Abstract
Kainate receptors (KARs) are heteromeric ionotropic glutamate receptors (GluRs) that play various roles in the regulation of synaptic transmission. The KAR subunits GluR5 and GluR6 exist under different splice variant isoforms in the C-terminal domain (GluR5a, GluR5b, GluR5c, GluR6a, GluR6b). The differential role of KAR subunit splice variants is presently unknown. In transfected COS-7 cells and neurons from wild-type and GluR5 × GluR6 mice, we have found that the subcellular localization and membrane delivery differed between these splice variants. GluR6a was highly expressed at the plasma membrane. GluR6b, GluR5a, and GluR5b were detected at lower levels in the plasma membrane and mainly colocalized with calreticulin in the endoplasmic reticulum (ER). GluR5c was strongly retained in the ER by an RXR motif. GluR6a acted as a key subunit splice variant promoting surface expression of ER-retained subunit splice variants when assembled in heteromeric KARs. Surface expression of GluR6a was independent of its PDZ (postsynaptic density-95/discs large/zona occludens-1) binding motif and was promoted by a stretch of four basic amino acid residues at its C terminus. Overall, splice variants and subunit composition of KARs regulate receptor trafficking from the endoplasmic reticulum to the plasma membrane.
Keywords: glutamate, kainate receptors, alternative splicing, intracellular trafficking, endoplasmic reticulum, hippocampal neuron
Introduction
Kainate receptors (KARs) share many structural, pharmacological, and biophysical properties with AMPA receptors, which mediate the majority of fast excitatory synaptic transmissions. KARs, however, appear to play a distinct role in synaptic transmission (Frerking and Nicoll, 2000; Kullmann, 2001; Lerma et al., 2001). At a presynaptic level, KARs act as modulators of transmitter release, at both GABA and glutamate synapses, inducing and regulating synaptic plasticity; at a postsynaptic level, KARs mediate a slow EPSC, which could play a role in the temporal integration of excitatory signals (Castillo et al., 1997; Vignes and Collingridge, 1997; Mulle et al., 1998; Frerking and Ohliger-Frerking, 2002) and in the regulation of neuronal excitability (Melyan et al., 2002).
KARs are hetero-oligomeric receptor channels composed of the subunits glutamate receptor (GluR) 5, GluR6, GluR7, KA1, and KA2. The subunit composition of KARs could be very diverse given the large combinatorial possibilities suggested by KAR subunit mRNA distribution at the tissue level (Wisden and Seeburg, 1993) and in single cells (Ruano et al., 1995; Bureau et al., 2000). GluR5, GluR6, and GluR7 function as homomeric recombinant receptor channels. KA1 and KA2 associate with these subunits to form heteromeric receptors with distinct pharmacological properties (Bettler and Mulle, 1995; Lerma et al., 2001). GluR5 and GluR6 also coassemble to generate recombinant and native receptors with novel functional properties (Cui and Mayer, 1999; Mulle et al., 2000; Paternain et al., 2000).
Complexity is increased by RNA editing and by the existence of splice variants for GluR5, GluR6, and GluR7 receptor subunits (Bettler and Mulle, 1995; Dingledine et al., 1999; Lerma et al., 2001). Apart from an alternatively spliced exon in the N-terminal domain of the GluR5 subunit, all splice variants differ in the sequence of their cytoplasmic C-terminal domain. This domain has now been extensively shown for AMPA and NMDA receptors to be the site of interactions of proteins involved in the trafficking and stabilization of receptors in the synaptic membrane, and in turn, to play a role in synaptic plasticity (Barry and Ziff, 2002; Carroll and Zukin, 2002). Splice variants in the C-terminal domains of GluR5 (GluR5a, GluR5b, and GluR5c) and GluR6 (GluR6a and GluR6b) have been identified (Bettler et al., 1990; Sommer et al., 1992; Gregor et al., 1993; Barbon and Barlati, 2000; Barbon et al., 2001; Jamain et al., 2002) (see Fig. 1A,B). GluR5b is 49 amino acids longer than GluR5a, which contains a stop codon 16 amino acids after the end of membrane domain IV. GluR5c contains an additional 29 amino acid cassette. GluR6a and GluR6b differ in their C-terminal domains by two distinct sequences of 54 and 15 amino acids, respectively, inserted 14 amino acids after membrane domain IV. The C-terminal ends of GluR5b, GluR5c, and GluR6a display postsynaptic density-95/discs large/zona occludens-1 (PDZ)-domain binding motifs (ETVA and ETMA, respectively). The idea that interaction of the different splice variants with distinct sets of intracellular partners such as PDZ domain-containing proteins (Garcia et al., 1998; Hirbec et al., 2003) or proteins of the cadherin-catenin complex (Coussen et al., 2002) accounts for the differential trafficking or activity of KARs has not yet been explored. Here we present the first report comparing the intracellular trafficking and surface expression of the different GluR5 and GluR6 subunit splice variants in heterologous cells and in neurons.
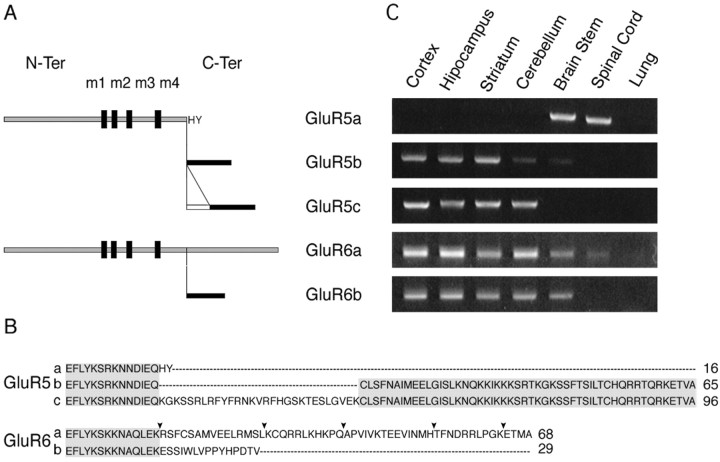
Figure 1.
C-terminal isoforms of GluR5 and GluR6 subunits and mRNA expression in various brain regions. A, Schematic representation of GluR5 and GluR6 C-terminal isoforms generated by alternative splicing. B, Clustal W alignment of GluR5 and GluR6 C-terminal splice variants (at the end of sequence, size in amino acid). Gray box corresponds to homologous regions between isoforms. Black arrowheads indicate the truncated forms used further. C, Specific RT-PCR products of each isoform in ethidium bromide agarose gels. Representative example from a set of four independent experiments is shown. All C-terminal isoforms are detected in the mouse nervous system (lung as a negative control).
Materials and Methods
RT-PCR experiments. RNA was prepared from tissues from adult C57BL/6 mice using the RNA NOW protocol (Ozyme), with 50-100 mg of starting material. The RT step was performed on 1 μg of total RNA with oligo-dT primers. RT products were amplified using splice variant-specific reverse primer as follows: GluR5 forward, GTGACGAGGGGTACCAAAGGGTGCTC; GluR5a reverse, TACGACACTTCAGTAATGCTG; GluR5b reverse, TCACGCCACAGTCTCTTTTCT; GluR5c reverse, ACCAAGGCTTTCTGTTTTGCT; GluR6 forward, ACTCCAAAGGCTATGGCGTTGG; GluR6a reverse, GGCACTTCAGGGACATTCTCAGCTC; GluR6b reverse, CTGGATGGTATGGTGGCACTAACC. PCR was performed using 40 cycles: 30 sec at 94°C; 30 sec at the optimal temperature (GluR5a, 47°C; GluR5b, 48°C; GluR5c, 50°C; GluR6a, 57°C; GluR6b, 58°C); 45 sec at 72°C. The size of the PCR products was as follows: GluR5a, 370 bp; GluR5b, 600 bp; GluR5c, 430 bp; GluR6a, 250 bp; GluR6b, 320 bp.
DNA constructs. Six consecutive c-myc epitopes were introduced after the predicted signal peptide of rat GluR5a(Q) and GluR6a(Q) cDNA (accession numbers: GluR5, M83560/P22756; GluR6a, NM_021956/NP_068775) and subcloned into pcDNA3 vector. The BstXI site located before membrane domain IV of the protein was mutated in the NheI restriction site by PCR, and an AflII restriction site was introduced after the stop codon. This cassette allowed exchanges for the different deleted GluR6 and GluR5 C-terminal domains. DNA coding for the last GluR6b (NM_175768/NP_786944) 15 amino acids was produced by PCR reaction and taken as a matrix together with GluR6a plasmid to produce by PCR the cassette containing the NheI and AflII restriction sites. cDNAs coding for GluR5b and GluR5c C-terminus domains were introduced by replacing the corresponding PstI-NotI fragments into myc-GluR5a cut by the same enzymes. Site-directed mutagenesis was performed using the QuickChange XL kit (Stratagene, Amsterdam, The Netherlands). Each tagged isoform (as well as truncation mutants of GluR6a) was expressed in human embryonic kidney (HEK) 293 cells, and their electrophysiological response to glutamate application was tested. Saturating concentrations of glutamate (10 mm) evoked fast activating inward currents for all tagged splice variants that were tested, except for GluR5c (supplemental data).
