Abstract
The cardiomyocyte cytoskeleton, including the sarcomeric contractile apparatus, forms a cohesive network with cellular adhesions at the plasma membrane and nuclear–cytoskeletal linkages (LINC complexes) at the nuclear envelope. Human cardiomyopathies are genetically linked to the LINC complex and A-type lamins, but a full understanding of disease etiology in these patients is lacking. Here we show that SUN2-null mice display cardiac hypertrophy coincident with enhanced AKT/MAPK signaling, as has been described previously for mice lacking A-type lamins. Surprisingly, in contrast to lamin A/C-null mice, SUN2-null mice fail to show coincident fibrosis or upregulation of pathological hypertrophy markers. Thus, cardiac hypertrophy is uncoupled from profibrotic signaling in this mouse model, which we tie to a requirement for the LINC complex in productive TGFβ signaling. In the absence of SUN2, we detect elevated levels of the integral inner nuclear membrane protein MAN1, an established negative regulator of TGFβ signaling, at the nuclear envelope. We suggest that A-type lamins and SUN2 play antagonistic roles in the modulation of profibrotic signaling through opposite effects on MAN1 levels at the nuclear lamina, suggesting a new perspective on disease etiology.
INTRODUCTION
The mammalian myocardium is composed of cardiomyocytes, which contain sarcomeres, the basic structural unit of muscle. Sarcomeres form a cohesive tissue-scale network of cell–cell adhesions at the intercalated disk (ICD) and cell–extracellular matrix adhesions at costameres in these cells. Embedded into the contractile network of cardiomyocytes is the nucleus, which is mechanically integrated into the cytoskeleton through nuclear envelope-spanning LINC (linker of nucleoskeleton and cytoskeleton) complexes, which consist of SUN domain proteins in the inner nuclear membrane and KASH domain proteins, Nesprins, or SYNEs in mammals, in the outer nuclear membrane (Chang et al., 2015). These complexes interface directly and indirectly with all components of the cytoskeleton on their cytoplasmic faces, and interact with the nuclear lamina on their nucleoplasmic faces (Chang et al., 2015). While it is well established that the formation and maintenance of cellular adhesions is dependent on proper cytoskeletal function (reviewed in Sequeira et al., 2014), our previous work (and that of others) implicates LINC complexes and the nuclear lamina as unexpected regulators of cellular adhesions at the plasma membrane (Mounkes et al., 2005; Frock et al., 2012; Stewart et al., 2015). In addition to their structural roles, adhesions and components of the sarcomere are mechanoresponsive and play an important role in the biochemical signaling processes that promote cardiac function, such as the normal hypertrophic growth that occurs during cardiac development (reviewed in McCain and Parker, 2011; Maillet et al., 2013). Thus, it is not surprising that defects in adhesion are strongly implicated in human cardiomyopathies (Sheikh et al., 2009; Delmar and McKenna, 2010; Israeli-Rosenberg et al., 2014).
The LINC complex and other components of the nuclear lamina, such as emerin and A-type lamins, are also genetically linked to various human myopathies, including dilated cardiomyopathy, arrhythmogenic cardiomyopathy, and syndromes with cardiac involvement such as Emery–Dreifuss muscular dystrophy (EDMD; Ostlund et al., 2001; Stroud et al., 2014). Strikingly, a large proportion of individuals with dilated cardiomyopathies exhibit lamin A/C point mutations, preceded in prevalence only by mutations in titin (Arbustini et al., 2002; Cahill et al., 2013). Point mutations in Sun2, one of the most widely expressed genes encoding a SUN domain–containing protein in mammals, have been found to act as genetic modifiers of cardiomyopathy, for example, exacerbating the severity of the disease in patients with mutations in the sarcomere component myosin binding protein C or other components of the nuclear lamina (Meinke et al., 2014). These point mutations in Sun2 reside either in the lamin A-binding region (M50T) or in the coiled-coil region, required for the trimerization of LINC complexes and Nesprin engagement (V378I; Sosa et al., 2012; Meinke et al., 2014). Further, SUN2 V378I and other point mutations in the coiled-coil region have been found in patients with EDMD or cardiomyopathy symptoms (Meinke et al., 2014). Despite the strong association between disruption of the LINC complex or nuclear lamina and human cardiac disease, a mechanistic understanding of the origin of these genetic diseases remains elusive. One prevailing model focuses on characteristic changes in nuclear morphology that accompany disruption of nuclear lamina components (Mounkes et al., 2005; Gupta et al., 2010; Zwerger et al., 2013), suggesting that nuclear fragility is the driving force behind disease etiology (reviewed in Davidson and Lammerding, 2014).
An additional (or alternative) mechanistic link has recently come to light from studies employing mouse models of lamin depletion (or a cardiomyopathy-linked lamin A/C mutation, H222P). Here, several studies have uncovered altered MAP kinase and AKT signaling as a driver of cardiac dysfunction (Muchir et al., 2007, 2012b; Wu et al., 2011; Choi et al., 2012); reversing this biochemical cascade by treatment with the TOR inhibitor rapamycin improved heart function (Choi et al., 2012; Liao et al., 2016). However, the mechanism(s) by which mutations in lamin A (or loss of lamin A) alter the AKT and TOR pathways remains unknown. Importantly, both the AKT and the profibrotic TGFβ pathways that drive cardiac hypertrophy and interstitial fibrosis are mechanoresponsive, largely through pathways sensitive to inputs from cellular adhesions and/or the state of the actomyosin cytoskeleton (Iijima et al., 2002; Balasubramanian and Kuppuswamy, 2003; Gomez et al., 2010; Young et al., 2014; Hinz, 2015; O’Connor et al., 2015; Varney et al., 2016). Models of LINC complex ablation provide an opportunity to test whether communication of mechanical signals from the cytoskeleton to the nuclear interior (Lammerding et al., 2004; Lee et al., 2007; Hale et al., 2008; Stewart-Hutchinson et al., 2008; Luxton et al., 2010; Folker et al., 2011; Khatau et al., 2012; Long et al., 2013; Myat et al., 2015; Stewart et al., 2015) contributes to myocardium function, although to date this avenue of investigation has been largely unexplored.
Importantly, suppressing MAPK signaling improves cardiac performance in the lamin A/C H222P mouse model, significantly ameliorating the coincident fibrosis (Muchir et al., 2007, 2012a; Wu et al., 2011; Choi et al., 2012). Moreover, fibrosis is a hallmark of the dysfunction characteristic of other laminopathic syndromes, including progeria and lipodystrophy (Olive et al., 2010; Le Dour et al., 2017). While the underlying mechanisms are poorly understood, it is interesting to note that MAN1, an integral inner nuclear membrane (INM) protein that requires A-type lamins to be retained at the INM (Ostlund et al., 2006), is a clear negative regulator of TGFβ signaling across a broad range of organisms (Raju et al., 2003; Lin et al., 2005; Ishimura et al., 2006). In addition to binding directly to R-SMADs, MAN1 has also been suggested to regulate SMAD signaling in a tissue stiffness-dependent manner (Chambers et al., 2018).
Here, to address how LINC complexes contribute to heart function, we have examined the consequences of disrupting SUN2-containing LINC complexes in the murine myocardium, and we find that this genetic perturbation induces cardiac hypertrophy. Sun2-/- mice display elevated AKT-mTOR and MAPK signaling in the myocardium, which we tie to increased integrin engagement at costameres. Surprisingly, these mice fail to induce expression of classic hypertrophy-associated genes, have a normal lifespan, lack fibrosis, and demonstrate down-regulation or unaltered levels of TGFβ target genes despite elevated levels of a transducer of this pathway, nuclear phospho-SMAD2. While lamin A/C is required for MAN1 targeting, we find that SUN2-null mice instead display elevated retention of MAN1 at the nuclear lamina. Taken together, these results suggest that A-type lamins and the LINC complex act in concert to regulate prohypertrophic signaling, but play antagonistic roles in driving fibrosis.
RESULTS
Mice deficient for Sun2 undergo cardiac hypertrophy
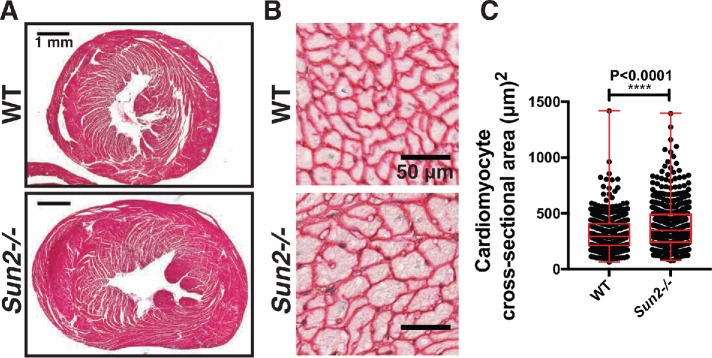
To assess the functional consequences of Sun2 loss in the murine myocardium, we obtained a previously reported Sun2-/- whole-body knockout mouse model (Lei et al., 2009). In wild-type (WT) left ventricular cardiomyocytes, SUN2 is localized to the nuclear envelope and is absent from Sun2-/- tissue (Supplemental Figure 1A); SUN1 expression is not substantially different in the hearts of Sun2-/- mice compared with WT (Supplemental Figure 1B). While we did not observe increases in spontaneous cardiac-associated deaths in aged mice (>1 yr), gross histology of hearts cut at the midventricular level revealed enlargement of Sun2-/- hearts in comparison with WT hearts at more than 1 yr of age (Figure 1A). These findings were recapitulated at the cellular level, as we observed significant enlargement of individual cardiomyocytes in the papillary muscle of Sun2-/- mice (Figure 1, B and C). These results suggest that Sun2-/- mice exhibit age-related cardiac hypertrophy at both the cellular and tissue levels.
FIGURE 1:
Sun2-/- murine hearts exhibit hypertrophy. (A) Paraffin-embedded hearts isolated from 13-mo-old WT and Sun2-/- mice were stained with Masson’s trichrome. Representative images show enlargement of the Sun2-/- heart in comparison with the WT; images of additional hearts are displayed in Supplemental Figure 1C. (B) Paraffin-embedded hearts from WT and Sun2-/- mice were stained with antibodies against laminin to reveal cardiomyocyte outlines. Cardiomyocytes from left ventricular papillary muscle are shown in cross section. Note the enlargement of Sun2-/- cells as compared with WT. (C) Quantification of left ventricular papillary muscle cardiomyocyte cross-sectional area, showing a greater population of enlarged cells in Sun2-/- than in WT heart. n ≥ 86 cells (86–198 cells) for each of three mice per genotype. Error bars indicate SDs. Statistical significance determined by unpaired, two-tailed t test.
