Abstract
Background
Expression patterns between males and females vary in every adult tissue, even in organs with no conspicuous dimorphisms such as the heart. While studies of male and female differences have traditionally focused on the influence of sex hormones, these do not account for all the differences at the molecular and epigenetic levels. We previously reported that a substantial number of genes were differentially expressed in male and female mouse embryonic stem (ES) cells and revealed dose-dependent enhancer activity in response to Prdm14, a key pluripotency factor expressed more highly in female ES cells. In this work, we investigated the role of Prdm14 in establishing sex-specific gene expression networks. We surveyed the sex-specific landscape in early embryogenesis with special reference to cardiac development. We generated sex-specific co-expression networks from mouse ES cells, examined the presence of sex-specific chromatin domains, and analyzed previously published datasets from different developmental time points to characterize how sex-biased gene expression waxes and wanes to evaluate whether sex-biased networks are detectable throughout heart development.
Results
We performed ChIP-seq on male and female mouse ES cells to determine differences in chromatin status. Our study reveals sex-biased histone modifications, underscoring the potential for the sex chromosome complement to prime the genome differently in early development with consequences for later expression biases. Upon differentiation of ES cells to cardiac precursors, we found sex-biased expression of key transcription and epigenetic factors, some of which persisted from the undifferentiated state. Using network analyses, we also found that Prdm14 plays a prominent role in regulating a subset of dimorphic expression patterns. To determine whether sex-biased expression is present throughout cardiogenesis, we re-analyzed data from two published studies that sampled the transcriptomes of mouse hearts from 8.5 days post-coitum embryos to neonates and adults. We found sex-biased expression at every stage in heart development, and interestingly, identified a subset of genes that exhibit the same bias across multiple cardiogenic stages.
Conclusions
Overall, our results support the existence of sexually dimorphic gene expression profiles and regulatory networks at every stage of cardiac development, some of which may be established in early embryogenesis and epigenetically perpetuated.
Electronic supplementary material
The online version of this article (10.1186/s13293-019-0259-1) contains supplementary material, which is available to authorized users.
Keywords: Sex chromosomes, Regulatory networks, Cardiogenesis, Sexual dimorphism, Differentiation
Background
It has long been acknowledged that clinical presentation of cardiovascular disease differs between men and women. Even in healthy adults, there are baseline sex differences in cardiovascular structure and function [1]. With the advent of sensitive sequencing technologies, a surprising amount of transcriptional and epigenomic variability has recently been shown between men and women in most adult tissues, including the heart [2–5]. Detailed studies in cardiomyocytes in humans, rats, and mice have also revealed sexual dimorphisms in transcriptome and function [6]. Most of these differences have been attributed to hormonal factors, yet results from many studies have shown that pathways other than hormones play an important role [7, 8]. For example, sex chromosomes contribute independently to sex biases in gene expression, although the specific sex chromosome-linked genes and their downstream targets have not been elucidated.
Genetic and epigenetic factors involved in normal cardiac development have been extensively characterized [9–14] and the transcriptional networks vital for cardiogenesis are well established. Generally, there has been no expectation in the developmental field that sex is relevant to early embryonic processes. Yet many congenital heart defects exhibit sex biases in presentation, mortality, and morbidity [15] and are primarily due to disruptions occurring before gonad formation. Furthermore, gestational insults, such as maternal undernutrition, are associated with sex-specific alterations in fetal heart development [16]. These imbalances have not been explained at either the genetic or developmental level and indicate that sex is an important biological variable during early embryogenesis.
In fact, sex-specific expression differences in early embryogenesis are widespread across the animal kingdom. Recent studies in non-mammalian model organisms have reported sex-biased expression at stages in which visible phenotypic differences between the sexes have not yet become apparent [17]. However, the question of whether this is also true of mammals has rarely been addressed.
One exception is the growing body of reports on mouse embryonic stem (ES) cells, which are self-renewing, pluripotent derivatives from pre-implantation embryos. Understanding the gene networks that control ES cells has been a major focus for many years [18–20] and recently, a surprising amount of sexual dimorphism in gene expression has been revealed in ES cells in both mice and humans [21–24]. Some expression differences were expected due to the presence of two active X chromosomes in females versus one in male cells. However, the majority of biases emanate from autosomal genes, including genes encoding dose-dependent transcription factors (TFs) and epigenetic and remodeling enzymes (EREs). This suggests that sex-specific gene networks are established by the sex chromosomes before X chromosome inactivation (XCI) occurs in female cells. Sophisticated network analyses have provided insight into the biology of organ development and can be deployed on the available data to address this possibility.
Evidence supports that sex-biased expression of regulatory factors in early embryogenesis establishes sex-specific epigenomic landscapes. Yet whether these differences are reversed by dosage compensation or are perpetuated during embryogenesis, with consequences for organogenesis and beyond, is unknown. Thus, it is necessary to characterize male and female transcriptomes over ontogeny in mammalian systems and to determine if they are connected to later adult phenotypes.
Here, we propose that the sex-biased expression of certain TFs and EREs in early development marks the genome with lasting effects across the lifespan [25]. We posit that while lineage specification decreases the range of sex-biased gene expression, sex-specific epigenetic marks persist and result in differential expression at later developmental stages [25]. We characterize candidates for these effects by elucidating co-expression and protein-protein interaction networks underlying the sex biases in male and female mouse ES cells. Our results highlight a co-expression module that is highly correlated with sex chromosome composition and identifies Prdm14, a sex-biased gene with higher expression in female ES cells, as a key regulator of sex biases in ES cells. Using heart development as a model process, we report sex-biased expression in male and female ES cells differentiated to cardiac precursors, in in vivo embryonic hearts and in adult cardiomyocytes. By focusing on transcriptional and epigenetic factors, we identify a subset of sex differences established in early embryogenesis that persist throughout lineage determination and cardiac organogenesis. Furthermore, we find evidence that Prdm14 regulates target genes that are sex-biased during cardiac development and, surprisingly, in the adult heart, when Prdm14 is no longer expressed.
Methods
Construction of weighted gene co-expression network and modules
We used a previously published RNA-sequencing (RNA-seq) dataset from six male (40, XY) and six female (40, XX) mouse ES cell lines (GSE90516) for network analysis [24]. We derived these cell lines from independent F1 hybrid blastocysts resulting from reciprocal crosses of mouse substrains C57BL/6 and CAST/EiJ by natural mating. Each cell line was maintained in ES cell culture medium (DMEM, 15% fetal calf serum, 1 mM sodium pyruvate, 2 mM l-glutamine, 1% non-essential aminoacids, 0.1 mM 2-mercaptoethanol and 1000 U/ml leukemia inhibitory factor) in 5% CO2 at 37°. The data was generated using HiSeq 2500 single end reads of 50 base pairs. We reported hundreds of coding and non-coding RNAs that were differentially expressed between male and female ES cell lines, after filtering for strain-specific effects [24].
To prevent bias, all aligned transcripts were used to establish a weighted gene co-expression network analysis (WGCNA), a widely used systems biology method that employs gene expression data to construct a scale-free network [26]. The WGCNA package in R, version 1.6, is available at https://cran.r-project.org/web/packages/WGCNA/index.html. For the analysis herein, Pearson’s correlation matrices were calculated for all pairs of genes evaluating the correlation coefficient between gene m and gene n such that Smn = |cor(m,n)|. Next, the Pearson’s correlation matrices were transformed into matrices defining connection strengths using the power function amn = power(Smn,β) = |Smn|β. In doing so, strong correlations are emphasized and the influence of weak correlation is reduced on an exponential scale. To obtain a scale-free network, we performed network topology analysis for thresholding powers from 1 to 20. The lowest power value for scale-free topology was 10, so β was set at 10.
The connectivity of pairs of genes was evaluated by calculating topology overlap (TO). TO is a robust indicator of the relationships among neighborhoods of genes. The TO was then used to perform hierarchical average linkage clustering to identify gene co-expression modules. Modules are branches of a hierarchical cluster tree defined using the top-down dynamic tree cut method [27] with a minimum module size of 50 genes. After module identification, a t test was used to calculate the p value of candidate genes. The gene significance (GS) was defined as the mediated p value of each gene (GS = lgP). From this, the module significance (MS) was defined using the average GS from all the genes within said module.
Transcription factor motif analysis
The set of genes within the module most highly correlated with cell sex, comprised of 1624 genes, was analyzed for known and de novo transcription factor motif binding sites. The parameters were set to cover the promoter using − 5000 to + 1000 bp of the transcriptional start site in the HOMER online software suite (http://homer.ucsd.edu/homer/) [28].
Ingenuity pathway analysis
We analyzed gene sets using the ingenuity systems pathways analysis (IPA) tool (Qiagen; Redwood City, CA). Datasets were subjected to IPA Core Analysis and then analyzed using IPA Upstream Regulator, Downstream Effects, and Canonical Pathways analytic tools. To capture regulatory networks, we focused on transcription factors and epigenetic and remodeling enzymes. The IPA output was exported as Microsoft Excel files to prepare the supplemental tables.
Chromatin immunoprecipitation and sequencing
Four low-passage (p7-9) independent mouse ES cell lines, two male lines (40, XY) and two female lines (40, XX), were grown on inactivated C57BL/6 mouse embryonic fibroblasts (MEFs). The MEFs are prepared from pooled embryos and include both male and female cells. ES cells were passaged at least twice prior to harvest to achieve high cell numbers for chromatin immunoprecipitation and sequencing (ChIP-seq). Cells were collected using 0.25% Trypsin + EDTA and MEF-depleted for 1 h at 37 °C in 5% carbon dioxide. After collecting the ES cells, residual MEFs were less than 1.5% of the final cell suspension. Because of their low numbers and the fact that they are a mixed population of male and female cells, any remaining MEFs are not expected to skew the results obtained from the ES cells. ES cells were cross-linked using formaldehyde at a final concentration of 1% followed by quenching with 1 M glycine. Sonication, immunoprecipitation, library construction, and sequencing were performed as previously described with minor modifications [29]. Briefly, three consecutive lysis buffers were used to ensure adequate nuclei release. Sonication was performed in a Q-Sonica cup horn sonication system using amplitude 70 with 30 s ON/OFF cycles for 10–15 min dependent on desired size and sonication efficiency of each sample. Samples were sonicated to a range of 100–500 base pairs. Sonicated chromatin was diluted in immunoprecipitation buffer. Two million cells were used for each immunoprecipitation (IP), with five consecutive IPs for each histone modification of interest. Ten percent of the initial sample volume per IP was set aside to serve as input control prior to addition of the appropriate antibody. Additional file 1: Table S1 provides the specifics for the antibodies used, with 2.5 μg of each antibody per IP. To isolate the antibody-bound fragments of interest, we used 50 μl of a 50/50 mix of Dynabeads TM Protein A (catalog # 10002D, lot # 00448844) and Protein G (catalog # 10003D, lot # 00486042) and an overnight incubation at 4 °C.
Beads were washed with RIPA buffer for five consecutive washes followed by a single wash with Tris-EDTA buffer. Complexes were eluted from the beads with 50 mM Tris, 10 mM EDTA, 1% (w/v) SDS, pH 8.0, and crosslinks were reversed. Concentration of resultant DNA was determined using Qubit according to manufacturer’s protocol. ChIP-seq libraries were prepared using DNA SMARTTM ChIP-Seq Kit. Sequencing was performed using Illumina HiSeq 2500 generating single end reads of 50 base pairs. Sequences were aligned to mouse genome assembly (mm9) using Bowtie2 v2.1.0 with default settings [30]. Using the Bedtools software suite for genome arithmetic [31], we determined the degree of enrichment of reads genome-wide. For data visualization, we used R software suite. To visualize enrichment patterns of histone modifications at promoters and enhancers, we used ngs.plot [32], using enhancer annotations from a previous report [33].
