in the indicated genotypes were determined by immunoblotting using anti-GFP and anti-Myc antibodies. Ponceau S staining of Rubisco is shown as a protein loading control. (D) RT-qPCR analysis of EXO70B1 expression. Total RNA was isolated from leaves of the 5-week-old Arabidopsis plants. The expression levels of EXO70B1 were normalized to the reference gene ACTIN2. Data represent the mean and standard deviation of three biological replicates. Three technical replicates for each biological sample were used. The lowercase letters indicate statistically significant differences (P < 0.05; one-way ANOVA). (E) In vitro self-ubiquitination assay of AvrPtoB. Recombinant GST-AvrPtoB was incubated in ubiquitination buffer in the presence or absence of ubiquitin, E1 (UBE1), or E2 (UBC8) at 30°C for 2 h. Recombinant GST-AvrPtoBF479A was used as a negative control. Reaction products were resolved using SDS-PAGE and subjected to immunoblot analysis with anti-GST and anti-Ub antibodies. Asterisk indicates the contaminating bands. (F) In vitro ubiquitination assay of EXO70B1 by AvrPtoB. Recombinant MBP-EXO70B1 was incubated in ubiquitination buffer in the presence or absence of ubiquitin, E1 (UBE1), E2 (UBC8), or E3 (AvrPtoB or AvrPtoBF479A). The reaction mixtures were subjected to immunoblot analysis with anti-MBP and anti-GST antibodies. Ubiquitinated MBP-EXO70B1 was detected by anti-MBP antibody. These experiments were repeated three times with similar results.
To further confirm that AvrPtoB mediates the degradation of EXO70B1, we next performed a protoplast transient expression assay. The EXO70B1-HA and GFP proteins were transiently co-expressed with AvrPtoB-Myc or AvrPtoBF479A-Myc in protoplasts prepared from Arabidopsis Col-0 leaves. After overnight transfection, protoplasts were incubated with or without MG132 for another 3 h. Total proteins were extracted and examined by immunoblotting analysis with anti-HA, anti-GFP, and anti-Myc antibodies. The level of EXO70B1-HA was decreased in the presence of AvrPtoB-Myc, but not AvrPtoBF479A-Myc ( Figure 3B ). The amount of EXO70B1-HA was increased in the presence of MG132 ( Figure 3B ), suggesting that the degradation of EXO70B1-HA by AvrPtoB-Myc in vivo in an E3-ligase-dependent manner is controlled by the 26S proteasome. The level of GFP, which was used to assess the efficiency of protoplast transfection, and Rubisco, which was shown as an equal protein loading control, remained unchanged regardless of the presence of MG132. Taken together, these data indicated that the degradation of EXO70B1 is regulated by AvrPtoB in a proteasome-dependent manner.
To further analyze whether AvrPtoB regulates the degradation of EXO70B1 in Arabidopsis, we introduced the 35S:AvrPtoB-Myc clone into the exo70B1-3 transgenic line, which contains a construct encoding functional EXO70B1-GFP in the exo70B1-3 mutant background (Zhao et al., 2015). Several independent stable transgenic lines were obtained, and the 35S:AvrPtoB-Myc #2 and #7 plants were chosen for further research. We extracted total proteins from 5-week-old Col-0, the exo70B1-3 transgenic line, and the 35S:AvrPtoB-Myc #2 and #7 plants and detected EXO70B1-GFP and AvrPtoB-Myc by immunoblotting analysis with anti-GFP and anti-Myc antibodies, respectively. The 35S:AvrPtoB-Myc #2 and #7 transgenic plants expressed AvrPtoB-Myc of the correct size, and the amount of EXO70B1-GFP was lower in the 35S:AvrPtoB-Myc transgenic plants compared with the controls ( Figure 3C ). We also tested the transcript levels of EXO70B1 and found that they were slightly increased in the 35S:AvrPtoB-Myc #2 and #7 plants ( Figure 3D ), indicating that AvrPtoB destabilizes EXO70B1 by post-translational regulation.
To investigate whether EXO70B1 is a substrate of AvrPtoB, we performed in vitro ubiquitination assays to analyze whether AvrPtoB can ubiquitinate EXO70B1. First, we used human E1 (UBE1) and Arabidopsis E2 (UBC8) to assess the in vitro E3 ligase activity of AvrPtoB. Recombinant full-length GST-AvrPtoB and GST-AvrPtoBF479A were incubated with ubiquitination buffer in the presence or absence of ubiquitin (Ub), UBE1, and UBC8 at 30°C for 2 h. The reaction mixture was analyzed by immunoblotting using anti-Ub and anti-GST antibodies. We observed GST-AvrPtoB E3 ligase activity as evidenced by high-molecular-mass smear ladders detected by both anti-Ub and anti-GST antibodies ( Figure 3E ), indicating that GST-AvrPtoB could self-ubiquitinate in our experimental conditions. In contrast, the E3-ligase-deficient mutation of AvrPtoB, GST-AvrPtoBF479A, did not have E3 ligase activity ( Figure 3E ).
Next, a bacterially expressed MBP-EXO70B1 recombinant protein was used for in vitro ubiquitination assays in the presence of all the other components required for the ubiquitination reaction. After 3 h of incubation, multiple ubiquitinated forms of MBP-EXO70B1 appeared in the presence of Ub, UBE1, UBC8, and GST-AvrPtoB. However, coincubation of MBP-EXO70B1 with GST-AvrPtoBF479A failed to produce any detectable high-molecular-mass smear ladders ( Figure 3F ), suggesting that the presence of an active E3 ligase domain is required for the ubiquitination of MBP-EXO70B1 by GST-AvrPtoB. Taken together, these data indicated that EXO70B1 is a substrate of AvrPtoB and is ubiquitinated by AvrPtoB.
Since EXO70B1 and EXO70B2 show the highest sequence similarity and both are required for PTI responses (Stegmann et al., 2012), we also analyzed whether AvrPtoB targets EXO70B2. With Pto as a positive control, we found that EXO70B2 also interacted with AvrPtoB ( Supplementary Figure S2A ). Next, with MBP-EXO70B1 as a positive control, no ubiquitination form of MBP-EXO70B2 was detected in the presence of all required ubiquitination components in our experimental condition ( Supplementary Figure S2B ). Taken together, these data indicated that EXO70B1, not EXO70B2, is a substrate of AvrPtoB.