Sun2-/- mice exhibit altered sarcomere structure and adhesion defects
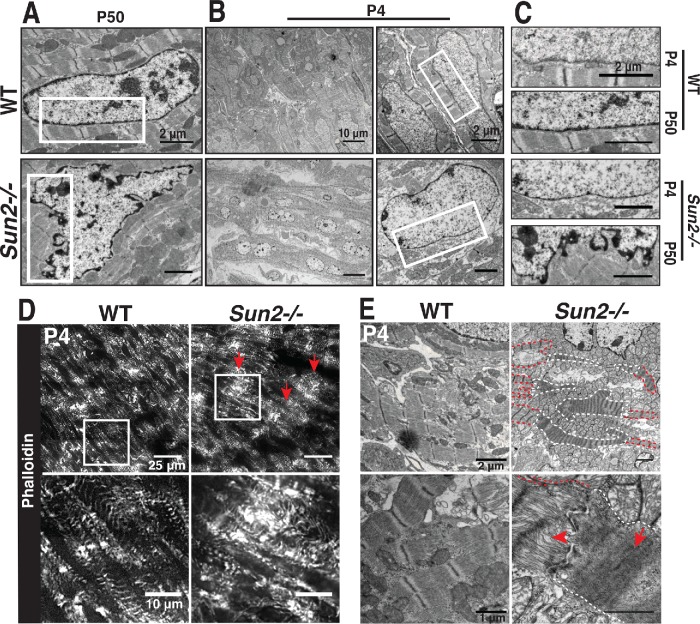
Cardiac dysfunction is often tied to changes in sarcomere structure. In particular, myofibril disarray has been linked to sarcomere mutations, many of which drive increased contractile function of the sarcomere at the cellular level (Michele et al., 1999; Lowey, 2002; Moore et al., 2012). Assessment of sarcomere organization in P50 mice by staining left ventricle (LV) cardiac tissue sections with phalloidin revealed the stereotypical banding of F-actin, corresponding to I-bands, in WT mice (Figure 2A). However, Sun2-/- tissue displayed actin bands of irregular width that did not align laterally between adjacent myofibrils, as well as regions with extensive actin disorganization (Figure 2A, arrowheads). At higher magnification by transmission electron microscopy (TEM), we find that while many regions of the Sun2-/- tissue still exhibited grossly intact myofibril structure (Figure 2B), these regions displayed misaligned and wavy Z-bands (Figure 2B, red lines) and M-bands (Figure 2B, arrowheads), loss of clearly defined I-bands (Figure 2B, arrows) and H-zones (Figure 2B, arrowheads), and reduced sarcomere length (Figure 2C). Focal regions of severe myofibril disarray with complete loss of sarcomere structure were also observed in the Sun2-/- LV (Figure 2B, bottom panels).
FIGURE 2:
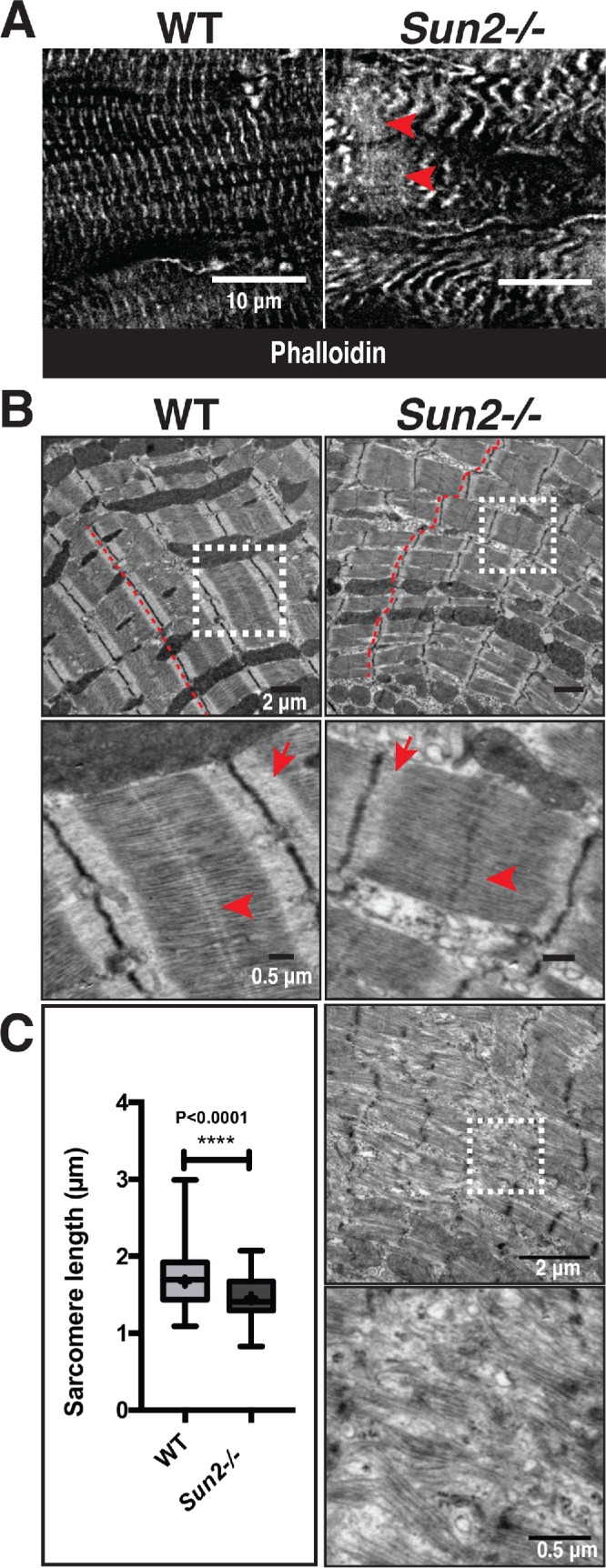
Sarcomere organization is perturbed in P50 Sun2-/- mice. (A) Frozen P50 WT and Sun2-/- cardiac left ventricle tissue was sectioned and stained with phalloidin to label F-actin in I-bands. While I-bands are evenly registered in WT tissue, the regularity of this pattern is lost in Sun2-/-. Note the variability in width of I-bands, their loss of alignment and increased wavy appearance, and focal loss of sarcomere structure (arrowheads). (B) Transmission electron micrographs (TEMs) of P50 WT and Sun2-/- left ventricle tissue. Classical sarcomere structure can be observed in WT mice, while Sun2-/- tissue exhibits misaligned and wavy Z-bands (compare red lines in WT vs. Sun2-/-) and loss of clearly defined I-bands (arrows). Focal regions of complete sarcomere disruption are also present in the Sun2-/ tissue (bottom two panels). (C) Sarcomere length, as measured from Z-band to Z-band, is reduced in P50 Sun2-/- tissue. n = 3 mice for each genotype. Statistical significance determined by unpaired, two-tailed t test.
Our previous work showing a requirement for SUN2 LINC complexes in intercellular adhesion organization and function (Stewart et al., 2015) inspired us to examine the structure of intercalated discs, the intercellular adhesions that connect adjacent cardiomyocytes. TEM analysis revealed discontinuous jagged intercalated discs with lacunae where sarcomeric cytoskeletal filaments failed to properly interface with intercalated disk components in P50 Sun2-/- tissue (Supplemental Figure 2A, arrows). Further, we observed reduced electron density along the intercalated disk membranes, suggesting that there was reduced adhesion protein accumulation at these sites (Supplemental Figure 2A, arrowheads). Immunofluorescence staining of intercalated disk components desmoplakin I/II, corresponding to desmosomes, and β-catenin, corresponding to adherens junctions, revealed discrete bands that ran perpendicular to the cardiomyocyte lateral membranes in WT tissue (Supplemental Figure 2, B and C). However, we frequently observed expansion of these normally discrete bands into several thinner, separated bands in the Sun2-/- tissue (Supplemental Figure 2, B and C, arrowheads), which mirrored the discontinuous appearance of the intercalated discs at the level of TEM (Supplemental Figure 2A). These results suggest that intercellular adhesion is structurally perturbed in the absence of SUN2 LINC complexes.
Sarcomere disorganization precedes changes in nuclear morphology in Sun2-/- mice
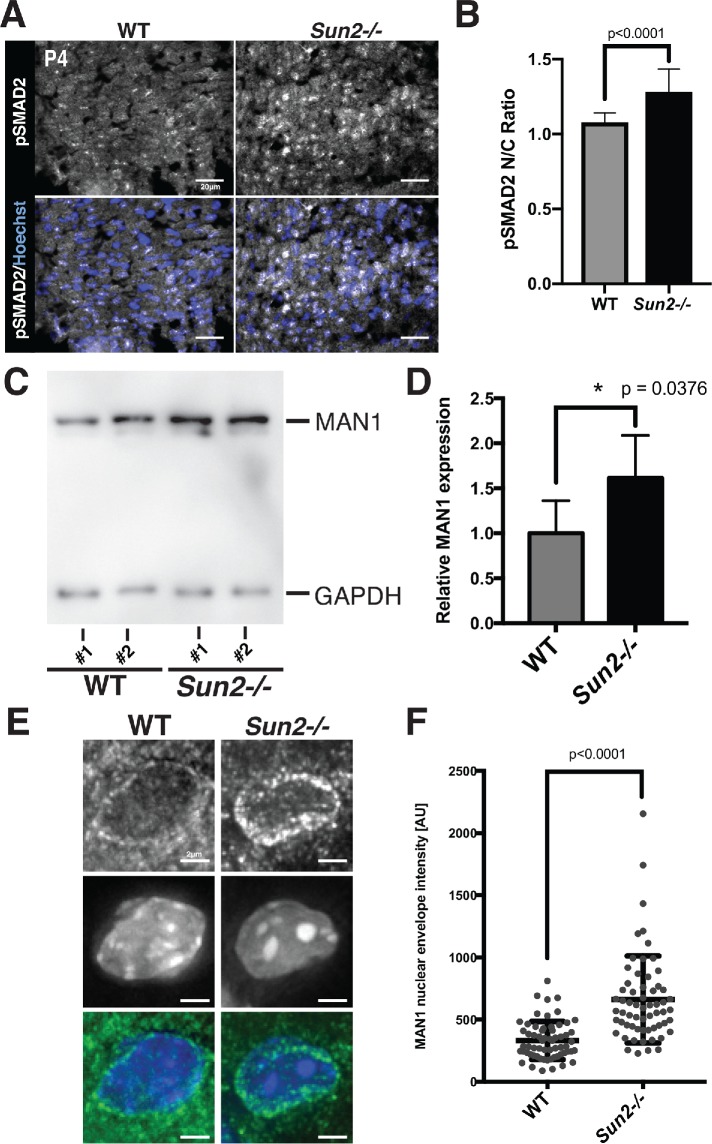
By P50 we observed extensive changes in sarcomere organization and cardiomyocyte adhesion in Sun2-/- mice. To address the likely foundational defect that drives these phenotypes, we next investigated early time points in mouse development. Further, as we observed altered nuclear morphology in P50 Sun2-/- cardiac tissue (Figure 3, A and C), which has been suggested to drive disease in laminopathies and nuclear envelopathies (Mounkes et al., 2005; Gupta et al., 2010; Zwerger et al., 2013), we also wished to investigate whether changes in the myocardium ultrastructure preceded or followed changes in nuclear shape. Morphological changes in nuclear shape were not present at P4 (Figure 3, B and C), suggesting that defects in nuclear morphology arise only after mature cardiac beating begins. We did observe that P4 Sun2-/- nuclei exhibited a somewhat rounder morphology than the more elongated WT nuclei (Figure 3B); however, we would argue that this observation likely reflects LINC complex-dependent tension normally being exerted by the cytoskeleton on the nucleus in WT cardiomyocytes, a phenomenon that may be lost upon ablation of LINC complexes (Zwerger et al., 2013; Hatch and Hetzer, 2016).
