Abstract
Synthetic fibres are used in place of the natural grass worldwide, for realizing playgrounds, soccer fields and even domestic gardens or recreational structures. An intensive use of artificial turf is currently observed in sports facilities, due to lower costs, higher sustainability in recycling of materials, and advantages related to athletic practice and performance. However, even if chemical and physical risks were studied, the microbiological component was not fully addressed, especially considering a comprehensive evaluation of the microbiota in synthetic vs natural playground surfaces. Here, we investigated the microbial community present on soccer fields, using Next Generation Sequencing and a 16S amplicon sequencing approach. Artificial and natural turfs show own ecosystems with different microbial profiles and a mean Shannon's diversity value of 2.176 and 2.475, respectively. The bacterial community is significantly different between facilities (ANOSIM: R = 0.179; p < 0.001) and surface materials (ANOSIM: R = 0.172; p < 0.005). The relative abundance of potentially pathogenic bacterial OTUs was higher in synthetic than in natural samples (ANOVA, F = 2.2). Soccer fields are characterized by their own microbiota, showing a different 16S amplicon sequencing signature between natural and artificial turfs.
Keywords: Bioinformatics, Genetics, Microbiology, Molecular biology, Toxicology
1. Introduction
Artificial turfs are surfaces of synthetic fibres more and more frequently used in place of the natural grass (Watterson, 2017; Fleming, 2011). Synthetic pitches are commercialized worldwide for realizing playgrounds, soccer, football or rugby fields and even in domestic gardens or recreational structures. Their successful diffusion is supported by lower costs, higher sustainability in materials reuse, water saving, and other advantages related to athletic practice and performance (Sánchez-Sánchez et al., 2018; Burillo et al., 2014). Starting from the 60s several synthetic materials were developed, and the technology was continuously and globally evolving. In recent years, several raw resources were used as a backing from jute to nylon or polypropylene, but these surfaces were considered too stiff and abrasive (Sandkuehler et al., 2010). The third generation of turfs was introduced in the 2000s and is characterized by long and less densely packed tufts fibres as well as an infill comprising elastomeric material, such as crumb rubber (Severn et al., 2011; Emery et al., 2016). The synthetic turfs are assembled with natural products such as cork or coir, a coconut-derived material, sand and crumb rubber as “soil” or “infills”, but not grass substitutes (Hongling et al., 2014). Therefore, a continuous progress in the field generated very different matrices that were extensively applied as pitches for different uses in place of natural grass.
The intensive diffusion of this technology in sport facilities raised several doubts on possible health risks, both injuries, chemical and microbiological hazards for users and environments (Watterson, 2017; Perkins et al., 2019). Recent studies have showed higher rates of abrasion injuries on artificial turf surfaces compared to natural grass playing fields (Twomey et al., 2018; Meyers, 2013; Williams et al., 2016). The debate about chemical hazards of artificial pitches and playgrounds is very current topic between manufacturers, suppliers, purchasers, workers and users of different playgrounds (Etrma, 2016; Moore, 2014). Several studies have identified potential hazards associated with synthetic turf such as heavy metals, volatile organic compounds, polycyclic aromatic hydrocarbons including benzopyrenes and phthalates (Anderson et al., 2006; Perkins et al., 2019; Celeiro et al., 2018). However, hazards associated with artificial turfs were not definitely confirmed and scientists, public health operators, consumers or environment agencies still continue to provide additional updates on studies or regulations (Macfarlane et al., 2015; USEPA, 2016; EHHI, 2016). The microbiological risk has been less investigated, even if several studies raised a possible association between turf burns and infections in injured athletes, identifying the synthetic turf as a possible source of pathogens, including community-acquired methicillin-resistant Staphylococcus aureus and other antibiotic resistant microorganisms (CDC, 2003; Kirkland and Adams, 2008; Cohen, 2008). It was suggested that the turf infill may represent a favourable niche for the accumulation and selection of bacteria species, especially if maintenance is not regularly and appropriately performed (Bass and Hintze, 2008).
Here, we report a description of the microbiological profile of synthetic vs natural turfs used on soccer fields, suggesting possible applications in safety and management issues.
2. Methods
2.1. Study design and sampling
We collected a total of 51 samples from 11 facilities in the area of Rome, involving 7 artificial and 5 natural FIFA-regulatory fields. Of these, 11 samples (collected from 3 of these facilities) were excluded because did not pass the quality controls due to low quantity or low quality in the isolated DNA. Flocked swabs (FS) with the active dying system (model 4N6FLOQSwabs, Copan) were used to obtain samples (n = 40). At each soccer field, three points were sampled: center mark (CM), penalty area (PA), corner arc (CA). Each site was swabbed for between 10 and 15 s. Moreover, grass/sand (GS) samples (n = 11), approximately 3–10 mg, were directly collected in the lateral play area. Due to low quantity or quality in DNA extraction, 11 samples were excluded (10 FS and 1 GS sample). Failed DNA isolation or amplification may have been due to multiple factors: e.g. low DNA concentrations, presence of PCR inhibitors (i.e. humic substances, phenolic compounds, pesticides, detergents, etc). Finally, we performed the study on 40 samples that were processable: 27 from artificial and 13 from natural soccer fields, respectively corresponding to 7 artificial and 3 natural soccer fields. Table 1 reports descriptive metadata collected for each sample, including date, location and climate in the time of sampling. Samples were transferred on ice to laboratory (within 24 h) and stored at 4 °C until to extraction.
Table 1.
