SUMMARY
The ability of neurons to identify correct synaptic partners is fundamental to the proper assembly and function of neural circuits. Relative to other steps in circuit formation such as axon guidance, our knowledge of how synaptic partner selection is regulated is severely limited. Drosophila Dpr and DIP IgSF cell-surface proteins bind heterophilically and are expressed in a complementary manner between synaptic partners in the visual system. Here, we show that in the lamina, DIP mis-expression is sufficient to promote synapse formation with Dpr-expressing neurons and that disrupting DIP function results in ectopic synapse formation. These findings indicate that DIP proteins promote synapses to form between specific cell types and that in their absence neurons synapse with alternative partners. We propose that neurons have the capacity to synapse with a broad range of cell types and that synaptic specificity is achieved by establishing a preference for specific partners.
eTOC blurb
Xu et al., show that, in the Drosophila visual system, DIP IgSF proteins are not necessary for synaptogenesis but regulate synaptic specificity by promoting synapses to form between specific cell types
INTRODUCTION
The formation of precise connections between neurons underlies the structural organization and function of the nervous system. In general, precise neural connectivity is established in a stepwise manner that serves to reduce the molecular complexity necessary for specifying neural connections. For example, events such as axon guidance (Huber et al., 2003; Kolodkin and Tessier-Lavigne, 2011; Tessier-Lavigne and Goodman, 1996), topographic positioning (Cang and Feldheim, 2013; Feldheim and O’Leary, 2010; Flanagan, 2006), and laminar innervation (Baier, 2013; Huberman et al., 2010; Sanes and Yamagata, 1999) target neural processes to specific locations thereby restricting the partners available for synapse formation. Over the past several decades progress has been made in identifying molecules that regulate these processes. However, within their local environment neurons still face the challenge of identifying correct synaptic partners amidst many alternatives (referred to here as synaptic specificity), and how this is achieved remains poorly understood. Based on landmark studies showing that regenerating neurons have the capacity to distinguish appropriate from inappropriate synaptic targets (Langley, 1895) [reviewed in (Sperry, 1963)], it was proposed that synaptic specificity is regulated by molecular determinants that mediate recognition between synaptic partners. A common interpretation of this idea is that recognition of the correct partners is necessary for synaptogenesis. However, few molecules have been shown to directly mediate selective interactions between synaptic partners [but see (Ashley et al., 2019; Duan et al., 2014; Hong et al., 2012; Krishnaswamy et al., 2015; Mosca et al., 2012; Venkatasubramanian et al., 2019; Xu et al., 2018)].
Recent biochemical, gene expression and protein expression studies have demonstrated that the members of two subfamilies of the immunoglobulin superfamily (IgSF), the Dpr (defective proboscis retraction) family (21 members) (Carrillo et al., 2015; Nakamura et al., 2002) and the dpr-interacting proteins (DIPs) (11 members) (Cosmanescu et al., 2018; Ozkan et al., 2013), form a complex protein interaction network (Carrillo et al., 2015; Cheng et al., 2019; Cosmanescu et al., 2018; Ozkan et al., 2013; Zinn and Ozkan, 2017) and are expressed in a complementary manner between synaptically coupled cell types during development in the Drosophila visual system (Carrillo et al., 2015; Tan et al., 2015). Based on these findings, Dpr-DIP interactions are proposed to play an instructive role in regulating synaptic specificity (Carrillo et al., 2015; Tan et al., 2015). Dprs and DIPs have 2 or 3 Ig domains in their extracellular regions, respectively, and they predominantly bind heterophilically with few exhibiting homophilic binding. Dpr-DIP complexes bear a striking resemblance to the complexes of mammalian IgSF proteins and Dpr/DIP proteins are homologous to the IgLON protein family in vertebrates (Cheng et al., 2018; Zinn and Ozkan, 2017). Dpr-DIP interactions play diverse roles in regulating the assembly of neural circuits in different regions of the Drosophila nervous system. DIP-γ and Dpr11 regulate the morphogenesis of synaptic terminals at the neuromuscular junction and regulate cell survival in the visual system (Carrillo et al., 2015; Xu et al., 2018); interactions between synaptic partners mediated by DIP-α and Dprs 6 and 10 have also been shown to regulate cell survival, control layer innervation, and synapse number and distribution in the visual system (Xu et al., 2018); binding between DIP-α and Dpr10 has further been shown to regulate terminal branching of motor neuron axons onto specific body wall and leg muscles, a process proposed to mediate synaptic specificity between motor axons and target muscles (Ashley et al., 2018; Venkatasubramanian et al., 2018); and multiple Dpr-DIP interactions are thought to mediate axon-axon fasciculation in the olfactory system (Barish et al., 2018). However, whether Dpr and DIP proteins act instructively to regulate synaptic specificity remains unclear.
To test the latter possibility, we have focused on the lamina of the Drosophila optic lobe (Fig. 1A–C) which comprises a highly stereotyped cellular and synaptic architecture that has been extensively characterized in electron microscopy (EM) studies (Meinertzhagen and O’Neil, 1991; Rivera-Alba et al., 2011). Within the lamina, the synaptic terminals of photoreceptors R1–R6 (R cells) and the neurites of lamina neurons L1–L5 (L cells) organize into cylindrical modules called cartridges (Fig. 1A and B). Each cartridge receives input from R cells that detect light from the same point in visual space (Braitenburg, 1967; Kirschfeld, 1967), neighboring cartridges processing information from neighboring points in visual space so as to establish a retinotopic map in the lamina. The core of each cartridge primarily comprises the main axons of L1 and L2, and their dendrites sandwiched between a ring of six R-cell axon terminals (Fig. 1B). By contrast, the main neurites of L3–L5 are located around the cartridge circumference, although L3 sends dendrites into the cartridge core. R cells repeatedly synapse en passant onto L1–L3 dendrites throughout each cartridge, but L1–L3 neither synapse reciprocally onto R cells nor synapse with each other (Fig. 1C). In the proximal lamina, near the base of each cartridge, L4 extends dendrites into the cores of both its own cartridge (Fig. 1A) and those of two neighbors and forms reciprocal connections with L2 (Fig. 1C). All L cells send axons into the underlying medulla neuropil where they synapse onto specific target cells.
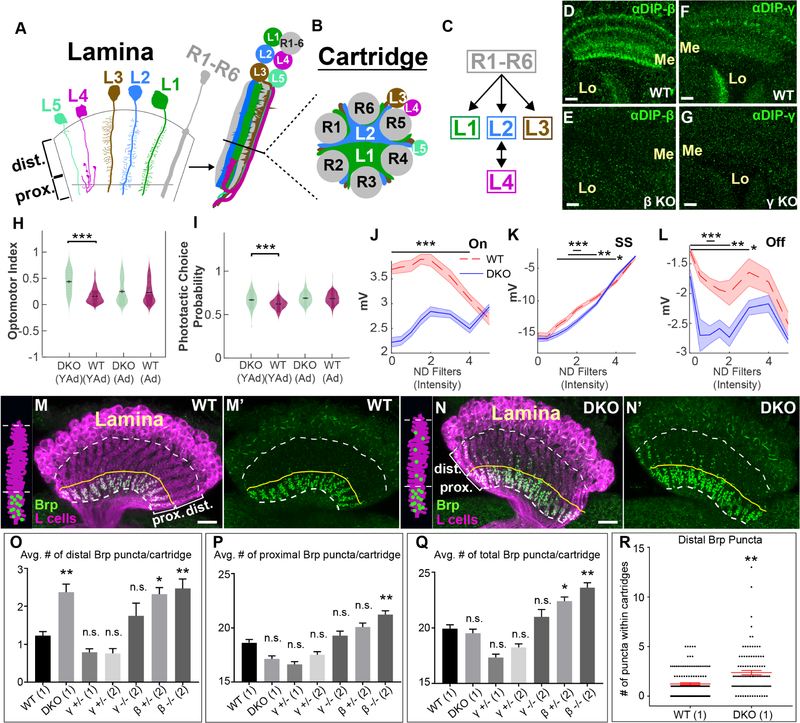
Figure 1. DIP proteins are required for visual function and proper synaptic connectivity.
(See also Figure S1)(A-C) Cellular and synaptic organization of the lamina.
(D-G) Confocal images showing DIP-β (D and E) or DIP-γ (F and G) immunolabeling (green) in the medulla (Me) and lobula (Lo) neuropils at 40–48 hours after puparium formation (h APF). Scale bar = 10μm.
(D and F) In wild type flies DIPs-β and γ are expressed in the medulla and lobula. n=6 brains and n=7 brains respectively.
(E and G) The expression of DIPs-β and γ in the medulla and lobula is severely reduced in flies homozygous for DIP-β (DIP-β1−95) (n=6 brains) or γ (DIP-γ1−67) null mutations (n=5 brains), respectively.
(H) Young adult (YAd; 1–2 day old) DKO flies (n=205) show enhanced tracking of a rotating optomotor stimuli compared to control flies (n=218). Adult flies (Ad; 13–15 days) showed no difference between DKO (n=163) and CTL (n=233) flies.
(I) Young adult (YAd; 1–2 day old) DKO flies (n=105) show an enhanced preference towards the lit arm of a phototactic choice y-maze compared to control flies (n=198). Adult flies (Ad; 13–15 days) showed no difference between DKO (n=168) and CTL (n=232) flies.
(J-L) ERG responses of 1–2 day old DKO (n=9) and control (n=7) flies by intensity for the On transient
(J), steady state response (K) and Off transient (L) components of the ERG in response to a 0.6 second flash of light (See figure S1F). Bars indicate statistical significance at the indicated intensities. Plots in J-L indicate mean of all measurements, shaded areas indicate the SEM).
(M-N’) Confocal images (longitudinal plane of the lamina cartridges) showing the distribution of Brp (green-smFPV5) expressed in L cells (magenta-LexAop-myr-tdTOM) in the laminas of wild type or DKO flies. Scale bar = 10μm. White dotted lines indicate the lamina neuropil. The yellow lines show the boundary between the distal and proximal lamina. We observed batch differences in the amount of Brp background signal in lamina neuron cell bodies (N, N’, O, O’).(M and M’) In wild type flies (n= 9 brains), Brp is restricted to the proximal lamina where L2 and L4 neurons are known to form synapses. (N and N’) In DKO flies (n= 7 brains), Brp is still localized to the proximal lamina, but ectopic Brp puncta are present in the distal lamina.
(O-Q) The average number of Brp puncta in the distal (O) or proximal (P) halves of lamina cartridges from different genotypes are shown. (Q) Indicates the average total number of Brp puncta within lamina cartridges. Results are from two different experiments (1/2). Statistical significance was established with respect to wild type flies (see methods section for a detailed description of statistical analyses). Data are represented as a mean +/− SEM.