FIGURE 3:
Nuclear morphology is normal at P4 but sarcomere organization is disrupted in Sun2-/- mice. (A–C) TEM of WT and Sun2-/- left ventricle tissue at P50 and P4. (A) At P50, nuclei in Sun2-/- tissue exhibit severe distortions of the nuclear envelope. (B) At P4, Sun2-/- nuclear envelope morphology is comparable to the morphology in WT tissue. Left panels display lower-magnification fields, illustrating the prevalence of morphologically similar nuclei in WT and Sun2-/- tissue. However, WT nuclei are more elongated than Sun2-/- nuclei. Right panels display higher magnification of single nuclei, highlighting the similarity in nuclear envelope structure in the two genotypes. (C) Insets from white boxed regions in A and B, illustrating the normal nuclear morphology at P4 and disruption by P50 in Sun2-/- tissue. (D) Frozen P4 WT and Sun2-/- cardiac left ventricle tissue was sectioned and stained with phalloidin. While nascent myofibrils in WT tissue display irregularity in I-band width and alignment, focal regions of Sun2-/- tissue exhibit severe sarcomere disruption (arrows). (E) TEM of P4 WT and Sun2-/- left ventricle tissue. Sarcomere structure is disrupted in Sun2-/- tissue, with cytoskeletal disorganization (arrowhead) and hypercontraction (arrow) visible in adjacent cells. Hypercontractile cells are outlined in white, while adjacent cells with more subtly disrupted sarcomere organization are outlined in red.
Given that the nuclei appeared intact in the Sun2-/- LV at P4, we next asked whether the P50 sarcomere defects (Figure 2) manifest before or after this stage in development; in addition, the ICDs are not yet mature at P4 (Hirschy et al., 2006; Wang et al., 2012), further allowing us to examine whether the sarcomere changes occurred before ICD formation. As many structures in the murine heart, including myofibrils and adhesions, continue to develop postnatally (Hirschy et al., 2006; Wang et al., 2012), phalloidin staining of P4 WT cardiac tissue revealed a less regular pattern of actin-rich I-bands than was observed at P50 (Figure 3D vs. Figure 2A). However, at this earlier age we again observed extensive disruptions in myofibril organization in Sun2-/- tissue in comparison with WT tissue (Figure 3D, arrows). These defects were further clarified following TEM of P4 LV myocardium, where Sun2-/- tissue featured ragged sarcomeres with a loss of clearly discernible I-bands (Figure 3E, arrowhead) and irregular Z-band spacing (Figure 3E, arrow). This indicates that sarcomere disarray is present during early postnatal cardiac development in mice lacking Sun2, before changes in nuclear morphology and ICD maturation.
We identified several regions in the P4 Sun2-/- tissue with extensive, striking reductions in the spacing between Z-bands, a phenotype previously linked to hypercontractility of the sarcomere (Lauritzen et al., 2009; Figure 3E, arrow). It appears that this effect was cell-autonomous, as individual cells with this hallmark of hypercontractility were located only sporadically through the examined tissue, while adjacent cells displayed more subtle changes in sarcomere structure (Figure 3E; compare cells outlined in white with adjacent cells outlined in red). In contrast to the P50 time point, we did not observe noticeable differences in intercellular adhesion in Sun2-/- mice during the 2-wk postnatal period when intercalated disk maturation occurred (Hirschy et al., 2006; Wang et al., 2012; Supplemental Figure 2D). This suggests that the sarcomere disarray in Sun2-/- mice precedes the alterations in intercellular adhesion, and that the intercalated disk defects observed by P50 may be a consequence of altered contractility and/or the sarcomere defects in these mice.
Sun2-/- mice exhibit increased integrin engagement and AKT/MAPK signaling
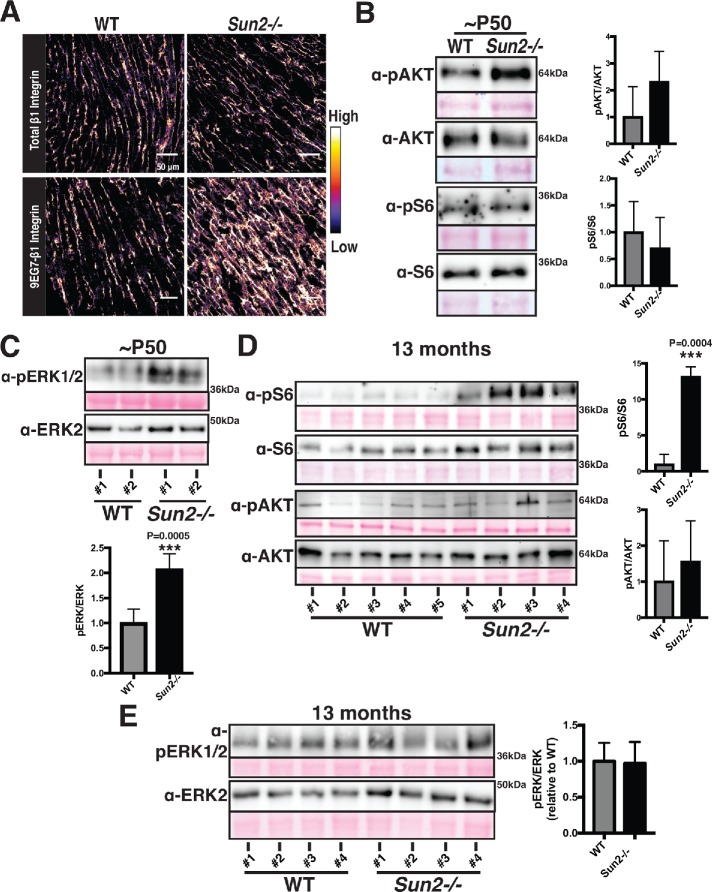
Given the established roles that costameres, the sites of cell–extracellular matrix adhesion, play in cardiac development, sarcomere organization, and integrin signaling upstream of the prohypertrophic AKT, mTOR, and MAPK pathways and their mechanoresponsive nature (MacKenna et al., 1998; Franchini et al., 2000; Laser et al., 2000; Balasubramanian and Kuppuswamy, 2003; Brancaccio et al., 2006; Dowling et al., 2008; Konieczny et al., 2008; McCain et al., 2012; Israeli-Rosenberg et al., 2014; Wilsbacher and Coughlin, 2015), we next examined costameres for functional changes in Sun2-/- mice. We employed a well-established antibody (9EG7) that recognizes a ligand-bound epitope of β1-integrin, thought to correspond to the active, engaged form of the protein (Lenter et al., 1993; Bazzoni et al., 1995; Su et al., 2016). While levels of total β1-integrin were similar in WT and Sun2-/- P50 tissue, the levels of active 9EG7 β1-integrin appeared substantially higher in the Sun2-/- tissue (Figure 4A; see additional examples in Supplemental Figure 2E). We did not observe an increase in active β1-integrin levels at P13 (Supplemental Figure 2F) or in 13-mo-old mice (Supplemental Figure 2G). These results suggest that integrin function at the costamere may be altered in the absence of SUN2 LINC complexes at a specific stage in murine cardiac development.
FIGURE 4:
Sun2-/- hearts display increased integrin engagement and AKT/MAPK signaling. (A) Frozen P50 WT and Sun2-/- cardiac left ventricle tissue was sectioned and stained with antibodies against total β1-integrin (top panels) or against the ligand-bound, active β1-integrin (9EG7) conformational epitope (bottom panels). Images were pseudocolored to depict the relative fluorescence intensity of the β1-integrin signal, with lighter colors indicative of higher intensity and darker colors indicative of lower intensity. Note the increased intensity of active β1-integrin in the Sun2-/- tissue. Images are representative of results for 3 mice per genotype (also see Supplemental Figure 2, E–G). (B–E) Representative immunoblots of ventricular lysate from three WT and three Sun2-/- mice at P50 or five WT and four Sun2-/- mice at 13 mo. Ponceau staining of total protein reveals even loading of samples. Plots to the right represent quantitative analysis from additional mice, represented as the mean ± SD for more than three WT or Sun2-/- mice. Full blots, used for quantifications, are shown in Supplemental Figure 3. For quantification, data are shown as a ratio of Sun2-/- to WT for the phosphorylated protein and total protein, expressed as a ratio. (B) Lysates from P50 mice were subjected to SDS–PAGE and immunoblotting with antibodies against phosphorylated AKT (pAKT), AKT, phosphorylated S6 (pS6), or S6, revealing elevated levels of pAKT in Sun2-/- tissue. Data are represented as the mean ± SD for three WT or Sun2-/- mice. (C) Lysates from P50 mice were subjected to SDS–PAGE and immunoblotting with antibodies against pERK1/2 and ERK2, revealing elevated levels of pERK1/2 in Sun2-/- tissue. Data are represented as the mean ± SD for three WT or Sun2-/- mice. (D) Lysates from 13-mo-old mice were subjected to SDS–PAGE and immunoblotting with antibodies against phosphorylated S6 (pS6), S6, phosphorylated AKT (pAKT), or AKT, revealing elevated levels of pS6 but similar levels of pAKT in Sun2-/- tissue vs. WT tissue. Data are represented as the mean ± SD for five WT or four Sun2-/- mice. (E) Lysates from 13-mo-old mice were subjected to SDS–PAGE and immunoblotting with antibodies against pERK1/2 and ERK2, revealing similar levels of pERK1/2 in Sun2-/- and WT tissues. Data are represented as the mean ± SD for three WT or Sun2-/- mice.