Descriptive metadata was collected for each sample, including date, location and climate in the time of sampling.
| ID sample | Sport Facilities | Sampling Time | Material | Sampling point |
|---|---|---|---|---|
| C1 | I | nov-16 | Synthetic turf | Center Mark |
| C2 | I | nov-16 | Synthetic turf | Corner Arc |
| C13 | I | nov-16 | Synthetic turf | Grass/sand play area |
| C3 | II | nov-16 | Synthetic turf | Center Mark |
| C4 | II | nov-16 | Synthetic turf | Corner Arc |
| C5 | II | nov-16 | Synthetic turf | Penalty Area |
| C14 | II | nov-16 | Synthetic turf | Grass/sand play area |
| C12 | II | nov-16 | Natural grass | Center Mark |
| C37 | II | Jan-18 | Natural grass | Center Mark |
| C38 | II | Jan-18 | Natural grass | Penalty Area |
| C39 | II | Jan-18 | Natural grass | Corner Arc |
| C49 | II | Jan-18 | Natural grass | Grass/sand play area |
| C6 | III | nov-16 | Synthetic turf | Center Mark |
| C7 | III | nov-16 | Synthetic turf | Corner Arc |
| C8 | III | nov-16 | Synthetic turf | Penalty Area |
| C15 | III | nov-16 | Synthetic turf | Grass/sand play area |
| C9 | IV | nov-16 | Synthetic turf | Center Mark |
| C10 | IV | nov-16 | Synthetic turf | Corner Arc |
| C11 | IV | nov-16 | Synthetic turf | Penalty Area |
| C16 | IV | nov-16 | Synthetic turf | Grass/sand play area |
| C28 | V | Jan-18 | Synthetic turf | Center Mark |
| C29 | V | Jan-18 | Synthetic turf | Penalty Area |
| C30 | V | Jan-18 | Synthetic turf | Corner Arc |
| C46 | V | Jan-18 | Synthetic turf | Grass/sand play area |
| C31 | VI | Jan-18 | Synthetic turf | Center Mark |
| C32 | VI | Jan-18 | Synthetic turf | Penalty Area |
| C33 | VI | Jan-18 | Synthetic turf | Corner Arc |
| C47 | VI | Jan-18 | Synthetic turf | Grass/sand play area |
| C34 | VII | Jan-18 | Synthetic turf | Center Mark |
| C35 | VII | Jan-18 | Synthetic turf | Penalty Area |
| C36 | VII | Jan-18 | Synthetic turf | Corner Arc |
| C48 | VII | Jan-18 | Synthetic turf | Grass/sand play area |
| C40 | VIII | Jan-18 | Natural grass | Center Mark |
| C41 | VIII | Jan-18 | Natural grass | Penalty Area |
| C42 | VIII | Jan-18 | Natural grass | Corner Arc |
| C50 | VIII | Jan-18 | Natural grass | Grass/sand play area |
| C43 | IX | Jan-18 | Natural grass | Center Mark |
| C44 | IX | Jan-18 | Natural grass | Penalty Area |
| C45 | IX | Jan-18 | Natural grass | Corner Arc |
| C51 | IX | Jan-18 | Natural grass | Grass/sand play area |
2.2. DNA extraction and purification
After sample collection, each swab was inserted into the semipermeable NAO Baskets and was broken inside at the breakpoint. The samples were pretreated with glass beads® (Sigma Aldrich, USA) and 200 μl of Lysozyme Solution® (Sigma Aldrich, USA), adapting the protocol as previous described (Valeriani et al., 2017, 2018a and b). In a second phase we followed the standard protocol procedure of GenElute® Bacterial Genomic DNA Kit (Sigma Aldrich, USA). For GS, the DNA extraction by pellets was performed with GenElute ® Bacterial Genomic DNA Kit, following the manufacturer's instruction. Sterile swabs were used as extraction and amplification controls.
2.3. 16S amplicon sequencing
Samples were prepared according to the “16S Metagenomic Sequencing Library Preparation” guide (Part# 15044223 rev. A; Illumina, San Diego, CA, USA). Briefly, the primers containing Illumina adapter and linker sequence and targeting the V1–V2 regions of bacterial 16S rRNA genes were used (Valeriani et al., 2018b; Wen et al., 2017). Three libraries with unique tags were generated for each sample as technical replicates. Each amplification reaction had a total volume of 25 μl containing 12.5 μl of KAPA HiFi HotStart ReadyMix (Roche, Pleasanton, CA), 5 μl of each primer (1 μlM), and 2 μl template DNA. Reactions were carried out on a Techne®TC-PLUS thermalcycler (VWR International, LLC, Radnor, USA). Thermal cycling conditions were as follows: an initial denaturation at 95 °C for 3 min, and 25 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, with a final extension at 72 °C for 5 min. Following amplification, 5 μl of PCR product from each reaction was used for agarose gel (1%) electrophoresis to confirm amplification. The cleaned libraries were quantified through DeNovix dsDNA fluorescence quantification kits (DeNovix Inc., DE, USA) and validated on Bioanalyzer DNA 1000 chip (Agilent, Santa Clara, CA. USA). The final concentration of libraries was prepared using the MiSeq Reagent Kit Preparation Guide (Illumina, San Diego, CA, USA). Briefly, the combined sample library was diluted to 4 nM, denatured with 0.2 N fresh NaOH, diluted to 4 pM by addition of Illumina HT1 buffer, and then mixed with an equal volume of 4 pM PhiX (sequencing control) (Illumina, San Diego, CA, USA). The library (600 μl) was loaded with read 1, read 2nd index sequencing primers on a 500-cycle (2 × 250 paired ends) reagent cartridge (Illumina), and run on a MiSeq desktop sequencer (Illumina San Diego, CA, USA).