Overexpression of AvrPtoB Leads to Stunted Phenotypes in Arabidopsis
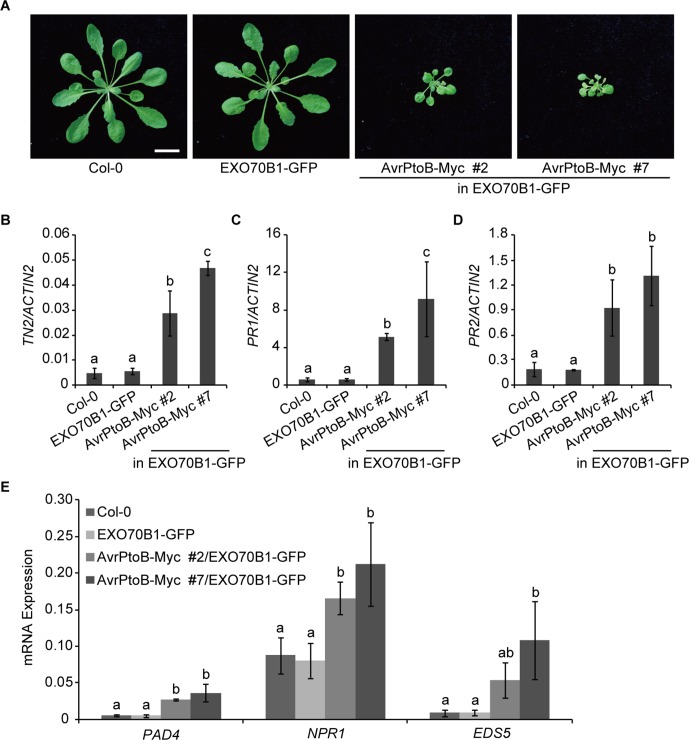
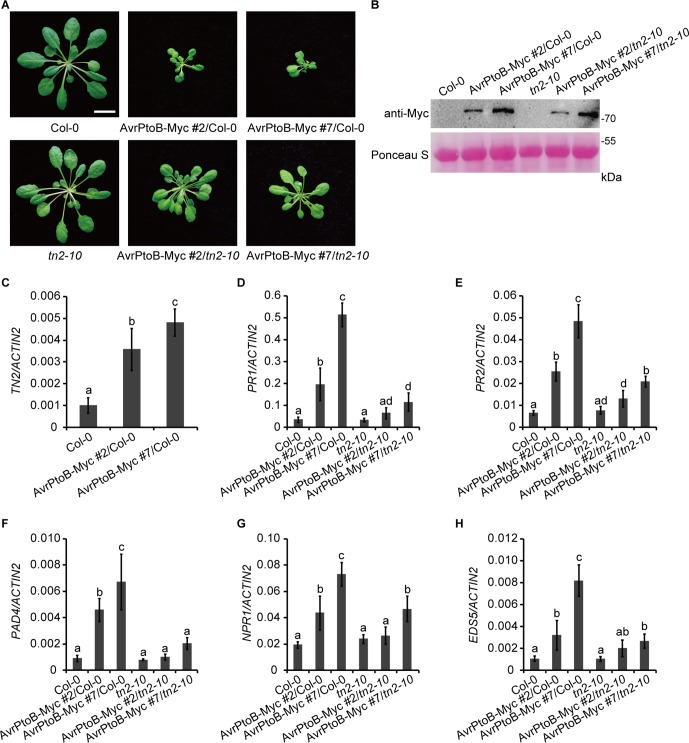
Further on, we analyzed the phenotypes of transgenic Arabidopsis plants that consistently express AvrPtoB. Compared with Col-0 and the exo70B1-3 transgenic line, 5-week-old 35S:AvrPtoB-Myc #2 and #7 transgenic plants displayed stunted phenotypes and growth defects ( Figure 4A ), which were similar to those in TN2 overexpressing plants (Li et al., 2009; Nandety et al., 2013). However, no obvious spontaneous cell death was observed in the 35S:AvrPtoB-Myc plants.
Figure 4.
Overexpression of AvrPtoB leads to stunted phenotypes in Arabidopsis. (A) Five-week-old plants of Col-0, the exo70B1-3 transgenic line (expressing EXO70B1-GFP in the exo70B1-3 mutant background), and two independent 35S:AvrPtoB-Myc transgenic Arabidopsis plants in the background of the exo70B1-3 transgenic line were photographed under short-day conditions. Bar = 1.0 cm. (B–E) RT-qPCR analysis of TN2 (B), PR1 (C), PR2 (D), and immune-related genes (E) expression. Total RNA was isolated from leaves of the 5-week-old Arabidopsis plants indicated in (A). The expression levels of indicated gene were normalized to the reference gene ACTIN2. Data represent the mean and standard deviation of three biological replicates. Three technical replicates for each biological sample were used. The lowercase letters indicate statistically significant differences (P < 0.05; one-way ANOVA). These experiments were repeated three times with similar results.
Next, we analyzed the transcript levels of TN2 in 5-week-old Col-0, the exo70B1-3 transgenic line, and the 35S:AvrPtoB-Myc #2 and #7 plants and found that the expression of TN2 was significantly up-regulated in the 35S:AvrPtoB-Myc #2 and #7 plants compared with the wild type and the exo70B1-3 transgenic line ( Figure 4B ). In exo70B1-3 mutant, the expression of PR1 and PR2 is constitutively up-regulated due to the activated TN2 (Zhao et al., 2015; Liu et al., 2017). Similarly, we found that the expression levels of PR1 and PR2 were also significantly up-regulated in the 35S:AvrPtoB-Myc #2 and #7 plants ( Figures 4C, D ). We further examined the expression of other immune-related genes, including PHYTOALEXIN DEFICIENT 4 (PAD4), NPR1, and ENHANCED DISEASE SUSCEPTIBILITY 5 (EDS5). The transcript levels of these genes were significantly up-regulated in the 35S:AvrPtoB-Myc #2 and #7 plants ( Figure 4E ). As shown in Figure 3C , AvrPtoB-Myc #7 accumulated much higher AvrPtoB-Myc protein levels and lower EXO70B1-GFP protein levels than did AvrPtoB-Myc #2, and the transcript levels of immune-related genes in AvrPtoB-Myc #7 were higher than those in AvrPtoB-Myc #2, which indicated that the stunted phenotypes and the up-regulation of immune-related genes in AvrPtoB-Myc plants are related to the low EXO70B1 protein levels. Taken together, these data indicated that overexpression of AvrPtoB leads to low accumulation of EXO70B1 and autoimmunity phenotypes, which may be caused by TN2-triggered plant immunity.