Hypertrophy can be driven by integrin signaling, including through the AKT and MAPK pathways (MacKenna et al., 1998; Franchini et al., 2000; Laser et al., 2000; Kim et al., 2003; Rota et al., 2005; Cittadini et al., 2006; Shiojima and Walsh, 2006; Catalucci et al., 2009; Maillet et al., 2013). Moreover, AKT activation has been linked to increased cardiac function and cardiomyocyte contractility (Kim et al., 2003; Rota et al., 2005; Cittadini et al., 2006; Catalucci et al., 2009). At the time point where we observe greater integrin engagement (P50), immunoblot analysis of WT and Sun2-/- myocardium reveals increased levels of phosphorylated AKT (Figure 4B) and phosphorylated ERK1/2 (Figure 4C) in Sun2-/- animals. Further, in hypertrophic 13-mo-old Sun2-/- mice we observed a dramatic increase in the level of phosphorylated ribosomal protein S6 (Figure 4D), a key target downstream of the AKT and mTOR pathways linked to the hypertrophic response (Shiojima and Walsh, 2006; Maillet et al., 2013). At this 13-mo time point, where Sun2-/- mice no longer display increased integrin engagement (Supplemental Figure 2G), we failed to observe a significant increase in AKT phosphorylation (Figure 4D) or pERK1/2 levels (Figure 4E).
Loss of SUN2 uncouples AKT- and MAPK-driven hypertrophy from cardiac fibrosis
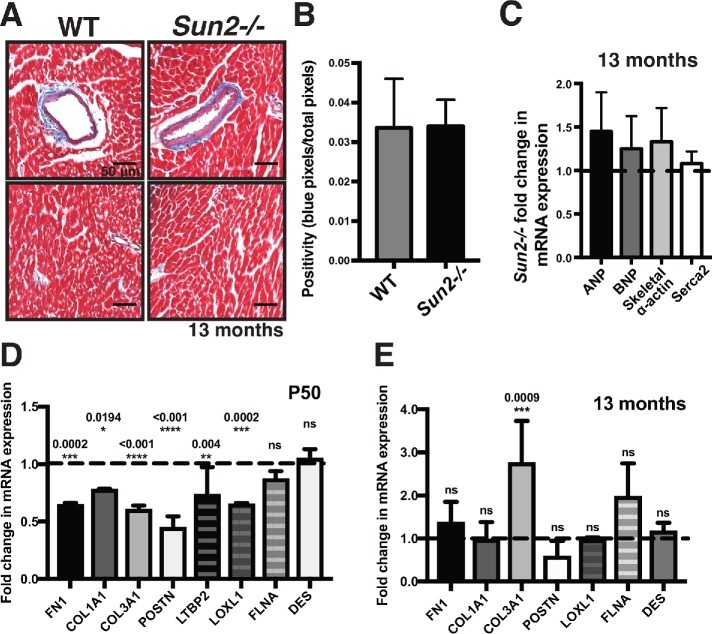
In the vast majority of disease contexts, cardiac hypertrophy occurs coincident with interstitial fibrosis (Ho et al., 2010), including in the laminopathic mouse models (Muchir et al., 2007, 2012b; Wu et al., 2011; Choi et al., 2012). Importantly, the consequences of disrupting lamin A function are quite severe, leading to death even before maturity is reached in the case of the lamin A/C-null model. Given the myriad cytological defects apparent in the myocardium of Sun2-/- mice, the gains in AKT and MAPK signaling, and cardiac hypertrophy, we were surprised that we failed to observe early death in the SUN2-null model. Remarkably, we found no increase in interstitial fibrosis in 13-mo-old Sun2-/- mice, as assessed by Masson’s trichrome staining of collagen and quantification with a color-based pixel count algorithm (Figure 5, A and B; see Materials and Methods). Moreover, pathological cardiac hypertrophy associated with fibrosis is typically accompanied by increased transcription of a number of fetal program genes involved in sarcomerogenesis, including involvement in the lamin A/C H222P mouse model (Kim et al., 2007; Taegtmeyer et al., 2010). However, quantitative PCR (qPCR) revealed no significant increases in the expression of ANP, BNP, skeletal α-actin, or Serca2 in Sun2-/- mice in comparison with WT mice (Figure 5C), suggesting that loss of SUN2 drives hallmarks of physiological rather than pathological hypertrophy (reviewed in Maillet et al., 2013). We next assessed the levels of gene targets of TGFβ, which is a master regulator of profibrotic signaling, at both P50 and at 13 mo. At P50 we observed a consistent down-regulation of TGFβ-associated profibrotic genes, including fibronectin (FN1) and the collagen genes COL1A1 and COL3A1 (Figure 5D). This does not appear to reflect a genomewide effect on the transcriptome, as a subset of TGFβ targets (FLNA) and nontargets (DES, which encodes Desmin) were unaltered (Figure 5D). At 13 mo, levels of the FN1 and COL1A1 transcripts were similar between WT and Sun2-/- mice (Figure 5E), consistent with the lack of fibrosis indicated by trichome staining (Figure 5, A and B), although we note that COL3A1 levels are significantly up-regulated in mice lacking SUN2.
FIGURE 5:
Sun2-/- hearts do not exhibit fibrosis and show reduced or unaltered expression of TGFβ target genes. (A) Images of trichrome-stained WT and Sun2-/- hearts are insets taken from Figure 1A. Sun2-/- tissue exhibits levels of collagen-positive staining (blue) equivalent to those in WT tissue surrounding blood vessels (top panels) and at interstitial sites (bottom panels). (B) Quantification of collagen-staining positiveness (the ratio of the number of blue pixels to total pixels; see Materials and Methods) in interstitial sites, showing equivalent levels of interstitial fibrosis in WT and Sun2-/- tissue. Unpaired, two-tailed t test failed to determine any difference to be significant. (C) qPCR analysis of ANP, BNP, skeletal α-actin, and Serca2 mRNA levels from 13-mo-old left ventricular total RNA. WT and Sun2-/- Ct values were normalized to GAPDH. Fold change of Sun2-/- mRNA levels was determined by calculating the 2ΔΔCt after taking the mean of WT ΔCt values from five mice. n = 4 Sun2-/- mice. Lack of statistical significance as determined by unpaired, two-tailed t test. (D) qPCR analysis of FN1 (fibronectin), COL1A1 (collagen type I), COL3A1 (collage type III), POSTN (periostin), LTBP2 (latent TGFβ-binding protein 2), LOXL1 (lysyl oxidase homologue 1), FLNA (filamin A), and DES (desmin) in P50 left ventricular total RNA, revealing similar or reduced levels of TGFβ transcriptional targets in Sun2-/- mice. WT and Sun2-/- Ct values were normalized to PPIA. Fold change of Sun2-/- mRNA levels was determined by calculating the 2ΔΔCt after taking the mean of WT ΔCt values from n = 2 WT or Sun2-/- mice. Statistical significance determined by one-way ANOVA followed by Dunnett’s multiple comparisons test. P values are indicated in the graph. (E) qPCR analysis of left ventricular total RNA from 13-mo-old animals as described in D.
Disrupted TGFβ signaling and increased retention of MAN1 at the nuclear envelope in SUN2-null mice
One of the major signaling pathways known to drive fibrosis in the heart is the TGFβ/SMAD cascade (Massagué and Wotton, 2000; Massagué, 2012; Travers et al., 2016). To determine the point at which the TGFβ signaling cascade (Massagué and Wotton, 2000; Massagué, 2012; Travers et al., 2016) is disrupted in the absence of SUN2, we performed immunofluorescence on P4 WT and Sun2-/- cardiac sections using antibodies against phosphorylated SMAD2 (pSMAD2; Figure 6, A and B). Surprisingly, Sun2-/- cardiac tissue exhibited a striking increase in the nuclear localization of pSMAD2 in comparison to WT tissue (Figure 6, A and B). These results raise the possibility that while the cytoplasmic aspects of TGFβ/SMAD signaling are increased in Sun2-/- cardiomyocytes, driving nuclear accumulation of pSMAD2, the nuclear aspects (transcription) are strongly abrogated in the absence of SUN2.
FIGURE 6:
MAN1 levels at the nuclear envelope are increased in the Sun2-/- myocardium. (A) Sun2-/- tissue exhibits qualitatively increased pSMAD2 nuclear localization. Frozen P4 WT and Sun2-/- cardiac left ventricle tissue was sectioned and stained with antibodies against pSMAD2 (gray) and counterstained with Hoechst 33342 (blue). Scale bar = 20 µm. (B) Enhanced pSMAD2 nuclear:cytoplasmic (N/C) ratio in Sun2-/- tissue. The relative maximum fluorescence intensity of the nuclear signal divided by the cytoplasmic signal for pSMAD2 in images obtained as in A. n = 503 cells for WT and n = 545 cells for Sun2-/-. Statistical significance determined by unpaired t test with Welch’s correction. (C) Lysates from WT and Sun2-/- P50 mice were subjected to SDS–PAGE and immunoblotting with antibodies against MAN1 and GAPDH (as a loading control). (D) Quantification of densitometry analysis for MAN1 normalized to GAPDH. Data are represented as the mean ± SD for four WT or Sun2-/- mice. (E) Representative fluorescence micrographs of MAN1 staining in frozen sections of the P4 myocardium from WT or Sun2-/- mice. (F) Peak fluorescence intensity of MAN1 at the nuclear envelope determined by line scan with local background subtraction. Plotted with the SD (n > 50 nuclei). Statistical significance determined by unpaired t test with Welch’s correction.
As MAN1 is established to be both a lamin A/C-binding protein and a nuclear-localized negative regulator of TGFβ, we next examined the levels and localization of MAN1 in the myocardium of Sun2-/- mice. AT P50, we found that total protein levels of MAN1 were moderately higher in Sun2-/- mice, as detected by immunoblotting (Figure 6, C and D). Further, in the myocardium of SUN2-null mice, we observed elevated levels of MAN1 by immunohistochemistry and quantitative analysis of fluorescence intensity across line profiles spanning the nuclear envelope (Figure 6, E and F). Thus, while upstream events that drive profibrotic pathways are likely stimulated in the absence of SUN2, mechanical signaling through the LINC complex may be required for nuclear pSMAD2 to act on its target genes, potentially by driving release of MAN1 from the nuclear lamina.
DISCUSSION
Here we demonstrate that a mouse model of SUN2 ablation develops cardiac hypertrophy, similarly to existing mouse models of A-type lamin dysfunction. Surprisingly, we observe that the SUN2-null mouse, unlike laminopathic mouse models, is protected from the involvement of interstitial fibrosis, suggesting that although the progrowth and profibrotic pathways may be driven by the same upstream factors, these two outcomes can be genetically uncoupled, potentially through distinct impacts on the targeting of MAN1 to the nuclear lamina. These findings raise the exciting possibility that future studies will reveal how modulating the nuclear lamina can be leveraged to disrupt tissue fibrosis.
