Abstract
The emerging role of cytoskeletal proteins in the cell nucleus has become a new frontier in cell biology. Actin and actin-binding proteins regulate chromatin and gene expression, but importantly they are beginning to be essential players in genome organization. These actin-based functions contribute to genome stability and integrity while affecting DNA replication and global transcription patterns. This is likely to occur through interactions of actin with nuclear components including nuclear lamina and subnuclear organelles. An exciting future challenge is to understand how these actin-based genome-wide mechanisms may regulate development and differentiation by interfering with the mechanical properties of the cell nucleus and how regulated actin polymerization plays a role in maintaining nuclear architecture. With a special focus on actin, here we summarize how cytoskeletal proteins operate in the nucleus and how they may be important to consolidate nuclear architecture for sustained gene expression or silencing.
CONNECTING GENE EXPRESSION AND GENOME ARCHITECTURE
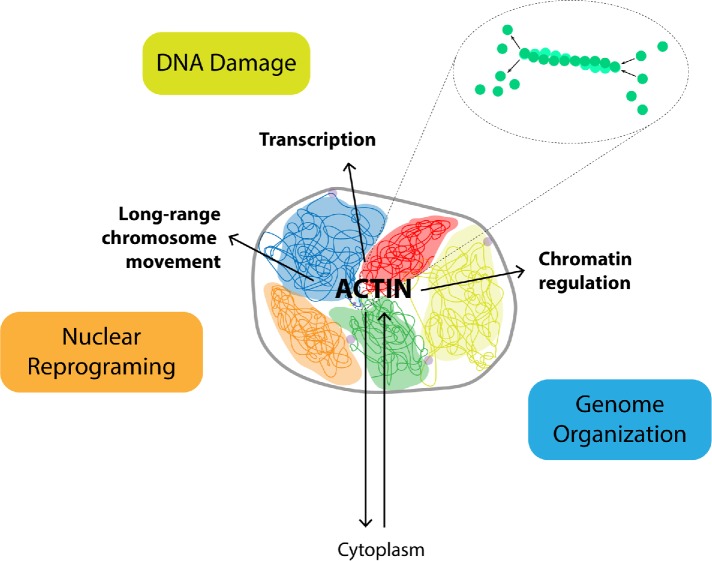
For a long period of time, the term “cytoskeletal proteins” has been synonymous with proteins strictly involved in cytoplasmic functions. Today this bias, with ancient biochemical roots dating back to the 1970s and 1980s, is finally coming to an end. The cell biology community is beginning to recognize that cytoskeletal proteins populate the cell nucleus and have fundamental functions also in this cellular compartment. Among the cytoskeletal proteins identified so far, actin is probably the best characterized. It has been associated with many nuclear functions together with well-characterized nuclear factors but also with myosin species and plenty of actin-regulating proteins, including actin-related proteins (ARPs) and those that control its polymerization state (Figure 1).
FIGURE 1:
Actin has been implicated in many nuclear tasks including chromatin regulation, transcription, and long-range chromosome movement. Recent evidence indicates that actin-based mechanisms impact cellular functions as different as DNA damage and nuclear reprogramming during differentiation. Because these functions are dependent on efficient organization of genomic architecture, we suggest a model where actin is a primary regulator of the functional architecture of the genome. We speculate that rounds of actin polymerization and depolymerization play an important role in facilitating interactions with different nuclear machinery.
