Abstract
In humans, insert mutations within the repetitive octapeptide region of the prion protein gene (Prnp) are often associated with familial spongiform encephalopathies. In this study, transgenic mice expressing bovine PrP (boTg mice) bearing an additional octapeptide insertion to the wild type (seven octapeptide repeats instead of six) showed an altered course of bovine spongiform encephalopathy (BSE) infection, reflected as reduced incubation times when compared with boTg mice expressing similar levels of the wild-type six-octapeptide protein. In both boTg mouse lines (bo6ORTg and bo7ORTg), incubation times were affected drastically depending on transgene expression levels and the inoculum used. In accordance with the lack of an interspecies barrier to BSE infection, we detected the typical signs of CNS spongiform degeneration by histopathological analysis and the presence of the bovine prion PrPres by Western blot or immunohistochemical analyses. When 7OR-PrPres was propagated in bo7ORTg mice, a similar earlier onset of clinical signs was observed compared with bo6ORTg mice. Proteins PrPC and PrPres containing seven octapeptides (7OR-PrPC and 7OR-PrPres) showed similar protease sensitivity and insolubility in nondenaturing detergents to homologous 6OR-PrPC and 6OR-PrPres. In addition, bo7ORTg mice showed a higher sensitivity than bo6ORTg mice for detecting prion infection in specimens previously diagnosed as negative by conventional biochemical techniques. In the absence of clinical signs of disease, 7OR-PrPres could be detected as early as 120 d after inoculation by immunohistochemical and Western blot analyses. These findings may help us improve the current mouse bioassays and understand the role of the octapeptide repeat region in susceptibility to disease.
Keywords: BSE transmission, transgenic mice, prion, octapeptide repeats, bioassay, scrapie, PrP
Introduction
Prion diseases, or transmissible spongiform encephalopathies (TSEs), form a class of neurodegenerative disorders characterized by the presence of an abnormal, protease-resistant (PrPres) isoform of prion protein in the brains of affected individuals (Prusiner, 1991). Prion diseases may be of an infectious, spontaneous, or inherited origin. Infectious TSEs have been described in animals and humans, the prototypes of which are scrapie in sheep and goats (Adams, 1986), bovine spongiform encephalopathy (BSE) in cattle (Prusiner, 1998), and kuru (Alpers and Rail, 1971) or variant Creutzfeldt-Jakob disease (vCJD) in humans (Will et al., 1996; Hill et al., 1997). Spontaneous and inherited TSEs have been mainly identified in humans and include CJD (Beck et al., 1969), Gertsmann-Sträussler-Scheinker disease (Bendheim, 1984; Collinge et al., 1989), and fatal familial insomnia (Manetto et al., 1992; Medori et al., 1992; Dormont, 1994; Goldfarb et al., 1994a).
In humans, inherited prion diseases have been associated with different point mutations (Palmer and Collinge, 1993; Ridley and Baker, 1993) and an increased number of octapeptide repeats within the PrP ORF (Owen et al., 1990; Goldfarb et al., 1991, 1993; Krasemann et al., 1995; Campbell et al., 1996; Cochran et al., 1996). In animals, the genetic instability of the corresponding region of tandem octapeptide coding repeats (ORs) has not been extensively explored, although some degree of polymorphism (five, six, and seven ORs) has been observed in cattle (Goldmann et al., 1991; Yoshimoto et al., 1992; Hunter et al., 1994; Neibergs et al., 1994; Walawski and Czarnik, 2003) and sheep (Goldmann et al., 1998). For instance, a recently identified seven OR polymorphism has been found to occur in at least 5% of Brown Swiss cows (Schlapfer et al., 1999).
Although pathogenic insertion mutations in the octapeptide repeat region (10, 12, and 13) have been described in humans (Goldfarb et al., 1991), no neuropathological disorders have been associated with natural cases of seven-octapeptide repeats in cattle (Schlapfer et al., 1999) or with extra PrP repeats in transgenic mice (Chiesa et al., 1998). Despite evidence that the region bearing the octarepeats is not essential for mediating prion pathogenesis and propagation, the repeat region may modulate the incubation time to disease (Fischer et al., 1996; Goldmann et al., 1998; Flechsig et al., 2000; Beck et al., 2001).
Whether a cow has five or six ORs within the PrP coding region, it is similarly susceptibly to BSE infection (Neibergs et al., 1994), although the natural susceptibility to BSE infection of cattle with the 7OR-PrP motif remains unknown. In this study, we show how bovine PrP transgenic mice with one extra octapeptide repeat insertion mutation (7OR-PrP) show reduced incubation and survival times after BSE prion inoculation compared with boTg mice (transgenic mice expressing bovine PrP) with the normal 6OR-PrP. This new bioassay model is more sensitive than the conventional boTg mouse model (6OR-PrP) in that it can detect minimal amounts of BSE prions, undetectable by conventional techniques. This finding may suggest a role for the octapeptide repeat gene region in the efficiency of prion transformation and propagation.
Materials and Methods
Plasmid constructs. The PrP ORF was isolated by PCR amplification from bovine DNA, using primers containing a XhoI restriction enzyme site adjacent to the translation start codon (5′-CCGCTCGAGGCCATCATGGTGAAAAGCCATATAG-3′) and stop codon (5′-CGGCTCGAGCTATCCTACTATGAG-3′). The 5′ primer also included Kozak sequences (Kozak, 1989). The resultant PCR fragment was subcloned into a T-tailed vector, and the insert was sequenced to confirm six copies of the octapeptide repeat sequence and no difference in the inferred amino acid sequence from the previously established bovine PrP gene sequence (GenBank accession number AF455119). The extra octapeptide repeat was introduced into the six-octapeptide repeat region of the bovine PrP gene as follows: 5′-CATGGAGGTGGCTGGGGCCAGCCC-3′ and 5′-CATGGGGCTGGCCCCAGCCACCTC-3′ primers were used to obtain a tandem of four artificial octarepeats containing ends compatible with the NcoI restriction enzyme site. The PrP ORF was cut with NcoI, thus eliminating three ORs, and the tandem of four artificial octarepeats with extreme ends compatible with NcoI was inserted (supplementary Fig. 1; available at www.jneurosci.org). The six (6OR) and seven (7OR) octarepeat PrP ORFs were excised from the T-tailed vector using the restriction enzyme XhoI and inserted into MoPrP.Xho (Borchelt et al., 1996), also digested with XhoI. This vector contains the murine PrP promoter, exon 1, intron 1, exon 2, and 3′ untranslated sequences.
Generating transgenic mice. MoPrP.Xho bovine transgenes (six ORs and seven ORs) containing mouse regulatory sequences were excised from the plasmid vector using the restriction endonuclease NotI to yield a 12 kb DNA fragment. Before microinjection, the fragment was purified using sodium chloride gradients as described previously (Fink, 1991) and resuspended in 10 mm Tris, pH 7.4, and 1 mm EDTA at a final concentration of 2-6 μg/ml. B6CBAf1 females mated with 129/Ola males carrying a null mutation in the endogenous PrP sequence (Manson et al., 1994) were superovulated to obtain 350 and 328 pronuclear stage ova, which were then microinjected with the 6OR or 7OR constructs, respectively. Homozygous transgenic lines were established in two steps. Founders were backcrossed to homozygous null animals muPrP-/- (Prnp-/-) to obtain homozygosity for the null mutation. Interbreeding within the same transgenic line yielded homozygosity for the bovine PrP constructs. Hybrids of B6CBAx129/Ola mice and Prnp-/- mice were used as control for all the infectivity studies.
