Abstract
Understanding how the blood transcriptome of human intracerebral hemorrhage (ICH) differs from ischemic stroke (IS) and matched controls (CTRL) will improve understanding of immune and coagulation pathways in both disorders. This study examined RNA from 99 human whole-blood samples using GeneChip® HTA 2.0 arrays to assess differentially expressed transcripts of alternatively spliced genes between ICH, IS and CTRL. We used a mixed regression model with FDR-corrected p(Dx) < 0.2 and p < 0.005 and |FC| > 1.2 for individual comparisons. For time-dependent analyses, subjects were divided into four time-points: 0(CTRL), <24 h, 24–48 h, >48 h; 489 transcripts were differentially expressed between ICH and CTRL, and 63 between IS and CTRL. ICH had differentially expressed T-cell receptor and CD36 genes, and iNOS, TLR, macrophage, and T-helper pathways. IS had more non-coding RNA. ICH and IS both had angiogenesis, CTLA4 in T lymphocytes, CD28 in T helper cells, NFAT regulation of immune response, and glucocorticoid receptor signaling pathways. Self-organizing maps revealed 4357 transcripts changing expression over time in ICH, and 1136 in IS. Understanding ICH and IS transcriptomes will be useful for biomarker development, treatment and prevention strategies, and for evaluating how well animal models recapitulate human ICH and IS.
Keywords: Alternative splicing, angiogenesis, intracerebral hemorrhage, ischemic stroke, T-cell receptors
Introduction
Intracerebral hemorrhages (ICHs) account for about 10%–15% of human strokes.1 ICH is a severe type of stroke,2 with worse functional outcomes and higher mortality rates compared to ischemic stroke (IS).1,3 Stroke diagnosis is based on patient history, neurological exam, and brain imaging. Differentiating ICH from IS can be challenging when imaging is not readily available. However, ruling out ICH is essential for early thrombolytic or endovascular therapy for IS to be initiated. Thus, understanding the specific pathways involved in the response to the acute brain injury caused by ICH and IS is essential for identifying potential diagnostic biomarkers, and novel targets for treatment and prevention.
Peripheral blood cells play major roles in clotting, atherosclerosis, and the acute brain injury caused by ICH and IS.4–9 Leukocytes interact with blood clots, platelets, atherosclerotic plaque and, through adhesion molecules, with endothelial cells and produce chemokines, cytokines and hormones.10,11 Peripheral leukocytes infiltrate injured brain and influence outcome.10–13 Though the peripheral immune response has been studied extensively in human IS,11,14–17 little is known about gene expression changes following ICH.16,18 In addition, little is known about the alternatively spliced transcriptome of peripheral blood cells following ICH and IS.14,16,18 Alternative splicing is the process by which a single gene produces multiple RNA transcripts and protein isoforms with different but related functions. It is one of the main means of regulating immune and clotting pathways and has been implicated in many diseases.19–21 Thus, we investigated peripheral blood at transcript-isoform level resolution following ICH and IS, and the changes that occur over time.
Our study reveals ICH has a different transcriptome architecture from IS and CTRL in the coding and non-coding transcriptome, involving differences in immune cell genes including T-cell receptors (TCRs) in ICH only, and in clotting, angiogenesis/vasculogenesis, macrophage, T-cell and cell death pathways in ICH and IS. The molecular differentiation of ICH and IS may be useful in biomarker development, understanding biological processes to identify novel targets for treatment or prevention, and for comparing to animal models of ICH and IS to determine which best recapitulate the immune response to ICH and IS in humans.
Materials and methods
Study subjects
Ninety-nine male (M) and female (F) patients with ICH (n = 33, 24 M/9 F), acute IS (n = 33, 24 M/9 F), and vascular risk factor (VRF) – matched control subjects (CTRL, n = 33, 24 M/9 F) were recruited between 2005 and 2013 from Universities of California at Davis and San Francisco, and at the University of Alberta, Canada. The protocol was approved by the UC Davis and UC San Francisco Institutional Review Boards and the University of Alberta Health Research Ethics Board and adheres to all federal and state regulations related to the protection of human research subjects, including The Common Rule, the principles of The Belmont Report, and Institutional Policies and procedures. Written informed consent was obtained from all participants or their proxy. ICH or IS diagnoses, made by board-certified neurologists, were confirmed by CT or MRI imaging.22 The 33 IS subjects were represented by 11 cardioembolic, 11 large-vessel, and 11 lacunar causes of IS. Five out of the 33 IS patients were treated with rtPA (recombinant tissue plasminogen activator) prior to the blood draw. CTRLs included subjects matched for VRFs and included stroke mimics, such as migraine and simple seizures. Samples were matched for age, race, sex, and VRFs, including hypertension, diabetes mellitus, hyperlipidemia, and smoking status. Exclusion criteria included previous stroke (for control subjects) and IS with hemorrhagic transformation.
Blood collection, RNA isolation
Blood collection in Paxgene tubes and RNA isolation were performed as previously described.16 There was a single blood draw per subject. Time after symptom onset for ICH and IS varied from 4.5 to 124.3 h (Supplementary Table 1). CTRL subjects were designated as time zero. Blood draw time was included as a covariate in statistical analyses.
Arrays and processing
The GeneChip® Human Transcriptome Array (HTA) 2.0 (Affymetrix, Santa Clara, CA) was used with details of the processing and quantification of expression provided in the Supplementary Methods.
Pairwise group comparisons
A mixed regression model was used for transcript-level analyses: Diagnosis (ICH, IS, CTRL, fixed effect), sex, age, time-since-event, and technical variation (scan batch, random effect). REML variance estimate suitable for mixed models was used.23 Transcripts with FDR-corrected p(Diagnosis) < 0.2, which also had p < 0.005 and |FC| > 1.2 for individual comparisons (ICH vs. CTRL, IS vs. CTRL, or ICH vs. IS), were considered significant. The Fisher’s least significant difference was used for individual contrasts.24
Time-dependent changes in transcript expression
For time-dependent analyses, VRF-matched controls (n = 33) were considered as time-point 0 (TP0), and ICH and IS subjects were binned separately for time since event into three time-points: < 24 h (TP1), 24–48 h (TP2) and >48 h (TP3). Mixed regression model on transcript-level data was used: diagnosis, sex, age, time-point, and an interaction of diagnosis × time-point. p < 0.005 and |FC| > 1.2 on the individual contrasts from the interaction term were considered significant.
Self-organizing maps analyses
To identify similar temporal profiles of expression following ICH and IS over the four time-points, we performed self-organizing map (SOM)25 and Learning and Neighborhood functions, as implemented in Partek Genomics Suite (Supplementary Methods).
Prediction of activation/inhibition of pathways and upstream transcriptional regulators
Pathway enrichment analysis was performed in ingenuity pathway analysis (IPA, Ingenuity Systems®).26 IPA was used to predict whether pathways were activated or inhibited and to identify potential upstream transcriptional regulators (Upstream Transcriptional Regulators Analysis, IPA, Supplementary Methods).
Results
Demographic and clinical characteristics
Demographic and clinical characteristics are in Supplementary Table 1. There was no significant difference in VRFs. The age of the subjects (years ± SD) was 62 ± 14.3 (ICH), 64.8 ± 13.0 (IS) and 63.5 ± 13.1 (CTRL) and was not significantly different between groups. Time since event between IS and ICH was not significant (p = 0.22) (Supplementary Table 1).
Transcriptome architecture differences between ICH and CTRL
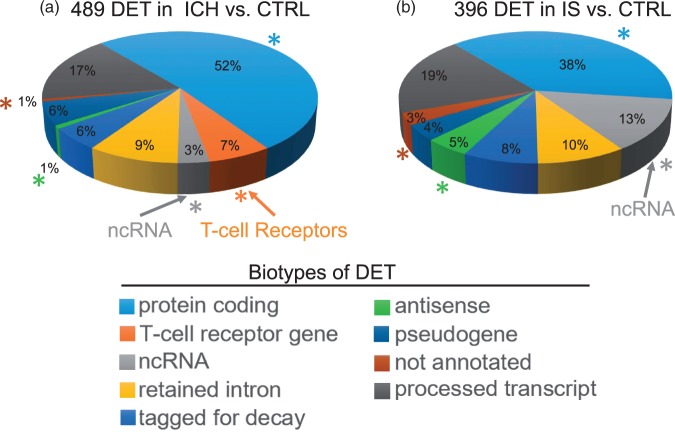
We identified 489 differentially expressed transcripts (DET) (394 genes) between ICH and CTRL (Figure 1(a), Supplementary Table 2(a), Supplementary Figure 2(a)). DET were predominantly protein coding (52%), of which 55% were down-regulated (Supplementary Table 2(a)); 3% were non-coding RNA (Figure 1(a)) and included microRNA, small-nuclear RNA, small-nucleolar RNA and long intergenic non-coding RNA (Supplementary Table 2(a)). Differences per biotype between the ICH versus CTRL and IS versus CTRL are illustrated in Figure 1(a) and (b).
Figure 1.
Transcriptome architecture of intracerebral hemorrhage (ICH, a) and ischemic stroke (IS, b) patients as compared to vascular risk-factor matched Controls (CTRL). The architecture is based upon differential expression of transcripts (DET) of alternatively spliced genes for different biological subtypes of RNA (Biotypes). Asterisks denote significant differences between the ICH and IS transcriptomes for the particular transcript biotype. The bright orange arrow points to T-Cell receptor genes (orange) and the light grey arrow points to non-coding RNA (ncRNA)(grey).