Hippocampal cultures. Primary cultures of hippocampal neurons were obtained from 1-d-old pups of C57BL/6 and GluR5 × GluR6 mutant mice (Mulle et al., 2000). Hippocampi were dissociated with trypsin followed by mechanical trituration and plated at 50,000 cells/cm2 in minimum essential medium (MEM)-Eagle supplemented with 0.5% d-glucose, 0.1 mg/ml transferrin, 25 μg/ml insulin, 2 mm Glutamax (Invitrogen), and 5 μg/ml gentamycin. B-27 (2%) (Sigma, St. Louis, MO) and 1 μm cytosine arabinoside were added 3 d after plating.
Transfection. COS-7 cells were transfected as described (Coussen et al., 2002). Hippocampal neurons (10-14 d old) were transfected using Lipofectamine 2000 transfection reagent (Invitrogen). Three micrograms of DNA in 500 μl of MEM-Eagle was mixed with 10 μl of Lipofectamine 2000 in 500 μl of MEM-Eagle and incubated at room temperature for 15 min. Cultured medium was then replaced by 1 ml of MEM-Eagle complemented with the transfection mix. After 1 hr of incubation, the transfection medium was replaced with 3 ml of growth medium. Experiments were performed 24 hr after transfection.
Immunostaining: double staining for surface expression experiments. Polyclonal anti-myc antibody (Ab) (10 μg/ml; catalog #06-549, Upstate Biotechnology, Lake Placid, NY) was incubated with the cells for 20 min at 20°C to limit antibody endocytosis. Cells were washed, fixed in paraformaldehyde (4%), and kept in PBS; 0.3% BSA and 0.05% saponin were added during the steps that require membrane permeabilization. Intracellular epitopes were detected using monoclonal 0.5 μg/ml anti-myc Ab (catalog #9E10; Roche, Hertforshire, UK) incubated for 1 hr at 20°C. ER staining was performed with polyclonal anti-calreticulin Ab (1:500; catalog #PA3-900, Upstate Biotechnology) for 1 hr at 20°C. Secondary Abs were incubated for 1 hr at 20°C and included Ab anti-rabbit Alexa 568- and 488-conjugated IgGs (catalog #A-11011 and #A-11001, Molecular Probes, Eugene, OR), anti-mouse Alexa 568- and 488-conjugated IgGs (10 μg/ml; catalog #A-11008 and #A-11004, Molecular Probes). The subcellular distribution of KAR subunit labeling was explored by sequential triple labeling. Neurons were incubated with monoclonal anti-myc Ab for 1 hr at 20°C; the primary antibody was then washed and revealed by the secondary Ab Alexa 568-conjugated IgGs in saturating conditions. Neurons were then permeabilized with saponin and incubated with monoclonal anti-microtubule-associated protein 2 (MAP2) Ab (1:500; catalog #HM-2, Sigma) and polyclonal anti-vesicular glutamate transporter 1 (VGlut1) Ab, (1:500) (Herzog et al., 2001) for 1 hr at 20°C. Anti-MAP2 and -VGlut1 antibodies were revealed by secondary Ab Cy5 (20 μg/ml; Jackson Immuno Research, West Grove, PA) and Alexa 488-conjugated IgGs respectively.
Surface biotinylation and glycosylation assays. COS-7 cells were washed two times with PBS, pH 8.0, incubated with 1 mg/ml of EZ-Link Sulfo-NHS-LC-Biotin (Pierce, Rockford, IL) in PBS for 30 min at 4°C, washed three times with PBS, and scraped in lysis buffer containing 25 mm HEPES, 150 mm NaCl, 1% Triton X-100, and a mix of protease inhibitors. After centrifugation, the supernatant was immunoprecipitated with 50 μl of Immunopure immobilized streptavidin beaded agarose overnight at 4°C and washed extensively. For deglycosylation experiments, endoglycosidase H (EndoH) and N-glycosidase F (PNG-F) were added to protein samples for 12 hr at 37°C. Quantification was performed using NIH Image J software.
Immunostaining data analysis. All images, except confocal images, were acquired on an Axiophot II microscope (Zeiss, Oberkochen, Germany) using a 63× objective, coupled to Quantix digital cooled CCD camera (Photometrix, Roper, NJ) driven by IPlab software (Scanalistics). The exposure settings corresponding to undersaturated acquisitions were kept the same for each condition. Images in the displayed figures are overexposed for a better demonstration but do not correspond to the analyzed images. The relative surface labeling value was analyzed as follows. Background was flattened in both intracellular and surface stainings, and positive signals were selected by intensity threshold calculated as the product of the remaining background by a constant (the estimation of the constant depended on the signal-to-noise ratio in each wavelength). Surface and intracellular labeled areas above threshold were measured (number of pixels: 11.8 nm2 per pixel). To determine the relative surface labeling for the aspecific signal attributable to the antibody, we calculated the ratio of surface staining of untranfected cells to intracellular staining of a neighbor transfected cell (i.e., with GluR5c in COS-7 cell, mean ± SEM = 0.9 ± 0.5%, and in neurons mean ± SEM = 5.4 ± 3.8%). Image analysis was performed using Metamorph Imaging software (Universal Imaging, West Chester, PA). Statistical significance was tested using a one-way or two-way ANOVA followed by a Wilcoxon nonparametric test. Confocal images were acquired on a Leica TCS SP2 microscope. The exposure settings and gain of laser were kept the same for each condition. To avoid cross-detection of Cy5 and Alexa 568, we used the sequential excitation-acquisition mode.
Results
We first verified that GluR5a, GluR5b, GluR5c, GluR6a, and GluR6b were all expressed in the mouse brain using RT-PCR from selected brain regions (Fig. 1C). GluR5a is detected in the brain stem and spinal cord. Both GluR5b and GluR5c are expressed in the cortex, the hippocampus, the striatum, and the cerebellum. GluR5c is not detected in the brain stem. The two C-terminal splice variants of GluR6 follow the same pattern of expression in all brain regions, except for the spinal cord, where GluR6b is not detected under our conditions.
Membrane delivery and subcellular localization of GluR5 and GluR6 C-terminal splice variants expressed as homo-oligomeric receptors
We tagged each GluR5 and GluR6 splice variant by insertion of six successive c-myc epitopes between the predicted signal peptide and the extracellular N terminus (Coussen et al., 2002). Myc-tagged GluR5 and GluR6 splice variants were transfected in COS-7 cells and labeled for surface receptors in live cells and for intracellular receptors after permeabilization (Fig. 2A). Surface labeling, distributed as clusters over all of the cell plasma membrane, was observed for all subunit isoforms, except for GluR5c (Fig. 2A). Qualitatively, the relative level of surface versus intracellular receptor labeling clearly varied among the GluR5 and GluR6 splice variants. Intracellular staining of GluR6a was concentrated and restricted to the center of the cell, in a compartment that might correspond to either the Golgi apparatus or a pericentriolar recycling compartment. In contrast, intracellular GluR6b and GluR5 isoforms were distributed in perinuclear and reticular structures likely corresponding to the ER. We have quantified the relative amount of surface-expressed receptors for each isoform (see Materials and Methods) (n = 10 cells in each condition) (Fig. 2B). GluR5a and GluR5b isoforms were detected at low levels at the plasma membrane (GluR5a: 9 ± 1%; GluR5b: 13 ± 3%), whereas the amount of surface staining for GluR5c was not different from background (GluR5c: 1.7 ± 0.3%; p < 0.001 vs GluR5a and GluR5b). Both GluR6 isoforms were detected at significantly higher levels than GluR5 isoforms (p < 0.05). GluR6a was detected at higher levels at the cell surface than GluR6b (GluR6a: 56 ± 3% of surface expression; GluR6b: 25 ± 5%; p < 0.005).
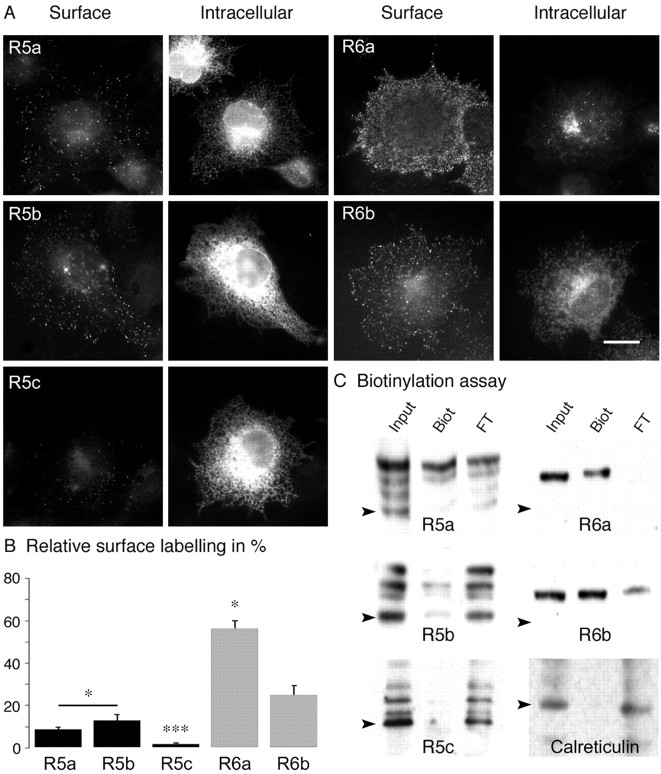
Figure 2.