The Autoimmunity Phenotypes in the AvrPtoB Overexpression Lines Are Partially Dependent on TN2
To investigate whether there is a direct link between the phenotypes of 35S:AvrPtoB-Myc plants and the accumulation of TN2, we crossed the 35S:AvrPtoB-Myc #2 and #7 plants with the tn2-10 mutant, a T-DNA insertion mutant of TN2. The tn2-10 mutation fully suppressed the spontaneous cell death and powdery mildew resistance in the exo70B1-3 mutant, indicating that tn2-10 is a loss-of-function mutation ( Supplementary Figures S3A–C ). The expression level of TN2 was up-regulated in the exo70B1-3 mutants, which is consistent with our early reports (Zhao et al., 2015; Liu et al., 2017), and was not detected in tn2-10 mutants and exo70B1-3 tn2-10 double mutants ( Supplementary Figure S3D ). We obtained stable 35S:AvrPtoB-Myc transgenic plants in the Col-0 (35S:AvrPtoB-Myc #2/Col-0 and 35S:AvrPtoB-Myc #7/Col-0) and tn2-10 mutant (35S:AvrPtoB-Myc #2/tn2-10 and 35S:AvrPtoB-Myc #7/tn2-10) background by crossing. We observed the stunted phenotypes of the 35S:AvrPtoB-Myc #2 and #7 plants in the Col-0 background, which was indistinguishable from the phenotypes of the 35S:AvrPtoB-Myc #2 and #7 plants in the background of the exo70B1-3 transgenic line. In contrast, the stunted phenotypes were partially rescued by the tn2-10 mutation ( Figure 5A ). The levels of AvrPtoB-Myc were similar in the 35S:AvrPtoB-Myc #2 and #7 plants in the Col-0 and tn2-10 background ( Figure 5B ), indicating that the stunted phenotype caused by the expression of AvrPtoB-Myc is partially dependent on TN2.
Figure 5.
The stunted phenotypes of the AvrPtoB overexpression lines are partially dependent on TN2. (A) Five-week-old Col-0, tn2-10, and two independent 35S:AvrPtoB-Myc transgenic Arabidopsis plants (35S:AvrPtoB-Myc #2 and 35S:AvrPtoB-Myc #7) in the Col-0 and tn2-10 background were photographed under short-day conditions. Bar = 1.0 cm. (B) The AvrPtoB-Myc fusion protein was examined by immunoblotting. Ponceau S staining of Rubisco was shown as a protein loading control. (C–H) RT-qPCR analysis of TN2 (C), PR1 (D), PR2 (E), PAD4 (F), NPR1 (G), and EDS5 (H) expression. Total RNA was isolated from leaves of the indicated 5-week-old Arabidopsis plants. The expression levels of indicated gene were normalized to the reference gene ACTIN2. Data represent the mean and standard deviation of three biological replicates. Three technical replicates for each biological sample were used. The lowercase letters indicate statistically significant differences (P < 0.05; one-way ANOVA). These experiments were repeated three times with similar results.
Consistent with the phenotypes, the transcript levels of TN2 were up-regulated in 35S:AvrPtoB-Myc #2/Col-0 and 35S:AvrPtoB-Myc #7/Col-0 plants ( Figure 5C ), and the up-regulation of PR1, PR2, PAD4, NPR1, and EDS5 expression levels was partially suppressed by the tn2-10 mutation ( Figures 5D–H ). Taken together, these data indicated that overexpression of AvrPtoB in Arabidopsis causes a decrease of EXO70B1 and activates TN2-triggered immunity.
AvrPtoB Overcomes EXO70B1-Mediated Resistance
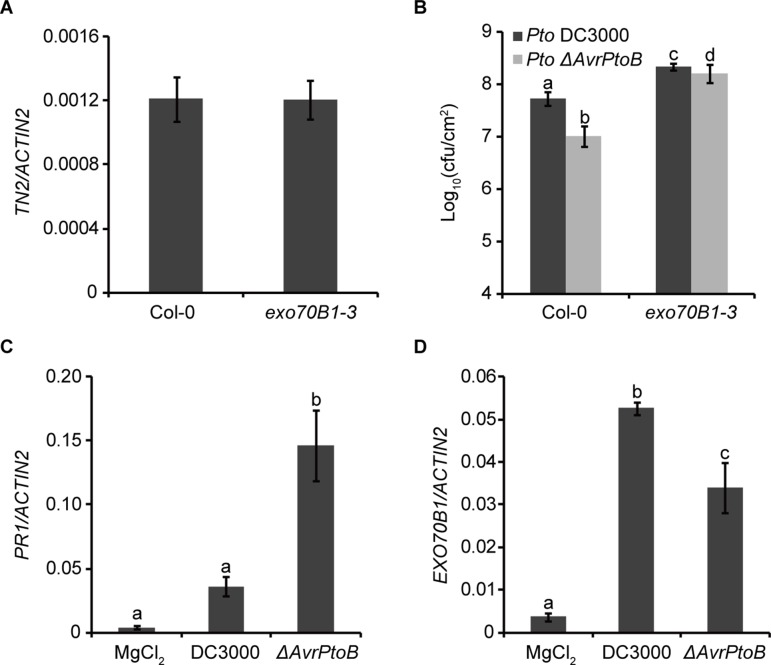
Previously, we showed that TN2 transcripts were up-regulated only in mature exo70B1-3 plants (Zhao et al., 2015), and the expression level of TN2 in 10-day-old exo70B1-3 seedlings was similar to that in Col-0 ( Figure 6A ). However, why the TN2 mRNA level did not change in young seedlings of exo70B1-3 mutant is still unclear. As the TN2 mRNA level did not change in young seedlings of exo70B1-3 mutant, the phenotypes of young exo70B1-3 seedlings would reflect the real role of EXO70B1 in resistance. Therefore, to assess whether degradation of EXO70B1 by AvrPtoB is important for the proliferation of Pto DC3000 in its host, we examined 10-day-old exo70B1-3 seedlings for responses to Pto DC3000 and the Pto DC3000 mutant strain lacking AvrPtoB (Pto ΔAvrPtoB). The virulence of the Pto ΔAvrPtoB strain was strongly reduced in Col-0 ( Figure 6B ), which is consistent with earlier reports (He et al., 2006; Gohre et al., 2008). In the exo70B1-3 mutant seedlings, bacterial numbers increased to similar extents for both Pto DC3000 strains, indicating that EXO70B1 contributes to bacterial resistance in Arabidopsis, and AvrPtoB enhances the virulence of Pto DC3000 by overcoming EXO70B1-mediated resistance.
Figure 6.