Screening of founders. DNA was prepared from tail biopsies as described previously (Hogan and Williams, 1981). MoPrP.Xho bovine transgenes were identified by a PCR assay using specific primers for the mouse PrP exon 2 and the bovine PrP ORF. The primers used were 5′-CCAGCCTCCACCACCATGTGGC-3′ and 5′-CATTCTGCCTTCCTAGTGGTACC-3′. The presence of PCR-amplified products of 315 and 339 nucleotides indicated the presence of MoPrP.Xho 6OR and 7OR bovine transgenes, respectively. The absence of murine PrP ORF in transgenic mice was confirmed by PCR using the following primers: 5′-ATGGCGAACCTTGGCTACTGGC-3′ and 5′-GATTATGGGTACCCCCTCCTTGG-3′.
Source of BSE inoculum; preparing brain homogenates. A pool of 49 BSE-infected cattle brains (TSE/08/59) supplied by the Veterinary Laboratory Agency (VLA) (New Haw, Addlestone, Surrey, UK) was used for some of the experimental inoculations and was denoted BSE1. The titer of this inoculum was ∼108 ID50 units per gram of bovine brainstem, as determined in the bo6ORTg110 mouse line (Castilla et al., 2003) (data not shown). For comparative infectivity studies, we also used material from one BSE-infected brainstem, also supplied by the VLA (RQ 225:PG1199/00), as inoculum BSE2. The BSE2 inoculum contained eight times more PrPres, as determined by immunoblot analysis (supplementary Fig. 2). We also used pooled brain homogenates from bo6ORTg mice generated in our laboratory (Castilla et al., 2003) and killed 120 (bo6ORTgBSE1120d) or 150 (bo6ORTgBSE1150d) days after inoculation with BSE1 in experimental infections. The bo6ORTgBSE1120d and bo6ORTgBSE1150d homogenates contained undetectable amounts of PrPres on conventional immunoblot analysis (data not shown). Finally, pooled infectious material from a first passage of BSE1 inoculum in a bo6ORTg110 mouse line (Castilla et al., 2003) or in a bo7ORTg006 mouse line generated in our laboratory (bo6ORTgBSE1 and bo7ORTgBSE1, respectively) were used in some experiments. In all the experiments, PBS was used as the negative inoculation control. Brain homogenates (10% wt/vol) were prepared in sterile Ca2+-free or Mg2+-free PBS by mechanical homogenization (OMNI International, Warrenton, VA). To minimize the risk of bacterial infection, all inocula were preheated for 10 min at 70°C before their use in mice.
Infection experiments. Groups of 4-13 mice (6-7 weeks old, weighing ∼20 gm) were inoculated in the right parietal lobe using a 25 gauge disposable hypodermic needle to deliver 20 μl of 10% brain homogenate to each animal. After inoculation, the mice were observed daily to check for clinical signs. Their neurological status was assessed twice weekly. A mouse was considered positive for prion disease when it showed two or three of 10 signs of neurological dysfunction (Scott et al., 1989, 1993). When disease progression was evident, animals were culled for ethical reasons, and their brains were harvested for analysis. These specimens were used for the histopathological determination of spongiform degeneration and to detect PrPres accumulation by immunohistochemistry or Western blotting.
Histopathology and immunohistochemistry. Brains were fixed by immersion in 10% buffered formalin for 1 week. The fixed brains were then cut into four pieces and immersed in 98% formic acid for 1 hr before routine processing and paraffin wax embedding. Five-micrometer-thick sections were cut and stained with hematoxylin and eosin for routine histopathological examination and immunohistochemical (IHC) techniques. The sections examined were taken from: (1) the medulla oblongata at the level of the obex and the pontine area; (2) the cerebellum; (3) the diencephalon, including thalamus; (4) the hippocampus; and (5) the cerebral cortex.
The avidin-biotin-peroxidase complex technique was used for the IHC detection of PrPres and GFAP. After dewaxing and dehydration, endogenous peroxidase was quenched by incubation with 3% hydrogen peroxide in methanol for 30 min at room temperature. Samples for PrPres labeling were rehydrated and pretreated with 98% formic acid for 15 min at room temperature, 4 m guanidine isothiocianate for 2 hr at 4°C, and proteinase K (4 μg/ml in Tris-HCl, pH 7.8; Roche, Basel, Switzerland) for 15 min at 37°C. Next, the tissue sections were rinsed in 0.01 m PBS, pH 7.4, and blocked with 10% NGS (Sigma, St. Louis, MO) for 30 min at room temperature. Sections were incubated overnight at 4°C with primary 6H4 monoclonal antibody (mAb) (Prionics, Schlieren, Switzerland) or 2A11 mAb (Brun et al., 2003) diluted 1:400 in PBS. Biotinylated goat anti-mouse IgG (Dako, Glostrup, Denmark) diluted 1:20 in PBS was used as secondary antibody, followed by the avidin-peroxidase complex (Vector Laboratories, Burlingame, CA). After developing with a 3,3′-diaminobenzidine tetrahydrochloride chromogenic substrate (Sigma), the slides were counterstained with Mayer′s hematoxylin for 1 min, dehydrated, and assembled.
Brain specimens for GFAP labeling were rehydrated without pretreatment and blocked as described above. The sections were incubated overnight at 4°C with primary anti-mouse GFAP polyclonal antibody (Dako) diluted 1:500 in PBS. Goat anti-rabbit IgG (Vector Laboratories) diluted 1:200 in PBS was used as the secondary antibody. The procedure was then continued as described above. PBS, nonimmune mouse serum, or nonimmune rabbit serum were used to replace specific primary antibodies to prepare negative controls.
Biochemical analysis of PrPC in bo6ORTg and bo7ORTg mice. Brain homogenates from bo6ORTg and bo7ORTg mice were solubilized in extraction buffer (0.5% NP-40, 1% sodium deoxycholate, and 10 mm EDTA in PBS, pH 7.4) in the presence of a protease inhibitor mixture (Complete; Roche). These samples were precleared by centrifugation at 10,000 × g for 5 min. The supernatant was then ultracentrifuged at 100,000 × g for 1 hr after the addition of Gdn-HCl at a final concentration of 1 and 3 m. Soluble proteins were precipitated by adding four volumes of cold methanol and centrifuging at 20,000 × g for 30 min. Insoluble fractions were exhaustively washed with 2% sarcosyl in PBS, pH 7.4, and subjected to an additional ultracentrifugation step. Insoluble and methanol-precipitated soluble proteins were subsequently analyzed by SDS-PAGE and Western blotting.
Western blot analysis. PrPC expression in transgenic mice was determined as follows. Brains from mice were homogenized in extraction buffer as above. These samples were precleared by centrifugation at 10,000 × g for 5 min and then either treated with 20 μg/ml proteinase K (Roche) at 37°C for 60 min or left untreated. An equal volume of 2× SDS reducing sample loading buffer was added to all samples, and each one was boiled for 5 min before loading on a SDS/12% polyacrylamide gel. mAbs 6H4, 2A11, and FH11 (Foster et al., 1996) were used at 1:5000, 1:1500, and 1:500 dilutions, respectively, for immunoblotting. Immunocomplexes were detected with HRP-conjugated anti-mouse IgG (Sigma). The immunoblots were developed under ECL (Amersham Biosciences, Piscataway, NJ).
Statistical analysis. We used SPSS 10.0 software (SPSS, Chicago, IL) to generate Kaplan-Meier curves and statistical data. The Student's t test for nonpaired data was used to compare the data obtained. After testing for normality (Kolmogorov-Smirnow test) and equal variance (Levene median test), incubation times were subjected to one-way ANOVA, followed by multiple pairwise comparison using Fisher's LSD test. The level of significance was set at a p < 0.05.