About 20% of cellular actin is present in the cell nucleus and a considerable fraction undergoes dynamic nucleocytoplasmic transport in and out of the nucleus. Maintenance of the nuclear actin levels by active nuclear transport by the importin/exportin/Ran system (Stuven et al., 2003; Dopie et al., 2012) are directly connected to nuclear function, and sufficient nuclear actin levels are required for transcription. Once in the cell nucleus, the general idea is that actin plays a direct role as part of several molecular machineries that regulate gene expression (Visa and Percipalle, 2010). In fact, there is biochemical evidence that actin is both physically associated and a bona fide component of many chromatin remodeling complexes, while at transcription sites actin binds to RNA polymerases and ribonucleoprotein complexes (Percipalle et al., 2001, 2003). In certain chromatin remodeling complexes, actin regulates the function of the ATPase subunit required for nucleosome remodeling (Olave et al., 2002; Kapoor and Shen, 2014), and can contribute to the recognition of extranucleosomal linker DNA as a subcomplex with actin-related proteins (Arp; Knoll et al., 2018). Biochemical association of actin with the RNA polymerase machineries has been reported by several studies (Fomproix and Percipalle, 2004; Hofmann et al., 2004; Hu et al., 2004; Philimonenko et al., 2004; Kukalev et al., 2005). We know that actin interacts with all three eukaryotic RNA polymerases and this interaction appears to be conserved through two of the subunits, Rbp6 and Rbp8, common to all three nuclear polymerases (Percipalle 2013). In the specific case of the RNA polymerase II machinery, actin binds to both inactive and active forms of the enzyme, and in particular, with the hyperphosphorylated form of the carboxyl-terminal domain (CTD; Hofmann et al., 2004; Kukalev et al., 2005). Interaction with the phosphorylated CTD is mediated by several factors including the heterogeneous nuclear ribonucleoprotein hnRNP U (SAF-A; Kukalev et al., 2005, Obrdlik et al., 2008). As transcription proceeds, a fraction of actin remains incorporated into nascent ribonucleoproteins in complex with heterogeneous nuclear ribonucleoproteins (hnRNPs) and accompanies the transcripts to polyribosomes (reviewed in Percipalle, 2009). The function of actin in pre-mRNPs and mature RNPs is still unclear. Some studies, however, have suggested a potential role for the RNP-associated actin in facilitating RNP assembly and disassembly at translation sites (Kotani et al., 2013). The wide role of actin is also supported by a recent mass spectrometry–based analysis of nuclear actin-interacting proteins, which revealed dynamic association of actin with proteins implicated in transcription preinitiation, elongation, and pre-mRNA processing (Viita et al., 2019). In addition, minigene splicing assays demonstrated a functional role for actin in alternative splicing, most likely by influencing the rate of transcription elongation (Viita et al., 2019). However, the molecular details are still unknown, and await high-resolution structures of actin in complex with the relevant proteins. Seeing the presence of actin in chromatin remodeling complexes, transcription apparatus, and nascent RNP, an interesting scenario is that actin functions as molecular links among the different machinery involved in gene expression from the gene to polyribosomes.
The next fascinating question is whether actin function in gene expression is restricted to the gene level or it impacts the architecture of the genome, seeing the importance of actin in long-range chromosome movement. In favor of a general function of actin in genome regulation, recent chromatin immunoprecipitation and deep sequencing studies have shown that actin binds across both mammalian and Drosophila genomes (Almuzzaini et al., 2016; Sokolova et al., 2018). Indeed, in mouse embryonic fibroblasts where both β-actin alleles are disrupted, we recently discovered that heterochromatin is reorganized. In the absence of β-actin, we revealed loss of HP1 (heterochromatin protein 1)-positive heterochromatin segregation at the nuclear envelope and loss of heterochromatin maintenance in the nuclear interior. These changes accompany alterations in nuclear size and extensive differential gene expression in comparison to wild-type cells (Xie et al., 2018a,b). Importantly, these phenotypes, both in terms of architecture and gene expression profiles, are rescued when β-actin is constitutively reintroduced in the cell nucleus in the knockout (KO) background (Xie et al., 2018a). We found that this nuclear β-actin pool regulates heterochromatin organization and gene expression by controlling deposition of Brg1, also known as SMARCA4 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin), across the genome (Xie et al., 2018a), impacting neurogenesis (Xie et al., 2018b). Brg1 protein is known to be regulated by actin and it is involved in nuclear reprogramming (Singhal et al. 2010). Although it cannot be excluded that some contribution to impaired heterochromatin organization and thus genome architecture may also derive from loss of the cytoplasmic β-actin pool, it is important to mention that these cells exhibit an intact cytoskeleton mainly due to the up-regulation of both α- and γ-actin cellular pools (Tondeleir et al., 2012; Xie et al., 2018a). In addition, in these mouse embryonic fibroblasts the up-regulated actin isoforms provide propulsive forces for cell migration, whereas β-actin retains a unique nuclear function that prevents myogenic differentiation by regulating global transcription (Tondeleir et al., 2012). It is therefore likely that in mouse embryonic fibroblasts it is the nuclear pool of β-actin that plays a direct role in the organization of the genome. In support of a unique role in gene expression, in vivo, the mouse model from which embryonic fibroblasts were generated is lethal at stage E10.5 when key developmental pathways kick in (Tondeleir et al., 2012; Xie and Percipalle, 2018). Prominent pleiotropic effects have been recently observed upon reduced amounts of β-actin levels in terms of cell shape, migration, proliferation, and gene expression impairing brain, heart, and kidney development (Cuvertino et al., 2017). Because cell shape, migration, and proliferation are results of changes in gene expression programs, it is still likely that even in this model system the primary β-actin function is still in the regulation of gene expression. On the other hand, cytoplasmic actin filaments in conjunction with components of the nuclear envelope may play an important role in genome organization through mechanosensing pathways (Shivashankar, 2019). Taken together, these observations indicate that nuclear β-actin is a key player in the architecture of the genome, although they raise numerous mechanistic questions, one of them being precisely the synergy between nuclear and cytoplasmic actin pools.