The earliest effects of SUN2 ablation on cardiac muscle manifest during development, consistent with the recent observation that the LINC complex nucleates developing sarcomeres at the nuclear envelope and promotes the maintenance of myofibril structure in Drosophila lateral transverse muscle (Auld and Folker, 2016), as well as the observation that overexpression of human Nesprin-1α2 in a zebrafish model leads to defects in cardiac development (Zhou et al., 2017). Although we cannot rule out a direct role for LINC complexes in organized sarcomere assembly during the development of mammalian cardiac muscle, we favor a model in which altered regulation of the contractile-adhesion network underlies the observed sarcomere defects both early and late in life in Sun2-/- mice. Indeed, integrin-based adhesions at costameres play essential roles in patterning the sarcomere during cardiac myofibrillogenesis (Rhee et al., 1994; Dabiri et al., 1997; Hirschy et al., 2006; Du et al., 2008). Such a model is consistent with our previous demonstration that the LINC complex can influence cell junctions (Stewart et al., 2015) and the apparent increase in the levels of β1-integrin in its active, ligand-bound conformation in Sun2-/- cardiac tissue (Figure 4A).
Further support for a potential foundational effect of increased contractility comes from our TEM analysis, which reveals that cardiomyocytes manifest with sarcomere defects reminiscent of those tied to hypercontractility (Lauritzen et al., 2009) shortly after birth. Interestingly, a small molecular inhibitor of sarcomere power output in cardiomyocytes can be protective in an HCM model, suggesting that increased cardiomyocyte contractility can drive cardiac hypertrophy (Green et al., 2016). Thus, altered contractile properties of Sun2-/- cardiomyocytes could drive the LV hypertrophy we observe. How could SUN2 LINC complexes influence sarcomere contractile function? Beyond its direct interactions with the actin cytoskeleton, the LINC complex is implicated as a regulator of RhoA activity (Thakar et al., 2017), interacts with other modulators of actin organization and function, such as the formin FHOD1 in fibroblasts (Kutscheidt et al., 2014), and is known to influence actin dynamics (Lammerding et al., 2004; Lee et al., 2007; Hale et al., 2008; Stewart-Hutchinson et al., 2008; Khatau et al., 2009; Luxton et al., 2010; Folker et al., 2011; Kim et al., 2012). However, it should be noted that while increased cardiac contractility at the organ level is frequently observed in HCM, cell autonomous hypercontractility is not always observed in these disease models (Moore et al., 2012). Recent work also suggests that the disease-associated D192G lamin A/C mutation may produce cytoskeletal and adhesion defects in neonatal rat cardiomyocytes (Lanzicher et al., 2015), while a human cardiomyopathy patient with the point mutation G382V in LMNA exhibited defective plakoglobin localization to ICDs in the right ventricle (Quarta et al., 2012). Whether cardiomyocytes in models of laminopathies exhibit altered adhesion function or contractile properties remains a critical future question.
Although it remains to be fully tested, we suggest that increased contractility in Sun2-/- mice could be responsible for the gain in β1-integrin engagement that we observe. Indeed, increased integrin activation can be driven by heightened intracellular actomyosin contractility, as has been shown for the integrin LFA-1 in migrating T-cells (Nordenfelt et al., 2016). Integrin engagement and subsequent activation of AKT signaling has also been shown to drive cardiac hypertrophy in mouse models (reviewed in Brancaccio et al., 2006; Sequeira et al., 2014), and AKT activation is linked to elevated cardiomyocyte contractility (Kim et al., 2003; Rota et al., 2005; Cittadini et al., 2006; Catalucci et al., 2009), suggesting a potential connection between contractility, active β1-integrin levels, and hypertrophy in Sun2-/- mice. Consistent with this model, we observe heightened levels of phosphorylated AKT at P50 in Sun2-/- mice (Figure 4B). Importantly, a similar transient AKT activation in adolescent mice followed by sustained AKT-mTOR downstream signaling has previously been described in the cardiac tissue of the lamin A/C H222P mutant mouse (Muchir et al., 2007, 2012a,b; Wu et al., 2011; Choi et al., 2012; Choi and Worman, 2013; Ramos et al., 2012). The mechanisms by which loss of A-type lamin function leads to increased AKT and MAPK signaling, observed in numerous mouse models (Muchir et al., 2007, 2012a,b; Wu et al., 2011; Choi et al., 2012; Choi and Worman, 2013; Ramos et al., 2012), has yet to be defined; our work suggests that further study of signaling from integrin-based adhesions may prove fruitful.
LMNA-/-, lamin A/C H222P, and the Sun2-/- mouse models all share an increase in SMAD2 phosphorylation and nuclear localization (Chatzifrangkeskou et al., 2016). In the context of lamin perturbation, this occurs coincident with the induction of TGFβ-associated gene expression and severe cardiac fibrosis (Chatzifrangkeskou et al., 2016). In contrast, Sun2-/- mice display hypertrophy without fibrosis, mimicking physiological hypertrophy, which is characterized by hypertrophy with a lack of fibrosis or reactivation of fetal program genes (Maillet et al., 2013). This is similar to what is observed during cardiac growth compensation in response to exercise and pregnancy (Frenzel et al., 1988; Gonzalez et al., 2007; Chung et al., 2012) or in models of AMP-insensitive “activated” AMPK mutations (Hinson et al., 2016). One model is that SUN2 is a required component of a pathway that “licenses” nuclear pSMAD2 to act on its target genes. Our observation that MAN1 accumulates at the nuclear lamina in the absence of SUN2 suggests that its repression of TGFβ/SMAD signaling is alleviated in contexts where the LINC complex is under tension.
Importantly, treatment of mouse models of lamin dysfunction with the TOR inhibitor rapamycin influences heart function positively and ameliorates cardiac hypertrophy and fibrosis (Muchir et al., 2007, 2012a,b; Wu et al., 2011; Choi et al., 2012; Choi and Worman, 2013; Ramos et al., 2012). As human laminopathy patients typically display left ventricular dilation coupled with fibrosis (Ostlund et al., 2001; Meinke et al., 2014; Stroud et al., 2014), identifying the mechanisms that drive profibrotic signaling remains a critical need. Further study of the nuclear aspect of this signaling cascade through modulating MAN1 may identify new approaches for intervening in pathological fibrosis.
MATERIALS AND METHODS
Mouse breeding and care
All animal care and experimental procedures were conducted in accord with requirements approved by the Institutional Animal Care and Use Committee of Yale University. Sun2-/- (strain B6;129S6-Sun2tm1Mhan/J) and C57BL/6 WT mice were obtained from Jackson ImmunoResearch Laboratories. Sun2-/- mice were previously generated through the replacement of exons 11–16 and part of exon 17 with a neomycin resistance cassette (Lei et al., 2009).
Mouse tissue isolation, histology, and immunofluorescence staining
Cardiac isolation.
WT and Sun2-/- murine hearts were isolated from P4 or P45/P47/P50 mice and perfused with PBS, and left ventricular tissue was either frozen in O.C.T. compound (Sakura, Tissue-Tek) at -80°C for cryosectioning or fixed in 10% neutral buffered Formalin and embedded in paraffin in collaboration with the Yale University Developmental Histology Facility in the Department of Pathology. Frozen samples were sectioned using a cryostat (CM3050S; Leica). For histological analyses, 10-µm sections were cut and stained with hematoxylin and eosin. For immunofluorescence, 6- or 8-µm sections were fixed in 4% formaldehyde or 4% paraformaldehyde at RT for 10 min.
Immunofluorescence staining.
For immunostaining with nonmouse primary antibodies, tissue sections were blocked in gelatin block (2.5% normal goat serum, 1% BSA, 2% gelatin, and 0.25% Triton X-100 in PBS) at RT for 1 h and incubated with the following primary antibodies overnight at 4°C: SUN2 (1:100; rabbit; Abcam ab124916), total β1-integrin (1:50; rat; Novus Biologicals NBP1-4323, clone KMI6), conformational epitope β1-integrin (1:50; rat; BD Pharmingen #550531, clone 9EG7), paxillin (1:100; rabbit; Abcam ab32084, clone Y113), or phosphorylated SMAD2 (1:100, rabbit, Cell Signaling #8828, clone D27F4). For immunostaining with the mouse primary antibodies desmoplakin I/II (1:50; mouse; Abcam ab16434, clone 2Q400), β-catenin (1:100; mouse; BD Transduction Labs #610153), and anti-MAN1 antibody (BBMAN1A22,44; kind gift from Brian Burke and Martina Maric, Institute of Medical Biology, Singapore), the Mouse-on-Mouse (M.O.M.) Immunodetection Kit blocking reagent and protein diluent (Vector Laboratories) were used according to the manufacturer’s instructions. Sections were subsequently washed in multiple changes of PBS and incubated with fluorescent dye–conjugated secondary antibodies (1:1000; mouse, rat, or rabbit; Alexa Fluor; Life Technologies) in gel block or M.O.M.-containing gel block when appropriate. Costaining with Hoechst 33342 (Thermo Fisher Scientific; 1:2000), Alexa Fluor 594-conjugated wheat germ agglutinin (Life Technologies; 1:2000), and/or Alexa Fluor 594-conjugated phalloidin (Invitrogen #A12381; 1:40) was performed when indicated. Sections were mounted using Fluoromount-G mounting medium (SouthernBiotech).
Transmission electron microscopy
TEM was performed in the Yale School of Medicine Center for Cellular and Molecular Imaging Electron Microscopy core facility. Left ventricular cardiac tissue from P4 and P45/P47 WT and Sun2-/- mice were isolated and processed for TEM; three mice were examined for each genotype at each time point. Tissue blocks were fixed in 2.5% glutaraldehyde/2% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, for 30 min at room temperature (RT) and 1.5 h at 4°C. The samples were rinsed in sodium cacodylate buffer and were postfixed in 1% osmium tetroxide for 1 h. The samples were rinsed and stained en bloc in aqueous 2% uranyl acetate for 1 h, followed by rinsing, dehydrating in an ethanol series to 100%, rinsing in 100% propylene oxide, infiltrating with EMbed 812 (Electron Microscopy Sciences) resin, and baking overnight at 60°C. Hardened blocks were cut using an ultramicrotome (UltraCut UC7; Leica). Ultrathin 60-nm sections were collected and stained using 2% uranyl acetate and lead citrate for transmission microscopy. Carbon-coated grids were viewed on a transmission electron microscope (Tecnai BioTWIN; FEI) at 80 kV. Images were taken using a charge-coupled device (CCD) camera (Morada; Olympus) and iTEM (Olympus) software.
Imaging and image analysis
Immunofluorescence imaging.