Protein-protein interaction networks
Protein-protein interaction networks (PPIs) were constructed with the STRING database using all differentially expressed genes between male and female ES cells (STRING version 10.5 [34]). To improve the quality of the resulting network, the “minimum required score” option was set to 0.7 and “text mining resources” were ignored. Analysis and graphing of the networks were performed in the Gephi software (version 0.9.2) [35]. Functional modules were detected using an algorithm to partition the network into communities of densely connected nodes [36]. Gene ontology (GO) analysis was performed using the ClueGO plug-in from Cytoscape [37]. GO terms were summarized using the REVIGO website (http://revigo.irg.hr/revigo.jsp) [38]. Network topology analysis and selection of important genes were done as previously described [39, 40].
ES cell differentiation
Two of the male and female ES cell lines from which ChIP-seq was performed were subjected to a standardized differentiation protocol that directs the stepwise differentiation of early embryonic cells into cardiac precursors [41], as assayed by marker gene analysis. Cells were cultured with leukemia inhibitory factor (LIF) on mouse embryonic fibroblasts (MEFs). Before differentiation, ES cells were dissociated, MEFs were eliminated as detailed above, and embryoid bodies were derived by hanging drop culture in medium without LIF. After 4 days, embryoid bodies were harvested and grown in medium containing Activin A, BMP4, and VEGF as monolayers until beating foci were observed. This optimized protocol yields > 75% cardiomyocytes [41]. At day 13 of initial LIF withdrawal, we picked beating foci from the plates and obtained RNA.
qPCR was performed to determine expression of pluripotency markers Nanog and Oct4 and cardiomyocyte markers Myh6 and Tnnt2 on cDNA generated using SuperScriptTM II (Invitrogen) and relative expression was assessed using PowerUp SYBR Green Master Mix (Thermo Fisher) and normalized to β-actin on the Applied Biosystems StepOnePlus Real-Time PCR System. RNA-seq was performed as previously described [24].
Meta-analysis of publicly available data
We leveraged existing expression datasets from across cardiac development in the mouse and stratified the data by sex when necessary. These collated data allowed us to determine whether there is dynamic sex-biased expression across cardiogenesis. Additional file 2: Table S2 details all the datasets examined herein.
Single cell data for 8.5, 9.5, and 10.5 days post coitum (dpc) embryonic [42] and neonatal mouse hearts [43] were downloaded and processed as follows: (1) if the fragments per kilobase of exon per million reads mapped (FPKM) was < 1, the gene was designated as not expressed; (2) genes with zero variance across all cells were removed. Cells were then sexed by determining the ratio of Xist to Eif2s3y, two oppositely biased genes, on a cell-by-cell basis. Cells with Xist/Eif2s3y ratios of at least 1.5 were considered as female and ratios below 1 were taken as male. t test analysis was performed on the samples from each stage, p values were used to calculate the false discovery rate (FDR), and genes with adjusted p value < 0.05 were selected as differentially expressed genes. Data for adult mouse hearts was already stratified by sex [44].
Transcription factor binding analysis
To detect recognition motifs of candidate transcription factors (TFs) in genes enriched in male or female cells, we used the genome-wide position matrix scanner from the Computational Cancer Genomics website (https://ccg.epfl.ch/pwmtools/pwmscan.php) with the JASPAR core vertebrate motif library (version 2018). We searched for Lef1 MA0768.1 and Zeb1 MA0103.3 motifs with a p value cutoff of 0.00001 with the Contra v3 tool (http://bioit2.irc.ugent.be/contra/v3/#/step/1) and uploaded the results as custom tracks in the UCSC browser.
Results
Defining a gene network associated with sex-biased gene expression
There is a substantial number of differentially expressed genes in male (40, XY) and female (40, XX) mouse embryonic stem (ES) cells, including transcription factors (TFs), and epigenetic and remodeling enzymes (EREs) [22–24]. Yet both male and female ES cells are pluripotent and can contribute to normal development. Thus, while overall pluripotency networks govern both XX and XY ES cells, we hypothesized that differentially expressed genes can shift network architecture or constitute subnetworks with distinct gene-gene correlations.
To determine whether the genes differentially expressed in XX and XY ES cells constitute sex-specific co-expression networks and identify genes with higher connectivity in each sex, we used normalized RNA-sequencing (RNA-seq) data from six male and six female mouse ES cell lines to perform weighted gene co-expression network analysis (WGCNA) [24, 27, 45] (see “Methods” section). Weighted gene co-expression network analysis allows partitioning of genes into modules that correlate with biological function and identifies the genes that are most likely to be crucial in regulating that function. WGCNA has been successfully applied to dissect the role of hormonal and sex chromosome effects in sex-biased co-expression networks in adult tissues [46].
Figure 1 shows the clustering dendrogram of co-expressed genes resulting from the WGCNA with the lowest power value for scale-free topology, β, set at 10. To avoid bias by pre-selecting genes with differential expression levels in male and female ES cells, we based the clustering on all aligned transcripts. Genes with similar patterns of expression were grouped into modules by hierarchical average linkage clustering using topological overlap [26]. The initial dynamic tree cut was further merged, to generate a subset of 11 distinct co-expression modules.
Fig. 1.
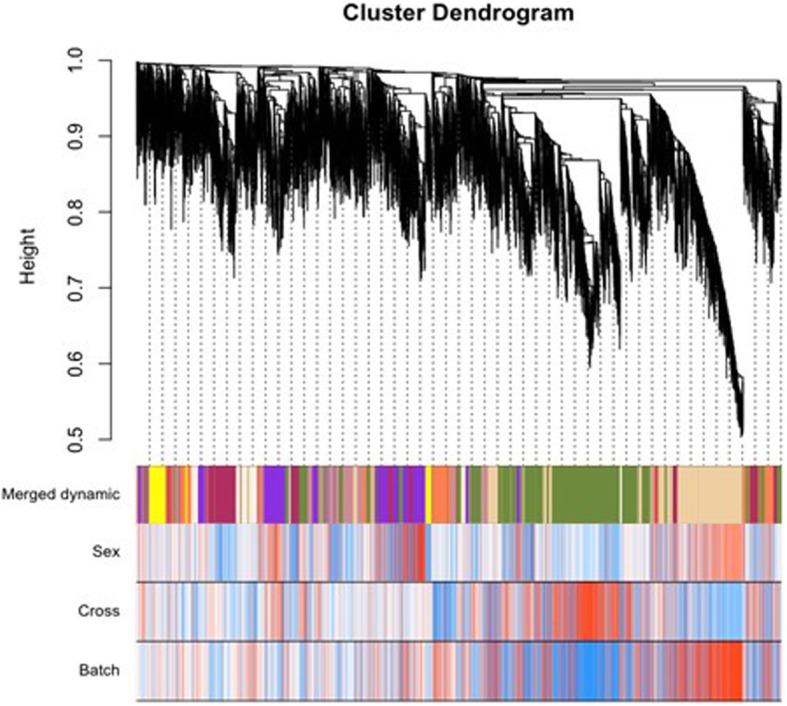
Weighted gene co-expression network analysis (WGCNA) for male and female ES cells. Expression modules were identified by weighted gene co-expression network analysis. Gene dendrograms display the co-expression modules identified by WGCNA from expression data from 6 male and 6 female ES cell lines and labeled by different colors. Dendrograms were generated by unsupervised hierarchical clustering of genes using topological overlap to identify co-expressed genes in modules. The significantly preserved modules are denoted by the striped colors in the bars below the dendrogram along the x-axis, referred to as the merged dynamic. The bars below the merged dynamic express correlation with sex, cross and RNA-seq batch. The y-axis shows the heights where the clusters merged
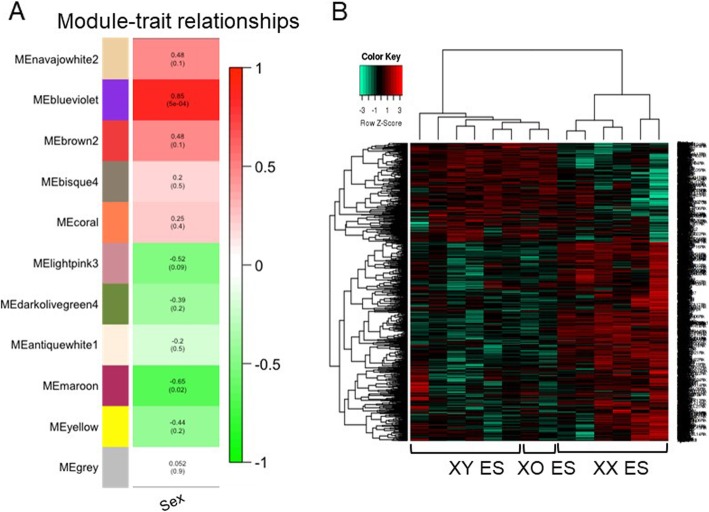
The first principal component of a given module is the module eigengene (ME), which represents the gene expression profile within that particular module. To understand the functional significance of the modules, we correlated the 11 MEs generated within the clustering dendrogram with traits of interest and isolated the most significant associations (Fig. 2a). According to the heatmap of module-trait correlations, sex showed a strong and independent association with a particular eigengene, the ME blue/violet (r = 0.85, p = 5e-04) and consisted of 1624 genes, including 84 TFs and 43 EREs (Additional file 3: Dataset S1).
Fig. 2.
Relationships of consensus modules (module eigengenes) with sex. a Each row in the table corresponds to a consensus module identified by distinct colors along the left y-axis. Each module eigengene (ME) was evaluated in relationship to sex. Numbers in the table report the correlation of the corresponding ME with sex, with the p values shown in parentheses. The degree of correlation, positive and negative is provided by the colored scale on the right y-axis. b Clustering of the mouse ES cell lines based upon the module eigengene, blue/violet. Heatmap showing separation of the lines by sex chromosome complement (XY, male; XX, female; XO, X chromosome monosomic) when the 1624 genes, contained within the blue/violet module from WGCNA were evaluated
To validate the gene cluster with a separate method, we produced a hierarchical clustering heatmap using the expression levels of the 1624 genes in the blue/violet module. Input of the genes contained within the blue/violet module into this separate pipeline did indeed show separation of the mouse ES cell lines by sex (Fig. 2b).
Distinct upstream regulators are associated with sex-biased functional pathways
To identify regulatory pathways for the genes in the blue/violet module (Fig. 2), i.e., the module best correlated with sex, we performed ingenuity pathway analysis independently on the XX- and XY-enriched TFs and EREs (Additional file 4: Dataset S2). We found that the top pathway for XX-enriched TFs and EREs was “DNA methylation and transcriptional repression” (p = 7.81 e−4), with Max and Mycn as the top upstream regulatory molecules. Analysis of the XY-enriched TFs and EREs from the blue/violet module identified “Jak1 in Interferon Signaling” as the top pathway (p = 2 e−3). Top upstream regulators were predicted to be Irf9 and Npc1.
Prdm14 motifs are enriched in promoters of sex-biased genes
We asked if the sex-biased clustering of the blue/violet genes was being driven by specific transcription factors and reflected sex-specific regulatory networks. To test this, we used HOMER to identify known transcription factor binding sites within the gene set in the blue/violet ME [28].