Results
Transgenic PrPC expression and phenotype of transgenic mouse lines
We obtained five different lines (founders) with seven OR PrPC (bo7ORPrPC) and two lines of heterozygous transgenic mice with six OR PrPC (bo6ORPrPC). All these lines also bore wild-type murine PrPC with five octarepeats (mu5ORPrPC). The expression of PrPC in these lines carrying both murine 5OR-PrPC and bovine 6OR-PrPC or 7OR-PrPC transgenes (PrP mu+/-bo+/-) was evaluated by subjecting brain homogenates to Western blot analysis using the 2A11 mAb (Fig. 1A), which is able to recognize both boPrPC and muPrPC. Western blotting with the 6H4 mAb yielded identical results (data not shown), thus only results using the 2A11 mAb are reported here. Although it was possible to distinguish between wild-type murine (5OR) and bovine (7OR) PrP according to their relative electrophoretic mobility, their expression was confirmed using the FH11 mAb, which does not recognize endogenous muPrP (Fig. 1B). Based on their expression levels, we then selected the lines bo7ORTg006 and bo7ORTg007 from the initial five heterozygous (PrP mu+/-bo+/-) Tg lines and bred them to homozygosity in a murine PrP null background (PrP mu-/-). Mouse lines bo6ORTg113 and bo6ORTg135 with six octarepeats were also bred to homozygosity in a murine PrP null background. This was achieved by crossing the selected lines with PrP null mice to give the transgene heterozygous lines (PrP mu-/-bo+/-). Then, by crossing heterozygous animals, we obtained the homozygous transgene genotype (PrP mu-/-bo+/+). The absence of the murine PrP gene in these animals was confirmed by PCR using specific primers (data not shown). Next, we established transgene expression levels in each homozygous mouse line by serial dilution of brain homogenates and compared these with PrPC levels in bovine brain homogenates. PrPC expression levels for the four lines bo7ORTg006, bo7ORTg007, bo6ORTg113, and bo6ORTg135 were found to be six-, four-, three-, and sixfold higher than PrPC levels in cattle, respectively (Fig. 1C). Furthermore, all the transgene heterozygous lines tested showed half the PrP expression levels detected in their homozygous counterparts (data not shown). Similar behavior and mean survival times were shown by the noninoculated mouse lines bo6ORTg (649 ± 49; mean days ± SEM) and bo7ORTg (631 ± 34).
Figure 1.
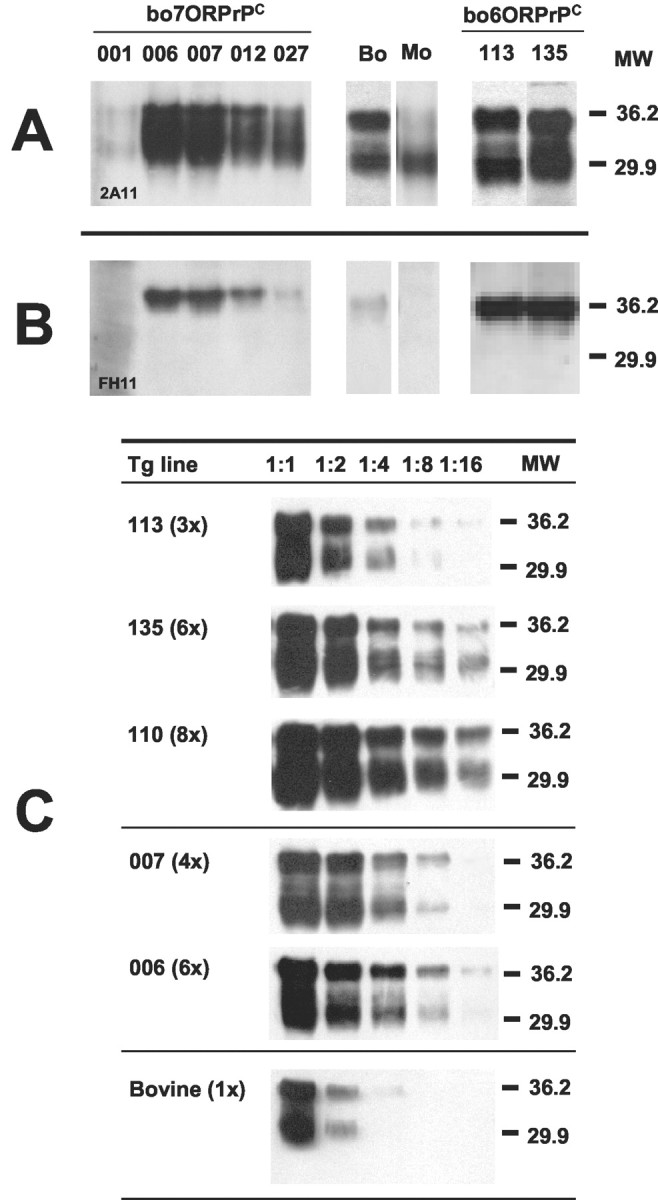
Expression of bovine 7OR-PrP and 6OR-PrP proteins in heterozygous (mu +/-bo +/-) boTg lines. Immunoblotting brain extracts from bo7ORTg mouse lines 001, 006, 007, 012, and 027 and bo6ORTg mouse lines 113 and 135 using mAb 2A11 (A) or FH11 (B). Serial dilutions of homozygous (mu-/-bo+/+) bo6ORTg (113, 135, and 110 mouse lines), bo7ORTg (007 and 006 mouse lines), and bovine brain homogenates were analyzed by Western blotting using mAb 2A11 (C). Bo, Cow brain extract; Mo, B6CBAx129/Ola brain extract. Equivalent amounts of total protein were loaded on each lane. Relative molecular mass is expressed in kilodaltons.
Susceptibility of bo6ORTg and bo7ORTg mice to BSE prions
Several BSE inocula (see Material and Methods) were used to compare the course of BSE infection in the selected mouse lines expressing bovine PrP with seven octarepeats (bo7ORTg006, bo7ORTg007) and in mice expressing similar levels of the wild-type six-octapeptide protein (bo6ORTg135, bo6ORTg113). After inoculation, incubation times (onset of clinical signs) and survival times were determined (Figs. 2, 3; Table 1). When disease progression was evident, the animals were culled, and their brains used for histopathological determination of spongiform degeneration (Fig. 4) and to detect PrPres accumulation by immunohistochemistry (Fig. 4) or Western blotting (Table 1; supplementary Fig. 3B).
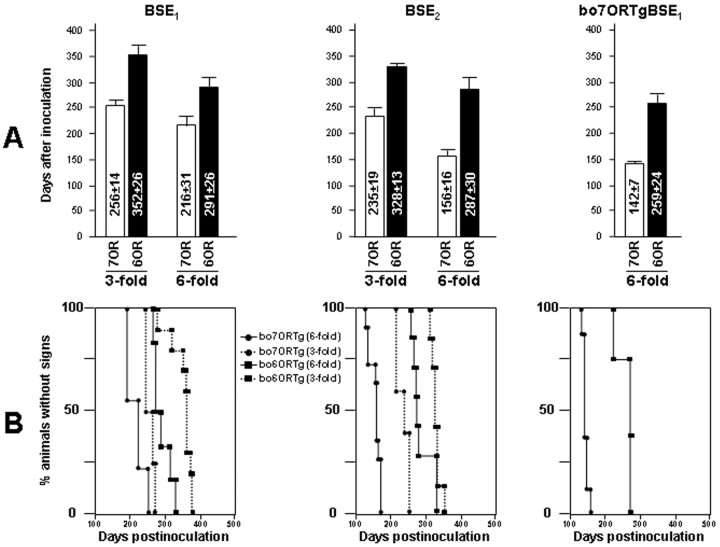
Figure 2.