Whole slides of hematoxylin- and eosin-stained or trichrome-stained cardiac sections were scanned using an Aperio digital pathology scanner at 40× magnification in collaboration with Yale University Pathology Digital Imaging in the Department of Pathology. Cardiac sections processed for immunofluorescence were imaged on a Leica SP5 confocal microscope or a widefield deconvolution microscope (DeltaVision; Applied Precision/GE Healthcare) with a CCD camera (CoolSNAP K4; Photometrics) and SoftWoRx software. All images acquired on the DeltaVision microscope were deconvolved using the Deconvolve tool (constrained iterative deconvolution) in SoftWoRx software. The DeltaVision microscope was equipped with an oil Plan Apochromat N 60×/1.42 NA objective (Olympus). In all cases, images were analyzed using Fiji software (ImageJ 1.48d; National Institutes of Health) as indicated.
Analysis of cardiomyocyte size.
Transverse midventricular sections of WT and Sun2-/- murine hearts were cut and stained with antibodies against laminin in collaboration with the Yale University Developmental Histology Facility in the Department of Pathology. The cross-sectional area of between 86 and 198 left ventricular papillary muscle cells was measured by manually outlining individual cells and using the measure tool in Fiji (ImageJ 1.51j; National Institutes of Health; Schindelin et al., 2012). All cells in a given field were measured and only fields with circular blood vessels—indicating that the adjacent cells were sectioned orthogonal to the plane—were included.
Analysis of fibrosis.
Transverse midventricular sections of WT and Sun2-/- murine hearts were cut and stained with Masson’s trichrome stain in collaboration with the Yale University Developmental Histology Facility in the Department of Pathology. Whole slides were scanned using an Aperio Digital Pathology Scanner at 40× magnification in collaboration with Yale University Pathology Digital Imaging in the Department of Pathology. The ImageScope 12.2 Positive Pixel Count algorithm (Aperio Technologies) was used to identify blue-colored collagen fibers in the tissue samples. Color hue and saturation values (hue value of 0.62, hue width of 0.40, and color saturation threshold of 0.005) were optimized before the analysis. Five or more fields of interstitial tissue were assessed for each of three mice per genotype.
Analysis of sarcomere length.
TEM micrographs of P50 WT and Sun2-/- ventricular tissue were acquired as described. The sarcomere length, defined as the distance from the center of one Z-band to the center of a successive Z-band, was measured in Fiji for between 94 and 235 sarcomeres for three mice of each genotype. All sarcomeres with visible Z-bands in a given field were measured.
Display of β1-integrin fluorescence intensity.
Images of WT and Sun2-/- ventricular tissue stained with antibodies against the ligand-bound β1-integrin conformation were acquired on the Leica SP5 microscope using the same acquisition settings between WT and Sun2-/- samples. WT and Sun2-/- images were scaled to one another and pseudocolored in Fiji using the Fire LUT, converting differences in pixel intensity to differences in display color; on this scale, lighter colors correspond to higher fluorescence intensity, while darker colors correspond to lower fluorescence intensity.
pSMAD2 nuclear:cytoplasmic intensity ratio measurements.
Using images as in Figure 6A, nuclei were segmented by thresholding the Hoechst signal, binarizing, and creating individual nuclear masks using the Analyze Particles function in ImageJ. The resulting masks were used to measure the nuclear pSMAD2 fluorescence intensity maxima following background subtraction. The masks were converted to a band of 1.5 pixels to measure perinuclear cytoplasmic intensity maxima. The nuclear:cytoplasmic intensity ratio was calculated as the nuclear intensity maximum divided by the cytoplasmic intensity maximum.
MAN1 fluorescence intensity measurements.
Using images as in Figure 6E, line profiles bisecting the nucleus of equal length were taken (>50 per sample) in ImageJ. The local intensity maxima at the nuclear envelope after subtraction of the mean intensity across the entire line profile was then calculated.
Western blotting
Following heart isolation and PBS perfusion, ventricular cardiac tissue was isolated from 13-mo-old WT and Sun2-/- mice or P45/P47 WT and Sun2-/- mice, minced, and homogenized using a Polytron PT 1200 E homogenizer (Kinematica AG) in radioimmunoprecipitation lysis buffer (50 mM Tris-Cl, pH 7.4, 1 mM EDTA, 1% NP-40 alternative, 0.1% Na deoxycholate, 0.1% SDS, 150 mM NaCl, protease inhibitor cocktail [Sigma-Aldrich]). The insoluble debris was removed by centrifugation for 30 min at 14,000 rpm in a 4°C tabletop microcentrifuge (Sorvall Legend Micro 21R; Thermo Fisher Scientific). Extracted protein samples were diluted in SDS–PAGE sample buffer and separated using a 7.5% polyacrylamide gel. The separated proteins were transferred to a nitrocellulose membrane using a transfer system (Bio-Rad Laboratories) and subjected to Ponceau S (Sigma) staining to observe total protein loading. Membranes were subsequently blocked in 10% nonfat milk (Omniblok; American Bioanalytical) or, if being blotted with phospho-antibodies, 5% BSA (AmericanBio) in TBST (TBS with Tween 20) for 1 h at RT and incubated with AKT (1:1000, rabbit, Cell Signaling 9272), pAKT (1:1000, rabbit, Cell Signaling 4060 clone D9E), S6 (1:5000, rabbit, Cell Signaling 2217 clone5G10), pS6 (1:5000, rabbit, Cell Signaling 2211 Ser235/236), ERK2 (1:200, rabbit, Santa Cruz SC-154), pERK1/2 (1:300, mouse, Cell Signaling 9101 Thr202/204), and MAN1/LEMD3 (1:1000, rabbit, ProSci 6603) primary antibodies at 4°C overnight. The membranes were washed for 15 min in TBST and then incubated for 1 h with HRP-conjugated secondary antibodies (1:5000; Pierce Antibodies; Thermo Fisher Scientific) in TBST at RT. After washes with TBST, the membranes were incubated with HRP chemiluminescent substrate (SuperSignal West Femto Chemiluminescent Substrate; Thermo Fisher Scientific) for 5 min and then imaged using a VersaDoc imaging system (Bio-Rad Laboratories).
qPCR
Following heart isolation and PBS perfusion, ventricular cardiac tissue was isolated from 13-mo-old WT and Sun2-/- mice or P54/P55 WT and Sun2-/- mice, minced, and homogenized in TRIzol (Invitrogen). Total RNA was isolated using the RNeasy kit (QIAGEN) according to manufacturer’s instructions. To remove genomic DNA, total RNA was digested with DNase I (Roche) according to manufacturer’s instructions and repurified using the RNeasy kit (QIAGEN). To generate cDNA, equal amounts of digested total RNA (1 μg) were added to SuperScript III Reverse Transcriptase (Invitrogen) reactions using Random Primer 9 (New England Biolabs). Quantitative real-time PCR was conducted with a Bio-Rad CFX96 Real-Time System using the iQ SYBR Green Supermix (Bio-Rad) for 39 cycles. Primers used in these experiments were ANP forward: CAAGATGCAGAAGCTGCTGG and reverse: GTGCTGCCTTGAGACCGAAGG; BNP forward: GTTTGGGCTGTAACGCACTGA and reverse: GAAAGAGACCCAGGCAGAGTCA; Skeletal α-actin forward: CATGAAGATCAAGATCATCGC and reverse: CTGGAAGGTGGACAGCGAGGC; Serca2 forward: GTGTGGCAGGAAAGAAATGC and reverse: CCAGGAACTATGTCTTTAGC; FN1 forward: TTCAAGTGTGATCCCCATGAAG and reverse: CAGGTCTACGGCAGTTGTCA; COL1A1 forward: CTGGCGGTTCAGGTCCAAT and reverse: TTCAGGCAATCCAGAGC; COL3A1 forward: CTGTAACATGGAAACTGGGGAAA and ereverse: CCATAGCTGAACTGAAAACCACC; POSTN forward: CCTGCCCTTATATGCTCTGCT and reverse: AAACATGGTCAATAGGCATCACT; LTBP2 forward: GCTCACCGGGAGAAATGTCTG and reverse: CAGGTTTGATACAGTGGTTGGT; LOXL1 forward: GAGTGCTATTGCGCTTCCC and reverse: GGTTGCCGAAGTCACAGGT; FLNA forward: GGCTACGGTGGGCTTAGTC and reverse: GTGGGACAGTAGGTGACCCT; DES forward: TACACCTGCGAGATTGATGC and reverse: ACATCCAAGGCCATCTTCAC; GAPDH forward: CGTAGACAAAATGGTGAAGGTCGG and reverse: AAGCAGTTGGTGGTGCAGGATG; and PPIA forward: GAGCTGTTTGCAGACAAAGTTC and reverse: CCCTGGCACATGAATCCTGG. PCR product levels were normalized to peptidylprolyl isomerase A (PPIA) or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA levels.
Supplementary Material
Acknowledgments
This work was supported by American Heart Association Predoctoral Fellowship #16PRE27460000 (R.M.S.), an Outstanding Early Investigator Award from the Ludwig Family Foundation (M.C.K.), and R01 GM129308-01 from the National Institutes of Health/National Institute of General Medical Sciences (M.C.K.). We thank Lawrence Young and Stuart Campbell of Yale University for valuable discussions; Morven Graham, Nicole Mikush, and Kevin Su for their technical expertise; and Martina Maric for the kind gift of the BBMAN1A22,44 antibody to MAN1 for staining tissue.
Abbreviations used:
- HCM
hypertrophic cardiomyopathy
- ICD
intercalated disk
- LINC
linker of nucleoskeleton and cytoskeleton
- LV
left ventricle
- qPCR
quantitative PCR
- TEM
transmission electron microscopy
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-07-0438) on May 15, 2019.
