Abstract
Purpose
We recently reported a new method accessing proteins from extracellular matrix by imaging mass spectrometry (ECM IMS). ECM IMS was evaluated for use in exploring breast tissue pathologies.
Experimental Design
A tissue microarray (TMA) was analyzed that had 176 cores of biopsies and lumpectomies spanning breast pathologies of inflammation, hyperplasia, fibroadenoma, invasive ductal carcinoma and invasive lobular carcinoma and normal adjacent to tumor (NAT). NAT was compared to subtypes by area under the receiver operating curve (ROC) >0.7. A lumpectomy was also characterized for collagen organization by microscopy and stromal protein distribution by IMS. LC-based high-resolution accurate mass (HRAM) proteomics was used to identify proteins from the lumpectomy.
Results
TMA analysis showed distinct spectral signatures reflecting a heterogeneous tissue microenvironment. Ninety-four peaks showed a ROC >0.7 compared to NAT; NAT had overall higher intensities. Lumpectomy analysis by IMS visualized a complex central tumor region with distal tumor regions. HRAM LC-based proteomics identified 39 stromal proteins. Accurate mass matches between image data and LC-based proteomics demonstrated a heterogeneous collagen type environment in the central tumor.
Conclusions
Data portray the heterogeneous stromal microenvironment of breast pathologies, including alteration of multiple collagen type patterns. ECM IMS is a promising new tool for investigating the stromal microenvironment of breast tissue including cancer.
Keywords: breast cancer, extracellular matrix, imaging mass spectrometry, proteomics, peptide imaging, formalin-fixed, paraffin-embedded tissue imaging, tissue imaging, MALDI imaging mass spectrometry
Introduction
Breast stroma is a highly organized composition of collagens and other extracellular matrix (ECM) proteins with a significant role in regulation of breast health.[1] Dense breast stroma has a significantly higher risk of developing cancer with tumors likely to form in regions of increased collagen.[2] Generally, in breast cancer progression, stroma composition becomes denser through excessive extrusion of ECM proteins. This is due to a combination of aberrant myoepithelial-luminal regulation of stroma and later altered regulation of fibrillary collagen by macrophage cells.[1, 3] Increased stroma density has been shown to be a predictor of localized recurrence after radiotherapy.[4] For metastatic cancers, stroma shows pathological distributions of increased collagen deposition near the tumor, linear collagen organization, and stiffening through crosslinking and post-translational modifications (PTMs).[5, 6] Breast cancer stromal regulation thus has use as a predictive risk factor, as a prognostic indicator, and in the diagnoses of tumor progression. However, a major limitation to understanding the role of stroma in breast cancer has been a lack of tools that can precisely define the biochemical aspects of stromal composition from increased stromal density to tumor metastasis.[1, 7, 8]
Microscopic imaging has been a main analytical tool for the study of breast stroma. Microscopy methods use an assortment of stains and physical properties to report on stromal collagen fiber organization, quantity, alignment, and length in relationship to breast cancer progression.[1, 7] Movat’s pentachrome or the trichrome stain histological stain reports collagen distribution in relationship to other tissue components such as elastin, glycosaminoglycans and muscle, but does not give information on fiber arrangement. The picrosirus red stain (PSR) as visualized with a circularly polarized light filter (CPLF) demonstrates collagen organization and fiber detail but cannot report on collagen type or potential PTMs.[9] The gold standard for quantitative assessment of collagen changes in breast cancer is second harmonic generation (SHG) microscopy.[6] SHG uses processes of light scattering and photon absorption to report nonlinear polarization properties of biological materials for high resolution images of collagen organization.[10] SHG observations termed tumor associated collagen signatures (TACS) relate collagen organization to breast tumor stage and prognosis in human and mouse models. [1, 6] These studies show that increased deposition of collagen occurs near the tumor in early tumor formation (TACS-1), forms linear fibers parallel to tumor boundaries as tumor increases (TACS-2) with multiple fibers oriented perpendicularly to the tumor border (TACS-3). TACS-3 is associated with metastasis and tumor cells are frequently observed migrating along the aligned collagen fibers. Both PSR-CPLF and SHG define that breast cancer has quantitative collagen metrics characteristic for staging[11]; however, these methods cannot report on biochemical content driving stromal expression that results in a tumor permissive environment.
MALDI IMS is an established imaging modality that uses mass spectrometry to probe molecular features of histology beyond what can be visualized by microscopy. MALDI IMS may be used to detect molecular patterns from hundreds to thousands of metabolites, lipids, peptides, intact proteins, including potential PTMs corresponding to histological features from thin tissue sections.[12] This highly multiplexed information is used to further characterize pathology by providing additional molecular details such as biomarkers or signatures of disease stage.[13, 14] MALDI IMS may be used for spatially localized proteomics analogous to LC-based proteomic strategies by spraying a thin molecular layer of trypsin onto the tissue to release tryptic peptides without delocalization.[15, 16] After digestion in a controlled temperature, high humidity chamber, the tissue is sprayed with a chemical matrix to facilitate peptide ionization. A laser is then systematically stepped across the tissue, desorbing and ionizing peptides at ordered, discrete locations for detection. Typically hundreds to thousands of peptides are detected from a single tissue section and each peptide can be visualized as a heatmap distribution across the tissue features. MALDI IMS of tryptic peptides in breast cancer tissues has shown that this imaging modality can define protein features of breast cancer including estrogen-receptor positive versus estrogen-receptor negative tumors[17], invasive ductal carcinoma[18], metastatic signatures[16], and stromal activation.[13] A disadvantage in using trypsin to access proteins by MALDI IMS is that this approach has limited reporting of collagen types and ECM content from inaccessibility due to the PTMs, especially of lysines, and/or the highly organized collagen suprastructures.