The organization of the genome in the nucleus of a eukaryotic cell is complex and dynamic. The architecture of the interphase nucleus is linked to the spatial arrangement of genes and gene clusters, the structure of chromatin, and the accessibility of regulatory DNA elements. Changes in genome architecture are beginning to be associated with differentiation. Therefore, a fascinating hypothesis is that nuclear actin by controlling the organization of the genome plays a primary role in cellular differentiation. As mentioned above, the β-actin mouse model generated by the Ampe lab is embryonic-lethal at stage E10.5 before the onset of key developmental pathways such as neurogenesis (Tondeleir et al., 2012). Further, in embryonic fibroblasts derived from the same mouse model transdifferentiated to neurons, we discovered that neurogenesis is affected due to loss of β-actin–dependent Brg1 genomic deposition at regulatory regions of genes involved in neuronal development, leading to higher heterochromatin content at these specific loci (Xie et al., 2018b). Ongoing work suggests that these mechanisms are conserved in other differentiation models (Percipalle, unpublished data). If β-actin plays an important role in regulating the genome, it is also likely that in the absence of β-actin we have a loss of genomic stability and, therefore, integrity. In this context, a similar mechanism could be envisaged because the chromatin remodeling complex BAF is also known to be recruited to sites of DNA damage (Kwon et al., 2015). A speculation is that impairment of actin-based mechanisms regulating genome architecture may, therefore, be a direct cause of disease.
One of the major questions is whether the function of actin in genome stability and integrity is linked to its ability to undergo changes in its polymerization state. In the next section we highlight recent evidence supporting the importance of actin polymerization in nuclear function.
NUCLEAR ACTIN POLYMERIZATION AND DEPOLYMERIZATION
Perhaps the most enigmatic aspect of nuclear actin has been its polymerization status (Figure 1). In most cases, canonical actin filaments cannot be detected in the cell nucleus, for example, with phalloidin staining, suggesting that in the nucleus, actin could be mostly monomeric or adopt specific conformations (reviewed in Grosse and Vartiainen, 2013). Earlier FRAP studies had already suggested the presence of also polymeric forms of nuclear actin (McDonald et al., 2006; Dopie et al., 2012), and the usage of novel, nuclear-targeted probes that recognize different forms of actin have revolutionized this field. First, the Mullins lab utilized known actin-binding domains, and demonstrated with an utrophin-based probe, UTR230-EN, short polymeric nuclear actin structures that were excluded from the chromatin-rich areas (Belin et al., 2013). The function of these structures is not known, but it was speculated that they could contribute to the viscoelastic nature of the nucleoplasm. The Grosse lab, on the other hand, utilized both nuclear-targeted LifeAct and nuclear actin chromobody (camelid nanobody) to demonstrate transient nuclear actin polymerization upon serum stimulation (Baarlink et al., 2013) as well as upon cell spreading or fibronectin-induced integrin signaling (Plessner et al., 2015). In both cases, the polymerization required formins mDia1/2 and was linked to activation of the MRTF-A transcription cofactor (Baarlink et al., 2013; Plessner et al., 2015), which regulates the activity of the transcription factor SRF in response to changes in actin monomer levels in both the cytoplasm and the nucleus (Vartiainen et al., 2007). Linker of nucleoskeleton and cytoskeleton (LINC) complex, as well as components of the nuclear lamina, were required for the cell spreading–induced nuclear actin polymerization (Plessner et al., 2015), implying mechanical sensing of the signal from integrin-mediated cell adhesions into the nucleus. Interestingly, also engagement of the T-cell antigen receptor (TCR) results in very fast, transient nuclear actin polymerization. TCR engagement leads to elevated intranuclear Ca2+-levels, which stimulates nuclear actin polymerization via N-WASP and Arp2/3 complex, with the formin Fmn2 also playing a role. Formation of this nuclear actin filament network was shown to be required, via an unknown mechanism, for the efficient expression of key effector cytokines, and essential for CD4+ T-cell helper function in vivo (Tsopoulidis et al., 2019). Beyond transcriptional activation, transient nuclear actin polymerization has also been reported in the early G1 phase of the cell cycle (Baarlink et al., 2017; Parisis et al., 2017). The Grosse lab demonstrated that this nuclear actin polymerization was required for nuclear expansion and consequently for chromatin decondensation after mitosis (Baarlink et al., 2017), while the Fisher lab suggested a role for nuclear actin dynamics in DNA replication (Parisis et al., 2017). The actin nucleator involved during early G1 has not been unambiguously demonstrated, although formins may play a role (Parisis et al., 2017). However, cofilin, an actin disassembly factor, is essential for the filament turnover (Baarlink et al., 2017). Taken together, these studies have unambiguously demonstrated that nuclear actin can polymerize, that the polymerization is very tightly regulated, and most importantly, that it plays essential roles in processes ranging from activation of specific transcriptional programs to cell cycle progression.
Nuclear actin polymerizes also in response to DNA damage, and it seems that many different actin-regulatory proteins contribute to the process. In Drosophila cells, double-strand breaks in heterochromatin relocalize to the nuclear periphery to repair sites. This takes place by directed motion along nuclear actin filaments, which are assembled by the Arp2/3 complex at the repair sites. Also nuclear myosins play a role here, and myosin activator Unc45 recruits them to repair sites through interactions with the repair complex proteins Smc5/6 (Caridi et al., 2018). Interestingly, nuclear myosin 1 has been linked to large rearrangements in chromosomal territories that take place during DNA damage (Fatakia et al., 2017), implying that an actomyosin motor could drive chromatin movements at various scales during DNA damage response. In mammalian cells, double-strand breaks in active genes cluster, and the clustering coincides with delayed repair. Interestingly, clustering seems to require the activity of formin family member Fmn2 (Aymard et al., 2017), which has previously been shown to induce polymerization of nuclear actin, together with Spire 1/2 proteins, in response to DNA damage (Belin et al., 2015). However, a subsequent study found that nuclear actin polymerization, mediated by WASp-Arp2/3 complex, is specifically required for clustering of DNA breaks undergoing homology-directed repair, whereas nonhomologous end-joining was not affected. Curiously, in this case nuclear actin did not seem to promote directed motion of the DNA breaks, but rather increased their general mobility, which was described as confined Brownian motion (Schrank et al., 2018). Phosphoinositides, which are important regulators of several actin-binding proteins in the cytoplasm, are also present in the cell nucleus, and accumulate to the sites of DNA damage, where they recruit mDia2 to induce nuclear actin polymerization, which is required for the recruitment of ATR kinase, an essential activator of the DNA damage checkpoint that leads to cell cycle arrest (Wang et al., 2017). In addition, recent work has shown that there is an important role for mDia2-mediated nuclear actin polymerization to confine centromere movement during CENP-A loading (Liu et al., 2018). Taken together, it is evident from the existing literature that nuclear actin does polymerize upon DNA damage, and that this polymerization is required for the efficient repair of the damage. However, several mechanisms have been suggested, and in the future, it will be important to understand whether these differences reflect true multifunctionality of nuclear actin upon DNA damage or stem from the different experimental systems and/or used methodology. Of note, in addition to movement of DNA repair sites, nuclear actin has also been linked to movement of genomic loci and even whole chromosomes. For instance, actin has been linked to movement of U2 snRNA locus toward Cajal bodies (Dundr et al., 2007), to movement of HSP70 transgene toward nuclear speckles during heat shock (Khanna et al., 2014), and together with myosins, implicated in the rearrangement of specific chromosomes upon serum withdrawal (Mehta et al., 2010). This indicates that analogous to its functions in the cytoplasm, one of the main functions of actin could be to mediate dynamic chromatin rearrangements.