2.4. Analysis of sequences and statistics
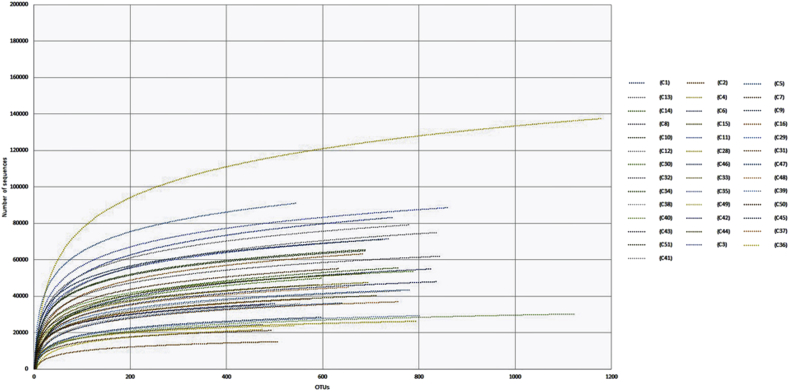
Raw sequence data was processed using an in-house pipeline which was built on the Galaxy platform and incorporated various software tools to evaluate the quality of the raw sequence data (e.g. FastQC, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). All data sets were rigorously screened to remove low quality reads (short reads >200 nt, zero-ambiguous sequences). Demultiplexing was performed to remove PhiX sequences and sort sequences; moreover, to minimize sequencing errors and ensure sequence quality, the reads were trimmed based on the sequence quality score using Btrim (Kong, 2011). OTUs were clustered at a 97% similarity level and final OTUs were generated based on the clustering results and taxonomic annotation of individual OTUs was based on representative sequences using RDP's 16S Classifier 2.5. Rarefaction curves were calculated for each sample (Fig. 1) as previously reported (Wen et al., 2017). Relative abundances of community members were determined with rarefied data and summarized at each taxonomic level (cut off 0.2%). The sequence reads were analyzed, also, in the cloud environment BaseSpace through the 16S Metagenomics app (version 1.0.1; Illumina®): the taxonomic database used was the Illumina-curated version (May 2013 release of the Greengenes Consortium Database). Microbiota phylogenic distribution was analyzed using Metagenassist (Arndt et al., 2012): input files were produced in the format of CSV files, including taxonomic profile file with taxonomic abundance for each sample and further file, containing metadata information (e.g. material of turf). Multivariate analysis, the PCoA and partial least square-discriminant analysis (PLS-DA) were performed in order to investigate the dissimilarity between groups, using METAGENassist platform. We performed feature selection using PLS-DA and 10-fold cross validation to tune algorithm parameters and to check model validity (in this analysis I facility is not considered because is compound by only three sampling points). The putative functional profiles based on the 16S community composition were investigated by automated taxonomic-to-phenotypic mapping using a METAGENassist platform and NCBI microbial taxonomy (Arndt et al., 2012). Based on Greengenes taxonomic assignment, pathogens were calculated considering those OTUs with a >97% sequence similarity respect to known human pathogens (NIH Human Microbiome Project; Knights et al., 2011). The result is presented as a heatmap. Agglomerative hierarchical clustering was performed by treating everyone as a separate cluster and then proceeds to combine them until all samples belong to a single cluster (the data are analyzed for human pathogen phenotype, Arndt et al., 2012). Beta-diversity clustering was analyzed using ANOSIM for categorical variables (material, facility and sampling point). Analysis of variance (ANOVA) was run to assess significant differences in relative abundance of OTUs on different categories. A PERMDISP test was run to assess the significance of beta-diversity distribution variation between sample types. To assess sequencing depth, alpha rarefaction plots were done in mothur (v 1.31.1) and R (version 3.1.3) using packages ‘ggplot2’ and ‘vegan’ (R Core team 2013).
Fig. 1.
Rarefaction curves calculated for each sample, showing an adequate and reliable sampling and sequencing effort for describing the bacterial community.
3. Results
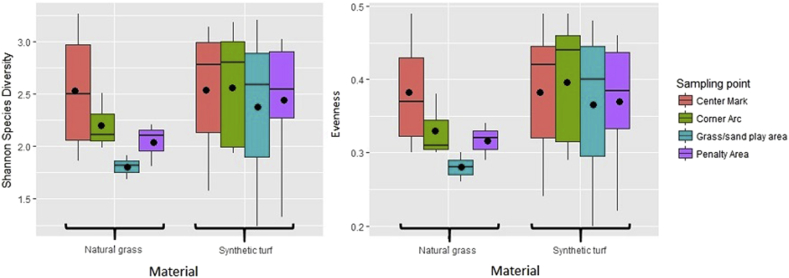
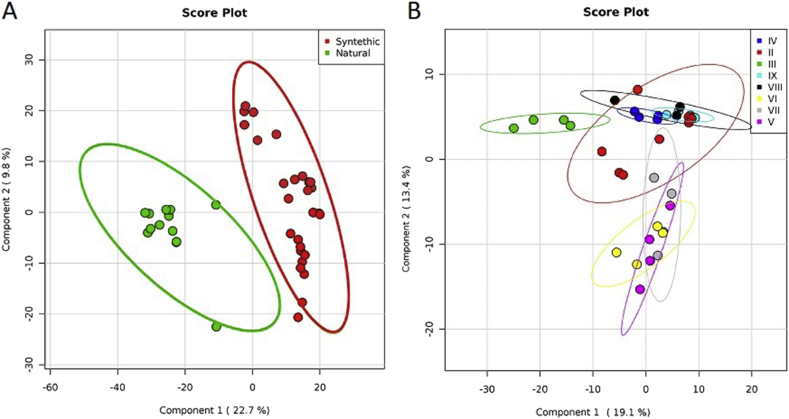
A total of 2,268,361 sequences were generated from 40 samples (Table 1 and detailed results of the amplicon sequencing analysis are shown in Table 2). The number of sequences for each sample ranged from 27,268 to 471,205 leading to the identification of 1181 OTUs defined at 97% identity. Rarefaction curves were calculated for each sample (Fig. 1), showing an adequate and reliable sampling and sequencing effort for describing the bacterial community (Wen et al., 2017). Interestingly, several reads resulted as unknown representing 21.8% and 6.69% in synthetic and natural turfs, respectively. We performed the analysis of the significance of alpha diversity measures between sample groups (Fig. 2). Overall, there was no specific trend detected in the richness and diversity of taxa between samples. Indeed, as determined by the Shannon species diversity and the Evenness index, the complexity of bacterial communities varied slightly between type of materials. The different sampling points collected from natural fields show very narrower confidence interval, except for the CM point. The mean Shannon's diversity values for the synthetic soccer fields exhibited slightly higher diversity level (Natural: H = 2.176 ± 0.456 and E = 0.33 ± 0.06; Synthetic H = 2.475 ± 0.161 and E = 0.38 ± 0.088). Bacterial community composition was significantly different between facilities (ANOSIM: R = 0.179; p < 0.001). Similar results were obtained using the Principal Component Analysis (PCA) to assess clustering (and potential separation) of facilities (Fig. 3a). The bacterial community pooled by the different types of materials was significantly different (R = 0.172; p < 0.005). The partial least square-discriminant analysis (PLS-DA) depicts a Pearson distance showing a separation between the natural and synthetic soccer field, suggesting a noticeable shift in community structures among the two groups of turfs (Fig. 3b). The analysis of similarities trough ANOSYM between several sampling points showed no diversification between the points in the same location within the field (e.g. all CM respect to all CA or PA; ANOSIM: R = -0.111; p > 0.6). However, when performing the analysis within the same facility we can detect a dissimilarity between each of the different locations (e.g. CM, CA or PA; ANOSIM: R = 0.513; p < 0.001), suggesting that the microbiota composition may consistently be influenced by both the facility and the location within the playground.