(R) Shows the number of distal Brp puncta within individual cartridges scored in wild type and DKO flies. Each star indicates a cartridge. +/− SEM is shown in red. (Statistical significance- *<.05, **<.005, ***<.0005)
Previous studies have characterized the mechanisms underlying targeting and positioning of neural processes to and within cartridges, respectively. Interactions between R cell axons (Clandinin and Zipursky, 2000; Langen et al., 2015), mediated by the cell surface molecules N-Cadherin (CadN) (Lee et al., 2001; Schwabe et al., 2013) and Flamingo (Chen and Clandinin, 2008; Lee et al., 2003; Schwabe et al., 2013), and CadN-dependent interactions between R cell axons and L cells (Prakash et al., 2005) target the axons of R cells that “view” the same point in visual space to the same cartridge. The receptor tyrosine phosphatases Lar (Clandinin et al., 2001) and PTP69D (Newsome et al., 2000) and the scaffold protein Liprin-α (Choe et al., 2006) are also required for R cell axon target specificity. Within cartridges, differential adhesion mediated by CadN optimally positions R cell and L cell neurites for synapse formation, with those cells forming the most connections (L1 and L2) occupying the cartridge core and cells forming fewer connections restricted to the periphery (Schwabe et al., 2014). In CadN mutant flies there was a drastic reduction of R cell synapses (Schwabe et al., 2014), indicating that neurite positioning within cartridges or CadN play a crucial role in synaptogenesis or maintenance of synapses. Despite these advances, the mechanisms that control synaptic specificity within cartridges remain unknown.
Using loss- and gain-of-function genetic approaches we have exploited the cell-type specificity of synapse formation within lamina cartridges to ask whether DIP proteins are necessary and sufficient for synaptic specificity. Our findings demonstrate that DIP proteins are necessary for proper visual function and support a role for DIP-β in promoting synapses to form between L4 and L2 neurons. When DIP-β function is disrupted L4 neurons form ectopic synapses. This suggests that L4 neurons have the capacity to synapse with multiple cell types but a preference for L2 neurons is established by DIP-β most likely through interactions with Dpr proteins. Our findings argue against the idea that specific interactions between correct synaptic partners are necessary for synapse formation and support a model whereby instead, such interactions establish a relative preference for synapses to form between specific cell types.
RESULTS
DIP proteins are necessary for proper synaptic connectivity and visual function
In the lamina, L4 and L2 neurons selectively form reciprocal connections in the proximal lamina (Meinertzhagen and O’Neil, 1991; Rivera-Alba et al., 2011). L2 abuts L1 extensively throughout the cartridge (Fig. 1B) yet is only pre-synaptic in the proximal region, where it synapses primarily onto L4, while L4 dendrites extend into the proximal cartridge core (Fig. 1A) and encounter both L1 and L2 neurites, yet primarily synapse onto L2. A previous study demonstrated that during synaptogenesis L4 dendritic branches strongly express the IgSF protein Kirre, and that disrupting kirre reduces the number of L4–L2 synapses (Luthy et al., 2014). Thus, Kirre is required for synapse formation or maintenance in this context. However, the mechanisms underlying the selectivity of synapse formation between L4 and L2 neurons remain unknown. Previously, through use of RNA-seq and GAL4 reporters both L4 and L2 were found to express a single DIP during pupal development, L4 expressing DIP-β and L2 expressing DIP-γ (Tan et al., 2015). DIP-β is known to bind to 7 different Dpr proteins in vitro (Carrillo et al., 2015; Cosmanescu et al., 2018; Ozkan et al., 2013), 6 of which have been shown to be expressed in L2 neurons (Tan et al., 2015). DIP-γ is known to bind 4 Dprs in vitro (Ozkan et al., 2013). While L4 was not found to express any of these Dprs at 40 hours after puparium formation (h APF) (Tan et al., 2015), we reasoned that L4 may express one or more of these during actual synapse formation, which occurs later in development (see below). Thus, we hypothesized that DIP-β-Dpr interactions, DIP-γ-Dpr interactions, or both promote selective synapse formation between L4 and L2 neurons. As DIPs-β and γ bind to many Dprs, to test this hypothesis we concentrated our efforts on addressing the functions of DIPs-β and γ.
Using the CRISPR/Cas9 system we generated early stop mutations near the translational start sites of DIPs-β and γ (see Methods section). DIP-β and γ immunolabeling were as a result eliminated in the optic lobes of flies homozygous for these mutations (Fig. 1D–G), demonstrating their effectiveness in disrupting DIP function. To determine if DIPs-β and γ are important for circuit formation in the visual system, we assessed whether disrupting these genes in combination (i.e. double KO [DKO]) caused deficits in visual function using behavioral and physiological assays (Werkhoven et al., 2019). We found that young adult (YAd) 1–2 day-old DKO flies were more responsive to apparent motion cues compared with control flies in the optomotor assay (Fig. 1H), and showed a stronger photopositive bias than control flies in the phototaxis assay (Fig. 1I). Interestingly, these effects were transient because adult (Ad) 13–15 day-old DKO and control flies performed similarly in both assays (Fig. 1H and I). In the phototaxis assay we also observed differences in average speed between adult DKO and control flies and in the number of trials triggered by both young adult and adult DKO and control flies, likely due to differences in activity (Fig. S1B and C). Our data indicate that DIP-β, γ or both are required for visually guided behavior. ERG recordings revealed significant differences between DKO and control flies for both On- and Off-transient responses, which are thought to correspond to the activities of L1 and L2 neurons (Coombe, 1986), and for the sustained component (SS) corresponding to the photoreceptor response (Heisenberg, 1971) (Fig. 1J–L, S1F). These differences varied as a function of light intensity and were most pronounced at intermediate light intensities (Fig. 1J–L). The phototactic bias was similarly largest at intermediate light intensities (Fig. S1D and E), suggesting that abnormal phototaxis in DKO flies might be caused by altered neural activity in the visual system. Taken together, these findings demonstrate that DIP-β, γ or both are necessary for proper visual function, motivating further investigation of their potential role in regulating connectivity between L4 and L2 neurons.
To test whether DIP proteins are necessary for synaptic specificity, we used synaptic tagging with recombination (STaR) (Chen et al., 2014; Peng et al., 2018) to label L4–L2 synapses selectively. Using STaR the active zone protein Bruchpilot (Brp) (Wagh et al., 2006) can be tagged in a cell type-specific manner depending on the expression of Flp recombinase (Golic and Lindquist, 1989), while being expressed from its native promoter within a bacterial artificial chromosome (BAC). Moreover, the cells that express tagged-Brp can also be made to express a fluorescent reporter through the LexA/LexAop system (Lai and Lee, 2006), providing a context in which to assess Brp localization. It has been shown that Brp puncta number correlates well with synapse number determined by EM (Chen et al., 2014). Since L4 and L2 are the only L cells that are pre-synaptic in the lamina and because they predominantly synapse with each other (Meinertzhagen and O’Neil, 1991; Rivera-Alba et al., 2011), selectively expressing Flp in L cells allows selective visualization of L4–L2 synapses in the proximal lamina (Fig. 1M and M’, S1A). In the absence of DIP function, we expected to observe a reduction in the number of Brp puncta in the proximal lamina, which would indicate a loss of L4–L2 synapses. However, in flies doubly mutant for DIPs-β and γ (DKO flies) the number of Brp puncta in the proximal lamina was qualitatively similar to that of wild type flies (Fig. 1N and N’). Interestingly, we observed abnormally large numbers of Brp puncta in the distal lamina of DKO flies compared with wild type flies, indicating that in the absence of DIP function additional synapses form at ectopic locations.
To quantify this phenotype, we imaged along the long axis of lamina cartridges using confocal microscopy and took Z-stacks of the laminas of wild type and DKO flies. Using a customized machine learning algorithm (see Methods section) we segmented individual cartridges and counted the number of Brp puncta within distal and proximal halves. As the proximal lamina in wild type flies, defined by the location of L4 dendrites and L4–L2 synapses, spans less than half of the lamina neuropil, the number of distal lamina synapses counted with this method may represent an underestimate. We found that the average number of Brp puncta in the distal halves of cartridges was significantly higher in DKO flies compared with wild type flies (Fig. 1O, R). A statistically significant difference in the number of Brp puncta in the proximal halves of cartridges from DKO flies relative to wild type flies was not observed (Fig. 1Q), but many synapses form in this region compared to the distal lamina. Additionally, no difference in the total number of Brp puncta within cartridges from DKO versus wild type flies was detected. Thus, in DKO flies L cell synapses still form in the lamina but some are abnormally distributed within the distal lamina. Together, these data indicate that DIP proteins are necessary for proper synaptic connectivity but are not required for synapse formation.
L4 neurons form ectopic synapses and have altered dendrite morphology in the absence of DIP-β function
To assess which L cell subtypes contribute ectopic Brp puncta in the distal lamina in the absence of DIP function, we used STaR to selectively label Brp in each L cell independently in wild type, control (β+/−; γ+/−) or DKO flies using cell type-specific GAL4 drivers (Tuthill et al., 2013) to control expression of FLP recombinase. These experiments revealed that L4 neurons (Fig. 2A, B, C) have an increased number of Brp puncta in the distal laminas of DKO flies compared with wild type flies (Fig. 2D). We did not observe differences in the number of Brp puncta in the proximal lamina or in the total number of puncta per cartridge (Fig. 2E and F). The increase in distal Brp puncta in L4 neurons is less than that observed when visualizing Brp in all L cells (compare Figs. 1O and 2D), indicating that other L cells may contribute to the phenotype. However, in the distal halves of the laminas of DKO flies increased numbers of Brp puncta were not detected in L1–L3, or L5 neurons (Fig. S2A–K) compared with control or wild type flies. As the GAL4 drivers for L1–L3 and L5 neurons turn on in the adult stage, it remains possible that limited expression of tagged-Brp in these neurons was insufficient to label all their pre-synaptic sites. By contrast, in experiments where Brp was labeled in L4 or all L cells tagged-Brp is activated in early pupal development or from the time the L cells are born, respectively.
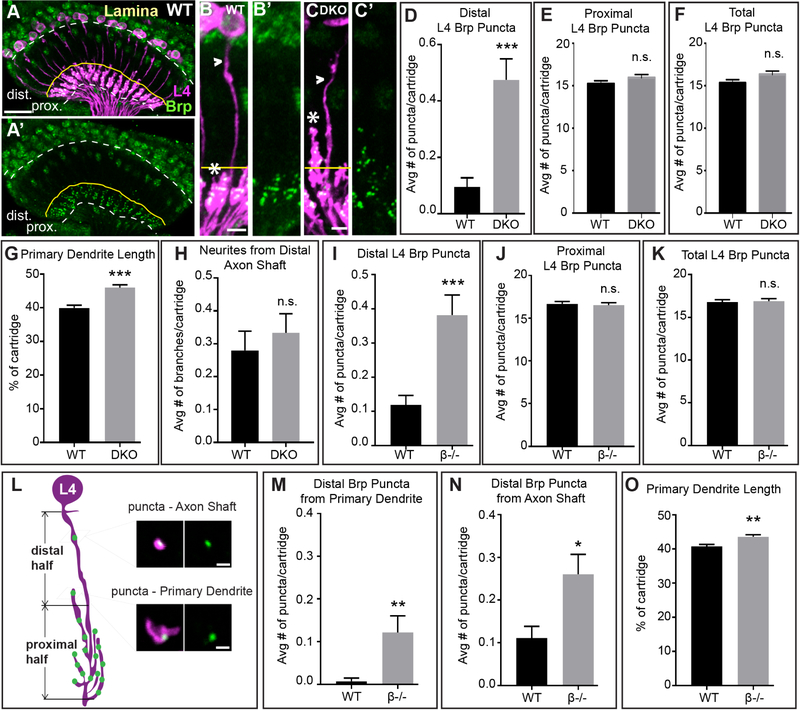
Figure 2. L4 neurons form ectopic synapses and have altered dendrite morphology in the absence of DIP-β function.
