SUMMARY
Tumors display reprogrammed metabolic activities that promote cancer progression. We currently possess a limited understanding of the processes governing tumor metabolism in vivo and of the most efficient approaches to identify metabolic vulnerabilities susceptible to therapeutic targeting. While much of the literature focuses on stereotyped, cell-autonomous pathways like glycolysis, recent work emphasizes heterogeneity and flexibility of metabolism between tumors and even within distinct regions of solid tumors. Metabolic heterogeneity is important because it influences therapeutic vulnerabilities and may predict clinical outcomes. This Review describes current concepts about metabolic regulation in tumors, focusing on processes intrinsic to cancer cells and on factors imposed upon cancer cells by the tumor microenvironment. We discuss experimental approaches to identify subtype-selective metabolic vulnerabilities in preclinical cancer models. Finally, we describe efforts to characterize metabolism in primary human tumors, which should produce new insights into metabolic heterogeneity in the context of clinically-relevant microenvironments.
INTRODUCTION
The field of cancer metabolism was founded on the principle that tumors share homogenous metabolic features that distinguish them from non-malignant tissues. Early studies by Otto Warburg in the 1920s quantified nutrient and gas exchanges in primary tissues and culminated in his view that a marked, irreversible suppression of respiration was not only a generalized characteristic of tumor metabolism, but held the key to cancer therapy (Warburg, 1956). The kernel of this view was Warburg’s observation that tumor tissue and cancer cells convert glucose to lactate under aerobic conditions, rather than oxidizing the glucose to CO2. Controversy ensued about the extent of respiratory impairment in tumors, with many lines of evidence arguing against permanent damage to respiratory mechanisms as a general feature of cancer (for an excellent discussion of this history, see (Koppenol et al., 2011)). Nevertheless, the concept of the Warburg effect (i.e. excessive aerobic glycolysis) as a hallmark of malignancy persists and is supported by the observation that oncogene expression is sufficient to stimulate various aspects of glycolysis in cultured cells (Elstrom et al., 2004; Flier et al., 1987; Shim et al., 1997).
It is increasingly clear that cancer cells and tumors have heterogeneous metabolic preferences and dependencies, and the recent application of advanced technologies including metabolomics, metabolic flux analysis and functional genomics supports this view. Metabolic phenotypes in tumors are both heterogeneous and flexible, and they result from the combined effects of many different factors, some intrinsic to the cancer cell (e.g., cell lineage; differentiation state; somatically-acquired mutations) and others imposed by the microenvironment (e.g. nutrient milieu; interactions with extracellular matrix and stromal cells). Understanding how metabolic phenotypes emerge and evolve will demand that we understand the influence of these factors and their relative impact on the tumor.
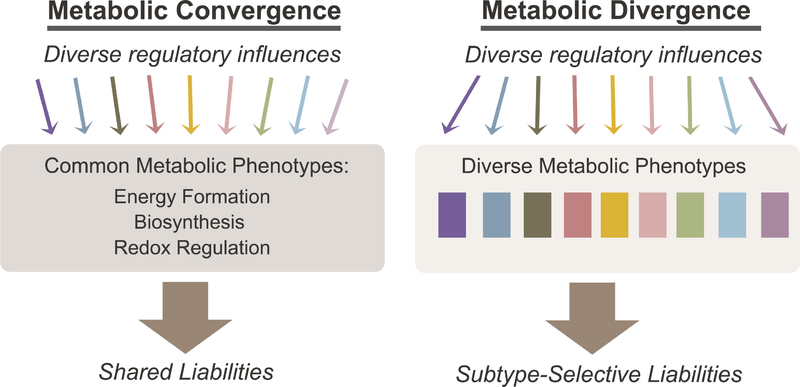
Cancer metabolism consists of both convergent and divergent metabolic phenotypes, and it is useful to consider what these properties can tell us about the mechanistic basis of metabolic reprogramming and implications for therapy (Figure 1). Convergent properties are stimulated by any of a large number of factors and thus are shared among diverse tumor types. These properties include widely studied pathways under oncogenic control, such as the enhanced ability to generate energy, synthesize macromolecules and maintain redox homeostasis (DeBerardinis and Chandel, 2016). Some convergent properties are considered hallmarks of malignancy (Hanahan and Weinberg, 2011). Glycolysis is a prime example of a convergent property, because it can be activated by many different oncogenic drivers or by hypoxia (Elstrom et al., 2004; Flier et al., 1987; Shim et al., 1997). Divergent properties occur when distinct factors stimulate heterogeneous pathways, leading to idiosyncratic or subtype-selective phenotypes. Examples of divergent pathways include (R)-2HG accumulation in tumors with IDH1 or IDH2 mutations, or metabolic preferences that differentiate tumors with distinct oncogenotypes (Dang et al., 2009; Shackelford et al., 2013; Ward et al., 2010). Interest in divergent pathways has risen markedly over the past decade, increasing our appreciation of metabolic heterogeneity in cancer (Boroughs and DeBerardinis, 2015). Understanding metabolic heterogeneity is important because it influences how we think about exploiting metabolic reprogramming to treat cancer. Therapies against convergent vs. divergent properties would have distinct challenges. In principle, treatments targeting convergent phenotypes would have broad utility, but in practice these core pathways are usually shared by non-malignant tissues, possibly narrowing the therapeutic index. In contrast divergent properties are by definition confined to subsets of cancers, and as a result targeting them may result in more acceptable toxicity profiles, although efficacy would be limited to distinct subsets.
Figure 1. Convergent and divergent metabolic properties in cancer.
Convergent metabolic properties (left) arise downstream of diverse regulatory influences. Convergent properties are observed frequently throughout cancer models and include core pathways such as those that allow cells to produce energy, build macromolecules and maintain redox balance. Divergent properties (right), in contrast, appear in distinct molecular subsets of cancer and contribute to metabolic heterogeneity. Convergent and divergent metabolic phenotypes may both give rise to metabolic liabilities, although the generality of these liabilities is predicted to differ according to the class.
This review considers how heterogeneous metabolic phenotypes arise in tumors. It also discusses the implications of metabolic heterogeneity on therapeutic vulnerabilities in cancer, and how new approaches are improving our ability to study metabolic heterogeneity in disease-relevant contexts.
METABOLIC HETEROGENEITY INDUCED BY GENETIC ALTERATIONS
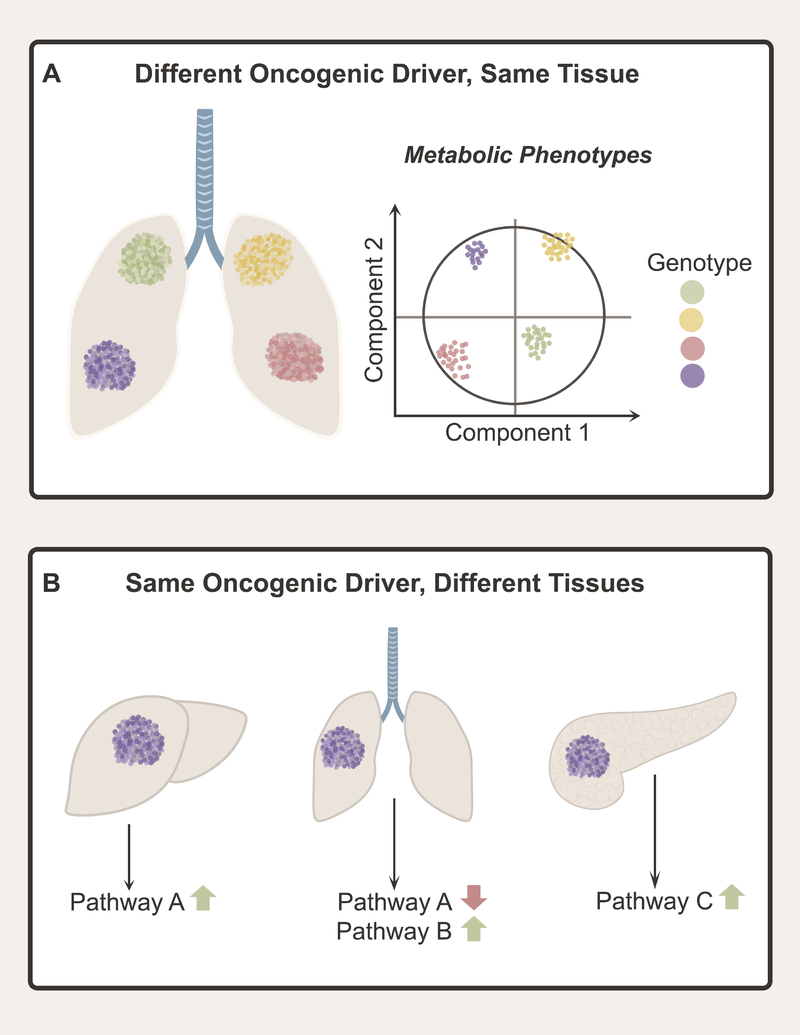
As analysis of cancer genetics and metabolism merged over the past two decades, it became clear that mutations in oncogenes and tumor suppressors can stimulate cell-autonomous metabolic reprogramming. Human lung adenocarcinoma provides an example of the prevalence of mutations that can perturb metabolism (Cancer Genome Atlas Research, 2014). In these tumors, all seven of the genes with the highest frequency of somatically-acquired mutations, TP53, KRAS, KEAP1, STK11, EGFR, NF1 and BRAF, have been reported to regulate metabolism either directly or indirectly. These mutations contribute to metabolic heterogeneity because divergent, cell-autonomous metabolic effects have been reported for specific mutations, such as the predilection of KEAP1-mutant tumors to resist oxidative stress and require glutamine catabolism (Romero et al., 2017). Thus, the oncogenotype can drive particular metabolic features in tumors arising in the same tissue (Figure 2A).
Figure 2. Oncogenic drivers and tissue of origin influence cancer metabolism.
(A) Different oncogenic drivers can produce divergent metabolic features, contributing to metabolic heterogeneity among tumors arising in the same tissue. (B) Cell and tissue of origin also contribute to metabolic heterogeneity. Different tissues display different native metabolic programs, and some aspects of these programs are retained in tumors arising in the tissue. As a result, tumors arising in different tissues may display markedly divergent metabolic phenotypes even if they contain the same oncogenic driver.