Notably, 7% of all DET in ICH were TCR genes: 33 transcripts, all were of the alpha type, and all were down-regulated in ICH compared to CTRL. Twenty-nine were from the TR_J gene type (for the Joining segment), one from the TR_C gene type (for the Constant segment), two from the TR_V gene type (for the Variable segment), and one TR_V Pseudogene (Supplementary Table 2(a)).
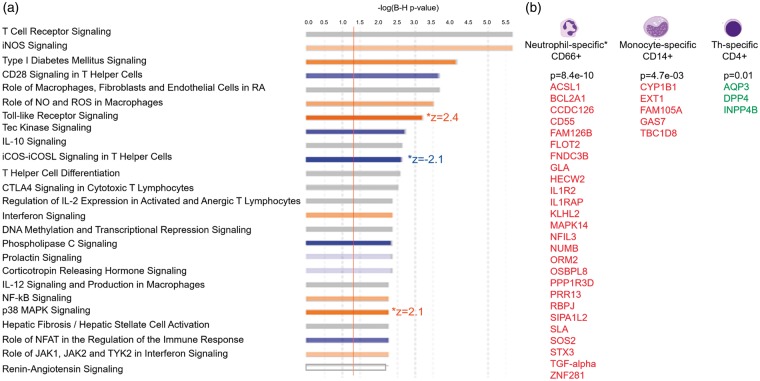
To identify blood cell-type specific DET following ICH, we overlapped our DET with genes reported to be cell-type specific in human peripheral blood.27 In ICH, the up-regulated transcripts showed significant overlap with CD66b(+) (Neutrophil)-specific genes (hypergeometric probability of overlap p = 8.4e-10), with CD14(+)(Monocytes) (p = 4.7e-03), and the down-regulated transcripts showed significant overlap with CD4(+)(T helper (Th) cells)-specific genes (p = 1.2e-02) (Figure 2(b)).
Figure 2.
Over-represented canonical pathways (a, top 25 pathways) and differentially expressed transcripts from cell type – specific genes (b) in ICH versus CTRL. (a) The X-axis represents negative log10 (Benjamini-Hochberg corrected p-value). −log10 (Benjamini-Hochberg corrected p-value) > 1.3 corresponds to Benjamini-Hochberg corrected p-value < 0.05. Orange bars represent predicted activation of the pathway, blue bars – predicted inhibition of the pathways, grey bars – direction cannot be predicted. Pathways having significant Z-scores for activation (Z > 2) or for inhibition (Z < −2) are denoted to the right of the pathway bar. (b) DET of cell-type specific genes in the 489 DET from ICH versus CTRL analysis. P-values represent the hypergeometric probability of overlap between our DET list (by gene symbol) and the list of cell-type specific genes from the HaemAtlas.27 Red – genes with upregulated DET (expressed higher in ICH than in CTRL); green – genes with down-regulated DET (expressed lower in ICH than in CTRL).
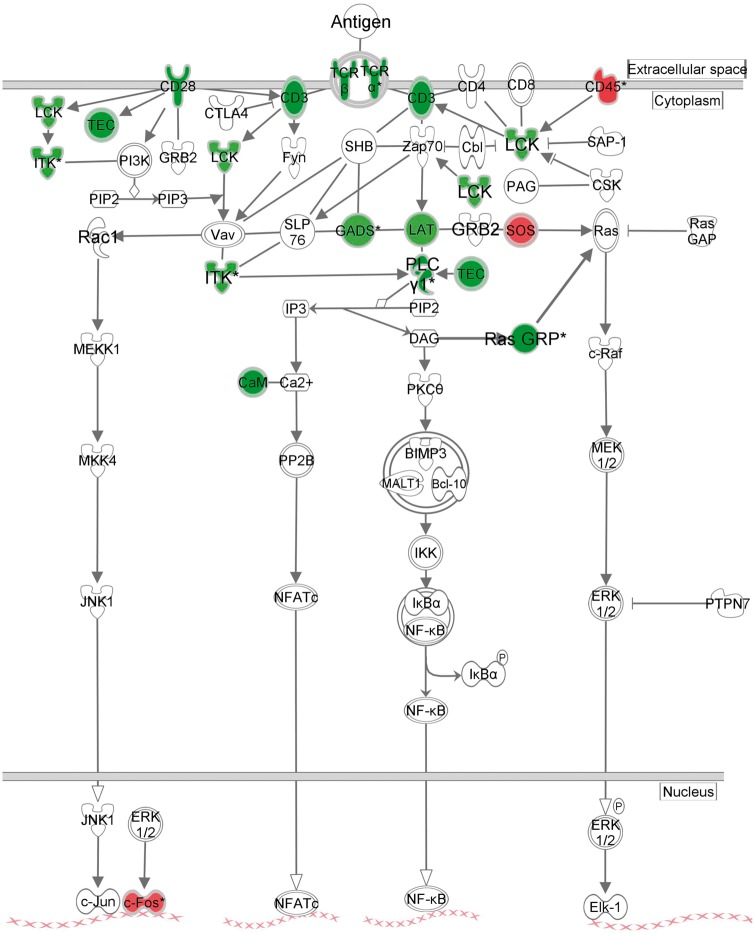
Functional analysis of the 489 ICH DET revealed they were enriched in 70 canonical pathways (Figure 2(a), Supplementary Table 3(a)). Most were involved in immune system function, including 30 cellular immune response, 20 cytokine, and 10 T-cell pathways – as well as TLR Signaling, iNOS and Production of NO and ROS in Macrophages, Leukocyte Extravasation Signaling, and Renin-Angiotensin Signaling. The most highly over-represented pathway, TCR Signaling, is presented in Figure 3. ICH DET from 36 genes were involved in angiogenesis and/or vasculogenesis (Supplementary Figure 1(a)).
Figure 3.
Ingenuity pathway analysis representation of the most significant pathway in ICH versus CTRL – T cell receptor signaling. Green – down-regulated transcript in ICH as compared to CTRL; Red – up-regulated transcript in ICH as compared to CTRL.
Transcriptome architecture differences between IS and CTRL
There were 63 DET (from 61 genes) between IS and CTRL (Supplementary Table 2(b), Supplementary Figure 2(b)). To increase the DET in the IS vs. CTRL subjects for pathway analysis so numbers of DET were similar to ICH versus CTRL, we selected IS versus CTRL DET with p < 0.005 and FC > |1.2|. This yielded 396 DET (from 379 genes) between IS and CTRL (Supplementary Table 2(c)). The transcriptome architecture for IS versus CTRL showed the DET were predominantly protein coding transcripts (38%), of which 59% were down-regulated (Figure 1(b)). Non-coding DET were also observed (13%) (Figure 1(b)), including miRNA (13), snRNA (14), snoRNA (2) and lincRNA (8), with 91% of these ncRNAs being upregulated in IS (Supplementary Table 2(c)).
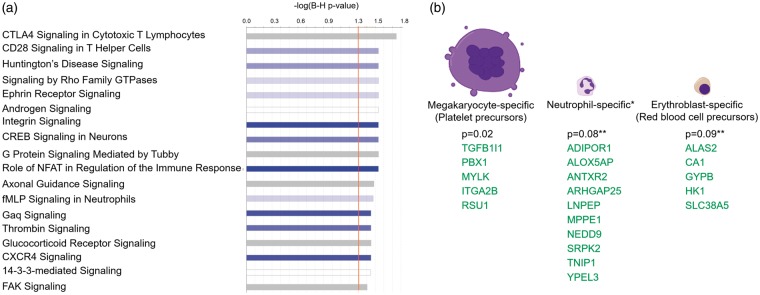
Functional analysis revealed the 396 IS DET were over-represented in 18 canonical pathways (Figure 4(a), Supplementary Table 3(b)), including immune pathways CTLA4 Signaling in Cytotoxic T Lymphocytes, CD28 Signaling in T-helper cells, NFAT Regulation of the Immune Response, fMLP Signaling in Neutrophils and CXCR4 Signaling; coagulation pathways such as Thrombin Signaling; and cell–cell and cell–extracellular matrix (ECM) interactions, such as Integrin and FAK Signaling pathways; 26 DET were from genes involved in angiogenesis (Supplementary Figure 1(b), p-value of overlap = 0.013). Downregulated transcripts showed a trend towards significance for over-representation in neutrophil-specific and erythroblast-specific genes (Figure 4(b)). The down-regulated transcripts showed a significant overlap with megakaryocyte-specific genes (p = 0.02) (Figure 4(b)).
Figure 4.
Over-represented canonical pathways (a) and differentially expressed transcripts from cell type – specific genes (b) in IS versus CTRL. A. The X-axis represents negative log10 (Benjamini-Hochberg corrected p-value). −log10 (Benjamini-Hochberg corrected p-value) > 1.3 corresponds to Benjamini-Hochberg corrected p-value < 0.05. Blue bars – predicted inhibition of the pathways (though no pathway passes Z < − 2 for significant inhibition), grey bars – direction of the pathway cannot be predicted. (b) DET of cell-type specific genes in the 396 DET from IS versus CTRL analysis. P-values represent the hypergeometric probability of overlap between our DET list (by gene symbol) and the list of cell-type specific genes from the HaemAtlas.27 Green – genes with down-regulated DET (expressed lower in IS than in CTRL).
