Abstract
Dividing cells must balance the maintenance of genome integrity with the generation of cytoskeletal forces that control chromosome position. In this study, we investigate how forces on astral microtubules impact the genome during cell division by using live-cell imaging of the cytoskeleton, chromatin, and DNA damage repair in budding yeast. Our results demonstrate that dynein-dependent forces on astral microtubules are propagated through the spindle during nuclear migration and when in excess can increase the frequency of double-stranded breaks (DSBs). Under these conditions, we find that homology-directed repair of DSBs is delayed, indicating antagonism between nuclear migration and the mechanism of homology-directed repair. These effects are partially rescued by mutants that weaken pericentric cohesion or mutants that decrease constriction on the nucleus as it moves through the bud neck. We propose that minimizing nuclear movement aids in finding a donor strand for homologous recombination.
INTRODUCTION
How cells balance the integrity of the genome with the generation of cytoskeletal-based forces is an important question. During cell division, the cytoskeleton generates forces to build the mitotic spindle, separate duplicated chromatids and move each copy of the genome to regions of the cytoplasm that will belong to the resulting daughter cells. Cells accomplish these tasks using spatially distinct microtubule networks. Within the spindle, microtubule-associated proteins (MAPs) and motors organize spindle microtubules to segregate chromosomes and stabilize the spindle. Outside the spindle, astral microtubules and associated proteins move each copy of the genome to precise locations in the dividing cell. How is genome integrity maintained when the cell undergoes such dramatic reorganization?
During cell division, the genome is exposed to many counteracting forces involved in building and positioning the mitotic spindle. A bipolar spindle is built through dynamic microtubules interacting with motor proteins, cross-linkers, and kinetochores on the chromosomes. These components within the spindle provide opposing forces that are thought to counterbalance each other to keep the spindle at an optimal length as microtubules form bioriented, tensile attachments to sister chromatids that are required for faithful chromosome segregation (McIntosh et al., 1969; Cottingham and Hoyt, 1997; Dumont and Mitchison, 2009). The core structural component of the spindle is microtubules. Outward spindle forces are provided by the combined action of Ase1/PRC1 proteins that selectively cross-link anti-parallel microtubules and kinesin-5 motors that slide these microtubules apart (Cole et al., 1994; Pellman et al., 1995; Winey et al., 1995; Kapitein et al., 2005). Inward forces are provided by kinesin-14 motors and microtubules that attach to the kinetochore complex built on centromeric DNA (Figure 1A; Bouck et al., 2008; Cheeseman and Desai, 2008). The attachment of each chromatid to microtubules from opposite spindle poles creates tension between sister chromatids due to the elastic properties of DNA and integrated proteins (Goshima and Yanagida, 2000; Tanaka et al., 2000; Pearson et al., 2001; Stephens et al., 2011). In addition, cohesin proteins hold sister chromatids together and are important for maintaining and sensing proper tension between chromatids (Stephens et al., 2011; Yong-Gonzales et al., 2012). While the balance of forces builds and maintains a bipolar spindle, how forces outside of the nucleus impact spindle dynamics is unclear.
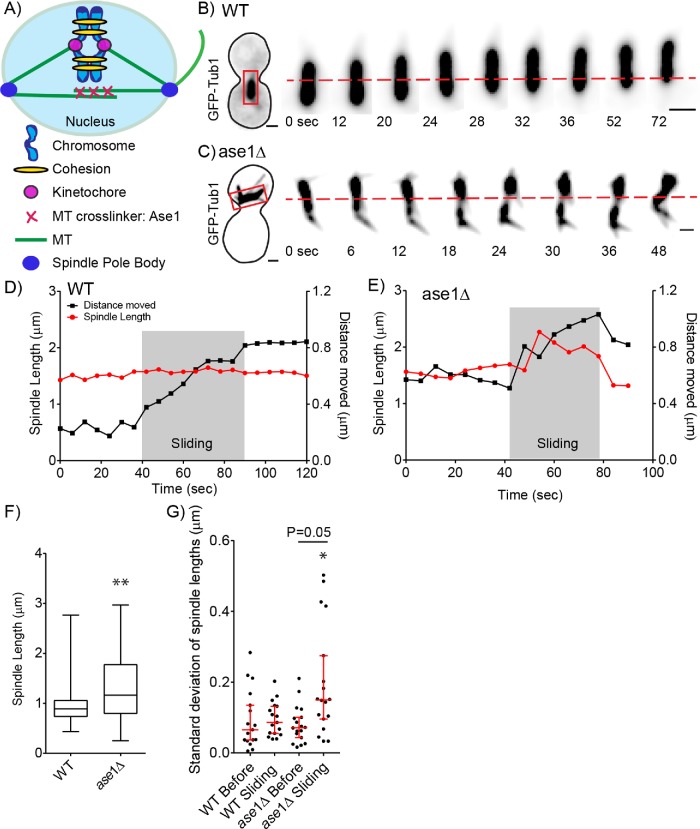
FIGURE 1:
Spindle length and variation increases during nuclear migration. (A) Schematic of spindle and components that contribute to the balance of forces in budding yeast. (B) Time-lapse image series of GFP-Tub1 in a WT cell. Each image is a maximum intensity projection from a confocal z-series. Spindle length during a sliding event was taken from the red boxed region. Time 0 is the spindle before movement. The red dashed line denotes the center of the spindle at time 0. Bar, 1 µm. (C) Time-lapse image series of GFP-Tub1 in an ase1∆ null mutant. Spindle length during a sliding event was taken from the red boxed region. The red dashed line denotes the center of the spindle at time 0. At time 0, the spindle is compact with the length at 0.98 µm. Bar, 1 µm. (D) Plot of spindle length over time for a WT cell during nuclear migration. On the left, the y-axis is spindle length in micrometers, and on the right, the y-axis is distance the spindle moves in micrometers. The x-axis is the duration in seconds. Red dots represent spindle length in micrometers and black dots represent distance the spindle moves in relation to the starting spindle position as 0. The gray box indicates the sliding event. The WT spindle moves a distance of 0.5 µm and the spindle length remains around 1.5 µm with a low SD of 0.02. (E) Plot of spindle length over time for the ase1∆ mutant. Red dots represent spindle length in micrometers and black dots represent the distance the spindle moves in relation to the starting spindle position as 0. The gray box indicates the sliding event. The ase1∆ mutant spindle moves 0.5 µm and the spindle length changes from 1.6 to 2.3 µm with a SD of 0.21. (F) Box and whisker plot of preanaphase spindle lengths. The median length for WT spindles is 0.89 µm with a SD of 0.28 in comparison to ase1∆ mutant containing significantly greater preanaphase spindle length with the median at 1.27 µm with a SD of 0.79; n = 400 cells. Asterisks denote a significant difference from WT. **p < 0.0001 determined by Mann–Whitney test. (G) Dot plot of SD of spindle lengths in micrometers. Each point represents the SD for the spindle lengths reached before or during a sliding event. Red bars represent the median and 95% CI; n = 17 sliding events. Asterisks denote a significant difference. *p < 0.05 determined by Mann–Whitney test.
Astral microtubules interact with the cell cortex to position the mitotic spindle within the cell. Dynein, kinesins, and bridging proteins that link to actin-based motors interact with astral microtubule plus ends to move the spindle (Carminati and Stearns, 1997; Cottingham and Hoyt, 1997; Miller and Rose, 1998; Adames and Cooper, 2000). Similar to maintaining the balance of forces within the spindle, forces from motor proteins must be balanced to regulate astral microtubule length and organization outside of the spindle. Altered astral microtubule stability results in aberrant spindle positioning (Huffaker et al., 1988; Gupta et al., 2006; Rankin and Wordeman, 2010; Tame et al., 2014; Estrem et al., 2017). How astral microtubule-based forces influence the organization and integrity of chromosomes is not understood.
Budding yeast provides an ideal model system to study how forces outside the nucleus impact the integrity of the genome during mitosis. Budding yeast exhibit a closed mitosis, where the nuclear envelope does not break down (Byers and Goetsch, 1975). The dividing cell moves the nucleus, containing the genome and spindle, through a cellular constriction between the mother and daughter, called the bud neck (Adames and Cooper, 2000; Yeh et al., 2000). This movement is accomplished by complementary pathways that first orient the spindle along the mother–daughter axis and then move the spindle into the bud neck. A bridging protein, Kar9, is recruited to the plus ends of astral microtubules growing toward the new daughter cell and interacts with type-V myosins that move along the actin network to orient the assembling spindle (Miller and Rose, 1998; Adames and Cooper, 2000; Yin et al., 2000; Hwang et al., 2003) Then prior to anaphase, the motor protein dynein attaches to a receptor at the cell cortex and generates pulling forces on astral microtubules to move the spindle and nucleus into the bud neck (Li et al., 1993; Adames and Cooper, 2000; Lee et al., 2005). As dynein moves toward the microtubule minus end at the spindle pole body, the yeast microtubule organizing center, it slides the astral microtubule along the cell cortex and pulls the spindle in the same direction as the microtubule. This process is called a sliding event. As the nucleus is pulled into the bud neck, the nuclear envelope is deformed from a circle into a bow tie shape (Byers and Goetsch, 1975; Cottingham and Hoyt, 1997). Dynein acts exclusively outside of the nucleus and its activity is restricted to positioning the spindle beginning in preanaphase and through anaphase (Yeh, 1995; Woodruff et al., 2009). It is unknown whether genome integrity is impacted during nuclear movement in mitosis.