Table 2.
Summary of NGS analysis after quality assessment of sequences.
| Samples | Number Reads (Passing filter) | Shannon (Species Diversity) | Number of Species (Identified) | Evenness |
|---|---|---|---|---|
| C1 | 250,451 | 2.784 | 746 | 0.42 |
| C2 | 170,372 | 2.882 | 684 | 0.44 |
| C13 | 218,213 | 2.927 | 778 | 0.44 |
| C3 | 260,439 | 3.078 | 861 | 0.46 |
| C4 | 203,130 | 3.121 | 795 | 0.47 |
| C5 | 95,282 | 3.011 | 779 | 0.45 |
| C14 | 143,017 | 2.855 | 598 | 0.45 |
| C6 | 145,551 | 2.908 | 824 | 0.43 |
| C7 | 27,268 | 2.803 | 495 | 0.45 |
| C8 | 69,188 | 2.583 | 590 | 0.40 |
| C15 | 133,129 | 2.589 | 688 | 0.40 |
| C9 | 53,454 | 3.142 | 597 | 0,49 |
| C10 | 108,481 | 3.182 | 690 | 0.49 |
| C11 | 173,499 | 3.025 | 740 | 0.46 |
| C16 | 91,324 | 3.203 | 758 | 0.48 |
| C12 | 218,172 | 3.268 | 836 | 0.49 |
| C28 | 471,205 | 2.145 | 1181 | 0.30 |
| C29 | 195,921 | 2.190 | 886 | 0.32 |
| C30 | 145,530 | 1.935 | 789 | 0.29 |
| C46 | 58,667 | 1.832 | 502 | 0.29 |
| C31 | 29,025 | 2.128 | 507 | 0.34 |
| C32 | 138,279 | 2.516 | 843 | 0.37 |
| C33 | 105,569 | 2.042 | 696 | 0.31 |
| C47 | 80,684 | 1.967 | 640 | 0.30 |
| C34 | 148,085 | 1.576 | 758 | 0.24 |
| C35 | 39,718 | 1.328 | 435 | 0.22 |
| C36 | 36,546 | 1.943 | 476 | 0.32 |
| C48 | 108,348 | 1.243 | 579 | 0.20 |
| C37 | 113,812 | 2.132 | 654 | 0,33 |
| C38 | 85,091 | 1.810 | 555 | 0.29 |
| C39 | 191,71 | 1.986 | 802 | 0.30 |
| C49 | 66,060 | 1.913 | 543 | 0.30 |
| C40 | 136,100 | 2.871 | 1125 | 0.41 |
| C41 | 85,696 | 2.107 | 761 | 0.32 |
| C42 | 105,308 | 2.115 | 837 | 0.31 |
| C50 | 111,098 | 1.688 | 633 | 0.26 |
| C43 | 66,734 | 1.864 | 481 | 0.30 |
| C44 | 83,162 | 2.209 | 714 | 0.34 |
| C45 | 58,294 | 2.511 | 727 | 0.38 |
| C51 | 105,428 | 1.819 | 690 | 0.28 |
Fig. 2.
A box plot presentation of the distribution of Shannon and Evenness indices of the four groups in natural and synthetic turf.
Fig. 3.
a) Principal coordinate analysis (PCoA) scatterplot of the normalized relative abundance of all samples, divided by type of material (red: synthetic and green: natural). Data are plotted at the genus-level classification. b) Partial least square-discriminant analysis (PLS-DA) depicts Pearson distance between different samples using phylogeny distribution based on 16S rRNA genes. Samples are, respectively, coloured according to sampling points.
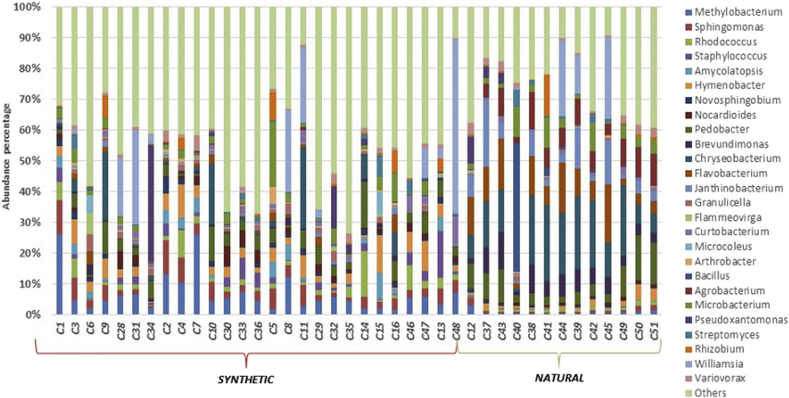
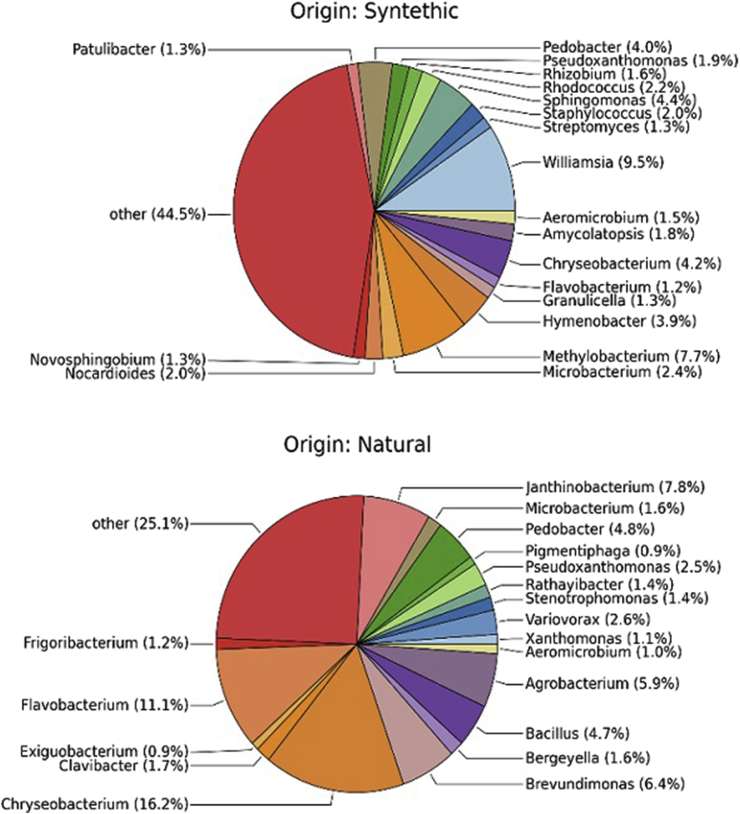
Regarding the observed genera, Chryseobacterium was the predominant one, accounting for 8.8% of total effective bacterial sequences in all samples, both synthetic and natural (Figs. 4 and 5). Other dominant genera include other environmental bacteria such as Flavobacterium (5%) and Pedobacter (3.6%). However, when samples were grouped by type of turf, synthetic kinds harbored a different microbiota (Fig. 5). In these samples median UniFrac distances were larger, and the distributions were wider, suggesting that the beta-diversity variance was significantly greater and probably influenced by anthropic and environmental contaminations. OTUs closely related to the genus Williamsia were significantly enriched on synthetic samples (9.5 % relative abundance versus <0.1 in natural turfs; p value < 0.001). The genera Methylobacterium, Sphingomonas, Hymenobacter and Rhodococcus were also abundant in synthetic turfs (respectively 7.7 %, 4.4 %, 3.9 %, 2.2% relative abundance). In particular, Hymenobacter and Rhodococcus are representative of a possible avian or soil contamination, respectively; supporting the hypothesis for a higher maintenance of pathogens and or anthropic/animal contaminations in synthetic turfs. Moreover, Staphylococcus, Nocardioides and Streptomyces