Comparing the transcriptome architectures of IS and ICH
Figure 1 compares the ICH (489 DET in ICH vs. CTRL) (a) and IS (396 DET in IS vs. CTRL) (b) transcriptomes. ICH had significantly more DET from TCR genes (7%) than IS (0%) (Figure 1). ICH had less DET in ncRNA genes (3%) than IS (13%). ICH had more protein coding genes (52%) than IS (38%) and less antisense DET (0.8%–1%) than IS (5%). When the biotype representation of the more stringent list of 63 DET in IS versus CTRL (Supplementary Figure 1(b)) was compared to the biotypes of the 489 DET in ICH versus CTRL (Figure 1(b) and Supplementary Figure 1(a)), TCR genes biotype was significantly different (p = 0.04), while the ncRNA biotype showed a trend towards significance (p = 0.08).
The upstream regulators predicted to contribute to the observed changes in the alternatively spliced transcriptome were different in ICH compared to IS (Supplementary Figure 3). For example, cytokine IFNB1 (interferon beta 1) was predicted to be inhibited in ICH, but not involved in IS. IL17A (interleukin 17 A), CSF3 (colony stimulating factor 3), IL1 (interleukin 1) and OSM (oncostatin M) were predicted to be activated in ICH but not in IS. Transcription regulators, such as MYC (v-myc avian myelocytomatosis viral oncogene homolog), VHL (von Hippel-Lindau tumor suppressor) and E2F1 (E2F transcription factor 1 [Cell cycle regulator E2F1 modulates angiogenesis via p53-dependent transcriptional control of VEGF]) were predicted to be inhibited in ICH (MYC approached significance for activation in IS) but not IS. The growth factors TGFA and HGF were predicted to be activated in ICH but not IS.
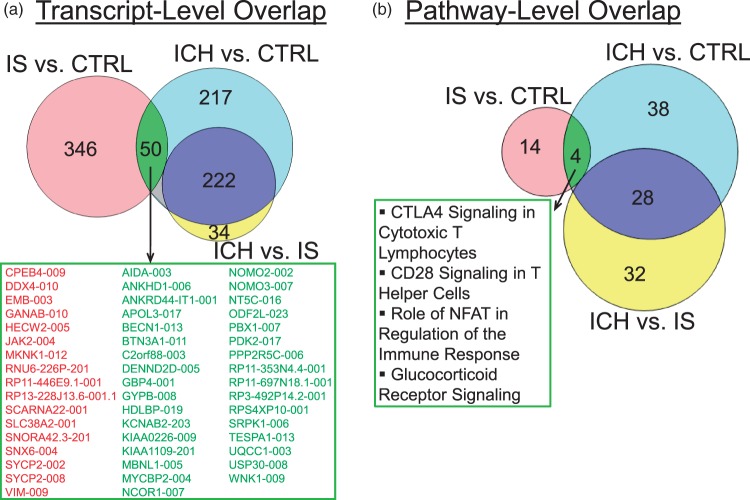
There were some common features of the ICH and IS transcriptomes. There was a significant overlap between the 396 DET in IS versus CTRL and the 489 DET in ICH versus CTRL (33 downregulated in IS and ICH, and 17 up-regulated in IS and ICH; hypergeometric probability of overlap p < 1e-16) (Figure 5(a)). These common transcripts included immune/inflammatory response genes such as JAK2, RUBCN and VIM (vimentin, implicated in neuroinflammation of brain28), and transcriptional activation of RNA genes, such as PBX1, NCOR1, MYCBP2, and PPP2R5C.
Figure 5.
Overlap of regulated transcripts (a) and pathways (b) in IS and ICH. For the indicated genes in the box in A, red are upregulated DET and green are down-regulated DET.
Four pathways overlapped between ICH and IS (Figure 5(b)). Even though these four pathways overlapped, most regulated transcripts were different. The only common transcripts from these four overlapping pathways were JAK2 (Janus kinase 2, upregulated in both ICH and IS), PPP2R5C (protein phosphatase 2 regulatory subunit B gamma, downregulated in both ICH and IS), as well as transcription factor PBX1 (PBX homeobox 1), which was downregulated in both ICH and IS, with much greater down-regulation in ICH (−17.2 fold) than IS (−1.7 fold).
Direct comparison of ICH and IS transcriptomes
A direct comparison between the ICH and IS transcriptomes identified 256 DET (from 212 genes) (Supplementary Figure 4, Supplementary Table 2(d)). The DET were over-represented in 55 pathways (Supplementary Figure 5(a), top 25 pathways, Supplementary Table 3(c)). These included cellular immune response (25 pathways, 8 of which were T-cell pathways), and/or cytokine signaling (13 pathways), and/or humoral immune response (3 pathways), growth factor signaling (such as HGF Signaling, ErbB Signaling) and cardiovascular signaling (renin-angiotensin signaling) (Supplementary Table 3(c)).
The DET expressed at higher levels in ICH than in IS showed significant overlap with neutrophil-specific genes (hypergeometric probability of overlap p = 6.4e-12) (ACSL1, BCL2A1, CD55, FAM126B, FLOT2, FNDC3B, GCA, IL1R2, IL1RAP, KLHL2, MAPK14, NFIL3, NUMB, PPP1R3D, RBPJ, SIPA1L2, SLA, STX3, TBC1D14, TGF-alpha), and with monocytes (p = 8.5e-03) (CYP1B1, EXT1, GAS7, TBC1D8). The transcripts expressed at lower levels in ICH than IS showed significant overlap with T helper-specific genes (p = 1.4e-04) (AQP3, DPP4, FHIT, INPP4B) (Supplementary Figure 5(b)).
Transcriptome changes within 24 h following ICH and IS
In the first 24 h, there were 2667 DET in ICH compared to 311 in IS (Supplementary Table 4(a) and (b)) with a specific architecture for each (Supplementary Figure 6(a)). The ICH transcriptome had higher proportion of DET from protein coding genes and pseudogenes. ICH had 55 DET from TCR genes compared to none for IS. ICH had less ncRNA and antisense RNA compared to IS (Supplementary Figure 6(a)); 18 DET overlapped between the ICH and IS (Supplementary Figure 6(b)). There were 135 pathways over-represented in ICH in the first 24 h, including 51 immune pathways (Supplementary Table 5(a)). The most over-represented pathways were TCR signaling (FDR corrected p-value = 2.1e-08), and 8 out of the top 10 pathways were T-cell pathways (FDR p = 2.1e-08–3.4e-04). There were also macrophage, B cell and natural killer cell immune pathways, and hypoxia and thrombin signaling were also associated with ICH.
There were 29 pathways over-represented in IS in the first 24 h (Supplementary Table 5(b)). These included nine immune pathways, thrombin signaling and cell–cell interactions.
The difference between the early ICH and IS transcriptomes from the CTRL transcriptome was also underscored by the fact that the 2667 DET in ICH and the 311 DET in IS could separate the ICH and IS from CTRL based on principal components analysis (Supplementary Figure 7(a) and (b)).
Dynamic changes in the transcriptome architecture in IS and ICH
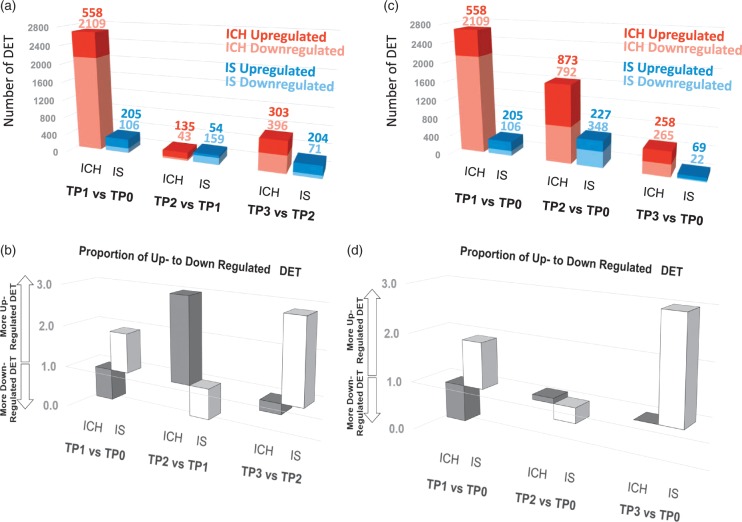
We next examined DET over time. We designated CTRL as time-point 0 (TP0), and examined DET at TP1 (<24 h), TP2 (24–48 h), and TP3 (>48 h). Overall, there were 4537 transcripts in ICH (from 3259 genes) that changed expression over time in ICH, while 1136 transcripts (from 1016 genes) changed in IS (p < 0.005 and FC > |1.2|) (Figure 6, Supplementary Table 8). ICH and IS showed different patterns of transcript regulation over time (Figure 6). For example, for each period, the proportion of up to down-regulated DET was opposite for ICH and IS (Figure 6(b) and (d)). The most pronounced transcriptomic response was in the acute phase (TP1 vs. TP0, <24 h) for both ICH (2,667 DET) and IS (311 DET) (Figure 6(a)). In terms of total number of up- and down-regulated DET, this was greatest at TP1 for ICH and decreased over time (Figure 6(c)). For IS, however, the total number of up- and down-regulated DET increased from TP1 to TP2 and then decreased from TP2 to TP3, with the greatest number of regulated DET at 24–48 h (Figure 6(c)).
Figure 6.
Dynamic changes in transcript expression in ICH and IS. TP0 indicates time-point 0 (CTRL), TP1 indicates time-point 1 (ICH or IS at < 24 h post event), TP2 indicates time-point 2 (ICH or IS between 24 h and 48 h post event) and TP3 indicates time-point 3 (ICH or IS > 48 h post event). A. number of DET between two consecutive time points. (b) Proportion of up- to down-regulated DET between two consecutive time points. (c) Number of DET between each ICH and IS time point and CTRL (as TP0). (d) Proportion of up- to down-regulated DET between each ICH and IS time point and CTRL (as TP0).