Cells use multiple checkpoints to ensure the integrity of chromosome segregation and the genome during cell division. Within the spindle, microtubules from opposing poles must attach to sister chromatids to satisfy the spindle assembly checkpoint and proceed into anaphase (Cohen-Fix et al., 1996; Schott and Hoyt, 1998; Biggins and Murray, 2001). Astral microtubules must move the spindle to a precise location to satisfy the spindle position checkpoint prior to mitotic exit (Pereira et al., 2000). For the genome, DNA damage surveillance is ongoing in the nucleus, ready to halt the cell cycle to repair damage. If DNA damage occurs in S or G2 of the cell cycle, the DNA damage checkpoint blocks progression into anaphase until the damage is repaired (Weinert and Hartwell, 1988). Homologous recombination is the major mechanism of DNA double-stranded break (DSB) repair to retain genome integrity from budding yeast to mammals (Orr-Weaver et al., 1981; Szostak et al., 1983; Jasin and Berg, 1988). Homologous recombination occurs in S and/or G2 of the cell cycle when a copy of the broken DNA strand is present and nearby (Symington et al., 2014). The DSB undergoes 5′ to 3′ resection by the Mre11-Rad50-Xrxs2 (MRX) complex, priming the leading strand for strand invasion when a region of homology is found (Ivanov et al., 1994; Mimitou and Symington, 2010). Rad52 is essential for homologous recombination in budding yeast due to its role in mediating the loading of Rad51 and annealing complementary single-stranded DNA (Game and Mortimer, 1974; Mortensen et al., 1996; New et al., 1998; Shinohara and Ogawa, 1998). Rad51 recombinase searches for the homologous sequence on the sister chromatid for the donor template (Sung, 1997). The rate-limiting step in the repair process is searching for a donor template (Lee et al., 2016). DSBs increase chromosomal mobility, which is regulated by many of the repair factors involved in homologous recombination (Dion et al., 2012; Miné-Hattab and Rothstein, 2012). How mobility is used to repair DNA damage is still an open question. One proposed mechanism is chromosomal mobility aids in the search to find the homologous donor strand of DNA (Lee et al., 2016; Lawrimore et al., 2017). Mine-Hattab and Rothstein (2012) showed that chromosome mobility in response to DNA damage depends on Rad51 and its upstream regulator Sae2, indicating that increased chromosomal movement contributes to repair by homologous recombination (Miné-Hattab and Rothstein, 2012). Consistent with this notion, DSB repair pathways appear to promote chromosome movement through a mechanism involving nuclear microtubules and kinesin-14/Kar3 motors, and this mechanism is required for efficient repair (Chung et al., 2015; Oshidari et al., 2018). Furthermore, Lawrimore et al. (2017) found microtubules and motor proteins outside of the nucleus contribute to chromosomal movement (Lottersberger et al., 2015; Lawrimore et al., 2017). While these studies demonstrate the importance of cytoskeletal forces in facilitating DSB repair, how these forces are regulated to promote efficient repair is unknown.
In this study, we test the hypothesis that the astral microtubule network outside of the nucleus is regulated to limit spindle movement and thereby protect the integrity of the genome. We use live-cell imaging of mutants that increase the frequency and extent of nuclear migration through the bud neck to examine how forces outside of the nucleus impact spindle stability, nuclear morphology, and the efficiency of DNA repair. Our results show that forces exerted on astral microtubules are transmitted through spindle microtubules and lead to distortions of pericentric DNA. Cells experiencing increased nuclear migration exhibit persistent Rad52 foci, indicative of delayed DSB repair during S and G2 phase. This repair defect can be rescued by cohesion mutants that loosen pericentric cohesion and allow more mobility between sister chromatids. In addition, we show extended nuclear deformations caused by dynein pulling the nucleus through the narrow bud neck lead to increased DNA damage and this damage is alleviated by mutants that widen the bud neck and relieve nuclear constriction. Our results demonstrate that aberrant forces generated by astral microtubules can damage the genome during mitosis and suggest proper regulation of astral microtubules may be coordinated with pathways that maintain genome integrity.
RESULTS
Forces from nuclear migration are transmitted through the spindle
We first tested the prediction that dynein-dependent forces on astral microtubules alter mitotic spindle organization by measuring changes in spindle length during nuclear migration. Wild-type (WT) spindles containing GFP-labeled microtubules (GFP-Tub1) were measured before and during nuclear migration. SD of spindle length was calculated by the square root of the variance, which is the average of the squared differences from the mean for that time period. We define nuclear migration as when an astral microtubule undergoes a dynein-dependent sliding event, pulling the spindle and nucleus forward. Figure 1B shows an example of nuclear migration in a WT cell. In this example, spindle length remains constant during migration, with an average length of 1.5 µm and a SD of 0.02 µm (Figure 1, B and D).
We reasoned that proteins that cross-link interpolar microtubules might resist spindle length changes when pulling forces are transmitted from astral microtubules. To test this prediction, we deleted ASE1, which cross-links interpolar microtubules in the spindle, and analyzed spindle length over time (Pellman et al., 1995; Juang et al., 1997). Figure 1C shows an example of nuclear migration in an ase1∆ mutant cell. At time 0, before the spindle moves, the spindle is compact with the length at 1.6 µm (Figure 1C). As the leading edge of the nucleus moves through the bud neck, the spindle elongates to a maximum length of 2.27 µm. When nuclear movement stops, the spindle shortens and returns to a length similar to the length observed before migration. This ase1∆ cell shows an average spindle length of 1.9 µm during migration with a SD of 0.21 µm (Figure 1, C and E).
By analyzing a population of cells, we find that ase1∆ mutant spindles exhibit a wider distribution of spindle lengths and a significantly greater mean spindle length compared with WT cells in preanaphase (Figure 1F). To distinguish whether ase1∆ spindles exhibit increased changes in length due to pulling forces from astral microtubules, we compared the SD of spindle lengths before and during nuclear migration (Figure 1G). We examined 17 nuclear migration events and measured spindle lengths over time, focusing on the length of the spindle immediately before and during the sliding event. Spindle length SD did not change significantly before and during sliding for WT cells (p = 0.95 determined by Mann–Whitney test) (Figure 1G). In contrast, spindle length SD did change significantly before and during sliding for ase1∆ mutant cells (p = 0.005 determined by Mann–Whitney test) (Figure 1G). Our results show that dynein-dependent pulling forces on astral microtubules normally cause the entire spindle to move without changing length. However, when cross-links between interpolar microtubules are disrupted, pulling forces on astral microtubules cause one pole to move away from the other and the spindle to change its length.
Nuclear migration increases tension on pericentric chromatin
The above results indicate that interpolar microtubule cross-links transmit astral microtubule pulling forces across the spindle during nuclear migration. We next asked whether chromosomes also experience pulling forces from astral microtubules. We used a previously characterized method to image pericentric chromatin by integrating a tetO array 2 kb away from the centromere on chromosome IV, and expressing tetR-GFP to label this region (Figure 2A) (Goshima and Yanagida, 2000; Brito et al., 2010; Fees et al., 2016). We refer to this as CENIV-GFP. By labeling the region proximal to a centromere, we have the resolution to see how this pericentric region is organized and positioned relative to the homologous region on its sister chromatid in the mitotic spindle. When the regions are not under tension by the pulling forces of spindle microtubules, the GFP-labeled CENIV appears as a single focus (Figure 2B) (Goshima and Yanagida, 2000; Tanaka et al., 2000; Pearson et al., 2001; Chacón et al., 2014). When sister chromatids are bioriented and under tension, the labeled pericentric regions can be resolved into two foci (Figure 2B) (Goshima and Yanagida, 2000; Pearson et al., 2001; Tanaka et al., 2000). In strains expressing labeled CENIV-GFP and differentially labeled spindle pole bodies (SPBs), we analyzed the CENIV foci number in all cells containing two SPBs separated by less than 1 µm to ensure that they were in preanaphase. We used this system to test the prediction that nuclear migration alters chromosome organization in the spindle; specifically, that pulling forces from nuclear migration increase tension on sister chromatids.
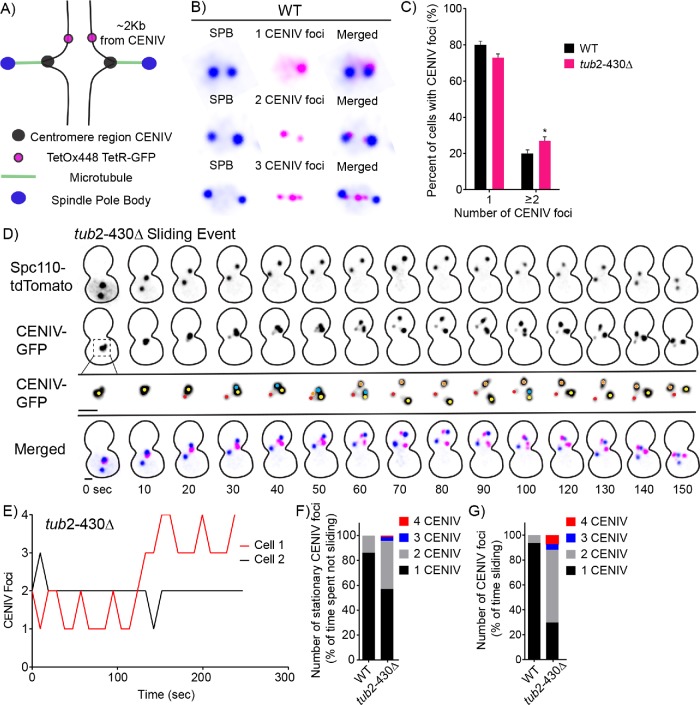
FIGURE 2:
Increased tension on pericentromeric chromatin during excessive nuclear migration mutant. (A) Schematic of CENIV TetR-GFP/TetO array. The tetOx448 array of repeats is located 2 kb away from the centromeric region on chromosome IV. In this system, we can visualize the region proximal to centromere with TetR-GFP and SPBs labeled with Spc110-tdTomato. (B) Representative images of WT cells with GFP-labeled pericentromeric region of chromosome IV and labeled SPBs in three different tension states. In the top panel, sister chromatids are not under tension in which the pericentromeric regions are combined into one CENIV-GFP foci. In the middle panel, the two sister chromatids are under tension, in which the pericentromeric regions can be resolved into two CENIV-GFP foci. In WT cells, on rare occasion, we see cells with three CENIV-GFP foci. (C) Frequency distribution for the percentage of preanaphase cells with one and two or more CENIV foci in WT and tub2-430∆ mutant strains. WT, n = 331; tub2-430∆, n = 531 cells. Error bars represent SEP. *p = 0.04 by Fisher’s exact test. (D) Time-lapse image series of a tub-430∆ mutant cell during a sliding event. Panel 1 displays SPBs, labeled by Spc110-tdTomato. Panels 2 and 3 display CENIV-GFP. Panel 2 matches the scale of panel 1. Panel 3 is an enlargement to visualize CENIV number. Colored dots denote and follow CENIV foci number. Panel 4 depicts the merged images. Blue is Spc110-tdTomato and pink is CENIV-GFP. Time is in seconds. Bar, 1 µm. (E) Representative life plots of CENIV-GFP foci transitions for two tub2-430∆ mutant cells. Cell 1 in red represents the CENIV foci depicted in D. (F) Stacked bar graph displaying the percentage of time CENIV-GFP spends with each foci number containing a stationary spindle for WT and tub2-430∆ strains. WT, n = 25; tub2-430∆, n = 23 cells. (G) Stacked bar graph displaying the percentage of time CENIV-GFP spends with each foci number when the spindle is sliding for WT and tub2-430∆ strains. WT, n = 25; tub2-430∆, n = 23 cells.