Onset of clinical signs after inoculating BSE in different bovine transgenic mice. In the columns (A) or Kaplan-Meier curves (B), incubation times are compared among bo6ORTg and bo7ORTg lines (showing threefold or sixfold the PrPC levels detected in bovine brain homogenates) inoculated with BSE1, BSE2, and first passage BSE1 in bo7ORTg (bo7ORTgBSE1). Values are expressed as days after inoculation ± SD.
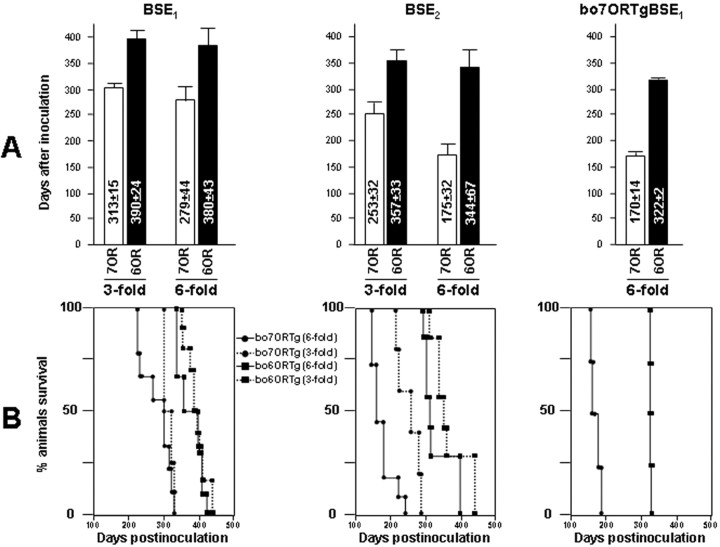
Figure 3.
Survival times after BSE inoculation in different bovine transgenic mice. In the columns (A) or Kaplan-Meier curves (B), survival times are compared among bo6ORTg and bo7ORTg (showing threefold or sixfold the PrPC levels detected in bovine brain homogenates) inoculated with BSE1, BSE2, and first passage BSE1 in bo7ORTg (bo7ORBSE1). Values are expressed as days after inoculation ± SD.
Table 1.
Susceptibility of 60R and 70R transgenic mice to BSE prions
|
|
|
Transgene expressiona |
Incubation time (days ± SEM) |
|
||
|---|---|---|---|---|---|---|
| Recipient |
Inoculum |
Illness |
Death |
Numberb
|
||
| Non-Tg (B6xCBAx1290la) | BSE1 | 1x | 614 = 26 | 656 ± 30 | 12 | |
| Non-Tg (prnp−/−) | BSE1 | 0x | 599 = 14 | 688 ± 35 | 8 | |
| bo6ORTg110+/+ | BSE1 | 8x | 272 ± 8 | 326 ± 18 | 13 | |
| bo6ORTg110+/− | BSE1 | 4x | 311 = 17 | 359 ± 15 | 11 | |
| bo6ORTg113+/+ | BSE1 | 3x | 352 = 10 | 410 ± 7 | 10 | |
| bo6ORTg135+/+ | BSE1 | 6x | 290 = 10 | 381 ± 18 | 6 | |
| bo7ORTg006+/+ | BSE1 | 6x | 216 ± 8 | 279 ± 14 | 9 | |
| bo7ORTg006+/− | BSE1 | 3x | 257 ± 7 | 313 ± 7 | 4 | |
| bo7ORTg007+/+ | BSE1 | 4x | 237 ± 4 | 286 ± 5 | 4 | |
| bo7ORTg007+/− | BSE1 | 2x | 325 = 10 | 376 ± 14 | 6 | |
| bo6ORTg110+/+ | BSE2 | 8x | 251 = 20 | 308 ± 5 | 5 | |
| bo6ORTg113+/+ | BSE2 | 3x | 328 ± 5 | 377 ± 8 | 7 | |
| bo6ORTg135+/+ | BSE2 | 6x | 257 = 11 | 324 ± 25 | 7 | |
| bo7ORTg006+/+ | BSE2 | 6x | 156 ± 5 | 175 ± 10 | 11 | |
| bo7ORTg006+/− | BSE2 | 3x | 235 ± 3 | 253 ± 14 | 5 | |
| bo7ORTg007+/+ | BSE2 | 4x | 218 ± 7 | 237 ± 7 | 4 | |
| bo7ORTg007+/− | BSE2 | 2x | 294 ± 5 | 337 ± 26 | 4 | |
| bo6ORTg110+/+ | bo7ORTgBSE1 | 8x | 205 ± 4 | 291 ± 18 | 6 | |
| bo6ORTg113+/+ | bo7ORTgBSE1 | 3x | 280 ± 6 | 319 ± 9 | 9 | |
| bo6ORTg135+/+ | bo7ORTgBSE1 | 6x | 259 = 12 | 323 ± 1 | 4 | |
| bo7ORTg006+/+ | bo7ORTgBSE1 | 6x | 142 ± 3 | 171 ± 5 | 8 | |
| bo7ORTg007+/+ | bo7ORTgBSE1 | 4x | 279 = 11 | 331 ± 20 | 8 | |
| bo6ORTg110+/+ | PBS inoculated | 8x | 490 ± 5 | 599 ± 27 | 4 | |
| bo6ORTg113+/+ | PBS inoculated | 3x | 491 ± 5 | 581 ± 39 | 4 | |
| bo6ORTg135+/+ | PBS inoculated | 6x | 461 = 10 | 607 ± 49 | 4 | |
| bo7ORTg006+/+ | PBS inoculated | 6x | 561 ± 36 | 589 ± 34 | 6 | |
| bo7ORTg007+/+
|
PBS inoculated |
4x |
>550 |
>550 |
6 |
|
+/+ or +/−, Homozygous or heterozygous for the bovine prnp gene. All transgenic animals are murine prnp−/−.
Relative to cattle PrP expression.
Number of inoculated animals.
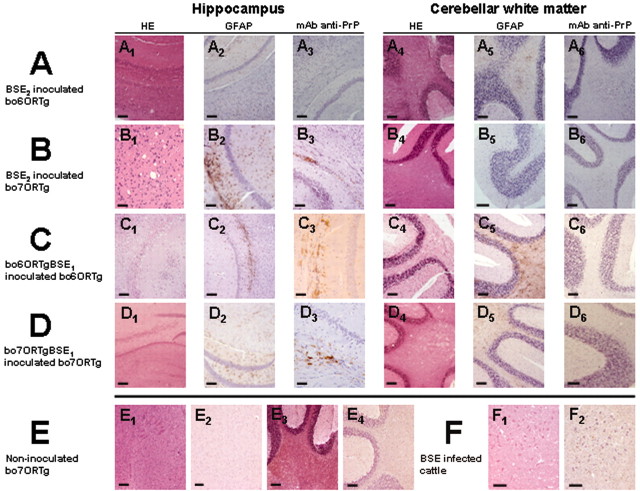
Figure 4.
A, B, Histopathological changes consisting in vacuolation in the deeper layers of the cerebral cortex and the hippocampus (1) and the cerebellar white matter (4); GFAP immundetection in the deeper layers of the cerebral cortex and the hippocampus (2) and cerebellar white matter (5) showing astrogliosis and astrocytosis; and PrPres immunodetection using mAb 2A11 in the deeper layers of the cerebral cortex and the hippocampus (3) and the cerebellar white matter (6) of bo6ORTg (A) or bo7ORTg (B) mice inoculated with BSE2 inoculum (∼200 d after inoculation). C, D, Histopathological changes consisting in vacuolation in the deeper layers of the cerebral cortex and the hippocampus (1) and the cerebellar white matter (4); GFAP immundetection in the deeper layers of the cerebral cortex and the hippocampus (2) and cerebellar white matter (5) showing astrogliosis and astrocytosis; and PrPres immunodetection using mAb 2A11 in the deeper layers of the cerebral cortex and the hippocampus (3) and the cerebellar white matter (6) of boORTg mice inoculated with the BSE1 inoculum passage one in bo6ORTg (C) or bo7ORTg mice inoculated with BSE1 inoculum passage once in bo7ORTg mice (D) (∼250 d after inoculation). E, F, Histopathology (1 and 3) and PrPres immunodetection (2 and 4) of bo6ORTg noninoculated mice as negative controls, showing no changes (E). Histopthological changes consisting in vacuolation (1) and PrPres immunodetection (2) in the brainstem of BSE-infected cattle as positive control (F). Scale bar, 50 μm.