HOMER motif analysis yielded significantly enriched TF motifs in the promoters of genes in the blue/violet module eigengene (Table 1). The transcription factor TEAD (TEA/ATTS domain) was the top and most significantly enriched motif (p value 1e-15). TEAD proteins are pivotal transcription factors implicated in development as well as in cancer [47]. Leukemia inhibitory factor, present in the culture medium, activates the Yes-associated protein (YAP) and TEA domain TEAD2 transcription factor pathway, which contributes to mouse ES cell maintenance of pluripotency and self-renewal. The stem cell factors Nanog and Oct3/4 are targets of the TEAD-pathway [48]. These pluripotency factors had similar expression levels among all male and female ES cell lines tested, and TEAD was not differentially expressed at the RNA level. However, it was previously reported that Tead1 and Tead2 are male-biased at the protein level [23]. Thus, further investigation is required to determine if these factors contribute to the sex-specific effects or whether they appear with the HOMER analysis due to their contribution to pluripotency per se.
Table 1.
HOMER motif analysis of the promoters of genes in the blue/violet module eigengene
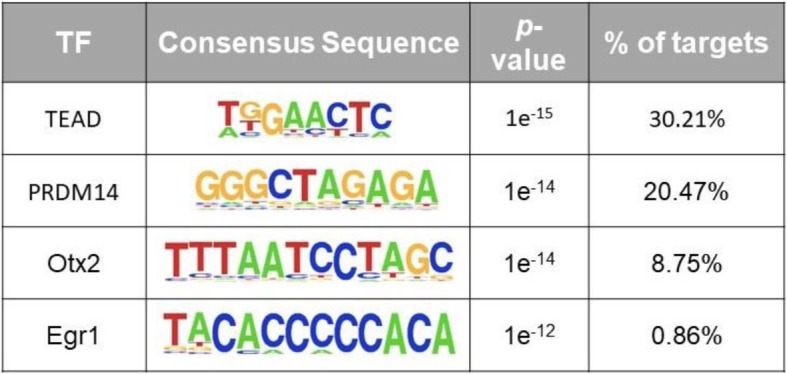
Genomic regions for − 5000 bp to + 1000 bp around the transcriptional start site were pulled for each of the 1624 genes contained within the blue/violet module eigengene and input into HOMER. Shown here are the four top hits and corresponding p values and percentage of coverage within target sequences
Interestingly, Prdm14 is a top hit and the second highest hit in HOMER (Table 1). Prdm14 is expressed more highly in female (XX) than in male (XY) ES cells, a bias that occurs independently of whether the ES cells are cultured in LIF/serum or 2i [22, 24] and is also seen at the protein level [23]. Prdm14 is a bi-functional TF with a cardinal role in ES cell pluripotency and in the establishment of primordial germ cells. Prdm14 can either activate or repress gene expression, depending on its interacting partners [49]. Recruitment of polycomb repressive complex 2 (PRC2) by Prdm14 results in transcriptional repression, whereas cooperation with estrogen-related receptor β (Esrrβ) activates target gene expression. However, the mechanisms by which Prdm14 selectively partners with its alternative co-factors, resulting in gene activation or repression, are not understood. Nevertheless, Prdm14 is a strong candidate for regulating gene expression differentially in male and female ES cells and establishing sex-biased epigenetic marks.
Prdm14 target genes encoding TFs have sex-biased expression
To identify downstream targets of Prdm14, we curated and compared publicly available expression profiles of ES cells depleted of Prdm14, focusing on TFs and EREs. Several studies have reported Prdm14 knockout or knockdown in ES cells, with inconsistent results, likely due to varying culture conditions, strains, and karyotypes [50–52]. Therefore, we focused on a report with siRNA-mediated knockdown of Prdm14 in wild-type female 129/Ola ES cells, with the caveat that culture conditions were 2i (versus LIF/serum in our lab) [50].
ES cells depleted of Prdm14 have a more “male-like” expression pattern, with upregulation of Foxi3, Sox11, Gata4, Dnmt3a, and Dnmt3l, which are highly expressed in wild-type male ES cells. Genes that are downregulated in Prdm14-depleted female ES cells, such as Mitf, Zeb1, and Prdm14 itself, are enriched in wild-type female ES cells. More than 10% of genes followed this pattern. This confirms that Prdm14 regulates a subset of genes, while also indicating that there are other factors involved in sex-biased expression.
Male and female ES cells exhibit sex biases in chromatin modifications
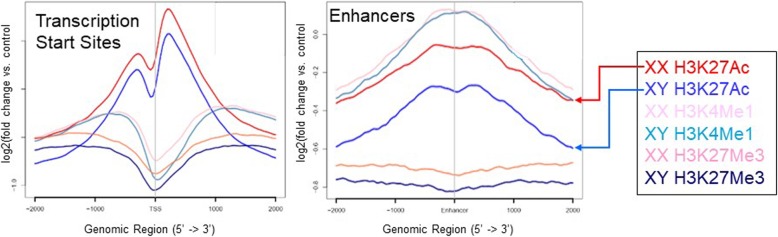
To determine if the differential transcriptomes between XX and XY ES cells are reflected in the chromatin structure, we performed chromatin immunoprecipitation and sequencing (ChIP-seq) on six early passage independent ES cell lines of each sex, i.e., the same cell lines for which we had reported sex-biased expression [24]. Antibodies against histone modifications H3K4Me1, H3K27Me3, and H3K27Ac were used to precipitate chromatin substrates with our standard protocol. The presence of H3K27Ac, indicating active chromatin, showed a significant difference between XX and XY ES cells at known enhancer regions (Fig. 3). This suggests that the main biases between XX and XY ES cells are established by TFs and EREs that bind and modify enhancer sequences.
Fig. 3.
Sex-biased chromatin modifications at regulatory sequences in ES cells. ChIP-Seq results on two XX (red, light pink, dark pink lines) and XY (blue, teal, and dark blue lines) ES cell lines for H3K27Ac, H3K27Me3, and H3K4Me1, respectively, are shown. IgG served as a control. NGS-plot was used to evaluate the enrichment of the histone modifications at transcriptional start sites and known enhancers. The plots depict the average profile of histone modifications at regions of interest, providing a quantitative view of the patterns for each ES cell line
To determine whether there was concordance between Prdm14 binding, gene expression biases, and differential chromatin modifications, we integrated available Prdm14 ChIP-seq data in ES cells [50] with our sex-specific chromatin studies for genes that respond to Prdm14 according to the knockdown studies.
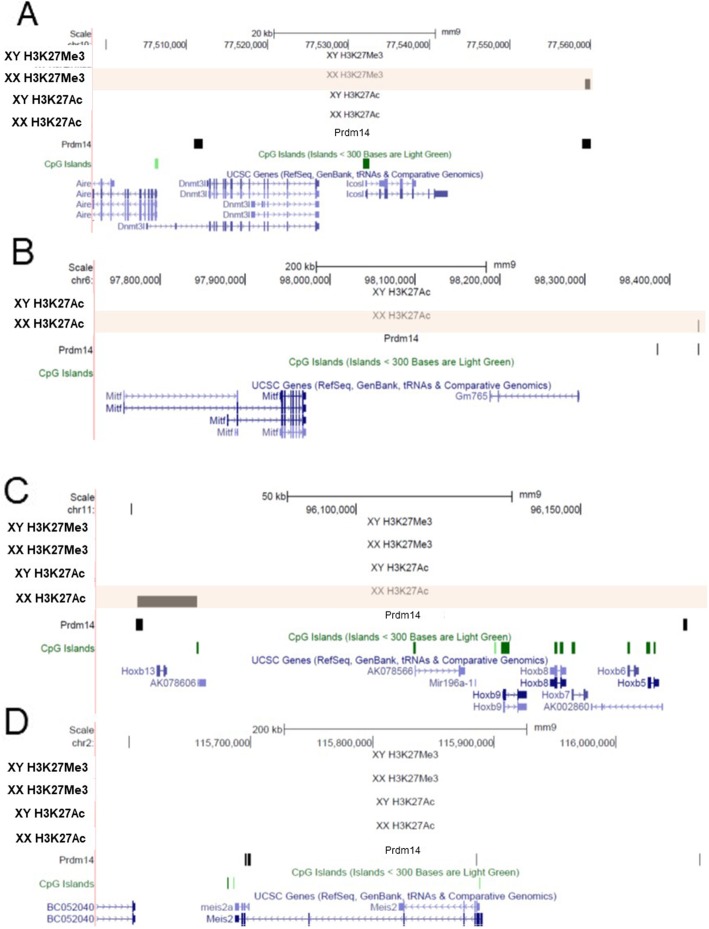
Our analysis identified three groups of differentially expressed genes: (1) genes that exhibited sex-biased chromatin modifications and Prdm14 binding, (2) genes with Prdm14 occupancy and no sex-specific histone modifications, and (3) genes with neither detectable sex-biased chromatin modification nor Prdm14 occupancy. For example, Dnmt3l, more highly expressed in male ES cells, has a Prdm14 binding site 40 kb downstream of the transcription start site, which is enriched in H3K27Me3, a repressive mark, in female ES cells (Fig. 4). One of the Prdm14 binding sites downstream of Mitf, more highly expressed in XX ES cells, has enrichment of H3K27Ac in those cells. Hoxb9 shows a similar pattern, with a Prdm14 binding site enriched in H3K27Ac in female ES cells which have higher expression. On the other hand, there are several Prdm14 binding sites upstream and in the promoter of Meis2, but we did not detect differential histone modifications in male and female ES cells, although it is more highly expressed in female cells. Genes such as Sohlh2 do not have apparent Prdm14 binding in their vicinity, indicating that they are regulated by other, as yet unknown TFs.
Fig. 4.
Differential H3K27Ac and H3K27Me3 enrichment in male and female ES cells. UCSC browser screen shots are shown with tracks denoting chromatin status designated as XY or XX. Black bars indicate presence of an enriched mark or Prdm14 binding in the corresponding track. Prdm14 occupancy track in ES cells was obtained from Ma et al. Browser shots for a Dnmt3l, b Mitf, c Hoxb9, and d Meis2
Protein-protein interaction network analysis of ES cell transcriptomes reveals overlap with Prdm14 target genes
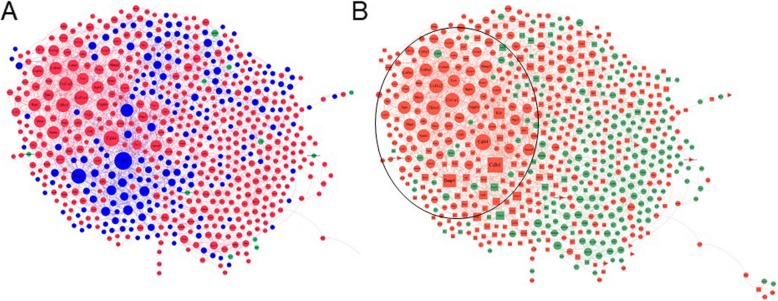
Data from the differentially expressed genes in male and female ES cells were used to construct a protein-protein interaction (PPI) network (Fig. 5a). Overlaying the information from the sex differences in gene expression shows that there are sex-biased modules within the global interaction network. We compared the genes from the blue/violet module eigengene from the WGCNA to the nodes of the PPI network. Two hundred twenty-five genes were shared between them (green nodes in Fig. 5b). Analysis of the network revealed six modules (Additional file 6: Figure S1), one of which contained the most important nodes based on topological analysis (degree, betweenness, and closeness centrality metrics in Additional file 5: Dataset 3). GO analysis of this module showed “blood vessel morphogenesis” and “Bmp signaling” as top terms (Additional file 7: Dataset S4).