First, we used single time-point imaging to measure the frequencies of preanaphase cells containing different numbers of CENIV-GFP foci. In WT preanaphase cells, 79% contain one CENIV-GFP focus, and 20% contain two or more CENIV-GFP foci (Figure 2C). To test our prediction that nuclear migration alters tension on pericentric chromatin, we used a mutant that exhibits excessive nuclear migration. We previously found that a β-tubulin mutant, tub2-430∆, causes hyperstable microtubules and increased interaction with dynein at the cell cortex, producing increased frequency and duration of spindle migration, and therefore nuclear migration (Estrem et al., 2017). In our single time-point experiment, the tub2-430∆ mutant exhibits an increase in cells containing two or more CENIV-GFP foci (27%) compared with WT cells (20%), consistent with previous studies (p = 0.04 by Fisher’s exact test; Figure 2C) (Fees et al., 2016). We then used time-lapse imaging to determine whether pericentric regions are specifically under higher tension during nuclear migration. Figure 2D shows a representative series of time-lapse images from a tub2-430∆ mutant cell. When the bipolar spindle moves through the bud neck, the CENIV-GFP changes from a single focus into multiple foci (Figure 2, D and E). Applying this analysis across a population of cells, we predict an increase in cells containing two CENIV foci during nuclear migration, when the spindle is moving through the cell. WT cells contain one CENIV foci 86% of the time and two CENIV foci 13% of the time while the spindle is stationary (Figure 2F). While the spindle is moving, WT cells contain one CENIV foci 93% of the time and two CENIV foci 6% of the time (Figure 2G). Thus, we do not see an increase in time spent in a two CENIV state during nuclear migration in WT cells. However, we find that the two CENIV state is more common in tub2-430∆ mutants, compared with WT controls (p < 0.0001 by Fisher’s exact test; Figure 2, F and G). Furthermore, the two CENIV-GFP state is specifically enriched in tub2-430∆ mutants during periods of spindle movement (p < 0.0001 by Fisher’s exact test; Figure 2, F and G). These data suggest that increased nuclear migration alters the tension on pericentric chromatin.
In addition to the two CENIV-GFP state, which is indicative of tension across sister chromatids, we also noticed that tub2-430∆ mutants transiently visit states with three or four CENIV-GFP foci. Our time-lapse imaging data indicate that these three and four CENIV-GFP states are transient, arise during spindle migration, and are specifically enriched in tub2-430∆ mutants (Figure 2, D–G). Consistent with this, we identified cells with three or four CENIV-GFP foci in a small portion of tub2-430∆ mutants (22/531 cells) and in a smaller portion of WT controls (5/331 cells) in our single time-point analysis. These results indicate that increased outside spindle forces can be transmitted to pericentric chromatin and alter its position and organization.
Mutants with increased nuclear migration contain a higher frequency of cells with DSBs
Our results demonstrate that forces that drive migration of the spindle and nucleus are transmitted to pericentric chromatin, and increased frequency and duration of migration disrupt normal chromatin organization. Next, we sought to determine whether increasing the movement of the spindle and nucleus damages the genome. We first tested the prediction that mutants that promote excessive nuclear migration might be highly sensitive to defects in DNA repair. A previous genome-wide screen identified negative genetic interactions between the tub2-430∆ mutant and null mutants that disrupt DNA repair (Aiken et al., 2014). Specifically, the identified interactions were significantly enriched for genes associated with the Gene Ontology term “response to DNA damage stimulus” (p = 0.003). These genes include the DNA recombinase RAD52, components of the Ctf18-RFC complex (CTF18, CTF8, DCC1), and components of the Cul8 RING ubiquitin ligase complex (MMS1 and MMS22) (Game and Mortimer, 1974; Prakash and Prakash, 1977; Kouprina et al., 1993; Shinohara and Ogawa, 1998). We confirmed a subset of these negative genetic interactions by using meiotic crosses to generate double mutants combining tub2-430∆ with rad52∆, ctf18∆, or mms22∆. In each case, double mutant progeny are either inviable or exhibit impaired growth, compared with single mutant controls (Supplemental Figure S1, A and B). These results support the prediction that cells experiencing excessive migration of the spindle and nucleus are highly sensitive to defects in DNA repair.
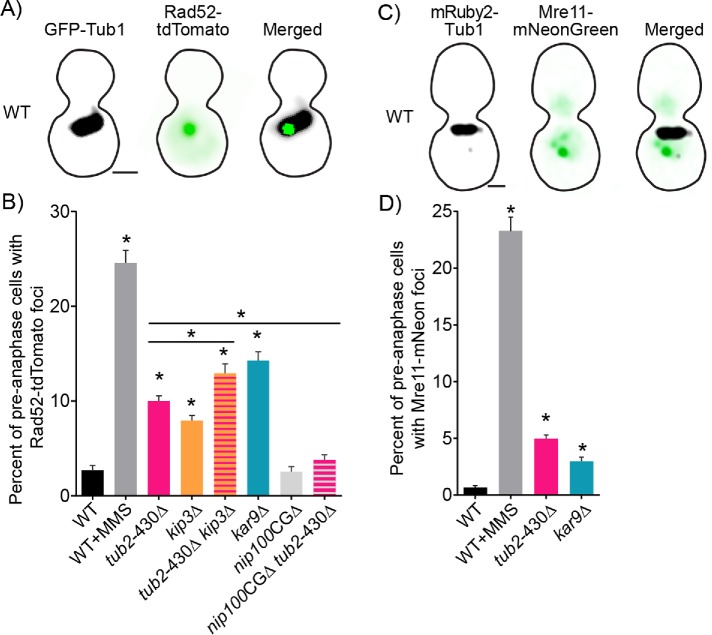
To measure DNA damage in cells, we visualized DSBs by fusing tdTomato to the C-terminus of the DNA repair factor Rad52 and expressing it from the native locus (Figure 3A). In the absence of DSBs, fluorescently labeled Rad52 is diffuse in the nucleus; but upon introducing DSBs, Rad52 forms a focus at the site of the DNA break and aids in homology-directed repair (Lisby et al., 2001) (Figure 3A). We validated that Rad52-tdTomato foci represent sites of DSB repair in three ways. First, we show that cells treated with the DNA alkylating agent methyl methanesulfonate (MMS) exhibit an increased frequency of Rad52-tdTomato foci, compared with untreated controls (Figure 3B). Second, we found that Rad52-tdTomato foci were almost always detected in WT cells during preanaphase, but not during G1 or late anaphase (Supplemental Figure S2A). This result is consistent with the known role of Rad52 in mediating repair when a duplicate copy of the genome is present, in S and G2 phases of the cell cycle (Orr-Weaver et al., 1981; Szostak et al., 1983; Lisby et al., 2003). Third, we compared our Rad52-tdTomato results to separate experiments using an integrated mNeonGreen fusion to Mre11, the nuclease subunit of the MRX complex, which functions at the initial repair step of the 5′ to 3′s resection of a DSB (Ivanov et al., 1994; Mimitou and Symington, 2010). Again, in the presence of DNA-damaging agent MMS, we saw a significantly increased frequency of preanaphase cells with Mre11-mNeonGreen foci (Figure 3, C and D).
FIGURE 3:
Mutants with increased nuclear migration contain a higher frequency of cells with DSBs. (A) Representative image of a WT cell expressing GFP-Tub1 and Rad52 labeled with tdTomato. Bar, 1 µm. (B) The percentage of cells containing Rad52 foci in preanaphase of the cell cycle. Percentages are calculated by number of cells containing Rad52 foci divided by number of cells in preanaphase of the cell cycle. WT, n = 2581; WT MMS, n = 972; tub2-430∆, n = 2653; kip3∆, n = 2294; tub2-430∆ kip3∆, n = 1762; kar9∆, n = 541; nip100-CG∆, n = 2696; tub2-430∆ nip100-CG∆, n = 2494 cells. Error bars represent SEP. Asterisks denote a significant difference from WT. *p < 0.001; determined by Fisher’s exact test. (C) Representative image of a WT cell expressing mRuby2-Tub1 and Mre11 labeled with mNeonGreen. Bar, 1 µm. (D) The percentage of cells containing Mre11 foci in preanaphase of the cell cycle. Percentages are calculated by number of cells containing Mre11 foci divided by number of cells in preanaphase of the cell cycle. WT, n = 1471; WT MMS, n = 794; tub2-430∆, n = 2178; kar9∆, n = 1448 cells. Error bars represent SEP. Asterisks denote a significant difference. *p < 0.001 determined by Fisher’s exact test.
We find that 10% of tub2-430∆ mutant cells contain Rad52-tdTomato foci compared with 3% of WT controls (p < 0.001; determined by Fisher’s exact test; Figure 3B). These Rad52-tdTomato foci were almost exclusively seen in tub2-430∆ cells in S and G2 phase, as determined by the morphology of spindles labeled with GFP-Tub1 (Supplemental Figure S2A). Since we saw this increase in Rad52 frequency specifically in preanaphase cells (i.e., cells in S and G2 phase), we focused our subsequent analysis on cells in preanaphase. In separate experiments, we found that combining the tub2-430∆ with the DNA repair mutants mec1∆ and sml1∆ produces an additive increase in the frequency of cells with Rad52 foci (Supplemental Figure S2B). Cells containing the mec1∆ sml1∆ null mutations contain more DNA damage in S and G2 due to collapsed replication forks and proceed through anaphase without delay even when containing DNA damage (Lisby et al., 2001; Weinert, 1998; Feng, 2017).
To determine whether the observed increase in DSBs in tub2-430∆ is a product of increased outside forces on the spindle, we independently analyzed two other mutants known to increase nuclear migration (Estrem et al., 2017). A null mutant of KIP3, a kinesin 8 in budding yeast, and a null mutant of KAR9 contain stable astral microtubules that promote increased frequency and duration of spindle migration (Gupta et al., 2006; Estrem et al., 2017) (Supplemental Figure S2, C and D). The tub2-430∆ and the kip3∆ mutants individually contain a significant increase, 10 and 8%, respectively, of preanaphase cells with Rad52 foci, compared with WT, at 3% of preanaphase cells (p < 0.0001 by Fisher’s exact test; Figure 3B). In addition, the tub2-430∆ and kip3∆ double mutant, which exhibits additive effects on microtubule stability and spindle movement (Estrem et al., 2017) (Supplemental Figure S2, C and D), exhibits Rad52 foci in 13% of cells, significantly increased compared with either of the single mutants (p < 0.0001 by Fisher’s exact test; Figure 3B). The kar9∆ single mutant exhibits the greatest frequency of cells with Rad52 foci at 14% of preanaphase cells and is significantly greater than either WT controls or tub2-430∆ mutants (p < 0.0001 by Fisher’s exact test; Figure 3B). The kar9∆ mutant only modifies astral microtubules and contains significantly more spindle movement compared with the tub2-430∆ mutant, which affects all microtubules (Supplemental Figure S2D); therefore, the increase in Rad52 foci correlates with forces specifically from astral microtubules that lead to increased frequency of nuclear migration.