AvrPtoB overcomes EXO70B1-mediated resistance. (A) TN2 transcript levels were examined by RT-qPCR. Total RNA was isolated from the indicated 10-day-old seedlings. The mRNA expression level was normalized to the reference gene ACTIN2. Data represent the mean and standard deviation of three biological replicates. Three technical replicates for each biological sample were used. This experiment was repeated three times with similar results. (B) AvrPtoB contributes to the virulence of Pto DC3000, which is dependent on its ability to disable the function of EXO70B1. Ten-day-old Col-0 and exo70B1-3 seedlings were infected with Pto DC3000 and Pto ΔAvrPtoB. The bacterial growth assays were performed 3 days after infection. Results represent the mean and standard deviation of three independent experiments each consisting four independent biological samples. cfu, colony-forming units. The lowercase letters indicate statistically significant differences (P < 0.05; nested ANOVA). (C) and (D) Pto DC3000 enhances the expression of EXO70B1. Leaves of soil-grown wild-type Col-0 Arabidopsis plants were hand infiltrated with Pto DC3000 or the Pto ΔAvrPtoB mutant at 108 cfu/ml and harvested at 24-h postinoculation. The transcript levels of PR1 and EXO70B1 were measured by RT-qPCR as described above. Data represent the mean and standard deviation of three biological replicates. Three technical replicates for each biological sample were used. These experiments were repeated three times with similar results. The lowercase letters indicate statistically significant differences (P < 0.05; one-way ANOVA).
To gain more insight into the function of EXO70B1 in plant immunity, we analyzed the transcripts of EXO70B1 as well as PR1 in Col-0 plants after inoculation with Pto DC3000 and Pto ΔAvrPtoB. Similar to the recent report, the PR1 transcript level was lower in plants inoculated with Pto DC3000 than with Pto ΔAvrPtoB (Chen et al., 2017) ( Figure 6C ). The expression levels of EXO70B1 were up-regulated after inoculation with both strains, and the up-regulation was enhanced in the presence of AvrPtoB ( Figure 6D ).
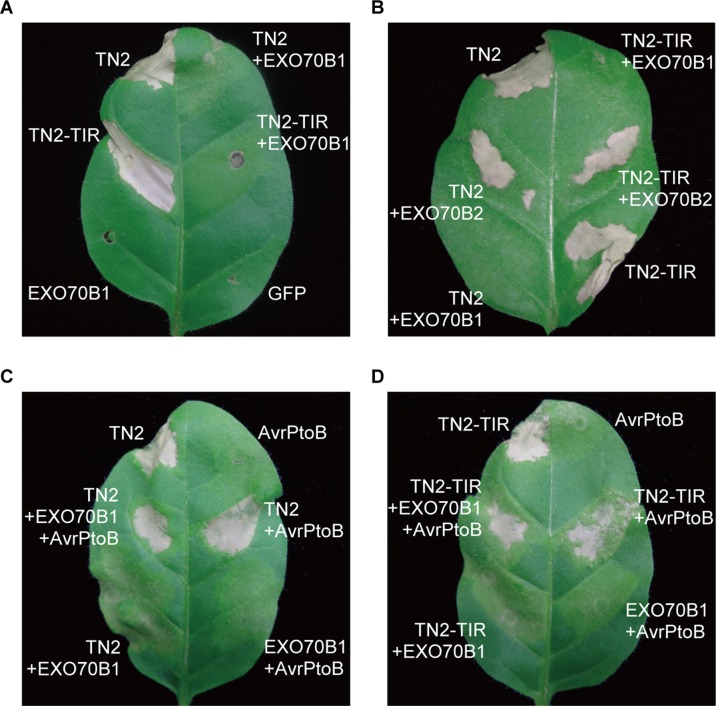
EXO70B1 Suppresses TN2-Triggered Cell Death in Tobacco
To further investigate the relationship between EXO70B1 and TN2, we expressed these proteins in N. tabacum. The expression of TN2 or TN2-TIR triggered a strong and rapid hypersensitive response (HR). This TN2- or TN2-TIR-triggered cell death was suppressed by co-expression with EXO70B1, not EXO70B2 ( Figures 7A, B ). TN2- or TN2-TIR-triggered cell death was also suppressed when we co-expressed TN2 or TN2-TIR with EXO70B1 using different Agrobacterium GV3101 concentrations, and proteins of the correct size were detected by immunoblotting analysis ( Supplementary Figure S4 ). These results indicated that EXO70B1 may maintain TN2 in an inactive status, and TN2 triggers cell death only in the absence of EXO70B1.
Figure 7.
EXO70B1 suppresses TN2- or TN2-TIR-triggered cell death in N. tabacum. (A) and (B) TN2- or TN2-TIR-triggered cell death was suppressed by EXO70B1, but not EXO70B2, in N. tabacum. (C) and (D) AvrPtoB rescues TN2- or TN2-TIR-triggered cell death, which is suppressed by EXO70B1, in N. tabacum. Agrobacterium GV3101 cells carrying different constructs were mixed prior to adding infiltration buffer. The concentration of each agrobacteria in the mix was brought to OD600 = 0.5 and infiltrated into 5-week-old N. tabacum leaves for transient expression. Cell death was observed 36 to 48 h later. Pictures were taken 4 days postinfiltration. These experiments were repeated three times with similar results.
To further study the role of AvrPtoB in TN2-triggered cell death, we co-expressed TN2 or TN2-TIR with EXO70B1 and AvrPtoB in N. tabacum. As shown in Figures 7C, D , AvrPtoB rescued TN2- or TN2-TIR-triggered cell death, which was suppressed by EXO70B1. Similar results were observed in N. benthamiana leaves ( Supplementary Figure S5 ). Taken together, these data indicated that EXO70B1 associates with TN2 and suppresses the activation of TN2, and TN2 is activated after the degradation of EXO70B1 by AvrPtoB in both N. tabacum and N. benthamiana.
Discussion
EXO70B1 is a component of the exocyst complex and belongs to the EXO70 protein family. In this study, we show that EXO70B1 is required for early immune signaling events, such as the ROS burst and callose deposition, triggered by flg22, elf18, and chitin ( Figure 1 ), which is consistent with the previous report on the requirement for EXO70B1 in flg22-triggered ROS burst (Stegmann et al., 2012). We also show that AvrPtoB, an effector of Pto DC3000, targets EXO70B1 to suppress plant immunity ( Figures 2 , 3 , and 6 ). Overexpression of AvrPtoB in Arabidopsis leads to autoimmunity, which is partially dependent of TN2 ( Figures 4 and 5 ). In addition, EXO70B1 inhibits the cell death triggered by TN2 ( Figure 7 and Supplementary Figure S4 ), suggesting that EXO70B1 associates with TN2 and may maintain TN2 in an inactive status.