Membrane delivery of homomeric receptors in COS-7 cells. A, Double staining of tagged subunits transfected in COS-7 cells: surface column represents live staining; intracellular column corresponds to staining of the same cells after permeabilization. Scale bar, 10 μm. B, Quantitative analysis of surface labeling for GluR5 and GluR6 isoforms (n = 10; R5a, R5b vs R6 isoforms, barred asterisk, p < 0.05; R5c vs R5a and R5b, ***p < 0.001; R6a vs R6b, *p < 0.005). C, Biotinylation assays in transfected COS-7 cells: “Input” corresponds to total proteins, “Biot” to the biotinylated fraction, and “FT” (flow through) to unbiotinylated intracellular proteins. For all panels, Western blotting was performed with polyclonal anti-myc antibody for KAR subunits and with an anti-calreticulin antibody. The amount of samples loaded on the gel in the three lanes was adjusted to be in the same range of optical density. Black arrowheads indicate the predicted molecular weight of tagged subunits and calreticulin.
To confirm that KAR subunits and alternative splice variants are differentially targeted to the plasma membrane of COS-7 cells, we used a biotinylation assay. GluR5a, GluR5b, GluR6a, and GluR6b were detected as surface-expressed proteins, consistent with fluorescence image analysis. No significant biotinylated fraction was detected for GluR5c. Unexpectedly, several bands were detected for GluR5a, GluR5b, and GluR5c. The same bands were detected when Western blots were reprobed with a C-terminus GluR5b/c antibody (data not shown), suggesting that the bottom bands do not correspond to degradation products. The top bands detected for GluR5 and GluR6 isoforms corresponded to higher molecular weights than predicted for non-glycosylated forms. The relative amount of biotinylated (surface) KARs relative to the total amount of receptors (input lane) was quantified by image analysis of the Western blots. The intensity of the different biotinylated bands was cumulated for a given splice variant. Quantification revealed differences between subunits and splice variants that were consistent with fluorescence measurements of cell surface staining (GluR5a: 12 ± 2%; GluR5b: 11 ± 3%; GluR5c: 2 ± 1%; GluR6a: 88 ± 3%; GluR6b: 39 ± 6%; n = 3). As a control, we verified that calreticulin (60 kDa), an intracellular protein, was not biotinylated in our assay (Fig. 2C).
The low expression of GluR5 splice variants at the plasma membrane of COS-7 cells could be caused by retention in the ER, which serves as a quality control checkpoint for properly folded and assembled multimeric receptors before export to the Golgi apparatus and to the plasma membrane (Ma and Jan, 2002). We observed that GluR5 splice variants colocalized with calreticulin, an ER resident chaperone protein (Leach et al., 2002) (Fig. 3A). GluR6b staining was colocalized with calreticulin in bright puncta that may correspond to ER exit sites (Fig. 3A) (Ellgaard and Helenius, 2003). Finally, no colocalization with calreticulin was observed for GluR6a.
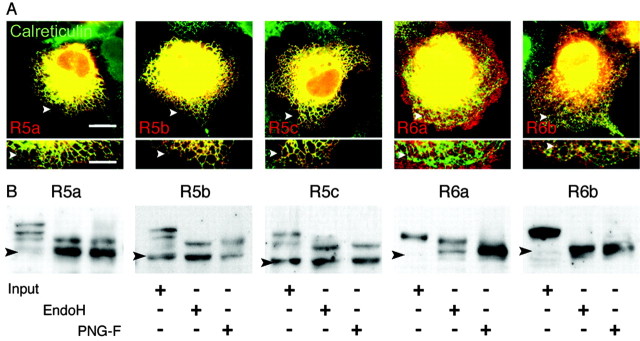
Figure 3.
Distribution of GluR5 and GluR6 isoforms in the ER. A, Transfected COS-7 cells were costained for total c-myc epitopes (red) and the ER resident protein calreticulin (green). Regions of interest are represented at a higher magnification under the corresponding cells; white arrowheads serve as a reference. GluR5 isoforms mainly overlap with the ER marker, and GluR6b partially colocalize in hot spots. No colocalization is observed for GluR6a. Scale bars: top images, 10 μm; boxes below, 2 μm. B, Western blot of protein extracts from transfected COS-7 cells blotted with an anti-myc antibody. Input lane represents the total protein sample; EndoH and PNG-F, respectively, correspond to digestion products. GluR5 isoforms are deglycosylated in the same manner by both enzyme treatments. GluR6a is partly resistant to EndoH, whereas GluR6b is sensitive to EndoH and PNG-F. Black arrowheads indicate the predicted molecular weights of tagged subunits.
We next examined the pattern of glycosylation of KAR subunit isoforms. In Western blots from total protein extracts of transfected COS-7 cells, several bands were detected for GluR5 isoforms (Fig. 3B, input lane). The molecular weight of the lower bands corresponded to the predicted molecular weight of non-glycosylated myc-GluR5 isoforms, i.e., 109 kDa for GluR5a, 115 kDa for GluR5b, and 118 kDa for GluR5c. The higher bands might correspond to glycosylated subunits. GluR5 carries eight predicted N-glycosylation sites (PubMed accession number Q60934). A single band was observed for GluR6 isoforms (Fig. 3B, input lane). This band corresponded to a higher molecular weight than predicted for non-glycosylated myc-GluR6 isoforms, i.e., 113 kDa for GluR6a and 108 kDa for GluR6b. GluR6 isoforms carry eight predicted N-glycosylation sites (PubMed P39087). We treated protein extracts with PNG-F, which removes N-glycans. GluR6 proteins treated with PNG-F migrated to the molecular weight predicted for non-glycosylated proteins. GluR5 isoforms treated with PNG-F displayed two bands, one of which corresponded to a non-glycosylated subunit. A second band with a slightly higher molecular weight was also observed, potentially associated with a different form of post-translational modification, independent of C-terminus alternative splicing. Resistance to EndoH is conferred by mannosidase 2 processing of the protein that occurs in the Golgi apparatus. GluR6a is resistant in part to the treatment by EndoH. Similar results were obtained with GluR6a containing KARs extracted from the brain of transgenic mice expressing myc-tagged GluR6a (Coussen et al., 2002) (see supplemental data). For all other isoforms, no Endo-H-resistant fraction could be detected under our experimental conditions (Fig. 3B), reflecting the low amount of proteins that have undergone the last phases of glycosylation and have trafficked to the plasma membrane. GluR5 surface (biotinylated) proteins were detected as several bands that may correspond to different glycosylated forms (Fig. 2C), suggesting that incompletely glycosylated GluR5 subunits might have trafficked to the plasma membrane. In keeping with this suggestion, glycosylation is not necessary for the proper function and expression of KARs in oocytes (Everts et al., 1997). Alternatively, GluR5 isoforms might exist under other different post-translational states. Overall, the low surface expression of homomeric GluR5 isoforms and of GluR6b can be explained by restricted export of the subunits from the ER, whereas GluR6a trafficks readily from the ER to the plasma membrane.
Membrane delivery and ER retention of GluR5 and GluR6 splice variants in hippocampal neurons
We next examined surface and intracellular receptors in cultured mouse hippocampal neurons transfected at 14 d in vitro. Both GluR5 and GluR6 isoforms were detected at the plasma membrane as puncta along neurites and neuronal cell bodies (Fig. 4A, “surface” column). Intracellular labeling of all isoforms was diffuse and observed in the soma and all along neuritic extensions (Fig. 4A, “intracellular” column). To quantify surface expression of GluR5 and GluR6 isoforms in hippocampal neurons, we used the same ratiometric measurement as in COS-7 cells (n = 20 cells for each isoform) (Fig. 4B). GluR5a and GluR5b displayed comparable levels of surface expression (GluR5a: 34 ± 7%; GluR5b: 33 ± 4%). GluR5c extracellular expression was weak, albeit significantly different from background (GluR5c: 18 ± 3%; p < 0.005). GluR6 isoforms displayed higher levels of surface expression than GluR5 (GluR6a: 77 ± 3%; GluR6b: 64 ± 2%). Intracellular GluR5 isoforms strongly colocalized with the ER marker calreticulin in neurites (Fig. 4C) as well as in the cell body (data not shown). We observed only limited colocalization of calreticulin with GluR6b and no colocalization with GluR6a. In hippocampal neurons as well as in COS-7 cells, differential membrane delivery of GluR5 and GluR6 isoforms could thus be explained by differential trafficking through the ER.