(see also Figure S2)
(A and A’) Confocal images (longitudinal plane of the lamina cartridges) showing the distribution of Brp (green-smGFPV5) expressed in L4 cells (magenta-LexAop-myr-tdTOM, 31C06AD (II), 34G07DBD (III) L4 split-GAL4 + UAS-FLP) in the lamina of wild type flies. Brp is restricted to the proximal lamina where L4 neurons are known to form synapses (n= 7 brains). White dotted lines indicate the lamina neuropil. The yellow lines show the boundary between the distal and proximal lamina. Scale bar = 10μm.
(B-C’) Confocal images of L4 neurons expressing Brp-smGFPV5 (STaR) in wild type (B and B’) and DKO flies (C and C’). In DKO flies primary dendrites extend into the distal lamina. Arrowheads indicate L4 axons and asterisks indicate primary dendrites. The yellow line approximates the boundary between the proximal and distal lamina. Scale bar = 5μm.
(D-F) The average number of Brp puncta in L4 neurons present within the distal (D) or proximal (E) halves of lamina cartridges, and the average total number of puncta within cartridges (F) in wild type or DKO flies. (distal: wild type n = 105 cartridges; 7 brains, DKO n = 120 cartridges; 8 brains; proximal and total: wild type n = 75, 5 brains, DKO n = 75 cartridges, 5 brains). Data are represented as a mean +/−SEM.
(G) L4 primary dendrite length calculated as percentage of total cartridge length in wild type (n=105 cartridges) and DKO (n = 120 cartridges) flies.
(H) Average number of neurites from L4 axons in the distal half of the lamina in wild type (n=75 cartridges) and DKO (n = 75 cartridges) flies. Data are represented as a mean +/− SEM.
(I-K) The average number of Brp puncta in L4 neurons present within the distal (I) or proximal (J) halves of lamina cartridges, and the average total number of puncta within cartridges (K) in wild type or in DIP-β KO flies. (wild type n = 135 cartridges; 9 brains, DIP-β KO n = 165 cartridges; 11 brains). Data are represented as a mean +/− SEM.
(L) Illustration of L4 morphology and presynaptic sites (Brp puncta) in the lamina of a DIP-β KO fly. Primary dendrites are mostly located in the proximal half of the lamina, with some extending into the distal half. Brp puncta are observed in the distal half from two sources – the axon shaft and primary dendrites. Scale bar = 1μm.
(M and N) The average number of distal Brp puncta from L4 primary dendrites (M) or L4 axon shafts
(N) in wild type (n=135 cartridges) and DIP-β KO (n=165 cartridges) flies.
(O) L4 primary dendrite length calculated as percentage of total cartridge length in wild type (n=135 cartridges) and DIP-β KO (n = 165 cartridges) flies.(Statistical significance- *<.05, **<.005, ***<.0005)
In DKO flies, L4 axons were morphologically indistinguishable from L4 axons in wild type flies (Fig. 2B and C, H). However, the primary dendrites of L4 neurons in DKO flies extended more distally within the cartridge core than did primary dendrites from wild type flies (Fig. 2B and C, G), even though the general spacing of primary L4 dendrites appeared normal (Fig. S2L and M).
To determine whether the synaptic and dendritic phenotypes in DKO flies resulted from disrupting DIP-β, γ, or both, we analyzed single knockout flies. We found that disrupting one or both copies of DIP-β caused an increase in the number of Brp puncta in L cells in the distal lamina similar to DKO flies (Fig. 1O), and an increase in the total number of Brp puncta per cartridge (Fig. 1Q). Disrupting one copy of DIP-β considerably reduced DIP-β immunolabeling in the optic lobe (Fig. S1G and H). When both copies of DIP-β were disrupted we also observed a significant increase in the number of Brp puncta L cells form in the proximal lamina (Fig. 1P). Cell type-specific STaR experiments revealed that, as in DKO flies, in DIP-β KO flies L4 neurons have increased numbers of Brp puncta in the distal lamina compared with wild type flies (Fig. 2I). In addition, primary dendrites in DIP-β KO flies extended further distally than in wild type flies (Fig. 2O) similar to what we observed in DKO flies (Fig. 2G). In DIP-β KO flies, the majority of distal Brp puncta were present on L4 axon shafts with only a subset formed on primary dendrites (Fig. 2L–N). Thus, ectopic synapse formation in the distal lamina under these conditions does not correlate with altered dendritic morphology. KO of DIP-γ did not result in a statistically significant difference in distal or proximal Brp puncta relative to wild type flies (Fig. 1O and P). Collectively, these findings indicate that disrupting DIP-β causes L4 neurons to form ectopic synapses and have altered dendritic morphology.
DIP-β is cell autonomously required in L4 neurons for proper synaptic connectivity
DIP-β is expressed in multiple cell types in the visual system (Cosmanescu et al., 2018) (Fig. 1D), and so to determine if synaptic defects observed in DIP-β KO flies result from the disruption of DIP-β in L4 neurons, as opposed to other neurons, we undertook conditional knockdown (cKD) experiments. We expressed DIP-β RNAi exclusively in L cells and visualized L cell synapses in the lamina using STaR. As DIP-β is normally expressed in L4 but no other L cells (Tan et al., 2015), disrupting DIP-β in all L cells is analogous to disrupting DIP-β in L4 neurons alone. Developmental analyses revealed that DIP-β immunolabeling becomes detectable in the proximal lamina in a pattern reminiscent of L4 dendritic processes at 72h APF (Fig. S3A–C’), and this labeling is eliminated in DIP-β KO flies (Fig. 3A and B). Expressing DIP-β RNAi in L cells (Fig. 3C–D’) or L4 neurons selectively (Fig. S3C–D”) also significantly reduced DIP-β immunolabeling in the proximal lamina demonstrating the efficacy of knockdown and showing that L4 dendrites are the primary source of DIP-β in this region. Additionally, when DIP-β was knocked down in L cells we observed increases in Brp puncta in the distal (Fig. 3E, G) and proximal (Fig. 3F) lamina similar to that observed in whole fly DIP-β mutants (Fig. 1O and P). These findings demonstrate that DIP-β is required in L4 neurons to establish normal synaptic connectivity.
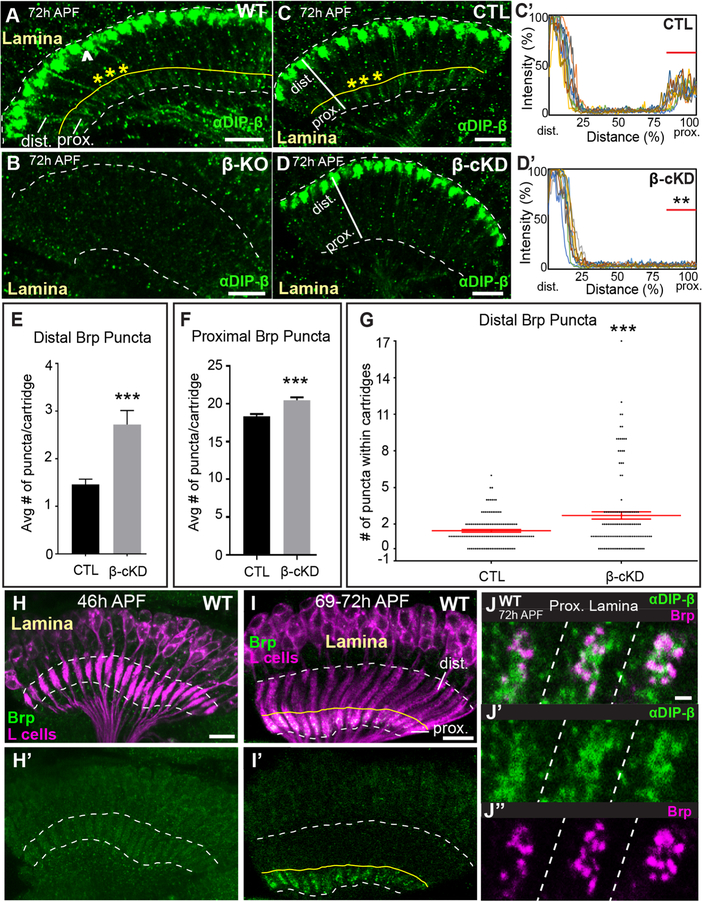
Figure 3. DIP-β in L4 neurons is necessary for proper synaptic connectivity.
(See also Figure S3) (A and B) Confocal images showing DIP-β immunolabeling (green) in the laminas of wild type (A) or DIP-β KO flies (B) at 72h APF. The dotted white lines demarcate the lamina neuropil. The yellow line in (A) indicates the boundary between the proximal and distal lamina. The arrowhead in (A) indicates DIP-β expression in non-L4 neurons likely to be LaWF2 neurons. Yellow asterisks in (A) indicate individual cartridges. Scale bar = 10μm.
(C-D’) DIP-β immunolabeling in the proximal lamina at 72h APF in control (UAS-β-RNAi only) and conditional knockdown flies (β-cKD, UAS-β-RNAi + 9B08-GAL4).
(C and D) Confocal images of DIP-β immunolabeling in the laminas of a control fly (C) or a β-cKD fly (D). The white lines show the region of lamina cartridges assessed in (C’ and D’). In (C), the yellow asterisks indicate individual cartridges. Scale bar = 10μm.
(C’ and D’) Quantification of DIP-β fluorescence intensity along the long axis of lamina cartridges (see white lines in (C and D)). Significantly reduced fluorescence intensity is observed in the proximal lamina (80–100% distance) of β-cKD flies (the proximal lamina is marked by red bar in C’ and D’) compared to control flies (n=3 cartridges per brain, n=10 brains per condition).
(E) The average number of Brp puncta in the distal halves of lamina cartridges in control (n= 120 cartridges; 6 brains) and β-cKD (n= 120 cartridges; 6 brains) flies. Data are represented as a mean +/−SEM.
(F) The average number of Brp puncta in the proximal halves of lamina cartridges in control (n= 120 cartridges; 6 brains) and β-cKD (n= 120 cartridges; 6 brains) flies. Data are represented as a mean +/−SEM.
(G) Total number of Brp puncta in the distal halves of cartridges in control (n= 120 cartridges; 6 brains) and β-cKD (n=120 cartridges; 6 brains) flies. Each dot represents a cartridge. Red bar indicates +/−SEM.
(H-I’) Confocal images in a longitudinal plane of lamina cartridges showing the developmental timing of Brp expression (green-smGFPV5) in L cells (magenta-myr-tdTOM) in the lamina. The dotted lines delineate the lamina neuropil. The yellow line in I and I’ indicates the boundary between the distal and proximal lamina. n=5 brains (46h and 69–72h APF). Scale bar = 10μm.
(J-J”) Co-labeling of DIP-β (green) and Brp (magenta-smGFPV5) at 72h APF in the proximal regions of three lamina cartridges separated by dotted lines. Scale bar =1μm.
