Abstract
L-type Ca2+ channels (LTCCs) drive the bulk of voltage-gated Ca2+ entry in vertebrate inner ear hair cells (HCs) and are essential for mammalian auditory processing. LTCC currents have been implicated in neurotransmitter release at the HC afferent active zone, the ribbon synapse. It is likely that LTCCs play a direct role in vesicle fusion; however, the subcellular localization of the channels in HCs has not been fully resolved. Via positional cloning, we show that mutations in a zebrafish LTCC encoding gene, cav1.3a, underlie the auditory-vestibular defects of gemini (gem) circler mutants. gem homozygous receptor mutant HCs display normal cell viability, afferent synaptogenesis, and peripheral innervation, yet exhibit strongly reduced extracellular potentials (∼50% of wild-type potentials). Apical FM1-43 uptake, however, is unaffected in gem mutant HCs, suggesting that mechanotransduction channels are functional. Using a Gem-specific antibody, we show that the bulk of Gem/Cav1.3a immunoreactivity in HCs is restricted to basally located focal spots. The number and location of focal spots relative to nerve terminals, and their remarkable ring-shaped structure, which is reminiscent of synaptic dense bodies, are consistent with Gem/Cav1.3a channels clustering at HC ribbon synapses.
Keywords: gemini, L-type calcium channels, deafness, hair cell, ribbon synapse, microphonics, synaptic transmission
Introduction
Ca2+ entry in sensory hair cells (HCs) of the vertebrate inner ear contributes to multiple essential steps in auditory processing, including mechanotransduction (transduction current and adaptation), frequency tuning, efferent modulation, and neurotransmitter release onto postsynaptic afferent terminals (Hudspeth, 1989; Lenzi and Roberts, 1994; Engel et al., 2002; Fuchs et al., 2003). Transmitter release by HCs is exceptionally fast and tonic (Moser and Beutner, 2000; Beutner et al., 2001; Glowatzki and Fuchs, 2002) and relies on a structurally specialized active zone, the ribbon synapse (RS). HC RSs are characterized by the presence of a sphere-shaped proteinaceous organelle (also known as dense body) adjacent to the presynaptic membrane. Dense bodies are ∼300 nm in diameter and are surrounded by a halo of tethered synaptic vesicles (Lenzi and von Gersdorff, 2001; Lenzi et al., 2002; Fuchs et al., 2003). Despite their essential role in photoreceptor neurotransmission (Allwardt et al., 2001; Altrock et al., 2003; Dick et al., 2003), the mechanism by which synaptic ribbons orchestrate sensory receptor neurotransmission remains unclear (Parsons and Sterling, 2003). Tonic transmission by HCs implies numerous and sustained vesicle fusion events at the active zone and therefore sets high demands on the capacity of HCs to produce elevated Ca2+ levels adjacent to RSs (Zenisek et al., 2003).
Ca2+ imaging and electrophysiological studies have indicated that the bulk of Ca2+ influx in HCs occurs in hotspots located in discrete regions of the basolateral membrane. These hotspots, which presumably correspond to Ca2+ channel clusters, might coincide with HC presynaptic zones (Roberts et al., 1990; Issa and Hudspeth, 1994; Tucker and Fettiplace, 1995; Martinez-Dunst et al., 1997; Zenisek et al., 2003). In support of this hypothesis, large intramembrane particles have been observed in the vicinity of RSs in a number that agrees with the predicted number of Ca2+ channels per HC (Roberts et al., 1990). Hence, the majority of the Ca2+ channels of the HC may localize near ribbons, thereby providing the amount of Ca2+ entry that is necessary for HC neurotransmission.
The Ca2+ influx that triggers transmitter release at HC RSs is likely to be controlled by voltage-gated channels and is blocked by L-type Ca2+ channel (LTCC) antagonists [e.g., dihydropyridines (DHPs)], suggesting that LTCCs participate in HC transmitter release (Zhang et al., 1999; Moser and Beutner, 2000; Spassova et al., 2001; Robertson and Paki, 2002). Consistent with this hypothesis, the predominant Ca2+ channel in avian HCs is an LTCC, the class D LTCC (Cav1.3, also known as α1D/CACNA1D) (Kollmar et al., 1997b), and mice carrying a targeted null allele of the encoding gene display profound congenital deafness (Platzer et al., 2000; Namkung et al., 2001). In a recent study, Brandt and colleagues (2003) demonstrated that HC exocytosis is reduced in dissected organs of Corti from Cav1.3-/- mice, suggesting a direct role in neurotransmission (Brandt et al., 2003). Cav1.3 immunoreactivity has been detected in a broad region of the basolateral membrane of chinchilla HCs (Lopez et al., 1999), and a recent report states a potential colocalization with the SNARE [SNAP (synaptosome-associated protein of 25 kDa) receptor] protein syntaxin, yet in a nonprecisely identified region of the HC membrane (Song et al., 2003). In HCs, a significant number of vesicle fusion events possibly involving syntaxin occur outside RSs (Zenisek et al., 2003). Hence, it remains to be shown whether Cav1.3 effectively localizes at HC RSs. In this report, we show that mutations in cav1.3a are responsible for auditory-vestibular defects in zebrafish gemini mutants, and we present the subcellular localization of this channel in HCs.
Materials and Methods
Zebrafish strains. The tc123d mutation was initially isolated in an N-ethyl-N-nitrosourea (ENU) large-scale mutagenesis screen for locomotion behavior-defective mutants (Granato et al., 1996). Additional studies established tc123d as the founder of the gemini (gem) complementation group of zebrafish vestibular (“circler”) mutants (Nicolson et al., 1998). The gem tn004 allele was recovered from an ENU-based noncomplementation screen after a survey of 321 genomes. The dominant-viable albino line, which produces embryos devoid of pigment, was used for in situ hybridization experiments at larval stages. Larvae were raised in E3 medium (in mm: 5 NaCl, 0.17 KCl, 0.33 CaCl2, and 0.33 MgCL2)at 28°C. All mutations were maintained in the heterozygous state in standard Tu background. The WIK polymorphic strain was used for genetic mapping.
Phalloidin, FM1-43, and acridine orange labeling. Five days postfertilization (dpf), larvae were fixed in 4% paraformaldehyde (PFA) at 4°C for 24 hr, permeabilized in 2% Triton X-100 and 4% PFA overnight at 4°C, and stained with Alexa Fluor-conjugated phalloidin (Molecular Probes, Eugene, OR) at a 1:100 dilution (2.5 mg/ml) in PBS and 0.1% Tween 20 (PBST) for an additional night at 4°C. Larvae were then washed at least four times for 20 min in PBST at room temperature in the dark and were mounted for confocal microscopy (see below). Staining of live larvae (5 dpf) with the styryl dye FM1-43 (Molecular Probes) were performed as described previously using a 20 sec exposure time (Seiler and Nicolson, 1999). For acridine orange labeling, live larvae (7 dpf) of given genotype were rinsed several times in E3, incubated in 5 μg/ml acridine orange (Sigma-Aldrich, St. Louis, MO) for 2 min, and rinsed again two to three times. FM1-43- and acridine orange-labeled larvae were examined under a fluorescence microscope using 10-20× objectives and a 63× water lens.
Electrophysiological recordings. Response currents in HCs were recorded from tail lateral line (LL) neuromasts (NMs) as extracellular potentials [“microphonics” (Flock, 1965)] essentially as described previously (Nicolson et al., 1998). Isolated tails were placed in Ringer's solution containing 100 nm tetrodotoxin, and a patch pipette (filled with Ringer's solution; 1-5 MΩ) was placed at the base of a hair bundle. Potentials were recorded with an Axoclamp 2B amplifier (Axon Instruments, Foster City, CA) in bridge mode, amplified 2000-fold, and digitized at 10 kHz. Mechanical stimulation of HCs was performed using a fluid jet (Denk and Webb, 1992). A micropipette (tip diameter, 50-150 μm) filled with Ringer's solution was oriented along the anteroposterior axis and placed 50-200 μm away from the NM. Fluid fluxes were generated by bidirectional sinusoidal modulations of the pressure within the pipette. Pressure intensity was adjusted under visual control to produce deflections of the hair bundle >10° to saturate HC responses. Data were recorded for 500 msec. After 150 msec, a fluid jet stimulus (20 Hz for 200 msec) was applied. At least 200 trials were averaged for each NM and bandpass filtered at 7-48 Hz.
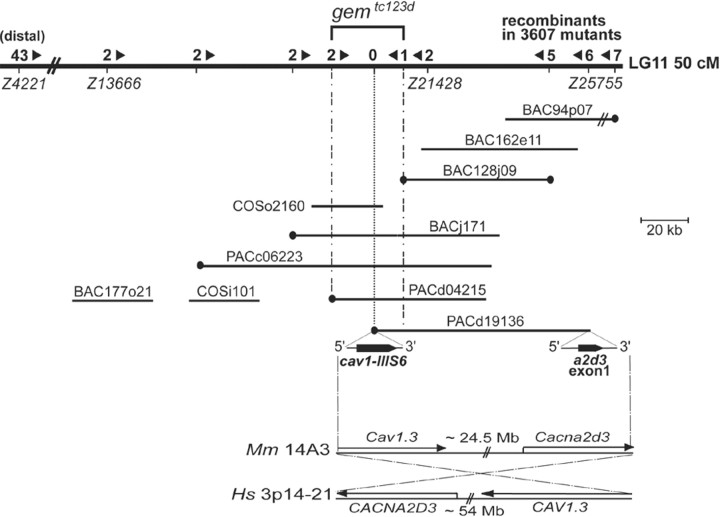
Positional cloning. Meiotic mapping lines (Tutc123d/WIK) were generated by crossing gemtc123d heterozygotes to homozygous WIK fish. Their progeny was scored for auditory-vestibular defects (e.g., acoustic startle reflex and circling behavior) at 5 dpf as described previously (Granato et al., 1996; Nicolson et al., 1998) (see mapping scheme at http://www.eb.tuebingen.mpg.de/dept3/geisler/). Larval genomic DNA was prepared and used for PCR as described previously (Geisler, 2002). DNA pools from 48 mutant and wild-type sibling larvae were used for bulked segregation analysis (Michelmore et al., 1991) whereby simple sequence length polymorphisms (SSLPs) from a low-density genome-wide scanning panel were used to test for linkage to tc123d. Z4190 [linkage group (LG) 11, 40.5 cM] was found syntenic to the gem locus. High-resolution genetic mapping was performed by genotyping 3607 individual mutant F2s (7214 meioses) with additional SSLPs in the area (http://zebrafish.mgh.harvard.edu/zebrafish/index.htm), the loci of which were confirmed by radiation hybrid (RH) mapping (Goodfellow T51 zebrafish RH panel; Invitrogen, Carlsbad, CA) (Kwok et al., 1998). Recombination events at SSLPs Z4221 and Z21428 defined an initial tc123d critical interval of 4 cM (∼3 Mb), with the nearest marker, Z21428, located at an estimated physical distance of ∼20 kb (0.03 cM; two recombinants per 3607 mutants) proximal to the mutation. This marker was used to screen P1 artificial chromosome [Resource Center for the German Genome Project (RZPD), Berlin, Germany], bacterial artificial chromosome (BAC) (BioCat, Heidelberg, Germany), yeast artificial chromosome (Research Genetics, Huntsville, AL), and cosmid (COS) (RZPD) PCR-able libraries for initiating the assembly of a physical contig through the gem locus. Clones were obtained from the RZPD and were purified using the Qiagen (Hilden, Germany) Large-Construct kit and the PSIψ Clone Big BAC DNA isolation kit (Princeton Separations, Adelphia, NJ). Clone ends were sequenced using standard methods and screened for polymorphic markers [SSLPs, simple sequence length polymorphisms, and single nucleotide polymorphisms (SNPs)]. Newly generated markers and novel SSLPs mapped to the area (such as Z13666, recombinants at which were used to close the contig distal to tc123d) were used to genotype recombinants, to establish clone overlaps, and for additional rounds of library screening. Chromosomal walk orientation was confirmed by PCR screening contiguous genomic clones with markers proximal to Z21428 (e.g., Z25755) (see Fig. 2). Clone inserts were sized by pulse-field gel electrophoresis as described previously (Geisler, 2002). Although we could not establish a tiling path covering the entire gem interval, the retention of two Z13666 crossovers at the distal end of PACd04215 and of one Z21428 recombinant at the distal end of BAC128j09 implied that the tc123d mutation resided in a small genomic region containing the LTCC-encoding distal end of PACd19136 (see Fig. 2). PACd19136 was shotgun sequenced at The Wellcome Trust Sanger Institute (Hinxton, Cambridge, UK).