Both the bo6ORTg and bo7ORTg mouse lines developed identical signs of CNS dysfunction after BSE inoculation. Incubation times were affected drastically depending on transgene expression levels and the inoculum used (Figs. 2, 3; Table 1). Thus, when we inoculated bo6ORTg and bo7ORTg mice with the “high PrPres” inocula (BSE2), reduced incubation and survival times were recorded compared with those shown by the same mouse lines inoculated with a “low PrPres” inoculum (BSE1) (Figs. 2, 3; Table 1). Moreover, the two homologous Tg mouse lines showed different incubation and survival times according to their respective boPrPC expression levels, irrespective of the inoculum used. For instance, the bo6ORTg135 line inoculated with BSE2 showed incubation and survival times of 287 ± 1 and 344 ± 25 d, respectively, whereas these times were 328 ± 5 and 357 ± 12 d for the bo6ORTg113 line, which expresses lower levels of the bovine prion protein (Table 1). Similarly, the bo7ORTg006 and bo7ORTg007 mouse lines showed incubation times of 156 ± 5 and 268 ± 7 d, respectively, and survival times of 175 ± 10 and 287 ± 8 d, respectively (Table 1). In addition, these differences in incubation and survival times showed good correlation with gene dosage (homozygous vs heterozygous) within a single transgenic mouse line (Table 1). To explore the effects of the number of octapeptide repeats, groups of bo6ORTg and bo7ORTg mice expressing identical amounts of PrPC were inoculated with the three different inocula (BSE1, BSE2, or bo7ORTgBSE1). In these experiments, the bo7ORTg mice consistently showed reduced incubation and survival times (Figs. 2, 3; Table 1). This reduction in incubation and survival times was enhanced in lines expressing higher amounts of boPrPC.
In line with the biochemical data, histopathological and immunohistological studies revealed PrPres deposition in brain sections from mice receiving injections of each of the three types of BSE inoculum. The histological pattern observed for the BSE inoculum or first passage BSE in both bo6ORTg or bo7ORTg mice (Fig. 4A-D) was indistinguishable from that of classic BSE in cattle (Fig. 4F), mainly vacuolization of the neuropil mostly in the brain stem, hippocampus (Fig. 4A1,B1,C1,D1), and cerebellar white matter (Fig. 4A4,B4,C4,D4). However, we observed several patterns indicating immunopositivity and correlating with Western blotting patterns for PrPres. The most common were fine granular and punctuate neuropil labeling and stellate labeling foci, which seemed to be associated with glial cells. However, we also observed granular staining in neuron cytoplasms and around the neurons, occasionally as plaque-like deposits (Fig. 4A3,A6,B3,B6,C3,C6,D3,D6). These labeling patterns were mostly observed in the neuropil of the rostral brain stem, in cerebellar nuclei (Fig. 4A6,B6,C6,D6), and in the deep layers of the cerebral cortex and the hypocampus (Fig. 4A3,B3,C3,D3) and were identical for the bo6ORTg and bo7ORTg lines, irrespective of the inoculum used. Severe vacuolization and PrPres deposition were accompanied by astrocytic gliosis, as observed by the IHC detection of GFAP in different structures of the encephalic area. Vacuoles were often enveloped by astrocytic prolongations, and astrocytosis was also observed as an enlarged astrocyte cytoplasm (Fig. 4A2,A5,B2,B5,C2,C5,D2,D5). No histopathological changes (Fig. 4E1,E3) in PrPres deposition (Fig. 4E2,E4) or in astrocyte reactions (data not shown) were observed in noninoculated mice used as negative controls.
Biochemical studies on PrPC and PrPres in bo6ORTg and bo7ORTg mice
We then went on to establish whether the addition of an extra octarepeat insertion mutation modifies the biochemical properties of the bo7OR-PrP protein in relation to the wild-type bo6OR-PrP protein. To this end, brain homogenates from six bo6ORTg and six bo7ORTg homozygous mice were solubilized in extraction buffer and ultracentrifuged at 100,000 × g for 1 hr, as described in Material and Methods. Subsequent Western blotting analysis of the soluble and insoluble proteins indicated identical biochemical behavior of the bo6OR-PrP and bo7OR-PrP proteins (supplementary Fig. 3A). In an additional set of experiments, we evaluated the different PrPres obtained after inoculating mice with BSE1, BSE2, bo6ORTgBSE1/BSE2, or bo7ORTgBSE1/BSE2 (i.e., BSE1 or BSE2 inocula passaged once in bo6ORTg or bo7ORTg mice), or first passage bo6ORTgBSE1 in bo6ORTg or bo7ORTg mice (i.e., second passage BSE1 in each mouse line). This analysis was performed by Western blotting detergent extracts of infected boTg mice brain homogenates after standard proteinase K treatment. As shown in supplementary Figure 3B, the band patterns were identical for the different BSE inocula and the bo6ORTg and bo7ORTg mice.
Comparative studies using brain homogenates from a silent carrier as the inoculum
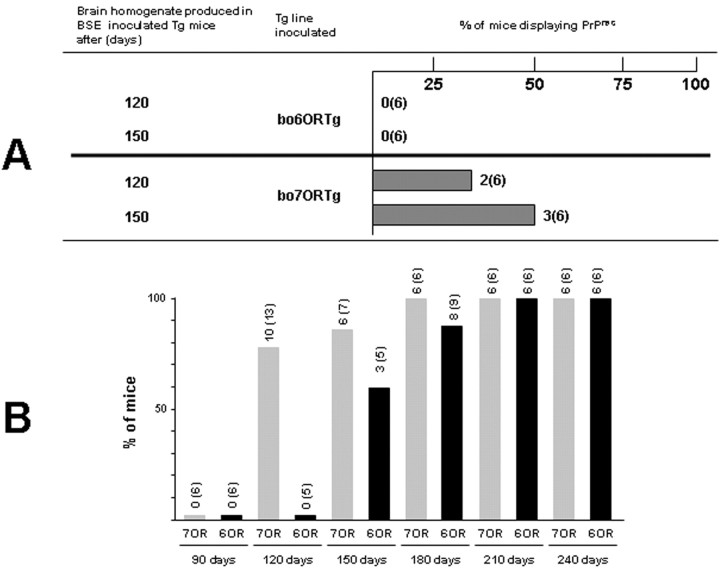
To confirm the increased capacity of bo7ORTg versus bo6ORTg mice to propagate bovine prions and, therefore, act as a more sensitive model for detecting the prions, we tried to detect infectivity in samples previously diagnosed as negative by conventional techniques. Both the 6OR and 7OR mouse lines were inoculated with bo6ORTgBSE1120d and bo6ORTgBSE1150d, corresponding to pooled brain homogenates from bo6ORTg mice killed at 120 or 150 d after inoculation with BSE1, respectively. These homogenates had been diagnosed as PrPres negative by conventional immunoblot analysis (data not shown). Three hundred thirty days after inoculation (sufficient time to detect PrPres in 100% of BSE1-inoculated bo6ORTg and bo7ORTg mice), PrPres could be detected by Western blotting in 33% (two of six) of the bo6ORTgBSE1120d-inoculated bo7ORTg mice and in 50% (three of six) of the bo6ORTgBSE1150d-inoculated bo7ORTg mice (Fig. 5A). In sharp contrast, no PrPres protein could be detected in any of the bo6ORTg mice (n = 6) (Fig. 5A). These findings were confirmed by IHC analysis (data not shown) and clearly indicate a role for the octapeptide region in modulating susceptibility to infection.