Clustering of DET based on similarity in their time-dependent profiles
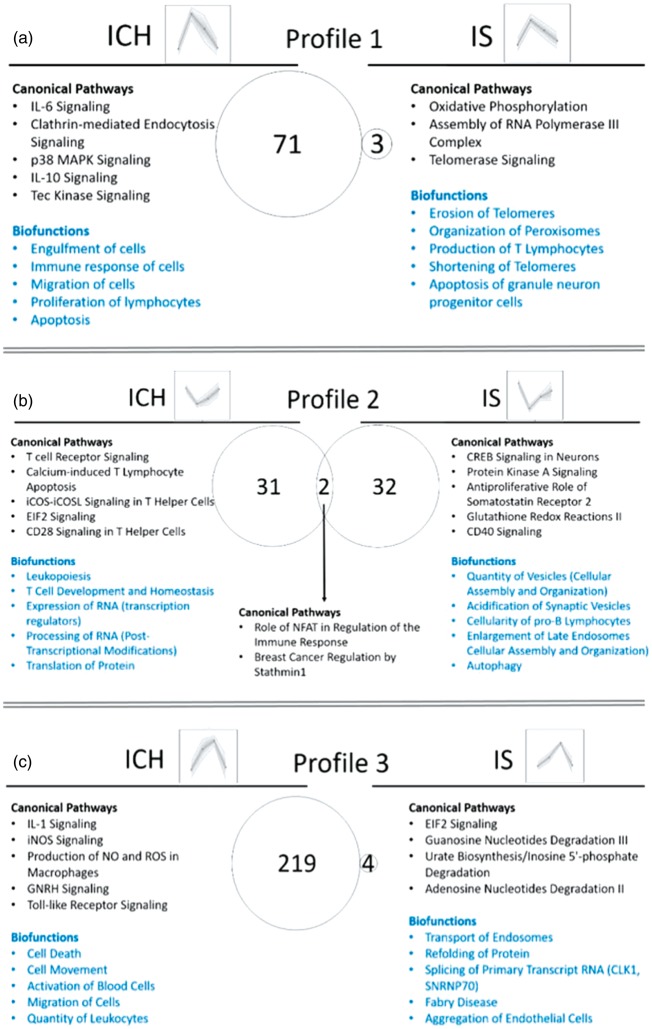
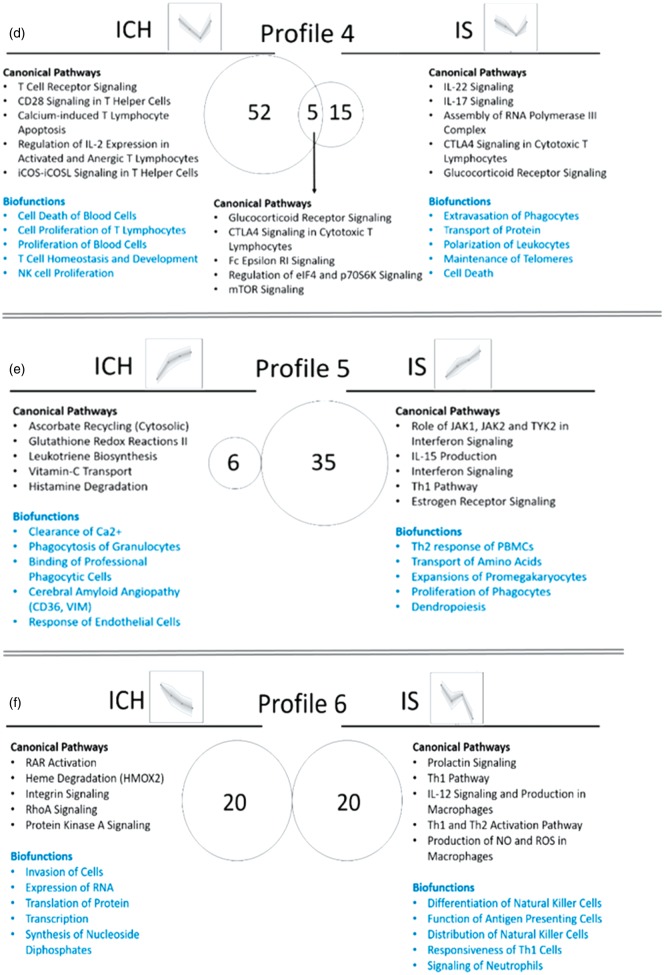
Time-dependent changes of DET were clustered using SOMs to identify transcripts with common expression profiles and to examine the biological functions and pathways for each profile. Though we started with 16 temporal profiles, similar profiles were grouped together to yield 10 temporal profiles. Supplementary Figure 8 shows eight profiles, and Supplementary Figure 9 – the other two profiles. The transcriptome architecture varied for each of the different time-dependent profiles for ICH and IS (Supplementary Figure 8). For example, transcripts encoding TCR genes were differentially expressed in profiles 2, 4, and 6 in ICH (bright orange pie chart segments and arrows in Supplementary Figure 8; asterisks show statistical significance for the difference between IS and ICH), while no TCR genes were regulated over time in IS. In addition, there were more ncRNAs (non-coding RNAs) in IS in profiles 1, 3, 5, 6 and 7, compared to the analogous profiles in ICH (light grey pie chart segments in Supplementary Figure 8).
Biological processes and pathways over-represented over time in ICH and IS
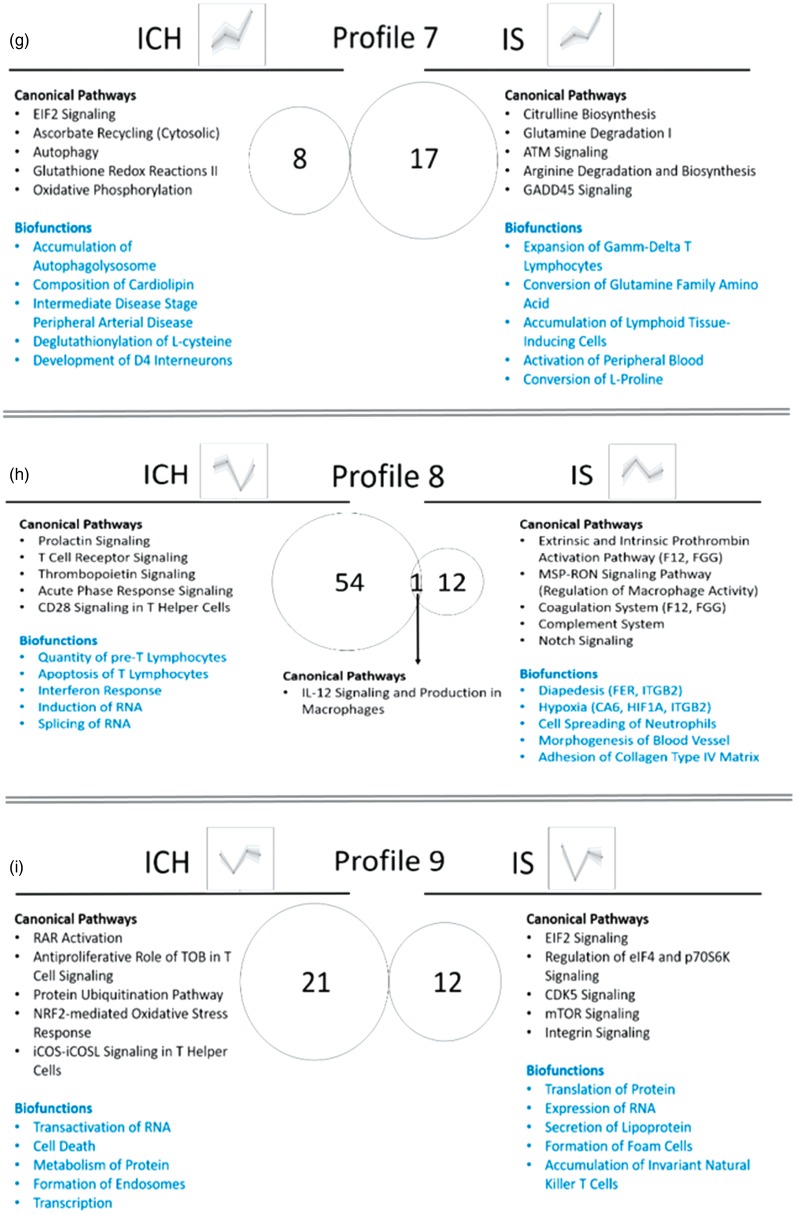
We next examined biological pathways and processes (Supplementary Table 7, Figure 7 – for profiles 1–9, Supplementary Figure 9 – for Profile 10) and Canonical Pathways and Biofunctions (p < 0.05) (Supplementary Table 8) that changed over time in ICH and IS. There were many changes including inflammatory pathways in neutrophils, macrophages, T-cells and natural killer (NK) cells; cell death, coagulation and metabolic pathways; and biofunctions involved in regulation of RNA transcription and splicing, post-transcriptional regulation and regulation of protein synthesis. Notably, DET for CD36 and VIM are in Profile 5 in ICH, both involved in cerebral amyloid angiopathy, one of the major causes of ICH. FER (FER tyrosine kinase) and ITGB2 (Integrin Subunit Beta 2) are in Profile 8 in IS and are involved in infiltration of immune cells to sites of inflammation. Coagulation Factor 12 (F12) and fibrinogen gamma chain (FGG) are in Profile 8 in IS, and are involved in extrinsic and intrinsic prothrombin activation pathways. HMOX2 is in Profile 6 in ICH and participates in heme degradation. Hypoxia-related genes CA6 (carbonic anhydrase 6) and HIF1A (hypoxia inducible factor 1 alpha subunit) are in Profile 8 in IS (Supplementary Table 8, Figure 7).