(Statistical significance- *<.05, **<.005, ***<.0005)
DIP-β localizes to L4 dendrites during synapse formation
To gain further insight into DIP-β function we assessed the timing of DIP-β expression in L4 neurons with respect to the formation of L4–L2 synapses and its subcellular localization during synapse formation visualized through immunolabeling and confocal microscopy. DIP-β immunolabeling was not detected in the lamina at 24h APF (Fig. S3A), but strong immunolabeling was detected in the most distal region of the lamina neuropil at 48h APF (Fig. S3B). This labeling likely represents DIP-β expressed in LaWF2 neurons (Tuthill et al., 2013). At 72h APF faint immunolabeling could be observed in the proximal lamina on L4 dendrites (Fig. 3A–D’, S3C–D”). To determine if the timing of DIP-β localization to L4 dendrites coincided with synapse formation, we used STaR to label Brp in L cells (Fig. S1A) and assessed Brp localization during pupal development in the laminas of wild type flies. We found that L4–L2 synapses form between 46–69h APF (Fig. 3H–I’), similar to the time when DIP-β becomes localized to L4 dendrites (between 48–72h APF) (Fig. 3A, C, S3A–C’). To shed light on whether DIP-β localizes to developing synapses, we simultaneously visualized DIP-β (immunolabeling) and Brp in L cells (STaR) at 72h APF using confocal microscopy (Fig. 3J–J”). We found that DIP-β was more diffusely distributed than Brp, but that some of the DIP-β protein appeared to be organized into clusters that overlapped with or were adjacent to Brp puncta. This was consistent between cartridges and across brains. Taken together, these findings show that DIP-β localizes to L4 dendrites during synapse formation between L4 and L2 neurons, consistent with a role for DIP-β in establishing connectivity between these neurons.
DIP mis-expression promotes synapse formation in Dpr-expressing lamina neurons
If Dpr-DIP interactions act instructively to control synaptic specificity, then they should be sufficient to promote synapse formation between specific cell types. To test this possibility, we exploited the cell type-specificity of synaptic connections between L cells and R cells in the lamina. Within each cartridge R cells synapse en passant onto L1–L3, but L1–L3 do not reciprocally synapse back onto R cells, nor do they synapse with each other (Meinertzhagen and O’Neil, 1991; Rivera-Alba et al., 2011) (Fig.1C). This specificity is striking given that processes of these neurons are densely packed within the cartridge and contact each other extensively. L cells express high levels of Dprs (Tan et al., 2015) and in general, both L cells and R cells express low levels of DIPs (Tan et al., 2015; Zhang et al., 2016). Thus, we hypothesized that if Dpr-DIP interactions promote synapse formation then mis-expressing DIPs in R cells should cause L cells to synapse onto R cells. Likewise, mis-expressing DIPs in L1–L3 should cause these cells to synapse with each other.
To test this hypothesis, we mis-expressed DIPs-γ and ε either together or independently and DIP-β independently in R cells or L cells and visualized L cell synapses in the lamina using STaR as in the DIP KO experiments. We chose these DIPs because they have broad Dpr binding specificities and are known to bind to Dprs expressed in L cells (Carrillo et al., 2015; Cosmanescu et al., 2018; Ozkan et al., 2013; Tan et al., 2015). In control flies, L cell synapses were restricted to the proximal lamina where L4 and L2 form reciprocal connections (Fig. 4A and A’). Strikingly, mis-expression of both DIPs-γ and ε (Fig. S4A and A’) or DIP-γ alone in R cells (Fig. 4B and B’) caused L cells to form streams of ectopic synapses throughout lamina cartridges. On average, 34% of lamina cartridges in young adult (1–2 day old) flies mis-expressing DIP-γ in R cells displayed clusters of ectopic L cell synapses in the distal lamina, while none of the cartridges in control flies showed this phenotype (Fig. 4D). While both DIPs-ε and γ were strongly expressed upon mis-expression in R cells (Fig. S4C and D), mis-expression of DIP-ε alone did not cause the formation of ectopic synapses (Figs. S4B and B’), nor did mis-expression of DIP-β (Fig. S4G and G’). Cross sections through the lamina of flies mis-expressing DIP-γ in R cells revealed the presence of fused cartridges in both control and mis-expression flies (Fig. S4E and F).
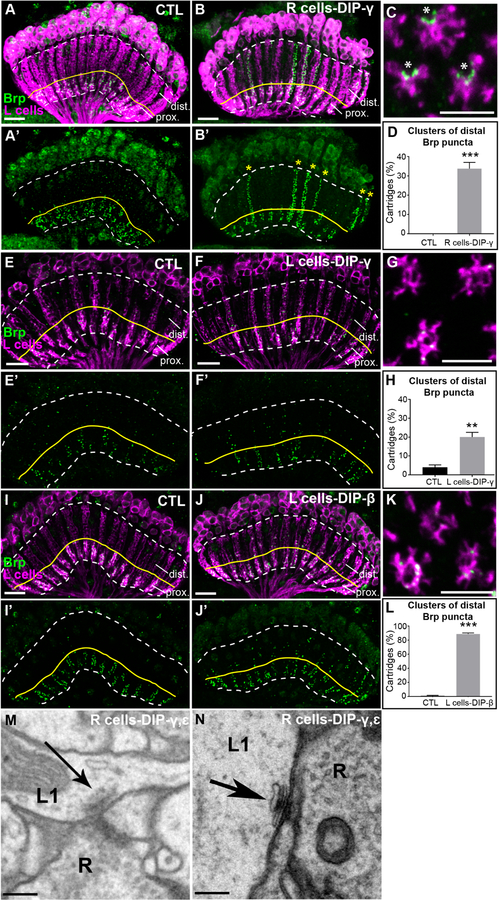
Figure 4. DIP mis-expression promotes synapse formation with Dpr-expressing lamina neurons.
(See also Figures S4 and S5, and Table S1)
(A-B’) Confocal images (longitudinal plane of lamina cartridges, 1–2 day old adults) showing the distribution of Brp (green-smFPV5) expressed in L cells (magenta-LexAop-myr-tdTOM) in the laminas of control (UAS-DIP-γ) flies or flies expressing DIP-γ in R cells (UAS-DIP-γ and GMR-GAL4). The white dotted line outlines the lamina and the yellow line separates the proximal (prox.) and distal lamina (dist.). Scale bar = 10μm.
(A and A’) Brp is restricted to the proximal lamina in control flies.
(B and B’) Streams of ectopic Brp puncta are detected throughout lamina cartridges (yellow stars in B’) when mis-expressing DIP-γ in R cells.
(C) A confocal image of a cross section through the lamina of a fly mis-expressing DIP-γ in R cells. Asterisks indicate the presumed positions of photoreceptor axons. Scale bar = 5μm.
(D) Quantification of the percentage of cartridges containing clusters of Brp puncta in the distal lamina in control flies (n= 11) and flies mis-expressing DIP-γ in R cells (n= 11). Clusters were defined as ≥3 consecutive z-stack slices containing distal Brp puncta. Only unmerged cartridges were considered in the quantification (n=25 cartridges/brain). Data are represented as a mean +/− SEM.
(E-F’) Mis-expression of DIP-γ in L cells (27G05-GAL4). Confocal images show the distribution of Brp (green-smFPV5) expressed in L cells (magenta-LexAop-myr-tdTOM) in the laminas of control flies (27G05-GAL4 alone) or experimental flies (27G05-GAL4 and UAS-DIP-γ). The region above the yellow line delineates the distal lamina (dist.). Scale bar = 10μm.
(E and E’) Brp is localized to the proximal lamina in control (27G05-GAL4) flies, occasionally with some puncta in the distal lamina. n=5 brains.
(F and F’) L cells form ectopic synapses in the distal regions of lamina cartridges (yellow stars in F’) upon mis-expression of DIP-γ in L cells. n= 5 brains.
(G) A confocal image of a cross section through the lamina of a fly mis-expressing DIP-γ in L cells. Scale bar = 5μm.
(H) Quantification of percentage of cartridges containing clusters of Brp puncta in the distal lamina in control flies (n=5) and flies mis-expressing DIP-γ in L cells (n=5). Clusters were defined as ≥5 distal Brp puncta within 5 consecutive z-stack slices of each cartridge (n=25 cartridges/brain). Data are represented as a mean +/− SEM.
(I and I’) Brp is localized to the proximal lamina in control (UAS-DIP-β) flies. n=5 brains.
(J and J’) In flies mis-expressing DIP-β in L cells (27G05-GAL4), ectopic synapses are present throughout lamina cartridges. n=7 brains.
(K) A confocal image of a cross section through the lamina of a fly mis-expressing DIP-β in L cells. Scale bar = 5μm.
(L) Quantification of percentage of cartridges containing clusters of Brp puncta in the distal lamina in control flies (n=5) and flies mis-expressing DIP-β in L cells (n=5). Clusters were defined as ≥5 distal Brp puncta within 5 consecutive z-stack slices of each cartridge (n=25 cartridges/brain). Data are represented as a mean +/− SEM.
(M and N) Putative L1-R cell synapses in flies mis-expressing DIPs-γ and ε in R cells identified by EM.
Scale bar = 200nm.
(Statistical significance- *<.05, **<.005, ***<.0005)
The presence of the GMR-GAL4 driver alone (R cell expression) was sufficient to induce cartridge fusion (Fig. S4E). Importantly, the induction of ectopic synapses does not correlate with cartridge fusion, as many unfused cartridges contained ectopic synapses (Fig. 4C and D, S4F). Analysis of cross sections also revealed that ectopic synapses frequently formed on the edges of L cell profiles consistent with the positions of R cell axon terminals (Fig. 4C). Mis-expression of DIPs-γ (Fig. 4E–H) or β (Fig. 4I–L) only in L cells also caused L cells to form ectopic synapses throughout lamina cartridges. Mis-expression of DIP-γ in L cells induced ectopic synapse formation in ~20% of lamina cartridges (Fig. 4H), while ~90% of cartridges contained clusters of Brp puncta in the distal lamina upon DIP-β mis-expression in L cells (Fig. 4L). Cross section views through cartridges revealed that ectopic Brp puncta were distributed throughout L cell processes within cartridges (as opposed to the edges of cartridges as in R cell mis-expression experiments) (compare Figs. 4C, G, K) consistent with L cell-L cell synapses. Together, these findings show that mis-expression of DIPs-γ and β causes L cells to form ectopic synapses in a predictable manner.
To visualize the morphologies and cellular constitution of ectopic synapses induced by DIP mis-expression we utilized EM. We cut the lamina of a fly mis-expressing both DIPs-γ and ε in R cells into 50–60 nm sections and imaged these using transmission electron microscopy (TEM). We then identified the synapses formed in the sections and assigned them to cellular profiles based on previously established criteria (Meinertzhagen, 1996; Meinertzhagen and O’Neil, 1991). We found that the positions of cell types within the cartridge were normal, with L1 and L2 always paired at the cartridge axis surrounded by R cell terminals (Fig. S5). In addition, the numbers of synapses formed by R cells(R cells pre-synaptic) was similar to those reported previously for the wild type, with the 6 R cell profiles together contributing 330 synapses (Table S1) (Meinertzhagen and O’Neil, 1991; Rivera-Alba et al., 2011). Thus, DIP-mis-expression did not significantly perturb the general cellular architecture of the cartridge, or synapse formation in R cells. We identified 86 L cell synapses within the cartridge (L cells pre-synaptic) (Table S1), ~3–4 times more L cell synapses than was previously reported for wild type cartridges (Meinertzhagen and O’Neil, 1991; Rivera-Alba et al., 2011). These were distributed throughout the cartridge, with 31 L cell synapses in the distal half (Table S1). In addition, we identified presynaptic sites formed by L1 (x12), L3 (x13), and L5 (x5) neurons which were previously not found to be pre-synaptic in the lamina (we also identified L2 and L4 presynaptic sites). In some cases, identified R cell profiles were adjacent to L cell presynaptic sites (Fig. 4M and N) consistent with L cell to R cell synapses, although a full reconstruction would be necessary to determine the degree to which L cell synapses form onto R cells upon DIP mis-expression. Together, these findings complement and support our confocal analyses and show that DIP-mis-expression promotes synapse formation in a manner predicted by Dpr expression.