Recently, we developed a MALDI IMS approach that reports on collagens and other stromal proteins from thin tissue sections.[19] The method, which we term ECM IMS, uses a bacterial matrix metalloproteinase (MMP), collagenase type III (COLase3), to target ECM proteins for digestion to peptides, which are then mapped as 2D distributions by MALDI IMS. Compared to mammalian MMPs which target specific collagens for degradation, bacterial MMPs are unbiased to collagen types and tissue origin.[19, 20] Previous evaluation of the COLase3 proteome by high resolution accurate mass (HRAM) LC-based proteomic methods identified that the method pulls down not only multiple collagen types, but also the collagen interactome. In the current work, we use the method to explore breast pathologies as a proof of concept towards breast cancer studies. We apply the method to a tissue microarray (TMA) defined for breast tissue pathologies including invasive cancers, as well as a lumpectomy characterized for collagen organization. Data are reported in the context of parallel HRAM proteomic studies done on the breast cancer tissue lumpectomy, coupled with protein interaction analysis. This approach provides a new tool for understanding stromal features of breast tissue including cancer progression.
Materials and Methods
Experimental
Materials
Acetonitrile, α-Cyano-4-hydroxycinnamic acid (CHCA), trifluoroacetic acid (TFA), and Trizma® base were purchased from Sigma-Aldrich (St. Louis, MO, USA). Collagenase type III (COLase3) (C. histolyticum) was purchased from StemCell Technologies (Cambridge, MA, USA). Xylenes, 200 proof ethanol, methanol, citraconic anhydride were purchased from Fisher Scientific (Pittsburgh, PA, USA).
Tissue Procurement
Tissue use was in accordance with protocols approved by the Medical University of South Carolina Institutional Review Board. The tissue microarray (TMA) was purchased from Biomax (Rockville MD, USA) as defined breast tissue pathology with 176 cases of breast pathologies from normal adjacent to tumor, inflammation, hyperplasia, fibroadenoma, invasive ductal carcinoma, intraductal carcinoma, and lobular carcininoma. The TMA was created in 2014 and used the WHO 2012 guidelines for grading. The Nottingham modification of the Scarff-Bloom-Richardson (NSBR) histological grading system was used for invasive breast cancer (IBC). Each core was further evaluated by a collaborating pathologist for tumor, stroma, glandular, inflammation and read to determine status of adenosis, inflammation, fibroadenoma and squamous cell carcinoma (rare). Tissue sections including the lumpectomy used for proteomics were procured via the Biorepository & Tissue Analysis Shared Resource at the Hollings Cancer Center, Medical University of South Carolina with Institutional Review Board (IRB) approval.
Tissue Preparation
The formalin-fixed, paraffin-embedded (FFPE) tissues and TMA were prepared as previously described.[19] Briefly, tissues were heated, dewaxed, antigen retrieved at pH 9, and sprayed with a thin molecular layer of collagenase type III (COLase3; StemCell Technologies) using an automated sprayer (M3 TM-Sprayer, HTXImaging, Chapel Hill, NC, USA). COLAse3 was sprayed onto tissues with the same automated sprayer using parameters of 45°C, 10 psi, 25 μL/min, 1200 velocity, and 15 passes with a 3.0 mm offset. Samples were digested in high humidity at 37.5°C for five hours followed automated spraying of CHCA matrix prepared as 7 mg/mL in 50% acetonitrile, 1% TFA with a spiked standard of 200 femtomole/microliter [Glu1]-Fibrinopeptide B human (Glufib) (Sigma-Aldrich), St. Louis, MO, USA). CHCA was sprayed onto tissues with the same automated sprayer using parameters of 77°C, 10 psi, 100 μL/min, 1300 velocity, and 10 passes with a 2.5 mm offset.
Imaging Mass Spectrometry
Samples were analyzed by MALDI-FT-ICR (solariX™ Legacy 7.0 Tesla, Bruker, Bremen, Germany) in positive ion mode, collecting 300 laser shots per pixel using the SmartWalk feature to raster the laser in a 25 μm diameter. Transients of 1 megaword were acquired in broadband mode over m/z 700–5000, with a calculated on-tissue mass resolution at full width half maximum of 81,000 at m/z 1400. Lockmass on Glufib peptide was maintained at 5 ppm during tissue imaging. Data were visualized in flexImaging 4.0 and analyzed by SCiLS Lab software 2017a both from Bruker Daltonics, Bremen, Germany. All images are shown normalized to total ion current. Image segmentation is reported using the Euclidean metric for segmentation. Extracted peak areas were exported from SCiLs and are shown visualized with MultiExperiment Viewer (http://www.tm4.org), a cloud-based freeware for large data.[21] Hierarchical clustering of peak areas used the Manhattan method with average linkage as a distance metric selection. Area under the receiver-operating curve (AUC) was used to explore the potential of peaks that discriminated between cancer types, subtypes and staging. In these studies, cancer types that had few cores (squamous, cores= 2; lobular in situ, cores=3) were removed from the analyses.
Proteomics
Sections of cancer tissue were treated as described previously[19], digesting with COLase3 overnight. Quantitation reported an average of 25 μg collected from 15 mm x 15mm x 5μm breast sections. From these, 4μg of COLAse3 peptides was dried down, cleaned up by solid phase extraction using a C18 Ziptip (EMD Millipore, Darmstadt, Germany) following the manufacturer’s protocol. Peptides were analyzed by data dependent acquisition (data exclusion enabled) on an Orbitrap Elite mass spectrometer equipped with a LC Packings U3000 nano LC system (Thermo Scientific). A single Fourier transform mass spectrometry survey scan acquired in the orbitrap followed by collision-induced dissociation MS/MS of the top 10 most intense ions in the ion trap. Tandem mass spectra were searched using both MASCOT (Version 2.4.01) and SEQUEST HT via Proteome Discoverer 1.4 (Thermo Scientific, Waltham, MA, USA) against a subset of human protein sequences downloaded on May 7, 2017 from UniProtKB (SwissProt) containing 1,783 entries (keywords used: collagen, elastin, aggrecan, gelatin, osteonectin, perlecan, plasminogen, and fibronectin). Search parameters were unspecified enzyme, precursor mass tolerances of ± 20 ppm, and fragment mass tolerances ± 0.8 Da. Methionine oxidation, asparagine and glutamine deamidation were used as variable modifications. Data were uploaded into Scaffold v4.8.1 (Proteomesoftware, Portland OR, USA) and a peptide probability of 99% was used to report peptide identifications. Protein localization was determined using Uniprot and Genecard. Accurate mass comparisons between image data and proteomic data were done by calculating accurate mass of a peptide using Protein Prospector version 5.22.0 (Baker, P.R. and Clauser, K.R. http://prospector.ucsf.edu, University of California at San Francisco, San Francisco CA, USA).