were detected in synthetic turfs (relative abundance: 2 %, 2 %, 1.3 %, respectively). Conversely, the natural surfaces were found to have a microbial community structure much more comparable to those present in phyllosphere environments, with the lowest median UniFrac distance between samples and a narrow distribution of these distances. Chryseobacterium was observed in all samples but more present on artificial mats (16.2 % relative abundance versus <0.1 in natural turf; p value < 0.001). In natural turfs, Flavobacterium, Janthinobacterium, Brevundimonas, Agrobacterium and Bacillus (relative abundance: 11.1 %, 7.8 %, 6.4 %, 5.9% and 4.7%, respectively) were also detected.
Fig. 4.
Summary of bacterial community abundance of the associated with type of material at each sampling point. To simplify community representation, OTUs less than 1% were discarded and count in the other. While each surface displays a unique community structure, surfaces were similar across all facilities.
Fig. 5.
Microbial community in synthetic and natural turfs. Representative diagram reporting data at genus level for all known genera. The subgroup “other” includes all genera below 1% abundance. Unknown genera represent 21.8% and 6.69% in synthetic and natural turfs, respectively (data not shown).
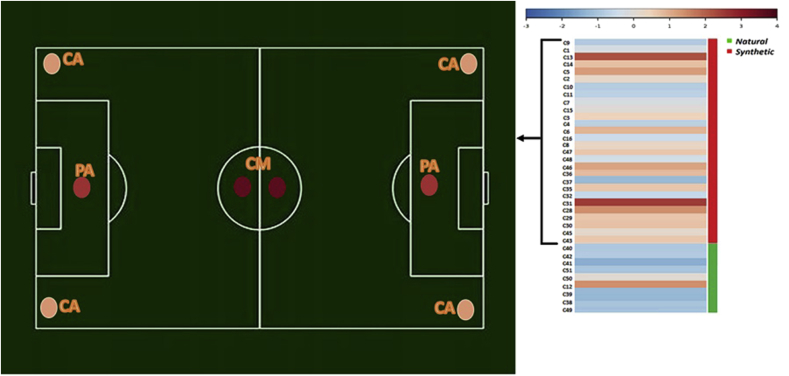
The analysis of potentially pathogenic genera resulted in a higher relative abundance of OTUs corresponding to opportunistic bacteria more in synthetic (23%) than in natural samples (13%) as summarized in Fig. 6. Indeed, samples from synthetic fields showed OTUs related to human associated bacteria, so that taxa that are commonly found in the human microbiome, were found more frequently, including members of the families Burkholderiaceae, Pseudoxantomonadaceae, Staphylococcaceae and genera Staphylococcus. Moreover, the relative abundance of potentially pathogenic bacteria was independent from the sampling point along the field but significantly varied between the different sport facilities (ANOVA, F = 2.2), suggesting a role for the general environment, anthropic contamination, everyday use and management of the playgrounds.
Fig. 6.
Heat map of pathogen percentage. Agglomerative hierarchical clustering was performed (the data are analyzed for human pathogen phenotype). Pathogen were calculated considering those OTUs with a >97% sequence similarity respect to known human pathogens (NIH Human Microbiome Project). In field soccer picture were only considered the synthetic turf samples and the average value for each sampling point. CM: center mark, PA: penalty area, CA: corner arc.
4. Discussion
The chemical and physical risks related to the use of synthetic fields have been investigated for several years, but the biological aspects were not yet fully clarified (CDC, 2003; Kirkland and Adams, 2008). The complexity of the novel materials used for synthetic fields can represent a condition for harboring specific bacteria communities. The role of infill structures was even associated with the accumulation of potential pathogenic organisms and the infectious risk after injuries (Kirkland and Adams, 2008; Cohen, 2008; Bass and Hintze, 2008). 16S amplicon sequencing analysis of synthetic and natural fields confirmed the presence of pathogens but also revealed the existence of a specific microflora (Fig. 5). Interestingly, unknown sequences were over three times more frequent in synthetic vs natural turfs (about 22% vs 7%, respectively), suggesting a specific unknown component present in artificial carpets respect to the well characterize microflora reported in soil and grass (Giampaoli et al., 2014; Nurulita et al., 2016).