Figure 5.
A, Comparative studies using brain homogenates from silent carriers as inoculum. Bo6ORTg and bo7ORTg mice were inoculated with bo6ORTgBSE1120d (120) and bo6ORTgBSE1150d (150) (two pools of brain homogenates from bo6ORTg mice killed at 120 or 150 d after inoculation with BSE1 inoculum). Both inocula contained amounts of PrPres undetectable by conventional immunoblot analysis. Tg mice were killed at 330 d after inoculation, and PrPres from brain samples were analyzed by Western blot after proteinase K (20 μg/ml) treatment. Columns indicate the percentage of Tg mice showing PrPres at the indicated time after inoculation. B, Comparative kinetics of PrPres detection after BSE2 inoculation in bo6ORTg and bo7ORTg mice. Bovine transgenic mice expressing 6OR-PrP or 7OR-PrP were inoculated with BSE2 inoculum and killed at the indicated time points, irrespective of the onset of neurological signs. PrPres in brain samples were analyzed by Western blotting after proteinase K (20 μg/ml) treatment. Columns indicate the percentage of Tg mice showing PrPres at the indicated time after inoculation.
Kinetics of prion detection
In addition to the susceptibility study, we performed a kinetic experiment to determine, in relative terms, the earliest time point at which the prions could be detected in both bo6ORTg and bo7ORTg mice. For this experiment, we used one of our previous mouse lines, bo6ORTg110 (Castilla et al., 2003), which expresses eight times the PrPC levels found in bovine brain homogenates, and the present bo7ORTg006 mouse line (expressing six times the level). The animals were inoculated with the BSE2 inoculum and killed 90, 120, 150, 180, 210, or 240 d after inoculation, after which PrPres was detected by Western blotting. These culling times were adhered to irrespective of the onset of neurological signs. As shown in Figure 5B, 120 d after inoculation over 75% of the bo7ORTg mice showed PrPres, whereas at this stage no protein could be detected in any of the bo6ORTg. Moreover, it was not until 150 d after inoculation that PrPres could be detected in 60% of the bo6ORTg mice, whereas six of seven (87%) of the bo7ORTg mice scored positive for the protein. These results were also confirmed by IHC analysis (data not shown).
Discussion
Spontaneous and inherited TSEs, characterized mainly in humans, are associated with the presence of an increased number of ORs within the PrP ORF (Goldfarb et al., 1991, 1993; Krasemann et al., 1995; Campbell et al., 1996; Cochran et al., 1996). Insertion mutation-derived spontaneous human TSEs show different phenotypes and behavior related to the number of additional octapeptide repeats within the PrP region. Thus, the number of ORs has been shown to affect the severity of symptoms and onset of clinical signs (Campbell et al., 1996). Although it is known that the region comprising the OR is not essential for mediating prion pathogenesis and propagation, it may modulate the incubation time to disease (Goldmann et al., 1998; Flechsig et al., 2000; Beck et al., 2001).
In the present study, we tried to evaluate the effect of one extra octarepeat insertion mutation (7OR-PrP) on the efficiency of prion transformation and propagation using a model of transgenic mice expressing bovine PrP. High expression levels of the prion protein have been related to a shorter incubation time for the prions to propagate in mice (Scott et al., 1997; Castilla et al., 2003). Thus, we used two boTg lines for each OR type: bo6ORTg113 showing a threefold relative expression level to cattle PrP expression (3x), bo6ORTg135 (6x), bo7ORTg007 (4x), and bo7ORTg006 (6x). The level of expression shown by these mouse lines also ensures the rapid propagation of the prion, but without reaching the pathogenicity threshold.
As expected, BSE transmission was highly effective in both bo6OR and bo7OR transgenic mice, in agreement with the lack of an effective species barrier. In both bo6OR and bo7OR transgenic mice, the incubation and survival times were clearly affected depending on transgene expression levels and the inoculum used.
Remarkably, the two types of transgenic mice (bo6OR and bo7OR) significantly differed in the time taken for clinical signs to appear and in survival time after BSE infection. Our results clearly demonstrate that bo7ORTg mice show an altered course of BSE infection, reflected as reduced incubation and survival times when compared with bo6ORTg mice expressing similar levels of the wild-type six-octapeptide protein These differences were confirmed using three different inocula (Figs. 2, 3; Table 1). In accordance with the lack of an interspecies barrier to BSE infection, we detected the typical signs of CNS spongiform degeneration by histopathological analysis (Fig. 4) and the presence of the bovine prion PrPres by Western blotting (supplementary Fig. 3B) or IHC analyses (Fig. 4). The histological patterns observed in both bo6ORTg or bo7ORTg mice for the different BSE inocula used were indistinguishable from that of classic BSE in cattle (Fig. 4).
Given these results were obtained in transgenic mouse lines with identical bovine PrP expression levels, we can attribute these differences to the presence of an extra OR in the transgene, suggesting that 7OR-PrP can be transformed into PrPres more efficiently than its homologous 6OR-PrP. It is known that bovines show three different polymorphisms (five, six, and seven ORs) in the PrP gene, and the statistics indicate that cattle expressing the five- or six-OR form of the PrP are similarly susceptible to BSE infection (Neibergs et al., 1994). However, to date there are no data on the susceptibility of cattle expressing the 7OR-PrP protein to BSE infection, a polymorphism occurring in at least 5% of Brown Swiss cows (Schlapfer et al., 1999).
To establish whether the addition of an extra octarepeat insertion mutation modifies the biochemical properties of the bo7OR-PrP protein in relation to the wild-type bo6OR-PrP protein, we determined the protease sensitivity and insolubility in nondenaturing detergents of both proteins. As shown in supplementary Figure 3A, PrPC and PrPres containing seven octapeptides (7OR-PrPC and 7OR-PrPres) showed similar protease sensitivity and insolubility in nondenaturing detergents to homologous 6OR-PrPC and 6OR-PrPres. An increased number of ORs has been related previously to enhanced aggregation and proteinase K (PK) resistance of the mutant PrP protein in the hamster (Priola and Chesebro, 1998). Nevertheless, despite the changes in the biochemical properties of PrP noted when there was a considerable difference in the number of ORs, hamster 7OR PrP molecules showed similar aggregation and protease resistance to six-OR PrP molecules. These data are consistent with our findings and with in vivo observations that increasing numbers of the repeat motifs lead to the earlier onset of familial CJD (Goldfarb et al., 1994b; Capellari et al., 1997). Indeed, the enhanced aggregation and protease resistance properties of the mutant PrPs could, in some way, affect the appearance of the disease associated with these mutations. It is clear that a greater understanding of PrP structure and the effects of adding repeats to its coding region is required. In effect, it has been proposed that the properties of PrP proteins with more than seven ORs in their coding region (toxicity, insolubility, and high aggregation capacity) could be directly related to an increase in the number of ORs. The mechanism through which extra copies of the OR may influence the aggregation properties of PrP is unclear. However, it has been suggested that the repeat region of the protein may itself act as a site promoting PrP self-aggregation or aggregation between PrP and other cell factors (Jarrett and Lansbury, 1993; Priola and Chesebro, 1998). This is consistent with the idea that PrP mutants with more than one extra copy in the OR region could more efficiently spontaneously generate PrPres in vivo. Based on our finding of identical biochemical properties of 6OR and 7OR PrP, we propose that the addition of a single OR could lead to a change in PrP structure of insufficient magnitude to confer the protein any discernible biochemical properties but might make the PrP molecules more prone to transformation to PrPres. This rationale would explain why our bovine 6OR- and 7OR-PrP show identical solubility and PK sensitivity but a marked difference in prion propagation in a murine background (Table 1).