DISCUSSION
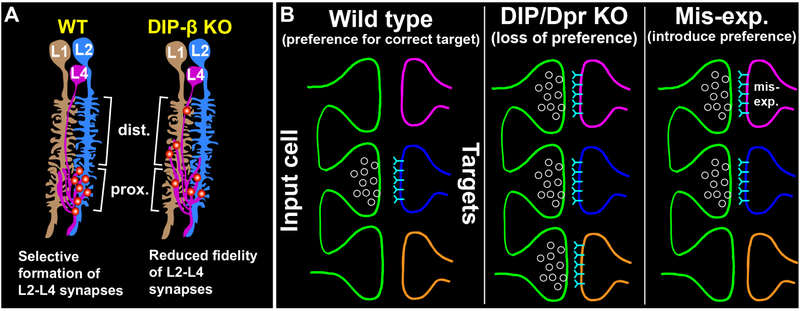
Neurobiologists have long thought that appropriate synaptic partners express complementary molecules that allow them to identify each other within a dense meshwork of alternative neurites through a lock-and-key mechanism. A common interpretation of this idea is that interactions between correct partners mediated by complementary adhesion or recognition molecules are necessary for synaptogenesis in an “all or nothing” process. Based on their heterophilic binding and matching expression in synaptically connected cell types, Dpr and DIP IgSF proteins have been proposed to play an instructive role in regulating synaptic specificity through a complementary binding mechanism. The findings we present here support a role for DIP proteins in instructing synaptic partner selection, most likely through interactions with Dpr proteins. However, rather than being necessary for synaptogenesis, we propose that DIP proteins regulate synaptic specificity by establishing a preference for synapses to form between specific cell types (see below). In this view, synaptic specificity reflects a relative preference for certain partners rather than an absolute or categorical recognition. In the absence of such preference neurons have the capacity to synapse with other cell types.
DIP proteins are necessary for correct visual function
Our findings demonstrate that DIP-β, γ or both are required for proper visual function. In DKO flies, we observed changes in two visually guided behaviors, the optomotor reflex and phototaxis, and also saw altered responses of photoreceptors and L1 and L2 neurons to light in electroretinogram recordings. For both optomotor and phototaxis assays the phenotype in young adult-DKO flies was not present in adult-DKO flies. One interesting possibility is that through experience DKO flies are able to compensate for the lack of DIP function. Whether the behavioral and physiological abnormalities observed in DKO flies are caused by the synaptic phenotypes reported here remains to be determined.
DIP-β is required for proper synaptic connectivity
We hypothesize that DIP-β regulates L4–L2 connectivity in multiple ways. First, DIP-β regulates the morphology of primary L4 dendrites. We hypothesize that interactions between DIP-β and L2 Dprs mediate adhesion between primary L4 dendrites and L2 processes in the cartridge core. We speculate that when this adhesion is reduced by disrupting DIP-β, L4 dendrites extend further distally and contact and synapse with alternative cell types (e.g. L1) in the distal lamina (Fig. 5A). However, as most ectopic synapses form on L4 axons in DIP-β KO flies, ectopic synapse formation does not strongly correlate with altered dendritic morphology.
Figure 5. Dpr-DIP interactions may regulate synaptic specificity by establishing a preference for synaptic partners.
(A) Working model of how DIP-β-Dpr interactions may regulate selective synapse formation between L4 and L2 neurons. In wild type flies, L4 and L2 selectively synapse with each other in the proximal lamina. In DIP-β KO flies, we propose that the fidelity of L4–L2 synapse formation is reduced and that L4 neurons form synapses with alternative cell types (e.g. L1) in both the distal and proximal lamina.
(B) General model of how Dpr-DIP interactions may regulate synaptic specificity. (Left panel) In a wild type background, Dpr-DIP interactions establish a preference for synapses to form between appropriate synaptic partners, potentially by concentrating synaptic machinery to specific cell-cell contacts. (Middle panel) When Dpr-DIP interactions are disrupted there is a reduced preference for the correct synaptic partner. Neurons have the capacity to synapse with other cell types. (Right panel) Inducing ectopic Dpr-DIP interactions introduces a preference for inappropriate synaptic partners.
Second, our mis-expression experiments support a synapse promoting function for DIP-β. As mis-expression of DIP-β in L cells but not R cells was sufficient to induce ectopic synapse formation in L cells, DIP-β may act presynaptically to promote synapse formation. We speculate that DIP-β in L4 neurons binds to Dprs in L2 neurons and promotes synapse formation onto L2 neurons by recruiting synaptic machinery to sites of L4–L2 contact. Thus, disrupting DIP-β may result in the accumulation of synaptic proteins at sites of contact with other cell types, leading to abnormal synapse formation in the proximal and distal lamina (Fig. 5A). Consistent with this, in DIP-β KO flies most of the ectopic pre-synaptic sites in the distal lamina form on L4 axons which are restricted to the cartridge periphery. It is likely that these ectopic synapses represent synapses with cell types other than L2, which occupies the cartridge core.
Our findings support a role for DIP-β in establishing L4–L2 connectivity by regulating dendrite morphology and by promoting synapse formation. As DIP proteins are primarily known to bind heterophilically with Dpr proteins (Carrillo et al., 2015; Cheng et al., 2019; Cosmanescu et al., 2018; Ozkan et al., 2013) (some DIPs bind homophilically (Cosmanescu et al., 2018)), and disrupting DIP-Dpr interactions in vivo has been shown to phenocopy the loss of DIPs or Dprs (Xu et al., 2018), it is likely that DIP-β functions in both contexts by interacting with cognate Dpr proteins expressed in L2 neurons. However, we cannot rule out Dpr-independent functions. To test this, it is crucial to identify Dprs expressed in L2 that bind DIP-β during synapse formation and test whether disrupting them, or the ability of DIP-β to bind them, phenocopies the loss of DIP-β in L4 neurons. We previously showed that L2 neurons express 6 of the 7 Dprs known to bind DIP-β at 40h APF, and at least 2 of these Dprs at 72h APF (Tan et al., 2015) (Table1). L1 neurons were found to express 3 of the 7 Dprs at 40h APF and at least 1 of these Dprs at 72h APF (Tan et al., 2015) (Table1). Thus, the preference for L4 neurons to synapse with L2 neurons over L1 neurons may be accounted for by the fact that L2 neurons express more Dprs that bind DIP-β than L1 neurons during synapse formation. Additionally, it will be important to determine if Dpr and DIP proteins cluster together at developing L4–L2 synapses and interact with synaptic proteins. Given that Dprs and DIPs lack obvious intracellular signaling motifs and that many are predicted to be linked to the plasma membrane through a lipid anchor (Cheng et al., 2018), if they interact with synaptic machinery this would be likely to occur through co-receptors.
Table1. The expression of Dprs that bind DIP-β in L1 and L2 neurons.
The expression of Dprs that bind DIP-β in L1 and L2 neurons at 40h and 72h APF as reported by (Tan et al., 2015). The binding affinity of DIP-β for specific Dprs is shown in parentheses, represented as dissociation constants (nm) determined by (Cosmanescu et al., 2018). The expression of Dprs 6, 10, 11, and 15 were assessed at the protein level. The expression of all other Dprs was examined at the mRNA level through RNA-seq.
| 40h APF | L1 | Dpr10 (54.9) | Dpr15 (22) | Dpr21 (1.83) | |||
| L2 | Dpr6 (19.4) | Dpr8 (1.52) | Dpr9 (4.07) | Dpr10 (54.9) | Dpr11 (94) | Dpr21 (1.83) | |
| 72h APF | L1 | Dpr10 (54.9) | |||||
| L2 | Dpr6 (19.4) | Dpr11 (94) |
DIP mis-expression changes the synaptic connections of L cells in a predictable manner
Our mis-expression experiments support a role for DIPs-β and γ in promoting synapse formation with Dpr-expressing neurons. Interestingly, since L cells and R cells already contact each other extensively within cartridges, DIP mis-expression is unlikely to promote synapse formation by forcing contact between these neurons. Rather, DIP mis-expression may make L cells competent to synapse onto R cells and each other. We hypothesize that mis-expressing DIP-γ in R cells or L cells and mis-expressing DIP-β in L cells promotes trans interactions with cognate Dprs expressed in L cells that go on to recruit synaptic machinery resulting in synapse formation. However it’s possible that DIP proteins promote synapse formation independent of Dprs. Experiments eliminating the ability of DIPs-γ or β to bind Dprs or disrupting the function of Dprs that bind DIP-γ or β in L cells are needed to determine whether specific Dpr-DIP interactions contribute to ectopic synapse formation in DIP mis-expression experiments.
It is unclear why the mis-expression phenotypes are non-uniform (not all cartridges contain ectopic synapses). It’s possible that synapse refinement contributes to the non-uniform pattern of ectopic synapse formation. For example, early in development many cartridges may contain ectopic synapses which then become pruned away over time.
Dpr-DIP interactions may regulate synaptic specificity by establishing a preference for synaptic partners
Synapse formation is robust [see also (Hassan and Hiesinger, 2015)]. In both vertebrates and invertebrates it has been shown that disrupting proteins known to regulate synapse organization (Chen et al., 2017; Mosca et al., 2012; Mosca and Luo, 2014; Robbins et al., 2010; Sudhof, 2017; Varoqueaux et al., 2006) or specificity (Krishnaswamy et al., 2015; Shen and Bargmann, 2003; Shen et al., 2004) does not prevent neurons from forming synapses. In addition, in the absence of appropriate partners neurons have the capacity to synapse with alternative partners (Bekkers and Stevens, 1991; Cash et al., 1992; Duan et al., 2014; Peng et al., 2018; Shen and Bargmann, 2003). The findings we report here are consistent with these observations. Collectively, our data support the idea that DIP proteins play an instructive role in establishing synaptic specificity but show that they are not necessary for synapse formation. Our working hypothesis is that interactions between DIP-β in L4 neurons and cognate Dprs in L2 neurons establish a preference for L4 to synapse onto L2 over other cell partnerships in the cartridge (Fig. 5A). In the absence of this preference L4 has the capacity to synapse with other cell types (e.g. L1). Similarly, mis-expression of DIP-γ in R cells or L cells and DIP-β in L cells establishes a preference for L cells to synapse with R cells or each other. Thus, we propose that when Dpr-DIP interactions are disrupted synapses still form but reflect a loss of preference for the correct partners, and that inducing ectopic Dpr-DIP interactions introduces incorrect preferences that promote synapses to form between incorrect partners (Fig. 5B). Similar models of synaptic specificity have been proposed in C. elegans and mice. In C. elegans, interactions between the IgSF proteins SYG-1, expressed in the HSNL neuron, and SYG-2, expressed in vulval epithelial cells (guidepost cells), restrict the subcellular location of synapse formation in HSNL, biasing HSNL to synapse with specific partners (Shen and Bargmann, 2003; Shen et al., 2004). An important difference between the SYG proteins and Dprs/DIPs is that Dpr-DIP interactions occur between synaptic partners rather than with guidepost cells. In the mouse retina, the IGSF protein Sdk2 regulates selective synapse formation between an interneuron and a retinal ganglion neuron after they have innervated the correct sublaminae (Krishnaswamy et al., 2015). It was proposed that Sdk2 and other similar molecules may bias synapses to form between specific cell types within sublaminae. Thus, our study together with previous studies suggests an evolutionarily shared strategy for establishing synaptic specificity.