Network Analysis
The upstream regulator networks were generated using Ingenuity Pathways Analysis (IPA) (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis). Protein identifications were uploaded into IPA to identify regulators of the protein dataset. Mechanistic networks were calculated using a p-value <1.0E-12 for inclusion in the network, filtering all predicted relationships between regulators by p-value ≤0.01.
Results and Discussion
Overview
A commercially prepared TMA and a breast cancer tissue were used to demonstrate evaluation of breast stroma by ECM IMS. The TMA was composed of 176 needle core biopsies and lumpectomies from individual cases associated with a spectrum of breast pathologies ranging from normal adjacent to invasive carcinomas (Table 1, Supplement Table 1). Data for each core includes estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) status (Supplemental Figure 1; Supporting Material). A tissue section of a lumpectomy was analyzed by microscopy, ECM IMS and HRAM proteomics to demonstrate obtainable stroma data in the context of collagen organization. HRAM-identified peptides were mapped by high mass accuracy in both the lumpectomy and the TMA data to illustrate known stromal patterns in the complex tumor microenvironment relative to pathological progression.
Table 1.
Characteristics of the TMA. Cores marked as squamous cell carcinoma (4 cores) were not used in statistical calculations.
| Age (female) | 46.7 ± 11.7 (range 19–82) | |
| Pathology diagnosis | Stage | Cores |
| Normal adjacent tissue | - | 16 |
| Plasma cell mastitis (inflammation) | - | 8 |
| Acute mastitis | - | 1 |
| Interstitial chronic inflammation | - | 7 |
| Adenosis with hyperplasia of breast duct | - | 7 |
| Hyperplasia (adenosis) | - | 7 |
| Atypical hyperplasia (grade II) | - | 1 |
| Fibroadenoma | - | 8 |
| Intraductal carcinoma | 0 | 19 |
| Intraductal carcinoma | IIA | 4 |
| Lobular carcinoma in situ | 0 | 3 |
| Invasive ductal carcinoma | I | 9 |
| Invasive ductal carcinoma | IIA | 39 |
| Invasive ductal carcinoma | IIB | 11 |
| Invasive ductal carcinoma | IIIA | 11 |
| Invasive ductal carcinoma | IIIB | 7 |
| Invasive lobular carcinoma | IIA | 6 |
| Invasive lobular carcinoma | IIIA | 3 |
| Invasive lobular carcinoma | IIB | 11 |
| Invasive lobular carcinoma | IIIB | 2 |
| Squamous cell carcinoma | I | 2 |
| Squamous cell carcinoma | IIB | 1 |
| Squamous cell carcinoma | IIIB | 1 |
Heuristic Analysis of the TMA
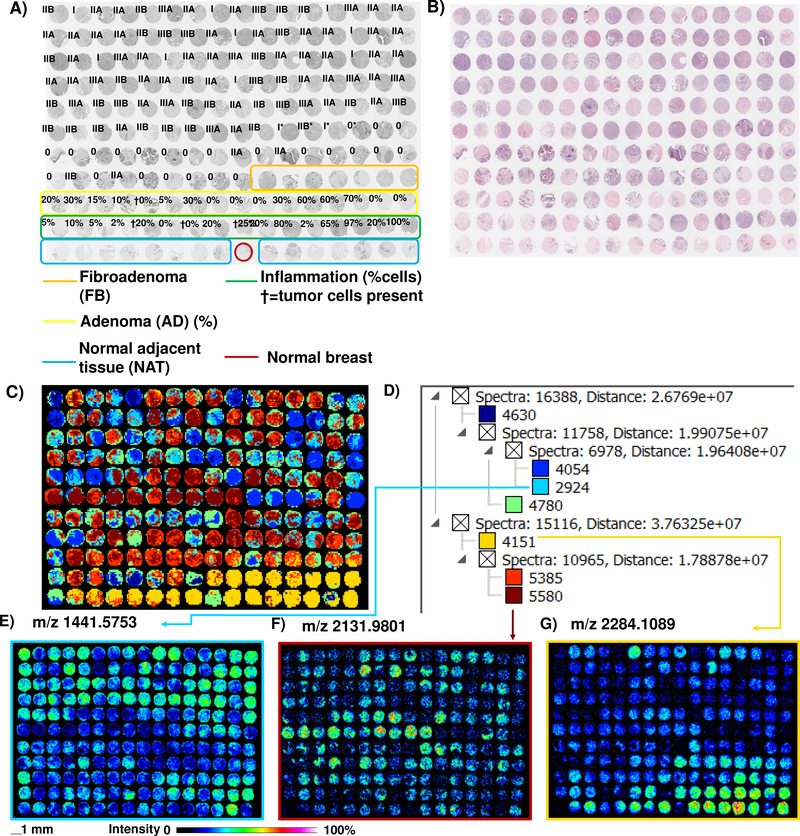
A goal of MALDI IMS is to identify novel molecular patterns of pathology; therefore we started our evaluation with exploratory approaches on unidentified peaks using image segmentation and data visualization. Image segmentation is a method that groups similar spectral features into regions, labeling and color coding them to identify molecular patterns across a tissue.[22] A total of 3,575 peaks found in various TMA cores were used to visualize overall spectral patterns linked to tissue features (Figure 1, Supplemental Figure 2). NAT was labeled as having distinct spectral features from adenomas, fibroadenoma, and cancer pathologies, as well as one normal breast tissue core derived from a patient without breast cancer. Interestingly, eight patient cores diagnosed as inflammation showed overlap with NAT tissue. Examination of inflammatory cell content in each core did not reveal a differences due to numbers of inflammatory cells (Supplemental Table 2, Supporting Material). Extracted peptide images from segments highlighted distinctive images related to breast pathologies (Figures 1E–G). Although image segmentation gave a good overview of potential differences in proteomes, it appeared that the patterns found by ECM imaging were more complex than could be described by regionalized spectral clustering compared to cellular content, tissue or tumor type.