REFERENCES
- Arbustini E, Pilotto A, Repetto A, Grasso M, Negri A, Diegoli M, Campana C, Scelsi L, Baldini E, Gavazzi A, et al (2002). Autosomal dominant dilated cardiomyopathy with atrioventricular block: a lamin A/C defect-related disease. J Am Coll Cardiol , 981–990. [DOI] [PubMed] [Google Scholar]
- Auld AL, Folker ES. (2016). Nucleus-dependent sarcomere assembly is mediated by the LINC complex. Mol Biol Cell , 2351–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Kuppuswamy D. (2003). RGD-containing peptides activate S6K1 through beta3 integrin in adult cardiac muscle cells. J Biol Chem , 42214–42224. [DOI] [PubMed] [Google Scholar]
- Bazzoni G, Shih DT, Buck CA, Hemler ME. (1995). Monoclonal antibody 9EG7 defines a novel beta 1 integrin epitope induced by soluble ligand and manganese, but inhibited by calcium. J Biol Chem , 25570–25577. [DOI] [PubMed] [Google Scholar]
- Brancaccio M, Hirsch E, Notte A, Selvetella G, Lembo G, Tarone G. (2006). Integrin signalling: the tug-of-war in heart hypertrophy. Cardiovasc Res , 422–433. [DOI] [PubMed] [Google Scholar]
- Cahill TJ, Ashrafian H, Watkins H. (2013). Genetic cardiomyopathies causing heart failure. Circ Res , 660–675. [DOI] [PubMed] [Google Scholar]
- Catalucci D, Latronico MVG, Ceci M, Rusconi F, Young HS, Gallo P, Santonastasi M, Bellacosa A, Brown JH, Condorelli G. (2009). Akt increases sarcoplasmic reticulum Ca2+ cycling by direct phosphorylation of phospholamban at Thr17. J Biol Chem , 28180–28187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers DM, Moretti L, Zhang JJ, Cooper SW, Chambers DM, Santangelo PJ, Barker TH. (2018). LEM domain-containing protein 3 antagonizes TGFβ-SMAD2/3 signaling in a stiffness-dependent manner in both the nucleus and cytosol. J Biol Chem , 15867–15886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Worman HJ, Gundersen GG. (2015). Accessorizing and anchoring the LINC complex for multifunctionality. J Cell Biol , 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzifrangkeskou M, Le Dour C, Wu W, Morrow JP, Joseph LC, Beuvin M, Sera F, Homma S, Vignier N, Mougenot N, et al (2016). ERK1/2 directly acts on CTGF/CCN2 expression to mediate myocardial fibrosis in cardiomyopathy caused by mutations in the lamin A/C gene. Hum Mol Genet , 2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JC, Muchir A, Wu W, Iwata S, Homma S, Morrow JP, Worman HJ. (2012). Temsirolimus activates autophagy and ameliorates cardiomyopathy caused by lamin A/C gene mutation. Sci Transl Med , 144ra102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JC, Worman HJ. (2013). Reactivation of autophagy ameliorates LMNA cardiomyopathy. Autophagy , 110–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E, Yeung F, Leinwand LA. (2012). Akt and MAPK signaling mediate pregnancy-induced cardiac adaptation. J Appl Physiol , 1564–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cittadini A, Monti MG, Iaccarino G, Di Rella F, Tsichlis PN, Di Gianni A, Strömer H, Sorriento D, Peschle C, Trimarco B, et al (2006). Adenoviral gene transfer of Akt enhances myocardial contractility and intracellular calcium handling. Gene Ther , 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabiri GA, Turnacioglu KK, Sanger JM, Sanger JW. (1997). Myofibrillogenesis visualized in living embryonic cardiomyocytes. Proc Natl Acad Sci USA , 9493–9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson PM, Lammerding J. (2014). Broken nuclei—lamins, nuclear mechanics, and disease. Trends Cell Biol , 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmar M, McKenna WJ. (2010). The cardiac desmosome and arrhythmogenic cardiomyopathies: from gene to disease. Circ Res , 700–714. [DOI] [PubMed] [Google Scholar]
- Dowling JJ, Gibbs E, Russell M, Goldman D, Minarcik J, Golden JA, Feldman EL. (2008). Kindlin-2 is an essential component of intercalated discs and is required for vertebrate cardiac structure and function. Circ Res , 423–431. [DOI] [PubMed] [Google Scholar]
- Du A, Sanger JM, Sanger JW. (2008). Cardiac myofibrillogenesis inside intact embryonic hearts. Dev Biol , 236–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker ES, Ostlund C, Luxton GWG, Worman HJ, Gundersen GG. (2011). Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc Natl Acad Sci USA , 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini KG, Torsoni AS, Soares PH, Saad MJ. (2000). Early activation of the multicomponent signaling complex associated with focal adhesion kinase induced by pressure overload in the rat heart. Circ Res , 558–565. [DOI] [PubMed] [Google Scholar]
- Frenzel H, Schwartzkopff B, Höltermann W, Schnürch HG, Novi A, Hort W. (1988). Regression of cardiac hypertrophy: morphometric and biochemical studies in rat heart after swimming training. J Mol Cell Cardiol , 737–751. [DOI] [PubMed] [Google Scholar]
- Frock RL, Chen SC, Da D-F, Frett E, Lau C, Brown C, Pak DN, Wang Y, Muchir A, Worman HJ, et al (2012). Cardiomyocyte-specific expression of lamin a improves cardiac function in Lmna-/- mice. PLoS One , e42918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez EW, Chen QK, Gjorevski N, Nelson CM. (2010). Tissue geometry patterns epithelial-mesenchymal transition via intercellular mechanotransduction. J Cell Biochem , 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez AMD, Osorio JC, Manlhiot C, Gruber D, Homma S, Mital S. (2007). Hypertrophy signaling during peripartum cardiac remodeling. Am J Physiol Heart Circ Physiol , H3008–H3013. [DOI] [PubMed] [Google Scholar]
- Green EM, Wakimoto H, Anderson RL, Evanchik MJ, Gorham JM, Harrison BC, Henze M, Kawas R, Oslob JD, Rodriguez HM, et al (2016). A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science , 617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Bilinska ZT, Sylvius N, Boudreau E, Veinot JP, Labib S, Bolongo PM, Hamza A, Jackson T, Ploski R, et al (2010). Genetic and ultrastructural studies in dilated cardiomyopathy patients: a large deletion in the lamin A/C gene is associated with cardiomyocyte nuclear envelope disruption. Basic Res Cardiol , 365–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CM, Shrestha AL, Khatau SB, Stewart-Hutchinson PJ, Hernandez L, Stewart CL, Hodzic D, Wirtz D. (2008). Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys J , 5462–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Hetzer MW. (2016). Nuclear envelope rupture is induced by actin-based nucleus confinement. J Cell Biol , 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson JT, Chopra A, Lowe A, Sheng CC, Gupta RM, Kuppusamy R, O’Sullivan J, Rowe G, Wakimoto H, Gorham J, et al (2016). Integrative analysis of PRKAG2 cardiomyopathy iPS and microtissue models identifies AMPK as a regulator of metabolism, survival, and fibrosis. Cell Rep , 3292–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B. (2015). The extracellular matrix and transforming growth factor-β1: tale of a strained relationship. Matrix Biol , 54–65. [DOI] [PubMed] [Google Scholar]
- Hirschy A, Schatzmann F, Ehler E, Perriard J-C. (2006). Establishment of cardiac cytoarchitecture in the developing mouse heart. Dev Biol , 430–441. [DOI] [PubMed] [Google Scholar]
- Ho CY, Lopez B, Coelho-Filho OR, Lakdawala NK, Cirino AL, Jarolim P, Kwong R, Gonzalez A, Colan SD, Seidman JG, et al (2010). Myocardial fibrosis as an early manifestation of hypertrophic cardiomyopathy. N Engl J Med , 552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima Y, Laser M, Shiraishi H, Willey CD, Sundaravadivel B, Xu L, McDermott PJ, Kuppuswamy D. (2002). c-Raf/MEK/ERK pathway controls protein kinase C-mediated p70S6K activation in adult cardiac muscle cells. J Biol Chem , 23065–23075. [DOI] [PubMed] [Google Scholar]
- Ishimura A, Ng JK, Taira M, Young SG, Osada S-I. (2006). Man1, an inner nuclear membrane protein, regulates vascular remodeling by modulating transforming growth factor beta signaling. Development , 3919–3928. [DOI] [PubMed] [Google Scholar]
- Israeli-Rosenberg S, Manso AM, Okada H, Ross RS. (2014). Integrins and integrin-associated proteins in the cardiac myocyte. Circ Res , 572–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatau SB, Bloom RJ, Bajpai S, Razafsky D, Zang S, Giri A, Wu PH, Marchand J, Celedon A, Hale CM, et al (2012). The distinct roles of the nucleus and nucleus–cytoskeleton connections in three-dimensional cell migration. Sci Rep , 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D. (2009). A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci USA , 19017–19022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-H, Khatau SB, Feng Y, Walcott S, Sun SX, Longmore GD, Wirtz D. (2012). Actin cap associated focal adhesions and their distinct role in cellular mechanosensing. Sci Rep , 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Porreca GJ, Song L, Greenway SC, Gorham JM, Church GM, Seidman CE, Seidman JG. (2007). Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science , 1481–1484. [DOI] [PubMed] [Google Scholar]
- Kim YK, Kim SJ, Yatani A, Huang Y, Castelli G, Vatner DE, Liu J, Zhang Q, Diaz G, Zieba R, et al (2003). Mechanism of enhanced cardiac function in mice with hypertrophy induced by overexpressed Akt. J Biol Chem , 47622–47628. [DOI] [PubMed] [Google Scholar]
- Konieczny P, Fuchs P, Reipert S, Kunz WS, Zeöld A, Fischer I, Paulin D, Schröder R, Wiche G. (2008). Myofiber integrity depends on desmin network targeting to Z-disks and costameres via distinct plectin isoforms. J Cell Biol , 667–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutscheidt S, Zhu R, Antoku S, Luxton GWG, Stagljar I, Fackler OT, Gundersen GG. (2014). FHOD1 interaction with nesprin-2G mediates TAN line formation and nuclear movement. Nat Cell Biol , 708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. (2004). Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest , 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzicher T, Martinelli V, Puzzi L, Del Favero G, Codan B, Long CS, Mestroni L, Taylor MRG, Sbaizero O. (2015). The cardiomyopathy lamin A/C D192G mutation disrupts whole-cell biomechanics in cardiomyocytes as measured by atomic force microscopy loading-unloading curve analysis. Sci Rep , 13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laser M, Willey CD, Jiang W, Cooper G, Menick DR, Zile MR, Kuppuswamy D. (2000). Integrin activation and focal complex formation in cardiac hypertrophy. J Biol Chem , 35624–35630. [DOI] [PubMed] [Google Scholar]
- Lauritzen F, Paulsen G, Raastad T, Bergersen LH, Owe SG. (2009). Gross ultrastructural changes and necrotic fiber segments in elbow flexor muscles after maximal voluntary eccentric action in humans. J Appl Physiol , 1923–1934. [DOI] [PubMed] [Google Scholar]