Figure 4.
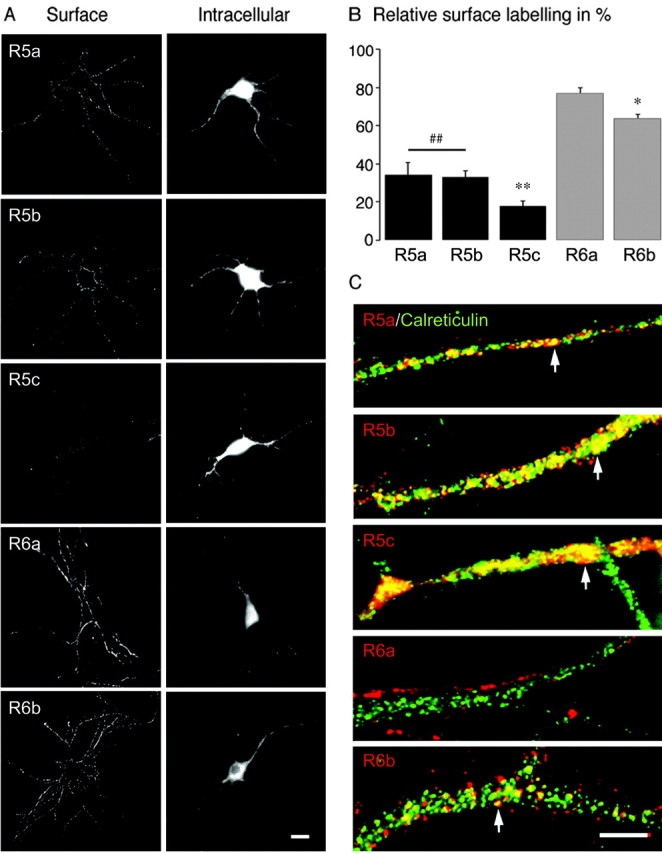
Subcellular distribution of GluR5 and GluR6 isoforms in cultured hippocampal neurons. A, Surface column represents surface staining; intracellular column shows anti-myc staining of the same cells after permeabilization. All isoforms were detected at the plasma membrane of transfected neurons, distributed as clusters in the cell body and along neuritic extensions. GluR5 isoforms are less able to be detected at the cell surface. Scale bar, 10 μm. B, Quantification of the relative surface labeling of each subunit (n = 20; R5a, R5b vs R6 isoforms, ##p < 0.005; R5c vs R5a and R5b, **p < 0.005; R6a vs R6b, *p < 0.05). C, GluR5 and GluR6 isoforms (red) costained with calreticulin (green). GluR5 isoforms and GluR6b colocalize in part with the ER marker (white arrowheads); no colocalization is observed for GluR6a. Scale bar, 2 μm.
GluR6a promotes heteromeric KAR trafficking to the plasma membrane
GluR5 and GluR6 subunits are known to be expressed in cultured hippocampal neurons (Ruano et al., 1995). These subunits could coassemble with the transfected KAR subunits and influence their trafficking. We thus repeated the experiments in cultured hippocampal neurons derived from mice deficient for the two major KAR subunits GluR5 and GluR6 (R5-/- × R6-/- mice) (Fig. 5) (Mulle et al., 2000). We found striking differences in the distribution of transfected tagged subunits in these neurons compared with wild-type neurons (Fig. 5A). GluR5a, GluR5b, and GluR5c are less able to be detected at the surface of R5-/- × R6-/- neurons compared with wild-type neurons (GluR5a: 17 ± 4%; GluR5b: 24 ± 4%; GluR5c: 9 ± 1%; p < 0.05 vs wild type; not significantly different from background for GluR5c) (Fig. 5B). Surface expression of GluR6b decreased mostly in neurons derived from R5-/-xR6-/- mice compared with neurons from wild-type mice (GluR6b: 23 ± 4%; p < 0.001) (Fig. 5). In contrast, GluR6a surface expression in neurons from R5-/- × R6-/- mice was comparable with that in neurons from wild-type mice (GluR6a: 69 ± 3%) (Fig. 5). Altogether, these results indicate that endogenously expressed KAR subunits are essential for the membrane delivery of GluR5c to the plasma membrane and that they positively regulate surface expression of GluR5a, GluR5b, and GluR6b without affecting GluR6a.
Figure 5.
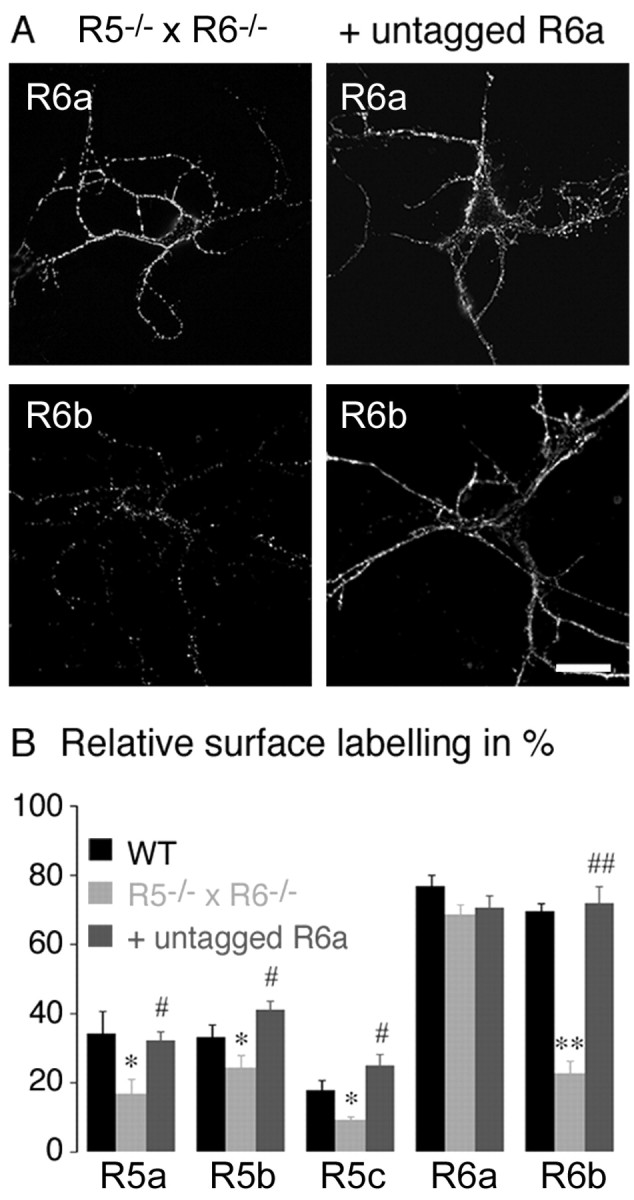
Membrane delivery of GluR5 and GluR6 subunits in R5-/- × R6-/- mice neurons and cotransfected with untagged GluR6a. A, Left column, Surface staining for tagged GluR6a and tagged GluR6b; right column, surface staining for tagged GluR6a and tagged GluR6b cotransfected with untagged GluR6a. Scale bar, 10 μm. B, Relative surface labeling for GluR5 and GluR6 isoforms in neurons from wild-type and R5-/- × R6-/- mice, and in neurons from R5 × R6 mice cotransfected with untagged GluR6a (n = 20; R5 × R6 vs wild type, **p < 0.001, *p < 0.05; R5-/- × R6-/-, cotransfection or not with untagged GluR6a, ##p < 0.001, #p < 0.05). WT, Wild type.
GluR6 is highly expressed in hippocampal pyramidal neurons in culture (Ruano et al., 1995), and GluR6a displays high levels of surface expression in all conditions tested. We thus reasoned that GluR6a, by coassembly with either one of the GluR5 isoforms or with GluR6b, could promote their delivery to the plasma membrane. We cotransfected hippocampal neurons from R5-/- × R6-/- mice with each tagged GluR5 and GluR6 isoform and with the untagged GluR6a subunit in a 1:1 ratio. In these conditions, the level of expression of each subunit isoform recovered the relative surface labeling that they displayed when expressed in neurons from wild-type mice (Fig. 5A,B). In neurons from R5-/- × R6-/- mice, the level of surface expression of the GluR5 isoforms and of GluR6b was significantly higher when these isoforms were cotransfected with GluR6a (GluR5a: 32 ± 3%; GluR5b: 41 ± 3%; GluR5c: 25 ± 3%; p < 0.05; GluR6b: 72 ± 5%; p < 0.001). Tagged GluR6a was detected at comparable levels when coexpressed with untagged GluR6a under the same conditions (GluR6a: 70 ± 4%). The low surface expression level of KAR subunits in neurons from R5-/- × R6-/- mice is thus fully rescued by coexpression with GluR6a. In COS-7 cells, GluR7a but not KA1 or KA2 also promoted surface expression of the ER retained subunit splice variant GluR5b (see supplemental data). It should be noted, however, that GluR7 does not appear to be expressed in cultured hippocampal neurons (Ruano et al., 1995). Overall, GluR6a appears to be a critical subunit for the targeting of heteromeric KARs to the neuronal plasma membrane.