STAR Methods text
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Matt Pecot (matthew_pecot@hms.harvard.edu).
Antibodies and fly lines generated in this study will be distributed upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Experimental model used in this study- Drosophila melanogaster
Flies were raised on standard cornmeal-agar based medium and maintained at 25°C with 50–60% humidity. Male and female flies were used at the following developmental stages: 24h APF (after puparium formation), 48h APF, 72h APF, young adult (1–2 days-old), adult (~2 weeks old).
METHOD DETAILS
Production of DIP antibodies
DIP-γ antigen: (aa22–393) full length except the predicted signal peptide and the TM domain (guinea pig)
GSTQNQHHESSSQLDPDPEFIGFINNVTYPAGREAILACSVRNLGKNKVGWLRASDQTVLALQGRVVT HNARISVMHQDMHTWKLKISKLRESDRGCYMCQINTSPMKKQVGCIDVQVPPDIINEESSADLAVQEG EDATLTCKATGNPQPRVTWRREDGEMILIRKPGSRELMKVESYNGSSLRLLRLERRQMGAYLCIASND VPPAVSKRVSLSVQFAPMVRAPSQLLGTPLGSDVQLECQVEASPSPVSYWLKGARTSNGFASVSTAS LESGSPGPEMLLDGPKYGITERRDGYRGVMLLVVRSFSPSDVGTYHCVSTNSLGRAEGTLRLYEIKLH PGASASNDDHLNYIGGLEEAARNAGRSNRTTWQ
DIP-β antigen: (88–470aa) full length except a few AAs of the predicted signal peptide and the TM domain (guinea pig)
NKISSVGAFEPDFVIPLENVTIAQGRDATFTCVVNNLGGHRVSGDGSSAPAKVAWIKADAKAILAIHEHVI TNNDRLSVQHNDYNTWTLNIRGVKMEDAGKYMCQVNTDPMKMQTATLEVVIPPDIINEETSGDMMVP EGGSAKLVCRARGHPKPKITWRREDGREIIARNGSHQKTKAQSVEGEMLTLSKITRSEMGAYMCIASN GVPPTVSKRMKLQVHFHPLVQVPNQLVGAPVLTDVTLICNVEASPKAINYWQRENGEMIIAGDRYALTE KENNMYAIEMILHIKRLQSSDFGGYKCISKNSIGDTEGTIRLYEMERPGKKILRDDDLNEVSKNEVVQKD TRSEDGSRNLNGRLYKDRAPDQHPASGSDQLLGRGTMR
DIP-ε antigen: (249–444aa)
VDFSPMVWIPHQLVGIPIGFNITLECFIEANPTSLNYWTRENDQMITESSKYKTETIPGHPSYKATMRLTI TNVQSSDYGNYKCVAKNPRGDMDGNIKLYMSSPPTTQPPPTTTTLRRTTTTAAEIALDGYINTPLNGNG IGIVGEGPTNSVIASGKSSIKYLSNLNEIDKSKQKLTGSSPKGFDWSKGKSSGSHG
Antigens and antibodies were produced at Genescript.
Immunohistochemistry
Fly brains were dissected in Schneider’s medium and fixed in 4% paraformaldehyde in phosphate buffered lysine for 25 min. After fixation, brains were quickly washed with phosphate buffer saline (PBS) with 0.5% Triton-X-100 (PBT) and incubated in PBT for at least 2 hr at room temperature. Next, brains were incubated in blocking buffer (10% NGS, 0.5% Triton-X-100 in PBS) overnight at 4°C. Brains were then incubated in primary antibody (diluted in blocking buffer) at 4°C for at least two nights. Follow ing primary antibody incubation, brains were washed with PBT three times, 1 hr per wash. Next, brains were incubated in secondary antibody (diluted in blocking buffer) at 4°C for at least two nights. Following secondary antibody incubation, brains were washed with PBT two times, followed by one wash in PBS, 1 hr per wash. Finally, brains were mounted in SlowFade Gold antifade reagent (Thermo Fisher Scientific, Waltham, MA).
Confocal imaging was accomplished using either a Leica SP8 laser scanning confocal microscope or a Zeiss LSM800 Laser Scanning Microscope.
Electron microscopy
The heads of 6-day old flies were dissected, immersed in a cacodylate-buffered paraformaldehyde and glutaraldehyde primary fixative, and processed for EM, as previously reported (Meinertzhagen and O’Neil, 1991; Meinertzhagen, 1996). Sections from Epon embedded specimens were cut serially at 60 nm, stained with 4% aqueous uranyl acetate and viewed with a FEI Tecnai 12 electron microscope operated at 80kV, and images collected with a Gatan 832 digital camera. A series of 500 consecutive sections in total was cut, 320 of which were imaged, aligned in Image J, and profiles identified and synapses marked manually.
Optomotor and phototaxis assays
Behavioral assays were performed using the MARGO platform as described previously (Werkhoven et al. 2019). Briefly, co-housed DKO and CTL flies were transferred to individual optomotor (Fig 1H) or phototactic y-maze (Fig 1I) arenas. In the optomotor assay, fly location was tracked and a rotating pinwheel stimuli was centered on the fly (angular speed of 320 deg/s, spatial frequency of 0.02 cycles/deg). Optomotor index was measured as the fraction of angular movement of the fly occuring in the direction of the stimuli, normalize between 1 (all movement in the direction of the stimuli) and −1 (all movement occurring in the direction opposing the stimuli). In the phototactic Y-maze assay, flies were placed in a y-maze with an LED at the end of each arm as their location was tracked. Intensity ranged from 940.3 nW to 11.35 mW as measured by an optical power meter (400nm, ThorLabs). As the fly approached the center of the arena, the LED appeared at the end of one of the non-occupied arms of the Y-maze. Phototactic choice probability was calculated by measuring the percentage of trials in which the fly chose the lit arm of the Y-maze.
Electroretinogram recordings
Electroretinogram recordings were performed on female flies using pairs of pulled glass electrodes (Sutter) filled with physiological drosophila saline (103mM NaCl, 3 mM KCl, 5mM TES,9 mM trehalos, 10mM glucose, 26mM NaHCO3, 1 mM NaH2PO4, 4mM MgCl2, 1.5 mM CaCl2). One electrode was placed on the surface of the cornea while the other was impaled in the thorax of the fly. Stimuli were generated by a white LED (ThorLabs) and delivered through the optics of the microscope. Stimuli were shaped by inserting neutral density filters to adjust the intensity of the light: intensity ranged from 33.5mW at ND0 to 475μW as measured by an optical power meter (400nm, ThorLabs). Data were acquired using an A-M Model 3000 extracellular amplifier, digitized at 20,000 Hz usingan Instrutech-ITC18 digitizer and acquired using Igor Pro 7. ERG waveforms were low pass filtered at 1000Hz and down-sampled to 500Hz for analysis—all ERG analysis was performed with custom MATLAB scripts. Differences between DKO and control flies were tested using a likelihood-ratio test of two linear mixed effects models (using the compare and fitlme() functions in MATLAB) on three measures of the ERG waveform, the steady state voltage (indicative of photoreceptor response, measured as the mean voltage in the final .25 seconds of the light step), and the on and off transient response (indicative of L1 and L2 response; the on response was measured as the maximum positive deflection within .1 seconds relative to the voltage before the stimulus, the off response was measured as the minimum voltage in the .1 seconds following light offset relative to the steady state voltage). For each measure, a model comparing light intensity, genotype, and their interaction was compared to the null hypothesis model with only light intensity. In both models an additional grouping variable was added to model effects for individual flies, such that H0: Yij=β0+β1NDi+β2Flyj+ eij and H1:Yij=β0+β1NDi+β2Genotypei+β3ND∗Genotypei+β2Flyj+ eij where irepresents each trial and j represents each fly.
Machine learning algorithm for tracing cartridges and puncta
Detecting individual cartridges was challenging due to the heterogeneity in intensity and texture within a dataset. We, therefore, trained a random forest model using 10–15 3D annotated training examples in the Matlab-based VoxelClassifier (https://hms-idac.github.io/VoxelClassifier/). In the training examples, background and cartridge pixels were annotated as separate classes. The features that were trained consisted of intensity derivatives, Laplacian of Gaussian kernels, steerable filters, and basic texture features such as standard deviation and entropy within a 5×5 or 11×11 neighborhood. The resulting probability class map for the cartridge (foreground class) was further processed by applying a gaussian filter with a sigma of similar radius as a typical cartridge and identifying regional maxima in each plane. These were dilated and skeletonized to form continuous filaments along the center of each cartridge. At each pixel along each filament, we employed region growing to the outer edge of each cartridge. This was repeated at each plane until the entire structure of the cartridge had been reconstructed throughout the dataset. To find puncta, we convolved a 3D Laplacian of Gaussian filter with a sigma of 2 that approximated the radius of each puncta, and identified regional maxima. This generated candidate spots on the puncta and background. To eliminate false positives, we set a robust threshold as 4 standard deviations above the median response and masked out spots that were not within a cartridge. The remaining puncta in each cartridge were counted and exported for further analysis. We used Imaris v8 (Bitplane) to verify the accuracy of cartridge and puncta segmentation.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of L2 and L4 cell numbers
L2 and L4 cell numbers were determined blind to genotype using cell-specific genetic labeling (described above). Each cartridge in the lamina contains dendrites from a single L2 neuron and the axon/neurite of a single L4 neuron. As cartridges are regularly spaced within the lamina, L2 dendrites and L4 axons within each cartridge can be identified in cross section views of the lamina. The percentage of cartridges containing L2 or L4 neurons was determined for each optic lobe scored. The percentages for lobes of the same genotype were pooled and the average percentage was determined.
Quantification of Brp puncta in the distal regions of lamina cartridges
Using confocal microscopy, we generated z-stacks of the lamina down the long axis of lamina cartridges. Within each z-stack (i.e. each optic lobe) 25 well labeled cartridges were identified and the number of Brp puncta in their distal halves was counted. The top (distal edge) and bottom (proximal edge) of each cartridge was determined by the first and last sections containing L cell processes (myrtd::TOM), respectively. The midpoint of each cartridge was then identified as the section in between the top and bottom sections. Brp puncta were counted in the sections distal to the midpoint of each cartridge. This stringent criterion was used to avoid counting L2–L4 synapses in the proximal lamina. It is likely that our quantification of distal Brp puncta is an underestimate of the number of ectopic synapses formed in the absence of DIP function. Genotypes were scored in a blind manner by three individuals, and their scores were averaged.
Quantification of DIP-β fluorescence signal
Using Zeiss image analysis software, we quantified DIP-β signal in conditional knockdown and control brains by measuring fluorescence signal through the cartridge trajectory (3 cartridges per brain). Signal intensity values and cartridge lengths were converted to percentages by setting the highest intensity within each cartridge as 100% intensity and the full length of the cartridge as 100% distance. Statistical analysis using unpaired t tests was performed after setting uniform intervals (using the spline function on matlab) of 0.01% distance.