Much of what we know about how the oncogenotype reprograms metabolism comes from isogenic models containing or lacking a mutation of interest. This approach isolates the metabolic impact of mutations one at a time and provides a basis for genotype-phenotype relationships in cancer metabolism. But the extensive molecular heterogeneity of human tumors may obscure the effects of mutations in any one gene, limiting the ability of isogenic models to uncover metabolic features that will translate to clinical studies. Most human tumors acquire hundreds of somatic mutations in coding regions (Martincorena and Campbell, 2015), many of which are predicted to impact metabolism. Studies in isogenic systems do not test whether mutations in single genes elicit metabolic effects robust enough to be detected across genetically-diverse tumors.
A few studies have begun to profile many tumors or cell lines derived from the same type of cancer to test whether single-gene effects drive recognizable metabolic features. A large study of metabolic heterogeneity in patient-derived melanoma xenografts concluded that every tumor had a unique metabolomic signature (Shi et al., 2017). Strikingly, each of over 180 tumor fragments analyzed in the study was most related to all other fragments descended from the same human tumor, even when the fragments arose from tumors in different mice. Oncogenic BRAF, a common driver mutation with clear metabolic effects in isogenic studies, was not sufficient to produce a recognizable metabolomic fingerprint in this genetically-diverse tumor panel. BRAF-mutant tumors did, however, have consistent features in more focused metabolic analyses, such as a high turnover of glycolytic intermediates during infusion with 13C-glucose.
Profiling panels of cancer cell lines can identify metabolic alterations referable to one mutation or a combination of mutations. Dependence on de novo serine biosynthesis can be directed by genomic amplification of the gene encoding phosphoglycerate dehydrogenase (PHGDH) (Locasale et al., 2011; Possemato et al., 2011). Unbiased profiling in several dozen lung cancer cell lines revealed that activation of the NRF2 transcriptional program, through KEAP1 loss and other mechanisms, also regulates activity and dependence on this pathway (DeNicola et al., 2015). Pancreatic ductal adenocarcinoma is dominated by oncogenic KRAS mutations, but metabolic profiling in cell lines revealed distinct metabolic subclasses correlating with expression of mesenchymal markers and sensitivity to specific metabolic inhibitors (Daemen et al., 2015). In small cell lung cancer (SCLC), analysis of 25 genetically-distinct cell lines revealed a metabolomic signature that distinguished cell lines containing or lacking the lineage oncoprotein ASCL1 (Huang et al., 2018). In this study, most ASCL1-low cell lines had high levels of MYC and expressed the guanosine biosynthetic enzymes inosine monophosphate dehydrogenase-1 and −2 (IMPDH1 and IMPDH2), both of which are MYC transcriptional targets. MYC expression was sufficient to enhance guanosine synthesis and to render SCLC cells sensitive to IMPDH inhibitors in culture and in vivo (Huang et al., 2018). An even broader study profiling over 900 cell lines from the Cancer Cell Line Encyclopedia revealed remarkable metabolic heterogeneity and uncovered novel associations among metabolites, gene expression signatures and therapeutic sensitivities, including hypermethylation of the gene encoding asparagine synthetase (ASNS) and sensitivity to L-asparaginase (Li et al., 2019). Altogether these studies show that even in cancer cell lines, divergent metabolic properties can point towards therapeutic vulnerabilities.
To fully understand the genetic basis of metabolic heterogeneity in cancer, we also need to consider the combinatorial effects of coincident mutations, because these combinations can change the clinical features of the tumor. Several studies have begun to do this, particularly in non-small cell lung cancer (NSCLC) where the large number of cell lines and mouse models make it feasible to detect combinatorial effects. Oncogenic KRAS has many effects on metabolism, but KRAS-mutant tumors contain other mutations that also modify metabolism. For example, KRAS stimulates macropinocytosis, which allows cancer cells to scavenge extracellular protein as a source of nutrition (Commisso et al., 2013). Other signaling pathways modify the ability of macropinocytosis to promote cell growth, with activation of Akt and mTORC1 addicting cells to free amino acids and impairing growth on extracellular protein (Palm et al., 2017; Palm et al., 2015). NSCLC cells with concurrent mutations in KRAS and STK11 develop a metabolomic signature of altered nitrogen metabolism distinct from cells containing oncogenic KRAS alone. The metabolic response to STK11 loss in the context of oncogenic KRAS includes addiction to the urea cycle enzyme carbamoyl-phosphate synthase-1 (CPS1) (Kim et al., 2017). Although STK11 loss is sufficient to drive CPS1 expression, only cells with concomitant mutations in STK11 and KRAS require CPS1 for survival. CPS1 dependence is related to its ability to sustain pools of pyrimidine nucleotides, which comprise a node of metabolic vulnerability in cells with this combination of mutations (Kim et al., 2017; Liu et al., 2013).
Genomic deletions also contribute to metabolic heterogeneity. These effects include bystander deletions of genes encoding metabolic enzymes that neighbor tumor suppressor genes. Homozygous deletion of CDKN2A, a tumor suppressor on human chromosome 9p21, usually involves co-deletion of the gene encoding methylthioadenosine phosphorylase (MTAP). Loss of MTAP results in elevated levels of methylthioadenosine (MTA), which partially inhibits the arginine methyltransferase PRMT5 and sensitizes cells to small-molecule PRMT5 inhibitors (Kryukov et al., 2016; Marjon et al., 2016). Thus, passenger deletion of a metabolic enzyme results in a therapeutic liability that can be exploited in pre-clinical cancer models.
EFFECT OF CELL AND TISSUE OF ORIGIN ON METABOLIC HETEROGENEITY
In mammals, homeostasis requires dispersion of metabolic functions among the organs. During fasting and catabolic stress, the liver disposes of excess nitrogen through the urea cycle and produces glucose and ketone bodies for use by other organs. Muscle, adipose tissue, and lungs synthesize and release glutamine, while the kidney catabolizes it to generate ammonia for acid-base balance and carbon for gluconeogenesis (Hensley et al., 2013; Stumvoll et al., 1999). Germline mutations in enzymes from these pathways alter homeostasis and result in systemic diseases. Expression profiling indicates that tumors tend to retain the metabolic network of the parental tissue, in part through tissue-specific epigenetic regulation (Gaude and Frezza, 2016; Hu et al., 2013). This may explain the finding that oncogenic drivers induce different metabolic phenotypes in tumors arising in different organs (Figure 2B). In mice, MYC stimulates glutamine catabolism in liver tumors but glutamine synthesis in the lung (Yuneva et al., 2012). Similarly, oncogenic KRAS has different effects on branched-amino acid (BCAA) metabolism in lung and pancreatic tumors (Mayers et al., 2016).
It follows that tumors located in the same tissue but derived from different cells of origin or acquiring distinct molecular and histological features would also display different metabolic properties. This appears to be the case in some human cancers. Gene expression signatures delineate clinically distinct subtypes of breast cancer (Perou et al., 2000), and these subtypes exhibit metabolic differences defined by lineage-specific gene expression patterns. Basal breast tumors express low levels of glutamine synthetase (glutamate-ammonia ligase, GLUL), and cells derived from these tumors are dependent on an exogenous glutamine supply. In contrast, luminal tumor cells express GLUL and resist glutamine deprivation in culture (Kung et al., 2011). Cell lines derived from triple-negative breast cancers, particularly basal and claudin-low tumors, consume glutamine to drive cystine import through the xCT glutamate-cystine antiporter. Withdrawing glutamine or inhibiting xCT suppresses growth of this subtype (Timmerman et al., 2013). Different subtypes of non-small cell lung cancer also exhibit distinct metabolic phenotypes and liabilities. Compared to human lung adenocarcinomas, squamous cell tumors express higher levels of the glucose transporter GLUT1, have higher uptake of the glucose analog FDG on clinical PET imaging, and display sensitivity to glucose uptake inhibitors in xenograft models (Goodwin et al., 2017).
EPIGENETIC REGULATION OF METABOLIC HETEROGENEITY
Many aspects of tumor heterogeneity result from epigenetic rather than genomic alterations. Epigenetic control of gene expression is fine-tuned by a balance between enzymes that “write” regulatory marks onto DNA and histones (e.g. DNA- and histone methyltransferases and histone acetyltransferases) and enzymes that “erase” these marks (e.g. histone-deacetylases, histone- and DNA-demethylases)(Allis and Jenuwein, 2016). Epigenetics contributes to cell fate determination and is a major driver of functional heterogeneity in mammalian tissues. Within tumors, cellular and clonal heterogeneity is partially determined by epigenetically-defined hierarchies. During oncogenesis, environmental stress including ROS accumulation can promote clonal expansion of cells with epigenetic abnormalities (Easwaran et al., 2014). Further alterations confer tumor-initiating capacity to a subset of these cells, which need to acquire metabolic properties consistent with long-term survival in the tumor microenvironment (Jones and Baylin, 2002; Machida, 2017).
Because metabolic intermediates serve as cofactors and substrates for epigenetic writers and erasers, the metabolic state influences epigenetics (Figure 3) (Kaelin and McKnight, 2013). DNA methyltransferases (DNMTs) catalyze the covalent addition of a methyl group to cytosine to form 5-methylcytosine, with highly methylated CpG islands generally associated with transcriptional repression (Edwards et al., 2017). Widespread DNA hypomethylation is a prominent feature of cancer genomes (Kulis and Esteller, 2010), while hypermethylation of regulatory regions is a mechanism to inactivate expression of tumor suppressor genes such as BRCA1 and RB (Esteller, 2002; Herman, 1999). Histone methyltransferases (HMTs) catalyze transfer of methyl groups to lysine and arginine in histone proteins (Hyun et al., 2017), resulting in either activation or repression of gene expression depending on the mark. Both DNMTs and HMTs utilize S-adenosylmethionine (SAM), an intermediate of the methionine cycle, as a methyl donor (Figure 3) (Ulrey et al., 2005). After donating its methyl group, SAM becomes S-adenosyl-homocysteine (SAH), which inhibits DNMTs and HMTs. Thus, alterations in the SAM/SAH ratio regulate methyltransferase activity (Williams and Schalinske, 2007). In cancer, enhanced flow through the methionine cycle produces an excess of SAM and contributes to hypermethylation of DNA and histones, and inappropriate gene silencing (Maddocks et al., 2016; Ulanovskaya et al., 2013).