FUTURE PERSPECTIVES
During the past decade, actin has emerged as an essential protein also in the cell nucleus, with functional roles in processes ranging from transcriptional regulation to maintenance of genomic integrity. Here, a future development is to obtain structural data to gain unambiguous evidence of the role of actin interactions with RNA polymerases across the gene. In terms of a potential role of actin at the genomic level, an interesting model that we have previously discussed is that an actin-based nucleoskeleton may regulate the functional architecture of the genome (Xie and Percipalle, 2018). This would be particularly important during the process of development and differentiation when changes in genome organization ensure that gene programs are either activated or repressed to achieve a specific cellular identity. We now have the possibility to address these exciting questions by applying chromosome conformation capture technologies coupled to deep sequencing in cells with compromised actin expression. An exciting development has also been the realization that nuclear actin polymerizes with the help of many proteins that regulate actin dynamics also in the cytoplasm, and that also the filamentous form of actin has pivotal roles in key nuclear processes. A critical challenge in the field is to uncover the molecular mechanisms involved. The multifunctional nature of actin both in the cytoplasm and in the nucleus poses a significant challenge to this, and necessitates the identification and comprehensive characterization of the nuclear binding partners for actin. Further development of imaging techniques, both superresolution light microscopy as well as perhaps CryoEM techniques for structural cell biology would facilitate ultrastructural analysis of nuclear actin filament networks.
Acknowledgments
This work was partly supported by grants from New York University Abu Dhabi, the Swedish Research Council (Vetenskapsrådet), and the Swedish Cancer Society (Cancerfonden) to P.P., as well as by grants from the Academy of Finland, the Jane and Aatos Erkko Foundation, the Sigrid Juselius Foundation, and the Helsinki Institute of Life Science to M.K.V.
Abbreviations used:
- ARPs
actin-related proteins
- BAF
Brahma-related gene-1 associated factor
- Brg1
Brahma-related gene-1
- CTD
carboxyl terminal domain
- FRAP
fluorescence recovery after photobleaching
- hnRNP
heterogeneous nuclear ribonucleoprotein
- HP1
heterochromatin-binding protein 1
- LINC
linker of nucleoskeleton and cytoskeleton
- SAF-A
scaffold attachment factor A
- SMARCA4
SWI/SNF-related matrix associated actin dependent regulator of chromatin
- SWI/SNF
SWItch/sucrose non-fermentable
Footnotes
REFERENCES
- Almuzzaini B, Sarshad AA, Rahmanto AS, Hansson ML, Von Euler A, Sangfelt O, Visa N, Farrants AK, Percipalle P. (2016). In β-actin knockouts, epigenetic reprogramming and rDNA transcription inactivation lead to growth and proliferation defects. FASEB J , 2860–2873. [DOI] [PubMed] [Google Scholar]
- Aymard F, Aguirrebengoa M, Guillou E, Javierre BM, Bugler B, Arnould C, Rocher V, Iacovoni JS, Biernacka A, Skrzypczak M, et al (2017). Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes. Nat Struct Mol Biol , 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baarlink C, Plessner M, Sherrard A, Morita K, Misu S, Virant D, Kleinschnitz EM, Harniman R, Alibhai D, Baumeister S, et al (2017). A transient pool of nuclear F-actin at mitotic exit controls chromatin organization. Nat Cell Biol , 1389–1399. [DOI] [PubMed] [Google Scholar]
- Baarlink C, Wang H, Grosse R. (2013). Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science , 864–867. [DOI] [PubMed] [Google Scholar]