Statistical Analysis of Brp puncta
To evaluate differences in the distal, proximal, and total number of Brp puncta between genotypes we fitted a general linear model with number of Brp puncta per cartridge as the response variable, and experiment identifier (2 levels) and genotype (7 levels) as factors (using the multicomp package of the R statistical computation software). We restricted the multiple comparison to contrasts WT vs. DKO, WT vs. γ +/−, WT vs. γ −/−, WT vs. β+/−, WT vs. β−/−, DKO vs. γ +/−, DKO vs. γ −/−, DKO vs/ β+/−, DKO vs. β−/−, γ +/− vs. γ −/−, γ +/− vs β+/−, γ +/− vs β−/− vs, β+/− vs γ −/−, β−/− vs β+/−, β−/− vs γ −/−, and reported the p-values of the individual contrasts.
Statistical analysis by figure
Figure 1
(H) Optomotor index, DKO (YAd) vs CTL (YAd): p = 4.9e-37. p-values were computed via rank sum test and were corrected for multiple comparisons.
(I) Phototactic choice probability, DKO (YAd) vs. CTL (YAd), p = 3.3e-09. p-values were computed via rank sum test and were corrected for multiple comparisons.
(J-L) p-values in J-L calculated via likelihood-ratio-test of linear mixed effects models (H0: No Genotype Effect, H1: Genotype effect)
(J) p <0.0005
(K) p <0.0005, p<0.005, p<0.05
(L) p <0.0005, p<0.005, p<0.05
(O) GLM: WT vs. DKO, p<0.001, WT vs. β+/−, p =0.01938, WT vs. β−/−, p= 0.00489;
(P) GLM: WT vs. β−/−, p= 0.00465;
(Q) GLM: WT vs. β+/−, p = 0.00684, WT vs. β−/−, p<0.001
(R) GLM: WT vs. DKO, p<0.001
Figure 2
(D) WT vs. DKO, p <0.0001, unpaired t test
(E) WT vs. DKO, p =0.1671, unpaired t test
(F) WT vs. DKO, p = 0.0677, unpaired t test
(G) WT vs. DKO, p <0.0001, unpaired t test
(H) WT vs. DKO, p=0.5190, unpaired t test
(I) WT vs. β−/−, p <0.0001, unpaired t test
(J) WT vs. β−/−, p = 0.7453, unpaired t test
(K) WT vs. β−/−, p = 0.7449, unpaired t test
(M) WT vs. β−/−, p = 0.0049, unpaired t test
(N) WT vs. β−/−, p = 0.0057, unpaired t test
(O) WT vs. β−/−, p = 0.0008, unpaired t test
Figure 3
(C’ and D’) p<0.005 from 80–83% distance; p<0.0001 from 84% −100% distance, unpaired t-tests for each 0.01% distance interval.
(E and G) p = 0.0001, unpaired t test
(F) p < 0.0001, unpaired t test
Figure 4
(D) CTL vs. R cells-DIP- γ, p < 0.0001, unpaired t test
(H) CTL vs. L cells-DIP- γ, p = 0.0014, unpaired t test
(L) CTL vs. L-cells-DIP- β, p < 0.0001, unpaired t test
Figure S1
(B) DKO (Ad) vs. CTL (Ad), p=2.2e-05. p-values were computed via rank sum test and were corrected for multiple comparisons.
(C) DKO (YAd) vs. CTL (YAd), p=6.7e-04 and DKO (Ad) vs CTL (Ad), p=2.2e-09. p-values were computed via rank sum test and were corrected for multiple comparisons.
(F) On transient WT vs DKO, (p=3.3e-36), steady state (SS) WT vs DKO, (p=3.6e-06), off transient (Off) WT vs DKO (p=2.2e-5).
Figure S2
(I) WT vs. DKO, p =0.2090, unpaired t test
(J) WT vs. DKO, p = 0.7053, unpaired t test
(K) WT vs. DKO, p = 0.3840, unpaired t test
Figure S3
(C” and D”) p<0.05 from 80–100% distance, unpaired t tests for each 0.01% distance interval.
DATA AND CODE AVAILABILITY
This study did not generate/analyze [datasets/code]
Supplementary Material
KEY RESOURCES TABLE
| Resource/Reagent | Source | Identifier |
|---|---|---|
| Experimental Models: Organisms/Strains | ||
| Strain (Drosophila melanogaster) 48A08AD (II), 66A01DBD (III) [L1 split-GAL4] |
Janelia Research Campus(Tuthill et al.,2013) | N/A |
| Strain (Drosophila melanogaster) 53G02AD (II), 29G11DBD (III) [L2 split-GAL4] | Janelia Research Campus(Tuthill et al.,2013) | N/A |
| Strain (Drosophila melanogaster) 64B03AD (II), 14B07DBD (III) [L3 split-GAL4] |
Janelia Research Campus(Tuthill et al.,2013) | N/A |
| Strain (Drosophila melanogaster) 31C06AD (II), 34G07DBD (III), [L4 split-GAL4] | Janelia Research Campus(Tuthill et al.,2013) | N/A |
| Strain (Drosophila melanogaster) 64D07AD (II), 37E10DBD (III) [L5 split-GAL4] |
Janelia Research Campus(Tuthill et al.,2013) | N/A |
| Strain (Drosophila melanogaster) 79C23S-GS-FRT-stop-FRT-smFPV5–2A-LexAVP16 | J. Peng (Peng et al., 2018) | N/A |
| Strain (Drosophila melanogaster) LexAop-myr::tdTomato (III) | Akin and Zipursky, 2016 | N/A |
| Strain (Drosophila melanogaster) UAS-Flp (II) | Bloomington Drosophila Stock Center | RRID:BDSC_4540 |
| Strain (Drosophila melanogaster) 27G05-FLPG5.PEST (attp5) | Janelia research campus (Peng et al., 2018) | N/A |
| Strain (Drosophila melanogaster) GMR-GAL4 (III) | Bloomington Drosophila Stock Center | RRID:BDSC_8121 |
| Strain (Drosophila melanogaster) 27G05-GAL4 (attp2) | Bloomington Drosophila Stock Center | RRID:BDSC_48703 |
| Strain (Drosophila melanogaster) UAS-β-RNAi (attp40) | Bloomington Drosophila Stock Center | RRID:BDSC_38310 |
| Strain (Drosophila melanogaster) 9B08GAL4 | (Pecot et al., 2013) | N/A |
| Strain (Drosophila melanogaster) DIP-β1−95 | This study | N/A |
| Strain (Drosophila melanogaster) DIP-γ1−67 | This study | N/A |
| Strain (Drosophila melanogaster) UAS-DIP-γ | This study | N/A |
| Strain (Drosophila melanogaster) UAS-DIP-ε | This study | N/A |
| Strain (Drosophila melanogaster) UAS-DIP-β | This study | N/A |
| Antibodies | ||
| Antibody Anti-V5 (mouse) 1:200 | Bio-Rad/AbD Serotec | Cat# MCA2892GA; RRID:AB_1658039 |
| Antibody Anti-DsRed (rabbit) 1:200 | Clontech Laboratories, Inc. | Cat# 632496; RRID:AB_10013483 |
| Antibody anti-chaoptin (mouse) 1:20 | Developmental Studies Hybridoma Bank | Cat# 24B10, RRID:AB_528161 |
| Antibody Anti-DIP-Beta (guinea pig) 1:300 | This study | N/A |
| Antibody Anti-DIP-Epsilon (rabbit) 1:500 | This study | N/A |
| Antibody Anti-DIP-Gamma (guinea pig) 1:400 | This study | N/A |
| Antibody Goat anti-Mouse IgG (H&L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 1:500 | Thermo Fisher Scientific | Cat# A11029 |
| Antibody Antibody, Alexa Fluor 647 1:500 | Thermo Fisher Scientific | Cat# A21245 |
| Antibody 647 Goat anti-Guinea Pig IgG (H&L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 1:500 | Thermo Fisher Scientific | Cat# A21450 |
| Software and Algorithms | ||
| Cartridge tracing algorithm | This paper | https://hms-idac.github.io/VoxelClassifier/ |
| MATLAB | Mathworks | https://www.mathworks.com/products/matlab.html |
| Prism | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| Fiji | Schindelin et al., 2012 | https://fiji.sc/ |
| R https://www.r-project.org/ | R core team (2013) | https://www.r-project.org/ |
Highlights.