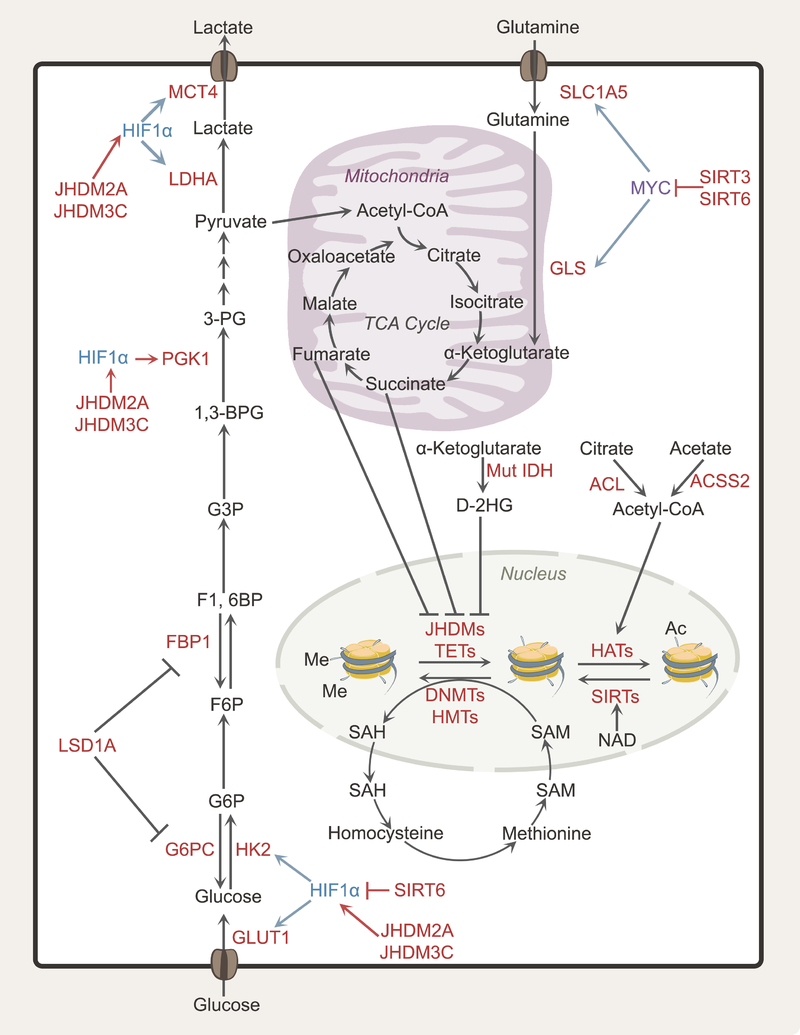
Figure 3. Epigenetics and metabolism in cancer cells.
Epigenetic regulation of gene expression contributes to metabolic heterogeneity because many metabolic enzymes and nutrient transporters are regulated by epigenetic modifications of histones (acetylation, methylation) and DNA (methylation). Conversely, these epigenetic modifications respond to the metabolic state of the cell. The relative abundances of SAM and SAH regulate DNA and histone methyltransferases, while the abundance of acetyl-CoA, the ratio of acetyl-CoA to free CoA and NAD levels can regulate histone acetylation. The TCA cycle intermediate α-KG affects demethylation of histones and DNA by acting as a co-substrate for JHDM histone demethylases and TET-family methylcytosine dioxygenases, respectively. The oncometabolites fumarate, succinate and 2-HG are dicarboxylates that compete with α-KG, interfering with TET/JHDM function. Abbreviations: HK2, hexokinase 2; MCT4, monocarboxylate transporter 4; SAM, S-adenosylmethionine; SAH, S-adenosyl-homocysteine; NAD, nicotinamide adenine dinucleotide; α-KG, alpha-ketoglutarate; 2-HG, 2-hydroxyglutarate; TET, 10–11 translocation enzyme; JHDM, Jumonji C domain-containing histone demethylase.
TCA cycle intermediates also influence the epigenetic landscape. DNA and histone demethylation is catalyzed by 10–11 translocation (TETs) enzymes and Jumonji C domain-containing histone demethylases (JHDMs), respectively; these enzymes are dioxygenases that require α-ketoglutarate (α-KG) as a co-substrate (Zhang et al., 2017a). Pathologically high levels of other dicarboxylates compete with α-KG and interfere with these and other dioxygenases (Figure 3) (Laukka et al., 2016; Lu et al., 2012; Smith et al., 2007). Several “oncometabolites,” metabolites that accumulate in a genetically-specified manner in tumors, operate through this mechanism. Fumarate and succinate accumulate in tumors lacking fumarate hydratase (FH) or components of the succinate dehydrogenase (SDH) complex, resulting in hypermethylation of DNA and histones (Xiao et al., 2012). Somatic mutations in isocitrate dehydrogenases-1 and −2 (IDH1 and IDH2) occur in gliomas, acute myeloid leukemias and other cancers (Losman and Kaelin, 2013). These mutant enzymes convert α-KG to (D)-2-hydroxyglutarate (D-2HG), a molecule that inhibits TETs and JHDMs histone- and DNA-modifying enzymes and interferes with expression of genes involved in differentiation (Lu et al., 2012). Some tumors accumulate L-2HG as a consequence of silencing of L-2HG dehydrogenase (L-2HGDH), the enzyme that keeps L-2HG levels low by converting it to α-KG. L-2HGDH expression is suppressed in clear cell renal cell carcinoma relative to adjacent kidney, and this is thought to contribute to altered epigenetics in these tumors (Shim et al., 2014). L-2HG also accumulates during hypoxia and impaired electron transport chain function, suggesting that this metabolite might commonly impact epigenetics in solid tumors (Mullen et al., 2014; Wise et al., 2011).
Many metabolic genes are under epigenetic control (Figure 3). Promoter hypomethylation in hexokinase 2 (HK2) in hepatocellular carcinoma and glioblastoma enhances its expression and confers heightened glycolytic flux. (Goel et al., 2003; Wolf et al., 2011). Fructose 1,6-bisphosphatase 1 (FBP1), a rate-limiting enzyme in gluconeogenesis, is silenced by promoter hypermethylation, inducing higher glycolytic rates in gastric, colon, liver and basal-like breast cancers (Chen et al., 2011; Dong et al., 2013). In clear cell renal cell carcinoma (ccRCC), re-expression of FBP1 suppresses tumor growth through both metabolic and non-metabolic effects, with the latter involving FBP1’s ability to impair HIF-dependent transcriptional activity in the nucleus (Li et al., 2014). In bladder cancer, cooperativity between HIF1α and JHDM2A results in promoter demethylation and enhanced expression of genes encoding transporters and enzymes in glycolysis, including GLUT1, HK2, PGK1, LDHA and MCT4 (Wan et al., 2017). JHDM3C-mediated epigenetic effects drive glycolytic gene expression in breast cancer (Luo et al., 2012). In hepatocellular carcinoma, Lysine Demethylase 1A (LSD1A) inhibits gluconeogenesis by suppressing transcription of glucose-6-phosphatase (G6PC) and FBP1 (Pan et al., 2013) and favors HIF-1α-dependent glycolytic metabolism (Figure 3) (Sakamoto et al., 2015).
Histone acetylation reduces the electronic interaction between histones and negatively charged DNA, thereby favoring DNA accessibility and gene transcription. The acetylation state is determined by the balance between histone acetyltransferases (HATs) and histone deacetylases (HDACs) that catalyze addition and removal of acetyl groups on lysine residues, respectively (Yang and Seto, 2007). Similar to methylation, this process is modulated by metabolic intermediates. HATs transfer the acetyl group from acetyl-CoA to lysine, producing ε-N-acetyl lysine. The abundance of acetyl-CoA and the ratio of acetyl-CoA to coenzyme A can regulate the extent of histone acetylation (Lee et al., 2014). In yeast, exogenously-provided acetate is sufficient to stimulate histone acetylation and growth (Cai et al., 2011). In mammalian cells, acetyl-CoA for histone acetylation is produced by at least two enzymes, acetyl-CoA synthetase 2 (ACSS2) and ATP-citrate lyase (ACL, Figure 3) (Pietrocola et al., 2015; Wellen et al., 2009). Cancer cells often over-express ACL, which contributes to the nuclear acetyl-CoA pool necessary for histone acetylation and gene expression (Zaidi et al., 2012).
Histone deacetylases of the sirtuin family also govern important aspects of metabolism in cancer (Figure 3). SIRT1 modulates lipid and glucose metabolism by deacetylating key metabolic regulators such as PGC1α and SREBP1 as well as the glycolytic enzyme phosphoglycerate mutase-1 (PGAM1) (Chang and Guarente, 2014; Hallows et al., 2012). SIRT3 increases mitochondrial fatty-acid oxidation by long-chain acyl coenzyme A dehydrogenase (LCAD) deacetylation, and tunes mitochondrial superoxide levels by activating manganese superoxide dismutase (MnSOD)(Tao et al., 2010). These enzymes use NAD as a substrate, cleaving it to produce nicotinamide as part of their catalytic mechanism. Tumor suppressor activity has been ascribed to several sirtuin family members via their ability to influence oncogenic transcriptional and metabolic programs. SIRT6 constrains aerobic glycolysis by functioning as a co-repressor of HIF1α and deacetylating H3K9 at the promoters of HIF1α target genes (Sebastian et al., 2012; Zhong et al., 2010). It also acts as a MYC co-repressor and inhibits ribosomal biogenesis and glutamine metabolism (Sebastian et al., 2012). SIRT3 suppresses glucose metabolism by inhibiting HIF1α-dependent transcription (Bell et al., 2011).
These processes are relevant to metabolic heterogeneity because epigenetic modifications are sensitive to changes in metabolite levels. In cultured mouse embryonic stem cells, DNA methylation and H3K27 histone methylation are sensitive to the ratio of α-ketoglutarate to succinate (that is, the substrate and product of α-ketoglutarate-dependent dioxygenases that erase epigenetic marks on histones and DNA)(Carey et al., 2015). These findings raise the question of whether regional metabolic differences within solid tumors bring about regional epigenetic effects. Preclinical data suggest that this may be the case. Glutamine-deprived core regions of melanomas in mice have markedly increased histone H3 methylation relative to glutamine-rich peripheral regions, consistent with suppressed HDM activity in the core. Inhibiting the conversion of glutamine to α-ketoglutarate in the peripheral regions enhances histone methylation (Pan et al., 2016). In these models, glutamine deprivation altered gene expression and resulted in resistance to BRAF inhibitors, suggesting that metabolically-determined epigenetic effects can impair therapeutic responses.