This is the first study using 16S amplicon sequencing to characterize the microbiota of soccer fields turfs. The observed data suggested a possible microbial signature own of synthetic vs natural fields. Therefore, athletes, workers and users are exposed to different microbial communities based on the composition of the carpet. The 16S amplicon sequencing approach allowed also the detection of sequences corresponding to common microbial indicators (e.g. Staphylococcus), in agreement with other studies performed in sport facilities using traditional culture-based methods (CDC, 2003; Kirkland and Adams, 2008; Cohen, 2008; Bass and Hintze, 2008). We would have expected synthetic turfs as an adverse environment for microorganisms, but the whole of the observed results showed no different trends in richness and biodiversity distribution of taxa respect to natural grass. The synthetic soccer fields exhibited even a moderately higher mean Shannon's values. A possible explanation can be found in the contamination of the infill with organic materials, but also in the presence of carbohydrates, amino acids, aliphatic and aromatic acids, fatty acids, that can be released and likely to be a driving force in the structure of the microbial biodiversity within the artificial niche (Nurulita et al., 2016; Prescott and Grayston, 2013; Xue and Huang, 2014; Krashevska et al., 2015). No major differences were observed in alfa-diversity index, but the dissimilarity was evident when using the beta-diversity indicators, supporting a 16S signature approach in characterizing synthetic turfs. Phyllosphere genera were observed in the natural turfs (e.g. Janthinobacterium, Agrobacterium, Variovorax, Pedobacter) as expected because of the presence of soil and grass (Simon et al., 2019; Hassani et al., 2018). Several of these bacteria were detected also in synthetic turfs (Figs. 4 and 5), even if less representatively. Being ubiquitous, they could easily contaminate the artificial carpets becoming part of their microbial community, as observed on the surfaces of other synthetic matrices. Interestingly, bacteria from different sources can be found in synthetic turfs, but not conversely in natural ones. Different synthetic materials already were shown to provide a cozy microenvironment to harbour bacteria from anthropic, animal (e.g. Staphylococcus, Streptomyces, Nocardioldes, Hymenobacter), or other natural sources (Williamsia, Chryseobacterium, Rhodococcus) (Mafu et al., 1990; Carniello et al., 2018; Sharma et al., 2018; Masoud, 2017). Therefore, a major factor driving beta-diversity variance in artificial surfaces may likely be due to contamination with human sweat or saliva as well as from the natural microflora in the surrounding area. This was not observed in natural turfs probably due to the competition driven by the rich endophytic microflora (Simon et al., 2019; Hassani et al., 2018; Mafu et al., 1990). Mesophilic bacteria, including pathogens, were detected more frequently in the penalty area and centre circle of synthetic turfs, even if the analysis of similarities for the several sampling points showed no changes in microflora profile. These results suggest that microbial communities fluctuate around a common biodiversity centroid, as already reported for other sport plants (Wood et al., 2015). However, within the same facility clear differences can be observed between different sampled areas. The whole of observed results suggests that in synthetic fields the microbial community structure is primarily defined by the anthropic contamination. Management, use, and maintenance of the facility may also play a major role in determining the microbial load and its composition. Infill materials can represent a potential source for bacterial grow posing putatively higher infection risks respect to natural fields, as previously reported for cased of cutaneous infections in soccer players using synthetic turfs (CDC, 2003; Kirkland and Adams, 2008; Cohen, 2008). The microbiota is not an absolute entity, but it represents the result of a complex interaction between the availability of natural microorganisms, the properties of that ecological niche and the influence of different environmental factors. In particular, turfs microbiota can be influenced by local factors (e.g. maintenance products e.g. detergents or pesticides, respectively for synthetic vs natural carpets) or other external factors (e.g. anthropic, animal or environmental pollutants). Some biological pollutants are traceable by microbiota analysis and were detected as possible contaminants in the synthetic turfs (e.g. bacteria of human, animal or soil origin). Otherwise, the microbiota itself can represent a promising approach for detecting traces of contaminants such as biological fluids, feces, plants or other contaminants in different materials and matrices (Valeriani et al., 2018a; Miletto and Lindow, 2015; Leung and Lee, 2016; Mucci et al., 2019). However, exposure to several chemical factors such as volatile organic compounds, particulate matter, polycyclic aromatic hydrocarbons as well as physical factors including ultraviolet radiation, temperature, humidity may represent interfering factors in microbiota formation and stability. A specific issue concerns the micro-conditions within the infill structure of different synthetic materials after exposure to different external factors. Our data focus on the 16S signature of the microbiota and are not so extended to address all the different raising issues. Results from samples coming from different areas, show the presence of a common core structure of the microbiota in synthetic turfs but cannot provide significative evidence for possible differences due to the complexity of the exposure to external factors. Further and more extended studies are required to address this issue in different playgrounds, starting also from the availability of the present report and available dataset. The 16S amplicon sequencing characterization of turf surfaces may represent a new marker for studying the biological component of synthetic turfs and finally improve management and hygiene in environments for sport and recreational facilities.
5. Conclusions
Soccer playgrounds surfaces are characterized by their own microbiota, showing a different 16S amplicon sequencing signature between natural and artificial turfs. For the first time we report a microbiota analysis of turfs commonly used for playing soccer, football, rugby or other sports as well as recreational and urban playgrounds. Synthetic soccer fields harbor a microflora from anthropic and environmental sources whereas the traditional natural grass carpets show a soil-related microbial community. Understanding the microbial component in different materials will eventually provide information on their ecology and on the potential health impact of exposed athletes or maintenance workers, both indirectly exposed and through possible injuries. In addition to the several studies addressing the physical and chemical risks related to the synthetic turfs, here we report some data on the biological component and on the application of high throughput sequencing on DNA samples from playground surfaces. Since synthetic fibers made to look like natural grass are often used also for other recreational or furnishing decoration purposes in private and public areas, further advances in the field may provide knowledge for risk assessment and tools for appropriate management and maintenance of synthetic turfs.
Declarations
Author contribution statement
Frederica Valeriani: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Lory Marika Margarucci: Analyzed and interpreted the data.