Figure 2.
Integrated genetic and physical map of the gem locus on zebrafish LG11 and syntenic relationships with mammalian genomes. A genomic region of ∼5 cM located at 50 cM from the top of LG11 is shown. The number of recombinants between tested markers and tc123d are shown above the chromosome line. Arrowheads indicate the direction to the mutation. The assembled physical contig of PAC, BAC, and COS clones across the mutant locus is shown underneath the LG line. The scale bar indicates clone insert sizes. BAC94p07 (60 kb) is not drawn to scale. Clone ends depicted by black circles contained Tu/WIK polymorphism(s) and were used as markers to genotype recombinants. The gem critical interval was defined by the retention of one proximal and two distal recombinants at markers d04215/Sp6 and 128j09/T7, respectively. Exons harbored by both ends of PACd19136 are magnified under the clone line, with their respective 5′-3′ orientations. Bottom, Conservation of the cav1/a2d3 synteny in zebrafish, mouse (Mm) (chromosome 14, A3 region; ∼24 Mb from the proximal telomere) and human (Hs) (chromosome 3, p14-21 region; ∼54 Mb from the proximal telomere) genomes.
Cloning of zebrafish cav1.3a and cav1.3b. Reverse-transcription (RT)PCR (RT-for-PCR kit; Clontech, Palo Alto, CA) and rapid amplification of cDNA ends (RACE) (SMART and MARATHON RACE cDNA amplification kits; Clontech) were used to assemble a full-length cav1.3a cDNA. At each step, RACE products were excised from agarose gels, extracted (Nucleotrap; Macherey-Nagel, Easton, PA), subcloned in the TA cloning pCRII-TOPO vector (Invitrogen), sequenced, and blasted to GenBank. The assembled cav1.3a 3′ end sequence [d19136 distal exon-3′ untranslated region (UTR)] was confirmed by long-range PCR (LRPCR) on cDNA (Advantage II PCR system; Clontech) and by alignments with PACd19136 shotgun reads. Surprisingly, the last rounds of cav1.3a 5′ rapid amplification of cDNA ends PCR-yielded products with an apparently premature, in-frame stop codon when compared with other vertebrate cav1.3 orthologs; predicted protein sequence upstream of this stop codon aligned well to higher vertebrate Cav1.3 N termini. However, the recent cloning of a cav1.3 ortholog from trout (Ramakrishnan et al., 2002) indicates that this apparent stop codon is conserved among teleosts and likely belongs to the 5′ UTR. Genscan (http://genes.mit.edu/GENSCAN.html), Promoter 2.0, and Netstart 1.0 (http://www.cbs.dtu.dk/services/) analyses of the zebrafish genome draft assembly (z06) contigs (http://www.ensembl.org/Daniorerio/) were further consistent with the gene being complete. Full-length cav1.3b was mainly deduced from Blast and Genscan searches of z06 followed by LR-PCRs on cDNA. Protein sequence alignments and phylogenetic analyses were performed using ClustalW and Phylip algorithms (http://clustalw.genome.ad.jp/). The domain structure of Cav1.3a and Cav1.3b proteins were deduced from SMART analysis (http://smart.embl-heidelberg.de) and from sequence comparisons with previously proposed mammalian, avian, and teleost Cav1.3 domains (Soldatov, 1992; Kollmar et al., 1997b; Ramakrishnan et al., 2002). Template cDNAs used in all PCRs above were obtained from 5 dpf larval total RNA extracts (Nucleospin RNAII; Macherey-Nagel) using standard or RACE first-strand cDNA synthesis RT reactions according to the instructions of the manufacturer. The adult expression pattern of cav1.3a was assessed by semiquantitative RT-PCR of total RNA pools from homogenates of various adult tissues. GenBank accession numbers are AY528224 (cav1.3a) and AY528225 (cav1.3b).
Mutational analysis. Total RNA extracts from at least three independent batches of tc123d and tn004 homozygotes and from their wild-type siblings served as templates for RT-PCR using primers amplifying overlapping fragments covering the entire cav1.3a ORF. PCR products were sequenced for mutational analysis. To distinguish natural SNPs from potential ENU-induced mutations, mutant and wild-type sibling sequences were compared using Lasergene software from DNASTAR (Madison, WI). The tc123d nonsense mutation occurs ∼400 bp downstream of the PACd19136 distal exon, consistent with the mapping data implicating this exon as being tightly linked to the mutation. The tn004 missense mutation was not found in all other tested zebrafish genetic backgrounds, including WIK, and no additional mutation other than silent or shared with other backgrounds, all yielding conservative amino acid polymorphisms, could be detected in cav1.3a transcripts of either mutant allele. Both mutations were confirmed by sequencing PCR products from reactions using genomic DNA as template.
In situ hybridization. cav1.3a (nucleotides 3734-4537) and cav1.3b (nucleotides 5653-6101) digoxygenin-labeled (Roche Diagnostics, Mannheim, Germany) sense and antisense riboprobes were transcribed in vitro and used for whole-mount in situ hybridization essentially as described previously (Hauptmann and Gerster, 1994). Five days postfertilization, albino larvae were treated 25 min in 10 μg/ml proteinase K to obtain efficient labeling of internal tissues, e.g., the pancreas. In ∼5% of stained larvae, strong staining was unexpectedly observed on the left side of specimens, i.e., opposite to the right location of the pancreas, and in place of the liver. We believe, however, that it is the pancreas and not the liver that was stained in these larvae and that these specimens reflected the low occurrence of heterotaxia observed in several fish species. In some experiments, stained larvae were Technovit embedded, sectioned, and labeled with nuclear fast red (Vector Laboratories, Burlingame, CA). No signals were detected with any of the cav1.3a and cav1.3b control sense probes.
Electron microscopy. Whole larvae (5 dpf) of given genotype were anesthetized with 0.02% 3-aminobenzoic acid ethyl ester and then fixed by immersion in 2.0% glutaraldehyde and 1.0% PFA in normal solution (in mm: 145 NaCl, 3 KCl, 1.8 CaCl2, and 10 HEPES, pH 7.2) overnight to several days at 4°C. Specimens were fixed with 1.0% OsO4 in H2O for 10 min on ice, followed by fixation and contrast with 1.0% uranyl acetate for 1 hr on ice, and then dehydrated with several steps in ethanol and embedded in Epon. Ultrathin sections of gemtc123d homozygous mutant specimens (n = 5) and wild-type siblings (n = 3) were stained with lead citrate and uranyl acetate and were examined using a Philips CM 10 electron microscope.
Antibodies. The Gem polyclonal antibody (pAb) Gem25.2 was generated by immunizing rabbits with the Cav1.3a-specific peptide NH2-(C)KSKKQGSSANTRPQR-COOH (zebrafish Cav1.3a amino acids 42-56) (SEQLAB, Göttingen, Germany). Crude Gem25.2 antiserum was used for immunohistochemistry (diluted 1:1000) because affinity-purified Gem25.2 eluates produced considerably weaker stainings. Preimmune antiserum did not produce a signal. In peptide competition experiments, the Gem25.2 antiserum (1:1000) was preadsorbed with 0.1 μg/ml peptide antigen (mass ratio of 10:1) for 2 hr at 37°C in PBS, the antigen-pAb mixture was centrifuged at 16,000 × g for 30 min, and the resulting supernatant was used for immunohistochemistry as described below. Staining with preadsorbed antiserum was done in parallel with labeling using crude antiserum. A mouse monoclonal anti-Synaptophysin Ab (Synaptic Systems, Göttingen, Germany), was used at a 1:10 dilution. Secondary Abs were Alexa Fluor (488, 546, 568) conjugated goat anti-rabbit IgG, goat anti-mouse IgG, and goat anti-mouse IgM Abs (all diluted 1:500) from Molecular Probes.
Whole-mount immunohistochemistry and confocal microscopy. Five days postfertilization larvae of given genotype were fixed overnight at 4°C in 4% PFA and were processed immediately for whole-mount immunohistochemistry (long-term storage in methanol or 4% PFA was detrimental to immunoreactivity). Specimens were permeabilized with 1.5% Triton X-100 overnight at 4°C in fixative, washed four times (20 min each rinse) in PBST, and blocked in 1% bovine serum albumin (BSA) in PBST for at least 1 hr at room temperature. Larvae were then incubated with the primary Abs of interest at appropriate dilutions (see above) in 1% BSA-PBST for at least 24 hr at 4°C, washed extensively in PBST, stained with appropriate combinations of Alexa Fluor-conjugated secondary Abs for a minimum of 4 hr at room temperature in the dark, washed again extensively in PBST, and mounted under thin glass fibers, on their sides, for confocal microscopy analyses (see below). Images of NM hair cells were generated with a Leica (Nussloch, Germany) DM IRB/E inverted confocal microscope using a 100× oil lens. Images are either single confocal plane sections (0.5-1 μm) or Z-stack projections of 16-32 sections scanned from basolateral to postsynaptic regions of HCs. The apicolateral region of HCs was systematically omitted from our projections because nonspecific Gem25.2 staining was observed in HC kinocilia and bases of stereocilia.