EFFECTS IMPOSED BY NUTRIENT LIMITATION IN THE MICROENVIRONMENT
Many non-cell autonomous factors in the tumor microenvironment (TME) influence metabolism and are themselves heterogeneous. These factors are difficult to replicate in culture, in part because the metabolic milieu of tumors in vivo is still poorly defined. The nutrient-rich composition of conventional culture medium differs markedly from mammalian plasma, with high levels of glucose, glutamine and essential amino acids designed to maximize cell proliferation. The metabolic composition of interstitial fluid from murine pancreatic tumors also differs from the plasma, with further depletion of glucose and some amino acids (Sullivan et al., 2019). Differences in nutrient availability impact metabolic activities and dependencies. Culture media formulations designed to recapitulate human plasma uncover new mechanisms of metabolic regulation and cancer cell growth. One such formulation revealed that physiological levels of uric acid, not usually present in commercial tissue culture medium, reduces pyrimidine synthesis by inhibiting uridine monophosphate synthase (Cantor et al., 2017). Sub-physiological levels of selenium in commercial medium results in oxidative damage and limits colony formation in culture (Vande Voorde et al., 2019). Culturing cancer cells in adult bovine serum reduces dependence on extracellular glutamine relative to conventional medium; this difference is the result of the low abundance of cystine in adult serum and reduced dependence on intracellular glutamate to support xCT-mediated cystine import (Muir et al., 2017). Thus, the nutrient milieu can override the effects of the oncogenotype on cell-intrinsic metabolic preferences.
Oxygen limitation also affects metabolism and can modify the impact of oncogenic drivers. In the presence of oxygen, MYC stimulates a transcriptional program that drives nutrient uptake, glycolysis, nucleotide synthesis and ribosome biogenesis. It induces mitochondrial biogenesis via Transcription Factor A, Mitochondrial (TFAM), which regulates mitochondrial transcription and DNA replication (Li et al., 2005). MYC also stimulates mitochondrial glutamine catabolism by regulating expression of glutamine transporters and glutaminase (Gao et al., 2009; Wise et al., 2008). Like MYC, hypoxia-inducible factor-1 (HIF-1) stimulates expression of glycolytic genes. But HIF-1 reduces mitochondrial metabolism through multiple mechanisms, including by suppressing MYC. HIF-1 activates transcription of MAX interactor 1, dimerization protein (MXI1), a MYC antagonist that attenuates MYC-induced mitochondrial biogenesis, and promotes proteasome-dependent degradation of MYC (Zhang et al., 2007). HIF-1 also inhibits pyruvate oxidation by inducing pyruvate dehydrogenase kinase 1 (PDK1), which phosphorylates and inactivates pyruvate dehydrogenase (Papandreou et al., 2006), and induces mitophagy (Zhang et al., 2008).
HIF-1 and MYC exert regional metabolic control in solid tumors. Cells located close to the blood supply tend to be under MYC influence, generating ATP aerobically and upregulating anabolic pathways to support proliferation (Stine et al., 2015). Cells located further from the blood supply experience lower O2 levels and induce HIF-1 (Gordan et al., 2007). These cells activate catabolic pathways like autophagy to provide energy and biosynthetic precursors. Lactate metabolism is thought to be subject to regional differences in oxygen availability, with hypoxic cancer cells consuming glucose and secreting lactate while cells in better-perfused regions import lactate and use it as a fuel (Sonveaux et al., 2008). These relationships may be clinically relevant, because heterogeneity of hypoxia and glucose uptake in human solid tumors predicts poor outcomes in some studies (Chicklore et al., 2013; Vaupel and Mayer, 2007).
INTERACTIONS WITH MATRIX AND TUMOR STROMA
Interactions between malignant cells and other components of the TME, including non-malignant cells and extracellular matrix (ECM) also contribute to metabolic heterogeneity. The tumor microenvironment contains blood vessels, cancer-associated fibroblasts (CAFs), lymphocytes and other immune cells, signaling molecules and ECM, whose functions and proportions are distinct from normal tissue (Figure 4). The microenvironment is temporally and spatially heterogeneous due to variations in blood flow, nutrient availability and cellular composition. In some contexts, cancer cells exploit the local nutrient milieu provided by non-malignant cells. Ovarian cancer cells metastasize to the omentum, a large abdominal organ rich in adipocytes. Co-culture experiments revealed transfer of fatty acids from adipocytes to ovarian carcinoma cells, stimulating fatty acid oxidation and growth in the cancer cells (Nieman et al., 2011). In pancreatic adenocarcinoma, stroma-associated pancreatic stellate cells secrete alanine, providing carbon and nitrogen for cancer cell proliferation (Sousa et al., 2016). Bone marrow stromal cells provide cysteine to promote glutathione (GSH) synthesis for chronic lymphocytic leukemia (CLL) cells; unlike many cancer cells, CLL cells cannot take up the abundant disulfide cystine, rendering them dependent on cysteine from neighboring stromal cells (Zhang et al., 2012).
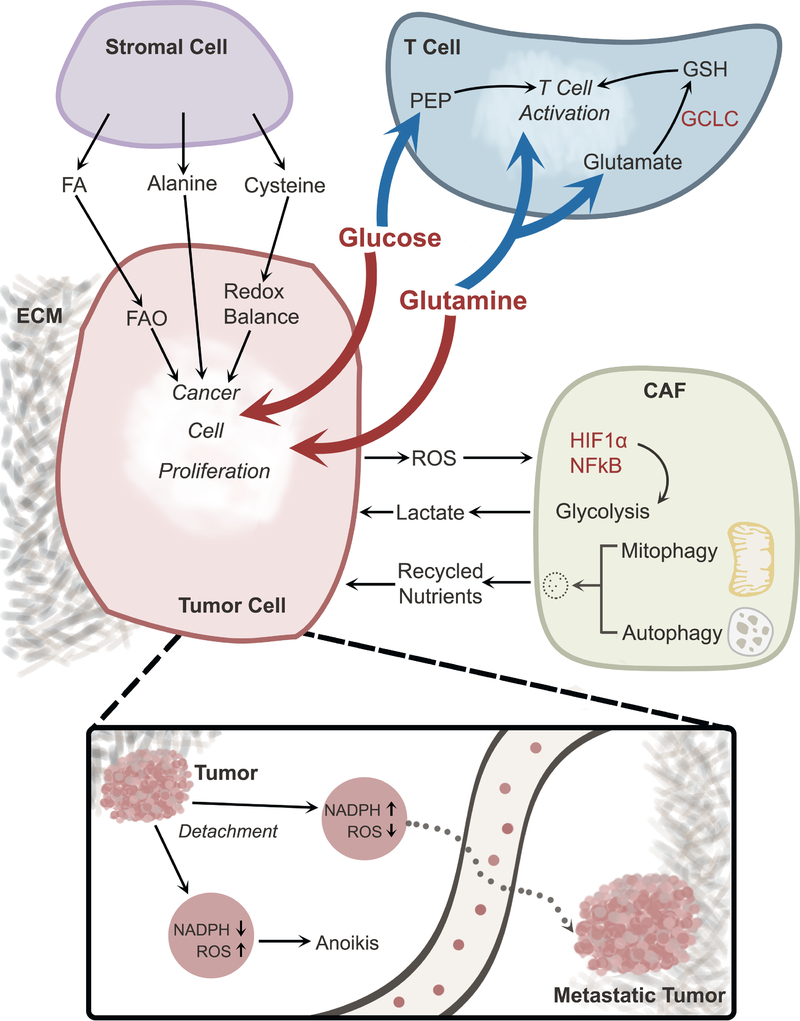
Figure 4. Metabolic cross-talk in the tumor microenvironment.
Interactions among cancer cells and other components of the TME contribute to metabolic heterogeneity. Cancer cells and T cells compete for nutrients (e.g., glucose and glutamine), and excessive consumption of these nutrients by cancer cells suppresses T cell activation. Other cells in the TME also engage in metabolic cross-talk with cancer cells. Stromal cells provide nutrients that support cancer cell proliferation. CAFs respond to oxidative stress imposed by cancer cells by activating HIF1α and NFκB, thereby stimulating glycolysis and secreting lactate, which may be taken up by cancer cells. Degradative processes like mitophagy and autophagy in CAFs also provide nutrients to cancer cells. Loss of ECM attachment suppresses NADPH production, resulting in ROS-mediated anoikis; this form of cell death can be overcome through oncogene-stimulated NADPH production by the pentose phosphate pathway. Reducing oxidative stress allows cells to survive in the detached state and promotes formation of distant metastases. Abbreviations, TME, tumor microenvironment; ECM, extracellular matrix; FA, fatty acid; FAO, fatty acid oxidation; CAF, cancer-associated fibroblast; ROS, reactive oxygen species; PEP, phosphoenolpyruvate; GSH, glutathione, reduced; GCLC, Glutamate-Cysteine Ligase.
CAFs interact extensively with cancer cell metabolism (Figure 4). Elevated ROS production by cancer cells activates HIF-1 and NFκB in CAFs. This stimulates aerobic glycolysis in CAFs, inducing them to produce and release lactate, which is taken up and consumed by cancer cells (Bonuccelli et al., 2010; Fiaschi et al., 2012; Zhang et al., 2015). Mechanistically, this metabolic coupling involves differential expression of monocarboxylate transporters (MCTs) in CAFs and cancer cells. Lactate is extruded from CAFs via MCT4 and then taken up by MCT1 in cancer cells. In some settings, cancer cells induce autophagy and mitophagy in CAFs, causing the CAFs to release recycled metabolites for cancer cell consumption (Martinez-Outschoorn et al., 2011). In prostate cancer, metabolic interactions between stromal and cancer cells are mediated in part by p62, a protein involved in numerous stress responses and signaling mechanisms (Moscat and Diaz-Meco, 2012). Loss of p62 in tumor stroma leads to impairment of mTORC1 and MYC-regulated nutrient metabolism, inducing oxidative stress and release of IL-6, which promotes cancer cell invasion and tumor growth (Valencia et al., 2014). Loss of stromal cell p62 also promotes ATF4-dependent asparagine synthesis and release into the microenvironment, providing a source of this amino acid for neighboring prostate cancer cells (Linares et al., 2017).