To confirm our results, we conducted independent experiments measuring the frequency of Mre11-mNeonGreen foci and again found a significant increase in cells containing foci in the tub2-430∆ and kar9∆ mutants (p < 0.001 by Fisher’s exact test; Figure 3D). Mre11 is recruited to the site of the DSBs earlier than Rad52 and stays bound for a shorter amount of time (Lisby et al., 2004; Supplemental Figure S3). Accordingly, the frequency of Mre11 foci may reflect the early stages of DSB repair, while Rad52 foci may reflect later stages. Interestingly, WT cells treated with MMS exhibit similar frequencies of Mre11 and Rad52 foci, while tub2-430∆ and kar9∆ cells exhibit higher frequencies of Rad52 foci (Figure 3, B and D). Together, these results indicate that cells experiencing increased nuclear migration also exhibit increased DSBs and may be particularly impacted during later stages of DSB repair.
Having established that increased nuclear migration results in increased DSBs, we asked whether suppressing nuclear migration could suppress the frequency of DSBs. The dynactin subunit Nip100/p150 contains a CAP-Gly domain, which is a microtubule-binding domain that promotes dynein activity (Kardon et al., 2009; Moore et al., 2009; Nirschl et al., 2016). Removing the CAP-Gly domain partially impairs dynein activity, and reduces the frequency and duration of nuclear migration in yeast (Moore et al., 2009; Supplemental Figure S2E). We find that nip100CAP-Gly∆ single mutants exhibit Rad52 foci in 3% of cells, similar to WT controls (Figure 3B). Double mutants combining nip100CAP-Gly∆ with tub2-430∆ show significantly fewer cells with Rad52 foci compared with tub2-430∆ single mutants (4%; p < 0.0001 by Fisher’s exact test) (Figure 3B). These results show that the level of nuclear migration activity plays a determining role in the prevalence of DSBs.
DSB repair is delayed during excessive nuclear migration
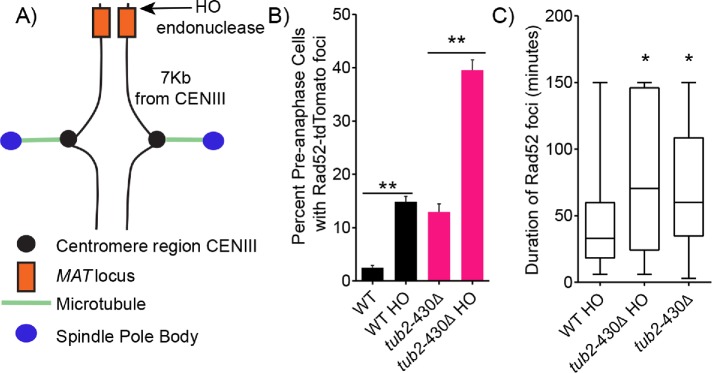
The increased frequency of cells containing Rad52 foci could be due to an increase in the generation of DSBs and/or a delay of DSB repair. To distinguish between these possibilities, we designed an experiment to measure the efficiency of DSB repair by comparing the lifetime of Rad52-tdTomato foci from formation to disappearance. For these experiments, we induced a DSB at a specific site on the genome by conditionally expressing the HO endonuclease. The HO endonuclease creates a DSB at the MAT locus, which is located on chromosome III, 7 kb from the centromere (Figure 4A) (Kostriken et al., 1983; Haber, 1992). This allowed us to generate a consistent frequency of Rad52 foci in WT and tub2-430∆ cells that is sufficient for our experiment. Expressing HO with a galactose-inducible promoter for 2 h in WT cells significantly increased the frequency of preanaphase cells containing Rad52 foci from 2% to 15% (Figure 4B). By comparison, inducing HO expression in tub2-430∆ cells significantly increased the frequency of preanaphase cells containing Rad52 foci from 13% to 40% (Figure 4B). We then measured the lifetime of Rad52 foci in WT and tub2-430∆ cells after HO induction by using time-lapse imaging of living cells. We find that Rad52 foci are significantly longer lived in the tub2-430∆ mutant cells, with a median lifetime of 70 min compared with 33 min in WT cells (p = 0.0006 by Mann–Whitney test) (Figure 4C). Rad52 foci lifetimes were not significantly different in tub2-430∆ cells containing HO-induced DSBs at the MAT locus compared with uninduced tub2-430∆ cells, in which DSBs are presumably at other locations on the genome (p = 0.2 by Mann–Whitney test) (Figure 4C). These results indicate that DSB repair is delayed in cells with increased spindle movement.
FIGURE 4:
Excessive nuclear migration mutants are slow to repair DSBs. (A) Schematic diagram of the MAT locus and where HO endonuclease cuts the DNA on chromosome III. (B) The percentage of cells containing Rad52 foci in preanaphase of the cell cycle. Percentages are calculated by number of cells containing Rad52 foci divided by number of cells in preanaphase of the cell cycle. WT, n = 1220; WT HO induced, n = 1220; tub2-430∆, n = 463; tub2-430∆ HO induced, n = 612 cells. Error bars represent SEP. Asterisks denote a significant difference. **p < 0.001; determined by Fisher’s exact test. (C) Box and whisker plot of duration of Rad52 foci in minutes. The center bar denotes the median, the box denotes the first and third quartiles, and whiskers are maxima and minima. WT and tub2-430∆ cells containing Rad52 foci induced with galactose for 2 h. WT HO, n = 71; tub2-430∆ HO n = 68; tub2-430∆ containing Rad52 foci containing glucose (uninduced for HO), n = 52 cells. Asterisks denote a significant difference. *p < 0.0003, determined by Mann–Whitney test.
Pericentric cohesion mutant decreases the frequency of DSBs in excessive nuclear migration
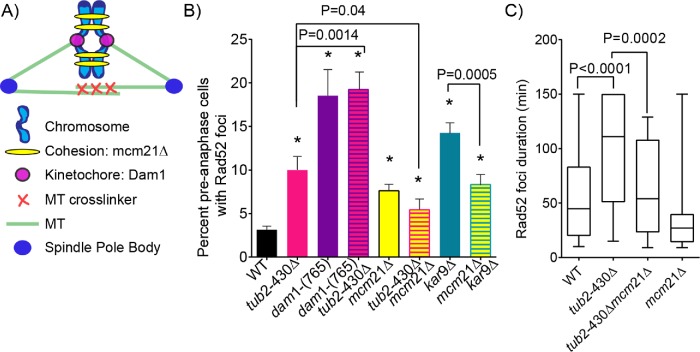
How might astral microtubule forces inhibit DSB repair? To investigate the role of tension within the spindle, we used previously characterized mutants that increase or decrease tension on pericentric chromatin and combined these with our nuclear migration mutants to assess effects on DSBs. First we tested the prediction that extraneous tension pulling sister chromatids apart would inhibit DSB repair due to altering the proximity and/or mobility of homologous chromosomes. Dam1 is part of the kinetochore complex that links centromeres to the plus ends of spindle microtubules (Hofmann et al., 1998). In dam1-765 mutants, kinetochores fail to attach to microtubule plus ends, and instead bind closer to minus ends, causing kinetochores to cluster near the spindle poles (Figure 5, A and B; Shimogawa et al., 2006). We found that 19% of dam1-765 mutant cells exhibit Rad52-tdTomato foci, which is significantly greater than either WT controls or nuclear migration mutants (p < 0.0001 determined by Fisher’s exact test) (Figure 5B). Double mutants combining tub2-430∆ with dam1-765 exhibit a frequency of Rad52-tdTomato foci similar to that seen in dam1-765 single mutants (19%; Figure 5B). Thus, increased tension across sister chromatids promotes a high level of DSBs, even when spindle migration is normal.
FIGURE 5:
Pericentric cohesion mutant decreases the frequency of DSBs in excessive nuclear migration mutants. (A) Schematic diagram of spindle components that make up and contribute to forces within the mitotic spindle. (B) The percentage of cells containing Rad52 foci in preanaphase of the cell cycle. Percentages are calculated by number of cells containing Rad52 foci divided by number of cells in preanaphase of the cell cycle. WT, n = 2581; tub2-430∆, n = 2653; tub2-430∆ mcm21∆, n = 276; mcm21∆, n = 1209; kar9∆, n = 891; mcm21∆ kar9∆, n = 621; dam1-(765), n = 135; dam1-(765) tub2-430∆, n = 135 cells. Error bars represent SEP. Asterisks denote a significant difference. *p < 0.05 determined by Fisher’s exact test. (C) Box and whisker plot of duration of Rad52 foci in minutes. The center bar denotes the median, box denotes the first and third quartiles, and whiskers are maxima and minima. The maximum duration is 150 min. Black bars represent the 95% CI. WT, n = 61; tub2-430∆, n = 35; tub2-430∆ mcm21∆, n = 42; mcm21∆, n = 18 cells; p values are determined by Mann–Whitney test.
If increasing tension between sister chromatids increases the prevalence of DSBs, then we predicted that decreasing tension might have the opposite effect. Pericentric cohesion couples sister chromatids and normally promotes the transmission of forces from kinetochore-microtubule attachments (Goshima and Yanagida, 2000). We used the mcm21∆ mutant to disrupt pericentric cohesion. The mcm21∆ mutation reduces cohesion from pericentric chromatin, decreasing tension between sister chromatids, and thereby increasing the number of preanaphase cells with two CENIV foci (Ng et al., 2009; Fees et al., 2016). In our experiment, mcm21∆ mutants alone exhibit a slight but significant increase in the frequency of cells with Rad52 foci, compared with WT controls (7% compared with 3%, respectively; p = 0.007 determined by Fisher’s exact test; Figure 5B). However, the mcm21∆ mutation suppressed DSBs when combined with mutants that increase spindle migration. Both tub2-430∆ mcm21∆ and kar9∆ mcm21∆ double mutants exhibit significantly fewer cells with Rad52-tdTomato foci (5% for tub2-430∆ mcm21∆; 8% for kar9∆ mcm21∆) compared with kar9∆ and tub2-430∆ single mutants alone (10% for tub2-430∆; 14% for kar9∆; p = 0.04 determined by Fisher’s exact test; Figure 5B). In addition, Rad52-tdTomato foci were significantly shorter-lived in tub2-430∆ mcm21∆ double mutant compared with tub2-430∆ single mutants (p < 0.0002 by Mann–Whitney test; Figure 5C). These data suggest that pericentric cohesion inhibits in DSB repair during excessive spindle migration.