TCR genes and pathways differentiate ICH from IS
There were 55 DET (ICH_TP1 vs. CTRL_TP0) from TCR genes in ICH that were not detected in IS (TCR genes from Supplementary Table 4). Supplementary Figure 10(a) and (b) show that expression values of these can differentiate ICH on principal components analysis from IS and CTRL, respectively. In addition, there were 107 transcripts from genes involved in TCR function (Supplementary Table 9) that were differentially expressed in ICH_TP1 versus CTRL_TP0 that could differentiate ICH from IS_TP1 (Supplementary Figure 11(a)) and from CTRL_TP0 (Supplementary Figure 11(b)) on principal components analysis, but they cannot differentiate IS from CTRL.
Cell count and CIBERSORT deconvoluted data
Complete blood count with or without differential was performed on some of the subjects as part of their medical work-up. There was no statistical significance (Mann–Whitney p-value < 0.05, 1000 iterations) in the numbers of their white blood cells, hemoglobin, hematocrit, platelets, and the percentage of their neutrophils, monocytes and lymphocytes (Supplementary Table 10). In addition, we performed CIBERSORT deconvolution gene expression analysis (see Supplementary Methods and Supplementary Table 11). Of the 22 human hematopoietic phenotypes investigated by the deconvolution algorithm, including seven T cell phenotypes, significant difference was predicted only in the T-cell CD4 memory resting phenotype in the ICH versus CTRL and ICH versus IS comparisons (lower in ICH than in IS and CTRL) (Supplementary Table 11). Indeed, the deconvolution analysis suggested a significantly smaller number of CD4+ resting T-cells in ICH versus CTRL and ICH versus IS, but no significant difference in γδT-Cells, CD8+ T-Cells, CD4+ memory activated T-Cells, CD4+ naïve T-Cells types, follicular helper T-Cells, regulatory T-Cells (Tregs), NK cells resting, NK cells activated, memory B cells, naïve B cells, and Plasma Cells.
Discussion
The data show alternatively spliced transcripts in blood of ICH patients differ from IS, with little overlap between the two. The time course and numbers of regulated transcripts differ in ICH compared to IS. There were many TCR transcripts differentially expressed in ICH not observed in IS, and a lesser percentage of non-coding RNA in ICH compared to IS. The data support the notion that alternative splicing is important in ICH and IS.29–32 The study is significant for identifying potential treatment targets for acute and subacute immune responses which may be involved in injury and repair, and to identify biomarkers that might be examined within the rtPA or endovascular therapy windows. Since little is known about the genomic response following ICH in human peripheral blood, we focused the discussion on ICH.
ICH immune response
There is a strong immune/inflammatory response after ICH, which includes microglial activation, leukocyte infiltration and production of inflammatory mediators.6 Indeed, HMGB1 Signaling, IL-6 Signaling, TLR Signaling and p38MAPK Signaling were predicted to be activated following ICH (Z > 2.0). HMGB1, a potent initiator of inflammation, belongs to the so-called danger-associated molecular pattern molecules (DAMPs) which are released from dying cells.7 DAMPs act through receptors, including toll-like receptors (TLRs) to induce inflammation. TLR Signaling was predicted to be activated in ICH. Transcripts from eight genes (including TLR2, TLR4, TLR5 and TLR8) were upregulated in ICH versus CTRL; and TLR1, TLR2, TLR4, TLR5 and TLR8 showed time-dependent changes in ICH. TLR4 signaling in resident microglia and peripheral infiltrating leukocytes has been implicated in ICH-induced brain injury.33 Increased expression of TLR2 and TLR4 in peripheral monocytes is associated with poor outcomes in ICH patients.34 Improved neurological function after experimental ICH is observed in TLR4-knockout mice,33,35 and the TLR2/4 heterodimer mediates inflammatory injury in ICH.36 Thus, TLRs are potential therapeutic targets in ICH.33,37
IL-6 (Interleukin 6) and p38 MAPK signaling pathways were also predicted to be activated in ICH. IL-6 is a regulator of acute-phase responses and a lymphocyte stimulating factor. Increased plasma IL-6 has been associated with increased risk for hemorrhage from cerebral arteriovenous malformations (AVMs).38 IL-6 induces expression of MMP-9 which degrades extracellular matrix in vasculature, damages endothelial cells, and increases risk for AVMs to rupture, particularly in carriers of a SNP in IL-6 (174 G > C).38 p38 MAPK, p38 mitogen-activated protein family of kinases are activated by inflammatory cytokines, death ligands, TGF-β-related polypeptides and oxidative stress, which results in enhanced transcriptional activity, protein synthesis and cell death. Transforming growth factor-β (TGF-β) signaling pathway was also activated in ICH (Z = 2.0, p = 8.73E-03). The TGF-β receptor complex is involved in hereditary hemorrhagic telangiectasia, which has a high rate of AVM and ICH.38 Our data also show TGF signaling in blood leukocytes modulates hemorrhagic transformation following IS.39,40 Thus, these DET and pathways may be therapeutic targets for ICH.
Neutrophils, monocytes and T-cells following ICH
Following ICH, the blood–brain barrier (BBB) is disrupted with associated edema and leukocyte extravasation.6,41 Leukocyte extravasation signaling was over-represented in ICH. This is the process by which leukocytes migrate from blood to tissue during injury/inflammation. Neutrophils are the first leukocyte type to enter the brain after ICH. Their role appears to be primarily deleterious in ICH, though some functions are beneficial.42 However, increased neutrophils or increased neutrophil to lymphocyte ratio in humans are associated with increased perihematomal edema,43 correlate with 90-day outcomes after ICH,44,45 ICH expansion46 and 30-day mortality after ICH.47 In addition, though neutrophils appear to remain in and around vessels following IS, neutrophils enter brain parenchyma and the clot following ICH.48 Neutrophils contribute to the ICH-induced brain injury by disrupting the BBB, producing reactive oxygen species, and proinflammatory molecules.9 Neutrophil depletion decreases BBB breakdown, axon injury and inflammation following experimental ICH.49
Monocytes also enter the brain after ICH and appear to have adverse effects early.41 Indeed, monocyte blood counts are associated with human 30-day case fatality after ICH.50 Monocytes also appear to contribute to tissue repair and hematoma phagocytosis later after ICH.41 Macrophages and dendritic cells dominate the inflammatory infiltrate by 12 h after experimental ICH.51 Studies suggest that monocyte entry is reduced and outcomes are improved after neutrophil depletion in experimental ICH.52 In our study, in ICH versus CTRL, we found a significant enrichment of up-regulated transcripts from CD66+ neutrophil- and CD14+ monocyte-specific genes, signifying the importance of differential transcript expression in these cell types following ICH.
A major finding was the large number of over-represented T-cell pathways in ICH versus CTRL, the majority of which were represented by down-regulated transcripts. In addition, there was significant overlap with CD4+ Th-specific genes in ICH versus CTRL, which were also down-regulated (Figure 2). Many TCR transcripts were also down-regulated in ICH (discussed below). iCOS-iCOSL signaling in T helper cells was predicted to be suppressed in ICH versus CTRL. This regulates activation of T-cells and effector T-cell functions and generates Th1 and Th2 responses. In IS, different T-cell subsets may play critical roles in brain injury and repair mechanisms.53 Less is known about the role of T lymphocytes following ICH.9 Lymphocytes have been detected in cerebrospinal fluid by 6 h following ICH.9 In a mouse model of ICH, CD4+ T cells were the predominant leukocyte type infiltrating brain one day post ICH, with numbers peaking at five days.54 Different T cell populations infiltrate the brain after ICH, including proinflammatory γδT cells and immunosuppressive Treg (regulatory T-Cells).9 Treg transfer attenuates neurological deficit after ICH55 and perihematomal inflammation in experimental models of ICH.56 Regulatory T cells decrease ICH injury by modulating microglia/monocytes via IL10/GSK3β/PTEN.57
Our data also show T-cell signaling pathways are important in ICH. There were several T-cell pathways in profiles such as 2, 3, 4, 8 and 9 (Figure 7). There were common pathways that were over-represented in different time-dependent profiles. For example, Profiles 3 and 4 in ICH, which had opposite direction (Profile 3 peaked at day 2, while Profile 4 dipped at day 2), had 35 common pathways, 10 of which were T cell pathways. These complex temporal profiles of T-Cell pathways underscore the importance of further investigating the transcriptome changes in isolated different subpopulations of T-cells following ICH in human. One possibility is that there is a decrease in T-cell numbers (or at least for some of the T-cell subtypes) in the peripheral blood following ICH, and/or they might be sequestered in the brain at the site of the hemorrhage/hematoma.
Figure 7.
Biological interpretation of the time-dependent profiles of ICH and IS (Profiles 1–9; Profile 10 is presented in Supplementary Figure 8). Venn diagram displays the number of canonical pathways over-represented in each of the time-dependent profiles. The top five relevant canonical pathways and top five relevant biofunctions are listed. (a) Profile 1; (b) Profile 2; (c) Profile 3; (d) Profile 4; (e) Profile 5; (f) Profile 6; (g) Profile 7; (h) Profile 8; (i) Profile 9.