The ECM also regulates metabolism, resulting in metabolic adaptations when cells lose matrix attachment (Figure 4, inset). In non-transformed breast epithelial cells, matrix detachment results in reduced glucose uptake, reduced NADPH production from the pentose phosphate pathway, and ROS-induced anoikis; this deleterious effect is overcome by oncogenic signaling, which permits cancer cells to maintain glucose metabolism after detachment (Schafer et al., 2009). Cancer cells capitalize on persistent pentose phosphate pathway flux to transmit reducing equivalents from the cytosol to the mitochondria, where ROS accumulation limits growth in the detached state (Jiang et al., 2016). These mechanisms to counteract ROS after loss of attachment are relevant to metastasis, because oxidative stress poses a major bottleneck to efficient formation of distant macrometastases in multiple models (Le Gal et al., 2015; Piskounova et al., 2015).
METABOLIC RELATIONSHIPS BETWEEN CANCER CELLS AND IMMUNE CELLS
Interactions between cancer cells and immune cells within the TME are important because immune cell function requires precise metabolic regulation and because manipulating the TME’s metabolic milieu might enhance the benefits of clinical immunotherapy. Preclinical studies suggest that metabolic heterogeneity within the TME influences local immune cell function and might contribute to immunotherapy treatment failures. Thus, there could be clinical value in defining the metabolic basis of effective cytotoxic immune responses against cancer cells and the factors limiting such responses in vivo.
In order to understand these relationships, we first need to understand the metabolic needs of immune cells, particularly T cells. Mitogen stimulation of T cells in culture induces metabolic changes similar to the constitutive metabolic state of cancer cell lines. A robust but self-limited activation of aerobic glycolysis was described in mitogen-stimulated lymphocytes as early as the 1970s and was among the first demonstrations of the Warburg effect outside of cancer (Wang et al., 1976). Rat thymocytes induce glutamine consumption, glutaminase activity and oxidation of both glutamine and glucose when stimulated to proliferate in culture (Brand, 1985). Inhibiting glucose metabolism prevents effector T cell functions (Cham et al., 2008; Macintyre et al., 2014). Inhibiting glutaminase impairs T cell proliferation and skews T cell differentiation towards the Th1 subset at the expense of Th17 cells (Johnson et al., 2018). In mice, T-cell specific impairment of electron transport chain complex III or IV reduces respiration and impairs differentiation of T cell subsets, resulting in lethal autoinflammation and ineffective regulatory T cell suppressor function in the case of complex III deficiency (Tarasenko et al., 2017; Weinberg et al., 2019).
Recent work indicates key roles for glutathione, serine and methionine metabolism in T cells. Deletion of Glutamate-Cysteine Ligase (GCLC), an enzyme required for glutathione biosynthesis, prevented T cells from achieving sustained states of activation, and reduced the manifestations of autoimmunity in vivo (Mak et al., 2017). Dietary serine insufficiency restricts T cell expansion in vivo by depleting the serine-dependent one-carbon pool for nucleotide biosynthesis (Ma et al., 2017). Restricting methionine access in antigen-stimulated T cells prevents them from carrying out epigenetic modifications that enable differentiation into effector cells (Sinclair et al., 2019).
Because cancer cells and activated T cells share metabolic features, competition for fuels in the TME could impact immune cell function. These competitive relationships likely involve multiple immune cell populations, but the effect of cancer cell glycolysis on T cell function provides a useful example (Figure 4). In models of rapid tumor growth in mice, tumor-infiltrating T-lymphocytes (TILs) display low levels of glucose uptake, possibly because cancer cells outcompete TILs for glucose (Chang et al., 2015). This renders the TILs hyporesponsive, even when confronted with antigenic tumor cells. TIL responsiveness is boosted by immune checkpoint inhibition, which enhances TIL glycolysis, or simply by injecting glucose into the mice. Glycolytic intermediates themselves may enhance T cell function within tumors. Phosphoenolpyruvate (PEP) sustains anti-tumor effector functions of TILs, but glucose limitation in the TME suppresses TIL PEP levels and reduces anti-tumor responses. Strikingly, over-expressing enzymes that increase PEP levels increases effector functions in tumor-specific CD4 and CD8 TILs, thereby restricting tumor growth (Ho et al., 2015). These reports suggest that therapies to increase glucose availability in the TME might enhance anti-tumor immunity and potentiate the effect of checkpoint blockade.
Several other metabolites in the TME have been reported to impact immune cell function. Lactate derived from cancer cells suppresses T cell and NK cell function (Brand et al., 2016; Fischer et al., 2007) and inhibits monocyte activation and differentiation of dendritic cells (Gottfried et al., 2006). It also induces M2 polarization of macrophages through a mechanism that involves HIF-1α and promotes tumor growth (Colegio et al., 2014). High intracellular arginine levels promote T cell survival and anti-tumor activity (Geiger et al., 2016), and therapies that deplete arginine from the TME induce the accumulation of immune-suppressor cells (Fletcher et al., 2015). Tryptophan catabolism through the kynurenine pathway blunts some aspects of both innate and adaptive immunity, suggesting that blocking the initial steps of tryptophan degradation might enhance the efficacy of immunotherapy (Munn and Mellor, 2013). Several inhibitors of this pathway have reached clinical trials, although the recent closure of a Phase III trial without positive results has tempered enthusiasm somewhat (Platten et al., 2019).
Macromolecular degradation and related pathways are also involved in immune cell function in the TME. Autophagy inhibitors are proposed as cancer therapeutics because of autophagy’s role as a survival mechanism during nutrient deprivation in cancer cells. But a genetic model of inducible autophagy inhibition in pancreatic cancer revealed both cancer cell-autonomous and non-autonomous mechanisms of tumor suppression (Yang et al., 2018). In this autochthonous model, autophagy inhibition led to tumor regression, but implanting autophagy-defective cancer cells into nude mice delayed tumor growth without inducing regression. Part of the difference was related to an enhanced inflammatory infiltrate, particularly macrophages, that contributed to tumor regression in the autochthonous model. Similarly, genetic deletion of any of several autophagy genes in the host resulted in reduced growth of autophagy-competent cancer cells in syngeneic models (DeVorkin et al., 2019). The mechanism involved enhanced production of CD8+ T-effector cells, and depleting these cells reversed the tumor suppressive effect of autophagy inhibition. Finally, the LC3-associated phagocytosis (LAP) pathway, which promotes macrophage scavenging of dead cells in the TME, sustains tumor growth in syngeneic models (Cunha et al., 2018). Inhibiting this pathway in macrophages promotes both innate and adaptive immune responses that suppress tumor growth.
Nutrient availability and regional competition for fuels are inconsistent within solid tumors. Local metabolic preferences of malignant cells may also be influenced by clonal heterogeneity, and this could further impact metabolic interactions among cancer cells, immune cells and other cells in the stroma. Immune cells are presumably subjected to variable metabolic pressures within single tumors, and this complexity is likely amplified in human tumors because of their size, mutational composition and variable exposure to environmental agents.
METABOLIC HETEROGENEITY IN HUMAN TUMORS
Efforts to study cancer metabolism directly in patients have begun to uncover common and heterogeneous metabolic features. A meta-analysis aggregating metabolomics data from over 100 different cohorts covering 18 tumor types revealed a small number of common features across multiple cancers, including elevated lactate in the tumor and reduced glutamine in the blood, and many other features that varied among different cancers (Goveia et al., 2016). Other studies focused on single cancer types have identified variable metabolite levels correlating with imaging features, therapy exposure or progression (Hakimi et al., 2016; Liu et al., 2016; Sreekumar et al., 2009; Zhang et al., 2017b). The emerging role of metabolomic phenotyping in human cancer was recently reviewed (Kaushik and DeBerardinis, 2018).
Isotope tracing can also be used to assess human tumor metabolism. In this approach, an isotope-labeled nutrient (e.g. 13C-glucose) is introduced shortly before or during procurement of a surgical tumor specimen. The labeled nutrient is taken up by the tumor and the label is distributed to metabolites as a consequence of metabolic activity in the tissue. The resulting 13C labeling features provide complementary information to metabolite abundance measurements made through metabolomics. Early studies in non-small cell lung cancer (Fan et al., 2009) and primary and metastatic brain tumors (Maher et al., 2012) emphasized the surprisingly extensive pyruvate oxidation in the TCA cycle. In lung tumors, labeling of TCA cycle intermediates from 13C-glucose exceeds metabolite labeling in the adjacent lung, with pyruvate carboxylase and pyruvate dehydrogenase contributing to enhanced TCA cycle activity in the tumors (Hensley et al., 2016; Sellers et al., 2015). These findings are interesting because many of the tumors were FDG-PET-avid on clinical imaging and had evidence of robust glycolytic intermediate labeling, indicating that oxidation in the TCA cycle is a major fate of glucose carbon even in “glycolytic” tumors. Regions of high proliferative cell content also had high levels of glucose oxidation, implying a role for the TCA cycle in tumor cell proliferation in human tumors (Hensley et al., 2016).
Other studies have reported metabolism of alternative fuels in human tumors. In gliomas and brain metastases, glucose oxidation accounts for only a minor fraction of total acetyl-CoA for the TCA cycle (Maher et al., 2012). Infusing 13C-acetate led to high levels of enrichment of TCA cycle intermediates, indicating that these tumors have the capacity to oxidize acetate from the circulation (Mashimo et al., 2014). In non-small cell lung cancer, labeling from 13C-glucose is heterogeneous among different tumors and different regions of the same tumor, indicating variable utilization of glucose as a fuel (Hensley et al., 2016). Comparing isotope labeling, gene expression and pre-surgical imaging features in the same regions revealed that variable levels of tissue perfusion correlate with fuel preferences in lung cancer (Hensley et al., 2016). Areas with the highest 13C enrichment in TCA cycle intermediates express genes related to glucose oxidation and have lower perfusion as assessed by dynamic contrast-enhanced magnetic resonance imaging prior to surgery. Areas with higher perfusion express genes related to lipid and amino acid metabolism and display lower 13C enrichment in the TCA cycle. One alternative carbon source accounting for heterogeneous metabolite labeling in the tumors is lactate. Over half of human non-small cell cancers transmit 13C from circulating 13C-lactate into TCA cycle metabolites (Faubert et al., 2017). This phenotype warrants further study, as it appears to correlate with early disease progression in lung cancer patients (Faubert et al., 2017).