Figure 1.
Image segmentation of a TMA of breast cancer progression after ECM IMS, done on 3,575 peaks. A) Pathologist grading on TMA. B) Hematoxylin and eosin stain. C) Image segmentation of peaks detected across the cores by MALDI IMS. The data shows distinct separation of noncancerous and cancer stages; cancer types are heterogenous. D) Spectral clusters associated with the image segmentation. Color corresponds to that of spectral cluster in C). See Supplemental Figure 1 and Supplemental Table 1 for further information on TMA Grades and Supplemental Table 2 for cellular and tissue composition.
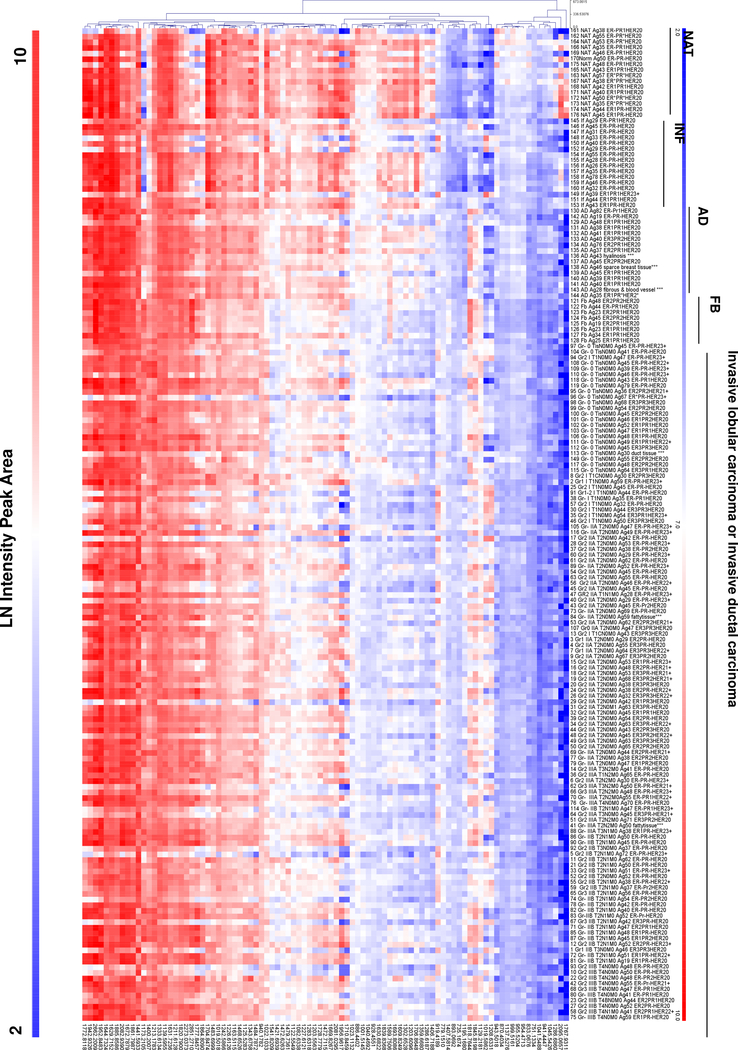
Area under the receiver-operating curve (AUC) was used to define discriminating peaks from the TMA. A total of 94 peaks distinguished tissue pathologies in comparison to NAT using AUC ≥0.7. The peak areas of the 94 discriminating peptides were extracted and assembled as a heatmap of peak areas (Figure 2). This suggested that NAT generally had higher expression values. This is consistent with stromal proteins actively regulating tumor status via the tissue microenvironment surrounding the tumor.[7, 23] Some m/z appeared to align with ER status in NAT, forming clusters of higher intensity. Evaluation of cores with no tumor and low inflammatory cells (<20% cell content) compared to cores with no tumor and high numbers of inflammatory cells (>20% content) revealed 20 peaks that could discriminate between low and high percentage inflammatory infiltrate (Table 2). Invasive subtypes showed dramatic decreases in a subpopulation of the peptides, with high variation overall. We were unable to assess stromal proteins based on survival status and the treatment regiment, since these parameters were unknown. There were comparisons that discriminated between receptor types at same stage or between stages. This was likely due to the much smaller cohorts of each stage that also included heterogeneous receptor status. Ongoing work investigates larger cohorts as well as contrasting active tumors with benign tumors such as fibroadenomas and phyllodes tumors. Comparison of Stage 0 tumors which are non-invasive without metastatic potential (n=22) and Stage I-III invasive tumors with metastatic potential (n=98) showed peaks trending towards discriminatory values (m/z 954.4944 AUC =0.67; m/z 1001.5136 AUC=0.65; m/z 1208.6345 AUC=0.67). Additionally, comparison of samples stratified by cancer type rather than stage showed discriminating peaks (Table 3). Comparisons made between intraductal carcinoma (cores=23), invasive lobular carcinoma (cores=22) and invasive ductal carcinoma (cores= 68) resulted in a total of 14 peaks that could distinguish between cancer types. Taken together, the analysis showed that the method may be used to define changes in breast pathologies.
Figure 2.
Peak intensity heat map of 94 peaks with area under the receiver operating curve >0.7. Data demonstrates that NAT has higher intensity patterns than most tissues. Fibroadenoma (FB) has a unique peak intensity pattern. Invasive type carcinomas have heterogeneous peak intensities, likely reflecting variations in receptor status and potentially patient age. Abbreviations: NAT- normal adjacent to tumor; INF- inflammation; AD- adenoma; FB- fibroadenoma.
Table 2.
Peptides discriminating between breast tissue with low inflammatory cells (<20%) and high inflammatory cells (>20%) using area under the receiver operating curve (AUC) ≥0.7.
| AUC | |
|---|---|
| 797.1228 | 0.737 |
| 940.1782 | 0.754 |
| 1173.2105 | 0.741 |
| 1218.5471 | 0.740 |
| 1333.6134 | 0.756 |
| 1430.6268 | 0.730 |
| 1460.6842 | 0.707 |
| 1479.7339 | 0.781 |
| 1483.7305 | 0.798 |
| 1483.7759 | 0.705 |
| 1541.8232 | 0.725 |
| 1628.7326 | 0.715 |
| 1629.7506 | 0.822 |
| 1644.7354 | 0.765 |
| 1656.7866 | 0.722 |
| 1704.8474 | 0.866 |
| 1706.8619 | 0.802 |
| 1731.7600 | 0.755 |
| 1885.8586 | 0.778 |
| 2284.1428 | 0.730 |
Table 3.