Subcellular localization of surface KARs in cultured neurons
The role of KARs in synaptic transmission depends on the subcellular localization of these receptors in specific membrane domains. No study has yet given a detailed description of the subcellular localization of KARs in hippocampal cultured neurons. Because of the lack of suitable N-terminal antibodies that would allow the localization of KARs in nonpermeabilized cells, we have not been able to study the distribution of native KAR subunits at the plasma membrane. Using transfection of tagged receptors, one must take into account the presence of endogenous subunits that can interfere with the trafficking of tagged KAR subunits through coassembly. By using neuronal cultures from R5-/- × R6-/- mice, we circumvent this problem, in great part, because these subunits are the major channel-forming subunits, although we cannot rule out a role for other subunits in the subcellular targeting of KARs. We examined the surface localization of transfected tagged GluR5a, GluR5b, GluR6a, and GluR6b in cultured hippocampal neurons. Triple labeling was performed to explore the distribution of individual tagged subunits in relation to axonal versus dendritic compartments and in relation to glutamatergic synapses. Dendrites were labeled with an antibody against MAP2. Glutamatergic synapses were labeled with an antibody against VGluT1 (Herzog et al., 2001). We found that GluR5a, GluR5b, GluR6a, and GluR6b were equally distributed at the surface of MAP2-positive and MAP2-negative neurites (Fig. 6). In addition, GluR5 and GluR6 splice variants did not colocalize with glutamatergic synapses (Fig. 6, large squares). For GluR6, however, we found clusters in close apposition with nontransfected MAP2-positive neurites, likely corresponding to presynaptic terminals. These results suggest that alternative splicing of GluR5 and GluR6 subunits might not be critical for the polarized targeting of KARs in cultured hippocampal neurons.
Figure 6.
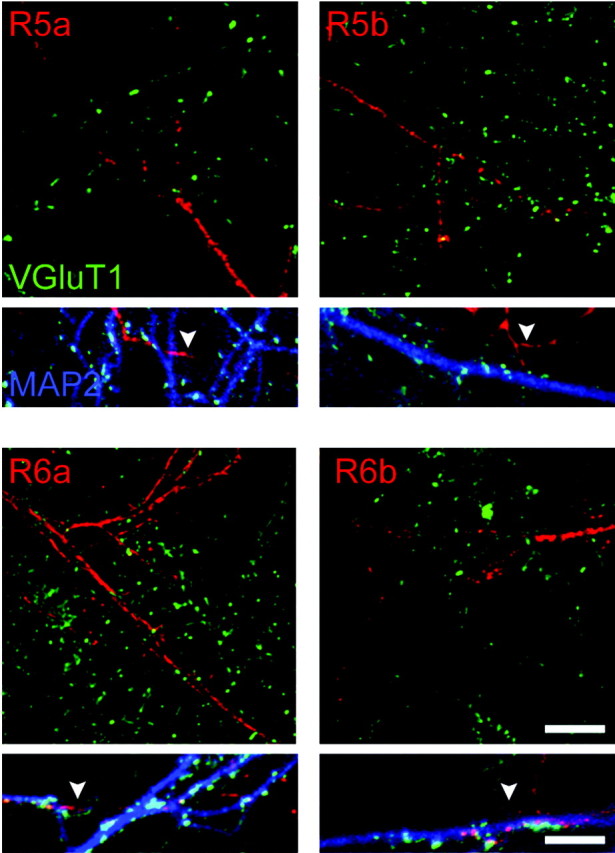
Subcellular distribution of KAR subunit isoforms. Costaining of surface KAR subunit isoforms with the dendritic marker MAP2 and the marker for glutamatergic synapses VGluT1 in neurons from R5-/- × R6-/- mice. The top images show the distribution of each subunit isoform (myc, red) (except GluR5c) in dendrites (costaining of myc and MAP2 indicating dendritic localization of KARs is not shown for clarity) in reference to glutamatergic synapses (VGluT1, green). None of these subunits colocalize with the synaptic marker VGluT1. The boxes below show triple labeling: MAP2 (blue), myc (red), and VGluT1 (green). White arrowheads indicate MAP2-negative extensions, which are likely axons. All subunits are detected on axons of transfected neurons. Scale bars: top images, 5 μm; boxes below, 2 μm.
Molecular determinants for GluR5c ER retention and GluR6a surface targeting
GluR5c differs from GluR5b by an insertion of 29 amino acids. This sequence contains an RXR motif (Fig. 7A) that was identified as an ER retention signal for both potassium channels and G-protein-coupled receptors (Zerangue et al., 1999; Margeta-Mitrovic et al., 2000). We thus examined whether this motif was responsible for ER retention of GluR5c in COS-7 cells and in neurons from R5-/- × R6-/- mice. We replaced the two arginines by alanine residues in myc-tagged GluR5c (GluR5c-ALA). At variance with GluR5c, GluR5c-ALA transfected in COS-7 cells was expressed at the plasma membrane (Fig. 7B) at levels comparable with GluR5b (relative amount of surface expressed receptors: GluR5c: 1.7 ± 0.3%; GluR5c-ALA: 12 ± 3%; GluR5b: 13 ± 3%). As for GluR5b, intracellular GluR5c-ALA was found in ER-like compartments. Similar results were obtained by transfecting GluR5c-ALA in hippocampal neurons derived from R5-/- × R6-/- mice (GluR5c: 9 ± 1%; GluR5c-ALA: 24 ± 2%; GluR5b: 24 ± 4%) (Fig. 7C). The RLR motif in GluR5c thus acts as an ER retention signal. In addition, it has been shown recently that other molecular determinants common to both GluR5b and GluR5c appear to account for the low surface expression levels of GluR5b and GluR5c-ALA (Ren et al., 2003b).
Figure 7.
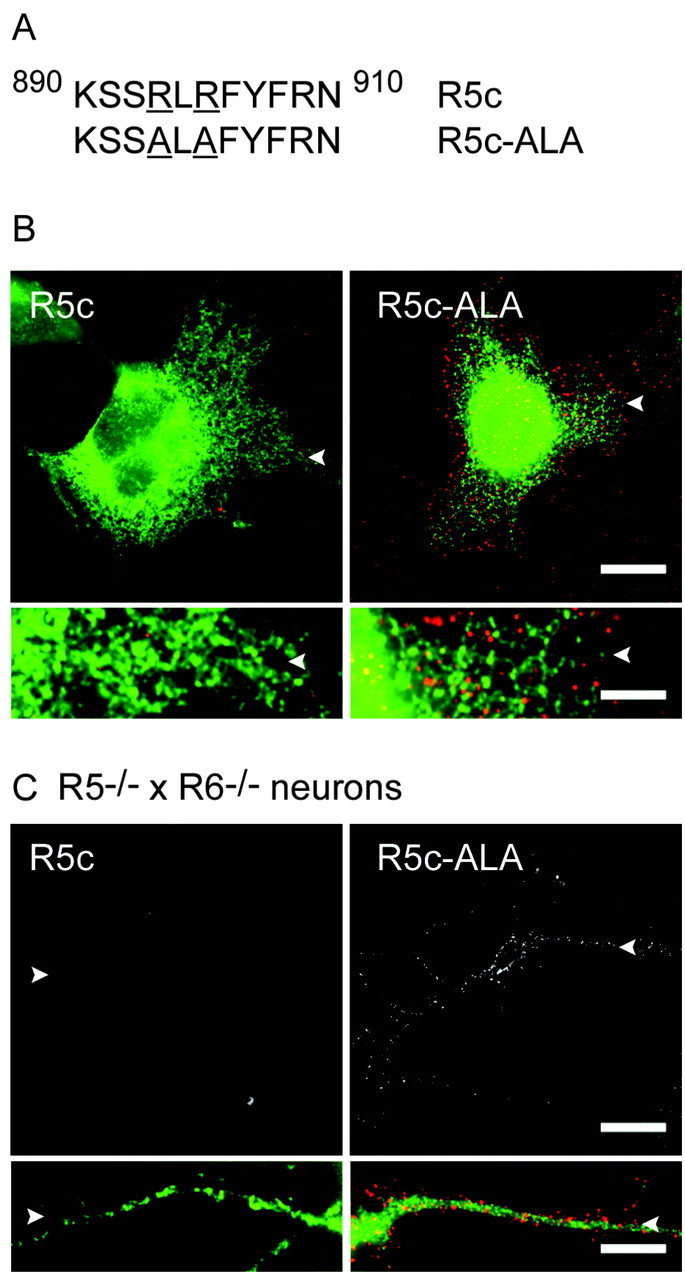
GluR5c carries an RXR-type ER retention motif. A, Sequence of GluR5c surrounding the RXR site (890-910) and its mutated form GluR5c-ALA. B, Double staining (surface in red and intracellular in green) of GluR5c and GluR5c-ALA in transfected COS-7 cells. Mutation of the RXR site in GluR5c allows the detection of surface-expressed receptors (square pictures and boxes below) without changing the ER-like distribution of intracellular labeling. C, Expression of GluR5c and GluR5c-ALA in cultured neurons from R5-/- × R6-/- mice. Only GluR5c-ALA is detected at the cell surface (in top images, surface staining; in boxes below, surface staining in red and intracellular labeling in green). Bottom images represent at a higher magnification a detail of the corresponding top image; white arrowheads serve as a reference. Scale bars: top images, 10 μm for top images; bottom images, 2 μm.