By applying the same 13C-glucose infusion protocol to multiple types of human cancer, Courtney et al. demonstrated metabolic heterogeneity among tumors at different anatomic sites (Courtney et al., 2018). Tumors in the brain and lung oxidized glucose, consistent with the studies cited above. However, ccRCCs had markedly reduced labeling of TCA cycle intermediates relative to adjacent, non-malignant kidney tissue. These tumors instead had elevated labeling of glycolytic intermediates compared to the kidney, consistent with the classical Warburg effect. The phenotype of prominent glycolysis and suppressed glucose oxidation may reflect the frequent loss of the von Hippel Lindau (VHL) tumor suppressor and chronic pseudohypoxic responses in ccRCC. These studies are valuable because they incorporate the incredible genetic and histological diversity of human cancer and thus have the potential to produce a realistic view of the clinically-relevant scope of metabolic heterogeneity. Over time, it is anticipated that these studies will report the effects of therapy and metastasis on metabolism, which could provide insights for metabolic interventions to suppress cancer progression.
CONCLUSIONS, CHALLENGES AND EMERGING QUESTIONS
The term “cancer metabolism” is often used to imply a common set of metabolic changes that accompany malignancy, but in reality tumors are metabolically heterogeneous. The causes of metabolic heterogeneity are multi-factorial, involving influences both intrinsic and extrinsic to the cancer cell. This makes it challenging to decode the metabolic phenotype of an individual tumor, and to understand when a particular phenotype predicts vulnerability to a therapy as opposed to a bystander effect. Classifying the many causes of metabolic reprogramming in malignant tissue is still a major challenge. But an even larger challenge is to define how different factors act in concert to determine the metabolic phenotype. It is worth taking these challenges on. The emergence of high-content data sets and informatics tools to analyze them, together with more informative metabolic phenotyping techniques, should make it possible to tackle the metabolic complexity of human cancer. As has been the case throughout nearly a century of research in cancer metabolism, understanding the processes by which metabolic properties become reprogrammed will likely continue to teach us about the basis of malignancy.
A practical consideration about how to move the field forward relates to which model systems to use. Although metabolic phenotyping of human cancers has become more prominent, we also need to define experimentally tractable model systems that will help test hypotheses arising from human studies and which can produce insights that can be translated to clinical studies with a high success rate. A combination of techniques will be needed. Genetically modified mice that provide the ability to study cancer initiation and progression in the context of a complex microenvironment will continue to be important. But systems that can recapitulate the genetic heterogeneity and TME of human tumors are also needed. Live cultures of freshly-prepared primary human tumor slices are one possibility (Fan et al., 2016). This approach is amenable to drug treatment, stable isotope tracing, regional heterogeneity analysis, and perhaps could be combined with physiological culture medium to reduce the metabolic artifacts of commercial medium.
Recent observations suggest that we are just scratching the surface on the complexity of non-cell autonomous drivers of metabolic heterogeneity. A fascinating development is the realization that local commensal microbiota impact tumor development in mice. In a genetic model of oncogenic KRAS-driven lung cancer, suppressing the microbial community in the lung reduced tumor burden (Jin et al., 2019). Given the complexity of the human microbiome and its relationship to chronic metabolic diseases, it will be interesting to study whether particular microorganisms promote local inflammation to contribute to cancer. To this end, a large number of clinical trials are examining the effect of manipulating the gut microbiome in cancer (McQuade et al., 2019). Finally, the metabolic health of the patient impacts cancer risk, as the incidence of many cancers is positively correlated with obesity. The mechanisms underlying these relationships are under investigation. But obesity also influences the effect of cancer therapy, positively in some cases. One retrospective, multiple-cohort study in melanoma found that obese men with metastatic melanoma had improved progression-free survival over non-obese peers after targeted therapies or immune checkpoint inhibitors (McQuade et al., 2018). Clearly there is much more to learn.
ACKNOWLEDGEMENTS
The rapid growth of research in cancer metabolism made it impossible to cite a large number of excellent studies relevant to the topic of this review. We are grateful to Katie Regan for help with the Figures. R.J.D. is supported by Howard Hughes Medical Institute, the National Cancer Institute (R35CA22044901) and the Robert L. Moody Faculty Scholar Award. J.K. is supported by the National Cancer Institute (1K22CA226676-01A1) and American Lung Association (LCD-614827). R.J.D. is an advisor for Agios Pharmaceuticals and holds shares in Agios Pharmaceuticals and Peloton Therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allis CD, and Jenuwein T (2016). The molecular hallmarks of epigenetic control. Nature Reviews Genetics 17, 487. [DOI] [PubMed] [Google Scholar]
- Bell EL, Emerling BM, Ricoult SJ, and Guarente L (2011). SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene 30, 2986–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonuccelli G, Whitaker-Menezes D, Castello-Cros R, Pavlides S, Pestell RG, Fatatis A, Witkiewicz AK, Vander Heiden MG, Migneco G, Chiavarina B, et al. (2010). The reverse Warburg effect: glycolysis inhibitors prevent the tumor promoting effects of caveolin-1 deficient cancer associated fibroblasts. Cell cycle (Georgetown, Tex.) 9, 1960–1971. [DOI] [PubMed] [Google Scholar]
- Boroughs LK, and DeBerardinis RJ (2015). Metabolic pathways promoting cancer cell survival and growth. Nature cell biology 17, 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A, Singer K, Koehl GE, Kolitzus M, Schoenhammer G, Thiel A, Matos C, Bruss C, Klobuch S, Peter K, et al. (2016). LDHA-Associated Lactic Acid Production Blunts Tumor Immunosurveillance by T and NK Cells. Cell metabolism 24, 657–671. [DOI] [PubMed] [Google Scholar]
- Brand K (1985). Glutamine and glucose metabolism during thymocyte proliferation. Pathways of glutamine and glutamate metabolism. Biochem J 228, 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, and Tu BP (2011). Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Molecular cell 42, 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. (2014). Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor JR, Abu-Remaileh M, Kanarek N, Freinkman E, Gao X, Louissaint A Jr., Lewis CA, and Sabatini DM (2017). Physiologic Medium Rewires Cellular Metabolism and Reveals Uric Acid as an Endogenous Inhibitor of UMP Synthase. Cell 169, 258–272.e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Finley LW, Cross JR, Allis CD, and Thompson CB (2015). Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518, 413–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cham CM, Driessens G, O’Keefe JP, and Gajewski TF (2008). Glucose deprivation inhibits multiple key gene expression events and effector functions in CD8+ T cells. 38, 2438–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Qiu J, O’Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ, et al. (2015). Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 162, 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, and Guarente L (2014). SIRT1 and other sirtuins in metabolism. Trends in endocrinology and metabolism: TEM 25, 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Zhang J, Li N, Qian Z, Zhu M, Li Q, Zheng J, Wang X, and Shi G (2011). Promoter hypermethylation mediated downregulation of FBP1 in human hepatocellular carcinoma and colon cancer. PloS one 6, e25564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicklore S, Goh V, Siddique M, Roy A, Marsden PK, and Cook GJR (2013). Quantifying tumour heterogeneity in 18F-FDG PET/CT imaging by texture analysis. European Journal of Nuclear Medicine and Molecular Imaging 40, 133–140. [DOI] [PubMed] [Google Scholar]
- Colegio OR, Chu N-Q, Szabo AL, Chu T, Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC, Phillips GM, et al. (2014). Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, et al. (2013). Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha LD, Yang M, Carter R, Guy C, Harris L, Crawford JC, Quarato G, Boada-Romero E, Kalkavan H, Johnson MDL, et al. (2018). LC3-Associated Phagocytosis in Myeloid Cells Promotes Tumor Immune Tolerance. Cell 175, 429–441 e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daemen A, Peterson D, Sahu N, McCord R, Du X, Liu B, Kowanetz K, Hong R, Moffat J, Gao M, et al. (2015). Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. 112, E4410–E4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. (2009). Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, and Chandel NS (2016). Fundamentals of cancer metabolism. Science advances 2, e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, et al. (2015). NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nature genetics 47, 1475–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVorkin L, Pavey N, Carleton G, Comber A, Ho C, Lim J, McNamara E, Huang H, Kim P, Zacharias LG, et al. (2019). Autophagy Regulation of Metabolism Is Required for CD8(+) T Cell Anti-tumor Immunity. Cell reports 27, 502–513 e505. [DOI] [PubMed] [Google Scholar]
- Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S, Lin Y, Yao J, Shi J, Kang T, et al. (2013). Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer cell 23, 316–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easwaran H, Tsai HC, and Baylin SB (2014). Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Molecular cell 54, 716–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JR, Yarychkivska O, Boulard M, and Bestor TH (2017). DNA methylation and DNA methyltransferases. Epigenetics & Chromatin 10, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, et al. (2004). Akt stimulates aerobic glycolysis in cancer cells. Cancer research 64, 3892–3899. [DOI] [PubMed] [Google Scholar]
- Esteller M (2002). CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene 21, 5427–5440. [DOI] [PubMed] [Google Scholar]
- Fan TW, Lane AN, and Higashi RM (2016). Stable Isotope Resolved Metabolomics Studies in Ex Vivo Tissue Slices. Bio-protocol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TW, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, and Miller DM (2009). Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM). Molecular cancer 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, Yang C, Do QN, Doucette S, Burguete D, et al. (2017). Lactate Metabolism in Human Lung Tumors. Cell 171, 358–371.e359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiaschi T, Marini A, Giannoni E, Taddei ML, Gandellini P, De Donatis A, Lanciotti M, Serni S, Cirri P, and Chiarugi P (2012). Reciprocal Metabolic Reprogramming through Lactate Shuttle Coordinately Influences Tumor-Stroma Interplay. 72, 5130–5140. [DOI] [PubMed] [Google Scholar]
- Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, Gottfried E, Schwarz S, Rothe G, Hoves S, et al. (2007). Inhibitory effect of tumor cell-derived lactic acid on human T cells. 109, 3812–3819. [DOI] [PubMed] [Google Scholar]