Comparison of breast cancer types using area under the receiver operating curve (AUC) ≥0.7. IDC- Invasive ductal carcinoma, ILC- Invasive lobular carcinoma.
| IDC vs Intraductal | IDC vs ILC | ILC vs Intraductal | |
|---|---|---|---|
| 919.4189 | 0.67 | 0.77 | 0.77 |
| 954.4963 | 0.70 | 0.60 | 0.62 |
| 984.1856 | 0.61 | 0.74 | 0.74 |
| 1001.5145 | 0.70 | 0.66 | 0.44 |
| 1019.5862 | 0.75 | 0.81 | 0.81 |
| 1125.5283 | 0.61 | 0.74 | 0.74 |
| 1139.5955 | 0.60 | 0.71 | 0.71 |
| 1173.2105 | 0.60 | 0.74 | 0.74 |
| 1441.5933 | 0.75 | 0.78 | 0.57 |
| 1675.7846 | 0.56 | 0.71 | 0.65 |
| 1772.8118 | 0.53 | 0.72 | 0.72 |
| 1861.8117 | 0.54 | 0.73 | 0.73 |
| 2663.2051 | 0.53 | 0.68 | 0.70 |
Breast Lumpectomy Analysis by Microscopy and ECM IMS
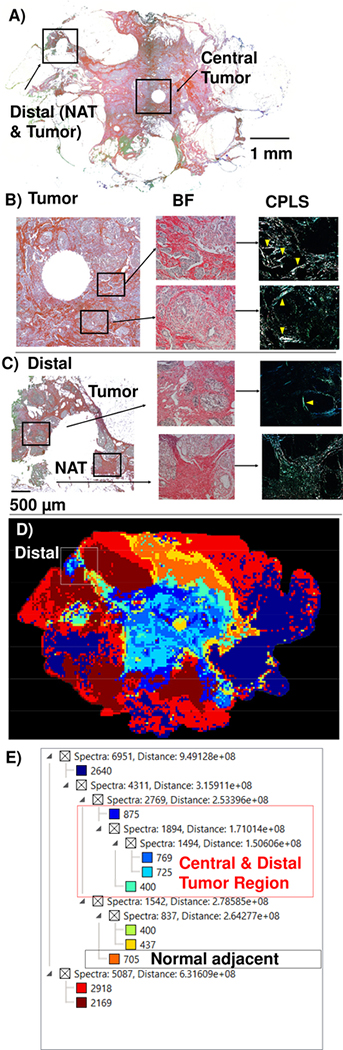
A lumpectomy was characterized by multiple methods to understand IMS data relative to collagen organization (Figure 3). Picrosirius red stain (PSR) viewed under a circularly polarized light filter on the breast tumor (Fig 3B) characterized the tumor collagen as having thick fibers with linear arrangements parallel to the tumor border, suggestive of a TACS-2 tumor type.[6] Examination of distal regions showed normal adjacent tissue (NAT) having homogenous distribution of short thin fibers (Fig 3C). In the distal region, parallel collagen deposition was also observed surrounding a region within 0.5 mm between NAT and surgical boundaries, suggesting the presence of a TACS-1 type tumor. A serial section analyzed by ECM IMS reported 2,210 peaks. Image segmentation demonstrated the tumor as a primary pattern distinguished from surrounding fatty tissue (Figure 3D, E). Fatty tissue patterns were heterogeneous. Corresponding with PSR staining studies, the NAT region showed a pattern matching primary tumor located between defined NAT and surgical marked tumor border. Further expansion of spectral clusters in the tumor region resulted in detailed structure emerging with complex patterning (Supplemental Figure 2). Overall, ECM IMS appeared to accurately report the breast tumor microenvironment correlating with that obtained by microscopy of collagen alignment. In particular, ECM IMS was able to highlight that a distal region of tumor existed between NAT and the surgical boundaries.
Figure 3.
Pathology of the lumpectomy. A) Overall lumpectomy shown with picrosirius red stain by brightfield highlighting regions in the central tumor or distal to tumor. Circles in the tumor and NAT region depicts location where a tissue core was collected by for other studies. B) Tumor region. Brightfield shows tumor structure and CPFL demonstrates linear organization of collagen near tumor boundaries. C) Distal region. Brightfield demonstrates tissue structure and CPFL highlights collagen organization. NAT shows homogenous distribution of short fibers. A tumor was found between the surgical border and NAT. This shows thick fibers parallel with tumor boundaries. D) Image segmentation shows the central tumor colored in royal blue. The distal region is correctly identified as tumorous. E) Spectral clusters of the image segmentation. Yellow arrows mark regions of collagen alignment. Abbreviations: NAT- normal adjacent to tumor; CPFL- circularly polarized light filter.