Results
Cellular basis of the gemini auditory-vestibular phenotype
Two large-scale chemical mutagenesis screens for locomotion and behavioral defects in 5 dpf zebrafish larvae led to the isolation of several mutants, including gem, exhibiting fully penetrant deafness and imbalance (“circling”) phenotypes despite normal larval morphology (Granato et al., 1996; Nicolson et al., 1998). The genes affected in circler mutants may encode proteins that are specifically required for HC function and are therefore of great interest for dissecting the molecular bases of inherited forms of nonsyndromic deafness in humans (for review, see Ashmore, 1998; Petit et al., 2001; Morton, 2002; Whitfield, 2002). The gem complementation group initially consisted of only one allele, tc123d (Nicolson et al., 1998). To facilitate the molecular characterization of the underlying gene, we conducted a small-scale noncomplementation screen and recovered an additional allele, tn004, from the inspection of ∼320 genomes. After several outcrosses, tc123d and tn004 homozygotes and tc123d/tn004 transheterozygotes exhibited identical behavioral phenotypes. Both mutations are larval lethal and segregate in a recessive pattern. In rare cases, we observed a weak semidominance associated with the tn004 allele (data not shown).
Initial analyses of gemtc123d lateral line NM HCs had shown that microphonic potentials were reduced by 70% compared with wild-type HCs, despite normal HC morphology (Nicolson et al., 1998). Microphonic potentials are a readout of the sum of stimulus-evoked transepithelial return currents that result from asymmetric ionic currents across the HC membrane (e.g., the transduction current in apical stereocilia and voltage- and time-dependent currents in the basolateral membrane) (Corey and Hudspeth, 1983). Initiation of the currents, however, depends on the presence of the transduction current that triggers depolarization (Corey and Hudspeth, 1983). Hence, the strongly affected yet detectable microphonics of tc123d HCs suggested that the mutation might generate a hypomorphic allele of a gene encoding an essential component of the transduction machinery of the HC (Nicolson et al., 1998). Alternatively, gem could encode a modulator of the transduction apparatus or a HC protein required downstream of transduction.
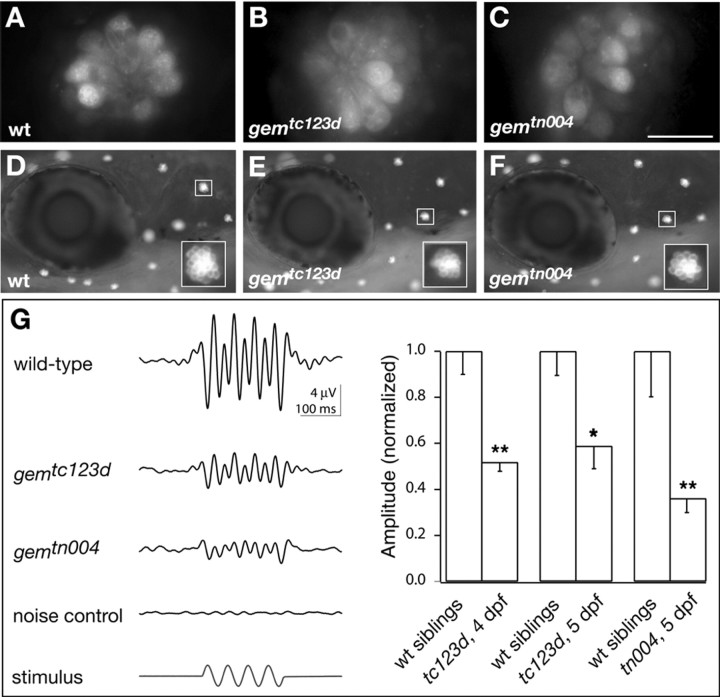
gem-homozygous HCs from both mutant alleles were examined at the cellular level (Fig. 1). Because a slight reduction in the number of HCs per sensory epithelium may account for a reduction in microphonic amplitude (Nicolson et al., 1998), we first examined whether gem mutations compromise early HC viability. We labeled live larvae (7 dpf) with the cell-death marker acridine orange. NM HC nuclei of gemtc123d and gemtn004 homozygotes were indistinguishable from those of wild-type larvae because no obvious labeling was observed (Fig. 1A-C). In contrast, the pyknotic nuclei of the HC degeneration mutant skylab (skb) (Nicolson et al., 1998) were brightly labeled at this stage (data not shown). These observations indicated that microphonic reduction in gem mutants does not arise from defects in HC survival. To investigate whether a functional defect in transduction could account for the reduction in microphonics, we labeled live gem larvae with the vital dye FM1-43 and assessed dye incorporation in NM HCs. FM1-43 uptake by zebrafish HCs is mediated by apical endocytosis, a mechanotransduction-dependent process. Zebrafish larval HCs in which transduction is abolished, such as mar mutants or nompC morphants, or zebrafish HCs exposed to blockers of the transduction channel do not take up the dye (Seiler and Nicolson, 1999; Sidi et al., 2003). Although an endocytic route of entry was also reported in guinea pig inner HCs (Griesinger et al., 2002), other studies suggest that FM1-43 may enter through transduction channels (Gale et al., 2001; Meyers et al., 2003). We compared FM1-43 uptake by gem HCs of either allele and found that it was normal (Fig. 1D-F), suggesting that transduction is not grossly affected in gem mutants.
Figure 1.
gem mutants have reduced HC extracellular receptor potentials. A-F, Acridine orange and FM1-43 labeling, respectively, of HCs in live zebrafish larvae (A-C, 7 dpf; D-F, 5 dpf) of indicated genotypes. Scale bar: (in C) A-C, 10 μm; D-F, 150 μm. G, Microphonic potentials are reduced in gem NMs. Left, Microphonic potentials in response to a saturating sinusoidal deflection of the hair bundle (20 Hz, 200 msec; see bottom trace) in wild-type (wt) and the two mutant gem alleles. In the noise control, the recording electrode and stimulus pipette were placed between NMs to assess the recording noise. Each trace was averaged over at least 200 trials and bandpass filtered between 7 and 48 Hz. The bottom trace shows the driver potential applied to the stimulus pipette obtained with the sinusoidal command function. Right, Statistical comparison of microphonic potentials in mutants and wild-type siblings. The amplitude of microphonic potentials was measured as the average of the four largest differences between adjacent positive and negative peaks during stimulation. Mean amplitudes were normalized to the mean amplitude in wild-type siblings and amounted to 52 ± 18% in tc123d at 4 dpf (mean ± SD; n = 12 mutants; n = 6 wild type), 59 ± 20% in tc123d at 5 dpf (n = 7 mutants; n = 4 wild type), and 36 ± 16% in tn004 at 5 dpf (n = 7 mutants; n = 5 wild type). The reduction of microphonic potential amplitude relative to the response of wild-type siblings was statistically significant in all cases (Wilcoxon rank-sum test). Significance levels are as follows: *p < 0.05; **p < 0.01.
We then examined microphonic potentials in gem mutants. Recordings were made extracellularly with a patch pipette while the stereocilia were deflected by a water jet from a stimulus pipette (for details, see Materials and Methods). Microphonic potentials often show pronounced response components at twice the stimulus frequency because NMs contain HCs with opposite preferred directions (Flock, 1965) (Fig. 1G, left). In tc123d homozygotes, we observed a reduction in microphonic amplitude (∼50% of wild type) but not a complete absence of microphonics (Fig. 1G, right), consistent with previous findings (Nicolson et al., 1998). tn004 mutant NM HCs showed a similar reduction in microphonic amplitude compared with wild type (Fig. 1G, right), indicating that the gem alleles we isolated are of similar strength.
gem encodes Cav1.3a
To understand how gem mutations impair HC microphonic potentials without compromising mechanotransduction activity, we sought to identify the underlying mutated gene. Bulk segregation analysis using a genome-wide scanning panel of microsatellite markers detected a significant linkage between the tc123d mutation and a marker located ∼50 cM distal to the LG11 proximal telomere. Because no obvious candidates had been mapped previously to this region, we undertook a positional cloning approach (for details, see Materials and Methods). The distal end of the genomic clone PACd19136, which physically mapped within the gem interval, contained an exon encoding an LTCC transmembrane segment (cav1-IIIS6) (Fig. 2). The proximal end of PACd19126 also contained coding sequence, with an exon encoding the N terminus of the α2δ3 subunit of LTCCs (also known as CACNA2D3), a gene that is linked to the CAV1.3 gene (also known as CACNA1D) in human and mouse genomes (Fig. 2, bottom). This conservation of synteny suggested that the cav1-IIIS6 exon we identified belonged to a zebrafish cav1.3 ortholog. In agreement with this notion, database searches indicated that the peptide with the closest homology to cav1-IIIS6 was transmembrane segment IIIS6 of chick Cav1.3 (also known as CHCACHA1D) (Kollmar et al., 1997b). In addition, cav1.3 was an excellent candidate for gem because Cav1.3 knock-out mice, similarly to gem mutant zebrafish, exhibit deafness (Platzer et al., 2000).
To investigate this hypothesis further, we conducted in situ hybridization experiments with probes derived from cav1 RACE fragments that were obtained using primers located in cav1-IIIS6. The RACE reactions yielded partial cDNAs that corresponded to at least two genes we tentatively named cav1.3a and cav1.3b. Indeed, radiation hybrid mapping revealed that a subset of RACE fragments (cav1.3b fragments) mapped to another chromosome (LG8, 100 cM distance from top). In contrast to zebrafish, mammalian genomes contain a single cav1.3 copy. Interestingly, we found that zebrafish cav1.3a and cav1.3b are expressed in mutually exclusive domains. Consistent with cav1.3a being a promising candidate for gem, it is strongly expressed in inner ear and LL sensory epithelia from 30 hours postfertilization (hpf) onward (Fig. 3A-F). In the sensory patches of the larval ear, the cristae and maculae, expression appeared to be restricted to HCs (Fig. 3E and data not shown). LL NMs exhibited a similar HC-specific expression (Fig. 3F). We did not observe cav1.3a expression in supporting cells (Fig. 3E,F) or in the auditory and LL ganglion cell bodies (Fig. 3D). Similarly to its mammalian ortholog (Williams et al., 1992; Iwashima et al., 1993; Takimoto et al., 1997), cav1.3a is also expressed in the CNS, pancreas, and retina. CNS expression is first detectable in discrete regions of the forebrain (telencephalon and thalamic and hypothalamic ventral diencephalon), midbrain, and ventral hindbrain (possibly branchial motor neurons) (Fig. 3A,B). From 48 hpf onward, cav1.3a expression spreads to most regions of the CNS (Fig. 3C,D). The onset of pancreatic expression occurs at ∼30 hpf (Fig. 3A,B), soon after the differentiation of the first insulin-expressing cells (Biemar et al., 2001). cav1.3a mRNA levels persist throughout development in pancreatic cells that presumably belong to the β-islet (Fig. 3G-I). Retinal expression initiates at 48 hpf and appears to be restricted to the inner plexiform layer, basal cells of the inner nuclear layer (amacrine cells), and the ganglion cell layer (Fig. 3H,J). In the adult zebrafish, cav1.3a is expressed in identical tissues and, in addition, at low levels in the heart (Fig. 3N). In contrast, no cav1.3b message was detected in HCs, CNS, pancreas, or heart at all stages examined (Fig. 3K,L and data not shown). Expression of cav1.3b in the eye was restricted to the outer plexiform and photoreceptor cell layers (Fig. 3M). Consistent with its expression in photoreceptors, cav1.3b message is also abundant in pinealocytes of the epiphysis from 30 hpf onward (Fig. 3K-M). Starting at 72 hpf, cav1.3b expression spreads to lateral cells of the pineal organ. These cells presumably belong to the habenular nuclei (Fig. 3L, inset) (Concha et al., 2003).