Gianluca Gianfranceschi, Antonello Ciccarelli, Filippo Tajani: Performed the experiments.
Nicolina Mucci, Maurizio Ripani: Conceived and designed the experiments.
Vincenzo Romano Spica: Conceived and designed the experiments; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- Anderson M.E., Kirkland K.H., Guidotti T.L., Rose C. A case study of tire crumb use on playgrounds: risk analysis and communication when major clinical knowledge gaps exist. Environ. Health Perspect. 2006;114:1–3. doi: 10.1289/ehp.7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt D., Xia J., Liu Y. METAGENassist: a comprehensive web server for comparative metagenomics. Nucleic Acids Res. 2012;40:88–95. doi: 10.1093/nar/gks497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J.J., Hintze D.W. Determination of microbial populations in a synthetic turf system. Skyline – Big Sky Undergrad. J. 2008;1(Suppl 1) http://www.synturf.org/images/Bass_and_Hintze_viewcontent.pdf Available at: [Google Scholar]
- Burillo P., Gallardo L., Felipe J.L., Gallardo A.M. Artificial turf surfaces: perception of safety, sporting feature, satisfaction and preference of football users. Eur. J. Sport Sci. 2014;14(1):437–447. doi: 10.1080/17461391.2012.713005. [DOI] [PubMed] [Google Scholar]
- Carniello V., Peterson B.W., van der Mei H.C., Busscher H.J. Physico-chemistry from initial bacterial adhesion to surface-programmed biofilm growth. Adv. Colloid Interface Sci. 2018;261:1–14. doi: 10.1016/j.cis.2018.10.005. [DOI] [PubMed] [Google Scholar]
- CDC, Centers for Disease Control and Prevention Centers for disease control and prevention. Methicillin-resistant Staphylococcus aureus infections among competitive sports participants. Colorado, Indiana, Pennsylvania, and Los Angeles county, 2000–2003. MMWR Morb. Mortal. Wkly. Rep. 2003;52:793–795. [PubMed] [Google Scholar]
- Celeiro M., Dagnac T., Llompart M. Determination of priority and other hazardous substances in football fields of synthetic turf by gas chromatography-mass spectrometry: a health and environmental concern. Chemosphere. 2018;195:201–211. doi: 10.1016/j.chemosphere.2017.12.063. [DOI] [PubMed] [Google Scholar]
- Cohen P.R. The skin in the gym: a comprehensive review of the cutaneous manifestations of community-acquired methicillin-resistant Staphylococcus aureus infection in athletes. Clin. Dermatol. 2008;26(1):16–26. doi: 10.1016/j.clindermatol.2007.10.006. [DOI] [PubMed] [Google Scholar]
- EHHI . 2016. Environment & Human Health, Inc. Reports to Promote Policy Changes.www.ehhi.org Available online: [Google Scholar]
- Emery J., Driscoll H.F., Barnes A., James D.M. Third generation artificial pitch quality in commercial football centers. Procedia Eng. 2016;147:860–865. [Google Scholar]
- ETRMA . 2016. Safety of Recycled Rubber Infill Material. Brussels.http://www.etrma.org/uploads/Modules/Documentsmanager/20161017_etrma_crumb-rubber_vf-2.pdf Available online: [Google Scholar]
- Fleming P. Artificial turf systems for sport surfaces: current knowledge and research needs. Proc. Inst. Mech. Eng. P J. Sport. Eng. Technol. 2011;225:43–63. [Google Scholar]
- Giampaoli S., Berti A., Di Maggio R.M., Pilli E., Valentini A., Valeriani F., Gianfranceschi G., Barni F., Ripani L., Romano Spica V. The environmental biological signature: NGS profiling for forensic comparison of soils. Forensic Sci. Int. 2014;240:41–47. doi: 10.1016/j.forsciint.2014.02.028. [DOI] [PubMed] [Google Scholar]
- Hassani M.A., Durán P., Hacquard S. Microbial interactions within the plant holobiont. Microbiome. 2018;6(1):58. doi: 10.1186/s40168-018-0445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongling Y., Baicun Z., Heng L., Weiguang G., Ting W., Weishan W. Nanosilica and olyethylene based artificial turf-abrasion resistance and mechanical properties. Procedia Eng. 2014;72:901–906. [Google Scholar]
- Kirkland E.B., Adams B.B. Methicillin-resistant Staphylococcus aureus and athletes. J. Am. Acad. Dermatol. 2008;59(3):494–502. doi: 10.1016/j.jaad.2008.04.016. [DOI] [PubMed] [Google Scholar]
- Knights D., Kuczynski J., Charlson E.S., Zaneveld J., Mozer M.C., Collman R.G. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods. 2011;8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y. Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics. 2011;98(2):152–153. doi: 10.1016/j.ygeno.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Krashevska V., Klarner B., Widyastuti R., Maraun M., Scheu S. Impact of tropical lowland rainforest conversion into rubber and oil palm plantations on soil microbial communities. Biol. Fertil. Soils. 2015;51:697–705. [Google Scholar]
- Leung M.H., Lee P.K. The roles of the outdoors and occupants in contributing to a potential pan-microbiome of the built environment: a review. Microbiome. 2016;4(1):21. doi: 10.1186/s40168-016-0165-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane R., Carrasco C., Alam Y., Archbold J. Toronto Public Health; Toronto, ON, Canada: 2015. Health Impact Assessment of the Use of Artificial Turf in Toronto. [Google Scholar]
- Mafu A.A., Roy D., Goulet J., Savoie L., Roy R. Efficiency of sanitizing agents for destroying Listeria monocytogenes on contaminated surfaces. J. Dairy Sci. 1990;73(12):3428–3432. doi: 10.3168/jds.S0022-0302(90)79040-6. [DOI] [PubMed] [Google Scholar]
- Masoud K. Williamsia spp. are emerging opportunistic bacteria. New Microb. New Infect. 2017;21:88–89. doi: 10.1016/j.nmni.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers M.C. Incidence, mechanisms, and severity of match-related collegiate women’s soccer injuries on fieldturf and natural grass surfaces: a 5-yearprospective study. Am. J. Sports Med. 2013;41(10):2409–2420. doi: 10.1177/0363546513498994. [DOI] [PubMed] [Google Scholar]