Proteomics
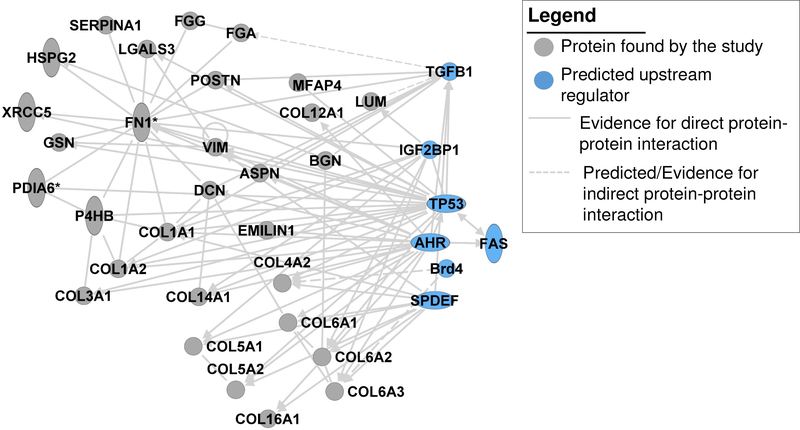
To increase our understanding of what types of peptides are detected from breast tissue by COLase3 digestion, we performed HRAM proteomics on the same breast cancer tissue used for IMS. This resulted in a total of 38 proteins with two or more unique peptides (Supplemental Table 3). A total of 34 (90%) proteins were annotated as localized to ECM, 13 of which were collagen types. Protein function changes according to cellular localization.[24] Several proteins had literature reporting differential localization (ENOA, PDIA1, PRDX5, TLN1), showing the potential for differential functions in breast cancer. Proteins were assessed for functional annotation and potential regulators using predictive and literature supported protein interactions (Table 3, Figure 4). Functional annotations showed that the proteins identified have a strong association with progressive processes of breast cancer, reflecting the heterogeneous tumor microenvironment (Table 4). Networked interactions identified FAS, AHR, Brd4, SPDEF, IGFBP2, TP53, COLQ and TGFB1 as among the main potential regulators of this distinct collection of proteins (Figure 4). Most interactors were known regulators of breast cancer. For instance, IGFBP2 has a pleitropic influence on tissue regulation and elevated levels are associated with aggressive breast cancer phenotypes.[25] Likewise, aryl hydrocarbon receptor (AhR) is a multipotent xenobiotic interactor that drives tumorigenic processes in advanced breast cancer.[26] Additionally, Brd4 is associated with drug resistant HER2+/ErbB2+ breast cancer types.[27] On the other hand, we were unable to find an association with breast cancer for COLQ (Collagen Like Tail Subunit of Asymmetric Acetylcholinesterase). Therefore, COLase3 proteomics on single FFPE tissue sections both accurately reports tissue regulators of breast cancer and may also predict new topics for breast cancer research. This suggests that COLase3 proteomics on larger cohorts may yield information on common regulators of specific breast cancer subtypes.
Figure 4.
Network interactions of proteins identified by LC-MS/MS after collagenase type III digestion (gray circles), including predicted upstream regulators (blue circles). Abbreviations: Collagen alpha-1(I) chain, COL1A1; Collagen alpha-2(I) chain, COL1A2; Collagen alpha-1(III) chain, COL3A1; Collagen alpha-2(IV) chain, COL4A2;Collagen alpha-1(V) chain, COL5A1; Collagen alpha-2(V) chain, COL5A2; Collagen alpha-1(VI) chain,COL6A1; Collagen alpha-2(VI) chain, COL6A2; Collagen alpha-3(VI) chain, COL6A3; Collagen alpha-1(XII) chain, COL12A1; Collagen alpha-1(XIV) chain, COL14A1; Collagen alpha-1(XVI) chain, COL16A1; Asporin, ASPN; Biglycan. BGN; Decorin, DCN; Dermatopontin, DPT; Elastin microfibrillar interacting protein 1, EMILIN1; Alpha-enolase, ENO1; Fibrinogen alpha chain, FGA; Fibrinogen gamma chain, FGG; Isoform 14 of Fibronectin, FN1; Gelsolin, GSN;Galectin-3, LGALS3; Prelamin-A/C, LMNA; Lumican, LUM; Microfibril-associated glycoprotein 4, MFAP4;Protein disulfide-isomerase, P4HB;Phosphatidylethanolamine-binding protein 1,PEBP1; Isoform 4 of Plectin, PLEC; Isoform 3 of Periostin, POSTN; Peroxiredoxin-5, mitochondrial, PRDX5; Prolargin, PRELP; Alpha-1-antitrypsin, SERPINA1;Transforming growth factor-beta-induced protein, TGFBI;Talin-1, TLN1;Tenascin-X, TNXB; Vimentin, VIM; X-ray repair cr,s-complementing protein 5, XRCC5.
Table 4.
Functional annotations associated with the proteome.
| Functional Annotation | p-value | Molecules |
|---|---|---|
| Cancer of secretory structure | 1.56E-06 | ASPN, BGN, COL12A1, COL14A1, COL1A1, COL1A2, COL3A1, COL4A2, COL5A1, COL5A2, COL6A1, COL6A2, COL6A3, EMILIN1, ENO1, FGA, FN1, GSN, HSPG2, LGALS3, LUM, PLEC, PRDX5, SERPINA1, TGFBI, VIM, XRCC5 |
| Stage I-II cancer | 7.06E-04 | COL1A2, FGG, FN1 |
| Incidence of malignant tumor | 1.05E-03 | DCN, LGALS3, TGFBI, XRCC5 |
| Invasive breast carcinoma | 1.06E-03 | BGN, COL1A2, FN1, POSTN |
| Metastasis | 5.92E-05 | COL4A2, COL6A3, FGG, FN1, LGALS3, PLEC, SERPINA1, TLN1, VIM |
Identified Peptide Expression Patterns by IMS
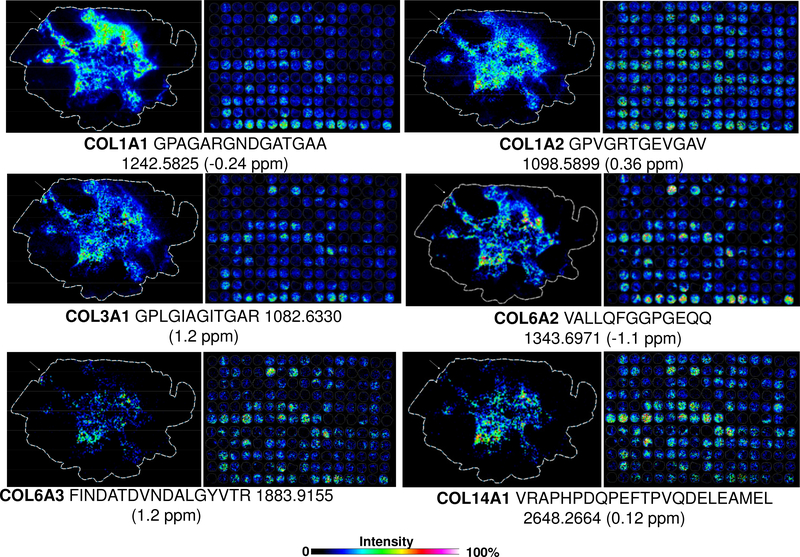
Peptides identified by HRAM LC-based proteomics were mapped by high mass accuracy on the TMA and lumpectomy data (Supplemental Table 4). There were some challenges with matching between electrospray ionization LC-based proteomics and MALDI IMS, likely due to differences in ionization. Figure 5 shows examples of specific collagen peptides mapped across the lumpectomy and the TMA. Many collagen peptides showed higher expression in NAT, again, corresponding to the stromal proteins regulating tumor status via the tissue surrounding the tumor.[7, 23] Many had partial expression in the distal TACS-1 type tumor tissue near surgical boundaries, but appeared with altered expression in the TMA. An exception was COL14A1, which had higher intensity in the mixed invasive tumor cores by TMA and appeared in the main tumor near the TACS-2 signatures in the lumpectomy. This expression pattern follows a previous report that collagen XIV is highly expressed in metastatic breast tissues by quantitative proteomic studies.[28] Interestingly, one of the two peptides previously used for MRM by Goto et al[28] partially matched with a COLase3 peptide ITGPPTELITSEVTARS (underscore highlights sequence homologous to the tryptic peptide found by Goto et al[28]), validating the ECM method and providing independent evidence of the role of collagen type XIV in advanced breast cancer. Further work is being done to determine the link between heterogeneous expression patterns shown on current TMA, along with additional TMAs that have highly defined patient populations. Overall, the ECM method illustrates targeted multiplexed targeted imaging of collagen sequences relevant to breast pathologies.