In vesicle trafficking, exocytosis plays fundamental roles by maintaining membrane integrity and contributing to membrane remodeling in response to altered environmental conditions (Ding et al., 2011). RIN4 recruits EXO70B1 to the PM, and AvrRpt2 can release both RIN4 and EXO70B1 to the cytoplasm, indicating that the localization of EXO70B1 at PM may be required for plant immunity (Sabol et al., 2017). The exo70B1-3 mutant was compromised in early responses to multiple PAMPs, which is likely due to impaired PRRs signaling. Future studies will need to determine whether EXO70B1-associated exocyst complex contributes to the recycling of important components in PTI signaling. Despite the importance of the exocyst complex in vesicle trafficking and immune signaling, very few effectors are reported to target the components of the exocyst complex. In rice, the effector Avr-Pii from the rice blast fungus Magnaporthe oryzae interacts with OsEXO70F2 and OsEXO70F3, and OsEXO70F3, not EXO70F2, is specifically involved in Pii-dependent resistance (Fujisaki et al., 2015). Similarly, in this study, we show that both EXO70B1 and EXO70B2 interact with AvrPtoB ( Figure 2 and Supplementary Figure S2 ), but only EXO70B1 is ubiquitinated by AvrPtoB ( Figure 3 and Supplementary Figure S2 ). These results indicated that AvrPtoB associates with both EXO70B1 and EXO70B2 as EXO70B1 and EXO70B2 show the highest sequence similarity, but AvrPtoB specifically ubiquitinates and mediates the degradation of EXO70B1 to suppress EXO70B1-related plant immune responses.
The amino acid residues 206–307 of AvrPtoB, also known as the NPR1-interacting domain (NID) (Chen et al., 2017), are necessary for the interaction between AvrPtoB and EXO70B1 ( Figure 2D ). This domain is sufficient for the interaction between AvrPtoB and most of its targets, including NPR1, MKK2 (MAP kinase kinase 2), Bti9 (AvrPtoB tomato-interacting protein 9), and CERK1 (Gimenez-Ibanez et al., 2009; Zeng et al., 2012; Wei et al., 2015; Chen et al., 2017). Besides the NID, the Fen-interacting domain (FID) of AvrPtoB, encoded by amino acid residues 308–387, is another important region for the interaction between AvrPtoB and its targets, including Fen, FLS2, and BAK1 (Rosebrock et al., 2007; Gohre et al., 2008). Among the AvrPtoB-targeted proteins, only Pto binds to the Pto-interacting domain (PID) between amino acids 121 and 200, and to the FID domain of AvrPtoB, all other targets of AvrPtoB bind to the 206–387 amino acid region (NID and FID) of AvrPtoB. However, no interaction was observed between the full-length AvrPtoB and EXO70B1 in Y2H assay maybe because of the AvrPtoB E3 ligase activity. It is likely in the early report that the E3 ligase-deficient AvrPtoB mutants, not wild-type AvrPtoB, interacted with Fen in Y2H assay (Rosebrock et al., 2007). Further study is also needed to investigate why the N-terminal region of AvrPtoB can target so many different proteins, including protein kinases, transcriptional coactivator NPR1, and the subunit of exocyst complex EXO70B1.
AvrPtoB targets a number of regulators of plant immunity and contributes to the virulence of Pto DC3000. However, there is no evidence that any targets of AvrPtoB are monitored by NLR proteins and trigger ETI in Arabidopsis when targeted. Here, we showed that AvrPtoB ubiquitinates and degrades EXO70B1, and overexpression of AvrPtoB in Arabidopsis leads to autoimmunity, which is similar to the phenotype of TN2 overexpression plants reported previously (Li et al., 2009; Nandety et al., 2013), but not to the phenotype of exo70B1 mutants ( Figure 4 ). The most like reason is that the TN2 transcripts were only up-regulated in older exo70B1-3 plants (Zhao et al., 2015), which is different from TN2 overexpression plants. Therefore, AvrPtoB overexpression phenotypes maybe due to the consistent up-regulation of TN2. Moreover, the autoimmunity phenotypes of the 35S:AvrPtoB-Myc transgenic plants are partially dependent on TN2 ( Figure 5 ). The reason why the tn2-10 mutation does not fully rescue the stunted phenotypes of the 35S:AvrPtoB-Myc transgenic plants may be because there are many targets of AvrPtoB in Arabidopsis and AvrPtoB may trigger other forms of NLR-mediated resistance. For instance, AvrPtoB also targets BAK1, and loss-of-function bak1 mutants show a cell death phenotype (Kemmerling et al., 2007), which could be caused by activation of an undefined NLR protein.
Pto DC3000, a bacterial strain carrying AvrPtoB, did not show a reduced fitness postinoculation to Col-0 ( Figure 6B ). The most likely explanation for this is that AvrPtoB targets lots of proteins functioning in plant immunity, and modifications of other targets by AvrPtoB may contribute to its virulence, which overcome TN2-mediated resistance. For instance, NPR1, which contributes to TN2-mediated resistance (Zhao et al., 2015), is also targeted by AvrPtoB (Chen et al., 2017), and the modification of NPR1 by AvrPtoB could interfere with TN2-mediated resistance. In addition, the amount of AvrPtoB secreted into plant cells by Pto DC3000 is limited, which is insufficient to trigger TN2-mediated resistance. Furthermore, several other effectors can interact with EXO70B1, and those effectors may modify EXO70B1 as well. The modification of EXO70B1 by other effectors may influence the ubiquitination and degradation of EXO70B1 mediated by AvrPtoB. Therefore, TN2-mediated resistance is not activated.
Ectopic expression of the full-length or truncated TIR domain of some NLR proteins in the absence of pathogens can trigger an HR in N. tabacum (Swiderski et al., 2009; Bernoux et al., 2011). In this study, we show that transient expression of TN2 or TN2-TIR alone triggers strong and rapid cell death in N. tabacum, which is suppressed by co-expression of EXO70B1, not EXO70B2 ( Figure 7 ), indicating that EXO70B1 specifically suppresses the activation of TN2, and which is consistent with results on TN2 interaction with EXO70B1, but not with EXO70B2 ( Supplementary Figure S2A ). Additionally, TN2 and TN2-TIR accumulated in the presence of EXO70B1 ( Supplementary Figure S4 ), indicating that EXO70B1 associates with TN2 and may maintain TN2 in an inactive status, rather than to degrade TN2 by EXO70B1-associated autophagy activity.