DIP-β is necessary for proper synaptic connectivity in the Drosophila visual system
DIPs-β and γ are sufficient to promote synapse formation in vivo
DIP IgSF proteins are necessary for proper visual function in Drosophila
ACKNOWLEDGEMENTS
We would like to thank Drs. Zipursky, Ginty, Kaeser and Chen for many helpful discussions and comments on the manuscript. This research was supported by the NIH/NINDS (National Institute of Neurological Disorders and Stroke) grants K01 K01NS094545 (M.Y.P.), and R01 1R01NS110713–01 (M.Y.P.) by the McKnight Foundation (Scholar Award) (M.Y.P.), and by a Whitehall Foundation Research Grant (M.Y.P.). C.X. was supported by an Alice and Joseph Brooks Postdoctoral Fellowship, and an Edward R. and Anne G. Lefler Center Postdoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Akin O, and Zipursky SL (2016). Frazzled promotes growth cone attachment at the source of a Netrin gradient in the Drosophila visual system. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley J, Sorrentino V, Lobb-Rabe M, Nagarkar-Jaiswal S, Tan L, Xu S, Xiao Q, Zinn K, and Carrillo RA (2019). Transsynaptic interactions between IgSF proteins DIP-alpha and Dpr10 are required for motor neuron targeting specificity. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley J, Sorrentino V, Nagarkar-Jaiswal S, Tan L, Xu S, Xiao Q, Zinn K, and Carrillo RA (2018). Transsynaptic interactions between IgSF proteins DIP-α and Dpr10 are required for motor neuron targeting specificity in Drosophila. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier H (2013). Synaptic laminae in the visual system: molecular mechanisms forming layers of perception. Annu Rev Cell Dev Biol 29, 385–416. [DOI] [PubMed] [Google Scholar]
- Barish S, Nuss S, Strunilin I, Bao S, Mukherjee S, Jones CD, and Volkan PC (2018). Combinations of DIPs and Dprs control organization of olfactory receptor neuron terminals in Drosophila. PLoS Genet 14, e1007560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkers JM, and Stevens CF (1991). Excitatory and inhibitory autaptic currents in isolated hippocampal neurons maintained in cell culture. Proc Natl Acad Sci U S A 88, 7834–7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braitenburg V (1967). Patterns of projection in the visual system of the fly. I. Retina-lamina projections. Exp Brain Res 3, 271–298. [DOI] [PubMed] [Google Scholar]
- Cang J, and Feldheim DA (2013). Developmental mechanisms of topographic map formation and alignment. Annu Rev Neurosci 36, 51–77. [DOI] [PubMed] [Google Scholar]
- Carrillo RA, Ozkan E, Menon KP, Nagarkar-Jaiswal S, Lee PT, Jeon M, Birnbaum ME, Bellen HJ, Garcia KC, and Zinn K (2015). Control of Synaptic Connectivity by a Network of Drosophila IgSF Cell Surface Proteins. Cell 163, 1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash S, Chiba A, and Keshishian H (1992). Alternate neuromuscular target selection following the loss of single muscle fibers in Drosophila. J Neurosci 12, 2051–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Jiang M, Zhang B, Gokce O, and Sudhof TC (2017). Conditional Deletion of All Neurexins Defines Diversity of Essential Synaptic Organizer Functions for Neurexins. Neuron 94, 611–625 e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PL, and Clandinin TR (2008). The cadherin Flamingo mediates level-dependent interactions that guide photoreceptor target choice in Drosophila. Neuron 58, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Akin O, Nern A, Tsui CY, Pecot MY, and Zipursky SL (2014). Cell-type-specific labeling of synapses in vivo through synaptic tagging with recombination. Neuron 81, 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Ashley J, Kurleto JD, Lobb-Rabe M, Park YJ, Carrillo RA, and Ozkan E (2019). Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Park Y, Kurleto JD, Jeon M, Zinn K, Thornton JW, and Ozkan E (2018). A new family of neural wiring receptors across bilaterians defined by phylogenetic, biochemical and structural evidence. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KM, Prakash S, Bright A, and Clandinin TR (2006). Liprin-alpha is required for photoreceptor target selection in Drosophila. Proc Natl Acad Sci U S A 103, 11601–11606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clandinin TR, Lee CH, Herman T, Lee RC, Yang AY, Ovasapyan S, and Zipursky SL (2001). Drosophila LAR regulates R1–R6 and R7 target specificity in the visual system. Neuron 32, 237–248. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, and Zipursky SL (2000). Afferent growth cone interactions control synaptic specificity in the Drosophila visual system. Neuron 28, 427–436. [DOI] [PubMed] [Google Scholar]
- Coombe PE (1986). The large monopolar cells L1 and L2 are responsible for ERG transients in Drosophila. Journal of Comparative Physiology A 159, 655–665. [Google Scholar]
- Cosmanescu F, Katsamba PS, Sergeeva AP, Ahlsen G, Patel SD, Brewer JJ, Tan L, Xu S, Xiao Q, Nagarkar-Jaiswal S, et al. (2018). Neuron-Subtype-Specific Expression, Interaction Affinities, and Specificity Determinants of DIP/Dpr Cell Recognition Proteins. Neuron 100, 1385–1400 e1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Krishnaswamy A, De la Huerta I, and Sanes JR (2014). Type II cadherins guide assembly of a direction-selective retinal circuit. Cell 158, 793–807. [DOI] [PubMed] [Google Scholar]
- Feldheim DA, and O’Leary DD (2010). Visual map development: bidirectional signaling, bifunctional guidance molecules, and competition. Cold Spring Harb Perspect Biol 2, a001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JG (2006). Neural map specification by gradients. Curr Opin Neurobiol 16, 59–66. [DOI] [PubMed] [Google Scholar]
- Golic KG, and Lindquist S (1989). The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59, 499–509. [DOI] [PubMed] [Google Scholar]
- Hassan BA, and Hiesinger PR (2015). Beyond Molecular Codes: Simple Rules to Wire Complex Brains. Cell 163, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg M (1971). Separation of receptor and lamina potentials in the electroretinogram of normal and mutant Drosophila. J Exp Biol 55, 85–100. [DOI] [PubMed] [Google Scholar]
- Hong W, Mosca TJ, and Luo L (2012). Teneurins instruct synaptic partner matching in an olfactory map. Nature 484, 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber AB, Kolodkin AL, Ginty DD, and Cloutier JF (2003). Signaling at the growth cone: ligand-receptor complexes and the control of axon growth and guidance. Annu Rev Neurosci 26, 509–563. [DOI] [PubMed] [Google Scholar]
- Huberman AD, Clandinin TR, and Baier H (2010). Molecular and cellular mechanisms of lamina-specific axon targeting. Cold Spring Harb Perspect Biol 2, a001743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschfeld K (1967). Die Projektion der optischen Umwelt auf das Raster der Rhabdomere im Komplexauge von Musca. Exp Brain Res 3, 248–270. [DOI] [PubMed] [Google Scholar]
- Kolodkin AL, and Tessier-Lavigne M (2011). Mechanisms and molecules of neuronal wiring: a primer. Cold Spring Harb Perspect Biol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy A, Yamagata M, Duan X, Hong YK, and Sanes JR (2015). Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature 524, 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, and Lee T (2006). Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci 9, 703–709. [DOI] [PubMed] [Google Scholar]
- Langen M, Agi E, Altschuler DJ, Wu LF, Altschuler SJ, and Hiesinger PR (2015). The Developmental Rules of Neural Superposition in Drosophila. Cell 162, 120–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley JN (1895). Note on Regeneration of Prae-Ganglionic Fibres of the Sympathetic. J Physiol 18, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Herman T, Clandinin TR, Lee R, and Zipursky SL (2001). N-cadherin regulates target specificity in the Drosophila visual system. Neuron 30, 437–450. [DOI] [PubMed] [Google Scholar]
- Lee RC, Clandinin TR, Lee CH, Chen PL, Meinertzhagen IA, and Zipursky SL (2003). The protocadherin Flamingo is required for axon target selection in the Drosophila visual system. Nat Neurosci 6, 557–563. [DOI] [PubMed] [Google Scholar]
- Luthy K, Ahrens B, Rawal S, Lu Z, Tarnogorska D, Meinertzhagen IA, and Fischbach KF (2014). The irre cell recognition module (IRM) protein Kirre is required to form the reciprocal synaptic network of L4 neurons in the Drosophila lamina. J Neurogenet 28, 291–301. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA (1996). Ultrastructure and quantification of synapses in the insect nervous system. J Neurosci Methods 69, 59–73. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, and O’Neil SD (1991). Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. J Comp Neurol 305, 232–263. [DOI] [PubMed] [Google Scholar]
- Mosca TJ, Hong W, Dani VS, Favaloro V, and Luo L (2012). Trans-synaptic Teneurin signalling in neuromuscular synapse organization and target choice. Nature 484, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca TJ, and Luo L (2014). Synaptic organization of the Drosophila antennal lobe and its regulation by the Teneurins. Elife 3, e03726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Baldwin D, Hannaford S, Palka J, and Montell C (2002). Defective proboscis extension response (DPR), a member of the Ig superfamily required for the gustatory response to salt. J Neurosci 22, 3463–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome TP, Asling B, and Dickson BJ (2000). Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127, 851–860. [DOI] [PubMed] [Google Scholar]
- Ozkan E, Carrillo RA, Eastman CL, Weiszmann R, Waghray D, Johnson KG, Zinn K, Celniker SE, and Garcia KC (2013). An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell 154, 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecot MY, Tadros W, Nern A, Bader M, Chen Y, and Zipursky SL (2013). Multiple interactions control synaptic layer specificity in the Drosophila visual system. Neuron 77, 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Santiago IJ, Ahn C, Gur B, Tsui CK, Su Z, Xu C, Karakhanyan A, Silies M, and Pecot MY (2018). Drosophila Fezf coordinates laminar-specific connectivity through cell-intrinsic and cell-extrinsic mechanisms. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Caldwell JC, Eberl DF, and Clandinin TR (2005). Drosophila N-cadherin mediates an attractive interaction between photoreceptor axons and their targets. Nat Neurosci 8, 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Alba M, Vitaladevuni SN, Mishchenko Y, Lu Z, Takemura SY, Scheffer L, Meinertzhagen IA, Chklovskii DB, and de Polavieja GG (2011). Wiring economy and volume exclusion determine neuronal placement in the Drosophila brain. Curr Biol 21, 2000–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins EM, Krupp AJ, Perez de Arce K, Ghosh AK, Fogel AI, Boucard A, Sudhof TC, Stein V, and Biederer T (2010). SynCAM 1 adhesion dynamically regulates synapse number and impacts plasticity and learning. Neuron 68, 894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, and Yamagata M (1999). Formation of lamina-specific synaptic connections. Curr Opin Neurobiol 9, 79–87. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe T, Borycz JA, Meinertzhagen IA, and Clandinin TR (2014). Differential adhesion determines the organization of synaptic fascicles in the Drosophila visual system. Curr Biol 24, 1304–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe T, Neuert H, and Clandinin TR (2013). A network of cadherin-mediated interactions polarizes growth cones to determine targeting specificity. Cell 154, 351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, and Bargmann CI (2003). The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell 112, 619–630. [DOI] [PubMed] [Google Scholar]
- Shen K, Fetter RD, and Bargmann CI (2004). Synaptic specificity is generated by the synaptic guidepost protein SYG-2 and its receptor, SYG-1. Cell 116, 869–881. [DOI] [PubMed] [Google Scholar]
- Sperry RW (1963). Chemoaffinity in the Orderly Growth of Nerve Fiber Patterns and Connections. Proc Natl Acad Sci U S A 50, 703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC (2017). Synaptic Neurexin Complexes: A Molecular Code for the Logic of Neural Circuits. Cell 171, 745–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Zhang KX, Pecot MY, Nagarkar-Jaiswal S, Lee PT, Takemura SY, McEwen JM, Nern A, Xu S, Tadros W, et al. (2015). Ig Superfamily Ligand and Receptor Pairs Expressed in Synaptic Partners in Drosophila. Cell 163, 1756–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier-Lavigne M, and Goodman CS (1996). The molecular biology of axon guidance. Science 274, 1123–1133. [DOI] [PubMed] [Google Scholar]
- Tuthill JC, Nern A, Holtz SL, Rubin GM, and Reiser MB (2013). Contributions of the 12 neuron classes in the fly lamina to motion vision. Neuron 79, 128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, Zhang W, Sudhof TC, and Brose N (2006). Neuroligins determine synapse maturation and function. Neuron 51, 741–754. [DOI] [PubMed] [Google Scholar]
- Venkatasubramanian L, Guo Z, Xu S, Tan L, Xiao Q, Nagarkar-Jaiswal S, and Mann RS (2018). Stereotyped Terminal Axon Branching of Leg Motor Neurons Mediated by IgSF Proteins DIP-α and Dpr10. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatasubramanian L, Guo Z, Xu S, Tan L, Xiao Q, Nagarkar-Jaiswal S, and Mann RS (2019). Stereotyped terminal axon branching of leg motor neurons mediated by IgSF proteins DIP-alpha and Dpr10. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Durrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, et al. (2006). Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron 49, 833–844. [DOI] [PubMed] [Google Scholar]
- Werkhoven Z, Rohrsen C, Qin C, Brems B, and de Bivort B (2019). MARGO (Massively Automated Real-time GUI for Object-tracking), a platform for high-throughput ethology. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Xiao Q, Cosmanescu F, Sergeeva AP, Yoo J, Lin Y, Katsamba PS, Ahlsen G, Kaufman J, Linaval NT, et al. (2018). Interactions between the Ig-Superfamily Proteins DIP-alpha and Dpr6/10 Regulate Assembly of Neural Circuits. Neuron 100, 1369–1384 e1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KX, Tan L, Pellegrini M, Zipursky SL, and McEwen JM (2016). Rapid Changes in the Translatome during the Conversion of Growth Cones to Synaptic Terminals. Cell Rep 14, 1258–1271. [DOI] [PubMed] [Google Scholar]
- Zinn K, and Ozkan E (2017). Neural immunoglobulin superfamily interaction networks. Curr Opin Neurobiol 45, 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze [datasets/code]