Decreasing nuclear deformation decreases the frequency of DSBs
Having established that forces from astral microtubules can stretch pericentric chromatin and delay repair, we next asked whether the movement of the nucleus within dividing cells deforms the genome and if this might also contribute to genome damage. Therefore, we investigated how movement of the spindle and nucleus impacts the three-dimensional structure of the genome. Previous studies in mammalian cells have shown that forcing cells to migrate through a narrow pore can increase DNA damage through several possible mechanisms, such as nuclear rupture, DNA herniation, mechanical shearing of DNA, and mislocalization of DNA repair factors within the nucleus (Shah et al., 2017; Xia et al., 2018). We did not find evidence of nuclear rupture in preanaphase budding yeast undergoing nuclear migration. We tested this possibility using time-lapse imaging of WT and tub2-430∆ mutants expressing nuclear-targeted GFP (NLS-GFP); but found no instances of NLS-GFP leaking out of the nucleus and into the cytoplasm in 45 cells analyzed (Supplemental Movies S1 and S2). We therefore asked whether the increased DSBs observed during nuclear migration in budding yeast was caused by the deformation of the nucleus and genome as it moves through the narrow bud neck region between the mother and daughter compartments. We first sought to assess the magnitude of deformation by imaging the genome using a fluorescent mNeonGreen fusion to the C-terminus of the core histone 2B protein, Htb2. Supplemental Movie 3 shows the fluorescently labeled genome in a cell transitioning from preanaphase into anaphase, as the spindle is pulled into the bud neck. The labeled genome maintains a largely circular shape in preanaphase (Figure 6A; see 0- to 12-min time points). At anaphase, the initial movement of the spindle into the bud neck is accompanied by a narrow extension of the genome, followed by the translocation of Htb2-mNeonGreen into two circular lobes in the mother and daughter (Supplemental Movies S3–S6; see 0- to 54-min time points).
FIGURE 6:
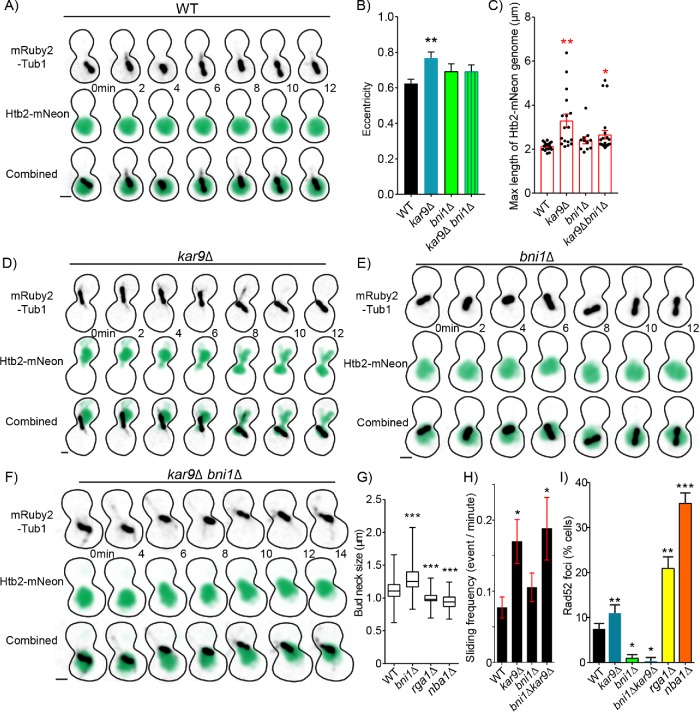
Nuclear deformation increases the frequency of DSBs. (A) Time-lapse image series of Htb2-mNeonGreen and mRuby-Tub1 in a WT cell in preanaphase. Each image is a maximum intensity projection from a confocal z-series. Bar, 1 µm. (B) Mean of maximum eccentricity reached in preanaphase. Error bars represent the standard error of the mean. WT, n = 19; kar9∆, n = 17; bni1∆, n = 11; kar9∆ bni1∆, n = 23 cells. Asterisks denote a significant difference from WT. **p < 0. 0.001 determined by t test. (C) Dot plot of maximum length reached in micrometers of Htb2-mNeon-labeled genome of cell in preanaphase. Red bars represent the mean ± standard error of the mean. WT, n = 19; kar9∆, n = 17; bni1∆, n = 11; kar9∆ bni1∆, n = 23 cells. Asterisks denote a significant difference from WT.*p < 0.05; **p < 0.001 determined by t test. (D) Time-lapse image series of Htb2-mNeonGreen and mRuby-Tub1 in a kar9∆ mutant cell in preanaphase. Each image is a maximum intensity projection from a confocal z-series. Bar, 1 µm. (E) Time-lapse image series of Htb2-mNeonGreen and mRuby-Tub1 in a bni1∆ mutant cell in preanaphase. Each image is a maximum intensity projection from a confocal z-series. Bar, 1 µm. (F) Time-lapse image series of Htb2-mNeonGreen and mRuby-Tub1 in a kar9∆, bni1∆ double mutant cell in preanaphase. Each image is a maximum intensity projection from a confocal z-series. Bar, 1 µm. (G) Box and whisker plot of bud neck sizes in micrometers. The center bar denotes the median, the box denotes the first and third quartiles, and whiskers are maxima and minima. WT, n = 354; bni1∆, n = 108; rga1∆, n = 154; nba1∆, n = 163 cells. Asterisks denote a significant difference from WT. ***p < 0.0001 determined by Mann–Whitney test. (H) Mean frequency of sliding events. Cells were arrested in 200 mM hydroxyurea for 2 h and scored for microtubule sliding events (see Materials and Methods). WT, n = 35; kar9∆, n = 23; bni1∆, n = 35; bni1∆ kar9∆, n = 16 cells. Asterisks denote a significant difference from WT. *p < 0.05 determined by t -test. (I) The percentage of cells containing Rad52 foci in preanaphase of the cell cycle. Percentages are calculated by the number of cells containing Rad52 foci divided by the number of cells in preanaphase of the cell cycle. WT, n = 597; kar9∆, n = 598; bni1∆, n = 736; bni1∆ kar9∆, n = 754; rga1∆, n = 253 nba1∆, n = 429 cells. Error bars represent SEP. Asterisks denote a significant difference from WT. *p < 0.05; **p < 0.001; ***p < 0.0001 determined by Fisher’s exact test.
Video S1.
GFP‐NLS and SPC110‐DsRed in a WT cell. Time‐lapse images of a WT cell expressing GFP‐NLS and SPC110‐DsRed were captured on a spinning‐disk confocal microscope at 10‐s intervals for 10 min. Each image represents a composite of 13 planes separated by 450 nm. The video plays at four frames per second. Strain: WT, yJM970.
Video S2.
GFP‐NLS and SPC110‐DsRed in a tub2‐430Δ cell. Time‐lapse images of a tub2‐430Δ cell expressing GFP‐NLS and SPC110‐DsRed were captured on a spinning‐disk confocal microscope at 10‐s intervals for 10 min. Each image represents a composite of 13 planes separated by 450 nm. The video plays at four frames per second. Strain: tub2‐430Δ, yJM971.
Video S3.
HTB2‐mNeonGreen and mRuby2‐Tub1 in a WT cell. Time‐lapse images of a WT cell expressing HTB2‐mNeonGreen and mRuby2‐Tub1 were captured on a spinning‐disk confocal microscope at 3‐minute intervals for 1 hour. Each image represents a composite of 13 planes separated by 450 nm. The video plays at four frames per second. Strain: WT, yJM3264.
Video S4.
HTB2‐mNeonGreen and mRuby2‐Tub1 in a kar9Δ cell. Time‐lapse images of a kar9Δ cell expressing HTB2‐mNeonGreen and mRuby2‐Tub1 were captured on a spinning‐disk confocal microscope at 3‐minute intervals for 1 hour. Each image represents a composite of 13 planes separated by 450 nm. The video plays at four frames per second. Strain: kar9Δ, yJM3405.
Video S5.
HTB2‐mNeonGreen and mRuby2‐Tub1 in a bni1Δ cell. Time‐lapse images of a bni1Δ cell expressing HTB2‐mNeonGreen and mRuby2‐Tub1 were captured on a spinning‐disk confocal microscope at 3‐minute intervals for 1 hour. Each image represents a composite of 13 planes separated by 450 nm. The video plays at four frames per second. Strain: bni1Δ, yJM3308.
Video S6.
HTB2‐mNeonGreen and mRuby2‐Tub1 in a kar9Δ bni1Δ cell. Time‐lapse images of a kar9Δ bni1Δ cell expressing HTB2‐mNeonGreen and mRuby2‐Tub1 were captured on a spinning‐disk confocal microscope at 3‐minute intervals for 1 hour. Each image represents a composite of 13 planes separated by 450 nm. The video plays at four frames per second. Strain: kar9Δ bni1Δ, yJM3411.
To test whether increased nuclear migration mutants deform the genome, we measured the eccentricity and diameter of Htb2-mNeonGreen over time in preanaphase cells (Figure 6, A and B). Eccentricity is defined as the deviation of an object from circularity. Preanaphase WT cells have an average maximum eccentricity of 0.6 ± 0.02 and a mean length across the longest extension of the nucleus of 2.1 ± 0.04 µm (mean ± SEM; Figure 6, B and C). By comparison, the increased nuclear migration mutant, kar9∆, has significantly increased average maximum eccentricity (0.8 ± 0.03; p = 0.001 by t test) and mean genome extension (3.3 ± 0.3 µm; p = 0.0004; Figure 6, A–D). These results are consistent with increased nuclear migration exacerbating the deformation of the genome.
To determine whether movement through the constricted space of the bud neck contributes to deformation of the genome and DNA damage we generated mutant cells with increased bud neck diameter. Bni1 is a formin that nucleates actin filaments to supply the formation of the bud (Lee et al., 1999). bni1∆ null mutants have a significantly wider bud neck with median diameter of 1.3 ± 0.1 µm compared with WT cells with median diameter of 1.1 ± 0.2 µm (median ± 95% CI; p < 0.0001 determined by Mann–Whitney test; Figure 6, E and G). We also compared the diameter of the bud neck in relation to the total diameter of the cell (measuring the long axis of the budding cell) to show the bni1∆ mutants have a wider bud neck in proportion to the total cell size (Supplemental Figure S4A). Widening the bud neck suppressed the deformation of the genome during nuclear migration. We find that preanaphase bni1∆ single mutant cells exhibit eccentricity and diameter of Htb2-mNeonGreen signal that is similar to WT controls (p = 0.14 and 0.05 by t test; Figure 6, B and C). In kar9∆ bni1∆ double mutants that increase nuclear migration activity and have wider bud necks, eccentricity, and genome diameter are also comparable to WT levels (p = 0.15; p = 0.05 determined by t test; Figure 6, B, C, F, and H).