To evaluate the infectivity of newly formed 7OR-PrPSc in bo7ORTg mice inoculated with BSE1, compared with that of 6OR-PrPSc, we inoculated groups of bo6ORTg and bo7ORTg mice showing identical PrPC expression levels. Consistently, the bo7ORTg mice showed the shortest incubation and survival times (Figs. 2, 3). The possibility that a passage inoculum in a murine context (boTgBSE1) promotes prion propagation is precluded by the fact that both inocula showed similar levels of PrPres (BSE2 as 6OR-PrPSc and bo7ORTgBSE1 as 7OR-PrPSc) and that bo6ORTgBSE1, a 6OR-PrPSc obtained from BSE1-inoculated bo6ORTg, showed similar results (data not shown). These infectivity data suggest that the 6OR-PrPSc and 7OR-PrPSc structures could also be similar.
The proposed ability of 7OR-PrP to be transformed and propagated more efficiently than its homologous 6OR-PrP could also explain the fact that the infectivity of inocula derived from asymptomatic animals was only detected in the bo7ORTg mice, whereas bo6ORTg mice were not able to detect infectivity in these inocula containing a minimal amount of BSE prions (Fig. 5A). These results clearly indicate a role for the octapeptide region in modulating susceptibility to infection.
In addition, our kinetic experiments designed to determine the earliest time point, in relative terms, at which the prions could be detected in bo6ORTg and bo7ORTg mice after BSE inoculation (Fig. 5B) demonstrated the bo7ORTg model to be a faster mouse bioassay for BSE detection. This finding is also consistent with the idea that 7OR-PrP is more efficiently transformed into PrPres than 6OR-PrP. Presumably, after 7OR-PrPSc production, the prion would be propagated quickly, reducing the time needed to detect PrPres by conventional methods. The higher sensitivity of the bo7ORTg mouse bioassay makes it suitable for assessing infectivity in different cow tissues, in particular those mostly consumed by humans. This mouse line may also have applications in checking drugs or medical products derived from cattle for prion contamination (e.g., vaccines containing bovine serum).
In summary, the addition of octapeptide repeats in the PrP coding region could give rise to a different PrP structure from that of the wild-type PrP, capable of acquiring properties according to the number of ORs added. The addition of a single OR would appear to generate a new 7OR-PrP structure that can be transformed more efficiently than its homologous 6OR-PrP but is insufficiently different for its biochemical properties to be affected.
Footnotes
This work was supported by a Spanish national grant (SC00-055). We thank J. C. Manson for supplying the Prnp -/- mice, D. R. Borchelt for supplying the MoPrP.Xho plasmid, and the Veterinary Laboratory Agency (Weybridge, UK) for supplying BSE brain material. We also acknowledge Fernando Rodríguez for critical reading of this manuscript.
Correspondence should be addressed to Dr. Juan María Torres Trillo, Centro de Investigación en Sanidad Animal, INIA, Valdeolmos, 28130 Madrid, Spain. E-mail: jmtorres@inia.es.
Copyright © 2004 Society for Neuroscience 0270-6474/04/242156-09$15.00/0
References
- Adams DH (1986) The nature of the scrapie agent. Med Hypotheses 20: 37-50. [DOI] [PubMed] [Google Scholar]
- Alpers M, Rail L (1971) Kuru and Creutzfeldt-Jakob disease: clinical and aetiological aspects. Proc Aust Assoc Neurol 8: 7-15. [PubMed] [Google Scholar]
- Beck E, Daniel PM, Matthews WB, Stevens DL, Alpers MP, Asher DM, Gajdusek DC, Gibbs Jr CJ (1969) Creutzfeldt-Jakob disease. The neuropathology of a transmission experiment. Brain 92: 699-716. [DOI] [PubMed] [Google Scholar]
- Beck JA, Mead S, Campbell TA, Dickinson A, Wientjens DP, Croes EA, Van Duijn CM, Collinge J (2001) Two-octapeptide repeat deletion of prion protein associated with rapidly progressive dementia. Neurology 57: 354-356. [DOI] [PubMed] [Google Scholar]
- Bendheim PE (1984) The human spongiform encephalopathies. Neurol Clin 2: 281-298. [PubMed] [Google Scholar]
- Borchelt DR, Davis J, Fischer M, Lee MK, Slunt HH, Ratovitsky T, Regard J, Copeland NG, Jenkins NA, Sisodia SS, Price DL (1996) A vector for expressing foreign genes in the brains and hearts of transgenic mice. Genet Anal 13: 159-163. [DOI] [PubMed] [Google Scholar]
- Brun A, Castilla J,. Ramírez MA, Prager K, Parra B, Salguero FJ, Shiveral D, Sánchez C, Sánchez-Vizcaíno JM, Douglas A, Torres, JM (2004) Proteinase-K enhanced immunoreactivity to the PrP specific monoclonal antibody 2A11. Neurosci Res 48: 75-83. [DOI] [PubMed] [Google Scholar]
- Campbell TA, Palmer MS, Will RG, Gibb WR, Luthert PJ, Collinge J (1996) A prion disease with a novel 96-base pair insertional mutation in the prion protein gene. Neurology 46: 761-766. [DOI] [PubMed] [Google Scholar]
- Capellari S, Vital C, Parchi P, Petersen RB, Ferrer X, Jarnier D, Pegoraro E, Gambetti P, Julien J (1997) Familial prion disease with a novel 144-bp insertion in the prion protein gene in a Basque family. Neurology 49: 133-141. [DOI] [PubMed] [Google Scholar]
- Castilla J, Gutiérrez Adán A, Brun A, Pintado B, Ramírez MA, Parra B, Doyle D, Rogers M, Salguero J, Sánchez C, Sánchez-Vizcaíno JM, Torres JM (2003) Early detection of PrPres in BSE-infected bovine PrP transgenic mice. Arch Virol 148: 677-691. [DOI] [PubMed] [Google Scholar]
- Chiesa R, Piccardo P, Ghetti B, Harris DA (1998) Neurological illness in transgenic mice expressing a prion protein with an insertional mutation. Neuron 21: 1339-1351. [DOI] [PubMed] [Google Scholar]
- Cochran EJ, Bennett DA, Cervenakova L, Kenney K, Bernard B, Foster NL, Benson DF, Goldfarb LG, Brown P (1996) Familial Creutzfeldt-Jakob disease with a five-repeat octapeptide insert mutation. Neurology 47: 727-733. [DOI] [PubMed] [Google Scholar]
- Collinge J, Harding AE, Owen F, Poulter M, Lofthouse R, Boughey AM, Shah T, Crow TJ (1989) Diagnosis of Gerstmann-Straussler syndrome in familial dementia with prion protein gene analysis. Lancet 2: 15-17. [DOI] [PubMed] [Google Scholar]
- Dormont D (1994) Human transmissible subacute spongiform encephalopathy. Bull Acad Natl Med 178: 887-903. [PubMed] [Google Scholar]
- Fink PS (1991) Using sodium chloride step gradients to fractionate DNA fragments. Biotechniques 10: 446, 448:450. [PubMed] [Google Scholar]
- Fischer M, Rulicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C (1996) Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J 15: 1255-1264. [PMC free article] [PubMed] [Google Scholar]