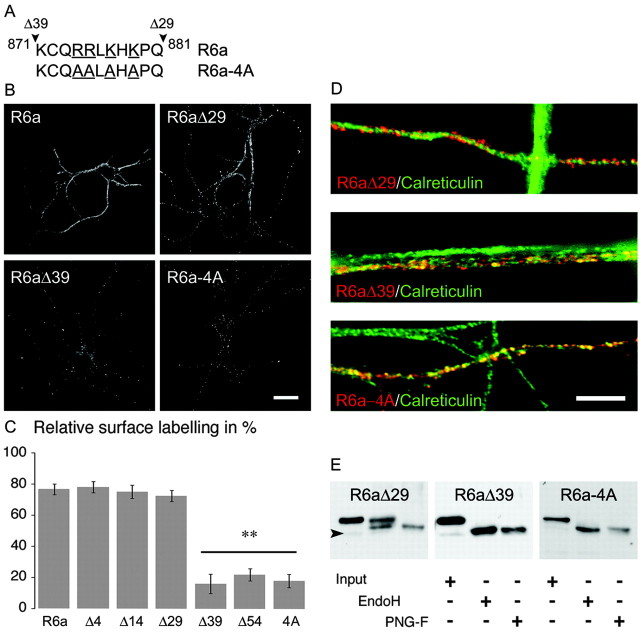
We examined whether GluR6a carries a membrane delivery motif by generating deletion mutants of the GluR6a C-terminal domain. The C-terminal end of GluR6a interacts with PDZ domain-containing proteins, such as postsynaptic density-95 (PSD-95), glutamate receptor interacting protein (GRIP), and protein intreacting with C kinase-1 (PICK1) (Garcia et al., 1998; Hirbec et al., 2003), as well as with proteins of the cadherin/catenin complex and CASK (calmodulin-dependent serine kinase) (Coussen et al., 2002). To investigate the potential role of these interacting proteins in the membrane delivery of GluR6a and to identify sequences responsible for forward trafficking of GluR6a, we have transfected COS-7 cells with different truncated forms of this subunit: GluR6aΔ4, truncated for the PDZ binding motif (ETMA); GluR6aΔ14, truncated for the last 14 amino acids essential for the interaction with β-catenin (Fig. 1B). The deletion of PDZ and cadherin-catenin interaction domains did not affect membrane delivery of GluR6a in COS-7 cells. In contrast, truncation of the last 39 amino acids of GluR6a critically decreased its surface expression (GluR6a: 66 ± 4%; GluR6aΔ4: 59 ± 5%; GluR6aΔ14: 64 ± 5%; GluR6aΔ29: 64 ± 6%; GluR6aΔ39: 5 ± 3%; GluR6aΔ39 vs others; p < 0.001). In hippocampal neurons from R5-/- × R6-/- mice, we obtained results similar to those described above for COS-7 cells (Fig. 8B,C). No significant difference was observed in surface expression among GluR6a, GluR6aΔ4, GluR6aΔ14, and GluR6aΔ29 (GluR6a: 77 ± 3%; GluR6aΔ4: 78 ± 4%; GluR6aΔ14: 75 ± 4%; GluR6aΔ29: 72 ± 4%; n = 20 cells) (Fig. 8B). Membrane delivery of GluR6a was clearly impaired when the last 39 or 54 amino acids were removed (GluR6aΔ39: 16 ± 6%; GluR6aΔ54: 22 ± 4%; GluR6aΔ39 vs others; p < 0.001). At variance with GluR6a or GluR6aΔ29, GluR6aΔ39 colocalized with calreticulin in neurites (Fig. 8D) and in cell bodies (data not shown) and is completely sensitive to EndoH (Fig. 8E), indicating that this truncated subunit is retained in the ER. These results strongly suggest that the GluR6a splice variant carries an essential protein sequence responsible for the high surface expression of KARs between amino acids 871 and 882 (Fig. 8A). This sequence contains four charged amino acid residues that we have mutated to alanines (GluR6a-4A). In COS-7 cells and in neurons derived from R5-/- × R6-/- mice, GluR6a-4A was detected less at plasma membrane than full-length GluR6a (Fig. 8) (COS-7 cells: GluR6a-4A: 5 ± 2%; R5-/- × R6-/- neurons: GluR6a-4A: 18 ± 4%; p < 0.001 compared with GluR6a). Intracellular GluR6a-4A is colocalized with the ER marker calreticulin (Fig. 8D) and is completely sensitive to EndoH (Fig. 8E). We have thus identified a group of four charged amino acid residues (RRXKXK) as an essential determinant for the high surface expression of GluR6a in the plasma membrane of COS-7 cells and neurons.
Figure 8.
GluR6a carries an ER export motif. A, Sequence of GluR6a composed between GluR6a Δ29 and GluR6a Δ39 (black arrowheads; amino acids 871-881) and mutation of the four underlined amino acids (GluR6a-4A). B, Surface expression of GluR6a, GluR6aΔ29, GluR6aΔ39, and GluR6a-4A in cultured neurons from R5-/- × R6-/- mice. Loss or mutation of the four basic amino acids (874-RRXKXK-879) critically decreases surface expression of GluR6a-containing KARs. Scale bar, 10 μm. C, Relative surface labeling of truncated and mutated isoforms in neurons from R5-/- × R6-/- mice (n = 20; R6aΔ39, R6aΔ54, and R6a-4A vs others, **p < 0.001). D, Costaining of GluR6aΔ29, GluR6aΔ39, and GluR6a-4A with calreticulin. Yellow puncta show that GluR6aΔ39 and GluR6a-4A localize in the ER. Scale bar, 2 μm. E, Western blot of protein extracts from transfected COS-7 cells blotted with an anti-myc antibody. Input lane is the total protein sample; EndoH and PNG-F, respectively, correspond to digestion products of corresponding enzymes. Like GluR6a, GluR6aΔ29 is partially resistant to EndoH, whereas GluR6aΔ39 and GluR6a-4A are completely deglycosylated by EndoH and PNG-F. Black arrowhead indicates the predicted molecular weights of tagged subunits.
Discussion
Regulation of the trafficking of glutamate receptors to the plasma membrane provides an essential mechanism for the control of synaptic strength (Barry and Ziff, 2002; Carroll and Zukin, 2002). Although recent progress has been made in the understanding of the trafficking of KARs (Coussen et al., 2002; Hirbec et al., 2003; Ren et al., 2003a,b), little is known about the role of the various subunit splice variants in the trafficking and function of KARs. Here we have compared intracellular trafficking and cell surface expression of splice variants for the two major channel-forming KAR subunits GluR5 and GluR6. We report that C-terminal splice variants of GluR5 and GluR6 differentially traffick from the ER to the plasma membrane. In cell lines and in cultured hippocampal neurons, we show that KAR subunit splice variants display no surface expression (GluR5c), low surface expression (GluR5a, GluR5b, GluR6b), or high surface expression (GluR6a). Splice variants with low surface expression are retained in the ER. We demonstrate that GluR5c is fully retained in the ER because of an “RXR” ER retention motif. In contrast, GluR6a appears as a key splice variant that readily targets to the plasma membrane. Furthermore, GluR6a promotes surface expression of heteromeric KARs containing ER retained splice variants. We identify a stretch of four basic amino acids as being responsible for the export of GluR6a from the ER to the plasma membrane.
Membrane delivery of GluR5 splice variants
At variance with GluR5a and GluR5b, which were detected (at low levels) at the plasma membrane, no surface expression was observed for GluR5c when expressed alone in cell lines or in neurons derived from mutant mice. In addition, no current was activated by glutamate when GluR5c was expressed in HEK 293 cells (data not shown). The C-terminal domain of GluR5c contains an RXR motif, which functions as an ER retention site in KATP channels (Zerangue et al., 1999), in GABAB(1) receptors (Margeta-Mitrovic et al., 2000; Pagano et al., 2001), in some NMDA receptor splice variants (Standley et al., 2000; Scott et al., 2001), and in the KA2 subunit (Schwarz et al., 2001; Ren et al., 2003a). Accordingly, mutation of RLR to ALA in the C-terminal domain of GluR5c yields KARs that are expressed at the plasma membrane in heterologous cells and in neurons. While this work was being completed, a study reported the identification of a novel ER retention signal in GluR5b (and thus GluR5c) that controls receptor trafficking (Ren et al., 2003b). At variance with the RXR signal, this novel retention signal does not fully prevent ER export but rather restricts the trafficking of GluR5b to the plasma membrane. Indeed, GluR5b is significantly expressed at the surface of heterologous cells and mutant cultured neurons (Ren et al., 2003b; this study). In contrast, single transmembrane constructs fused to the C terminus of GluR5b (Ren et al., 2003b) are not detected at the plasma membrane, indicating that assembly of KARs as multimeric receptors includes key steps in the regulation of ER sorting. Using full-length receptor subunits, the trafficking of GluR5a and GluR5b appeared to be undistinguishable. Thus, although GluR5a lacks the retention signal found in GluR5b, the level of surface expression of GluR5a is similar to that of GluR5b. A possible explanation is that GluR5b also carries an ER export signal that is silenced by its ER retention site, whereas GluR5a passively trafficks from the ER to the plasma membrane.