Figure 3.
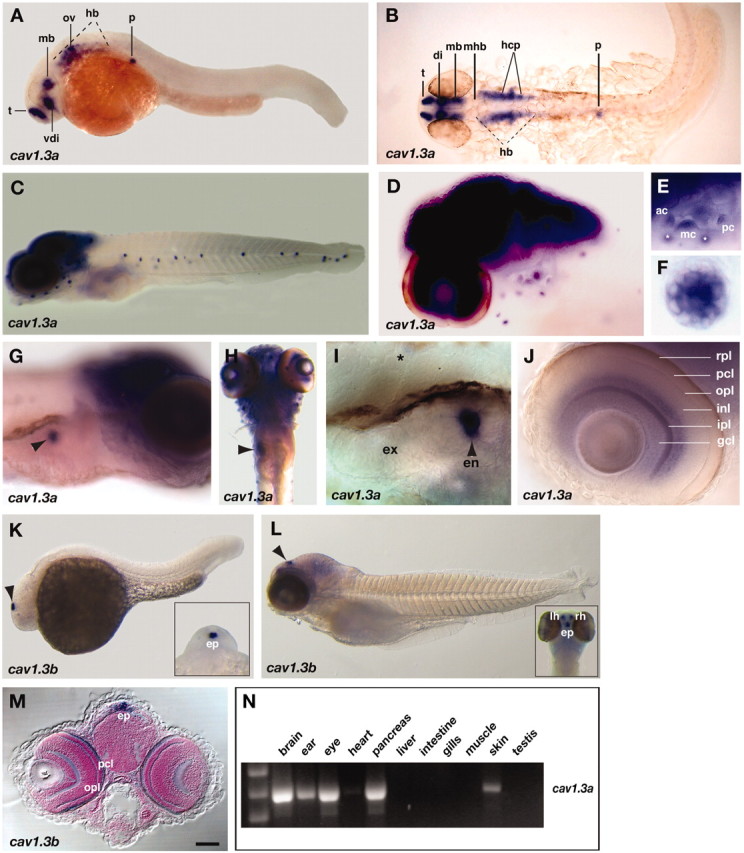
cav1.3a and cav1.3b mRNA expression patterns in the developing and adult zebrafish. mRNA (blue) was detected at embryonic (A, B, 30 hpf; K, 24 hpf) and larval (C-J, L, M, 5 dpf) stages by whole-mount in situ hybridization with cav1.3a (A-J) and cav1.3b (K-M) antisense riboprobes. A, C-G, I-L, Lateral views with anterior to the left and to the right, respectively. B, Dorsal view (anterior to the right) of the same embryo as in A; the embryo was flat mounted. H, Ventral view, anterior to the top. M, Frontal view of a larva cross-sectioned at the level of the epiphysis. E, F, I, J, Close-ups of the following organs (5 dpf): E, ear (focus is on the three cristae; white asterisks denote anterior LL NMs that are not in focus); F, anterior LL NM; I, bean-shaped pancreas. A black asterisk marks expression in a neighboring NM; J, eye. G, H, Arrows mark expression in the pancreas. K, L, Arrows mark expression in the epiphysis organ. K, L, Insets, Anterior dorsal views of the same specimens. t, Telencephalon; vdi, ventral diencephalon; mb, midbrain; ov, otic vesicle; hb, hindbrain; p, pancreas; di, diencephalon; mhb, midbrain-hindbrain boundary; hcp, HC primordium; ac, anterior crista; mc, medial crista, pc, posterior crista; ex, exocrine; en, endocrine; rpl, retinal pigmented epithelial layer; pcl, photoreceptor cell layer; ipl, inner plexiform layer; opl, outer plexiform layer; inl, inner nuclear layer; gcl, ganglion cell layer; ep, epiphysis; lh, left habenula; rh, right habenula. Scale bar: (in M) A, B, 100 μm; C, H, 150 μm; D, 80 μm; E, 65 μm; F, 5 μm; G, 60 μm; I, J, 20 μm; K, 125 μm; L, 200 μm; M, 90 μm. N, A cav1.3a-specific band is detected by RT-PCRs of total RNA from adult brain, ear, eye, pancreas, heart (low levels), and testis tissue homogenates.
Our genetic and physical mapping results, together with in situ expression analyses, established cav1.3a as an excellent candidate for gem. We therefore assembled a full-length cav1.3a cDNA for mutational analysis. This cDNA was derived from one of the numerous cav1.3a splice variants we isolated during RACE experiments and may represent the longest form expressed in larval zebrafish. The 6548 kb cav1.3a message encodes a 2082 amino acid protein, the sequence and domain structure of which are highly similar and identical, respectively, to that of other vertebrate Cav1.3 proteins (Soldatov, 1992; Kollmar et al., 1997b) (Fig. 4A) (for details, see supplemental Fig. 1, available at www.neurosci.org). Phylogenetic analyses clearly identified Cav1.3a as the closest relative to higher vertebrate Cav1.3 proteins, with Cav1.3b representing a distant member of the same class (Fig. 4C) (supplemental Fig. 1, available at www.jneurosci.org).
Figure 4.
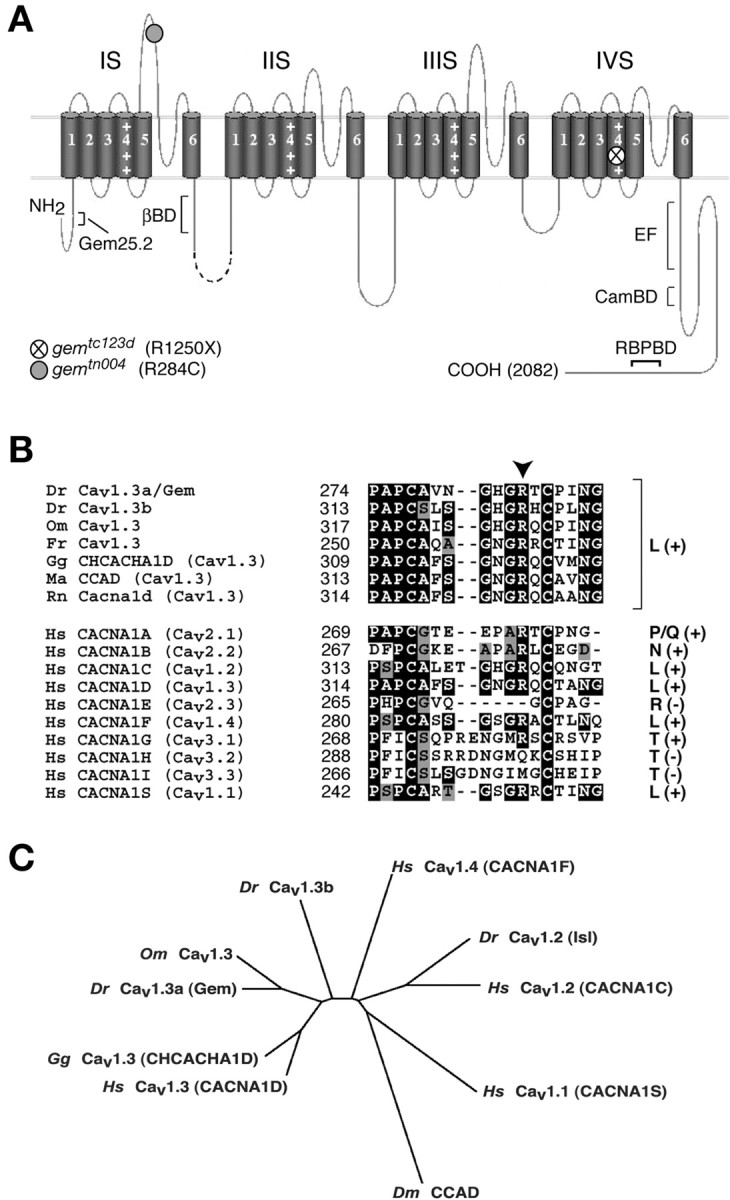
gem encodes the Cav1.3a isoform of zebrafish LTCCs. A, Topographic diagram of the Cav1.3a secondary structure. The channel core consists of four transmembrane domains subdivided into six hydrophobic segments (1-6) and a pore (P) region, with each transmembrane domain connected to the next by an intracellular loop of variable length. gem mutant lesions are indicated by circles. Positively charged residues in voltage-sensor domains (I-IV S4) are illustrated by + signs. The position of the Gem25.2 epitope, the Ca2+-binding helix-loop-helix unit (EF) hand, and other functionally characterized domains are indicated. BD, Binding domain; βBD, β subunit binding domain; CamBD, calmodulin binding domain; RBPbd, RIM binding protein binding domain (for details, see supplemental Fig. 1, available at www.jneurosci.org). B, Alignments of the IS5-IP region of a variety of voltage-gated Ca2+ channels. Top, 1.3 isoforms from various species. Bottom, Various isoforms of human Cav channels. Corresponding channel classes (L-, P/Q-, N-, R-, and T-type) are indicated at the right. + or - signs indicated in parentheses stand for conservation or nonconservation, respectively, of the Arg residue mutated in gemtn004 (arrowhead). C, Phylip unrooted dendogram deduced from Clustal alignments of the shown proteins, indicating that cav1.3a is the true ortholog of higher vertebrate cav1.3 genes. Cav1.3b appears to belong to the 1.3 class, suggesting that it is a true paralogue of cav1.3a that arose through duplication of a cav1.3 common ancestor and that subsequently diverged. wt, Wild type; Dr, Danio rerio; Hs, Homo sapiens; Gg, Gallus gallus; Om, Oncorhynchus mykiss (trout); Fr, Fugu rubripes; Rn, Rattus norvegicus; Ma, Mesocricetus auratus; Dm, Drosophila melanogaster.
In cav1.3a cDNAs from gemtc123d homozygotes, we identified a C → T transition at nucleotide 3970 exchanging an Arg codon (CGA, R1250) for a stop codon (TGA) (Fig. 4A). This nonsense mutation affects a hydrophilic residue of transmembrane segment IVS4 and disrupts the integrity of the last transmembrane domain while also removing the C-terminal tail (Fig. 4A). In gemtn004 transcripts, we detected a C → T transition at nucleotide 1072 substituting an Arg (CGC, R284) for a Cys (TGC) residue in the IS5-IP region (Fig. 4A). This missense mutation affects an amino acid conserved in all known vertebrate Cav1.3 channels, as well as in most voltage-gated Ca2+ channel classes across species (Fig. 4B). The presence of deleterious mutations in cav1.3a transcripts from both gem alleles provided strong evidence that cav1.3a is the gem gene. In addition, sequencing and gene prediction analyses of PACd19136 revealed that no gene other than cav1.3a resides within the gem critical interval. The sole other coding sequences detected on the clone (α2δ3 exon 1 and an opsin gene) reside 3′ to the BAC128j09 recombination breakpoint. Altogether, these results demonstrated that gem encodes Cav1.3a.