Figure 5.
Identified collagen peptides linked to the lumpectomy and tissue microarray by high mass accuracy. White arrow indicates distal region associated as tumor tissue by imaging mass spectrometry, situated between normal adjacent to tumor tissue and primary tumor. Abbreviations: Collagen alpha-1(I) chain, COL1A1; Collagen alpha-2(I) chain, COL1A2; Collagen alpha-1(III) chain, COL3A1; Collagen alpha-2(VI) chain, COL6A2; Collagen alpha-3(VI) chain, COL6A3; Collagen alpha-1(XIV) chain, COL14A1.
Conclusion
In the current study, we demonstrated a novel IMS method to access stromal proteins of breast cancer progression. The use of collagenase type III to access ECM peptides produced several thousand peaks from both TMA and breast tissue biopsies. Heuristic analysis of the peaks depicted progressive breast cancer processes and reported a heterogeneous stromal microenvironment associated with breast cancer. HRAM LC-MS/MS proteomics on COLase3 peptides reported known functions and regulators of breast cancer. Specific peptides linked to COL14A1 were found to have higher levels in invasive type tumors, which correlated closely with previous reports. Larger cohorts with well-defined patient data are needed to assess if the method can distinguish a stromal signature based on receptor status. Significant work remains in targeted identification of COLase3 peptides for informative image distribution linked to subtype status. Post translationally modified peptides are of interest and work is being done to enhance detection of these peptides. Continual efforts query different breast cancer subtypes using COLase3 to expand databases of cancer relevant stromal proteins. This work demonstrates that ECM IMS is a valid technique capable of accurately probing the breast stroma microenvironment. Integration of this method with current microscopy techniques will enhance our understanding of how stromal proteins regulate tumor expansion. This may lead to new therapeutic avenues for resistant and aggressive breast cancers.
Supplementary Material
Statement of Clinical Relevance.
Stromal proteins, which are primarily extracellular matrix (ECM) proteins, play a significant role in regulating all types of breast cancer progression, but tools that can report molecular details of ECM proteins are limited. We examine a new imaging mass spectrometry method that targets and reports ECM proteins using a commercially produced tissue microarray (TMA) containing 176 patient samples of breast tissue. Breast pathologies include benign disease such as inflamed breast tissue, fibroadenoma, and non-invasive and invasive breast cancer. This new approach provides an approach to understand ECM protein regulation in breast pathologies.
Acknowledgements
The authors appreciate the discussion on LC-MS/MS proteomic topics with Jennifer Bethard. PMA is supported in part by the National Institute of General Medical Sciences (P20 GM103542), National Cancer Institute (1R21CA207779), and by pilot research funding from an American Cancer Society Institutional Research Grant awarded to the Hollings Cancer Center, Medical University of South Carolina. Additional support was provided by the South Carolina Centers of Economic Excellence SmartState program to RRD. The Orbitrap Elite used in this research was funded by S10 D010731 to LEB. Microscopy data supported by the Cell & Molecular Imaging Shared Resource, Hollings Cancer Center, Medical University of South Carolina (P30 CA138313) and the Shared Instrumentation Grant S10 OD018113. The MUSC Mass Spectrometry Facility and SCW are supported by the Office of the Provost and the South Carolina COBRE in Oxidants, Redox Balance and Stress Signaling (GM 103542). There are no Conflicts of Interest.