- Belin BJ, Cimini BA, Blackburn EH, Mullins RD. (2013). Visualization of actin filaments and monomers in somatic cell nuclei. Mol Biol Cell , 982–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin BJ, Lee T, Mullins RD. (2015). DNA damage induces nuclear actin filament assembly by Formin-2 and Spire-(1/2) that promotes efficient DNA repair. Elife , e07735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caridi CP, D’Agostino C, Ryu T, Zapotoczny G, Delabaere L, Li X, Khodaverdian VY, Amaral N, Lin E, Rau AR, Chiolo I. (2018). Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature , 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvertino S, Stuart HM, Chandler KE, Roberts NA, Armstrong R, Bernardini L, Bhaskar S, Callewaert B, Clayton-Smith J, Hernando Davalillo C, et al (2017). ACTB loss-of-function mutations result in a pleiotropic developmental disorder. Am J Hum Genet , 1021–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopie J, Skarp KP, Kaisa Rajakyla E, Tanhuanpaa K, Vartiainen MK. (2012). Active maintenance of nuclear actin by importin 9 supports transcription. Proc Natl Acad Sci USA , E544–E552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. (2007). Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol , 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatakia SN, Kulashreshtha M, Mehta IS, Rao BJ. (2017). Chromosome territory relocation paradigm during DNA damage response: some insights from molecular biology to physics. Nucleus , 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomproix N, Percipalle P. (2004). An actin-myosin complex on actively transcribing genes. Exp Cell Res , 140–148. [DOI] [PubMed] [Google Scholar]
- Grosse R, Vartiainen MK. (2013). To be or not to be assembled: progressing into nuclear actin filaments. Nat Rev Mol Cell Biol , 693–697. [DOI] [PubMed] [Google Scholar]
- Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, Kysela K, Goodrich JA, Lessard JL, Hope TJ, et al (2004). Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol , 1094–1101. [DOI] [PubMed] [Google Scholar]
- Hu P, Wu S, Hernandez N. (2004). A role for β-actin in RNA polymerase III transcription. Genes Dev , 3010–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor P, Shen XT. (2014). Mechanisms of nuclear actin in chromatin-remodeling complexes. Trends Cell Biol , 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna N, Hu Y, Belmont AS. (2014). HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr Biol , 1138–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll KR, Eustermann S, Niebauer V, Oberbeckmann E, Stoehr G, Schall K, Tosi A, Schwarz M, Buchfellner A, Korber P, Hopfner KP. (2018). The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling. Nat Struct Mol Biol , 823–832. [DOI] [PubMed] [Google Scholar]
- Kotani T, Yasuda K, Ota R, Yamashita M. (2013). Cyclin B1 mRNA translation is temporally controlled through formation and disassembly of RNA granules. J Cell Biol , 1041–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukalev A, Nord Y, Palmberg C, Bergman T, Percipalle P. (2005). Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat Struct Mol Biol , 238–244. [DOI] [PubMed] [Google Scholar]
- Kwon SJ, Park JH, Park EJ, Lee SA, Lee HS, Kamg SW, Kwon J. (2015). ATM-mediated phosphorylation of the chromatin remodeling enzyme BRG1 modulates DNA double-strand break repair. Oncogene , 303–313. [DOI] [PubMed] [Google Scholar]
- Liu C, Zhu R, Mao Y. (2018). Nuclear actin polymerized by mDia2 confines centromere movement during CENP-A loading. iScience , 314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ. (2006). Nucleoplasmic β-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol , 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta IS, Amira M, Harvey AJ, Bridger JM. (2010). Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts. Genome Biol , R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrdlik A, Kukalev A, Louvet E, Farrants AK, Caputo L, Percipalle P. (2008). The histone acetyltransferase PCAF associates with actin and hnRNP U for RNA polymerase II transcription. Mol Cell Biol , 6342–6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olave IA, Reck-Peterson SL, Crabtree GR. (2002). Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem , 755–781. [DOI] [PubMed] [Google Scholar]