Since widening the bud neck suppresses genome deformation during nuclear migration, we predicted that widening the bud neck might also suppress DNA DSBs. Consistent with this prediction, we found that 1.0% of preanaphase bni1∆ mutant cells exhibit Rad52 foci, which is significantly decreased compared with WT controls (7%; p = 0.02 by Fisher’s exact test; Figure 6I). Furthermore, widening the bud neck also suppresses DNA DSBs in mutants that increase nuclear migration. The bni1∆ kar9∆ double mutant cells exhibit a lower frequency of Rad52 foci (0.3%) compared with kar9∆ single mutants (11%; p < 0.0001 by Fisher’s exact test; Figure 6I).
We reasoned that since widening the bud neck suppresses DSBs, then decreasing bud neck size would increase DNA DSBs. We tested this prediction using two mutants, rga1∆ and nba1∆, that exhibit significantly smaller bud neck width with median diameter of 0.98 ± 0.1 µm and 0.93 ± 0.1 µm compared with WT cells (median ± 95% CI; p < 0.0001 determined by Mann–Whitney test; Figure 6G)(Meitinger et al., 2014). Consistent with our prediction, we found that 21% and 35% of preanaphase rga1∆ and nba1∆ mutant cells contain Rad52 foci, which is significantly increased compared with WT controls (7%; p < 0.0001 by Fisher’s exact test; Figure 6I). These results demonstrate that the deformation of the nucleus as it is pulled into the bud neck contributes to the prevalence of DNA DSBs.
DISCUSSION
The movement of chromosomes is precisely coordinated in mitosis, but how forces outside of the mitotic spindle might impact genome integrity is unclear. Our study demonstrates that forces from astral microtubules are transmitted to the genome, and increasing these forces promotes DNA damage during cell division. We propose that nuclear movement must be coordinated with DNA replication and repair to minimize genome damage.
Our results suggest a model where enhanced nuclear migration increases tension within the spindle and delays homology-directed repair of DSBs. We found the frequency of DNA damage was increased in excessive nuclear migration mutants and they are twice as slow to repair DSBs (Figures 3B and 4D). We predict excessive nuclear migration alters the mobility of the genome, inhibiting DNA repair. Genome mobility is an important part of DNA repair and aids in searching for homology regions (Miné-Hattab and Rothstein, 2012). Cohesion confines the area that pericentric chromatin loops can sample (Stephens et al., 2011), and cells are known to relax cohesion tethers around the centromere through phosphorylation in response to DNA damage, indicating that the relaxation of cohesion around the centromere may be a requirement for Rad51 to search for regions of homology (Wu and Yu, 2012; Strecker et al., 2016). Accordingly, increasing tension around the centromere by excessive pulling forces might inhibit the search for homologous donor strands that are required for homology-directed repair. Our strongest evidence in support of this model is the finding that decreasing pericentric cohesion in mutants with increased nuclear migration suppresses Rad52 foci and foci lifetimes are shorter, indicating these mutants have better repair efficiency than the nuclear migration mutants alone (Figure 5D).
Although our results indicate that forces from astral microtubules can impair the efficiency of homology-directed repair, it is unclear whether these forces also generate DSBs directly. Our analysis of pericentric chromatin in Figure 2 provides evidence that excessive nuclear migration increases tension across sister chromatids, which could potentially cause DSBs. Also, during spindle movement, we see CENIV-GFP foci separate into as many as four CENIV-GFP foci (Figure 2, D, E, and G). Whether these represent DSBs or stretching of pericentric chromatin is not clear. Nevertheless, these results reveal that forces from astral microtubules are increasing tension on regions of DNA close to kinetochore attachment sites. In addition, we found increasing tension within the spindle, through the dam1-765 mutant, was sufficient to increase the frequency of cells containing Rad52 foci, even when astral microtubule forces are normal (Figure 5E). Taken together, these results indicate that increasing microtubule-based forces from inside the spindle or from outside the spindle can increase the prevalence of DNA damage. Whether these forces cause DSBs, delay repair of normally occurring damage, or both will require further study.
How might excessive nuclear migration inhibit timely homology-directed repair? Our results from dividing yeast bear some resemblance to observations of DNA damage in migrating mammalian cells. Inducing mammalian interphase cells to migrate through environments with pore sizes smaller than the diameter of the nucleus (e.g., 3-µm pores and 5-µm nuclei) promotes DNA damage (Denais et al., 2016; Shah et al., 2017). Under these conditions, cells first display nuclear deformities that can lead to rupture of the nuclear envelope due to mechanical forces from actin on the nucleus (Denais et al., 2016; Raab et al., 2016; Thiam et al., 2016). Repeatedly forcing cells through a 3-µm pore results in even greater increased DNA mutation and chromosome copy number changes (Irianto et al., 2017). For comparison, budding yeast nuclei are on average 2 µm wide and move through a bud neck that is 1.1 µm wide, which is an even smaller ratio of nucleus to constriction size than in the mammalian cell experiments. While we do not observe nuclear rupture during mitosis in budding yeast, we do find that widening the bud neck suppresses nuclear deformation and the frequency of DSBs, while decreasing bud neck size increases the frequency of DSBs (Figure 6I). Therefore, our results suggest that the accumulation of DNA damage during nuclear migration in budding yeast may be causally linked to the deformation of the nucleus, rather than nuclear envelope rupture. Recent evidence from human cancer cells demonstrates that high membrane curvature causes the mislocalization of repair factors (Xia et al., 2018). While additional mechanisms could contribute to the increase in DSB frequency, repair factor mislocalization could contribute to the delay in repair we see in nuclear migration mutants. The apparently antagonistic relationship between bud neck size and genome integrity also raises the interesting evolutionary question of why budding yeast build a bud neck that is approximately half the diameter of the G2 nucleus. Perhaps the narrow diameter of the neck could act as a size exclusion filter that normally prevents the movement of a G2 nucleus, with its duplicated genome, into the bud, but permits the movement of a single copy-sized genome into the bud at anaphase.
Several lines of evidence suggest that the activation of dynein-dependent spindle movement may normally be regulated by pathways that monitor the integrity and replication of the genome. During cell division, dynein moves the spindle and nucleus into the bud neck around the time of anaphase onset (Yeh, 1995, 2000). Inducing DNA damage with HO endonuclease or DNA-damaging drugs causes cells to arrest at the G2 to M transition and exhibit low levels of nuclear movement (Dotiwala et al., 2007; Bierle et al., 2015). During this arrest, dynein activity appears to be inhibited by DNA damage checkpoint signaling, because null mutants lacking RAD53 or CHK1 function exhibit increased nuclear migration (Dotiwala et al., 2007). How might DNA damage response pathways inhibit dynein? We know that the timing of dynein activation is regulated through its activating partner dynactin, which is recruited to the plus ends of astral microtubules prior to anaphase onset (Moore et al., 2008; Woodruff et al., 2009). The protein She1 inhibits dynein by preventing dynactin from localizing to the microtubule until G2 phase in the cell cycle and by directly inhibiting dynein motility (Woodruff et al., 2009; Bergman et al., 2012; Markus et al., 2012; Ecklund et al., 2017). In addition, our previous findings and our results here demonstrate that astral microtubule stability also regulates dynein activity, suggesting another possible mechanism for limiting dynein-dependent spindle movement to a narrow window of the cell cycle (Estrem et al., 2017). Further studies will be needed to determine whether signaling pathways responding to DNA damage or replication stress impinge on these mechanisms to regulate dynein activity, and whether these mechanisms are conserved across species.
MATERIALS AND METHODS
Yeast strains and manipulation
Chemicals and reagents were from Fisher Scientific (Pittsburgh, PA) and Sigma-Aldrich (St. Louis, MO), unless stated otherwise. General yeast manipulation, media, and transformation were performed by standard methods (Amberg et al., 2005). Fluorescent tag fusions to Rad52, Mre11, Spc110, Htb2, and Myo1 are integrated at the native loci (Sheff and Thorn, 2004). The mNeonGreen fluor was provided by Allele Biotechnology and Pharmaceuticals (San Diego, CA) (Shaner et al., 2013). GFP-Tub1 and mRuby-Tub1 fusions were integrated and expressed ectopically to the native TUB1 gene (Song and Lee, 2001; Markus et al., 2015). The mutant alleles of TUB2 and NIP100 were generated at the native chromosomal loci (Moore et al., 2008; Aiken et al., 2014). Deletion mutants were generated by conventional PCR-mediated methods (Petracek and Longtine, 2002). Strains and plasmids are listed in Supplemental Tables 1 and 2.
Live-cell imaging
Cells were grown overnight in rich media (2% glucose, 2% peptone, and 1% yeast extract) at 30°C then diluted into synthetic media (2% glucose; CSM from Sunrise Science Products, #1001, San Francisco, CA) and grown to log phase at 30°C. Cells were grown asynchronously to early log phase in nonfluorescent media and adhered to slide chambers coated with concanavalin A (Fees et al., 2017). Slide chambers were sealed with VALAP (Vaseline, lanolin, and paraffin at 1:1:1).
Images were collected on a Nikon Ti-E microscope equipped with a 1.45 NA 100 × CFI Plan Apo objective, piezo electric stage (Physik Instrumente, Auburn, MA), spinning disk confocal scanner unit (CSU10; Yokogawa), 488- and 561-nm lasers (Agilent Technologies, Santa Clara, CA), and an EMCCD camera (iXon Ultra 897; Andor Technology, Belfast, UK) using NIS Elements software (Nikon). During acquisition, the temperature of the stage was 25°C. z-Series consisted of 13 images separated by 450 nm.
Analysis of preanaphase spindle length and sliding
Spindle lengths were measured in cells expressing an integrated fusion of GFP to the N-terminus of α-tubulin (GFP-Tub1) in asynchronous populations. All images were collected under the same microscope setting of 92% 488 laser power and exposure of 50 ms. Thirteen z-slices were taken at 500-nm intervals. Image stacks were collapsed into two-dimensional (2D) image projections based on maximum pixel intensity. Spindle lengths were measured along the long axis of the spindle in large, budded cells before anaphase onset. According to Winey et al. (1995), spindle lengths of 1.5–6 µm are considered anaphase B spindle length in WT cells (Winey and O’Toole, 2001). To analyze changes in preanaphase spindle length, we measured spindle lengths below 3 µm to include mutant spindle lengths. We use additional criteria to exclude anaphase spindles, noting spindle lengths between 1.5 and 3 µm that did not consistently elongate over time. A custom MATLAB program was used to define the spindle using 2D projected images and applying a fluorescence intensity threshold. Then the program measured the length along the long axis of the spindle and tracked the spindle centroid over time. Spindle displacement was calculated from centroids using Pythagorean theorem relative to the first frame. A sliding event was defined as three or more frames of directed movement, meaning spindle movement in the same direction within a 45° angle constraint. Change in spindle length was calculated by the absolute value of the difference between spindle lengths at each time point and the mean spindle length of the entire sliding event or the same duration before the sliding event. SD of spindle length was calculated by the square root of the variance, which is the average of the squared differences from the mean for that time period. WT and ase1∆ spindle lengths did not fit a normal distribution and therefore we used Mann–Whitney test to determine p values. For all livecell imaging experiments, cells were analyzed from at least three separate experiments performed on different days.