Membrane delivery of GluR6 splice variants
GluR6a and GluR6b differentially traffick to the plasma membrane. GluR6a surface expression level is significantly higher than GluR6b in both COS-7 cells and in neurons derived from R5-/- × R6-/- mice. These isoforms are found in distinct intracellular compartments. Although GluR6a is not detected in the ER, GluR6b is sensitive to deglycosylation by EndoH and partly colocalizes with calreticulin in hot spots that may correspond to ER exit sites (Ellgaard and Helenius, 2003). The differences between GluR6a and GluR6b likely arise from a sequence downstream of the common 14 amino acids sequence. By sequential truncation of the C-terminal domain of GluR6a followed by site-directed mutagenesis, we identified a critical forward trafficking motif that promotes surface expression of GluR6 receptors at the plasma membrane of heterologous cells and neurons. This motif consists of four basic amino acids (RRXKXK). Mutation of these four amino acids induced low surface expression and retention of GluR6a in the ER. Studies of the trafficking of potassium channels of the Kir family and of NMDA receptors have revealed the presence of various ER export motifs (Ma et al., 2001; Ma and Jan, 2002; Mu et al., 2003) that are different from the RRXKXK motif identified for GluR6a.
GluR6a promotes surface expression of heteromeric receptors
GluR6a facilitates membrane delivery of other subunit splice variants that are retained mostly in the ER when expressed as homomeric receptors. This effect is dominant for GluR6b, because GluR6a-GluR6b heteromers display surface expression levels equivalent to homomeric GluR6a. Heteromerization of GluR5c with GluR6a counterbalances, at least in part, ER retention, possibly by masking this site, as shown for heteromeric GluR6-KA2 receptors (Ren et al., 2003a). In wild-type cultures, GluR5 isoforms are targeted more efficiently to the plasma membrane than in cultures derived from R5-/- × R6-/- mice. The most likely reason for this difference is the presence of an endogenous GluR6a that could associate with the transfected subunit and facilitate membrane delivery of the other subunit splice variants. Consistent with this hypothesis, coexpression of GluR6a with GluR6b or GluR5 isoforms in mutant neurons rescues membrane delivery of KARs to levels observed in wild-type neurons.
In our immunofluorescence measurements, however, the relative surface expression of GluR5-GluR6a heteromers is lower than for GluR6a alone. A likely explanation is that our measurement takes into account both homomeric myc-GluR5 and heteromeric myc-GluR5-GluR6 receptors. If homomeric myc-GluR5 receptors compose a large part of intracellular receptors, the relative surface expression of heteromeric myc-GluR5-GluR6 receptors would be underestimated. Another possibility is that ER retention signals in GluR5b and GluR5c isoforms tend to retain associated subunits in the ER; however, although no ER retention site has been described for GluR5a, the level of surface expression of GluR5a in the presence of GluR6a is similar to that of GluR5b. Our data thus suggest that even in the presence of GluR6a, the level of GluR5-containing KARs at the plasma membrane is limited to a certain level. The degree of oligomerization of ionotropic glutamate receptors depends on N-terminal extracellular domains (Ayalon and Stern-Bach, 2001) that differ between GluR5 and GluR6. One can speculate that a quality control mechanism limits the exit of GluR5-containing heteromers from the ER.
In summary, intracellular trafficking and surface expression of KARs is finely regulated by a balance between different categories of sites present in the C-terminal domain of various splice variants. These sites either promote or restrict trafficking of KARs from the ER to the plasma membrane.
Scaffold proteins and membrane delivery of KARs
Scaffold proteins, such as PDZ domain-containing proteins, may associate with receptors and channels and either facilitate or inhibit ER to Golgi trafficking of channels and receptors (Ma and Jan, 2002). PDZ domain proteins such as PSD-95, PICK1, GRIP, and syntenin are interactors of the KAR subunits GluR5b, GluR5c, and GluR6a (Garcia et al., 1998; Hirbec et al., 2003). Several lines of evidence suggest that the interaction of KAR subunits with PDZ domain-containing proteins does not affect the targeting of KAR subunits to the cell surface in our conditions, as also shown for KA2 and GluR5b (Ren et al., 2003a,b). Most importantly, the deletion of PDZ domain interaction sites in GluR6a does not inhibit targeting to the cell surface in COS-7 cells and in cultured neurons derived from R5-/- × R6-/- mice. In addition, the three splice variants GluR5b, GluR5c, and GluR6a, which interact with PDZ domain proteins, show very contrasting levels of surface expression in cultured hippocampal neurons that express PDZ domain proteins such as PSD-95. Thus, in the present conditions, interaction of GluR5 and GluR6 isoforms with PDZ domain proteins does not participate in the regulated trafficking from the ER to the plasma membrane, as is the case for NMDA receptors (Standley et al., 2000; Scott et al., 2001). These interactions, however, appear to play a role in the stabilization of KARs at postsynaptic sites in neurons where such KARs are present (Hirbec et al., 2003).
Functional significance: splice variants in native receptors
Heteromeric receptors probably compose most native receptors, as indicated by biochemical experiments and electrophysiological experiments comparing wild-type and KAR subunit-deficient mice (Mulle et al., 2000; Lerma et al., 2001; Coussen et al., 2002; Contractor et al., 2003). In hippocampal interneurons, KARs are likely heteromers composed of both GluR5 and GluR6 subunits (Mulle et al., 2000; Paternain et al., 2000). In these heteromeric KARs, association with GluR6a would facilitate trafficking to the plasma membrane. Our RT-PCR experiments indicate that various splice variants for each of the GluR5 or GluR6 subunits are expressed in the same brain regions, raising the possibility that a single neuron could express various isoforms of the same subunit. Native KARs could thus be composed of a combination of various splice variants. A still unresolved question is whether the various functions subserved by KARs arise from various KARs with different compositions. Although clear functional differences exist between GluR5 and GluR6 subunits (Lerma et al., 2001), an electrophysiological survey of recombinant KARs has not yet provided evidence for functional divergence between the subunit splice variants. The function of KARs is highly dependent on their subcellular localization at either presynaptic or postsynaptic sites. Additional work will be needed to understand how the interaction of the C-terminal domain of KAR splice variants with distinct subsets of intracellular proteins might finely regulate membrane expression in different neuronal domains that pertain to specific functions of KARs.
Footnotes
This work was supported by grants from the Centre National de la Recherche Scientifique, the Ministère de la Recherche of France, the Conseil Régional d'Aquitaine, and the European Commission (contract QLRT-2000-02089). We thank Steve Heinemann and the Salk Institute for making mutant mice available, Agnès Hémar and Rodrigo Cunha for helpful discussion on this manuscript, and Françoise Rossignol for technical help.
Correspondence should be addressed to Christophe Mulle, Laboratoire “Physiologie Cellulaire de la Synapse,” Centre National de la Recherche Scientifique Unité Mixte de Recherche 5091, Institut François Magendie, Université Bordeaux 2, rue C. Saint-Saëns, 33077 Bordeaux Cedex, France. E-mail: mulle@u-bordeaux2.fr.