Abbreviations
- ECM
extracellular matrix protein
- PTM
post-translational modification
- MALDI IMS matrix
assisted laser desorption /ionization imaging mass spectrometry
- HRAM
high resolution accurate mass
References
- [1].Conklin MW, Keely PJ, Cell Adhesion & Migration 2012, 6, 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Boyd NF, Dite GS, Stone J, Gunasekara A, English DR, McCredie MRE, Giles GG, Tritchler D, Chiarelli A, Yaffe MJ, New England Journal of Medicine 2002, 347, 886; T. Li, L. Sun, N. Miller, T. Nicklee, J. Woo, L. Hulse-Smith, M.-S. Tsao, R. Khokha, L. Martin, N. Boyd, Cancer Epidemiology and Prevention Biomarkers 2005, 14, 343. [DOI] [PubMed] [Google Scholar]
- [3].Quail DF, Joyce JA, Nature Medicine 2013, 19, 1423; S. Ingthorsson, B. Hilmarsdottir, J. Kricker, M. K. Magnusson, T. Gudjonsson, Current Molecular Biology Reports 2015, 1, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Park CC, Rembert J, Chew K, Moore D, Kerlikowske K, International Journal of Radiation Oncology 2009, 73, 75; Y.-S. Huang, J. L.-Y. Chen, C.-S. Huang, S.-H. Kuo, F.-S. Jaw, Y.-H. Tseng, W.-C. Ko, Y.-C. Chang, Breast Cancer Research 2016, 18, 120. [DOI] [PubMed] [Google Scholar]
- [5].Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, Cell 2009, 139, 891; D. M. Gilkes, P. Chaturvedi, S. Bajpai, C. C. Wong, H. Wei, S. Pitcairn, M. E. Hubbi, D. Wirtz, G. L. Semenza, Cancer Research 2013, 73, 3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Conklin MW, Eickhoff JC, Riching KM, Pehlke CA, Eliceiri KW, Provenzano PP, Friedl A, Keely PJ, The American Journal of Pathology 2011, 178, 1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fang M, Yuan J, Peng C, Li Y, Tumor Biology 2014, 35, 2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Giussani M, Merlino G, Cappelletti V, Tagliabue E, Daidone MG, 2015, 35, 3. [DOI] [PubMed] [Google Scholar]
- [9].Puchtler H, Waldrop FS, Valentine LS, zur Pathologie Beiträge 1973, 150, 174; L. C. U. Junqueira, G. Bignolas, R. R. Brentani, The Histochemical Journal 1979, 11, 447; P. Whittaker, R. A. Kloner, D. R. Boughner, J. G. Pickering, Basic Research in Cardiology 1994, 89, 397. [DOI] [PubMed] [Google Scholar]
- [10].Campagnola PJ, Lewis A, Loew LM, Biophysical Journal 1999, 77, 3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Drifka CR, Loeffler AG, Mathewson K, Mehta G, Keikhosravi A, Liu Y, Lemancik S, Ricke WA, Weber SM, Kao WJ, Journal of Histochemistry & Cytochemistry 2016, 64, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Angel PM, Caprioli RM, Biochemistry 2013, 52, 3818; J. L. Norris, R. M. Caprioli, Chemical Reviews 2013, 113, 2309; R. R. Drake, T. W. Powers, E. E. Jones, E. Bruner, A. S. Mehta, P. M. Angel, Advances in Cancer Research 2017, 134, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dekker TJA, Balluff BD, Jones EA, Schöne C. d. D., Schmitt M, Aubele M, Kroep JR, Smit VT, Tollenaar RAEM, Mesker WE, Journal of proteome research 2014, 13, 4730. [DOI] [PubMed] [Google Scholar]
- [14].Gemoll T, Strohkamp S, Schillo K, Thorns C, Habermann JK, Oncotarget 2015; C. Marquardt, T. Tolstik, C. Bielecki, R. Kaufmann, A. C. Crecelius, U. S. Schubert, U. Settmacher, A. Stallmach, O. Dirsch, Zeitschrift für Gastroenterologie 2015, 53, 33; M. Kriegsmann, R. Casadonte, J. Kriegsmann, H. Dienemann, P. Schirmacher, J. H. Kobarg, K. Schwamborn, A. Stenzinger, A. Warth, W. Weichert, Molecular & Cellular Proteomics 2016, 15, 3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Groseclose MR, Andersson M, Hardesty WM, Caprioli RM, Journal of Mass Spectrometry 2007, 42, 254. [DOI] [PubMed] [Google Scholar]
- [16].Casadonte R, Kriegsmann M, Zweynert F, Friedrich K, Bretton G, Otto M, Deininger SO, Paape R, Belau E, Suckau D, Proteomics 2014, 14, 956. [DOI] [PubMed] [Google Scholar]
- [17].Sanders ME, Dias EC, Xu BJ, Mobley JA, Billheimer D, Roder H, Grigorieva J, Dowsett M, Arteaga CL, Caprioli RM, Journal of Proteome Research 2008, 7, 1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Seeley EH, Caprioli RM, Proceedings of the National Academy of Sciences 2008, 105, 18126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Angel PM, Comte-Walters S, Ball LE, Talbot K, Brockbank KGM, Mehta AS, Drake RR, Journal of Proteome Research 2017, doi: 10.1021/acs.jproteome.7b00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Toyoshima T, Matsushita O, Minami J, Nishi N, Okabe A, Itano T, Connective Tissue Research 2001, 42, 281. [DOI] [PubMed] [Google Scholar]
- [21].Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J, Methods in Enzymology 2006, 411, 134. [DOI] [PubMed] [Google Scholar]
- [22].Deininger S. r.-O., Ebert MP, Futterer A, Gerhard M, Röcken C, Journal of Proteome Research 2008, 7, 5230; T. Alexandrov, Bmc Bioinformatics 2012, 13, S11; H. Thiele, S. Heldmann, D. Trede, J. Strehlow, S. Wirtz, W. Dreher, J. Berger, J. Oetjen, J. H. Kobarg, B. Fischer, Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics 2014, 1844, 117. [DOI] [PubMed] [Google Scholar]
- [23].Golaraei A, Kontenis L, Cisek R, Tokarz D, Done SJ, Wilson BC, Barzda V, Biomedical Optics Express 2016, 7, 4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Scott MS, Calafell SJ, Thomas DY, Hallett MT, PLoS Computational Biology 2005, 1, e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dean SJR, Perks CM, Holly JMP, Bhoo-Pathy N, Looi L-M, Mohammed NAT, Mun K-S, Teo S-H, Koobotse MO, Yip C-H, American Journal of Clinical Pathology 2014, 141, 323; S. W. Yau, W. J. Azar, M. A. Sabin, G. A. Werther, V. C. Russo, Journal of Cell Communication and Signaling 2015, 9, 125. [DOI] [PubMed] [Google Scholar]
- [26].Thompson PA, Khatami M, Baglole CJ, Sun J, Harris SA, Moon E-Y, Al-Mulla F, Al-Temaimi R, Brown DG, Colacci AM, Carcinogenesis 2015, 36, S232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zambrano JN, Neely BA, Yeh ES, Pharmacological research 2017, 119, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Goto R, Nakamura Y, Takami T, Sanke T, Tozuka Z, PloS One 2015, 10, e0130760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.