Fig. 5.
Protein-protein interaction networks. a PPIs were constructed from differentially expressed genes from male and female ES cells. The networks includes sex-biased modules highlighted by red (female-enriched) and blue (male-enriched) nodes. b PPI network compared to genes in the blue/violet module from the WGCNA analysis. The most important module (based on topological analysis) is encircled. The common genes are green, unique genes are orange; squares represent male-biased and circles female-biased genes
Prdm14 was contained in the module with the most important nodes and showed connections to Dazl, Tcl1, Wnt3, Cdx2, Dnmt3b, Prdm6, Bmp4, Lin28a, Lefty2, T, and Gata4. Strikingly, many of the nodes in this PPI module, such as Zeb1, Lefty1, Gata4, Dusp6, and Sox11, are direct Prdm14 transcriptional targets in ES cells.
Male and female cardiac precursors also show sex biases in gene expression
Thus far, we showed that there are sex-specific expression and protein-protein interaction networks in ES cells. Upon differentiation of female ES cells, one of the two X chromosomes is inactivated, a massive epigenetic event that equalizes most of the X-linked genes between males and females. This transition mirrors the in vivo process of blastocyst implantation, during which female embryos undergo X chromosome inactivation (XCI).
To determine whether some sex-biased expression differences were perpetuated after XCI and during the beginning stages of lineage determination, we subjected two male and female ES cell lines to an optimized differentiation protocol to generate cardiac precursors and performed RNA-seq (Fig. 6). At day 13 after LIF withdrawal, ES cells have differentiated to cardiac precursors corresponding to 8.5–9.5 days post coitum (dpc) cardiac progenitors in vivo. RT-PCR confirmed that stem cell markers such as Nanog and Oct4 were downregulated, whereas markers of cardiac differentiation, such as Tnnt2 and Myh6, were upregulated in both sexes, as previously reported (Additional file 8: Figure S2) [10, 53].
Fig. 6.
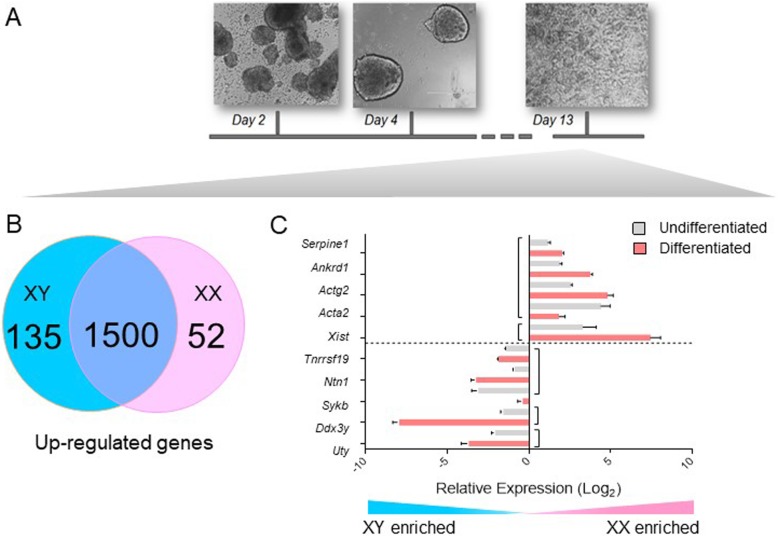
Differentiation of male and female ES cells into cardiac precursors. Top, images resulting from differentiation of ES cell lines according to standard protocol, with beating cardiac precursor cells at day 13 of LIF withdrawal. Below left, comparison of up-regulated genes between XX and XY ES cell lines indicating common and sex-specifically expressed RNAs (q < 0.01). Below right, expression of a subset of sex-biased genes expressed before and after differentiation of ES cells as analyzed by qRT-PCR in undifferentiated ES cells (gray) and derived cardiac precursors (coral). Error bars represent S.E.M. of duplicate experiments with three replicates each
We compared transcriptomes between differentiated male and female cell lines and found 157 genes that were differentially expressed at a FDR < 0.01 (Additional file 9: Dataset S5). The Xist non-coding RNA, which is involved in X chromosome inactivation, was more highly expressed in female cells, as expected. The male cells showed higher expression of 2 Y chromosome-linked genes, Ddx3y and Uty (Kdm6c). Interestingly, four TFs were more highly expressed in male cells, Ferd3l, Pou3f3, Six6, and St18. Ferd3l and Pou3f3 have nearby Prdm14 binding sites in undifferentiated ES cells, although we did not detect differential histone modifications in their vicinity (Additional file 10: Figure S3). Overall, this data shows that although the number of genes exhibiting sex differences diminishes during lineage determination, some biases persist. ChIP-seq data for cardiac precursors derived from male and female ES cells are needed to determine which epigenetic differences also persist after differentiation.
Sex biases in cardiac expression exist in early cardiac developmental stages in vivo
To elucidate how sex biases in gene expression vary during cardiac development, we collated and analyzed single cell transcriptional profiles from mouse embryonic hearts at 8.5, 9.5, and 10.5 dpc [42] and post-natal day 1 (p1) (Additional file 2: Table S2) [43]. Single cell data was downloaded and sexed (Additional file 11: Dataset S6). We found that there were hundreds of sex-biased genes at every stage. Some of these were stage-specific and some were common to two or more time points. For example, Lef1 was more highly expressed in male than female ES cells, and the same was true for 8.5 dpc and p1 hearts. Tbx20 was also enriched in male ES cells, cardiac precursors, and in 10.5 dpc and p1 hearts.
The majority of genes with sex-biased expression were male-biased at every stage. The number of female-enriched genes peaked dramatically at 9.5 dpc and decreased thereafter. At 8.5 dpc, only three X-linked genes, including Xist, were female-biased, whereas 19 X-linked genes were male-biased. At 10.5, eight X-linked genes were more highly expressed in females, including Xist, Tsix, and three genes that had not been characterized as escapees. More than 30 X-linked genes showed male-biased expression, indicating that some genes are not dosage compensated by X chromosome inactivation, at least at this stage in this tissue.
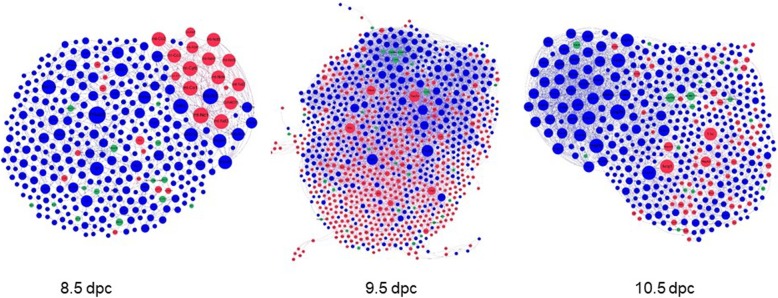
Protein-protein interaction networks were constructed with the sex-stratified expression data from 8.5, 9.5, and 10.5 dpc hearts (Fig. 7, Additional file 12: Figure S4, Additional file 5: Dataset S3). Sex biases in specific modules varied across the developmental stages, suggesting a highly dynamic but constant pattern of sexual dimorphism at the molecular level.
Fig. 7.
Protein-protein interaction networks in early cardiac development. PPIs were constructed from differentially expressed genes in 8.5, 9.5, and 10.5 dpc hearts as assayed by single-cell RNA-seq. Networks include sex-biased modules highlighted by red (female-enriched) and blue (male-enriched) nodes (based on data from Li, G. et al.)
Adult male and female hearts have sex-specific pathways
To explore whether there are expression differences in male and female adult C57BL/6 mouse hearts, we inspected recently published transcriptomic data across 17 tissues, stratified by sex [44]. Strikingly, 908 and 148 genes exhibited expression biases in adult male and female hearts, respectively, again showing that male-biased genes are more numerous. Interestingly, 38 X-linked genes were male-biased, suggesting male-specific regulation of these genes.
We investigated whether TFs that were sex-biased in adult hearts showed Prdm14 binding in ES cells. We found that Nkx2.5, Lef1, Id2, Ikzf3, and Srebf2 had Prdm14 occupancy in or near their promoter regions (Additional file 13: Figure S5), suggesting that their sex differences could have been established in early development. Differential histone modifications, however, were not apparent in ES cells in these regions.
We used Ingenuity Pathway Analysis to identify enrichment of biological network components in the sex-specific gene signatures in the adult heart. The top canonical pathways differed between male and female cardiac cells (Additional file 14: Datasets S7 and S8). Cardiovascular disease was the top disease association and cardiovascular system development and function was one of the top networks in importance for females, but surprisingly, cancer was the top disease association as well as the top network for males. Regulatory component analysis predicted distinct upstream regulatory factors for the male and female expression patterns. For example, Tp53, Nr3c2, and Tbx5 were among the top transcriptional regulators for female cells, whereas Ncor1 and Smad3 were identified for male cells.
Conserved sex-biased expression between mouse and human hearts
We compared sex-biased genes in adult heart ventricles between mouse and human. Differentially expressed genes between male and female human hearts were obtained from DeMeo et al., in which expression from the GTEx portal was stratified by sex [4]. There are 70 and 328 genes that are enriched in females and males, respectively, in both mouse and human (Additional file 15: Dataset 9). Among these are TFs Bhlhe40, Tcf15, Npas3, and Mafa, which are enriched in female hearts. Males show higher levels of Ehf, Etv1, Foxk1, Ikzf2, Meis2, and Tbx20, among others and of EREs Hat1, Cdyl, and Rad54l2.
There is sex-biased expression at specific developmental time points for important cardiac regulators
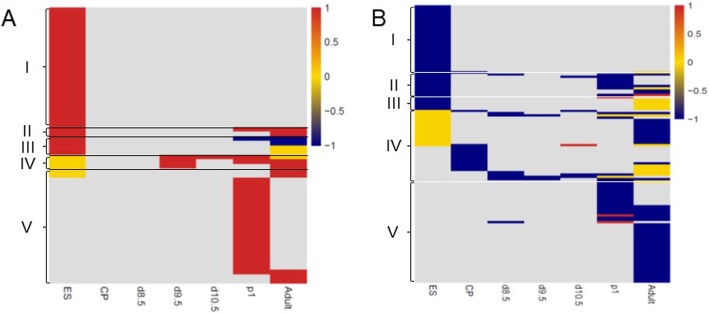
To query temporal changes in sex-biased expression profiles, we compared differential expression from ES cells (our data), embryonic and neonatal hearts, and adult cardiac myocytes [44]. Figure 8 and Tables 2 and 3 show sex-biased expression of TFs and EREs at each stage of heart development. Several different patterns can be visualized. Some genes encoding TFs and EREs are only expressed at one stage and others across several stages. For the latter group, there are subsets of genes that either maintain, acquire, lose, or even reverse their bias. A distinct group of genes, for example, Carhsp1 (male-biased) and Bhlhe40 (female-biased), display sex differences before gonad formation and the appearance of sex hormones. Our data also reveals sex disparities in expression that only become apparent in neonates and adults, suggesting that these respond, at least in part, to hormonal differences.
Fig. 8.
Expression of sex-biased transcription and epigenetic factors throughout development. Schematic heatmap representation of Tables 2 (a) and 3 (b) indicating expression and sex biases of transcription and epigenetic factors at each time point. Data was compiled from a female and b male ES cells, derived cardiac precursors (CP), hearts from 8.5, 9.5, and 10.5 days post coitum (dpc) embryos, neonates (p1), and adult mice (Ad). Each row is a specific transcription or epigenetic factor, with a total of 60 for females and 61 for males; the color denotes expression detected and enrichment in XX (red), XY (blue), or not biased (yellow). Group I: biased in ES cells, not expressed thereafter; groups II, III: biased in ES cells and same (II) or different (III) bias at other stages; group IV: biased after implantation but before gonadogenesis; group V: biased only after gonadogenesis
Table 2.