The genome of the Arabidopsis ecotype Col-0 encodes about 50 atypical NLR proteins, including TIR, TIR-X, and TIR-NBS (Meyers et al., 2002), some of which have been reported to play roles in plant immunity. For instance, RESPONSE TO HOPBA1 (RBA1), a TIR-only protein, functions as a pathogen sensor and recognizes the HopBA1 effector directly (Nishimura et al., 2017). TN13 is a MODIFIER OF SNC1, 6 (MOS6) interaction partner required for resistance against Pto DC3000 lacking AvrPto and AvrPtoB (Roth et al., 2017). However, how truncated NLR proteins activate disease resistance is not clear. It is proposed that the truncated NLR proteins might not function alone and instead play a role in immunity with full-length NLR proteins (Nandety et al., 2013). Future studies are needed to determine whether overexpression of AvrPtoB triggered autoimmunity and TN2-activated resistance rely on full-length NLR proteins.
Conclusion
Our study demonstrated that EXO70B1, a subunit of the exocyst complex, plays an important role in immune responses triggered by multiple PAMPs. Pto DC3000 secretes AvrPtoB into plant cells to target EXO70B1, which may contribute to the trafficking of plasma membrane proteins involved in PAMP-triggered immunity. EXO70B1 associates with and maintains TN2, a truncated NLR protein, in an inactive status. Although the role of EXO70B1 in plant immunity and how EXO70B1 inhibits TN2 activation need further study, our results highlight that the Arabidopsis exocyst component is a target of effectors and may be guarded by a truncated NLR protein.
Data Availability
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT5G03540 (At EXO70A1), At5g58430 (At EXO70B1), At1g07000 (At EXO70B2), At1g17615 (At TN2), At2g14610 (At PR1), AT3g57260 (At PR2), At3g52430 (At PAD4), At1g64280 (At NPR1), At4g39030 (At EDS5), At3g18780 (At ACTIN2), AAF76306 (Sl Pto), AAO57459 (AvrPto), AY074795 (AvrPtoB), AAO54130 (Hop H1), AAO53599 (Hop K1), AF458392 (Hop O1-1), and AF458403 (Hop Y1).
Author Contributions
DT and WW conceived and initiated the research and designed the experiments; WW, NL, CG, and LR performed the experiments; WW, NL, CG, and DT analyzed the data; DT and WW wrote the paper.
Funding
The work was supported by grants from the National Natural Science Foundation of China (31761133017) and National Science Fund for Distinguished Young Scholars of China (31525019) to DT.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Jian-Min Zhou for Pto ΔAvrPtoB strain, Dr. Gitta Coaker for N. tabacum seeds, Dr. Shuhua Yang for pSuper1300 vectors, and Dr. Qi Xie for anti-Ub antibody.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01027/full#supplementary-material
References
- Abramovitch R. B., Janjusevic R., Stebbins C. E., Martin G. B. (2006). Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc. Natl. Acad. Sci. U.S.A. 103, 2851–2856. 10.1073/pnas.0507892103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramovitch R. B., Kim Y. J., Chen S., Dickman M. B., Martin G. B. (2003). Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 22, 60–69. 10.1093/emboj/cdg006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam L., Somerville S. C. (1996). Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana . Plant J. 9, 341–356. 10.1046/j.1365-313X.1996.09030341.x [DOI] [PubMed] [Google Scholar]
- Bernoux M., Ve T., Williams S., Warren C., Hatters D., Valkov E., et al. (2011). Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9, 200–211. 10.1016/j.chom.2011.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogyo M., Mcmaster J. S., Gaczynska M., Tortorella D., Goldberg A. L., Ploegh H. (1997). Covalent modification of the active site threonine of proteasomal β subunits and the Escherichia coli homolog HslV by a new class of inhibitors. Proc. Natl. Acad. Sci. U.S.A. 94, 6629–6634. 10.1073/pnas.94.13.6629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Chen J., Li M., Chang M., Xu K., Shang Z., et al. (2017). A bacterial type III effector targets the master regulator of salicylic acid signaling, NPR1, to subvert plant immunity. Cell Host Microbe 22, 777–788. 10.1016/j.chom.2017.10.019 [DOI] [PubMed] [Google Scholar]
- Cheng W., Munkvold K. R., Gao H., Mathieu J., Schwizer S., Wang S., et al. (2011). Structural analysis of Pseudomonas syringae AvrPtoB bound to host BAK1 reveals two similar kinase-interacting domains in a type III Effector. Cell Host Microbe 10, 616–626. 10.1016/j.chom.2011.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nurnberger T., Jones J. D., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. 10.1038/nature05999 [DOI] [PubMed] [Google Scholar]
- Clough S., Bent A. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cui H., Tsuda K., Parker J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. 10.1146/annurev-arplant-050213-040012 [DOI] [PubMed] [Google Scholar]
- Cvrckova F., Zarsky V. (2013). Old AIMs of the exocyst evidence for an ancestral association of exocyst subunits with autophagy-associated Atg8 proteins. Plant Signal Behav. 8, e27099. 10.4161/psb.27099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres M., Mansfield J. W., Grabov N., Brown I. R., Ammouneh H., Tsiamis G., et al. (2006). Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis . Plant J. 47, 368–382. 10.1111/j.1365-313X.2006.02798.x [DOI] [PubMed] [Google Scholar]
- Ding Z., Galvan-Ampudia C. S., Demarsy E., Langowski L., Kleine-Vehn J., Fan Y., et al. (2011). Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis . Nat. Cell Biol. 13, 447–452. 10.1038/ncb2208 [DOI] [PubMed] [Google Scholar]
- Du Y., Mpina M. H., Birch P. R., Bouwmeester K., Govers F. (2015). Phytophthora infestans RXLR effector AVR1 interacts with exocyst component Sec5 to manipulate plant immunity. Plant Physiol. 169, 1975–1990. 10.1104/pp.15.01169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxbury Z., Ma Y., Furzer O. J., Huh S. U., Cevik V., Jones J. D., et al. (2016). Pathogen perception by NLRs in plants and animals: parallel worlds. BioEssays 38, 769–781. 10.1002/bies.201600046 [DOI] [PubMed] [Google Scholar]