To explore the possibility of a lower number of T-cells in the peripheral blood, we examined whether there were cell number differences between the ICH, IS and CTRL groups. As noted in the results, there were no significant differences in the white blood cell count, platelet count, and in the total lymphocyte, monocyte and neutrophil percent at the time of the blood draw used for the transcriptomic study. The deconvolution analysis,58 however, suggested a significantly smaller number of CD4+ resting cells in ICH versus CTRL and ICH versus IS. Thus, it is possible that decreases of circulating CD4+ T cells might account for the decreases of T cell receptor and signaling genes in this study. This needs to be examined in the future.
TCR transcripts can differentiate ICH from IS
Transcripts from TCR genes were only differentially expressed in ICH, but not in IS as compared to CTRL; 33 DET from TCR genes were differentially expressed (all down-regulated) between ICH and CTRL, and the expression of many TCR transcripts was time-dependent with 55 DET from TCR genes being down-regulated within 24 h post ICH.
TCR are antigen receptors on T cells. They recognize MHC (HLA) associated antigenic peptides that are presented by antigen presenting cells. TCR consists of two distinct units: an antigen-recognizing unit comprised either of TCR α-β heterodimer, or γ-δ heterodimer, and a signal transduction unit comprised the CD3 complex. TCR α-β heterodimer recognizes processed antigens which are presented as peptides by the HLA proteins, whereas TR γ-δ recognizes nonpeptidic antigens. The α, β, γ and δ chains are encoded by genes located in four major loci: the TR α (TRA), TR β (TRB), TR γ (TRG) and TR δ (TRD). Four TCR gene types: variable (V), diversity (D) (for TRB and TRD), joining (J) and constant (C) genes contribute to the TCR chain synthesis. After somatic recombination, the variable domain at the N-terminal end of each TCR chain engages in a V-(D)-J rearrangement. The remainder of the chain, or constant region, is encoded by a C gene. Thus, there is large diversity of TCRs that is a result of the combinatorial and junctional diversity.59 In our data, we had many transcripts from TCR genes that were differentially expressed between ICH and CTRL, all down-regulated in ICH, and most all were from the α, J (joining) type. Time-dependent profiles 2,4 and 6 in ICH (Figure 7) also contained TCR transcripts. In addition, during the acute response to ICH (within 24 h), there were 55 TCR transcripts, all down-regulated in ICH compared to CTRL. TCR genes and genes from T-cell associated pathways were able to differentiate ICH from IS on a principal components analysis. IS could not be differentiated from CTRL based on TCR genes, signifying they may be ICH-specific biomarkers. These data suggest TCR genes might be used as novel biomarkers to differentiate ICH from IS which needs to be confirmed in future studies.
Angiogenesis/vasculogenesis following ICH
A significant enrichment of vasculogenesis and angiogenesis-associated genes was found in ICH versus CTRL (transcripts from 36 genes). Both processes were predicted to have tendency towards being activated (positive Z-scores, but smaller than 2.0 – the threshold for significant activation). Previous studies suggest upregulation of angiogenesis following ICH, leading to remodeling that may improve function in animal models and patients.60,61 Circulating endothelial progenitor cells show therapeutic promise in animal ICH models.61 Thus, the differentially expressed transcripts from genes implicated in angiogenesis or vasculogenesis provided here may represent potential human therapeutic targets. Thus, following ICH, just as occurs in IS, there is vascular re-modeling which is likely modulated by the peripheral immune system.
Other genes and pathways
There were many splicing regulators that were differentially expressed in ICH. For example, several FYN transcripts, which is a Src protein tyrosine kinase family member, involved in intracellular signaling of various processes including T-cell development and activation,62 were differentially expressed over time following ICH. Transcripts from SRSF1, SFSF7, SFSF8 and SRSF11 were differentially expressed over time in ICH. SRSF genes (serine and arginine rich splicing factors) are implicated in regulating alternative splicing in immune cells.31 Transcripts from TIA-1 (TIA1 cytotoxic granule associated RNA binding protein, formerly known as T-cell restricted intracellular antigen 1) had time-dependent expression in ICH and IS. TIA-1 is involved in alternative pre-RNA splicing and regulation of mRNA translation and is a silencing translator of TNFα.31 These data underscore the potential importance of alterative splicing in the pathophysiology of ICH.
The expression of CD36 and VIM transcripts increased over time in ICH (Profile 5, Figure 7). CD36 and VIM are implicated in cerebral amyloid angiopathy (CAA) which is one of the major causes of ICH (12–15% in elderly).63 CAA develops as a result of deposition of amyloid-β (Aβ) in cerebral blood vessels and leads to damaged endothelial cells, disrupted blood–brain barrier, and disrupted vascular function. CD36, which is an innate immune receptor involved in Aβ trafficking, has been implicated in CAA. CD36 was suggested to promote vascular amyloid deposition and cerebrovascular damage, leading to neurovascular dysfunction and cognitive deficits.64 In addition, CD36 is a phagocytosis scavenger receptor found on monocytes, macrophages, microglia and plays a role in resolution of the clot following ICH. Stimulation of CD36 with agonists of PPARγ, RXR and Nrf2 improves hematoma resolution and improves behavioral outcomes in experimental ICH.65,66
VIM is a cytoskeletal protein involved in the immune response and in LDL transport. It is found on the surface of numerous cells, including platelets, apoptotic neutrophils and T cells, activated macrophages, vascular endothelial cells and brain microvascular endothelial cells.67–71 VIM is expressed on the surface of vascular endothelial cells (so-called superficial VIM) following infection with dengue virus (causing hemorrhagic disease) and has been proposed to be involved in binding of the virus.72 VIM has also been implicated in neuroinflammation, cerebral ischemia, and in CAA.28,73–75 Our data underscore the need to investigate the role of CD36, VIM and the many other genes in ICH patients.
There were two DET in RNF213 (Ring Finger Protein 213) that changed with ICH. Variants in this gene are associated with Moyamoya disease which is associated with ICH that results from progressive internal carotid artery occlusions and proliferation of lenticulostriate penetrator arteries.76
Non-coding transcriptome in ICH
miRNA-21 was upregulated in ICH versus CTRL and had time-dependent expression as well. There was significant overlap between the DET of ICH versus CTRL and miR-21 targets (15 genes overlapped), and the majority (11 of 15 genes) were down-regulated as would be expected, since mRNA levels have opposite direction of the levels of the targeting miRNA. miRNA-21 plays a role in angiogenesis by promoting survival, migration and tube formation of endothelial cells, which simultaneously inhibits tissue inhibitor of metalloproteinase-3 (TIMP3) expression and promotes matrix metalloproteinase-2 (MMP2) and matrix metalloproteinase-9 (MMP9) expression and secretion.77 Other pathways regulated by miRNAs regulated following ICH include an influx of macrophages (miR-181),78 B-cell development (miR-132),79 expansion of B-1a lymphocytes (miR-210),79 proliferation of lymphocytes (miR-132, miR-181, miR-21, miR-210),79 Th1 and Th2 pathways (miR-21),80 and angiogenesis/vasculogenesis (miR-21, miR-210, miR-132, miR-181).81 miR-181 regulates inflammatory responses in monocytes and macrophages,78 acts as antigen sensitivity “rheostat” during T-cell development,82 and influences the outcome of cerebral ischemia in vitro and in vivo.83 Analyses of the combined mRNA-miRNA and other ncRNA expression networks will shed more light on the involvement of these regulatory RNAs in the peripheral immune response to ICH and their involvement of angiogenesis/vasculogenesis.
Limitations
Sample sizes were relatively small, requiring validation in future studies. Since VRFs were matched between ICH and IS subjects, it is likely they were on similar medications – though this will have to be studied in detail in the future. Investigating individual blood cell types in the future will help delineate their unique contribution to the immune response and pathophysiology of ICH and IS that could only be inferred from the whole blood studies here. Future validation of individual DET in a larger, independent cohort and with alternative expression methods, such as qRT-PCR or targeted RNA-Seq is needed. In addition, even though we did not observe subgrouping based on biological sex and rtPA administration (Supplementary Figure 13), how the transcriptome changes following ICH and IS in relation to these and other factors will need to be investigated in future studies. This is not a prospective study. Since no pre-IS and pre-ICH samples are available for the IS and ICH subjects, the initiation of the differential expression cannot be pinpointed exactly. Nevertheless, examining the immune response over time provides us with a better understanding of the acute and subacute changes following ICH and IS, as well as the processes involved in injury and repair. In addition, differences in the cell numbers of specific cell subtypes may drive some of the observed differences. That is why future studies should focus on isolated cell subtypes, particularly T-cells, neutrophils and monocytes to further investigate the findings.
Supplemental Material
Supplemental material for The intracerebral hemorrhage blood transcriptome in humans differs from the ischemic stroke and vascular risk factor control blood transcriptomes by Boryana Stamova, Bradley P Ander, Glen Jickling, Farah Hamade, Marc Durocher, Xinhua Zhan, Da Zhi Liu, Xiyuan Cheng, Heather Hull, Alan Yee, Kwan Ng, Natasha Shroff and Frank R Sharp in Journal of Cerebral Blood Flow & Metabolism
Supplemental Material
Supplemental material, Supplementary Tables for The intracerebral hemorrhage blood transcriptome in humans differs from the ischemic stroke and vascular risk factor control blood transcriptomes by Boryana Stamova, Bradley P Ander, Glen Jickling, Farah Hamade, Marc Durocher, Xinhua Zhan, Da Zhi Liu, Xiyuan Cheng, Heather Hull, Alan Yee, Kwan Ng, Natasha Shroff and Frank R Sharp in Journal of Cerebral Blood Flow & Metabolism
Supplemental Material
Supplemental material, Supplementary Figures 1 to 13 for The intracerebral hemorrhage blood transcriptome in humans differs from the ischemic stroke and vascular risk factor control blood transcriptomes by Boryana Stamova, Bradley P Ander, Glen Jickling, Farah Hamade, Marc Durocher, Xinhua Zhan, Da Zhi Liu, Xiyuan Cheng, Heather Hull, Alan Yee, Kwan Ng, Natasha Shroff and Frank R Sharp in Journal of Cerebral Blood Flow & Metabolism
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: These studies were supported by American Heart Association grants to Stamova and Liu, an American Heart Association Fellow to Faculty award to Jickling, and National Institutes of Health/National Institute of Neurological Disorders and Stroke grants to Sharp, Jickling, Stamova and Ander (NS079153, NS075035, NS097000, NS101718).