The Gem25.2 antiserum specifically detects Gem/Cav1.3a channels as focal spots localized near the basal region of NMs
To investigate the role of Gem/Cav1.3a in HCs, we sought to localize the channel by whole-mount immunohistochemistry. We generated rabbit antisera directed against various potentially antigenic peptides of zebrafish Cav1.3a and assessed their immunoreactivity in intact 5 dpf zebrafish larvae using confocal microscopy. One of these antisera, Gem25.2, produced strong immunoreactivity in inner ear and NM sensory neuroepithelia and was selected for additional analyses. The Gem25.2 antigen is a Cav1.3a-specific peptide located in the N-terminal tail (KSKKQGSSANTRPQR, residues 42-56) (Fig. 4A) (supplemental Fig. 1, available at www.jneurosci.org).
Figure 5A shows a typical ventrolateral side view of Gem25.2 immunoreactivity in a zebrafish anterodorsal NM and underlying LL nerve, with the dorsal-most plane coinciding with the basal surface of HCs. Labeling of the LL nerve and HC basal membranes is obvious. Notably, strong immunoreactivity occurs in discrete regions of the basal surface of the NM that may coincide with the interface between LL axons and HCs. We hereafter refer to these distinct sites of immunoreactivity as “focal spots.” No or negligible labeling was detected within the basal cytoplasm of HCs.
Figure 5.
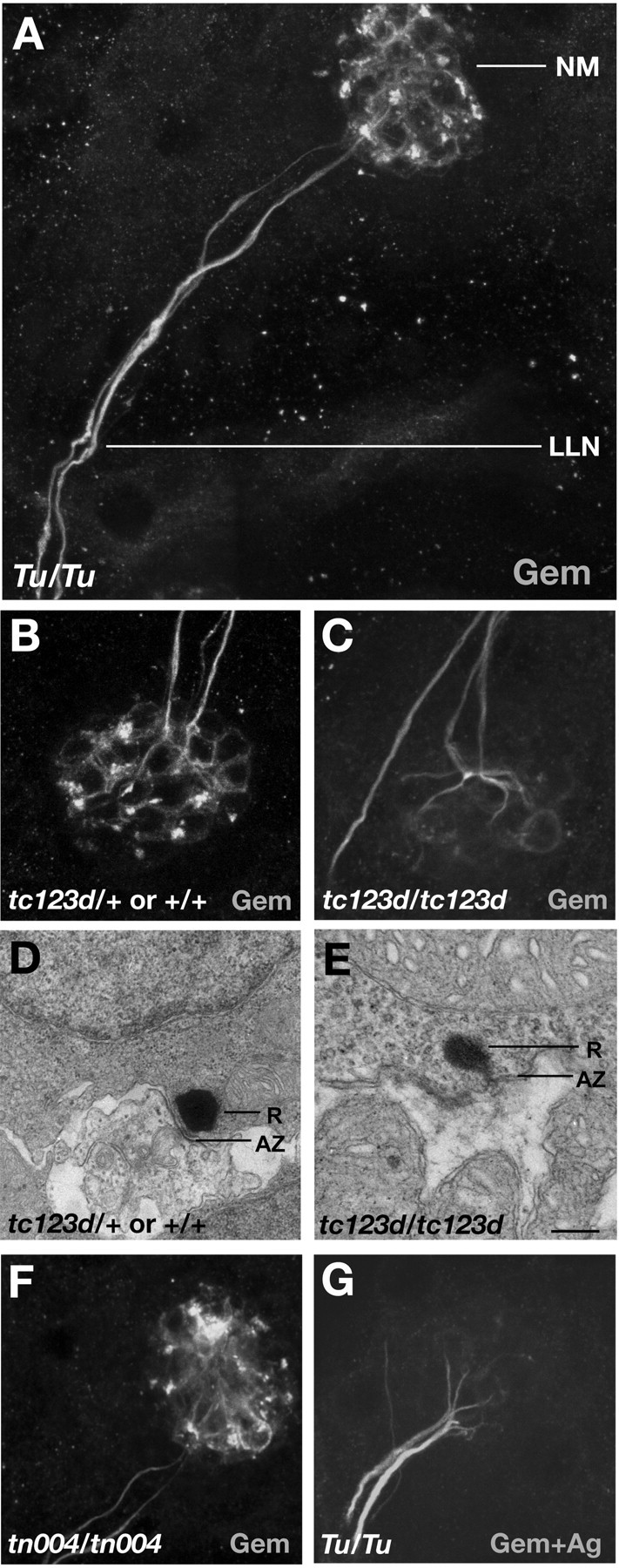
Specific immunoreactivity of the Gem25. 2 antiserum in the basal region of NMs. Images are Z-stack projections of 16 confocal sections with the basal surface of NMs and LL nerve as apical-most and distal-most section planes, respectively. Genotypes and Ab solutions are indicated in left and right bottom corners, respectively. A, Ventrolateral view of Gem25.2 labeling of an anterodorsal NM and underlying LL nerve in a 5 dpf intact zebrafish wild-type larva. B, C, Gem25.2 labeling of tc123d sibling (B) and mutant (C) NMs and LL nerves (images are projections, as in A). D, E, TEM sections of tc123d sibling and mutant HC RSs. The ribbon (R) and underlying active zone (AZ) are depicted. F, Gem25.2 labeling of the interface between agemtn004 mutant NM and underlying LL nerve. G, Labeling produced by the Gem25.2 antiserum preincubated with its target antigen. LLN, Lateral line nerve. Scale bar: (in E) A, 5 μm; B, C, F, G, 3 μm; D, E, 300 nm.
We tested the specificity of Gem25.2 by peptide competition and by examining immunoreactivity in gem mutants. During preliminary binding to its antigen, the Gem25.2 antiserum still produced strong labeling of the LL nerve. In contrast, overall labeling of HC basal membranes and immunoreactive focal spots was blocked by peptide competition (Fig. 5G). These results suggest that, in contrast to HC membrane labeling and focal spots, LL axon staining is nonspecific. A similar result was obtained when gemtc123d mutants and wild-type siblings were analyzed. gem sibling specimens produced analogous immunoreactivity to that shown in Figure 5A, whereas tc123d mutants resembled specimens stained with Gem25.2 preincubated with the antigen (Fig. 5C). Axon labeling was retained in tc123d mutants, but staining of HC basal membranes was weaker and/or more diffuse. Most striking was the complete absence of focal spots (Fig. 5B,C). In contrast to the tc123d nonsense mutation, the missense tn004 mutation did not affect any features of Gem25.2 immunoreactivity (Fig. 5F).
RT-PCR and in situ hybridization indicated that cav1.3a mRNA levels are normal in tc123d homozygotes (data not shown). This result excludes that the absence of Gem25.2 focal spots in tc123d mutants results from a nonsense-mediated decay of the transcript. In addition, the Gem25.2 epitope is not affected by the tc123d C-terminal truncation (Fig. 4A). The following possibilities could explain the absence of Gem25.2 focal spots in tc123d mutants: (1) tc123d-truncated Cav1.3a channels are unstable and degraded, (2) the conformation of the truncated channels compromises plasma membrane integration and/or immunoreactivity of the antigen, (3) tc123d channels lack a critical C-terminal site required for membrane localization and/or clustering efficiency, or (4) an epistatic effect of the mutation, such as abnormal synaptogenesis or synaptic degeneration, removes not only Cav1.3a channels but also other components of the cellular structure(s) underlying the focal spots. Only the latter hypothesis is compatible with the possibility that Gem25.2-immunoreactive focal spots are nonspecific. In transmission electron microscopic (TEM) sections of tc123d mutant NMs, however, ribbons are present near active zones, such as in HCs of wild-type siblings (Fig. 5D,E). Hence, peptide competition and mutant labeling experiments strongly suggest that Gem25.2 specifically recognizes Cav1.3a.
Focal spots of Gem/Cav1.3a immunoreactivity localize within the basal membrane of HCs
The spatial distribution of Gem25.2 focal spots reveals that Gem/Cav1.3a preferentially localizes in discrete regions of the basal area of NMs that may coincide with the interface between HCs and underlying axons. We first assessed whether the Gem25.2 focal spots occur within or outside the HC membrane (e.g., in adjacent axons or NM support cells). We performed double-labeling experiments using fluorescently labeled phalloidin to visualize the submembranous actin cytoskeleton of HCs. Figure 6 shows a side view (lateral section) of a dorsoanterior NM. The outlines of HCs and underlying supporting cells can be visualized in a bright-field image of the section (Fig. 6A). As predicted, incubation with phalloidin yielded strong staining of the actin-rich stereociliary bundles but also uniformly labeled the HC walls (Fig. 6C). In the two HCs that are in focus, Gem25.2 immunoreactivity can be found all along the basolateral membrane, where it appears rather uniform and diffuse, and in discrete regions of the basal membrane, which are strongly labeled and presumably correspond to the Gem25.2 focal spots (Fig. 6B). This staining clearly overlapped with that of phalloidin (Fig. 6D). No obvious Gem25.2 labeling was detected outside or underneath HCs (Fig. 6B,D). Therefore, Gem/Cav1.3a channels localize within specific areas of the HC basal membrane.
Figure 6.
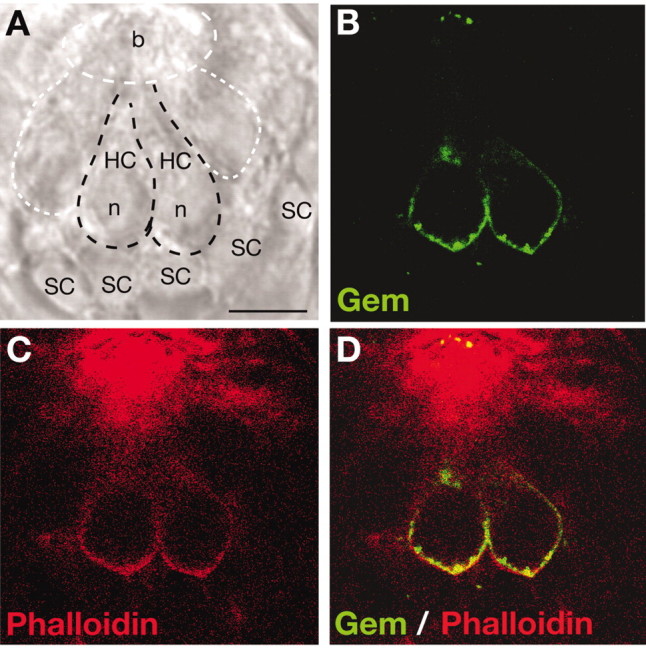
Gem/Cav1.3a focal spots localize within the HC basal membrane. A-D, Lateral view(single confocal section, 0.5 μm thick) of an anterodorsal NM of a 5 dpf wild-type zebrafish larva double-labeled with Gem25.2 and phalloidin, a marker of actin filaments. A, Bright-field view of the NM and underlying supporting cells. Black dashed regions delineate two HCs that are in focus. White-dashed regions indicate two HCs that are not in focus and the surface of the NM in which HC stereociliary bundles reside (this region coincides with strong phalloidin staining in C). B, C, Fluorescent images of the same section showing sites of Gem25.2 immunoreactivity (B) and phalloidin labeling (C). D, Merged image of B and C, showing the position of Gem25.2-immunoreactive zones relative to the phalloidin-labeled cortical actin. Scale bar, 3.5 μm. b, Bundles; n, nucleus; SC, supporting cell.