Female-biased expression of transcription and epigenetic factors
| Transcription factors and epigenetic remodelling enzymes | ES | CP | 8.5 | 9.5 | 10.5 | p1 | Adult | Chr | Name |
|---|---|---|---|---|---|---|---|---|---|
| Hmgb3 | XX | X | High mobility group box 3 | ||||||
| Dmrtb1 | XX | 4 | DMRT-like family B with proline-rich C-terminal, 1 | ||||||
| Hoxb9 | XX | 11 | Homeobox B9 | ||||||
| Hoxc8 | XX | 15 | Homeobox C8 | ||||||
| Klf8 | XX | X | Kruppel-like factor 8 | ||||||
| Mitf | XX | 6 | Melanogenesis associated transcription factor | ||||||
| Prdm14 | XX | 1 | PR domain containing 14 | ||||||
| Rhox1 | XX | X | Reproductive homeobox 1 | ||||||
| Sohlh2 | XX | 3 | Spermatogenesis and oogenesis specific basic helix-loop-helix 2 | ||||||
| Spic | XX | 13 | Serine (or cysteine) peptidase inhibitor, clade B, member 6c | ||||||
| Tbx15 | XX | 3 | T-box 15 | ||||||
| Zeb1 | XX | 18 | Zinc finger E-box binding homeobox 1 | ||||||
| Zfp182 | XX | X | Zinc finger protein 182 | ||||||
| Zfp275 | XX | X | Zinc finger protein 275 | ||||||
| Zfp449 | XX | X | Zinc finger protein 449 | ||||||
| Zfp59 | XX | 7 | Zinc finger protein 59 | ||||||
| Zxdb | XX | X | Zinc finger, X-linked, duplicated B | ||||||
| Mecp2 | XX | X | Methyl CpG binding protein 2 | ||||||
| Nkap | XX | X | NFKB activating protein | ||||||
| Ogt | XX | X | O-linked N-acetylglucosamine (GlcNAc) transferase | ||||||
| Top2b | XX | 14 | Topoisomerase (DNA) II beta | ||||||
| Trim16 | XX | 11 | Tripartite motif-containing 16 | ||||||
| Zmym3 | XX | X | Zinc finger, MYM-type 3 | ||||||
| Apobec2 | XX | 17 | Apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 | ||||||
| Baz2b | XX | 2 | Bromodomain adjacent to zinc finger domain, 2B | ||||||
| Prrx1 | XX | XX | 1 | Paired related homeobox 1 | |||||
| Kdm6a | XX | XX | XX | X | Lysine (K)-specific demethylase 6A | ||||
| Lbh | XX | XX | 17 | Limb-bud and heart | |||||
| Meis2 | XX | XY | 2 | Meis homeobox 2 | |||||
| Zfp9 | XX | XY | 6 | Zinc finger protein 9 | |||||
| Aff2 | XX | XX | nb | X | AF4/FMR2 family, member 2 | ||||
| Atrx | XX | XY | nb | X | ATRX, chromatin remodeler | ||||
| Arid1b | nb | XX | XX | nb | 17 | AT rich interactive domain 1B (SWI-like) | |||
| Bhlhe40 | nb | XX | XX | 6 | Basic helix-loop-helix family, member e40 | ||||
| Zfp51 | nb | XX | XX | 17 | Zinc finger protein 51 | ||||
| Klf15 | nb | XX | 6 | Kruppel-like factor 15 | |||||
| Mafk | nb | XX | XX | 5 | v-Maf musculoaponeurotic fibrosarcoma oncogene family, protein K (avian) | ||||
| Aff3 | nb | XX | XX | XX | XY | AF4/FMR2 Family Member 3 | |||
| Heyl | XX | 4 | Hairy/enhancer-of-split related with YRPW motif-like | ||||||
| Hopx | XX | 5 | HOP homeobox | ||||||
| Dlx1 | XX | 2 | Distal-less homeobox 1 | ||||||
| Rsl1 | XX | 13 | Regulator of sex limited protein 1 | ||||||
| Zbtb45 | XX | 7 | Zinc finger and BTB domain containing 45 | ||||||
| Zfp282 | XX | 6 | Zinc finger protein 282 | ||||||
| Zfp472 | XX | 17 | Zinc finger protein 472 | ||||||
| Zfp498 | XX | 5 | Zinc finger and SCAN domain containing 25 | ||||||
| Zfp758 | XX | 17 | Zinc finger protein 758 | ||||||
| Zkscan14 | XX | 5 | Zinc finger with KRAB and SCAN domains 14 | ||||||
| Zkscan6 | XX | 11 | Zinc finger with KRAB and SCAN domains 6 | ||||||
| Nat10 | XX | 2 | N-acetyltransferase 10 | ||||||
| Senp3 | XX | 11 | SUMO/sentrin specific peptidase 3 | ||||||
| Setmar | XX | 6 | SET domain without mariner transposase fusion | ||||||
| Sirt7 | XX | 11 | sirtuin 7 | ||||||
| Suv39h2 | XX | 2 | suppressor of variegation 3-9 2 | ||||||
| Bard1 | XX | 1 | BRCA1 associated RING domain 1 | ||||||
| Cbx2 | XX | 11 | Chromobox 2 | ||||||
| Cbx7 | XX | 15 | Chromobox 7 | ||||||
| Rcc1 | XX | 4 | Regulator of chromosome condensation 1 | ||||||
| Nkx2-5 | XX | XX | 17 | NK2 homeobox 5 | |||||
| Irf4 | XX | 13 | Interferon regulatory factor 4 | ||||||
| Cecr2 | XX | 6 | CECR2, histone acetyl-lysine reader |
ES embryonic stem cells; CP cardiac precursors; 8.5, 9.5, 10.5 days post coitum (dpc) embryonic hearts (single cell); p1 neonatal hearts; Chr chromosome; empty cells expression not detected; nb expressed but not biased
Table 3.
Male-biased expression of transcription and epigenetic factors
| Transcription factors and epigenetic and remodelling enzymes | ES | CP | 8.5 | 9.5 | 10.5 | p1 | Adult | Chr | Name |
|---|---|---|---|---|---|---|---|---|---|
| Egr4 | XY | 6 | Early growth response 4 | ||||||
| Cdx1 | XY | 18 | Caudal type homeobox 1 | ||||||
| E2f7 | XY | 10 | E2F transcription factor 7 | ||||||
| Eomes | XY | 9 | Eomesodermin | ||||||
| Evx1 | XY | 6 | Even-skipped homeobox 1 | ||||||
| Foxi3 | XY | 6 | Forkhead box I3 | ||||||
| Foxp4 | XY | 17 | Forkhead box P4 | ||||||
| Insm1 | XY | 2 | Insulinoma-associated 1 | ||||||
| Lin28a | XY | 4 | Lin-28 homolog A (C. elegans) | ||||||
| Mesp1 | XY | 7 | Mesoderm posterior 1 | ||||||
| Mixl1 | XY | 1 | Mix1 homeobox-like 1 (Xenopus laevis) | ||||||
| Nr6a1 | XY | 2 | Nuclear receptor subfamily 5, group A, member 1 | ||||||
| Pou2f3 | XY | 9 | POU domain, class 2, transcription factor 3 | ||||||
| Sox11 | XY | 12 | SRY (sex determining region Y)-box 11 | ||||||
| Sp5 | XY | 9 | Per-hexamer repeat gene 4 | ||||||
| Sp8 | XY | 12 | Trans-acting transcription factor 8 | ||||||
| T | XY | 17 | Brachyury, T-box transcription factor T | ||||||
| Tcf7 | XY | 11 | Transcription factor 7, T cell specific | ||||||
| Wiz | XY | 17 | Widely-interspaced zinc finger motifs | ||||||
| Ybx2 | XY | 11 | Y box protein 2 | ||||||
| Zglp1 | XY | 9 | Zinc finger, GATA-like protein 1 | ||||||
| Dnmt3b | XY | 2 | DNA methyltransferase 3B | ||||||
| Dnmt3l | XY | 10 | DNA (cytosine-5-)-methyltransferase 3-like | ||||||
| Phc1 | XY | 6 | Polyhomeotic 1 | ||||||
| Plac8 | XY | 5 | Placenta-specific 8 | ||||||
| Tdrd5 | XY | 1 | Tudor domain containing 5 | ||||||
| Bahcc1 | XY | 11 | BAH domain and coiled-coil containing 1 | ||||||
| Kdm6c | XY | XY | XY | XY | XY | XY | Y | Ubiq.transcribed tetratricopeptide repeat gene, Y chr | |
| Bcl6b | XY | XY | nb | 11 | B cell CLL/lymphoma 6, member B | ||||
| Irf8 | XY | XY | nb | 8 | Interferon regulatory factor 8 | ||||
| Arid1a | XY | XY | 4 | AT rich interactive domain 1A (SWI-like) | |||||
| Lef1 | XY | XY | XY | XY | 3 | Lymphoid enhancer binding factor 1 | |||
| Tbx20 | XY | XY | XY | XY | 9 | T-box 20 | |||
| Smarcd1 | XY | XY | 15 | SWI/SNF related, matrix assoc., actin-dep. reg. of chromatin d1 | |||||
| Zbtb7a | XY | XY | 10 | Zinc finger and BTB domain containing 7a | |||||
| Hif3a | XY | XY | XY | 7 | hypoxia inducible factor 3, alpha subunit | ||||
| Nfkb2 | XY | XY | XY | 19 | nuclear factor of κ light polypeptide enhancer B cells 2, p49/p100 | ||||
| Pbx2 | XY | XY | nb | 17 | Pre B cell leukemia homeobox 2 | ||||
| Prdm6 | XY | XY | 18 | PR domain containing 6 | |||||
| Id2 | XY | XY | 12 | Inhibitor of DNA binding 2 | |||||
| Dot1l | XY | XY | XX | 10 | DOT1-like, histone H3 methyltransferase (S. cerevisiae) | ||||
| Zfp296 | XY | XX | 7 | Zinc finger protein 296 | |||||
| Gata4 | XY | nb | 14 | GATA binding protein 4 | |||||
| Mycn | XY | nb | 12 | v-myc avian myelocytomatosis related og, neuroblastoma derived | |||||
| Hdac5 | XY | nb | 11 | Histone deacetylase 5 | |||||
| Nfxl1 | XY | nb | 5 | Nuclear transcription factor, X-box binding-like 1 | |||||
| Pax6 | nb | XY | 2 | Paired box 6 | |||||
| Brd4 | nb | XY | nb | 17 | Bromodomain containing 4 | ||||
| Carhsp1 | nb | XY | XY | XY | 16 | Calcium regulated heat stable protein 1 | |||
| Csde1 | nb | XY | XY | nb | nb | 3 | Cold shock domain containing E1, RNA binding | ||
| Mef2d | nb | XY | 3 | Myocyte enhancer factor 2D | |||||
| Atf5 | nb | XY | 7 | activating transcription factor 5 | |||||
| Bach2 | nb | XY | 4 | BTB and CNC homology, basic leucine zipper transcription factor 2 | |||||
| Cdc5l | nb | XY | 17 | Cell division cycle 5-like (S. pombe) | |||||
| Crebzf | nb | XY | 7 | CREB/ATF bZIP transcription factor | |||||
| Gm13139 | nb | XY | 4 | Zinc finger protein 991 | |||||
| Hmgb1 | nb | XY | 2 | Predicted gene, 21596 | |||||
| Hmgb2 | nb | XY | 8 | High mobility group box 2 | |||||
| Klf10 | nb | XY | 15 | Kruppel-like factor 10 | |||||
| Nfic | nb | XY | 10 | Nuclear factor I/C | |||||
| Nfkb1 | nb | XY | 3 | Nuclear factor of κ light polypeptide enhancer in B cells 1, p105 | |||||
| Pias2 | nb | XY | 18 | Protein inhibitor of activated STAT 2 |
ES embryonic stem cells; CP cardiac precursors; 8.5, 9.5, 10.5 days post coitum (dpc) embryonic hearts (single cell); p1 neonatal hearts; Chr chromosome; empty cells expression not detected; nb expressed but not biased
Thirty-six genes have conserved sex-biased expression in ES cells and adult hearts. Of those, six genes are more highly expressed in females at both stages, four of which are X-linked. Interestingly, only one of the X-linked genes has been previously described as escaping X chromosome inactivation (XCI) (Kdm6a) [54]. Thirty genes are male-biased in both ES cells and adult cardiomyocytes, including the three transcription factors Nfkb2, Lef1, Id2, and the epigenetic enzymes Uty and Prdm6.