- Frye C. A., Innes R. W. (1998). An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10, 947–956. 10.1105/tpc.10.6.947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisaki K., Abe Y., Ito A., Saitoh H., Yoshida K., Kanzaki H., et al. (2015). Rice Exo70 interacts with a fungal effector, AVR-Pii, and is required for AVR-Pii-triggered immunity. Plant J. 83, 875–887. 10.1111/tpj.12934 [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S., Hann D. R., Ntoukakis V., Petutschnig E., Lipka V., Rathjen J. P. (2009). AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19, 423–429. 10.1016/j.cub.2009.01.054 [DOI] [PubMed] [Google Scholar]
- Gohre V., Spallek T., Haweker H., Mersmann S., Mentzel T., Boller T., et al. (2008). Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18, 1824–1832. 10.1016/j.cub.2008.10.063 [DOI] [PubMed] [Google Scholar]
- Gu Y., Zavaliev R., Dong X. (2017). Membrane trafficking in plant immunity. Mol. Plant 10, 1026–1034. 10.1016/j.molp.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Chancey S. T., Tian F., Ge Z., Jamir Y., Alfano J. R. (2005). Pseudomonas syringae type III chaperones ShcO1, ShcS1, and ShcS2 facilitate translocation of their cognate effectors and can substitute for each other in the secretion of HopO1-1. J. Bacteriol. 187, 4257–4269. 10.1128/JB.187.12.4257-4269.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman D. S., Vinatzer B. A., Sarkar S. F., Ranall M. V., Kettler G., Greenberg J. T. (2002). A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae . Science 295, 1722–1726. 10.1126/science.295.5560.1722 [DOI] [PubMed] [Google Scholar]
- He P., Chintamanani S., Chen Z., Zhu L., Kunkel B. N., Alfano J. R., et al. (2004). Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J. 37, 589–602. 10.1111/j.1365-313X.2003.01986.x [DOI] [PubMed] [Google Scholar]
- He P., Shan L., Lin N. C., Martin G. B., Kemmerling B., Nurnberger T., et al. (2006). Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125, 563–575. 10.1016/j.cell.2006.02.047 [DOI] [PubMed] [Google Scholar]
- Hong D., Jeon B. W., Kim S. Y., Hwang J. U., Lee Y. (2016). The ROP2-RIC7 pathway negatively regulates light-induced stomatal opening by inhibiting exocyst subunit Exo70B1 in Arabidopsis . New Phytol. 209, 624–635. 10.1111/nph.13625 [DOI] [PubMed] [Google Scholar]
- Jones J. D., Dangl J. L. (2006). The plant immune system. Nature 444, 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kato H., Saito T., Ito H., Komeda Y., Kato A. (2014). Overexpression of the TIR-X gene results in a dwarf phenotype and activation of defense-related gene expression in Arabidopsis thaliana . J. Plant Physiol. 171, 382–388. 10.1016/j.jplph.2013.12.002 [DOI] [PubMed] [Google Scholar]
- Kemmerling B., Schwedt A., Rodriguez P., Mazzotta S., Frank M., Qamar S. A., et al. (2007). The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr. Biol. 17, 1116–1122. 10.1016/j.cub.2007.05.046 [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Lin N. C., Martin G. B. (2002). Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109, 589–598. 10.1016/S0092-8674(02)00743-2 [DOI] [PubMed] [Google Scholar]
- Kulich I., Pecenkova T., Sekeres J., Smetana O., Fendrych M., Foissner I., et al. (2013). Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic 14, 1155–1165. 10.1111/tra.12101 [DOI] [PubMed] [Google Scholar]
- Li Y., Pennington B., Hua J. (2009). Multiple R-like genes are negatively regulated by BON1 and BON3 in Arabidopsis . Mol. Plant-Microbe Interact. 22, 840–848. 10.1094/MPMI-22-7-0840 [DOI] [PubMed] [Google Scholar]
- Liu N., Hake K., Wang W., Zhao T., Romeis T., Tang D. (2017). Calcium-dependent protein kinase5 associates with the truncated NLR protein TIR-NBS2 to contribute to exo70B1-mediated immunity. Plant Cell 29, 746–759. 10.1105/tpc.16.00822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Wu S., Gao X., Zhang Y., Shan L., He P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 496–501. 10.1073/pnas.0909705107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Caldwell K. S., Wroblewski T., Wright M. E., Michelmore R. W. (2009). Proteolysis of a negative regulator of innate immunity is dependent on resistance genes in tomato and Nicotiana benthamiana and induced by multiple bacterial effectors. Plant Cell 21, 2458–2472. 10.1105/tpc.107.056044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B. C., Morgante M., Michelmore R. W. (2002). TIR-X and TIR-NBS proteins: two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J. 32, 77–92. 10.1046/j.1365-313X.2002.01404.x [DOI] [PubMed] [Google Scholar]
- Nandety R. S., Caplan J. L., Cavanaugh K., Perroud B., Wroblewski T., Michelmore R. W., et al. (2013). The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol. 162, 1459–1472. 10.1104/pp.113.219162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathalie L.-C., Bouhidel K. (2014). Plasma membrane protein trafficking in plant–microbe interactions: a plant cell point of view. Front. Plant Sci. 5, 735. 10.3389/fpls.2014.00735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M. T., Anderson R. G., Cherkis K. A., Law T. F., Liu Q. L., Machius M., et al. (2017). TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis . Proc. Natl. Acad. Sci. U.S.A. 114, e2053–e2062. 10.1073/pnas.1620973114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., Debroy S., Lee Y. H., Pumplin N., Jones J., He S. Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313, 220–223. 10.1126/science.1129523 [DOI] [PubMed] [Google Scholar]
- Robatzek S. (2007). Vesicle trafficking in plant immune responses. Cell Microbiol. 9, 1–8. 10.1111/j.1462-5822.2006.00829.x [DOI] [PubMed] [Google Scholar]
- Rosebrock T. R., Zeng L., Brady J. J., Abramovitch R. B., Xiao F., Martin G. B. (2007). A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448, 370–374. 10.1038/nature05966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C., Ludke D., Klenke M., Quathamer A., Valerius O., Braus G. H., et al. (2017). The truncated NLR protein TIR-NBS13 is a MOS6/IMPORTIN-alpha3 interaction partner required for plant immunity. Plant J. 92, 808–821. 10.1111/tpj.13717 [DOI] [PubMed] [Google Scholar]