The position, fine structure, and number of Gem25.2 focal spots are consistent with Gem/Cav1.3a channels clustering at HC RSs
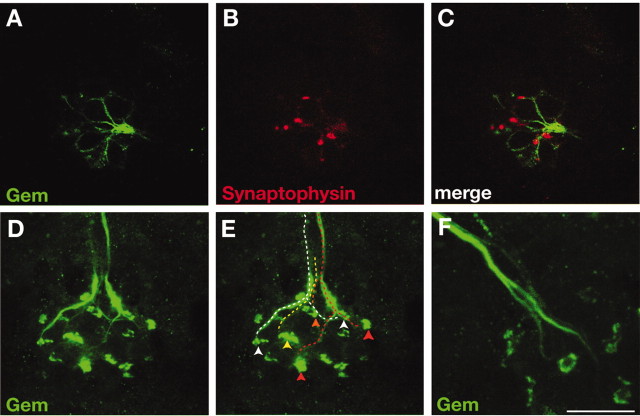
The basal-most localization of Gem25.2 focal spots in HCs suggests that Gem/Cav1.3a channels may localize at RSs. In TEM sections of vertebrate HCs, RSs are typically found apposing afferent terminals (Fig. 5D,E) (see Fig. 9A,B) (Lenzi et al., 2002; Fuchs et al., 2003). Hence, we wanted to establish the location of the Gem25.2 focal spots relative to afferent postsynaptic zones. We first tested whether the nonspecific yet strong labeling of LL axons by Gem25.2 correlated with afferent and/or efferent axons. Synaptophysin is one of the SNARE proteins that, although detected at photoreceptor ribbons (Von Kriegstein et al., 1999), is completely absent at HC RSs (Safieddine and Wenthold, 1999). It is, however, present in efferent terminals in which it contributes to vesicle fusion (Safieddine and Wenthold, 1999; Waka et al., 2003). We therefore used synaptophysin as a specific marker for efferent synapses. As shown in Figure 7A-C, anti-synaptophysin immunoreactivity did not coincide with Gem25.2-labeled axon endings, implying that the nonspecifically labeled axons are LL afferents. We then determined the position of Gem25.2-labeled afferent terminals relative to the Gem25.2-labeled HC focal spots. We could identify most contacts between the HCs of each NM and underlying afferents (Fig. 7D,E). Strikingly, these contacts always coincided with focal spots (Fig. 7E). Hence, Gem/Cav1.3a channels reside in the position that is expected for HC RS-specific Ca2+ channels.
Figure 9.
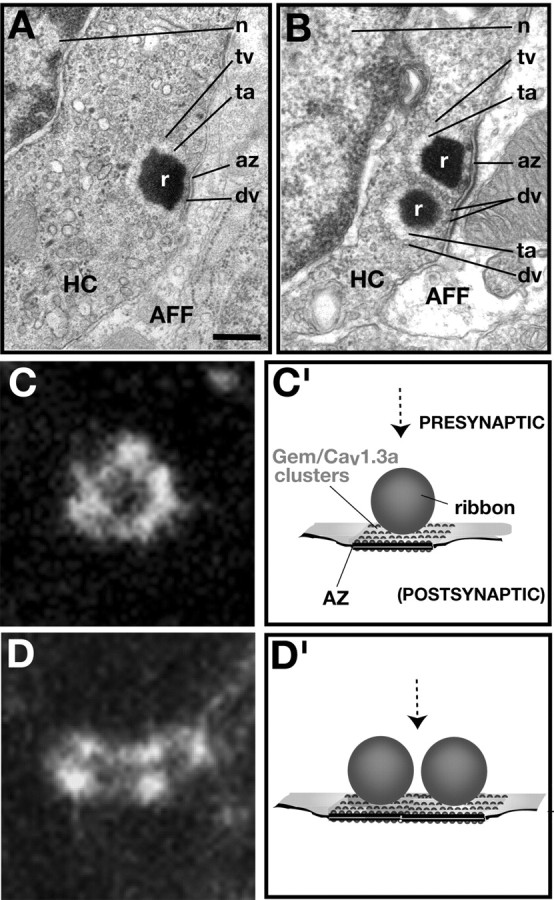
A model for Gem/Cav1.3a localization in zebrafish HCs. A, B, TEM sections of 5 dpf wild-type HC afferent terminals showing the presence of either one (A) or two (B) ribbons at the RS. C, D, Gem25.2 focal spots at high magnification. Note their circular or elliptic shape and their punctated aspect, suggesting a clustering of single or groups of Gem/Cav1.3a channels adjacent to ribbons. C′, D′, Drawings of HC RSs from a lateral view. The dashed arrow in each panel indicates the view (dorsal) from which the drawings are related to C and D, respectively. Scale bar: (in A) A, 375 nm; B, 300 nm; C, D, 500 nm. AFF, Afferent terminal; az, active zone; fv, fusing vesicle; n, nucleus; r, ribbon; ta, tethering apparatus; tv, tethered vesicle.
Figure 7.
Gem25.2 focal spots appose lateral line afferent terminals and have a ring-like morphology. A-C, The efferent marker synaptophysin does not colocalize with Gem25.2 nonspecific labeling of LL fibers. Images are Z-stack projections of serial confocal sections (0.5 μm thick) through the region just basal to an anterodorsal NM of a 5 dpf wild-type zebrafish larva double labeled with Gem25.2 (green) and synaptophysin (red) antibodies. D-F, Nonspecifically labeled afferent fibers project to Gem25.2-positive focal spots. D, Projection of 16 lateral sections (0.5 μm thick) through an anterodorsal NM in a region encompassing LL nerve axons and the basal surface of HCs. E, Same image as in D illustrating all clearly distinguishable axons and their target focal spot (depicted by dashed lines and arrowheads of identical colors). F, Projection of three lateral sections (0.5 μm thick) through a different anterodorsal NM, seen at twofold higher magnification than that in D and E and showing the branching of two individual axons near their target focal spots. Note the ring-shaped structure of the focal spots. Scale bars: (in C) A-C, 3 μm; (in F) D, E, 3.5 μm; F, 1.8 μm.
Examination of the positions of focal spots relative to afferent terminals at high magnification revealed that focal spots were organized in a stereotypical shape resembling that of a ring (Figs. 7F, 8B-D, insets). The rings of Gem25.2 immunoreactivity appeared to be formed by individual dots linked one to another by more diffuse staining (Figs. 8B-D, insets, 9C,D), suggesting that the rings are in fact formed by clusters of individual and/or groups of Gem/Cav1.3a channels. We further noticed a striking correlation between the spherical-like structure and diameter (∼300 nm) of the nonlabeled, central area of Gem25.2 focal spots and the spherical form and diameter that are characteristic of HC synaptic ribbons (Fig. 5D,E). Hence, the fine structure of Gem25.2 focal spots is also in strong agreement with the notion that Gem/Cav1.3a channels reside at HC RSs.
Figure 8.
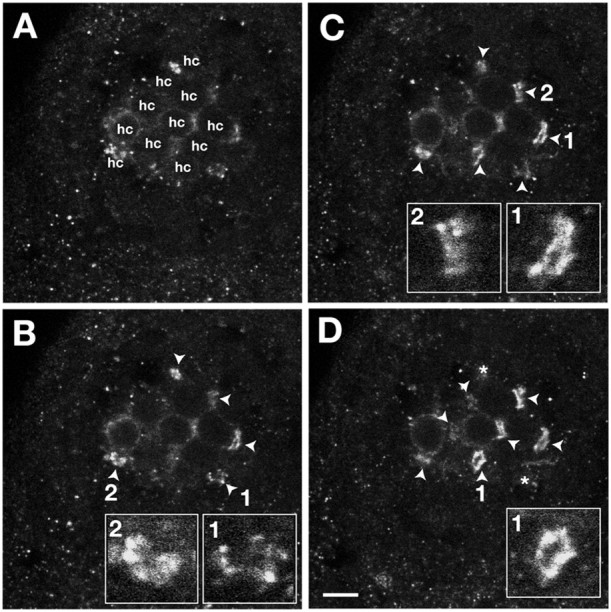
Number and fine structure of Gem25.2-immunoreactive rings in basal membranes of NM HCs. A-D, Ventral views of four serial confocal sections (0.3 μm thick) through an anterodorsal NM of a 5 dpf wild-type zebrafish larva labeled with Gem25.2. The apical-most section (A) coincides with the mediolateral membrane of the HCs of the NMs; Gem25.2-immunoreactive rings are not yet in focus. The basal-most section(D) is through the basal-most surface of the NM; deeper sections do not contain any additional Gem25.2 rings. The number of HCs was counted in focal planes in which the cell outlines were clearly visible (denoted hc in A). Focal spots in focus are marked by arrowheads and then by asterisks as they leave the focal plane (to mark their former position). In this experiment, we counted 11 HCs and nine focal spots. Insets in B-D are close-ups of individual focal spots. Scale bar, 3 μm.
One other implication of Gem/Cav1.3a localizing at HC RSs is that the number of channel clusters per HC should be similar to the number of ribbons per HC. Examination of TEM sections suggests that 5 dpf HCs contain at least two RSs, often occurring in pairs (Fig. 9B). We counted focal spots in 20 NMs from 10 different specimens (one representative NM is shown as an example in Fig. 8) and obtained an overall ratio of 0.8 focal spots per HC (n = 177 focal spots; n = 218 HCs). The number of focal spots is therefore lower than expected. Less than perfect immunoreactivity, masking effects of background or overall membrane staining, and/or the occurrence of RSs in pairs may account for this difference. Nearly 20% of Gem25.2 clusters had an elliptic rather than a spherical shape (Figs. 8C, insets, 9D). In contrast to rings of Gem25.2 immunoreactivity that are suggestive of the presence of a single ribbon (Fig. 9A,C′), the elliptic patterns of immunoreactivity may correspond to those 5 dpf HC RSs composed of two ribbons (Fig. 9B,D′). Although we could not establish a strict numerical agreement between focal spots and RSs, these experiments nevertheless indicated that the number of Gem25.2 focal spots is in the range of that of HC RSs. On the basis of position, fine structure, and number of Gem25.2 focal spots, we conclude that Gem/Cav1.3a channels cluster at HC RSs.
Discussion
In this study, we identified the molecular defects associated with the gem deafness-imbalance phenotype as point mutations in a Ca2+ channel encoding gene, cav1.3a. The gem gene encodes one of the two class 1.3 pore-forming subunits of zebrafish LTCCs and is strongly expressed in inner ear and NM HCs. Similarly to Cav1.3 knock-out mice (Platzer et al., 2000), deafness in gem arises from HC-specific defects. Our analysis of the distribution of Gem/Cav1.3a proteins in wild-type and mutant HCs strongly favors loss of Ca2+ influx at RSs as the basis for the gem mutant phenotype. Together, our results support the notion that the unusual biophysical properties of Cav1.3 LTCCs (Kollmar et al., 1997a,b) are of evolutionary and functional relevance to the unique property of vertebrate HCs to produce fast and sustained transmitter release (Fuchs et al., 2003).