- Fletcher M, Ramirez ME, Sierra RA, Raber P, Thevenot P, Al-Khami AA, Sanchez-Pino D, Hernandez C, Wyczechowska DD, Ochoa AC, et al. (2015). l-Arginine depletion blunts antitumor T-cell responses by inducing myeloid-derived suppressor cells. Cancer research 75, 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flier JS, Mueckler MM, Usher P, and Lodish HF (1987). Elevated levels of glucose transport and transporter messenger RNA are induced by ras or src oncogenes. Science (New York, N.Y.) 235, 1492–1495. [DOI] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. (2009). c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458, 762–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaude E, and Frezza C (2016). Tissue-specific and convergent metabolic transformation of cancer correlates with metastatic potential and patient survival. Nature Communications 7, 13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger R, Rieckmann JC, Wolf T, Basso C, Feng Y, Fuhrer T, Kogadeeva M, Picotti P, Meissner F, Mann M, et al. (2016). L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell 167, 829–842.e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A, Mathupala SP, and Pedersen PL (2003). Glucose metabolism in cancer. Evidence that demethylation events play a role in activating type II hexokinase gene expression. The Journal of biological chemistry 278, 15333–15340. [DOI] [PubMed] [Google Scholar]
- Goodwin J, Neugent ML, Lee SY, Choe JH, Choi H, Jenkins DMR, Ruthenborg RJ, Robinson MW, Jeong JY, Wake M, et al. (2017). The distinct metabolic phenotype of lung squamous cell carcinoma defines selective vulnerability to glycolytic inhibition. Nat Commun 8, 15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, and Simon MC (2007). HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer cell 12, 108–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottfried E, Kunz-Schughart LA, Ebner S, Mueller-Klieser W, Hoves S, Andreesen R, Mackensen A, and Kreutz M (2006). Tumor-derived lactic acid modulates dendritic cell activation and antigen expression. Blood 107, 2013–2021. [DOI] [PubMed] [Google Scholar]
- Goveia J, Pircher A, Conradi LC, Kalucka J, Lagani V, Dewerchin M, Eelen G, DeBerardinis RJ, Wilson ID, and Carmeliet P (2016). Meta-analysis of clinical metabolic profiling studies in cancer: challenges and opportunities. EMBO Mol Med 8, 1134–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi AA, Reznik E, Lee CH, Creighton CJ, Brannon AR, Luna A, Aksoy BA, Liu EM, Shen R, Lee W, et al. (2016). An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer cell 29, 104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Yu W, and Denu JM (2012). Regulation of glycolytic enzyme phosphoglycerate mutase-1 by Sirt1 protein-mediated deacetylation. The Journal of biological chemistry 287, 3850–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, and Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L, et al. (2016). Metabolic Heterogeneity in Human Lung Tumors. Cell 164, 681–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley CT, Wasti AT, and DeBerardinis RJ (2013). Glutamine and cancer: cell biology, physiology, and clinical opportunities. The Journal of clinical investigation 123, 3678–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG (1999). Hypermethylation of tumor suppressor genes in cancer. Seminars in cancer biology 9, 359–367. [DOI] [PubMed] [Google Scholar]
- Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, et al. (2015). Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell 162, 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Locasale JW, Bielas JH, O’Sullivan J, Sheahan K, Cantley LC, Heiden MGV, and Vitkup D (2013). Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nature Biotechnology 31, 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Ni M, Chalishazar MD, Huffman KE, Kim J, Cai L, Shi X, Cai F, Zacharias LG, Ireland AS, et al. (2018). Inosine Monophosphate Dehydrogenase Dependence in a Subset of Small Cell Lung Cancers. Cell metabolism 28, 369–382.e365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun K, Jeon J, Park K, and Kim J (2017). Writing, erasing and reading histone lysine methylations. Experimental &Amp; Molecular Medicine 49, e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, Terada LS, Adams ND, McCabe MT, Pietrak B, et al. (2016). Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 532, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, Ameh S, Sandel D, Liang XS, Mazzilli S, et al. (2019). Commensal Microbiota Promote Lung Cancer Development via gammadelta T Cells. Cell 176, 998–1013 e1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MO, Wolf MM, Madden MZ, Andrejeva G, Sugiura A, Contreras DC, Maseda D, Liberti MV, Paz K, Kishton RJ, et al. (2018). Distinct Regulation of Th17 and Th1 Cell Differentiation by Glutaminase-Dependent Metabolism. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, and Baylin SB (2002). The fundamental role of epigenetic events in cancer. Nature reviews. Genetics 3, 415–428. [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr., and McKnight SL (2013). Influence of metabolism on epigenetics and disease. Cell 153, 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik AK, and DeBerardinis RJ (2018). Applications of metabolomics to study cancer metabolism. Biochim Biophys Acta Rev Cancer 1870, 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hu Z, Cai L, Li K, Choi E, Faubert B, Bezwada D, Rodriguez-Canales J, Villalobos P, Lin YF, et al. (2017). CPS1 maintains pyrimidine pools and DNA synthesis in KRAS/LKB1-mutant lung cancer cells. Nature 546, 168–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, and Dang CV (2011). Otto Warburg’s contributions to current concepts of cancer metabolism. Nature reviews. Cancer 11, 325–337. [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Wilson FH, Ruth JR, Paulk J, Tsherniak A, Marlow SE, Vazquez F, Weir BA, Fitzgerald ME, Tanaka M, et al. (2016). MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science (New York, N.Y.) 351, 1214–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis M, and Esteller M (2010). DNA methylation and cancer. Advances in genetics 70, 27–56. [DOI] [PubMed] [Google Scholar]
- Kung HN, Marks JR, and Chi JT (2011). Glutamine synthetase is a genetic determinant of cell type-specific glutamine independence in breast epithelia. PLoS Genet 7, e1002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laukka T, Mariani CJ, Ihantola T, Cao JZ, Hokkanen J, Kaelin WG Jr., Godley LA, and Koivunen P (2016). Fumarate and Succinate Regulate Expression of Hypoxia-inducible Genes via TET Enzymes. The Journal of biological chemistry 291, 4256–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gal K, Ibrahim MX, Wiel C, Sayin VI, Akula MK, Karlsson C, Dalin MG, Akyurek LM, Lindahl P, Nilsson J, et al. (2015). Antioxidants can increase melanoma metastasis in mice. Science translational medicine 7, 308re308. [DOI] [PubMed] [Google Scholar]
- Lee JV, Carrer A, Shah S, Snyder NW, Wei S, Venneti S, Worth AJ, Yuan ZF, Lim HW, Liu S, et al. (2014). Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell metabolism 20, 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Qiu B, Lee DS, Walton ZE, Ochocki JD, Mathew LK, Mancuso A, Gade TP, Keith B, Nissim I, et al. (2014). Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature 513, 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, and Dang CV (2005). Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Molecular and cellular biology 25, 6225–6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ning S, Ghandi M, Kryukov GV, Gopal S, Deik A, Souza A, Pierce K, Keskula P, Hernandez D, et al. (2019). The landscape of cancer cell line metabolism. Nature medicine 25, 850–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares JF, Cordes T, Duran A, Reina-Campos M, Valencia T, Ahn CS, Castilla EA, Moscat J, Metallo CM, and Diaz-Meco MT (2017). ATF4-Induced Metabolic Reprograming Is a Synthetic Vulnerability of the p62-Deficient Tumor Stroma. Cell metabolism 26, 817–829 e816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Romero IL, Litchfield LM, Lengyel E, and Locasale JW (2016). Metformin Targets Central Carbon Metabolism and Reveals Mitochondrial Requirements in Human Cancers. Cell metabolism 24, 728–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Marks K, Cowley GS, Carretero J, Liu Q, Nieland TJ, Xu C, Cohoon TJ, Gao P, Zhang Y, et al. (2013). Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer. Cancer discovery 3, 870–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. (2011). Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature genetics 43, 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losman JA, and Kaelin WG Jr. (2013). What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes & development 27, 836–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. (2012). IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Chang R, Zhong J, Pandey A, and Semenza GL (2012). Histone demethylase JMJD2C is a coactivator for hypoxia-inducible factor 1 that is required for breast cancer progression. Proceedings of the National Academy of Sciences of the United States of America 109, E3367–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma EH, Bantug G, Griss T, Condotta S, Johnson RM, Samborska B, Mainolfi N, Suri V, Guak H, Balmer ML, et al. (2017). Serine Is an Essential Metabolite for Effector T Cell Expansion. Cell metabolism 25, 482. [DOI] [PubMed] [Google Scholar]
- Machida K (2017). Pluripotency transcription factors and Metabolic reprogramming of mitochondria in tumor-initiating stem-like cells. Antioxidants & redox signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, Anderson SM, Abel ED, Chen BJ, Hale LP, et al. (2014). The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell metabolism 20, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks OD, Labuschagne CF, Adams PD, and Vousden KH (2016). Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells. Molecular cell 61, 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher EA, Marin-Valencia I, Bachoo RM, Mashimo T, Raisanen J, Hatanpaa KJ, Jindal A, Jeffrey FM, Choi C, Madden C, et al. (2012). Metabolism of [U-13 C]glucose in human brain tumors in vivo. NMR in biomedicine 25, 1234–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak TW, Grusdat M, Duncan GS, Dostert C, Nonnenmacher Y, Cox M, Binsfeld C, Hao Z, Brustle A, Itsumi M, et al. (2017). Glutathione Primes T Cell Metabolism for Inflammation. Immunity 46, 675–689. [DOI] [PubMed] [Google Scholar]
- Marjon K, Cameron MJ, Quang P, Clasquin MF, Mandley E, Kunii K, McVay M, Choe S, Kernytsky A, Gross S, et al. (2016). MTAP Deletions in Cancer Create Vulnerability to Targeting of the MAT2A/PRMT5/RIOK1 Axis. Cell reports 15, 574–587. [DOI] [PubMed] [Google Scholar]
- Martincorena I, and Campbell PJ (2015). Somatic mutation in cancer and normal cells. Science (New York, N.Y.) 349, 1483–1489. [DOI] [PubMed] [Google Scholar]
- Martinez-Outschoorn UE, Pavlides S, Howell A, Pestell RG, Tanowitz HB, Sotgia F, and Lisanti MP (2011). Stromal-epithelial metabolic coupling in cancer: integrating autophagy and metabolism in the tumor microenvironment. The international journal of biochemistry & cell biology 43, 1045–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo T, Pichumani K, Vemireddy V, Hatanpaa KJ, Singh DK, Sirasanagandla S, Nannepaga S, Piccirillo SG, Kovacs Z, Foong C, et al. (2014). Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell 159, 1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR, Lau AN, Ji BW, Dixit PD, Hosios AM, et al. (2016). Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science (New York, N.Y.) 353, 1161–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade JL, Daniel CR, Helmink BA, and Wargo JA (2019). Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol 20, e77–e91. [DOI] [PubMed] [Google Scholar]