Analysis of CENIV foci number
To assess the tension state between sister kinetochores, we imaged the pericentric regions of chromosome IV labeled with a TetO array. Strains containing TetOx448 array integrated 2 kb from centromere on chromosome IV, along with a TetR fusion to GFP, were derived from Brito et al. (2010) (Goshima and Yanagida, 2000; Fees et al., 2016). For single time-point images, we analyzed cells containing two Spc110-tdTomato foci less than 1.8 µm to ensure cells were in preanaphase (Fees et al., 2016). From this preanaphase cell population, we categorized CENIV-GFP foci number into bins of one or two or more CENIV-GFP foci per cell. The number of cells in each category was divided by total cells counted to achieve the percentage of cells containing each number of CENIV foci.
To assess how three and four CENIV-GFP foci arise, we imaged cells over time. Cells containing Spc110-tdTomato and CENIV-GFP were imaged at 10 s intervals for 5 min. We separated time periods of when spindles were moving from time periods while spindles were stationary. Moving spindles were classified as three or more frames of Spc110-tdTomato movement within a 45° angle constraint. The CENIV-GFP foci number was then recorded for each time point. To calculate the percentage of time a cell spent with each CEN foci number, we divided the total time a cell contained one, two, three, or four CENIV-GFP foci by the duration of total spindle movement or the total duration containing a stationary spindle. For example, we collected the total time cells contained two CENIV-GFP foci during spindle migration and divided that duration by the total time of all observed migrating spindles.
Analysis of rad52 and mre11 foci
To assess DSB repair, we imaged living cells expressing either labeled Rad52 or Mre11, along with labeled microtubules to allow for determination of cell cycle state, based on spindle morphology. Two-color images of living cells expressing GFP-Tub1 and Rad52-tdTomato were collected by confocal microscopy, as described above. First, each cell was categorized into three different phases of the cell cycle based on spindle morphology and bud size. The microtubule network and bud size were visualized by GFP–Tub1-labeling microtubules and diffuse fluorescence in the cytoplasm, respectively. Cells without a bud and without a linear, bipolar spindle were characterized as G1. Cells with a bud and a short bipolar spindle (<1.8 µm) were characterized as M cells (combining S phase and G2/M phase). Cells with an elongated bipolar spindle greater than 1.8 µm were characterized as anaphase cells (Winey et al., 1995; Estrem et al., 2017). After cell cycle characterization, each cell was analyzed for the presence of a Rad52 focus. Two-dimensional image projections were run through a custom-built ImageJ macro to identify the presence of Rad52 foci in each cell. Using our standard imaging conditions, the average fluorescence intensity of Rad52-tdTomato foci is 2204 ± 94 a.u. (mean ± 95% CI). Therefore, we designed an ImageJ macro to identify foci above the lower 95% CI of the mean (i.e., 2100 a.u.). We also set the area of a Rad-52 focus must be above the confocal resolution limit of at least 285 nm. Similar settings were defined for Rad52-YFP and Mre11-mNeonGreen.
We measured the duration of Rad52 foci lifetime by time-lapse imaging of living cells expressing GFP-Tub1 and Rad52-tdTomato. The lifetime of a Rad52 focus was calculated by the last frame in which the foci was detected above our fluorescence intensity threshold (2100 a.u.) subtracted by the first frame the focus was detected above the threshold, multiplied by the time interval of 3 min. Owing to the variability of long duration time-lapse imaging, the maximum foci duration was set at 150 min. Therefore, any foci duration over 150 min was changed to 150 min to include long-lasting foci in the data set.
HO endonuclease assay
We induced a DSB by conditionally expressing the HO endonuclease. The HO endonuclease creates DSBs at the MAT locus, which is located on chromosome III, 7 kb from the centromere (Haber, 1992). Cells were transformed with a plasmid in which the HO gene is carried on a single-copy plasmid under control of the GAL10 promoter (Wu and Haber, 1996; Dotiwala et al., 2007). Overexpression was achieved by first growing cells to early log phase in media containing 2% raffinose, then adding galactose to 2% and returning to a 30°C shaking incubator for 2 h. Overexpression was confirmed by the increase in Rad52 foci in WT controls compared with uninduced cells.
Bud neck size analysis
Bud neck diameter and cell size were measured in 2D maximum intensity projections of cells expressing eGFP fused to the type II myosin (Myo1-EGFP), which localizes to the cytokinetic ring (Bezanilla et al., 2000). Cell size was measured from the distal tip of the bud to the distal tip of the mother based on the signal from free GFP-tubulin subunits in the cytoplasm.
Htb2 imaging and analysis
To assess genome shape changes during mitosis, cells expressing differentially labeled histone H2B (Htb2-mNeonGreen) and microtubules (mRuby-Tub1) were imaged at 2-min intervals for 1 h. Eccentricity was calculated by the taking the distance from the centroid to the ellipsoid focus (centroid of the H2B fluorescence distribution) over the distance from that focus to the vertex. The value calculated is between 0 and 1, with closer to 0 representing more circularity and closer to 1 deviating from circularity. To measure the width of the genome in the bud neck during anaphase, we identified anaphase cells based on spindle morphology and measured the width of Htb2-mNeon Green signal along the axis of the bud neck.
Drug treatment
To validate that Rad52-tdTomato and Mre11-mNeonGreen foci represent sites of DSB repair, we induced DNA damage with methyl methanesulfonate (MMS). MMS is a DNA-alkylating agent that causes reactive oxygen species production that leads to multiple mechanisms of DNA damage, including double stranded breaks (Rowe et al., 2008). Cells expressing GFP-Tub1 and Rad52-tdTomato or Mre110-mNeonGreen were grown to early log phase in synthetic media at 30°C and then treated with 0.03% MMS for 2 h before imaging.
Supplementary Material
Acknowledgments
We thank Colby Fees for providing MATLAB code used for data analysis and Steven Markus for sharing yeast strains. This work was supported by National Institutes of Health grant R01 GM112893 to J.K.M. C.E. was supported by the University of Colorado Anschutz Medical Campus (Pre-doctoral Training Program in Molecular Biology grant NIH-T32-GM008730, Butcher Innovation Award provided by Chancellor Elliman, and the Bolie Scholarship award by the Molecular Biology program).
Abbreviations used:
- DSB
double-stranded break
- GFP-Tub1
GFP-labeled microtubules
- MAP
microtubule-associated protein
- MMS
methyl methanesulfonate
- SPBs
pindle pole bodies
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-12-0808) on May 8, 2019.
REFERENCES
- Adames NR, Cooper JA. (2000). Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol , 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiken J, Sept D, Costanzo M, Boone C, Cooper JA, Moore JK. (2014). Genome-wide analysis reveals novel and discrete functions for tubulin carboxy-terminal tails. Curr Biol , 1295–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Burke D, Strathern JN, Burke D, Cold Spring Harbor Laboratory (2005). Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Bergman ZJ, Xia X, Amaro IA, Huffaker TC. (2012). Constitutive dynein activity in she1 mutants reveals differences in microtubule attachment at the yeast spindle pole body. Mol Biol Cell , 2319–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M, Wilson JM, Pollard TD. (2000). Fission yeast myosin-II isoforms assemble into contractile rings at distinct times during mitosis. Curr Biol , 397–400. [DOI] [PubMed] [Google Scholar]
- Bierle LA, Reich KL, Taylor BE, Blatt EB, Middleton SM, Burke SD, Stultz LK, Hanson PK, Partridge JF, Miller ME. (2015). DNA damage response checkpoint activation drives KP1019 dependent pre-anaphase cell cycle delay in S. cerevisiae. PLoS One , 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Murray AW. (2001). The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev , 3118–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck DC, Joglekar AP, Bloom KS. (2008). Design features of a mitotic spindle: balancing tension and compression at a single microtubule kinetochore interface in budding yeast. Annu Rev Genet , 335–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito IL, Yu H-G, Amon A. (2010). Condensins promote coorientation of sister chromatids during meiosis I in budding yeast. Genetics , 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. (1975). Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol , 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. (1997). Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex J Cell Biol , 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón JM, Mukherjee S, Schuster BM, Clarke DJ, Gardner MK. (2014). Pericentromere tension is self-regulated by spindle structure in metaphase. J Cell Biol , 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. (2008). Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol , 33–46. [DOI] [PubMed] [Google Scholar]
- Chung DKC, Chan JNY, Strecker J, Zhang W, Ebrahimi-Ardebili S, Lu T, Abraham KJ, Durocher D, Mekhail K. (2015). Perinuclear tethers license telomeric DSBs for a broad kinesin- and NPC-dependent DNA repair process. Nat Commun , 7742. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. (1996). Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev , 3081–3093. [DOI] [PubMed] [Google Scholar]
- Cole DG, Saxton WM, Sheehan KB, Scholey JM. (1994). A “slow” homotetrameric kinesin-related motor protein purified from Drosophila embryos. J Biol Chem , 22913–22916. [PMC free article] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. (1997). Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J Cell Biol , 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais CM, Gilbert RM, Isermann P, McGregor AL, Te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J. (2016). Nuclear envelope rupture and repair during cancer cell migration. Science , 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM. (2012). Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat Cell Biol , 502–510. [DOI] [PubMed] [Google Scholar]
- Dotiwala F, Haase J, Arbel-Eden A, Bloom K, Haber JE. (2007). The yeast DNA damage checkpoint proteins control a cytoplasmic response to DNA damage Proc Natl Acad Sci USA , 11358–11363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S, Mitchison TJ. (2009). Force and length in the mitotic spindle. Curr Biol , R749–R761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecklund KH, Morisaki T, Lammers LG, Marzo MG, Stasevich TJ, Markus SM. (2017). She1 affects dynein through direct interactions with the microtubule and the dynein microtubule-binding domain. Nat Commun , 2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrem C, Fees CP, Moore JK. (2017). Dynein is regulated by the stability of its microtubule track. J Cell Biol , 2047–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fees CP, Aiken J, O’Toole ET, Giddings TH, Moore JK. (2016). The negatively charged carboxy-terminal tail of β-tubulin promotes proper chromosome segregation. Mol Biol Cell , 1786–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fees CP, Estrem C, Moore JK. (2017). High-resolution imaging and analysis of individual astral microtubule dynamics in budding yeast. J Vis Exp , 55610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W. (2017). Mec1/ATR, the program manager of Nucleic Acids Inc. Genes (Basel) , 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game JC, Mortimer RK. (1974). A genetic study of X-ray sensitive mutants in yeast. Mutat Res , 281–292. [DOI] [PubMed] [Google Scholar]
- Goshima G, Yanagida M. (2000). Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell , 619–633. [DOI] [PubMed] [Google Scholar]
- Gupta ML, Carvalho P, Roof DM, Pellman D. (2006). Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol , 913–923. [DOI] [PubMed] [Google Scholar]
- Haber JE. (1992). Mating-type gene switching in Saccharomyces cerevisiae Trends Genet , 446–452. [DOI] [PubMed] [Google Scholar]