The effects of gem mutations on HC microphonic potentials and channel localization
In seminal studies of bullfrog saccular HCs, at least four currents were proposed to contribute to the generation of the HC microphonic potential: the transduction current, a Ca2+-dependent K+ (BK) current, and two voltage-dependent currents, one for K+ (Kv current) and one for Ca2+ (the Cav current presently studied) (Corey and Hudspeth, 1983; Lewis and Hudspeth, 1983). In gemtc123d homozygous HCs, the microphonics are reduced by ∼50%. gemtn004 mutant HCs show a similar reduction (up to ∼60%). In general, zebrafish mutant HCs with a comparable reduction in microphonics show readily visible defects in FM1-43 uptake (Seiler and Nicolson, 1999). HCs carrying weak alleles of sputnik/cadherin23, for instance, display a microphonic defect similar to that seen in gem mutants and are obviously affected in their ability to take up FM1-43. However, FM1-43 uptake appears normal in gem mutants, suggesting that mechanotransduction channels are functional. Our uptake experiments may not detect subtle differences in transduction efficiency. Normal transduction currents were also reported in isolated HCs from Cav1.3-/- mice (Brandt et al., 2003). The reduction in microphonic potentials may be attributed to the loss of Gem/Cav1.3a channel activity and possibly to the loss of the downstream BK current. Consistent with this notion, mouse Cav1.3-/- HCs lack BK currents, and BK channels appear to be absent (Brandt et al., 2003). In gem mutant HCs, lack of BK currents may perturb cell repolarization after stimulus-induced depolarization, an effect that will reduce the driving force on the transduction current. Alternatively, the lack of Gem/Cav1.3a channel activity may lead to secondary defects, which in turn affect the microphonic potentials.
The gemtc123d mutation is predicted to truncate the channel within the fourth transmembrane domain (IVS), thereby removing two transmembrane segments (IVS5 and IVS6) and one pore helix (IVP). Although the predicted truncated channel retains the epitope recognized by the Gem25.2 Ab, we observed strongly reduced immunoreactivity in basolateral HC membranes of tc123d homozygotes. tc123d may therefore be a function-null allele, if not a protein-null allele, of gem/cav1.3a. In contrast, both the levels and localization of Cav1.3a produced by the gemtn004 missense allele appear normal (Fig. 5F). This observation indicates that tn004-encoded Cav1.3a channels are nonfunctional, suggesting a crucial role for the mutated arginine residue in channel conductance (R284, corresponding to R324 in human Cav1.3). Consistent with this hypothesis, the amino acid invariance at this position extends from class 1.3 L-type Ca2+ channels to most vertebrate voltage-gated Ca2+ channels (R-type and several T-type channels being exceptions) (Fig. 4B). In addition, residues in the IS5-IS6 region had been postulated previously to play a critical role in Cav1-mediated LTCC conductance (Dirksen et al., 1997). Additional studies will help establish the precise role of the tn004-affected residue in Cav1.3a activity.
Gem/Cav1.3a as a HC ribbon-specific Ca2+ channel
Voltage-gated Ca2+ entry is important for HC transmitter release (Roberts et al., 1990; Beutner et al., 2001). Whether the contribution of the Cav1.3 isoform of LTCCs to vesicle fusion is direct or indirect, however, has remained unclear. Although capacitance measurements and afferent nerve recordings of DHP-treated HC preparations from mouse and frog strongly suggest an involvement of LTCCs in HC neurotransmission (Zhang et al., 1999; Spassova et al., 2001; Robertson and Paki, 2002), these studies suffer the inherent caveat of possible nonspecific effects of the drug. However, a recent study of inner ear HCs isolated from Cav1.3-/- mice has shown that capacitance measurements in response to depolarization are reduced (Brandt et al., 2003). Moreover, many studies are consistent with LTCCs localizing at sensory receptor RSs (Roberts et al., 1990; Issa and Hudspeth, 1994; Tucker and Fettiplace, 1995; Martinez-Dunst et al., 1997; Nachman-Clewner et al., 1999; Hibino et al., 2002; Zenisek et al., 2003). High-resolution subcellular localization of Cav1.3 in HCs, however, has not been reported. Recent immunohistochemistry studies detected Cav1.3 proteins either in a broad region of the HC basolateral membrane (Lopez et al., 1999) or as colocalized with the SNARE protein syntaxin (Song et al., 2003). However, the precise position of the immunoreactivity relative to afferent presynaptic terminals was not assessed. Our immunolocalization data indicate that (1) Gem channels preferentially localize in discrete regions of the basal membrane of HCs, (2) the number of discrete regions per HC is in the range of the number of RSs per HC, (3) the discrete regions map to the sites of neurotransmission, that is, at the interface with afferent endings, and (4) the pattern of immunoreactivity in these discrete regions is reminiscent of the spherical structure of HC ribbons, suggesting that single or groups of Gem/Cav1.3a channels cluster in the HC membrane near the ribbon (Fig. 9C′,D′). Altogether, these observations strongly suggest that Gem/Cav1.3a is present at the HC RS.
Etiology for the gem deafness phenotype and concluding remarks
Our characterization of the zebrafish circler mutant gemini provides a valuable, nonmammalian genetic model for dissecting the function and evolution of Cav1.3 in vertebrate HCs. The results from our phenotypic, electrophysiological, and immunolocalization analyses, considered in the light of recent analyses of murine Cav1.3-/- HCs (Platzer et al., 2000; Brandt et al., 2003; Glueckert et al., 2003), lead us to propose that absence of Ca2+ influx at HC RSs, and therefore a likely defect in transmitter release, accounts for the gem deafness phenotype. Future studies using gem mutants may further help to assign a direct role for Cav1.3a channels in HC transmission and may increase our understanding of the means by which this Ca2+ channel underlies the unusual kinetics and throughput of HC neurotransmission.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft and Sonderforschungsbereich 430 (U.S., T.N.),the Max-Planck Gesellschaft(S.S.,E.B.-N.), Association pour la Recherche contre le Cancer, and Fondation pour la Recherche Medicale (S.S.). We thank A. Hruscha and O. Heindrich at the Max-Planck-Institut für Entwicklungsbiologie for technical support and C. Seiler and C. Söllner for helpful comments during the course of this work.
Correspondence should be addressed to Teresa Nicolson, Oregon Hearing Research Center and Vollum Institute, Mail Code L335A, 3181 Southwest Sam Jackson Park Road, Portland, OR 97239. E-mail: nicolson@ohsu.edu.
Copyright © 2004 Society for Neuroscience 0270-6474/04/244213-11$15.00/0
References
- Allwardt BA, Lall AB, Brockerhoff SE, Dowling JE (2001) Synapse formation is arrested in retinal photoreceptors of the zebrafish nrc mutant. J Neurosci 21: 2330-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altrock WD, tom Dieck S, Sokolov M, Meyer AC, Sigler A, Brakebusch C, Fassler R, Richter K, Boeckers TM, Potschka H, Brandt C, Loscher W, Grimberg D, Dresbach T, Hempelmann A, Hassan H, Balschun D, Frey JU, Brandstatter JH, Garner CC, Rosenmund C, Gundelfinger ED (2003) Functional inactivation of a fraction of excitatory synapses in mice deficient for the active zone protein bassoon. Neuron 37: 787-800. [DOI] [PubMed] [Google Scholar]
- Ashmore J (1998) Mechanosensation: swimming round in circles. Curr Biol 8: R425-R427. [DOI] [PubMed] [Google Scholar]
- Beutner D, Voets T, Neher E, Moser T (2001) Calcium dependence of exocytosis and endocytosis at the cochlear inner hair cell afferent synapse. Neuron 29: 681-690. [DOI] [PubMed] [Google Scholar]
- Biemar F, Argenton F, Schmidtke R, Epperlein S, Peers B, Driever W (2001) Pancreas development in zebrafish: early dispersed appearance of endocrine hormone expressing cells and their convergence to form the definitive islet. Dev Biol 230: 189-203. [DOI] [PubMed] [Google Scholar]
- Brandt A, Striessnig J, Moser T (2003) Cav1.3 channels are essential for development and presynaptic activity of cochlear inner hair cells. J Neurosci 23: 10832-10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha ML, Russell C, Regan JC, Tawk M, Sidi S, Gilmour DT, Kapsimali M, Sumoy L, Goldstone K, Amaya E, Kimelman D, Nicolson T, Grunder S, Gomperts M, Clarke JD, Wilson SW (2003) Local tissue interactions across the dorsal midline of the forebrain establish CNS laterality. Neuron 39: 423-438. [DOI] [PubMed] [Google Scholar]
- Corey DP, Hudspeth AJ (1983) Analysis of the microphonic potential of the bullfrog's sacculus. J Neurosci 3: 942-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leon M, Wang Y, Jones L, Perez-Reyes E, Wei X, Soong TW, Snutch TP, Yue DT (1995) Essential Ca2+-binding motif for Ca2+-sensitive inactivation of L-type Ca2+ channels. Science 270: 1502-1506. [DOI] [PubMed] [Google Scholar]
- Denk W, Webb WW (1992) Forward and reverse transduction at the limit of sensitivity studied by correlating electrical and mechanical fluctuations in frog saccular hair cells. Hear Res 60: 89-102. [DOI] [PubMed] [Google Scholar]
- Dick O, tom Dieck S, Altrock WD, Ammermuller J, Weiler R, Garner CC, Gundelfinger ED, Brandstatter JH (2003) The presynaptic active zone protein bassoon is essential for photoreceptor ribbon synapse formation in the retina. Neuron 37: 775-786. [DOI] [PubMed] [Google Scholar]
- Dirksen RT, Nakai J, Gonzalez A, Imoto K, Beam KG (1997) The S5-S6 linker of repeat I is a critical determinant of L-type Ca2+ channel conductance. Biophys J 73: 1402-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmetsch RE, Pajvani U, Fife K, Spotts JM, Greenberg ME (2001) Signaling to the nucleus by an L-type calcium channel-calmodulin complex through the MAP kinase pathway. Science 294: 333-339. [DOI] [PubMed] [Google Scholar]
- Engel J, Michna M, Platzer J, Striessnig J (2002) Calcium channels in mouse hair cells: function, properties and pharmacology. Adv Otorhinolaryngol 59: 35-41. [DOI] [PubMed] [Google Scholar]
- Flock A (1965) Transducing mechanisms in the lateral line canal organ receptors. Cold Spring Harb Symp Quant Biol 30: 133-145. [DOI] [PubMed] [Google Scholar]
- Fuchs PA, Glowatzki E, Moser T (2003) The afferent synapse of cochlear hair cells. Curr Opin Neurobiol 13: 452-458. [DOI] [PubMed] [Google Scholar]
- Gale JE, Marcotti W, Kennedy HJ, Kros CJ, Richardson GP (2001) FM1-43 dye behaves as a permeant blocker of the hair-cell mechanotransducer channel. J Neurosci 21: 7013-7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler R (2002) Mapping and cloning. In: Zebrafish: a practical approach, Chap 7 (Nuesslein-Volhard C, Dahm R, eds). New York: Oxford University.