- McQuade JL, Daniel CR, Hess KR, Mak C, Wang DY, Rai RR, Park JJ, Haydu LE, Spencer C, Wongchenko M, et al. (2018). Association of body-mass index and outcomes in patients with metastatic melanoma treated with targeted therapy, immunotherapy, or chemotherapy: a retrospective, multicohort analysis. Lancet Oncol 19, 310–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J, and Diaz-Meco MT (2012). p62: a versatile multitasker takes on cancer. Trends in biochemical sciences 37, 230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir A, Danai LV, Gui DY, Waingarten CY, Lewis CA, and Vander Heiden MG (2017). Environmental cystine drives glutamine anaplerosis and sensitizes cancer cells to glutaminase inhibition. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AR, Hu Z, Shi X, Jiang L, Boroughs LK, Kovacs Z, Boriack R, Rakheja D, Sullivan LB, Linehan WM, et al. (2014). Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell reports 7, 1679–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn DH, and Mellor AL (2013). Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends in immunology 34, 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB, Hotamisligil GS, et al. (2011). Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nature medicine 17, 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, Araki J, King B, DeMatteo RG, and Thompson CB (2017). Critical role for PI3-kinase in regulating the use of proteins as an amino acid source. Proceedings of the National Academy of Sciences of the United States of America 114, E8628–E8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, Park Y, Wright K, Pavlova NN, Tuveson DA, and Thompson CB (2015). The Utilization of Extracellular Proteins as Nutrients Is Suppressed by mTORC1. Cell 162, 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Mao C, and Wang YX (2013). Suppression of gluconeogenic gene expression by LSD1-mediated histone demethylation. PloS one 8, e66294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M, Reid MA, Lowman XH, Kulkarni RP, Tran TQ, Liu X, Yang Y, Hernandez-Davies JE, Rosales KK, Li H, et al. (2016). Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nature cell biology 18, 1090–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, and Denko NC (2006). HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell metabolism 3, 187–197. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. (2000). Molecular portraits of human breast tumours. Nature 406, 747–752. [DOI] [PubMed] [Google Scholar]
- Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, and Kroemer G (2015). Acetyl coenzyme A: a central metabolite and second messenger. Cell metabolism 21, 805–821. [DOI] [PubMed] [Google Scholar]
- Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, Leitch AM, Johnson TM, DeBerardinis RJ, and Morrison SJ (2015). Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 527, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten M, Nollen EAA, Rohrig UF, Fallarino F, and Opitz CA (2019). Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nature reviews. Drug discovery 18, 379–401. [DOI] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. (2011). Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, Karakousi TR, Ellis DC, Bhutkar A, Sanchez-Rivera FJ, et al. (2017). Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nature medicine 23, 1362–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A, Hino S, Nagaoka K, Anan K, Takase R, Matsumori H, Ojima H, Kanai Y, Arita K, and Nakao M (2015). Lysine Demethylase LSD1 Coordinates Glycolytic and Mitochondrial Metabolism in Hepatocellular Carcinoma Cells. Cancer research 75, 1445–1456. [DOI] [PubMed] [Google Scholar]
- Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, and Brugge JS (2009). Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 461, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, et al. (2012). The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151, 1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers K, Fox MP, Bousamra M 2nd, Slone SP, Higashi RM, Miller DM, Wang Y, Yan J, Yuneva MO, Deshpande R, et al. (2015). Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. The Journal of clinical investigation 125, 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, Wei L, Fishbein MC, Czernin J, Mischel PS, et al. (2013). LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer cell 23, 143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Tasdogan A, Huang F, Hu Z, Morrison SJ, and DeBerardinis RJ (2017). The abundance of metabolites related to protein methylation correlates with the metastatic capacity of human melanoma xenografts. Science advances 3, eaao5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EH, Livi CB, Rakheja D, Tan J, Benson D, Parekh V, Kho EY, Ghosh AP, Kirkman R, Velu S, et al. (2014). L-2-Hydroxyglutarate: an epigenetic modifier and putative oncometabolite in renal cancer. Cancer discovery 4, 1290–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, and Dang CV (1997). c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proceedings of the National Academy of Sciences of the United States of America 94, 6658–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair LV, Howden AJ, Brenes A, Spinelli L, Hukelmann JL, Macintyre AN, Liu X, Thomson S, Taylor PM, Rathmell JC, et al. (2019). Antigen receptor control of methionine metabolism in T cells. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EH, Janknecht R, and Maher IIILJ (2007). Succinate inhibition of a-ketoglutarate-dependent enzymes in a yeast model of paraganglioma. Human Molecular Genetics 16, 3136–3148. [DOI] [PubMed] [Google Scholar]
- Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, De Saedeleer CJ, Kennedy KM, Diepart C, Jordan BF, et al. (2008). Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. The Journal of clinical investigation 118, 3930–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying H, et al. (2016). Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, et al. (2009). Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457, 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Stine ZE, Walton ZE, Altman BJ, Hsieh AL, and Dang CV (2015). MYC, Metabolism, and Cancer. Cancer discovery 5, 1024–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumvoll M, Perriello G, Meyer C, and Gerich J (1999). Role of glutamine in human carbohydrate metabolism in kidney and other tissues. Kidney international 55, 778–792. [DOI] [PubMed] [Google Scholar]
- Sullivan MR, Danai LV, Lewis CA, Chan SH, Gui DY, Kunchok T, Dennstedt EA, Vander Heiden MG, and Muir A (2019). Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, et al. (2010). Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Molecular cell 40, 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasenko TN, Pacheco SE, Koenig MK, Gomez-Rodriguez J, Kapnick SM, Diaz F, Zerfas PM, Barca E, Sudderth J, DeBerardinis RJ, et al. (2017). Cytochrome c Oxidase Activity Is a Metabolic Checkpoint that Regulates Cell Fate Decisions During T Cell Activation and Differentiation. Cell metabolism 25, 1254–1268.e1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman LA, Holton T, Yuneva M, Louie RJ, Padro M, Daemen A, Hu M, Chan DA, Ethier SP, van ‘t Veer LJ, et al. (2013). Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer cell 24, 450–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulanovskaya OA, Zuhl AM, and Cravatt BF (2013). NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nature chemical biology 9, 300–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrey CL, Liu L, Andrews LG, and Tollefsbol TO (2005). The impact of metabolism on DNA methylation. Human Molecular Genetics 14, R139–R147. [DOI] [PubMed] [Google Scholar]
- Valencia T, Kim JY, Abu-Baker S, Moscat-Pardos J, Ahn CS, Reina-Campos M, Duran A, Castilla EA, Metallo CM, Diaz-Meco MT, et al. (2014). Metabolic reprogramming of stromal fibroblasts through p62-mTORC1 signaling promotes inflammation and tumorigenesis. Cancer cell 26, 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Voorde J, Ackermann T, Pfetzer N, Sumpton D, Mackay G, Kalna G, Nixon C, Blyth K, Gottlieb E, and Tardito S (2019). Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Sci Adv 5, eaau7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaupel P, and Mayer A (2007). Hypoxia in cancer: significance and impact on clinical outcome. Cancer and Metastasis Reviews 26, 225–239. [DOI] [PubMed] [Google Scholar]
- Wan W, Peng K, Li M, Qin L, Tong Z, Yan J, Shen B, and Yu C (2017). Histone demethylase JMJD1A promotes urinary bladder cancer progression by enhancing glycolysis through coactivation of hypoxia inducible factor 1alpha. Oncogene 36, 3868–3877. [DOI] [PubMed] [Google Scholar]
- Wang T, Marquardt C, and Foker J (1976). Aerobic glycolysis during lymphocyte proliferation. Nature 261, 702–705. [DOI] [PubMed] [Google Scholar]
- Warburg O (1956). On the origin of cancer cells. Science (New York, N.Y.) 123, 309–314. [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. (2010). The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer cell 17, 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SE, Singer BD, Steinert EM, Martinez CA, Mehta MM, Martinez-Reyes I, Gao P, Helmin KA, Abdala-Valencia H, Sena LA, et al. (2019). Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature 565, 495–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, and Thompson CB (2009). ATP-citrate lyase links cellular metabolism to histone acetylation. Science (New York, N.Y.) 324, 1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KT, and Schalinske KL (2007). New Insights into the Regulation of Methyl Group and Homocysteine Metabolism. The Journal of Nutrition 137, 311–314. [DOI] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, et al. (2008). Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proceedings of the National Academy of Sciences of the United States of America 105, 18782–18787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, and Thompson CB (2011). Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proceedings of the National Academy of Sciences of the United States of America 108, 19611–19616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf A, Agnihotri S, Munoz D, and Guha A (2011). Developmental profile and regulation of the glycolytic enzyme hexokinase 2 in normal brain and glioblastoma multiforme. Neurobiology of disease 44, 84–91. [DOI] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, et al. (2012). Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes & development 26, 1326–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Herter-Sprie G, Zhang H, Lin EY, Biancur D, Wang X, Deng J, Hai J, Yang S, Wong KK, et al. (2018). Autophagy Sustains Pancreatic Cancer Growth through Both Cell-Autonomous and Nonautonomous Mechanisms. Cancer discovery 8, 276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, and Seto E (2007). HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26, 5310. [DOI] [PubMed] [Google Scholar]
- Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Mates JM, Alonso FJ, Wang C, Seo Y, et al. (2012). The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell metabolism 15, 157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi N, Swinnen JV, and Smans K (2012). ATP-citrate lyase: a key player in cancer metabolism. Cancer research 72, 3709–3714. [DOI] [PubMed] [Google Scholar]
- Zhang D, Wang Y, Shi Z, Liu J, Sun P, Hou X, Zhang J, Zhao S, Zhou BP, and Mi J (2015). Metabolic reprogramming of cancer-associated fibroblasts by IDH3alpha downregulation. Cell reports 10, 1335–1348. [DOI] [PubMed] [Google Scholar]
- Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, and Semenza GL (2008). Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. The Journal of biological chemistry 283, 10892–10903. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, and Semenza GL (2007). HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer cell 11, 407–420. [DOI] [PubMed] [Google Scholar]
- Zhang J, Pavlova NN, and Thompson CB (2017a). Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine. The EMBO journal 36, 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Trachootham D, Liu J, Chen G, Pelicano H, Garcia-Prieto C, Lu W, Burger JA, Croce CM, Plunkett W, et al. (2012). Stromal control of cystine metabolism promotes cancer cell survival in chronic lymphocytic leukaemia. Nature cell biology 14, 276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Udayakumar D, Cai L, Hu Z, Kapur P, Kho EY, Pavia-Jimenez A, Fulkerson M, de Leon AD, Yuan Q, et al. (2017b). Addressing metabolic heterogeneity in clear cell renal cell carcinoma with quantitative Dixon MRI. JCI Insight 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. (2010). The histone deacetylase Sirt6 regulates glucose homeostasis via HIF1αlpha. Cell 140, 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]