- Hofmann C, Cheeseman IM, Goode BL, Mcdonald KL, Barnes G, Drubin DG. (1998). Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function J Cell Biol , 1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker TC, Thomas JH, Botstein D. (1988). Diverse effects of β-tubulin mutations on microtubule formation and function. J Cell Biol , 1997–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E, Kusch J, Barral Y, Huffaker TC. (2003). Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J Cell Biol , 483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Xia Y, Pfeifer CR, Athirasala A, Ji J, Alvey C, Tewari M, Bennett RR, Harding SM, Liu AJ, et al. (2017). DNA damage follows repair factor depletion and portends genome variation in cancer cells after pore migration. Curr Biol , 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE. (1994). Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol , 3414–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, Berg P. (1988). Homologous integration in mammalian cells without target gene selection. Genes Dev. , 1353–1363. [DOI] [PubMed] [Google Scholar]
- Juang YL, Huang J, Peters JM, McLaughlin ME, Tai CY, Pellman D. (1997). APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science , 1311–1314. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJG, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. (2005). The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature , 114–118. [DOI] [PubMed] [Google Scholar]
- Kardon JR, Reck-Peterson SL, Vale RD. (2009). Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc Natl Acad Sci USA , 5669–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostriken R, Strathern JN, Klar AJS, Hicks JB, Heffron F. (1983). A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell , 167–174. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Tsouladze A, Koryabin M, Hieter P, Spencer F, Larionov V. (1993). Identification and genetic mapping ofCHL genes controlling mitotic chromosome transmission in yeast. Yeast , 11–19. [DOI] [PubMed] [Google Scholar]
- Lawrimore J, Barry TM, Barry RM, York AC, Friedman B, Cook DM, Akialis K, Tyler J, Vasquez P, Yeh E, et al. (2017). Microtubule dynamics drive enhanced chromatin motion and mobilize telomeres in response to DNA damage. Mol Biol Cell , 1701–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L, Klee SK, Evangelista M, Boone C, Pellman D. (1999). Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J Cell Biol , 947–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WL, Kaiser MA, Cooper JA. (2005). The offloading model for dynein function: differential function of motor subunits. J Cell Biol , 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-S, Wang RW, Chang H-H, Capurso D, Segal MR, Haber JE. (2016). Chromosome position determines the success of double-strand break repair. Proc Natl Acad Sci USA , E146–E154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Yeh E, Hays T, Bloom K. (1993). Disruption of mitotic spindle orientation in a yeast dynein mutant Proc Natl Acad Sci USA , 10096–10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH. (2001). Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA , 8276–8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R. (2003). Colocalization of multiple DNA double-strand breaks at a single Rad52 repair center. Nat Cell Biol , 572–577. [DOI] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. (2004). Choreography of the DNA damage response. Cell , 699–713. [DOI] [PubMed] [Google Scholar]
- Lottersberger F, Karssemeijer RA, Dimitrova N, de Lange T. (2015). 53BP1 and the LINC complex promote microtubule-dependent DSB mobility and DNA repair. Cell , 880–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus SM, Kalutkiewicz KA, Lee WL. (2012). Astral microtubule asymmetry provides directional cues for spindle positioning in budding yeast. Exp Cell Res , 1400–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus SM, Omer S, Baranowski K, Lee WL. (2015). Improved plasmids for fluorescent protein tagging of microtubules in Saccharomyces cerevisiae. Traffic , 773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Helper PK, van Wie DG. (1969). Model for mitosis. Nature , 659–663. [Google Scholar]
- Meitinger F, Khmelinskii A, Morlot S, Kurtulmus B, Palani S, Andres-Pons A, Hub B, Knop M, Charvin G, Pereira G. (2014). A memory system of negative polarity cues prevents replicative aging. Cell , 1056–1069. [DOI] [PubMed] [Google Scholar]
- Miller RK, Rose MD. (1998). Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol , 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. (2010). Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. , 3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miné-Hattab J, Rothstein R. (2012). Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol , 510–517. [DOI] [PubMed] [Google Scholar]
- Moore JK, Li J, Cooper JA. (2008). Dynactin function in mitotic spindle positioning. Traffic , 510–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Sept D, Cooper JA. (2009). Neurodegeneration mutations in dynactin impair dynein-dependent nuclear migration Proc Natl Acad Sci USA , 5147–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. (1996). DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci USA , 10729–10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. (1998). Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature , 407–410. [DOI] [PubMed] [Google Scholar]
- Ng TM, Waples WG, Lavoie BD, Biggins S. (2009). Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol Biol Cell , 3818–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirschl JJ, Magiera MM, Lazarus JE, Janke C, Holzbaur ELF. (2016). α-Tubulin tyrosination and CLIP-170 phosphorylation regulate the initiation of dynein-driven transport in neurons. Cell Rep , 2637–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW, Rothstein RJ. (1981). Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci USA , 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshidari R, Strecker J, Chung DKC, Abraham KJ, Chan JNY, Damaren CJ, Mekhail K. (2018). Nuclear microtubule filaments mediate non-linear directional motion of chromatin and promote DNA repair. Nat Commun , 2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Salmon ED, Bloom K. (2001). Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol , 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellman D, Bagget M, Tu H, Fink GR. (1995). Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. J Cell Biol , 1373–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Höfken T, Grindlay J, Manson C, Schiebel E. (2000). The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell , 1–10. [PubMed] [Google Scholar]
- Petracek ME, Longtine MS. (2002). PCR-based engineering of yeast genome. Methods Enzymol , 445–469. [DOI] [PubMed] [Google Scholar]
- Prakash L, Prakash S. (1977). Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics , 33–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M, Gentili M, de Belly H, Thiam H-R, Vargas P, Jimenez AJ, Lautenschlaeger F, Voituriez R, Lennon-Dumenil A-M, Manel N, et al. (2016). ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science , 359–362. [DOI] [PubMed] [Google Scholar]
- Rankin KE, Wordeman L. (2010). Long astral microtubules uncouple mitotic spindles from the cytokinetic furrow. J Cell Biol , 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe LA, Degtyareva N, Doetsch PW. (2008). DNA damage-induced reactive oxygen species (ROS) stress response in Saccharomyces cerevisiae. Natl Inst Heal , 1167–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott EJ, Hoyt MA. (1998). Dominant alleles of Saccharomyces cerevisiae CDC20 reveal its role in promoting anaphase. Genetics , 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Wolf K, Lammerding J. (2017). Bursting the bubble—nuclear envelope rupture as a path to genomic instability? Trends Cell Biol , 546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, et al. (2013). A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods , 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff MA, Thorn KS. (2004). Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast , 661–670. [DOI] [PubMed] [Google Scholar]
- Shimogawa MM, Graczyk B, Gardner MK, Francis SE, White EA, Ess M, Molk JN, Ruse C, Niessen S, Yates JR, et al. (2006). Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr Biol , 1489–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. (1998). Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature , 404–407. [DOI] [PubMed] [Google Scholar]
- Song S, Lee KS. (2001). A novel function of Saccharomyces cerevisiae CDC5 in cytokinesis. J Cell Biol , 451–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Haase J, Vicci L, Taylor RM, Bloom K. (2011). Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J Cell Biol , 1167–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker J, Gupta GD, Zhang W, Bashkurov M, Landry MC, Pelletier L, Durocher D. (2016). DNA damage signalling targets the kinetochore to promote chromatin mobility. Nat Cell Biol , 281–290. [DOI] [PubMed] [Google Scholar]
- Sung P. (1997). Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem , 28194–28197. [DOI] [PubMed] [Google Scholar]
- Symington LS, Rothstein R, Lisby M. (2014). Mechanisms and regulation of mitotic recombination in saccharomyces cerevisiae. Genetics , 795–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. (1983). The double-strand-break repair model for recombination. Cell , 25–35. [DOI] [PubMed] [Google Scholar]
- Tame M, Raaijmakers J, Van Den Broek B, Lindqvist A, Jalink K, Medema RH. (2014). Cell cycle astral microtubules control redistribution of dynein at the cell cortex to facilitate spindle positioning. Cell Cycle , 1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Fuchs J, Loidl J, Nasmyth K. (2000). Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat Cell Biol , 492–499. [DOI] [PubMed] [Google Scholar]
- Thiam HR, Vargas P, Carpi N, Crespo CL, Raab M, Terriac E, King MC, Jacobelli J, Alberts AS, Stradal T, et al. (2016). Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat Commun , 10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. (1998). DNA damage and checkpoint pathways: molecular anatomy and interactions with repair. Cell , 555–558. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. (1988). The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science , 317–22. [DOI] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, McDonald KL, McIntosh JR. (1995). Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol , 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, O’Toole ET. (2001). The spindle cycle in budding yeast. Nat Cell Biol , E23–E27. [DOI] [PubMed] [Google Scholar]
- Woodruff JB, Drubin DG, Barnes G. (2009). Dynein-driven mitotic spindle positioning restricted to anaphase by She1p inhibition of dynactin recruitment dynein is a minus-end-directed microtubule motor important for mitotic spindle positioning. Mol Biol Cell , 3003–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Haber JE. (1996). A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell , 277–285. [DOI] [PubMed] [Google Scholar]
- Wu N, Yu H. (2012). The Smc complexes in DNA damage response. Cell Biosci , 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Ivanovska IL, Zhu K, Smith L, Irianto J, Pfeifer CR, Alvey CM, Ji J, Liu D, Cho S, et al. (2018). Nuclear rupture at sites of high curvature compromises retention of DNA repair factors. J Cell Biol , 3796–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E. (1995). Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol , 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Yang C, Chin E, Maddox P, Salmon ED, Lew DJ, Bloom K. (2000). Dynamic positioning of mitotic spindles in yeast: Role of microtubule motors and cortical determinants. Mol Biol Cell , 3949–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Pruyne D, Huffaker TC, Bretscher A. (2000). Myosin V orientates the mitotic spindle in yeast. Nature , 1013–1015. [DOI] [PubMed] [Google Scholar]
- Yong-Gonzales V, Hang LE, Castellucci F, Branzei D, Zhao X. (2012). The Smc5-Smc6 complex regulates recombination at centromeric regions and affects kinetochore protein sumoylation during normal growth. PLoS One , e51540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.