- Parisis N, Krasinska L, Harker B, Urbach S, Rossignol M, Camasses A, Dewar J, Morin N, Fisher D. (2017). Initiation of DNA replication requires actin dynamics and formin activity. EMBO J , 3212–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P. (2009). The long journey of actin and actin-associated proteins from genes to polysomes. Cell Mol Life Sci , 2151–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P. (2013). Co-transcriptional nuclear actin dynamics. Nucleus , 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, Fomproix N, Kylberg K, Miralles F, Bjorkroth B, Daneholt B, Visa N. (2003). An actin-ribonucleoprotein interaction is involved in transcription by RNA polymerase II. Proc Natl Acad Sci USA , 6475–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, Zhao J, Pope B, Weeds A, Lindberg U, Daneholt B. (2001). Actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from the gene to polysomes. J Cell Biol , 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philimonenko VV, Zhao J, Iben S, Dingová H, Kyselá K, Kahle M, Zentgraf H, Hofmann WA, de Lanerolle P, Hozák P, Grummt I. (2004). Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol , 1165–1172. [DOI] [PubMed] [Google Scholar]
- Plessner M, Melak M, Chinchilla P, Baarlink C, Grosse R. (2015). Nuclear F-actin formation and reorganization upon cell spreading. J Biol Chem , 11209–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrank BR, Aparicio T, Li Y, Chang W, Chait BT, Gundersen GG, Gottesman ME, Gautier J. (2018). Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature , 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivashankar GV. (2019). Mechanical regulation of genome architecture and cell-fate decisions. Curr Opin Cell Biol , 115–121. [DOI] [PubMed] [Google Scholar]
- Singhal N, Graumann J, Wu G, Araúzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Schöler HR. (2010). Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell , 943–955. [DOI] [PubMed] [Google Scholar]
- Sokolova M, Moore HM, Prajapati B, Dopie J, Merilainen L, Honkanen M, Matos RC, Poukkula M, Hietakangas V, Vartiainen MK. (2018). Nuclear actin is required for transcription during Drosophila oogenesis. iScience , 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuven T, Hartmann E, Gorlich D. (2003). Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J , 5928–5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondeleir D, Lambrechts A, Müller M, Jonckheere V, Doll T, Vandamme D, Bakkali K, Waterschoot D, Lemaistre M, Debeir O, et al (2012). Cells lacking β-actin are genetically reprogrammed and maintain conditional migratory capacity. Mol Cell Proteomics , 255–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsopoulidis N, Kaw S, Laketa V, Kutscheidt S, Baarlink C, Stolp B, Grosse R, Fackler OT. (2019). T cell receptor–triggered nuclear actin network formation drives CD4+ T cell effector functions. Sci Immunol , eaav1987. [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, Treisman R. (2007). Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science , 1749–1752. [DOI] [PubMed] [Google Scholar]
- Viita T, Kyheröinen S, Prajapati B, Virtanen J, Frilander MJ, Varjosalo M, Vartiainen MK. (2019). Nuclear actin interactome analysis links actin to KAT14 histone acetyl transferase and mRNA splicing. J Cell Sci , jcs226852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N, Percipalle P. (2010). Nuclear functions of actin. Cold Spring Harb Perspect Biol , a000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Hariharan A, Bastianello G, Toyama Y, Shivashankar GV, Foiani M, Sheetz MP. (2017). DNA damage causes rapid accumulation of phosphoinositides for ATR signaling. Nat Commun , 2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Almuzzaini B, Drou N, Kremb S, Yousif A, Farrants AÖ, Gunsalus K, Percipalle P. (2018a). β-actin-dependent global chromatin organization and gene expression programs control cellular identity. FASEB J , 1296–1314. [DOI] [PubMed] [Google Scholar]
- Xie X, Jankauskas R, Mazari AMA, Drou N, Percipalle P. (2018b). β-actin regulates a heterochromatin landscape essential for optimal induction of neuronal programs during direct reprograming. PLoS Genet , e1007846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Percipalle P. (2018). An actin-based nucleoskeleton involved in gene regulation and genome organization. Biochem Biophys Res Commun , 378–386. [DOI] [PubMed] [Google Scholar]