- Ryu M. Y., Cho S. K., Kim W. T. (2010). The Arabidopsis C3H2C3-type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid-dependent response to drought stress. Plant Physiol. 154, 1983–1997. 10.1104/pp.110.164749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabol P., Kulich I., Zarsky V. (2017). RIN4 recruits the exocyst subunit EXO70B1 to the plasma membrane. J. Exp. Bot. 68, 3253–3265. 10.1093/jxb/erx007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo D. H., Ahn M. Y., Park K. Y., Kim E. Y., Kim W. T. (2016). The N-terminal UND motif of the Arabidopsis U-Box E3 ligase PUB18 is critical for the negative regulation of ABA-mediated stomatal movement and determines its ubiquitination specificity for exocyst subunit Exo70B1. Plant Cell 28, 2952–2973. 10.1105/tpc.16.00347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L., He P., Li J., Heese A., Peck S. C., Nurnberger T., et al. (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor–signaling complexes and impede plant immunity. Cell Host Microbe 4, 17–27. 10.1016/j.chom.2008.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Shen Q., Qi Y., Yan H., Nie H., Chen Y., et al. (2013). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis . Plant Cell 25, 1143–1157. 10.1105/tpc.112.107904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M., Anderson R. G., Ichimura K., Pecenkova T., Reuter P., Zarsky V., et al. (2012). The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis . Plant Cell 24, 4703–4716. 10.1105/tpc.112.104463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M., Anderson R. G., Westphal L., Rosahl S., Mcdowell J. M., Trujillo M. (2013). The exocyst subunit Exo70B1 is involved in the immune response of Arabidopsis thaliana to different pathogens and cell death. Plant Signal Behav. 8, e27421. 10.4161/psb.27421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li L., Macho A. P., Han Z., Hu Z., Zipfel C., et al. (2013). Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342, 624–628. 10.1126/science.1243825 [DOI] [PubMed] [Google Scholar]
- Swiderski M. R., Birker D., Jones J. (2009). The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol. Plant-Microbe Interact. 20, 156–165. 10.1094/MPMI-22-2-0157 [DOI] [PubMed] [Google Scholar]
- Tang D., Wang G., Zhou J. M. (2017). Receptor kinases in plant–pathogen interactions: more than pattern recognition. Plant Cell 29, 618–637. 10.1105/tpc.16.00891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez A. C., Oney M., Huot B., Xu S., He S. Y. (2017). Diverse mechanisms of resistance to Pseudomonas syringae in a thousand natural accessions of Arabidopsis thaliana . New Phytol. 214, 1673–1687. 10.1111/nph.14517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C. F., Kvitko B. H., Shimizu R., Crabill E., Alfano J. R., Lin N. C., et al. (2007). A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana . Plant J. 51, 32–46. 10.1111/j.1365-313X.2007.03126.x [DOI] [PubMed] [Google Scholar]
- Wei H. L., Chakravarthy S., Mathieu J., Helmann T. C., Stodghill P., Swingle B., et al. (2015). Pseudomonas syringae pv. tomato DC3000 type III secretion effector polymutants reveal an interplay between HopAD1 and AvrPtoB. Cell Host Microbe 17, 752–762. 10.1016/j.chom.2015.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Liu S., Zhao Y., Wang W., Kong Z., Tang D. (2015). Enhanced disease resistance4 associates with clathrin heavy chain2 and modulates plant immunity by regulating relocation of EDR1 in Arabidopsis . Plant Cell 27, 857–873. 10.1105/tpc.114.134668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhou J. M. (2013). Receptor-like kinases in plant innate immunity. J. Integr. Plant Biol. 55, 1271–1286. 10.1111/jipb.12123 [DOI] [PubMed] [Google Scholar]
- Xiang T., Zong N., Zhang J., Chen J., Chen M., Zhou J. (2011). BAK1 is not a target of the Pseudomonas syringae effector AvrPto. Mol. Plant-Microbe Interact. 24, 100–107. 10.1094/MPMI-04-10-0096 [DOI] [PubMed] [Google Scholar]
- Xiang T., Zong N., Zou Y., Wu Y., Zhang J., Xing W., et al. (2008). Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18, 74–80. 10.1016/j.cub.2007.12.020 [DOI] [PubMed] [Google Scholar]
- Xiao F., He P., Abramovitch R. B., Dawson J. E., Nicholson L. K., Sheen J., et al. (2007). The N-terminal region of Pseudomonas type III effector AvrPtoB elicits Pto-dependent immunity and has two distinct virulence determinants. Plant J. 52, 595–614. 10.1111/j.1365-313X.2007.03259.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Sanz-Burgos A. P., Guo H. S., Garcia J. A., Gutierrez C. (1999). Grab proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol. Biol. 39, 647–656. 10.1023/A:1006138221874 [DOI] [PubMed] [Google Scholar]
- Yu F., Lou L., Tian M., Li Q., Ding Y., Cao X., et al. (2016). ESCRT-I component VPS23A affects ABA signaling by recognizing ABA receptors for endosomal degradation. Mol. Plant 9, 1570–1582. 10.1016/j.molp.2016.11.002 [DOI] [PubMed] [Google Scholar]
- Zeng L., Velasquez A. C., Munkvold K. R., Zhang J., Martin G. B. (2012). A tomato LysM receptor-like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J. 69, 92–103. 10.1111/j.1365-313X.2011.04773.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Li W., Xiang T., Liu Z., Laluk K., Ding X., et al. , , et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7, 290–301. 10.1016/j.chom.2010.03.007 [DOI] [PubMed] [Google Scholar]
- Zhang J., Shao F., Li Y., Cui H., Chen L., Li H., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1, 175–185. 10.1016/j.chom.2007.03.006 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang Y., Liu J., Ding Y., Wang S., Zhang X., et al. (2017). Temperature-dependent autoimmunity mediated by chs1 requires its neighboring TNL gene SOC3 . New Phytol. 213, 1330–1345. 10.1111/nph.14216 [DOI] [PubMed] [Google Scholar]
- Zhao T., Rui L., Li J., Nishimura M. T., Vogel J. P., Liu N., et al. (2015). A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet. 11, e1004945. 10.1371/journal.pgen.1004945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wu G., Shi H., Tang D. (2019). Receptor-like kinase 902 associates with and phosphorylates brassinosteroid-signaling KINASE1 to regulate plant immunity. Mol. Plant 12, 59–70. 10.1016/j.molp.2018.10.008 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J. D., Boller T., et al. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760. 10.1016/j.cell.2006.03.037 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E. J., Jones J. D., Felix G., et al. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767. 10.1038/nature02485 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT5G03540 (At EXO70A1), At5g58430 (At EXO70B1), At1g07000 (At EXO70B2), At1g17615 (At TN2), At2g14610 (At PR1), AT3g57260 (At PR2), At3g52430 (At PAD4), At1g64280 (At NPR1), At4g39030 (At EDS5), At3g18780 (At ACTIN2), AAF76306 (Sl Pto), AAO57459 (AvrPto), AY074795 (AvrPtoB), AAO54130 (Hop H1), AAO53599 (Hop K1), AF458392 (Hop O1-1), and AF458403 (Hop Y1).