Copyright © 2004 Society for Neuroscience 0270-6474/04/242506-10$15.00/0
References
- Ayalon G, Stern-Bach Y (2001) Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions. Neuron 31: 103-113. [DOI] [PubMed] [Google Scholar]
- Barbon A, Barlati S (2000) Genomic organization, proposed alternative splicing mechanisms, and RNA editing structure of GRIK1. Cytogenet Cell Genet 88: 236-239. [DOI] [PubMed] [Google Scholar]
- Barbon A, Vallini I, Barlati S (2001) Genomic organization of the human GRIK2 gene and evidence for multiple splicing variants. Gene 274: 187-197. [DOI] [PubMed] [Google Scholar]
- Barry MF, Ziff EB (2002) Receptor trafficking and the plasticity of excitatory synapses. Curr Opin Neurobiol 12: 279-286. [DOI] [PubMed] [Google Scholar]
- Bettler B, Mulle C (1995) AMPA and kainate receptors. Neuropharmacology 34: 123-139. [DOI] [PubMed] [Google Scholar]
- Bettler B, Boulter J, Hermans-Borgmeyer I, O'Shea-Greenfield A, Deneris ES, Moll C, Borgmeyer U, Hollmann M, Heinemann S (1990) Cloning of a novel glutamate receptor subunit, GluR5: expression in the nervous system during development. Neuron 5: 583-595. [DOI] [PubMed] [Google Scholar]
- Bureau I, Dieudonne S, Coussen F, Mulle C (2000) Kainate receptor-mediated synaptic currents in cerebellar Golgi cells are not shaped by diffusion of glutamate. Proc Natl Acad Sci USA 97: 6838-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll RC, Zukin RS (2002) NMDA-receptor trafficking and targeting: implications for synaptic transmission and plasticity. Trends Neurosci 25: 571-577. [DOI] [PubMed] [Google Scholar]
- Castillo PE, Malenka RC, Nicoll RA (1997) Kainate receptors mediate a slow postsynaptic current in hippocampal CA3 neurons. Nature 388: 182-186. [DOI] [PubMed] [Google Scholar]
- Contractor A, Sailer AW, Darstein M, Maron C, Xu J, Swanson GT, Heinemann SF (2003) Loss of kainate receptor-mediated heterosynaptic facilitation of mossy-fiber synapses in KA2-/- mice. J Neurosci 23: 422-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussen F, Normand E, Marchal C, Costet P, Choquet D, Lambert M, Mege RM, Mulle C (2002) Recruitment of the kainate receptor subunit glutamate receptor 6 by cadherin/catenin complexes. J Neurosci 22: 6426-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Mayer ML (1999) Heteromeric kainate receptors formed by the coassembly of GluR5, GluR6, and GluR7. J Neurosci 19: 8281-8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF (1999) The glutamate receptor ion channels. Pharmacol Rev 51: 7-61. [PubMed] [Google Scholar]
- Ellgaard L, Helenius A (2003) Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol 4: 181-191. [DOI] [PubMed] [Google Scholar]
- Everts I, Villmann C, Hollmann M (1997) N-Glycosylation is not a prerequisite for glutamate receptor function but is essential for lectin modulation. Mol Pharmacol 52: 861-873. [DOI] [PubMed] [Google Scholar]
- Frerking M, Nicoll RA (2000) Synaptic kainate receptors. Curr Opin Neurobiol 10: 342-351. [DOI] [PubMed] [Google Scholar]
- Frerking M, Ohliger-Frerking P (2002) AMPA receptors and kainate receptors encode different features of afferent activity. J Neurosci 22: 7434-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia EP, Mehta S, Blair LA, Wells DG, Shang J, Fukushima T, Fallon JR, Garner CC, Marshall J (1998) SAP90 binds and clusters kainate receptors causing incomplete desensitization. Neuron 21: 727-739. [DOI] [PubMed] [Google Scholar]
- Gregor P, O'Hara BF, Yang X, Uhl GR (1993) Expression and novel subunit isoforms of glutamate receptor genes GluR5 and GluR6. NeuroReport 4: 1343-1346. [DOI] [PubMed] [Google Scholar]
- Herzog E, Bellenchi GC, Gras C, Bernard V, Ravassard P, Bedet C, Gasnier B, Giros B, El Mestikawy S (2001) The existence of a second vesicular glutamate transporter specifies subpopulations of glutamatergic neurons. J Neurosci 21: RC181(1-6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirbec H, Francis JC, Lauri SE, Braithwaite SP, Coussen F, Mulle C, Dev KK, Couthino V, Meyer G, Isaac JT, Collingridge GL, Henley JM (2003) Rapid and differential regulation of AMPA and kainate receptors at hippocampal mossy fibre synapses by PICK1 and GRIP. Neuron 37: 625-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Betancur C, Quach H, Philippe A, Fellous M, Giros B, Gillberg C, Leboyer M, Bourgeron T (2002) Linkage and association of the glutamate receptor 6 gene with autism. Mol Psychiatry 7: 302-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullmann DM (2001) Presynaptic kainate receptors in the hippocampus: slowly emerging from obscurity. Neuron 32: 561-564. [DOI] [PubMed] [Google Scholar]
- Leach MR, Cohen-Doyle MF, Thomas DY, Williams DB (2002) Localization of the lectin, ERp57 binding, and polypeptide binding sites of calnexin and calreticulin. J Biol Chem 277: 29686-29697. [DOI] [PubMed] [Google Scholar]
- Lerma J, Paternain AV, Rodriguez-Moreno A, Lopez-Garcia JC (2001) Molecular physiology of kainate receptors. Physiol Rev 81: 971-998. [DOI] [PubMed] [Google Scholar]
- Ma D, Jan LY (2002) ER transport signals and trafficking of potassium channels and receptors. Curr Opin Neurobiol 12: 287-292. [DOI] [PubMed] [Google Scholar]
- Ma D, Zerangue N, Lin YF, Collins A, Yu M, Jan YN, Jan LY (2001) Role of ER export signals in controlling surface potassium channel numbers. Science 291: 316-319. [DOI] [PubMed] [Google Scholar]
- Margeta-Mitrovic M, Jan YN, Jan LY (2000) A trafficking checkpoint controls GABA(B) receptor heterodimerization. Neuron 27: 97-106. [DOI] [PubMed] [Google Scholar]
- Melyan Z, Wheal HV, Lancaster B (2002) Metabotropic-mediated kainate receptor regulation of IsAHP and excitability in pyramidal cells. Neuron 34: 107-114. [DOI] [PubMed] [Google Scholar]
- Mu Y, Otsuka T, Horton A, Scott DB, Ehlers MD (2003) Activity-dependent mRNA splicing controls ER export and synaptic delivery of NMDA receptors. Neuron 40: 581-594. [DOI] [PubMed] [Google Scholar]
- Mulle C, Andreas S, Pérez-Otaño I, Dickinson-Anson H, Castillo PE, Bureau I, Maron C, Gage FH, Mann JR, Bettler B, Heinemann SF (1998) Altered synaptic physiology and reduced susceptibility to kainate induced seizures in GluR6-deficient mice. Nature 392: 601-604. [DOI] [PubMed] [Google Scholar]
- Mulle C, Sailer A, Swanson GT, Brana C, O'Gorman S, Bettler B, Heinemann SF (2000) Subunit composition of kainate receptors in hippocampal interneurons. Neuron 28: 475-484. [DOI] [PubMed] [Google Scholar]
- Pagano A, Rovelli G, Mosbacher J, Lohmann T, Duthey B, Stauffer D, Ristig D, Schuler V, Meigel I, Lampert C, Stein T, Prezeau L, Blahos J, Pin J, Froestl W, Kuhn R, Heid J, Kaupmann K, Bettler B (2001) C-terminal interaction is essential for surface trafficking but not for heteromeric assembly of GABA(b) receptors. J Neurosci 21: 1189-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paternain AV, Herrera MT, Nieto MA, Lerma J (2000) GluR5 and GluR6 kainate receptor subunits coexist in hippocampal neurons and coassemble to form functional receptors. J Neurosci 20: 196-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Riley NJ, Garcia EP, Sanders JM, Swanson GT, Marshall J (2003a) Multiple trafficking signals regulate kainate receptor KA2 subunit surface expression. J Neurosci 23: 6608-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Riley NJ, Needleman LA, Sanders JM, Swanson GT, Marshall J (2003b) Cell surface expression of GluR5 kainate receptors is regulated by an endoplasmic reticulum retention signal. J Biol Chem, 278: 52700-52709. [DOI] [PubMed] [Google Scholar]
- Ruano D, Lambolez B, Rossier J, Paternain AV, Lerma J (1995) Kainate receptor subunits expressed in single cultured hippocampal neurons: molecular and functional variants by RNA editing. Neuron 14: 1009-1017. [DOI] [PubMed] [Google Scholar]
- Schwarz MK, Pawlak V, Osten P, Mack V, Seeburg PH, Kohr G (2001) Dominance of the lurcher mutation in heteromeric kainate and AMPA receptor channels. Eur J Neurosci 14: 861-868. [DOI] [PubMed] [Google Scholar]
- Scott DB, Blanpied TA, Swanson GT, Zhang C, Ehlers MD (2001) An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J Neurosci 21: 3063-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Burnashev N, Verdoorn TA, Keinänen K, Sakmann B, Seeburg PH (1992) A glutamate receptor with high affinity for domoate and kainate. EMBO J 11: 1651-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standley S, Roche KW, McCallum J, Sans N, Wenthold RJ (2000) PDZ domain suppression of an ER retention signal in NMDA receptor NR1 splice variants. Neuron 28: 887-898. [DOI] [PubMed] [Google Scholar]
- Vignes M, Collingridge GL (1997) The synaptic activation of kainate receptors. Nature 388: 179-182. [DOI] [PubMed] [Google Scholar]
- Wisden W, Seeburg P (1993) A complex mosaic of high-affinity kainate receptors in rat brain. J Neurosci 13: 3582-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerangue N, Schwappach B, Jan YN, Jan LY (1999) A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron 22: 537-548. [DOI] [PubMed] [Google Scholar]