- Le Dour C, Wu W, Béréziat V, Capeau J, Vigouroux C, Worman HJ. (2017). Extracellular matrix remodeling and transforming growth factor-β signaling abnormalities induced by lamin A/C variants that cause lipodystrophy. J Lipid Res , 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JSH, Hale CM, Panorchan P, Khatau SB, George JP, Tseng Y, Stewart CL, Hodzic D, Wirtz D. (2007). Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys J , 2542–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K, Zhang X, Ding X, Guo X, Chen M, Zhu B, Xu T, Zhuang Y, Xu R, Han M. (2009). SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc Natl Acad Sci USA , 10207–10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenter M, Uhlig H, Hamann A, Jenö P, Imhof B, Vestweber D. (1993). A monoclonal antibody against an activation epitope on mouse integrin chain beta 1 blocks adhesion of lymphocytes to the endothelial integrin alpha 6 beta 1. Proc Natl Acad Sci USA , 9051–9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CY, Anderson SS, Chicoine NH, Mayfield JR, Academia EC, Wilson JA, Pongkietisak C, Thompson MA, Lagmay EP, Miller DM, et al (2016). Rapamycin reverses metabolic deficits in lamin A/C-deficient mice. Cell Rep , 2542–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Morrison JM, Wu W, Worman HJ. (2005). MAN1, an integral protein of the inner nuclear membrane, binds Smad2 and Smad3 and antagonizes transforming growth factor-beta signaling. Hum Mol Genet , 437–445. [DOI] [PubMed] [Google Scholar]
- Long JB, Bagonis M, Lowery LA, Lee H, Danuser G, Van Vactor D. (2013). Multiparametric analysis of CLASP-interacting protein functions during interphase microtubule dynamics. Mol Cell Biol , 1528–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowey S. (2002). Functional consequences of mutations in the myosin heavy chain at sites implicated in familial hypertrophic cardiomyopathy. Trends Cardiovasc Med , 348–354. [DOI] [PubMed] [Google Scholar]
- Luxton GWG, Gomes ER, Folker ES, Vintinner E, Gundersen GG. (2010a). Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science , 956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenna DA, Dolfi F, Vuori K, Ruoslahti E. (1998). Extracellular signal-regulated kinase and c-Jun NH2-terminal kinase activation by mechanical stretch is integrin-dependent and matrix-specific in rat cardiac fibroblasts. J Clin Invest , 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet M, van Berlo JH, Molkentin JD. (2013). Molecular basis of physiological heart growth: fundamental concepts and new players. Nat Rev Mol Cell Biol , 38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. (2012). TGFβ signalling in context. Nat Rev Mol Cell Biol , 616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, Wotton D. (2000). Transcriptional control by the TGF-beta/Smad signaling system. Embo J , 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCain ML, Lee H, Aratyn-Schaus Y, Kléber AG, Parker KK. (2012). Cooperative coupling of cell-matrix and cell-cell adhesions in cardiac muscle. Proc Natl Acad Sci USA , 9881–9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCain ML, Parker KK. (2011). Mechanotransduction: the role of mechanical stress, myocyte shape, and cytoskeletal architecture on cardiac function. Pflugers Arch , 89–104. [DOI] [PubMed] [Google Scholar]
- Meinke P, Mattioli E, Haque F, Antoku S, Columbaro M, Straatman KR, Worman HJ, Gundersen GG, Lattanzi G, Wehnert M, et al (2014). Muscular dystrophy-associated SUN1 and SUN2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genet , e1004605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michele DE, Albayya FP, Metzger JM. (1999). Direct, convergent hypersensitivity of calcium-activated force generation produced by hypertrophic cardiomyopathy mutant alpha-tropomyosins in adult cardiac myocytes. Nat Med , 1413–1417. [DOI] [PubMed] [Google Scholar]
- Moore JR, Leinwand L, Warshaw DM. (2012). Understanding cardiomyopathy phenotypes based on the functional impact of mutations in the myosin motor. Circ Res , 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounkes LC, Kozlov SV, Rottman JN, Stewart CL. (2005). Expression of an LMNA-N195K variant of A-type lamins results in cardiac conduction defects and death in mice. Hum Mol Genet , 2167–2180. [DOI] [PubMed] [Google Scholar]
- Muchir A, Pavlidis P, Decostre V, Herron AJ, Arimura T, Bonne G, Worman HJ. (2007). Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery–Dreifuss muscular dystrophy. J Clin Invest , 1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchir A, Reilly SA, Wu W, Iwata S, Homma S, Bonne G, Worman HJ. (2012a). Treatment with selumetinib preserves cardiac function and improves survival in cardiomyopathy caused by mutation in the lamin A/C gene. Cardiovasc Res , 311–319. [DOI] [PubMed] [Google Scholar]
- Muchir A, Wu W, Choi JC, Iwata S, Morrow J, Homma S, Worman HJ. (2012b). Abnormal p38α mitogen-activated protein kinase signaling in dilated cardiomyopathy caused by lamin A/C gene mutation. Hum Mol Genet , 4325–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myat MM, Rashmi RN, Manna D, Xu N, Patel U, Galiano M, Zielinski K, Lam A, Welte MA. (2015). Drosophila KASH-domain protein Klarsicht regulates microtubule stability and integrin receptor localization during collective cell migration. Dev Biol , 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordenfelt P, Elliott HL, Springer TA. (2016). Coordinated integrin activation by actin-dependent force during T-cell migration. Nat Commun , 13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor JW, Riley PN, Nalluri SM, Ashar PK, Gomez EW. (2015). Matrix rigidity mediates TGFβ1-Induced epithelial-myofibroblast transition by controlling cytoskeletal organization and MRTF-A localization. J Cell Physiol , 1829–1839. [DOI] [PubMed] [Google Scholar]
- Olive M, et al (2010). Cardiovascular pathology in Hutchinson–Gilford progeria: correlation with the vascular pathology of aging. Arterioscler. Thromb Vasc Biol , 2301–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund C, Bonne G, Schwartz K, Worman HJ. (2001). Properties of lamin A mutants found in Emery–Dreifuss muscular dystrophy, cardiomyopathy and Dunnigan-type partial lipodystrophy. J Cell Sci , 4435–4445. [DOI] [PubMed] [Google Scholar]
- Ostlund C, Sullivan T, Stewart CL, Worman HJ. (2006). Dependence of diffusional mobility of integral inner nuclear membrane proteins on A-type lamins. Biochemistry , 1374–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarta G, Syrris P, Ashworth M, Jenkins S, Zuborne Alapi K, Morgan J, Muir A, Pantazis A, McKenna WJ, Elliott PM. (2012). Mutations in the lamin A/C gene mimic arrhythmogenic right ventricular cardiomyopathy. Eur Heart J , 1128–1136. [DOI] [PubMed] [Google Scholar]
- Raju GP, Dimova N, Klein PS, Huang H-C. (2003). SANE, a novel LEM domain protein, regulates bone morphogenetic protein signaling through interaction with Smad1. J Biol Chem , 428–437. [DOI] [PubMed] [Google Scholar]
- Ramos FJ, Chen SC, Garelick MG, Dai DF, Liao CY, Schreiber KH, MacKay VL, An EH, Strong R, Ladiges WC, et al (2012). Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci Transl Med , 144ra103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee D, Sanger JM, Sanger JW. (1994). The premyofibril: evidence for its role in myofibrillogenesis. Cell Motil Cytoskeleton , 1–24. [DOI] [PubMed] [Google Scholar]
- Rota M, Boni A, Urbanek K, Padin-Iruegas ME, Kajstura TJ, Fiore G, Kubo H, Sonnenblick EH, Musso E, Houser SR, et al (2005). Nuclear targeting of Akt enhances ventricular function and myocyte contractility. Circ Res , 1332–1341. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira V, Nijenkamp LLAM, Regan JA, van der Velden J. (2014). The physiological role of cardiac cytoskeleton and its alterations in heart failure. Biochim Biophys Acta , 700–722. [DOI] [PubMed] [Google Scholar]
- Sheikh F, Ross RS, Chen J. (2009). Cell–cell connection to cardiac disease. Trends Cardiovasc Med , 182–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiojima I, Walsh K. (2006). Regulation of cardiac growth and coronary angiogenesis by the Akt/PKB signaling pathway. Genes Dev , 3347–3365. [DOI] [PubMed] [Google Scholar]
- Sosa BA, Rothballer A, Kutay U, Schwartz TU. (2012). LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell , 1035–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RM, Zubek AE, Rosowski KA, Schreiner SM, Horsley V, King MC. (2015). Nuclear–cytoskeletal linkages facilitate cross talk between the nucleus and intercellular adhesions. J Cell Biol , 403–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. (2008). Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res , 1892–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud MJ, Banerjee I, Veevers J, Chen J. (2014). Linker of nucleoskeleton and cytoskeleton complex proteins in cardiac structure, function, and disease. Circ Res , 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Xia W, Li J, Walz T, Humphries MJ, Vestweber D, Cabañas C, Lu C, Springer TA. (2016). Relating conformation to function in integrin α5β;1. Proc Natl Acad Sci USA , E3872–E3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taegtmeyer H, Sen S, Vela D. (2010). Return to the fetal gene program: a suggested metabolic link to gene expression in the heart. Ann NY Acad Sci , 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakar K, May CK, Rogers A, Carroll CW. (2017). Opposing roles for distinct LINC complexes in regulation of the small GTPase RhoA. Mol Biol Cell , 182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers JG, Kamal FA, Robbins J, Yutzey KE, Blaxall BC. (2016). Cardiac fibrosis: the fibroblast awakens. Circ Res , 1021–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varney SD, Betts CB, Zheng R, Wu L, Hinz B, Zhou J, Van De Water L. (2016). Hic-5 is required for myofibroblast differentiation by regulating mechanically dependent MRTF-A nuclear accumulation. J Cell Sci , 774–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Lin JL-C, Wu K-H, Wang D-Z, Reiter RS, Sinn HW, Lin C-I, Lin C-JJ. (2012). Xin proteins and intercalated disc maturation, signaling and diseases. Front Biosci (Landmark Ed) , 2566–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsbacher LD, Coughlin SR. (2015). Analysis of cardiomyocyte development using immunofluorescence in embryonic mouse heart. J Vis Exp e52644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Muchir A, Shan J, Bonne G, Worman HJ. (2011). Mitogen-activated protein kinase inhibitors improve heart function and prevent fibrosis in cardiomyopathy caused by mutation in lamin A/C gene. Circulation , 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JL, Kretchmer K, Ondeck MG, Zambon AC, Engler AJ. (2014). Mechanosensitive kinases regulate stiffness-induced cardiomyocyte maturation. Sci Rep , 6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Li C, Zhou B, Sun H, Koullourou V, Holt I, Puckelwartz MJ, Warren DT, Hayward R, Lin Z, et al (2017). Novel nesprin-1 mutations associated with dilated cardiomyopathy cause nuclear envelope disruption and defects in myogenesis. Hum Mol Genet , 2258–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerger M, Jaalouk DE, Lombardi ML, Isermann P, Mauermann M, Dialynas G, Herrmann H, Wallrath LL, Lammerding J. (2013). Myopathic lamin mutations impair nuclear stability in cells and tissue and disrupt nucleo-cytoskeletal coupling. Hum Mol Genet , 2335–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.