Some genes expressed at early stages are still expressed in neonates or adults but lose their sex differences or even exhibit reversals in sex biases. In females, the X-linked Aff2 and Atrx lose their bias, which likely reflects dosage compensation after X chromosome inactivation in female cells. However, Meis2 and Zfp9 switch to male-biased expression in adults (Table 2). Seven male-biased genes, including Irf8, Pbx2, Gata4, and Hdac5, which exhibit higher expression in male ES cells, become equally expressed in adult hearts of both sexes. Dot1l and Zfp296 reverse their bias and are more highly expressed in females at later stages (Table 3).
We also find several genes that are not expressed differentially in male and female ES cells and later acquire a sex bias. These are good candidates for genes regulated by hormonal factors, although Esr1, the only estrogen receptor expressed in the heart, is not differentially expressed between males and females. RNA encoding the androgen receptor is also not sex-biased in the adult heart, suggesting that hormonal regulation depends on other co-factors and/or differential chromatin environment of the target genes.
To further explore the role TFs expressed in early development have at later stages, we identified binding sites for Lef1 and Zeb1 within the regulatory regions of genes differentially expressed between male and female cardiomyocytes. Lef1 is enriched in male ES cells, 8.5 dpc embryonic hearts, and in neonatal and adult hearts. Genes that harbored Lef1 binding motifs included other TFs that are also male-biased in ES cells, such as Mixl1, Mesp1, Irf8, and Tbx20, but also genes that are only later expressed in the adult heart, such as Gata5 and Foxo6, which are also male-enriched (Fig. 9a). Zeb1 is enriched in female ES cells and not detected at later stages, but its cognate motifs are present in genes that are female-biased in adult heart, such as Cecr2 and Nkx2-5 (Fig. 9b). These results suggest that TFs expressed in early development may determine sex-biased gene expression at later stages.
Fig. 9.
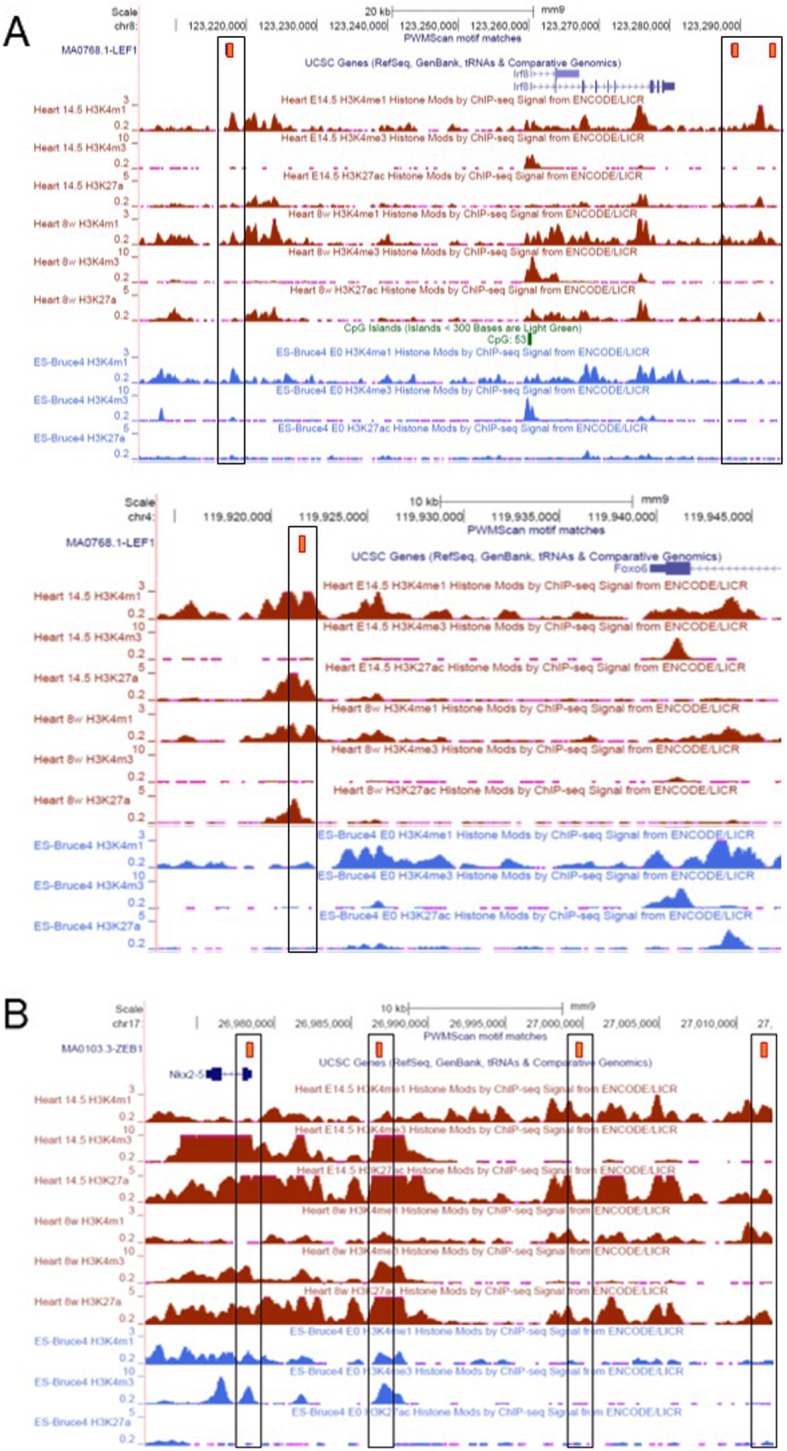
UCSC browser screenshots of genes regulated by sex-biased transcription factors (TFs). Custom tracks show TF binding sites for (a) Lef1 (male-biased) and (b) Zeb1 (female-biased) for genes that share the same bias with the TFs, with the binding sites denoted as orange bars. Also shown are the histone modification profiles for ES cells and 14.5 dpc and adult hearts, highlighting active histone marks coinciding with TF binding sites
Discussion
This study challenges the expectation that sex biases in gene regulation are non-existent during early mammalian development. Although sex determination has been traditionally associated with processes leading to distinct reproductive systems in males and females, we show that sex biases appear soon after fertilization and may have sex-specific repercussions during organogenesis, some of which persist in adults. In the absence of time-series experiments from pre-implantation embryos throughout lineage determination and organogenesis, we capitalized on our own data and a series of previously published RNA-seq datasets.
Gene co-expression network analysis identifies Prdm14 as a key determinant of sex-biased gene expression in ES cells
Previous reports have identified thousands of genes differentially expressed in male and female ES cells and pre-implantation embryos in rodents, bovine, primates, and humans [21–24, 54–59]. In this work, we asked whether sexual dimorphism is detectable at the molecular level of gene expression and enriched in protein-protein interaction networks in early development. Both WGCNA and PPI networks revealed that important modules associated with sex are enriched in genes with Prdm14 cognate binding sites and are Prdm14 target genes.
Prdm14 is important for pluripotency in ES cells [49, 52] and is a key regulator of primordial germ cell specification [60, 61]. In contrast to other PRDM family members, Prdm14 does not exhibit histone methyltransferase, but has been shown to partner with enzymes that catalyze post-translational modification of histones [49]. In fact, as seen in our ChIP-seq data, male and female ES cells have differential chromatin modifications, some of which are associated to Prdm14 occupancy at regulatory sequences. In addition, Prdm14 binding is found in promoters or neighboring regions of genes that are not expressed in ES cells. Thus, epigenetic marks established in pre-implantation stages can potentially result in sex-biased gene expression later in development.
Prdm14 expression is downregulated after differentiation of male and female ES cells and after implantation in vivo. However, female ES cells are developmentally delayed relative to male cells due to the process of X chromosome inactivation (XCI) [56]. Consequently, they are exposed to higher Prdm14 levels for a longer period, which could lead to the establishment of female-specific epigenetic marks. In fact, we previously reported that a Prdm14-responsive enhancer exhibited higher activity in female ES cells, strongly suggesting that Prdm14 target gene levels are dosage-sensitive [24]. In addition, it is possible that a subset of genes regulated by Prdm14 is distinct in male and female ES cells. This is also true of any dose-dependent TF or ERE with sex-biased expression. Therefore, future ChIP-seq studies for TFs and chromatin modifications performed in a sex-stratified manner should allow us to distinguish between these possibilities.
Our studies also show that there are factors in addition to Prdm14 that regulate sex-biased gene expression. For example, X-linked genes that are expressed from the two active X chromosomes, such as Atrx, Kdm6a, and Klf8, are strong candidates for involvement in sex-biased expression. However, autosomal factors, such as Lef1 and Zeb1, could be involved as well. In theory, TFs that are not sex-biased could also be important for differential gene expression and according to our network analysis (WGCNA), there are a host of other TFs and EREs that are significantly correlated with sex such as Arid3b, Smad4, Jarid2, and Kdm8. For regulatory factors that are not sex-biased per se, their cognate sites could present different accessibility in male and female cells or there could be differential availability of their co-factors.
Differentiated ES cells exhibit sex-biased gene expression
Differentiation of male and female ES cell lines into cardiac precursors drastically changed the transcriptional profile of the cells, but we still detected sex-biased expression. Most X-linked genes were expressed equally between male and female cells due to the process of XCI, but unexpectedly, four were more highly expressed in male cells, suggesting that there is male-specific regulation of some X-linked genes. Some of the sex differences in gene expression could represent the slight developmental delay of the female cells. Yet some expression differences observed in ES cells persist in the adult heart, suggesting that these are independent of developmental stage and are integrating bona fide sex-specific regulatory networks.
While the protocol we used for differentiation of ES cells into cardiac precursors has been derived from the extensive knowledge on cardiogenesis in vivo [41], the in vitro derivation of cardiac progenitors lacks other factors, such as spatial context, that are important for proper organ formation. For example, during heart development in vivo, multiple cell types, including transient populations, interact in three dimensions and receive input from surrounding tissues. However, single-cell analyses of early cardiac stages have pinpointed that cardiac progenitors derived from ES cells have a transcriptome corresponding to 9.5 dpc single-cell cardiomyocytes[43], a stage in which fibroblasts are not yet apparent. Thus, differentiated ES cells serve as a close approximation of the early stages of heart development.