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions
BS and FRS designed the study and wrote the manuscript. BS performed the data analysis and results interpretation. BPA and MD assisted in data analysis. GJ and FRS oversaw subject selection and diagnosis. FH, BPA, MD, HH, NS, XZ, DL, XC, BS, FRS, GJ, AY, KN participated in the conduct and discussions of the studies, and read and reviewed the manuscript.
Supplementary material
Supplementary material for this paper can be found at the journal website: http://journals.sagepub.com/home/jcb
References
- 1.Benjamin EJ, Blaha MJ, Chiuve SE, et al. Heart disease and stroke statistics-2017 update: a report from the American Heart Association. Circulation 2017; 135: e146–e603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qureshi AI, Mendelow AD, Hanley DF. Intracerebral haemorrhage. Lancet 2009; 373: 1632–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Asch CJ, Luitse MJ, Rinkel GJ, et al. Incidence, case fatality, and functional outcome of intracerebral haemorrhage over time, according to age, sex, and ethnic origin: a systematic review and meta-analysis. Lancet Neurol 2010; 9: 167–176. [DOI] [PubMed] [Google Scholar]
- 4.Macrez R, Ali C, Toutirais O, et al. Stroke and the immune system: from pathophysiology to new therapeutic strategies. Lancet Neurol 2011; 10: 471–480. [DOI] [PubMed] [Google Scholar]
- 5.Ziai WC. Hematology and inflammatory signaling of intracerebral hemorrhage. Stroke 2013; 44: S74–S78. [DOI] [PubMed] [Google Scholar]
- 6.Keep RF, Hua Y, Xi G. Intracerebral haemorrhage: mechanisms of injury and therapeutic targets. Lancet Neurol 2012; 11: 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamorro A, Meisel A, Planas AM, et al. The immunology of acute stroke. Nat Rev Neurol 2012; 8: 401–410. [DOI] [PubMed] [Google Scholar]
- 8.Iadecola C, Anrather J. The immunology of stroke: from mechanisms to translation. Nat Med 2011; 17: 796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mracsko E, Veltkamp R. Neuroinflammation after intracerebral hemorrhage. Front Cell Neurosci 2014; 8: 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharp FR, Jickling GC, Stamova B, et al. Molecular markers and mechanisms of stroke: RNA studies of blood in animals and humans. J Cereb Blood Flow Metab 2011; 31: 1513–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharp FR, Jickling GC. Whole genome expression of cellular response to stroke. Stroke 2013; 44: S23–S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agnihotri S, Czap A, Staff I, et al. Peripheral leukocyte counts and outcomes after intracerebral hemorrhage. J Neuroinflammation 2011; 8: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackenzie JM, Clayton JA. Early cellular events in the penumbra of human spontaneous intracerebral hemorrhage. J Stroke Cerebrovasc Dis 1999; 8: 1–8. [DOI] [PubMed] [Google Scholar]
- 14.Meller R, Pearson AN, Hardy JJ, et al. Blood transcriptome changes after stroke in an African American population. Ann Clin Transl Neurol 2016; 3: 70–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asano S, Chantler PD, Barr TL. Gene expression profiling in stroke: relevance of blood-brain interaction. Curr Opin Pharmacol 2016; 26: 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dykstra-Aiello C, Jickling GC, Ander BP, et al. Altered expression of long noncoding RNAs in blood after ischemic stroke and proximity to putative stroke risk loci. Stroke 2016; 47: 2896–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore DF, Li H, Jeffries N, et al. Using peripheral blood mononuclear cells to determine a gene expression profile of acute ischemic stroke: a pilot investigation. Circulation 2005; 111: 212–221. [DOI] [PubMed] [Google Scholar]
- 18.Sang M, Wang X, Zhang H, et al. Gene expression profile of peripheral blood mononuclear cells in response to intracerebral hemorrhage. DNA Cell Biol 2017; 36: 647–654. [DOI] [PubMed] [Google Scholar]
- 19.Gamazon ER, Stranger BE. Genomics of alternative splicing: evolution, development and pathophysiology. Hum Genet 2014; 133: 679–687. [DOI] [PubMed] [Google Scholar]
- 20.Poulos MG, Batra R, Charizanis K, et al. Developments in RNA splicing and disease. Cold Spring Harb Perspect Biol 2011; 3: a000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet 2016; 17: 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yew KS, Cheng E. Acute stroke diagnosis. Am Fam Physician 2009; 80: 33–40. [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson WA., Jr The problem of negative estimates of variance components. Ann Math Stat. 33: 273–289. [Google Scholar]
- 24.Tamhane AC, Dunlop DD. Statistics and data analysis: From elementary to intermediate, Upper Saddle River, NJ: Prentice Hall, 2000. [Google Scholar]
- 25.Kohonen T. Self-organizing maps, 3rd ed. Berlin, Heidelberg, New York: Springer.
- 26.Stamova B, Green PG, Tian Y, et al. Correlations between gene expression and mercury levels in blood of boys with and without autism. Neurotox Res 2011; 19: 31–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watkins NA, Gusnanto A, de Bono B, et al. A HaemAtlas: characterizing gene expression in differentiated human blood cells. Blood 2009; 113: e1–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang SX, Slinn J, Aylsworth A, et al. Vimentin participates in microglia activation and neurotoxicity in cerebral ischemia. J Neurochem 2012; 122: 764–774. [DOI] [PubMed] [Google Scholar]
- 29.Pan Q, Shai O, Lee LJ, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008; 40: 1413–1415. [DOI] [PubMed] [Google Scholar]
- 30.Lynch KW. Consequences of regulated pre-mRNA splicing in the immune system. Nat Rev Immunol 2004; 4: 931–940. [DOI] [PubMed] [Google Scholar]
- 31.Martinez NM, Lynch KW. Control of alternative splicing in immune responses: many regulators, many predictions, much still to learn. Immunol Rev 2013; 253: 216–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luco RF, Allo M, Schor IE, et al. Epigenetics in alternative pre-mRNA splicing. Cell 2011; 144: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Y, Wang Y, Wang J, et al. Inflammation in intracerebral hemorrhage: from mechanisms to clinical translation. Prog Neurobiol 2014; 115: 25–44. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez-Yanez M, Brea D, Arias S, et al. Increased expression of Toll-like receptors 2 and 4 is associated with poor outcome in intracerebral hemorrhage. J Neuroimmunol 2012; 247: 75–80. [DOI] [PubMed] [Google Scholar]
- 35.Sansing LH, Harris TH, Welsh FA, et al. Toll-like receptor 4 contributes to poor outcome after intracerebral hemorrhage. Ann Neurol 2011; 70: 646–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang YC, Zhou Y, Fang H, et al. Toll-like receptor 2/4 heterodimer mediates inflammatory injury in intracerebral hemorrhage. Ann Neurol 2014; 75: 876–889. [DOI] [PubMed] [Google Scholar]
- 37.Xi G, Strahle J, Hua Y, et al. Progress in translational research on intracerebral hemorrhage: is there an end in sight? Prog Neurobiol 2014; 115: 45–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mouchtouris N, Jabbour PM, Starke RM, et al. Biology of cerebral arteriovenous malformations with a focus on inflammation. J Cereb Blood Flow Metab 2015; 35: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jickling GC, Ander BP, Stamova B, et al. RNA in blood is altered prior to hemorrhagic transformation in ischemic stroke. Ann Neurol 2013; 74: 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jickling GC, Liu D, Stamova B, et al. Hemorrhagic transformation after ischemic stroke in animals and humans. J Cereb Blood Flow Metab 2014; 34: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keep RF, Zhou N, Xiang J, et al. Vascular disruption and blood-brain barrier dysfunction in intracerebral hemorrhage. Fluids Barriers CNS 2014; 11: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao X, Ting SM, Liu CH, et al. Neutrophil polarization by IL-27 as a therapeutic target for intracerebral hemorrhage. Nat Commun 2017; 8: 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gusdon AM, Gialdini G, Kone G, et al. Neutrophil-lymphocyte ratio and perihematomal edema growth in intracerebral hemorrhage. Stroke 2017; 48: 2589–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao C, Hu X, Wang J, et al. Admission neutrophil count and neutrophil to lymphocyte ratio predict 90-day outcome in intracerebral hemorrhage. Biomark Med 2017; 11: 33–42. [DOI] [PubMed] [Google Scholar]
- 45.Lattanzi S, Cagnetti C, Provinciali L, et al. Neutrophil-to-lymphocyte ratio predicts the outcome of acute intracerebral hemorrhage. Stroke 2016; 47: 1654–1657. [DOI] [PubMed] [Google Scholar]