- Glowatzki E, Fuchs PA (2002) Transmitter release at the hair cell ribbon synapse. Nat Neurosci 5: 147-154. [DOI] [PubMed] [Google Scholar]
- Glueckert R, Wietzorrek G, Kammen-Jolly K, Scholtz A, Stephan K, Striessnig J, Schrott-Fischer A (2003) Role of class D L-type Ca2+ channels for cochlear morphology. Hear Res 178: 95-105. [DOI] [PubMed] [Google Scholar]
- Granato M, van Eeden FJ, Schach U, Trowe T, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Nusslein-Volhard C (1996) Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development 123: 399-413. [DOI] [PubMed] [Google Scholar]
- Griesinger CB, Richards CD, Ashmore JF (2002) FM1-43 reveals membrane recycling in adult inner hair cells of the mammalian cochlea. J Neurosci 22: 3939-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptmann G, Gerster T (1994) Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet 10: 266. [DOI] [PubMed] [Google Scholar]
- Hibino H, Pironkova R, Onwumere O, Vologodskaia M, Hudspeth AJ, Lesage F (2002) RIM binding proteins (RBPs) couple Rab3-interacting molecules (RIMs) to voltage-gated Ca2+ channels. Neuron 34: 411-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudspeth AJ (1989) How the ear's works work. Nature 341: 397-404. [DOI] [PubMed] [Google Scholar]
- Issa NP, Hudspeth AJ (1994) Clustering of Ca2+ channels and Ca2+-activated K+ channels at fluorescently labeled presynaptic active zones of hair cells. Proc Natl Acad Sci USA 91: 7578-7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashima Y, Pugh W, Depaoli AM, Takeda J, Seino S, Bell GI, Polonsky KS (1993) Expression of calcium channel mRNAs in rat pancreatic islets and downregulation after glucose infusion. Diabetes 42: 948-955. [DOI] [PubMed] [Google Scholar]
- Kollmar R, Montgomery LG, Fak J, Henry LJ, Hudspeth AJ (1997a) Predominance of the alpha1D subunit in L-type voltage-gated Ca2+ channels of hair cells in the chicken's cochlea. Proc Natl Acad Sci USA 94: 14883-14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmar R, Fak J, Montgomery LG, Hudspeth AJ (1997b) Hair cell-specific splicing of mRNA for the alpha1D subunit of voltage-gated Ca2+ channels in the chicken's cochlea. Proc Natl Acad Sci USA 94: 14889-14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok C, Korn RM, Davis ME, Burt DW, Critcher R, McCarthy L, Paw BH, Zon LI, Goodfellow PN, Schmitt K (1998) Characterization of whole genome radiation hybrid mapping resources for non-mammalian vertebrates. Nucleic Acids Res 26: 3562-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzi D, Roberts WM (1994) Calcium signalling in hair cells: multiple roles in a compact cell. Curr Opin Neurobiol 4: 496-502. [DOI] [PubMed] [Google Scholar]
- Lenzi D, von Gersdorff H (2001) Structure suggests function: the case for synaptic ribbons as exocytotic nanomachines. BioEssays 23: 831-840. [DOI] [PubMed] [Google Scholar]
- Lenzi D, Crum J, Ellisman MH, Roberts WM (2002) Depolarization redistributes synaptic membrane and creates a gradient of vesicles on the synaptic body at a ribbon synapse. Neuron 36: 649-659. [DOI] [PubMed] [Google Scholar]
- Lewis RS, Hudspeth AJ (1983) Voltage- and ion-dependent conductances in solitary vertebrate hair cells. Nature 304: 538-541. [DOI] [PubMed] [Google Scholar]
- Lopez I, Ishiyama G, Ishiyama A, Jen JC, Liu F, Baloh RW (1999) Differential subcellular immunolocalization of voltage-gated calcium channel alpha1 subunits in the chinchilla cristae ampullaris. Neuroscience 92: 773-782. [DOI] [PubMed] [Google Scholar]
- Martinez-Dunst C, Michaels RL, Fuchs PA (1997) Release sites and calcium channels in hair cells of the chick's cochlea. J Neurosci 17: 9133-9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers JR, MacDonald RB, Duggan A, Lenzi D, Standaert DG, Corwin JT, Corey DP (2003) Lighting up the senses: FM1-43 loading of sensory cells through nonselective ion channels. J Neurosci 23: 4054-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88: 9828-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton CC (2002) Genetics, genomics and gene discovery in the auditory system. Hum Mol Genet 11: 1229-1240. [DOI] [PubMed] [Google Scholar]
- Moser T, Beutner D (2000) Kinetics of exocytosis and endocytosis at the cochlear inner hair cell afferent synapse of the mouse. Proc Natl Acad Sci USA 97: 883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman-Clewner M, St. Jules R, Townes-Anderson E (1999) L-type calcium channels in the photoreceptor ribbon synapse: localization and role in plasticity. J Comp Neurol 415: 1-16. [PubMed] [Google Scholar]
- Namkung Y, Skrypnyk N, Jeong MJ, Lee T, Lee MS, Kim HL, Chin H, Suh PG, Kim SS, Shin HS (2001) Requirement for the L-type Ca2+ channel alpha(1D) subunit in postnatal pancreatic beta cell generation. J Clin Invest 108: 1015-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson T, Rusch A, Friedrich RW, Granato M, Ruppersberg JP, Nusslein-Volhard C (1998) Genetic analysis of vertebrate sensory hair cell mechanosensation: the zebrafish circler mutants. Neuron 20: 271-283. [DOI] [PubMed] [Google Scholar]
- Parsons TD, Sterling P (2003) Synaptic ribbon. Conveyor belt or safety belt? Neuron 37: 379-382. [DOI] [PubMed] [Google Scholar]
- Petit C, Levilliers J, Hardelin JP (2001) Molecular genetics of hearing loss. Annu Rev Genet 35: 589-646. [DOI] [PubMed] [Google Scholar]
- Platzer J, Engel J, Schrott-Fischer A, Stephan K, Bova S, Chen H, Zheng H, Striessnig J (2000) Congenital deafness and sinoatrial node dysfunction in mice lacking class D L-type Ca2+ channels. Cell 102: 89-97. [DOI] [PubMed] [Google Scholar]
- Pragnell M, De Waard M, Mori Y, Tanabe T, Snutch TP, Campbell KP (1994) Calcium channel beta-subunit binds to a conserved motif in the I-II cytoplasmic linker of the alpha 1-subunit. Nature 368: 67-70. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan NA, Green GE, Pasha R, Drescher MJ, Swanson GS, Perin PC, Lakhani RS, Ahsan SF, Hatfield JS, Khan KM, Drescher DG (2002) Voltage-gated Ca2+ channel Ca(V)1.3 subunit expressed in the hair cell epithelium of the sacculus of the trout Oncorhynchus mykiss: cloning and comparison across vertebrate classes. Brain Res Mol Brain Res 109: 69-83. [DOI] [PubMed] [Google Scholar]
- Roberts WM, Jacobs RA, Hudspeth AJ (1990) Colocalization of ion channels involved in frequency selectivity and synaptic transmission at presynaptic active zones of hair cells. J Neurosci 10: 3664-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D, Paki B (2002) Role of L-type Ca2+ channels in transmitter release from mammalian inner hair cells. II. Single-neuron activity. J Neurophysiol 87: 2734-2740. [DOI] [PubMed] [Google Scholar]
- Safieddine S, Wenthold RJ (1999) SNARE complex at the ribbon synapses of cochlear hair cells: analysis of synaptic vesicle- and synaptic membrane-associated proteins. Eur J Neurosci 11: 803-812. [DOI] [PubMed] [Google Scholar]
- Seiler C, Nicolson T (1999) Defective calmodulin-dependent rapid apical endocytosis in zebrafish sensory hair cell mutants. J Neurobiol 41: 424-434. [PubMed] [Google Scholar]
- Sidi S, Friedrich RW, Nicolson T (2003) NompC TRP channel required for vertebrate sensory hair cell mechanotransduction. Science 301: 96-99. [DOI] [PubMed] [Google Scholar]
- Soldatov NM (1992) Molecular diversity of L-type Ca2+ channel transcripts in human fibroblasts. Proc Natl Acad Sci USA 89: 4628-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Nie L, Rodriguez-Contreras A, Sheng ZH, Yamoah EN (2003) Functional interaction of auxiliary subunits and synaptic proteins with Ca(v)1.3 may impart hair cell Ca2+ current properties. J Neurophysiol 89: 1143-1149. [DOI] [PubMed] [Google Scholar]
- Spassova M, Eisen MD, Saunders JC, Parsons TD (2001) Chick cochlear hair cell exocytosis mediated by dihydropyridine-sensitive calcium channels. J Physiol (Lond) 535: 689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto K, Li D, Nerbonne JM, Levitan ES (1997) Distribution, splicing and glucocorticoid-induced expression of cardiac alpha 1C and alpha 1D voltage-gated Ca2+ channel mRNAs. J Mol Cell Cardiol 29: 3035-3042. [DOI] [PubMed] [Google Scholar]
- Tucker T, Fettiplace R (1995) Confocal imaging of calcium microdomains and calcium extrusion in turtle hair cells. Neuron 15: 1323-1335. [DOI] [PubMed] [Google Scholar]
- Von Kriegstein K, Schmitz F, Link E, Sudhof TC (1999) Distribution of synaptic vesicle proteins in the mammalian retina identifies obligatory and facultative components of ribbon synapses. Eur J Neurosci 11: 1335-1348. [DOI] [PubMed] [Google Scholar]
- Waka N, Knipper M, Engel J (2003) Localization of the calcium channel subunits Cav1.2 (alpha1C) and Cav2.3 (alpha1E) in the mouse organ of Corti. Histol Histopathol 18: 1115-1123. [DOI] [PubMed] [Google Scholar]
- Whitfield TT (2002) Zebrafish as a model for hearing and deafness. J Neurobiol 53: 157-171. [DOI] [PubMed] [Google Scholar]
- Williams ME, Feldman DH, McCue AF, Brenner R, Velicelebi G, Ellis SB, Harpold MM (1992) Structure and functional expression of alpha 1, alpha 2, and beta subunits of a novel human neuronal calcium channel subtype. Neuron 8: 71-84. [DOI] [PubMed] [Google Scholar]
- Zenisek D, Davila V, Wan L, Almers W (2003) Imaging calcium entry sites and ribbon structures in two presynaptic cells. J Neurosci 23: 2538-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SY, Robertson D, Yates G, Everett A (1999) Role of L-type Ca2+ channels in transmitter release from mammalian inner hair cells I. Gross sound-evoked potentials. J Neurophysiol 82: 3307-3315. [DOI] [PubMed] [Google Scholar]
- Zuhlke RD, Pitt GS, Deisseroth K, Tsien RW, Reuter H (1999) Calmodulin supports both inactivation and facilitation of L-type calcium channels. Nature 399: 159-162. [DOI] [PubMed] [Google Scholar]