- Flechsig E, Shmerling D, Hegyi I, Raeber AJ, Fischer M, Cozzio A, von Mering C, Aguzzi A, Weissmann C (2000) Prion protein devoid of the octapeptide repeat region restores susceptibility to scrapie in PrP knockout mice. Neuron 27: 399-408. [DOI] [PubMed] [Google Scholar]
- Foster JD, Wilson M, Hunter N (1996) Immunolocalisation of the prion protein (PrP) in the brains of sheep with scrapie. Vet Rec 139: 512-515. [DOI] [PubMed] [Google Scholar]
- Goldfarb LG, Brown P, McCombie WR, Goldgaber D, Swergold GD, Wills PR, Cervenakova L, Baron H, Gibbs Jr CJ, Gajdusek DC (1991) Transmissible familial Creutzfeldt-Jakob disease associated with five, seven, and eight extra octapeptide coding repeats in the PRNP gene. Proc Natl Acad Sci USA 88: 10926-10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb LG, Brown P, Little BW, Cervenakova L, Kenney K, Gibbs Jr CJ, Gajdusek DC (1993) A new (two-repeat) octapeptide coding insert mutation in Creutzfeldt-Jakob disease. Neurology 43: 2392-2394. [DOI] [PubMed] [Google Scholar]
- Goldfarb LG, Brown P, Cervenakova L, Gajdusek DC (1994a) Molecular genetic studies of Creutzfeldt-Jakob disease. Mol Neurobiol 8: 89-97. [DOI] [PubMed] [Google Scholar]
- Goldfarb LG, Brown P, Cervenakova L, Gajdusek DC (1994b) Genetic analysis of Creutzfeldt-Jakob disease and related disorders. Philos Trans R Soc Lond B Biol Sci 343: 379-384. [DOI] [PubMed] [Google Scholar]
- Goldmann W, Hunter N, Martin T, Dawson M, Hope J (1991) Different forms of the bovine PrP gene have five or six copies of a short, G-C-rich element within the protein-coding exon. J Gen Virol 72: 201-204. [DOI] [PubMed] [Google Scholar]
- Goldmann W, Chong A, Foster J, Hope J, Hunter N (1998) The shortest known prion protein gene allele occurs in goats, has only three octapeptide repeats and is non-pathogenic. J Gen Virol 79: 3173-3176. [DOI] [PubMed] [Google Scholar]
- Hill AF, Desbruslais M, Joiner S, Sidle KC, Gowland I, Collinge J, Doey LJ, Lantos P (1997) The same prion strain causes vCJD and BSE. Nature 389: 448-450, 526. [DOI] [PubMed] [Google Scholar]
- Hogan B, Williams J (1981) Integration of foreign genes into the mammalian germ line: genetic engineering enters a new era. Nature 294: 9-10. [DOI] [PubMed] [Google Scholar]
- Hunter N, Goldmann W, Smith G, Hope J (1994) Frequencies of PrP gene variants in healthy cattle and cattle with BSE in Scotland. Vet Rec 135: 400-403. [DOI] [PubMed] [Google Scholar]
- Jarrett JT, Lansbury Jr PT (1993) Seeding “one-dimensional crystallization” of amyloid: a pathogenic mechanism in Alzheimer's disease and scrapie? Cell 73: 1055-1058. [DOI] [PubMed] [Google Scholar]
- Kozak M (1989) The scanning model for translation: an update. J Cell Biol 108: 229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann S, Zerr I, Weber T, Poser S, Kretzschmar H, Hunsmann G, Bodemer W (1995) Prion disease associated with a novel nine octapeptide repeat insertion in the PRNP gene. Brain Res Mol Brain Res 34: 173-176. [DOI] [PubMed] [Google Scholar]
- Manetto V, Medori R, Cortelli P, Montagna P, Tinuper P, Baruzzi A, Rancurel G, Hauw JJ, Vanderhaeghen JJ, Mailleux P (1992) Fatal familial insomnia: clinical and pathologic study of five new cases. Neurology 42: 312-319. [DOI] [PubMed] [Google Scholar]
- Manson JC, Clarke AR, Hooper ML, Aitchison L, McConnell I, Hope J (1994) 129/Ola mice carrying a null mutation in PrP that abolishes mRNA production are developmentally normal. Mol Neurobiol 8: 121-127. [DOI] [PubMed] [Google Scholar]
- Medori R, Montagna P, Tritschler HJ, LeBlanc A, Cortelli P, Tinuper P, Lugaresi E, Gambetti P (1992) Fatal familial insomnia: a second kindred with mutation of prion protein gene at codon 178. Neurology 42: 669-670. [DOI] [PubMed] [Google Scholar]
- Neibergs HL, Ryan AM, Womack JE, Spooner RL, Williams JL (1994) Polymorphism analysis of the prion gene in BSE-affected and unaffected cattle. Anim Genet 25: 313-317. [DOI] [PubMed] [Google Scholar]
- Owen F, Poulter M, Shah T, Collinge J, Lofthouse R, Baker H, Ridley R, McVey J, Crow TJ (1990) An in-frame insertion in the prion protein gene in familial Creutzfeldt-Jakob disease. Brain Res Mol Brain Res 7: 273-276. [DOI] [PubMed] [Google Scholar]
- Palmer MS, Collinge J (1993) Mutations and polymorphisms in the prion protein gene. Hum Mutat 2: 168-173. [DOI] [PubMed] [Google Scholar]
- Priola SA, Chesebro B (1998) Abnormal properties of prion protein with insertional mutations in different cell types. J Biol Chem 273: 11980-11985. [DOI] [PubMed] [Google Scholar]
- Prusiner SB (1991) Molecular biology of prion diseases. Science 252: 1515-1522. [DOI] [PubMed] [Google Scholar]
- Prusiner SB (1998) The prion diseases. Brain Pathol 8: 499-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley RM, Baker HF (1993) Genetics of human prion disease. Dev Biol Stand 80: 15-23. [PubMed] [Google Scholar]
- Schlapfer I, Saitbekova N, Gaillard C, Dolf G (1999) A new allelic variant in the bovine prion protein gene (PRNP) coding region. Anim Genet 30: 386-387. [DOI] [PubMed] [Google Scholar]
- Scott M, Foster D, Mirenda C, Serban D, Coufal F, Walchli M, Torchia M, Groth D, Carlson G, DeArmond SJ (1989) Transgenic mice expressing hamster prion protein produce species- specific scrapie infectivity and amyloid plaques. Cell 59: 847-857. [DOI] [PubMed] [Google Scholar]
- Scott M, Groth D, Foster D, Torchia M, Yang SL, DeArmond SJ, Prusiner SB (1993) Propagation of prions with artificial properties in transgenic mice expressing chimeric PrP genes. Cell 73: 979-988. [DOI] [PubMed] [Google Scholar]
- Scott MR, Safar J, Telling G, Nguyen O, Groth D, Torchia M, Koehler R, Tremblay P, Walther D, Cohen FE, DeArmond SJ, Prusiner SB (1997) Identification of a prion protein epitope modulating transmission of bovine spongiform encephalopathy prions to transgenic mice. Proc Natl Acad Sci USA 94: 14279-14284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walawski K, Czarnik U (2003) Prion octapeptide-repeat polymorphism in Polish Black-and-White cattle. J Appl Genet 44: 191-195. [PubMed] [Google Scholar]
- Will RG, Ironside JW, Zeidler M, Cousens SN, Estibeiro K, Alperovitch A, Poser S, Pocchiari M, Hofman A, Smith PG (1996) A new variant of Creutzfeldt-Jakob disease in the UK. Lancet 347: 921-925. [DOI] [PubMed] [Google Scholar]
- Yoshimoto J, Iinuma T, Ishiguro N, Horiuchi M, Imamura M, Shinagawa M (1992) Comparative sequence analysis and expression of bovine PrP gene in mouse L-929 cells. Virus Genes 6: 343-356. [DOI] [PubMed] [Google Scholar]