- 46.Morotti A, Phuah CL, Anderson CD, et al. Leukocyte count and intracerebral hemorrhage expansion. Stroke 2016; 47: 1473–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang F, Hu S, Ding Y, et al. Neutrophil-to-lymphocyte ratio and 30-day mortality in patients with acute intracerebral hemorrhage. J Stroke Cerebrovasc Dis 2016; 25: 182–187. [DOI] [PubMed] [Google Scholar]
- 48.Zhao X, Sun G, Zhang H, et al. Polymorphonuclear neutrophil in brain parenchyma after experimental intracerebral hemorrhage. Transl Stroke Res 2014; 5: 554–561. [DOI] [PubMed] [Google Scholar]
- 49.Moxon-Emre I, Schlichter LC. Neutrophil depletion reduces blood-brain barrier breakdown, axon injury, and inflammation after intracerebral hemorrhage. J Neuropathol Exp Neurol 2011; 70: 218–235. [DOI] [PubMed] [Google Scholar]
- 50.Adeoye O, Walsh K, Woo JG, et al. Peripheral monocyte count is associated with case fatality after intracerebral hemorrhage. J Stroke Cerebrovasc Dis 2014; 23: e107–e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hammond MD, Ai Y, Sansing LH. Gr1+ Macrophages and dendritic cells dominate the inflammatory infiltrate 12 hours after experimental intracerebral hemorrhage. Transl Stroke Res 2012; 3: s125–s131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sansing LH, Harris TH, Kasner SE, et al. Neutrophil depletion diminishes monocyte infiltration and improves functional outcome after experimental intracerebral hemorrhage. Acta Neurochir Suppl 2011; 111: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu L, Jian Z, Stary C, et al. T cells and cerebral ischemic stroke. Neurochem Res 2015; 40: 1786–1791. [DOI] [PubMed] [Google Scholar]
- 54.Mracsko E, Javidi E, Na SY, et al. Leukocyte invasion of the brain after experimental intracerebral hemorrhage in mice. Stroke 2014; 45: 2107–2114. [DOI] [PubMed] [Google Scholar]
- 55.Yang Z, Yu A, Liu Y, et al. Regulatory T cells inhibit microglia activation and protect against inflammatory injury in intracerebral hemorrhage. Int Immunopharmacol 2014; 22: 522–525. [DOI] [PubMed] [Google Scholar]
- 56.Mao LL, Yuan H, Wang WW, et al. Adoptive regulatory T-cell therapy attenuates perihematomal inflammation in a mouse model of experimental intracerebral hemorrhage. Cell Mol Neurobiol 2017; 37: 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou K, Zhong Q, Wang YC, et al. Regulatory T cells ameliorate intracerebral hemorrhage-induced inflammatory injury by modulating microglia/macrophage polarization through the IL-10/GSK3beta/PTEN axis. J Cereb Blood Flow Metab 2017; 37: 967–979. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015; 12: 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Janeway CAJ, Travers P, Walport M, et al. Immunology: The immune system in health and disease, 5th ed New York: Garland Science, 2001. [Google Scholar]
- 60.Lei C, Lin S, Zhang C, et al. Effects of high-mobility group box1 on cerebral angiogenesis and neurogenesis after intracerebral hemorrhage. Neuroscience 2013; 229: 12–19. [DOI] [PubMed] [Google Scholar]
- 61.Pias-Peleteiro J, Campos F, Castillo J, et al. Endothelial progenitor cells as a therapeutic option in intracerebral hemorrhage. Neural Regen Res 2017; 12: 558–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palacios EH, Weiss A. Function of the Src-family kinases, Lck and Fyn, in T-cell development and activation. Oncogene 2004; 23: 7990–8000. [DOI] [PubMed] [Google Scholar]
- 63.Mehndiratta P, Manjila S, Ostergard T, et al. Cerebral amyloid angiopathy-associated intracerebral hemorrhage: pathology and management. Neurosurg Focus 2012; 32: E7. [DOI] [PubMed] [Google Scholar]
- 64.Park L, Zhou J, Zhou P, et al. Innate immunity receptor CD36 promotes cerebral amyloid angiopathy. Proc Natl Acad Sci U S A 2013; 110: 3089–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao X, Sun G, Zhang J, et al. Hematoma resolution as a target for intracerebral hemorrhage treatment: role for peroxisome proliferator-activated receptor gamma in microglia/macrophages. Ann Neurol 2007; 61: 352–362. [DOI] [PubMed] [Google Scholar]
- 66.Zhao XR, Gonzales N, Aronowski J. Pleiotropic role of PPARgamma in intracerebral hemorrhage: an intricate system involving Nrf2, RXR, and NF-kappaB. CNS Neurosci Ther 2015; 21: 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moisan E, Girard D. Cell surface expression of intermediate filament proteins vimentin and lamin B1 in human neutrophil spontaneous apoptosis. J Leukoc Biol 2006; 79: 489–498. [DOI] [PubMed] [Google Scholar]
- 68.Boilard E, Bourgoin SG, Bernatchez C, et al. Identification of an autoantigen on the surface of apoptotic human T cells as a new protein interacting with inflammatory group IIA phospholipase A2. Blood 2003; 102: 2901–2909. [DOI] [PubMed] [Google Scholar]
- 69.Mor-Vaknin N, Punturieri A, Sitwala K, et al. Vimentin is secreted by activated macrophages. Nat Cell Biol 2003; 5: 59–63. [DOI] [PubMed] [Google Scholar]
- 70.Xu B, deWaal RM, Mor-Vaknin N, et al. The endothelial cell-specific antibody PAL-E identifies a secreted form of vimentin in the blood vasculature. Mol Cell Biol 2004; 24: 9198–9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Podor TJ, Singh D, Chindemi P, et al. Vimentin exposed on activated platelets and platelet microparticles localizes vitronectin and plasminogen activator inhibitor complexes on their surface. J Biol Chem 2002; 277: 7529–7539. [DOI] [PubMed] [Google Scholar]
- 72.Yang J, Zou L, Yang Y, et al. Superficial vimentin mediates DENV-2 infection of vascular endothelial cells. Sci Rep 2016; 6: 38372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kindy MS, Bhat AN, Bhat NR. Transient ischemia stimulates glial fibrillary acid protein and vimentin gene expression in the gerbil neocortex, striatum and hippocampus. Brain Res Mol Brain Res 1992; 13: 199–206. [DOI] [PubMed] [Google Scholar]
- 74.Xiong H, Callaghan D, Jones A, et al. ABCG2 is upregulated in Alzheimer’s brain with cerebral amyloid angiopathy and may act as a gatekeeper at the blood-brain barrier for Abeta(1-40) peptides. J Neurosci 2009; 29: 5463–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Levin EC, Acharya NK, Sedeyn JC, et al. Neuronal expression of vimentin in the Alzheimer’s disease brain may be part of a generalized dendritic damage-response mechanism. Brain Res 2009; 1298: 194–207. [DOI] [PubMed] [Google Scholar]
- 76.Fujimura M, Bang OY, Kim JS. Moyamoya disease. Front Neurol Neurosci 2016; 40: 204–220. [DOI] [PubMed] [Google Scholar]
- 77.Hu J, Ni S, Cao Y, et al. The angiogenic effect of microRNA-21 targeting TIMP3 through the regulation of MMP2 and MMP9. PLoS One 2016; 11: e0149537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xie W, Li M, Xu N, et al. MiR-181a regulates inflammation responses in monocytes and macrophages. PLoS One 2013; 8: e58639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol 2016; 16: 279–294. [DOI] [PubMed] [Google Scholar]
- 80.Murugaiyan G, Garo LP, Weiner HL. MicroRNA-21, T helper lineage and autoimmunity. Oncotarget 2015; 6: 9644–9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Landskroner-Eiger S, Moneke I, Sessa WC. miRNAs as modulators of angiogenesis. Cold Spring Harb Perspect Med 2013; 3: a006643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li QJ, Chau J, Ebert PJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 2007; 129: 147–161. [DOI] [PubMed] [Google Scholar]
- 83.Ouyang YB, Lu Y, Yue S, et al. miR-181 regulates GRP78 and influences outcome from cerebral ischemia in vitro and in vivo. Neurobiol Dis 2012; 45: 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material for The intracerebral hemorrhage blood transcriptome in humans differs from the ischemic stroke and vascular risk factor control blood transcriptomes by Boryana Stamova, Bradley P Ander, Glen Jickling, Farah Hamade, Marc Durocher, Xinhua Zhan, Da Zhi Liu, Xiyuan Cheng, Heather Hull, Alan Yee, Kwan Ng, Natasha Shroff and Frank R Sharp in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, Supplementary Tables for The intracerebral hemorrhage blood transcriptome in humans differs from the ischemic stroke and vascular risk factor control blood transcriptomes by Boryana Stamova, Bradley P Ander, Glen Jickling, Farah Hamade, Marc Durocher, Xinhua Zhan, Da Zhi Liu, Xiyuan Cheng, Heather Hull, Alan Yee, Kwan Ng, Natasha Shroff and Frank R Sharp in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, Supplementary Figures 1 to 13 for The intracerebral hemorrhage blood transcriptome in humans differs from the ischemic stroke and vascular risk factor control blood transcriptomes by Boryana Stamova, Bradley P Ander, Glen Jickling, Farah Hamade, Marc Durocher, Xinhua Zhan, Da Zhi Liu, Xiyuan Cheng, Heather Hull, Alan Yee, Kwan Ng, Natasha Shroff and Frank R Sharp in Journal of Cerebral Blood Flow & Metabolism