- Miletto M., Lindow S.E. Relative and contextual contribution of different sources to the composition and abundance of indoor air bacteria in residences. Microbiome. 2015;3:61. doi: 10.1186/s40168-015-0128-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. Rubber and Plastics News; Akron, OH, USA: 2014. Industry Defends Use of Crumb Rubber in Artificial Surfaces. [Google Scholar]
- Mucci N., Gianfranceschi G., Cianfanelli C., Santucci S., Romano Spica V., Valeriani F. Can air microbiota be a novel marker for public health? A sampling model and preliminary data from different environments. Aerobiologia. 2019 [Epub ahead of print] [Google Scholar]
- Nurulita Y., Adetutu E.M., Kadali K.K., Zul D., Mansur A.A., Ball A.S. The assessment of the impact of oil palm and rubber plantations on the biotic and abiotic properties of tropical peat swamp soil in Indonesia. Int. J. Agric. Sustain. 2016;13:150–166. [Google Scholar]
- Perkins A.N., Inayat-Hussain S.H., Deziel N.C., Johnson C.H., Ferguson S.S., Garcia-Milian R., Thompson D.C.-, Vasiliou V. Evaluation of potential carcinogenicity of organic chemicals in synthetic turf crumb rubber. Environ. Res. 2019;49:163–172. doi: 10.1016/j.envres.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott C.E., Grayston S.J. Tree species influence on microbial communities in litter and soil: current knowledge and research needs. For. Ecol. Manag. 2013;309:19–27. [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2013. A Language and Environment for Statistical Computing.http://www.R-project.org [Google Scholar]
- Sánchez-Sánchez J., García-Unanue J., Gallardo A.M., Gallardo L., Hexaire P., Felipe J.L. Effect of structural components, mechanical wear and environmental conditions on the player–surface interaction on artificial turf football pitches. Mater. Des. 2018;140:172–178. [Google Scholar]
- Sandkuehler P., Torres E., Allgeuer Th. Polyolefin Materials and technology in artificial turf I: yarn developments. Sport. Technol. 2010;3(3):52–58. [Google Scholar]
- Severn K., Fleming P., Clarke J.D., Carre M.J. Science of synthetic turf surfaces: investigating traction behaviour. Proc. Inst. Mech. Eng. P J. Sport. Eng. Technol. 2011;225:147–158. [Google Scholar]
- Sharma R., Sharma K., Sawhney R. Evidence of variable bacterial colonization on coloured elastomeric ligatures during orthodontic treatment: an intermodular comparative study. J Clin Exp Dent. 2018;10(3):271–278. doi: 10.4317/jced.54610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J.C., Marchesi J.R., Mougel C., Selosse M.A. Host-microbiota interactions: from holobiont theory to analysis. Microbiome. 2019;7(1):5. doi: 10.1186/s40168-019-0619-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey D.M., Petrass L.A., Fleming P., Lenehan K. Abrasion injuries on artificial turf: a systematic review. J. Sci. Med. Sport. 2018;1440–2440(18):31127. doi: 10.1016/j.jsams.2018.11.005. [DOI] [PubMed] [Google Scholar]
- USEPA, United States Environmental Protection Agency, 2017 . USEPA/CDC/USCPSC 2017; USEPA; Washington, DC, USA: 2016. Federal Research Action Plan on Recycled Tire Crumb Used on Playing Fields and Playgrounds: Status Report. [Google Scholar]
- Valeriani F., Agodi A., Casini B., Cristina M.L., D'Errico M.M., Gianfranceschi G., Liguori G., Liguori R., Mucci N., Mura I., Pasquarella C., Piana A., Sotgiu G., Privitera G., Protano C., Quattrocchi A., Ripabelli G., Rossini A., Spagnolo A.M., Tamburro M., Tardivo S., Veronesi L., Vitali M., Romano Spica V. Potential testing of reprocessing procedures by real-time polymerase chain reaction: a multicenter study of colonoscopy devices. Am. J. Infect. Contr. 2018;46(2):159–164. doi: 10.1016/j.ajic.2017.08.008. [DOI] [PubMed] [Google Scholar]
- Valeriani F., Cianfanelli C., Gianfranceschi G., Santucci S., Romano Spica V., Mucci N. Monitoring biodiversity in libraries: a pilot study and perspectives for indoor air quality. J Prev Med Hyg. 2017;58(3):238–251. [PMC free article] [PubMed] [Google Scholar]
- Valeriani F., Crognale S., Protano C., Gianfranceschi G., Orsini M., Vitali M., Romano Spica V. Metagenomic analysis of bacterial community in a travertine depositing hot spring. New Microbiol. 2018;41(2):126–135. [PubMed] [Google Scholar]
- Watterson A. Artificial turf: contested terrains for precautionary public health with particular reference to Europe? Int. J. Environ. Res. Public Health. 2017;14:1050. doi: 10.3390/ijerph14091050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C., Wu L., Qin Y., Van Nostrand J.D., Ning D., Sun B., Zhou J. Evaluation of the reproducibility of amplicon sequencing with Illumina MiSeq platform. PLoS One. 2017;12(4) doi: 10.1371/journal.pone.0176716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S., Trewartha G., Kemp S.P., Michell R., Stokes K.A. The influence of an artificial playingsurface on injury risk and perceptions of muscle soreness in elite Rugby Union. Scand. J. Med. Sci. Sport. 2016;26(1):101–108. doi: 10.1111/sms.12402. [DOI] [PubMed] [Google Scholar]
- Wood M., Gibbons S.M., Lax S., Eshoo-Anton T.W., Owens S.M., Kennedy S., Gilbert J.A., Hampton-Marcell J.T. Athletic equipment microbiota are shaped by interactions with human skin. Microbiome. 2015;3:25. doi: 10.1186/s40168-015-0088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D., Huang X. Changes in soil microbial community structure with planting years and cultivars of tree peony (Paeonia suffruticosa) World J. Microbiol. Biotechnol. 2014;30:389397. doi: 10.1007/s11274-013-1457-3. [DOI] [PubMed] [Google Scholar]