Sex-biased gene expression exists at every stage during cardiac development
To determine whether the sex biases in differentiated ES cells are present in vivo, we inspected previously published data from specific stages during heart development. Single-cell assessment of transcriptional profiles in early stages of cardiogenesis has allowed detailed analysis of the step-wise specification of cardiac progenitors, but the available data are not stratified by sex. For each sample, we genotyped for sex and re-analyzed these data and observed sex-biased expression across all the available stages of heart development. We also observed short bursts of sex-biased expression of regulatory factors at single stages, raising the question of whether these are capable of encoding persistent dimorphisms. In addition, we show that some genes equalize their expression, while others become biased in the opposite direction, which raises important questions on the mechanisms by which these events occur.
We recognize several caveats in this study. First, compiling datasets from different reports presents challenges because of the different experimental designs. Our own data is from ES cells in culture subjected to a directed differentiation protocol that only partially recapitulates the complex processes in vivo. Second, the single-cell RNA-seq data from embryonic and neonatal hearts, while useful for distinguishing cell populations, is necessarily incomplete. Currently, single-cell RNA-seq only detects a fraction of the transcriptome, with a bias towards high expression transcripts, which excludes many TFs that are expressed at relatively low levels.
Systems-level analyses have yielded valuable information on the correlations between congenital heart disease and their developmental origins [62, 63]. Transcriptomic data for early developmental stages is sparse, however. Nevertheless, the currently available datasets reveal sex-biased expression at every stage and suggest novel hypotheses for future mechanistic studies. Our analyses also open questions on how the fluctuations in sex-biased expression are regulated, how they are reflected in epigenetic differences between male and female cells, and how widely these occur in other tissues during embryogenesis. Our data also serves as a platform to identify the role of sex hormones in countering or compounding sex biases. Future studies will enable dissection of the effects of sex chromosomes and hormonal influence on sexual dimorphism. Ultimately, expanding developmental studies will allow us to connect early sexual dimorphism to the sex biases that occur in adult health and disease.
Conclusions
The ability to profile transcriptomes has heightened interest in sex-biased gene expression, especially after recent reports that show substantial differences between males and females in humans and other animal models, even in organ systems that are overtly identical [4, 5, 44, 64]. The focus on adult tissues reflects a broadly held assumption that sex-biased expression is unimportant during early embryogenesis, during which critical lineage decisions are made, and that sex-specific selection only operates after the reproductive interests of the sexes have diverged [65]. In non-mammalian species, however, there is evidence that sex biases at the transcriptomic level occur throughout development [17, 66]. Here, we address a major gap in developmental studies by detecting sex-biased expression during mouse cardiac development. Our data strongly suggest that some of the differences in transcriptomic profiles in adult hearts may be established epigenetically before the appearance of sex hormones. Our observations open the field to explore the timing and extent of sex-specific transcriptional and epigenetic profiles in other organ systems and their relevance to sexual dimorphisms in adult health and disease.
Additional files
Table S1. List of antibodies used for ChIP-seq (XLSX 9 kb)
Table S2. Accession and metadata of all samples analyzed in this study (XLSX 10 kb)
Dataset S1. Genes in blue/violet module eigengene (XLSX 29 kb)
Dataset S2. Ingenuity Pathway Analysis Upstream Regulators (XLSX 9 kb)
Dataset S3. Topology analysis for protein-protein interaction networks (XLSX 141 kb)
Figure S1. Modularity in male and female ES cell protein-protein interaction networks (JPG 50 kb)
Dataset S4. Gene ontology for protein-protein interaction networks (XLSX 15 kb)
Figure S2. qPCR of markers before and after ES cell differentiation (JPG 38 kb)
Dataset S5. Differentially expressed genes in male and female cardiac precursors (XLSX 10 kb)
Figure S3. Male-enriched genes after differentiation of ES cells. UCSC browser screen shots with tracks denoting Prdm14 occupancy (track obtained from Ma et al.) (JPG 41 kb)
Dataset S6. Sex Biases in Single Cell Data (XLSX 108 kb)
Figure S4. Modularity in protein-protein interaction networks (JPG 114 kb)
Figure S5. Differentially expressed genes in adult male and female hearts (JPG 113 kb)
Datasets S7 and S8. Ingenuity Pathway Analysis for sex-biased genes in adult heart (XLSX 19 kb)
Dataset S9. Sex-biased Genes Shared between Human and Mouse Adult Hearts (XLSX 13 kb)
Acknowledgments
We thank Rachael Werner for technical support and Joanne Thorvaldsen for comments and suggestions. This project was supported by a Temple University Bridge Grant (to N.E.).
Abbreviations
- EREs
Epigenetic and remodeling enzymes
- ES cells
Embryonic stem cells
- FPKM
Fragments per kilobase of exon per million reads mapped
- GS
Gene significance
- LIF
Leukemia inhibitory factor
- ME
Module eigengene
- MS
Module significance
- PPIs
Protein-protein interaction networks
- TFs
Transcription factors
- TO
Topology overlap
- WGCNA
Weighted gene co-expression network analysis
- XCI
X chromosome inactivation
Authors’ contributions
NE conceived and designed the experiments. DFD generated the data. RK and RJK performed the data analysis. NE and RJK interpreted the data. JM contributed bioinformatic support. NE wrote the manuscript. NE, JM and RK generated the figures. All authors gave final approval of the published version. All authors are accountable for all aspects of the work.
Funding
This study was funded by a Temple University Bridge Grant.
Availability of data and materials
Data generated has been deposited in GEO: GSE90516.
Data from other reports and their supplementary information files was also analyzed:
Li G, Xu A, Sim S, Priest JR, Tian X, Khan T, Quertermous T, Zhou B, Tsao PS, Quake SR et al: Transcriptomic Profiling Maps Anatomically Patterned Subpopulations among Single Embryonic Cardiac Cells. Dev Cell 2016, 39(4):491-507. GSE76118
DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W et al: Single-Cell Resolution of Temporal Gene Expression during Heart Development. Dev Cell 2016, 39(4):480-490. Obtained from the author.
Li B, Qing T, Zhu J, Wen Z, Yu Y, Fukumura R, Zheng Y, Gondo Y, Shi L: A Comprehensive Mouse Transcriptomic BodyMap across 17 Tissues by RNA-seq. Sci Rep 2017, 7(1):4200. PRJNA375882
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Daniel F. Deegan and Reza Karbalaei contributed equally to this work.
Contributor Information
Daniel F. Deegan, Email: dandeegan14@temple.edu
Reza Karbalaei, Email: tuk35072@temple.edu.
Jozef Madzo, Email: jmadzo@temple.edu.
Rob J. Kulathinal, Email: robkulathinal@temple.edu
Nora Engel, Phone: 215-707-7611, Email: noraengel@temple.edu.
References
- 1.Blenck CL, Harvey PA, Reckelhoff JF, Leinwand LA. The importance of biological sex and estrogen in rodent models of cardiovascular health and disease. Circ Res. 2016;118(8):1294–1312. doi: 10.1161/CIRCRESAHA.116.307509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singmann P, Shem-Tov D, Wahl S, Grallert H, Fiorito G, Shin SY, Schramm K, Wolf P, Kunze S, Baran Y, et al. Characterization of whole-genome autosomal differences of DNA methylation between men and women. Epigenetics Chromatin. 2015;8:43. doi: 10.1186/s13072-015-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isensee J, Ruiz Noppinger P. Sexually dimorphic gene expression in mammalian somatic tissue. Gender medicine. 2007;4(Suppl B):S75–S95. doi: 10.1016/s1550-8579(07)80049-0. [DOI] [PubMed] [Google Scholar]
- 4.Chen C-Y, Lopes-Ramos C, Kuijjer ML, Paulson JN, Sonawane AR, Fagny M, Platig J, Glass K, Quackenbush J, DeMeo DL: sexual dimorphism in gene expression and regulatory networks across human tissues. bioRxiv 2016. [DOI] [PMC free article] [PubMed]
- 5.Gershoni M, Pietrokovski S. The landscape of sex-differential transcriptome and its consequent selection in human adults. BMC Biol. 2017;15(1):7. doi: 10.1186/s12915-017-0352-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trexler CL, Odell AT, Jeong MY, Dowell RD, Leinwand LA. Transcriptome and functional profile of cardiac myocytes is influenced by biological sex. Circ Cardiovasc Genet. 2017;10(5):1-11. [DOI] [PMC free article] [PubMed]
- 7.Isensee J, Witt H, Pregla R, Hetzer R, Regitz-Zagrosek V, Noppinger PR. Sexually dimorphic gene expression in the heart of mice and men. J Mol Med (Berl) 2008;86(1):61–74. doi: 10.1007/s00109-007-0240-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Y, Fuscoe JC, Zhao C, Guo C, Jia M, Qing T, Bannon DI, Lancashire L, Bao W, Du T, et al. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun. 2014;5:3230. doi: 10.1038/ncomms4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srivastava D. Making or Breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151(1):206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain R, Epstein JA. Competent for commitment: you've got to have heart! Genes Dev. 2018;32(1):4–13. doi: 10.1101/gad.308353.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steimle JD, Moskowitz IP. TBX5: A key regulator of heart development. Curr Top Dev Biol. 2017;122:195–221. doi: 10.1016/bs.ctdb.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bardot E, Calderon D, Santoriello F, Han S, Cheung K, Jadhav B, Burtscher I, Artap S, Jain R, Epstein J, et al. Foxa2 identifies a cardiac progenitor population with ventricular differentiation potential. Nat Commun. 2017;8:14428. doi: 10.1038/ncomms14428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azhar M, Ware SM. Genetic and developmental basis of cardiovascular malformations. Clin Perinatol. 2016;43(1):39–53. doi: 10.1016/j.clp.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somerville J. The Denolin lecture: the woman with congenital heart disease. Eur Heart J. 1998;19(12):1766–1775. doi: 10.1053/euhj.1998.1204. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins KJ, Correa A, Feinstein JA, Botto L, Britt AE, Daniels SR, Elixson M, Warnes CA, Webb CL. American Heart Association Council on Cardiovascular Disease in the Y: Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115(23):2995–3014. doi: 10.1161/CIRCULATIONAHA.106.183216. [DOI] [PubMed] [Google Scholar]
- 17.Mank JE, Nam K, Brunstrom B. Ellegren H: ontogenetic complexity of sexual dimorphism and sex-specific selection. Mol Biol Evol. 2010;27(7):1570–1578. doi: 10.1093/molbev/msq042. [DOI] [PubMed] [Google Scholar]
- 18.Dunn SJ, Martello G, Yordanov B, Emmott S, Smith AG. defining an essential transcription factor program for naive pluripotency. Science. 2014;344(6188):1156–1160. doi: 10.1126/science.1248882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H, Ang YS, Sevilla A, Lemischka IR, Ma'ayan A. Construction and validation of a regulatory network for pluripotency and self-renewal of mouse embryonic stem cells. PLoS Comput Biol. 2014;10(8):e1003777. doi: 10.1371/journal.pcbi.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Som A, Harder C, Greber B, Siatkowski M, Paudel Y, Warsow G, Cap C, Scholer H, Fuellen G. The PluriNetWork: an electronic representation of the network underlying pluripotency in mouse, and its applications. PLoS One. 2010;5(12):e15165. doi: 10.1371/journal.pone.0015165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe R, Gemma C, Rakyan VK, Holland ML. Sexually dimorphic gene expression emerges with embryonic genome activation and is dynamic throughout development. BMC Genomics. 2015;16:295. doi: 10.1186/s12864-015-1506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marks H, Kerstens HH, Barakat TS, Splinter E, Dirks RA, van Mierlo G, Joshi O, Wang SY, Babak T, Albers CA, et al. Dynamics of gene silencing during X inactivation using allele-specific RNA-seq. Genome Biol. 2015;16:149. doi: 10.1186/s13059-015-0698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi J, Clement K, Huebner AJ, Webster J, Rose CM, Brumbaugh J, Walsh RM, Lee S, Savol A, Etchegaray JP, et al. DUSP9 modulates DNA hypomethylation in female mouse pluripotent stem cells. Cell Stem Cell. 2017;20(5):706–719. doi: 10.1016/j.stem.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Werner R, Schultz B, Huhn J, Jelinek J, Madzo J, Engel N. Sex chromosomes drive gene expression and regulatory dimorphisms in mouse embryonic stem cells. Biol Sex Differ. 2017;8:28. doi: 10.1186/s13293-017-0150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engel N. Sex Differences in Early Embryogenesis: inter-chromosomal regulation sets the stage for sex-biased gene networks: the dialogue between the sex chromosomes and autosomes imposes sexual identity soon after fertilization. BioEssays. 2018;40(9):e1800073. doi: 10.1002/bies.201800073. [DOI] [PubMed] [Google Scholar]
- 26.Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol. 2008;4(8):e1000117. doi: 10.1371/journal.pcbi.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medvedeva YA, Fridman MV, Oparina NJ, Malko DB, Ermakova EO, Kulakovskiy IV, Heinzel A, Makeev VJ. Intergenic, gene terminal, and intragenic CpG islands in the human genome. BMC Genomics. 2010;11:48. doi: 10.1186/1471-2164-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lupey-Green LN, Moquin SA, Martin KA, McDevitt SM, Hulse M, Caruso LB, Pomerantz RT, Miranda JL, Tempera I. PARP1 restricts Epstein Barr Virus lytic reactivation by binding the BZLF1 promoter. Virology. 2017;507:220–230. doi: 10.1016/j.virol.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B: Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics 2010, Chapter 11:Unit 11 17. [DOI] [PMC free article] [PubMed]
- 31.Quinlan Aaron R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Current Protocols in Bioinformatics. 2014;47(1):11.12.1-11.12.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen Li, Shao Ningyi, Liu Xiaochuan, Nestler Eric. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15(1):284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bastian M. H S, Jacomy M. : Gephi: an open source software for exploring and manipulating networks. International AAAI Conference on Weblogs and Social Media 2009.
- 36.Blondel VD. Fast unfolding of communities in large networks. J Stat Mech. 2008;arXiv:0803.0476. [Google Scholar]
- 37.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6(7):e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karbalaei R, Allahyari M, Rezaei-Tavirani M, Asadzadeh-Aghdaei H, Zali MR. Protein-protein interaction analysis of Alzheimer`s disease and NAFLD based on systems biology methods unhide common ancestor pathways. Gastroenterol Hepatol Bed Bench. 2018;11(1):27–33. [PMC free article] [PubMed] [Google Scholar]
- 40.Karbalaei R, Piran M, Rezaei-Tavirani M, Asadzadeh-Aghdaei H, Heidari MH. A systems biology analysis protein-protein interaction of NASH and IBD based on comprehensive gene information. Gastroenterol Hepatol Bed Bench. 2017;10(3):194–201. [PMC free article] [PubMed] [Google Scholar]
- 41.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8(2):228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 42.Li G, Xu A, Sim S, Priest JR, Tian X, Khan T, Quertermous T, Zhou B, Tsao PS, Quake SR, et al. Transcriptomic profiling maps anatomically patterned subpopulations among single embryonic cardiac cells. Dev Cell. 2016;39(4):491–507. doi: 10.1016/j.devcel.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W, et al. Single-cell resolution of temporal gene expression during heart development. Dev Cell. 2016;39(4):480–490. doi: 10.1016/j.devcel.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li B, Qing T, Zhu J, Wen Z, Yu Y, Fukumura R, Zheng Y, Gondo Y, Shi L. A comprehensive mouse transcriptomic bodymap across 17 Tissues by RNA-seq. Sci Rep. 2017;7(1):4200. doi: 10.1038/s41598-017-04520-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arnold AP, van Nas A, Lusis AJ. Systems biology asks new questions about sex differences. Trends Endocrinol Metab. 2009;20(10):471–476. doi: 10.1016/j.tem.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Nas A, Guhathakurta D, Wang SS, Yehya N, Horvath S, Zhang B, Ingram-Drake L, Chaudhuri G, Schadt EE, Drake TA, et al. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology. 2009;150(3):1235–1249. doi: 10.1210/en.2008-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pobbati AV, Hong W. Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol Ther. 2013;14(5):390–398. doi: 10.4161/cbt.23788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pijuan-Galito S, Tamm C, Anneren C. Serum inter-alpha-inhibitor activates the Yes tyrosine kinase and YAP/TEAD transcriptional complex in mouse embryonic stem cells. J Biol Chem. 2014;289(48):33492–33502. doi: 10.1074/jbc.M114.580076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakaki F, Saitou M. PRDM14: a unique regulator for pluripotency and epigenetic reprogramming. Trends Biochem Sci. 2014;39(6):289–298. doi: 10.1016/j.tibs.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 50.Ma Z, Swigut T, Valouev A, Rada-Iglesias A, Wysocka J. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat Struct Mol Biol. 2011;18(2):120–127. doi: 10.1038/nsmb.2000. [DOI] [PubMed] [Google Scholar]
- 51.Yamaji M, Ueda J, Hayashi K, Ohta H, Yabuta Y, Kurimoto K, Nakato R, Yamada Y, Shirahige K, Saitou M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell. 2013;12(3):368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 52.Grabole N, Tischler J, Hackett JA, Kim S, Tang F, Leitch HG, Magnusdottir E, Surani MA. Prdm14 promotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation. EMBO Rep. 2013;14(7):629–637. doi: 10.1038/embor.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keller GM. In vitro differentiation of embryonic stem cells. Curr Opin Cell Biol. 1995;7(6):862–869. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- 54.Berletch JB, Deng X, Nguyen DK, Disteche CM. Female bias in Rhox6 and 9 regulation by the histone demethylase KDM6A. PLoS Genet. 2013;9(5):e1003489. doi: 10.1371/journal.pgen.1003489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burgoyne PS, Thornhill AR, Boudrean SK, Darling SM, Bishop CE, Evans EP. The genetic basis of XX-XY differences present before gonadal sex differentiation in the mouse. Philos Trans R Soc Lond B Biol Sci. 1995;350(1333):253–260. doi: 10.1098/rstb.1995.0159. [DOI] [PubMed] [Google Scholar]
- 56.Schulz EG, Meisig J, Nakamura T, Okamoto I, Sieber A, Picard C, Borensztein M, Saitou M, Bluthgen N, Heard E. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell. 2014;14(2):203–216. doi: 10.1016/j.stem.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 57.Bermejo-Alvarez P, Rizos D, Rath D, Lonergan P, Gutierrez-Adan A. Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. Proc Natl Acad Sci U S A. 2010;107(8):3394–3399. doi: 10.1073/pnas.0913843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen G, Schell JP, Benitez JA, Petropoulos S, Yilmaz M, Reinius B, Alekseenko Z, Shi L, Hedlund E, Lanner F, et al. Single-cell analyses of X chromosome inactivation dynamics and pluripotency during differentiation. Genome Res. 2016. [DOI] [PMC free article] [PubMed]
- 59.Petropoulos S, Edsgard D, Reinius B, Deng Q, Panula SP, Codeluppi S, Reyes AP, Linnarsson S, Sandberg R, Lanner F. Single-cell RNA-Seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell. 2016;167(1):285. doi: 10.1016/j.cell.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, Yamanaka K, Ohinata Y, Saitou M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet. 2008;40(8):1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- 61.Magnusdottir E, Surani MA. How to make a primordial germ cell. Development. 2014;141(2):245–252. doi: 10.1242/dev.098269. [DOI] [PubMed] [Google Scholar]
- 62.Lage K, Greenway SC, Rosenfeld JA, Wakimoto H, Gorham JM, Segre AV, Roberts AE, Smoot LB, Pu WT, Pereira AC, et al. Genetic and environmental risk factors in congenital heart disease functionally converge in protein networks driving heart development. Proc Natl Acad Sci U S A. 2012;109(35):14035–14040. doi: 10.1073/pnas.1210730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lage K, Mollgard K, Greenway S, Wakimoto H, Gorham JM, Workman CT, Bendsen E, Hansen NT, Rigina O, Roque FS, et al. Dissecting spatio-temporal protein networks driving human heart development and related disorders. Mol Syst Biol. 2010;6:381. doi: 10.1038/msb.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16(8):995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rice WR, Chippindale AK. Sexual recombination and the power of natural selection. Science. 2001;294(5542):555–559. doi: 10.1126/science.1061380. [DOI] [PubMed] [Google Scholar]
- 66.Perry JC, Harrison PW, Mank JE. The ontogeny and evolution of sex-biased gene expression in Drosophila melanogaster. Mol Biol Evol. 2014;31(5):1206–1219. doi: 10.1093/molbev/msu072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of antibodies used for ChIP-seq (XLSX 9 kb)
Table S2. Accession and metadata of all samples analyzed in this study (XLSX 10 kb)
Dataset S1. Genes in blue/violet module eigengene (XLSX 29 kb)
Dataset S2. Ingenuity Pathway Analysis Upstream Regulators (XLSX 9 kb)
Dataset S3. Topology analysis for protein-protein interaction networks (XLSX 141 kb)
Figure S1. Modularity in male and female ES cell protein-protein interaction networks (JPG 50 kb)
Dataset S4. Gene ontology for protein-protein interaction networks (XLSX 15 kb)
Figure S2. qPCR of markers before and after ES cell differentiation (JPG 38 kb)
Dataset S5. Differentially expressed genes in male and female cardiac precursors (XLSX 10 kb)
Figure S3. Male-enriched genes after differentiation of ES cells. UCSC browser screen shots with tracks denoting Prdm14 occupancy (track obtained from Ma et al.) (JPG 41 kb)
Dataset S6. Sex Biases in Single Cell Data (XLSX 108 kb)
Figure S4. Modularity in protein-protein interaction networks (JPG 114 kb)
Figure S5. Differentially expressed genes in adult male and female hearts (JPG 113 kb)
Datasets S7 and S8. Ingenuity Pathway Analysis for sex-biased genes in adult heart (XLSX 19 kb)
Dataset S9. Sex-biased Genes Shared between Human and Mouse Adult Hearts (XLSX 13 kb)
Data Availability Statement
Data generated has been deposited in GEO: GSE90516.
Data from other reports and their supplementary information files was also analyzed:
Li G, Xu A, Sim S, Priest JR, Tian X, Khan T, Quertermous T, Zhou B, Tsao PS, Quake SR et al: Transcriptomic Profiling Maps Anatomically Patterned Subpopulations among Single Embryonic Cardiac Cells. Dev Cell 2016, 39(4):491-507. GSE76118
DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W et al: Single-Cell Resolution of Temporal Gene Expression during Heart Development. Dev Cell 2016, 39(4):480-490. Obtained from the author.
Li B, Qing T, Zhu J, Wen Z, Yu Y, Fukumura R, Zheng Y, Gondo Y, Shi L: A Comprehensive Mouse Transcriptomic BodyMap across 17 Tissues by RNA-seq. Sci Rep 2017, 7(1):4200. PRJNA375882